Abstract
ELMODs are a family of three mammalian paralogues that display GTPase-activating protein (GAP) activity toward a uniquely broad array of ADP-ribosylation factor (ARF) family GTPases that includes ARF-like (ARL) proteins. ELMODs are ubiquitously expressed in mammalian tissues, highly conserved across eukaryotes, and ancient in origin, being present in the last eukaryotic common ancestor. We described functions of ELMOD2 in immortalized mouse embryonic fibroblasts (MEFs) in the regulation of cell division, microtubules, ciliogenesis, and mitochondrial fusion. Here, using similar strategies with the paralogues ELMOD1 and ELMOD3, we identify novel functions and locations of these cell regulators and compare them to those of ELMOD2, allowing the determination of functional redundancy among the family members. We found strong similarities in phenotypes resulting from deletion of either Elmod1 or Elmod3 and marked differences from those arising in Elmod2 deletion lines. Deletion of either Elmod1 or Elmod3 results in the decreased ability of cells to form primary cilia, loss of a subset of proteins from cilia, and accumulation of some ciliary proteins at the Golgi, predicted to result from compromised traffic from the Golgi to cilia. These phenotypes are reversed upon activating mutant expression of either ARL3 or ARL16, linking their roles to ELMOD1/3 actions.
INTRODUCTION
The ARF family of regulatory GTPases is large, with ∼30 mammalian members that are highly conserved throughout eukaryotic evolution, 16 of which are predicted to have been present in the last eukaryotic common ancestor (Vargova et al., 2021). ARFs and ARLs regulate a range of essential cellular processes including roles in membrane traffic, primary cilia, mitochondria, tubulin biogenesis, and microtubule dynamics (Cherfils, 2014; Jackson and Bouvet, 2014; Sztul et al., 2019; Casalou et al., 2020; Fisher et al., 2020). A technical challenge in elucidating mechanisms of such actions is the fact that specificity of GTPase actions is often lost in in vitro assays using purified components, as they fail to replicate the intricate cellular contexts under which they drive biological functions. Other challenges include that GTPases frequently act from multiple sites yet localize to each site only transiently. This holds true for the GTPases along with the guanine nucleotide exchange factors (GEFs) that activate them and the GTPase-activating proteins (GAPs) that both inactivate the GTPase and frequently serve effector functions. As a result, we have only fragmentary data on the substrate (GTPase) specificity for any ARF GAP or ARF GEF as well as the cellular functions of each. There are 28 known or predicted ARF GAPs, 24 of which were identified based on the presence of an ARF GAP domain (Kahn et al., 2008). Of the ARF GAPs that lack the consensus ARF GAP domain, three mammalian proteins share an ELMO domain (ELMOD1–3) with broad specificity for GTPases in the ARF family (East et al., 2012), while the sole outlier, RP2, possesses GAP activity specific to ARL2 and ARL3 (Veltel et al., 2008). Those ARF GAPs that share the ARF GAP domain have been tested using in vitro assays and found to act with various degrees of specificity toward one or more of the six mammalian ARFs (ARF1–6; though typically only ARF1 and ARF6 have been tested), but few have been tested against any of the 22 ARF-like (ARL) GTPases. In efforts to gain insights into signaling by the ARLs, we purified ELMOD2 as an ARL2 GAP and found that all three of the human ELMOD proteins act on several different ARLs as well as ARFs (East et al., 2012; Ivanova et al., 2014). Thus, this broad specificity in in vitro GAP assays suggests the possibility that ELMODs act in an even broader set of signaling pathways than do the ARF GAPs.
The ELMO (cell EnguLfment and MOtility) family of proteins consists of six members in humans that share an ELMO domain and are equally divided into structurally and phylogenetically distinct subgroups, termed ELMOs and ELMODs. The ELMODs are ancient proteins that span the entire diversity of eukaryotes and date back to the last eukaryotic common ancestor (East et al., 2012). In contrast, the ELMOs are a more recent evolutionary family, found only in metazoans and fungi, and are predicted to have emerged from the ELMODs (East et al., 2012). While the ELMODs are GAPs with broad substrate specificities toward ARFs and ARLs, the ELMOs primarily function in complex with DOCK proteins as unconventional GEFs for RHO/RAC GTPases (which act at the leading edge of migrating cells) and lack GAP activity (Reddien and Horvitz, 2004; Laurin and Cote, 2014). In mouse embryonic fibroblasts (MEFs), ELMOD2 serves critical roles in the regulation of cytokinesis, ciliation, microtubule stability, mitochondrial fusion, ciliogenesis, anchoring of rootlets to centrosomes, and lipid metabolism at lipid droplets (Suzuki et al., 2015; Newman et al., 2017; Zhou et al., 2017; Schiavon et al., 2019; Turn et al., 2020, 2021). The effects of ELMOD2 deletion on mitochondrial fusion and microtubules are linked to ARL2 (Schiavon et al., 2019; Turn et al., 2020), while effects on cell division/abscission are specifically suppressed by expression of activated ARF6 (Turn et al., 2020), demonstrating that one ELMOD can regulate multiple pathways through distinct GTPases. Because ELMOD2 is a member of a three-gene family in mammals, each of which shares a common functional domain. We sought to compare these actions to those of other ELMODs. This was to pave the way for defining the cellular functions of each of the family members and also to assess the extent of functional redundancy that may suppress the appearance of even more deficits in cells when any one member is mutated or deleted.
Mutations in ELMOD2 are linked to familial idiopathic pulmonary fibrosis as well as antiviral response in both mice and humans (Hodgson et al., 2006; Pulkkinen et al., 2010). In contrast, ELMOD1 mutations and ELMOD3 mutations are linked to deafness, autism, and intellectual disability in humans and mice (Johnson et al., 2012; Jaworek et al., 2013; Li et al., 2018; Miryounesi et al., 2019). Hearing deficits linked to mutation of either Elmod1 or Elmod3 in mice are also linked to defects in stereocilia, or actin-based apical projections of inner ear hair cells. For stereocilia to form, the hair cell must first project a primary cilium (in the context of hair cells, called the “kinocilium”), which plays a critical role in signaling for proper stereocilia morphology and orientation. Almost all mammalian cells generate at least one cilium, motile (e.g., flagellum) or nonmotile (primary cilia and kinocilia), and some cells (particularly epithelial) display multiciliation. Cilia are primarily present in cells that have exited the cell cycle at least in part because centrosomes are involved in formation of the basal body, the associated distal appendages, the ciliary pocket, and the microtubule-based axoneme that extends and pushes out the nascent cilium. The early steps of ciliogenesis, termed licensing, can be induced in cell culture via serum starvation. This initiates the recruitment of CEP164 to the mother centriole followed by recruitment of TTBK2 and later “uncapping” or loss of CP110 (Tsang and Dynlacht, 2013; Yadav et al., 2016). The release of CP110 is followed by ciliary vesicle docking and axoneme extension (Yadav et al., 2016). For additional details of ciliogenesis, consult the following reviews: Bernabe-Rubio and Alonso, 2017; Reiter and Leroux, 2017; and Chen et al., 2021. Once formed, cilia are dependent on three known protein complexes to engineer the entry and export of ciliary proteins as well as movement along the axoneme: intraflagellar transport complex A (IFT-A, retrograde traffic), IFT complex B (IFT-B, anterograde traffic), and the BBSome (Sedmak and Wolfrum, 2010; Liem et al., 2012; Lechtreck, 2015; Nachury, 2018; Kobayashi et al., 2021). Despite characterizations of their actions as multisubunit protein complexes, there is evidence that one of the 16 components of the IFT-B complex, IFT20, acts independently of the complex at the Golgi to promote traffic to the ciliary base (Follit et al., 2006; Keady et al., 2011). Knockout (KO) of IFT20 causes a more severe phenotype than does KO of the IFT-A core subunit IFT140 (Crouse et al., 2014).
Much less is known about routes that proteins take to get to or through the ciliary base, but there are likely several different routes and regulators as well as differences between membrane and soluble proteins. Soluble proteins <∼100 kDa (9 nm diameter) can simply diffuse into cilia (Nachury and Mick, 2019). On the other hand, membrane proteins are synthesized on the rough endoplasmic reticulum (ER) membrane, traffic through the Golgi, and are directed to the basal body through incompletely understood processes. Notable among the proteins implicated in regulating import and/or retention of ciliary proteins are four ARF family GTPases: ARL3, ARL6, ARL13B, and ARL16 (Fisher et al., 2020). ARL13B interacts with INPP5E, a PIP 5′-phosphatase, and PDE6D (aka PDE6δ or Pr/BPδ), a transporter of prenylated cargoes that includes INPP5E (Humbert et al., 2012; Qiu et al., 2020; Fujisawa et al., 2021). ARL13B possesses GEF activity for ARL3, which in turn can bind directly to PDE6D, resulting in the release of its cargo (Miertzschke et al., 2014; Gotthardt et al., 2015; Ivanova et al., 2017; Hanke-Gogokhia et al., 2018). While ARL13B can be palmitoylated on cysteine residues near the N-terminus and thereby become anchored in the membrane, it is not clear how ARL3 is retained in cilia, though its local activation by ARL13B may regulate its localization in cilia. We also have recently identified defects in ciliogenesis and ciliary protein content in cells deleted for ARL16 (Dewees et al., 2021). Even less well understood are the regulators of these ciliary GTPases, as to date very little is known about the ARF GEFs and GAPs that work in primary cilia.
Here we describe the use of a number of approaches, relying heavily on CRISPR/Cas9-edited MEFs, to begin defining the cellular functions of Elmod1 and Elmod3 and comparing the degree of functional overlap/specificity among the family members (Schiavon et al., 2019; Turn et al., 2020, 2021). ELMOD1 and ELMOD3 studies to date have focused on in vitro biochemical activities and analyses of mutations in mammals, so the cellular functions remain largely uncharacterized. We use MEFs as our model system to allow detailed comparisons to earlier studies of Elmod2 KO in isogenic cell lines. We include descriptions of organelle morphologies and pathways that appear unaltered in these KO lines as a broad scan for unexpected, potential functions, as such data also provide supportive evidence of the specificity of the defects identified.
RESULTS
CRISPR/Cas9 KOs of Elmod1, Elmod3, or both in immortalized MEFs
In efforts to identify the cellular functions of ELMOD1 and ELMOD3, we used CRISPR/Cas9 in MEFs to introduce frameshift mutations in Elmod1, Elmod3, or both Elmod1/Elmod3. We previously used the same procedure to generate Elmod2 KO MEFs and described multiple resulting phenotypes.(Schiavon et al., 2019; Turn et al., 2020, 2021) We used two or more guides to target exons (see Supplemental Figure S1, A and B, and Materials and Methods for details) and identified at least two clones from each guide to obtain lines with null alleles. We summarize the alleles for all cell lines in Supplemental Figure S1C. We predict that all 17 lines result in the loss of functional proteins or null alleles, as these frameshifts are targeted upstream of the sole functional (ELMOD) domain. Thus, we refer to these lines as KO and double KO (DKO) (Elmod1/Elmod3 DKO) lines. There are currently no antibodies specific to ELMOD1 or ELMOD3 with the requisite sensitivity to detect endogenous proteins in MEFs, so further confirmation by immunoblotting was not possible. We also generated clones with no mutations in the targeted region. These are retained as additional controls, as they went through the same transfection, selection, and cloning process as the KO and DKO lines. We refer to such lines as CRISPR WT (CWT) to distinguish them from the parental WT lines. The use of more than one guide, multiple alleles, and independent clones helps alleviate concerns regarding off-target effects, alternative splicing, and downstream initiation. As an additional protection against off-target effects, we also performed rescue experiments in which we transiently reexpressed the disrupted gene to test for reversal of key phenotypes.
Many organelles and processes appear unaltered in cells deleted for Elmod1, Elmod3, or DKOs
Because ELMODs display in vitro GAP activity with a number of different ARF family GTPase substrates, known to regulate a wide array of cellular processes, we screened the KO lines using markers of different organelles and processes to identify gross changes. For example, we previously reported (East et al., 2012) localization of ELMOD1 at nuclear speckles, but no evidence of changes in nuclear speckles were evident in any of these KO lines (Supplemental Figure S2A). Hoechst staining revealed no evidence of changes in nuclear size, number, or morphology (Supplemental Figure S2B). Because we first purified ELMODs as ARL2 GAPs, we looked at processes previously reported to be regulated by ARL2 but found no evident changes in mitochondrial morphology (Supplemental Figure S2C), microtubule networks (Supplemental Figure S2D), or centrosome numbers (Turn et al., 2020, 2021). Phalloidin staining for actin was not overtly altered among Elmod1 or Elmod3 KO cell lines (Supplemental Figure S2E) when compared with wild-type (WT) cells. Because we earlier identified Golgi as a site of action of overexpressed ELMOD1 in HeLa cells (East et al., 2012), we looked at markers of the cis-Golgi (GM130; Supplemental Figure S2F), trans-Golgi network (TGN) (Golgin-97; Supplemental Figure S2G), TGN/endosomes (BIG2/ARFGEF2; Supplemental Figure S2G), and recycling endosomes (RAB11-FIP1, RAB11-FIP3, or RAB11-FIP5) and again found no evidence of gross morphological alterations. Because KO of Elmod2 caused a number of readily identified phenotypes, we also examined those in Elmod1/3 KO cells. Cell cycle was not obviously altered based on propidium iodide staining and DNA content analysis using flow cytometry (Supplemental Figure S3A). Neither were ciliary rootlets altered, as they were intact and appeared similar in size around centrosomes that were not significantly more separated than centrosomes in WT cells (Supplemental Figure S3, B and C), in contrast to what was observed in Elmod2 KOs (Turn et al., 2021). Cold and nocodazole sensitivity of microtubules are obvious in MEFs lacking ELMOD2 (Turn et al., 2020), but neither phenotype was evident in lines deleted for Elmod1, Elmod3, or both. Number and size of focal adhesions also are unaltered from WT in Elmod1/3 KO and DKO cells (Supplemental Figure S4).
ELMOD1 and ELMOD3 are localized in ciliary compartments of mouse photoreceptor cells
Previous data from our lab revealed novel functions for ELMOD2 as a negative regulator of ciliogenesis as its deletion caused increases in ciliation. Motivated by this and the genetic data linking both ELMOD1 and ELMOD3 to defects in stereocilia, we investigated potential roles for ELMOD1 and ELMOD3 in cilia. To begin to test this model, we searched for novel localizations for ELMOD1 and ELMOD3. ELMODs are low-abundance proteins, with ELMOD1 estimated to be present <0.01% of total cell protein in HeLa cells (East et al., 2012). Despite evidence of the expression of all three transcripts in MEFs from RNA-seq data (though levels of the mRNAs were so low as to challenge accurate quantification, Ct ≥ 35), they are absent from a database of more than 8400 proteins identified in MEFs by mass spectrometry (unpublished observation), presumably due to low protein expression. Neither commercial nor in-house ELMOD1 and ELMOD3 antibodies yielded specific signal by immunofluorescence in MEFs or in immunoblots of total MEF lysates. We next turned to the well-studied model for proteins implicated in cilia, retinal photoreceptor cells, to identify specific localization of each protein.
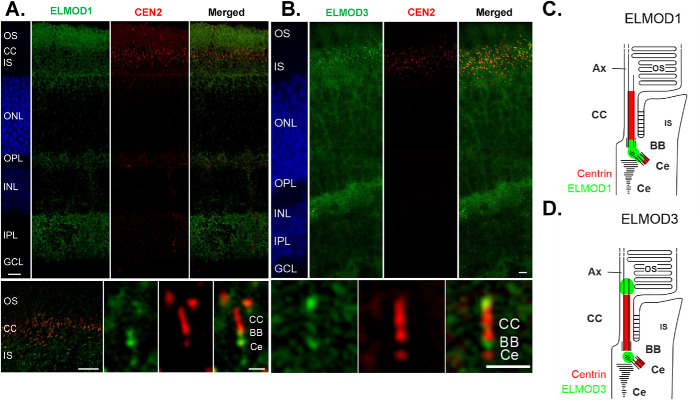
Mouse retinas were prepared and processed as described previously (Turn et al., 2021) and in Materials and Methods. In cryosections through the retina of the eGFP-Centrin mouse, eGFP-Centrin was visualized to identify the connecting cilium and basal body. Counterstaining with 4′,6-diamidino-2-phenylindole (DAPI) was used to demonstrate the nuclear layers. ELMOD1 (Figure 1A) and ELMOD3 (Figure 1B) were found in the plexiform layers of the retina (Figure 1, IPL), where a high density of synapses localize, though each of the ELMODs is more prominent in the photoreceptor region (Figure 1, IS, CC, OS). Higher magnification of this region revealed that both ELMOD1 and ELMOD3 were present at the base of the connecting cilium (Figure 1, A and B, bottom panels), which links the outer segments (=modified primary cilium) and inner segments of photoreceptor cells and resembles an elongated transition zone (TZ) of primary cilia (Turn et al., 2021). There, ELMOD1 staining was found in the basal body and the daughter centriole, as well as in between the basal body and the daughter centriole (Figure 1, A, bottom panel, and C). In contrast, we found ELMOD3 not only in the linkage between the basal body and the daughter centriole but also in an extension of the connecting cilium toward the axoneme of the photoreceptor outer segment (Figure 1, B, bottom panel, and D), which is consistent with the previously reported localization of ELMOD2 (Turn et al., 2021).
FIGURE 1:
ELMOD1 and ELMOD3 show overlapping, but also distinctive, localization patterns in mouse retinal photoreceptor cells. Retinas of eGFP-CETN2 mice were processed and analyzed with a deconvolution microscope, as described in Materials and Methods, to determine ELMOD1 and ELMOD3 localization. (A, large top panel) ELMOD1 immunolabeling of cryosections through the retina revealed a prominent staining in a region of the photoreceptor cells comprising the outer segment (OS), the connecting cilium (CC, red, Cen 2), and the inner segment (IS). In addition, fade staining was observed in the outer and inner plexiform layer (OPL, IPL), while other retinal layers, the outer and inner nuclear layer (ONL, INL, blue DAPI staining) and ganglion cell layer (GCL), did not show substantial staining. (Bottom panel) Higher magnification of photoreceptor region (left) and of the ciliary part of a photoreceptor cell (right) revealed ELMOD1 localization at the base of the connecting cilium (CC) in the basal body (BB) and the adjacent daughter centriole (Ce) (both red) as well as within the bridge in between. (B, large top panel) ELMOD3 immunolabeling in a transgenic eGFP-CETN2 mouse retina revealed staining at the region containing photoreceptor cells and both plexiform layers (OPL, IPL) (bottom panel). Higher magnification of the ciliary part of a photoreceptor cell showed that ELMOD3 was restricted to a localization between the BB and Ce at the base of the CC. In addition, ELMOD3 was stained at the base of the outer segment, a compartment above the CC. (C, D) Scheme of ELMOD1 and ELMOD3 photoreceptor cells. Scale = A and B, top panels: 15 µm; bottom panels (higher magnifications), 5 µm.
In conclusion, ELMOD1 and ELMOD3 show distinct localization patterns but overlap in their localization at the base of the connecting cilium. Both the base and the tip of the connecting cilium are associated with numerous signaling pathways. Interestingly ELMOD1, ELMOD2, and ELMOD3 all show joint localization around the connecting cilium. This could indicate a common site of action for the three proteins at the base and for ELMOD2 and ELMOD3 at the tip of the connecting cilium.
Both Elmod1 KOs and Elmod3 KOs cause decreased ciliogenesis at a late step in licensing
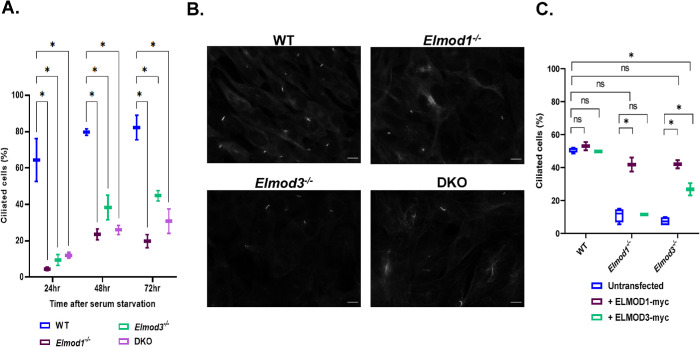
Given the evidence for ELMOD1 and ELMOD3 localization to basal bodies in retinal cells and roles of ELMOD2 in cilia, we next examined whether primary cilia form at normal rates and with typical protein content in Elmod1 and Elmod3 KO lines. Using acetylated tubulin (Ac Tub) antibody to mark the axonemes, we monitored the percentage of ciliated cells after 24, 48, and 72 h of serum starvation (0.5% fetal bovine serum [FBS]). We found that ciliation was strongly decreased in Elmod1 and Elmod3 single KO MEFs, with <10% of cells on average having a cilium after 24 hr of serum starvation, compared with >60% in WT controls (Figure 2, A and B). While the percentage of ciliated cells increased at later times of serum starvation in both WT populations and each of the KO clones, the latter never approached levels seen in WT cells (Figure 2A). In the course of this work, we visually inspected hundreds of cells in biological replicates from multiple clones for each genotype and observed no obvious, consistent differences in ciliary length.
FIGURE 2:
Elmod1 and/or Elmod3 KO in MEFs cause decreased ciliation. Cloned lines of WT, Elmod1 KO, Elmod3 KO, and DKO MEFs were serum starved for either 24, 48, or 72 h to induce ciliogenesis before staining for Ac Tub (to mark cilia) as described in Materials and Methods. (A) The percentages of cells with cilia were scored (two cell lines per genotype, 100 cells each) at each time point. The experiment was performed in duplicate (N = 2), and results were graphed as box-and-whisker plots. (B) Representative images of WT and KO cells were collected at 60× magnification by wide-field imaging 72 h after serum starvation. Scale bar = 10 µm. (C) Plasmids directing expression of ELMOD1-myc, ELMOD3-myc, or GFP were transiently transfected into WT, Elmod1 KO, or Elmod3 KO lines (two each). One day later, they were serum starved for 24 h and then stained and scored for cilia as in panel A. Only cells with evident protein expression (myc positive) were scored. Experiments were scored in duplicate (N = 2), 100 cells per replicate. Results were graphed as box-and-whisker plots using GraphPad. Statistical significance was assessed via one-way ANOVA, performing multiple comparisons. ns = not significant, *p < 0.05.
We performed rescue experiments in which we transiently expressed either ELMOD1-myc or ELMOD3-myc proteins in KO, DKO, and WT MEFs and scored ciliation after 24 h of serum starvation. Expression of ELMOD1-myc in Elmod1 KO lines was sufficient to bring ciliation percentages close to those of parental WT controls, while expression of ELMOD1-myc in WT cells had no significant effect on ciliation (Figure 2C). Because we scored all myc-positive cells, even those staining weakly, there may be a lower limit of expression required to bring ciliation percentages fully up to those seen in WT cells. Expression of ELMOD3-myc was also tested and found capable of reversing the decreased ciliation in Elmod3 deleted cells, though only about halfway back to levels seen in WT cells (Figure 2C). Interestingly, expression of ELMOD1-myc also rescued ciliation in cells deleted for Elmod3 (Figure 2C). Indeed, the effect of ELMOD1-myc expression on restoration of ciliation in Elmod3 KO lines was comparable to that seen in ELMOD1-myc rescue of Elmod1 KOs and of ELMOD3-myc in Elmod3 KO cells (Figure 2C). In contrast, expression of ELMOD3-myc in Elmod1 KOs failed to restore ciliation (Figure 2C). We compared the relative levels of ELMOD1-myc and ELMOD3-myc expression by immunoblotting for the myc tag in total cell lysates of WT cells 24 h after transient expression of each protein (Supplemental Figure S5). This revealed that ELMOD1-myc is expressed to considerably higher levels than is ELMOD3-myc (Supplemental Figure S5), perhaps explaining its greater potency in rescue. Unfortunately, the low levels of endogenous ELMOD protein expression in MEFs prohibited quantitative comparisons between endogenous and exogenous protein expression. We currently interpret these findings as more consistent with ELMOD1 and ELMOD3 acting on a common pathway but at distinct steps, as explained in more detail in the Discussion.
Having identified that loss of ELMOD1 and ELMOD3 leads to ciliation defects, we next sought to monitor the early steps required for licensing of ciliogenesis in mutant MEFs. We previously showed that ELMOD2 regulates the earliest stages of ciliogenesis, so we predicted that ELMOD1 and/or ELMOD3 may also be acting early in ciliogenesis. We examined recruitment of CEP164 (normally to nascent distal appendages) and loss of CP110 or “uncapping,” both of which are critical for licensing of axoneme growth (Graser et al., 2007; Humbert et al., 2012; Schmidt et al., 2012; Cajanek and Nigg, 2014; Lo et al., 2019). We found no differences in the degree of either CEP164 recruitment (Supplemental Figure S5A) or CP110 loss (Supplemental Figure S5B) in any of the mutant MEF lines. We also stained cells for CEP290, a marker of the TZ at the base of cilia, and also found no changes in the extent of its recruitment (Supplemental Figure S6C). Together, we interpret these results as evidence that the ciliation defect seen in Elmod1/3 KO lines is downstream of the uncapping of CP110 at the distal appendages and of TZ construction involving CEP290.
KO of Elmod1 or Elmod3 alters the protein composition in cilia
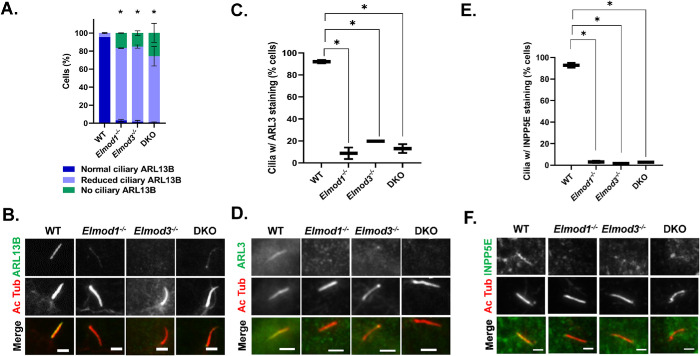
We next sought to determine whether loss of ELMOD1 or ELMOD3 also had effects on ciliary protein composition. To monitor the protein content of cilia, we first examined ARL13B, which is bound to the ciliary membrane through N-terminal palmitoylation and localizes along the entire ciliary length (Cevik et al., 2010; Larkins et al., 2011; Mariani et al., 2016; Roy et al., 2017; Fisher et al., 2020). We observed reduced ciliary ARL13B staining in both KO and the DKO lines, revealing compromised import (or increased export) in cells that lack ELMOD1 and/or ELMOD3 (Figure 3, A and B). In WT cells, all Ac Tub staining overlapped with ARL13B staining, indicating that WT cilia contain ARL13B, with little variation in the intensity of staining. In contrast, each of the Elmod1, Elmod3, and DKO lines examined displayed a large majority of Ac Tub–positive cilia in which staining of ARL13B was clearly reduced and was even undetectable in ∼15–20% of cilia (Figure 3A). These differences were so stark, at least in part due to the strong staining of ciliary ARL13B in WT cells, that the quantification of pixel intensities was unnecessary. In DKO cells, more cilia (∼26%) were devoid of ARL13B, though the difference in levels from the single KO lines was not statistically significant. These findings were further confirmed by quantification of ARL13B pixel intensity (Supplemental Figure S11).
FIGURE 3:
Elmod1 or Elmod3 KO causes loss of ARL13B, ARL3, and INPP5E from cilia. Cells (two lines per genotype, from different guides) were grown to ∼80% confluence before inducing ciliation via serum starvation for 72 h and then staining for Ac Tub and either ARL3, ARL13B, or INPP5E. (A) ARL13B levels were binned as normal (readily identifiable before confirming with the Ac Tub channel), reduced (identifiable as ciliary only upon confirmation with Ac Tub staining), or absent (no staining evident that overlaps with Ac Tub). Scoring was performed on 100 cells for each of two cell lines in replicate (N = 2), and error bars represent SEM. (B) Cells treated as in A, stained for Ac Tub and ARL13B, were collected by wide-field microscopy at 100×, and representative images are shown. Note that in these images the Elmod1 KO and DKO cells were scored as reduced, as faint staining is evident, while in the Elmod3 KO cell it is not. (C) Ciliary ARL3 was scored in cells treated as described in panel A. Owing to the weaker overall staining of ARL3, scoring was binned as either present or absent from cilia, identified via Ac Tub staining. Scoring was performed on 100 cells for each of two cell lines in replicate (N = 2), and box-and-whisker plots from the scoring are shown; error bars = minimum and maximum. (D) Representative images from cells stained for ARL3 and Ac Tub are shown. (E) INPP5E and Ac Tub staining was performed as described in Materials and Methods, and scoring of INPP5E presence in cilia was performed as described in panel C, being binned as either present or absent. (F) Representative images from cells stained for Ac Tub and INPP5E are shown. Statistical significance for all data was determined via one-way ANOVA, performing multiple comparisons, using GraphPad Prism Software. *p < 0.05. For all images shown, scale bar = 10 µm.
ARL13B can act as a GEF for ARL3 (Miertzschke et al., 2014; Gotthardt et al., 2015; Ivanova et al., 2017). Import of ARL3 into cilia is thought to be mediated by simple diffusion, while its retention may be ARL13B-dependent (Kosling et al., 2018; Gigante et al., 2020). We examined ARL3 localization in cilia and found it to be strongly decreased in Elmod1 and Elmod3 KO lines (Figure 3, C and D). Because staining of ARL3 in cilia is not as intense as that of ARL13B, we scored in a binary manner, “normal” versus “reduced,” with the latter being scored only when unambiguously below levels seen in most WT cells. Despite this conservative approach to scoring, the loss of ARL3 staining was quite strong, with more than 80% of cilia having reduced ARL3 staining, compared with WT cells where fewer than 10% displayed lower ciliary ARL3. Thus, both ARL13B and ARL3 are strongly reduced in cilia, due to either decreased import or increased export (failed retention).
Among other actions in cells, ciliary ARL3 binds the prenylated cargo transporter PDE6D, causing it to release its cargo at that site (Gotthardt et al., 2015; Fansa et al., 2016; Dyson et al., 2017). Perhaps the best known prenylated cargo involved is the lipid phosphatase INPP5E, active in modifying the lipid composition of cilia (Zhang et al., 2007; Bielas et al., 2009; Thomas et al., 2014; Garcia-Gonzalo et al., 2015; Stephen and Ismail, 2016; Alkanderi et al., 2018; Kosling et al., 2018; Ukhanov et al., 2022). Once in cilia, INPP5E is thought to bind directly to ARL13B, which aids in its retention (Humbert et al., 2012; Qiu et al., 2020; Fujisawa et al., 2021). Thus, we also analyzed the ciliary content of INPP5E in Elmod1, Elmod3, and DKO cells. We found a near complete loss of INPP5E staining in each of the KO lines (Figure 3, E and F). The reduction in INPP5E in cilia may be an indirect result from the loss of ARL13B or ARL3, or it may result from other defects (see below).
Cilia are required to transduce vertebrate Hedgehog (Hh) signaling, which is regulated by both ARL13B and INPP5E (Huangfu et al., 2003; Huangfu and Anderson, 2005; Caspary et al., 2007; Garcia-Gonzalo et al., 2015; Constable et al., 2020). GLI3 is one of three transcription factors that localize to cilia and increase at the ciliary tip in response to Hh stimulation (Haycraft et al., 2005). We examined ciliary GLI3 in the KO lines and found no differences in GLI3’s ciliary staining or enrichment at the ciliary tip (Supplemental Figure S7). We also stained for markers of the intraflagellar transport complexes IFT-A (IFT140) and IFT-B (IFT88) and found no differences between WT and KOs in their staining in cilia (Supplemental Figure S8). Taken together, these data show that ciliary protein content is altered in cells lacking Elmod1 and/or Elmod3, but it appears to be a select group of proteins with previously identified ciliary links that are reduced in cilia.
INPP5E and IFT140 accumulate in the Golgi in Elmod1/3 and DKO MEFs
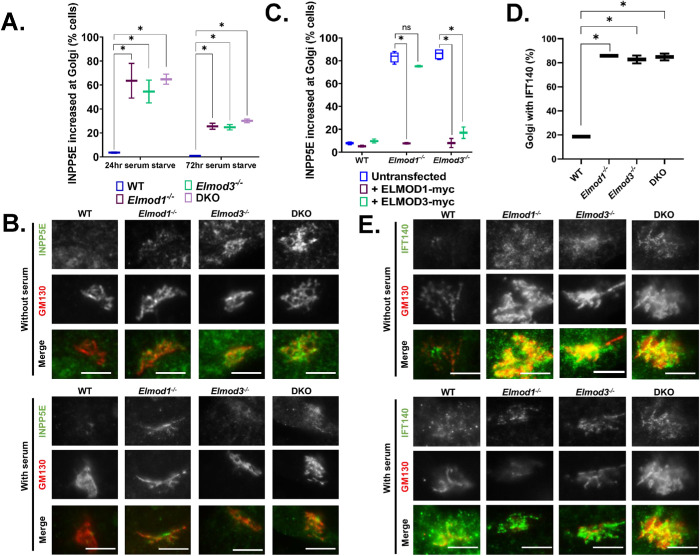
As a C-terminal isoprenylated protein, INPP5E traffics throughout the cell as cargo of the PDE6D transporter and is deposited onto membranes upon either ARL2 or ARL3 binding to PDE6D (Zhang et al., 2012; Gotthardt et al., 2015; Fansa et al., 2016; Hanke-Gogokhia et al., 2016; Stephen and Ismail, 2016; Wright et al., 2016; Kosling et al., 2018). Thus, the absence of INPP5E from cilia might be explained by the lack of ARL3, if required for dislocation from PDE6D, or lack of retention offered by binding to ARL13B (Humbert et al., 2012; Qiu et al., 2020). While staining ELMOD1/3 KO lines for INPP5E, we noticed an increase in INPP5E staining intensity in serum-starved cells (24 h) that colocalized with the Golgi marker GM130 (Figure 4, A and B). Under these conditions, INPP5E staining at the Golgi in WT cells was weak and usually absent. In contrast, the presence of INPP5E at the Golgi is evident in well over half of all Elmod1, Elmod3, and DKO cell lines after 24 h of serum starvation (Figure 4, A and B). Note that INPP5E staining at cilia was performed using paraformaldehyde (PFA) as fixative, while its presence at the Golgi was evident only after methanol fixation. Interestingly, by 72 h this effect was reduced, with only ∼25–30% of Elmod1, Elmod3, or DKO cells displaying INPP5E at the Golgi. This reduction over time suggests a transience to this phenomenon that may result from a pulse of new INPP5E synthesis in response to serum starvation or a redistribution within the cell. A more detailed time course was not investigated at this time but may be an interesting future direction to pursue.
FIGURE 4:
INPP5E and IFT140 accumulate at the Golgi in Elmod1 or Elmod3 KO lines. Cells were serum starved for 24 or 72 h, followed by staining for GM130 and either INPP5E (A–C) or IFT140 (D–F). (A) The presence of INPP5E colocalizing with GM130 was scored for each genotype, using two different clones of each and scoring 100 cells in duplicate experiments (N = 2). Results were graphed as box-and-whisker plots via GraphPad Prism. *p < 0.05, calculated via one-way ANOVA with multiple comparisons. (B) Representative images of WT and KO cells serum starved for 72 h and stained for GM130 and INPP5E were collected at 100× magnification with wide-field microscopy. Scale bar = 10 µm. (C) WT, Elmod1, and Elmod3 KO cells were transfected with plasmids directing the expression of ELMOD1-myc or ELMOD3-myc. Twenty-four hours later, the cells were serum starved for 24 h before fixing and staining for GM130 and INPP5E. The increased abundance of IFT140 at the Golgi was scored as in panel A and graphed as box-and-whisker plots. *p < 0.05, calculated via one-way ANOVA with multiple comparisons. (D) The presence of IFT140 colocalizing with GM130 was scored for each genotype, using two clones of each KO line and one WT line, scoring 100 cells in duplicate experiments (N = 2). Results were plotted as box-and-whisker plots. *p < 0.05, calculated via one-way ANOVA with multiple comparisons. (E) Images of IFT140 at the Golgi, using GM130 as marker, in each line. Cells were serum starved for 24 h and stained for GM130 and IFT140.
We performed rescue experiments using transient expression of either ELMOD1-myc or ELMOD3-myc. Expression of either protein had no impact on the percentages of cells displaying staining of INPP5E at the Golgi in WT cells (Figure 4C). Expression of either protein in lines deleted for that protein reduced the staining of INPP5E at the Golgi to near WT levels (Figure 4C). As we observed for rescue of overall ciliation percentages, expression of ELMOD1-myc reversed the increased levels of INPP5E in both Elmod1 and Elmod3 KO lines, while ELMOD3-myc expression rescued this phenotype only in Elmod3 KOs (Figure 4C).
The increased localization of INPP5E at the Golgi in Elmod1 and Elmod3 KO lines also prompted us to look for the presence of other ciliary proteins at the Golgi. We found that IFT140, a core component of IFT-A, is also increased at the Golgi in Elmod1/3 KO lines compared with WT cells (Figure 4, D and E). In marked contrast, we did not detect any increased staining for other IFT proteins at the Golgi, including IFT81, IFT88, and IFT144 (unpublished data). Thus, while the presence of IFT140 at the Golgi is increased in KO lines, it appears to be there independently of the IFT-A complex. The increased Golgi staining of IFT140 was also evident in KO lines without serum starvation (Figure 4E), suggesting a block or delay in traffic that is independent of induced ciliogenesis.
In the process, we also observed strong IFT140 staining that was typically adjacent to, but rarely overlapped with, the Golgi marker (GM130) and was more restricted in space. Costaining of IFT140 and rootletin in WT cells demonstrates that IFT140 is present at ciliary rootlets and that this likely is the source of the strong staining near the Golgi (Figure 4E). IFT140 staining of rootlets was unaltered from that seen in WT cells in any of the Elmod1/3 KO lines (Supplemental Figure S9).
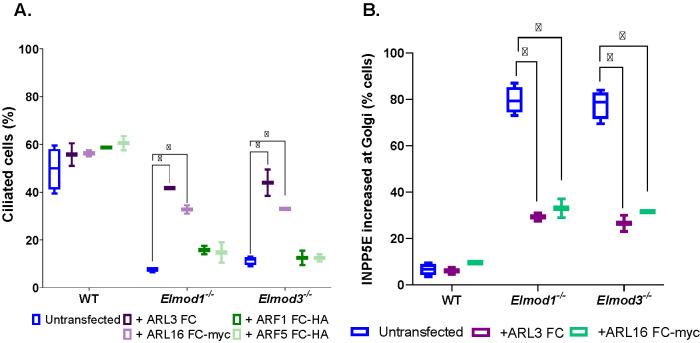
Expression of either activated ARL3 or activated ARL16 rescues ciliation defects and accumulation of INPP5E and IFT140 at the Golgi in Elmod1 KO or Elmod3 KO lines
ARF GAPs have dual functions in cells: both to terminate/dampen signaling from specific GTPases to which they bind and to propagate the downstream signal, most commonly via recruitment of other proteins (East and Kahn, 2011; Sztul et al., 2019). In efforts to identify the GTPase(s) acting in a GAP-sensitive pathway, it has proven to be fruitful to test for rescue of phenotypes resulting from deletion of the GAP using activated mutants of different GTPases. Because the commonly used “Q to L” mutants (corresponding to Q71 in ARF1 or Q61 in HRAS) are often quite toxic in cells, we use “fast cycling” mutants that become activated independently of an ARF GEF and, unlike the Q to L mutants, can still cycle between active and inactive conformations and thereby propagate the signal without locking up the pathway (Santy, 2002; Turn et al., 2020, 2021). Transient transfections were used to express fast-cycling mutants of ARF1 (ARF1[T161A]-HA), ARF5 (ARF5[T161A]-HA), ARL3 (ARL3[L161A]-myc), or ARL16 (ARL16[R153A]-myc) in WT and KO MEFs. Expression of either activated ARL3-myc or ARL16-myc restored ciliation percentages near WT levels (Figure 5A), functionally linking ARL3 and ARL16 to the actions of ELMOD1 and ELMOD3 in ciliation. In contrast, neither ARF1[T161A]-HA nor ARF5[T161A]-HA expression had any effect on ciliation of WT, Elmod1, or Elmod3 KO cells (Figure 5A), supporting the conclusion that there is specificity to the GTPases acting with ELMOD1/3 in ciliation. Finally, we examined whether the expression of the same two activated ARF family members, ARL3 and ARL16, that restored ciliation also reversed the accumulation of INPP5E at the Golgi. We found that each of these GTPases partially reversed the increased staining of INPP5E at the Golgi (Figure 5B). Thus, deficiencies in ciliation correlate well with increased INPP5E localization at the Golgi, and each is reversed either by expression of deleted ELMOD or by activated ARL3 or ARL16, suggesting functional linkage between these two lesions.
FIGURE 5:
The ciliation defect in Elmod1 or Elmod3 KO cells can be reversed upon transient expression of activated ARL3 or ARL16. (A) WT, Elmod1 KO, and Elmod3 KO lines were transfected with plasmids directing expression of fast-cycling mutants of ARF1-HA, ARF5-HA, ARL3, or ARL16-myc. The next day, cells were serum starved for 24 h and then fixed and stained for Ac Tub and the corresponding tag. (B) The same procedure was carried out as described for panel A, except that only fast-cycling mutants of ARL3 and ARL16 were examined and that staining was for the expressed protein and INPP5E. Only those cells expressing exogenous proteins were scored. Two lines for each genotype were scored, 100 cells each, and the experiment was repeated in duplicate (N = 2). The box-and-whisker plot shows the averages. *p < 0.05, calculated via one-way ANOVA with multiple comparisons.
DISCUSSION
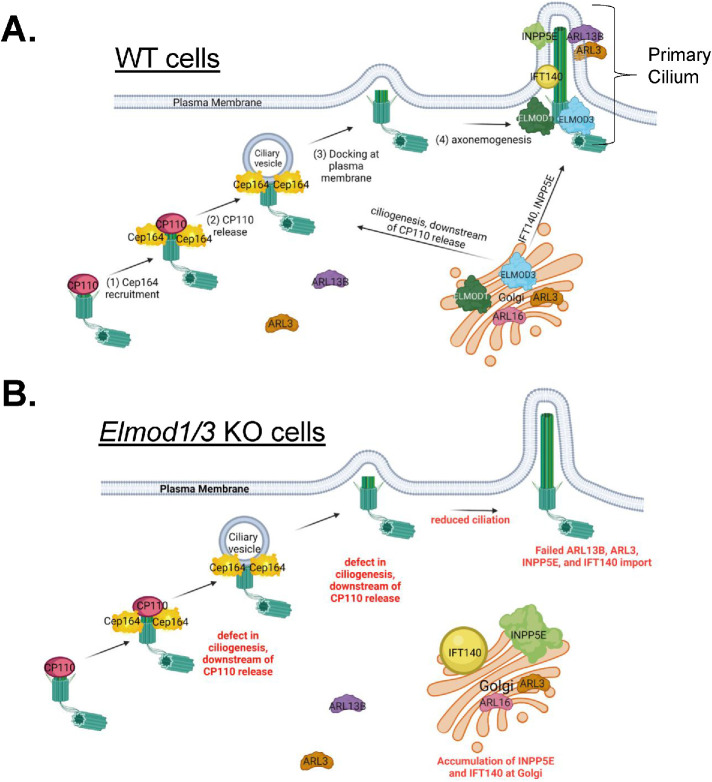
We developed KO MEF lines to define cellular roles of the ARF GAPs ELMOD1 and ELMOD3 in mammalian cells. We found that Elmod1 and Elmod3 KOs have decreased ciliation, loss of import, or retention of several ciliary proteins that are important in signaling and accumulation of some of the same proteins at the Golgi. We also identified likely roles for ARL16 and ARL3 in ELMOD1 and ELMOD3 function. In several respects, these results parallel studies of the other paralogue, ELMOD2, in that ELMOD1 and ELMOD3 appear to act at more than one place and in concert with more than one GTPase within a given cell (Schiavon et al., 2019; Turn et al., 2020, 2021). However, ELMOD1 and ELMOD3 play roles distinct from those described for ELMOD2, most notably with opposite effects on ciliation, as summarized in Table 1. We propose a model whereby ELMOD1 and ELMOD3 act at cilia to regulate ciliogenesis and protein import and also at the Golgi to regulate the export of specific cargoes (INPP5E and IFT140) that may strongly influence ciliary generation and protein content (Figure 6). These findings clearly call for future studies to better define molecular mechanisms of all three ELMODs in ciliogenesis, ciliary protein recruitment, Golgi traffic, and related processes in multiple tissues and cell types.
TABLE 1:
Summary of phenotypes found in MEFs deleted for Elmod1 KO, Elmod2 KO, Elmod3 KO, and Elmod1/Elmod3 DKO MEFs using CRISPR/Cas9.
| Phenotype | Elmod1 KO | Elmod3 KO | DKO | Elmod2 KO |
|---|---|---|---|---|
| Mitochondrial morphology (HSP60 staining) | Normal | Normal | Normal | Fragmented |
| Centrosome number (γ-tubulin staining) | Normal | Normal | Normal | Supernumerary |
| DNA content (propidium iodide staining) | Normal | Normal | Normal | Polyploid |
| Nuclear number (Hoechst staining) | Normal | Normal | Normal | Multinucleated |
| Rootlet morphology (Rootletin staining) | Normal | Normal | Normal | Fragmented |
| Microtubule sensitivity (cold/nocodazole) | Normal | Normal | Normal | Increased |
| Ciliation (% of cells) (Ac Tub staining) | Decreased | Decreased | Decreased | Increased |
| Multiciliation) (Ac Tub staining) | Normal | Normal | Normal | Multiciliated |
| Ciliogenesis licensing (Cep164, CP110 staining) | Normal | Normal | Normal | Increased (CEP164↑ CP110↓) |
| ARL13B/ARL3 in cilia (ARL13B/ARL3 staining) | Decreased | Decreased | Decreased | Normal |
| INPP5E in cilia (INPP5E staining) | Decreased | Decreased | Decreased | ND |
| lFT140 at basal body (IFT140 staining) | Normal | Normal | Normal | ND |
| IFT140 at Golgi (IFT140 staining) | Increased | Increased | Increased | ND |
The top of the chart indicates the cell lines, the leftmost column lists the phenotypes assayed and markers used, and phenotypes are indicated under each cell line as normal (no change from WT) or other. ND, Not determined.
FIGURE 6:
Model for ELMOD1 and ELMOD3 function as regulators of (A) ciliogenesis and (B) traffic of key ciliary cargoes. We propose that ELMOD1 and ELMOD3 are acting in concert and in at least three processes to regulate ciliogenesis and the traffic of key ciliary cargoes from the Golgi to cilia. With respect to ciliogenesis, we propose that ELMOD1 and ELMOD3 are regulating ciliogenesis from the basal body, after the release of CP110 from distal appendages. ELMOD1 and ELMOD3 are also required for ARL13B in cilia, which in turn aids in the ciliary retention of ARL3. These first two actions may be closely linked in space, though neither ARL13B nor ARL3 is required for ciliogenesis, so we consider them separate at this time. We believe that ELMOD1 and ELMOD3 also act from the Golgi to regulate export of INPP5E and IFT140, though through distinct mechanisms, because export of INPP5E requires PDE6D while export of IFT140 does not. Finally, we speculate that ELMOD1 and ELMOD3 can also act from endosomes, perhaps directly on ARL16, and that in their absence ARL16 is strongly increased on endosomal membranes. Perhaps this would result in its depletion from other sites, including the Golgi, causing delays or defects in the export of specific proteins from the Golgi. Figure was created using BioRender.
The diversity in sites of action and effects on different processes makes dissection of molecular mechanisms for ELMOD1 and ELMOD3 a challenge, as recently highlighted (Sztul et al., 2019). To begin to address such questions, we looked at localization of ELMOD1 and ELMOD3 in cultured mammalian cells and retinal tissue. We reported earlier that endogenous ELMOD1 is overwhelmingly soluble by cell fractionation of normal rat kidney (NRK) cells (East et al., 2012). Unfortunately, neither homemade nor commercial antibodies directed against ELMOD1 and ELMOD3 were sensitive enough to identify localizations of endogenous proteins in MEFs. However, ELMOD1 is at the basal body and centriole of mouse retinal cells (Figure 1, A and C). ELMOD3 staining overlapped with that of ELMOD1 at the base of the connecting cilium of the centriole in photoreceptor cells (Figure 1, B and D). The overlap between localizations of ELMOD1 and ELMOD3 in mouse retinal cells is consistent with previously reported localizations of ELMOD1 in hair cells at the cell apex (Krey et al., 2018) and ELMOD3 in stereocilia and kinocilia in the organ of Corti and inner ear (Jaworek et al., 2013). The localizations of ELMOD3 are also quite similar to what we reported earlier for ELMOD2 in mouse retina (Turn et al., 2021). Previous work revealed that endogenous ELMOD3 localizes to stereocilia and kinocilia in both inner and outer hair cells (Jaworek et al., 2013) and that green fluorescent protein (GFP)-ELMOD3 recruits to stereocilia in inner ear explants and to cortical actin structures in a confluent monolayer of Madin–Darby canine kidney (MDCK) cells (Li et al., 2018). ELMOD3 has been implicated as a target of the cilia-related RFX2 transcription factor (Chung et al., 2014) and was also localized to basal bodies in a related, high-throughput screen (Tu et al., 2018). Thus, the locations of ELMOD1 and ELMOD3 in cells and tissues are incompletely understood and may show tissue specificity. We fully expect the ELMODs to act at additional sites, likely in all cells, for example, at subcortical actin, as suggested by expression of ELMOD3-GFP in MDCK cells (Li et al., 2019). On the basis of our data, though, we predict that these players are commonly found at the basal body/TZ, where they may act on aspects of ciliogenesis and regulated protein import/export. Interestingly, we also note strong staining of ELMOD1/3 at the inner plexiform layer of the retina, which is rich in synapses. These findings may lay the groundwork for future studies designed to look at potential roles for ELMODs at synapses in neural tissues.
We describe three cilia-related consequences from loss of either ELMOD1 or ELMOD3 that likely represent distinct actions of these ARF GAPs: decreased ciliation, loss of several proteins from cilia, and increased accumulation of some of the same proteins at the Golgi. We found that the defect in ciliation occurs at a step downstream of “uncapping” of the mother centrosome. Potential later steps where they might act include the expansion of the ciliary pocket or extension of the axoneme, particularly as ARF family GTPases play multiple roles in vesicular membrane traffic (Donaldson and Jackson, 2011), and ARL2 in regulation of microtubules (Francis et al., 2017a,b). In contrast, the other paralogue ELMOD2 is required quite early in the process, upstream of CEP164 recruitment, and its absence results in increases both in the percentage of cells that ciliate as well as in multiciliation, neither of which were phenotypes found in either ELMOD1 or ELMOD3 KOs. Furthermore, ELMOD2 localizes to rootlets, and its deletion causes striking fragmentation of ciliary rootlets with centrosome separation (Turn et al., 2021). In contrast, neither Elmod1 nor Elmod3 KOs show such effects on rootlet integrity and centrosome separation (Supplemental Figure S3, B and C). Together, these data point to a combination of discrete and overlapping functions between members of the ELMOD family.
We also discovered changes in the ciliary content of at least three proteins in cells lacking ELMOD1 and/or ELMOD3: ARL13B, ARL3, and INPP5E. Note that this is not a universal defect in all ciliary protein traffic, as such factors as Ac Tub, IFT88, IFT140, and GLI3 show normal localization, as do the regulators of early ciliogenesis (i.e., CEP164 and CP110). The proteins that are lost are not all transported to cilia by a common mechanism, so a single site of action seems unlikely. ARL3, at ∼20 kDa, is small enough to diffuse across the TZ, and its abundance in cilia may be increased as a result of its binding/activation by ARL13B with its ARL3 GEF activity (Gotthardt et al., 2015). If this is the case, then the loss of ARL3 may be simply secondary to the decreased ciliary abundance of ARL13B. INPP5E is a farnesylated cargo that is trafficked to cilia by the transporter PDE6D, which releases cargoes upon binding to activated ARL3 (Kosling et al., 2018). However, ciliary targeting of INPP5E is dependent on ARL13B and not ARL2 or ARL3 (Humbert et al., 2012; Fujisawa et al., 2021). Thus, the decreased ciliary accumulation of INPP5E may also be an indirect result from the loss of ARL13B. How ARL13B gets to and is retained at cilia is less clear, though a role for Ahi1 acting at the TZ to affect ARL13B levels has been shown, as have links to Tulp3 and RPGRIP1L (Andreu-Cervera et al., 2019; Hwang et al., 2019; Munoz-Estrada and Ferland, 2019). ARL13B becomes membrane associated after palmitoylation of cysteines near its N-terminus and likely acts more like a membrane protein than the others (Cevik et al., 2010; Mariani et al., 2016; Roy et al., 2017). While mutations in ARL13B can cause decreased ciliation and shortened cilia, none of the other proteins with altered localization in Elmod1/3 KO lines is required for growth of a cilium of normal length (Jonassen et al., 2012). Thus, we posit that the traffic defect(s) in Elmod1 and Elmod3 KOs, whether at the Golgi or at the ciliary base, represent a lesion distinct from that which decreases ciliogenesis overall, while the decrease in percentage of ciliated cells after serum starvation in Elmod1/3 KO lines may be common to the losses observed in Arl13b null cells (Caspary et al., 2007; Larkins et al., 2011; Mariani et al., 2016).
INPP5E colocalizes extensively with β-COP at the Golgi in 3T3-L1 adipocytes (Kong et al., 2006) and Tera-1 cells (Kong et al., 2000). In contrast, in RPE-hTERT cells, INPP5E was found at the ciliary axoneme with only minimal nonciliary staining (Bielas et al., 2009). INPP5E is cytosolic and at the plasma membrane in macrophages (Horan et al., 2007). Thus, while farnesylated INPP5E traffics throughout the cell while bound to PDE6D, just when and where these two first encounter one another are unknown. The accumulation of INPP5E and intraflagellar transport (IFT)140 at the Golgi in Elmod1 and Elmod3 KO cells may have physiologically important consequences to IFT-A assembly and functions as well as the regulation of phosphatidylinositol phosphates in multiple membranes, making clean dissection of molecular mechanisms challenging, but worthy of further study. That is, the increased abundance of INPP5E at the Golgi and its loss from cilia are expected to cause alterations in the lipid composition of membranes at each compartment, with likely indirect consequences to localization and actions of any number of other proteins.
The finding that IFT140 and INPP5E accumulate at the Golgi in both Elmod1 and Elmod3 KO cells could explain their loss from cilia. We can detect INPP5E staining at the Golgi in WT MEFs, though it is clearly much fainter and less consistently present at the Golgi than in Elmod1/3 KO lines. This is consistent with a model in which IFT140 normally traffics through the Golgi, despite the lack of an identifiable membrane-binding motif. Little is known about how IFT140 is moved through cells and assembles into the core IFT-A complex. In contrast to INPP5E, IFT140 has not previously been shown to localize to Golgi. Future studies into how IFT140 is trafficked will provide critical new insight into the mechanisms by which IFT-A import occurs—an understudied question.
These findings should raise awareness of the potential for proteins acting at the Golgi to have consequences in ciliary biology. A clear precedent has been set by studies of IFT20, which localizes to the basal body as well as the Golgi, where it works with GMAP210 in the sorting and traffic of proteins to ciliary membranes (Follit et al., 2006, 2008). The actions of IFT20 and IFT140 were compared, and while each was found to be critical for membrane protein traffic to cilia, IFT20 was found to act at the Golgi while IFT140 acted from the plasma membrane (Crouse et al., 2014). In this case, though, it was likely the IFT-A complex and not IFT140 acting alone that was under study, as IFT140 deletion would remove both pools. Interestingly, IFT20 has more recently been found to play a role in integrin recycling, cell migration, and focal adhesion dynamics, potentially linking these membrane dynamics to ciliary protein traffic (Su et al., 2020). Open questions include how IFT140 associates with membranes (as it lacks a transmembrane domain as well as classic acylation and prenylation motifs), as well as how and where it assembles into the IFT-A core complex.
ELMOD1 and ELMOD3 display differences in specific activities against ARFs and ARLs in the in vitro GAP assay (Ivanova et al., 2014) yet almost identical phenotypes when deleted in MEFs. The simplest model to emerge from these results is that they act in a common pathway, though most likely on different steps. This is also supported by the findings that the DKO lines do not display stronger phenotypes than either KO alone. That expression of ELMOD1-myc rescues the loss of ciliation in both Elmod1 and Elmod3 KO lines is also consistent with them sharing a common pathway and possibly with ELMOD1 acting downstream of ELMOD3, though the fact that ELMOD1-myc is expressed to higher levels than ELMOD3-myc may provide an alternate explanation for this finding. The partial rescue of Elmod3 KO lines by ELMOD3-myc, and perhaps even its inability to reverse the ciliation defect in Elmod1 KO lines, may result from lower levels of expression. Finally, we cannot exclude the possibility that while both proteins act on a common step or in a common pathway, they may have different specific activities as GAPs and thus require different levels of expression for full rescue.
The finding that Elmod1 KO and Elmod3 KO ciliary defects are rescued by expression of either activated ARL3 or ARL16 is also consistent with ELMOD1/3 sharing a common pathway and strongly implicates these two GTPases in that pathway. The differences in functions identified for ELMOD1 and ELMOD3 versus ELMOD2 (Table 1) likely result from their functional interactions with different ARF family GTPases: with ELMOD2 acting with ARL2 and ARF6 (Schiavon et al., 2019; Turn et al., 2020, 2021) and ELMOD1/3 acting with ARL3 and ARL16. A broader sampling of the 30 mammalian ARF family GTPases would be required to gain a more complete picture of the specificities for each of these GAPs/effectors in each pathway. ELMODs also have highly divergent N- and C-termini that are not thought to be involved in GTPase binding or hydrolysis. Yet, they may confer distinct spatial regulation or binding partners in cells. We interpret rescue of phenotypes resulting from KO of an ARF GAP by an activating mutant of an ARF family GTPase as evidence of their interaction in a common pathway. Work from Cardenas-Rodriguez et al. (2021) showed that, in zebrafish, mutations in Cep290 can result in increased expression of a number of proteins, including ARL3, ARL13B, and UNC119 and that exogenous expression of these same proteins can reverse ciliary phenotypes. We have not yet searched for changes in gene expression in response to KO of Elmods in MEFs.
Our findings can also be considered in light of the evolutionary history of the ELMOD family. ELMODs are ancient proteins, with between one and six genes present in a broad spectrum of eukaryotes; mammals express three, ELMOD1–3. While mammalian ELMODs exhibit some shared specificities for GTPases (Ivanova et al., 2014), there are also some striking differences in the actions of ELMOD2 from those of ELMOD1 and ELMOD3 in MEFs, and likely all cells. The lack of changes in centrosome numbers and separation, rootlet integrity, cell cycle, and microtubule sensitivities in Elmod1/3 KO cells is in marked contrast to what we recently reported in Elmod2 KO lines (Turn et al., 2020, 2021) (Table 1). The absence of changes in both CEP164 recruitment and release of CP110 is also opposite to those changes observed in ELMOD2 KO lines (Table 1). Thus, ELMOD1/3 versus ELMOD2 play roles in ciliogenesis but appear to act at distinct steps in the pathway, and their actions lead to opposite effects on ciliogenesis. Interestingly, a recent study in Arabidopsis identified roles for ELMODs in pollen aperture formation, with two paralogues apparently working together and another appearing to act in opposition to those two (Zhou et al., 2021), perhaps analogous to what we report here. From our earlier phylogenetic analyses of the ELMODs, we concluded that there was at least one, and likely two, ELMOD present in the last eukaryotic common ancestor (East et al., 2012). We speculate that the differences in functions ascribed to ELMOD2 versus ELMOD1/3 may represent a very early divergence in function within the ELMOD family in the evolution of eukaryotes.
Our characterization of ELMOD1 and/or ELMOD3 in mammalian cells has revealed a number of novel functions for these two ARF GAPs and also prompts new questions worth exploring. Our results give us a better understanding of roles for Elmod1 or Elmod3 in primary cilia function that may be disrupted in the context of disease. Our findings may provide the foundation for understanding how Elmod1 or Elmod3 mutations lead to cellular defects that propagate disease, such as hearing impairment and autism. We focused on the use of MEFs, in large part to allow direct comparisons to comparable studies on ELMOD2 but recognize the possibility of differences between cell and tissue types. While we found that ciliation was compromised in Elmod1/3 KO lines downstream of CP110 cap release, it is quite possible that detailed study, for example, using electron microscopy, may reveal defects in ciliary vesicle recruitment, axoneme extension, or other specific steps in ciliogenesis. Such studies are technically challenging and time consuming due to the small size of cilia, incomplete ciliation of most cell types, and need for serial sectioning. The finding that the increased abundance of INPP5E at the Golgi was higher after 24 h of serum starvation than after 72 h is suggestive of a transience to this phenotype, though it was evident in both regular (10%) and low (0.5%) serum conditions. One possible explanation for this may be that export of INPP5E and IFT140 from the Golgi is normally occurring at very close to maximal rates and that loss of either ELMOD1 or ELMOD3 impairs that process, resulting in their accumulation. Finally, testing a more complete set of fast-cycling mutants of the 30 mammalian ARF family GTPases for rescue of the phenotypes identified here would allow for a far better understanding of the specificities for GTPases by these GAPs in the pathways under study. We are currently working to generate such a complete collection of activating mutations, but this too will require more time and study.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Reagents, antibodies, and plasmids
All chemicals used were purchased from commercial sources. The following commercial antibodies were used in these studies: SC35 (1:500; Abcam; ab11826), HSP60 (1:1000; Stressgen; ADI-SPA-807), α-tubulin (1:1000; Sigma; T9026), γ-tubulin (1:5000; Sigma T6557 or Abcam ab11317), GM-130 (1:1,000; BD/Transduction; 610823), Golgin97 (1:500; Proteintech; 12640-1-ap), BIG2 (1:200; EMD Millipore; MABS1246), Rootletin (1:500; EMD Millipore; ABN1686), centrin clone 20H5 (1:1000; Sigma; 04-1624), Ac Tub (1:1000; Sigma; T5192), α-myc (1:1000; Invitrogen R950-25 or Abcam ab9132), CEP164 (1:100; Santa Cruz; sc-515403), CP110 (1:100; Proteintech; 66448-1-ig), CEP290 (1:100; Proteintech; 22490-1-ap), ARL13B (1:1000; Proteintech; 17711-1-AP), INPP5E (1:100; Proteintech; 17797-1-ap), Gli3 (1:1000; R&D Systems; AF3690), β-COP (1:2000; ThermoFisher; PA1-061), IFT140 (1:500; Proteintech; 17460-1-AP), IFT88 (1:500; Proteintech; 13967-1-ap), α-HA (1:1000; Covance; MMS-101P), β1-integrin 1 (1:200; Sigma; MAB1997), active integrin clone 9EG7 (1:200; BD-Pharmingen; 553715). The following antibodies were generously provided from other labs: polyclonal, rabbit antibodies raised against FIP1 (1:500) and FIP5 (1:500) were from Rytis Prekeris (University of Colorado), and sheep anti-FIP3 (1:500) was from Jim Goldenring (Vanderbilt University). The Kahn lab generated ARL3 rabbit polyclonal antibody, which we use at 1:1000 dilution (Cavenagh et al., 1994).
Plasmids directing expression of mouse ELMOD1-myc or ELMOD3-myc were codon optimized for expression and synthesized by GeneArt and later moved into the pcDNA3.1 vector. Fast-cycling mutants of GTPases were generated by site-directed mutagenesis of the residue corresponding to T161 in ARF6 (Santy, 2002), followed by sequencing of the complete open reading frame to confirm the mutation and lack of extraneous changes.
Cell culture, transfections, and induction of ciliation
MEFs used in this study were originally obtained from the American Type Culture Collection (ATCC CRL-2991) and were maintained in DMEM with 10% FBS (Atlanta Biologicals; S11150) and 2 mM glutamine. Antibiotics were not used in routine culture to minimize chances of mycoplasma contamination, which was monitored by DNA staining. In any experiment in which comparisons were planned between lines with different genotypes, attention was paid to ensure comparable feeding/plating schedules and cell densities were used to minimize the likelihood that such variables may confound the data or their interpretations.
Transient transfections of WT or KO MEFs for rescue experiments were performed using jetOPTIMUS (VWR; 76299-634). Cells seeded at 90% confluence were transfected with a ratio of 4 µg DNA:4 µl jetOPTIMUS transfection reagent: 400 µl jetOPTIMUS buffer adhering to the protocol provided by the manufacturer. The DNA/jetOPTIMUS mixture was added dropwise to each respective well, and samples were returned to 37°C to incubate for 24 h. Cells were maintained in normal growth serum (i.e., 10% FBS in DMEM). The next day, cells were replated as needed for different experiments.
Induction of ciliation involved switching to low-FBS (0.5%) medium 1 d after plating and allowing ciliation to progress for 24–72 h before cells were fixed. Ciliation is also increased at higher cell densities, so cells were typically seeded at ∼80–90% density on day 0, with attention that all cell lines in the experiment were seeded at the same density. Cells in low serum proliferate very slowly, if at all, so densities approached confluence without overcrowding that would challenge scoring.
CRISPR/Cas9 genome editing
Genome editing in MEFs was performed as previously described (Schiavon et al., 2019; Turn et al., 2020, 2021). Benchling software (www.benchling.com/academic/) was used to design four 20-nucleotide(nt) guides. To facilitate expression from the U6 promoter, a “G” was substituted for the first nucleotide for each guide RNA. Primers were purchased from IDT based on the following templates: 5′-CACC(N20)-3′ and 5′-AAAC(NR20)-3′, where N20 and NR20 refer to the 20-nt protospacer sequence and its reverse complement, respectively. The guides used to generate KO lines in MEFs were as follows.
Elmod1 guide 1: CACCGGATGCGGAAACTCACCGGA
Elmod1 guide 2: CACCGTTTGCTACGGCACCAAACC
Elmod3 guide 2: CACCGATGCCATGGTTCGTCAGCT
Elmod3 guide 3: CACCGCCCATTGGTTTCTGCCGTC
Complementary oligos were annealed and cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 vector (Addgene plasmid #62988) at the BbsI sites. Guides were targeted near the N-terminus of the protein and upstream of the ELMOD domain to optimize the likelihood of null alleles. We generated at least two different clones from at least two different guides, each with unique frame shifting mutations on both alleles, to protect against both off-target effects and the potential for use of downstream initiation of protein synthesis, alternative splicing, or other confounding changes (Smits et al., 2019). Two ELMOD1 guides were used to generate four independent ELMOD1 KO clones (two from each guide), while two ELMOD3 guides, targeting exons 5 and 8, were used to generate 13 clones (three from one guide and 10 from the other) deleted for ELMOD3. We also generated two ELMOD1/ELMOD3 DKO lines by using one of the ELMOD3 guides (guide 2) transfected into one of our ELMOD1 KO lines (KO #2; Supplemental Figure S1C).
Low-passage MEFs were grown to 90% confluence in six-well dishes, transfected via Lipofectamine 2000 at a 1:3 ratio of DNA to Lipofectamine reagent with 4 µg of DNA, and then replated onto 10 cm plates for growth overnight. Puromycin (3 μg/ml; Sigma #P8833) was added the next day and maintained for 4 d to enrich for transfected cells. Individual clones were isolated via limited dilution in 96-well plates, followed by expansion, cryopreservation, and sequencing of genomic DNA after PCR amplification of the region surrounding the targeted site to identify frame shifting mutations that propagate early stop codons. After initial testing of phenotypes and finding consistencies between KO lines, we chose two from each gene for more detailed analyses, though often examined every KO line available.
During the cloning of the Elmod3 KO lines, but more so the DKO lines, we noted an apparent decrease in cell attachment to plates, evident as rounded cells after plating at low cell density. Prior treatment of plates with fibronectin alleviated this problem. To assess possible changes in cell attachment in the cloned lines, both the number and size of focal adhesions were quantified, and no differences were observed (Supplemental Figure S9, A and B). No defects in the actin cytoskeleton were noted in ELMOD1 or ELMOD3 KO lines, as detected by phalloidin staining (Figure S10). Thus, an attachment defect was not evident in the cell lines described here or in cloning of KO lines for multiple other genes studied in our lab. Thus, the reasons for possible attachment problems are unknown and were not pursued further at this time.
Flow cytometry analysis for DNA content
As previously described (Turn et al., 2020), unsynchronized cells were prepared for flow cytometry by trypsinizing the cells, taking up the cells in ice-cold phosphate-buffered saline (PBS), washing with ice-cold PBS, and fixing the cells with ice-cold 70% ethanol dropwise while simultaneously vortexing to reduce the risk of cell clumping. Both supernatant and adherent cells were collected to ensure that we had a full representation of the cell population. The day of flow cytometry, cells were spun down, washed with ice-cold phosphate citrate buffer (0.1 M citric acid in PBS, pH 7.8) two times, treated with RNase A for 15 min (100 µg/ml; Sigma; R5125), and treated with propidium iodide for 45 min (50 µg/ml; Sigma; P4170) to stain for DNA content. Cells were passed through a cell strainer and run on a FACSymphony A3. The G1 peak of WT cells was set at a 50,000 voltage for each run, and these settings were used to acquire all subsequent samples run that day to ensure that we accurately track 2N, 4N, and >4N peaks. At least 10,000 cells were collected per sample. FloJo software was used to plot data shown.
Microscopy
Before imaging, cells were plated onto fibronectin-coated 18 mm glass coverslips (#1.5; Fisher Scientific; 12-545-81), according to the specified experimental conditions, and processed for immunocytochemistry. Images were collected via wide-field microscopy on our Olympus IX81 microscope with Slidebook software at 60× and 100× magnification (UPIanFI; 1.30 NA oil). The same acquisition settings were used to ensure accurate comparisons (e.g., same sample preparation, magnification, gain, offset, laser power). Data were processed using FIJI imaging software, making sure to apply the same image processing techniques to each sample of a given data set (e.g., cropping, magnification, brightness, contrast, background subtraction). In the case of retinal tissue, samples were imaged using a Leica DM6000B deconvolution microscope (Leica Microsystems, Bensheim, Germany) or a Leica SP8 laser scanning confocal microscope (Leica Microsystems, Bensheim, Germany). Data were processed via Adobe Photoshop CS (Adobe Systems, San Jose, CA).
Immunofluorescence
The following protocols were used based on the antigen we were targeting (described below):
Methanol fixation protocol.
The methanol fixation protocol was mostly for centrosomal/centriolar markers to provide cleaner imaging without cell background/preserve structure. Cells plated on coverslips were fixed with methanol for 10 min at –20°C before being washed 4× with PBS. Cells were blocked for 1 h at room temperature with 10% FBS in PBS and incubated overnight with primary antibody diluted in 10% FBS in PBS at 4°C. Cells were washed with PBS 4× before being incubated with secondary antibody (1:500 dilution in 10% FBS in PBS) for 1 h at room temperature in the dark. Cells were washed with PBS 2×, stained with 1:5000 Hoechst in PBS for 4 min, and washed 2× with PBS before being mounted overnight with a 1:9 ratio of PPD (p-phenylenediamine dihydrochloride; ACROS Organics; 624-18-0) to MOWIOL 4-88 Reagent (CALBIOCHEM; 475904) mounting medium.
PFA fixation protocol.
The PFA fixation protocol was to preserve membranes/as a good general fixation protocol for cilia. Cells were fixed with prewarmed (37°C) 4% PFA in PBS for 15 min. Cells were washed 2× with PBS and permeabilized with 0.1% Triton X-100 for 10 min. Cells were blocked with 1% bovine serum albumin (BSA) in PBS for 1 h at room temperature and incubated with primary antibody diluted in 1% BSA in PBS overnight at 4°C. Cells were washed 4× with PBS, incubated with secondary antibody diluted in 1% BSA in PBS for 1 h at room temperature, and washed and mounted on slides as described above.
Note that use of the same antibody but with different staining protocols may yield differences. For example, INPP5E at cilia is optimally visualized after PFA fixation, while its presence at the Golgi is most prominent after methanol fixation. Variations in staining of cells after different fixation or permeabilization protocols using one antibody typically arise as a result of changes in protein structure and epitope exposure, retention/loss of binding partners that also may alter epitope availability, or washout of one pool of antigen that may have masked the staining of another pool. The last is common for proteins that are largely soluble but with transient localization to membrane compartments, as is common for ARF family GTPases and their regulators.
Immunoblotting
Cells were collected and cell pellets were lysed in PBS with 1% Triton X-100 on ice. After 15 min, cells were spun in a microfuge at 14,000 × g for 30 min to remove insoluble material. Proteins (40 µg) were loaded into lanes of an 11% polyacrylamide gel and resolved at 60 mA. Proteins were later transferred onto nitrocellulose filters overnight at 20 mV. The next day, filters were stained briefly with Ponceau S to confirm equal transfer and then put into Blotto blocking buffer (5% dry milk in PBS) for 1 h before the addition of primary antibody overnight. Filters were washed 3 × 10 min in PBST (PBS with 1% Tween 20) before adding secondary antibody and incubating for 1 h, followed by 3 × 10 min washes in PBST. Imaging was performed using a Bio-Rad imager.
Scoring of cell phenotypes
For all phenotypes described above, experiments were performed in triplicate and scored in at least duplicate, 100 cells per sample. All scoring was performed blinded, checking the genotypes of the samples only after all data were collected. For phenotypes such as ciliation, nucleation, and centrosome/centriole counts, scoring was binned based on the number of each organelle present/the number of the organelles positive for that marker. For markers in which it was related to a degree of localization in cilia (e.g., ARL13B), we binned them as either present (visible even without checking the Ac Tub channel), reduced (present, but noticeable only upon switching to the Ac Tub channel), or absent (cannot be detected even upon switching to the Ac Tub channel). For all ciliary phenotyping, Ac Tub was used as a costain to detect cilia and draw accurate comparisons. For centrosomal/basal body scoring, γ-tubulin was used as the standard comparison point. Finally, for Golgi staining/localization, either GM130 or Golgin97 was used to mark Golgi.
Sample preparation and staining of mouse retinas
Animals.
Transgenic eGFP-CETN2 mice (Higginbotham et al., 2004) were kept on a 12-h light–dark schedule at 22°C, with free access to food and water. Animal health was monitored on a regular basis, and all other procedures complied with the German Law on Animal Protection and the Institute for Laboratory Animal Research Guide for the Care and Use of Laboratory Animals at the Johannes Gutenberg University.
Immunohistochemistry of retinal sections .
Mouse retinas were dissected from enucleated eyeballs and cryofixed in melting isopentane and cryosectioned as previously described (Wolfrum, 1991; Karlstetter et al., 2014). Cryosections (10 µm thick) were placed on poly-l-lysine–precoated coverslips and incubated with 0.01% Tween 20 in PBS for 20 min. After washing, sections were flooded with blocking solution (0.5% cold-water fish gelatin plus 0.1% ovalbumin in PBS) and incubated for at least 30 min followed by an overnight incubation with primary antibodies at 4°C in blocking solution (Trojan et al., 2008). Washed cryosections were then incubated with secondary antibodies conjugated to Alexa 488 or Alexa 568 (Invitrogen) in blocking solution and with DAPI (Sigma-Aldrich) to stain the DNA of nuclei for 1.5 h at room temperature in the dark. After three washes in PBS, specimens were mounted in MOWIOL 4.88 (Hoechst) and imaged using a Leica DM6000B microscope (Leica).
Statistics
All experiments were scored at least in duplicate and performed at least in triplicate, using at least two clones from at least two different guides for each gene targeted (ELMOD1 or ELMOD3). Unless otherwise stated, at least 100 cells were scored per sample, the standard sample size both in our lab and in the field. Furthermore, we performed power analysis to ensure that our sample size was sufficient to assess statistical significance. Having scored 100 cells per each four replicates for each genotype, we determined that we were 90–100% powered to detect changes in ciliation rate (64.25 ± 16.1 in WT vs. 12.00 ± 7.16). Error bars in the bar graphs represent the SEM, and box-and-whisker plots indicate the range of the data along with the median and upper/lower quartiles. One-way or two-way analysis of variance (ANOVA) tests were used to determine whether there were significant differences between test groups. The presence of an asterisk in a figure indicates statistical significance: *p < 0.05, as also indicated in figure legends. We consider the individual lines as biological replicates. Therefore, if we report that a sample has an N = 4, this indicates that four different lines were scored in (at least) duplicate, and the averages of those duplicates are presented in the graphs.
Reagents and model systems
All cells used in these experiments were generated from the same immortalized MEF line, obtained from the ATCC (ATCC CRL-2991). All CRISPR KO lines were genotyped at the time of creation using sequencing as our measure for reagent authentication, and we maintain these lines at low passage and using proper cell culture techniques to ensure that all lines stay a clean monogenic population. As the need arises, we resequence the population to ensure that we are using the correct cell lines. All experiments were performed blinded to ensure rigor and reproducibility. Once published, all reagents described in this article will be made readily available.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health: R35GM122568 to R.A.K., R35GM122549 to T.C., R01GM127361 to J.E.C.; 1F31CA236493-02 to R.E.T., and 1F31HD096815-03 to S.I.D.; the Foundation Fighting Blindness (FFB PPA-0717-0719-RAD) to U.W.; the German Research Council/DFG in the framework of SPP SPP2127—Gene and Cell based therapies to counteract neuro-retinal degeneration WO548/9-1 to U.W.; and the joint training program between the Emory University School of Medicine and the Xiangya School of Medicine, Central South University (Y.H.). We thank colleagues for their generous sharing of key reagents, in particular Rytis Prekeris and Jim Goldenring for antibodies and plasmids. This research was supported in part by the Emory University Integrated Cellular Imaging (ICI) Microscopy and Pediatrics Flow Cytometry Cores. We thank the Emory Biochemistry Department and Laney Graduate School for their ongoing support and shared resources provided.
Abbreviations used:
- Ac Tub
acetylated tubulin
- CWT
CRISPR-WT (CRISPR-treated cells without edits in the targeted region)
- DKO
double knockout (of both ELMOD1 and ELMOD3)
- FBS
fetal bovine serum
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- IFT
intraflagellar transport
- KO
knockout
- MEF
mouse embryonic fibroblast
- PCM
pericentriolar material
- PFA
paraformaldehyde
- SHH
Sonic Hedgehog
- Smo
Smoothened
- TZ
transition zone
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-09-0443) on November 24, 2021.
REFERENCES
- Alkanderi S, Molinari E, Shaheen R, Elmaghloob Y, Stephen LA, Sammut V, Ramsbottom SA, Srivastava S, Cairns G, Edwards N, et al. (2018). ARL3 mutations cause Joubert syndrome by disrupting ciliary protein composition. Am J Hum Genet 103, 612–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu-Cervera A, Anselme I, Karam A, Laclef C, Catala M, Schneider-Maunoury S (2019). The ciliopathy gene Ftm/Rpgrip1l controls mouse forebrain patterning via region-specific modulation of Hedgehog/Gli signaling. J Neurosci 39, 2398–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabe-Rubio M, Alonso MA (2017). Routes and machinery of primary cilium biogenesis. Cell Mol Life Sci 74, 4077–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas SL, Silhavy JL, Brancati F, Kisseleva MV, Al-Gazali L, Sztriha L, Bayoumi RA, Zaki MS, Abdel-Aleem A, Rosti RO, et al. (2009). Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat Genet 41, 1032–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajanek L, Nigg EA (2014). Cep164 triggers ciliogenesis by recruiting Tau tubulin kinase 2 to the mother centriole. Proc Natl Acad Sci USA 111, E2841–E2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas-Rodriguez M, Austin-Tse C, Bergboer JGM, Molinari E, Sugano Y, Bachmann-Gagescu R, Sayer JA, Drummond IA (2021). Genetic compensation for cilia defects in cep290 mutants by upregulation of cilia-associated small GTPases. J Cell Sci 134, 10.1242/jcs.258568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalou C, Ferreira A, Barral DC (2020). The role of ARF family proteins and their regulators and effectors in cancer progression: a therapeutic perspective. Front Cell Dev Biol 8, 10.3389/fcell.2020.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV (2007). The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell 12, 767–778. [DOI] [PubMed] [Google Scholar]
- Cavenagh MM, Breiner M, Schurmann A, Rosenwald AG, Terui T, Zhang C, Randazzo PA, Adams M, Joost HG, Kahn RA (1994). ADP-ribosylation factor (ARF)-like 3, a new member of the ARF family of GTP-binding proteins cloned from human and rat tissues. J Biol Chem 269, 18937–18942. [PubMed] [Google Scholar]
- Cevik S, Hori Y, Kaplan OI, Kida K, Toivenon T, Foley-Fisher C, Cottell D, Katada T, Kontani K, Blacque OE (2010). Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J Cell Biol 188, 953–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Kelley RA, Li T, Swaroop A (2021). Primary cilia biogenesis and associated retinal ciliopathies. Semin Cell Dev Biol 110, 70–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J (2014). Arf GTPases and their effectors: assembling multivalent membrane-binding platforms. Curr Opin Struct Biol 29, 67–76. [DOI] [PubMed] [Google Scholar]
- Chung M-I, Kwon T, Tu F, Brooks ER, Gupta R, Meyer M, Baker JC, Marcotte EM, Wallingford JB (2014). Coordinated genomic control of ciliogenesis and cell movement by RFX2. eLife 3, e01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constable S, Long AB, Floyd KA, Schurmans S, Caspary T (2020). The ciliary phosphatidylinositol phosphatase Inpp5e plays positive and negative regulatory roles in Shh signaling. Development 147, dev183301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse JA, Lopes VS, Sanagustin JT, Keady BT, Williams DS, Pazour GJ (2014). Distinct functions for IFT140 and IFT20 in opsin transport. Cytoskeleton (Hoboken) 71, 302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewees SI, Vargova R, Hardin KR, Turn RE, Devi S, Linnert J, Wolfrum U, Caspary T, Elias M, Kahn R (2021). Phylogenetic profiling and cellular analyses of ARL16 reveal roles in traffic of IFT140 and INPP5E. bioRxiv, 10.1101/2021.10.14.464442:2021.2010.2014.464442. 10.1101/2021.10.14.464442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL (2011). ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol 12, 362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson JM, Conduit SE, Feeney SJ, Hakim S, DiTommaso T, Fulcher AJ, Sriratana A, Ramm G, Horan KA, Gurung R, et al. (2017). INPP5E regulates phosphoinositide-dependent cilia transition zone function. J Cell Biol 216, 247–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East MP, Bowzard JB, Dacks JB, Kahn RA (2012). ELMO domains, evolutionary and functional characterization of a novel GTPase-activating protein (GAP) domain for Arf protein family GTPases. J Biol Chem 287, 39538–39553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East MP, Kahn RA (2011). Models for the functions of Arf GAPs. Semin Cell Dev Biol 22, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fansa EK, Kosling SK, Zent E, Wittinghofer A, Ismail S (2016). PDE6delta-mediated sorting of INPP5E into the cilium is determined by cargo-carrier affinity. Nat Commun 7, 11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Kuna D, Caspary T, Kahn RA, Sztul E (2020). ARF family GTPases with links to cilia. Am J Physiol Cell Physiol 319, C404–C418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, San Agustin JT, Xu F, Jonassen JA, Samtani R, Lo CW, Pazour GJ (2008). The golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex. PLoS Genet 4, e1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ (2006). The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell 17, 3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis JW, Goswami D, Novick SJ, Pascal BD, Weikum ER, Ortlund EA, Griffin PR, Kahn RA (2017a). Nucleotide binding to ARL2 in the TBCDARL2beta-tubulin complex drives conformational changes in beta-tubulin. J Mol Biol 429, 3696–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis JW, Newman LE, Cunningham LA, Kahn RA (2017b). A trimer consisting of the tubulin-specific chaperone D (TBCD), regulatory GTPase ARL2, and beta-tubulin is required for maintaining the microtubule network. J Biol Chem 292, 4336–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa S, Qiu H, Nozaki S, Chiba S, Katoh Y, Nakayama K (2021). ARL3 and ARL13B GTPases participate in distinct steps of INPP5E targeting to the ciliary membrane. Biol Open, 10.1242/bio.058843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Phua SC, Roberson EC, Garcia G 3rd, Abedin M, Schurmans S, Inoue T, Reiter JF (2015). Phosphoinositides regulate ciliary protein trafficking to modulate Hedgehog signaling. Dev Cell 34, 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigante ED, Taylor MR, Ivanova AA, Kahn RA, Caspary T (2020). ARL13B regulates Sonic hedgehog signaling from outside primary cilia. eLife 9, e50434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt K, Lokaj M, Koerner C, Falk N, Giessl A, Wittinghofer A (2015). A G-protein activation cascade from Arl13B to Arl3 and implications for ciliary targeting of lipidated proteins. eLife 4, e11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Clech MLe, Nigg EA (2007). Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol 179, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke-Gogokhia C, Frederick JM, Zhang H, Baehr W (2018). Binary function of ARL3-GTP revealed by gene knockouts. Adv Exp Med Biol 1074, 317–325. [DOI] [PubMed] [Google Scholar]
- Hanke-Gogokhia C, Zhang H, Frederick JM, Baehr W (2016). The function of Arf-like proteins ARL2 and ARL3 in photoreceptors. Adv Exp Med Biol 854, 655–661. [DOI] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK (2005). Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet 1, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham H, Bielas S, Tanaka T, Gleeson JG (2004). Transgenic mouse line with green-fluorescent protein-labeled Centrin 2 allows visualization of the centrosome in living cells. Transgenic Res 13, 155–164. [DOI] [PubMed] [Google Scholar]
- Hodgson U, Pulkkinen V, Dixon M, Peyrard-Janvid M, Rehn M, Lahermo P, Ollikainen V, Salmenkivi K, Kinnula V, Kere J, et al. (2006). ELMOD2 is a candidate gene for familial idiopathic pulmonary fibrosis. Am J Hum Genet 79, 149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan KA, Watanabe K, Kong AM, Bailey CG, Rasko JE, Sasaki T, Mitchell CA (2007). Regulation of FcgammaR-stimulated phagocytosis by the 72-kDa inositol polyphosphate 5-phosphatase: SHIP1, but not the 72-kDa 5-phosphatase, regulates complement receptor 3 mediated phagocytosis by differential recruitment of these 5-phosphatases to the phagocytic cup. Blood 110, 4480–4491. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV (2005). Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci USA 102, 11325–11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV (2003). Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83–87. [DOI] [PubMed] [Google Scholar]
- Humbert MC, Weihbrecht K, Searby CC, Li Y, Pope RM, Sheffield VC, Seo S (2012). ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc Natl Acad Sci USA 109, 19691–19696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SH, Somatilaka BN, Badgandi H, Palicharla VR, Walker R, Shelton JM, Qian F, Mukhopadhyay S (2019). Tulp3 regulates renal cystogenesis by trafficking of cystoproteins to cilia. Curr Biol 29, 790–802.e795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova AA, Caspary T, Seyfried NT, Duong DM, West AB, Liu Z, Kahn RA (2017). Biochemical characterization of purified mammalian ARL13B protein indicates that it is an atypical GTPase and ARL3 guanine nucleotide exchange factor (GEF). J Biol Chem 292, 11091–11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova AA, East MP, Yi SL, Kahn RA (2014). Characterization of recombinant ELMOD (cell engulfment and motility domain) proteins as GTPase-activating proteins (GAPs) for ARF family GTPases. J Biol Chem 289, 11111–11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CL, Bouvet S (2014). Arfs at a glance. J Cell Sci 127, 4103–4109. [DOI] [PubMed] [Google Scholar]
- Jaworek TJ, Richard EM, Ivanova AA, Giese AP, Choo DI, Khan SN, Riazuddin S, Kahn RA, Riazuddin S (2013). An alteration in ELMOD3, an Arl2 GTPase-activating protein, is associated with hearing impairment in humans. PLoS Genet 9, e1003774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Longo-Guess CM, Gagnon LH (2012). Mutations of the mouse ELMO domain containing 1 gene (Elmod1) link small GTPase signaling to actin cytoskeleton dynamics in hair cell stereocilia. PLoS One 7, e36074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen JA, SanAgustin J, Baker SP, Pazour GJ (2012). Disruption of IFT complex A causes cystic kidneys without mitotic spindle misorientation. J Am Soc Nephrol 23, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Bruford E, Inoue H, Logsdon JM Jr, Nie Z, Premont RT, Randazzo PA, Satake M, Theibert AB, Zapp ML, Cassel D (2008). Consensus nomenclature for the human ArfGAP domain-containing proteins. J Cell Biol 182, 1039–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlstetter M, Sorusch N, Caramoy A, Dannhausen K, Aslanidis A, Fauser S, Boesl MR, Nagel-Wolfrum K, Tamm ER, Jagle H, et al. (2014). Disruption of the retinitis pigmentosa 28 gene Fam161a in mice affects photoreceptor ciliary structure and leads to progressive retinal degeneration. Hum Mol Genet 23, 5197–5210. [DOI] [PubMed] [Google Scholar]
- Keady BT, Le YZ, Pazour GJ (2011). IFT20 is required for opsin trafficking and photoreceptor outer segment development. Mol Biol Cell 22, 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ishida Y, Hirano T, Katoh Y, Nakayama K (2021). Cooperation of the IFT-A complex with the IFT-B complex is required for ciliary retrograde protein trafficking and GPCR import. Mol Biol Cell 32, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong AM, Horan KA, Sriratana A, Bailey CG, Collyer LJ, Nandurkar HH, Shisheva A, Layton MJ, Rasko JE, Rowe T, Mitchell CA (2006). Phosphatidylinositol 3-phosphate [PtdIns3P] is generated at the plasma membrane by an inositol polyphosphate 5-phosphatase: endogenous PtdIns3P can promote GLUT4 translocation to the plasma membrane. Mol Cell Biol 26, 6065–6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong AM, Speed CJ, O’Malley CJ, Layton MJ, Meehan T, Loveland KL, Cheema S, Ooms LM, Mitchell CA (2000). Cloning and characterization of a 72-kDa inositol-polyphosphate 5-phosphatase localized to the Golgi network. J Biol Chem. 275, 24052–24064. [DOI] [PubMed] [Google Scholar]
- Kosling SK, Fansa EK, Maffini S, Wittinghofer A (2018). Mechanism and dynamics of INPP5E transport into and inside the ciliary compartment. Biol Chem 399, 277–292. [DOI] [PubMed] [Google Scholar]
- Krey JF, Dumont RA, Wilmarth PA, David LL, Johnson KR, Barr-Gillespie PG (2018). ELMOD1 stimulates ARF6-GTP hydrolysis to stabilize apical structures in developing vestibular hair cells. J Neurosci 38, 843–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins CE, Aviles GD, East MP, Kahn RA, Caspary T (2011). Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell 22, 4694–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin M, Cote JF (2014). Insights into the biological functions of Dock family guanine nucleotide exchange factors. Genes Dev 28, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF (2015). IFT–cargo interactions and protein transport in cilia. Trends Biochem Sci 40, 765–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Feng Y, Chen A, Li T, Huang S, Liu J, Liu X, Liu Y, Gao J, Yan D, et al. (2019). Elmod3 knockout leads to progressive hearing loss and abnormalities in cochlear hair cell stereocilia. Hum Mol Genet 28, 4103–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Sun J, Ling J, Li J, He C, Liu Y, Chen H, Men M, Niu Z, Deng Y, et al. (2018). ELMOD3, a novel causative gene, associated with human autosomal dominant nonsyndromic and progressive hearing loss. Hum Genet 137, 329–342. [DOI] [PubMed] [Google Scholar]
- Liem KF Jr, Ashe A, He M, Satir P, Moran J, Beier D, Wicking C, Anderson KV (2012). The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. J Cell Biol 197, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CH, Lin IH, Yang TT, Huang YC, Tanos BE, Chou PC, Chang CW, Tsay YG, Liao JC, Wang WJ (2019). Phosphorylation of CEP83 by TTBK2 is necessary for cilia initiation. J Cell Biol 218, 3489–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani LE, Bijlsma MF, Ivanova AI, Suciu SK, Kahn RA, Caspary T (2016). Arl13b regulates Shh signaling from both inside and outside the cilium. Mol Biol Cell 27, 3780–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miertzschke M, Koerner C, Spoerner M, Wittinghofer A (2014). Structural insights into the small G protein Arl13B and implications for Joubert syndrome. Biochem J 457, 301–311. [DOI] [PubMed] [Google Scholar]
- Miryounesi M, Bahari S, Salehpour S, Alipour N, Ghafouri-Fard S (2019). ELMO domain containing 1 (ELMOD1) gene mutation is associated with mental retardation and autism spectrum disorder. J Mol Neurosci 69, 312–315. [DOI] [PubMed] [Google Scholar]
- Munoz-Estrada J, Ferland RJ (2019). Ahi1 promotes Arl13b ciliary recruitment, regulates Arl13b stability and is required for normal cell migration. J Cell Sci 132, jcs230680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV (2018). The molecular machines that traffic signaling receptors into and out of cilia. Curr Opin Cell Biol 51, 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Mick DU (2019). Establishing and regulating the composition of cilia for signal transduction. Nat Rev Mol Cell Biol 20, 389–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman LE, Schiavon CR, Zhou C, Kahn RA (2017). The abundance of the ARL2 GTPase and its GAP, ELMOD2, at mitochondria are modulated by the fusogenic activity of mitofusins and stressors. PLoS One 12, e0175164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulkkinen V, Bruce S, Rintahaka J, Hodgson U, Laitinen T, Alenius H, Kinnula VL, Myllarniemi M, Matikainen S, Kere J (2010). ELMOD2, a candidate gene for idiopathic pulmonary fibrosis, regulates antiviral responses. FASEB J 24, 1167–1177. [DOI] [PubMed] [Google Scholar]
- Qiu H, Fujisawa S, Nozaki S, Katoh Y, Nakayama K (2020). Interaction of INPP5E with ARL13B is essential for its ciliary membrane retention but dispensable for its ciliary entry. Biol Open, 10.1242/bio.057653. 10.1242/bio.057653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR (2004). The engulfment process of programmed cell death in Caenorhabditis elegans. Annu Rev Cell Dev Biol 20, 193–221. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Leroux MR (2017). Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K, Jerman S, Jozsef L, McNamara T, Onyekaba G, Sun Z, Marin EP (2017). Palmitoylation of the ciliary GTPase Arl13b is necessary for its stability and its role in cilia formation. J Biol Chem 292, 17703–17717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santy LC (2002). Characterization of a fast cycling ADP-ribosylation factor 6 mutant. J Biol Chem 277, 40185–40188. [DOI] [PubMed] [Google Scholar]
- Schiavon CR, Turn RE, Newman LE, Kahn RA (2019). ELMOD2 regulates mitochondrial fusion in a mitofusin-dependent manner, downstream of ARL2. Mol Biol Cell 30, 1198–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KN, Kuhns S, Neuner A, Hub B, Zentgraf H, Pereira G (2012). Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J Cell Biol 199, 1083–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedmak T, Wolfrum U (2010). Intraflagellar transport molecules in ciliary and nonciliary cells of the retina. J Cell Biol 189, 171–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits AH, Ziebell F, Joberty G, Zinn N, Mueller WF, Clauder-Munster S, Eberhard D, Falth Savitski M, Grandi P, Jakob P, et al. (2019). Biological plasticity rescues target activity in CRISPR knock outs. Nat Methods 16, 1087–1093. [DOI] [PubMed] [Google Scholar]
- Stephen LA, Ismail S (2016). Shuttling and sorting lipid-modified cargo into the cilia. Biochem Soc Trans 44, 1273–1280. [DOI] [PubMed] [Google Scholar]
- Su S, Begum S, Ezratty EJ (2020). An IFT20 mechanotrafficking axis is required for integrin recycling, focal adhesion dynamics, and polarized cell migration. Mol Biol Cell 31, 1917–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Murakami T, Cheng J, Kano H, Fukata M, Fujimoto T (2015). ELMOD2 is anchored to lipid droplets by palmitoylation and regulates adipocyte triglyceride lipase recruitment. Mol Biol Cell 26, 2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztul E, Chen PW, Casanova JE, Cherfils J, Dacks JB, Lambright DG, Lee FS, Randazzo PA, Santy LC, Schurmann A, et al. (2019). ARF GTPases and their GEFs and GAPs: concepts and challenges. Mol Biol Cell 30, 1249–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Wright KJ, Corre SL, Micalizzi A, Romani M, Abhyankar A, Saada J, Perrault I, Amiel J, Litzler J, et al. (2014). A homozygous PDE6D mutation in Joubert syndrome impairs targeting of farnesylated INPP5E protein to the primary cilium. Hum Mutat 35, 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojan P, Krauss N, Choe HW, Giessl A, Pulvermuller A, Wolfrum U (2008). Centrins in retinal photoreceptor cells: regulators in the connecting cilium. Prog Retin Eye Res 27, 237–259. [DOI] [PubMed] [Google Scholar]
- Tsang WY, Dynlacht BD (2013). CP110 and its network of partners coordinately regulate cilia assembly. Cilia 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu F, Sedzinski J, Ma Y, Marcotte EM, Wallingford JB (2018). Protein localization screening in vivo reveals novel regulators of multiciliated cell development and function. J Cell Sci 131, jcs206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turn RE, East MP, Prekeris R, Kahn RA (2020). The ARF GAP ELMOD2 acts with different GTPases to regulate centrosomal microtubule nucleation and cytokinesis. Mol Biol Cell 31, 2070–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turn RE, Linnert J, Gigante ED, Wolfrum U, Caspary T, Kahn RA (2021). Roles for ELMOD2 and Rootletin in ciliogenesis. Mol Biol Cell 32, 800–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukhanov K, Uytingco C, Green W, Zhang L, Schurmans S, Martens JR (2022). INPP5E controls ciliary localization of phospholipids and the odor response in olfactory sensory neurons. J Cell Sci 135, jcs258364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargova R, Wideman JG, Derelle R, Klimes V, Kahn RA, Dacks JB, Elias M (2021). A eukaryote-wide perspective on the diversity and evolution of the ARF GTPase protein family. Genome Biol Evol 13, evab157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltel S, Kravchenko A, Ismail S, Wittinghofer A (2008). Specificity of Arl2/Arl3 signaling is mediated by a ternary Arl3-effector-GAP complex. FEBS Lett 582, 2501–2507. [DOI] [PubMed] [Google Scholar]
- Wolfrum U (1991). Distribution of F-actin in the compound eye of the blowfly, Calliphora erythrocephala (Diptera, Insecta). Cell Tissue Res 263, 399–403. [Google Scholar]
- Wright ZC, Singh RK, Alpino R, Goldberg AF, Sokolov M, Ramamurthy V (2016). ARL3 regulates trafficking of prenylated phototransduction proteins to the rod outer segment. Hum Mol Genet 25, 2031–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav SP, Sharma NK, Liu C, Dong L, Li T, Swaroop A (2016). Centrosomal protein CP110 controls maturation of the mother centriole during cilia biogenesis. Development 143, 1491–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Constantine R, Frederick JM, Baehr W (2012). The prenyl-binding protein PrBP/delta: A chaperone participating in intracellular trafficking. Vision Res 75, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li S, Doan T, Rieke F, Detwiler PB, Frederick JM, Baehr W (2007). Deletion of PrBP/delta impedes transport of GRK1 and PDE6 catalytic subunits to photoreceptor outer segments. Proc Natl Acad Sci USA 104, 8857–8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CX, Shi LY, Li RC, Liu YH, Xu BQ, Liu JW, Yuan B, Yang ZX, Ying XY, Zhang D (2017). GTPase-activating protein Elmod2 is essential for meiotic progression in mouse oocytes. Cell Cycle 16, 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Amom P, Reeder SH, Lee BH, Helton A, Dobritsa AA (2021). Members of the ELMOD protein family specify formation of distinct aperture domains on the Arabidopsis pollen surface. eLife, 10, e71061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.