Abstract
This work reviews nuclear compartments, defined broadly to include distinct nuclear structures, bodies, and chromosome domains. It first summarizes original cytological observations before comparing concepts of nuclear compartments emerging from microscopy versus genomic approaches and then introducing new multiplexed imaging approaches that promise in the future to meld both approaches. I discuss how previous models of radial distribution of chromosomes or the binary division of the genome into A and B compartments are now being refined by the recognition of more complex nuclear compartmentalization. The poorly understood question of how these nuclear compartments are established and maintained is then discussed, including through the modern perspective of phase separation, before moving on to address possible functions of nuclear compartments, using the possible role of nuclear speckles in modulating gene expression as an example. Finally, the review concludes with a discussion of future questions for this field.
The Oxford Language definition of compartment is “a separate section of a structure in which certain items can be kept separate from others.” In addition to their diffuse localization throughout the nucleoplasm, many proteins and RNAs concentrate within distinct nuclear bodies, or within less distinct, but still spatially concentrated, condensates. The cumulative volume of all these nuclear bodies and condensates, still unknown, likely occupies a significant fraction of the total interchromosomal nuclear space, suggesting that a large portion of the genome lies within small distances from multiple nuclear compartments with distinct functional properties. Meanwhile, different types of chromatin domains position differentially but specifically near these different nuclear nonchromosomal compartments, while also compacting to form discrete chromosome structures that may themselves function as a distinct kind of nuclear compartment. A classic example would be the inactive mammalian X chromosome, which is positioned preferentially adjacent to either the nuclear or nucleolar periphery while also compacting into a condensed “Barr body” (Barr and Bertram 1951; Barr and Carr 1962; Belmont et al. 1986; Zhang et al. 2007; Rego et al. 2008). Recently, the Barr body has been proposed to function as a phase-separated condensate that would exclude specific proteins and macromolecular complexes based on additional molecular properties beyond simply size (Cerase et al. 2019; Pandya-Jones et al. 2020).
Here, I first survey over 100 years of cytology, describing this multitude of nuclear bodies and structures. I then discuss previous imaging approaches to studying chromosome nuclear compartmentalization and compare this with modern genomic methods for describing the same. Briefly, I review recent new imaging approaches that promise to meld genomic and imaging approaches. This is followed by bringing in the modern perspective of phase separation to the discussion of nuclear compartmentalization. I then address the currently poorly understood question of how these nuclear compartments are established and maintained, before discussing how to approach the functions of nuclear compartments, using recent experiments examining the possible contribution of nuclear speckles to gene regulation as an example. Finally, I comment on challenges in addressing the role of nuclear compartments in nuclear genome organization and function before concluding with a discussion of outstanding questions in the field.
AN INCOMPLETE SURVEY OF NUCLEAR BODIES AND NUCLEAR COMPARTMENTS
The Usual Suspects
One definition of nuclear bodies describes them as “nonmembrane-bound structures that can be visualized as independent domains by transmission electron microscopy without antibody labeling” (Spector 2006). But decades before the invention of transmission electron microscopy (TEM), four major nuclear bodies/compartments—the nuclear periphery/lamina, nucleoli, nuclear speckles, and Cajal bodies—were either inferred from (nuclear envelope and lamina) or specifically stained and visualized by light microscopy (nucleoli, nuclear speckles, and Cajal bodies) (Lafarga et al. 2009).
Nucleolus
The nucleolus is the largest nuclear body and the “factory” for ribosome transcription, representing ∼60% of total transcription within the nucleus, and assembly (Schöfer and Weipoltshammer 2018). In tumors, larger nucleoli correlate with higher tumor growth and ribosomal RNA (rRNA) synthesis rates (Derenzini et al. 2000). Numbers and intranuclear positioning of nucleoli vary from a centrally located, single nucleolus to multiple nucleoli located at noncentral locations in different cell types. Although appearing interiorly located in many typical light microscopy images (Fig. 1A), nucleoli typically are attached either directly to the nuclear lamina or indirectly through association with peripheral heterochromatin; frequently, these attachments are associated with invaginations of the nuclear envelope (Bourgeois et al. 1979; Bourgeois and Hubert 1988). TEM revealed the nucleolus's characteristic structure (Fig. 1E,F): a large granular component (GC) within which are embedded dense fibrillar components (DFCs) surrounding fibrillar centers (FCs) (Bernhard et al. 1952; Yasuzumi et al. 1958; Smetana and Busch 1964). With cell stress, FC/DFCs locate more toward the nucleolar exterior (Frottin et al. 2019; Latonen 2019). A consensus has emerged that transcription occurs at the interface between the DFC and FC, and possibly also within the DFCs, with initial rRNA processing occurring in the DFC followed by posttranscriptional processing in the GC (Raška et al. 2006; Boisvert et al. 2007; Schöfer and Weipoltshammer 2018). In many cell types, condensed chromatin coats some of the nucleolar periphery.
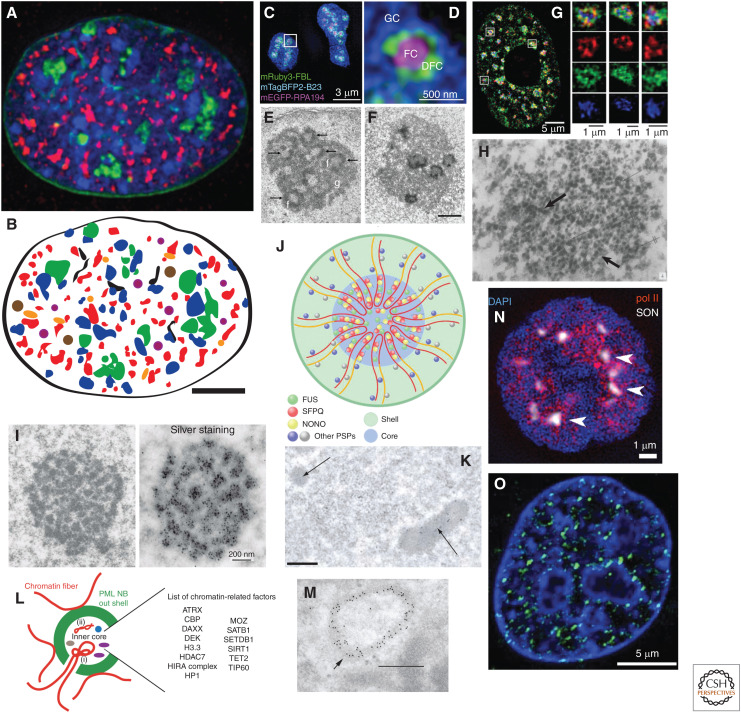
Figure 1.
Varied nuclear bodies occupy significant fraction of cell nucleus. (A) Mouse NIH 3T3 fibroblast cell stained with DAPI (blue) to highlight DNA-dense bodies, including chromocenters, and expressing fluorescently labeled proteins to identify nuclear lamina (green, lamin B1), nucleoli (green, fibrillarin), and nuclear speckles (Magoh, red) (Zhao et al. 2020). (Image courtesy of Dr. Pankaj Chaturvedi.) (B) Same nucleus as A was used to outline nuclear lamina (black), nucleoli (green), and nuclear speckles (red) and then superimpose typical numbers for NIH 3T3 cells of additional nuclear bodies: Cajal bodies (brown), paraspeckles (orange), and promyelocytic leukemia (PML) bodies (purple). Scale bar, 5 μm (A,B). (C,D) Nucleoli stained for granular compartment (GC) (blue, B23), fibrillar center (FC) (purple, RPA194), and dense fibrillar component (DFC) (fibrillarin, FBL). Scale bars, 3 µm (C); 500 nm (D). (Panels C and D from Figure 1C in Yao et al. 2019; reprinted, with permission, from Elsevier © 2019.) (E,F) Electron microscopy (EM) visualization of nucleoli showing GC (g), DFC (arrows), FC (f). Conventional uranyl and lead staining were used in E, and silver staining for NOC proteins used in F. (Panels E and F from Penzo et al. 2019; reprinted under the Creative Commons Attribution License CC BY 4.0.) (G) 3D SIM microscopy showing nuclear speckles with partial spatial separation between anti-“SC35” (actually anti-SRRM2, see text) staining (blue) and MALAT1 (red) and U2 RNA (green). (Continued) (Panel G from Fei et al. 2017; reprinted, with permission, from Company of Biologists © 2017.) (H) EM visualization of interchromatin granule cluster (IGC)/nuclear speckle in rat adrenal cortex cell; granules sometimes appear in linear chains (arrows). (Panel H from Monneron and Bernhard 1969; reprinted, with permission, from Elsevier © 1969.) (I) EM visualization of large Cajal (accessory) bodies in neuronal cells showing “coiled thread” internal structure using conventional (left) or silver (right) staining. Scale bar, 200 nm. (Panel I from Lafarga et al. 2017; reprinted, with permission, from Taylor & Francis © 2017.) (J) Model of paraspeckles showing proposed scaffolding role of NEAT1_2 long noncoding RNA (lncRNA) (red and yellow lines) recruiting/binding different paraspeckle proteins over 5′ and 3′ ends (outer shell regions) versus internal sequences (core region). (Panel J from McCluggage and Fox 2021; reprinted, with permission, from John Wiley and Sons © 2017.) (K) EM visualization of round or elliptical paraspeckles (arrows). Scale bar, 0.5 μm. (Panel K is from Fox and Lamond 2010; image courtesy of Sylvie Souquere and Gerard Pierron, Villejuif, France.) (L) Model for PML body structure showing PML protein outer shell and an inner cavity containing many PML proteins, including chromatin factors and, in some cases, chromatin. (Panel L from Corpet et al. 2020; reprinted, with permission, from Oxford University Press © 2020.) (M) EM visualization of PML body showing immunogold-labeled PML protein outer shell. Scale bar, 0.5 μm. (Panel M from Lallemand-Breitenbach and de Thè 2018; reprinted, with permission, from Elsevier © 2018.) (N) Immunostained nucleus from human K562 cell showing RNA Pol II CTD Ser5p foci (red) clustered around nuclear speckles (SON, white) as well as other nuclear interior regions but depleted from periphery of nucleus counterstained for DNA with DAPI (blue). (Panel N from Chen et al. 2018b; reprinted under the Creative Commons License CC BY-NC-SA 4.0.) (O) Live-cell imaging of Mediator (MED1) condensates (green) visualized in mouse embryonic stem cell (mESC) nucleus stained with Hoechst for DNA (blue). (Panel O from Sabari et al. 2018; reprinted, with permission, from The American Association for the Advancement of Science © 2018.)
On reformation of the nucleus in G1 phase of the cell cycle, many components of the GC colocalize within a number of “prenucleolar bodies” (PNBs) (Hernandez-Verdun 2011), which sometimes localize nearer to the nuclear periphery, and also show chromatin juxtaposed to the PNB periphery (Ochs et al. 1985; Zatsepina et al. 1997). Originally, PBNs were conceptualized as being precursors to nucleolar assembly, with PNBs imagined as migrating and fusing with chromosome nucleolar organizing regions (NORs) early in G1 phase to form the nucleolus. The advent of live-cell imaging using green fluorescent protein (GFP) instead revealed GC components diffusing at different rates out of PNBs and accumulating within reforming nucleoli, with different GC components shifting from PNBs to nucleoli with different kinetics during G1 phase (Dundr et al. 2000; Hernandez-Verdun 2011). PNBs also appear to be sites of resumed processing of precursor rRNAs present during mitosis (Carron et al. 2012).
Although nucleoli are sites of rRNA transcription, they also have been implicated in a multitude of additional processes ranging from gene silencing to assembly and regulation of other non-ribosomal nucleoproteins (RNPs), including telomerase, regulation of protein activity, including p53, and more generally in biological processes, such as development, organismal aging, and stress responses (Boulon et al. 2010; Pederson 2011; Tiku and Antebi 2018; Iarovaia et al. 2019; Latonen 2019; Weeks et al. 2019).
Nuclear Speckles
Ramon y Cajal in 1910 described “hyaline grumes” as a distinct nuclear body using a modified silver-staining procedure (Lafarga et al. 2009). These nuclear speckles were rediscovered in the mid-twentieth century using TEM as interchromatin granule clusters (IGCs) consisting of ∼20–25-nm-diameter RNP particles (granules) lying between and adjacent to chromatin domains (Swift 1959; Bernhard and Granboulan 1963; Monneron and Bernhard 1969). These granules frequently align forming linear chains or apparent rods (Fig. 1H). They were rediscovered yet again as bodies enriched in splicing factors, snRNAs, polyA-RNAs, and immunostaining by an antibody to a phosphorylated epitope tied to several splicing factors, including SC35 (SRSF2) (Fig. 1A,G), but recently revealed to primarily immunostain a different protein, SRRM2 (Ilik et al. 2020). In fibroblasts, nuclear speckles are largely excluded from near the nuclear lamina and concentrated within the equatorial plane of the nuclear interior (Fig. 1A; Carter et al. 1993); exclusion from the nuclear periphery and concentration toward the nuclear interior appears common in many cell types. A recent tyramide signal amplification (TSA)-proximity-labeling proteomics study revealed both SRRM2 and SON, with similar levels of enrichment, as the most highly enriched proteins in nuclear speckles (Dopie et al. 2020). Double SRMM2 and SON RNAi knockdown caused significant dissolution of nuclear speckle staining using multiple speckle markers, suggesting that either protein is sufficient for nuclear speckle formation (Ilik et al. 2020). Immunoelectron microscopy staining tied these variably named structures (nuclear speckles, polyA-islands, SC-35 islands) to the previously described IGCs, and, in turn, these IGCs to the hyaline grumes identified by Cajal through specific silver-staining protocols (Lafarga et al. 2009; Spector and Lamond 2011).
Local concentrations of splicing factors, visualized by light microscopy, are still conflated with “nuclear speckles”, a name now reserved for the light microscopy analog of the IGCs visualized by electron microscopy (EM). Whereas all IGCs show accumulation of various splicing factors, not all splicing factor local accumulations represent IGCs, as clearly revealed by analysis of transgene arrays (Hochberg-Laufer et al. 2019).
Nuclear speckles form rapidly after mitosis, beginning in late telophase (Ferreira et al. 1994; Thiry 1995; Tripathi and Parnaik 2008). Analogous perhaps to PNBs, discrete bodies containing the nuclear speckle protein MFAB1 appear in the reforming telophase nucleus even while the SON and SRRM2 proteins are still cytoplasmic in mitotic interchromatin granules (MIGs) (Dopie et al. 2020). Whether these MFAB1 bodies nucleate nuclear speckles or instead accumulate nuclear speckle proteins that then diffuse and concentrate into nuclear speckles remains unknown. Even after SC35 staining (SRRM2), nuclear speckles first appear in late telophase/early G1 nuclei; SR proteins accumulate first near NORs before then accumulating through an apparent RNA Pol II transcription-dependent mechanism in nuclear speckles (Bubulya et al. 2004).
Nuclear speckles have alternatively been proposed to serve as storage sites for factors involved in RNA Pol II transcription and RNA processing or as gene expression “hubs” for a subset of highly active genes (Hall et al. 2006; Spector and Lamond 2011). These two models are not mutually exclusive. RNA processing factors were proposed to transit from nuclear speckles to active genes and then recycle back to nuclear speckles to be “recharged” for another cycle of RNA processing; this cycling between nuclear speckle and adjacent transcription sites was proposed to be linked to cycles of posttranscriptional modifications, particularly phosphorylation and dephosphorylation, providing “recharging” of these factors for another RNA processing cycle (Misteli and Spector 1997). A unified model then was proposed in which the positioning of a subset of active genes adjacent to nuclear speckles would facilitate this recycling of RNA processing factors from nuclear speckles to transcription, thus supporting high rates of gene expression for these nuclear-speckle-adjacent genes (Hall et al. 2006).
Cajal Bodies
Similar to nuclear speckles, Cajal bodies were first identified as a distinct nuclear body in neurons by Cajal using combinations of histochemical stains or silver-staining procedures (Lafarga et al. 2009). They were rediscovered and named by electron microscopists as “coiled bodies,” ∼0.2–2 µm in diameter (Cioce and Lamond 2005), owing to their appearance after heavy-metal staining (Fig. 1I; Monneron and Bernhard 1969). They were then identified again through their immunostaining against the marker protein, coilin (Andrade et al. 1991; Raška et al. 1991; Lafarga et al. 2009). Cajal bodies were previously known as nucleolar accessory bodies because of their location close to nucleoli in some cell types. Several Cajal bodies (up to ∼10) per nucleus are present in cells associated with high transcriptional activity and/or growth rates, including rapidly dividing embryonic cells, cancer cells, and neurons, but present in fewer numbers or even less than one Cajal body per cell in nontransformed cell types (Ogg and Lamond 2002; Cioce and Lamond 2005; Strzelecka et al. 2010; Machyna et al. 2013).
Cajal body formation is dependent both on the presence of coilin and SMN; Cajal bodies are enriched in RNPs and factors involved in RNP maturation, including spliceosomal snRNPs, scaRNPs, snoRNPs, and the telomerase RNP. Spliceosomal snRNPs are both assembled and recycled within Cajal bodies, and thus Cajal bodies have been proposed as sites of accelerated assembly and modification of multiple small RNA-containing RNPs (Machyna et al. 2013; Meier 2017). Cajal bodies associate with active gene loci, most notably the tandem U2 snRNA gene locus (Machyna et al. 2013; Sawyer et al. 2016).
Nuclear Pores and Nuclear Lamina
Both nuclear pores and the nuclear lamina have been reviewed extensively elsewhere (de Leeuw et al. 2018; Lin and Hoelz 2019; Briand and Collas 2020; Cho and Hetzer 2020; also see Miroshnikova and Wickström 2021; Pawar and Kutay 2021). TEM visualized the nuclear lamina as a fibrous layer lying between the inner nuclear envelope and the peripheral, condensed chromatin (Fawcett 1966). The intermediate filament lamin proteins—lamins A and C, both encoded by the LMNA gene, lamin B1, and lamin B2 in mammalian cells—comprise the major constituents of the nuclear lamina. Recent superresolution light microscopy reveals that these various lamins are concentrated differentially within spatially distinct meshworks, separated by several hundred nanometers, within the lamina (Shimi et al. 2015; Xie et al. 2016). Previous TEM tomography had shown local attachments of chromatin to the nuclear periphery underlying regions of high lamin B concentration within this meshwork (Belmont et al. 1993). A small fraction of lamin A is nucleoplasmic, where it may play diverse roles separate from its function at the nuclear lamina (Briand and Collas 2020). Mutations in lamin A are associated with ∼15 diseases collectively termed laminopathies, including one type of premature aging disease (Hutchinson-Gilford progeria syndrome), Emery-Dreifuss and other muscle dystrophies, lipodystrophies, and peripheral neuropathies (Kang et al. 2018; Osmanagic-Myers and Foisner 2019; Briand and Collas 2020).
The nuclear lamina contains hundreds of additional proteins, many of which interact directly or indirectly with lamins (Wilson and Foisner 2010; Mehus et al. 2016; Wong et al. 2021). This includes inner nuclear membrane (INM) proteins as well as proteins concentrated near the lamina and peripheral chromatin. INM proteins notably include LEM-domain proteins (Wilson and Foisner 2010), which in mammals include LAP2 (α, β, and other isoforms), emerin, MAN1, and LEM2/NET25. The INM also includes lamin B receptor (LBR), which combines chromatin-binding and sterol reductase domains, SUN domain proteins that link to KASH domain proteins in the outer nuclear membrane that interact directly with cytoskeleton, centrosome, and organelle proteins (de Leeuw et al. 2018), as well as many other transmembrane proteins that may provide cell-specific links of specific chromosome regions to the nuclear periphery (Robson et al. 2016). These proteins together with lamins interact with chromatin as well as various transcription factors, chromatin modifying, and signaling proteins. At least in some postmitotic cell types, lamin A/C and LBR together anchor peripheral chromatin to the nuclear lamina; their knockout results in “inverted nuclei” with peripheral chromatin now located in the nuclear interior (Solovei et al. 2013).
Proposed functions of the nuclear lamina include imparting mechanical stability to the nucleus, the anchoring of specific chromatin domains (lamin-associated domains [LADs]) to the nuclear periphery (discussed later in this article), repression of gene activity of these LADs and possibly maintenance of epigenetic gene silencing, and cell signaling (Wilson and Foisner 2010; van Steensel and Belmont 2017; de Leeuw et al. 2018; Briand and Collas 2020).
The nuclear envelope is perforated by thousands of nuclear pores. These large protein complexes were first visualized by early EM as ∼30 nm holes in the nuclear membrane (Callan and Tomlin 1950). Early observations on sectioned nuclei visualized ∼150 nm diameter annuli and already made observations of chromatin-free channels extending from these nuclear pores well into the nuclear interior (Watson 1959). The physiological relevance of nuclear pores to transport in and out of the nucleus was realized immediately and thus they have been a focus of extensive biochemical, molecular, structural, and biophysical research ever since. Cryo-EM reconstructions now provide the highest resolution imaging of intact nuclear pore structure, which includes a central inner pore ring between outer and inner rings, all with eightfold symmetry, plus filaments extending into the cytoplasm on one side and a “fish-trap”-shaped nuclear basket extending into the nucleoplasm (Lin and Hoelz 2019).
Molecular cloning of nuclear pore proteins, or nucleoporins, revealed a subset of NUPs with intrinsically disordered, FG repeats that together create the permeability barrier of the nuclear pore and interact with exportin and importin transport factors (Beck and Hurt 2017; Lin and Hoelz 2019). Biophysical studies suggest the local concentration of these FG repeat NUPs in the nuclear pore channel induce either a liquid–liquid phase separation or hydrogel accounting for this barrier. This type of barrier, involving a high frequency of low-affinity interactions, may account for the combined high selectivity plus high rates of nuclear pore traffic (Schmidt and Görlich 2016; Frey et al. 2018; Celetti et al. 2020). The nuclear basket, comprised largely of the protein TPR, has been implicated in helping to maintain a chromatin-free channel facing the nucleoplasmic side of the nuclear pore. The loss of peripheral heterochromatin during oncogene-induced cell senescence was correlated with increased nuclear pore density and reversed by knockdown of TPR (Boumendil et al. 2019).
Beyond nuclear import and export of proteins and RNPs, nuclear pores and NUPs likely have additional functions. Both gene activation and transcriptional memory and gene silencing have been linked to direct contacts of genes with nuclear pores across a wide range of species from yeast to human (Randise-Hinchliff and Brickner 2018; Cho and Hetzer 2020). Additionally, a subset of NUPs shuttle between nuclear pores and the nuclear interior and contribute to gene regulation (Cho and Hetzer 2020). DNA break repair has been linked as well to chromosome movement to and contact with nuclear pores (Seeber and Gasser 2017). Conversely, certain nuclear compartmentalization, including association with the nuclear lamina, may restrain chromosome movement and the available molecular pathways for DNA repair (Lemaître et al. 2014; Schep et al. 2021).
Given these diverse functions, nuclear pores and/or NUPs have been implicated in a wide range of biological processes ranging from control of differentiation and cell identity, viral infection, cancer, cell senescence and organismal premature and pathological aging, and neurodegenerative diseases (Boumendil et al. 2019; Cho and Hetzer 2020).
Other Well-Known Suspects
Many additional nuclear bodies have been described through imaging of the distribution of specific proteins and/or RNAs. In some cases, TEM has recognized these as distinct domains that can be recognized subsequently without antibody staining. Like Cajal bodies, some of these bodies may be present in only a small fraction of cells and a subset of cell types—for example, cancer cells, rapidly growing normal cell types, and/or metabolically active cells such as neurons.
Paraspeckles
Paraspeckles were first recognized through immunostaining against a protein, PSPC1 (Paraspeckle Protein 1), identified in a nucleolus proteomics screen (Fox et al. 2002; Fox and Lamond 2010). Unexpectedly, PSPC1 localized in distinct nuclear bodies away from nucleoli but near or adjacent to nuclear speckles; however, PSPC1 did localize in perinucleolar “caps” in early G1 nuclei before the onset of significant transcription (Fox et al. 2002, 2005). Core paraspeckle proteins include the three members—PSF/SFPQ, NONO/P54NRB, and PSPC1—of the DBHS family and RNA-binding motif protein 14 (RMB14) (Fig. 1J; Nakagawa et al. 2018). In most mammalian cultured cells, ∼5–20 paraspeckles are present, appearing as ellipsoidal, ∼0.5–1 μm diameter bodies (Fig. 1K; Clemson et al. 2009; Fox and Lamond 2010). Paraspeckles are now known to be nucleated by the NEAT1_2 long noncoding RNA (lncRNA) (Fig. 1J; Hutchinson et al. 2007; Clemson et al. 2009) and to concentrate certain nuclear-retained mRNAs containing long 3′UTRs with A-I edited stretches of inverted repeats (Chen and Carmichael 2009). Regulated cleavage of the 3′UTR of such mRNAs, as described first for Ctn mRNA, can lead to rapid nuclear export and a rapid increase in protein expression (Prasanth et al. 2005). Whereas normal paraspeckles are not required for Ctn nuclear retention, they do regulate the nuclear compartmentalization of Ctn, may modulate its A-I editing, and have been reported to regulate the export of other structured RNAs (Anantharaman et al. 2016). In mice, only a few tissues normally contain paraspeckles, but they can be induced under special conditions, including after various types of cell stresses (McCluggage and Fox 2021). Paraspeckles have been proposed to act in gene regulation through protein and RNA sequestration, thereby possibly playing a role in modulating various stress responses, including the hypoxic response, the circadian rhythm, and cell proliferation, among other pathways, and may play a role in miRNA processing (Pisani and Baron 2019).
PML Bodies
PML (promyelocytic leukemia) protein bodies are enigmatic nuclear compartments (Fig. 1L) implicated in a wide range of cell responses and processes, with PML body number and size regulated by various cellular stresses, including viral infections, and implicated in chromatin remodeling, DNA repair, apoptosis, cell senescence, stem cell renewal, antiviral activity, and inhibition of neurodegenerative diseases (Lallemand-Breitenbach and de Thé 2018; Corpet et al. 2020). They were discovered originally through immunostaining of the PML tumor suppressor gene product, and then connected to heterogenous-type spherical objects visualized previously by EM (Fig. 1M; Lallemand-Breitenbach and de Thé 2010). PML bodies are round, ∼100–1000 nm in diameter, and are present in most mammalian cells at copy numbers of ∼1–30 (5–15 in typical cell lines). They are nucleated through a spherical shell assembly of PML protein subunits and appear to transiently recruit a wide range of seemingly unrelated proteins, including the histone chaperone DAXX, the HIRA H3.3-specific histone chaperone complex, SETDB1, the transcriptional coactivator CBP, and PTEN, perhaps at least in part through SUMO conjugation of PML and recruited proteins. Protein sequestration within PML bodies and enhanced protein modifications and/or degradation within PML bodies may all be related to PML function. PML bodies have been observed to interact with particular gene loci, including the MHC gene cluster (Shiels et al. 2001; Gialitakis et al. 2010), the TP53 locus (Sun et al. 2003), active histone genes in S-phase, and transcriptionally active genes in general (Wang et al. 2004; Corpet et al. 2020). In at least one case, this gene association was correlated with transcriptional memory, in which repeated gene induction is associated with a faster response and a higher level of gene induction (Gialitakis et al. 2010). Specialized PML bodies are associated with telomeres undergoing alternative lengthening of telomeres (ALTs) (Lallemand-Breitenbach and de Thé 2018; Corpet et al. 2020).
Perinucleolar Compartment (PNC)
These appear as ∼250–4000 nm diameter caps, containing 80–180 nm electron-dense threads as visualized by EM, on the nucleolar surface in many cancer cells but not nontransformed cells or tissues (Pollock et al. 2011). Their prevalence and number per nucleus correlates with metastatic potential of primary tumor cells and inversely correlates with patient survival. PNCs contain a subset of RNA Pol III transcribed small, noncoding RNAs and RNA-binding proteins associated with RNA processing of RNA Pol II transcribed transcripts plus nucleolin, a nucleolar-localized protein involved in rRNA processing. In mice, metarrestin, a drug selected for its ability to disassemble PNCs inhibited metastatic development and extended survival in several cancer models (Frankowski et al. 2018).
Histone Locus Body (HLB)
These bodies appear similar to Cajal bodies, sharing some components, but appear specifically adjacent to histone genes and contain many additional components related to histone gene transcription and processing including coactivator NPAT and FLASH, involved in 3′ end processing of histone transcripts (Yang et al. 2009; Machyna et al. 2013).
Cleavage Bodies
Numbering one to several per nucleus, these are bodies ranging from ∼0.3 to 1 μm in diameter and enriched in factors involved in 3′ cleavage and polyadenylation of nascent transcripts; they often are found in spatial proximity or overlap with Cajal bodies, HLBs, and GEMs, which are bodies containing the SMN protein (Li et al. 2006). Note that GEMs, HLBs, and cleavage bodies frequently lie adjacent to each other or overlap; they have been proposed to represent “sub-Cajal” bodies that are precursors to Cajal bodies (Machyna et al. 2013), perhaps as multilayered condensates that then fuse into one merged condensate, analogous to what has been proposed for the different nucleolar compartments (Lafontaine et al. 2021).
Less Conventional Suspects
A number of nuclear structures do not meet the traditional definition of nuclear bodies that can be recognized by EM without specific staining but fit the definition of nuclear compartments that concentrate factors. One example would be “transcription factories,” defined by immunostaining against RNA Pol II carboxy-terminal domain (CTD) amino acid repeats phosphorylated at specific sites (Ser2, Ser5) in both fixed and living cells (Fig. 1N; Xie and Pombo 2006; Uchino et al. 2021). Several hundred to thousands of punctate foci, ∼80–130 nm in diameter, are present per nucleus (Jackson et al. 1998; Cook 1999; Eskiw and Fraser 2011), localizing adjacent to many active gene loci (Osborne et al. 2004, 2007; Ferrai et al. 2010). They were speculated to represent small clusters of RNA Pol II through which DNA is “reeled” during transcriptional elongation (Iborra et al. 1996; Jackson et al. 1998; Cook 1999). Their functional significance—as storage sites for initiating or elongating RNA Pol II polymerases or actual active polymerases engaged on DNA—remains unknown, but they are found clustered around nuclear speckles and other active nuclear regions (Chen and Belmont 2019), show increased numbers adjacent to more active Hsp70 genes (Kim et al. 2020), and show high contact frequencies with many highly active gene loci (Takei et al. 2021).
Condensates of subunits of the transcriptional Mediator complex are a second example of these unconventional nuclear compartments that do not form a nuclear body or structure identifiable by TEM without immunostaining. Mediator aggregates form both in vitro and in vivo (Fig. 1O; Cho et al. 2018; Guo et al. 2019) and superenhancers in mouse embryonic stem cell (mESC) nuclei frequently associate adjacent to these condensates (Sabari et al. 2018). In vitro, Mediator subunit condensates excluded RNA Pol II with phosphorylated CTDs but were miscible with condensates formed from splicing factors (Guo et al. 2019). Thus, RNA Pol II CTD phosphorylation was proposed to transfer RNA Pol II engaged genes from Mediator condensates involved in transcriptional initiation to condensates enriched in splicing factors involved in transcriptional elongation (Guo et al. 2019).
Additional examples would include specialized chromatin domains—for example, chromocenters and polycomb bodies formed in certain species and/or cell types by coalescence of pericentric, constitutive heterochromatin or polycomb-silenced regions, respectively. Both form long-distance contacts in both cis and trans with other chromatin regions correlating with their gene silencing (Csink and Henikoff 1996; Dernburg et al. 1996; Brown et al. 1999; Bantignies et al. 2011; Pirrotta and Li 2012).
More broadly, chromatin domains in general, and entire chromosomes such as the mammalian inactive X chromosome, are increasingly being thought of as discrete compartments that may interact in cis and trans with other similar chromatin compartments as discussed below.
CHROMATIN COMPARTMENTS—INSIGHTS FROM IMAGING
Early Cytology
Key features of metazoan nuclear chromosome were first recognized roughly a century ago. Folding of chromatin into largely discrete, localized interphase chromosomes territories, as reviewed elsewhere (Cremer and Cremer 2010), was first inferred by early cytologists. Decades later, the concept of chromosome territories was resurrected by the Cremer laboratory's observation that microirradiation of local nuclear regions caused DNA damage and repair in only a small number of interphase chromosomes (Cremer and Cremer 2010). With the continued evolution of fluorescence in situ hybridization and light microscopy, chromosome territories were then directly visualized by whole chromosome “paints.” Similarly, the existence in many species and cell types of a variant “Rabl” interphase chromosome configuration, in which centromeres and telomeres localize to opposite poles of the nuclear periphery, was observed by early cytologists examining chromosomes exiting and reentering mitosis (Rabl 1885). Recent Hi-C and molecular analysis has revealed that this Rabl configuration likely has appeared in multiple species through convergent evolution driven by condensin II reduced activity through mutations in condensin II subunits (Hoencamp et al. 2021).
The original definition of heterochromatin as chromosome regions that remain condensed after mitosis during interphase was made by Emil Heitz in the 1920s (Heitz 1928; Passarge 1979). Heitz defined heterochromatin as chromosome regions that remained condensed throughout most of the cell cycle, loosening only briefly before mitosis, and condensing again before the mitotic condensation of euchromatin, the chromatin that did decondense during interphase (Heitz 1928; Brown 1966; Passarge 1979). Heitz later made the association between heterochromatin and gene-poor chromosome regions. Subsequently, heterochromatin was divided into constitutive heterochromatin, which remains heterochromatic in all developmental stages and all tissues, and facultative heterochromatin, which does not. Constitutive heterochromatin is found in many species flanking centromeres, near telomeres, adjacent to the NOR, in sex chromosomes, and scattered in blocks throughout the euchromatin chromosome arms.
In the 1960s, TEM revealed the tight apposition of a chromatin layer adjacent to the nuclear lamina as well as adjacent at the nucleolar periphery and in the nuclear interior (e.g., Fawcett 1966). This early TEM also led to the textbook labeling of heterochromatin and euchromatin in which the darkly stained chromatin after heavy metal staining is considered heterochromatin, whereas the lightly stained regions with apparent finely fibrillar and granular texture is considered euchromatin.
This textbook model is almost assuredly incorrect. Most “heterochromatin” regions visualized by EM would fall under the original euchromatin definition of chromosome regions that decondense during interphase, forming “chromomeres,” or granular-type structures, and “chromonema,” or fiber-type structures, viewed by histological staining and light microscopy and later by EM (Zatsepina et al. 1983; Belmont et al. 1989). Notably, Sklar and Whitock used light microscopy of living cells containing polytene chromosomes to show the fixation-induced appearance of structure in the nuclear “sap” during fixation and then showed that a similar “euchromatin” TEM-staining pattern filled the chromosome-free nucleoplasm in Drosophila salivary gland nuclei between the clearly distinguished polytene chromosomes and also in mammalian liver nuclei after centrifuging chromatin to the opposing half of the nucleus (Skaer and Whytock 1976; Skaer and Whytock 1977). Meanwhile, multiple methods suggest that chromatin in many somatic cell types exists in large-scale domains that are likely comparable to the chromomeres visualized by early cytologists; these methods include a variety of alternative DNA staining, sample preparation, and light and electron microscopy imaging approaches (Belmont et al. 1989; Olins et al. 1989; Testillano et al. 1991; Derenzini et al. 1993; Bohrmann and Kellenberger 1994; Biggiogera et al. 1996; Bazett-Jones and Hendzel 1999; Nozaki et al. 2017; Hoffman et al. 2020; Miron et al. 2020). Thus, the textbook “heterochromatin” in heavy-metal stained EM images likely represents the bulk of genomic DNA, with smaller differences between active and inactive genomic regions than suggested by the typically used labeling of “heterochromatin” and “euchromatin.” Further confirmation of the existence of stable folding of early DNA replicating, euchromatin into chromonema fibers extending over micron distances comes from combined live-cell visualization and immunogold staining of engineered chromosome regions (Kireev et al. 2008; Hu et al. 2009; Deng et al. 2016).
Emergence of Radial and Binary Models of Nuclear Compartmentalization by Imaging
Distinct, large blocks of heterochromatin, as defined by Heitz, were seen to preferentially associate with the nuclear and nucleolar periphery. This included the Barr body, the mammalian inactive X chromosome that was visualized associated with the nucleolar periphery in neurons and the nuclear periphery, and/or nucleolus in other cell types (Barr and Bertram 1951; Belmont et al. 1986; Zhang et al. 2007). The development of immunostaining, labeled nucleotides, and in situ hybridization (ISH) methods rapidly advanced our appreciation for distinct spatial nuclear compartmentalization to the entire genome.
Early fluorescence in situ hybridization (FISH) experiments exploiting repetitive DNA probes showed preferential association of telomeres and centromeres with the nuclear periphery and nucleoli as a function of cell type and cell-cycle stage and proliferation (Vourc'h et al. 1993; Solovei et al. 2004). The later development of whole chromosome FISH paints revealed the gene-poor human chromosome 18 frequently juxtaposed to the nuclear periphery and more peripheral than the more centrally located, and nucleolar-associated, gene-rich chromosome 19 (Croft et al. 1999). Analysis of all human chromosomes revealed varying distributions relative to the periphery versus nuclear center, with a general dependence both on chromosome size and on gene density, with increasing gene density/activity associated with a more central location (Cremer et al. 2003; Bolzer et al. 2005). This was observed in multiple cell types, although some cell types, especially with flatter nuclei, also showed a dependence on chromosome size, with smaller chromosomes more central and larger chromosomes more peripheral (Bolzer et al. 2005). Analysis of individual gene locations by many laboratories also revealed a correlation between gene activity and radial positioning of gene loci (Takizawa et al. 2008; Bickmore 2013); this relationship between gene activity and radial positioning more recently has been attributed to the gene density and activity of ∼Mbp chromosome regions (Kölbl et al. 2012).
These results led to a radial positioning model of genome organization with more active chromosomal loci located more interiorly and more silent chromosomal loci located more peripherally in the nucleus. Notably, this is a statistical, correlative model and there is a large variability in radial positioning for any particular chromosome locus, as well as a large variability in radial positioning among different genes with similar expression levels (Takizawa et al. 2008). Bias in radial positioning could simply be the indirect result of association of chromosome loci with different nuclear compartments, which themselves are distributed with a radial bias (Chen et al. 2018b; Misteli 2020).
Meanwhile, other imaging observations led instead to an approximately binary model of genome nuclear organization. LINE-1 repeats, enriched in gene-poor, mitotic chromosome G-bands, concentrate in a thin rim adjacent to the nuclear and nucleolar periphery, whereas Alu repeats, enriched in gene-rich, mitotic chromosome R-bands, distribute through much of the nuclear interior (Fig. 2A; Korenberg and Rykowski 1988; Bolzer et al. 2005; Solovei et al. 2009; Lu et al. 2021). Similarly, labeling of DNA replication revealed two main replication patterns—early replicating DNA distributed over most of the nuclear interior but excluded from the nuclear and nucleolar periphery versus middle to late replicating distributed similarly to the LINE-1 repeats in a rim adjacent to the nuclear and nucleolar periphery (Fig. 2B)—plus a minor, late-replicating pattern in a small number of large domains distributed in the nuclear interior (O'Keefe et al. 1992). Whereas the first two patterns were each estimated to occupy several hours of S-phase, the late replicating stage, thought to correspond to constitutive heterochromatin, was estimated to occur in a shorter time window (Dimitrova and Gilbert 1999). Later, visualization of the redistribution of LADs after mitosis revealed their concentration at both the nuclear and nucleolar periphery (Fig. 2C; Kind et al. 2013).
Figure 2.
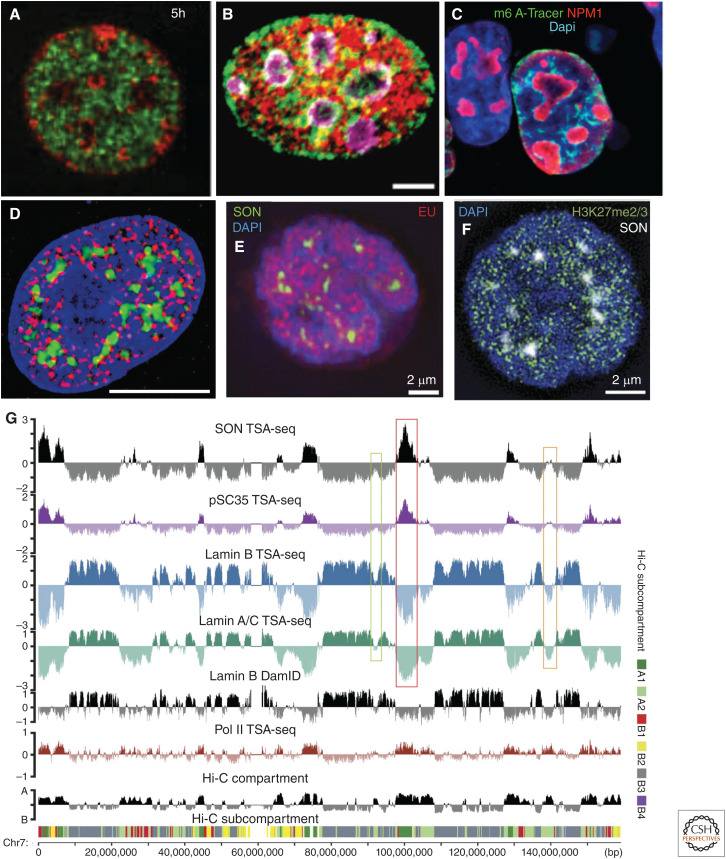
Both imaging and sequencing-based genomics methods suggest binary model for nuclear genome organization as a first approximation to a more complex organization. (A–C) An approximately binary nuclear genome organization revealed by imaging L1 repeat enriched chromatin, late-replicating DNA, and lamina-associated domains (LADs) largely at the nuclear and nucleolar peripheries with B1/Alu repeat enriched chromatin, early-replicating DNA, and intervening LADs (iLADs) in the nuclear interior. (A) Mouse embryonic fibroblast, early (green) versus late (red) DNA replication pulse labeling (5 h chase between early and late labeling). (Panel A from Wu et al. 2006; reprinted, with permission, from The Rockefeller Press © 2006.) (Continued) (B) Mouse C2C12 cell. L1 (green) or B repeat (red) FISH with nucleoli stained by nucleolin (purple). Scale bar, 5 μm. (Panel B from Lu et al. 2021; reprinted under the terms of the Creative Commons CC BY license.) (C) LADs, whose DNA was methylated by contact with lamin B1 fused to Dam methylase in the preceding interphase, stochastically redistribute early in the next interphase to the nuclear lamina, periphery of the nucleoli (red, NPM1), and nuclear interior (blue, DAPI), as visualized by the binding of the m6A-Tracer protein (green) that binds methylated DNA. (Panel C from Kind et al. 2013; reprinted, with permission, from Elsevier © 2013.) (D–F) Signs of a more complex nuclear genome organization emerge after staining for nuclear speckles and various marks of active versus repressive chromatin. (D) Hyperacetylated histones (red) are distributed nonuniformly within nuclear interior (DNA, blue), including concentrations adjacent to nuclear speckles (green). Scale bar, 10 μm. (Panel D from Hendzel et al. 1998; reprinted, with permission, from the American Society for Cell Biology © 1998.) (E,F) Local concentrations of EU pulse-labeling of nascent transcripts (red) revealing transcriptionally active chromosome regions dispersed nonuniformly through nuclear interior (DAPI, blue), including surrounding nuclear speckles (SON, green) (E); in contrast, repressive H3K27me3 mark (green) for facultative heterochromatin also is present in foci distributed throughout most of the nucleus from the nuclear periphery to the edge of nuclear speckles (F). (Panels E and F from Chen et al. 2018b; reprinted under the Creative Commons License CC BY-NC-SA 4.0.) (G) Genome browser view showing how largely binary division of nuclear genome organization based on lamin B1 DamID, Hi-C compartment (EV1) score, or RNA Pol II CTD Ser5p TSA-seq is further subdivided into chromosomal regions with varying distances to nuclear speckles or from nuclear lamina, as seen by varying location and heights/depths of SON/SC35 TSA-seq peaks/valleys, varying depths of lamin A/C and B TSA-seq valleys, as well as Hi-C subcompartments (note correlation of A1 subcompartment with SON/SC35 TSA-seq peaks and varied localization of B1 [enriched in H3K27me3 mark] along chromosome). (Panel G from Chen et al. 2018b; reprinted under the Creative Commons License CC BY-NC-SA 4.0.)
Both the radial genome positioning and binary models of genome organization clearly represent approximations. Indeed, beyond the binary division of early and late DNA replication patterns, a third, very late DNA replication pattern shows large, condensed, and likely heterochromatic regions in the nuclear interior. Chromocenters and the Y chromosome, representing constitutive heterochromatin, as well as the facultative heterochromatic Barr body can be found in the nuclear interior as well as the nuclear and nucleolar periphery. More prevalent, however, are the hundreds to thousands of foci located in the nuclear interior with inactive chromatin marks, for example, H3K27me3 (Fig. 2F) or H3K9me3, interspersed with foci located in the nuclear interior with active chromatin marks (Fig. 2D) or even foci of nascent RNA (Fig. 2E).
In an early, unusually insightful study, Shopland and colleagues FISH labeled a 4.3 Mbp chromosome region, painting several, 400–1000 kbp gene-poor domains green and interspersed, 280–890 kbp gene-rich domains red (Shopland et al. 2006). As expected, a “barber-striped” pattern along a linear interphase chromosome trajectory was observed, but only in ∼20% of G1 chromosomes. Instead, alternate conformations emerged with red domains and green domains self-segregated with like domains. This included clusters of red adjacent to the nuclear periphery with green domains located more interior, clusters of green surrounded by red domains, and zigzag patterns in which linear arrays of red domains abutted a parallel array of green domains. Thus, gene-poor versus gene-rich chromatin domains, segregated at this light microscopy resolution through apparent preferential interactions of like-domains with each other, independent of treatment with the transcriptional inhibitor, DRB. In contrast, a chromosome paint of a uniformly active, ∼4 Mbp, gene-rich region showed a higher percentage of striped patterns and decreased percentages of alternative clustered or zigzag patterns (Shopland et al. 2006).
An analogous self-organization of active versus inactive DNA sequence was revealed in the folding of engineered, large BAC (bacterial artificial chromosome) transgene arrays 10s to 100s of Mbp consisting of multiple copies of single, ∼200 kbp BACs (Sinclair et al. 2010). The plasmid backbone and a 10 kb 256mer lac operator repeat inserted within the human DNA BAC inserts from multiple BACs came together into separate heterochromatin foci enriched in the histome modification H3K9me3 and the architectural heterochromatin protein HP1—the vector backbone in one set of clusters and the lac operator repeat in another set of clusters. In contrast, active gene sequences and polycomb-repressed regions within α-globin BAC transgene arrays arrange toward the periphery of the BAC transgene array “territory” but in separate apparent clusters. Nascent transgene RNA and RNA Pol II concentrated in an outer rim surrounding the BAC transgene “territory.” The segregation of H3K27me3-modified, polycomb-repressed α-globin genes from the GFP-lac repressor stained lac operator repeats was maintained even in mitotic chromosomes. This tendency of self-sorting and self-association of like sequences was particularly enhanced in undifferentiated mESCs (Sinclair et al. 2010).
This observed clustering of ectopic lac operator and vector backbone repeats, described above, may be revealing mechanisms acting on endogenous genomic repeats to shape chromosome folding. Indeed, homotypic clustering of L1 and Alu repeats driven by repeat RNA transcription was proposed recently to contribute to nuclear compartmentalization (Lu et al. 2021). More generally, computer polymer-folding simulations have suggested that this self-association of active with active and inactive with inactive chromatin regions, together with their affinity for different nuclear compartments, is likely a major driver of nuclear genome folding, as reviewed recently (Misteli 2020).
A different question is how compartmentalization of DNA into large-scale chromatin domains might be stable during interphase progression and particularly during DNA replication. Similar to the transcription “factory” model, discussed previously, combined live-cell imaging plus light and electron microscopy imaging of pulse-chased replicated DNA suggested large-scale chromatin domains remain condensed during DNA replication, with DNA instead pulled out of these chromatin domains and into adjacent PCNA-enriched “replication foci” and then “snapping back” into the chromatin domain after replication (Deng et al. 2016).
GENOMIC ANALYSIS OF GENOME NUCLEAR ORGANIZATION
Binary Compartment Model Based on Genome-Wide Mapping of Genomes
More recently developed genome-wide mapping methods are complementary to the imaging methods that generated these initial conceptual frameworks for genome organization. The first genomic method to suggest an approximately binary division of the genome was the measurement of DNA replication timing (Schübeler et al. 2002; White et al. 2004; Hiratani et al. 2008). A two-fraction assay using early versus late pulse-labeling suggested a largely binary division of the genome into early versus late replicating domains with transition zones, possibly corresponding to single, elongating replication forks, connecting early with late regions. In Drosophila, constitutive heterochromatin regions were, as in imaging studies, detected as replicating even later (Schübeler et al. 2002).
The subsequent development of molecular proximity mapping methods revealed a striking division of the genome into discrete domains. One early genome-wide proximity-mapping method was DamID, which relies on methylation of DNA regions that interact with a protein of interest (van Steensel and Henikoff 2000; van Steensel et al. 2001). DamID showed differential molecular interaction of genome regions with nuclear lamina proteins such as lamin B1 (Fig. 2G) or emerin (Guelen et al. 2008; Peric-Hupkes et al. 2010). Thus, DamID mapping provided an approximately binary division of the genome into LADs and intervening domains (iLADs), with small genomic regions connecting the LADs and iLADs. Constitutive (cLADs) are LADs in most or all cell lines tested, while facultative LADs (fLADs) convert to iLADs in some cell lines or during differentiation (Peric-Hupkes et al. 2010; Meuleman et al. 2013; Robson et al. 2016).
The development of chromosome capture conformation (3C) methods, and ultimately Hi-C, led to the third independent suggestion of a similar binary genome division (Lieberman-Aiden et al. 2009). Hi-C interaction maps across chromosomes showed higher than expected cross-linking between particular genome regions that were classified as “A” or “B” “compartments”: Both A and B compartments across a chromosome interacted at a higher frequency with other like compartments than expected from the average decrease in interaction frequency observed as a function of genomic distance. This division into A and B compartments emerges as principal component 1 in a principal component analysis (PCA) of Hi-C interaction frequencies, measuring the division of the genome into A (positive eigenvector 1, EV1) and B (negative EV1) compartments (Fig. 2G, “compartment score”).
This genome binary division into LADs versus iLADs closely parallels B and A compartments (Fig. 2G; van Steensel and Belmont 2017), and, with the addition of transition zones, late and early DNA replicating domains (Ryba et al. 2010). LAD/B/late genomic regions have lower gene density, lower transcriptional activity, and epigenetic marks associated with gene silencing, whereas iLADs/A/early regions have higher gene density, higher transcriptional activity, and epigenetic marks associated with active chromatin (de Wit and van Steensel 2009; Peric-Hupkes and van Steensel 2010; van Steensel and Belmont 2017; Zhao et al. 2017).
Beyond A and B Compartments
Improved Hi-C methods combined with much greater read depth further divided the original Hi-C A and B compartments into A1 and A2 active and B1, B2, and B3 major subcompartments in the G12878 lymphoblastoid cell line (Fig. 2G; Rao et al. 2014). Whereas the B1 subcompartment was enriched in epigenetic markers related to polycomb silencing, B2 and B3 subcompartments overlapped extensively with LADs. These subcompartments are assumed to correspond to spatially distinct active (A1 and A2) and repressive (B1, B2, and B3) nuclear compartmentalization. The B2 subcompartment regions are enriched on smaller chromosomes and acrocentric chromosomes containing NORs, associating the B2 subcompartment with a more nucleolar localization.
Sequencing of residual DNA associated with biochemically purified nucleoli better identify nucleolar-associated domains (NADs) (Németh et al. 2010; van Koningsbruggen et al. 2010; Bizhanova and Kaufman 2021). NADs show extensive overlap with LADs, with B2 Hi-C subcompartment genomic regions overrepresented relative to B3 regions. Overlap between LADs and NADs was expected from live-cell imaging experiments showing a stochastic shuffling of LADs between association with the nuclear lamina versus nucleolar periphery after mitosis (Kind et al. 2013). NAD-seq revealed additional H3K27me3-enriched “type 2” NADs, distinct from LADs, as confirmed by FISH (Vertii et al. 2019).
Several newer, non-3C-related genomic methods, have suggested further assignment of Hi-C-defined compartments to specific nuclear bodies, such as nucleoli and nuclear speckles.
SPRITE (split-pool recognition of interactions by tag extension), using a series of dilutions and pooling of complexes, adds unique sequencing barcodes to multiple DNA and RNA fragments associated with the same individual complexes produced by sonicating chemically cross-linked cells (Quinodoz et al. 2018). In this approach, high-throughput DNA sequencing of sequencing libraries produced from large numbers of complexes can produce two-way “interaction frequencies” analogous to Hi-C, but corresponding instead to DNA/RNA fragments colocalizing in the same complex. But SPRITE also yields frequencies of simultaneous “interactions” from larger numbers of fragments all colocalizing in the same complex. In mESCs, SPRITE identified “active” and “repressive” ∼1 Mbp regions defined through their interaction frequencies with a small number of either highly active or inactive “hubs”—chromosome regions with unusually high numbers of interchromosomal contacts in sonicated complexes. Subsequent FISH validation showed that the hub contact frequency of SPRITE-defined active chromosome regions correlated with the frequency of colocalization of these regions with nuclear speckles; instead, the hub contact frequency SPRITE-defined repressive chromosome regions correlated with the colocalization of these regions with nucleoli (Quinodoz et al. 2018).
More recently, RD-SPRITE, an improved version of SPRITE with greatly improved RNA detection, has provided more direct mapping of DNA sequences interacting with specific RNAs enriched in different nuclear bodies and also mapped imprinted chromosomal domains showing domain-wide interactions with specific lncRNAs associated with their silencing (Quinodoz et al. 2020).
A different mapping method, MARGI, ligates nearby RNA and DNA fragments in cross-linked nuclei to provide a sequencing readout of RNA colocalizing near DNA sequences (Chen et al. 2018a). Cross-linking of ncRNAs (snRNAs and MALAT1) enriched in nuclear speckles revealed large domains corresponding approximately to the entire Hi-C A compartment. However, given that MARGI reads out molecular-scale interactions, this colocalization may simply reflect the known local enrichment of snRNAs and Malat1 at actively transcribing gene bodies, rather than proximity of the chromosome region to nuclear speckles (Engreitz et al. 2014; Chen and Belmont 2019).
Genomic regions interacting with nuclear speckles were measured more directly using a new genomics method, TSA-seq (Chen et al. 2018b). TSA uses indirect immunofluorescence using a secondary antibody coupled to horse-radish peroxidase (HRP). HRP catalyzes the generation of tyramide (phenol)–biotin free radicals. The sustained generation of tyramide–biotin free radicals combined with their diffusion from the site of generation, and an approximately constant probability over time and space in their quenching, creates an exponentially decreasing free-radical concentration gradient that can be used to measure the distance of a DNA region from the staining target.
Nuclear speckle and lamin TSA-seq showed that in K562 erythroleukemia cells the previously identified A1 Hi-C subcompartment corresponded to the ∼20% of the genome closest to nuclear speckles and far from the nuclear lamina, the A2 Hi-C subcompartment instead localized at intermediate distance to nuclear speckles, whereas the inactive B2 and B3 Hi-C subcompartments were distant from nuclear speckles and close to the nuclear lamina (Fig. 2G; Chen et al. 2018b).
TSA-seq suggested several additional concepts deviating further from both the binary and radial models of genome organization. Speckle-associated domains (SPADs), corresponding to the ∼5% of the genome closest to nuclear speckles, were near deterministically located adjacent to nuclear speckles (∼95% or more of alleles). These SPADs plus flanking LADs, Mbps distant to these SPADs, defined anchor points for predicting several Mbp chromosome trajectories extending from nuclear lamina to nuclear speckles and back. More generally, gene-dense expression “hot zones” are located at the apexes of predicted chromosome trajectories projecting from the nuclear lamina variable distances into the nuclear interior. Inferred distances to either the nuclear lamina or nuclear speckles were proposed to represent better metrics for describing genome organization than radial distance to the nuclear center (Chen et al. 2018b).
Further division of the genome into multiple states with varying nuclear spatial localization was achieved for K562 cells by combining TSA-seq, DamID, and Hi-C data using a hidden Markov random field model, SPIN (spatial position inference of the nuclear genome), to identify multiple states, each predicted to share a distinctive nuclear localization (Wang et al. 2021). SPIN further divided LADs into lamina-associated, two near-lamina, and lamina-like states and divided iLADs into speckle-associated, three interior active, and two interior repressive states, each with distinctive histone modifications, DNA replication timing, and gene expression levels.
APEX (enhanced ascorbic peroxidase [APX]), a related proximity labeling method implemented typically in live cells (Rhee et al. 2013), recently has been applied to mapping genome organization relative to PML bodies (Kurihara et al. 2020). APEX uses expression of a fusion protein between an engineered, monomeric ascorbate peroxidase and a cellular protein localizing in the target cellular compartment; labeling of chromatin by the phenol–biotin free radical is subsequently detected by ChIP-seq. Although a region of the Y chromosome was mapped consistently near PML bodies, no other chromosomal region was detectable as preferentially lying near PML bodies. The APEX tagged PML protein mapped locally to a large number of promoter and enhancer regions, at well below the expected diffusion radius of the phenol–biotin free radical. This labeling may be an artifact caused by APEX labeling of DNA-specific regulatory proteins inside of PML bodies followed by their rapid diffusion out of the PML bodies and their subsequent binding to distant regulatory DNA sequences (Kurihara et al. 2020).
Finally, the ligation-independent genome architecture mapping (GAM) genomic method notably detects a significantly higher number of long-distance and trans chromosomal interactions as compared with Hi-C (Beagrie et al. 2017, 2020). GAM involves sequencing DNA from thin sections cut randomly from many cell nuclei; DNA sequences colocalizing within nuclear space are more likely to be present within randomly sampled nuclear cross-sections. Long-distance, “multi-way” interactions involving simultaneous colocalization of different DNA sequences detected by GAM were suggested to be the consequence of chromosomal interactions with nuclear bodies such as nuclear speckles and/or “transcription factories” or other condensates associated with active gene expression (Beagrie et al. 2017).
BACK TO THE FUTURE—MELDING GENOMICS WITH MULTIPLEXED NUCLEIC ACID AND PROTEIN IMAGING
All sequence-based, genomic mapping approaches, and especially those performed on ensembles of cells, face the challenge of relating their results and predictions to actual nuclear and chromosomal structures. New, multiplexed FISH approaches using bar-coded oligonucleotide probes are promising to bring genomics to single-cell imaging (Cardozo Gizzi et al. 2019; Mateo et al. 2019; Nguyen et al. 2020). Two new studies point to a future in which near genome-wide coverage of chromosome loci, and eventually chromosome trajectories, will be visualized relative to immunostained nuclear structures and bodies. Both mapped ∼1000–3000 chromosome loci, plus large numbers of nascent pre-RNAs, relative to the nuclear lamina, nucleoli, and nuclear speckles in IMR90 human fibroblasts (Su et al. 2020) and to the same nuclear compartment markers plus additional chromatin marks in mESCs (Takei et al. 2021), using multiple rounds of FISH hybridization, combinatorial labeling, and decoding schemes.
Chromosome loci showed variable contact frequencies between nuclear lamina, nucleoli, and nuclear speckles that correlated with previous genomic data. Both studies showed increased and decreased gene expression levels for loci associated with nuclear speckles and the lamina, respectively. Takei et al. identified chromosomal loci with unusually high contact frequency with specific nuclear structures, including specific chromatin marks, RNA Pol II foci, heterochromatin defined by DNA staining, or nuclear bodies (lamina, nucleoli, nuclear speckles) (Fig. 3A). Su et al. described reduced percentages of transcriptionally active gene alleles with detectable nascent transcripts for alleles associated with the nuclear lamina, whereas slightly higher active allele frequencies were observed for gene alleles associated with nuclear speckles. Takei et al. inferred a prepositioning of highly active genes near nuclear speckles and/or regions of high active chromatin marks, regardless of on/off status of alleles in particular cells.
Figure 3.
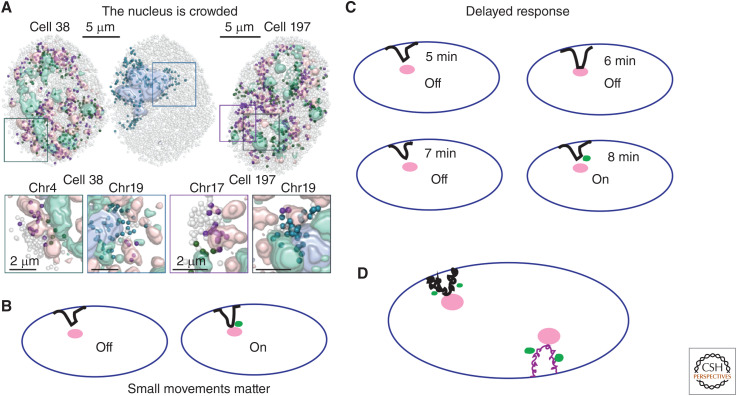
Challenges for the future. (A) The nucleus is crowded with many internal nuclear bodies and structures as revealed by multiplexed imaging of DNA loci relative to nuclear speckles (pink), nucleoli (blue), or H3K9me3-enriched heterochromatin, including chromocenters, in mouse embryonic stem cells (mESCs). A number of specific chromosome loci appear as “fixed” relative to these nuclear structures, meaning they show statistically unusually high frequency of colocalization for particular structures—pink dots for nuclear speckle-associated, green dots for H3K9me3-associated, and blue dots for nucleolar-associated—as compared with chromosome loci that do not show elevated association frequencies with any of these structures (gray dots). (Panel A from Takei et al. 2021; reprinted, with permission, from Nature Publishing © 2021.) (B) Small movements matter: relative movements closer or further to specific nuclear bodies/structures, even of several hundred nm, can be highly correlated with changes in gene expression. (C) Delayed response: greatly complicating analysis of the possible causal relationship between nuclear localization and changes in DNA functional output is that the change in output—i.e., transcription (nascent transcripts represented as green dot)—may show a delayed response. Here, a gene locus starts at 5 min after gene induction a small distance from a nuclear speckle, touches the speckle at 6 min, and in a delayed response, turns on to higher levels at 8 min when it is again away from the nuclear speckle. Examination in fixed cells would instead lead to the inference of a high level of transcription even without nuclear speckle contact. (D) An integrated view: Traditionally, our field has focused solely on individual chromosome loci and their nuclear position. However, movements of chromosome loci toward or away from specific nuclear bodies/structures could be associated with coordinated changes in nuclear localization, large-scale chromatin compaction, and even biochemical changes to flanking chromatin regions, possibly Mbps in size. Here, an active chromosome movement of a speckle-associated chromosomal locus toward a nuclear speckle, followed by its attachment to the speckle, combined with the anchoring of a neighboring LAD to the nuclear lamina could differentially alter the chromatin compaction of the intervening several Mbp of DNA between these two chromosome loci, possibly even leading to differential gene expression (nascent transcripts, green dots) as a function of differential chromosomal stretching.
THE ELEPHANT IN THE ROOM: NUCLEAR COMPARTMENTS, PHASE SEPARATION, AND CONDENSATES
Liquid–liquid phase separation (LLPS) has gathered momentum as a major paradigm change in cell biology today (Hyman et al. 2014; Shin and Brangwynne 2017). LLPS refers to the demixing of liquids into spatially separate phases, analogous to the separation of oil and water. Many nuclear bodies show at least a subset of LLPS characteristics; these include fusion, fission, dripping, rounding, viscoelastic behavior, and rapid diffusion of proteins within bodies and exchange of these proteins out of the bodies, but with reflection at the nucleoplasmic/body boundary. Moreover, many nuclear body proteins, either individually or in a mixture with other proteins or RNAs, form liquid phase-separated droplets in vitro. An excellent example in which LLPS properties play a likely role in establishing nuclear body structure and function is the nucleolus, as reviewed recently (Lafontaine et al. 2021).
As a result, LLPS is commonly invoked as a universal mechanism for the organizing principle of nearly all nuclear bodies. Yet, the “elephant in the room” that many are reluctant to discuss is that there are few instances where “classic” liquid-like behavior, involving the phase-separation of pure liquid states, has been shown definitively in vivo (McSwiggen et al. 2019). If one defines condensates as local concentrations of subunits in the absence of a separating lipid membrane enclosure, then condensates would include many additional types of matter besides liquids such as liquid crystals, gels, and solids; condensates would also include the concentrated binding of proteins to localized RNA aggregates or condensed chromatin domains (Boeynaems et al. 2018; Lyon et al. 2021). Moreover, some characteristics of LLPS may be shared by these other types of condensates, although a given cellular body might transition between liquid and gel or gel and solid. Moreover, these transitions might occur either uniformly or heterogeneously within the body (Boeynaems et al. 2018; Lyon et al. 2021).
Indeed, by two criteria for a true liquid-like condensate—the absence of internal structure and the surface tension induced rounding of the condensate—several nuclear bodies clearly deviate. Cajal bodies show a distinctive coiled structure (Fig. 1I); paraspeckles are now being modeled as block copolymer micelles with a characteristic cylindrical shape and an ordered interior organized by the nucleating and required lncRNA NEAT1_2 (Fig. 1J; Yamazaki et al. 2021); PML bodies show an outer core shell formed by the PML protein (Fig. 1M; Yamazaki et al. 2021); IGCs/nuclear speckles show nonround shapes formed by clusters of 20–25 nm RNP granules, which often align linearly as chains of granules (Fig. 1H; Bernhard and Granboulan 1963). Moreover, the actual functional nuclear speckle likely includes the interchromatin granules together with additional proteins and RNAs, including the lncRNA MALAT1, which fill space between these granules and/or surround the outside of the granule cluster, as visualized by superresolution light microscopy (Fig. 1G; Fei et al. 2017).
These features of many nuclear bodies deviate from the typically round foci formed by in vivo overexpression of many nuclear body proteins that frequently contain intrinsically disordered regions (IDRs) favoring LLPS. Indeed, some nuclear body proteins appear under normal physiological conditions to be in a nonliquid condensate state, which may be poised to undergo LLPS during cell-cycle progression or after physiological perturbation. For example, MFAP1 and PRPF38A reenter nuclei and form round, droplet-like bodies ∼10–20 min before the entry of the nuclear speckle putative scaffolding proteins SON and SRRM2 and their colocalization with nuclear speckles (Dopie et al. 2020; Ilik et al. 2020). These MFAP1/PRPF38A bodies either represent nucleation sites for the reforming nuclear speckles or instead are local condensates of proteins that later will concentrate in nuclear speckles forming elsewhere. RNA Pol II inhibition during interphase causes the same MFAP1 and PRPF38A proteins to exit nuclear speckles and form round bodies adjacent to and away from nuclear speckles (Dopie et al. 2020). Meanwhile, RNA Pol II inhibition also causes nuclear speckle rounding, as expected for a transition to a more liquid-like condensate.
For these reasons, more complex models beyond LLPS likely will be required to understand the full range of behaviors shown by many nuclear bodies.
This also holds true for chromatin domains, which recently have been suggested to form via LLPS (Maeshima et al. 2020). In vitro, nucleosome oligomers form aggregates as a function of polycation concentration, and similar drop-like structures were observed after injection into nuclei of live cells (Gibson et al. 2019). This aggregation of nucleosomes can be enhanced by binding of HP1 through an HP1-induced change in the histone octamer structure (Sanulli et al. 2019).
However, numerous mechanical measurements and live-cell imaging experiments instead have shown elastic behavior of whole chromosomes, nuclear chromatin, and interphase chromosome regions. The first suggestion of chromosome regions behaving with liquid-like properties was for HP1-enriched Drosophila and mouse chromocenters, based on the demonstrated formation of HP1 droplets in vitro and observation of fusion and fission of groups of pericentric heterochromatin regions in live cells (Larson et al. 2017; Strom et al. 2017). Recent studies, however, now show that the HP1/DNA “droplets” behave more like a cross-linked gel against short-duration mechanical impulses, with mobile HP1 acting as a cross-linker of immobile DNA fragments (Keenen et al. 2021). In vivo, HP1 is not essential to the maintenance of chromocenter compaction and the concentration of HP1 in the nucleoplasm and chromocenter shows concentration-dependent behavior different than expected for LLPS (Erdel et al. 2020). Meanwhile, in vivo experiments reveal that whereas proteins mix rapidly within chromocenters, DNA from individual pericentric heterochromatin regions do not, suggesting a model in which solid/elastic chromosome structures serve as “scaffolds” on which LLPS might occur for chromatin-associated proteins (Strickfaden et al. 2020). Meanwhile, older experiments using photoactivation fluorescently labeled DNA had clearly established very non-liquid-like and stable interphase positioning of chromatin through cell-cycle progression through much of interphase (Walter et al. 2003).
More complex models beyond LLPS—likely combining polyelectrolyte electrostatic interactions, fiber–fiber interactions, protein and ncRNA cross-linking, and active enzymatic processes such as cohesin and condensin-mediated loop extrusion—will be needed to understand how elastic chromosomes can coexist with clearly visualized compartmentalization of different heterochromatin and euchromatic chromosome regions.
ESTABLISHMENT, MAINTENANCE, AND CHANGES IN NUCLEAR COMPARTMENTALIZATION
Although an extensive literature exists describing the reformation after mitosis of major nuclear bodies and compartments, we still have a surprisingly poor grasp of the larger picture of how these different compartments are established relative to each other and the forces and mechanisms that drive these rearrangements. In some cell types, nucleoli have been described as fusing and moving to the nuclear center during cell-cycle progression (Amenta 1961; González and Nardone 1968; Savino et al. 2001). More broadly, what forces lead to a single, centrally located nucleolus in one cell type (e.g., certain neurons [Manuelidis 1984]) versus multiple, scattered nucleoli in other cell types? What controls the variable sizes and numbers of nuclear speckles in different cell types, their restriction to the nuclear equatorial plane in fibroblasts, and their exclusion from the nuclear periphery and concentration in the nuclear interior in multiple cell types? What forces and mechanisms effect changes in nuclear compartments during cell-cycle progression or cell differentiation?
Both Cajal bodies and nuclear speckles are mobile, in some cases moving up to several microns through the nucleus at velocities up to ∼1 µm/min (Platani et al. 2000; Kim et al. 2019). In response to several cell stresses (heat shock, heavy metal, transcriptional inhibition) or during entry into prophase, nuclear speckles show a “follow-the-leader” movement with smaller nuclear speckles moving in DNA-depleted channels to fuse with larger speckles; new nuclear speckles then nucleate and move along a similar path to fuse with the same nuclear speckle (Kim et al. 2019).
In the case of chromosomal compartmentalization, early pulse-chase experiments showed that the differential spatial localization of early versus late replicating chromosomal regions to the nuclear interior versus periphery, respectively, is established during the first few hours of G1 phase (Ferreira et al. 1997). These and related experiments (Walter et al. 2003) suggested the idea that chromosome position becomes relatively fixed early in G1 phase and that cells need to passage through mitosis to rearrange their chromosome compartmentalization.
More recently, Hi-C of cells synchronized in their progression from mitosis into G1 showed establishment of A/B compartments within the first few hours of G1, agreeing with earlier microscopy work (Abramo et al. 2019; Zhang et al. 2019). However, changes in Hi-C A/B compartmentalization occur both during differentiation (Miura et al. 2019) or after physiological stimulation within a single cell cycle (Amat et al. 2019; Zhou et al. 2019).
Indeed, more than 70 years ago, the specific movement of the inactive X chromosome away from the edge of the nucleolus into the nuclear interior and then back again was observed during the several week period of wound healing of crushed motor neurons, permanently arrested in the cell cycle in G1 (Barr and Bertram 1951). In addition, several examples of stereotyped, long-range chromosome movements without an intervening mitosis or change in differentiation have been reported (Chuang and Belmont 2007).
Recent development of a new DamID method, pA-DamID, with greatly increased time resolution has suggested progressive changes in the magnitude of subsets of lamin pA-DamID LAD signals during cell-cycle progression, suggesting that a set of LADs at the ends of chromosome arms decrease their interactions with the nuclear lamina after early G1, whereas a different set of LADs toward the middle of the chromosomes increase their interactions (van Schaik et al. 2020). Similar results were independently reported using lamin cTSA-seq, a modified form of TSA-seq in which chromatin pulldown replaces DNA pulldown (Tran et al. 2021).
As an early model system to study chromosome movements tied to changes in gene expression, inducible tethering of the acidic activation domain of VP16 resulted in the directional movement of a peripherally located plasmid transgene over distances up to several microns and at average velocities of ∼0.4 μm/min (Chuang et al. 2006). These movements toward the nuclear interior were directly or indirectly related to actin and nuclear myosin 1c. Similar, actin-dependent movements were reported after induction of a transgene array of U2 genes toward Cajal bodies (Dundr et al. 2007).
More recently, a directional, linear movement, in some cases up to 4 μm and at velocities of 1–2 μm/min, was visualized for a large plasmid HSPA1A transgene array from the nuclear periphery to nuclear speckles after heat shock (Khanna et al. 2014). Similar linear, directional movements were seen for BAC Hsp70 (HSPA1A/HSPA1B/HSPA1L) transgenes toward nuclear speckles after heat shock (Khanna et al. 2014; Kim et al. 2020).
The most detailed molecular dissection of the possible mechanism of interphase chromosome movements toward specific nuclear compartments has been done in budding yeast in the context of the INO1 gene movement toward nuclear pores in response to transcriptional activation (Wang et al. 2020). Based on these studies, a model has been proposed for long-range, processive chromosome movement by a one-headed myosin recruited by the Put3 transcription factor binding to a DNA “zipcode.” This model suggests that binding of the one-headed myosin to F-actin is stabilized by a second, chaperone-dependent interaction of INO80, recruited to nearby H2A.Z-modified nucleosomes, with F-actin. The motor activity of the one-headed myosin, at least an in vitro actin filament gliding assay, was also dependent on the presence of the Hsp90 chaperone and a cochaperone (Wang et al. 2020).
FUNCTION OF NUCLEAR COMPARTMENTALIZATION
The extensive compartmentalization of the nucleus is evident, but what function does it serve? Two traditional explanations of nuclear compartmentalization have been proposed. The first is the idea of compartments increasing local concentrations of key components for enzymatic reactions and/or assemblies of macromolecular complexes. The second is the idea of compartments acting as local sites for storage or sequestration of components whose activities are normally elsewhere. With the new paradigm of condensates and phase separation, these older models have been further refined to include the idea that condensates may act as selective filters to pass through and/or concentrate certain classes of proteins or RNAs while excluding others.
However, experimentally probing the function of nuclear bodies and compartmentalization has been challenging. As just one example, many of the same proteins responsible for rRNA transcription, processing, and assembly into ribosomes are implicated in phase separation and formation of nucleolar structure. Thus, experiments manipulating the levels of these same proteins to perturb nucleolar assembly and then assaying effects on ribosome synthesis would be difficult to interpret. Indeed, although principles of phase separation derived from equilibrium thermodynamics are instructive, normal nucleolar assembly and structure is driven by the nonequilibrium transcription, RNA processing, and assembly of rRNA into ribosomes (Lafontaine et al. 2021), and it is challenging to uncouple the functional output of nucleoli from their organization. Moreover, release of the many additional proteins and RNAs that concentrate in nucleoli by induced nucleolar disassembly would lead to pleiotropic effects potentially causing significant indirect effects that again would complicate interpretation of experimental results.
Similarly, recent simultaneous knockdown of SON and SRRM2 was shown to “dissolve” nuclear speckles, raising the concentrations of their components in the nucleoplasm (Ilik et al. 2020). But both SON and SRRM2 bind near promoters and gene bodies at most active genes, including away from nuclear speckles. Indeed, most nuclear speckle proteins are present and likely playing important functional roles at or near genes away from these speckles. Thus, manipulation of nuclear speckles followed by assaying effects on gene expression also are complicated by the likely indirect effects induced by changing the concentrations of speckle components in the nucleoplasm.
One promising new approach has been to induce nucleation of condensates locally using optogentic tools (Kichuk et al. 2021). At the very least, this approach allows exploration and testing of proposed models for the effects of condensates on biochemical and molecular processes. To the extent that these induced condensates approximate the true behaviors of the more complex, actual nuclear bodies, these optogenetic approaches should provide a productive means to probe the functions of nuclear bodies (Zhu et al. 2019; Wei et al. 2020; Zhang et al. 2020a; Ma et al. 2021).
Nuclear Speckles as a Model to Probe Compartment Structure and Function Relationship
Numerous reviews have focused on the function of various nuclear compartments and structures. Here, we focus on recent experiments in nuclear speckles that have used temporal ordering of events, in addition to perturbation of nuclear bodies, as a tool for probing nuclear body function.
TSA-seq mapping reveals several features of speckles: First, ∼5% of the genome is near deterministically positioned within <500 nm of nuclear speckles; second, inferred average distances to nuclear speckles are largely conserved between different cell types, with ∼90% of the genome not statistically different in position in pairwise cell line comparisons; and third, small-to-moderate shifts of the remaining 10% of the genome are highly correlated with changes in gene expression, with shifts relatively closer (further) to speckles associated with a strong bias toward significantly increased (decreased) gene expression (Chen et al. 2018b).
As described previously, mechanism(s) exist to move specific transgenes—for example, HSPA1A—directionally to nuclear speckles in just several minutes (Khanna et al. 2014). A more direct link between nuclear speckle proximity and gene expression was suggested by the temporal sequence between contact of these HSPA1A plasmid transgenes with nuclear speckles and HSPA1A gene expression, as revealed by live-cell imaging. Increased nascent HSPA1A transcripts appeared always after first contact, but never before contact with nuclear speckles. The time lag between first appearance of the nascent transcript signal was several minutes shorter if the transgene array first contacted a larger versus smaller nuclear speckle. Contact with a smaller speckle led to an initial increase in nuclear speckle size above a certain size before the appearance of a nascent transcript signal above background. Later, both nascent transcript signals and nuclear speckle size increased in parallel. This increased size and coordinate growth of both speckles and nascent transcript signals is intriguing in light of the proposed model, described previously, of cycles of PTMs for splicing factors trafficking between speckle to gene and then back to speckle (Misteli and Spector 1997; Hall et al. 2006). Blocking movement to nuclear speckles by blocking actin polymerization resulted in reduced HSPA1A heat-shock induction of plasmid transgenes away from nuclear speckles but no reduction in expression for plasmid transgenes associated with nuclear speckles (Khanna et al. 2014).
Using HSPA1B BAC transgene arrays, which more completely recapitulate the heat-shock-induced expression behavior of the endogenous locus, produced similar results and showed the phenomenon of “gene expression amplification” (Kim et al. 2020). Although the BAC transgenes showed 100% induction within several minutes after heat shock, regardless of speckle association, transgenes in contact with nuclear speckles showed a several-fold increased production of nascent transcripts, which may be at least partially explained by reduced exosome degradation of nascent transcripts near nuclear speckles. This was also seen for the endogenous Hsp70 locus and several HSPA1A flanking genes at both the endogenous locus and the BAC transgenes at normal temperature. Again, live-cell imaging showed that these BAC transgenes did not show jumps in nascent transcript production until after nuclear speckle contact; reduced gene expression occurred within several minutes of loss of contact with nuclear speckles (Kim et al. 2020).
In light of the observed movement of plasmid and BAC transgenes toward nuclear speckles after heat shock, it was surprising that about half of all heat-shock genes, including HSPA1A, are near 100% localized adjacent to nuclear speckles even before heat shock, as revealed by TSA-seq and DNA FISH in multiple human cell lines (Zhang et al. 2020b). The remaining half of expressed heat-shock gene loci are near or at moderate distance (HSPH1A) from nuclear speckles but move closer after heat shock. The HSPH1 gene locus was the furthest of all heat-shock loci from nuclear speckles at 37°C (∼50% genomic percentile) and showed the slowest induction kinetics. FISH experiments revealed a gene amplification as well after nuclear speckle contact for the endogenous HSPH1A gene (Zhang et al. 2020b).
Finally, comparing four cell types, no large differences in relative nuclear speckle position were observed (Zhang et al. 2020b). Genes that are positioned closer to nuclear speckles in one cell type are positioned just a few hundred nanometers in average distance further from nuclear speckles in other cell types. This suggests a level of “prewiring” of gene loci relative to nuclear speckles such that genes move only small distances to or from nuclear speckles during differentiation or physiological stimuli (Zhang et al. 2020b).
Such a prewiring is further suggested by recent analysis of p53-responsive genes (Alexander et al. 2021). Approximately half of ∼20 surveyed p53-responsive genes moved closer to nuclear speckles after p53 induction and, interestingly, these were the genes that were already positioned close to nuclear speckles even before p53 activation. This induced association with nuclear speckles was dependent on a different region of the p53 protein from the known trans-activating or DNA-binding domains of p53. FISH analysis again revealed a gene expression amplification phenomenon, with faster and higher induction of increased gene expression for alleles within ∼0.5 μm of nuclear speckles. Perturbation of nuclear speckles by SON knockdown perturbed this increased gene expression observed with nuclear speckle proximity. TSA-seq revealed several hundred inferred p53-responsive genes that moved closer to nuclear speckles after p53 activation and again showed increased expression, and these were the ∼30% of p53-responsive genes that were prepositioned near nuclear speckles before p53 activation (Alexander et al. 2021).
These p53 experiments suggest that the phenomenon of gene expression amplification may be seen for large numbers of genes beyond heat-shock loci. Further experiments should reveal whether this is a general phenomenon of stress-responsive genes, as well as genes moving closer to nuclear speckles as a function of other physiological stimuli and/or development.
A very different theory has been proposed recently in which nuclear speckles rather than simply serving as storage sites for RNA processing factors instead act to actively regulate the nucleoplasmic levels of key RNA processing and splicing factors (Hasenson and Shav-Tal 2020). This produces an “action-at-a-distance” regulation in which changing nucleoplasmic levels of limiting factors by concentration or release by nuclear speckles can change the RNA processing and alternative splicing of genes throughout the nucleus. This model was prompted by observations from live-cell imaging of a transgene array with transgenes containing varying numbers of intron/exon junctions (Hochberg-Laufer et al. 2019). Accumulation of nascent RNA transcripts near the site of transcription was observed only for transgenes containing larger numbers of introns and this was modeled as a delay in splicing occurring after transcription but before release of the nascent transcripts into the nucleoplasm. This delay in release of nascent transcripts could be rescued either by overexpression of a subset of splicing factors or, much more interestingly, after “dissolving” of nuclear speckles by overexpression of the Clk1 kinase, which has been proposed to cause release of splicing factors from nuclear speckles after their phosphorylation (Hochberg-Laufer et al. 2019).
In summary, these recent experiments suggest a closer than previously appreciated relationship between nuclear speckles and regulation of gene expression. In retrospect, this close relationship had been concealed by what appears to be a very precisely controlled yet dynamic prepositioning of many endogenous genes close to nuclear speckles. Likewise, we may be underappreciating the role of nuclear speckles in buffering nucleoplasmic levels of factors involved in RNA processing, and how variations in nuclear speckle protein fluxes and dynamics between cell types may have global influence on patterns of gene expression. Dissecting the mechanisms underlying this prepositioning of gene loci near nuclear speckles, as well as the dynamics of nuclear speckle components, may be required to appreciate the full extent of this influence of nuclear speckles on gene expression.
FUTURE CHALLENGES AND QUESTIONS
The nucleus—and more specifically the interchromosomal space—is crowded (Fig. 3A). Immunostaining and fluorescent microscopy may give us a “tunnel vision” picture owing to the difficulty of visualizing more than three or four components simultaneously. On the other hand, genome-wide mapping methods are an oversimplification of reality because they only present the average over millions of cells and do not take into account cell-to-cell variability. We now realize that any given interphase chromosome domain is likely within a short distance to multiple nuclear “locales” with distinctive functional properties (Fig. 3B). New imaging approaches have made this conceptualization concrete with their visualization of many chromosome loci appearing “fixed” relative to different nuclear interior “environments” defined by proximity to specific nuclear bodies and/or chromatin marks (Fig. 3A; Su et al. 2020; Takei et al. 2021). Newly identified phase-separated nuclear condensates, in addition to previously described nuclear bodies, are only adding to this complexity. Given the number of nuclear proteins with IDRs prone to phase separate above a critical concentration, we likely are still just seeing only the tip of the iceberg in terms of further subdivision of nuclear interchromosomal space. Meanwhile, chromatin domains themselves are assuming characteristics of compartmentalized units in terms of limiting diffusion of large, macromolecular complexes involved in DNA functions.
Previous models emphasizing nuclear radial gene positioning or binary division into iLAD/early replicating/A or LAD/late replicating/B compartments can now be seen as underlying fundamental, yet oversimplified, features of nuclear genome organization. Statistical trends of radial positioning are likely the indirect result of the organization of chromosomes relative to specific nuclear bodies and compartments that themselves are distributed as a function of radial position. Meanwhile, the apparent binary division of the genome conceals an emerging reality that both the nuclear interior, and likely the nuclear periphery, are further divided into functionally distinct locales. Different alleles of the same chromosome locus may distribute among different but related spatial regions and move between them over physiological timescales. For example, recent high-throughput, multiplexed imaging revealed a maximum of ∼40% of LAD alleles in actual contact with the nuclear lamina in IMR90 human fibroblasts, with most LAD alleles not associated with the nuclear lamina but instead localizing near the nucleolar periphery or elsewhere in the nuclear interior (Su et al. 2020). Still unknown is what fraction of LAD alleles will localize to euchromatic versus heterochromatic local environments. Similarly, a large fraction of certain speckle-associated chromosome loci localizes very near nuclear speckles, but what fraction of these speckle-associated alleles instead localize away from nuclear speckles but close to other types of nuclear local environments associated with active gene expression?
Not only do “small distances matter”—separating nuclear compartments with distinct functional properties—but chromosomal domains may be “prepositioned” near the appropriate nuclear compartments required for their full and/or more robust activation or silencing. Hi-C defined nuclear compartments are ∼Mbp in size and contain many genes showing uncorrelated changes in gene activity. Therefore, it was inferred that smaller units of genome organization such as TADs and sub-TADs would be more intimately connected to gene regulation. However, a newer generation of genomic methods are showing statistically significant changes in positioning of genomic regions that can be comparable or even smaller in size than TADs. In the case of nuclear speckles, these small shifts in relative positions strongly correlate with changes in gene expression (Zhang et al. 2020b; Alexander et al. 2021).
Complicating matters further, functional consequences of small nuclear movements can be masked by subsequent movements and delayed functional outcomes of contact with nuclear compartments (Fig. 3C). This was seen over a timescale of minutes in the case of Hsp70 transgenes contacting nuclear speckles but then turning on even after nuclear speckle detachment (Khanna et al. 2014) and also taking several minutes to turn off after detaching from a nuclear speckle (Kim et al. 2020). But time delays between nuclear compartment contact and functional output might extend in other contexts to entire cell cycles or longer. For example, in fission yeast, several cell cycles were required for full loss of epigenetic silencing of a gene after knockdown of a protein required for its anchoring to the nuclear periphery (Holla et al. 2020).
Finally, we may need a more integrated view of nuclear organization in which we not only examine the location of specific chromosomal regions, and the possible functional consequence of these localizations, but also extend our analysis to larger chromosome trajectories and examine the possible coupling between changes in chromosome positioning within the nucleus simultaneous with changes in chromatin conformation and biochemical marks (Fig. 3D). Relative movements of one chromosome region within the nucleus might not only cause coordinated movements of ∼Mbp-scale flanking regions but also lead to changes in chromosome stretching and chromatin compaction, which have been shown to change gene expression (Tajik et al. 2016; Sun et al. 2020). This could create an “action-at-a-distance” mechanism by which there might be coordinated changes in nuclear position, chromatin folding, and gene expression over Mbp-linked chromosomal regions.
Thus, new multiplexed imaging approaches to visualize multi-Mbp chromosome trajectories, guided by genome-wide genomics methods providing readouts of nuclear position, DNA topology, and chromatin packing, will need to be complemented by live-cell imaging to establish temporal ordering of events linking nuclear position, chromosome folding, biochemical modifications, and, finally, functional output.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) Grants R01 GM58460, U01DK127422 and UM1HG011593. We would like to thank the NIH Common Fund, the Office of Strategic Coordination, and the Office of the NIH Director for funding the 4D Nucleome Program, which has supported the latter two grants. We thank Dr. Pankaj Chaturvedi, University of Illinois, Urbana, for the image in Figure 1A.
Footnotes
Editors: Ana Pombo, Martin W. Hetzer, and Tom Misteli
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abramo K, Valton AL, Venev SV, Ozadam H, Fox AN, Dekker J. 2019. A chromosome folding intermediate at the condensin-to-cohesin transition during telophase. Nat Cell Biol 21: 1393–1402. 10.1038/s41556-019-0406-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander KA, Coté A, Nguyen SC, Zhang L, Gholamalamdari O, Agudelo-Garcia P, Lin-Shiao E, Tanim KMA, Lim J, Biddle N, et al. 2021. P53 mediates target gene association with nuclear speckles for amplified RNA expression. Mol Cell 81: 1666–1681.e6. 10.1016/j.molcel.2021.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amat R, Böttcher R, Le Dily F, Vidal E, Quilez J, Cuartero Y, Beato M, de Nadal E, Posas F. 2019. Rapid reversible changes in compartments and local chromatin organization revealed by hyperosmotic shock. Genome Res 29: 18–28. 10.1101/gr.238527.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amenta PS. 1961. Fusion of nucleoli in cells cultured from the heart of Triturus viridescens. Anat Rec 139: 155–165. 10.1002/ar.1091390207 [DOI] [PubMed] [Google Scholar]
- Anantharaman A, Jadaliha M, Tripathi V, Nakagawa S, Hirose T, Jantsch MF, Prasanth SG, Prasanth KV. 2016. Paraspeckles modulate the intranuclear distribution of paraspeckle-associated Ctn RNA. Sci Rep 6: 34043. 10.1038/srep34043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. 1991. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med 173: 1407–1419. 10.1084/jem.173.6.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. 2011. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144: 214–226. 10.1016/j.cell.2010.12.026 [DOI] [PubMed] [Google Scholar]
- Barr ML, Bertram EG. 1951. The behavior of nuclear structures during depletion and restoration of Nissl material in motor neurons. J Anat 85: 171–181. [PMC free article] [PubMed] [Google Scholar]
- Barr ML, Carr DH. 1962. Correlations between sex chromatin and sex chromosomes. Acta Cytol 6: 34–45. [PubMed] [Google Scholar]
- Bazett-Jones DP, Hendzel MJ. 1999. Electron spectroscopic imaging of chromatin. Methods 17: 188–200. 10.1006/meth.1998.0729 [DOI] [PubMed] [Google Scholar]
- Beagrie RA, Scialdone A, Schueler M, Kraemer DC, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR, et al. 2017. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543: 519–524. 10.1038/nature21411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagrie RA, Thieme CJ, Annunziatella C, Baugher C, Zhang Y, Schueler M, Kramer DC, Chiariello AM, Bianco S, Kukalev A, et al. 2020. Multiplex-GAM: genome-wide identification of chromatin contacts yields insights not captured by Hi-C. bioRxiv 10.1101/2020.07.31.230284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Hurt E. 2017. The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol 18: 73–89. 10.1038/nrm.2016.147 [DOI] [PubMed] [Google Scholar]
- Belmont AS, Bignone F, Ts'o PO. 1986. The relative intranuclear positions of Barr bodies in XXX non-transformed human fibroblasts. Exp Cell Res 165: 165–179. 10.1016/0014-4827(86)90541-0 [DOI] [PubMed] [Google Scholar]
- Belmont AS, Braunfeld MB, Sedat JW, Agard DA. 1989. Large-scale chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma 98: 129–143. 10.1007/BF00291049 [DOI] [PubMed] [Google Scholar]
- Belmont AS, Zhai Y, Thilenius A. 1993. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J Cell Biol 123: 1671–1685. 10.1083/jcb.123.6.1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard W, Granboulan N. 1963. The fine structure of the cancer cell nucleus. Exp Cell Res 9: 19–53. 10.1016/0014-4827(63)90243-X [DOI] [PubMed] [Google Scholar]
- Bernhard W, Haguenau F, Oberling C. 1952. L'ultrastructure du nucléole de quelques cellules animales, révélée par le microscope électronique. Experientia 8: 58–59. 10.1007/BF02139019 [DOI] [PubMed] [Google Scholar]
- Bickmore WA. 2013. The spatial organization of the human genome. Annu Rev Genomics Hum Genet 14: 67–84. 10.1146/annurev-genom-091212-153515 [DOI] [PubMed] [Google Scholar]
- Biggiogera M, Courtens JL, Derenzini M, Fakan S, Hernandez-Verdun D, Risueno MC, Soyer-Gobillard MO. 1996. Osmium ammine: review of current applications to visualize DNA in electron microscopy. Biol Cell 87: 121–132. 10.1111/j.1768-322X.1996.tb00974.x [DOI] [PubMed] [Google Scholar]
- Bizhanova A, Kaufman PD. 2021. Close to the edge: heterochromatin at the nucleolar and nuclear peripheries. Biochim Biophys Acta Gene Regul Mech 1864: 194666. 10.1016/j.bbagrm.2020.194666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, et al. 2018. Protein phase separation: a new phase in cell biology. Trends Cell Biol 28: 420–435. 10.1016/j.tcb.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohrmann B, Kellenberger E. 1994. Immunostaining of DNA in electron microscopy: an amplification and staining procedure for thin sections as alternative to gold labeling. J Histochem Cytochem 42: 635–643. 10.1177/42.5.7512586 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. 2007. The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585. 10.1038/nrm2184 [DOI] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Müller S, Eils R, Cremer C, Speicher MR, et al. 2005. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 3: e157. 10.1371/journal.pbio.0030157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. 2010. The nucleolus under stress. Mol Cell 40: 216–227. 10.1016/j.molcel.2010.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumendil C, Hari P, Olsen KCF, Acosta JC, Bickmore WA. 2019. Nuclear pore density controls heterochromatin reorganization during senescence. Genes Dev 33: 144–149. 10.1101/gad.321117.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois CA, Hubert J. 1988. Spatial relationship between the nucleolus and the nuclear envelope: structural aspects and functional significance. Int Rev Cytol 111: 1–52. 10.1016/S0074-7696(08)61730-1 [DOI] [PubMed] [Google Scholar]
- Bourgeois CA, Hemon D, Bouteille M. 1979. Structural relationship between the nucleolus and the nuclear envelope. J Ultrastruct Res 68: 328–340. 10.1016/S0022-5320(79)90165-5 [DOI] [PubMed] [Google Scholar]
- Briand N, Collas P. 2020. Lamina-associated domains: peripheral matters and internal affairs. Genome Biol 21: 85. 10.1186/s13059-020-02003-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SW. 1966. Heterochromatin. Science 151: 417–425. 10.1126/science.151.3709.417 [DOI] [PubMed] [Google Scholar]
- Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG. 1999. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell 3: 207–217. 10.1016/S1097-2765(00)80311-1 [DOI] [PubMed] [Google Scholar]
- Bubulya PA, Prasanth KV, Deerinck TJ, Gerlich D, Beaudouin J, Ellisman MH, Ellenberg J, Spector DL. 2004. Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J Cell Biol 167: 51–63. 10.1083/jcb.200404120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan HG, Tomlin SG. 1950. Experimental studies on amphibian oocyte nuclei. I: Investigation of the structure of the nuclear membrane by means of the electron microscope. Proc R Soc Lond B Biol Sci 137: 367–378. 10.1098/rspb.1950.0047 [DOI] [PubMed] [Google Scholar]
- Cardozo Gizzi AM, Cattoni DI, Fiche JB, Espinola SM, Gurgo J, Messina O, Houbron C, Ogiyama Y, Papadopoulos GL, Cavalli G, et al. 2019. Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms. Mol Cell 74: 212–222.e5. 10.1016/j.molcel.2019.01.011 [DOI] [PubMed] [Google Scholar]
- Carron C, Balor S, Delavoie F, Plisson-Chastang C, Faubladier M, Gleizes PE, O'Donohue MF. 2012. Post-mitotic dynamics of pre-nucleolar bodies is driven by pre-rRNA processing. J Cell Sci 125: 4532–4542. [DOI] [PubMed] [Google Scholar]
- Carter KC, Bowman D, Carrington W, Fogarty K, McNeil JA, Fay FS, Lawrence JB. 1993. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science 259: 1330–1335. 10.1126/science.8446902 [DOI] [PubMed] [Google Scholar]
- Celetti G, Paci G, Caria J, VanDelinder V, Bachand G, Lemke EA. 2020. The liquid state of FG-nucleoporins mimics permeability barrier properties of nuclear pore complexes. J Cell Biol 219: e201907157. 10.1083/jcb.201907157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerase A, Armaos A, Neumayer C, Avner P, Guttman M, Tartaglia GG. 2019. Phase separation drives X-chromosome inactivation: a hypothesis. Nat Struct Mol Biol 26: 331–334. 10.1038/s41594-019-0223-0 [DOI] [PubMed] [Google Scholar]
- Chen Y, Belmont AS. 2019. Genome organization around nuclear speckles. Curr Opin Genet Dev 55: 91–99. 10.1016/j.gde.2019.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 35: 467–478. 10.1016/j.molcel.2009.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Yan Z, Li S, Huang N, Huang X, Zhang J, Zhong S. 2018a. RNAs as proximity-labeling media for identifying nuclear speckle positions relative to the genome. iScience 4: 204–215. 10.1016/j.isci.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang Y, Wang Y, Zhang L, Brinkman EK, Adam SA, Goldman R, van Steensel B, Ma J, Belmont AS. 2018b. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J Cell Biol 217: 4025–4048. 10.1083/jcb.201807108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho UH, Hetzer MW. 2020. Nuclear periphery takes center stage: the role of nuclear pore complexes in cell identity and aging. Neuron 106: 899–911. 10.1016/j.neuron.2020.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, Cisse II. 2018. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361: 412–415. 10.1126/science.aar4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Belmont AS. 2007. Moving chromatin within the interphase nucleus-controlled transitions? Semin Cell Dev Biol 18: 698–706. 10.1016/j.semcdb.2007.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. 2006. Long-range directional movement of an interphase chromosome site. Curr Biol 16: 825–831. 10.1016/j.cub.2006.03.059 [DOI] [PubMed] [Google Scholar]
- Cioce M, Lamond AI. 2005. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol 21: 105–131. 10.1146/annurev.cellbio.20.010403.103738 [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726. 10.1016/j.molcel.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PR. 1999. The organization of replication and transcription. Science 284: 1790–1795. 10.1126/science.284.5421.1790 [DOI] [PubMed] [Google Scholar]
- Corpet A, Kleijwegt C, Roubille S, Juillard F, Jacquet K, Texier P, Lomonte P. 2020. PML nuclear bodies and chromatin dynamics: catch me if you can! Nucleic Acids Res 48: 11890–11912. 10.1093/nar/gkaa828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M. 2010. Chromosome territories. Cold Spring Harb Perspect Biol 2: a003889. 10.1101/cshperspect.a003889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer M, Küpper K, Wagler B, Wizelman L, von Hase J, Weiland Y, Kreja L, Diebold J, Speicher MR, Cremer T. 2003. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J Cell Biol 162: 809–820. 10.1083/jcb.200304096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 145: 1119–1131. 10.1083/jcb.145.6.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csink AK, Henikoff S. 1996. Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature 381: 529–531. 10.1038/381529a0 [DOI] [PubMed] [Google Scholar]
- de Leeuw R, Gruenbaum Y, Medalia O. 2018. Nuclear lamins: thin filaments with major functions. Trends Cell Biol 28: 34–45. 10.1016/j.tcb.2017.08.004 [DOI] [PubMed] [Google Scholar]
- Deng X, Zhironkina OA, Cherepanynets VD, Strelkova OS, Kireev II, Belmont AS. 2016. Cytology of DNA replication reveals dynamic plasticity of large-scale chromatin fibers. Curr Biol 26: 2527–2534. 10.1016/j.cub.2016.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenzini M, Farabegoli F, Trerè D. 1993. Localization of DNA in the fibrillar components of the nucleolus: a cytochemical and morphometric study. J Histochem Cytochem 41: 829–836. 10.1177/41.6.8315275 [DOI] [PubMed] [Google Scholar]
- Derenzini M, Trerè D, Pession A, Govoni M, Sirri V, Chieco P. 2000. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J Pathol 191: 181–186. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Broman KW, Fung JC, Marshall WF, Philips J, Agard DA, Sedat JW. 1996. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell 85: 745–759. 10.1016/S0092-8674(00)81240-4 [DOI] [PubMed] [Google Scholar]
- de Wit E, van Steensel B. 2009. Chromatin domains in higher eukaryotes: insights from genome-wide mapping studies. Chromosoma 118: 25–36. 10.1007/s00412-008-0186-0 [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. 1999. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell 4: 983–993. 10.1016/S1097-2765(00)80227-0 [DOI] [PubMed] [Google Scholar]
- Dopie J, Sweredoski MJ, Moradian A, Belmont AS. 2020. Tyramide signal amplification mass spectrometry (TSA-MS) ratio identifies nuclear speckle proteins. J Cell Biol 219: e201910207. 10.1083/jcb.201910207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Misteli T, Olson MO. 2000. The dynamics of postmitotic reassembly of the nucleolus. J Cell Biol 150: 433–446. 10.1083/jcb.150.3.433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. 2007. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 179: 1095–1103. 10.1083/jcb.200710058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. 2014. RNA–RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell 159: 188–199. 10.1016/j.cell.2014.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, Rademacher A, Vlijm R, Tünnermann J, Frank L, Weinmann R, Schweigert E, Yserentant K, Hummert J, Bauer C, et al. 2020. Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol Cell 78: 236–249 e237. 10.1016/j.molcel.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskiw CH, Fraser P. 2011. Ultrastructural study of transcription factories in mouse erythroblasts. J Cell Sci 124: 3676–3683. 10.1242/jcs.087981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett DW. 1966. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am J Anat 119: 129–145. 10.1002/aja.1001190108 [DOI] [PubMed] [Google Scholar]
- Fei J, Jadaliha M, Harmon TS, Li ITS, Hua B, Hao Q, Holehouse AS, Reyer M, Sun Q, Freier SM, et al. 2017. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J Cell Sci 130: 4180–4192. 10.1242/jcs.206854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrai C, Xie SQ, Luraghi P, Munari D, Ramirez F, Branco MR, Pombo A, Crippa MP. 2010. Poised transcription factories prime silent uPA gene prior to activation. PLoS Biol 8: e1000270. 10.1371/journal.pbio.1000270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira JA, Carmo-Fonseca M, Lamond AI. 1994. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J Cell Biol 126: 11–23. 10.1083/jcb.126.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira J, Paolella G, Ramos C, Lamond AI. 1997. Spatial organization of large-scale chromatin domains in the nucleus: a magnified view of single chromosome territories. J Cell Biol 139: 1597–1610. 10.1083/jcb.139.7.1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lamond AI. 2010. Paraspeckles. Cold Spring Harb Perspect Biol 2: a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. 2002. Paraspeckles: a novel nuclear domain. Curr Biol 12: 13–25. 10.1016/S0960-9822(01)00632-7 [DOI] [PubMed] [Google Scholar]
- Fox AH, Bond CS, Lamond AI. 2005. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell 16: 5304–5315. 10.1091/mbc.e05-06-0587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankowski KJ, Wang C, Patnaik S, Schoenen FJ, Southall N, Li D, Teper Y, Sun W, Kandela I, Hu D, et al. 2018. Metarrestin, a perinucleolar compartment inhibitor, effectively suppresses metastasis. Sci Transl Med 10: eaap8307. 10.1126/scitranslmed.aap8307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Rees R, Schünemann J, Ng SC, Funfgeld K, Huyton T, Görlich D. 2018. Surface properties determining passage rates of proteins through nuclear pores. Cell 174: 202–217.e9. 10.1016/j.cell.2018.05.045 [DOI] [PubMed] [Google Scholar]
- Frottin F, Schueder F, Tiwary S, Gupta R, Körner R, Schlichthaerle T, Cox J, Jungmann R, Hartl FU, Hipp MS. 2019. The nucleolus functions as a phase-separated protein quality control compartment. Science 365: 342–347. 10.1126/science.aaw9157 [DOI] [PubMed] [Google Scholar]
- Gialitakis M, Arampatzi P, Makatounakis T, Papamatheakis J. 2010. γ Interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol Cell Biol 30: 2046–2056. 10.1128/MCB.00906-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, Rosen MK. 2019. Organization of chromatin by intrinsic and regulated phase separation. Cell 179: 470–484.e21. 10.1016/j.cell.2019.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González SG, Nardone RM. 1968. Cyclic nucleolar changes during the cell cycle. I: Variations in number, size, morphology and position. Exp Cell Res 50: 599–615. 10.1016/0014-4827(68)90422-9 [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951. 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall'Agnese A, Hannett NM, Spille JH, Afeyan LK, Zamudio AV, Shrinivas K, et al. 2019. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572: 543–548. 10.1038/s41586-019-1464-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Smith KP, Byron M, Lawrence JB. 2006. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol 288A: 664–675. 10.1002/ar.a.20336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenson SE, Shav-Tal Y. 2020. Speculating on the roles of nuclear speckles: how RNA-protein nuclear assemblies affect gene expression. Bioessays 42: 2000104. 10.1002/bies.202000104 [DOI] [PubMed] [Google Scholar]
- Heitz E. 1928. Das heterochromatin der moose. I Jahrb Wiss Bot 69: 762–818. [Google Scholar]
- Hendzel MJ, Kruhlak MJ, Bazett-Jones DP. 1998. Organization of highly acetylated chromatin around sites of heterogeneous nuclear RNA accumulation. Mol Biol Cell 9: 2491–2507. 10.1091/mbc.9.9.2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D. 2011. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2: 189–194. 10.4161/nucl.2.3.16246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schübeler D, Gilbert DM. 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6: e245. 10.1371/journal.pbio.0060245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg-Laufer H, Neufeld N, Brody Y, Nadav-Eliyahu S, Ben-Yishay R, Shav-Tal Y. 2019. Availability of splicing factors in the nucleoplasm can regulate the release of mRNA from the gene after transcription. PLoS Genet 15: e1008459. 10.1371/journal.pgen.1008459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoencamp C, Dudchenko O, Elbatsh AMO, Brahmachari S, Raaijmakers JA, van Schaik T, Sedeño Cacciatore A, Contessoto VG, van Heesbeen R, van den Broek B, et al. 2021. 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science 372: 984–989. 10.1126/science.abe2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DP, Shtengel G, Xu CS, Campbell KR, Freeman M, Wang L, Milkie DE, Pasolli HA, Iyer N, Bogovic JA, et al. 2020. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 367: eaaz5357. 10.1126/science.aaz5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holla S, Dhakshnamoorthy J, Folco HD, Balachandran V, Xiao H, Sun LL, Wheeler D, Zofall M, Grewal SIS. 2020. Positioning heterochromatin at the nuclear periphery suppresses histone turnover to promote epigenetic inheritance. Cell 180: 150–164.e15. 10.1016/j.cell.2019.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Kireev I, Plutz MJ, Ashourian N, Belmont AS. 2009. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J Cell Biol 185: 87–100. 10.1083/jcb.200809196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. 2007. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8: 39. 10.1186/1471-2164-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Jülicher F. 2014. Liquid–liquid phase separation in biology. Annu Rev Cell Dev Biol 30: 39–58. 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- Iarovaia OV, Minina EP, Sheval EV, Onichtchouk D, Dokudovskaya S, Razin SV, Vassetzky YS. 2019. Nucleolus: a central hub for nuclear functions. Trends Cell Biol 29: 647–659. 10.1016/j.tcb.2019.04.003 [DOI] [PubMed] [Google Scholar]
- Iborra FJ, Pombo A, Jackson DA, Cook PR. 1996. Active RNA polymerases are localized within discrete transcription “factories’ in human nuclei. J Cell Sci 109: 1427–1436. 10.1242/jcs.109.6.1427 [DOI] [PubMed] [Google Scholar]
- Ilik IA, Malszycki M, Lubke AK, Schade C, Meierhofer D, Aktas T. 2020. SON and SRRM2 are essential for nuclear speckle formation. eLife 9: e60579. 10.7554/eLife.60579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Iborra FJ, Manders EM, Cook PR. 1998. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol Biol Cell 9: 1523–1536. 10.1091/mbc.9.6.1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SM, Yoon MH, Park BJ. 2018. Laminopathies; mutations on single gene and various human genetic diseases. BMB Rep 51: 327–337. 10.5483/BMBRep.2018.51.7.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenen MM, Brown D, Brennan LD, Renger R, Khoo H, Carlson CR, Huang B, Grill SW, Narlikar GJ, Redding S. 2021. HP1 proteins compact DNA into mechanically and positionally stable phase separated domains. eLife 10: e64563. 10.7554/eLife.64563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna N, Hu Y, Belmont AS. 2014. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr Biol 24: 1138–1144. 10.1016/j.cub.2014.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichuk TC, Carrasco-Lopez C, Avalos JL. 2021. Lights up on organelles: optogenetic tools to control subcellular structure and organization. Wiley Interdiscip Rev Syst Biol Med 13: e1500. [DOI] [PubMed] [Google Scholar]
- Kim J, Han KY, Khanna N, Ha T, Belmont AS. 2019. Nuclear speckle fusion via long-range directional motion regulates speckle morphology after transcriptional inhibition. J Cell Sci 132: jcs226563. 10.1242/jcs.226563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Venkata NC, Hernandez Gonzalez GA, Khanna N, Belmont AS. 2020. Gene expression amplification by nuclear speckle association. J Cell Biol 219: e201904046. 10.1083/jcb.201904046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. 2013. Single-cell dynamics of genome-nuclear lamina interactions. Cell 153: 178–192. 10.1016/j.cell.2013.02.028 [DOI] [PubMed] [Google Scholar]
- Kireev I, Lakonishok M, Liu W, Joshi VN, Powell R, Belmont AS. 2008. In vivo immunogold labeling confirms large-scale chromatin folding motifs. Nat Methods 5: 311–313. 10.1038/nmeth.1196 [DOI] [PubMed] [Google Scholar]
- Kölbl AC, Weigl D, Mulaw M, Thormeyer T, Bohlander SK, Cremer T, Dietzel S. 2012. The radial nuclear positioning of genes correlates with features of megabase-sized chromatin domains. Chromosome Res 20: 735–752. 10.1007/s10577-012-9309-9 [DOI] [PubMed] [Google Scholar]
- Korenberg JR, Rykowski MC. 1988. Human genome organization: alu, lines, and the molecular structure of metaphase chromosome bands. Cell 53: 391–400. 10.1016/0092-8674(88)90159-6 [DOI] [PubMed] [Google Scholar]
- Kurihara M, Kato K, Sanbo C, Shigenobu S, Ohkawa Y, Fuchigami T, Miyanari Y. 2020. Genomic profiling by ALaP-seq reveals transcriptional regulation by PML bodies through DNMT3A exclusion. Mol Cell 78: 493–505.e8. 10.1016/j.molcel.2020.04.004 [DOI] [PubMed] [Google Scholar]
- Lafarga M, Casafont I, Bengoechea R, Tapia O, Berciano MT. 2009. Cajal's contribution to the knowledge of the neuronal cell nucleus. Chromosoma 118: 437–443. 10.1007/s00412-009-0212-x [DOI] [PubMed] [Google Scholar]
- Lafarga M, Tapia O, Romero AM, Berciano MT. 2017. Cajal bodies in neurons. RNA Biol 14: 712–725. 10.1080/15476286.2016.1231360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. 2021. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol 22: 165–182. 10.1038/s41580-020-0272-6 [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, de Thé H. 2010. PML nuclear bodies. Cold Spring Harb Perspect Biol 2: a000661. 10.1101/cshperspect.a000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, de Thé H. 2018. PML nuclear bodies: from architecture to function. Curr Opin Cell Biol 52: 154–161. 10.1016/j.ceb.2018.03.011 [DOI] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. 2017. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547: 236–240. 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latonen L. 2019. Phase-to-phase with nucleoli—stress responses, protein aggregation and novel roles of RNA. Front Cell Neurosci 13: 151. 10.3389/fncel.2019.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître C, Grabarz A, Tsouroula K, Andronov L, Furst A, Pankotai T, Heyer V, Rogier M, Attwood KM, Kessler P, et al. 2014. Nuclear position dictates DNA repair pathway choice. Genes Dev 28: 2450–2463. 10.1101/gad.248369.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Roy K, Katyal S, Sun X, Bléoo S, Godbout R. 2006. Dynamic nature of cleavage bodies and their spatial relationship to DDX1 bodies, cajal bodies, and gems. Mol Biol Cell 17: 1126–1140. 10.1091/mbc.e05-08-0768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DH, Hoelz A. 2019. The structure of the nuclear pore complex (an update). Annu Rev Biochem 88: 725–783. 10.1146/annurev-biochem-062917-011901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JY, Chang L, Li T, Wang T, Yin Y, Zhan G, Han X, Zhang K, Tao Y, Percharde M, et al. 2021. Homotypic clustering of L1 and B1/Alu repeats compartmentalizes the 3D genome. Cell Res 31: 613–630. 10.1038/s41422-020-00466-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon AS, Peeples WB, Rosen MK. 2021. A framework for understanding the functions of biomolecular condensates across scales. Nat Rev Mol Cell Biol 22: 215–235. 10.1038/s41580-020-00303-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Gao Z, Wu J, Zhong B, Xie Y, Huang W, Lin Y. 2021. Co-condensation between transcription factor and coactivator p300 modulates transcriptional bursting kinetics. Mol Cell 81: 1682–1697 e1687. 10.1016/j.molcel.2021.01.031 [DOI] [PubMed] [Google Scholar]
- Machyna M, Heyn P, Neugebauer KM. 2013. Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA 4: 17–34. 10.1002/wrna.1139 [DOI] [PubMed] [Google Scholar]
- Maeshima K, Tamura S, Hansen JC, Itoh Y. 2020. Fluid-like chromatin: toward understanding the real chromatin organization present in the cell. Curr Opin Cell Biol 64: 77–89. 10.1016/j.ceb.2020.02.016 [DOI] [PubMed] [Google Scholar]
- Manuelidis L. 1984. Active nucleolus organizers are precisely positioned in adult central nervous system cells but not in neuroectodermal tumor cells. J Neuropathol Exp Neurol 43: 225–241. 10.1097/00005072-198405000-00002 [DOI] [PubMed] [Google Scholar]
- Mateo LJ, Murphy SE, Hafner A, Cinquini IS, Walker CA, Boettiger AN. 2019. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568: 49–54. 10.1038/s41586-019-1035-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluggage F, Fox AH. 2021. Paraspeckle nuclear condensates: global sensors of cell stress? Bioessays 43: 2000245. 10.1002/bies.202000245 [DOI] [PubMed] [Google Scholar]
- McSwiggen DT, Mir M, Darzacq X, Tjian R. 2019. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev 33: 1619–1634. 10.1101/gad.331520.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehus AA, Anderson RH, Roux KJ. 2016. BioID identification of lamin-associated proteins. Methods Enzymol 569: 3–22. 10.1016/bs.mie.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT. 2017. RNA modification in cajal bodies. RNA Biol 14: 693–700. 10.1080/15476286.2016.1249091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. 2013. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res 23: 270–280. 10.1101/gr.141028.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron E, Oldenkamp R, Brown JM, Pinto DMS, Xu CS, Faria AR, Shaban HA, Rhodes JDP, Innocent C, de Ornellas S, et al. 2020. Chromatin arranges in chains of mesoscale domains with nanoscale functional topography independent of cohesin. Sci Adv 6: eaba8811. 10.1126/sciadv.aba8811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Miroshnikova YA, Wickström SA. 2021. Mechanical forces in nuclear organization. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. 2020. The self-organizing genome: principles of genome architecture and function. Cell 183: 28–45. 10.1016/j.cell.2020.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL. 1997. Protein phosphorylation and the nuclear organization of pre-mRNA splicing. Trends Cell Biol 7: 135–138. 10.1016/S0962-8924(96)20043-1 [DOI] [PubMed] [Google Scholar]
- Miura H, Takahashi S, Poonperm R, Tanigawa A, Takebayashi SI, Hiratani I. 2019. Single-cell DNA replication profiling identifies spatiotemporal developmental dynamics of chromosome organization. Nat Genet 51: 1356–1368. 10.1038/s41588-019-0474-z [DOI] [PubMed] [Google Scholar]
- Monneron A, Bernhard W. 1969. Fine structural organization of the interphase nucleus in some mammalian cells. J Ultrastruct Res 27: 266–288. 10.1016/S0022-5320(69)80017-1 [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Yamazaki T, Hirose T. 2018. Molecular dissection of nuclear paraspeckles: towards understanding the emerging world of the RNP milieu. Open Biol 8: 180150. 10.1098/rsob.180150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Péterfia B, Solovei I, Cremer T, Dopazo J, Längst G. 2010. Initial genomics of the human nucleolus. PLoS Genet 6: e1000889. 10.1371/journal.pgen.1000889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HQ, Chattoraj S, Castillo D, Nguyen SC, Nir G, Lioutas A, Hershberg EA, Martins NMC, Reginato PL, Hannan M, et al. 2020. 3D mapping and accelerated super-resolution imaging of the human genome using in situ sequencing. Nat Methods 17: 822–832. 10.1038/s41592-020-0890-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki T, Imai R, Tanbo M, Nagashima R, Tamura S, Tani T, Joti Y, Tomita M, Hibino K, Kanemaki MT, et al. 2017. Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol Cell 67: 282–293.e7. 10.1016/j.molcel.2017.06.018 [DOI] [PubMed] [Google Scholar]
- Ochs RL, Lischwe MA, Shen E, Carroll RE, Busch H. 1985. Nucleologenesis: composition and fate of prenucleolar bodies. Chromosoma 92: 330–336. 10.1007/BF00327463 [DOI] [PubMed] [Google Scholar]
- Ogg SC, Lamond AI. 2002. Cajal bodies and coilin—moving towards function. J Cell Biol 159: 17–21. 10.1083/jcb.200206111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe RT, Henderson SC, Spector DL. 1992. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J Cell Biol 116: 1095–1110. 10.1083/jcb.116.5.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins AL, Moyer BA, Kim SH, Allison DP. 1989. Synthesis of a more stable osmium ammine electron-dense DNA stain. J Histochem Cytochem 37: 395–398. 10.1177/37.3.2465337 [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36: 1065–1071. 10.1038/ng1423 [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. 2007. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol 5: e192. 10.1371/journal.pbio.0050192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanagic-Myers S, Foisner R. 2019. The structural and gene expression hypotheses in laminopathic diseases—not so different after all. Mol Biol Cell 30: 1786–1790. 10.1091/mbc.E18-10-0672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya-Jones A, Markaki Y, Serizay J, Chitiashvili T, Mancia Leon WR, Damianov A, Chronis C, Papp B, Chen CK, McKee R, et al. 2020. A protein assembly mediates Xist localization and gene silencing. Nature 587: 145–151. 10.1038/s41586-020-2703-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarge E. 1979. Emil heitz and the concept of heterochromatin: longitudinal chromosome differentiation was recognized fifty years ago. Am J Hum Genet 31: 106–115. [PMC free article] [PubMed] [Google Scholar]
- *.Pawar S, Kutay U. 2021. The diverse cellular functions of inner nuclear membrane proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a040477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. 2011. The nucleolus. Cold Spring Harb Perspect Biol 3: a000638. 10.1101/cshperspect.a000638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzo M, Montanaro L, Treré D, Derenzini M. 2019. The ribosome biogenesis—cancer connection. Cells 8: 55. 10.3390/cells8010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, van Steensel B. 2010. Role of the nuclear lamina in genome organization and gene expression. Cold Spring Harb Symp Quant Biol 75: 517–524. 10.1101/sqb.2010.75.014 [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613. 10.1016/j.molcel.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V, Li HB. 2012. A view of nuclear Polycomb bodies. Curr Opin Genet Dev 22: 101–109. 10.1016/j.gde.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani G, Baron B. 2019. Nuclear paraspeckles function in mediating gene regulatory and apoptotic pathways. Noncoding RNA Res 4: 128–134. 10.1016/j.ncrna.2019.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Swedlow JR, Lamond AI. 2000. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol 151: 1561–1574. 10.1083/jcb.151.7.1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock C, Daily K, Nguyen VT, Wang C, Lewandowska MA, Bensaude O, Huang S. 2011. Characterization of MRP RNA-protein interactions within the perinucleolar compartment. Mol Biol Cell 22: 858–866. 10.1091/mbc.e10-09-0768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. 2005. Regulating gene expression through RNA nuclear retention. Cell 123: 249–263. 10.1016/j.cell.2005.08.033 [DOI] [PubMed] [Google Scholar]
- Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, et al. 2018. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174: 744–757.e24. 10.1016/j.cell.2018.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz SA, Bhat P, Ollikainen N, Jachowicz JW, Banerjee AK, Chovanec P, Blanco MR, Chow A, Markaki Y, Plath K, et al. 2020. RNA promotes the formation of spatial compartments in the nucleus. bioRxiv 10.1101/2020.08.25.267435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl C. 1885. Uber zelltheilung. Morphol Jahrbuch 10: 214–330. [Google Scholar]
- Randise-Hinchliff C, Brickner JH. 2018. Nuclear pore complex in genome organization and gene expression in yeast. In Nuclear pore complexes in genome organization, function and maintenance (ed. D'Angelo M), pp. 87–109. Springer, Berlin. [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raška I, Andrade LE, Ochs RL, Chan EK, Chang CM, Roos G, Tan EM. 1991. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res 195: 27–37. 10.1016/0014-4827(91)90496-H [DOI] [PubMed] [Google Scholar]
- Raška I, Shaw PJ, Cmarko D. 2006. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol 18: 325–334. 10.1016/j.ceb.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Rego A, Sinclair PB, Tao W, Kireev I, Belmont AS. 2008. The facultative heterochromatin of the inactive X chromosome has a distinctive condensed ultrastructure. J Cell Sci 121: 1119–1127. 10.1242/jcs.026104 [DOI] [PubMed] [Google Scholar]
- Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. 2013. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339: 1328–1331. 10.1126/science.1230593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson MI, de Las Heras JI, Czapiewski R, Lê Thành P, Booth DG, Kelly DA, Webb S, Kerr ARW, Schirmer EC. 2016. Tissue-specific gene repositioning by muscle nuclear membrane proteins enhances repression of critical developmental genes during myogenesis. Mol Cell 62: 834–847. 10.1016/j.molcel.2016.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770. 10.1101/gr.099655.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. 2018. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361: eaar3958. 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL, Griffin PR, Gross JD, Narlikar GJ. 2019. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575: 390–394. 10.1038/s41586-019-1669-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino TM, Gébrane-Younés J, De Mey J, Sibarita JB, Hernandez-Verdun D. 2001. Nucleolar assembly of the rRNA processing machinery in living cells. J Cell Biol 153: 1097–1110. 10.1083/jcb.153.5.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer IA, Sturgill D, Sung MH, Hager GL, Dundr M. 2016. Cajal body function in genome organization and transcriptome diversity. Bioessays 38: 1197–1208. 10.1002/bies.201600144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep R, Brinkman EK, Leemans C, Vergara X, van der Weide RH, Morris B, van Schaik T, Manzo SG, Peric-Hupkes D, van den Berg J, et al. 2021. Impact of chromatin context on Cas9-induced DNA double-strand break repair pathway balance. Mol Cell 81: 2216–2230.e10. 10.1016/j.molcel.2021.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, Görlich D. 2016. Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem Sci 41: 46–61. 10.1016/j.tibs.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Schöfer C, Weipoltshammer K. 2018. Nucleolus and chromatin. Histochem Cell Biol 150: 209–225. 10.1007/s00418-018-1696-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M. 2002. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet 32: 438–442. 10.1038/ng1005 [DOI] [PubMed] [Google Scholar]
- Seeber A, Gasser SM. 2017. Chromatin organization and dynamics in double-strand break repair. Curr Opin Genet Dev 43: 9–16. 10.1016/j.gde.2016.10.005 [DOI] [PubMed] [Google Scholar]
- Shiels C, Islam SA, Vatcheva R, Sasieni P, Sternberg MJ, Freemont PS, Sheer D. 2001. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J Cell Sci 114: 3705–3716. 10.1242/jcs.114.20.3705 [DOI] [PubMed] [Google Scholar]
- Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD. 2015. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell 26: 4075–4086. 10.1091/mbc.E15-07-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. 2017. Liquid phase condensation in cell physiology and disease. Science 357: eaaf4382. 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- Shopland LS, Lynch CR, Peterson KA, Thornton K, Kepper N, Hase J, Stein S, Vincent S, Molloy KR, Kreth G, et al. 2006. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol 174: 27–38. 10.1083/jcb.200603083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair P, Bian Q, Plutz M, Heard E, Belmont AS. 2010. Dynamic plasticity of large-scale chromatin structure revealed by self-assembly of engineered chromosome regions. J Cell Biol 190: 761–776. 10.1083/jcb.200912167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaer RJ, Whytock S. 1976. The fixation of nuclei and chromosomes. J Cell Sci 20: 221–231. 10.1242/jcs.20.1.221 [DOI] [PubMed] [Google Scholar]
- Skaer RJ, Whytock S. 1977. Chromatin-like artifacts from nuclear sap. J Cell Sci 26: 301–310. 10.1242/jcs.26.1.301 [DOI] [PubMed] [Google Scholar]
- Smetana K, Busch H. 1964. Studies on the ultrastructure of the nucleoli of the Walker tumor and rat liver. Cancer Res 24: 537–557. [PubMed] [Google Scholar]
- Solovei I, Schermelleh L, Düring K, Engelhardt A, Stein S, Cremer C, Cremer T. 2004. Differences in centromere positioning of cycling and postmitotic human cell types. Chromosoma 112: 410–423. 10.1007/s00412-004-0287-3 [DOI] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctôt C, Kösem S, Peichl L, Cremer T, Guck J, Joffe B. 2009. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137: 356–368. 10.1016/j.cell.2009.01.052 [DOI] [PubMed] [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. 2013. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152: 584–598. 10.1016/j.cell.2013.01.009 [DOI] [PubMed] [Google Scholar]
- Spector DL. 2006. Snapshot: cellular bodies. Cell 127: 1071.e1–1071.e2. 10.1016/j.cell.2006.11.026 [DOI] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. 2011. Nuclear speckles. Cold Spring Harb Perspect Biol 3: a000646. 10.1101/cshperspect.a000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden H, Tolsma TO, Sharma A, Underhill DA, Hansen JC, Hendzel MJ. 2020. Condensed chromatin behaves like a solid on the mesoscale in vitro and in living cells. Cell 183: 1772–1784 e1713. 10.1016/j.cell.2020.11.027 [DOI] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. 2017. Phase separation drives heterochromatin domain formation. Nature 547: 241–245. 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka M, Oates AC, Neugebauer KM. 2010. Dynamic control of cajal body number during zebrafish embryogenesis. Nucleus 1: 96–108. 10.4161/nucl.1.1.10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su JH, Zheng P, Kinrot SS, Bintu B, Zhuang X. 2020. Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell 182: 1641–1659.e26. 10.1016/j.cell.2020.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Durrin LK, Krontiris TG. 2003. Specific interaction of PML bodies with the TP53 locus in Jurkat interphase nuclei. Genomics 82: 250–252. 10.1016/S0888-7543(03)00075-2 [DOI] [PubMed] [Google Scholar]
- Sun J, Chen J, Mohagheghian E, Wang N. 2020. Force-induced gene up-regulation does not follow the weak power law but depends on H3K9 demethylation. Sci Adv 6: eaay9095. 10.1126/sciadv.aay9095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift H. 1959. Studies on nuclear fine structure. Brookhaven Symp Biol 12: 134–152. [PubMed] [Google Scholar]
- Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, Singh R, Khanna N, Belmont AS, Wang N. 2016. Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater 15: 1287–1296. 10.1038/nmat4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y, Yun J, Zheng S, Ollikainen N, Pierson N, White J, Shah S, Thomassie J, Suo S, Eng CL, et al. 2021. Integrated spatial genomics reveals global architecture of single nuclei. Nature 590: 344–350. 10.1038/s41586-020-03126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Meaburn KJ, Misteli T. 2008. The meaning of gene positioning. Cell 135: 9–13. 10.1016/j.cell.2008.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testillano PS, Sanchez-Pina MA, Olmedilla A, Ollacarizqueta MA, Tandler CJ, Risueño MC. 1991. A specific ultrastructural method to reveal DNA: the NAMA-Ur. J Histochem Cytochem 39: 1427–1438. 10.1177/39.10.1719069 [DOI] [PubMed] [Google Scholar]
- Thiry M. 1995. Behavior of interchromatin granules during the cell cycle. Eur J Cell Biol 68: 14–24. [PubMed] [Google Scholar]
- Tiku V, Antebi A. 2018. Nucleolar function in lifespan regulation. Trends Cell Biol 28: 662–672. 10.1016/j.tcb.2018.03.007 [DOI] [PubMed] [Google Scholar]
- Tran JR, Adam SA, Goldman RD, Zheng Y. 2021. Dynamic nuclear lamina-chromatin interactions during G1 progression. bioRxiv 10.1101/2021.01.03.425156 [DOI] [Google Scholar]
- Tripathi K, Parnaik VK. 2008. Differential dynamics of splicing factor SC35 during the cell cycle. J Biosci 33: 345–354. 10.1007/s12038-008-0054-3 [DOI] [PubMed] [Google Scholar]
- Uchino S, Ito Y, Sato Y, Handa T, Ohkawa Y, Tokunaga M, Kimura H. 2021. Visualizing transcription sites in living cells using a genetically encoded probe specific for the elongating form of RNA polymerase II. bioRxiv 10.1101/2021.04.27.441582 [DOI] [Google Scholar]
- van Koningsbruggen S, Gierliński M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. 2010. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell 21: 3735–3748. 10.1091/mbc.e10-06-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik T, Vos M, Peric-Hupkes D, Hn Celie P, van Steensel B. 2020. Cell cycle dynamics of lamina-associated DNA. EMBO Rep 21: e50636. 10.15252/embr.202050636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Belmont AS. 2017. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169: 780–791. 10.1016/j.cell.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Henikoff S. 2000. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol 18: 424–428. 10.1038/74487 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Henikoff S. 2001. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet 27: 304–308. 10.1038/85871 [DOI] [PubMed] [Google Scholar]
- Vertii A, Ou J, Yu J, Yan A, Pagès H, Liu H, Zhu LJ, Kaufman PD. 2019. Two contrasting classes of nucleolus-associated domains in mouse fibroblast heterochromatin. Genome Res 29: 1235–1249. 10.1101/gr.247072.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourc'h C, Taruscio D, Boyle AL, Ward DC. 1993. Cell cycle-dependent distribution of telomeres, centromeres, and chromosome-specific subsatellite domains in the interphase nucleus of mouse lymphocytes. Exp Cell Res 205: 142–151. 10.1006/excr.1993.1068 [DOI] [PubMed] [Google Scholar]
- Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. 2003. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol 160: 685–697. 10.1083/jcb.200211103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shiels C, Sasieni P, Wu PJ, Islam SA, Freemont PS, Sheer D. 2004. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J Cell Biol 164: 515–526. 10.1083/jcb.200305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Kolhe JA, Gioacchini N, Baade I, Brieher WM, Peterson CL, Freeman BC. 2020. Mechanism of long-range chromosome motion triggered by gene activation. Dev Cell 52: 309–320.e5. 10.1016/j.devcel.2019.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Zhang R, van Schaik T, Zhang L, Sasaki T, Peric-Hupkes D, Chen Y, Gilbert DM, van Steensel B, et al. 2021. SPIN reveals genome-wide landscape of nuclear compartmentalization. Genome Biol 22: 36. 10.1186/s13059-020-02253-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ML. 1959. Further observations on the nuclear envelope of the animal cell. J Biophys Biochem Cytol 6: 147–156. 10.1083/jcb.6.2.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks SE, Metge BJ, Samant RS. 2019. The nucleolus: a central response hub for the stressors that drive cancer progression. Cell Mol Life Sci 76: 4511–4524. 10.1007/s00018-019-03231-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei MT, Chang YC, Shimobayashi SF, Shin Y, Strom AR, Brangwynne CP. 2020. Nucleated transcriptional condensates amplify gene expression. Nat Cell Biol 22: 1187–1196. 10.1038/s41556-020-00578-6 [DOI] [PubMed] [Google Scholar]
- White EJ, Emanuelsson O, Scalzo D, Royce T, Kosak S, Oakeley EJ, Weissman S, Gerstein M, Groudine M, Snyder M, et al. 2004. DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. Proc Natl Acad Sci 101: 17771–17776. 10.1073/pnas.0408170101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KL, Foisner R. 2010. Lamin-binding proteins. Cold Spring Harb Perspect Biol 2: a000554. 10.1101/cshperspect.a000554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong X, Cutler JA, Hoskins VE, Gordon M, Madugundu AK, Pandey A, Reddy KL. 2021. Mapping the micro-proteome of the nuclear lamina and lamina-associated domains. Life Sci Alliance 4: e202000774. 10.26508/lsa.202000774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Singh PB, Gilbert DM. 2006. Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J Cell Biol 174: 185–194. 10.1083/jcb.200601113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie SQ, Pombo A. 2006. Distribution of different phosphorylated forms of RNA polymerase II in relation to cajal and PML bodies in human cells: an ultrastructural study. Histochem Cell Biol 125: 21–31. 10.1007/s00418-005-0064-2 [DOI] [PubMed] [Google Scholar]
- Xie W, Chojnowski A, Boudier T, Lim JS, Ahmed S, Ser Z, Stewart C, Burke B. 2016. A-type lamins form distinct filamentous networks with differential nuclear pore complex associations. Curr Biol 26: 2651–2658. 10.1016/j.cub.2016.07.049 [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Yamamoto T, Yoshino H, Souquere S, Nakagawa S, Pierron G, Hirose T. 2021. Paraspeckles are constructed as block copolymer micelles. EMBO J 40: e107270. 10.15252/embj.2020107270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z. 2009. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol Cell 36: 267–278. 10.1016/j.molcel.2009.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao RW, Xu G, Wang Y, Shan L, Luan PF, Wang Y, Wu M, Yang LZ, Xing YH, Yang L, et al. 2019. Nascent pre-rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus. Mol Cell 76: 767–783.e11. 10.1016/j.molcel.2019.08.014 [DOI] [PubMed] [Google Scholar]
- Yasuzumi G, Sawada T, Sugihara R, Kiriyama M, Sugioka M. 1958. Electron microscope researches on the ultrastructure of nucleoli in animal tissues. Z Zellforsch Mikrosk Anat 48: 10–23. 10.1007/BF00496710 [DOI] [PubMed] [Google Scholar]
- Zatsepina OV, Poliakov V, Chentsov Iu S. 1983. Electron microscopic study of the chromonema and chromomeres in mitotic and interphase chromosomes. Tsitologiia 25: 123–129. [PubMed] [Google Scholar]
- Zatsepina OV, Dudnic OA, Todorov IT, Thiry M, Spring H, Trendelenburg MF. 1997. Experimental induction of prenucleolar bodies (PNBs) in interphase cells: interphase PNBs show similar characteristics as those typically observed at telophase of mitosis in untreated cells. Chromosoma 105: 418–430. 10.1007/BF02510478 [DOI] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT. 2007. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129: 693–706. 10.1016/j.cell.2007.03.036 [DOI] [PubMed] [Google Scholar]
- Zhang H, Emerson DJ, Gilgenast TG, Titus KR, Lan Y, Huang P, Zhang D, Wang H, Keller CA, Giardine B, et al. 2019. Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature 576: 158–162. 10.1038/s41586-019-1778-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhao R, Tones J, Liu M, Dilley RL, Chenoweth DM, Greenberg RA, Lampson MA. 2020a. Nuclear body phase separation drives telomere clustering in ALT cancer cells. Mol Biol Cell 31: 2048–2056. 10.1091/mbc.E19-10-0589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang Y, Chen Y, Gholamalamdari O, Wang Y, Ma J, Belmont AS. 2020b. TSA-seq reveals a largely conserved genome organization relative to nuclear speckles with small position changes tightly correlated with gene expression changes. Genome Res 31: 151–264. 10.1101/gr.266239.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao PA, Rivera-Mulia JC, Gilbert DM. 2017. Replication domains: genome compartmentalization into functional replication units. In DNA replication: from old principles to new discoveries (ed. Masai H, Foiani M), pp. 229–257. Springer, Singapore. [DOI] [PubMed] [Google Scholar]
- Zhao B, Chaturvedi P, Zimmerman DL, Belmont AS. 2020. Efficient and reproducible multigene expression after single-step transfection using improved BAC transgenesis and engineering toolkit. ACS Synth Biol 9: 1100–1116. 10.1021/acssynbio.9b00457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Gerrard DL, Wang J, Li T, Yang Y, Fritz AJ, Rajendran M, Fu X, Stein G, Schiff R, et al. 2019. Temporal dynamic reorganization of 3D chromatin architecture in hormone-induced breast cancer and endocrine resistance. Nat Commun 10: 1522. 10.1038/s41467-019-09320-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Richardson TM, Wacheul L, Wei MT, Feric M, Whitney G, Lafontaine DLJ, Brangwynne CP. 2019. Controlling the material properties and rRNA processing function of the nucleolus using light. Proc Natl Acad Sci 116: 17330–17335. 10.1073/pnas.1903870116 [DOI] [PMC free article] [PubMed] [Google Scholar]