Abstract
We have developed two sets of Campylobacter shuttle vectors containing either the gfp (green fluorescent protein), yfp (yellow fluorescent protein), or cfp (cyan fluorescent protein) reporter gene. In one set, the reporter gene is fused to a consensus Campylobacter promoter sequence (Pc). The other set contains a pUC18 multicloning site upstream of the reporter gene, allowing the construction of transcriptional fusions using known promoters or random genomic fragments. C. jejuni cells transformed with the Pc fusion plasmids are strongly fluorescent and easily visualized on chicken skin, on plant tissue, and within infected Caco-2 cells. In each C. jejuni strain tested, these plasmids were maintained over several passages in the absence of antibiotic selection. Also, in many C. jejuni strains, >91% of the cells transformed with the Pc fusion plasmids remained fluorescent after several days. Experiments with yellow fluorescent and cyan fluorescent C. jejuni transformants suggest that aggregates containing two or more strains of C. jejuni may be present in an enrichment broth culture. Colonies arising from these aggregates would be heterologous in nature; therefore, isolation of a pure culture of C. jejuni, by selecting single colonies, from an environmental sample may not always yield a single strain.
The gram-negative bacterium Campylobacter jejuni is a commensal organism in a wide variety of animals, including cattle and swine, and in birds, such as poultry (10, 36). In humans, however, C. jejuni is a leading cause of acute inflammatory enteritis. Inflammation and diarrhea are thought to be due to the adherence of C. jejuni to the colonic mucosa and the invasion of human epithelial cells (48). Some complications associated with this enteritis, such as Guillain-Barré syndrome (20, 22, 51), a disease of the peripheral nervous system, are potentially fatal. While many cases of C. jejuni enteritis are caused by contaminated meat, untreated water, or raw milk (14), the primary route of infection is through improperly handled or undercooked poultry (36, 37). C. jejuni has been detected on raw chicken carcasses, and close to 100% of retail broiler chickens are contaminated with this bacterium (37).
In order to understand the pathogenicity of C. jejuni, it is important to examine the adherence of the organism to both poultry carcasses and human gastrointestinal epithelia, as well as the invasion of human epithelial cells. Several different methods have been developed for detecting C. jejuni adherence and invasion, including indirect immunofluorescence (21, 24), electron microscopy (11, 26, 35), radiolabeling of bacteria (15, 26, 35), Giemsa staining (17), acridine orange-crystal violet staining (16, 21, 23), and Nomarsky differential interference contrast-UV incident-light microscopy (6). While these methods successfully detect adherent and internalized C. jejuni, they have several limitations which decrease their versatility in in vivo studies. Many of the procedures listed above are destructive, time-consuming, or difficult to perform or cannot be used on fresh tissue. The signal from radiolabeled C. jejuni cells or C. jejuni cells stained with DNA intercalating dyes (e.g., Hoechst or DAPI [4′, 6-diamidino-2-phenylindole]) will become diluted over time due to cell division. The fluorescent labeling of bacterial membrane proteins using fluorescein isothiocyanate is subject to similar dilution. In addition, because these fluorescent compounds are extrinsic, they can be irreversibly photobleached. Finally, the differentiation of individual Campylobacter strains is not possible using fluorescent antibody conjugates due to the lack of fine specificity of the antibodies.
An ideal method for studying adherence and invasion uses an intrinsic tag, in which both extracellular and intercellular bacterial cells can be detected in real time without the necessity of a secondary substrate. Bacteria can be tagged by transforming the cells with plasmids that contain a constitutively expressed reporter gene fusion. Several reporter genes (e.g., lacZ, luxAB, and cat) have been shown to function in C. jejuni (1, 33, 34, 49). However, these reporter genes require the addition of an exogenous substrate. The green fluorescent protein (GFP) of Aequorea victoria, encoded by the reporter gene gfp, fluoresces in the absence of any added cofactor or substrate (7) and can be expressed in a wide variety of bacterial species (44). GFP is very stable and resistant to photobleaching (7). The intrinsic fluorescence and stability of GFP permit the nondestructive visualization of gfp-containing cells, even in complex environments. Additionally, several GFP alleles exist in which the emission spectrum of the protein has been shifted, resulting in a blue fluorescent protein (BFP) (fluorescence maximum wavelength [λmax] = 440 nm), a cyan fluorescent protein (CFP) (fluorescence λmax = 477 nm), or a yellow fluorescent protein (YFP) (fluorescence λmax = 527 nm) (42).
Plasmids are available that contain gfp fused to a strong or constitutively expressed promoter (9, 18, 29, 31, 32); however, these plasmids are not suitable for use in C. jejuni because the origins of replication and antibiotic resistance genes (e.g., bla and neo) present on these plasmids do not function in Campylobacter (19, 27, 50). Shuttle vectors that function in C. jejuni and other Campylobacter species have been constructed (34, 46, 47, 50). These vectors contain a Campylobacter-derived chloramphenicol or kanamycin resistance gene and two replication regions, the Escherichia coli ColE1 replicon and the replication region from a Campylobacter coli plasmid (41), allowing these vectors to be maintained in both C. jejuni and E. coli.
For experiments in vivo, the promoter fused to gfp on the marker plasmid should be strong and insensitive to most regulatory signals. Many E. coli promoters, including several strong, well-characterized promoters (e.g., Plac), do not function in C. jejuni (39, 49). Several C. jejuni promoters have been characterized (1, 2, 40); however, many are environmentally or nutritionally regulated. Recently, Wösten et al. compared several C. jejuni promoters and determined a putative promoter consensus sequence (49). Presumably, gfp fused to this consensus sequence would be constitutively expressed at a high level in the absence of any added protein-binding sites.
In this report we describe the construction of two sets of C. jejuni shuttle vectors containing the gfp, yfp, or cfp reporter gene. One set of plasmids contains the pUC18 polylinker upstream of the reporter gene. The other set contains a constitutively expressed transcriptional fusion consisting of a Campylobacter consensus promoter sequence fused to the reporter gene. These plasmids are maintained over several passages in the absence of antibiotic selection. Additionally, in many transformed C. jejuni strains, >91% of the cells remained fluorescent after being subcultured for 7 to 10 days. The construction and use of these plasmids in attachment and invasion studies are discussed.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and chemicals.
C. jejuni ATCC 43446 (Table 1) was obtained from P. Guerry, Naval Medical Research Institute (NMRI), Bethesda, Md., and was isolated from human feces. C. jejuni RM1221 (Table 1) was isolated from a 1 M NaCl wash of a store-bought chicken carcass. C. jejuni D781 (Table 1) was originally isolated from a chicken cloaca and was obtained from R. Meinersmann, USDA Agricultural Research Service, Athens, Ga. Spontaneous streptomycin-resistant (Smr) mutants of RM1221 were isolated by plating 109 to 1010 CFU of RM1221 per ml on brucella agar (BA) amended with 100 μg of streptomycin per ml (BA-SM). After 48 h, several colonies appeared; three of these putative Smr RM1221 colonies were restreaked twice on BA-SM. Smr colonies from these plates were designated RM1221S.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| ATCC 43446 | C. jejuni, serotype O:19 | P. Guerry; isolated from human feces |
| RM1221 | C. jejuni | This study; isolated from a chicken carcass |
| RM1221S | Smr mutant of RM1221 | This study |
| D781 | C. jejuni, serotype O:2,33 | R. Meinersmann; isolated from a chicken cloaca |
| 43446gfp | ATCC 43446(pWM1007) | This study |
| 43446yfp | ATCC 43446(pWM1008) | This study |
| 43446cfp | ATCC 43446(pWM1009) | This study |
| 1221gfp | RM1221(pWM1007) | This study |
| 1221yfp | RM1221(pWM1008) | This study |
| 1221cfp | RM1221(pWM1009) | This study |
| 781gfp | D781(pWM1007) | This study |
| 781yfp | D781(pWM1008) | This study |
| 781cfp | D781(pWM1009) | This study |
| Plasmids | ||
| pRK2013 | IncP Kmr Tra RK2+ ΔrepRK2 repE1+ | 12 |
| pMW10 | Kmr Mob+repB lacZ oriV | 49 |
| pWM1001 | Kmr; pMW10 ΔlacZΩ[(T1)4-gfp-T1] | This study |
| pWM1007 | Kmr; pMW10 ΔlacZΩ[(T1)4-Pc-gfp-T1] | This study |
| pWM1008 | Kmr; pMW10 ΔlacZΩ[(T1)4-Pc-yfp-T1] | This study |
| pWM1009 | Kmr; pMW10 ΔlacZΩ[(T1)4-Pc-cfp-T1] | This study |
| pWM1011 | Kmr; pMW10 ΔlacZΩ[(T1)4-yfp-T1] | This study |
| pWM1012 | Kmr; pMW10 ΔlacZΩ[(T1)4-cfp-T1] | This study |
C. jejuni was routinely cultured at 42°C under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) on BA supplemented with 0.025% FeSO4 · 7H2O, 0.025% sodium metabisulfite (anhydrous), and 0.025% sodium pyruvate (anhydrous).
Restriction and DNA-modifying enzymes were obtained from New England Biolabs (Beverly, Mass.) or Roche Molecular Biochemicals (Indianapolis, Ind.). Oligonucleotides were purchased from Oligos Etc. (Wilsonville, Oreg.) or Integrated DNA Technologies, Inc. (Coralville, Iowa). All chemicals (unless specified otherwise) were purchased from Sigma Chemicals (St. Louis, Mo.) or Fisher Scientific (Pittsburgh, Pa.). Alexa Fluor 546 conjugates, Alexa Fluor 568 conjugates, BODIPY 650/665 conjugates, and SYPRO Red protein gel stain were purchased from Molecular Probes (Eugene, Oreg.).
Preparation of polyclonal anti-Campylobacter antiserum.
C. jejuni strain RM1221 was grown on BA. A suspension of 2 × 107 CFU/ml in sterile phosphate-buffered saline (PBS) was prepared, and 0.5 ml was injected into the ear vein of a New Zealand White rabbit. The rabbit was immunized again at 4 and 8 weeks with 2 × 108 CFU of the same strain (per ml) that had been stored at −20°C and then thawed. Serum was collected and fractionated by precipitation with saturated ammonium sulfate (50% [vol/vol] final), followed by DEAE column chromatography. The fractions containing predominantly immunoglobulin G (IgG) were pooled and designated anti-Cj IgG.
Culture of Caco-2 cells.
Caco-2 cells, derived from a human colonic carcinoma, were obtained from the American Type Culture Collection (ATCC HTB-37). The cells were routinely cultured in Eagle's minimum essential medium with Earle's salts and amended with 2 mM l-glutamine, 20% fetal bovine serum, nonessential amino acids (0.1 mM [final concentration]), and sodium pyruvate (1 mM [final concentration]).
Bacterial transformation.
Plasmid DNA was mobilized into the C. jejuni strain RM1221S by triparental mating using a green, yellow, or cyan fluorescent E. coli DH5α transformant as the donor strain and DH5α(pRK2013) as the helper strain. Overnight RM1221S cultures were restreaked onto fresh BA-SM medium and grown for 9 h as described above. After 9 h, cells were removed and resuspended in PBS to an optical density at 600 nm (OD600) of 1.0. Overnight cultures of the donor and helper E. coli strains were subcultured into fresh Luria broth and grown to an OD600 of ca. 1.2. Cells were mixed at a ratio of either 1:1:10 or 1:1:100 (donor/helper/recipient), spotted onto BA plates, and incubated overnight at 42°C under microaerophilic conditions. The mating spots were then resuspended in PBS and centrifuged for 40 s at 3,000 × g to pellet the E. coli cells. The supernatant was removed and plated onto BA amended with 100 and 200 μg of streptomycin and kanamycin, respectively, per ml. The plates were examined after 2 to 3 days for the appearance of fluorescent C. jejuni colonies.
Construction of the gfp plasmids pWM1001 and pWM1007.
Plasmid pWM1001 was constructed by creating a unique PspOMI site in pMW10 (49) and then inserting the NotI-ended gfp promoter-probe cassette from pNH18/8 (30) into that PspOMI site. To create the PspOMI restriction site, oligonucleotide BspEco (Table 2) was first self-annealed to form a double-stranded (ds) adaptor. pMW10 was digested with EcoRI, and the 7,170-bp fragment, containing the origin of replication and Kmr gene, was ligated to the ds adaptor. To generate a constitutively expressed gfp fusion, a ds adaptor was synthesized that contains a promoter sequence based on the C. jejuni consensus promoter of Wösten et al. (49). The promoter sequence contains most of the consensus elements as described (49); however, minor modifications were made in the nonconsensus bases in order to minimize RNA secondary structure. The sequence of the ds adaptor, between the BamHI- and EcoRI-compatible ends, is 5′ GTTATTTTAAGTCTTAGTTTAGTTTTTTTGGTATAATTA 3′. This adaptor was ligated into BamHI-, EcoRI-digested pWM1001 to create the plasmid pWM1007 (Fig. 1).
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence |
|---|---|
| BspEco | 5′ AAT TGG GGC CCC 3′ |
| YFP1 | 5′ ACT TGT CAC TAC TTT CGG TTA TGG TCT TCA ATG CTT TGC AAG ATA CCC AGA TC 3′ |
| YFP2 | 5′ AAC CAT TAC CTG TCC TAC CAA TCT GCC CTT TCG 3′ |
| YCFP5′ | 5′ TGC CAT GGC CCA CCC TCG TGA CCA 3′ |
| CFP3′ | 5′ GGT TTC GAA AGG GCG GAC TGG GTG CTC AG 3′ |
FIG. 1.
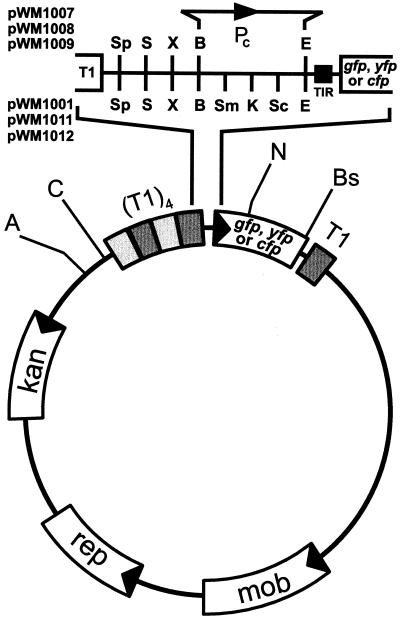
gfp, yfp, and cfp plasmids. Each plasmid contains a Campylobacter-derived kanamycin resistance gene (kan) and the mob (mob) and repB (rep) genes required for replication in Campylobacter. Additionally, each plasmid contains a Campylobacter origin of replication and the pBR322 ColE1 origin of replication (not shown). (T1)4, four tandem copies of the T1 terminator from the E. coli rrnB1 operon (5); Pc, consensus Campylobacter promoter (present only in plasmids pWM1007-9); TIR, translation initiation region (8) containing a phage T7 translational enhancer and a consensus Shine-Dalgarno region; T1, single rrnB1 terminator; unique restriction sites: A, AgeI; B, BamHI; Bs, BstBI; C, ClaI; E, EcoRI; N, NcoI; S, SalI; Sp, SphI; X, XbaI. Additional unique restriction sites (Sm, SmaI; K, KpnI; and Sc, SacI) are present in some plasmids (pWM1001, pWM1011, and pWM1012) between the BamHI and EcoRI sites in the MCS.
Construction of yfp plasmids pWM1008 and pWM1011.
pWM1007 was mutagenized using the GeneEditor in vitro site-directed mutagenesis system (Promega, Madison, Wis.) with oligonucleotides YFP1 and YFP2 (Table 2) as the mutagenesis oligonucleotides. The manufacturer's suggested protocol was followed with two exceptions: no antibiotic selection oligonucleotides were used and the BMH71-18mutS and JM109 transformants were not incubated with the antibiotic selection mix; instead, the JM109 transformants were examined on agar plates with an MZ-FLIII fluorescence stereomicroscope (Leica Microsystems Heidelberg GmbH, Heidelberg, Germany) equipped with a 41017 Endow GFP filter set (Chroma Technology Corp, Brattleboro, Vt.). Yellow fluorescent mutant colonies were isolated from among the background of green fluorescent colonies, and the plasmid DNA that was purified from these transformants was designated pWM1008 (Fig. 1). A ClaI-, EcoRI-ended fragment from pWM1001 was ligated to ClaI-, EcoRI-digested pWM1008 to create the plasmid pWM1011.
Construction of the cfp plasmids pWM1009 and pWM1012.
A fragment of the cfp allele from pECFP (Clontech, Palo Alto, Calif.) was amplified using the primer set YCFP5′-CFP3′ (Table 2). PCRs were carried out at 30 cycles of 1 min at 95°C, 2 min at 50°C, and 3 min at 72°C. The amplified product was ligated to pCR2.1 (Invitrogen, Carlsbad, Calif.). A NcoI-, BstBI-ended cfp fragment from this plasmid was ligated to NcoI-, BstBI-digested pWM1007 to create plasmid pWM1009 (Fig. 1). A ClaI-, EcoRI-ended fragment from pWM1001 was ligated to ClaI-, EcoRI-digested pWM1009 to create the plasmid pWM1012.
Plasmid and fluorescence stability in vitro.
C. jejuni cells transformed with pWM1007, pWM1008, or pWM1009 were plated onto BA amended with 200 μg of kanamycin per ml (BA-KM) and grown for 48 h as described above. Cells were then streaked onto BA medium and grown under microaerophilic conditions at 42°C. Cells were removed after 24 h, resuspended in PBS, diluted 10−4, 10−5, and 10−6, and plated on BA and BA-KM media. The colonies on each plate were counted after 24 h; the nonfluorescent colonies were counted using an MZ-FLIII fluorescence stereomicroscope. This procedure was repeated for 5 to 10 days. BA is slightly fluorescent at CFP excitation and emission wavelengths, thereby complicating detection of the cyan fluorescent C. jejuni colonies. To increase contrast, the culture medium was amended with 0.4% bacteriological charcoal.
Fluorescence microscopy of C. jejuni cells and colonies.
Bacterial colonies were photographed using a Leica MZ-FLIII fluorescence stereomicroscope equipped with a DKC-5000 charge-coupled device (CCD) camera (Sony Medical Systems, Park Ridge, N.J.); individual cells were photographed using a Leica DM-RB epifluorescence microscope (Leica Microsystems) equipped with a Sony DKC-5000 CCD camera. The GFP, YFP, and CFP filter sets used were 41017 Endow GFP, 41028 Yellow GFP, and 31044 v2. Cyan GFP, respectively, and were obtained from Chroma Technology Corp.
Confocal laser scanning microscopy (CLSM).
Confocal microscopy was performed with a TCS NT confocal microscope (Leica Microsystems). An Ar laser (excitation wavelength [λexc] = 488 nm) was used to excite GFP and a Kr laser (λexc = 568 nm) was used to excite red autofluorescent chloroplasts, Alexa Fluor 546-stained molecules, or Alexa Fluor 568-stained cells. A He-Ne laser (λexc = 633 nm) was used to excite BODIPY 650/665-stained actin filaments or SYPRO Red-stained tissue. GFP fluorescence was detected with the BP525/50 filter set and assigned the color green. Red autofluorescent or fluorescent Alexa Fluor emissions were detected with the LP590 (two-color images) or BP600/30 (three-color images) filter set and assigned the color red. Fluorescent BODIPY and SYPRO emissions were detected with the LP645 filter set and assigned the color blue. Two- or three-color images were obtained by overlaying images from individual channels using the TCS NT software (version 1.6.551; Leica Microsystems).
Adherence of green fluorescent C. jejuni cells to chicken breast skin.
781gfp cells were grown overnight on BA-KM, transferred to 35 ml of brucella broth amended with 200 μg of kanamycin per ml (BB-KM), and grown overnight in a vented tissue culture flask. The cells were centrifuged at 8,000 × g for 8 min and resuspended in PBS at an OD600 of ca. 0.8. A sample of fresh chicken breast skin was first stained for 15 min with SYPRO Red protein gel stain, washed three times with PBS, and incubated for 40 min at room temperature with the 781gfp cell suspension. The skin was then washed three times with PBS, placed on a glass slide with 50% glycerol in PBS, and observed under the CLSM.
Adherence and recovery of yellow and cyan fluorescent C. jejuni cells from chicken breast skin.
Strains 1221yfp, 1221cfp, 781yfp, and 781cfp were grown overnight in BB-KM and centrifuged as described above. The cells were resuspended in PBS, and the OD600 for each suspension was measured. The suspensions were then diluted in PBS to a final concentration of 106 CFU/ml. Two mixtures containing 1221yfp and 781cfp (1:1) and 1221cfp and 781yfp (1:1) were prepared and added (5 ml/g of tissue) to samples of fresh chicken breast skin. The skin was incubated for 1 h at room temperature and washed for 5 min twice with PBS. Adherent C. jejuni cells on the chicken skin were recovered by stomaching for 2 min in Preston selective enrichment broth (PSEB) (Preston medium [43] supplemented with 10% lysed horse blood) (5 ml/g of tissue). The PSEB, containing the recovered C. jejuni, was sonicated for 30 s, diluted, and plated on CCDA medium (3) amended with 200 μg of kanamycin per ml (CCDA-KM). As a control, each mixture was diluted and plated directly onto CCDA-KM. Additionally, overnight broth cultures were adjusted to equivalent OD600 values, mixed as described above, diluted, and plated onto BA-KM amended with 0.4% bacteriological charcoal.
Adherence of C. jejuni cells to cilantro.
Strain 1221gfp was grown overnight on BA-KM and grown under microaerophilic conditions as described above. An inoculum suspension was prepared by resuspending cells from the plate at a final concentration of 109 cells/ml in 0.5 mM PBS. A fully developed cilantro leaf was inoculated by immersing it in the Campylobacter suspension. The inoculated leaf was placed on a humid filter paper in a closed petri dish and incubated for 1 h at 37°C. The leaf was rinsed gently three times in 0.5 mM PBS, mounted in Aqua-Poly/Mount (Polysciences, Warrington, Pa.), and examined under CLSM.
Detection of internalized C. jejuni in Caco-2 cells.
Caco-2 cells were seeded onto round, glass coverslips, placed into a 24-well tissue culture dish, and grown overnight, as described above, to a cell density of ca. 105 cells per well. The coverslips in three wells were removed; the adherent cells were trypsinized and counted on a hemocytometer to determine the average number of cells per coverslip. 781gfp cells were grown in BB-KM as described above. The cells were examined by epifluorescence microscopy and determined to be fluorescent and primarily spiral shaped. The C. jejuni cells were centrifuged, resuspended in PBS, and inoculated into the wells containing Caco-2 cells at a multiplicity of infection of 200. After incubation of the inoculated Caco-2 cells at 37°C in a 5% CO2 incubator for 2 h, the tissue culture medium was removed and the cells were washed three times with 1 ml of Earle's balanced salt solution. Three sets of coverslips were prepared.
In the first set, the C. jejuni cells and the Caco-2 cell tubulin were visualized. The cells were fixed with 3.7% formalin for 10 min. After fixation, the Caco-2 cells on the coverslips were washed three times with PBS, treated with 1% glycine in PBS for 10 min, washed three times with PBS, permeabilized with 0.1% Triton X-100 in PBS for 5 min, and again washed three times with PBS. Following permeabilization, the Caco-2 cells were treated with 1% bovine serum albumin (BSA) in PBS (BSA-PBS) for 40 min to reduce nonspecific binding. The Caco-2 cells were then incubated with 50 μl of mouse monoclonal, anti-bovine α-tubulin (200 μg of stock solution per ml in BSA-PBS, 2 mM NaN3, diluted to 0.5 μg/ml in BSA- PBS; Molecular Probes Inc.) at 37°C for 1 h. The antibody conjugate was removed and the cells were washed three times with PBS. The coverslips were incubated in the dark for 1 h with 50 μl of Alexa Fluor 546 goat anti-mouse IgG (heavy and light chain) conjugate (2 mg of stock solution per ml in 0.1 M NaPO4, 0.1 M NaCl, 2 mM NaN3 [pH 7.5] diluted to 5 μg/ml in BSA-PBS), washed three times with PBS, and mounted on a slide. The C. jejuni and Caco-2 cells were observed under CLSM.
Fresh tissue culture medium, amended with 200 μg of gentamicin per ml, was added to the second and third set of coverslips, and the Caco-2 cells were incubated for 2 h at 37°C as described above. The gentamicin was then removed by gently washing the cells three times with 1 ml of Earle's balanced salt solution. The second set of coverslips was used to determine the average number of internalized C. jejuni bacteria per Caco-2 cell. Internalized C. jejuni cells were quantified by lysing the Caco-2 cells with 0.1% Triton X-100 for 15 min at room temperature, and then plating serial dilutions of the lysate on BA-KM and CCDA-KM. It was determined previously that Caco-2 cells were insensitive to gentamicin at 200 μg/ml and that C. jejuni cells were not affected by a 15-min incubation in 0.1% Triton X-100.
To visualize antibody-labeled C. jejuni cells and actin filaments in the third set, phalloidin and a polyclonal antiserum raised against C. jejuni were used. The Caco-2 cells on the remaining coverslips were fixed, permeabilized, and blocked as described above. The coverslips were incubated with anti-Cj IgG (10 μg/ml in BSA-PBS) for 1 h and then washed three times with BSA-PBS. Alexa Fluor 568 goat anti-rabbit IgG (heavy and light chain) conjugate (5 μg/ml in BSA-PBS) was added. After incubation for 1 h, the antibody conjugate was removed by washing the coverslips three times in BSA-PBS. Actin filaments were stained by incubating the Caco-2 cells for 20 min with BODIPY 650/665-conjugated phalloidin (0.3 μM in BSA-PBS) for 20 min. After incubation, the coverslips were washed three times with PBS and mounted with ProLong Antifade (Molecular Probes Inc.) on microscope slides. The cells were observed under CLSM.
Nucleotide sequence accession numbers.
The nucleotide sequences of the plasmids pWM1001, pWM1007, pWM1008, pWM1009, pWM1011, and pWM1012 have been submitted to the GenBank nucleotide database under the accession numbers AF292555, AF292556, AF292557, AF292558, AF292559, and AF292560, respectively.
RESULTS
Construction of gfp, yfp, and cfp shuttle plasmids.
Our goal was to construct two sets of shuttle plasmids for transforming Campylobacter jejuni. The first set would contain either gfp, yfp, or cfp downstream of a multicloning site (MCS), into which uncharacterized genomic DNA fragments or DNA fragments containing a known promoter sequence could be cloned. The second set would contain the same reporter genes fused to a strong, constitutively expressed promoter. Campylobacter cells transformed with any plasmid from this second set would be intrinsically labeled and would presumably fluoresce under all environmental conditions. To construct a gfp shuttle plasmid suitable for use in C. jejuni, we modified an existing Campylobacter shuttle plasmid by inserting a gfp-containing promoter probe cassette. The Campylobacter plasmid that we used was pMW10 (49). This plasmid, derived from Campylobacter coli plasmid pIP1433 (41), contains a Campylobacter origin of replication, the repB and mob genes necessary for replication and mobilization, a Campylobacter-derived kanamycin resistance gene, and the ColE1 origin of replication from pBR322. The promoter-probe cassette that we selected was the gfp cassette from pNH18/8 (30). This cassette can be excised from pNH18/8 by NotI and contains, from left to right, four copies of the E. coli rrnB1 T1 terminator (5), the pUC18 MCS, an optimized translation initiation region (8), the gfp gene, and a single T1 terminator. The four upstream terminators effectively reduce background GFP fluorescence by shielding the gfp gene from transcription initiating outside the cassette (30). Strong promoters may interfere with plasmid replication (38); the downstream terminator minimizes any potential interference (4, 38). The lacZ gene in pMW10 and the restriction sites present 5′ of lacZ were replaced by the promoter-probe cassette from pNH18/8 to create the plasmid pWM1001.
A strong promoter, one that could be reasonably predicted to be insensitive to all regulatory signals, was required to create a set of plasmids that could be used to tag C. jejuni cells. We first constructed an artificial, constitutive, promoter sequence by annealing together two oligonucleotides. The promoter (Pc) contained in this DNA fragment is based on the consensus promoter sequence of Wösten et al. (49). The −35, −16, and −10 motifs were left intact; nonconserved bases were optimized in order to minimize potential RNA secondary structure. This promoter fragment was ligated into the MCS of pWM1001 to create the plasmid pWM1007.
To construct Pc-yfp and Pc-cfp marker plasmids, pWM1007 was mutagenized using in vitro site-directed mutagenesis (see Materials and Methods). A yellow fluorescent variant of pWM1007 was isolated and designated pWM1008. This mutagenesis procedure, however, did not result in any cyan fluorescent mutants. Therefore, a cyan fluorescent variant of pWM1007 was constructed by amplifying a segment of the cfp gene of pECFP (Clontech) and ligating it into pWM1007. The resulting Pc-cfp fusion plasmid was designated pWM1009. Finally, promoterless versions of pWM1008 and pWM1009 were constructed by replacing the MCS-Pc region in these two plasmids with the MCS from pWM1001, creating the plasmids pWM1011 and pWM1012, respectively.
Characterization of shuttle plasmids.
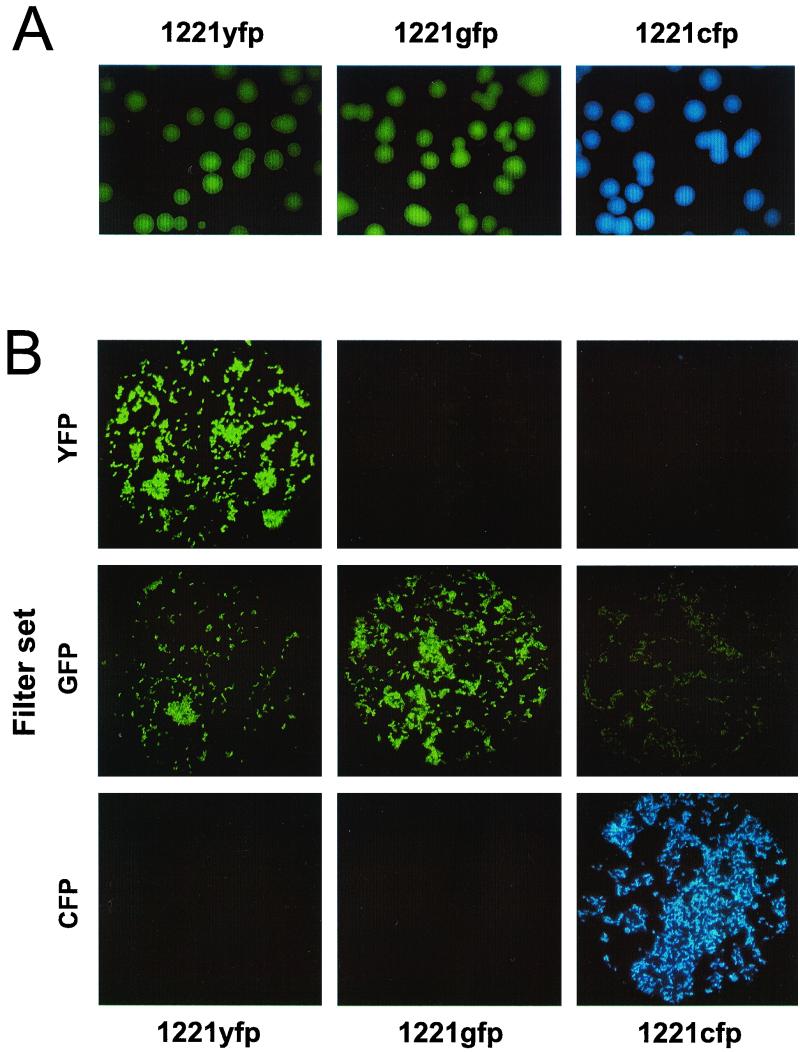
To verify that the gfp-, yfp-, and cfp-containing plasmids would function in C. jejuni, the C. jejuni strain RM1221 (Table 1) was transformed with either pWM1007 (1221gfp), pWM1008 (1221yfp), or pWM1009 (1221cfp). Electroporation of RM1221 with E. coli-derived DNA was not successful. Therefore, we first mobilized the shuttle plasmids into a Smr variant of RM1221 (RM1221S) by triparental mating. Although the mating efficiency was low, DNA isolated from the RM1221S transformants could then be electroporated into wild-type RM1221 at a high frequency. DNA isolated from RM1221 could also be used to electroporate other C. jejuni strains, such as ATCC 43446 (Table 1); however, many other C. jejuni strains (10 out of 15 tested) could not be so transformed, suggesting a possible restriction barrier (data not shown). Colonies and individual cells of 1221gfp, 1221yfp, and 1221cfp are shown in Fig. 2. Individual C. jejuni cells were photographed with either their cognate filter set (i.e., 1221yfp with the yfp filter set) or the other two filter sets (Fig. 2B). 1221yfp, 1221gfp, and 1221cfp cells are strongly fluorescent when viewed with the yfp, gfp, or cfp filter set, respectively. No fluorescence is seen when 1221gfp or 1221cfp cells are viewed with the yfp filter set; also, no fluorescence is seen when 1221yfp or 1221gfp cells are visualized with the cfp filter set. However, there is considerable background fluorescence when 1221yfp or 1221cfp cells are viewed with the gfp filter set.
FIG. 2.
Fluorescence of C. jejuni RM1221 transformed with Pc-gfp, Pc-yfp or Pc-cfp fusion plasmids. RM1221 was transformed with either pWM1007 (1221gfp), pWM1008 (1221yfp), or pWM1009 (1221cfp). (A) Colonies of 1221gfp, 1221yfp, or 1221cfp. All three strains were grown on BA-KM amended with bacteriological charcoal. Colonies were photographed using a Leica MZ-FLIII fluorescence stereomicroscope equipped with appropriate filters and a Sony DKC-5000 CCD camera. (B) Individual 1221gfp, 1221yfp, or 1221cfp cells visualized using the gfp, yfp, and cfp filter sets. Cells were visualized using a Leica DM-RB epifluorescence microscope equipped with a Sony DKC-5000 CCD camera.
Many additional C. jejuni strains transformed with pWM1007, pWM1008, or pWM1009 were equally fluorescent (data not shown). Additionally, E. coli, Salmonella enterica serovar Newport, Enterobacter sp., and Pantoea agglomerans cells transformed with these plasmids are very fluorescent (data not shown) suggesting that these plasmids can be used to tag multiple enteric taxa.
Plasmid and fluorescence stability in C. jejuni.
One set of gfp, yfp, and cfp plasmids (pWM1007, pWM1008, and pWM1009, respectively [Fig. 1]) contains a constitutively expressed transcriptional fusion that can be used to intrinsically tag C. jejuni cells. The successful use of these plasmids requires that the fluorescence of Campylobacter cells that have been transformed with these vectors be stable, in the absence of antibiotic selection, over any experimental time course. Fluorescence can be lost over time in two ways: the plasmid can be lost from the cell or the reporter gene can become inactive, either through deletion of the reporter gene or the accumulation of deleterious point mutations.
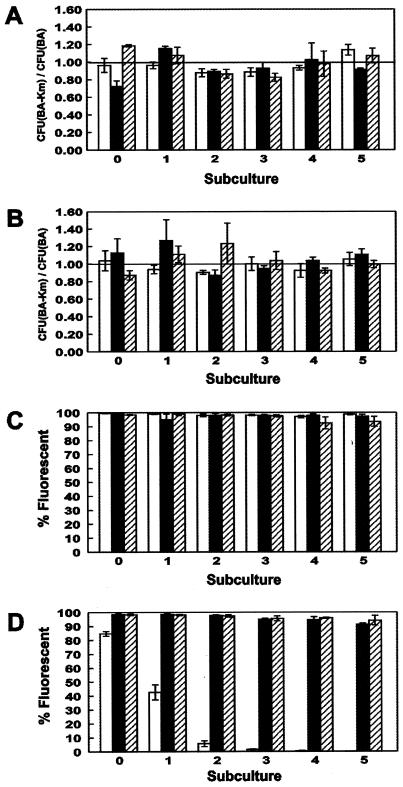
The stability of the shuttle plasmids in vitro was quantified by determining the ratio of colonies on media with (BA-KM) and without (BA) kanamycin. After each subculture, the ratio of the number of colonies on BA-KM to that on BA was around 1.0 (Fig. 3A and B), indicating that the shuttle plasmids are stably maintained in C. jejuni in the absence of antibiotic selection. A previous experiment with 43446gfp and 1221gfp showed that pWM1007 is maintained after least 10 passages (data not shown).
FIG. 3.
Plasmid and fluorescence stability of pWM1007, pWM1008, and pWM1009 in C. jejuni ATCC 43446 and RM1221. Subculture 0 represents cells grown on BA-KM; subcultures 1 through 5 represent cells grown on BA. Error bars indicate the standard error of the mean. (A and B) Stability of pWM1007 (white bars), pWM1008 (black bars), or pWM1009 (striped bars) in RM1221 (A) and ATCC 43446 (B). Each data point represents the average ratio from three replicate sets of plates. (C and D) Stability of gfp (white bars), yfp (black bars), or cfp (striped bars) in transformed RM1221 (C) and ATCC 43446 (D) on BA-KM. Each data point represents the average value from three replicate sets of plates.
The percentage of fluorescent cells was calculated after each subculture. Over 90% of the 1221gfp, 1221yfp, or 1221cfp cells remained fluorescent after five subcultures (Fig. 3C). However, although >91% of the 43446yfp or 43446cfp cells remained fluorescent after five subcultures, the fluorescence of 43446gfp cells was rapidly lost over time and was negligible after four subcultures (Fig. 3D). A previous experiment with 43446gfp revealed that >91% of the cells remained fluorescent after 10 days (data not shown). Analysis of plasmid DNA isolated from nonfluorescent colonies of 43446gfp indicated that the loss of fluorescence is due to deletion of gfp (data not shown), presumably caused by recombination between the T1 terminators 5′ and 3′ of the reporter gene (30). We have transformed several additional strains of C. jejuni with the three shuttle plasmids. Fluorescence appears to be stable in all of the C. jejuni strains tested in vitro with the exception of 43446gfp (data not shown).
To assess the stability of the shuttle plasmids in vivo, 24-day-old White Leghorn chickens were gavaged with 107 1221yfp, 1221cfp, 781yfp, or 781cfp cells alone or in combination. At various intervals, a chicken from each group was sacrificed and the enteric tissue was removed. Sections of enteric tissue were incubated in PSEB for 18 h under normal growth conditions. The PSEB was then diluted and plated on CCDA. One hundred percent of the recovered colonies were fluorescent 19 days after inoculation (data not shown).
Visualization of transformed C. jejuni cells on poultry, on plant surfaces, and after invasion of Caco-2 cells.
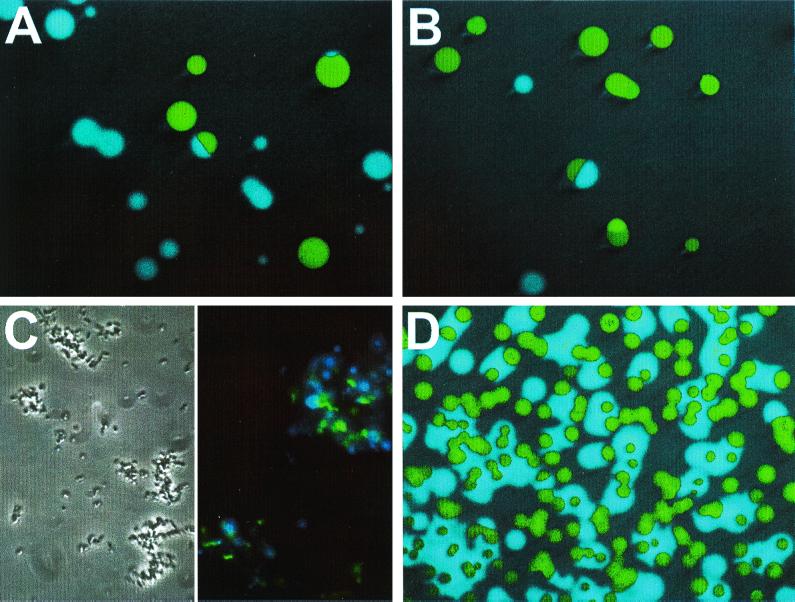
To assess the versatility of the fusion plasmids for use in fluorescence microscopy, we inoculated chicken breast skin and a cilantro (Coriandrum sativum L.) leaf with green fluorescent C. jejuni to verify that there is sufficient fluorescence emitted by the bacteria to visualize individual cells against these backgrounds. Spiral forms of 781gfp cells are clearly visible on chicken skin counterstained with SYPRO Red (Fig. 4A). Green or yellow fluorescent C. jejuni was also visible on chicken skin counterstained with other dyes (e.g., SYPRO Orange [data not shown]). Green fluorescent 1221gfp cells were easily visualized against the red autofluorescence of the epidermis of a cilantro leaf (Fig. 4B). Green fluorescent C. jejuni was also visible on leaves of spinach and red leaf lettuce contained in commercial Spring Mix (data not shown).
FIG. 4.
Representative CLSM images of fluorescent C. jejuni associated with various tissues or internalized within Caco-2 cells. Images were magnified 630× (A, B, and D) or 1,000× (C). Scale bars represent 20 μm. (A) Extended focus image of 781gfp cells attached to chicken breast skin. (B) Projection of 35 x-y sections showing 1221gfp cells associated with a physical lesion on a cilantro leaf. (C) 781gfp internalized within Caco-2 cells. (D) Extended focus image of 781gfp internalized within Caco-2 cells. The tissue sample was scanned in the z plane in 0.5- to 1-μm sections. Arrows indicate internalized C. jejuni cells (red) detected by the polyclonal sera and Alexa Fluor 568 goat anti-rabbit IgG.
To determine whether tagged C. jejuni cells could be visualized after internalization into Caco-2 cells and if internalized cells could be detected by a C. jejuni polyclonal antiserum, gfp-transformed C. jejuni cells were used in invasion assays with cultured Caco-2 cells. There was no decrease in internalization compared to D781 when 781gfp, 781yfp, or 781cfp cells were incubated with Caco-2 cells (data not shown), indicating that neither the presence of the plasmid nor the fluorescent protein, per se, affected invasion. Spiral, green fluorescent 781gfp cells were readily visible after internalization in Caco-2 cells stained with antitubulin antibodies (Fig. 4C). 781gfp cells were also visible in Caco-2 cells after a 2-h treatment with gentamicin (200 μg/ml), added to kill extracellular Campylobacter (Fig. 4D). In this experiment, the Caco-2 cells were permeabilized, after incubation with C. jejuni, and then probed with an anti-C. jejuni polyclonal antiserum and an Alexa Fluor 568-labeled secondary antibody conjugate. A small percentage of the internalized cells were detected by the antiserum (Fig. 4D). However, the polyclonal antiserum and secondary antibody conjugate detected 100% of the 781gfp cells on a slide mount (data not shown), suggesting either that epitope expression had been altered in internalized C. jejuni or that only a fraction of the internalized 781gfp cells were accessible to the antisera. Highly green fluorescent cells labeled with the Alexa 568 antibody conjugate should appear yellow in overlay; however, no yellow cells were visible. Control experiments indicated that treating green fluorescent C. jejuni cells with antibodies greatly reduced their fluorescence (data not shown). When the gain in the green channel was increased, faint green fluorescent C. jejuni cells were detected at the same location as the red cells; therefore, these red cells do indeed represent green fluorescent Campylobacter cells that were detected by the antisera.
Aggregation and colony morphology of yellow and cyan fluorescent C. jejuni transformants.
The C. jejuni strains RM1221 and D781 were isolated from chicken (Table 1). To determine if these two strains adhere differently to chicken breast skin, two mixtures were prepared: a 1:1 mixture of 1221yfp and 781cfp and a 1:1 mixture of 1221cfp and 781yfp. These mixtures were inoculated onto, and recovered from, a sample of chicken skin. Two- to threefold more cyan fluorescent colonies were seen when 1221cfp and 781yfp cells were mixed (Fig. 5A). Also, two- to threefold more yellow fluorescent colonies were seen when 1221yfp and 781cfp cells were mixed (Fig. 5B). However, similar results were obtained when the inocula were plated directly onto CCDA-KM (data not shown), suggesting that the apparent difference between the number of RM1221 and D781 transformants is due to a discrepancy in the original cell concentrations and is not due to differences in the adherence of the two strains to chicken skin.
FIG. 5.
Representative microscopic images of yellow and cyan C. jejuni colonies and cells following inoculation of chicken breast skin or after growth in vitro. (A, B, and D) Colonies were photographed using a Leica MZ-FLIII fluorescence stereomicroscope equipped with the appropriate filters and a Sony DKC-5000 CCD camera. Individual cells (C) were visualized using a Leica DM-RB epifluorescence microscope equipped with a Hamamatsu C4742-95 cooled CCD camera (Hamamatsu Photonics K.K., Hamamatsu, Japan). Panels A, B, and D and the right half of panel C represent a composite of the same field photographed with the yfp and cfp filter sets. The image obtained with the yfp filter set was overlaid onto the image obtained with the cfp filter set using Corel Photo-Paint9 (Corel Corp., Ottawa, Canada). (A and B) Colonies of 1221cfp and 781yfp (A) or 1221yfp and 781cfp (B). Cells were inoculated onto a sample of chicken skin, removed, and grown on CCDA amended with 200 μg of kanamycin per ml. (C) Bright-field (left half) and epifluorescent (right half) images of 781yfp and 1221cfp aggregates. Mixtures (1:1) of 781yfp and 1221cfp cells were grown for 24 h in BB, vortexed vigorously, and observed as a wet mount. The aggregates in the left half of panel C are not the same as those in the right half. (D) Colonies of 1221cfp and 781yfp. Cells were grown for 24 h in BB-KM and plated on BA-KM amended with 0.4% bacteriological charcoal.
Over 6% of the colonies on CCDA-KM are sectored (data not shown). In some instances, half of the colony is cyan and the other half is yellow (Fig. 5A and B). Other colonies which show different degrees of sectoring can also be seen (Fig. 5A, upper right). As it is unlikely that a single point mutation could convert a yellow fluorescent cell into a cyan fluorescent cell, such sectored colonies are probably the result of aggregate formation in the broth cultures or PBS suspensions. Bright-field examination of a 1:1 broth culture mixture of RM1221 and D781 transformants indicates that a large proportion of the C. jejuni cells in the mixture are contained in aggregates, even after vigorous vortexing of the mixture (Fig. 5C, left half). Some aggregates contain both yellow fluorescent and cyan fluorescent cells (Fig. 5C, right half); other aggregates contain only yellow fluorescent cells or only cyan fluorescent cells (data not shown).
When a broth culture mixture of 1221cfp and 781yfp is plated on BA-KM (Fig. 5D), the cyan fluorescent 1221cfp transformants appear to have a much different colony morphology than the yellow fluorescent D781 transformants; several yellow fluorescent colonies are completely encompassed by cyan fluorescent cells. Similarly, mixtures of 1221yfp and 781cfp plated on the same medium result in cyan fluorescent colonies surrounded by yellow fluorescent cells (data not shown). However, neither the sectored colonies nor the colonies in which the D781 transformants are enveloped by the RM1221 transformants can be distinguished from the other colonies under bright-field examination (data not shown).
DISCUSSION
We have constructed two sets of Campylobacter shuttle vectors which contain either the gfp, yfp, or cfp reporter gene. These plasmids confer kanamycin resistance and have two origins of replication, the ColE1 origin from pBR322 and a Campylobacter-derived origin, which allow these vectors to be maintained in both C. jejuni and several enteric taxa. In one set of plasmids, the reporter gene is promoterless. A MCS from pUC18, present upstream of the reporter gene, provides several unique restriction sites into which known promoter sequences or random genomic fragments can be inserted. These vectors are particularly valuable in conjunction with a fluorescence-activated cell sorter where cells containing transcriptional fusions that are induced by certain environmental stimuli can be separated from the larger population (44, 45).
In the other set of plasmids, the reporter gene is fused to an artificial Campylobacter promoter sequence (Pc). This sequence contains the −35, −16, and −10 motifs that have been proposed to constitute the ς70 promoter in Campylobacter (49). As the promoter motifs were constructed to be consensus at each position and the nonconserved bases surrounding the motifs were adjusted to minimize potential RNA secondary structure, this promoter was predicted to be very strong; indeed, cells tagged with these Pc fusion plasmids are extremely fluorescent (Fig. 2, 4, and 5). Additionally, although the Pc promoter was based on a compilation of several putative Campylobacter promoter sequences (49), this promoter can function in several enteric taxa. No binding sites for regulatory proteins (e.g., Cap and Fur) were inserted, and no promoters for alternate ς factors (i.e., ς28 and ς54) are present; therefore, this promoter should be constitutive and insensitive to many environmental and nutritional regulatory signals. However, the effect of other regulatory signals, such as DNA supercoiling, on transcription from the Campylobacter promoter has not been addressed.
While both sets of plasmids are stable in the absence of antibiotic selection in C. jejuni and in most laboratory strains of E. coli (e.g., DH5α) in vitro, they are not maintained, under the same conditions, in some enteric strains (data not shown). This is probably because rop is absent, as in the pUC plasmids, leading to a higher copy number (28), which may be detrimental to some strains. Also, in some strains transformed with the Pc fusion plasmids, fluorescence may be lost over time. This loss of fluorescence is due to the deletion of the reporter gene, presumably following recombination between the rrnB1 T1 terminators (30), and is independent of the presence of kanamycin in the culture medium (data not shown). We have transformed multiple strains of C. jejuni with all three fusion plasmids; only the strain 43446gfp shows any fluorescence instability in vitro (Fig. 3C). Fluorescence is stable in 43446yfp and 43446cfp (Fig. 3C). However, fluorescence was also initially stable in 43446gfp; therefore, it is possible that 43446yfp and 43446cfp may eventually show the same instability. If so, then the high rate of homologous recombination in these three strains may reflect a difference in the genotype of ATCC 43446 as compared to other C. jejuni strains. Additional C. jejuni strains may show a similar loss of fluorescence over time; therefore, although fluorescence is stable in most transformed C. jejuni strains, it would be advisable to test fluorescence stability in all mobilized strains before using them in situ. Also, although low fluorescence stability in vitro might suggest low stability in vivo, high fluorescence stability in vitro does not always reflect high stability in vivo. However, fluorescence stability would only have an impact when the transformants undergo multiple rounds of cell division. Where fluorescence is unstable, the proportion of the nonfluorescent subpopulation would increase with successive generations. For some experiments, such as attachment to chicken skin, the effect of fluorescence stability would be minimal. In other cases, where the time course of an experiment is measured in days or weeks, fluorescence stability would be much more important. In one such experiment, 24-day-old White Leghorn chickens were gavaged with RM1221 and D781 transformants. One hundred percent of the Campylobacter colonies recovered from enteric tissue 19 days after inoculation, in the absence of antibiotic treatment (data not shown), were fluorescent, suggesting that under certain conditions fluorescence is stable in vivo. Other animal or plant hosts or other conditions in which these transformants might be used have not been tested.
The multiple fluorescent proteins encoded by these vectors permit the design of experiments in which the same sample is inoculated simultaneously with two or more tagged strains. In one such experiment, a sample of chicken breast skin could be inoculated with a mixture containing a yellow and a cyan fluorescent C. jejuni strain. Differences in the adherence of the two strains would be reflected in the number of yellow and cyan fluorescent colonies present after recovery and plating. To test whether two strains adhere differently to chicken skin, we coinoculated chicken skin with fluorescent D781 and RM1221 transformants. While no difference was seen in the adherence of D781 or RM1221 to chicken skin, a large number of sectored colonies were detected, in which a single colony contained both yellow and cyan fluorescent cells. These colonies are not due to the conversion of one fluorescent phenotype to the other, but probably originate from aggregates of C. jejuni cells in the sample (Fig. 5C). A colony which is half yellow and half cyan (Fig. 5A and B) might result from a yellow fluorescent cell and a cyan fluorescent cell. Other colonies, where one fluorescent phenotype represents a small percentage of the total colony (Fig. 5A, upper right), suggest aggregates containing a much larger number of cells. Over 6% of the colonies on CCDA-KM are sectored; as an aggregate of yellow or cyan fluorescent C. jejuni would result in a yellow or cyan flourescent colony, respectively, this figure probably represents the minimum number of colonies that resulted from an aggregate. These aggregates, containing yellow fluorescent cells, cyan fluorescent cells, or a mixture of the two, were present even though the samples were sonicated or vortexed vigorously before plating (Fig. 5C). Therefore, standard plate counts of C. jejuni may significantly underestimate the number of cells in a sample. Of greater concern is the fact that since RM1221 and D781 cells were able to form aggregates, any colony that arose from such an aggregate would not represent a single strain of C. jejuni. Thus, it may be difficult to obtain a pure culture of C. jejuni from an environmental sample (e.g., poultry) via enrichment, plating, and isolation using single colony picks.
Additionally, RM1221 transformants appear to have a much different colony morphology on BA-KM than the D781 transformants (Fig. 5D). This difference, however, is not seen when the cells are plated on CCDA (Fig. 5A and B). The alteration in colony morphology on CCDA may reflect reduced motility. Sodium deoxycholate, present in CCDA at a concentration of 0.1%, has been shown to inhibit the motility of Proteus mirabilis and E. coli (13).
The fusion plasmids described in this paper represent a significant addition to the molecular tools available to study Campylobacter adherence and invasion. C. jejuni cells transformed with the gfp vector pWM1007 are clearly visible on chicken skin (Fig. 4A) and on leaf surfaces (Fig. 4B). Also, transformed C. jejuni are readily detected after internalization into Caco-2 cells (Fig. 4C and D), in agreement with the observations reported by Konkel et al. on the binding of gfp-tagged C. jejuni to INT-407 cells (25). The ability to intrinsically tag Campylobacter cells with gfp, yfp, or cfp fusion plasmids obviates other procedures that are more destructive or time-consuming or that require the addition of an exogenous substrate. Since GFP is extremely stable and is constitutively expressed in cells transformed with these fusion plasmids, cells thus tagged can be monitored over the course of several days after inoculation without the decay of fluorescence or the dilution of fluorescence due to cell division. These plasmids also offer an improvement in the detection of internalized Campylobacter in contrast to those procedures that use polyclonal or monoclonal antisera and immunofluorescence to detect internalized C. jejuni (21, 24). Whereas our C. jejuni-polyclonal antiserum detected close to 100% of the cells in vitro (data not shown), only a fraction of the C. jejuni cells were detected by the antiserum after invasion (Fig. 4D). Relying solely on immunofluorescence analyses might therefore underestimate the proportion of internalized cells.
ACKNOWLEDGMENTS
We thank R. Meinersmann, P. Guerry, and M. Wösten for providing strains and plasmids.
This work was funded by the U.S. Department of Agriculture Agricultural Research Service CRIS project 5325-42000-022.
REFERENCES
- 1.Alm R A, Guerry P, Trust T J. The Campylobacter sigma 54 flaB flagellin promoter is subject to environmental regulation. J Bacteriol. 1993;175:4448–4455. doi: 10.1128/jb.175.14.4448-4455.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baillon M L, van Vliet A H, Ketley J M, Constantinidou C, Penn C W. An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J Bacteriol. 1999;181:4798–4804. doi: 10.1128/jb.181.16.4798-4804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolton F J, Hutchinson D N, Coates D. Blood-free selective medium for isolation of Campylobacter jejuni from feces. J Clin Microbiol. 1984;19:169–171. doi: 10.1128/jcm.19.2.169-171.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosius J. Toxicity of an overproduced foreign gene product in Escherichia coli and its use in plasmid vectors for the selection of transcription terminators. Gene. 1984;27:161–172. doi: 10.1016/0378-1119(84)90137-9. [DOI] [PubMed] [Google Scholar]
- 5.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 6.Bukholm G, Kapperud G. Expression of Campylobacter jejuni invasiveness in cell cultures coinfected with other bacteria. Infect Immun. 1987;55:2816–2821. doi: 10.1128/iai.55.11.2816-2821.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 8.Cheng X, Patterson T A. Construction and use of lambda PL promoter vectors for direct cloning and high level expression of PCR amplified DNA coding sequences. Nucleic Acids Res. 1992;20:4591–4598. doi: 10.1093/nar/20.17.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen B B, Sternberg C, Molin S. Bacterial plasmid conjugation on semi-solid surfaces monitored with the green fluorescent protein (GFP) from Aequorea victoria as a marker. Gene. 1996;173:59–65. doi: 10.1016/0378-1119(95)00707-5. [DOI] [PubMed] [Google Scholar]
- 10.Cover T L, Blaser M J. The pathobiology of Campylobacter infections in humans. Annu Rev Med. 1989;40:269–285. doi: 10.1146/annurev.me.40.020189.001413. [DOI] [PubMed] [Google Scholar]
- 11.de Melo M A, Gabbiani G, Pechere J C. Cellular events and intracellular survival of Campylobacter jejuni during infection of HEp-2 cells. Infect Immun. 1989;57:2214–2222. doi: 10.1128/iai.57.7.2214-2222.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Mello A, Yotis W W. The action of sodium deoxycholate on Escherichia coli. Appl Environ Microbiol. 1987;53:1944–1946. doi: 10.1128/aem.53.8.1944-1946.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle M P, Jones D M. Food-borne transmission and antibiotic resistance of Campylobacter jejuni. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 45–48. [Google Scholar]
- 15.Everest P H, Goossens H, Butzler J P, Lloyd D, Knutton S, Ketley J M, Williams P H. Differentiated Caco-2 cells as a model for enteric invasion by Campylobacter jejuni and C. coli. J Med Microbiol. 1992;37:319–325. doi: 10.1099/00222615-37-5-319. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez H, Eller G, Freymuller E, Vivanco T. Detection of Campylobacter jejuni invasion of HEp-2 cells by acridine orange-crystal violet staining. Mem Inst Oswaldo Cruz. 1997;92:509–511. doi: 10.1590/s0074-02761997000400012. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez H, Trabulsi L R. Invasive and enterotoxic properties in Campylobacter jejuni and Campylobacter coli strains isolated from humans and animals. Biol Res. 1995;28:205–210. [PubMed] [Google Scholar]
- 18.Gage D J, Bobo T, Long S R. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa) J Bacteriol. 1996;178:7159–7166. doi: 10.1128/jb.178.24.7159-7166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerry P, Yao R, Alm R A, Burr D H, Trust T J. Systems of experimental genetics for Campylobacter species. Methods Enzymol. 1994;235:474–481. doi: 10.1016/0076-6879(94)35163-5. [DOI] [PubMed] [Google Scholar]
- 20.Hahn A F. Guillain-Barre syndrome. Lancet. 1998;352:635–641. doi: 10.1016/S0140-6736(97)12308-X. [DOI] [PubMed] [Google Scholar]
- 21.Hu L, Kopecko D J. Campylobacter jejuni 81–176 associates with microtubules and dynein during invasion of human intestinal cells. Infect Immun. 1999;67:4171–4182. doi: 10.1128/iai.67.8.4171-4182.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes R A, Hadden R D, Gregson N A, Smith K J. Pathogenesis of Guillain-Barre syndrome. J Neuroimmunol. 1999;100:74–97. doi: 10.1016/s0165-5728(99)00195-2. [DOI] [PubMed] [Google Scholar]
- 23.Konkel M E, Cieplak W., Jr Altered synthetic response of Campylobacter jejuni to cocultivation with human epithelial cells is associated with enhanced internalization. Infect Immun. 1992;60:4945–4949. doi: 10.1128/iai.60.11.4945-4949.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konkel M E, Hayes S F, Joens L A, Cieplak W., Jr Characteristics of the internalization and intracellular survival of Campylobacter jejuni in human epithelial cell cultures. Microb Pathog. 1992;13:357–370. doi: 10.1016/0882-4010(92)90079-4. [DOI] [PubMed] [Google Scholar]
- 25.Konkel M E, Joens L A, Mixter P F. Molecular characterization of Campylobacter jejuni virulence determinants. In: Nachamkin I, Blaser M J, editors. Campylobacter. 2nd ed. Washington, D.C.: American Society for Microbiology; 2000. pp. 217–240. [Google Scholar]
- 26.Konkel M E, Mead D J, Hayes S F, Cieplak W., Jr Translocation of Campylobacter jejuni across human polarized epithelial cell monolayer cultures. J Infect Dis. 1992;166:308–315. doi: 10.1093/infdis/166.2.308. [DOI] [PubMed] [Google Scholar]
- 27.Labigne-Roussel A, Harel J, Tompkins L. Gene transfer from Escherichia coli to Campylobacter species: development of shuttle vectors for genetic analysis of Campylobacter jejuni. J Bacteriol. 1987;169:5320–5323. doi: 10.1128/jb.169.11.5320-5323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacatena R M, Banner D W, Castagnoli L, Cesareni G. Control of initiation of pMB1 replication: purified Rop protein and RNA I affect primer formation in vitro. Cell. 1984;37:1009–1014. doi: 10.1016/0092-8674(84)90435-5. [DOI] [PubMed] [Google Scholar]
- 29.Matthysse A G, Stretton S, Dandie C, McClure N C, Goodman A E. Construction of GFP vectors for use in gram-negative bacteria other than Escherichia coli. FEMS Microbiol Lett. 1996;145:87–94. doi: 10.1111/j.1574-6968.1996.tb08561.x. [DOI] [PubMed] [Google Scholar]
- 30.Miller W G, Leveau J H J, Lindow S E. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol Plant-Microbe Interact. 2000;13:1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- 31.Miller W G, Lindow S E. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene. 1997;191:149–153. doi: 10.1016/s0378-1119(97)00051-6. [DOI] [PubMed] [Google Scholar]
- 32.Ouahrani-Bettache S, Porte F, Teyssier J, Liautard J P, Kohler S. pBBR1-GFP: a broad-host-range vector for prokaryotic promoter studies. BioTechniques. 1999;26:620–622. doi: 10.2144/99264bm05. [DOI] [PubMed] [Google Scholar]
- 33.Park S F. The use of hipO, encoding benzoylglycine amidohydrolase (hippuricase), as a reporter of gene expression in Campylobacter coli. Lett Appl Microbiol. 1999;28:285–290. doi: 10.1046/j.1365-2672.1999.00524.x. [DOI] [PubMed] [Google Scholar]
- 34.Purdy D, Park S F. Heterologous gene expression in Campylobacter coli: the use of bacterial luciferase in a promoter probe vector. FEMS Microbiol Lett. 1993;111:233–237. doi: 10.1111/j.1574-6968.1993.tb06391.x. [DOI] [PubMed] [Google Scholar]
- 35.Russell R G, Blake D C., Jr Cell association and invasion of Caco-2 cells by Campylobacter jejuni. Infect Immun. 1994;62:3773–3779. doi: 10.1128/iai.62.9.3773-3779.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skirrow M B, Blaser M J. Clinical and epidemiologic considerations. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 3–8. [Google Scholar]
- 37.Stern N J. Reservoirs for Campylobacter jejuni and approaches for intervention in poultry. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 49–60. [Google Scholar]
- 38.Stueber D, Bujard H. Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1982;1:1399–1404. doi: 10.1002/j.1460-2075.1982.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor D E. Genetics of Campylobacter and Helicobacter. Annu Rev Microbiol. 1992;46:35–64. doi: 10.1146/annurev.mi.46.100192.000343. [DOI] [PubMed] [Google Scholar]
- 40.Thies F L, Karch H, Hartung H P, Giegerich G. The ClpB protein from Campylobacter jejuni: molecular characterization of the encoding gene and antigenicity of the recombinant protein. Gene. 1999;230:61–67. doi: 10.1016/s0378-1119(99)00054-2. [DOI] [PubMed] [Google Scholar]
- 41.Trieu-Cuot P, Gerbaud G, Lambert T, Courvalin P. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 1985;4:3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsien R, Prasher D. Molecular biology and mutation of green fluorescent protein. In: Chalfie M, Kain S, editors. Green fluorescent protein: properties, applications, and protocols. Chichester, England: John Wiley and Sons Ltd.; 1998. pp. 97–118. [Google Scholar]
- 43.Uyttendaele M, Debevere J. Evaluation of Preston medium for detection of Campylobacter jejuni in vitro and in artificially and naturally contaminated poultry products. Food Microbiol (London) 1996;13:115–122. [Google Scholar]
- 44.Valdivia R H, Cormack B P, Falkow S. The uses of green fluorescent protein in prokaryotes. In: Chalfie M, Kain S, editors. Green fluorescent protein: properties, applications, and protocols. Chichester, England: John Wiley and Sons Ltd.; 1998. pp. 121–138. [Google Scholar]
- 45.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Taylor D E. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene. 1990;94:23–28. doi: 10.1016/0378-1119(90)90463-2. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Taylor D E. Natural transformation in Campylobacter species. J Bacteriol. 1990;172:949–955. doi: 10.1128/jb.172.2.949-955.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wooldridge K G, Ketley J M. Campylobacter-host cell interactions. Trends Microbiol. 1997;5:96–102. doi: 10.1016/S0966-842X(97)01004-4. [DOI] [PubMed] [Google Scholar]
- 49.Wösten M M, Boeve M, Koot M G, van Nuene A C, van der Zeijst B A. Identification of Campylobacter jejuni promoter sequences. J Bacteriol. 1998;180:594–599. doi: 10.1128/jb.180.3.594-599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao R, Alm R A, Trust T J, Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene. 1993;130:127–130. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]
- 51.Yuki N. Pathogenesis of Guillain-Barre and Miller Fisher syndromes subsequent to Campylobacter jejuni enteritis. Jpn J Infect Dis. 1999;52:99–105. [PubMed] [Google Scholar]