ABSTRACT
The outer membrane (OM) of Gram-negative bacteria is an essential organelle that acts as a formidable barrier to antibiotics. Increasingly prevalent resistance to existing drugs has exacerbated the need for antibiotic discovery efforts targeting the OM. Acylated proteins, known as lipoproteins, are essential in every pathway needed to build the OM. The central role of OM lipoproteins makes their biogenesis a uniquely attractive therapeutic target, but it also complicates in vivo identification of on-pathway inhibitors, as inhibition of OM lipoprotein biogenesis broadly disrupts OM assembly. Here, we use genetics to probe the eight essential proteins involved in OM lipoprotein maturation and trafficking. We define a biological signature consisting of three simple assays that can characteristically identify OM lipoprotein biogenesis defects in vivo. We find that several known chemical inhibitors of OM lipoprotein biogenesis conform to the biological signature. We also examine MAC13243, a proposed inhibitor of OM lipoprotein biogenesis, and find that it fails to conform to the biological signature. Indeed, we demonstrate that MAC13243 activity relies entirely on a target outside of the OM lipoprotein biogenesis pathway. Hence, our signature offers simple tools to easily assess whether antibiotic lead compounds target an essential pathway that is the hub of OM assembly.
KEYWORDS: antibiotics, cell envelope, lipoproteins, outer membrane, stress response
INTRODUCTION
Since the advent of antibiotics, treatment of infection has been a race against time. Once antibiotics are introduced clinically, bacteria often quickly develop resistance. Antibiotic discovery efforts, with an emphasis on novel bacterial targets, are essential to the continuation of the current medical treatment model for curing infections. Resistance among Gram-negative pathogens is particularly concerning, as discovery of novel antibiotic classes targeting these bacteria has proved to be especially difficult (1). Gram-negative bacteria, such as Escherichia coli, have an outer membrane (OM) that acts as a selective permeability barrier against extracellular onslaughts, such as host immune factors and antibiotics (2). Thus, the OM is a prime antibiotic target, both because it is essential and because it is a protective barrier, leading many recent antibiotic discovery efforts to focus on OM biogenesis (3, 4).
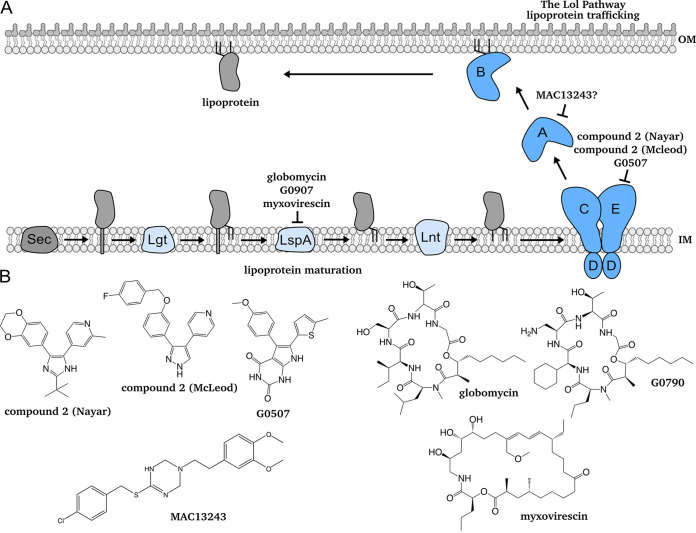
The OM is an asymmetric lipid bilayer. The inner leaflet consists of phospholipids, while the outer leaflet primarily consists of lipopolysaccharide (LPS) (5). Construction of the OM requires specialized machinery, particularly because highly hydrophobic proteins and lipids must, somehow, cross an aqueous periplasm (Fig. 1) (6). Three machines are largely responsible for OM biogenesis: the lipopolysaccharide transport (Lpt) machine shuttles LPS to the OM (7), the β-barrel assembly machine (Bam) folds β-barrel proteins into the OM (8), and the localization of lipoprotein (Lol) pathway traffics lipoproteins to the OM (9). Notably, Bam, Lpt, and Lol require at least one essential OM lipoprotein component: BamD, LptE, and LolB, respectively (10–12). Thus, OM lipoprotein biogenesis, comprised of the lipoprotein maturation and trafficking pathways, is key to construction and integrity of the OM.
FIG 1.
Lipoprotein maturation and trafficking and outer membrane (OM) lipoprotein biogenesis inhibitors. (A) Lipoproteins exit the cytoplasm via the Sec translocon, where they are tethered by their signal sequence in the inner membrane (IM). Before they are trafficked to the OM, lipoproteins must be modified by a series of lipoprotein maturation enzymes in the IM. Lipoproteins undergo sequential modifications by Lgt, LspA, and Lnt. Modified, triacylated lipoproteins are extracted by LolCDE. LolA receives lipoproteins from LolC, shielding their hydrophobic acyl chains as it traffics them across the aqueous periplasm. At the OM, LolB receives and inserts lipoproteins. This work focuses on two known compounds that inhibit lipoprotein maturation and trafficking: globomycin inhibits LspA, while compound 2 inhibits LolCDE. (B) Chemical structures of known and proposed inhibitors of OM lipoprotein biogenesis, as depicted in A.
All lipoproteins are synthesized in the cytoplasm then translocated across the cytoplasmic membrane. Lipoproteins destined for the OM must undergo a series of sequential modifications in the inner membrane (IM) before they are trafficked to the OM (Fig. 1) (13, 14). First, the enzyme Lgt transfers a diacylglyceryl moiety from phosphatidylglycerol to an invariant cysteine of a target lipoprotein (15, 16). Next, the type II signal peptidase LspA cleaves the signal peptide (17). Finally, the acyltransferase Lnt adds a third acyl chain from phosphatidylethanolamine to the now N-terminal cysteine, producing a mature lipoprotein (18, 19). Lipoprotein maturation enzymes are highly conserved and essential among Gram-negative bacteria. However, some species can remain viable without lnt in laboratory conditions (20, 21).
A mature lipoprotein is trafficked to the OM if it contains residues specifying an OM localization signal, which varies across species (22–24). An ATP-binding cassette (ABC) transporter (LolCDE in E. coli) extracts mature, OM-targeted lipoproteins from the IM (25). Then, the chaperone LolA receives lipoproteins from LolC, shielding their hydrophobic acyl chains from the aqueous periplasm (26, 27). Finally, the OM lipoprotein LolB receives lipoproteins from LolA and inserts them into the OM (26). Many clinically important species produce LolB, although some Gram-negative species lack a clear homolog (9). A LolAB-independent trafficking mechanism also exists, although it alone cannot support viability in wild-type E. coli (28).
As OM assembly relies on lipoproteins, OM lipoprotein biogenesis is a crucial target for novel antibacterials. This pathway requires up to eight essential and conserved proteins, offering an array of potential therapeutic targets. In fact, a recent CRISPRi screen of the essential genes of Vibrio cholerae found that depletion of genes in the Lol trafficking pathway caused a more severe decrease in viability than any other essential genes (29).
Lipoproteins play an essential role in OM assembly, complicating unambiguous identification of OM lipoprotein biogenesis inhibitors in vivo. Inhibitors of OM lipoprotein biogenesis will wreak widespread havoc on β-barrel assembly, LPS transport, and cell wall biosynthesis. Lipoprotein trafficking and OM biogenesis are so entwined that lipoprotein trafficking inhibitors have emerged from screens designed to identify inhibitors of cell wall synthesis (30) and activators of σE, a monitor of β-barrel assembly (31).
In vivo target validation of new compounds active against essential pathways remains challenging. No protocol to validate inhibition of lipoprotein maturation or trafficking factors exists. In this work, we define a unique biological signature for target validation of OM lipoprotein biogenesis inhibitors in E. coli. Our signature consists of three biological effects that, collectively, are hallmarks of defective OM lipoprotein biogenesis: (i) increased OM permeability, (ii) toxicity of the major OM lipoprotein Lpp, and (iii) activation of the Cpx envelope stress response by a sensory OM lipoprotein, NlpE. We validate this signature using genetic depletions and chemical inhibitors (compound 2, globomycin) of essential steps in OM lipoprotein biogenesis. We then demonstrate the utility of our signature by examining MAC13243, a proposed LolA inhibitor, and find that MAC13243 fails to fulfill our biological signature. Finally, using genetics, we confirm that MAC13243 bioactivity is independent of LolA.
RESULTS
Depletion of OM lipoprotein biogenesis factors causes OM permeability.
To establish a biological signature of lipoprotein maturation or trafficking inhibition, we used our current understanding of OM assembly to develop assays that report on OM lipoprotein biogenesis defects. Since at least one lipoprotein is essential to each OM biogenesis machine, we hypothesized that disrupting OM lipoprotein biogenesis would cause OM assembly defects that affect its antibiotic barrier function.
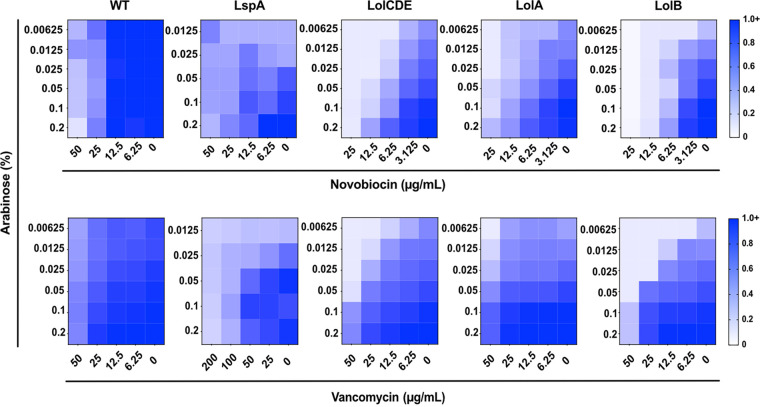
To assess OM barrier integrity when OM lipoprotein biogenesis is limited, we used a series of E. coli strains in which expression of OM lipoprotein biogenesis genes (lspA, lolCDE, lolA, and lolB) depends on arabinose induction. We chose to analyze LspA, LolCDE, and LolA as chemical inhibitors of each have been identified and characterized (30–37). We included LolB in our characterization as the response to its depletion is well characterized (38). Growth in media lacking arabinose depletes these essential proteins. We used checkerboard assays to measure sensitivity to three large scaffold antibiotics, which cannot pass through an intact OM, in response to depletion of OM lipoprotein biogenesis factors. Each antibiotic had a distinct target: novobiocin (a hydrophobic DNA gyrase inhibitor; Fig. 2), vancomycin (a hydrophilic cell wall biosynthesis inhibitor; Fig. 2), and rifampicin (a hydrophobic RNA polymerase inhibitor; Fig. S1).
FIG 2.
Depletion of lipoprotein maturation or trafficking factors causes outer membrane permeability. Strains in which LspA, LolCDE, LolA, or LolB were under an arabinose-dependent promoter were grown in decreasing concentrations of inducer and increasing concentrations of two large scaffold antibiotics, novobiocin and vancomycin. Depletion of any OM lipoprotein biogenesis factor tested caused increased sensitivity to large scaffold antibiotics. Arabinose did not affect the sensitivity of wild type (WT) to large scaffold antibiotics. Data are from three independent experiments. Averaged density (A600nm) values of antibiotic-treated cultures relative to mock-treated control (set as 1.0) are shown.
Depletion of lipoprotein maturation or trafficking factors causes outer membrane permeability. Strains in which the only copy of lspA, lolCDE, lolA, or lolB was under an arabinose inducible promoter were grown with increasing concentrations of inducer and the large scaffold antibiotics rifampicin and erythromycin. Depletion increased outer membrane (OM) permeability, as measured by sensitivity to large scaffold antibiotics. Data are from three independent experiments; averaged density (A600nm) values of antibiotic-treated cultures relative to mock-treated control (set as 1.0) are shown. Download FIG S1, TIF file, 0.3 MB (308.7KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
As we depleted each protein, sensitivity to large scaffold antibiotics increased (Fig. 2 and Fig. S1). We observed variation in the extent of antibiotic sensitivity caused by depletion of each OM lipoprotein biogenesis factor, likely reflecting differing levels of depletion achievable with each construct. Nonetheless, depleting OM lipoprotein biogenesis increased OM permeability to antibiotics. Interestingly, recent work found that novobiocin has an additional target in the Lpt machine, LptB (39). As LPS transport is integral to OM integrity, this could exacerbate the permeabilization seen upon depletion of OM lipoprotein biogenesis factors. Additionally, the permeabilizing effect was compound specific, as decreasing induction of lspA, lolCDE, lolA, or lolB did not sensitize cells to erythromycin (a hydrophobic macrolide inhibitor of translation; Fig. S1). Selective permeability caused by OM assembly mutants was previously observed and remains poorly understood (40). Our data confirm that defects in OM lipoprotein biogenesis weaken the integrity of the OM barrier.
Loss of Lpp alleviates OM lipoprotein biogenesis defects.
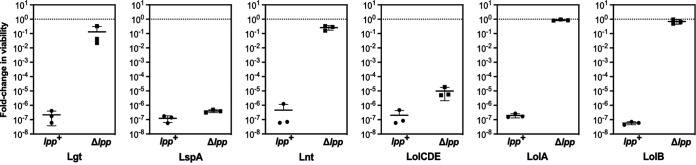
In addition to disrupting OM construction, OM lipoprotein biogenesis defects cause IM mislocalization of OM-targeted lipoproteins, which can be toxic (28). One such example is the OM lipoprotein Lpp, which covalently cross-links to the cell wall from the OM, providing important architectural stability to the cell envelope (41–43). When Lpp is not trafficked efficiently, it accumulates in the IM and errantly cross-links to peptidoglycan. Lpp cross-linking from the IM is lethal (44). Hence, although lpp is not essential, efficient OM lipoprotein biogenesis of Lpp is essential. We reasoned that Δlpp would prevent toxicity, increasing viability when OM lipoprotein biogenesis is limited. We assessed the viability of LspA-, LolCDE-, LolA-, or LolB-depleted strains in the presence or absence of lpp using arabinose-inducible constructs (Fig. 3). Each gene is essential in both lpp+ and Δlpp backgrounds; therefore, inducer-independent growth in these strains relies on leaky expression of the gene construct. Tight regulation of the LspA-depletion strain was previously demonstrated, and thus, viability of the LspA strain was measured in the presence of a low level of inducer (34). Viability of all other constructs was measured in the absence of inducer.
FIG 3.
Deletion of lpp protects against lipoprotein maturation and trafficking defects. Relative viability of strains with arabinose-dependent expression of OM lipoprotein biogenesis proteins (Lgt, LspA, Lnt, LolCDE, LolA, or LolB). Viable counts per mL of culture were enumerated in the presence of 0.2% arabinose or in the absence of arabinose and used to determine the fold change in viability when inducer is absent. For LspA strains, the comparison was made between arabinose replete conditions (0.2%) and arabinose deplete (0.002%) conditions. Data represent three independent experiments and the mean.
Depletion of any OM lipoprotein biogenesis factor severely reduced viability of wild-type E. coli, as expected for essential genes. In all instances, Δlpp improved viability without inducer. While Δlpp caused striking increases in viability in Lgt-, Lnt-, LolA-, and LolB-depleted cells, we measured only modest increases in viability in LspA- and LolCDE-depleted cells. The variation in the alleviation of toxicity in Δlpp strains likely reflects the dissimilar levels of depletion achievable with each gene construct, with little leaky expression of LspA or LolCDE. Additionally, the LspA construct is expressed from a p15a medium copy number plasmid whose instability likely plays a role in the relatively tight depletion that we observe (34). Importantly, Δlpp did not improve viability when essential components of the Bam and Lpt machines (BamD and LptE) were depleted (Fig. S2). On the contrary, there was a modest decrease in viability of the BamD- and LptE-depletion strains in the absence of Lpp. Therefore, Δlpp does not alleviate cell envelope defects in other essential OM assembly pathways. Rather, our data show that Δlpp specifically improves the viability of cells when OM lipoprotein biogenesis is depleted.
Deletion of lpp does not protect against general OM biogenesis defects. Relative viability of strains with arabinose-dependent expression of bamD or lptE. Viable counts per mL of culture were enumerated in the presence of 0.2% arabinose or in the absence of arabinose and used to determine the fold change in viability when inducer is absent. Data represent three independent experiments and the mean. Download FIG S2, TIF file, 0.1 MB (84.8KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Depletion of OM lipoprotein biogenesis causes NlpE-dependent activation of Cpx.
A series of stress responses monitor OM and cell envelope integrity (45). Among these is Cpx, a two-component system comprised of the histidine kinase CpxA and the response regulator CpxR (46). Together, CpxAR respond to OM perturbations and various other cellular signals (47). Cpx was recently shown to alleviate stress caused by defects in late steps of lipoprotein trafficking (28, 48, 49). We hypothesized that defective OM lipoprotein biogenesis would similarly activate Cpx, marking a signature of OM lipoprotein biogenesis stress.
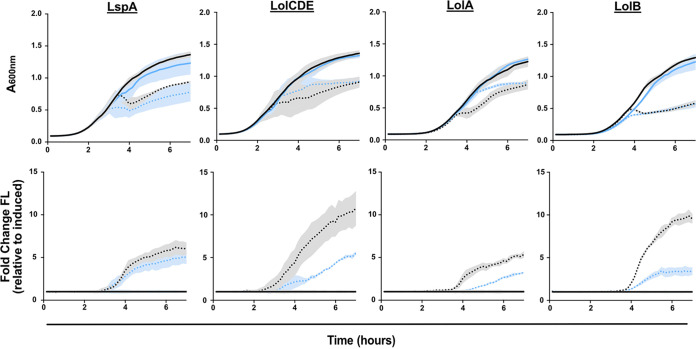
To assess Cpx activation when OM lipoprotein biogenesis is defective, we used a reporter plasmid carrying a transcriptional green fluorescent protein (gfp) fusion to the promoter of the CpxAR-regulated gene cpxP (PcpxP-gfp). The plasmid was introduced into the LspA-, LolCDE-, LolA-, and LolB-depletion strains. We monitored GFP fluorescence as each OM lipoprotein biogenesis factor was depleted during subculture without inducer (Fig. 4). As expected, depletion of each OM lipoprotein biogenesis factor reduced growth. As growth slowed, we detected strong increases in fluorescence from PcpxP-gfp (Fig. 4), indicating activation of Cpx.
FIG 4.
Depletion of lipoprotein maturation or trafficking causes NlpE-dependent activation of the Cpx stress response. Strains carrying PcpxP-gfp and inducible LspA, LolCDE, LolA, or LolB were grown with (solid) or without (dots) inducer (0.2% arabinose). Culture density (A600nm; top) and fluorescence (FL) were measured to calculate fluorescence per cell (fluorescence/A600nm; bottom). Strains were tested in the presence (black) or absence (blue) of nlpE. Data are average ± SD; n = 3.
As a variety of stimuli activates Cpx, we wanted to test whether the observed rapid and potent Cpx activation was specific to OM lipoprotein biogenesis defects. Recent work proposed that Cpx activation in response to defects in late OM lipoprotein biogenesis is due to mislocalization of the OM sensor lipoprotein NlpE to the IM (48, 49). We reasoned that if the observed Cpx activation was caused by sensing OM lipoprotein biogenesis defects, the early, strong Cpx activation would be NlpE-dependent. Hence, we deleted nlpE from our depletion strains and monitored expression from PcpxP-gfp. Growth of all strains was similar in the presence and absence of nlpE. Importantly, deletion of nlpE decreased fluorescence upon depletion of LolCDE, LolA, or LolB, indicating NlpE-dependent Cpx activation. We did not observe clear NlpE-dependent Cpx activation in the LspA strain, likely a product of the construct’s tight repression and instability causing rapid, widespread OM defects that cause generalized activation of Cpx and other cell envelope stress responses. However, as depletion of LolCDE, LolA, or LolB causes NlpE-dependent Cpx activation, we conclude that this is a strong indicator of OM lipoprotein biogenesis limitation.
Stress responses with OM lipoprotein sensors are activated by limited OM lipoprotein biogenesis.
Two other envelope stress responses monitor OM defects: Rcs and σE. Rcs monitors defects through the OM lipoprotein RcsF (50), while σE directly detects misfolded β-barrel proteins. We reasoned that OM lipoprotein biogenesis defects would lead to early activation of Rcs but would not activate σE, as no lipoprotein is involved in the σE response. To assess Rcs and σE activation, we introduced reporter plasmids carrying a transcriptional gfp fusion to the Rcs-responsive osmB promoter (PosmB-gfp) or the σE-dependent micA promoter (PmicA-gfp) (Fig. S3) (51, 52). We also constructed a control plasmid expressing GFP from a housekeeping RpoD-dependent promoter (PrpoD-gfp) to control for artifactual increases in fluorescence (Fig. S3). We introduced each plasmid into the LspA-, LolCDE-, LolA-, and LolB-depletion strains and measured growth and GFP fluorescence during depletion. We observed increases in fluorescence from PosmB-gfp when OM lipoprotein biogenesis factors were depleted, indicating Rcs activation. Conversely, depletion did not strongly activate PmicA-gfp, with the exception of LspA. The activation of PmicA-gfp in the LspA-depletion background illustrates the generalized OM defects caused by tight depletion of the LspA protein. None of the depletion strains caused strong activation of the control reporter PrpoD-gfp. Thus, we propose that the specific activation of Cpx and Rcs is a strong indicator of OM lipoprotein biogenesis inhibition, while an absence of σE activation is important for discrimination between specific OM lipoprotein defects and generalized cell envelope defects.
Depletion of lipoprotein maturation or trafficking activates the Rcs stress response but does not activate general OM stress responses. Inducible LspA, LolCDE, LolA, or LolB strains carrying PosmB-gfp, PmicA-gfp, or PrpoD-gfp were grown with (black solid) or without (blue dots) inducer. Culture density (A600nm) and fluorescence were measured to calculate fluorescence per cell (fluorescence/A600nm). Data are average ± SD; n = 3. Download FIG S3, TIF file, 0.7 MB (739.2KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Chemical inhibitors of OM lipoprotein biogenesis conform to the biological signature.
Genetic depletions allowed us to establish three signature hallmarks of defects in OM lipoprotein biogenesis: (i) OM permeabilization, (ii) Lpp toxicity, and (iii) NlpE-specific activation of Cpx. We next tested our signature using chemical inhibition of the OM lipoprotein biogenesis pathway. Several compounds targeting OM lipoprotein biogenesis have been described. Three compounds target LspA: globomycin (36, 37), G0790 (35), and myxovirescin (53) (Fig. 1). We chose globomycin as a representative LspA inhibitor, as G0790 is an analog of globomycin and as mutations in Lpp confer resistance to globomycin, G0790, and myxovirescin, indicating functional overlap in all three compounds. Multiple compounds targeting LolCDE have also been described, and all three compounds share structural similarities (3, 30–32) (Fig. 1). Mutations in LolCDE confer pan-resistance to each of these compounds (30–32). We chose the pyridine-imidazole “compound 2 (McLeod)” as a representative inhibitor of LolCDE function (32).
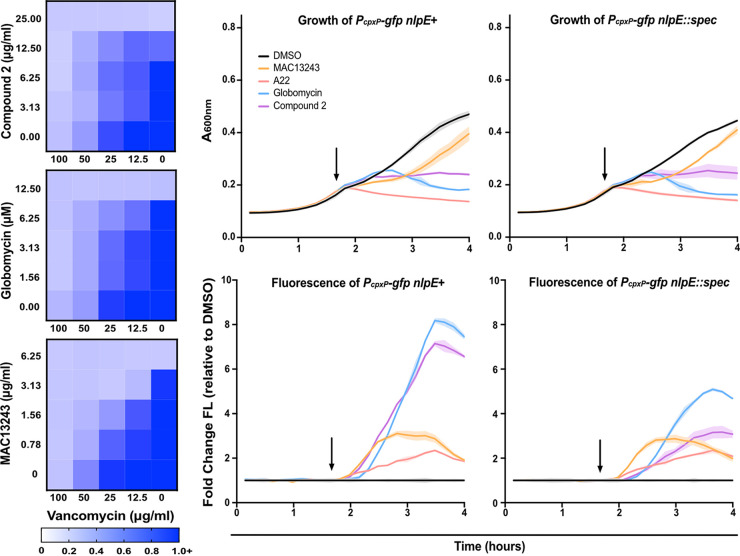
To probe OM permeability, we assessed sensitivity to large scaffold antibiotics upon treatment with globomycin or compound 2 using checkerboard assays. As globomycin and compound 2 poorly penetrate E. coli, we tested a ΔtolC strain in which antibiotic efflux is inactivated. A ΔtolC strain should allow cellular accumulation of both compounds. Treatment with globomycin or compound 2 sensitized cells to vancomycin, indicating decreased integrity of the OM barrier (Fig. 5). Thus, both compounds satisfied the first criterium of the proposed biological signature.
FIG 5.
Chemical inhibitors of lipoprotein maturation or trafficking fit the expected profile of OM lipoprotein biogenesis inhibition. (Left) OM permeability was assessed in efflux defective mutants (ΔtolC) by treatment with increasing concentrations of vancomycin and compound 2 (top), globomycin (middle), or MAC13243 (bottom). Data are from three independent experiments. Averaged density (A600nm) values of antibiotic-treated cultures relative to mock-treated control (set as 1.0) are shown. (Right) Culture density (A600nm; top) and fluorescence were used to calculate fluorescence per cell (bottom) in ΔtolC strains with native nlpE or with a chromosomal deletion of nlpE (nlpE::spec). Cells were treated after 100 min (arrow), with DMSO (black), MAC13243 (orange), A22 (red), globomycin (blue), or compound 2 (purple). Data are average ± SD; n = 3.
We next tested whether Δlpp was protective against treatment with either inhibitor. In agreement with previous work, Δlpp increased the MIC of globomycin and compound 2 (Table 1) (32, 54). Therefore, both compounds fulfill the second criterium of our signature.
TABLE 1.
Deletion of lpp increases resistance to OM lipoprotein biogenesis inhibitors
| Strain | Globomycin (μM) | Compound 2 (μg/mL) | A22 (μg/mL) | MAC13243 (μg/mL) |
|---|---|---|---|---|
| lpp+ | 12.5 | 10 | 2.5 | 2.5 |
| Δlpp | >50 | >40 | 2.5 | 2.5 |
OM, outer membrane.
Finally, we tested stress-response activation upon treatment with both inhibitors. Strains were treated with globomycin or compound 2 after 100 min of growth. Both compounds inhibited growth similarly. Almost immediately after treatment with either compound, we detected a strong increase in fluorescence from a PcpxP-gfp reporter (Fig. 5). Deletion of nlpE delayed and strongly reduced GFP fluorescence upon treatment with either compound. We found that globomycin induced fluorescence from Rcs-activated PosmB-gfp, consistent with prior observations (Fig. S4). Treatment with compound 2 also increased fluorescence of PosmB-gfp (Fig. S4). However, both globomycin and compound 2 caused little activation of the σE-dependent micA promoter (PmicA-gfp) (Fig. S4). Thus, both globomycin and compound 2 satisfy all three of our criteria and fully conform to our biological signature. Collectively, our data demonstrate that OM lipoprotein biogenesis defects, whether induced genetically or with chemical inhibitors, produce a distinctive biological signature of OM lipoprotein biogenesis inhibition.
Chemical inhibitors of OM lipoprotein biogenesis cause Rcs activation but do not activate σE or RpoD. Stress response activation after treatment (arrow) with DMSO (black), MAC13243 (orange), A22 (red), globomycin (blue), or compound 2 (purple) was assessed in a ΔtolC background. Culture density (A600nm, top) and fluorescence were used to calculate fluorescence per cell (A600nm/Fluorescence) in strains carrying PosmB-gfp, PmicA-gfp, or PrpoD-gfp. Data are average ± SD; n = 3. Download FIG S4, TIF file, 0.2 MB (193.6KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proposed LolA inhibitor MAC13243 does not fit the biological signature.
Recent work proposed that MAC13243 inhibits LolA (33). Indeed, MAC13243 permeabilizes E. coli to large scaffold antibiotics (55), and LolA over-production protects against MAC13243 (33). Curiously, MAC13243 degrades into a thiourea compound closely related to A22, an inhibitor of the essential cytoskeletal protein MreB (56–58). In vitro, MAC13243 and A22 interact with purified LolA (58). However, clear in vivo LolA inhibition has yet to be demonstrated. We hypothesized that, if they target LolA in vivo, both MAC13243 and A22 would fit our biological signature.
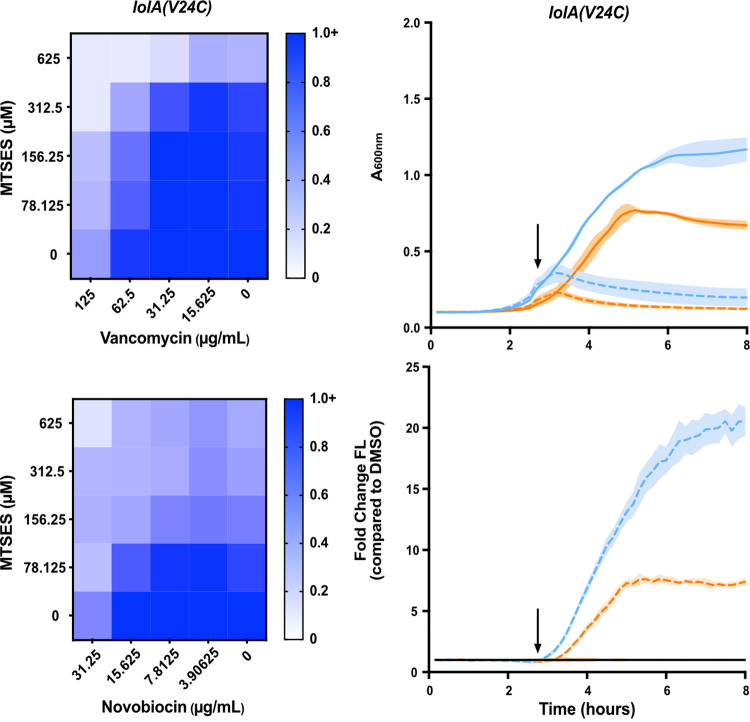
As a control for LolA inhibition, we also designed an allele-specific system for inhibiting LolA. First, we introduced a V24C substitution in LolA. The V24 residue is proposed to be important to lipoprotein binding by LolA (26). A plasmid carrying lolA(V24C)-complemented deletion of native lolA, indicating that the mutation does not reduce LolA activity. To inhibit LolA(V24C), we treated cells with the thiol-reactive compound 2-[(methylsulfonyl)thio]-ethanesulfonic acid (MTSES). Previous work illustrated that, despite the potential effect of MTSES on any thiol group in the cell, introduction of cysteines at key sites causes protein-specific sensitivity to MTSES (59, 60). We reasoned that introduction of a cysteine at the V24 residue would provide an MTSES target within a region known to be functionally important to LolA and would potentially impair LolA. Indeed, lolA(V24C) was more sensitive to MTSES than lolA+ (Fig. S5). Treatment with MTSES caused only minor growth defects in the lolA+ strain, yet the same treatment was lethal in the lolA(V24C) strain (Fig. S5). Hence, MTSES allowed us to semiselectively inhibit LolA in strains producing the V24C variant.
2-[(Methylsulfonyl)thio]-ethanesulfonic acid (MTSES) inhibits the activity of LolA(V24C) and resistance to MTSES increases in the absence of Lpp. (A) Growth (A600nm) of strains carrying either lolA+ (black) or lolA(V24C) (blue) was measured upon treatment with MTSES. In early log phase (arrow), strains were treated with 0.5 mM MTSES (dotted) or vehicle control (1% DMSO) (solid). Data are average ± SD; n = 3. (B) MICs of strains carrying either lolA+ or lolA(V24C) were assessed in the presence (lpp+) and absence (Δlpp) of Lpp. Cultures were grown in 24-well plates (n = 3). Download FIG S5, TIF file, 0.2 MB (165.3KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We tested MAC13243, A22, and our allele-specific MTSES inhibitor system using our biological signature. Treatment with MAC13243 increased sensitivity to novobiocin, vancomycin, and rifampicin, indicating increased OM permeability (Fig. 5). This is in keeping with previous observations that MAC13243 permeabilizes E. coli to vancomycin (55) and observations that A22 permeabilizes E. coli to novobiocin (61). Increasing concentrations of MTSES also increased sensitivity of LolA(V24C) to large scaffold antibiotics (Fig. 6; Fig. S6).
FIG 6.
An allele specific inhibitor of LolA causes OM permeability and activation of the Cpx stress response. (Left) OM permeability to vancomycin and novobiocin was assessed in a strain carrying lolA(V24C) upon treatment with increasing concentrations of 2-[(methylsulfonyl)thio]-ethanesulfonic acid (MTSES). Data are from three independent experiments. Averaged density (A600nm) values of antibiotic-treated cultures relative to mock-treated control (set as 1.0) are shown. (Right) Growth (A600nm) of strains carrying lolA(V24C) and a Cpx reporter plasmid (PcpxP-gfp) was measured. Strains either had native nlpE (blue) or were ΔnlpE::spec (orange). In early log phase (arrow), strains were treated with 0.5 mM MTSES (dotted) or vehicle control (1% DMSO) (solid). Fluorescence was measured and normalized to A600nm to calculate fluorescence per cell. Reporter values were normalized to a DMSO-treated control. Data are average ± SD; n = 3.
An allele specific inhibitor of LolA causes OM permeability to antibiotics. OM permeability to vancomycin, novobiocin, rifampicin, and erythromycin was assessed in lolA+ strains upon treatment with increasing amounts of MTSES (top). OM permeability to rifampicin and erythromycin were also assessed in a strain carrying lolA(V24C) upon increasing concentrations of MTSES (bottom). Data are from three independent experiments; averaged density (A600nm) values of antibiotic-treated cultures relative to mock-treated control (set as 1.0) are shown. Download FIG S6, TIF file, 0.2 MB (248.8KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Since Δlpp vastly improves viability when LolA is depleted, we expected Δlpp would make E. coli more tolerant to a compound targeting LolA. Indeed, Δlpp increased the MIC of MTSES in the lolA(V24C) strain (Fig. S5). However, Δlpp had no effect on the MIC of MAC13243 or A22 (Table 1). This suggests that toxic mislocalization of Lpp is not a significant contributor to the lethality of MAC13243 or A22.
Next, we evaluated Cpx activation upon chemical inhibition of LolA. Treatment of lolA(V24C) with MTSES caused rapid growth arrest and strong activation of Cpx, just as we observed upon LolA depletion. Moreover, this activation was clearly NlpE-dependent (Fig. 6; Fig. S7). Treatment with MAC13243 or A22 also caused rapid growth inhibition and strong Cpx activation (Fig. 5). However, Cpx activation in response to MAC13243 or A22 treatment was entirely NlpE independent (Fig. 5). Analysis of Rcs activation showed that MTSES induced Rcs in a nonallele-specific manner (Fig. S8). Both MAC13243 and A22 caused delayed Rcs activation (Fig. S4).
MTSES treatment activates the Cpx stress response. Cpx activation upon treatment with MTSES was assessed in a lolA+ background. Growth (A600nm) of the lolA+ strains carrying a Cpx reporter plasmid (PcpxP-gfp) was measured. Strains were either nlpE+ (blue) or nlpE::spec (orange). In early log phase (arrow), strains were treated with DMSO (1%) (solid) or 0.5 mM MTSES (dotted). Fluorescence per cell was calculated using fluorescence normalized to A600nm. Reported values are normalized to a DMSO-treated control. Data are average ± SD; n = 3. Download FIG S7, TIF file, 0.2 MB (166.6KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MTSES treatment causes activation of the Rcs stress response. Cpx activation upon treatment with MTSES was assessed in lolA+ (blue) and lolA(V24C) (orange) strains carrying PosmB-gfp, PmicA-gfp, or PrpoD-gfp. Growth (A600nm) and fluorescence were measured after treatment with DMSO (1%) (solid) or 0.5 mM MTSES (dotted) in early log phase. Fluorescence per cell was calculated by normalizing fluorescence to A600nm. Reported fluorescence values are normalized to a DMSO-treated control. Cultures were grown in 96-well, black-walled plates. Data are average ± SD; n = 3. Download FIG S8, TIF file, 0.2 MB (227.7KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Thus, MAC13243 and A22 fail to meet the biological signature of OM lipoprotein biogenesis inhibition. Deletion of lpp does not alleviate lethal effects of either compound, and both compounds activate Cpx in an NlpE-independent manner. Our data suggest that treatment with these compounds does not appreciably inhibit OM lipoprotein biogenesis, suggesting LolA is not inhibited in vivo.
MAC13243 activity is LolA independent.
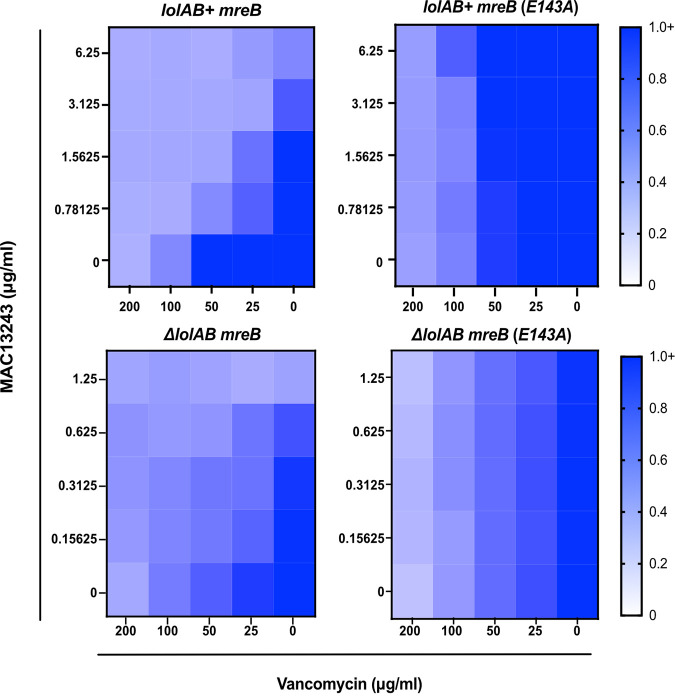
We sought to conclusively assess if the biological activity of MAC13243 occurs through inhibition of LolA in vivo. While lolA is essential in wild-type E. coli, genetic conditions exist under which both lolA and lolB can be deleted (28). As LolA and LolB work in concert, we examined the activity of MAC13243 in a strain lacking both LolA and LolB (ΔlolAB). We expected that MAC13243 would affect cells that produce LolA and LolB (lolAB+) but would not show activity in cells that lack the proposed LolA target (ΔlolAB). Surprisingly, we observed that MAC13243 causes OM permeabilization even in the absence of LolA (Fig. 7). Thus, the OM-permeabilizing effect of MAC13243 is not dependent on LolA inhibition.
FIG 7.
MAC13243 activity is independent of LolA. To test MAC13243 activity on LolA, OM permeability of strains in which chromosomal lolAB (lolAB+) are present or absent (ΔlolAB) was assessed. Strains carried either wild-type mreB or mreB(E143A) and were assessed upon treatment with increasing concentrations of MAC13243 and vancomycin. Data are from three independent experiments. Averaged density (A600nm) values of antibiotic-treated cultures relative to mock-treated control (set as 1.0) are shown.
The inhibition of MreB by A22 is well characterized. Since MAC13243 is chemically similar to A22, we examined whether the permeabilizing effect of MAC13243 relied on MreB inhibition. An E143A substitution in MreB confers resistance to A22, likely by preventing its binding (62). Interestingly, an mreB(E143A) allele also increased resistance to MAC13243. Moreover, in a mreB(E143A) background, MAC13243 did not permeabilize the OM to large scaffold antibiotics (Fig. 7). Hence, the activity of MAC13243 was entirely dependent on a susceptible MreB protein and independent of the presence of LolA in the cell. Collectively, our data strongly argue that the in vivo target of MAC13243 is MreB, not LolA.
DISCUSSION
OM lipoprotein biogenesis is an attractive antibiotic target, as it is required for OM construction and integrity. However, there is currently no protocol for validating lipoprotein maturation or trafficking inhibitors. Herein, we establish a 3-fold biological signature of OM lipoprotein biogenesis limitation: (i) permeabilization of the OM to large scaffold antibiotics, (ii) toxicity of Lpp, and (iii) NlpE-dependent activation of Cpx. This signature can be used to validate OM lipoprotein biogenesis inhibitors in vivo. Indeed, known inhibitors fully conform to this signature.
The first parameter of our biological signature is OM permeabilization. Prior work firmly established that mutations in the Bam and Lpt machines permeabilize the OM to antibiotics (40). This property has been exploited for genetic analysis of Bam and Lpt. Our data now show that the same chemical genetic logic extends to OM lipoprotein biogenesis. Increased OM permeability is arguably the least discerning parameter in our biological signature, since permeability can be expected in response to defects in OM assembly, cell wall synthesis, or antibiotic efflux. As such, we see OM permeability as a primary classifier, which, if not satisfied, can exclude compounds that do not target OM lipoprotein biogenesis.
The second parameter of our biological signature relies on increased viability in the absence of Lpp. Notably, we show that Δlpp alleviates defects in any stage of OM lipoprotein biogenesis yet does not alleviate defects in other OM assembly pathways (Bam and Lpt). The covalent linkage between OM-localized Lpp and cell wall peptidoglycan serves an important role in cell envelope architecture (42, 43). However, when Lpp cross-links from the IM, it is lethal to the cell (44). Defects at any stage in OM lipoprotein biogenesis should cause Lpp to accumulate in the IM, and deletion of lpp prevents lethal toxicity. In fact, lpp mutations alleviate temperature sensitivity of E. coli or Salmonella lgt mutations (63, 64) and confer resistance to globomycin or LolCDE-targeting chemical inhibitors (32, 54).
Loss-of-function lpp mutations can be isolated with high frequency in the laboratory, suggesting a ready genetic route for resistance to novel therapeutics targeting OM lipoprotein biogenesis. Yet it is unclear that similar lpp mutations could be isolated in a clinical context. The absence of Lpp dysregulates the cell envelope architecture, which leads to excessive OM blebbing and hypersensitivity to detergents frequently encountered by enteric bacteria, such as bile salts (65). Indeed, Δlpp mutants survive poorly in mammalian hosts and are highly sensitive to complement-mediated immune clearance in serum (66–70). Therefore, although lpp is not essential in the laboratory, there is strong evidence to suggest that lpp is essential for infection. It is highly unlikely that lpp mutations could arise inside patients or animals treated with OM lipoprotein biogenesis inhibitors. Similarly, Acinetobacter baumannii mutants that no longer produce lipooligosaccharide can be readily isolated in the lab following colistin selection, but no such mutants have been recovered clinically from colistin-treated patients (71, 72).
A recent study described a macrocyclic peptide (G2824) that inhibits Lgt activity and is bactericidal to E. coli (73). Notably, Δlpp did not confer resistance to G2824. In fact, Δlpp-sensitized bacteria to G2824. This is unexpected in light of our data showing that Δlpp significantly increased viability of Lgt-depleted E. coli and other studies reporting that lpp mutations alleviate the effects of defective lgt alleles (63, 64). G2824 has two reported activities: it impedes lipoprotein modification by Lgt, and it prevents Lpp attachment to peptidoglycan. This dual activity suggests G2824 may have multiple targets in vivo. Both of the inhibited reactions, Lgt modification and Lpp attachment to peptidoglycan by the L,D-transpeptidases LdtABC, rely on cysteine residues. If G2824 has affinity for cysteines in the periplasm, it would interfere with both lipoprotein maturation and Lpp-peptidoglycan attachment, as reported. This hypothesis requires testing, but such a generalized activity of G2824 in the cell envelope would explain why Δlpp sensitizes E. coli treated with G2824. The absence of Lpp causes severe envelope disruption that is exacerbated by inhibiting transpeptidases and cysteine-dependent periplasmic reactions (74).
The final parameter of our biological signature of OM lipoprotein biogenesis inhibition is NlpE-dependent activation of Cpx. Recent work revealed that NlpE acts as a real-time sensor of lipoprotein stress (38, 48). When lipoprotein trafficking is disrupted, NlpE becomes trapped in the IM, where it signals to CpxA. In keeping with this model, we found that depletion or chemical inhibition of OM lipoprotein biogenesis causes NlpE-dependent activation of Cpx. As lipoprotein trafficking is just one of the stressors to which CpxAR responds, general cell envelope defects likely still activate Cpx, yet they do so independently of NlpE. Although LspA depletion only caused NlpE-independent Cpx activation, globomycin, a well-studied LspA inhibitor, caused clear NlpE-dependent Cpx activation. The conformity of globomycin to our biological signature indicates the usefulness of our assay for validation of OM lipoprotein biogenesis inhibitors. NlpE allows rapid, robust activation of Cpx, speaking to its imperative role reacting to OM lipoprotein biogenesis stress and its usefulness as a criterium in the biological signature of OM lipoprotein biogenesis inhibition.
In addition to Cpx activation, our data indicate that Rcs activation is a strong indicator of OM lipoprotein biogenesis inhibition. Stress response activation is, thus, a powerful tool for the identification and validation of OM biogenesis inhibition. However, as measuring the NlpE-dependence of Cpx activation provides direct assessment of OM lipoprotein biogenesis, we found it to be the most informative parameter for identification of OM lipoprotein biogenesis inhibitors. Together, these three criteria can be used as a powerful tool for in vivo validation of OM lipoprotein biogenesis inhibitors, although careful consideration should be given when using them for comparing efficacy. Differences in the ability to penetrate the OM among a set of inhibitors could lead to a false perception of potency against a given target. We see the greatest utility of the signature as a simple procedure for validating OM lipoprotein biogenesis inhibitors, not as a tool for comparing their potency.
Finally, our results offer an essential conclusion to an ongoing discussion of the true target of MAC13243 in vivo. MAC13243 was originally discovered using overexpression of the essential genes of E. coli (33). Overexpression of LolA protected against treatment with MAC13243 (33). Later studies found that MAC13243 degrades under aqueous conditions into S-(4-chlorobenzyl)isothiourea, a close analog of the known MreB inhibitor A22 (58). In vitro, MAC13243, its S-(4-chlorobenzyl)isothiourea derivative, and A22 were all suggested to bind purified LolA (58). Given this evidence, MAC13243 has been embraced in the field as a LolA inhibitor (55, 75–77). Our results, however, indicate that neither MAC13243 nor A22 conform to the expected signature of an OM lipoprotein biogenesis inhibitor. As Δlpp offers no protection and Cpx activation is NlpE-independent in response to MAC13243 or A22 treatment, we suggest that neither compound appreciably impedes LolA activity in vivo. MAC13243 and A22 only conform to one criterium of our signature: OM permeabilization. Interestingly, we found that MAC13243 still causes OM permeabilization in the absence of LolA. Conversely, OM permeabilization does require a susceptible allele of mreB. Therefore, the OM permeability caused by MAC13243 is likely a result of defects in the Rod system caused by MreB inhibition, as previously suggested (78).
Comparing otherwise isogenic lolAB+ and ΔlolAB strains, we detected an increase in sensitivity to vancomycin. Hence, the loss of the LolAB trafficking pathway caused additional antibiotic sensitivity. We would expect that a compound that inhibits LolA should similarly sensitize to vancomycin. However, we failed to see any sensitization to vancomycin, even at high concentrations of MAC13243, in either mreB+ or mreB(E143A). Collectively, our data strongly argue against any in vivo activity of MAC13243 against LolA. Recent evidence also supports this conclusion. Overexpression of an inhibitor’s target can confer resistance to some inhibitors. This was the interpretation originally used to explain how LolA overexpression provides resistance to MAC13243. However, recent work found that LolA overexpression triggers activation of Rcs, explaining the protective effect of LolA overexpression against MAC13243 (78). Inactivation of Rcs abolished the protective effect of LolA overexpression. Thus, it is Rcs activation, not LolA overexpression, that is protective. Given our evidence, MAC13243 should be classified as an MreB-inhibiting compound, since it has no apparent activity against LolA in vivo.
We propose that the described biological signature discerns between those compounds that specifically inhibit OM lipoprotein biogenesis and those that interfere with the closely related processes of OM or cell envelope assembly. In an age of increasing antibiotic resistance, discovery efforts are imperative, yet lead compound target validation in vivo, especially for compounds targeting essential proteins and processes, remains challenging. Several groups have recently established clever methods to act as roadmaps for discovery and validation of on-pathway inhibitors of a variety of cellular pathways, including β-barrel assembly and cell elongation (78, 79). Our biological signature of OM lipoprotein biogenesis adds to this suite of resources, providing an invaluable tool for rapid validation of inhibitors of OM lipoprotein biogenesis.
MATERIALS AND METHODS
Strain construction.
Strains and plasmids used are provided in Table S1 and S2, respectively. Oligonucleotides used in constructing strains and plasmids are also provided in Table S2. Strains were grown in Lennox Broth (LB) supplemented with ampicillin (Amp; 125 mg/L), spectinomycin (Spec; 50 mg/L), kanamycin (Kan; 25 mg/L), or arabinose (Ara; 0.2% wt/vol) as needed. The tolC and lpp kanamycin-resistant deletion-insertion mutants were obtained from the Keio collection (80). The ΔlolA::kan, ΔlolB::kan, ΔnlpE::spec, and ΔlolCDE::cam alleles were previously described (28, 81). Deletion-insertion mutations and the complementing constructs of lspA, lnt, and lgt have also been previously described (34, 82, 83). A22-resistant mreB(E143A) was previously described (62). Strains were constructed by standard P1vir transduction of antibiotic resistance-marked alleles or by standard plasmid transformations.
Strains used in this study. Download Table S1, DOCX file, 0.03 MB (28.9KB, docx) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids and oligonucleotides used in this study. Download Table S2, DOCX file, 0.02 MB (23KB, docx) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Checkerboard assays.
Overnight cultures were diluted to optical density at 600 nm (OD600) = 0.1, then further diluted 1:1,000 into fresh broth. For LspA-, LolCDE-, LolA-, and LolB-depletion strains, 60 μL of subculture was added to each well of a 96-well microtiter plate. Next, varying amounts of arabinose (diluted in LB broth) were added in a volume of 20 μL. Finally, varying concentrations of antibiotic (diluted in LB broth) were added in a volume of 20 μL. Plates were sealed with Breathe-Easy Gas Permeable Film (Sigma Z380059) and incubated overnight at 37°C. For checkerboard assays using MTSES, subcultures were prepared as described above in 60 μL LB with 0.2% arabinose. To each well, varying MTSES amounts (20 μL) and varying antibiotic amounts (20 μL) were added. Plates were incubated for 48 h at 30°C. For MAC13243 checkerboard assays, cultures were prepared as described above in 60 μL volume (without arabinose). Varying amounts of MAC13243 (in 20 μL) and antibiotic (in 20 μL) were added to each well, and plates were incubated for 2 days at 30°C. Checkerboard assays using globomycin and compound 2 were prepared as described above but scaled down to 40 μL final volume in a 384-well microtiter plate and incubated overnight at 37°C. In all cases, A600nm was read using a Synergy H1 microplate reader (Biotek).
MTSES growth curves and GFP reporter assays.
Overnight cultures were diluted 1:1,000 into LB broth supplemented with arabinose (0.02%) and Kan, where appropriate. Aliquots of 1.96 mL seeded each well of a 24-well microtiter plate. Plates were sealed with breathable film and incubated at 37°C with shaking in a Synergy H1 measuring A600nm. After 180 min, MTSES or vehicle control DMSO was added in a volume of 40 μL. Plates were then returned to the plate reader.
Cell viability assays.
Overnight cultures of depletion strains (lpp+ or Δlpp) were grown in LB supplemented with 0.2% arabinose. Dilutions of the saturated culture were plated onto LB with arabinose and LB alone (Lgt, Lnt, LolCDE, LolA, and LolB depletions) or 0.02% arabinose (LspA depletion). Plates were incubated at 37°C overnight. Viable counts were enumerated as CFU per mL of culture. The ratio of viable cells in the presence or absence of arabinose was calculated.
MIC assays.
Cultures (5 × 104 cells/mL) were seeded (98 μL) into wells of a 96-well microtiter plate. Two-fold serial dilutions of antibiotic or chemical compound were added in a volume of 2 μL. Plates were incubated overnight at 37°C.
GFP reporter plasmids and cloning.
Cpx, Rcs, and RpoD GFP transcriptional reporters were constructed by amplifying the promoter regions of cpxP, osmB, and rpoD using the primers listed in Table S2. Amplicons were used in Gibson assembly reactions with pUA66 (51) to generate plasmids. A Strep II affinity tag was introduced to the C-terminus of LolA using a PCR site-directed insertion strategy. The V24C substitution was introduced by PCR site-directed mutagenesis. We confirmed that V24C is produced at WT-equivalent levels using α-strep II immunoblotting.
GFP reporter assays.
Overnight cultures of GFP reporter strains were subcultured into fresh LB, and 198 μL was seeded into black 96-well microtiter plates. Varying amounts of inducer or compound were added in a volume of 2 μL. Plates were grown at 37°C with shaking in a Synergy H1 plate reader (Biotek), and A600nm and GFP fluorescence was measured every 10 min. The amount of GFP per cell was calculated as a ratio of fluorescence to A600nm.
ACKNOWLEDGMENTS
This work was supported by grant 1R35GM133509 (to M.G.), fellowship F31AI147589 (to K.M.L.), and training grant T32AI106699 (to H.C.S.).
We thank Daniel Wall (University of Wyoming), Nienke Buddelmeijer (Institut Pasteur), and Timothy Meredith (Pennsylvania State University) for providing LspA-, Lgt-, and Lnt-depletion strains, respectively. We thank Benjamin Bratton (Vanderbilt University Medical Center) for providing mreB alleles. We also thank Sarah McLeod (Entasis Therapeutics) for provinding Compound 2 (McLeod). We are grateful to Kerrie May, William Shafer, and all members of the Grabowicz laboratory for helpful discussions and critical review of the manuscript.
We declare no conflicts of interest.
Contributor Information
Marcin Grabowicz, Email: marcin.grabowicz@emory.edu.
M. Stephen Trent, University of Georgia
REFERENCES
- 1.Lewis K. 2020. The science of antibiotic discovery. Cell 181:29–45. doi: 10.1016/j.cell.2020.02.056. [DOI] [PubMed] [Google Scholar]
- 2.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehman KM, Grabowicz M. 2019. Countering Gram-negative antibiotic resistance: recent progress in disrupting the outer membrane with novel therapeutics. Antibiotics 8:163. doi: 10.3390/antibiotics8040163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacNair CR, Tsai CN, Brown ED. 2020. Creative targeting of the Gram‐negative outer membrane in antibiotic discovery. Ann N Y Acad Sci 1459:69–85. doi: 10.1111/nyas.14280. [DOI] [PubMed] [Google Scholar]
- 5.Kamio Y, Nikaido H. 1976. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase C and cyanogen bromide activated dextran in the external medium. Biochemistry 15:2561–2570. [DOI] [PubMed] [Google Scholar]
- 6.May KL, Silhavy TJ. 2017. Making a membrane on the other side of the wall. Biochim Biophys Acta Mol Cell Biol Lipids 1862:1386–1393. doi: 10.1016/j.bbalip.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. 2016. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomasek D, Kahne D. 2021. The assembly of β-barrel outer membrane proteins. Curr Opin Microbiol 60:16–23. doi: 10.1016/j.mib.2021.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grabowicz M. 2018. Lipoprotein transport: greasing the machines of outer membrane biogenesis. BioEssays 40:1700187. doi: 10.1002/bies.201700187. [DOI] [PubMed] [Google Scholar]
- 10.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol 61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. 2008. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci USA 105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka K, Matsuyama SI, Tokuda H. 2001. Deletion of lolB, encoding an outer membrane lipoprotein, is lethal for Escherichia coli and causes accumulation of lipoprotein localization intermediates in the periplasm. J Bacteriol 183:6538–6542. doi: 10.1128/JB.183.22.6538-6542.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda A, Matsuyama S, Hara T, Nakayama J, Nagasawa H, Tokuda H. 2002. Aminoacylation of the N-terminal cysteine is essential for Lol-dependent release of lipoproteins from membranes but does not depend on lipoprotein sorting signals. J Biol Chem 277:43512–43518. doi: 10.1074/jbc.M206816200. [DOI] [PubMed] [Google Scholar]
- 14.Buddelmeijer N. 2015. The molecular mechanism of bacterial lipoprotein modification—how, when and why? FEMS Microbiol Rev 39:246–261. doi: 10.1093/femsre/fuu006. [DOI] [PubMed] [Google Scholar]
- 15.Sankaran K, Wu HC. 1994. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J Biol Chem 269:19701–19706. doi: 10.1016/S0021-9258(17)32077-X. [DOI] [PubMed] [Google Scholar]
- 16.Inouye S, Franceschini T, Sato M, Itakura K, Inouye M. 1983. Prolipoprotein signal peptidase of Escherichia coli requires a cysteine residue at the cleavage site. EMBO J 2:87–91. doi: 10.1002/j.1460-2075.1983.tb01386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokunaga M, Loranger JM, Wu HC. 1984. A distinct signal peptidase for prolipoprotein in Escherichia coli. J Cell Biochem 24:113–120. doi: 10.1002/jcb.240240203. [DOI] [PubMed] [Google Scholar]
- 18.Gupta S, Wu HC. 1991. Identification and subcellular localization of apolipoprotein N-acyltransferase in Escherichia coli. FEMS Microbiology Lett 78:37–42. [DOI] [PubMed] [Google Scholar]
- 19.Noland CL, Kattke MD, Diao J, Gloor SL, Pantua H, Reichelt M, Katakam AK, Yan D, Kang J, Zilberleyb I, Xu M, Kapadia SB, Murray JM. 2017. Structural insights into lipoprotein N-acylation by Escherichia coli apolipoprotein N-acyltransferase. Proc Natl Acad Sci USA 114:E6044–E6053. doi: 10.1073/pnas.1707813114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gwin CM, Prakash N, Christian Belisario J, Haider L, Rosen ML, Martinez LR, Rigel NW. 2018. The apolipoprotein N-acyl transferase Lnt is dispensable for growth in Acinetobacter species. Microbiology (Reading) 164:1547–1556. doi: 10.1099/mic.0.000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LoVullo ED, Wright LF, Isabella V, Huntley JF, Pavelka MS. 2015. Revisiting the Gram-negative lipoprotein paradigm. J Bacteriol 197:1705–1715. doi: 10.1128/JB.02414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuda K, Matsuyama S, Tokuda H. 2002. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc Natl Acad Sci USA 99:7390–73895. doi: 10.1073/pnas.112085599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka S, Narita S, Tokuda H. 2007. Characterization of the Pseudomonas aeruginosa (italicized) Lol system as a lipoprotein sorting mechanism. J Biol Chem 282:13379–13384. doi: 10.1074/jbc.M611840200. [DOI] [PubMed] [Google Scholar]
- 24.Grabowicz M. 2019. Lipoproteins and their trafficking to the outer membrane. EcoSal Plus 8:2. doi: 10.1128/ecosalplus.esp-0038-2018. [DOI] [PubMed] [Google Scholar]
- 25.Yakushi T, Masuda K, Narita S, Matsuyama S, Tokuda H. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat Cell Biol 2:212–218. doi: 10.1038/35008635. [DOI] [PubMed] [Google Scholar]
- 26.Okuda S, Tokuda H. 2009. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc Natl Acad Sci USA 106:5877–5882. doi: 10.1073/pnas.0900896106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan E, Greene NP, Crow A, Koronakis V. 2018. Insights into bacterial lipoprotein trafficking from a structure of LolA bound to the LolC periplasmic domain. Proc Natl Acad Sci USA 115:E7389–E7397. doi: 10.1073/pnas.1806822115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grabowicz M, Silhavy TJ. 2017. Redefining the essential trafficking pathway for outer membrane lipoproteins. Proc Natl Acad Sci USA 114:4769–4774. doi: 10.1073/pnas.1702248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caro F, Place NM, Mekalanos JJ. 2019. Analysis of lipoprotein transport depletion in Vibrio cholerae using CRISPRi. Proc Natl Acad Sci USA 116:17013–17010. doi: 10.1073/pnas.1906158116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nayar AS, Dougherty TJ, Ferguson KE, Granger BA, McWilliams L, Stacey C, Leach LJ, Narita S, Tokuda H, Miller AA, Brown DG, McLeod SM. 2015. Novel antibacterial targets and compounds revealed by a high-throughput cell wall reporter assay. J Bacteriol 197:1726–1734. doi: 10.1128/JB.02552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nickerson NN, Jao CC, Xu Y, Quinn J, Skippington E, Alexander MK, Miu A, Skelton N, Hankins J.v, Lopez MS, Koth CM, Rutherford S, Nishiyama M. 2018. A novel inhibitor of the LolCDE ABC transporter essential for lipoprotein trafficking in Gram-negative bacteria. Antimicrob Agents Chemother 62:e02151-17. doi: 10.1128/AAC.02151-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLeod SM, Fleming PR, MacCormack K, McLaughlin RE, Whiteaker JD, Narita S, Mori M, Tokuda H, Miller AA. 2015. Small-molecule inhibitors of Gram-negative lipoprotein trafficking discovered by phenotypic screening. J Bacteriol 197:1075–1082. doi: 10.1128/JB.02352-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pathania R, Zlitni S, Barker C, Das R, Gerritsma DA, Lebert J, Awuah E, Melacini G, Capretta FA, Brown ED. 2009. Chemical genomics in Escherichia coli identifies an inhibitor of bacterial lipoprotein targeting. Nat Chem Biol 5:849–856. doi: 10.1038/nchembio.221. [DOI] [PubMed] [Google Scholar]
- 34.Xiao Y, Wall D. 2014. Genetic redundancy, proximity, and functionality of lspA, the target of antibiotic TA, in the Myxococcus xanthus producer strain. J Bacteriol 196:1174–1183. doi: 10.1128/JB.01361-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pantua H, Skippington E, Braun M-G, Noland CL, Diao J, Peng Y, Gloor SL, Yan D, Kang J, Katakam AK, Reeder J, Castanedo GM, Garland K, Komuves L, Sagolla M, Austin CD, Murray J, Xu Y, Modrusan Z, Xu M, Hanan EJ, Kapadia SB. 2020. Unstable mechanisms of resistance to inhibitors of Escherichia mBio 11:e02018-20. doi: 10.1128/mBio.02018-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inukai M, Nakajima M, Osawa M, Haneishi T, Arai M. 1978. Globomycin, a new peptide antibiotic with spheroplast-forming activity: Isolation and physico-chemical and biological characterization. J Antibiot (Tokyo) 31:421–425. doi: 10.7164/antibiotics.31.421. [DOI] [PubMed] [Google Scholar]
- 37.Vogeley L, el Arnaout T, Bailey J, Stansfeld PJ, Boland C, Caffrey M. 2016. Structural basis of lipoprotein signal peptidase II action and inhibition by the antibiotic globomycin. Science 351:876–880. doi: 10.1126/science.aad3747. [DOI] [PubMed] [Google Scholar]
- 38.May KL, Lehman KM, Mitchell AM, Grabowicz M. 2019. A stress response monitoring lipoprotein trafficking to the outer membrane. mBio 10:e00618-19. doi: 10.1128/mBio.00618-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.May JM, Owens TW, Mandler MD, Simpson BW, Lazarus MB, Sherman DJ, Davis RM, Okuda S, Massefski W, Ruiz N, Kahne D. 2017. The antibiotic novobiocin binds and activates the ATPase that powers lipopolysaccharide transport. J Am Chem Soc 139:17221–17224. doi: 10.1021/jacs.7b07736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz N, Falcone B, Kahne D, Silhavy TJ. 2005. Chemical conditionality: a genetic strategy to probe organelle assembly Cell 121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 41.Braun V, Rehn K. 1969. Chemical characterization, spatial distribution and function of a lipoprotein (Murein-Lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem 10:426–438. doi: 10.1111/j.1432-1033.1969.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 42.Mathelié-Guinlet M, Asmar AT, Collet J-F, Dufrêne YF. 2020. Lipoprotein Lpp regulates the mechanical properties of the E. coli cell envelope. Nat Commun 11:1789. doi: 10.1038/s41467-020-15489-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mandela E, Stubenrauch CJ, Ryoo D, Hwang H, Cohen EJ, Torres VL, Deo P, Webb CT, Huang C, Schittenhelm RB, Beeby M, Gumbart J, Lithgow T, Hay ID. 2022. Adaptation of the periplasm to maintain spatial constraints essential for cell envelope processes and cell viability. Elife 11:e73516. doi: 10.7554/elife.73516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yakushi T, Tajima T, Matsuyama S, Tokuda H. 1997. Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. J Bacteriol 179:2857–2862. doi: 10.1128/jb.179.9.2857-2862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell AM, Silhavy TJ. 2019. Envelope stress responses: balancing damage repair and toxicity. Nat Rev Microbiol 17:417–428. doi: 10.1038/s41579-019-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hews CL, Cho T, Rowley G, Raivio TL. 2019. Maintaining integrity under stress: envelope stress response regulation of pathogenesis in Gram-negative bacteria. Front Cell Infect Microbiol 9:313–325. doi: 10.3389/fcimb.2019.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogt SL, Raivio TL. 2012. Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol Lett 326:2–11. doi: 10.1111/j.1574-6968.2011.02406.x. [DOI] [PubMed] [Google Scholar]
- 48.Delhaye A, Laloux G, Collet JF. 2019. The lipoprotein NlpE Is a Cpx sensor that serves as a sentinel for protein sorting and folding defects in the Escherichia coli envelope. J Bacteriol 201:e00611-18. doi: 10.1128/jb.00611-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.May KL, Lehman KM, Mitchell AM, Grabowicz M. 2019. A stress response monitoring lipoprotein trafficking to the outer membrane. mBio 10:e00618-19. doi: 10.1128/mBio.00618-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shiba Y, Miyagawa H, Nagahama H, Matsumoto K, Kondo D, Matsuoka S, Matsumoto K, Hara H. 2012. Exploring the relationship between lipoprotein mislocalization and activation of the Rcs signal transduction system in Escherichia coli. Microbiology (Reading) 158:1238–1248. doi: 10.1099/mic.0.056945-0. [DOI] [PubMed] [Google Scholar]
- 51.Mutalik VK, Nonaka G, Ades SE, Rhodius VA, Gross CA. 2009. Promoter strength properties of the complete sigma E regulon of Escherichia coli and Salmonella enterica. J Bacteriol 191:7279–7287. doi: 10.1128/JB.01047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konovalova A, Mitchell AM, Silhavy TJ. 2016. A lipoprotein/β-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. Elife 5:e15276. doi: 10.7554/elife.15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao Y, Gerth K, Müller R, Wall D. 2012. Myxobacterium-produced antibiotic TA (Myxovirescin) inhibits type ii signal peptidase. Antimicrob Agents Chemother 56:2014–2021. doi: 10.1128/AAC.06148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zwiebel LJ, Inukai M, Nakamura K, Inouye M. 1981. Preferential selection of deletion mutations of the outer membrane lipoprotein gene of Escherichia coli by globomycin. J Bacteriol 145:654–656. doi: 10.1128/jb.145.1.654-656.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muheim C, Götzke H, Eriksson AU, Lindberg S, Lauritsen I, Nørholm MHH, Daley DO. 2017. Increasing the permeability of Escherichia coli using MAC13243. Sci Rep 7:17629. doi: 10.1038/s41598-017-17772-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwai N, Nagai K, Wachi M. 2002. Novel S-benzylisothiourea compound that induces spherical cells in Escherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2. Biosci Biotechnol Biochem 66:2658–2662. doi: 10.1271/bbb.66.2658. [DOI] [PubMed] [Google Scholar]
- 57.Bean GJ, Flickinger ST, Westler WM, Mccully ME, Sept D, Weibel DB, Amann KJ. 2009. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry 48:4852–4857. doi: 10.1021/bi900014d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barker CA, Allison SE, Zlitni S, Nguyen ND, Das R, Melacini G, Capretta AA, Brown ED. 2013. Degradation of MAC13243 and studies of the interaction of resulting thiourea compounds with the lipoprotein targeting chaperone LolA. Bioorg Med Chem Lett 23:2426–2431. doi: 10.1016/j.bmcl.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 59.Butler EK, Davis RM, Bari V, Nicholson PA, Ruiz N. 2013. Structure-function analysis of MurJ reveals a solvent-exposed cavity containing residues essential for peptidoglycan biogenesis in Escherichia coli. J Bacteriol 195:4639–4649. doi: 10.1128/JB.00731-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sham L-T, Butler EK, Lebar MD, Kahne D, Bernhardt TG, Ruiz N. 2014. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345:220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor PL, Rossi L, de Pascale G, Wright GD. 2012. A forward chemical screen identifies antibiotic adjuvants in Escherichia coli. ACS Chem Biol 7:1547–1555. doi: 10.1021/cb300269g. [DOI] [PubMed] [Google Scholar]
- 62.Ouzounov N, Nguyen JP, Bratton BP, Jacobowitz D, Gitai Z, Shaevitz JW. 2016. MreB orientation correlates with cell diameter in Escherichia coli. Biophys J 111:1035–1043. doi: 10.1016/j.bpj.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gan K, Gupta SD, Sankaran K, Schmid MB, Wu HC. 1993. Isolation and characterization of a temperature-sensitive mutant of Salmonella typhimurium defective in prolipoprotein modification. J Biol Chem 268:16544–16550. doi: 10.1016/S0021-9258(19)85453-4. [DOI] [PubMed] [Google Scholar]
- 64.Gan K, Sankaran K, Williams MG, Aldea M, Rudd KE, Kushner SR, Wu HC. 1995. The umpA gene of Escherichia coli encodes phosphatidylglycerol:prolipoprotein diacylglyceryl transferase (lgt) and regulates thymidylate synthase levels through translational coupling. J Bacteriol 177:1879–1882. doi: 10.1128/jb.177.7.1879-1882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asmar AT, Collet JF. 2018. Lpp, the Braun lipoprotein, turns 50—major achievements and remaining issues. FEMS Microbiology Lett 365:1–8. doi: 10.1093/femsle/fny199. [DOI] [PubMed] [Google Scholar]
- 66.Phan M-D, Peters KM, Sarkar S, Lukowski SW, Allsopp LP, Moriel DG, Achard MES, Totsika M, Marshall VM, Upton M, Beatson SA, Schembri MA. 2013. The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet 9:e1003834. doi: 10.1371/journal.pgen.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sha J, Agar SL, Baze WB, Olano JP, Fadl AA, Erova TE, Wang S, Foltz SM, Suarez G, Motin VL, Chauhan S, Klimpel GR, Peterson JW, Chopra AK. 2008. Braun lipoprotein (Lpp) contributes to virulence of Yersiniae: potential role of Lpp in inducing bubonic and pneumonic plague. Infect Immun 76:1390–1409. doi: 10.1128/IAI.01529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fadl AA, Sha J, Klimpel GR, Olano JP, Niesel DW, Chopra AK. 2005. Murein lipoprotein is a critical outer membrane component involved in Salmonella enterica serovar typhimurium systemic infection. Infect Immun 73:1081–1096. doi: 10.1128/IAI.73.2.1081-1096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diao J, Bouwman C, Yan D, Kang J, Katakam AK, Liu P, Pantua H, Abbas AR, Nickerson NN, Austin C, Reichelt M, Sandoval W, Xu M, Whitfield C, Kapadia SB. 2017. Peptidoglycan association of murein lipoprotein is required for KpsD-dependent group 2 capsular polysaccharide expression and serum resistance in a uropathogenic Escherichia coli isolate. mBio 8:e00603-17. doi: 10.1128/mbio.00603-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sha J, Fadl AA, Klimpel GR, Niesel DW, Popov VL, Chopra AK. 2004. The two murein lipoproteins of Salmonella enterica serovar typhimurium contribute to the virulence of the organism. Infect Immun 72:3987–4003. doi: 10.1128/IAI.72.7.3987-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moffatt JH, Harper M, Harrison P, Hale JDF, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, Adler B, Nation RL, Li J, Boyce JD. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boll JM, Crofts AA, Peters K, Cattoir V, Vollmer W, Davies BW, Trent MS. 2016. A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide-deficient Acinetobacter baumannii. Proc Natl Acad Sci USA 113:E6228–E6237. doi: 10.1073/pnas.1611594113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diao J, Komura R, Sano T, Pantua H, Storek KM, Inaba H, Ogawa H, Noland CL, Peng Y, Gloor SL, Yan D, Kang J, Katakam AK, Volny M, Liu P, Nickerson NN, Sandoval W, Austin CD, Murray J, Rutherford ST, Reichelt M, Xu Y, Xu M, Yanagida H, Nishikawa J, Reid PC, Cunningham CN, Kapadia SB. 2021. Inhibition of Escherichia coli lipoprotein diacylglyceryl transferase is insensitive to resistance caused by deletion of Braun’s lipoprotein. J Bacteriol 203:e0014921. doi: 10.1128/jb.00149-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Collet J-F, Cho SH, Iorga BI, Goemans C.v. 2020. How the assembly and protection of the bacterial cell envelope depend on cysteine residues. J Biol Chem 295:11984–11994. doi: 10.1074/jbc.REV120.011201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boags A, Samsudin F, Khalid S. 2019. Details of hydrophobic entanglement between small molecules and Braun’s lipoprotein within the cavity of the bacterial chaperone LolA. Sci Rep 9:3717. doi: 10.1038/s41598-019-40170-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pandeya A, Ojo I, Alegun O, Wei Y. 2020. Periplasmic targets for the development of effective antimicrobials against Gram-negative bacteria. ACS Infect Dis 6:2337–2354. doi: 10.1021/acsinfecdis.0c00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pedebos C, Smith IPS, Boags A, Khalid S. 2021. The hitchhiker’s guide to the periplasm: unexpected molecular interactions of polymyxin B1 in E. coli. Structure 29:444–456.e2. doi: 10.1016/j.str.2021.01.009. [DOI] [PubMed] [Google Scholar]
- 78.Buss JA, Baidin V, Welsh MA, Flores-Kim J, Cho H, Wood BM, Uehara T, Walker S, Kahne D, Bernhardt TG. 2019. Pathway-directed screen for inhibitors of the bacterial cell elongation machinery. Antimicrob Agents Chemother 63:e01530-18. doi: 10.1128/AAC.01530-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Konovalova A, Grabowicz M, Balibar CJ, Malinverni JC, Painter RE, Riley D, Mann PA, Wang H, Garlisi CG, Sherborne B, Rigel NW, Ricci DP, Black TA, Roemer T, Silhavy TJ, Walker SS. 2018. Inhibitor of intramembrane protease RseP blocks the σE response causing lethal accumulation of unfolded outer membrane proteins. Proc Natl Acad Sci USA 115:E6614–E6621. doi: 10.1073/pnas.1806107115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K‐12 in‐frame, single‐gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Snyder WB, Davis LJ, Danese PN, Cosma CL, Silhavy TJ. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J Bacteriol 177:4216–4223. doi: 10.1128/jb.177.15.4216-4223.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pailler J, Aucher W, Pires M, Buddelmeijer N. 2012. Phosphatidylglycerol::prolipoprotein diacylglyceryl transferase (Lgt) of Escherichia coli has seven transmembrane segments, and its essential residues are embedded in the membrane. J Bacteriol 194:2142–2151. doi: 10.1128/JB.06641-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Armbruster KM, Meredith TC. 2017. Identification of the Lyso-form N -Acyl intramolecular transferase in low-GC firmicutes. J Bacteriol 199:e00099-17. doi: 10.1128/jb.00099-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Depletion of lipoprotein maturation or trafficking factors causes outer membrane permeability. Strains in which the only copy of lspA, lolCDE, lolA, or lolB was under an arabinose inducible promoter were grown with increasing concentrations of inducer and the large scaffold antibiotics rifampicin and erythromycin. Depletion increased outer membrane (OM) permeability, as measured by sensitivity to large scaffold antibiotics. Data are from three independent experiments; averaged density (A600nm) values of antibiotic-treated cultures relative to mock-treated control (set as 1.0) are shown. Download FIG S1, TIF file, 0.3 MB (308.7KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Deletion of lpp does not protect against general OM biogenesis defects. Relative viability of strains with arabinose-dependent expression of bamD or lptE. Viable counts per mL of culture were enumerated in the presence of 0.2% arabinose or in the absence of arabinose and used to determine the fold change in viability when inducer is absent. Data represent three independent experiments and the mean. Download FIG S2, TIF file, 0.1 MB (84.8KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Depletion of lipoprotein maturation or trafficking activates the Rcs stress response but does not activate general OM stress responses. Inducible LspA, LolCDE, LolA, or LolB strains carrying PosmB-gfp, PmicA-gfp, or PrpoD-gfp were grown with (black solid) or without (blue dots) inducer. Culture density (A600nm) and fluorescence were measured to calculate fluorescence per cell (fluorescence/A600nm). Data are average ± SD; n = 3. Download FIG S3, TIF file, 0.7 MB (739.2KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Chemical inhibitors of OM lipoprotein biogenesis cause Rcs activation but do not activate σE or RpoD. Stress response activation after treatment (arrow) with DMSO (black), MAC13243 (orange), A22 (red), globomycin (blue), or compound 2 (purple) was assessed in a ΔtolC background. Culture density (A600nm, top) and fluorescence were used to calculate fluorescence per cell (A600nm/Fluorescence) in strains carrying PosmB-gfp, PmicA-gfp, or PrpoD-gfp. Data are average ± SD; n = 3. Download FIG S4, TIF file, 0.2 MB (193.6KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
2-[(Methylsulfonyl)thio]-ethanesulfonic acid (MTSES) inhibits the activity of LolA(V24C) and resistance to MTSES increases in the absence of Lpp. (A) Growth (A600nm) of strains carrying either lolA+ (black) or lolA(V24C) (blue) was measured upon treatment with MTSES. In early log phase (arrow), strains were treated with 0.5 mM MTSES (dotted) or vehicle control (1% DMSO) (solid). Data are average ± SD; n = 3. (B) MICs of strains carrying either lolA+ or lolA(V24C) were assessed in the presence (lpp+) and absence (Δlpp) of Lpp. Cultures were grown in 24-well plates (n = 3). Download FIG S5, TIF file, 0.2 MB (165.3KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
An allele specific inhibitor of LolA causes OM permeability to antibiotics. OM permeability to vancomycin, novobiocin, rifampicin, and erythromycin was assessed in lolA+ strains upon treatment with increasing amounts of MTSES (top). OM permeability to rifampicin and erythromycin were also assessed in a strain carrying lolA(V24C) upon increasing concentrations of MTSES (bottom). Data are from three independent experiments; averaged density (A600nm) values of antibiotic-treated cultures relative to mock-treated control (set as 1.0) are shown. Download FIG S6, TIF file, 0.2 MB (248.8KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MTSES treatment activates the Cpx stress response. Cpx activation upon treatment with MTSES was assessed in a lolA+ background. Growth (A600nm) of the lolA+ strains carrying a Cpx reporter plasmid (PcpxP-gfp) was measured. Strains were either nlpE+ (blue) or nlpE::spec (orange). In early log phase (arrow), strains were treated with DMSO (1%) (solid) or 0.5 mM MTSES (dotted). Fluorescence per cell was calculated using fluorescence normalized to A600nm. Reported values are normalized to a DMSO-treated control. Data are average ± SD; n = 3. Download FIG S7, TIF file, 0.2 MB (166.6KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MTSES treatment causes activation of the Rcs stress response. Cpx activation upon treatment with MTSES was assessed in lolA+ (blue) and lolA(V24C) (orange) strains carrying PosmB-gfp, PmicA-gfp, or PrpoD-gfp. Growth (A600nm) and fluorescence were measured after treatment with DMSO (1%) (solid) or 0.5 mM MTSES (dotted) in early log phase. Fluorescence per cell was calculated by normalizing fluorescence to A600nm. Reported fluorescence values are normalized to a DMSO-treated control. Cultures were grown in 96-well, black-walled plates. Data are average ± SD; n = 3. Download FIG S8, TIF file, 0.2 MB (227.7KB, tif) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains used in this study. Download Table S1, DOCX file, 0.03 MB (28.9KB, docx) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids and oligonucleotides used in this study. Download Table S2, DOCX file, 0.02 MB (23KB, docx) .
Copyright © 2022 Lehman et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.