Summary
Background
Streptococcus dysgalactiae subspecies equisimilis (SDSE) has emerged as an important cause of severe invasive infections including streptococcal toxic shock syndrome (STSS). The present study aimed to identify genes involved in differences in invasiveness between STSS and non-invasive SDSE isolates.
Methods
STSS and non-invasive SDSE isolates were analysed to identify csrS/csrR mutations, followed by a comparative analysis of genomic sequences to identify mutations in other genes. Mutant strains were generated to examine changes in gene expression profiles and altered pathogenicity in mice.
Findings
Of the 79 STSS-SDSE clinical isolates, 15 (19.0%) harboured csrS/csrR mutations, while none were found in the non-invasive SDSE isolates. We identified a small RNA (sRNA) that comprised three direct repeats along with an inverted repeat and was transcribed in the same direction as the sagA gene. The sRNA was referred to as srrG (streptolysin S regulatory RNA in GGS). srrG mutations were identified in the STSS-SDSE strains and were found to be associated with elevated expression of the streptolysin S (SLS) gene cluster and enhanced pathogenicity in mice.
Interpretation
The csrS/csrR and srrG mutations that increased virulence gene expression in STSS-SDSE isolates were identified, and strains carrying these mutations caused increased lethality in mice. A significantly higher frequency of mutations was observed in STSS-SDSE isolates, thereby highlighting their importance in STSS.
Funding
Japan Agency for Medical Research and Development, the Japan Society for the Promotion of Science (JSPS), and the Ministry of Health, Labor, and Welfare of Japan.
Keywords: Streptococcus dysgalactiae subspecies equisimilis, Streptococcus toxic shock syndrome, Mutation, Invasive infections, Bacterial infection
Abbreviations: SDSE, Streptococcus dysgalactiae subspecies equisimilis; STSS, streptococcal toxic shock syndrome; sRNA, small RNA; SLS, streptolysin S; GGS, group G streptococci; GAS, group A streptococci; ST, sequence type; FDR, false discovery rate; FC, fold change; DEGs, differentially expressed genes
Research in context.
Evidence before this study
Streptococcus dysgalactiae subspecies equisimilis (SDSE) strains are an important cause of severe invasive infections, including systemic disease characterised by streptococcal toxic shock syndrome. SDSE infections range from pharyngitis and mild skin and soft-tissue conditions including wound infection, erysipelas, and cellulitis, to life-threatening necrotising fasciitis and streptococcal toxic shock syndrome, thereby reflecting the clinical picture of S. pyogenes. PubMed searches for articles in any language with the terms “Streptococcus dysgalactiae subsp. equisimilis” AND “streptococcal toxic shock syndrome” from database inception to January 12, 2022, retrieved 29 articles, none of which pertained to differences in virulence between isolates from mild infection and streptococcal toxic shock syndrome (STSS). Identification of differences between STSS and non-invasive SDSE isolates is essential for developing effective treatment and vaccines.
Added value of this study
We report factors involved in differences in invasiveness between STSS and non-invasive SDSE clinical isolates. The STSS-SDSE isolates harboured mutations in at least one of the three genes, at varied sites between individual strains. The mutations resulted in elevated expression of virulence genes, as well as increased pathogenicity in animal studies.
Implications of all the available evidence
“Deleterious mutations” that destroy their own genes occur in SDSE, which concomitantly result in elevated expression of virulence genes and severe invasive infections including STSS. A proper comprehension of the mechanisms via which these virulence factors operate is crucial, especially in the case of SLS that plays an important role in SDSE induced STSS and was upregulated by these mutations.
Alt-text: Unlabelled box
Introduction
Group G streptococci (GGS), common among the normal flora of human skin, pharynx, and gastrointestinal tract,1 are virulent and pyogenic β-haemolytic streptococci classified as Streptococcus dysgalactiae subspecies equisimilis (SDSE) and cause pharyngitis, skin and soft tissue infections, bacteraemia, and endocarditis.2 Although SDSE is distinct from S. pyogenes (group A streptococci (GAS)), a substantial overlap of clinical symptoms and virulence factors exists between the two.2,3
S. pyogenes-caused streptococcal toxic shock syndrome (STSS) emerged as a serious concern in the 1980s, with fulminant onset of rapidly progressing symptoms including necrotising fasciitis, disseminated intravascular coagulopathy, and rapid multi-organ failure leading to shock and death. GGS identified as SDSE are also known to cause STSS, with the first case being reported in Minneapolis, United States of America, in 1996, followed by several reports.4, 5, 6, 7, 8, 9 Increasingly, SDSE isolates have been recovered from severe invasive streptococcal infections; SDSE has emerged as an important cause of severe invasive infections.2,10,11
Since SDSE emm genes show polymorphisms similar to S. pyogenes,12 gene sequence analysis has been utilised for epidemiologic analysis. To date, >30 emm types are recognised among SDSEs (http://www2a.cdc.gov/ncidod/biotech/strepblast.asp), with stG6792 being the most prevalent STSS-SDSE isolate in Japan.9
Our studies on STSS-GAS isolates revealed high frequency mutations in regulators including csrS (covS), csrR (covR), and/or rgg, enhancing virulence gene expression.13,14 While SDSE possesses csrS/csrR, it lacks the genomic region that harbours rgg.15 Watanabe et al. (2013) reported that only two (2.4%) of 83 invasive infection causing SDSE strains were csrS-defective.16 In contrast to S. pyogenes strains, csrS/csrR mutation frequency in invasive disease-causing SDSE isolates is low.13,14,17 Further, differences in the proportion of csrS/csrR mutations between STSS-SDSE isolates and non-invasive SDSE isolates are unknown. Therefore, the importance of mutations in these and other SDSE genes in the context of STSS remains unknown. This study aimed to determine the csrS/csrR sequence in SDSE isolates to identify mutations with potential roles in STSS development. Our findings revealed the presence of csrS/csrR mutations in almost a quarter of examined STSS-SDSE strains. Additionally, a mutation was identified in srrG in an STSS-SDSE clinical isolate.
Methods
Study design and bacteria
STSS- and non-invasive infection-causing SDSE isolates (Appendix 1), which are of the stG6792 genotype and sequence type (ST) 17, were obtained from pathogen collections at the National Institute of Infectious Diseases, and prefectural Public Health Institutes located in Fukushima, Tokyo, Kanagawa, Osaka, Yamaguchi, and Oita, Japan. Diagnostic criteria for STSS were as described by the Working Group on Severe Streptococcal Infections (1993).18 In total, 79 SDSE isolates were collected between 1995 and 2020 from sterile body sites of patients with STSS as part of standard patient care and 34 non-invasive patient isolates were collected between 2003 and 2020 (Appendix 1). The present study analysed differences between clinical isolates of STSS and non-invasive infections caused by SDSE. This study followed the Strengthening the Reporting of Genetic Association Studies reporting guideline.19
Ethics statement
This study complied with guidelines of the Declaration of Helsinki and was approved by institutional individual ethics committees for the use of human subjects (the National Institute of Infectious Diseases Ethic Review Board for Human Subjects [Approval number: 19]) and animal experiments (the National Institute of Infectious Diseases Animal Experiments Committee [Approval number: 120073]). Animal experiments adhered to guidelines of the Guide for Animal Experiments, National Institute of Infectious Diseases, Japan and complied with the U.K. Animals (Scientific Procedures) Act, 1986, associated guidelines of the EU directive 2010/63/EU, and National Institutes of Health guide for the care and use of laboratory animals (NIH publication no. 8023, revised 1978).
Plasmids, primers, and culture conditions
The plasmids and primers used in this study are described in Appendix 2. Escherichia coli DH5α was used as the host for plasmid construction and was grown in either liquid Luria-Bertani medium with shaking or on agar plates at 37°C. SDSE were cultured in either Todd-Hewitt broth (Becton Dickinson, Tokyo, Japan) supplemented with 0.5% yeast extract (THY media), brain heart infusion broth (Becton Dickinson) without agitation, or on Columbia agar supplemented with 5% sheep blood (Becton Dickinson). Cultures were grown at 37°C in air supplemented with 5% CO2. The following antibiotics were added to media as per requirement: kanamycin (Km), 25 μg/mL for E. coli and 200 μg/mL for SDSE; and spectinomycin (Sp), 50 μg/mL for E. coli and SDSE. SDSE growth was turbidimetrically monitored at 600 nm using a MiniPhoto 518R (Taitec, Tokyo, Japan).
DNA sequencing
Nucleotide sequences were determined by automated sequencing performed on the Applied Biosystems 3130xl Genetic Analyser (Applied Biosystems, Tokyo, Japan). The srrG sequence have been submitted to DDBJ and assigned DDBJ accession number LC685842
Construction of the sagA-phoZF transcriptional fusion plasmid
Plasmids that could measure sagA transcriptional levels were constructed using the pABG5 plasmid,20 that contains the phoZF reporter gene. When expressed, the reporter gene produces alkaline phosphatase that is secreted and can be easily measured. The promoter of the phoZF gene, but not the Shine-Dalgarno (SD) sequence of pABG5 was deleted while engineering the construct referred to as pABG0. The sic promoters of STSS and non-invasive SDSE isolates were amplified by PCR using the following primers: upstream primers, sagp-F0_Bm and sagp-F3_Bm, and downstream primer, sagp-R_Ec. PCR products were digested with BamH1 and EcoR1 to facilitate cloning into the BamHI-EcoRI sites located upstream of the phoZF SD region in pABG0, which resulted in generation of the fusion plasmids pABGsagA80, pABGsagA960, pABGsagA80-0, and pABGsagA960-0.
Construction of srrG, GGS_0543 and GGS_1256 complementation plasmids
Potential srrG, GGS_0543, and GGS_1256 mutations found in STSS-SDSE isolates, were mapped by amplifying these genes in the non-invasive isolate SDSE80 by PCR. The srrG gene was amplified using the primers sRNA_F_Bm and sRNA_R_Ml. The GGS_0543 gene was amplified using the primers vicR-F and vicR-R. The GGS_1256 gene was amplified using the primers tetR-F and tetR-R. The resulting PCR products were cloned into pAT18,21 thereby yielding pATsrrG, pAT-0543, and pAT-1256, respectively.
Construction of pATsrrGΔTTAAAGA plasmids
The deleted-srrG gene was amplified using the primers sRNA_F_Bm and sRNA_R_Ml. We used NIH960 genomic DNA as a template. PCR products were cloned into pAT18,21 thereby yielding pATsrrGΔTTAAAGA.
Construction of deletion mutants
Construction of the csrS mutant
The csrS deletion mutant was generated by amplifying three DNA fragments by PCR. A DNA fragment containing the 5ʹ end of csrS and the adjacent upstream region was amplified from the non-invasive isolate, SDSE80, using the primers csrS-del1 and csrS-del2. A fragment containing the 3ʹ end of csrS and the adjacent downstream region was amplified from SDSE80 using the primers csrS-del3 and csrS-del4. A DNA fragment containing pSET4s22 (pSET4s-PCR) was amplified using the primers pSET4s-PCR1 and pSET4s-PCR2. The three PCR products were circularised using the In-Fusion HD cloning kit (Takara Bio, Shiga, Japan), as per the manufacturer's instructions to create the plasmid pSET4scsrS. This plasmid was introduced into SDSE80 by electroporation, and transformants were selected on spectinomycin agar at 28°C. Cells with chromosomal integration of pSET4scsrS were selected by the growth of transformants in the presence of spectinomycin at 37°C. The plasmid integrated strain was serially passaged in liquid culture at 28°C without spectinomycin selection to facilitate excision of the plasmid, thereby leaving behind the desired mutation in the chromosome. The replacement of the native csrS gene by the csrS deleted mutant allele was verified by PCR, and the resultant strains were named SDSE80csrS.
Construction of the srrG mutant
The srrG deletion mutant was generated by amplifying two DNA fragments using PCR. A DNA fragment containing the mutated srrG gene was amplified from the NIH960 STSS isolate using the primers srrG-mut1 and srrG-mut2. A second DNA fragment containing pSET4s22 (pSET4s-PCR) was amplified using the primers pSET4s-PCR1 and pSET4s-PCR2. The two PCR products were circularised using the In-Fusion HD cloning kit (Takara Bio, Shiga, Japan), as per the manufacturer's instructions to create the plasmid pSET4ssrrG. This plasmid was introduced into the non-invasive strain SDSE80 by electroporation, and transformants were selected as described above. The replacement of the native srrG gene by the srrG disruption mutant was verified by PCR and sequencing, and the resultant strain was named SDSE80srrG.
Quantitative RT-PCR analysis
Total RNA was extracted from 79 STSS isolates and 34 non-invasive isolates. Quantitative RT-PCR was subsequently performed to assess sagA transcriptional abundance in an attempt to examine the expression profiles of virulence genes in SDSE isolates. The expression level of SDSE80 strain is shown as 1, and relative quantification was performed. Briefly, SDSE was grown to late-log phase (OD600 = 0.8–1.0) at 37°C in air supplemented with 5% CO2, and total RNA was extracted using the RNeasy Protect Bacteria Mini Kit (QIAGEN), according to the manufacturer's instructions. Complementary DNA was synthesised using the PrimeScript RT reagent Kit (Perfect Real Time) (Takara Bio), as per the manufacturer's instructions. Transcript levels were determined using the ABI PRISM Sequence Detection System 7500 (Applied BioSystems) and Premix Ex Taq (Perfect Real Time) (Takara Bio). For real-time amplification, template equivalent to 5 ng of total RNA was utilised. Measurements were performed in triplicate, and a reverse transcription negative blank of each sample as well as a no template blank served as negative controls. gyrA was used as an internal control. The primers and probes are described in Appendix 2.
Animals
Sixty-five male 5‒6-week-old outbred ddY mice and forty SKH1 hairless mice, weighing 20‒25 g, were procured from SLC and the Jackson Laboratory Japan, Inc. and maintained under specific pathogen-free conditions. Groups of five mice were allocated to cages and maintained in the Animal Experiment Facility at the National Institute of Infectious Diseases with a 0800–2000 h lighting cycle, under standard practices for mice husbandry.
phoZF alkaline phosphatase activity assay
The reporter gene phoZF utilised in this study encodes a chimeric protein comprising the N-terminal domain of protein F, and the C-terminal domain of Enterococcus faecalis alkaline phosphatase (PhoZF). Secreted PhoZF was detected in the supernatant, and ALP activity was measured as previously described.23
Genome comparisons
The whole genome sequences of the STSS isolate, NIH960, and the non-invasive isolates, SDSE77 and SDSE80, were determined by paired-end sequencing on an Illumina Hiseq2000 sequencing system. Comparisons between the same were made using a data mining service (Macrogen Japan, Tokyo, Japan). The S. dysgalactiae subsp. equisimilis RE378 sequence (GenBank accession no. AP011114) was utilised as the reference genome sequence. The whole-genome sequencing raw data are accessible under BioProject no. PRJDB13169.
RNA-seq
Total RNA was extracted using the RNeasy Protect Bacteria Mini Kit (Qiagen, Tokyo, Japan) and sequenced using the Illumina NovaSeq platform in paired-end mode with a read length of 150 bp. Approximately 3 Gb of raw reads were generated for each sample. RNA-sequence sample preparation, sequencing, and data mining were carried out by Genewiz Japan. Genes with a false discovery rate (FDR) below 0.005 and a fold change (FC) lower or higher than two were considered differentially expressed genes (DEGs). The raw RNA-seq data were submitted to DDBJ/EMBL-EBI/GenBank under BioProject accession no. PRJDB13168.
SDSE infection in a mouse model
The model has been previously described.13 A total of 6 × 107 SDSE CFUs were injected intra-peritoneally into 5‒6-week-old male ddY outbred mice (15 mice/SDSE strain). The number of surviving mice was compared statistically among SDSE80, SDSEsrrG, and SDSEcsrS using the Kaplan-Meier log-rank test. For the subcutaneous infection model, 5‒6-week-old male SKH1 hairless mice (10 mice/SDSE strain) were injected with 3 × 107 SDSE CFUs under anaesthesia, followed by daily measurement of the lesion area. In all cases, mice were randomly allocated to the different experimental groups. One group was set to 5 mice for statistical processing. Two or three experiments were performed for reproducibility. We observed frequently and euthanised without waiting for death if clinical symptoms such as hair growth, eye closure, and akinesia could predict death. Mice that reached the humanitarian endpoint and mice that had completed the observation period were euthanised by CO2 asphyxiation.
Histopathology
Tissues from SDSE infected mice were fixed in 10% formalin in PBS. Paraffin-embedded sections were stained with haematoxylin and eosin (Sapporo General Pathology Laboratory Co. Ltd., Hokkaido, Japan).
Statistical analysis
Statistical analysis was performed using Ekuseru-Toukei 2012 (Social Survey Research Information Co., Ltd., Tokyo, Japan). Survival curves were assessed using the Kaplan-Meier estimator; differences in the survival trends among SDSE80, SDSEsrrG, and SDSEcsrS were evaluated using the log-rank Mantel–Cox test. Differences in mutation frequency between STSS isolates and non-invasive isolates were compared using the χ2 test. Blood cell count and lesion size were compared between SDSE80 and SDSEsrrG or SDSEcsrS using the z-test. Statistical significance was defined as p < 0.05.
Role of the funding source
The funding body played no role in the study design, data collection, data analysis, data interpretation, or writing of this report. All authors had complete access to the study data and bear final responsibility for the decision to submit the study for publication.
Results
csrS/csrR mutation frequency STSS clinical isolates
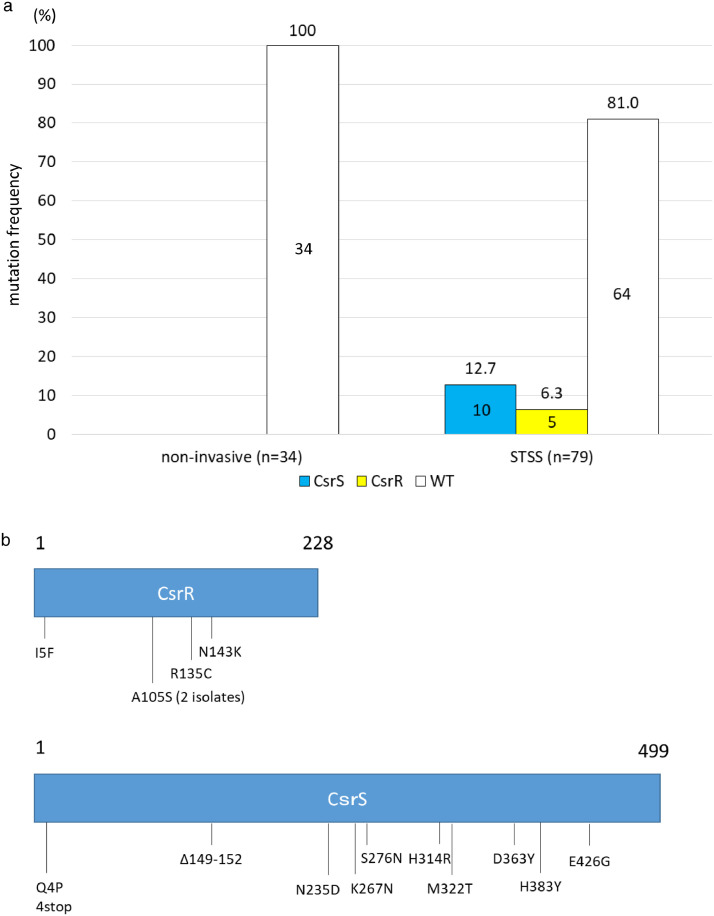
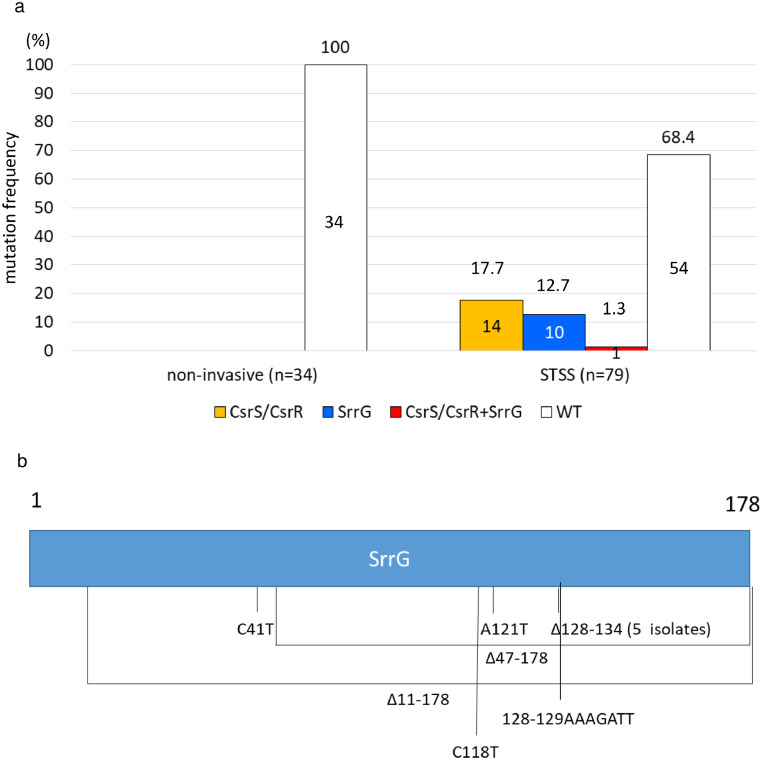
We previously reported several csrS/csrR mutations in STSS-GAS clinical isolates.13 Watanabe et al.16 reported that 2.4% (2/83) of 83 invasive-SDSE isolates were csrS-defective, and only four of 83 isolates caused STSS. Therefore, we sequenced csrS and csrR from 79 STSS clinical isolates (stG6792 genotype and ST17) collected from sterile body sites and 34 non-invasive clinical isolates (stG6792 genotype and ST17) obtained from non-sterile body sites, to evaluate mutation frequencies of mutations in these genes in STSS-causing SDSE. In total, 15 (19.0%) STSS-SDSE isolates harboured deletions or point mutations at various sites in csrS/csrR (Figure 1a, 1b, and Appendix 1)), rendering their products non-functional.16 On account of the lower frequency of csrS/csrR mutations in SDSE isolates than in GAS isolates in the context of SDSE, the remaining 64 (81.0%) STSS-SDSE isolates may be presumed to have mutations in genes other than csrS and csrR. In contrast, none of the non-invasive-SDSE (0/34) isolates carried mutations in these genes (p = 0.0064 by χ2 analysis) (Figure 1a). Although we found a higher frequency of csrS/csrR mutations among STSS-SDSE isolates than among non-invasive-SDSE isolates, they were not observed in all STSS isolates, suggesting that these isolates harbour mutations in other regulatory genes.
Figure 1.
CsrS and CsrR mutation frequency and positions.
(a) The CsrS and CsrR mutation frequency in S. dysgalactiae subsp. equisimilis isolates from 79 STSS and 34 non-invasive infections. STSS isolates (19.0%: CsrS, 12.7%; CsrR, 6.3%) carried CsrS/CsrR mutations, whereas non-invasive isolates did not harbour any mutations in these genes. WT indicated intact CsrS/CsrR. Numbers above bars indicate the percentages, and number in the boxes indicate the number of isolates. (b) CsrS/CsrR amino acid sequence variations. Labels refer to amino acid positions, and numbers above boxes indicate amino acid number. 1 is the start codon and 228 and 499 are penultimate amino acids to the stop codons of CsrR and CsrS, respectively.
Selection of STSS isolates with significantly elevated virulence gene expression
Quantitative RT-PCR was performed for 16 of 64 STSS isolates, which were randomly selected, without mutations in csrS and csrR and three non-invasive isolates that functioned as negative controls to assess the expression of two virulence genes, namely sagA and slo, that are elevated in csrS/csrR mutant strains.16,24 The selection criterion was >2-fold expression of at least one virulence gene in the STSS-SDSE isolate over that in the negative controls (Appendix 3). qRT-PCR analysis identified two isolates with elevated sagA expression that met this criterion.
Mutation spectrum of STSS and non-invasive SDSE isolates
The genomic sequences of one STSS-SDSE isolate (NIH960) with elevated sagA expression and two non-invasive isolates (SDSE77 and SDSE80) were compared to identify mutations enhancing sagA expression. Total DNA was isolated for DNA library construction and whole genome sequencing to analyse single nucleotide polymorphisms (SNPs) and insertion/deletion (InDel) mutations that may be associated with elevated sagA expression in STSS-SDSE isolates. This led to the identification of 509 non-synonymous SNPs, 1110 synonymous SNPs, and 209 SNPs that were present in the inter-genic regions of the STSS-SDSE isolates and absent in the non-invasive isolates, apart from seven inter-genic InDel mutations and eight frameshift InDel mutations. Among these, we identified mutations in two regulators, namely GGS_0543 and GGS_1256 in RE378 (Accession number AP011114), and a deletion mutation upstream of sagA. The intact GGS_0543 and GGS_1256 genes from SDSE80 were inserted into pAT18 to generate pAT-0543 or pAT-1256, respectively for complementation testing. The elevated sagA mRNA levels in NIH960 were not reduced to levels similar to those in SDSE80 due to lack of complementation post introduction of these plasmids (Appendix 4).
Promoter analysis of sagA
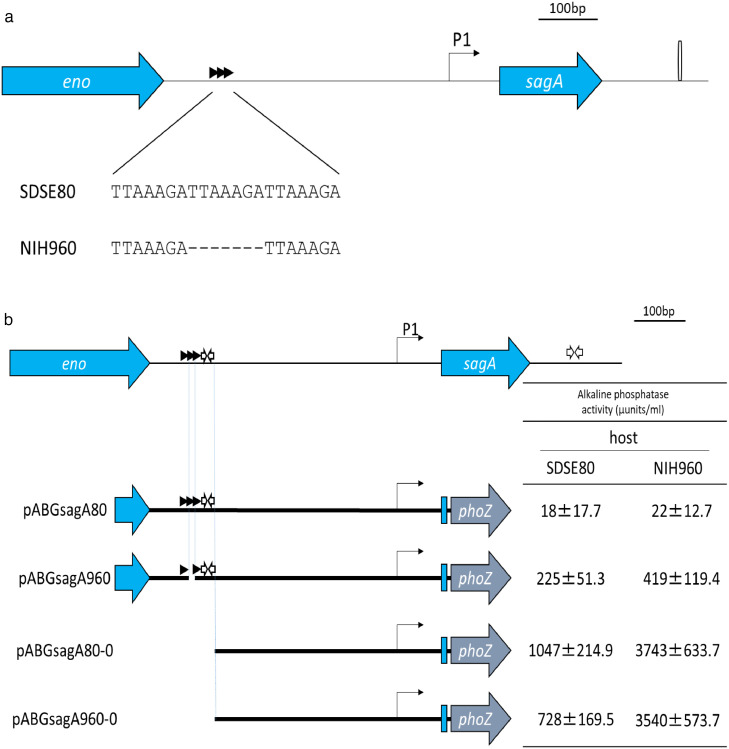
SDSE80 contains three direct TTAAAGA repeats upstream of sagA, one of which is absent in NIH960 (Figure 2a). The effect of this deletion on sagA transcription was assessed by alkaline phosphatase reporter assays. Briefly, we introduced either the sagA-phoZF transcriptional fusion plasmid pABGsagA960 containing the upstream region of sagA derived from NIH960, or the pABGsagA80 plasmid containing the upstream region of sagA derived from SDSE80, into the SDSE80 isolate (Figure 2b) and subsequently measured alkaline phosphatase activity. Higher activity was noted in the strain harbouring pABGsagA960 with the deletion than in the one harbouring pABGsagA80 (Figure 2b). Therefore, elevated sagA expression is attributable to the deletion in NIH960. In contrast, no observable differences in expression levels of sagA promoters (pABGsagA80-0 and pABGsagA960-0), which did not contain the deleted region, were seen in SDSE80 and NIH960.
Figure 2.
sagA upstream sequence and analysis of sagA expression levels.
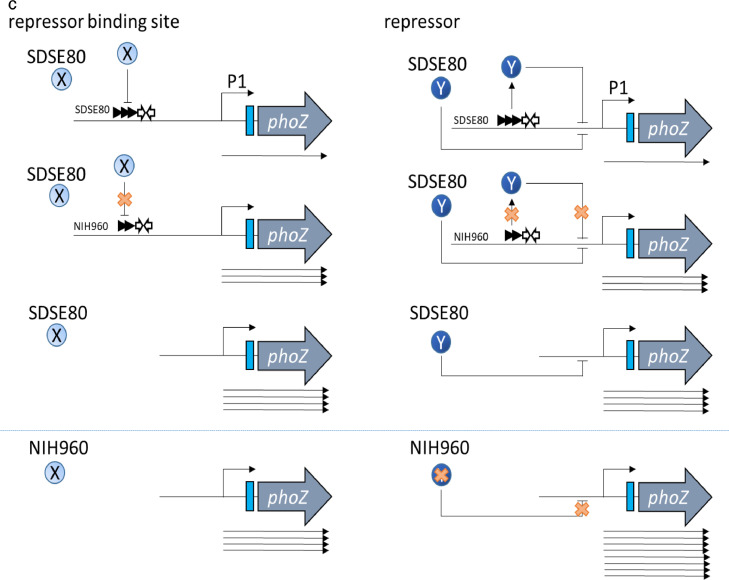
(a) The SDSE80 upstream sequence comprises 3 direct TTAAAGA repeats, while NIH960 comprises 2 direct repeats. (b) Truncation analysis of sagA promoter. Horizontal lines indicate DNA regions that were PCR-amplified and inserted into the promoter probe vector, pABG0.28 Alkaline phosphatase enzyme activity was measured in either SDSE80 or NIH960 harbouring one of the resulting plasmids. Mean activity of each strain is representative of 3 independent experiments. (c) Enhanced sagA expression consequent to the deletion of a direct repeat. Left: Expression levels of the SDSE80 and NIH960 sagA gene promoter regions without the deleted region do not change on being introduced into NIH960 if the region containing direct repeats represents a repressor binding region. Right: Expression levels of sagA promoter region without the deleted region are enhanced on being introduced into NIH960 if the region containing direct repeats encodes a repressor on account of the repressor on the NIH960 chromosome being destroyed. Horizontal lines indicate DNA regions that were inserted into the promoter probe vector, pABG0. Arrows beneath the horizontal lines represent RNA transcripts. Arrows above the horizontal lines represent direct repeats. White arrows above the horizontal lines represent inverted repeats. SDSE80 and NIH960 indicate the host. X and Y indicate repressors. X and Y beneath the host name indicate repressors encoded by chromosomal DNA. Cross marks indicate loss of function. P1 indicates the expected transcriptional start site as per the promoter sequence.
The above-mentioned observation could be explained either by the region encompassing the deletion being a repressor-binding region or by this region encoding a repressor. In the former case, the deletion would interfere with repressor binding; in the latter, the mutated gene product would be non-functional. Both scenarios would result in elevated sagA expression. This was investigated by introducing the pABG80-0 or pABG960-0 plasmid into NIH960 cells and subsequently performing reporter assays. If the deleted region represented a repressor binding region, expression levels of SDSE80 and NIH960 sagA promoter regions without the deletion would not change on introduction into NIH960 because the promoter regions did not show a difference in the sequence of the cloning region between pABG80-0 and pABG960-0 (Figure 2c). An alternate model might state that if the region binds a repressor, the introduction of the mutated promoter section could cause additional sequestering of the repressor away from the chromosomal promoter, further increasing sagA expression. This hypothesis hinges on dose-dependent repression by the repressor (Figure 2c). In this study, introduction of the pABG80-0 or pABG960-0 plasmids into NIH960 cells resulted in elevated sagA expression compared to that observed post their introduction into SDSE80 (Figure 2b), indicating that the region encompassing the deletion encodes a repressor.
Analysis of transcripts encoded by the region containing direct repeats
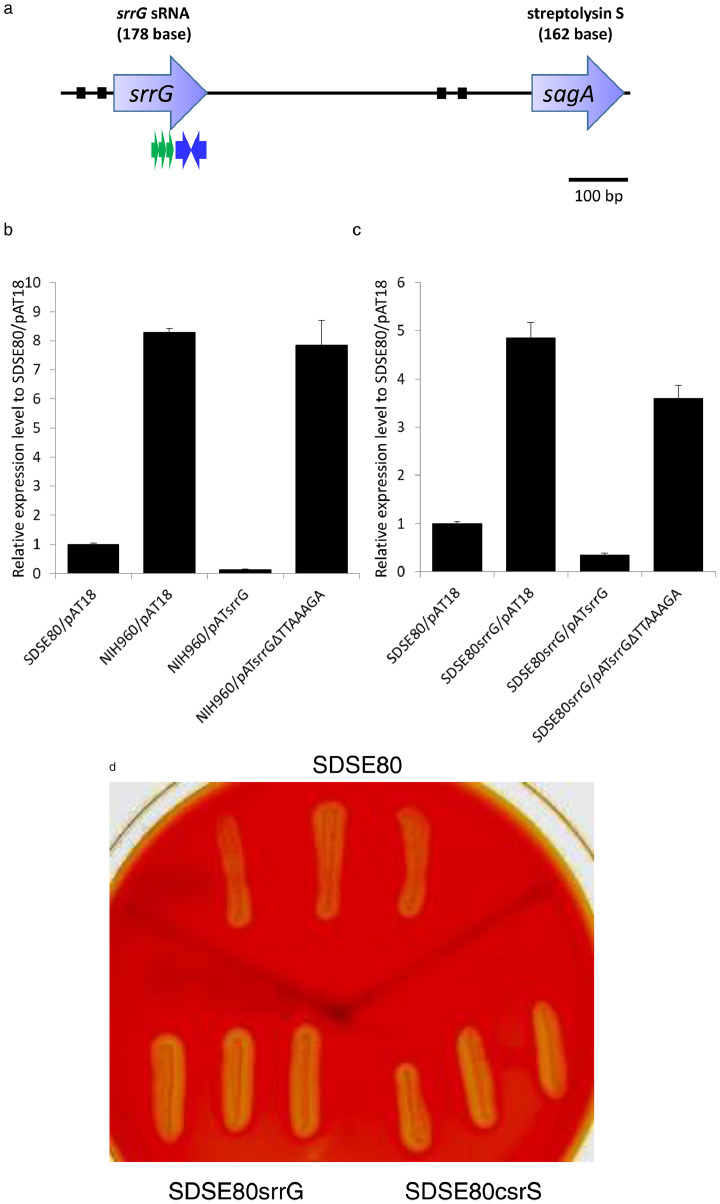
An ORF search in this region to identify possible transcripts revealed no clear ORF. Consequently, the ends of the transcripts encoded by the region containing the direct repeats were determined by 5ʹ-RACE and 3ʹ-RACE. This revealed the presence of a 178-bp transcript beginning 713-bp upstream and ending 536-bp upstream from the sagA start codon (Figure 3a), with the RNA being transcribed in the same direction as sagA. Additionally, an inverted repeat (AAAGCTTGGGA-N3-TCCCAAGCTTT) located 3ʹ to the direct repeats was identified. This small RNA (sRNA) was referred to as srrG (streptolysin S regulatory RNA in GGS).
Figure 3.
SrrG sRNA sequence and its effect on sagA expression.
(a) Position of SrrG sRNA. The putative −10 and/or −35 promoter sequences are indicated by black boxes and the putative rho-independent (intrinsic) terminators are highlighted by inverted blue arrows. The green arrow indicates the direct repeats region. The transcriptional product of SrrG was determined by 5′ and 3′ RACE. (b–c) sagA expression post introduction of SrrG sRNA into (b) NIH960 and (c) SDSE80srrG was determined by RT-PCR. sagA expression in each strain was analysed by RT-PCR. Columns represent the relative sagA mRNA levels in each strain. The expression level of SDSE80/pAT18 strain is shown as 1. Error bars displayed as mean +/- standard deviation (±S.D.) (n = 4). (d) Haemolysis in the srrG and csrS mutants after 5 hours using Columbia agar plates with 5 % sheep blood.
srrG mutation results in enhanced sagA expression in STSS isolates
The increase in sagA mRNA abundance in NIH960 was complemented by the introduction of pATsrrG, a pAT18 plasmid containing the intact SDSE80 srrG, but not by that of pATsrrGΔTTAAAGA, a plasmid with a deleted TTAAATA sequence in the srrG gene. This resulted in a reversal of elevated sagA mRNA levels in NIH960, to those observed in SDSE80 post introduction of pATsrrG (Figure 3b). Further, a srrG mutant strain, SDSE80srrG, was created by introducing a srrG deletion mutation derived from NIH960 into a non-invasive isolate prior to performing complementation tests and haemolytic phenotype. sagA mRNA levels in SDSE80srrG were lower than those in SDSE80 post introduction of pATsrrG, but not with pATsrrGΔTTAAAGA (Figure 3c), indicating that elevated sagA expression in NIH960 was consequent to deletion of direct repeats in the srrG sRNA. Additionally, the srrG mutant was found to be more haemolytic than SDSE80 (Figure 3d).
Screening of srrG-regulated genes via transcriptome analysis
The ability of srrG to regulate transcription of various genes was tested by RNA-seq, followed by a comparative analysis of mRNA levels between wild-type SDSE80 and SDSE80srrG. The results identified 21 transcripts with differential abundance that fulfilled the FC criterion in the mutant in comparison to the wild type (FC: |log2FC| > 1, p-value < 0.005, and FDR: q-value < 0.005 [Appendix 5]). From the heat map of FCs of differential expression patterns (DEPs) across SDSE80 and SDSE80srrG, we observed that most of the DEPs in SDSE80 and SDSE80srrG were similar (Appendix 6a). The full list of DEGs is available in Appendix 5. We subjected DEGs from both SDSE80 and SDSE80srrG to Gene Ontology analysis (Appendix 6b) and pathway analysis (Appendix 6c). The two strains had almost the same biological process regulations. Transcripts of known putative function with notable differential abundance between the mutant and wild type included sagA encoding SLS and other associated genes (GGS_0679 through GGS_0687; FCs in the mutant over wild-type, log2FC: 1.11-2.19). Oppositely the transcript encoding the M protein demonstrated differential abundance (log2FC: -7.42). Thus, transcription of the sagA operon is influenced by the srrG mutation.
srrG mutation frequency in STSS isolates
srrG was sequenced in 79 STSS-SDSE clinical isolates collected from sterile body sites and 34 non-invasive clinical isolates obtained from non-sterile body sites to evaluate the frequency of STSS-causing srrG mutations in SDSE. In total, 11 (13.9%) STSS-SDSE isolates harboured srrG mutations, including deletions, insertions, or point variations, resulting in loss of function as evident by the elevated sagA expression (Figure 4a, b and Appendix 1). In contrast, no non-invasive isolate carried a mutation. STSS isolates possessed a significantly higher frequency of srrG mutations compared to non-invasive isolates (p = 0.022 by χ2 analysis).
Figure 4.
SrrG mutation frequency and positions.
(a) SrrG and/or CsrS/CsrR mutation frequency among S. dysgalactiae subsp. equisimilis isolates from 79 STSS and 34 non-invasive infections. Twenty-five (31.6%) of STSS isolates harboured SrrG and/or the CsrS/CsrR mutations, while no non-invasive disease isolate carried a mutation in the srrG gene. WT indicates intact SrrG and CsrS/CsrR. Numbers above bars indicate the percentages, and number in the boxes indicate the number of isolates. (b) SrrG sequence variations. Labels refer to nucleotide positions, and numbers above boxes indicate nucleotide number.
Further, 31.6% STSS-SDSE isolates harboured csrS/R and/or srrG mutations, which is significantly higher than 0% of non-invasive isolates harbouring mutations in these genes (p = 0.00020 by χ2 analysis) (Figure 4a).
Importance of srrG mutation in pathogenesis of invasive infections in mouse models
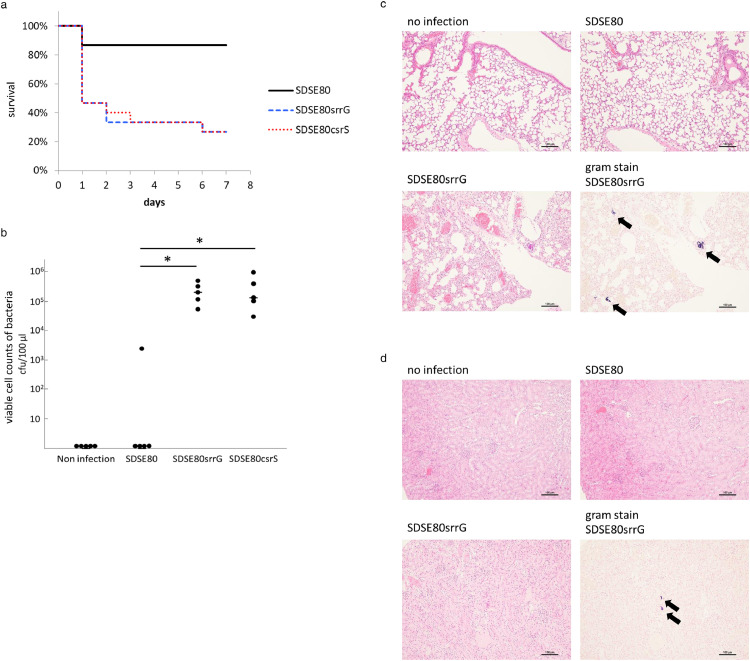
The histopathology and lethality of SDSE80srrG and SDSE80 infections were compared in vivo by injecting them intra-peritoneally in a mouse model. The srrG mutant strain demonstrated significantly higher lethality than the SDSE80 strain (p < 0.01 by a log-rank test) (Figure 5a), indicating that srrG mutated strains are more virulent than SDSE80 strains that cause non-invasive infections. SDSE80srrG-infected mice developed bacteraemia 21 h post intra-peritoneal injection, whereas bacteraemia was only occasionally detected in SDSE80-infected mice (p < 0.05 by z-test) (Figure 5b). Histopathological examination of SDSE80srrG-infected mice revealed bacterial colonies and inflammatory cell infiltration in the vicinity of pulmonary bronchi and blood vessels (Figure 5c). Further, bacterial colonies were also observed in the kidneys of these animals (Figure 5d). In contrast however, no significant pathological changes were observed in SDSE80-infected mice (Figure 5d).
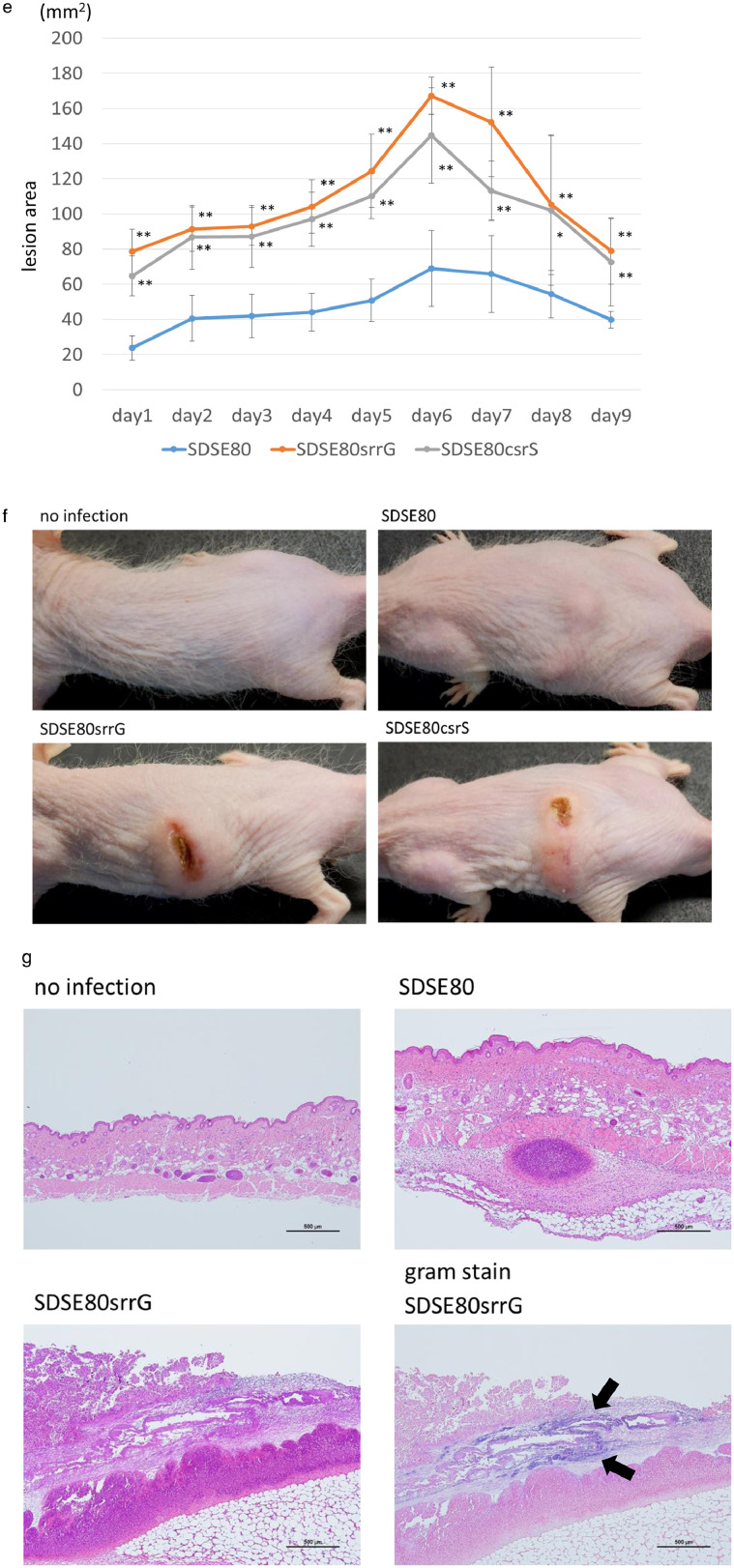
Figure 5.
SrrG and CsrS mutations enhance the lethality and histopathology of SDSE in in vivo mouse models.
(a) Survival curves of mice infected with each strain. Mice were intra-peritoneally inoculated with 6 × 107 CFUs of each SDSE and survival was observed up to 7 days post-infection. Differences in mortality were statistically significant (p < 0.01 by a log-rank test), as determined by a log-rank test. Survival curves were generated from 3 independent experiments using a total of 15 ddY mice per strain. (b) Counts of SDSE in 100 μL blood of each male ddY mouse. Blood was withdrawn 21 h post injection and bacterial counts were determined by plating on agar. The (–) bar represents median values, *(p < 0.05 by z-test). (c–d) Histopathological changes in the lung (c) and kidneys (d) of SDSE infected mice. Tissues were extracted 21 h post intra-peritoneal injection with 3 × 107 CFUs of SDSE. Black arrows indicate clusters of bacteria in lungs (c) and kidneys (d). The scale bar indicates 100 μm. (e-g) Lesion areas of subcutaneous infection in hairless mice injected with SDSE. (e) 3 × 107 CFUs of SDSE was injected subcutaneously, and lesion area were measured daily post infection (n = 5). Lesion areas in mice infected with srrG or csrS mutant strains were significantly larger than in those infected with wild type SrrG strains (SDSE80) (* p < 0.05; ** p < 0.01 using the z-test). Error bars displayed as mean +/- SD. Representative gross (f), and microscopic histopathological findings (g) in mice infected with SDSE. (g) Black arrows indicate clusters of bacteria. The scale bar indicates 500 μm.
Subcutaneous SDSE80srrG infection in a soft-tissue infection mouse model revealed significantly larger lesions compared to those observed on SDSE80 infection (p < 0.01 by z-test) (Figure 5e); Figures 5f and 5g display the representative gross and microscopic pathological findings. Examination of lesions caused by SDSEsrrG revealed indurated zones surrounded by oedema (Figure 5f) that progressed (Figure 5e) to concentrated ulcers and necrosis, which did not penetrate underlying muscle tissue (Figure 5f). SDSE80-infected animals, however, did not develop necrotic lesions. Tissues biopsied 48 h post-infection with either SDSE80 or SDSEsrrG revealed infiltration by inflammatory cells (Figure 5g). Gram staining of subcutaneous tissue sections of SDSE80srrG-infected mice infected with revealed Gram-positive cocci; the same was not observed in SDSE80-infected mice. This indicates in vivo virulence is higher for the srrG mutated isolate than for the wild type srrG isolate.
Comparison of virulence between csrS and srrG mutations
Since we identified a srrG mutation that conferred increased virulence and identified csrS/csrR mutations in 15 (19.0%) SDSE-STSS isolates (Figure 1a), we compared effects of csrS and srrG mutations in the context of virulence gene expression and in vivo lethality. The regulatory role of csrS/csrR in transcription of virulence genes was analysed by RNA-seq followed by a comparative analysis of mRNA levels between wild-type SDSE80 and SDSE80csrS. In total, 138 transcripts exhibited differential abundance as per the criteria previously described. Transcripts of known putative function with notable differential abundance between the mutant and the wild type included sagA and other associated genes (GGS_0679 through GGS_0687). Additionally, transcripts encoding M protein (stG6792), C5a peptidase (scpA, scpB), streptolysin O (slo), NADase (nga), C3-degrading proteinase (cppA), IL-8 protease like protein (cepA), laminin binding protein (lmb), complement inhibitor protein (sic), immunoglobulin G-binding protein (spg), fibronectin-binding protein (GGS_0161), and collagen adhesion protein (GGS_0158) also demonstrated differential abundance (Appendix 5). This indicates that sagA operon transcription is influenced by mutations in csrS/csrR and srrG.
The csrS mutant, as well as the srrG mutant, are more haemolytic than SDSE80 (Figure 3d). In vivo pathogenic effects of csrS and srrG mutations were compared in mice. Compared to SDSE80 infection, intra-peritoneal SDSE80csrS infection of mice caused earlier and higher mortality (p < 0.01 by a log-rank test), elevated blood bacterial counts (p < 0.05 by z-test), larger lesions (p < 0.05 by z-test) and marked changes in gross presentation. In contrast, there was almost no difference in lethality, blood bacterial counts, lesion size, and gross presentation compared to those seen with SDSE80srrG infection (Figure 5a, b, e, f).
Discussion
SDSE-caused STSS is a serious health concern globally. The present study identified a high frequency of causative negative regulatory mutations in STSS-SDSE clinical isolates in comparison with non-invasive-SDSE clinical isolates in a certain molecular clone, including an srrG mutant affecting multiple organs and enhancing lethality in a mouse model. Here, we reveal mutations in srrG and csrS/csrR, which negatively regulate virulence genes, in STSS-SDSE clinical isolates.
Watanabe et al. (2013) reported that 2.4% (2/83 isolates) of SDSE-invasive isolates carried csrS mutations.16 We examined whether STSS isolates causing more severe invasive infections than strains analysed in the above-mentioned study harboured csrS/csrR mutations; 19.0% of STSS-SDSE isolates carried loss-of-function csrS/csrR mutations (Figure 1a), indicating that they are more prominent in STSS-SDSE isolates than in mildly invasive isolates.
Higher sagA expression in STSS-SDSE isolates than in non-invasive isolates (Appendix 1 and Figure 3b) is mediated by srrG mutations. The loss of srrG function due to these mutations primarily affected the sagA operon (Appendix 5). The expression of the protein related to the secretion and modification of SagA25 was increased, as well as the SagA protein, which functions as a toxin, resulting in severe multi-organ damage in mice (Figure 5b–g). Therefore, SDSE strains with srrG mutations may be reasonably expected to cause severe invasive infections including STSS in humans.
srrG encoded a 178-base sRNA comprising direct repeats and an inverted repeat (Figure 3a). Although it negatively regulates sagA operon expression, no sequence homology was found between srrG and the peripheral region of sagA. Reporter assays revealed that srrG acts in trans (Figure 2b); however, its regulatory mechanism remains unknown and requires further investigation.
Analysis of srrG and csrS/csrR sequences of STSS-SDSE isolates revealed different mutations between strains (Figure 1b, 4b and Appendix 1). This, however, does not signify that STSS is caused by spread of clonal mutants, as indicated by our findings on mutations in different STSS-GAS isolates. In an invasive infection model using GAS, half of the strains in the skin lesions were mutated and all strains in the blood were mutant strains.26 In addition, strains with and without mutations in CsrS survived in lesions, but only strains with mutations in blood, spleen, and liver were confirmed to survive.27 Considering several factors including, a model in which mutation occurs after entering the body, no mutation being found in strains isolated from the pharynx (Figure 4a), significant mutations present in STSS isolates, which were isolates from a sterile site (Figure 4a), the mutation position differing in each strain (Figure 1a, 4a), and the mutant strains showing growth in organs in animal experiments (Figure 4), it is suggested that SDSE may mutate in vivo, spread in the body, and cause severe invasive infections, including STSS, as observed for STSS-GAS isolates.
Some strains with the same mutations show significant differences in the expression of sagA (e.g. NIH960, 11.46; NIH2805, 38.24). The expression of sagA in NIH2805 is similar to that in NIH2182 (46.43), which has mutations in both SrrG and CsrS (Appendix 1). In NIH2805, csrS/csrR is intact, and therefore, it may have another mutation that replaces csrS/csrR. As a result, the expression of sagA may be higher than that in others. Further research is needed to investigate this possibility.
csrS/csrR and srrG mutation frequency in STSS-SDSE isolates (31.6%) was significantly higher than that in non-invasive isolates (0%), suggesting that mutations in regulators of pathogenic genes may play important roles in STSS development. However, a larger proportion of these isolates (68.4%) harboured no csrS/csrR or srrG mutations and may carry mutations in other regulators. As mutations in an invasive infection may occur after entering the body,27 strains with mutation and those without mutations may be mixed 26; not all strains may have mutation after invasion, or a strain without mutations may be selected on isolation. Host factors may also contribute to clinical severity of infections caused by strains without mutations in these three genes. For instance, certain human leukocyte antigen class II haplotypes are associated with the risk of high disease severity in GAS infections,28 and the importance of both host and environmental factors has been previously reported.29 Further, 75% of STSS-SDSE patients in Japan have at least one or more underlying conditions,6 which may also contribute to STSS risk.
sagA is negatively regulated by both CsrS and SrrG16 (Figure 3 and Appendix 1). Humar et al.4 reported that SLS is an important virulence factor for severe invasive infections. We found increased SLS expression in STSS-SDSE isolates carrying csrS and/or srrG mutations, thereby further supporting the role of SLS as an important effector in severe invasive infections caused by SDSE. SLS-type gene clusters have been identified in Clostridium botulinum, Clostridium sporogenes, Staphylococcus aureus, Listeria monocytogenes, Streptococcus iniae, S. anginosus, and S. pyogenes.30, 31, 32, 33 As these pathogens commonly cause invasive infections, the over-expression of toxins, including SLS-like toxins, may contribute to the severity of invasive infections.
In conclusion, the present study identified csrS/csrR and srrG mutations that cause elevated expression of virulence genes in STSS-SDSE isolates. The significantly higher frequency of mutations in STSS-SDSE isolates and the enhanced lethality associated with these mutations highlight their importance in STSS onset and pathogenesis.
Contributors
TI, MO, and YA wrote the manuscript. TI, MO, and YA planned and supervised the study as well as verified underlying data. TI, HO, KC, YK, TY, RO, YD, MS, JI, MO, and YA performed the experiments and analysed data. TI, HO, KC, YK, TY, RO, YD, MS, and JI either contributed to the data or characterised samples. All authors interpreted the results, had complete access to the study data, critically reviewed all drafts of the manuscript, approved this final version of the manuscript, and bear final responsibility for the decision to submit it for publication.
Data sharing statement
Raw bacterial load data of the tested samples can be obtained from the corresponding author upon request.
Declaration of interests
We declare no competing interests.
Acknowledgments
This work was supported by the Ministry of Health, Labor, and Welfare of Japan [grant number 19HA1001], a Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science (JSPS) [grant number 21K07034] and the Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development (AMED) [grant number JP22fk0108130].
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104133.
Appendix. Supplementary materials
References
- 1.Gaviria JM, Bisno AL., Group C, streptococci G. Oxford University Press; New York: 2000. Streptococcal Infections: Clinical Aspects, Microbiology, and Molecular Pathogenesis; pp. 238–254. In D. L. Stevens and E. L. Kaplan (eds.) [Google Scholar]
- 2.Brandt CM, Spellerberg B. Human infections due to Streptococcus dysgalactiae subspecies Equisimilis. Clin Infect Dis. 2009;49:766–772. doi: 10.1086/605085. [DOI] [PubMed] [Google Scholar]
- 3.Loubinoux J, Plainvert C, Collobert G, et al. Adult invasive and noninvasive infections due to Streptococcus dysgalactiae subsp. Equisimilis in France from 2006 to 2010. J Clin Microbiol. 2013;51:2724–2727. doi: 10.1128/JCM.01262-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humar D, Datta V, Bast DJ, Beall B, De Azavedo JC, Nizet V. Streptolysin S, necrotizing infections produced by group G streptococcus. Lancet. 2002;359:124–129. doi: 10.1016/S0140-6736(02)07371-3. [DOI] [PubMed] [Google Scholar]
- 5.Wagner JG, Schlievert PM, Assimacopoulos AP, Stoehr JA, Carson PJ, Komadina K. Acute group G streptococcal myositis associated with streptococcal toxic shock syndrome: case report and review. Clin Infect Dis. 1996;23:1159–1161. doi: 10.1093/clinids/23.5.1159. [DOI] [PubMed] [Google Scholar]
- 6.Ikebe T, Murayama S, Saitoh K, et al. Surveillance of severe invasive group G streptococci infections and molecular typing of isolates in Japan. Epidemiol Infect. 2004;132:145–149. doi: 10.1017/s0950268803001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashikawa S, Iinuma Y, Furushita M, et al. Characterization of group C and G streptococcal strains that cause streptococcal toxic shock syndrome. J Clin Microbiol. 2004;42:186–192. doi: 10.1128/JCM.42.1.186-192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikebe T, Oguro Y, Ogata K, et al. Surveillance of severe invasive group G streptococcal infections during 2002–2008 in Japan. Jpn J Infect Dis. 2010;63:372–375. [PubMed] [Google Scholar]
- 9.Ikebe T, Okuno R, Sasaki M, et al. Molecular characterization and antibiotic resistance of Streptococcus dysgalactiae subspecies equisimilis isolated from patients with streptococcal toxic shock syndrome. J Infect Chemother. 2018;24:117–122. doi: 10.1016/j.jiac.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Broyles LN, Beneden CV, Beall B, et al. Population-based study of invasive disease due to beta-hemolytic streptococci of groups other than A and B. Clin Infect Dis. 2009;48:706–712. doi: 10.1086/597035. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi T, Sunaoshi K, Sunakawa K, et al. Clinical aspects of invasive infections with Streptococcus dysgalactiae ssp. equisimilis in Japan: differences with respect to Streptococcus pyogenes and Streptococcus agalactiae infections. Clin Microbiol Infect. 2010;16:1097–1103. doi: 10.1111/j.1469-0691.2009.03047.x. [DOI] [PubMed] [Google Scholar]
- 12.Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–958. doi: 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikebe T, Ato M, Matsumura T, et al. Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikebe T, Matsumura T, Nihonmatsu H, et al. Spontaneous mutations in Streptococcus pyogenes isolates from patients with streptococcal toxic shock syndrome play a role in virulence. Sci Rep. 2016;6:28761. doi: 10.1038/srep28761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimomura Y, Okumura K, Murayama SY, et al. Complete genome sequencing and analysis of a Lancefield group G Streptococcus dysgalactiae subsp. equisimilis strain causing streptococcal toxic shock syndrome (STSS) BMC Genomics. 2011;12:17. doi: 10.1186/1471-2164-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe S, Shimomura Y, Ubukata K, Kirikae T, Miyoshi-Akiyama T. Concomitant regulation of host tissue-destroying virulence factors and carbohydrate metabolism during invasive diseases induced by group G streptococci. J Infect Dis. 2013;208(9):1482–1493. doi: 10.1093/infdis/jit353. [DOI] [PubMed] [Google Scholar]
- 17.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of streptococci reveals a mutation that modulates the global phenotype and disease specificity. PLoS Pathog. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Working Group for Severe Streptococcal Infections Defining group A streptococcal toxic shock syndrome: rationale and consensus definition. JAMA. 1993;269:390–391. [PubMed] [Google Scholar]
- 19.Little J, Higgins JP, Ioannidis JP, et al. Strengthening the reporting of genetic association studies (STREGA) – an extension of the STROBE statement. Genet Epidemiol. 2009;33:581–598. doi: 10.1002/gepi.20410. [DOI] [PubMed] [Google Scholar]
- 20.Alexander B, Parsonage GD, Ross RP, Caparon MG. The RofA binding site in Streptococcus pyogenes is utilised in multiple transcriptional pathways. J Bacteriol. 2000;182:1529–1540. doi: 10.1128/jb.182.6.1529-1540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene. 1991;102:99–104. doi: 10.1016/0378-1119(91)90546-n. [DOI] [PubMed] [Google Scholar]
- 22.Takamatsu D, Osaki M, Sekizaki T. Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid. 2001;46:140–148. doi: 10.1006/plas.2001.1532. [DOI] [PubMed] [Google Scholar]
- 23.Ikebe T, Endoh M, Watanabe H. Increased expression of the ska gene in emm49-genotyped Streptococcus pyogenes strains isolated from patients with severe invasive streptococcal infections. Jpn J Infect Dis. 2005;58:272–275. [PubMed] [Google Scholar]
- 24.Hasegawa T, Matsumoto M, Hata N, Yano H, Isaka M, Tatsuno I. Homologous role of CovRS two-component regulatory system in NAD+ -glycohydrolase activity in Streptococcus dysgalactiae subsp. equisimilis as in Streptococcus pyogenes. APMIS. 2019;127:87–92. doi: 10.1111/apm.12914. [DOI] [PubMed] [Google Scholar]
- 25.Nizet V. Streptococcal beta-hemolysins: genetics and role in disease pathogenesis. Trends Microbiol. 2002;10:575–580. doi: 10.1016/s0966-842x(02)02473-3. [DOI] [PubMed] [Google Scholar]
- 26.Cole JN, McArthur JD, McKay FC, et al. Trigger for group A streptococcal M1T1 invasive disease. FASEB J. 2006;20:1745–1747. doi: 10.1096/fj.06-5804fje. [DOI] [PubMed] [Google Scholar]
- 27.Walker MJ, Hollands A, Sanderson-Smith ML, et al. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med. 2007;13:981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- 28.Kotb M, Norrby-Teglund A, McGeer A, et al. Immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat Med. 2002;8:1398–1404. doi: 10.1038/nm1202-800. [DOI] [PubMed] [Google Scholar]
- 29.Factor SH, Levine OS, Schwartz B, et al. Invasive group A streptococcal disease: risk factors for adults. Emerg Infect Dis. 2003;9:970–977. doi: 10.3201/eid0908.020745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalez DJ, Lee SW, Hensler ME, et al. Clostridiolysin S, a post-translationally modified biotoxin from Clostridium botulinum. J Biol Chem. 2010;285:28220–28228. doi: 10.1074/jbc.M110.118554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cotter PD, Draper LA, Lawton EM, et al. Listeriolysin S, a novel peptide hemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SW, Mitchell DA, Markley AL, et al. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc Natl Acad Sci USA. 2008;105:5879–5884. doi: 10.1073/pnas.0801338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asam D, Mauerer S, Walheim E, Spellerberg B. Identification of β-haemolysin-encoding genes in Streptococcus anginosus. Mol Oral Microbiol. 2013;28:302–315. doi: 10.1111/omi.12026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.