SUMMARY
Giant congenital melanocytic nevi are NRAS-driven proliferations that may cover up to 80% of the body surface. Their most dangerous consequence is progression to melanoma. This risk often triggers preemptive extensive surgical excisions in childhood, producing lifelong severe challenges. We present preclinical models including multiple genetically engineered mice and xenografted human lesions, which enabled testing locally applied pharmacologic agents to avoid surgery. The murine models permitted identification of proliferative vs senescent nevus phases, and treatments targeting both. These nevi recapitulated histologic and molecular features of human giant congenital nevi including risk of melanoma transformation. Cutaneously delivered MEK, PI3K and c-KIT inhibitors or pro-inflammatory squaric acid dibutylester (SADBE) achieved major regressions. SADBE triggered innate immunity that ablated detectable nevocytes, fully prevented melanoma, and regressed human giant nevus xenografts. These findings reveal nevus mechanistic vulnerabilities and suggest opportunities for topical interventions that may alter therapeutic options for children with congenital giant nevi.
Keywords: Congenital melanocytic nevus, melanoma, hapten, Nras, mole, prevention and topical
In Brief
Large congenital nevi, or highly pigmented regions of skin, can present major cosmetic and psychosocial issues and have a significant chance of turning into malignant melanoma. However, current treatment methods provide only partial removal and can lead to scarring. Here, congenital nevus mouse models were developed and used to identify topical therapies that were highly effective at clearing nevi and protecting against melanoma formation.
Graphical Abstract
Introduction
Congenital melanocytic nevi are benign proliferations of nevomelanocytes that are present at birth. Large or giant congenital nevi occur in approximately 1 in 20,000 births (Chien et al., 2010; Hashmi et al., 2009). The estimated rate of conversion from benign nevus to malignant melanoma is approximately 2% and 12% in patients with large and giant congenital nevi, respectively (Bett, 2005; Kinsler et al., 2009; Vourc’h-Jourdain et al., 2013; Zaal et al., 2005). Other side effects of congenital melanocytic nevi include weakened skin integrity within the lesions, as well as major cosmetic and psychosocial ramifications due to their appearance (Koot et al., 2000; Price and Schaffer, 2010; Tromberg et al., 2005).
The current available treatments for patients with congenital melanocytic nevi include surgical excision, dermatome shaving, pigment-specific laser and others (Ibrahimi et al., 2012; Viana et al., 2013). However, removal of a complete lesion by these approaches is often impossible. The development of alternative therapies that avoid the shortcomings of the current treatments (such as extensive scarring) while minimizing the lifetime risk of melanoma, are needed.
Activating NRAS mutations are the sole recurrent somatic mutation found in up to 80–95% of large and giant congenital nevi (Bauer et al., 2007). The majority of NRAS mutations are Q61K or Q61R amino acid substitutions, resulting in a constitutively active NRAS protein (Bauer et al., 2007; Charbel et al., 2014; Kinsler et al., 2013). Shakhova and colleagues (Shakhova et al., 2012) have demonstrated that transgenic mutant Nras targeting melanocytes via the tyrosinase promoter can cause Sox10 dependent giant congenital nevi in mice, in agreement with several groups who used tyr::Nras models to demonstrate and study nevi or progression to melanoma (Ackermann et al., 2005; Pawlikowski et al., 2015; Shakhova et al., 2012).
In this study, we describe the development and characterization of murine congenital nevus models that accurately recapitulate key features of human giant congenital nevi, including risk of progression to invasive melanoma. We utilized these preclinical models to test a collection of localized drug treatments and observed that MEK-targeted local therapies exhibit regressive activity. We additionally observed a profound clearance of the lesions following topical squaric acid dibutyl ester (SADBE)-based immunotherapy, an approach which also offered 100% protection against melanoma formation within treated skin of prospectively followed mice. The activity of SADBE was shown to be mediated by macrophages. Therefore, it was possible to confirm SADBE efficacy in xenografts of human resected giant nevi established in immunodeficient (SHO) mice which retain macrophage function. These data suggest that an immune-based approach that utilizes topical SADBE may hold therapeutic potential for human giant congenital nevi.
RESULTS
Melanocyte-specific NrasQ61R driven congenital nevus models recapitulate genetic and phenotypic features of human giant congenital nevi
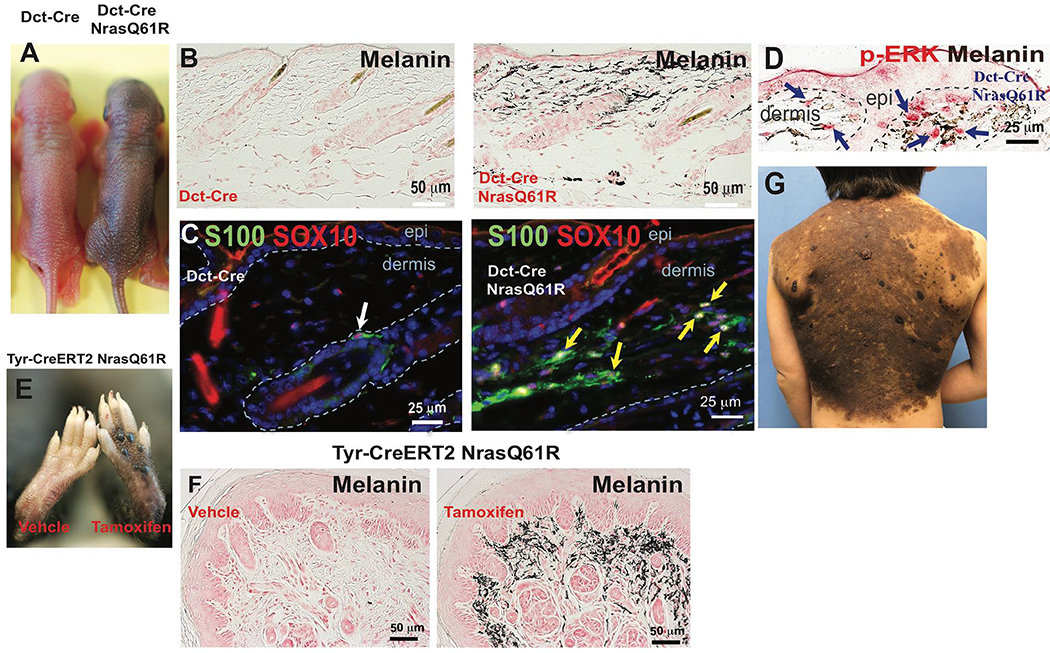
We generated and then characterized both constitutive and inducible murine giant congenital nevus models, and compared their features to those of corresponding human lesions. Melanocyte-specific NrasQ61R mutant mice were generated by crossing LSL-NrasQ61R knock-in mice (Burd et al., 2014) with transgenic mice expressing homozygous Cre recombinase under the control of melanocyte-specific promoters, including dopachrome tautomerase (Dct) and tyrosinase (Tyr). Newborn mice harboring the Dct-Cre-mediated NrasQ61R mutation displayed cutaneous hyperpigmentation, and retained their congenital pigmented lesions throughout life, without spontaneous regression (Figures 1A and S1A). Abundant ectopic dermal melanin was detected in mutant skin (Figures 1B and S1B), and the melanin producing dermal melanocytes stained positively for the NrasQ61R mutation (Figure S1C). Melanocytic markers such as S100, SOX10, MITF and phospho-ERK were also present within dermal nevus cells (Figures 1C, 1D, and S1D–F). When lesions from Nras mutant mice were juxtaposed with human giant congenital nevi (e.g., Figures 1G and S2) multiple comparable histologic features were detected. Similar to the human giant congenital nevi, the mutant mouse nevi were adjacent to adnexal structures while sparing other structures, distributed within papillary and reticular dermis extending into superficial subcutaneous fat, and combined with variably pigmented spindle shaped nevus cells. In addition, superficial aggregates of nevus cells formed nests, and penetrated along collagen bundles parallel to the long axis of the epidermis.
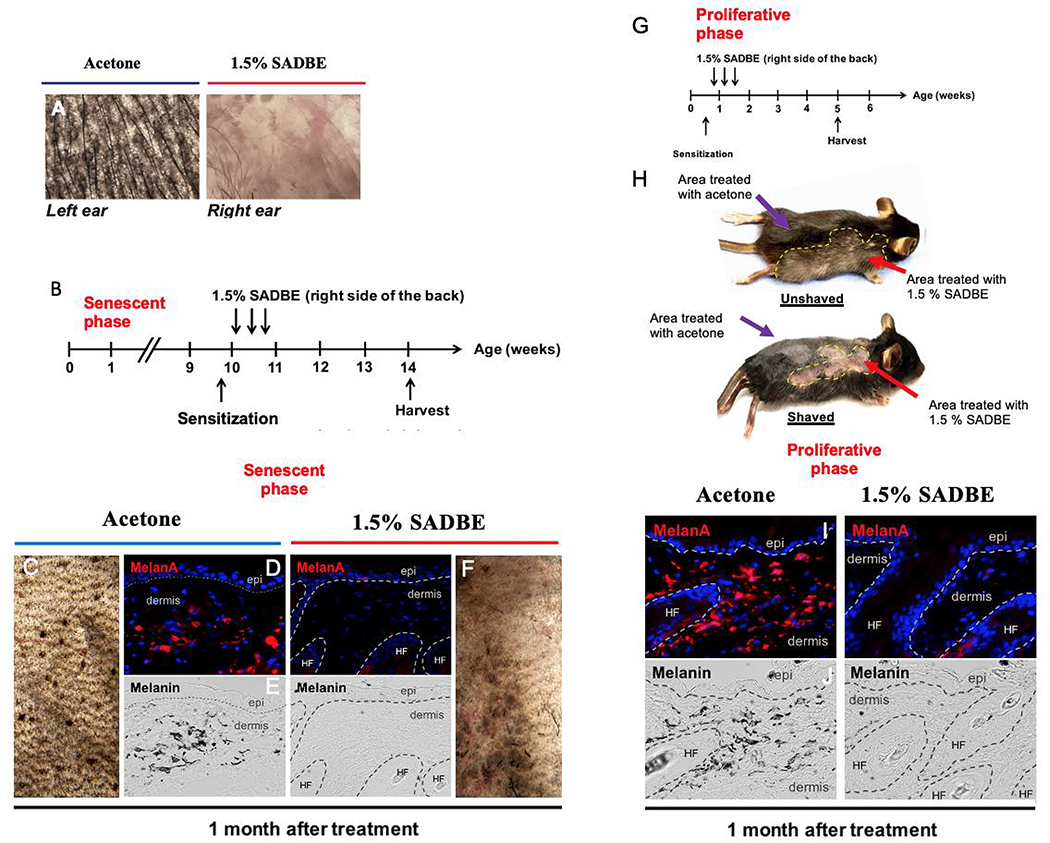
Figure 1. Melanocyte-specific NrasQ61R mutant mice recapitulates histologic and molecular features of human giant congenital melanocytic nevi.
(A) Features of melanocytic nevi in 3-day-old Dct-Cre NrasQ61R. (B) Melanin of adult Dct-Cre NrasQ61R mice was detected by Fontana-Masson staining. (C-D) Immunostains. Paraffin sections were immunostained for S100 (C, green), SOX10 (C, red), or p-ERK (D, red). (E) Features of melanocytic nevi in tamoxifen-induced Tyr-CreERT2 NrasQ61R mice. (F) Melanin in tamoxifen-induced Tyr-CreERT2 NrasQ61R mice was detected by Fontana-Masson staining. (G) The clinical appearance of a human giant congenital nevus is shown in. White arrow in (C) indicates normal signal in hair follicle. Yellow arrows in (C) indicate ectopic signals in the dermis of nevus skin. Blue arrows in (D) indicate positive signals. See also supplementary Figures 1 and 2.
In order to assess proliferative vs senescent phases of nevus development, melanocyte-specific expression of inducible NrasQ61R mutation was activated by topical 4-hydroxytamoxifen (tamoxifen) treatment in neonatal offspring of LSL-NrasQ61R knock-in mice crossed with tamoxifen-inducible Tyr-CreERT2 mice. 2 weeks after induction skin hyperpigmentation was observed, driven by ectopic dermal melanocytes which developed in tamoxifen-treated areas of skin (Figures 1E–F and S1G–H). We also generated mice by using constitutive Tyr-Cre-mediated NrasQ61R expression. The combination of these alleles was lethal in most offspring. In addition to skin hyperpigmentation, the few surviving Tyr-Cre NrasQ61R mutants displayed hydrocephalus (Figure. S1I) and other neurologic abnormalities, such as neurocutaneous melanosis, (Figure S1J–N) which are in line with similar murine and human Nras mutations expressed in mice and in children (Kinsler et al., 2013; Pedersen et al., 2013; Shah, 2010; Shakhova et al., 2012).
NrasQ61R mutation-driven congenital nevi exhibit proliferative and senescent phases
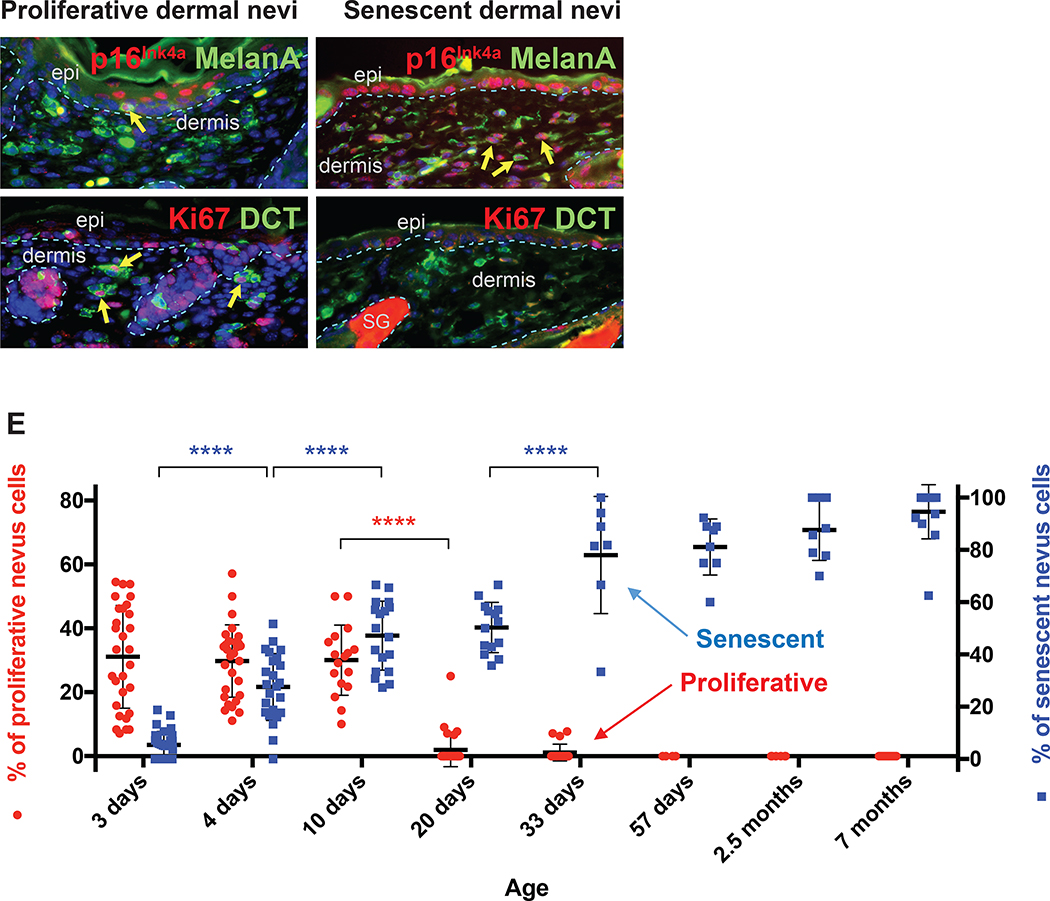
Since the therapeutic efficacies of various treatments may depend on whether nevus cells are proliferating or arrested/senescent, we quantified the proportion of dermal melanocytic cells in Dct-Cre NrasQ61R mice, at various timepoints relative to date of birth, measuring proliferative and senescence markers. The results indicated that neonatal nevus cells are negative for p16Ink4a (Figure 2A) and positive for Ki67 (Figure 2B), consistent with a proliferative phase. In contrast, adult nevus cells are positive for p16Ink4a (Figure 2C), but not Ki67 (Figure 2D) consistent with growth arrest and senescence (Gray-Schopfer et al., 2006; Tran et al., 2012). Quantification of these data revealed that NrasQ61R mutation-driven congenital nevi are initially proliferative and become senescent over approximately 3 weeks (Figure 2E, note left and right Y axes), a feature that has also been observed in humans (Gerami and Paller, 2013; Leech et al., 2004; Phadke et al., 2011).
Figure 2. NrasQ61R mutation-driven congenital nevi are initially proliferative and subsequently become senescent.
(A, B) Representative images of proliferative nevi. (C, D) Representative images of senescent nevi. Paraffin sections were immunostained for p16Ink4a (A and C, red), MelanA/MART1 (A and C, green), Ki67 (B and D, red), or DCT (B and D, green). Yellow arrows indicate double-positive cells, and white arrows indicate single-positive cells. Dorsal skin samples from Dct-Cre NrasQ61R/Q61R mutant mice were harvested at the ages indicated in E. Samples from three mice of each age group, with five to ten of non-adjacent samples per mouse, were quantified. Nevus cell proliferation and senescence were measured by immunofluorescence staining for Ki67 and p16Ink4a, respectively. Quantification of the above staining revealed a statistically significant decrease in proliferative nevus cell between 10 and 20 days of age (E, red). The percentage of senescent nevus cells gradually increases between age groups until mice reach approximately 1 month of age (E, blue). Red and blue colored dots indicate mean percentages of nevus cells that are proliferative (left Y-axis) or senescent (right Y-axis), respectively. The bars represent the standard error of the mean (SEM). P-values were calculated using one-way ANOVA with Tukey’s multiple comparison test (****P<0.0001).
NrasQ61R- driven nevi transform into invasive melanoma
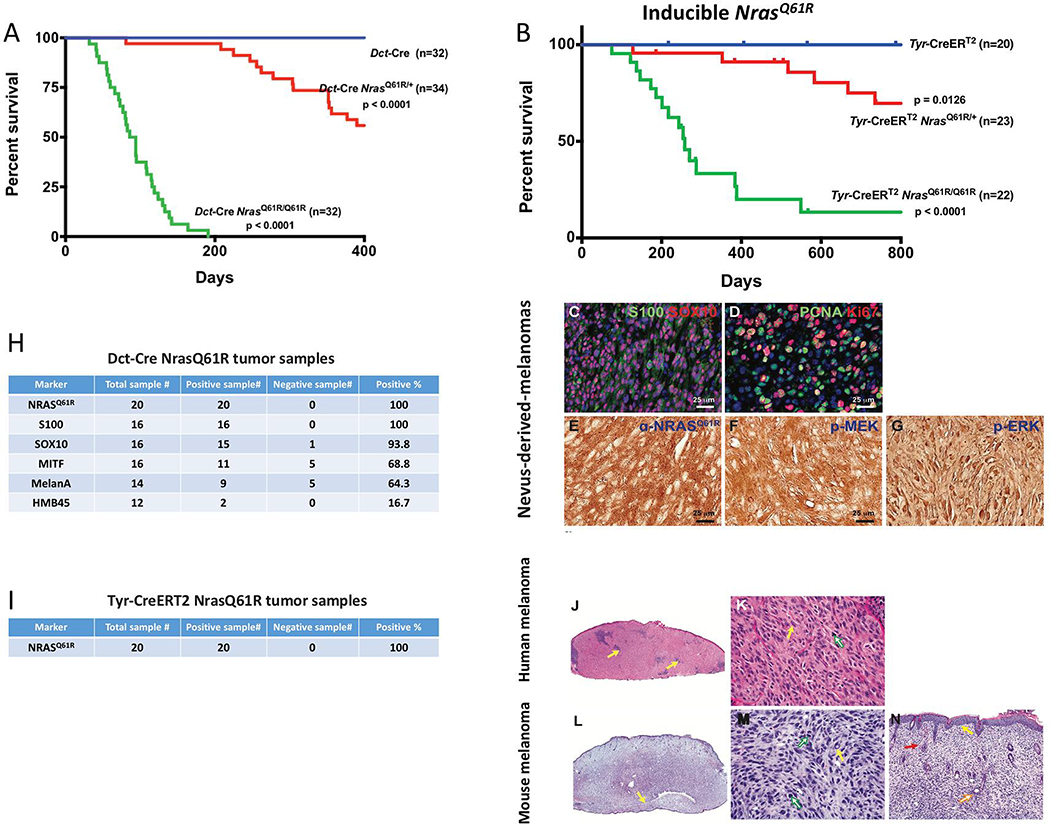
Human giant congenital nevi carry a significant risk of melanomagenesis (Bajaj et al., 2009; Krengel et al., 2006; Shah, 2010). We therefore assessed the risk of melanoma development in our NrasQ61R mutation-driven murine nevus models. Tumor-free survival curves for Nras mutant animals displayed significant rates of melanoma formation in Dct-Cre NrasQ61R mice (Figure 3A) and Tyr-CreERT2 NrasQ61R mice (Figure 3B) compared with control mice. In humans, melanomas derived from giant congenital nevi are typically heterozygous for mutant NRAS (Kinsler et al., 2013; Price, 2016). Consistent with this, we observed significant latency and sporadic frequency of spontaneous melanoma formation in NrasQ61R/+ heterozygous mice activated by either constitutive (Dct-Cre) or inducible (Tyr-CreERT2) Cre alleles (Figure 3A–B, red lines), thus resembling another feature of human giant congenital nevi. Two copies of NrasQ61R produced melanoma transformation with a shorter latency and a higher penetrance in both the constitutive (Dct-Cre) and inducible (Tyr-CreERT2) backgrounds (Figure 3A–B, green lines). Furthermore, we found that Dct-Cre NrasQ61R mutation-driven melanomas were positive for melanocytic markers, including S100, SOX10 (Figure 3C), and MITF (Figure 3H) and highly proliferative (Figure 3D). The activating NrasQ61R epitope was found in both models (Figure 3E and 3I) and 3 out of 3 melanomas arising on heterozygous NrasQ61R mice retained the wild type Nras allele (Figure S3). The activation of MEK/ERK signaling in Dct-Cre NrasQ61R nevus-derived melanomas was confirmed by immunofluorescence (Figure 3F and 3G). Dct-Cre NrasQ61R mutation-driven murine melanomas (Figure 3L–N) also showed similar histologic features when compared to those of human melanomas (Figure 3J–K).
Figure 3. Melanocyte-specific NrasQ61R activating mutation in mice results in high incidence of melanoma with shared histologic features of human melanoma.
(A) Dct-Cre NrasQ61R/Q61R mutant (green, n=35), Dct-Cre NrasQ61R/+ mutant (red, n=34), and Dct-Cre control (blue, n=32) mice were examined to compare the incidence of NrasQ61R mutation-driven melanoma formation. Survival curves were plotted using the Kaplan-Meier method and analyzed by the log-rank test. The tumor-free survival rates were significantly different between the control and Dct-Cre NrasQ61R/Q61R groups (P<0.0001), and between the control and Dct-Cre NrasQ61R/+ groups (P<0.0001). (B) The incidence of NrasQ61R mutation-driven melanomas was measured in tamoxifen-induced Tyr-CreERT2 NrasQ61R/Q61R mutant (green, n=22), Tyr-CreERT2 NrasQ61R/+ mutant (red, n=23), and Tyr-CreERT2 control (blue, n=20) mice. Statistical significance was defined as P<0.05. (C-G) Dct-Cre NrasQ61R tumor sections were immunostained for S100 (C, green), SOX10 (C, red), PCNA (D, green), Ki67 (D, red), NrasQ61R (E, brown), p-MEK (F, brown), or p-ERK (G, brown). (H) Melanocytic markers, including S100, SOX10, MITF, MelanA/MART-1, and HMB45, were assessed by immunostaining in Dct-Cre NrasQ61R tumor samples, and the percentage of positive samples for each marker was calculated. (I) Tyr-CreERT2 NrasQ61R tumor samples were examined using an antibody against NrasQ61R and the percentage of NrasQ61R-positive samples was calculated. (J-K) H&E stained histological images of human melanomas. (L-N) H&E stained histological images of NrasQ61R mutation-driven murine melanomas. are compared with those of. Histologic images at low and high magnifications are shown (J, L, 20x) (K, M, 400x; N, 100x). The infiltrate of human primary malignant melanomas replaces the entire dermis as a sheet, punctuated by nidi of inflammation (J, yellow arrows). The nuclear-to-cytoplasmic ratios, hyperchromatic nuclei, and sometimes multiple nucleoli are shown (K, yellow arrow). The atypical mitotic figure is visible (K, green arrow). The infiltration of murine melanoma can be observed in the ear as a sheet of tumor cells, encircling the cartilage (L, yellow arrow). Severe pleomorphism of the murine melanoma cells, with large nuclear-to-cytoplasmic ratios, hyperchromatic nuclei, and intranuclear vacuoles (M, yellow arrow), and mitotic activity, (M, green arrows) is observed. The murine melanoma abuts the epidermis, with no grenz zone (N, yellow arrow), and invades the hair follicle adventitia (N, orange arrow). The extravasation of red blood cells (N, red arrow), a common finding in human melanoma, is also detected. See also supplemental Figures 3 and 4.
RNA- sequencing was performed on nevi containing pinna and tumors from Dct-Cre NrasQ61R/+ and Tyr-CreERT2NrasQ61R/Q61R mice and compared to normal pinna. Pigmentation/melanocytic genes were somewhat upregulated with larger upregulation in homozygous NrasQ61R/Q61R nevi (Figure S4A), likely reflecting the relative mixture of nevus cells within other skin cells. Previously published human CMN expression data(Martins da Silva et al., 2019) showed heterogeneous expression patterns with some showing only weak melanocytic genes (e.g., MITF, SOX10) while others showing higher melanocytic genes expression (Figure S4B). In addition, there is evidence of upregulated interferon-α response as well as G2M checkpoint in the mouse CMN models compared to normal skin (Figure S4C). Up-regulated transcription factors, such as hematopoietic factor Gfi1b (fold change = 33.3, q-val < 3.7 10−2), regulatory T-cell-specific factor Foxp3 (fold change = 3.7, q-val<1.5 10−2) and lymphoid specific transcription factor Spi-b (fold change=3.0, q-val<4.5 10−2) also indicate immune infiltration in heterozygous nevi. Gfi and Foxp3 are included among the top 20 Lisa-predicted TF regulators of the differentially expressed genes, along with other immune related factors, Stat1, Stat3, Runx1, Irf4, RelA and Tbx21 (Figure S4D). In tumors, both models exhibited heterogeneity in expression patterns, likely reflecting variable second genetic events which followed the initiating NrasQ61R genomic event. Ngfr and Sox10 were upregulated in Dct-Cre NrasQ61R/+ tumors (Figure S4E).
Interestingly, Sox9 was down regulated in tumors from both models, similar to SOX9 in human melanoma (Yang et al., 2019). The interferon-α response was upregulated in the mouse tumors (Figure S4F). Human CMN samples (Martins da Silva et al., 2019) expressed a number of pathways in common with the murine CMNs, including upregulated interferon-α and downregulated oxidative phosphorylation and adipogenesis (Figure S4G). We therefore utilized these murine models of NrasQ61Rmutation-driven melanocytic nevi in subsequent experiments to preclinically evaluate therapeutic strategies to regress nevi and prevent progression to melanoma.
Signaling inhibitors can regress NrasQ61R mutation-driven melanocytic nevi
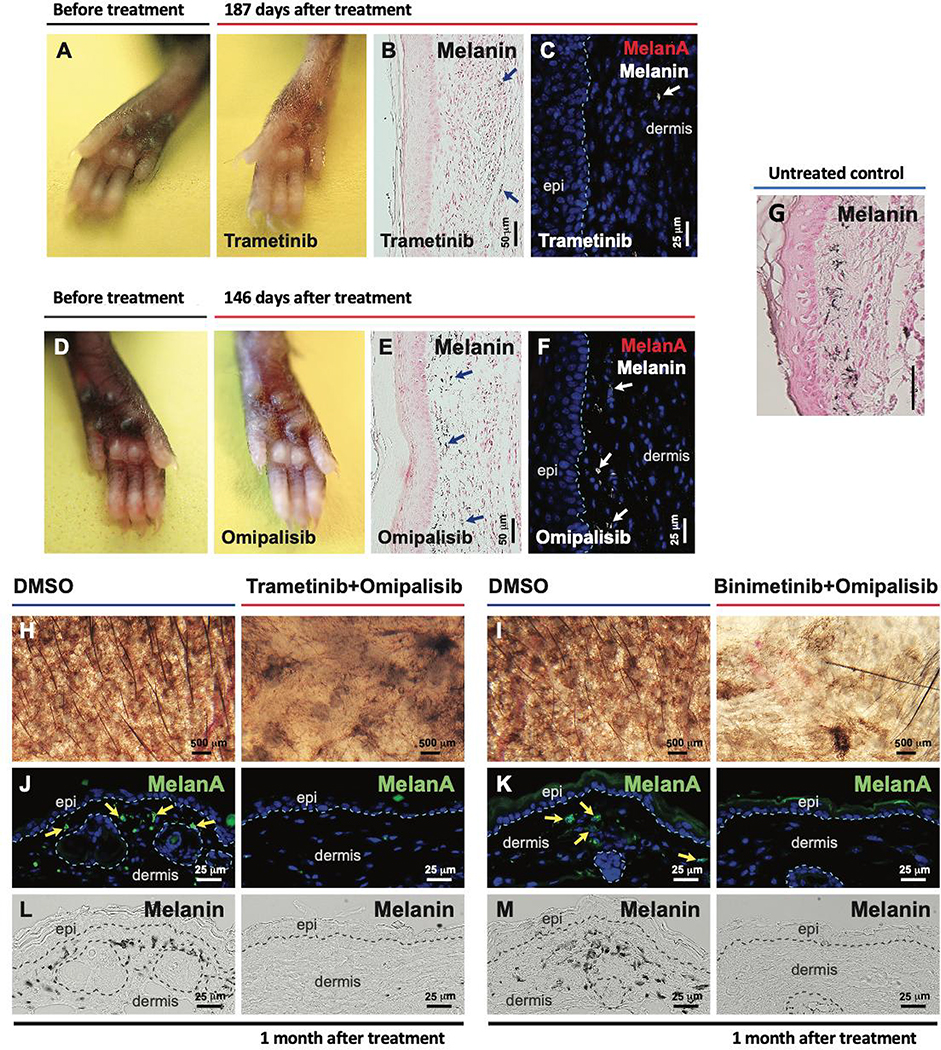
We hypothesized that MAPK or PI3K pathway inhibitors (either alone or in combination) may lead to regression of NrasQ61R mutation-driven giant congenital nevi, as evidence supporting this for MEK inhibitors has been previously reported (Kinsler et al., 2017; Mir et al., 2019; Pawlikowski et al., 2015; Rouille et al., 2019). We tested MEK and/or PI3K inhibitors applied as direct intralesional injection into paws (skin is thicker) or topical application to ears (Table S1). Local administration of these agents to 2-month-old tamoxifen-induced Tyr-CreERT2 NrasQ61R mice led to a significant loss of nevus cells and hypopigmentation (Figures 4 and S5). In topical single-drug therapies, the MEK inhibitor, binimetinib, induced clear hypopigmentation (Figure S5B) and the PI3K inhibitor, omipalisib, caused moderate hypopigmentation in the dermis (Figure S5C). Subcutaneous injection of the MEK inhibitors such as trametinib (Figure 4A–B), and binimetinib (Figure S5 A–B), or of the PI3K inhibitor, omipalisib (Figure 4D–E, Figure S5C), resulted in hypopigmentation and almost complete loss of melanocytes (Figures 4C and 4F), compared to untreated control Figure 4G). These data suggest that localized treatment of post-senescent nevi with inhibitors targeting kinases downstream of RAS may cause the selective loss of nevus cells albeit not necessarily complete ablation. Drukker et al. have shown activated AKT pathway activity in giant nevi (Drukker et al., 2013). We therefore tested various kinase-targeting combinations. When applied to right ear skin and compared to contralateral ears treated with vehicle control, we observed that topical combinations of trametinib and omipalisib (Figure 4G, 4I and 4K) or binimetinib and omipalisib (Figure 4H, 4J and 4L) led to strong melanocyte regression and dramatic depigmentation, demonstrating that the effects were localized and not systemic. MEK- or PI3K-targeted local therapies alone or in combination, may thus regress NrasQ61R driven nevi. The c-KIT inhibitor, imatinib, has inhibitory effects on human melanocytes and infrequently cause skin hypopigmentation (Cerchione et al., 2009; Hemesath et al., 1998; Nishimura et al., 2002; Tsao et al., 2003; Wehrle-Haller, 2003; Wu et al., 2000; Yoshida et al., 2001). We found that single-drug treatment with imatinib resulted in measurable depigmentation (Figure S5K). Moreover, a topical combination of binimetinib with imatinib led to more profound melanocyte regressions (Figure S5F and S5G) and depigmentation (Figure S5D and S5E). This combination induced immune responses (Figure S5H–J), as evident by the increased inflammatory cell recruitment (CD11c+, CD163+, and CD16+) to treated lesions.
Figure 4. Local therapy with small molecule inhibitors of the NRAS signaling pathway regress NrasQ61R-driven melanocytic nevi.
(A-F) NrasQ61R mutation-driven nevi were induced in Tyr-CreERT2 LSL-NrasQ61R newborn mice with tamoxifen over the first postnatal week, and 2 months later local treatments of senescent nevi were initiated with MEK and/or PI3K inhibitors or DMSO vehicle control. Trametinib (A-C, 0.1 μg /μl) or omipalisib (D-F, 0.03 μg /μl) was subcutaneously injected into the pigmented paws of tamoxifen-induced Tyr-CreERT2 NrasQ61R/+ heterozygous mutants in 10 μl of 10% DMSO, three times per week for two weeks. Paws were visualized 187 days after treatment with Trametinib (A) or 146 days after treatment with omipalisib (D). Melanin (B, and E, blue arrows) was detected by Fontana-Masson staining. Tissue sections were stained for MelanA/MART1 (C, F). White arrows (in C and F) indicate dermal melanin detected by bright-field microscopy. (G) Representative image of melanin detected by Fontana-Masson staining of non-treated tamoxifen-induced Tyr-CreERT2 NrasQ61R/+ mice from postnatal day 16, 2 weeks after tamoxifen induction. (H-M) Topical combinatorial therapies. Topical combinatorial therapies with the PI3K inhibitor, omipalisib (10 μg/ μl), plus the MEK inhibitor, trametinib (H, J, L, 2 μg/ μl) or binimetinib (I, K, M, 10 μg/ μl), were applied to the pigmented ears of tamoxifen-induced Tyr-CreERT2 NrasQ61R/Q61R mutant mice. All topical agents were administered in 5 μl DMSO, five times per week for 3 weeks (‘Trametinib+Omipalisib’ and their vehicle control treatments) or for 6 weeks (‘Binimetinib+Omipalisib’ and their vehicle control treatments). Ear skin tissues were harvested 1 month after the final drug treatment, and nevus regression was examined. Images were obtained with a stereomicroscope, and the dark brown particles indicate melanin (H, I). Tissue sections were stained for MelanA/MART1 (J, K, green). Yellow arrows indicate positive signals. Dermal melanin detected by bright-field microscopy is shown (L, M, black). We used five mice per treatment group, and representative images are shown. See also supplemental Figure 5.
Topical squaric acid dibutylester induces local inflammation and nevus cell destruction
We chose to test the efficacy of haptens, as these are low molecular weight chemicals that become antigenic when bound to a carrier molecule, and consequently elicit a well-known ‘contact hypersensitivity’-like response. (Chipinda et al., 2011; Vocanson et al., 2009). To determine whether topical hapten-based immunotherapy is capable of regressing congenital nevi, we tested SADBE, as a single-agent topical immunotherapy. SADBE was chosen for this study because it is already clinically used by dermatologists as a topical agent for alopecia areata and warts, and because it has been described to induce hypopigmentation or vitiligo (Sakai et al., 2020; Silverberg et al., 2000; Valsecchi et al., 1989). Two months after tamoxifen induction of nevi in ear skin of Tyr-CreERT2 NrasQ61R neonatal mice, the NrasQ61R mutation-driven nevus mice were sensitized by applying 2% SADBE to the right side of the shaved abdomen. Subsequent treatment of the right ear with topical 1.5% SADBE occurred three times over the following week. This short-term single agent therapy resulted in major regression of congenital nevi (Figure 5A). We next tested the efficacy of SADBE single-agent on both senescent and proliferative giant congenital nevi in dorsal skin of Dct-Cre NrasQ61R mutant mice as depicted in Figure 5B and G, respectively. When senescent nevus lesions were treated with SADBE beginning at 2 months of age, we observed skin depigmentation in areas treated with SADBE (Figure 5C and 5F). This regression was confirmed by the disappearance of both melanocytes and melanin (Figure 5D and 5E). To study susceptibility of proliferative nevi, SADBE treatments were initiated within a week of birth as depicted in Figure 5G, prior to the loss of proliferative nevus features (Figure 2E). Nevus regression was again noted (Figure 5H, lower panel red arrow) as well as subsequent leukotrichia (white hair) in SADBE-treated areas (Figure 5H, upper panel red arrow). In contrast, melanocytic nevi remained in areas treated with vehicle control (Figure 5H, purple arrows). Disappearance of both melanocytes and melanin was verified in treated lesions (Figure 5I and 5J).
Figure 5. Single-agent local immunotherapy with topical SADBE results in significant regression of both neonatal proliferative and postnatal senescent congenital nevi.
(A) SADBE (1.5%, topical) was applied to the pigmented right ears of tamoxifen-induced Tyr-CreERT2 NrasQ61R/Q61R mutant mice 2 months after tamoxifen induction, whereas the pigmented left ears of the same animals were treated with acetone vehicle. Ear tissue images were obtained with a stereomicroscope, and dark brown particles indicate melanin. (B) Treatment scheme of adult Dct-Cre NrasQ61R/Q61R. Single-drug local therapy with 1.5% SADBE was administered topically to the right back. Acetone was applied topically to the left back of each SADBE-treated mouse as a vehicle control. All treatments were carried out three times per week (every other day), and the treated skin tissues were harvested 1 month after the first treatment to assess regression of melanocytic nevi (C, F). Tissue sections were immunostained for MelanA/MART1 (D, I). Melanin (black) detected by bright-field microscopy is shown in (E, J). (G) Treatment scheme of neonatal Dct-Cre NrasQ61R/Q61R mice. (H) Representative images of treated Dct-Cre NrasQ61R/Q61R mice unshaved (upper panel) and shaved (lower panel) are shown. White hairs were detectable in treated areas (H, upper panel red arrow). Regression of melanocytic nevi was detectable in treated areas (H, lower panel red arrow). The purple arrows in (H) indicate unchanged nevi in the nevus lesion treated with acetone vehicle. Yellow dashed lines in H demarcate the region treated with 1.5% SADBE. Stippled white lines in (D, E, I, J) separate the epidermis (epi) and hair follicles (HF) from the dermis.
Topical squaric acid dibutylester regression of nevi involves recruitment and M1-like polarization of macrophages.
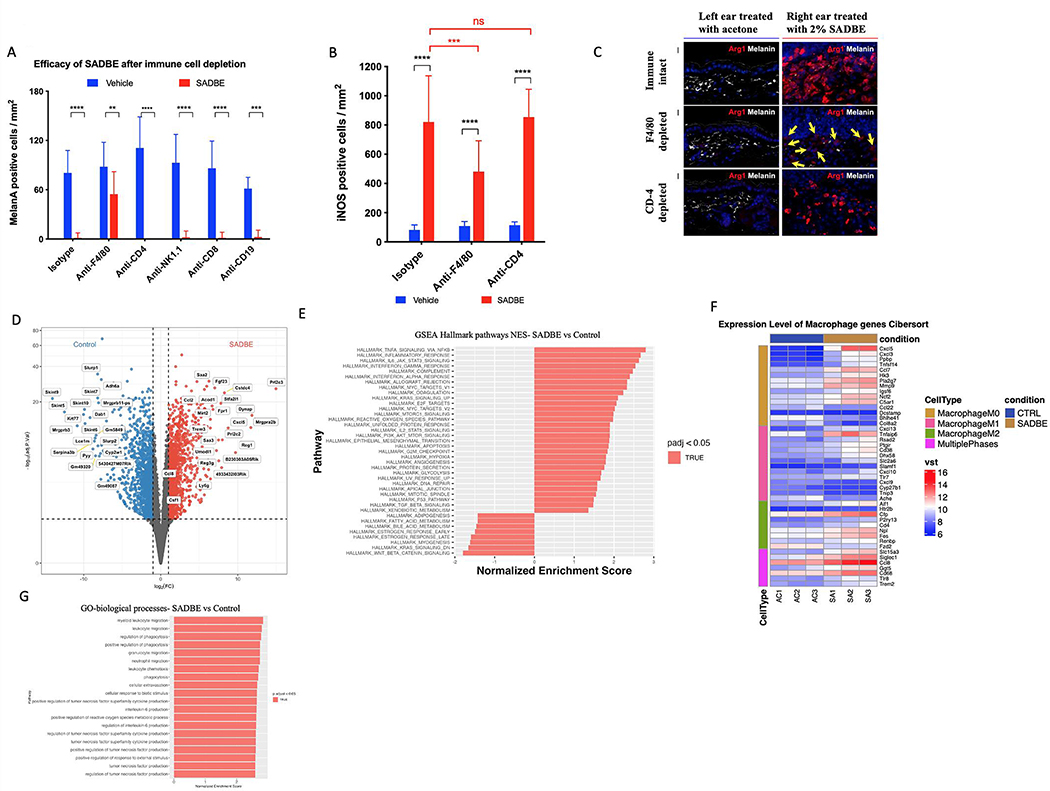
In order to assess immune cell requirements for the SADBE dependent regression of nevi, depletion of specific immune populations was performed on 2-month-old Dct-Cre NrasQ61R mice through IP injection of monoclonal antibodies directed at distinct immune lineages. Skin melan A positive nevus cells were quantified after SADBE treatment and compared to vehicle control (Figure 6A). Depletion of macrophages via anti-F4/80 significantly inhibited the ability of SADBE to clear melan A positive cells.; however, depletion of CD4+ T-cells, NK1.1+ NK cells CD8+ T-cells, and CD19+ B-cells, did not diminish the regression efficacy of SADBE. Depletion of tissue-resident cells (such as macrophages) is typically incomplete compared with other hematopoietic lineages, but the diminished efficacy of SADBE despite incomplete depletion (Figure 6B) strongly suggests that these cells play a key functional role in nevus clearance. Cells expressing the pro-inflammatory macrophage (M1) marker iNOS were also strongly recruited by SADBE (Figure 6B), as were anti-inflammatory Arg1+ cells (Figure 6C). Furthermore, CD4+ T-cell depletion enhanced macrophage recruitment, with modestly larger increases in pro-inflammatory (and decreased anti-inflammatory Arg1+) populations and slightly higher melanin clearance (Figure 6C). Thus, SADBE appears to attract both inflammatory and anti-inflammatory macrophages. To independently assess the effect of SADBE on the presence of markers for these cell populations, Dct-Cre NrasQ61R mice were treated with either SADBE or vehicle control (3 per treatment or control groups) for 72h, and RNA-Seq was carried out. Differential gene expression analysis revealed macrophage-related cytokines among top upregulated genes (Figure 6D). GSEA analysis showed TNFα signaling via NFKB as the top upregulated pathway (Figure 6E). SADBE was shown to highly induce genes of multiple macrophage subtypes, which included pro-inflammatory M1-like polarization genes (Figure 6F) and GO biological processes analysis showed migration and regulation of phagocytosis as enriched pathways (Figure 6G).
Figure 6. Antibody mediated depletion of inflammatory cell lineages in mice and RNAseq reveal macrophage recruitment by SADBE.
(A) Quantification of MelanA positive cells per mm2 in Dct-Cre NrasQ61R mice after immunodepletion by IP injection with 750 μg of anti F4/80, anti-CD-4, anti-NK1.1, anti-CD-8 and anti-CD-19. (B) Quantification of INOS positive cells per mm2 in Dct-Cre NrasQ61R mice after immunodepletion by IP injection with 750 μg of anti F4/80 and anti-CD-4. (C) Arg1 staining (red) of ear skin sections following depletion with anti-CD-4, anti-F4/80, isotype control and 2% SADBE treatment for 1 week. Yellow arrows indicate negative signals. Melanin detected by bright-field microscopy is shown in white. (D) Differential gene expression analysis (E) GSEA hallmark pathways analysis (F) Heat map of macrophage marker groups according to macrophage phase and (G) GO biological processes analysis. Dct-Cre NrasQ61R mice were treated with SADBE (2%) or vehicle control (acetone) on dorsal skin (3 mice per treatment or control group). Whole skin RNA-Seq was performed 72h after treatment. Macrophage-associated genes for M0, M1, and M2 phases were collected from the hematopoietic gene signature set LM22.
Topical squaric acid dibutylester decreases nevus cell numbers in human CMN xenografts.
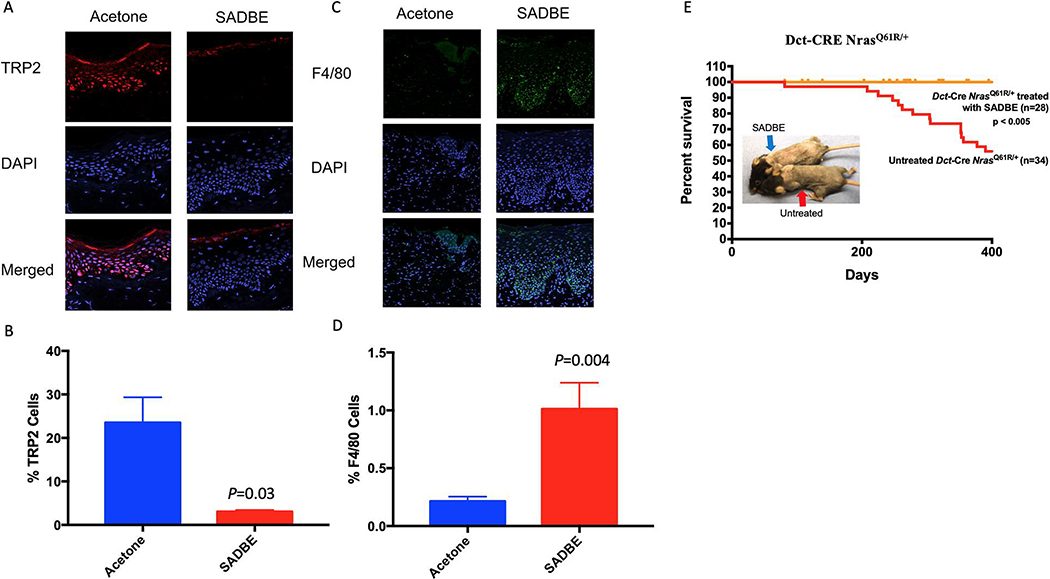
These lineage-ablation and expression studies suggested that SADBE’s clearance of nevus cells was not dependent upon adaptive immune cell types, but rather required macrophage function. We therefore engrafted human discarded resected CMN tissue onto immunodeficient SHO mice (which lack adaptive immunity, but retain at least partial macrophage function) (Bancroft and Kelly, 1994; Charles River) in order to test responses to SADBE. Eight human CMN xenografts were thus generated from a single giant nevus specimen. We utilized the epidermal region of the lesion that contained a significant epidermal nevus component for engraftment, thereby enhancing vascular supply and immune access. Following a 4-month recovery period, the grafts were subjected to biweekly topical treatment with 1% SADBE (or vehicle control) with dosing guided by potential sensitivity of the graft, followed by harvesting and staining for nevus cells using the melanocytic antibody anti-TRP2. SADBE treatments in these xenografted mice led to thickening of the stratum corneum (superficial dead cell layer of xenografts, Figure S6A), which contained significantly accumulated pigment, thus obscuring underlying cellular pigmentation changes. Stratum corneum thickening was not observed in mouse models (above) or in humans that were treated with SADBE for warts or Alopecia Areata. Immunofluorescence staining and automated quantification of underlying nevus cells (TRP2+) revealed a striking melanocyte decrease of ~90% (P=0.03) in human giant nevus xenografts following SADBE treatment as compared to vehicle controls (Figure 7A–B). To further examine whether SADBE treatments of human-to-mouse xenografts were inducing macrophage recruitment (as in the murine model), specimens from two weeks following initiation of topical SADBE treatment were stained with the macrophage marker F4/80. Figure 7C–D, depict the results of F4/80+ immunostaining and its quantification, and reveal significant recruitment of mouse macrophages (F4/80+) into the CMN tissue (vehicle 0.21 +/− 0.03, SADBE 1.01 +/− 0.22, P=0.004). These results corroborate nevus cell regressions within human tissue, showing that SADBE exhibits the ability to induce macrophage recruitment together with the significant clearance of human CMN cells within the murine SHO model.
Figure 7. SADBE decreases melanocyte numbers in CMN xenografts and prevents melanoma formation in mice.
(A) Immunofluorescent staining of CMN tissue xenografts treated with topical application of 1% SADBE or acetone control for 8 months: DAPI (blue) TRP2 (red) Merged (purple). (B) Quantification of the percentage of TRP2 positive cells out of total cells in the epidermis. (C) Immunofluorescent staining of CMN tissue xenografts treated with topical application of 1% SADBE or acetone control for 2 weeks: DAPI (blue) F4/80 (green) Merged (cyan). (D) Quantification of F4/80 positive cells as percent of total cells. (E) Effect of SADBE on melanoma-genesis and mouse survival in Dct-Cre NrasQ61R/+ mice. Mice were sensitized with a 2% SADBE in acetone treatment of the right side of the shaved abdomen. Three days later, sensitized mice were treated with 1.5% SADBE over the shaved dorsal (back) skin (treated and untreated controls indicated by arrows). Treatments with SADBE were carried out every other day (total of three treatments), and tumor formation was monitored. Untreated control tumor incidences are the same (historical) control Dct-CRE NrasQ61R/+ mice in Figure 3A. In a population where anatomic location was tracked, dorsal back specific melanomas occurred in 13 of 21 (61.9%) control mice. See also supplemental Figure 6.
SADBE prevents melanoma formation in NrasQ61R mice
To more stringently test whether SADBE could visually regress melanocytic nevus cells, pigmentation, and also prevent melanoma formation, we treated a cohort of 29 Dct-Cre NrasQ61R/+ mice with SADBE over dorsal trunk skin and observed their long-term melanoma incidence for up to 400 days. As shown in Figure 7E, pretreatment with SADBE fully protected against melanoma formation within the treated skin (dorsal half of the trunk) during the observation period. Importantly, six melanomas did form in the SADBE treated mice; however, all of these tumors were in untreated areas of the skin, where depigmentation was also not observed (Figure 7E, blue arrow), further confirming the localized activity of the regressive treatment. Finally, to determine whether lower doses of SADBE may be capable of minimizing toxicity while maintaining efficacy at regressing CMN cells in the Dct-NrasQ61R model, we tested the lower dose 0.5% SADBE 3 times per week for 6 total doses (2 weeks) rather than 1.5% for 3 doses. We observed similar depigmentation with the lower dose of SADBE (Figure S6B), suggesting that additional dose/schedule options should be pursued before future application of this strategy.
Discussion
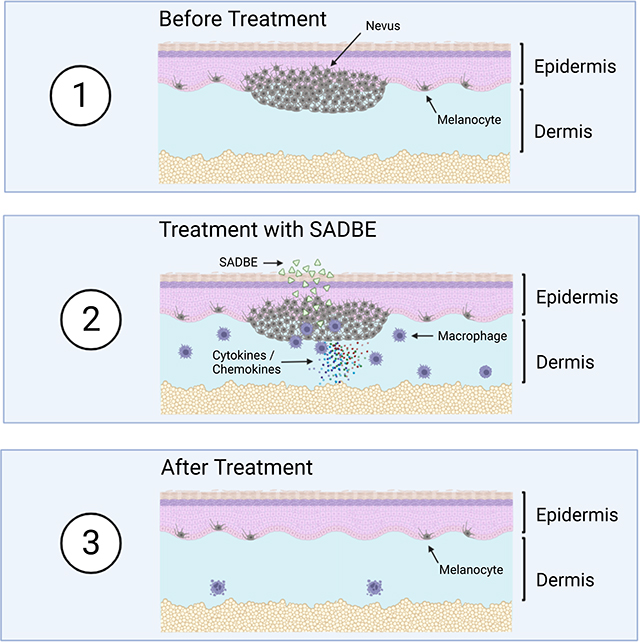
We report the development of a collection of engineered murine models for giant congenital nevi, which permit us to distinguish proliferative from senescent phases of these lesions. These preclinical models complement previously reported Nras-transgenic models (Ackermann et al., 2005; Shakhova et al., 2012) and permitted the identification of multi-pronged therapeutic approaches for non-surgical regression of the lesions and prevention of melanoma formation. These data suggest potentially valuable treatment strategies for children with giant congenital nevi morbidity which otherwise lack effective nonsurgical therapies. Along with alleviating the cosmetic and psychosocial effects of giant congenital melanocytic nevi (Koot et al., 2000; Price and Schaffer, 2010; Tromberg et al., 2005), a driving purpose of this study is to diminish the increased melanoma risk arising from these lesions, as was strongly observed following one week (3 treatments) of topical SADBE in our mouse models. It is important to note that the melanoma risk in patients with CMN is not fully confined to cutaneous locations, as deeper melanocytic populations may exist, due to differences in the developmental states in which the nevus formed (oncogenic mutation). Nonetheless, cutaneous melanoma-genesis is a significant challenge in these patients, and likely contributes importantly to current parental decisions to seek debilitating surgical resections. The plausibility of exploiting the pro-inflammatory hapten SADBE is supported by its active use in several unrelated clinical contexts (Happle et al., 1980; Micali et al., 2000), as well as anecdotal reports of vitiligo-like depigmentation in humans where the agent was used topically for other indications such as alopecia areata (Sakai et al., 2020; Valsecchi et al., 1989). While SADBE achieved melanocyte clearance, we emphasize the value in further interrogating kinase targeting agents which, despite requiring further development as local-cutaneous drugs, offer potential advantages of specific molecular targeting.
Mechanistically, melanocyte clearance utilizes recruitment of macrophages into the skin. Other studies in mice (Feng et al., 2017; Gupta et al., 2006; Zoller et al., 2004) and a human case report (Herman G et al 2004) have demonstrated macrophage recruitment by haptens suggesting that the macrophage recruitment observed here is unlikely to be particular to nevi or SADBE. Our RNA-seq data of SADBE treated mice revealed multiple macrophage cytokines, such as CCL2 (Fuentes et al., 1995; Rollins, 1991) and CCL7 (Xuan et al., 2015) as well as the monocyte recruiting chemokine CCL8 (Ji et al., 2019) and macrophage differentiating cytokine CSF-1 (Jones and Ricardo, 2013), which may contribute to macrophage recruitment to the treated area. Furthermore, the TNFα signaling pathway was found to be highly upregulated after SADBE treatment. TNFα can elevate cytotoxic activity of macrophages(Salim et al., 2016). Interestingly, TNFα is also able to affect both the growth and melanogenesis of melanocytes (Englaro et al., 1999; Hu et al., 2002). In addition, increased levels of TNFα were found in vitiligo lesions, and exposure of melanocytes to TNFα was reported to downregulate MITF-M and induce apoptosis (Singh et al., 2021). TNFα is produced predominantly by macrophages (Riches et al., 1996). Therefore, TNFα induction represents a candidate mechanism of melanocyte/nevocyte reduction either by elevating cytotoxic activity of macrophages or by inducing melanocyte apoptosis. Since our GO analysis revealed phagocytosis as an enriched pathway, this process may participate in nevus clearance. Our functional data demonstrated that macrophage depletion impeded SADBE’s efficacy at nevus regression, and we believe future studies could further clarify exactly which macrophage functions contribute to this activity. In our studies, manipulations favoring pro-inflammatory macrophages enhanced melanocyte clearance, suggesting that future strategies to modulate this macrophage population might further refine the selectivity of the treatment. Interestingly, our LISA analysis and GSEA pathway analysis revealed many transcription factors involved in immune system and interferon-α pathway activity to be highly regulated both in mouse and human CMN. This could be as a result of the senescent characteristics of the nevi which can induce the Senescence-Associated Secretory Phenotype (Coppé et al., 2010). Given clinical anecdotes of melanocyte loss after SADBE in settings independent of CMN, these findings suggest potential applications for other conditions where melanocyte clearance may be beneficial, such as cases where distinct driver oncogenes drive melanocytic neoplasia. Collectively, this study provides preclinical genetically-driven and human-engrafted congenital nevus models which show translational potential for future human clinical testing.
Limitations of the study
In this proof-of-principle study, several murine models of Congenital Giant Nevi are developed, and various treatments are tested to regress the lesions. However, there are several limitations:
While genetically matching the NRASQ61R gene driving many human CMNs, the nevi are present within mouse skin which differs structurally from human skin. For example, interfollicular dorsal skin in mice does not normally contain melanocytes, and ear skin, which does contain epidermal melanocytes in mouse, is particularly thin, lacking barrier features of human skin.
A variety of kinase-targeting agents demonstrated significant regressions of the CMN cells in the mouse CMN lesions. However, these agents have not been developed for either topical or injected cutaneous delivery. Therefore, the feasibility of delivering these agents into human skin awaits further study.
SADBE efficiently regresses CMN cells in several of the genetically engineered mouse CMN models. However, it is unknown how the relative dosing of SADBE in mouse will apply to humans, a question which requires additional evaluation.
SADBE also regressed ~90% of human CMNs within resected human CMN specimens, xenografted onto immunodeficient mice, and recruited macrophages into the CMN xenografts. However, the immunodeficient background of the xenograft recipients remains an imperfect predictor of in-vivo responses in immune-intact human skin.
RESOURCE AVALIABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Dr. David Fisher, M.D., Ph.D. (DFISHER3@mgh.harvard.edu).
Materials Availability
Mouse lines generated in this study will be distributed upon request to other investigators under a Material Transfer Agreement. All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
All the Software packages and methods used in this study have been properly detailed and referenced under the Software and algorithms listed in Key resources table. All RNA-seq data has been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-TRP2/DCT | Santa Cruz Biotechnology | Cat#sc-74439; RRID:AB_1130818 |
| Mouse monoclonal anti-SOX10 | Santa Cruz Biotechnology | Cat# sc-365692; RRID:AB_10844002 |
| Mouse monoclonal anti-MITF | Leica Biosystems | Cat# NCL-L-MITF; RRID:AB_564111 |
| Mouse monoclonal anti-Ki67 | Leica Biosystems | Cat# KI67-MM1-CE; RRID:AB_563841 |
| Rabbit polyclonal anti-S100 | DAKO | Cat# Z031129-2; RRID:AB_2315306 |
| Mouse monoclonal anti-HMB45 | DAKO | Cat# M0634; RRID:AB_2335682 |
| Rabbit monoclonal anti-Phospho-p44/42 MAPK | Cell Signaling Technology | Cat# 4370; RRID:AB_2315112 |
| Rabbit monoclonal anti-Phospho-MEK1/2 (Ser221) (166F8) | Cell Signaling Technology | Cat# 2338; RRID:AB_490903 |
| Mouse monoclonal anti-PCNA | Cell Signaling Technology | Cat# 2586; RRID:AB_2160343 |
| Rabbit monoclonal anti-CD8 | Cell Signaling Technology | Cat# 98941; RRID:AB_2756376 |
| Rabbit polyclonal anti-CD163 | Biorbyt | Cat# orb13303; RRID:AB_10749283 |
| Mouse monoclonal anti-CD161 | Novus Biologicals | Cat# NBP2-14844; RRID: N/A |
| Rabbit polyclonal anti-NRASQ61R | Abcam | Cat# ab227658; RRID: N/A |
| Mouse monoclonal anti-p16INK4A | Abcam | Cat# ab54210; RRID:AB_881819 |
| Rabbit monoclonal anti-MelanA | Abcam | Cat #ab210546; RRID:AB_2889292 |
| Hamster monoclonal anti-CD11c | Abcam | Cat# ab33483; RRID:AB_726084 |
| Rat monoclonal anti-CD3 | Abcam | Cat# ab11089; RRID:AB_2889189 |
| Rabbit monoclonal anti-CD4 | Abcam | Cat# ab183685; RRID:AB_2686917 |
| Rat monoclonal InVivoMAb anti-mouse CD4 | BioXCell | Cat# BE0003-1; RRID:AB_1107636 |
| Rat monoclonal InVivoMAb anti-mouse CD8α | BioXCell | Cat# BE0061; RRID:AB_1125541 |
| Rat monoclonal InVivoMAb rat IgG2b isotype control, anti-keyhole limpet hemocyanin | BioXCell | Cat# BE0090; RRID:AB_1107780 |
| Rat monoclonal InVivoMab anti-mouse F4/80 | BioXCell | Cat# BE0206; RRID:AB_10949019 |
| Rabbit polyclonal anti-TRP2 | Abcam | Cat# ab74073; RRID:AB_1524517 |
| Rat monoclonal anti-F4/80 | Abcam | Cat# ab16911; RRID:AB_443548 |
| Goat polyclonal Alexa Fluor 488-AffiniPure Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch Labs | Cat# 111-545-144; RRID:AB_2338052 |
| Donkey polyclonal Alexa Fluor® 594 AffiniPure Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch Labs | Cat# 711-585-152; RRID:AB_2340621 |
| Rabbit monoclonal anti-SOX10 | Abcam | Cat# ab155279; RRID:AB_2650603 |
| Mouse monoclonal anti-HMB-45 | Abcam | Cat# ab732; RRID:AB_305844 |
| Bacterial and Viral Strains | ||
| Biological samples | ||
| De-identified nevus tissue | Mass General Brigham Healthcare | IRB Protocol 2017P000992 |
| Chemicals, peptides, and recombinant proteins | ||
| 4-hydroxytamoxifen | Sigma-Aldrich | Cat#H6278; CAS: 68392-35-8 |
| Binimetinib (MEK162) | Selleckchem | Cat#S7007; CAS 606143-89-9 |
| Trametinib (GSK1120212) | Selleckchem | Cat# S2673; CAS 871700-17-3 |
| Omipalisib (GSK2126458) | Selleckchem | Cat# S2658; CAS 1086062-66-9 |
| Imatinib (STI571) | Selleckchem | Cat# S2475; CAS 152459-95-5 |
| Squaric acid dibutylester (SADBE) | Sigma-Aldrich | Cat# 339792; CAS 2892-62-8 |
| 16% Paraformaldehyde Aqueous Solution, EM Grade, Ampoule 10 ML | Electron Microscopy Sciences | Cat# 50-980-487 |
| Hematoxylin 2 | Epredia | Cat# 7231 |
| Nuclear Fast Red | Abcam | Cat #ab246831 |
| RNAlater | Life Technologies | Cat# AM7020 |
| EQUISUL-SDT® (SULFADIAZINE/TRIMETHOPRIM) | Aurora Pharmaceuticals | Cat# SC-395895Rx |
| Citrate Buffer (pH 6.0), Concentrate | Life Technologies | Cat# 005000 |
| Goat Serum | Sigma Aldrich | Cat# G9023 |
| DAPI | Sigma Aldrich | Cat# D9542; CAS:28718-90-3 |
| Fluoromount G | Southern Biotech | Cat# 0100-01 |
| Critical commercial assays | ||
| RNeasy Plus Universal Kit | Qiagen | Cat# 73404 |
| Agilent RNA 6000 Nano Kit | Agilent Technologies | Cat# G2938-90034 |
| Fontana-Masson Stain Kit (Melanin Stain) | Abcam | Cat# ab150669 |
| RNA FFPE Miniprep kit | Zymo Research | Cat# R1009 |
| Agilent RNA 6000 Pico Kit | Agilent Technologies | Cat# G2938-90046 |
| Quant-it™ RiboGreen RNA Assay Kit and RiboGreen RNA Reagent | ThermoFisher Scientific | Cat# R11490 |
| Illumina Ribo-Zero Plus rRNA Depletion Kit | Illumina | Cat# 20037135 |
| QIAamp DNA FFPE Tissue Kit | Qiagen | Cat# 56404 |
| High Sensitivity RNA ScreenTape Analysis | Agilent Technologies | Cat# 5067-5579 |
| Illumina® Stranded mRNA Prep, Ligation (96 Samples) | Ilumina | Cat# 20040534 |
| Deposited data / Publicly available data | ||
| Raw and analyzed CMN Model RNA Seq Data | This Paper | GEO: |
| Raw and analyzed DCT SADBE RNA Seq Data | This Paper | GEO: |
| LM22 (leukocyte gene signature matrix) | Stanford University CIBERSORT | Newman et al. 2015 |
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| Mouse: Lox-stop-Lox (LSL)-NrasQ61R: C57Bl/6 Nrastm1.1Nesh | Burd et al., 2014 Sourced from Drs. Christin Burd and Norman Sharpless, University of North Carolina | MGI:5645358 |
| Mouse: Dct-Cre: C57BL/6 Dcttm1(Cre)Bee | Guyonneau et al., 2004 Sourced from Dr. Friedrich Beermann, ISREC, Lausanne, Switzerland | MGI:2387116 |
| Mouse: Tyr-Cre: C57BL/6 Tg(Tyr-cre)1Lru | Delmas et al., 2003 Source from Dr. Lionel Larue, Institut Curie, Paris, France | MGI:3573939 |
| Mouse: Tyr-CreERT2: C57B1/6 Tg(Tyr-cre/ERT2)13Bos | Bosenberg et al., 2006 Sourced from Dr. Marcus Bosenberg, Yale University | MGI:3641203 |
| Mouse:SHO: Crl:SHO-PrkdcscidHrhr | Charles River Laboratories | RRID: IMSR_CRL:474 |
| Oligonucleotides | ||
| Primer DCT Forward: TTGAGAGGAGAGGAAAGGGC | Guyonneau et al., 2004 | N/A |
| Primer DCT Reverse: CACGCCATCCAAGGTCATGC | Guyonneau et al., 2004 | N/A |
| Primer CRE Reverse: CATTGCTGTCACTTGGTCGT | Guyonneau et al., 2004 | N/A |
| Primer NRAS Q61R: GCAAGAGGCCCGGCAGTACCTA | Burd et al., 2014 | N/A |
| Primer NRAS 1: AGACGCGGAGACTTGGCGAGC | Burd et al., 2014 | N/A |
| Primer NRAS 2: GCTGGATCGTCAAGGCGCTTTTCC | Burd et al., 2014 | N/A |
| Primer Tyr::CRE<ERT2> 1: CAGGGTGTTATAAGCAATCCC | Bosenberg et al., 2006 | N/A |
| Primer Tyr::CRE<ERT2> 2: CCTGGAAAATGCTTCTGTCCG | Bosenberg et al., 2006 | N/A |
| Primer Tyr::CRE Forward: GTCACTCCAGGGGTTGCTGG | Delmas et al., 2003 | N/A |
| Primer Tyr::CRE Reverse: CCGCCGCATAACCAGTGA | Delmas et al., 2003 | N/A |
| Recombinant DNA | ||
| Software and algorithms | ||
| GraphPad Random Treatment Assigner | GraphPad | https://www.graphpad.com/quickcalcs/randomize1/ |
| Zeiss Zen Image Software, Version 2.3 | Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html |
| ImageJ | National Institutes of Health (NIH) | https://imagej.nih.gov/ij/ |
| NIS Elements imaging software | Nikon Instruments Inc | https://www.microscope.healthcare.nikon.com/products/software/nis-elements |
| DESeq2 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Prism 7 | Graphpad | https://www.graphpad.com/support/prism-7-updates/ |
| DRAGEN FastQC | Illumina | https://support.illumina.com/content/dam/illumina-support/help/Illumina_DRAGEN_Bio_IT_Platform_v3_7_1000000141465/Content/SW/Informatics/Dragen/FASTQC_Intro_fDG.htm |
| Salmon v 1.8.0 | Patro et al., 2017 | https://github.com/COMBINE-lab/Salmon |
| NDP.view2 v 2.8.24 | Hamamatsu | https://www.hamamatsu.com/us/en/product/life-science-and-medical-systems/digital-slide-scanner/U12388-01.html |
| R package: fgsea | Korotkevich et al., 2021 | https://doi.org/10.1101/060012 |
| R package: clusterProfiler | Yu et al., 2012 | https://doi.org/10.1089/omi.2011.0118 |
| Other | ||
EXPERIEMTNAL MODEL AND SUBJECT DETAILS
Ethics Statement
Mouse studies and procedures were approved by the Institutional Animal Care and Use Committee of Massachusetts General Hospital and conducted strictly in accordance with the approved animal handling protocol.
Mice
Mice were assigned randomly to the different experimental groups by covariate adaptive randomization using an online calculator (www.graphpad.com/quickcalcs/randomize1/). All mice were matched by gender and age, as indicated in each experiment. All mice were group-housed, fed ProLab IsoPro RMH 300 Irradiated feed, and kept at 68–73 °F. Mice were genotyped according to the protocol obtained from their respective publications or distributers. Primers can be found in the Key Resources Table.
Generation of melanocyte-specific NrasQ61R mutant mice
Melanocyte-specific NrasQ61R expression was induced by crossing LSL-NrasQ61R mice (Burd et al., 2014) (generated by Drs. Christin Burd and Norman Sharpless, University of North Carolina) with melanocyte-targeted Cre recombinase transgenic mice containing either Dct promoter-driven Cre (Dct-Cre mice (Guyonneau et al., 2004), kindly provided by Dr. Friedrich Beermann, ISREC, Lausanne, Switzerland), Tyr promoter-driven Cre Tyr-Cre mice (Delmas et al., 2003), kindly provided by Dr. Lionel Larue, Institut Curie, Paris, France), or inducible Tyr promoter-driven CreERT2 (Tyr-CreERT2 mice) (Bosenberg et al., 2006), kindly provided by Dr. Marcus Bosenberg, Yale University). To obtain a melanocyte-specific inducible Nras mutation, two-day-old female LSL-NrasQ61R newborn mice harboring Tyr-CreERT2 were painted topically with 25 mg/ml 4-hydroxytamoxifen (Sigma-Aldrich, #H6278) dissolved in dimethyl sulfoxide (DMSO), once daily for 1 week.
METHOD DETAILS
Animal studies and analysis of melanoma incidence
We measured the incidence of NrasQ61R mutation-driven melanoma formation in both Dct-Cre NrasQ61R mutant and tamoxifen-induced Tyr-CreERT2 NrasQ61R mutant mice. These experiments were performed with > 20 mice per group (both genders, classified by ages), and their survival curves were calculated by the log-rank test at a two-sided significance level of P<0.05.
Quantification of proliferative and senescent phases
Dorsal skin samples from Dct-Cre NrasQ61R/Q61R mutant mice were harvested at the ages indicated. Samples from three mice of each age group, with five to ten non-adjacent samples per mouse, were quantified. Nevus cell proliferation and senescence were measured by immunofluorescence staining for Ki67 (Leica Biosystems, #KI67-MM1-CE) and p16Ink4a (Abcam, # ab54210), respectively and quantification of Ki67+MelanA+ double-positive proliferative nevus cells or p16+MelanA+ double-positive senescent nevus cells was calculated as a percentage of the total number of MelanA+ positive nevus cells.
Quantification of Melan A or iNOS positive cells
Ear skin samples from three mice of each study group, with three to five non-adjacent samples per mouse, were quantified. The number of MelanA-positive nevus cells (Figure 6A) or iNOS-positive cells (Figure 6B) was measured and calculated per mm2 unit area.
Treatment procedures for therapeutic agents
Selected therapeutic agents or vehicle controls were applied on NrasQ61R mutation-driven nevus lesions by either direct intralesional injection (paws) or topical application (ears and dorsal skin). Agents tested included: (1) small molecules targeting NRAS signaling pathways: binimetinib (MEK162) (Sellechkchem, #S7007), trametinib (GSK1120212) (Selleckchem, #S2673), and omipalisib (GSK2126458) (Selleckchem, #S2658); (2) the melanocyte-selective toxic agent, imatinib (STI571) (Selleckchem, #S2475); SADBE (squaric acid dibutylester) (Sigma-Aldrich, #339792). Details are provided in Table S1 For all topical hapten-based immunotherapies. Mice were sensitized with 2% SADBE in acetone which was applied to the right side of the shaved abdomen. Three days later, sensitized mice were challenged with the same drug applied to the right ear or dorsal skin. An identical quantity of acetone (without SADBE) was administered topically to the left ear or dorsal skin as control. All drug treatments in mice were performed under isoflurane inhalation anesthesia. Mice were examined for signs of toxicity and for depigmentation during each treatment. Animals were euthanized in accordance with a set schedule, and the treated lesions were collected and processed for assessment of melanocyte destruction. Each sample was compared to the vehicle control from the same mouse, and five mice per treatment group were typically used excluding 0.5% SADBE 3 times per week for 2 weeks, where 3 mice per treatment group were used (Figure S4B).
Histology, staining, and imaging
Skin samples were taken from euthanized mice, fixed in 4% paraformaldehyde (Electron Microscopy Sciences, #50–980-487) overnight at 4°C, and embedded in paraffin using standard procedures. 5 μm sections were prepared from tissue blocks and stained with hematoxylin and eosin (H&E) (Epredia, #7231) for histologic analysis. Fontana-Masson silver stain (Abcam, #ab150669) was used to detect the melanin pigments following the provided protocol with one modification by leaving the slides in Ammoniacal Silver solution for 40 minutes rather than the recommended 30 minutes for melanin, and stained sections were counterstained with Nuclear Fast Red (Abcam, #ab246831). Stained images were captured using a Hamamatsu Nanozoomer, and analyzed using NDP.view2 software (Hamamatsu).
For immunostaining, sections were subjected to heat-induced antigen retrieval and incubated with primary antibodies against TRP2/DCT (1:100) or SOX10 (1:50) (Santa Cruz Biotechnology, #sc-74439, #sc-365692); MITF (1:50) or Ki67 (1:50) (Leica Biosystems, #NCL-L-MITF, #KI67-MM1-CE); S100 (1:2000) or HMB-45 (1:100) (DAKO, #Z031129–2, #M0634); p-p44/42 MAPK (1:50), p-MEK1/2 (1:50), PCNA (1:2000), or CD8 (1:400) (Cell Signaling Technology, #4370, #2338, #2586); CD163 (1:100) (Biorbyt, #orb13303); CD161 (1:50) (Novus Biologicals, #NBP2–14844); NRASQ61R (1:100), p16INK4A (1:1000), MelanA (1:500), CD11c (1:100), CD3 (1:100), or CD4 (1:500) (Abcam, #ab227658, #ab54210, #ab210546, #ab33483, #ab33483, #ab11089, #ab183685). Stained images were captured using a Zeiss AxioObserver Z1 microscope and an AxioCam MRC digital camera for brightfield or Photometrics CoolSNAP HQ2 interline CCD camera for fluorescence, and Ziess Zen software, version 2.3 (Carl Zeiss Microscopy LLC, Thornwood, NY, USA). Immunofluorescence-stained sections were counterstained with 4’,6-diamidino-2-phenylindole (DAPI) (Sigma Aldrich, #D9542), and ImageJ software (National Institutes of Health [NIH]) (Schneider et al., 2012) was used for image processing and analysis. Skin tissue images were obtained with a Nikon SMZ1500 stereomicroscope, a Nikon DS-Ri2 microscope camera, and NIS Elements imaging software, version 4.30 (Nikon Instruments Inc., Melville, NY, USA).
Nras loss of heterozygosity
DNA from 5–10 5um sections taken from paraffin blocks, was extracted using the QIAamp DNA FFPE Tissue Kit (QIAGEN, #56404). PCR reactions were run with approximately 200ng of DNA following the established genotyping test PCR for the DCT:CRE, NRAS mice. PCR products were run on a QIAxcel to visualize the bands and determine estimated concentrations. Toe DNA samples were collected during routine genotyping of pups to use as controls.
In vivo depletion of immune cells
750 μg of the following antibodies were IP injected on day 7, 4, and 2 prior to sensitization with SADBE, and 300 μg on days 0 (sensitization with 2% SADBE), 3, 5, 7, 10: InVivoMAb anti-mouse CD4_GK1.5 clone (BioXCell, #BE0003–1), InVivoMAb anti-mouse CD8α _2.43 clone (BioXCell, #BE0061), InVivoMAb anti-rat IgG2b isotype control; anti-keyhole limpet hemocyanin_LTF-2 clone (BioXCell, #BE0090), and InVivoMAb anti-mouse F4/80_CI:A3–1 clone (BioXCell, #BE0206).
RNA-Sequencing of SADBE treated samples
Whole skin sections were harvested from treated areas of Dct-Cre NrasQ61R/+ mice 72 hours after treatment with 2% SADBE or vehicle control (acetone) and stored in RNAlater (Ambion/Life Technologies, #AM7020). Total RNA was isolated from skin using the RNeasy Plus Universal Kit (Qiagen, #73404) with disruption and homogenization using the TissueLyser II system (Qiagen). All extracted RNA samples possessed RIN > 8.0 by Agilent TapeStation System with High Sensitivity RNA Screen Tape (Agilent Technologies, #5067–55779) and proceeded for NGS library construction with Illumina Stranded mRNA Prep, Ligation kit (Illumina, #20040534). Each amplified, then bead-cleaned library was assessed for its quality and quantity by Agilent TapeStation with High Sensitivity D1000 ScreenTape and Qubit fluorometer 4.0.(Invitrogen), respectively. Each library was normalized at 4 nM, then pooled, denatured, and loaded onto the Illumina NextSeq550 at 2×74 base-pair specification. The generated FastQ files were processed under DRAGEN FastQC bioinformatic pipeline (Illumina) to assess the overall quality of the data for adapter contamination or GC content.
RNA-Sequencing mouse model comparison
FFPE blocks of tumor and control tissues(n=18) were cut at 8 um thickness and RNA was extracted with RNA FFPE Miniprep kit (Zymo Research, #R1009) following manufacturer recommendations. Extracted RNA quality was assessed by the Agilent Bioanalyzer system with Eukaryote Total RNA Pico assay (Agilent Technologies, #G2938–90046). The overall quantity was measured by Nanodrop 2000 Spectrophotometer followed by Quant-it RiboGreen RNA Assay (ThermoFisher Scientific, #R11490) to optimize inputs for NGS library construction. The maximum input of 1 ug was processed for library construction with Stranded total RNA with ligation with Ribozero plus kit (Illumina, #20037135). Each amplified then bead-cleaned library was assessed for its quality and quantity by Agilent TapeStation with High Sensitivity D1000 ScreenTapes and Qubit fluorometer 4.0. (Invitrogen, Waltham, MA), respectively. Each library was normalized at 4 nM, then pooled, denatured, and loaded onto the Illumina NextSeq550 targeting 80 million reads per sample at 1×76 base-pair specification. The generated FastQ files were processed under DRAGEN FastQC bioinformatic pipeline (Illumina) to assess the overall quality of the data for adapter contamination or GC content.
RNA-Sequencing Analysis
We performed RNAseq and differential expression analysis for in-house mouse RNAseq datasets and one publicly available human nevi cohort. Read alignment was preformed using STAR (Dobin et al., 2013), followed by transcript quantification using Salmon (Patro et al., 2017), and differential analysis using DESeq2(Love et al., 2014) that compares SADBE-treated samples and control samples. The set of differentially expressed genes were determined using absolute LogFoldChange>1 and FDR<0.01. To investigate macrophage-related genes, we obtained macrophage gene lists from LM22 (Newman et al., 2015) which includes markers for macrophage states M0, M1, and M2. We also obtained from TIDE (Jiang et al., 2018) the gene correlation scores with tumor M2 macrophage levels. For the systematic (Jiang et al., 2018) pathway analysis on differentially expressed genes, we performed gene-set enrichment analysis using R packages fgsea(Korotkevich et al., 2021) and clusterProfiler(Yu et al., 2012). The above analysis was performed on both mouse and human RNAseq datasets using corresponding genomic references in each species; specifically, for the mouse set with untreated nevi, normal, and melanoma, differential expression was carried out using protein-coding genes to facilitate pathway interpretation between nevi and normal skin samples. After obtaining differential gene lists, we applied LISA (Qin et al., 2020) to infer transcriptional regulators using public ChIP-seq profiles.
Transplantation of CMN xenografts
Discarded de-identified nevus tissue was obtained under IRB approved protocol 2017P000992 (Mass General Brigham Healthcare). Nevi were transplanted onto the back of SHO (Charles River Laboratories, MA, USA) 62-day-old female mice according to previous published work (Schiferle et al., 2021). Briefly, using a DermaBlade Shave Biopsy Instrument (AccuTec Blades Inc., #72–0001-0000) under sterile conditions, subcutaneous dermal fat was removed from the dermal side of the giant nevus to enhance vascularization and grafting efficiency. Mice were anesthetized and placed on a heating pad. The surgical site was sterilized with 70% ethanol. Nevi were transplanted onto recipient mice using surgical grade sutures and were bandaged using Vaseline gauze and elastic adhesive bandage. Mice were placed on sulfadiazine/trimethoprim antibiotics (Equisul-SDT Oral Suspension) (Aurora Pharmaceuticals, #sc-395895Rx) after transplantation. Bandages were removed 5 to 7 days after surgery and graft sutures were removed 14 days post-surgery.
SADBE treatment of xenografts
8 xenografts from one donor were implanted on 4 SHO 62-day-old female mice and 2 xenografts from an additional donor were implanted on 1 SHO mouse. For melanocyte cell count, topical application of 1% SADBE using a cotton tip was performed twice a week for 8 months on 3 of the xenografts, 5 of the xenografts from the same donor received acetone control. Treatments did not induce recognized side effects; behavior remained normal and morphologic structure of the skin remained intact with minimal surrounding erythema. For macrophage cell counts, 2 additional xenografts from a different donor were treated with 1% SADBE or acetone control for 2 weeks after which the xenografts were harvested for further F4/80 macrophage staining.
Imaging of CMN xenografts
Formalin–fixed paraffin-embedded CMN xenografts (5 μm) were labeled with the following antibodies: TRP2 antibody (1:100) (Abcam, #ab74073) and F4/80 antibody (1:100) (Abcam, #ab16911). Deparaffinization of slides was performed with 100% xylene for 2×5 min, followed by 100, 90, 70, 50 and 30% ethanol for 2 min each. Slides were washed in PBS for 5 min. Permeabilization was conducted in 0.2% triton-X for 5 min and then the slides were washed 2×3 min in 0.01% PBS-tween 20. Antigen retrieval was performed by incubating the slides with citrate buffer (pH = 6) at 95 °C for 20 minutes. Sections were washed with 0.01% PBS-tween 20 for 3×3 min and blocked for 1 hour with PBS containing 10% normal goat serum and 5% BSA. Slides were incubated overnight at 4 °C with primary antibodies diluted in blocking buffer. Sections were washed with 0.01% PBS-tween 20 for 3×3 min, followed by secondary antibody incubation [1:500; goat anti-Rabbit IgG (H+L) Alexa Fluor® 488-AffiniPure (Jackson ImmunoResearch Laboratories, #111–545-144), or 1:500 Alexa Fluor® 594 AffiniPure Donkey Anti-Rabbit IgG (H+L) (Jackson ImmunoResearch Laboratories, #711–585-152)] for 2 hours at room temperature. Slides were washed 3×3 min with 0.01% PBS-tween 20, incubated with DAPI (Sigma-Aldrich, #D9542) for 5 minutes for nuclei staining and coverslipped using Fluoromount G (Southern Biotech, #0100–01). Slides were placed at 4°C for 48 hours before imaging. Photomicrographs were obtained under either×20 or×40 objective using a Ziess Axio Photo Observer Microscope or Nikon SMZ1500 stereomicroscope. ImageJ (National Institutes of Health [NIH]) was used to identify cells based on size, circularity, nuclear presence, and intensity. Positive cells were counted in both the epidermis and dermis of each specimen for F4/80 and throughout epidermis for TRP2. In each area, the number of positive cells was counted and reported in relationship to the total number of DAPI-positive cells. Thus, results are expressed as a percentage of positive cells for each marker analyzed per total cells. Quantifications were done on multiple sections from each CMN xenograft, typically 3–20 images per sample.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed with GraphPad Prism 7 software, and the evaluations were conducted by using a Student’s t-test, one-way ANOVA with Tukey’s multiple comparison test, or log-rank test for survival analysis. A probability value of P<0.05 was considered statistically significant (*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001).
Supplementary Material
Supplemental Table S1. Therapeutic agents. Related to Figures 4–7.
Concentrations and protocols of the agents used for localized treatments.
Figure S1. Additional melanocyte-specific NrasQ61R mutation-driven congenital nevus models mimic genetic and phenotypic features of human giant congenital nevi, related to Figure 1
(A) Features of melanocytic nevi of adult Dct-Cre NrasQ61R mouse. (B-F) Features of melanocytic nevi of 3-day-old Dct-Cre NrasQ61R mouse. (A)Skin hyperpigmentation was observed in melanocyte-specific NrasQ61R mutant mice, but not in control mice. (B) Melanin from 3-day-old Dct-Cre NrasQ61R mice was detected by Fontana-Masson staining. (C-F) Immunostaining of paraffin sections. Paraffin sections were immunostained for (mutant-specific) NrasQ61R (C, dark brown), DCT (D, red in upper panel, green in lower panel), MITF (D, red in lower panel), S100 (E, green), SOX10 (E, red), or p-ERK (F, red). Red arrows in (C) indicate follicular melanin detected by bright-field microscopy, blue arrows in (C) indicate positive signals. Phosphorylated ERK (yellow arrows in F) was detectable in dermal nevus cells showing melanin deposition (white, F). (G-H) Features of melanocytic nevi of tamoxifen-induced Tyr-CreERT2 NrasQ61R mutant mice were observed (G, tail, H, ear). (I) Surviving Tyr-Cre NrasQ61R/+ heterozygous mutant mouse was compared with control littermates at 1 month of age. The mutant mouse developed hydrocephalus and had growth retardation. (J-L) Hyperpigmentation. Skin hyperpigmentation in Tyr-Cre NrasQ61R/+ mice was observed in the snout/perioral region (J), paws (K), and tail (L). (M-N) Ectopic pigmentation was compared in the parietal-temporal lobe (M, upper image) and the area of the superior and interior colliculi (N, lower image). Yellow arrows in (N) indicate ectopic melanocytes in the meninges of the mutant brain, but not in control tissue.
Figure S2. Comparison of mouse vs human giant congenital nevi, related to Figure 1
(A–D) Hematoxylin and eosin (H&E) stained histological images of human congenital nevi. (E-H)Hematoxylin and eosin (H&E) stained histological images of NrasQ61R mutation-driven murine congenital nevi. Histologic images at low magnifications (A,C and D, 200x) and high magnification (B, E, G, and H, 400x; F: 600x) are presented. Yellow arrows in (A) indicate nevus cells. Red arrow in (A) indicates nevi exhibiting perivascular distribution. Green arrow (A) indicates nevi associated with the adventitia of the hair follicle. Yellow arrows in (B) indicate nevus cells having hyperchromatic nuclei. Green arrow in (E) indicates nevus cells distributed around some adnexal structures. Yellow arrow in (F) indicates nevus cells with small hyperchromatic nuclei. Red arrow in (F) indicates spindle shaped nevus cells. Yellow arrow in (C) indicates nevus cells displaying increased cytoplasm. Green arrow in (C) indicates large nests of pigmentation. Red arrow in (C) indicates melanocytes infiltrating the reticular dermis. Yellow arrow in (G) indicates nevus cells having hyperchromatic nuclei and a round cytoplasm. Green arrow in (G) indicates a superficial aggregate of nevus cells forming a nest.
Figure S3. Loss of heterozygosity determination in Nras-driven mice melanoma, related to Figure 3
3 heterozygous Dct-Cre NrasQ61R/+ tumors and control tissue were tested using PCR primers that discriminate between the wildtype (WT), mutant (Q61R), and recombined mutant (Q61R+CRE) alleles. All three tumors retained the wildtype allele.
Figure S4. RNA-sequencing of mouse nevi and melanoma compared to normal mouse skin and human CMN, related to Figure 3.
(A) Heat map of melanocytic genes in nevi Nras mouse models. (B) Heat map of melanocytic genes in human CMN samples. (C) GSEA hallmark pathway analysis of nevi from Nras mouse models in comparison with normal skin. (D) Top 20 Lisa-predicted transcription factors based on top 200 significantly up-regulated genes from the Nevi vs normal skin comparison. (E) Heat map of melanocytic genes in melanoma Nras mouse models. (F) GSEA hallmark pathway analysis of melanoma from Nras mouse models in comparison with normal skin. (G) GSEA hallmark pathway analysis of human CMN samples in comparison with human healthy skin samples from the same cohort.
Figure S5. Kinase inhibitor treatments to induce nevus regression, related to Figure 4
The MEK inhibitor, binimetinib (A, 0.6 μg/μl), or the c-KIT inhibitor, imatinib (K, 10 μg/μl), was subcutaneously injected into the pigmented paws of tamoxifen-induced Tyr-CreERT2 NrasQ61R/Q61R mice in 10 μl of 10% DMSO three times per week for two weeks. Depigmentation was then assessed 2 weeks (A, K) and 6 weeks (A) after the final drug treatment. (B, C) Pigmented right ears of tamoxifen-induced Tyr-CreERT2 NrasQ61R mutant mice were topically treated with the MEK inhibitor, binimetinib (B, 10 μg /μl) or the PI3K inhibitor, omipalisib (C, 10 μg /μl), five times per week for 2 months; the vehicle control, DMSO, was applied at the same time points to the pigmented left ear of the same mouse. All agents were administered topically in 5 μl DMSO. Ear skin tissues were harvested 15 weeks after the final drug treatment, and nevus regression was assessed. Melanin detected by bright-field microscopy is shown (B, C, black). Five mice per each treatment group were used, and representative images are shown. (D-J) Topical combinatorial therapy with binimetinib (5 μg/μl) plus imatinib (50 μg/μl) was applied to the pigmented ears of tamoxifen-induced Tyr-CreERT2 NrasQ61R/Q61R mutant mice. Topical agents were administered in 5 μl DMSO, four times per week for 7 weeks. Shortly thereafter ear skin tissues were collected and analyzed. Photographic images of ear tissues are shown (D). Melanin is shown in images of hematoxylin and eosin (H&E) stained ear skin (E, brown). Red arrows in (E) indicate dermal melanin detected by bright-field microscopy. Ear skin sections were stained with DCT (F, red), SOX10 (G, red), CD11c (H, red), CD163 (I, red), or CD16 (J, red) antibodies. Blue arrows in (F, I, J) and yellow arrows in (G and H) indicate positive signals. Melanin detected by bright-field microscopy is shown as dark brown in (F, I, J) and white in (G, H). We used five mice per treatment group, and representative images are shown.
Figure S6. Fontana Masson of xenografts and efficacy of low dose SADBE in mice, related to Figure 7
(A) Fontana Masson staining of human xenografts treated with SADBE (1%) compared to acetone control. (B) SADBE (0.5%, topical) was applied to the pigmented right ears of tamoxifen-induced Dct-Cre NrasQ61R/+ mutant 3 times a week for 2 weeks, and the pigmented left ears of the same animals were treated with acetone vehicle. Ear tissue images were obtained with a stereomicroscope, and dark brown particles indicate melanin.
LIFE SCIENCE TABLE WITH EXAMPLES FOR AUTHOR REFERENCE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Snail | Cell Signaling Technology | Cat#3879S; RRID: AB_2255011 |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat#T9026; RRID: AB_477593 |
| Rabbit polyclonal anti-BMAL1 | This paper | N/A |
| Bacterial and virus strains | ||
| pAAV-hSyn-DIO-hM3D(Gq)-mCherry | Krashes et al., 2011 | Addgene AAV5; 44361-AAV5 |
| AAV5-EF1a-DIO-hChR2(H134R)-EYFP | Hope Center Viral Vectors Core | N/A |
| Cowpox virus Brighton Red | BEI Resources | NR-88 |
| Zika-SMGC-1, GENBANK: KX266255 | Isolated from patient (Wang et al., 2016) | N/A |
| Staphylococcus aureus | ATCC | ATCC 29213 |
| Streptococcus pyogenes: M1 serotype strain: strain SF370; M1 GAS | ATCC | ATCC 700294 |
| Biological samples | ||
| Healthy adult BA9 brain tissue | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat#UMB1455 |
| Human hippocampal brain blocks | New York Brain Bank | http://nybb.hs.columbia.edu/ |
| Patient-derived xenografts (PDX) | Children’s Oncology Group Cell Culture and Xenograft Repository | http://cogcell.org/ |
| Chemicals, peptides, and recombinant proteins | ||
| MK-2206 AKT inhibitor | Selleck Chemicals | S1078; CAS: 1032350-13-2 |
| SB-505124 | Sigma-Aldrich | S4696; CAS: 694433-59-5 (free base) |
| Picrotoxin | Sigma-Aldrich | P1675; CAS: 124-87-8 |
| Human TGF-β | R&D | 240-B; GenPept: P01137 |
| Activated S6K1 | Millipore | Cat#14-486 |
| GST-BMAL1 | Novus | Cat#H00000406-P01 |
| Critical commercial assays | ||
| EasyTag EXPRESS 35S Protein Labeling Kit | PerkinElmer | NEG772014MC |
| CaspaseGlo 3/7 | Promega | G8090 |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202-1012 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE63473 |
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Nanog STILT inference | This paper; Mendeley Data | http://dx.doi.org/10.17632/wx6s4mj7s8.2 |
| Affinity-based mass spectrometry performed with 57 genes | This paper; Mendeley Data | Table S8; http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Experimental models: Cell lines | ||
| Hamster: CHO cells | ATCC | CRL-11268 |
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Norbert Perrimon | FlyBase: FBtc0000181 |
| Human: Passage 40 H9 ES cells | MSKCC stem cell core facility | N/A |
| Human: HUES 8 hESC line (NIH approval number NIHhESC-09-0021) | HSCI iPS Core | hES Cell Line: HUES-8 |
| Experimental models: Organisms/strains | ||
| C. elegans: Strain BC4011: srl-1(s2500) II; dpy-18(e364) III; unc-46(e177)rol-3(s1040) V. | Caenorhabditis Genetics Center | WB Strain: BC4011; WormBase: WBVar00241916 |
| D. melanogaster: RNAi of Sxl: y[1] sc[*] v[1]; P{TRiP.HMS00609}attP2 | Bloomington Drosophila Stock Center | BDSC:34393; FlyBase: FBtp0064874 |
| S. cerevisiae: Strain background: W303 | ATCC | ATTC: 208353 |
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/3J | The Jackson Laboratory | JAX: 006494 |
| Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J | The Jackson Laboratory | RRID: IMSR_JAX:008471 |
| Zebrafish: Tg(Shha:GFP)t10: t10Tg | Neumann and Nuesslein-Volhard, 2000 | ZFIN: ZDB-GENO-060207-1 |
| Arabidopsis: 35S::PIF4-YFP, BZR1-CFP | Wang et al., 2012 | N/A |
| Arabidopsis: JYB1021.2: pS24(AT5G58010)::cS24:GFP(-G):NOS #1 | NASC | NASC ID: N70450 |
| Oligonucleotides | ||
| siRNA targeting sequence: PIP5K I alpha #1: ACACAGUACUCAGUUGAUA | This paper | N/A |
| Primers for XX, see Table SX | This paper | N/A |
| Primer: GFP/YFP/CFP Forward: GCACGACTTCTTCAAGTCCGCCATGCC | This paper | N/A |
| Morpholino: MO-pax2a GGTCTGCTTTGCAGTGAATATCCAT | Gene Tools | ZFIN: ZDB-MRPHLNO-061106-5 |
| ACTB (hs01060665_g1) | Life Technologies | Cat#4331182 |
| RNA sequence: hnRNPA1_ligand: UAGGGACUUAGGGUUCUCUCUAGGGACUUAGGGUUCUCUCUAGGGA | This paper | N/A |
| Recombinant DNA | ||
| pLVX-Tight-Puro (TetOn) | Clonetech | Cat#632162 |
| Plasmid: GFP-Nito | This paper | N/A |
| cDNA GH111110 | Drosophila Genomics Resource Center | DGRC:5666; FlyBase:FBcl0130415 |
| AAV2/1-hsyn-GCaMP6- WPRE | Chen et al., 2013 | N/A |
| Mouse raptor: pLKO mouse shRNA 1 raptor | Thoreen et al., 2009 | Addgene Plasmid #21339 |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| ICS algorithm | This paper; Mendeley Data | http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Other | ||
| Sequence data, analyses, and resources related to the ultra-deep sequencing of the AML31 tumor, relapse, and matched normal | This paper | http://aml31.genome.wustl.edu |
| Resource website for the AML31 publication | This paper | https://github.com/chrisamiller/aml31SuppSite |
PHYSICAL SCIENCE TABLE WITH EXAMPLES FOR AUTHOR REFERENCE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| QD605 streptavidin conjugated quantum dot | Thermo Fisher Scientific | Cat#Q10101MP |
| Platinum black | Sigma-Aldrich | Cat#205915 |
| Sodium formate BioUltra, ≥99.0% (NT) | Sigma-Aldrich | Cat#71359 |
| Chloramphenicol | Sigma-Aldrich | Cat#C0378 |
| Carbon dioxide (13C, 99%) (<2% 18O) | Cambridge Isotope Laboratories | CLM-185-5 |
| Poly(vinylidene fluoride-co-hexafluoropropylene) | Sigma-Aldrich | 427179 |
| PTFE Hydrophilic Membrane Filters, 0.22 μm, 90 mm | Scientificfilters.com/Tisch Scientific | SF13842 |
| Critical commercial assays | ||
| Folic Acid (FA) ELISA kit | Alpha Diagnostic International | Cat# 0365-0B9 |
| TMT10plex Isobaric Label Reagent Set | Thermo Fisher | A37725 |
| Surface Plasmon Resonance CM5 kit | GE Healthcare | Cat#29104988 |
| NanoBRET Target Engagement K-5 kit | Promega | Cat#N2500 |
| Deposited data | ||
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Structure of compound 5 | This paper; Cambridge Crystallographic Data Center | CCDC: 2016466 |
| Code for constraints-based modeling and analysis of autotrophic E. coli | This paper | https://gitlab.com/elad.noor/sloppy/tree/master/rubisco |
| Software and algorithms | ||
| Gaussian09 | Frish et al., 2013 | https://gaussian.com |
| Python version 2.7 | Python Software Foundation | https://www.python.org |
| ChemDraw Professional 18.0 | PerkinElmer | https://www.perkinelmer.com/category/chemdraw |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| Other | ||
| DASGIP MX4/4 Gas Mixing Module for 4 Vessels with a Mass Flow Controller | Eppendorf | Cat#76DGMX44 |
| Agilent 1200 series HPLC | Agilent Technologies | https://www.agilent.com/en/products/liquid-chromatography |
| PHI Quantera II XPS | ULVAC-PHI, Inc. | https://www.ulvac-phi.com/en/products/xps/phi-quantera-ii/ |
Highlights.
Models of melanocyte inducible NrasQ61R mimic human congenital melanocytic nevi.
Locally delivered MEK, PI3K and c-KIT inhibitors are able to regress the nevi.
SADBE regresses nevi in mice and human CMN xenografts, and prevents melanoma in mice.
SADBE induces inflammation and recruits macrophages that lead to nevus clearance.
Acknowledgements:
The authors gratefully acknowledge the original sources of key mouse strains by Dr. Lionel Larue, Institut Curie, Paris, France (Tyr promoter-driven Cre), Dr. Friedrich Beermann, Swiss Institute for Experimental Cancer Research, Epalinges, Switzerland (Dct-CRE), and Dr. Marcus Bosenberg, Yale University (Tyr-CreERT2, tamoxifen inducible). The authors gratefully acknowledge support from NIH: (D.E.F.) R01AR072304, (D.E.F.) R01AR043369; (D.E.F.&X.S.L.) P01CA163222; (D.E.F.) R01CA222871; (D.E.F.) the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation; and (J.L.C.) the MGH Summer Research Trainee Program. The Graphical Abstract was created with biorender.com.
DECLARATION OF INTERESTS: D.E.F. has a financial interest in Soltego, a company developing salt inducible kinase inhibitors for topical skin-darkening treatments that might be used for a broad set of human applications. The interests of D.E.F. were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
C.L.C. has a financial interest in 4Immune, a company developing cell therapy treatments that can be used for a broad set of human applications. The interests of C.L.C were reviewed and are managed by Mass General Brigham in accordance with their conflict of interest policies.
X.S.L. is a cofounder, board member, SAB member, and consultant of GV20 Oncotherapy and its subsidiaries; stockholder of BMY, TMO, WBA, ABT, ABBV, and JNJ; and received research funding from Takeda, Sanofi, Bristol Myers Squibb, and Novartis. M.C.M. discloses consulting relationship with Novartis, Advisory Board with BioCoz and Caliber ID, and author Royalties with Wiley & Sons.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackermann J, Frutschi M, Kaloulis K, McKee T, Trumpp A, and Beermann F (2005). Metastasizing melanoma formation caused by expression of activated N-RasQ61K on an INK4adeficient background. Cancer Res 65, 4005–4011. 10.1158/0008-5472.CAN-04-2970. [DOI] [PubMed] [Google Scholar]
- Bajaj MS, Khuraijam N, Sen S, and Pushker N (2009). Congenital melanocytoma manifesting as proptosis with multiple cutaneous melanocytic nevi and oculodermal melanosis. Arch Ophthalmol 127, 937–939. 10.1001/archophthalmol.2009.146. [DOI] [PubMed] [Google Scholar]
- Bancroft GJ, and Kelly JP (1994). Macrophage activation and innate resistance to infection in SCID mice. Immunobiology 191, 424–431. 10.1016/S0171-2985(11)80448-1. [DOI] [PubMed] [Google Scholar]
- Bauer J, Curtin JA, Pinkel D, and Bastian BC (2007). Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. J Invest Dermatol 127, 179–182. 10.1038/sj.jid.5700490. [DOI] [PubMed] [Google Scholar]
- Bett BJ (2005). Large or multiple congenital melanocytic nevi: occurrence of cutaneous melanoma in 1008 persons. J Am Acad Dermatol 52, 793–797. 10.1016/j.jaad.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Bosenberg M, Muthusamy V, Curley DP, Wang Z, Hobbs C, Nelson B, Nogueira C, Horner JW 2nd, Depinho R, and Chin L (2006). Characterization of melanocyte-specific inducible Cre recombinase transgenic mice. Genesis 44, 262–267. 10.1002/dvg.20205. [DOI] [PubMed] [Google Scholar]
- Burd CE, Liu W, Huynh MV, Waqas MA, Gillahan JE, Clark KS, Fu K, Martin BL, Jeck WR, Souroullas GP, et al. (2014). Mutation-specific RAS oncogenicity explains NRAS codon 61 selection in melanoma. Cancer Discov 4, 1418–1429. 10.1158/2159-8290.Cd-14-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchione C, Fabbricini R, Pane F, and Luciano L (2009). Vitiligo-like lesions in an adult patient treated with Imatinib mesylate. Leuk Res 33, e104–105. 10.1016/j.leukres.2008.12.023. [DOI] [PubMed] [Google Scholar]
- Charbel C, Fontaine RH, Malouf GG, Picard A, Kadlub N, El-Murr N, How-Kit A, Su X, Coulomb-L’Hermine A, Tost J, et al. (2014). NRAS mutation is the sole recurrent somatic mutation in large congenital melanocytic nevi. J Invest Dermatol 134, 1067–1074. 10.1038/jid.2013.429. Charles River. https://www.criver.com/products-services/find-model/scid-hairless-outbred-sho-mouse?region=3646. [DOI] [PubMed] [Google Scholar]
- Chien JC, Niu DM, Wang MS, Liu MT, Lirng JF, Chen SJ, and Hwang B (2010). Giant congenital melanocytic nevi in neonates: report of two cases. Pediatr Neonatol 51, 61–64. 10.1016/S1875-9572(10)60012-5. [DOI] [PubMed] [Google Scholar]
- Chipinda I, Hettick JM, and Siegel PD (2011). Haptenation: chemical reactivity and protein binding. J Allergy (Cairo) 2011, 839682. 10.1155/2011/839682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé JP, Desprez PY, Krtolica A, and Campisi J (2010). The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol 5, 99–118. 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas V, Martinozzi S, Bourgeois Y, Holzenberger M, and Larue L (2003). Cre-mediated recombination in the skin melanocyte lineage. Genesis 36, 73–80. 10.1002/gene.10197. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drukker L, Margulis A, Chaouat M, Levitzki R, Maiorenko E, and Ben Bassat H (2013). Changes of PI3K/AKT/BCL2 signaling proteins in congenital Giant Nevi: melanocytes contribute to their increased survival and integrity. J Recept Signal Transduct Res 33, 359–366. 10.3109/10799893.2013.838785. [DOI] [PubMed] [Google Scholar]
- Englaro W, Bahadoran P, Bertolotto C, Busca R, Derijard B, Livolsi A, Peyron JF, Ortonne JP, and Ballotti R (1999). Tumor necrosis factor alpha-mediated inhibition of melanogenesis is dependent on nuclear factor kappa B activation. Oncogene 18, 1553–1559. 10.1038/sj.onc.1202446. [DOI] [PubMed] [Google Scholar]
- Feng J, Yang P, Mack MR, Dryn D, Luo J, Gong X, Liu S, Oetjen LK, Zholos AV, Mei Z, et al. (2017). Sensory TRP channels contribute differentially to skin inflammation and persistent itch. Nat Commun 8, 980. 10.1038/s41467-017-01056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes ME, Durham SK, Swerdel MR, Lewin AC, Barton DS, Megill JR, Bravo R, and Lira SA (1995). Controlled recruitment of monocytes and macrophages to specific organs through transgenic expression of monocyte chemoattractant protein-1. J Immunol 155, 5769–5776. [PubMed] [Google Scholar]
- Gerami P, and Paller AS (2013). Making a mountain out of a molehill: NRAS, mosaicism, and large congenital nevi. J Invest Dermatol 133, 2127–2130. 10.1038/jid.2013.146. [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer VC, Cheong SC, Chong H, Chow J, Moss T, Abdel-Malek ZA, Marais R, Wynford-Thomas D, and Bennett DC (2006). Cellular senescence in naevi and immortalisation in melanoma: a role for p16? Br J Cancer 95, 496–505. 10.1038/sj.bjc.6603283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Freyschmidt-Paul P, Vitacolonna M, Kiessling S, Hummel S, Hildebrand D, Marhaba R, and Zoller M (2006). A chronic contact eczema impedes migration of antigen-presenting cells in alopecia areata. J Invest Dermatol 126, 1559–1573. 10.1038/sj.jid.5700328. [DOI] [PubMed] [Google Scholar]
- Guyonneau L, Murisier F, Rossier A, Moulin A, and Beermann F (2004). Melanocytes and pigmentation are affected in dopachrome tautomerase knockout mice. Mol Cell Biol 24, 3396–3403. 10.1128/mcb.24.8.3396-3403.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happle R, Kalveram KJ, Buchner U, Echternacht-Happle K, Goggelmann W, and Summer KH (1980). Contact allergy as a therapeutic tool for alopecia areata: application of squaric acid dibutylester. Dermatologica 161, 289–297. 10.1159/000250380. [DOI] [PubMed] [Google Scholar]
- Hashmi GS, Ahmed SS, and Khan S (2009). Congenital giant melanocytic nevi. Rare Tumors 1, e9. 10.4081/rt.2009.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath TJ, Price ER, Takemoto C, Badalian T, and Fisher DE (1998). MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature 391, 298–301. 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- Hu D-N, McCormick S, and Mehta S (2002). Effects of Tumor Necrosis Factor-alpha on the Growth and Melanogenesis of Human Uveal Melanocytes In Vitro. Investigative Ophthalmology & Visual Science 43, 1089–1089. [Google Scholar]
- Ibrahimi OA, Alikhan A, and Eisen DB (2012). Congenital melanocytic nevi: where are we now? Part II. Treatment options and approach to treatment. J Am Acad Dermatol 67, 515 e511–513; quiz 528–530. 10.1016/j.jaad.2012.06.022. [DOI] [PubMed] [Google Scholar]
- Ji J, Wang P, Zhou Q, Zhu L, Zhang H, Zhang Y, Zheng Z, Bhatta AK, Zhang G, and Wang X (2019). CCL8 enhances sensitivity of cutaneous squamous cell carcinoma to photodynamic therapy by recruiting M1 macrophages. Photodiagnosis Photodyn Ther 26, 235–243. 10.1016/j.pdpdt.2019.03.014. [DOI] [PubMed] [Google Scholar]
- Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, Li Z, Traugh N, Bu X, Li B, et al. (2018). Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 24, 1550–1558. 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CV, and Ricardo SD (2013). Macrophages and CSF-1: implications for development and beyond. Organogenesis 9, 249–260. 10.4161/org.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsler VA, Birley J, and Atherton DJ (2009). Great Ormond Street Hospital for Children Registry for congenital melanocytic naevi: prospective study 1988–2007. Part 1-epidemiology, phenotype and outcomes. Br J Dermatol 160, 143–150. 10.1111/j.1365-2133.2008.08849.x. [DOI] [PubMed] [Google Scholar]
- Kinsler VA, O’Hare P, Jacques T, Hargrave D, and Slater O (2017). MEK inhibition appears to improve symptom control in primary NRAS-driven CNS melanoma in children. Br J Cancer 116, 990–993. 10.1038/bjc.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsler VA, Thomas AC, Ishida M, Bulstrode NW, Loughlin S, Hing S, Chalker J, McKenzie K, Abu-Amero S, Slater O, et al. (2013). Multiple congenital melanocytic nevi and neurocutaneous melanosis are caused by postzygotic mutations in codon 61 of NRAS. J Invest Dermatol 133, 2229–2236. 10.1038/jid.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koot HM, de Waard-van der Spek F, Peer CD, Mulder PG, and Oranje AP (2000). Psychosocial sequelae in 29 children with giant congenital melanocytic naevi. Clin Exp Dermatol 25, 589–593. 10.1046/j.1365-2230.2000.00712.x. [DOI] [PubMed] [Google Scholar]
- Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov M, and Sergushichev A (2021). Fast gene set enrichment analysis.
- Krengel S, Hauschild A, and Schafer T (2006). Melanoma risk in congenital melanocytic naevi: a systematic review. Br J Dermatol 155, 1–8. 10.1111/j.1365-2133.2006.07218.x. [DOI] [PubMed] [Google Scholar]
- Leech SN, Bell H, Leonard N, Jones SL, Geurin D, McKee PH, and Lawrence CM (2004). Neonatal giant congenital nevi with proliferative nodules: a clinicopathologic study and literature review of neonatal melanoma. Arch Dermatol 140, 83–88. 10.1001/archderm.140.1.83. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins da Silva V, Martinez-Barrios E, Tell-Marti G, Dabad M, Carrera C, Aguilera P, Brualla D, Esteve-Codina A, Vicente A, Puig S, et al. (2019). Genetic Abnormalities in Large to Giant Congenital Nevi: Beyond NRAS Mutations. J Invest Dermatol 139, 900–908. 10.1016/j.jid.2018.07.045. [DOI] [PubMed] [Google Scholar]
- Micali G, Nasca MR, Tedeschi A, Dall’Oglio F, and Pulvirenti N (2000). Use of squaric acid dibutylester (SADBE) for cutaneous warts in children. Pediatr Dermatol 17, 315–318. 10.1046/j.1525-1470.2000.01762.x. [DOI] [PubMed] [Google Scholar]
- Mir A, Agim NG, Kane AA, Josephs SC, Park JY, and Ludwig K (2019). Giant Congenital Melanocytic Nevus Treated With Trametinib. Pediatrics 143. 10.1542/peds.2018-2469. [DOI] [PubMed] [Google Scholar]
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, and Alizadeh AA (2015). Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12, 453–457. 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura EK, Jordan SA, Oshima H, Yoshida H, Osawa M, Moriyama M, Jackson IJ, Barrandon Y, Miyachi Y, and Nishikawa S (2002). Dominant role of the niche in melanocyte stem-cell fate determination. Nature 416, 854–860. 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419. 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlikowski JS, Brock C, Chen SC, Al-Olabi L, Nixon C, McGregor F, Paine S, Chanudet E, Lambie W, Holmes WM, et al. (2015). Acute Inhibition of MEK Suppresses Congenital Melanocytic Nevus Syndrome in a Murine Model Driven by Activated NRAS and Wnt Signaling. J Invest Dermatol 135, 2902. 10.1038/jid.2015.230. [DOI] [PubMed] [Google Scholar]
- Pedersen M, Kusters-Vandevelde HVN, Viros A, Groenen P, Sanchez-Laorden B, Gilhuis JH, van Engen-van Grunsven IA, Renier W, Schieving J, Niculescu-Duvaz I, et al. (2013). Primary melanoma of the CNS in children is driven by congenital expression of oncogenic NRAS in melanocytes. Cancer Discov 3, 458–469. 10.1158/2159-8290.CD-12-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadke PA, Rakheja D, Le LP, Selim MA, Kapur P, Davis A, Mihm MC Jr., and Hoang MP (2011). Proliferative nodules arising within congenital melanocytic nevi: a histologic, immunohistochemical, and molecular analyses of 43 cases. Am J Surg Pathol 35, 656–669. 10.1097/PAS.0b013e31821375ea. [DOI] [PubMed] [Google Scholar]
- Price HN (2016). Congenital melanocytic nevi: update in genetics and management. Curr Opin Pediatr 28, 476–482. 10.1097/mop.0000000000000384. [DOI] [PubMed] [Google Scholar]
- Price HN, and Schaffer JV (2010). Congenital melanocytic nevi-when to worry and how to treat: Facts and controversies. Clin Dermatol 28, 293–302. 10.1016/j.clindermatol.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Qin Q, Fan J, Zheng R, Wan C, Mei S, Wu Q, Sun H, Brown M, Zhang J, Meyer CA, and Liu XS (2020). Lisa: inferring transcriptional regulators through integrative modeling of public chromatin accessibility and ChIP-seq data. Genome Biol 21, 32. 10.1186/s13059-020-1934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riches DW, Chan ED, and Winston BW (1996). TNF-alpha-induced regulation and signalling in macrophages. Immunobiology 195, 477–490. 10.1016/s0171-2985(96)80017-9. [DOI] [PubMed] [Google Scholar]
- Rollins BJ (1991). JE/MCP-1: an early-response gene encodes a monocyte-specific cytokine. Cancer Cells 3, 517–524. [PubMed] [Google Scholar]
- Rouille T, Aractingi S, Kadlub N, Fraitag S, How-Kit A, Daunay A, Hivelin M, Moguelet P, Picard A, Fontaine RH, and Guegan S (2019). Local Inhibition of MEK/Akt Prevents Cellular Growth in Human Congenital Melanocytic Nevi. J Invest Dermatol 139, 2004–2015 e2013. 10.1016/j.jid.2019.03.1156. [DOI] [PubMed] [Google Scholar]
- Sakai K, Fukushima S, Mizuhashi S, Jinnin M, Makino T, Inoue Y, and Ihn H (2020). Effect of topical immunotherapy with squaric acid dibutylester for alopecia areata in Japanese patients. Allergol Int 69, 274–278. 10.1016/j.alit.2019.10.008. [DOI] [PubMed] [Google Scholar]
- Salim T, Sershen CL, and May EE (2016). Investigating the Role of TNF-alpha and IFN-gamma Activation on the Dynamics of iNOS Gene Expression in LPS Stimulated Macrophages. PLoS One 11, e0153289. 10.1371/journal.pone.0153289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiferle EB, Cheon SY, Ham S, Son HG, Messerschmidt JL, Lawrence DP, Cohen JV, Flaherty KT, Moon JJ, Lian CG, et al. (2021). Rejection of benign melanocytic nevi by nevus-resident CD4(+) T cells. Sci Adv 7. 10.1126/sciadv.abg4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah KN (2010). The risk of melanoma and neurocutaneous melanosis associated with congenital melanocytic nevi. Semin Cutan Med Surg 29, 159–164. 10.1016/j.sder.2010.06.007. [DOI] [PubMed] [Google Scholar]
- O., Zingg D, Schaefer SM, Hari L, Civenni G, Blunschi J, Claudinot S, Okoniewski M, Beermann F, Mihic-Probst D, et al. (2012). Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nat Cell Biol 14, 882–890. 10.1038/ncb2535. [DOI] [PubMed] [Google Scholar]
- Silverberg NB, Lim JK, Paller AS, and Mancini AJ (2000). Squaric acid immunotherapy for warts in children. J Am Acad Dermatol 42, 803–808. 10.1067/mjd.2000.103631. [DOI] [PubMed] [Google Scholar]
- Singh M, Mansuri MS, Kadam A, Palit SP, Dwivedi M, Laddha NC, and Begum R (2021). Tumor Necrosis Factor-alpha affects melanocyte survival and melanin synthesis via multiple pathways in vitiligo. Cytokine 140, 155432. 10.1016/j.cyto.2021.155432. [DOI] [PubMed] [Google Scholar]
- Tran SL, Haferkamp S, Scurr LL, Gowrishankar K, Becker TM, Desilva C, Thompson JF, Scolyer RA, Kefford RF, and Rizos H (2012). Absence of distinguishing senescence traits in human melanocytic nevi. J Invest Dermatol 132, 2226–2234. 10.1038/jid.2012.126. [DOI] [PubMed] [Google Scholar]
- Tromberg J, Bauer B, Benvenuto-Andrade C, and Marghoob AA (2005). Congenital melanocytic nevi needing treatment. Dermatol Ther 18, 136–150. 10.1111/j.1529-8019.2005.05012.x. [DOI] [PubMed] [Google Scholar]
- Tsao AS, Kantarjian H, Cortes J, O’Brien S, and Talpaz M (2003). Imatinib mesylate causes hypopigmentation in the skin. Cancer 98, 2483–2487. 10.1002/cncr.11812. [DOI] [PubMed] [Google Scholar]
- Valsecchi R, Pansera B, Rossi A, and Cainelli T (1989). [Pigmentation abnormalities in the course of topical immunotherapy of alopecia areata]. G Ital Dermatol Venereol 124, 31–32. [PubMed] [Google Scholar]
- Viana AC, Gontijo B, and Bittencourt FV (2013). Giant congenital melanocytic nevus. An Bras Dermatol 88, 863–878. 10.1590/abd1806-4841.20132233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocanson M, Hennino A, Rozieres A, Poyet G, and Nicolas JF (2009). Effector and regulatory mechanisms in allergic contact dermatitis. Allergy 64, 1699–1714. 10.1111/j.1398-9995.2009.02082.x. [DOI] [PubMed] [Google Scholar]
- Vourc’h-Jourdain M, Martin L, Barbarot S, and aRed (2013). Large congenital melanocytic nevi: therapeutic management and melanoma risk: a systematic review. J Am Acad Dermatol 68, 493–498 e491–414. 10.1016/j.jaad.2012.09.039. [DOI] [PubMed] [Google Scholar]
- Wehrle-Haller B (2003). The role of Kit-ligand in melanocyte development and epidermal homeostasis. Pigment Cell Res 16, 287–296. [DOI] [PubMed] [Google Scholar]
- Wu M, Hemesath TJ, Takemoto CM, Horstmann MA, Wells AG, Price ER, Fisher DZ, and Fisher DE (2000). c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev 14, 301–312. [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Qu Q, Zheng B, Xiong S, and Fan GH (2015). The chemotaxis of M1 and M2 macrophages is regulated by different chemokines. J Leukoc Biol 97, 61–69. 10.1189/jlb.1A0314-170R. [DOI] [PubMed] [Google Scholar]
- Yang X, Liang R, Liu C, Liu JA, Cheung MPL, Liu X, Man OY, Guan XY, Lung HL, and Cheung M (2019). SOX9 is a dose-dependent metastatic fate determinant in melanoma. J Exp Clin Cancer Res 38, 17. 10.1186/s13046-018-0998-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Kunisada T, Grimm T, Nishimura EK, Nishioka E, and Nishikawa SI (2001). Review: melanocyte migration and survival controlled by SCF/c-kit expression. J Investig Dermatol Symp Proc 6, 1–5. 10.1046/j.0022-202x.2001.00006.x. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287. 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaal LH, Mooi WJ, Klip H, and van der Horst CM (2005). Risk of malignant transformation of congenital melanocytic nevi: a retrospective nationwide study from The Netherlands. Plast Reconstr Surg 116, 1902–1909. 10.1097/01.prs.0000189205.85968.12. [DOI] [PubMed] [Google Scholar]
- Zoller M, Freyschmidt-Paul P, Vitacolonna M, McElwee KJ, Hummel S, and Hoffmann R (2004). Chronic delayed-type hypersensitivity reaction as a means to treat alopecia areata. Clin Exp Immunol 135, 398–408. 10.1111/j.1365-2249.2003.02380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1. Therapeutic agents. Related to Figures 4–7.
Concentrations and protocols of the agents used for localized treatments.
Figure S1. Additional melanocyte-specific NrasQ61R mutation-driven congenital nevus models mimic genetic and phenotypic features of human giant congenital nevi, related to Figure 1
(A) Features of melanocytic nevi of adult Dct-Cre NrasQ61R mouse. (B-F) Features of melanocytic nevi of 3-day-old Dct-Cre NrasQ61R mouse. (A)Skin hyperpigmentation was observed in melanocyte-specific NrasQ61R mutant mice, but not in control mice. (B) Melanin from 3-day-old Dct-Cre NrasQ61R mice was detected by Fontana-Masson staining. (C-F) Immunostaining of paraffin sections. Paraffin sections were immunostained for (mutant-specific) NrasQ61R (C, dark brown), DCT (D, red in upper panel, green in lower panel), MITF (D, red in lower panel), S100 (E, green), SOX10 (E, red), or p-ERK (F, red). Red arrows in (C) indicate follicular melanin detected by bright-field microscopy, blue arrows in (C) indicate positive signals. Phosphorylated ERK (yellow arrows in F) was detectable in dermal nevus cells showing melanin deposition (white, F). (G-H) Features of melanocytic nevi of tamoxifen-induced Tyr-CreERT2 NrasQ61R mutant mice were observed (G, tail, H, ear). (I) Surviving Tyr-Cre NrasQ61R/+ heterozygous mutant mouse was compared with control littermates at 1 month of age. The mutant mouse developed hydrocephalus and had growth retardation. (J-L) Hyperpigmentation. Skin hyperpigmentation in Tyr-Cre NrasQ61R/+ mice was observed in the snout/perioral region (J), paws (K), and tail (L). (M-N) Ectopic pigmentation was compared in the parietal-temporal lobe (M, upper image) and the area of the superior and interior colliculi (N, lower image). Yellow arrows in (N) indicate ectopic melanocytes in the meninges of the mutant brain, but not in control tissue.
Figure S2. Comparison of mouse vs human giant congenital nevi, related to Figure 1
(A–D) Hematoxylin and eosin (H&E) stained histological images of human congenital nevi. (E-H)Hematoxylin and eosin (H&E) stained histological images of NrasQ61R mutation-driven murine congenital nevi. Histologic images at low magnifications (A,C and D, 200x) and high magnification (B, E, G, and H, 400x; F: 600x) are presented. Yellow arrows in (A) indicate nevus cells. Red arrow in (A) indicates nevi exhibiting perivascular distribution. Green arrow (A) indicates nevi associated with the adventitia of the hair follicle. Yellow arrows in (B) indicate nevus cells having hyperchromatic nuclei. Green arrow in (E) indicates nevus cells distributed around some adnexal structures. Yellow arrow in (F) indicates nevus cells with small hyperchromatic nuclei. Red arrow in (F) indicates spindle shaped nevus cells. Yellow arrow in (C) indicates nevus cells displaying increased cytoplasm. Green arrow in (C) indicates large nests of pigmentation. Red arrow in (C) indicates melanocytes infiltrating the reticular dermis. Yellow arrow in (G) indicates nevus cells having hyperchromatic nuclei and a round cytoplasm. Green arrow in (G) indicates a superficial aggregate of nevus cells forming a nest.
Figure S3. Loss of heterozygosity determination in Nras-driven mice melanoma, related to Figure 3
3 heterozygous Dct-Cre NrasQ61R/+ tumors and control tissue were tested using PCR primers that discriminate between the wildtype (WT), mutant (Q61R), and recombined mutant (Q61R+CRE) alleles. All three tumors retained the wildtype allele.
Figure S4. RNA-sequencing of mouse nevi and melanoma compared to normal mouse skin and human CMN, related to Figure 3.
(A) Heat map of melanocytic genes in nevi Nras mouse models. (B) Heat map of melanocytic genes in human CMN samples. (C) GSEA hallmark pathway analysis of nevi from Nras mouse models in comparison with normal skin. (D) Top 20 Lisa-predicted transcription factors based on top 200 significantly up-regulated genes from the Nevi vs normal skin comparison. (E) Heat map of melanocytic genes in melanoma Nras mouse models. (F) GSEA hallmark pathway analysis of melanoma from Nras mouse models in comparison with normal skin. (G) GSEA hallmark pathway analysis of human CMN samples in comparison with human healthy skin samples from the same cohort.
Figure S5. Kinase inhibitor treatments to induce nevus regression, related to Figure 4
The MEK inhibitor, binimetinib (A, 0.6 μg/μl), or the c-KIT inhibitor, imatinib (K, 10 μg/μl), was subcutaneously injected into the pigmented paws of tamoxifen-induced Tyr-CreERT2 NrasQ61R/Q61R mice in 10 μl of 10% DMSO three times per week for two weeks. Depigmentation was then assessed 2 weeks (A, K) and 6 weeks (A) after the final drug treatment. (B, C) Pigmented right ears of tamoxifen-induced Tyr-CreERT2 NrasQ61R mutant mice were topically treated with the MEK inhibitor, binimetinib (B, 10 μg /μl) or the PI3K inhibitor, omipalisib (C, 10 μg /μl), five times per week for 2 months; the vehicle control, DMSO, was applied at the same time points to the pigmented left ear of the same mouse. All agents were administered topically in 5 μl DMSO. Ear skin tissues were harvested 15 weeks after the final drug treatment, and nevus regression was assessed. Melanin detected by bright-field microscopy is shown (B, C, black). Five mice per each treatment group were used, and representative images are shown. (D-J) Topical combinatorial therapy with binimetinib (5 μg/μl) plus imatinib (50 μg/μl) was applied to the pigmented ears of tamoxifen-induced Tyr-CreERT2 NrasQ61R/Q61R mutant mice. Topical agents were administered in 5 μl DMSO, four times per week for 7 weeks. Shortly thereafter ear skin tissues were collected and analyzed. Photographic images of ear tissues are shown (D). Melanin is shown in images of hematoxylin and eosin (H&E) stained ear skin (E, brown). Red arrows in (E) indicate dermal melanin detected by bright-field microscopy. Ear skin sections were stained with DCT (F, red), SOX10 (G, red), CD11c (H, red), CD163 (I, red), or CD16 (J, red) antibodies. Blue arrows in (F, I, J) and yellow arrows in (G and H) indicate positive signals. Melanin detected by bright-field microscopy is shown as dark brown in (F, I, J) and white in (G, H). We used five mice per treatment group, and representative images are shown.
Figure S6. Fontana Masson of xenografts and efficacy of low dose SADBE in mice, related to Figure 7
(A) Fontana Masson staining of human xenografts treated with SADBE (1%) compared to acetone control. (B) SADBE (0.5%, topical) was applied to the pigmented right ears of tamoxifen-induced Dct-Cre NrasQ61R/+ mutant 3 times a week for 2 weeks, and the pigmented left ears of the same animals were treated with acetone vehicle. Ear tissue images were obtained with a stereomicroscope, and dark brown particles indicate melanin.
Data Availability Statement
All the Software packages and methods used in this study have been properly detailed and referenced under the Software and algorithms listed in Key resources table. All RNA-seq data has been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-TRP2/DCT | Santa Cruz Biotechnology | Cat#sc-74439; RRID:AB_1130818 |
| Mouse monoclonal anti-SOX10 | Santa Cruz Biotechnology | Cat# sc-365692; RRID:AB_10844002 |
| Mouse monoclonal anti-MITF | Leica Biosystems | Cat# NCL-L-MITF; RRID:AB_564111 |
| Mouse monoclonal anti-Ki67 | Leica Biosystems | Cat# KI67-MM1-CE; RRID:AB_563841 |
| Rabbit polyclonal anti-S100 | DAKO | Cat# Z031129-2; RRID:AB_2315306 |
| Mouse monoclonal anti-HMB45 | DAKO | Cat# M0634; RRID:AB_2335682 |
| Rabbit monoclonal anti-Phospho-p44/42 MAPK | Cell Signaling Technology | Cat# 4370; RRID:AB_2315112 |
| Rabbit monoclonal anti-Phospho-MEK1/2 (Ser221) (166F8) | Cell Signaling Technology | Cat# 2338; RRID:AB_490903 |
| Mouse monoclonal anti-PCNA | Cell Signaling Technology | Cat# 2586; RRID:AB_2160343 |
| Rabbit monoclonal anti-CD8 | Cell Signaling Technology | Cat# 98941; RRID:AB_2756376 |
| Rabbit polyclonal anti-CD163 | Biorbyt | Cat# orb13303; RRID:AB_10749283 |
| Mouse monoclonal anti-CD161 | Novus Biologicals | Cat# NBP2-14844; RRID: N/A |
| Rabbit polyclonal anti-NRASQ61R | Abcam | Cat# ab227658; RRID: N/A |
| Mouse monoclonal anti-p16INK4A | Abcam | Cat# ab54210; RRID:AB_881819 |
| Rabbit monoclonal anti-MelanA | Abcam | Cat #ab210546; RRID:AB_2889292 |
| Hamster monoclonal anti-CD11c | Abcam | Cat# ab33483; RRID:AB_726084 |
| Rat monoclonal anti-CD3 | Abcam | Cat# ab11089; RRID:AB_2889189 |
| Rabbit monoclonal anti-CD4 | Abcam | Cat# ab183685; RRID:AB_2686917 |
| Rat monoclonal InVivoMAb anti-mouse CD4 | BioXCell | Cat# BE0003-1; RRID:AB_1107636 |
| Rat monoclonal InVivoMAb anti-mouse CD8α | BioXCell | Cat# BE0061; RRID:AB_1125541 |
| Rat monoclonal InVivoMAb rat IgG2b isotype control, anti-keyhole limpet hemocyanin | BioXCell | Cat# BE0090; RRID:AB_1107780 |
| Rat monoclonal InVivoMab anti-mouse F4/80 | BioXCell | Cat# BE0206; RRID:AB_10949019 |
| Rabbit polyclonal anti-TRP2 | Abcam | Cat# ab74073; RRID:AB_1524517 |
| Rat monoclonal anti-F4/80 | Abcam | Cat# ab16911; RRID:AB_443548 |
| Goat polyclonal Alexa Fluor 488-AffiniPure Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch Labs | Cat# 111-545-144; RRID:AB_2338052 |
| Donkey polyclonal Alexa Fluor® 594 AffiniPure Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch Labs | Cat# 711-585-152; RRID:AB_2340621 |
| Rabbit monoclonal anti-SOX10 | Abcam | Cat# ab155279; RRID:AB_2650603 |
| Mouse monoclonal anti-HMB-45 | Abcam | Cat# ab732; RRID:AB_305844 |
| Bacterial and Viral Strains | ||
| Biological samples | ||
| De-identified nevus tissue | Mass General Brigham Healthcare | IRB Protocol 2017P000992 |
| Chemicals, peptides, and recombinant proteins | ||
| 4-hydroxytamoxifen | Sigma-Aldrich | Cat#H6278; CAS: 68392-35-8 |
| Binimetinib (MEK162) | Selleckchem | Cat#S7007; CAS 606143-89-9 |
| Trametinib (GSK1120212) | Selleckchem | Cat# S2673; CAS 871700-17-3 |
| Omipalisib (GSK2126458) | Selleckchem | Cat# S2658; CAS 1086062-66-9 |
| Imatinib (STI571) | Selleckchem | Cat# S2475; CAS 152459-95-5 |
| Squaric acid dibutylester (SADBE) | Sigma-Aldrich | Cat# 339792; CAS 2892-62-8 |
| 16% Paraformaldehyde Aqueous Solution, EM Grade, Ampoule 10 ML | Electron Microscopy Sciences | Cat# 50-980-487 |
| Hematoxylin 2 | Epredia | Cat# 7231 |
| Nuclear Fast Red | Abcam | Cat #ab246831 |
| RNAlater | Life Technologies | Cat# AM7020 |
| EQUISUL-SDT® (SULFADIAZINE/TRIMETHOPRIM) | Aurora Pharmaceuticals | Cat# SC-395895Rx |
| Citrate Buffer (pH 6.0), Concentrate | Life Technologies | Cat# 005000 |
| Goat Serum | Sigma Aldrich | Cat# G9023 |
| DAPI | Sigma Aldrich | Cat# D9542; CAS:28718-90-3 |
| Fluoromount G | Southern Biotech | Cat# 0100-01 |
| Critical commercial assays | ||
| RNeasy Plus Universal Kit | Qiagen | Cat# 73404 |
| Agilent RNA 6000 Nano Kit | Agilent Technologies | Cat# G2938-90034 |
| Fontana-Masson Stain Kit (Melanin Stain) | Abcam | Cat# ab150669 |
| RNA FFPE Miniprep kit | Zymo Research | Cat# R1009 |
| Agilent RNA 6000 Pico Kit | Agilent Technologies | Cat# G2938-90046 |
| Quant-it™ RiboGreen RNA Assay Kit and RiboGreen RNA Reagent | ThermoFisher Scientific | Cat# R11490 |
| Illumina Ribo-Zero Plus rRNA Depletion Kit | Illumina | Cat# 20037135 |
| QIAamp DNA FFPE Tissue Kit | Qiagen | Cat# 56404 |
| High Sensitivity RNA ScreenTape Analysis | Agilent Technologies | Cat# 5067-5579 |
| Illumina® Stranded mRNA Prep, Ligation (96 Samples) | Ilumina | Cat# 20040534 |
| Deposited data / Publicly available data | ||
| Raw and analyzed CMN Model RNA Seq Data | This Paper | GEO: |
| Raw and analyzed DCT SADBE RNA Seq Data | This Paper | GEO: |
| LM22 (leukocyte gene signature matrix) | Stanford University CIBERSORT | Newman et al. 2015 |
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| Mouse: Lox-stop-Lox (LSL)-NrasQ61R: C57Bl/6 Nrastm1.1Nesh | Burd et al., 2014 Sourced from Drs. Christin Burd and Norman Sharpless, University of North Carolina | MGI:5645358 |
| Mouse: Dct-Cre: C57BL/6 Dcttm1(Cre)Bee | Guyonneau et al., 2004 Sourced from Dr. Friedrich Beermann, ISREC, Lausanne, Switzerland | MGI:2387116 |
| Mouse: Tyr-Cre: C57BL/6 Tg(Tyr-cre)1Lru | Delmas et al., 2003 Source from Dr. Lionel Larue, Institut Curie, Paris, France | MGI:3573939 |
| Mouse: Tyr-CreERT2: C57B1/6 Tg(Tyr-cre/ERT2)13Bos | Bosenberg et al., 2006 Sourced from Dr. Marcus Bosenberg, Yale University | MGI:3641203 |
| Mouse:SHO: Crl:SHO-PrkdcscidHrhr | Charles River Laboratories | RRID: IMSR_CRL:474 |
| Oligonucleotides | ||
| Primer DCT Forward: TTGAGAGGAGAGGAAAGGGC | Guyonneau et al., 2004 | N/A |
| Primer DCT Reverse: CACGCCATCCAAGGTCATGC | Guyonneau et al., 2004 | N/A |
| Primer CRE Reverse: CATTGCTGTCACTTGGTCGT | Guyonneau et al., 2004 | N/A |
| Primer NRAS Q61R: GCAAGAGGCCCGGCAGTACCTA | Burd et al., 2014 | N/A |
| Primer NRAS 1: AGACGCGGAGACTTGGCGAGC | Burd et al., 2014 | N/A |
| Primer NRAS 2: GCTGGATCGTCAAGGCGCTTTTCC | Burd et al., 2014 | N/A |
| Primer Tyr::CRE<ERT2> 1: CAGGGTGTTATAAGCAATCCC | Bosenberg et al., 2006 | N/A |
| Primer Tyr::CRE<ERT2> 2: CCTGGAAAATGCTTCTGTCCG | Bosenberg et al., 2006 | N/A |
| Primer Tyr::CRE Forward: GTCACTCCAGGGGTTGCTGG | Delmas et al., 2003 | N/A |
| Primer Tyr::CRE Reverse: CCGCCGCATAACCAGTGA | Delmas et al., 2003 | N/A |
| Recombinant DNA | ||
| Software and algorithms | ||
| GraphPad Random Treatment Assigner | GraphPad | https://www.graphpad.com/quickcalcs/randomize1/ |
| Zeiss Zen Image Software, Version 2.3 | Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html |
| ImageJ | National Institutes of Health (NIH) | https://imagej.nih.gov/ij/ |
| NIS Elements imaging software | Nikon Instruments Inc | https://www.microscope.healthcare.nikon.com/products/software/nis-elements |
| DESeq2 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Prism 7 | Graphpad | https://www.graphpad.com/support/prism-7-updates/ |
| DRAGEN FastQC | Illumina | https://support.illumina.com/content/dam/illumina-support/help/Illumina_DRAGEN_Bio_IT_Platform_v3_7_1000000141465/Content/SW/Informatics/Dragen/FASTQC_Intro_fDG.htm |
| Salmon v 1.8.0 | Patro et al., 2017 | https://github.com/COMBINE-lab/Salmon |
| NDP.view2 v 2.8.24 | Hamamatsu | https://www.hamamatsu.com/us/en/product/life-science-and-medical-systems/digital-slide-scanner/U12388-01.html |
| R package: fgsea | Korotkevich et al., 2021 | https://doi.org/10.1101/060012 |
| R package: clusterProfiler | Yu et al., 2012 | https://doi.org/10.1089/omi.2011.0118 |
| Other | ||