Abstract
While most bones fully self-heal, certain diseases require bone allograft to assist with fracture healing. Bone allografts offer promise as treatments for such fractures due to their osteogenic properties. However, current bone allografts made of decellularized bone extracellular matrix (ECM) have high failure rates, and thus grafts which improve fracture healing outcomes are needed. Understanding specific changes to the ECM proteome during normal fracture healing would enable the identification of key proteins that could be used enhance osteogenicity of bone allograft. Here, we performed a timeseries analysis of the fracture callus in mice to investigate proteomic and mineralization changes to the ECM at key stages of fracture healing. We found that changes to the ECM proteome largely coincide with the distinct phases of fracture healing. Basement membrane proteins (AGRN, COL4, LAMA), cartilage proteins (COL2A1, ACAN), and collagen crosslinking enzymes (LOXL, PLOD, ITIH) were initially upregulated, followed by bone specific proteoglycans and collagens (IBSP, COL1A1). Various tissue proteases (MMP2, 9, 13, 14; CTSK, CTSG, ELANE) were expressed at different levels throughout fracture healing. These changes coordinated with mineralization of the fracture callus, which increased steeply during the initial stages of healing. Interestingly the later timepoint was characterized by a response to wound healing and high expression of clotting factors (F2, 7, 9, 10). We identified ELANE and ITIH2 as tissue remodeling enzymes having no prior known involvement with fracture healing. This data can be further mined to identify regenerative proteins for enhanced bone graft design.
INTRODUCTION:
Fracture repair undergoes a series of dynamic events in which the injured bone’s ECM becomes remodeled from an initial fibrous tissue into mineralized bone1. The fracture healing timeline in mice can be divided into several histological stages as illustrated in Figure 1, including an inflammatory response and hematoma (days 1-3 post-injury); second is the formation of a fibro-vascular soft callus (days 4-7); during the osteogenic phase, the soft callus becomes mineralized into a hard callus (days 7-14), and last is the remodeling phase in which the fracture callus is absorbed and remodeled to form normal bone (days 14-28 and beyond. Figure 1)2. Fracture healing can further be described by two distinct modes of repair: intramembranous ossification (IO) and endochondral ossification (EO)3. In IO, osteoprogenitors undergo osteogenic differentiation along the cortical bone to directly form bone4,5. During EO, progenitors undergo chondrogenesis within the fracture gap to create a cartilage template, which subsequently undergoes vascularization and mineralization of the template. Chondrocytes undergo apoptosis and/or transform to osteoblasts that form the new bone6-10. These processes occur simultaneously throughout fracture healing although EO is the predominant mechanism11.
Figure 1:
Schematic of injury, and workflow for proteomics. Mouse tibia underwent a three-point bending fracture. The fracture callus was harvested at days 7, 10, 14, and 21 post-injury, and was subjected to mass spectrometry and FTIR. The ECM proteome was then analyzed by bioinformatic analysis. (A-D) 4x and (E-H) 10x magnification images of the fracture callus showing Safranin-O staining, which stains bone turquoise blue, cartilage tissue red, and nuclei purple. At 7 days post-injury, fibrous repair tissue (Fib) occupies the injury site (A, E). By 10 days, cells within the injury site undergo chondrogenesis (Cart) to create the soft cartilage callus (B, F); some chondrocytes undergo hypertrophy in which they enlarge in size and secrete angiogenic factors and proteases (F). At 14 days post-injury, the fracture callus is a mix of cartilage tissue that is undergoing ossification (Bone) following vascularization (C, G). By day 21, the fracture callus is ossified, and occupied by mineralized bone tissue (D, H). Scale bars = 1mm
In recent years, the soft callus ECM generated during EO has attracted attention as a potential treatment to improve fracture healing outcomes. In cases of fracture healing, such an ECM graft could be used to increase healing rates and improve the quality of the healed bone12-15. The current gold standard graft is decellularized bone allograft which has problems with low osteointegration and osteoinduction16. While EO grafts have been investigated for this purpose, a deeper understanding of the ECM components of such grafts and how they change over time could lead to improved graft treatments that enhance fracture healing outcomes15.
In general, the ECM is the non-cellular network of macromolecules and minerals which provides mechanical and biochemical support for cells within a tissue. ECM components vary in composition among tissues and undergo substantial changes during injury17. Cartilage of the soft callus is composed mainly of collagen type II (COL2A1) and aggrecan (ACAN) which forms a tightly crosslinked ECM network that retains water and resists compressive forces18. During EO fracture healing, chondrocytes within the cartilage soft callus undergo hypertrophy to secrete angiogenic and neurotrophic signals which initiates invasion of the neurovascular bundle19-23. Proteases, including MMP2, MMP9 and MMP13, play an important role in remodeling the soft callus for subsequent phases bone formation24-28. Once remodeled, the mature bone ECM is composed mainly of calcium phosphate hydroxyapatite (~60%), collagen type I (COL1A1), and bone-specific glycoproteins and proteoglycans that aid in tissue mineralization29. While histological, cellular, and transcriptional hallmarks of fracture healing have been established, an understanding of how the fracture callus ECM proteome changes over time during normal bone healing is lacking8,30.
Following injury, bones are one of the few tissues that can fully regenerate to a pre-injured state. While this is the case for the majority of fractures, co-morbidities such as diabetes, obesity, smoking, osteoporosis and aging can result in impaired healing in up to 50% of these fractures. As such this presents a significant burden to patients suffering from non-healing fractures and adds substantial cost to the healthcare system31-34. In this work we investigate changes to the fracture callus ECM proteome that occur throughout normal fracture healing. A more complete understanding of healthy bone regeneration following fracture could lead to treatments that can induce bone healing in diseased states and improved biomimetic ECM grafts for fracture healing35.
METHODS:
Animal model of fracture healing:
An overview of the procedure is outlined in Figure 1. All murine studies were approved by the UCSF Institutional Animal Care and Use Committee and a standardized protocol for anesthesia, aseptic technique and post-operative analgesics was followed. Adult (10-14 weeks old) C57/BL6/J male mice (Jackson Laboratory) were obtained or bred for the study. To generate fractures, mice received a standardized, closed fracture in the mid-diaphysis of the tibia using a custom-built apparatus designed to deliver a reproducible three-point bending fracture by controlling the weight (460 g) and distance (14 cm) of the force. Fractures were not stabilized to promote robust endochondral repair as previously detailed36. Animals were allowed to ambulate freely post-operatively and pain was managed per the IACUC approved protocol. At days 7, 10, 14, or 21 animals were euthanized by CO2 asphyxiation, the fracture callus was harvested from the injury site and was subjected to mass spectrometry or FTIR analysis as described below. One tibia sample was used as a normal control.
Mass spectrometry proteomics:
An equal amount of fracture callus (5 mg) was harvested from the injury site of each animal at days 7 (n = 2), 10 (n = 2), 14 (n = 3), or 21 (n = 3) post fractures, and samples were processed for mass spectrometric analysis (LC–MS/MS) as mentioned previously37. The experiments were carried out using the EASY n-LC II system coupled to the LTQ Orbitrap Velos Pro mass spectrometer (Thermo Scientific, CA, USA). The same amount of peptides mixtures (1 μg) were separated and eluted using a 90 min 3–25% solvent B (99.9% acetonitrile and 0.1% formic acid) over solvent A (99.9% water and 0.1% formic acid) gradient, at 300 nL/min. The MS was operated in a Top12 data-dependent acquisition. Proteome Discoverer 1.4 (Thermo Scientific) and Mascot 2.5.1 (Matrix Science, UK) were used for protein inference in UniProtKB/ SwissProt FASTA database. Variable (methionine oxidation, asparagine and glutamine deamidation) and fixed (cysteine carbamidomethylation) modifications were taken into account. The peptide false discovery rate was set below 0.05. Label-free relative quantification analysis was performed with Sieve 2.1 (Thermo Scientific) and a spectral abundance alteration of at least 1.5-fold (over the control group) corroborated with a p-value < 0.05 was considered a significant protein level variation. Precursor ion intensities were normalized using the total ion current algorithm. Proteins with more than 3 missing average quantification ratios (AQR) and those with a median AQR between 0.67 and 1.5 were excluded from the analysis. Protein Center 3.41 (Thermo Scientific) was used for disparities evaluation of the identified proteins.
Fracture callus mineral composition and crystallinity
Chemical composition of the fracture callus was examined with Fourier Transform Infrared (FTIR) spectroscopy (n = 3/timepoint). Samples were desiccated through an ethanol series followed by exposure in a desiccant chamber. For each sample, a homogenized powder mixture was created of 1% tissue by weight in potassium bromide (KBr; Thermo Electron Corporation). The powder mixture was compressed using a manual die to create a pellet for FTIR spectroscopy. Spectroscopy was performed on a benchtop interferometer system (Nexus 870, Thermo Electron Corporation). Spectra were acquired using 256 scans at a spectral resolution of 4 cm−1. A background scan was recorded immediately following each sample scan to facilitate background correction. Following acquisition, the spectra were analyzed using chemical imaging software (Isys, Spectral Dimensions, Inc.). Spectra were baseline adjusted and the integrated areas of the amide I (1595-1720 cm−1), ν1 ν3 phosphate (PO4, 895-1215 cm−1), and carbonate (CO32−, 840-890 cm−1) bands were calculated. Mineral-to-matrix (PO4/amide I), carbonate-to-phosphate (CO32−/PO4), and carbonate-to-matrix (CO32−/amide I) ratios were calculated from integrated areas of the respective peaks. Additionally, peak heights were measured at specific wavenumbers to determine additional spectroscopic parameters. The ratio of 1030 cm−1 to 1020 cm−1 represents the ratio of stoichiometric apatite to non-stoichiometric apatite, a measure of crystallinity. Finally, the ratio of 1660 cm−1 to 1690 cm−1 represents the proportion of non-reducible to reducible crosslinks in the collagen, indicative of collagen maturity.
Safranin-O histology:
Safranin-O histology was performed as previously described15. Briefly, tibiae were harvested and fixed in 4% paraformaldehyde for 24 hours at 4°C. Tibias were then decalcified in 19% EDTA (pH 7.4) for 14 days at 4°C and then processed for paraffin or frozen histology. Histology staining to visualize bone and cartilage tissues included: Modified Milligan's Trichrome (bone = blue), Safranin-O/fast green (cartilage = red), or Hall and Brunt Quadruple stain (HBQ, bone = red, cartilage = blue).
Bioinformatics and statistical analyses:
Statistical analyses and graphs were made using the R statistical package38. InteractiVenn was utilized for visualization of protein identifications among the different groups39. Statistical comparison between two groups was performed with two sample equal variance t-test, and comparison among groups was performed with one-way equal variance ANOVA (using matrixTests ‘R’ package). FDR-adjusted p-values (q-value) < 0.05 were considered significant. Differentially expressed proteins (DEPs) between two groups were determined as having fold change > ∣2∣ and q-value < 0.0540. GO enrichment analysis was performed using g:Profler with Bonferroni correction and a non-redundant term list was generated using REVIGO41,42. To detect and visualize possible interaction networks of differentially abundant proteins, we used the STRING, KEGG, and PANTHER databases43,44.
RESULTS:
Clustering and top significant ECM proteins
In total, 251 ECM or ECM-related proteins were identified, with 181 expressed at significantly different levels across timepoints (q-value < 0.05). Analysis of all identified proteins showed clustering of samples according to timepoint, and most of these proteins decreased in expression over time (Figure 2A), indicating that ECM expression is upregulated early in fracture healing and decreases as time progresses. Samples also grouped by timepoint according to their principal components (Figure 2B). An in-depth analysis of the top 25 of these differentially expressed proteins by manual interpretation (ST 1) revealed that most of these ECM proteins are proteoglycans (HSPG, CSPG4); cartilage collagens (COL2A1, COL9A1 & 3, COL11A1); enzymes involved in collagen crosslinking and stability (PLOD1-3, LOXL2, P4HA1); and major components of the ECM (LAMA5, FBLN2, NID2, FN1). GO enrichment (Supplemental Figure 2A) revealed terms involved in ECM, collagen, and proteoglycan production and organization; endochondral bone morphogenesis and cartilage development; and skeletal system development. Further, expression of all top 25 proteins decreased over time as the fractures healed (Supplemental Figure 3).
Figure 2:
(A) Unsupervised clustering of the ECM proteome showing grouping of samples (x-axis) according to timepoint by protein expression (y-axis). (B) Principal component analysis showing separation of samples by timepoint.
Proteins upregulated early in fracture healing
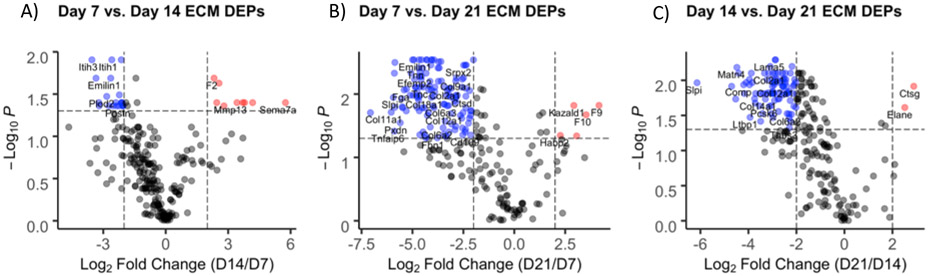
To further investigate ECM proteins involved in fracture healing, differentially expressed proteins (DEPs) were identified by comparing fold change (FC > ∣2∣) and significance values (q-value < 0.05) between timepoints. As with cluster analysis, volcano plots showed that expression of most proteins was highest at earlier timepoints (Figure 3). There were no DEPs between days 7 and 10 (not shown); 24 proteins downregulated and 9 upregulated comparing days 14 vs. 7 (Figure 3A); 137 proteins downregulated and 9 upregulated comparing days 21 vs. 7 (Figure 3B); and 130 proteins downregulated and 4 upregulated comparing days 21 vs. 14 (Figure 3C).
Figure 3:
Volcano plots representing differentially expressed ECM proteins between (A) days 14 and 7, (B) days 21 and 7, and (C) days 21 and 14. Red and blue labeled proteins are expressed at higher and lower levels, respectively, in day 14 vs. 7 (A), day 21 vs. 7 (B), and day 21 vs. 14.
Compared to day 14, DEPs upregulated at day 7 are primarily involved in wound healing (FN1, POSTN); collagen crosslinking (PLOD1-3, LOXL2-3); and elastic fiber formation (EMILIN1, EFEMP2, LTBP2) (Supplemental Table 3). GO enrichment of these proteins revealed biological process (BP) terms involved in extracellular matrix organization, cell adhesion, protein hydroxylation, and hyaluronan metabolic process; molecular function (MF) terms involved in binding calcium ions, integrins, carbohydrates, and glycosaminoglycans; and cellular component (CC) terms in the extracellular region, cell periphery, and elastic fibers (Supplemental Figure 4B). Most (22/24) of these proteins were also downregulated at day 21 (Supplemental Table 3). Top DEPs downregulated at day 21 compared to both day 7 (Supplemental Table 3) and day 14 (Supplemental Table 5) include major cartilage collagens (COL2, COL11, COL9) and glycoproteins (COMP); the hypertrophic cartilage protein COL10A1; basement membrane collagens (COL4, COL15, COL8); and proteins involved in wound healing and coagulation (FGA, POSTN, FGB). GO enrichment of these DEPs demonstrated a diverse range of BP terms involved in tissue growth including: developmental process, biogenesis, ECM organization, cell adhesion and locomotion, response to wounding, and hemostasis, as well as skeletal system development (Supplemental Figures 4B & 5B). MF terms included binding of ECM, calcium, glycosaminoglycans, collagen, and fibronectin. These results indicate a strong upregulation of ECM proteins expressed at day 7 that are largely involved in cartilage generation and stabilization as well as hemostasis and the regulation of wound healing following injury. Additionally, Panther Pathways for genes upregulated at day 7 include the blood coagulation, inflammation, integrin signaling pathway, and protein binding, consistent with hemostasis, resulting cell motility seen in a general response to injury at this timepoint.
Proteins upregulated later in fracture healing
GO enrichment of DEPs upregulated at the later day 14 and day 21 timepoints generally showed a response to wounding and ossification. GO BP terms for the day 14 DEPs include proteolysis, collagen catabolic process, and positive regulation of coagulation and hemostasis, while MF terms include hydrolase and endopeptidase activity, and collagen and fibronectin binding (Supplemental Figure 4A). These DEPs include cartilage remodeling enzymes (MMP13, CTSK); coagulation factors (F2, F7, F9, F10); and an insulin growth factor involved in bone regeneration (KAZALD1) (Supplemental Table 3). The day 21 upregulated DEP BP terms also include response to wounding, hemostasis, and regulation of blood coagulation, as well as ECM organization, ossification, and biomineralization. MF terms include the binding of calcium ions and growth factors, hydrolase and endopeptidase activity, and ECM components conferring tensile strength (Supplemental Figure 5A). These DEPs include the major bone collagen proteins (COL1A1, COL1A2); and the continued upregulation of coagulation factors (F2, F7, F9, F10) and the insulin growth factor which stimulates bone growth (KAZALD1) (ST 5).
GO terms for upregulated day 21 DEPs include anti-fungal and anti-bacterial responses for BP, and glycosaminoglycan and ECM binding and endopeptidase activity for MF in the ECM region (Supplemental Figure 6A). According to these data, the fracture callus continues to undergo response to wounding and an antibacterial and immune response, as well as biomineralization of tissue at these later timepoints. These DEPs include a mineralization phosphoprotein (MEPE); and tissue remodeling enzymes (ELANE, CTSG). Panther Pathway analysis of DEPs upregulated at later timepoints include angiogenesis, blood coagulation, and plasminogen activating cascade indicating that these proteins are involved in creating a vascular template and in different parts of the coagulation pathway.
STRING and KEGG network analysis
STRING and KEGG network analysis (Supplemental Figure 7-12) indicate strong ECM production and organization at day 7, and include both GAG and collagen biosynthesis and crosslinking, as well as ECM organization, focal adhesion, and complement activation. Interestingly the day 14 timepoint was characterized by complement and coagulation cascades, formation of a fibrin clot, apoptosis, and platelet and neutrophil degranulation, suggesting a strong homeostasis and innate immune response at this timepoint. Collagen and GAG biosynthesis, and the tissue remodeling enzymes collagenase 3, peptidase, and cathepsin K and lysosome, phagosome, and lysine degradation were also networks enriched at the day 14 timepoint, indicating both the production and digestion of tissues. The later day 21 timepoint was characterized by focal adhesions, protein digestion and absorption, and the tissue remodeling enzymes listed above indicating that the fracture callus is still undergoing active remodeled at this timepoint.
Analysis of canonical bone healing proteins
Figure 4 shows the expression level trends of select proteins commonly found in bone (Figure 4A) and cartilage tissue as it undergoes endochondral bone formation (Figure 4B). Expression patterns of these proteins are largely consistent with the literature, and follow the temporal pattens seen histologically (Figure 1), with high initial expression of a fibro-cartilage template, followed by its mineralization and remodeling. COL1A1 (the most prominent organic component of bone), DMP1 (an ECM phosphoprotein critical for proper bone mineralization and osteoblast differentiation), and MEPE (a calcium binding phosphoprotein that regulates mineralization) are all increasingly expressed as the fracture heals. Interestingly, IBSP, MGP (both major structural protein of bone critical for matrix mineralization), and BGN (prominent proteoglycan involved in collagen assembly) expression levels peak around day 14 and are expressed at lower levels by day 21 and in the tibia. For cartilage proteins, COL2A1 (major collagen of cartilage), COL9A1 (major crosslinker of COL2A1), COL10A1 (hypertrophic chondrocyte collagen), and the glycoproteins and glycosaminoglycans CSPG4, COMP, and ACAN (all prominent cartilage ECM proteins) are all expressed at high levels early in fracture healing and then drop in expression at the later timepoints. The enzymes PLOD2, LOXL2 (both critical for collagen and elastin crosslinking and stability), and ITIH2 (protease inhibitor involved in ECM stabilization by binding to hyaluronic acid) are all highly expressed at the early day 7 timepoint, while MMP13 (enzyme secreted by hypertrophic chondrocyte and involved in endochondral tissue remodeling) and CTSK (enzyme released by osteoclasts and involved in bone tissue remodeling) peak in expression at the day 14 timepoint45,46. For tissue proteases, we identified MMP2 (binds denatured type IV and V collagen and elastin), MMP9 (degrades type IV and V collagen), MMP13 (cleaves type II collagen, and to a lesser extent other fibrillar collagens, fibronectin, aggrecan), and MMP14 (activates MMP2; degrades collagens) (Supplemental Figure 13). These proteases were expressed at different levels throughout the fracture healing process, indicating unique roles in tissue digestion for each. Interestingly ELANE (elastase enzyme that hydrolyzes many ECM proteins) and CTSG were highly expressed at the day 21, indicating active remodeling at this later timepoint.
Figure 4:
Expression levels (mean +/− sd) of select proteins as they relate to endochondral bone formation. The tibia sample is included as a reference point. (A) includes proteins commonly found in cartilage tissue; (B) includes enzymes important for the transition from cartilage to bone, and (C) are proteins commonly found in bone tissue. The x-axis is time in days (day 7, day 10, day 14, and day 21) or tibia, and the y-axis is the log2 transformed protein expression levels. All proteins are expressed at significantly different levels (q-value < 0.05; except MEPE, q-value = 0.13).
Mineralization of the fracture callus coincides with respective proteins
FTIR spectroscopic analysis found an increasing mineral-to-matrix ratio (min/mat) from day 7 to day 14, after which mineral-to-matrix ratio was stable through day 21 (Figure 5). These results indicate that complete mineralization was achieved by day 14, after which mineral content relative to collagen content remained constant during any subsequent remodeling during the 14 to 21 day period. This increase coincided with increased expression of major bone proteins involved in mineralization (COL1A1, IBSP, and DMP1)
Figure 5:
FTIR results showing the mineral/matrix ratios across the different times since fracture. Y-axis values represent absorbance/absorbance (unitless). There was a significant increase in mineralization from day 10 to day 14, after which mineralization plateaued.
DISCUSSION:
In this study, we investigated changes to the fracture callus ECM proteome as it progressed from a soft fibro-cartilage callus to a mineralized hard callus. Using a tissue digestion method that has been highly optimized for the ECM, we identified 251 proteins, most of which were present at all timepoints40. In general, expression patterns of canonical endochondral fracture healing proteins were consistent with the current literature, with high expression of cartilage (COL2A1, ACAN, HSPG) proteins initially, followed by hypertrophic cartilage proteins and remodeling enzymes (COL10A1, MMP13), and finally mature bone proteins (COL1A1, DMP1) at later timepoints. GO terms of our top 25 differentially expressed ECM proteins were related to endochondral bone morphogenesis, cartilage development, and skeletal system development, further indicating a successful ECM proteomics strategy in this injury.
While most fractures will self-heal, endogenous bone regeneration can be insufficient in a wide variety of clinical scenarios that lead to impaired or delayed healing (diabetes, obesity, smoking, osteoporosis, aging) or in scenarios where bone formation needs to span a large gap (critical sized bone defects, spinal fusions, opening wedge osteotomies). In these cases, insufficient ECM is produced and bone grafting is often used to facilitate fracture healing47. Unfortunately, current bone allografts have low osteogenicity and poor integration with host tissue, and can result in slow healing times48. Because of this, endochondral grafts that mimic the endochondral bone formation process observed during healthy bone healing have been investigated13,49,50. While these grafts have been used successfully in cases of spinal fusion51,52 and nonunion53, they remain poorly characterized. Understanding the ECM protein components within the endochondral fracture callus of successfully healed bone could lead to the identification of specific proteins involved in pro-regenerative fracture healing, and therefore graft design for improved fracture healing.
This strategy has been used in designing synthetic peptides that enhance bone healing by mimicking ECM components. For example, the synthetic peptide B2A was designed to mimic the positive BMP-2 receptor modulator54 while P-15 was designed to mimic collagen and enhance osteoblastogenesis. Such synthetic peptides are examples of successful treatments for bone healing inspired from the fracture callus ECM55 and have been shown to treat delayed union fractures in patients56. Building upon this strategy, our data could be further mined for peptides or proteins associated with a pro-regenerative bone healing response.
In addition to hallmark collagen proteins, our early timepoint also had high expression of basement membrane proteins and elastin, indicating strong production of connective tissue proteins at day 7. The matricellular proteins (TNN, TNC, COMP) were also identified as DEPs with higher expression early in fracture healing (day 7). COMP has an established role in cartilage matrix organization, and while recent studies suggest involvement bone integrity57 or osteogenesis58, there are no known studies to date on its role in fracture repair. Similarly, there are limited publications about tenanscins (TNN, TNC, TNW) during fracture repair. TNN and/or TNC were only recently reported to have expression by chondrocytes and osteoblasts in vitro, a role in mechanosensation59,60, and responsiveness to canonical signaling programs required for fracture repair (Wnt and BMP).61 Taken together these data highlight how the fracture matrix proteomics may identify key functional processes that have previously been underappreciated.
Tissue proteases which are important for bone remodeling were expressed throughout healing, indicating prominent roles for different proteases at different timepoints62. MMPs 2 and 14 were expressed early, while MMPs 9 and 13 and CTSK were expressed during the conversion of cartilage to bone, and CTSG was observed at high levels at day 21. ELANE (neutrophil elastase) was expressed at highest levels at day 21 and the tibia with no previously reported role in fracture healing. ITIH2 (serine protease inhibitor) was expressed at high levels at our early time point, yet its involvement in fracture healing is unknown. Differential expression of these proteases are likely important for healthy fracture healing, and their timely expression may help with graft design for enhanced fracture healing.
We also identified ECM enzymes with critical roles in fracture healing: PLOD2 and LOXL2 (both catalyze collagen crosslinking and stability) were highly expressed early in fracture healing, while IBSP and MEPE (both involved in tissue mineralization) had higher expression at later timepoints. Deficiencies in these proteins result in bone pathologies and delays in fracture healing63,64,65-67, and these are examples of proteins that could be harnessed in an intelligently designed graft for functionally enhanced fracture repair.
Limitations of our study include the mass spectrometry data dependent acquisition strategy used, which identifies high abundance targets and ignores those at lower abundance. Nonetheless, we identified a high number of ECM proteins whose expression patterns align with the literature. While we report a small sample size (n=2, 3) in this pilot study, we observed clear separation of samples based on their timepoints using principal components as well as with unsupervised clustering. Regarding sex dimorphism, there are known differences in fracture healing between males and females, and recent research indicates that male mice display more rapid fracture healing, have more prominent soft callus formation, and higher bone mineral density68. These results suggest differences in ECM remodeling between males and females, and future studies should include females. While clinical fractures are immobilized, which generates IO healing as well as EO, here we use an EO-biased model that allowed us to investigate this predominant mechanism of fracture healing. Future directions for this work include investigating earlier and later timepoints to capture the early inflammatory and late remodeling responses.
Our work aimed to investigate a timeseries analysis of the fracture callus ECM proteome in order to understand important changes that occur during healthy bone healing. This data can be further mined to identify peptides and proteins involved in a pro-regenerative healing response for better bone graft design.
Supplementary Material
ACKNOWLEDGEMENTS:
This publication was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through UCSF-CTSI Grant Number UL1 TR000004 (Award # A119683, PI Bahney). This project was also supported the UCSF Resource Allocation Program Grant Funding (Bahney). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or University. We would also like to thank Kelsey M O’Hara (SPRI) for her assistance staining the fracture calli and assembling the images into figure 1 and Gina Baldoza (UCSF) for grants administration.
REFERENCES
- 1.Bahney CS et al. Cellular biology of fracture healing. Journal of orthopaedic research : official publication of the Orthopaedic Research Society 37, 35–50, doi: 10.1002/jor.24170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahney CS et al. Cellular biology of fracture healing. Journal of Orthopaedic Research® 37, 35–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marsell R & Einhorn TA The biology of fracture healing. Injury 42, 551–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Percival CJ & Richtsmeier JT Angiogenesis and intramembranous osteogenesis. Developmental Dynamics 242, 909–922 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sfeir C, Ho L, Doll BA, Azari K & Hollinger JO in Bone regeneration and repair 21–44 (Springer, 2005). [Google Scholar]
- 6.Hu DP et al. Cartilage to bone transformation during fracture healing is coordinated by the invading vasculature and induction of the core pluripotency genes. Development 144, 221–234, doi: 10.1242/dev.130807 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahney CS, Hu DP, Miclau III T & Marcucio RS The multifaceted role of the vasculature in endochondral fracture repair. Frontiers In endocrinology 6, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bais M et al. Transcriptional analysis of fracture healing and the induction of embryonic stem cell–related genes. PloS one 4, e5393 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimitriou R, Jones E, McGonagle D & Giannoudis PV Bone regeneration: current concepts and future directions. BMC medicine 9, 66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeung Tsang K, Wa Tsang S, Chan D & Cheah KS The chondrocytic journey in endochondral bone growth and skeletal dysplasia. Birth defects research. Part C, Embryo today : reviews 102, 52–73, doi: 10.1002/bdrc.21060 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Wong SA et al. Microenvironmental Regulation of Chondrocyte Plasticity in Endochondral Repair-A New Frontier for Developmental Engineering. Frontiers in bioengineering and biotechnology 6, 58, doi: 10.3389/fbioe.2018.00058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calciolari E & Donos N Proteomic and transcriptomic approaches for studying bone regeneration in health and systemically compromised conditions. PROTEOMICS–Clinical Applications 14, 1900084 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Dennis SC, Berkland CJ, Bonewald LF & Detamore MS Endochondral ossification for enhancing bone regeneration: converging native extracellular matrix biomaterials and developmental engineering in vivo. Tissue Engineering Part B: Reviews 21, 247–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheehy EJ, Vinardell T, Buckley CT & Kelly DJ Engineering osteochondral constructs through spatial regulation of endochondral ossification. Acta Biomater 9, 5484–5492, doi: 10.1016/j.actbio.2012.11.008 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Bahney CS et al. Stem Cell–Derived Endochondral Cartilage Stimulates Bone Healing by Tissue Transformation. Journal of Bone and Mineral Research 29, 1269–1282, doi: 10.1002/jbmr.2148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothrauff BB & Tuan RS Decellularized bone extracellular matrix in skeletal tissue engineering. Biochemical Society transactions, doi: 10.1042/bst20190079 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Lin X, Patil S, Gao Y-G & Qian A The Bone Extracellular Matrix in Bone Formation and Regeneration. Frontiers in Pharmacology 11, doi: 10.3389/fphar.2020.00757 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Carvajal ZY et al. Cartilage tissue engineering: the role of extracellular matrix (ECM) and novel strategies. (IntechOpen, 2013). [Google Scholar]
- 19.Maes C et al. Placental growth factor mediates mesenchymal cell development, cartilage turnover, and bone remodeling during fracture repair. The Journal of clinical investigation 116, 1230–1242, doi: 10.1172/JCI26772 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivera KO et al. Local injections of beta-NGF accelerates endochondral fracture repair by promoting cartilage to bone conversion. Sci Rep 10, 22241, doi: 10.1038/s41598-020-78983-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z et al. Fracture repair requires TrkA signaling by skeletal sensory nerves. The Journal of clinical investigation 129, 5137–5150, doi: 10.1172/JCI128428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zelzer E et al. VEGFA is necessary for chondrocyte survival during bone development. Development 131, 2161–2171, doi: 10.1242/dev.01053 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Gerber HP et al. VEGF couples hypertrophic cartilage remodeling, ossification and angiogenesis during endochondral bone formation. Nat Med 5, 623–628, doi: 10.1038/9467 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Inada M et al. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc Natl Acad Sci U S A 101, 17192–17197, doi:0407788101 [pii] 10.1073/pnas.0407788101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behonick DJ et al. Role of matrix metalloproteinase 13 in both endochondral and intramembranous ossification during skeletal regeneration. PloS one 2, e1150, doi: 10.1371/journal.pone.0001150 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colnot C, Thompson Z, Miclau T, Werb Z & Helms JA Altered fracture repair in the absence of MMP9. Development 130, 4123–4133 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X et al. MMP9 regulates the cellular response to inflammation after skeletal injury. Bone 52, 111–119, doi: 10.1016/j.bone.2012.09.018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieu S et al. Impaired remodeling phase of fracture repair in the absence of matrix metalloproteinase-2. Disease models & mechanisms 4, 203–211, doi: 10.1242/dmm.006304 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlesinger PH et al. Cellular and extracellular matrix of bone, with principles of synthesis and dependency of mineral deposition on cell membrane transport. American Journal of Physiology-Cell Physiology 318, C111–C124, doi: 10.1152/ajpcell.00120.2019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calciolari E & Donos N Proteomic and Transcriptomic Approaches for Studying Bone Regeneration in Health and Systemically Compromised Conditions. PROTEOMICS – Clinical Applications 14, 1900084, doi: 10.1002/prca.201900084 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Gibon E, Lu L & Goodman SB Aging, inflammation, stem cells, and bone healing. Stem cell research & therapy 7, 44, doi: 10.1186/s13287-016-0300-9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.General, O. o. t. S. Bone health and osteoporosis: a report of the Surgeon General. (2004). [PubMed]
- 33.Moseley KF Type 2 diabetes and bone fractures. Curr Opin Endocrinol Diabetes Obes 19, 128–135, doi: 10.1097/MED.0b013e328350a6e1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams SA et al. Economic burden of osteoporotic fractures in US managed care enrollees. Am J Manag Care 26, e142–e149 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Carvalho MS, Poundarik AA, Cabral JMS, da Silva CL & Vashishth D Biomimetic matrices for rapidly forming mineralized bone tissue based on stem cell-mediated osteogenesis. Scientific reports 8, 14388, doi: 10.1038/s41598-018-32794-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le A, Miclau T, Hu D & Helms J Molecular aspects of healing in stabilized and non-stabilized fractures. Journal of Orthopaedic Research 19, 78–84 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Uyy E et al. Endoplasmic reticulum chaperones are potential active factors in thyroid tumorigenesis. Journal of proteome research 15, 3377–3387 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Team, R. C. R: A language and environment for statistical computing. (2013).
- 39.Heberle H, Meirelles GV, da Silva FR, Telles GP & Minghim R InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16, 169, doi: 10.1186/s12859-015-0611-3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCabe MC et al. Evaluation and Refinement of Sample Preparation Methods for Extracellular Matrix Proteome Coverage. Molecular & cellular proteomics : MCP 20, 100079, doi: 10.1016/j.mcpro.2021.100079 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raudvere U et al. g: Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic acids research 47, W191–W198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Supek F, Bošnjak M, Škunca N & Šmuc T REVIGO summarizes and visualizes long lists of gene ontology terms. PloS one 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szklarczyk D et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic acids research 43, D447–D452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M & Tanabe M KEGG: integrating viruses and cellular organisms. Nucleic acids research 49, D545–d551, doi: 10.1093/nar/gkaa970 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kosaki N et al. Impaired bone fracture healing in matrix metalloproteinase-13 deficient mice. Biochemical and biophysical research communications 354, 846–851 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Walia B, Lingenheld E, Duong L, Sanjay A & Drissi H. A novel role for cathepsin K in periosteal osteoclast precursors during fracture repair. Annals of the New York Academy of Sciences 1415, 57–68, doi: 10.1111/nyas.13629 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Holmes D Closing the gap. Nature 550, S194–S195 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Baldwin P et al. Autograft, Allograft, and Bone Graft Substitutes: Clinical Evidence and Indications for Use in the Setting of Orthopaedic Trauma Surgery. J Orthop Trauma 33, 203–213, doi: 10.1097/bot.0000000000001420 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Fu R, Liu C, Yan Y, Li Q & Huang RL Bone defect reconstruction via endochondral ossification: A developmental engineering strategy. J Tissue Eng 12, 20417314211004211, doi: 10.1177/20417314211004211 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson EM, Matsiko A, Farrell E, Kelly DJ & O'Brien FJ Recapitulating endochondral ossification: a promising route to in vivo bone regeneration. Journal of tissue engineering and regenerative medicine 9, 889–902, doi: 10.1002/term.1918 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Dang ABC et al. Repurposing Human Osteoarthritic Cartilage as a Bone Graft Substitute in an Athymic Rat Posterolateral Spinal Fusion Model. Int J Spine Surg 12, 735–742, doi: 10.14444/5092 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sielatycki JA et al. Autologous chondrocyte grafting promotes bone formation in the posterolateral spine. JOR Spine 1, e1001, doi: 10.1002/jsp2.1001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Freeman FE & McNamara LM Endochondral Priming: A Developmental Engineering Strategy for Bone Tissue Regeneration. Tissue engineering. Part B, Reviews, doi: 10.1089/ten.TEB.2016.0197 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Lin X et al. B2A peptide induces chondrogenic differentiation in vitro and enhances cartilage repair in rats. Journal of Orthopaedic Research 30, 1221–1228, doi: 10.1002/jor.22078 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ho-Shui-Ling A et al. Bone regeneration strategies: Engineered scaffolds, bioactive molecules and stem cells current stage and future perspectives. Biomaterials 180, 143–162, doi: 10.1016/j.biomaterials.2018.07.017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomar F, Orozco R, Villar JL & Arrizabalaga F P-15 small peptide bone graft substitute in the treatment of non-unions and delayed union. A pilot clinical trial. International orthopaedics 31, 93–99 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coustry F et al. Mutant cartilage oligomeric matrix protein (COMP) compromises bone integrity, joint function and the balance between adipogenesis and osteogenesis. Matrix biology : journal of the International Society for Matrix Biology 67, 75–89, doi: 10.1016/j.matbio.2017.12.014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ishida K et al. Cartilage oligomeric matrix protein enhances osteogenesis by directly binding and activating bone morphogenetic protein-2. Bone 55, 23–35, doi: 10.1016/j.bone.2013.03.007 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Morgan JM, Wong A, Yellowley CE & Genetos DC Regulation of tenascin expression in bone. Journal of cellular biochemistry 112, 3354–3363, doi: 10.1002/jcb.23265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kimura H, Akiyama H, Nakamura T & de Crombrugghe B Tenascin-W inhibits proliferation and differentiation of preosteoblasts during endochondral bone formation. Biochemical and biophysical research communications 356, 935–941, doi: 10.1016/j.bbrc.2007.03.071 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Imhof T et al. Pivotal Role of Tenascin-W (-N) in Postnatal Incisor Growth and Periodontal Ligament Remodeling. Front Immunol 11, 608223, doi: 10.3389/fimmu.2020.608223 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cassuto J, Folestad A, Göthlin J, Malchau H & Kärrholm J Concerted actions by MMPs, ADAMTS and serine proteases during remodeling of the cartilage callus into bone during osseointegration of hip implants. Bone Rep 13, 100715, doi: 10.1016/j.bonr.2020.100715 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lv F et al. Novel Mutations in PLOD2 Cause Rare Bruck Syndrome. Calcified tissue international 102, 296–309, doi: 10.1007/s00223-017-0360-6 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Leal GF et al. Expanding the Clinical Spectrum of Phenotypes Caused by Pathogenic Variants in PLOD2. Journal of Bone and Mineral Research 33, 753–760, doi: 10.1002/jbmr.3348 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Alam I et al. SIBLING family genes and bone mineral density: association and allele-specific expression in humans. Bone 64, 166–172, doi: 10.1016/j.bone.2014.04.013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Estrada K et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 44, 491–501, doi: 10.1038/ng.2249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Surakka I et al. MEPE loss-of-function variant associates with decreased bone mineral density and increased fracture risk. Nature communications 11, 4093, doi: 10.1038/s41467-020-17315-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haffner-Luntzer M, Fischer V & Ignatius A Differences in Fracture Healing Between Female and Male C57BL/6J Mice. Frontiers in Physiology 12, doi: 10.3389/fphys.2021.712494 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.