Abstract
Chestnut blight, caused by the fungus Cryphonectria parasitica, has been effectively controlled with double-stranded RNA hypoviruses in Europe for over 40 years. The marked reduction in the virulence of C. parasitica by hypoviruses is a phenomenon known as hypovirulence. This virus-fungus pathosystem has become a model system for the study of biological control of fungi with viruses. We studied variation in tolerance to hypoviruses in fungal hosts and variation in virulence among virus isolates from a local population in Italy. Tolerance is defined as the relative fitness of a fungal individual when infected with hypoviruses (compared to being uninfected); virulence is defined for each hypovirus as the reduction in fitness of fungal hosts relative to virus-free hosts. Six hypovirus-infected isolates of C. parasitica were sampled from the population, and each hypovirus was transferred into six hypovirus-free recipient isolates. The resulting 36 hypovirus-fungus combinations were used to estimate genetic variation in tolerance to hypoviruses, in hypovirus virulence, and in virus-fungus interactions. Four phenotypes were evaluated for each virus-fungus combination to estimate relative fitness: (i) sporulation, i.e., the number of asexual spores (conidia) produced; (ii) canker area on field-inoculated chestnut trees, (iii) vertical transmission of hypoviruses into conidia, and (iv) conidial germination. Two-way analysis of variance (ANOVA) revealed significant interactions (P < 0.001) between viruses and fungal isolates for sporulation and canker area but not for conidial germination or transmission. One-way ANOVA among hypoviruses (within each fungal isolate) and among fungal isolates (within each hypovirus) revealed significant genetic variation (P < 0.01) in hypovirus virulence and fungal tolerance within several fungal isolates, and hypoviruses, respectively. These interactions and the significant genetic variation in several fitness characters indicate the potential for future evolution of these characters. However, biological control is unlikely to break down due to evolution of tolerance to hypoviruses in the fungus because the magnitudes of tolerance and interactions were relatively small.
Despite widespread release over many years, the efficacy of biological control agents in populations of target pests has remained relatively stable (22, 25). This stability contrasts markedly to the situation with chemical pesticides, where numerous target pests have evolved resistance to a wide range of pesticides (19). In a recent review, Holt and Hochberg (25) summarized some of the hypotheses that have been proposed to explain this dichotomy. These hypotheses include inadequate relevant genetic variation in target pests, genetic constraints (trade-offs) between resistance (or tolerance) to a biological control agent and fitness, weak selection, temporally varying selection, and coevolutionary dynamics. Few of these hypotheses have been addressed experimentally in biological control systems, although genetic variation in resistance to parasitoids (6, 22) and genetic trade-offs (27, 45) have been documented. However, the paucity of experimental data bearing on these questions has precluded general predictions about the stability of biological control systems (25); this lack of data is particularly evident in biological control systems involving fungal plant pathogens.
To assess the potential erosion in efficacy of a biological control interaction, the fitnesses of both host and pathogen need to be determined in relation to each other. In this report, we compare the fitness of viruses and their fungal hosts in a system that has been exploited for biological control. Hypovirulence is a phenomenon found in the chestnut blight fungus, Cryphonectria parasitica (Murrill) Barr (formerly Endothia parasitica). Biological control of chestnut blight results when C. parasitica is infected with double-stranded RNA (dsRNA) viruses in the family Hypoviridae (for reviews, see references 2, 18, 20, 21, 23, 30, 35, and 44); these viruses are referred to in the vernacular as hypoviruses (23). Virus-infected individuals are markedly less virulent to their chestnut hosts, producing superficial cankers and rarely killing trees. In Europe, where biological control of chestnut blight has been most successful, Cryphonectria hypovirus 1 (CHV1) (23) is the only hypovirus that has been found (1); therefore, we limit our discussion to this virus. Infection with CHV1 reduces the fitness of C. parasitica in several ways, the most obvious being reductions in asexual sporulation and canker growth and almost complete inhibition of female sexual fertility (9, 11, 46). Reductions in fungal fitness caused by virus infection, therefore, could exert strong selection on C. parasitica for tolerance to viruses and could reduce the effectiveness of biological control.
To study the evolutionary stability of a biological control system, interactions between host and pathogen genotypes also must be assessed. Previous work on C. parasitica has shown that the effects of hypoviruses on fungal fitness vary among fungal genotypes (7, 9, 10, 31, 39, 41). These earlier studies were done with relatively few isolates that were collected from geographically separated populations. In this study, we were interested in interactions of viruses and fungi within a local population in order to investigate the stability of biological control at a geographic scale at which these hosts and pathogens interact in nature. Studies of the population structure of C. parasitica (33) and its hypoviruses (Y.-C. Liu and M. G. Milgroom, unpublished data) have demonstrated that gene flow is restricted among fungal and viral populations in Italy. Therefore, local populations are the most relevant for studying the evolution of these interactions and for inferring the potential for evolutionary change in this system.
The specific objectives of this study were to determine if there is genetic variation in tolerance to hypoviruses within a population of C. parasitica, whether there is variation in virulence toward C. parasitica in the corresponding hypovirus population, and whether there are significant interactions among fungus and virus isolates. We defined tolerance to hypoviruses in terms of the relative fitness of a fungal genotype when infected with hypoviruses and when uninfected. Although some evolutionary biologists have used a similar definition for resistance in animals to pathogens and parasites, we use the term tolerance as it has been used in studies of plant-pathogen or plant-herbivore interactions (16, 26, 40, 43). Finally, we define virulence quantitatively for each hypovirus as the reduction in fitness of fungal hosts relative to virus-free hosts. Although the definitions of these terms may differ from their use in virology, we have attempted to be consistent with terminology used in the ecological and evolutionary literature.
MATERIALS AND METHODS
Random samples of fungi and hypoviruses.
We used 158 isolates of C. parasitica previously sampled from a chestnut coppice forest in Bergamo, Italy (8). All isolates were screened for the presence of hypoviruses with an immunoblot procedure using a monoclonal antibody specific for dsRNA (36, 42). Six hypovirus-infected and six hypovirus-free isolates were randomly selected based on immunoblot results. Presence (and absence) of dsRNA in the sampled isolates was confirmed using a modification (36) of the CF11 cellulose column purification procedure of Morris and Dodds (34). The identity of the sampled dsRNAs as CHV1 was determined by hybridization as described previously (36, 37) and by nucleotide sequencing (Liu and Milgroom, unpublished). The hypovirus-free isolates were HPC21, HPC33, HPC36, PC13, PC61, and PC72; the hypovirus-infected isolates were HPC27, HPC29, HPC39, PC29, PC76 and PC89. Hypovirus isolates derived from these C. parasitica isolates will be referred to by their isolate names enclosed in brackets; e.g., [HPC27] denotes the isolate of CHV1 originally found in C. parasitica isolate HPC27. Although all hypovirus isolates had a high degree of nucleotide sequence similarity, each isolate was unique (Liu and Milgroom, unpublished). In addition to randomly sampled virus isolates from Bergamo, we used a standard laboratory virus isolate, CHV1-EP713 from C. parasitica isolate UEP1 (38), provided by N. K. Van Alfen.
Transmission of hypoviruses among isolates.
We transferred hypoviruses from each of the six infected donor isolates, plus CHV1-EP713, into each of the six virus-free recipient isolates through hyphal anastomoses, resulting in 42 hypovirus-fungus combinations. All isolates of C. parasitica infected with CHV1 had a white phenotype in culture, while uninfected strains were yellow-orange (4, 9, 21). We used this phenotype to follow the transmission of hypoviruses between isolates that were cocultured on potato dextrose agar (PDA; Difco Laboratories, Detroit, Mich.) (4, 29). We first attempted direct donor-to-recipient transmission of hypoviruses. If direct transmission failed due to vegetative incompatibility barriers (3, 29), hypoviruses were transferred indirectly via isolates in different vegetative compatibility types as described previously (3, 28, 29). Hypovirus transmission was confirmed by CF11 column purification of dsRNA from recipient isolates as described above.
Fitness of hypovirus-infected isolates and hypoviruses.
Fitness of hypovirus-infected fungal isolates was assessed in four ways. First, sporulation, i.e., asexual production of spores (conidia), was estimated in vitro. Second, canker growth was estimated in field experiments. Third, the abundance of fungal stromata, which contain pycnidia (and sometimes perithecia), was assayed in a sample of cankers in field experiments. Fourth, the effect of virus infection on spore germination was investigated. The fitness of each hypovirus was estimated as the total number of infected conidia produced by each host isolate.
Sporulation, germination, and virus transmission into conidia.
We grew all 42 hypovirus-fungus combinations, plus the six hypovirus-free (recipient) isolates as controls (48 total treatments), in 9-cm-diameter plastic petri dishes containing 25 ml of PDA under cool white fluorescent light with a 12-h light/12-h dark photoperiod at 25°C for 14 to 21 days. Conidia were washed from the surface of each plate with 10 ml of sterile water and counted with a hemocytometer. All isolates were cultured, and spores were counted, at four different times to estimate sporulation. We estimated conidial germination during two repetitions of these experiments. Conidial suspensions were spread on PDA, allowed to germinate for 24 h, and then examined under a dissecting microscope at 100×. We observed 100 conidia from each isolate and identified the growth of germ tubes. To estimate vertical transmission into conidia, we sampled 120 conidia from each of the 42 virus-infected isolates, 60 with germ tubes (fast germinators) and 60 without germ tubes (slow germinators), transferred them to PDA, and incubated them for a 1 week. The presence of hypovirus in each isolate was determined by comparing its cultural morphology with that of the parental hypovirus-free isolates grown from conidia under identical conditions. Viral fitness was estimated by determining the total number of virus-infected conidia produced, calculated as total number of conidia (sporulation) times germination rate times vertical transmission rate.
Field inoculations.
The 48 virus-fungus combinations were inoculated on 3-year-old Castanea sativa stems located in a 1-ha chestnut coppice forest in Ambivere, Bergamo, Italy. Two inoculation sites were used on each of 120 stems of similar size (6- to 7-cm diameter at 1-m height). Each virus-fungus combination was inoculated onto five trees. Inoculations were done by inserting agar mycelium plugs into circular wounds made to the depth of the cambium with an 8-mm-diameter corkborer. Mock inoculations were made on three trees by inserting sterile agar plugs. The two wounds on each stem were at least 1 m apart and on opposite sides of the stem. Isolates were inoculated on 21 May 1997, and the areas of the resulting cankers were measured on 1 July 1998 (1-year rating) and 17 February 1999 (2-year rating). The area (square centimeters) of each canker was estimated by measuring the length (L) and width (W) on the perpendicular axes of each canker and applying the formula for an ellipse (πLW/4).
Cankers that developed after field inoculations were examined for the presence of stromata. On 12 January 2000, bark samples (ca. 2 by 4 cm) were removed from cankers where stromata were present or near the inoculation wound in cankers when stromata were not clearly visible. Samples were collected from 168 of the 210 inoculations for virus-infected isolates and 22 of the 30 inoculations for virus-free isolates. If no stromata were visible, samples were surface disinfested in 70% ethanol for 30 s and 0.5% sodium hypochlorite for 1 min, rinsed in sterile water, and incubated in a moist chamber for 2 weeks for further examination.
The recovery of virus-infected isolates was attempted from two replicates of each of the 42 virus-fungus combinations after incubation in moist chambers. Pieces of stromata or actively growing mycelium were transferred onto PDA. One isolate from each canker was tested for vegetative compatibility (8) with the original isolate inoculated. The recovered isolate was also grown on PDA to observe whether it had the white colony phenotype diagnostic of virus infection.
Data analysis.
Each measure of fitness was analyzed separately using analysis of variance (ANOVA) with a general linear model in Minitab 12 (Minitab Inc., State College, Pa.). Experiments were initially designed as randomized complete block designs with time of experiment as the blocking factor. Time of experiment was found to be nonsignificant in all analyses; the designs were collapsed into two-factor experiments for sporulation and canker size and three-factor experiments for conidial germination and virus transmission into conidia. Fungal isolate and hypovirus isolate were the two factors common to all experiments; the third factors in the conidial germination and vertical transmission experiments were infection (hypovirus-free versus hypovirus-infected isolates) and germination time (conidia which germinated in less than 24 h [fast germinators] versus conidia which germinated after 24 h [slow germinators]), respectively. Residual analysis of ANOVA using untransformed sporulation and canker area data indicated nonconstancy of error variance. Subsequent ANOVAs of sporulation and canker area were performed with natural logarithm-transformed data, which stabilized the error variances. Germination and transmission data, expressed as proportions, were arcsine square-root transformed to stabilize variances. Comparisons between fungal isolates infected with control hypovirus CHV1-EP713 and sampled hypoviruses were made using t tests with pairing by fungal isolate.
RESULTS
Sporulation.
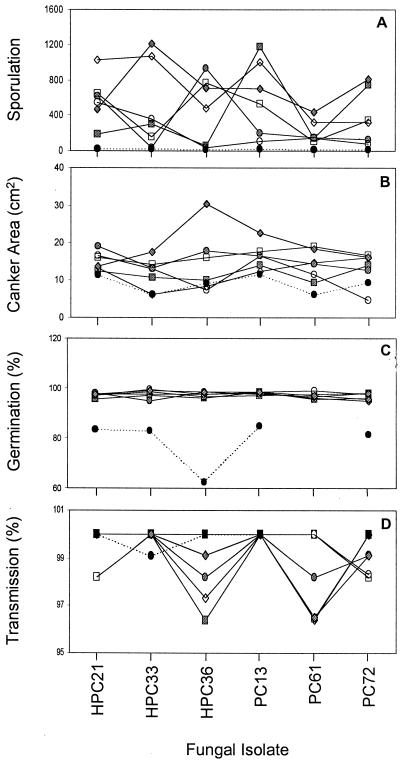
Hypovirus infection significantly reduced the sporulation of hypovirus-infected isolates relative to the hypovirus-free isolates (Table 1). Interactions between fungal and hypovirus isolates were highly significant (P < 0.001) (Fig. 1A; Table 2); therefore, we analyzed simple effects. One-way ANOVA among hypoviruses (within each fungal isolate) revealed that variation among hypoviruses was highly significant (P < 0.01) for four fungal isolates (HPC33, HPC36, PC13, and PC72) but not for isolates HPC21 and PC61 (Table 3). Variation among fungal isolates (within each hypovirus isolate) was significant (P < 0.01) for all hypoviruses except [PC89] (Table 4). Control hypovirus CHV1-EP713 had a significantly greater effect on sporulation than the Bergamo hypoviruses (P < 0.01) (Table 1).
TABLE 1.
Effects of virus infection on sporulation, canker area, and conidial germination of six isolates of C. parasitica from Bergamo, Italy
| Fitness measure | Virus isolate | Determinationa for fungal isolate:
|
|||||
|---|---|---|---|---|---|---|---|
| HPC21 | HPC33 | HPC36 | PC13 | PC61 | PC72 | ||
| Sporulation (conidia/0.5 μl) | Noneb | 2,300 (650) | 3,100 (600) | 1,700 (300) | 2,500 (1,300) | 2,800 (1,600) | 3,100 (1,600) |
| [HPC27] | 1,000 (330) | 1,100 (310) | 480 (330) | 1,000 (490) | 320 (96) | 320 (160) | |
| [HPC29] | 190 (230) | 300 (170) | 60 (50) | 1,200 (680) | 140 (140) | 750 (160) | |
| [HPC39] | 610 (490) | 34 (15) | 930 (740) | 200 (190) | 150 (140) | 130 (160) | |
| [PC29] | 650 (320) | 160 (80) | 770 (160) | 530 (290) | 110 (67) | 340 (420) | |
| [PC76] | 550 (250) | 360 (330) | 34 (17) | 110 (65) | 150 (46) | 82 (49) | |
| [PC89] | 470 (410) | 1,200 (660) | 710 (280) | 700 (94) | 440 (210) | 810 (460) | |
| [UEP1]c | 20 (8) | 20 (4) | 13 (3) | 24 (13) | 14 (5) | 17 (5) | |
| Canker area (cm2) | None | 141 (34) | 181 (43) | 129 (52) | 125 (64) | 136 (67) | 159 (56) |
| [HPC27] | 13 (4) | 6 (2) | 8 (2) | 12 (3) | 15 (6) | 16 (6) | |
| [HPC29] | 12 (4) | 11 (4) | 10 (3) | 14 (2) | 9 (2) | 14 (3) | |
| [HPC39] | 19 (2) | 13 (5) | 18 (7) | 17 (6) | 14 (3) | 13 (3) | |
| [PC29] | 16 (4) | 14 (2) | 16 (5) | 18 (5) | 19 (7) | 17 (4) | |
| [PC76] | 17 (3) | 13 (4) | 7 (1) | 16 (3) | 11 (1) | 5 (1) | |
| [PC89] | 14 (1) | 17 (5) | 31 (17) | 23 (4) | 18 (5) | 16 (5) | |
| [UEP1] | 11 (4) | 6 (2) | 9 (1) | 12 (3) | 6 (2) | 9 (2) | |
| Germination (% of total) | None | 99 (1) | 99 (1) | 98 (2) | 98 (2) | 98 (2) | 99 (2) |
| [HPC27] | 98 (1) | 98 (2) | 98 (1) | 98 (1) | 96 (0) | 95 (0) | |
| [HPC29] | 96 (1) | 97 (0) | 96 (1) | 99 (1) | 96 (1) | 96 (1) | |
| [HPC39] | 98 (3) | 95 (0) | 99 (1) | 98 (1) | 97 (2) | 98 (1) | |
| [PC29] | 97 (0) | 99 (1) | 97 (1) | 97 (0) | 98 (1) | 98 (0) | |
| [PC76] | 97 (1) | 100 (1) | 98 (1) | 99 (1) | 99 (0) | 98 (2) | |
| [PC89] | 98 (1) | 99 (0) | 99 (1) | 99 (1) | 97 (3) | 96 (1) | |
| [UEP1] | 84 (4) | 83 (6) | 63 (15) | 85 (6) | NT | 82 (1) | |
| Vertical transmission (% of fast germinators containing hypovirus) | [HPC27] | 100 (0) | 100 (0) | 100 (0) | 98 (2) | 99 (1) | 100 (0) |
| [HPC29] | 97 (1) | 100 (0) | 96 (1) | 100 (0) | 100 (0) | 100 (0) | |
| [HPC39] | 99 (1) | 100 (0) | 100 (0) | 100 (0) | 100 (0) | 100 (0) | |
| [PC29] | 100 (0) | 98 (3) | 100 (0) | 100 (0) | 100 (0) | 100 (0) | |
| [PC76] | 99 (1) | 100 (0) | 100 (0) | 100 (0) | 97 (0) | 100 (0) | |
| [PC89] | 98 (2) | 96 (1) | 98 (3) | 99 (1) | 98 (2) | 97 (1) | |
| [UEP1] | 100 (0) | 100 (0) | 99 (1) | 100 (0) | NT | 100 (0) | |
Mean, with standard deviation in parentheses (four replications for sporulation, five replications for canker area, and two replications for germination and transmission). NT, not tested.
Virus-free fungal isolate.
Control hypovirus CHV1-EP713 (from isolate UEP1 [38]).
FIG. 1.
Interactions between hypoviruses and fungal isolates plotted by fungal isolate. (A) Sporulation (conidia/0.5 μl); (B) canker area (square centimeters) 1 year postinoculation; (C) percent germination; (D) percent transmission into conidia for fast germinators. Each solid line represents a different hypovirus isolate, and the dotted line (black circles) represents the control hypovirus, CHV1-EP713. Each data point represents the mean response of four independent experiments for sporulation and germination, five replicate inoculations for canker area, and two independent experiments for transmission. Data for C. parasitica isolate PC61 infected with CHV1-EP713 are missing for germination and transmission.
TABLE 2.
ANOVA of sporulation, canker area, conidial germination, transmission, and virus fitness of C. parasitica isolates from Bergamo, Italy, infected with hypoviruses
| Expt | Source | df | Mean square | F | P |
|---|---|---|---|---|---|
| Sporulation | Fungus | 5 | 2.93 | 0.79 | 0.57 |
| Virus | 5 | 10.52 | 2.82 | 0.04 | |
| Fungus × virus | 25 | 3.73 | 5.38 | <0.001 | |
| Error | 108 | 0.69 | |||
| Total | 143 | ||||
| Canker areaa | Fungus | 5 | 0.48 | 1.16 | 0.36 |
| Virus | 5 | 1.76 | 4.24 | 0.01 | |
| Fungus × virus | 25 | 0.41 | 5.02 | <0.001 | |
| Error | 144 | 0.08 | |||
| Total | 179 | ||||
| Conidial germination | Infectionb | 1 | 0.13 | 29.31 | <0.001 |
| Fungus | 5 | 0.005 | 0.86 | 0.52 | |
| Virus | 5 | 0.002 | 0.31 | 0.90 | |
| Fungus × virus | 25 | 0.006 | 1.32 | 0.17 | |
| Error | 107 | 0.004 | |||
| Total | 143 | ||||
| Vertical transmission | Germinationc | 1 | 0.010 | 0.55 | 0.46 |
| Fungus | 5 | 0.046 | 2.02 | 0.11 | |
| Virus | 5 | 0.023 | 1.02 | 0.43 | |
| Fungus × virus | 25 | 0.023 | 1.30 | 0.18 | |
| Error | 107 | 0.017 | |||
| Total | 143 | ||||
| Virus fitnessd | Fungus | 5 | 14.99 | 0.79 | 0.56 |
| Virus | 5 | 51.86 | 2.75 | 0.04 | |
| Fungus × virus | 25 | 94.47 | 5.45 | <0.001 | |
| Error | 108 | 74.82 | |||
| Total | 143 |
Measured 1 year after inoculation.
Hypovirus-free versus hypovirus-infected conidia.
Fast versus slow germinators.
Measured as sporulation × germination × vertical transmission.
TABLE 3.
One-way ANOVA of hypovirus virulence within each C. parasitica isolate from Bergamo, Italy, measured as sporulation, canker area, conidial germination, vertical transmission, and virus fitness
| Source | df | HPC21
|
HPC33
|
HPC36
|
PC13
|
PC61
|
PC72
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MSd | F | P | MS | F | P | MS | F | P | MS | F | P | MS | F | P | MS | F | P | ||
| Sporulation | 5 | 2.79 | 2.42 | 0.08 | 7.07 | 16.92 | <0.001 | 8.59 | 18.10 | <0.001 | 4.34 | 7.18 | 0.001 | 1.87 | 2.26 | 0.09 | 4.48 | 6.65 | 0.001 |
| Error | 18 | 1.15 | 0.42 | 0.48 | 0.61 | 0.83 | 0.67 | ||||||||||||
| Canker areaa | 5 | 0.15 | 2.56 | 0.05 | 0.67 | 6.36 | 0.001 | 1.29 | 11.89 | <0.001 | 0.21 | 3.79 | 0.011 | 0.34 | 3.63 | 0.01 | 1.17 | 16.07 | <0.001 |
| Error | 24 | 0.06 | 0.11 | 0.11 | 0.06 | 0.09 | 0.07 | ||||||||||||
| Germinationb | 5 | 0.002 | 0.68 | 0.66 | 0.007 | 4.23 | 0.054 | 0.002 | 2.88 | 0.12 | 0.001 | 1.05 | 0.468 | 0.003 | 1.60 | 0.29 | 0.003 | 1.95 | 0.22 |
| Error | 6 | 0.004 | 0.002 | 0.001 | 0.001 | 0.002 | 0.002 | ||||||||||||
| Vertical transmission | 5 | 0.006 | 1.23 | 0.40 | 0.01 | 4.75 | 0.042 | 0.014 | 4.91 | 0.04 | 0.003 | 0.81 | 0.581 | 0.010 | 2.73 | 0.13 | 0.008 | 23.90 | 0.001 |
| Error | 6 | 0.005 | 0.003 | 0.003 | 0.004 | 0.004 | 0.0003 | ||||||||||||
| Virus fitnessc | 5 | 2.90 | 2.51 | 0.07 | 7.11 | 17.01 | <0.001 | 8.74 | 18.42 | <0.001 | 4.32 | 7.14 | 0.001 | 1.84 | 2.22 | 0.10 | 4.35 | 6.45 | 0.001 |
| Error | 18 | 1.15 | 0.42 | 0.48 | 0.61 | 0.83 | 0.67 | ||||||||||||
Measured 1 year after inoculation.
Fast germinators only.
Measured as sporulation × germination × vertical transmission.
MS, mean square.
TABLE 4.
One-way ANOVA of fungal resistance within each CHV1 isolate from Bergamo, Italy, measured as sporulation, canker area, conidial germination, vertical transmission and virus fitness
| Source | df | [HPC27]
|
[HPC29]
|
[HPC39]
|
[PC29]
|
[PC76]
|
[PC89]
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MSd | F | P | MS | F | P | MS | F | P | MS | F | P | MS | F | P | MS | F | P | ||
| Sporulation | 5 | 1.47 | 5.27 | 0.004 | 6.57 | 6.37 | 0.001 | 5.56 | 4.38 | 0.01 | 2.89 | 5.90 | 0.002 | 3.96 | 11.0 | <0.001 | 1.13 | 1.55 | 0.23 |
| Error | 18 | 0.28 | 1.03 | 1.27 | 0.49 | 0.36 | 0.73 | ||||||||||||
| Canker areaa | 5 | 0.65 | 5.39 | 0.002 | 0.16 | 2.16 | 0.09 | 0.14 | 1.66 | 0.18 | 0.04 | 0.44 | 0.816 | 1.25 | 24.52 | <0.001 | 0.32 | 3.88 | 0.01 |
| Error | 24 | 0.12 | 0.08 | 0.09 | 0.08 | 0.05 | 0.08 | ||||||||||||
| Germinationb | 5 | 0.002 | 1.56 | 0.30 | 0.003 | 3.66 | 0.07 | 0.004 | 0.90 | 0.54 | 0.001 | 4.11 | 0.06 | 0.004 | 1.84 | 0.24 | 0.003 | 1.92 | 0.23 |
| Error | 6 | 0.002 | 0.001 | 0.004 | <0.001 | 0.002 | 0.002 | ||||||||||||
| Vertical transmission | 5 | 0.003 | 0.81 | 0.58 | 0.017 | 43.88 | <0.001 | 0.001 | 1.00 | 0.49 | 0.003 | 1.00 | 0.49 | 0.010 | 8.67 | 0.01 | 0.005 | 0.57 | 0.72 |
| Error | 6 | 0.004 | <0.001 | 0.001 | 0.003 | 0.001 | 0.009 | ||||||||||||
| Virus fitnessc | 5 | 1.46 | 5.26 | 0.004 | 6.71 | 6.51 | 0.001 | 5.55 | 4.37 | 0.01 | 2.91 | 5.94 | 0.002 | 3.94 | 10.94 | <0.001 | 1.10 | 1.51 | 0.24 |
| Error | 18 | 0.28 | 1.03 | 1.27 | 0.49 | 0.36 | 0.73 | ||||||||||||
Measured 1 year after inoculation.
Fast germinators only.
Measured as sporulation × germination × vertical transmission.
MS, mean square.
Canker size.
Hypovirus infection significantly reduced the size of cankers on chestnut trees in the field for both the 1-year and 2-year ratings (data from the 1-year rating are shown in Table 1). Interactions between fungal isolates and hypovirus isolates were highly significant (P < 0.001) (Fig. 1B; Table 2); therefore, we analyzed simple effects. One-way ANOVAs among hypoviruses (within each fungal isolate) revealed that variation among hypoviruses was significant (P < 0.05) for all isolates except HPC21, for which variation was marginally significant (P = 0.054) (Table 3). Variation among fungal isolates (within each hypovirus isolate) was significant (P < 0.05) for three hypoviruses, [HPC27], [PC76], and [PC89] (Table 4). Control hypovirus CHV1-EP713 had a significantly greater effect on reducing canker area compared to the Bergamo hypoviruses (P < 0.01) (Table 1). No cankers developed on trees mock inoculated with water agar plugs.
Presence of asexual sporulation and perithecia in field inoculations.
Stromata were observed on 21 of 22 cankers examined in the laboratory that were induced by virus-free isolates; 11 of these had perithecia. In contrast, none of the 168 cankers induced by virus-infected isolates had stromata or perithecia when examined in the laboratory immediately after collection from the field. After a 2-week incubation in moist chambers, we observed in 54% of cankers a few small pycnidia covering less than 5% of the bark surface area. Virus-infected isolates vegetatively compatible with the isolates originally used for inoculation were recovered from 66% of the healed cankers. No statistical analyses were attempted on these data because neither asexual nor sexual sporulation was evident on any cankers caused by virus-infected isolates without incubation in moist chambers.
Conidial germination.
Conidial germination rates after 24 h incubation were greater than 95% for all hypovirus-infected isolates (Table 1). Hypovirus infection reduced overall conidial germination by approximately 1% relative to the uninfected isolates (Table 1); this difference, though statistically significant, was small (Fig. 1C; Table 2). ANOVA revealed no significant variation among hypoviruses, fungal isolates, or their interactions (Table 2). Control hypovirus CHV1-EP713 had a significantly greater effect on germination compared to the Bergamo hypoviruses (P < 0.01) (Table 1).
Vertical transmission.
Hypoviruses were transmitted into >95% of conidia of all fungal isolates (Table 1). ANOVA revealed no significant variation among hypoviruses, fungal isolates, or their interactions (Fig. 1D; Table 2). No significant differences in rates of transmission were detected between conidia germinating by 24 h (fast germinators) and conidia germinating after 24 h (slow germinators) (Table 2). Transmission rates for control hypovirus CHV1-EP713 were significantly greater than those for the Bergamo hypoviruses for the fast germinators (P < 0.01) (Table 1) but not for the slow germinators (P > 0.05).
Hypovirus fitness.
The total number of virus-infected conidia produced in vitro was used as the sole estimate of hypovirus fitness. This number was calculated as total sporulation times the germination rate times the proportion of conidia containing hypoviruses (vertical transmission). Because germination and vertical transmission were nearly 100% for all virus-fungus combinations, hypovirus fitness is virtually identical to results for sporulation (Tables 2 to 4).
DISCUSSION
We found significant genetic variation in hypovirus virulence, in tolerance to hypoviruses, and in interactions among fungus and virus isolates from a local population in Italy. Hypovirus infection had large effects on fungal fitness, reducing sporulation and canker growth, consistent with previous reports (9, 24, 41). In contrast, hypovirus infection had a very minor effect on conidial germination, and hypoviruses were transmitted at high rates into conidia. Variation in tolerance was expressed as significant (α = 0.05) differences in sporulation among fungal isolates when infected with five of six hypovirus isolates and as significant differences in canker area for three of six hypoviruses. From these data we also could estimate variation in virulence among hypoviruses, defined in terms of the reduction in fitness of fungal isolates caused by virus infection. Hypovirus virulence within each fungal isolate varied significantly in four of six fungal isolates for sporulation and for five of six fungal isolates for canker area.
CHV1-EP713 (originally isolated from France), used as a control in this study, had more severe phenotypic effects on C. parasitica than did hypovirus isolates from Bergamo. CHV1-EP713 was also found to be more virulent than CHV1-Euro7 from Italy (7). Nucleotide sequences of the virus isolates from Bergamo that we used in this study were more similar to the sequence of CHV1-Euro7 than that of CHV1-EP713 (Liu and Milgroom, unpublished), which are representative of two distinct CHV1 groups in Europe (1, 7). However, we are less concerned with variation between different virus groups than with variation within local populations for making inferences about the potential evolution of this system.
Hypoviruses were vertically transmitted into conidia of C. parasitica at high rates in this population. Transmission rates were greater than 95% for all virus-fungus combinations, and no evidence was obtained for any variation in rate due to hypovirus or fungal isolate. In contrast, vertical transmission rates of North American hypoviruses and other dsRNAs into conidia of C. parasitica vary widely (0 to 100%) (11–13, 31, 41). Few data are available on transmission rates of CHV1 into conidia of C. parasitica. One study of a hypovirus from Italy (presumably CHV1) in an American C. parasitica isolate found that the hypovirus was transmitted to 43 to 77% of conidia (41), which is considerably lower than the rates observed in our study. It is not possible to determine whether variation in vertical transmission rates is a function of virus or fungus genotypes in these previous studies because the same virus isolates were not studied in the same host isolates.
The variation in tolerance to hypoviruses observed in this study indicates that C. parasitica has some potential to evolve higher levels of tolerance to hypoviruses in the Bergamo population. Predicting evolutionary trends may be complicated by the significant hypovirus-fungus interactions detected for two of four fungal fitness measures, demonstrating that some hypovirus genotypes are better adapted to particular fungal genotypes, or vice versa. Although we found statistically significant variation in virulence, in tolerance among fungal isolates, and in interactions between the two, the magnitude of variation was not large enough relative to the marked effects of hypoviruses on fungal fitness to signal an erosion of biological control with hypovirulence. Sporulation, both in vitro and in field inoculations, and canker size were both greatly reduced by hypovirus infection, regardless of the significant variation observed. Variation in resistance or susceptibility to biological control agents has been observed in several other systems but has not resulted in decreased effectiveness of control (22, 25, 27, 45). Henter and Via (22) proposed three hypotheses to explain this lack of response: (i) that fitness costs or trade-offs were associated with resistance, (ii) that negative correlations existed between susceptibility to the parasitoid being studied and responses to other potential selective agents, and (iii) that significant interactions occurred between parasitoid and insect genotypes, resulting in frequency-dependent selection. Experimental evidence for fitness costs associated with parasitoid or parasite resistance has been obtained for some insects (14, 15, 27, 45). It is possible that tolerance to hypoviruses in C. parasitica similarly imposes fitness costs on tolerant isolates in the absence of hypovirus infection. Selection experiments such as those performed by Kraaijeveld and Godfray (27) may prove useful in testing whether there are fitness costs in the C. parasitica-hypovirus interaction.
Assessing the evolutionary stability in the hypovirus-C. parasitica system is complicated by the fact that virus fitness was measured solely in terms of vertical transmission into conidia. With strict vertical transmission, virus fitness is totally confounded with sporulation, a measure of fungal fitness. Therefore, selection should favor virus genotypes that have the least effect on sporulation while simultaneously favoring fungal isolates that are more tolerant to virus infection (barring trade-offs, etc., as discussed above), hence producing more spores. Both of these trajectories would point toward the erosion of biological control. However, this scenario is oversimplified because the evolution of virulence in this system will depend on both vertical and horizontal transmission of viruses, and we know little about the horizontal transmission of hypoviruses among fungal individuals in the field. Horizontal transmission is likely to be inversely correlated to the diversity of vegetative compatibility types (29, 32); however, we are aware no estimates of the relative contributions of horizontal and vertical transmission outside the laboratory. This information is highly significant in the context of stability of biocontrol because, in contrast to vertical transmission, high levels of horizontal transmission theoretically favor greater pathogen virulence (5, 17). Studies designed to estimate horizontal transmission in nature (for example, tracking the transmission of specific hypovirus genotypes or estimating gene flow among vegetative compatibility types) are needed to predict the evolutionary direction of this system.
ACKNOWLEDGMENTS
Yir-Chung Liu and Tobin L. Peever contributed equally to all aspects of this research.
We thank Mario Intropido, Antonio De Martino, and Luca Rancati for assistance with the field inoculations.
This research was funded by USDA NRI competitive grant 95-3703-1708 and McIntire-Stennis project NYC-153553.
REFERENCES
- 1.Allemann C, Hoegger P, Heiniger U, Rigling D. Genetic variation of Cryphonectria hypoviruses (CHV1) in Europe, assessed using RFLP markers. Mol Ecol. 1999;8:843–854. doi: 10.1046/j.1365-294x.1999.00639.x. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostakis S L. Biological control of chestnut blight. Science. 1982;215:466–471. doi: 10.1126/science.215.4532.466. [DOI] [PubMed] [Google Scholar]
- 3.Anagnostakis S L. Conversion to curative morphology in Endothia parasitica and its restriction by vegetative compatibility. Mycologia. 1983;75:777–780. [Google Scholar]
- 4.Anagnostakis S L, Day P R. Hypovirulence conversion in Endothia parasitica. Phytopathology. 1979;69:1226–1229. [Google Scholar]
- 5.Bull J J. Virulence. Evolution. 1994;48:1423–1437. doi: 10.1111/j.1558-5646.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 6.Carton Y, Capy P, Nappi A J. Genetic variability of host-parasite relationship traits: utilization of isofemale lines in a Drosophila simulans parasitic wasp. Genet Selection Evol. 1989;21:437–446. [Google Scholar]
- 7.Chen B, Nuss D L. Infectious cDNA clone of hypovirus CHV1-Euro7: a comparative virology approach to investigate virus-mediated hypovirulence of the chestnut blight fungus Cryphonectria parasitica. J Virol. 1999;73:985–992. doi: 10.1128/jvi.73.2.985-992.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortesi P, Milgroom M G, Bisiach M. Distribution and diversity of vegetative compatibility types in subpopulations of Cryphonectria parasitica in Italy. Mycol Res. 1996;100:1087–1093. [Google Scholar]
- 9.Elliston J E. Characteristics of dsRNA-free and dsRNA-containing strains of Endothia parasitica in relation to hypovirulence. Phytopathology. 1985;75:151–158. [Google Scholar]
- 10.Elliston J E. Further evidence for two cytoplasmic hypovirulence agents in a strain of Endothia parasitica from western Michigan. Phytopathology. 1985;75:1405–1413. [Google Scholar]
- 11.Elliston J E. Preliminary evidence for two debilitating cytoplasmic agents in a strain of Endothia parasitica from western Michigan. Phytopathology. 1985;75:170–173. [Google Scholar]
- 12.Enebak S A, Hillman B I, MacDonald W L. A hypovirulent isolate of Cryphonectria parasitica with multiple, genetically unique dsRNA segments. Mol Plant-Microbe Interact. 1994;7:590–595. [Google Scholar]
- 13.Enebak S A, MacDonald W L, Hillman B I. Effect of dsRNA associated with isolates of Cryphonectria parasitica from the central Appalachians and their relatedness to other dsRNAs from North America and Europe. Phytopathology. 1994;84:528–534. [Google Scholar]
- 14.Fellowes M D E, Kraaijeveld A R, Godfray H C J. Association between feeding rate and parasitoid resistance in Drosophila melanogaster. Evolution. 1999;53:1302–1305. doi: 10.1111/j.1558-5646.1999.tb04544.x. [DOI] [PubMed] [Google Scholar]
- 15.Fellowes M D E, Kraaijeveld A R, Godfray H C J. Trade-off associated with selection for increased ability to resist parasitoid attack in Drosophila melanogaster. Proc R Soc Lond B. 1998;265:1553–1558. doi: 10.1098/rspb.1998.0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fornoni J, Núñez-Farfán J. Evolutionary ecology of Datura stramonium: genetic variation and costs for tolerance to defoliation. Evolution. 2000;54:789–797. [PubMed] [Google Scholar]
- 17.Frank S A. Models of parasite virulence. Q Rev Biol. 1996;71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- 18.Fulbright D W, Weidlich W H, Haufler K Z, Thomas C S, Paul C P. Chestnut blight and recovering American chestnut trees in Michigan. Can J Bot. 1983;61:3164–3171. [Google Scholar]
- 19.Georghiou P. Pesticide resistance: strategies and tactics for management. Washington, D.C.: National Academy Press; 1986. pp. 14–43. [Google Scholar]
- 20.Griffin G J. Chestnut blight and its control. Hortic Rev. 1986;8:291–336. [Google Scholar]
- 21.Heiniger U, Rigling D. Biological control of chestnut blight in Europe. Annu Rev Phytopathol. 1994;32:581–599. [Google Scholar]
- 22.Henter H J, Via S. The potential for coeveolution in a host-parasitoid system. I. Genetic variation within an aphid population in susceptibility to a parasitic wasp. Evolution. 1995;49:427–438. doi: 10.1111/j.1558-5646.1995.tb02275.x. [DOI] [PubMed] [Google Scholar]
- 23.Hillman B I, Fulbright D W, Nuss D L, Van Alfen N K. Hypoviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M P, Summers M D, editors. Sixth Report of the International Committee for the Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. pp. 261–264. [Google Scholar]
- 24.Hillman B I, Shapira R, Nuss D L. Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology. 1990;80:950–956. [Google Scholar]
- 25.Holt R D, Hochberg M E. When is biological control evolutionarily stable (or is it)? Ecology. 1997;78:1673–1683. [Google Scholar]
- 26.Juenger T, Bergelson J. The evolution of compensation to herbivory in scarlet gilia, Ipomospsis aggregata: herbivore-imposed natural selection and the quantitative genetics of tolerance. Evolution. 2000;54:764–777. doi: 10.1111/j.0014-3820.2000.tb00078.x. [DOI] [PubMed] [Google Scholar]
- 27.Kraaijeveld A R, Godfray H C J. Trade-off between parasitoid resistance and larval competitive ability in Drosophila melanogaster. Nature. 1997;389:278–280. doi: 10.1038/38483. [DOI] [PubMed] [Google Scholar]
- 28.Kuhlman E G, Bhattacharyya H, Nash B L, Double M L, MacDonald W L. Identifying hypovirulent isolates of Cryphonectria parasitica with broad conversion capacity. Phytopathology. 1984;74:676–682. [Google Scholar]
- 29.Liu Y-C, Milgroom M G. Correlation between hypovirus transmission and the number of vegetative incompatibility (vic) genes different among isolates from a natural population of Cryphonectria parasitica. Phytopathology. 1996;86:79–86. [Google Scholar]
- 30.MacDonald W L, Fulbright D W. Biological control of chestnut blight: use and limitations of transmissible hypovirulence. Plant Dis. 1991;75:656–661. [Google Scholar]
- 31.Melzer M S, Dunn M, Zhou T, Boland G J. Assessment of hypovirulent isolates of Cryphonectria parasitica for potential in biological control of chestnut blight. Can J Plant Pathol. 1997;19:69–77. [Google Scholar]
- 32.Milgroom M G. Populations of fungi and their viruses. In: Worrall J, editor. Structure and dynamics of fungal populations. London, United Kingdom: Chapman & Hall; 1999. pp. 283–305. [Google Scholar]
- 33.Milgroom M G, Cortesi P. Analysis of population structure of the chestnut blight fungus based on vegetative incompatibility genotypes. Proc Natl Acad Sci USA. 1999;96:10518–10523. doi: 10.1073/pnas.96.18.10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris T J, Dodds J A. Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology. 1979;69:854–858. [Google Scholar]
- 35.Nuss D L. Biological control of chestnut blight: an example of virus-mediated attenuation of fungal pathogenesis. Microbiol Rev. 1992;56:561–576. doi: 10.1128/mr.56.4.561-576.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peever T L, Liu Y-C, Milgroom M G. Diversity of hypoviruses and other double-stranded RNAs in Cryphonectria parasitica in North America. Phytopathology. 1997;87:1026–1033. doi: 10.1094/PHYTO.1997.87.10.1026. [DOI] [PubMed] [Google Scholar]
- 37.Peever T L, Liu Y-C, Wang K, Hillman B I, Foglia R, Milgroom M G. Incidence and diversity of double-stranded RNAs infecting the chestnut blight fungus, Cryphonectria parasitica, in China and Japan. Phytopathology. 1998;88:811–817. doi: 10.1094/PHYTO.1998.88.8.811. [DOI] [PubMed] [Google Scholar]
- 38.Powell W A, Van Alfen N K. Differential accumulation of poly(A)+ RNA between virulent and double-stranded RNA-induced hypovirulent strains of Cryphonectria (Endothia) parasitica. Mol Cell Biol. 1987;7:3688–3693. doi: 10.1128/mcb.7.10.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rigling D, Heiniger U, Hohl H. Reduction in laccase activity in dsRNA-containing hypovirulent strains of Cryphonectria (Endothia) parasitica. Phytopathology. 1989;79:219–223. [Google Scholar]
- 40.Rosenthal J P, Kotanen P M. Terrestrial plant tolerance to herbivory. Trends Ecol Evol. 1994;9:145–148. doi: 10.1016/0169-5347(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 41.Russin J S, Shain L. Disseminative fitness of Endothia parasitica containing different agents for cytoplasmic hypovirulence. Can J Bot. 1985;65:54–57. [Google Scholar]
- 42.Schönborn J, Oberstrab J, Breyel E, Tittgen J, Schumacher J, Lukacs N. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 1991;19:2993–3000. doi: 10.1093/nar/19.11.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simms E L, Triplett J. Costs and benefits of plant responses to disease: resistance and tolerance. Evolution. 1994;48:1973–1985. doi: 10.1111/j.1558-5646.1994.tb02227.x. [DOI] [PubMed] [Google Scholar]
- 44.Van Alfen N K, Jaynes R A, Anagnostakis S L, Day P R. Chestnut blight: biological control by transmissible hypovirulence in Endothia parasitica. Science. 1975;189:890–891. doi: 10.1126/science.189.4206.890. [DOI] [PubMed] [Google Scholar]
- 45.Yan G, Severson D W, Christensen B M. Costs and benefits of mosquito refractoriness to malaria parasites: implications for genetic variability of mosquitoes and genetic control of malaria. Evolution. 1997;51:441–450. doi: 10.1111/j.1558-5646.1997.tb02431.x. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L, Baasiri R A, Van Alfen N K. Viral repression of fungal pheromone precursor gene expression. Mol Cell Biol. 1998;18:953–959. doi: 10.1128/mcb.18.2.953. [DOI] [PMC free article] [PubMed] [Google Scholar]