Abstract
Severe congenital neutropenia (SCN) is a life-threatening marrow failure disorder, usually caused by heterozygous mutations in ELANE. Potential genetic treatment strategies include biallelic knockout or gene correction via homology-directed repair (HDR). Such strategies, however, involve the potential loss of the essential function of the normal allele product or limited coverage of diverse monogenic mutations within the patient population, respectively. As an alternative, we have developed a novel CRISPR-based monoallelic knockout strategy that precisely targets the heterozygous sites of single-nucleotide polymorphisms (SNPs) associated with most ELANE mutated alleles. In vitro studies demonstrate that patients’ unedited hematopoietic CD34+ cells have significant abnormalities in differentiation and maturation, consistent with the hematopoietic defect in SCN patients. Selective knockout of the mutant ELANE allele alleviated these cellular abnormalities and resulted in about 50%–70% increase in normally functioning neutrophils (p < 0.0001). Genomic analysis confirmed that ELANE knockout was specific to the mutant allele and involved no off-targets. These results demonstrate the therapeutic potential of selective allele editing that may be applicable to SCN and other autosomal dominant disorders.
Keywords: ELANE, neutropenia, CRISPR, gene editing, allele-specific editing, neutrophils
Graphical abstract
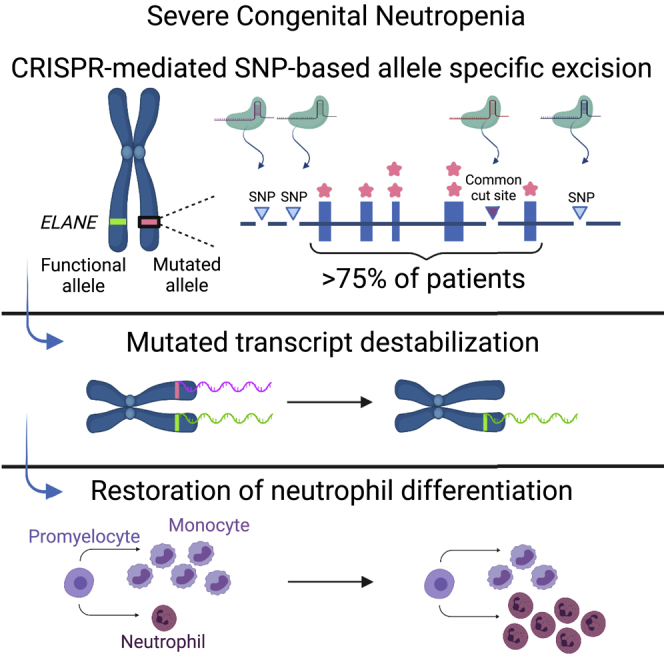
Dale, Emmanuel, and colleagues use highly precise CRISPR methodologies to knock out a mutant ELANE allele in severe congenital neutropenia (SCN), while keeping the wild-type allele intact. Knockout promotes neutrophil differentiation. This strategy involves targeting heterozygous sites of SNPs adjacent to ELANE’s mutations and could potentially treat the majority of SCN patients.
Introduction
Severe congenital neutropenia (SCN) is a rare, life-threatening hematopoietic disorder characterized by a paucity of mature neutrophils.1,2 SCN patients typically present within the first 6 months of life with recurrent, occasionally life-threatening infections attributable to a selective defect in neutrophil production.3
The most frequent causes of SCN (45%–88% according to the inclusion criteria and cohort used in the report) are more than 100 different autosomal dominant mutations in the ELANE gene encoding neutrophil elastase (NE).1,3, 4, 5, 6, 7, 8 The molecular pathways that underlie ELANE-associated neutropenia are complex and not fully understood.4,9 One possible mechanism is the misfolding of mutant NE, which is improperly processed, resulting in aberrant intracellular localization. Cytoplasmic aggregation of mutant NE induces endoplasmic reticulum (ER) stress and the unfolded protein response, eventually leading to maturation arrest, i.e., a robust marrow that produces only a few metamyelocytes, bands, and mature neutrophils.1,10
Currently, SCN patients can be treated effectively, long term, with daily injections of granulocyte colony-stimulating factor (G-CSF).11 However, such patients continue to be at risk of evolution to myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML).1,6,12, 13, 14, To date, the only alternative treatment available is allogeneic hematopoietic stem cell transplantation (HSCT),15 which requires matched donors and can lead to graft-versus-host disease and serious fatal infections.16,17 Thus, there is a great interest in new and more efficient treatments.
Biallelic knockout of ELANE alleles (both mutant and wild type) has been suggested by some groups as a potential treatment for ELANE-mediated SCN. Studies have shown that complete ELANE knockout in SCN patient-derived HSCs resolves the maturation arrest and results in normal neutrophil differentiation in vitro.18, 19, 20 However, removal of both ELANE alleles dampens the levels of NE, a protein highly involved in host immune defenses21, 22, 23, 24, 25, 26, 27, 28 and recently reported to attenuate tumor growth.29
Recently, Tran et al. used the CRISPR-Cas9 system and a DNA donor template to initiate homology-directed repair (HDR) of a mutated ELANE gene.30 Although promising, this approach poses several challenges.31, 32, 33 HDR-based gene editing targets specific mutations. Given the numerous mutations associated with SCN,1,4 mutation-specific targeting approaches are relevant to only a minor portion of the patient population.
In addition, successful gene correction in hematopoietic stem cells (HSCs) is often achieved in vitro, but some studies have reported a limited long-term reconstitution of edited HSCs in vivo.34
In this study we present a CRISPR-based potential gene therapy that provides specific knockout of an ELANE mutant allele while preserving the functional wild-type allele. Monoallelic knockout in SCN patient-derived HSCs, using an optimized high-fidelity nuclease, resolves maturation arrest and restores neutrophil differentiation. The target sites for such a unique knockout strategy are single-nucleotide polymorphisms (SNPs) that are frequently heterozygous in the SCN patient population and are linked to the majority of ELANE mutations. Thus, this approach could provide a feasible treatment to more than about 75% of ELANE-mediated SCN patients and potentially serve as a model for treatment of other autosomal dominant disorders.
Results
Representative ELANE mutations and respective therapeutic strategies
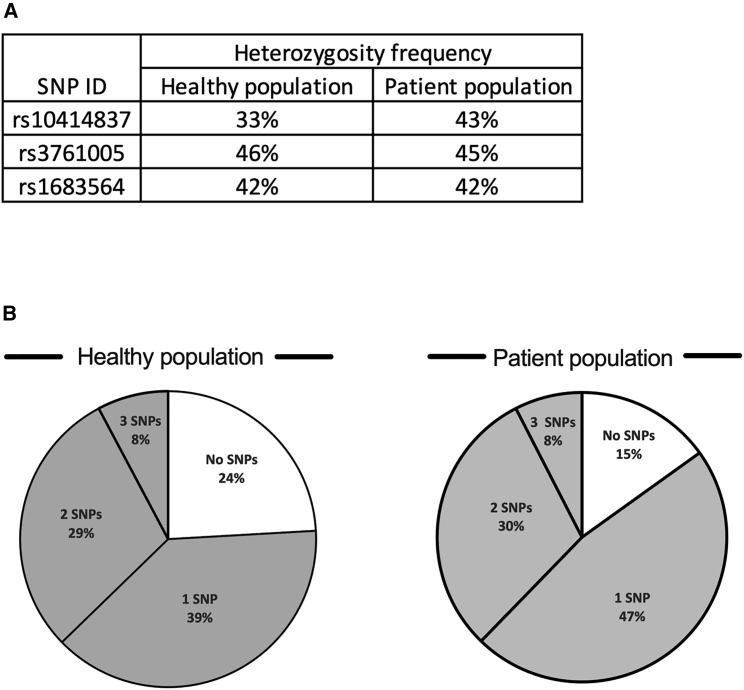
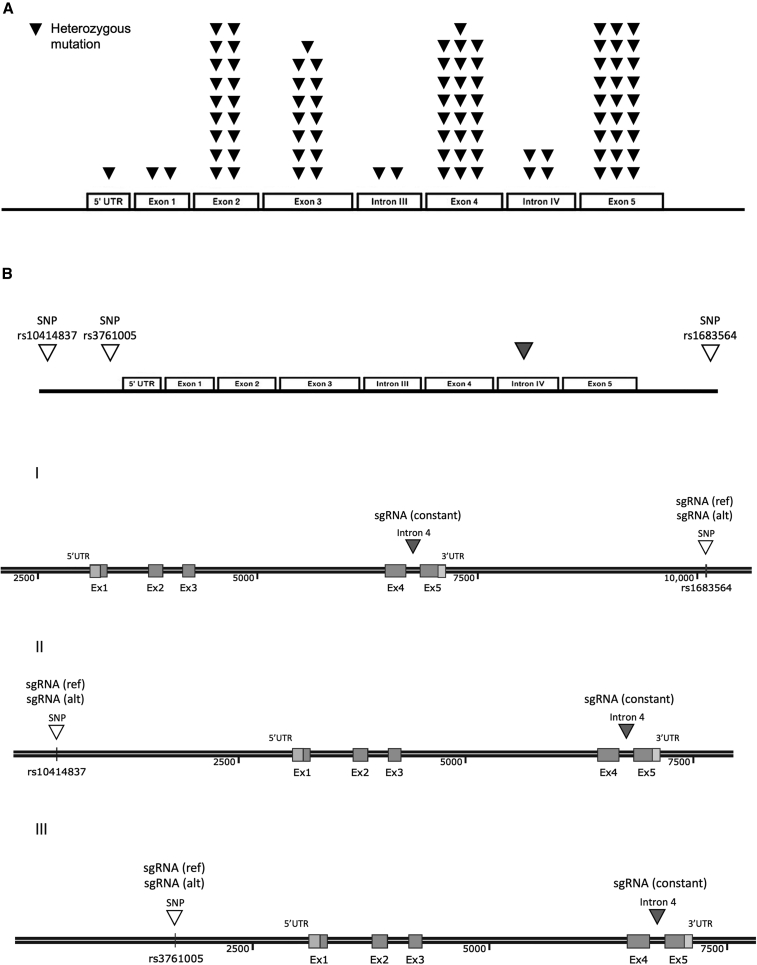
SCN is associated with numerous heterozygous mutations in ELANE. The current study presents a novel approach for knockout of the mutated ELANE allele by targeting heterozygous sites of SNPs that are adjacent to the majority of ELANE-mediated SCN mutations, instead of independently editing each pathogenic mutation. Of hundreds of potential SNPs in the ELANE gene, we identified three SNPs (termed herein rs1683564, rs10414837, and rs3761005), retrieved from the healthy database of the 1000 Genomes Project Consortium, that are frequently heterozygous in the healthy population. Analysis of the healthy and patient populations revealed similar heterozygosity frequencies between the two populations for each of the SNPs (Figure 1A). About 76%–85% of each of the populations (healthy and patient, respectively) was heterozygous in at least one of the SNPs, indicating the applicability of our strategy to more than about 75% of the SCN patient population (Figure 1B and Table S1). For each of the three SNPs, located along the abundant clusters of ELANE mutations (Figure 2A and Figure 2B, top), we developed an editing ribonucleoprotein (RNP) composition that includes two different guides and an optimized CRISPR-associated nuclease termed OMNI A1 V10 (discovery and engineering of the nuclease are described in the supplemental information). One guide (termed herein sgRNA constant) is common to all three compositions and cuts both ELANE alleles at intron 4. The editing at the intron site did not affect exons 4 and 5 or any of the regulatory elements within the intron (Figure S1). The second guide targets the heterozygous form of one of the three SNPs and therefore cuts only one allele. The composition that targets SNP rs1683564 excises exon 5 and the entire 3′ UTR, causing the degradation of the destabilized mRNA transcript (Figure 2B [I]). The compositions that target either SNP rs10414837 or SNP rs3761005 (Figure S2) lead to excision of most of the coding region and the promotor, thus preventing the transcription of the mutated allele (Figure 2B [II and III]). The current study is focused on composition I, targeting the rs1683564 SNP. The more prevalent form of rs1683564 SNP is cytosine and is referred to herein as the reference (ref) allele. Adenosine is the less prevalent form of the SNP and is referred to herein as the alternative (alt) allele. Prior to treatment, patient cells are genotyped to determine if the mutation and the SNP are on the same allele or different alleles in a process termed linkage determination (Figure S3). If the pathogenic mutation is linked to the reference allele, a nuclease-guide RNA composition including a guide (sgRNA(ref)) that targets the cytosine form of the SNP is chosen (termed herein RNP(ref)). If the pathogenic mutation is linked to the alternative allele, a composition including a guide (sgRNA(alt)) that targets the adenosine form of the SNP is chosen (termed herein RNP(alt)). The guides differ by only one nucleotide and when used with the sgRNA constant result in the same editing outcome (Figure S4).
Figure 1.
Heterozygosity frequency and coverage of patient and healthy populations by the three SNPs
(A) Heterozygosity frequency of each of the three chosen SNPs in the healthy (left) and patient (right) populations. Heterozygosity frequency was similar among the healthy and patient populations for each of the three SNPs. rs10414837, p = 0.126, odds ratio = 0.653; rs3761005, p = 0.9615, odds ratio = 1.014; rs1683564, p = 0.9475, odds ratio = 1.019; all analyzed by chi-square. (B) Pie chart presenting the percentages of the population being heterozygous for at least one of the three chosen SNPs (gray) in the healthy (left) or patient (right) population. The healthy and patient populations are covered similarly by the three SNPs (more than 75%, p = 0.1285, odds ratio = 0.56, chi-square).
Figure 2.
Schematic of representative ELANE mutations and suggested therapeutic strategies
(A) Linear representation of ELANE’s five exons and four introns showing locations of representative heterozygous mutations associated with SCN depicted as black inverted triangles. Based on Makaryan et al.4 (B) Schematics of three identified SNPs (white inverted triangles) associated with the majority of ELANE mutations and a common cut site (gray inverted triangle), based on which three allele-specific sgRNA guides and a constant guide were designed representing three monoallelic excision strategies.
Allele specificity and excision efficiency using OMNI A1 V10 nuclease in an SNP-based knockout strategy
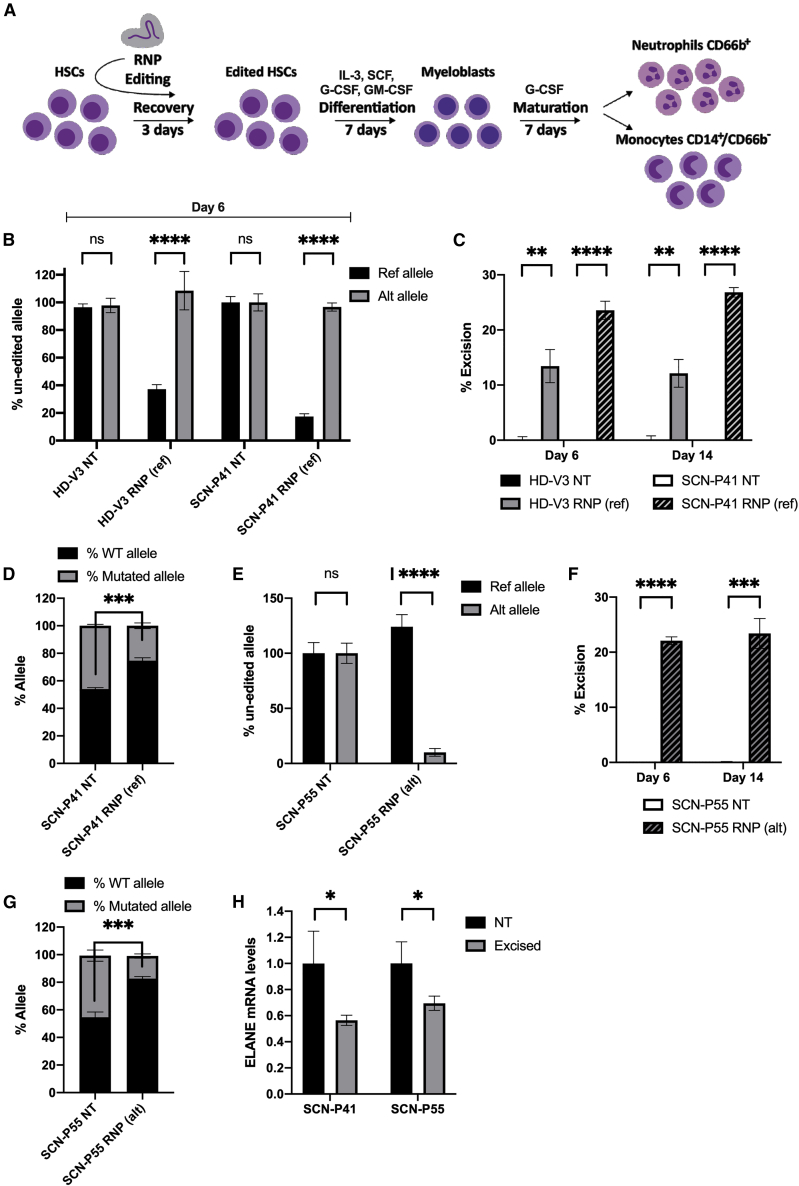
Unlike most CRISPR-associated editing strategies that cut the target gene in both alleles, our approach is directed at knockout of the mutated allele while keeping the wild-type functional allele intact. To demonstrate the feasibility of our monoallelic editing strategy, HSCs heterozygous to SNP rs1683564, taken from healthy donors and SCN patients, were excised using either RNP(ref) or RNP(alt) (according to their linkage) or left non-treated (NT). HSCs recovered for 3 days in CD34+ expansion medium; then were cultured for 7 days in the presence of IL-3, SCF, GM-CSF, and G-CSF for proliferation and myeloid progenitor differentiation; and subsequently were stimulated with G-CSF for a further 7 days for neutrophil differentiation (Figure 3A).
Figure 3.
Allele specificity and excision efficiency of OMNI A1 V10 nuclease compositions
(A) Scheme depicting experimental workflow. HSCs from healthy donors and SCN patients were electroporated with RNPs or left non-treated followed by 3 days of recovery in CD34+ expansion medium. Cells were then subjected to differentiation by culturing for 7 days with IL-3, SCF, GM-CSF, and G-CSF for proliferation and myeloid progenitor differentiation, followed by a 7-day culture in G-CSF for neutrophil differentiation. (B) Bar graph representing percentages of un-edited reference (black) and alternative (gray) alleles at day 6 of differentiation in HSCs taken from either a healthy donor (HD-V3) or an SCN patient (SCN-P41) and treated with RNP(ref) composition or left non-treated (NT), as measured by ddPCR (n = 3 groups of cells from HD-V3 healthy/SCN-P41 patient donors). Statistical significance is indicated by ∗∗∗∗p < 0.0001; ns, not statistically significant. (C) Bar graph representing percentages of excision at days 6 and 14 of differentiation in HSCs taken from either a healthy donor (HD-V3) (non-treated [NT, black] or RNP(ref)-treated [gray]) or an SCN patient (SCN-P41) (NT [white] or RNP(ref)-treated [hatched]), as measured by ddPCR (n = 3 groups of cells from HD-V3 healthy/SCN-P41 patient donors). Statistical significance is indicated by ∗∗p < 0.01, ∗∗∗∗p < 0.0001. (D) Bar graph representing percentages of wild-type (black) and mutated (gray) alleles in cDNA taken from SCN-P41 patient HSCs that were either RNP(ref)-treated or left NT, as measured by NGS targeting the mutation site (n = 3 groups of cells from patient SCN-P41). Statistical significance is indicated by ∗∗∗p < 0.001. (E) Bar graph representing percentages of unedited reference (black) and alternative (gray) alleles at day 6 of differentiation in HSCs taken from an SCN patient (SCN-P55) treated with RNP(alt) composition or left NT, as measured by ddPCR (n = 3 groups of cells from patient donor SCN-P55). Statistical significance is indicated by ∗∗∗∗p < 0.0001; ns, not statistically significant. (F) Bar graph representing percentages of excision at days 6 and 14 of differentiation in HSCs taken from an SCN patient (SCN-P55): NT (white) or RNP(alt)-treated (hatched), as measured by ddPCR (n = 3 groups of cells from patient donor SCN-P55). Statistical significance is indicated by ∗∗∗p < 0.001,∗∗∗∗p < 0.0001. (G) Bar graph representing percentages of wild-type (black) and mutated (gray) alleles in cDNA taken from patient SCN-P55 HSCs that were either RNP(alt)-treated or left NT, as measured by NGS targeting the mutation site (n = 3 groups of cells from patient SCN-P55). Statistical significance is indicated by ∗∗∗p < 0.001. (H) Bar graph representing ELANE mRNA levels in day 6 differentiated HSCs of patients SCN-P41 and SCN-P55 that were either NT (black) or RNP(ref)/RNP(alt)-treated, respectively (excised, gray). Data are presented relative to the NT group (n = 3 groups of cells from patients SCN-P41 and SCN-P55). Statistical significance is indicated by ∗p < 0.05. Bars represent mean values with standard deviation.
A fraction of the cells was harvested at days 6 and 14 of differentiation for genomic DNA or RNA extraction. Allele specificity was determined at day 6 of differentiation by two competitive probes binding either the alternative allele or the reference allele (for probes’ specificity see supplemental information and Figure S5). To test the RNP(ref) composition we used HSCs from SCN patient P41 (SCN-P41) harboring a mutation on the reference allele and HSCs from healthy donor V3 (HD-V3), both heterozygous to the reference form of the SNP. Digital Droplet PCR (ddPCR) revealed that editing using RNP(ref) was specific, as only about 40% and about 20% of the reference allele remained intact in HSCs of HD-V3 and SCN-P41, respectively (Figure 3B). The alternative allele, however, was not affected by the RNP(ref) composition, as indicated by its high levels, which were similar to those of NT cells (Figure 3B). These results demonstrated that treatment with the RNP(ref) composition leads to allele-specific editing.
Next, we evaluated excision efficiency at days 6 and 14 of neutrophil differentiation. Excision was determined by amplification of two regions in ELANE using two differently labeled probes, one for exon 1, which is not affected by the current excision strategy, and a second probe for exon 5, which is degraded upon excision (Figure 2B [I]). The ratio between the signals of the two probes was translated to excision efficiency. Treatment with RNP(ref) resulted in about 13% excision in HSCs of HD-V3 and about 25% excision in HSCs of SCN-P41 (Figure 3C). Given the high specificity of the nuclease (Figure 3B), this excision correlated to about 50% of the cell population that had undergone excision at the reference allele in HSCs of SCN-P41. RNP(ref) treatment also involved about 6% inversion events in HSCs of SCN-P41 as measured by EvaGreen staining (Figures S6A and S6B). Of note, other experiments evaluating RNP(ref)-mediated excision levels in HSCs from healthy donors showed higher excision levels than reported above that were comparable to those measured in HSCs of SCN-P41 (Figure S7). In addition, next-generation sequencing (NGS) analysis of cDNA from SCN-P41 cells targeting exon 4 harboring a mutation showed a 1:1 ratio between wild-type and mutated alleles in NT cells. RNP(ref) treatment shifted the wild-type-to-mutated allele ratio to 3:1 by differentially reducing the mutated allele transcript, thereby enriching the wild-type allele (Figure 3D).
To evaluate the specificity and efficiency of the RNP(alt) composition, we used HSCs from SCN patient P55 (SCN-P55) harboring a mutation on the alternative allele and heterozygous to the alternative form of the SNP. RNP(alt)-based treatment resulted in editing of about 90% of the alternative allele (only 10% remained intact), whereas the reference allele was kept intact at day 6 of differentiation (Figure 3E). Excision levels in SCN-P55 HSCs were about 23% at days 6 and 14 of neutrophil differentiation (Figure 3F), indicating that about 46% of the cell population had undergone excision at the alternative allele. RNP(alt) treatment resulted in 7.6% inversion events in SCN-P55 HSCs as measured by EvaGreen staining (Figure S6C). Specificity and excision efficiency of RNP(alt)-edited HSCs of healthy donors were tested and found comparable to those obtained in patient-derived cells (Figure S7). In addition, NGS analysis of cDNA from SCN-P55 cells targeting exon 5 harboring a mutation showed enrichment of the wild-type allele (Figure 3G). ELANE mRNA levels were decreased following excision in cells from patients SCN-P41 and SCN-P55 (Figure 3H). In view of the increased wild-type:mutant allele ratio obtained following excision (Figures 3D and 3G), the reduced mRNA levels were mainly a result of the degradation of the mutated transcript.
Next, we confirmed that RNP(alt)-based excision had occurred in a sub-population of HSCs (CD34+/CD90+ cells) that is considered essential for multilineage engraftment and hematopoietic reconstitution35 (Figure S8). An unbiased survey (GUIDE-seq) of whole-genome off-target cleavage using OMNI A1 V10 nuclease and each of constant guide (SgRNA(constant)), reference guide (SgRNA(ref)), and alternative guide (SgRNA(alt)) resulted in no identified off-targets (≤4 mismatches) (Figures S9A–S9C and Table S2). In addition, in silico off-target analysis was performed for each of the guides and identified a few potential off-targets. None of these off-targets were validated by a rhAmpSeq analysis done on edited HSCs from patients SCN-P41 and SCN-P55 (Figure S9D), demonstrating the high fidelity of the nuclease compositions. Taken together, the results provided above present an active, highly accurate nuclease that can target the mutant allele while preserving the intact wild-type functional allele.
OMNI A1 V10-facilitated editing boosts neutrophil differentiation and maturation in vitro
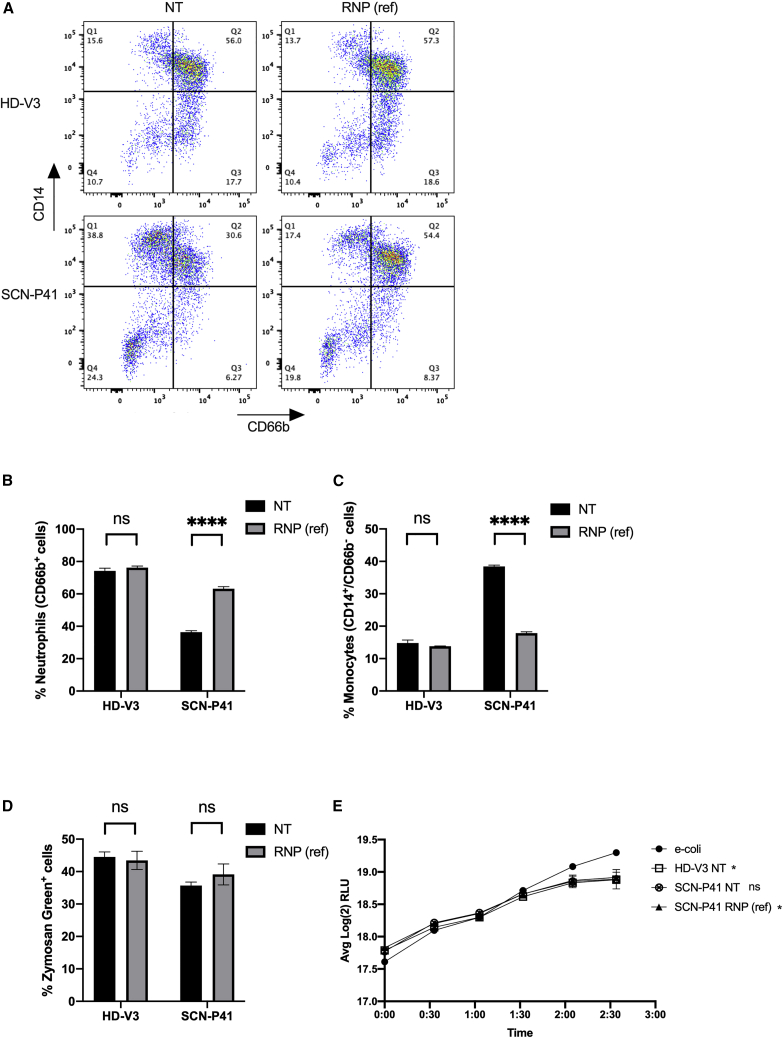
To demonstrate the functional outcome of our SNP-based single-allelic editing approach we evaluated neutrophil differentiation and maturation capacities of RNP(ref)-edited and NT healthy and patient-derived HSCs in vitro, at day 14 of differentiation. Flow cytometric analysis showed that about 74% of the HD-V3 HSCs, subjected to the differentiation protocol differentiated into neutrophils (CD66b+; two right quarters [Q2 + Q3] of the dot plot), compared with only 36% of the SCN-P41-derived HSCs. In contrast, only about 15% of HD-V3 HSCs differentiated into monocytes (CD14+/CD66b−; upper left quarter [Q1] of the dot plot) compared with 38% of the SCN-P41-derived HSCs. These observations are consistent with the characteristic hematopoietic defect in SCN patients (Figures 4A–4C; NT: HD-V3 versus SCN-P41). SCN-P41-derived HSCs treated with RNP(ref) showed a 74% increase in neutrophils (CD66b+ cells) and a 2-fold reduction in the monocytic subset (CD14+/CD66b−) (Figures 4A–4C; SCN-P41: NT versus RNP(ref)). A similar increase in neutrophil count was observed in SCN-P41-derived edited HSCs by flow cytometric analysis of CD11b+/CD15+ cells (Figures S10A and S10B). RNP(ref)-mediated editing in HSCs of HD-V3 did not affect the monocytic and neutrophilic subsets, supporting the safety of this composition (Figures 4A–4C; HD-V3: NT versus RNP(ref)). After demonstrating that our allele-specific editing approach significantly improved the cellular abnormalities associated with SCN, we next assessed neutrophilic functions in RNP(ref)-treated and NT healthy (V3) and patient (P41) HSC-derived neutrophils. In vitro phagocytic capacity was tested by measurement of phagosomal uptake of zymosan green particles by neutrophils from the different groups. Flow cytometric analysis revealed equivalent levels of phagocytosis in RNP(ref)-treated and NT neutrophils from both HD-V3 and SCN-P41 (Figure 4D). In addition, anti-bacterial killing capacity was examined by incubating neutrophils derived from NT healthy (HD-V3) and patient (SCN-P41) HSCs and RNP(ref)-treated patient HSCs (SCN-P41, RNP(ref)) with E. coli bacteria expressing the bacterial luciferase gene and tracking real-time changes in light emission, expressed as relative light units (RLUs). Healthy NT, RNP(ref)-treated, and NT patient-derived neutrophils exhibited efficient bacterial killing, as indicated by a 23%–25% decrease in bacterial unit (RLU) relative to the bacteria-only control (Figure 4E). The SCN-P41 NT group showed reduced bacterial units, although not statistically different from the bacteria-only control group.
Figure 4.
RNP(ref)-facilitated editing boosts neutrophil differentiation and maturation in vitro
(A) Representative fluorescence-activated cell sorting (FACS) plots of non-treated (NT, left) and RNP(ref)-treated (right) healthy donor (HD-V3, top) and patient SCN-P41 (bottom ) differentiated HSCs, analyzed for neutrophilic (CD66b+) and monocytic (CD14+/CD66b−) subsets. (B) Quantitative analysis of respective FACS data for percentages of neutrophils (CD66b+ cells) in healthy (HD-V3) and SCN patient (SCN-P41) differentiated HSCs that were NT (black) or treated with RNP(ref) (gray) (n = 3 groups of cells from HD-V3 healthy/SCN-P41 patient donors). Statistical significance is indicated by ∗∗∗∗p < 0.0001; ns, not statistically significant. (C) Quantitative analysis of respective FACS data for percentages of monocytes (CD14+/CD66b− cells) in healthy (HD-V3) and SCN patient (SCN-P41) differentiated HSCs that were NT (black) or treated with RNP(ref) (gray) (n = 3 groups of cells from HD-V3 healthy/SCN-P41 patient donors). Statistical significance is indicated by ∗∗∗∗p < 0.0001; ns, not statistically significant. (D) Quantification of percentages of zymosan green uptake by healthy (HD-V3) and SCN patient (SCN-P41) differentiated HSCs that were NT (black) or treated with RNP(ref) (gray). ns, not statistically significant. (E) Graph depicts real-time change in light emission, in relative light units (RLUs), from 200,000 luciferase-expressing bacterial cells incubated with differentiated neutrophils from healthy NT (HD-V3; square), patient NT (SCN-P41; crossed circle), or patient RNP(ref)-treated (SCN-P41, RNP(ref); triangle) HSCs compared with bacterial cells-only control (e-coli; circle). Statistical significance for each of the groups versus the E. coli control at the last time point presented, when RLU levels reached plateau, is indicated by ∗p < 0.05; ∗∗p < 0.01; ns, not statistically significant. Bars represent mean values with standard deviation.
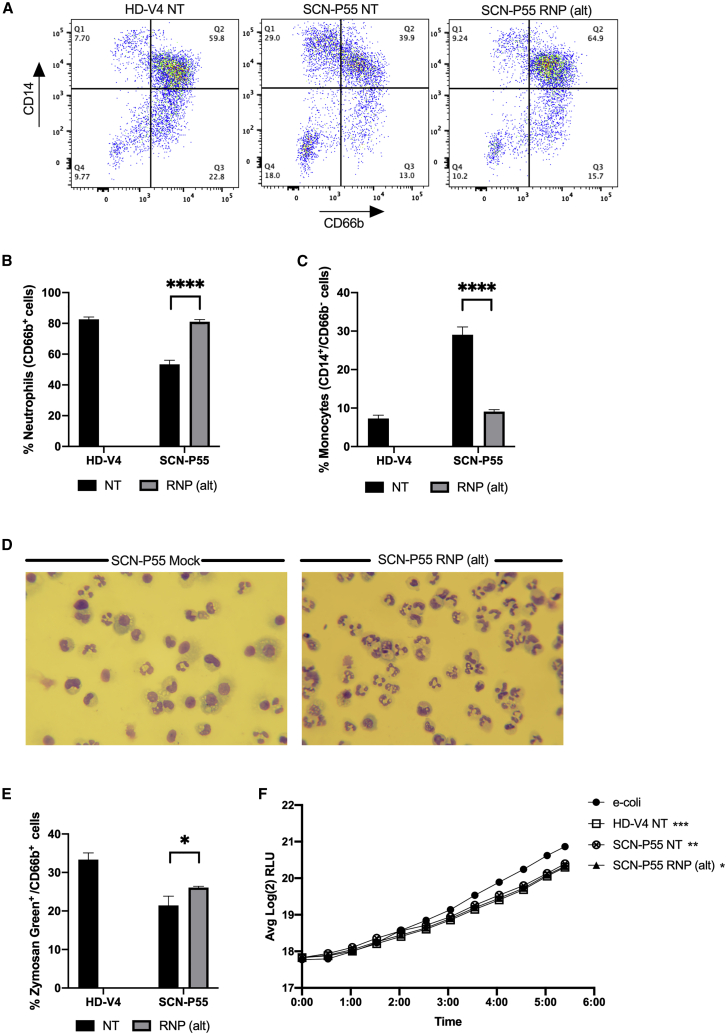
An experiment using the RNP(ref) composition was conducted on cells from another patient (SCN-P42) and gave similar excision, differentiation, and functional results, including histological staining demonstrating the restoration of neutrophil differentiation (Figure S11). Thus, specific knockout of the ELANE mutated allele by RNP(ref) composition ameliorated the aberrant phenotype of attenuated differentiation toward neutrophils while preserving essential neutrophil functions. A similar analysis was performed on the RNP(alt) composition. Flow cytometric analysis showed that about 83% of HD-V4-derived HSCs differentiated into neutrophils (CD66b+; two right quarters [Q2 + Q3] of the dot plot) and about 7% differentiated into monocytes (CD14+/CD66b−; upper left quarter [Q1] of the dot plot). SCN-P55-derived HSCs showed a lower differentiation toward neutrophils (about 53%) and a higher differentiation to monocytes (about 29%) (Figures 5A–5C; NT: HD-V4 versus SCN-P55), representing a typical SCN hematopoietic defect. SCN-P55-derived HSCs treated with RNP(alt) presented a 1.5-fold increase in neutrophils (CD66b+ cells) and a 3-fold reduction in the monocytic subset (CD14+/CD66b−) (Figures 5A–5C; SCN-P41: NT versus RNP(alt)). A similar increase in neutrophil subset was observed in SCN-P55-derived edited HSCs by flow cytometric analysis of CD11b+/CD15+ cells (Figures S10C and S10D). Diff-Quik staining of SCN-P55-derived HSCs treated with RNP(alt) or electroporated without a nuclease composition (SCN-P55 Mock) revealed higher numbers of cells with classical polymorphonuclear neutrophilic morphology in the RNP(alt)-treated group compared with the mock group (Figure 5D). Flow cytometric analysis of zymosan green particles revealed slightly higher levels of phagocytosis in RNP(alt)-treated neutrophils compared with NT neutrophils from SCN-P55 (Figure 5E). In addition, healthy NT (HD-V4), patient NT (SCN-P55), and RNP(alt)-treated (SCN-P55, RNP(alt)) neutrophils exhibited efficient bacterial killing, as indicated by a significant 30% decrease in bacterial unit (RLU) relative to the bacteria-only control (Figure 5F). A classical pathogen-killing mechanism of neutrophils is the formation of neutrophil extracellular traps (NETs). We therefore confirmed that our excision strategy does not compromise NETosis capacity (Figure S12).
Figure 5.
RNP(alt)-mediated editing promotes HSC differentiation toward functional neutrophils in vitro
(A) Representative FACS plots of non-treated healthy donor (HD-V4 NT, left), NT SCN patient (SCN-P55, middle), and RNP(alt)-treated SCN patient (right) differentiated HSCs, analyzed for neutrophilic (CD66b+) and monocytic (CD14+/CD66b−) subsets. (B) Quantitative analysis of respective FACS data for percentages of neutrophils (CD66b+ cells) in differentiated HSCs from NT healthy donor (HD-V4; black) and SCN patient (SCN-P55) either NT (black) or treated with RNP(alt) (gray) (n = 3 groups of cells from HD-V4 healthy/SCN-P55 patient donors). Statistical significance is indicated by ∗∗∗∗p < 0.0001. (C) Quantitative analysis of respective FACS data for percentages of monocytes (CD14+/CD66b− cells) in differentiated HSCs from NT healthy donor (HD-V4; black) and SCN patient (SCN-P55) either NT (black) or treated with RNP(alt) (gray) (n = 3 groups of cells from HD-V4 healthy/SCN-P55 patient donors). Statistical significance is indicated by ∗∗∗∗p < 0.0001. (D) Diff-Quik staining of P55 SCN patient-derived differentiated HSCs treated with RNP(alt) or electroporated without a nuclease composition (SCN-P55 Mock). Microphotographs were taken on a Leitz Laborlux S polarizing light microscope at 400× magnification using a Nikon DSLR digital camera. (E) Quantification of percentages of zymosan green uptake by differentiated HSCs from NT healthy donor (HD-V4; black) and SCN patient (SCN-P55) either NT (black) or treated with RNP(alt) (gray) (n = 3 groups of cells from HD-V4 healthy/SCN-P55 patient donors). Statistical significance is indicated by ∗p < 0.05. (F) Graph depicts real-time change in light emission in relative light units (RLUs) from 200,000 luciferase-expressing bacterial cells incubated with differentiated neutrophils from healthy NT (HD-V4; square) HSCs, patient NT (SCN-P55; crossed circle), and patient RNP(alt)-treated (SCN-P55, RNP(alt); triangle) HSCs compared with bacterial cells-only control (e-coli; circle). Statistical significance for each of the groups versus E. coli control at the last time point presented, when RLU levels reached plateau, is indicated by ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Bars represent mean values with standard deviation.
An experiment using the RNP(alt) composition was conducted on cells from another patient (SCN-P12) and showed similar excision, differentiation, and functional results (Figure S13). Moreover, another experiment performed on patient cells (SCN-P56) harboring a mutation on exon 2, located upstream of the mutations found in previous patients (exons 4 and 5), showed similar results (Figure S14). The results described herein indicate that a single-allelic knockout of an ELANE mutated allele, using RNP(ref) and RNP(alt) nuclease compositions, is effective and safe both in boosting neutrophil differentiation and in maintaining essential neutrophilic core functions.
Discussion
The prognosis of most SCN patients has been dramatically improved following the introduction of G-CSF therapy, owing to the increase in absolute neutrophil counts and a reduced incidence of infections.1 Nonetheless, patients on long-term G-CSF treatment remain at risk of hematological complications, especially those who respond poorly to this treatment or require high daily doses of G-CSF.6,12, 13, 14 G-CSF therapy induces compensatory mechanisms of granulopoiesis, but does not treat the etiological roots of the disease. Moreover, the full functions of neutrophils in SCN patients on G-CSF therapy may not be completely restored, which may account for the complications still observed in treated patients.1,12,13,36
The most frequent causes of SCN are heterozygous mutations in the ELANE gene encoding NE.4,37,38 ELANE mutations are characterized by their dominant nature, resulting in the development of neutropenia despite the presence of one intact wild-type ELANE allele.8,37,39,40 The pathophysiological pathways that underlie neutrophil maturation arrest may vary depending on which elastase domain was affected by the mutation.41,42 However, a common possible etiology relates to the production of an abnormal NE that cannot be folded, secreted, or degraded, resulting in its intracellular accumulation and mislocalization. These events induce ER stress and the unfolded protein response, leading to increased apoptosis of neutrophil precursors.9,43, 44, 45 Moreover, co-expression of the mutant and wild-type forms of NE within the same cell results in inhibition of the wild-type NE activity.41 Thus, a desired therapy, as proposed in the current study, would be one that specifically removes the mutated allele, thereby preventing the destructive cascade of events initiated by abnormal NE, but at the same time preserves the wild-type ELANE allele, intact and functional.
Recently, Nasri et al. demonstrated that CRISPR-Cas9-mediated ELANE gene deletion in cells from SCN patients harboring ELANE mutations increased neutrophil differentiation and maturation in vitro. The study reported that the resulting neutrophils retained phagocytic, oxidative, and chemotactic functions.19 However, such an approach could be counterproductive, as it completely eliminates expression of NE, which has been reported to mediate a non-redundant role in innate immune defense against pathogens. Deficiency in NE results in increased susceptibility to sepsis and death following infection in gram-negative bacteria.23,24 NE targets bacterial virulence proteins, modulates inflammatory cytokines, and degrades bacterial outer-membrane proteins.21,25, 26, 27,46,47 In addition, NE has an essential fungicidal activity.22
Another mechanism by which neutrophils kill bacteria, fungi, and parasites is the formation of NETs. Upon pathogen recognition, NE is released from the neutrophil’s granules and translocates to the nucleus, where it degrades histones and promotes chromatin decondensation. The chromatin expands, leading to cell rupture and the release of web-like chromatin fibers that trap and kill pathogens.48 Papayannopoulos et al. demonstrated that chromatin decondensation is blocked by specific pharmacological inhibitors of NE and that NE knockout animals do not form NETs, and their neutrophils exhibit condensed nuclei, in an in vivo mouse model of lung infection.48 Interestingly, our allele-specific excision strategy, which keeps the wild-type NE intact, resulted in preservation of NETosis capacity. Taken together, the abundant published experimental data unequivocally attribute to NE essential non-redundant roles. Therefore, strategies aimed at eliminating NE must be reevaluated, considering its beneficial functions.
Further support for the need to preserve functional NE comes from a recent study identifying the anti-cancer properties of this protein. The study reported that NE attenuates primary tumor growth and produces a CD8+ T-cell-mediated abscopal effect to attack distant metastases. NE selectively induces DNA damage and promotes apoptosis in cancer cells while sparing non-cancer cells. Interestingly, this effect is attenuated in vivo due to the presence of serine protease inhibitors that limit NE activity, resulting in protection and prosperity of cancer cells.29 Thus, complete removal of NE could have detrimental consequences. In alignment with this finding, there are no examples of healthy individuals carrying a biallelic ELANE knockout,49 while naturally occurring single-allele knockout exists in healthy people.50 Notably, allele-specific knockout is a desired therapeutic goal, not only in SCN, but also in other autosomal dominant disorders, as was recently demonstrated for Huntington’s disease.51
In view of the importance of keeping a functional copy of ELANE, we suggest a unique approach for targeting SCN-related ELANE mutated alleles while sparing the wild-type allele. Such an approach employs a composition, consisting of a CRISPR-associated nuclease (OMNI A1 V10) and two single guide RNAs, that specifically excises a fraction of the mutated allele. Our results demonstrated editing at the site of the SNP in about 80%–90% of the mutant alleles in HSCs from patients harboring a mutation on either the reference or the alternative allele. In contrast, the complementary wild-type allele was maintained intact at about 97%–100%, indicating allele-specific editing took place. NGS analysis of cDNA for the mutation site in excised patient cells showed an enrichment in the wild-type allele compared with non-excised cells, further supporting the knockout of the mutant allele alongside preservation of the wild-type copy. In addition, OMNI A1 V10 showed high fidelity without traceable off targets. These unique features of our nuclease may provide an advantage and should be explored in the context of other indications that are dominant, dominant negative, and compound heterozygous, covering most genetic disorders that other technologies cannot address.
The OMNI A1 V10 composition showed excision efficiency of about 25%. Notably, given the high allele specificity of this nuclease composition, it is estimated that about 50% of the cell population had undergone excision at the mutant allele. With respect to the functional aspects, ELANE monoallelic excision significantly enhanced neutrophil differentiation in vitro. It also reduced the aberrant numbers of monocytes, consistent with the hematopoietic defect of SCN patients. Excised neutrophils showed normal phagocytic and bacterial killing capacities, indicating that the editing did not impair core neutrophilic functions and is therefore safe. Notably, analysis of phagocytosis out of the total CD66b+ population showed no differences between patient-derived edited and NT neutrophils (Figure S15), supporting the claim that our editing strategy rescues the impaired differentiation arrest in patient-derived cells rather than affecting the function of neutrophils.
In principle, an alternative approach for restoring neutrophil differentiation in SCN-derived HSCs could be templated HDR for correction of ELANE mutations.30 However, in most cases, HDR-based gene correction necessitates tailor-made repair strategies for specific mutations. Given the numerous mutations associated with SCN,1, 2, 3, 4 this approach would be clinically unattainable for some of the patients. In contrast, the editing approach demonstrated in this study is based on targeting SNPs that are frequently heterozygous in the SCN patient population and are linked to the majority of ELANE mutations, rather than addressing each mutation independently. Patients’ cells were genotyped to determine SNP-mutation linkage. Then, a relevant composition was chosen based on the SNP-mutation localization to either the reference or the alternative allele. By using only three editing strategies (one for each identified SNP, each employing two different compositions directed to either the reference or the alternative allele), the approach described herein could provide a therapeutic solution to more than about 75% of the ELANE-mediated SCN patient population through autologous HSC monoallelic editing and subsequent transplantation.
Thus, the current study presents a novel CRISPR-based strategy of specific monoallelic knockout. Such a strategy was found to be efficient, functional, accurate, and safe, thereby providing an alternative therapeutic route for SCN. The technology disclosed herein could potentially offer new therapeutic opportunities for other genetic disorders. Of note, the development of a clinical composition requires additional studies to meet the highest safety and efficacy standards. For example, an in vivo engraftment study using an immunocompromised mouse model is needed to support the long-term engraftment and multilineage reconstitution potential of our edited cells. Moreover, although the GUIDE-seq and in silico analyses provided in our study did not depict any off-targets, additional biochemical assays for assessment of off-targets and editing-mediated translocations are necessary.
Materials and methods
Human HSC isolation
HSCs were isolated from SCN patients’ bone marrow and healthy donors’ mobilized peripheral blood.
Heterozygosity frequency of SNPs in the healthy and patient populations
Variant call files encompassing the ELANE gene region (±3 kb of ELANE) were downloaded from the 1000 Genomes Project Consortium (phase 3). Genotypes from 2,407 unrelated individuals were analyzed. Three SNPs were chosen (rs3761005, rs1683564, and rs10414837 polymorphisms) to optimize the population coverage for allele-specific ELANE knockout. Fifty-three patients’ samples were sequenced for the pathogenic mutation and the three chosen SNPs. The heterozygosity frequency of each of the three chosen SNPs and the percentage of the population being heterozygous for at least one of them were calculated for both the healthy and the patient populations.
CRISPR-associated OMNI-A1-V10 ELANE gene editing
An RNP system at a molar ratio of 1:2.5 (OMNI-A1-V10 nuclease:sgRNA) was used. Human CD34+ cells were electroporated using the CA-137 program (Lonza 4D, Nucleofector).
Digital Droplet PCR (excision, allele specificity)
Excision and allele specificity were measured using Digital Droplet PCR on genomic DNA. For excision reaction, amplification of two regions in ELANE, exon 1 and exon 5, was performed, using two different probes, FAM(X1) and HEX(X1), respectively. The ratio between the probe signals was translated to excision efficiency. For allele specificity, a FAM probe (binding the alternative allele) and a HEX probe (binding the reference allele) were used (FAM + HEX). The ratio between the two probes was normalized to endogenous genes.
Assessment of mutated:wild-type allele ratio
cDNAs were mapped using NGS targeting exons 4 and 5 harboring S126L and R220Q mutations in patients 41 and 55, respectively. The relative ratio of mutated:wild-type alleles in treated cells was calculated and compared with that of NT cells.
ELANE mRNA expression levels estimate
RNA was purified from day 6 differentiated patient HSCs excised with either RNP(ref) or RNP(alt), mock treated or not treated, using the RNeasy Mini Kit (Qiagen no. 74104). cDNA was prepared using a High-Capacity RNA-to-cDNA kit (Applied Biosystems, no. 4387406). ELANE expression levels were measured by ddPCR and normalized to GAPDH mRNA levels. Data are presented relative to ELANE levels in mock or NT samples.
Differentiation assay
Edited and NT HSCs were subjected to a differentiation protocol adopted from Nasri et al.19 On day 14, cells were analyzed by flow cytometry for monocytic (CD14+/CD66b−) and neutrophilic (CD66b+), (CD11b+/CD15+) subsets.
Bacterial-killing assay
Day 13 differentiated HSCs were evaluated for their bacterial-killing capacity as described in Atosuo and Lilius,52 using luciferase-expressing bacteria.53 RLUs were measured over 5 h. The last time point presented is when RLU levels reached plateau. Wells without differentiated HSCs (E. coli only) and with the phagocytosis inhibitor cytochalasin D (data not shown) served as controls.
Phagocytosis assay
Phagocytosis capacity was evaluated using the EZCell phagocytosis assay kit (Green Zymosan) (BioVision, cat. no. K397). Cells were analyzed by flow cytometry for internalization of opsonized fluorescent zymosan green particles.
Further details on fluorescence-activated cell sorting (FACS) antibodies, probes, sequences, assays, protocols, and additional methods are provided in the supplemental information.
Acknowledgments
This study was supported by research funding from EmendoBio, Inc., and the National Institute of Health (1RO1HL151629 and 5R24AI049393) to D.C.D. and Gil Harari, PhD (CEO, MediStat Ltd), assisted with statistical analysis. The work presented in this article was done in Seattle, Washington, USA.
Author contributions
P.S., V.M., Y.D., A.H., R.E., and D.C.D. designed the research. P.S., Y.D., L.P., D.E., E.S., O.H., and T.P. performed research, analyzed, and interpreted data. L.R., T.B., and M.G. designed the engineered nuclease; M.A. produced the nuclease. P.S., V.M., L.P., E.S., R.E., and D.C.D. commented on the manuscript. A.L.D. collected and interpreted data, performed statistical analysis, and wrote the manuscript.
Declaration of interests
L.P., L.R., T.B., D.E., E.S., O.H., M.A., A.L.D., A.H., and R.E. are employed by EmendoBio, Inc., which is the owner of the editing technology reported in this research. Y.D. and M.G. are former employees of EmendoBio, Inc. P.S., V.M., and T.P. receive research funding from EmendoBio, Inc. D.C.D. is a consultant and receives honoraria and research funding from EmendoBio, Inc., Amgen, and X4 Pharmaceuticals.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2022.06.002.
Contributor Information
Rafi Emmanuel, Email: rafi@emendobio.com.
David C. Dale, Email: dcdale@uw.edu.
Supplemental information
Raw data from an unbiased survey (GUIDE-seq) of whole-genome off-target cleavage using OMNI A1 V10 nuclease and each of constant guide (SgRNA(constant)), reference guide (SgRNA(ref)), and alternative guide (SgRNA(alt)).
References
- 1.Skokowa J., Dale D.C., Touw I.P., Zeidler C., Welte K. Severe congenital neutropenias. Nat. Rev. Dis. Primer. 2017;3 doi: 10.1038/nrdp.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dale D.C., Link D.C. The many causes of severe congenital neutropenia. N. Engl. J. Med. 2009;360:3–5. doi: 10.1056/nejmp0806821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janeway K.A. In: Comprehensive Pediatric Hospital Medicine. Zaoutis L.B., Chiang V.W., editors. Mosby; 2007. Chapter 118 - neutropenia; pp. 738–743. [Google Scholar]
- 4.Makaryan V., Zeidler C., Bolyard A.A., Skokowa J., Rodger E., Kelley M.L., Boxer L.A., Bonilla M.A., Newburger P.E., Shimamura A., et al. The diversity of mutations and clinical outcomes for ELANE-associated neutropenia. Curr. Opin. Hematol. 2015;22:3–11. doi: 10.1097/moh.0000000000000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellanné-Chantelot C., Clauin S., Leblanc T., Cassinat B., Rodrigues-Lima F., Beaufils S., Vaury C., Barkaoui M., Fenneteau O., Maier-Redelsperger M., et al. Mutations in the ELA2 gene correlate with more severe expression of neutropenia: a study of 81 patients from the French Neutropenia Register. Blood. 2004;103:4119–4125. doi: 10.1182/blood-2003-10-3518. [DOI] [PubMed] [Google Scholar]
- 6.Welte K., Zeidler C., Dale D.C. Severe congenital neutropenia. Semin. Hematol. 2006;43:189–195. doi: 10.1053/j.seminhematol.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Skokowa J., Germeshausen M., Zeidler C., Welte K. Severe congenital neutropenia: inheritance and pathophysiology. Curr. Opin. Hematol. 2007;14:21–28. doi: 10.1097/00062752-200701000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Dale D.C., Person R.E., Bolyard A.A., Aprikyan A.G., Bos C., Bonilla M.A., Boxer L.A., Kannourakis G., Zeidler C., Welte K., et al. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood. 2000;96:2317–2322. doi: 10.1182/blood.v96.7.2317. [DOI] [PubMed] [Google Scholar]
- 9.Horwitz M.S., Duan Z., Korkmaz B., Lee H.-H., Mealiffe M.E., Salipante S.J. Neutrophil elastase in cyclic and severe congenital neutropenia. Blood. 2007;109:1817–1824. doi: 10.1182/blood-2006-08-019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makaryan V., Kelley M.L., Fletcher B., Bolyard A.A., Aprikyan A.A., Dale D.C. Elastase inhibitors as potential therapies for ELANE-associated neutropenia. J. Leukoc. Biol. 2017;102:1143–1151. doi: 10.1189/jlb.5a1016-445r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dale D.C., Bonilla M.A., Davis M.W., Nakanishi A.M., Hammond W.P., Kurtzberg J., Wang W., Jakubowski A., Winton E., Lalezari P., JAkubowski A. A randomized controlled phase III trial of recombinant human granulocyte colony-stimulating factor (Filgrastim) for treatment of severe chronic neutropenia. Blood. 1993;81:2496–2502. doi: 10.1182/blood.v81.10.2496.bloodjournal81102496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg P.S., Alter B.P., Bolyard A.A., Bonilla M.A., Boxer L.A., Cham B., Fier C., Freedman M., Kannourakis G., Kinsey S., et al. The incidence of leukemia and mortality from sepsis in patients with severe congenital neutropenia receiving long-term G-CSF therapy. Blood. 2006;107:4628–4635. doi: 10.1182/blood-2005-11-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg P.S., Zeidler C., Bolyard A.A., Alter B.P., Bonilla M.A., Boxer L.A., Dror Y., Kinsey S., Link D.C., Newburger P.E., et al. Stable long-term risk of leukaemia in patients with severe congenital neutropenia maintained on G-CSF therapy. Br. J. Haematol. 2010;150:196–199. doi: 10.1111/j.1365-2141.2010.08216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donadieu J., Leblanc T., Bader Meunier B., Barkaoui M., Fenneteau O., Bertrand Y., Maier-Redelsperger M., Micheau M., Stephan J.L., Phillipe N., et al. French Severe Chronic Neutropenia Study Group Analysis of risk factors for myelodysplasias, leukemias and death from infection among patients with congenital neutropenia. Experience of the French Severe Chronic Neutropenia Study Group. Haematologica. 2005;90:45–53. [PubMed] [Google Scholar]
- 15.Rotulo G.A., Beaupain B., Rialland F., Paillard C., Nachit O., Galambrun C., Gandemer V., Bertrand Y., Neven B., Dore E., et al. HSCT may lower leukemia risk in ELANE neutropenia: a before–after study from the French Severe Congenital Neutropenia Registry. Bone Marrow Transplant. 2020;55:1614–1622. doi: 10.1038/s41409-020-0800-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fioredda F., Iacobelli S., van Biezen A., Gaspar B., Ancliff P., Donadieu J., Aljurf M., Peters C., Calvillo M., Matthes-Martin S., et al. Stem cell transplantation in severe congenital neutropenia: an analysis from the European Society for Blood and Marrow Transplantation. Blood. 2015;126:1885–1892. doi: 10.1182/blood-2015-02-628859. quiz 1970. [DOI] [PubMed] [Google Scholar]
- 17.Zeidler C., Welte K., Barak Y., Barriga F., Bolyard A.A., Boxer L., Cornu G., Cowan M.J., Dale D.C., Flood T., et al. Stem cell transplantation in patients with severe congenital neutropenia without evidence of leukemic transformation. Blood. 2000;95:1195–1198. [PubMed] [Google Scholar]
- 18.Rao S., Brito-Frazao J., Serbin A.V., Yao Q., Luk K., Wu Y., Zeng J., Ren C., Watkinson R., Armant M., et al. Gene editing ELANE in human hematopoietic stem and progenitor cells reveals disease mechanisms and therapeutic strategies for severe congenital neutropenia. Blood. 2019;134:3. doi: 10.1182/blood-2019-131073. [DOI] [Google Scholar]
- 19.Nasri M., Ritter M., Mir P., Dannenmann B., Aghaallaei N., Amend D., Makaryan V., Xu Y., Fletcher B., Bernhard R., et al. CRISPR/Cas9-mediated ELANE knockout enables neutrophilic maturation of primary hematopoietic stem and progenitor cells and induced pluripotent stem cells of severe congenital neutropenia patients. Haematologica. 2020;105:598–609. doi: 10.3324/haematol.2019.221804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao S., Yao Y., Soares de Brito J., Yao Q., Shen A.H., Watkinson R.E., Kennedy A.L., Coyne S., Ren C., Zeng J., et al. Dissecting ELANE neutropenia pathogenicity by human HSC gene editing. Cell Stem Cell. 2021;28:833–845.e5. doi: 10.1016/j.stem.2020.12.015. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirche T.O., Benabid R., Deslee G., Gangloff S., Achilefu S., Guenounou M., Lebargy F., Hancock R.E., Belaaouaj A. Neutrophil elastase mediates innate host protection against Pseudomonas aeruginosa. J. Immunol. 2008;181:4945–4954. doi: 10.4049/jimmunol.181.7.4945. Baltim. Md 1950. [DOI] [PubMed] [Google Scholar]
- 22.Tkalcevic J., Novelli M., Phylactides M., Iredale J.P., Segal A.W., Roes J. Impaired immunity and enhanced resistance to endotoxin in the absence of neutrophil elastase and cathepsin G. Immunity. 2000;12:201–210. doi: 10.1016/s1074-7613(00)80173-9. [DOI] [PubMed] [Google Scholar]
- 23.Belaaouaj A. Neutrophil elastase-mediated killing of bacteria: lessons from targeted mutagenesis. Microbes Infect. 2002;4:1259–1264. doi: 10.1016/s1286-4579(02)01654-4. [DOI] [PubMed] [Google Scholar]
- 24.Belaaouaj A., McCarthy R., Baumann M., Gao Z., Ley T.J., Abraham S.N., Shapiro S.D. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat. Med. 1998;4:615–618. doi: 10.1038/nm0598-615. [DOI] [PubMed] [Google Scholar]
- 25.Reeves E.P., Lu H., Jacobs H.L., Messina C.G.M., Bolsover S., Gabella G., Potma E.O., Warley A., Roes J., Segal A.W. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature. 2002;416:291–297. doi: 10.1038/416291a. [DOI] [PubMed] [Google Scholar]
- 26.Weinrauch Y., Drujan D., Shapiro S.D., Weiss J., Zychlinsky A. Neutrophil elastase targets virulence factors of enterobacteria. Nature. 2002;417:91–94. doi: 10.1038/417091a. [DOI] [PubMed] [Google Scholar]
- 27.Benabid R., Wartelle J., Malleret L., Guyot N., Gangloff S., Lebargy F., Belaaouaj A. Neutrophil elastase modulates cytokine expression. J. Biol. Chem. 2012;287:34883–34894. doi: 10.1074/jbc.m112.361352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., Weinrauch Y., Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 29.Cui C., Chakraborty K., Tang X.A., Zhou G., Schoenfelt K.Q., Becker K.M., Hoffman A., Chang Y.-F., Blank A., Reardon C.A., et al. Neutrophil elastase selectively kills cancer cells and attenuates tumorigenesis. Cell. 2021;184:3163–3177.e21. doi: 10.1016/j.cell.2021.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran N.T., Graf R., Wulf-Goldenberg A., Stecklum M., Strauß G., Kühn R., Kocks C., Rajewsky K., Chu V.T. CRISPR-Cas9-Mediated ELANE mutation correction in hematopoietic stem and progenitor cells to treat severe congenital neutropenia. Mol. Ther. J. Am. Soc. Gene Ther. 2020;28:2621–2634. doi: 10.1016/j.ymthe.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeWitt M.A., Magis W., Bray N.L., Wang T., Berman J.R., Urbinati F., Heo S.-J., Mitros T., Muñoz D.P., Boffelli D., et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl. Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo C.Y., Long J.D., Campo-Fernandez B., de Oliveira S., Cooper A.R., Romero Z., Hoban M.D., Joglekar A.V., Lill G.R., Kaufman M.L., et al. Site-specific gene editing of human hematopoietic stem cells for X-linked hyper-IgM syndrome. Cell Rep. 2018;23:2606–2616. doi: 10.1016/j.celrep.2018.04.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavel-Dinu M., Wiebking V., Dejene B.T., Srifa W., Mantri S., Nicolas C.E., Lee C., Bao G., Kildebeck E.J., Punjya N., et al. Gene correction for SCID-X1 in long-term hematopoietic stem cells. Nat. Commun. 2019;10:1634. doi: 10.1038/s41467-019-09614-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romero Z., Lomova A., Said S., Miggelbrink A., Kuo C.Y., Campo-Fernandez B., Hoban M.D., Masiuk K.E., Clark D.N., Long J., et al. Editing the sickle cell disease mutation in human hematopoietic stem cells: comparison of endonucleases and homologous donor templates. Mol. Ther. 2019;27:1389–1406. doi: 10.1016/j.ymthe.2019.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radtke S., Pande D., Cui M., Perez A.M., Chan Y.-Y., Enstrom M., Schmuck S., Berger A., Eunson T., Adair J.E., Kiem H.P. Purification of human CD34+CD90+ HSCs reduces target cell population and improves lentiviral transduction for gene therapy. Mol. Ther. - Methods Clin. Dev. 2020;18:679–691. doi: 10.1016/j.omtm.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donini M., Fontana S., Savoldi G., Vermi W., Tassone L., Gentili F., Zenaro E., Ferrari D., Notarangelo L.D., Porta F., et al. G-CSF treatment of severe congenital neutropenia reverses neutropenia but does not correct the underlying functional deficiency of the neutrophil in defending against microorganisms. Blood. 2007;109:4716–4723. doi: 10.1182/blood-2006-09-045427. [DOI] [PubMed] [Google Scholar]
- 37.Horwitz M.S., Corey S.J., Grimes H.L., Tidwell T. ELANE mutations in cyclic and severe congenital neutropenia: genetics and pathophysiology. Hematol. Oncol. Clin. North Am. 2013;27:19–41. doi: 10.1016/j.hoc.2012.10.004. vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Germeshausen M., Deerberg S., Peter Y., Reimer C., Kratz C.P., Ballmaier M. The spectrum of ELANE mutations and their implications in severe congenital and cyclic neutropenia. Hum. Mutat. 2013;34:905–914. doi: 10.1002/humu.22308. [DOI] [PubMed] [Google Scholar]
- 39.Dale D.C., Makaryan V. In: GeneReviews®. Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J., Mirzaa G., Amemiya A., editors. University of Washington, Seattle; 1993. ELANE-related Neutropenia. [PubMed] [Google Scholar]
- 40.Briars G.L., Parry H.F., Ansari B.M. Dominantly inherited severe congenital neutropenia. J. Infect. 1996;33:123–126. doi: 10.1016/s0163-4453(96)93081-9. [DOI] [PubMed] [Google Scholar]
- 41.Li F.-Q., Horwitz M. Characterization of mutant neutrophil elastase in severe congenital neutropenia. J. Biol. Chem. 2001;276:14230–14241. doi: 10.1074/jbc.m010279200. [DOI] [PubMed] [Google Scholar]
- 42.Aprikyan A.A.G., Kutyavin T., Stein S., Aprikian P., Rodger E., Liles W.C., Boxer L.A., Dale D.C. Cellular and molecular abnormalities in severe congenital neutropenia predisposing to leukemia. Exp. Hematol. 2003;31:372–381. doi: 10.1016/s0301-472x(03)00048-1. [DOI] [PubMed] [Google Scholar]
- 43.Grenda D.S., Murakami M., Ghatak J., Xia J., Boxer L.A., Dale D., Dinauer M.C., Link D.C. Mutations of the ELA2 gene found in patients with severe congenital neutropenia induce the unfolded protein response and cellular apoptosis. Blood. 2007;110:4179–4187. doi: 10.1182/blood-2006-11-057299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Köllner I., Sodeik B., Schreek S., Heyn H., von Neuhoff N., Germeshausen M., Zeidler C., Krüger M., Schlegelberger B., Welte K., Beger C. Mutations in neutrophil elastase causing congenital neutropenia lead to cytoplasmic protein accumulation and induction of the unfolded protein response. Blood. 2006;108:493–500. doi: 10.1182/blood-2005-11-4689. [DOI] [PubMed] [Google Scholar]
- 45.Nayak R.C., Trump L.R., Aronow B.J., Myers K., Mehta P., Kalfa T., Wellendorf A.M., Valencia C.A., Paddison P.J., Horwitz M.S., et al. Pathogenesis of ELANE-mutant severe neutropenia revealed by induced pluripotent stem cells. J. Clin. Invest. 2015;125:3103–3116. doi: 10.1172/jci80924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia R., Gusmani L., Murgia R., Guarnaccia C., Cinco M., Rottini G. Elastase is the only human neutrophil granule protein that alone is responsible for in vitro killing of Borrelia burgdorferi. Infect. Immun. 1998;66:1408–1412. doi: 10.1128/iai.66.4.1408-1412.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.López-Boado Y.S., Espinola M., Bahr S., Belaaouaj A. Neutrophil serine proteinases cleave bacterial Flagellin, abrogating its host response-inducing activity. J. Immunol. 2004;172:509–515. doi: 10.4049/jimmunol.172.1.509. [DOI] [PubMed] [Google Scholar]
- 48.Papayannopoulos V., Metzler K.D., Hakkim A., Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J. Cell Biol. 2010;191:677–691. doi: 10.1083/jcb.201006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.ELANE | gnomAD v2.1.1 | gnomAD. https://gnomad.broadinstitute.org/gene/ENSG00000197561?dataset=gnomad_r2_1
- 50.Horwitz M.S., Laurino M.Y., Keel S.B. Normal peripheral blood neutrophil numbers accompanying ELANE whole gene deletion mutation. Blood Adv. 2019;3:2470–2473. doi: 10.1182/bloodadvances.2019000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oikemus S.R., Pfister E., Sapp E., Chase K.O., Kennington L.A., Hudgens E., Miller R., Zhu L.J., Chaudhary A., Mick E.O., et al. Allele-specific knockdown of mutant HTT protein via editing at coding region SNP heterozygosities. Hum. Gene Ther. 2021 doi: 10.1089/hum.2020.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Atosuo J.T., Lilius E.-M. The real-time-based assessment of the microbial killing by the antimicrobial compounds of neutrophils. Sci. World J. 2011;11:2382–2390. doi: 10.1100/2011/376278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karsi A., Lawrence M.L. Broad host range fluorescence and bioluminescence expression vectors for Gram-negative bacteria. Plasmid. 2007;57:286–295. doi: 10.1016/j.plasmid.2006.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw data from an unbiased survey (GUIDE-seq) of whole-genome off-target cleavage using OMNI A1 V10 nuclease and each of constant guide (SgRNA(constant)), reference guide (SgRNA(ref)), and alternative guide (SgRNA(alt)).