Abstract
Studies conducted using murine arthritis models have indicated that the use of in vitro-transcribed messenger RNA (IVT mRNA) is an effective therapeutic approach for joint diseases. However, the use of IVT mRNA in human synovial cells has not been widely studied. Recently, the outbreak of the novel coronavirus disease has accelerated the development of innovative mRNA vaccines, such as those containing a modified nucleic acid, N1-methylpseudouridine-5′-triphosphate (m1ψ). IVT mRNA is an attractive tool for biological experiments and drug discovery. To verify the protein expression from IVT mRNA in vitro, primary cultured fibroblast-like synoviocytes (FLS) and MH7A human synovial fibroblast cells were transfected with enhanced green fluorescent protein (EGFP) mRNA with or without m1ψ incorporation. EGFP was detected using western blotting and fluorescence microscopy. A multiplex assay was performed to comprehensively understand IVT mRNA-induced immunogenicity. Gene expression levels were measured using reverse transcription polymerase chain reaction. In both MH7A cells and FLS, cells transfected with EGFP mRNA containing m1ψ generated higher levels of EGFP than those transfected with unmodified EGFP or control mRNAs. The multiplex assay of the FLS culture supernatant and reverse transcription polymerase chain reaction for FLS revealed that both concentration and expression of IL-6, TNF-α, and CXCL10 were upregulated by unmodified EGFP mRNA, whereas they were suppressed by EGFP mRNA with m1ψ. Overall, m1ψ incorporation enhanced protein expression and decreased the expression of cytokines. These findings may contribute to arthritis research.
Keywords: IVT mRNA, Synoviocyte, Fibroblast-like synoviocyte, EGFP, Multiplex assay
Introduction
Studies conducted using murine arthritis models have indicated that the use of in vitro-transcribed messenger RNA (IVT mRNA) is an effective therapeutic approach for joint diseases (Aini et al. 2016; Mokuda et al. 2019). IVT mRNA is synthesized in vitro from template DNA. When an IVT mRNA penetrates the cell membrane and enters the cytoplasm, it instantly initiates the production of the foreign protein using intracellular ribosomes. Unlike plasmid DNA and viral vectors, IVT mRNA can function without entering the nucleus and is degraded through the endogenous metabolic pathway after protein production (Sahin et al. 2014). Therapeutic strategies using IVT mRNA are attractive and have been examined in vivo. However, the in vitro-use of IVT mRNA in human synoviocytes has been poorly studied (Mokuda et al. 2015).
IVT mRNA introduced into the cytoplasm stimulates intracellular innate immune responses through pattern-recognition receptors (PRRs) (Sahin et al. 2014). Exogenous single-stranded RNA (ssRNA) is known to react with Toll-like receptor 7 (TLR7), TLR8, protein kinase R (PKR) and retinoic acid-inducible gene-I (RIG-I) (Anderson et al. 2010; Lund et al. 2004; Diebold et al. 2004; Heil et al. 2004; Kato et al. 2006; Pichlmair et al. 2006). In addition, when IVT mRNA forms a hairpin or partially forms double-stranded RNA (dsRNA), it may also be recognized by PRRs, such as TLR3, RIG-I, PKR, 2′–5′-oligoadenylate synthetase (OAS)/ribonuclease L and melanoma differentiation-associated protein 5 (MDA5) (Yoneyama et al. 2004; Kato et al. 2008; Li et al. 2009; Anderson et al. 2011). It has been reported that the incorporation of modified nucleic acids, such as pseudouridine-5′-triphosphate (ψ), into IVT mRNA is an effective method to reduce immunogenicity (Anderson et al. 2011; Karikó et al. 2005, 2008). The advantages of using ψ and 5-methylcytidine-5′-triphosphate have also been reported (Sullenger and Nair 2016; Mokuda et al. 2019).
Recently, the novel coronavirus disease 2019 pandemic, which has been ongoing since December 2019, has led to the development of innovative mRNA vaccines such as BNT162b2 and mRNA-1273, containing N1-methylpseudouridine-5′-triphosphate (m1ψ) (Polack et al. 2020; Baden et al. 2021). These vaccines seldom have negative effects on the human body, at least in the short term (within approximately 1 year), indicating that the use of m1ψ is safe. The incorporation of m1ψ into IVT mRNA has been highlighted as a useful method for enhancing protein expression (Andries et al. 2015; Svitkin et al. 2017; Van der Jeught et al. 2018; Foster et al. 2019; Hadas et al. 2019; Parr et al. 2020; Nelson et al. 2020; Nance and Meier 2021); m1ψ has a unique benefit for therapeutic methods using IVT mRNA.
In this study, we aimed to evaluate the efficiency of gene expression and immunogenicity of exogenous m1ψ-incorporated IVT mRNA using a human fibroblast cell line and primary cultured fibroblast-like synoviocytes (FLS). We demonstrate that lipofection of m1ψ-incorporated IVT mRNA is a potential in vitro gene transfer technique for FLS.
Materials and methods
Ethics
This study was approved by the clinical ethics committee of Hiroshima University Hospital (approval number: E-668; approval date: February 1, 2017). The methods were performed in accordance with the approved guidelines. We collected synovial tissues from patients with rheumatoid arthritis (RA) who fulfilled the classification criteria of the American College of Rheumatology 1987 (Arnett et al. 1988). Synovial tissues were harvested from patients with RA who underwent total joint replacement or synovectomy after they provided informed consent and signed a written consent form.
Preparation of MH7A cells and primary cultured FLS
The human synovial cell line, MH7A, was isolated from the knee joint of a patient with RA (Miyazawa et al., Kissei Pharmaceutical Co., Ltd., 1998). MH7A cells were obtained from the Riken Cell Bank (Ibaraki, Japan). These cells were cultured in RPMI1640 (FUJIFILM Wako Pure Chemical Co., Osaka, Japan) supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA), l-glutamine (300 mg/L), pyruvate (110 mg/L), HEPES (5958 mg/L), and penicillin/streptomycin (FUJIFILM Wako Pure Chemical Co.). MH7A cells were used in the experiments and were starved in serum-free RPMI1640 for 24 h before measuring protein expression. For FLS preparation, synovial tissues from three patients with RA were minced and incubated with 1 mg/mL collagenase/dispase (Roche, Indianapolis, IN, USA) in phosphate-buffered saline (pH 7.2, FUJIFILM Wako Pure Chemical Co.) for 1 h at 37 °C. The synovial cells were then filtered, washed, and cultured. During incubation, the supernatant was replaced frequently to withdraw any nonadherent cells. Adherent FLS were then subcultured at 1:3 after reaching 80% confluence and were passaged. FLS were cultured in Dulbecco’s modified Eagle medium (DMEM; FUJIFILM Wako Pure Chemical Co.) supplemented with 10% fetal bovine serum (Sigma-Aldrich) and penicillin/streptomycin (FUJIFILM Wako Pure Chemical Co.). FLS were used in the experiments at passages 3–4 and were starved in DMEM containing 0.5% fetal bovine serum for 24 h before measuring mRNA and protein expression. Both cell types were incubated at 37 °C under 5% CO2.
Preparation of IVT mRNA and transfection into the cultured cells
To prepare templates for IVT mRNA with a cap structure and poly A sequence, the following plasmids were previously constructed in our laboratory: pcDNA3-A124 and pcDNA3-enhanced green fluorescent protein (EGFP)-A124 vectors, containing both untranslated regions and poly A contiguous sequences (Holtkamp et al. 2006; Mokuda et al. 2015, 2019). Template plasmids were linearized, and in vitro transcription was performed using the HiScribe T7 ARCA mRNA Kit (New England Biolabs, Ipswich, MA, USA) to generate IVT mRNA according to the manufacturer’s instructions. m1ψ was purchased from TriLink BioTechnologies (San Diego, CA, USA). Components of the mixture except for the template plasmids, reaction buffer, and T7 polymerase were as follows: (1) For unmodified IVT mRNA: 4 mM cap analog (anti-reverse cap analog; 3′-O-Me-m7G(5′)ppp(5′)G), 1.25 mM ATP, 1 mM GTP, 1.25 mM CTP, and 1.25 mM UTP; (2) For IVT mRNA (m1ψ): 4 mM cap analog (anti-reverse cap analog), 1.25 mM ATP, 1 mM GTP, 1.25 mM CTP, 1.25 mM UTP, and 2.5 mM m1ψ. After in vitro transcription at 37 °C for 1 h, IVT mRNA was purified using a Monarch RNA Cleanup Kit (New England Biolabs). IVT mRNA was then transfected into cells using Lipofectamine MessengerMAX (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Briefly, mRNA was mixed with Lipofectamine reagent at a ratio of 1:3 (pmol mRNA: µL Lipofectamine) in serum-free DMEM. The complex was allowed to form for 20 min at 25 °C. At 24 h after transfection, both cells and their culture supernatants were harvested for the subsequent experiments.
Western blotting
The plated cells were washed with phosphate-buffered saline before collection. Proteins from the cultured cells were processed using SuperSep Ace 15% precast gel (FUJIFILM Wako Pure Chemical Co.) and transferred on to a polyvinylidene fluoride membrane. The membranes were probed with anti-GFP (0.5 µg/mL, chicken polyclonal; Genscript, Piscataway, NJ, USA) and anti-β-actin (1 µg/mL, mouse monoclonal, clone. AC-15; Sigma-Aldrich) antibodies. Horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA) were then added. Horseradish peroxidase activity was detected using ECL prime reagents (Cytiva, Tokyo, Japan), followed by imaging with Image Quant LAS 500 (Cytiva).
Measurement of EGFP fluorescence intensity
To observe the fluorescence of live cells, the culture medium was replaced with FluoroBrite DMEM (Thermo Fisher Scientific), and the cells were then imaged under the green channel of a ZOE Fluorescent Cell Imager (Bio-Rad Laboratories, Hercules, CA, USA).
To measure the fluorescence intensity in multi-well plates, the transfected cells were plated in a 96-well black plate (FUJIFILM Wako Pure Chemical Co.). The culture supernatant was replaced with 100 μL of FluoroBrite DMEM before fluorescence detection. Fluorescence was measured using a SpectraMax iD3 system (Molecular Devices, Sunnyvale, CA, USA). The excitation and emission wavelengths used for EGFP detection were 460 and 535 nm, respectively. Subsequently, to compensate for the fluorescence affected by cell viability, the obtained fluorescence intensity was divided by the intensity of resorufin fluorescence, as described below.
Measurement of cell viability
To measure the viability of the cultured cells, the transfected cells plated in 96-well black plates were examined using a CellTiter-Blue Cell Viability Assay kit (Promega, Madison, WI, USA). Briefly, 20 μL of CellTiter-Blue Reagent (resazurin) was added to 100 μL of the medium in each well and incubated at 37 °C for 1 h. Resazurin is reduced to resorufin by the aerobic respiration of metabolically active cells. The fluorescence of resorufin was measured using a SpectraMax iD3 system (Molecular Devices) at the excitation and emission wavelengths of 550 and 595 nm, respectively.
Immunocytochemistry staining
Because fibroblasts harvested from human adult specimens often exhibit autofluorescence, which is the natural emission from biological structures, such as lipofuscin (Georgakopoulou et al. 2013), it is difficult to detect FLS using immunocytochemistry with Alexa Fluor 488. FLS were cultured on cover slips placed in a six-well tissue culture plate. After transfection with IVT mRNA, the cells were incubated for 24 h. The cells were fixed with a mixture of acetone and methanol for 20 min at 25 °C, and then blocked with 5% Blocking One P reagent (Nacalai Tesque, Kyoto, Japan) diluted with Tris-buffered saline for 1 h at 25 °C. The cells were incubated overnight at 4 °C with the primary antibodies against the GFP tag (8 µg/mL; rabbit polyclonal; Proteintech, Rosemont, IL, USA), and then with Alexa Fluor 647-conjugated anti-rabbit IgG antibodies (4 µg/mL; host: donkey; Abcam, Cambridge, UK) for 1 h at 25 °C. The cells on the coverslips were mounted on glass slides with VECTASHIELD mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector, Burlingame, CA, USA). Anti-GFP antibody signals were detected using a digital microscope VHX-7000 (KEYENCE, Osaka, Japan).
Multiplex assay using fluorescence-activated cell sorting (FACS)
The concentrations of interleukin-1β (IL-1β), IL-6, IL-8, IL-10, IL-12p70, interferon-α2 (IFN-α2), IFN-β, IFN-λ1, IFN-λ2/3, IFN-γ, tumor necrosis factor-α (TNF-α), C-X-C motif chemokine ligand 10 (CXCL10), and granulocyte macrophage colony-stimulating factor (GM-CSF) in the supernatant of cultured cells were measured using a LEGENDplex Multi-Analyte Flow Assay Kit, Human Anti-Virus Response Panel (Biolegend, San Diego, USA). The assay was performed in a V-bottom plate according to the manufacturer’s protocol. Data were acquired using a Cytoflex flow cytometer (Beckman Coulter, Brea, CA, USA) and analyzed using the Legendplex Data Analysis V7.1 software (Biolegend). The sensitivity of the assay, indicated in parentheses, was as follows: IL-1β (19.3 pg/mL), IL-6 (11.7 pg/mL), IL-8 (5.7 pg/mL), IL-10 (15.4 pg/mL), IL-12p70 (3.7 pg/mL), IFN-α2 (2.9 pg/mL), IFN-β (105.7 pg/mL), IFN-λ1 (47.7 pg/mL), IFN-λ2/3 (270.5 pg/mL), IFN-γ (39.8 pg/mL), TNF-α (16.9 pg/mL), CXCL10 (16.3 pg/mL), and GM-CSF (15.6 pg/mL).
Reverse transcription-quantitative polymerase chain reaction
Total RNA was extracted and purified from the cultured cells using the TRIzol reagent (Life Technologies, Carlsbad, CA, USA) and a Direct-Zol RNA kit (Zymo Research, Irvine, CA, USA), followed by cDNA synthesis using a PrimeScript RT Reagent Kit (Takara Bio, Kusatsu, Japan). Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed using Brilliant II SYBR Green QPCR Master Mix (Agilent Technologies, Santa Clara, CA, USA) on a CFX Connect Real-Time PCR Detection System (Bio-Rad Laboratories). The upstream and downstream primer sequences were as follows: human IL6, 5′-GCAGAAAAAGGCAAAGAATC-3′ and 5′-CTACATTTGCCGAAGAGC-3′; human TNF, 5′-AGGCAGTCAGATCATCTTC-3′ and 5′-TTATCTCTCAGCTCCACG-3′; and human CXCL10, 5′-AAAGCAGTTAGCAAGGAAAG-3′ and 5′-TCATTGGTCACCTTTTAGTG-3′. The upstream and downstream primer sequences for human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were 5′-CTTTTGCGTCGCCAG-3′ and 5′- TTGATGGCAACAATATCCAC-3′, respectively.
Statistical analysis
All statistical analyses were performed using Student’s t-test. All graphs show the results of one representative experiment from several individual experiments. The results were analyzed and processed using the GraphPad Prism 9 software (GraphPad, Inc., La Jolla, CA, USA).
Results
IVT mRNA induction in the MH7A cell line
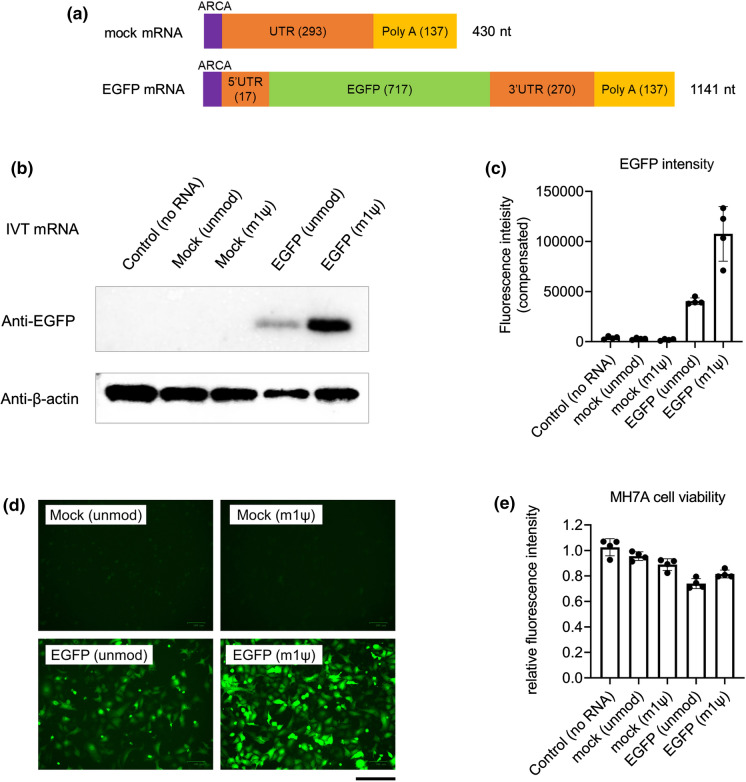
Transfection of IVT mRNA in vigorously dividing cells is easy, similar to plasmid transfection. Thus, we first introduced EGFP mRNA into MH7A cells, which has a high proliferation ability. We prepared the following four types of IVT mRNAs: mock mRNA, with or without m1ψ-incorporation, and EGFP mRNA, with or without m1ψ-incorporation. Mock mRNA, which cannot produce any protein, had approximately 400 nucleotides (nt) and EGFP mRNA had approximately 1100 nt (Fig. 1a). MH7A cells were collected at 24 h after mRNA transfection. EGFP expression in EGFP mRNA (m1ψ)-transfected cells was higher than that in unmodified EGFP mRNA-transfected cells or in the control groups (mock mRNA-transfected) as evident from the western blotting analysis, fluorescence microplate measurement, and fluorescence microscopy (Fig. 1b–d). The transfection efficiency and viability of MH7A cells were also examined. Live cell EGFP fluorescence was detected in most EGFP mRNA (m1ψ)-transfected cells (Fig. 1d). The percentages of EGFP fluorescence-emitting cells transfected with unmodified EGFP mRNA and EGFP mRNA (m1ψ) were 73.1 ± 2.0 and 84.1 ± 2.0 (%; mean ± SEM; N = 3), respectively. The viability of these cells was higher than 80% (Fig. 1e). However, MH7A cells transfected with EGFP mRNA (unmodified) were less viable compared with those in other groups.
Fig. 1.
EGFP mRNA transfection in MH7A human fibroblasts. MH7A cells were transfected with in vitro-transcribed mRNA (IVT mRNA) and harvested at 24 h post transfection. a Scheme of the IVT mRNA design. b Western blotting of EGFP. c Intensity of fluorescence from MH7A cells in 96-well plates (n = 4, mean ± SEM). d Images of MH7A captured using a fluorescence microscope. Scale bar, 0.2 mm. e Cell viability was measured using the CellTiter-Blue Cell Viability Assay kit (n = 4, mean ± SEM). ARCA, anti-reverse cap analog; EGFP, enhanced green fluorescent protein; UTR, untranslated region
IVT mRNA induction in primary cultured FLS
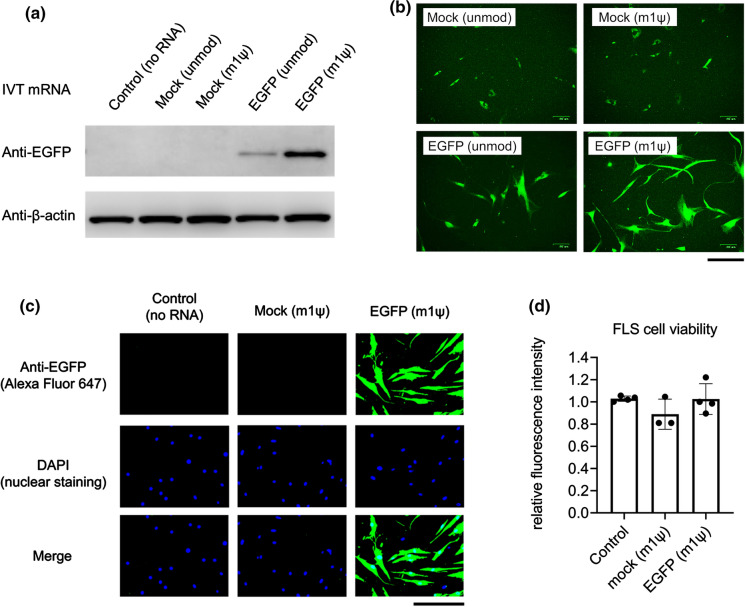
We examined the expression level of EGFP in transfected primary cultured FLS. As shown in Fig. 2a (western blotting) and Fig. 2b (fluorescence microscopy), EGFP expression in EGFP mRNA (m1ψ)-transfected FLS was higher than that in the negative control groups or in unmodified EGFP mRNA-transfected FLS. Unexpectedly, FLS produced green auto-fluorescence in the control groups. Therefore, it was difficult to accurately evaluate the intensity of EGFP fluorescence, which was detected using either fluorescence microplate measurement or fluorescence microscopy. We also performed immunocytochemistry using Alexa Fluor 647 to calculate the transfection efficiency. The percentage of EGFP expressing cells transfected with EGFP mRNA (m1ψ) was 85.2 ± 3.5 (%; mean ± SEM; N = 3) (Fig. 2c), and the viability of these cells was more than 90% (Fig. 2d).
Fig. 2.
EGFP mRNA transfection in primary human fibroblast-like synoviocytes (FLS). FLS were transfected with in vitro-transcribed mRNA (IVT mRNA) and harvested at 24 h after transfection. a Western blotting of EGFP. b Images of FLS captured using fluorescence microscopy. Scale bar, 0.2 mm. c Immunocytochemical staining for EGFP. Scale bar, 0.2 mm. d Measurement of cell viability using the CellTiter-Blue Cell Viability Assay kit (n = 3–4, mean ± SEM)
These findings indicate that IVT mRNA containing m1ψ can induce adequate expression of foreign protein in primary culture FLS, with acceptable cell viability.
Comprehensive analysis of IVT mRNA-stimulated production of cytokines and chemokines in FLS
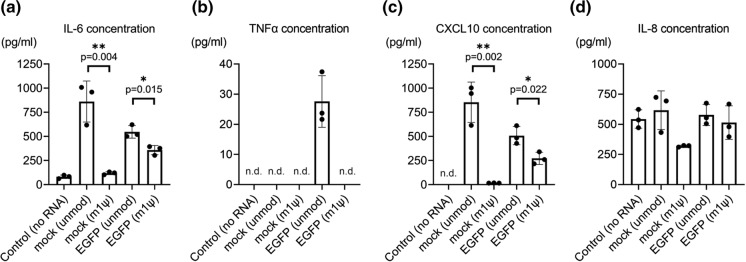
Introduced IVT mRNA is considered to be recognized by some PRRs and induces post-viral infection-like responses, which can activate two main cascades, namely, nuclear factor-kappa B (NF-κB)-mediated signaling and interferon-regulatory factor (IRF)-mediated signaling (Sahin et al. 2014; Nance and Meier 2021). To clarify these responses against IVT mRNA, we used a multiplex assay to measure 13 different proteins, IL-1β, IL-6, IL-8, IL-10, IL-12p70, IFN-α2, IFN-β, IFN-λ1, IFN-λ2/3, IFN-γ, TNF-α, CXCL10, and GM-CSF, in the culture supernatants from 0 to 24 h after transfection. As a result, IL-6, TNF-α, CXCL10, and IL-8 were detected, whereas the other nine proteins were below the detection limits. Unmodified mock mRNA increased the protein levels of IL-6 and CXCL10. Moreover, unmodified EGFP mRNA increased the protein levels of IL-6, TNF-α, and CXCL10 (Fig. 3a–c). Both mock and EGFP mRNAs incorporated with m1ψ downregulated this elevation. IL-8 showed high endogenous excretion in the no RNA control group, and it did not increase in the presence of unmodified IVT mRNA (Fig. 3d).
Fig. 3.
Multiplex assay of cytokines and chemokines in in vitro-transcribed mRNA (IVT mRNA)-transfected fibroblast-like synoviocytes (FLS) culture supernatant. The protein concentrations in the FLS culture supernatant were measured using a LEGENDplex Multi-Analyte Flow Assay kit. The samples contained humoral factors 24 h after transfection (n = 3). a IL-6 (pg/mL), b TNF-α (pg/mL), c CXCL10 (pg/mL), and d IL-8 (pg/mL) were detected. IL-1β, IFN-λ1, IL-12p70, IFN-α2, N-IFN-λ2/3, GM-CSF, IFN-β, IL-10, and IFN-γ were below detectable limits. Data are presented as mean ± SEM. t-test, *p < 0.05, **p < 0.01
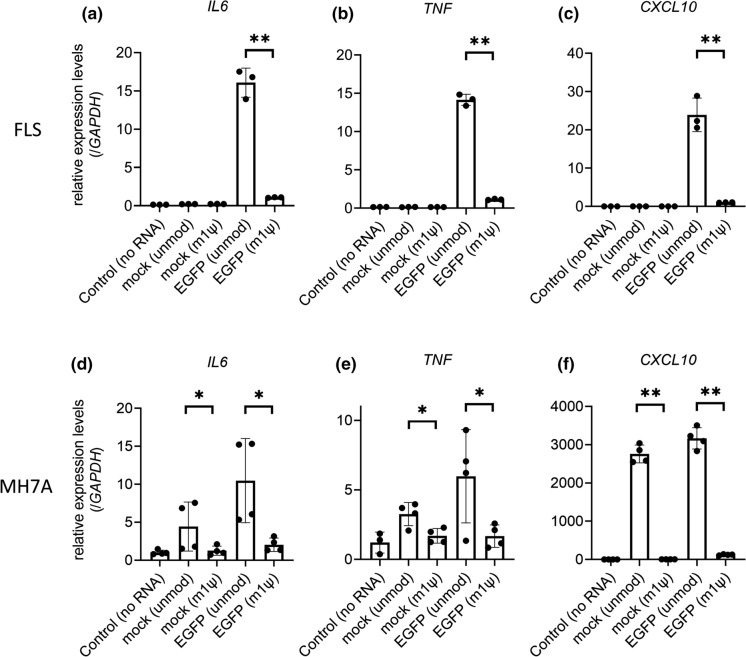
These measured protein concentrations reflected the 24-h accumulation of the secreted proteins after transfection. We also performed RT-qPCR of transfected FLS at 24 h post-transfection and found that the expression of three candidate genes (IL6, TNF, and CXCL10) from unmodified EGFP mRNA-transfected FLS was considerably higher than that in other groups, including that in m1ψ-incorporated IVT mRNA-transfected samples (Fig. 4a–c). In addition, we examined them using MH7A cells and found that expression levels of IL6, TNF, and CXCL10 in MH7A cells transfected with unmodified mock- and EGFP mRNAs were considerably higher than those in m1ψ-incorporated groups (Fig. 4d–f).
Fig. 4.
Gene Expression levels of humoral factors in in vitro-transcribed mRNA (IVT mRNA)-transfected fibroblast-like synoviocytes (FLS) and MH7A cells. RT-qPCR of IVT mRNA-transfected cells showed changes in the expression of three candidate genes. a–c FLS (n = 3): a IL6, b TNF, and c CXCL10. Gene expression levels were normalized to those of GAPDH. d–f MH7A (n = 4): d IL6, e TNF, and f CXCL10. Gene expression levels were normalized to those of GAPDH. The samples were harvested at 24 h after transfection. Data are presented as mean ± SEM. t-test, *p < 0.05, **p < 0.01
In summary, unmodified IVT mRNA simultaneously elevated the levels of both NF-κB-mediated pro-inflammatory cytokines, IL-6 and TNF-α, and an IRF-mediated chemokine, CXCL10 in cultured synovial cells. In addition, m1ψ-incorporation into IVT mRNA suppressed this elevation.
Discussion
From 1993 to 2000, IVT mRNA was developed for use in vaccination approach against cancer and infectious diseases, mainly because it does not undergo mutagenesis and can transiently produce various exogenous proteins (Martinon et al. 1993; Conry et al. 1995; Boczkowski et al. 1996; Mandl et al. 1998; Koido et al. 2000; Schirrmacher et al. 2000). In contrast, IVT mRNA itself can induce unfavorable immunogenicity. When IVT mRNA is used as a vaccine, these immune-stimulatory responses may act as adjuvants and promote adaptive immune responses against the introduced exogenous protein (Sander et al. 2011). In contrast, when IVT mRNA is treated as a gene replacement therapy, additive immune responses are not necessary, and it may have disadvantages. Joint diseases might be good targets for IVT mRNA gene replacement therapy. Cartilage protective genes, such as WWP2, might contribute to RA or osteoarthritis because of which articular cartilage-specific treatment has not been established (Mokuda et al. 2019).
Inflammation exacerbates the pathophysiology of joint diseases, and human RA-derived FLS reportedly produce high levels of IL-6 and CXCL10 (Bartok and Firestein 2010; Kuranobu et al. 2020; Yukawa et al. 2020). Therefore, it is desirable that IVT mRNA has low immunogenicity when developing a therapeutic strategy involving gene replacement for arthritis. To verify the usefulness of m1ψ-incorporated IVT mRNA for cultured cells, we utilized primary cultured human FLS. To date, transfection of both unmodified IVT mRNA and plasmid DNA using lipofection in FLS has been technically difficult, and these molecules must be introduced using electroporation or viral vectors. Our results support the notion that m1ψ-modified IVT mRNA-lipofection method is convenient, facilitates high protein production, and as noted below, has low immunogenicity.
To avoid immune responses induced by IVT mRNA, it is necessary to block the interaction between these ssRNAs, which partially form double strands, and PRRs, such as TLR3, TLR7, TLR8, RIG-I, PKR, OAS/ribonuclease L, and MDA5 (Anderson et al., 2011; Alexopoulou et al. 2001; Heil et al. 2004; Karikó et al. 2005; Kato et al. 2006). One strategy for this is to use modified nucleic acids, such as m1ψ. Some structural mechanisms of m1ψ for immunogenicity suppression have already been advocated (Nance and Meier 2021). Incorporation of m1ψ into an mRNA could block the formation of secondary structures, such as hairpins, which are recognized by RIG-I and TLR3. Moreover, ssRNA is sensed by TLR7. m1ψ alters the hydrogen bonding face and disrupts the interaction of immune sensors, such as TLR7, even in the absence of dsRNA. However, the biological functions of m1ψ-incorporated IVT mRNA in cultured human cells are not well understood. Most studies have indicated that this mRNA increases the expression efficiency; however, its ability to suppress innate immune responses has not been evaluated in detail. Particularly, there are only a few comprehensive studies on cytokine production induced by IVT mRNA-stimulated cells at the protein level. Kormann et al. reported both elevated TNFα and IL-8 concentrations in human monocyte culture supernatant stimulated by IVT mRNA (Kormann et al. 2011). In the present study, we analyzed the comprehensive immune response induced by IVT mRNA. IL-6, TNF-α, and CXCL10 were particularly noticeable among the 13 proteins tested. Measurement of the protein concentrations and mRNA expression revealed that m1ψ incorporation can repress the immunogenicity of IVT mRNA. The following findings of in vitro experiments have been reported: expressions of both IFN-β and C–C motif chemokine ligand 5 (CCL5) are suppressed in luciferase IVT mRNA (m1ψ)-transfected human A549 cells; expressions of both IFN-α and IFN-β are downregulated in luciferase IVT mRNA (m1ψ)-transfected rat cardiomyocytes; and expression of CXCL10 is downregulated in human erythropoietin IVT mRNA (m1ψ)-transfected human monocyte-derived macrophages in vitro (Andries et al. 2015; Hadas et al. 2019; Nelson et al. 2020). These results are consistent with our results.
We also generated IVT mRNA of two different lengths: approximately 400 nt (mock) and approximately 1100 nt (EGFP). RIG-I recognizes dsRNA less than 1000 nt in length and with a terminal 5′-triphosphate, and MDA5 recognizes dsRNA more than 1000 nt in length and with a terminal 5′-triphosphate (Hornung et al. 2006; Kato et al. 2008). Different responses against these two PRRs may affect the gene expression pattern. Our results show that 24-h accumulated protein levels of IL-6 and CXCL10 in EGFP mRNA (m1ψ)-transfected FLS were higher than those in cells transfected with mock mRNA (m1ψ), although the gene expression levels of IL6 and CXCL10 at 24 h post-transfection were considerably low in these two groups of FLS. Thus, immunogenicity in FLS was completely suppressed for IVT mRNA having a length less than 1000 nt, whereas reactivity to IVT mRNA, more than 1000 nt in length, was retained within 24 h after transfection. The experimental methods should be further developed to eliminate the immunogenicity induced by IVT mRNA. Moreover, TNF-α production in FLS transfected with unmodified mock mRNA was not detected. We presume that the failure to detect TNF-α production in FLS transfected with mock mRNA (short form, about 400 nt) might be because of the fact that the RIG-I response is not required for TNF-α production.
The relationship between inflammatory cytokines and IVT mRNA-induced translation by ribosomes is also unknown. We also investigated the expression levels of ribosomal proteins. In our preliminary data, expression levels of ribosomal proteins did not alter dramatically under inflammatory conditions (data not shown). Further investigations are needed in this regard.
In conclusion, we demonstrate that m1ψ incorporation enhances exogenous protein expression and suppresses immunogenicity in primary human cells. Thus, m1ψ-incorporated IVT mRNA will be useful for primary culture experiments, including those in arthritis research.
Acknowledgements
We used MH7A cells with the MTA from KISSEI Pharmaceutical Co., Ltd.. We would like to thank Shigeru Miyaki (Hiroshima University) for the preparation of experimental devices.
Author contribution
SM, HW, HK, MI, and KA performed the experiments and analyzed the data. SM, HW, SH, and ES planned the experiments and wrote the manuscript.
Funding
This research was funded by JSPS KAKENHI (Grant Number 19K18499), Mitsubishi Foundation, Takeda Science Foundation, Mochida Memorial Foundation for Medical and Pharmaceutical Research, Japanese Respiratory Foundation Grant, Japan Rheumatism Foundation, The Japan College of Rheumatology Grant for Promoting Research for Early RA, and The Nakatomi Foundation to S.M.
Data availability
The source data for most of the figures are available from the authors.
Declarations
Conflict of interest
The authors declare no conflicts of interest associated with the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sho Mokuda, Email: sho-mokuda@hiroshima-u.ac.jp.
Hirofumi Watanabe, Email: watahir@hiroshima-u.ac.jp.
Hiroki Kohno, Email: hirkohno@hiroshima-u.ac.jp.
Michinori Ishitoku, Email: m141093i@hiroshima-u.ac.jp.
Kei Araki, Email: arakikei@hiroshima-u.ac.jp.
Shintaro Hirata, Email: shirata@hiroshima-u.ac.jp.
Eiji Sugiyama, Email: esugiyam@hiroshima-u.ac.jp.
References
- Aini H, Itaka K, Fujisawa A, et al. Messenger RNA delivery of a cartilage-anabolic transcription factor as a disease-modifying strategy for osteoarthritis treatment. Sci Rep. 2016;6:18743. doi: 10.1038/srep18743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Anderson BR, Muramatsu H, Jha BK, et al. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BR, Muramatsu H, Nallagatla SR, et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andries O, Mc Cafferty S, De Smedt SC, et al. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J Control Release. 2015;217:337–344. doi: 10.1016/J.JCONREL.2015.08.051. [DOI] [PubMed] [Google Scholar]
- Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/ART.1780310302. [DOI] [PubMed] [Google Scholar]
- Baden LR, El-Sahly HM, Essink B, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403–416. doi: 10.1056/NEJMOA2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartok B, Firestein GS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev. 2010;233:233–255. doi: 10.1111/J.0105-2896.2009.00859.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczkowski D, Nair SK, Snyder D, Gilboa E. Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J Exp Med. 1996;184:465–472. doi: 10.1084/jem.184.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conry RM, LoBuglio AF, Wright M, et al. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995;55:1397–1400. [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, et al. Innate Antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/SCIENCE.1093616. [DOI] [PubMed] [Google Scholar]
- Foster JB, Choudhari N, Perazzelli J, et al. Purification of mRNA encoding chimeric antigen receptor is critical for generation of a robust T-Cell response. Hum Gene Ther. 2019;30:168–178. doi: 10.1089/HUM.2018.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulou EA, Tsimaratou K, Evangelou K, et al. Specific lipofuscin staining as a novel biomarker to detect replicative and stress-induced senescence. A method applicable in cryo-preserved and archival tissues. Aging. 2013;5:37–50. doi: 10.18632/aging.100527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadas Y, Sultana N, Youssef E, et al. Optimizing modified mRNA in vitro synthesis protocol for heart gene therapy. Mol Ther Methods Clin Dev. 2019;14:300–305. doi: 10.1016/J.OMTM.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/SCIENCE.1093620. [DOI] [PubMed] [Google Scholar]
- Holtkamp S, Kreiter S, Selmi A, et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–4017. doi: 10.1182/BLOOD-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, et al. 5’-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/SCIENCE.1132505. [DOI] [PubMed] [Google Scholar]
- Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/J.IMMUNI.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Karikó K, Muramatsu H, Welsh FA, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16:1833–1840. doi: 10.1038/MT.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/JEM.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/NATURE04734. [DOI] [PubMed] [Google Scholar]
- Koido S, Kashiwaba M, Chen D, et al. Induction of antitumor immunity by vaccination of dendritic cells transfected with MUC1 RNA. J Immunol. 2000;165:5713–5719. doi: 10.4049/jimmunol.165.10.5713. [DOI] [PubMed] [Google Scholar]
- Kormann MSD, Hasenpusch G, Aneja MK, et al. expression of therapeutic proteins after delivery of chemically modified mrNA in mice. Nat Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- Kuranobu T, Mokuda S, Oi K, et al. Activin A expressed in rheumatoid synovial cells downregulates TNFα-induced CXCL10 expression and osteoclastogenesis. Pathobiology. 2020;87:198–207. doi: 10.1159/000506260. [DOI] [PubMed] [Google Scholar]
- Li X, Lu C, Stewart M, et al. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Arch Biochem Biophys. 2009;488:23–33. doi: 10.1016/J.ABB.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Lund JM, Alexopoulou L, Sato A, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl CW, Aberle JH, Aberle SW, et al. In vitro-synthesized infectious RNA as an attenuated live vaccine in a flavivirus model. Nat Med. 1998;4:1438–1440. doi: 10.1038/4031. [DOI] [PubMed] [Google Scholar]
- Martinon F, Krishnan S, Lenzen G, et al. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur J Immunol. 1993;23:1719–1722. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- Mokuda S, Miyazaki T, Ito Y, et al. The proto-oncogene survivin splice variant 2B is induced by PDGF and leads to cell proliferation in rheumatoid arthritis fibroblast-like synoviocytes. Sci Rep. 2015;5:9795. doi: 10.1038/srep09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokuda S, Nakamichi R, Matsuzaki T, et al. Wwp2 maintains cartilage homeostasis through regulation of Adamts5. Nat Commun. 2019;10:2429. doi: 10.1038/s41467-019-10177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance KD, Meier JL. Modifications in an emergency: the role of N1-methylpseudouridine in COVID-19 vaccines. ACS Cent Sci. 2021;7:748–756. doi: 10.1021/ACSCENTSCI.1C00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J, Sorensen EW, Mintri S, et al. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci Adv. 2020;6:eaaz6893. doi: 10.1126/SCIADV.AAZ6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr CJC, Wada S, Kotake K, et al. N 1-Methylpseudouridine substitution enhances the performance of synthetic mRNA switches in cells. Nucleic Acids Res. 2020;48:E35. doi: 10.1093/NAR/GKAA070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5ʹ-phosphates. Science. 2006;314:997–1001. doi: 10.1126/SCIENCE.1132998. [DOI] [PubMed] [Google Scholar]
- Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMOA2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U, Karikó K, Türeci Ö. MRNA-based therapeutics-developing a new class of drugs. Nat Rev Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- Sander LE, Davis MJ, Boekschoten MV, et al. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474:385–392. doi: 10.1038/NATURE10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirrmacher V, Förg P, Dalemans W, et al. Intra-pinna anti-tumor vaccination with self-replicating infectious RNA or with DNA encoding a model tumor antigen and a cytokine. Gene Ther. 2000;7:1137–1147. doi: 10.1038/sj.gt.3301220. [DOI] [PubMed] [Google Scholar]
- Sullenger BA, Nair S. From the RNA world to the clinic. Science. 2016;352:1417–1420. doi: 10.1126/science.aad8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Cheng YM, Chakraborty T, et al. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 2017;45:6023–6036. doi: 10.1093/NAR/GKX135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Jeught K, De KS, Bialkowski L, et al. Dendritic cell targeting mRNA lipopolyplexes combine strong antitumor T-cell immunity with improved inflammatory safety. ACS Nano. 2018;12:9815–9829. doi: 10.1021/ACSNANO.8B00966. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/NI1087. [DOI] [PubMed] [Google Scholar]
- Yukawa K, Mokuda S, Kohno H, et al. Serum CXCL10 levels are associated with better responses to abatacept treatment of rheumatoid arthritis. Clin Exp Rheumatol. 2020;38:956–963. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The source data for most of the figures are available from the authors.