Significance
Behçet’s disease (BD) is a systemic vasculitis, and its pathogenesis is elusive. Limited understanding of the immune disturbances in BD hindered the identification of therapeutic targets. Here, we performed single-cell and bulk RNA sequencing to reveal the comprehensive landscape of cellular and molecular changes in BD blood. We observed the mobilization of monocytes, especially a monocyte subset (C1q-high monocytes) in BD. Further assays revealed the proinflammatory features and clinical relevance of this subset. Activated interferon-γ (IFN-γ) signaling in BD patients induced C1q-high monocyte expansion, which was recovered by tofacitinib treatment. Our findings provide comprehensive understanding of BD immunopathogenesis, highlighting the proinflammatory contribution and therapeutic potential of C1q-high monocytes.
Keywords: Behçet’s disease, single-cell analysis, monocytes, interferon
Abstract
Behçet’s disease (BD) is a chronic vasculitis characterized by systemic immune aberrations. However, a comprehensive understanding of immune disturbances in BD and how they contribute to BD pathogenesis is lacking. Here, we performed single-cell and bulk RNA sequencing to profile peripheral blood mononuclear cells (PBMCs) and isolated monocytes from BD patients and healthy donors. We observed prominent expansion and transcriptional changes in monocytes in PBMCs from BD patients. Deciphering the monocyte heterogeneity revealed the accumulation of C1q-high (C1qhi) monocytes in BD. Pseudotime inference indicated that BD monocytes markedly shifted their differentiation toward inflammation-accompanied and C1qhi monocyte–ended trajectory. Further experiments showed that C1qhi monocytes enhanced phagocytosis and proinflammatory cytokine secretion, and multiplatform analyses revealed the significant clinical relevance of this subtype. Mechanistically, C1qhi monocytes were induced by activated interferon-γ (IFN-γ) signaling in BD patients and were decreased by tofacitinib treatment. Our study illustrates the BD immune landscape and the unrecognized contribution of C1qhi monocytes to BD hyperinflammation, showing their potential as therapeutic targets and clinical assessment indexes.
Behçet’s disease (BD) is a systemic inflammatory disorder affecting blood vessels and commonly manifests as recurrent oral/genital ulceration and skin lesions, while patients with exacerbated BD exhibit multiorgan involvement such as uveitis and gastrointestinal, neurological, vascular, and cardiac symptoms, which cause significant morbidity and mortality (1–3). BD displays geographical variations in prevalence, with a higher incidence (20 to ∼602 cases per 100,000 population) in Silk Road countries spanning from China in the east to Turkey in the Mediterranean area (2–6). The atypical symptoms and regional differences in disease prevalence pose diagnostic and therapeutic challenges for clinicians. Thus, the identification of laboratory indices to improve empirical judgment and BD therapies is needed.
Accumulating evidence has shown that immunological abnormalities are pivotal in BD development, although genetic variants and environmental stimuli are also important triggers (7). Patients with BD exhibit aberrant and excessive activation of both innate and adaptive immunity (corresponding to the features of autoinflammatory or autoimmune diseases) (7, 8), including the overproduction of proinflammatory cytokines (interferon-γ [IFN-γ], interleukin 6 [IL-6], and tumor necrosis factor α [TNF-α]) (9) and skewed T-hepler (Th) 1 and Th17 cell activation (10, 11). Intense efforts into understanding BD pathogenesis have focused on specific immune cells based on select cell-surface markers (12–14). However, a comprehensive and unbiased depiction of immune disturbances in BD remains to be elucidated.
In previous BD studies, the transcriptomic profiling at the bulk level lacked the resolution to capture cellular heterogeneity and was limited in the ability to identify the pathogenetic contribution of certain cell subtypes (15–17). Recently, single-cell RNA sequencing (scRNA-seq) technology has shown unprecedented value in discovering pathophysiological immune changes at higher resolution (18–23). However, the molecular characteristics of diverse immune cell subsets associated with BD at single-cell resolution and how these features contribute to exaggerated inflammation in BD patients have not been previously described.
To address these questions in an unbiased manner, we applied scRNA-seq to first delineate the immune landscape in peripheral blood mononuclear cells (PBMCs) that usually serve as an attractive source for understanding the pathogenesis of diverse diseases with abnormal immunophenotypes (24–26). Monocytes with significant expansion and transcriptional changes were observed in BD. Further investigations demonstrated that the unappreciated C1q-high (C1qhi) monocyte subtype exhibited proinflammatory characteristics and significant clinical relevance in BD. Overall, our study offers a comprehensive view of immune dysfunction in BD and highlights the proinflammatory contribution of C1qhi monocytes to BD immunopathogenesis, which might facilitate the development of clinical assessments and targeted therapies for BD.
Results
The Single-Cell Landscape of Peripheral Mononuclear Cells in BD.
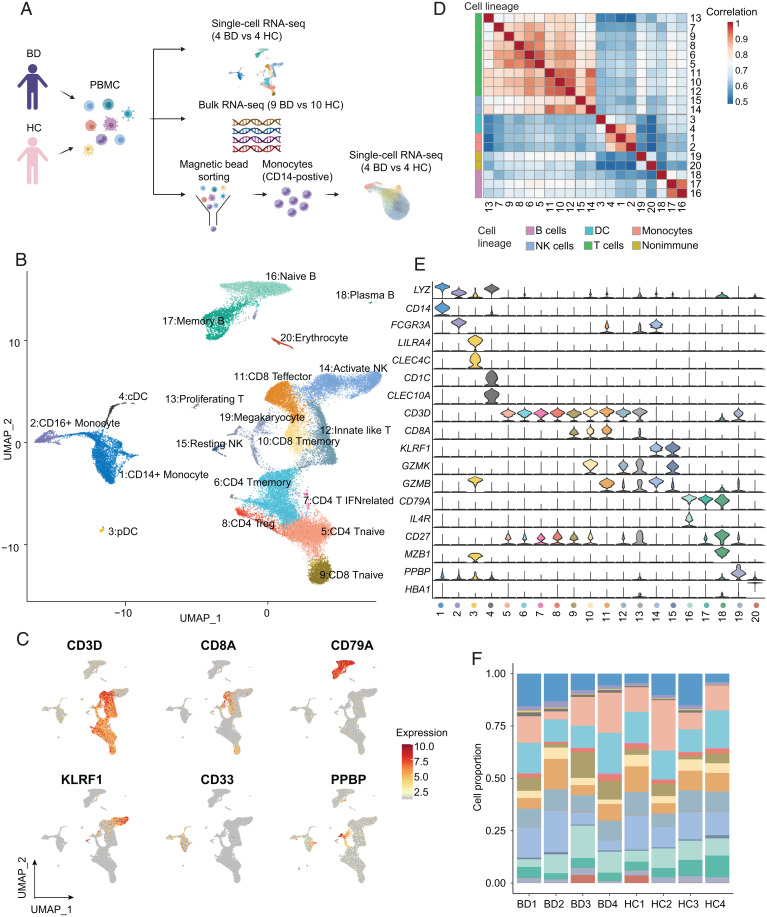
We performed droplet-based scRNA-seq (10X Genomics) to delineate the immune landscape of PBMCs from four treatment-naive BD patients and four healthy controls (HCs) (Fig. 1A and Dataset S1). A total of 36,190 high-quality cells of 41,889 cells were retained for downstream analysis after removing cells with a low sequence depth and a high ratio of mitochondrial genes (SI Appendix, Fig. S1A); the analyzed cells included 17,678 (48.8%) from BD patients and 18,512 (51.2%) from HCs, and an average of 1,096 genes per cell was monitored (SI Appendix, Fig. S1 B–D). After batch effect correction, dimension reduction, and graph-based clustering, we identified 20 distinctive clusters (Fig. 1B) within five major cell lineages, including myeloid cells (CD33+), T cells (CD3D+), natural killer (NK) cells (KLRF1+), B cells (CD79A+), and nonimmune cell lineages (PPBP+) (Fig. 1C). As expected, unsupervised hierarchical clustering of all cell clusters confirmed the major cell lineages (Fig. 1D).
Fig. 1.
High-resolution mapping of PBMCs in BD patients and healthy donors. (A) Schematic workflow of the experimental strategy. (B) UMAP visualization of 20 unique cell clusters (colors) in PBMCs from four BD patients and four HCs. (C) UMAP plot showing the expression of canonical markers of the main cell lineages by color. (D) Heatmap showing Spearman correlation (colors) between the log-normalized average expression of all clusters. Cell clusters are ordered by hierarchical clustering and annotated with the corresponding cell lineage (color-coded bar). (E) Violin plots showing the expression of canonical markers that recognize each cell cluster. (F) Proportions of cell clusters among total cells in individual patients. QC, quality control; UMAP, uniform manifold approximation and projection.
The high sensitivity of scRNA-seq allowed us to further map these clusters to immune cell subsets. Specifically, we identified four myeloid cell subtypes according to the expression of canonical markers and cell type–specific gene sets (27) (Fig. 1E and SI Appendix, S1 E and F and Dataset S2), including CD14+ monocytes (LYZ+CD14+), CD16+ monocytes (LYZ+FCGR3A+), conventional dendritic cells (cDCs; CD1C+CLEC10A+), and plasmacytoid dendritic cells (pDCs; LILRA4+CLEC4C+). We also identified nine T cell subtypes, namely, naive CD4 T cells (CD4 Tnaive; CD3D+ CCR7+), memory CD4 T cells (CD4 Tmemory; CD3D+ CD40LG+), IFN-related CD4 T cells (CD4 T IFNrelated; CD3D+ ISG15+), regulatory T cells (Tregs; CD3D+ FOXP3+), innate-like T cells (innate lymphoid cells; CD3D+ CD127+ KLRG1+ SLC4A10+), memory CD8 T cells (CD8 Tmemory; CD8A+ GZMK+), effector CD8 T cells (CD8 Teffector; CD8A+ GZMB+), and proliferating T cells (proliferating T; CD3D+ MKI67+) (SI Appendix, Fig. S1H), which were confirmed by evaluating T cell signature gene sets (27) (SI Appendix, Fig. S1G). In addition, other subpopulations were annotated mainly according to unique marker genes, including resting NK cells/CD56brightCD16− NK (NCAM1high CD16− KLRF1+ GZMK+), active NK cells/CD56dimCD16+ NK (NCAM1dim CD16− KLRF1+ GZMB+), naive B cells (CD79A+ IL4R+), memory B cells (CD79A+ CD27+), plasma B cells (CD79A+ MZB1+), megakaryocytes (PPBP+), and erythrocytes (HBA1+) (Fig. 1E and SI Appendix, Fig. S1I). All clusters contained cells from multiple BD patients, suggesting the absence of significant batch effects (Fig. 1F and SI Appendix, Fig. S1J). Thus, we delineated the peripheral immune landscape in BD at single-cell resolution.
Monocytes Contribute to Systematic Immune Aberrations in BD.
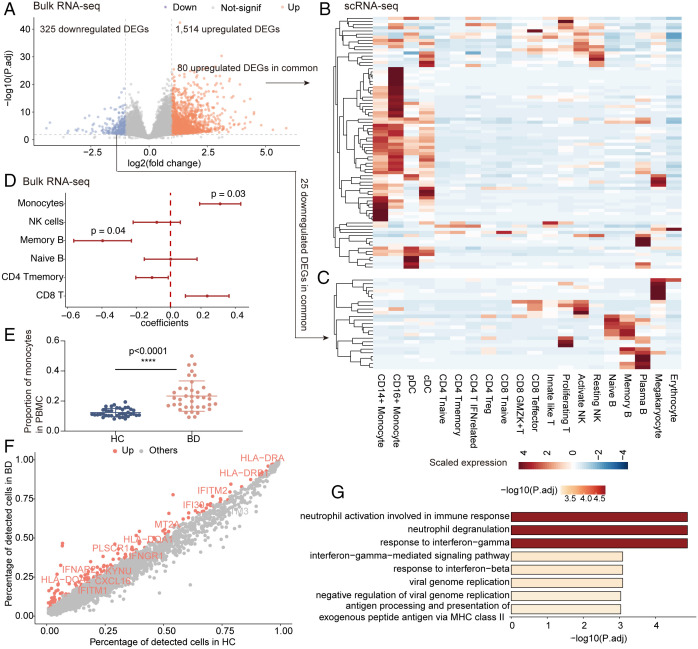
Aiming to decipher the changes in immune cell composition in BD, we conducted integrative analyses of scRNA-seq data with bulk RNA-seq data (9 BD patients and 10 HCs) to increase patient numbers. Principal component analysis of bulk RNA-seq data using all quantified genes revealed that the first principal component clearly distinguished BD patients from HCs (SI Appendix, Fig. S2A), indicating dramatic differences in the genome-wide profiles of BD patients compared with HCs. We then identified 1,514 up-regulated and 325 down-regulated differentially expressed genes (DEGs) in BD from bulk RNA-seq data (Fig. 2A). Upon mapping these DEGs to cell types identified in scRNA-seq data, we found that the up-regulated DEGs were relatively enriched in monocytes and DCs (Fig. 2B), while the down-regulated genes were enriched in B cells (Fig. 2C). However, the fold changes in these DEGs between BD patients and HCs did not reveal a pronounced enrichment in specific cell types (SI Appendix, Fig. S2B). This result implies that BD-associated variations in the bulk transcriptome are more likely to be impacted by changes in cellular composition than by intracellular changes. We next deconvoluted the immune cell subtypes in bulk RNA-seq data by CIBERSORT (28) with the LM22 signature matrix, which distinguishes human hematopoietic populations isolated from peripheral blood (28). Consistently, we found a higher proportion of monocytes in BD patients than in HCs (Fig. 2D), which was further confirmed by complete blood count data (23.3 ± 10.1% versus 12.4 ± 2.8%, P < 0.0001; Fig. 2E) and scRNA-seq data (SI Appendix, Fig. S2C), suggesting an aberrant role of monocytes in BD. Additionally, smaller memory B cell populations were observed in BD patients than in HCs (Fig. 2D), as previously reported (29).
Fig. 2.
Aberrant monocytes in PBMCs of BD patients. (A) Volcano plot showing the DEGs in bulk RNA-seq data comparing the BD and HC groups (criteria: P < 0.01 and absolute log2 [fold change] >1). Red, significant up-regulation; purple, significant down-regulation; gray, not significant (not-signif). (B and C) Heatmap visualizing the row-scaled average expression of common DEGs in cell types identified in scRNA-seq data, with up-regulated DEGs in B and down-regulated DEGs in C. Common DEGs are defined as the overlap between DEGs in bulk RNA-seq data in A and highly variable genes in scRNA-seq data. (D) Forest plots showing the BD-association cell types. Coefficients, 95% CIs, and P values were derived from the linear mixed models between condition (BD/HC) and cell proportions, with age as the covariate. The cell proportions in bulk RNA-seq data (9 BD patients and 10 HCs) were deconvolved by CIBERSORT using the default LM22 signature. (E) The proportions of monocytes in total PBMCs of BD patients and HCs based on complete blood counts (n = 38 per group). The data are summarized as the mean ± SD. (F) Scatter plot showing the percentage of detected genes (dots) in HC (x axis) and BD (y axis) monocytes from scRNA-seq data. The colored genes indicate significantly up-regulated genes in BD monocytes, and the labeled genes are those within the response to the IFN-γ pathway in the Gene Ontology (GO) database. (G) Bar plot showing the top enriched GO pathways of the significantly up-regulated genes in F. The Wilcoxon test (A and D) and independent-sample t test (E) were applied. ****P < 0.0001. P.adj, adjusted P value.
Next, we investigated intracellular biological differences associated with BD development in the major cell lineages using scRNA-seq data. We identified DEGs that distinguished BD from HC in individual cell lineages and determined the enriched biological pathways. The bacterial and viral infection pathways were enriched in most lineages (SI Appendix, Fig. S2D), supporting the hypothesis that infectious agents trigger BD pathogenesis (30, 31). In T cells, Th1 cell differentiation was among the top enriched pathways (SI Appendix, Fig. S2D), suggesting Th1-mediated hyperactivity in BD (32). In monocytes, the top pathways were the IFN-γ response, processing and presentation of exogenous antigens, and neutrophil activation (Fig. 2 F and G and Dataset S3). Moreover, the up-regulated DEGs in monocytes from scRNA-seq data were shared with the DEGs from the bulk RNA-seq data (SI Appendix, Fig. S2E), further confirming the significant contribution of monocytes to the global transcriptome aberrations in BD. Taken together, the integrated analysis of bulk RNA-seq data systematically identified BD-associated immune disturbances and indicated the prominent mobilization of monocytes in BD.
Identification of Monocyte Heterogeneity.
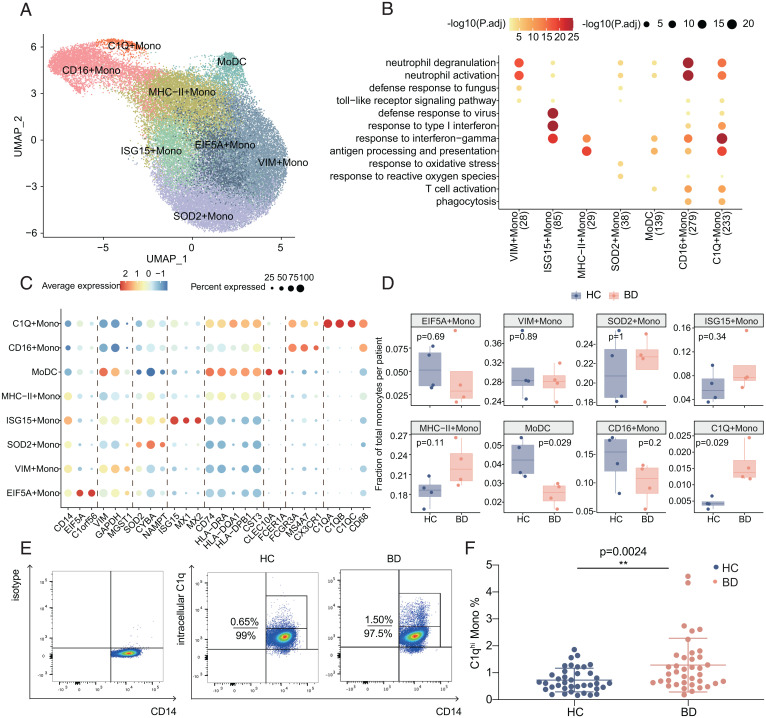
To increase cell-level resolution and dissect monocyte heterogeneity, we performed scRNA-seq of magnetic bead–sorted monocytes (CD14+) from PBMCs of four BD patients and four HCs. We retained 39,385 monocytes that passed rigorous filtering, including 17,321 cells (44%) from HCs and 22,064 cells (56%) from BD patients. By graph-based clustering, we captured the profiles of eight clusters (Fig. 3A and Dataset S4), which were not confounded by either a specific patient or condition (SI Appendix, Fig. S3 A and B).
Fig. 3.
Heterogeneity of monocyte subtypes in BD. (A) UMAP visualization of sorted monocytes (CD14+) from blood samples of BD patients (n = 4) and HCs (n = 4), colored by the identified monocyte subtypes. (B) Representative Gene Ontology (GO) pathways enriched by the highly expressed genes in monocyte subtypes. Both the color and size of the individual dots indicate the −log10–transformed Benjamini-Hochberg–corrected P value. (C) Dot plot displaying the expression of highly expressed genes for eight cell subtypes. The size of each circle corresponds to the percentage of cells in the subtype expressing the gene, and the color represents the average expression. (D) Boxplot comparing the differences in cell proportions between BD patients (red) and HCs (blue) in the sorted monocyte scRNA-seq data. The data are summarized as the mean ± SD. (E and F) Representative flow cytometry plot (E) and summary of the proportions of C1qhi monocytes (F) in peripheral monocytes from active BD patients and HCs (n = 38 per group). The data are summarized as the mean ± SD. The Wilcoxon test (D) and independent-sample t test (F) were applied. **P < 0.01. P.adj, adjusted P value; UMAP, uniform manifold approximation and projection.
The heterogeneity of monocytes corresponds to diverse functional specialization (33). We then compared the functional phenotypes of all eight subtypes by assessing the highly expressed genes and corresponding enriched pathways (Fig. 3 B and C). Five of the eight monocyte subtypes maintained higher levels of CD14 (Fig. 3C). Specifically, VIM-high monocytes (VIM Monos) overexpressed S100A family genes (S100A8, S100A9, and S100A12), which are markers of human myeloid–derived suppressor cells (34). Although the expression profile of SOD2-high monocytes (SOD2 Monos) resembled that of VIM Monos (SI Appendix, Fig. S3C), SOD2 Monos were enriched in the response to oxidative stress and overexpressed redox-related genes (SOD2, CYBA, and NAMPT) that are associated with impaired fibrinogen function in BD patients (35). EIF5A-high monocytes (EIF5A Monos) involved cell proliferation genes (EIF5A and C1orf56); ISG15-high monocytes (ISG15 Monos) showed preferential expression of IFN-inducible genes (ISG15, MX1, and MX2); and major histocompatibility complex (MHC)-II–high monocytes (MHC-II Monos) were enriched for genes in antigen presentation pathways, such as those encoding MHC class II chains (HLA-DRA, HLA-DQA1, HLA-DPB1, and CD74). Among the three subtypes with lower CD14 expression (Fig. 3C), CD16-high monocytes (CD16 Monos) presented nonclassical monocyte markers (FCGR3A, MS4A7, and CX3CR1), and monocyte-derived DCs (MoDCs) showed overexpression of DC markers (CLEC10A, FCER1A, CST3, and CD74) with enrichment of antigen presentation pathways and the DC-specific gene signature (Fig. 3 B and C and SI Appendix, Fig. S3D). C1qhi monocytes (C1Q Monos) highly expressed components of complement 1q (C1q genes, including C1QA, C1QB, and C1QC) and macrophage markers (CD68) (Fig. 3C) and also exhibited a macrophage-like phenotype (SI Appendix, Fig. S3D).
We next investigated BD-associated differences in monocyte composition. Comparing the relative cell proportions in the BD and HC groups, we observed a notable increase in C1Q Monos in BD patients and a significant decrease in MoDCs (Fig. 3D). Using flow cytometry, we observed the existence of this subtype and validated that the C1qhi monocyte population (C1Q Monos in scRNA-seq data) was increased in active BD patients (1.28 ± 0.99% versus 0.73 ± 0.44%, P = 0.0024; Fig. 3 E and F). Altogether, analysis of the scRNA-seq data for sorted monocytes revealed the heterogeneity of circulating monocytes and the corresponding diversification of monocyte functions and significantly increased C1qhi monocytes in BD patients.
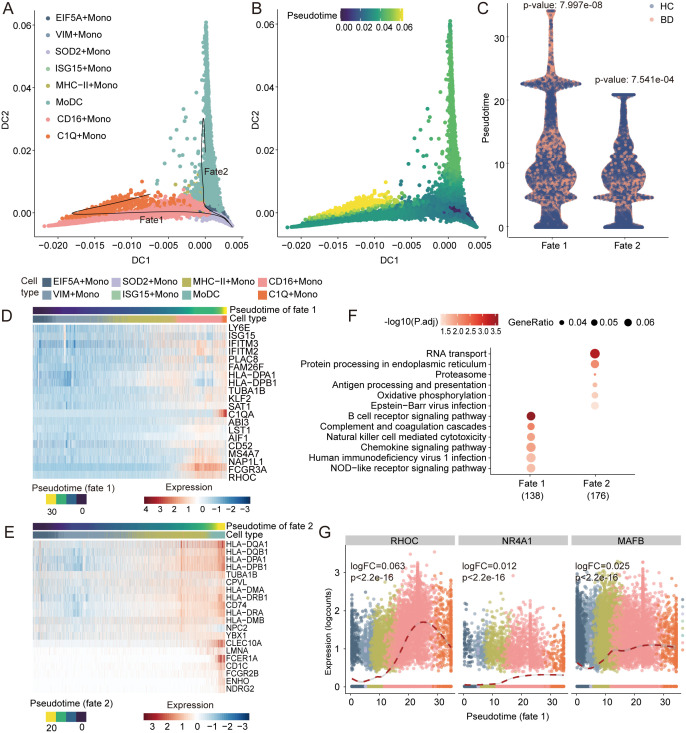
Differentiation Trajectories Toward C1qhi Monocytes Are Accompanied by Inflammatory Pathways.
To explore the transitional relationships across monocyte subtypes, we determined the pseudotemporal order and reconstructed the differentiation trajectory using a diffusion map (36), TSCAN (37), and Slingshot algorithms (38). The pseudotime trajectory axes branched from MHC-II Monos into two termini: fate 1, C1Q Monos, and fate 2, MoDCs (Fig. 4 A and B). These two fates corresponded to the roles of monocytes in replenishing macrophages and DCs (39) and were confirmed by Monocle 3 (40) (SI Appendix, Fig. S4A). Interestingly, a significantly imbalanced distribution of two conditions appeared along with the two fates, with BD monocytes aggregating at the end of fate 1 but more HC monocytes accumulating at the end of fate 2 (Fig. 4C). This result corresponded with the increased C1Q Monos and decreased MoDCs signatures in BD (Fig. 3D). The remarkable shift in monocyte differentiation toward C1Q Monos, but not MoDCs, suggests the role of C1Q Monos in modulating exaggerated inflammation in BD.
Fig. 4.
Trajectory inference of monocyte differentiation. (A and B) Visualization of monocyte trajectories using the first two diffusion components inferred by the diffusion map algorithm, colored by monocyte subtype (A) and pseudotime (B). Each dot represents a cell, and the black lines in A show Slingshot trajectories. (C) Comparison of the pseudotime differences between BD patients (red) and HCs (blue) within fate 1 and fate 2. The Wilcoxon test was used for statistical analysis. (D and E) Heatmap showing the gene expression (normalized log count, shown by color) of correlated genes along fate 1 (D) and fate 2 (E), annotated by the corresponding pseudotime and cell type. (F) Pathway enrichment of pseudotime-correlated genes for the two fates. The top Kyoto Encyclopedia of Genes and Genomes terms are shown colored by the Benjamini-Hochberg–corrected P values. The dot size indicates the ratio of enriched genes in the pathway. (G) Expression of known TFs driving macrophage development along fate 1, colored by monocyte subtypes. Dashed line shows the smoothed fit of TF’s expression along fate 1. Labeled are P value and log fold-change (logFC) of the TF along fate 1. P.adj, adjusted P value.
Next, we investigated the correlated gene expression profiles and pathways underlying the two differentiation trajectories (Dataset S5). We observed the gradual up-regulation of genes encoding inflammatory cytokine-related proteins (ISG15, TNFRSF1B, and S100A11) and complement components (C1q genes) along fate 1 (Fig. 4D). In contrast, fate 2 showed variations in antigen presentation genes, including MHC-II genes (HLA-DR, HLA-DP, and HLA-DQ), the Fc fragment of the immunoglobulin E receptor (FCER1A and FCER2B), and DC markers (CLEC10A, CD1C, and CD74) (Fig. 4E). Pathway enrichment analyses identified fate 1 was in conjunction with inflammation-related pathways (41–43), such as chemokine signaling pathway, nucleotide oligomerization domain (NOD)-like receptor signaling pathway, and NK cell–mediated cytotoxicity, while fate 2 was associated with antigen presentation–related pathways. These results indicated functional divergence of cells adopting the two fates. As expected, phagocytosis and antigen presentation were enhanced along fate 1 and fate 2, respectively (SI Appendix, Fig. S4 B and C). In addition, the expression of transcription factors (TFs) engaged in the monocyte-to-macrophage transition (RHOC, NR4A1, and MAFB) gradually increased in fate 1, which ended with the macrophage-like subtypes (C1Q Monos), but not in fate 2, which concluded with the MoDCs subtype (Fig. 4G and SI Appendix, Fig. S4D). Altogether, monocytes in BD patients preferentially differentiate toward a C1Q Monos–ended and inflammation-accompanied trajectory, which suggests the proinflammatory characteristics of C1Q Monos.
C1Q Monos Contribute to Hyperinflammation in BD.
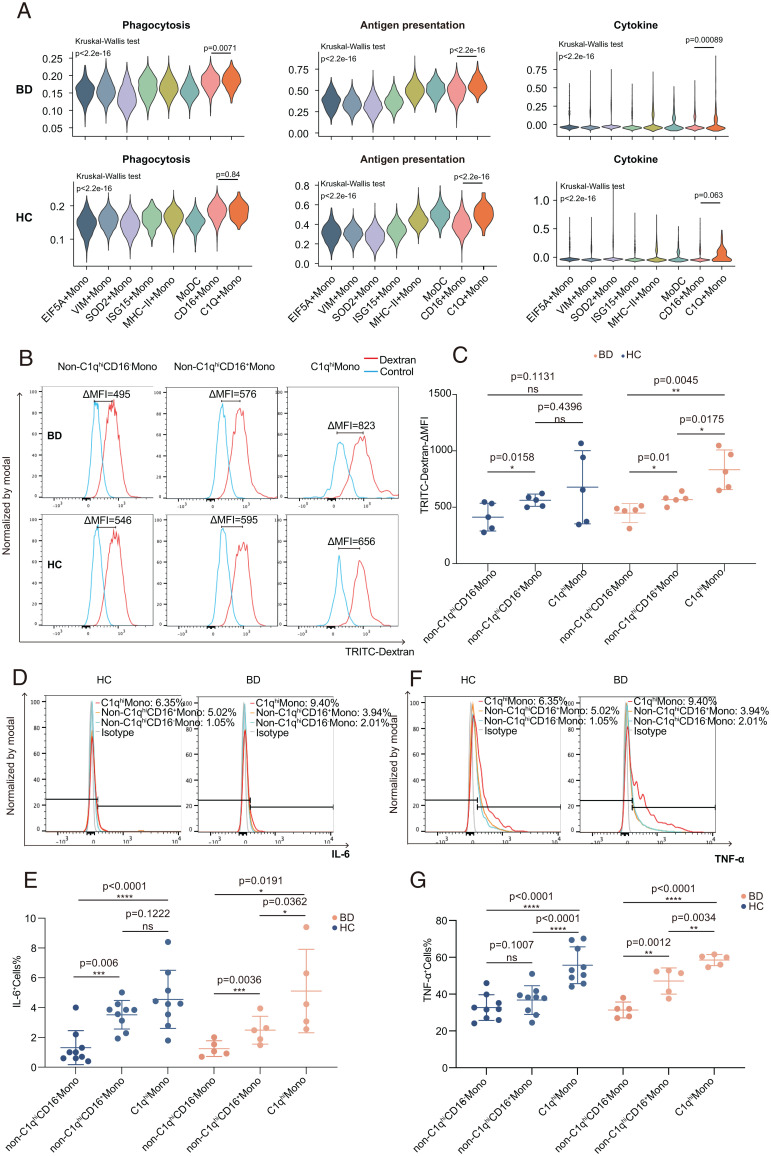
To test whether C1Q Monos manifest proinflammatory properties, we analyzed three key inflammation-regulatory capacities of monocytes, including phagocytosis, antigen presentation, and cytokine secretion. C1Q Monos indicated greater functional capacity than other subtypes (Fig. 5A), as evidenced by the overexpression of BD-associated proinflammatory cytokines (TNF and IL6) (7, 44), FcR activators (LYN and HCK), recruiters of cytoskeleton remodelers (PIK3CG, WASP2, and VASP), and MHC-II components (SI Appendix, Fig. S5 A–C).
Fig. 5.
C1qhi monocytes exhibited proinflammatory properties. (A) Violin plots showing the average expression levels of monocyte functional gene sets (SI Appendix, Materials and Methods) among all monocyte subtypes (colors). Top: BD patients; Bottom, HCs. Groupwise P values were calculated via the Kruskal-Wallis test, and pairwise P values were calculated via the Wilcoxon test. (B and C) Representative histograms (B) and summary (C) of the phagocytosis capability of C1qhi monocytes, non-C1qhiCD16+ monocytes, and non-C1qhiCD16− monocytes from BD patients (n = 5) and HCs (n = 5). The data are summarized as the mean ± SD. (D–G) Representative histograms (Top) and summary (Bottom) of the flow cytometry analysis results showing the significantly higher production of IL-6 (n = 5 in BD and n = 9 in HC; D and E) and TNF-α (n = 5 in BD and n = 9 in HC; F and G) in C1qhi monocytes than in non-C1qhiCD16+ monocytes and non-C1qhiCD16− monocytes after LPS stimulation. The data are summarized as the mean ± SD. Paired t tests (C, E, and G) were applied. TRITC-dextran, tetramethylrhodamine isothiocyanate-dextran; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. ns, not significant.
Next, we examined the phagocytic capabilities by incubating monocytes with fluorescence-labeled dextran. BD C1qhi monocytes engulfed more dextran than CD16+ monocytes (Δmean fluorescence intensity [MFI]: 832.8 ± 175.4 versus 567.8 ± 43.5, P = 0.0175) and CD16− monocytes (ΔMFI: 832.8 ± 175.4 versus 448 ± 84.6, P = 0.0045; Fig. 5 B and C and SI Appendix, Fig. S5D). We further detected proinflammatory cytokine production in lipopolysaccharide (LPS)-stimulated monocytes and observed that BD C1qhi monocytes produced significantly higher levels of IL-6 and TNF-α than CD16+ monocytes (IL-6: 5.1 ± 2.8% versus 2.5 ± 0.9%, P = 0.0362; TNF-α: 58.5 ± 3.0% versus 47.1 ± 7.1%, P = 0.0034) or CD16− monocytes (IL-6: 5.1 ± 2.8% versus 1.2 ± 0.5%, P = 0.0191; TNF-α: 58.5 ± 3.0% versus 31.4 ± 4.4%, P < 0.0001; Fig. 5 D–G). Collectively, our functional assays confirmed the proinflammatory characteristics of C1qhi monocytes, providing insights into the pathogenicity of C1qhi monocytes in the hyperinflammatory responses of BD patients.
Activated IFN-γ Signaling Stimulates the Expansion of C1qhi Monocytes in BD.
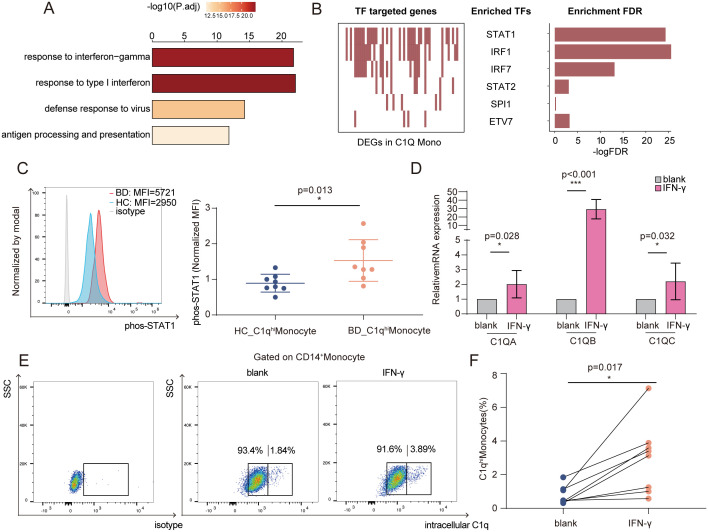
To understand the underlying mechanisms of C1qhi monocyte expansion in BD, we first analyzed DEGs in C1qhi monocytes between BD and HC and identified 254 up-regulated genes and 115 down-regulated genes in BD (SI Appendix, Fig. S6A). The up-regulated genes were prominently enriched in pathways of response to IFN-γ (Fig. 6A), corresponding to the global overexpression of IFN-γ–inducible genes in C1qhi monocytes (SI Appendix, Fig. S6A) and the DEG enrichment in total monocytes (Fig. 2G). Additionally, the overexpression of these IFN-induced genes (ISG15, LYE6, IFITM2, and IFITM3) was accompanied by preferential differentiation into C1qhi monocytes (Fig. 4D). Next, we used SCENIC tools (45) to predict which TFs modulate these DEGs and noted the marked enrichment of TFs that regulate IFN-γ response pathways, including STAT1 and IRF1 (Fig. 6B and SI Appendix, Fig. S6B); the expression of these TFs was also elevated in C1qhi monocytes from BD patients (SI Appendix, Fig. S6C). STAT1 phosphorylation was validated to be significantly increased in C1qhi monocytes from BD patients (normalized MFI: 1.5 ± 0.7 versus 0.9 ± 0.3, P = 0.013; Fig. 6C).
Fig. 6.
Increased IFN-γ levels led to C1qhi monocyte expansion in BD. (A) Bar plot showing the top four enriched pathways of significantly up-regulated genes in C1qhi monocytes from BD patients. (B) Top TFs predicted as regulators of DEGs (BD versus HC) in C1qhi monocytes by the SCENIC algorithm. Each red rectangle in the left panel represents a gene targeted by the TF. The significant TFs are shown in the middle panel, and the right panel shows the FDR value for the TF calculated by the hypergeometric test for the TF-targeted genes and all other DEGs. (C) Representative histograms (left) and quantification (right) of phosphorylated STAT1 levels (normalized MFI values) in C1qhi monocytes from the BD and HC groups (n = 8 per group). (D) Monocytes were stimulated with IFN-γ for 6 h, and relative mRNA expression (C1q genes) was measured using qRT-PCR (n = 5). (E and F) Representative flow cytometry plot (E) and graph (F) displaying the proportions of C1qhi monocytes after 24 h of IFN-γ stimulation (n = 8). The paired t test (F) and independent-samples t test (C and D) were applied. *P < 0.05; ***P < 0.001. FDR, false discovery rate; P.adj, adjusted P value; SSC, side scatter; phos-STAT1, phosphorylated signal transducer and activator of transcription 1.
The enriched IFN-related pathways in BD C1qhi monocytes inspired us to ask whether IFNs can stimulate C1qhi monocytes. We found that IFN-γ treatment significantly increased the expression of C1q genes in HC monocytes (fold change: 2.0 ± 0.9 for C1QA, P = 0.028; 29.2 ± 11.5 for C1QB, P < 0.0001; 2.2 ± 1.2 for C1QC, P = 0.032; Fig. 6D) and increased the proportion of C1qhi monocytes (3.0 ± 2.1% versus 0.8 ± 0.5%, P = 0.013; Fig. 6 E and F), which was significantly higher than IFN-α or IFN-β (SI Appendix, Fig. S6 D–F)
Meanwhile, we observed markedly increased IFN-γ concentrations in serum from BD patients (14.6 ± 14.8 pg/mL versus 4.012 ± 3.2 pg/mL, P < 0.0001; SI Appendix, Fig. S6G). By examining the main IFN-γ–producing cells, we found a significant increase in Th1 cells (IFN-γ–positive CD4 cells) among CD4+ T cells (12.9 ± 4.7% versus 8.15 ± 2.6%, P = 0.038; SI Appendix, Fig. S6 H and I). To gain insights into the regulatory relationships among these cell clusters, we inferred putative interactions between C1qhi monocytes and T/NK cells (SI Appendix, Fig. S7A and Dataset S6). Incoming interactions from T/NK cells to C1qhi monocytes were increased mainly in CD4+ T cells from BD patients but were reduced in CD8+ T cells and NK cells, in line with the increased IFN-γ production by CD4+ T cells. Additionally, the increased number of outgoing interactions from C1qhi monocytes to T/NK cells indicates intense signalings were sent from C1qhi monocytes in BD patients. Proinflammatory characteristics were also shown in BD-associated interactions, including CD40-CD40LG (46), HLA-DPB1-TNFSF13B (47), CXCL10-CXCR3 (48), and TNFRSF10B-TNFSF10 (49) (SI Appendix, Fig. S7 B and C and Dataset S6). These analyses reveal frequent crosstalk between C1qhi monocytes and CD4+ T cells in BD patients.
IFN-γ is a major effector in the pathogenesis of numerous inflammatory and autoimmune diseases (50, 51). We evaluated whether C1qhi monocytes are also present in patients with cancer or other IFN-γ–related immune diseases. Through an integrative analysis, we observed that the C1qhi monocytes in BD showed similar transcriptomic patterns to the C1QA monocytes found in rheumatoid arthritis (SDY998) (52) (SI Appendix, Fig. S7D). We also observed C1qhi monocytes in other immune diseases, such as systemic lupus erythematosus (Gene Expression Omnibus [GEO] database accession No. GSE135779) (53), and Kawasaki disease (GSE168732) (54) (SI Appendix, Fig. S7D), but not in blood cancers, including acute myeloid leukemia (GSE116256) (55), acute lymphocytic leukemia (GSE132509) (56), and chronic lymphocytic leukemia patients (GSE111014) (57) (SI Appendix, Fig. S7E and Dataset S7). Altogether, our data suggest the unrecognized mechanism by which IFN-γ promotes the generation of C1qhi monocytes in BD, and this mechanism may also be involved in the pathogenesis of other immune diseases.
C1qhi Monocytes Distinguish BD Patients From Healthy Donors and Respond to Treatment.
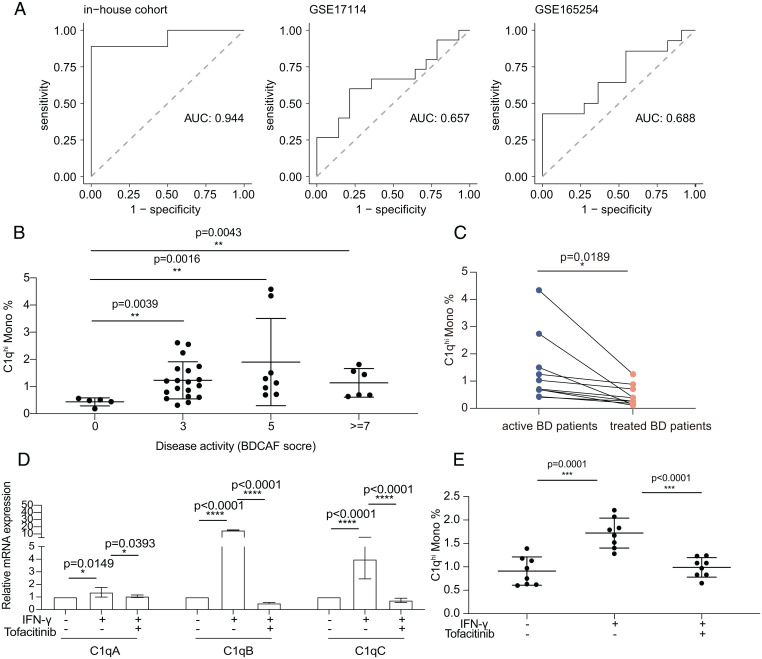
To explore the potential clinical value of C1Q Monos, we mined the predictive power of highly expressed genes in C1Q Monos in our in-house cohort (n = 19) and three published cohorts with active BD patients (GSE17114, n = 29; GSE165254, n = 25; and GSE70403, n = 58). The top five expressed genes in C1Q Monos were found to significantly explain variance and predict BD patients with an area under the curve above 0.7 in the three cohorts (Fig. 7A). Consistently, these five genes showed significantly higher relative expressions than randomly selected genes in the HC-corrected gene profiles of BD patients in the GSE70403 cohort (SI Appendix, Fig. S8A). Among the top five genes, C1q genes (C1QA, C1QB, and C1QC) were significantly up-regulated in the bulk RNA-seq analysis of PBMCs (SI Appendix, Fig. S8B), and C1q protein levels were also markedly increased in serum from BD patients (108.9 ± 18.4 μg/mL versus 82.0 ± 26.8 μg/mL, P = 0.03; SI Appendix, Fig. S8C). These results reveal C1Q Monos could distinguish active BD patients from HCs in diverse cohorts.
Fig. 7.
C1qhi monocyte distinguished BD patients from HCs and responded to BD treatment. (A) Receiver operating characteristic curve of the mean expression of the top five highly expressed genes (C1QB, C1QA, C1QC, FCGR3A, and HES4) in C1Q Monos to distinguish BD patients from HCs using in-house bulk RNA-seq data (n = 19), GSE17114 (n = 29), and GSE165254 (n = 25). (B) Proportion of C1qhi monocytes in circulating monocytes across BD patients (n = 38) with different disease activity indexes (BDCAF scores). Higher BDCAF scores mean more severe disease states. The data are summarized as the mean ± SD. (C) Proportion of C1qhi monocytes in peripheral monocytes from active and immunosuppressant-treated BD patients (n = 10). (D and E) IFN-γ–stimulated C1qhi monocytes were decreased after tofacitinib treatment, detected by qRT-PCR (D, at mRNA level) and flow cytometry (E, at protein level). Proportion of C1qhi monocytes in peripheral HC monocytes is shown in y axis (E). The data are summarized as the mean ± SD. The paired t test (C) and independent-samples t test (D and E) were applied. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. AUC, area under the curve.
We investigated BD-associated genes identified by genome-wide association studies across monocyte subtypes and found that C1Q Monos had the highest average expression of these genes (SI Appendix, Fig. S8D). Furthermore, C1qhi monocytes were positively correlated with the erythrocyte sedimentation rate (a clinical diagnostic parameter measuring the degree of BD inflammation) (r = 0.46, P = 0.004; SI Appendix, Fig. S8E) and showed higher proportions in BD patients with high disease activity scores (estimated by Behçet’s Disease Current Activity Form [BDCAF] scores) (Fig. 7B). In brief, these results reveal the significant association of C1qhi monocytes with disease activity, indicating their potential as clinical parameters to identify BD patients in the active inflammation stage.
Furthermore, we explored whether C1qhi monocytes could respond to drug treatment in BD. The immunosuppressive therapies significantly decreased C1qhi monocytes within the same patient after treatment (1.4 ± 1.2% versus 0.5 ± 0.4%, P = 0.0189; Fig. 7C). The Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway signaling is a canonical pathway that IFN-γ utilizes to activate STAT1 TF (51). Having observed the prominent activation of STAT1 and IFN-γ signaling in C1qhi monocytes (Fig. 6C), we next tested whether C1qhi monocytes respond to tofacitinib (a drug suppressing JAK-STAT signaling), which was a promising intervention for BD in our recent clinical studies (58). Intriguingly, tofacitinib treatment significantly decreased IFN-γ–stimulated C1qhi monocytes at both transcriptional (fold change: 1.4 ± 0.4 versus 1.1 ± 0.1 for C1QA, P = 0.0393; 15.2 ± 0.6 versus 0.5 ± 0.1 for C1QB, P < 0.0001; 4.0 ± 1.6 versus 0.7 ± 0.2 for C1QC, P < 0.0001, Fig. 7D), and protein levels (1.7 ± 0.3% versus 1.0 ± 0.2%, P < 0.0001; Fig. 7E and SI Appendix, Fig. S8F). Altogether, these results provide insights into the potential of C1qhi monocytes in monitoring treatment efficacy and indicate that targeting C1qhi monocytes could be an effective strategy for BD therapies.
Discussion
BD shares common features with autoinflammatory and autoimmune diseases (2, 5, 7). Our findings focused on revealing the unexplored immune landscape in BD at single-cell resolution, especially the markedly increased monocyte population. Among monocyte subtypes, the increased C1qhi monocytes manifested proinflammatory features and significant clinical relevance. This subtype was induced by IFN-γ in BD serum but was recovered by tofacitinib treatment. Briefly, our study delineates a single-cell view of circulating blood in BD patients, increasing the understanding of how certain subtypes contribute to BD hyperinflammatory status and revealing potential targets for BD therapies and clinical assessment.
Monocytes serve as a bridge between innate and adaptive immunity owing to their versatile functions, including antigen presentation, phagocytosis, and cytokine secretion (59). Impaired monocyte function is emerging in multiple autoinflammatory and autoimmune diseases (60–64). Our study found increased monocytes in BD blood, which may contribute to aberrant monocyte infiltration in the BD lesions, such as intestinal ulcers, blood vessels and the brain (65–67). Monocytes are a heterogeneous population of cells with functional variation (33), our previous study revealed the dysfunction of monocyte subsets in BD (50). Here, we leveraged the high resolution of single-cell technologies to redefine monocytes into eight subtypes, thereby gaining insight into the pathogenic role of monocyte subtypes in BD.
Our experiments showed the expansion of C1qhi monocytes in BD patients, as well as the enhanced phagocytosis and overproduction of proinflammatory cytokines. C1q also promoted the phagocytic capabilities of human monocytes and macrophages (68, 69) and stimulated the release of inflammatory cytokines (TNF-α and IL-6) from monocytes in RA patients (70). Consistently, trajectory inference revealed that BD monocytes preferentially differentiated into C1qhi monocytes but not monocyte-derived DCs, as suggested by previous studies showing that C1q expression increased in inflammatory monocytes (71) and inhibited DC differentiation (72). Moreover, C1q plays a central role in host defense as a pattern recognition molecule that recognizes both self and nonself ligands (73–76), indicating that proinflammatory C1qhi monocytes might help BD patients defend against pathogens that are known as critical triggers of BD development (7, 31).
It remains elusive how IFN-γ signaling mediates the immune disturbances in BD, although IFN-γ levels were significantly increased in BD patients (77–79). Our study discovered that C1qhi monocytes respond to IFN-γ activation in BD patients. As a critical transcriptional modulator of IFN-γ (51), STAT1 was also prominently activated in C1qhi monocytes, similar to the activation of STAT1 in CD14+ monocytes (80) and active macrophages (81) in other immune diseases. Additionally, monocytes treated with IFN-γ showed increased C1qhi monocytes, as suggested by the previous finding that exogenous IFN-γ induced C1q messenger RNA (mRNA) synthesis in murine macrophages (82). As Th1 cells are the main IFN-γ–producing cells among CD4+ T cells, our findings of increased IFN-γ secretion and Th1 cell differentiation support the hypothesis of activated Th1-dominated immunity in BD (79). Overall, these results suggest a model wherein overactive Th1 cells in BD patients increase the serum concentration of IFN-γ, which induces the expansion of C1qhi monocytes. As C1qhi monocytes were also identified in other immune diseases, our findings may provide insight into their pathogenic role in not only BD but also numerous IFN-γ–involved diseases.
A critical unmet need in BD is the identification of therapeutic targets and laboratory indexes to aid in clinical assessment (3). We found that C1qhi monocytes expansion was tightly correlated with BD disease activity. Furthermore, the marker genes of C1qhi monocytes significantly predicted BD patients in multiple cohorts, and C1q protein levels were increased in BD serum. Of note, C1qhi monocytes responded to both immunosuppressive agents and tofacitinib, which our recent study reported as an effective and safe drug for BD patients (58). These data implied that C1qhi monocytes and their markers could be potential clinical parameters to assess therapeutic efficacy and that targeting this subset would bring therapeutic benefits to BD patients.
There are several limitations of our current study. First, due to the low prevalence of BD, the limited number of BD patients in the scRNA-seq data may introduce statistical bias. Hence, we verified our findings by combining both experiments and additional analyses of public cohorts. Second, we merely focused on immune cells from circulating blood, and further study of BD lesions will improve understanding of the immune landscape of BD. Third, functional phenotypes and markers of the other monocyte subtypes will also need to be confirmed in the future.
In conclusion, our study delineates the immune landscape of BD at single-cell resolution, identifies the discriminative markers and transitional dynamics of monocyte subtypes, and pinpoints the proinflammatory contribution of C1qhi monocytes in BD. Our data provide a valuable scRNA-seq resource for BD-related studies and shed light on potential targets for targeted therapies and clinical assessment of BD.
Materials and Methods
Peripheral blood samples were collected from 52 BD patients who fulfilled the 2013 International Criteria for BD (83) and 56 HCs (Dataset S1). Sixteen samples were used for scRNA-seq(four BD and four HC samples for individual scRNA-seq data), and 19 samples were used for bulk RNA-seq (9 BD and 10 HC). The study was approved by the ethical committee of the Peking Union Medical College Hospital (JS-3418). All subjects provided written informed consent.
Detailed methods are provided in SI Appendix .
Supplementary Material
Acknowledgments
We thank all patients for their consent to participate in the study and thank Chenran Xu for his constructive suggestions. This work was supported by grants from the National Natural Science Foundation of China (Grant Nos.: 82171800, 81871299, and 82030017), the National Key Research and Development Project of China (Grant Nos.: 2019YFA0801500 and 2020YFC2008003), and the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (Grant Nos.: 2021-1-I2M-050 and 2019-RC-HL-006).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. Y.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2204289119/-/DCSupplemental.
Data Availability
The raw data and metadata of bulk RNA-seq and scRNA-seq data have been deposited in the GEO database (accession Nos. GSE198533 and GSE198616). The scripts used for all analyses in this study are available at GitHub (https://github.com/wangxiaoman618/BD_project.git). All other data are included in the manuscript and supporting information.
References
- 1.Tascilar K., et al. , Vascular involvement in Behçet’s syndrome: A retrospective analysis of associations and the time course. Rheumatology (Oxford) 53, 2018–2022 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Greco A., et al. , Behçet’s disease: New insights into pathophysiology, clinical features and treatment options. Autoimmun. Rev. 17, 567–575 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Hatemi G., Seyahi E., Fresko I., Talarico R., Hamuryudan V., One year in review 2020: Behçet’s syndrome. Clin. Exp. Rheumatol. 38 (suppl. 127), 3–10 (2020). [PubMed] [Google Scholar]
- 4.Leonardo N. M., McNeil J., Behcet’s disease: Is there geographical variation? A review far from the silk road. Int. J. Rheumatol. 2015, 945262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yazici H., Seyahi E., Hatemi G., Yazici Y., Behçet syndrome: A contemporary view. Nat. Rev. Rheumatol. 14, 107–119 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Pineton de Chambrun M., Wechsler B., Geri G., Cacoub P., Saadoun D., New insights into the pathogenesis of Behçet’s disease. Autoimmun. Rev. 11, 687–698 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Tong B., Liu X., Xiao J., Su G., Immunopathogenesis of Behcet’s disease. Front. Immunol. 10, 665 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hedayatfar A., Behçet’s disease: Autoimmune or autoinflammatory? J. Ophthalmic Vis. Res. 8, 291–293 (2013). [PMC free article] [PubMed] [Google Scholar]
- 9.Ben Ahmed M., Houman H., Miled M., Dellagi K., Louzir H., Involvement of chemokines and Th1 cytokines in the pathogenesis of mucocutaneous lesions of Behçet’s disease. Arthritis Rheum. 50, 2291–2295 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Koarada S., et al. , Increased entry of CD4+ T cells into the Th1 cytokine effector pathway during T-cell division following stimulation in Behcet’s disease. Rheumatology (Oxford) 43, 843–851 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Hamzaoui K., Borhani Haghighi A., Ghorbel I. B., Houman H., RORC and Foxp3 axis in cerebrospinal fluid of patients with neuro-Behçet’s disease. J. Neuroimmunol. 233, 249–253 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Parlakgul G., et al. , Expression of regulatory receptors on γδ T cells and their cytokine production in Behcet’s disease. Arthritis Res. Ther. 15, R15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasan M. S., Ryan P. L., Bergmeier L. A., Fortune F., Circulating NK cells and their subsets in Behçet’s disease. Clin. Exp. Immunol. 188, 311–322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clemente Ximenis A., et al. , In vitro evaluation of γδ T cells regulatory function in Behçet’s disease patients and healthy controls. Hum. Immunol. 77, 20–28 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Okuzaki D., et al. , Microarray and whole-exome sequencing analysis of familial Behçet’s disease patients. Sci. Rep. 6, 19456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tulunay A., et al. , Activation of the JAK/STAT pathway in Behcet’s disease. Genes Immun. 16, 170–175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verrou K. M., et al. , Distinct transcriptional profile of blood mononuclear cells in Behcet’s disease: Insights into the central role of neutrophil chemotaxis. Rheumatology (Oxford) 60, 4910–4919 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Arazi A., et al. ; Accelerating Medicines Partnership in SLE Network, The immune cell landscape in kidneys of patients with lupus nephritis. Nat. Immunol. 20, 902–914 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Der E., et al. ; Accelerating Medicines Partnership Rheumatoid Arthritis and Systemic Lupus Erythematosus (AMP RA/SLE) Consortium, Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat. Immunol. 20, 915–927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valenzi E., et al. , Single-cell analysis reveals fibroblast heterogeneity and myofibroblasts in systemic sclerosis-associated interstitial lung disease. Ann. Rheum. Dis. 78, 1379–1387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng S., et al. , A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 184, 792–809.e23 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Zhang L., et al. , Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 181, 442–459.e29 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Yu X.-X., et al. , Sequential progenitor states mark the generation of pancreatic endocrine lineages in mice and humans. Cell Res. 31, 886–903 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Böttcher C., et al. , Multi-parameter immune profiling of peripheral blood mononuclear cells by multiplexed single-cell mass cytometry in patients with early multiple sclerosis. Sci. Rep. 9, 19471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao Z. X., Miller J. S., Zheng S. G., An updated advance of autoantibodies in autoimmune diseases. Autoimmun. Rev. 20, 102743 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Ren X., et al. , COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell 184, 1895–1913.e1819 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leruste A., et al. , Clonally expanded T cells reveal immunogenicity of rhabdoid tumors. Cancer Cell 36, 597–612.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Newman A. M., et al. , Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bashford-Rogers R. J. M., et al. , Analysis of the B cell receptor repertoire in six immune-mediated diseases. Nature 574, 122–126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeuchi M., et al. , Dense genotyping of immune-related loci implicates host responses to microbial exposure in Behçet’s disease susceptibility. Nat. Genet. 49, 438–443 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mehmood N., Low L., Wallace G. R., Behçet’s disease: Do microbiomes and genetics collaborate in pathogenesis? Front. Immunol. 12, 648341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leccese P., Alpsoy E., Behçet’s disease: An overview of etiopathogenesis. Front. Immunol. 10, 1067 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guilliams M., Mildner A., Yona S., Developmental and functional heterogeneity of monocytes. Immunity 49, 595–613 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Zhao F., et al. , S100A9 a new marker for monocytic human myeloid-derived suppressor cells. Immunology 136, 176–183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becatti M., et al. , Neutrophil activation promotes fibrinogen oxidation and thrombus formation in Behçet disease. Circulation 133, 302–311 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Angerer P., et al. , destiny: Diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Ji Z., Ji H., TSCAN: Pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis. Nucleic Acids Res. 44, e117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Street K., et al. , Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coillard A., Segura E., In vivo differentiation of human monocytes. Front. Immunol. 10, 1907 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao J., et al. , The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elavazhagan S., et al. , Granzyme B expression is enhanced in human monocytes by TLR8 agonists and contributes to antibody-dependent cellular cytotoxicity. J. Immunol. 194, 2786–2795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lazennec G., Richmond A., Chemokines and chemokine receptors: New insights into cancer-related inflammation. Trends Mol. Med. 16, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong Y., Kinio A., Saleh M., Functions of NOD-like receptors in human diseases. Front. Immunol. 4, 333 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Z. Y., Chen S. L., Shen N., Lu Y., Cytokines and Behcet’s disease. Autoimmun. Rev. 11, 699–704 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Aibar S., et al. , SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grewal I. S., Flavell R. A., The role of CD40 ligand in costimulation and T-cell activation. Immunol. Rev. 153, 85–106 (1996). [DOI] [PubMed] [Google Scholar]
- 47.Steri M., et al. , Overexpression of the cytokine BAFF and autoimmunity risk. N. Engl. J. Med. 376, 1615–1626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee E. Y., Lee Z. H., Song Y. W., CXCL10 and autoimmune diseases. Autoimmun. Rev. 8, 379–383 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Sullivan G. P., et al. , TRAIL receptors serve as stress-associated molecular patterns to promote ER-stress-induced inflammation. Dev. Cell 52, 714–730.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Li C., et al. , Aberrant monocyte subsets in patients with Behçet’s disease. Clin. Immunol. 225, 108683 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Hu X., Ivashkiv L. B., Cross-regulation of signaling pathways by interferon-γ: Implications for immune responses and autoimmune diseases. Immunity 31, 539–550 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang F., et al. ; Accelerating Medicines Partnership Rheumatoid Arthritis and Systemic Lupus Erythematosus (AMP RA/SLE) Consortium, Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 20, 928–942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nehar-Belaid D., et al. , Mapping systemic lupus erythematosus heterogeneity at the single-cell level. Nat. Immunol. 21, 1094–1106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z., et al. , Single-cell RNA sequencing of peripheral blood mononuclear cells from acute Kawasaki disease patients. Nat. Commun. 12, 5444 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Galen P., et al. , Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell 176, 1265–1281.e24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caron M., et al. , Single-cell analysis of childhood leukemia reveals a link between developmental states and ribosomal protein expression as a source of intra-individual heterogeneity. Sci. Rep. 10, 8079 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rendeiro A. F., et al. , Chromatin mapping and single-cell immune profiling define the temporal dynamics of ibrutinib response in CLL. Nat. Commun. 11, 577 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J., et al. , A pilot study of tofacitinib for refractory Behçet’s syndrome. Ann. Rheum. Dis. 79, 1517–1520 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Stuart T., et al. , Comprehensive integration of single-cell data. Cell 177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narasimhan P. B., Marcovecchio P., Hamers A. A. J., Hedrick C. C., Nonclassical monocytes in health and disease. Annu. Rev. Immunol. 37, 439–456 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Puchner A., et al. , Non-classical monocytes as mediators of tissue destruction in arthritis. Ann. Rheum. Dis. 77, 1490–1497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wildenberg M. E., et al. , Increased frequency of CD16+ monocytes and the presence of activated dendritic cells in salivary glands in primary Sjögren syndrome. Ann. Rheum. Dis. 68, 420–426 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Ma W.-T., Gao F., Gu K., Chen D.-K., The role of monocytes and macrophages in autoimmune diseases: A comprehensive review. Front. Immunol. 10, 1140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bentham J., et al. , Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 47, 1457–1464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hirohata S., Histopathology of central nervous system lesions in Behçet’s disease. J. Neurol. Sci. 267, 41–47 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Hirohata S., Kikuchi H., Histopathology of the ruptured pulmonary artery aneurysm in a patient with Behçet’s disease. Clin. Exp. Rheumatol. 27 (suppl. 53), S91–S95 (2009). [PubMed] [Google Scholar]
- 67.Nara K., et al. , Involvement of innate immunity in the pathogenesis of intestinal Behçet’s disease. Clin. Exp. Immunol. 152, 245–251 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bobak D. A., Frank M. M., Tenner A. J., C1q acts synergistically with phorbol dibutyrate to activate CR1-mediated phagocytosis by human mononuclear phagocytes. Eur. J. Immunol. 18, 2001–2007 (1988). [DOI] [PubMed] [Google Scholar]
- 69.Bobak D. A., Gaither T. A., Frank M. M., Tenner A. J., Modulation of FcR function by complement: Subcomponent C1q enhances the phagocytosis of IgG-opsonized targets by human monocytes and culture-derived macrophages. J. Immunol. 138, 1150–1156 (1987). [PubMed] [Google Scholar]
- 70.Wu X. Y., et al. , Complement C1q synergizes with PTX3 in promoting NLRP3 inflammasome over-activation and pyroptosis in rheumatoid arthritis. J. Autoimmun. 106, 102336 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Schmiedel B. J., et al. , Impact of genetic polymorphisms on human immune cell gene expression. Cell 175, 1701–1715.e1716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hosszu K. K., Santiago-Schwarz F., Peerschke E. I. B., Ghebrehiwet B., Evidence that a C1q/C1qR system regulates monocyte-derived dendritic cell differentiation at the interface of innate and acquired immunity. Innate Immun. 16, 115–127 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Son M., Diamond B., Santiago-Schwarz F., Fundamental role of C1q in autoimmunity and inflammation. Immunol. Res. 63, 101–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Païdassi H., et al. , C1q binds phosphatidylserine and likely acts as a multiligand-bridging molecule in apoptotic cell recognition. J. Immunol. 180, 2329–2338 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gaboriaud C., Frachet P., Thielens N. M., Arlaud G. J., The human c1q globular domain: Structure and recognition of non-immune self ligands. Front. Immunol. 2, 92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bohlson S. S., Fraser D. A., Tenner A. J., Complement proteins C1q and MBL are pattern recognition molecules that signal immediate and long-term protective immune functions. Mol. Immunol. 44, 33–43 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Aridogan B. C., et al. , Serum levels of IL-4, IL-10, IL-12, IL-13 and IFN-gamma in Behçet’s disease. J. Dermatol. 30, 602–607 (2003). [DOI] [PubMed] [Google Scholar]
- 78.Ahn J. K., Yu H. G., Chung H., Park Y. G., Intraocular cytokine environment in active Behçet uveitis. Am. J. Ophthalmol. 142, 429–434 (2006). [DOI] [PubMed] [Google Scholar]
- 79.El-Asrar A. M. A., et al. , Cytokine profiles in aqueous humor of patients with different clinical entities of endogenous uveitis. Clin. Immunol. 139, 177–184 (2011). [DOI] [PubMed] [Google Scholar]
- 80.Puccetti A., et al. , Gene expression profiling in Behcet’s disease indicates an autoimmune component in the pathogenesis of the disease and opens new avenues for targeted therapy. J. Immunol. Res. 2018, 4246965 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu X., et al. , Sensitization of IFN-γ Jak-STAT signaling during macrophage activation. Nat. Immunol. 3, 859–866 (2002). [DOI] [PubMed] [Google Scholar]
- 82.Kolosov M., Kolosova I., leu R. W., Stimulation of macrophage synthesis of complement C1q by interferon-γ mediated by endogenous interferon-α/β. J. Interferon Cytokine Res. 16, 245–249 (1996). [DOI] [PubMed] [Google Scholar]
- 83.International Team for the Revision of the International Criteria for Behçet’s Disease (ITR-ICBD), The International Criteria for Behçet’s Disease (ICBD): A collaborative study of 27 countries on the sensitivity and specificity of the new criteria. J. Eur. Acad. Dermatol. Venereol. 28, 338–347 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data and metadata of bulk RNA-seq and scRNA-seq data have been deposited in the GEO database (accession Nos. GSE198533 and GSE198616). The scripts used for all analyses in this study are available at GitHub (https://github.com/wangxiaoman618/BD_project.git). All other data are included in the manuscript and supporting information.