Abstract
Purpose of Review
Increases in ambient levels of air pollutants have been linked to lung inflammation and remodeling, processes that lead to the development and exacerbation of allergic asthma. Conventional research has focused on the role of CD4+ T helper 2 (TH2) cells in the pathogenesis of air pollution-induced asthma. However, much work in the past decade has uncovered an array of air pollution-induced non-TH2 immune mechanisms that contribute to allergic airway inflammation and disease.
Recent Findings
In this article, we review current research demonstrating the connection between common air pollutants and their downstream effects on non-TH2 immune responses emerging as key players in asthma, including PRRs, ILCs, and non-TH2 T cell subsets. We also discuss the proposed mechanisms by which air pollution increases immune-mediated asthma risk, including pre-existing genetic risk, epigenetic alterations in immune cells, and perturbation of the composition and function of the lung and gut microbiomes.
Summary
Together, these studies reveal the multifaceted impacts of various air pollutants on innate and adaptive immune functions via genetic, epigenetic, and microbiome-based mechanisms that facilitate the induction and worsening of asthma.
Keywords: Asthma, Air pollution, Epigenetics, Microbiome, T cells, Innate lymphoid cells
Introduction
Asthma is a heterogeneous lung disease that currently affects approximately 25 million people in the USA [1]. This chronic pulmonary disease is characterized by reversible airway obstruction, bronchial hyperresponsiveness, pulmonary inflammation, and increased airway secretions [2]. Maladaptive pulmonary immune responses to allergens and/or other environmental exposures are thought to give rise to asthma [2, 3]. Epithelial inflammation stemming from these exposures leads to a cascade of immunological events, including activation and priming of antigen-presenting cells (APCs), polarization and clonal expansion of naïve T cells, and secretion of cytokines and chemokines that recruit eosinophils and neutrophils into the airspace [4, 5]. These responses lead to structural alterations of the lung, including mucus hypersecretion, increased goblet cell numbers, and peribronchiolar fibrosis [6]. These pathologies and cellular mechanisms have been characterized in both rodent models and patient populations. However, the exact mechanisms by which these maladaptive pulmonary immune responses arise are still being defined.
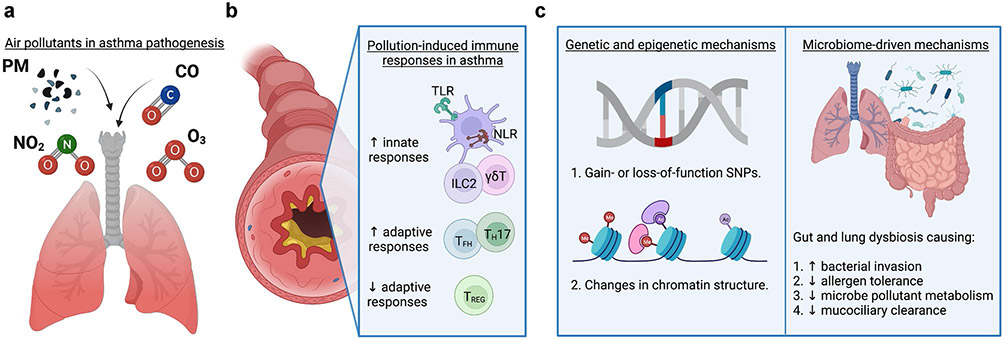
Both epidemiological and laboratory studies have reported that air pollution can increase susceptibility to and severity of asthma [7•]. Air pollution exposure can directly and indirectly stimulate the innate and adaptive immune responses that are known to drive asthma pathogenesis (Fig. 1a-b). A major component of air pollution is particulate matter (PM), ranging from coarse PM with diameter ≤ 10 μm (PM10) that tends to deposit in the upper airway, fine PM ≤ 2.5 μm (PM2.5) that can deposit in the central and peripheral airways and alveoli, and ultrafine PM ≤ 0.1 μm (UFPs) that transiently affects respiratory tissues along the whole tract [8, 9]. Diesel exhaust particles (DEP) include a combination of PM with organic compounds such as polycyclic aromatic hydrocarbons (PAHs), sulfate, nitrate, and other trace elements [10]. In addition to PM, air pollution also contains a significant portion of noxious gasses, which include carbon monoxide (CO), nitrogen dioxide (NO2), and ozone (O3) [11]. Several studies in recent years have shown that these air pollutants alone can elicit an immune response that can potentially influence asthma pathogenesis. Therefore, understanding how air pollution induces pathogenic immune responses in the lung may provide novel therapeutic targets to prevent or treat asthma.
Fig. 1.
Effects of air pollution on novel immune-mediated mechanisms of asthma pathogenesis. a Pollutants that induce or exacerbate asthma by altering immune-mediated responses. b Air pollution induced non-TH2 immune responses, leading to increased susceptibility to or severity of asthma. c Mechanisms of pollutant-driven asthma pathogenesis, including genetic risk, epigenetic alterations, and changes in the lung/gut microbiome. Created with BioRender.com
This review will provide insight into emerging investigations of the relationships between air pollution exposure and asthma pathogenesis. Specific emphasis will be placed on novel pulmonary immune mechanisms known to be altered with exposure to select air pollutants. Within this scope, the role of genetics and epigenetics in these immunological and inflammatory mechanisms, and how the microbiome (both in the lung and gut) can modulate these responses, will also be explored. This overview will highlight what is known about these novel mechanisms and propose further areas of study to better examine the impact of air pollution on innate and adaptive immune responses that contribute to asthma.
Air Pollution Modulates Asthma Through the Classical TH2 Immune Response
Typically, the immune response driving sensitization in asthma begins with airway inflammation leading to the activation of APCs that interact with naïve CD4+ T lymphocytes using major histocompatibility complex (MHC) molecules loaded with antigen. A variety of environmental antigens polarize naïve CD4+ T cells toward the TH2 cell fate that is characteristic of the classical asthma phenotype [12]. TH2 cells are the canonical T cell population implicated in eosinophilic asthma and produce several pro-inflammatory cytokines including IL-4, IL-5, and IL-13 that affect downstream immune responses [13-15]. The presence of these TH2 cytokines in the environment leads to immunoglobulin E (IgE) class switching and cytokine production by B cells that drive many of the pathologies associated with subsequent allergen challenge [16]. Consequently, activation and cross-linking of the IgE-FcεRI complex on effector cells (i.e., mast cells and basophils) lead to the release of vasoactive soluble mediators such as prostaglandins, leukotrienes, and histamine, resulting in bronchial mucosa edema, mucous production, and smooth muscle constriction [13, 16-19]. The combination of these immune responses and subsequent inflammatory factors leads to airway hyperresponsiveness (AHR) and airway obstruction associated with asthma, as well as other inflammatory airway diseases.
Though there have been extensive investigations in both laboratory models and human subjects defining how air pollution exposure alters TH2–driven asthma [20•, 21-23], there has been much less progress in understanding the inflammatory processes that initiate asthma and/or other T cell subsets known to alter the asthmatic phenotype. While a range of innate and adaptive immune response mechanisms has been described beyond TH2 cells, this review will focus on how air pollution alters other innate and adaptive immune responses in asthma, including diverse T cell subsets, pattern recognition receptors (PRRs), and innate lymphoid cell (ILC) populations (Fig. 1b).
Novel Roles of Non-TH2 T Cell Subsets in Asthma
It has long been appreciated that the adaptive immune response plays a critical role in asthma development and exacerbation. The traditional understanding of asthma pathogenesis implicates TH2 cells as the main drivers of eosinophilic allergic asthma. However, recent studies have noted a role for non-TH2 subsets, including T helper 17 (TH17), T follicular helper (TFH), regulatory T (TREG), and gamma delta T (γδT cells in asthma. Additionally, emerging evidence has also shown that air pollution affects the polarization and function of these cells in allergic airway disease. Below we review the recent literature describing the impact of air pollution on non-TH2 T cell subsets and their roles in asthma pathogenesis.
TH17 Cells
TH17 cells are well-known to play a pro-inflammatory role in the body through the production of IL-17, which contributes to the pathogenesis of many autoimmune and inflammatory diseases like rheumatoid arthritis, psoriasis, multiple sclerosis, inflammatory bowel disease, and asthma [24]. In contrast to type-2-mediated eosinophilic asthma, many studies support a role for TH17 cells in driving neutrophilic asthma, which has been observed in a subset of asthmatics that are resistant to corticosteroid treatment [25]. A balance between pro-inflammatory TH17 cells and immunosuppressive TREG cells has been commonly described in the literature to play a significant role in asthma [26-28]. In mice, Zhou et al. found that PM2.5 exposure in a murine model of asthma increased the TH17/TREG ratio and symptoms associated with asthma exacerbations [29]. Increases in TH17 cells and effector cytokines such as IL-17A, IL-17F, and IL-23 have also been noted with DEP co-exposure with house dust mite (HDM) antigen [30]. Consistent with this, asthmatic children exposed to DEP also had significant increases in serum IL-17A [30]. These findings support the idea that air pollution exposure may contribute to the heterogeneity of asthma, potentially through TH17 polarization and expansion.
TFH Cells
TFH cells express the lineage-defining transcription factor B cell lymphoma-6 protein (BCL-6), as well as surface C-X-C chemokine receptor 5 (CXCR5), programmed death protein 1 (PD-1), and the inducible T cell co-stimulator (ICOS) [31, 32]. CXCR5 directs trafficking of TFH cells to the B cell follicle of secondary lymphoid organs, such as the tonsil, spleen, and lymph nodes. Here, TFH cells activate B cells, leading to the formation of germinal centers, affinity maturation, and productive antibody-mediated immunity and humoral immune responses [32, 33]. Within the context of asthma pathogenesis, class switching of IgE, the most common isotype implicated in allergic asthma, can be positively or negatively regulated by the TFH–produced cytokines IL-4 and IL-21, respectively [34•, 35]. Thus, TFH cells have an indirect role in mediating allergic asthma development via regulation of B cell and IgE responses.
TFH cells are also known to display functional plasticity via their differentiation into TFH1, TFH2, TFH13, TFH17, and regulatory (TFR) cells; these subsets are specific for various immune responses, with transcription factors and phenotypes similar to their TH1, TH2, and TH17 counterparts [32, 34•]. TFH2 cells are of particular interest in asthma because they secrete IL-4, similar to TH2 cells, and thus initiate and support IgE production in allergic disease [36-39]. Furthermore, TFH13 cells, which produce IL-4 and IL-13 to regulate antibody class switching and IgE affinity, are significantly increased in patients with allergic rhinitis and asthma [34•, 40].
Given the relatively recent acknowledgement of the role of TFH2 cells in asthma [41] and the recent discovery of TFH13 cells [34•], the impacts of air pollution on TFH cells and asthma remain unknown. However, there is data suggesting that TFH cells are not exempt from the effects of air pollution on asthma exacerbation. Ma et al. found that in response to PM2.5 exposure, CD4+ and CD8+ T cells exhibited a macrophage-dependent production of cytokines including IL-21, which could suggest the possibility of increased TFH differentiation, though a more targeted investigation is needed [42].
TREG Cells
TREG cells play an important role in the negative regulation of immune cells that cause allergic and autoimmune responses. There are two main subsets of TREG cells: “natural” TREG (nTREG) cells, which develop in the thymus, and “induced” TREG (iTREG) cells, which develop in the periphery [43]. Once they develop, nTREG cells express the lineage-defining transcription factor forkhead box P3 (Foxp3), whereas iTREG cells upregulate Foxp3 expression following polarization [44, 45]. To perform their regulatory functions, TREG cells secrete immunosuppressive cytokines such as IL-10 and TGF-β [46], suppress APC activation [47], and sequester IL-2 [48], among other effector functions. These effects dampen the innate immune response [49] and suppress proliferation of differentiating immune cells like T cells that have been linked to asthma pathogenesis [50]. Additionally, TREG cells have been shown to play a role in inhibiting the proximal pathways of allergic sensitization and IgE production in response to allergen exposure [44, 51].
Studies have shown an association between ambient air pollution, impaired TREG function, and increased morbidity in people with asthma [52, 53]. A 2010 study comparing asthmatic and non-asthmatic children from Fresno, CA (poor air quality, with PM concentrations exceeding the federal annual standard by over 40%) to those from Stanford, CA (good air quality compared to Fresno) found that the more severe Fresno asthma group had reduced TREG cell immunosuppression, chemotaxis, and function [52]. This was also noted in another cohort in the 2015–2018 Nutrition in Early Life and Asthma (NELA) birth cohort, where García-Serna et al. found that levels of TREG cells were decreased in newborn cord blood exposed to transient NO2 or PM10 pollution in utero [54], corroborating similar data found in newborns and children [55]. Despite this, it is still unclear how air pollutants alter TREG numbers and function in the context of asthma.
γδT Cells
γδT cells are unconventional, innate-like T cells with T cell receptors (TCRs) made of γ and δ chains (instead of conventional α and β chains) that provide a bridge between innate and adaptive immune functions. Composed of only 5% of peripheral T cells, γδT cells can be divided into subsets that function analogous to conventional TH1, TH2, TH17, TREG, and TFH cells [56]. Thus, the contribution of γδT cells to asthma pathogenesis is complex. Although the overall proportion of γδT cells is lower in asthmatic patients [57], murine asthma models show that TH2-like and TH17-like γδT cells are increased in the blood and bronchoalveolar lavage (BAL), respectively [58]. Pulmonary γδT cells contribute to asthma pathogenesis by producing IL-4 [57], which enhances allergen-induced late airway responses and inflammation [59]. However, γδT cells have also been noted to inhibit AHR production via interferon gamma (IFN-γ) secretion and suppression of IgE production [60]. Additionally, IL-17-producing γδT cells play a dual role in asthma by altering AHR [61]. Recent publications have described an association between air pollution and γδT cell function. For example, O3 has been shown to increase total γδT and IL-13+ γδT cells within the lungs of obese mice compared to wildtype mice [62]. Additionally, murine lung injury models have also shown that O3 and PM2.5 can increase the number of IL-17A–secreting cells, the majority of which are γδT cells, thus promoting lung fibrosis and inflammation [63, 64]. Further studies are needed to more fully understand how γδT cell subsets are impacted by air pollution in the context of asthma.
Innate Lymphoid Cells
In addition to the various T cell immune responses contributing to asthma, innate responses to air pollution have also been shown to play important roles in this chronic lung disease. An emerging area of innate immunity that has been implicated in asthma pathogenesis is the contribution of ILCs. ILCs are a family of non-T, non-B lymphocytes that have conserved effector cell function and are present in mucosal and lymphoid tissues. ILCs survey tissues for pathogens and damage to rapidly and efficiently respond in an antigen-independent manner [65]. They play a critical role in tissue homeostasis, resistance to infection, control of the composition of commensal microbiota, and pathology at mucosal surfaces [66, 67]. ILCs are composed of five subfamilies based on surface expression markers and effector function: the two cytotoxic ILC subfamilies (natural killer (NK) cells and lymphoid tissue-inducer cells (LTi)) and the three helper ILC subfamilies (type 1 ILCs (ILC1s), ILC2s, and ILC3s) [65, 68, 69]. The helper ILCs parallel CD4+ T helper cells: ILC1s, ILC2s, and ILC3s function like TH1, TH2, and TH17 cells, respectively.
In asthma pathogenesis, ILC2s are the most widely studied subfamily. ILC2s function most like the asthma-mediating TH2 cells through production of IL-4, IL-5, and IL-13, which promote eosinophil recruitment, macrophage polarization, mast cell activation, goblet cell mucus production, and smooth muscle contraction [65, 70-72]. ILC2s also represent the most common resident and migratory ILC population in the asthmatic lung, with their frequency and activation increased during asthma [73, 74]. Furthermore, in a chronic murine model of allergic asthma, lung ILC2s, and not T or B cells, were required for disease maintenance [75]. It should be noted that ILC2 frequency decreases with inhaled corticosteroid treatment [76, 77]. Recent studies have unveiled novel ILC2 populations, including regulatory IL-10+ ILC2s [78-81] and novel ILC2-to-ILC1 or ILC2-to-ILC3 plasticity [68, 82], indicating that there may be multiple roles that other ILC subsets play in asthma pathogenesis.
As the role of ILCs begins to emerge in the context of allergic asthma, several studies have shown that air pollution, specifically PM and O3, can alter ILC functions in the lung. Recent studies have reported that air pollution can reduce IFN-γ production and the cytotoxicity of ILC1s. Additionally, O3 can stimulate lung ILC2s by increasing IL-33 levels and ILC2–specific activation, proliferation, and airway inflammation [62, 83-87]. Lastly, recent laboratory studies found that co-exposure of DEP with an allergen (e.g., HDM), but not DEP or HDM alone, leads to marked increases in IL-25 and IL-33 and moderate increases in ILC2 levels [88]. This suggests that DEPs may work synergistically with allergens to induce lung inflammation. These findings of ILCs have also been noted in human studies where PM10 exposure levels had a positive correlation with the frequency of ILC2s, particularly in severe asthmatics, whereas ILC1s correlated with O3, NO2, and CO exposure [89]. Taken together, these data identify an emerging role for ILC modulation by air pollution, which may contribute to how these environmental exposures alter the susceptibility and heterogeneity of asthma.
Pattern Recognition Receptors in Asthma
Beyond ILCs, critical components of the innate inflammatory response involved in initiating asthmatic responses are pattern recognition receptors (PRRs), which recognize Pathogen-Associated Molecular Patterns (PAMPs) and Damage-Associated Molecular Patterns (DAMPs) [90]. Primarily found on APCs, such as dendritic cells (DCs) and macrophages, PRRs include four main types: (1) toll-like receptors (TLRs), (2) nucleotide-binding oligomerization domain (NOD)-leucine rich repeats (LRR)-containing receptors (NLRs), (3) retinoic acid-inducible gene 1-like receptors (RLR), and (4) C-type lectin receptors (CLRs). Due to their ability to quickly sense environmental triggers, including air pollutants, TLRs and NLRs play major roles in the pathogenesis of lung inflammation and allergic asthma. It is through PRR signaling that many of the innate and adaptive immune responses downstream of pollutant exposure begin to unfold, as we detail in the sections that follow.
Toll-Like Receptors
TLRs are transmembrane receptors of the innate immune system located on the cell surface or in endosomes that are important for initiating adaptive immune responses. Environmental allergens are known to activate the TLRs of the lung airway epithelia, leading to allergic and asthmatic disease [91]. TLR2 and TLR4, which recognize gram-negative and -positive bacteria, respectively, are the most well-studied in the development and exacerbation of allergic asthma. TLR2 and/or TLR4 activation is known to drive exacerbation of acute and chronic inflammation associated with asthma and allergic disease by promoting neutrophil, eosinophil, TH2, and TH17 activation [92-95]. Deletion of TLR4 and downstream signaling molecules, including MyD88, leads to a reduction of asthma-related inflammation, as evidenced by various laboratory studies [96-99]. Additionally, TLR2 directly activates lung type 2 innate lymphoid cells (ILC2s), which are a source of IL-5 and IL-13 in allergic airway inflammation [100]. However, recent studies have noted a nonredundant role for other TLRs in allergic asthma. For instance, TLR9 has been shown to prevent ILC2–driven AHR [101]. At the same time, TLR9 mediates airway inflammation by activating NLRP3 inflammasome and increasing oxidative stress [102]. Interestingly, severe asthmatics have been found to have decreased expression of TLR5 and TLR7 [103], suggesting that severe asthmatics may suffer from insufficient TLR signaling during bacterial or viral infections, leading to asthma exacerbation.
Given the role of TLRs in allergic asthma, determining whether air pollutants modulate TLR signaling in a way that increases allergic asthma burden is of great interest. Data from both in vivo and in vitro laboratory studies have shown that air pollution alone (O3 and/or PM) can induce TLR2– and TLR4–dependent cytokine production [104-106] and AHR [107-112]. Although it is unclear how these pollutants can induce pulmonary inflammation and/or dysfunction, it is thought that this could be driven by the DAMPs generated in the airspace following exposure or because of the PAMPs; the exposures can carry into the lung. For example, PM2.5 has been found to contain lipotoeic acid (LTA) and lipopolysaccharide (LPS, or endotoxin), and when compared to LPS alone, PM2.5 and LPS co-exposure resulted in an enhanced TLR2/TLR4/MyD88–driven allergic airway inflammation and eosinophilia [113, 114•, 115]. Thus, the interplay between air pollution and TLR–induced asthma is an evolving field, and as functional and nonfunctional TLR variants and their downstream signaling networks are further described, there will be greater insight into how these PRRs contribute to air pollution-induced asthma exacerbation.
NOD-Like Receptors
Unlike the transmembrane TLRs, NLRs are cytosolic innate immune receptors that sense intracellular microbial products. The five NLR subfamilies are NLRA, NLRC, NLRC, NLRP, and NLRX. The most studied NLRs include nucleotide-binding oligomerization domain-containing protein 1 (NOD1) and NOD2 in the NLRC subfamily, and NOD–, LRR–, and pyrin domain-containing protein 3 (NLRP3) in the NLRP subfamily [116]. Many of these NLRs have been implicated in the onset and/or progression of asthma, with much of the research focused on the role of NLRP3–driven activation of the inflammasome in asthma [78, 117-124]. For instance, increased sputum NLRP3 expression correlates with neutrophilic inflammation and asthma severity [125]. However, other NLRs are beginning to be recognized for their contributions in the onset and/or exacerbation of allergic asthma. For example, NOD1 gene variants [126, 127] and dysregulated expression of NOD1 isoforms [128] have been shown to alter asthma pathogenesis. Additionally, NOD2 ligands can lead to TH2 activation and increase asthmatic inflammation [129-131], although the exact role of this NLR in the immune response driving allergic asthma is still debated [132].
Recent studies have noted that the exacerbation of asthmatic responses by air pollutants may be through NLR–driven mechanisms. NLRP3 is known to be activated by environmental oxidants, including O3 [133, 134] and PM [135-137]. It has been proposed that NLRs are activated by these air pollutants via reactive oxygen species (ROS)–induced mitochondrial dysfunction [133, 138] and extracellular release of intracellular DAMPs such as the nuclear high mobility group box 1 (HMGB1) protein [139, 140]. Additionally, PM exposure was found to activate the sterol regulatory element-binding protein 1 (SREBP1)/Pirin (PIR) axis via Sirtuin1 (SIRT1) inhibition [141], which in turn activates the NLRP3 inflammasome, leading to acute and chronic lung inflammation. This NLRP3 activation has also been noted with other ambient particle exposures such as DEP, leading to airway inflammation and mucus secretion [142, 143]. Taken together, these data show that air pollution initiation of NLR–based signaling may be a novel and emerging area in the study of asthma pathogenesis.
Mechanisms by which Air Pollutants Influence Asthma Pathogenesis and Exacerbation
It is clear that air pollutants, either alone or in conjunction with allergens, alter innate and adaptive immune responses as discussed above. However, the biological processes of how pollutants induce these immune changes have yet to be fully established. Three candidate mechanisms have been recently uncovered that strongly suggest that air pollution modulates immune cell functions via (1) interaction with genetic risk, (2) epigenetic changes in immune cells that alter gene expression, and (3) altered composition and function of the lung and gut microbiome (Fig. 1c). Below, we briefly describe the current understanding of these novel mechanisms by which air pollution alters the immune response, thus increasing susceptibility and/or severity of asthma.
Air Pollution and Genetic Risk Predisposing to Asthma Development
Despite the environmental impacts on respiratory disease pathogenesis, asthma remains a notably heterogenous disease with substantial genetic contributions [144]. Thus, identifying the genes and genetic variants involved in asthma is of great interest for comprehensive prevention and management of this disease. Several studies have identified several genes contributing to an individuals’ genetic risk for the development and severity of asthma, including IL-13, TNF, ADAM33, IL-4RA, DPP10, PHF11, NPSR1, HLA-G, CYFIP2, IRAK3, COL6A5, OPN3/CHML, and TBXA2R [144]. More recently, genome-wide association studies (GWAS) have been used to identify disease associated with over 500,000 single nucleotide polymorphisms (SNPs), or specific gene variants across populations that lead to increased or decreased risk [145]. For example, SNPs in NLRs including NOD1 have been shown to either protect against or induce asthma [126, 127]. Additionally, NLRP3 and Caspase 1 (CASP1) polymorphisms have been associated with either increased or decreased asthma risk in a population of Brazilian children [146•]. Beyond PRRs, other studies have identified asthma-associated SNPs in ILC2 gene regulatory elements that could increase disease risk [147•], though the contributions of specific SNPs must be further defined. SNPs that are protective against asthma have also been identified in TH17 cell functioning pathways [148]. Together, genetic variation and SNPs highly influence susceptibility to or exacerbation of asthma.
Currently, there are emerging studies that have defined a direct interaction between air pollution and genetic risk for developing asthma [149]. Recent studies have reported that SNPs in certain PRRs interact with air pollution to induce asthma [150, 151]. In mice exposed to O3, specific polymorphisms in the TLR4 and TLR5 altered asthma pathogenesis [152, 153••]. Gain-of-function SNPs in NLRP1 have been found to activate its inflammasome following air pollution exposure, leading to high IgE levels and asthma exacerbation [154]. Additionally, a recent genome-wide interaction study (GWIS) analyzing the impact of NO2 air pollution on childhood asthma identified SNPs in the novel loci B4GALT5 and in SNPs previously associated with lung disease, ADCY2 and DLG2 [155]. Even though these data are still being generated, thus far it seems that air pollution plays a pervasive role in asthma pathogenesis in those with increased genetic risk. Additional GWAS studies will provide an unbiased approach for understanding which individuals and populations have a higher genetic risk for asthma, as well as reveal how air pollution contributes to this genetic risk.
Air Pollution and Epigenetic Changes Inducing Asthma
Beyond the mere presence or absence of asthma-related genes in an individual, asthma induction and severity are also influenced by dynamically regulated gene expression, referred to as epigenetics [156]. These epigenetic processes include chromatin remodeling, biochemical changes to DNA and histones—such as methylation (typically leading to reduced DNA accessibility) and acetylation (typically leading to increased DNA accessibility)—and RNA interference, among others. Ultimately, these changes result in altered gene transcription, transcriptional responsiveness to stimuli, or translational availability of gene transcripts [157]. Further still, some of these epigenetic changes and their resulting traits seem to demonstrate transgenerational inheritance [158-160], adding to the complexity of epigenetic contributions in health and disease.
The data connecting specific epigenetic changes to asthma are plentiful [161-164], to the extent that they give rise to an emerging paradigm that asthma is an “epigenetic disease” of the immune system. For example, several studies link DNA methylation of immunosuppressive TREG genes to the exacerbation of asthma [165, 166]. Meanwhile, altered methylation in loci linked to IL-4, IL-13, IL-5RA, ZPBP2, RUNX3, TIGIT, and ALOX15 has also been associated with asthma [167-171]. Histone modifications also play a significant role in asthma-related gene expression, as the permissive modifications histone H3 lysine K4 (H3K4) trimethylation and histone hyperacetylation have been connected to increased T cell activation and airway remodeling [172-174]. A large number of noncoding microRNAs (miRNAs), which block or alter mRNA translation, have also been heavily implicated in asthmatic phenotypes and responses to therapy [175-178]. In addition, an interesting human study of the Isle of Wight birth cohort shows that specific DNA methylation changes associated with asthma persist from F0 to either F2 or F3 generations of asthma patients [179, 180].
There is now a large body of work demonstrating epigenetic changes that occur in response to environmental pollutants that are associated with asthma pathogenesis [181••, 182-184]. From these studies, we now know that PM, O3, and other air pollutants can contribute to airway inflammation via epigenetic enzyme perturbation (namely the ten-eleven translocation (TET) 1–3 and DNA methyltransferase 3 (DNMT3) A-B enzymes) [185], DNA methylation [52, 166, 186-190], histone modifications [191], and miRNA regulation [192-195]. It is possible that some of these components impair the expression of epigenetically acting enzymes responsible for maintaining or altering the epigenetic landscape [196, 197]. With exposure to air pollution, many of these epigenetic changes have been noted in DCs [198, 199], PRRs [200, 201], ILCs [147•, 202], and various T cell subsets [52, 173, 186, 203-205, 206••, 207]. Notably, there is an increasing body of literature connecting urban lifestyle [208] or specific pollutant exposures, such as DEP, concentrated urban air particles (CAP) [205, 209-217, 218••], and black carbon particles [219, 220], to aberrant epigenetic signatures seen in asthma. In fact, DEP and CAP can transmit an asthma risk phenotype to F2 and F3 generations, an effect which seems to be linked to epigenome-wide methylation aberrations [199].
Whether these epigenetic changes seen in humans and mice directly cause asthma is yet to be determined. Future causality studies may confirm a paradigm of “asthma as an epigenetic disease,” which would shift our understanding of asthma from the current “inflammatory disease of the airways” consensus definition. As novel tools emerge for targeted manipulation of the epigenome, which have only recently been developed within the last decade [221-234], this promises new avenues toward understanding how air pollution increases risk for asthma development.
Air Pollution and Microbiome Changes Inducing Asthma
In addition to genetic and epigenetic changes increasing risk for asthma, recent studies have highlighted that the commensal bacteria (termed the “microbiome”) in the lung and gut can influence asthma incidence and severity. Given the widespread colonization of these commensal microorganisms within the human body, the microbiome interacts with and alters the metabolic and effector functions of nearby and distant cells [235]. Collectively, the microbiome of the lung and gut is composed of bacteria, archaea, viruses, and fungi, which have been shown to play critical roles in the training and development of the host’s innate and adaptive immunity [236]. However, changes in the microbiome, whether through antibiotic use, diet change, or other environmental perturbations, can lead to immune responses that drive diseases such as food allergy, inflammatory bowel disease, rheumatoid arthritis, metabolic diseases, neurodegeneration, and asthma [237-242].
The association of an altered microbiome in the lung and asthma was initially reported by Hilty et al. [243], which has since been confirmed by multiple additional studies of the respiratory microbiome in humans [244-253] and mice [254, 255]. In a healthy adult human lung, the main phyla present are Bacteroidetes, Firmicutes, Actinobacteria, and Fusobacteria [246, 256], while the healthy gut normally contains thousands of microbiota species, especially from the Bacteroidetes, Firmicutes, Actinomycetes, and Verrucomicrobia phyla [257, 258]. Both laboratory and clinical studies of the lung and gut microbiome have shown that there are increases in the Proteobacteria phyla in asthma [243, 246, 253, 259-261], accompanied by decreases in beneficial Firmicutes and Bacteroidetes [247, 256, 262•]. In addition to reduced microbiome diversity, there also seems to be increased levels of the pathogenic Haemophilus and Moraxella bacteria in neutrophilic asthmatics compared to eosinophilic asthmatics [263•, 264]. Mechanistically, this altered microbiome contributes to asthma development in part through its impact on several innate and adaptive immune responses, including TLR signaling [265••, 266], NLRP3 signaling [118, 267•], and T cell responses [265••, 268, 269]. However, given the heterogeneity of asthma, the contributions of an altered lung and/or gut microbiome to an asthmatic phenotype are still widely unknown.
Recently, air pollution has also been shown to affect the composition and function of the microbiome [270]. A few studies have demonstrated that air pollution decreases airway microbiome diversity, which was associated with decreased lung function [271••, 272-274]. However, reports conflict on whether air pollution increases [272, 274-276] or decreases [277-279] microbiome alpha diversity. Furthermore, it has yet to be described if a specific taxa are enriched or depleted after air pollution exposure, nor is there a consistent pattern or predictability associated with these changes [39, 272, 274-276, 278-280]. Still, it is likely that the altered gut and lung microbiome following air pollution modulates asthma risk. Polluted air often contains pathogenic bacteria and thus may influence airway inflammation via direct introduction of harmful microbiota into the respiratory tract [66]. It has also been observed that air pollutants’ alteration of microbial components is associated with changes in asthma risk [281]. For example, altered microbiome composition changes the levels of tolerogenic short-chain fatty acids (SCFAs) produced by commensal bacteria; loss of these SCFAs, which normally reduce allergen sensitivity, airway inflammation, and asthma risk in infants [252, 282, 283] and mice [284-287], can lead to peripheral immune cell dysregulation [288] and increased risk of developing allergic asthma [289]. Beyond SCFAs, microbial LPS attached to air pollutants can further stimulate TLRs and activate downstream ROS and PAH–sensing pathways [3, 256]. However, this relationship is not simple. Higher levels of endotoxin in dust extract from homes of the Amish, a population known to be exposed to high microbial levels on their traditional farms and to have low asthma prevalence, have been shown to correlate with protection against innate and adaptive airway inflammation in children [290••] and in murine experimental asthma models [291]. The hygiene hypothesis and its proposed updates attempt to explain how exposure to some types of microbes in early life assists in immune system development [292]. Beyond microbial components themselves, new data are emerging showing the role of commensal bacteria in immune cell homeostasis and the subsequent perturbation of effector function following PM exposure [293•]. Taken together, these data provide a foundation for further investigations of how air pollution induces asthma by altering the microbiome.
The role of the microbiome in human disease remains an ever-expanding field requiring intensive biomedical characterization. Only a small number of studies on the direct effects of air pollution on the microbiome have been published in the last decade with a great deal of variability in their geographic populations, tissue sampling, and pollution-measuring methodologies. Thus, examining the effects of specific air pollution components in well-controlled animal studies will help to define their effects. Because the microbiome may potentially metabolize inhaled pollutants and modulate downstream immune responses [294], it would also be critical to thoroughly define the host homeostatic functions that the microbiome perform, including epigenetic alterations. With the integration of metagenomics and personalized medicine into clinical diagnosis and treatment becoming increasingly more mainstream, gut and lung microbiome diversity may be used to predict risk of allergic asthma. Finally, as the contribution of the microbiome to asthma is more precisely defined, it may be likely that prebiotics, probiotics, or targeted antibiotics could be designed for asthma prevention and treatment to offset damage from air pollution.
Conclusion
Given the established association between air pollutant exposure and asthma development and exacerbation, we have compiled the latest knowledge on the genetic, epigenetic, and microbiome-driven immune mechanisms by which asthma is worsened. The available literature has focused on TLRs and NLRs in the context of air pollution and asthma, though future work should explore the roles that the RLR and CLR PRRs play in asthma pathogenesis in the presence of air pollution. Given the collective contributions of different T cell subsets, increased research on the impact of air pollution on the functional plasticity of T cells and their subsequent effect on asthma phenotype and severity is also warranted. Further improvements in accessibility to epigenetic tools and microbiome diversity screens for physicians may ultimately help to predict who is at greatest risk for allergic and asthmatic disease.
Acknowledgements
Drs. Fedulov, Oldfield, and Wiscovitch-Russo are supported by NIEHS R01 ES030227. Dr. Oestreich is supported by NIAID R01 AI134972. Ms. Tuazon is supported by The Ohio State University Susan Huntington Dean's Distinguished University Fellowship. Drs. Gowdy and Dunigan-Russell are supported by NIEHS R01 ES028829.
Footnotes
Conflict of Interest Brita Kilburg-Basnyat reports being employed at Arcus Biosciences, Inc. as of 1/18/22. None of the work for this review paper was performed while employed by the biopharmaceutical company and the treatments Arcus is pursuing are oncology-related. Lauren Oldfield reports personal fees from Replay Holdings LLC, outside the submitted work. Jasmine Tuazon, Rosana Wiscovitch-Russo, Katelyn Dunigan-Russell, Alexey Fedulov, Kenneth Oestreich, and Kymberly Gowdy declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Centers for Disease Control and Prevention. 2019 National Health Interview Survey data [Internet]. U.S. Dep. Heal. Hum. Serv. 2020. [cited 2022 Jan 17]. Available from: https://www.cdc.gov/asthma/nhis/2019/data.htm [Google Scholar]
- 2.Chabra R, Gupta M. Allergic and environmental induced asthma [Internet]. StatPearls. Treasure Island (FL): StatPearls Publishing; 2021. Available from: https://pubmed.ncbi.nlm.nih.gov/30252274/ [Google Scholar]
- 3.Glencross DA, Ho TR, Camiña N, Hawrylowicz CM, Pfeffer PE. Air pollution and its effects on the immune system. Free Radic Biol Med. 2020;151:56–8. [DOI] [PubMed] [Google Scholar]
- 4.Holtzman MJ. Asthma as a chronic disease of the innate and adaptive immune systems responding to viruses and allergens. J Clin Invest. 2012;122:2741–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holgate ST. Innate and adaptive immune responses in asthma. Nat Med. 2012;18:673–83. [DOI] [PubMed] [Google Scholar]
- 6.Shen Y, Huang S, Kang J, Lin J, Lai K, Sun Y, et al. Management of airway mucus hypersecretion in chronic airway inflammatory disease: Chinese expert consensus (english edition). Int J COPD. 2018;13:399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.•. Liu Y, Pan J, Zhang H, Shi C, Li G, Peng Z, et al. Short-term exposure to ambient air pollution and asthma mortality. Am J Respir Crit Care Med. 2019;200:24–32. (COMMENT: Case crossover study of 4454 individuals who died from asthma in Hubei province, China, showing short-term exposure to PM2.5, NO2, and O3 was positively associated with asthma mortality.)
- 8.Vallero D. Fundamentals of air pollution, fifth edition. Fundam. Air Pollution, Fifth Ed. 2014. [Google Scholar]
- 9.U.S. EPA. Integrated science assessment for particulate matter (Final Report). Washington, D.C.; 2016. [Google Scholar]
- 10.Wichmann HE. Diesel exhaust particles. Inhal Toxicol. 2007;19:241–4. [DOI] [PubMed] [Google Scholar]
- 11.Tiotiu AI, Novakova P, Nedeva D, Chong-Neto HJ, Novakova S, Steiropoulos P, et al. Impact of air pollution on asthma outcomes. Int J Environ Res Public Health. 2020;17:6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammad H, Lambrecht BN. The basic immunology of asthma. Cell. 2021;184:1469–85. [DOI] [PubMed] [Google Scholar]
- 13.Novak N, Bieber T. Allergic and nonallergic forms of atopic diseases. J Allergy Clin Immunol. 2003;112:252–62. [DOI] [PubMed] [Google Scholar]
- 14.Walker JA, McKenzie ANJ. TH2 cell development and function. Nat Rev Immunol. 2018;18:121–33. [DOI] [PubMed] [Google Scholar]
- 15.Zhu J. T helper cell differentiation, heterogeneity, and plasticity. Cold Spring Harb Perspect Biol. 2018;10:a030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holgate ST, Polosa R. Treatment strategies for allergy and asthma. Nat Rev Immunol. 2008;8:218–30. [DOI] [PubMed] [Google Scholar]
- 17.Bax HJ, Keeble AH, Gould HJ. Cytokinergic IgE action in mast cell activation. Front Immunol. 2012;3:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poynter ME. Airway epithelial regulation of allergic sensitization in asthma. Pulm Pharmacol Ther. 2012;25:438–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Froidure A, Shen C, Pilette C. Dendritic cells revisited in human allergic rhinitis and asthma. Allergy Eur J Allergy Clin Immunol. 2016;71:137–48. [DOI] [PubMed] [Google Scholar]
- 20.•. Li N, Lewandowski RP, Sidhu D, Holz C, Jackson-Humbles D, Eiguren-Fernandez A, et al. Combined adjuvant effects of ambient vapor-phase organic components and particulate matter potently promote allergic sensitization and Th2-skewing cytokine and chemokine milieux in mice: the importance of mechanistic multi-pollutant research. Toxicol Lett [Internet]. 2021;356:21–32. Available from: https://www.sciencedirect.com/science/article/pii/S0378427421009000 (COMMENT: Letter showing the TH2-skewing effects of concurrent air pollution exposures; findings are likely relevant for other non-TH2 immune responses.)
- 21.Michaeloudes C, Abubakar-Waziri H, Lakhdar R, Raby K, Dixey P, Adcock IM, et al. Molecular mechanisms of oxidative stress in asthma. Mol Aspects Med. 2021;101026. [DOI] [PubMed] [Google Scholar]
- 22.Sun N, Niu Y, Zhang R, Huang Y, Wang J, Qiu W, et al. Ozone inhalation induces exacerbation of eosinophilic airway inflammation and Th2-skew immune response in a rat model of AR. Biomed Pharmacother. 2021;137:111261. [DOI] [PubMed] [Google Scholar]
- 23.Davidson EJ, Yang IV. Role of epigenetics in the development of childhood asthma. Curr Opin Allergy Clin Immunol. 2018;18:132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tesmer LA, Lundy SK, Sarkar S, Fox DA. Th17 cells in human disease. Immunol Rev. 2008;223:87–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei Q, Liao J, Jiang M, Liu J, Liang X, Nong G. Relationship between Th17-mediated immunity and airway inflammation in childhood neutrophilic asthma. Allergy, Asthma Clin Immunol. 2021;17:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Q Polarization of immune cells in the pathologic response to inhaled particulates. Front Immunol. 2020;11:1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iman M, Rezaei R, Azimzadeh Jamalkandi S, Shariati P, Kheradmand F, Salimian J. Th17/Treg immunoregulation and implications in treatment of sulfur mustard gas-induced lung diseases. Expert Rev Clin Immunol. 2017;13:1173–88. [DOI] [PubMed] [Google Scholar]
- 28.Hu Y, Chen Z, Zeng J, Zheng S, Sun L, Zhu L, et al. Th17/Treg imbalance is associated with reduced indoleamine 2,3 dioxygenase activity in childhood allergic asthma. Allergy, Asthma Clin Immunol. 2020;16:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou J, Xu J, Geng F, Peng L, Ye X, Yang D, et al. Childhood co-exposure of cold stress and PM2.5 aggravates the susceptibility and severity of asthma in adulthood of mice. Environ Toxicol. 2021;36:177–84. [DOI] [PubMed] [Google Scholar]
- 30.Brandt EB, Kovacic MB, Lee GB, Gibson AM, Acciani TH, Le Cras TD, et al. Diesel exhaust particle induction of IL-17A contributes to severe asthma. J Allergy Clin Immunol. 2013;132:1194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao Y, Chen CL, Yu D, Liu Z. Roles of follicular helper and regulatory T cells in allergic diseases and allergen immunotherapy. Allergy Eur J Allergy Clin Immunol. 2021;76:456–70. [DOI] [PubMed] [Google Scholar]
- 32.Olatunde AC, Hale JS, Lamb TJ. Cytokine-skewed Tfh cells: functional consequences for B cell help. Trends Immunol. 2021;42:536–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.•. Gowthaman U, Chen JS, Zhang B, Flynn WF, Lu Y, Song W, et al. Identification of a T follicular helper cell subset that drives anaphylactic IgE. Science (80- ). 2019;365:eaaw6433. (COMMENT: Suggests TFH populations may be more heavily involved in asthma pathogenesis and exacerbation than previously appreciated.)
- 35.Wade-Vallance AK, Allen CD. Intrinsic and extrinsic regulation of IgE B cell responses. Curr Opin Immunol. 2021;72:221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui YJ, Chen GH, Wang JL, Ma L, Guo XL, Liao JX, et al. Alterations of CD4+CXCR5+Tfh cells and its transcription regulatory factors in children with asthma. Chinese J Contemp Pediatr. 2014;16:1215–9. [PubMed] [Google Scholar]
- 37.Gong F, Zhu HY, Zhu J, Dong QJ, Huang X, Jiang DJ. Circulating CXCR5 + CD4 + T cells participate in the IgE accumulation in allergic asthma. Immunol Lett. 2018;197:9–14. [DOI] [PubMed] [Google Scholar]
- 38.Yao Y, Chen CL, Wang N, Wang ZC, Ma J, Zhu RF, et al. Correlation of allergen-specific T follicular helper cell counts with specific IgE levels and efficacy of allergen immunotherapy. J Allergy Clin Immunol. 2018;142:321–4. [DOI] [PubMed] [Google Scholar]
- 39.Yao Y, Wang ZC, Yu D, Liu Z. Role of allergen-specific T-follicular helper cells in immunotherapy. Curr Opin Allergy Clin Immunol. 2018;18:495–501. [DOI] [PubMed] [Google Scholar]
- 40.Clement RL, Daccache J, Mohammed MT, Diallo A, Blazar BR, Kuchroo VK, et al. Follicular regulatory T cells control humoral and allergic immunity by restraining early B cell responses. Nat Immunol. 2019;20:1360–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong F, Qian C, Zhu H, Zhu J, Pan Y, Dong Q, et al. Circulating follicular T-helper cell subset distribution in patients with asthma. Allergy Asthma Proc. 2016;37:154–61. [DOI] [PubMed] [Google Scholar]
- 42.Ma QY, Huang DY, Zhang HJ, Wang S, Chen XF. Exposure to particulate matter 2.5 (PM2.5) induced macrophage-dependent inflammation, characterized by increased Th1/Th17 cytokine secretion and cytotoxicity. Int Immunopharmacol. 2017;50:139–45. [DOI] [PubMed] [Google Scholar]
- 43.Schmitt EG, Williams CB. Generation and function of induced regulatory T cells. Front Immunol. 2013;4:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aron JL, Akbari O. Regulatory T cells and type 2 innate lymphoid cell-dependent asthma. Allergy Eur J Allergy Clin Immunol. 2017;72:1148–55. [DOI] [PubMed] [Google Scholar]
- 45.Roncarlo MG, Gregori S. Is FOXP3 a bona fide marker for human regulatory T cells? Eur J Immunol. 2008;38:925–7. [DOI] [PubMed] [Google Scholar]
- 46.Shevyrev D, Tereshchenko V. Treg heterogeneity, function, and homeostasis. Front Immunol. 2020;10:3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levings MK, Sangregorio R, Roncarolo MG. Human CD25+CD4+ T regulatory cells suppress naive and memory T cell proliferation and can be expanded in vitro without loss of function. J Exp Med. 2001;193:1295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chinen T, Kannan AK, Levine AG, Fan X, Klein U, Zheng Y, et al. An essential role for IL-2 receptor in regulatory T cell function. Nat Immunol. 2016;17:1322–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okeke EB, Uzonna JE. The pivotal role of regulatory T cells in the regulation of innate immune cells. Front Immunol. 2019;10:680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao S, Wang C. Regulatory T cells and asthma. J Zhejiang Univ Sci B. 2018;19:663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lan F, Zhang N, Bachert C, Zhang L. Stability of regulatory T cells in T helper 2–biased allergic airway diseases. Allergy Eur J Allergy Clin Immunol. 2020;75:1918–26. [DOI] [PubMed] [Google Scholar]
- 52.Nadeau K, McDonald-Hyman C, Noth EM, Pratt B, Hammond SK, Balmes J, et al. Ambient air pollution impairs regulatory T-cell function in asthma. J Allergy Clin Immunol. 2010;126:845–52. [DOI] [PubMed] [Google Scholar]
- 53.Rouadi PW, Idriss SA, Naclerio RM, Peden DB, Ansotegui IJ, Canonica GW, et al. Immunopathological features of air pollution and its impact on inflammatory airway diseases (IAD). World allergy organ J. 2020;13:100467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.García-Serna AM, Hernández-Caselles T, Jiménez-Guerrero P, Martín-Orozco E, Pérez-Fernández V, Cantero-Cano E, et al. Air pollution from traffic during pregnancy impairs newborn’s cord blood immune cells: the NELA cohort. Environ Res. 2021;198:110468. [DOI] [PubMed] [Google Scholar]
- 55.Baiïz N, Slama R, Béné MC, Charles MA, Kolopp-Sarda MN, Magnan A, et al. Maternal exposure to air pollution before and during pregnancy related to changes in newborn’s cord blood lymphocyte subpopulations. The EDEN study cohort. BMC Pregnancy Childbirth. 2011;11:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pang DJ, Neves JF, Sumaria N, Pennington DJ. Understanding the complexity of γδ T-cell subsets in mouse and human. Immunology. 2012;136:283–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zarobkiewicz MK, Wawryk-Gawda E, Kowalska W, Janiszewska M, Bojarska-Junak A. γδ T lymphocytes in asthma: a complicated picture. Arch Immunol Ther Exp (Warsz). 2021;69:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Belkadi A, Dietrich C, Machavoine F, Victor JR, Leite-de-Moraes M. γδ T cells amplify Blomia tropicalis-induced allergic airway disease. Allergy Eur J Allergy Clin Immunol. 2019;74:395–8. [DOI] [PubMed] [Google Scholar]
- 59.Tamura-Yamashita K, Endo J, Isogai S, Matsuoka K, Yonekawa H, Yoshizawa Y. γδ T cell is essential for allergen-induced late asthmatic response in a murine model of asthma. J Med Dent Sci. 2008;55:113–20. [PubMed] [Google Scholar]
- 60.Huang Y, Aydintug MK, Loomis J, MacLeod MK, McKee AS, Kirchenbaum G, et al. Antigen-specific regulation of IgE antibodies by non-antigen–specific γδ T cells. J Immunol. 2013;190:913–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kinyanjui MW, Shan J, Nakada EM, Qureshi ST, Fixman ED. Dose-dependent effects of IL-17 on IL-13–induced airway inflammatory responses and airway hyperresponsiveness. J Immunol. 2013;190:3859–68. [DOI] [PubMed] [Google Scholar]
- 62.Mathews JA, Krishnamoorthy N, Kasahara DI, Cho Y, Wurmbrand AP, Ribeiro L, et al. IL-33 drives augmented responses to ozone in obese mice. Environ Health Perspect. 2017;125:246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cong LH, Li T, Wang H, Wu YN, Wang SP, Zhao YY, et al. IL-17A-producing T cells exacerbate fine particulate matter-induced lung inflammation and fibrosis by inhibiting PI3K/Akt/mTOR-mediated autophagy. J Cell Mol Med. 2020;24:8532–44. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Che L, Jin Y, Zhang C, Lai T, Zhou H, Xia L, et al. Ozone-induced IL-17A and neutrophilic airway inflammation is orchestrated by the caspase-1-IL-1 cascade. Sci Rep. 2016;6:18680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wirtz S, Schulz-Kuhnt A, Neurath MF, Atreya I. Functional contribution and targeted migration of group-2 innate lymphoid cells in inflammatory lung diseases: being at the right place at the right time. Front Immunol. 2021;12:688879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mao K, Baptista AP, Tamoutounour S, Zhuang L, Bouladoux N, Martins AJ, et al. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature. 2018;554:255–9. [DOI] [PubMed] [Google Scholar]
- 67.Colonna M. Innate lymphoid cells: diversity, plasticity, and unique functions in immunity. Immunity. 2018;48:1104–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krabbendam L, Bal SM, Spits H, Golebski K. New insights into the function, development, and plasticity of type 2 innate lymphoid cells. Immunol Rev. 2018;286:74–85. [DOI] [PubMed] [Google Scholar]
- 69.Tynecka M, Radzikowska U, Eljaszewicz A. IL-10-producing innate lymphoid cells: did we find a missing piece of the puzzle? Allergy. 2021;76:3849–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kindermann M, Knipfer L, Atreya I, Wirtz S. ILC2s in infectious diseases and organ-specific fibrosis. Semin Immunopathol. 2018;40:379–92. [DOI] [PubMed] [Google Scholar]
- 71.Schulz-Kuhnt A, Wirtz S, Neurath MF, Atreya I. Regulation of human innate lymphoid cells in the context of mucosal inflammation. Front Immunol. 2020;11:1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Licona-Limón P, Kim LK, Palm NW, Flavell RA. TH2, allergy and group 2 innate lymphoid cells. Nat Immunol. 2013;14:536–42. [DOI] [PubMed] [Google Scholar]
- 73.Smith SG, Chen R, Kjarsgaard M, Huang C, Oliveria JP, O’Byrne PM, et al. Increased numbers of activated group 2 innate lymphoid cells in the airways of patients with severe asthma and persistent airway eosinophilia. J Allergy Clin Immunol. 2016;137:75–86.e8. [DOI] [PubMed] [Google Scholar]
- 74.Bartemes KR, Kephart GM, Fox SJ, Kita H. Enhanced innate type 2 immune response in peripheral blood from patients with asthma. J Allergy Clin Immunol. 2014;134:671–678.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Christianson CA, Goplen NP, Zafar I, Irvin C, Good JT, Rollins DR, et al. Persistence of asthma requires multiple feedback circuits involving type 2 innate lymphoid cells and IL-33. J Allergy Clin Immunol. 2015;136:59–68.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jia Y, Fang X, Zhu X, Bai C, Zhu L, Jin M, et al. IL-13+ Type 2 innate lymphoid cells correlate with asthma control status and treatment response. Am J Respir Cell Mol Biol. 2016;55:675–83. [DOI] [PubMed] [Google Scholar]
- 77.Nagakumar P, Puttur F, Gregory LG, Denney L, Fleming L, Bush A, et al. Pulmonary type-2 innate lymphoid cells in paediatric severe asthma: phenotype and response to steroids. Eur Respir J. 2019;54:1801809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim RY, Rae B, Neal R, Donovan C, Pinkerton J, Balachandran L, et al. Elucidating novel disease mechanisms in severe asthma. Clin Transl Immunol. 2016;5:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seehus CR, Kadavallore A, La TBD, Yeckes AR, Wang Y, Tang J, et al. Alternative activation generates IL-10 producing type 2 innate lymphoid cells. Nat Commun. 2017;8:1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang S, Xia P, Chen Y, Qu Y, Xiong Z, Ye B, et al. Regulatory innate lymphoid cells control innate intestinal inflammation. Cell. 2017;171:201–216.e18. [DOI] [PubMed] [Google Scholar]
- 81.Golebski K, Layhadi JA, Sahiner U, Steveling-Klein EH, Lenormand MM, Li RCY, et al. Induction of IL-10-producing type 2 innate lymphoid cells by allergen immunotherapy is associated with clinical response. Immunity. 2021;54:291–307.e7. [DOI] [PubMed] [Google Scholar]
- 82.Bal SM, Golebski K, Spits H. Plasticity of innate lymphoid cell subsets. Nat. Rev. Immunol 2020. [DOI] [PubMed] [Google Scholar]
- 83.Yang Q, Ge MQ, Kokalari B, Redai IG, Wang X, Kemeny DM, et al. Group 2 innate lymphoid cells mediate ozone-induced airway inflammation and hyperresponsiveness in mice. J Allergy Clin Immunol. 2016;132:571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Estrella B, Naumova EN, Cepeda M, Voortman T, Katsikis PD, Drexhage HA. Effects of air pollution on lung innate lymphoid cells: review of in vitro and in vivo experimental studies. Int J Environ Res Public Health. 2019;16:2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumagai K, Lewandowski RP, Jackson-Humbles DN, Buglak N, Li N, White K, et al. Innate lymphoid cells mediate pulmonary eosinophilic inflammation, airway mucous cell metaplasia, and type 2 immunity in mice exposed to ozone. Toxicol Pathol. 2017;45:692–704. [DOI] [PubMed] [Google Scholar]
- 86.Kumagai K, Lewandowski R, Jackson-Humbles DN, Li N, Van Dyken SJ, Wagner JG, et al. Ozone-induced nasal type 2 immunity in mice is dependent on innate lymphoid cells. Am J Respir Cell Mol Biol. 2016;54:782–91. [DOI] [PubMed] [Google Scholar]
- 87.Harkema JR, Wagner JG. Innate lymphoid cell–dependent airway epithelial and inflammatory responses to inhaled ozone: a new paradigm in pathogenesis. Toxicol Pathol. 2019;47:993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Grove KC, Provoost S, Hendriks RW, McKenzie ANJ, Seys LJM, Kumar S, et al. Dysregulation of type 2 innate lymphoid cells and TH2 cells impairs pollutant-induced allergic airway responses. J Allergy Clin Immunol. 2017;139:246–257.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim J, Kim YC, Ham J, Sohn KH, Lee SY, Chung DH, et al. The effect of air pollutants on airway innate immune cells in patients with asthma. Allergy Eur J Allergy Clin Immunol. 2020;75:2372–6. [DOI] [PubMed] [Google Scholar]
- 90.Amarante-Mendes GP, Adjemian S, Branco LM, Zanetti LC, Weinlich R, Bortoluci KR. Pattern recognition receptors and the host cell death molecular machinery. Front Immunol. 2018;9:2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bauer RN, Diaz-Sanchez D, Jaspers I. Effects of air pollutants on innate immunity: the role of toll-like receptors and nucleotide-binding oligomerization domain-like receptors. J Allergy Clin Immunol. 2012;129:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Eisenbarth SC, Piggott DA, Huleatt JW, Visintin I, Herrick CA, Bottomly K. Lipopolysaccharide-enhanced, toll-like receptor 4-dependent T helper cell type 2 responses to inhaled antigen. J Exp Med. 2002;196:1645–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mcalees JW, Whitehead GS, Harley ITW, Cappelletti M, Rewerts CL, Holdcroft AM, et al. Distinct Tlr4-expressing cell compartments control neutrophilic and eosinophilic airway inflammation. Mucosal Immunol. 2015;8:863–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lafferty EI, Qureshi ST, Schnare M. The role of toll-like receptors in acute and chronic lung inflammation. J Inflamm. 2010;7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ferreira DS, Annoni R, Silva LFF, Buttignol M, Santos ABG, Medeiros MCR, et al. Toll-like receptors 2, 3 and 4 and thymic stromal lymphopoietin expression in fatal asthma. Clin Exp Allergy. 2012;42:1459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Whitehead GS, Hussain S, Fannin R, Trempus CS, Innes CL, Schurman SH, et al. TLR5 activation exacerbates airway inflammation in asthma. Lung. 2020;198:289–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilson RH, Maruoka S, Whitehead GS, Foley JF, Flake GP, Sever ML, et al. The toll-like receptor 5 ligand flagellin promotes asthma by priming allergic responses to indoor allergens. Nat Med. 2012;18:1705–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cook DN. Role of environmental adjuvants in asthma development. Curr Allergy Asthma Rep. 2020;20:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Johnson AN, Harkema JR, Nelson AJ, Dickinson JD, Kalil J, Duryee MJ, et al. MyD88 regulates a prolonged adaptation response to environmental dust exposure-induced lung disease. Respir Res. 2020;21:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ishii T, Muroi M, Horiguchi K, Tanamoto K ichi, Nagase T, Yamashita N. Activation through toll-like receptor 2 on group 2 innate lymphoid cells can induce asthmatic characteristics. Clin Exp Allergy. 2019;49:1624–32. [DOI] [PubMed] [Google Scholar]
- 101.Thio CLP, Lai ACY, Chi PY, Webster G, Chang YJ. Toll-like receptor 9–dependent interferon production prevents group 2 innate lymphoid cell–driven airway hyperreactivity. J Allergy Clin Immunol. 2019;144:682–97. [DOI] [PubMed] [Google Scholar]
- 102.Zhao CC, Xie QM, Xu J, Yan XB, Fan XY, Wu HM. TLR9 mediates the activation of NLRP3 inflammasome and oxidative stress in murine allergic airway inflammation. Mol Immunol. 2020;125:24–31. [DOI] [PubMed] [Google Scholar]
- 103.Shikhagaie MM, Andersson CK, Mori M, Kortekaas Krohn I, Bergqvist A, Dahl R, et al. Mapping of TLR5 and TLR7 in central and distal human airways and identification of reduced TLR expression in severe asthma. Clin Exp Allergy. 2014;44:184–96. [DOI] [PubMed] [Google Scholar]
- 104.Shoenfelt J, Mitkus RJ, Zeisler R, Spatz RO, Powell J, Fenton MJ, et al. Involvement of TLR2 and TLR4 in inflammatory immune responses induced by fine and coarse ambient air particulate matter. J Leukoc Biol. 2009;86:303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Becker S, Fenton MJ, Soukup JM. Involvement of microbial components and toll-like receptors 2 and 4 in cytokine responses to air pollution particles. Am J Respir Cell Mol Biol. 2002;27:611–8. [DOI] [PubMed] [Google Scholar]
- 106.Tighe RM, Wheeler J, Hollingsworth JW. Air pollution and immune function. In: Nadadur S, Hollingsworth J, editors. Air Pollut Heal Eff [Internet]. London: Springer; 2015. p. 289–321. Available from: 10.1007/978-1-4471-6669-6_11 [DOI] [Google Scholar]
- 107.Mumby S, Chung KF, Adcock IM. Transcriptional effects of ozone and impact on airway inflammation. Front Immunol. 2019;10:1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hernandez ML, Lay JC, Harris B, Esther CR, Brickey WJ, Bromberg PA, et al. Atopic asthmatic subjects but not atopic subjects without asthma have enhanced inflammatory response to ozone. J Allergy Clin Immunol. 2010;126:537–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fakhrzadeh L, Laskin JD, Laskin DL. Ozone-induced production of nitric oxide and TNF-α and tissue injury are dependent on NF-κB p50. Am J Physiol - Lung Cell Mol Physiol. 2004;287:L279–85. [DOI] [PubMed] [Google Scholar]
- 110.Kleeberger SR, Reddy S, Zhang LY, Jedlicka AE. Genetic susceptibility to ozone-induced lung hyperpermeability. Role of toll-like receptor 4. Am J Respir Cell Mol Biol. 2000;22:620–7. [DOI] [PubMed] [Google Scholar]
- 111.Connor AJ, Laskin JD, Laskin DL. Ozone-induced lung injury and sterile inflammation. Role of toll-like receptor 4. Exp Mol Pathol. 2012;92:229–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Williams AS, Leung SY, Nath P, Khorasani NM, Bhavsar P, Issa R, et al. Role of TLR2, TLR4, and MyD88 in murine ozone-induced airway hyperresponsiveness and neutrophilia. J Appl Physiol. 2007;103:1189–95. [DOI] [PubMed] [Google Scholar]
- 113.He M, Ichinose T, Ren Y, Song Y, Yoshida Y, Arashidani K, et al. PM2.5-rich dust collected from the air in Fukuoka, Kyushu, Japan, can exacerbate murine lung eosinophilia. Inhal Toxicol. 2015;27:287–99. [DOI] [PubMed] [Google Scholar]
- 114.•. Fonceca AM, Zosky GR, Bozanich EM, Sutanto EN, Kicic A, McNamara PS, et al. Accumulation mode particles and LPS exposure induce TLR-4 dependent and independent inflammatory responses in the lung. Respir Res. 2018;19:15. (COMMENT: Highlights the combined impact of bacterial-derived LPS and ambient air pollution on TLR4-driven airway inflammation)
- 115.He M, Ichinose T, Yoshida Y, Arashidani K, Yoshida S, Takano H, et al. Urban PM2.5 exacerbates allergic inflammation in the murine lung via a TLR2/TLR4/MyD88-signaling pathway. Sci Rep. 2017;7:11027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tsang MSM, Hou T, Chan BCL, Wong CK. Immunological roles of NLR in allergic diseases and its underlying mechanisms. Int J Mol Sci. 2021;22:1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wood LG, Li Q, Scott HA, Rutting S, Berthon BS, Gibson PG, et al. Saturated fatty acids, obesity, and the nucleotide oligomerization domain–like receptor protein 3 (NLRP3) inflammasome in asthmatic patients. J Allergy Clin Immunol. 2019;143:305–15. [DOI] [PubMed] [Google Scholar]
- 118.Donovan C, Liu G, Shen S, Marshall JE, Kim RY, Alemao CA, et al. The role of the microbiome and the NLRP3 inflammasome in the gut and lung. J Leukoc Biol. 2020;108:925–35. [DOI] [PubMed] [Google Scholar]
- 119.Kim RY, Pinkerton JW, Gibson PG, Cooper MA, Horvat JC, Hansbro PM. Inflammasomes in COPD and neutrophilic asthma. Thorax. 2015;70:1199–201. [DOI] [PubMed] [Google Scholar]
- 120.Pinkerton JW, Kim RY, Robertson AAB, Hirota JA, Wood LG, Knight DA, et al. Inflammasomes in the lung. Mol Immunol. 2017;86:44–55. [DOI] [PubMed] [Google Scholar]
- 121.Hansbro PM, Kim RY, Starkey MR, Donovan C, Dua K, Mayall JR, et al. Mechanisms and treatments for severe, steroid-resistant allergic airway disease and asthma. Immunol Rev. 2017;278:41–62. [DOI] [PubMed] [Google Scholar]
- 122.Wadhwa R, Dua K, Adcock IM, Horvat JC, Kim RY, Hansbro PM. Cellular mechanisms underlying steroid-resistant asthma. Eur Respir Rev. 2019;28:190096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Simpson JL, Phipps S, Baines KJ, Oreo KM, Gunawardhana L, Gibson PG. Elevated expression of the NLRP3 inflammasome in neutrophilic asthma. Eur Respir J. 2014;43:1067–76. [DOI] [PubMed] [Google Scholar]
- 124.Theofani E, Semitekolou M, Morianos I, Samitas K, Xanthou G. Targeting NLRP3 inflammasome activation in severe asthma. J Clin Med. 2019;8:1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rossios C, Pavlidis S, Hoda U, Kuo CH, Wiegman C, Russell K, et al. Sputum transcriptomics reveal upregulation of IL-1 receptor family members in patients with severe asthma. J Allergy Clin Immunol. 2018;141:560–70. [DOI] [PubMed] [Google Scholar]
- 126.Eder W, Klimecki W, Yu L, Von Mutius E, Riedler J, Braun-Fahrländer C, et al. Association between exposure to farming, allergies and genetic variation in CARD4/NOD1. Allergy Eur J Allergy Clin Immunol. 2006;61:1117–24. [DOI] [PubMed] [Google Scholar]
- 127.Hysi P, Kabesch M, Moffatt MF, Schedel M, Carr D, Zhang Y, et al. NOD1 variation, immunoglobulin E and asthma. Hum Mol Genet. 2005;14:935–41. [DOI] [PubMed] [Google Scholar]
- 128.Girardin SE, Jéhanno M, Mengin-Lecreulx D, Sansonetti PJ, Alzari PM, Philpott DJ. Identification of the critical residues involved in peptidoglycan detection by Nod1. J Biol Chem. 2005;280:38648–56. [DOI] [PubMed] [Google Scholar]
- 129.Duan W, Mehta AK, Magalhaes JG, Ziegler SF, Dong C, Philpott DJ, et al. Innate signals from Nod2 block respiratory tolerance and program TH2-driven allergic inflammation. J Allergy Clin Immunol. 2010;126:1284–93.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Besnard AG, Guillou N, Tschopp J, Erard F, Couillin I, Iwakura Y, et al. NLRP3 inflammasome is required in murine asthma in the absence of aluminum adjuvant. Allergy Eur J Allergy Clin Immunol. 2011;66:1047–57. [DOI] [PubMed] [Google Scholar]
- 131.Bruchard M, Rebé C, Derangère V, Togbé D, Ryffel B, Boidot R, et al. The receptor NLRP3 is a transcriptional regulator of TH2 differentiation. Nat Immunol. 2015;16:859–70. [DOI] [PubMed] [Google Scholar]
- 132.Allen IC, Jania CM, Wilson JE, Tekeppe EM, Hua X, Brickey WJ, et al. Analysis of NLRP3 in the development of allergic airway disease in mice. J Immunol. 2012;188:2884–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xu M, Wang L, Wang M, Wang H, Zhang H, Chen Y, et al. Mitochondrial ROS and NLRP3 inflammasome in acute ozone-induced murine model of airway inflammation and bronchial hyperresponsiveness. Free Radic Res. 2019;53:780–90. [DOI] [PubMed] [Google Scholar]
- 134.Li F, Xu M, Wang M, Wang L, Wang H, Zhang H, et al. Roles of mitochondrial ROS and NLRP3 inflammasome in multiple ozone-induced lung inflammation and emphysema. Respir Res. 2018;19:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hirota JA, Hirota SA, Warner SM, Stefanowicz D, Shaheen F, Beck PL, et al. The airway epithelium nucleotide-binding domain and leucine-rich repeat protein 3 inflammasome is activated by urban particulate matter. J Allergy Clin Immunol. 2012;129:1116–25.e6. [DOI] [PubMed] [Google Scholar]
- 136.Hirota JA, Gold MJ, Hiebert PR, Parkinson LG, Wee T, Smith D, et al. The nucleotide-binding domain, leucine-rich repeat protein 3 inflammasome/IL-1 receptor I axis mediates innate, but not adaptive, immune responses after exposure to particulate matter under 10 μm. Am J Respir Cell Mol Biol. 2015;52:96–105. [DOI] [PubMed] [Google Scholar]
- 137.Chan YL, Wang B, Chen H, Ho KF, Cao J, Hai G, et al. Pulmonary inflammation induced by low-dose particulate matter exposure in mice. Am J Physiol - Lung Cell Mol Physiol. 2019;317:L424–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yu J, Nagasu H, Murakami T, Hoang H, Broderick L, Hoffman HM, et al. Inflammasome activation leads to Caspase-1-dependent mitochondrial damage and block of mitophagy. Proc Natl Acad Sci U S A. 2014;111:15514–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang T, Sun L, Wang T, Liu C, Zhang H, Zhang C, et al. Gestational exposure to PM2.5 leads to cognitive dysfunction in mice offspring via promoting HMGB1-NLRP3 axis mediated hippocampal inflammation. Ecotoxicol Environ Saf. 2021;223:112617. [DOI] [PubMed] [Google Scholar]
- 140.Chen Y, Li G, Liu Y, Werth VP, Williams KJ, Liu ML. Translocation of endogenous danger signal HMGB1 from nucleus to membrane microvesicles in macrophages. J Cell Physiol. 2016;231:2319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tien CP, Chen CH, Lin WY, Liu CS, Liu KJ, Hsiao M, et al. Ambient particulate matter attenuates Sirtuin1 and augments SREBP1-PIR axis to induce human pulmonary fibroblast inflammation: molecular mechanism of microenvironment associated with COPD. Aging (Albany NY). 2019;11:4654–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ko JW, Shin NR, Je-Oh L, Jung TY, Moon C, Kim TW, et al. Silica dioxide nanoparticles aggravate airway inflammation in an asthmatic mouse model via NLRP3 inflammasome activation. Regul Toxicol Pharmacol. 2020;112:104618. [DOI] [PubMed] [Google Scholar]
- 143.Im Kim D, Song MK, Lee K. Diesel exhaust particulates enhances susceptibility of LPS-induced acute lung injury through upregulation of the IL-17 cytokine-derived TGF-β1/collagen i expression and activation of NLRP3 inflammasome signaling in mice. Biomolecules. 2021;11:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hall R, Hall IP, Sayers I. Genetic risk factors for the development of pulmonary disease identified by genome-wide association. Respirology. 2019;24:204–14. [DOI] [PubMed] [Google Scholar]
- 145.Ntontsi P, Photiades A, Zervas E, Xanthou G, Samitas K. Genetics and epigenetics in asthma. Int J Mol Sci. 2021;22:2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.•. Queiroz G de A, da Silva RR, Pires A de O, Costa R dos S, Alcântara-Neves NM, da Silva TM, et al. New variants in NLRP3 inflammasome genes increase risk for asthma and Blomia tropicalis-induced allergy in a Brazilian population. Cytokine X. 2020;2:100032. (COMMENT: Comprehensive assessment of NLRP3 and CASP1 SNPs linked to pediatric asthma.)
- 147.•. Stadhouders R, Li BWS, de Bruijn MJW, Gomez A, Rao TN, Fehling HJ, et al. Epigenome analysis links gene regulatory elements in group 2 innate lymphocytes to asthma susceptibility. J Allergy Clin Immunol. 2018;142:1793–807. (COMMENT: Identified epigenetic changes in ILC2s, not found in TH2 cells, related to asthma risk.)
- 148.Abdollahi E, Tavasolian F, Momtazi-Borojeni AA, Samadi M, Rafatpanah H. Protective role of R381Q (rs11209026) polymorphism in IL-23R gene in immune-mediated diseases: a comprehensive review. J Immunotoxicol. 2016;13:286–300. [DOI] [PubMed] [Google Scholar]
- 149.Morales E, Duffy D. Genetics and gene-environment interactions in childhood and adult onset asthma. Front Pediatr. 2019;7:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gill MA. The role of dendritic cells in asthma. J Allergy Clin Immunol. 2012;129:889–901. [DOI] [PubMed] [Google Scholar]
- 151.Gaurav R, Agrawal DK. Clinical view on the importance of dendritic cells in asthma. Expert Rev Clin Immunol. 2013;9:899–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hussain S, Johnson CG, Sciurba J, Meng X, Stober VP, Liu C, et al. TLR5 participates in the TLR4 receptor complex and promotes MyD88-dependent signaling in environmental lung injury. Elife. 2020;9:e50458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.••. Schurman SH, Bravo MA, Innes CL, Jackson WB, McGrath JA, Miranda ML, et al. Toll-like receptor 4 pathway polymorphisms interact with pollution to influence asthma diagnosis and severity. Sci Rep. 2018;8:12713. (COMMENT: Connects asthma diagnosis and severity to TLR4 SNPs and geographic air pollutant exposure.)
- 154.Leal VNC, Genov IR, Mallozi MC, Solé D, Pontillo A. Polymorphisms in inflammasome genes and risk of asthma in Brazilian children. Mol Immunol. 2018;93:64–7. [DOI] [PubMed] [Google Scholar]
- 155.Gref A, Merid SK, Gruzieva O, Ballereau S, Becker A, Bellander T, et al. Genome-wide interaction analysis of air pollution exposure and childhood asthma with functional follow-up. Am J Respir Crit Care Med. 2017;195:1373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kabesch M, Tost J. Recent findings in the genetics and epigenetics of asthma and allergy. Semin Immunopathol. 2020;42:43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Choi BY, Han M, Kwak JW, Kim TH. Genetics and epigenetics in allergic rhinitis. Genes (Basel). 2021;12:2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liberman N, Wang SY, Greer EL. Transgenerational epigenetic inheritance: from phenomena to molecular mechanisms. Curr Opin Neurobiol. 2019;59:189–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shukla A, Bunkar N, Kumar R, Bhargava A, Tiwari R, Chaudhury K, et al. Air pollution associated epigenetic modifications: transgenerational inheritance and underlying molecular mechanisms. Sci Total Environ. 2019;656:760–77. [DOI] [PubMed] [Google Scholar]
- 160.Fitz-James MH, Cavalli G. Molecular mechanisms of transgenerational epigenetic inheritance. Nat Rev Genet. 2022;Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 161.Alexis NE, Huang YC, Rappold AG, Kehrl H, Devlin R, Peden DB. Patients with asthma demonstrate airway inflammation after exposure to concentrated ambient particulate matter. Am J Respir Crit Care Med. 2014;190:235–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kumar RK, Hitchins MP, Foster PS. Epigenetic changes in childhood asthma. Dis Model Mech. 2009;2:549–53. [DOI] [PubMed] [Google Scholar]
- 163.DeVries A, Vercelli D. Epigenetic mechanisms in asthma. Ann Am Thorac Soc. 2016;13(Suppl 1):S48–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Edris A, Dekker HT, Melen E, Lahousse L. Epigenome-wide association studies in asthma: a systematic review. Clin Exp Allergy. 2019;49:953–68. [DOI] [PubMed] [Google Scholar]
- 165.Li J, Sha J, Sun L, Zhu D, Meng C. Contribution of regulatory T cell methylation modifications to the pathogenesis of allergic airway diseases. J Immunol Res. 2021;2021:5590217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Runyon RS, Cachola LM, Rajeshuni N, Hunter T, Garcia M, Ahn R, et al. Asthma discordance in twins is linked to epigenetic modifications of T cells. PLoS ONE. 2012;7:23226205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li JY, Zhang Y, Lin XP, Ruan Y, Wang Y, Wang CS, et al. Association between DNA hypomethylation at IL13 gene and allergic rhinitis in house dust mite-sensitized subjects. Clin Exp Allergy. 2016;46:298–307. [DOI] [PubMed] [Google Scholar]
- 168.Yang IV, Pedersen BS, Liu AH, O’Connor GT, Pillai D, Kattan M, et al. The nasal methylome and childhood atopic asthma. J Allergy Clin Immunol. 2017;139:1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yang IV, Pedersen BS, Liu A, O’Connor GT, Teach SJ, Kattan M, et al. DNA methylation and childhood asthma in the inner city. J Allergy Clin Immunol. 2015;136:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Arathimos R, Suderman M, Sharp GC, Burrows K, Granell R, Tilling K, et al. Epigenome-wide association study of asthma and wheeze in childhood and adolescence. Clin Epigenetics. 2017;9:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Naumova AK, Al Tuwaijri A, Morin A, Vaillancout VT, Madore AM, Berlivet S, et al. Sex- and age-dependent DNA methylation at the 17q12-q21 locus associated with childhood asthma. Hum Genet. 2013;132:811–22. [DOI] [PubMed] [Google Scholar]
- 172.Rowell E, Wilson CB. Programming perpetual T helper cell plasticity. Immunity. 2009;30:7–9. [DOI] [PubMed] [Google Scholar]
- 173.Harb H, Raedler D, Ballenberger N, Böck A, Kesper DA, Renz H, et al. Childhood allergic asthma is associated with increased IL-13 and FOXP3 histone acetylation. J Allergy Clin Immunol. 2015;136:200–2. [DOI] [PubMed] [Google Scholar]
- 174.Cheng Q, Shang Y, Huang W, Zhang Q, Li X, Zhou Q. p300 mediates the histone acetylation of ORMDL3 to affect airway inflammation and remodeling in asthma. Int Immunopharmacol. 2019;76:105885. [DOI] [PubMed] [Google Scholar]
- 175.Elbehidy RM, Youssef DM, El-Shal AS, Shalaby SM, Sherbiny HS, Sherief LM, et al. MicroRNA-21 as a novel biomarker in diagnosis and response to therapy in asthmatic children. Mol Immunol. 2016;71:107–14. [DOI] [PubMed] [Google Scholar]
- 176.Panganiban RPL, Pinkerton MH, Maru SY, Jefferson SJ, Roff AN, Ishmael FT. Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. Am J Clin Exp Immunol. 2012;1:154–65. [PMC free article] [PubMed] [Google Scholar]
- 177.Korde A, Ahangari F, Haslip M, Zhang X, Liu Q, Cohn L, et al. An endothelial microRNA-1–regulated network controls eosinophil trafficking in asthma and chronic rhinosinusitis. J Allergy Clin Immunol. 2020;145:550–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wasti B, Liu SK, Xiang XD. Role of epigenetics in the pathogenesis, treatment, prediction, and cellular transformation of asthma. Mediators Inflamm. 2021;2021:9412929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Arshad SH, Karmaus W, Zhang H, Holloway JW. Multi-generational cohorts in asthma and allergy. J Allergy Clin Immunol. 2017;132:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Everson TM, Zhang H, Lockett GA, Kaushal A, Forthofer M, Ewart SL, et al. Epigenome-wide association study of asthma and wheeze characterizes loci within HK1. Allergy, Asthma Clin Immunol. 2019;15:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.••. Gruzieva O, Xu CJ, Yousefi P, Relton C, Merid SK, Breton CV, et al. Prenatal particulate air pollution and DNA methylation in newborns: an epigenome-wide meta-analysis. Environ Health Perspect. 2019;127:57012. (COMMENT: Highly relevant study in the field of transgenerational epigenetic effects of pollution on asthma risk.)
- 182.Panni T, Mehta AJ, Schwartz JD, Baccarelli AA, Just AC, Wolf K, et al. Genome-wide analysis of DNA methylation and fine particulate matter air pollution in three study populations: KORA F3, KORA F4, and the normative aging study. Environ Health Perspect. 2016;124:983–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Breton CV, Marutani AN. Air pollution and epigenetics: recent findings. Curr Environ Heal reports. 2014;1:35–45. [Google Scholar]
- 184.Ji H, Khurana Hershey GK. Genetic and epigenetic influence on the response to environmental particulate matter. J Allergy Clin Immunol. 2012;129:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Miller C, Schladweiler M, Dye J, Hazari M, Kodavanti U, Gilmour I. The early epigenetic response to ozone: impacts on DNA methylation and hydroxymethylation [Internet]. Durham, NC; 2016. Available from: https://cfpub.epa.gov/si/si_public_record_report.cfm?Lab=NHEERL&direntryid=334199 [Google Scholar]
- 186.Zhong J, Karlsson O, Wang G, Li J, Guo Y, Lin X, et al. B vitamins attenuate the epigenetic effects of ambient fine particles in a pilot human intervention trial. Proc Natl Acad Sci U S A. 2017;114:3503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.White AJ, Kresovich JK, Keller JP, Xu Z, Kaufman JD, Weinberg CR, et al. Air pollution, particulate matter composition and methylation-based biologic age. Environ Int. 2019;132:105071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rider CF, Carlsten C. Air pollution and DNA methylation: effects of exposure in humans. Clin Epigenetics. 2019;11:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li R, Zhou R, Zhang J. Function of PM2.5 in the pathogenesis of lung cancer and chronic airway inflammatory diseases. Oncol Lett. 2018;15:7506–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yang SI. Particulate matter and childhood allergic diseases. Korean J Pediatr. 2019;62:22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zheng Y, Sanchez-Guerra M, Zhang Z, Joyce BT, Zhong J, Kresovich JK, et al. Traffic-derived particulate matter exposure and histone H3 modification: a repeated measures study. Environ Res. 2017;153:112–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fry RC, Rager JE, Bauer R, Sebastian E, Peden DB, Jaspers I, et al. Air toxics and epigenetic effects: ozone altered microRNAs in the sputum of human subjects. Am J Physiol - Lung Cell Mol Physiol. 2014;306:L1129–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Fuentes N, Roy A, Mishra V, Cabello N, Silveyra P. Sex-specific microRNA expression networks in an acute mouse model of ozone-induced lung inflammation. Biol Sex Differ. 2018;9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Clay CC, Maniar-Hew K, Gerriets JE, Wang TT, Postlethwait EM, Evans MJ, et al. Early life ozone exposure results in dysregulated innate immune function and altered microRNA expression in airway epithelium. PLoS One. 2014;9:e90401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bhargava A, Shukla A, Bunkar N, Shandilya R, Lodhi L, Kumari R, et al. Exposure to ultrafine particulate matter induces NF-KB mediated epigenetic modifications. Environ Pollut. 2019;252:39–50. [DOI] [PubMed] [Google Scholar]
- 196.Somineni HK, Zhang X, Biagini Myers JM, Kovacic MB, Ulm A, Jurcak N, et al. Ten-eleven translocation 1 (TET1) methylation is associated with childhood asthma and traffic-related air pollution. J Allergy Clin Immunol. 2016;137:797–805.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhou W, Tian D, He J, Wang Y, Zhang L, Cui L, et al. Repeated PM2.5 exposure inhibits BEAS-2B cell P53 expression through ROS-Akt-DNMT3B pathway-mediated promoter hypermethylation. Oncotarget. 2016;7:20691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Mikhaylova L, Zhang Y, Kobzik L, Fedulov AV. Link between epigenomic alterations and genome-wide aberrant transcriptional response to allergen in dendritic cells conveying maternal asthma risk. PLoS One. 2013;8:e70387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gregory DJ, Kobzik L, Yang Z, McGuire CC, Fedulov AV. Transgenerational transmission of asthma risk after exposure to environmental particles during pregnancy. Am J Physiol - Lung Cell Mol Physiol. 2017;313:L395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bind M-A, Lepeule J, Zanobetti A, Gasparrini A, Baccarelli AA, Coull BA, et al. Air pollution and gene-specific methylation in the Normative Aging Study. Epigenetics. 2014;9:448–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bind MA, Baccarelli A, Zanobetti A, Tarantini L, Suh H, Vokonas P, et al. Air pollution and markers of coagulation, inflammation, and endothelial function: associations and epigene-environment interactions in an elderly cohort. Epidemiology. 2012;23:332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Verma M, Michalec L, Sripada A, McKay J, Sirohi K, Verma D, et al. The molecular and epigenetic mechanisms of innate lymphoid cell (ILC) memory and its relevance for asthma. J Exp Med. 2021;218:e20201354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Prunicki M, Stell L, Dinakarpandian D, de Planell-Saguer M, Lucas RW, Hammond SK, et al. Exposure to NO2, CO, and PM2.5 is linked to regional DNA methylation differences in asthma. Clin Epigenetics. 2018;10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hew KM, Walker AI, Kohli A, Garcia M, Syed A, Mcdonald-Hyman C, et al. Childhood exposure to ambient polycyclic aromatic hydrocarbons is linked to epigenetic modifications and impaired systemic immunity in T cells. Clin Exp Allergy. 2015;45:238–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sun L, Fu J, Lin SH, Sun JL, Xia L, Lin CH, et al. Particulate matter of 2.5 μm or less in diameter disturbs the balance of TH17/regulatory T cells by targeting glutamate oxaloacetate transaminase 1 and hypoxia-inducible factor 1α in an asthma model. J Allergy Clin Immunol. 2020;145:402–14. [DOI] [PubMed] [Google Scholar]
- 206.••. Prunicki M, Cauwenberghs N, Lee J, Zhou X, Movassagh H, Noth E, et al. Air pollution exposure is linked with methylation of immunoregulatory genes, altered immune cell profiles, and increased blood pressure in children. Sci Rep. 2021;11:4067. (COMMENT: Demonstrated that changes in air pollutants altered DMGs of TREG cells and T helper subsets.)
- 207.Strickland DH, Holt PG. T regulatory cells in childhood asthma. Trends Immunol. 2011;32:420–7. [DOI] [PubMed] [Google Scholar]
- 208.Yang IV, Pedersen BS, Liu A, O'Connor GT, Teach SJ, Kattan M, et al. DNA methylation and childhood asthma in the inner city. J Allergy Clin Immunol. 136:25769910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Liu J, Ballaney M, Al-alem U, Quan C, Jin X, Perera F, et al. Combined inhaled diesel exhaust particles and allergen exposure alter methylation of T helper genes and IgE production in vivo. Toxicol Sci. 2008;102:76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.de FC Lichtenfels AJ, Van Der Plaat DA, de Jong K, van Diemen CC, Postma DS, Nedeljkovic, et al. Long-term air pollution exposure, genome-wide DNA methylation and lung function in the Life-Lines Cohort Study. Env Heal Perspect. 2018;126:027004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Xia M, Viera-Hutchins L, Garcia-Lloret M, Noval Rivas M, Wise P, McGhee SA, et al. Vehicular exhaust particles promote allergic airway inflammation through an aryl hydrocarbon receptor-notch signaling cascade. J Allergy Clin Immunol. 2015;136:441–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Miousse IR, Chalbot MC, Pathak R, Lu X, Nzabarushimana E, Krager K, et al. In vitro toxicity and epigenotoxicity of different types of ambient particulate matter. Toxicol Sci. 2015;148:473–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, et al. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Env Heal Perspect. 2009;117:217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Salam MT, Byun HM, Lurmann F, Breton CV, Wang X, Eckel SP, et al. Genetic and epigenetic variations in inducible nitric oxide synthase promoter, particulate pollution, and exhaled nitric oxide levels in children. J Allergy Clin Immunol. 2012;129:232–9.e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sofer T, Baccarelli A, Cantone L, Coull B, Maity A, Lin X, et al. Exposure to airborne particulate matter is associated with methylation pattern in the asthma pathway. Epigenomics. 2013;5:147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Brunst KJ, Leung YK, Ryan PH, Khurana Hershey GK, Levin L, Ji H, et al. Forkhead box protein 3 (FOXP3) hypermethylation is associated with diesel exhaust exposure and risk for childhood asthma. J Allergy Clin Immunol. 2013;131:592–4.e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Jiang R, Jones MJ, Sava F, Kobor MS, Carlsten C. Short-term diesel exhaust inhalation in a controlled human crossover study is associated with changes in DNA methylation of circulating mononuclear cells in asthmatics. Part Fibre Toxicol. 2014;11:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.••. Zhang X, Chen X, Weirauch MT, Zhang X, Burleson JD, Brandt EB, et al. Diesel exhaust and house dust mite allergen lead to common changes in the airway methylome and hydroxymethylome. Env Epigenet. 2018;4:dvy020. (COMMENT: Genome-wide methylation study of patient cohorts exposed to air pollution (NO2 and PM2.5).)
- 219.Lovinsky-Desir S, Jung KH, Jezioro JR, Torrone DZ, de Planell-Saguer M, Yan B, et al. Physical activity, black carbon exposure, and DNA methylation in the FOXP3 promoter. Clin Epigenetics. 2017;9:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Jung KH, Lovinsky-Desir S, Yan B, Torrone D, Lawrence J, Jezioro J, et al. Effect of personal exposure to black carbon on changes in allergic asthma gene methylation measured 5 days later in urban children: importance of allergic sensitization. Clin Epigenetics. 2017;9:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Gregory DJ, Zhang Y, Kobzik L, Fedulov AV. Specific transcriptional enhancement of inducible nitric oxide synthase by targeted promoter demethylation. Epigenetics. 2013;8:1205–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Gregory DJ, Mikhaylova L, Fedulov AV. Selective DNA demethylation by fusion of TDG with a sequence-specific DNA-binding domain. Epigenetics. 2012;7:344–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Huang YH, Su J, Lei Y, Brunetti L, Gundry MC, Zhang X, et al. DNA epigenome editing using CRISPR-Cas SunTag-directed DNMT3A. Genome Biol. 2017;18:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Jeffries MA. Epigenetic editing: how cutting-edge targeted epigenetic modification might provide novel avenues for autoimmune disease therapy. Clin Immunol. 2018;196:49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ganesan A, Arimondo PB, Rots MG, Jeronimo C, Berdasco M. The timeline of epigenetic drug discovery: from reality to dreams. Clin Epigenetics. 2019;11:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Cano-Rodriguez D, Rots MG. Epigenetic editing: on the verge of reprogramming gene expression at will. Curr Genet Med Rep. 2016;4:170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Chen H, Kazemier HG, De Groote ML, Ruiters MHJ, Xu GL, Rots MG. Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res. 2014;42:1563–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol. 2013;31:1137–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Xu X, Tao Y, Gao X, Zhang L, Li X, Zou W, et al. A CRISPR-based approach for targeted DNA demethylation. Cell Discov. 2016;2:16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Rivenbark AG, Stolzenburg S, Beltran AS, Yuan X, Rots MG, Strahl BD, et al. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics. 2012;7:350–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Siddique AN, Nunna S, Rajavelu A, Zhang Y, Jurkowska RZ, Reinhardt R, et al. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a-Dnmt3L single-chain fusion protein with increased DNA methylation activity. J Mol Biol. 2013;425:479–91. [DOI] [PubMed] [Google Scholar]
- 232.Lu A, Wang J, Sun W, Huang W, Cai Z, Zhao G, et al. Reprogrammable CRISPR/dCas9-based recruitment of DNMT1 for site-specific DNA demethylation and gene regulation. Cell Discov. 2019;5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Vojta A, Dobrinic P, Tadic V, Bockor L, Korac P, Julg B, et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44:5615–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Xiong T, Meister GE, Workman RE, Kato NC, Spellberg MJ, Turker F, et al. Targeted DNA methylation in human cells using engineered dCas9-methyltransferases. Sci Rep. 2017;7:6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Zhu W, Wu Y, Liu H, Jiang C, Huo L. Gut–lung axis: microbial crosstalk in pediatric respiratory tract infections. Front Immunol. 2021;12:741233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Méndez CS, Bueno SM, Kalergis AM. Contribution of gut microbiota to immune tolerance in infants. J Immunol Res. 2021;2021:7823316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhang M, Sun K, Wu Y, Yang Y, Tso P, Wu Z. Interactions between intestinal microbiota and host immune response in inflammatory bowel disease. Front Immunol. 2017;8:942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Maeda Y, Takeda K. Host–microbiota interactions in rheumatoid arthritis. Exp Mol Med. 2019;51:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Belizário JE, Faintuch J, Garay-Malpartida M. Gut microbiome dysbiosis and immunometabolism: new frontiers for treatment of metabolic diseases. Mediators Inflamm. 2018;2018:2037838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Main BS, Minter MR. Microbial immuno-communication in neurodegenerative diseases. Front Neurosci. 2017;11:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;7:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127:372–381.e1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, Arron JR, et al. The airway microbiome in patients with severe asthma: associations with disease features and severity. J Allergy Clin Immunol. 2015;136:874–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol. 2013;131:346–52. e1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107:129–34. [DOI] [PubMed] [Google Scholar]
- 249.Van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol. 2011;128:948–55.e1-3. [DOI] [PubMed] [Google Scholar]
- 250.Vebø HC, Sekelja M, Nestestog R, Storrø O, Johnsen R, Øien T, et al. Temporal development of the infant gut microbiota in immunoglobulin E-sensitized and nonsensitized children determined by the GA-map infant array. Clin Vaccine Immunol. 2011;18:1326–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Penders J, Thijs C, Van Den Brandt PA, Kummeling I, Snijders B, Stelma F, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA birth cohort study. Gut. 2007;56:661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152. [DOI] [PubMed] [Google Scholar]
- 253.Bisgaard H, Li N, Bonnelykke K, Chawes BLK, Skov T, Paludan-Müller G, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128:646–52.e1-5. [DOI] [PubMed] [Google Scholar]
- 254.Roggenbuck M, Anderson D, Barfod KK, Feelisch M, Geldenhuys S, Sørensen SJ, et al. Vitamin D and allergic airway disease shape the murine lung microbiome in a sex-specific manner. Respir Res. 2016;17:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Remot A, Descamps D, Noordine ML, Boukadiri A, Mathieu E, Robert V, et al. Bacteria isolated from lung modulate asthma susceptibility in mice. ISME J. 2017;11:1061–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Soumana IH, Carlsten C. Air pollution and the respiratory microbiome. J Allergy Clin Immunol. 2021;148:67–9. [DOI] [PubMed] [Google Scholar]
- 257.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Shi CY, Yu CH, Yu WY, Ying HZ. Gut-lung microbiota in chronic pulmonary diseases: evolution, pathogenesis, and therapeutics. Can J Infect Dis Med Microbiol. 2021;2021:9278441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Robinson LB, Chen Arroyo AJ, Dantas MAS, Espinola JA, Sullivan AF, Camargo CA. Prenatal exposure to acid-suppressant medications and the risk of recurrent wheeze at 3 years of age in children with a history of severe bronchiolitis. J Allergy Clin Immunol Pract. 2019;7:2422–2424.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Durack J, Boushey HA, Lynch SV. Airway microbiota and the implications of dysbiosis in asthma. Curr Allergy Asthma Rep. 2016;16:52. [DOI] [PubMed] [Google Scholar]
- 261.Durack J, Huang YJ, Nariya S, Christian LS, Mark Ansel K, Beigelman A, et al. Bacterial biogeography of adult airways in atopic asthma. Microbiome. 2018;6:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.•. Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun. 2018;9:141. (COMMENT: Associates failed maturation of gut microbiota in 1-year-old children with increased pediatric asthma risk at age 5. Notable for its implications for preventing pediatric asthma via protection from gut dysbiosis.)
- 263.•. Taylor SL, Leong LEX, Choo JM, Wesselingh S, Yang IA, Upham JW, et al. Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. J Allergy Clin Immunol. 2018;141:94–103.e15. (COMMENT: With much microbiome work focusing on the gut, this study shows how airway microbiome is associated with adult asthma risk.)
- 264.Logotheti M, Agioutantis P, Katsaounou P, Loutrari H. Microbiome research and multi-omics integration for personalized medicine in asthma. J Pers Med. 2021;11:1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.••. Liu J, Tu C, Yu J, Chen M, Tan C, Zheng X, et al. Maternal microbiome regulation prevents early allergic airway diseases in mouse offspring. Pediatr Allergy Immunol. 2020;31:962–73. (COMMENT: Integrated microbiome interaction with immune mechanisms and epigenetic alterations leading to asthma development.)
- 266.Losol P, Kim S, Ahn S, Lee S, Choi J, Kim Y, et al. Genetic variants in the TLR-related pathway and smoking exposure alter the upper airway microbiota in adult asthmatic patients. Allergy. 2021;76:3217–20. [DOI] [PubMed] [Google Scholar]
- 267.•. Huang C, Wang J, Zheng X, Chen Y, Zhou R, Wei H, et al. Commensal bacteria aggravate allergic asthma via NLRP3/IL-1β signaling in post-weaning mice. J Autoimmun. 2018;93:104–13. (COMMENT: Evidence for a role of commensal bacteria in asthma pathogenesis by promoting NLRP3 expression in immune cells.)
- 268.Di Gangi A, Di Cicco ME, Comberiati P, Peroni DG. Go with your gut: the shaping of T-cell response by gut microbiota in allergic asthma. Front Immunol. 2020;11:1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Liu D, Tan Y, Bajinka O, Wang L, Tang Z. Th17/IL-17 Axis regulated by airway microbes get involved in the development of asthma. Curr Allergy Asthma Rep. 2020;20:11. [DOI] [PubMed] [Google Scholar]
- 270.Daniel S, Pusadkar V, McDonald J, Mirpuri J, Azad RK, Goven A, et al. Traffic generated emissions alter the lung microbiota by promoting the expansion of Proteobacteria in C57Bl/6 mice placed on a high-fat diet. Ecotoxicol Environ Saf. 2021;213:112035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.••. Wang L, Cheng H, Wang D, Zhao B, Zhang J, Cheng L, et al. Airway microbiome is associated with respiratory functions and responses to ambient particulate matter exposure. Ecotoxicol Environ Saf. 2019;13(Suppl 5):S438–46. (COMMENT: One of the first studies connecting PM exposure to airway microbiome perturbation and associated respiratory dysfunction.)
- 272.Hosgood HD, Mongodin EF, Wan Y, Hua X, Rothman N, Hu W, et al. The respiratory tract microbiome and its relationship to lung cancer and environmental exposures found in rural china. Environ Mol Mutagen. 2019;60:617–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Li X, Sun Y, An Y, Wang R, Lin H, Liu M, et al. Air pollution during the winter period and respiratory tract microbial imbalance in a healthy young population in Northeastern China. Environ Pollut. 2019;246:972–9. [DOI] [PubMed] [Google Scholar]
- 274.Li J, Hu Y, Liu L, Wang Q, Zeng J, Chen C. PM2.5 exposure perturbs lung microbiome and its metabolic profile in mice. Sci Total Environ. 2020;721:137432. [DOI] [PubMed] [Google Scholar]
- 275.Li N, He F, Liao B, Zhou Y, Li B, Ran P. Exposure to ambient particulate matter alters the microbial composition and induces immune changes in rat lung. Respir Res. 2017;18:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Yu G, Gail MH, Consonni D, Carugno M, Humphrys M, Pesatori AC, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Fitch MN, Phillippi D, Zhang Y, Lucero JA, Pandey RS, Liu J, et al. Effects of inhaled air pollution on markers of integrity, inflammation, and microbiota profiles of the intestines in Apolipoprotein E knockout mice. Environ Res. 2020;181:108913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Wang W, Zhou J, Chen M, Huang X, Xie X, Li W, et al. Exposure to concentrated ambient PM2.5 alters the composition of gut microbiota in a murine model. Part Fibre Toxicol. 2018;15:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Fouladi F, Bailey MJ, Patterson WB, Sioda M, Blakley IC, Fodor AA, et al. Air pollution exposure is associated with the gut microbiome as revealed by shotgun metagenomic sequencing. Environ Int. 2020;138:105604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Mutlu EA, Comba IY, Cho T, Engen PA, Yazıcı C, Soberanes S, et al. Inhalational exposure to particulate matter air pollution alters the composition of the gut microbiome. Environ Pollut. 2018;240:817–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Howard E, Orhurhu V, Huang L, Guthrie B, Phipatanakul W. The impact of ambient environmental exposures to microbial products on asthma outcomes from birth to childhood. Curr Allergy Asthma Rep. 2019;19:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Roduit C, Frei R, Ferstl R, Loeliger S, Westermann P, Rhyner C, et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy Eur J Allergy Clin Immunol. 2019;74:799–809. [DOI] [PubMed] [Google Scholar]
- 283.Stiemsma LT, Arrieta MC, Dimitriu PA, Cheng J, Thorson L, Lefebvre DL, et al. Shifts in Lachnospira and Clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma. Clin Sci. 2016;130:2199–207. [DOI] [PubMed] [Google Scholar]
- 284.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Di Yu, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–66. [DOI] [PubMed] [Google Scholar]
- 286.Thorburn AN, McKenzie CI, Shen S, Stanley D, MacIa L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015;6:7320. [DOI] [PubMed] [Google Scholar]
- 287.Zaiss MM, Rapin A, Lebon L, Dubey LK, Mosconi I, Sarter K, et al. The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity. 2015;43:998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Yip W, Hughes MR, Li Y, Cait A, Hirst M, Mohn WW, et al. Butyrate shapes immune cell fate and function in allergic asthma. Front Immunol. 2021;12:628453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Komlósi ZI, van de Veen W, Kovács N, Szűcs G, Sokolowska M, O’Mahony L, et al. Cellular and molecular mechanisms of allergic asthma. Mol Aspects Med. 2021;100995. [DOI] [PubMed] [Google Scholar]
- 290.••. Hrusch CL, Stein MM, Gozdz J, Holbreich M, von Mutius E, Vercelli D, et al. T-cell phenotypes are associated with serum IgE levels in Amish and Hutterite children. J Allergy Clin Immunol. 2019;144:1391–1401.e10. (COMMENT: Defines how T cell phenotypes change with different environmental microbiota exposure in Amish/Hutterite communities, affecting asthma outcomes.)
- 291.Stein MM, Hrusch CL, Gozdz J, Igartua C, Pivniouk V, Murray SE, et al. Innate immunity and asthma risk in Amish and Hutterite farm children. N Engl J Med. 2016;48:51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Strachan DP. Family size, infection and atopy: the first decade of the “hygiene hypothesis”. Thorax. 2000;55(Suppl 1):S2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.•. Yang C, Kwon D Il, Kim M, Im SH, Lee YJ. Commensal microbiome expands Tγδ17 cells in the lung and promotes particulate matter-induced acute neutrophilia. Front Immunol. 2021;12:645741. (COMMENT: Shows the microbiome affecting γδ T cell subsets in lung inflammation.)
- 294.Adar SD, Huffnagle GB, Curtis JL. The respiratory microbiome: an underappreciated player in the human response to inhaled pollutants? Ann Epidemiol. 2016;26:355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]