Summary
Serological markers are a promising tool for surveillance and targeted interventions for Plasmodium vivax malaria. P. vivax is closely related to the zoonotic parasite P. knowlesi, which also infects humans. P. vivax and P. knowlesi are co-endemic across much of South East Asia, making it important to design serological markers that minimize cross-reactivity in this region. To determine the degree of IgG cross-reactivity against a panel of P. vivax serological markers, we assayed samples from human patients with P. knowlesi malaria. IgG antibody reactivity is high against P. vivax proteins with high sequence identity with their P. knowlesi ortholog. IgG reactivity peaks at 7 days post-P. knowlesi infection and is short-lived, with minimal responses 1 year post-infection. We designed a panel of eight P. vivax proteins with low levels of cross-reactivity with P. knowlesi. This panel can accurately classify recent P. vivax infections while reducing misclassification of recent P. knowlesi infections.
Keywords: malaria, Plasmodium vivax, Plasmodium knowlesi, antibodies, serosurveillance, serological exposure markers, antibody cross-reactivity, malaria elimination, species cross-reactivity
Graphical abstract
Highlights
-
•
Serology is promising for actionable surveillance for Plasmodium vivax malaria
-
•
P. knowlesi infections induce cross-reactive IgG antibodies to P. vivax proteins
-
•
Cross-reactive IgG antibodies are short-lived following infection
-
•
Misclassification due to cross-reactivity can be reduced through antigen selection
Longley et al. characterize cross-reactivity against Plasmodium vivax proteins in people infected with the closely related parasite species Plasmodium knowlesi. They propose an approach to maintain sensitivity of P. vivax serological exposure markers while reducing misclassification due to cross-reactivity with P. knowlesi.
Introduction
To accelerate toward malaria elimination, new tools and interventions are needed. Malaria is caused by the parasite Plasmodium, with most of the disease in humans caused by P. falciparum, P. vivax, P. malariae, P. ovale, and P. knowlesi. P. falciparum and P. vivax account for the largest burdens of disease. In many co-endemic regions outside Africa, infection with P. vivax has become the predominant cause of malaria as the overall level of transmission has declined.1 Thus, although standard malaria control tools (such as those targeting the mosquito vector and case management) have had a significant impact on reducing transmission of P. falciparum, they have not had the same level of effect on P. vivax. This is most likely due to several distinct biological features of P. vivax infections that promote transmission. These include a cryptic endosplenic life cycle leading to a large hidden splenic reservoir of P. vivax parasites,2,3 which sustains a high prevalence of low-density asymptomatic blood stage infections4 that are still capable of onward transmission.5 Furthermore, an additional life cycle stage in the liver, known as the hypnozoite, can remain arrested or dormant for months to years following the initial mosquito bite-induced infection, before being reactivated by currently unknown signals to re-join the life cycle. Hepatic hypnozoites are major reservoirs for transmission in communities, with up to 80% of detected blood-stage infections attributed to hypnozoite relapse rather than new mosquito bite-induced infections.6, 7, 8
To address the unique challenge to malaria elimination posed by P. vivax, we have developed a surveillance tool: P. vivax serological exposure markers. By measuring total IgG antibody responses to a carefully selected panel of eight P. vivax proteins, we can classify individuals as recently exposed to a blood-stage P. vivax infection within the past 9 months with 80% sensitivity and 80% specificity.9 Due to the frequency of P. vivax relapses, with most occurring within 6–9 months following primary infection,10 we hypothesize that our P. vivax serological exposure markers can be used to indirectly identify hypnozoite carriers—individuals who are more likely to go on to develop recurrent P. vivax infections and sustain transmission. Our serological markers can thus be used as an effective surveillance tool for identifying clusters of P. vivax infections for efficient targeting of resources11 and for designing public health interventions relying on “serological testing and treatment” (seroTAT).12 SeroTAT can be used to identify hypnozoite carriers who can then be treated with appropriate anti-malarial drugs (including an anti-liver-stage component such as primaquine or tafenoquine) as part of an elimination campaign.9
The major species of Plasmodium causing disease in humans have different geographic distributions13 and different levels of relatedness.14 It is therefore important to characterize potential cross-reactivity against our P. vivax serological exposure markers in individuals who have had recent exposure to other Plasmodium infections. We identified no patterns of cross-reactivity with recent P. falciparum exposure in our original cohort studies,9 with the caveat that few individuals had P. falciparum infections in those observational cohorts (in Thailand, Brazil, and the Solomon Islands). In the limited number of prior studies assessing P. vivax- P. falciparum cross-reactive antibody responses,15 mixed results were observed with both species-specific16 and cross-reactive antibodies detected.17,18 However, even if cross-reactivity between our P. vivax proteins in individuals with recent P. falciparum infections becomes evident, this is not necessarily of programmatic concern. Similar risk factors for exposure in co-endemic regions are present, and there is potential benefit in treating individuals with P. falciparum infections with anti-hypnozoite drugs19 (due to the high risk of P. vivax recurrence following P. falciparum infections,20, 21, 22 and for the beneficial effect of primaquine clearing P. falciparum gametocytes, the sexual form of the parasite23). These benefits do need to be considered in the context of the risks of primaquine treatment in glucose-6-phosphate dehydrogenase (G6PD)-deficient individuals.24 However, continued improvements in rapid G6PD testing alleviate some of these concerns,25 along with a recent finding of higher G6PD activity levels in people with malaria compared with those without.26 P. vivax is more closely related to the zoonotic parasite P. knowlesi than to P. falciparum,14 and serological cross-reactivity has been reported.27, 28, 29 P. vivax and P. knowlesi are co-endemic in humans across much of Southeast Asia.30 Risk-factors for exposure to P. knowlesi and P. vivax differ,31 as do spatial clusters of infections,29 treatment,32 and risk mitigation strategies.31 For these reasons, and to optimize the utility of serological markers in Southeast Asia, it is of particular importance to characterize cross-reactivity against the P. vivax serological exposure markers in individuals with recent P. knowlesi infections.
Here, we aimed to assess cross-reactivity against a panel of P. vivax serological exposure markers (including our previously identified top combination of eight markers9) in samples from two clinical cohorts of patients with confirmed P. knowlesi malaria. We assessed whether our algorithm would classify these P. knowlesi patients as P. vivax exposed or not and whether we could improve our serological markers for use in P. vivax-P. knowlesi co-endemic areas by selecting proteins with absent or lower levels of cross-reactivity.
Results
Comparison of P. vivax and P. knowlesi protein sequences
We hypothesized that potential antibody cross-reactivity could relate to the level of sequence identity, and thus, we first constructed a pipeline to identify and then compare the sequences of our expanded panel of 21 P. vivax serological exposure markers, selected on the basis of our prior work9 (see STAR Methods for further details), with their identified P. knowlesi orthologs. Two different analytical approaches were used: (1) by accessing the list of orthologs on PlasmoDB for each P. vivax protein and (2) through an NCBI BlastP search of the protein sequence construct. P. knowlesi orthologs were found for 17 of 21 P. vivax proteins via PlasmoDB (using either the original macaque H strain33 or the later human red blood cell-adapted A1H1 line34) and for 20/21 by NCBI BlastP (any strain) (Table 1). Using both methods each of the 21 P. vivax proteins had at least one identified P. knowlesi ortholog. When multiple orthologs were found, the top hit was selected by highest identity. The similarity and identity values for the hits on PlasmoDB were obtained by aligning the full-length protein sequences provided in the database using the EMBOSS Needle protein alignment tool;35 the identity values for the NCBI BlastP hits were obtained from the BlastP search using only the protein construct sequence. The sequence identity of the full-length protein sequences (search strategy 1) ranged from 11% to 19% for MSP7F (A1H1 line or H strain, respectively) to 83.4% for MSP8. A similar trend was found using the specific protein construct and NCBI BlastP (search strategy 2); however, the percent identity was often higher using this method (Table 1).
Table 1.
Sequence comparison of the P. vivax proteins with their P. knowlesi orthologs
| P. vivax Protein | PlasmoDB code | # Hits P | Top Hit P | Synteny P | Similarity (%) P | Identity (%) P | Top Hit N | Identity (%) N |
|---|---|---|---|---|---|---|---|---|
| MSP8a | PVX_097625 | 1 | PKNH_1031500 | Y | 91.2 | 83.4 | AFL93300.1 | 84.52 |
| Pv-fam-aa | PVX_112670 | 24 | PKNH_1300800 | Y | 72.8 | 65.8 | OTN66803.1 | 83.11 |
| MSP1-19a | PVX_099980 | 1 | PKNH_0728900 | Y | 74.4 | 64.2 | AZL87433.1 | 81.31 |
| RAMA∗ | PVX_087885 | 0 | NA | NA | NA | NA | OTN68698.1 | 73.61 |
| RIPR | PVX_095055 | 1 | PKNH_0817000 | Y | 82.9 | 71.9 | XP_002258810.1 | 73.43 |
| Pv-fam-aa | PVX_096995 | 24 | PKNH_0200900 | Y | 50.9 | 41.1 | OTN66147.1 | 70.28 |
| PTEX150 | PVX_084720 | 1 | PKNH_0422900 | Y | 83.9 | 73.2 | OTN66840.1 | 72.02 |
| DBPII Sal 1 | PVX_110810 | 3 | PKNH_0623500 | Y | 64.7 | 52.1 | OTN68355.1 | 68.72 |
| DBPII AH | AAY34130.1b | 3 | PKNH_0623500 | Y | 64.7 | 51.9 | OTN68355.1 | 68.21 |
| Sexual-stage antigen s16 | PVX_000930 | 1 | PKNH_0304300 | Y | 78.6 | 67.1 | OTN68464.1 | 65.45 |
| TRAP | PVX_082735 | 1 | PKNH_1265400 | Y | 81.4 | 69.1 | AAG24613.1 | 64.23 |
| MSP3b | PVX_097680 | 15 | PKNH_1457400 | N | 42 | 28.6 | OTN63878.1 | 62.5 |
| MSP5 | PVX_003770 | 2 | PKNH_0414200 | Y | 65.4 | 49.9 | AAT77929.1 | 46.26 |
| Hypothetical | PVX_097715 | 0 | NA | NA | NA | NA | ANP24393.1 | 40.65 |
| EBP region IIa | KMZ83376.1b | 0 | NA | NA | NA | NA | QPL17772.1 | 35 |
| RBP2b1986–2653 | PVX_094255 | 2 | PKNH_0700200 | N | 52.2 | 29.1 | OTN67427.1 | 34.53 |
| MSP7F | PVX_082670 | 15 | PKNH_0726500 | N | 30.1 | 19 | AND94835.1 | 27.53 |
| RBP2b161–1454a | PVX_094255 | 2 | PKNH_0700200 | N | 52.2 | 29.1 | OTN67427.1 | 26.1 |
| RBP2a | PVX_121920 | 2 | PKNH_0700200 | N | 46.7 | 25.4 | OTN67427.1 | 25.77 |
| MSP7L | PVX_082700 | 0 | NA | NA | NA | NA | AND94835.1 | 24.29 |
| MSP3aa | PVX_097720 | 15 | PKNH_1457400 | N | 45.8 | 31.6 | NA | NA |
Full-length protein sequences were compared using the orthologs listed in PlasmoDB (H or A1H1 strain). The H strain and the A1H1 line gave identical results except for MSP7F; the H strain results are listed: A1H1 gave 11.5% identity and 18.8% similarity. Protein construct sequences (Table S1) were compared using NCBI BlastP (any strain) to identify the top hits. “P” denotes PlasmoDB pipeline, and “N” denotes NCBI pipeline. NA, no matches. Proteins are ordered by highest percent sequence identity using NCBI BlastP.
Top eight P. vivax protein serological-exposure marker.
GenBank IDs.
P. vivax protein-specific IgG antibody response in P. knowlesi clinical case samples
To assess potential cross-reactivity against the P. vivax proteins, we measured longitudinal total IgG antibody responses in patients with PCR-confirmed P. knowlesi monoinfection, enrolled in two clinical trials, denoted ACTKNOW (recruited over 2012–201436) and PACKNOW (recruited over 2016–201837,38) (Table 2). The two sample sets were initially analyzed separately, given that the risk of prior P. vivax infection was lower at the time of the PACKNOW trial.39 There were no differences in demographic variables such as age, sex, and self-reported history of malaria infections between the patients in the two trials (Table 2). The high proportion of males in both studies is typical of the existing evidence for P. knowlesi clinical presentation and epidemiology.31,40,41
Table 2.
Characteristics of patients in two P. knowlesi clinical trial cohorts used in the present study
| ACTKNOW (n = 99) | PACKNOW (n = 41) | |
|---|---|---|
| PCR-confirmed Plasmodium spp. | P. knowlesi | P. knowlesi |
| Collection year | 2012–2014 | 2016–2018 |
| Age (years), median (range)a | 34 (3–78) | 36 (20–70) |
| Gender, number femaleb (percent) | 29 (29.3%) | 6 (14.6%) |
| Self-reported fever days, median (range)a | 5 (0–14) | 4 (3–14) |
| Self-reported previous malaria infection, number yesb (percent) | 34 (34.3%) | 17 (41.5%) |
| Parasitaemia (parasites/μL), median (range)a | 2857 (56–43,721) | 2690 (37–185,553) |
| Treatment administered | Randomized to artesunate-mefloquine (n = 52) or chloroquine (n = 47) | Artemether-lumefantrine |
| Serology details | ||
| Sample size at enrollment | 99 | 41 |
| Follow-up timepoints | 3: days 0, 7, and 28 | 5: days 0, 7, 14, 28, and 365 |
| Total number of samples assessed | 296c | 166d |
Statistical difference between cohorts assessed by Mann-Whitney test, not significant.
Statistical difference between cohorts assessed by Fisher exact test, not significant.
One sample missing serology data at day 0.
Samples were not available at all time points for all patients.
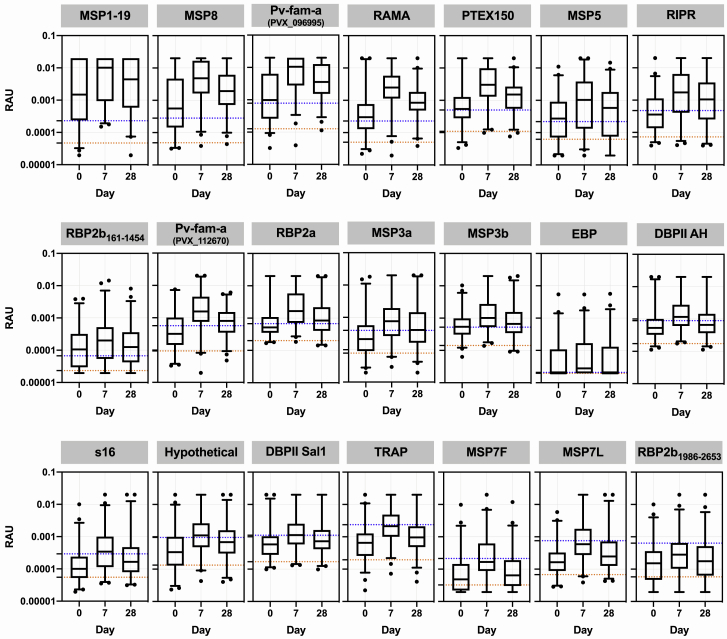
We first assessed potential cross-reactivity against the P. vivax proteins by using 99 patients from the ACTKNOW P. knowlesi clinical trial cohort, with plasma samples available at the time of diagnosis of P. knowlesi infection (day 0) and follow-up at day 7 and day 28. Median parasitemia prior to treatment (day 0) was 2,857 parasites/μL (IQR 648–7,177), with all subjects negative for asexual-stage parasites and 95% negative for sexual stages by the day 7 time point.36 Antibody responses were compared with malaria-naive negative-control samples from the non-endemic cities of Melbourne (Australia), Bangkok (Thailand), and Rio de Janeiro (Brazil) (n = 369), with a protein-specific seropositivity cut-off defined as the average of the negative controls plus two times the standard deviation. We observed an increase in the median IgG antibody response against all P. vivax proteins at day 7 compared with day 0, with IgG levels declining from the peak response by day 28 (Figure 1). For 17 of 21 P. vivax proteins, median IgG levels at day 7 (the peak of the response) were above the seropositivity cut-off. Of those 17 proteins, for five the median IgG level had dropped below the sero-positivity cut-off by day 28. For all P. vivax proteins there were at least some patients with responses above the sero-positivity cut-off at all three time points (see Figure S1 for the change in IgG levels at the individual level over time). We explored whether acquisition of cross-reactive IgG antibodies at day 0 was related to delayed presentation to the clinic (and therefore delayed enrollment into the study), using duration of fever as a surrogate for delayed presentation/duration of infection. There was no correlation between day 0 IgG antibodies and duration of fever for 19 of the 21 P. vivax proteins; there was a weak negative correlation for both DBPII constructs (Spearman correlation, AH r = −0.25, Sal1 r = −0.2, p < 0.05; see Figure S2). The highest levels of potential cross-reactivity were observed for MSP1-19, MSP8, Pv-fam-a (PlasmoDB: PVX_096,995) and RAMA, with median IgG levels at day 7 greater than 10-fold compared with the seropositivity cut-off. All four of these proteins are used in the classification algorithm based on a combination of antibody responses to eight proteins, with MSP1-19 and RAMA in particular having an important contribution to the classification performance based on a variable-importance plot.9
Figure 1.
IgG antibody levels against 21 P. vivax proteins in patients with clinical P. knowlesi infections
IgG levels were measured against the 21 P. vivax proteins using a multiplexed antibody assay. Individual patients (n = 99) had longitudinal samples obtained and run at the time of diagnosis of P. knowlesi infection (day 0), and days 7 and 28 following enrollment (ACTKNOW cohort). Day 0 has data from 98 samples. Results are expressed as relative antibody units (RAU). All samples were run in singlicate. Proteins are ordered by highest level of median IgG at day 7 compared with the seropositivity cut-off. Dashed lines indicate the malaria-naive negative-control samples (n = 369, MSP3b n = 213): orange, average of the negative control samples; blue, seropositivity cut-off (average plus 2× standard deviation). The box plots indicate the median, 25th, and 75th percentiles with the whiskers showing the 2.5 and 97.5 percentiles. Dots are outliers.
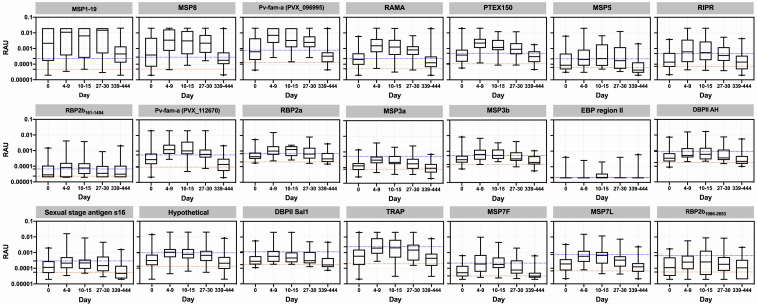
To confirm the results of the ACTKNOW study and to determine the longevity of the cross-reactive antibody response over 1 year (median 369 days; IQR 365–394) post-P. knowlesi infection, we measured IgG antibody levels against the 21 P. vivax proteins in the PACKNOW clinical trial cohort samples (n = 41 patients with up to five time points available, recruited over 2016–2018, at a time of near-elimination of P. vivax in Sabah, Malaysia). These patients were all treated with artemether-lumefantrine. Median parasitemia prior to treatment (day 0) was 2,690 parasites/μL (IQR 572–14,317), with all subjects negative for asexual-stage parasites by the day 7 time point.37,38 There was no significant difference in parasitemia at day 0 between the ACTKNOW and PACKNOW subjects (median 2,857 versus 2,690, respectively; see Table 2). Like the ACTKNOW cohort, we observed a peak in the total IgG response to all 21 P. vivax proteins at days 4–15 post enrollment, with responses generally declining by the day 27–30 time point (Figure 2). Total IgG antibodies against the protein P. vivax MSP1-19 were an exception, with elevated levels maintained at the day 27–30 time point and at 1 year. By 1 year after P. knowlesi infection, the median P. vivax total IgG antibodies had declined to below the seropositivity cut-off for all other proteins (Figure 2). There were fewer P. vivax proteins (n = 11/21), with a peak median total IgG response above the seropositivity cut-off in the PACKNOW samples (collected across 2016–2018) compared with the ACTKNOW samples (collected across 2012–2014; n = 17/21), although this was not statistically significant (Fisher exact test, p = 0.1). There was a significantly higher magnitude total IgG response to 9 of 21 P. vivax proteins in the ACTKNOW compared with PACKNOW cohorts at day 7 (Mann-Whitney U test; see Figure S3). The top four P. vivax proteins with the highest levels of cross-reactivity evident were the same as in the ACTKNOW cohort, being MSP1-19, MSP8, Pv-fam-a (PlasmoDB: PVX_096995), and RAMA (median IgG levels at day 7 more than 6-fold higher than the seropositivity cut-off).
Figure 2.
IgG antibody levels against 21 P. vivax proteins in patients up to 1 year post-clinical P. knowlesi infections
IgG levels were measured against the 21 P. vivax proteins using a multiplexed antibody assay. Samples were obtained and run at the time of P. knowlesi infection (day 0) (n = 41), days 4–9 (n = 35), days 10–15 (n = 15), days 27–30 (n = 33), and days 339–444 (n = 42) following enrollment (PACKNOW cohort). Results are expressed as RAU. All samples were run in singlicate. Proteins are ordered as per Figure 1. Dashed lines indicate the malaria-naive negative-control samples (n = 369, MSP3b n = 213): orange, average of the negative control samples; blue, seropositivity cut-off (average plus 2× standard deviation). The box plots indicate the median, 25th and 75th percentiles with the whiskers showing the 2.5 and 97.5 percentiles. Dots are outliers.
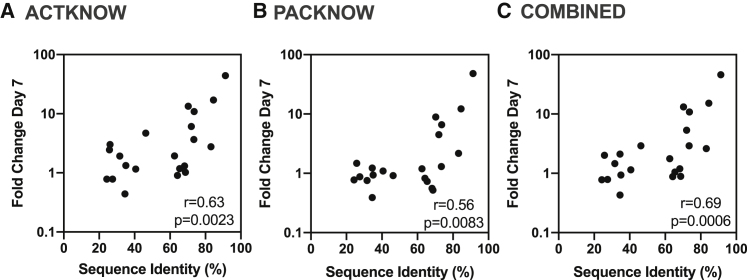
There was a statistically significant correlation between the fold change comparing the median IgG level at the peak at day 7 with the seropositivity cut-off and the percentage sequence identity of the P. knowlesi and P. vivax orthologs for both the ACTKNOW and PACKNOW cohorts (Figures 3A and 3B) (Spearman correlation, r = 0.63, p = 0.0023 and r = 0.56, p = 0.0083, respectively) and using median IgG antibody data from both patient cohorts combined (r = 0.69, p = 0.0006; Figure 3C).
Figure 3.
Correlation between the peak anti-P. vivax IgG level at day 7 and the percent sequence identity of the P. vivax and P. knowlesi orthologs
(A–C) The median IgG level at day 7 (the peak of the response) was divided by the seropositivity cut-off to generate the fold change at the peak compared with the background. The percentage sequence identity was calculated for the protein construct sequence using NCBI BlastP, or the PlasmoDB method when required (see Table 1). A Spearman’s correlation was performed to determine the relationship of the fold change with the sequence identity using data from all 21 P. vivax proteins, for the (A) ACTKNOW r = 0.63, p = 0.0023, (B) PACKNOW cohorts r = 0.56, p = 0.0083, and (C) ACTKNOW and PACKNOW combined (median antibody level of n = 134 P. knowlesi patients at day 7 divided by the seropositivity cut-off) r = 0.69, p = 0.0006.
Classification of recent exposure to P. vivax blood-stage infections in the P. knowlesi cohorts
Since high levels of cross-reactivity (>6- to 10-fold increase over the seropositivity cut-off) were evident among four of our top eight P. vivax proteins in the two series of P. knowlesi clinical case samples (MSP1-19, MSP8, Pv-fam-a (PlasmoDB: PVX_096995), RAMA), we next used these data in our previously developed classification algorithm to determine whether any of the samples from patients with current or recent P. knowlesi infections were classified as recently exposed to P. vivax. The top eight P. vivax proteins used in the classification algorithm were: MSP1-19, MSP8, Pv-fam-a (PlasmoDB: PVX_096995), RAMA, RBP2b161–1454, Pv-fam-a (PlasmoDB: PVX_112670), MSP3a, and EBP. We utilized a balanced sensitivity and specificity target of 79%. The Random Forest classification algorithm was trained using existing datasets from prospective longitudinal cohorts in Brazil, Thailand, and the Solomon Islands.9 In these studies, the prevalence of P. vivax infection was assessed monthly via qPCR for the year-long duration of the cohort, and antibody responses were measured at the final time point. This allowed a detailed characterization of the association between antibody responses and the time since previous P. vivax infection. We ran the algorithm on both the ACTKNOW and PACKNOW cohorts at all time points for which we had data, and the proportion of patients classified as positive by the algorithm varied between 17% and 82% (Table 3). The trend was for the highest numbers of P. knowlesi patients to be classified as recently exposed to P. vivax at the day 7 time point in both cohorts (82% ACTKNOW, 77% PACKNOW), in line with the highest peak IgG activity. The lowest proportion of P. knowlesi patients classified as positive was for the PACKNOW cohort at day 365 (7/42, 17%).
Table 3.
Output from P. vivax classifier using IgG antibody responses from (1) top eight P. vivax serological exposure markers and (2) two adjusted panels of eight P. vivax serological exposure markers with low levels of P. knowlesi-cross-reactivity
| Samples | Total N | Classified positive legacy 8, number (%) |
Classified positive modified 8, with RBP2b1986–2653, number (%) |
Classified positive modified 8, with RBP2b161–1454, number (%) |
|---|---|---|---|---|
| ACTKNOW | ||||
| Day 0 | 98 | 47 (48.0%) | 26 (26.5%) | 36 (36.7%) |
| Day 7 | 99 | 81 (81.8%) | 69 (69.7%) | 67 (67.7%) |
| Day 28 | 99 | 68 (68.7%) | 43 (43.4%) | 49 (49.5%) |
| PACKNOW | ||||
| Day 0 | 41 | 21 (51.2%) | 12 (29.3%) | 12 (29.3%) |
| Day 7 | 35 | 27 (77.1%) | 28 (80.0%) | 25 (71.4%) |
| Day 14 | 15 | 11 (73.3%) | 10 (66.7%) | 8 (53.3%) |
| Day 28 | 33 | 20 (60.6%) | 19 (57.8%) | 14 (42.4%) |
| Day 365 | 42 | 7 (16.7%) | 3 (7.1%) | 4 (9.5%) |
A classification of previous exposure was taken when predicted probability was greater than a cut off corresponding to the respective sensitivity and specificity targets of 79% and 79% (legacy), 72.4% and 70.8% (RBP2b1986–2653), or 78.7% and 76.9% (RBP2b161-–1454).
For the ACTKNOW cohort, of the 99 patients, 46 were classified positive by the algorithm at all three time points; 20 were classified positive at day 7 and day 28, 14 at day 7 only, two at day 28 only, and one at day 0 and day 7. Sixteen patients were not classified as positive at any time point. Although there was variability in the available plasma samples post-enrollment for the PACKNOW cohort, nine patients were not classified positive at any time point, and all those who were positive at a later time point (day 28 or day 365) were previously positive for at least one of the earlier time points (i.e., day 0, 7, or 14). The exceptions were one patient who was positive at only day 28 and one patient positive only at day 365 (the latter had notably high antibody levels at day 365, reaching equivalent of a 1/50 dilution of the positive-control pool against four of eight antigens).
Design and application of an adjusted P. vivax-specific serological marker panel
In both the ACTKNOW and PACKNOW P. knowlesi clinical cohorts, more than one-half the patients at day 7 and day 28 post-infection were classified as recently exposed to P. vivax blood-stage parasites by using our existing algorithm with our set of eight P. vivax serological exposure markers. This indicated that our current serological exposure marker panel was not P. vivax specific. We hypothesized that using proteins with low levels of cross-reactivity in the P. knowlesi-infected patients would reduce misclassification. We thus designed a modified panel of eight P. vivax serological exposure markers by selecting the proteins with the lowest levels of cross-reactivity in the P. knowlesi clinical samples (as determined by the median IgG level at day 7 from the ACTKNOW sample set compared with the background, as described in Figure 3). These proteins were the following: RBP2b1986–2653, MSP7L, MSP7F, TRAP, DBPII Sal1, hypothetical (PlasmoDB: PVX_097715), sexual-stage antigen s16, and EBP (all with a fold change of < 2). DBPII AH was not selected, given that this is a variant of the DBP region II that was already included. We re-trained the algorithm on our existing Thai, Brazilian, and Solomon Islands datasets9 for these eight P. vivax proteins. Youden’s J was used to determine the best trade-off balancing sensitivity and specificity, assuming equal importance of false positives and false negatives in the classification process. The algorithm trained with the modified panel of eight P. vivax proteins achieved 72.4% sensitivity and 70.8% specificity respectively, for classifying recent P. vivax infections within the prior 9 months using the existing Thai, Brazilian, and Solomon Islands datasets (Figure S4A). When applied to confirmed P. knowlesi clinical cases, we identified a reduced number of 210 of 462 (45%) samples being classed as recently exposed to P. vivax (Table 3) (compared with 281 of 462 (61%) using the original panel). Misclassification was still high at day 7 post-P. knowlesi infection (69.7% ACTKNOW and 80% PACKNOW) but less than 58% at day 28 and 7% at day 365. The greatest reduction in numbers and proportions of P. knowlesi patients classified positive by the algorithm was for the ACTKNOW cohort.
Although the modified combination of eight P. vivax proteins reduced overall misclassification of P. knowlesi clinical cases, the sensitivity and specificity for correctly classifying recent P. vivax infections (72.4%, 70.8%) was substantially lower than the original eight (80% for both9). We hypothesized that this was because the modified panel did not include the top classifier RBP2b161–1454 (see Figure S4 and Longley et al., 20209). As this protein also had relatively low levels of cross-reactivity in the P. knowlesi samples at day 7 (fold change 2.9 compared with the seropositivity cut-off), we replaced RBP2b1986–2653 in the modified panel with RBP2b161–1454. This improved classification of recent P. vivax infections in the Thai, Brazilian and Solomon Islands datasets to 78.7% sensitivity and 76.9% specificity (Figure S4B), which is more comparable with the original classifier. Applying this trained classification algorithm to the P. knowlesi sample sets resulted in 215 of 462 (47%) samples being misclassified (Table 3), lower than when the original algorithm (281) was used and similar to the 210 misclassified when RBP2b1986–2653 was used in the modified panel (modified algorithm RBP2b1986–2653 vs RBP2b161–1454, p = 0.79, Fisher exact test). At day 7 post-P. knowlesi infection 68%–71% were classified as recently exposed to P. vivax, reducing to 42%-50% at day 28 and 10% at day 365. Those patients positive at day 365 were also classified positive at earlier time points.
Association of peak antibody levels with age and P. knowlesi parasitemia
Antibody levels are commonly positively associated with age (due to both being a proxy for exposure and age itself) and sometimes with the antigenic input as measured by blood-stage parasitemia. We further explored whether the cross-reactive antibodies identified at day 7 were associated with either age or P. knowlesi parasitemia (as recorded at enrollment) through regression analyses. This analysis was performed using peak antibody levels at day 7 from both cohorts (n = 134). There was a weak positive association with age (range 3–78 years) for 11 of 21 P. vivax antigens (Table 4). In prior studies of P. knowlesi malaria, age was associated with parasitemia;41,42 thus, we also adjusted the age analyses for parasitemia, and the findings remained the same (Table 4). Parasitemia ranged from 56 to 185,553 parasites/μL, and there was no association between P. knowlesi parasitemia and peak antibody levels at day 7 for 19 of 21 P. vivax antigens (Table S2). For DBPII AH there was a negative association between parasitemia and antibody levels, whereas for MSP7F there was a positive association between parasitemia and antibody levels. There were also higher antibody levels in females compared with males for four proteins (MSP5, MSP3b, MSP3a, and DBPII AH; Table S2); three of these also had a significant positive association with age, which remained after adjustment for sex. No pattern was observed between these associations with the ranking of antigens by cross-reactivity (Table 4).
Table 4.
Associations between peak IgG anti-P. vivax antibody levels at day 7 in both P. knowlesi cohorts combined with age
| Protein | Fold Δ IgG | Age (unadjusted) |
Age (adjusted) |
||
|---|---|---|---|---|---|
| Coefficient (95% CI) | p value | Coefficient (95% CI) | p value | ||
| MSP1-19 | 43.98 | 0.012 (0.0052–0.02) | 0.001 | 0.012 (0.0052–0.020) | 0.001 |
| MSP8 | 17.07 | 0.005 (−0.0012–0.011) | 0.11 | 0.005 (−0.0012–0.011) | 0.111 |
| Pv-fam-a (PVX_096,995) | 13.39 | 0.0047 (−0.00062–0.01) | 0.083 | 0.0047 (−0.00064–0.01) | 0.084 |
| RAMA | 10.86 | 0.0046 (−0.0021–0.011) | 0.175 | 0.0046 (−0.0021–0.011) | 0.175 |
| PTEX150 | 6.08 | 0.013 (0.0053–0.020) | 0.001 | 0.013 (0.0053–0.02) | 0.001 |
| MSP5 | 4.70 | 0.021 (0.013–0.029) | <0.0001 | 0.021 (0.013–0.029) | <0.0001 |
| RIPR | 3.66 | 0.014 (0.0073–0.021) | <0.0001 | 0.014 (0.0073–0.0211) | <0.0001 |
| RBP2b161–1454 | 3.00 | 0.0077 (0.0012–0.014) | 0.02 | 0.0077 (0.0011–0.014) | 0.021 |
| Pv-fam-a (PVX_112,670) | 2.76 | −0.0048 (−0.01–0.0036) | 0.068 | −0.0048 (−0.01–0.00038) | 0.069 |
| RBP2a | 2.45 | 0.012 (0.0071–0.018) | <0.0001 | 0.012 (0.007–0.018) | <0.0001 |
| MSP3b | 1.93 | 0.0064 (0.0014–0.011) | 0.013 | 0.0064 (0.0013–0.012) | 0.014 |
| MSP3a | 1.92 | 0.015 (0.0088–0.021) | <0.0001 | 0.015 (0.0088–0.021) | <0.0001 |
| EBP | 1.33 | 0.012 (0.0072–0.016) | <0.0001 | 0.012 (0.0072–0.016) | <0.0001 |
| DBPII AH | 1.31 | 0.0044 (−0.00086–0.0097) | 0.1 | 0.0044 (−0.0008-0.0096) | 0.099 |
| S16 | 1.18 | 0.0061 (0.00020–0.012) | 0.043 | 0.0061 (0.00018–0.012) | 0.044 |
| Hypothetical | 1.16 | 0.0038 (−0.0028–0.010) | 0.258 | 0.0038 (−0.0028–0.01) | 0.26 |
| DBPII Sal1 | 1.02 | 0.0051 (−0.001–0.011) | 0.102 | 0.0051 (−0.001–0.011) | 0.102 |
| TRAP | 0.90 | 0.007 (0.0014–0.013) | 0.015 | 0.007 (0.014–0.013) | 0.015 |
| MSP7L | 0.78 | 0.0057 (−0.00012–0.012) | 0.055 | 0.0057 (−0.00013–0.0115) | 0.055 |
| MSP7F | 0.78 | 0.0066 (−0.0029–0.013) | 0.06 | 0.0066 (−0.0025–0.013) | 0.059 |
| RBP2b1986–2653 | 0.44 | 0.0042 (−0.0063–0.012) | 0.266 | 0.0042 (−0.0033–0.118) | 0.272 |
Regression analyses were performed univariably and adjusted with parasitemia (n = 134). Antigens are ordered by the fold change in the peak antibody level at day 7 in the ACTKNOW cohort compared with the seropositivity cut-off based on the negative-control samples (=Fold Δ IgG). CI, confidence interval.
Discussion
P. vivax serological markers, which reflect recent exposure to blood-stage P. vivax parasites and can indirectly identify hypnozoite carriers, could play an important role in accelerating malaria elimination programs. Application of seroTAT (the treatment of individuals identified as recently exposed) paves the way for reducing the large transmission reservoir caused by P. vivax hypnozoites while resulting in substantially less over-treatment compared with one alternative, mass drug administration. In this study, we aimed to identify whether cross-reactive antibodies against our P. vivax serological markers were present in patients with recent P. knowlesi infections. Using serial plasma samples from two cohorts of patients with P. knowlesi malaria, we found that cross-reactive antibodies are induced against most of the P. vivax proteins assessed. A statistically significant association was seen with the level of sequence identity between the P. vivax and P. knowlesi orthologs. The peak cross-reactive antibody response was at day 7 following infection, which is the same timing as the peak P. knowlesi-antibody response reported previously using a similar sample set from Sabah, Malaysia.28 This is also in line with the timing of peak IgG antibody responses reported for other species such as P. falciparum and P. vivax (within the first 2 weeks post-treatment).43, 44, 45 Antibodies against the P. vivax proteins had reduced in magnitude by day 28 post-infection, and the median IgG levels were below the seropositivity cut-off for all but one P. vivax protein (MSP1-19) at 1 year post-P. knowlesi infection.
The two cohorts of P. knowlesi clinical case samples used here were obtained during different time periods, 2012–2014 and 2016–2018. The incidence of P. vivax infections in Sabah, Malaysia, in 2012–2014 was modest, and transmission had declined even further by 2016–2018.39 Therefore, the risk that the P. knowlesi-infected patients had recent past exposure to P. vivax was low in both cohorts (notably, 14 patients were retrospectively excluded from the ACTKNOW study due to P. vivax infections detected by PCR, whereas only one was excluded from the PACKNOW study; see STAR Methods). The number of P. vivax proteins with antibody levels above the seropositive cut-off at the peak of the response was higher in the ACTKNOW cohort in 2012–2014 at 80.95% (17/21) compared with 52.38% (11/21) in the PACKNOW cohort in 2016–2018 (this difference was not statistically significant). There was also a significantly higher magnitude of the IgG response in the ACTKNOW cohort compared with the PACKNOW cohort for 9 of 21 P. vivax proteins at day 7. There was no significant difference in age between the two cohorts or in parasitemia at enrollment (Table 2). Overall, there were still anti-P. vivax antibody responses present in the later PACKNOW cohort; thus, our interpretation of these responses as cross-reactive is by far the most likely explanation, given the evidence (rather than recent past exposure to P. vivax infections). Furthermore, the P. vivax protein RBP2b is the top serological exposure marker and is highly accurate at predicting recent P. vivax infections even when used alone.9 The lack of antibody responses detected against RBP2b in the P. knowlesi-clinical cohorts suggests that it is unlikely that these patients also had recent P. vivax infections.
Our findings are in line with, but extend, past research. An extensive study of P. vivax-P. knowlesi antibody cross-reactivity to 19 blood-stage proteins was recently published,27 which included four of the proteins we assessed in the current study (MSP1, MSP8, DBP, and RAMA). Those authors generated rabbit antibodies against the 19 P. vivax proteins and found that they recognized P. knowlesi blood-stage parasites through immunofluorescence assays and were able to inhibit invasion of P. knowlesi into erythrocytes in vitro.27 Naturally acquired antibodies from individuals living in endemic areas were also analyzed, with evidence of cross-reactive antibodies from either of P. vivax or P. knowlesi patients against P. vivax or P. knowlesi recombinant proteins (building upon the authors’ prior work on one P. vivax protein, apical asparagine-rich protein46). Of relevance to our study, they found significant levels of reactivity in P. knowlesi patient sera against P. vivax MSP8,27 which is supported by our current data. The 19 P. vivax proteins assessed by Muh and colleagues had high levels of sequence identity with their P. knowlesi orthologs (>58.2%),27 with the exception of two RBP proteins (1a and 1b, 22.3% and 26.9%, respectively), and this could explain the universal cross-reactivity they identified. In contrast, our panel of P. vivax proteins had a large spread of sequence identity with their P. knowlesi orthologs, with nine proteins having less than 50% sequence identity, which enabled us to demonstrate the positive relationship between sequence identity and antibody cross-reactivity. A lower level of sequence identity comes with a decrease in the likelihood of cross-reactive continuous linear epitopes, which is in support of our finding. Further work would need to be undertaken to determine how lower sequence identity affects discontinuous conformational epitopes. Overall, we found that the presence and/or level of cross-reactivity is antigen specific. Our work further extends existing findings by demonstrating that cross-reactive antibody responses in P. knowlesi patients against P. vivax proteins are short-lived.
The most cross-reactive protein in our panel was MSP1-19 (>40-fold increase at day 7 compared with seropositivity cut-off). This magnitude of response is equivalent to that reached against a P. knowlesi-specific protein (PkSERA antigen 3) in a subset of these same P. knowlesi patient samples (50-fold at day 7 compared with day 0).28 Previous assessment of antibody responses against MSP1-19 in P. falciparum, P. vivax, P. malariae, and P. ovale (but notably did not include assessment of P. knowlesi) indicated a highly species-specific response.16 However, high-levels of cross-reactivity for MSP1-19 between P. vivax and P. knowlesi (>81% sequence identity between the Sal1 and H strains, respectively) have been reported in other studies in line with our work.27,28 P. vivax MSP1-19 was a vaccine candidate,47 but has not progressed to clinical trials.48 There remains interest in this candidate,49,50 and the high levels of cross-reactivity in our study with P. knowlesi suggest that potential cross-species immunity could be possible.15 Recent work has used P. knowlesi as a model for screening inhibitory activity of P. vivax antibodies.51 Polyclonal antibodies against the P. vivax proteins DBP, two 6-cysteine proteins (Pv12, Pv14), and the GPI-anchored micronemal antigen (GAMA) were able to inhibit invasion of wild-type P. knowlesi, demonstrating that cross-species functional immunity is possible (at least in vitro). No inhibition was evident against wild-type P. knowlesi when antibodies against P. vivax MSP3a and MSP7L were used, which is consistent with our findings of minimal cross-reactivity in P. knowlesi patients for these proteins. In addition to MSP1-19, we observed high levels of potential cross-reactive antibody responses against the P. vivax proteins MSP8, Pv-fam-a (PlasmoDB: PVX_096,995), and RAMA. These antigens could thus be considered for assessment of growth-inhibitory activity in the P. knowlesi model51 and considered as potential targets for cross-species protection. This is an important consideration, given the currently limited landscape of vaccine development specifically for P. knowlesi.52 In some previously co-endemic areas of Southeast Asia, the rise in incidence of symptomatic P. knowlesi malaria has followed the decline in P. vivax malaria, supporting the idea for development of a P. vivax vaccine that provides cross-reactive immunity against P. knowlesi.52 To date, there has been limited assessment of these three P. vivax proteins as vaccine candidates. Unfortunately, several of the most promising P. vivax vaccine candidates (including RBP2b, EBP, and MSP3a53) have low sequence identity with their P. knowlesi orthologs and limited evidence of cross-reactivity in our study. Although not an objective of our current study, we identified interesting associations between infection-related variables and antibodies against P. vivax DBPII, including shorter fever duration and lower parasitemia in P. knowlesi patients with higher antibodies. This suggests some effect of pre-existing immunity leading to a better ability to control the infection and thus warrants further attention.
All four of the most highly cross-reactive proteins are within our top eight panel of P. vivax serological exposure markers.9 However, levels of antibodies against our top serological marker, RBP2b (N- and C-terminal constructs), were low. When we applied our previously trained classification algorithm to the current P. knowlesi clinical datasets, 77%–82% of P. knowlesi patients at the peak antibody time point (day 7) in each cohort were classified as recently exposed to P. vivax blood-stage parasites in the past 9 months. This was reduced to 61%–69% at day 28 and 17% at day 365. As mentioned, at the time of the PACKNOW cohort (2016–2018), local P. vivax transmission in Sabah, Malaysia, had approached elimination, and nearly all malaria was from P. knowlesi.39 Thus, in this cohort there is less risk that the anti-P. vivax antibody responses measured are due to past exposure to P. vivax, and a lower proportion of P. knowlesi patients were accordingly classified as recently exposed compared with the earlier ACTKNOW cohort. However, 77% of PACKNOW patients at day 7 were classified as recently exposed to P. vivax, and thus, the substantial cross-reactivity identified against four of our top eight proteins has resulted in this (likely) misclassification. It is important to consider this level of misclassification in areas endemic for both P. vivax and P. knowlesi in the context of how the serological exposure markers will be applied.
Multiple-use cases for P. vivax serological markers exist.12 For an intervention, such as seroTAT, even with potential misclassification due to recent exposure to P. knowlesi, the serological markers would still outperform mass drug administration in terms of specificity. Hence, in co-endemic areas both the adjusted panel and the legacy panel would likely have significant benefit for accelerating elimination efforts. For surveillance purposes, such as identifying clusters of infections or levels of residual transmission to better target the limited resources of malaria control programs, it will be important to define whether the recent exposure was due to P. vivax or P. knowlesi. We therefore assessed performance of an adjusted panel of eight P. vivax serological markers, which were selected due to low levels of cross-reactivity in the P. knowlesi clinical cases. Suboptimal performance was initially observed, but after substitution of one fragment of RBP2b1986–2653 for the top-performing RBP2b161–1454, this modified panel of eight markers was still able to classify recent P. vivax infections with 78.7% sensitivity and 76.9% specificity while reducing the misclassification of recent P. knowlesi infections to 68%–71% at the peak day 7 time point. By day 28, this was <50% and importantly at 1 year post-P. knowlesi infection, 10%. This provides proof of principle that an adjusted panel of serological markers could be applied for use in co-endemic areas, but further optimization would be needed to completely reduce the risk of misclassifying recent P. knowlesi infections. This residual cross-reactivity at 1 year may assume greater importance for sero-surveillance purposes, as the proportion of all Plasmodium infections due to P. knowlesi rises with P. vivax elimination strategies in co-endemic areas.52 It would be useful to have plasma samples collected between 1 and 12 months post-P. knowlesi infection to better understand the dynamics of the cross-reactive antibody response. In addition, while we used a large and population-diverse panel of negative controls to generate seropositivity cut-offs and for training the algorithm, it would be beneficial to screen population-matched malaria-naive controls from a malaria-free area of Malaysia.
An alternative approach would be to include a panel of P. knowlesi-specific markers into the P. vivax serological assay, which could be cross-referenced to inform on recent P. knowlesi exposure. Recently a panel of recombinant P. knowlesi proteins has been developed for use as serological tools, with care being taken in their design to avoid regions of sequence with high levels of identity with other Plasmodium species.28 While further validation of species specificity is required, at least one protein (P. knowlesi serine repeat antigen 3 antigen 2) was able to identify P. knowlesi exposed individuals.28 Further work could also assess the use of other Ig isotypes or IgG subclasses detected against the P. vivax serological exposure markers, instead of total IgG, as another avenue for reducing misclassification at later time points post-P. knowlesi exposure, given the possible variability in their longevity.43,54 It may also be possible to design smaller P. vivax protein fragments of the top markers that have very low sequence identity with their P. knowlesi orthologs.
Our study provides important evidence of antibody cross-reactivity against P. vivax proteins in patients with P. knowlesi infections that are relatively short-lived and provides an approach for reducing the potential misclassification of Plasmodium species by serology in co-endemic areas. These data will be useful when P. vivax sero-surveillance results in settings co-endemic for P. knowlesi are being interpreted. It will be important to expand upon our findings to assess potential cross-reactivity with other zoonotic55 and non-zoonotic Plasmodium species, even though for the latter the levels are expected to be lower due to the further divergence in genetic relatedness with other species.14 Further assessment of recently developed P. knowlesi markers of exposure28 in P. vivax-endemic areas is warranted to confirm limited cross-reactivity for those specific proteins between the two species. We have demonstrated that it is possible to reduce cross-reactivity (i.e., improve specificity) without a loss in sensitivity through careful selection of antigens. The adjusted panel is thus well suited for use in co-endemic areas, particularly for seroTAT, although further improvements could be made to reduce cross-reactivity due to recent P. knowlesi infections. For sero-surveillance purposes, further work needs to be undertaken with a focus on either improving antigen selection and optimization for low levels of cross-reactivity or by supplementing the P. vivax panel with P. knowlesi antigens. For implementation of P. vivax serological exposure markers for seroTAT, further development of the assay into a point-of-care device is ongoing, along with protein optimization to potentially improve the sensitivity and specificity of the assay. We have demonstrated that cross-reactivity is a parameter that can be optimized and will result in a reduction in over-treatment. Alongside these laboratory and product development goals, a modeling approach is being applied to determine the benefits and risks, the ideal balance of sensitivity and specificity, and the schedule and frequency of screening and treatment required for effectively reducing P. vivax transmission.56
Limitations of the study
The P. knowlesi-infected individuals came from Sabah, Malaysia, where low levels of P. vivax transmission remains. Therefore, despite the low risk, a limitation of our study is that we cannot exclude the possibility of past P. vivax exposure. A limitation for understanding antibody kinetics or decay over time was that we did not have samples available at time points between 1 and 12 months post-P. knowlesi infection. Our Random Forest classification algorithm was trained on prior datasets that used a similar but different laboratory assay set-up, i.e., non-magnetic beads and a Luminex-200 instrument versus magnetic beads and a MAGPIX instrument. Data from these two assay platforms are well correlated,57 but potentially an improvement could be seen if the algorithm were retrained on magnetic-bead data and with inclusion of a population-matched negative-control sample set such as samples from a non-endemic area of Malaysia.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE-conjugated Donkey F(ab)2 anti-human IgG | Jackson Immunoresearch | JIR 709-116-098 RRID:AB_2340519 |
| Chemicals, peptides, and recombinant proteins | ||
| 15 P. vivax proteins expressed in WGCF | CellFree Sciences | |
| 2 P. vivax proteins expressed in E. coli | Wai-Hong Tham, WEHI | |
| 1 P vivax protein expressed in E. coli | Julie Healer, WEHI | |
| 3 P. vivax proteins expressed in E. coli | Chetan Chitnis, Institut Pasteur | |
| Magnetic COOH beads | BioRad | 171,506(xxx) |
| sulfo-N-hydroxysuccinimide (S-NHS) | Sigma | 56485 |
| N-ethyl-N′-(3-(dimethylamino)propyl)carbodiimide (EDC) | Sigma | 3449 |
| Bovine Serum Albumin (BSA) | Sigma | A7906 |
| Software and algorithms | ||
| PlasmoDB | Amos et al., 2022 | plasmodb.org/ |
| NCBI BlastP | Camacho et al., 2009 | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| EMBOSS Needle | Needleman et al., 1970 | https://www.ebi.ac.uk/Tools/psa/emboss_needle/ |
| Random Forest classification algorithm | Longley et al., 2020 | https://github.com/MWhite-InstitutPasteur/Pvivax_sero_dx |
| Random Forest classification algorithm RShiny App | Chotirat et al., 2021 | https://gitlab.pasteur.fr/tobadia/pvserotat-rshiny-app |
| Other | ||
| MAGPIX Instrument | Luminex | https://www.luminexcorp.com/magpix-system/#overview |
Resource availability
Lead contact
Further information and requests for resources and reagents should be direct to and will be fulfilled by the lead contact, Dr Rhea Longley (Longley.r@wehi.edu.au).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Ethical approvals
Samples from two clinical trials conducted in Sabah, Malaysia, were used for this study (ACTKNOW36 and PACKNOW37,38). Ethical approvals for these studies were obtained from the Human Research Ethics Committee at the Menzies School of Health Research, Darwin, Australia (approval numbers 2012-1815 and 16–2544) and the Medical Research and Ethics Committee of the Ministry of Health Malaysia, Malaysia (approval numbers NMRR-12-89-11,005 and NMRR-16-29,088). All patients gave written informed consent or an attending relative gave informed consent. Ethical approval was provided by the Human Research Ethics Committee at the Walter and Eliza Hall Institute of Medical Research for use of the Malaysian and negative control samples in Melbourne (approval number 14/02).
Study samples: P. knowlesi patients and negative controls
Longitudinal plasma samples were collected from patients enrolled in two P. knowlesi clinical trials conducted in Sabah, Malaysia, denoted as ACTKNOW and PACKNOW, and four panels of malaria-naïve negative controls.
The ACTKNOW study recruited patients over 2012–2014, with the aim of comparing artesunate-mefloquine versus chloroquine for the treatment of acute uncomplicated P. knowlesi malaria36 (clinicaltrials.gov registration number NCT01708876). Patients were recruited from three hospitals (Kudat, Kota Marudu, Pitas) and were eligible for inclusion if they had microscopic diagnosis of P. knowlesi monoinfection, were more than 1 year of age, more than 10kg, had a negative P. falciparum rapid diagnostic test, and had a fever (≤37.5°C) or history of fever in the past 48 h. Patients who were not confirmed by PCR to have P. knowlesi monoinfection were retrospectively excluded.36 Of those excluded, 14 were due to P. vivax infections detected upon PCR. The duration of self-reported history of fever (days) was recorded. Patients were treated as previously described,36 with approximately half randomised to receive chloroquine and half to receive artemisinin-based combination therapy. Plasma samples were available for the current study from 99 patients at day 0 (time of enrollment), day 7 and day 28. Data is presented from 98 samples at day 0 for 20/21 P. vivax proteins due to one sample failing quality control of the antibody data, and the incorrect sample being repeated by mistake. See Table 2 for patient demographic details include age and gender.
The PACKNOW study recruited patients over 2016–2018, with the aim of comparing regularly dosed paracetamol versus no paracetamol on renal function in P. knowlesi malaria37,38 (clinicaltrials.gov registration number NCT03056391). All patients received antimalarial drug treatment with artemether-lumefantrine. Patients were recruited from four hospitals (Queen Elizabeth Hospital, Keningau, Ranau, Kota Marudu) and were eligible for inclusion if they had a microscopic diagnosis of P. knowlesi monoinfection, fever (≥38°C) on admission or during the preceding 48 h, and were more than 5 years of age. Patients were enrolled within 18 h of commencing antimalarial treatment.37,38 Patients who were not confirmed by PCR to have P. knowlesi monoinfection were retrospectively excluded, using a validated PCR assay.58 In contrast to the ACTKNOW study, only one participant was excluded due to P. vivax infection detected by PCR (unpublished data). Plasma samples were available for the current study from 41 patients at day 0 (time of enrollment), 7, 14, 28 and 365 (these exact time-points were not always available, exact days post-enrolment are stated in the results and relevant figure legends). See Table 2 for patient demographic details include age and gender.
For both the ACTKNOW and PACKNOW cohorts, all patients were PCR-negative for P. vivax co-infection at enrollment, however it was unknown whether the P. knowlesi patients had past exposure to P. vivax parasites. P. vivax has been endemic in Sabah, Malaysia, but the number of malaria cases due to P. vivax has substantially declined from 2009–2017 (whilst the number of cases due to P. knowlesi has risen).39 In line, the number of patients retrospectively excluded from these studies due to P. vivax infection detected by PCR was 14 in ACTKNOW and only 1 in PACKNOW. All patient data variables are provided in Data S1.
We utilised four panels of malaria-naïve negative control plasma samples as previously described.9 Briefly, this included 102 individuals from the Volunteer Blood Donor Registry in Melbourne, Australia; 100 individuals from the Australian Red Cross, Melbourne, Australia; 72 individuals from the Thai Red Cross, Bangkok, Thailand; and 96 individuals from the Rio de Janeiro State Blood Bank, Rio de Janeiro, Brazil.
Method details
P. vivax proteins
21 P. vivax proteins were selected for assessment of antibody responses due to their ability to classify individuals as recently exposed to P. vivax parasites within the prior 9-month, as previously described.9 These included eight P. vivax proteins that, together, can classify recent P. vivax infections with 80% sensitivity and 80% specificity. The remainder of the proteins were selected due to their individual ability to accurately classify recent P. vivax infections, as described,11 or to provide further information on particular proteins (i.e. DBPII Sal1 was included to complement DBPII AH, and MSP3b was included to complement MSP3a). Details on the 21 P. vivax proteins, including the sequence construct and expression system, are provided in Table S1. In this manuscript we have referred to proteins by their annotation, with the PlasmoDB59 (plasmodb.org/) code in brackets if necessary. Proteins were expressed as previously described9 either by 1) dialysis-based refeeding wheat germ cellfree methods with purification by affinity matrix using a His-tag or 2) E. coli (Table S2). All constructs were based on the reference Sal1 strain except for DBPII where two strains were used (Sal1 and AH). Notations for MSP7 were from Garzón-Ospina 201660 and MSP3 from Kuamsab 2020.61
Sequence comparisons between the P. vivax proteins with their P. knowlesi orthologs were performed by identifying the orthologs in PlasmoDB or through NCBI BlastP.62 Only H strain and A1H1 line orthologs were selected using PlasmoDB whereas NCBI BlastP included all P. knowlesi strains. Sequence identity and similarity percentages were obtained using the EMBOSS Needle protein alignment tool (https://www.ebi.ac.uk/Tools/psa/emboss_needle/)35 or NCBI Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) directly. PlasmoDB comparisons were made between full-length protein sequences whereas NCBI Blast used the protein sequences of expression constructs as the query sequence.
Total IgG antibody assay
We used a multiplexed assay based on Luminex xMAP technology to measure total IgG antibody responses to the 21 P. vivax proteins. This assay was run on a MAGPIX instrument, as previously described.57 Briefly, the 21 P. vivax proteins were coupled to individual sets of internally labeled magnetic COOH beads at previously determined optimal concentrations (see Table S1) following standard methods.57 The magnetic COOH beads were first pre-activated in 100mM monobasic sodium phosphate using 50 mg/mL sulfo-NHS and 50 mg/mL EDC, then the P. vivax proteins were added in the amounts provided in Table S1 and incubated either for 2 h at room temperature or overnight at 4°C. The coupled beads were washed and stored in 1x PBS with 0.1% BSA, 0.02% Tween 20 and 0.05% sodium-azide (pH 7.4) at 4°C until use.
Antigen-specific total IgG was detected in plasma samples by incubating 500 coupled beads of each antigen per well with plasma diluted at 1:100 in PBT (1x PBS with 1% BSA and 0.05% Tween 20), followed by the addition of 0.5 mg/mL PE-conjugated Donkey F(ab)2 anti-human IgG (JIR 709-116-098). On each plate, a 2-fold serial dilution from 1/50 to 1/25,600 of a positive control hyperimmune plasma pool (generated from adults from PNG, a non-knowlesi-endemic region) was included. At least 15 beads per region were then acquired and read on a MAGPIX instrument as per the manufacturer’s instructions, with results expressed as the mean fluorescent intensity (MFI). Several datapoints were excluded for the negative control panels for MSP3b due to not passing quality control, final n = 213 (MSP3b only). All serology data is available in Data S2 and S3. Each sample was run in singlicate.
Existing datasets of total IgG antibodies against the same P. vivax proteins in three observational cohort studies (in P. vivax-endemic areas) were also used.9 These data were generated as previously described9 using a similar multiplexed assay, but with non-magnetic beads and run on a Luminex-200 instrument. Briefly, yearlong cohort studies were conducted in Thailand (Kanchanaburi and Ratchaburi provinces), and in non-knowlesi endemic Brazil (Manaus) and the Solomon Islands (Ngella) across 2013–2014. Each site enrolled between 928 and 1,274 individuals with blood samples taken every month for qPCR detection of P. vivax infections and plasma stored for antibody measurements. This enabled total IgG antibody responses measured at the final visit of the yearlong cohorts to be related to the time since prior P. vivax infection. These existing datasets are available here: https://github.com/MWhite-InstitutPasteur/Pvivax_sero_dx.
Quantification and statistical analysis
Statistical analysis
The raw MFI results were converted to relative antibody units (RAU) using protein-specific standard curve data, as previously described.57 A five-parameter logistic function was used to obtain an equivalent dilution value (expressed as the RAU) compared to the PNG control plasma, with extrapolation one step beyond the lowest dilution resulting in a range of values from 1.95 × 10−5 to 0.02 RAU. The standard curve conversion was performed in R version 4.1.1. Spearman’s r correlations were performed to correlate sequence identity with relative cross-reactivity levels. Spearman r values < 0.3 were considered weak, 0.3–0.7 moderate, and >0.7 strong correlations. Mann-Whitney U tests were used to compare antibody levels between various patient variables, or to compare antibody levels across cohorts. Fisher’s exact tests were used to compare categorical variables or outcomes. Correlations, Mann-Whitney U tests and Fisher’s exact tests were performed in Prism version 9. Linear regression analyses to assess associations between peak antibody levels with age, gender and parasitaemia were performed in Stata version 12. Antibody levels were first log10 transformed to better fit a normal distribution.
Classification of recent P. vivax infections within the prior 9-month was performed using a Random Forest classification algorithm. The algorithm used antibody data against the top eight P. vivax serological exposure markers that we had previously identified,9 and was trained with four existing datasets from our previous work (Thailand n = 826, Brazil n = 925, Solomon Islands n = 754 and negative controls n = 274, using non-magnetic beads and a Luminex-200 instrument).9 The top eight proteins were: MSP1-19, Pv-fam-a (PVX_096995), Pv-fam-a (PVX_112670), RAMA, MSP8, MSP3a, RBP2b161-1454, and EBP. A diagnostic target of 79% specificity and 79% sensitivity was selected.
Two modified sets of eight proteins were also tested in the classification algorithm, based on low cross-reactivity in the P. knowlesi clinical patient samples. A random forest classification algorithm was trained, using existing antibody data generated against the modified sets of eight proteins in our Thai, Brazilian and Solomon Islands studies (using non-magnetic beads and a Luminex-200 instrument).9 The classification algorithm was cross-validated using 1,000 randomly sampled, disjoint training and testing subsets, and the data presented in a receiver operating characteristic curve (Figure S4), with credible intervals corresponding to percentiles calculated among replicates. At every iteration, 2/3 of each cohort was used for training and 1/3 for testing. This newly modified and trained random forest algorithm was subsequently used to classify the samples from the P. knowlesi clinical cohorts as recently exposed to P. vivax blood-stage parasites in the prior 9-months, or not, with the optimal balanced diagnostic target that was achievable as per Youden’s J.
Additional resources
ACTKNOW clinical trial registry number NCT01708876: https://clinicaltrials.gov/ct2/show/NCT01708876?term=NCT01708876&draw=2&rank=1.
PACKNOW clinical trial registry number NCT03056391: https://clinicaltrials.gov/ct2/show/NCT03056391?term=NCT03056391&draw=2&rank=1.
Acknowledgments
We acknowledge the efforts of the original study teams for collecting the P. knowlesi clinical samples. We thank the following organisations for donation of malaria-naive control plasma: the Australian and Thai Red Cross; the Rio de Janeiro State Blood Bank (with support from Andre M. Siqueira); and the Volunteer Blood Donor Registry at WEHI. We thank Shazia Ruybal for support generating an automated quality control platform through R, based on prior work by Connie Li-Wai-Suen, which was also used for the standard curve conversion. We thank Sarah Miller for assistance with ethical approvals. We would like to thank the Director-General, Ministry of Health, Malaysia, for permission to publish this manuscript. We also acknowledge the Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS. Funding: WEHI Innovation Fund (R.L., I.M.). Clinical Trials Funding: Malaysian Ministry of Health (grant number BP00500420), the Asia Pacific Malaria Elimination Network (108-07), and the Australian National Health and Medical Research Council (NHMRC; 1037304, 1045156, 115680). NHMRC Fellowships to N.M.A. #1135820, M.J.G. #1138860, and R.L. #1173210. NHMRC grants #1092789, #1134989, #1132975 and #1043345 (I.M.). M.J.G. was also supported by the Australian Centre for International Agricultural Research (Grant# LS-2019-116).
Author contributions
Conceptualization: R.L., M.G., N.A., and I.M. Methodology – clinical trials & control samples: R.L., M.G., K.P., J.S., G.R., B.B., D.C., T.W., N.A., and I.M. Methodology – protein expression: E.T., T.T., M.H., C.E.C., J.H., and W.H.T. Methodology – antibody assays & analytics: R.L., K.S., R.M., N.N., T.O., and M.W. Investigation – laboratory: R.L., K.S., S.H., and R.M. Investigation – analytics: R.L., K.S., T.O., and M.W. Visualization: R.L., K.S., T.O., and M.W. Supervision: N.A. and I.M. Writing – original draft: R.L. Writing – reviewing & editing: R.L., M.G., K.S., S.H., K.P., N.N., T.O., R.M., E.T., T.T., M.H., C.E.C., D.C., G.R., K.T., C.D., J.H., W.H.T., J.S., M.W., B.B., T.W., N.A., and T.M.
Declaration of interests
R.L., M.W., T.T., and and I.M. are inventors on filed patent PCT/US17/67926 on a system, method, apparatus, and diagnostic test for P. vivax. M.H. was an employee of the company CellFree Sciences Co., Ltd.
Inclusion and diversity
While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: June 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100662.
Supplemental information
Data and code availability
-
•
All data are available in the main text or the supplemental information. Data S1 contains patient variables, Data S2 contains IgG antibody data from the P. knowlesi cohorts and Data S3 IgG antibody data from the malaria-naïve controls.
-
•
Existing code is available at https://github.com/MWhite-InstitutPasteur/Pvivax_sero_dx and https://gitlab.pasteur.fr/tobadia/pvserotat-rshiny-app.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- 1.Auburn S., Cheng Q., Marfurt J., Price R.N. The changing epidemiology of Plasmodium vivax: insights from conventional and novel surveillance tools. PLoS Med. 2021;18:e1003560. doi: 10.1371/journal.pmed.1003560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kho S., Qotrunnada L., Leonardo L., Andries B., Wardani P.A.I., Fricot A., Henry B., Hardy D., Margyaningsih N.I., Apriyanti D., et al. Hidden biomass of intact malaria parasites in the human spleen. N. Engl. J. Med. 2021;384:2067–2069. doi: 10.1056/NEJMc2023884. [DOI] [PubMed] [Google Scholar]
- 3.Kho S., Qotrunnada L., Leonardo L., Andries B., Wardani P.A.I., Fricot A., Henry B., Hardy D., Margyaningsih N.I., Apriyanti D., et al. Evaluation of splenic accumulation and colocalization of immature reticulocytes and Plasmodium vivax in asymptomatic malaria: a prospective human splenectomy study. PLoS Med. 2021;18:e1003632. doi: 10.1371/journal.pmed.1003632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreira C.M., Abo-Shehada M., Price R.N., Drakeley C.J. A systematic review of sub-microscopic Plasmodium vivax infection. Malar. J. 2015;14:360. doi: 10.1186/s12936-015-0884-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiattibutr K., Roobsoong W., Sriwichai P., Saeseu T., Rachaphaew N., Suansomjit C., Buates S., Obadia T., Mueller I., Cui L., et al. Infectivity of symptomatic and asymptomatic Plasmodium vivax infections to a Southeast Asian vector, Anopheles dirus. Int. J. Parasitol. 2017;47:163–170. doi: 10.1016/j.ijpara.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson L.J., Wampfler R., Betuela I., Karl S., White M.T., Li Wai Suen C.S.N., Hofmann N.E., Kinboro B., Waltmann A., Brewster J., et al. Strategies for understanding and reducing the Plasmodium vivax and Plasmodium ovale hypnozoite reservoir in Papua New Guinean children: a randomised placebo-controlled trial and mathematical model. PLoS Med. 2015;12:e1001891. doi: 10.1371/journal.pmed.1001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Commons R.J., Simpson J.A., Watson J., White N.J., Price R.N. Estimating the proportion of Plasmodium vivax recurrences caused by relapse: a systematic review and meta-analysis. Am. J. Trop. Med. Hyg. 2020;103:1094–1099. doi: 10.4269/ajtmh.20-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adekunle A.I., Pinkevych M., McGready R., Luxemburger C., White L.J., Nosten F., Cromer D., Davenport M.P. Modeling the dynamics of Plasmodium vivax infection and hypnozoite reactivation in vivo. PLoS Negl. Trop. Dis. 2015;9:e0003595. doi: 10.1371/journal.pntd.0003595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Longley R.J., White M.T., Takashima E., Brewster J., Morita M., Harbers M., Obadia T., Robinson L.J., Matsuura F., Liu Z.S.J., et al. Development and validation of serological markers for detecting recent Plasmodium vivax infection. Nat. Med. 2020;26:741–749. doi: 10.1038/s41591-020-0841-4. [DOI] [PubMed] [Google Scholar]
- 10.White N.J. Determinants of relapse periodicity in Plasmodium vivax malaria. Malar. J. 2011;10:297. doi: 10.1186/1475-2875-10-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chotirat S., Nekkab N., Kumpitak C., Hietanen J., White M.T., Kiattibutr K., Sa-angchai P., Brewster J., Schoffer K., Takashima E., et al. Application of 23 novel serological markers for identifying recent exposure to Plasmodium vivax parasites in an endemic population of western Thailand. Front. Microbiol. 2021;12:643501. doi: 10.3389/fmicb.2021.643501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenhouse B., Daily J., Guinovart C., Goncalves B., Beeson J., Bell D., Chang M.A., Cohen J.M., Ding X., Domingo G., et al. Priority use cases for antibody-detecting assays of recent malaria exposure as tools to achieve and sustain malaria elimination. Gates Open Res. 2019;3:131. doi: 10.12688/gatesopenres.12897.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO . World Health Organisation; 2021. World Malaria Report. [Google Scholar]
- 14.Loy D.E., Liu W., Li Y., Learn G.H., Plenderleith L.J., Sundararaman S.A., Sharp P.M., Hahn B.H. Out of Africa: origins and evolution of the human malaria parasites Plasmodium falciparum and plasmodium vivax. Int. J. Parasitol. 2017;47:87–97. doi: 10.1016/j.ijpara.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitran C.J., Yanow S.K. The case for exploiting cross-species epitopes in malaria vaccine design. Front. Immunol. 2020;11:335. doi: 10.3389/fimmu.2020.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Priest J.W., Plucinski M.M., Huber C.S., Rogier E., Mao B., Gregory C.J., Candrinho B., Colborn J., Barnwell J.W. Specificity of the IgG antibody response to Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, and Plasmodium ovale MSP1(19) subunit proteins in multiplexed serologic assays. Malar. J. 2018;17:417. doi: 10.1186/s12936-018-2566-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodberry T., Minigo G., Piera K.A., Hanley J.C., de Silva H.D., Salwati E., Kenangalem E., Tjitra E., Coppel R.L., Price R.N., et al. Antibodies to Plasmodium falciparum and Plasmodium vivax merozoite surface protein 5 in Indonesia: species-specific and cross-reactive responses. J. Infect. Dis. 2008;198:134–142. doi: 10.1086/588711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diggs C.L., Sadun E.H. Serological cross reactivity between plasmodium vivax and plasmodium falciparum as determined BY a modified fluorescent antibody test. Exp. Parasitol. 1965;16:217–223. doi: 10.1016/0014-4894(65)90046-9. [DOI] [PubMed] [Google Scholar]
- 19.Lacerda M.V.G., Bassat Q. Primaquine for all: is it time to simplify malaria treatment in co-endemic areas? Lancet Infect. Dis. 2019;19:10–12. doi: 10.1016/s1473-3099(18)30612-1. [DOI] [PubMed] [Google Scholar]
- 20.Looareesuwan S., White N.J., Chittamas S., Bunnag D., Harinasuta T. High rate of Plasmodium vivax relapse following treatment of falciparum malaria in Thailand. Lancet. 1987;330:1052–1055. doi: 10.1016/s0140-6736(87)91479-6. [DOI] [PubMed] [Google Scholar]
- 21.Douglas N.M., Nosten F., Ashley E.A., Phaiphun L., van Vugt M., Singhasivanon P., White N.J., Price R.N. Plasmodium vivax recurrence following falciparum and mixed species malaria: risk factors and effect of antimalarial kinetics. Clin. Infect. Dis. 2011;52:612–620. doi: 10.1093/cid/ciq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hossain M.S., Commons R.J., Douglas N.M., Thriemer K., Alemayehu B.H., Amaratunga C., Anvikar A.R., Ashley E.A., Asih P.B.S., Carrara V.I., et al. The risk of Plasmodium vivax parasitaemia after P. falciparum malaria: an individual patient data meta-analysis from the WorldWide Antimalarial Resistance Network. PLoS Med. 2020;17:e1003393. doi: 10.1371/journal.pmed.1003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eziefula A.C., Bousema T., Yeung S., Kamya M., Owaraganise A., Gabagaya G., Bradley J., Grignard L., Lanke K.H., Wanzira H., et al. Single dose primaquine for clearance of Plasmodium falciparum gametocytes in children with uncomplicated malaria in Uganda: a randomised, controlled, double-blind, dose-ranging trial. Lancet Infect. Dis. 2014;14:130–139. doi: 10.1016/s1473-3099(13)70268-8. [DOI] [PubMed] [Google Scholar]
- 24.Ashley E.A., Recht J., White N.J. Primaquine: the risks and the benefits. Malar. J. 2014;13:418. doi: 10.1186/1475-2875-13-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ley B., Winasti Satyagraha A., Rahmat H., von Fricken M.E., Douglas N.M., Pfeffer D.A., Espino F., von Seidlein L., Henriques G., Oo N.N., et al. Performance of the Access Bio/CareStart rapid diagnostic test for the detection of glucose-6-phosphate dehydrogenase deficiency: a systematic review and meta-analysis. PLoS Med. 2019;16:e1002992. doi: 10.1371/journal.pmed.1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ley B., Alam M.S., Kibria M.G., Marfurt J., Phru C.S., Ami J.Q., Thriemer K., Auburn S., Jahan N., Johora F.T., et al. Glucose-6-phosphate dehydrogenase activity in individuals with and without malaria: analysis of clinical trial, cross-sectional and case-control data from Bangladesh. PLoS Med. 2021;18:e1003576. doi: 10.1371/journal.pmed.1003576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muh F., Kim N., Nyunt M.H., Firdaus E.R., Han J.H., Hoque M.R., Lee S.K., Park J.H., Moon R.W., Lau Y.L., et al. Cross-species reactivity of antibodies against Plasmodium vivax blood-stage antigens to Plasmodium knowlesi. PLoS Negl. Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herman L.S., Fornace K., Phelan J., Grigg M.J., Anstey N.M., William T., Moon R.W., Blackman M.J., Drakeley C.J., Tetteh K.K.A. Identification and validation of a novel panel of Plasmodium knowlesi biomarkers of serological exposure. PLoS Negl. Trop. Dis. 2018;12:e0006457. doi: 10.1371/journal.pntd.0006457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fornace K.M., Herman L.S., Abidin T.R., Chua T.H., Daim S., Lorenzo P.J., Grignard L., Nuin N.A., Ying L.T., Grigg M.J., et al. Exposure and infection to Plasmodium knowlesi in case study communities in Northern Sabah, Malaysia and Palawan, the Philippines. PLoS Negl. Trop. Dis. 2018;12:e0006432. doi: 10.1371/journal.pntd.0006432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuenca P.R., Key S., Jumail A., Surendra H., Ferguson H.M., Drakeley C.J., Fornace K. Epidemiology of the zoonotic malaria Plasmodium knowlesi in changing landscapes. Adv. Parasitol. 2021;113:225–286. doi: 10.1016/bs.apar.2021.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Grigg M.J., Cox J., William T., Jelip J., Fornace K.M., Brock P.M., von Seidlein L., Barber B.E., Anstey N.M., Yeo T.W., Drakeley C.J. Individual-level factors associated with the risk of acquiring human Plasmodium knowlesi malaria in Malaysia: a case-control study. Lancet Planet. Health. 2017;1:e97–e104. doi: 10.1016/s2542-5196(17)30031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barber B.E., Grigg M.J., William T., Yeo T.W., Anstey N.M. The treatment of Plasmodium knowlesi malaria. Trends Parasitol. 2017;33:242–253. doi: 10.1016/j.pt.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Pain A., Böhme U., Berry A.E., Mungall K., Finn R.D., Jackson A.P., Mourier T., Mistry J., Pasini E.M., Aslett M.A., et al. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature. 2008;455:799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benavente E.D., de Sessions P.F., Moon R.W., Grainger M., Holder A.A., Blackman M.J., Roper C., Drakeley C.J., Pain A., Sutherland C.J., et al. A reference genome and methylome for the Plasmodium knowlesi A1-H.1 line. Int. J. Parasitol. 2018;48:191–196. doi: 10.1016/j.ijpara.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 36.Grigg M.J., William T., Menon J., Dhanaraj P., Barber B.E., Wilkes C.S., von Seidlein L., Rajahram G.S., Pasay C., McCarthy J.S., et al. Artesunate-mefloquine versus chloroquine for treatment of uncomplicated Plasmodium knowlesi malaria in Malaysia (ACT KNOW): an open-label, randomised controlled trial. Lancet Infect. Dis. 2016;16:180–188. doi: 10.1016/s1473-3099(15)00415-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooper D.J., Plewes K., Grigg M.J., Rajahram G.S., Piera K.A., William T., Chatfield M.D., Yeo T.W., Dondorp A.M., Anstey N.M., Barber B.E. The effect of regularly dosed paracetamol versus no paracetamol on renal function in Plasmodium knowlesi malaria (PACKNOW): study protocol for a randomised controlled trial. Trials. 2018;19:250. doi: 10.1186/s13063-018-2600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooper D.J., Grigg M.J., Plewes K., Rajahram G.S., Piera K.A., William T., Menon J., Koleth G., Edstein M.D., Birrell G.W., et al. The effect of regularly dosed acetaminophen versus no acetaminophen on renal function in Plasmodium knowlesi malaria (PACKNOW): a randomised controlled trial. Clin. Infect. Dis. 2022:ciac152. doi: 10.1093/cid/ciac152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cooper D.J., Rajahram G.S., William T., Jelip J., Mohammad R., Benedict J., Alaza D.A., Malacova E., Yeo T.W., Grigg M.J., et al. Plasmodium knowlesi malaria in Sabah, Malaysia, 2015-2017: ongoing increase in incidence despite near-elimination of the human-only Plasmodium species. Clin. Infect. Dis. 2020;70:361–367. doi: 10.1093/cid/ciz237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh B., Daneshvar C. Human infections and detection of Plasmodium knowlesi. Clin. Microbiol. Rev. 2013;26:165–184. doi: 10.1128/cmr.00079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grigg M.J., William T., Barber B.E., Rajahram G.S., Menon J., Schimann E., Piera K., Wilkes C.S., Patel K., Chandna A., et al. Age-related clinical spectrum of Plasmodium knowlesi malaria and predictors of severity. Clin. Infect. Dis. 2018;67:350–359. doi: 10.1093/cid/ciy065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barber B.E., Grigg M.J., William T., Piera K.A., Boyle M.J., Yeo T.W., Anstey N.M. Effects of aging on parasite biomass, inflammation, endothelial activation, microvascular dysfunction and disease severity in Plasmodium knowlesi and Plasmodium falciparum malaria. J. Infect. Dis. 2017;215:1908–1917. doi: 10.1093/infdis/jix193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogier E., Nace D., Dimbu P.R., Wakeman B., Pohl J., Beeson J.G., Drakeley C., Tetteh K., Plucinski M. Framework for characterizing longitudinal antibody response in children after Plasmodium falciparum infection. Front. Immunol. 2021;12:617951. doi: 10.3389/fimmu.2021.617951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yman V., White M.T., Asghar M., Sundling C., Sondén K., Draper S.J., Osier F.H.A., Färnert A. Antibody responses to merozoite antigens after natural Plasmodium falciparum infection: kinetics and longevity in absence of re-exposure. BMC Med. 2019;17:22. doi: 10.1186/s12916-019-1255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Z.S.J., Sattabongkot J., White M., Chotirat S., Kumpitak C., Takashima E., Harbers M., Tham W.H., Healer J., Chitnis C.E., et al. Naturally acquired antibody kinetics against Plasmodium vivax antigens in people from a low malaria transmission region in western Thailand. BMC Med. 2022;20:89. doi: 10.1186/s12916-022-02281-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muh F., Ahmed M.A., Han J.H., Nyunt M.H., Lee S.K., Lau Y.L., Kaneko O., Han E.T. Cross-species analysis of apical asparagine-rich protein of Plasmodium vivax and Plasmodium knowlesi. Sci. Rep. 2018;8:5781. doi: 10.1038/s41598-018-23728-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosa D.S., Iwai L.K., Tzelepis F., Bargieri D.Y., Medeiros M.A., Soares I.S., Sidney J., Sette A., Kalil J., Mello L.E., et al. Immunogenicity of a recombinant protein containing the Plasmodium vivax vaccine candidate MSP1(19) and two human CD4+ T-cell epitopes administered to non-human primates (Callithrix jacchus jacchus) Microb. Infect. 2006;8:2130–2137. doi: 10.1016/j.micinf.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 48.De S.L., Ntumngia F.B., Nicholas J., Adams J.H. Progress towards the development of a P. vivax vaccine. Expert Rev. Vaccines. 2021;20:97–112. doi: 10.1080/14760584.2021.1880898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kale S., Yadav C.P., Rao P.N., Shalini S., Eapen A., Srivasatava H.C., Sharma S.K., Pande V., Carlton J.M., Singh O.P., Mallick P.K. Antibody responses within two leading Plasmodium vivax vaccine candidate antigens in three geographically diverse malaria-endemic regions of India. Malar. J. 2019;18:425. doi: 10.1186/s12936-019-3066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCaffery J.N., Fonseca J.A., Singh B., Cabrera-Mora M., Bohannon C., Jacob J., Arévalo-Herrera M., Moreno A. A multi-stage Plasmodium vivax malaria vaccine candidate able to induce long-lived antibody responses against blood stage parasites and robust transmission-blocking activity. Front. Cell. Infect. Microbiol. 2019;9:135. doi: 10.3389/fcimb.2019.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ndegwa D.N., Kundu P., Hostetler J.B., Marin-Menendez A., Sanderson T., Mwikali K., Verzier L.H., Coyle R., Adjalley S., Rayner J.C. Using Plasmodium knowlesi as a model for screening Plasmodium vivax blood-stage malaria vaccine targets reveals new candidates. PLoS Pathog. 2021;17:e1008864. doi: 10.1371/journal.ppat.1008864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anstey N.M., Grigg M.J. Zoonotic malaria: the better you look, the more you find. J. Infect. Dis. 2019;219:679–681. doi: 10.1093/infdis/jiy520. [DOI] [PubMed] [Google Scholar]
- 53.França C.T., White M.T., He W.Q., Hostetler J.B., Brewster J., Frato G., Malhotra I., Gruszczyk J., Huon C., Lin E., et al. Identification of highly-protective combinations of Plasmodium vivax recombinant proteins for vaccine development. Elife. 2017;6:e28673. doi: 10.7554/eLife.28673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ssewanyana I., Rek J., Rodriguez I., Wu L., Arinaitwe E., Nankabirwa J.I., Beeson J.G., Mayanja-Kizza H., Rosenthal P.J., Dorsey G., et al. Impact of a rapid decline in malaria transmission on antimalarial IgG subclasses and avidity. Front. Immunol. 2020;11:576663. doi: 10.3389/fimmu.2020.576663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yap N.J., Hossain H., Nada-Raja T., Ngui R., Muslim A., Hoh B.P., Khaw L.T., Kadir K.A., Simon Divis P.C., Vythilingam I., et al. Natural human infections with Plasmodium cynomolgi, P. Inui, and 4 other simian malaria parasites, Malaysia. Emerg. Infect. Dis. 2021;27:2187–2191. doi: 10.3201/eid2708.204502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Obadia T., Nekkab N., Robinson L., Drakeley C., Mueller I., White M.T. Developing sero-diagnostic tests to facilitate Plasmodium vivax Serological Test-and-Treat approaches: modelling the balance between public health impact and overtreatment. BMC Med. 2022;20:98. doi: 10.1186/s12916-022-02285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mazhari R., Brewster J., Fong R., Bourke C., Liu Z.S.J., Takashima E., Tsuboi T., Tham W.H., Harbers M., Chitnis C., et al. A comparison of non-magnetic and magnetic beads for measuring IgG antibodies against Plasmodium vivax antigens in a multiplexed bead-based assay using Luminex technology (Bio-Plex 200 or MAGPIX) PLoS One. 2020;15:e0238010. doi: 10.1371/journal.pone.0238010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nuin N.A., Tan A.F., Lew Y.L., Piera K.A., William T., Rajahram G.S., Jelip J., Dony J.F., Mohammad R., Cooper D.J., et al. Comparative evaluation of two commercial real-time PCR kits (QuantiFast™ and abTES™) for the detection of Plasmodium knowlesi and other Plasmodium species in Sabah, Malaysia. Malar. J. 2020;19:306. doi: 10.1186/s12936-020-03379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amos B., Aurrecoechea C., Barba M., Barreto A., Basenko E.Y., Bażant W., Bażant W., Belnap R., Blevins A.S., Böhme U., et al. VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 2022;50 doi: 10.1093/nar/gkab929. D898–D911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garzón-Ospina D., Forero-Rodríguez J., Patarroyo M.A. Evidence of functional divergence in MSP7 paralogous proteins: a molecular-evolutionary and phylogenetic analysis. BMC Evol. Biol. 2016;16:256. doi: 10.1186/s12862-016-0830-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuamsab N., Putaporntip C., Pattanawong U., Jongwutiwes S. Insights into the molecular diversity of Plasmodium vivax merozoite surface protein-3γ (pvmsp3γ), a polymorphic member in the msp3 multi-gene family. Sci. Rep. 2020;10:10977. doi: 10.1038/s41598-020-67222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data are available in the main text or the supplemental information. Data S1 contains patient variables, Data S2 contains IgG antibody data from the P. knowlesi cohorts and Data S3 IgG antibody data from the malaria-naïve controls.
-
•
Existing code is available at https://github.com/MWhite-InstitutPasteur/Pvivax_sero_dx and https://gitlab.pasteur.fr/tobadia/pvserotat-rshiny-app.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.