Significance
Despite decades of progress in pancreas biology, the molecular features of pancreatic endocrine progenitors remain elusive, and how different hormone-producing islet endocrine cells choose their fate is largely unknown. A longstanding view suggests that ductal epithelium harbors bipotent progenitor cells, which are the origin of endocrine progenitors. Here, we provide evidence for the absence of a bipotent progenitor and suggest direct development of endocrine lineage from duct cells. Our results also suggest that chromatin “priming” in duct cells prior to Neurog3 expression is not required for endocrine differentiation.
Keywords: pancreas, Neurog3, ATAC-seq, scRNA-seq
Abstract
Delineating gene regulatory networks that orchestrate cell-type specification is a continuing challenge for developmental biologists. Single-cell analyses offer opportunities to address these challenges and accelerate discovery of rare cell lineage relationships and mechanisms underlying hierarchical lineage decisions. Here, we describe the molecular analysis of mouse pancreatic endocrine cell differentiation using single-cell transcriptomics, chromatin accessibility assays coupled to genetic labeling, and cytometry-based cell purification. We uncover transcription factor networks that delineate β-, α-, and δ-cell lineages. Through genomic footprint analysis, we identify transcription factor–regulatory DNA interactions governing pancreatic cell development at unprecedented resolution. Our analysis suggests that the transcription factor Neurog3 may act as a pioneer transcription factor to specify the pancreatic endocrine lineage. These findings could improve protocols to generate replacement endocrine cells from renewable sources, like stem cells, for diabetes therapy.
More than 400 million people are living with diabetes worldwide. Diabetes results from loss or dysfunction of hormone-producing endocrine islet cells in the pancreas, whose principal roles include regulation of circulating glucose levels. Advances in tissue engineering to replace nonfunctioning endocrine cells have sustained interest in understanding the molecular mechanisms of pancreatic endocrine cell differentiation (1).
A key event during endocrine pancreas development is expression of the transcription factor (TF) Neurog3 in select pancreatic duct cells (2). Neurog3 specifies endocrine progenitor cells, which differentiate into hormone-producing cells that delaminate from the duct and aggregate to form pancreatic islets (3–5). Several distinct endocrine cell types aggregate within pancreatic islets, including insulinpos β-cells, glucagonpos α-cells, somatostatinpos δ-cells, ghrelinpos ε-cells, and PPYpos γ-cells. Mice lacking pancreatic Neurog3 fail to develop endocrine islet cells (2, 6–8) In one model based on lineage tracing (9, 10), Neurog3pos cells are postulated to originate from a “bipotent progenitor” with potential to generate either ducts or islets (11).
Emerging single-cell technologies are revolutionizing developmental biology by enabling quantitative molecular analysis of transient, rare cell types in developing organs, especially lineage progenitor cells. Recently, several groups used single-cell RNA sequencing (scRNA-seq) to catalog dynamic transcriptome changes during mouse pancreas development and endocrine cell differentiation (12–17). Some studies provide evidence for the existence of endocrine progenitor subtypes, which may be biased toward specific hormone (17–19). While these reports contributed substantially to our understanding of endocrine pancreas development, they did not report the specification of the crucial islet δ-cell lineage (20) or investigate chromatin conformation changes by overcoming cell labeling ambiguities related to Neurog3-green fluorescent protein (GFP) cells (21).
To address these unmet needs, we used an integrative approach that combined cell-surface-marker-based sorting, genetic labeling, chromatin analysis, and single-cell assays to elucidate molecular mechanisms underlying gene expression changes during endocrine pancreas differentiation. By establishing pseudotime trajectories for hormone lineages, including islet δ-cells, we identified unique combinations of TFs guiding differentiation of the β-, α-, and δ-lineages. Chromatin accessibility analysis using Assay for Accessible Chromatin (ATAC-seq) (22) unexpectedly revealed extensive similarities between duct cells and those that activate Neurog3. We discovered genomic regions that undergo substantial transformation during development and identified enriched motifs in open chromatin specific to differentiation stages. We also applied powerful genomic footprint analysis to identify TF activity in open chromatin regions and found evidence of specific TF footprints linked to their associated motifs. Our analysis suggests a revised model for endocrine pancreas development by providing evidence for direct development of this lineage from duct cells and the absence of a bipotent progenitor.
Our results demonstrate the feasibility of using a combined scRNA-seq and ATAC-seq analysis to map gene regulatory networks that define pancreatic cell lineages. We anticipate our findings and those from similar works should foster efforts aiming to direct the development of renewable cell sources, like stem cells, for tissue replacement and regeneration.
Results
Single-Cell Transcriptomic Analysis of Endocrine Pancreas Development.
To understand gene expression dynamics during pancreatic endocrine cell differentiation, we performed scRNA-seq on cells isolated from mouse embryonic day 15.5 (E15.5) and E17.5 pancreas. We used the Neurog3-eGFP knock-in and Neurog3-Cre,Rosa-mTmG mice combined with cell-surface markers to isolate specific populations from the embryonic pancreas (see Methods) (21,23,24). We followed the SMART-Seq2 (25) protocol to sequence messenger RNAs (mRNAs) from single cells sorted into 96-well plates by fluorescence-activated cell sorting (FACS; SI Appendix, Fig. S1 A–D). Using this strategy, we collected and sequenced a total of 604 cells: 461 from E15.5 cells and 143 from E17.5 cells.
After initial read processing to count transcripts for each gene in each cell (Dataset S1), we used Monocle2 (26), a single-cell analysis tool, for downstream cell clustering and trajectory analysis (SI Appendix, Fig. S2). Unsupervised clustering organized cells based on transcriptome similarity, revealing a recognizable sequence of pancreatic endocrine cell differentiation (Fig. 1A and SI Appendix, Fig. S1 C and D). This developmental process included a progenitor cluster expressing high levels of Neurog3, a transitioning early endocrine cell cluster, a definitive endocrine cluster marked by high levels of Chga expression, and a cluster of exocrine cells marked by Cpa1 expression (Fig. 1B). We also found a small cluster of mesenchymal cells (14 cells, <3% of total cells), which were excluded from further analysis.
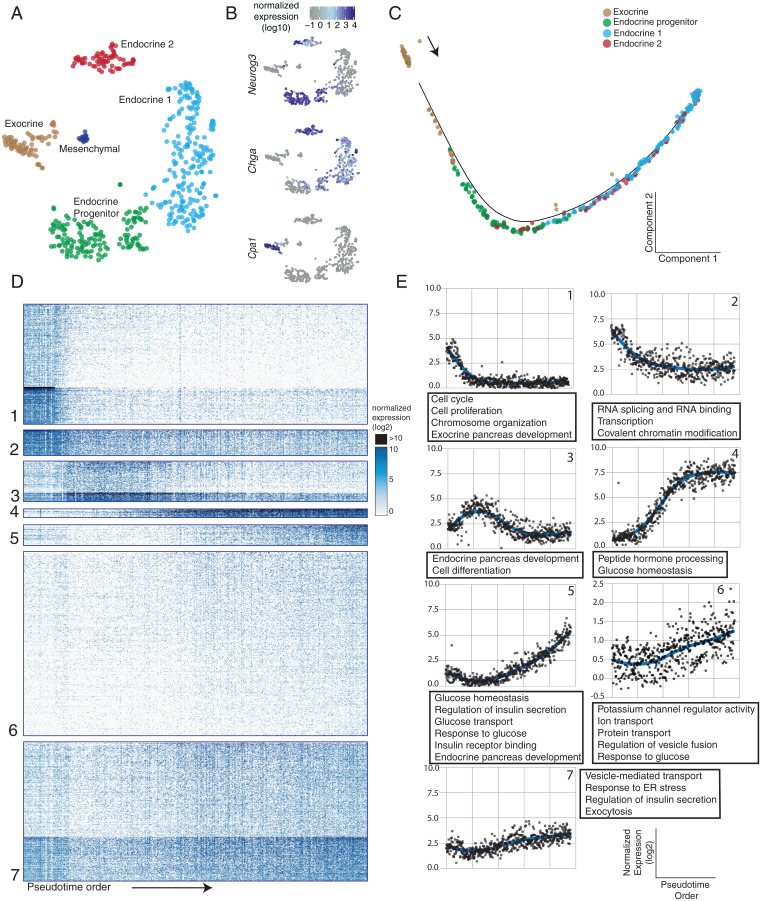
Fig. 1.
(A) t-SNE plot showing single-cell clusters, colored by cluster. Each dot is a single cell. Cluster names are indicated on the graph. (B) Marker gene expression levels overlaid onto the t-SNE plot: Cpa1 (exocrine), Neurog3 (endocrine progenitor), and Chga (pan-endocrine). (C) Alignment of single cells onto a pseudotime trajectory beginning with duct cells and ending with hormone-producing endocrine cells. Colors represent the clusters in A. (D) Heat map representation of >2,500 differentially expressed genes during pancreatic endocrine cell differentiation, organized into different clusters. Rows represent genes, and columns represent single cells ordered by the pseudotime order. (E) Graphs representing expression trends per cluster determined by fitting a LOESS curve of average gene expression per cluster, plotted over pseudotime. Each point represents the average expression of genes within each cluster for a single cell along pseudotime. Associated GO terms are listed in the text boxes.
To delineate gene expression programs involved in endocrine cell development, we aligned cells in a pseudotime trajectory based on quantitative gene expression profiles that change continuously in differentiating cells. This analysis placed all cells on a single trajectory that corroborated the known progression of duct cells into Neurog3pos progenitors, followed by hormone-expressing endocrine cells (Fig. 1C). We found more than 2,500 genes whose expression changed significantly along this pseudotime trajectory (q value < 0.05). Then, k-means analysis partitioned these differentially expressed genes into distinct gene clusters (Fig. 1D and Dataset S2). To better visualize the gene expression trends in each cluster, we used LOESS (or locally weighted) smoothing along pseudotime (Fig. 1E and SI Appendix, Methods). Gene Ontology (GO) term analysis identified enriched biological process terms in these clusters relevant to pancreatic differentiation (false discovery rate [FDR] < 0.2; Fig. 1E and Dataset S3) (3, 5).
Cluster 1 included genes that are expressed at high levels at the start of the pseudotime trajectory, then decline significantly or are extinguished as cells differentiate into the endocrine lineages. These genes included known regulators of multipotent pancreatic progenitor or exocrine cells (Ptf1a, Hes1, Notch1, Rbpj), the cell cycle (Mki67, Ccna2, Cdk1), and factors involved in maintenance of chromosome organization or covalent chromatin modifications (Smc4, Ezh2, and Ctcf). Cluster 2 genes had a similar trend, although their expression remained detectable in endocrine cells. These include genes regulating RNA binding and splicing, translation initiation, and ribonucleoprotein complexes. Cluster 3 genes are mainly expressed in endocrine progenitor cells and trend similarly with Neurog3 expression, including Pax4, Tox3, and Cbfa2t3. Most Cluster 3 transcripts were detectable only transiently in progenitor cells, then extinguished in endocrine cells. Cluster 3 was associated with GO terms related to cell differentiation and endocrine pancreas development (Dataset S3). Clusters 4, 5, and 6 contained genes whose expression increased following the Neurog3 induction. Cluster 4 genes included Chga, Pcsk2, Pax6, Iapp, Neurod1, and Isl1 and were turned on shortly after Neurog3 expression peaked, in early endocrine cells that still lack mRNAs encoding the principal islet hormones. Clusters 5 and 6 genes include the hormones Ins1, Ins2, Ppy, Sst, and Gcg, whose expressions peak in endocrine cells. These clusters also included genes involved in vesicle-mediated transport, ion transport, response to endoplasmic reticulum (ER) stress, regulation of insulin secretion, and exocytosis. Cluster 7 contained genes enriched with functions in the mitochondrial respiratory chain complex, proton transport, and ATP synthesis. Taken together, pancreatic endocrine cell specification involves highly dynamic gene regulatory programs, encompassing multiple groups of gene families with distinct functions.
Analysis of Pancreatic Endocrine Progenitors.
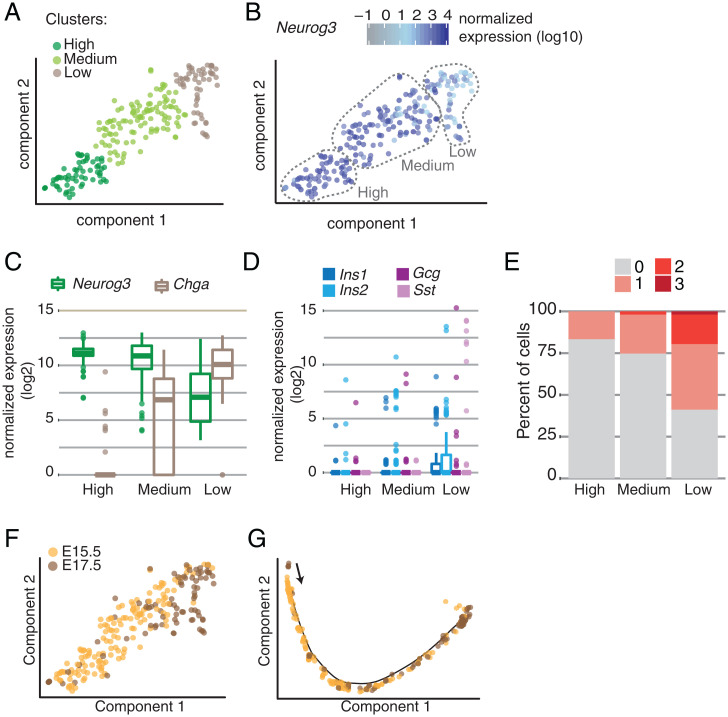
Prior studies reported the existence of distinct Neurog3pos endocrine progenitor subtypes (17–19). To investigate the heterogeneity in Neurog3pos progenitor cells, we focused on the cells expressing Neurog3 transcript in our dataset and visualized them using the t-SNE (t-distributed stochastic neighbor embedding) method. This analysis identified three clusters based on Neurog3 transcript abundance—designated as hi, med and lo—though none of the clusters split into visually distinct groups on the t-SNE projection (Fig. 2A). The Neurog3hi cells had the highest Neurog3 levels compared to other clusters (Fig. 2B), likely the result of increased Neurog3 transcription that occurs during the secondary transition of endocrine differentiation (7). Fewer than 10% of the Neurog3hi cells had detectable Chga expression (Fig. 2C). In comparison, Neurog3med and Neurog3lo cells had lower Neurog3 transcript levels, while Chga mRNA levels were increased (Fig. 2C). Thus, the observed “transcriptional heterogeneity” in Neurog3pos cells is likely a direct reflection of advancing development. Moreover, these data argue against a model where endocrine progenitor cells randomly develop from cells with heterogeneous Neurog3 levels. When we analyzed the expression of individual hormone genes, we found that the number of cells expressing Ins1, Ins2, Gcg, or Sst increased as cells transitioned from Neurog3hi to Neurog3lo progenitors, with Sst appearing only in the Neurog3lo cluster (Fig. 2D). Additionally, we investigated the number of cells simultaneously expressing one, two, or three of these hormone genes and found that the number of cells coexpressing multiple hormone genes increased as Neurog3 expression decreased. For instance, none of the Neurog3hi cells were polyhormonal, whereas 18% of Neurog3lo cells expressed two and 2% expressed all three hormone genes (Fig. 2E).
Fig. 2.
(A) t-SNE plot showing Neurog3-expressing cell subsets. Each dot is a single cell, colored by clusters or by (B) Neurog3 expression. (C) Box plots show normalized Neurog3 and Chga expression in each cluster. (D) Box plots show normalized hormone transcripts detected in each cluster. (E) Stacked bar plot showing the percent of cells within each cluster expressing zero, one, two, or three hormone transcripts (Ins1 or Ins2, Gcg, Sst). (F) t-SNE projection of Neurog3-expressing cells colored by the embryonic day they were isolated. (G) Pseudotime trajectory of Neurog3-expressing cells colored by the embryonic day they were isolated.
To investigate whether there is transcriptional heterogeneity in Neurog3pos endocrine progenitors isolated from different developmental stages, we examined all Neurog3pos cells by incorporating the embryonic stage information onto the clusters (Fig. 2F). We did not observe distinct clustering of E15.5 and E17.5 Neurog3pos endocrine progenitors; rather, the cells were arranged coincident with their developmental stage (Fig. 2F). When temporally ordering Neurog3pos cells via pseudotime analysis, the continuous developmental progression was apparent in a single trajectory without any branching (Fig. 2G). Taken together, in our dataset, we did not find evidence for lineage biases or subtypes in endocrine progenitors isolated from different embryonic time points. We found that nascent endocrine cells may transiently coexpress mRNAs encoding multiple hormones in an intermediate “polyhormonal” state preceding branch specification.
Single-Cell Trajectories Defining Endocrine Cell-Type Specification.
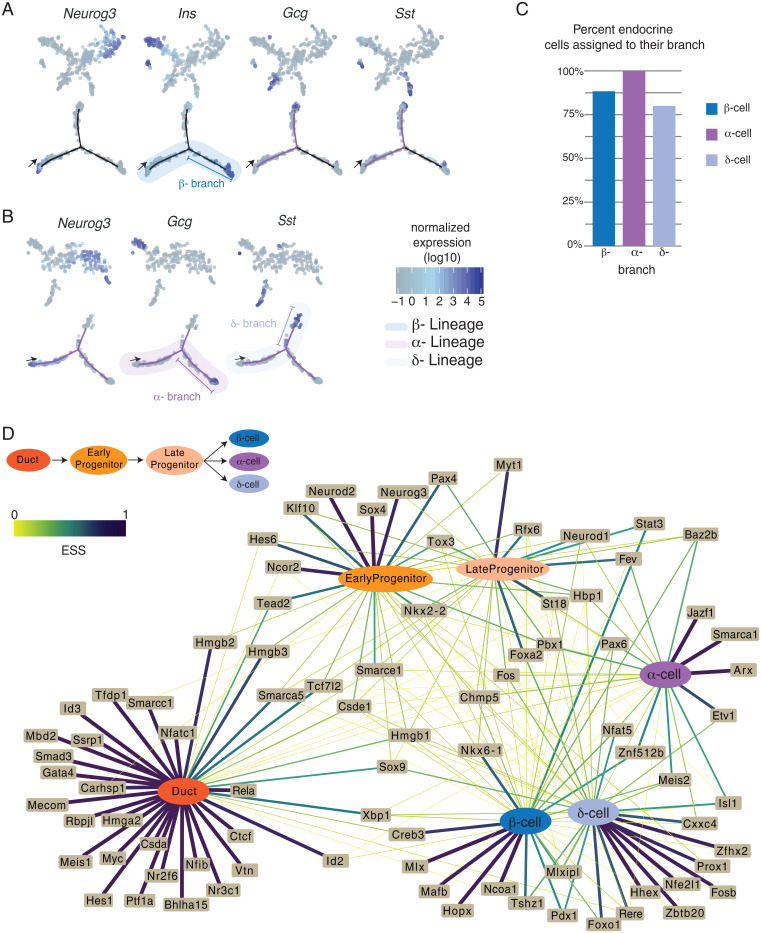
While Neurog3 is necessary and sufficient to establish the pancreatic endocrine lineage, the mechanisms underlying subsequent endocrine lineage diversification are not well established. Other studies using single-cell approaches successfully delineated β- and α-cell branches of islet endocrine cell differentiation but failed to identify a distinct branch for δ-cell specification (13, 15–19). In our data, an unsupervised approach including all cells also did not yield trajectories defining individual hormone lineages (Fig. 1C). We reasoned that when all cells are included, the substantial shift in gene expression programs at the onset of Neurog3 activation might hinder the discovery of less-pronounced differences in the initial β-, α-, and δ-cell lineage decisions. To circumvent this issue, we focused analysis on cells after Neurog3 peak expression (SI Appendix, Fig. S3) and performed semisupervised clustering with marker gene information (26). Briefly, endocrine progenitors and β-, α-, and δ-cells were preassigned based on marker genes before attempting clustering. A prior study used a similar approach to resolve mixed hematopoietic lineages (27). We then performed iterative rounds of trajectory analysis, sequentially removing cells already assigned to an endocrine cell branch in each iteration, until all branches were identified (Fig. 3 A and B). This approach successfully partitioned β-, α-, and δ-cells into nearly exclusive, specific branches (Fig. 3C), suggesting that expert curation can overcome some limitations of trajectory analysis (also see Discussion).
Fig. 3.
(A) (Top) t-SNE plots showing semisupervised clustering of single cells, first iteration to resolve β-lineage. Each dot is a single cell, colored by marker gene expression. (Bottom) Trajectory of cells beginning at the arrow; each dot is a single cell and is colored by marker gene expression. High expression of Ins1 and Ins2 is seen in cells at the end of the β-branch. (B) (Top) t-SNE plots showing semisupervised clustering of single cells, second iteration to resolve the α- and δ-lineages. Each dot is a single cell, colored by marker gene expression. (Bottom) Trajectory of cells beginning at the arrow; each dot is a single cell and is colored by marker gene expression. High expression of Gcg is seen on the α-branch, and Sst is seen in cells at the end of the δ-branch. (C) Bar graph indicating the percent of endocrine cells that were assigned in the appropriate branch: 88% for β-, 100% for α-, and 80% for δ-cells (D) Network showing the relationship between TF expression and cell state. The edges represent the expression specificity of TFs in each state. Thickness and color of the edges directly correspond to the expression specificity scores (ESSs) (SI Appendix, Methods). ESS values range from 0 to 1, where ESS = 1 means TF is exclusively expressed in that cell type, and ESS = 0 means no expression. Ubiquitous expression is ESS = 0.166.
TF Networks Regulating Islet Cell Lineage Gene Expression.
To reveal the gene expression changes underlying distinct trajectories of endocrine cell specification, we performed differential gene expression analysis among cells assigned to the β-, α-, and δ-lineages. We defined the lineages as beginning from the duct cells and ending with hormone-expressing endocrine cells (Fig. 3 A and B and Dataset S4). We focused our analysis on TFs due to their well-established role in determining cell fates. This analysis revealed 145 TFs whose expression changed significantly during endocrine cell differentiation (SI Appendix, Fig. S4). We visualized how these TFs may be regulating distinct lineages by constructing a network based on TF expression patterns in each cell type (duct, β-, α- and δ-cells) or state (early progenitor, late progenitor; Dataset S5, also see SI Appendix, Methods for details). For instance, Hes1 was detected in duct cells and, thus, was connected to the node representing the duct cell.
Topological examination of the TF expression–cell-state interaction network revealed three network patterns. In network pattern 1, we found TFs highly specific to a single lineage. For example, 92% of cells in the β-cell lineage express Nkx6-1, and 71% of α-cells express Arx. Nkx6-1 is thought to repress transcription of Arx, which specifies the α-cell lineage; conversely, Arx is postulated to repress transcription of Nkx6-1, which specifies the β-cell lineage (28). We found that Smarca1 is highly specific to the α-cell lineage, and this is consistent with recent reports of Smarca1 activation during α-cell development, prior to Gcg expression (13, 17). Smarca1 is an ATP-dependent chromatin remodeler, which can be selectively recruited to cell-type-specific enhancer elements (29). A second TF, Etv1, is a Neurog3 target (30), and in our data we find Etv1 is highly specific to the fetal α-cell lineage, indicating this TF has a functional role in α-cell development. In our network, we confirmed that Hhex is specific to the δ-lineage (31) and found additional factors. Zbtb20 has increased expression in δ-cells relative to β- and α-cells and, to our knowledge, has not been reported before. Instead, Zbtb20 was recently identified as a TF upregulated in the α-cell lineage (17). Because the δ-lineage was not defined in this report, it is possible that the uncategorized δ-cells aligned with the α-lineage instead. Other TFs that are highly specific to the δ-cell lineage, but with no known functions, include Zfhx2, Rere, and Cxxc4.
In network pattern 2, we found TFs that are expressed in multiple cell types or states. For instance, the high mobility group proteins Hmgb2, Hmgb3, and Tead2, a YAP (Yes-associated protein 1) signaling factor, are initially expressed in duct cells and continue to be expressed in early Neurog3pos progenitors. We also found known TFs—including Isl1, Rfx6, Pax6, and Meis2—in the β-, α-, and δ-cell lineages. In line with a prior report, almost all endocrine cells in the β-, α-, and δ- lineages appear to pass through a Fevpos stage after Neurog3 expression (13). In this network, Fev is most specific to late progenitors. After islet cells transit through a Fevpos stage, Fev expression rapidly declines in the β-cell lineage but remains at detectable levels in α- and δ-cells (SI Appendix, Fig. S4).
Network pattern 3 includes TFs that follow an ON-OFF-ON pattern as cells differentiate from duct to progenitors to endocrine lineages. For example, Xbp1 is abundant in duct cells, but its levels decrease in early and late Neurog3pos progenitors then increase in β-, α-, and δ-cells. In mice, loss of Xbp1 results in hyperglycemia (32), abnormal zymogen granules, and aplasia of acinar cells (33). Xbp1 is an essential regulator of the unfolded protein response and ER stress (34). Similarly, Creb3 and Id2 follow the ON-OFF-ON pattern. These TFs were recently reported to be associated with ER and oxidative stress response programs in human islet β-cells (35).
Chromatin Accessibility Dynamics during Islet Endocrine Cell Differentiation.
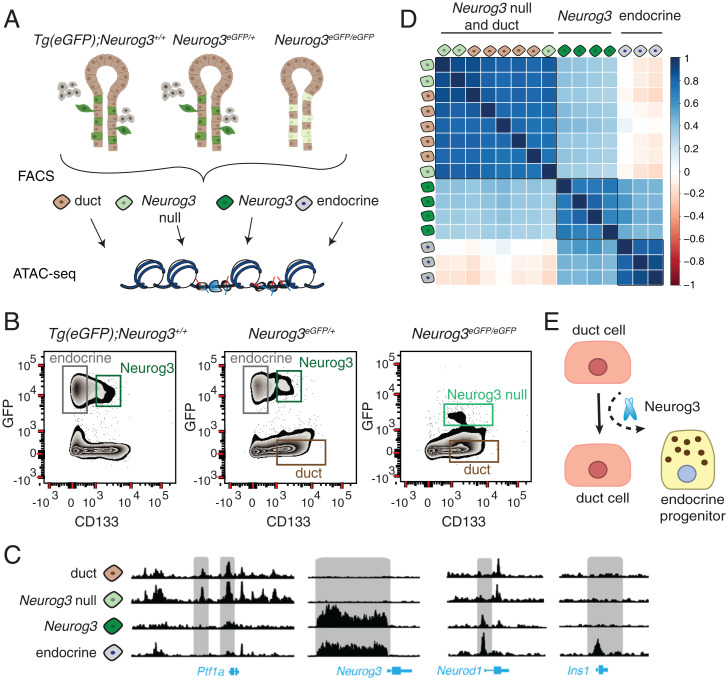
To investigate chromatin accessibility changes during endocrine cell differentiation, we performed ATAC-seq (22) on purified populations of duct, endocrine progenitor, and endocrine cells isolated from E15.5 pancreas using the Neurog3-eGFP knock-in mice (21) (Fig. 4A and SI Appendix, Table S1). In these mice, the coding region of Neurog3 is replaced by an eGFP cassette, thereby regulating eGFP production from the endogenous Neurog3 cis-regulatory element, including the promoter. As reported previously, heterozygous Neurog3eGFP/+ animals form a complete endocrine pancreas with no discernable phenotypes (21). However, in homozygous Neurog3eGFP/eGFP animals, eGFPpos cells lack Neurog3 and fail to differentiate further into the endocrine lineage.
Fig. 4.
(A) ATAC-seq workflow used in this study. (B) Representative FACS plots showing sorted cell populations and gating strategy. Three mouse genotypes were used to collect four types of cell populations from E15.5 embryos. (C) ATAC-seq reads obtained from different cell populations visualized on the genome browser near the gene loci: Ptf1a, Neurog3, Neurod1, Ins1. (D) Pearson’s correlation matrix showing the similarity of ATAC-seq samples. A value of 1 indicates high correlation, 0 indicates no correlation, and −1 indicates anticorrelation. The samples are colored as in A. (E) A parsimonious model for endocrine pancreas differentiation, where Neurog3 functions as a pioneer factor to shift cells from the default ductal lineage to the endocrine lineage.
To achieve requisite specificity needed for experiments involving purification of Neurog3-expressing cells, we managed two concerns not addressed in prior studies (19, 36). First, since Neurog3 protein stability is transient and short-lived compared to eGFP (37), we needed methods to discriminate between eGFPpos Neurog3pos progenitors and eGFPpos Neurog3neg endocrine cells that have ceased to express Neurog3. We achieved this using modified cell-sorting strategies (24) (see Methods). Second, to address possible concerns about Neurog3 gene dosage effects on endocrine cell differentiation, we used mice that are wild type (Tg[eGFP]; Neurog3) (38), heterozygous, or homozygous null for Neurog3 (Fig. 4B). This enabled direct comparison of chromatin states in endocrine progenitor cells with varying Neurog3 gene dosage. Specifically, we analyzed four distinct cell populations in different genetic backgrounds: 1) Neurog3pos hormoneneg cells (Neurog3); 2) eGFPpos Neurog3-null cells (Neurog3 null); 3) hormonepos islet cells (endocrine); and 4) duct cells (duct) (Figs. 4 A and B and SI Appendix, Table S1). In total, we performed ATAC-seq on 15 primary pancreatic cell samples.
After aligning sequencing reads, we visually inspected loci near genes essential for pancreas development like Ptf1a, Neurog3, Neurod1, and Ins1 (Fig. 4C). ATAC-seq revealed substantial reorganization of chromatin accessibility in regions near these and other genes (see below) during differentiation from duct cells to Neurog3pos endocrine progenitor cells and endocrine cells. For instance, open chromatin “control regions” in the Ptf1a locus were detected in wild-type duct cells and Neurog3-null cells; the accessibility of this chromatin was then eliminated as duct cells transitioned into endocrine progenitors, a “closed” state also maintained in endocrine cells (39). Neurog3 is thought to bind its own promoter (40), and, consistent with this view, we observed opening of the Neurog3 promoter in Neurog3 cells, coinciding with Neurog3 expression (Fig. 4C). In Neurod1 locus, an established Neurog3 target, promoter-proximal chromatin was closed in duct cells but became accessible in Neurog3pos endocrine progenitors. In the Ins1 locus, chromatin in control regions remained closed until cells committed to the endocrine lineage. Thus, cell purification combined with ATAC-seq generated high-quality chromatin maps that corresponded to distinct differentiation stages.
To investigate the similarity in chromatin states between ATAC-seq samples, we calculated pairwise Pearson correlation coefficients and organized samples by clustering (Fig. 4D). This analysis revealed three groups that corresponded to duct cells, Neurog3pos progenitors, and endocrine cells. Chromatin profiles of cells isolated either from wild-type or heterozygous Neurog3 mice were similar. Unexpectedly, Neurog3-null cells clustered with wild-type duct cells (Fig. 4D). If ductal epithelia harbored bipotent cells that could become either endocrine progenitors or duct cells, we expected to see a distinct clustering of Neurog3-null from duct cells. Thus, cells that activated Neurog3 transcription in the ductal epithelium but could not differentiate into endocrine lineage have chromatin that is indistinguishable from duct cells. This suggests that chromatin “priming” in duct cells prior to expression of Neurog3 is not required for endocrine differentiation. Furthermore, Neurog3 might be a pioneer TF whose functions include the capacity to initiate nucleosome displacement or conformational changes in inaccessible chromatin (Fig. 4E) (41).
Differentially Accessible Chromatin Regions Reveal Cis-Regulatory Elements that Mediate Endocrine Lineage Specification.
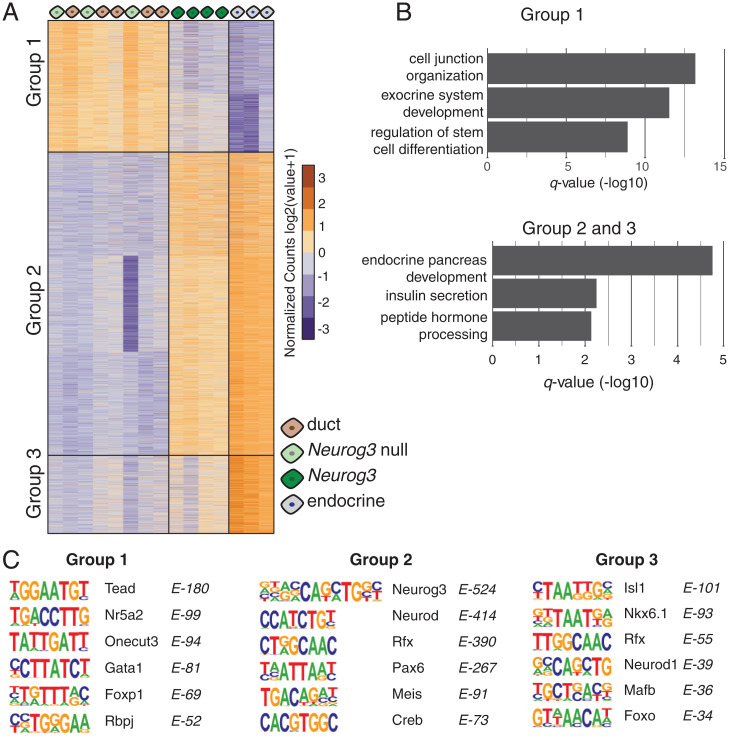
To identify differentially accessible chromatin regions in our sorted cell types, we analyzed the ATAC-seq signal at every peak across all samples using the DESeq algorithm (42). From a total of 116,942 ATAC-seq peaks, we found 10,687 that have significant accessibility changes between samples (FDR < 0.001). The k-means clustering of differentially open peaks revealed three main groups of genomic regions that represent the open chromatin profiles of distinct cell states (Fig. 5A and Dataset S6). In group I, we observed 2,754 accessible regions in duct cells (either wild type or Neurog3 null) that switch to a closed state in Neurog3pos progenitors and remain closed in endocrine cells. Using the GREAT algorithm (Genomic Regions Enrichment of Annotations Tool) (43), we found that these regions were associated with genes that have established roles in exocrine pancreas cell development, gland development, and cell proliferation like Fgfr, Smad, Ptf1a, Hes1, and Notch signaling (Fig. 5B). Group II includes 6,312 and group III includes 1,621 accessible regions (Fig. 5A). Based on the ATAC-seq signal, we observed that these regions are closed in duct cells, are open in Neurog3pos progenitors, and remain in open state in endocrine cells. The regions in group III have significantly stronger ATAC-seq signal in endocrine cells compared to endocrine progenitors, suggesting that other regulatory factors independent of Neurog3 might be enhancing the accessibility in these regions once the cells begin producing hormones. GREAT analysis linked chromatin from groups II and III to genes known to regulate endocrine pancreas differentiation, or cardinal features of islet function including peptide hormone processing, and regulation of calcium ion-dependent exocytosis (Fig. 5B).
Fig. 5.
(A) Heat map showing differentially open chromatin regions. Each column is an ATAC-seq sample, and each row is an open chromatin region, organized by k-means clustering. Three groups of open regions were identified and indicated on the graph. (B) Bar graphs show significant GO terms associated with open regions identified in A. (C) Position weight matrices of enriched TF motifs found in each of the three open chromatin groups.
To discover TF motifs within these dynamic chromatin regions, we performed TF motif enrichment analysis using the HOMER algorithm (Hypergeometric Optimization of Motif EnRichment) (44). Consistent with the GREAT analysis, we found overrepresented motifs (Fig. 5C) of exocrine lineage specific factors like Tead, Rbpj, and Nr5a2 in accessible chromatin regions of duct cells in group I. In contrast, our analysis of regions in group II identified Neurog3, NeuroD, Rfx, and Pax motifs— all known regulators of endocrine pancreas development. Likewise, the analysis of group III regions yielded enriched TF motifs of lineage markers of β- and α-cells, including Mafb and Isl1. Thus, by combining cell sorting, mouse genetics, and ATAC-seq, we identified developmentally resolved chromatin states and found sequence motifs enriched for regulators of pancreas development, demonstrating the sensitivity and specificity of our approach.
Identifying TF Occupancy in Regulatory Genomic Regions during Endocrine Cell Differentiation.
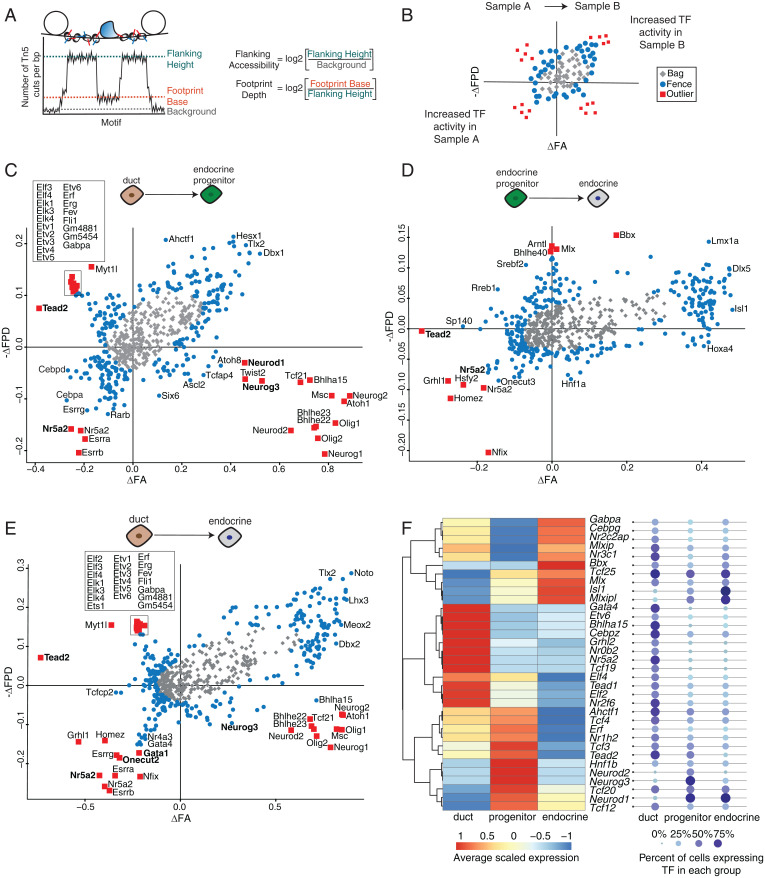
Chromatin accessibility assays, like ATAC-seq and Dnase-seq, enable identification of TF occupancy sites where DNA is protected from enzymatic cleavage or transposition due to TF binding, leaving a “TF footprint” (22, 45). We envisioned that an integrative approach combining TF footprint and single-cell gene expression profiles could uncover TF activity during endocrine pancreas differentiation. We used the BaGFoot algorithm (Bivariate Genomic Footprinting) to identify changes in TF occupancy between two cell states using our ATAC-seq samples (46). BaGFoot calculates two parameters for each TF motif: 1) footprint depth (FPD), the relative protection of DNA at the TF motif site; and 2) flanking accessibility (FA), the quantification of accessible chromatin near the TF motif (Fig. 6A). TF binding dynamics is expected to affect these two parameters genome-wide; thus, by comparing the FPD and FA between two samples, we can infer changes in TF activity. For instance, a motif with a deep FPD and high FA would indicate strong protection at the motif site. These results are represented in “bagplots,” which are analogous to “box and whisker” plots (Fig. 6B, also see SI Appendix, Methods).
Fig. 6.
(A) Cartoon describing how FPD and FA are calculated from ATAC-seq data. (B) Guide to interpret pairwise comparisons using a bagplot. (C–E) Bagplots displaying TFs with upregulated activity when comparing two samples. Outliers are marked by red squares, TFs in the fence are marked by blue circles, and TFs in the bag are marked by gray diamonds. Bolded TFs correspond to the de novo motifs found in the HOMER analysis. (F) Heat map showing average expression levels of outlier TFs in duct, progenitor, or endocrine cells. TFs are ordered by hierarchical clustering, and expression levels are scaled to each row. Each TF is detected in at least 25% of cells in each group.
We calculated the FPD and FA values for more than 650 curated TF motifs using our ATAC-seq data. Pairwise comparison of footprint signatures in duct cells and Neurog3pos progenitors or duct cells and endocrine cells revealed changes in TF activity. Consistent with the HOMER-based motif analysis, we found strong footprint signals for Gata and Onecut TFs and nuclear receptors in duct cells. In endocrine cells, we detected footprints for homeobox TFs including Isl1, Hnf1a, and Pou TFs (Fig. 6 C–E and Dataset S7). Comparison of Neurog3pos progenitors and endocrine cells revealed relatively modest TF activity changes (Fig. 6D). Similar to the findings above, the most significant changes in TF footprint activity occur during the transition from ductal to endocrine progenitor state, supporting the view that activation of Neurog3 is the main driver of changes in chromatin accessibility and gene expression.
We also calculated the FA and FPD scores of the TF motifs we derived de novo from our ATAC-seq motif enrichment analysis (Fig. 5C). These motifs displayed increased FA or FPD in the appropriate cell type (indicated in bold; Fig. 6 C–E and Dataset S7), independently validating the TF occupancy at these sequences.
While FPD and FA are often correlated, some TFs exhibited only increased FA without a detectable footprint, likely due to distinct DNA binding kinetics— for instance, those TFs with high OFF rates (46, 47). TFs matching this profile were basic helix–loop–helix factors including Neurog3, Neurod1, and Ascl2 in endocrine progenitors. In addition, some motifs were found in the second quadrant, displaying deeper FPD but decreased FA in endocrine or Neurog3pos progenitor samples compared to duct cells. This profile is consistent with repressor TFs, whose DNA binding activity leads to decreased accessibility surrounding the motif. We found that Tead factors and ETS (Erythroblast Transformation Specific) family TFs—including Etv6, Elf2/4, and Erf—were included in this group (Fig. 6 C and E).
Paralogous TFs often bind similar DNA motifs, resulting in nearly identical footprint scores. For instance, Neurog3 motif could also be recognized by Neurog1 or Neurog2 (Fig. 6 C and E). Thus, footprint analysis alone cannot determine which TF family member might be occupying the regulatory sequences in a particular cell type. Integrating BaGFoot results with single-cell expression data overcomes this limitation. We found more than 50 TFs whose expression correlates with a matching footprint (Fig. 6F and SI Appendix, Fig. S5 and Dataset S8). Among the TFs whose expression was detected in at least 25% of the cells within each group (Fig. 6F), we confirmed the activity of known regulators—for instance, Nr5a2 and Gata4 in duct cells (48, 49). In addition, we found footprints of several relatively less-studied nuclear receptor TFs (Nr2f6, Nr3c1), and we identified a Nuclear Factor 1 family TF, Nfix, that has increased activity in Neurog3pos progenitor cells (Fig. 6D and SI Appendix, Fig. S5). Taken together, footprint and expression analysis predicted dozens of regulators whose roles have not been previously explored in endocrine cell development and provided quantitative evidence of selective TF occupancy in different pancreatic cell types.
Discussion
Here, we established an integrative approach combining cell purification, genetic labeling, single-cell transcriptomics, chromatin accessibility assessment, and TF footprint analysis to elucidate molecular mechanisms underlying pancreatic endocrine cell specification. We show that mouse pancreatic endocrine cell development is a dynamic process involving a network of TFs whose expression is selectively tuned to define specific hormone lineages. We were able to delineate gene expression changes leading to δ-cell specification and nominate unrecognized factors that could regulate δ-cell function. We demonstrate that in developing pancreatic epithelial cells, chromatin undergoes substantial reorganization upon Neurog3 induction. In remodeled genomic regions during development, we identified enriched TF motifs and footprints that correspond to TF activity in specific cell types.
Two prior studies (17, 19) postulated that the Neurog3pos progenitors exhibit heterogeneity and temporal lineage biases. In our study, using the same mouse models and embryonic stages, we did not find evidence for such bias even though our gene expression results aligned well with differential gene expression reported by Scavuzzo and colleagues. Thus, differences in our findings may reflect interpretation of alternative analytical approaches, rather than primary data. Similar views about the challenges in single-cell analysis and biological interpretation were discussed in recent reviews (50, 51).
Using an iterative, semisupervised clustering approach, we successfully identified branching points that specify three hormone lineages, including β-, α-, and δ-cell lineages. In our dataset, we found only 13 pancreatic polypeptide (PP) cells, which did not provide sufficient statistical power to permit a PP-branch identification. Due to the known regulatory role of TFs, we focused on differentially expressed TFs between these lineages. We identified known, as well as previously understudied, pancreatic TFs that may have roles in islet endocrine cell specification. Based on analysis of TF expression in specific developmental timelines, we suggest that pancreatic lineage specification is governed by a network of TFs with dynamic, overlapping expression profiles. For instance, while Neurog3 is necessary for the endocrine lineage, it needs to be turned off to permit further differentiation of endocrine cell lineages. We speculate that this may explain the low efficiency observed in direct reprogramming approaches when a handful of lineage-specific TFs are constitutively overexpressed to force nonislet cells toward a β-cell fate (52, 53). Our focused analysis of Neurog3pos cells revealed that the pan-endocrine state precedes specific endocrine lineages. This may explain why the interconversion of hormone cell types does not require Neurog3 (54, 55). Consistent with this view, a recent report showed that NEUROG3 binds genomic regions regulating pan-endocrine genes but not regions regulating specific hormone expression (56). Likely reflecting the sensitivity of SMART-seq assays used here, we found that the early endocrine cells are polyhormonal as defined by their transcriptome. These results are also reminiscent of reports of polyhormonal cells generated during the in vitro differentiation experiments using human embryonic stem cells or adult tissues with endoderm origin (14, 57–60).
Chromatin accessibility is thought to be a better predictor of cell identity than transcriptome analysis, with changes in chromatin states often preceding changes in gene expression (61). By taking advantage of established cell markers and genetic models, we were able to dissect the chromatin accessibility changes during endocrine cell differentiation at unprecedented resolution. The unexpected similarity between duct cells and those that activate Neurog3 forced us to re-evaluate extant endocrine cell development models. For example, our findings provide evidence that pancreatic “trunk cells,” previously postulated to be oligopotent progenitors, may simply be duct cells that default to the ductal lineage in the absence of Neurog3 (Fig. 4E). Comparison of Neurog3pos cells from heterozygous (Neurog3+/eGFP) and homozygous wild-type (Tg(eGFP); Neurog3+/+) mice showed that a single, wild-type Neurog3 allele is sufficient to drive global chromatin reconfiguration in the pancreatic endocrine lineage. It is likely that in individual ductal epithelium cells, Neurog3 concentration needs to reach a critical threshold to compete with histone proteins for DNA binding (11, 62).
Using a TF footprint algorithm, we provide quantitative, cell-type-specific TF occupancy profiles at nucleotide resolution in pancreatic duct, endocrine progenitor, and endocrine cell regulatory DNA. To our knowledge, this is the most comprehensive analysis of TF activity correlated with gene expression during pancreas development in any organism. TF-regulatory DNA interactions form the basis of gene regulatory networks, which are central to determining and maintaining cell-type-specific transcription, cell fate, and function. Further delineation of gene regulatory networks defining pancreatic cell lineages will be crucial for understanding pancreas disorders and has the potential to improve gene therapy approaches using CRISPR-guided synthetic engineering to generate cells and tissues (63). Expanding these strategies to human pancreas or in vitro differentiation efforts using emerging single-cell technologies that query chromatin and gene expression profiles (64) could offer new approaches to investigating the pathogenesis of type 1 and type 2 diabetes.
Materials and Methods
Tissue Processing and FACS.
Pancreata were dissected from E15.5 and E17.5 embryos and checked for GFP using a fluorescence dissecting microscope. Details of mouse models can be found in SI Appendix, Materials and Methods, section A. GFPpos pancreata were then digested with Tryp-LE express (ThermoFisher, 12605-010) for 5 min at 37 °C, with regular pipet agitation to disrupt tissue. The digestion reaction was stopped by adding FACS buffer, which contains Ca2+ and Mg2+ free PBS (phosphate buffered saline) supplemented with 2% bovine serum albumin and 10 mM EGTA (ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid). The cell suspension was filtered to remove debris using a cell 70-μm cell strainer (BD Biosciences). Red blood cells were eliminated from dissociated cells using a lysis buffer (BioLegend). Cells were then stained with Aqua live/dead viability dye (Thermo Fisher) to exclude dead cells during sorting. Cells were incubated with a blocking solution containing FACS buffer and goat immunoglobulin G (Jackson Labs, 1:20 dilution) prior to staining with cell-surface antibodies. After blocking, antibody staining was performed on ice for 30 min using the following antibodies: biotin mouse anti-CD133 (13A4, 1:100; eBioscience) and Streptavidin-APC (1:200; eBioscience). We also used CD45-PE-Cy7 (eBioscience) to label and exclude leukocytes. We previously showed that CD133 labels Neurog3pos endocrine progenitors and duct cells (24). By contrast, hormonepos islet cells that no longer produce Neurog3 are CD133neg. After exclusion of CD45pos cells, the following gating strategies defined pancreas cell subpopulations: GFPposCD133neg cells were considered “endocrine,” GFPposCD133poscells were “Neurog3pos” or “Neurog3 null” if obtained from null animals, and GFPnegCD133pos cells were considered “duct” (24, 30). Representative gates are shown in Fig. 4B. Note that the GFP intensity of Neurog3-null cells is reduced. In wild type cells, Neurog3 normally enhances its own expression through an autoregulatory “positive feedback loop.” In null cells, this mechanism is likely absent (21, 40, 65).
scRNA-seq.
scRNA-seq libraries were generated using the SMART-Seq2 method as described (25). Dissociated cells were sorted directly into 96-well plates containing lysis buffer with ERCC (External RNA Controls Consortium) RNA spike-in controls (ThermoFisher). The details about the sorted cell populations, genotypes, and associated plate codes are available in the Gene Expression Omnibus (GEO) metadata file linked to this study (GSE146006). The lysis reaction was followed by reverse transcription with template-switch using an LNA-modified (locked nucleic acid) template switch oligos to generate complementary DNA (cDNA). After preamplification, DNA was purified and analyzed on an automated Fragment Analyzer (Advanced Analytical). A cDNA fragment profile corresponding to each single cell was individually inspected, and only wells with successful amplification products (concentration higher than 0.06 ng/ul) and no detectable RNA degradation were selected for final library preparation. Tagmentation assays and barcoded sequencing libraries were prepared using Nextera XT kit (Illumina) according to the manufacturer’s instructions. Barcoded libraries were pooled and subjected to 75-bp paired-end sequencing on the Illumina NextSeq instrument. Details of scRNA-Seq analysis are in SI Appendix, Materials and Methods, sections B–F.
ATAC-seq Assays and Data Processing.
Three mouse genotypes were used for ATAC-seq analysis: Tg-eGFP; Neurog3+/+, Neurog3eGFP/+, and Neurog3eGFP/eGFP. From these animals, different cell populations were isolated as described in the Tissue Processing and FACS section (also see SI Appendix, Table S1). ATAC-seq was performed following the protocol in Buenrostro et al. (22). On average, 10,000 sorted cells were used for each ATAC-seq assay. Sorted cells were pelleted at 300 g and washed once with PBS. Nuclei were isolated, followed by the transposition reaction. Transposed DNA fragments were purified using the Qiagen MinElute kit and amplified six to eight cycles using the Nextera (Illumina) PCR primers. Libraries were sequenced as 50 bp paired-end on HiSeq2000 platform. ATAC-seq data processing and genome alignment were performed with PEPATAC (version 0.8.2), a pipeline developed to analyze ATAC-seq samples (66). PEPATAC begins by trimming adapters using skewer (version 0.2.2) with the parameters “-f sanger -t 8 -m pe”. Trimmed fastq files were then mapped to the mm10 genome with bowtie2 (67) and the parameter “–very-sensitive.” Lastly, peaks were called using MACS2 (68) with “-q 0.01 –shift 0 –nomodel.” At the end of PEPATAC processing, 42 to 88 million reads aligned to the mouse genome, and 15,377 to 55,676 peaks per sample were detected. These peak regions were then merged using BedTools (69) to generate a nonoverlapping consensus peak list for downstream analysis. ATAC-seq fragments corresponding to the peaks were quantified by using the annotatePeaks.pl function in the HOMER suite, a genome analysis tool (v.4.10) (44). DESeq (42) was used to find regions with significantly different ATAC-seq counts by running a generalized linear model with the modelFormula set to “count∼condition” and “count∼1.” Accordingly, DESeq calculates P values and FDR. Peaks passing the FDR threshold < 0.001 were considered differentially open regions (DORs) between cell types (∼10,600 DORs). Pearson correlation coefficient method was used to determine the similarity between ATAC-seq samples based on DORs. The results were visualized using the R package ggcorrplot with hierarchical clustering. DORs and samples were clustered by Cluster 3.0 tool using the k-means method (70). ATAC-seq fragment counts were further normalized by log2 transformation after shifting values +1 for visualization in TreeView (71). To assign DORs to regulatory domains and putative target genes, we used the GREAT algorithm (v3.0.0) (43) with default settings. GREAT also outputs enriched GO terms associated with these regions. For the GO term enrichment analysis, DORs were used as test regions against whole genome (mm10) as background. Additional details about TF enrichment and footprint analysis are in SI Appendix, Materials and Methods, sections G–I.
Supplementary Material
Acknowledgments
We thank P. Batista and the members of the Arda and Kim laboratories for discussions and suggestions. This work was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (ZIA BC011798), by funds from a Juvenile Diabetes Research Foundation Advanced Postdoctoral Fellowship (3-APF-2016-172-A-N) to H.E.A.; by the National Institute of General Medical Sciences (R35-GM128636) to N.C.S.; and by National Institutes of Diabetes and Digestive and Kidney Diseases to S.K.K. (5R01 DK128932). Computational resources of the NIH High Performance Cluster Biowulf cluster supported the analysis in this work (https://hpc.nih.gov).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201267119/-/DCSupplemental.
Data Availability
The data discussed in this publication have been deposited in National Center for Biotechnology, Gene Expression Omnibus (GEO) (72) and are accessible through GEO Series accession numbers GSE146006 (73) and GSE65794 (74).
References
- 1.Siehler J., Blöchinger A. K., Meier M., Lickert H., Engineering islets from stem cells for advanced therapies of diabetes. Nat. Rev. Drug Discov. 20, 920–940 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Gradwohl G., Dierich A., LeMeur M., Guillemot F., neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. U.S.A. 97, 1607–1611 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arda H. E., Benitez C. M., Kim S. K., Gene regulatory networks governing pancreas development. Dev. Cell 25, 5–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benitez C. M., Goodyer W. R., Kim S. K., Deconstructing pancreas developmental biology. Cold Spring Harb. Perspect. Biol. 4, a012401 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastidas-Ponce A., Scheibner K., Lickert H., Bakhti M., Cellular and molecular mechanisms coordinating pancreas development. Development 144, 2873–2888 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Gu G., Dubauskaite J., Melton D. A., Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129, 2447–2457 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Schwitzgebel V. M., et al. , Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development 127, 3533–3542 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Smith S. B., Watada H., German M. S., Neurogenin3 activates the islet differentiation program while repressing its own expression. Mol. Endocrinol. 18, 142–149 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Kopinke D., et al. , Lineage tracing reveals the dynamic contribution of Hes1+ cells to the developing and adult pancreas. Development 138, 431–441 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solar M., et al. , Pancreatic exocrine duct cells give rise to insulin-producing β cells during embryogenesis but not after birth. Dev. Cell 17, 849–860 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Bankaitis E. D., Bechard M. E., Wright C. V. E., Feedback control of growth, differentiation, and morphogenesis of pancreatic endocrine progenitors in an epithelial plexus niche. Genes Dev. 29, 2203–2216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bastidas-Ponce A., et al. , Massive single-cell mRNA profiling reveals a detailed roadmap for pancreatic endocrinogenesis. Development 146, dev.173849 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Byrnes L. E., et al. , Lineage dynamics of murine pancreatic development at single-cell resolution. Nat. Commun. 9, 3922 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krentz N. A. J., et al. , Single-cell transcriptome profiling of mouse and hESC-derived pancreatic progenitors. Stem Cell Reports 11, 1551–1564 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu W.-L., et al. , Deciphering pancreatic islet β cell and α cell maturation pathways and characteristic features at the single-cell level. Cell Metab. 25, 1194–1205.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Sharon N., et al. , A peninsular structure coordinates asynchronous differentiation with morphogenesis to generate pancreatic islets. Cell 176, 790–804.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu X.-X., et al. , Defining multistep cell fate decision pathways during pancreatic development at single-cell resolution. EMBO J. 38, e100164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J., et al. , Neurog3-independent methylation is the earliest detectable mark distinguishing pancreatic progenitor identity. Dev. Cell 48, 49–63.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scavuzzo M. A., et al. , Endocrine lineage biases arise in temporally distinct endocrine progenitors during pancreatic morphogenesis. Nat. Commun. 9, 3356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arrojo E Drigo R., et al. , Structural basis for delta cell paracrine regulation in pancreatic islets. Nat. Commun. 10, 3700 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C. S., Perreault N., Brestelli J. E., Kaestner K. H., Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 16, 1488–1497 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L., A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Sugiyama T., Rodriguez R. T., McLean G. W., Kim S. K., Conserved markers of fetal pancreatic epithelium permit prospective isolation of islet progenitor cells by FACS. Proc. Natl. Acad. Sci. U.S.A. 104, 175–180 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picelli S., et al. , Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Qiu X., et al. , Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsson A., et al. , Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537, 698–702 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaffer A. E., et al. , Nkx6.1 controls a gene regulatory network required for establishing and maintaining pancreatic Beta cell identity. PLoS Genet. 9, e1003274 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vierbuchen T., et al. , AP-1 transcription factors and the BAF complex mediate signal-dependent enhancer selection. Mol. Cell 68, 1067–1082.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benitez C. M., et al. , An integrated cell purification and genomics strategy reveals multiple regulators of pancreas development. PLoS Genet. 10, e1004645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J., McKenna L. B., Bogue C. W., Kaestner K. H., The diabetes gene Hhex maintains δ-cell differentiation and islet function. Genes Dev. 28, 829–834 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee A.-H., Heidtman K., Hotamisligil G. S., Glimcher L. H., Dual and opposing roles of the unfolded protein response regulated by IRE1α and XBP1 in proinsulin processing and insulin secretion. Proc. Natl. Acad. Sci. U.S.A. 108, 8885–8890 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hess D. A., et al. , Extensive pancreas regeneration following acinar-specific disruption of Xbp1 in mice. Gastroenterology 141, 1463–1472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hetz C., The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13, 89–102 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Xin Y., et al. , Pseudotime ordering of single human β-cells reveals states of insulin production and unfolded protein response. Diabetes 67, 1783–1794 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Xu C.-R., et al. , Dynamics of genomic H3K27me3 domains and role of EZH2 during pancreatic endocrine specification. EMBO J. 33, 2157–2170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White P., May C. L., Lamounier R. N., Brestelli J. E., Kaestner K. H., Defining pancreatic endocrine precursors and their descendants. Diabetes 57, 654–668 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Gu G., et al. , Global expression analysis of gene regulatory pathways during endocrine pancreatic development. Development 131, 165–179 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Masui T., et al. , Transcriptional autoregulation controls pancreatic Ptf1a expression during development and adulthood. Mol. Cell. Biol. 28, 5458–5468 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ejarque M., et al. , Neurogenin3 cooperates with Foxa2 to autoactivate its own expression. J. Biol. Chem. 288, 11705–11717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaret K. S., Mango S. E., Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev. 37, 76–81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anders S., Huber W., Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLean C. Y., et al. , GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maurano M. T., et al. , Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baek S., Goldstein I., Hager G. L., Bivariate genomic footprinting detects changes in transcription factor activity. Cell Rep. 19, 1710–1722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corces M. R., et al. , The chromatin accessibility landscape of primary human cancers. Science 362, eaav1898 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hale M. A., et al. , The nuclear hormone receptor family member NR5A2 controls aspects of multipotent progenitor cell formation and acinar differentiation during pancreatic organogenesis. Development 141, 3123–3133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xuan S., et al. , Pancreas-specific deletion of mouse Gata4 and Gata6 causes pancreatic agenesis. J. Clin. Invest. 122, 3516–3528 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiselev V. Y., Andrews T. S., Hemberg M., Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 20, 273–282 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Tritschler S., et al. , Concepts and limitations for learning developmental trajectories from single cell genomics. Development 146, dev170506 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Hickey R. D., et al. , Generation of islet-like cells from mouse gall bladder by direct ex vivo reprogramming. Stem Cell Res. (Amst.) 11, 503–515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li W., et al. , In vivo reprogramming of pancreatic acinar cells to three islet endocrine subtypes. eLife 3, e01846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chakravarthy H., et al. , Converting adult pancreatic islet α cells into β cells by targeting both Dnmt1 and Arx. Cell Metab. 25, 622–634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Furuyama K., et al. , Diabetes relief in mice by glucose-sensing insulin-secreting human α-cells. Nature 567, 43–48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schreiber V., et al. , Extensive NEUROG3 occupancy in the human pancreatic endocrine gene regulatory network. Mol. Metab. 53, 101313 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galivo F., et al. , Reprogramming human gallbladder cells into insulin-producing β-like cells. PLoS One 12, e0181812 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee J., et al. , Expansion and conversion of human pancreatic ductal cells into insulin-secreting endocrine cells. eLife 2, e00940 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petersen M. B. K., et al. , Single-cell gene expression analysis of a human ESC model of pancreatic endocrine development reveals different paths to β-Cell differentiation. Stem Cell Reports 9, 1246–1261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Veres A., et al. , Charting cellular identity during human in vitro β-cell differentiation. Nature 569, 368–373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corces M. R., et al. , Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klemm S. L., Shipony Z., Greenleaf W. J., Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Bevacqua R. J., et al. , CRISPR-based genome editing in primary human pancreatic islet cells. Nat. Commun. 12, 2397 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma S., et al. , Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 183, 1103–1116.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang S., et al. , Myt1 and Ngn3 form a feed-forward expression loop to promote endocrine islet cell differentiation. Dev. Biol. 317, 531–540 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith J. P., et al. , PEPATAC: An optimized pipeline for ATAC-seq data analysis with serial alignments. NAR Genom. Bioinform. 3, lqab101 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feng J., Liu T., Qin B., Zhang Y., Liu X. S., Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Hoon M. J. L., Imoto S., Nolan J., Miyano S., Open source clustering software. Bioinformatics 20, 1453–1454 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Saldanha A. J., Java Treeview--extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Edgar R., Domrachev M., Lash A. E., Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.E. Duvall, H. Arda, S. Kim, Data from “Single cell transcriptome analysis of endocrine pancreas development in mice.” https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146006. Accessed 8 June 2022. [Google Scholar]
- 74.H. Arda, S. Kim, E. Duvall, Data from “Open chromatin landscape of pancreatic endocrine progenitors.” https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65794. Accessed 1 April 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in National Center for Biotechnology, Gene Expression Omnibus (GEO) (72) and are accessible through GEO Series accession numbers GSE146006 (73) and GSE65794 (74).