Abstract
Dramatic nuclear reorganization occurs during early development to convert terminally differentiated gametes to a totipotent zygote, which then gives rise to an embryo. Aberrant epigenome resetting severely impairs embryo development and even leads to lethality. How the epigenomes are inherited, reprogrammed, and reestablished in this critical developmental period has gradually been unveiled through the rapid development of technologies including ultrasensitive chromatin analysis methods. In this review, we summarize the latest findings on epigenetic reprogramming in gametogenesis and embryogenesis, and how it contributes to gamete maturation and parental-to-zygotic transition. Finally, we highlight the key questions that remain to be answered to fully understand chromatin regulation and nuclear reprogramming in early development.
Gametogenesis and embryogenesis are essential to give rise to a new life and to repopulate species that use sexual reproduction. Sperm and oocytes arise from primordial germ cells (PGCs) through spermatogenesis and oogenesis, respectively. After fertilization, the two terminally differentiated gametes fuse and form a totipotent zygote (Zhou and Dean 2015). During embryo cleavage, maternal proteins and mRNAs are gradually degraded, while the zygotic genome is transcriptionally activated, collectively leading to the maternal-to-zygotic transition (MZT) (Fig. 1; Schultz 2002; Walser and Lipshitz 2011). Zygotic genome activation ([ZGA], also known as embryonic genome activation) often takes place in two waves (for review, see Lee et al. 2014). During minor ZGA in mouse embryos, the transcription of a small subset of genes is accompanied by promiscuous, genome-wide transcription, which occurs from the S phase of a one-cell embryo until the onset of “major ZGA” in mid- to late two-cell embryos, when thousands of genes are activated (Bouniol et al. 1995; Nothias et al. 1996; Aoki et al. 1997; Hamatani et al. 2004; Wang et al. 2004; Zeng et al. 2004; Abe et al. 2015). Concomitantly, RNA polymerase II (Pol II) undergoes dynamic chromatin engagement during the minor-to-major ZGA transition (Bellier et al. 1997; Liu et al. 2020). In human, embryonic transcripts are first observed from the two-cell to the four-cell stage, and the major ZGA occurs around the eight-cell stage (Tesarik et al. 1987; Braude et al. 1988; Dobson et al. 2004). The totipotent zygote then develops to blastocyst containing the pluripotent inner cell mass (ICM), that subsequently produces germ layers during gastrulation and, ultimately, the whole organism (Chazaud and Yamanaka 2016). During this process, epigenetic regulation plays critical roles to ensure precise spatiotemporal gene regulation (Bird 2002; Kouzarides 2007) and successful early development (Burton and Torres-Padilla 2014; Eckersley-Maslin et al. 2018; Xu and Xie 2018). Aberrant epigenetic reprogramming leads to severe developmental defects and diseases (Greenberg and Bourc'his 2019; Jambhekar et al. 2019). However, it has long been enigmatic how the epigenomes are globally reprogrammed in mammalian early development at the molecular level. Furthermore, the role of the epigenome in early development remains elusive, with mechanistic studies often hindered by the limited amount of biological material that can be obtained from embryos. Recent advances in low-input or single-cell chromatin analysis begin to illuminate specific aspects of chromatin regulation during gametogenesis and embryogenesis. Here, we summarize the dynamics of the epigenomes in gametogenesis and early development, their functions and the possible mechanisms underlying the drastic changes. Last, we highlight crucial questions that remain unanswered.
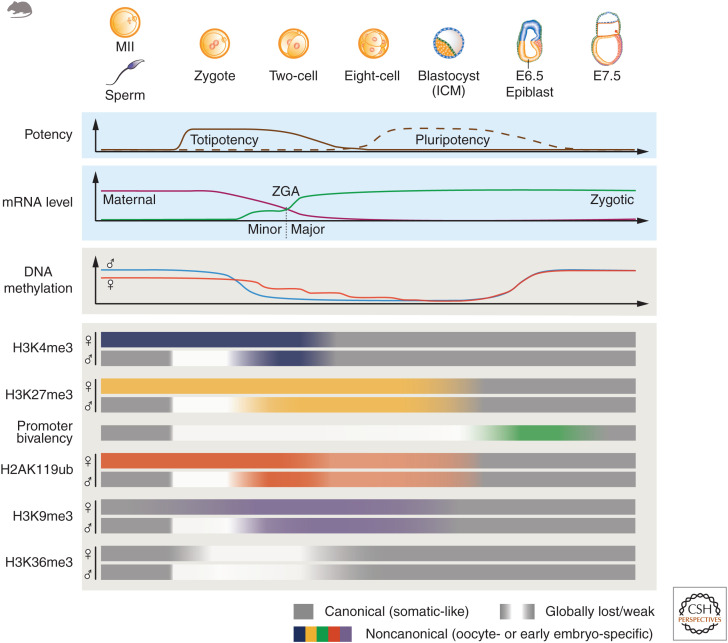
Figure 1.
Transcription and epigenetic dynamics from gametes to embryos in mouse. After fertilization, the terminally differentiated gametes are converted into a totipotent zygote. Inner cell mass (ICM) cells from blastocysts are pluripotent and can give rise to all embryonic tissues. The accumulated maternal mRNAs undergo degradation after fertilization, while minor and major zygotic genome activation (ZGA) emerge around the one-cell and two-cell stages, respectively. Parental genomes undergo global DNA demethylation starting from the zygote stage and the reestablishment of embryonic DNA methylome occurs in postimplantation embryos. For histone modifications, sperm largely display canonical distributions (similar to somatic cells), while oocytes and early embryos often acquire noncanonical patterns. H3K4me3 and H3K27me3 form noncanonical broad domains in oocytes, which are transiently inherited to the maternal allele of embryos (except for promoter H3K27me3). The sperm H3K4me3 and H3K27me3 quickly attenuate after fertilization. Weak and extremely large domains of H3K4me3 (at gene-rich regions) and H3K27me3 (at gene deserts) are de novo established on the paternal genome in zygotes. Concomitant with ZGA, both maternal and paternal H3K4me3 are reprogrammed to the canonical patterns, while the distal H3K27me3 domains persist until blastocyst, before being transformed into a canonical state in postimplantation embryos. Bivalency at developmental genes exists in mature gametes but is missing in preimplantation embryos. In E6.5 epiblast, H3K27me3 and unusually strong H3K4me3 form “super bivalency” (green), which is subsequently attenuated at E7.5. In mouse sperm, H2AK119ub resides at the promoters of canonical Polycomb targets. In mouse oocytes, it forms broad distal domains in H3K27me3 marked PMDs, and also occurs at active promoters associated with H3K4me3. Upon fertilization, H2AK119ub undergoes drastic changes, with global resetting of sperm signals but brief inheritance of oocyte H2AK119ub in one-cell embryos. H2AK119ub then rapidly remodels (mainly from the paternal allele but also partially from the maternal allele) and adopts similar patterns between the two alleles from the two-cell stage. After implantation, H2AK119ub and H3K27me3 become colocalized again at developmental gene promoters. H3K9me3 marks both a subset of promoters and LTRs in gametes. After fertilization, H3K9me3 is globally reset in an allele-specific manner, and the asymmetrical distribution of H3K9me3 persists to blastocyst. H3K36me3 enriches at actively expressed gene bodies in gametes. After fertilization, H3K36me3 is globally removed and is then reestablished upon ZGA. The enrichment of epigenetic modification is indicated by the widths and shades of the bars.
EPIGENOME REPROGRAMMING DURING EARLY DEVELOPMENT
DNA Methylation
5-methylcytosine (5mC) is a prevalent epigenetic mark in the mammalian genome, existing predominantly in CpG dinucleotides. It is essential for development by regulating genomic imprinting, transposon silencing, X chromosome inactivation (XCI), and gene repression (Bird 2002; Smith and Meissner 2013). In mammals, DNA methylation is established by de novo methyltransferases DNMT3A and DNMT3B, and maintained by DNMT1 through cell division, while active DNA demethylation is facilitated by the ten-eleven translocation (TET) methylcytosine dioxygenase family (Kohli and Zhang 2013; Pastor et al. 2013; Li and Zhang 2014). During germline development, PGCs first undergo extensive genome-wide DNA demethylation (Guibert et al. 2012; Seisenberger et al. 2012; Vincent et al. 2013), followed by sex-specific de novo DNA methylation (Fig. 1; Stewart et al. 2016). The proper establishment of DNA methylome is essential for spermatogenesis. In mouse, loss of DNMT3A or DNMT3L (a DNMT3A/B cofactor) causes catastrophic meiotic arrest in spermatogonia (Walsh et al. 1998; Bourc'his and Bestor 2004). Similarly, the mutation of DNMT3C, a male fetal germ cell-specific DNA methyltransferase, leads to derepression of young transposable elements and infertility (Barau et al. 2016). DNA methylation, however, appears to be dispensable for mouse oogenesis, as Dnmt3a or Dnmt3l knockout does not cause apparent oocyte developmental defects (Smallwood et al. 2011; Kobayashi et al. 2012; Shirane et al. 2013), although oocyte methylation is critical for early embryogenesis. This is perhaps not surprising since the de novo DNA methylation only occurs in late-stage oocytes (Stewart et al. 2016; Sendžikaite and Kelsey 2019; Yan et al. 2021). Notably, the mouse oocyte has a distinct DNA methylome in which only transcribed regions are highly methylated (Kobayashi et al. 2012; Shirane et al. 2013). This is attributed to the notion that Pol II can recruit SETD2 for H3K36me3 deposition, which further leads to the recruitment of DNMT3A (Dhayalan et al. 2010; Baubec et al. 2015; Dukatz et al. 2019). A major function of such transcription-dependent DNA methylation is to establish maternal imprints often by exploiting oocyte-specific promoters upstream of the imprinting control regions (ICRs) (Chotalia et al. 2009; Bartolomei and Ferguson-Smith 2011; Brind'Amour et al. 2018; Xu et al. 2019). Meanwhile, restricting DNA methylation to transcribed regions in mouse oocytes may also avoid undesired methylation of the paternal imprints. Intriguingly, while DNMT1 is dispensable for the establishment of the oocyte DNA methylome (Shirane et al. 2013), its activity needs to be strictly harnessed by STELLA, which sequestrates UHRF1 (a key cofactor of DNMT1) (Li et al. 2018b). Deletion of Stella results in aberrant accumulation of DNA methylation by DNMT1 genome wide, which interferes with ZGA (Li et al. 2018b). Notably, in the mouse male germ cells, NSD1 and H3K36me2 assume similar roles as SETD2 and H3K36me3 to establish the bulk DNA methylome (Shirane et al. 2020).
After fertilization, the parental genomes undergo global DNA demethylation (except for ICRs and some retrotransposons) through both active and passive mechanisms (Fig. 1; Smith et al. 2012; Guo et al. 2014; Shen et al. 2014; Wang et al. 2014). DNA methylation at ICRs controls allele-specific expression of a small subset of genes that play important roles in placenta, postnatal development, and brain functions (for review, see Tucci et al. 2019). Notably, de novo DNA methylation, although weak, also occurs amid global demethylation, and is subjected to TET-mediated DNA demethylation (Amouroux et al. 2016). During mouse somatic cell nuclear transfer (SCNT), such de novo methylation is suggested to impede ZGA and cloning efficiency (Gao et al. 2018b). In mouse, the global DNA methylation is quickly reestablished after implantation (Smith et al. 2017; Zhang et al. 2018b). The extraembryonic tissues, such as the placenta, are relatively hypomethylated compared to embryonic lineages, presumably forming epigenetic barriers (Schroeder et al. 2015; Smith et al. 2017; Zhang et al. 2018b). Similar findings were made in cultured human embryos from day 5 to day 14 (Zhou et al. 2019). Nevertheless, despite the detailed characterization, the function of postfertilization global DNA methylation resetting remains elusive. A plausible hypothesis is that it equalizes the two distinct gametic methylomes (except for ICRs). The demethylation may also be a prerequisite for the emergence of extraembryonic tissues. Another intriguing possibility is that the postfertilization global demethylation may facilitate the elongation of telomeres in early embryos, as observed in mouse embryonic stem cells (mESCs) (Gonzalo et al. 2006; Dan et al. 2017). Future studies are warranted to test these models.
HISTONE MODIFICATIONS
The posttranslational modifications of histones are fundamental epigenetic regulators that control many crucial cellular processes, including transcription, DNA repair, and DNA replication (Kouzarides 2007; Lawrence et al. 2016). Mounting evidence also shows that they play key roles in regulating spatial-temporal gene expression during gametogenesis and early embryo development.
H3K4me3
Although most histones in mature spermatozoa are replaced with protamine, the residual histones are still extensively modified (Gold et al. 2018). For example, H3K4me3, a hallmark of permissive promoters that can recruit chromatin remodelers and transcriptional machinery (Ruthenburg et al. 2007), remains preferentially enriched at CG-rich promoters in sperm as in most other cell types (Hammoud et al. 2009, Erkek et al. 2013). In growing oocytes, H3K4me3 is similarly restricted to active promoters. However, it becomes accumulated as broad domains at both promoters and distal regions in more mature oocytes (Fig. 1; Dahl et al. 2016; Liu et al. 2016b; Zhang et al. 2016; Hanna et al. 2018b). Such noncanonical H3K4me3 (ncH3K4me3) domains found in oocytes occur almost exclusively in partially methylated DNA domains (PMDs), which are essentially nontranscribed regions (Dahl et al. 2016; Zhang et al. 2016; Hanna et al. 2018b). The fully DNA-methylated regions are protected against ncH3K4me3 by DNA methylation in conjunction with H3K36me3, as SETD2 deficiency results in the loss of H3K36me3 and DNA methylation, and ectopic invasion of H3K4me3 (Hanna et al. 2018b; Xu et al. 2019). Notably, the ncH3K4me3 domains are deposited mainly by MLL2 (KMT2B) (Hanna et al. 2018b), which is also responsible for depositing H3K4me3 at poised promoters in mESCs (Hu et al. 2013; Denissov et al. 2014) and postimplantation embryos (Xiang et al. 2020) in a transcription-independent manner. Importantly, ablation of MLL2 leads to anovulation and oocyte death (Andreu-Vieyra et al. 2010). Intriguingly, either deficiency of Mll2 (Andreu-Vieyra et al. 2010) or overexpression of the H3K4me3 demethylase Kdm5b (Zhang et al. 2016) disrupts genome silencing in late-stage full-grown oocytes (FGOs). Although the underlying mechanism is currently unknown, such repressive function contrasts with that of H3K4me3 at active promoters. For example, the CXXC finger protein 1 (CFP1) is a component of the histone H3K4me3 methyltransferase SETD1A/B, which functions at constitutively active promoters (Lee and Skalnik 2005; Clouaire et al. 2012). Loss of CFP1 causes global down-regulation of H3K4me3 and transcription, as well as the failure of mouse oocyte maturation (Yu et al. 2017; Sha et al. 2018). These data are in line with the notion that different H3K4me3 methyltransferase complexes have distinct targets (Hu et al. 2017; Douillet et al. 2020).
Upon fertilization, the paternal H3K4me3 quickly diminishes (Lepikhov and Walter 2004; Lepikhov et al. 2008; Zhang et al. 2016), which may be linked to the extensive protamine-histone exchange (Burton and Torres-Padilla 2014). It is subsequently reestablished in late one-cell mouse embryos, but in a noncanonical pattern, forming weak but extremely large (up to several Mbs) domains in gene-rich regions (Zhang et al. 2016). By contrast, the maternal H3K4me3 is briefly inherited to zygotes. Both maternal and paternal H3K4me3 are then reprogrammed to the canonical patterns at the two-cell stage in a ZGA-dependent manner (Fig. 1; Dahl et al. 2016; Liu et al. 2016b; Zhang et al. 2016). This is accompanied by a decrease of global H3K4me3 in immunofluorescence, executed by two lysine demethylases KDM5A/5B that are activated during ZGA (Dahl et al. 2016). Knocking down these enzymes leads to embryonic lethality (Dahl et al. 2016). Intriguingly, ncH3K4me3 is absent in human germinal vesicle (GV) oocytes (Xia et al. 2019). Instead, de novo broad domains of H3K4me3 occur in CpG-rich, accessible regions in human pre-ZGA embryos, although its function remains unknown (Xia et al. 2019). Therefore, diverse mechanisms regulate H3K4me3 in mouse and human embryos.
H3K27me3
Polycomb repressive complex 2 (PRC2) is responsible for the deposition of the histone modification H3K27me3. Both PRC2 and H3K27me3 are evolutionarily conserved epigenetic regulators, which frequently repress key developmental genes and thus help maintain cell identity (Margueron and Reinberg 2011; Di Croce and Helin 2013). H3K27me3 is also enriched at developmental gene promoters in sperm (Hammoud et al. 2009, Brykczynska et al. 2010; Erkek et al. 2013). Similar to H3K4me3, widespread distal H3K27me3 domains are also found in PMDs in mouse oocytes (Figs. 1 and 2A; Liu et al. 2016b; Zheng et al. 2016). Despite its prevalence, oocyte H3K27me3 seems to be nonessential for oogenesis, since the oocyte-specific knockout of EED, a core component of PRC2 complex, does not cause notable oocyte developmental defects (Inoue et al. 2018; Prokopuk et al. 2018). However, knockout of Ring1a/Ring1b in oocytes, the key components of Polycomb repressive complex 1 (PRC1) that catalyzes H2AK119 monoubiquitination (H2AK119ub), leads to massive gene derepression, developmental defects in oocytes, and subsequent one-cell arrest after fertilization (Posfai et al. 2012; Du et al. 2020). These data are in agreement with the notion that PRC1 and its modification H2AK119ub are the major transcription repressors, while PRC2 and H3K27me3 facilitate such repression (Fursova et al. 2019; Blackledge et al. 2020; Tamburri et al. 2020). Upon fertilization, oocyte H3K27me3 at promoters of developmental genes is mostly lost, including at Hox gene promoters (Fig. 2A; Liu et al. 2016b; Zheng et al. 2016). The paternal H3K27me3 is quickly erased in zygotes, followed by gradual de novo formation of megabase-size broad domains of H3K27me3 in gene deserts (Zheng et al. 2016), although their functions remain unclear.
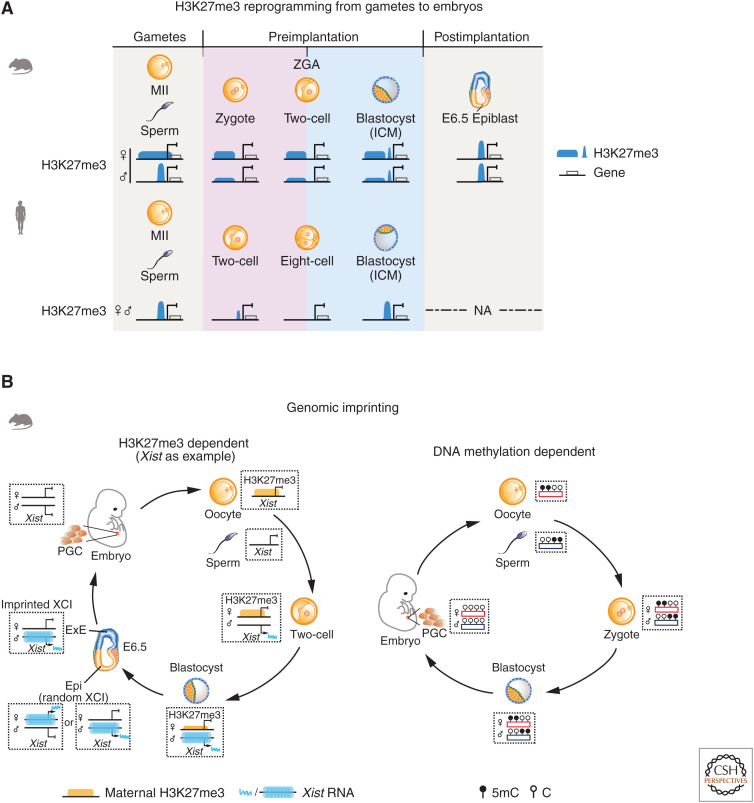
Figure 2.
The H3K27me3 reprogramming from gametes to embryos and genomic imprinting. (A) The schematics show the dynamics of H3K27me3 (blue) in gametes and early embryos in mouse and human. In mouse, broad domains of H3K27me3 from oocytes are transiently inherited to embryos until blastocyst (except for promoter H3K27me3). The sperm H3K27me3 is quickly removed after fertilization. Canonical H3K27me3 is established at promoters in blastocyst and in postimplantation embryos. By contrast, in both human oocytes and sperm, H3K27me3 exhibits canonical distributions. After fertilization, H3K27me3 is globally removed before zygotic genome activation (ZGA) and is reestablished as early as the morula stage. (NA) Data not available. (B) (Left) A schematic model showing H3K27me3-dependent imprinting (Xist as an example) in mouse. Oocyte-inherited H3K27me3 domains repress maternal Xist in mouse female embryos, leaving the paternal copy specifically expressed and initiates the imprinted X chromosome inactivation (XCI). In postimplantation embryos, the oocyte-inherited H3K27me3 domain at the Xist locus is lost. Epiblast (Epi) undergoes random XCI, while the imprinted XCI persists in the extraembryonic lineages, presumably due to additional epigenetic repression mechanisms. The inactive X chromosome is reactivated in the primordial germ cells (PGCs). (Right) A schematic model showing DNA methylation-dependent imprinting in mouse. Oocyte and sperm establish distinct DNA methylome and differentially methylated imprinting control regions (ICRs) or imprints during the gametogenesis. Imprints can survive the global DNA demethylation during preimplantation and is well maintained thereafter, directing allelic gene expression. They are, however, erased in PGCs, paving the way for imprint establishment for the next generation. (ICM) Inner cell mass.
Intriguingly, the distal domains of H3K27me3 from oocytes can persist until blastocyst stage (Zheng et al. 2016). These maternally inherited H3K27me3 domains were proven to function as an imprinting mark and dictate a small set of paternal-specific genes in early mouse embryos, similar to DNA methylation-mediated imprinting (Fig. 2B; Inoue et al. 2017a; Chen and Zhang 2020). These genes include Xist (Inoue et al. 2017b), the master regulator of XCI (Lee 2005). In female mouse embryos, the maternal Xist locus is coated with a broad H3K27me3 domain inherited from oocytes, leaving the paternal copy specifically expressed, which initiates “imprinted XCI” (Fig. 2B; Inoue et al. 2017b). Ablation of Eed in oocytes leads to random X-inactivation in female embryos (Inoue et al. 2018; Harris et al. 2019). Besides Xist, a small subset of genes with roles in placental development, such as Slc38a4, Sfmbt2, and Grab1, is also imprinted by oocyte-derived H3K27me3 (Inoue et al. 2017a). Intriguingly, while the oocyte-inherited H3K27me3 is lost in postimplantation embryos (Zheng et al. 2016; Inoue et al. 2017b; Chen et al. 2019b), the imprinted states of these genes (Platr20, Slc38a4, Gm32885, Gab1, Phf17, and Smoc1) are preserved in extraembryonic tissues by maternal-specific de novo DNA methylation (Chen et al. 2019b; Hanna et al. 2019), suggesting that DNA methylation and H3K27me3 act together to safeguard their imprinting. The importance of such H3K27me3-mediated imprinting was supported by data from the SCNT embryos, where oocyte H3K27me3-mediated imprinting is apparently missing (Matoba et al. 2018). Artificially forcing the monoallelic expression of these H3K27me3-controlled imprinted genes by allele-specific gene deletion can substantially improve mouse SCNT development (Wang et al. 2020). Interestingly, such H3K27me3-mediated imprinting appears to be missing in human. In human oocytes, H3K27me3 is strongly enriched at developmental gene promoters in a canonical pattern (Xia et al. 2019). Global H3K27me3 is erased after fertilization by the eight-cell stage, when ZGA happens. Embryonic H3K27me3 is subsequently restored at developmental genes in morulae and blastocysts (Fig. 2A; Xia et al. 2019; Zhang et al. 2019b). These results are consistent with the reported absence of imprinted XCI in human (Okamoto et al. 2011; Petropoulos et al. 2016). Mechanistically, in contrast with their abundant presence in mouse, the PRC2 core components, EED and SUZ12, are not expressed in human oocytes or pre-ZGA embryos, and are only activated after ZGA (Saha et al. 2013; Xia et al. 2019). These findings highlight the divergence of epigenetic regulation in early development among different species.
Bivalent H3K4me3 and H3K27me3
The “bivalent marks” refer to the co-occupancy of H3K4me3 and H3K27me3 at the promoters, often at developmental genes (Azuara et al. 2006; Bernstein et al. 2006). By bearing both permissive and repressive modifications, the bivalent marks are postulated to poise silenced key developmental genes for rapid reactivation upon cell differentiation. The bivalent marks widely exist in gametes, somatic cells, and PGCs (Vastenhouw and Schier 2012; Sachs et al. 2013; Hammoud et al. 2014). However, they are briefly absent in preimplantation embryos in mouse and human (Liu et al. 2016b; Zheng et al. 2016; Xia et al. 2019), with the underlying mechanisms and functions being unclear. One interesting possibility is that preimplantation embryos possess totipotency and pluripotency, while bivalent marks predominantly function during cell differentiation. Consistently, by E6.5, H3K27me3 and unusually strong H3K4me3 are restored at developmental gene promoters in the mouse epiblast, forming “super bivalency” (Fig. 1; Xiang et al. 2020). The “strong H3K4me3” is then quickly attenuated in fate-committed lineages and further decreases in somatic cells. Genes marked by “super-bivalency” also show strong spatial interactions as determined by Hi-C analysis (Xiang et al. 2020). In mESCs, such strong interactions rely on PRC1 and can help H3K27me3 spreading to regions away from “nucleation sites” (Schoenfelder et al. 2015; Oksuz et al. 2018; Kraft et al. 2020). Mechanistically, KMT2B (MLL2) is responsible for the “strong H3K4me3” at E6.5 but becomes partially dispensable by E8.5, as Kmt2b mutant embryos fail to activate a subset of developmental genes and die at E10.5 (Glaser et al. 2006; Xiang et al. 2020). As bivalent promoters are often intermediately accessible and enriched for paused Pol II (Stock et al. 2007; Brookes et al. 2012; Tee et al. 2014), it is speculated that “super-bivalency” may allow developmental genes to rapidly respond to developmental cues upon swift cell-fate decision during early lineage specification (Xiang et al. 2020). In addition, a subset of bivalent promoters is regulated by DPPA2/DPPA4 in mESCs, which safeguards promoter bivalency and prevents DNA methylation (Eckersley-Maslin et al. 2020; Gretarsson and Hackett 2020). Their roles in bivalency and differentiation in embryos are interesting topics for future studies.
H2AK119ub
H2AK119ub, deposited by PRC1, preferentially colocalizes with H3K27me3 at the promoters of developmental genes in most cell types including mouse sperm (Schoenfelder et al. 2015; Chen et al. 2021; Mei et al. 2021; Zhu et al. 2021). Similar to H3K27me3, H2AK119ub in mouse oocytes also exhibits a noncanonical distribution by forming broad domains in PMDs (Chen et al. 2021; Mei et al. 2021; Zhu et al. 2021). Interestingly, it also covers a fraction of active promoters marked by H3K4me3 (Chen et al. 2021; Mei et al. 2021; Zhu et al. 2021). H2AK119ub and H3K27me3 undergo distinct reprogramming dynamics in mouse early embryos. Upon fertilization, H2AK119ub, but not H3K27me3, is retained at promoters of developmental genes, and acute depletion of H2AK119ub in one-cell embryos causes derepression of developmental genes during ZGA and four-cell arrest, suggesting that H2AK119ub protects preimplantation embryo development from precocious expression of developmental genes when H3K27me3 is absent (Chen et al. 2021). Distal maternal H3K27me3 is highly stable and persists to blastocyst, which shows distinct pattern from that of the paternal allele. By contrast, H2AK119ub is much more dynamic after fertilization and the two alleles already adopt similar distributions by the two-cell stage (Chen et al. 2021; Mei et al. 2021; Zhu et al. 2021). Finally, H2AK119ub and H3K27me3 become colocalized again by exhibiting canonical patterns after implantation (Figs. 1 and 2A; Chen et al. 2021; Mei et al. 2021).
Interestingly, H2AK119ub has a complex interplay with H3K27me3 at maternal H3K27me3-controlled imprinted genes. Acute removal of H2AK119ub in mouse one-cell embryos does not affect H3K27me3-mediated imprinting at least by the four-cell stage (Chen et al. 2021), suggesting that H2AK119ub is dispensable for the maintenance of these H3K27me3 imprints. However, oocyte-specific knockout of Pcgf1 and Pcgf6, two key components of variant PRC1, leads to partial loss of H3K27me3 imprinting in both FGOs and morula (Mei et al. 2021), implying that H2AK119ub is involved in the establishment of some H3K27me3-mediated imprinting. Genes that lose H3K27me3 upon the Pcgf1 and Pcgf6 mutations also tend to be derepressed in FGOs (Mei et al. 2021). These findings highlight distinct roles and intricate cross talks between H2AK119ub and H3K27me3 in mouse early embryos, where H2AK119ub is more dynamic and can exert immediate repression, while H3K27me3 is relatively more stable and may serve as silencing memories.
H3K36me2/3
Methylation of H3K36 plays critical roles in transcription fidelity, RNA splicing, DNA replication, and DNA repair (Wagner and Carpenter 2012; Wozniak and Strahl 2014). H3K36me3 often enriches at actively expressed gene bodies, while H3K36me2 occupies both genic and intergenic regions (Wagner and Carpenter 2012; Wozniak and Strahl 2014; Weinberg et al. 2019). Both marks can recruit DNMT3A/B and subsequently specify patterns of DNA methylation (Dhayalan et al. 2010; Baubec et al. 2015; Dukatz et al. 2019; Weinberg et al. 2019). In mouse oocytes, DNA methylation is deposited to actively transcribed regions guided by H3K36me3, and a major function for such H3K36me3-guided DNA methylation is to establish maternal imprints (Chotalia et al. 2009; Bartolomei and Ferguson-Smith 2011; Kelsey and Feil 2013; Xu et al. 2019). Meanwhile, DNA methylation and H3K36me3 may also work synergistically to protect transcribing regions from H3K4me3 and H3K27me3, as observed in Setd2-deficient oocytes (Hanna et al. 2018b; Xu et al. 2019). As a result, Setd2-deficient oocytes show developmental defects and subsequent one-cell embryo arrest after fertilization (Xu et al. 2019). Spindle exchange experiments showed that such early arrest was mainly caused by cytosolic defect, as 54% of embryos reconstructed with wild-type (WT) cytoplasm and Setd2-deficient chromatin can develop to blastocyst. These embryos, however, eventually died after implantation presumably due to the loss of imprints (Xu et al. 2019). By contrast, in male germline, it is NSD1 and its catalytic product H3K36me2, but not SETD2 and H3K36me3, that direct the majority of de novo DNA methylation and protect transcribing regions from the spreading of H3K27me3 (Shirane et al. 2020). Male mice with ablated Nsd1 are infertile with hypogonadism and cannot generate mature spermatozoa (Shirane et al. 2020). These results indicate dimorphic mechanisms in DNA methylation establishment during gametogenesis. Why oocyte and sperm employ different H3K36 methylation to guide de novo DNA methylation awaits further investigation. After fertilization, H3K36me3 from both alleles is largely lost, before it reappears concomitant with ZGA (Fig. 1; Xu et al. 2019).
H3K9me3
H3K9me3 is a repressive mark that often decorates constitutive heterochromatin (Becker et al. 2016; Allshire and Madhani 2018). Disrupting H3K9me3 writers or erasers causes severe phenotypes in both oocytes and embryos. For example, mouse oocytes with deficient SETDB1, an H3K9 methyltransferase, suffer impaired meiosis and failure in silencing ERVK, ERVL, and MaLR retrotransposons (Eymery et al. 2016; Kim et al. 2016). Zygotic Setdb1-null embryos are lethal around implantation with defects in ICM growth (Dodge et al. 2004). Double knockout of H3K9 methyltransferase SUV39H1/2 leads to prenatal lethality in mice (Peters et al. 2001). Meanwhile, excessive H3K9me3 also needs to be curbed as the ablation of KDM4A, an H3K9me3 demethylase in oocytes, causes aberrant H3K9me3 invasion to the broad H3K4me3 domains, subsequently leading to impaired activation of genes and transposable elements during ZGA and preimplantation lethality (Sankar et al. 2020). H3K9me3 regulates mouse embryo development at least in three major aspects: facilitating the remodeling of heterochromatin, repressing repeats, and regulating lineage specification.
Mouse preimplantation embryos are featured with immature heterochromatin characterized by the absence of classic constitutive heterochromatin markers, such as HP1α and H4K20me3 (Wongtawan et al. 2011). The H3K9me3 methyltransferase SUV39H1 protein is not detected until the eight-cell stage (Burton et al. 2020). De novo H3K9me3 occurs in one-cell embryos (Wang et al. 2018; Burton et al. 2020), as ChIP-seq analyses confirmed that H3K9me3 undergoes a global resetting on both alleles after fertilization (Fig. 1; Wang et al. 2018). On the paternal genome, de novo H3K9me3 is mediated by SUV39H2, but is nonrepressive and instead bookmarks promoters for compaction at later stages (Burton et al. 2020). Such nonrepressive, immature heterochromatin appears to be essential for early development, as forced precocious expression of Suv39h1 restores constitutive heterochromatin but impairs embryonic development especially beyond the morula stage (Burton et al. 2020). Improper reprogramming of H3K9me3 in SCNT embryos results in insufficient gene activation in somatic H3K9me3-marked regions and poor development to blastocyst (Matoba et al. 2014). Such preimplantation developmental defects can be rescued by overexpression of the H3K9me3 demethylases Kdm4d or Kdm4b, or by knockdown of H3K9 methyltransferase Suv39h1/2 in donor cells in mouse (Matoba et al. 2014; Liu et al. 2016a). Similar findings were made in pig (Ruan et al. 2018), bovine (Liu et al. 2018a), monkey (Liu et al. 2018b), and human (Chung et al. 2015), suggesting that H3K9me3 is an evolutionarily conserved epigenetic barrier for cell-fate reprogramming.
After fertilization, a large portion of LTRs (long terminal retrotransposons) are activated (Peaston et al. 2004), possibly because of the relief of global epigenetic repression. The subsequent repression of LTRs coincides with the gradual reestablishment of H3K9me3 (Wang et al. 2018) and requires the histone chaperone chromatin assembly factor 1 (CAF-1) (Hatanaka et al. 2015; Wang et al. 2018). Knocking down the methyltransferases Suv39h1/2 leads to increased expression of LTRs in morula but not at the two-cell stage (Hatanaka et al. 2015; Burton et al. 2020), reinforcing the notion that H3K9me3 is repressive only in late-stage embryos.
Whereas H3K9me3 is primarily present in constitutive heterochromatin and transposons, it is also observed at lineage-specific genes, where it acts as a barrier for cell-fate changes by preventing precocious activation of lineage-incompatible genes (Becker et al. 2016). For example, the distributions of promoter H3K9me3 differ significantly between intra- and extraembryonic tissues of E6.5 and E7.5 embryos (Wang et al. 2018). Uncommitted germ-layer cells transiently employ high levels of H3K9me3 to repress genes involved in lineage specification, which are subsequently removed during differentiation to allow tissue-specific gene expression (Nicetto et al. 2019). Consequently, perturbation of H3K9me3 leads to aberrant lineage-specific gene expression (Nicetto et al. 2019). For example, mouse livers with endoderm-specific Setdb1/Suv39h1/2 triple-knockout resulted in the failed induction of hepatocyte-specific genes and aberrant expression of inappropriate lineage genes, concomitant with the loss of H3K9me3 and heterochromatin (Nicetto et al. 2019).
TRANSPOSABLE ELEMENTS
Transposable elements (TEs) occupy almost half of the mouse and human genome (Lander et al. 2001; de Koning et al. 2011). It has been increasingly evident that these elements, once thought as “junk DNA,” drive genetic innovation during evolution by introducing regulatory elements (such as promoters and enhancers) and transcription factor-binding sites (Rebollo et al. 2012; Chuong et al. 2017; Jangam et al. 2017; Bourque et al. 2018). MERVL, a member of endogenous retroviruses (ERVs), is actively transcribed during minor ZGA in mouse embryos (Svoboda et al. 2004), and was shown to regulate mouse preimplantation development (Huang et al. 2017). MERVL is reported to initiate hundreds of two-cell-specific transcripts in two-cell-like cells (2CLC), a rare and transient cell population in mESCs (Macfarlan et al. 2012; Genet and Torres-Padilla 2020). Notably, overexpressing DUX, a transcription factor specifically expressed during minor ZGA, can activate MERVL transcription and induce 2CLCs in mESCs through the DUX-miR-344-ZMYM2 pathway (De Iaco et al. 2017; Hendrickson et al. 2017; Whiddon et al. 2017; Yang et al. 2020). DUX itself is activated by two upstream transcription factors DPPA2/DPPA4 (Eckersley-Maslin et al. 2019) and negative elongation factor A (NELFA) (Hu et al. 2020). Apart from DUX, ZSCAN4, a nonconventional transcription factor (Falco et al. 2007; Ko 2016; Srinivasan et al. 2020), can also activate MERVL (Zhang et al. 2019a), in addition to its role in telomere elongation and genome integrity in mESCs (Ko 2016). However, ZSCAN4, in contrast to DUX, fails to do so in the absence of DPPA2/DPPA4 in mESCs. It was proposed that ZSCAN4 is not the driving factor but instead is a stabilizer to initiate the ZGA (De Iaco et al. 2019; Eckersley-Maslin et al. 2019). Notably, the Dux-knockout embryos show only minor defects of ZGA and MERVL transcription, and the majority of them can develop to term (Chen and Zhang 2019; Guo et al. 2019; De Iaco et al. 2020). DPPA2/DPPA4 double-null mice undergo normal embryogenesis but die perinatally (Nakamura et al. 2011). Therefore, these data suggest that additional ZGA regulators exist in embryos.
Long interspersed element-1 (LINE1), a subclass of non-LTR transposons, also plays a vital role in mouse embryo development (Beraldi et al. 2006; Jachowicz et al. 2017; Percharde et al. 2018). These elements are abundantly transcribed in mouse oocytes and fertilized embryos, before undergoing repression after the two-cell stage (Fadloun et al. 2013; Jachowicz et al. 2017). LINE1 knockdown in mouse zygotes was shown to impair ZGA and lead to two-cell stage arrest (Percharde et al. 2018). Interestingly, LINE1 transcription, but not its RNA, contributes to the embryo development by decondensing chromatin (Jachowicz et al. 2017). Precocious silencing or prolonged activation of LINE1 hinders chromatin relaxation or compaction, respectively, both resulting in embryo developmental defects (Beraldi et al. 2006; Jachowicz et al. 2017). Intriguingly, LINE1 RNA could also repress Dux expression in mESCs along with nucleolin and KAP1/TRIM28 (Percharde et al. 2018). It remains to be determined whether this is also the case in early embryos where Dux also needs to be shut down in a timely manner (Guo et al. 2019). Therefore, transposon elements are integral to the transcriptional circuitry in early development. Their exact roles in ZGA, totipotency, pluripotency, and lineage commitment await further investigation.
CHROMATIN ACCESSIBILITY
Accessible chromatin usually marks cis-regulatory elements, such as promoters, enhancers, insulators, where trans-acting factors interact with DNA and regulate gene expression (Klemm et al. 2019). Genome-wide accessible chromatin landscapes in mouse gametes and preimplantation were profiled with DNase I sequencing (liDNase-seq) (Lu et al. 2016; Inoue et al. 2017a) or transposase accessible chromatin-sequencing (ATAC-seq) (Wu et al. 2016; Jung et al. 2017, Lu et al. 2016; Wu et al. 2016, Jung et al. 2017, Gao et al. 2018a; Li et al. 2018a). Interestingly, a significant fraction of open chromatin appears to be specific for pre-ZGA embryos, and is lost upon genome activation (Wu et al. 2016, Lu et al. 2016; Wu et al. 2016, Jung et al. 2017; Gao et al. 2018a; Li et al. 2018a; Jung et al. 2019).
3D GENOME
In eukaryotes, the genome is packaged into a multilayer, higher-order chromatin configuration (Bickmore 2013; Gibcus and Dekker 2013; Gorkin et al. 2014; Bonev and Cavalli 2016; Misteli 2020). The chromosomes are nonrandomly distributed in interphase nucleus, occupying different chromosome territories. At the megabase scale, the genome is spatially partitioned into transcriptionally active compartment A, which preferentially resides near the nuclear speckles, and repressive compartment B, which is often associated with nuclear lamina or nucleolus (Lieberman-Aiden et al. 2009; van Steensel and Belmont 2017; Chen et al. 2018; Quinodoz et al. 2018). Topologically associating domains (TADs) exist as self-interacting domains usually with lengths of several hundred kilobase in mammals (Gibcus and Dekker 2013; Gorkin et al. 2014; Rowley and Corces 2018). They often arise via ATP-dependent loop extrusion, as cohesin extrudes chromatin until it is stopped by CCCTC-binding factor (CTCF) or other cis-acting factors (Sanborn et al. 2015; Fudenberg et al. 2016; Rao et al. 2017; Schwarzer et al. 2017; Vian et al. 2018; Davidson et al. 2019; Kim et al. 2019). At an even finer scale, the interactions among regulatory elements such as promoters and enhancers are crucial in regulating appropriate gene expression (Gorkin et al. 2014; Pombo and Dillon 2015; Schoenfelder and Fraser 2019). With the development of low-input or single-cell technologies to analyze chromatin organization, the dynamics of chromatin structure during gametogenesis and early embryo development begin to be unveiled, as summarized below.
Genome Organization in Spermatogenesis
To transport the haploid paternal genome to the offspring, the male germ cells undergo an orchestrated developmental process from spermatogonia to mature spermatozoa, which could be briefly divided into three major phases: mitosis (the proliferation and differentiation of spermatogonia), meiosis (the formation of haploid cells), and postmeiosis spermiogenesis (when the genome is subjected to compaction accompanied with histone-protamine exchange) (Handel and Schimenti 2010). In the meiosis I prophase, the homologous chromosomes undergo pairing (during the leptotene and zygotene stage), synapse (at the pachytene stage) and desynapse (at the onset of the diplotene stage) (Zickler and Kleckner 2015). Interestingly, TADs and compartments A/B dissolve in both monkey and mouse pachytene spermatocytes, when the homologous chromosomes parallelly align along the synaptonemal complex with chromatin loops extruding from the central axis (Alavattam et al. 2019; Patel et al. 2019; Vara et al. 2019; Wang et al. 2019b; Luo et al. 2020). At the same time, transcription-correlated local compartments (refined compartments A/B) (or point interactions/transcription hubs) emerge. These data raise a possibility that the chromatin loops facilitated by synaptonemal complex (Zickler and Kleckner 2015) may be incompatible with compartments A/B and chromatin loops mediated by CTCF (Nuebler et al. 2018). In line with this idea, refined compartments A/B are attenuated in synaptonemal complex mutants (Sycp2), with conventional TADs and compartmentalization partially restored (Wang et al. 2019b). Furthermore, conventional compartments and TADs also reappear in mature spermatozoa (Fig. 3; Battulin et al. 2015; Du et al. 2017; Jung et al. 2017; Ke et al. 2017; Alavattam et al. 2019; Wang et al. 2019b; Luo et al. 2020), although this was not detected in a separate study using a different sperm collection method (Vara et al. 2019). Interestingly, human mature spermatozoa have no TADs, which is attributed to CTCF silencing (Fig. 3; Chen et al. 2019a). Zebrafish spermatozoa also do not have conventional TADs, but possess unique “hinge-like” chromatin domains arranged similarly as mitotic chromosomes (Wike et al. 2021). These data show that extensive chromosome reorganization occurs during spermatogenesis and distinct mechanisms underline chromatin condensation in mature sperm in different species.
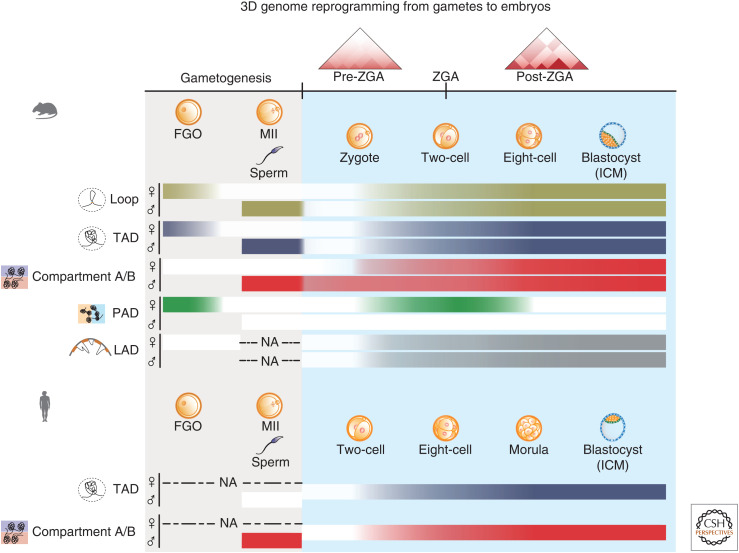
Figure 3.
3D genome dynamics in gametes and early embryos. The schematics showing the dynamics of higher-order chromatin structure from gametes to embryos in mouse and human. In mice, full-grown oocytes (FGOs) exhibit cohesin-independent, H3K27me3-marked compartmental domains (Polycomb-associating domains [PADs]). By contrast, topologically associating domains (TADs) become weak and compartments A/B are undetectable in FGOs. Lamina-associated domains (LADs) are also undetected in FGOs. In MII oocytes, chromatin adopts a mitotic-like chromatin structure, where compartments A/B, PADs, TADs, and loops are all lost. By contrast, mouse mature sperm shows canonical A/B compartmentalization, TADs, and loops. After fertilization, chromatin structure is dramatically dispersed, which shows weakened compartments, TADs, and loops. The genome structure is then gradually reestablished during preimplantation development. PADs briefly reappear specifically on the maternal allele from one-cell to eight-cell stages. LADs (gray) are de novo established after fertilization. Human embryos also exhibit diminished higher-order structure after fertilization, followed by consolidation of TADs and compartments A/B after zygotic genome activation (ZGA). Notably, human sperm has no typical TADs possibly due to the lack of CTCF protein. The strength of TADs, compartmentalization (compartments A/B and PADs/iPADs), and LADs are indicated by the shades of the bars. (NA) Data not available, (ICM) inner cell mass.
Genome Organization in Oogenesis
Oocytes are arrested at the diplotene stage for an extended time when the meiotic recombination is completed and the synaptonemal complex is largely dissembled (Handel and Schimenti 2010). Upon hormone stimulation, some oocytes undergo follicle growth, which start to accumulate proteins and RNAs for embryonic development, ultimately becoming FGOs (Bachvarova 1985; Li et al. 2010; Veselovska et al. 2015). TADs and chromatin loops can be detected in FGOs and, similar to those in soma, depend on SCC1, a key component of cohesin (Flyamer et al. 2017; Du et al. 2020; Silva et al. 2020). A meiotic-specific cohesin component, REC8, is required for the bivalent chromosome cohesion (Tachibana-Konwalski et al. 2010; Silva et al. 2020). Moreover, H3K27me3-marked compartmental domains (termed Polycomb-associating domains [PADs]) with strong interdomain interactions emerge in FGOs, concomitant with the loss of conventional compartments A/B (Flyamer et al. 2017; Du et al. 2020) and lamina-associated domains (LADs) (Fig. 3; van Steensel and Belmont 2017; Borsos et al. 2019). PADs require the PRC1 core components RING1A and RING1B, which catalyze H2AK119ub, but not the PRC2 protein EED and cohesin. Ring1a/Ring1b double knockout also partially derepresses the genes located in PADs in mouse oocytes, raising the possibility that PADs may play a role in gene silencing (Du et al. 2020). These data are in line with the notion that PRC1 is essential for both gene repression and Polycomb target interactions (Schuettengruber et al. 2017; Fursova et al. 2019; Blackledge et al. 2020; Boyle et al. 2020; Tamburri et al. 2020). Upon releasing oocytes from the diplotene arrest, PADs quickly disassemble (Du et al. 2020). When these cells are subsequently rearrested at metaphase II, the genome structure adopts a mitotic-like chromatin structure that lacks TADs, PADs, and compartments (Fig. 3; Du et al. 2017; Ke et al. 2017; Oomen and Dekker 2017). The mechanisms that lead to the formation of PADs in FGOs is currently unknown. It also remains unclear whether the collapse of chromatin around nucleolus in late-stage oocytes (Zuccotti et al. 1995) may contribute to the detachment of genome from the lamina and the emergence of PADs.
Conserved Relaxed Chromatin Organization after Fertilization
After fertilization, a prominent feature of chromatin of mouse zygotes is that it is extremely dispersed and lacks apparent condensed organization, as revealed by electron spectroscopic imaging (Ahmed et al. 2010). Consistently, low-input or single-cell Hi-C analyses show that chromatin adopts a highly dispersed or relaxed state in the zygotes and two-cell embryos, with loops, TADs, and compartments all substantially weakened compared with those in later-stage embryos (Fig. 3; Du et al. 2017; Flyamer et al. 2017; Gassler et al. 2017; Ke et al. 2017; Collombet et al. 2020). Remarkably, the extensively relaxed chromatin organization after fertilization is evolutionarily conserved, as similar findings were also made in fly (Hug et al. 2017; Ogiyama et al. 2018), fish (Kaaij et al. 2018; Nakamura et al. 2021; Wike et al. 2021), pig (Li et al. 2020), and human (Chen et al. 2019a). How chromatin relaxation occurs and whether it regulates early development remain a mystery. Such chromatin relaxation is also found in two-cell mouse SCNT embryos, implying that the ooplasm has a potent ability to relax chromatin organization (Chen et al. 2020; Zhang et al. 2020). Intriguingly, mimicking chromatin relaxation by pre-depletion of cohesin in the donor cells promotes the SCNT development, and this is in part via facilitating minor ZGA (Zhang et al. 2020). Further investigations are warranted to fully unravel the function of the highly dispersed chromatin state in early embryos.
Progressive Maturation of Chromatin Structure in Preimplantation Embryos
Chromatin structure reestablishment appears to be an unusually slow and prolonged process during early embryogenesis. The consolidation of TADs and the segregation of chromatin compartments are not completed until the eight-cell stage in mouse (Du et al. 2017; Ke et al. 2017), and around the morula stage in human (Fig. 3; Chen et al. 2019a). The maturation of TADs appears to require ZGA in human, presumably because CTCF is lowly expressed prior to ZGA but is highly induced during ZGA (Chen et al. 2019a). This is, however, not the case in mouse and fly embryos, where ZGA is dispensable for TAD and compartment A/B maturation (Du et al. 2017; Hug et al. 2017; Ke et al. 2017), indicating that additional mechanisms exist to help relax chromatin architecture in early development. The maturation of chromatin organization also includes equalizing chromatin organization between the two alleles. In mouse zygotes, the chromatin compartment is clearly visible for the paternal genome, while the maternal genome is poorly segregated (Du et al. 2017; Flyamer et al. 2017; Ke et al. 2017). The mechanisms underlying such differences remain unknown, although it is clear that the maternal and paternal genomes are independently segregated (Duffié and Bourc'his 2013) using two bipolar spindles in zygotes (Reichmann et al. 2018), as also captured by Hi-C (Du et al. 2017). The differential compartment reprogramming lasts until the eight-cell stage (Fig. 3; Du et al. 2017; Ke et al. 2017).
Besides TADs and compartments, LADs also undergo drastic reprogramming during mouse early embryogenesis (Borsos et al. 2019). They emerge as early as in the mouse zygote, similar to compartments, where the maternal genome exhibits poorly defined LADs, which then gradually are reinforced at the two-cell stage (Fig. 3; Du et al. 2017; Ke et al. 2017; Borsos et al. 2019). Furthermore, LADs in the two-cell embryos are in a noncanonical state, as many LADs colocalize with compartment A instead of compartment B, and LADs/inter-LADs show smaller differences of AT contents (Borsos et al. 2019). These observations likely reflect the transitionary nature of chromatin organization at this stage. Intriguingly, the ectopic expression of Kdm5b, the H3K4me3 demethylase, erases the paternal LADs in one-cell embryos (Borsos et al. 2019). How H3K4me3 regulates paternal LADs remains unknown. Notably, PADs briefly reappear on the maternal allele from one-cell to eight-cell stages (Fig. 3), concomitant with the brief inheritance of oocyte H3K27me3 domains (Zheng et al. 2016; Collombet et al. 2020; Du et al. 2020; Chen et al. 2021; Mei et al. 2021). Unlike what happens in oocytes, PADs in early embryos depend on PRC2 (Du et al. 2020). These observations indicate that histone modifications may also contribute to the reprogramming of chromatin architecture in embryos, and chromatin architecture matures at multidimensions in early development.
OUTSTANDING QUESTIONS
The recent exciting progress in understanding epigenetic reprogramming during early mammalian development inevitably raises many fundamental questions, as we discuss below.
-
Why do mammalian oocytes adopt noncanonical epigenomes?
Mouse oocytes adopt noncanonical epigenomes including DNA hypomethylated nontranscribing regions, broad domains of key histone modifications, and unique 3D genome structures. Why do mammalian oocytes, in contrast to sperm and soma, display so many unique features (Fig. 4)? This is particularly intriguing given zebrafish oocytes show relatively conventional epigenomes (Jiang et al. 2013; Potok et al. 2013; Zhang et al. 2018a; Zhu et al. 2019). One possibility lies in the fact that mammals require imprinting. To establish imprints, mammalian oocytes exploit a transcription-dependent mechanism to establish a partially methylated genome, which contrasts the hypermethylated sperm genome (Stewart et al. 2016; Sendžikaite and Kelsey 2019). It is, however, unclear why oocytes use such a dramatically distinct DNA methylome despite the fact that only a tiny portion are true imprints. It warrants further investigation if the remaining differential methylomes are simply “passenger marks” brought by the transcription-dependent de novo DNA methylation. Alternatively, they may provide a pool of candidate imprints during evolution, especially considering the existence of species-specific imprints (Bartolomei and Ferguson-Smith 2011).
Likewise, the broad domains of histone marks in mouse oocytes, which contrast with those in sperm, set the stage for noncanonical H3K27me3 imprinting (Inoue et al. 2017a,Xia et al. 2019). Several other factors may also contribute to such noncanonical patterning of histone marks. First, the prolonged prophase-I arrest of oocytes, which lacks DNA replication and cell division, provides ample time for the deposition of histone marks and the incorporation of histone variants (the deposition of which is DNA replication-independent). For example, H3.3 plays a critical role in oocytes as the deletion of H3.3 impairs oocyte competence, concomitant with aberrant chromatin accessibility, histone modifications, and ectopic gene activation (Lin et al. 2014; Nashun et al. 2015; Smith et al. 2020). Second, the lack of DNA methylation in nontranscribing regions may allow opportunistic deposition of opposing histone marks, as evidenced in the Dnmt3a/3b-double-knockout or Setd2-null oocytes (Hanna et al. 2018b; Xu et al. 2019). Nevertheless, how human oocytes manage to avoid these broad domains of histone marks (such as H3K4me3) (Xia et al. 2019) remains unclear. Last, the noncanonical histone marks may be related to oocyte-specific functions. For example, noncanonical H3K4me3 appears to be required for the transcription silencing of oocytes (Zhang et al. 2016; Hanna et al. 2018b).
-
Why do early mammalian embryos require genome-wide resetting rather than region-specific refining?
Embryos undergo extensive genome-wide epigenome reprogramming upon fertilization, which is rarely observed for other biological processes. A natural hypothesis is that the global resetting can efficiently convert the epigenomes from gametes to embryos. Nevertheless, neither the global chromatin relaxation nor genome-wide erasure of repressive epigenetic marks was reported in iPSC reprogramming, an equally dramatic cell-fate conversion event (Papp and Plath 2013). First, one possibility is that the remarkable difference between fertilization and iPSC conversion may reflect the distinct developmental potency for fertilized embryos (“totipotency”) and iPS cells (“pluripotency”). Perhaps the emergence of totipotency requires a globally permissive, naive chromatin state. Second, the global demethylation in early embryos may contribute to the establishment of hypomethylated extraembryonic tissues (Schroeder et al. 2015; Smith et al. 2017; Zhang et al. 2018b). Last, a third possible purpose of the dramatic reprogramming in early embryos may be to equalize the distinct epigenomes of two alleles inherited from sperm and oocytes, which arise during the establishment of imprints. As supporting evidence, zebrafish, which lack imprinting, do not undergo global DNA demethylation (Macleod et al. 1999; Jiang et al. 2013; Potok et al. 2013), but instead present locus-specific DNA methylation remodeling.
Notably, zebrafish gametes show canonical patterns of histone modifications, which are, however, rapidly lost during early embryogenesis, before being restored at regulatory elements around ZGA (Vastenhouw et al. 2010; Zhang et al. 2014, Murphy et al. 2018; Zhu et al. 2019). The differences between DNA methylation and histone modification reprogramming in zebrafish may be related to the fact that histone marks lack an efficient maintenance mechanism, compared with DNA methylation, as their post-DNA replication recovery is usually much slower than DNA methylation (Budhavarapu et al. 2013; Almouzni and Cedar 2016). Therefore, histone marks are perhaps more difficult to be maintained during the extremely rapid embryo cleavage in early development, until the cell cycle slows down around ZGA (Kane and Kimmel 1993).
-
How do embryos achieve successful MZT?
MZT is a key process in early embryos, which transfers the developmental control from maternally inherited gene products to zygotically synthesized factors (Schultz 2002; Walser and Lipshitz 2011). Mounting data begin to reveal the existence of a “primitive” chromatin state prior to ZGA featured with widespread active marks, immature heterochromatin, and relaxed chromatin structure (Fig. 4; Xia and Xie 2020). The mechanisms underlying the noncanonical setting of the epigenome, and their roles in MZT are fascinating questions and are discussed below.
First, fertilized mouse embryos are featured by compromised heterochromatin, as manifested by the lack of chromocenters and the rapid decrease or resetting of heterochromatin epigenetic marks such as H3K9me3, H3K64me3, H4K20me3, H3K27me3, and DNA methylation (Probst et al. 2007; Aguirre-Lavin et al. 2012; Burton and Torres-Padilla 2014). The loss of heterochromatin may be important for lengthening telomeres (Ko 2016) and for eliminating cell reprogramming barriers to release zygotic genome from a constrained environment to become a totipotent state (Zhou and Dean 2015; Becker et al. 2016). Nevertheless, it presumably also creates challenges and stresses to the embryos. For example, the global loss of DNA demethylation and repressive H3K9me3 can lead to derepression of repeats that threaten genome stability (Gerdes et al. 2016; Rodriguez-Terrones and Torres-Padilla 2018) and trigger immune responses (Grow et al. 2015). However, these repeats are also exploited by embryos for transcription regulation through creating new or alternative promoters, providing or disrupting TF-binding sites, regulating chromatin accessibility, and silencing nearby genes (Gerdes et al. 2016; Rodriguez-Terrones and Torres-Padilla 2018). Meanwhile, it is perhaps not surprising that embryos employ alternative mechanisms to withstand the detrimental effects from the loss of heterochromatin. For instance, in one-cell mouse embryos, the maternally provided PRC1 and corresponding H2AK119ub temporarily assume the function of H3K9me3 in the paternal genome at pericentromeric heterochromatin to inhibit pericentric satellite expression (Puschendorf et al. 2008; Tardat et al. 2015). Similarly, while H3K27me3 is erased from its canonical targets (developmental genes) in mouse preimplantation embryos (Zheng et al. 2016), H2AK119ub maintains these developmental genes nevertheless silenced (Chen et al. 2021).
Second, despite the largely silenced transcriptional state, pre-ZGA embryos show hyperpermissive chromatin featuring widespread transcription-independent active marks and open chromatin. For example, the paternal genomes of mouse one-cell embryos transiently acquire H3K4me3 domains in gene-rich regions (Zhang et al. 2016). Likewise, human pre-ZGA embryos exhibit broad domains of H3K4me3 at CG-rich promoters and distal regions (Xia et al. 2019). In zebrafish, widespread de novo H3K27ac occupies promoters throughout the genome prior to ZGA (Zhang et al. 2018a; Sato et al. 2019). The transcription-independent active histone marks likely reflect a “primitive” state of the genome and perhaps also prepares chromatin for coming ZGA. In support of this hypothesis, histone acetylation precedes and is required for transcription elongation in zebrafish early embryos (Zhang et al. 2018a; Chan et al. 2019; Sato et al. 2019). Such hyperpermissive chromatin is likely contributed by the loss of heterochromatic marks. In addition, one-cell embryos either inherit or acquire high levels of histone variants, including H3.3, H2A.X, and TH2A (Burton and Torres-Padilla 2014; Shinagawa et al. 2014; Funaya and Aoki 2017). For example, the paternal genome incorporates H3.3-packaged nucleosomes through protamine-histone exchange in one-cell mouse embryos (van der Heijden et al. 2005; Torres-Padilla et al. 2006; Santenard et al. 2010). Consequently, embryos deficient for maternal Hira, which is responsible for H3.3, suffer impaired male pronucleus formation, DNA replication, and rRNA transcription (Loppin et al. 2005; Inoue and Zhang 2014; Lin et al. 2014). The abundant histone variants may also contribute to the high mobility and dispersed state of zygotic chromatin (Ooga et al. 2016). Such hyperpermissive chromatin, however, needs to be curbed upon ZGA to ensure the fidelity of transcription programs. For example, exogenous promoters can fire without enhancers in the one-cell, but not two-cell, mouse embryos, indicating a progressively repressive chromatin environment (Wiekowski et al. 1991; Majumder et al. 1993). Repression of LINE-1 may also contribute to chromatin compaction beyond the two-cell stage (Jachowicz et al. 2017). In addition, transcription activity, in particular during minor ZGA, may play a critical role in this process, as transcription inhibition prevents ZGA-associated transition of open chromatin and active histone marks (Wu et al. 2016, Zhang et al. 2016; Li et al. 2018a).
Last, early embryos employ a highly conserved relaxed 3D chromatin organization featured by the loss of TADs, loops, and compartments (Zheng and Xie 2019). It remains unclear whether the long-range enhancer–promoter interactions are also lost and reset after fertilization. If so, one intriguing question is how embryos execute transcription programs without long-distance enhancer–promoter interactions and the facilitating TADs and loops when ZGA starts. It is possible that embryos may primarily use genes or enhancers that do not require long-distance interactions, which could help prevent premature firing of developmental genes and cell-fate commitment programs. Currently, the enhancer–promoter regulatory network at the beginning of life still remains largely unknown and awaits further investigations.
-
How do early embryos establish an embryonic epigenome?
The establishment of embryonic epigenome could be divided into two waves. Embryonic active epigenetic marks are often established around ZGA in mouse and human, with H3K4me3 occupying active promoters (Dahl et al. 2016; Zhang et al. 2016; Xia et al. 2019), and H3K27ac and open chromatin appearing at both active promoters and putative enhancers (Liu et al. 2016b; Lu et al. 2016; Wu et al. 2016, Gao et al. 2018a; Li et al. 2018a; Xia et al. 2019). ZGA is required for this process, as transcription inhibition blocks the redistribution of these active marks (Dahl et al. 2016; Wu et al. 2016, Zhang et al. 2016; Xia et al. 2019). These data paint a picture in which transcription facilitates chromatin state transitions, which presumably in turn potentiate transcription, forming a positive-feedback loop. By contrast, the establishment of repressive epigenetic marks (such as H3K27me3, H3K9me3, and DNA methylation) occurs later, often around implantation (Liu et al. 2016b; Zheng et al. 2016; Smith et al. 2017; Wang et al. 2018; Zhang et al. 2018b; Nicetto et al. 2019). Why does the reestablishment of repressive marks occur at a slower pace compared to active marks? One possibility is that the absence of these repressive marks at earlier stages may be a prerequisite to ensure the embryos to retain totipotency. Consistently, the removal of H3K9me3 by the depletion of H3K9 methyltransferase SETDB1 and its binding partner KAP1 promotes the transition from mESC to two-cell-like cells, during which cells partially regain totipotent potentials (Wu et al. 2020). These two waves of embryonic epigenome establishment may ensure the coordination of step-wise developmental events that include ZGA, totipotency emergence, and cell-fate determination (Fig. 4). At the gene level, diverse signaling pathways and transcription factors are often engaged in regulating the accurate embryonic epigenome establishment. For example, WNT and FGF signaling pathways are reported to modulate the global hypomethylation and CGI methylation in the extraembryonic lineages (Smith et al. 2017). Distinct transcription factors are expected to function in the lineage-specific distribution of DNA methylation and histone marks at regulatory elements based on the motif-enrichment analysis (Smith et al. 2017; Wang et al. 2018; Zhang et al. 2018b; Xiang et al. 2020). Future studies on the establishment of embryonic epigenome will undoubtedly broaden our understanding of the arise of totipotency, pluripotency, and lineage specification during embryo development.
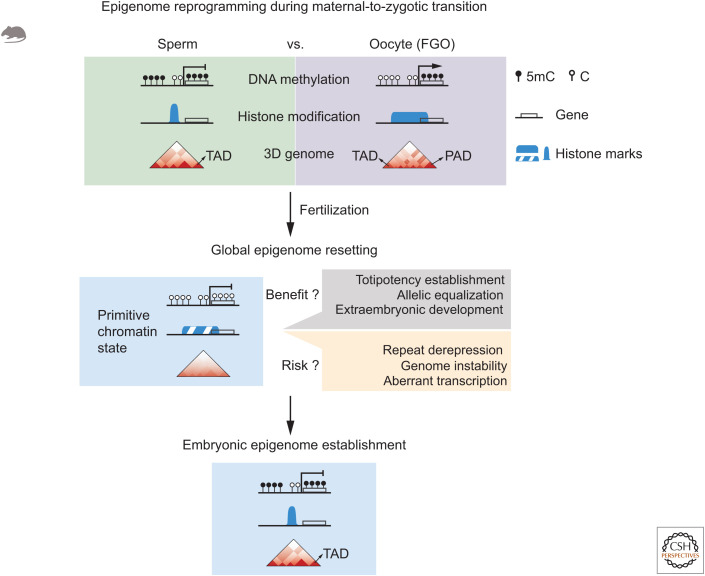
Figure 4.
Epigenome reprogramming during maternal-to-zygotic transition (MZT). Mouse oocytes, but not sperm, adopt noncanonical epigenomes including DNA hypomethylated nontranscribing regions, broad domains of key histone modifications, and unique 3D genome structures. After fertilization, the embryos undergo global epigenome resetting, featured by the global loss of repressive epigenetic marks (such as DNA methylation), widespread presence of active histone marks, and global chromatin relaxation. The global resetting might be beneficial for the establishment of totipotency, allelic epigenome equalization, and extraembryonic tissue development. Meanwhile, it may also expose embryos for risks of repeat derepression, genome instability, and aberrant transcription. After zygotic genome activation (ZGA), the embryos stepwise establish embryonic epigenomes to restore DNA methylation, histone marks, and higher-order chromatin structure. (FGO) Full-grown oocyte, (TAD) topologically associating domain, (PAD) Polycomb-associating domain.
CONCLUDING REMARKS
Tremendous advances in ultrasensitive chromatin analysis technologies have shown the epigenome reprogramming modes in mammalian gametogenesis and early development, and the mechanisms underlying these fascinating processes. Despite these breakthroughs, more questions arise as expected. First, with the emerging detailed maps of the epigenome, how and why these reprogramming events occur remain elusive. For example, what are the molecular mechanisms and the potential functions of the relaxed chromatin structure after fertilization? What are the key factors or events that trigger ZGA in mammals? Addressing these questions would bring the second major challenge in the field, which is the urgent need to develop innovative tools. For instance, CRISPR-based screening is a powerful tool to identify the key transcription regulators in cell lines (Alda-Catalinas et al. 2020). Yet, genome-wide screening is still a daunting task for early embryos. This is further complicated by the presence of abundant maternal proteins that can be highly stable in oocytes and early embryos. Easy-to-use protein degradation methods, such as PROTAC (Burslem and Crews 2020), ARMeD (Ibrahim et al. 2020), or Trim-away (Clift et al. 2017), are needed for the loss-of-function studies to investigate factors of interests. The emerging new in vitro models, such as blastoid and gastruloid (for review, see Baillie-Benson et al. 2020), could simulate embryonic development and be potentially used to screen and identify key regulators in embryonic development. Ultrasensitive and single-cell technologies, preferentially with the ability to probe multi-omics, should be instrumental in deciphering the spatiotemporal chromatin dynamics in early embryos. For example, TFs play essential roles in governing transcription circuitry especially during lineage specification. Although improved ChIP and ChIC-based methods (uliChIP, itChIP, CUT&RUN, CUT&Tag, ChIL-seq, CoBATCH, ACT-seq, etc.) (Brind'Amour et al. 2015; Skene and Henikoff 2017; Ai et al. 2019; Carter et al. 2019; Harada et al. 2019; Kaya-Okur et al. 2019; Wang et al. 2019a) have been invented, these methods rely on highly sensitive antibodies that are not always accessible. As another example, enhancer–promoter interactions are still difficult to study in early development because of the sparse Hi-C data from limited research materials. Improved chromatin conformation capture methods to enrich the regulatory chromatin interactions at high resolution is urgently needed. The Oligopaint-based DNA FISH (fluorescence in situ hybridization) and CRISPR-based live-cell imaging methods that can tag numerous genomic loci simultaneously (Kempfer and Pombo 2020) may facilitate decoding how dynamic chromatin conformation contributes to gene expression during embryogenesis. Single-cell, multi-omics methods would be essential to delineate the heterogeneity of cells during early development. Thirdly, epigenome reprogramming studies across species have revealed remarkable differences between mice and human, and even between monkey and human (Hanna et al. 2018a; Chen et al. 2019a; Wang et al. 2019b; Xia et al. 2019). Applying the research framework to other species will shed light on the evolution of epigenetic reprogramming and facilitate studies for key events that are found in human but not in mouse.
ACKNOWLEDGMENTS
We thank the laboratory members for the comments and help during the preparation of the manuscript. The review included only selected studies and we apologize to all the authors whose work was not cited due to space limitations. This work was supported by the National Key R&D Program of China (Grant No. 2019YFA0508901 to W.X.), the National Natural Science Foundation of China (Grant Nos. 31988101, 31830047, and 31725018 to W.X.), the THU-PKU Center for Life Sciences (W.X.), the Beijing Municipal Commission of Science and Technology (Grant No. Z181100001318006 to W.X.). W.X. is an HHMI International Research Scholar. Z.D. is supported by THU-PKU Center for Life Sciences postdoctoral fellowships.
Footnotes
Editors: Ana Pombo, Martin W. Hetzer, and Tom Misteli
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Abe K, Yamamoto R, Franke V, Cao MJ, Suzuki Y, Suzuki MG, Vlahovicek K, Svoboda P, Schultz RM, Aoki F. 2015. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3′ processing. EMBO J 34: 1523–1537. 10.15252/embj.201490648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre-Lavin T, Adenot P, Bonnet-Garnier A, Lehmann G, Fleurot R, Boulesteix C, Debey P, Beaujean N. 2012. 3D-FISH analysis of embryonic nuclei in mouse highlights several abrupt changes of nuclear organization during preimplantation development. BMC Dev Biol 12: 30. 10.1186/1471-213X-12-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP. 2010. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS ONE 5: e10531. 10.1371/journal.pone.0010531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai S, Xiong H, Li CC, Luo Y, Shi Q, Liu Y, Yu X, Li C, He A. 2019. Profiling chromatin states using single-cell itChIP-seq. Nat Cell Biol 21: 1164–1172. 10.1038/s41556-019-0383-5 [DOI] [PubMed] [Google Scholar]
- Alavattam KG, Maezawa S, Sakashita A, Khoury H, Barski A, Kaplan N, Namekawa SH. 2019. Attenuated chromatin compartmentalization in meiosis and its maturation in sperm development. Nat Struct Mol Biol 26: 175–184. 10.1038/s41594-019-0189-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alda-Catalinas C, Bredikhin D, Hernando-Herraez I, Santos F, Kubinyecz O, Eckersley-Maslin MA, Stegle O, Reik W. 2020. A single-cell transcriptomics CRISPR-activation screen identifies epigenetic regulators of the zygotic genome activation program. Cell Syst 11: 25–41.e9. 10.1016/j.cels.2020.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Madhani HD. 2018. Ten principles of heterochromatin formation and function. Nat Rev Mol Cell Biol 19: 229–244. 10.1038/nrm.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G, Cedar H. 2016. Maintenance of epigenetic information. Cold Spring Harb Perspect Biol 8: a019372. 10.1101/cshperspect.a019372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D'Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, et al. 2016. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat Cell Biol 18: 225–233. 10.1038/ncb3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu-Vieyra CV, Chen R, Agno JE, Glaser S, Anastassiadis K, Stewart AF, Matzuk MM. 2010. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. Plos Biol 8: e1000453. 10.1371/journal.pbio.1000453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM. 1997. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 181: 296–307. 10.1006/dbio.1996.8466 [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. 2006. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538. 10.1038/ncb1403 [DOI] [PubMed] [Google Scholar]
- Bachvarova R. 1985. Gene expression during oogenesis and oocyte development in mammals. Dev Biol (NY 1985) 1: 453–524. [DOI] [PubMed] [Google Scholar]
- Baillie-Benson P, Moris N, Martinez Arias A. 2020. Pluripotent stem cell models of early mammalian development. Curr Opin Cell Biol 66: 89–96. 10.1016/j.ceb.2020.05.010 [DOI] [PubMed] [Google Scholar]
- Barau J, Teissandier A, Zamudio N, Roy S, Nalesso V, Hérault Y, Guillou F, Bourc'his D. 2016. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354: 909–912. 10.1126/science.aah5143 [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Ferguson-Smith AC. 2011. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol 3: a002592. 10.1101/cshperspect.a002592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battulin N, Fishman VS, Mazur AM, Pomaznoy M, Khabarova AA, Afonnikov DA, Prokhortchouk EB, Serov OL. 2015. Comparison of the three-dimensional organization of sperm and fibroblast genomes using the Hi-C approach. Genome Biol 16: 77. 10.1186/s13059-015-0642-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, Schübeler D. 2015. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520: 243–247. 10.1038/nature14176 [DOI] [PubMed] [Google Scholar]
- Becker JS, Nicetto D, Zaret KS. 2016. H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet 32: 29–41. 10.1016/j.tig.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellier S, Chastant S, Adenot P, Vincent M, Renard JP, Bensaude O. 1997. Nuclear translocation and carboxyl-terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. EMBO J 16: 6250–6262. 10.1093/emboj/16.20.6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraldi R, Pittoggi C, Sciamanna I, Mattei E, Spadafora C. 2006. Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol Reprod Dev 73: 279–287. 10.1002/mrd.20423 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Bickmore WA. 2013. The spatial organization of the human genome. Annu Rev Genomics Hum Genet 14: 67–84. 10.1146/annurev-genom-091212-153515 [DOI] [PubMed] [Google Scholar]
- Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21. 10.1101/gad.947102 [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Fursova NA, Kelley JR, Huseyin MK, Feldmann A, Klose RJ. 2020. PRC1 catalytic activity is central to polycomb system function. Mol Cell 77: 857–874.e9. 10.1016/j.molcel.2019.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Cavalli G. 2016. Organization and function of the 3D genome. Nat Rev Genet 17: 661–678. 10.1038/nrg.2016.112 [DOI] [PubMed] [Google Scholar]
- Borsos M, Perricone SM, Schauer T, Pontabry J, de Luca KL, de Vries SS, Ruiz-Morales ER, Torres-Padilla ME, Kind J. 2019. Genome–lamina interactions are established de novo in the early mouse embryo. Nature 569: 729–733. 10.1038/s41586-019-1233-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouniol C, Nguyen E, Debey P. 1995. Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Exp Cell Res 218: 57–62. 10.1006/excr.1995.1130 [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99. 10.1038/nature02886 [DOI] [PubMed] [Google Scholar]
- Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, Imbeault M, Izsvák Z, Levin HL, Macfarlan TS, et al. 2018. Ten things you should know about transposable elements. Genome Biol 19: 199. 10.1186/s13059-018-1577-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S, Flyamer IM, Williamson I, Sengupta D, Bickmore WA, Illingworth RS. 2020. A central role for canonical PRC1 in shaping the 3D nuclear landscape. Genes Dev 34: 931–949. 10.1101/gad.336487.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braude P, Bolton V, Moore S. 1988. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 332: 459–461. 10.1038/332459a0 [DOI] [PubMed] [Google Scholar]
- Brind'Amour J, Liu S, Hudson M, Chen C, Karimi MM, Lorincz MC. 2015. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun 6: 6033. 10.1038/ncomms7033 [DOI] [PubMed] [Google Scholar]
- Brind'Amour J, Kobayashi H, Richard Albert J, Shirane K, Sakashita A, Kamio A, Bogutz A, Koike T, Karimi MM, Lefebvre L, et al. 2018. LTR retrotransposons transcribed in oocytes drive species-specific and heritable changes in DNA methylation. Nat Commun 9: 3331. 10.1038/s41467-018-05841-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, et al. 2012. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10: 157–170. 10.1016/j.stem.2011.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schübeler D, Stadler MB, Peters AH. 2010. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 17: 679–687. 10.1038/nsmb.1821 [DOI] [PubMed] [Google Scholar]
- Budhavarapu VN, Chavez M, Tyler JK. 2013. How is epigenetic information maintained through DNA replication? Epigenetics Chromatin 6: 32. 10.1186/1756-8935-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burslem GM, Crews CM. 2020. Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell 181: 102–114. 10.1016/j.cell.2019.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Torres-Padilla ME. 2014. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat Rev Mol Cell Biol 15: 723–735. 10.1038/nrm3885 [DOI] [PubMed] [Google Scholar]
- Burton A, Brochard V, Galan C, Ruiz-Morales ER, Rovira Q, Rodriguez-Terrones D, Kruse K, Le Gras S, Udayakumar VS, Chin HG, et al. 2020. Heterochromatin establishment during early mammalian development is regulated by pericentromeric RNA and characterized by non-repressive H3K9me3. Nat Cell Biol 22: 767–778. 10.1038/s41556-020-0536-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B, Ku WL, Kang JY, Hu G, Perrie J, Tang Q, Zhao K. 2019. Mapping histone modifications in low cell number and single cells using antibody-guided chromatin tagmentation (ACT-seq). Nat Commun 10: 3747. 10.1038/s41467-019-11559-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SH, Tang Y, Miao L, Darwich-Codore H, Vejnar CE, Beaudoin JD, Musaev D, Fernandez JP, Benitez MDJ, Bazzini AA, et al. 2019. Brd4 and P300 confer transcriptional competency during zygotic genome activation. Dev Cell 49: 867–881.e8. 10.1016/j.devcel.2019.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y. 2016. Lineage specification in the mouse preimplantation embryo. Development 143: 1063–1074. 10.1242/dev.128314 [DOI] [PubMed] [Google Scholar]
- Chen Z, Zhang Y. 2019. Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat Genet 51: 947–951. 10.1038/s41588-019-0418-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhang Y. 2020. Maternal H3K27me3-dependent autosomal and X chromosome imprinting. Nat Rev Genet 21: 555–571. 10.1038/s41576-020-0245-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang Y, Wang Y, Zhang L, Brinkman EK, Adam SA, Goldman R, van Steensel B, Ma J, Belmont AS. 2018. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J Cell Biol 217: 4025–4048. 10.1083/jcb.201807108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ke Y, Wu K, Zhao H, Sun Y, Gao L, Liu Z, Zhang J, Tao W, Hou Z, et al. 2019a. Key role for CTCF in establishing chromatin structure in human embryos. Nature 576: 306–310. 10.1038/s41586-019-1812-0 [DOI] [PubMed] [Google Scholar]
- Chen Z, Yin Q, Inoue A, Zhang C, Zhang Y. 2019b. Allelic H3K27me3 to allelic DNA methylation switch maintains noncanonical imprinting in extraembryonic cells. Sci Adv 5: eaay7246. 10.1126/sciadv.aay7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Zhu Q, Li C, Kou X, Zhao Y, Li Y, Xu R, Yang L, Yang L, Gu L, et al. 2020. Chromatin architecture reorganization in murine somatic cell nuclear transfer embryos. Nat Commun 11: 1813. 10.1038/s41467-020-15607-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Djekidel MN, Zhang Y. 2021. Distinct dynamics and functions of H2AK119ub1 and H3K27me3 in mouse preimplantation embryos. Nat Genet 53: 551–563. 10.1038/s41588-021-00821-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, James K, Dean W, Kelsey G. 2009. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev 23: 105–117. 10.1101/gad.495809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YG, Matoba S, Liu Y, Eum JH, Lu F, Jiang W, Lee JE, Sepilian V, Cha KY, Lee DR, et al. 2015. Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells. Cell Stem Cell 17: 758–766. 10.1016/j.stem.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet 18: 71–86. 10.1038/nrg.2016.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D, McEwan WA, Labzin LI, Konieczny V, Mogessie B, James LC, Schuh M. 2017. A method for the acute and rapid degradation of endogenous proteins. Cell 171: 1692–1706.e18. 10.1016/j.cell.2017.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, Andrews R, Lee JH, Skalnik D, Bird A. 2012. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev 26: 1714–1728. 10.1101/gad.194209.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombet S, Ranisavljevic N, Nagano T, Varnai C, Shisode T, Leung W, Piolot T, Galupa R, Borensztein M, Servant N, et al. 2020. Parental-to-embryo switch of chromosome organization in early embryogenesis. Nature 580: 142–146. 10.1038/s41586-020-2125-z [DOI] [PubMed] [Google Scholar]
- Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, Li G, Kuan S, Li B, Lee AY, et al. 2016. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537: 548–552. 10.1038/nature19360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J, Rousseau P, Hardikar S, Veland N, Wong J, Autexier C, Chen T. 2017. Zscan4 inhibits maintenance DNA methylation to facilitate telomere elongation in mouse embryonic stem cells. Cell Rep 20: 1936–1949. 10.1016/j.celrep.2017.07.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters JM. 2019. DNA loop extrusion by human cohesin. Science 366: 1338–1345. 10.1126/science.aaz3418 [DOI] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. 2017. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 49: 941–945. 10.1038/ng.3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Coudray A, Duc J, Trono D. 2019. DPPA2 and DPPA4 are necessary to establish a 2C-like state in mouse embryonic stem cells. EMBO Rep 20: e47382. 10.15252/embr.201847382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Verp S, Offner S, Grun D, Trono D. 2020. DUX is a non-essential synchronizer of zygotic genome activation. Development 147: dev177725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. Plos Genet 7: e1002384. 10.1371/journal.pgen.1002384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, Singh S, Anastassiadis K, Stunnenberg HG, Stewart AF. 2014. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development 141: 526–537. 10.1242/dev.102681 [DOI] [PubMed] [Google Scholar]
- Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. 2010. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem 285: 26114–26120. 10.1074/jbc.M109.089433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L, Helin K. 2013. Transcriptional regulation by polycomb group proteins. Nat Struct Mol Biol 20: 1147–1155. 10.1038/nsmb.2669 [DOI] [PubMed] [Google Scholar]
- Dobson AT, Raja R, Abeyta MJ, Taylor T, Shen S, Haqq C, Pera RA. 2004. The unique transcriptome through day 3 of human preimplantation development. Hum Mol Genet 13: 1461–1470. 10.1093/hmg/ddh157 [DOI] [PubMed] [Google Scholar]
- Dodge JE, Kang YK, Beppu H, Lei H, Li E. 2004. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol 24: 2478–2486. 10.1128/MCB.24.6.2478-2486.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillet D, Sze CC, Ryan C, Piunti A, Shah AP, Ugarenko M, Marshall SA, Rendleman EJ, Zha DD, Helmin KA, et al. 2020. Uncoupling histone H3K4 trimethylation from developmental gene expression via an equilibrium of COMPASS, Polycomb and DNA methylation. Nat Genet 52: 615–625. 10.1038/s41588-020-0618-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zheng H, Huang B, Ma R, Wu J, Zhang X, He J, Xiang Y, Wang Q, Li Y, et al. 2017. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547: 232–235. 10.1038/nature23263 [DOI] [PubMed] [Google Scholar]
- Du Z, Zheng H, Kawamura YK, Zhang K, Gassler J, Powell S, Xu Q, Lin Z, Xu K, Zhou Q, et al. 2020. Polycomb group proteins regulate chromatin architecture in mouse oocytes and early embryos. Mol Cell 77: 825–839.e7. 10.1016/j.molcel.2019.11.011 [DOI] [PubMed] [Google Scholar]
- Duffié R, Bourc'his D. 2013. Parental epigenetic asymmetry in mammals. Curr Top Dev Biol 104: 293–328. 10.1016/B978-0-12-416027-9.00009-7 [DOI] [PubMed] [Google Scholar]
- Dukatz M, Holzer K, Choudalakis M, Emperle M, Lungu C, Bashtrykov P, Jeltsch A. 2019. H3k36me2/3 binding and DNA binding of the DNA methyltransferase DNMT3A PWWP domain both contribute to its chromatin interaction. J Mol Biol 431: 5063–5074. 10.1016/j.jmb.2019.09.006 [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Alda-Catalinas C, Reik W. 2018. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat Rev Mol Cell Bio 19: 436–450. 10.1038/s41580-018-0008-z [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin M, Alda-Catalinas C, Blotenburg M, Kreibich E, Krueger C, Reik W. 2019. Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev 33: 194–208. 10.1101/gad.321174.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Parry A, Blotenburg M, Krueger C, Ito Y, Franklin VNR, Narita M, D'Santos CS, Reik W. 2020. Epigenetic priming by Dppa2 and 4 in pluripotency facilitates multi-lineage commitment. Nat Struct Mol Biol 27: 696–705. 10.1038/s41594-020-0443-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkek S, Hisano M, Liang CY, Gill M, Murr R, Dieker J, Schübeler D, van der Vlag J, Stadler MB, Peters AH. 2013. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol 20: 868–875. 10.1038/nsmb.2599 [DOI] [PubMed] [Google Scholar]
- Eymery A, Liu ZC, Ozonov EA, Stadler MB, Peters AHFM. 2016. The methyltransferase Setdb1 is essential for meiosis and mitosis in mouse oocytes and early embryos. Development 143: 2767–2779. [DOI] [PubMed] [Google Scholar]
- Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E, Carninci P, Torres-Padilla ME. 2013. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat Struct Mol Biol 20: 332–338. 10.1038/nsmb.2495 [DOI] [PubMed] [Google Scholar]
- Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. 2007. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol 307: 539–550. 10.1016/j.ydbio.2007.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, Tachibana-Konwalski K. 2017. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544: 110–114. 10.1038/nature21711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2016. Formation of chromosomal domains by loop extrusion. Cell Rep 15: 2038–2049. 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funaya S, Aoki F. 2017. Regulation of zygotic gene activation by chromatin structure and epigenetic factors. J Reprod Dev 63: 359–363. 10.1262/jrd.2017-058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fursova NA, Blackledge NP, Nakayama M, Ito S, Koseki Y, Farcas AM, King HW, Koseki H, Klose RJ. 2019. Synergy between variant PRC1 complexes defines polycomb-mediated gene repression. Mol Cell 74: 1020–1036.e8. 10.1016/j.molcel.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Wu K, Liu Z, Yao X, Yuan S, Tao W, Yi L, Yu G, Hou Z, Fan D, et al. 2018a. Chromatin accessibility landscape in human early embryos and its association with evolution. Cell 173: 248–259.e15. 10.1016/j.cell.2018.02.028 [DOI] [PubMed] [Google Scholar]
- Gao R, Wang C, Gao Y, Xiu W, Chen J, Kou X, Zhao Y, Liao Y, Bai D, Qiao Z, et al. 2018b. Inhibition of aberrant DNA re-methylation improves post-implantation development of somatic cell nuclear transfer embryos. Cell Stem Cell 23: 426–435.e5. 10.1016/j.stem.2018.07.017 [DOI] [PubMed] [Google Scholar]
- Gassler J, Brandão HB, Imakaev M, Flyamer IM, Ladstätter S, Bickmore WA, Peters JM, Mirny LA, Tachibana K. 2017. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J 36: 3600–3618. 10.15252/embj.201798083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genet M, Torres-Padilla ME. 2020. The molecular and cellular features of 2-cell-like cells: a reference guide. Development 147: dev189688. 10.1242/dev.189688 [DOI] [PubMed] [Google Scholar]
- Gerdes P, Richardson SR, Mager DL, Faulkner GJ. 2016. Transposable elements in the mammalian embryo: pioneers surviving through stealth and service. Genome Biol 17: 100. 10.1186/s13059-016-0965-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. 2013. The hierarchy of the 3D genome. Mol Cell 49: 773–782. 10.1016/j.molcel.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF. 2006. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 133: 1423–1432. 10.1242/dev.02302 [DOI] [PubMed] [Google Scholar]
- Gold HB, Jung YH, Corces VG. 2018. Not just heads and tails: the complexity of the sperm epigenome. J Biol Chem 293: 13815–13820. 10.1074/jbc.R117.001561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. 2006. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 8: 416–424. 10.1038/ncb1386 [DOI] [PubMed] [Google Scholar]
- Gorkin DU, Leung D, Ren B. 2014. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell 14: 762–775. 10.1016/j.stem.2014.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MVC, Bourc'his D. 2019. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol 20: 590–607. 10.1038/s41580-019-0159-6 [DOI] [PubMed] [Google Scholar]
- Gretarsson KH, Hackett JA. 2020. Dppa2 and Dppa4 counteract de novo methylation to establish a permissive epigenome for development. Nat Struct Mol Biol 27: 706–716. 10.1038/s41594-020-0445-1 [DOI] [PubMed] [Google Scholar]
- Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, et al. 2015. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522: 221–225. 10.1038/nature14308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibert S, Forne T, Weber M. 2012. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res 22: 633–641. 10.1101/gr.130997.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Li X, Liang D, Li T, Zhu P, Guo H, Wu X, Wen L, Gu TP, Hu B, et al. 2014. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15: 447–459. 10.1016/j.stem.2014.08.003 [DOI] [PubMed] [Google Scholar]
- Guo M, Zhang Y, Zhou J, Bi Y, Xu J, Xu C, Kou X, Zhao Y, Li Y, Tu Z, et al. 2019. Precise temporal regulation of Dux is important for embryo development. Cell Res 29: 956–959. 10.1038/s41422-019-0238-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS. 2004. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 6: 117–131. 10.1016/S1534-5807(03)00373-3 [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460: 473–478. 10.1038/nature08162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Low DH, Yi C, Carrell DT, Guccione E, Cairns BR. 2014. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15: 239–253. 10.1016/j.stem.2014.04.006 [DOI] [PubMed] [Google Scholar]
- Handel MA, Schimenti JC. 2010. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 11: 124–136. 10.1038/nrg2723 [DOI] [PubMed] [Google Scholar]
- Hanna CW, Demond H, Kelsey G. 2018a. Epigenetic regulation in development: is the mouse a good model for the human? Hum Reprod Update 24: 556–576. 10.1093/humupd/dmy021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna CW, Taudt A, Huang J, Gahurova L, Kranz A, Andrews S, Dean W, Stewart AF, Colomé-Tatché M, Kelsey G. 2018b. MLL2 conveys transcription-independent H3K4 trimethylation in oocytes. Nat Struct Mol Biol 25: 73–82. 10.1038/s41594-017-0013-5 [DOI] [PubMed] [Google Scholar]
- Hanna CW, Pérez-Palacios R, Gahurova L, Schubert M, Krueger F, Biggins L, Andrews S, Colomé-Tatché M, Bourc'his D, Dean W, et al. 2019. Endogenous retroviral insertions drive non-canonical imprinting in extra-embryonic tissues. Genome Biol 20: 225. 10.1186/s13059-019-1833-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Maehara K, Handa T, Arimura Y, Nogami J, Hayashi-Takanaka Y, Shirahige K, Kurumizaka H, Kimura H, Ohkawa Y. 2019. A chromatin integration labelling method enables epigenomic profiling with lower input. Nat Cell Biol 21: 287–296. 10.1038/s41556-018-0248-3 [DOI] [PubMed] [Google Scholar]
- Harris C, Cloutier M, Trotter M, Hinten M, Gayen S, Du Z, Xie W, Kalantry S. 2019. Conversion of random X-inactivation to imprinted X-inactivation by maternal PRC2. eLife 8: e44258. 10.7554/eLife.44258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka Y, Inoue K, Oikawa M, Kamimura S, Ogonuki N, Kodama EN, Ohkawa Y, Tsukada Y, Ogura A. 2015. Histone chaperone CAF-1 mediates repressive histone modifications to protect preimplantation mouse embryos from endogenous retrotransposons. Proc Natl Acad Sci 112: 14641–14646. 10.1073/pnas.1512775112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. 2017. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 49: 925–934. 10.1038/ng.3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Garruss AS, Gao X, Morgan MA, Cook M, Smith ER, Shilatifard A. 2013. The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat Struct Mol Biol 20: 1093–1097. 10.1038/nsmb.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Gao X, Cao K, Morgan MA, Mas G, Smith ER, Volk AG, Bartom ET, Crispino JD, Di Croce L, et al. 2017. Not all H3K4 methylations are created equal: Mll2/COMPASS dependency in primordial germ cell specification. Mol Cell 65: 460–475.e6. 10.1016/j.molcel.2017.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Tan DEK, Chia G, Tan H, Leong HF, Chen BJ, Lau MS, Tan KYS, Bi X, Yang D, et al. 2020. Maternal factor NELFA drives a 2C-like state in mouse embryonic stem cells. Nat Cell Biol 22: 175–186. 10.1038/s41556-019-0453-8 [DOI] [PubMed] [Google Scholar]
- Huang Y, Kim JK, Do DV, Lee C, Penfold CA, Zylicz JJ, Marioni JC, Hackett JA, Surani MA. 2017. Stella modulates transcriptional and endogenous retrovirus programs during maternal-to-zygotic transition. eLife 6: e22345. 10.7554/eLife.22345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug CB, Grimaldi AG, Kruse K, Vaquerizas JM. 2017. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169: 216–228.e19. 10.1016/j.cell.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Ibrahim AFM, Shen L, Tatham MH, Dickerson D, Prescott AR, Abidi N, Xirodimas DP, Hay RT. 2020. Antibody RING-mediated destruction of endogenous proteins. Mol Cell 79: 155–166.e9. 10.1016/j.molcel.2020.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. 2014. Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat Struct Mol Biol 21: 609–616. 10.1038/nsmb.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Jiang L, Lu F, Suzuki T, Zhang Y. 2017a. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547: 419–424. 10.1038/nature23262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Jiang L, Lu F, Zhang Y. 2017b. Genomic imprinting of Xist by maternal H3K27me3. Genes Dev 31: 1927–1932. 10.1101/gad.304113.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Chen Z, Yin Q, Zhang Y. 2018. Maternal Eed knockout causes loss of H3K27me3 imprinting and random X inactivation in the extraembryonic cells. Genes Dev 32: 1525–1536. 10.1101/gad.318675.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachowicz JW, Bing X, Pontabry J, Bošković A, Rando OJ, Torres-Padilla ME. 2017. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat Genet 49: 1502–1510. 10.1038/ng.3945 [DOI] [PubMed] [Google Scholar]
- Jambhekar A, Dhall A, Shi Y. 2019. Roles and regulation of histone methylation in animal development. Nat Rev Mol Cell Biol 20: 625–641. 10.1038/s41580-019-0151-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangam D, Feschotte C, Betrán E. 2017. Transposable element domestication as an adaptation to evolutionary conflicts. Trends Genet 33: 817–831. 10.1016/j.tig.2017.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Zhang J, Wang JJ, Wang L, Zhang L, Li G, Yang X, Ma X, Sun X, Cai J, et al. 2013. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell 153: 773–784. 10.1016/j.cell.2013.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YH, Sauria MEG, Lyu X, Cheema MS, Ausio J, Taylor J, Corces VG. 2017. Chromatin states in mouse sperm correlate with embryonic and adult regulatory landscapes. Cell Rep 18: 1366–1382. 10.1016/j.celrep.2017.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YH, Kremsky I, Gold HB, Rowley MJ, Punyawai K, Buonanotte A, Lyu X, Bixler BJ, Chan AWS, Corces VG. 2019. Maintenance of CTCF- and transcription factor-mediated interactions from the gametes to the early mouse embryo. Mol Cell 75: 154–171.e5. 10.1016/j.molcel.2019.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaaij LJ, van der Weide RH, Ketting RF, de Wit E. 2018. Systemic loss and gain of chromatin architecture throughout zebrafish development. Cell Rep 24: 1–10.e4. 10.1016/j.celrep.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. 1993. The zebrafish midblastula transition. Development 119: 447–456. 10.1242/dev.119.2.447 [DOI] [PubMed] [Google Scholar]
- Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S. 2019. CUT&tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10: 1930. 10.1038/s41467-019-09982-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Xu Y, Chen X, Feng S, Liu Z, Sun Y, Yao X, Li F, Zhu W, Gao L, et al. 2017. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell 170: 367–381.e20. 10.1016/j.cell.2017.06.029 [DOI] [PubMed] [Google Scholar]
- Kelsey G, Feil R. 2013. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos T R Soc B 368: 20110336. 10.1098/rstb.2011.0336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempfer R, Pombo A. 2020. Methods for mapping 3D chromosome architecture. Nat Rev Genet 21: 207–226. 10.1038/s41576-019-0195-2 [DOI] [PubMed] [Google Scholar]
- Kim J, Zhao H, Dan J, Kim S, Hardikar S, Hollowell D, Lin K, Lu Y, Takata Y, Shen J, et al. 2016. Maternal Setdb1 is required for meiotic progression and preimplantation development in mouse. PLoS Genet 12: e1005970. 10.1371/journal.pgen.1005970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H. 2019. Human cohesin compacts DNA by loop extrusion. Science 366: 1345–1349. 10.1126/science.aaz4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm SL, Shipony Z, Greenleaf WJ. 2019. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 20: 207–220. 10.1038/s41576-018-0089-8 [DOI] [PubMed] [Google Scholar]
- Ko MS. 2016. Zygotic genome activation revisited: looking through the expression and function of Zscan4. Curr Top Dev Biol 120: 103–124. 10.1016/bs.ctdb.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, et al. 2012. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. Plos Genet 8: e1002440. 10.1371/journal.pgen.1002440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. 2013. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502: 472–479. 10.1038/nature12750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Kraft K, Yost KE, Murphy S, Magg A, Long Y, Corces MR, Granja JM, Mundlos S, Cech TR, Boettiger A, et al. 2020. Polycomb-mediated genome architecture enables long-range spreading of H3K27 methylation. bioRxiv 10.1101/2020.1107.1127.223438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lawrence M, Daujat S, Schneider R. 2016. Lateral thinking: how histone modifications regulate gene expression. Trends Genet 32: 42–56. 10.1016/j.tig.2015.10.007 [DOI] [PubMed] [Google Scholar]
- Lee JT. 2005. Regulation of X-chromosome counting by Tsix and Xite sequences. Science 309: 768–771. 10.1126/science.1113673 [DOI] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG. 2005. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem 280: 41725–41731. 10.1074/jbc.M508312200 [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ. 2014. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol 30: 581–613. 10.1146/annurev-cellbio-100913-013027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepikhov K, Walter J. 2004. Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev Biol 4: 12. 10.1186/1471-213X-4-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepikhov K, Zakhartchenko V, Hao R, Yang F, Wrenzycki C, Niemann H, Wolf E, Walter J. 2008. Evidence for conserved DNA and histone H3 methylation reprogramming in mouse, bovine and rabbit zygotes. Epigenetics Chromatin 1: 8. 10.1186/1756-8935-1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Zhang Y. 2014. DNA methylation in mammals. Cold Spring Harb Perspect Biol 6: a019133. 10.1101/cshperspect.a019133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zheng P, Dean J. 2010. Maternal control of early mouse development. Development 137: 859–870. 10.1242/dev.039487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Guo F, Gao Y, Ren Y, Yuan P, Yan L, Li R, Lian Y, Li J, Hu B, et al. 2018a. Single-cell multi-omics sequencing of human early embryos. Nat Cell Biol 20: 847–858. 10.1038/s41556-018-0123-2 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Z, Chen J, Liu W, Lai W, Liu B, Li X, Liu L, Xu S, Dong Q, et al. 2018b. Stella safeguards the oocyte methylome by preventing de novo methylation mediated by DNMT1. Nature 564: 136–140. 10.1038/s41586-018-0751-5 [DOI] [PubMed] [Google Scholar]
- Li F, Wang D, Song R, Cao C, Zhang Z, Wang Y, Li X, Huang J, Liu Q, Hou N, et al. 2020. The asynchronous establishment of chromatin 3D architecture between in vitro fertilized and uniparental preimplantation pig embryos. Genome Biol 21: 203. 10.1186/s13059-020-02095-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. 2014. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell 30: 268–279. 10.1016/j.devcel.2014.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WQ, Liu XY, Wang CF, Gao YW, Gao R, Kou XC, Zhao YH, Li JY, Wu Y, Xiu WC, et al. 2016a. Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell Discov 2: 16010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang C, Liu W, Li J, Li C, Kou X, Chen J, Zhao Y, Gao H, Wang H, et al. 2016b. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537: 558–562. 10.1038/nature19362 [DOI] [PubMed] [Google Scholar]
- Liu X, Wang YZ, Gao YP, Su JM, Zhang JC, Xing XP, Zhou C, Yao KZ, An QL, Zhang Y. 2018a. H3k9 demethylase KDM4E is an epigenetic regulator for bovine embryonic development and a defective factor for nuclear reprogramming. Development 145: dev158261. 10.1242/dev.158261 [DOI] [PubMed] [Google Scholar]
- Liu Z, Cai YJ, Wang Y, Nie YH, Zhang CC, Xu YT, Zhang XT, Lu Y, Wang ZY, Poo MT, et al. 2018b. Cloning of macaque monkeys by somatic cell nuclear transfer. Cell 174: 245–245. 10.1016/j.cell.2018.01.036 [DOI] [PubMed] [Google Scholar]
- Liu B, Xu Q, Wang Q, Feng S, Lai F, Wang P, Zheng F, Xiang Y, Wu J, Nie J, et al. 2020. The landscape of RNA Pol II binding reveals a stepwise transition during ZGA. Nature 587: 139–144. 10.1038/s41586-020-2847-y [DOI] [PubMed] [Google Scholar]
- Loppin B, Bonnefoy E, Anselme C, Laurençon A, Karr TL, Couble P. 2005. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437: 1386–1390. 10.1038/nature04059 [DOI] [PubMed] [Google Scholar]
- Lu F, Liu Y, Inoue A, Suzuki T, Zhao K, Zhang Y. 2016. Establishing chromatin regulatory landscape during mouse preimplantation development. Cell 165: 1375–1388. 10.1016/j.cell.2016.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Wang X, Jiang H, Wang R, Chen J, Chen Y, Xu Q, Cao J, Gong X, Wu J, et al. 2020. Reorganized 3D genome structures support transcriptional regulation in mouse spermatogenesis. iScience 23: 101034. 10.1016/j.isci.2020.101034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. 10.1038/nature11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod D, Clark VH, Bird A. 1999. Absence of genome-wide changes in DNA methylation during development of the zebrafish. Nat Genet 23: 139–140. 10.1038/13767 [DOI] [PubMed] [Google Scholar]
- Majumder S, Miranda M, DePamphilis ML. 1993. Analysis of gene expression in mouse preimplantation embryos demonstrates that the primary role of enhancers is to relieve repression of promoters. EMBO J 12: 1131–1140. 10.1002/j.1460-2075.1993.tb05754.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The polycomb complex PRC2 and its mark in life. Nature 469: 343–349. 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Liu YT, Lu FL, Iwabuchi KA, Shen L, Inoue A, Zhang Y. 2014. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 159: 884–895. 10.1016/j.cell.2014.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Wang H, Jiang L, Lu F, Iwabuchi KA, Wu X, Inoue K, Yang L, Press W, Lee JT, et al. 2018. Loss of H3K27me3 imprinting in somatic cell nuclear transfer embryos disrupts post-implantation development. Cell Stem Cell 23: 343–354.e5. 10.1016/j.stem.2018.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei H, Kozuka C, Hayashi R, Kumon M, Koseki H, Inoue A. 2021. H2AK119ub1 guides maternal inheritance and zygotic deposition of H3K27me3 in mouse embryos. Nat Genet 53: 539–550. 10.1038/s41588-021-00820-3 [DOI] [PubMed] [Google Scholar]
- Misteli T. 2020. The self-organizing genome: principles of genome architecture and function. Cell 183: 28–45. 10.1016/j.cell.2020.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium; Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. 10.1038/nature01262 [DOI] [PubMed] [Google Scholar]
- Murphy PJ, Wu SF, James CR, Wike CL, Cairns BR. 2018. Placeholder nucleosomes underlie germline-to-embryo DNA methylation reprogramming. Cell 172: 993–1006.e13. 10.1016/j.cell.2018.01.022 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Nakagawa M, Ichisaka T, Shiota A, Yamanaka S. 2011. Essential roles of ECAT15-2/Dppa2 in functional lung development. Mol Cell Biol 31: 4366–4378. 10.1128/MCB.05701-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura R, Motai Y, Kumagai M, Wike CL, Nishiyama H, Nakatani Y, Durand NC, Kondo K, Kondo T, Tsukahara T, et al. 2021. CTCF looping is established during gastrulation in medaka embryos. Genome Res 31: 968–980. 10.1101/gr.269951.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashun B, Hill PWS, Smallwood SA, Dharmalingam G, Amouroux R, Clark SJ, Sharma V, Ndjetehe E, Pelczar P, Festenstein RJ, et al. 2015. Continuous histone replacement by Hira is essential for normal transcriptional regulation and de novo DNA methylation during mouse oogenesis. Mol Cell 60: 611–625. 10.1016/j.molcel.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicetto D, Donahue G, Jain T, Peng T, Sidoli S, Sheng L, Montavon T, Becker JS, Grindheim JM, Blahnik K, et al. 2019. H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science 363: 294–297. 10.1126/science.aau0583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothias JY, Miranda M, DePamphilis ML. 1996. Uncoupling of transcription and translation during zygotic gene activation in the mouse. EMBO J 15: 5715–5725. 10.1002/j.1460-2075.1996.tb00955.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA. 2018. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci 115: E6697–E6706. 10.1073/pnas.1717730115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiyama Y, Schuettengruber B, Papadopoulos GL, Chang JM, Cavalli G. 2018. Polycomb-dependent chromatin looping contributes to gene silencing during Drosophila development. Mol Cell 71: 73–88.e5. 10.1016/j.molcel.2018.05.032 [DOI] [PubMed] [Google Scholar]
- Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf JP, Renard JP, Duranthon V, et al. 2011. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472: 370–374. 10.1038/nature09872 [DOI] [PubMed] [Google Scholar]
- Oksuz O, Narendra V, Lee CH, Descostes N, LeRoy G, Raviram R, Blumenberg L, Karch K, Rocha PP, Garcia BA, et al. 2018. Capturing the onset of PRC2-mediated repressive domain formation. Mol Cell 70: 1149–1162.e5. 10.1016/j.molcel.2018.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooga M, Fulka H, Hashimoto S, Suzuki MG, Aoki F. 2016. Analysis of chromatin structure in mouse preimplantation embryos by fluorescent recovery after photobleaching. Epigenetics 11: 85–94. 10.1080/15592294.2015.1136774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oomen ME, Dekker J. 2017. Epigenetic characteristics of the mitotic chromosome in 1D and 3D. Crit Rev Biochem Mol Biol 52: 185–204. 10.1080/10409238.2017.1287160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Plath K. 2013. Epigenetics of reprogramming to induced pluripotency. Cell 152: 1324–1343. 10.1016/j.cell.2013.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. 2013. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 14: 341–356. 10.1038/nrm3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel L, Kang R, Rosenberg SC, Qiu YJ, Raviram R, Chee S, Hu R, Ren B, Cole F, Corbett KD. 2019. Dynamic reorganization of the genome shapes the recombination landscape in meiotic prophase. Nat Struct Mol Biol 26: 164–174. 10.1038/s41594-019-0187-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. 2004. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7: 597–606. 10.1016/j.devcel.2004.09.004 [DOI] [PubMed] [Google Scholar]
- Percharde M, Lin CJ, Yin Y, Guan J, Peixoto GA, Bulut-Karslioglu A, Biechele S, Huang B, Shen X, Ramalho-Santos M. 2018. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 174: 391–405.e19. 10.1016/j.cell.2018.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AHFM, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337. 10.1016/S0092-8674(01)00542-6 [DOI] [PubMed] [Google Scholar]
- Petropoulos S, Edsgärd D, Reinius B, Deng Q, Panula SP, Codeluppi S, Reyes AP, Linnarsson S, Sandberg R, Lanner F. 2016. Single-cell RNA-seq reveals lineage and x chromosome dynamics in human preimplantation embryos. Cell 167: 285. 10.1016/j.cell.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, Dillon N. 2015. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol 16: 245–257. 10.1038/nrm3965 [DOI] [PubMed] [Google Scholar]
- Posfai E, Kunzmann R, Brochard V, Salvaing J, Cabuy E, Roloff TC, Liu ZC, Tardat M, van Lohuizen M, Vidal M, et al. 2012. Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev 26: 920–932. 10.1101/gad.188094.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potok ME, Nix DA, Parnell TJ, Cairns BR. 2013. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 153: 759–772. 10.1016/j.cell.2013.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Santos F, Reik W, Almouzni G, Dean W. 2007. Structural differences in centromeric heterochromatin are spatially reconciled on fertilisation in the mouse zygote. Chromosoma 116: 403–415. 10.1007/s00412-007-0106-8 [DOI] [PubMed] [Google Scholar]
- Prokopuk L, Stringer JM, White CR, Vossen R, White SJ, Cohen ASA, Gibson WT, Western PS. 2018. Loss of maternal EED results in postnatal overgrowth. Clin Epigenetics 10: 95. 10.1186/s13148-018-0526-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschendorf M, Terranova R, Boutsma E, Mao X, Isono K, Brykczynska U, Kolb C, Otte AP, Koseki H, Orkin SH, et al. 2008. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet 40: 411–420. 10.1038/ng.99 [DOI] [PubMed] [Google Scholar]
- Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, et al. 2018. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174: 744–757.e24. 10.1016/j.cell.2018.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, St Hilaire BG, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171: 305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. 2012. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet 46: 21–42. 10.1146/annurev-genet-110711-155621 [DOI] [PubMed] [Google Scholar]
- Reichmann J, Nijmeijer B, Hossain MJ, Eguren M, Schneider I, Politi AZ, Roberti MJ, Hufnagel L, Hiiragi T, Ellenberg J. 2018. Dual-spindle formation in zygotes keeps parental genomes apart in early mammalian embryos. Science 361: 189–193. 10.1126/science.aar7462 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Terrones D, Torres-Padilla ME. 2018. Nimble and ready to mingle: transposon outbursts of early development. Trends Genet 34: 806–820. 10.1016/j.tig.2018.06.006 [DOI] [PubMed] [Google Scholar]
- Rowley MJ, Corces VG. 2018. Organizational principles of 3D genome architecture. Nat Rev Genet 19: 789–800. 10.1038/s41576-018-0060-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan DG, Peng JY, Wang XS, Ouyang Z, Zou QJ, Yang Y, Chen FB, Ge WK, Wu H, Liu ZM, et al. 2018. XIST derepression in active X chromosome hinders pig somatic cell nuclear transfer. Stem Cell Reports 10: 494–508. 10.1016/j.stemcr.2017.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. 2007. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30. 10.1016/j.molcel.2006.12.014 [DOI] [PubMed] [Google Scholar]
- Sachs M, Onodera C, Blaschke K, Ebata KT, Song JS, Ramalho-Santos M. 2013. Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep 3: 1777–1784. 10.1016/j.celrep.2013.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B, Home P, Ray S, Larson M, Paul A, Rajendran G, Behr B, Paul S. 2013. EED and KDM6B coordinate the first mammalian cell lineage commitment to ensure embryo implantation. Mol Cell Biol 33: 2691–2705. 10.1128/MCB.00069-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci 112: E6456–E6465. 10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar A, Lerdrup M, Manaf A, Johansen JV, Gonzalez JM, Borup R, Blanshard R, Klungland A, Hansen K, Andersen CY, et al. 2020. KDM4A regulates the maternal-to-zygotic transition by protecting broad H3K4me3 domains from H3K9me3 invasion in oocytes. Nat Cell Biol 22: 380–388. 10.1038/s41556-020-0494-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla ME. 2010. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol 12: 853–862. 10.1038/ncb2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Hilbert L, Oda H, Wan Y, Heddleston JM, Chew TL, Zaburdaev V, Keller P, Lionnet T, Vastenhouw N, et al. 2019. Histone H3K27 acetylation precedes active transcription during zebrafish zygotic genome activation as revealed by live-cell analysis. Development 146: dev179127. 10.1242/dev.179127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Fraser P. 2019. Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet 20: 437–455. 10.1038/s41576-019-0128-0 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sugar R, Dimond A, Javierre BM, Armstrong H, Mifsud B, Dimitrova E, Matheson L, Tavares-Cadete F, Furlan-Magaril M, et al. 2015. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat Genet 47: 1179–1186. 10.1038/ng.3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DI, Jayashankar K, Douglas KC, Thirkill TL, York D, Dickinson PJ, Williams LE, Samollow PB, Ross PJ, Bannasch DL, et al. 2015. Early developmental and evolutionary origins of gene body DNA methylation patterns in mammalian placentas. PLoS Genet 11: e1005442. 10.1371/journal.pgen.1005442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. 2017. Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171: 34–57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Schultz RM. 2002. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update 8: 323–331. 10.1093/humupd/8.4.323 [DOI] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering CH, Mirny L, et al. 2017. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551: 51–56. 10.1038/nature24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. 2012. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 48: 849–862. 10.1016/j.molcel.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendžikaite G, Kelsey G. 2019. The role and mechanisms of DNA methylation in the oocyte. Essays Biochem 63: 691–705. 10.1042/EBC20190043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha QQ, Dai XX, Jiang JC, Yu C, Jiang Y, Liu J, Ou XH, Zhang SY, Fan HY. 2018. CFP1 coordinates histone H3 lysine-4 trimethylation and meiotic cell cycle progression in mouse oocytes. Nat Commun 9: 3477. 10.1038/s41467-018-05930-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Inoue A, He J, Liu Y, Lu F, Zhang Y. 2014. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell 15: 459–471. 10.1016/j.stem.2014.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa T, Takagi T, Tsukamoto D, Tomaru C, Huynh LM, Sivaraman P, Kumarevel T, Inoue K, Nakato R, Katou Y, et al. 2014. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell 14: 217–227. 10.1016/j.stem.2013.12.015 [DOI] [PubMed] [Google Scholar]
- Shirane K, Toh H, Kobayashi H, Miura F, Chiba H, Ito T, Kono T, Sasaki H. 2013. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. Plos Genet 9: e1003439. 10.1371/journal.pgen.1003439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirane K, Miura F, Ito T, Lorincz MC. 2020. NSD1-deposited h3k36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nat Genet 52: 1088–1098. 10.1038/s41588-020-0689-z [DOI] [PubMed] [Google Scholar]
- Silva MCC, Powell S, Ladstätter S, Gassler J, Stocsits R, Tedeschi A, Peters JM, Tachibana K. 2020. Wapl releases Scc1-cohesin and regulates chromosome structure and segregation in mouse oocytes. J Cell Biol 219: e201906100. 10.1083/jcb.201906100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6: e21856. 10.7554/eLife.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G. 2011. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet 43: 811–814. 10.1038/ng.864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. 2013. DNA methylation: roles in mammalian development. Nat Rev Genet 14: 204–220. 10.1038/nrg3354 [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu HC, Gnirke A, Regev A, Meissner A. 2012. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484: 339–344. 10.1038/nature10960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Shi J, Gu H, Donaghey J, Clement K, Cacchiarelli D, Gnirke A, Michor F, Meissner A. 2017. Epigenetic restriction of extraembryonic lineages mirrors the somatic transition to cancer. Nature 549: 543–547. 10.1038/nature23891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Jiang Z, Susor A, Ming H, Tait J, Conti M, Lin C-J. 2020. The H3.3 chaperone Hira complex orchestrates oocyte developmental competence. bioRxiv 10.1101/2020.1105.1125.114124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Nady N, Arora N, Hsieh LJ, Swigut T, Narlikar GJ, Wossidlo M, Wysocka J. 2020. Zscan4 binds nucleosomal microsatellite DNA and protects mouse two-cell embryos from DNA damage. Sci Adv 6: eaaz9115. 10.1126/sciadv.aaz9115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart KR, Veselovska L, Kelsey G. 2016. Establishment and functions of DNA methylation in the germline. Epigenomics 8: 1399–1413. 10.2217/epi-2016-0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435. 10.1038/ncb1663 [DOI] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Anger M, Bernstein E, Hannon GJ, Schultz RM. 2004. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev Biol 269: 276–285. 10.1016/j.ydbio.2004.01.028 [DOI] [PubMed] [Google Scholar]
- Tachibana-Konwalski K, Godwin J, van der Weyden L, Champion L, Kudo NR, Adams DJ, Nasmyth K. 2010. Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes Dev 24: 2505–2516. 10.1101/gad.605910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburri S, Lavarone E, Fernández-Pérez D, Conway E, Zanotti M, Manganaro D, Pasini D. 2020. Histone H2AK119 mono-ubiquitination is essential for polycomb-mediated transcriptional repression. Mol Cell 77: 840–856.e5. 10.1016/j.molcel.2019.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Albert M, Kunzmann R, Liu Z, Kaustov L, Thierry R, Duan S, Brykczynska U, Arrowsmith CH, Peters AH. 2015. Cbx2 targets PRC1 to constitutive heterochromatin in mouse zygotes in a parent-of-origin-dependent manner. Mol Cell 58: 157–171. 10.1016/j.molcel.2015.02.013 [DOI] [PubMed] [Google Scholar]
- Tee WW, Shen SS, Oksuz O, Narendra V, Reinberg D. 2014. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell 156: 678–690. 10.1016/j.cell.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesařík J, Kopečný V, Plachot M, Mandelbaum J. 1987. Ultrastructural and autoradiographic observations on multinucleated blastomeres of human cleaving embryos obtained by in-vitro fertilization. Hum Reprod 2: 127–136. 10.1093/oxfordjournals.humrep.a136496 [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. 2006. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol 50: 455–461. [DOI] [PubMed] [Google Scholar]
- Tucci V, Isles AR, Kelsey G, Ferguson-Smith AC; Erice Imprinting Group. 2019. Genomic imprinting and physiological processes in mammals. Cell 176: 952–965. 10.1016/j.cell.2019.01.043 [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Dieker JW, Derijck AA, Muller S, Berden JH, Braat DD, van der Vlag J, de Boer P. 2005. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech Dev 122: 1008–1022. 10.1016/j.mod.2005.04.009 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Belmont AS. 2017. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169: 780–791. 10.1016/j.cell.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vara C, Paytuví-Gallart A, Cuartero Y, Le Dily F, Garcia F, Salvà-Castro J, Gómez HL, Julià E, Moutinho C, Aiese Cigliano R, et al. 2019. Three-dimensional genomic structure and cohesin occupancy correlate with transcriptional activity during spermatogenesis. Cell Rep 28: 352–367. e359. 10.1016/j.celrep.2019.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Schier AF. 2012. Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol 24: 374–386. 10.1016/j.ceb.2012.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. 2010. Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464: 922–926. 10.1038/nature08866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselovska L, Smallwood SA, Saadeh H, Stewart KR, Krueger F, Maupetit-Méhouas S, Arnaud P, Tomizawa S, Andrews S, Kelsey G. 2015. Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol 16: 209. 10.1186/s13059-015-0769-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vian L, Pękowska A, Rao SSP, Kieffer-Kwon KR, Jung S, Baranello L, Huang SC, El Khattabi L, Dose M, Pruett N, et al. 2018. The energetics and physiological impact of cohesin extrusion. Cell 173: 1165–1178.e20. 10.1016/j.cell.2018.03.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JJ, Huang Y, Chen PY, Feng S, Calvopiña JH, Nee K, Lee SA, Le T, Yoon AJ, Faull K, et al. 2013. Stage-specific roles for tet1 and tet2 in DNA demethylation in primordial germ cells. Cell Stem Cell 12: 470–478. 10.1016/j.stem.2013.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. 2012. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 13: 115–126. 10.1038/nrm3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser CB, Lipshitz HD. 2011. Transcript clearance during the maternal-to-zygotic transition. Curr Opin Genet Dev 21: 431–443. 10.1016/j.gde.2011.03.003 [DOI] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20: 116–117. 10.1038/2413 [DOI] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. 2004. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell 6: 133–144. 10.1016/S1534-5807(03)00404-0 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang J, Duan J, Gao X, Zhu W, Lu X, Yang L, Zhang J, Li G, Ci W, et al. 2014. Programming and inheritance of parental DNA methylomes in mammals. Cell 157: 979–991. 10.1016/j.cell.2014.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CF, Liu XY, Gao YW, Yang L, Li C, Liu WQ, Chen C, Kou XC, Zhao YH, Chen JY, et al. 2018. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat Cell Biol 20: 620–631. 10.1038/s41556-018-0093-4 [DOI] [PubMed] [Google Scholar]
- Wang Q, Xiong H, Ai S, Yu X, Liu Y, Zhang J, He A. 2019a. CoBATCH for high-throughput single-cell epigenomic profiling. Mol Cell 76: 206–216.e7. 10.1016/j.molcel.2019.07.015 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang H, Zhang Y, Du Z, Si W, Fan S, Qin D, Wang M, Duan Y, Li L, et al. 2019b. Reprogramming of meiotic chromatin architecture during spermatogenesis. Mol Cell 73: 547–561.e6. 10.1016/j.molcel.2018.11.019 [DOI] [PubMed] [Google Scholar]
- Wang LY, Li ZK, Wang LB, Liu C, Sun XH, Feng GH, Wang JQ, Li YF, Qiao LY, Nie H, et al. 2020. Overcoming intrinsic H3K27me3 imprinting barriers improves post-implantation development after somatic cell nuclear transfer. Cell Stem Cell 27: 315–325.e5. 10.1016/j.stem.2020.05.014 [DOI] [PubMed] [Google Scholar]
- Weinberg DN, Papillon-Cavanagh S, Chen HF, Yue Y, Chen X, Rajagopalan KN, Horth C, McGuire JT, Xu XJ, Nikbakht H, et al. 2019. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 573: 281–286. 10.1038/s41586-019-1534-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiddon JL, Langford AT, Wong CJ, Zhong JW, Tapscott SJ. 2017. Conservation and innovation in the DUX4-family gene network. Nat Genet 49: 935–940. 10.1038/ng.3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, DePamphilis ML. 1991. Regulation of gene expression in preimplantation mouse embryos: effects of the zygotic clock and the first mitosis on promoter and enhancer activities. Dev Biol 147: 403–414. 10.1016/0012-1606(91)90298-H [DOI] [PubMed] [Google Scholar]
- Wike CL, Guo Y, Tan M, Nakamura R, Shaw DK, Díaz N, Whittaker-Tademy AF, Durand NC, Lieberman Aiden E, Vaquerizas JM, et al. 2021. Chromatin architecture transitions from zebrafish sperm through early embryogenesis. Genome Res 31: 981–994. 10.1101/gr.269860.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongtawan T, Taylor JE, Lawson KA, Wilmut I, Pennings S. 2011. Histone H4K20me3 and HP1α are late heterochromatin markers in development, but present in undifferentiated embryonic stem cells. J Cell Sci 124: 1878–1890. 10.1242/jcs.080721 [DOI] [PubMed] [Google Scholar]
- Wozniak GG, Strahl BD. 2014. Hitting the “mark”: interpreting lysine methylation in the context of active transcription. Bba-Gene Regul Mech 1839: 1353–1361. [DOI] [PubMed] [Google Scholar]
- Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, et al. 2016. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534: 652–657. 10.1038/nature18606 [DOI] [PubMed] [Google Scholar]
- Wu J, Xu J, Liu B, Yao G, Wang P, Lin Z, Huang B, Wang X, Li T, Shi S, et al. 2018. Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature 557: 256–260. 10.1038/s41586-018-0080-8 [DOI] [PubMed] [Google Scholar]
- Wu K, Liu H, Wang Y, He J, Xu S, Chen Y, Kuang J, Liu J, Guo L, Li D, et al. 2020. SETDB1-mediated cell fate transition between 2C-like and pluripotent states. Cell Rep 30: 25–36.e6. 10.1016/j.celrep.2019.12.010 [DOI] [PubMed] [Google Scholar]
- Xia W, Xie W. 2020. Rebooting the epigenomes during mammalian early embryogenesis. Stem Cell Reports 15: 1158–1175. 10.1016/j.stemcr.2020.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Xu J, Yu G, Yao G, Xu K, Ma X, Zhang N, Liu B, Li T, Lin Z, et al. 2019. Resetting histone modifications during human parental-to-zygotic transition. Science 365: 353–360. 10.1126/science.aaw5118 [DOI] [PubMed] [Google Scholar]
- Xiang Y, Zhang Y, Xu Q, Zhou C, Liu B, Du Z, Zhang K, Zhang B, Wang X, Gayen S, et al. 2020. Epigenomic analysis of gastrulation identifies a unique chromatin state for primed pluripotency. Nat Genet 52: 95–105. 10.1038/s41588-019-0545-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu QH, Xie W. 2018. Epigenome in early mammalian development: inheritance, reprogramming and establishment. Trends Cell Biol 28: 237–253. 10.1016/j.tcb.2017.10.008 [DOI] [PubMed] [Google Scholar]
- Xu Q, Xiang Y, Wang Q, Wang L, Brind'Amour J, Bogutz AB, Zhang Y, Zhang B, Yu G, Xia W, et al. 2019. SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nat Genet 51: 844–856. 10.1038/s41588-019-0398-7 [DOI] [PubMed] [Google Scholar]
- Yan R, Gu C, You D, Huang Z, Qian J, Yang Q, Cheng X, Zhang L, Wang H, Wang P, et al. 2021. Decoding dynamic epigenetic landscapes in human oocytes using single-cell multi-omics sequencing. Cell Stem Cell S1934-5909: 00169. [DOI] [PubMed] [Google Scholar]
- Yang F, Huang X, Zang R, Chen J, Fidalgo M, Sanchez-Priego C, Yang J, Caichen A, Ma F, Macfarlan T, et al. 2020. DUX-miR-344-ZMYM2-mediated activation of MERVL LTRs induces a totipotent 2C-like state. Cell Stem Cell 26: 234–250.e7. 10.1016/j.stem.2020.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Fan X, Sha QQ, Wang HH, Li BT, Dai XX, Shen L, Liu J, Wang L, Liu K, et al. 2017. CFP1 regulates histone H3K4 trimethylation and developmental potential in mouse oocytes. Cell Rep 20: 1161–1172. 10.1016/j.celrep.2017.07.011 [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. 2004. Transcript profiling during preimplantation mouse development. Dev Biol 272: 483–496. 10.1016/j.ydbio.2004.05.018 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Vastenhouw NL, Feng J, Fu K, Wang C, Ge Y, Pauli A, van Hummelen P, Schier AF, Liu XS. 2014. Canonical nucleosome organization at promoters forms during genome activation. Genome Res 24: 260–266. 10.1101/gr.157750.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, Ming J, Wu X, Zhang Y, Xu Q, et al. 2016. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537: 553–557. 10.1038/nature19361 [DOI] [PubMed] [Google Scholar]
- Zhang B, Wu X, Zhang W, Shen W, Sun Q, Liu K, Zhang Y, Wang Q, Li Y, Meng A, et al. 2018a. Widespread enhancer dememorization and promoter priming during parental-to-zygotic transition. Mol Cell 72: 673–686.e6. 10.1016/j.molcel.2018.10.017 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiang Y, Yin Q, Du Z, Peng X, Wang Q, Fidalgo M, Xia W, Li Y, Zhao ZA, et al. 2018b. Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat Genet 50: 96–105. 10.1038/s41588-017-0003-x [DOI] [PubMed] [Google Scholar]
- Zhang W, Chen F, Chen R, Xie D, Yang J, Zhao X, Guo R, Zhang Y, Shen Y, Goke J, et al. 2019a. Zscan4c activates endogenous retrovirus MERVL and cleavage embryo genes. Nucleic Acids Res 47: 8485–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Chen Z, Yin Q, Zhang D, Racowsky C, Zhang Y. 2019b. Maternal-biased H3K27me3 correlates with paternal-specific gene expression in the human morula. Genes Dev 33: 382–387. 10.1101/gad.323105.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Wu DY, Zheng H, Wang Y, Sun QR, Liu X, Wang LY, Xiong WJ, Wang Q, Rhodes JDP, et al. 2020. Analysis of genome architecture during SCNT reveals a role of cohesin in impeding minor ZGA. Mol Cell 79: 234–250.e9. 10.1016/j.molcel.2020.06.001 [DOI] [PubMed] [Google Scholar]
- Zheng H, Xie W. 2019. The role of 3D genome organization in development and cell differentiation. Nat Rev Mol Cell Biol 20: 535–550. 10.1038/s41580-019-0132-4 [DOI] [PubMed] [Google Scholar]
- Zheng H, Huang B, Zhang B, Xiang Y, Du Z, Xu Q, Li Y, Wang Q, Ma J, Peng X, et al. 2016. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol Cell 63: 1066–1079. 10.1016/j.molcel.2016.08.032 [DOI] [PubMed] [Google Scholar]
- Zhou LQ, Dean J. 2015. Reprogramming the genome to totipotency in mouse embryos. Trends Cell Biol 25: 82–91. 10.1016/j.tcb.2014.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Wang R, Yuan P, Ren Y, Mao Y, Li R, Lian Y, Li J, Wen L, Yan L, et al. 2019. Reconstituting the transcriptome and DNA methylome landscapes of human implantation. Nature 572: 660–664. 10.1038/s41586-019-1500-0 [DOI] [PubMed] [Google Scholar]
- Zhu W, Xu X, Wang X, Liu J. 2019. Reprogramming histone modification patterns to coordinate gene expression in early zebrafish embryos. BMC Genomics 20: 248. 10.1186/s12864-019-5611-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Yu J, Rong Y, Wu Y-W, Li Y, Zhang L, Pan Y, Fan H-Y, Shen L. 2021. Genomewide decoupling of H2AK119ub1 and H3K27me3 in early mouse development. Sci Bull 10.1016/j.scib.2021.06.010 [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. 2015. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 7: a016626. 10.1101/cshperspect.a016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccotti M, Piccinelli A, Rossi PG, Garagna S, Redi CA. 1995. Chromatin organization during mouse oocyte growth. Mol Reprod Dev 41: 479–485. 10.1002/mrd.1080410410 [DOI] [PubMed] [Google Scholar]