Summary
In the TRIDENT-2 study, all pregnant women in the Netherlands are offered genome-wide non-invasive prenatal testing (GW-NIPT) with a choice of receiving either full screening or screening solely for common trisomies. Previous data showed that GW-NIPT can reliably detect common trisomies in the general obstetric population and that this test can also detect other chromosomal abnormalities (additional findings). However, evidence regarding the clinical impact of screening for additional findings is lacking. Therefore, we present follow-up results of the TRIDENT-2 study to determine this clinical impact based on the laboratory and perinatal outcomes of cases with additional findings. Between April 2017 and April 2019, additional findings were detected in 402/110,739 pregnancies (0.36%). For 358 cases, the origin was proven to be either fetal (n = 79; 22.1%), (assumed) confined placental mosaicism (CPM) (n = 189; 52.8%), or maternal (n = 90; 25.1%). For the remaining 44 (10.9%), the origin of the aberration could not be determined. Most fetal chromosomal aberrations were pathogenic and associated with severe clinical phenotypes (61/79; 77.2%). For CPM cases, occurrence of pre-eclampsia (8.5% [16/189] vs 0.5% [754/159,924]; RR 18.5), and birth weight <2.3rd percentile (13.6% [24/177] vs 2.5% [3,892/155,491]; RR 5.5) were significantly increased compared to the general obstetric population. Of the 90 maternal findings, 12 (13.3%) were malignancies and 32 (35.6%) (mosaic) pathogenic copy number variants, mostly associated with mild or no clinical phenotypes. Data from this large cohort study provide crucial information for deciding if and how to implement GW-NIPT in screening programs. Additionally, these data can inform the challenging interpretation, counseling, and follow-up of additional findings.
Keywords: NIPS, NIPT, cfDNA, common trisomies, fetal trisomy, first tier test, genome-wide, prenatal screening, rare autosomal trisomies, confined placental mosaicism
Introduction
The rising awareness regarding fetal testing, together with technological advancements, has boosted the market demand for non-invasive prenatal testing (NIPT), making this test an important component of prenatal care worldwide.1 Within this market, NIPT providers are expanding the test beyond screening for the common aneuploidies (trisomy 21 [Down syndrome], 18 [Edwards syndrome], and 13 [Patau syndrome])2, 3, 4 to genome-wide screening.1,5, 6, 7
Genome-wide NIPT (GW-NIPT) allows for the detection of chromosomal aberrations other than trisomies 21, 18, and 13 (additional findings). Although both the accuracy and scope of GW-NIPT continue to improve, the origin of the cell-free DNA (cfDNA) (maternal and placental rather than fetal) providing the basis for NIPT poses an inherent limitation to the test. A chromosomal abnormality detected by GW-NIPT does not always have to be fetal, and therefore requires cytogenetic follow-up testing to confirm its origin (see recommendations in web resources).8 In some cases, false-positive results have been attributed to a chromosomal abnormality that is confined to the placenta, while the fetus has a normal chromosome complement. This condition is known as confined placental mosaicism (CPM). GW-NIPT can also detect maternal aberrations including undiagnosed cancer. The possibility of an additional finding introduces a level of ambiguity to a test meant to detect severe fetal chromosomal aberrations and fuels the debate about the risks of false positives, parental anxiety, and a potential increase in (invasive) diagnostic procedures. In addition, due to the lack of data demonstrating the clinical relevance of reporting additional findings in a general obstetric population, GW-NIPT is currently considered controversial.9, 10, 11, 12
In 2014, the Dutch NIPT Consortium, a national partnership of professionals and other stakeholders involved in public prenatal care, was granted a governmental license to introduce NIPT in the Dutch prenatal screening program. This implementation study was called the Trial by Dutch Laboratories for Evaluation of Non-invasive Prenatal Testing (TRIDENT). The aim of the TRIDENT study is to determine whether and how NIPT should be offered within the national prenatal screening program in the Netherlands. The first phase (TRIDENT-1) offered NIPT as a second-tier screening test to women with an elevated risk for trisomy 21, 18, or 13 based on the first trimester combined test (FCT) or medical history (e.g., a previous child with a trisomy).13 TRIDENT-1 resulted in high NIPT uptake and a vast reduction of invasive tests, supporting the offer of NIPT to women with an increased risk for fetal trisomy.14 Additionally, results of a small cohort within the TRIDENT-1 study highlighted the potential clinical utility of screening for chromosomal aberrations other than the common trisomies by GW-NIPT in high-risk pregnancies.7 Phase 2 (TRIDENT-2) of the NIPT implementation study was initiated in 2017. NIPT as a first-tier screening test for trisomies 21, 18, and 13 and as an alternative to the FCT became available to the general obstetric population. A unique aspect of the TRIDENT-2 study is that women opting for NIPT can choose a test aimed at the analysis of the common trisomies only or a genome-wide test that also reports other autosomal chromosomal aberrations. The results of the first year of the TRIDENT-2 study were presented previously, focusing on the test uptake and test characteristics.5 GW-NIPT was shown to be a reliable and highly accurate screening test for the detection of common trisomies 21, 18, and 13 in the general obstetric population. In addition, the study showed the ability of GW-NIPT to detect other and less common chromosomal aberrations, together with the origin of these additional findings.5
We now present detailed cytogenetic and clinical follow-up data for all cases with additional findings within the first two years of the TRIDENT-2 study. With these new data, we determine not only the origin but also the implications of the detected chromosomal aberration for fetal and/or maternal health (clinical impact), filling a long-standing knowledge gap in the current literature.
Material and methods
Study design and population
The TRIDENT-2 study is a nationwide implementation study of NIPT in the Dutch prenatal screening program for trisomies 21, 18, and 13.5 In this study, NIPT is offered as a first-tier screening test to all pregnant women. High-risk pregnancies (based on medical history but not on maternal age alone) were not included as they were the subject of prenatal screening within the TRIDENT-1 study.7 After pre-test counseling by a certified obstetric care provider, women opting for NIPT could choose a test aimed at the analysis of chromosomes 21, 18, and 13 with or without the reporting of aberrations of other autosomal chromosomes. Sex chromosomes were not analyzed. Pregnant women paid an out-of-pocket fee of 175 euros (190 USD) for the NIPT in TRIDENT-2.15
Women with a NIPT result indicative of an additional finding were referred to one of the eight regional centers for prenatal diagnostics for a post-test counseling session by an experienced clinical geneticist and for subsequent obstetric follow-up care and genomic testing, according to national guidelines. Baseline characteristics of the study cohort were obtained from medical records and the national prenatal screening database (Peridos).
TRIDENT-2 was licensed by the Minister of Health, Welfare, and Sport (1017420-153371-PG) and approved by the Medical Center Ethics Committee of Erasmus MC, Rotterdam (MEC-2018-1685). All women consented to the use of their data for research purposes. More information about the inclusion and exclusion criteria for the TRIDENT-2 study, pre- and post-test counseling, and the advice for follow-up of additional findings can be found in a previous study5 and in the supplemental methods.
Laboratory analysis and bioinformatics of NIPT
Peripheral blood samples were collected at 11 weeks of pregnancy and sent to one of the three clinical genetic laboratories (Amsterdam UMC location VUMC, Rotterdam Erasmus MC, and Maastricht UMC+) performing NIPT within the TRIDENT-2 study. In the first year of TRIDENT-2, the protocol was as follows: blood was centrifuged at 1,600 × g for 10 min at 4°C without brakes and plasma was aspired. After a second centrifuge step for 10 min at 5,600 × g without brakes, cell-free DNA was isolated from the plasma through the use of QIAsymphony Circulating DNA Kits (QIAgen) followed by library preparation for genome-wide shallow sequencing (0.2×; 51 bp single-end) on the Illumina HiSeq4000 or the NextSeq500 sequencer (Illumina). In the second year, the VeriSeq NIPT Solution v.1.0.9, which involves 36-bp paired-end sequencing on a NextSeq500, was used according to the specifications of the supplier (Illumina). Bioinformatic analysis was performed using the WISECONDOR (v.2.0.1) algorithm with a resolution of approximately 10–15 Mb at the sequencing depth used. Individual z-scores per 1 Mbp bin size and Stouffer’s Z score over multiple 1 Mbp bins were calculated. The Z score cut-off of 3 was employed for calling trisomies and subchromosomal aberrations based on the sliding window approach. GRCh37 was used as reference genome. In cases where parents opted for targeted testing of trisomy 21, 18, and 13, a filter was applied to reveal only the results of WISECONDOR analysis of chromosomes 21, 18, and 13 and to mask other autosomes before the results were made available for interpretation. Chromosome 19 analysis by WISECONDOR is not reliable, because of a shortage of reference bins, and is therefore excluded from analysis.16 Sex chromosomes were not analyzed within the TRIDENT-2 study.
Additional findings
Additional findings were divided into three categories according to the type of chromosomal aberration.
-
1.
Rare autosomal trisomies (RATs): trisomies other than those involving chromosomes 21, 18, and 13. This group also includes combinations of two RATs or a RAT with a structural chromosomal aberration (combined events).
-
2.
Structural chromosomal aberrations (SAs): chromosomal abnormalities resulting from a loss and/or gain of part of a chromosomal segment. This group also includes combinations of two SAs (combined events).
-
3.
Complex profiles: chromosomal aberrations consisting of multiple losses and gains of whole, or parts, of chromosomes.
Follow-up investigations
Depending on the type of chromosomal aberration, invasive testing (chorionic villus sampling or amniocentesis) was performed and/or maternal blood was tested. The type of invasive procedure depended on the type of chromosomal aberration, gestational age, personal preferences, and the presence or absence of ultrasound abnormalities. Chorion villus biopsies and amniotic fluid cells were investigated with genomic arrays, fluorescent in situ hybridization (FISH), and/or conventional karyotyping. Uniparental disomy (UPD) studies were performed by single-nucleotide polymorphism (SNP) array or by polymorphic microsatellite repeat analysis, for cases in which NIPT showed a high risk for trisomy 6, 7, 11, 14, 15, or 20.
Postnatal genetic testing of the fetus/infant in some cases involved cytogenetic investigation of umbilical cord blood/biopsies, saliva, oral mucosa, and/or skin and organ biopsies. This included cases in which parents had refrained from prenatal invasive testing, cases where an abnormal NIPT result was caused by a maternal copy number variant (CNV) to test the infant’s carrier status, and cases of proven fetal chromosomal mosaicism, to study the distribution of abnormal cells in different fetal tissues. In some cases, additional placental studies by array and/or FISH were performed to confirm the placental origin of a genetic aberration. Alternatively, NIPT was occasionally repeated after the delivery to see whether the aberration was no longer present in the maternal blood, which would be indicative of its placental origin. Another reason to repeat NIPT after the delivery was to confirm a maternal origin of the chromosomal aberration. This could be to demonstrate an acquired chromosome abnormality (e.g., 5q/20q loss) or to find evidence that the abnormal NIPT result was caused by uterine fibroids. Additionally, in cases of a suspected maternal malignancy, extensive cytogenetic, oncologic, and/or hematologic follow-up investigations of the pregnant women were performed.17 In some cases, invasive testing was performed as well.
A structural anomaly scan was offered to all women with an additional finding.
Classification of additional findings
The origin of additional findings was classified according to criteria described previously.7
-
•
Fetal aberrations: chromosomal anomalies confirmed in the fetus by invasive diagnostic testing during pregnancy or in cord blood or other tissue postpartum.
-
•
Placental aberrations: chromosomal abnormalities not confirmed in the fetus but detected in chorionic villi (CV) during pregnancy or in placental biopsies after birth (confirmed CPM). Additionally, chromosomal aberrations typically involved in CPM (RATs) were considered cases of assumed CPM, based on large chorionic villi cytogenetic studies showing that trisomies other than 21, 18, and 13 are rarely confirmed in the fetus.8 This includes cases in which no cytogenetic testing was performed in fetus and placenta and cases where the test result in placenta and/or fetus was normal. Additionally, if placental biopsies tested normal, a placental aberration was considered not to be ruled out due to placental site variation.18
-
•
Maternal aberrations: chromosomal aberrations confirmed in maternal genomic DNA (e.g., [mosaic] trisomies and CNVs) or (assumed) acquired chromosomal aberrations most likely originating from a maternal malignant or benign tumor.
-
•
Unresolved: chromosomal aberrations of unknown origin, despite (multiple) cytogenetic follow-up tests, or because parents refrained from any follow-up testing.
Adverse perinatal outcomes
The following clinical data on adverse perinatal outcomes were retrieved from medical records (for more details, see supplemental methods).
-
1.
Maternal pregnancy complications: hypertensive disorders and gestational diabetes.
-
2.
Adverse pregnancy outcomes: (spontaneous) preterm birth (<37 weeks of gestation), delivery by emergency caesarean section, a birth weight <2.3rd or between 2.3rd–10th according to the Dutch reference curves,19,20 or postpartum hemorrhage of ≥1,000 mL.
-
3.
Adverse neonatal outcomes: 5-min Apgar score < 7, umbilical artery pH < 7.05, admission to neonatal intensive care unit (NICU), intra-uterine fetal demise (IUFD), neonatal death, or major congenital structural abnormalities detected by the structural anomaly scan, visual inspection at birth, autopsy, or at longer-term follow-up. Structural anomalies were classified as either major or minor according to the guidelines of European Surveillance of Congenital Anomalies (see EUROCAT in web resources).
Longer-term follow-up
Medical data on psychomotor and physical development of children born alive were obtained from the parents when the children were 6–24 months old, in cases of fetal or placental origin of the additional finding. The data were requested via a structured telephone interview and/or e-mail. In cases of abnormal development, the results of follow-up tests and examinations were requested via the physicians involved.
Definition of clinical impact
An additional finding was defined as having clinical impact if the chromosomal aberration was pathogenic and associated with a severe clinical phenotype in the fetus or the mother, or when the risk for an adverse perinatal outcome was increased in case of placental origin.
Reference population and statistical analysis
Descriptive statistics were used to describe maternal age, maternal BMI, gestational age, and birthweight. Adverse perinatal outcomes were compared to data from the Dutch national obstetric outcome registration (data from PERINED, accessed August 12, 2021). Prevalence of major congenital structural abnormalities was compared to the prevalence recorded in the Dutch EUROCAT registry (see web resources). Categorical data were compared using the chi-square test or Fisher exact test, depending on the outcome frequency. Relative risks are reported with 95% Katz log confidence intervals, where adjusted log intervals (i.e., addition of 0.5 'success' to each sample in the contingency table) were computed for contingency tables with at least one zero cell.21 These intervals have adequate coverage in samples of at least 75 cases.22 Statistical testing was carried out using R software, v.3.3.1 (R Project for Statistical Computing). A p value < 0.05 was considered statistically significant. For subgroup analyses, categorical data were compared using the Fisher exact test with the Bonferroni corrected significance level of 0.0125.
Results
Population
Between April 2017 and April 2019, NIPT results were provided for 149,318 pregnancies, resulting in a nationwide NIPT uptake of 43.2%, based on 345,413 pregnancies at 12 weeks of gestation in 2017–2018.23 110,739 (74.2%) chose genome-wide analysis and the remainder preferred targeted analysis of chromosomes 21, 18, and 13. A common aneuploidy was reported in 730 of the 149,318 (0.49%) pregnancies (trisomy 21: n = 503, 0.34%; trisomy 18: n = 118, 0.08%; and trisomy 13: n = 109, 0.07%). An additional finding was found in 402/110,739 (0.36%) cases, involving 196 (0.18%) RATs, 188 (0.17%) SAs, and 18 (0.02%) complex profiles. This implies that 1 in every 275 women opting for GW-NIPT received an abnormal result indicative of an additional finding. The performance of GW-NIPT for the detection of trisomy 21, 18, 13, and additional findings is given in Table S1. Table 1 presents the characteristics of the study cohort. For about half the women, it was their first pregnancy (48.5%). The mean maternal age of the women (32.9 years, interquartile range 30–36, median 33) was slightly higher than the Dutch average age of 31.4 years (source: Statistics Netherlands), and the mean gestational age was 12.4 weeks (interquartile range 11.6–12.9, median 12) at the time of blood draw. Table 2 shows the origin of the additional findings in relation to clinical outcomes.
Table 1.
Characteristics of women with additional findings
| Characteristics | Total (n = 402) | RATs (n = 196) | SAs (n = 188) | Complex profiles (n = 18) |
|---|---|---|---|---|
| Nulliparous | 48.5 (195) | 51.5 (101) | 45.7 (86) | 44.4 (8) |
| Maternal age (years) at NIPT blood draw | 33.0 (30.0–36.0) | 32.0 (29.0–36.0) | 33.0 (31.0–36.0) | 33.5 (32.0–37.0) |
| Maternal BMIa (kg/m2) at NIPT blood draw | 23.0 (21.0–25.6) | 22.7 (20.8–25.9) | 23.1 (21.4–25.4) | 23.3 (21.9–24.7) |
| GA at NIPT blood draw (weeks) | 12.0 (11.6–12.9) | 12.0 (11.4–13.0) | 12.0 (11.6–12.9) | 11.9 (11.7–12.9) |
| Non-smoker | 86.3 (347) | 82.7 (162) | 89.9 (169) | 88.9 (16) |
| Spontaneous conception | 86.3 (347) | 85.7 (168) | 87.2 (164) | 83.3 (15) |
BMI, body mass index; GA, gestational age; NIPT, non-invasive prenatal testing; RATs, rare autosomal trisomies; SAs, structural chromosomal aberrations. Table shows proportion (frequency) or median (interquartile range, 25th–75th percentile); percentages are calculated from the total number of cases within each group (column). There was a trivial amount of missing values (1%–3%) for the variables parity, smoking behavior, and method of conception.
The body-mass index is the weight in kilograms divided by the square of the height in meters.
Table 2.
The origin and outcomes of additional findings detected by genome-wide NIPT (n = 402)
| Outcomea |
Pregnancy Outcome |
||
|---|---|---|---|
| LB | TOP | IUFD | |
| Rare autosomal trisomies (n = 196) | |||
| Fetal origin (n = 15) | |||
| Pathogenic (n = 9) | 3 | 5 | 1 |
| VUS/benign (n = 6) | 4 | 2 | 0 |
| (Assumed) CPM origin (n = 179) | |||
| Adverse perinatal outcome yes (n = 94) | 86 | 3 | 5 |
| Adverse perinatal outcome no (n = 85) | 81 | 4 | 0 |
| Maternal origin (n = 2) | |||
| Pathogenic (n = 2) | 2 | 0 | 0 |
| Structural chromosomal aberrations (n = 188) | |||
| Fetal origin (n = 64) | |||
| Pathogenic (n = 52) | 4 | 48 | 0 |
| VUS/benign (n = 12) | 11 | 0 | 1 |
| CPM origin (n = 8) | |||
| Adverse perinatal outcome yes (n = 5) | 5 | 0 | 0 |
| Adverse perinatal outcome no (n = 3) | 3 | 0 | 0 |
| Maternal origin (n = 73) | |||
| Acquired (n = 23) | 22 | 0 | 1 |
| Pathogenic (n = 30) | 29 | 1 | 0 |
| VUS/benign (n = 20) | 20 | 0 | 0 |
| Unresolved origin (n = 43) | – | – | – |
| Complex (n = 18) | |||
| CPM origin (n = 2) | |||
| Adverse perinatal outcome yes (n = 1) | 1 | 0 | 0 |
| Adverse perinatal outcome no (n = 1) | 1 | 0 | 0 |
| Maternal origin (n = 15) | |||
| Acquired (n = 15) | 14 | 0 | 1 |
| Unresolved origin (n = 1) | – | – | – |
CPM, confined placental mosaicism; IUFD, intra-uterine fetal demise; LB, live born; NIPT, non-invasive prenatal testing; TOP, termination of pregnancy; VUS, variants of uncertain clinical significance.
Adverse perinatal outcome: at least one of the following adverse outcomes: 5-min Apgar score < 7, umbilical artery pH < 7.05, admission to neonatal intensive care unit (NICU), intra-uterine fetal demise (IUFD), neonatal death, major congenital structural abnormalities, (spontaneous) preterm birth (<37 weeks of gestation), delivery by emergency caesarean section, a birthweight below the 10th centile,19,20 postpartum hemorrhage of ≥1,000 mL, pregnancy-induced hypertension, pre-eclampsia, HELLP syndrome, or gestational diabetes.
Follow-up and outcomes
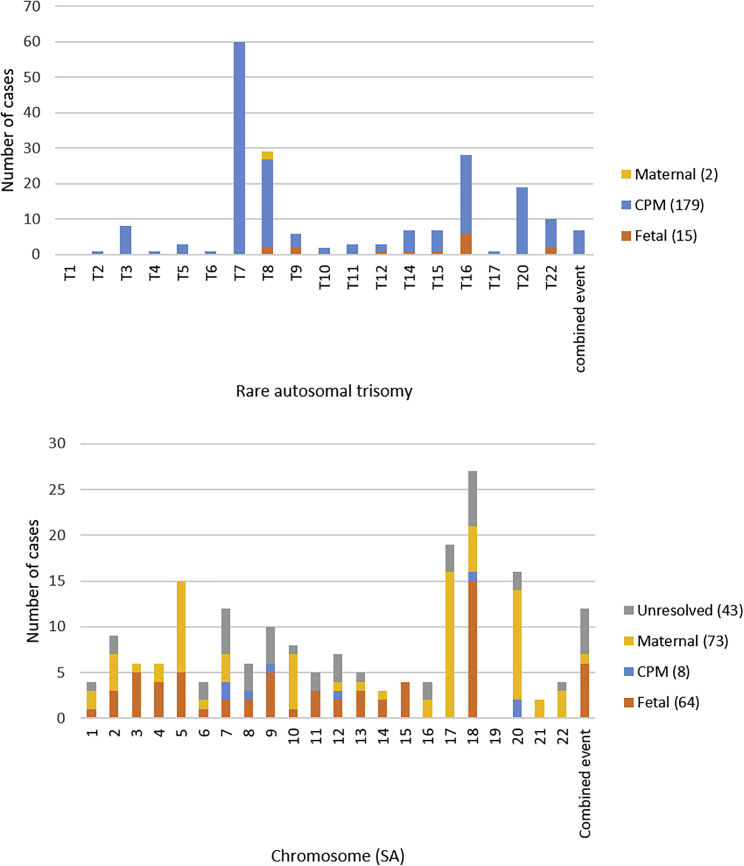
Available follow-up data are presented in Table 3. In 44/402 (10.9%) of the cases (SAs: 43/188, complex: 1/18), the origin remained unknown. Of the remaining 358 additional findings, the origin of the chromosomal aberration was proven to be fetal in 79/358 (22.1%) cases (RATs: 15/196, SAs: 64/145), (assumed) CPM in 189/358 (52.8%) cases (RATs: 179/197, SAs: 8/145, complex: 2/17), and maternal in 90/358 (25.1%) cases (RATs: 2/196, SAs: 73/145, complex: 15/17). The origin per RAT and SA is given in Figure 1.
Table 3.
Cytogenetic and clinical follow-up of women with additional findings
| GW-NIPT result | Number of cases, n | Available cytogenetic follow-up, n (%) |
Available clinical follow-up, n (%) |
||||
|---|---|---|---|---|---|---|---|
| Invasive testing | Fetus/neonatea | Pregnant womanb | Placenta biopsies | Structural anomaly scan | Postnatal outcome | ||
| RATs | 196 | 147 (75) | 165 (84) | 82 (42) | 60 (31) | 189 (96) | 193 (98) |
| SAs | 188 | 128 (68) | 140 (74) | 156 (83) | 26 (14) | 156 (83) | 183 (97) |
| Complex profiles | 18 | 11 (61) | 11 (61) | 17 (94) | 3 (17) | 18 (100) | 18 (100) |
| Total | 402 | 286 (71) | 316 (79) | 255 (63) | 89 (22) | 363 (90) | 394 (98) |
GW-NIPT, genome-wide non-invasive prenatal testing; RATs, rare autosomal trisomies; SAs, structural chromosomal aberrations. Percentages are calculated from the total number of cases within each group (row).
Prenatally and/or postnatally tested.
Cytogenetic testing of the pregnant women consisted of cytogenetic investigation of maternal blood, saliva, oral mucosa, and/or hair roots.
Figure 1.
Origin of additional findings per chromosome
Rare autosomal trisomies (upper panel) and structural chromosome aberrations (lower panel). CPM, confined placental mosaicism; SA, structural aberration; T, trisomy.
Fetal chromosomal aberrations
Of the fetal chromosomal aberrations, 61/79 (77.2%) were pathogenic (Table S2). The remainder were benign variants (10/79; 12.7%) or variants of uncertain clinical significance (VUSs) (8/79; 10.1%). Pathogenic fetal aberrations therefore accounted for 61/358 (17.0%) of additional findings with known origin.
Pathogenic aberrations included eight mosaic RATs, one maternal UPD15 (Prader-Willi syndrome), and 52 SAs, all associated with severe clinical phenotypes. In 53/61 (86.9%) cases, parents opted for termination of pregnancy. A structural anomaly scan was performed in 26 of these cases and major structural fetal anomalies were detected in 42.3% (11/26). In 8/61 (13.1%) cases, the pregnancy was not terminated. These were mostly cases of mosaic chromosomal aberrations, or aberrations with variable expression or incomplete penetrance inherited from an asymptomatic parent. In cases of mosaic trisomy 16 (3/8), IUFD occurred once, and the two other cases had adverse perinatal outcomes. The other 5/8 cases had no adverse perinatal outcomes during follow-up until 1–2 years of age.
VUSs/benign variants (18/79; 22.8%) included cases of fetal (segmental) UPD of a non-imprinted chromosome (n = 8) and CNVs (n = 10). 15/18 (83.3%) were live born, of which five had an adverse neonatal outcome. Major congenital anomalies were detected in 2/18 cases. In one pregnancy, IUFD occurred, and two were terminated for reasons other than the fetal chromosomal aberration.
(Assumed) CPM
In 189 cases, a confined placental origin of the chromosomal aberration was confirmed (n = 59; 31.2%) or assumed (n = 130; 68.8%) (Tables S3, S4, and S5). The vast majority involved RATs (n = 179; 94.7%), and the remainder were SAs (n = 8; 4.2%) and complex profiles (n = 2; 1.1%).
An IUFD occurred in 5/189 (2.6%) cases (trisomy 7, 8, 15, 16, 22) between 15 and 20 weeks of gestation. In 7/189 (3.7%) cases, pregnancy was terminated because of Turner or Klinefelter syndrome (NIPT result was indicative of a RAT; sex chromosomal aberrations were found after invasive testing), structural fetal anomalies, or social reasons. The remaining 177/189 (93.7%) pregnancies resulted in live borns.
Of (assumed) CPM cases, 52.9% (100/189) had adverse perinatal outcomes (Table 4). Compared to the general obstetric population, pregnancies with (assumed) CPM had a significantly increased risk for:
-
•
pre-eclampsia (8.5% [16/189] vs 0.5% [754/159,924]; relative risk [RR] 18.5, 95% CI 11.6–29.4, p < 0.001),
-
•
preterm birth (12.4% [22/177] vs 5.8% [8,784/150,471]; RR 2.2, 95% CI 1.5–3.2, p < 0.001),
-
•
birth weight <2.3rd percentile (13.6% [24/177] vs 2.5% [3,892/155,491]; RR 5.5, 95% CI 3.8–8.0, p < 0.001),
-
•
birth weight 2.3rd–10th percentile (13.0% [23/177] vs 7.4% [11,500/155,491]; RR 1.8, 95% CI 1.2–2.6, p = 0.005),
-
•
induction of labor (30.5% [54/177] vs 23.1% [34,756/150,471]; RR 1.3, 95% CI 1.1–1.7, p = 0.02),
-
•
planned caesarean section (12.4% [22/177] vs 7.8% [11,713/150,471]; RR 1.6, 95% CI 1.1–2.4, p = 0.02),
-
•
NICU admission (6.2% [11/177] vs 3.1% [4,879/157,391]; RR 2.1 95% CI 1.2–3.7, p = 0.02).
Table 4.
Characteristics and outcomes of pregnancies with CPM detected by GW-NIPT
| CPM (n = 189) | RR (95% CI) | p value | Estimated population percentagesa | |
|---|---|---|---|---|
| Characteristics | ||||
| Nulliparous | 50.3 (95) | – | – | – |
| Maternal age (years) at NIPT blood draw | 32.0 (29.0–36.0) | – | – | – |
| Maternal BMI (kg/m2) at NIPT blood draw | 22.7 (20.8–25.7) | – | – | – |
| GA (weeks) at NIPT blood draw | 12.0 (11.6–13.0) | – | – | – |
| Non-smoker | 82.5 (156) | – | – | – |
| Spontaneous conception | 85.7 (162) | – | – | – |
| Ultrasound | ||||
| Ultrasound structural abnormalities | 5.8 (11) | – | – | – |
| Major | 2.6 (5) | – | – | – |
| Minor | 3.2 (6) | – | – | – |
| Maternal pregnancy complications | ||||
| Pregnancy induced hypertension | 5.3 (10) | 1.0 (0.6–1.9) | 0.98 | 5.3 |
| Pre-eclampsia | 8.5 (16) | 18.5 (11.6–29.4) | <0.001 | 0.5 |
| HELLP syndrome | 2.1 (4) | – | – | – |
| Gestational diabetes | 4.2 (8) | 1.1 (0.6–2.1) | 0.96 | 4.2 |
| Termination of pregnancy | 3.7 (7) | – | – | – |
| Pregnancy outcomes | ||||
| GA (weeks) at birth | 39.3 (37.7, 40.3) | – | – | – |
| sPTB (<37 weeks GA) | 4.5 (8) | 1.4 (0.7–2.7) | 0.42 | 3.4 |
| Preterm birth | 12.4 (22) | 2.2 (1.5–3.2) | <0.001 | 5.8 |
| Birth weight, g | 3,175 (2,740, 3,560) | – | – | – |
| Birth weight <2.3rd centile | 13.6 (24) | 5.5 (3.8–8.0) | <0.001 | 2.5 |
| Birth weight 2.3rd–10th centile | 13.0 (23) | 1.8 (1.2–2.6) | 0.005 | 7.4 |
| Birth weight >10th centile | 71.8 (127) | 0.8 (0.7–0.9) | <0.001 | 90.1 |
| Onset of labor: Spontaneous | 55.4 (98) | 0.8 (0.7–0.9) | <0.001 | 69.1 |
| Onset of labor: Induction | 30.5 (54) | 1.3 (1.1–1.7) | 0.02 | 23.1 |
| Onset of labor: Planned caesarean section | 12.4 (22) | 1.6 (1.1–2.4) | 0.02 | 7.8 |
| Delivery: Spontaneous vaginal delivery | 71.8 (127) | 0.9 (0.8–1.0) | 0.07 | 77.4 |
| Delivery: Vaginal instrumental | 6.8 (12) | 1.0 (0.6–1.7) | 0.80 | 7.3 |
| Delivery: Elective caesarean section | 9.0 (16) | 1.2 (0.8–1.9) | 0.53 | 7.8 |
| Delivery: Emergency caesarean section | 10.7 (19) | 1.5 (1.0–2.2) | 0.11 | 7.5 |
| Postpartum hemorrhage (>1,000 mL) | 7.9 (15) | 1.3 (0.8–2.1) | 0.33 | 6.2 |
| Postpartum hemorrhage (>500 mL) | 18.0 (34) | – | – | – |
| Neonatal outcomes | ||||
| 5-min Apgar score <7 | 1.1 (2) | 0.8 (0.2–2.8) | 0.77 | 1.7 |
| Umbilical artery pH < 7.05b | 0.0 (0) | – | – | – |
| NICU admission | 6.2 (11) | 2.1 (1.2–3.7) | 0.02 | 3.1 |
| Neonatal death | 0.0 (0) | 0.6 (0.0–9.7) | 1.00 | 0.5 |
| Intra-uterine fetal demise | 2.6 (5) | – | – | – |
| Congenital structural abnormalitiesc | 11.6 (22) | – | – | – |
| Major | 4.2 (8) | 1.8 (0.9–3.5) | 0.15 | 2.5 |
| Minor | 7.4 (14) | – | – | – |
| Composite perinatal outcomesd | ||||
| Composite perinatal (neonatal) | 12.2 (23) | – | – | – |
| Composite perinatal (neonatal/sPTB/birth weight <p10) | 36.0 (68) | – | – | – |
| Composite (neonatal/pregnancy) | 46.6 (88) | – | – | – |
| Composite (neonatal/pregnancy/maternal) | 52.9 (100) | – | – | – |
Table shows proportion (frequency), or median (interquartile range, 25th–75th percentile). Baseline characteristics were not available for the general Dutch obstetric population. Percentages may not add up to 100% due to missing values. The numbers were calculated based on the outcomes of all cases or only the live borns, depending on the outcome measure. BMI, body mass index; CI, confidence interval; CPM, confined placental mosaicism; GA, gestational age; GW-NIPT, genome-wide non-invasive prenatal testing, HELLP, hemolysis, elevated liver enzymes, and low platelets; NICU, neonatal intensive care unit; RR, relative risk; sPTB, spontaneous preterm birth.
Data from the Dutch national obstetric outcome registration Perined. Incidence of major congenital structural abnormalities was compared to the incidence recorded in the Dutch EUROCAT registry. See web resources for URLs.
If the umbilical artery pH data were missing, but 5-min Apgar score was more than 7 and the neonate was not admitted to NICU, the neonatal outcome was classified as normal.
Total number of cases with congenital abnormalities detected on ultrasound and/or at birth or at the longer-term (including cases of TOP and IUFD).
Composite perinatal (neonatal): at least one of the following adverse outcomes: 5-min Apgar score <7, umbilical artery pH < 7.05, admission to neonatal intensive care unit (NICU), intra-uterine fetal demise (IUFD), neonatal death, and major congenital structural abnormalities. Composite perinatal (pregnancy): at least one of the following adverse outcomes: (spontaneous) preterm birth (<37 weeks of gestation), delivery by emergency caesarean section, a birthweight below the 10th centile,19,20 and postpartum hemorrhage of ≥1,000 mL. Composite perinatal (maternal): at least one of the following adverse outcomes: pregnancy induced hypertension, pre-eclampsia, HELLP syndrome, and gestational diabetes.
When excluding cases of (assumed) CPM trisomy 16 for which the association with adverse perinatal outcomes is well known,11,12 the risk for pre-eclampsia, preterm birth, birth weight <2.3rd percentile, birth weight between the 2.3rd and 10th percentile, and an onset of labor by planned caesarean section remained significantly increased (Table S6).
Exploratory analysis CPM trisomy 7, 8, 16, and 20
Figure S1 and Tables S7, S8, S9, and S10 show outcomes for the four most commonly detected subgroups of CPM; trisomies 7, 8, 16, and 20.
-
•
CPM trisomy 7 showed a significant increased risk for a birth weight <2.3rd percentile (11.9% [7/59] vs 2.5% [3,892/155,491]; relative risk [RR] 5.0, 95% CI 2.6–9.8, p < 0.001).
-
•
CPM trisomy 8 was significantly associated with an induction of labor (47.8% [11/23] vs 23.1% [34,756/150,471]; RR 2.1, 95% CI 1.4–3.2, p = 0.005).
-
•
CPM trisomy 16 showed a significant increased risk for pre-eclampsia (27.3% [6/22] vs 0.5% [754/159,924]; RR 70.7, 95% CI 37.6–132.9, p < 0.001), preterm birth (26.3% [5/19] vs 5.8% [8,784/150,471]; RR 4.8, 95% CI 2.4–9.8, p = 0.004), birth weight <2.3rd percentile (52.6% [10/19] vs 2.5% [3,892/155,491]; RR 21.5, 95% CI 14.2–32.5, p < 0.001), and induction of labor (68.4% [13/19] vs 23.1% [34,756/150,471]; RR 3.0, 95% CI 2.2–4.0, p < 0.001).
-
•
CPM trisomy 20 was significantly associated with pre-eclampsia (10.5% [2/19] vs 0.5% [754/159,924]; RR 27.2, 95% CI 8.5–86.7, p = 0.004) and with an onset of labor by planned caesarean section (38.9% [7/18] vs 7.8% [11,713/150,471]; RR 5.2, 95% CI 3.0–9.1, p < 0.001).
Maternal chromosomal aberrations
A maternal origin of the chromosomal aberration was identified in 90/358 (25.1%) cases (Table S11). These chromosomal aberrations were subdivided into acquired aberrations (38/90; 42.2%), constitutional pathogenic aberrations (32/90; 35.6%), and VUS/benign aberrations (20/90; 22.2%).
The group of acquired chromosomal aberrations included 15 complex profiles; in 12 cases the aberrations originated from a maternal malignancy, in two cases from uterine fibroids, and in one case it was caused by familial Mediterranean fever. Additionally, structural chromosomal aberrations were found in eight women with fibroids, including two cases with a chromosome 7q deletion known to be associated with these benign tumors.24 Furthermore, chromosome 5q (n = 7) and 20q (n = 8) deletions, both known to be associated with myeloid neoplasms, were found in 15 cases. None of these women were diagnosed with myeloid neoplasm. Follow-up diagnostic fetal testing occurred in 6/15, all with normal results.
The largest subgroup of women with a constitutional pathogenic chromosomal aberration had a deletion of peripheral myelin protein 22 (PMP22) (n = 10) or its reciprocal duplication (n = 6). This gene is associated with hereditary neuropathy with liability to pressure palsies (HNPP) and Charcot-Marie-Tooth (CMT1A), respectively. Of these women, 13/16 (81.3%) were symptomatic. None of the women with a PMP22 deletion/duplication opted for invasive testing. Additionally, one case with a 22q11.2 deletion and two cases with the reciprocal 22q11.21 duplication were reported, one of each had symptoms related to the syndrome. A mosaic pathogenic CNV was detected in 11 asymptomatic women, and mosaic trisomy 8 was found in two phenotypically normal women. None of these were inherited by their offspring.
Finally, 13 women had chromosomal aberrations which were found to be variants of unknown significance or benign CNVs (n = 7). In 5/20, follow-up diagnostic testing of the fetus or cytogenetic testing on cord blood was performed with a normal result.
Unresolved
Forty-four of 402 (10.9%) additional findings were classified as unresolved, as the origin of the abnormal NIPT result could not be determined despite follow-up investigations in all but three cases. Most cases, 42/44 (95.5%), were born alive (Table S12). In 2/44 (4.5%) cases, major structural congenital abnormalities were detected. A relation between these abnormalities and the NIPT result could not be established with follow-up postnatal cytogenetic testing or based on current literature. One pregnancy was terminated on social grounds and one ended in an IUFD (1st trimester), both without any follow-up testing of the fetus.
Discussion
The nationwide implementation of NIPT in the Netherlands (TRIDENT-2 study) offered all pregnant women the choice of testing for common trisomies 21, 18, and 13 only or genome-wide testing. Previous studies showed that GW-NIPT is a reliable and robust screening test for common trisomies.5, 6, 7 Additionally, these studies showed the ability of GW-NIPT to detect chromosomal aberrations other than the common trisomies and their possible origins (fetal, placental, and maternal). In order to explore the clinical impact of screening for chromosomal aberrations other than common trisomies on fetal and/or maternal health, we collected detailed cytogenetic and clinical follow-up data from a large cohort of women who received additional findings detected by GW-NIPT.
Between April 2017 and April 2019, NIPT results were provided for 149,318 pregnancies, with 74.2% of pregnant women choosing to have genome-wide chromosomal analysis. Additional findings were detected in 1 out of every 275 performed GW-NIPT and accounted for 35.5% (402/1,132) of all abnormal NIPT results within the TRIDENT-2 cohort. These additional findings included 196 rare autosomal trisomies (RATs), 188 structural aberrations (SAs), and 18 complex profiles. Genetic follow-up testing showed a fetal, (assumed) placental, or (assumed) maternal origin of chromosomal aberrations in 22.1%, 52.8%, and 25.1% of cases, respectively. PPVs differed largely between RATs and SAs (7.7% vs 44.1%), which is in line with previous research.5,6
Of all fetal chromosomal aberrations, 77.2% were pathogenic. The frequency of these pathogenic abnormalities is comparable to that of the common fetal trisomy 13 or 18,5 for which screening is offered in many countries.1 In the majority of cases, knowledge of gene content and function was sufficiently available in current literature and online databases to predict the clinical phenotype associated with the detected chromosome aberration. In 86.9%, parents opted for termination of pregnancy. Furthermore, in more than half of the pathogenic chromosomal abnormalities, no malformations were detected on a second trimester structural anomaly scan. This outcome is in line with previous reports25,26 showing that not all (severe) diseases can be detected by ultrasound.
Screening with GW-NIPT resulted not only in the reporting of pathogenic CNVs associated with severe phenotypes, but also in the incidental reporting of fetal VUSs or benign CNVs. Reporting VUSs or benign CNVs should be avoided as this leads to unnecessary follow-up testing and possible decisions based on uncertain information. Genetic variants detected with NIPT were cautiously interpreted by accredited clinical laboratory geneticists and clinical geneticists. Reporting fetal VUSs or benign CNVs was mainly due to technical limitations of NIPT regarding the precise determination of the size and breakpoints of CNVs, and to discrepancies that existed between the NIPT result and the final fetal karyotype in cases of SAs.27 With advancing insights, the reporting of abnormal NIPT results has already improved over the years. This will only further improve with the increasing availability of data and experience of the specialists involved.
About half of the additional findings were (assumed) CPM. The majority (94.7%) were RATs. Previous studies have assessed the association between CPM and adverse pregnancy outcomes.28 These were, however, mostly small cohort studies of high-risk pregnancies. More importantly, the reported findings of these studies were conflicting, except for the significant association between CPM trisomy 16 and adverse pregnancy outcomes. We now demonstrate that CPM is associated with adverse perinatal outcomes in an unselected population of pregnant women. The risks for pre-eclampsia and low birth weight were significantly increased compared to the general obstetric population. These associations were not limited to CPM trisomy 16 as suggested before,11,12 but remained significant when trisomy 16 cases were excluded. In contrast to CPM trisomy 16, we did not find an increased risk for fetal structural anomalies for the other CPM cases. We argue that in case of CPM, women should be offered tailored perinatal obstetric care, including the monitoring of placental functioning and fetal growth, and the advice to use acetylsalicylic acid to lower the risk for pre-eclampsia. We hypothesize that if CPM is present, this might result in abnormal placentation, impairing placental function and fetal development. Our explanatory analyses on the larger subgroups of CPM cases (trisomy 7, 8, 16, and 20) show that the risk of adverse pregnancy outcomes differs per chromosome; however, more data are necessary to be able to draw final conclusions for each individual RAT, as the subgroups were still relatively small.
A maternal origin was confirmed in 25.1% of cases with additional findings. About 40% of the maternal findings were (assumed) acquired chromosomal aberrations, the others were constitutional CNVs. In accordance with other studies, acquired complex profiles associated with maternal malignancies were detected, but rare.29,30 The clinical details of the (suspected) malignancies within TRIDENT-2 are presented in a separate paper.17 The clinical impact of the 5q and 20q deletions is still unclear and a long-term follow-up study is ongoing to look into the possible association with myeloid neoplasms.31 More than half of the women with constitutional chromosomal aberrations had disease-related symptoms not previously diagnosed that could now be explained (mostly HNPP and CMT1A cases). None of the women with mosaic pathogenic chromosomal aberrations associated with well-known abnormal phenotypes had clinical symptoms, probably due to a (low) level and organ distribution of the mosaicism. However, their offspring is at risk for the syndrome and invasive testing can be offered.
Finally, in 10.9% of cases, the origin of the abnormal NIPT result remained unresolved, despite follow-up testing in the fetus and the mother in most cases. The type of these chromosomal aberrations did not provide any clues regarding their origin. Complete follow-up testing requires analysis of all tissues that may contribute to the cfDNA fraction of maternal plasma, which was not always feasible. This mainly concerns availability of placentas postnatally. Other plausible explanations for aberrant NIPT findings could be an unnoticed vanishing twin, fetal or maternal low-grade chromosomal mosaicism, or a technical artifact. Currently, the advice for pregnancies with an additional finding of unknown origin is to offer parents close obstetric surveillance. This advice was based on the results of the TRIDENT-1 study, in which an increased risk for obstetric pathology and fetal anomalies was found in these cases.7 This advice will stand, until new evidence emerges from further research that specifically focuses on the unresolved cases.
Screening for additional findings has inevitably led to an increased number of invasive tests performed (0.3%; 286/110,739). However, this increase should be weighed against the clinical impact of additional findings. In general, follow-up strategies for additional findings can be discussed on a case-by-case basis. These strategies may consist of invasive fetal testing, investigation of maternal DNA, ultrasound follow-up, the use of acetylsalicylic acid, or a combination of these. The strategy should be determined based on the type of chromosomal aberration involved. Based on our data, refraining from invasive testing can be considered in cases where NIPT is indicative of trisomy 7, as all 60 cases turned out to be (assumed) CPM. We have insufficient data to draw such conclusions for the other additional findings.
We recognize that with GW-NIPT screening, pre- and post-test counseling is more challenging because of the variety of chromosome aberrations that can be detected. We therefore argue that access to appropriately trained counsellors and clinical (laboratory) geneticists, national recommendations, and society guidelines is crucial for the responsible implementation of GW-NIPT screening. Additionally, we see the necessity of investigating parental experiences with GW-NIPT in terms of stress and anxiety associated with opting for prenatal screening and possible choices after abnormal test results. This is an important topic within the TRIDENT-2 study. Finally, we would like to emphasize that an essential part of the evaluation of screening tools and screening programs is a cost-effectiveness analysis. Therefore, the cost-effectiveness of GW-NIPT as compared to other prenatal screening tests should be investigated in future research.
This research is unique in the size and detail of the cytogenetic and clinical follow-up of cases with chromosomal aberrations other than the common trisomies detected by GW-NIPT. Professional societies have been expressing their need for objective data assessing the clinical utility of GW-NIPT for years. This study adds substantially to that need. However, this research also has some limitations. First, although we present a large dataset of pregnancies with additional findings, the number of cases for each individual chromosomal aberration (specifically, the RATs) is still too low to provide individual chances for a true fetal origin (PPV) and advice on specific follow-up testing. Second, the governmental license for the TRIDENT-2 study did not allow the analysis of the sex chromosomes. This decision was made partly because of ethical concerns for sex selection, and partly because of discussions on the lack of clinical utility for screening for sex chromosomal abnormalities and the low positive predictive value. We are aware that analysis of the sex chromosomes, including gender determination, is sometimes included in the offer of prenatal screening in other countries (mainly offered by commercial laboratories).1,32 With this study, we cannot present data on sex chromosomal aneuploidies which could have been informative for screening policy in other countries. Third, we made no distinction between confirmed CPM cases and assumed CPM cases when describing the results. In the follow-up protocol for additional findings, the examination of placentas postpartum was recommended in some cases (details in supplemental methods) but was not mandatory. As a result, in many cases the placenta has not been examined. It is known from previous studies that mosaicism can be lost in the placenta over gestation or can be localized.18 Therefore, a normal placental biopsy at term does not rule out CPM with certainty. Additionally, for the majority of assumed CPM cases (80%) in our cohort, a fetal origin was ruled out with invasive testing during pregnancy or cytogenetic testing on umbilical cord blood after delivery. For the remaining cases we rely on the knowledge about the low chance of a true fetal origin in case of a RAT.8 Fourth, we recognize that the reported incidence of pre-eclampsia in the general Dutch obstetric population is low (0.5%) compared to the estimates of pre-eclampsia incidence in large systematic reviews (3%–8%).33,34 Although the quality of data from the Dutch national obstetric outcome registration (Perined) is generally very high (data delivery by healthcare providers is mandatory and is done according to national standards), the large difference could indicate an underreporting of pre-eclampsia in this database. However, the conclusion of a significantly increased risk for pre-eclampsia for pregnancies complicated with CPM holds true, even if the true rate of PE in the general obstetric population would be significantly higher than the incidence reported in the national outcome registration (Perined). Finally, in ten cases we assumed that fibroids caused the abnormal NIPT result. No cytogenetic tests were performed on the fibroids to confirm the association. Uterine fibroids are the most common pelvic tumors of the female genital tract.35 It is known that placental estrogens and progesterone, and an array of endocrine and paracrine factors, affect fibroid blood supply, growth rate, and the risk of degeneration along the gestational and postpartum periods. It is estimated that about 11% of all pregnant women have fibroids, and that 40%–50% of fibroids show karyotypically detectable chromosomal abnormalities.35,36 Chromosomal abnormalities known to be associated with fibroids include specific deletions of chromosome 7q,24,36 consistent with the NIPT finding of two women in our sample with fibroids. For the remaining cases, there was no typical chromosomal aberration detected, but in most cases, a fetal, maternal, and placental origin was ruled out.
In conclusion, about one in every 275 women in a general obstetric population opting for GW-NIPT receives an additional finding. The majority of additional findings identified by GW-NIPT have clinical impact. Most fetal chromosomal aberrations are pathogenic and associated with severe clinical phenotypes. (Assumed) CPM is significantly associated with adverse perinatal outcomes, requiring tailored obstetric care. The clinical impact of maternal findings is predominantly limited to maternal malignancies. Our data provide crucial information for the decision whether and how to implement GW-NIPT in screening programs, and can inform the challenging interpretations and counseling of additional findings.
Acknowledgments
The authors would like to thank all participating women for their contribution to the TRIDENT-2 study. We also thank all who have contributed to the organization and execution of the study, including health professionals, laboratory staff, and other supporting staff. Staff members of RIVM/CvB, the Regional Centers for Prenatal Screening, Peridos the online national digital registration system for prenatal screening, and EUROCAT are acknowledged for their collaboration. Beau van Hulst and Maurits van Prooyen Schuurman are acknowledged for their contribution to the data collection for this study, and Sandra van ‘t Padje is acknowledged for her work as functional manager of the NIPT Consortium. This work was supported by a grant from the Netherlands Organisation for Health Research and Development (ZonMw, No. 543002001).
Declaration of interests
E.A.S. was one of the applicants of the ZonMW grant and is a member of the GenQA advisory board (unpaid) and the associate editor of Extracellular Vesicles and Circulating Nucleic Acids. D.V.O. is a member of the project group NIPT additional findings and Project group NIPT Laboratories of the National Institute for Public Health & Environment (RIVM) – Center for Population Screening (CvB). L.H. was one of the applicants of the ZonMW grant. M.N.B. is a member of the project group NIPT additional findings, of the RIVM-CvB (unpaid). C.J.B. is a member of the project group NIPT additional findings, of the RIVM-CvB (unpaid). M.J.P. is a member of the Project group NIPT Quality, and Project group NIPT additional findings, all of the RIVM-CvB (unpaid). M.V.E.M. is a member of the Program Committee Prenatal Screening, Project group NIPT Quality, and Project group NIPT Laboratories, all of the RIVM-CvB (unpaid). Part of the ZonMW grant was paid to his institution for TRIDENT-2 bio-informatic research. R.-J.H.G. was one of the applicants of the ZonMW grant, and he is a member of the Program Committee Prenatal Screening, Project group NIPT Quality, and Project group NIPT additional findings, all of the RIVM-CvB (unpaid).
Published: June 2, 2022; corrected online June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.04.018.
Web resources
About NIPT and the TRIDENT studies, https://www.meerovernipt.nl
English information leaflet about prenatal screening for Down's, Edwards' and Patau's syndromes, https://www.pns.nl/documenten/information-about-prenatal-screening-for-down-syndrome-edwards-syndrome-and-pataus
EUROCAT, Complete EUROCAT Guide 1.4 and Reference Documents (v.01.12.2020), https://eu-rd-platform.jrc.ec.europa.eu/eurocat/data-collection/guidelines-for-data-registration_en.
EUROCAT prevalence report (accessed August 12, 2021), https://eu-rd-platform.jrc.ec.europa.eu/eurocat/eurocat-data/prevalence/export_en
Online national digital registration system for prenatal screening Peridos, https://www.peridos.nl
PERINED, accessed August 2021, http://www.peristat.nl
Recommendations for further investigation in case of suspicion for Trisomy 21, 18 and 13 and additional findings detected with NIPT (in Dutch), 2018, https://www.nvog.nl/wp-content/uploads/2018/06/Protocol-vervolgonderzoek-bij-afwijkende-NIPT-versie-1-dd-06062018-DEF.pdf
Supplemental information
Table S8. Characteristics and outcomes of pregnancies with CPM trisomy 8 detected by GW-NIPT
Data and code availability
All data collected for this study are present in the paper or supplemental information or are available under a data use agreement and subject to the limitations of the informed consent document.
References
- 1.Gadsboll K., Petersen O.B., Gatinois V., Strange H., Jacobsson B., Wapner R., Vermeesch J.R., Group N.I.-m.S., Vogel I. Current use of noninvasive prenatal testing in Europe, Australia and the USA: a graphical presentation. Acta Obstet. Gynecol. Scand. 2020;99:722–730. doi: 10.1111/aogs.13841. [DOI] [PubMed] [Google Scholar]
- 2.Gil M.M., Accurti V., Santacruz B., Plana M.N., Nicolaides K.H. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet. Gynecol. 2017;50:302–314. doi: 10.1002/uog.17484. [DOI] [PubMed] [Google Scholar]
- 3.Norton M.E., Jacobsson B., Swamy G.K., Laurent L.C., Ranzini A.C., Brar H., Tomlinson M.W., Pereira L., Spitz J.L., Hollemon D., et al. Cell-free DNA analysis for noninvasive examination of trisomy. N. Engl. J. Med. 2015;70:483–484. doi: 10.1097/01.ogx.0000470657.58577.f2. [DOI] [PubMed] [Google Scholar]
- 4.Taylor-Phillips S., Freeman K., Geppert J., Agbebiyi A., Uthman O.A., Madan J., Clarke A., Quenby S., Clarke A. Accuracy of non-invasive prenatal testing using cell-free DNA for detection of Down, Edwards and Patau syndromes: a systematic review and meta-analysis. BMJ Open. 2016;6:e010002. doi: 10.1136/bmjopen-2015-010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Meij K.R.M., Sistermans E.A., Macville M.V.E., Stevens S.J.C., Bax C.J., Bekker M.N., Bilardo C.M., Boon E.M.J., Boter M., Diderich K.E.M., et al. TRIDENT-2: national implementation of genome-wide non-invasive prenatal testing as a first-tier screening test in The Netherlands. Am. J. Hum. Genet. 2019;105:1091–1101. doi: 10.1016/j.ajhg.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Den Bogaert K., Lannoo L., Brison N., Gatinois V., Baetens M., Blaumeiser B., Boemer F., Bourlard L., Bours V., De Leener A., et al. Outcome of publicly funded nationwide first-tier noninvasive prenatal screening. Genet. Med. 2021;23:1137–1142. doi: 10.1038/s41436-021-01101-4. [DOI] [PubMed] [Google Scholar]
- 7.Van Opstal D., van Maarle M.C., Lichtenbelt K., Weiss M.M., Schuring-Blom H., Bhola S.L., Hoffer M.J.V., Huijsdens-van Amsterdam K., Macville M.V., Kooper A.J.A., et al. Origin and clinical relevance of chromosomal aberrations other than the common trisomies detected by genome-wide NIPS: results of the TRIDENT study. Genet. Med. 2018;20:480–485. doi: 10.1038/gim.2017.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Opstal D., Srebniak M.I. Cytogenetic confirmation of a positive NIPT result: evidence-based choice between chorionic villus sampling and amniocentesis depending on chromosome aberration. Expert Rev. Mol. Diagn. 2016;16:513–520. doi: 10.1586/14737159.2016.1152890. [DOI] [PubMed] [Google Scholar]
- 9.Bekker M.N., Henneman L., Macville M.V.E., Sistermans E.A., Galjaard R.J.H. Benefit vs potential harm of genome-wide prenatal cfDNA testing requires further investigation and should not be dismissed based on current data. Ultrasound Obstet. Gynecol. 2020;55:695–696. doi: 10.1002/uog.22030. [DOI] [PubMed] [Google Scholar]
- 10.Benn P. Expanding non-invasive prenatal testing beyond chromosomes 21, 18, 13, X and Y. Clin. Genet. 2016;90:477–485. doi: 10.1111/cge.12818. [DOI] [PubMed] [Google Scholar]
- 11.Grati F.R., Ferreira J., Benn P., Izzi C., Verdi F., Vercellotti E., Dalpiaz C., D'Ajello P., Filippi E., Volpe N., et al. Outcomes in pregnancies with a confined placental mosaicism and implications for prenatal screening using cell-free DNA. Genet. Med. 2020;22:309–316. doi: 10.1038/s41436-019-0630-y. [DOI] [PubMed] [Google Scholar]
- 12.Jani J.C., Gil M.M., Benachi A., Prefumo F., Kagan K.O., Tabor A., Bilardo C.M., Di Renzo G.C., Nicolaides K.H. Genome-wide cfDNA testing of maternal blood. Ultrasound Obstet. Gynecol. 2020;55:13–14. doi: 10.1002/uog.21945. [DOI] [PubMed] [Google Scholar]
- 13.van Schendel R.V., van El C.G., Pajkrt E., Henneman L., Cornel M.C. Implementing non-invasive prenatal testing for aneuploidy in a national healthcare system: global challenges and national solutions. BMC Health Serv. Res. 2017;17:670. doi: 10.1186/s12913-017-2618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oepkes D., Page-Christiaens G.C., Bax C.J., Bekker M.N., Bilardo C.M., Boon E.M.J., Schuring-Blom G.H., Coumans A.B.C., Faas B.H., Galjaard R.H., et al. Trial by Dutch laboratories for evaluation of non-invasive prenatal testing. Part I-clinical impact. Prenat Diagn. 2016;36:1083–1090. doi: 10.1002/pd.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gezondheidsraad . Den Haag; 2016. Wet op het bevolkingsonderzoek: NIPT als eerste test voor de syndromen van Down, Patau en Edwards. Contract No.: publicatienr. 2016/10. [Google Scholar]
- 16.Straver R., Sistermans E.A., Holstege H., Visser A., Oudejans C.B.M., Reinders M.J.T. WISECONDOR: detection of fetal aberrations from shallow sequencing maternal plasma based on a within-sample comparison scheme. Nucleic Acids Res. 2014;42:e31. doi: 10.1093/nar/gkt992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heesterbeek C.J., Aukema S.M., Galjaard R.J.H., Boon E.M.J., Srebniak M.I., Bouman K., Faas B.H.W., Govaerts L.C.P., Hoffer M.J.V., den Hollander N.S., et al. Noninvasive prenatal test results indicative of maternal malignancies: a nationwide genetic and clinical follow-up study. J. Clin. Oncol. 2022:JCO2102260. doi: 10.1200/jco.21.02260. [DOI] [PubMed] [Google Scholar]
- 18.Schuring-Blom G.H., Keuzer M., Jakobs M.E., Van den Brande D.M., Visser H.M., Wiegant J., Hoovers J.M.N., Leschot N.J. Molecular cytogenetic analysis of term placentae suspected of mosaicism using fluorescence in situ hybridization. Prenat Diagn. 1993;13:671–679. doi: 10.1002/pd.1970130803. [DOI] [PubMed] [Google Scholar]
- 19.Hoftiezer L., Hof M.H.P., Dijs-Elsinga J., Hogeveen M., Hukkelhoven C.W., Hukkelhoven C., van Lingen R.A. From population reference to national standard: new and improved birthweight charts. Am. J. Obstet. Gynecol. 2019;220:383.e1–383.e17. doi: 10.1016/j.ajog.2018.12.023. [DOI] [PubMed] [Google Scholar]
- 20.Hoftiezer L., Hukkelhoven C.W.P.M., Hogeveen M., Straatman H.M.P.M., van Lingen R.A. Defining small-for-gestational-age: prescriptive versus descriptive birthweight standards. Eur. J. Pediatr. 2016;175:1047–1057. doi: 10.1007/s00431-016-2740-8. [DOI] [PubMed] [Google Scholar]
- 21.Katz D.B.J., Baptista J., Azen S.P., Pike M.C. Obtaining confidence intervals for the risk ratio in cohort studies. Biometrics. 1978;34:469–474. doi: 10.2307/2530610. [DOI] [Google Scholar]
- 22.Fagerland MW L.S., Laake P. Recommended confidence intervals for two independent binomial proportions. Stat. Methods Med. Res. 2011;0:1–31. doi: 10.1177/0962280211415469. [DOI] [PubMed] [Google Scholar]
- 23.Scientific Center for Quality of Healthcare (IQ healthcare). Professionalsmonitor 2020 Prenatale screening op down-, edwards- en patausyndroom en het Structureel Echoscopisch Onderzoek.
- 24.Vanharanta S., Wortham N.C., Laiho P., Sjoberg J., Aittomaki K., Arola J., Tomlinson I.P., Karhu A., Arango D., Aaltonen L.A. 7q deletion mapping and expression profiling in uterine fibroids. Oncogene. 2005;24:6545–6554. doi: 10.1038/sj.onc.1208784. [DOI] [PubMed] [Google Scholar]
- 25.Srebniak M.I., Joosten M., Knapen M.F.C.M., Arends L.R., Polak M., van Veen S., Go A.T.J.I., Van Opstal D. Frequency of submicroscopic chromosomal aberrations in pregnancies without increased risk for structural chromosomal aberrations: systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2018;73:517–519. doi: 10.1097/01.ogx.0000546163.62393.50. [DOI] [PubMed] [Google Scholar]
- 26.Baena N., De Vigan C., Cariati E., Clementi M., Stoll C., Caballin M.R., Guitart M., Group E.W. Prenatal detection of rare chromosomal autosomal abnormalities in Europe. Am. J. Med. Genet. A. 2003;118A:319–327. doi: 10.1002/ajmg.a.10104. [DOI] [PubMed] [Google Scholar]
- 27.Van Opstal D., van Veen S., Joosten M., Diderich K.E.M., Govaerts L.C.P., Polak J., van Koetsveld N., Boter M., Go A.T., Papatsonis D.N.M., et al. Placental studies elucidate discrepancies between NIPT showing a structural chromosome aberration and a differently abnormal fetal karyotype. Prenat Diagn. 2019;39:1016–1025. doi: 10.1002/pd.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eggenhuizen G.M., Go A., Koster M.P.H., Baart E.B., Galjaard R.J. Confined placental mosaicism and the association with pregnancy outcome and fetal growth: a review of the literature. Hum. Reprod. Update. 2021;27:885–903. doi: 10.1093/humupd/dmab009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amant F., Verheecke M., Wlodarska I., Dehaspe L., Brady P., Brison N., Van Den Bogaert K., Dierickx D., Vandecaveye V., Tousseyn T., et al. Presymptomatic identification of cancers in pregnant women during noninvasive prenatal testing. JAMA Oncol. 2015;1:814–819. doi: 10.1001/jamaoncol.2015.1883. [DOI] [PubMed] [Google Scholar]
- 30.Bianchi D.W., Chudova D., Sehnert A.J., Bhatt S., Murray K., Prosen T.L., Garber J.E., Wilkins-Haug L., Vora N.L., Warsof S., et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA. 2015;314:162–169. doi: 10.1001/jama.2015.7120. [DOI] [PubMed] [Google Scholar]
- 31.Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., Le Beau M.M., Bloomfield C.D., Cazzola M., Vardiman J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 32.Kornman L., Palma-Dias R., Nisbet D., Scott F., Menezes M., da Silva Costa F., McLennan A. Non-invasive prenatal testing for sex chromosome aneuploidy in routine clinical practice. Fetal Diagn. Ther. 2018;44:85–90. doi: 10.1159/000479460. [DOI] [PubMed] [Google Scholar]
- 33.Abalos E., Cuesta C., Grosso A.L., Chou D., Say L. Global and regional estimates of preeclampsia and eclampsia: a systematic review. Eur. J. Obstet. Gynecol. Reprod. Biol. 2013;170:1–7. doi: 10.1016/j.ejogrb.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Hutcheon J.A., Lisonkova S., Joseph K.S. Epidemiology of pre-eclampsia and the other hypertensive disorders of pregnancy. Best Pract. Res. Clin. Obstet. Gynaecol. 2011;25:391–403. doi: 10.1016/j.bpobgyn.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Coutinho L.M., Assis W.A., Spagnuolo-Souza A., Reis F.M. Uterine fibroids and pregnancy: how do they affect each other? Reprod. Sci. 2021 doi: 10.1007/s43032-021-00656-6. [DOI] [PubMed] [Google Scholar]
- 36.Medikare V., Kandukuri L.R., Ananthapur V., Deenadayal M., Nallari P. The genetic bases of uterine fibroids; a review. J. Reprod. Infertil. 2011;12:181–191. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S8. Characteristics and outcomes of pregnancies with CPM trisomy 8 detected by GW-NIPT
Data Availability Statement
All data collected for this study are present in the paper or supplemental information or are available under a data use agreement and subject to the limitations of the informed consent document.