Abstract
Coronavirus disease (COVID-19) has affected people for over two years. Moreover, the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants has raised concerns regarding its accurate diagnosis. Here, we report a colorimetric DNAzyme reaction triggered by loop-mediated isothermal amplification (LAMP) with clustered regularly interspaced short palindromic repeats (CRISPR), referred to as DAMPR assay for detecting SARS-CoV-2 and variants genes with attomolar sensitivity within an hour. The CRISPR-associated protein 9 (Cas9) system eliminated false-positive signals of LAMP products, improving the accuracy of DAMPR assay. Further, we fabricated a portable DAMPR assay system using a three-dimensional printing technique and developed a machine learning (ML)-based smartphone application to routinely check diagnostic results of SARS-CoV-2 and variants. Among blind tests of 136 clinical samples, the proposed system successfully diagnosed COVID-19 patients with a clinical sensitivity and specificity of 100% each. More importantly, the D614G (variant-common), T478K (delta-specific), and A67V (omicron-specific) mutations of the SARS-CoV-2 S gene were detected selectively, enabling the diagnosis of 70 SARS-CoV-2 delta or omicron variant patients. The DAMPR assay system is expected to be employed for on-site, rapid, accurate detection of SARS-CoV-2 and its variants gene and employed in the diagnosis of various infectious diseases.
Keywords: SARS-CoV-2, variants, smartphone, CRISPR-Cas9, machine learning
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread to nearly every corner of the globe, causing widespread societal instability following its initial outbreak in December 2019.1 Globally, as of June 2022, there have been 535,863,950 confirmed cases of coronavirus disease 2019 (COVID-19) with 6,314,972 confirmed deaths.2 The pandemic is yet to be controlled, despite the administration of a total of 11,864,214,773 vaccine doses according to the WHO.2 The confirmed cases of COVID-19 in USA and China are 85,981,213 and 886,865, respectively.2
The large scale of the pandemic has ironically facilitated the advance of diagnostic techniques and systems significantly during the past two years. From the beginning of the early spread of COVID-19 until now, molecular diagnostic methods represented by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) have been primarily used for the detection of SARS-CoV-2.3 In addition, immunoassays also have been complementarily employed to aid in the simple diagnosis, rapid screening, vaccination monitoring, and prognosis of patients.4 Moreover, various research works have led to the development of several types of innovative diagnostic methods to inhibit the spread of SARS-CoV-2, including next-generation sequencing (NGS) and lateral-flow immunochromatographic assay.5,6 Although the recent development of vaccines for COVID-19 and subsequent mass inoculation drives conducted appeared to have brought the pandemic under control, the emergence of SARS-CoV-2 variant strains such as delta and omicron and the consequent rise in breakthrough infected cases have once more prompted concerns regarding the accurate diagnosis of such strains.7
Molecular diagnostic tests have been vital in controlling infectious disease outbreaks.8 In the case of the current COVID-19 pandemic, the qRT-PCR method has cemented its status as the gold standard for the detection of SARS-CoV-2.9 However, the PCR method is somewhat inconvenient as it requires sample transport, huge and centralized instruments, and highly trained personnel.10 Consequently, there have been calls for the development of on-site, rapid, sensitive, and selective molecular diagnostic methods, and several approaches have been developed including reverse transcription loop-mediated isothermal amplification (RT-LAMP),11,12 clustered regularly interspaced short palindromic repeats (CRISPR)-based diagnostics,13−15 and nanomaterials-utilized diagnostics.16 Among them, RT-LAMP is considered the most representative molecular diagnostic technology for point of care testing (POCT) because of its high efficiency, isothermal amplification, and good tolerance to crudely processed biological samples.17 Since April 26, 2021, Seoul National University in the Republic of Korea has conducted RT-LAMP testing on all members to render the university a “COVID-19 safe zone”,18 thereby demonstrating the practicality of the RT-LAMP method. However, the LAMP technique suffers from the problem of false-positive amplification owing to product cross-contamination.19,20 Moreover, the changes in nucleic acids of SARS-CoV-2 variants may affect the utility of current LAMP diagnostic assays. Consequently, the development of an advanced LAMP method enabling the detection of SARS-CoV-2 and variants by eliminating the false-positive signals and maintaining the high amplifying efficiency has garnered attention.
The CRISPR–CRISPR-associated protein (Cas) system is considered a promising genetic engineering technique in molecular biology because of its highly specific target recognition ability.21 Recently, CRISPR-Cas systems have been employed for both genetic engineering and disease diagnostics, resulting in state-of-the-art diagnostic technologies combining various sensing techniques.22 The rapidly evolving area of CRISPR-based diagnostics is based on properties such as specificity, programmability, and ease of use,23 which has inspired us to develop an on-site, rapid, and accurate detection system for SARS-CoV-2 and variants by integrating the RT-LAMP technique with the CRISPR-Cas9 system.
We report a colorimetric DNAzyme reaction triggered by LAMP with CRISPR (DAMPR) assay system for the detection of SARS-CoV-2 and its variants. The advantages of the proposed system are as follows: (1) The DAMPR assay can detect three genes of SARS-CoV-2 simultaneously with attomolar sensitivity (1.08 aM for ORF1 gene, 0.92 aM for N gene, and 1.37 aM for S gene), reducing the false-negative signals. (2) The adoption of the Cas9/guide RNA (gRNA) system eliminates the false-positive signals by double-checking the amplified LAMP products. (3) It can be applied to the detection of SARS-CoV-2 variants using specifically designed Cas9/gRNA complexes. (4) Colorimetric signals provided by the DAMPR assay render naked-eye detection of viruses possible. Furthermore, a machine learning (ML) model was trained with smartphone images after the assay, enabling the semiquantitative detection of SARS-CoV-2 and variants without complicated optical devices. (5) The portable DAMPR assay system was fabricated using a three-dimensional (3D) printing technique, and the end-user-friendly smartphone application was developed as well. The system and application facilitate the on-site, rapid, and accurate detection of SARS-CoV-2 and variants within an hour. (6) The developed system was examined using SARS-CoV-2 genes, viral lysates, virus-spiked human nasopharyngeal aspirates, and clinical samples, thereby proving its sensing capability. (7) A total of 136 clinical samples were blindly tested using the DAMPR assay system, exhibiting clinical sensitivity and specificity of 100% each. Furthermore, the DAMPR assay system identified 70 patients infected with SARS-CoV-2 delta or omicron variants, indicating that the developed DAMPR assay system can play a key role in POCT, where a range of viruses must be efficiently assayed.
Results
Principle of DAMPR Assay
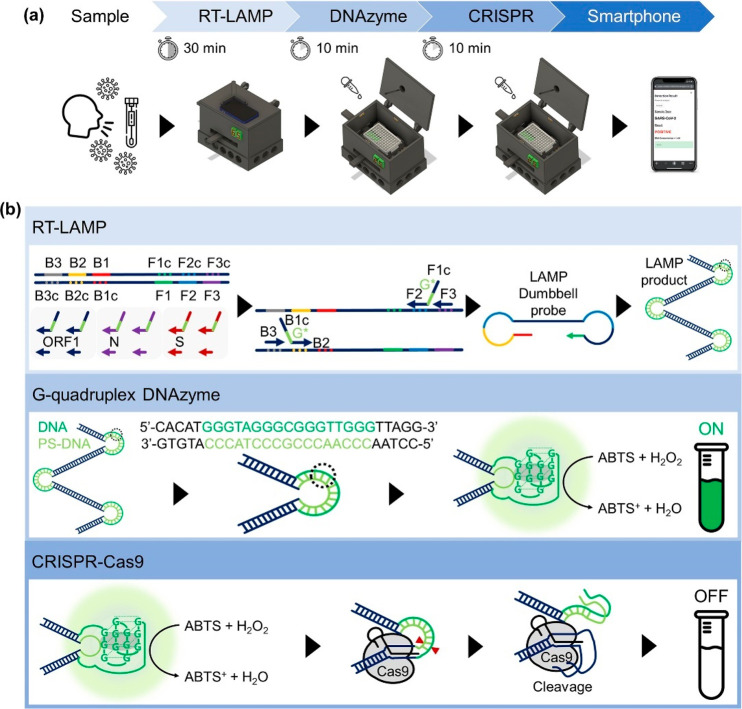
The schematic of the procedure of DAMPR assay is presented in Figure 1a. The assay was developed to use the nasopharyngeal aspirates or sputum samples in virus transport media directly, as these clinical samples have been frequently used for the diagnosis of COVID-19 patients.24 First, the collected samples in the media were added to a 96-well microplate with RT-LAMP mixture and subsequently reacted for 30 min at 65 °C, followed by the addition of DNAzyme reagents to the plate and incubation for 10 min at 25 °C. Subsequently, a photograph of the well plate was acquired using a smartphone on the DAMPR assay system. To double-check amplified LAMP products, CRISPR-Cas9 and gRNA complexes were treated and incubated for 10 min, and the photograph was acquired again. Finally, the diagnostic result of DAMPR assay can be obtained via the smartphone application (positive or negative results for the sample and the concentration level of SARS-CoV-2 in the sample) after uploading the photographs (Movie S1).
Figure 1.
(a) Procedure of DAMPR assay for SARS-CoV-2 detection. (b) Principle of DAMPR assay including RT-LAMP, G-quadruplex DNAzyme, and CRISPR-Cas9 reactions.
The detailed principle of DAMPR assay is shown in Figure 1b. The DAMPR assay begins with an RT-LAMP reaction employing pairs of inner and outer primers. The forward inner primer (FIP) and backward inner primer (BIP) were designed comprising three functional domains: target recognition site at 3′ end (F2 and B2), G-quadruplex complementary sequence with phosphorothioate (PS) modification at the link (G*), and target complementary site at 5′ end (F1c and B1c). The upper panel of Figure 1b shows that four types of primers and a strand-displacing DNA polymerase amplify the target gene. The RT-LAMP reaction produces dumbbell structured intermediate amplicons to facilitate subsequent rounds of amplification through the extension on the loops and additional annealing of primers. Subsequently, several G-quadruplex DNA/PS-DNA duplexes are acquired, which can be denatured easily and thus become active G-quadruplex DNAzymes, because PS-modification of DNA renders the base stacking interaction weak within the double-stranded form and, consequently, reduces the melting temperature (Tm) of the DNA/PS-DNA duplex.25−28 The produced G-quadruplex DNAzymes interact with hemin, turning on the colorimetric signal in the presence of H2O2 and 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) considering their peroxidase mimicking activity (middle panel of Figure 1b).29,30 This DNAzyme reaction indicates that the SARS-CoV-2 can be identified via the naked eye. Finally, Cas9/gRNA complexes are introduced to double-check the false-positive products during the RT-LAMP reaction (lower panel of Figure 1b). As gRNAs are specifically designed for ORF1, N, and S genes, the Cas9/gRNA complexes recognize the protospacer adjacent motif (PAM) (5′-NGG) sequence within the correctly amplified LAMP products and cleave the G-quadruplex DNAzyme. This induces the decrease of colorimetric signal, enabling the rechecking of the amplified target sequences via the naked eye. Shortly after, when RT-LAMP and DNAZyme reactions occur accurately, the colorimetric signal decreases after the CRISPR-Cas9 reaction. In contrast, on the generation of false-positive LAMP products, no CRISPR-Cas9 cleavage occurs, thereby maintaining the colorimetric signal.
Optimization and Evaluation of DAMPR Assay
The DAMPR assay was optimized via the examination of the reaction conditions including primers, gRNAs, reaction temperature and time, and concentrations of hemin, ABTS, and Cas9/gRNA complex. The primers for RT-LAMP were selected on the basis of the ORF1, S, and N gene sequences of SARS-CoV-2 (Figure 2a). Further, the primers comprising DNA and PS-DNA were compared using real-time fluorescence curves and absorbance spectra of LAMP reactions. Both primers exhibited similar gene amplification efficiencies in the fluorescence curves, implying that the PS-DNA and normal DNA primers are comparable (Figure S1a). However, in the absorbance spectra, the PS-DNA primers exhibit higher signals than DNA primers (Figure S1b), indicating that DNAzymes can be produced more efficiently using PS-DNA primers. The number of primers (four and six) was also compared similarly (Figure S1c,d). Considering the optimization results, four types of primers comprising PS-DNA were selected for the DAMPR assay (Table S1). Further, the gRNAs targeted the sequences of the SARS-CoV-2 ORF1, N, or S genes and G-quadruplex DNAzyme (Figure 2a and Figure S2a,c,e), and their optimum sequences were chosen by comparing the decrease of absorbance signals after CRISPR reactions (Figure S2b,d,f). The gRNA sequences for DAMPR assay are presented in Table S2. The reaction temperature and time for G-quadruplex DNAzyme and CRISPR-Cas9 reactions were set to 25 °C and 10 min, respectively, considering the experimental results (Figure S3a,b). A 0.8 μM hemin and 0.3 mM ABTS were determined to be the optimal conditions for DNAzyme reaction (Figure S3c), and 10× Cas9/gRNA ribonucleoprotein (RNP) to target gene was chosen for CRISPR-Cas9 reaction (Figure S3d).
Figure 2.
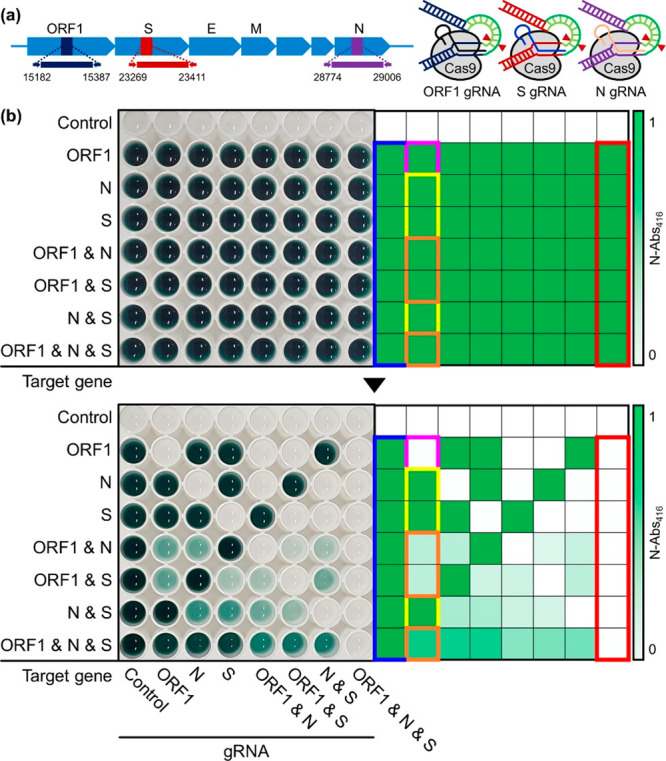
(a) Schematic illustration of target ORF1, S, and N genes of SARS-CoV-2 and Cas9/gRNA complexes for DAMPR assay. (b) Photograph of well plate and corresponding heat map after the detection of SARS-CoV-2 genes using RT-LAMP and DNAzyme reactions (upper). Photograph of the same well plate and corresponding heat map after double-checking of SARS-CoV-2 genes using Cas9/gRNA complexes (lower).
Following the optimization of DAMPR assay, ORF1, S, and N genes within SARS-CoV-2 were detected multiple times with various wells to verify the feasibility of the assay. The upper panel of Figure 2b shows a photograph of the well plate after the detection of SARS-CoV-2 genes by RT-LAMP and DNAzyme reactions, while the tested target genes are presented on the left side of the photograph. The target genes had concentrations of 1 nM. All wells except the control were dark green in color, which was clearly visible, indicating feasible naked-eye detection of SARS-CoV-2 genes. Further, the corresponding heat map for the normalized absorbance at 416 nm (N-Abs416) implies that the LAMP and DNAzyme reactions occurred in the presence of target genes. The lower panel of Figure 2b is a photograph of the same well plate following the addition and incubation of CRISPR-Cas9 systems, while the added gRNAs are mentioned at the bottom of the photograph. The corresponding heat map for the N-Abs416 indicates the decrease of colorimetric signals after the CRISPR-Cas9 reactions. All absorbance spectra for Figure 2b are presented in Figure S4. After the addition of all types of Cas9/gRNA complexes, the dark green color disappeared completely (red boxes in Figure 2b), whereas in the absence of Cas9/gRNA complexes, it is preserved (blue boxes in Figure 2b). The colorimetric signals decrease only after the addition of the Cas9/gRNA complexes corresponding to the amplified target genes. Consider the second column of the well plate as an example, wherein, as shown in the upper image of Figure 2b, all wells in the column provided dark color signals following the detection of each target gene by RT-LAMP and DNAzyme reactions. After the treatment of the Cas9/ORF1 gRNA complex, the color signals of the wells changed individually. The color signal was completely reduced in the ORF1 gene only amplified well (magenta boxes in Figure 2b), owing to the complete cleavage of the amplified target gene by the Cas9/ORF1 gRNA complex. The lack of complementary target genes for the Cas9/ORF1 gRNA complex resulted in no change in the N and/or S genes-detected wells (yellow boxes in Figure 2b). However, the dark green signals decreased when ORF1 and N and/or S genes coexisted in the wells (orange boxes in Figure 2b). Moreover, the N-Abs416 decreased more in ORF1 and N or S genes-amplified wells, while less decrease in N-Abs416 was observed in all three target genes-amplified wells as half of the LAMP products were cleaved by the Cas9/ORF1 gRNA complex in ORF1 and N or S genes-amplified wells, while one-third were cleaved by the complex in all target-amplified well. This implies that the Cas9/ORF1 gRNA complex cleaved the corresponding target gene precisely. Thus, the DAMPR assay detects ORF1, N, and S genes of SARS-CoV-2 and also double-checks the amplified genes, reducing the false-negative and false-positive signals.
Abs416 was measured as a function of the target gene concentration to estimate the sensitivity of DAMPR assay (Figure 3). The experimental results indicate that the absorbance intensity increases with an increase in the concentrations of the target gene up to 1 nM. In the range 1 aM–1 nM, an excellent linear relationship (R2 = 0.9987, R2 = 0.9938, and R2 = 0.9921 for ORF1, N, and S genes, respectively) exists and the limit of detection (LOD) was calculated as 1.08 aM (10 copies/sample), 0.92 aM (9 copies/sample), and 1.37 aM (13 copies/sample) for ORF1, N, and S genes, respectively, according to 3σ/slope (σ is the standard deviation at the lowest concentration of the linear range) (blue data in Figure 3b,d,f). This value is lower than or comparable to previously reported SARS-CoV-2 assays (Table S3). Meanwhile, red data in Figure 3b,d,f show that Abs416 decreases after Cas9/gRNA reaction through the whole concentrations. Despite a lower concentration than 100 aM, Abs416 reduced distinguishably after Cas9/gRNA addition, implying that the double-checking mechanism of DAMPR assay can operate in a wide concentration range of samples. The images in Figure 3b,d,f are representative well plates after LAMP and DNAzyme reactions (ON) and CRISPR reaction (OFF) with various concentrations of target genes. The real-time fluorescence curves for the detection of three SARS-CoV-2 genes are also provided in Figure S5a,b.
Figure 3.
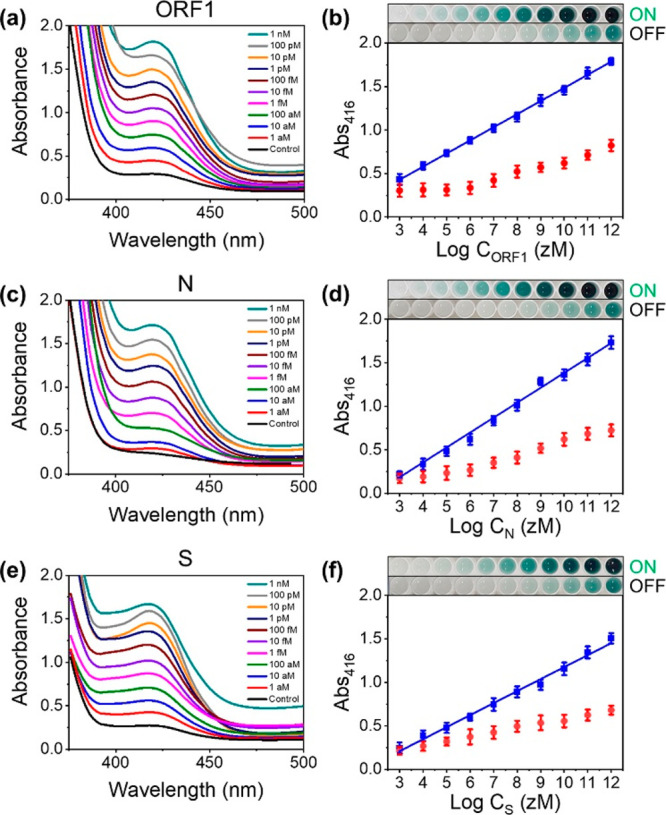
(a, c, and e) Absorbance spectra after detection of SARS-CoV-2 (a) ORF1, (c) N, and (e) S genes using RT-LAMP and DNAzyme reactions. The concentrations of genes were varied from 1 aM to 1 nM. (b, d, and f) Plots of Abs416 as a function of logarithmic concentration of SARS-CoV-2 (b) ORF1, (d) N, and (f) S genes (n = 6, error bar = standard deviation). Blue data were obtained after the detection of SARS-CoV-2 genes using RT-LAMP and DNAzyme reactions. Red data were obtained after double-checking of SARS-CoV-2 genes using Cas9/gRNA complexes. Photographs are well plates after RT-LAMP and DNAzyme reactions (ON) and CRISPR reaction (OFF). The left wells are control samples.
The specificity of DAMPR assay was examined using several types of viral genes (SARS-CoV-2, influenza A virus subtype H1N1, H3N2, H5N2, H1N2, and H3H8, and respiratory syncytial virus A (RSV A)). As shown in Figure S5c, significant enhancement of the absorbance signal was observed only in the presence of SARS-CoV-2, clearly demonstrating that DAMPR assay has excellent specificity corresponding for the target.
Further, we attempted to detect SARS-CoV-2 lysates by DAMPR assay. SARS-CoV-2 was provided by the Korea National Institute of Health and cultured in Biosafety Level 3 (BL-3) laboratory of Korea Research Institute of Bioscience and Biotechnology (KRIBB). SARS-CoV-2 lysates (1, 10, 50, 100, 150, 200, and 300 plaque-forming unit (PFU)/mL) were added to a well plate and DAMPR assay was performed (Figure S6a). The colorimetric signals indicate that the virus is detectable at a low concentration of 1 PFU/mL. In addition, SARS-CoV-2-spiked human nasopharyngeal aspirates and sputum samples (n = 5), obtained from the patients negatively diagnosed for COVID-19 by qRT-PCR, were tested as well. Figure S6b indicates that DAMPR assay provides positive results for all samples, demonstrating the assay detection of SARS-CoV-2 directly in the human fluid samples. Figure S7 indicates that DAMPR assay could detect SARS-CoV-2 without RNA extraction and preamplification processes.
Development of DAMPR Assay System
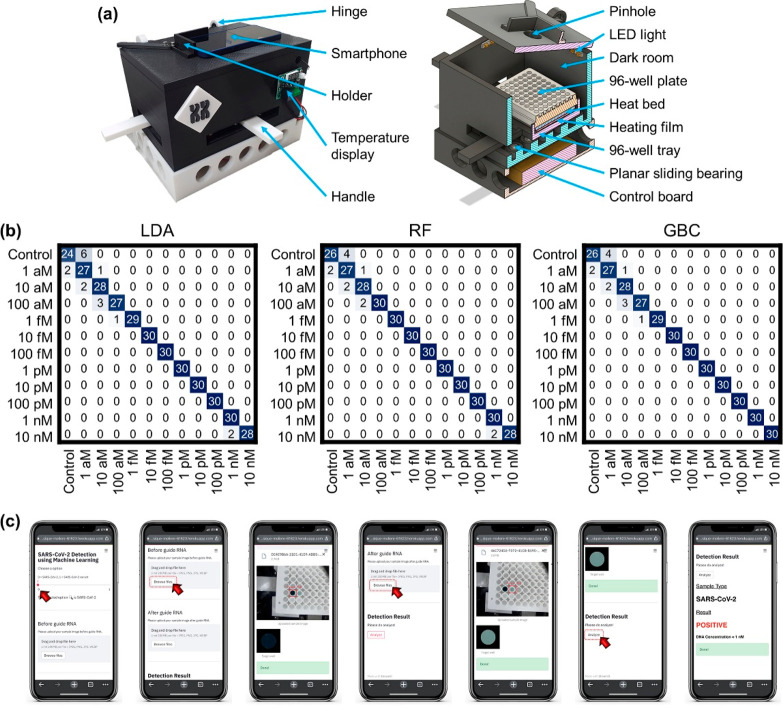
Following the evaluation of DAMPR assay, we strived to construct a portable DAMPR assay system. The left image of Figure 4a shows the portable DAMPR assay system fabricated via a 3D printer. A hinge structure was applied to open and close the system, thereby enabling well plate places in dark room. A holder to fix a smartphone was attached at the top. The system front housed the temperature controller, and two handles allowed the control of the well plate, thereby enabling the easy capture of photographs without opening the system. Further, the right image of Figure 4a illustrates the inside structure of the system. Pinhole, led light, and dark room allowed for capturing photographs under constant conditions, enhancing the accuracy of DAMPR assay results. A reaction plate can be placed on a tray employing a heat bed and heating film for temperature control. Under the 96-well tray, a planar sliding bearing system exists for the movement of the tray. At the system bottom, a control board was inserted. The constructed DAMPR assay system weighs ∼900 g and is operated via battery or external power supply. Further, the system is 170 × 250 × 207 mm3 in width, depth, and height, respectively. Overall, an adult can handle the DAMPR assay system, allowing for the on-site detection of SARS-CoV-2.
Figure 4.
(a) Photograph of portable DAMPR assay system (left) and inside structure illustration of portable DAMPR assay system (right). (b) Confusion matrices for the detection of SARS-CoV-2 gene by LDA, RF, and GBC classifiers, respectively. (c) User manual of smartphone application for SARS-CoV-2 detection by DAMPR assay system. The exampled diagnostic result is positive.
In the DAMPR assay system, no optical instruments are allowed for the measurement of absorbance in the system. Instead, a smartphone was adopted to analyze the diagnostic results of DAMPR assay. Further, to acquire accurate diagnostic results of SARS-CoV-2, ML techniques were employed. The linear discriminant analysis (LDA), random forest (RF), and gradient boosting classifier (GBC) classifiers based on ML analysis quantified the concentration of the SARS-CoV-2 gene by learning the 300 sample images for each concentration. The accuracy, precision, recall, and F1 score of the classifiers were computed to assess their performance (Tables S4 and S5). According to a comparison of the LDA, RF, and GBC classifiers on the DAMPR assay, the RF had the best result with 99.38% accuracy. Consequently, for clear understanding, confusion matrices based on the correlation between the true and prediction labels are shown in Figure 4b, which enable visualization of the performance of the LDA, RF, and GBC, respectively. Similarly, the RF classifier showed the best correlation between true and prediction labels; we, therefore, applied the RF classifier to the DAMPR assay system.
Finally, the custom-designed smartphone application was developed for the public to conveniently and rapidly check SARS-CoV-2 diagnostic results and viral concentration (Figure 4c, Figure S8, and Movie S1). Prior to transferring the image to the server, image processing techniques were embedded in the application to determine the region of interest (ROI). For classification, the processed image is sent to the server via the Firebase with the result displayed on the smartphone screen following concentration level classification. The criteria for determining SARS-CoV-2 Positive/Negative/False Positive results are as follows: (1) If the estimated concentration level in the first photograph is greater than 1 aM and level in the second photograph is lower than that in the first, the result is considered positive (Figure 4c). (2) If the estimated concentration level in the first photograph is 0 and the level observed in the second photograph is the same as that in the first, the sample is considered negative (Figure S8a). (3) Finally, if the concentration level in the first photograph is greater than 1 aM, accompanied by a similar or higher level in the second photograph, the sample is considered false-positive (Figure S8b). The difference in concentration level between the first and second photographs was determined on the basis of the experimental results shown in Figure 3. With the development of the portable DAMPR assay system and user-friendly smartphone application, it is anticipated that SARS-CoV-2 can be detected on-site simply within an hour.
Diagnosis of COVID-19 Patients by Using DAMPR Assay System
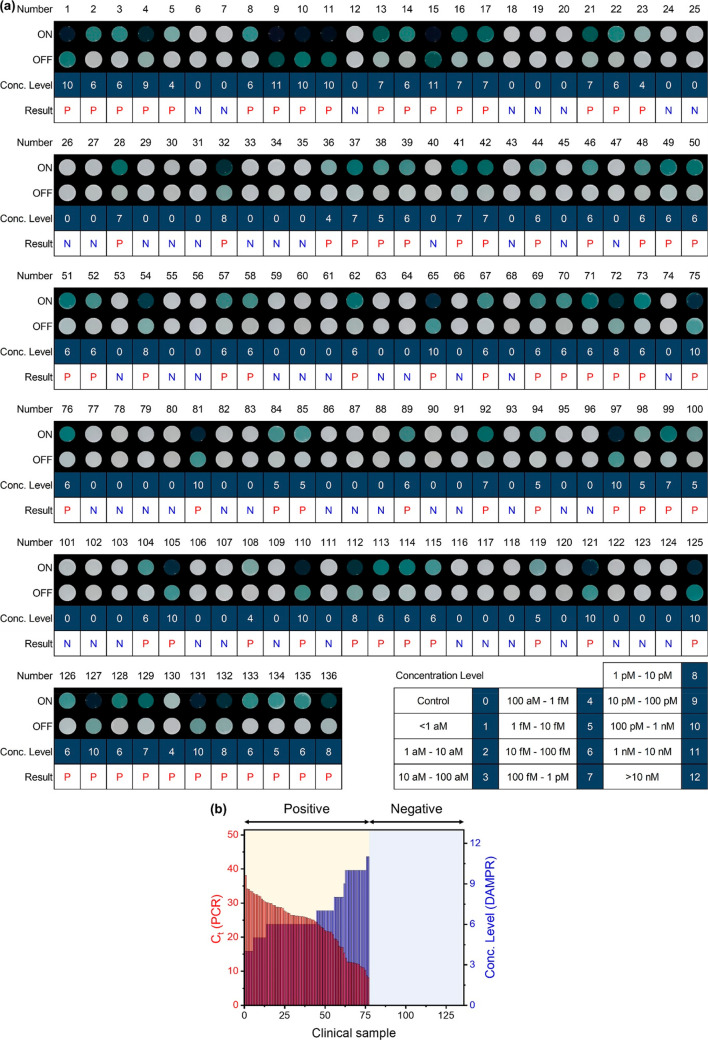
The DAMPR assay system was applied to diagnose COVID-19 patients. A total of 136 samples were acquired from Gyeongsang National University College of Medicine and Yonsei University Health Service Center, Severance Hospital and were directly used for DAMPR assay in blind conditions. The diagnostic results were compared with qRT-PCR results (Table S6). Figure 5a shows DAMPR assay results for the 136 clinical samples in this study, with patient number (Number), processed image for detection (ON), processed image for double-checking (OFF), the concentration level of SARS-CoV-2 (Conc. Level), and diagnostic result (Result). The image in Figure 5a was processed for ML analysis. Seventy-seven samples were diagnosed as positive for SARS-CoV-2, while the remaining 59 were negative. This result perfectly matched with the diagnostic results based on qRT-PCR, indicating 100% clinical sensitivity and specificity of the DAMPR assay system. Furthermore, the estimated viral concentration levels by DAMPR assay system inversely agree well with the cycle threshold (Ct) values of the qRT-PCR results (R = −0.9785). Figure 5b is a comparative plot of Ct value (PCR) and Conc. Level (DAMPR) as a function of the clinical sample, wherein the samples are classified as positive and negative. In the case of the negative samples, both DAMPR and PCR results provided no values, whereas for the positive samples, the concentration levels of SARS-CoV-2 estimated by the DAMPR assay system are inversely correlated to the Ct values obtained by qRT-PCR. This indicates that the portable DAMPR assay system can detect SARS-CoV-2 semiquantitatively. The DAMPR assay system is, thus, expected to provide positive or negative results for SARS-CoV-2 along with an accurate concentration level of the virus. Patients with high viral concentrations may become superspreaders and are critical for the prevention of viral spreads.31 As a result, the DAMPR assay system could be effective for both screening unknown COVID-19 patients and preventing possible viral superspreaders.
Figure 5.
(a) Diagnostic results of 136 clinical samples using DAMPR assay system. Result tables consist of patient number (Number), processed image after LAMP and DNAzyme reactions (ON), processed image after CRISPR reaction (OFF), concentration level of SARS-CoV-2 (Conc. Level), and diagnostic result (Result). Number of concentration level is noted at right-bottom. Diagnostic result is written P (red) for positive and N (blue) for negative. (b) Comparative plot of Ct value (PCR) and Conc. Level (DAMPR) as a function of 136 clinical samples.
Detection of SARS-CoV-2 Variants by Using v-DAMPR Assay System
Since the emergence of the COVID-19 pandemic, SARS-CoV-2 has mutated over time, resulting in variations in the population of circulating viral strains.32 Currently, alpha, beta, gamma, delta, and omicron variants of SARS-CoV-2 have been classified as variants of concern (VOCs), while epsilon, zeta, eta, theta, iota, kappa, lambda, and mu are variants of interest (VOIs) by the World Health Organization (WHO).33,34 As the SARS-CoV-2 variants are expected to exhibit higher transmission rates and resistivity against vaccines than wild-type (WT) SARS-CoV-2, the detection of SARS-CoV-2 variants is important.35
D614G mutation of SARS-CoV-2 spike (S) protein is considered an important parameter in both viral spread and vaccine efficacy, owing to its ability to induce the increased affinity of the virus with angiotensin-converting enzyme 2 (ACE2) receptor, enhancement of viral replication in the upper respiratory tract, and increased susceptibility of the virus to neutralization by antibodies (Figure 6a).36,37 As the D614G mutation has been found in all SARS-CoV-2 VOCs and VOIs, we attempted to detect the D614G mutation of SARS-CoV-2 by using the variant-specific DAMPR assay (v-DAMPR assay). Furthermore, we sought to detect the delta (T478K)- and omicron (A67V)-specific variants, respectively, because these two variants have become dominant among SARS-CoV-2 variants. Figure 6b shows the sequences of WT, D614G, T478K, and A67V SARS-CoV-2. A single mutation in the S gene of the SARS-CoV-2 D614G variant can stimulate the formation of glycine instead of aspartic acid in the S protein. Also, the SARS-CoV-2 T478K variant can stimulate the formation of lysine instead of threonine in S protein and the SARS-CoV-2 A67V variant can stimulate the formation of valine instead of alanine in S protein. Considering these sequence variations, we designed the v-DAMPR assay to recognize the SARS-CoV-2 variants. Briefly, the RT-LAMP and DNAzyme reactions are processed in a manner similar to the DAMPR assay. At this stage, both WT and mutant SARS-CoV-2 provide highly increased color signals. Following the photographing, the Cas9/mutation-specific gRNA complex is treated. For samples including SARS-CoV-2 variants, the color signal of the well plate decreases following the CRISPR-Cas9 reaction. For WT SARS-CoV-2 present in the sample, dark green color is preserved following the CRISPR reaction. The v-DAMPR assay can recognize SARS-CoV-2 on the basis of the first color development and subsequently identify whether the detected virus is WT or a variant on the basis of the second color change. Previous studies have concluded that the PAM-adjacent sequences, defined as the sequences of 6–12 bp immediately upstream of the PAM site, are critical for the target recognition ability of the CRISPR-Cas9 system.38 As D614G, T478K, and A67V gRNA can recognize the mutation within the PAM-adjacent sequences, the v-DAMPR assay identifies the mutations efficiently.
Figure 6.
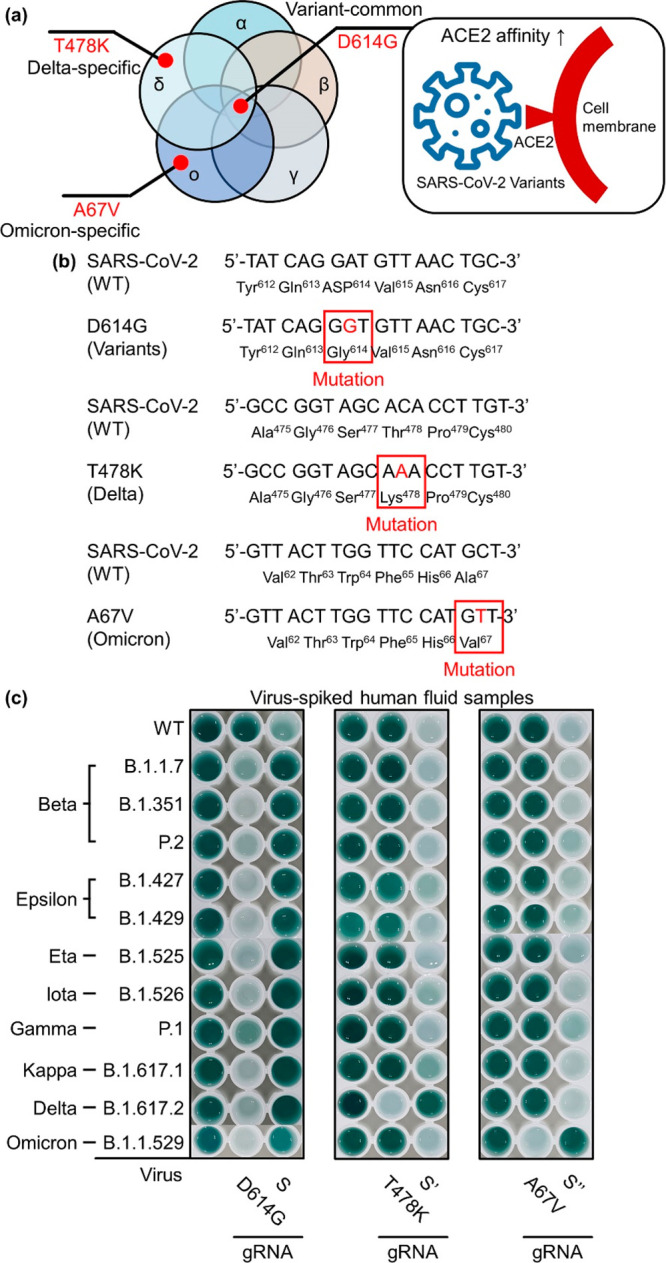
(a) D614G, T478K, and A67V mutations of SARS-CoV-2 S protein. (b) Sequences of WT, D614G, T478K, and A67V SARS-CoV-2 S protein. (c) Photograph of well plates after detection of WT and SARS-CoV-2 variants (beta, epsilon, eta, iota, gamma, kappa, delta, and omicron) using v-DAMPR assay from viral lysate-spiked human nasopharyngeal aspirate samples (102 PFU/mL).
Figure 6c represents the v-DAMPR assay results following the detection of WT SARS-CoV-2 and its variants (beta, epsilon, eta, iota, gamma, kappa, delta, and omicron) in human fluid samples with the concentration of the viruses at 102 PFU/mL. All SARS-CoV-2 strains exhibited a dark green color after LAMP and DNAzyme reactions. Furthermore, following the Cas9/mutation-specific gRNA reaction, the color of SARS-CoV-2 variants became sparse but that of WT SARS-CoV-2 was maintained. In detail, when D614G gRNA was used, all SARS-CoV-2 variants exhibited weak signals because this mutation is common in SARS-CoV-2 variants. In contrast, upon the reaction with the Cas9/S gRNA complex, the signal of only the WT virus decreased, while those of mutant viruses were unchanged. When T478K (delta-specific) gRNA was used, only the delta variant of SARS-CoV-2 provided sparse color. In the control experiment using Cas9/S′ gRNA complex, only the color of delta SARS-CoV-2 was unchanged. Finally, when A67V (omicron-specific) gRNA was employed, the signals of the omicron variant of SARS-CoV-2 decreased significantly. In the presence of S″ gRNA, SARS-CoV-2 viruses except for omicron variant showed decreased signals. This verifies that the v-DAMPR assay can specifically detect the various SARS-CoV-2 variants.
For the on-site detection of SARS-CoV-2 variants, the v-DAMPR assay was applied to a portable system and a smartphone application was developed in a manner similar to that described before. On uploading two photographs acquired from the v-DAMPR assay system, the diagnostic result for SARS-CoV-2 variants and concentration level are informed routinely (Figure S9 and Movie S2). The developed v-DAMPR assay system was employed for the diagnosis of 10 patients infected with SARS-CoV-2 variants (1–10 in Figure 7a) and 10 clinical samples obtained from the WT SARS-CoV-2-infected patients (11–20 in Figure 7a). In this case, the D614G gRNA was used to determine the presence of SARS-CoV-2 variants. The first color signals turned on for all samples (ON), implying successful detection of SARS-CoV-2, whereas the second color signals decreased for SARS-CoV-2 variant samples but were maintained for the WT virus samples (OFF). In addition, we tried to diagnose 50 patients infected with the delta variant of SARS-CoV-2. In this experiment, the T478K gRNA was employed. As shown in Figure 7b, the dark color was obtained for 50 samples (ON), and the color became sparse after reaction with Cas9/T478K gRNA (OFF). The smartphone application suggested positive results for all samples. Lastly, 10 patients infected with the omicron variant of SARS-CoV-2 were diagnosed by the v-DAMPR assay system with A67V gRNA. These samples showed strong first color signals and weak second signals, indicating the identification of the SARS-CoV-2 omicron variant (Figure 7c). The estimated concentration levels of SARS-CoV-2 by v-DAMPR assay system (Conc. Level) are consistent with the Ct values of qRT-PCR for the same clinical samples (Table S7). Thus, the diagnostic results confirm that the v-DAMPR assay system can accurately identify the patients infected with SARS-CoV-2 variants. Since the RT-LAMP, DNAzyme, and CRISPR-Cas9 reactions contributed synergistically to the sensitive and selective detection of SARS-CoV-2, the developed DAMPR assay system including smartphone application realized on-site, rapid, and simple detection of SARS-CoV-2 and variants.
Figure 7.
(a–c) Diagnostic results of 80 clinical samples using v-DAMPR assay system. Result tables consist of patient number (Number), processed image after LAMP and DNAzyme reactions (ON), processed image after Cas9/gRNA reaction (OFF), concentration level of SARS-CoV-2 (Conc. Level), and diagnostic result (Result). Number of concentration level is noted at right-bottom. Diagnostic result is written P (red) for SARS-CoV-2 variants and N (blue) for WT SARS-CoV-2. (a) Clincal samples from 1 to 20 were analyzed by v-DAMPR assay system with D614G gRNA for the detection of SARS-CoV-2 variants, (b) samples from 21 to 70 with T478K for SARS-CoV-2 delta variant, and (c) samples from 71 to 80 with A67V for SARS-CoV-2 omicron variant.
Discussion
In general, the RT-LAMP technique assessed the color change of the pH-sensitive dye by utilizing the principle that extensive DNA synthesis lowers the pH of the reaction solution. However, according to a prior study, biological samples such as nasopharyngeal swab eluates might reduce the pH when applied directly to the LAMP reagents, resulting in false-positive results.39,40 Thus, the RT-LAMP amplification product should be detected directly. Consequently, we employed a DNAzyme reaction induced by RT-LAMP amplification products. In addition, PS-DNA primers facilitate the DNAzyme reaction by lowering the Tm of the DNA/PS-DNA LAMP products and, consequently, enable the detection of the target genes by the naked eye. The comparison of DAMPR assay with previous colorimetric SARS-CoV-2 detection methods is presented in Figure S10.
The recent molecular diagnostic approaches have achieved low LOD values of a few copies/sample or aM, and the DAMPR assay exhibits attomolar sensitivity for ORF1, N, and S genes of SARS-CoV-2 (Table S3). The high sensitivity of molecular diagnostic methods has enabled the precise detection of genes with a small volume of samples, thereby improving the capability of the disease diagnosis greatly. However, enhancement in the sensitivity of methods has prompted concerns for false signals. For the accurate detection of target genes, the DAMPR assay was designed to detect ORF1, N, and S genes, whereas previous technologies usually detected one or two genes.41,42 This contributes to reducing the false-negative signals. Furthermore, the double-checking system by Cas9/gRNA complexes eliminated false-positive signals in DAMPR assay, enabling accurate diagnosis of the clinical samples without false signals (Figure S11).
The estimated minimum number of sample size was 49 patients and 49 controls when the parameters required to estimate the sample size was as below: type I error = 5%, power = 80%, sensitivity in null hypothesis = 50%, sensitivity in alternative hypothesis = 70% (clinically important difference = 20%), and disease prevalence = 50%).43 On fixing parameters including type I error, power, and disease prevalence, only clinically important differences increase, while the required minimum sample size decreases. Therefore, a total of 216 study subjects (157 patients and 59 controls) are sufficient to evaluate the diagnostic performance of the DAMPR assay system in the current situation, with 100% sensitivity and specificity.
The identification of SARS-CoV-2 VOCs and VOIs is critical because these variants could exhibit higher transmission rates and even resistivity against vaccines.44 To facilitate on-site rapid identification of SARS-CoV-2 variants, v-DAMPR assay, capable of detecting D614G, T478K, and A67V mutations of SARS-CoV-2, was developed. The single nucleotide polymorphism (SNP) recognition nature of CRISPR-Cas9 facilitated the development of v-DAMPR assay. Currently, the SARS-CoV-2 VOCs and VOIs have been found to possess D614G mutation in the S gene.45 Therefore, the positive result of v-DAMPR assay system with D614G gRNA indicates that the tested sample includes SARS-CoV-2 VOCs or VOIs. Further, v-DAMPR assay system with T478K and A67V gRNAs could successfully identify the delta and omicron variants of SARS-COV-2. The applicability of v-DAMPR assay to recent variants of viruses proves that the current assay system might be useful in responding to future emerging infectious viruses.
The DAMPR assay can be performed almost everywhere owing to its smartphone-based portable model. The diagnostic result and concentration level of SARS-CoV-2 can be checked using just two pictures acquired during the DAMPR assay. Moreover, the DAMPR assay system can detect several tens of samples. The blind test of 136 clinical samples was completed within 3 h using the DAMPR assay system. Furthermore, the detectable sample number of DAMPR assay system can be expanded easily and owing to the daily tested numbers for COVID-19; this diagnostic system is expected to be capable of massive sample detection.
The consistency of signal output interpretation was enhanced via an automated mobile phone application, allowing for reporting the diagnostic result and the concentration of the target gene. The smartphone application allows for the identification of individuals who may be viral supercarriers and, therefore, superspreaders. The previous literature has suggested that 80–90% of infections are caused by 10–20% of infected individuals.46,47 Therefore, identifying viral supercarriers within presymptomatic and asymptomatic stages of the disease in the community is vital. The DAMPR assay system may be useful to determine a majority of infected individuals within asymptomatic stages of the disease. Further, according to the previous result, the samples of SARS-CoV-2 positive patients with asymptomatic stages have a mean Ct value of 32, corresponding to a sample with a viral RNA load of approximately 1 × 105 copies/mL;48 as the LOD of the DAMPR assay system covers this range of asymptomatic patients, it allows the assessment/prediction of transmission of the virus by screening viral supercarriers by the DAMPR assay system.
We blindly tested a sample of 136 SARS-CoV-2 patients (Table S6). Among them, 126 samples (No. 1–126) are nasopharyngeal swabs and 10 samples (No. 127–136) are sputum. For the diagnostic test of laboratory-developed or commercial platforms for SARS-CoV-2, the FDA-approved sample types are nasopharyngeal swabs and sputum.49,50 Although there is a difference in the collection method between the nasopharyngeal aspirate sample and the sputum sample, there is no significant difference in the subsequent processing, accuracy, and sensitivity between two samples.51 Therefore, we performed the DAMPR assay on these two samples and could detect SARS-CoV-2 successfully.
However, the current DAMPR assay system has the opportunity for improvement. Despite the detection of SARS-CoV-2 and its variants in many clinical samples, a completely automated approach for massive analysis is required. Further, the ML-based smartphone application can be improved by preparing and training more photographs, thereby enhancing the accuracy of the diagnostic result. The DAMPR assay must be widened to the precise analysis of SARS-CoV-2 variants through the design of LAMP primers and gRNA sequences. In the future, we aim to improve the DAMPR assay system and hope that this system will be extensively used for the diagnosis of infectious diseases in the real world.
Conclusions
In summary, we developed a colorimetric DNAzyme reaction triggered by LAMP with CRISPR, referred to as DAMPR assay for detecting SARS-CoV-2 and variants genes, eliminating the false-positive signals by the double-checking system. Further, the portable assay system and the smartphone application were also developed for POCT diagnostics of COVID-19. Using the assay system, a total of 216 clinical samples were diagnosed with 100% sensitivity and specificity. We anticipate that the DAMPR assay can be expanded to various biomedical diagnostics.
Methods
Materials
All nucleic acids used in this study were synthesized from Integrated DNA Technologies, Inc. (Coralville, IA) and Bioneer Co. (Daejeon, Korea). Hemin, NaCl, dimethyl sulfoxide (DMSO), 2-[4-(2-hydroxyethyl)piperazin-1-yl] ethanesulfonic acid (HEPES), ABTS, tosyl phenylalanyl chloromethyl ketone (TPCK) trypsin, Triton X-100, and H2O2 were purchased from Sigma-Aldrich (St. Louis, MO). A WarmStart LAMP Kit (DNA and RNA), Cas9 nuclease (Streptococcus pyogenes), Luna Universal one-step RT-qPCR, and 10× NEBuffer r3.1 were purchased from New England Biolabs Inc. (Beverly, MA). Thermo Fisher Scientific provided Dulbecco’s modified Eagle’ medium (DMEM), antibiotic-antimycotic, and 10× tris-borate ethylenediaminetetraacetic acid (EDTA) (TBE) buffer (Waltham, MA). Biotium’s GelRed nucleic acid dye (41003) was used (Hayward, CA). LPS Solution provided the tris(2-carboxyethyl)phosphine (TCEP) (Daejeon, Korea). Dynebio provided the EDTA solution (Seongnam-si, Gyeonggi-do, Korea). Hemin stock solution (1 mM) was made in DMSO and kept at −20 °C in the dark.
SARS-CoV-2 strains (BetaCoV/Korea/KCDC03/2020, hCoV-19/Korea/KDCA51463/2021; GR, B.1.1.7, hCoV-19/Korea/KDCA55905/2021; GH, B.1.351, hCoV-19/Korea/KDCA72731/2021; GR, P.2 lineage, hCoV-19/Korea/KDCA49671/2021; GH, B.1.427, hCoV-19/Korea/KDCA59777/2021; GH, B.1.429, hCoV-19/Korea/KDCA79765/2021; G, B.1.525, hCoV-19/Korea/KDCA49671/2021; GH, B.1.526, hCoV-19/Korea/KDCA95637/2021; GR, P.1, hCoV-19/Korea/KDCA105288/2021; G, B.1.617.1, hCoV-19/Korea/KDCA119861/2021; G, B.1.617.2, hCoV-19/Korea/KDCA447321/2021; GRA, B.1.1529) were provided by the National Culture Collection for Pathogens (NCCP), which is operated by the Korea National Institute of Health. The strains were cultured as described previously.52,53 All experiments involving the use of SARS-CoV-2 were performed at the Korea Centers for Disease Control and Prevention (KCDC)-approved BL-3 facility of KRIBB in accordance with institutional biosafety requirements. The genomic RNA of the viruses (SARS-CoV-2, influenza A virus subtype H1N1, H3N2, H5N2, H1N2, and H3H8, and RSV A) was provided by the NCCP.
Kapton film heater (12 V, 3W, Ø22), aluminum heat exchanger 1T (138 × 96), 12 V digital temperature control switch module (SZH-AT024), control board (SMPS KO-120W12 V), and led module (MW3HT217ZN-70) were purchased from DEVICE MART (Incheon, Korea).
DAMPR Assay
First, the 10× primer stocks (16 μM FIP, 16 μM BIP, 2 μM FIP, and 2 μM BIP in nuclease-free water) were prepared. All components were thawed and, subsequently, used at 25 °C. They were then placed on ice, vortexed briefly to allow mixing, and centrifuged to collect reaction materials. LAMP reaction mixture (25 μL) containing 12.5 μL of WarmStart LAMP 2× Master Mix, 2.5 μL of 10× primer stocks, 9 μL of distilled water (DW), and 1 μL of the sample were prepared and incubated at 65 °C for 30 min. Subsequently, 7 μL of DW, 8 μL of 10 μM hemin, 10 μL of 3 mM ABTS, and 10 μL of 6 mM H2O2 were added to the RT-LAMP reaction product and incubated at 25 °C for 10 min. Following G-quadruplex DNAzyme reaction, the CRISPR solution comprising 28 μL of DW, 10 μL of 10× NEBuffer r3.1 (100 mM NaCl, 50 mM tris-HCl, 10 mM MgCl2, 100 μg/mL recombinant albumin, pH 7.9), 1 μL of 1 μM gRNA, and 1 μL of 1 μM Cas9 was added to the product and incubated at 25 °C for 10 min.
An android phone was used to capture the images of reaction products, and the absorbance signals were analyzed with a Tecan Infinite M200 pro microplate reader from 375 to 500 nm (Mannedorf, Switzerland).
To prepare PCR amplification products (ORF1, N, and S genes of SARS-CoV-2), qRT-PCR was performed on a C1000 thermal cycler (Biorad, CA) in a 50 μL solution containing genomic RNA (10 ng), with primer set (0.4 μM each), 1× Luna Universal one-step reaction mix, and 1× Luna WarmStart RT enzyme mix. qRT-PCR was programmed for 10 min at 55 °C for reverse transcription and for 1 min at 95 °C for initial denaturation, followed by 45 cycles of 10 s at 95 °C and 30 s at 60 °C, following which, the qRT-PCR products were confirmed via agarose gel electrophoresis, purified using a NucleoSpin Gel and PCR cleanup (Macherey-Nagel, Düren, Germany), and their concentrations determined using a nanodrop ND-1000 spectrophotometer (Wilmington, DE). The used primers were ORF1 FP and BP, N FP and BP, and S FP and BP, respectively.
Ninety microliters of virus samples were mixed with 10 μL of TCEP/EDTA (final concentrations of 100 and 1 mM, respectively) and heated at 50 °C for 5 min and 64 °C for 5 min to make viral lysates. The viral lysate samples were detected by DAMPR assay as described above. Also, the viral lysate-spiked human nasopharyngeal aspirates and sputum samples were detected by DAMPR assay as described above.
For the RNA extraction, 100 μL of SARS-CoV-2 lysate (104 PFU/mL) was applied to a QIAamp Viral RNA kit (Qiagen, Germany) following the manufacturer’s protocol. One microliter of the extracted sample was used for DAMPR assay.
Development of DAMPR Assay System
The DAMPR assay system was designed employing Autodesk’s Fusion 360 3D CAD software. The housing and components were printed using a Fused Deposition Modeling (FDM) 3D printer (Thing & Thinks, Incheon, Korea). Black poly(lactic acid) (PLA) filament was chosen for printing the housing, thereby minimizing the reflectance during photography. To perform the RT-LAMP reaction at 65 °C, a heating bed was fabricated using an aluminum plate and a film heater, which spread the heat evenly to the entire 96-well plate. Regarding the heating bed, a 1 mm thick aluminum plate was cut to 11 cm × 8 cm, and 11 Kapton film heaters with Ø22 mm were attached behind the aluminum plate. The film heaters were soldered in parallel, powered from a 12 V power supply and connected to a digital temperature controller with a temperature sensor. The set point for the temperature controller circuit, which can handle the temperature up to 110 °C in a closed-loop by attaching the temperature sensor under the heating bed, was programmed to 65 °C, and the temperature was maintained at a constant. To prevent thermal deformation of the structure printed by the 3D printer, a 1.5 cm high iron spacer was configured in the form of support, which prevented the heated bed from getting attached to the well tray. The tray could be moved by attaching a handle to capture all wells of the microplate by smartphone, and a planar sliding bearing was applied between the housing and the tray to smoothen the movement by reducing frictional force. Moreover, as external light does not enter the interior space, the housing is manufactured in the form of a dark room such that external light does not penetrate into the interior space. Further, the LED lights were soldered in series with a 300 Ω resistor, allowing passage of a current of 30 mA when connected to a 12 V power supply. LED lights were attached to the top of the four walls in the dark room to realize a constant light source. A hinge structure was applied to the system such that a 96-well plate could be placed in dark room. A holder was attached to fix a smartphone at the top of the system, ensuring a consistent focal distance was maintained and the nonuniformity of the lighting condition was resolved.
As shown in Figure 1a, RT-LAMP and G-quadruplex DNAzyme reactions are processed in a well plate of the DAMPR assay system. Following the reactions, the first photograph is acquired. Thereafter, CRISPR-Cas9 reaction is completed and the second photograph is obtained. Subsequently, two images acquired during the DAMPR assay are transmitted to the ML-based colorimetric program developed in Python through the web-based smartphone application (http://classique-moliere-61823.herokuapp.com), wherein the ROI where the color change occurs during the DAMPR assay being extracted. Several image processing methods can be applied to extract the ROI from the photograph, including thresholding, binarizing, masking, and contour detection. Subsequently, the extracted ROI is masked with the original image to extract the features for ML-based classifier, which receives the color features of ROI as input and outputs the concentration level of SARS-CoV-2. Further, comparing the outputs of two images, the final diagnostic result is displayed in the end-user application. The current platform works with any operating system (Windows, Mac OSX, or Linux) that supports Python over 3.8.
The smartphone app was designed with a simple and user-friendly interface capable of streamlit-based image processing. It was deployed on Heroku, an example of Platform as a Service (PAAS). Heroku runs our application within dynos smart containers on a reliable, fully managed runtime environment. The smartphone application has the ability to crop the desired ROI in the target image. After the image is captured using the camera in the application, the desired ROI defined its edge and measured its diameter employing the Hough Circle Algorithm. This algorithm has been used to determine an arbitrary shape in images and to measure its diameter in pixels. From the cropped image, the same color features used in the classifier trained using ML were extracted and inputted to the learned classifier, and then, the viral concentration level was estimated.
The SARS-CoV-2 diagnostic results on the application are determined as follows. If the concentration level in the first photograph is greater than 1 aM and the level in the second photograph is lower than that in the first, the result is positive. If the concentration level in the first photograph is 0 and the level observed in the second photograph is the same as that in the first, the result is negative. Finally, if the concentration level in the first photograph is greater than 1 aM, accompanied by a similar or higher level in the second photograph, the result is false-positive.
Training ML Model
With the smartphone-based platform, the best LDA, RF, and GBC classifiers were determined. After image processing, the classifiers were trained by extracting color features. To analyze the effect of color spaces on concentration levels, the RGB values of the image were first transformed to HSV and L*a*b*. Subsequently, for each R, G, B, H, S, V, L*, a*, b* color channel, mean values were determined. In the Python programming language, a total of nine characteristics were used to train the classifiers. In machine learning, k-fold cross-validation is a common technique for evaluating classifier performance that divides the data set into k equal folds.54 The k −1 number of folds is used to train the model, while the remaining fold is used to test the trained model. This procedure is performed k times. As a result, the various fold is used as a training and testing set in each repeat, resulting in k different accuracy values, with the average of these values used to calculate the classifier’s overall accuracy. In this case, k is set to 10, which has been reported to be adequate for avoiding problems caused by large bias and variance.55
Diagnosis of COVID-19 Patients by Using DAMPR Assay System
A total of 216 samples were acquired from Gyeongsang National University College of Medicine and Yonsei University Health Service Center, Severance Hospital. The nasopharyngeal aspirates and sputum samples were collected from patients by using flocked nasopharyngeal swabs and, thereafter, placed into the virus transport media (3 mL, Copan Diagnostics Inc., Murrieta, CA). All samples were stored at −70 °C until use. The protocol for this study was reviewed and approved by the Institutional Review Board of Gyeongsang National University College of Medicine, Jinju, Korea (IRB approval number: 2020-10-002) and Yonsei University Health Service Center, Severance Hospital, Seoul, Korea (IRB approval number: 4-2020-0465). Also, written informed consent was acquired from each subject. For the diagnosis of COVID-19 patients by using DAMPR assay, the clinical samples were directly used and the assay procedures were the same as described above except for the final concentration of Cas9/gRNA complexes (100 nM).
For comparison of DAMPR assay with qRT-PCR, qRT-PCR was performed on a C1000 thermal cycler in a 50 μL solution containing clinical sample (1 μL), a primer set (0.4 μM each), 1× Luna Universal one-step reaction mix, and 1× Luna WarmStart RT enzyme mix. qRT-PCR was programmed for 10 min at 55 °C for reverse transcription and for 1 min at 95 °C for initial denaturation, followed by 45 cycles of 10 s at 95 °C and 30 s at 60 °C. The employed primers for SARS-CoV-2 are ORF1 FP and BP, N FP and BP, and S FP and BP, respectively.
Detection of SARS-CoV-2 Variants by the v-DAMPR Assay System
For the detection of WT and D614G SARS-CoV-2 by the v-DAMPR assay system, two types of CRISPR-Cas9 systems were employed: first, the Cas9/S gRNA complex that recognizes the S gene of WT SARS-CoV-2 and, second, the Cas9/D614G gRNA complex that recognizes the S gene of D614G SARS-CoV-2. For the detection of WT and T478K SARS-CoV-2, two CRISPR-Cas9 systems were employed: first, the Cas9/S′ gRNA complex and, second, the Cas9/T478K gRNA complex. For the detection of WT and A67V SARS-CoV-2, two CRISPR-Cas9 systems were employed: first, the Cas9/S″ gRNA complex and, second, the Cas9/A67V gRNA complex. The assay procedures were the same as above, except for the employed gRNAs, which are different. For the diagnosis of COVID-19 patients infected with SARS-CoV-2 variants by using v-DAMPR assay, the clinical sample was directly used, and the assay procedures followed were the same as mentioned before.
The smartphone application for SARS-CoV-2 variants was developed as described above. The diagnostic results of the application are determined as follows. If the concentration level in the first photograph is greater than 1 aM and the level in the second photograph is lower than that in the first, the sample is positive for variants. If the concentration level in the first photograph is 0 and the level observed in the second photograph is the same as that in the first, the sample is negative for variants. Finally, if the concentration level in the first photograph is greater than 1 aM, accompanied by a similar or higher level in the second photograph, the sample is positive but not for variants.
For comparison of v-DAMPR assay with qRT-PCR, the results were provided from Gyeongsang National University College of Medicine.
Acknowledgments
This research was supported by National R&D Programs through National Research Foundation (NRF) of Korea funded by Ministry of Science and ICT (MSIT) of Korea (NRF-2021M3E5E3080379, NRF-2021M3E5E3080382, NRF-2021M3H4A1A02051048, NRF-2018M3A9E2022821, NRF-2021M3E5E3080844, NRF-2022R1C1C1008815, and NRF-2020R1A2C1010453), Global Frontier Program through Center for BioNano Health-Guard funded by MSIT of Korea (H-GUARD_2014M3A6B2060507 and H-GUARD_2013M3A6B2078950), R&D Program through National Research Council of Science & Technology (NST) of Korea funded by MSIT of Korea (No. CRC21021-100), Technology Development Program for Biological Hazards Management in Indoor Air through Korea Environment Industry & Technology Institute (KEITI) funded by Ministry of Environment (ME) of Korea (2021003370003), K-Sensor Technology Development Program funded by Ministry of Trade, Industry, and Energy (MOTIE) of Korea (RS-2022-00154855), Nanomedical Devices Development Program of National Nano Fab Center (CSM2105M101), and KRIBB Research Initiative Program (1711134081).
Glossary
Abbreviations Used
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- LAMP
loop-mediated isothermal amplification
- CRISPR
clustered regularly interspaced short palindromic repeats
- Cas9
CRISPR-associated protein 9
- ML
machine learning
- COVID-19
coronavirus disease 2019
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- NGS
next-generation sequencing
- gRNA
guide RNA
- ABTS
2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)
- PAM
protospacer adjacent motif
- LOD
limit of detection
- LDA
linear discriminant analysis
- RF
random forest
- GBC
gradient boosting classifier
- VOC
variants of concern
- VOI
variants of interest
- ACE2
angiotensin-converting enzyme 2
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.2c04840.
Figures of primer optimization, gRNA optimization, DNAzyme and CRISPR reactions optimization, absorbance spectra, real-time fluorescence curves during detection of SARS-CoV-2 genes using RT-LAMP reactions, plot of Ct value as a function of logarithmic concentration of SARS-CoV-2 genes, plot of Abs416 as a function of viral gene, real-time fluorescence curves during detection of SARS-CoV-2 lysates and control using RT-LAMP reactions before and after RNA extraction, plot of Abs416 as a function of RNA extraction, user manuals of smartphone application for SARS-CoV-2 detection by DAMPR assay system, user manuals of smartphone application for SARS-CoV-2 variants detection by v-DAMPR assay system, comparison of DAMPR assay with the previous colorimetric SARS-CoV-2 detection methods, and comparison of DAMPR assay with the previously integrated assays of LAMP and CRISPR-Cas systems and tables of primer sequences used in this study, gRNA sequences used in this study, comparison of this work with previous reported SARS-CoV-2 assays, evaluation of LDA, RF, and GBC classifiers for DAMPR assay system in terms of precision, recall, and F1 score, accuracy of LDA, RF, and GBC classifiers for DAMPR assay system, diagnostic results of 136 clinical samples by qRT-PCR, and diagnostic results of 80 clinical samples by qRT-PCR (PDF)
Movie of smartphone application for SARS-CoV-2 detection using DAMPR assay system (MP4)
Movie of smartphone application for SARS-CoV-2 omicron variant detection using v-DAMPR assay system (MP4)
Author Contributions
◆ J.S. and B.C. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Hu B.; Guo H.; Zhou P.; Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) . Coronavirus Disease (COVID-19) Pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed 2021-12).
- Liu R.; Han H.; Liu F.; Lv Z.; Wu K.; Liu Y.; Feng Y.; Zhu C. Positive Rate of RT-PCR Detection of SARS-CoV-2 Infection in 4880 Cases from One Hospital in Wuhan, China, from Jan to Feb 2020. Clin. Chim. Acta 2020, 505, 172–175. 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D.; Liu L.; Zhang M.; Hu Y.; Yang Q.; Guo J.; Dai Y.; Xu Y.; Cai Y.; Chen X.; Huang K.; Zhang Z. Evaluations of the Serological Test in the Diagnosis of 2019 Novel Coronavirus (SARS-CoV-2) Infections during the COVID-19 Outbreak. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 2271–2277. 10.1007/s10096-020-03978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. S.; Sathe L.; Munugala C.; Jones E. M.; Gasperini M.; Lubock N. B.; Yarza F.; Thompson E. M.; Kovary K. M.; Park J.; Marquette D.; Kay S.; Lucas M.; Love T.; Sina Booeshaghi A.; Brandenberg O. F.; Guo L.; Boocock J.; Hochman M.; Simpkins S. W.; et al. Massively Scaled-up Testing for SARS-CoV-2 RNA via next-Generation Sequencing of Pooled and Barcoded Nasal and Saliva Samples. Nat. Biomed. Eng. 2021, 5, 657–665. 10.1038/s41551-021-00754-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos I.; Gruson D.; Kabamba B.; Dahma H.; Van den Wijngaert S.; Reza S.; Carbone V.; Vandenberg O.; Gulbis B.; Wolff F.; Rodriguez-Villalobos H. Evaluation of Two Automated and Three Rapid Lateral Flow Immunoassays for the Detection of Anti-SARS-CoV-2 Antibodies. J. Clin. Virol. 2020, 128, 104413. 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B.; Fischer W. M.; Gnanakaran S.; Yoon H.; Theiler J.; Abfalterer W.; Hengartner N.; Giorgi E. E.; Bhattacharya T.; Foley B.; Hastie K. M.; Parker M. D.; Partridge D. G.; Evans C. M.; Freeman T. M.; de Silva T. I.; McDanal C.; Perez L. G.; Tang H.; Moon-Walker A.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland C. A.; Kiechle F. L. Point-of-Care Molecular Diagnostic Systems — Past, Present and Future. Curr. Opin. Microbiol. 2005, 8, 504–509. 10.1016/j.mib.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Wang J.; Cai K.; Zhang R.; He X.; Shen X.; Liu J.; Xu J.; Qiu F.; Lei W.; Wang J.; Li X.; Gao Y.; Jiang Y.; Xu W.; Ma X. Novel One-Step Single-Tube Nested Quantitative Real-Time PCR Assay for Highly Sensitive Detection of SARS-CoV-2. Anal. Chem. 2020, 92, 9399–9404. 10.1021/acs.analchem.0c01884. [DOI] [PubMed] [Google Scholar]
- Kubista M.; Andrade J. M.; Bengtsson M.; Forootan A.; Jonák J.; Lind K.; Sindelka R.; Sjöback R.; Sjögreen B.; Strömbom L.; Ståhlberg A.; Zoric N. The Real-Time Polymerase Chain Reaction. Mol. Aspects. Med. 2006, 27, 95–125. 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Lalli M. A.; Langmade J. S.; Chen X.; Fronick C. C.; Sawyer C. S.; Burcea L. C.; Wilkinson M. N.; Fulton R. S.; Heinz M.; Buchser W. J.; Head R. D.; Mitra R. D.; Milbrandt J. Rapid and Extraction-Free Detection of SARS-CoV-2 from Saliva by Colorimetric Reverse-Transcription Loop-Mediated Isothermal Amplification. Clin. Chem. 2021, 67, 415–424. 10.1093/clinchem/hvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.; Wang X.; Han L.; Chen T.; Wang L.; Li H.; Li S.; He L.; Fu X.; Chen S.; Xing M.; Chen H.; Wang Y. Multiplex Reverse Transcription Loop-Mediated Isothermal Amplification Combined with Nanoparticle-Based Lateral Flow Biosensor for the Diagnosis of COVID-19. Biosens. Bioelectron. 2020, 166, 112437. 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. S.; Hsieh K.; Chen L.; Kaushik A.; Trick A. Y.; Wang T. Digital CRISPR/Cas-Assisted Assay for Rapid and Sensitive Detection of SARS-CoV-2. Adv. Sci. 2021, 8, 2003564. 10.1002/advs.202003564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri R.; Tang Z.; Dong M.; Liu T.; Kshirsagar A.; Guan W. CRISPR-Based Detection of SARS-CoV-2: A Review from Sample to Result. Biosens. Bioelectron. 2021, 178, 113012. 10.1016/j.bios.2021.113012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.; Tian D.; Liu Y.; Lin Z.; Lyon C. J.; Lai W.; Fusco D.; Drouin A.; Yin X.; Hu T.; Ning B. Ultra-Sensitive and High-Throughput CRISPR-p Owered COVID-19 Diagnosis. Biosens. Bioelectron. 2020, 164, 112316. 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra P.; Alafeef M.; Dighe K.; Frieman M. B.; Pan D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627. 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolone S. N.; Tree M. O.; Conway M. J.; Chancellor M. B.; Lamb L. E. Reverse Transcription-Loop-Mediated Isothermal Amplification (RT-LAMP) Assay for Zika Virus and Housekeeping Genes in Urine, Serum, and Mosquito Samples. J. Vis. Exp. 2018, 139, e58436. 10.3791/58436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoul National University . Frequently Asked Questions about SNU Rapid COVID-19 Test. https://en.snu.ac.kr/snunow/snu_media/news?md=v&bbsidx=132529 (accessed 2021-06).
- Aoki M. N.; de Oliveira Coelho B.; Góes L. G. B.; Minoprio P.; Durigon E. L.; Morello L. G.; Marchini F. K.; Riediger I. N.; do Carmo Debur M.; Nakaya H. I.; Blanes L. Colorimetric RT-LAMP SARS-CoV-2 Diagnostic Sensitivity Relies on Color Interpretation and Viral Load. Sci. Rep. 2021, 11, 1–10. 10.1038/s41598-021-88506-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K.; Mage P. L.; Csordas A. T.; Eisenstein M.; Tom Soh H. Simultaneous Elimination of Carryover Contamination and Detection of DNA with Uracil-DNA-Glycosylase-Supplemented Loop-Mediated Isothermal Amplification (UDG-LAMP). Chem. Commun. 2014, 50, 3747–3749. 10.1039/c4cc00540f. [DOI] [PubMed] [Google Scholar]
- Song J.; Kim S.; Kim H. Y.; Hur K. H.; Kim Y.; Park H. G. A Novel Method to Detect Mutation in DNA by Utilizing Exponential Amplification Reaction Triggered by the CRISPR-Cas9 System. Nanoscale 2021, 13, 7193–7201. 10.1039/D1NR00438G. [DOI] [PubMed] [Google Scholar]
- Guo L.; Sun X.; Wang X.; Liang C.; Jiang H.; Gao Q.; Dai M.; Qu B.; Fang S.; Mao Y.; Chen Y.; Feng G.; Gu Q.; Wang R. R.; Zhou Q.; Li W. SARS-CoV-2 Detection with CRISPR Diagnostics. Cell Discov. 2020, 6, 1–4. 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski M. M.; Abudayyeh O. O.; Gootenberg J. S.; Zhang F.; Collins J. J. CRISPR-Based Diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. 10.1038/s41551-021-00760-7. [DOI] [PubMed] [Google Scholar]
- Hong K. H.; Lee S. W.; Kim T. S.; Huh H. J.; Lee J.; Kim S. Y.; Park J.-S.; Kim G. J.; Sung H.; Roh K. H.; Kim J.-S.; Kim H. S.; Lee S.-T.; Seong M.-W.; Ryoo N.; Lee H.; Kwon K. C.; Yoo C. K. Guidelines for Laboratory Diagnosis of Coronavirus Disease 2019 (COVID-19) in Korea. Ann. Lab. Med. 2020, 40, 351–360. 10.3343/alm.2020.40.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczkowska M.; Guga P.; Stec W. J. Stereodefined Phosphorothioate Analogues of DNA: Relative Thermodynamic Stability of the Model PS-DNA/DNA and PS-DNA/RNA Complexes. Biochemistry 2002, 41, 12483–12487. 10.1021/bi026225z. [DOI] [PubMed] [Google Scholar]
- LaPlanche L. A.; James T. L.; Powell C.; Wilson W. D.; Uznanski B.; Stec W. J.; Summers M. F.; Zon G. Phosphorothioate-Modified Oligodeoxyribonucleotides. III. NMR and UV Spectroscoptc Studies of the R p - R p, S p - S p, and R p - S p Duplexes, [d(GG s AATTCC)] 2, Derived from Diastereomeric O -Ethyl Phosphorothioates. Nucleic Acids Res. 1986, 14, 9081–9093. 10.1093/nar/14.22.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.; Kim H. Y.; Kim S.; Jung Y.; Park H. G. Self-Priming Phosphorothioated Hairpin-Mediated Isothermal Amplification. Biosens. Bioelectron. 2021, 178, 113051. 10.1016/j.bios.2021.113051. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Yao C.; Wang Y.; Yang C. A G-Quadruplex DNAzyme-Based LAMP Biosensing Platform for a Novel Colorimetric Detection of Listeria Monocytogenes. Anal. Methods 2018, 10, 848–854. 10.1039/C7AY02908J. [DOI] [Google Scholar]
- Fu R.; Jeon K.; Jung C.; Park H. G. An Ultrasensitive Peroxidase DNAzyme-Associated Aptasensor That Utilizes a Target-Triggered Enzymatic Signal Amplification Strategy. Chem. Commun. 2011, 47, 9876–9878. 10.1039/c1cc13122b. [DOI] [PubMed] [Google Scholar]
- Kong D.-M.; Xu J.; Shen H.-X. Positive Effects of ATP on G-Quadruplex-Hemin DNAzyme-Mediated Reactions. Anal. Chem. 2010, 82, 6148–6153. 10.1021/ac100940v. [DOI] [PubMed] [Google Scholar]
- Beldomenico P. M. Do Superspreaders Generate New Superspreaders? A Hypothesis to Explain the Propagation Pattern of COVID-19. J. Infect. Dis. 2020, 96, 461–463. 10.1016/j.ijid.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) . Tracking SARS-CoV-2 Variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed 2021-11).
- Harvey W. T.; Carabelli A. M.; Jackson B.; Gupta R. K.; Thomson E. C.; Harrison E. M.; Ludden C.; Reeve R.; Rambaut A.; Peacock S. J.; Robertson D. L. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. Nat. Rev. Microbiol. 2021, 19, 409–424. 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N. G.; Jarvis C. I.; Edmunds W. J.; Jewell N. P.; Diaz-Ordaz K.; Keogh R. H. Increased Mortality in Community-Tested Cases of SARS-CoV-2 Lineage B.1.1.7. Nature 2021, 593, 270–274. 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.; Thao T. T. N.; Hoffmann D.; Taddeo A.; Ebert N.; Labroussaa F.; Pohlmann A.; King J.; Steiner S.; Kelly J. N.; Portmann J.; Halwe N. J.; Ulrich L.; Trüeb B. S.; Fan X.; Hoffmann B.; Wang L.; Thomann L.; Lin X.; Stalder H.; et al. SARS-CoV-2 Spike D614G Change Enhances Replication and Transmission. Nature 2021, 592, 122–127. 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
- Plante J. A.; Liu Y.; Liu J.; Xia H.; Johnson B. A.; Lokugamage K. G.; Zhang X.; Muruato A. E.; Zou J.; Fontes-Garfias C. R.; Mirchandani D.; Scharton D.; Bilello J. P.; Ku Z.; An Z.; Kalveram B.; Freiberg A. N.; Menachery V. D.; Xie X.; Plante K. S.; et al. Spike Mutation D614G Alters SARS-CoV-2 Fitness. Nature 2021, 592, 116–121. 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Liu Y.; Xia H.; Zou J.; Weaver S. C.; Swanson K. A.; Cai H.; Cutler M.; Cooper D.; Muik A.; Jansen K. U.; Sahin U.; Xie X.; Dormitzer P. R.; Shi P.-Y. BNT162b2-Elicited Neutralization of B.1.617 and Other SARS-CoV-2 Variants. Nature 2021, 596, 273–275. 10.1038/s41586-021-03693-y. [DOI] [PubMed] [Google Scholar]
- O’Geen H.; Henry I. M.; Bhakta M. S.; Meckler J. F.; Segal D. J. A Genome-Wide Analysis of Cas9 Binding Specificity Using ChIP-Seq and Targeted Sequence Capture. Nucleic Acids Res. 2015, 43, 3389–3404. 10.1093/nar/gkv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao Thi V. L.; Herbst K.; Boerner K.; Meurer M.; Kremer L. P.; Kirrmaier D.; Freistaedter A.; Papagiannidis D.; Galmozzi C.; Stanifer M. L.; Boulant S.; Klein S.; Chlanda P.; Khalid D.; Barreto Miranda I.; Schnitzler P.; Kräusslich H.-G.; Knop M.; Anders S. A Colorimetric RT-LAMP Assay and LAMP-Sequencing for Detecting SARS-CoV-2 RNA in Clinical Samples. Sci. Transl. Med. 2020, 12, eabc7075. 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawattanapaiboon K.; Pasomsub E.; Prombun P.; Wongbunmak A.; Jenjitwanich A.; Mahasupachai P.; Vetcho P.; Chayrach C.; Manatjaroenlap N.; Samphaongern C.; Watthanachockchai T.; Leedorkmai P.; Manopwisedjaroen S.; Akkarawongsapat R.; Thitithanyanont A.; Phanchana M.; Panbangred W.; Chauvatcharin S.; Srikhirin T. Colorimetric Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) as a Visual Diagnostic Platform for the Detection of the Emerging Coronavirus SARS-CoV-2. Analyst 2021, 146, 471–477. 10.1039/D0AN01775B. [DOI] [PubMed] [Google Scholar]
- Yan C.; Cui J.; Huang L.; Du B.; Chen L.; Xue G.; Li S.; Zhang W.; Zhao L.; Sun Y.; Yao H.; Li N.; Zhao H.; Feng Y.; Liu S.; Zhang Q.; Liu D.; Yuan J. Rapid and Visual Detection of 2019 Novel Coronavirus (SARS-CoV-2) by a Reverse Transcription Loop-Mediated Isothermal Amplification Assay. Clin. Microbiol. Infect. 2020, 26, 773–779. 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.; Qian C.; Pang Y.; Li M.; Yang Y.; Ma H.; Zhao M.; Qian F.; Yu H.; Liu Z.; Ni T.; Zheng Y.; Wang Y. OpvCRISPR: One-Pot Visual RT-LAMP-CRISPR Platform for SARS-Cov-2 Detection. Biosens. Bioelectron. 2021, 172, 112766. 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujang M. A.; Adnan T. H. Requirements for Minimum Sample Size for Sensitivity and Specificity Analysis. J. Clin. Diagn. Res. 2016, 210, YE01–YE06. 10.7860/JCDR/2016/18129.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K.; Tzou P. L.; Nouhin J.; Gupta R. K.; de Oliveira T.; Kosakovsky Pond S. L.; Fera D.; Shafer R. W. The Biological and Clinical Significance of Emerging SARS-CoV-2 Variants. Nat. Rev. Genet. 2021, 22, 757–773. 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono S.; Zhang Y.; Ode H.; Sano K.; Tan T. S.; Imai K.; Miyoshi K.; Kishigami S.; Ueno T.; Iwatani Y.; Suzuki T.; Tokunaga K. SARS-CoV-2 D614G Spike Mutation Increases Entry Efficiency with Enhanced ACE2-Binding Affinity. Nat. Commun. 2021, 12, 848. 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.; Martin M. A.; Harel N.; Tirosh O.; Kustin T.; Meir M.; Sorek N.; Gefen-Halevi S.; Amit S.; Vorontsov O.; Shaag A.; Wolf D.; Peretz A.; Shemer-Avni Y.; Roif-Kaminsky D.; Kopelman N. M.; Huppert A.; Koelle K.; Stern A. Full Genome Viral Sequences Inform Patterns of SARS-CoV-2 Spread into and within Israel. Nat. Commun. 2020, 11, 5518. 10.1038/s41467-020-19248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam D. C.; Wu P.; Wong J. Y.; Lau E. H. Y.; Tsang T. K.; Cauchemez S.; Leung G. M.; Cowling B. J. Clustering and Superspreading Potential of SARS-CoV-2 Infections in Hong Kong. Nat. Med. 2020, 26, 1714–1719. 10.1038/s41591-020-1092-0. [DOI] [PubMed] [Google Scholar]
- Ra S. H.; Lim J. S.; Kim G.; Kim M. J.; Jung J.; Kim S.-H. Upper Respiratory Viral Load in Asymptomatic Individuals and Mildly Symptomatic Patients with SARS-CoV-2 Infection. Thorax 2021, 76, 61–63. 10.1136/thoraxjnl-2020-215042. [DOI] [PubMed] [Google Scholar]
- Wong S. C. Y.; Tse H.; Siu H. K.; Kwong T. S.; Chu M. Y.; Yau F. Y. S.; Cheung I. Y. Y.; Tse C. W. S.; Poon K. C.; Cheung K. C.; Wu T. C.; Chan J. W. M.; Cheuk W.; Lung D. C. Posterior Oropharyngeal Saliva for the Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2020, 71, 2939–2946. 10.1093/cid/ciaa797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K. K.; Lu L.; Yip C. C.; Poon R. W.; Fung A. M.; Cheng A.; Lui D. H.; Ho D. T.; Hung I. F.; Chan K.-H.; Yuen K.-Y. Additional Molecular Testing of Saliva Specimens Improves the Detection of Respiratory Viruses. Emerg. Microbes. Infect. 2017, 6, 1–7. 10.1038/emi.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-I.; Kim S.-G.; Kim S.-M.; Kim E.-H.; Park S.-J.; Yu K.-M.; Chang J.-H.; Kim E. J.; Lee S.; Casel M. A. B.; Um J.; Song M.-S.; Jeong H. W.; Lai V. D.; Kim Y.; Chin B. S.; Park J.-S.; Chung K.-H.; Foo S.-S.; Poo H.; et al. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell host & Microbe 2020, 27, 704–709. 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha H.; Kim H.; Joung Y.; Kang H.; Moon J.; Jang H.; Park S.; Kwon H.-J.; Lee I.-C.; Kim S.; Yong D.; Yoon S.-W.; Park S.-G.; Guk K.; Lim E.-K.; Park H. G.; Choo J.; Jung J.; Kang T. Surface-Enhanced Raman Scattering-Based Immunoassay for Severe Acute Respiratory Syndrome Coronavirus 2. Biosens. Bioelectron. 2022, 202, 114008. 10.1016/j.bios.2022.114008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J.; Kwon H.-J.; Yong D.; Lee I.-C.; Kim H.; Kang H.; Lim E.-K.; Lee K.-S.; Jung J.; Park H. G.; Kang T. Colorimetric Detection of SARS-CoV-2 and Drug-Resistant PH1N1 Using CRISPR/DCas9. ACS Sens. 2020, 5, 4017–4026. 10.1021/acssensors.0c01929. [DOI] [PubMed] [Google Scholar]
- Elmaz F.; Büyükçakır B.; Yücel Ö.; Mutlu A. Y. Classification of Solid Fuels with Machine Learning. Fuel 2020, 266, 117066. 10.1016/j.fuel.2020.117066. [DOI] [Google Scholar]
- James G.; Witten D.; Hastie T.; Tibshirani R.. An Introduction to Statistical Learning; Springer: New York, 2013; Vol. 112, p 18. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.