Abstract
We studied the secretion of recombinant human insulin-like growth factor 1 (rhIGF-1) from transformed yeast cells. The hIGF-1 gene was fused to the mating factor α prepro- leader sequence under the control of the constitutive ACT1 promoter. We found that the inactivation of the GAS1 gene in the host strain led to a supersecretory phenotype yielding a considerable increase, from 8 to 55 mg/liter, in rhIGF-1 production.
Human insulin-like growth factor 1 (hIGF-1, or somatomedin C) is a protein of 70 amino acids purified from human serum (21). hIGF-1 is a hormone of vast pharmaceutical interest, since it is responsible for the regulation of growth and differentiation of various cell types (10). Native recombinant hIGF-1 (rhIGF-1) has been expressed mainly in Escherichia coli (14, 22), mammalian tissue cultures (2), and budding yeast (3, 8), with productions ranging between 2 and 8 mg/liter. In yeast cells, different homologous leader sequences have been tested to promote secretion of rhIGF-1 (24); however, secretion can be achieved only using the prepro-α-factor sequence, or, even if in smaller quantities, by fusion between the SUC2 or PHO5 signal peptide and the pro-α-factor (pαFL) (5). Probably only pαFL confers on the hIGF-1 molecule an optimal conformation that is crucial for translocation (5). Nevertheless, native rhIGF-1 represents only 10 to 20% of the total rhIGF-1 production; most of the rhIGF-1 is inactive, essentially composed of dimers or multimers. Such forms are due to the formation of incorrect intra- and intermolecular disulfide bonds (4, 7, 23). In this report we describe an integrated approach to increasing the secretion and/or production in Saccharomyces cerevisiae of monomeric correctly folded rhIGF-1 using an expression system based on the prepro-α-factor leader sequence. A single change in the genetic background of the host strain allowed us to considerably increase the total secreted rhIGF-1.
Production of native rhIGF-1 from recombinant yeast cells.
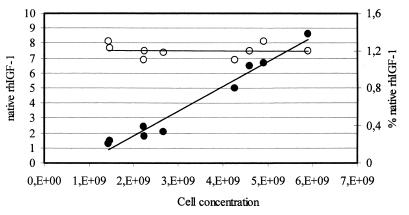
For the production of rhIGF-1 from transformed budding yeast cells, we used the expression vector p539/12 (13) and the yeast strain GcP3 (MATa pep4-3 prb1-1122 ura3-52 leu2 gal2 cir°) as the host. On the plasmid p539/12, which bears the entire 2μm sequence, the hIGF-1 gene is fused with the prepro-α-factor sequence under the control of the constitutive ACT1 yeast promoter. The GcP3 host is [cir°]. This combination—GcP3[p539/12]—allows a stable amplification of the plasmid at a very high copy number per cell (100 to 200) during growth on both mineral-selective and rich complex media (references 1, 6, 11, and 28 and data not shown). To obtain high productions of the recombinant biomass, we cultured the recombinant host in a fed-batch stirred tank bioreactor, following a simple protocol where the addition of a solution of fresh glucose and other nutrients was controlled based on the ethanol concentration (20). We ran many different fed-batch tests, changing the composition of the feed medium. We obtained productions of monomeric native rhIGF-1 from about 2 to 3 mg/liter (glucose [50%, wt/vol] used as feed) to 8.6 mg/liter (glucose [50%, wt/vol], hydrolyzed casein [1.3%, wt/vol], and other mineral elements [20] used as feed). Figure 1 plots the production of monomeric native rhIGF-1 against the cell concentration during the time course of different fed-batch tests. Cell concentration was determined with a Coulter Counter (27). Monomeric and total rhIGF-1 were measured by high-pressure liquid chromatography as described by Gellerfors et al. (13); the total concentration of secreted proteins was determined using the Bio-Rad DC protein assay kit. Interestingly, for all the tests the native rhIGF-1 represented 1.2% of the total secreted proteins and 10% of the total rhIGF-1 produced. Therefore, these data seem to indicate that the production of rhIGF-1 is simply related to the amount of biomass of the recombinant strain. Western analysis of total protein extracts did not reveal any intracellular rhIGF-1 accumulation (data not shown).
FIG. 1.
Production of recombinant native rhIGF-1 (●) (milligrams per liter) plotted against the cell concentration (cells/milliliter) during the time course of different fed-batch tests. The percentage of native rhIGF-1 on the total amount of secreted proteins is also reported ( ). Experimental results are the averages of at least two independent fed-batch tests.
Finally, neither the use of a stronger promoter (the inducible UASGAL1–10) nor an attempt to lower the local product concentration in the endoplasmic reticulum (by using a single-copy centromeric vector) gave interesting results (data not shown).
Development of a supersecretory phenotype.
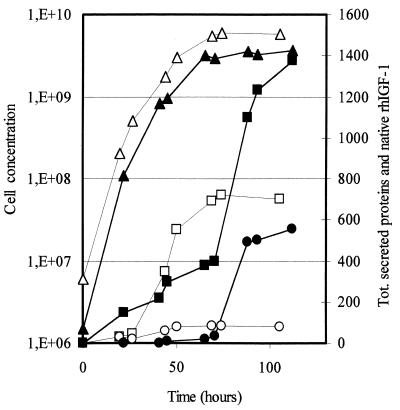
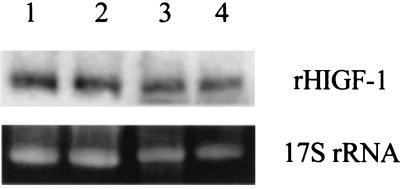
The GAS1 yeast gene codes for a glycoprotein anchored to the plasma membrane by a glycosylphosphatidylinositol whose function is necessary for the correct assembly of cell wall polymers in S. cerevisiae. gas1Δ mutants show morphogenetic defects due to changes in the composition and organization of cell wall constituents (see reference 19 for a review). To test the effects (if any) of such a mutation on rhIGF-1 secretion, the GAS1 gene was inactivated in the GcP3[p539/12] transformed strain by the one-step gene replacement procedure (25), yielding the strain GcP3Δ12[p539/12]. The inactivation of the GAS1 gene in the GcP3[p539/12] strain affected the morphology and the growth rate, as already reported for other strains (18, 26). The main effect on rhIGF-1 accumulation was particularly evident in fed-batch fermentations, where the production phase increased from about 50 h to about 115 h of fermentation. Figure 2 compares the time course fed-batch fermentations of the GcP3[p539/12] and GcP3Δ12[p539/12] strains carried out at pH 5.7 using glucose (50%, wt/vol), hydrolyzed casein (1.3%, wt/vol) and other mineral elements (20) as feed. A much higher production of total (535 to 540 mg/liter compared to 80 to 85 mg/liter) and native (55.6 mg/liter compared to 8.6 mg/liter) rhIGF-1 was obtained. The fraction of native rhIGF-1 on the total secreted proteins (4% compared to 1.2%) also noticeably increased, while the ratio of native to total rhIGF-1 did not change. More importantly, GAS1 gene inactivation had a strong effect on the global secretion. This was particularly evident during the last stage of fermentation. This effect could be ascribed to the modification of the genetic background of the host strain. The absence of cytoplasmic markers (i.e., aldolase and triosephosphate isomerase) in the medium rules out the possibility of a massive cellular lysis during fermentation (data not shown). To clarify the reason for the different rhIGF-1 secretion levels, we compared the amounts of rhIGF-1 mRNA expressed in the GcP3[p539/12] and GcP3Δ12[p539/12] strains: total RNA, prepared according to the method of selective precipitation with LiCl (9), was subjected to Northern analysis. Hybridization was performed as previously described (17) using a nonradioactive single-strand RNA probe, anti-hIGF-1, generated by in vitro transcription (Dig RNA labeling kit; Boehringer Mannheim). Despite the different production of both total and native rhIGF-1, we did not detect a significant difference between the rhIGF-1 mRNA of the two strains (Fig. 3), indicating that the enhanced secretion from the GcP3Δ12[p539/12] strain is not the result of upregulated transcription. Interestingly, also for the GcP3Δ12[p539/12] transformed cells, we never observed intracellular accumulation of rhIGF-1. These two considerations taken together seem to indicate that the strong improvement obtained from the mutant GcP3Δ12 host could be determined simply by using a faster rhIGF-1 secretion process. In fact, a higher secretion rate could allow a better accumulation of the heterologous protein in the medium, outflanking the eventual intracellular degradation of the protein. Supporting this interpretation, we observed an increase of the total secreted proteins from the mutant GcP3Δ12 (Fig. 2). These increased secretion levels could be related to a decreased cell wall binding of proteins, among them rhIGF-1. In this respect, gas1Δ mutants showed a changed composition and organization of cell wall constituents (19), and increased global secretion levels associated with alterations in the cell wall architecture already have been observed both in S. cerevisiae (12) and in the fungus Trichoderma reesei (16). Finally, a similar correlation between the secretion rate and the levels of a secreted heterologous protein have been recently reported by other authors; Katakura et al. (15) showed that replacement of Trp19 with Tyr in the bovine β-lactoglobulin yielded an improved secretion level (six times more) from recombinant S. cerevisiae cells. Similar to the results shown in this study, the higher secretion levels are related not to upregulation of specific transcription but to a higher secretion rate.
FIG. 2.
Native rhIGF-1 secretion during a typical fed-batch fermentation of GcP3[p539/12]- and GcP3Δ12[p539/12]-transformed cells. Open symbols, GcP3[p539/12]; closed symbols; GcP3Δ12[p539/12]; ▴ and Δ, cell concentration, in cells per milliliter; █ and □, total proteins secreted, in milligrams per liter; ● and ○, native rhIGF-1, in milligrams per liter (original data have been multiplied by 10).
FIG. 3.
Northern analysis of rhIGF-1 expression in gas1Δ cells. Total RNA was extracted from the mutant GcP3Δ12 (gas1∷LEU2) (lanes 1 and 2) and its isogenic strain GcP3 (lanes 3 and 4), expressing rhIGF-1. Cells were collected at two different cellular densities. About the same amount of RNA (10 μg) was loaded on each lane. The ethidium bromide-stained gel of the autoradiogram is shown in the lower panel. Densitometric quantification of mRNA was performed by a computer program, and mRNA loading was normalized using the 17S rRNA.
In conclusion, our results show that a careful choice of the host strain enables noticeably increased production and yield of a heterologous protein.
REFERENCES
- 1.Armstrong K A, Som T, Volkert F C, Rose A, Broach J R. Propagation and expression of genes in yeast using 2-micron circle vectors. In: Barr P J, Brake A J, Valenzuela P, editors. Yeast genetic engineering. London, England: Butterworths; 1989. pp. 165–192. [PubMed] [Google Scholar]
- 2.Bayne M L, Cascieri M A, Kelder B, Applebaum J, Chicchi G, Shapiro J A, Pasleau F, Kopchick J J. Expression of a synthetic gene encoding human insulin-like growth factor-I in cultured mouse fibroblasts. Proc Natl Acad Sci USA. 1987;84:2638–2642. doi: 10.1073/pnas.84.9.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayne M L, Applebaum J, Chicchi G G, Hayes N S, Green B G, Cascieri M A. Expression, purification and characterization of recombinant human insulin-like growth factor I in yeast. Gene. 1988;66:235–244. doi: 10.1016/0378-1119(88)90360-5. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhuri B, Helliwell S B, Priestle J P. A Lys27 to Glu27mutation in the human insulin-like growth factor-I prevents disulphide linked dimerization and allows secretion of BiP when expressed in yeast. FEBS Lett. 1991;294:213–216. doi: 10.1016/0014-5793(91)81432-8. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhuri B, Steube K, Stephan C. The pro-region of the yeast preproα-factor is essential for membrane translocation of human insulin-like growth factor-I in vivo. Eur J Biochem. 1992;206:793–800. doi: 10.1111/j.1432-1033.1992.tb16986.x. [DOI] [PubMed] [Google Scholar]
- 6.Dobson M J, Futcher A B, Cox B S. Control of recombination within and between DNA plasmids of Saccharomyces cerevisiae. Curr Genet. 1980;2:193–200. doi: 10.1007/BF00435685. [DOI] [PubMed] [Google Scholar]
- 7.Elliott S, Fagin K D, Narhi L O, Miller J A, Jones M, Koski R, Peters M, Hsieh P, Sachdev R, Rosenfeld R D, Rhode M F, Arakawa T. Yeast-derived recombinant human insulin-like growth factor-I: production, purification, and structural characterization. J Protein Chem. 1990;9:95–104. doi: 10.1007/BF01024990. [DOI] [PubMed] [Google Scholar]
- 8.Ernst J F. Improved secretion of heterologous proteins by Saccharomyces cerevisiae: effects of promoter substitution in alpha-factor fusions. DNA. 1986;5:483–492. doi: 10.1089/dna.1.1986.5.483. [DOI] [PubMed] [Google Scholar]
- 9.Federoff H J, Cohen J D, Eccleshall T R, Needleman R B, Buchferer B A, Giacalone J, Marmur J. Isolation of a maltase structural gene from Saccharomyces carlsbergensis. J Bacteriol. 1982;149:1064–1070. doi: 10.1128/jb.149.3.1064-1070.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Froesch E R, Hussain M A, Schmid C, Zapf J. Insulin-like growth factor I: physiology, metabolic effects, and clinical uses. Diabetes Metab Rev. 1996;12:195–215. doi: 10.1002/(SICI)1099-0895(199610)12:3<195::AID-DMR164>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 11.Futcher A B, Cox B S. Copy number and the stability of 2-μm circle-based artificial plasmids of Saccharomyces cerevisiae. J Bacteriol. 1984;157:283–290. doi: 10.1128/jb.157.1.283-290.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaynor E C, Mondesert G, Grimme S J, Reed S I, Orlean P, Emr S D. MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast. Mol Biol Cell. 1999;10:627–648. doi: 10.1091/mbc.10.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gellerfors P, Axelsson K, Helander A, Johansson S, Kenne L, Lindqvist S, Pavlu B, Skottner A, Fryklund L. Isolation and characterization of a glycosylated form of human insulin-like growth factor I produced in Saccharomyces cerevisiae. J Biol Chem. 1989;264:11444–11449. [PubMed] [Google Scholar]
- 14.Joly J C, Leung W S, Swartz J R. Overexpression of Escherichia colioxidoreductases increases recombinant insulin-like growth factor-I accumulation. Proc Natl Acad Sci USA. 1998;95:2773–2777. doi: 10.1073/pnas.95.6.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katakura Y, Ametani A, Totsuka M, Nagafuchi S, Kaminogawa S. Accelerated secretion of mutant β-lactoglobulin in Saccharomyces cerevisiaeresulting from a single amino acid substitution. Biochim Biophys Acta. 1999;1432:302–312. doi: 10.1016/s0167-4838(99)00099-0. [DOI] [PubMed] [Google Scholar]
- 16.Kruszewska J S, Butterweck A H, Kurzątkowski W, Migdalski A, Kubicek C P, Palamarczyk G. Overexpression of the Saccharomyces cerevisiae mannosylphosphodolichol synthase-encoding gene in Trichoderma reeseiresults in an increased level of protein secretion and abnormal cell ultrastructure. Appl Environ Microbiol. 1999;65:2382–2387. doi: 10.1128/aem.65.6.2382-2387.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popolo L, Cavadini P, Vai M, Alberghina L. Transcript accumulation of the GGP1gene, encoding a yeast GPI-anchored glycoprotein, is inhibited during arrest in the G1 phase and during sporulation. Curr Genet. 1983;24:382–387. doi: 10.1007/BF00351845. [DOI] [PubMed] [Google Scholar]
- 18.Popolo L, Vai M, Gatti E, Porello S, Bonfante P, Balestrini R, Alberghina L. Physiological analysis of mutants indicates involvement of the Saccharomyces cerevisiaeGPI-anchored protein gp115 in morphogenesis and cell separation. J Bacteriol. 1993;175:1879–1885. doi: 10.1128/jb.175.7.1879-1885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popolo L, Vai M. The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochim Biophys Acta. 1999;1426:385–400. doi: 10.1016/s0304-4165(98)00138-x. [DOI] [PubMed] [Google Scholar]
- 20.Porro D, Martegani E, Tura A, Ranzi B M. Development of a pH controlled fed-batch system for budding yeast. Res Microbiol. 1991;142:535–539. doi: 10.1016/0923-2508(91)90186-e. [DOI] [PubMed] [Google Scholar]
- 21.Rinderknecht E, Humbel R E. The amino acid sequence of human insulin-like growth factor I and its structural homology with proinsulin. J Biol Chem. 1978;253:2769–2776. [PubMed] [Google Scholar]
- 22.Schulz M-F, Buell G, Schmid E, Movva R, Selzer G. Increased expression in Escherichia coliof a synthetic gene encoding human somatomedin C after gene duplication and fusion. J Bacteriol. 1987;169:5385–5392. doi: 10.1128/jb.169.12.5385-5392.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steube K, Chaudhuri B, Marki W, Merryweather J P, Heim J. α-Factor-leader-directed secretion of recombinant human-insulin-like growth factor-I from Saccharomyces cerevisiae. Eur J Biochem. 1991;198:651–657. doi: 10.1111/j.1432-1033.1991.tb16063.x. [DOI] [PubMed] [Google Scholar]
- 24.Taussig R, Carlson M. Nucleotide sequence of the yeast SUC2gene for invertase. Nucleic Acids Res. 1983;11:1943–1954. doi: 10.1093/nar/11.6.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vai M, Gatti E, Lacanà E, Popolo L, Alberghina L. Isolation and deduced amino acid sequence of the gene encoding gp115, a yeast glycophospholipid-anchored protein. J Biol Chem. 1991;266:12242–12248. [PubMed] [Google Scholar]
- 26.Vai M, Orlandi I, Cavadini P, Alberghina L, Popolo L. Candida albicans homologue of GGP1/GAS1 gene is functional in Saccharomyces cerevisiaeand contains the determinants for glycosylphosphatidylinositol attachment. Yeast. 1996;12:361–368. doi: 10.1002/(SICI)1097-0061(19960330)12:4%3C361::AID-YEA920%3E3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 27.Vanoni M, Vai M, Popolo L, Alberghina L. Structural heterogeneity in populations of the budding yeast Saccharomyces cerevisiae. J Bacteriol. 1983;156:1282–1291. doi: 10.1128/jb.156.3.1282-1291.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walmsley R M, Gardner D C, Oliver S G. Stability of a cloned gene in chemostat culture. Mol Gen Genet. 1983;194:361–365. doi: 10.1007/BF00392175. [DOI] [PubMed] [Google Scholar]