Abstract
STUDY QUESTION
Do polymorphisms in the anti-Müllerian hormone (AMH) promoter have an effect on AMH levels in patients with polycystic ovary syndrome (PCOS)?
SUMMARY ANSWER
We have identified a novel AMH promoter polymorphism rs10406324 that is associated with lower serum AMH levels and is suggested to play a role in the mechanism of regulation of AMH gene expression in women.
WHAT IS KNOWN ALREADY
Follicle number is positively correlated with serum AMH levels, reflected by elevated AMH levels in women with PCOS. In addition, it is suggested that AMH production per follicle is higher in women with PCOS than in normo-ovulatory women, implying an altered regulation of AMH in PCOS.
STUDY DESIGN, SIZE, DURATION
A discovery cohort of 655 PCOS women of Northern European ancestry and both an internal and external validation PCOS cohort (n = 458 and n = 321, respectively) were included in this study. Summary-level data of an AMH genome-wide association study meta-analysis including 7049 normo-ovulatory women was included as a control cohort. A genetic approach was taken through association analysis and in silico analysis of the associated variants in the AMH promoter. In vitro analysis was performed to investigate the functional mechanisms.
PARTICIPANTS/MATERIALS, SETTING, METHODS
All common two-allelic single-nucleotide polymorphisms (SNPs) in the region Chr19:2 245 353–2 250 827 bp (Build 37) were selected for the analysis. Linear regression analyses were performed to determine the association between SNPs in the AMH promoter region and serum AMH levels. For the in silico analysis, the webtools ‘HaploReg’ v4.1 for ENCODE prediction weight matrices and ‘atSNP’ were used. In vitro analysis was performed using KK1 cells, a mouse granulosa cell line and COV434 cells, a human granulosa tumor cell line. Cells were transfected with the reference or the variant human AMH promoter reporter construct together with several transcription factors (TFs). Dual-Glo® Luciferase Assay was performed to measure the luciferase activity.
MAIN RESULTS AND THE ROLE OF CHANCE
Polymorphism rs10406324 was significantly associated with serum AMH levels in all three PCOS cohorts. Carriers of the minor allele G had significantly lower log-transformed serum AMH levels compared to non-carriers (P = 8.58 × 10−8, P = 1.35 × 10−3 and P = 1.24 × 10−3, respectively). This result was validated in a subsequent meta-analysis (P = 3.24 × 10−12). Interestingly, rs10406324 was not associated with follicle count, nor with other clinical traits. Also, in normo-ovulatory women, the minor allele of this variant was associated with lower serum AMH levels (P = 1.04 × 10−5). These findings suggest that polymorphism rs10406324 plays a role in the regulation of AMH expression, irrespective of clinical background. In silico analysis suggested a decreased binding affinity of the TFs steroidogenenic factor 1, estrogen-related receptor alpha and glucocorticoid receptor to the minor allele G variant, however in vitro analysis did not show a difference in promoter activity between the A and G allele.
LIMITATIONS, REASONS FOR CAUTION
Functional analyses were performed in a mouse and a human granulosa cell line using an AMH promoter reporter construct. This may have limited assessment of the impact of the polymorphism on higher order chromatin structures. Human granulosa cells generated from induced pluripotent stem cells, combined with gene editing, may provide a method to elucidate the exact mechanism behind the decrease in serum AMH levels in carriers of the −210 G allele. We acknowledge that the lack of follicle number in the external validation and the control cohort is a limitation of the paper. Although we observed that the association between rs10406324 and AMH levels was independent of follicle number in our discovery and internal validation PCOS cohorts, we cannot fully rule out that the observed effects on serum AMH levels are, in part, caused by differences in follicle number.
WIDER IMPLICATIONS OF THE FINDINGS
These results suggest that variations in serum AMH levels are not only caused by differences in follicle number but also by genetic factors. Therefore, the genetic context should be taken into consideration when assessing serum AMH levels in women. This may have clinical consequences when serum AMH levels are used as a marker for the polycystic ovarian morphology phenotype.
STUDY FUNDING/COMPETING INTEREST(S)
No external funding was used. J.S.E.L. has received consultancy fees from the following companies: Ferring, Roche Diagnostics and Ansh Labs and has received travel reimbursement from Ferring. J.A.V. has received royalties from AMH assays, paid to the institute/lab with no personal financial gain. The other authors declare no competing interests.
TRIAL REGISTRATION NUMBER
N/A.
Keywords: AMH, PCOS, SNP, genetics, ovary
Introduction
Polycystic ovary syndrome (PCOS) is a very heterogeneous disease, diagnosed based on the presence of at least two of the following three criteria: (i) ovulatory dysfunction (OD); (ii) clinical and/or biochemical hyperandrogenism (HA) and (iii) polycystic ovarian morphology (PCOM) (Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group, 2004; Dewailly et al., 2014). With a prevalence of 10–15% in the European population, PCOS is the most common endocrine disorder in females of reproductive age (Bozdag et al., 2016; Neven et al., 2018). Using the diagnostic criteria, four different phenotypes can be identified: (A) OD, HA and PCOM; (B) OD and HA; (C) HA and PCOM; and (D) OD and PCOM (Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group, 2004). Of these four phenotypes, Phenotypes A and B are considered the most severe phenotypes, presenting with more pronounced menstrual dysfunction and the highest risk for metabolic syndrome (Lizneva et al., 2016).
In addition to the three main characteristics, patients with PCOS have significantly increased levels of anti-Müllerian hormone (AMH) (Pigny et al., 2003; Laven et al., 2004; Park et al., 2010). AMH is a member of the transforming growth factor β family and is expressed by granulosa cells of growing gonadotropin-independent follicles, mainly the preantral and small antral follicles (Cate et al., 1986; Weenen et al., 2004). In women with PCOS, the increased number of antral follicles, causing the PCOM phenotype, is strongly correlated with the elevated circulating AMH levels. Hence, serum AMH levels are postulated as a diagnostic marker for PCOS as an alternative to follicle count (Dewailly, 2016; Lie Fong et al., 2017). In the ovary, AMH plays a role in folliculogenesis and the selection of the dominant follicle. During this selection process, the sensitivity of follicles for FSH increases and the follicles become gonadotrophin-dependent for further growth. AMH inhibits this FSH-dependent follicle growth. Thus, increased AMH levels reduce FSH sensitivity, thereby contributing to the accumulation of small antral follicles and OD in PCOS (Visser et al., 2006; Garg and Tal, 2016).
Because of the proposed role of AMH in the PCOS etiology, previous genetic studies have focused on variants in the AMH gene and its specific type II receptor (AMHR2). In our previous study, we studied the association of the AMH single-nucleotide polymorphism (SNP) rs10407022 (Ile49Ser) and the AMHR2 polymorphism rs2002555 with the PCOS phenotype (Kevenaar et al., 2008). Although these SNPs were not risk alleles for PCOS, carriers of the AMH 49Ser allele significantly less often had PCOM, which is in line with the decreased bioactivity of the AMH 49Ser variant observed in vitro. Two recent studies (Gorsic et al., 2017, 2019) identified several heterozygous PCOS-specific polymorphisms in the AMH and the AMHR2 genes and subsequent in vitro assays showed that these polymorphisms have impaired AMH bioactivity and a dominant negative effect on AMH signaling. These studies strongly support the hypothesis that AMH plays an important role in the PCOS phenotype. While these studies focused on the functional role of AMH in the PCOS etiology, potential changes in the regulation of AMH gene expression in PCOS have not been addressed.
An altered regulation of AMH gene expression in PCOS has been hypothesized. Several studies have shown that an increased serum AMH level in PCOS is not only explained by increased number of follicles but also by increased AMH expression per follicle (Bhide et al., 2015; Alebic et al., 2018). Indeed, AMH as well as AMHR2 gene expression are increased and granulosa cells from PCOS patients compared to normo-ovulatory women (Catteau-Jonard et al., 2008; Pierre et al., 2017). In addition, whereas in normo-ovulatory women AMH expression is absent in gonadotropin-dependent follicles after follicle maturation, in PCOS patients this suppression was not observed (Kedem et al., 2013; Kristensen et al., 2019). Surprisingly, despite its specific expression pattern and the clinical use of serum AMH levels, still little is known about the transcriptional regulation of AMH, in general but also in patients with PCOS. Currently, only a few transcription factors (TFs) have been identified to play a role in the regulation of AMH expression in the ovary, including GATA4, FOXL2 and steroidogenenic factor 1 (SF1) (Rey et al., 2003; Jin et al., 2016).
Hence, the aim of this study is to gain more insight into the regulation of AMH gene expression in PCOS patients. We hypothesized that polymorphisms in the AMH promoter region may affect serum AMH levels. Therefore, we investigated whether SNPs in the human AMH promoter were associated with serum AMH levels in PCOS patients. In addition, we have performed in silico and in vitro analyses to investigate the functional mechanisms of the associated SNP.
Materials and methods
Subjects and phenotyping
Discovery PCOS cohort
In total, 655 women of reproductive age and European ancestry from the previously described Rotterdam PCOS cohort were included in this study (Day et al., 2018). The Rotterdam PCOS cohort, the CyclusOLigoAmennorroe (COLA) study, was approved by the institutional review board (Medical Ethics Committee) of the Erasmus Medical Center (04-263). PCOS was diagnosed based on the Rotterdam criteria requiring at least two out of the following three diagnostic criteria: (i) oligo- or amenorrhea, defined as chronic menstrual cycle interval >35 days or >188 days, respectively; (ii) clinical HA, defined as modified Ferriman Gallwey (FG) score above 5 and/or biochemical HA, defined as elevated (>4.5) free androgen index [testosterone × 100/SHBG] (Bui et al., 2015); and (iii) polycystic ovaries, defined as 12 or more follicles of 2–9 mm in each ovary and/or an ovarian volume >10 ml (Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group, 2004). Serum AMH levels were measured with the picoAMH assay [Ansh Labs®, Webster, TX, USA]. Serum testosterone was measured using radioimmunoassays (Diagnostic Products Corporation). Serum FSH and LH were measured using the Siemens Immulite 2000XPi, and total follicle count (TFC) and ovarian volume were assessed as described earlier (van Santbrink et al., 1997; Kevenaar et al., 2008).
Internal replication PCOS cohort
Internal replication was carried out in a follow-up sample of the discovery PCOS cohort. A total of 458 women of European ancestry, diagnosed based on the Rotterdam PCOS criteria, were included. The diagnostic criteria for biochemical HA was adjusted to a cutoff value of 2.9 for patients entering the dataset after 20 August 2012, due to the use of UPLC-MS/MS instead of radioimmunoassay for the measurement of serum testosterone (Bui et al., 2015). Serum AMH levels were measured with the picoAMH assay [Ansh Labs®, Webster, TX, USA].
External replication PCOS cohort
External replication was carried out in an independent cohort. A total of 321 women from the Chicago PCOS cohort were included. The Chicago PCOS cohort was approved by the Northwestern IRB (#STU00008096). Patients were included when they met the 1990 National Institutes of Health (NIH) criteria for PCOS, requiring the presence of both clinical and/or biochemical HA and OD. Cases fulfilling these criteria also meet the Rotterdam criteria for PCOS. Serum AMH levels were measured with the picoAMH assay [Ansh Labs®, Webster, TX, USA].
Genotyping
All genotyping specifications for the PCOS cohorts have been described earlier in a large genome-wide association study (GWAS) meta-analysis in PCOS patients (Day et al., 2018). Samples used in this study were selected from the Rotterdam PCOS cohort based on the presence of AMH levels measured by the picoAMH assay [Ansh Labs®, Webster, TX, USA] (Day et al., 2018). All two-allelic SNPs in the region Chr19:2 245 353–2 250 827 bp (Build 37, hg19) were selected. This region covers 3969 bp of the human AMH promoter and 1505 bp downstream of the protein coding region of AMH, covering the first three exons of the AMH protein. Only SNPs with a minor allele frequency (MAF) >1% and imputation quality (r2) >0.75 were included for the analysis. The quality control check analysis was described previously (Day et al., 2018). Linkage disequilibrium (LD) between SNPs was calculated using the Ensembl library 1000GENOMES:phase_3:CEU (Zerbino et al., 2018).
Control cohort
Summary-level data from an AMH GWAS meta-analysis including data of 7049 normo-ovulatory premenopausal women (median age ranged from 15.3 to 48 years across included GWAS) was included as a control study. The data included in this GWAS meta-analysis and the performed data analyses have been described previously (Verdiesen et al., 2022). All studies included in this GWAS meta-analysis converted AMH levels to pmol/l and transformed AMH levels using rank-based inverse normal transformation to guarantee a normal distribution.
Deleteriousness prediction
Deleteriousness of the associated variant was assessed using the online tool Combined Annotation Dependent depletion v1.2 (CADD) (Rentzsch et al., 2019). CADD is an annotation tool that predicts the potential functional consequences of an SNP. The CADD C-score evaluates the severity of the deleteriousness and a CADD C-score above 15 was considered deleterious.
TF binding motif analysis
Predicted TF binding scores were calculated using the in silico webtools ‘HaploReg’ v4.1 for ENCODE prediction weight matrices (Kheradpour and Kellis, 2014; Zerbino et al., 2018) and ‘atSNP’, a package for affinity testing for the impact of SNPs on TF binding motifs in the human genome (Zuo et al., 2015; Shin et al., 2019). The significance of changes in TF binding sides was assessed by comparing the difference in binding affinity of the TF for motifs containing either the reference allele or the variant allele. The results are presented as log odds scores. According to the atSNP protocol, a P-value below 0.0001 was considered statistically significant (Zuo et al., 2015). TFs without known expression in ovarian tissues (normalized expression <1) were excluded from the analysis (Uhlén et al., 2015).
Targeted mutagenesis
The pGL3 luciferase reporter containing the -643/-1 bp human AMH promoter (a gift from Prof. Dr. J. Bae) was used (Park et al., 2014). Site-directed mutagenesis of the AMH promoter reporter was performed using the Quick change site-directed mutagenesis kit to introduce the polymorphism rs10406324 (−210 A>G) using the following primers: Forward primer 5′—CAA GGA CGG CAT GTT GAC AC—3′ and Reverse primer 5′—GTG TCA ACA TGC CGT CCT TG—3′ [Integrated DNA technologies]. To generate the AMH promoter with the mutated proximal SF1 binding site the following primers were used: Forward primer 5′—GTC CCC CAA TTT CGC GGC AG—3′ and reverse primer 5′—CTG CCG CGA AAT TGG GGG AC—3′ (Campbell et al., 2008).
Cell culture and transfections
KK1 cells, a mouse granulosa cell line (a gift from Dr. I. Huhtaniemi), and COV434 cells, a human tumor granulosa cell line (Sigma-Aldrich, European Collection of Authenticated Cell Cultures), were cultured in a humidified incubator at 37°C with 5% CO2, using Dulbecco’s Modified Eagle Medium F-12 Nutrient Mixture (DMEM/F-12) + GlutaMAX [Gibco, Invitrogen, Breda, The Netherlands] with 10% Fetal Calf Serum [FCS; Life Technologies Europe BV] and 1% Penicillin-Streptomycin [P/S; Gibco] (Kananen et al., 1995).
KK1 and COV434 cells were seeded at 1 × 105–1.5 × 105 cells per well in 24-well or 48-well plates. After 24 h, cells were transfected with the reference or variant human AMH promoter reporter construct (167 ng/well) together with 83 ng/well of one the following constructs: (i) pCMV5-SF1-FL (a kind gift from Prof. Dr. J. Bae); (ii) phGR-SB (a gift from Andrew Cato, Addgene plasmid #133409) (Heck et al., 1994); or (iii) pCMV-flag-ERRα (a gift from Toren Finkel, Addgene plasmid #10975) (Ichida et al., 2002). Cells cotransfected with the glucocorticoid receptor (GR) plasmid were stimulated with vehicle or 5 nM dexamethasone for 24 h. The renilla luciferase reporter construct (83 ng/well) [pnull-RL, Promega Benelux BV, Leiden, The Netherlands] served as an internal control to normalize for transfection efficiency.
Cells were lysed 24–48 h after transfection and luciferase activity was measured using the Dual-Glo® Luciferase Assay according to the manufacturer’s protocol (Promega) using the Victor X4 luminometer (PerkinElmer, Waltham, MA, USA).
Statistical analysis
Baseline characteristics of the PCOS patients from the discovery and replication cohorts are presented as median and mean with SEM or indicated otherwise. Serum AMH levels were log-transformed prior to the analysis. Continuous variables were compared using an independent t-test or Mann–Whitney test, when the data were not normally distributed. Normality was tested using a Shapiro–Wilk test. Categorical variables were compared using a chi-squared test.
Linear regression analyses were performed to determine the association between SNPs in the AMH promoter region and serum AMH levels. In the discovery cohort and the internal replication cohort, these analyses were adjusted for age, BMI and TFC as these clinical parameters have previously been shown to correlate with AMH levels (Moy et al., 2015; Christiansen et al., 2016; Kruszynska and Slowinska-Srzednicka, 2017). For the regression analysis, the genetic data were added as a continuous term, assuming an additive genetic model. The threshold for the Wald test of significance was set at a P < 8.51 x 10−3, based on matrix spectral decomposition with an effective number of independent variables (VEffLi) of six (Li and Ji, 2005). This P-value correction method adjusts for the strength of LD between all polymorphisms included in the analysis. In the external replication cohort, the analysis was adjusted for age and BMI only, as data on follicle count were not available.
To determine the influence of the SNP on the PCOS phenotype, we compared phenotypic characteristics of PCOS patients who carried either one or two copies of the minor allele of the significantly associated SNP with PCOS patients homozygous for the major allele. For continuous variables, analysis of covariate variances AN(C)OVA were performed and chi-squared tests were used for categorical variables. AMH, testosterone, follicle count, FSH and LH levels were adjusted for age and were log-transformed together with BMI to approximate a normal distribution. Minor allele frequencies were compared to the Rotterdam Study population, an ongoing prospective cohort study in of the general population, by performing a chi-squared test (Ikram et al., 2020). Meta-analysis was performed using a fixed-effects model with rmeta and a sample-size weighted meta-analysis using METAL (Willer et al., 2010). The analyses were performed using Rstudio version 3.6.2 (http://www.rstudio.com/).
For the functional studies, unpaired t-test was performed to analyze the difference between group means using Prism software (GraphPad Prism 5.1 Software, Inc. San Diego, CA, USA). A P-value <0.05 was considered statistically significant.
Results
Clinical characteristics of the PCOS cohorts
Baseline characteristics of the discovery cohort and both replication cohorts are listed in Table I. In the internal validation cohort, PCOS patients had significantly lower testosterone levels compared to the discovery cohort. This can be explained by different techniques used over time for the measurement of testosterone. In the discovery cohort, a radioimmunoassay was used to measure testosterone levels, whereas in the validation cohort UPLC-MS/MS was used. Moreover, the percentage women with polycystic ovaries in the discovery cohort was slightly, but significantly lower than in the internal validation cohort (96.3% vs 98.9%, respectively).
Table I.
Clinical characteristics of the patient population in the discovery and replication cohorts.
| Discovery cohort | Internal validation cohort | External validation cohort |
P-value |
||
|---|---|---|---|---|---|
| (n = 655) | (n = 457) | (n = 321) | Discovery vs internal validation cohort | Discovery vs external validation cohort | |
| median | median | median | |||
| (mean ± SEM) | (mean ± SEM) | (mean ± SEM) | |||
| Age, years | 28.9 | 28.6 | 28.0 | 0.61 | 0.01 |
| (28.8 ± 0.19) | (28.7 ± 0.21) | (28.0 ± 0.26) | |||
| BMI, kg/m2 | 24.4 | 23.7 | 35.5 | 0.12 | 2.2 × 10−16 |
| (26.0 ± 0.23) | (25.5 ± 0.28) | (35.7 ± 0.47) | |||
| AMH, ng/ml | 8.17 | 8.42 | 10.7 | 0.88 | 3.41 × 10−5 |
| (9.82 ± 0.27) | (9.45 ± 0.27) | (12.1 ± 0.45) | |||
| Total follicle count, n | 37.0 | 39.0 | – | 0.03 | – |
| (42.3 ± 0.84) | (45.0 ± 1.03) | ||||
| Testosterone, pmol/l | 1.70 | 1.34 | – | 8.97 × 10−12 | – |
| (1.86 ± 0.03) | (1.53 ± 0.04) | ||||
| FSH | 5.80 | 5.70 | – | 0.85 | – |
| (5.83 ± 0.10) | (5.89 ± 0.13) | ||||
| LH | 7.90 | 7.00 | – | 0.06 | – |
| (9.76 ± 0.34) | (9.55 ± 0.49) | ||||
| Ovulatory dysfunction, n (%) | 647 (98.8%) | 446 (97.6%) | 321 (100%) | 0.21 | – |
| Hyperandrogenism, n (%) | 337 (52.4%) | 221 (48.5%) | 321 (100%) | 0.34 | – |
| Polycystic ovaries, n (%) | 631 (96.3%) | 452 (98.9%) | – | 0.01 | – |
AMH, anti-Müllerian hormone. Values are expressed as median (mean ± SEM) or as percentages. AMH, testosterone, follicle count, FSH and LH levels were age-adjusted. P-values were obtained by performing the Mann–Whitney U test.
In the external replication cohort, the PCOS patients were younger and had significantly higher BMI and serum AMH levels, compared to the discovery cohort. Furthermore, in line with the NIH criteria, all patients were hyperandrogenic compared to only 52.4% in the discovery cohort.
Identification and association analysis of an AMH promoter SNP
A total of 24 SNPs were identified in the Chr19:2 245 353–2 250 827 bp (hg19) region of the AMH gene. There were 13 SNPs excluded, because their imputation quality was below the threshold of 0.75. The remaining 11 SNPs were included for linear regression analyses; of these, eight SNPs were located in the promoter region and three SNPs were in exon 1 of the AMH gene (Table II). Minor allele frequencies were similar between the discovery cohort and the Rotterdam study cohort, a prospective cohort study of the general population (Ikram et al., 2020).
Table II.
Characteristics of 11 SNPs located in the AMH gene region of the discovery cohort.
| SNP | Observed MAF | Rotterdam study MAF | Position | Location | Major allele | Minor allele | P-value |
|---|---|---|---|---|---|---|---|
| rs116548172 | 0.02 | 0.02 | 2245457 | Promoter | A | G | 0.69 |
| rs45521740 | 0.06 | 0.05 | 2245622 | Promoter | G | A | 0.70 |
| rs139821116 | 0.02 | 0.02 | 2246154 | Promoter | A | G | 0.11 |
| rs4806833 | 0.08 | 0.09 | 2246403 | Promoter | A | G | 0.78 |
| rs2074861 | 0.16 | 0.16 | 2246652 | Promoter | A | G | 0.97 |
| rs4807216 | 0.15 | 0.15 | 2248683 | Promoter | T | C | 0.99 |
| rs3761018 | 0.04 | 0.05 | 2248882 | Promoter | A | G | 0.22 |
| rs10406324 | 0.04 | 0.04 | 2249112 | Promoter | A | G | 0.65 |
| rs10407022 | 0.18 | 0.18 | 2249477 | First exon | T | G | 0.99 |
| rs61736572 | 0.04 | 0.05 | 2249583 | First exon | G | A | 0.28 |
| rs61736575 | 0.04 | 0.05 | 2249634 | First exon | G | A | 0.25 |
MAF, minor allele frequency; SNP, single-nucleotide polymorphism. Eight SNPs are located in the human AMH promoter; three SNPs (rs10407022, rs61736572 and rs61736575) are located in exon 1 of the AMH gene. Position is based on build GRCh37/hg19. The Rotterdam study (RS) is an ongoing population based cohort in Rotterdam, The Netherlands. MAF are based on RS phase 1 (n = 7983). P-values were obtained by performing a chi-squared test and comparing the observed MAF with the Rotterdam study MAF.
As indicated in Table III, only SNP rs10406324 (−210 A>G) was significantly associated with log-transformed serum AMH levels. Carriers of the minor allele G had significantly lower log-transformed serum AMH levels compared to non-carriers (estimate −0.53, SE 0.10, P = 8.58 × 10−8). Each extra copy of the G allele resulted in a decrease of the log-transformed serum AMH levels by 0.53.
Table III.
Linear regression model analyses of the 11 SNPs.
| SNP | Effect allele | Effect allele frequency | Estimate | Standard error | P-value |
|---|---|---|---|---|---|
| rs116548172 | G | 0.02 | −0.17 | 0.11 | 0.12 |
| rs45521740 | A | 0.06 | −0.04 | 0.09 | 0.57 |
| rs139821116 | G | 0.02 | −0.16 | 0.12 | 0.16 |
| rs4806833 | G | 0.08 | −0.15 | 0.07 | 0.03 |
| rs2074861 | G | 0.16 | −0.08 | 0.05 | 0.12 |
| rs4807216 | C | 0.15 | −0.08 | 0.05 | 0.12 |
| rs3761018 | G | 0.04 | 0.15 | 0.11 | 0.17 |
| rs10406324 | G | 0.04 | −0.53 | 0.10 | 8.58 × 10−8 |
| rs10407022 | G | 0.18 | −0.04 | 0.05 | 0.47 |
| rs61736572 | A | 0.04 | 0.15 | 0.11 | 0.17 |
| rs61736575 | A | 0.04 | 0.14 | 0.11 | 0.17 |
SNP, single-nucleotide polymorphism. Linear regression models included log-transformed serum AMH levels as dependent variable and age, BMI, follicle count and genotype (continuous term) as independent variables. An additive genetic model was assumed. P-values are obtained by performing Wald test of significance. A Veffli corrected P-value below <0.000851 was considered as significant.
We also performed an allele carrier model analysis. In accordance with the regression analysis, carriers of the rs10406324 G allele had significantly lower serum AMH levels corrected for age compared to non-carriers (respectively 10.0 ± 0.29 ng/ml vs 7.69 ± 0.86 ng/ml, P = 1.19 × 10−3) (Table IV).
Table IV.
Characteristics of the carrier allele model analysis in the discovery cohort.
| Discovery cohort | |||
|---|---|---|---|
| AA | AG + GG | P-value | |
| n = 602 | n = 53 | ||
| median | median | ||
| (mean ± SEM) | (mean ± SEM) | ||
| AMH, ng/ml | 8.37 | 5.57 | 1.19 × 10−3 |
| (10.0 ± 0.29) | (7.69 ± 0.86) | ||
| Age, years | 28.8 | 29.8 | 0.53 |
| (28.8 ± 0.20) | (29.2 ± 0.65) | ||
| BMI, kg/m2 | 25.9 | 25.9 | 0.18 |
| (25.9 ± 0.24) | (26.9 ± 0.79) | ||
| Total follicle number | 37.0 | 42.0 | 0.25 |
| (42.2 ± 0.89) | (43.6 ± 2.41) | ||
| Testosterone, pmol/l | 1.70 | 1.60 | 0.32 |
| (1.87 ± 0.03) | (1.76 ± 0.12) | ||
| FSH | 5.80 | 6.20 | 0.92 |
| (5.83 ± 0.10) | (5.82 ± 0.28) | ||
| LH | 7.90 | 7.10 | 0.54 |
| (9.75 ± 0.36) | (9.89 ± 1.12) | ||
| LH/FSH | 1.47 | 1.34 | 0.46 |
| (1.77 ± 0.05) | (1.64 ± 0.14) | ||
| Ovulatory dysfunction, n (%) | 595 (98.8%) | 52 (98.1%) | 1 |
| Hyperandrogenism, n (%) | 309 (51.3%) | 28 (52.8%) | 0.95 |
| Polycystic ovaries, n (%) | 579 (96.2%) | 52 (98.1%) | 0.74 |
AMH, anti-Müllerian hormone. Values are expressed as median (mean ± SEM) or as percentages. P-values were obtained by performing ANCOVA with age as the covariate.
Replication of the AMH promoter SNP rs10406324
We have replicated the AMH promoter polymorphism rs10406324 using an internal and an independent external replication PCOS cohort. In both cohorts, PCOS patients carrying the rs10406324 G allele had significantly lower serum AMH levels compared to non-carriers (estimate −0.39, SE 0.12, P = 1.35 × 10−3 and estimate −0.60, SE 0.18, P = 1.24 × 10−3, respectively). In addition, allele carrier model analysis in the replication cohorts also confirmed that carriers of the G allele had significantly lower log-transformed serum AMH levels compared to non-carriers (9.70 ± 0.29 ng/ml vs 5.89 ± 0.62 ng/ml, P = 1.59 × 10−4 and 12.4 ± 0.47 ng/ml vs 8.22 ± 1.51 ng/ml, P = 6.45 × 10−3, respectively) (Table V). The MAF of polymorphism rs10406324 in the discovery cohort did not significantly differ between the internal and external replication cohorts (P = 0.035 and P = 0.042, respectively). Comparison with the MAF of the polymorphism rs10406324 reported in Northern and Western European populations also did not show any difference with our PCOS cohort (1000 Genomes Project Consortium et al., 2015). This suggests that the prevalence of the variant rs10406324 in women with PCOS is similar to the Western European population.
Table V.
Characteristics of the carrier model analysis in both replication cohorts.
| Internal validation cohort |
External validation cohort |
|||||
|---|---|---|---|---|---|---|
| AA | AG + GG | P-value | AA | AG + GG | P-value | |
| n = 427 | n = 30 | n = 300 | n = 21 | |||
| median (mean ± SEM) | median (mean ± SEM) | median (mean ± SEM) | median (mean ± SEM) | |||
| AMH, ng/ml | 8.92 | 5.26 | 1.59 × 10−4 | 10.9 | 5.73 | 6.45 × 10−3 |
| (9.70 ± 0.29) | (5.89 ± 0.62) | (12.4 ± 0.47) | (8.22 ± 1.51) | |||
| Age, years | 28.7 | 27.2 | 0.52 | 28.0 | 26.0 | 0.08 |
| (28.7 ± 0.21) | (28.1 ± 1.00) | (28.1 ± 0.27) | (26.3 ± 0.91) | |||
| BMI, kg/m2 | 23.5 | 25.2 | 0.11 | 35.5 | 35.5 | 0.63 |
| (25.4 ± 0.29) | (27.0 ± 0.98) | (35.8 ± 0.5) | (34.9 ± 1.6) | |||
| Total follicle number | 40.0 | 36.5 | 0.16 | – | – | – |
| (45.3 ± 1.08) | (39.0 ± 2.78) | |||||
| Testosterone, pmol/l | 1.35 | 1.31 | 0.44 | – | – | – |
| (1.53 ± 0.04) | (1.55 ± 0.19) | |||||
| FSH | 5.80 | 5.30 | 0.44 | – | – | – |
| (5.94 ± 0.14) | (5.22 ± 0.32) | |||||
| LH | 7.20 | 5.58 | 0.05 | – | – | – |
| (9.76 ± 0.52) | (6.55 ± 0.81) | |||||
| LH/FSH | 1.34 | 1.19 | 0.09 | |||
| (1.70 ± 0.07) | (1.25 ± 0.13) | |||||
| Ovulatory dysfunction, n (%) | 416 (97.4%) | 30 (100%) | 0.78 | – | – | – |
| Hyperandrogenism, n (%) | 208 (48.7%) | 13 (43.3%) | 0.70 | – | – | – |
| Polycystic ovaries, n (%) | 423 (99.1%) | 29 (96.7%) | 0.76 | – | – | – |
AMH, anti-Müllerian hormone. Values are expressed as median (mean ± SEM) or as percentages. P-values were obtained by performing ANCOVA with age as the covariate.
Effect of the AMH promoter SNP rs10406324 in normo-ovulatory women
To determine whether the observed association was PCOS-specific, we next assessed the association of polymorphism rs10406324 with serum AMH levels in a relatively large AMH GWAS analysis including normo-ovulatory women. Similar to the observation in the PCOS cohorts, normo-ovulatory women carrying the minor allele G had significantly lower circulating AMH levels compared to non-carriers (estimate = −0.16, SE = 0.04, P = 1.04 × 10−5). In this cohort, the MAF of polymorphism rs10406324 was 0.04, which is similar to the MAF observed in our PCOS cohorts.
Meta-analysis
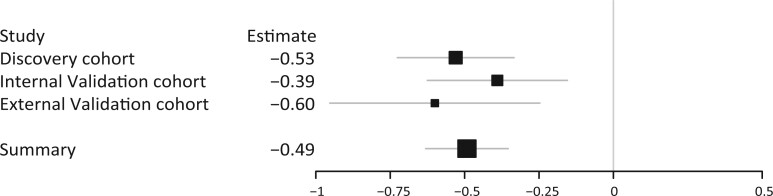
A fixed effect model of all three PCOS cohorts combined showed that carriers of the G allele have significantly lower serum AMH levels (estimate = −0.49, SE = 0.07, P = 3.24 × 10−12) (Fig. 1). A meta-analysis using a random effect model was not performed, as the P-value for heterogeneity was not significant (P = 0.54). Meta-analysis using a sample-size weighted model of all PCOS cohorts and the control cohort combined resulted in a P-value of P = 5.16 × 10−12.
Figure 1.
Forrest plot of the fixed-effects meta-analysis. Forrest plot of the three different PCOS cohorts together with the summary statistic. The size of the squares reflect the weight of the cohort in the combined analysis. The large square represents the overall summary effect and the gray lines represent the CI. PCOS, polycystic ovary syndrome.
Phenotypic analysis of the AMH promoter SNP rs10406324
In order to explain the observed association between the AMH promoter SNP rs10406324 and AMH levels, we investigated its association with phenotypic characteristics of PCOS. In our discovery PCOS cohort, neither follicle count nor any of the other quantitative traits (age, BMI, testosterone, FSH, LH, LH/FSH, OD, HA and PCOM) were significantly different between carriers and non-carriers of the minor allele G (Table IV). Also in both replication cohorts, no additional associations were observed (Table V).
Next, we determined whether the association of rs10406324 with AMH levels was PCOS phenotype specific. Phenotypes B and C were excluded from further analysis as these groups only contained one heterozygous carrier of the minor allele G. The MAF of rs10406324 stratified by phenotype was 0.04 in both Phenotypes A and D (P = 0.18) (Table VI). Comparing Phenotypes A and D showed that patients with Phenotype A had the highest serum AMH levels (11.3 ± 0.47 ng/ml vs 8.90 ± 0.31 ng/ml, respectively, P = 5.90 × 10−5). Moreover, only within Phenotype A, carriers of the minor allele had significantly lower serum AMH levels compared to non-carriers (P = 1.46 × 10−4 versus P = 0.11) (Table VI). Since Phenotypes A and D differ in the presence of HA, we also performed a stratification by presence of HA, comparing patients in Phenotype D with patient in Phenotypes A, B and C together. This yielded similar results, with significantly lower serum AMH levels in carriers of the minor allele G only in patients with HA (P = 4.55 × 10−3 versus P = 0.11). Stratification between lean (<25 kg/m2) and obese (>30 kg/m2) patients did not yield significant differences (data not shown).
Table VI.
Stratification by PCOS phenotype in the discovery cohort.
| Phenotype | AMH | N. | MAF | AA | AG + GG | P-value |
|---|---|---|---|---|---|---|
| (ng/ml) | AA/AG + GG | AMH (ng/ml) | AMH (ng/ml) | |||
| median (mean ± SEM) | median (mean ± SEM) | median (mean ± SEM) | ||||
| A (OD/HA/PCOM) | 9.36 | 279/26 | 0.04 | 9.49 | 6.84 | 1.46 × 10−4 |
| (11.3 ± 0.47) | (11.6 ± 0.49) | (8.64 ± 1.44) | ||||
| B (OD/HA) | 2.77 | 23/1 | 0.02 | 2.65 | 4.4 | – |
| (3.51 ± 0.64) | (3.48 ± 0.67) | |||||
| C (HA/PCOM) | 8.08 | 7/1 | 0.06 | 8.91 | 1.54 | – |
| (6.93 ± 1.42) | (7.70 ± 1.37) | |||||
| D (OD/PCOM) | 7.36 | 393/25 | 0.04 | 7.69 | 5.57 | 0.11 |
| (8.90 ± 0.31) | (9.05 ± 0.32) | (7.07 ± 1.02) |
AMH, anti-Müllerian hormone; HA, biochemical hyperandrogenism; MAF, minor allele frequency; OD, ovulatory dysfunction; PCOM, polycystic ovarian morphology. Comparison of the AMH levels between carriers and non-carriers of the SNP rs10406324 stratified by PCOS phenotype. Values are expressed as median (mean ± SEM) or as percentages or indicated otherwise. P-values are obtained by performing ANCOVA with age as the covariate.
TF binding motif prediction
The SNP rs10406324 was not in strong LD (r2 > 0.75) with any other SNP in a region of 50kb surrounding the polymorphism, enhancing the probability of it being a causal SNP. The CADD C-score to evaluate the severity of the deleteriousness of the SNP was 8.61, meaning that the SNP rs10406324 is not predicted to be deleterious (Rentzsch et al., 2019).
In addition, motif binding affinity analysis was performed to predict whether the polymorphism rs10406324 introduces or changes a TF binding site (TFBS) in the AMH promoter. According to this analysis, the variant is predicted to reduce the binding affinity of SF1, GR and estrogen-related receptor alpha (ESRRA). Binding of SF1 is predicted to decrease in log odds from 11.6 to 8.6, binding of GR is predicted to decrease from −18.6 to −24.6, and binding of ESRRA is predicted to decrease from −12.2 to −35.2. The predicted change of binding affinity of ESRRA was borderline significant (P = 0.0004), however since ESRRA is expressed in ovarian tissue and has been shown to bind the SF1 TFBS (Bonnelye et al., 1997), we decided to include this TF in the subsequent analyses.
Functional analysis of SF1, GR and ESRRA on hAMH promoter activity
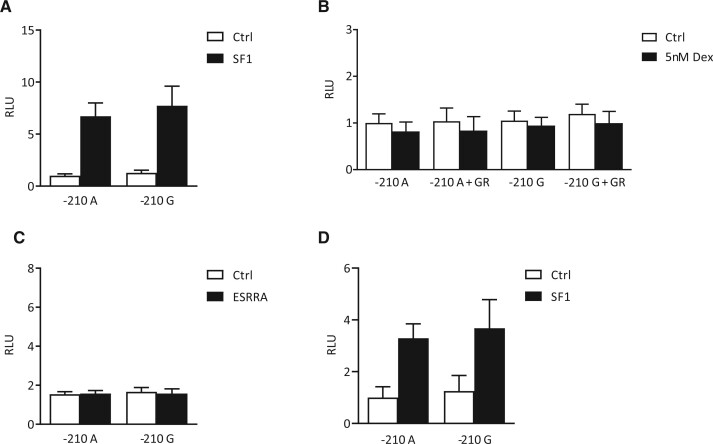
To study the effect of the SNP rs10406324 in vitro, the variant was introduced into a human AMH promoter reporter construct. Transfection of the hAMH promoter constructs into KK1 and COV434 cells did not show differences in basal activity between the −210 G variant compared to the −210 A variant (P = 0.41 and P = 0.76, respectively) (Figure 2A and 2D). Co-transfection with SF1 increased the activity of both the −210 A and the −210 G AMH promoter reporter to a similar extent (P = 0.68 and P = 0.74, respectively) (Figure 2A and 2D).
Figure 2.
Luciferase reporter assay of the effect of the polymorphism rs10406324 on human AMH promoter activity. −210 A, human AMH promoter construct containing −210 A allele; −210 G, human AMH promoter construct containing −210 G allele; dex, dexamethasone; ESRRA, estrogen-related receptor alpha; GR, glucocorticoid receptor; RLU, relative luciferase unit; SF1, steroidogenic factor 1. Luciferase reporter assay to compare the effect of SF1 in KK1 cells [A], GR with and without stimulation of 5 nM dexamethasone in KK1 cells [B], ESRRA binding in KK1 cells [C] and the effect of SF1 in COV434 cells [D] on the activity of the human AMH promoter containing the −210 A allele and the human AMH promoter containing the −210 G allele. Values are expressed by luminescence rate. P-values were obtained by performing an unpaired t-test. All comparisons between −210 A and −210 G were non-significant (P > 0.05).
The AMH promoter is known to contain two SF1 binding sites and based on previously published studies, the SNP rs10406324 is located in the distal TFBS of SF1 (Jin et al., 2016; Schteingart et al., 2019). Therefore, we mutated the proximal SF1 binding site, to investigate the effect of rs10406324 on SF1-induced AMH promoter activity after binding only the distal site. In the presence of a mutated proximal SF1 binding site, basal and SF1-induced −210 A and −210 G AMH promoter activity also did not differ (data not shown). Likewise, co-transfection with GR with or without dexamethasone stimulation and co-transfection with ESRRA in KK1 cells did not show any differences between the −210 A and the −210 G AMH promoter activity (P = 0.73, P = 0.71 and P = 0.98, respectively) (Figure 2B and 2C).
Discussion
We report a novel AMH promoter polymorphism rs10406324 (−210 A>G) to be associated with serum AMH levels in patients with PCOS. Carriers of the minor allele G have significantly lower serum AMH levels in both linear regression model analysis as well as in carrier allele model analysis. These results were confirmed in an internal as well as an external replication cohort and in the subsequent meta-analysis. Importantly, the number of antral follicles, assessed by ultrasound, did not differ between carriers and non-carriers of polymorphism rs10406324. This strongly suggests that genetic differences in the AMH promoter may lead to a differential regulation of AMH expression and contribute to observed variations in serum AMH levels in otherwise phenotypically similar PCOS patients.
Previous genetic studies have also identified polymorphisms located in the AMH gene in relation to AMH levels. A GWAS meta-analysis on serum AMH levels in 3344 normo-ovulatory women of European ancestry did not find any novel polymorphisms besides the known variant rs16991615 in the MCM8 gene (Ruth et al., 2019). However, a more recent large GWAS meta-analysis on circulating AMH levels in a cohort of 7049 normo-ovulatory women of European ancestry, did find three novel signals of which one was located in the AMH gene (rs10417628) (Verdiesen et al., 2022). However, the rs10417628 variant has been shown to display loss of immunoreactivity, depending on the AMH ELISA used (Hoyos et al., 2020), which could explain the observed association. Both of these large GWAS studies on AMH levels in normo-ovulatory women did not report the rs10406324 polymorphism as being genome-wide significant (Ruth et al., 2019; Verdiesen et al., 2022). However, when specifically looking up this polymorphism in the GWAS summary statistics of Verdiesen et al. (2022), we observed that, similar to PCOS patients, normo-ovulatory women carrying the minor allele G have lower AMH levels compared to non-carriers. This suggests that polymorphism rs10406324 is an AMH level regulating variant in women, irrespective of the clinical background.
Recent studies hypothesize that PCOS may constitute different genetic diseases, showing different clinical phenotypes (Dapas et al., 2020). Particularly for Phenotypes A and D, a different genetic background may explain the presence or absence of HA. In our study, stratification by phenotype did not show any difference in MAF for rs10406324, thus this SNP is not a risk allele for a particular phenotype. However, only within Phenotype A, carriers and non-carriers had significantly different AMH levels, with carriers having lower levels. Unfortunately, Phenotypes B and C could not be analyzed as the number of carriers were too low. However, stratification by HA yielded similar results with the highest serum AMH levels in non-carriers. These results suggest that in PCOS, factors that lead to HA may contribute to the regulation of AMH expression.
The effect of androgens on AMH expression in granulosa cells has been investigated in several studies, yielding contradictory results (Zhang et al., 2016; Pierre et al., 2017; Dilaver et al., 2019; Dewailly et al., 2020). The only study performed in luteinized granulosa cells of women with PCOS showed that AMH mRNA expression was increased after stimulation with 5α-DHT in women with PCOS, but not in normo-ovulatory women (Pierre et al., 2017). The exact mechanism for this differential effect of androgens on AMH expression remains to be elucidated. However, these studies suggest that in PCOS, the transcriptional regulation of the AMH promoter is altered. Aberrant expression of TFs and/or epigenetic changes could be potential mechanisms explaining this difference. Binding sites for several transcriptions factors, such as SF1, GATA4, SOX9, WT-1 and FOXL2 have been identified in the human AMH promoter (Shen et al., 1994; Watanabe et al., 2000; Rey et al., 2003; Jin et al., 2016; Schteingart et al., 2019). Especially an intact distal SF1 binding site has been shown to be crucial for human AMH promoter activity using a Sertoli cell line (Schteingart et al., 2019). Expression quantitative trait locus data, available in the Genotype-Tissue Expression (GTEx) project (The GTEx Consortium, 2015), for the variant rs10406324 is scarce. An associated with AMH expression in several tissues is reported, but not in known AMH expressing tissues (The GTEx Consortium, 2015). Our in silico analysis showed that rs10406324 is located at the distal SF1 binding site. A decrease in binding affinity of SF1 to this binding motif was predicted in the presence of the minor allele G. However, subsequent in vitro results did not show a significant difference in basal or SF1-induced AMH promoter activity using promoter constructs containing either the major or minor allele of the SNP rs10406324.
Moreover, our in silico analysis predicted that the SNP rs10406324 may impact the binding of the TFs GR and ESRRA. A role for these TFs in the regulation of AMH expression has not been studied yet. Interestingly, ESRRA, also known as estrogen-related receptor 1 (ERR-1), is known to have affinity for the SF1 binding site (Bonnelye et al., 1997). However, in our in vitro study, ESRRA did not have a direct effect on AMH promoter activity.
It is plausible that the functional effects could not be demonstrated as both cell lines may lack the proper follicle-stage specific environment. Furthermore, the use of AMH promoter constructs may limit the assessment of the impact of the polymorphism on higher order structures of the chromatin. It is plausible that the polymorphism impacts the mode of action of transcription on the 3D genome organization (Kim and Shendure, 2019). Human granulosa cells generated from induced pluripotent stem cells, combined with gene editing, may provide a more accurate model to study the functional mechanism of the polymorphism. Alternatively, it is possible that polymorphism rs10406324 is not the lead SNP affecting regulation of AMH expression. In our study, we have not detected other variants in LD with the polymorphism within a range of 50kb. However, variants further up- or downstream that may induce the observed effects cannot be ruled out.
To conclude, we have identified an AMH promoter polymorphism rs10406324 (−210 A>G) that is associated with serum AMH levels in women. Carriers of the minor allele G had significantly lower serum AMH levels. Importantly, the observed association was independent of total follicle number and other PCOS parameters. These results suggest that variations in serum AMH levels are not only caused by differences in follicle number but also by genetic factors. This should be taken into account when serum AMH levels are considered as a marker for the PCOM phenotype. Results of future GWAS studies on serum AMH levels will therefore be of interest to identify additional genetic variants that influence AMH expression.
Data availability
The data underlying this article cannot be shared publicly due to the privacy of individuals who participated in the study. The data will be shared on reasonable request to the corresponding author.
Authors’ roles
L.M.E.M., Y.V.L., J.S.E.L. and J.A.V. were involved in the initial study design. L.B. and A.G.U. (discovery cohort and internal validation cohort), R.M.G.V. (control cohort), R.K.S. and A.D. (external validation cohort) and A.Mc.L. (in vitro analysis) were involved in the acquisition of (part of) the data. L.M.E.M. carried out the analyses and drafted the article. All authors were involved in the interpretation of the results, revising and approving the manuscript.
Funding
No external funding was received for this study.
Conflict of interest
J.S.E.L. has received consultancy fees from the following companies: Ferring, Roche Diagnostics and Ansh Labs and has received travel reimbursement from Ferring. J.A.V. has received royalties from AMH assays, paid to the institute/lab with no personal financial gain.
Contributor Information
Loes M E Moolhuijsen, Department of Internal Medicine, Erasmus MC, University Medical Center, Rotterdam, the Netherlands.
Yvonne V Louwers, Division of Reproductive Endocrinology and Infertility, Department of Obstetrics and Gynaecology, Erasmus MC, University Medical Center, Rotterdam, the Netherlands.
Anke McLuskey, Department of Internal Medicine, Erasmus MC, University Medical Center, Rotterdam, the Netherlands.
Linda Broer, Department of Internal Medicine, Erasmus MC, University Medical Center, Rotterdam, the Netherlands.
Andre G Uitterlinden, Department of Internal Medicine, Erasmus MC, University Medical Center, Rotterdam, the Netherlands.
Renée M G Verdiesen, Division of Molecular Pathology, The Netherlands Cancer Institute—Antoni van Leeuwenhoek Hospital, Amsterdam, the Netherlands; Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht University, Utrecht, the Netherlands.
Ryan K Sisk, Division of Endocrinology, Metabolism, and Molecular Medicine, Department of Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
Andrea Dunaif, Division of Endocrinology, Diabetes and Bone Disease, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Joop S E Laven, Division of Reproductive Endocrinology and Infertility, Department of Obstetrics and Gynaecology, Erasmus MC, University Medical Center, Rotterdam, the Netherlands.
Jenny A Visser, Department of Internal Medicine, Erasmus MC, University Medical Center, Rotterdam, the Netherlands.
References
- Alebic MS, Stojanovic N, Dewailly D.. Discordance between serum anti-Mullerian hormone concentrations and antral follicle counts: not only technical issues. Hum Reprod 2018;33:1141–1148. [DOI] [PubMed] [Google Scholar]
- Bhide P, Dilgil M, Gudi A, Shah A, Akwaa C, Homburg R.. Each small antral follicle in ovaries of women with polycystic ovary syndrome produces more antimullerian hormone than its counterpart in a normal ovary: an observational cross-sectional study. Fertil Steril 2015;103:537–541. [DOI] [PubMed] [Google Scholar]
- Bonnelye E, Vanacker JM, Dittmar T, Begue A, Desbiens X, Denhardt DT, Aubin JE, Laudet V, Fournier B.. The ERR-1 orphan receptor is a transcriptional activator expressed during bone development. Mol Endocrinol 1997;11:905–916. [DOI] [PubMed] [Google Scholar]
- Bozdag G, Mumusoglu S, Zengin D, Karabulut E, Yildiz BO.. The prevalence and phenotypic features of polycystic ovary syndrome: a systematic review and meta-analysis. Hum Reprod 2016;31:2841–2855. [DOI] [PubMed] [Google Scholar]
- Bui HN, Sluss PM, Hayes FJ, Blincko S, Knol DL, Blankenstein MA, Heijboer AC.. Testosterone, free testosterone, and free androgen index in women: reference intervals, biological variation, and diagnostic value in polycystic ovary syndrome. Clin Chim Acta 2015;450:227–232. [DOI] [PubMed] [Google Scholar]
- Campbell LA, Faivre EJ, Show MD, Ingraham JG, Flinders J, Gross JD, Ingraham HA.. Decreased recognition of SUMO-sensitive target genes following modification of SF-1 (NR5A1). Mol Cell Biol 2008;28:7476–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate RL, Mattaliano RJ, Hession C, Tizard R, Farber NM, Cheung A, Ninfa EG, Frey AZ, Gash DJ, Chow EP.. Isolation of the bovine and human genes for Mullerian inhibiting substance and expression of the human gene in animal cells. Cell 1986;45:685–698. [DOI] [PubMed] [Google Scholar]
- Catteau-Jonard S, Jamin SP, Leclerc A, Gonzales J, Dewailly D, di Clemente N.. Anti-Mullerian hormone, its receptor, FSH receptor, and androgen receptor genes are overexpressed by granulosa cells from stimulated follicles in women with polycystic ovary syndrome. J Clin Endocrinol Metab 2008;93:4456–4461. [DOI] [PubMed] [Google Scholar]
- Christiansen SC, Eilertsen TB, Vanky E, Carlsen SM.. Does AMH reflect follicle number similarly in women with and without PCOS? PLoS One 2016;11:e0146739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapas M, Lin FTJ, Nadkarni GN, Sisk R, Legro RS, Urbanek M, Hayes MG, Dunaif A.. Distinct subtypes of polycystic ovary syndrome with novel genetic associations: an unsupervised, phenotypic clustering analysis. PLoS Med 2020;17:e1003132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day F, Karaderi T, Jones MR, Meun C, He C, Drong A, Kraft P, Lin N, Huang H, Broer L. et al. Large-scale genome-wide meta-analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnosis criteria. PLoS Genet 2018;14:e1007813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewailly D. Diagnostic criteria for PCOS: Is there a need for a rethink? Best Pract Res Clin Obstet Gynaecol 2016;37:5–11. [DOI] [PubMed] [Google Scholar]
- Dewailly D, Barbotin AL, Dumont A, Catteau-Jonard S, Robin G.. Role of anti-Mullerian hormone in the pathogenesis of polycystic ovary syndrome. Front Endocrinol (Lausanne) 2020;11:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewailly D, Lujan ME, Carmina E, Cedars MI, Laven J, Norman RJ, Escobar-Morreale HF.. Definition and significance of polycystic ovarian morphology: a task force report from the Androgen Excess and Polycystic Ovary Syndrome Society. Hum Reprod Update 2014;20:334–352. [DOI] [PubMed] [Google Scholar]
- Dilaver N, Pellatt L, Jameson E, Ogunjimi M, Bano G, Homburg R, D Mason H, Rice S.. The regulation and signalling of anti-Mullerian hormone in human granulosa cells: relevance to polycystic ovary syndrome. Hum Reprod 2019;34:2467–2479. [DOI] [PubMed] [Google Scholar]
- Garg D, Tal R.. The role of AMH in the pathophysiology of polycystic ovarian syndrome. Reprod Biomed Online 2016;33:15–28. [DOI] [PubMed] [Google Scholar]
- 1000 Genomes Project ConsortiumAuton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsic LK, Dapas M, Legro RS, Hayes MG, Urbanek M.. Functional genetic variation in the anti-mullerian hormone pathway in women with polycystic ovary syndrome. J Clin Endocrinol Metab 2019;104:2855–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsic LK, Kosova G, Werstein B, Sisk R, Legro RS, Hayes MG, Teixeira JM, Dunaif A, Urbanek M.. Pathogenic anti-mullerian hormone variants in polycystic ovary syndrome. J Clin Endocrinol Metab 2017;102:2862–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck S, Kullmann M, Gast A, Ponta H, Rahmsdorf HJ, Herrlich P, Cato AC.. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J 1994;13:4087–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyos LR, Visser JA, McLuskey A, Chazenbalk GD, Grogan TR, Dumesic DA.. Loss of anti-Mullerian hormone (AMH) immunoactivity due to a homozygous AMH gene variant rs10417628 in a woman with classical polycystic ovary syndrome (PCOS). Hum Reprod 2020;35:2294–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida M, Nemoto S, Finkel T.. Identification of a specific molecular repressor of the peroxisome proliferator-activated receptor gamma Coactivator-1 alpha (PGC-1alpha). J Biol Chem 2002;277:50991–50995. [DOI] [PubMed] [Google Scholar]
- Ikram MA, Brusselle G, Ghanbari M, Goedegebure A, Ikram MK, Kavousi M, Kieboom BCT, Klaver CCW, de Knegt RJ, Luik AI. et al. Objectives, design and main findings until 2020 from the Rotterdam Study. Eur J Epidemiol 2020;35:483–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Won M, Park SE, Lee S, Park M, Bae J.. FOXL2 is an essential activator of SF-1-induced transcriptional regulation of anti-mullerian hormone in human granulosa cells. PLoS One 2016;11:e0159112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kananen K, Markkula M, Rainio E, Su JG, Hsueh AJ, Huhtaniemi IT.. Gonadal tumorigenesis in transgenic mice bearing the mouse inhibin alpha-subunit promoter/simian virus T-antigen fusion gene: characterization of ovarian tumors and establishment of gonadotropin-responsive granulosa cell lines. Mol Endocrinol 1995;9:616–627. [DOI] [PubMed] [Google Scholar]
- Kedem A, Hourvitz A, Yung Y, Shalev L, Yerushalmi GM, Kanety H, Hanochi M, Maman E.. Anti-Mullerian hormone (AMH) downregulation in late antral stages is impaired in PCOS patients. A study in normo-ovulatory and PCOS patients undergoing in vitro maturation (IVM) treatments. Gynecol Endocrinol 2013;29:651–656. [DOI] [PubMed] [Google Scholar]
- Kevenaar ME, Laven JS, Fong SL, Uitterlinden AG, de Jong FH, Themmen AP, Visser JA.. A functional anti-mullerian hormone gene polymorphism is associated with follicle number and androgen levels in polycystic ovary syndrome patients. J Clin Endocrinol Metab 2008;93:1310–1316. [DOI] [PubMed] [Google Scholar]
- Kheradpour P, Kellis M.. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res 2014;42:2976–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Shendure J.. Mechanisms of interplay between transcription factors and the 3D genome. Mol Cell 2019;76:306–319. [DOI] [PubMed] [Google Scholar]
- Kristensen SG, Kumar A, Kalra B, Pors SE, Botkjaer JA, Mamsen LS, Colmorn LB, Fedder J, Ernst E, Owens L. et al. Quantitative differences in TGF-beta family members measured in small antral follicle fluids from women with or without PCO. J Clin Endocrinol Metab 2019;104:6371–6384. [DOI] [PubMed] [Google Scholar]
- Kruszynska A, Slowinska-Srzednicka J.. Anti-mullerian hormone (AMH) as a good predictor of time of menopause. Prz Menopauzalny 2017;16:47–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laven JS, Mulders AG, Visser JA, Themmen AP, De Jong FH, Fauser BC.. Anti-Mullerian hormone serum concentrations in and anovulatory women of reproductive age. J Clin Endocrinol Metab 2004;89:318–323. [DOI] [PubMed] [Google Scholar]
- Li J, Ji L.. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 2005;95:221–227. [DOI] [PubMed] [Google Scholar]
- Lie Fong S, Laven JSE, Duhamel A, Dewailly D.. Polycystic ovarian morphology and the diagnosis of polycystic ovary syndrome: redefining threshold levels for follicle count and serum anti-Mullerian hormone using cluster analysis. Hum Reprod 2017;32:1723–1731. [DOI] [PubMed] [Google Scholar]
- Lizneva D, Suturina L, Walker W, Brakta S, Gavrilova-Jordan L, Azziz R.. Criteria, prevalence, and phenotypes of polycystic ovary syndrome. Fertil Steril 2016;106:6–15. [DOI] [PubMed] [Google Scholar]
- Moy V, Jindal S, Lieman H, Buyuk E.. Obesity adversely affects serum anti-mullerian hormone (AMH) levels in Caucasian women. J Assist Reprod Genet 2015;32:1305–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neven ACH, Laven J, Teede HJ, Boyle JA.. A summary on polycystic ovary syndrome: diagnostic criteria, prevalence, clinical manifestations, and management according to the Latest International Guidelines. Semin Reprod Med 2018;36:5–12. [DOI] [PubMed] [Google Scholar]
- Park AS, Lawson MA, Chuan SS, Oberfield SE, Hoeger KM, Witchel SF, Chang RJ.. Serum anti-mullerian hormone concentrations are elevated in oligomenorrheic girls without evidence of hyperandrogenism. J Clin Endocrinol Metab 2010;95:1786–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Suh DS, Lee K, Bae J.. Positive cross talk between FOXL2 and antimullerian hormone regulates ovarian reserve. Fertil Steril 2014;102:847–855.e1. [DOI] [PubMed] [Google Scholar]
- Pierre A, Taieb J, Giton F, Grynberg M, Touleimat S, El Hachem H, Fanchin R, Monniaux D, Cohen-Tannoudji J, di Clemente N. et al. Dysregulation of the anti-mullerian hormone system by steroids in women with polycystic ovary syndrome. J Clin Endocrinol Metab 2017;102:3970–3978. [DOI] [PubMed] [Google Scholar]
- Pigny P, Merlen E, Robert Y, Cortet-Rudelli C, Decanter C, Jonard S, Dewailly D.. Elevated serum level of anti-mullerian hormone in patients with polycystic ovary syndrome: relationship to the ovarian follicle excess and to the follicular arrest. J Clin Endocrinol Metab 2003;88:5957–5962. [DOI] [PubMed] [Google Scholar]
- Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M.. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 2019;47:D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey R, Lukas-Croisier C, Lasala C, Bedecarras P.. AMH/MIS: what we know already about the gene, the protein and its regulation. Mol Cell Endocrinol 2003;211:21–31. [DOI] [PubMed] [Google Scholar]
- Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril 2004;81:19–25. [DOI] [PubMed] [Google Scholar]
- Ruth KS, Soares ALG, Borges MC, Eliassen AH, Hankinson SE, Jones ME, Kraft P, Nichols HB, Sandler DP, Schoemaker MJ. et al. Genome-wide association study of anti-Mullerian hormone levels in pre-menopausal women of late reproductive age and relationship with genetic determinants of reproductive lifespan. Hum Mol Genet 2019;28:1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schteingart HF, Picard JY, Valeri C, Marshall I, Treton D, di Clemente N, Rey RA, Josso N.. A mutation inactivating the distal SF1 binding site on the human anti-Mullerian hormone promoter causes persistent Mullerian duct syndrome. Hum Mol Genet 2019;28:3211–3218. [DOI] [PubMed] [Google Scholar]
- Shen WH, Moore CC, Ikeda Y, Parker KL, Ingraham HA.. Nuclear receptor steroidogenic factor 1 regulates the mullerian inhibiting substance gene: a link to the sex determination cascade. Cell 1994;77:651–661. [DOI] [PubMed] [Google Scholar]
- Shin S, Hudson R, Harrison C, Craven M, Keleş S.. atSNP Search: a web resource for statistically evaluating influence of human genetic variation on transcription factor binding. Bioinformatics 2019;35:2657–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A. et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- van Santbrink EJ, Hop WC, Fauser BC.. Classification of normogonadotropic infertility: polycystic ovaries diagnosed by ultrasound versus endocrine characteristics of polycystic ovary syndrome. Fertil Steril 1997;67:452–458. [DOI] [PubMed] [Google Scholar]
- Verdiesen RMG, van der Schouw YT, van Gils CH, Verschuren WMM, Broekmans FJM, Borges MC, Goncalves Soares AL, Lawlor DA, Eliassen AH, Kraft P. et al. Genome-wide association study meta-analysis identifies three novel loci for circulating anti-Mullerian hormone levels in women. Hum Reprod 2022;37:1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser JA, de Jong FH, Laven JS, Themmen AP.. Anti-Mullerian hormone: a new marker for ovarian function. Reproduction 2006;131:1–9. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Clarke TR, Lane AH, Wang X, Donahoe PK.. Endogenous expression of Mullerian inhibiting substance in early postnatal rat sertoli cells requires multiple steroidogenic factor-1 and GATA-4-binding sites. Proc Natl Acad Sci USA 2000;97:1624–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weenen C, Laven JS, Von Bergh AR, Cranfield M, Groome NP, Visser JA, Kramer P, Fauser BC, Themmen AP.. Anti-mullerian hormone expression pattern in the human ovary: potential implications for initial and cyclic follicle recruitment. Mol Hum Reprod 2004;10:77–83. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR.. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG. et al. Ensembl 2018. Nucleic Acids Res 2018;46:D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang SF, Zheng JD, Zhao CB, Zhang YN, Liu LL, Huang JH.. Effects of testosterone on the expression levels of AMH, VEGF and HIF-1alpha in mouse granulosa cells. Exp Ther Med 2016;12:883–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo C, Shin S, Keleş S.. atSNP: transcription factor binding affinity testing for regulatory SNP detection. Bioinformatics 2015;31:3353–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article cannot be shared publicly due to the privacy of individuals who participated in the study. The data will be shared on reasonable request to the corresponding author.