Abstract
Analogues and derivatives of natural nucleosides/nucleotides are considered among the most successful bioactive species of drug-like compounds in modern medicinal chemistry, as they are well recognized for their diverse and efficient pharmacological activities in humans, especially as antivirals and antitumors. Coronavirus disease 2019 (COVID-19) is still almost incurable, with its infectious viral microbe, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), continuing to wreak devastation around the world. This global crisis pushed all involved scientists, including drug discoverers and clinical researchers, to try to find an effective and broad-spectrum anti-COVID-19 drug. Didanosine (2′,3′-dideoxyinosine, DDI) is a synthetic inosine/adenosine/guanosine analogue and highly active antiretroviral therapeutic agent used for the treatment of human immunodeficiency virus infection and acquired immunodeficiency syndrome (HIV/AIDS). This potent reverse-transcriptase inhibitor is characterized by proven strong pharmacological effects against the viral genome, which may successfully take part in the effective treatment of SARS-CoV-2/COVID-19. Additionally, targeting the pivotal SARS-CoV-2 replication enzyme, RNA-dependent RNA polymerase (RdRp), is a very successful tactic to combat COVID-19 irrespective of the SARS-CoV-2 variant type because RdRps are broadly conserved among all SARS-CoV-2 strains. Herein, the current study proved for the first time, using the in vitro antiviral evaluation, that DDI is capable of potently inhibiting the replication of the novel virulent progenies of SARS-CoV-2 with quite tiny in vitro anti-SARS-CoV-2 and anti-RdRp EC50 values of around 3.1 and 0.19 μM, respectively, surpassing remdesivir together with its active metabolite (GS-441524). Thereafter, the in silico computational interpretation of the biological results supported that DDI strongly targets the key pocket of the SARS-CoV-2 RdRp main catalytic active site. The ideal pharmacophoric characteristics of the ligand DDI make it a typical inhibiting agent of SARS-CoV-2 multiplication processes (including high-fidelity proofreading), with its elastic structure open for many kinds of derivatization. In brief, the present results further uphold and propose the repurposing potentials of DDI against the different types of COVID-19 and convincingly motivate us to quickly launch its extensive preclinical/clinical pharmacological evaluations, hoping to combine it in the COVID-19 therapeutic protocols soon.
1. Introduction
After more than 26 months, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) still remains a major global threat and concerns to humans since the first emergence of the virus in Wuhan, China.1 The dangerous disease that results from this virus, coronavirus disease 2019 (COVID-19), is still rapidly ongoing everywhere in the world with officially affirmed infections and deaths reaching more than 444 and 6 million, respectively, as a result of this worldwide pandemic.2 The growing evolution of very spreadable and resistant novel lineages/variants of the COVID-19 virus, especially in 2021, has created the need for the scientific and health communities in all the nations to search for efficient medicines/vaccines that will be successful in resisting and inhibiting this irritating virus, together with finding drugs that possess the capacities to abolish or neutralize all, or most of, the serious to very serious effects of COVID-19 on human bodies; thus finding effective and comprehensive (e.g., dual-action) anti-SARS-CoV-2/anti-COVID-19 medicinal agents will be quite advantageous for this currently resistant-to-treatment infection and all or most of its health sequelae.3 Several new and repurposed natural and/or synthetic compounds are, for the time being, under broad international and multinational investigations (including preclinical studies and human clinical trials) in order to be pharmacologically evaluated as effective anti-COVID-19 drugs (e.g., nirmatrelvir, molnupiravir, remdesivir and its active metabolites GS-441524/GS-443902, cordycepin, favipiravir and its active derivative cyanorona-20, hydroxychloroquine and its brother chloroquine, CoViTris2020 and its 1,3,4-oxadiazole family members taroxaz-104/ChloViD2020, teriflunomide and its prodrug leflunomide, ivermectin, umifenovir (also known as Arbidol), and colchicine), but most of them did not prove successful broad-spectrum effectiveness in the long run until now (i.e., the end outcomes of many of these global investigations are not declared to date).4−23
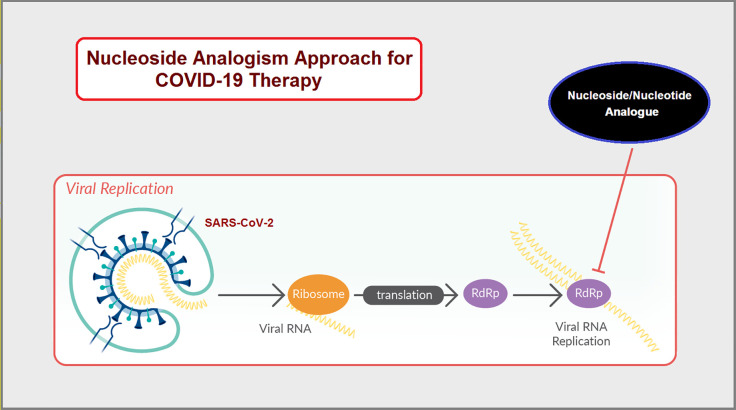
Tactical nucleos(t)ide analogism is certainly among the favorable therapeutic choices in drug molecule design for pharmaceutical chemists to stop coronavirus multiplication inside the tissues of the human body.6−12,24 In this anti-COVID-19 therapeutic tactic, the used nucleoside/nucleotide analogue makes use of its close similarity with the normal natural nucleos(t)ides to misguide and deceive the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp is the nonstructural protein complex 12/7/8 “nsp12/nsp7/nsp8 or simply nsp12/7/8”; nsp12 is the polymerase that binds to the two major proteinous cofactors, nsp7 and nsp8; it is a very important enzyme in the replication as well as transcription of the coronavirus genome, and as a consequence, the strong inhibition/hindrance of the performance of this enzyme will severely deteriorate SARS-CoV-2 replication) through incorporation and combination mainly in the growing viral genetic strands instead of the real/correct naturally occurring nucleos(t)ides, resulting in reduplicated excess, inappropriate, and ambiguous coding along with premature termination of mRNA synthesis and, at the end, formation of vague RNA strands; these pseudostrands make abnormal noninfectious and inactive (ineffective) viral particles, hence no further replication and reproduction of the virus occurs (Figure 1).24 Some of the previously mentioned used and experimental anti-COVID-19 medicines/compounds, such as molnupiravir, remdesivir, and cyanorona-20, as well as their active metabolites, β-d-N4-hydroxycytidine (NHC), GS-441524, and favipiravir (Figure 2), count on this smart mechanism in their effective inhibitory activities against SARS-CoV-2.5−8,10−12 One of them, the synthetic drug molnupiravir, which is a prodrug of the synthetic active nucleoside derivative NHC, is now finally approved for medical use in mild-to-moderate (MtoM) COVID-19 cases in some countries.5 With the progressive advent of more malicious/resistant new strains of SARS-CoV-2, searching for more potent and broad-spectrum synthetic anti-COVID-19 drugs became a must.6−12
Figure 1.
Illustrative exemplification of the nucleoside/nucleotide analogism maneuver tactic employed for the strong inhibition of SARS-CoV-2 replication.
Figure 2.
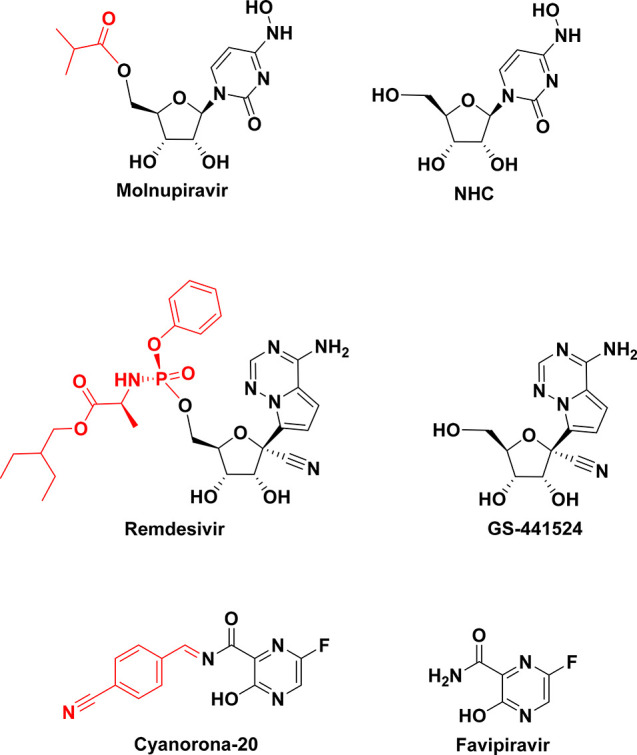
Chemical structures of the three pairs of anti-COVID-19 compounds: molnupiravir/NHC, remdesivir/GS-441524, and cyanorona-20/favipiravir.
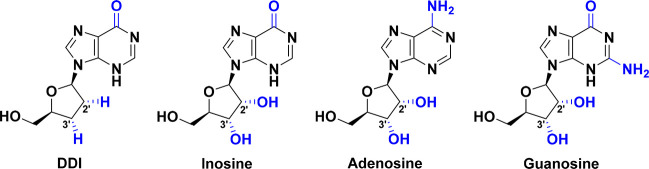
Didanosine (DDI) is a synthetic dideoxynucleoside compound which is a purine 2′,3′-dideoxyribonucleoside.25 Chemically, DDI is 2′,3′-dideoxyinosine (2,3-ddI), which is named per IUPAC as 9-[(2R,5S)-5-(hydroxymethyl)oxolan-2-yl]-3H-purin-6-one (Figure 3).25 The DDI molecule is present in either of two isomeric forms of the keto tautomer via intramolecular conversion in aqueous media (Figure 4).25,26 This synthetic nucleoside analogue can be seen as an inosine analogue (in which the two hydroxyl groups at the 2′ and 3′ positions on the ribose sugar moiety are replaced by two hydrogens, i.e., in which its β-d-ribofuranosyl ring is 2′,3′-dideoxygenated), adenosine analogue (in which the two hydroxyl groups at the 2′ and 3′ positions on the ribose sugar moiety are replaced by two hydrogens, along with conversion of the adenine base to hypoxanthine base), or guanosine analogue (in which the two hydroxyl groups at the 2′ and 3′ positions on the ribose sugar moiety are replaced by two hydrogens, along with conversion of the guanine base to hypoxanthine base) (Figure 3). DDI was first recognized as the major active metabolite of its prodrug, 2′,3′-dideoxyadenosine (DDA), and is responsible for most of the potent bioactivities of this prodrug.27,28 Being a purine nucleoside analogue, antimetabolite, and reverse transcriptase inhibitor, DDI is used as an antiviral agent combined with other agents in the treatment of human immunodeficiency virus (HIV) infection and its fatal disease, acquired immunodeficiency syndrome (AIDS).27−29 DDI is very effective against HIV type 1 (HIV-1) because it is specifically a potent HIV-1 reverse transcriptase inhibitor.27−29 The absence of a hydroxyl determinant at the 3′ position of the ribose moiety in the DDI molecule prevents and blocks the formation of the vital phosphodiester linkages that are essentially required for the completion of nucleic acid chains in DNA and RNA.9,30 Additionally, it was recently proven and reported that reduction (i.e., absence of hydroxyl group(s) and replacement of it/them by mainly hydrogens) at the 3′ carbon or both the 2′ and 3′ carbons of the ribose moiety of any designed nucleoside analogue is necessary for a strong anti-SARS-CoV-2 effect.9,24,30,31 DDI is primarily a potent inhibitory agent of HIV replication, serving as an efficient chain terminator of viral DNA by binding to reverse transcriptase.27−29 One of the major metabolic pathways of DDI inside the human body is its intracellular phosphorylation (mainly after amination) to another active nucleoside analogue, DDA triphosphate (DDA-TP), which is supposed to be one of the main active metabolites of DDI.32
Figure 3.
Chemical structures of DDI, inosine, adenosine, and guanosine.
Figure 4.
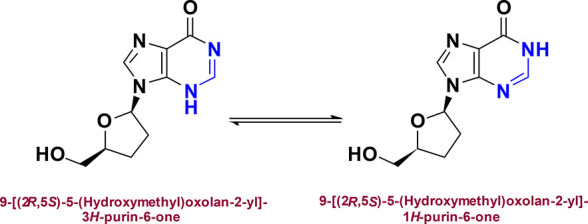
Isomeric structures of the predominant keto tautomer of DDI in aqueous media.
Interestingly, the DDI molecule has the ideal characteristics to become a potentially successful anti-SARS-CoV-2 nucleoside analogue, and it even surpasses almost all of the investigational and approved natural and synthetic nucleoside analogues in some needed anticoronaviral and anti-COVID-19 properties, therefore a strong rationale for DDI repurposing against COVID-19 infections can be established in the following 20 points.25−40 First, DDI has smaller molecular weight (236.23 Da) and volume (199.28 Å3), relative to most of the other nucleosides and nucleoside analogues, which are very favorable from the biological and pharmacokinetic points of view. Second, the relative similarity in chemical structure and molecular size of DDI to natural purine nucleos(t)ides gives its constructure significant ability to reasonably fit within the known active site pocket of the SARS-CoV-2 RdRp. Third, the absence of a 3′-hydroxyl group in the DDI molecule will result in forced termination (i.e., impairment) of the RdRp critical catalytic reactions in SARS-CoV-2 replication/transcription processes. Fourth, the previous third point will result also in mutations of the very large coronaviral genomes that encode the high-fidelity 3′–5′ exonuclease enzyme (SARS-CoV-2 proofreading 3′-to-5′ exoribonuclease (ExoN) or nonstructural protein 14 (nsp14)) involved and needed in SARS-CoV-2 genomic proofreading processes (the activity of nsp14 is enhanced by its activator the cofactor nonstructural protein 10 or nsp10); the correct proofreading function is very important for the increase of replication fidelity by removing mismatched nucleotides, thus the produced mutated/disabled nsp14 (if some are newly formed), i.e., nsp14-like protein, will have disrupted roles, leading to a decrease in replication fidelity of the coronaviral genome. Fifth, on the other hand, the deficiency of a 2′-hydroxyl group in the DDI molecule (the offending nucleoside/nucleotide analogue) may disable the activity of the already-present coronaviral nsp14/10 complex, which normally requires a 2′-hydroxyl group at the 3′ end of the growing RNA strand, and this will mostly lead to premature chain termination and continuous disability of the exonuclease to excise and remove the offending nucleotide analogue. Sixth, DDI has previously been shown to block the polymerases of some other resistant RNA viruses (such as HIV-1), even those with different polymerase types (e.g., reverse transcriptase), thus it has a significant potential to also inhibit the SARS-CoV-2 RdRp. Seventh, the DDI molecule displays inhibitory affinity and selectivity for SARS-CoV-2 RdRp significantly higher than that for human cellular DNA and RNA polymerases (the findings and facts that are reported and proven in the current research study). Eighth, the spatial configuration of the DDI molecule is typically the most convenient conformational form required for molecular positioning inside the key cavity of the SARS-CoV-2 RdRp main active site and interacting with the amino acid residues of this cavity (this fact is also proven in the current research study). Ninth, the structural analogism of DDI with the natural endogenous nucleosides adenosine/inosine/guanosine makes the human biological system incapable of identifying and distinguishing this molecule; that is, various enzymes significantly fail to discriminate it from the endogenous adenosine/inosine/guanosine; through this disguise tactic, given DDI can be efficiently engaged in inhibiting a variety of specific biochemical pathways/reactions that contribute to the continuation of SARS-CoV-2 multiplication and infection, for example, it may give rise to potent poly(A) polymerase blockade (i.e., strong polyadenylation inhibition), intense shortening of poly(A) tails, persistent destabilization of mRNAs, potent adenine/guanine biosynthesis impairment, and also premature terminus of protein synthesis. Tenth, DDI is an adenosine kinase (ADK) inhibitor which potently interferes with the ADK activity (ADK is the main regulatory enzyme of adenosine biosynthesis; e.g., ADK phosphorylates cytokinin nucleosides to keep a sufficient pool of bioactive cytokinins through this nucleoside–nucleotide interconversion, and cytokinin availability, in turn, significantly increases the human cells susceptibility to the coronaviral infection). This action will greatly increase the probable anti-SARS-CoV-2 activity of DDI. Eleventh, DDI has a potential interleukin 2 (IL-2) receptor alpha (IL-2Rα; IL-2 is one of the major immunogenic/inflammatory mediators responsible for the severe immunogenic cytokine storm and extensive inflammation of the COVID-19 status) antagonistic effect, and this action will greatly increase the expected comprehensive anti-COVID-19 activities of DDI. Twelfth, DDI is computationally predicted to be a SARS-CoV-2 helicase (nonstructural protein 13 or nsp13) inhibitor (the coronaviral helicase is a vital replication enzyme that is mainly responsible for catalyzing the unwinding of duplex oligonucleotides into single strands in a nucleoside-5′-triphosphate (NTP)-dependent modality, thus inhibiting this enzyme will also inhibit SARS-CoV-2 multiplication). Thirteenth, DDI is expected to be a very beneficial medication for the acute lung fibrosis caused by COVID-19 infection (by matching single-cell RNA sequencing data in a bioinformatics-based technique). Fourteenth, the pathogenesis molecular mechanisms used by the two viruses HIV-1 and SARS-CoV-2 are significantly similar, increasing the potential that DDI may also succeed in clinically inhibiting SARS-CoV-2. Fifteenth, DDI and its nucleotidic triphosphate form (DDI-TP) are already FDA-approved drugs known to have relatively reasonable levels of toxicity and are more likely to be well tolerated by COVID-19 patients. Sixteenth, the structural similarity with three major cellular nucleosides, inosine, adenosine, and guanosine (respectively), not one, renders DDI bioacting on most adenosine receptors and quite analogous to these important nucleosides in most of their bioactivities that may engage in comprehensive anti-COVID-19 treatment in humans. Seventeenth, the structural analogism to adenosine/inosine/guanosine molecules gives strong triple camouflage abilities to the DDI molecule, rendering it capable of inhibiting and impairing some of the bioactions of the adenosine/inosine/guanosine molecules that may aggravate the COVID-19 status. Eighteenth, DDI is an ideal drug-like molecule as it completely complies with Lipinski’s rule of five (Ro5) without any violations. Nineteenth, DDI is a biologically compatible molecule with better abilities to efficiently pass through the biomembranes in comparison to adenosine, inosine, and guanosine molecules due to the comparatively stronger lipophilic characteristics that result from a lack of the two hydrophilic hydroxyl groups directly attached to the ribose moiety (i.e., the DDI molecule is less hydrophilic than adenosine, inosine, and guanosine molecules). Twentieth, DDI has highly balanced lipophilic/hydrophilic properties, as it has a moderate log P value of −0.95.
Most of the aforementioned diverse points about DDI are extremely needed in the comprehensive battle against SARS-CoV-2 infection (i.e., in the rational design of an effective multitarget anticoronaviral agent). This makes DDI a very promising candidate as an anti-COVID-19 drug with multiple mechanisms of action. Using only hypothetical bioinformatics and in silico computational approaches, few recently published theoretical studies shed light on the possible use of DDI against COVID-19.35,37,38 Herein, in this new study, we report for the first time that DDI can successfully inhibit the coronaviral multiplication (i.e., can act as an effective SARS-CoV-2 replication inhibitor) by considerably lowering the number of SARS-CoV-2 copies reproduced, which is proven to be caused by principally blocking the genomic RNA biosynthesis mediated mainly via the SARA-CoV-2 RdRp, using the strategy of nucleoside similarity (as formerly explained). In this efficient mechanism of anti-RdRp action, the nucleoside-like DDI molecule is first easily phosphorylated to its mono-, di-, and triphosphate ester forms (i.e., its endogenous nucleotide analogues) intracellularly, and then the highly active nucleotide analogue DDI-TP could be readily incorporated into RNA instead of the naturally occurring chemicosimilar purine ribonucleotides, adenosine triphosphate (ATP) and guanosine triphosphate (GTP); this consequently inhibits and closes transcription elongation and creation of coronaviral RNA strands in all steps (i.e., acts as an RNA elongation inhibitor because of the hydroxyl moiety deficiency at the 3′ position of the molecule, and this one-hydroxyl group absence greatly antagonizes and blocks the SARS-CoV-2 RdRp action through the prevention of phosphodiester linkage formation, which is required for proper completion of nucleic acid chains as formerly demonstrated), affording imperfect impaired premature RNAs in the growing viral mRNA strands and genomes, and eventually, this vague coding leads to substantial inhibition of SARS-CoV-2 different copying (replication/transcription) processes and creation of inefficient and nonviral (i.e., non-SARS-CoV-2) particles instead of the active and pernicious SARS-CoV-2 particles (Figure 5). This current work preclinically evaluated and proved the potential potent anti-SARS-CoV-2/anti-COVID-19 activities of DDI based on two validated in vitro bioassays, anti-SARS-CoV-2 assay and anti-RdRp assay (along with an in silico molecular docking interpretation of the biological evaluation). Considering all of the preceding encouraging literature data along with the very promising biological evaluation outcomes of the present research, DDI can be repositioned to in vivo studies to assess its defensive and inhibiting activities specifically on SARS-CoV-2 particle infestation (as an anti-RNA-virus agent) in mammals and to clinical trials to assess its preventative and inhibiting activities comprehensively on COVID-19 status progression as a whole (as an anti-COVD-19-condition agent) in COVID-19 human patients.
Figure 5.
Representation of the presently proven mode of strong anti-SARS-CoV-2 action of DDI (anti-RdRp activities).
2. Results and Discussion
2.1. Evaluation of the In Vitro Anti-SARS-CoV-2 and Cytotoxic Bioactivities of DDI
Table 1 displays the obtained data from both the experimental in vitro anti-SARS-CoV-2 and cytotoxicity bioassays in detail. The used SARS-CoV-2 variant in the anticoronaviral bioassay is the new strain VOC-202012/01, which is one of the most virulent/resistant strains of the virus. The demonstrated data interestingly showed the considerably greater antiviral effectiveness of DDI on the recently appearing variants and lineages of SARS-CoV-2 compared to that of each one of the two reference (positive control) drugs remdesivir and GS-441524 (please note that the placebo drug, the solvent DMSO, showed extremely negligible activities). DDI was found to clearly impair and inhibit the entire SARS-CoV-2 replication/transcription in the used Vero E6 cells with an EC50 value significantly smaller than that of the 100 μM stock concentration. Importantly, DDI (EC50 = 3.1 μM) was found to be around 6.8 and 5 times as potent as the reference drugs remdesivir (EC50 = 21 μM) and GS-441524 (EC50 = 15.60 μM), respectively, in relation to the tested in vitro anti-VOC-202012/01/anti-SARS-CoV-2 activity. In accordance with the cytotoxicity evaluation test, in vitro CC50 of DDI is considerably greater than 100 μM, thus this nucleoside analogue is assumed to have very favorable high clinical selectivity index (SI) (SIDDI > 32.26, whereas remdesivir and GS-441524 references have narrower SIs, SIremdesivir > 4.76 and SIGS-441524 > 6.41), indicating the specific/selective anti-RNA capabilities (RNA-disrupting activities) of the DDI molecule on the new SARS-CoV-2 genomic RNA rather than the known human genome. DDI displayed a significantly low value of the concentration that causes 100% in vitro suppression of the viral VOC-202012/01 cytopathic effects (CPEIC100 = 8.95 μM), which is smaller than the corresponding values of both remdesivir (CPEIC100 = 25.17 μM) and GS-441524 (CPEIC100 = 17.40 μM). In harmony with its potent activity against the infectious coronaviral VOC-202012/01 strain, DDI also exhibited very small required concentration for 50% in vitro decrease in the number of coronaviral RNA copies of the VOC-202012/01 strain (3.47 μM), which is comparatively smaller than the respective values of the two agents remdesivir and GS-441524 (22.92 and 16.04 μM, respectively). The EC90 value for DDI, which is preferable for in vivo/clinical studies, was also small and consistent with the EC50 values (being not that far from the EC50 values indicates the significant potency of DDI), as seen in Table 1.
Table 1. Anti-SARS-CoV-2 (Anti-COVID-19) Activities and Cytotoxicity of the Target Repurposed Drug DDI (Using the Two Reference Agents Remdesivir and GS-441524 as the Positive Control Drugs and the Placebo Solvent DMSO as the Negative Control Drug) against SARS-CoV-2 VOC-202012/01 Strain (in Vero E6 Cells).
| suppression
of SARS-CoV-2 replication in
vitro (anti-VOC-202012/01 bioactivities) (μM) |
||||||
|---|---|---|---|---|---|---|
| categorization | compound | CC50a (μM) | 100% CPE inhibitory concentration (CPEIC100)b | 50% decrease in infectious virus (EC50)c | 50% decrease in viral RNA copy (EC50)d | 90% decrease in infectious virus (EC90)e |
| repurposed medication | DDI | >100 | 8.95 ± 0.52 | 3.10 ± 0.14 | 3.47 ± 0.15 | 17.80 ± 0.69 |
| reference drugs | remdesivir | >100 | 25.17 ± 2.51 | 21.00 ± 1.97 | 22.92 ± 1.99 | >100 |
| GS-441524 | >100 | 17.40 ± 1.83 | 15.60 ± 0.76 | 16.04 ± 0.81 | 93.36 ± 4.70 | |
| placebo solvent | DMSO | >100 | >100 | >100 | >100 | >100 |
CC50 or 50% cytotoxic concentration is the concentration of the assayed compound which kills half of the cells in an uninfected cell culture. CC50 was estimated with sequentially diluted compounds in Vero E6 cells at 48 h postincubation utilizing CellTiter-Glo luminescent cell viability assay (Promega).
CPEIC100 or 100% CPE inhibitory concentration is the least concentration of the assayed compound which causes 100% inhibition of the cytopathic effects (CPE) of SARS-CoV-2 VOC-202012/01 virus in Vero E6 cells under increasing concentrations of the assayed compound at 48 h postinfection. Compounds were sequentially diluted from 100 μM concentration.
EC50 or 50% effective concentration is the concentration of the assayed compound which is needed for 50% decrease in infectious SARS-CoV-2 VOC-202012/01 virus particles in vitro. EC50 is estimated by infectious virus yield in culture supernatant at 48 h postinfection (log10 TCID50/mL).
EC50 or 50% effective concentration is the concentration of the assayed compound which is needed for 50% decrease in SARS-CoV-2 VOC-202012/01 viral RNA copies in vitro. EC50 is estimated by viral RNA copies number in culture supernatant at 48 h postinfection (log10 RNA copies/mL).
EC90 or 90% effective concentration is the concentration of the assayed compound which is needed for 90% decrease in infectious SARS-CoV-2 VOC-202012/01 virus particles in vitro. EC90 is estimated by infectious virus yield in culture supernatant at 48 h postinfection (log10 TCID90/mL).
It was unexpectedly noted that DDI inhibits the coronaviral particles in a mixed mode, rapid-onset mode then time-dependent mode, as it runs to its maximal effectiveness on the coronavirus within 12–24 h of starting treatment. Although DDI has a short plasma half-life which does not exceed 2 h (DDI has peak plasma concentrations that appear at 0.5–1.5 h due to rapid absorption and high bioavailability), but it has a much longer intracellular life duration (this is mainly because its net anti-SARS-CoV-2 activities do not just rely on its own molecule but further on almost all of its active nucleos(t)idic metabolites which reside inside the human cells in considerable concentrations for longer periods of time than their parent DDI molecule).25 This observation reflects the expected double clinical anti-SARS-CoV-2 action of DDI, which comprises both the rapid effect (first action) and the sustained effect (i.e., prolonged anticoronaviral activities, second action) against COVID-19, which will be very advantageous and required in almost all patients with COVID-19 (because it will cover the therapeutic regimens and protocols of the diverse types of COVID-19 cases). As previously mentioned, a major portion of DDI molecules would be biometabolized through in vivo metabolic phosphorylation to another active form, DDI-TP (one of its nucleotidic forms). This sort of cellular metabolic transformation, specifically, would not impede the potential anti-COVID-19 actions of DDI and would not result in any therapeutic problem for DDI clinical usage, as it is a beneficial chemical metabolism which converts the nucleoside analogue to the more biocompatible form, the nucleotide analogue (the more needed form), with no hydroxylation of the 2′ and 3′ positions of the DDI molecule (as formerly pointed out, the nucleotide analogue DDI-TP could be actively merged into the coronaviral RNA and, accordingly, inhibit or stop the transcription elongation and RNA synthesis and impair mRNA translation due to the lack of essential hydroxyl groups at the 2′/3′ carbons of the ribose moiety). Again, the metabolic resemblance with the biosimilar nucleoside (inosine, adenosine, and guanosine) molecules significantly helps the DDI molecule to deceive the biological system of humans and perform its intentional therapeutic roles in COVID-19 treatment. The current results of this bioassay are in excellent agreement with almost all of the proposed points of the study rationale previously presented and discussed in section 1.
2.2. Evaluation of the In Vitro Anti-RdRp Bioactivity of DDI
This robust cell-based test, the in vitro anti-SARS-CoV-2 RdRp bioassay, was recently developed using Gaussia luciferase (Gluc) as the reporter to determine the anticoronaviral RdRp activity of mainly nucleoside analogues (nucleotide prodrugs) with no need for synthesizing the active nucleotidic triphosphate forms of the nucleoside analogues (or of the other non-triphosphorylated nucleotidic analogues) as for the cell-free assays.41 Additionally, it was confirmed beyond doubt, through the findings of this new assay, that the exonuclease activity of SARS-CoV-2 nsp14 significantly enhances the SARS-CoV-2 RdRp resistance to the inhibitors/blockers of the nucleos(t)ide analogue class (one of the main factors that increases resistance and severe pathogenicity of SARS-CoV-2 is its ability to encode the nsp14 exoribonuclease, which is capable of excising mistaken mutagenic nucleotides misincorporated by the low-fidelity nsp12 into the growing viral RNA strands, causing resistance to nucleos(t)ide analogue therapeutics, i.e., leading to little anticoronaviral activities of most of the existing nucleos(t)ide analogue remedies), thus nsp14 was included and fixed in the protocol of this screening assay for candidate anti-SARS-CoV-2 RdRp agents (unlike the classic analytical cell-free assay).41−43
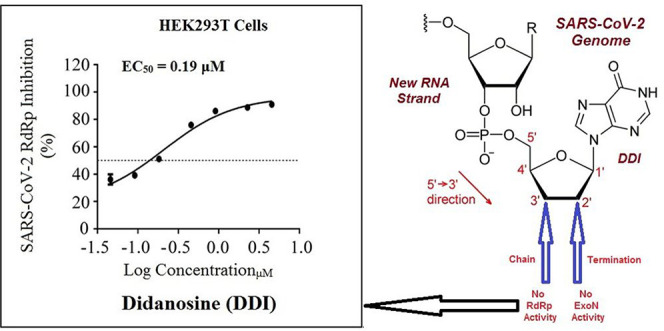
Table 2 presents the detailed resulting values from the current in vitro anti-SARS-CoV-2 RdRp bioassay. Herein, we focus mainly on the two major protein complexes that control the SARS-CoV-2 replication processes, nsp12/7/8 polymerase complex and nsp14/10 exoribonuclease complex. This assay greatly simulates the corresponding natural replication processes that occur for the SARS-CoV-2 particles, as it functionally mimics the RNAs synthetic processes driven by the in vivo SARS-CoV-2 RdRp.44 The obtained data demonstrated that DDI successfully suppressed SARS-CoV-2 RdRp activity with a very small promising EC50 value of 0.19 μM (Figure 6), which is slightly increased in the presence of SARS-CoV-2 exoribonuclease (the wild-type) to about 0.31 μM. Mutations in the exoribonuclease (i.e., the mutated type; e.g., D90A/E92A mutations of the active catalytic residues in nsp14 as in our current case) reinforced the anti-RdRp activity of DDI to an excellent EC50 value of 0.24 μM (i.e., lower than that obtained in the presence of the normal wild-type). The two potent reference agents, remdesivir and GS-441524, showed significantly higher values, revealing the obvious superiority of DDI. It is clearly observed from the values in Table 2 that as much as the EC50 values of the nucleoside analogue against the polymerase alone and against the polymerase in the presence of the exoribonuclease are close to each other, the more potent this nucleoside analogue inhibitor is (i.e., as more expected for this investigated nucleoside analogue to be an ideally effective RdRp inhibitor). From the results, we can also conclude that an ideal potent SARS-CoV-2 RdRp inhibitor should have a ratio of EC50(polymerase+exoribonuclease)/EC50(polymerase) that is very close to 1 and less than 2. As this ratio decreases, the compound has greater potential to succeed at inhibiting the SARS-CoV-2 replication more perfectly. DDI presented the highest resistance, among the tested compounds, to the coronaviral exoribonuclease activity in HEK293T cells. The promising abilities of DDI to block the polymerase nsp12 and combat the exoribonuclease nsp14 (the enzyme that is responsible for increasing the resistance of the major targeted enzyme the SARS-CoV-2 RdRp to the various nucleotide analogue inhibitors) interestingly support the repurposing potentials of DDI. It is worth mentioning that DDI and molnupiravir are the only synthetic nucleoside analogues that have such unique anti-SARS-CoV-2 activities against the resistant/different SARS-CoV-2 variants in very significant values to date.41
Table 2. Anti-SARS-CoV-2 RdRp Activities (along with Respective Ratios) of the Target Repurposed Drug DDI (Using the Two Reference Agents Remdesivir and GS-441524 as the Positive Control Drugs and the Placebo Solvent DMSO as the Negative Control Drug) in HEK293T Cells, Expressed as EC50 Values in μMa.
| inhibition
of SARS-CoV-2 RdRp in vitro (EC50 in μM)b |
respective
ratios of EC50 |
|||||
|---|---|---|---|---|---|---|
| categorization | compound | nsp12 | nsp12 + nsp14 | nsp12 + nsp14mutant | (nsp12 + nsp14)/nsp12 | (nsp12 + nsp14mutant)/nsp12 |
| repurposed medication | DDI | 0.19 ± 0.02 | 0.31 ± 0.03 | 0.24 ± 0.02 | 1.63 | 1.26 |
| reference drugs | remdesivir | 1.11 ± 0.06 | 2.00 ± 0.09 | 1.52 ± 0.08 | 1.80 | 1.37 |
| GS-441524 | 1.04 ± 0.05 | 1.95 ± 0.09 | 1.46 ± 0.07 | 1.88 | 1.40 | |
| placebo solvent | DMSO | >100 | >100 | >100 | NAc | NA |
Please note that, in this table, nsp12 refers to nsp12/7/8 complex, nsp14 refers to nsp14/10 complex, and nsp14mutant refers to nsp14mutant/10 complex.
EC50 or 50% effective concentration is the concentration of the assayed compound which is needed for 50% decrease in the COVID-19 polymerase (SARS-CoV-2 RdRp) activity in vitro. EC50 is expressed in μM.
NA means not available (i.e., it was not determined).
Figure 6.
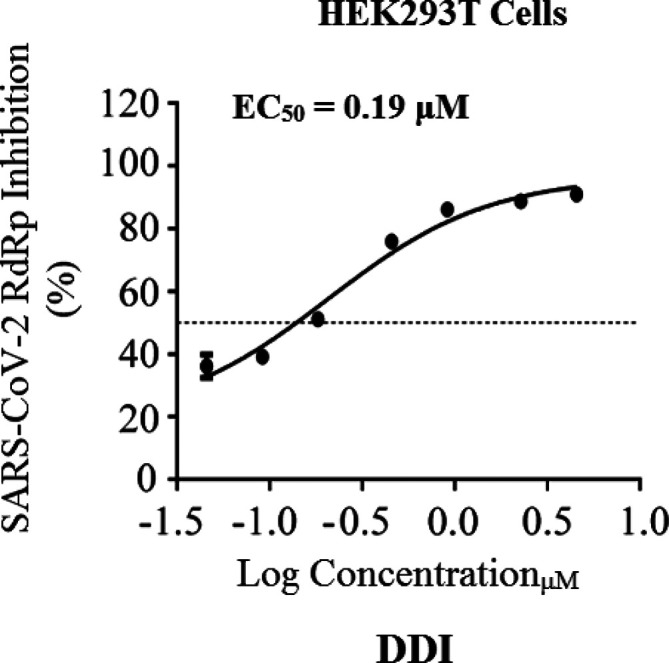
Dose-dependent inhibition CoV-Gluc by DDI. The used HEK293T cells were accurately transfected with CoV-Gluc, nsp12, nsp7, and nsp8 plasmid DNAs at the standard ratio of 1:10:30:30; then 12 h after transfection, cells were reseeded in 96-well plates (104/well) and treated with sequentially diluted DDI. After 24 h of continuous incubation, Gluc activities in supernatants were measured. Results are exhibited herein as the mean of three independent determinations.
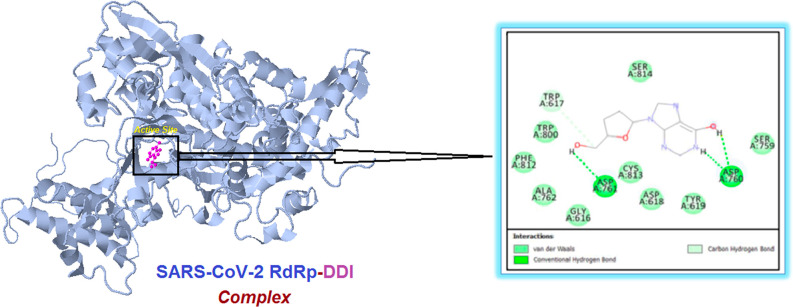
A simple illustrative and informative complementary molecular docking study of DDI in SARS-CoV-2 RdRp was done, utilizing a docking web server, following the previous promising biological results.45 The obtained results of this prevalidated in silico evaluation clearly exposed the very sturdy intermolecular interactions with the most important amino acid residues of the primary active site of the SARS-CoV-2 RdRp, confirming that the DDI molecule interacts actively with the molecule of RdRp protein (Figure 7). These strong inhibiting interactions were mirrored in the resulting comparatively low (highly negative) net binding energy that is around −10.3 kcal/mol at the best pose of docking, along with constructing a very stable RdRp–DDI complex. Interestingly, it was discovered that the DDI molecule attacks and binds to both critical catalytic amino acids Asp760 and Asp761 of the active site of SARS-CoV-2 RdRp through both robust hydrogen bonds/hydrophobic interactions (Figure 7). In addition, the DDI molecule was found to attach to some amino acid residues that are also catalytic or neighboring/very neighboring to the catalytic residues in the active site of RdRp, e.g., Trp617, Asp618, Tyr619, Ser759, Ala762, and Trp800 residues (Figure 7). These supplemental intermolecular interactions are mediated by means of various interactions, such as nonbonding and hydrophobic interactions (Figure 7). These current findings concerning the potent SARS-CoV-2 RdRp-binding properties of DDI are also in perfect agreement with almost all of the research rationale points previously proposed and explained in section 1.
Figure 7.
Illustration of the molecular docking output displaying the best foreseen binding mode of the DDI molecule (represented in magenta color) with the active site residues (present inside the small black rectangle and amplified in the right panel) of the SARS-CoV-2 RdRp macromolecule (represented in cyan color) using the COVID-19 Docking Server procedure.
3. Conclusions and Future Recommendations
While the world waits for mass COVID-19 vaccination, there is an immediate need for successful medications as available short-term postinfection weapons to fight the SARS-CoV-2 infection, in general, and to complement or boost the vaccine doses’ effectiveness against the attack of the mutated and newer variants of SARS-CoV-2, specifically. In this context, the drug repositioning strategy is an approach able to guarantee positive results quickly. In this regard, it is biologically well known that many nucleoside precursors and nucleoside-mimicking analogues may effectively block the growth and multiplication of viruses, providing efficient first-choice drugs for diverse viral diseases, including COVID-19 infection. Consequently, the current preclinical work aims to start the repurposing journey of DDI, an FDA-approved synthetic anti-HIV-1 inosine nucleoside analogue, against the resistant SARS-CoV-2 strains and COVID-19. The anticipated comprehensive nature of DDI in COVID-19 therapy principally comes from two practically proven paths. First, it could effectively inhibit the replication and permanence of the COVID-19-causing microbial virus itself with considerably potent broad-scope activities (including actions against the most contemporary strains of SARS-CoV-2), which are reaching a final EC50 of about 3.1 μM on the complete SARS-CoV-2 particles and a specific EC50 of about 0.19 μM on only the SARS-CoV-2 RdRp enzyme (DDI molecule strongly hits the two important catalytic amino acids, Asp760/Asp761, of the main active pocket of the SARS-CoV-2 RdRp). This anti-SARS-CoV-2 effect of DDI is because of the chemical mimicking (analogism) of the DDI molecule with the three naturally occurring human biomolecules, inosine, adenosine, and guanosine, with the lack of the two hydroxyls at the 2′ and 3′ positions on the ribose moiety (i.e., pseudoribosyl sugar residue). During viral replication, DDI and DDI-TP significantly compete with adenosine/guanosine and ATP/GTP, respectively, at their active site of SARS-CoV-2 RdRp, and consequently, this low-fidelity SARS-CoV-2 RdRp misincorporates DDI-TP (fake or faulty nucleotide) at the 3′ end of the RNA being synthesized (i.e., into the growing and newly synthesized RNA strands) in place of real/native ATP and GTP. Because the ribose of DDI-TP is a dideoxyribose (no 3′-hydroxyl group), RdRp will be unable to continue the replication process and chain extension is terminated as there is now no suitable substrate for chain extension to occur if the faulty nucleotide DDI-TP molecule is not completely removed by the exoribonuclease, which in turn will be unable to excise the DDI-TP molecule (in order to correct and recomplete the replication process) because of the absence of the other hydroxyl of the two adjacent hydroxyls of the ribose of DDI-TP (no 2′-hydroxyl group). Consequently, everything will stop as a result of these two synergistic effects of DDI, and the opposite strand of the coronaviral RNA is not going to get synthesized completely so RNA replication will also stop. Moreover, when the SARS-CoV-2 RdRp enzyme endeavors to copy the resultant unstable incomplete DDI-TP-containing RNA, it either interprets it incorrectly or fails to interpret it at all. Note that in normal conditions, as previously explained, when the primary copying/proofreading functions of SARS-CoV-2 RdRp are disturbed because of the mistaken addition of an incorrect nucleotide at the 3′ end of the RNA strand synthesized by this polymerase, the enzymatic activity of SARS-CoV-2 3′-5′ exonuclease removes and excises this nucleotide, giving the SARS-CoV-2 RdRp a second chance to add the correct nucleotide; these complementary excision and correcting actions of the proofreading function of the exonuclease could not be perfectly performed if the molecule of the added incorrect nucleotide lacks the two adjacent hydroxyls of the ribose moiety or at least the one at the 2′ position, which is the case with DDI, as clearly represented in the designed explanatory model of Figure 8. This hampered interpretation as well as the resulting flaws in the viral genetic code result in a large number of alterations (i.e., mutations) in all downstream coronaviral copies that are a lot more than the COVID-19 virus can handle and repair (in virology, this is known as the viral lethal mutagenesis/error catastrophe theory). Second, it could reduce and alleviate the significant health consequences of COVID-19, which are primarily related to the SARS-CoV-2 infection of the patients’ respiratory and cardiovascular systems, as this attack typically causes acute immunologic and inflammatory biological disturbances (see several and ample advantages of DDI use in the comprehensive COVID-19 therapy in section 1).
Figure 8.
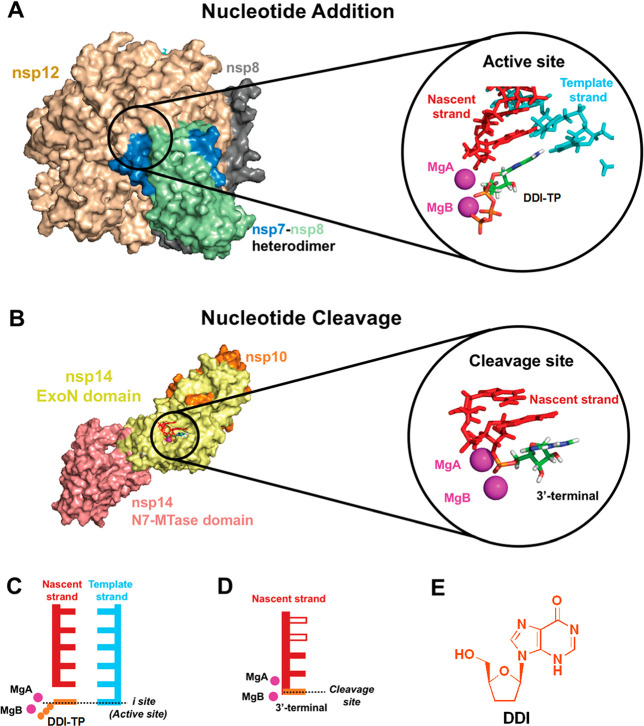
Newly designed structural model (A–D) of SARS-CoV-2 RdRp and ExoN (or 3′-to-5′ exoribonuclease) for investigating and explaining the inhibitory mechanisms of DDI against both enzymes, i.e., the dual inhibitory mode of action of DDI in coronaviral multiplication. (A) 3D model of the protein complex nsp12–nsp7–nsp8 for nucleotide addition. The known active site of this polymerase is circled and amplified in the right panel. The nascent and template coronaviral RNA strands are colored in red and cyan, respectively. DDI-TP molecule (in orange) is bound in the active site pocket, and two Mg2+ ions are displayed as magenta spheres. (B) 3D model of the protein complex nsp14–nsp10, containing the ExoN domain for precise nucleotide cleavage (this domain plays an important RNA proofreading role for resisting and preventing the coronaviral lethal mutagenesis). The ExoN cleavage site pocket is circled and amplified in the right panel. Three nucleotides are modeled, involving the 3′-terminal site utilized for modeling DDI-TP. Magenta spheres portray two Mg2+ ions required for cleavage. (C) Cartoon model of the active site in SARS-CoV-2 RdRp (inhibited/blocked by DDI-TP). (D) Cartoon model of the cleavage site in SARS-CoV-2 ExoN (inhibited/blocked by DDI-TP). The three terminal nucleotides utilized in this model are portrayed by color-filled rectangles. The ones which are not involved in this model are portrayed by empty rectangles. (E) Chemical structure of DDI in the original nucleoside form.
On the other hand, it has been previously argued that the use or the repurposing of exogenous dideoxynucleoside analogues (ddNs) and dideoxynucleoside triphosphate analogues (i.e., dideoxynucleotide analogues or ddNTPs), such as DDI, will not be adequately efficient against the SARS-CoV-2 because the RdRp is relatively more specific/selective for nucleoside triphosphates (NTPs), i.e., nucleotides, than for dideoxynucleotides (ddNTPs). However, another ddN analogue, which is the synthetic dideoxycytidine analogue lamivudine (3TC), has been proposed and in some instances demonstrated to be effective against the SARS-CoV-2 RdRp activity.24,31 Furthermore, in one of these previous reports, not only 3TC but also DDI have been included in the list of ddNTP analogues suggested to be suitable to try against the SARS-CoV-2 RdRp.31 Taken together, the present and previous results show that, although not being NTP analogues, the ddNTP analogues such as DDI or 3TC may exert very effective and unique biological inhibition of both principal activities, the RNA polymerase and RNA proofreading activities, of the SARS-CoV-2 RdRp, as a result of the lack of both essential adjacent hydroxyls, the 2′ and 3′ hydroxyls, in the ribose moiety of these analogues. Therefore, one of the novelties of this paper is that DDI is presented as one of the first dideoxy analogues of the natural nucleoside/nucleotide substrates (ddNs and ddNTPs) of the SARS-CoV-2 RdRp proposed as a potent anti-SARS-CoV-2/anti-COVID-19 agent.
In light of the comprehensive findings of the current research, it is proven that the DDI molecule interestingly outperforms a number of many other under-investigation anti-SARS-CoV-2 agents, especially those effective only on the spike (S) protein, in being acting as a nonselective/nonspecific anti-SARS-CoV-2 agent, i.e., able to act on almost all SARS-CoV-2 variants and lineages, as its anti-SARS-CoV-2 bioactivities do not rely upon the possible blocking effect on the changeable S protein which is mutated from one SARS-CoV-2 strain to another, but they depend mainly on the proven inhibitory actions on the fixed RdRp enzyme which has almost the same amino acid sequences in all SARS-CoV-2 strains to date (DDI possesses general broad-spectrum anti-SARS-CoV-2 properties that are likely to be effective against all SARS-CoV-2 strains, including the Omicron variant, irrespective of the characteristic mutations of the S protein in each specific strain). Intriguingly, the clear preponderance, in nearly all anti-COVID-19 properties and items, of DDI over the potent anticoronaviral agents, e.g., molnupiravir, remdesivir, and GS-441524, supports DDI candidacy as a superior prospective COVID-19 comprehensive therapeutic agent. It is also worth noting that the DDI molecule possesses more than 10 highly reactable atoms fit for several chemical reactions; therefore, hundreds of diverse potential derivatives/analogues with required improved pharmacokinetic and/or pharmacodynamic traits could be effectively planned and synthesized according to the biocompatible chemical scaffold of this invaluable molecule that belongs to the nucleos(t)ide analogue class of antiviral medicinal drugs. Finally, but certainly not least, the scientific community is strongly encouraged to pursue the DDI repurposing path against COVID-19 by conducting broad global preclinical and clinical investigations (as a suggestion, it is preferred to formulate and use DDI in the form of a nasal/oral inhaler for more rapid, direct, and targeted delivery and action, along with avoiding its weak acid stability in the stomach) to thoroughly assess the final efficacy and safety of DDI for use in the treatment and prevention of all forms of COVID-19 infections in a comprehensive manner.
4. Materials and Methods
4.1. Specifications of the Assayed Chemicals and Molecular Docking
DDI (2′,3′-dideoxyinosine, CAS registry number 69655-05-6) and remdesivir (GS-5734, CAS registry number 1809249-37-3) were purchased from Biosynth Carbosynth (Carbosynth Ltd., Berkshire, U.K.) (for DDI, product code ND02929, purity ≥98%; for remdesivir, product code AG170167, purity ≥98%), whereas the other reference compound GS-441524 (CAS registry number 1191237-69-0) was purchased from MedChemExpress (MCE, MedChemExpress LLC, New Jersey, U.S.A.) (catalog number HY-103586, purity 99.77%). The ultrapure solvent dimethyl sulfoxide (DMSO, CAS registry number 67-68-5) was purchased from a local distributor, El-Gomhouria Company For Drugs (El-Gomhouria Co. For Trading Drugs, Chemicals & Medical Supplies, Mansoura Branch, Egypt) (purity ≥99.9% “anhydrous”). The simple and direct illustrative molecular docking of DDI in SARS-CoV-2 RdRp was performed utilizing a validated web server called COVID-19 Docking Server. This server uses AutoDock Vina 1.2.0 software as the docking engine; the 3D structure of the SARS-CoV-2 RdRp (nsp12/7/8) protein cocrystallized in a complex with RNA as well as the triphosphate form of remdesivir (RTP) was obtained from the Protein Data Bank (PDB) database with the code of 7BV2, employing the validated remdesivir docking protocol as the comparison protocol for the ligand DDI.45
4.2. In Vitro Anti-SARS-CoV-2 and Cytotoxic Bioactivities Assay of DDI
This reliable in vitro anti-COVID-19 assay (including the cytotoxicity test) depends primarily on Rabie’s verified techniques.11,12,15−18 All operations were implemented in a biosafety level 3 (BSL-3) laboratory of a specialized research facility. The tested new variant of SARS-CoV-2 virus, the first variant of concern from December 2020 (VOC-202012/01), was isolated from the fresh nasopharyngeal aspirate and throat swab of a 31-year-old male COVID-19 patient, employing Vero E6 cells (ATCC CRL-1586), on October 20, 2021. The starting titer of the stock virus (107.25 TCID50/mL) was made after three serial passes in Vero E6 cells in infection media (Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 4.5 g/L d-glucose, 100 mg/L sodium pyruvate, 2% fetal bovine serum (FBS), 100 000 U/L penicillin–streptomycin, and 25 mM N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid (HEPES)). The tested drugs are as described and specified in section 4.1. Preliminary pilot assays were first done with the main purpose to find the best concentration of DDI, remdesivir, and GS-441524 to adequately begin the in vitro anti-COVID-19/cytotoxicity tests. As a result, the stocks of the tested compounds were precisely produced by dissolving each of the three compounds in the solvent DMSO to get a 100 μM concentration of each compound. In addition, DMSO was utilized as a negative control to ensure that the study is placebo-controlled. To estimate the anti-SARS-CoV-2 activities of the target compound, DDI, as compared to those of each of the two positive control drugs, remdesivir and GS-441524, as well as those of the negative control solvent, DMSO, Vero E6 cells were first treated with the four compounds diluted in infection media for 1 h before infection by the new strain of the SARS-CoV-2 at MOI = 0.02. During the 2 h incubation period, all of the four assayed compounds were kept with the virus inoculum. After incubation, the inoculum was withdrawn, and the cells were completely fed with infection media containing the diluted test compounds. Supernatants were collected immediately after 48 h of incubation at 37 °C to determine virus loads using the TCID50 or quantitative real-time RT-PCR (qRT-PCR) (TaqMan Fast Virus 1-Step Master Mix) assays. Virus loads in these assays were fitted in logarithm scale (log10 TCID50/mL, log10 TCID90/mL, and log10 viral RNA copies/mL), not in linear scale, under increasing concentrations of the tested compounds. Four-parameter logistic regression (GraphPad Prism) was employed to fit the dose–response curves and obtain the EC50 and EC90 of the tested compounds, which inhibit viral replication of the SARS-CoV-2 (CPEIC100 was also determined for each compound). Cytotoxicity of each compound of the four tested compounds was assessed as well in Vero E6 cells utilizing the CellTiter-Glo luminescent cell viability assay (Promega). Final outcomes were accurately represented as the mean ± standard deviation (SD) from at least three separate trials. Statistical analysis was carried out using SkanIt 4.0 research edition software (Thermo Fisher Scientific) and Prism V5 software (GraphPad). At p < 0.05, all of the results were judged to be statistically significant.
4.3. In Vitro Anti-RdRp Assay (SARS-CoV-2-RdRp-Gluc Reporter Assay) of DDI
First, the used cells, 293T cells (ATCC CRL-3216), were kept in DMEM (Gibco) with 10% (v/v) FBS (Gibco), and then they were cultured at 37 °C in a CO2-humidified atmosphere (5%). HEK293T cells were transfected employing Vigofect transfection reagents (Vigorous) according to the manufacturer’s precise instructions. The required plasmid DNAs, antibodies, and reagents were purchased and treated exactly as in the literature procedures.41 The tested drugs are as described and specified in section 4.1. Also, Western blotting (for the collected transfected HEK293T cells), real-time RT-PCR (for the extracted total RNA of transfected HEK293T cells), and cell viability test (using cell counting kit-8 (CCK8), Beyotime) were exactly performed as the typical procedures of the literature.41 The method of the validated newly designed in vitro SARS-CoV-2-RdRp-Gluc reporter assay was performed according to the original procedures of its designers, Zhao and colleagues (please note that HEK293T cells were transfected in this assay with CoV-Gluc, nsp12, nsp7, and nsp8 plasmid DNAs in the ratio of 1:10:30:30, and with CoV-Gluc, nsp12, nsp7, nsp8, nsp10, and nsp14 plasmid DNAs in the ratio of 1:10:30:30:10:90).41 Typically as instructed in this original method, a stock of coelenterazine-h was dissolved in absolute ethyl alcohol to a concentration of 1.022 mM/L.41 Right before each assay, the stock was diluted in phosphate-buffered saline to 16.7 μM and incubated in the dark for 30 min at room temperature.41 For the luminescence assay, 10 μL of supernatant was added to each well of a white and opaque 96-well plate, then 60 μL of 16.7 μM coelenterazine-h was injected, and the respective luminescence was measured for 0.5 s utilizing the Berthold Centro XS3 LB 960 microplate luminometer.41 Final outcomes were accurately represented as the mean ± SD from at least three separate trials. Statistical analysis was carried out using SkanIt 4.0 research edition software (Thermo Fisher Scientific) and Prism V5 software (GraphPad). At p < 0.05, all of the results were judged to be statistically significant.
Acknowledgments
This novel discovery did not receive any external financing. The author expresses his gratitude and sincere appreciation to any person who sincerely assisted to render this novel research and work coming out to light.
The author declares no competing financial interest.
References
- Hui D. S.; I Azhar E.; Madani T. A.; Ntoumi F.; Kock R.; Dar O.; Ippolito G.; Mchugh T. D.; Memish Z. A.; Drosten C.; Zumla A.; Petersen E. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health — The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Map, available from Johns Hopkins Coronavirus Research Center, https://coronavirus.jhu.edu/map.html (accessed 2022-03-05).
- Chitalia V. C.; Munawar A. H. A painful lesson from the COVID-19 pandemic: the need for broad-spectrum, host-directed antivirals. J. Transl. Med. 2020, 18, 390. 10.1186/s12967-020-02476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase E. Covid-19: Pfizer’s paxlovid is 89% effective in patients at risk of serious illness, company reports. BMJ 2021, 375, n2713. 10.1136/bmj.n2713. [DOI] [PubMed] [Google Scholar]
- Imran M.; Kumar Arora M.; Asdaq S. M. B.; Khan S. A.; Alaqel S. I.; Alshammari M. K.; Alshehri M. M.; Alshrari A. S.; Mateq Ali A.; Al-shammeri A. M.; Alhazmi B. D.; Harshan A. A.; Alam M. T.; Abida Discovery, Development, and Patent Trends on Molnupiravir: A Prospective Oral Treatment for COVID-19. Molecules 2021, 26, 5795. 10.3390/molecules26195795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moirangthem D. S.; Surbala L. Remdesivir (GS-5734) in COVID-19 Therapy: The Fourth Chance. Curr. Drug Targets 2021, 22, 1346–1356. 10.2174/1389450121999201202110303. [DOI] [PubMed] [Google Scholar]
- Yan V. C.; Muller F. L. Advantages of the Parent Nucleoside GS-441524 over Remdesivir for Covid-19 Treatment. ACS Med. Chem. Lett. 2020, 11, 1361–1366. 10.1021/acsmedchemlett.0c00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunotte L.; Zheng S.; Mecate-Zambrano A.; Tang J.; Ludwig S.; Rescher U.; Schloer S. Combination Therapy with Fluoxetine and the Nucleoside Analog GS-441524 Exerts Synergistic Antiviral Effects against Different SARS-CoV-2 Variants In Vitro. Pharmaceutics 2021, 13, 1400. 10.3390/pharmaceutics13091400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie A. M. Potent Inhibitory Activities of the Adenosine Analogue Cordycepin on SARS-CoV-2 Replication. ACS Omega 2022, 7, 2960–2969. 10.1021/acsomega.1c05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q.; Yang M.; Liu D.; Chen J.; Shu D.; Xia J.; Liao X.; Gu Y.; Cai Q.; Yang Y.; Shen C.; Li X.; Peng L.; Huang D.; Zhang J.; Zhang S.; Wang F.; Liu J.; Chen L.; Chen S.; Wang Z.; Zhang Z.; Cao R.; Zhong W.; Liu Y.; Liu L. Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study. Engineering 2020, 6, 1192–1198. 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie A. M. Discovery of (E)-N-(4-cyanobenzylidene)-6-fluoro-3-hydroxypyrazine-2-carboxamide (cyanorona-20): the first potent and specific anti-COVID-19 drug. Chem. Pap. 2021, 75, 4669–4685. 10.1007/s11696-021-01640-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rabie A. M. Cyanorona-20: The first potent anti-SARS-CoV-2 agent. Int. Immunopharmacol. 2021, 98, 107831. 10.1016/j.intimp.2021.107831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip A.; Ahn J.; Zhou Y.; Goy A. H.; Hansen E.; Pecora A. L.; Sinclaire B. A.; Bednarz U.; Marafelias M.; Sawczuk I. S.; Underwood J. P. III; Walker D. M.; Prasad R.; Sweeney R. L.; Ponce M. G.; La Capra S.; Cunningham F. J.; Calise A. G.; Pulver B. L.; Ruocco D.; Mojares G. E.; Eagan M. P.; Ziontz K. L.; Mastrokyriakos P.; Goldberg S. L. Hydroxychloroquine in the treatment of outpatients with mildly symptomatic COVID-19: a multi-center observational study. BMC Infect. Dis. 2021, 21, 72. 10.1186/s12879-021-05773-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Cao R.; Zhang L.; Yang X.; Liu J.; Xu M.; Shi Z.; Hu Z.; Zhong W.; Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie A. M. Two antioxidant 2,5-disubstituted-1,3,4-oxadiazoles (CoViTris2020 and ChloViD2020): successful repurposing against COVID-19 as the first potent multitarget anti-SARS-CoV-2 drugs. New J. Chem. 2021, 45, 761–771. 10.1039/D0NJ03708G. [DOI] [Google Scholar]
- Rabie A. M. CoViTris2020 and ChloViD2020: a striking new hope in COVID-19 therapy. Mol. Diversity 2021, 25, 1839–1854. 10.1007/s11030-020-10169-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rabie A. M. Potent toxic effects of Taroxaz-104 on the replication of SARS-CoV-2 particles. Chem.-Biol. Interact. 2021, 343, 109480. 10.1016/j.cbi.2021.109480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie A. M. Discovery of Taroxaz-104: The first potent antidote of SARS-CoV-2 VOC-202012/01 strain. J. Mol. Struct. 2021, 1246, 131106. 10.1016/j.molstruc.2021.131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H.; Sarma P.; Bhattacharyya A.; Sharma S.; Chhimpa N.; Prajapat M.; Prakash A.; Kumar S.; Singh A.; Singh R.; Avti P.; Thota P.; Medhi B. Efficacy and safety of dihydroorotate dehydrogenase (DHODH) inhibitors “leflunomide” and “teriflunomide” in Covid-19: A narrative review. Eur. J. Pharmacol. 2021, 906, 174233. 10.1016/j.ejphar.2021.174233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie A. M. Teriflunomide: A possible effective drug for the comprehensive treatment of COVID-19. Curr. Res. Pharmacol. Drug Discovery 2021, 2, 100055. 10.1016/j.crphar.2021.100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L.; Druce J. D.; Catton M. G.; Jans D. A.; Wagstaff K. M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020, 178, 104787. 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Cao R.; Zhang H.; Liu J.; Xu M.; Hu H.; Li Y.; Zhao L.; Li W.; Sun X.; Yang X.; Shi Z.; Deng F.; Hu Z.; Zhong W.; Wang M. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discovery 2020, 6, 28. 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif J.-C.; Bouabdallaoui N.; L’Allier P. L.; Gaudet D.; Shah B.; Pillinger M. H.; Lopez-Sendon J.; da Luz P.; Verret L.; Audet S.; Dupuis J.; Denault A.; Pelletier M.; Tessier P. A.; Samson S.; Fortin D.; Tardif J.-D.; Busseuil D.; Goulet E.; Lacoste C.; Dubois A.; Joshi A. Y; Waters D. D; Hsue P.; Lepor N. E.; Lesage F.; Sainturet N.; Roy-Clavel E.; Bassevitch Z.; Orfanos A.; Stamatescu G.; Grégoire J. C.; Busque L.; Lavallée C.; Hétu P.-O.; Paquette J.-S.; Deftereos S. G.; Levesque S.; Cossette M.; Nozza A.; Chabot-Blanchet M.; Dubé M.-P.; Guertin M.-C.; Boivin G. Colchicine for community-treated patients with COVID-19 (COLCORONA): a phase 3, randomised, double-blinded, adaptive, placebo-controlled, multicentre trial. Lancet Respir. Med. 2021, 9, 924–932. 10.1016/S2213-2600(21)00222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien M.; Anderson T. K.; Jockusch S.; Tao C.; Li X.; Kumar S.; Russo J. J.; Kirchdoerfer R. N.; Ju J. Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, a Key Drug Target for COVID-19. J. Proteome Res. 2020, 19, 4690–4697. 10.1021/acs.jproteome.0c00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didanosine, PubChem CID: 135398739, available from PubChem, https://pubchem.ncbi.nlm.nih.gov/compound/135398739 (accessed 2022-01-31).
- Martins F. T.; Legendre A. O.; Honorato S. B.; Ayala A. P.; Doriguetto A. C.; Ellena J. Solvothermal Preparation of Drug Crystals: Didanosine. Cryst. Growth Des. 2010, 10, 1885–1891. 10.1021/cg9015959. [DOI] [Google Scholar]
- Cooney D. A.; Ahluwalia G.; Mitsuya H.; Fridland A.; Johnson M.; Hao Z.; Dalal M.; Balzarini J.; Broder S.; Johns D. G. Initial studies on the cellular pharmacology of 2′,3′-dideoxyadenosine, an inhibitor of HTLV-III infectivity. Biochem. Pharmacol. 1987, 36, 1765–1768. 10.1016/0006-2952(87)90235-8. [DOI] [PubMed] [Google Scholar]
- Ahluwalia G.; Cooney D. A.; Mitsuya H.; Fridland A.; Flora K. P.; Hao Z.; Dalal M.; Broder S.; Johns D. G. Initial studies on the cellular pharmacology of 2′,3′-dideoxyinosine, an inhibitor of HIV infectivity. Biochem. Pharmacol. 1987, 36, 3797–3800. 10.1016/0006-2952(87)90440-0. [DOI] [PubMed] [Google Scholar]
- Mitsuya H.; Broder S. Inhibition of the in vitro infectivity and cytopathic effect of human T-lymphotrophic virus type III/lymphadenopathy-associated virus (HTLV-III/LAV) by 2′,3′-dideoxynucleosides. Proc. Natl. Acad. Sci. U. S. A. 1986, 83, 1911–1915. 10.1073/pnas.83.6.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch S.; Tao C.; Li X.; Anderson T. K.; Chien M.; Kumar S.; Russo J. J.; Kirchdoerfer R. N.; Ju J. A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19. Antiviral Res. 2020, 180, 104857. 10.1016/j.antiviral.2020.104857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Trejo J. J.; Ortega R.; Zarco-Zavala M. Putative Repurposing of Lamivudine, a Nucleoside/Nucleotide Analogue and Antiretroviral to Improve the Outcome of Cancer and COVID-19 Patients. Front. Oncol. 2021, 11, 664794. 10.3389/fonc.2021.664794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kewn S.; Hoggard P. G.; Henry-Mowatt J. S.; Veal G. J.; Sales S. D.; Barry M. G.; Back D. J. Intracellular Activation of 2′,3′-Dideoxyinosine and Drug Interactions in Vitro. AIDS Res. Hum. Retroviruses 1999, 15, 793–802. 10.1089/088922299310692. [DOI] [PubMed] [Google Scholar]
- Molinspiration Property Engine (v2021.10), available from Molinspiration Cheminformatics, https://www.molinspiration.com/cgi-bin/properties (accessed 2022-01-25).
- Mclaren C.; Datema R.; Knupp C. A.; Buroker R. A. Didanosine. Antiviral Chem. Chemother. 1991, 2, 321–328. 10.1177/095632029100200601. [DOI] [Google Scholar]
- Cava C.; Bertoli G.; Castiglioni I. In Silico Discovery of Candidate Drugs against Covid-19. Viruses 2020, 12, 404. 10.3390/v12040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliji S.; Lacatus G.; Sunter G. The interaction between geminivirus pathogenicity proteins and adenosine kinase leads to increased expression of primary cytokinin-responsive genes. Virology 2010, 402, 238–247. 10.1016/j.virol.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alakwaa F. M. Repurposing Didanosine as a Potential Treatment for COVID-19 Using Single-Cell RNA Sequencing Data. mSystems 2020, 5, e00297-20. 10.1128/mSystems.00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta Y.; Maciorowski D.; Mathur R.; Pearce C. M.; Ilc D. J.; Husein H.; Bharti A.; Becker D.; Brijesh R.; Bradfute S. B.; Durvasula R.; Kempaiah P. Revealing SARS-CoV-2 Functional Druggability Through Multi-Target Cadd Screening of Repurposable Drugs. Preprints 2020, 2020050199. 10.20944/preprints202005.0199.v1. [DOI] [Google Scholar]
- Borgio J. F.; Alsuwat H. S.; Al Otaibi W. M.; Ibrahim A. M.; Almandil N. B.; Al Asoom L. I.; Salahuddin M.; Kamaraj B.; AbdulAzeez S. State-of-the-art tools unveil potent drug targets amongst clinically approved drugs to inhibit helicase in SARS-CoV-2. Arch. Med. Sci. 2020, 16, 508–518. 10.5114/aoms.2020.94567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illanes-Álvarez F.; Márquez-Ruiz D.; Márquez-Coello M.; Cuesta-Sancho S.; Girón-González J. A. Similarities and differences between HIV and SARS-CoV-2. Int. J. Med. Sci. 2021, 18, 846–851. 10.7150/ijms.50133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.; Guo S.; Yi D.; Li Q.; Ma L.; Zhang Y.; Wang J.; Li X.; Guo F.; Lin R.; Liang C.; Liu Z.; Cen S. A cell-based assay to discover inhibitors of SARS-CoV-2 RNA dependent RNA polymerase. Antiviral Res. 2021, 190, 105078. 10.1016/j.antiviral.2021.105078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. C.; Blanc H.; Vignuzzi M.; Denison M. R. Coronaviruses Lacking Exoribonuclease Activity Are Susceptible to Lethal Mutagenesis: Evidence for Proofreading and Potential Therapeutics. PLoS Pathog. 2013, 9, e1003565. 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F.; Subissi L.; Silveira De Morais A. T.; Le N.; Sevajol M.; Gluais L.; Decroly E.; Vonrhein C.; Bricogne G.; Canard B.; Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E162–E171. 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen H. S.; Kokic G.; Farnung L.; Dienemann C.; Tegunov D.; Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584, 154–156. 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- COVID-19 Docking Server, https://ncov.schanglab.org.cn (accessed 2022-01-26).