Abstract
The purpose of this study is to investigate hyperspectral microscopic imaging and deep learning methods for automatic detection of head and neck squamous cell carcinoma (SCC) on histologic slides. Hyperspectral imaging (HSI) cubes were acquired from pathologic slides of 18 patients with SCC of the larynx, hypopharynx, and buccal mucosa. An Inception-based two-dimensional convolutional neural network (CNN) was trained and validated for the HSI data. The automatic deep learning method was tested with independent data of human patients. This study demonstrated the feasibility of using hyperspectral microscopic imaging and deep learning classification to aid pathologists in detecting SCC on histologic slides.
Keywords: Hyperspectral microscopic imaging, head and neck cancer, histology, digital pathology, convolutional neural network, deep learning, classification
1. PURPOSE
Head and neck cancer is the sixth most common cancer worldwide [1-3]. Squamous cell carcinoma (SCC) takes more than 90% of the cancers of the upper aerodigestive tract of the head and neck region [2]. Surgical resection has long been the major and standard treatment approach for head and neck SCC [4, 5]. During tumor resection surgery, surgeons rely on intraoperative pathologist consultation (IPC) using frozen section microscopic analysis to determine the surgical margin . However, IPC is a time-consuming task, as it requires skilled pathologists to manually examine the morphological features of the specimens, and it is subject to error of pathologists’ histological interpretation [6-11]. A fast and automatics method for cancer detection in the histological slides can facilitate this process and potentially improve the surgical outcome.
Computer-aided pathology (CAP) has been an emerging trend, which aims to improve the reproducibility and objectivity of pathological diagnosis as well as to save time in the routine examination [12]. Machine learning techniques, particularly deep learning models, have been playing an important role in CAP [13]. Many studies of deep learning-based cancer detection in whole-slide digitalized histological images have been investigated, and most of them were carried out in RGB images. Halicek et al. [14, 15] used a patch-based Inception-v4 convolutional neural network (CNN) for head and neck cancer detection and localization in digitalized whole slide image (WSI) of hematoxylin and eosin (H&E) stained histological slides. The network achieved an AUC of 0.916 for SCC and 0.954 for thyroid cancer. Wang et al. [16] proposed using a transfer learning CNN and patch aggregation strategy for colorectal cancer diagnosis in weakly labeled pathological images, with a resulted average AUC of 0.981. Sitnik et al. [17] employed UNet++ architecture for pixel-level detection of colon cancer metastasis in the liver and achieved 0.81 balanced accuracy. Mavuduru et al [18] used U-Net architecture for head and neck SCC detection in digitalized WSI and achieved an average AUC of 0.89.
Hyperspectral imaging is an optical imaging technique that has been increasingly used in biomedical applications. It captures the spatial and spectral information of the imaged tissue, revealing the chemical composition and morphological features in a single image modality. Therefore, it is a promising technology to aid the histopathological analysis of tissue samples. Hyperspectral microscopic imaging (HSMI) has been investigated in some recent studies. Ortega et al. [19] implemented automatic breast cancer cell detection in hyperspectral histologic images with an average testing AUC of 0.90. The comparison between the classification results using HSI and RGB suggested that HSI outperforms RGB. Nakaya et al. [20] used a support vector machine classifier and the average spectra of nuclei extracted from hyperspectral images for colon cancer detection. Xu et al. (Xu 2020, microalgae) used principal components analysis (PCA) and peak ratio algorithms for dimensionality reduction and feature extraction, followed by a support vector machine model for the detection of microalgae. Ishikawa et al.[21] proposed a pattern recognition method named hyperspectral analysis of pathological slides based on stain spectrum (HAPSS) to process the spectra of the histologic hyperspectral images and used an SVM classifier for pancreatic tumor nuclei detection, which obtained a maximal accuracy of 0.94. However, many of these studies were based on a nucleus level, and they were usually coupled with feature-based machine learning approaches for image analysis.
Our previous study investigated the feasibility and usefulness of HSI for head and neck SCC nuclei detection in histologic slides [22]. We compared the classification performance of a CNN trained with HSI and pseudo-RGB patches of the nuclei, respectively. The CNN trained with HSI got an average AUC of 0.94 and an average accuracy of 0.82, which were slightly higher than the AUC and accuracy using RGB. The results demonstrate that the morphological features of nuclei play a dominant role in classification and the extra spectral information in hyperspectral images is able to improve the diagnostic outcome. The study proved the advantage of utilizing HSI as an automated tool in pathology since it can potentially save time and provide an accurate diagnosis with both the spatial and spectral information in the image. However, this method mainly works in the regions where a large number of nuclei exist, such as the cancer nest. It becomes less efficient when applied to the whole slide.
In this study, we investigate the feasibility of using patch-based CNN for the detection of head and neck SCC in the whole hyperspectral images of H&E stained histologic slides. Different from the previous study that merely extracted patches of nuclei, this work uses patches generated from the whole image, thus the morphological and spectral information of the whole imaged area is utilized. We also compared the classification performance using HSI and RGB images.
2. METHODS
2.1. Histologic Slides from Head and Neck SCC Patients
The histologic slides used in this study are a subset of our head and neck cancer dataset, which has been reported in our previous studies [15]. Eighteen histologic slides of larynx, hypopharynx, and buccal mucosa from 18 different head and neck cancer (SCC, HPV-negative) patients were utilized. The tissue of each slide was resected at the tumor-normal margin, containing both cancerous and normal tissue. The slides were digitalized at 40× magnification, and the cancerous regions in the digitalized histology images were manually annotated by a board-certificated pathologist, which were used as the ground truth.
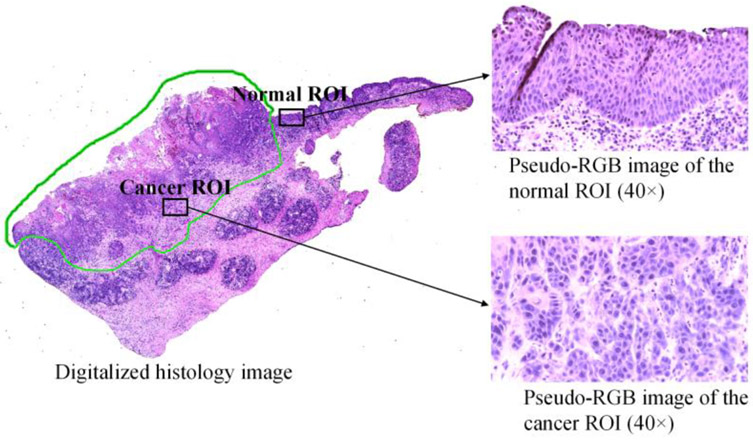
Within the annotated cancerous region of each slide, we carefully selected several regions of interest (ROIs) to scan. Considering the possible inter-pathologist variation of histologic diagnosis, we imaged the tissue that is away from the edge of the annotation to avoid any interface area. As nuclei carry important cancer-related information, the imaged regions should contain adequate nuclei. Therefore, all selected cancerous ROIs were at or close to the cancer nest, where a mass of cells extends to the surrounding area of cancerous growth. For the normal ROIs, we selected from the 2nd to 4th layer of healthy stratified squamous epithelium, because that is where the SCC mostly origin from. For each slide, we chose at least 3 regions of interest for cancerous tissue and 3 ROIs for normal tissue. For the two slides that did not contain any healthy epithelium, we only selected and imaged cancerous ROIs for them. In total, we collected 121 hyperspectral images, where 70 are cancerous and 51 are normal. Each image corresponds to one selected ROI. Figure 1 shows an example of a histologic slide of tumor-normal margin tissue as well as the normal and cancerous ROIs selected from the slide. The image of the whole slide on the left is the digitalized histology image, the green annotation indicates the cancerous region in the slide; the two pseudo-RGB images of the selected ROIs were synthesized from the acquired hyperspectral images using a customized transformation function, which will be described in Section 2.2.
Figure 1.
Selection of the regions of interest in the histologic slide for quantitative testing: digitalized histology image of the whole slide as well as synthesized-RGB images of the selected normal and cancerous ROIs (Imaged at 40×).
2.2. Hyperspectral images and data preparation
The H&E stained histologic slides were imaged using a customized hyperspectral microscopic imaging system with an objective lens of 40× magnification. The system contained an optical microscope, a customized compact hyperspectral camera, and a high-precision motorized stage, as presented in our previous paper [22]. The camera could scan the field of view (FO V) through the translation of the imaging sensor inside of the camera, thus no relative motion between the camera and the slide was needed. The motorized stage in our system, different from the one that is usually employed in a line-scanning hyperspectral microscopic system, was used to carefully select the ROI in the slide. Although we did not focus on acquiring hyperspectral WSIs of the slides, the developed system was able to fulfill that by capturing images of different regions and stitching them to make a WSI. The light source was a 100-Watt halogen lamp that was installed with the microscope. The HSI camera acquired hyperspectral images in the visible wavelength range from 467 nm to 900 nm. The image size was 3600 pixel × 2048 pixel × 146 bands with a spatial resolution of approximately 139 nm/pixel. The spatial size of the field of view was about 500 × 284 μm. However, we found that the NIR wavelength bands in the images carried some noise and the spectral difference between cancerous tissue and normal tissue was less distinctive compared to the visible wavelength bands, so we only used the 78 bands from 467 nm to 720 nm.
Because of the slight variance of thickness in the histological slide, the microscope needed to be focused for each ROI. After the image acquisition of each ROI, a white reference image of a blank area on the slide was captured immediately with the same illumination setting and focus. The dark reference image was acquired automatically by the camera along with the acquisition of each hyperspectral image. All hyperspectral images were calibrated with the corresponding white and dark reference images to obtain the normalized transmittance of the slide, as Equation (1) shows.
| (1) |
where Transmittance (λ) is the normalized transmittance for wavelength λ, Iraw (λ) is the intensity value in the raw hyperspectral image, Iwhite(λ) and Idark(λ) are the intensity values in the white and dark reference images, respectively.
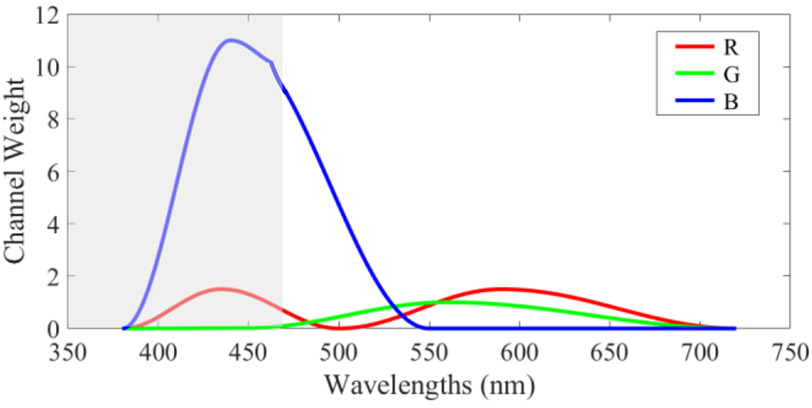
After calibration, we synthesized pseudo-RGB images from hyperspectral images for better visualization and data selection. We utilized a series of cosine functions to make up the HSI-to-RGB transformation function, which had a similar shape with the spectral response of human eye perception to colors [23], as Figure 2 shows. Due to the limited wavelength range of our hyperspectral camera, the spectral information of the slide from 380 to 470 nm was unavailable. In order to balance the three channels, we increased the channel weight of red (R) within 380-500 nm, and the channel weight of blue (B) within 380-550 nm. The colors that we see through our eyes are influenced by both the color perception of the human eye and the spectral signature of the light source. However, the image calibration shown by equation (1) removed the influence of light source from the hyperspectral images. Therefore, in order to have the synthesized RGB images as real as possible, we took the light source into consideration and slightly increased the channel weight of red within 500-720 nm, because the irradiance of the halogen light source is higher in the red-color wavelength range. After applying tins customized transformation, the image was multiplied with a constant of 2 to adjust the brightness.
Figure 2.
Customized transformation to synthesize a pseudo-RGB image from a hyperspectral image, modified from the human eye spectral response for red (R), green (G), and blue (B). The grey area shows the unavailable wavelength range of our hyperspectral camera.
Then, we generated patches from all hyperspectral images using a sliding window of 101 pixels. To ensure that the patches contain enough histologic features for classification, we set the patch size to 404×404×78. Each pair of adjacent patches shared ¾ overlapped regions. Then, all patches were 4 times down-sampled in the spatial dimensions to 101×101×78, corresponding to a 10× magnification. It is worth noting that we also tried using a smaller patch size of 51×51 pixels (after down-sampling) in order to have more patches for training, but the results turned out to be less satisfying. The same sliding window was applied to the pseudo-RGB images to generate RGB patches.
Because the spatial dimension of the hyperspectral images was 3600×2048, each image should be cut into 544 patches in total. However, patches from the edge of tissue contained much blank background pixels, and including them in the dataset would reduce the efficiency of classification. Therefore, these patches were removed. Since the background pixels in our synthesized-RGB images were saturated, and the images were in double-precision format, the sum of three color channels (R, G, and B) of these pixels should be no smaller than 3. Thus, we calculated the number of the background pixels in each patch by setting a threshold of 3 to the sum of three color channels of the RGB patch. If the number of background pixels was more than 50% of the total pixels (10201 pixels) in an RGB patch, both the HSI patch and the RGB patch were removed.
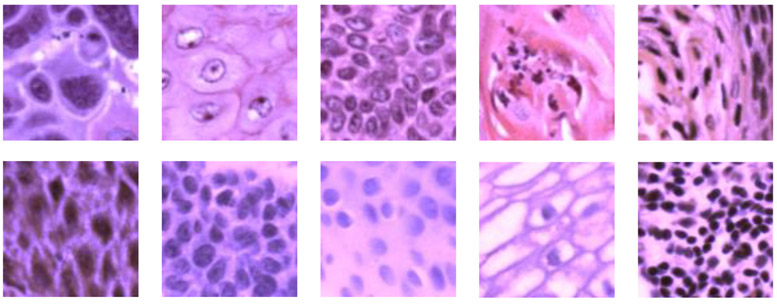
Figure 3 shows some of the extracted cancerous and normal patches in pseudo-RGB. Some typical histological features of SCC could be observed in the cancerous patches, including enlarged nuclei, keratinizing SCC, basaloid SCC, and regions with chronic inflammation. Because the normal ROIs were selected from the stratified epithelium, most normal patches only show the features of different layers in the epithelium and the region under the epithelium with inflammation. The number of images and patches generated from each patient are summarized in Table 1.
Figure 3.
Pseudo-RGB histological patches (101 × 101 pixels) showing anatomical diversity. First row: Patches extracted from cancerous ROIs with various histological features. Second row: Patches generated from images of normal ROIs, showing features at different layers of the stratified epithelium as well as the region under epithelium with inflammation.
Table 1.
Summary of the hyperspectral histologic dataset.
| Patient # |
Organ | Number of images |
Number of patches |
|||
|---|---|---|---|---|---|---|
| Cancerous | Normal | Cancerous | Normal | Total | ||
| 62 | Larynx | 3 | 3 | 1632 | 968 | 2600 |
| 68 | Hypopharynx | 4 | 4 | 2021 | 1939 | 3960 |
| 74 | Larynx | 3 | 3 | 1632 | 1618 | 3250 |
| 110 | Larynx | 5 | 0 | 2720 | 0 | 2720 |
| 120 | Buccal mucosa | 6 | 5 | 3104 | 2696 | 5800 |
| 127 | Larynx | 4 | 3 | 2157 | 1443 | 3600 |
| 134 | Larynx | 3 | 3 | 1088 | 712 | 1800 |
| 137 | Larynx | 3 | 3 | 1632 | 788 | 2420 |
| 146 | Buccal mucosa | 7 | 3 | 3808 | 1432 | 5240 |
| 149 | Buccal mucosa | 3 | 0 | 1080 | 0 | 1080 |
| 154 | Larynx | 3 | 3 | 1080 | 1602 | 2682 |
| 161 | Larynx | 4 | 3 | 2176 | 1024 | 3200 |
| 166 | Larynx | 4 | 3 | 2136 | 1624 | 3760 |
| 172 | Larynx | 5 | 3 | 2718 | 1442 | 4160 |
| 174 | Larynx | 4 | 3 | 2176 | 1564 | 3740 |
| 184 | Larynx | 3 | 3 | 1605 | 1625 | 3230 |
| 187 | Larynx | 3 | 3 | 1632 | 1308 | 2940 |
| 188 | larynx | 3 | 3 | 1632 | 948 | 2580 |
| Total | 70 | 51 | 36029 | 22733 | 58762 | |
2.3. Convolutional neural network classification
The generated patches from hyperspectral images were used to train, validate and test an Inception-based two-dimensional CNN. The CNN architecture was modified from the original Inceptio-v4 network [24] to be adapted to our patch size. Each convolutional layer in the network was followed by a BatchNormalization layer [25] with a momentum of 0.997 as well as ReLU activation. A 40% dropout was applied for each convolutional layer. The CNN architecture and the output size of each layer/block are shown in Table 2.
Table 2.
CNN architecture.
| Layer/Block | Output size |
|---|---|
| Input | 101×101×78 |
| Conv2D, ‘valid’ | 50×50×80 |
| Conv2D, ‘valid’ | 48×48×86 |
| Conv2D, ‘valid’ | 23×23×92 |
| Inception A block ×4 | 23×23×384 |
| Reduction A block | 11×11×1024 |
| Inception B block ×7 | 11×11×1024 |
| Reduction B block | 5×5×1536 |
| Inception C block ×3 | 5×5×1536 |
| Average Pool | 1×1×1536 |
| Flatten | 1536 |
| Dense (‘sigmoid’) | 1 |
The CNN was implemented using Keras [26] with a Tensorflow backend on a Titan XP NVIDIA GPU. The optimizer was Adadelta [27] with an initial learning rate of 1. The network was trained with a batch size of 16. The activation function of the output layer was sigmoid, and the loss function was binary cross-entropy. During network training, we used the callback function to monitor the validation accuracy and saved every checkpoint with improved validation accuracy. After training, we used the checkpoint that had the highest validation accuracy to validate and test on the whole image.
2.4. Data partition
In this work, all image patches were split into training, validation, and testing groups to evaluate the network. To evaluate the proposed method for real clinical application, it is optimal to test the network on as many independent patients as possible. However, due to the small size of our dataset, leaving many patients out for testing might cause the inadequacy of training data. Finally, we decided to carried out 6-fold cross-validation and then test the network on an independent group of 3 different patients.
Except for the three testing patients, all other patients have been used for training in different folds. Because the validation data should have both classes of labels (tumor and normal), 12 patients were divided into 6 groups and took turns to be used for validation in 6 different folds. Patient #110 and #149 were excluded from being used as validation data because of the absence of normal images, while #146 was not used due to the extremely unbalanced label classes. We randomly included 2 patients for validation in each fold to avoid bias to one specific patient, but for patient #120 who has the most images, we manually selected patient #137 with a relatively small number of image patches to be included in the same fold. The three testing patients (#74, #154, #172) were also randomly selected. The data of one patient has never been used in the training, validation, or testing group at the same time.
2.5. Post-processing
During the training process, we used the image patches as described in section 2.2 for network training and validation accuracy monitoring. After the training was done and the checkpoint was selected, we generated new validation patches and testing patches of 404×404×78 from the whole images in the validation group and testing group using a sliding window of 204 pixels. The new patches were also reshaped to 101×101×78. Each image could generate 144 patches in total. All patches were used for classification regardless of how much blank area was included, in order to evaluate the classification performance in whole images. After classification, we reconstructed the probability map of each image. For the overlapped area between adjacent patches, we calculated the average probability. The optimal threshold was determined according to the receiver operating characteristic (ROC) curve and applied to the probability maps to generate binary classification masks. Then, we calculated the evaluation metrics using the reconstructed binary masks. We used the same method as stated in section 2.2 to exclude the background pixels in the reconstructed binary masks, so that the classification labels of the large blank areas in the reconstructed binary masks were not used for metrics calculation. In other words, we only calculated the evaluation metrics using the tissue pixels.
2.6. Evaluation metrics
In this study, we use overall accuracy, sensitivity, and specificity to evaluate the classification performance, as shown in Equation (2-4). Overall accuracy is defined as the ratio of the number of all the correctly labeled patches to the total number of patches in the testing group. Sensitivity and specificity are calculated from true positive (TP), true negative (TN), false positive (FP), and false negative (FN), where positive corresponds to tumor and negative to normal. Sensitivity is the ratio of TP to the sum of TP and FN, and it measures the portion of cancerous tissue that is correctly labeled; specificity is the ratio of TN to the sum of TN and FP, and it represents the ability of the classifier to identify true normal tissue.
| (2) |
| (3) |
| (4) |
3. RESULTS
3.1. Validation results
Because we did not use all patients for testing, the validation results of 12 patients served as an important evaluation of the network. Therefore, we report the validation performance of each patient as Table 3 shows. The patch-based 2D-CNN achieved an average validation accuracy of 0.73, as well as 0.78 sensitivity and 0.62 specificity. Generally, the networks had a better ability to detect cancer than normal tissue in the hyperspectral histologic images.
Table 3.
Validation results per patient.
| Patient # | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| 62 | 0.66 | 0.51 | 0.92 |
| 68 | 0.90 | 0.96 | 0.80 |
| 120 | 0.82 | 0.70 | 0.95 |
| 127 | 0.86 | 0.97 | 0.71 |
| 134 | 0.72 | 0.99 | 0.33 |
| 137 | 0.88 | 0.99 | 0.63 |
| 161 | 0.52 | 1.00 | 0 |
| 166 | 0.57 | 0 | 1.00 |
| 174 | 0.75 | 0.93 | 0.42 |
| 184 | 0.59 | 0.47 | 0.67 |
| 187 | 0.68 | 0.90 | 0.33 |
| 188 | 0.84 | 0.96 | 0.71 |
| Average | 0.73 | 0.78 | 0.62 |
Because most patients had more tumor images than normal images, the two classes of data in the training group were unbalanced, especially when some patients with balanced data were left out for validation or testing and the two patients with only tumor data were used for training. This could be one possible reason for higher sensitivity than specificity in the validation results. On the other hand, in some slides, the epithelium was very thin, and inflamed tissue under the epithelium with full of lymphocytes and barely any nuclei were included in the images. Because tissue inflammation happens in both the normal and cancerous tissue, our networks classified a portion of the inflamed tissue in the normal images as tumor, which caused the low specificity of a few patients especially #134, #137. In the future, we hope to solve it by using a larger training dataset with more patient data and more images of normal tissue. In addition, images of patient #161 and #166, which were validated in the same fold, were all classified as one type. If exclude these two patients, the average validation accuracy, sensitivity, and specificity are 0.77, 0.84, and 0.65.
In order to compare the classification performance of HSI and RGB and investigate the usefulness of the extra spectral information in HSI, we used synthesized-RGB patches to train the same network as HSI. The hyperparameters and data partition for the CNN of synthesized-RGB were exactly the same as that of HSI. However, the validation results using synthesized-RGB patches were much worse than HSI. For 9 validation patients except #62, #68, and #120, almost all images of each patient were classified as one class, while the validation performance of the 3 patients was also worse than HSI. Therefore, we did not report the results of synthesized-RGB here. It is worth noting that one of our previous work [14] implemented an inception-based CNN with a very large dataset (102 SCC patients) of digitalized whole-slide histologic images and got an average area under the curve (AUC) of 0.916. The unsatisfying performance of synthesized-RGB in this study is probably due to the small size of the dataset. Nevertheless, the comparison between HSI and synthesized-RGB proved that the spectral information in HSI could improve the distinction ability.
3.2. Testing results
The testing results of 3 patients using HSI are shown in Table 4. The network was able to discriminate between cancerous and normal tissue in the testing data. The average testing accuracy, sensitivity, and specificity were 0.73, 0.77, and 0.69, respectively. For patient #74, the misclassified normal tissue were mainly from the first layer of epithelium with sparse nuclei, as well as the basement membrane with dense nuclei. For both patient #154 and #172, the misclassified regions were mainly around the keratinizing pearl in the slides where very few nuclei existed.
Table 4.
Testing results per patient.
| Patient # | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| 74 | 0.59 | 0.99 | 0.18 |
| 154 | 0.79 | 0.58 | 0.94 |
| 172 | 0.82 | 0.75 | 0.96 |
| Average | 0.73 | 0.77 | 0.69 |
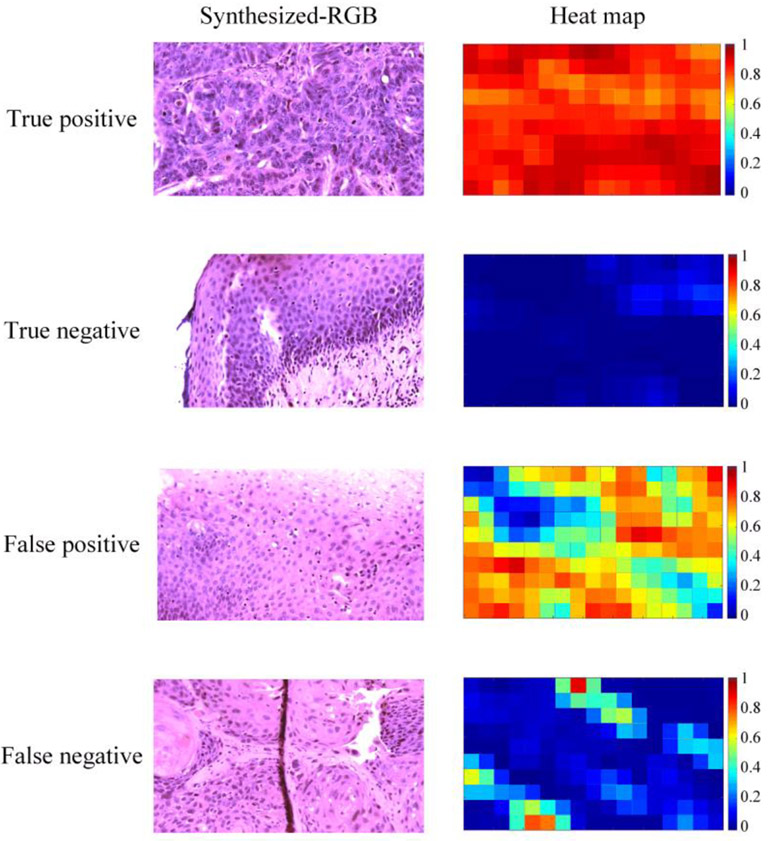
In general, tumor images with obvious histologic features such as enlarged nuclei and keratinizing SCC were mostly well classified. Normal images that were acquired at the second and third layers of the stratified epithelium, where normal-sized nuclei were evenly distributed, also got good results. We looked into the images that had less satisfying results, and the main reasons include the small number of nuclei in the ROI, image out of focus in some regions, and the overexposure in some regions where the slide became very thin. In addition, images of normal tissues with very thin epithelium, dense nuclei, and many lymphocytes tended to be classified as cancer. Figure 4 shows some images and heat maps of true positive, true negative, false positive, and false negative.
Figure 4.
Synthesized-RGB images and heat maps of true positive, true negative, false positive, and false negative. First row: a correctly classified whole image of cancerous tissue with keratinizing SCC and enlarged nuclei. Second row: a correctly classified whole image of normal epithelium, note that the small region on the top right comer was out of focus, which influenced the classification probability. Third row: normal epithelium with false positives due to fewer nuclei (top right) and dense nuclei (bottom left). Fourth row: tumor tissue with false negatives, probably due to the less distinct histologic features in the field of view.
4. DISCUSSION & CONCLUSION
In this preliminary study, we used a modified Inception-based CNN for head and neck cancer detection in the whole hyperspectral images of histologic slides. H&E stained histological slides of larynx, hypopharynx, and buccal mucosa from 18 different patients were included in this study. Patches generated from the whole hyperspectral image of the slides were used to train, validate, and test the CNN. Synthesized-RGB patches were used to train and validate the same networks but received a worse performance than HSI. The comparison between HSI and synthesized-RGB has proven the usefulness of the extra spectral information in HSI for this application. Because the automated hyperspectral whole-slide imaging system was not available, we manually selected and scanned ROIs in the slides, which resulted in a relatively small dataset in this study. The normal ROIs were mainly selected from the healthy stratified epithelium, which had a limited representation of histological features in normal tissue. Despite this study had certain limitations, it demonstrates the potential of hyperspectral microscopic imaging to provide a fast pathological diagnosis. In the future, we will refine the hyperspectral microscopic imaging system and develop automated scanning and stitching, so that we can conduct cancer detection in larger regions or even the whole slide. We also plan to include a larger dataset of histologic slides for the study of deep learning methods. With the automated system, the proposed method could potentially aid intraoperative pathologist consultation for whole-slide cancer detection and improve surgical outcomes.
ACKNOWLEDGEMENTS
This research was supported in part by the U.S. National Institutes of Health (NIH) grants (R01CA156775, R01CA204254, R01HL140325, and R21CA231911) and by the Cancer Prevention and Research Institute of Texas (CPRIT) grant RP190588.
Footnotes
DISCLOSURES
The authors have no relevant financial interests in this article and no potential conflicts of interest to disclose. Informed consent was obtained from all patients in accordance with Emory Institutional Review Board policies under the Head and Neck Satellite Tissue Bank (HNSB, IRB00003208) protocol.
REFERENCES
- [1].Vigneswaran N, and Williams MD, “Epidemiologic trends in head and neck cancer and aids in diagnosis,” Oral and Maxillofacial Surgery Clinics, 26(2), 123–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Leemans CR, Braakhuis BJ, and Brakenhoff RH, “The molecular biology of head and neck cancer,” Nature reviews cancer, 11(1), 9–22 (2011). [DOI] [PubMed] [Google Scholar]
- [3].Wissinger E, Griebsch I, Lungershausen J, Foster T, and Pashos CL, “The economic burden of head and neck cancer: a systematic literature review,” Pharmacoeconomics, 32(9), 865–882 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Haddad RI, and Shin DM, “Recent advances in head and neck cancer,” New England Journal of Medicine, 359(11), 1143–1154 (2008). [DOI] [PubMed] [Google Scholar]
- [5].Argiris A, Karamouzis MV, Raben D, and Ferris RL, “Head and neck cancer,” The Lancet, 371(9625), 1695–1709 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hinni ML, Ferlito A, Brandwein-Gensler MS, Takes RP, Silver CE, Westra WH, Seethala RR, Rodrigo JP, Corry J, and Bradford CR, “Surgical margins in head and neck cancer: a contemporary review,” Head & neck, 35(9), 1362–1370 (2013). [DOI] [PubMed] [Google Scholar]
- [7].Meier JD, Oliver DA, and Varvares MA, “Surgical margin determination in head and neck oncology: current clinical practice. The results of an International American Head and Neck Society Member Survey,” Head & Neck: Journal for the Sciences and Specialties of the Head and Neck, 27(11), 952–958 (2005). [DOI] [PubMed] [Google Scholar]
- [8].Sutton D, Brown J, Rogers SN, Vaughan E, and Woolgar J, “The prognostic implications of the surgical margin in oral squamous cell carcinoma,” International journal of oral and maxillofacial surgery, 32(1), 30–34 (2003). [DOI] [PubMed] [Google Scholar]
- [9].Kerker FA, Adler W, Brunner K, Moest T, Wurm MC, Nkenke E, Neukam FW, and von Wilmowsky C, “Anatomical locations in the oral cavity where surgical resections of oral squamous cell carcinomas are associated with a close or positive margin—a retrospective study,” Clinical oral investigations, 22(4), 1625–1630 (2018). [DOI] [PubMed] [Google Scholar]
- [10].Gandour-Edwards RF, Donald PJ, and Wiese DA, “Accuracy of intraoperative frozen section diagnosis in head and neck surgery: experience at a university medical center,” Head & neck, 15(1), 33–38 (1993). [DOI] [PubMed] [Google Scholar]
- [11].Ortega S, Halicek M, Fabelo H, Camacho R, Plaza M. d. I. L., Godtliebsen F, Callicó GM, and Fei B, “Hyperspectral Imaging for the Detection of Glioblastoma Tumor Cells in H&E Slides Using Convolutional Neural Networks,” Sensors, 20(7), 1911 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Madabhushi A, and Lee G, “Image analysis and machine learning in digital pathology: Challenges and opportunities,” Medical Image Analysis 33:170–175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nam S, Chong Y, Jung CK, Kwak T-Y, Lee JY, Park J, Rho MJ, and Go H, “Introduction to digital pathology and computer-aided pathology,” Journal of pathology and translational medicine, 54(2), 125–134 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Halicek M, Shahedi M, Little JV, Chen AY, Myers LL, Sumer BD, Fei B, Tomaszewski JE, and Ward AD, "Detection of squamous cell carcinoma in digitized histological images from the head and neck using convolutional neural networks", Proc. SPIE 10956, 109560K (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Halicek M, Shahedi M, Little JV, Chen AY, Myers LL, Sumer BD, and Fei B, “Head and Neck Cancer Detection in Digitized Whole-Slide Histology Using Convolutional Neural Networks,” Scientific Reports, 9(1), 14043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang K-S, Yu G, Xu C, Meng X-H, Zhou J, Zheng C, Deng Z, Shang L, Liu R, and Su S, “Accurate Diagnosis of Colorectal Cancer Based on Histopathology Images Using Artificial Intelligence,” bioRxiv, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sitnik D, Kopriva I, Aralica G, Pačić A, Hadžija MP, and Hadžija M, "Transfer learning approach for intraoperative pixel-based diagnosis of colon cancer metastasis in a liver from hematoxylin-eosin stained specimens." Proc. SPIE 11320, 113200A (2020). [Google Scholar]
- [18].Mavuduru A, Halicek M, Shahedi M, Little J, Chen A, Myers L, and Fei B, "Using a 22-layer U-Net to perform segmentation of squamous cell carcinoma on digitized head and neck histological images" Proc. SPIE 11320, 113200C (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ortega S, Halicek M, Fabelo H, Guerra R, Lopez C, Lejeune M, Godtliebsen F, Callico G, and Fei B, "Hyperspectral imaging and deep learning for the detection of breast cancer cells in digitized histological images." Proc. SPIE 11320, 113200V (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nakaya D, Tsutsumiuchi A, Satori S, Saegusa M, Yoshida T, Yokoi A, Kanoh M, Tomaszewski JE, and Ward AD, "Digital pathology with hyperspectral imaging for colon and ovarian cancer." 10956, 109560X. [Google Scholar]
- [21].Ishikawa M, Okamoto C, Shinoda K, Komagata H, Iwamoto C, Ohuchida K, Hashizume M, Shimizu A, and Kobayashi N, “Detection of pancreatic tumor cell nuclei via a hyperspectral analysis of pathological slides based on stain spectra,” Biomedical Optics Express, 10(9), (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ma L, Halicek M, Zhou X, Dormer J, and Fei B, "Hyperspectral microscopic imaging for automatic detection of head and neck squamous cell carcinoma using histologic image and machine learning." Proc. SPIE 11320, 113200W (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vos JJ, “Colorimetric and photometric properties of a 2° fundamental observer,” Color Research & Application, 3(3), 125–128 (1978). [Google Scholar]
- [24].Szegedy C, Ioffe S, Vanhoucke V, and Alemi AA, "Inception-v4, inception-resnet and the impact of residual connections on learning," Proceedings of the AAAI Conference on Artificial Intelligence, 31(1), (2017) [Google Scholar]
- [25].Ioffe S, and Szegedy C, “Batch normalization: Accelerating deep network training by reducing internal covariate shift,” arXiv preprint arXiv:1502.03167, (2015). [Google Scholar]
- [26].Ketkar N, "Introduction to Keras", Deep Learning with Python, Springer; (2017), pp. 97–111. [Google Scholar]
- [27].Zeiler MD, “Adadelta: an adaptive learning rate method,” arXiv preprint arXiv:1212.5701, (2012). [Google Scholar]