The omicron SARS-CoV-2 variant of concern (B.1.1.529) was first reported in South Africa in mid-November, 2021. Early data indicated that infection with omicron (around 99% BA.1 lineage during this period) was associated with a lower risk of hospitalisation and lower risk of severe illness, once hospitalised, compared with delta (B.1.617.2) variant infection.1 Recently, the BA.2 lineage has increased in many areas globally, including South Africa, associated with increases in case numbers in some settings. In South Africa, the BA.2 lineage was first detected on Nov 17, 2021. From week 49 of 2021 (starting Dec 5, 2021), the proportion of BA.2 lineage began to increase, making up 84% (27 of 32) of all sequenced samples by week 5 of 2022 (week ending Feb 5, 2022).2 Replacement of BA.1 by BA.2 occurred in a period when SARS-CoV-2 case numbers were declining from the fourth wave peak in South Africa and was associated with a brief increase in case numbers in children of school-going age and slowing of the rate of decline compared with previous waves. The BA.1 lineage contains a 69–70 amino acid deletion in the spike protein, which is associated with S-gene target failure (SGTF) when tested using the TaqPath COVID‑19 PCR test (Thermo Fisher Scientific, Waltham, MA, USA). At the time of this study, BA.2 lacks this deletion, hence infections with BA.2 are S-gene positive on this assay.
Similar to BA.1, BA.2 is associated with substantial loss in neutralising activity in individuals infected with wild-type SARS-CoV-2 or recipients of mRNA vaccines.3 BA.2 has also been associated with increased transmissibility compared with BA.1,4 and in England was shown to have an increased growth rate compared with BA.1.5 However, data are lacking on the clinical severity of the BA.2 lineage compared with BA.1. We aimed to assess the severity of BA.2 infections compared with BA.1 in South Africa.
Using previously described methods,1 we performed individual-level data linkage for national data from three sources: (1) national COVID-19 case data, (2) SARS-CoV-2 laboratory test data for public sector laboratories and one large private sector laboratory, and (3) DATCOV, which is an active surveillance system for COVID-19 hospital admissions in South Africa (including both incidental and attributable admissions). Case and test data were obtained on Jan 29, 2022, and DATCOV data on Feb 10, 2022. In this analysis, restricted to tests performed on the TaqPath COVID‑19 assay, S-gene positive and SGTF infections were considered proxies for omicron lineages BA.2 and BA.1, respectively. Among 680 555 COVID-19 cases identified during the study period, the test used was known in 282 298 (41·5%) cases, and among these, 133 665 (47·3%) were diagnosed using the TaqPath COVID‑19 PCR test.
Two multivariable logistic regression models were generated to assess risk factors for (1) hospitalisation and (2) severe disease among hospitalised individuals (subset of individuals in model 1), comparing S-gene positive infections (proxy for BA.2) with SGTF infections (proxy for BA.1). We controlled for factors associated with hospitalisation (age, sex, presence of comorbidity, province, health-care sector, and previous SARS-CoV-2 infection) and factors associated with severity (age, presence of comorbidity, sex, province, health-care sector, number of days between the dates of specimen collection and hospital admission, known previous SARS-CoV-2 infection, and SARS-CoV-2 vaccination status) based on important predictors of outcome in South Africa6, 7 in the respective models. Data on comorbidities and SARS-CoV-2 vaccination were available only for hospitalised individuals. Cases were censored to those with a specimen collected before Jan 20, 2022, to allow for at least 3 weeks of follow-up. Severity analysis was restricted to admissions that had already accumulated outcomes, and all patients still in hospital were excluded. Severe disease was defined (based on a modification of WHO recommendations8) as a hospitalised patient meeting at least one of the following criteria: admitted to the intensive care unit, received any level of oxygen treatment, ventilated, received extracorporeal membrane oxygenation, experienced acute respiratory distress syndrome, or died.
Ethical approval was obtained from the Human Research Ethics Committee (Medical) of the University of the Witwatersrand for the collection of COVID-19 case and test data as part of essential communicable disease surveillance (M210752), and for the DATCOV surveillance programme (M2010108).
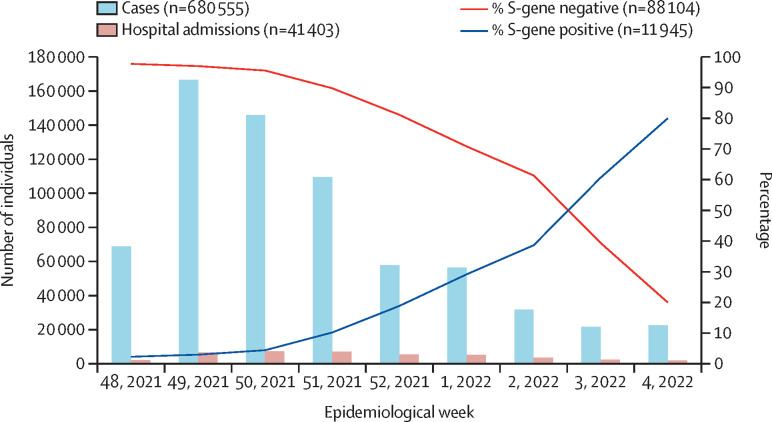
From Dec 1, 2021, to Jan 29, 2022, 680 555 SARS-CoV-2 infections were reported. From week 49 of 2021 (starting Dec 5, 2021) to week 4 of 2022 (ending Jan 29, 2022), the proportion of S-gene positive infections increased from 3% (931 of 31 271) to 80% (2425 of 3031; figure ). Among 95 470 samples tested using the TaqPath COVID-19 PCR assay, 3·6% of individuals with S-gene positive infection (BA.2 proxy) were hospitalised compared with 3·4% with SGTF infection (BA.1 proxy; appendix pp 1–2).
Figure.
Number of cases detected, hospital admissions, and percentage of S-gene positive and S-gene target failure (S-gene negative) infections among tests performed on the TaqPath assay by epidemiological week, Dec 5, 2021, to Jan 29, 2022
By multivariable analysis, after controlling for factors associated with hospitalisation, the odds of being admitted to hospital did not differ between individuals with S-gene positive (BA.2 proxy) infection compared with SGTF (BA.1 proxy) infection (adjusted odds ratio [aOR] 0·96 [95% CI 0·85–1·09]; appendix pp 1–2). In addition to geographical factors, hospital admission was associated with female sex (aOR 1·14 [1·06–1·22]) as well as young age (<5 years, aOR 7·49 [6·02–9·32]) and older age (40–59 years, aOR 1·39 [1·16–1·66] and ≥60 years, aOR 4·97 [4·12–5·94]) compared with individuals aged 19–24 years. Individuals in the private health-care sector were less likely to be admitted to hospital than those in the public sector (aOR 0·63 [0·58–0·68]).
Among hospitalised individuals diagnosed between Dec 1, 2021, and Jan 20, 2022, after controlling for factors associated with severe disease, the odds of severe disease did not differ between individuals with S-gene positive infection and individuals with SGTF infection (aOR 0·91 [95% CI 0·68–1·22]; appendix pp 3–5). The odds of severe disease was higher among individuals with a comorbidity (aOR 1·52 [1·25–1·84]), and among individuals aged 40–59 years (aOR 2·09 [1·33–3·31]) and 60 years or older (aOR 4·36, [2·77–6·85]), compared with individuals aged 19–24 years. The odds of severe disease were lower in children aged 5–12 years (compared with individuals aged 19–24 years), in females, and in individuals who had received at least one dose of SARS-CoV-2 vaccine. The distribution of the clinical severity components did not differ for individuals with S-gene positive infection compared with SGTF infection (appendix p 6).
Limitations of our study include restriction to samples tested with the TaqPath COVID-19 PCR assay, biasing data geographically, and that we used S-gene positive infection as a proxy for BA.2 lineage infection. Some misclassification could have occurred with other non-omicron variants, but these made up less than 2% of all detected viruses in December, 2021, and January, 2022. There could be a lag in hospitalisation and severe outcomes leading to underestimation of severe illness. To address this issue, we analysed data from hospitalised patients with known outcomes and censored cases to ensure at least 3 weeks of follow-up. We only had vaccination information for hospitalised patients, and this was based on self-report. Reinfection is probably also under-ascertained, as less than 10% of SARS-CoV-2 cases were diagnosed during the first and second waves in South Africa.9
We found a similar proportion of individuals were hospitalised and developed severe illness, given hospitalisation, for individuals infected with BA.1 compared with BA.2, during the omicron-dominated fourth wave in South Africa. These data are reassuring as they suggest that although BA.2 might have a competitive advantage over BA.1 in some settings, the clinical profile of illness remains similar. South Africa might differ from other settings in having a high level of previous immunity after natural infection,10 and data evaluating BA.2 severity are needed from other settings.
Acknowledgments
CC has received grant support from the South African Medical Research Council, UK Foreign, Commonwealth and Development Office, Wellcome Trust, US Centers for Disease Control and Prevention (CDC), and Sanofi Pasteur. NW has received grant support from Sanofi Pasteur and the Bill & Melinda Gates Foundation. AvG has received grant support from the US CDC, Africa Centres for Disease Control and Prevention, African Society for Laboratory Medicine, South African Medical Research Council, WHO Regional Office for Africa, The Fleming Fund, and Wellcome Trust. RW (a member of the DATCOV-Gen author group) declares personal shareholding in Adcock Ingram Holdings, Dischem Pharmacies, Discovery, Netcare, and Aspen Pharmacare Holdings. All other authors declare no competing interests. This study was funded by the South African Medical Research Council with funds received from the National Department of Health. Sequencing activities for the National Institute for Communicable Diseases are supported by a conditional grant from the South African National Department of Health as part of the emergency COVID-19 response; a cooperative agreement between the National Institute for Communicable Diseases of the National Health Laboratory Service and the US CDC; the African Society of Laboratory Medicine and Africa Centers for Disease Control and Prevention through a sub-award from the Bill & Melinda Gates Foundation; the UK Foreign, Commonwealth and Development Office and the Wellcome Trust; and the UK Department of Health and Social Care, managed by the Fleming Fund and performed under the auspices of the SEQAFRICA project. This research was also supported by The Coronavirus Aid, Relief, and Economic Security Act through the US CDC and the COVID International Task Force funds through the CDC under the terms of a subcontract with the African Field Epidemiology Network. Screening for SGTF at the University of Cape Town was supported by the Wellcome Centre for Infectious Diseases Research in Africa, which is supported by the Wellcome Trust. The findings and conclusions in this Correspondence are those of the authors and do not necessarily represent the official position of the funding agencies. The funders played no role in the writing of the Correspondence or the decision to submit for publication. Data used in this Correspondence are available upon reasonable request to cherylc@nicd.ac.za. Acknowledgments and contributor details for this Correspondence are listed in the appendix (p 7). Members of the DATCOV-Gen author group are listed in the appendix (p 8). AvG and CC contributed equally.
Supplementary Material
References
- 1.Wolter N, Jassat W, Walaza S, et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet. 2022;399:437–446. doi: 10.1016/S0140-6736(22)00017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Network for Genomic Surveillance in South Africa (NGS-SA) SARS-CoV-2 sequencing update. Feb 11, 2022. https://www.nicd.ac.za/wp-content/uploads/2022/02/Update-of-SA-sequencing-data-from-GISAID-11-Feb-2022.pdf
- 3.Iketani S, Liu L, Guo Y, et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyngse FP, Kirkeby CT, Denwood M, et al. Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: evidence from Danish households. medRxiv. 2022 doi: 10.1101/2022.01.28.22270044. published online Jan 30. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.UK Health Security Agency SARS-CoV-2 variants of concern and variants under investigation in England: technical briefing 35. Jan 28, 2022. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1050999/Technical-Briefing-35-28January2022.pdf
- 6.Jassat W, Cohen C, Tempia S, et al. Risk factors for COVID-19-related in-hospital mortality in a high HIV and tuberculosis prevalence setting in South Africa: a cohort study. Lancet HIV. 2021;8:e554–e567. doi: 10.1016/S2352-3018(21)00151-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies M-A, Kassanjee R, Rousseau P, et al. Outcomes of laboratory-confirmed SARS-CoV-2 infection in the Omicron-driven fourth wave compared with previous waves in the Western Cape Province, South Africa. Trop Med Int Health. 2022;27:564–573. doi: 10.1111/tmi.13752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO COVID-19 clinical management: living guidance. Jan 25, 2021. https://apps.who.int/iris/handle/10665/338882
- 9.Kleynhans J, Tempia S, Wolter N, et al. SARS-CoV-2 seroprevalence in a rural and urban household cohort during first and second waves of infections, South Africa, July 2020–March 2021. Emerg Infect Dis. 2021;27:3020–3029. doi: 10.3201/eid2712.211465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen C, Kleynhans J, von Gottberg A, et al. SARS-CoV-2 incidence, transmission, and reinfection in a rural and an urban setting: results of the PHIRST-C cohort study, South Africa, 2020–21. Lancet Infect Dis. 2022;22:821–834. doi: 10.1016/S1473-3099(22)00069-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.