Summary
Polygenic risk scores (PRSs) quantify the contribution of multiple genetic loci to an individual’s likelihood of a complex trait or disease. However, existing PRSs estimate this likelihood with common genetic variants, excluding the impact of rare variants. Here, we report on a method to identify rare variants associated with outlier gene expression and integrate their impact into PRS predictions for body mass index (BMI), obesity, and bariatric surgery. Between the top and bottom 10%, we observed a 20.8% increase in risk for obesity (p = 3 × 10−14), 62.3% increase in risk for severe obesity (p = 1 × 10−6), and median 5.29 years earlier onset for bariatric surgery (p = 0.008), as a function of expression outlier-associated rare variant burden when controlling for common variant PRS. We show that these predictions were more significant than integrating the effects of rare protein-truncating variants (PTVs), observing a mean 19% increase in phenotypic variance explained with expression outlier-associated rare variants when compared with PTVs (p = 2 × 10−15). We replicated these findings by using data from the Million Veteran Program and demonstrated that PRSs across multiple traits and diseases can benefit from the inclusion of expression outlier-associated rare variants identified through population-scale transcriptome sequencing.
Keywords: polygenic risk scores, rare variants, transcriptomics, BMI, obesity, complex disease
Introduction
A major goal of complex disease genetics is predicting an individual’s disease risk. Recent efforts have aimed at summarizing genome-wide risk for multiple traits and diseases via polygenic risk scores (PRSs),1, 2, 3, 4, 5, 6 which are derived by summing genome-wide common genetic variants associated with a given trait or disease. PRSs have demonstrated stratification of genetic disease risk, but there remains substantial unexplained variability in these predictions.7 One potential explanation for this variability is the presence of rare variants with large phenotypic effects that are unaccounted for in PRS models.2
Despite the well-known contributions of specific, rare genetic variants to complex traits and diseases,8,9 rare variants in aggregate have been difficult to robustly characterize and integrate into PRS predictions because of their abundance in the genome, poor interpretability, and sample size constraints. Currently, beyond single risk loci such as APOE in Alzheimer disease,10 BRCA1 in breast cancer,11 and LDLR in familial hypercholesterolemia,12 the majority of DNA-sequencing-based approaches focus only on rare, protein-truncating variants (PTVs).13 New approaches that aggregate multiple rare variants have provided opportunity to improve risk prediction,14 however these have further focused only on missense and PTV rare variant burden. To extend to other impactful variants, recent studies have focused on individuals with outlier gene expression demonstrating enrichments of multiple classes of rare variants beyond missense and PTVs,15, 16, 17, 18 finding that such variants can have large effects on traits and diseases.19,20
Given the known large phenotypic effects of rare variants associated with outlier gene expression—and that these variants are not currently included in existing PRSs—we sought to test whether this subset of rare variants in aggregate can aid in genetic risk prediction. We developed an approach that integrates outlier-associated rare variants from population-scale, transcriptome sequencing in the GTEx project (v8), and demonstrated improved disease and trait prediction in the UK Biobank (UKB)21 and Million Veteran Program (MVP).
Subjects and methods
GTEx v8 data
Rare SNV calls passing quality control and mapped to the hg38 genome build were obtained from GTEx (v8) whole-genome sequencing (WGS) data (see data and code availability). Using the software bedtools22 (--window flag), variants were linked to genes if falling within the gene body, 10 kb upstream of transcription start site or 10 kb downstream of the transcription end site. The 10 kb window was chosen on the basis of prior studies that demonstrated significant rare variant enrichment for outliers compared to non-outliers with this window size.15,19,23 We restricted variants to those mapping to genes within autosomes. Using the software Vcfanno,24 variants were intersected with gnomAD (version r2.0.2, with liftover to the hg38 genome build)25 and CADD26 databases to obtain the minor allele frequency (MAF) and CADD score, respectively, for each variant. MAFs were calculated across all individuals in gnomAD and were retained for variants with gnomAD MAF ≤ 1%. Variant effect annotations were obtained with Variant Effect Predictor (VEP) (version 88).27
RNA sequencing (RNA-seq) data were obtained from GTEx (v8) (see data and code availability). To identify GTEx outlier gene expression samples, normalized gene expression values (TPM) were processed across 49 GTEx v8 tissues, limited to autosomal genes annotated as protein coding or lincRNA. A minimum expression filter was applied per gene (≥20% individuals with TPM > 0.1 and read count > 6); genes not passing this filter were removed. Expression values were PEER28 factor corrected (with 15 factors for tissues with ≤150 samples, 30 for tissues with <250 samples, 45 for tissues with <350 samples, and 60 for tissues with ≥350 samples) and adjusted for the lead cis-eQTL per gene for a given tissue as well as genotype principal components of ancestry 1–3 and sex. Finally, residuals were scaled and centered to generate expression Z scores. Individuals exhibiting global patterns of outlier gene expression for a given tissue were removed from the final corrected expression matrix for that tissue. Global outlier is defined as any individual who has a gene expression abs(Z score) ≥ 2 in more than 100 genes in a given tissue.
UKB data
UKB Phase 2 genome-wide association study (GWAS) summary statistics were obtained from the Neale Lab server (see data and code availability). For the continuous trait BMI, we selected the inverse-rank-normalized (IRNT), both-sexes version (file name: “21001_irnt.gwas.imputed_v3.both_sexes.tsv.bgz”). We selected a further 1,963 GWASs for permutation testing by using the following filtering criteria: both-sexes; IRNT version for all continuous traits; any ordinal trait (n_cases = NA); or binary traits with n_cases ≥ 1,000. As described more fully in the Neale Lab server documentation, the variants included in each GWAS had been filtered for imputation score > 0.8, UKB MAF > 0.1%, and Hardy-Weinberg equilibrium p value > 1 × 10−10. Additionally, we removed all variants flagged as low confidence (low_confidence_variant = TRUE).
All other phenotypic and genetic data were obtained from the data instance approved under UKB application #24983 (see data and code availability). Based on the information provided in protocol 44532, Stanford University IRB review has determined that the research does not involve human subjects as defined in 45 CFR 46.102(f) or 21 CFR 50.3(g). All participants of UK Biobank provided written informed consent. Individual-level values for weight (UKB data field #21002) and BMI (UKB data field #21001) were downloaded from the relevant phenotype file. We averaged (using median) overall observations per individual for anyone with multiple observations of the same phenotype. Additional phenotypic and demographic data included age, sex, genotype-derived principal components 1–10, genotyping array, and comparative body size at age 10 (UKB data field #1687). To compute age at bariatric surgery, we used OPCS-4 records for procedure codes G28.1 (partial gastrectomy and anastomosis of stomach to duodenum), G28.2 (partial gastrectomy and anastomosis of stomach to transposed jejunum), G28.3 (partial gastrectomy and anastomosis of stomach to jejunum NEC), G28.4 (sleeve gastrectomy and duodenal switch), G28.5 (sleeve gastrectomy NEC), G31.2 (bypass of stomach by anastomosis of stomach to duodenum), G32.1 (bypass of stomach by anastomosis of stomach to transposed jejunum), G33.1 (bypass of stomach by anastomosis of stomach to jejunum NEC), and G71.6 (duodenal switch) combined with an approximate date of birth from the fields month of birth (UKB data field #52) and year of birth (UKB data field #34). We followed the same procedure to obtain an approximate age when ICD-10 code E66 (obesity) was first reported in the medical record (UKB data field #130792). We further used age as directly reported in the relevant file for diagnosis of stroke (UKB data field #4056) and diagnosis of pulmonary embolism (UKB data field #4022).
Individual-level genotypes for outlier-associated and matched control rare variants were obtained from UKB genotyping callset version 3. Variants in this callset were mapped to the hg19 genome build; therefore, we used the software CrossMap29 to convert genome coordinates from hg19 to hg38 given that GTEx (v8) is mapped to hg38. We restricted variants to the high-confidence set included in the UKB GWAS files described above.
UKB validation cohort
We defined a non-overlapping UKB cohort separate from the individuals included in the GWAS described above (see UKB data). Using the self-identified non-British White labels that were reported in the UKB metadata, we first inferred a larger cohort of predicted non-British White individuals by using the first and second genotype principal components. All individuals without a self-reported ethnic identity that were within +/−3 SD of the calculated mean principal component 1 (PC1) and principal component 2 (PC2) values were inferred to be non-British White. All self-reported non-British White individuals that fell out of this range were excluded. We found that the BMI PRS distribution of this non-British White cohort did not differ significantly from a normal distribution (Shapiro-Wilk normality test; p = 0.2774), suggesting that the PRS generalizes well to this cohort. We further obtained the plate and well information for all individuals included in the UKB GWAS described above by using the “european_samples.tsv” file available from the Neale Lab server (see data and code availability), as well as for all individuals in our non-British White cohort. We removed any individuals who appeared in the intersecting set.
Million Veteran Program (MVP) validation cohort
DNA extracted from individual blood samples were genotyped with a customized Affymetrix Axiom biobank array, the MVP 1.0 Genotyping Array. The array was enriched for both common and rare genetic variants of clinical significance in different ethnic backgrounds. Quality-control procedures used to assign ancestry, remove low-quality samples and variants, and perform genotype imputation to the 1000 Genomes reference panel were previously described.30 Individuals related more than second-degree cousins were excluded.
We conducted HARE (harmonized ancestry and race/ethnicity) analysis by using race/ethnicity information from MVP participants.31 Genotyped MVP participants are assigned into one of the four HARE groups (Hispanic, non-Hispanic White, non-Hispanic Black, and non-Hispanic Asian) and “other.” The analysis is based on a machine-learning algorithm, which integrates race/ethnicity information from MVP baseline survey and high-density genetic variation data. Trans-ethnic, and ethnicity-specific principal-component analyses were performed with flashPCA.32 BMI was calculated as average BMI with all measurements within a 3-year window around the date of MVP enrollment (i.e., 1.5 years before/after the date of enrollment), excluding height measurements that were >3 in or weight measurements that were >60 lbs from the average of each participant.
Genetic association with BMI in the MVP cohort was examined among 217,980 non-Hispanic White participants. Given differences between MVP and UKB in genotyping array and imputation, we could include 57,686 outlier-associated variants in the independent outlier gene count (IOGC) score calculation for MVP—65.6% of the total outlier-associated variant set. As a result of data access restrictions, we were unable to calculate PRSs in MVP.
Identifying rare variants associated with GTEx gene expression outliers and non-outliers
To link rare variants to expression outliers, we used the processed RNA-seq data (see GTEx v8 data) for each tissue and identified individuals passing a defined absolute Z score expression level for a given gene (abs(Z score) ≥ 2). We also identified the set of individuals with non-outlier gene expression, defined as abs(Z score) < 1 for a given gene and tissue. We retained only the genes with at least one outlier individual. From the set of variants identified in outlier individuals, we removed variants that were observed in any individual not passing the defined absolute Z score threshold in any tissue. We further removed any variants linked to inconsistent outlier directions in the same gene (e.g., under-expression in one outlier individual and over-expression in another). For the set of variants identified in non-outlier individuals, we defined a corresponding set of matching variants on the basis of the CADD score and gnomAD MAF of outlier variants in the same gene and tissue. For CADD, we required a match within a +/−1 window. For gnomAD MAF, we required a match within 0.1% of outlier variant gnomAD MAF (for example, for an outlier variant with gnomAD MAF of 0.3%, the match window would be 0.2%–0.4%). We subsequently confirmed there was no difference in local linkage disequilibrium (LD) for outlier and matched non-outlier variants. For analyses integrating PRSs, we further subset the list of outlier-associated and non-variants mapping to genes containing ≥1 PRS variant.
Annotating missense variants and PTVs in UKB
To identify missense variants, we used annotations available on the Neale Lab server (see data and code availability) by using the “consequence” column available in the “variants.tsv.bgz” file. We annotated predicted rare protein-truncating SNVs in the UKB by using the imputed genotype callset (version 3). We restricted variants to the high-confidence variant set described above (see UKB data) and further retained only those variants with a rate of missingness < 1% and UKB MAF < 1%. We performed variant annotation by using the Ensembl VEP27 (April 2017 version) with the LOFTEE plugin25 using the hg19 genome build. We considered the following predicted consequences as protein-truncating SNVs: frameshift variant, splice acceptor variant, splice donor variant, stop lost, stop gained, and start lost. Finally, repeating the same process as used for outlier-associated and matched non-outlier variants, we restricted variants to those mapping to genes with ≥1 PRS variant. We checked for overlap between this set of PTVs and outlier-associated variants, finding that 138 variants overlapped in both sets.
Calculating PRSs
We computed PRSs by using publicly available PRS weights obtained from The Polygenic Score (PGS) Catalog (see data and code availability): body mass index (PGS Catalog ID: PGS000027). Scores were calculated with the software plink (version 2.0) (--score flag, including “sum” modifier). Scores were scaled to generate PRS Z scores.
GWAS effect size permutation test
We performed a permutation test (n permutations = 1,000) to assess how often randomly drawn outlier-associated variants had larger GWAS effect sizes than matched non-outlier variants across genes. For each GWAS, the input data are files containing outlier-associated and non-outlier variants along with GWAS effect size, gene ID, outlier direction (under-expression/over-expression), and tissue. Additionally, for GWAS of traits and disease where we also integrate PRS information, we subset outlier genes to those with ≥1 PRS variant. For each tissue/gene/outlier-direction tuple, we randomly select one outlier-associated variant and one matched non-outlier variant. We then identify the variant (i.e., outlier-associated/non-outlier) with the largest GWAS absolute effect size. Summarizing across all tuples, we construct a 2 × 2 contingency table to compute the odds of observing an outlier-associated variant with a larger absolute effect size than non-outlier variant across genes. To generate a null distribution, we repeated this analysis for matched non-outlier variants only, comparing two randomly chosen non-outlier variants per tuple.
GTEx cis-eQTL slope and phenotype risk concordance with outlier-associated rare variants
Significant GTEx cis-eQTL summary statistics were obtained from the GTEx Portal (“∗.v8.signif_variant_gene_pairs.txt” file suffix; see data and code availability). For each tissue, we selected genes with ≥1 outlier-associated variant and ≥1 cis-eQTL variant and filtered for SNVs only and merged variants with UKB GWAS summary statistics. For genes with >1 cis-eQTL in a given tissue, the variant with the smallest UKB GWAS p value was retained. We removed genes where cis-eQTLs had either risk or protective GWAS effects but the same eQTL slope direction. Outlier-associated variants were then compared on the slope and GWAS effect direction of gene-level summarized cis-eQTL results (e.g., for cis-eQTL variants with a positive median cis-eQTL slope and GWAS risk direction, we assessed if outlier-associated variants for the same gene were overexpressed and had a GWAS risk direction). We stratified results by cis-eQTL GWAS p value, number of cis-eQTL tissues per gene, and outlier expression Z score.
Outlier-associated variant effect enrichment in BMI PRS outliers
To investigate enrichments in outlier-associated variant effects in individuals whose observed BMI differs substantially from their PRS-group prediction, we first calculated BMI Z scores separately across deciles of PRS risk. We then assessed enrichment across variant effect subtypes for IOGC top and bottom decile individuals who fall close or far from mean PRS-decile BMI. Enrichments were composed of individuals with respect to their IOGC score direction—for bottom 10% IOGC individuals, we defined BMI Z score < 0 and > −0.5 as close to mean and Z score < −2.5 as far from mean. Similarly, for top 10% IOGC individuals, BMI Z score > 0 and < 0.5 was considered close to mean and Z score > 2.5 far from mean.
Quantifying effects of IOGC score on phenotype prediction
Outlier-associated, non-outlier, and predicted protein-truncating variants were written to a separate file and input to the software plink33 (version 2.0; using --extract flag) to identify UKB individuals in the validation cohort who are heterozygous or homozygous for each variant. We then used the relevant UKB GWAS (e.g., BMI) effect estimate to assign effect directions to each outlier variant (i.e., risk/protective). As noted above (see UKB validation cohort), the UKB cohort used to estimate variant effect direction is non-overlapping with the cohort we used to calculate IOGC scores.
To compute IOGC scores for each individual in our UKB validation cohort, we first count the number of unique effect directions per gene. Per individual, we convert the beta effect estimate per variant to an integer by using a sign function,
where βk is the UKB GWAS beta coefficient for variant k. In practice, effect sizes of zero are not generally observed, so we expect to see only values of −1 or 1. Following this step, we take the distinct values per gene (i.e., remove duplicates); because our goal is to use outlier variants to quantify putative outlier gene expression, this step prevents double counting. As such, if we denote the vector of sgn(βk) for variants linked to a given i gene as s, then
We define si for each gene with at least one outlier variant, then take a sum over genes to yield the IOGC score. We split the genotypes of individual j into vectors gij for each θi and compute:
Linear regression was used for quantitative phenotypes and logistic regression for binary phenotypes. All statistical analyses were performed with R (version 3.6.0). Plots were generated with ggplot2 (version 3.3.0).34
Results
Identification of rare variants associated with gene expression outliers
To identify candidate, large-effect rare variants, we focused on rare variants associated with gene expression outliers that could also be tested for their effects on complex traits in the UKB. We first intersected the set of SNVs with gnomAD25 MAF > 0 and ≤ 1% identified in GTEx v8 with high-quality imputed variants in the UKB (subjects and methods; Figure 1A). From a starting set of 8,150,921 unique rare SNVs, we identified 1,773,318 (21.8%) variants that overlapped with those in UKB. From this intersecting set, we compared the set of variants found in GTEx outlier to non-outlier individuals to isolate the subset of rare variants present in gene expression outlier individuals only (subjects and methods). Variants were subsequently annotated to a gene if they fell within the gene body or +/−10 kb around the gene. Following this approach, we identified 90,898 unique outlier-associated variants for 15,871 genes.
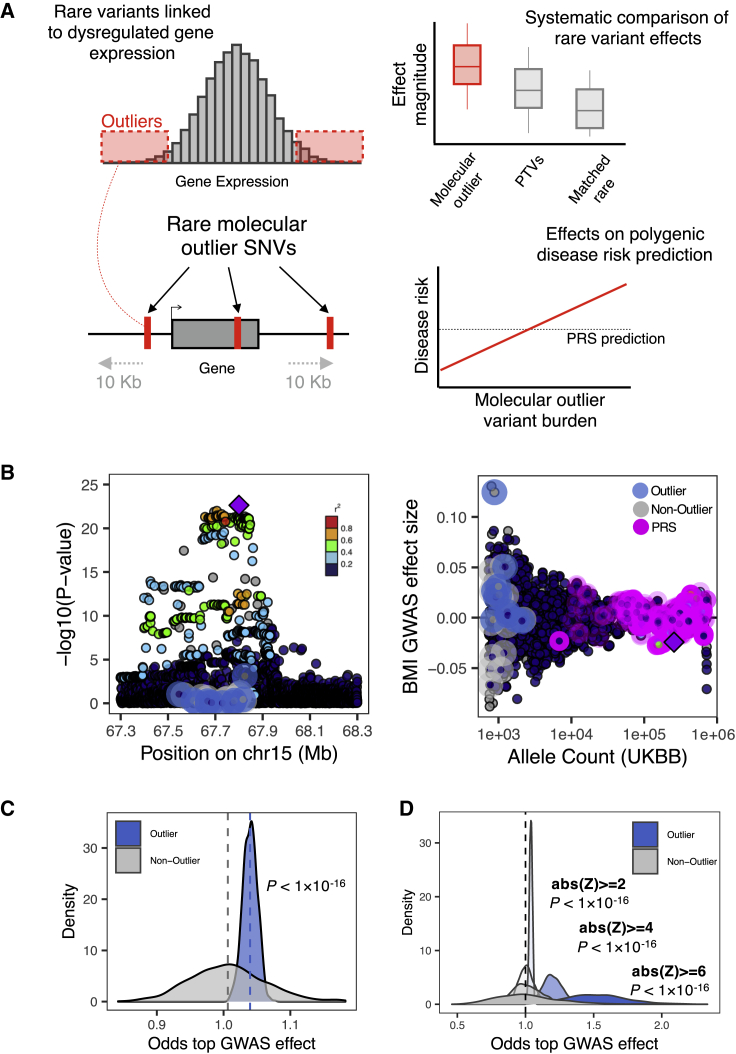
Figure 1.
Phenotypic effects of rare outlier-associated variants across genes
(A) Rare SNVs were identified in gene expression outlier individuals across 49 GTEx v8 tissues. The phenotypic effects of these variants were systematically compared with protein-truncating variants and matched rare non-outlier variants and jointly modeled with PRS estimates.
(B) Example gene locus (MAP2K5) containing a common variant genome-wide significant hit for BMI illustrates the large phenotypic effect of an outlier-associated variant: (left) showing distribution of −log10(p values) for UKB BMI GWAS for all outlier (blue halo) and non-outlier (gray halo) variants linked to the gene; (right) associated effect sizes, stratified by UKB allele count and highlighting variants included in a PRS for BMI (pink halo). Points colored by LD (1000 Genomes phase 3, European samples) relative to lead variant in gene locus (purple diamond).
(C) Distribution of odds estimates from permutation testing to assess how often randomly drawn outlier-associated variants had larger BMI GWAS effect sizes than matched non-outlier variants across genes (blue shading). This process was repeated for randomly selected non-outlier variants only (gray). p values obtained with a Wilcoxon test.
(D) Distribution of odds from permutation testing (permutation testing method as detailed in B), across more-stringent outlier Z scores. p values obtained with a Wilcoxon test.
We observed that individuals were often carriers for multiple outlier-associated rare variants highlighting the potential for rare variants in aggregate to contribute to a polygenic phenotype. Within the UKB, we observed that each individual had an average of 288 (SD = 53) outlier-associated rare variants. In comparison, each individual had on average 25 (SD = 3) rare PTVs (subjects and methods).
Rare expression outlier-associated variants impact BMI
To assess the degree to which multiple rare variants across genes can impact a polygenic trait, we focused on BMI from the UKB. We first observed that outlier-associated rare variants had the potential for large BMI effects; for example, an outlier-associated rare variant linked to the gene MAP2K5 had a BMI effect size in the top 0.05% of all variants with a locus centered on the top genome-wide significant hit (Figure 1B) and was further in the top 0.003% across all variants genome-wide.
We next systematically assessed whether outlier-associated rare variants had larger effect sizes on BMI than non-outlier rare variants (subjects and methods). We observed a median odds of 1.04 when comparing outlier versus non-outlier variants and a median odds of 1.00 when comparing non-outlier variants to themselves (Wilcoxon test, p < 1 × 10−16) (Figure 1C). We repeated this approach for increasing outlier expression Z score thresholds, observing progressively increased odds of outlier variants with larger BMI effects (median odds (BMI GWAS): abs(Z) > 4 = 1.20; abs(Z) > 6 = 1.51) but not when comparing non-outlier variants only (median odds (BMI GWAS): abs(Z) > 4 = 1.00; abs(Z) > 6 = 1.00) (Wilcoxon test, p < 1 × 10−16 for both comparisons) (Figure 1D).
We subsequently ran the permutation test across 1,963 traits and diseases including GWAS meta-categories (cancer, non-cancer diseases, treatment/medication) released in UKB Phase 2 GWAS (subjects and methods). We observed a median odds of 1.03 (SD = 0.02) across all disease and traits when comparing outlier with non-outlier variants, an effect that further increased with increasing outlier expression Z score thresholds (Figure S1). This indicates that outlier-associated rare variants are modestly enriched for rare variants with impacts on multiple traits and diseases and that the degree of outlier influences the magnitude of this effect.
To validate whether rare, outlier-associated variant effects on BMI were consistent in allelic series with common effects, we tested the consistency of their effect directions on BMI with common cis-eQTL variants. We compared the BMI GWAS effect direction between cis-eQTLs and outlier-associated variants at each locus (as an example, positive cis-eQTL slope and over-expression outliers both leading to increased BMI risk) (subjects and methods). We stratified results by cis-eQTL variant BMI p value and outlier-associated variant Z score and observed an overall mean concordance of 69% (binomial test, p = 0.001) (Figure S2).
IOGC score improves genetic risk prediction
By demonstrating that multiple rare variants can contribute to a polygenic trait, such as BMI, we next assessed whether we could construct a score to aggregate their impacts in combination with established PRS predictions. We first calculated BMI and obesity PRS for a subset of UKB individuals (n = 96,606) and observed the expected gradients in mean BMI and weight increasing by PRS deciles (Figure S3). We then used a linear regression model to assess change in BMI given an individual’s PRS, sex, age, first ten components of genetic ancestry, genotyping array, and a novel score that quantifies the total outlier-associated, rare variant burden per individual, computed by subtracting total protective from total risk outlier-associated variants collapsed to gene level (subjects and methods). We refer to this score as the independent outlier gene count (IOGC) score. We observed significant coefficient estimates for 10/15 features in the model (Figure S4). Further, as PRS was also included in the model, subsequent analyses focused on the additional predictive benefit of the IOGC score.
We observed that each standard deviation (SD) increase in IOGC score (mean absolute change in net outlier-associated gene count = 14.5 genes) was associated with a mean rate of change in BMI of 0.139 kg/m2 (linear regression, p < 1 × 10−16) (Figure 2A). We observed similar predictive power for obesity (BMI ≥ 30 kg/m2), diagnostic history of obesity (ICD-10 code E66), and severe obesity (BMI ≥ 40 kg/m2) as a function of IOGC (Figures 2B and 2C; Figures S5A and S5B; Table S1). We investigated whether the degree of outlier gene expression integrated into the IOGC score affected change in BMI, observing an increase in IOGC score coefficient at more extreme Z score thresholds (abs(Z) ≥ 2: linear regression r = 0.009, p < 1 × 10−16; abs(Z) ≥ 3: r = 0.015, p = 2 × 10−6; abs(Z) ≥ 4: r = 0.016, p = 0.02) (Figure S5C), and that the IOGC score subset to under-expression outlier-associated variants had slightly larger impacts than over-expression outlier-associated variants (under-expression outlier: linear regression r = 0.011, p = 3 × 10−12; over-expression outlier: r = 0.009, p = 2 × 10−12) (Figure S5D). We also see evidence that IOGC is transferable across ancestries (Figures S5E–S5G), however we expect that increased study of non-Europeans will enrich the discovery of outlier-associated rare variants.
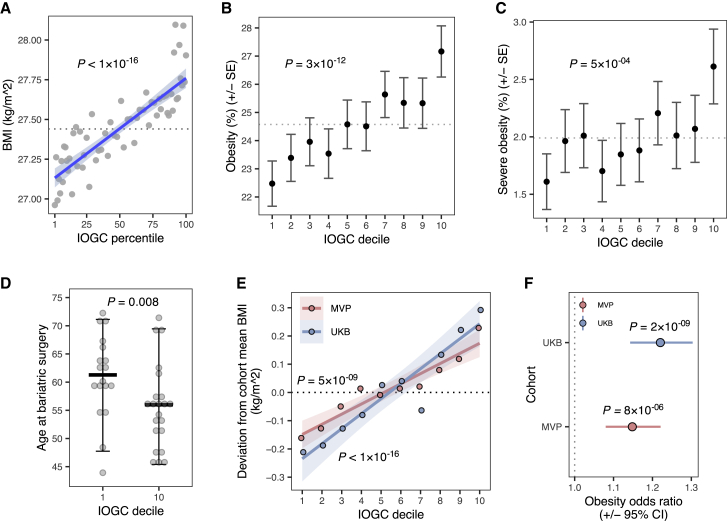
Figure 2.
Increasing burden of outlier rare variants is associated with a significant deviation in body mass index and obesity and earlier age of onset for bariatric surgery
(A) Mean BMI across 96,606 UKB individuals binned by IOGC score percentile (gray points). Linear regression fit is displayed in blue. Dashed line indicates cohort average. p value obtained from linear regression.
(B and C) Mean rates of obesity (BMI ≥ 30 kg/m2) (B) and severe obesity (BMI ≥ 40 kg/m2) (C), across deciles of IOGC score. Dashed line indicates cohort averages. Error bars indicate standard error of the mean. p values obtained from logistic regression.
(D) Age at time of bariatric surgery for individuals in top and bottom 10% of IOGC score. Crossbars indicate median, error bars indicate 90% of data range. p value obtained from Wilcoxon test.
(E) Deviation from cohort mean BMI in UKB and MVP as a function of IOGC score decile. p values obtained from linear regression.
(F) Risk for obesity (BMI ≥ 30 kg/m2) in UKB and MVP comparing individuals stratified to top and bottom 10% of IOGC score. Error bars indicate 95% confidence interval. p values obtained from Fisher's exact test.
Given the trajectories for obesity risk associated with IOGC score, we hypothesized that IOGC could impact the time course for obesity-related medical interventions such as bariatric surgery. For individuals with evidence of bariatric surgery (n = 159; subjects and methods), we calculated age at time of procedure and observed a median 5.29 years earlier onset among individuals in the top decile of IOGC score compared to bottom decile (median age at bariatric surgery: decile 1 = 61.28; decile 10 = 55.98; Wilcoxon test, p = 0.008; Figure 2D) and a median 6.16 years earlier medical diagnosis of obesity (ICD-10 code E66) (median age at diagnosis: decile 1 = 54.30; decile 10 = 48.14; Wilcoxon test, p = 0.009). Among the individuals in the top decile of IOGC who had a medical history of bariatric surgery, 50% would not have been considered high-risk from their PRS alone (defined as PRS Z score < 1). Among individuals classified as severely obese (BMI ≥ 40 kg/m2), we observed suggestive evidence of an earlier age of onset for comorbidities, including stroke and pulmonary embolism, as a function of IOGC score (Figure S6).
We further observed that the IOGC score was predictive for effects that manifest from childhood. We identified a subset of individuals in UKB (n = 45,840) who provided self-reported information on being “plumper” or “thinner” than average at age 10 (UKB data field #1687). We tested the association of IOGC score with childhood body size (subjects and methods) and observed a modest increase in the likelihood across each decile of IOGC of having a “plumper” comparative body size at 10 (logistic regression, p = 0.003); comparing low and high deciles of IOGC score (10%, 90%), we observed a mean “plumper” body size at age 10 in IOGC decile 1 of 30.41% and 33.98% in IOGC decile 10.
We investigated whether IOGC from UKB replicated in a large-scale external cohort. We calculated IOGC in MVP by using the intersecting set of outlier-associated variants available in both cohorts (N variants = 57,686) (subjects and methods). We observed that IOGC was significantly associated with BMI in MVP (linear regression, r = 0.007, p = 5 × 10−9; Figure 2E). We also observed replication of effects of IOGC on risk for obesity when comparing individuals in the top decile of IOGC score compared to bottom decile (Fisher’s exact test, MVP: odds ratio = 1.15 [CI 1.08–1.22], p = 8 × 10−6; UKB: odds ratio = 1.22 [CI 1.15–1.30], p = 2 × 10−9; subjects and methods; Figure 2F).
To assess whether rare variants used in calculating the IOGC were driving effects on BMI in excess of the addition of random rare variants, we compared IOGC results for outlier-associated variants and matched, non-outlier variants (subjects and methods). We observed an overall 33.3% increase in incremental R2 when using outlier-associated compared with non-outlier variants (mean incremental R2: outlier = 0.047%; non-outlier = 0.036%; Wilcoxon test, p < 1 × 10−16). This effect was pronounced even for smaller matched subsets of outlier and non-outlier variants (Figure S7A) as well as matched rare missense variants and PTVs in intersecting genes (subjects and methods; Figure S7B). We repeated this test for the subset of outlier-associated variants at an expression outlier threshold abs(Z score) ≥ 3, observing a greater than 3-fold increase in incremental R2 compared to matched non-outlier variants (mean incremental R2: outlier = 0.016%; non-outlier = 0.005%; Wilcoxon test, p < 1 × 10−16), highlighting the larger phenotypic effects of rare variants linked to more-extreme gene expression outliers.
IOGC aggregates multiple classes of large-impact rare variants beyond PTVs
We investigated the composition of outlier-associated variants contributing to IOGC-based prediction. We compared rare variants within individuals with significant uncaptured variance between their PRS prediction and their observed BMI (subjects and methods). We observed that individuals far from their predicted PRS mean were significantly enriched for missense, regulatory, 3′ and 5′ UTR outlier-associated variants, and outlier-associated variants proximal (+/−1 kb) to the transcription start site (TSS) (Figure 3A) and with further evidence of contributions from splice outlier-associated variants (Fisher’s exact test; missense: odds ratio = 1.11 [CI 0.99–1.23], p = 0.05; regulatory: odds ratio = 1.06 [CI 1.02–1.10], p = 0.006; 3′ UTR: odds ratio = 1.09 [CI 1.01–1.17], p = 0.04; 5′ UTR: odds ratio = 1.19 [CI 1.03–1.37], p = 0.02; TSS: 1.14 [CI 1.05–1.24], p = 0.002). We repeated this process with matched non-outlier variants (subjects and methods) and observed no significant enrichments.
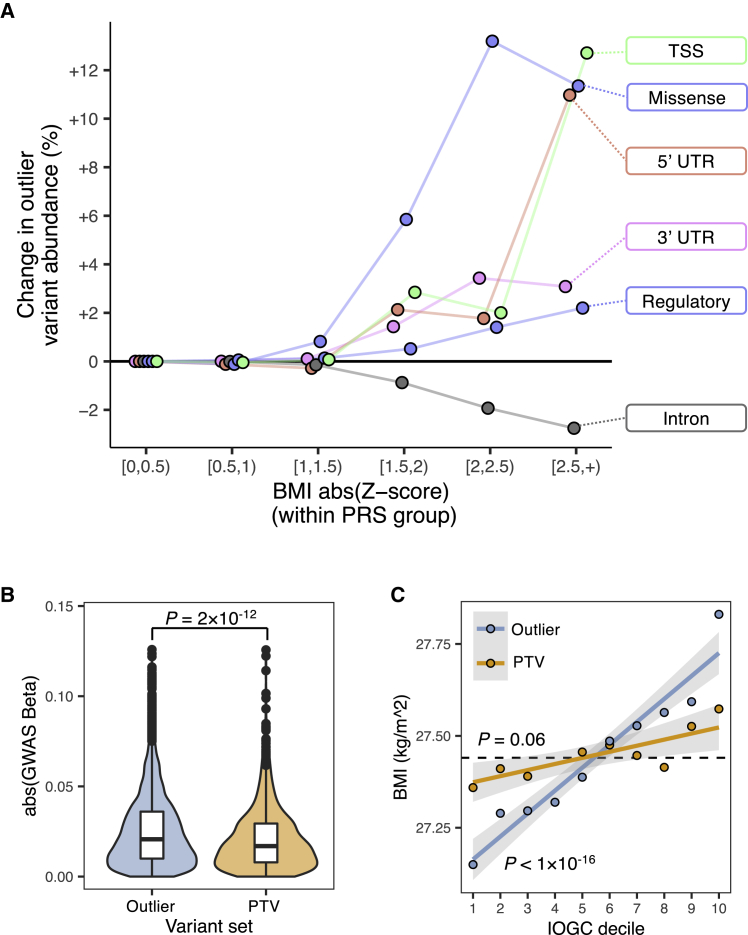
Figure 3.
Integrating expression outlier information highlights multiple classes of large-effect rare variants beyond PTVs
(A) Difference in outlier-associated variant burden across categories of variant effects as a functional of observed BMI Z score computed within PRS decile groups. Difference is with respect to outlier-associated variant abundance within bin [0,0.5). Individuals who have an observed BMI that deviates substantially from their PRS prediction are enriched for multiple classes of functional outlier-associated variants.
(B) BMI GWAS effect sizes for outlier-associated variants and PTVs. p value obtained from Wilcoxon test.
(C) BMI across deciles of IOGC score for outlier-associated variants and PTVs. p value obtained from linear regression.
We further sought to establish the relative predictive power of outlier-associated variants compared to the aggregated effects of rare PTVs. From the same high-confidence UKB imputed rare variant set as described above, we identified 1,509 rare PTVs mapped to 1,354 BMI PRS genes, comprising 344 frameshift indels, 192 splice acceptor, 317 splice donor, 78 start lost, 525 stop gained, and 53 stop lost variants (methods). Restricting to the intersecting set of genes with at least one outlier-associated variant and one PTV (n genes = 1,016), we observed that outlier-associated variants had overall larger BMI effects than PTVs (Wilcoxon test, p = 2 × 10−12) (Figure 3B). We calculated IOGC scores by using the full set of PTVs and observed no significant effect on BMI (linear regression, p = 0.06) (Figure 3C). In a permutation test using variants within the intersecting gene set (n genes = 1,016), we observed a mean increase of 19% in incremental R2 comparing IOGC by using outlier-associated variants or PTVs (mean incremental R2: outlier-associated variants = 0.0029%; PTVs = 0.0024%; Wilcoxon test, p = 2 × 10−15) (Figure S8).
Discussion
Integration of rare variants within PRSs provides an opportunity to improve prediction of genetic traits and diseases.11,35,36 We have demonstrated that a high burden of rare variants identified by their association with outlier gene expression can lead to substantial deviations in PRS-predicted phenotype. Furthermore, by integrating these rare variants into genetic risk prediction using the IOGC score, we demonstrated improvements in predicting risk for obesity beyond what was achievable with common variant-based PRSs. For example, we observed that IOGC could account for observed instances of PRS lower-risk individuals with increased risk for severe-obesity-related medical interventions, such as bariatric surgery.
The power of this approach is enabled by identifying expression outlier-associated rare variants in GTEx; these variants represent strong candidates for corresponding phenotypic effects and are not limited to protein-coding variant effects alone.19 However, given that this cohort is limited to 714 individuals, it is certain that many expression outlier-associated rare variants remain to be identified. Future large-scale RNA-seq studies in population biobanks, catalogs of expression outlier-associated rare variants, and personal -omics will only increase the efficacy of this approach. Furthermore, we could recover only a subset of outlier-associated rare variants in UKB because of limitations in rare variant imputation; recently released WGS in the UKB will recover more expression outlier-associated rare variants including ultra-rare variants, indels, and structural variants (SVs).37, 38, 39 Notably however, by using only the subset of outlier-associated rare variants between GTEx and UKB, we demonstrated improved power for genetic risk prediction beyond what could be achieved by integrating the effects of multiple rare PTVs alone.
Our approach has the opportunity for multiple future methodological improvements. The IOGC score assesses the number of genes with outlier-associated variants and does not use their effect size weights because of statistical imprecision as a result of current cohort sizes. A number of other rare variant burden testing approaches are available.40 A recent study by Lali et al.14 showed that it is possible to construct an individual rare variant score without collapsing to genes; however, this was limited to variants mapping to only a subset of the full set of genes containing PRS variants. By including outlier-associated rare variants in these models, we would expect opportunities to increase the power of these approaches. Further, in this work, we focused only on gene expression outlier-associated rare variants. Our own work has shown that splicing outlier-associated rare variants are also abundant and can contribute to complex traits.19 Future studies may benefit from integrating a broader collection of outlier-associated rare variant effects or outliers from additional multi-omics phenotypes.
Combined, our study demonstrates utility for the prediction of polygenic traits and diseases with both gene expression outlier-associated rare variants and PRS.
Acknowledgments
We thank Jonathan Pritchard, Hakhamanesh Mostafavi, and other members of the Pritchard Lab at Stanford University for helpful comments on this manuscript. C.S. was supported by NIH grant T32LM012409. N.M.F. and M.G.D. were supported by NSF Graduate Research Fellowships (grant number 1656518) and, for N.M.F., the Stanford Center for Computational, Evolutionary and Human Genomics. M.A. is supported by NLM training grant T15LM007033. X.L. is supported by the NNSF of China (grant number 31970554) and National Key R&D Program of China (grant numbers 2021YFA0805200 and 2019YFC1315804). M.J.G. was supported by a Stanford Graduate Fellowship. Y.T. was supported by a Funai Overseas Scholarship from the Funai Foundation for Information Technology. M.A.R. was supported by NIH grants U01HG009080 and R01HG010140. S.B.M. is supported by NIH grants U01HG009431, R01HL142015, R01HG008150, R01AG066490, U01HG009080, R01HG011432, R01MH125244, and U01AG072573. This research was conducted with the UK Biobank Resource under application number 24983, “Generating effective therapeutic hypotheses from genomic and hospital linkage data.” This work used supercomputing resources provided by the Stanford Genetics Bioinformatics Service Center supported by NIH grant number S10OD023452. This research was supported by funding from the Department of Veterans Affairs Office of Research and Development, Million Veteran Program (MVP) grant numbers I01-BX003340, I01-BX003362, and I01-BX004821. A list of MVP core investigators appears in the supplemental information. This publication does not represent the views of the NIH, VA, or the US Government. The funders had no role in study design, data collection and analysis, decision to publish, or manuscript preparation.
Author contributions
Conceptualization, C.S. and S.B.M.; Methodology, C.S. and S.B.M.; Software, C.S., N.M.F., and Q.H.; Formal Analysis, C.S., N.M.F., Q.H., M.G.D., Y.T., A.S.R., M.R.K., J.M.J., X.L., and M.J.G.; Investigation, C.S., N.M.F., M.G.D., A.S.R., and S.B.M.; Data Curation, T.L.A., M.A., and M.A.R.; Resources, Q.H., J.H., T.L.A., C.J.O., Y.V.S., C.K., A.R., and M.A.R.; Writing - Original Draft, C.S. and S.B.M.; Writing - Review & Editing, C.S., N.M.F., M.G.D., A.S.R., M.A., Q.H., J.H., T.L.A., C.J.O., Y.V.S., M.A.R., C.K., A.R., and S.B.M.; Funding Acquisition, M.A.R. and S.B.M.
Declaration of interests
S.B.M. is a consultant for Myome Inc, Tenaya Therapeutics, and BioMarin. C.J.O. is an employee of Novartis Institute for Biomedical Research. S.B.M. and C.S. report a patent application related to this work.
Published: May 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.04.015.
Contributor Information
Craig Smail, Email: csmail@cmh.edu.
Stephen B. Montgomery, Email: smontgom@stanford.edu.
Supplemental information
Data and code availability
GTEx (v8) RNA-seq and WGS data are available from dbGaP (dbGaP: phs000424.v8.p2). GTEx (v8) eQTL summary statistics were obtained from the GTEx Portal available at https://gtexportal.org/home/datasets. UK Biobank (UKB) data were obtained under application number 24983 (PI: Dr. Manuel Rivas). UKB Phase 2 GWAS summary statistics were obtained from the Neale Lab server available at http://www.nealelab.is/uk-biobank. Polygenic risk score (PRS) for body mass index was obtained from PGS Catalog available at https://www.pgscatalog.org/. Gene annotation data were obtained from GENCODE (version 19) available at https://www.gencodegenes.org/human/release_19.html. Allele frequency data were obtained from gnomAD (version r2.0.2) available at https://console.cloud.google.com/storage/browser/gnomad-public/release/2.0.2/. The code generated during this study is available at https://github.com/csmail/iogc.
References
- 1.Khera A.V., Chaffin M., Wade K.H., Zahid S., Brancale J., Xia R., Distefano M., Senol-Cosar O., Haas M.E., Bick A., et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell. 2019;177:587–596.e9. doi: 10.1016/j.cell.2019.03.028.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin A.R., Daly M.J., Robinson E.B., Hyman S.E., Neale B.M. Predicting polygenic risk of psychiatric disorders. Biol. Psychiatry. 2019;86:97–109. doi: 10.1016/j.biopsych.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elliott J., Bodinier B., Bond T.A., Chadeau-Hyam M., Evangelou E., Moons K.G.M., Dehghan A., Muller D.C., Elliott P., Tzoulaki I. Predictive accuracy of a polygenic risk score–enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA. 2020;323:636–645. doi: 10.1001/jama.2019.22241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khera A.V., Chaffin M., Aragam K.G., Haas M.E., Roselli C., Choi S.H., Natarajan P., Lander E.S., Lubitz S.A., Ellinor P.T., et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y.D., Hurson A.N., Zhang H., Choudhury P.P., Easton D.F., Milne R.L., Simard J., Hall P., Michailidou K., Dennis J., et al. Assessment of polygenic architecture and risk prediction based on common variants across fourteen cancers. Nat. Commun. 2020;11:3353. doi: 10.1038/s41467-020-16483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riveros-Mckay F., Weale M.E., Moore R., Selzam S., Krapohl E., Sivley R.M., Tarran W.A., Sørensen P., Lachapelle A.S., Griffiths J.A., et al. Integrated polygenic tool substantially enhances coronary artery disease prediction. Circ. Genom Precis Med. 2021;14:e003304. doi: 10.1161/circgen.120.003304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torkamani A., Wineinger N.E., Topol E.J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 8.Yang J., Bakshi A., Zhu Z., Hemani G., Vinkhuyzen A.A.E., Lee S.H., Robinson M.R., Perry J.R.B., Nolte I.M., van Vliet-Ostaptchouk J.V., et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet. 2015;47:1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mancuso N., Rohland N., Rand K.A., Tandon A., Quinque D., Mallick S., Stram A., Sheng X., Easton D.F., Eeles R.A., et al. The contribution of rare variation to prostate cancer heritability. Nat. Genet. 2016;48:30–35. doi: 10.1038/ng.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leonenko G., Baker E., Stevenson-Hoare J., Sierksma A., Fiers M., Williams J., de Strooper B., Escott-Price V. Identifying individuals with high risk of Alzheimer’s disease using polygenic risk scores. Nat. Commun. 2021;12:4506. doi: 10.1038/s41467-021-24082-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuchenbaecker K.B., McGuffog L., Barrowdale D., Lee A., Soucy P., Dennis J., Domchek S.M., Robson M., Spurdle A.B., Ramus S.J., et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J. Natl. Cancer Inst. 2017;109:djw302. doi: 10.1093/jnci/djw302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel A.P., Wang M., Fahed A.C., Mason-Suares H., Brockman D., Pelletier R., Amr S., Machini K., Hawley M., Witkowski L., et al. Association of rare pathogenic DNA variants for familial hypercholesterolemia, hereditary breast and ovarian cancer syndrome, and lynch syndrome with disease risk in adults according to family history. JAMA Netw. Open. 2020;3:e203959. doi: 10.1001/jamanetworkopen.2020.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akbari P., Gilani A., Sosina O., Kosmicki J.A., Khrimian L., Fang Y.-Y., Persaud T., Garcia V., Sun D., Li A., et al. Sequencing of 640,000 exomes identifies GPR75 variants associated with protection from obesity. Science. 2021;373:eabf8683. doi: 10.1126/science.abf8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lali R., Chong M., Omidi A., Mohammadi-Shemirani P., Le A., Cui E., Paré G. Calibrated rare variant genetic risk scores for complex disease prediction using large exome sequence repositories. Nat. Commun. 2021;12:5852. doi: 10.1038/s41467-021-26114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X., Kim Y., Tsang E.K., Davis J.R., Damani F.N., Chiang C., Hess G.T., Zappala Z., Strober B.J., Scott A.J., et al. The impact of rare variation on gene expression across tissues. Nature. 2017;550:239–243. doi: 10.1038/nature24267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J., Akinsanmi I., Arafat D., Cradick T.J., Lee C.M., Banskota S., Marigorta U.M., Bao G., Gibson G. A burden of rare variants associated with extremes of gene expression in human peripheral blood. Am. J. Hum. Genet. 2016;98:299–309. doi: 10.1016/j.ajhg.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X., Battle A., Karczewski K.J., Zappala Z., Knowles D.A., Smith K.S., Kukurba K.R., Wu E., Simon N., Montgomery S.B. Transcriptome sequencing of a large human family identifies the impact of rare noncoding variants. Am. J. Hum. Genet. 2014;95:245–256. doi: 10.1016/j.ajhg.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng Y., Wang G., Yang E., Ji G., Brinkmeyer-Langford C.L., Cai J.J. Aberrant gene expression in humans. Plos Genet. 2015;11:e1004942. doi: 10.1371/journal.pgen.1004942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferraro N.M., Strober B.J., Einson J., Abell N.S., Aguet F., Barbeira A.N., Brandt M., Bucan M., Castel S.E., Davis J.R., et al. Transcriptomic signatures across human tissues identify functional rare genetic variation. Science. 2020;369:eaaz5900. doi: 10.1126/science.aaz5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonder M.J., Smail C., Gloudemans M.J., Frésard L., Jakubosky D., D’Antonio M., Li X., Ferraro N.M., Carcamo-Orive I., Mirauta B., et al. Identification of rare and common regulatory variants in pluripotent cells using population-scale transcriptomics. Nat. Genet. 2021;53:313–321. doi: 10.1038/s41588-021-00800-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montgomery S.B., Lappalainen T., Gutierrez-Arcelus M., Dermitzakis E.T. Rare and common regulatory variation in population-scale sequenced human genomes. PLoS Genet. 2011;7:e1002144. doi: 10.1371/journal.pgen.1002144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen B.S., Layer R.M., Quinlan A.R. Vcfanno: fast, flexible annotation of genetic variants. Genome Biol. 2016;17:118. doi: 10.1186/s13059-016-0973-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karczewski K.J., Francioli L.C., MacArthur D.G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1530/ey.17.14.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stegle O., Parts L., Piipari M., Winn J., Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao H., Sun Z., Wang J., Huang H., Kocher J.-P., Wang L. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics. 2014;30:1006–1007. doi: 10.1093/bioinformatics/btt730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klarin D., Damrauer S.M., Cho K., Sun Y.V., Teslovich T.M., Honerlaw J., Gagnon D.R., DuVall S.L., Li J., Peloso G.M., et al. Genetics of blood lipids among ∼300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 2018;50:1514–1523. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang H., Hui Q., Lynch J., Honerlaw J., Assimes T.L., Huang J., Vujkovic M., Damrauer S.M., Pyarajan S., Gaziano J.M., et al. Harmonizing genetic ancestry and self-identified race/ethnicity in genome-wide association studies. Am. J. Hum. Genet. 2019;105:763–772. doi: 10.1016/j.ajhg.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abraham G., Qiu Y., Inouye M. FlashPCA2: principal component analysis of Biobank-scale genotype datasets. Bioinformatics. 2017;33:2776–2778. doi: 10.1093/bioinformatics/btx299. [DOI] [PubMed] [Google Scholar]
- 33.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickham H. Springer; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 35.Tansey K.E., Rees E., Linden D.E., Ripke S., Chambert K.D., Moran J.L., McCarroll S.A., Holmans P., Kirov G., Walters J., et al. Common alleles contribute to schizophrenia in CNV carriers. Mol. Psychiatry. 2015;21:1085–1089. doi: 10.1038/mp.2015.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fahed A.C., Wang M., Homburger J.R., Patel A.P., Bick A.G., Neben C.L., Lai C., Brockman D., Philippakis A., Ellinor P.T., et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat. Commun. 2020;11:3635. doi: 10.1038/s41467-020-17374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hernandez R.D., Uricchio L.H., Hartman K., Ye C., Dahl A., Zaitlen N. Ultrarare variants drive substantial cis heritability of human gene expression. Nat. Genet. 2019;51:1349–1355. doi: 10.1038/s41588-019-0487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eyre-Walker A. Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proc. Natl. Acad. Sci. U S A. 2010;107:1752–1756. doi: 10.1073/pnas.0906182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng J., de Vlaming R., Wu Y., Robinson M.R., Lloyd-Jones L.R., Yengo L., Yap C.X., Xue A., Sidorenko J., McRae A.F., et al. Signatures of negative selection in the genetic architecture of human complex traits. Nat. Genet. 2018;50:746–753. doi: 10.1038/s41588-018-0101-4. [DOI] [PubMed] [Google Scholar]
- 40.Povysil G., Petrovski S., Hostyk J., Aggarwal V., Allen A.S., Goldstein D.B. Rare-variant collapsing analyses for complex traits: guidelines and applications. Nat. Rev. Genet. 2019;20:747–759. doi: 10.1038/s41576-019-0177-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GTEx (v8) RNA-seq and WGS data are available from dbGaP (dbGaP: phs000424.v8.p2). GTEx (v8) eQTL summary statistics were obtained from the GTEx Portal available at https://gtexportal.org/home/datasets. UK Biobank (UKB) data were obtained under application number 24983 (PI: Dr. Manuel Rivas). UKB Phase 2 GWAS summary statistics were obtained from the Neale Lab server available at http://www.nealelab.is/uk-biobank. Polygenic risk score (PRS) for body mass index was obtained from PGS Catalog available at https://www.pgscatalog.org/. Gene annotation data were obtained from GENCODE (version 19) available at https://www.gencodegenes.org/human/release_19.html. Allele frequency data were obtained from gnomAD (version r2.0.2) available at https://console.cloud.google.com/storage/browser/gnomad-public/release/2.0.2/. The code generated during this study is available at https://github.com/csmail/iogc.