ABSTRACT
Antimicrobial resistance (AMR) represents a significant source of morbidity and mortality worldwide, with expectations that AMR-associated consequences will continue to worsen throughout the coming decades. Since resistance to antibiotics is encoded in the microbiome, interventions aimed at altering the taxonomic composition of the gut might allow us to prophylactically engineer microbiomes that harbor fewer antibiotic resistant genes (ARGs). Diet is one method of intervention, and yet little is known about the association between diet and antimicrobial resistance. To address this knowledge gap, we examined diet using the food frequency questionnaire (FFQ; habitual diet) and 24-h dietary recalls (Automated Self-Administered 24-h [ASA24®] tool) coupled with an analysis of the microbiome using shotgun metagenome sequencing in 290 healthy adult participants of the United States Department of Agriculture (USDA) Nutritional Phenotyping Study. We found that aminoglycosides were the most abundant and prevalent mechanism of AMR in these healthy adults and that aminoglycoside-O-phosphotransferases (aph3-dprime) correlated negatively with total calories and soluble fiber intake. Individuals in the lowest quartile of ARGs (low-ARG) consumed significantly more fiber in their diets than medium- and high-ARG individuals, which was concomitant with increased abundances of obligate anaerobes, especially from the family Clostridiaceae, in their gut microbiota. Finally, we applied machine learning to examine 387 dietary, physiological, and lifestyle features for associations with antimicrobial resistance, finding that increased phylogenetic diversity of diet was associated with low-ARG individuals. These data suggest diet may be a potential method for reducing the burden of AMR.
KEYWORDS: antibiotic resistance, bioinformatics, diet, diversity, fiber, food, gut microbes, human health, machine learning, metagenomes, microbiome, nutrition
INTRODUCTION
Antimicrobial resistance (AMR) represents a serious global health threat to humans and animals, with many reports predicting AMR as a major cause of death worldwide by 2050 (1). Contributing to the growing epidemic are the overuse of antibiotics in farming practices and unnecessary clinical prescription, such as in the treatment for sore throat due to viral infection (2–4). Additional factors, ranging from poor sanitation to the inadequate adherence of antibiotic treatment regimens to climate change, likely also play a role (5–7). A lack of investment into new antibiotic therapies has compounded the AMR problem; the World Health Organization acknowledges that the current clinical pipeline for novel antimicrobial therapeutics remains inadequate to overcome AMR (8). Consequently, researchers estimated that in the year 2019, there were 4.95 million (95% uncertainty interval, 3.62 to 6.57) deaths worldwide in which AMR was likely an associated factor (9).
In humans, antimicrobial resistance is harbored in the microbiome, where microbes carry genetically encoded strategies to survive contact with antibiotics. The collection of these genes is known as the human resistome. The human resistome varies, in part, as a function of lifestyle. For example, individuals living in rural societies (i.e., nonindustrialized societies) with no access to modern medicine (10, 11), and even ancient humans (12), show evidence of abundant and diverse reservoirs of antibiotic resistant genes (ARGs) within fecal metagenomes even though these populations were not exposed to modern antibiotics. Regardless, the composition and diversity of ARGs in industrialized populations are significantly different from nonindustrialized populations (10). One explanation for differences in antimicrobial resistance profiles could be differences in diet. The dietary changes that occurred in tandem with industrialization may have resulted in alterations to the microbiome, expanding the niche for antibiotic-resistant bacteria (13, 14). For example, low dietary fiber intake (a hallmark of diets in industrialized countries) reduces the substrate availability for microbes that convert fiber to short-chain fatty acids (SCFAs) and changes the compositions of the microbiome. Additionally, while ARGs are abundant within taxa in both the Firmicutes and Proteobacteria phyla (these phyla have undergone recent name changes to Bacillota and Pseudomonadota, respectively), proteobacteria are predominantly Gram-negative, facultative anaerobes that are sensitive to low pH. Therefore, this group is likely to be selected against in the presence of a higher fiber diet.
In a prior study with dairy cows, we demonstrated that the colostrum consumed by the calves was the most likely source of antimicrobial resistance genes and that these ARGs could then be suppressed by increasing fiber in the diet (15). This conclusion suggests that the diet can be both a source of AMR and a solution to reduce AMR. However, there have been no studies to fully characterize AMR in humans and its relationship to diet.
In the current study, we sought to better understand which dietary and lifestyle factors are predictive of antibiotic resistance in healthy U.S. adults. Prior to data generation, we outlined several hypotheses surrounding diet and AMR. Specifically, our primary hypothesis was that an increased intake of dietary fiber would be associated with reduced ARG abundance in human fecal metagenomes. Our secondary hypotheses were that increased habitual animal protein intake and saturated fat intake would be associated with increased ARG abundance while a higher Healthy Eating Index (HEI) would be associated with decreased ARG abundance. Finally, we hypothesized that lower stool pH, as a proxy for fermentation in the colon, would be associated with a lower abundance of ARGs. Beyond these directed hypotheses, we utilized machine learning approaches on a variety of diet, physiological, and lifestyle features to assess whether the abundance of antibiotic genes is correlated with variables outside the scope of our directed hypotheses. The present study aims to address a critical knowledge gap regarding the impact of diet on antimicrobial resistance.
RESULTS
Participant characteristics.
Participants with stool selected for metagenomic sequencing (n = 290) varied by age, sex, and body mass index (BMI) as shown in Table 1. While these participants identified mostly as white (n = 205), there were also Hispanic (n = 38), black (n = 13), and Asian (n = 32) ethnicities represented. Most participants were born in the United States (n = 241); however, several were born in other countries (n = 49). Both sexes were represented nearly equally, with 140 males and 150 females. The cohort reflects the multiethnic population in Davis, California, and surrounding areas.
TABLE 1.
General characteristics of each ARG cluster for all individualsa
| Characteristic | Data by cluster |
||
|---|---|---|---|
| Low-ARG | Medium-ARG | High-ARG | |
| Avg age (y [range]) | 42.7 (19–66) | 39.5 (18–65) | 41.4 (20–64) |
| BMI (kg/m2 [range]) | 27.7 (19.7–43.9) | 27.3 (18.2–43.2) | 26.5 (18.0–38.0) |
| No. of males | 33 | 72 | 35 |
| No. of females | 40 | 72 | 38 |
| Ethnicity (n) | |||
| Asian | 4 | 15 | 13 |
| Black | 4 | 6 | 3 |
| Hispanic | 12 | 18 | 8 |
| White | 52 | 104 | 49 |
| Born in United States (n) | |||
| Yes | 60 | 125 | 56 |
| No | 13 | 19 | 17 |
N = 290.
Metagenomic sequencing statistics.
The number of sequence reads per sample and average read length before and after the preprocessing stages of the bioinformatics pipeline are shown in Table S4 in the supplemental material. Even after we removed reads that mapped to the human genome or failed quality checking or were duplicates, there were 27.2 (standard error of the mean [SEM], 0.22) million unique reads per sample that were then mapped to an antimicrobial resistance gene database. We discovered previously that few paired-end reads can be merged using standard library preparation protocols, which is problematic when mapping to amino acid sequences (16). Therefore, we adjusted the insert library size to enable overlapping reads (see Materials and Methods). Most reads overlapped (79.20% ± 1.35%), resulting in merged reads of 259 ± 4.79 bp long (SEM calculated using million as the read number unit).
Sequencing read loss due to QC steps. Download Table S4, XLSX file, 0.01 MB (10.5KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The composition and abundance of ARGs vary significantly in a healthy cohort.
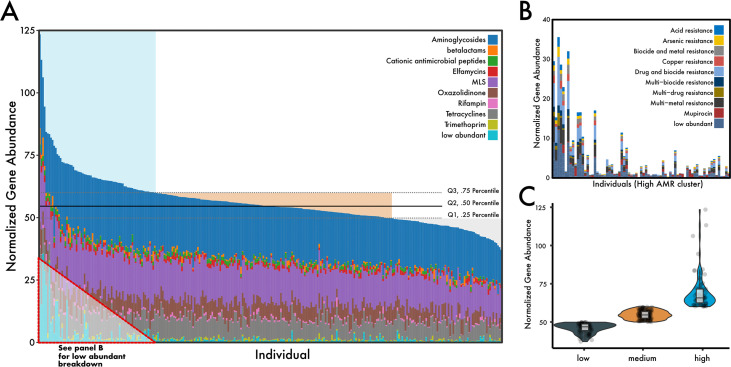
The total abundance of antimicrobial resistance genes, post-normalization, ranged from 37 to 123 reads per kb per genome equivalent (RPKG). Aminoglycoside resistance was the most common mechanism of antimicrobial resistance within the cohort, with an average abundance of 21.4 ± 3.34 RPKG (coefficient of variation, 15.6%), followed by resistance to macrolide-lincosamide-streptogramin (MLS) (14.7 ± 2.68 RPKG), and tetracyclines (7.84 ± 2.51 RPKG) (Fig. 1A; see Fig. S2A in the supplemental material). Individuals with high ARG abundance generally had a higher diversity of AMR mechanisms found in their resistomes (Fig. S2B). Similarly, a group of low-abundant mechanisms that included metal and multidrug resistance was increased in individuals with a high abundance of ARGs (Fig. 1B) but absent or reduced in individuals with lower ARG abundance profiles.
FIG 1.
Distribution of antibiotic resistance mechanisms in a healthy U.S. cohort. (A) Stacked bar chart of the top nine most abundant ARG mechanisms across 290 individuals, normalized to genome equivalents (see Materials and Methods). Horizontal quartile lines are drawn based on total ARG abundance and represent our clustering strategy. (B) A zoom-in of the low-abundant mechanisms that are found increasingly throughout the high-ARG group. (C) Total ARG abundance violin plot summary between ARG clusters. Points represent the sum of ARG abundance within individuals.
(A) ARG composition (top) and taxonomic assignments (bottom, using CAT) of most abundant contigs to which ARG reads map. (B) Alpha diversity (evenness, richness, and Shannon diversity) of ARG genes across different ARG clusters. Download FIG S2, TIF file, 2.8 MB (2.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
For classification purposes, we used quartiles to cluster individuals into three groups based on their total antimicrobial gene abundance (Fig. 1C). The low-ARG cluster represented 73 individuals in quartile 1, who had an average ARG abundance of 46.0 ± 3.1 RPKG. The medium-ARG cluster represented 144 individuals between quartile 2 and 3 three who had an average ARG abundance of 54.7 ± 2.7 RPKG. Finally, the high-ARG cluster represented 73 individuals who had an average ARG abundance of 69.2 ± 11.3 RPKG. The composition of AMR genes between these groups appeared to have substantial variation (permutational multivariate analysis of variance [PERMANOVA], R2 = 0.19, P = 0.001); however, the assumption of dispersions was not met (permutational analysis of multivariate dispersions [PERMDISP], P < 0.001). The lack of dispersion homogeneity between the ARG clusters was likely not a function of sample size differences; indeed, after subsampling ARG clusters to equal the number of individuals (n = 73), significant differences remained (PERMDISP, P < 0.001) and high-ARG individuals appeared more disperse in ordination than in the other clusters (data not shown).
Differences in ARG abundance are associated with differences in the gut microbiome.
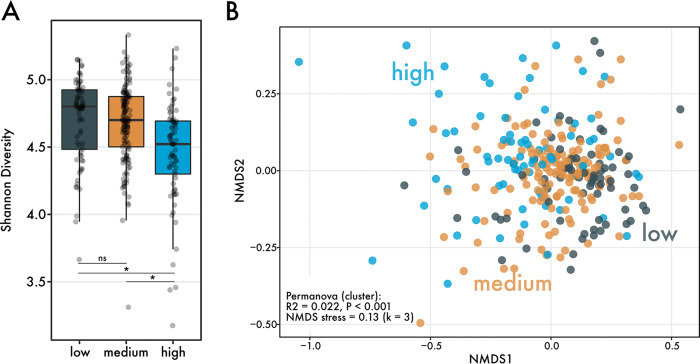
Next, we asked whether differences in ARG abundance were associated with differences in gut microbiome diversity and composition, potentially indicating a shared microbial community structure or function within high-ARG individuals. The three ARG clusters differed significantly in the Shannon diversity index, a metric of alpha diversity (Fig. 2A) (Kruskal-Wallis, P < 0.05). Post hoc tests revealed that individuals within the high-ARG group harbored significantly less diverse gut microbiomes than those in low- and medium-ARG groups. The average number of Kraken-classified species in high-ARG (post-rarefaction) was 2,305 (SD, 317), which is over 100 species less than that for medium-ARG (2,408 ± 229) and low-ARG clusters (2,423 ± 235).
FIG 2.
Differences in microbiome diversity between ARG clusters. (A) Shannon diversity between ARG clusters (n = 290 individuals). Significance was determined using the Kruskal-Wallis test and a post hoc Dunn test, with P values correcting using the Benjamini-Hochberg method. (B) Nonmetric multidimensional scaling of Bray-Curtis distances between individuals’ microbiomes. Points are colored by ARG cluster. Beta diversity differences were determined using a PERMANOVA, which revealed significant differences in the community composition between the clusters.
We also investigated whether the composition of individuals’ microbiomes vary between ARG clusters. Between low-, medium-, and high-ARG clusters, the taxonomic composition of the microbiome differed significantly (Fig. 2B) (PERMANOVA, R2 = 0.022, P < 0.001; PERMDISP, P > 0.05). To determine which taxa were driving these compositional differences, we performed a linear discriminant analysis that revealed high-ARG microbiomes were distinguished by significantly more reads mapping to bacterial families Streptococcoceae and Enterobacteriaceae (see Fig. S3 in the supplemental material). Very few families were distinguishing for medium-ARG microbiomes, notably species of Clostridiaceae, in addition to reads mapping to the Elusimicrobiaceae family. Finally, low-ARG microbiomes were characterized by a greater abundance of mostly anaerobic microbes, especially many different species from the family Clostridiaceae.
LDA analyses identifying bacterial families that vary in abundance between ARG clusters. Download FIG S3, TIF file, 2.7 MB (2.7MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
High fiber and low protein intake correlate with low antimicrobial resistance.
Similar to the microbiome, we asked whether aspects of diet related to our directed hypotheses, such as fiber and protein consumption, varied between ARG clusters. Our results indicate that habitual fiber intake and soluble fiber intake (calorie adjusted) measured using the food frequency questionnaire (FFQ) were significantly different between ARG clusters (Table 2). Post hoc tests show that significant differences in fiber intake were driven by differences between low-ARG and medium-ARG clusters. Calorie-adjusted fiber intake was highest in low-ARG individuals compared with individuals in medium- and high-ARG clusters. We also examined differences in protein intake between ARG clusters. Habitual protein intake, especially from beef and pork sources, was significantly lower in low-ARG individuals than that in individuals with more abundant antibiotic-resistant genes.
TABLE 2.
Tests of directed hypothesesa
| Dietary component | Data source | Data by cluster |
Transformation, test |
P value of: |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Low (n = 63) | Medium (n = 126) | High (n = 57) | Low vs medium | Low vs high | Medium vs high | All | |||
| Avg fiber intake_tnfs | ASA24 | 26.67 ± 10.8 (1.35)b | 24.96 ± 13.53 (1.2)b | 26.42 ± 12.31 (1.63)b | sqrt(x), ANOVA, Tukey | 0.423 | 0.982 | 0.58 | 0.376 |
| Fiber from food and supplements | ASA24 | 12.29 ± 4.63 (0.58) c | 10.81 ± 4.55 (0.4) c | 12.1 ± 5.21 (0.69) c | yeojohnson, ANOVA, Tukey | 0.079 | 0.983 | 0.146 | 0.045 |
| Total fiber intakec | FFQ | 25.94 ± 11.34 (1.42)b | 24.85 ± 12.18 (1.08)b | 28.41 ± 11.96 (1.58)b | yeojohnson, ANOVA, Tukey | 0.655 | 0.494 | 0.084 | 0.101 |
| FFQ | 13.05 ± 3.92 (0.49) c | 11.7 ± 3.82 (0.34) c | 12.59 ± 3.77 (0.5) c | arcsinh, ANOVA, Tukey | 0.034 | 0.798 | 0.214 | 0.03 | |
| Soluble fiber intake | FFQ | 7.2 ± 3.2 (0.4)b | 6.82 ± 3.73 (0.33)b | 7.73 ± 3.32 (0.44)b | yeojohnson, ANOVA, Tukey | 0.517 | 0.645 | 0.097 | 0.107 |
| FFQ | 3.6 ± 1.07 (0.13) c | 3.17 ± 1.05 (0.09) c | 3.44 ± 1.13 (0.15) c | Log10(x), ANOVA, Tukey | 0.015 | 0.616 | 0.236 | 0.016 | |
| Habitual protein intake from animals | FFQ | 46.76 ± 24.52 (3.06) b | 54.48 ± 25.4 (2.25) b | 55.58 ± 27.35 (3.62) b | arcsinh, ANOVA, Tukey | 0.029 | 0.042 | 0.951 | 0.019 |
| Saturated fatty acid intake | FFQ | 26.45 ± 11.33 (1.42)b | 29 ± 12.66 (1.12)b | 30.13 ± 12.49 (1.65)b | BoxCox, ANOVA, Tukey | 0.295 | 0.168 | 0.802 | 0.166 |
| Total energy intake | FFQ | 2008.95 ± 734.73 (91.84)d | 2135.41 ± 820.54 (72.81)d | 2288.85 ± 825.32 (109.32)d | arcsinh, ANOVA, Tukey | 0.599 | 0.12 | 0.379 | 0.141 |
| Beef/pork consumption, including cured meat | FFQ | 3.28 ± 2.16 (0.27) e | 4.06 ± 2.3 (0.2) e | 4.12 ± 2.5 (0.33) e | arcsinh, ANOVA, Tukey | 0.017 | 0.077 | 0.985 | 0.018 |
| FFQ | 1.62 ± 0.83 (0.1) f | 1.93 ± 0.78 (0.07) f | 1.81 ± 0.82 (0.11) f | yeojohnson, ANOVA, Tukey | 0.027 | 0.369 | 0.606 | 0.036 | |
| Beef/pork consumption, excluding cured meat | FFQ | 1.11 ± 0.91 (0.11) e | 1.44 ± 1.13 (0.1) e | 1.47 ± 1.18 (0.16) e | boxcox, ANOVA, Tukey | 0.041 | 0.157 | 0.969 | 0.045 |
| FFQ | 0.53 ± 0.32 (0.04)f | 0.67 ± 0.42 (0.04)f | 0.61 ± 0.37 (0.05)f | yeojohnson, ANOVA, Tukey | 0.045 | 0.465 | 0.602 | 0.056 | |
| HEI total score | ASA24 | 64.33 ± 13.29 (1.66) | 61.53 ± 14.1 (1.25) | 63.57 ± 12 (1.59) | None, ANOVA, Tukey | 0.364 | 0.949 | 0.607 | 0.343 |
| Stool pH | Not applicable | 7.08 ± 0.54 (0.07) | 6.97 ± 0.61 (0.05) | 6.87 ± 0.52 (0.07) | None, ANOVA, Tukey | 0.431 | 0.121 | 0.539 | 0.143 |
| BMI | Not applicable | 27.64 ± 5.4 (0.67) | 27.2 ± 5 (0.44) | 26.1 ± 4.08 (0.54) | yeojohnson, ANOVA, Tukey | 0.837 | 0.258 | 0.422 | 0.27 |
Shows statistics regarding the hypotheses made about diet and other physiological metrics and their relationship with antimicrobial resistance. These comparisons were made across a subset of the same individuals for which we had data on every dietary/physiological feature used in this analysis (n = 246 individuals). Rows with features that were significantly different between ARG clusters are bolded. Values displayed in parenthetical are formated as Mean ± standard deviation (standard error of mean).
Units are g.
Units are g/1,000 kcal.
Units are kcal.
Units are oz eq.
Units are oz eq/1,000 kcal.
While we hypothesized that the healthy eating index (HEI) and stool pH would vary between ARG clusters, we found no significant differences in these variables. The HEI of low-ARG individuals trended higher (healthier) than that of the other clusters; however, stool pH, a proxy for SCFA metabolism, paradoxically also trended higher in low-ARG. Taken together, individuals with a lower abundance of antibiotic resistance genes consumed diets higher in fiber and lower in animal protein, but these patterns in nutrient intake were not reflected in HEI differences between ARG clusters.
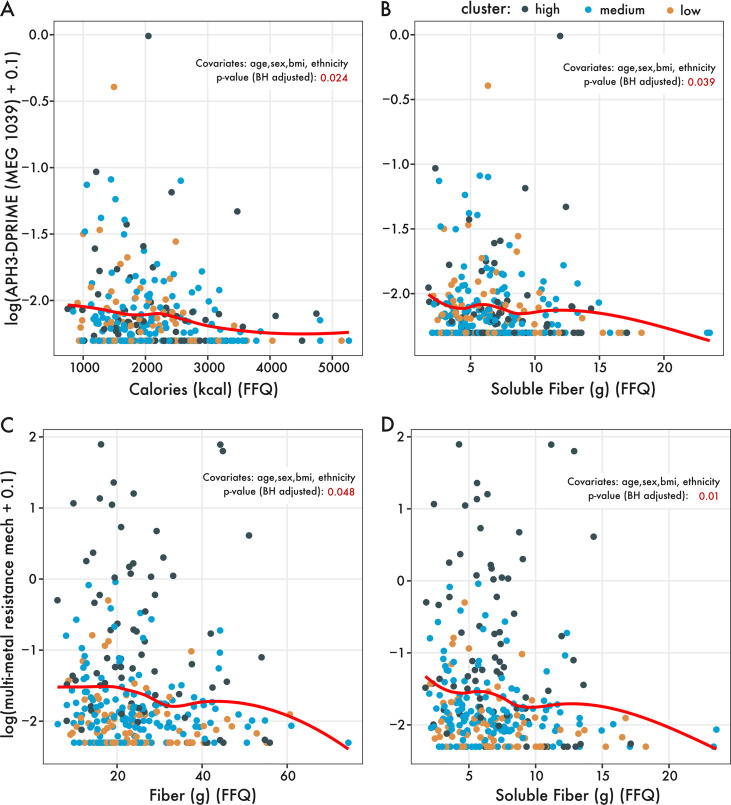
Next, we determined whether there were specific AMR mechanisms or genes that varied with diet. Using ordinal logistic regression, we analyzed the relationship between antimicrobial resistance mechanisms or genes with variables from our directed hypotheses (i.e., dietary fiber, protein, and HEI). Notably, after correcting for known covariates (age, sex, BMI, and ethnicity), the gene aminoglycoside-O-phosphotransferase (aph3-dprime) varied significantly with soluble fiber and habitual calorie intake, decreasing in abundance with increasing soluble fiber and calorie intake (Fig. 3A and B; see Fig. S4A in the supplemental material). Associations at the AMR mechanism level revealed several dietary associations with multimetal resistance. Specifically, multimetal resistance varied significantly with dietary fiber and soluble fiber intake after we corrected for covariates (Fig. 3B and C, Fig. S4B). In summary, increasing fiber intake appears to be associated with decreased abundances of specifically aminoglycoside-O-phosphotransferases and genes involved in multimetal resistance.
FIG 3.
Proportional odds logistic regression identifies relationships between dietary nutrients from directed hypotheses and ARG genes or mechanisms. The abundance of MEG 1039, an aminoglycoside-O-phosphotransferase (aph3-dprime), varied significantly with (A) total calorie intake and (B) soluble fiber intake (n = 290 individuals). Likewise, aggregated at the mechanism level, multimetal resistance significantly varied with (C) dietary fiber intake and (D) soluble fiber intake. The red regression line is based on the geom_smooth() function using the locally estimated scatterplot smoothing (LOESS) method from the R package ggplot2. The P values of the POLR regression are included in red, noting which covariates were included in the POLR analysis.
(A) Regression analysis of the association between habitual calorie intake and the gene aminoglycoside-O-phosphotransferase (aph3-dprime) (no covariate adjustment). (B) The association between soluble fiber intake and multimetal resistance (no covariate adjustment). Download FIG S4, TIF file, 1.1 MB (1.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Machine learning identifies fiber and diet diversity as predictive features distinguishing ARG profiles.
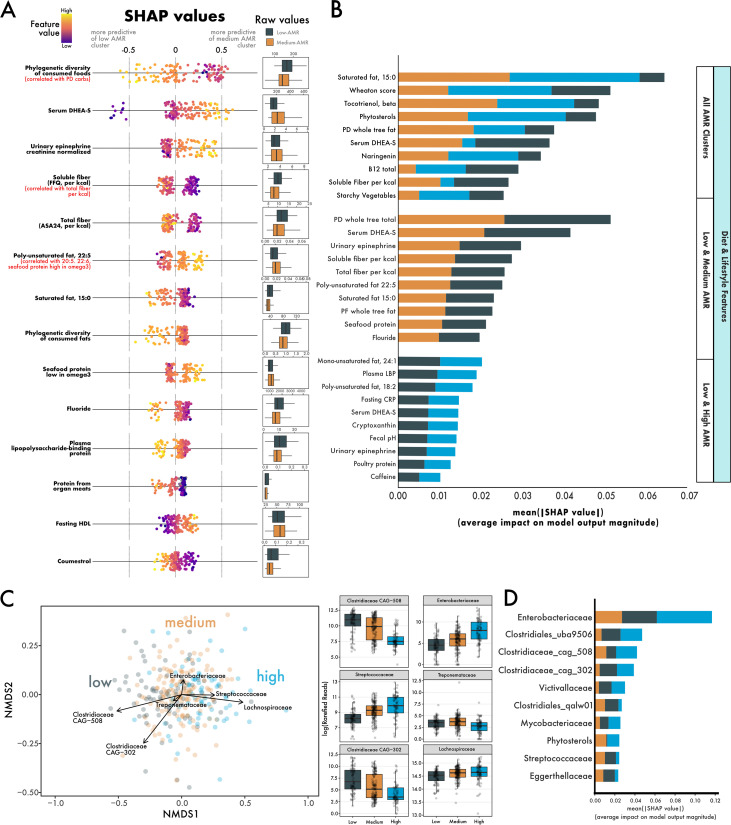
Finally, we leveraged machine learning and our large number of diet, lifestyle, and microbial measurements to identify other potential factors differentiating ARG clusters. We analyzed 387 dietary, lifestyle, and physiological features from a subset of 187 individuals (92 males and 95 females) aged 19 to 66 years (mean, 42.2 ± 13.8). A random forest model comparing the diet and lifestyle between low- and medium-ARG individuals performed better based on the area under the receiver operating characteristic curve (ROC AUC) score than models comparing between low- and high-ARG clusters or comparisons between all three clusters (see Table S3 in the supplemental material). The phylogenetic diversity of food intake (i.e., how many different types of foods were consumed) was the most explanatory feature between low- and medium-ARG clusters, with low-ARG individuals having more diverse diets than medium-ARG individuals (Fig. 4A). While other features were distinguishing between low- and medium-ARG, a higher intake of dietary fiber (ASA, calorie adjusted) and soluble fiber (FFQ, calorie adjusted) were particularly indicative of low-ARG individuals (Fig. 4A).
FIG 4.
Using machine learning to identify aspects of diet, lifestyle, and the microbiome that predict ARG abundance. (A) SHAP values depicting the predictive power of a random forest using diet and lifestyle features to distinguish individuals in the low-ARG and medium-ARG groups. High levels of diversity in diet (which was correlated with diversity in carbohydrates) was most predictive of low-ARG individuals. (B) Features, sorted by their mean absolute SHAP value, which distinguish all ARG clusters, low-ARG (gray, n = 45) and medium-ARG (gold, n = 95), and low-ARG and high-ARG (blue, n = 47). (C) A nonmetric multidimensional scaling of Bray-Curtis distances between individuals’ microbiomes, with overlaid vectors of bacterial families identified as strong predictors of ARG cluster membership. Box plots show the log-transformed abundance of the families between ARG clusters. (D) A random forest using both microbiome and diet/lifestyle data uses mainly microbiome features in distinguishing ARG clusters.
Machine learning model performance metrics. Download Table S3, XLSX file, 0.01 MB (11.2KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Furthermore, we investigated differences between low- and high-ARG and between all three clusters together (Fig. 4B). One feature shared between these comparisons was plasma dehydroepiandrosterone sulfate (DHEA-S), driven by low values in low-ARG individuals (see Fig. S5 in the supplemental material). The predictive strength of features used to distinguish low- from high-ARG individuals was relatively weaker (Fig. 4B), with the largest mean absolute SHapley Additive exPlanations (SHAP) value being approximately 0.02, which is less than half as much as comparisons between low- versus medium-ARG or all ARG clusters.
(A) Significantly decreased DHEA-S levels in low-ARG compared with that of medium- and high-ARG. (B) Most measured DHEA-S values fall within the normal clinical range. Solid lines indicate lower bounds of normal DHEA-S for age and sex. Dashed lines indicate upper bounds. Download FIG S5, TIF file, 1.4 MB (1.4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Next, to support results from the linear discriminant analysis on microbiome data, we used a random forest model to identify bacterial families that differentiate ARG clusters. The abundances of taxa such as Clostridium CAG-508, Enterobacteriaceae, and Streptococcaceae stratified linearly between low-, medium-, and high-ARG clusters (Fig. 4C). Random forest models made using microbiome data performed better than diet/lifestyle data at classifying ARG clusters (Table S3). Indeed, after we combined microbiome, diet, and lifestyle data, nearly all the top predictive features were microbe abundances (Fig. 4D).
DISCUSSION
In the present study, we analyzed the association between healthy individuals’ resistome, diet, and microbiome. While studies in animals suggest that nutrient intake can alter antimicrobial resistance (15), a knowledge gap exists regarding how diet might influence the abundance and composition of antibiotic resistance genes in humans. This knowledge gap is particularly prevalent for adults, as several studies have investigated resistome development in infants (17–20). We leveraged a large and diverse cohort of healthy individuals who provided habitual and recent dietary information, coupled with physiological or microbiome measurements from blood, urine, and stool. With this cohort, we specifically sought to answer how aspects of diet vary as a function of total abundance of antibiotic resistance genes. Using traditional statistics and machine learning, we show that increased fiber and diet diversity covary with ARG abundance, with individuals consuming increased fiber and more diverse diets having lower total abundances of antibiotic resistance genes. We also show strong signatures of antimicrobial resistance in the gut microbiome. Our machine learning model indicates that the abundance of bacterial families such as Enterobacteriaceae and Streptococcaceae are both strong predictors of AMR. Our results provide a framework for future studies which might seek to reduce the burden of antimicrobial resistance through interventional means, such as diet.
We shotgun sequenced the gut microbiomes of 290 healthy individuals revealing, somewhat surprisingly, a large diversity (in both abundance and composition) of antibiotic resistance genes. The diversity of antimicrobial resistance mechanisms increased within increasing total ARG abundance (Fig. S2B), even after normalizing for sequence depth. Within the cohort, the most common antimicrobial resistance mechanism was aminoglycoside resistance, followed by resistance to macrolide-lincosamide-streptogramin and tetracyclines, which are all clinically important classes of antibiotics for humans (Fig. 1A). Studies have shown aminoglycoside resistance to be widespread in different environments, including wastewater, livestock farms, and human fecal samples (21). Within human gut metagenomes specifically, aminoglycoside resistance is extremely prevalent in individuals from both industrialized and nonindustrialized societies; however, its abundance is greater within industrialized societies (22). The prevalence of aminoglycoside resistance, especially within individuals from industrialized environments, likely results from widespread coselection. Indeed, in a study examining antibiotic exposure in pigs, aminoglycoside-O-phosphotransferases (incidentally, a gene in our study significantly inversely correlated with soluble fiber intake [Fig. 3A]) increased in abundance even when aminoglycoside antibiotics were not administered to the pigs (23). Furthermore, in a study examining the distribution of ARGs worldwide, it was shown that the resistomes of individuals from the United States skewed higher in abundance of aminoglycosides, which differed from individuals from Mongolia (24). Once encoded in the microbiome, aminoglycoside resistance genes are promiscuously transferable elements, finding home in phylogenetically disparate genomes (25).
We suspected that differences in the resistomes between individuals would reflect differences in the microbiome. Both the diversity and composition of the microbiome differed significantly between low-, medium-, and high-ARG clusters (Fig. 2). Recent work has shown that the density of an antibiotic gene network increases as microbiome diversity decreases (26). The authors speculated this trend might be due to selection preserving connected organisms or that decreased diversity promotes more opportunities for connectedness. Notwithstanding, we show that an increased abundance of genes involved in antimicrobial resistances was significantly associated with decreased alpha diversity (Fig. 2A). Moreover, the composition of the microbiome differed significantly between ARG clusters. Notably, individuals in the high-ARG cluster had an increased abundance of Streptococacceae and Enterobacteriaceae (Fig. 4C, Fig. S3). These results were supported by machine learning, which identified these bacterial families as highly predictive of the high-ARG cluster (Fig. 4D).
Enterobacteriaceae in particular has been shown to bloom transiently in response to a 5-day antibiotic cocktail regimen (27). Interestingly, this bloom of Enterobacteriaceae follows an increase in redox potential, suggesting species within Enterobacteriaceae might utilize their diverse set of respiration pathways to establish themselves as pioneer species after antibiotic treatment (27). Conversely, low abundances of facultative anaerobes (i.e., Streptococacceae and Enterobacteriaceae) and higher abundances of strict anaerobes, such as those from the family Clostrideaceae, were more predictive of low-ARG individuals (Fig. 4D). Central to both of these observations is the role of oxygen in the gut (reviewed in reference 28). Studies have shown that antibiotics reduce taxa in the gut responsible for the production of short-chain fatty acids (which include members of Clostrideaceae) (29), which in turn disrupts peroxisome proliferator-activated receptor-γ (PPAR-γ) signaling (30). Proper PPAR-γ signaling directs colonocytes to oxidize short-chain fatty acids, a process which requires enough oxygen such that a gradient is maintained and the lumen remains anaerobic. Disrupted PPAR-γ signaling leads to altered colonocyte metabolism, releasing oxygen and other terminal electron acceptors into the gut, potentially expanding a niche for facultative anaerobes (28). Taken together, antibiotic treatment depletes obligate anaerobes in the gut, which diminishes the production of SCFAs and shifts colonocyte metabolism toward one that favors blooms of inflammation-associated facultative anaerobes. Our findings that strict anaerobes are more predictive of low-ARG and facultative anaerobes are more predictive of high-ARG support this relationship between antibiotics and oxygen in the gut.
Our study addresses a critical knowledge gap surrounding the role of diet and antimicrobial resistance. While microbial fermentation biproducts (i.e., SCFAs) impact colonocyte metabolism and microbiome composition, a key piece missing thus far is how dietary fiber, the substrate for SCFA production, associates with antimicrobial resistance profiles. In dairy cows, an increase in complex carbohydrates was associated with a decrease in antibiotic resistance gene abundance (15). We hypothesized that increased dietary fiber would be correlated inversely with antibiotic resistance gene abundance. Our results show that both total fiber and soluble fiber consumption were highest in individuals in the low-ARG cluster (Table 2). Additionally, fiber correlated inversely with aph3-dprime gene abundance (involved in aminoglycoside resistance) and multimetal resistance mechanisms (Fig. 3). We do note our findings regarding metal resistance associating with lower fiber intake may, in part, reflect our increased ability to detect metal resistance genes; a primary objective of the second iteration of the MEGARes database (v2.0) was the inclusion of metal resistance and biocide genes (31). Finally, between low-ARG and medium-ARG clusters, our machine learning model identified higher recent fiber intake (ASA24) and soluble fiber (FFQ) as predictive of low-ARG individuals (Fig. 4A). Puzzlingly, fecal pH, a proxy for SCFA metabolism, trended higher in low-ARG individuals. One explanation may be that the rapid absorption of SCFAs across the gut epithelial fails to contribute to pH lowering in fecal samples. These data are supported by in vitro work showing no positive correlations between fecal SCFA abundance and acid production by the gut microbiota (32). Indeed, others have shown that modifying diet with large increases in the substrates necessary for SCFA production has no significant impact on fecal SCFAs measured (33). To that end, future work should aim to measure serum SCFAs or some other proxy for SCFA production, such as transcriptional profiles of colonocyte SCFA transporters.
Moreover, results relating to our directed hypotheses surrounding protein and antimicrobial resistance were generally supported; decreased protein intake correlated with decreased ARG abundance. These results support a study examining community-acquired urinary tract infections in an elderly population, which showed that more antibiotic-resistant bacterial isolates were recovered from individuals with increased chicken and pork intake (32). However, while antimicrobial resistance in animals raised for food is increasing globally (34), the directionality of AMR transfer between humans and animals, or even whether one exists at all, remains controversial and inconclusive (35, 36). Several studies have shown specific transfer of antibiotic-resistant bacteria between humans and farm animals (37–40). Yet, with the exception of a few countries (e.g., Switzerland [41]), the surveillance needed to understand the magnitude or even directionality of this transfer (human to animal or animal to human) is currently lacking. Many of the systems in place to identify foodborne pathogens use pulsed-field gel electrophoresis (PFGE) or multilocus sequence typing (MLST), which do not have the resolving power to detect bacterial strain-level transmission during surveillance efforts (42).
Some aspects of diet and lifestyle identified by the random forest as predictive of low-ARG individuals have generally been associated with health. For example, increased phylogenetic diversity of diet (which was highly cocorrelated with phylogenetic diversity of dietary carbohydrates) was increased in low-ARG individuals. This finding is consistent with a recent study examining diet and the microbiome in a cohort of 1,112 individuals, which showed that a diet high in plant diversity or a diet with a high healthy eating index was correlated strongly with gut microbiome composition (43). Furthermore, our machine learning models found fecal calprotectin, a marker of gut inflammation, increased with increasing ARG abundance. Notably, however, the random forest model was poor at distinguishing low-ARG from high-ARG when using diet and lifestyle features (Fig. 4B). We suspect that we are missing features that may distinguish low-ARG from high-ARG more confidently, such as antibiotic usage history. Repeated antibiotic exposure can shift the gut microbiome to an altered state that is different from the pre-antibiotic baseline (44). Multiple perturbations yielding altered steady states in ecological systems have been discussed (45), the insights of which may appropriately describe the effects of repeated antibiotic exposure on the microbiome.
While our machine learning approach largely supported our primary hypothesis, some features identified by the random forest model were more enigmatic in their relationship with AMR. Dehydroepiandrosterone sulfate (DHEA-S), an androgen precursor with diverse health benefits (46–48), was used in all models as a valuable predictor of ARG cluster membership. While the hormone was low in low-ARG individuals (Fig. S5A), it should be noted that nearly all individuals had measured values within the normal clinical range (Fig. S5B). Although DHEA-S levels are known to fluctuate between sexes and throughout life (49), we do not suspect that the models are using DHEA-S as a proxy for age or sex since neither age nor sex were important for distinguishing ARG clusters. One study examining hormone profiles as a function of diet in pre- and post-menopausal women found that vegetarians (who consumed more fiber than their omnivorous counterparts) contained significantly lower levels of plasma DHEA-S and a generally beneficial metabolic profile (50). Moreover, increased levels of DHEA-S have been shown to reduce the protein levels of PPAR-γ in adipocytes in vitro, an effect that may extend to gut colonocytes, disrupting the oxygen balance discussed previously (51). The presence of DHEA has even been shown to increase the antibiotic resistance of Staphylococcus aureus in vitro (52). While it remains difficult to say with any certainty if decreased levels of DHEA-S are biologically meaningful in the context of diet and AMR, our work, combined with existing studies, suggest that DHEA-S may fluctuate as a function of diet. Furthermore, the higher levels of DHEA-S we observed in medium-ARG and high-ARG individuals may contribute directly and indirectly to increased antimicrobial resistance.
An important caveat of our study is its observational nature; while our results might inspire some important and testable hypotheses, presently, we are unable to untangle any casual relationships between diet and antimicrobial resistance. Furthermore, we are not able to assess the impact of previous antibiotic use or other treatments which may influence the establishment or detection of antibiotic resistance genes. Finally, we acknowledge that the inherent temporal variability found in the microbiome is not captured in this study, and longitudinal efforts would allow us to examine questions such as the intraindividual stability of the resistome.
In conclusion, we report that a diverse cohort of healthy individuals harbor significant variability in their resistomes. We show that ARG diversity was associated with diversity in diet and the microbiome. Specifically, we showed that individuals with lower abundances of antibiotic resistance genes consumed more diverse diets that were richer in fiber and limited in animal protein. We suspect that increased fiber likely drives the composition of the gut toward a more obligate anaerobe state, reducing footholds for facultative anaerobes, which are known harbors of inflammation and antibiotic resistance. Critical next steps include assessing SCFA metabolism more directly to better determine whether high-fiber diets contribute to microbial communities intrinsically lower in antibiotic-resistant genes. Ultimately, future work may use research-based dietary guidelines to reduce the incidence of antimicrobial resistance, thus lifting an immense burden on health care systems worldwide.
MATERIALS AND METHODS
Participants.
Healthy adults, aged 18 to 65 y, male or female, with a BMI of 18 to 44 kg/m2 living near Davis, California, were recruited in the USDA Nutritional Phenotyping Study, which is a cross-sectional observational trial in which the fecal microbiome, among other outcomes, was assessed in the context of self-reported diet, physical activity, and physiological status (53). Men and women were recruited to fill nine bins within sex, to balance BMI and age, using three BMI categories (<25, 25 to 29, and 30 to 44 kg/m2) within each age category (18 to 33, 34 to 49, and 50 to 65 y). Participants were excluded if they had high blood pressure (systolic blood pressure of >140 mm Hg or diastolic blood pressure of >90 mm Hg) when measured on-site or if they had any active chronic disease requiring daily medication, including, but not limited to, diabetes mellitus, cardiovascular disease, cancer, gastrointestinal disorders, kidney disease, liver disease, bleeding disorders, asthma, autoimmune disorders, hypertension, or osteoporosis. Participants were excluded from fecal metagenome sequencing if their dietary record was considerably incomplete or if the extracted metagenomic DNA was low in concentration (<100 ng/μL) or quality (A260/280 of <1.78; A260/230 of <1.72). In total, 290 individuals were included in the current study. The study is registered on ClinicalTrials.gov (identifier NCT02367287) and received ethical approval from the University of California Davis Institutional Review Board.
Stool collection and processing.
Participants were instructed to collect a single stool sample in a Ziploc bag enclosed in a hard, plastic container with a lid, and immediately place it in a cooler containing ice packs. The cooler was brought to the research center as soon as possible for same-day processing. Stool was homogenized in a Stomacher for 3 min and flash frozen on dry ice before being broken into aliquots and stored at −70°C until DNA isolation. Stool consistency was technician scored and pH analyzed as described previously (54, 55).
DNA extraction, library preparation, and sequencing.
DNA was isolated with the ZymoBiomics DNA miniprep kit (Zymo Research) from approximately 100 mg of homogenized stool per the manufacturer’s protocol. Beta-mercaptoethanol was added to the DNA binding buffer at a 0.5% (vol/vol) concentration. Five cycles of 1 min bead beating at maximum speed (6.5 m/s) followed by 5 min cooling on ice were performed using a FastPrep-24 homogenizer (MP Biomedicals) for a total of 5 min of bead beating. DNA was eluted into 50 μL DNase/RNase-free water. The majority of the DNA preparations (>95%) had A260/280 and A260/230 ratios greater than 1.80, and the lowest A260/280 and A260/230 ratios of sequenced samples were 1.78 and 1.72, respectively. DNA was quantified with the Qubit double-stranded DNA (dsDNA) broad-range (BR) assay (Thermo Fisher) and then diluted to 100 ng/μL. Representative samples were resolved on an agarose gel to confirm that DNA was intact and RNA free prior to library construction.
Whole-genome shotgun sequencing library preparation, library quality control (QC), quantification, and pooling were performed by DNA Technologies & Expression Analysis Core Laboratory at University of California Davis Genome Center. Library insert sizes were 250 to 280 bp, and exact size selection of DNA fragments was performed with a PippinHT instrument (Sage Science). Ninety-six to 98 libraries were combined in a single pool for each sequencing batch sequenced in a 2 × 150-bp format on the NovaSeq6000 (Illumina) platform with an S4 flow cell to result in an approximately 20-bp overlap in reads.
Metagenomic sequence analysis.
BMTagger (56) was used to remove reads aligning to the human genome version GRCh38.p13 (57) from all samples. Afterward, Trimmomatic version 0.33 (58) was used to remove TruSeq paired-end adapters and reads were trimmed as described previously (59) with a sliding window of 4 bp, a minimum average quality of 15, and a minimum length of 99 bp. Duplicated reads were then removed using FastUniq version 1.1 (60) with default settings. The resulting paired-end reads were assembled using FLASH version 1.2.11 (61) with the overlapping length set between 10 bp to 100 bp and the mismatch ratio set at 0.1. Community-wide taxonomy profiling was performed with Kraken2 (62) and aligned to a prebuilt custom database (release 95 [13.07.2020]) using Sturo (63). Kraken2mpa.py from KrakenTools was used to format Kraken outputs for downstream analysis. Taxa which had only one sequence read of support were dropped. Sequence depth was normalized by permuted rarefaction using the R package EcolUtils (64), to a depth of 5,833,371 reads per sample averaged across 5 permutations.
Antimicrobial-resistant gene analysis.
Paired-end deduplicated reads were mapped against the MEGARes 2.0 database (31) using BWA version 0.7.16a (65) to determine antimicrobial drug, biocide, and metal resistance genes. For ARGs with significant correlations with diet, we checked whether the ARG required single nucleotide polymorphism (SNP) confirmations as advised by MEGARes. The resulting alignment files were then analyzed with the resistome analyzer (https://github.com/cdeanj/resistomeanalyzer) to calculate the abundances of AMR at the gene level. To normalize the raw ARG counts to reads per kb per genome equivalent [RPKG; RPKG = raw counts/(gene length × genome equivalent)], genome equivalents based on the total paired-end, deduplicated reads for each sample were estimated using MicrobeCensus version 1.1.1 (66).
MEGAHIT version 1.29 (67) was used to assemble paired-end deduplicated reads into contigs. These contigs were then used as references to align ARG-mapped short reads using BWA-MEM (65). Next, contigs containing ARG reads were retained for taxonomy identification using the Contig Annotation Tool (CAT) version 5.0.3 and the preprepared database version 2020-06-18 (68). Sequence manipulation, including sam-to-fastq file conversion and read filtering, was performed using the BBMap toolkit version 37.68 (69).
Dietary assessment.
Recent dietary intake was assessed using the Automated Self-Administered 24-h (ASA24) Dietary Assessment Tool, versions 2014 and 2016 (70), as described previously (54). The average intake of dietary components was calculated using the average across all at-home 24-h recalls that passed quality control, as we have described previously (71). Habitual diet was estimated using the 2014 Block FFQ (NutritionQuest, Berkeley, CA) and quality checked manually by a registered dietitian as described previously (55). Diet quality was estimated using the healthy eating index (HEI) (72). The phylogenetic diversity of food and macronutrients were calculated as described previously (73).
Markers of physiological stress load.
Total physiological stress load was estimated using allostatic load (AL) score, which was calculated as reported previously (54, 74). Briefly, AL was calculated using urinary cortisol, norepinephrine, and epinephrine levels (corrected for urinary creatinine levels); resting systolic and diastolic blood pressure; overnight-fasted waist-to-hip ratio, fasting serum levels of high sensitivity C-reactive protein (hsCRP), cholesterol, and HDL cholesterol; fasting plasma levels of dehydroepiandrosterone sulfate (DHEA-S); and whole blood hemoglobin A1c (HbA1c). Stress load and AL score are positively correlated (75). AL may be more predictive of a broad range of stress-related conditions (76).
Assessment of physical activity.
Energy expenditure from physical activity was monitored as described previously (54). Briefly, individuals were monitored using a Respironics Actical accelerometer for a period of approximately 7 d between two study visits. Time spent performing physical activity at sedentary, light, moderate, and vigorous activities were averaged based on all days for which a minimum of 12 h of activity was recorded.
Blood collection.
Blood was drawn in the morning after a 12-h overnight fast (water being allowed to maintain hydration) following consumption of a standard meal the evening before (53). Plasma was collected from blood tubes that used sodium heparin or EDTA as anticoagulants that were centrifuged at 1300 × g at 4°C for 10 min. Aliquots of plasma were prepared immediately and stored in Cryo-Store vials at −80°C for further analysis.
Complete blood count (CBC) with differential.
During this 4-year recruitment period (June 2015 through July 2019), the CBC analyses were performed using whole blood (treated with EDTA as an anticoagulant) at the University of California, Davis Pathology Laboratory using a Beckman Coulter LH750/780 (prior to October 2016) or a Beckman Coulter DxH800 automated hematology analyzer, with the exception that 12 samples early in the study (prior to August 14, 2015) were analyzed on an Abbott Cell-Dyn 322 analyzer at the Western Human Nutrition Research Center (WHNRC).
Plasma C-reactive protein (CRP) measurement.
The concentration of CRP in plasma was measured using the Vplex Vascular Injury Panel 1 kit from samples diluted 1:1,000 with the MSD Sector Imager 2400 instrument (Meso Scale Discovery, Rockville, MD). Three levels of lyophilized controls were used on each plate to assess plate-to-plate variation. Mean concentration (mg/L) of duplicate wells was used for analysis.
Plasma lipopolysaccharide-binding protein (LBP) measurement.
Plasma LBP was measured from fasting heparin plasma stored at −70°C after being thawed at room temperature and centrifuged at 10,000 × g for 2 min to clarify. Samples were then diluted down to a final concentration of 1/800 and run per enzyme-linked immunosorbent assay (ELISA) kit instructions (Abnova; catalog [cat] number KA0448).
Fecal calprotectin and myeloperoxidase (MPO) measurement.
Fecal calprotectin was measured using homogenized stool specimens stored at −70°C that were first thawed at 4°C for 1 h and then brought to room temperature in small batches. Fifteen milligrams of a fecal sample were extracted to result in a 1/100 dilution with the IDK extract stool sample preparation system (Immundiagnostik) following kit instructions. Stool extracts were divided into aliquots and kept frozen at −20°C for up to 4 days. Calprotectin (Immundiagnostik; cat number K6927) and MPO (Immundiagnostik; cat number KR6630) ELISAs were run per instructions after diluting the extracts as recommended for each kit.
Statistical methods.
Individuals were divided into three ARG groups based on quartiles of total ARG abundance (sum total of all antibiotic-resistant genes). For example, low-ARG individuals were individuals with the 25th percentile or lower (up to quartile 1 [Q1]) of total ARG abundance. Medium-ARG individuals had total ARG abundance between the 25th and the 75th percentiles (Q1 to Q3). Finally, high-ARG individuals were above quartile 3, containing the most ARGs by total abundance. This clustering strategy was implemented, in part, to isolate the high-ARG and low-ARG individuals, to help reduce noise in ML classification. We suspect this strategy also reasonably reflects the natural distribution of ARGs in a population (i.e., a large group of individuals with middling ARG abundance, with smaller groups at the tails). Indeed, this distribution has been seen in a much larger (n = 2,037) cohort examining ARGs in healthy individuals (24). Furthermore, we examined other grouping strategies, such as using three equal groups (tertiles) and k-means clustering on ARG composition (data not shown). Tertile groups performed poorly in ML classification. While there was large overlap between the k-means grouping and the ARG grouping presented here, we ultimately used the quartile strategy because our questions were about the abundance of ARGs and not the ARG composition that k-means was basing the clusters on. The Shannon diversity metric, used to assess alpha diversity of antibiotic resistance genes and microbiome taxa, was calculated using the Vegan package (v.2.5-7) (77) in R (v.4.0.1) (78). Testing differences in the Shannon index between ARG clusters was done using the nonparametric Kruskal-Wallis test in base R, followed by a post hoc pairwise Dunn test from the Dunn package (v.1.3.5) (79), using the Benjamini-Hochberg method (80) to correct for multiple comparisons. Nonmetric multidimensional scaling (NMDS) ordination on Bray-Curtis distances of the microbiome was done using data from Kraken outputs and the metaMDS() function in Vegan. Overlaid vectors of bacterial families in ordination space was done using the envfit() feature from Vegan. To test for compositional differences between ARG clusters, we used a permutation multivariate analysis of variance (PERMANOVA) from adonis() function in Vegan, using 999 permutations. The equality of dispersions between ARG clusters, an assumption of PERMANOVA, was tested using the betadisper() function in Vegan. Differences in abundances of bacterial families between ARG clusters was determined using linear discriminant analysis (LDA) implemented by the program linear discriminant analysis effect size (LEfSe) (81) on the Galaxy Hutlab server (https://huttenhower.sph.harvard.edu/galaxy/), using default parameters. Comparisons based on our directed hypotheses between ARG clusters were done using an analysis of variance (ANOVA), and pairwise post hoc tests were done using the Tukey honestly significant difference (HSD) test on a subset of 246 individuals which had complete data for these comparisons. Homogeneity of variances between groups was confirmed using the Levene test from the Car package (82), and normality of residuals was assessed using the Shapiro-Wilk test. Nonnormal variables were transformed using the bestNormalize package (83) in R. Summary statistics are presented as mean ± standard deviation unless otherwise stated.
ARG abundance was also analyzed as a continuous outcome using proportional odds logistic regression (POLR) (84), which we use here as a semiparametric regression method as described in reference 85. P values were adjusted for multiple testing across ARGs using the Benjamini-Hochberg false discovery rate (FDR) controlling method (80) on a subset of 288 individuals for which we had sufficient ARG abundance data. POLR analyses were conducted using R v.4.0.1 (06 June 2020) (78). POLR models were fitted using the function polr in the R package MASS (v.7.3-53) (86). Regressions were done both crudely and covariate adjusted for age, sex, and BMI, which have been shown to impact microbiome composition (87, 88). ggPlot2 (89) was used to plot figures, and postprocessing was done in Affinity Designer (v.1.10.4) (Serif Europe Ltd.).
Machine learning analyses.
Machine learning models were built using the scikit-learn v.0.23.2 (90) implementation of a random forest classifier in python 3.7 (91). All models based on diet and lifestyle parameters (including stress-related AL and AL components) were built using a subset of 387 features (see Table S1 in the supplemental material) collected as part of the USDA Nutritional Phenotyping Study. To preserve as many samples as possible, some features with missing values were dropped from further analysis, which resulted in 187 individuals from all 3 ARG groups used in ML analysis (a comparison of characteristics between the initial cohort of 290 individuals and the machine learning subset cohort can be found in Fig. S1 in the supplemental material). The mikropml package in R (92) was used to one-hot encode categorical variables and drop features with zero variance. If features were correlated at a Spearman rho of 0.8, one of the correlated features was kept and the rest were dropped prior to random forest modeling (see Table S2 in the supplemental material). However, highly predictive features that were cocorrelated with dropped features were indicated within figures. Pairwise model comparisons were carried out, specifically low-ARG versus medium-ARG, low-ARG versus high-ARG, and a multiclass comparison between all ARG clusters. The data set was split using 70% of samples for training and 30% for testing. Within the training split, 2,000 iterations of hyperparameter tuning (n_estimators, min_sample_split, max_features, and max_samples) were done using RandomizedSearchCV. Hyperparameter tuning used 5-fold-stratified cross-validation, and models were evaluated by their balanced accuracy score. The model was assessed on the test data using confusion matrices, receiver operator curve AUC, accuracy, F1 (macro averaged), and Cohen’s kappa (Table S3). The optimum model was fit to the entire training data set and then evaluated using the testing set. The predictive power of features for the model was determined using the SHapley Additive exPlanations (SHAP) package (93) as described previously (55). For reproducibility, the random state for each step of the pipeline was set to the same value.
Dietary, lifestyle, and physiology features and their abbreviations used. Download Table S1, TXT file, 0.02 MB (18.3KB, txt) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Machine learning features which were correlated at Spearman rho of 0.80. Download Table S2, TXT file, 0.00 MB (5.8KB, txt) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Cohort characteristic differences between the initial (n = 290) and the ML subset cohort (n = 187). Download FIG S1, TIF file, 1.2 MB (1.2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data availability.
Metagenomes are deposited in NCBI Sequence Read Archive (SRA) under the study accession SRP354271. Requests for nonmetagenomic data from the USDA ARS WHNRC Nutritional Phenotyping Study used in this analysis should be made via an email to the senior WHNRC author on the publication of interest. Requests will be reviewed quarterly by a committee consisting of the study investigators. Scripts for processing raw sequence data can be found on GitHub (https://github.com/dglemay/ARG_metagenome). To aid in the reproducibility of statistical analyses and visualizations, a separate GitHub repository contains a docker container with all R packages used (https://github.com/aoliver44/ARGs_and_Diet). Machine learning scripts can be found at the same GitHub link, along with pickle files of the models used to generate these results. A docker container for the machine learning analysis in python is also provided for reproducibility.
ACKNOWLEDGMENTS
We thank Eduardo Cervantes, Ellen Bonnel, and the USDA Nutritional Phenotyping team for assisting in data collection, the USDA Bioanalytical Support Lab for sample management and stool processing and Sarah Spearman for assistance with fecal sample analysis. We thank Jules Larke and Elizabeth Chin for thoughtful discussions related to machine learning and Rachel Waymack for edits for clarity.
This research was primarily supported by USDA Agricultural Research Service grants 2032-51530-026-00D and 2032-51530-022-00-D and by the 2019 USDA-ARS Antimicrobial Resistance Funding (number 0426572). A.O. was supported by an appointment to the Research Participation Program at the Agricultural Research Service, United States Department of Agriculture, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and ARS. This research used resources provided by the SCINet project of the USDA Agricultural Research Service, ARS project number 0500-00093-001-00-D.
The clinical trial is registered online at https://www.clinicaltrials.gov/ (identifier NCT02367287).
Contributor Information
Danielle G. Lemay, Email: danielle.lemay@usda.gov.
Melinda M. Pettigrew, Yale School of Public Health
REFERENCES
- 1.Wellcome Trust. 2016. The review on antimicrobial resistance. Tackling drug-resistant infections globally: final report and recommendations. UK Government, Wellcome Trust, London, UK. [Google Scholar]
- 2.Van Boeckel TP, Glennon EE, Chen D, Gilbert M, Robinson TP, Grenfell BT, Levin SA, Bonhoeffer S, Laxminarayan R. 2017. Reducing antimicrobial use in food animals. Science 357:1350–1352. doi: 10.1126/science.aao1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pouwels KB, Dolk FCK, Smith DRM, Robotham JV, Smieszek T. 2018. Actual versus “ideal” antibiotic prescribing for common conditions in English primary care. J Antimicrob Chemother 73:19–26. doi: 10.1093/jac/dkx502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roope LSJ, Smith RD, Pouwels KB, Buchanan J, Abel L, Eibich P, Butler CC, Tan PS, Sarah Walker A, Robotham JV, Wordsworth S. 2019. The challenge of antimicrobial resistance: what economics can contribute. Science 364:eaau4679. doi: 10.1126/science.aau4679. [DOI] [PubMed] [Google Scholar]
- 5.Macfadden DR, Mcgough SF, Fisman D, Santillana M, Brownstein JS. 2018. Antibiotic resistance increases with local temperature. Nat Clim Chang 8:510–514. doi: 10.1038/s41558-018-0161-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boonyasiri A, Tangkoskul T, Seenama C, Saiyarin J, Tiengrim S, Thamlikitkul V. 2014. Prevalence of antibiotic resistant bacteria in healthy adults, foods, food animals, and the environment in selected areas in Thailand. Pathog Glob Health 108:235–245. doi: 10.1179/2047773214Y.0000000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vikesland P, Garner E, Gupta S, Kang S, Maile-Moskowitz A, Zhu N. 2019. Differential drivers of antimicrobial resistance across the world. Acc Chem Res 52:916–924. doi: 10.1021/acs.accounts.8b00643. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization. 2019. Antibacterial agents in clinical development: an analysis of the antibacterial clinical development pipeline. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 9.Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, Han C, Bisignano C, Rao P, Wool E, Johnson SC, Browne AJ, Chipeta MG, Fell F, Hackett S, Haines-Woodhouse G, Kashef Hamadani BH, Kumaran EAP, McManigal B, Agarwal R, Akech S, Albertson S, Amuasi J, Andrews J, Aravkin A, Ashley E, Bailey F, Baker S, Basnyat B, Bekker A, Bender R, Bethou A, Bielicki J, Boonkasidecha S, Bukosia J, Carvalheiro C, Castañeda-Orjuela C, Chansamouth V, Chaurasia S, Chiurchiù S, Chowdhury F, Cook AJ, Cooper B, Cressey TR, Criollo-Mora E, Cunningham M, Darboe S, Day NPJ, De Luca M, Dokova K, et al. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, Knight R, Manjurano A, Changalucha J, Elias JE, Dominguez-Bello MG, Sonnenburg JL. 2017. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357:802–806. doi: 10.1126/science.aan4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcón Ó, Lander O, McDonald J, Cox M, Walter J, Oh PL, Ruiz JF, Rodriguez S, Shen N, Song SJ, Metcalf J, Knight R, Dantas G, Dominguez-Bello MG. 2015. The microbiome of uncontacted Amerindians. Sci Adv 1:e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wibowo MC, Yang Z, Borry M, Hübner A, Huang KD, Tierney BT, Zimmerman S, Barajas-Olmos F, Contreras-Cubas C, García-Ortiz H, Martínez-Hernández A, Luber JM, Kirstahler P, Blohm T, Smiley FE, Arnold R, Ballal SA, Pamp SJ, Russ J, Maixner F, Rota-Stabelli O, Segata N, Reinhard K, Orozco L, Warinner C, Snow M, LeBlanc S, Kostic AD. 2021. Reconstruction of ancient microbial genomes from the human gut. Nature 594:234–239. doi: 10.1038/s41586-021-03532-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonnenburg ED, Sonnenburg JL. 2019. The ancestral and industrialized gut microbiota and implications for human health. Nat Rev Microbiol 17:383–390. doi: 10.1038/s41579-019-0191-8. [DOI] [PubMed] [Google Scholar]
- 14.Sonnenburg JL, Sonnenburg ED. 2019. Vulnerability of the industrialized microbiota. Science 366:eaaw9255. doi: 10.1126/science.aaw9255. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Taft DH, Maldonado-Gomez MX, Johnson D, Treiber ML, Lemay DG, DePeters EJ, Mills DA. 2019. The fecal resistome of dairy cattle is associated with diet during nursing. Nat Commun 10:4406. doi: 10.1038/s41467-019-12111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treiber ML, Taft DH, Korf I, Mills DA, Lemay DG. 2020. Pre-and post-sequencing recommendations for functional annotation of human fecal metagenomes. BMC Bioinformatics 21:74. doi: 10.1186/s12859-020-3416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lebeaux RM, Coker MO, Dade EF, Palys TJ, Morrison HG, Ross BD, Baker ER, Karagas MR, Madan JC, Hoen AG. 2021. The infant gut resistome is associated with E. coli and early-life exposures. BMC Microbiol 21:201. doi: 10.1186/s12866-021-02129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Stokholm J, Brejnrod A, Vestergaard GA, Russel J, Trivedi U, Thorsen J, Gupta S, Hjelmsø MH, Shah SA, Rasmussen MA, Bisgaard H, Sørensen SJ. 2021. The infant gut resistome associates with E. coli, environmental exposures, gut microbiome maturity, and asthma-associated bacterial composition. Cell Host Microbe 29:975–987.e4. doi: 10.1016/j.chom.2021.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Rahman SF, Olm MR, Morowitz MJ, Banfield JF. 2018. Machine learning leveraging genomes from metagenomes identifies influential antibiotic resistance genes in the infant gut microbiome. mSystems 3:e00123-17. doi: 10.1128/mSystems.00123-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pärnänen K, Karkman A, Hultman J, Lyra C, Bengtsson-Palme J, Larsson DGJ, Rautava S, Isolauri E, Salminen S, Kumar H, Satokari R, Virta M. 2018. Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat Commun 9:3891. doi: 10.1038/s41467-018-06393-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Yang Y, Ma L, Ju F, Guo F, Tiedje JM, Zhang T. 2015. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J 9:2490–2502. doi: 10.1038/ismej.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brito IL, Yilmaz S, Huang K, Xu L, Jupiter SD, Jenkins AP, Naisilisili W, Tamminen M, Smillie CS, Wortman JR, Birren BW, Xavier RJ, Blainey PC, Singh AK, Gevers D, Alm EJ. 2016. Mobile genes in the human microbiome are structured from global to individual scales. Nature 535:435–439. doi: 10.1038/nature18927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, Stedtfeld RD, Sul WJ, Stedtfeld TM, Chai B, Cole JR, Hashsham SA, Tiedje JM, Stanton TB. 2012. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci USA 109:1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu Q, Wang J, Yan Y, Roy B, Chen Y, Shang X, Dou T, Han L. 2020. Metagenomic analysis reveals the distribution of antibiotic resistance genes in a large-scale population of healthy individuals and patients with varied diseases. Front Mol Biosci 7:590018. doi: 10.3389/fmolb.2020.590018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellabaan MMH, Munck C, Porse A, Imamovic L, Sommer MOA. 2021. Forecasting the dissemination of antibiotic resistance genes across bacterial genomes. Nat Commun 12:2435. doi: 10.1038/s41467-021-22757-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kent AG, Vill AC, Shi Q, Satlin MJ, Brito IL. 2020. Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi-C. Nat Commun 11:4379. doi: 10.1038/s41467-020-18164-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reese AT, Cho EH, Klitzman B, Nichols SP, Wisniewski NA, Villa MM, Durand HK, Jiang S, Midani FS, Nimmagadda SN, O'Connell TM, Wright JP, Deshusses MA, David LA. 2018. Antibiotic-induced changes in the microbiota disrupt redox dynamics in the gut. Elife 7:e35987. doi: 10.7554/eLife.35987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litvak Y, Byndloss MX, Bäumler AJ. 2018. Colonocyte metabolism shapes the gut microbiota. Science 362:eaat9076. doi: 10.1126/science.aat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, Weir TL, Ehrentraut SF, Pickel C, Kuhn KA, Lanis JM, Nguyen V, Taylor CT, Colgan SP. 2015. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17:662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Byndloss MX, Olsan EE, Rivera-Chávez F, Tiffany CR, Cevallos SA, Lokken KL, Torres TP, Byndloss AJ, Faber F, Gao Y, Litvak Y, Lopez CA, Xu G, Napoli E, Giulivi C, Tsolis RM, Revzin A, Lebrilla CB, Bäumler AJ. 2017. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357:570–575. doi: 10.1126/science.aam9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doster E, Lakin SM, Dean CJ, Wolfe C, Young JG, Boucher C, Belk KE, Noyes NR, Morley PS. 2020. MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res 48:E561–E569. doi: 10.1093/nar/gkz1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmes ZC, Silverman JD, Dressman HK, Wei Z, Dallow EP, Armstrong SC, Seed PC, Rawls JF, David LA. 2020. Short-chain fatty acid production by gut microbiota from children with obesity differs according to prebiotic choice and bacterial community composition. mBio 11:e00914-20. doi: 10.1128/mBio.00914-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliver A, Chase AB, Weihe C, Orchanian SB, Riedel SF, Hendrickson CL, Lay M, Sewall JM, Martiny JBH, Whiteson K. 2021. High-fiber, whole-food dietary intervention alters the human gut microbiome but not fecal short-chain fatty acids. mSystems 6:e00115-21. doi: 10.1128/mSystems.00115-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Boeckel TP, Pires J, Silvester R, Zhao C, Song J, Criscuolo NG, Gilbert M, Bonhoeffer S, Laxminarayan R. 2019. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 365:eaaw1944. doi: 10.1126/science.aaw1944. [DOI] [PubMed] [Google Scholar]
- 35.Muloi D, Ward MJ, Pedersen AB, Fèvre EM, Woolhouse MEJ, van Bunnik BAD. 2018. Are food animals responsible for transfer of antimicrobial-resistant Escherichia coli or their resistance determinants to human populations? A systematic review. Foodborne Pathog Dis 15:467–474. doi: 10.1089/fpd.2017.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woolhouse M, Ward M, van Bunnik B, Farrar J. 2015. Antimicrobial resistance in humans, livestock and the wider environment. Philos Trans R Soc Lond B Biol Sci 370:20140083. doi: 10.1098/rstb.2014.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mather AE, Reid SWJ, Maskell DJ, Parkhill J, Fookes MC, Harris SR, Brown DJ, Coia JE, Mulvey MR, Gilmour MW, Petrovska L, de Pinna E, Kuroda M, Akiba M, Izumiya H, Connor TR, Suchard MA, Lemey P, Mellor DJ, Haydon DT, Thomson NR. 2013. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science 341:1514–1517. doi: 10.1126/science.1240578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward MJ, Gibbons CL, McAdam PR, van Bunnik BAD, Girvan EK, Edwards GF, Fitzgerald JR, Woolhouse MEJ. 2014. Time-scaled evolutionary analysis of the transmission and antibiotic resistance dynamics of Staphylococcus aureus clonal complex 398. Appl Environ Microbiol 80:7275–7282. doi: 10.1128/AEM.01777-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spoor LE, McAdam PR, Weinert LA, Rambaut A, Hasman H, Aarestrup FM, Kearns AM, Larsen AR, Skov RL, Fitzgerald JR. 2013. Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. mBio 4:e00356-13. doi: 10.1128/mBio.00356-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowder BV, Guinane CM, Ben Zakour NL, Weinert LA, Conway-Morris A, Cartwright RA, Simpson AJ, Rambaut A, Nübel U, Fitzgerald JR. 2009. Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc Natl Acad Sci USA 106:19545–19550. doi: 10.1073/pnas.0909285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thanner S, Drissner D, Walsh F. 2016. Antimicrobial resistance in agriculture. mBio 7:467–474. doi: 10.1128/mBio.02227-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker CT, Huynh S, Alexander A, Oliver AS, Cooper KK. 2021. Genomic characterization of Salmonella typhimurium DT104 strains associated with cattle and beef products. Pathogens 10:529. doi: 10.3390/pathogens10050529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DA, Leeming E, Gibson R, Le Roy C, Al Khatib H, Francis L, Mazidi M, Mompeo O, Valles-Colomer M, Tett A, Beghini F, Dubois L, Bazzani D, Thomas AM, Mirzayi C, Khleborodova A, Oh S, Hine R, Bonnett C, Capdevila J, Danzanvilliers S, Giordano F, Geistlinger L, Waldron L, Davies R, Hadjigeorgiou G, Wolf J, Ordovás JM, Gardner C, Franks PW, Chan AT, Huttenhower C, Spector TD, Segata N. 2021. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med 27:321–332. doi: 10.1038/s41591-020-01183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dethlefsen L, Relman DA. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA 108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paine RT, Tegner MJ, Johnson EA. 1998. Compounded perturbations yield ecological surprises. Ecosystems 1:535–545. doi: 10.1007/s100219900049. [DOI] [Google Scholar]
- 46.Krysiak R, Frysz-Naglak D, Okopień B. 2008. Current views on the role of dehydroepiandrosterone in physiology, pathology and therapy. Pol Merkur Lekarski 24:66–71. [PubMed] [Google Scholar]
- 47.Genazzani AD, Lanzoni C, Genazzani AR. 2012. Might DHEA be considered a beneficial replacement therapy in the elderly? Drugs Aging 24:173–185. doi: 10.2165/00002512-200724030-00001. [DOI] [PubMed] [Google Scholar]
- 48.Giunta S, Sergio G. 2008. Exploring the complex relations between inflammation and aging (inflamm-aging): anti-inflamm-aging remodelling of inflamm- aging, from robustness to frailty. Inflamm Res 57:558–563. doi: 10.1007/s00011-008-7243-2. [DOI] [PubMed] [Google Scholar]
- 49.Young DG, Skibinski G, Mason JI, James K. 1999. The influence of age and gender on serum dehydroepiandrosterone sulphate (DHEA-S), IL-6, IL-6 soluble receptor (IL-6 sR) and transforming growth factor beta 1 (TGF-β1) levels in normal healthy blood donors. Clin Exp Immunol 117:476–481. doi: 10.1046/j.1365-2249.1999.01003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karelis AD, Fex A, Filion ME, Adlercreutz H, Aubertin-Leheudre M. 2010. Comparison of sex hormonal and metabolic profiles between omnivores and vegetarians in pre- and post-menopausal women. Br J Nutr 104:222–226. doi: 10.1017/S0007114510000619. [DOI] [PubMed] [Google Scholar]
- 51.Kajita K, Ishizuka T, Mune T, Miura A, Ishizawa M, Kanoh Y, Kawai Y, Natsume Y, Yasuda K. 2003. Dehydroepiandrosterone down-regulates the expression of peroxisome proliferator-activated receptor γ in adipocytes. Endocrinology 144:253–259. doi: 10.1210/en.2002-220039. [DOI] [PubMed] [Google Scholar]
- 52.Plotkin BJ, Konakieva MI. 2017. Attenuation of antimicrobial activity by the human steroid hormones. Steroids 128:120–127. doi: 10.1016/j.steroids.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 53.Baldiviez LM, Keim NL, Laugero KD, Hwang DH, Huang L, Woodhouse LR, Burnett DJ, Zerofsky MS, Bonnel EL, Allen LH, Newman JW, Stephensen CB. 2017. Design and implementation of a cross-sectional nutritional phenotyping study in healthy US adults. BMC Nutr 3:79. doi: 10.1186/s40795-017-0197-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lemay DG, Baldiviez LM, Chin EL, Spearman SS, Cervantes E, Woodhouse LR, Keim NL, Stephensen CB, Laugero KD. 2021. Technician-scored stool consistency spans the full range of the bristol scale in a healthy US population and differs by diet and chronic stress load. J Nutr 151:1443–1452. doi: 10.1093/jn/nxab019. [DOI] [PubMed] [Google Scholar]
- 55.Chin EL, Van Loan M, Spearman SS, Bonnel EL, Laugero KD, Stephensen CB, Lemay DG. 2021. Machine learning identifies stool pH as a predictor of bone mineral density in healthy multiethnic US adults. J Nutr 151:3379–3390. doi: 10.1093/jn/nxab266. [DOI] [PubMed] [Google Scholar]
- 56.Rotmistrovsky K, Agarwala R. 2011. BMTagger: Best Match Tagger for removing human reads from metagenomics datasets. https://ftp.ncbi.nlm.nih.gov/pub/agarwala/bmtagger/.
- 57.Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen HC, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D, Fulton RS, Kremitzki M, Magrini V, Markovic C, McGrath S, Steinberg KM, Auger K, Chow W, Collins J, Harden G, Hubbard T, Pelan S, Simpson JT, Threadgold G, Torrance J, Wood JM, Clarke L, Koren S, Boitano M, Peluso P, Li H, Chin CS, Phillippy AM, Durbin R, Wilson RK, Flicek P, Eichler EE, Church DM. 2017. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res 27:849–864. doi: 10.1101/gr.213611.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taft DH, Liu J, Maldonado-Gomez MX, Akre S, Huda MN, Ahmad SM, Stephensen CB, Mills DA. 2018. Bifidobacterial dominance of the gut in early life and acquisition of antimicrobial resistance. mSphere 3:e00441-18. doi: 10.1128/mSphere.00441-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu H, Luo X, Qian J, Pang X, Song J, Qian G, Chen J, Chen S. 2012. FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS One 7:e52249. doi: 10.1371/journal.pone.0052249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De La Cuesta-Zuluaga J, Ley RE, Youngblut ND. 2020. Struo: a pipeline for building custom databases for common metagenome profilers. Bioinformatics 36:2314–2315. doi: 10.1093/bioinformatics/btz899. [DOI] [PubMed] [Google Scholar]
- 64.Salazar G. 2000. EcolUtils: utilities for community ecology analysis. 0.1.
- 65.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nayfach S, Pollard KS. 2015. Average genome size estimation improves comparative metagenomics and sheds light on the functional ecology of the human microbiome. Genome Biol 16:51. doi: 10.1186/s13059-015-0611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 68.Von Meijenfeldt FAB, Arkhipova K, Cambuy DD, Coutinho FH, Dutilh BE. 2019. Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol 20:217. doi: 10.1186/s13059-019-1817-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bushnell B. 2015. BBMap. https://sourceforge.net/projects/bbmap/.
- 70.Epidemiology and Genomics Research Program. 2016. Automated Self-Administered 24-Hour (ASA24®) Dietary Assessment Tool. NIH, Bethesda, MD. [Google Scholar]
- 71.Bouzid YY, Arsenault JE, Bonnel EL, Cervantes E, Kan A, Keim NL, Lemay DG, Stephensen CB. 2021. Effect of manual data cleaning on nutrient intakes using the Automated Self-Administered 24-Hour Dietary Assessment Tool (ASA24). Curr Dev Nutr 5:nzab005. doi: 10.1093/cdn/nzab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.U.S. Department of Health and Human Services. 2015. 2015–2020 Dietary guidelines for Americans, eighth edition. Department of Health and Human Services, Washington, DC. [Google Scholar]
- 73.Kable ME, Chin EL, Storms D, Lemay DG, Stephensen CB. 2022. Tree-Based analysis of dietary diversity captures associations between fiber intake and gut microbiota composition in a healthy US adult cohort. J Nutr 152:779–788. doi: 10.1093/jn/nxab430. [DOI] [PubMed] [Google Scholar]
- 74.Soltani H, Keim NL, Laugero KD. 2018. Diet quality for sodium and vegetables mediate effects of whole food diets on 8-week changes in stress load. Nutrients 10:1606. doi: 10.3390/nu10111606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mcewen BS. 2004. Protection and damage from acute and chronic stress: allostasis and allostatic overload and relevance to the pathophysiology of psychiatric disorders. Ann N Y Acad Sci 1032:1–7. doi: 10.1196/annals.1314.001. [DOI] [PubMed] [Google Scholar]
- 76.Juster R-P, McEwen BS, Lupien SJ. 2010. Allostatic load biomarkers of chronic stress and impact on health and cognition. Neurosci Biobehav Rev 35:2–16. doi: 10.1016/j.neubiorev.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 77.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2019. vegan: community Ecology Package. R package version. https://CRAN.R-project.org/package=vegan.
- 78.Zhang Z, Ersoz E, Lai C-Q, Todhunter RJ, Tiwari HK, Gore M, Bradbury PJ, Yu J, Arnett Dk Ordovas JM, Buckler ES, Cho RJ, Mindrinos M, Richards DR, Sapolsky RJ, Anderson M, Drenkard E, Dewdney J, Reuber TL, Stammers M, Federspiel N, Theologis A, Yang WH, Hubbell E, Au M, Chung EY, Lashkari D, Lemieux B, Dean C, Lipshutz RJ, Ausubel FM, Davis RW, Oefner PJ, Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES, Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES, Lenné JM, Takan JP, Mgonja MA, Manyasa EO, Kaloki P, Wanyera N, Okwadi J, et al. 2014. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 79.Dinno A. 2017. dunn.test: Dunn’s test of multiple comparisons using rank sums, version 1.3.5.
- 80.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 81.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fox J, Weisberg S. 2019. An {R} companion to applied regression, third edition. Sage Publications, Thousand Oaks, CA. [Google Scholar]
- 83.Peterson RA. 2021. Finding optimal normalizing transformations via bestNormalize. R J 13:310–329. doi: 10.32614/rj-2021-041. [DOI] [Google Scholar]
- 84.McCullagh P. 1980. Regression models for ordinal data. J R Stat Soc Ser B 42:109–142. doi: 10.1111/j.2517-6161.1980.tb01109.x. [DOI] [Google Scholar]
- 85.Liu Q, Shepherd BE, Li C, Harrell FE. 2017. Modeling continuous response variables using ordinal regression. Stat Med 36:4316–4335. doi: 10.1002/sim.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Venables WN, Ripley BD. 2002. Modern applied statistics with S. Springer, New York, NY. [Google Scholar]
- 87.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, Tito RY, Chaffron S, Rymenans L, Verspecht C, De Sutter L, Lima-Mendez G, D'hoe K, Jonckheere K, Homola D, Garcia R, Tigchelaar EF, Eeckhaudt L, Fu J, Henckaerts L, Zhernakova A, Wijmenga C, Raes J. 2016. Population-level analysis of gut microbiome variation. Science 352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 88.Park J, Kato K, Murakami H, Hosomi K, Tanisawa K, Nakagata T, Ohno H, Konishi K, Kawashima H, Chen YA, Mohsen A, Zhong Xiao J, Odamaki T, Kunisawa J, Mizuguchi K, Miyachi M. 2021. Comprehensive analysis of gut microbiota of a healthy population and covariates affecting microbial variation in two large Japanese cohorts. BMC Microbiol 21:151. doi: 10.1186/s12866-021-02215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wickham H. 2016. ggplot2: elegant graphics for data analysis. J R Stat Soc Ser A Stat Soc 174:245. [Google Scholar]
- 90.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830. [Google Scholar]
- 91.van Rossum G, Drake FL. 2009. Python 3 reference manual. CreateSpace, Scotts Valley, CA. [Google Scholar]
- 92.Topçuoğlu B, Lapp Z, Sovacool K, Snitkin E, Wiens J, Schloss P. 2021. mikropml: user-friendly R package for supervised machine learning pipelines. J Open Source Softw 6:3073. doi: 10.21105/joss.03073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lundberg SM, Lee S-I. 2017. A unified approach to interpreting model predictions. Adv Neural Inf Process Syst 16:426–430. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequencing read loss due to QC steps. Download Table S4, XLSX file, 0.01 MB (10.5KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(A) ARG composition (top) and taxonomic assignments (bottom, using CAT) of most abundant contigs to which ARG reads map. (B) Alpha diversity (evenness, richness, and Shannon diversity) of ARG genes across different ARG clusters. Download FIG S2, TIF file, 2.8 MB (2.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
LDA analyses identifying bacterial families that vary in abundance between ARG clusters. Download FIG S3, TIF file, 2.7 MB (2.7MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(A) Regression analysis of the association between habitual calorie intake and the gene aminoglycoside-O-phosphotransferase (aph3-dprime) (no covariate adjustment). (B) The association between soluble fiber intake and multimetal resistance (no covariate adjustment). Download FIG S4, TIF file, 1.1 MB (1.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Machine learning model performance metrics. Download Table S3, XLSX file, 0.01 MB (11.2KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(A) Significantly decreased DHEA-S levels in low-ARG compared with that of medium- and high-ARG. (B) Most measured DHEA-S values fall within the normal clinical range. Solid lines indicate lower bounds of normal DHEA-S for age and sex. Dashed lines indicate upper bounds. Download FIG S5, TIF file, 1.4 MB (1.4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Dietary, lifestyle, and physiology features and their abbreviations used. Download Table S1, TXT file, 0.02 MB (18.3KB, txt) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Machine learning features which were correlated at Spearman rho of 0.80. Download Table S2, TXT file, 0.00 MB (5.8KB, txt) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Cohort characteristic differences between the initial (n = 290) and the ML subset cohort (n = 187). Download FIG S1, TIF file, 1.2 MB (1.2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
Metagenomes are deposited in NCBI Sequence Read Archive (SRA) under the study accession SRP354271. Requests for nonmetagenomic data from the USDA ARS WHNRC Nutritional Phenotyping Study used in this analysis should be made via an email to the senior WHNRC author on the publication of interest. Requests will be reviewed quarterly by a committee consisting of the study investigators. Scripts for processing raw sequence data can be found on GitHub (https://github.com/dglemay/ARG_metagenome). To aid in the reproducibility of statistical analyses and visualizations, a separate GitHub repository contains a docker container with all R packages used (https://github.com/aoliver44/ARGs_and_Diet). Machine learning scripts can be found at the same GitHub link, along with pickle files of the models used to generate these results. A docker container for the machine learning analysis in python is also provided for reproducibility.