Abstract
Human cytomegalovirus (HCMV) is a ubiquitous pathogen belongs to betaherpesvirus subfamily. RNA2.7 is a highly conserved long non-coding RNA accounting for more than 20% of total viral transcripts. In our study, functions of HCMV RNA2.7 were investigated by comparison of host cellular transcriptomes between cells infected with HCMV clinical strain and RNA2.7 deleted mutant. It was demonstrated that RNA polymerase II (Pol II)-dependent host gene transcriptions were significantly activated when RNA2.7 was removed during infection. A 145 nt-in-length motif within RNA2.7 was identified to inhibit the phosphorylation of Pol II Serine-2 (Pol II S2) by reducing the interaction between Pol II and phosphorylated cyclin-dependent kinase 9 (pCDK9). Due to the loss of Pol II S2 phosphorylation, cellular DNA pre-replication complex (pre-RC) factors, including Cdt1 and Cdc6, were significantly decreased, which prevented more cells from entering into S phase and facilitated viral DNA replication. Our results provide new insights of HCMV RNA2.7 functions in regulation of host cellular transcription.
Keywords: Human cytomegalovirus (HCMV), RNA2.7, RNA polymerase II (Pol II), Cyclin-dependent kinase 9 (CDK9), Phosphorylation
Highlights
-
•
HCMV RNA2.7 inhibits the phosphorylation of Pol II Serine-2.
-
•
RNA2.7 reduces the interactions between Pol II and pCDK9.
-
•
RNA2.7 regulates cell cycle by preventing cells from entering into S phase.
1. Introduction
Human cytomegalovirus (HCMV) is a ubiquitous human pathogen belongs to the betaherpesvirus subfamily and encodes more than 200 open reading frames (ORFs) (Stern-Ginossar et al., 2012). Following primary infection, HCMV usually establishes lifelong latent infection with episodes of reactivation, mainly in the immune compromised host. The infection or reactivation of HCMV in immune compromised patients, such as transplant recipients and acquired immune deficiency syndrome (AIDS) patients, can cause severe diseases (Cheeran et al., 2009; Drew, 1992; Razonable and Eid, 2009).
Long non-coding RNAs (lncRNAs) are roughly defined as RNA molecules of more than 200 bases in length without protein-coding capacity. Many studies have illuminated that lncRNAs could regulate gene expression at transcriptional level or posttranscriptional level (Jarroux et al., 2017; Wei et al., 2017). Accumulating evidences indicate that viruses also produce lncRNAs, which may play important roles in a variety of biological and pathological processes (Park and Miller, 2018; Pijlman et al., 2008; Rennekamp and Lieberman, 2011; Saayman et al., 2014; Vallery et al., 2018).
High-resolution transcriptome mapping of HCMV showed that most viral transcripts during infection were viral lncRNAs (Gatherer et al., 2011; Rossetto et al., 2013), including RNA1.2, RNA2.7, RNA4.9 and RNA5.0. HCMV RNA2.7 is a viral lncRNA of 2.7-kb in length, which was previously termed as beta2.7 (McDonough and Spector, 1983). RNA2.7 was abundant at early time of infection, accounting for more than 20% of total viral transcripts (Greenaway and Wilkinson, 1987). Recent studies have verified that RNA2.7 can prevent cell apoptosis by interacts with mitochondrial complex I during HCMV infection to maintain high levels of energy production in infected cells (Poole et al., 2016; Reeves et al., 2007). However, most functions of HCMV RNA2.7 involved in HCMV infection still remain unclear.
In this study, functions of HCMV RNA2.7 were investigated by comparison of host cellular transcriptomes between cells infected with HCMV clinical strain and RNA2.7 deleted mutant. A 145 nt-in-length motif in RNA2.7 was identified to inhibit RNA Polymerase II (Pol II) phosphorylation by reducing the contactions between Pol II and phosphorylated cyclin-dependent kinase 9 (pCDK9). In addition, evidences were presented that cellular DNA pre-replication complex (pre-RC) factors were decreased due to the repression of Pol II phosphorylation, which might influence cell cycle progression to facilitate viral DNA replication. Our findings might provide a new understanding of RNA2.7 function by which HCMV could keep survival and productive infection in host cells.
2. Materials and methods
2.1. Cell culture
Human embryonic lung fibroblast (HELF) cells were obtained from Shanghai Institutes for Biological Sciences (Cat#GNHu41). Cells were maintained in minimal essential medium (MEM) (Gibco, Carlsbad, CA, USA, Cat#11575032) supplemented with 10% fetal bovine serum (FBS) (Gibco, Cat#10100147), 100 units/mL penicillin and streptomycin.
HEK293 cells were maintained in Dulbecco's modification of Eagle's medium (DMEM) (Gibco, Cat#12320032) supplemented with 10% FBS, 100 units/mL penicillin and streptomycin.
2.2. Virus preparation
Bacterial artificial chromosome of a characterized HCMV clinical low-passage isolate HAN (BAC HAN) has been constructed and labeled with green fluorescent protein (GFP) as previously described (Zhao et al., 2016). BAC HAN stock was prepared on HELF cells and the virus titer was determined by standard median tissue culture infective dose (TCID50) assays.
2.3. Construction of HCMV RNA2.7 deleted mutant
RNA2.7 deleted mutant was constructed from BAC HAN by homologous recombination. A fragment encoding kanamycin resistance gene was amplified from the previously described plasmid pGEM-oriV/Kan using forward and reverse RNA2.7Del primers (forward: 5′-AGATCGCTGCTGCTCCGGCGTTCTCCAGAAGCCCCGGCGGGCGAATCGGCCTGTCTCTTATACACATCTCAACCATC-3′; reverse: 5′-GCATGCAAACTTCTCATTTATTGTGTCTACTACTCTGTGTTGCTACAGGGAGCTGTCTCTTATACACATCTCAACCCTG-3′; sequence homologous to RNA2.7 flanking regions was underlined). Purified PCR product was electro-transferred into competent bacteria DY380 that contained BAC HAN construct. Colonies were picked and the deletion of HCMV RNA2.7 was approved by PCR and sequencing directly.
BAC HANΔRNA2.7 plasmid was extracted from the identified bacteria DY380 colony using Nucleo BondXtra Midi Kit (Macherey-Nagel, Duren, Germany, Cat#740412) following the manufacturer's protocol and was transfected into HELF cells by electroporation. The cells were then maintained under standard cell culture conditions until cytopathic effects (CPEs) and GFP signal were clearly apparent. The reconstituted virus was named as HANΔRNA2.7, and the virus stock was prepared as described previously.
2.4. Validation of HCMV RNA2.7 deletion
HELF cells growing in 6-well plates were infected with HAN or HANΔRNA2.7 at an MOI of 1.0 for 72 h. Total RNAs were extracted from cells using Trizol reagent (ThermoFisher, Wilmington, DE, USA, Cat#15596026) according to the protocol, dissolved in 40 μL RNase free water and were then treated by TURBO DNA-free™ Kit (ThermoFisher, Cat#AM1907). RNA preparations were estimated by electrophoresis on 1% agarose gel and were quantified by ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). Total RNAs were reverse-transcribed using the SuperScript III First-strand synthesis system (ThermoFisher, Cat#18080051).
Forward and reverse RNA2.7 reverse transcript (RT) primers were designed and used to amplify RNA2.7 from the cDNAs. Transcript of HCMV UL83 was amplified as a control. The primer sequences used in PCR are shown in Supplementary Table S2. Products were analyzed by electrophoresis on a 1.5% agarose gel containing ethidium bromide and were visualized under ultraviolet light.
The transcriptions of genes neighbor to RNA2.7 (RL1, RL6, RL8A and RL9A) and selected viral immediate-early (UL123), early (UL44 and UL55) and late (UL83 and UL99) genes were quantified using QuantiTect SYBR Green-based PCR Kits (ThermoFisher, Cat#4309155) on Applied Biosystems 7300 Fast Real-Time PCR System (Applied Biosystem, Foster City, CA, USA). Primers for quantitative PCR are listed in Supplementary Table S2. Detections were performed in biological triplicates and the relative transcription levels of viral genes were normalized to that of housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in corresponding samples using 2−ΔΔCTmethods.
DNA of HAN or HANΔRNA2.7 infected HELF cells was isolated using QIAamp DNA Mini Kit (QIAGEN, Dusseldorf, Germany, Cat#51306). The viral genomes were assessed with restriction endonucleases EcoR I (Takara, Ostu, Shiga, Japan, #1040S) and Spe I (Takara, #1086S), respectively. Products were analyzed by electrophoresis on a 0.3% agarose gel containing ethidium bromide and were visualized under ultraviolet light.
2.5. Transcriptomic microarray assays and statistical analysis
HELF cells growing in 100 mm plates were infected with HAN or HANΔRNA2.7 at an MOI of 1.0. Total RNAs was extracted and purified at 72 h post-infection (hpi) using RNeasy Mini Kit (QIAGEN, Cat#74104) following the manufacturer's instruction and was checked to inspect RNA integration by an Agilent Bioanalyzer 2100 (Agilent technologies, California, USA). Microarray assays were performed at Shanghai Bio Corporation (National Engineering Center for Biochip in Shanghai, China) using GeneChipTM Human Genome U133 Plus 2.0 Assays with 54675 probes representing 29255 genes/transcripts (ThermoFisher). Raw data were processed and normalized by Expression Console software (ThermoFisher, version 1.1.2). The transcriptomic microarray analysis was done with three biological replicates. Genes with empirical Bayes-adjusted P values less than 0.05 and fold change greater than 2.0 were considered differentially transcribed.
Differentially transcribed genes were functionally categorized using ToppGene (https://toppgene.cchmc.org). Activities of transcriptional regulators were predicted using Ingenuity Pathway Analysis software (QIAGEN Ingenuity Systems).
2.6. Western blots of Pol II and CDK9
HELF cells infected with HAN or HANΔRNA2.7 at an MOI of 1.0 were harvested and re-suspended using mammalian protein extraction reagent M-PER (ThermoFisher, Cat#78501) with protease and phosphatase inhibitor cocktail (Abcam, Cambridge, MA, USA, #ab201111). Lysates were separated by SDS-PAGE. Blotted PVDF membranes were incubated with antibodies against Pol II (Millipore, #05-623-Z) (Millipore, Billerica, MA, USA), Pol II S2 (Abcam, #ab5095), Pol II S5 (Abcam, #ab5131), pCDK9 (Abcam, #ab79178), IE1 (ThermoFisher, MA1-7596) and GAPDH (Abcam, #8245), followed by anti-IgG-HRP with ECL Western blot reagent. Protein relative densities were quantified using ImageJ software 1.44p (NIH). GAPDH was used as a quantitative control.
2.7. Vector construction and transfection
HCMV RNA2.7 sequence was amplified from cDNA using PrimerSTAR Max DNA Polymerase Mix (TaKaRa, #R045A). A series of RNA2.7 sequences in different lengths were obtained using different primers. The primers used in the vector construction are listed in Supplementary Table S2. PCR products were inserted into pcDNA3.1 (−) vector (ThermoFisher, #79520) and the recombinants were transformed into TOP10 (TianGenBioTech, Bejing, China, #CB104). All constructs were verified by sequencing.
HELF cells were prepared in 6-well plates. A total of 2 μg vectors per well were transfected into the cells using Attractene Transfection reagent (QIAGEN, #301005) after cell preparation. Cells without transfection or transfected with pcDNA3.1 (−) vector were used as a negative control. Proteins were extracted for Western blot analysis or CoIP at 48 h after transfection.
2.8. Co-immunoprecipitation (CoIP)
HELF or HEK293 cells growing in 100 mm plates were infected with different HCMV strains at an MOI of 1.0 or transfected with vectors transcribing RNA2.7C2c. PureProteomeTMProteinA/G Mix Magnetic Beads (Millipore, #LSKMAGAG10) were coated with anti-Pol II antibody (Millipore, #05-623-Z) and were incubated with lysates at 4 °C overnight. The captured protein complex was eluted with 60 μL SDS-PAGE sample loading buffer (Beyotime, Nantong, China, #P0015) and was then heated at 70 °C for 10 min. After the beads were removed, the supernatants were loaded on 8% SDS-PAGE gel. Blotted PVDF membranes were incubated with antibodies against Pol II and pCDK9, followed by peroxidase-conjugated goat anti-mouse or rabbit IgG (ZSGB-BIO, Guangzhou, China, #ZB-2305 or #ZB-2301) with ECL Western blot reagent (ThermoFisher). Protein relative densities were quantified using ImageJ software 1.44p (NIH). Pol II captured was measured as a quantitative reference to calculate the relative amount of pCDK9 binding to Pol II.
2.9. RNA immunoprecipitation (RIP)
A total of 1 × 107 HELF cells were infected with HAN at an MOI of 1.0 for 72 h. Cells were treated with alfa-amanitin (MedChem Express, Cambridge, MA, USA, #23109-05-9) 2 h before cell collection at a final concentration of 1 μg/mL. RIP experiments were performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, #10007D). Cells were pelleted and re-suspended with RIP lysis buffer plus protease and RNase inhibitors. The cell lysates were incubated with rabbit anti-Pol II antibody (Millipore, #05-623-25UG) coated beads overnight according to the protocol. Rabbit IgG was used as a control. After treating with proteinase K, the immunoprecipitated RNAs were extracted by RNeasyMinElutte Cleanup Kit (QIAGEN, #74104) and reversely transcribed using PrimeScript RT Master Mix (TaKaRa, #RR037A). The abundance of RNA2.7 was detected using primers RNA2.7D forwards and reverse by RT-PCR. UL123 (IE1) mRNA and GAPDH mRNA were reversed and amplified as negative control. Primer information is listed in Supplementary Table S2.
2.10. Dose-dependent assay
HEK293 cells growing in 6-well plates were transfected with vectors transcribing RNA2.7 (4 μg per well) or RNA2.7C2c (4, 2 and 1 μg per well). Cell lysates were separated by SDS-PAGE. Blotted PVDF membranes were incubated with antibodies against Pol II (Millipore, #05-623-Z), Pol II S2 (Abcam, #ab5095), pCDK9 (Abcam, #ab79178) and GAPDH (Abcam, #ab8245), followed by anti-IgG-HRP with ECL Western blot reagent (ThermoFisher). Protein relative densities were quantified using ImageJ software 1.44p (NIH). GAPDH was used as a quantitative control.
2.11. RNA electrophoretic mobility shift assay (RNA EMSA)
Nucleoprotein was extracted from 1 × 107 HELF cells using NE-PER Nuclear and Cytoplasmic Extraction Reagents (ThermoFisher, #78833) according to its instrument. Pol II C-terminal domain (CTD) protein was purified using PureProteomeTMProtein A/G Mix Magnetic Beads (Millipore, #LSKMAGAG10) and anti-Pol II CTD antibody (Millipore, #05-623-Z). The purified Pol II CTD protein was eluted with 60 μL native elution buffer (0.2 mol/L Glycine-HCl, PH 2.5) and was then neutralized by adding 5 μL neutralization buffer (1 mol/L Tris-HCl, PH 8.5).
RNA2.7C2c sequence containing T7 promoter was generated by PCR using Phusion High-Fidelity PCR Master Mix (NEB, Ipswich, MA, USA, #M0531S). PCR primers (T7RNA2.7C2c Forward and Reverse) were listed in Supplementary Table S2. PCR products were cloned and sequenced. The 145 nt-in-length RNA2.7C2c probe and competitive RNA2.7C2c were transcribed using T7 RNA Polymerase (NEB, #M0251S). RNA2.7C2c probe was labeled with biotin by adding biotin-16-UTP (ThermoFisher, #AM8452) into transcription reaction. After DNase treatment, probes were purified using RNeasy Mini Kit (QIAGEN, #74104) and were kept at −80 °C till use.
RNA EMSA was carried out with LightShift®Chemiluminescent RNA EMSA Kit (ThermoFisher, #20148). To relax RNA folding, probes were heated at 80 °C for 5 min and were then placed on ice before use. Binding reactions were assembled with 2 nmol/L RNA2.7C2c probe and 4 μg nucleoprotein or purified Pol II CTD protein with gradient concentrations in binding buffer, 5% glycerol and 25 nmol/L DTT in 20 μL. In addition, unlabeled RNA2.7C2c was used as specific competitor. Anti-Pol II antibody (Millipore, #05-623-Z) was added for super shift. After 20 min incubation at room temperature, the samples were loaded onto 4% polyacrylamide gel in 0.5x TBE buffer and run for 90 min at 100 V at 4 °C. Samples were transferred to Nylon membrane at 400 mA for 60 min and were crosslinked using a commercial ultraviolet light crosslinking instrument. Biotin-labeled probes were detected with Chemilumiescent Nucleic Acid Detection Module (ThermoFisher, #89880).
2.12. Propidium iodide staining
To synchronize cells in G0, HELF cells were grown in 25T-flasks in serum-free medium for 24 h. Quiescent cells were infected with HAN or HANΔRNA2.7 at an MOI of 1 for 72 h. Four hours before harvest, HANΔRNA2.7 infected cells were treated with 5,6-dichloro-1-b-D-ribofuranosyl benzimidazole (DRB) at a final concentration of 200 nmol/L. Mock-infected cells were measured as control. Cells were washed twice in ice cold PBS and were then harvested using cell dissociation buffer before being centrifuged. Cell pellets were resuspended in 70% ethanol and were kept at −80 °C overnight. The fixed cells were then washed in PBS and resuspended in PI/RNase Staining buffer (ThermoFisher, #550825). Cells were incubated for 30 min at room temperature, washed twice in PBS, resuspended in 0.5 mL PBS and finally analyzed by flow cytometry. Single cells were gated and percentages of cells in the G0/G1 phases of the cell cycle were calculated using winMDi.
2.13. Quantitative PCR and Western blots of pre-RC components
Quiescent cells were infected with HAN or HANΔRNA2.7 at an MOI of 1.0 for 72 h. DRB was added into the supernatant 4 h before cell collection at a final concentration of 200 nmol/L. Mock-infected cells were used as controls.
Transcriptions of MCM2, MCM4, MCM5, Cdt1 and Cdc6 were quantified from the cDNA using QuantiTect SYBR Green-based PCR Kits (QIAGEN) on Applied Biosystems 7300 Fast Real-Time PCR System (Applied Biosystem). Primer details are listed in Supplementary Table S2. The reaction conditions were as follows: an initial denaturation at 95 °C for 2 min, then 40 cycles of annealing/extension at 60 °C for 30 s followed by a final denaturation at 95 °C for 15 s. Detections were performed in biological triplicates and relative transcription levels of MCM2, MCM4, MCM5, Cdt1 and Cdc6 were normalized to that of housekeeping gene GAPDH in corresponding samples using 2−ΔΔCT method.
Lysates were separated by SDS-PAGE. Blotted PVDF membranes were incubated with antibodies against Pol II S2 (Abcam, #ab5095), Cdt1 (Abcam, #ab202067), Cdc6 (Abcam, #ab109315) and GAPDH, followed by peroxidase-conjugated goat anti-mouse or rabbit IgG (ZSGB-BIO, #ZB-2305 or #ZB-2301).
2.14. Virus titer assay
HELF cells were plated into 6-well plates (8 × 105 cells/well), synchronized in serum-free medium for 24 h and were then infected with HAN or HANΔRNA2.7 at an MOI of 0.1 or 3.0. After 2 h of incubation, medium was removed, the cells were rinsed with phosphorylate-buffered saline for three times, and fresh DMEM was added. Supernatant samples were collected at different time points and were stored at −80 °C until they were assayed. Infectious virus titers were determined by TCID50. HELF cells were resuspended in growth medium at a concentration of 2 × 105 cells/mL, and were seeded into 96-well plates (100 μL per well). On the following day, supernatant sample after 10-fold serial dilutions were added into each well in a row of the 96-well plates of cells and were cultured for approximately 14 days. GFP-positive wells were counted and the corresponding titer number was found using the TCID50 chart. An average titer was derived from three independent experiments.
2.15. siRNA design and transfection
siRNAs specific to Cdt1 and Cdc6 (RiboBio, Guangzhou, China) were diluted with RNase free water to a final concentration of 20 μmol/L. All siRNAs were stored as aliquots at −80 °C to avoid multiple freeze thaw cycles. The siRNA sequences are as follows: siRNA to CDT1: GCACCAGGAGGTCAGATTA; siRNA to Cdc6: GTGTGAGACTATTCAAGCA.
HELF cells were prepared in 6-well plates with 70% confluence. Transfection of siRNAs was carried out using HiPerfect Transfection Reagent (QIAGEN, #301704) according to its instrument. The final concentration of siRNA in each well was 100 nmol/L. MEM with 10% FBS was changed by MEM with 2% FBS at 24 h post transfection for viral infection.
2.16. Quantitative PCR of viral DNA
siRNAs transfected HELF cells were infected with HAN or HANΔRNA2.7 at an MOI of 1.0. DNA samples were extracted at 2, 24 and 96 hpi, respectively. Quantitative PCR was carried out for HCMV UL83 and GAPDH using QuantiTect SYBR Green-based PCR Kits (QIAGEN, #204243) on Applied Biosystems 7300 Fast Real-Time PCR System (Applied Biosystems). Primer details are listed in Supplementary Table S2. The reaction conditions were as follows: an initial denaturation at 95 °C for 2 min, then 40 cycles of annealing/extension at 60 °C for 30 s then denaturation at 95 °C for 15 s. Detections were performed in biological triplicates and the relative level of viral DNA was normalized to that of housekeeping gene GAPDH in corresponding samples using 2−ΔΔCTmethods.
2.17. Statistical analysis
Statistical analyses were performed using Excel and GraphPad Prism 5.0. Statistically significant differences were calculated using unpaired 2-tailed Student's t-test. In all cases, P value less than 0.05 was considered significant.
3. Results
3.1. Host cellular transcriptomes are differential in cells infected with different HCMV constructs
HCMV RNA2.7 is a transcript from the antisense strand of HCMV genome between RL1 and RL6 (Supplementary Fig. S1A). To study the functions of HCMV RNA2.7, an RNA2.7 deleted mutant (HANΔRNA2.7) was constructed by homologous recombination based on a previously constructed HCMV bacterial artificial chromosome (BAC) HAN, which was the first characterized HCMV clinical strain in China (Supplementary Fig. S1B) (Zhao et al., 2016). The viral genomes were assessed with restriction endonucleases and reverse-transcription PCR respectively (Supplementary Fig. S1C, S1D). The transcriptions of flanking genes (RL1, RL6, RL8A and RL9A) and selected viral immediate-early (UL123), early (UL44 and UL55) and late (UL83 and UL99) genes were measured at 72 h post infection (hpi) (Supplementary Fig. S1E). The transcriptions of viral genes were not interrupted by the deletion of RNA2.7. Based on these results, HANΔRNA2.7 strain could be applied, as an RNA2.7 knocked out HCMV stock in our further study.
To address the effects of RNA2.7 on host gene transcriptions, cells infected with HAN or HANΔRNA2.7 were harvested at 72 hpi and were subjected to whole genome transcriptomic analysis. In total, transcriptions of 2,520 cellular genes were substantially altered by infection with HAN compared with mock infected cells, while transcriptions of 4,286 cellular genes were changed by HANΔRNA2.7 infection (Fig. 1A). After sorting according to relative transcription levels, four patterns were identified. Transcriptions of 509 genes were only affected by HAN infection and were designated as HAN specific transcriptional genes. Transcriptions of 54 cellular genes were inversely regulated between HAN and HANΔRNA2.7 infections, while 1,957 genes were similarly regulated by both strains. A total of 2,275 genes were HANΔRNA2.7 specific, among which more than 60% (1,455/2,275) genes were up-regulated. In general, host genes were transcribed more actively in response to HANΔRNA2.7 infection compared with HAN infection, suggesting either an activation of gene transcription or a lack of suppression at transcriptional level due to deletion of RNA2.7.
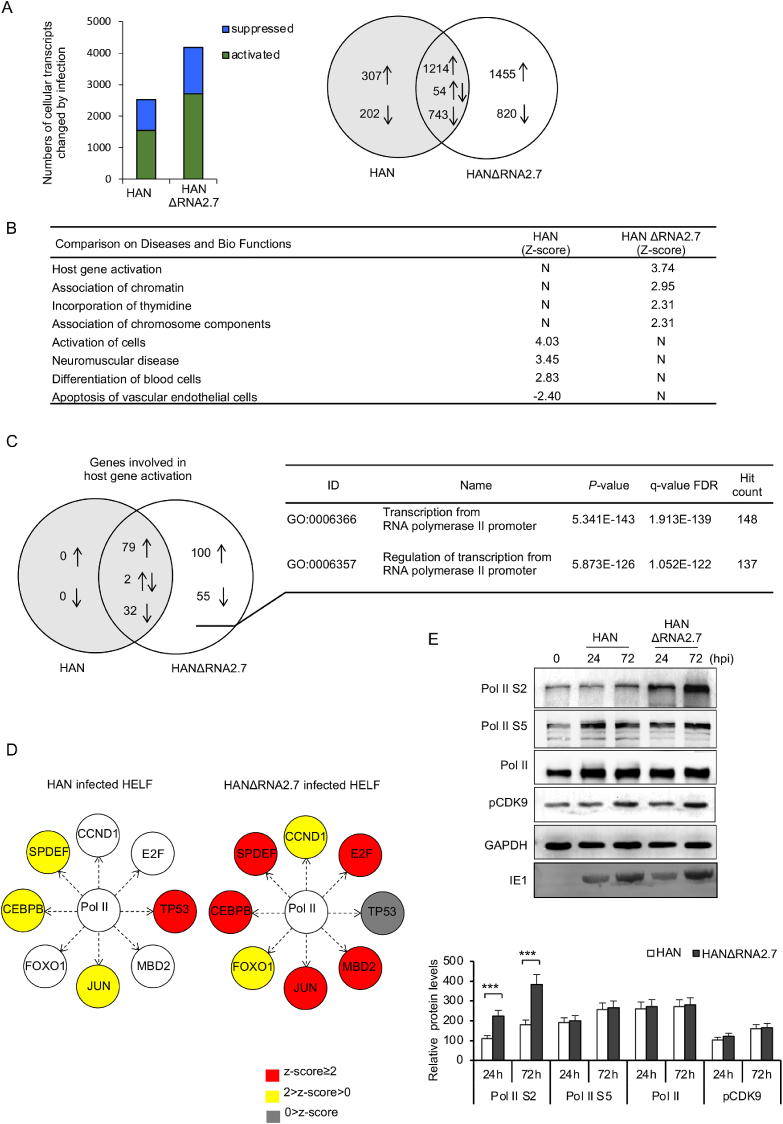
Fig. 1.
Repression of cellular Pol II-dependent transcription by HCMV RNA2.7. A General alteration of genome transcription in cells infected with HAN or HANΔRNA2.7 (MOI: 1.0). Cellular transcripts changed by infection are sorted in Venn diagram. B Comparison results on diseases and biofunctions of HAN or HANΔRNA2.7 altered transcripts. Their effects are indicated with Z-score: an active effect is indicated with Z-score > 2.0; a suppressive effect is indicated with Z-score < −2.0. C Venn diagram showing genes involved in host gene activation. HANΔRNA2.7 specific genes were analyzed and categorized into transcription from Pol II promoter. D Activation of Pol II-dependent regulators. Activities of transcriptional regulators were modified by RNA2.7. The effects are indicated with Z-score: Z-score > 2.0 indicates a significantly transcriptional activation of downstream genes; 2.0 > Z-score > 0 indicates a transcriptional activation of downstream genes; 0 > Z-score indicates a transcriptional repression of downstream genes. E Western blot analysis for pCDK9, Pol II and its two phosphorylated forms (Pol II S2 and Pol II S5) in cells infected with HAN or HANΔRNA2.7 at different time points. IE1 and GAPDH were measured as controls. Data are presented as mean ± SEM. Statistical analysis was performed by Student's t-test. ∗∗∗P < 0.0001.
3.2. HCMV RNA2.7 inhibits host Pol II S2 phosphorylation during infection
Host transcripts altered by HAN or HANΔRNA2.7 infection were compared based on diseases and biofunctions by Z-scores. A Z-score larger than 2.0 always indicates an activation, while a Z-score less than −2.0 indicates a repression. Results with top different Z-scores between HAN and HANΔRNA2.7 infected cells are listed in Fig. 1B. In HAN infected cells, genes involved in the apoptosis of vascular endothelial cells were indicated to be repressed with Z-score of −2.40. On the other hand, genes involved in host gene activation were indicated to be significantly activated in HANΔRNA2.7 infected cells (Z-score = 3.74). Genes involved in host gene activation were then analyzed and detailed information was listed in Supplementary Table S1. No gene was grouped specific to HAN infection for host gene activation, except for 113 genes whose transcriptions were both altered in cells infected with HAN and HANΔRNA2.7. Transcriptions of 155 genes associated with host gene activation were influenced specifically by HANΔRNA2.7 infection (Fig. 1C). It was suggested that RNA2.7 might suppress the host gene activation, and the suppression could be eliminated by deletion of RNA2.7.
The genes associated with host gene activation were further analyzed for gene ontology. It was predicted that 148 genes out of the 155 genes specific to HANΔRNA2.7 infection (95.48%) were categorized into transcription from Pol II promoter. Meanwhile, the activities of Pol II-dependent regulators such as JUN, FOXO1, CEBPB, SPDEF, CCND1, E2F and MBD2 were evaluated with Z-scores in cells infected with HAN or HANΔRNA2.7 (Fig. 1D). It was found that, the Z-scores of these regulators were increased when RNA2.7 was removed during HCMV infection. It was indicated that transcriptions of their downstream host genes were significantly activated in HANΔRNA2.7 infected cells. Based on these results, HCMV RNA2.7 was indicated to play a role in the regulation of host Pol II-dependent transcription, suggesting an effective regulation of RNA2.7 on quantity or activity of Pol II during HCMV infection.
Pol II has been known to control most eukaryotic gene transcriptions including mRNA precursors and non-coding RNAs (Lee et al., 2004). It is a multiprotein complex composed of 12 subunits. Pol II CTD is a component of the largest subunit and contains multiple repeats of the heptapeptide consensus sequence YSPTSPS. Transcription through Pol II extensively depends on the phosphorylation of serine-2 (S2) and serine-5 (S5) in CTD.
To investigate the effects of HCMV RNA2.7 on Pol II activity, HELF cells were infected with HAN or HANΔRNA2.7. Total proteins were extracted at 0, 24 and 72 hpi, respectively. Protein levels of Pol II and its two phosphorylated forms, Pol II S2 and Pol II S5, were measured. As shown in Fig. 1E, Pol II S2 was significantly increased in cells infected with HANΔRNA2.7 compared to that in HAN infected cells. The increase of Pol II S2 could be detected early at 24 hpi. However, total Pol II and Pol II S5 proteins were not significantly different between cells infected with HAN or HANΔRNA2.7. It was indicated that the prominent effect of RNA2.7 on Pol II function is to inhibit Pol II S2 phosphorylation during HCMV infection.
3.3. HCMV RNA2.7 reduces interaction between pCDK9 and Pol II
Phosphorylation of Pol II S2 promotes Pol II to overcome transcriptional blocks and increases the efficiency of 3′-end processing during elongation. Moreover, pCDK9 is an important enzyme mediating Pol II S2 phosphorylation. Therefore, protein levels of pCDK9 were also measured and compared between cells infected with HAN or HANΔRNA2.7, while no difference was found between them (Fig. 1E). The inhibition of Pol II S2 phosphorylation by RNA2.7 was not due to suspected quantity changes of pCDK9 proteins.
Thus, the effect of RNA2.7 on the interaction between pCDK9 and Pol II was investigated. Pol II and its binding proteins were immunoprecipitated using anti-Pol II antibody in HELF cells infected with different strains. Pol II-bound pCDK9 was measured in the captured proteins by Western blot. Immunoprecipitated Pol II protein was measured as a reference for calculation. The amount of pCDK9 binding to Pol II was increased by more than 47.6% in HANΔRNA2.7 infected cells than that in HAN infected cells (Fig. 2A). It was illustrated that RNA2.7 could reduce the binding of pCDK9 to Pol II.
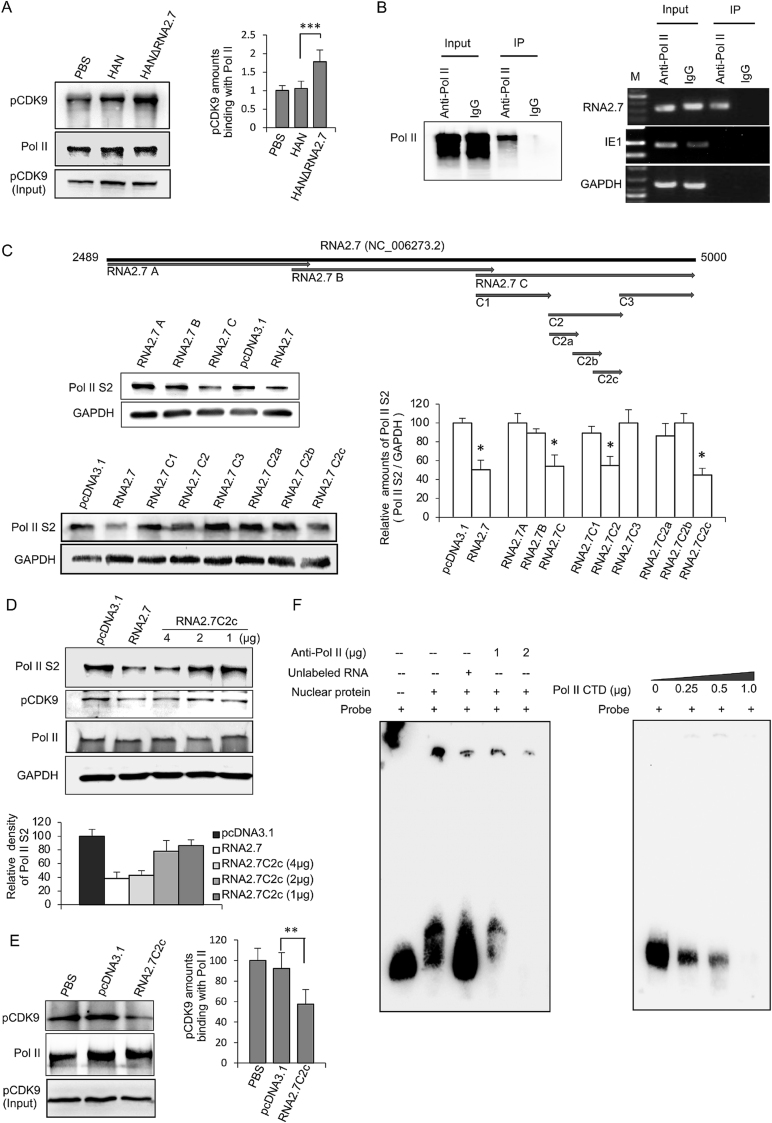
Fig. 2.
Inhibition of Pol II S2 phosphorylation by HCMV RNA2.7. A Co-immunoprecipitation and Western blot analysis of Pol II binding pCDK9 proteins in cells infected with HAN or HANΔRNA2.7. The relative amounts of pCDK9 binding to Pol II are quantified by densitometry with Pol II as a reference. Data are presented as mean ± SEM. B Interaction between RNA2.7 and Pol II. Pol II binding RNAs were immunopricipitated. Captured RNA was reverse transcribed and amplified using RNA2.7 specific primer. IE1 and GAPDH cDNA was amplified as negative control. M: DNA ladder marker (DL2000 for RNA2.7 and GAPDH; DL500 for IE1). C Effect of RNA2.7C2c on inhibiting Pol II S2 phosphorylation. A series of vectors transcribing RNA2.7 gene with different lengths were constructed and transfected into HEK293 cells. By Western blot analysis, RNA2.7C2c was identified to inhibit Pol II S2 phosphorylation functionally. The amounts of Pol II S2 proteins are quantified by densitometry. Data are presented as mean ± SEM. D RNA2.7C2c showing dose-dependent effect on inhibition of Pol II S2 phosphorylation. E Effect of RNA2.7C2c on interaction between Pol II and pCDK9. Vector transcribing RNA2.7C2c was transfected into HEK293 cells. Cells transfected with pcDNA3.1 were used as a negative control. By co-immunoprecipitation and Western blot analysis, RNA2.7C2c was verified to block the interaction between pCDK9 and Pol II protein. The relative amounts of pCDK9 binding to Pol II are quantified by densitometry with Pol II as a reference. Data are presented as mean ± SEM. F Confirmation of interaction between RNA2.7C2c and Pol II protein by RNA EMSA. RNA2.7C2c probe and competitive RNA2.7C2c were 145 nt in length. RNA2.7C2c probe was labeled with biotin. Pol II CTD proteins were purified from nucleoprotein using anti Pol II CTD antibody. Statistical analysis was performed by Student's t-test. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.0001. CTD, C-terminal domain.
To explore the relationship between RNA2.7 and Pol II, RNA immunoprecipitation (RIP) was performed to immunoprecipitate Pol II binding RNAs in infected cells. To exclude the disturbance of the binding between the undergoing transcriptional RNA2.7 and Pol II, alfa-amanitin, which can block all undergoing Pol II-dependent transcription processes, was added into cells 2 h before immunoprecipitation. Captured RNA was reverse-transcribed and amplified using RNA2.7 specific primers. Transcript of HCMV UL123 was amplified as negative control. RNA2.7 was confirmed in captured Pol II binding RNAs, which indicated a directly physical interaction between Pol II protein and RNA2.7 (Fig. 2B).
3.4. A 145 nt-in-length motif within RNA2.7 binds to Pol II and inhibits Pol II S2 phosphorylation
To investigate the functional motif of RNA2.7 mediating the inhibition of Pol II S2 phosphorylation, a series of vectors producing RNA2.7 fragments in different lengths were constructed and transfected into HELF cells (Fig. 2C). Pol II S2 proteins in transfected cells were detected and compared at 48 h post transfection. Cells transfected with pcDNA3.1 was used as a reference. A 145 nt-in-length motif (RNA2.7C2c) located in the 3′ terminus of RNA2.7 was identified to functionally inhibit Pol II S2 phosphorylation. Compared to reference value, the level of endogenous Pol II S2 protein was decreased by 54.9% in cells transfected with vector transcribing RNA2.7C2c. Moreover, the inhibition of Pol II S2 phosphorylation by RNA2.7C2c also showed a dose-dependent effect (Fig. 2D).
The effect of RNA2.7C2c to prevent pCDK9 binding with Pol II was further confirmed. HELF cells were transfected with pcDNA3.1 or vectors producing RNA2.7C2c. Pol II and its binding proteins were co-immunoprecipitated using anti-Pol II antibody. Compared to cells transfected with pcDNA3.1, the amount of pCDK9 binding to Pol II was decreased by about 42.6% in cells transfected with RNA2.7C2c vector (Fig. 2E).
To address the interaction between PoI II and RNA2.7C2c, RNA EMSA was carried out using biotin-labeled RNA2.7C2c RNA probes. A band appeared in nucleoproteins after incubation with RNA2.7C2c probes (Fig. 2F). Adding either competitive RNA or anti-Pol II antibody in the reaction systems could weaken the band. Although no classic super-shift band was obtained after incubation with Pol II antibody, the strip was more weakened when more anti-Pol II antibody was added. No interaction between pCDK9 and RNA2.7C2c was found by RNA EMSA using anti-pCDK9 antibody (Supplementary Fig. S2). Additionally, biotin-labeled RNA2.7C2c RNA probes were incubated with purified Pol II CTD proteins and were then detected by RNA EMSA. Along with the increase of amounts of Pol II CTD proteins added into incubation, the quantity of detected RNA probes was decreased correspondingly (Fig. 2F). The results confirmed that RNA2.7C2c is a functional motif of RNA2.7 to inhibit Pol II S2 phosphorylation by a physical binding to Pol II protein directly.
3.5. Inhibition of Pol II S2 phosphorylation alters cell cycle progression
Pathway analysis was carried out for cellular transcripts altered by HAN or HANΔRNA2.7 infection according to their known or suggested functions. Pathways involved in cell cycle regulation were significantly influenced in HANΔRNA2.7 infected cells (P < 0.05) (Supplementary Fig. S3A). To investigate whether the effects of RNA2.7 on host cell cycle control was associated with the inhibition of Pol II S2 phosphorylation, cell cycles were analyzed for cells infected with different HCMV strains. Chemical DRB, which is a specific inhibitor of Pol II S2 phosphorylation, was used for rescue. HELF cells were pretreated using DRB at a final concentration of 200 nmol/L before HANΔRNA2.7 infection. Compared with cells infected with HAN, less cells (73.39% vs 79.81%, P = 0.0485) were arrested at G0/G1 phase when infected with HANΔRNA2.7 (Fig. 3A). With a rescue of inhibition of Pol II S2 phosphorylation by DRB, the cell population in G0/G1 phases of HANΔRNA2.7 infected cells was significantly increased from 73.39% to 91.07% (P = 0.0002). These results indicate that RNA2.7 had an effect on preventing cells entering into S phase, which was similar to DRB by inhibiting Pol II S2 phosphorylation.
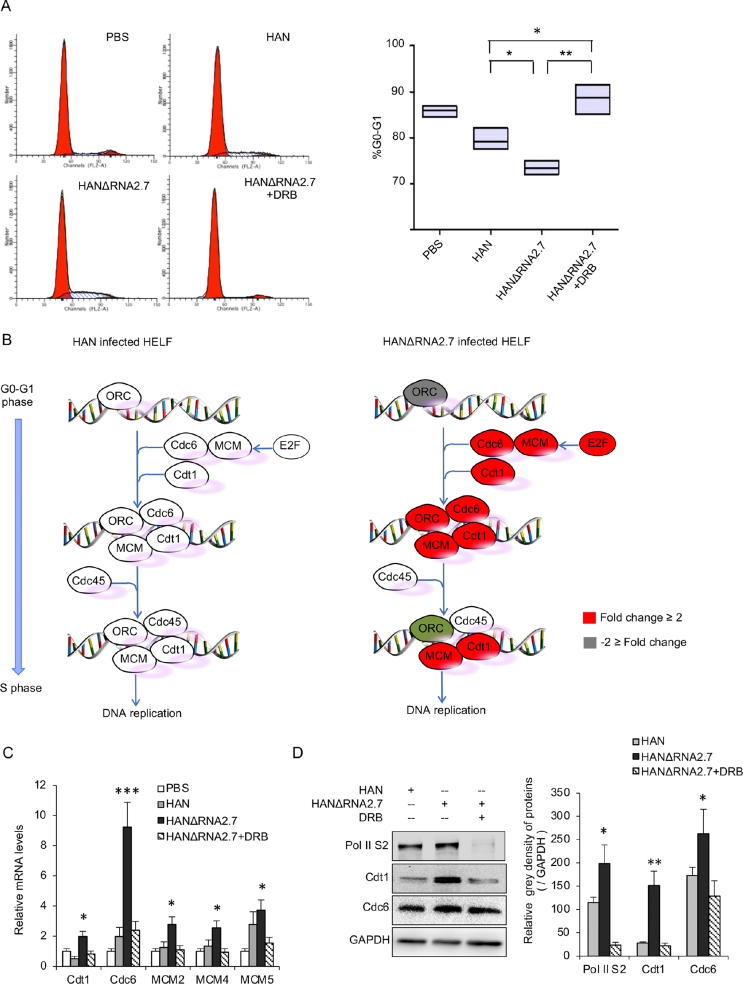
Fig. 3.
Regulation of host cell cycle by HCMV RNA2.7. A Block of host cells entry into S phase by RNA2.7. HELF cells were infected with HAN or HANΔRNA2.7 (MOI = 1.0). DRB was added 4 h before infection with a final concentration of 200 nmol/L and was used as a positive control. Cells were stained and analyzed by flow cytometry. Percentages of cells in the G0/G1 phases of the cell cycle were calculated and presented as mean ± SEM. B Model of pre-RC formation for cellular DNA replication. Components involved in pre-RC formation were up-regulated in HANΔRNA2.7 infected cells and were facilitated cellular DNA replication. Gene that mRNA fold change more than 2 is indicated with red color. Gene that mRNA fold change less than −2 is indicated with green color. C Effects of RNA2.7 on genes facilitating pre-RC formation. Transcriptions of MCM2, MCM4, MCM5, Cdt1 and Cdc6 were increased in HANΔRNA2.7 infected cells, and could be decreased by inhibiting Pol II S2 phosphorylation. Data are presented as mean ± SEM. D Effects of RNA2.7 on expressions of Cdt1 and Cdc6. Protein levels of Cdt1 and Cdc6 were increased in HANΔRNA2.7 infected cells, and could be decreased by inhibiting Pol II S2 phosphorylation. Data are presented as mean ± SEM. Statistical analysis was performed by Student's t-test. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.0001. DRB, 5,6-dichloro-1-b-D-ribofuranosyl benzimidazole; pre-RC, pre-replication complex.
Pre-RC has been well known to mainly promote cellular DNA replication and drive cells from G0/G1 phase into S phase. Genes involved in the formation of pre-RC, such as MCMs, Cdt1 and Cdc6, are products of Pol II-dependent transcription. The results of transcriptomic assay indicated that transcriptions of MCMs, Cdt1 and Cdc6 increased in HANΔRNA2.7 infected cells compared to those in HAN infected cells (Fig. 3B and Supplementary Fig. S3B). These phenomena were validated using real-time PCR and Western blots respectively. The results showed that mRNA levels of MCMs, Cdt1 and Cdc6 were increased in HANΔRNA2.7 infected cells at 72 hpi compared to those in HAN infected cells, and the differences could be eliminated by DRB treatment (Fig. 3C). The same changes were also observed for Cdt1 and Cdc6 proteins (Fig. 3D). It was speculated that RNA2.7 could reduce the factors of pre-RC and prevent host cells entering S phase through the inhibition of Pol II S2 phosphorylation.
3.6. Reduction of Cdt1 and Cdc6 proteins facilitates viral DNA replication
HELF cells were infected with HAN or HANΔRNA2.7 at low and high MOI (MOI = 0.1 or 3.0), respectively. Viral DNA replications were measured within nine days. A slight but significant difference of virus growth curves was observed between HAN and HANΔRNA2.7 strains at an MOI of 3.0. It was found that HAN strain replicates more productively than HANΔRNA2.7 strain (Fig. 4A).
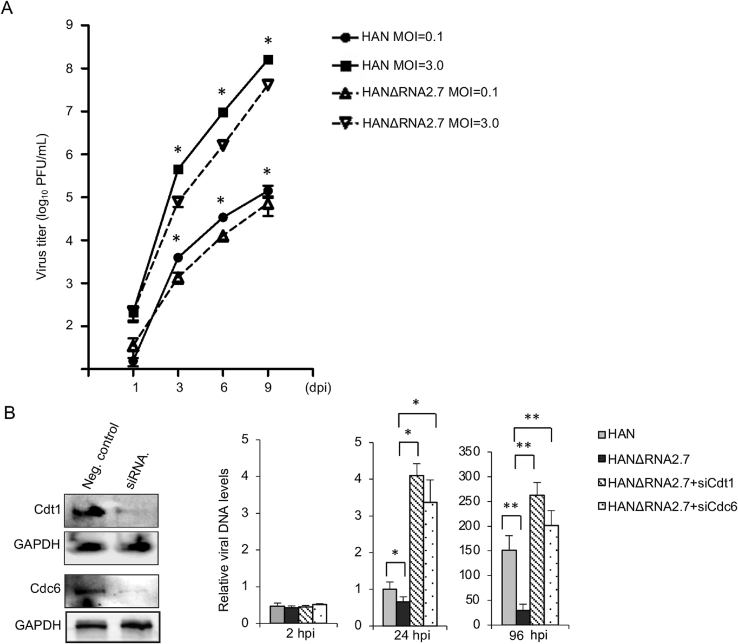
Fig. 4.
Effects of RNA2.7 on viral DNA replication. A Growth curves of HAN and HANΔRNA2.7 in HELF cells. Growth curves were performed in quiescent HELF cells infected with HAN or HANΔRNA2.7 at an MOI of 0.1 and 3. Supernatant samples were collected at 1, 3, 6 and 9 dpi. Titers were determined by TCID50. HAN is indicated with solid lines, and HANΔRNA2.7 is indicated with dashed lines. Data are averages from three independent experiments, and standard errors are indicated. B Up-regulation of viral replication by Cdt1 and Cdc6 knockdown. Evaluated siRNA specific to Cdt1 and Cdc6 were transfected into HELF cells. Cells transfected with siRNAs specific to Cdt1 or Cdc6 were infected with HANΔRNA2.7 (MOI = 1.0). Cells transfected with siRNA negative control were infected with HAN or HANΔRNA2.7. Viral DNA levels were quantified at 2, 24 and 96 hpi. Relative viral DNA levels are presented as mean ± SEM. Statistical analysis was performed by Student's t-test. ∗P < 0.05; ∗∗P < 0.01. TCID50, standard median tissue culture infective dose.
As known, viral DNA replication is intimately linked to host cell cycle control. To study whether the reduction of Cdt1 and Cdc6 proteins by RNA2.7 could influence HCMV DNA replication, Cdt1 and Cdc6 specific siRNAs were designed and transfected into HELF cells. Cdt1 or Cdc6 knockdown cells were infected with HAN or HANΔRNA2.7 at an MOI of 1.0. Relative viral DNA levels were measured at 24 and 96 hpi, respectively. The results showed that the deletion of RNA2.7 from genome caused a decrease of viral DNA replication by 33.63% at 24 hpi and 70.41% at 96 hpi. The supression of viral DNA replication could be reversed by Cdt1 or Cdc6 knockdown. The knockdown of Cdt1 or Cdc6 in HANΔRNA2.7 infected cells resulted in a major increase with fold changes of 8.88 or 6.81 in viral DNA level at 96 hpi (Fig. 4B). The results implied that HCMV RNA2.7 could facilitate viral DNA replication through repressing Cdt1 and Cdc6 expressions.
4. Discussion
Some researchers have been carried out to investigate the functions of RNA2.7. Potential cellular protein partners that might interact with RNA2.7 were directly screened among cDNA library in lambda phage using RNA probe (Rossetto et al., 2013). NADH ubiquinone oxidoreductase, a subunit of mitochondrial complex I was identified to interact with RNA2.7. The interaction of RNA2.7 with mitochondrial complex I could inhibit rotenone-induced apoptosis in neuronal cells (Reeves et al., 2007). However, the functions of RNA2.7 are still far from clear.
Viruses can extensively manipulate cellular gene expressions to maintain intracellular conditions benefiting viral survival and propagation. Transcription is the first step of gene expression and Pol II is an important enzyme participating in eukaryotic transcriptions (Schier and Taatjes, 2020). From our microarray results, an extensive activation of cellular transcription was observed in cells infected with RNA2.7-deleted mutant. The activated transcriptions were mainly categorized into Pol II-dependent transcriptions. Moreover, without RNA2.7, regulators of Pol II-dependent transcriptions were more activated during HCMV lytic infection. Generally, our results indicate a repression of cellular Pol II-dependent transcription caused by HCMV RNA2.7 during infection.
Diverse mechanisms of many viruses have been reported to hijack Pol II-dependent transcriptions to shut down host gene expression and facilitate virus productive infection. For example, herpes simplex virus I (HSV-1) have evolved various mechanisms to repress host Pol II transcriptional activities, including altering positioning of Pol II on host genes and inhibiting Pol II S2 phosphorylation (Birkenheuer et al., 2018; Fraser and Rice, 2005, 2007; Spencer et al., 1997; Zaborowska et al., 2014). Human Kaposi's sarcoma-associated virus (KSHV) ORF24 encoded protein makes a direct protein-protein contact with Pol II CTD domain to direct viral late gene transcription (Castañeda et al., 2020). HCMV UL79 has been confirmed to benefit accumulation of viral transcription by interaction with Pol II during late stages of infection (Peng et al., 2014). In our results, HCMV RNA2.7 was also found to inhibit Pol II S2 phosphorylation without altering pCDK9 levels. A 145 nt-in-length motif (RNA2.7C2c) in RNA2.7 was further identified to inhibit Pol II S2 phosphorylation by blocking the interaction between pCDK9 and Pol II. The inhibition was independent to the virions. It is well known that Pol II S2 phosphorylation is involved in the elongation of transcription. It is speculated that HCMV RNA2.7 might play a role in preventing overproduction of host components, which was disadvantageous to viral replication. Our findings provide more evidences for host Pol II functional modification by HCMV.
The interaction between RNA2.7C2c and Pol II was validated using RNA EMSA. Regrettably, no super-shift band was obtained by adding anti-Pol II antibody into RNA EMSA system. This might be due to the large molecular weight of RNA2.7C2c-Pol II complex that prevented them entering gel in electrophoresis. However, the detected quantity of biotin-labeled RNA2.7C2c reduced gradually along with the increasing input of purified Pol II correspondingly, which confirmed that there was a directly interaction between RNA2.7C2c and Pol II. Based on our results above, it is speculated that the interaction between RNA2.7 and Pol II may occupy or change the functional structure of Pol II for pCDK9 binding, and may competitively inhibit Pol II S2 phosphorylation.
Compared to HAN infected cells, more cells were found to be driven into S phase for cellular DNA replication in HANΔRNA2.7 infected cells. It is established that HCMV infection can lead to cell cycle arrest at some points (Kalejta and Shenk, 2002; Spector, 2015). Cell cycle progression is regulated by a wide variety of factors. Previous study has showed that the HCMV-mediated disruption of cell cycle control occurred at multiple aspects of gene expression, including gene transcription. For example, HCMV pp71 and UL97 can affect cell cycle progression by promoting the releasing of E2F/DP to activate gene transcription (McElroy et al., 2000). The assembly of pre-RC at DNA replication origins during G1 phase of the cell cycle initiates cellular G1-S-phase transition. MCMs, Cdt1 and Cdc6 are key components involved in pre-RC formation (Nishitani et al., 2001; Randell et al., 2006; Speck et al., 2005; Wohlschlegel et al., 2000). The binding of Cdt1 and Cdc6 to the complex allows recruitment of MCMs (Blow and Dutta, 2005). The transcriptions of MCMs, Cdt1 and Cdc6 were activated in HANΔRNA2.7 infected cells, and the activation could be eliminated by DRB treatment before infection. It was speculated that the higher expressions of Cdt1 and Cdc6 in HANΔRNA2.7 infected cells might be associated with the loss of inhibition of Pol II S2 phosphorylation by RNA2.7.
Cellular DNA replication may compete against viral DNA replication, since more molecules are used for cellular DNA replication and less is available for viral DNA replication. Arresting host cells at G0/G1 phase is essential to initiate HCMV gene expression at the time of infection (Fortunato et al., 2002). It has been reported that depletion of pre-RC factors in HCMV infected cell could promote viral replication (Biswas et al., 2003; Braun et al., 2012; Qian et al., 2010; Wiebusch et al., 2003). Cdt1 and Cdc6 levels were significantly increased in HANΔRNA2.7 infected cells. HCMV DNA replication levels decreased correspondingly and the suppression could be inverted by transfection with siRNA targeting Cdt1 and Cdc6. In previous study, the growth kinetics of Toledo and its RNA2.7-deleted mutant were compared in human foetal foreskin fibroblasts at an MOI of 0.01 (McSharry et al., 2003). The virus replication levels of Toledo were slightly higher than mutant strain, but removal of RNA2.7 could not cause growing defect of Toledo strain. Sinclair et al. found that the virus titers of Toledo were decreased when RNA2.7 was deleted in U373 cells (Reeves et al., 2007). We also measured the growth curves of HAN and HANΔRNA2.7 in HELF at low and high MOI (MOI = 0.1 or 3.0), respectively. There was a slight but significant difference of virus growth between HAN and HANΔRNA2.7 at high MOI, which could present viral replication abilities more relevantly without spreading.
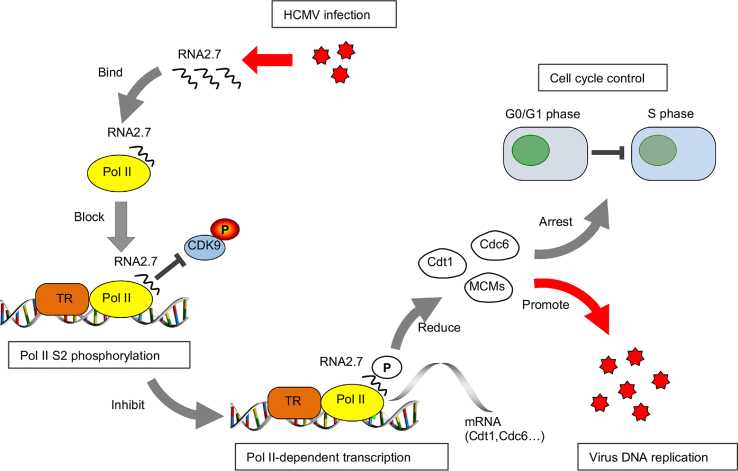
Altogether, our results suggest a potential pathway by which RNA2.7 plays a role at the host transcriptional regulation (Fig. 5): HCMV RNA2.7 binds to Pol II and inhibits Pol II S2 phosphorylation by blocking the interaction between pCDK9and Pol II. The loss of S2-phosphorylated Pol II leads to lower level expressions of host gene, including MCMs, Cdt1 and Cdc6. The reduction of pre-RC factors prevents host cells driven into S phase for cellular DNA replication so as to facilitate viral DNA replication. We have tried to investigate the functions of RNA2.7 by comparison between HCMV and RNA2.7-deleted HCMV infections, while detection of a revertant of RNA2.7-deleted HCMV infection would make our evidences much stronger by excluding unexpected genomic mutation occurred during BAC mutagenesis. Generally in our view, the effect of HCMV RNA2.7 on Pol II phosphorylation works more like a buffer. In cells infected with HCMV, RNA2.7 stabilizes the Pol II phosphorylation at normal levels, which will prevent host aggressive response to lytic infection.
Fig. 5.
Model for the mechanism of HCMV RNA2.7 in cell cycle control. RNA2.7 binds to Pol II and inhibits Pol II S2 phosphorylation by blocking interactions between pCDK9 and Pol II. The inhibition of Pol II S2 phosphorylation decreases the levels of MCMs, Cdt1 and Cdc6, and disturbs the formation of pre-RC which leads to host cell cycle arrest at G0/G1 phase and facilitates viral DNA replication.
HCMV has both lytic and latent phases in its life cycle like other human herpesviruses. RNA4.9 was proposed to play a role in transcriptional repression of viral IE gene expression for viral latent infection (Rossetto et al., 2013). Similar to RNA4.9, RNA2.7 was detected in latent CD14+ and CD34+ cells at relatively high levels. Similarly, it is prospected that HCMV RNA2.7 may play a role in intracellular modulation in some aspects for the progress of latency or reactivation, such as cellular transcription and cell cycle progression. More work on RNA2.7 functions for HCMV latency and reactivation is needed in further study.
5. Conclusions
HCMV RNA2.7 is a highly conserved virus-encoded lncRNA. Here we presented evidences to confirm function of RNA2.7 on inhibition of RNA Pol II S2 phosphorylation and its effects on cell cycle control. The repression of Pol II activity might reduce host responses to infection and benefit viral replication. Our findings provided a new understanding of RNA2.7 function by which HCMV could keep survival and productive infection in host cells.
Data availability
The transcriptomic microarray data are available in the NCBI's Gene Expression Omnibus database (accession number GSE73954).
Ethics statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author contributions
Yujing Huang: methology, investigation, data curation, writing-original draft, editing. Xin Guo: methology, investigation, software. Jing Zhang: methology, validation. Jianming Li: validation. Mingyi Xu: software, visualization. Qing Wang: validation. Zhongyang Liu: methology, investigation. Yanping Ma: data curation. Ying Qi: recourses. Qiang Ruan: funding acquisition, project administration, supervision.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (82071664).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2022.02.011.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Birkenheuer C., Danko C., Baines J. Herpes simplex virus 1 dramatically alters loading and positioning of RNA polymerase II on host genes early in infection. J. Virol. 2018;92 doi: 10.1128/JVI.02184-17. e02184-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas N., Sanchez V., Spector D. Human cytomegalovirus infection leads to accumulation of geminin and inhibition of the licensing of cellular DNA replication. J. Virol. 2003;77:2369–2376. doi: 10.1128/JVI.77.4.2369-2376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J., Dutta A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T., Poole E., Sinclair J. Depletion of cellular pre-replication complex factors results in increased human cytomegalovirus DNA replication. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda A., Didychuk A., Louder R., McCollum C., Davis Z., Nogales E., Glaunsinger B. The gammaherpesviral TATA-box-binding protein directly interacts with the CTD of host RNA Pol II to direct late gene transcription. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeran M., Lokensgard J., Schleiss M. Neuropathogenesis of congenital cytomegalovirus infection: disease mechanisms and prospects for intervention. Clin. Microbiol. Rev. 2009;22:99–126. doi: 10.1128/CMR.00023-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew W. Cytomegalovirus infection in patients with AIDS. Clin. Infect. Dis. 1992;14:608–615. doi: 10.1093/clinids/14.2.608-a. [DOI] [PubMed] [Google Scholar]
- Fortunato E., Sanchez V., Yen J., Spector D. Infection of cells with human cytomegalovirus during S phase results in a blockade to immediate-early gene expression that can be overcome by inhibition of the proteasome. J. Virol. 2002;76:5369–5379. doi: 10.1128/JVI.76.11.5369-5379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser K., Rice S. Herpes simplex virus type 1 infection leads to loss of serine-2 phosphorylation on the carboxyl-terminal domain of RNA polymerase II. J. Virol. 2005;79:11323–11334. doi: 10.1128/JVI.79.17.11323-11334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser K., Rice S. Herpes simplex virus immediate-early protein ICP22 triggers loss of serine 2-phosphorylated RNA polymerase II. J. Virol. 2007;81:5091–5101. doi: 10.1128/JVI.00184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatherer D., Seirafian S., Cunningham C., Holton M., Dargan D., Baluchova K., Hector R., Galbraith J., Herzyk P., Wilkinson G., Davison A. High-resolution human cytomegalovirus transcriptome. Proc. Natl. Acad. Sci. U. S. A. 2011;108:19755–19760. doi: 10.1073/pnas.1115861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenaway P., Wilkinson G. Nucleotide sequence of the most abundantly transcribed early gene of human cytomegalovirus strain AD169. Virus Res. 1987;7:17–31. doi: 10.1016/0168-1702(87)90055-4. [DOI] [PubMed] [Google Scholar]
- Jarroux J., Morillon A., Pinskaya M. History, discovery, and classification of lncRNAs. Adv. Exp. Med. Biol. 2017;1008:1–46. doi: 10.1007/978-981-10-5203-3_1. [DOI] [PubMed] [Google Scholar]
- Kalejta R., Shenk T. Manipulation of the cell cycle by human cytomegalovirus. Front. Biosci. 2002;7:d295–306. doi: 10.2741/kalejta. [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K., Lee S., Baek S., Kim V. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy A., Dwarakanath R., Spector D. Dysregulation of cyclin E gene expression in human cytomegalovirus-infected cells requires viral early gene expression and is associated with changes in the Rb-related protein p130. J. Virol. 2000;74:4192–4206. doi: 10.1128/jvi.74.9.4192-4206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough S., Spector D. Transcription in human fibroblasts permissively infected by human cytomegalovirus strain AD169. Virology. 1983;125:31–46. doi: 10.1016/0042-6822(83)90061-2. [DOI] [PubMed] [Google Scholar]
- McSharry B., Tomasec P., Neale M., Wilkinson G. The most abundantly transcribed human cytomegalovirus gene (beta 2.7) is non-essential for growth in vitro. J. Gen. Virol. 2003;84:2511–2516. doi: 10.1099/vir.0.19298-0. [DOI] [PubMed] [Google Scholar]
- Nishitani H., Taraviras S., Lygerou Z., Nishimoto T. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J. Biol. Chem. 2001;276:44905–44911. doi: 10.1074/jbc.M105406200. [DOI] [PubMed] [Google Scholar]
- Park R., Miller G. Epstein-barr virus-induced nodules on viral replication compartments contain RNA processing proteins and a viral long noncoding RNA. J. Virol. 2018;92 doi: 10.1128/JVI.01254-18. e01254-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng Y., Campbell J., Lenschow D., Yu D. Human cytomegalovirus pUL79 is an elongation factor of RNA polymerase II for viral gene transcription. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijlman G., Funk A., Kondratieva N., Leung J., Torres S., van der Aa L., Liu W., Palmenberg A., Shi P., Hall R., Khromykh A. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Poole E., Kuan W., Barker R., Sinclair J. The human cytomegalovirus non-coding Beta2.7 RNA as a novel therapeutic for Parkinson's disease--Translational research with no translation. Virus Res. 2016;212:64–69. doi: 10.1016/j.virusres.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Qian Z., Leung-Pineda V., Xuan B., Piwnica-Worms H., Yu D. Human cytomegalovirus protein pUL117 targets the mini-chromosome maintenance complex and suppresses cellular DNA synthesis. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell J., Bowers J., Rodríguez H., Bell S. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol. Cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Razonable R., Eid A. Viral infections in transplant recipients. Transplantation. 2009;100:479–501. [PubMed] [Google Scholar]
- Reeves M., Davies A., McSharry B., Wilkinson G., Sinclair J. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science. 2007;316:1345–1348. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- Rennekamp A., Lieberman P. Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt. J. Virol. 2011;85:2837–2850. doi: 10.1128/JVI.02175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto C., Tarrant-Elorza M., Pari G. Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14 (+) monocytes and CD34 (+) cells. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman S., Ackley A., Turner A., Famiglietti M., Bosque A., Clemson M., Planelles V., Morris K. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol. Ther. 2014;22:1164–1175. doi: 10.1038/mt.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier A., Taatjes D. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 2020;34:465–488. doi: 10.1101/gad.335679.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C., Chen Z., Li H., Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat. Struct. Mol. Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. Human cytomegalovirus riding the cell cycle. Med. Microbiol. Immunol. 2015;204:409–419. doi: 10.1007/s00430-015-0396-z. [DOI] [PubMed] [Google Scholar]
- Spencer C., Dahmus M., Rice S. Repression of host RNA polymerase II transcription by herpes simplex virus type 1. J. Virol. 1997;71:2031–2040. doi: 10.1128/jvi.71.3.2031-2040.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N., Weisburd B., Michalski A., Le V., Hein M., Huang S., Ma M., Shen B., Qian S., Hengel H., Mann M., Ingolia N., Weissman J. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallery T., Withers J., Andoh J., Steitz J. Kaposi’s sarcoma-associated herpesvirus mRNA accumulation in nuclear foci is influenced by viral DNA replication and viral noncoding polyadenylated nuclear RNA. J. Virol. 2018;92 doi: 10.1128/JVI.00220-18. e00220-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Huang K., Yang C., Kang C. Non-coding RNAs as regulators in epigenetics (Review) Oncol. Rep. 2017;37:3–9. doi: 10.3892/or.2016.5236. [DOI] [PubMed] [Google Scholar]
- Wiebusch L., Asmar J., Uecker R., Hagemeier C. Human cytomegalovirus immediate-early protein 2 (IE2)-mediated activation of cyclin E is cell-cycle-independent and forces S-phase entry in IE2-arrested cells. J. Gen. Virol. 2003;84:51–60. doi: 10.1099/vir.0.18702-0. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel J., Dwyer B., Dhar S., Cvetic C., Walter J. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- Zaborowska J., Baumli S., Laitem C., O'Reilly D., Thomas P., O'Hare P., Murphy S. Herpes Simplex Virus 1 (HSV-1) ICP22 protein directly interacts with cyclin-dependent kinase (CDK)9 to inhibit RNA polymerase II transcription elongation. PLoS One. 2014;9 doi: 10.1371/journal.pone.0107654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Shen Z., Liu Z., Zeng W., Cheng S., Ma Y., Rayner S., Yang B., Qiao G., Jiang H., Gao S., Zhu H., Xu F., Ruan Q., Luo M. Identification and BAC construction of Han, the first characterized HCMV clinical strain in China. J. Med. Virol. 2016;88:859–870. doi: 10.1002/jmv.24396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptomic microarray data are available in the NCBI's Gene Expression Omnibus database (accession number GSE73954).