ABSTRACT
Downstream next-generation sequencing (NGS) of the syphilis spirochete Treponema pallidum subspecies pallidum (T. pallidum) is hindered by low bacterial loads and the overwhelming presence of background metagenomic DNA in clinical specimens. In this study, we investigated selective whole-genome amplification (SWGA) utilizing multiple displacement amplification (MDA) in conjunction with custom oligonucleotides with an increased specificity for the T. pallidum genome and the capture and removal of 5′-C-phosphate-G-3′ (CpG) methylated host DNA using the NEBNext Microbiome DNA enrichment kit followed by MDA with the REPLI-g single cell kit as enrichment methods to improve the yields of T. pallidum DNA in isolates and lesion specimens from syphilis patients. Sequencing was performed using the Illumina MiSeq v2 500 cycle or NovaSeq 6000 SP platform. These two enrichment methods led to 93 to 98% genome coverage at 5 reads/site in 5 clinical specimens from the United States and rabbit-propagated isolates, containing >14 T. pallidum genomic copies/μL of sample for SWGA and >129 genomic copies/μL for CpG methylation capture with MDA. Variant analysis using sequencing data derived from SWGA-enriched specimens showed that all 5 clinical strains had the A2058G mutation associated with azithromycin resistance. SWGA is a robust method that allows direct whole-genome sequencing (WGS) of specimens containing very low numbers of T. pallidum, which has been challenging until now.
IMPORTANCE Syphilis is a sexually transmitted, disseminated acute and chronic infection caused by the bacterial pathogen Treponema pallidum subspecies pallidum. Primary syphilis typically presents as single or multiple mucocutaneous lesions and, if left untreated, can progress through multiple stages with various clinical manifestations. Molecular studies often rely on direct amplification of DNA sequences from clinical specimens; however, this can be impacted by inadequate samples due to disease progression or timing of patients seeking clinical care. While genotyping has provided important data on circulating strains over the past 2 decades, WGS data are needed to better understand strain diversity, perform evolutionary tracing, and monitor antimicrobial resistance markers. The significance of our research is the development of an SWGA DNA enrichment method that expands the range of clinical specimens that can be directly sequenced to include samples with low numbers of T. pallidum.
KEYWORDS: DNA enrichment, Treponema pallidum, metagenomics, syphilis, whole-genome sequencing
INTRODUCTION
Syphilis, caused by Treponema pallidum subspecies pallidum (here referred to as T. pallidum), is steadily increasing in the United States. In 2019, 38,992 cases (11.9 per 100,000 people) of primary and secondary (P&S) syphilis and 1,870 cases (48.5 per 100,000 live births) of congenital syphilis were reported to the CDC, representing a 167.2% increase in P&S syphilis rates since 2010 and a 291.1% increase in congenital syphilis since 2015 (1). Penicillin has been the drug of choice for treating all stages of syphilis; however, azithromycin has been used as an alternative to penicillin for treating early syphilis or contacts of syphilis cases worldwide. Macrolide-resistant T. pallidum strains associated with two mutations (A2508G, A2509G) in the 23S rRNA genes have since been reported worldwide (2–5). While macrolides are not recommended as first-line treatment in many countries, periodic monitoring is useful to determine the prevalence of resistant strains (6).
Molecular studies on contemporary T. pallidum strains in the United States remains challenging due to the limited number of whole genomes sequenced directly from clinical specimens. Strain diversity has been gleaned from molecular epidemiology studies, which are based on a few genetic loci but may not be representative of the entire T. pallidum genome (7–10). In addition, studies have relied primarily on strains propagated in rabbits or DNA amplified directly from clinical specimens, because T. pallidum cannot be grown on routine laboratory medium. However, advances have been made with in vitro tissue culture and the propagation of T. pallidum in rabbits from cryopreserved genital lesion specimens, which may make routine culture directly from clinical specimens a possibility in the near future (11–13).
Metagenomic shotgun sequencing approaches have made significant advances in recent years, with sequence data being used for pathogen detection, whole-genome-based typing, and antimicrobial resistance marker detection, in addition to phylogenetic analyses (14–16). However, direct whole-genome sequencing (WGS) of T. pallidum from clinical specimens and rabbit isolates can be problematic due to bacterial genomic DNA (gDNA) being outweighed by either human or rabbit DNA, respectively. Several DNA enrichment methods have been described for T. pallidum, including RNA bait capture techniques, methyl-directed enrichment using the restriction nuclease DpnI, and pooled whole-genome amplification (17–22). These methods have generated T. pallidum-specific WGS data from over 700 metagenomic samples; however, sequencing specimens with low numbers of T. pallidum remains challenging. Therefore, new approaches that would enable sequencing of samples with low bacterial loads are needed. Host DNA removal by 5′-C-phosphate-G-3′ (CpG) methylated capture and selective whole-genome amplification (SWGA), which allows selective amplification of gDNA of the species of interest compared to host DNA, have been successfully used for enriching bacterial gDNA in metagenomic samples (23–27); however, these methods have not been applied to T. pallidum.
In this study, we describe a DNA enrichment method based on selective whole-genome amplification (SWGA) using multiple displacement amplification (MDA) and custom primers that enables WGS of clinical specimens with very low genomic copies of T. pallidum. We also investigated an alternative method, the NEBNext Microbiome DNA enrichment kit, that uses CpG methylated capture of host DNA followed by MDA with the REPLI-g single cell kit.
RESULTS
Real-time qPCR on clinical specimens and spiked samples.
A total of 15 clinical specimens were included in this study (Table 1). DNAs were extracted using standard or large-scale extraction protocols, and T. pallidum gDNA (copies/μL of extract) was estimated using quantitative PCR (qPCR) targeting the polA gene. As shown in Table 1, 3 specimens had >100 T. pallidum gDNA copies/μL and the remaining 12 specimens had <32 copies/μL. PCR amplification of the human RNase P gene (RNP) was used to estimate the concentration of human gDNA in each sample (28). RNP threshold cycle (CT) values in the specimens ranged from 22.61 (highest concentration of RNP) to 38.32 (lowest concentration of RNP) (Table 1).
TABLE 1.
Clinical and laboratory data for specimens and the T. pallidum isolatea
| Sample/isolate IDb | Collection yr | Gender | Sexual orientationc | Syphilis stage | Site of lesion | Antibody titer (assay)d | qPCR (T. pallidum gDNA in extract) (copies/μL) | RNPCT | Extraction method | Reference or source |
|---|---|---|---|---|---|---|---|---|---|---|
| CDC-SF003 | 2017 | Male | MSM | Primary | Penis | 1:4 (VDRL) | 9,680 | NA | Standard | 11 |
| EUHM-001 | 2019 | Male | MSM | Secondary | Neck | 1:128 (RPR) | <1 | 29.59 ± 0.20 | Standard | This study |
| EUHM-002 | 2019 | Male | MSM | Secondary | Perianal | 1:256 (RPR) | <1 | 28.15 ± 0.04 | Standard | This study |
| EUHM-003 | 2019 | Male | MSM | Secondary | Penis | 1:32 (RPR) | <1 | 29.35 ± 0.08 | Standard | This study |
| EUHM-004 | 2019 | Male | MSM | Primary | Penis | 1:4 (RPR) | 106.7 ± 6.5 | 25.48 ± 0.04 | Standard | This study |
| EUHM-005 | 2019 | Male | MSM | Secondary | Penis | 1:64 (RPR) | <1 | 33.34 ± 0.03 | Standard | This study |
| EUHM-006 | 2019 | Male | MSM | Primary | Penis | 1:16 (RPR) | <1 | 31.62 ± 0.03 | Standard | This study |
| EUHM-007 | 2019 | Male | MSM | Secondary | Hand | 1:64 (RPR) | <1 | 38.32 ± 0.1 | Standard | This study |
| EUHM-008 | 2019 | Male | MSM | Secondary | Scrotum | 1:64 (RPR) | 0.9 ± 0.1 | 31.00 ± 0.1 | Standard | This study |
| EUHM-009 | 2019 | Male | MSM | Secondary | Scrotum | 1:64 (RPR) | <1 | 33.24 ± 0.14 | Standard | This study |
| EUHM-010 | 2019 | Male | MSM | Secondary | Scrotum | 1:128 (RPR) | <1 | 31.27 ± 0.08 | Standard | This study |
| EUHM-011 | 2019 | Male | MSM | Primary | Penis | 1:32 (RPR) | <1 | 32.87 ± 0.21 | Standard | This study |
| EUHM-012 | 2019 | Male | MSM | Primary | Penis | 1:8 (RPR) | 31.5 ± 0.5 | 22.61 ± 0.08 | Large scale | This study |
| EUHM-013 | 2020 | Male | MSM | Secondary | Penis | 1:64 (RPR) | 122 ± 1.2 | 31.38 ± 0.21 | Large scale | This study |
| EUHM-014 | 2020 | Male | MSM | Secondary | NA | 1:16 (RPR) | 103 ± 6.7 | 24.06 ± 0.1 | Large scale | This study |
| STLC-001 | 2020 | Male | MSW | Primary | Penis | NR (RPR) | 28.8 ± 3.1 | 25.57 ± 0.07 | Standard | This study |
NA, not available; NR, nonreactive.
EUHM, Emory University Hospital, Atlanta, GA; STLC, St. Louis County STD Clinic, St. Louis. MO.
MSM, men who have sex with men; MSW, men who have sex with women.
VDRL, Venereal Disease Research Laboratory test; RPR, rapid plasma reagin test.
To determine the limit of detection (LoD) for each enrichment method, spiked samples were composed of a 10-fold dilution series of T. pallidum Nichols DNA against a background of a constant concentration of human DNA. An RNPCT of 25.48, corresponding to the lowest CT value among specimens extracted with the standard protocol, was targeted as the cutoff for all the spiked samples. Based on qPCR, the copy number of the undiluted spiked sample was estimated at 11,066.17 ± 364.60 gDNA copies/μL with an average RNPCT value of 25.07 ± 0.04 (Table S1). The dilution series generated from this spiked sample averaged 1,016 ± 9.41 down to 1.57 ± 0.12 T. pallidum gDNA copies/μL. The RNPCT values averaged 25.02 ± 0.03 among all samples, with no significant difference observed among the RNPCT values for each dilution in the series (P > 0.3; Table S1).
Copy number, RNPCT, and sequencing results for NEB+MDA- and SWGA-enriched spiked samples. Download Table S1, CSV file, 0.00 MB (4.4KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
NEBNext microbiome enrichment with MDA.
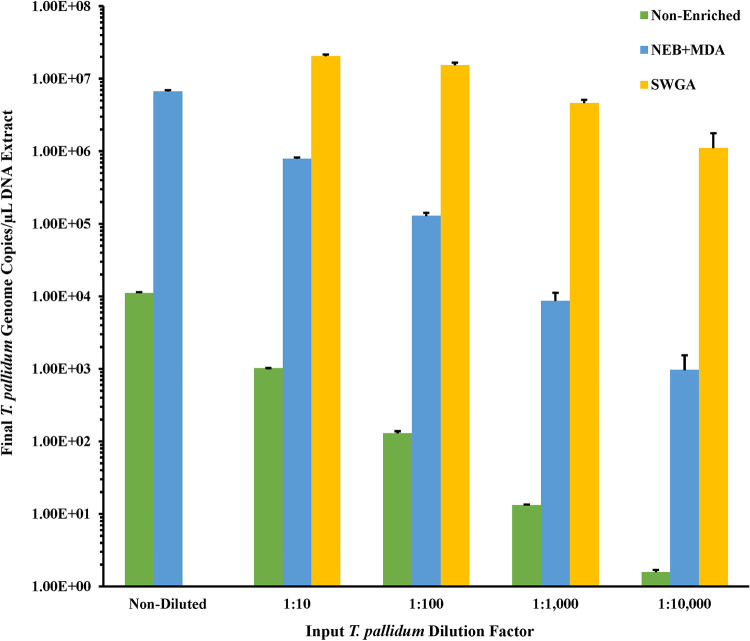
To determine the minimum T. pallidum copy number input required to generate adequate sequencing coverage, each of the replicate 10-fold-diluted spiked samples were enriched with the New England Biolabs (NEB) microbiome enrichment kit with subsequent REPLI-g single-cell MDA (here referred to as NEB+MDA). We observed an increase in T. pallidum gDNA, determined by qPCR, in all spiked samples after enrichment (Fig. 1; Table S1). The enriched spiked samples indicated an average of 6.67 × 106 ± 2.74 × 105 to 964 ± 574.23 gDNA copies/μL in the undiluted to 1:10,000 diluted samples, resulting in a 482 to 995.09× enrichment (Table S1). Upon comparing the average RNPCT of each dilution in the series, the enriched samples indicated 29.28 ± 1.07 to 31.42 ± 0.45 for the neat (undiluted) to 1:10,000 dilution (Table S1). The RNPCT values of each enriched sample in the dilution series were not significantly different from one another, with an average RNPCT of 30.64 ± 0.33 for all dilutions in the series (P = 0.22).
FIG 1.
T. pallidum gDNA copies/μL for the 10-fold dilution series spiked samples enriched by the NEB+MDA or SWGA. Spiked samples were composed of a 10-fold dilution series of T. pallidum Nichols DNA and a constant concentration of human DNA. T. pallidum genome DNA (copies/μL of DNA extract) in samples pre- and postenrichment was estimated using PCR targeting the polA gene and are shown in the bar graph. The y axis has been log10-scaled for depiction of the nonenriched dilution series. Error bars represent the standard error among three replicate enriched T. pallidum samples.
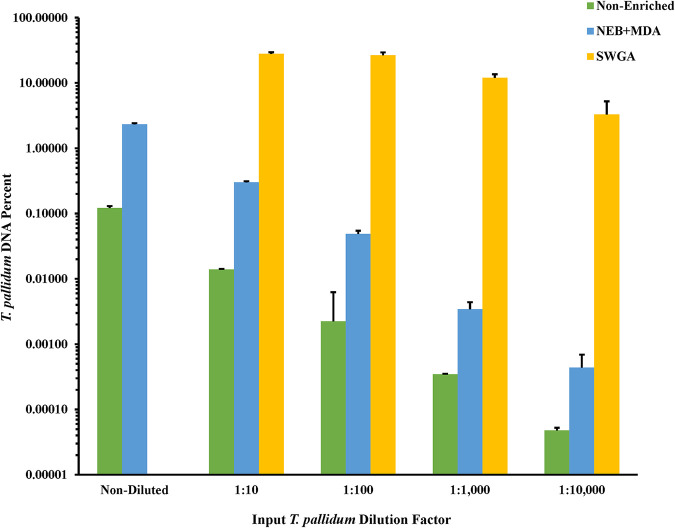
After enrichment with NEB+MDA, the average percentage of T. pallidum DNA in the total DNA ranged from 2.33% ± 0.10 to 0.0004% ± 0.0003% for the neat to 1:10,000 dilution samples, resulting in up to a 26.12-fold increase in the percentage of T. pallidum DNA among all enriched replicates (Fig. 2). Apart from enriched samples from the 1:100 and 1:1,000 dilutions, we observed that increasing the T. pallidum input gDNA copy number 10-fold resulted in a significant increase in the total DNA belonging to T. pallidum postenrichment (P = 0.06 and P < 0.05, respectively).
FIG 2.
Relative percent T. pallidum Nichols DNA in total DNA for nonenriched and NEB+MDA- and SWGA-enriched spiked samples. Spiked samples were composed of a 10-fold dilution series of T. pallidum Nichols DNA and a constant concentration of human DNA. The percent T. pallidum DNA in total DNA was calculated based on the input DNA concentration and gDNA copies/μL (nonenriched) and the DNA concentration and gDNA copies/μL for the Nichols-spiked samples postenrichment (NEB+MDA or SWGA). Genome copies were estimated from measured T. pallidum polA copies/μL of DNA extract. The y axis is log10-scaled for depiction of the nonenriched dilution series. Error bars represent the standard error among three replicate samples.
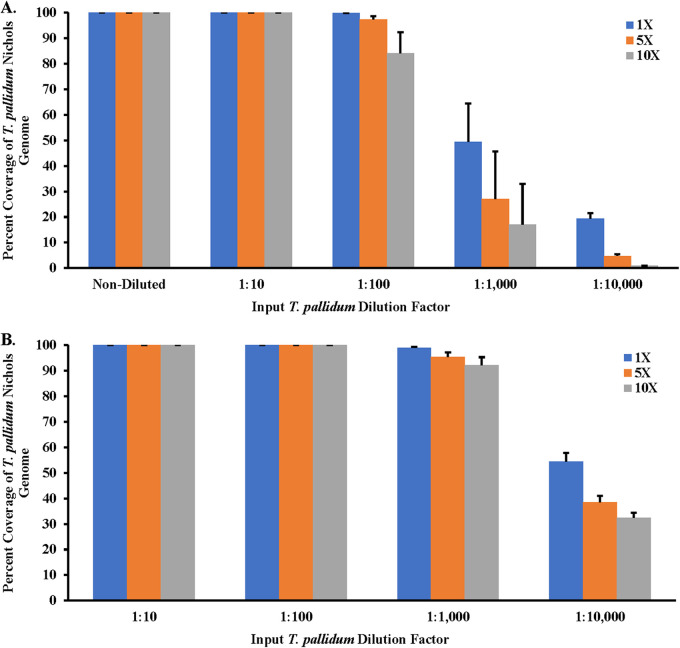
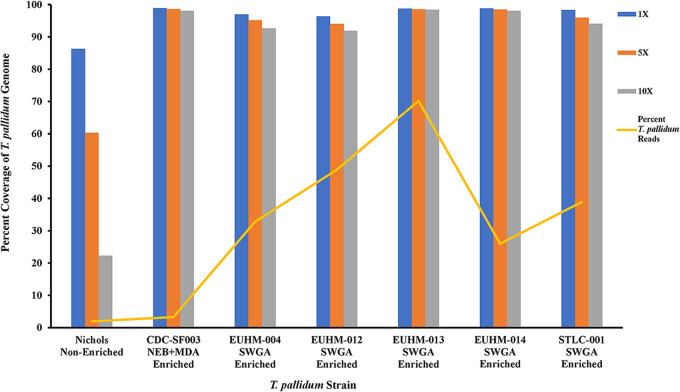
To confirm if the increase in T. pallidum gDNA copies correlated with the increase in genome coverage, sequencing data derived from samples enriched by NEB+MDA were mapped against the T. pallidum Nichols reference genome (GenBank version number NC_021490.2). The genome sequencing data showed 0.01% to 10.52% of the quality-controlled reads binned as T. pallidum, along with a mean read mapping depth to the T. pallidum Nichols reference genome ranging from 0.51× to 501.75×. An average percentage coverage exceeding 97.29% across the Nichols reference genome with at least 5 reads mapped per site (5×) for the neat to 1:100 diluted samples, was observed among the NEB+MDA enriched samples (Fig. 3A; Table S1 and Fig. S1). At the same time, for a higher coverage of at least 10 reads mapped per nucleotide (10×), the 1:100 diluted samples had an average percentage coverage of 84.14%, while neat and 1:10 dilution samples covered at 99.99% across the reference genome. A sharp decline in coverage was observed in the enriched 1:1,000 and 1:10,000 diluted samples. With the quality control (QC) criteria for efficiency set at ≥90% at ≥5× read depth, samples sequenced post-NEB+MDA enrichment had an LoD of 129 T. pallidum gDNA copies/μL (Fig. 3A; Table S1 and Fig. S1). A comparison of all the genome data generated from the serially diluted samples and the nonenriched T. pallidum Nichols control using the NEB+MDA protocol revealed no genetic variants, verifying that single nucleotide polymorphisms (SNPs), inversions, or deletions were not introduced during whole-genome amplification. NEB+MDA was subsequently used to enrich CDC-SF003, a recently propagated clinical isolate (11), with 2.39 × 106 ± 1.35 × 105 gDNA copies/μL of DNA extract observed postenrichment. We observed that 1.06% of the total DNA belonged to T. pallidum postenrichment, and 3.29% of the host-removed quality-controlled sequencing reads were classified as T. pallidum. Sequencing indicated a 98.60% coverage across the T. pallidum SS14 reference genome (GenBank version number NC_021508.1) at 5× read depth with a mean mapping depth of 46.43× (Fig. 4 and Table 2; Fig. S2).
FIG 3.
Percent coverage of sequencing reads of enriched T. pallidum Nichols spiked samples. Treponemal reads with at least 1 read mapped per site (1×) against the T. pallidum subsp. pallidum Nichols reference genome (GenBank version number NC_000919.1) and percent coverage of the T. pallidum genome are shown. (A) Sequencing reads of samples enriched using the NEB+MDA method. (B) Sequencing reads of samples enriched using SWGA. All samples were sequenced using the Illumina NovaSeq 6000 platform. Error bars represent the standard error between the mapped reads derived from three replicate enriched Nichols samples.
FIG 4.
Percent coverage of a nonenriched T. pallidum Nichols isolate control containing 1,063.1 ± 45.22 T. pallidum genomic copies/μL of DNA extract, NEB+MDA-enriched clinical isolate CDC-SF003, and SWGA-enriched clinical specimens sequenced using the Illumina MiSeq v2 (500- cycle) platform. The percentages of T. pallidum reads are derived from down-selected T. pallidum reads. Prefiltered reads for Nichols-CDC were mapped to the Nichols reference genome (GenBank version number NC_000919.1). The prefiltered reads in all clinical isolates and specimens were mapped against the SS14 reference genome (GenBank version number NC_021508.1).
TABLE 2.
Sequencing percent coverage for the Nichols isolate, clinical isolate CDC-SF003, and clinical specimens across the T. pallidum reference genome
| Sample | Enrichment methoda | Clonal complex | T. pallidum gDNA copies/μL post enrichmentc | Raw read pairs | Nonhost read pairs | Total read pairs after QC | Read pairs classified as T. pallidum | Total read pairs classified as T. pallidum (%) | Mean read depthd | Genome covered ≥1× (%)d | Genome covered ≥5× (%)d | Genome covered ≥10× (%)d |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nichols_CDCb | Nonenriched | Nichols-like | 1,063.1 ± 45.22 | 4,053,500 | 3,645,649 | 3,588,414 | 70,299 | 1.96 | 6.33 | 86.26 | 60.30 | 22.28 |
| CDC-SF003 | NEB+MDA | SS14-like | 2,394,930 ± 135,210 | 5,798,777 | 3,988,173 | 3,949,036 | 129,998 | 3.29 | 46.44 | 98.87 | 98.60 | 98.01 |
| EUHM-004 | SWGA | Nichols-like | 6,367,089.5 ± 240,811.5 | 6,102,826 | 4,440,618 | 4,280,401 | 1,403,645 | 32.79 | 370.39 | 96.99 | 95.13 | 92.67 |
| EUHM-012 | SWGA | Nichols-like | 2,140,753 ± 28,192 | 10,350,274 | 5,870,287 | 5,716,082 | 2,793,693 | 48.87 | 639.86 | 96.34 | 93.98 | 91.89 |
| EUHM-013 | SWGA | SS14-like | 5,159,716 ± 220,318.5 | 11,975,324 | 11,966,460 | 11,838,431 | 8,308,234 | 70.18 | 2,503.96 | 98.72 | 98.56 | 98.37 |
| EUHM-014 | SWGA | Nichols-like | 2,573,508 ± 221,900.5 | 11,250,518 | 9,266,926 | 9,059,022 | 2,355,426 | 26.00 | 930.87 | 98.79 | 98.49 | 98.04 |
| STLC-001 | SWGA | SS14-like | 7,420,534 ± 719,765 | 11,293,960 | 7,770,834 | 7,721,767 | 3,004,631 | 38.91 | 1,133.43 | 98.32 | 95.94 | 94.10 |
All sequencing was performed using Illumina’s MiSeq v2 (500 cycle) platform.
Nonenriched T. pallidum Nichols isolate used as MiSeq control.
Based on T. pallidum polA calculated copies/μL.
Calculated after quality assessment and T. pallidum selection of reads.
Coverage plots for the T. pallidum Nichols undiluted and 10-fold dilution series enriched using the NEB+MDA. Quality-controlled T. pallidum reads were mapped to the Nichols reference genome (GenBank version number NC_000919.1), and the log10 of the coverage at each mapped nucleotide position is shown on the x axis. Samples were sequenced using the Illumina NovaSeq 6000 platform. Download FIG S1, TIF file, 1.9 MB (1.9MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Coverage plots for a nonenriched T. pallidum Nichols isolate control containing 1,063.1 ± 45.22 T. pallidum genomic copies/μL of DNA extract, NEB+MDA-enriched clinical isolate CDC-SF003 and the SWGA-enriched Emory- and St. Louis-derived clinical specimens. T. pallidum Nichols reference isolate quality-controlled T. pallidum reads were mapped to the Nichols reference genome (GenBank version number NC_000919.1). An input number of 1,000 genomic copies/μL T. pallidum was used for the Nichols nonenriched and SWGA enriched samples. Quality-controlled T. pallidum reads for the Emory- and St. Louis-derived specimens and CDC-SF003 were mapped to the SS14 reference genome (GenBank version number NC_021508.1). The log10 of the coverage at each mapped nucleotide position is shown on the x axis. All samples were sequenced using the Illumina MiSeq v2 (500-cycle) platform. Download FIG S2, TIF file, 1.5 MB (1.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
SWGA enrichment of T. pallidum Nichols.
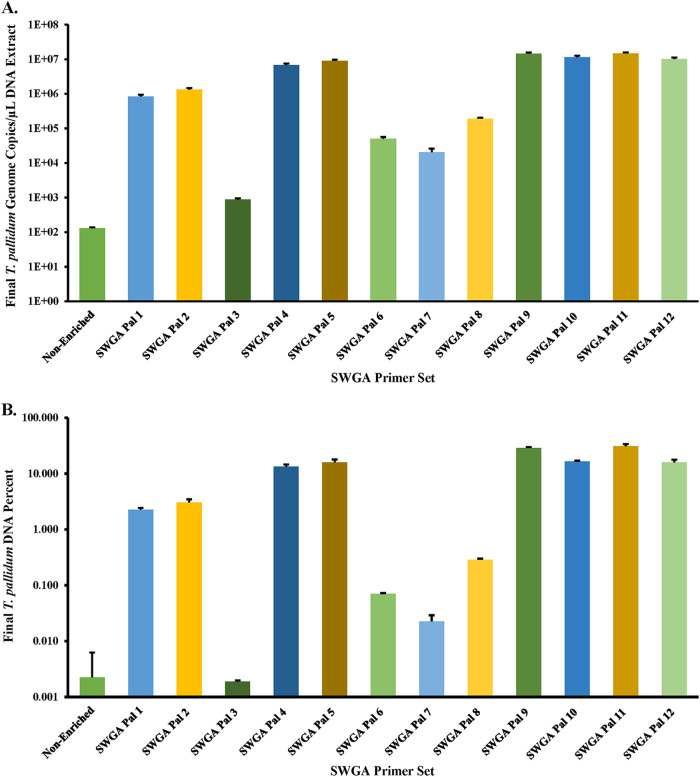
While NEB+MDA was useful for enriching samples with >129 T. pallidum gDNA copies/μL, we sought an alternative method for enriching clinical specimens with lower T. pallidum gDNA copies (Table 1). Selective whole-genome amplification (here referred to as SWGA) was chosen based on its success in other bacteria (25–27). To determine an optimal primer set for enriching T. pallidum during SWGA, a total of 12 primer sets were tested using Equiphi29 MDA (Thermo Fisher Scientific, Waltham, MA; Tables S2 and S3) and the 1:100 diluted Nichols DNA sample (~129 gDNA copies/μL) (Table S1; Table S4). The efficacy of the SWGA primer sets varied, resulting in DNA enrichment differences between 6.86 to 1.16 × 105 times (Fig. 5A) with 7 of the 12 primer sets (SWGA Pal 2, 4, 5, 9, 10, 11, and 12) and a >10,000-fold increase in gDNA copy number. SWGA Pal 9 and Pal 11 gave the highest enrichment at 1.13 × 105 and 1.16 × 105 times, respectively (Table S4). The difference observed between Pal 9 and Pal 11 in the T. pallidum gDNA copy number and relative percent DNA belonging to T. pallidum were not significantly different, and Pal 11 was selected for testing the SWGA limit of detection (P > 0.1; Fig. 5; Table S4).
FIG 5.
SWGA primer set validation. (A) T. pallidum gDNA copies/μL for the Nichols spiked sample (1:100 diluted) enriched with each SWGA primer set (1 to 12). (B) Relative percent T. pallidum DNA for the Nichols spiked sample (1:100 dilution) enriched with each SWGA primer set. Percent T. pallidum DNA was calculated based on the input DNA concentration and gDNA copies/μL for the Nichols mock samples post-SWGA enrichment. The spiked sample contained purified human gDNA, and the T. pallidum genome copies were derived from qPCR-measured T. pallidum polA copies/μL of DNA extract. The input T. pallidum gDNA copies/μL of DNA is displayed as nonenriched. The y axis is log10-scaled in each panel for depiction of the relative percent T. pallidum post-enrichment with each primer set. Error bars represent the standard error among three replicate Nichols samples.
List of primers and probes used in this study. Download Table S2, DOCX file, 0.02 MB (23KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Binding distribution of the swga Toolkit-generated primer sets to the T. pallidum and Homo sapiens genomes. Download Table S3, DOCX file, 0.02 MB (23.2KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
SWGA primer validation preenrichment and postenrichment PCR results. Download Table S4, CSV file, 0.00 MB (2.2KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To determine the SWGA Pal 11 primer set’s LoD and enrichment for T. pallidum, SWGA was performed in triplicate on the 10-fold dilution series, except for the neat sample. We observed a marked increase in T. pallidum gDNA copy number in every dilution in the series postenrichment (Fig. 1; Table S1). The gDNA copy number ranged from 1.11 × 106 ± 6.68 × 105 for the 1:10,000 dilution to 2.04 × 107 ± 1.20 × 107 in the 1:10 dilution (Table S1). Compared to the input copy number, this was a 2.01 × 104-fold to 5.53 × 105-fold increase in the enriched samples, from 1:10 to 1:10,000 dilution, respectively. Upon comparing the average RNPCT of each dilution in the series, the SWGA-enriched samples indicated a 29.36 ± 0.37 to 28.65 ± 0.16 increase for the 1:10 to 1:10,000 dilution, respectively (Table S1). The average RNPCT values at each 10-fold increase in T. pallidum concentration were not significantly different from one another (P > 0.1); however, by increasing the T. pallidum gDNA input 100-fold, we observed a significant decrease in RNP concentration (P < 0.03).
After enrichment with SWGA, we observed that dilutions ranging from 1:10 to 1:10,000 held 27.93% ± 1.57% to 3.29% ± 1.93% of the total DNA belonging to T. pallidum, respectively (Fig. 2). This reflected up to a 1.63 × 105-fold increase in the relative T. pallidum among all replicate SWGA-enriched samples compared to unenriched samples. All samples were significantly increased in their relative T. pallidum DNA compared to their respective inputs (P < 0.0001). While there were observed deviations in the percent DNA between replicates, the 1:10,000 diluted replicates still yielded a 28.40-fold ± 17.71-fold increase in DNA belonging to T. pallidum post-SWGA compared to the nonenriched neat dilution (P < 0.0001).
Genome sequencing data derived from the SWGA-enriched Nichols samples showed 0.98% to 78.05% of the quality-controlled reads binned as T. pallidum, along with a mean mapping depth to the T. pallidum Nichols reference genome ranging from 65.82 to 4.89 × 103. An average percent coverage of 98.67% ± 0.005% for the 1:10 diluted samples down to 96.15% ± 0.082% in the 1:1,000 diluted samples across the Nichols genome at 5× read depth was observed among the SWGA-enriched samples. A sharp decline in coverage was observed at the 1:10,000 dilution (Fig. 3B; Table S1 and Fig. S3). Comparing NEB+MDA and SWGA for enrichment, we observed that SWGA consistently produced higher relative T. pallidum DNA in all 10-fold diluted samples (P < 0.01). In addition, there was a sharp decline in coverage observed in the 1:1,000 diluted samples enriched by NEB+MDA, while this drop was not observed in samples enriched by SWGA, which still held >95% coverage at 5× read depth. As observed with NEB+MDA, no SNPs, inversions, or deletions were introduced during enrichment with SWGA. SWGA was subsequently used for enriching clinical specimens in this study.
Coverage plots for the T. pallidum Nichols dilution series by SWGA. Plots range from the lowest concentration T. pallidum Nichols mock samples (1:10,000) to the highest concentration (1:10) enriched using SWGA. Quality-controlled T. pallidum reads were mapped to the Nichols reference genome (GenBank version number NC_000919.1), and the log10 of the coverage at each mapped nucleotide position is shown on the x axis. Samples were sequenced using the Illumina NovaSeq 6000 platform. Download FIG S3, TIF file, 1.8 MB (1.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
SWGA enrichment of clinical strains.
SWGA on specimen EUHM-004 gave an average T. pallidum gDNA of 6.37 × 106 ± 2.24 × 105 copies/μL, with 5.56% of the total DNA belonging to T. pallidum (Table 2). Next-generation sequencing (NGS) on the MiSeq v2 (500-cycle) platform revealed 95.13% coverage across the T. pallidum genome at 5× read depth (Fig. 4; Table 2 and Fig. S2). Due to the overall low copy number obtained by standard DNA extraction, three specimens (EUHM-012, EUHM-013, and EUHM-014) were extracted using a large-scale method, yielding 31.5 ± 0.5, 122 ± 1.15, and 103 ± 6.55 T. pallidum gDNA copies/μL of extract, respectively (Table 1). For EUHM-012, we observed an average of 2.14 × 106 ± 2.82 × 104 T. pallidum gDNA copies/μL, with 1.72% of the total DNA belonging to T. Pallidum postenrichment by SWGA (Table 2). Sequencing indicated a 93.98% coverage across the T. pallidum genome at 5× read depth (Fig. 4; Table 2 and Fig. S2).
Compared to EUHM-012, EUHM-013 had a higher T. pallidum gDNA copy number/μL at 5.16 × 106 ± 2.20 × 105, with 15.48% of the total DNA belonging to T. pallidum postenrichment by SWGA (Table 2). The sequencing data correlated with the qPCR data, indicating a 98.56% coverage across the T. pallidum genome at 5× read depth (Fig. 4 and Table 2; Fig. S2). We also observed that EUHM-014 held an increased T. pallidum gDNA copy number post-SWGA, with 2.57 × 106 ± 2.21× 105 copies/μL and 4.72% of the total DNA belonging to T. pallidum (Table 2). Upon sequencing, we observed 98.49% coverage across the T. pallidum genome at 5× read depth (Fig. 4 and Table 2; Fig. S2). The T. pallidum gDNA copy number for specimen STLC-001 was 7.42 × 106 ± 7.20 × 105 copies/μL, with 8.34% of the total DNA belonging to T. pallidum (Table 2). The sequencing coverage was 95.94% at 5× read depth, where 38.91% of the quality-controlled reads binned as T. pallidum along with a mean read depth coverage of 1,133.43× (Fig. 4 and Table 2; Fig. S2).
Phylogenetic analysis and characterization of genotypic macrolide resistance.
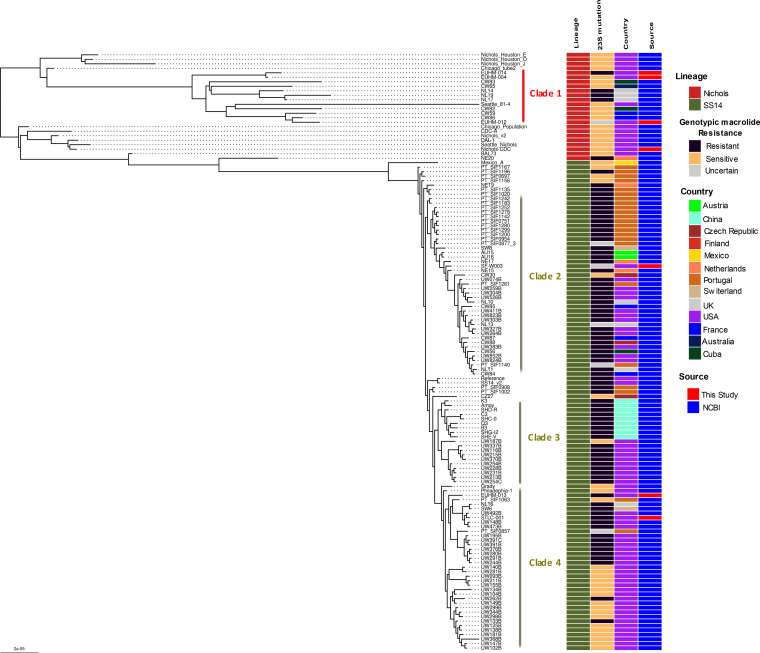
A whole-genome phylogenetic tree was constructed using the genomes derived from the 5 clinical specimens and 2 isolates along with 126 high-quality published T. pallidum genome sequences as of May 2021 (18–21, 29–31; see Table S5). Phylogenetic analysis revealed the presence of two dominant lineages (Nichols-like and Street-14 [SS14]-like), of which most strains belonged to the SS14-like lineage. We identified a total of four monophyletic clades within this phylogenetic tree with ≥30 bootstrap support (Fig. 6). Three of the clinically derived genomes from Atlanta, Georgia, EUHM-004 (2019) EUHM-012 (2019), and EUHM-014 (2020), belonged to the Nichols-like lineage (clade 1; n = 12; Fig. 6). Interestingly, the other nine Nichols-like genomes in clade 1 were recent clinically derived genomes from Cuba (n = 2; 2015 to 2016), Australia (n = 1; 2014), France (n = 2; 2012 to 2013), and the United Kingdom (n = 3; 2016) and, together with the specimens from Atlanta, were distinct from the original Nichols strain isolated in 1912 and sent to different North American labs as in vivo-derived clones. The three clinical specimens from Atlanta (EUHM-004, EUHM-012, and EUHM-014) and three clinically derived genomes from the United Kingdom isolated in 2016 (NL14, NL19, and NL17) carried the 23S rRNA A2058G mutation, which confers macrolide resistance, suggesting a recent acquisition of this antibiotic resistance variant in the Nichols-like lineage.
FIG 6.
Maximum likelihood global phylogenetic tree of the 7 T. pallidum strains sequenced in this study along with 122 high-quality (with 5× read depth covering >90% of the genome) publicly available T. pallidum genomes. The two major lineages, Nichols-like and SS14-like, are highlighted along with the presence of genotypic mutation responsible for macrolide resistance and country of origin.
Even though previous phylogenomic analyses indicated that the SS14-lineage showed a polyphyletic structure, our phylogenetic analysis with a greater number of genomes showed the presence of 3 monophyletic clades (clades 2, 3, and 4) (18, 20; Fig. 6). Clades 2 and 4 contained genomes clustered within the previously reported SS14Ω-A subcluster, which also contained two clades corresponding to the clades 2 and 4 detected in this study, and contained genomes derived from Europe and North America, while clade 3 was like subcluster SS14Ω-B and composed of Chinese- and North American-derived T. pallidum genomes (18). The rabbit-propagated isolate, CDC-SF003 (San Francisco, USA; 2017) sequenced in this study clustered within clade 2, while the EUHM-013 (Atlanta, USA; 2020) and STLC-001 (St. Louis, USA; 2020) genomes clustered within clade 4. Sequence analysis showed that all 3 strains carried the A2058G antimicrobial resistance variant for macrolide resistance. Macrolide resistance strains were widespread among the SS14 lineage, with a higher proportion among the genomes in clades 2 and 3 than those in clade 4 genomes. The A2058G point mutation identified in 4 patient specimens and isolate CDC-SF003 was verified by real-time PCR testing of gDNA and SWGA-enriched samples (data not shown). There was inadequate sample of the fifth specimen to confirm the mutation by real-time PCR testing.
All the Nichols-like genomes derived from the NEB+MDA and SWGA 10-fold dilution series that contained T. pallidum reads mapped to ≥90% of the genome with at least 5× read depth formed a tight monophyletic clade (bootstrap support of 88/100) and clustered with the lab-derived Nichols-Houston-J genome (bootstrap support of 100/100), indicating that genomes generated from both methods are adequate to capture genetic variants required to perform a high-resolution phylogenetic analysis (Fig. S4).
Maximum-likelihood global phylogenetic tree with all the genomes sequenced from all the enriched Nichols dilution series with NEB+MDA and SWGA that generated at least 5× read depth covering at least 90% of the genome. All highlighted samples were sequenced using the Illumina NovaSeq 6000 platform. Download FIG S4, TIF file, 2.8 MB (2.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
WGS of T. pallidum is often challenging due to low bacterial loads or the difficulty of obtaining adequate samples for testing. In this study, we sought to develop a method for performing WGS from rabbit-propagated isolates and clinical specimens containing lower T. pallidum numbers, leading us to investigate CpG capture and SWGA.
During the testing of the 10-fold dilution series of Nichols spiked samples, we observed increases in T. pallidum gDNA copy numbers and relative T. pallidum percent DNA in the neat to 1:1,000 dilutions enriched by NEB+MDA compared to the nonenriched inputs. There was no significant difference in the relative human RNPCT from dilution to dilution. While the neat to 1:100 dilution spiked samples demonstrated an average percent coverage exceeding 97.29% at 5× read depth across the Nichols strain genome, genomic coverage for 1:1,000 to 1:1,0000 diluted spiked samples was poor. We observed that an input of >129 T. pallidum gDNA copies/μL can generate >95% coverage at 5× read depth from the Nichols strain post-NEB+MDA enrichment. Post-NEB+MDA enrichment of isolate CDC-SF003 demonstrated >98% coverage at 5× read depth across the T. pallidum genome. Variant analysis correlated with real-time PCR detection of the mutations associated with macrolide resistance in CDC-SF003. In addition, phylogenetics revealed that this strain belonged to the SS14 lineage, which correlated with its enhanced CDC typing method (ECDCT) strain type, 14d9f, as previously reported (11). While this enrichment method yielded good results with isolates, the fact that most clinical specimens collected in this study had fewer than 100 T. pallidum gDNA copies/μL extract led us to consider an alternative method.
We observed that samples enriched by SWGA using multiple primer sets exhibited a 10,000-fold increase in T. pallidum genome copy number, with Pal 9 and 11 producing the highest relative percent T. pallidum DNA at 29% and 31%, respectively. While we chose to work with Pal 11 as the optimal set, Pal 9 could also be a good alternative for enriching syphilis specimens. Further testing using Pal 11 showed that the limit of detection was increased compared to that of the T. pallidum enrichment obtained with NEB+MDA, with significant increases in both T. pallidum gDNA copy number and percent T. pallidum across the 10-fold dilution series of spiked samples. Coverage across the T. pallidum genome exceeded 95% at 5× read depth for all diluted samples, apart from the 1:10,000 diluted samples. Interestingly, we observed that increasing the T. pallidum DNA input 100-fold resulted in a significant decrease in the presence of RNP postenrichment. Our data show that >14 T. pallidum gDNA copies/μL can generate at least 95% coverage at 5× read depth with the Nichols strain using the SWGA enrichment method, which translated well to the clinical specimens tested. While there was a decrease in coverage in one of the clinical specimens at 94.44% with 5× read depth compared to the 98.62% coverage at 5× read depth observed in the 1:100 diluted Nichols isolates, this could be primarily due to the improved capabilities of the NovaSeq 6000 platform compared to the MiSeq v2 (500-cycle) platform used for sequencing. Another possible reason for the variation in coverage could be the lower T. pallidum input copy number in the clinical specimens.
The genomes derived directly from the 5 clinical specimens using SWGA were phylogenetically associated with the representative lineages (either Nichols-like or SS14-like) and provided high levels of within-lineage strain resolution, which is ideal for effective tracking of various strains circulating within a geographical area and outbreak investigations. In addition, the NGS methods described here can be used for macrolide resistance marker detection. As observed with NEB+MDA enrichment, azithromycin mutation detection performed on the SWGA-enriched specimens matched the results obtained by real-time PCR, indicating that all clinical specimens contained the A2058G mutation. SWGA-based enrichment also enabled sequencing of specimens within the range of detection limits for real-time PCR assays, suggesting that our NGS workflow can be adapted for T. pallidum detection in metagenomic samples.
In terms of expense, both methods are cost-effective for enriching T. pallidum gDNA, and while SWGA is less expensive than NEB+MDA, sequencing reagents are the true limiting factor for WGS. With the recent advancements in large-scale sequencing platforms, overall sequencing costs can be further reduced. While NovaSeq 6000 has a much higher potential for multiplex sequencing, our data show compatibility of these enrichments for both the NovaSeq 6000 and MiSeq platforms.
While we successfully enriched T. pallidum whole genomes in clinical specimens, the success of SWGA is limited by the constraint on primer size, which may reduce the selectivity for the target genome. Phi29 functions best between 30°C and 35°C, and ramp-down incubations have been shown as an effective means of utilizing larger primers with increased melting temperatures (26, 27, 32, 33). To help alleviate the constraints on primer size, we utilized a thermostable phi29 mutant which has a much higher optimal temperature at 45°C (34) compared to the 30°C to 35°C functional range of the phi29 polymerase (26, 27). This higher optimal temperature permits the use of longer oligonucleotides in the SWGA reaction, potentially increasing the selectivity for the T. pallidum genome. The phi29 mutant has also been shown to be more efficient, with a 3-h exhaustion time compared to the 8 to 16 h required for the wild-type phi29 (34). In addition, the genome sequence data generated from NEB+MDA- and SWGA-enriched samples revealed that no SNPs, inversions, or deletions were introduced during whole-genome amplification.
Our results show that SWGA is a more sensitive, less cumbersome, and faster method for enriching clinical specimens compared to NEB+MDA, allowing for WGS of T. pallidum with a minimum input of 14 gDNA copies/μL. In addition, the sequencing data generated are of sufficient quality to enable phylogenetic analyses and detection of mutations associated with azithromycin resistance. While NEB+MDA was unsuitable for the clinical specimens in this study, our data suggest that it can be used for DNA extracts containing >129 T. pallidum gDNA copies/μL.
MATERIALS AND METHODS
Specimen collection, T. pallidum strains used for WGS, and real-time qPCR.
Specimens used in this study were collected from men presenting with lesions of primary or secondary syphilis to the Emory Infectious Diseases Clinic, Emory University Hospital Midtown (EUHM) in Atlanta, GA, and the St. Louis County STD Clinic (STLC) in St. Louis, MO (Table 1). Patients were diagnosed with syphilis based on clinical presentation and serology testing. A total of 14 swab specimens were collected in Aptima Multitest storage medium (Hologic, Inc., Marlborough, MA) at the Emory Infectious Diseases Clinic, and 1 specimen was from the St. Louis County STD Clinic (Table 1). All specimens were stored at −80°C until shipment on dry ice to the CDC. The T. pallidum Nichols reference strain was used for initial optimization and validation of the two enrichment methods. A recent rabbit-propagated isolate, CDC-SF003, was also included for testing (Table 1; 11). Prior to study commencement, local institutional review board (IRB) approvals were obtained from Emory University and St. Louis County Department of Public Health, and the project was approved at CDC.
DNA was extracted from specimens and rabbit testis extracts using the QIAamp DNA minikit (Qiagen, Germantown, MD) following the manufacturer’s recommendations. To increase our chances of successfully sequencing three specimens, large-scale DNA extraction was carried out on 1.5 mL of specimen using the QIAamp DNA minikit with slight modifications for upscaling (Table 1). Briefly, proteinase K was added at 0.1× total sample volume. AL buffer and ethanol were added at 1× total sample volume. Each sample was processed through a single column and eluted in 100 μL AE buffer (Qiagen). DNA samples were tested with a real-time quantitative duplex PCR (qPCR) targeting the polA gene of T. pallidum and the human Rnase P gene (RNP) using a Rotor-Gene 6000 instrument (Qiagen) as previously described with modifications (11, 28); see (Text S1).
NEBNext microbiome enrichment and multiple displacement amplification (MDA).
Initially, DNA concentrations of extracts from clinical specimens and rabbit-propagated strains were measured using the Qubit double-stranded DNA (dsDNA) high-sensitivity (HS) assay (Thermo Fisher Scientific, Waltham, MA). Capture and removal of CpG methylated host DNA from samples were carried out using the NEBNext microbiome DNA enrichment kit (New England Biolabs [NEB], Ipswich, MA), following the manufacturer’s recommendations with modifications (New England Biolabs). For all samples tested, 250 ng of DNA was subjected to two rounds of bead capture using NEB and enriched treponemal gDNA was purified using AMPure XP beads (Beckman Coulter, Indianapolis, IN). Enriched DNA samples were stored at −20°C until multiple displacement amplification (MDA) was performed. MDA was carried out using the REPLI-g single cell kit following the manufacturer’s recommendations with slight modifications (Qiagen). MDA reaction mixtures were incubated at 30°C for 16 h. Following amplification, the polymerase was inactivated at 65°C for 10 min, and samples were purified with AMPure XP beads and eluted with 100 μL 1× AE buffer (Qiagen). For enrichment by MDA, no template controls were included to confirm the absence of T. pallidum.
A 10-fold dilution series on the Nichols strain was used to determine the limit of detection (LoD) for enrichment (see Text S1) with NEB+MDA followed by sequencing on an Illumina NovaSeq 6000 instrument. After DNA extraction, each dilution in the series was enriched by NEB+MDA, gDNA copy numbers were estimated by polA qPCR, and sequencing was performed in triplicate. Enriched samples were diluted 1:10 prior to measurement of RNP amplification. The LoD was set at the minimum genome copy number required to generate a ≥5× read depth with ≥95% genome coverage compared to the reference genome.
Selective whole-genome amplification (SWGA) primer design, validation, and enrichment.
Primers with an increased affinity to T. pallidum were identified using the swga toolkit as previously described with slight modifications (https://www.github.com/eclarke/swga; 26; see Text S1). Eight primer sets (SWGA Pal 1 to 8), and 4 additional primer sets (SWGA Pal 9 to 12) generated by combining primers in the initial set (Table S1), were chosen for SWGA using EquiPhi29 DNA polymerase (Thermo Fisher Scientific, Waltham, MA). To account for the 3′–5′ exonuclease activity of the phi29 polymerase, all SWGA primers were generated with phosphorothioate bonds between the last two nucleotides at the 3′ end (Table S1). Each of the 12 primer sets was tested in triplicate against the spiked sample diluted to an estimated 100 T. pallidum gDNA copies/μL (see Text S1).
Supplemental Materials and Methods. Download TEXT S1, DOCX file, 0.03 MB (30.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Prior to SWGA enrichment, samples were denatured for 5 min at 95°C by adding 2.5 μL DNA to 2.5 μL reaction buffer, containing custom primers, and then placed immediately on ice until the Equiphi29 master mix, prepared as per the manufacturer’s recommendations, was added. Whole-genome amplification was carried out following the manufacturer’s recommendations with modifications (Thermo Fisher Scientific; 34). The reaction contained EquiPhi29 master mix, with EquiPhi29 reaction buffer at a final concentration of 1×, with each primer at a final concentration of 4 μM, and nuclease-free H2O was added to a final reaction volume of 20 μL. Reaction tubes were gently mixed by pulse vortexing and incubated at 45°C for 3 h. The reaction was stopped by inactivating the DNA polymerase at 65°C for 15 min. All reactions were purified using AMPure XP beads and eluted in 100 μL AE buffer (Qiagen). No template controls were included to confirm the absence of contaminate T. pallidum DNA.
The relative percent T. pallidum in each sample was calculated as shown in Fig. S1. SWGA Pal 11 was chosen for testing the LoD for downstream genome sequencing post-SWGA enrichment using the 10-fold dilution series, excluding the undiluted (neat) spiked sample. All enriched samples were validated by polA real-time qPCR in triplicate.
Sequencing and genome analysis of T. pallidum strains.
Libraries were prepared using the NEBNext Ultra DNA library preparation kit for NovaSeq and the NEBNext Ultra II FS DNA library preparation kit for MiSeq sequencing following the manufacturer’s recommendations (New England Biolabs, Ipswich, MA). For the validation experiments, sequencing was carried out on the Nichols reference strain using the NovaSeq 6000 platform following the manufacturer’s recommendations (Illumina, San Diego, CA). Sequencing of isolate CDC-SF003 and swab specimens was carried out using the MiSeq v2 (500-cycle) platform following the manufacturer’s recommendations (Illumina).
Sequenced genomic reads were first filtered from the raw sequencing data set using Bowtie 2 v2.2.9 (35), which removed any contaminating human sequences using the h19 human reference genome and rabbit reference genome for rabbit-propagated clinical isolates (36). Cutadapt v1.8.3 was used to trim specified primers and adaptors and to filter out reads below Phred quality scores of 15 and read lengths below 50 bp (37). PCR duplicates were removed using Clumpify (sourceforge.net/projects/bbmap/) with the dedupe=t option to prevent biased coverage of genomic regions. Treponemal reads were selected using K-SLAM, a k-mer-based metagenomics taxonomic profiler, which used a database containing all bacterial and archaeal reference nucleotide sequences (38). The presence of T. pallidum sequences was also confirmed using MetaPhlan2 (39). Prefiltered treponemal reads were mapped against either the T. pallidum subsp. pallidum SS14 reference genome (GenBank version number NC_021508.1) or the T. pallidum subsp. pallidum Nichols reference genome (GenBank version number NC_000919.1) using the Burrows-Wheeler Aligner MEM algorithm (BWA MEM) v2.12.0 (MapQ, ≥20), followed by consensus sequence generation and estimation of sequencing depth and mapping statistics using SAMtools (options “depth” and “mpileup”) and bcftools v1.9. The prefiltered treponemal reads were also used to generate de novo short-read assemblies using SPAdes v3.7.0 with the “careful” option (40–42).
To assess whether there were introductions of amplification-induced SNPs, inversions, or deletions for both NEB+MDA and SWGA, we compared the sequencing data generated from all three replicates for each of the 10-fold dilution series for both protocols to the control genome generated directly from an unamplified DNA sample containing 10,000 gDNA copies/μL of T. pallidum Nichols. The same Nichols DNA was used for the dilution series and the unamplified sample. Essentially, the sequencing reads of the Nichols control genome were de novo assembled using SPAdes v3.7.0 as described above, and the resultant contigs were arranged using ABACAS in reference to the T. pallidum Nichols reference genome (GenBank version number NC_021490.2) to generate a circular pseudochromosome (43). All T. pallidum reads from all the NEB and SWGA 10-fold diluted samples were mapped against the control T. pallidum Nichols pseudochromosome using Snippy v4.3.8 and checked for any genetic variants in the form of SNPs, insertions, or deletions introduced during multiple amplification steps.
Phylogenetic analyses.
We used a reference mapping approach by mapping the filtered treponemal reads to a custom version of the T. pallidum SS14 reference genome (GenBank version number NC_021508.1) by masking around 30,000 nucleotide positions belonging to 12 repetitive tpr genes A to L, along with arp and TPANIC_0470 genes using BEDTools v2.17.0 maskfasta (44). Full-length whole-genome alignment was generated using Snippy v4.3.8 (https://github.com/tseemann/snippy), which identifies variants using FreeBayes v1.0.2 with a minimum 5× read coverage and 90% read concordance at a locus for each SNP (45). Regions of increased density of homoplasious SNPs introduced by possible recombination events were predicted iteratively and masked using Gubbins (46). The final phylogenetic tree was reconstructed using RAxML on the recombination-removed alignment using the GTR+GAMMA model of nucleotide substitution with a majority-rule consensus (MRE) convergence criterion to reconstruct an ascertainment bias-corrected (Stamatakis method) maximum-likelihood (ML) phylogeny (47).
Apart from the genomes sequenced in this study, 126 high-quality (with at least 5× read depth covering >90% of the genome) T. pallidum genomes deposited in the NCBI Sequence Read Archive (SRA) under BioProject numbers PRJEB20795 and PRJNA508872 were also included (18, 20). A second phylogenetic tree was also reconstructed by including all the genomes sequenced from the 10-fold dilution series for both NEB+MDA- and SWGA-enriched samples. Genomic sequencing data from samples included in the phylogenetic analyses covered at least 90% of the reference genome with 5× read depth. Variant calls for the A2058G and A2059G macrolide resistance mutations using genomic data were validated with a previously described real-time PCR assay (5).
Statistical analyses.
Statistical analyses were performed in R (R Foundation for Statistical Computing, Vienna, Austria) using the R companion software RStudio (Boston, MA). Statistical significance was determined by analysis of variance (ANOVA) and Tukey post hoc multiple-comparison tests. T. pallidum percent DNA was normalized through log10 conversions. Quantitative data are presented as means ± standard error. Differences were considered statistically significant if P was <0.05.
Data availability.
All sequencing data associated with this study were submitted to the National Center for Biotechnology Information Sequence Read Archive (SRA) under the BioProject number PRJNA744275.
T. pallidum strains included in the phylogenetic tree. Download Table S5, CSV file, 0.00 MB (5KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ACKNOWLEDGMENTS
We thank Teressa Burns at the Emory University Hospital Midtown; Tamara Jones from the St. Louis County STD Clinic; Yetty Fakile, Kevin Pettus, and Jack Cartee at CDC’s Division of STD Prevention; the veterinary staff at the CDC’s Comparative Medicine Branch; Mark Itsko at CDC’s Division of Bacterial Diseases; and Nikhat Sulaiman and Justin Lee at CDC’s Division of Scientific Resources for their assistance, consults, and support throughout this study.
This work was made possible through CDC’s Division of STD Prevention with support from the Advanced Molecular Detection (AMD) program and, in part, by an appointment (C.M.T.) to the Research Participation Program at the CDC, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and CDC.
Allan Pillay and Ellen N. Kersh conceived the study. Allan Pillay, Charles M. Thurlow, Cheng Chen, and Lilia Ganova-Raeva designed the study. Charles M. Thurlow and Allan Pillay designed the enrichment protocols. Charles M. Thurlow designed the SWGA-specific custom primer sets used during this study and performed all enrichment experiments. Charles M. Thurlow, Allan Pillay, Samantha S. Katz, Lara Pereira, Alyssa Debra, Kendra Vilfort, Yongcheng Sun, Kai-Hua Chi, and Damien Danavall performed the laboratory experiments and assisted with specimen collection. Kimberly Workowski, Stephanie E. Cohen, Hilary Reno, and Susan S. Philip collected clinical specimens and patient data. Mark Burroughs, Mili Sheth, and Charles M. Thurlow performed Illumina sequencing. Sandeep J. Joseph performed the bioinformatic analyses of the genomic data and the phylogenetic analysis and contributed to the generation of tables and figures. Charles M. Thurlow and Sandeep J. Joseph performed data analysis. Charles M. Thurlow wrote and prepared the manuscript with oversight by Allan Pillay and contributions from Sandeep J. Joseph and Weiping Cao, which was reviewed by all authors for revisions.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. We declare that there are no competing interests.
Contributor Information
Charles M. Thurlow, Email: cthurlow@cdc.gov.
Allan Pillay, Email: apillay@cdc.gov.
Maria L. Marco, University of California, Davis
REFERENCES
- 1.CDC. 2020. Sexually transmitted disease surveillance 2019. US Department of Health and Human Services, Atlanta, GA. [Google Scholar]
- 2.Lukehart SA, Godornes C, Molini BJ, Sonnett P, Hopkins S, Mulcahy F, Engelman J, Mitchell SJ, Rompalo AM, Marra CM, Klausner JD. 2004. Macrolide resistance in Treponema pallidum in the United States and Ireland. N Engl J Med 351:154–158. doi: 10.1056/NEJMoa040216. [DOI] [PubMed] [Google Scholar]
- 3.Tipple C, Taylor GP. 2015. Syphilis testing, typing, and treatment follow-up: a new era for an old disease. Curr Opin Infect Dis 28:53–60. doi: 10.1097/QCO.0000000000000124. [DOI] [PubMed] [Google Scholar]
- 4.Smajs D, Pastekova L, Grillova L. 2015. Macrolide resistance in the syphilis spirochete, Treponema pallidum ssp. pallidum: can we also expect macrolide-resistant yaws strains? Am J Trop Med Hyg 93:678–683. doi: 10.4269/ajtmh.15-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C-Y, Chi K-H, Pillay A, Nachamkin E, Su JR, Ballard RC. 2013. Detection of the A2058G and A2059G 23S rRNA gene point mutations associated with azithromycin resistance in Treponema pallidum by use of a TaqMan real-time multiplex PCR assay. J Clin Microbiol 51:908–913. doi: 10.1128/JCM.02770-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Workowski KA, Bolan GA, Centers for Disease Control and Prevention. 2015. Sexually transmitted diseases treatment guidelines, 2015. MMWR Recomm Rep 64:1–137. [PMC free article] [PubMed] [Google Scholar]
- 7.Marra C, Sahi S, Tantalo L, Godornes C, Reid T, Behets F, Rompalo A, Klausner JD, Yin Y, Mulcahy F, Golden MR, Centurion-Lara A, Lukehart SA. 2010. Enhanced molecular typing of Treponema pallidum: geographical distribution of strain types and association with neurosyphilis. J Infect Dis 202:1380–1388. doi: 10.1086/656533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillay A, Lee M-K, Slezak T, Katz SS, Sun Y, Chi K-H, Morshed M, Philip S, Ballard RC, Chen CY. 2019. Increased discrimination of Treponema pallidum strains by subtyping with a 4-component system incorporating a mononucleotide tandem repeat in rpsA. Sex Transm Dis 46:e42–e45. doi: 10.1097/OLQ.0000000000000935. [DOI] [PubMed] [Google Scholar]
- 9.Katz K, Pillay A, Ahrens K, Kohn R, Hermanstyne K, Bernstein K, Ballard R, Klausner J. 2010. Molecular epidemiology of syphilis: San Francisco, 2004–2007. Sex Transm Dis 37:660–663. doi: 10.1097/OLQ.0b013e3181e1a77a. [DOI] [PubMed] [Google Scholar]
- 10.Grillová L, Bawa T, Mikalová L, Gayet-Ageron A, Nieselt K, Strouhal M, Sednaoui P, Ferry T, Cavassini M, Lautenschlager S, Dutly F, Pla-Díaz M, Krützen M, González-Candelas F, Bagheri HC, Šmajs D, Arora N, Bosshard PP. 2018. Molecular characterization of Treponema pallidum subsp. pallidum in Switzerland and France with a new multilocus sequence typing scheme. PLoS One 13:e0200773. doi: 10.1371/journal.pone.0200773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereira LE, Katz SS, Sun Y, Mills P, Taylor W, Atkins P, Thurlow CM, Chi K-H, Danavall D, Cook N, Ahmed T, Debra A, Philip S, Cohen S, Workowski KA, Kersh E, Fakile Y, Chen CY, Pillay A. 2020. Successful isolation of Treponema pallidum strains from patients’ cryopreserved ulcer exudate using the rabbit model. PLoS One 15:e0227769. doi: 10.1371/journal.pone.0227769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edmondson DG, Hu B, Norris SJ. 2018. Long-term in vitro culture of the syphilis spirochete Treponema pallidum subsp. pallidum. mBio 9:e01153-18. doi: 10.1128/mBio.01153-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edmondson DG, DeLay BD, Kowis LE, Norris SJ. 2021. Parameters affecting continuous in vitro culture of Treponema pallidum strains. mBio 12:e03536-20. doi: 10.1128/mBio.03536-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bachmann NL, Rockett RJ, Timms VJ, Sintchenko V. 2018. Advances in clinical sample preparation for identification and characterization of bacterial pathogens using metagenomics. Front Public Health 6:363. doi: 10.3389/fpubh.2018.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorburn F, Bennett S, Modha S, Murdoch D, Gunson R, Murcia PR. 2015. The use of next generation sequencing in the diagnosis and typing of respiratory infections. J Clin Virol 69:96–100. doi: 10.1016/j.jcv.2015.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lefterova MI, Suarez CJ, Banaei N, Pinsky BA. 2015. Next-generation sequencing for infectious disease diagnosis and management: a report of the Association for Molecular Pathology. J Mol Diagn 17:623–634. doi: 10.1016/j.jmoldx.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Beale MA, Marks M, Cole MJ, Lee M-K, Pitt R, Ruis C, Balla E, Crucitti T, Ewens M, Fernández-Naval C, Grankvist A, Guiver M, Kenyon CR, Khairullin R, Kularatne R, Arando M, Molini BJ, Obukhov A, Page EE, Petrovay F, Rietmeijer C, Rowley D, Shokoples S, Smit E, Sweeney EL, Taiaroa G, Vera JH, Wennerås C, Whiley DM, Williamson DA, Hughes G, Naidu P, Unemo M, Krajden M, Lukehart SA, Morshed MG, Fifer H, Thomson NR. 2021. Global phylogeny of Treponema pallidum lineages reveals recent expansion and spread of contemporary syphilis. Nat Microbiol 6:1549–1560. doi: 10.1038/s41564-021-01000-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beale MA, Marks M, Sahi SK, Tantalo LC, Nori AV, French P, Lukehart SA, Marra CM, Thomson NR. 2019. Genomic epidemiology of syphilis reveals independent emergence of macrolide resistance across multiple circulating lineages. Nat Commun 10:3255. doi: 10.1038/s41467-019-11216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinto M, Borges V, Antelo M, Pinheiro M, Nunes A, Azevedo J, Borrego MJ, Mendonça J, Carpinteiro D, Vieira L, Gomes JP. 2016. Genome-scale analysis of the non-cultivable Treponema pallidum reveals extensive within-patient genetic variation. Nat Microbiol 2:16190. doi: 10.1038/nmicrobiol.2016.190. [DOI] [PubMed] [Google Scholar]
- 20.Grillová L, Oppelt J, Mikalová L, Nováková M, Giacani L, Niesnerová A, Noda AA, Mechaly AE, Pospíšilová P, Čejková D, Grange PA, Dupin N, Strnadel R, Chen M, Denham I, Arora N, Picardeau M, Weston C, Forsyth RA, Šmajs D. 2019. Directly sequenced genomes of contemporary strains of syphilis reveal recombination-driven diversity in genes encoding predicted surface-exposed antigens. Front Microbiol 10:1691–1691. doi: 10.3389/fmicb.2019.01691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arora N, Schuenemann VJ, Jäger G, Peltzer A, Seitz A, Herbig A, Strouhal M, Grillová L, Sánchez-Busó L, Kühnert D, Bos KI, Davis LR, Mikalová L, Bruisten S, Komericki P, French P, Grant PR, Pando MA, Vaulet LG, Fermepin MR, Martinez A, Centurion Lara A, Giacani L, Norris SJ, Šmajs D, Bosshard PP, González-Candelas F, Nieselt K, Krause J, Bagheri HC. 2016. Origin of modern syphilis and emergence of a pandemic Treponema pallidum cluster. Nat Microbiol 2:16245. doi: 10.1038/nmicrobiol.2016.245. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, Šmajs D, Hu Y, Ke W, Pospíšilová P, Hawley KL, Caimano MJ, Radolf JD, Sena A, Tucker JD, Yang B, Juliano JJ, Zheng H, Parr JB. 2021. Analysis of Treponema pallidum strains from China using improved methods for whole-genome sequencing from primary syphilis chancres. J Infect Dis 223:848–853. doi: 10.1093/infdis/jiaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feehery GR, Yigit E, Oyola SO, Langhorst BW, Schmidt VT, Stewart FJ, Dimalanta ET, Amaral-Zettler LA, Davis T, Quail MA, Pradhan S. 2013. A method for selectively enriching microbial DNA from contaminating vertebrate host DNA. PLoS One 8:e76096. doi: 10.1371/journal.pone.0076096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thoendel M, Jeraldo PR, Greenwood-Quaintance KE, Yao JZ, Chia N, Hanssen AD, Abdel MP, Patel R. 2016. Comparison of microbial DNA enrichment tools for metagenomic whole genome sequencing. J Microbiol Methods 127:141–145. doi: 10.1016/j.mimet.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leichty AR, Brisson D. 2014. Selective whole genome amplification for resequencing target microbial species from complex natural samples. Genetics 198:473–481. doi: 10.1534/genetics.114.165498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke EL, Sundararaman SA, Seifert SN, Bushman FD, Hahn BH, Brisson D. 2017. swga: a primer design toolkit for selective whole genome amplification. Bioinformatics 33:2071–2077. doi: 10.1093/bioinformatics/btx118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itsko M, Retchless AC, Joseph SJ, Norris Turner A, Bazan JA, Sadji AY, Ouédraogo-Traoré R, Wang X. 2020. Full molecular typing of Neisseria meningitidis directly from clinical specimens for outbreak investigation. J Clin Microbiol 58:e01780-20. doi: 10.1128/JCM.01780-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chi KH, Danavall D, Taleo F, Pillay A, Ye T, Nachamkin E, Kool JL, Fegan D, Asiedu K, Vestergaard LS, Ballard RC, Chen CY. 2015. Molecular differentiation of Treponema pallidum subspecies in skin ulceration clinically suspected as yaws in Vanuatu using real-time multiplex PCR and serological methods. Am J Trop Med Hyg 92:134–138. doi: 10.4269/ajtmh.14-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Čejková D, Zobaníková M, Chen L, Pospíšilová P, Strouhal M, Qin X, Mikalová L, Norris SJ, Muzny DM, Gibbs RA, Fulton LL, Sodergren E, Weinstock GM, Šmajs D. 2012. Whole genome sequences of three Treponema pallidum ssp. pertenue strains: yaws and syphilis treponemes differ in less than 0.2% of the genome sequence. PLoS Negl Trop Dis 6:e1471. doi: 10.1371/journal.pntd.0001471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pětrošová H, Pospíšilová P, Strouhal M, Čejková D, Zobaníková M, Mikalová L, Sodergren E, Weinstock GM, Šmajs D. 2013. Resequencing of Treponema pallidum ssp. pallidum strains Nichols and SS14: correction of sequencing errors resulted in increased separation of syphilis treponeme subclusters. PLoS One 8:e74319. doi: 10.1371/journal.pone.0074319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun J, Meng Z, Wu K, Liu B, Zhang S, Liu Y, Wang Y, Zheng H, Huang J, Zhou P. 2016. Tracing the origin of Treponema pallidum in China using next-generation sequencing. Oncotarget 7:42904–42918. doi: 10.18632/oncotarget.10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sundararaman SA, Plenderleith LJ, Liu W, Loy DE, Learn GH, Li Y, Shaw KS, Ayouba A, Peeters M, Speede S, Shaw GM, Bushman FD, Brisson D, Rayner JC, Sharp PM, Hahn BH. 2016. Genomes of cryptic chimpanzee Plasmodium species reveal key evolutionary events leading to human malaria. Nat Commun 7:11078. doi: 10.1038/ncomms11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowell AA-O, Loy DE, Sundararaman SA, Valdivia H, Fisch K, Lescano AG, Baldeviano GC, Durand S, Gerbasi V, Sutherland CJ, Nolder D, Vinetz JM, Hahn BH, Winzeler EA. 2017. Selective whole-genome amplification is a robust method that enables scalable whole-genome sequencing of Plasmodium vivax from unprocessed clinical samples. mBio 8:e02257-16. doi: 10.1128/mBio.02257-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Povilaitis T, Alzbutas G, Sukackaite R, Siurkus J, Skirgaila R. 2016. In vitro evolution of phi29 DNA polymerase using isothermal compartmentalized self replication technique. Protein Eng Des Sel 29:617–628. doi: 10.1093/protein/gzw052. [DOI] [PubMed] [Google Scholar]
- 35.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, Hunt SE, Cole CG, Coggill PC, Rice CM, Ning Z, Rogers J, Bentley DR, Kwok PY, Mardis ER, Yeh RT, Schultz B, Cook L, Davenport R, Dante M, Fulton L, Hillier L, Waterston RH, McPherson JD, Gilman B, Schaffner S, Van Etten WJ, Reich D, Higgins J, Daly MJ, Blumenstiel B, Baldwin J, Stange-Thomann N, Zody MC, Linton L, Lander ES, Altshuler D, International SNP Map Working Group. 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 37.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 38.Ainsworth D, Sternberg MJE, Raczy C, Butcher SA. 2017. k-SLAM: accurate and ultra-fast taxonomic classification and gene identification for large metagenomic data sets. Nucleic Acids Res 45:1649–1656. doi: 10.1093/nar/gkw1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. 2015. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 40.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997. doi: 10.48550/arXiv.1303.3997. [DOI]
- 41.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Assefa S, Keane TM, Otto TD, Newbold C, Berriman M. 2009. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics 25:1968–1969. doi: 10.1093/bioinformatics/btp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv 1207.3907. doi: 10.48550/arXiv.1207.3907. [DOI]
- 46.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Copy number, RNPCT, and sequencing results for NEB+MDA- and SWGA-enriched spiked samples. Download Table S1, CSV file, 0.00 MB (4.4KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Coverage plots for the T. pallidum Nichols undiluted and 10-fold dilution series enriched using the NEB+MDA. Quality-controlled T. pallidum reads were mapped to the Nichols reference genome (GenBank version number NC_000919.1), and the log10 of the coverage at each mapped nucleotide position is shown on the x axis. Samples were sequenced using the Illumina NovaSeq 6000 platform. Download FIG S1, TIF file, 1.9 MB (1.9MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Coverage plots for a nonenriched T. pallidum Nichols isolate control containing 1,063.1 ± 45.22 T. pallidum genomic copies/μL of DNA extract, NEB+MDA-enriched clinical isolate CDC-SF003 and the SWGA-enriched Emory- and St. Louis-derived clinical specimens. T. pallidum Nichols reference isolate quality-controlled T. pallidum reads were mapped to the Nichols reference genome (GenBank version number NC_000919.1). An input number of 1,000 genomic copies/μL T. pallidum was used for the Nichols nonenriched and SWGA enriched samples. Quality-controlled T. pallidum reads for the Emory- and St. Louis-derived specimens and CDC-SF003 were mapped to the SS14 reference genome (GenBank version number NC_021508.1). The log10 of the coverage at each mapped nucleotide position is shown on the x axis. All samples were sequenced using the Illumina MiSeq v2 (500-cycle) platform. Download FIG S2, TIF file, 1.5 MB (1.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
List of primers and probes used in this study. Download Table S2, DOCX file, 0.02 MB (23KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Binding distribution of the swga Toolkit-generated primer sets to the T. pallidum and Homo sapiens genomes. Download Table S3, DOCX file, 0.02 MB (23.2KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
SWGA primer validation preenrichment and postenrichment PCR results. Download Table S4, CSV file, 0.00 MB (2.2KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Coverage plots for the T. pallidum Nichols dilution series by SWGA. Plots range from the lowest concentration T. pallidum Nichols mock samples (1:10,000) to the highest concentration (1:10) enriched using SWGA. Quality-controlled T. pallidum reads were mapped to the Nichols reference genome (GenBank version number NC_000919.1), and the log10 of the coverage at each mapped nucleotide position is shown on the x axis. Samples were sequenced using the Illumina NovaSeq 6000 platform. Download FIG S3, TIF file, 1.8 MB (1.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Maximum-likelihood global phylogenetic tree with all the genomes sequenced from all the enriched Nichols dilution series with NEB+MDA and SWGA that generated at least 5× read depth covering at least 90% of the genome. All highlighted samples were sequenced using the Illumina NovaSeq 6000 platform. Download FIG S4, TIF file, 2.8 MB (2.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Supplemental Materials and Methods. Download TEXT S1, DOCX file, 0.03 MB (30.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
T. pallidum strains included in the phylogenetic tree. Download Table S5, CSV file, 0.00 MB (5KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
All sequencing data associated with this study were submitted to the National Center for Biotechnology Information Sequence Read Archive (SRA) under the BioProject number PRJNA744275.
T. pallidum strains included in the phylogenetic tree. Download Table S5, CSV file, 0.00 MB (5KB, csv) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.