Abstract
Over the past 30 years, functional magnetic resonance imaging (fMRI) has emerged as a powerful tool to non-invasively study the activity and function of the human brain. But along with the potential of fMRI to shed light on neurological, psychiatric, and psychological processes, there are methodological challenges and criticisms. We herein provide an fMRI primer designed for a diverse audience, from the neuroimaging novice to the experienced user. Part 1: Overview: “What is fMRI and what can it tell us?”. Part 2: Basic fMRI principles: MR physics, the BOLD signal, and components of a typical scan session. Part 3: Basic fMRI experimental design: why timing is critical, and common sources of noise in the signal. Part 4: Basic fMRI analysis methods: software, the three stages of data analysis (preprocessing, individual, and group level), and a survey of advanced topics and methods including connectivity, machine learning, and assessing statistical significance. Part 5: Criticism, crises, and opportunities related to power of studies, computing requirements, logistical and interpretational challenges, and methodological debate (assessing causality, circular correlations, and open science best practices). We trust that the novice will gain an understanding of the appropriate uses and limitations of fMRI, and for the experienced user, a concise update on current issues and methodological advances.
Part 1: The Big Picture
Since it first became possible to conduct functional magnetic resonance imaging (fMRI) scans on humans more than 30 years ago [1–3], the popularity of neuroimaging has grown steadily. Most researchers have probably seen colorful maps of the brain in journal articles. The aim of this article is to fill in knowledge gaps for everyone from neuroimaging novices, to experienced users wanting to refresh on fundamentals.
Here, we hope to provide a diverse audience a friendly introduction to fMRI, and a table of common fMRI terminology (Tables 1 & 2). We address what fMRI is, briefly how it works, important methodological considerations, and caveats. We highlight recent advances in the field, as well as criticisms and limitations, to facilitate interpretation of the fMRI literature.
Table 1.
Magnetic Resonance Imaging Terms
| Term/Acronym | Meaning |
|---|---|
| Artifact | An irregularity in the magnetic field caused by head movement, bony cavities, or the presence of metal, which leads to loss of integrity of signal. |
| Atlas | A brain template used to define regions and/or networks, e.g., the MNI Atlas, Talairach Atlas, Yeo Atlas. |
| Block fMRI | Consecutive blocks of task stimuli presented in series (e.g., for 30s), interspersed with rest periods. |
| BOLD | Blood Oxygenation Level Dependent signal; measures magnitude of activation using contrast between oxygenated and deoxygenated blood. |
| Cluster | A contiguous part of a statistical brain map after some method of significance testing (e.g., “we identified a significant cluster in the left insula”). |
| Connectivity | Estimates of the dynamic temporal relationship between brain cells, areas, regions or networks. |
| Event-related | Task-based design, individual task trials interspersed with variable inter-trial interval |
| FWE/FDR | Family wise error/false discovery rate: multiple testing considerations to correct for the ~50,000 voxels in a brain. |
| GLM | General linear model - most common statistical approach for fMRI data, implemented in all major packages (FSL, AFNI, SPM). |
| HRF | Hemodynamic response function - the shape and lag used to model the BOLD response. |
| ITI | Inter-trial interval - the delay between successive task trials. |
| Mask | To extract signal from a discrete region or network for analysis, a mask is created, which is a 3D map of which voxels should be included in analysis. |
| Network | An interacting, topologically organized set of brain regions. |
| RF pulse | Radiofrequency pulse - the perturbational signal emitted by the scanner. |
| ROI | Region of interest; a predefined brain region used for analysis of discrete, usually anatomically-defined brain regions. |
| rsfMRI | Resting state fMRI - participant rests in the scanner with eyes open or closed. |
| SNR | Signal-to-noise ratio, an index of data quality; e.g., “there is low SNR in that region due to an artifact near the orbital sinus”. |
| Slice | A full acquisition in one plane; e.g., “36 sagittal slices”. |
| tesla | Unit of measurement for magnet strength; abbreviated “T” (e.g., a 3T scanner). |
| Time series autocorrelation | Degree of similarity in activation between two regions while accounting for the time lapse between them. |
| TE | Time to echo - the time between administration of the magnetic pulse and the corresponding echo from the perturbed atomic nuclei in the brain. |
| TR | Time to repetition - the time between magnetic pulses (typically ~2s). |
| Volume | A single complete acquisition (whole brain/part of the brain) (e.g., “336 volumes”). |
| Voxel | A 3D pixel (e.g., “the region contained 200 voxels”). |
Table 2.
Data Analysis Principles
| Preprocessing Step | Purpose | Best Practices/Guidelines |
|---|---|---|
| Brain extraction | Remove skull from image | Visually check skull removal for all images (affects registration) |
| Global signal regression | Normalize BOLD values to participant mean | Much debate over whether this helps or hurts measurement |
| High pass filtering | Removes low-frequency “BOLD drift“ artifacts | Typical value is 100–120 seconds |
| Linear detrending | Corrects for how BOLD signal “drifts” upwards over time | Use linear trend correction |
| Motion correction | Estimate and regress out effects of inadvertent head motion | Roll, pitch and yaw displacements; can add their temporal derivatives also |
| Physiological denoising | Record cardiac, respiratory, and other physiological signals during the scan, and then filter out the corresponding noise from fMRI data | Essential for rsFMRI, helpful for task fMRI |
| Prewhitening | Remove time series autocorrelation to make estimation more valid and efficient | Useful in most cases, but not when trials <50 or TR >30s |
| Reconstruction | Convert “raw” data into a 4D (i.e., including changes over time) brain image | Varies by scanner/software |
| Registration/normalization | Warp individual participant brain to a generic standard template | Check fits carefully - many issues originate here! |
| Slice timing correction | Corrects for differences in acquisition of slices | Only applicable for certain acquisitions; check with MR physicist |
| Spatial smoothing | Improve estimation by blurring nearby signals together | 5–8mm is standard; 10–12 may be justified; 15+ is generally not advised |
What is fMRI and What is it Good For?
There are multiple options for non-invasive neuroimaging. While this paper focuses on magnetic resonance imaging (MRI), other methods include electroencephalography (EEG), magnetoencephalography (MEG), positron emission tomography (PET), and functional near-infrared spectroscopy (fNIRS). EEG, MEG, and fNIRS are more affordable and portable options for neuroimaging, but they are currently limited in their ability to accurately image subcortical (deep) brain structures [4]. Structural methods include diffusion tensor imaging (DTI) tracks diffusion of water molecules to reveal white matter tracts in the brain, which can be used in the diagnosis of various neurological disorders [5]. Functional MRI (fMRI) is one of the more expensive options but offers the most comprehensive coverage of regional brain activity. Functional MRI involves collecting dynamic images of blood flow in the brain, which can be used to characterize activation of brain regions, or connectivity (i.e., temporal correlations) between regions [6]. Scanning approaches to measurement of activation include using arterial spin labeling (ASL), which quantifies blood perfusion in the brain, and measuring the blood oxygen level dependent (BOLD) signal, in which the degree of oxygen utilization is proportional to the level of neuronal activity, or (less commonly) a combination of the two. There are three primary modalities of data collection in fMRI: task-dependent, passive responses to stimuli, and resting state. Task-dependent fMRI involves having participants complete behavioral tasks in the scanner, ranging from simple motor tasks (e.g., finger tapping) to responding to complex social scenarios [7]. This approach measures task-specific changes in brain activation or connectivity [8.]. Stimuli can also be presented without requiring a cognitive or behavioral response. In this case, the natural, event-related responses in the brain (passive responses) are captured. Resting state fMRI involves having participants rest quietly in the scanner, and can be used as a “baseline” measure of brain function in the absence of a task [9].
A Brief History of fMRI
Magnetic resonance imaging (MRI) was developed before functional magnetic resonance imaging (fMRI), first being conducted in humans in 1977. Although both methods utilize the same instrumentation and principles of physics, it is important to distinguish MRI from fMRI. MRI uses a broad category of imaging approaches to visualize the structure and function of the human body, whereas fMRI is a derivative of MRI involving the measurement of brain function specifically. Functional MRI was first performed in humans in the early 1990s [1–3,15]. The discovery that the paramagnetic properties of deoxygenated blood could be used to generate a proxy of brain activity via the BOLD signal was a major breakthrough for neuroimaging [2,15,16]. Early experiments involved mapping brain responses to stimuli e.g., flashing checkerboards, to activate the visual system [17] or finger tapping to induce motor-related activation [18]. Recent examples of studies have used computational models to characterize how brain regions become activated during genital stimulation and orgasm in men and women [22–25]. The number of fMRI papers published each year has grown steadily since its inception, with as many as 2,000 fMRI-related articles being published each month [26], as evidence that, despite its inherent limitations, fMRI has gained wide acceptance and can provide valuable insights into brain function,
fMRI Studies in Sexual Medicine
A google scholar search of “sexual response and fMRI” yields over 50,000 results. The following is a brief summary of the main themes of this literature. Functional MRI has been applied to a variety of phenomena related to sexuality, using multiple strategies. By recording spontaneous non-task-related (i.e.“resting state”) brain activity, fMRI has been used to analyze correlation with premenstrual syndrome, phases of the menstrual cycle (luteal vs follicular) and menopausal transition; also, erectile dysfunction, premature ejaculation, diverse sexualities (sexual orientation, sexual preferences, gender identity), prostatitis and pelvic pain syndrome. Alternatively, event-related fMRI of brain activity has been used in the following ways: to analyze brain regions that respond to genital stimulation (by self or by partner) in men and women during arousal and orgasm; to investigate the effect of erotogenic visual stimulation on brain activity related to gender, gender dysphoria, androgen insensitivity syndrome, sexual orientation (gay, straight, bisexual), gender identity (cis/trans), and paraphilias; effects of hormones (e.g., contraceptive, kisspeptin), side effects on sexual function of medication (e.g., amisulpride, reboxetine) and recreational substances (e.g., methamphetamine); to record physiological responses (e.g., erection, lubrication) and/or affective responses (e.g., sexual desire, hypoactive sexual desire disorder, subjective arousal, post-orgasmic resolution phase) to erotogenic stimulation; and to assess brain activity correlates of sexual disorders (e.g., pedophilia, compulsive sexual behavior, premature ejaculation, and vulvodynia). Refer to Table 4 for representative examples of the application of fMRI in each domain.
Table 4.
Central nervous system correlates of human sexuality and representative recent citations.
| Healthy sexual function | Diverse sexualities | Physiological aspects of sexuality | Sexual pathology and dysfunction |
|---|---|---|---|
| Sexual desire [161] | Sex differences [160, 155, 52] | Menstrual cycle [136, 141] | Pelvic pain syndromes [154, 167, 137] |
| Arousal [185, 144, 169] | Gender Identity (cis/trans, gender dysphoria) [149, 179, 177] | Pharmacology (e.g., medications, recreational drugs, side effects) [158, 180] | Pedophilia [166, 143] |
| Orgasm [182, 159] | Sexual orientation (heterosexual, homosexual, Bisexual) [149, 160, 179] | Urologic [181, 174] | Maladaptive behavior (e.g. risky sexual behavior, compulsive sexual behavior, excessive masturbation, pornography addiction) [150, 156, 170] |
| Sexual satisfaction [186, 183] | Sexual preferences (BDSM, kink) | Mapping genital sensory projections to the brain [23, 25, 131, 140, 144, 146] | Disorders of the nervous system (e.g. spinal cord injury) [24, 157, 182] |
| Hormonal factors [163, 177, 172, 147] | Erectile dysfunction Premature ejaculation [171, 184, 168] |
Part 2: How it Works
Basic MR Physics
Magnetic resonance imaging is widely used in clinical and research settings to generate three-dimensional images of the anatomy and function of the human body. The strength of the magnetic field of the scanner is measured in units of tesla (T). Most modern scanners are either 1.5T or 3T, with higher tesla machines being mostly reserved for research use (e.g., 7T, 10.5T). The basic premise of MRI is that different bodily tissues vary in their fat, water, and/or oxygen content, and these tissue types can be distinguished from each other by generating image contrast, e.g., contrast between grey and white matter in the brain [15]. The details of how this is done are beyond the scope of this article; however, in brief, the method capitalizes on the inherent property of atomic nuclei to receive and transmit radio frequency (RF) energy and align themselves along a magnetic field [27]. Initially aligned through a strong magnetic field, an RF pulse delivered by the MRI machine perturbs the nuclei so that they momentarily change their orientation (alignment/synchronization). Sensors then measure the time it takes for the nuclei to return to equilibrium, i.e., relaxation; the original orientation (in structural imaging) or desynchronization (fMRI). This return to equilibrium releases energy that can be captured in various ways by manipulating two key parameters: the time to repetition (TR: time between RF pulses) and the time to echo (TE: the time between the RF pulse and the corresponding emission of RF energy in the responding nuclei). Varying these parameters leads to different types of image contrast. The primary types of image contrast are referred to as T1- and T2-weighted images, referring to different relaxation properties induced by varying the TR and TE parameters [15,28–30]. In T1-weighted images, a short TE and TR is used, producing an image in which the cerebrospinal fluid is dark. In T2-weighted images, longer TE and TR values produce an image in which the cerebrospinal fluid is bright white. The type of images used for fMRI are T2-weighted [31], which involve the long TE values and capture perturbations in the magnetic field that are produced from deoxygenated blood [2].
What is the BOLD Signal?
The BOLD signal is measurable because of the paramagnetic properties of deoxygenated blood [2,16,32]. An increase in neuronal activity generates a demand of those neurons for increased oxygen. This oxygen utilization creates a local deoxygenation of the blood [32]. The consequent local change in magnetic property of iron in the blood produces a measurable local perturbation in the magnetic field [16,32]. This process is referred to as the hemodynamic response, which peaks 4–6 seconds from the time of increased activity of the neurons.
Blood flow and neuronal activation relate to one another in a complex process called neurovascular coupling, [16,32]. The exact biological nature of the mechanisms that underlie neurovascular coupling have not been elucidated despite being the basis of the BOLD signal, but may be related to the metabolic demands of neural activity [136]. Proposed cellular components of the mechanism include astrocytes [34,35], pericytes, and interneurons, as well as vascular endothelium [32,36], but the extent to which each component contributes to the BOLD signal remains unknown. Without fully understanding the cellular and molecular mechanisms, we remain limited in our ability to understand brain-behavior links at the cellular level in humans. The evidence that we do have comes from fMRI scans collected concurrently with invasive electrophysiological single-neuron recording in monkeys and rodents [35,37,38].
A common misconception about fMRI is that relative increases and decreases in BOLD response correspond to excitatory or inhibitory neuronal dynamics. Activation identified using fMRI does not distinguish between activation of a group of excitatory neurons versus activation of a group of inhibitory neurons. What appears to “increase” or “decrease” is relative to what is being used as the baseline condition (e.g. rest, active control, etc.). For example, if a participant’s mind wanders during a rest period, areas that increase activation during daydreaming may appear to show a reduction in activation after the switch to task performance. While it is tempting to interpret such “deactivations” as inhibition, it can also be explained by blood flow demands to that region reflecting passive disengagement. Activation in one brain region can represent an inhibitory process; decreases in activation observed in a region could occur in a group of inhibitory neurons, so that the disinhibition results in activation of the neurons to which they project.
At a higher level, there are consistent relationships among human behavior, patterns of activation in groups of neurons, and the BOLD signal, thus making meaningful links between movement, perception or cognition and the BOLD signal [12]. There are some considerations about these correlations. First, individual neurons within an active group release different excitatory and inhibitory neurotransmitters, thereby producing a net effect. The net effect may actively increase or decrease a function to the same degree. Thus, the overall direction of BOLD activity should be interpreted with caution. Second, vasculature in the brain is affected by factors such as aging and disease [39,40]. The variability among participants introduced into fMRI data from non-neural sources (e.g., vasculature integrity, overall health, diet) remains perhaps the biggest challenge in functional brain imaging - but some statistical solutions have been proposed, as well as using multiple types of imaging methods on each participant, such as by capitalizing on properties of multi-echo scans, in which a combination of TE parameters is used as a means of reducing fMRI signal “noise” [41,42].
Part 3: fMRI Experimental Design: Basics
The measurement underlying fMRI is made possible by the observation that deoxygenated blood can be discerned from oxygenated blood in localized regions of the brain, producing an image contrast [2,16,32]. Thus, the BOLD signal is a proxy for neuronal dynamics. Since this measurement is indirect (as opposed to direct invasive single-neuron recording), the signal is susceptible to contamination from non-physiological factors, or physiological factors not of interest to the question at hand (see Common Confounds section). It is also a slow signal (relative to the millisecond precision of neuronal action potential activity, EEG or MEG), with the stimulus-induced signal peaking 4–6s after stimulus onset and not returning back to baseline for as long as 12–20 seconds [40; Figure 1]. Consequently, experiments must be designed carefully to accommodate the temporal dynamics of the signal.
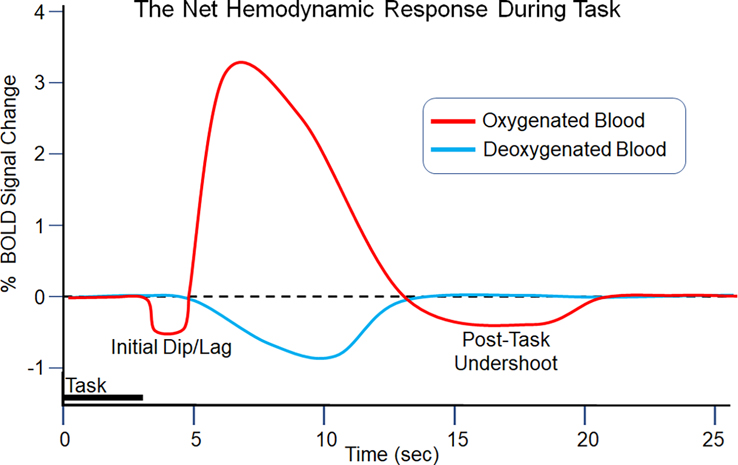
Figure 1.
The Hemodynamic Response Function (HRF). This figure depicts a model of the average blood oxygenation level (red) and deoxygenation level (blue) in a hypothetical region of the brain. Although the features of the signal are generally agreed upon – the initial dip, followed by a delayed peak, and then a dip back below baseline – the specific parameters are known to vary among individuals (e.g., older vs younger adults) and between brain regions (e.g., those with different levels of vascularity). This type of HRF form should be considered a model that approximates the form of the “true” HRF.
The focal location of changes in the BOLD signal is measured in cubic elements called voxels (3D pixels) that are time-bound [Table 1]. Each voxel holds brain space of a specified volume, such as three cubic millimeters. An active region of the brain is often composed of hundreds of voxels [33].
Why Timing is Critical
Experimental design is also of particular importance in fMRI given the intensive labor and high financial costs involved. Timing is critical because an improperly timed protocol can result in uninterpretable results. Each of the three main types of fMRI (resting state fMRI (rsfmRI), task-based fMRI, and passive response) can be a part of the same research design.
Resting state fMRI refers to having the participant lie quietly in the scanner, eyes open or closed, while spontaneous BOLD fluctuations are measured [44]. Often participants are asked to fixate their gaze on a crosshair pictured on a screen in order to standardize the stimulus input across participants. The advantage of resting state fMRI is that it requires little from the participant except to lie still, and is therefore well suited for clinical populations, children, and others who may be unable to provide behavioral responses in the scanner.
In task-based fMRI, participants perform a behavioral task during the scan. The earliest and simplest fMRI tasks involved having the participant tap their finger to activate the motor cortex, or view flashing checkerboards to stimulate the visual system. Current task-based fMRI has become more sophisticated, involving tasks that might test attention abilities, responses to movie or song clips, or sensory self-stimulation of different bodily and genital regions to map the brain pathways that underlie these responses [23,25].
For task-based fMRI, the main research design options are event-related and block design [Figure 3]. Event-related (ER) design refers to presenting stimuli with a variable inter-trial interval of sufficiently long duration to allow BOLD signal for multiple conditions to be separately extracted and analyzed. This avoids BOLD signals arising from one condition overlapping onto other conditions [8; Figure 3]. Event-related designs are appropriate for tasks that involve inherent unpredictability, such as both correct and incorrect participant responses. In such cases, event-related designs inevitably decrease the signal-to-noise ratio, and therefore require more total trials and a longer scan; however, they enable analysis of individual trial responses that cannot be predicted in advance.
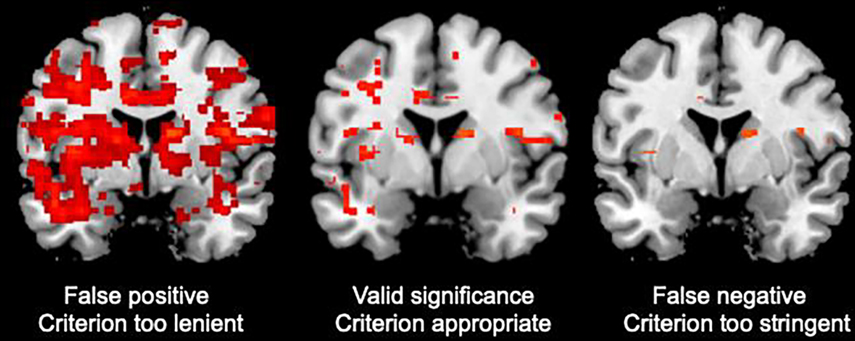
Figure 3.
Hypothetical examples of overly permissive thresholding, appropriate thresholding, and overly stringent thresholding.
Block designs use repeated trials of the same type toward achieving a “steady state” of BOLD activation (e.g., viewing a series of 10 images for 3 seconds each; see Figure 3). Block designs aggregate signal across multiple trials, which leads to higher signal-to-noise ratios [52]. While block designs have higher predictability, they are not appropriate for all research questions.
Hybrid approaches have also been applied, such as mixed block and event trial structures, or having some trials with standardized, naturalistic stimuli such as photographs or movies [46,53]. The presentation of naturalistic stimuli allows investigators to record the response of the brain to stimuli in the absence of behavioral or cognitive tasks or instructions (i.e., passive response). It is also important to incorporate adequate rest periods between trials both to allow the BOLD signal to return to baseline before transitioning from one task condition to the next and to give participants a break so that cognitive fatigue is reduced. Some researchers consider the brain network reconfigurations between task and rest to be important and should not be ignored [54,55].
Common Sources of Noise in the fMRI Signal
The BOLD signal is an indirect measure of brain activity with no established “truth” regarding what the signal should look like in a given area for a given person [54]. It is therefore particularly important to identify and control for known sources of noise. Noise refers to aspects of the signal that are not true brain activity, but instead factors that cause distortions in estimation of the physiological BOLD signal. Signal-to-noise ratio (SNR) is a way to quantify data quality and can be calculated in a variety of ways [52]. The most common artifacts that reduce SNR are produced by motion [56], loss of signal near bony areas such as the sinuses and ears [57], and cardiac and respiratory activity [42] - but even factors such as MRI scanner design characteristics can impact fMRI measurement stability [58].
Head motion during scanning is a pernicious issue that has inspired a number of creative solutions. Motion of even just half a millimeter is enough to inflate statistical estimates [56,59]. Imagine how in a photograph taken of a moving person, it might be hard to identify where the eyes and mouth are if there is blurring of the face. On a brain image, motion can similarly lead to “smearing” of signal that can look indistinguishable from true activation. Small amounts of truly random motion from participants shifting in the scanner can usually be dealt with; however, task-linked motion (such as nodding every time a participant pushes a joystick forward), or frequent and excessive motion, will likely confound results [13] and such subjects’ data may need to be removed from analysis. Many techniques, statistical and otherwise, have been developed to mitigate motion effects, including: regressing out 6 or more estimates of brain displacement [42,56], visualizing spikes in BOLD values that correspond to spikes in motion and removing those values, i.e., “scrubbing” [60], behavioral training programs that teach participants motion boundaries [59], and custom head restraints that physically restrict motion [22; see https://caseforge.com/, UC Berkeley, CA).
Signal loss near the nasal sinuses and ear canal occurs because the scanner’s magnetic field becomes distorted when passing through these empty cavities that are surrounded by bone [57]. This results in signal “dropout” in which an area of the brain that is visible on an anatomical scan appears “missing” in the functional scan. Similar dropout can be seen if participants have metal dental work.
These dropouts can clearly be seen in one of the common functional imaging scans (an echo planar imaging EPI scan) in which a single shot of RF allows a complete image to be captured [61]. These dropout regions unfortunately cannot be recovered; however, signal loss can be mitigated by optimizing scan parameters, such as with a spiral scan, which captures the readout of signal both going into the brain and leaving the brain [57,61,62]. It is also beneficial to use a more sensitive head coil [63]. A 32-channel head coil, compared to an 8-channel head coil, has been found to be more sensitive and more homogeneous throughout data collection for brain volume, white matter connectivity, and functional connectivity in posterior areas [63]. The degree of dropout can also be estimated using standard quality-control measures. When extracting activation from brain regions prone to signal dropout, it is important to exclude zero-value voxels when estimating average time series of a region. Best practices dictate defining a priori how signal dropout will be addressed, such as by customizing analysis parameters to accommodate the unique dropout patterns for each participant [21]. Researchers interested in the function of regions prone to dropout, such as the medial prefrontal cortex and temporal cortices, should pay particular attention to this issue.
Additionally, cardiac, vascular, and respiratory signals contribute to fluctuations in BOLD signal [42]. Respiratory rhythms can lock with task onsets in a stereotyped way that leads to greater blood oxygenation during inhalation [64]. Vascular structural changes with age also impact the BOLD signal [39,65]. Confounds are a particular challenge for resting state scans [66] in which there is no behavioral measure to use to anchor analysis of BOLD hemodynamics. This has led to a variety of sophisticated methods such as independent component analysis [66], and tools designed for the rsfMRI user [Table 3]. Some researchers propose collecting longer durations of resting state data [67] and using methods to increase the signal-to-noise ratio [68,69]. Another hybrid approach is passive viewing of neutral photographs or movies to promote more consistency among participants [46,53] based on phase-locking of eye gaze and associated respiration and heart rate. Enhanced imaging sequences such as multi-band and multi-echo can also improve data quality, as explained above [48,49,51].
Table 3.
List of tools for rsfMRI and task fMRI data analysis.
Part 4: A Brief Review of Analysis Methods
There are a variety of excellent resources that address how to perform fMRI analysis [see Supplemental Material]. The present paper will focus on broader aspects of the purpose of each stage of analysis, and the main considerations for evaluating parameters and analysis approaches. The data analysis plan should be chosen before data are collected, and it is imperative to choose the appropriate type of analysis, and the appropriate sequence of analysis steps (i.e., pipeline), to avoid mismodeling [70]. Mismodeling occurs when researchers choose the inappropriate type of analysis or parameters for their data, build a predictive model based on such analysis, and then make claims based on the model. Any conclusions drawn from a mismodel should be viewed with skepticism. All details of data collection and analysis should be reported in a manuscript, enabling other investigators to verify the method, and replicate the analysis [71].
Which Software(s)?
A major challenge for neuroimagers is the vast array of analysis packages, each with its own set of researcher degrees of freedom in terms of which parameters to select. It is vexing that using different software can lead to minor, and even major differences in findings [72]. These challenges underscore the importance of transparency of reporting [73,74], data and code sharing [72,75,76], and prespecified analysis plans [75].
The three most popular data analysis suites are FMRIB’s Software Library or FSL [77–79], Statistical Parametric Mapping [SPM; 80], and Analysis of Functional NeuroImaging [AFNI; 81]. Many of these programs are also partially or wholly integrated into statistical computing environments such as R [82] which uses the fslr package [83], AFNI’s 3dLME tool [84], or MATLAB [85] in the case of SPM [72,80].
Typically, researchers become expert in the package used by whoever trained them. However, increasingly, workflow managers such as NiPype [86] and fMRIPrep [87] string together the most optimal functions across multiple packages. Depending on the researcher’s specific question (88), application of a workflow manager may be the optimal analytic method. A workflow manager is any system of automated processes that allows more time and resources to be spared during data collection and analysis. Traditionally, much of the research process was done manually, which is very time consuming. We recommend that novice neuroimagers start by finding the workflow used in previous, recently published studies most similar to theirs, and consulting with a collaborator who has relevant methodological expertise for their opinion (often scanning centers have such a person on staff). Choosing the wrong analysis parameters can lead to false positives [76], reduced sensitivity to actual effects [89], or replicability issues. Careful attention must be paid to the choice of software and settings [72].
Preprocessing
Preprocessing refers to the steps taken to prepare the “raw” unprocessed BOLD data for single-subject and eventually group analysis. Table 2 outlines the typical steps of preprocessing and their purpose. A full explanation of the “why” and “how” of preprocessing is beyond the scope of this paper [but see 88]. In sum, there are a series of techniques designed to improve statistical estimation given the known noisiness of the BOLD signal. Changing these initial parameters can potentially have major consequences in subsequent analyses, particularly for resting state connectivity [42,90,91]. While leaving them to software defaults is likely acceptable in many cases, they are important and often overlooked considerations. For example, incomplete brain “extraction” (i.e., leaving parts of the skull in the images) can lead to poor registration between the subject’s brain and a generic, standard template brain, which might then lead to erroneous estimation of the location of the BOLD signal in the brain.
It is therefore critical to visually inspect data after each phase of preprocessing, and the most current pipelines include data reports with many visualizations intended for this purpose [86,87,92]. When evaluating an fMRI publication, it is helpful to check that the appropriate preprocessing steps have been carried out and that the parameters used are in the typical range (or well justified if they deviate from defaults). In some cases, it should also be demonstrated that results are robust to minor preprocessing changes (particularly for rsfMRI). This can be shown by producing replicable results even with the use of slightly different pipelines.
Anatomical scans are typically used to aid in “registration” or “normalization” of fMRI images. The blurry, low resolution functional images are matched to the sharper anatomical images via a warping procedure, and this warped image is then matched to a generic, standard template (e.g., the “Talairach brain atlas”, which is based on the average of a large sample of brains) to enable combining images in a common space across individuals [93].
First Level/Individual Subject Analysis
Just as in the case of behavioral or survey data, individual BOLD activations need to be summarized before they can be included in analyses aimed at understanding group, task, or experimental condition-based differences. The two most common ways to analyze brain activation are as changes in magnitude of the BOLD response or the connectivity strength [94]. Magnitude of the BOLD response across the whole brain, or a selected region of the brain, can be compared for one experimental condition relative to another for each participant, to see which regions of the brain show more or less BOLD activity when engaged in a particular behavior. Connectivity analysis involves extracting time series from brain regions and computing a measure of the relationship among them, such as a correlation coefficient or partial correlation. The relationships can also be built as models with dynamic causal modeling [DCM; 20,95] or Bayesian graph analysis [96,97]. Dynamic causal models describe the correlated dynamics between two brain regions, and how the relationship is modulated by experimental manipulation or other brain activity [20]. These models are based on a predetermined distribution of data from past empirical evidence, so this method requires expertise [20]. Bayesian network models are graphical representations of probabilistic associations between active voxels [96]. The values from first-level analysis are then typically summarized statistically into a set of values per person, and further testing is carried out at the group level.
Group Models
Parametric Approaches
Group level comparisons that aggregate across participants have been the primary method in neuroimaging for comparing experimental conditions, groups, or both. The most ubiquitous approach is the general linear model [GLM; 98], which is implemented in all the major software packages (FSL, SPM, AFNI, etc.). This approach is parametric, which means that the distribution of the data is known and is based on a fixed set of parameters. It is also univariate, meaning single variable, i.e., level of BOLD activity. This method quantifies the linear relationship between each voxel’s activation and a design matrix indicating which conditions occurred at specific points in the time-course. It is considered a linear model because it is represented by the following equation: Y=Xβ+ϵ, in which each individual in the group has their own unique slope and Y-intercept [98]. Briefly, voxels that increase or decrease their activation when engaged in a task, relative to a baseline condition or rest, are identified using statistical testing. These clusters of activation contain a range of activation values, which are then subjected to a thresholding procedure to determine statistical significance when multiple comparisons are accounted for to control the rate of false positives [33,99].
A number of issues have been raised with parametric analysis approaches. These models involve assumptions about data distributions that do not actually hold true in many, if not most, cases of group fMRI data. This may lead to inflated error rates that are closer to 30–40% versus the 5% error expected when a result has a p value of less than 0.05.
Nonparametric approaches
To address these limitations, nonparametric approaches have been adapted for use with fMRI. Nonparametric means that the data distribution is treated as unknown and the parameters are not fixed. These methods include using alternative forms of correlation such as the Spearman ranked correlation, Wilcoxon ranked correlation, or permutation testing. Permutation testing refers to calculating a test statistic by quantifying all possible combinations of the data points. This process builds a sampling distribution of the expected null hypothesis which can then be used to infer statistical significance. This approach can be computationally demanding for fMRI data, although tools exist to make it easier (see Table 3).
Bayesian Approaches
Frequentist statistics test an hypothesis without incorporating any probability of whether that hypothesis is correct. Bayesian statistics, on the other hand, incorporate probabilities in the form of prior information or priors. These priors can be known in advance, or estimated from the data using sampling procedures in a similar manner as permutation testing. Bayesian approaches can increase power, such as in the case of hierarchical Bayesian analysis, in which information about the distribution of the group is used to constrain estimates at the individual level, a process called “shrinkage” for how it “shrinks” the distribution by pulling in outliers. Examples of these approaches for neuroimaging data include the IMaGES algorithm [97,102].
Advanced modeling approaches
More sophisticated methods have recently become available that make it easier to analyze repeated measures or nested designs, such as longitudinal studies with multiple visits, or treatment studies that have responder and non-responder groups. For example, the 3dLME function in AFNI uses the linear mixed effects [lme4, 100] package in R to take in first-level statistical maps of either activation or connectivity. The user can then make predictions about group differences while taking into account individual variations. This provides additional flexibility, especially for longitudinal designs, clinical trials with pre-post measurements, intervention studies, and other more complex study designs.
Connectivity analysis is based on the assumption that if the levels of BOLD activity in two different brain regions are correlated in time, then those two brain regions are functionally connected in some way. Whereas the general linear model assesses changes in BOLD magnitude, connectivity analysis quantifies relationships between time series in sets of regions. A full discussion of the range of methods, interpretations, and pitfalls of connectivity analysis is beyond the scope of this paper [but see 97,101–104]. Connectivity analysis involves different assumptions, and reveals information that differs from GLM. Most approaches summarize these temporal dynamics using a summary measure for each participant (e.g., correlation of BOLD activity levels between region A and region B over X time points). These summaries are then aggregated at the group level for standard statistical testing (e.g., paired t-test or multi-level model).
Additional methods include multi-variate pattern analysis (MVPA), which identifies nonlinear patterns of activation in brain regions in response to a stimulus [108,109]. “Representational similarity analysis” measures the relative similarity/dissimilarity of the multivariate pattern evoked by different stimuli [53]. Another method is independent component analysis (ICA), which separates each piece of the BOLD signal into spatial-temporal pieces for analysis [110]. Other methods involve extracting a summary measure, such as the average connectivity in a network, or a change in parameter estimates, and subject them to further testing using approaches such as machine learning classification to delineate group memberships [108], or canonical correlation to link between groups of brain variables and sets of other measures, such as behavior or self-report [111].
Assessing Statistical Significance
The accurate assessment of statistical significance of fMRI findings is perhaps the most debated issue in neuroimaging analysis. The multiple comparison problem states that given the tens of thousands of voxels in the brain, statistically testing each one as if it is independent of any other, does not accurately capture both the temporal and the spatial relationships among voxels, especially adjacent voxels. This also introduces chance into the analysis, in which case some voxels are false positives instead of true activations. Pitfalls of doing this incorrectly were exemplified with the now-infamous “dead salmon experiment” [113], in which researchers performed an fMRI analysis on the brain of a dead salmon and identified significantly activated voxels when using a relaxed criterion of statistical significance [45,114].
The false discovery rate (FDR) is a method of conceptualizing the rate of type I errors in null hypothesis testing when conducting multiple comparisons [115,116]. Each of the major software packages offers slightly different options to correct for multiple comparisons. The most conservative method is voxel-wise FDR, which treats each voxel as independent and penalizes for the full number of tests conducted. However, this method risks a high rate of false negatives because it ignores the spatial relationship among adjacent voxels [115, Figure 3].
Cluster thresholding, on the other hand, takes into account these spatial relationships by not treating voxels as independent but instead identifying “clusters.” In order to determine cluster threshold, criteria must be set for the clusters, and then the statistical program calculates the likelihood that adjacent voxels belong to a cluster. This is done by incorporating the z-value of the voxels; higher z-values correspond to greater activation. The threshold is designed to “cut off” voxels below a certain z-value and only include for analysis those that are above the value. Once those candidate voxels have been identified, then the cluster probability is calculated - the likelihood that a cluster of that size and magnitude occurred due to chance.[72].
The challenge of cluster thresholding is that there is typically a tradeoff between power and accuracy. Methods for selecting the appropriate cluster size and z-score are highly debated, and there is as yet no consensus on the appropriate set of criteria. However, generally a cluster threshold of z=3.29 is considered acceptably stringent. Allowing more results (voxels) to be included by using a less stringent threshold may avoid false negatives at the expense of accepting false positives [33,115]. Especially in precision medicine applications, there are issues with replicability and generalizability when thresholding values are not standardized. At worst this can facilitate “p hacking” of results (repeated testing until significance is achieved without correcting for multiple testing). The scientific consequences of a false positive from p-hacking are generally considered greater because it is likely to influence other researchers more than a false negative. False negatives, on the other hand, are harder to publish (other than in a registered report) and this can lead to bias in published effects aka the “file drawer effect” Note also that many sexual medicine studies are pilot studies in understudied or rare populations and topics, and such studies should be clearly differentiated from fully-powered studies.
A recent approach that strikes an acceptable balance between power and precision is threshold-free cluster enhancement [TFCE; 117]. Rather than prescribing a minimum Z-score, this method estimates which sets of voxels are clusters by looking at both strength of signal in a voxel as well as the signal in other adjacent voxels. This leads to an enhancement of signal but preserves the location of the true local maxima. The TFCE method relies on permutation testing, which is a statistical test to determine the distribution of the observed data under the null hypothesis. Within that distribution lies the maximum TFCE score across voxels. Then, this is compared to the TFCE image to assess significance of a TFCE statistical map. The 95th percentile in the permuted null distribution can then be used to threshold the TFCE image to give inference at the p<0.05 (corrected) level [117].
Probabilistic TFCE [117] further improves on this method by eliminating the need to conduct permutation testing (which is time consuming and computationally demanding). Instead, it incorporates the probability that a voxel with a given value X is significant, given its proximity to other voxels with values close to X. Clusters that are larger, more contiguous, and consistent in their values are therefore given higher confidence whereas those that are spatially non-contiguous and high in variability are assigned lower confidence. This increases the power to detect true effects, without requiring the overly punitive FDR or the degrees of freedom of setting criteria for cluster thresholding. However, like most cluster-thresholding techniques, small important brain regions (e.g., in the hypothalamus), or distributed networks of activation, may be overlooked in favor of larger (e.g., cortical) regions.
Part 5: Criticisms, Crises, and Opportunities
Because there are a number of challenges specific to fMRI, there have been numerous calls for methodological improvements and best practices [118]. The best methods to identify robust, reproducible, and valid results is perhaps the most debated issue in neuroimaging analysis (second only to “what is the BOLD signal?”). Problems with earlier studies included insufficient control of false positives [115], significant differences between software packages and code bugs [72], along with different preprocessing pipelines and regression techniques [74,91]. Even using scanners from different vendors [58] creates a lack of generalizability and replicable findings [76]. Additionally, physiological confounds [42], low power [45], and overfitting of models to data [119] have led some researchers to question the usefulness and viability of fMRI [115]. Overfitting occurs when a selected model is too stringent based on the original dataset it was built from. This limits its accuracy and use with future datasets [119]. However, most of these doubts stem from research that lacks merit and standardization, which is a fixable problem for the field.
Underpowered Studies
Statistical “power” refers to the ability to detect a true effect. Low power most commonly results from small sample sizes. Recent progress has been made with large-scale scanning initiatives that overcome low power by scanning hundreds of participants such as in the Human Connectome Project [120]. Power can also be affected by statistical package parameters and superficial data collection, which may limit “orthogonalization” [121]. Orthogonalization means that all research variables are separated mathematically, and function independently of each other. It is imperative to account for all aspects of variables and data, starting with research design and ending with the statistical package suite. Additional means to increase power include choosing the appropriate number of stimuli based on the number of participants [122], behavioral prescreening [123], and proper operationalization of variables [124]. Note that the high cost of neuroimaging can make achieving appropriate power challenging; when properly powered studies are not possible, research should be clearly labelled as pilot, exploratory, or preliminary findings. It is also recommended to compute a power analysis based on your research design. This can be simulated with the tidyverse package [125] in R.
Computing Resources Needed to Efficiently Process Data
A well-powered laptop can typically handle a standard analysis workflow on a modest size data set, such as 8-minute scans for <25 participants; however, if running a more computationally intensive workflow, better resources are required. For larger sample sizes, it is recommended that a dedicated multi-core computer be used -- either a standalone workstation or a cluster computing system, available at most major universities [126]. This allows parallelization of analyses - meaning that the workflow is broken into smaller discrete jobs which run at the same time on different computing cores. Some programs, like FSL, will automatically parallelize jobs; for others this may require indicating the parameter in the workflow design.
Logistical Limitations
Another challenge is that running a well-powered fMRI study is expensive in terms of scan costs, training, time, and staff labor. There are also limitations to the types of tasks that can be done by participants in the scanner, given the need to keep the head still, the requirement that additional physiological or behavioral equipment must be MRI compatible (not containing ferrous metal elements), and the confined nature of the scanner. Some participants, such as those who have a pacemaker, or implanted iron-containing metal (e.g., shrapnel, spinal fusion devices) or even some tattoos, cannot safely be scanned because the magnet will interfere with device function or heat up the metal. This limits the generalizability of results to full populations.
Interpretational Challenges
The results of fMRI should be interpreted with a clear understanding of the constraints of the method. The lay public find maps of “brain blobs” highly convincing, and indeed they suggest that we can easily measure biological underpinnings of complex processes. However, there are important fallacies of interpretation and misuses of methods that have been identified in the literature [114].
An example of a logical fallacy is reverse inference. Reverse inference is an error of interpretation, in which a brain region that increases its activation in response to stimulus X is described as “performing” that function [127]. As we now know, many functions of the brain are orchestrated by networks, and some functions cannot be attributed to single regions. To take a simple example, imagine a study in which you show a person in the scanner a photo of a peanut butter and jelly sandwich and a photo of jelly, and average their response to each. If you took the “jelly” response and subtracted it from the “peanut butter and jelly sandwich” response, you would be left with the “peanut butter sandwich” region of the brain. Reverse inference says “yes”; however, this may not be the full story -- the regions that appear to be “selective” for peanut butter sandwich might actually code features such as the color, memories of the taste, fear if one has a peanut allergy, etc.
Past studies have made similar, oversimplified inferences of brain functions based on flawed data collection [127]. This introduced doubt into the field, but also allowed investigators to reflect on and raise research standards. Linking brain activation to specific behavior patterns or internal experiences requires sophisticated experimental design. All conditions must be kept constant, except the key variable being manipulated. Allen et al. [25] provide a sound example of using the subtraction method to distinguish rectum stimulation from prostate stimulation-induced activations in the sensory cortex of men. The stimulating probe was first pressed against just the rectal wall, then more forcefully against the rectal wall to stimulate the prostate also. The sensory cortical response to the former was subtracted from the latter, revealing a unique response to the prostate stimulation. If robust research principles are used, we can make stronger claims about brain function and the human experience.
The Causality Question
A thorny issue in connectivity analysis is the question of causal inference [102,104]. Since most studies of causality infer relationships based on sequential response (cause precedes effect) it logically follows that it might be possible to estimate such relationships from temporal dynamics in fMRI. However, this has proven to be a complex and controversial undertaking, due largely to the signal-to-noise issues inherent in neuroimaging as well as conceptual and statistical concerns [104]. Causal modeling typically requires a priori hypotheses about brain regions that may be active in the scanner. It is not always possible to know which brain networks will be relevant, and lack of precision in this process can defeat the premise of hypothesis testing [128]. However, brain network analysis can also provide useful information. The information should be viewed as inferential and not definitive [129]. There are important steps to take when assessing presumptive causal evidence [106]. Causal modeling graphs in conjunction with other methods may provide stronger evidence. The basic approach is to analyze a response evoked by a stimulus. One such method is “transcranial magnetic stimulation (TMS)”, which temporarily perturbs regions of the brain to evoke a region-specific response [130]. Another method is applying a sensory stimulus and mapping the responsive brain regions [131]. These methods provide direct evidence of brain function similar to single-neuron or EEG evoked responses, especially when combined with results from additional imaging methods [130,132,133].
Circular Correlations
Some high correlations have been labeled as circular, unscientific and biased. This occurs when non-independent region of interest (ROI) analyses are used. For example, after conducting and thresholding a whole-brain analysis, the researcher picks the most active area, extracts its signal and correlates it with an individual difference measure, and claims a high correlation between a signal in Region X and that individual difference. This method leads to bias because the two analyses are not independent -- the selection process (whole brain analysis) biases the subsequent ROI analysis (aka cherry picking). In an influential paper, Vul et al. [134] argued that true correlations 0.8 and above are rare, despite many papers claiming to have found such high correlations. The majority of publications also did not share their data and code openly, and were reluctant to freely share the information with Vul et al. According to Poldrack and Mumford [99], it is possible to find high correlations, but researchers must use sound principles to make such claims.
The solution to this is to use independent ROIs for analysis - predefined brain regions based on anatomical, meta-analytic, or other methods before any analyses are conducted. Behavioral data and fMRI data also depend on the reliability of measures, the use of multiple comparison corrections (to avoid the “dead salmon error”), and appropriate modeling choices [99]. Open science best practices, such as preregistration of hypotheses and methods before data collection and sharing of data and code, can also contribute to the replicability of findings. If these principles are implemented, the chances of circular correlations being published will be greatly reduced.
Circular analysis, also termed, “double dipping”, refers to choices made in analysis that are contingent on results of a previous analysis, and therefore biased. Button [75] found that 42% of randomly sampled articles (n=134) from five journals re-used previously published data to make new claims. This is problematic because if the original data collection was biased, the impact “voodoo” gets recycled from one analysis to the next. This highlights the importance of proper hypothesis testing and replication of findings across laboratories.
Potential Solutions to Methodological Limitations
Some researchers propose an open-science approach, in which data, protocols, and code are shared to promote transparency, rigor, and reproducibility. This can also help researchers to standardize research practices [73, 74], and to choose appropriate experimental designs for research questions. Sharing code repositories, analysis pipelines [72], significant and nonsignificant results [76], and all raw data [75] could improve research quality. As a preventative step, preregistering hypotheses and predefining analysis plans ensures that researchers do not engage in excessive post hoc hypothesis testing or “p-hacking” [75].
Methodological improvements are continually evolving to meet the needs of researchers, including faster, quieter and more precise imaging strategies (i.e., “sequences”) [30,61,49,51,135]. The fMRI of the future will likely be more efficient and with higher average signal-to-noise ratios than what is currently available.
Summary
Functional MRI is a tool with clinical as well as research potential, but it is also a nascent, technically complex method whose basic premise rests on a number of scientific unknowns. It is important to be an informed consumer of the fMRI literature. If one is conducting neuroimaging experiments, a healthy skepticism is warranted, along with keeping up with constant technological advances (and challenges). A list of resources for performing fMRI analysis while cognizant of its pitfalls can be found in the Supplemental Methods. We hope that this paper has reduced some of the mystery of how colorful brain maps are generated and highlighted both the pros and cons of the method.
Supplementary Material
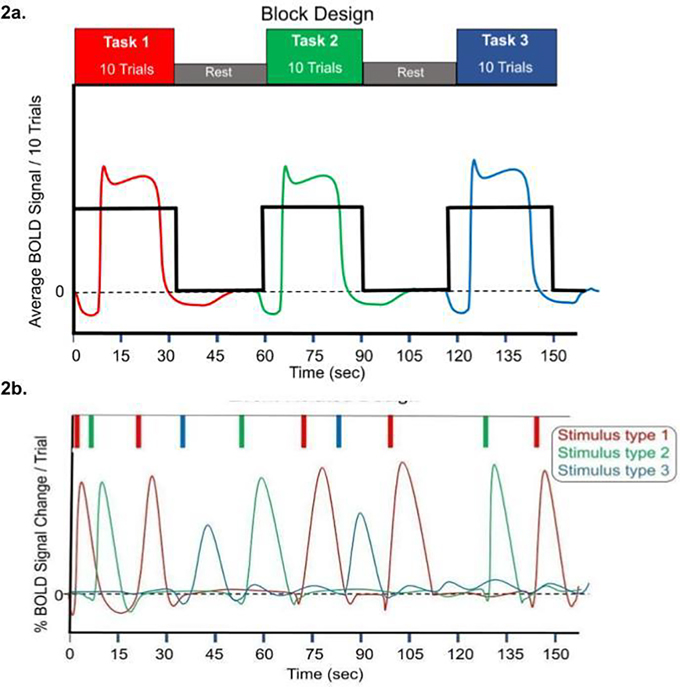
Figure 2.
a. Block Design schematic. A hypothetical average BOLD response is shown for three blocks of stimulus presentation, which consist of ten trials of three different tasks presented over a 30s time period, followed by 30s of rest. For each period of stimulus presentation, the summed HRF exhibits an initial dip, peak, then plateaus during the majority of the stimulus presentation period, finally dipping below baseline, and eventually stabilizing back at zero before the next block.2b. Event-related design. Three types of stimuli are presented in an interspersed manner with varying temporal delay. If the events are adequately spaced, HRF modeling can be applied to estimate effects of individual trials or stimulus types. ‘
Acknowledgements
This work was supported, in part, by the Intramural Research Program of the NIH, National Center for Complementary and Integrative Health. We would like to thank Dr. Gary Glover of the Radiology Department at Stanford University for providing us with pictures of the Lucas Center for Neurobiological Imaging’s MRI equipment, as well as Halee Staggs and Eli Ventura for their input on the manuscript.
Footnotes
Disclosure Statement
There are no conflicts of interest surrounding this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Colleen Mills-Finnerty, Palo Alto Veterans Health Care System, Palo Alto, CA, USA; Stanford University, Department of Psychiatry and Behavioral Sciences, Palo Alto, CA, USA.
Eleni Frangos, National Center for Complementary and Integrative Health, National Institutes of Health, Bethesda, MD, USA.
Kachina Allen, Southern Cross University, Psychology Department, Bilinga, QLD, AUS.
Barry Komisaruk, Rutgers University, Psychology Department, Newark, NJ, USA.
Nan Wise, Rutgers University, Psychology Department, Newark, NJ, USA.
References
- 1.Bandettini PA, Wong EC, Hinks RS, et al. Time course EPI of human brain function during task activation. Magn Reson Med. 1992;25:390–7. [DOI] [PubMed] [Google Scholar]
- 2.Ogawa S, Lee TM, Kay AR, et al. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc Natl Acad Sci USA. 1990;87:9868–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogawa S, Tank DW, Menon R, et al. Intrinsic signal changes accompanying sensory stimulation: Functional brain mapping with magnetic resonance imaging. Proc Natl Acad Sci USA. 1992;89:5951–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeung MK, Chan AS. A systematic review of the application of functional near-infrared spectroscopy to the study of cerebral hemodynamics in healthy aging. Neuropsychol Rev. 2020;31:139–66. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Shao Z, Xu Y, et al. Disrupted frontostriatal connectivity in primary insomnia: a DTI study. Brain Imaging and Behavior. 21. [DOI] [PubMed] [Google Scholar]
- 6.Shine JM, Koyejo O, Bell PT, et al. Estimation of dynamic functional connectivity using multiplication of temporal derivatives. Neuroimage. 2015;122:399–407. [DOI] [PubMed] [Google Scholar]
- 7.Hildebrandt MK, Jauk E, Lehmann K, et al. Brain activation during social cognition predicts everyday perspective-taking: a combined fMRI and ecological momentary assessment study of the social brain. Neuroimage. 2021;227:117624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Reilly JXO, Woolrich MW, Behrens TEJ, et al. Tools of the trade: psychophysiological interactions and functional connectivity. Soc Cogn Affect Neurosci. 2012;7:604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Zuo X, He Y. Graph-based network analysis of resting-state functional MRI. Front Syst Neurosci. 2010;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komisaruk BR, Rodriguez del Cerro MC. Human sexual behavior related to pathology and activity of the brain. Handb Clin Neurol. 2015;130:109–19. [DOI] [PubMed] [Google Scholar]
- 11.Jones SE, Lempka SF, Gopalakrishnan R, et al. Functional magnetic resonance imaging correlates of ventral striatal deep brain stimulation for poststroke pain. Neuromodulation. 2021;24(2):259–64. [DOI] [PubMed] [Google Scholar]
- 12.D’Esposito M Are individual differences in human brain organization measured with functional MRI meaningful? Proc Natl Acad Sci USA. 2019;116:22432–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubois J, Adolphs R. Building a science of individual differences from fMRI. Trends Cogn Sci. 2016;20:425–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung MH, Martins B, Privratsky A, et al. Individual differences in rate of acquiring stable neural representations of tasks in fMRI. PLoS ONE. 2018;13:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosen BR, Belliveau JW, Aronen HJ, et al. Susceptibility contrast imaging of cerebral blood volume: human experience. Magn Reson Med. 1991;22:293–9. [DOI] [PubMed] [Google Scholar]
- 16.Gauthier CJ, Fan AP. BOLD signal physiology: models and applications. Neuroimage. 2019;187:116–27. [DOI] [PubMed] [Google Scholar]
- 17.Dale AM, Buckner RL. Selective averaging of rapidly presented individual trials using fMRI. Hum Brain Mapp. 1997;5:329–40. [DOI] [PubMed] [Google Scholar]
- 18.Wexler BE, Fulbright RK, Lacadie CM, et al. An fMRI study of the human cortical motor system response to increasing functional demands. Magn Reson Imaging. 1997;15:385–96. [DOI] [PubMed] [Google Scholar]
- 19.Kriegeskorte N, Douglas PK. Cognitive computational neuroscience. Nat Neurosci. 2018;21:1148–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephan KE, Penny WD, Moran RJ, et al. Ten simple rules for dynamic causal modeling. Neuroimage. 2010;49:3099–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mills-Finnerty C, Hanson C, Khadr M, et al. Computations and connectivity underlying aversive counterfactuals. Brain Connect. 2020;10:467–478. [DOI] [PubMed] [Google Scholar]
- 22.Wise NJ, Frangos E, Komisaruk BR. Brain activity unique to orgasm in women: an fMRI analysis. J Sex Med. 2017;14(11):1380–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wise NJ, Frangos E, Komisaruk BR. Activation of sensory cortex by imagined genital stimulation: an fMRI analysis. Socioaffect Neurosci Psychol. 2016;6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komisaruk BR, Whipple B, Crawford A et al. Brain activation during vaginocervical self-stimulation and orgasm in women with complete spinal cord injury: fMRI evidence of mediation by the Vagus nerves. Brain Res. 2004;1024:77–88. [DOI] [PubMed] [Google Scholar]
- 25.Allen K, Wise N, Frangos E, Komisaruk B. (2020) Male urogenital system mapped onto the sensory cortex: Functional magnetic resonance imaging evidence. J Sex Med; 17:603–13. [DOI] [PubMed] [Google Scholar]
- 26.Marinsek N 30 years of trends in the MRI and fMRI literatures. 2017. Available at: https://nikkimarinsek.com/blog/fmri-bursts
- 27.Vassiliou VS, Cameron D, Prasad SK, et al. Magnetic resonance imaging: physics basics for the cardiologist. JRSM Cardiovasc Dis. 2018;7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brant-Zawadzki M, Gillan GD, Nitz WR. MP RAGE: a three-dimensional, T1-weighted, gradient-echo sequence - initial experience in the brain. Radiol. 1992;182:769–75. [DOI] [PubMed] [Google Scholar]
- 29.McPhee KC, Wilman AH. T2 quantification from only proton density and T2-weighted MRI by modelling actual refocusing angles. Neuroimage. 2015;118:642–50. [DOI] [PubMed] [Google Scholar]
- 30.Feinberg DA, Setsompop K. Ultra-fast MRI of the human brain with simultaneous multi-slice imaging. J Magn Reson. 2013;229:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chavhan GB, Babyn PS, Thomas B, et al. Principles, techniques, and applications of T2*-based MR imaging and its special applications. Radiographics. 2009;29:1433–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hillman EMC. Coupling mechanism and significance of the BOLD signal: a status report. Annu Rev Neurosci. 2014;37:161–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo CW, Krishnan A, Wager TD. Cluster-extent based thresholding in fMRI analyses: pitfalls and recommendations. Neuroimage. 2014;91:412–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishra A, Reynolds JP, Chen Y, et al. Astrocytes mediate neurovascular signaling to capillary pericytes but not to arterioles. Nat Neurosci. 2016;19:1619–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M, He Y, Sejnowski TJ, et al. Brain-state dependent astrocytic Ca2+ signals arecoupled to both positive and negative BOLD-fMRI signals. Proc Natl Acad Sci USA. 2018;115:E1647–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Özbay PS, Chang C, Picchioni D, et al. Contribution of systemic vascular effects to fMRI activity in white matter. Neuroimage. 2018;176:541–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Logothetis NK, Pfeuffer J. On the nature of the BOLD fMRI contrast mechanism. Mag Reson Imaging. 2004;22:1517–31. [DOI] [PubMed] [Google Scholar]
- 38.Musall S, Kaufman MT, Juavinett AL,et al. Single-trial neural dynamics are dominated by richly varied movements. Nat Neurosci. 2019;22:1677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsvetanov KA, Henson RN, Jones PS, et al. The effects of age on resting-state BOLD signal variability is explained by cardiovascular and neurovascular factors. Psychophysiology. 2020;00:e13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Handwerker DA, Gonzalez-Castillo J, D’Esposito M, et al. The continuing challenge of understanding and modeling hemodynamic variation in fMRI. Neuroimage. 2012;62:1017–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsvetanov KA, Henson RNA, Rowe JB. Separating vascular and neuronal effects of age on fMRI BOLD signals. Philos Trans R Soc Lond B Biol Sci. 2020;376:20190631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caballero-Gaudes C, Reynolds RC. Methods for cleaning the BOLD fMRI signal. Neuroimage. 2017;154:128–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dawson DA, Cha K, Lewis LB, et al. Evaluation and calibration of functional network modeling methods based on known anatomical connections. Neuroimage. 2013;67:331–43. [DOI] [PubMed] [Google Scholar]
- 44.Brodoehl S, Witte OW, Klingner CM. Measuring eye states in functional MRI. BMC Neurosci. 2016;17:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cremers HR, Wager TD, Yarkoni T. The relation between statistical power and inference in fMRI. PLoS One. 2017;12:e0184923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun J, Hu X, Huang X, et al. Inferring consistent functional interaction patterns from natural stimulus FMRI data. Neuroimage. 2012;61:987–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H, Han X, Jin M, et al. Cerebral blood flow alterations in hemodialysis patients with and without restless legs syndrome: an arterial spin labeling study. Brain Imaging Behav. 2020;15:401–09. [DOI] [PubMed] [Google Scholar]
- 48.Xu J, Moeller S, Auerbach EJ, et al. Evaluation of slice accelerations using multiband echo planar imaging at 3 T. Neuroimage. 2013;83:991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paasonen J, Laakso H, Pirttimäki T, et al. Multi-band SWIFT enables quiet and artefact-free EEG-fMRI and awake fMRI studies in rat. Neuroimage. 2020;206:116338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kundu P, Voon V, Balchandani P, et al. Multi-echo fMRI: a review of applications in fMRI denoising and analysis of BOLD signals. Neuroimage. 2017;154:59–80. [DOI] [PubMed] [Google Scholar]
- 51.Diekhoff S, Uludag K, Sparing R, et al. Functional localization in the human brain: gradient-echo, spin-echo, and arterial spin-labeling fMRI compared with neuronavigated TMS. Hum Brain Mapp. 2011;32:341–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welvaert M, Rosseel Y. On the definition of signal-to-noise ratio and contrast-to-noise ratio for FMRI data. PLoS ONE, 2013;8(11):e77089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finn ES, Glerean E, Khojandi AY, et al. Idiosynchrony: from shared responses to individual differences during naturalistic neuroimaging. Neuroimage. 2020;215:116828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salehi M, Greene AS, Karbasi A, et al. There is no single functional atlas even for a single individual: functional parcel definitions change with task. Neuroimage. 2020;208:116366. [DOI] [PubMed] [Google Scholar]
- 55.Deco G, Tononi G, Boly M, et al. Rethinking segregation and integration: contributions of whole-brain modelling. Nat Rev Neurosci. 2015;16:430–9. [DOI] [PubMed] [Google Scholar]
- 56.Siegel JS, Mitra A, Laumann TO, et al. Data quality influences observed links between functional connectivity and behavior. Cereb Cortex. 2017;27:4492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glover GH, Law CS. Spiral-in/out BOLD fMRI for increased SNR and reduced susceptibility artifacts. Magn Reson Med. 2001;46:515–22. [DOI] [PubMed] [Google Scholar]
- 58.Badhwar AP, Collin-Verreault Y, Orban P, et al. Multivariate consistency of resting-state fMRI connectivity maps acquired on a single individual over 2.5 years, 13 sites and 3 vendors. Neuroimage. 2020;205:116210. [DOI] [PubMed] [Google Scholar]
- 59.Krause F, Benjamins C, Eck J, et al. Active head motion reduction in magnetic resonance imaging using tactile feedback. Hum Brain Mapp. 2019;40:4026–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li W, Qiao L, Zhang L, et al. Functional brain network estimation with time series self-scrubbing. IEEE J Biomed Health Inform. 2019;23: 2494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Glover GH. Spiral imaging in fMRI. Neuroimage. 2012;62:706–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glover GH. Simple analytic spiral K-space algorithm. Magn Reson Med. 1999;42:412–5. [DOI] [PubMed] [Google Scholar]
- 63.Panman JL, To YY, van der Ende EL, et al. Bias introduced by multiple head coils in MRI research: an 8 channel and 32 channel coil comparison. Front Neurosci. 2019;15;13:729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gitelman DR, Penny WD, Ashburner J, et al. Modeling regional and psychophysiologic interactions in fMRI: the importance of hemodynamic deconvolution. Neuroimage. 2003;19:200–7. [DOI] [PubMed] [Google Scholar]
- 65.Kannurpatti SS, Motes MA, Rypma B, et al. Neural and vascular variability and the fMRI-BOLD response in normal aging. Magn Reson Imaging. 2010;28:466–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cole DM, Smith SM, Beckmann CF. Advances and pitfalls in the analysis and interpretation of resting-state FMRI data. Front Syst Neurosci. 2010;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stark CE, Squire LR. When zero is not zero: the problem of ambiguous baseline conditions in fMRI. Proc Natl Acad Sci USA. 2001;98:12760–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mejia AF, Nebel MB, Shou H, et al. Improving reliability of subject-level resting-state fMRI parcellation with shrinkage estimators. Neuroimage. 2015;112:14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Caparelli EC, Ross TJ, Gu H, et al. Factors affecting detection power of blood oxygen-level dependent signal in resting-state functional magnetic resonance imaging using high-resolution echo-planar imaging. Brain Connect. 2019;9(8):638–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Loh JM, Lindquist MA, Wager TD. Residual analysis for detecting mis-modeling in fMRI. Stat Sin. 2008;18:1421–48. [Google Scholar]
- 71.Soch J, Haynes JD, Allefeld C. How to avoid mismodelling in GLM-based fMRI data analysis: cross-validated Bayesian model selection. NeuroImage, 2016;141:469–89. [DOI] [PubMed] [Google Scholar]
- 72.Bowring A, Maumet C, Nichols TE. Exploring the impact of analysis software on task fMRI results. Hum Brain Mapp. 2019;40:3362–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fallani FDV, Richiardi J, Chavez M, et al. Graph analysis of functional brain networks: practical issues in translational neuroscience. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hallquist MN, Hillary FG. Graph theory approaches to functional network organization in brain disorders: a critique for a brave new small-world. Netw Neurosci. 2019;3:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Button KS. Double-dipping revisited. Nat Neurosci. 2019;22:688–90. [DOI] [PubMed] [Google Scholar]
- 76.Carp J The secret lives of experiments: methods reporting in the fMRI literature. Neuroimage. 2012;63:289–300. [DOI] [PubMed] [Google Scholar]
- 77.Woolrich MW, Jbabdi S, Patenaude B, et al. Bayesian analysis of neuroimaging data in FSL. Neuroimage. 2009;45:173–86. [DOI] [PubMed] [Google Scholar]
- 78.Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:208–19. [DOI] [PubMed] [Google Scholar]
- 79.Jenkinson M, Beckmann CF, Behrens TE, et al. FSL. Neuroimage. 2012;62:782–90. [DOI] [PubMed] [Google Scholar]
- 80.Frackowiak RSJ, Friston KJ, Frith CD, et al. Human Brain Function. Academic Press; USA. 1997. [Google Scholar]
- 81.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comp Biomed Res. 1996;29:162–73. [DOI] [PubMed] [Google Scholar]
- 82.R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2020. Available from: https://www.R-project.org/ [Google Scholar]
- 83.Muschelli J, Sweeney E, Lindquist M, et al. fslr: connecting the FSL software with R. 2018. F1000Res. 2018;7:599. [PMC free article] [PubMed] [Google Scholar]
- 84.Chen G, Saad ZS, Britton JC, et al. Linear mixed-effects modeling approach to fMRI group analysis. Neuroimage. 2013;73:176–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.The Math Works. MATLAB. Version 2020a. [Google Scholar]
- 86.Gorgolewski K, Burns CD, Madison C, et al. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework. Front Neuroinform. 2011;5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Esteban O, Markiewicz CJ, Blair RW, et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat Methods. 2019;16:111–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Churchill NW, Spring R, Afshin-pour B, et al. An automated, adaptive framework for optimizing preprocessing pipelines in task-based functional MRI. PLoS One. 2015;10:e0131520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Noble S, Scheinost D, Constable RT. Cluster failure or power failure? Evaluating sensitivity in cluster-level inference. Neuroimage. 2020;209:116468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aquino KM, Fulcher BD, Parkes L, et al. Identifying and removing widespread signal deflections from fMRI data: rethinking the global signal regression problem. NeuroImage. 2020;212:116614. [DOI] [PubMed] [Google Scholar]
- 91.Hallquist MN, Hwang K, Luna B. The nuisance of nuisance regression: spectral misspecification in a common approach to resting-state fMRI preprocessing reintroduces noise and obscures functional connectivity. Neuroimage. 2013;82:208–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Power JD. A simple but useful way to assess fMRI scan qualities. Neuroimage. 2016;154:150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang D, Du Y, Cheng H, et al. Groupwise spatial normalization of fMRI data based on multi-range functional connectivity patterns. Neuroimage. 2013;82:355–72. [DOI] [PubMed] [Google Scholar]
- 94.Lindquist MA, Meng J, Atlas LY, et al. Modeling the hemodynamic response function in fMRI: efficiency, bias and mis-modeling. Neuroimage. 2009;45:S187–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Friston KJ, Harrison L, Penny W. Dynamic causal modelling. Neuroimage. 2003;19:1273. [DOI] [PubMed] [Google Scholar]
- 96.Chen R, Herskovitz E. Graphical model based functional analysis of fMRI images. Neuroimage. 2007;35:635–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ramsey JD, Hanson SJ, Glymour C. Multi-subject search correctly identifies causal connections and most causal directions in the DCM models of the Smith et al. simulation study. Neuroimage. 2011;58:838–48. [DOI] [PubMed] [Google Scholar]
- 98.Monti MM. Statistical analysis of fMRI time-series: a critical review of the GLM approach. Front Hum Neurosci. 2011;5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Poldrack RA, Mumford J. Independence in ROI analysis: where is the voodoo? Soc Cogn Affect Neurosci. 2009;4:208–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bates D, Mächler M, Bolker B, et al. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48. [Google Scholar]
- 101.Smith SM. The future of FMRI connectivity. Neuroimage. 2012;62:1257–66. [DOI] [PubMed] [Google Scholar]
- 102.Ramsey JD, Hanson SJ, Hanson C, et al. Six problems for causal inference from fMRI. Neuroimage. 2010;49:1545–58. [DOI] [PubMed] [Google Scholar]
- 103.Goldenberg D, Galván A. The use of functional and effective connectivity techniques to understand the developing brain. Dev Cogn Neurosci. 2015;12:155–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reid AT, Headley DB, Mill RD, et al. Advancing functional connectivity research fromassociation to causation. Nat Neurosci. 2019;22:1751–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lohmann G, Erfurth K, Müller K, et al. Critical comments on dynamic causal modelling. Neuroimage. 2012;59:2322–9. [DOI] [PubMed] [Google Scholar]
- 106.Bielczyk NZ, Uithol S, van Mourik T, et al. Disentangling causal webs in the brain using functional magnetic resonance imaging: a review of current approaches. Netw Neurosci. 2019;3:237–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mills-Finnerty C, Hanson C, Hanson S.J. Brain network response underlying decisions about abstract reinforcers. Neuroimage. 2014;103:48–54. [DOI] [PubMed] [Google Scholar]
- 108.Haynes J-D. A primer on pattern-based approaches to fMRI: principles, pitfalls, and perspectives. Neuron. 2015;87:257–70. [DOI] [PubMed] [Google Scholar]
- 109.Li Y, Richardson RM, Ghuman AS. Multi-connection pattern analysis: decoding the representational content of neural communication. Neuron. 2016;63:902–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Salimi-Khorshidi G, Douaud G, Beckmann CF, et al. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. Neuroimage. 2014;90:449–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhuang X, Yang Z, Sreenivasan KR, et al. Multivariate group-level analysis for task fMRI data with canonical correlation analysis. Neuroimage. 2019;194:25–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Madan CR. Creating 3D visualizations of MRI data: a brief guide. F1000Res. 2015;4:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bennett CM, Miller MB, Wolford GL. Neural correlates of interspecies perspective taking in the post-mortem atlantic salmon: an argument for multiple comparisons correction. Neuroimage. 2009. Jul;47(1):S125. [Google Scholar]
- 114.Lyon L Dead salmon and voodoo correlations: should we be sceptical about functional MRI? Brain. 2017;140:e53. [DOI] [PubMed] [Google Scholar]
- 115.Eklund A, Nichols TE, Knutsson H. Cluster failure: why fMRI inferences for spatial extent have inflated false-positive rates. Proc Natl Acad Sci USA. 2016;113:7900–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kessler D, Angstadt M, Sripada CS. Reevaluating “cluster failure” in fMRI using nonparametric control of the false discovery rate. Proc Natl Acad Sci. 2017;114:E3372–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Spisák T, Spisák Z, Zunhammer M, et al. Probabilistic TFCE: a generalized combination of cluster size and voxel intensity to increase statistical power. Neuroimage. 2019;185:12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mills-Finnerty C Five best practices for fMRI research. J Reprod Neurosci. 2021;2. [Google Scholar]
- 119.Hosseini M, Powell M, Collins J, et al. I tried a bunch of things: the dangers of unexpected overfitting in classification of brain data. Neurosci Biobehav Rev. 2020;119:456–67. [DOI] [PubMed] [Google Scholar]
- 120.Connectome Coordination Facility. About the CCF (CCF overview). 2020.
- 121.Mumford JA, Poline J-B, Poldrack RA. Orthogonalization of regressors in fMRI models.PLoS One. 2015;10:e0126255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Westfall J, Kenny DA, Judd CM. Statistical power and optimal design in experiments in which samples of participants respond to samples of stimuli. J Exp Psychol Gen. 2014;143:2020–45. [DOI] [PubMed] [Google Scholar]
- 123.de Haas B How to enhance the power to detect brain-behavior correlations with limited resources. Front Hum Neurosci. 2018;12:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Smith PL, Little DR. Small is beautiful: in defense of the small-N design. Psychon Bull Rev. 2018;25:2083–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wickham H, Averick M, Bryan J, et al. Welcome to the tidyverse. J Open Source Softw. 2019;4:1686. [Google Scholar]
- 126.Russpoldrack.org. Computing models for a neuroimaging lab. 2019.
- 127.Ritchie JB, Kaplan DM, Klein C. Decoding the brain: neural representation and the limits of multivariate pattern analysis in cognitive neuroscience. Br J Philos Sci. 2019;70:581–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ryan O, Bringmann LF, Schuurman NK. The challenge of generating causal hypotheses using network models. PsyArXiv. 2019:1–29. [Google Scholar]
- 129.Deuker L, Bullmore ET, Smith M, et al. Reproducibility of graph metrics of human brain functional networks. Neuroimage. 2009;47:1460–8. [DOI] [PubMed] [Google Scholar]
- 130.Hallam GP, Whitney C, Hymers M, et al. Charting the effects of TMS with fMRI: modulation of cortical recruitment within the distributed network supporting semantic control. Neuropsychologia. 2016;93:40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Komisaruk BR, Wise N, Frangos E, et al. Women’s clitoris, vagina, and cervix mapped on the sensory cortex: fMRI evidence. J Sex Med. 2011;8:2822–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Peters JC, Reithler J, Graaf TA, et al. Concurrent human TMS-EEG-fMRI enables monitoring of oscillatory brain state-dependent gating of cortico-subcortical network activity. Commun Biol. 2020;3:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bleyenheuft Y, Dricot L, Gilis N, et al. Capturing neuroplastic changes after bimanual intensive rehabilitation in children with unilateral spastic cerebral palsy: a combined DTI, TMS and fMRI pilot study. Res Dev Disabil 2015:43–44:136–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vul E, Harris C, Winkielman P, et al. Puzzlingly high correlations in fMRI studies of emotion, personality, and social cognition. Perspect Psychol Science. 2009;4(3):274–290. [DOI] [PubMed] [Google Scholar]
- 135.Solana AB, Menini A, Sacolick LI, et al. Quiet and distortion-free, whole brain BOLD fMRI using T2 -prepared RUFIS. Magn Reson Med. 2016;75:1402–12. [DOI] [PubMed] [Google Scholar]
- 136.Mann K, Deny S, Ganguli S, Clandinin TR. Coupling of activity, metabolism and behaviour across the Drosophila brain. Nat 2021. 5937858 [Internet]. 2021 Apr 28 [cited 2021 Oct 21];593(7858):244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.