Abstract
Introduction
We aimed to provide cut points for the automated Elecsys Alzheimer's disease (AD) cerebrospinal fluid (CSF) biomarkers.
Methods
Cut points for Elecsys amyloid beta 42 (Aβ42), total tau (t‐tau), hyperphosphorylated tau (p‐tau), and t‐tau/Aβ42 and p‐tau/Aβ42 ratios were evaluated in Mayo Clinic Study of Aging (n = 804) and Mayo Clinic Alzheimer's Disease Research Center (n = 70) participants.
Results
The t‐tau/Aβ42 and p‐tau/Aβ42 ratios had a higher percent agreement with normal/abnormal amyloid positron emission tomography (PET) than the individual CSF markers. Reciever Operating Characteristic (ROC)‐based cut points were 0.26 (0.24–0.27) for t‐tau/Aβ42 and 0.023 (0.020–0.025) for p‐tau/Aβ42. Ratio cut points derived from other cohorts performed as well in our cohort as our own did. Individual biomarkers had worse diagnostic properties and more variable results in terms of positive and negative percent agreement (PPA and NPA).
Conclusion
CSF t‐tau/Aβ42 and p‐tau/Aβ42 ratios are very robust indicators of AD. For individual biomarkers, the intended use should determine which cut point is chosen.
Keywords: Alzheimer's disease, biomarkers, CSF, cut point
1. INTRODUCTION
Cerebrospinal fluid (CSF) amyloid beta 1‐42 (Aβ42), total tau (t‐tau), and hyperphosphorylated tau (p‐tau) are well‐established biomarkers for Alzheimer's disease (AD). 1 They measure soluble forms of the proteins that are the pathological hallmarks of AD. Despite ample research, use of CSF biomarkers is still hampered by technical issues, mainly between‐laboratory variation and lot‐to‐lot variation. 2 Differences in analytical procedures are a major cause of between‐laboratory variation. 3 Automated assays, such as the Elecsys assays by Roche, aim to standardize and control the analytic process and thus provide more reliable measurements. 4 , 5 , 6
RESEARCH IN CONTEXT
Systematic review: Using PubMed, we found seven prior publications in which cut points for the Elecsys amyloid beta 42 (Aβ42), t‐tau, p‐tau, t‐tau/Aβ42 ratio, or p‐tau/Aβ42 ratio were determined. None of these studies focused on defining cut points for use in various contexts.
Interpretation: We defined cut points for various clinical and scientific uses. Best overall agreement with normal/abnormal amyloid positron emission tomography (PET) was reached by using either the t‐tau/Aβ42 or p‐tau/Aβ42 ratio (Youden index: 0.26 and 0.023). When assessing abnormality using individual cerebrospinal fluid (CSF) biomarkers, we propose a dynamic biomarker view is best implemented by using a combination of a mixture modeling cut point for Aβ42 (1026 pg/mL) and the cut points from an Reciever Operating Characteristic (ROC) curve‐based analysis discriminating cognitively impaired abnormal amyloid PET versus cognitively unimpaired normal amyloid PET for t‐tau and p‐tau (Youden index: 238 and 22 pg/mL, respectively).
Future directions: The cut points provided need to be further evaluated for their diagnostic and prognostic performances.
HIGHLIGHTS
Agreement between amyloid positron emission tomography (PET) and cerebrospinal fluid (CSF) ratios of p‐tau/amyloid beta 42 (Aβ42) and t‐tau/Aβ42 ratios is excellent.
Ratio cut points derived from other cohorts perform as well in our cohort as our own ratio cut points.
For individual biomarkers, positive precent agreement (PPA) and negative percent agreement (NPA) are more variable between different cut points.
Although the AD pathophysiological process is a continuum, both clinicians and researchers use cut points to define who has AD and who does not. The gold standard for this classification should be pathologic evidence of AD, but this is often insufficiently available and has its own challenges. 7 Arguably, the best alternative is to define cut points based on their utility in different situations. For example, therapeutic trials that select participants based on the presence of amyloid pathophysiology may want to use a more sensitive cut point, thus ensuring inclusion of every person who might have AD. However, a clinician seeing a patient may want a more conservative (specific) cut point to ensure that a diagnosis of AD is only communicated when one can be certain this is the true cause of the clinical syndrome.
Several prior publications have reported cut points for the Elecsys assays. Three used amyloid positron emission tomography (PET) binding as the reference standard, 8 , 9 , 10 one used Innotest enzyme‐linked immunosorbent assay (ELISA) (Fujirebio, Gent) in CSF, and one used both. 11 , 12 More recently, p‐tau and t‐tau cut points were determined based on the distinction between high‐ and low‐risk mild cognitive impairment (MCI). 13 However, none of these studies examined the utility of multiple references to tailor their results to different uses of the proposed cut points.
In this study, we aimed to define cut points for Elecsys Aβ42, t‐tau, and p‐tau, and the t‐tau/Aβ42 and p‐tau/Aβ42 ratios. We used multiple approaches to assess how sensitive cut points were to different statistical methodologies and to facilitate using these cut points in various ways. In addition, we integrated previously published cut‐points into our results for comparison.
2. METHODS
2.1. Participants
We included participants from the Mayo Clinic Study of Aging (MCSA, n = 804) and the Mayo Clinic Alzheimer Disease Research Center (ADRC, n = 70), both in Rochester, Minnesota. The MCSA is a longitudinal, population‐based study of residents of Olmsted County, Minnesota that was established in 2004. Details of the study design and conduct of the study are reported elsewhere. 14 , 15 Briefly, MCSA participants are evaluated clinically every 15 months. Each evaluation includes separate assessments by a study coordinator, a physician, and a psychometrist. Final clinical diagnoses were established by consensus using previously published criteria. 16 , 17 Starting November 2007, MCSA participants were recruited to undergo a lumbar puncture. The ADRC is a clinic‐based study in which patients who present to a behavioral neurologist are invited to participate. The present analyses included MCSA and ADRC participants who underwent lumbar puncture and were either cognitively unimpaired (CU, n = 727), had mild cognitive impairment (MCI, n = 105), or had AD dementia (AD dementia, n = 42). These individuals make up the CSF cohort. A subset (n = 524) also underwent 11C Pittsburgh Compound B PET imaging ( 11C PiB PET) within 1 year of the lumbar puncture and are labeled the CSF + PET subset.
2.2. Standard protocol approvals, registrations, and patient consents
The institutional review boards of the Mayo Clinic and the Olmsted Medical Center approved all study protocols, and written informed consent was obtained from all participants.
2.3. CSF analyses
CSF samples were obtained by lumbar puncture from the L3 and L4 intervertebral space. Lumbar puncture was performed early in the morning after fasting. CSF was collected and transported to the lab in polypropylene tubes where it was aliquoted in 0.5 mL polypropylene tubes and stored at −80°C until use without any additional freeze‐thaw cycles. Aβ42, t‐tau, and hyperphosphorylated tau‐181 (p‐tau) were analyzed using Elecsys β‐amyloid(1‐42) CSF, Elecsys Total‐Tau CSF, and Elecsys Phospho‐Tau (181P) CSF electrochemiluminescence immunoassays (Roche Diagnostics International Ltd, Rotkreuz, Switzerland) 4 , 5 , 6 ; these assays are approved for clinical use in countries accepting the Conformité Européenne (CE) mark. In the United States, these immunoassays are for investigational purposes and are not yet approved for clinical use. The quality control process consisted of an initial method validation including precision and accuracy analysis prior to starting the analysis and the use of Elecsys PreciControl samples to monitor quality during the trial as described previously. 18
2.4. PET imaging
In the CSF + PET subset, amyloid PET imaging was performed with 11C Pittsburg Compound B (or PiB). Imaging consisted of four, 5‐minute dynamic frames acquired from 40‐60 minutes after injection. Amyloid PET was analyzed using an in‐house fully automated image processing pipeline. 19 Image voxel values were extracted from automatically labeled regions of interest (ROIs). A “global” amyloid PET standardized uptake value ratio (SUVR) was calculated as the voxel‐number weighted average of the median uptake across the following target regions: prefrontal, orbitofrontal, parietal, temporal, anterior cingulate, and posterior cingulate/precuneus divided by the median uptake in a cerebellar crus reference region. Target regions were white matter and gray matter sharpened to exclude CSF voxels; the reference region was gray matter sharpened. Partial volume correction was not used. Centiloid values were converted from the SUVR values as recommended in Klunk et al. 20 , 21
2.5. Statistical analyses
Associations between individual CSF biomarkers and the t‐tau/Aβ42 and p‐tau/Aβ42 ratios versus amyloid PET were assessed using Spearman rank correlations, which are non‐parametric and do not assume an underlying linear relationship.
We used mixture modeling and Receiver Operating Characteristic (ROC) methods to define multiple cut points for each CSF biomarker to be applied in different contexts and evaluated the performance of each. Univariate mixture modeling was based on the premise that our sample consisted of two subgroups: one with AD and one without. CSF values were log‐transformed for the mixture modeling. These mixture model cut points were defined as the point where the posterior probability was 50% assuming equal prior prevalence.
Using the CSF + PET subset, ROC curves were generated with normal versus abnormal amyloid PET (A− vs A+) as the primary reference for Aβ42 and the t‐tau/Aβ42 and p‐tau/Aβ42 ratios. ROC cut points were defined based on both the Youden index, which maximizes sensitivity plus specificity, and also on the cut point that would result in 90% positive percent agreement (PPA). A− versus A+ was determined using mixture modeling of log‐transformed SUVR values in the CSF + PET subset.
Because ample evidence suggests CSF p‐tau and t‐tau change later in the disease course of AD than amyloid, the most appropriate reference for creating cut points for t‐tau and p‐tau is arguably not A− versus A+. Nor is a purely clinically defined reference that does not take into account amyloid abnormality appropriate. Therefore, for p‐tau and t‐tau we used a reference of CU A− versus cognitively impaired (CI) participants (those with MCI or AD dementia) A+ as the primary reference. ROC cut points were again defined based on the Youden index and 90% PPA from these curves. Bootstrapping was used to estimate 95% confidence intervals for all of the mixture modeling and ROC‐based cut points.
For each biomarker, we performed a sensitivity analysis using the alternative (non‐primary) reference. In this sensitivity analysis for Aβ42, t‐tau/Aβ42, and p‐tau/Aβ42 ratios, the reference was CU A− versus CI A+, and for t‐tau and p‐tau, it was A− versus A+. The samples, methods, and references used for defining the different cut points are summarized in the Supplemental Table 1.
For the mixture model cut points and the cut points derived from the ROC analyses with A+ versus A− as the reference, we summarized the agreement between normal/abnormal CSF and normal/abnormal amyloid PET. For the cut points derived from the ROC analyses with CI A+ versus CU A−, we summarized the agreement between normal/abnormal CSF and CU A−/CI A+. In addition, we summarized the agreement for previously published cut‐points in the same manner, taking our primary reference for each of the biomarkers as outcome (A+ vs A− for Aβ42 and the ratios and CU A− vs CI A+ for p‐tau and t‐tau).
Although amyloid PET is the best surrogate marker we have for pathological evidence of AD during life, pathology itself is the real gold standard. In view of this difference, we use the terms overall percent agreement (OPA) instead of concordance, positive percent agreement (PPA) instead of sensitivity, and negative percent agreement (NPA) instead of specificity. We also report Cohen's kappa, which can be interpreted as a measure of percent agreement adjusted for agreement due to chance.
All statistical analyses were done using the R language and environment for statistical computing version 3.6.2.
3. RESULTS
Participants in the CSF cohort had a median age of 73 (interquartile range 64‐79). Males (502, 57%) were slightly overrepresented. The age and sex distribution was similar for the CSF + PET subset (71 [63–78] years, 56% male). For further descriptions of the cohort see Table 1.
TABLE 1.
Demographic characteristics by clinical diagnosis within the CSF cohort and CSF + PET subset
| CSF cohort | CSF + PET subset | |||||||
|---|---|---|---|---|---|---|---|---|
| Characteristic | All | CU | MCI | AD dementia | All | CU | MCI | AD dementia |
| Number of participants | 874 | 727 | 105 | 42 | 524 | 416 | 69 | 39 |
| Cohort study, no. (%) | ||||||||
| MCSA | 804 (92%) | 715 (98%) | 84 (80%) | 5 (12%) | 475 (91%) | 414 (100%) | 54 (78%) | 7 (18%) |
| ADRC | 70 (8%) | 12 (2%) | 21 (20%) | 37 (88%) | 49 (9%) | 2 (0%) | 15 (22%) | 32 (82%) |
| Age (years) | ||||||||
| Median (IQR) | 73 (64, 79) | 72 (63, 79) | 77 (71, 83) | 72 (65, 80) | 71 (63, 78) | 70 (61, 77) | 77 (68, 83) | 72 (64, 80) |
| Range | 32 to 95 | 32 to 95 | 51 to 92 | 50 to 91 | 32 to 95 | 32 to 95 | 51 to 94 | 51 to 88 |
| Male sex, no. (%) | 502 (57%) | 410 (56%) | 66 (63%) | 26 (62%) | 296 (56%) | 229 (55%) | 43 (62%) | 24 (62%) |
| Education (years), median (IQR) | 14 (12, 16) | 14 (12, 16) | 14 (12, 16) | 16 (12, 16) | 14 (12, 16) | 14 (12, 16) | 14 (12, 17) | 16 (12, 16) |
| APOE ε4 carrier, no. (%) | 262 (30%) | 191 (26%) | 41 (39%) | 30 (71%) | 171 (33%) | 117 (28%) | 27 (39%) | 27 (69%) |
| Short Test of Mental Status, Median (IQR) | 35 (33, 37) | 36 (34, 37) | 32 (29, 33) | 26 (21, 28) | 35 (33, 37) | 36 (34, 37) | 32 (29, 34) | 25 (20, 28) |
| Aβ42 (pg/mL), median (IQR) | 1050 (716, 1493) | 1107 (792, 1538) | 847 (602, 1223) | 479 (351, 585) | 1026 (676, 1444) | 1099 (794, 1515) | 764 (490, 1183) | 510 (403, 607) |
| t‐tau (pg/mL), median (IQR) | 220 (168, 289) | 213 (166, 270) | 246 (190, 345) | 318 (254, 467) | 217 (168, 284) | 208 (163, 266) | 244 (187, 302) | 361 (265, 530) |
| p‐tau (pg/mL) median (IQR) | 19 (14, 25) | 18 (14, 23) | 22 (16, 31) | 29 (25, 41) | 19 (14, 25) | 18 (14, 23) | 21 (16, 27) | 32 (26, 47) |
| t‐tau/Aβ42, median (IQR) | 0.18 (0.14, 0.31) | 0.17 (0.14, 0.25) | 0.25 (0.16, 0.54) | 0.79 (0.53, 1.08) | 0.19 (0.14, 0.34) | 0.17 (0.14, 0.25) | 0.26 (0.17, 0.58) | 0.81 (0.52, 1.07) |
| p‐tau/Aβ42, Median (IQR) | 0.015 (0.012, 0.027) | 0.015 (0.012, 0.021) | 0.021 (0.014, 0.049) | 0.069 (0.049, 0.108) | 0.016 (0.012, 0.030) | 0.015 (0.012, 0.021) | 0.021 (0.015, 0.054) | 0.071 (0.049, 0.111) |
| Amyloid PET | ||||||||
| SUVR, median (IQR) | 1.40 (1.33, 1.69) | 1.38 (1.32, 1.50) | 1.60 (1.37, 2.24) | 2.43 (2.23, 2.63) | ||||
| Abnormal, no. (%) | 146 (28%) | 75 (18%) | 35 (51%) | 36 (92%) | ||||
CU = Cognitively Unimpaired, MCI = Mild Cognitive Impairment, AD = Alzheimer's Disease, MSCA = Mayo Clinic Study of Aging, ADRC = Alzheimer's Disease Research Center, IQR = Inter‐quartile Range, AB42 = amyloid‐beta42, t‐tau = total tau, p‐tau = hyperphosphorylated tau, SUVR = Standard Uptake Value Ratio.
3.1. Correlations
We found moderate correlations between amyloid PET and CSF Aβ42 (rank correlation [rho] = −0.53, P < .001), and t‐tau (rho = 0.43, P < .001), and p‐tau (rho = 0.48, P < .001) (Figure 1). Correlation between amyloid PET and t‐tau/Aβ42 and p‐tau/Aβ42 was higher (rho = 0.70, P < .001 for both).
FIGURE 1.
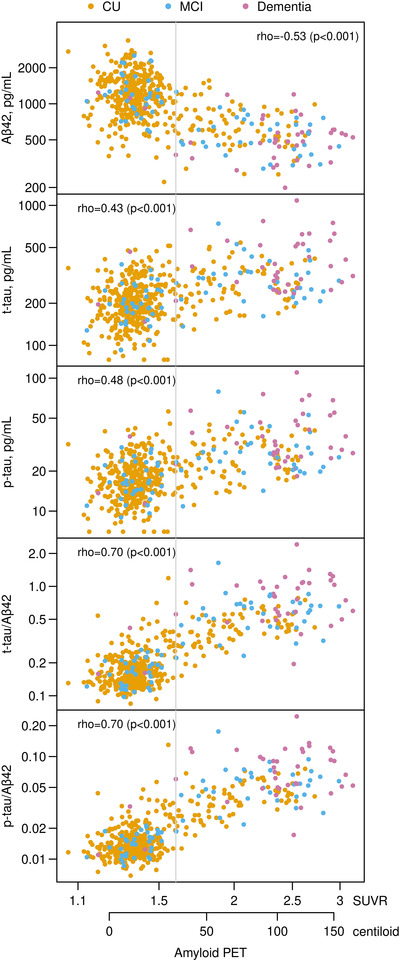
Scatter plots of the cerebrospinal fluid (CSF) biomarkers versus amyloid Positron Emission Tomography (PET) with rank correlations. Individual points are colored by clinical diagnosis CU = cognitively unimpaired, MCI = Mild Cognitive Impairment). The gray vertical line represents the amyloid PET cut point of 1.6 Standard Uptake Value Ratio (SUVR, 32 centiloid)
3.2. Gaussian mixture modeling for amyloid PET
Amyloid PET was amenable to mixture modeling in our cohort (Supplemental Figure 1). The mixture‐based cut point between the two groups based on equal prior prevalence was an SUVR of 1.60 (95% confidence interval [ 1.55–1.67) or centiloid of 32 (95% confidence interval 28‐38). A+ was therefore defined as an SUVR ≥1.60 and A− as an SUVR < 1.60.
3.3. CSF Aβ42
The CSF Aβ42 distribution was unimodal, roughly symmetric, and did not clearly suggest a mixture of two underlying groups. However, mixture modeling is not necessarily inappropriate in this context, since a unimodal distribution may result from a mixture of two groups whose modes are relatively close. The cut point resulting from this method was 1026 pg/mL (95% confidence interval 952‐1542) (Figure 2). The ROC curve with amyloid PET‐based A+ versus A− as the reference had an area under the curve (AUC) of 0.89. The cut point at which 90% PPA was reached was 954 (95% confidence interval 883‐1185). The Youden index resulted in a cut point of 893 (95% confidence interval 791‐953). The OPA ranged from 73% (for the mixture model) to 79% (for the Youden index) and the PPA was higher than NPA for each cut point. These cut points did not indicate strong agreement with the amyloid PET cut point (kappa 0.46–0.54). All previously published cut points for Aβ42 had very similar PPA compared to our results (91–92% vs 86–92%). However, most of the published cut points were a little higher at ≈1100 pg/mL. Those had lower NPAs (58–59%) compared to our results (66–76%).
FIGURE 2.
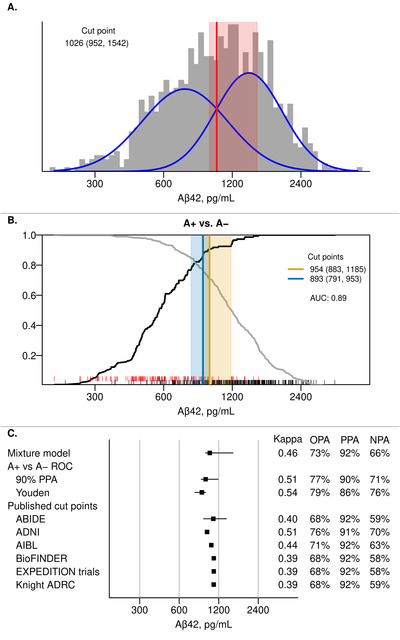
CSF Aβ42 cut points. Panel A shows a histogram of Aβ42 in the CSF cohort with estimated probability density curves of the two groups identified through univariate mixture modeling (blue). The optimal cut point with 95% bootstrap confidence interval is shown in red. Panel B shows the positive percent agreement (PPA, black line) and negative percent agreement (NPA, gray line) for Aβ42 from an Receiving Operating Characteristic (ROC) analysis using A+ versus A− as the reference within the CSF + PET subset. A+ was defined as amyloid PET ≥1.60 SUVR (32 centiloid). The cut point (95% bootstrap confidence interval) that results in 90% PPA is shown in orange and the Youden method cut point is shown in blue. Rug plots at the bottom of each panel indicate Aβ42 values for A+ (red) and A− (black) individuals. Panel C summarizes the Aβ42 cut points from Panels A and B as well as performance of previously published cut points in our cohort. 8 , 9 , 10 , 11 , 12 These cut points were: 1092 pg/mL for Alzheimer's Biomarkers In Daily PracticE (ABIDE), 977 pg/mL for Alzheimer's Disease Neuroimaging Initiative (ADNI), 1054 pg/mL for the Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing (AIBL), 1100 pg/mL for Biomarkers For Identifying Neurodegenerative Disorders Early and Reliably (BioFINDER), 1100 pg/mL for the EXPEDITION trials, and 1098 pg/mL for the Knight Alzheimer's Disease Research Center (ADRC). 8 , 9 , 10 , 11 , 12 Kappa, overall percent agreement (OPA), positive percent agreement (PPA), and negative percent agreement (NPA) are reported with amyloid PET (A+ vs A−) as reference.
We also show the ROC cut points for CSF Aβ42 using the non‐primary reference of CI A+ participants and CU A− participants (Supplemental Figure 1), The 90% PPA cut point was 820 (95% CI 743‐979) and the Youden cut point was 817 (95% CI 676‐906); both reached an OPA of 84%.
3.4. CSF t‐tau/Aβ42 ratio
The t‐tau/Aβ42 ratio suggested a mixture distribution and its general shape was similar to that of amyloid PET (Figure 3). The mixture model indicated a cut point of 0.23 (95% confidence interval 0.21–0.24). The PET based A+ versus A− ROC curve had an AUC of 0.97. The 90% PPA cut point based on this curve was 0.28 (95% confidence interval 0.25–0.30). The Youden‐index resulted in a cut point of 0.26 (95% confidence interval 0.24–0.27). The mixture model‐based cut point had exceptionally high PPA (97%), but somewhat lower NPA (86%), with an OPA of 89%. More balanced results were reached with the 0.26 and 0.28 cut points (OPA 92% with PPA and NPA > 90% for both). The t‐tau/Aβ42 ratio cut points provided comparatively good agreement with the amyloid PET cut point (kappa 0.75–0.81). Three of four previously published cut points performed the same as our ROC‐based cut points, 9 , 10 whereas the Knight ADRC cut point performed similar to our mixture modeling cut point. 8
FIGURE 3.
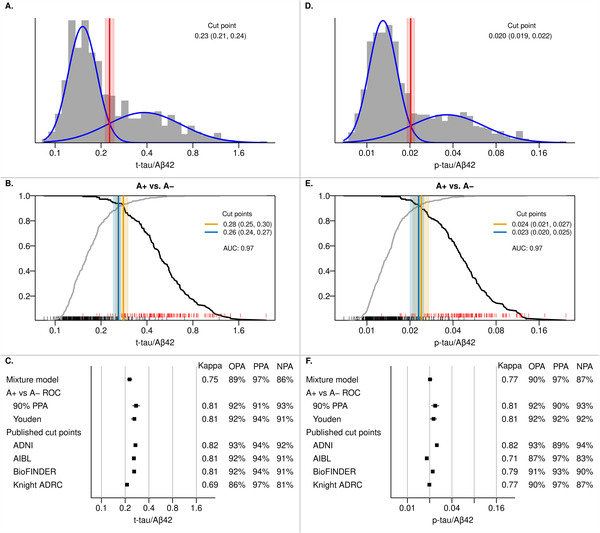
CSF T‐tau/Aβ42 and P‐tau/Aβ42 cut points. Panels A and D show the histograms of t‐tau/Aβ42 and p‐tau/Aβ42 in the CSF cohort with estimated probability density curves of the two groups identified through univariate mixture modeling (blue). The optimal cut point with 95% bootstrap confidence interval is shown in red. Panels B and E show the positive percent agreement (PPA, black line) and negative percent agreement (NPA, gray line) for t‐tau/Aβ42 and p‐tau/Aβ42, respectively, from an ROC analysis using A+ versus A− as the reference within the CSF + PET subset. A+ was defined as amyloid PET ≥1.60 SUVR (32 centiloid). The cut point (95% bootstrap confidence interval) that results in 90% PPA is shown in orange and the Youden method cut point is shown in blue. RUG plots at the bottom of each panel indicate CSF values for A+ (red) and A− (black) individuals. Panels C and F summarize the t‐tau/Aβ42 and p‐tau/Aβ cut points, respectively, from the previous panels and the performance of previously published cut‐points in our cohort. 8 , 9 , 10 For t‐tau/Aβ42, these cut points were 0.27 for ADNI, 0.258 for AIBL, 0.26 for BioFINDER, and 0.211 for the Knight ADRC. 8 , 9 , 10 For p‐tau/Aβ42, these cut points were 0.025 for ADNI, 0.0183 for AIBL, 0.022 for BioFINDER, and 0.0198 for the Knight ADRC. Kappa, overall percent agreement (OPA), PPA, and NPA with amyloid PET as reference (A+ vs A−) are reported.
The analyses using the non‐primary reference (CI A+ vs CU A−) are shown in Supplemental Figure 2 and resulted in higher cut points. The 90% PPA cut point was 0.35 (95% confidence interval 0.26–0.42, OPA 95%). The Youden‐based cut point was 0.32 (95% confidence interval 0.26–0.40, OPA 94%).
3.5. CSF p‐tau/Aβ42 ratio
The p‐tau/Aβ42 ratio distribution was similar to that of t‐tau/Aβ42 and resulted in a mixture model cut point of 0.020 (95% confidence interval 0.019–0.022) (Figure 3). The amyloid PET based A+ versus A− ROC curve resulted in an AUC of 0.97. The resulting 90% PPA cut point was 0.024 (95% confidence interval 0.021–0.027) and the cut point based on the Youden index was 0.023 (95% CI 0.020–0.025). OPAs for all cut points were between 90% and 92%. Similar to results for the t‐tau/Aβ42 ratio, the mixture model resulted in a high PPA (97%) but somewhat lower NPA (87%). The most balanced result was reached when using the Youden index: PPA and NPA were both 92% for the 0.023 cut point. Kappas for the p‐tau/Aβ42 ratio cut points ranged from 0.77 to 0.81. Cut points from other cohorts varied some but performed very similar to either our mixture model or the ROC‐based cut points. 8 , 9 , 10
Cut points based on the non‐primary reference of CI A+ and CU A− are also shown in Supplemental Figure 2 and resulted in higher (more conservative) cut points with excellent agreement: OPA was 95% and 94% for the 90% PPA and Youden index based cut points of 0.031 (95% confidence interval 0.023–0.038) and 0.026 (95% confidence interval 0.023–0.030), respectively.
3.6. CSF t‐tau
Figure 4 depicts our primary analyses for t‐tau and p‐tau. Like CSF Aβ42, the t‐tau and p‐tau distributions were roughly unimodal and symmetric. Mixture modeling resulted in a cut point of 258 pg/mL (95% confidence interval 227‐350) for t‐tau. The CI A+ versus CU A− analysis had an AUC of 0.84 and resulted in a 90% PPA cut point of 213 (95% confidence interval 191‐230) and the Youden cut point was 238 (95% confidence interval 214‐265). OPAs were between 64% and 76% for these methods. The mixture model cut point resulted in the highest NPA (78%, PPA 70%), whereas the 90% PPA cut point resulted in the highest PPA (90% by definition, NPA 59%). Kappa values were low (0.28–0.37). Kappa values and OPAs of previously published cut points were in the same range as those resulting from our analyses. However, we found large differences in PPA (ranging from 51% to 90%) and NPA (ranging from 59% to 87%) when individual cut points from other cohorts were evaluated.
FIGURE 4.
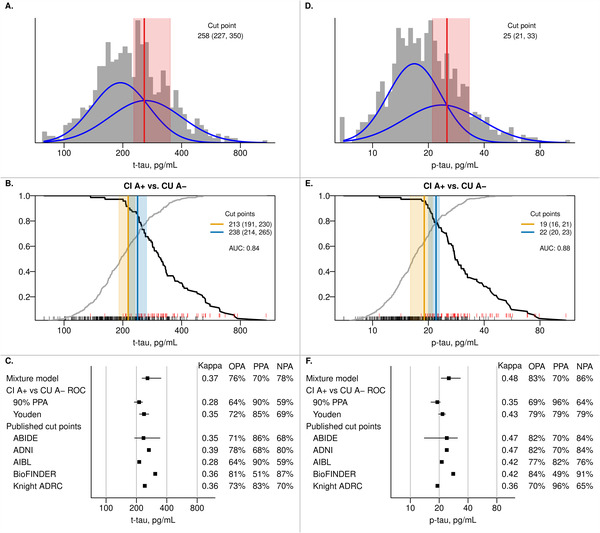
CSF t‐tau and p‐tau cut points. Panels A and D show the histograms of t‐tau and p‐tau in the cerebrospinal fluid (CSF) cohort with estimated probability density curves of the two groups identified through univariate mixture modeling (blue). The optimal cut point with 95% bootstrap confidence interval is shown in red. Panels B and E show the positive percent agreement (PPA, black line) and negative percent agreement (NPA, gray line) for t‐tau and p‐tau, respectively, from an receiver‐operating characteristic (ROC) analysis using cognitively impaired (CI) A+ versus cognitively unimpaired (CU) A− as the reference within the CSF + PET subset (see Methods – Statistical Analysis for more information). A+ was defined as amyloid PET ≥1.60 SUVR (32 centiloid). The cut point (95% bootstrap confidence interval) that results in 90% PPA is shown in orange and the Youden method cut point is shown in blue. RUG plots at the bottom of each panel indicate CSF values for CI A+ (red) and CU A− (black) individuals. Panels C and F summarize the t‐tau and p‐tau cut points, respectively, from the previous panels as well as performance of previously published cut points in our cohort. For t‐tau, these cut points were 235 pg/mL for ABIDE, 266 pg/mL for ADNI, 213 pg/mL for AIBL, 307 pg/mL for BioFINDER, and 242 pg/mL for the Knight ADRC. 8 , 10 , 12 , 13 For p‐tau, these were 24 pg/mL for ABIDE, 24 pg/mL for ADNI, 21.3 pg/mL for AIBL, 28 pg/mL for BioFINDER, and 19.2 pg/mL for the Knight ADRC. 8 , 10 , 12 , 13 Kappa, overall percent agreement (OPA), PPA, and NPA with CI A+ versus CU A− as the reference are reported.
When using the non‐primary reference of A+ versus A−, the 90% PPA cut point was 185 (95% confidence interval 169‐202) and the Youden index resulted in a cut point of 223 (95% confidence interval 207‐257). Agreement between these cut points and PET based A+ versus A− classification was poor with OPAs of 56% and 68%, respectively (Supplemental Figure 2).
3.7. CSF p‐tau
Mixture modeling resulted in a cut point for p‐tau of 25 pg/mL (95% confidence interval 21‐33) (Figure 4). From the ROC CI A+ versus CU A− analysis, the AUC was 0.88, the 90% PPA cut point was 19 pg/mL (95% confidence interval 16‐21), and the Youden index resulted in a cut point of 22 (95% confidence interval 20‐23). OPAs for the mixture model (78%) and the Youden index (79%) were very similar, with the mixture model favoring NPA (86% with a PPA of 58%) and the Youden index giving a more balanced result (PPA 79%, NPA 79%). Kappa values were also low for p‐tau (0.35–0.44). As was the case for t‐tau, kappa values and OPAs of previously published cut points were similar to those from our primary analyses, but individual cut points covered a wide range of PPA (ranging from 49% to 96%) and NPA (from 65% to 91%) values.
When using A+ versus A− as the non‐primary reference, the cut point using 90% PPA was 17 pg/mL (95% confidence interval 15‐18, OPA 64%) (Supplemental Figure 2). The cut point based on the Youden index was 21 (95% confidence interval 18‐23, OPA 74%).
4. DISCUSSION
In the current study, we provide several possible cut points for the CSF Elecsys AD markers. We confirm prior evidence that when the goal of using this assay is in high agreement with normal/abnormal amyloid PET imaging, the t‐tau/Aβ42 or p‐tau/Aβ42 ratios clearly provide better results than any of the individual CSF biomarkers. 8 , 9 All of the ratio cut points provide good to very good agreement with very little difference between the different results. Based on the theoretical basis of the Youden index, cut points resulting from this method may be preferred (0.26 for the t‐tau/Aβ42 ratio and 0.023 for the p‐tau/Aβ42 ratio). Notably, ratio cut‐points from other large cohorts (especially ADNI, AIBL, and BioFINDER for t‐tau/Aβ42 and ADNI and BioFINDER for p‐tau/Aβ42) perform as well in our cohort as our own Youden index–based cut points. 9 , 10 These findings strongly support the generalizability of the ratio cut‐off points defined in this study.
Many factors could have induced larger differences. The most important of these is pre‐analytical variability. Automated assays like the Elecsys assays aim to reduce analytical variability. However, despite efforts to promote uniform sample handling, considerable pre‐analytical variability still exists. 22 , 23 Aside from a few key elements that were consistent across all studies mentioned above and our own (use of polypropylene tubes, fewer than two freeze‐thaw cycles), pre‐analytical factors do not seem to influence results in such a way that they lead to greatly different cut points. Another factor worth mentioning that could have influenced cut points when taking amyloid PET as the reference is the manner in which the amyloid PET cut point is determined. For the current study, we used mixture modeling to determine an amyloid PET cut point, whereas in both ADNI and BioFINDER, amyloid PET visual read was used. In addition, the amyloid PET cut point we used in the current manuscript corresponds to a centiloid value of 32 (95% CI 28‐38), which is higher than the centiloid values that were previously published for the MCSA and those that were suggested based on a comparison between amyloid PET and neuropathology in a multicenter study. 24 , 25 Using a lower (centiloid) value may have resulted in different cut point recommendations. However, the similarity between our cut points for the CSF ratios with amyloid PET A+ versus A− as the reference and those from other cohorts is striking. 8 , 9 , 10 The methodological differences described appear to have little effect on the cut points found, indicating their robustness across a variety of factors.
Any CSF assay measures concentrations present in the CSF at a single time point. This is the result of release in, and clearance from, CSF of that specific protein, but likely also from more general CSF dynamics (such as CSF production and clearance). 18 This is different from PET imaging, which measures cumulative deposition of the protein of interest. Using CSF ratios may effectively correct for the effect that CSF dynamics have on inter‐individual biomarker concentration differences. This may be one of the factors that contribute to the ratios performing better than Aβ42 alone when amyloid PET is used as reference. Another factor may be that CSF Aβ42 becomes abnormal prior to amyloid PET imaging, whereas CSF p‐tau and t‐tau become abnormal later. The ratio could be reflecting these disparate dynamics and therefore have better overall agreement with amyloid PET imaging.
Creating reliable cut points for individual CSF biomarkers is more challenging as indicated by the considerably worse diagnostic properties. Still, unique information may be provided by each individual biomarker, 26 , 27 which may be lost when using the ratios. In addition, staging according to a dynamic biomarker view, such as the one most recently incorporated in the AT(N) framework requires separate use of the individual biomarkers. 28 , 29 However, going forward, it will be important to carefully assess how well the CSF biomarkers conform to the AT(N) framework.
For CSF Aβ42, our cut points varied between 893 pg/mL for the ROC‐based Youden index and 1026 pg/mL for the Gaussian mixture modeling. Mixture models do not require a reference group and provide a cut‐off that corresponds to a post‐test probability of 50%, which can be thought of as rather lenient. Others have shown that Aβ42 cut points based on mixture modeling have better prognostic performance than lower ones derived from categorical comparisons between CU and AD dementia subjects, likely due to the increased detection of early amyloid pathology. 30 With this purpose in mind, 1026 pg/mL would be the optimal choice in our cohort. However, in a diagnostic setting one might want to minimize the risk of false‐positive results and ensure a higher NPA (at the cost of lower PPA). For example, a cut point of 893 pg/mL would correctly classify an additional 10% of those with a normal amyloid PET result as normal (a 10% increase in NPA to 76%) at the cost of wrongly classifying an additional 6% of those with an abnormal amyloid PET as normal (a 6% reduction in PPA to 86%).
Some of the previously published cut points for Aβ42 (977 pg/mL for ADNI and 1054 pg/mL for AIBL) are so similar to our own that they could be used interchangeably. 9 , 10 For cut points closer to 1100 pg/mL (ABIDE, BioFINDER, EXPEDITION trials, and Knight ADRC), NPA decreases somewhat without any increase in PPA making such cut points a less optimal choice based on our data. 8 , 9 , 12
Prior evidence from the MCSA and another publication considering multiple cohorts suggests that there are two types of individuals with elevated tau in CSF within the normal aging/preclinical AD population. 18 , 31 One type appears to be part of a group of participants in which t‐tau and p‐tau are highly positively correlated with Aβ42. The other type of individuals with elevated tau is associated with low Aβ42 and may thus be more likely to be part of the AD pathophysiological spectrum. With these findings in mind, and for the purpose of identifying useful cut points for individual biomarkers, we aimed to create a cut point that would most accurately reflect the difference between normal and abnormal p‐tau or t‐tau within the second type by comparing amyloid PET‐based CU A− with CI A+ (MCI/AD dementia) participants. The resulting cut points were somewhat lower than those resulting from Gaussian mixture modeling, but similar to the cut points resulting from the A+ versus A− analysis (without taking cognitive status into account). A dynamic biomarker model may be best served by using the ROC (Youden‐index) based cut points of 238 pg/mL for t‐tau and 22 pg/mL for p‐tau.
Diagnostic properties of cut points for Elecsys t‐tau and p‐tau are generally worse and vary more greatly than those for the ratios. This finding is emphasized when comparing our results to previously published cut points. 8 , 10 , 12 , 13 For p‐tau, cut points from ABIDE and ADNI performed similarly to our mixture model cut point and the AIBL cut point to our Youden‐index based cut point. 9 , 10 , 12 These similarities are striking, because each of these studies used different reference standards. A high PPA (96%) is reached by using either our 90% PPA cut point or the cut point from the Knight ADRC, but NPAs are quite low for both (64% and 65%). Conversely, the cut point from BioFINDER results in a very high NPA (91%) with very low PPA (49%), meaning 51% of CI participants with abnormal amyloid PET in our cohort would not be classified as having abnormal p‐tau if this cut point were to be implemented. Similar phenomena are seen for t‐tau. Here, cut points from ABIDE, ADNI, and the Knight ADRC perform similarly to either our Gaussian mixture modeling or Youden index–based cut points, whereas those from AIBL and BioFINDER either favor higher PPA or NPA at the expense of a considerably lower NPA or PPA, respectively. 8 , 9 , 10 , 12 Specifically for t‐tau and p‐tau, considering the “trade‐offs” between false positives and false negatives is important.
The strengths of the current study include the large sample size with a large subset of participants who had amyloid PET data available. In addition, CSF analyses were done using a single reagent lot in samples that were stored in polypropylene tubes and thawed no more than once. The small number of AD dementia cases should be considered a limitation of the study. Black and Latino participants are under‐represented in the MCSA. This could have influenced results, especially for the individual biomarkers. However, using the more robust p‐tau/Aβ42 may ameliorate differences between ethnic groups. 32 In addition, 16% of our samples were above the prescribed upper detection limit of 1700 pg/mL for the Aβ42 assay. Although this is well within the normal range, extrapolated values above this limit could have influenced the parameter estimates in the Gaussian mixture models. Other methods were rank based and unaffected by this truncation.
CONFLICT OF INTEREST
Dr. A.C. van Harten, H.J. Wiste, S.D. Weigand, Dr. W Kremers, Dr. R.B. Dyer, and Dr. A. Algeciras‐Schimnich report no disclosures. Dr. Mielke served as a consultant to Eli Lilly and Lysosomal Therapeutics, Inc. She receives research support from the National Institutes of Health (NIH) (R01 AG49704, P50 AG44170, U01 AG06786, RF1 AG55151), Department of Defense (W81XWH‐15‐1), and unrestricted research grants from Biogen, Roche, and Lundbeck. Dr. Eichenlaub is an employee of Roche Diagnostics International Ltd, Rotkreuz, Switzerland. Dr. Knopman serves on a Data Safety Monitoring Board for Lundbeck Pharmaceuticals and for the DIAN study; is an investigator in clinical trials sponsored by Biogen, Lilly Pharmaceuticals and the Alzheimer's Disease Cooperative Study; and receives research support from the NIH. Dr. C. Jack Jr. has provided consulting services for Eli Lilly Co. He receives research funding from the NIH (R01 AG011378, U01 HL096917, U01 AG024904, RO1 AG041851, R01 AG037551, R01 AG043392, U01 AG006786) and the Alexander Family Alzheimer's Disease Research Professorship of the Mayo Foundation. Dr. Petersen is a consultant for Roche, Inc., Merck, Inc., Biogen, Inc., Genentech, Inc. Eisai, Inc., and GE Healthcare. The Elecsys β‐Amyloid (1‐42) CSF assay, the Elecsys Phospho‐Tau (181P) CSF assay, and the Elecsys Total‐Tau CSF assay are not approved for clinical use in the United States.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
ELECSYS is a trademark of Roche. All other product names and trademarks are the property of their respective owners. The authors thank Ekaterina Manuilova for her review of the manuscript. Funding was provided by the following foundations: NIA: U01 AG006786, P50 AG016574, GHR Foundation, and Mayo Foundation for Medical Education and Research. Roche Diagnostics provided the assay kits for CSF biomarker analysis.
van Harten AC, Wiste HJ, Weigand SD, et al. Detection of Alzheimer's disease using Elecsys amyloid beta 1‐42, p‐tau, and t‐tau assays. Alzheimer's Dement. 2022;18:635–644. 10.1002/alz.12406
REFERENCES
- 1. Molinuevo JL, Ayton S, Batrla R, et al. Current state of Alzheimer's fluid biomarkers. Acta Neuropathologica. 2018;136:821‐853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mattsson N, Andreasson U, Persson S, et al. CSF biomarker variability in the Alzheimer's Association quality control program. Alzheimer's & Dementia. 2013;9:251‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Verwey NA, van der Flier WM, Blennow K, et al. A worldwide multicentre comparison of assays for cerebrospinal fluid biomarkers in Alzheimer's disease. Ann Clin Biochem. 2009;46:235‐240. Internet]]. [DOI] [PubMed] [Google Scholar]
- 4. Bittner T, Zetterberg H, Teunissen CE, et al. Technical performance of a novel, fully automated electrochemiluminescence immunoassay for the quantitation of β‐amyloid (1‐42) in human cerebrospinal fluid. Alzheimers Dement. 2016;12:517‐526. [DOI] [PubMed] [Google Scholar]
- 5. Lifke V, Kollmorgen G, Manuilova E, et al. Elecsys®Total‐Tau and Phospho‐Tau (181P) CSF assays: analytical performance of the novel, fully automated immunoassays for quantification of tau proteins in human cerebrospinal fluid. Clin Biochem. 2019;72:30‐38. [DOI] [PubMed] [Google Scholar]
- 6. Rozga M, Bittner T, Höglund K, Blennow K. Accuracy of cerebrospinal fluid Aβ1‐42 measurements: evaluation of pre‐analytical factors using a novel Elecsys immunosassay. Clin Chem Lab Med. 2017;55:1545‐1554. [DOI] [PubMed] [Google Scholar]
- 7. Scheltens P, Rockwood K. How golden is the gold standard of neuropathology in dementia?. Alzheimers Dement. 2011;7:486‐489. [DOI] [PubMed] [Google Scholar]
- 8. Schindler SE, Gray JD, Gordon BA, et al. Cerebrospinal fluid biomarkers measured by Elecsys assays compared to amyloid imaging. Alzheimers Dement. 2018;14:1460‐1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hansson O, Seibyl J, Stomrud E, et al. CSF biomarkers of Alzheimer's disease concord with amyloid‐β PET and predict clinical progression: a study of fully automated immunoassays in BioFINDER and ADNI cohorts. Alzheimers Dement. 2018;14:1470‐1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doecke JD, Ward L, Burnham SC, et al. Elecsys CSF biomarker immunoassays demonstrate concordance with amyloid‐PET imaging. Alzheimers Res Ther. 2020;12:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shaw Leslie M., Waligorska Teresa, Fields Leona, Korecka Magdalena, Figurski Michal, Trojanowski John Q., Eichenlaub Udo, Wahl Simone, Quan Marian, Pontecorvo Michael J., Lachno D. Richard, Talbot Jayne A., Andersen Scott W., Siemers Eric R., Dean Robert A. (2018) Derivation of cutoffs for the Elecsys® amyloid β (1–42) assay in Alzheimer's disease. Alzheimer's & Dementia: Diagnosis, Assessment & Disease Monitoring, 10 (1), 698–705. 10.1016/j.dadm.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Willemse EAJ, van Maurik IS, Tijms BM, et al. Diagnostic performance of Elecsys immunoassays for cerebrospinal fluid Alzheimer's disease biomarkers in a nonacademic, multicenter memory clinic cohort: the ABIDE project. Alzheimers Dement (Amst). 2018;10:563‐572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blennow K, Shaw LM, Stomrud E, et al. Predicting clinical decline and conversion to Alzheimer's disease or dementia using novel Elecsys Aβ(1‐42). pTau and tTau CSF immunoassays Sci Rep. 2019;9:19024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roberts Rosebud O., Geda Yonas E., Knopman David S., Cha Ruth H., Pankratz V. Shane, Boeve Bradley F., Ivnik Robert J., Tangalos Eric G., Petersen Ronald C., Rocca Walter A. (2008) The Mayo Clinic Study of Aging: Design and Sampling, Participation, Baseline Measures and Sample Characteristics. Neuroepidemiology, 30 (1), 58–69. 10.1159/000115751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petersen R. C., Roberts R. O., Knopman D. S., Geda Y. E., Cha R. H., Pankratz V. S., Boeve B. F., Tangalos E. G., Ivnik R. J., Rocca W. A. (2010) Prevalence of mild cognitive impairment is higher in men: The Mayo Clinic Study of Aging. Neurology, 75 (10), 889–897. 10.1212/wnl.0b013e3181f11d85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med. 2004;256:183‐194. [DOI] [PubMed] [Google Scholar]
- 17. McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging‐Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263‐269. Internet]]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Harten Argonde C., Wiste Heather J., Weigand Stephen D., Mielke Michelle M., Kremers Walter K., Eichenlaub Udo, Batrla‐Utermann Richard, Dyer Roy B., Algeciras‐Schimnich Alicia, Knopman David S., Jack Clifford R., Petersen Ronald C. (2020) CSF biomarkers in Olmsted County. Neurology, 95 (3), e256–e267. 10.1212/wnl.0000000000009874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jack CR, Wiste HJ, Weigand SD, et al. Defining imaging biomarker cut points for brain aging and Alzheimer's disease. Alzheimers Dement. 2017;13:205‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klunk WE, Koeppe RA, Price JC, et al. The Centiloid Project: standardizing quantitative amyloid plaque estimation by PET. Alz Dem. 2015;11:1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwarz CG, Tosakulwong N, Senjem ML, et al. Considerations for performing level‐2 centiloid transformations for Amyloid PET SUVR values. Sci Rep. 2018;8(1):7421‐7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hansson O, Mikulskis A, Fagan AM, et al. The impact of preanalytical variables on measuring cerebrospinal fluid biomarkers for Alzheimer's disease diagnosis: a review. Alzheimers Dement. 2018;14:1313‐1333. [DOI] [PubMed] [Google Scholar]
- 23. Teunissen CE, Tumani H, Engelborghs S, Mollenhauer B. Biobanking of CSF: international standardization to optimize biomarker development. Clin Biochem. 2014;47:288‐292. [DOI] [PubMed] [Google Scholar]
- 24. Lowe VJ, Lundt ES, Albertson SM, et al. Neuroimaging correlates with neuropathologic schemes in neurodegenerative disease. Alzheimers Dement. 2019;15:927‐939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. La Joie R, Ayakta N, Seeley WW, et al. Multisite study of the relationships between antemortem [11C]PIB‐PET Centiloid values and postmortem measures of Alzheimer's disease neuropathology. Alzheimers Dement. 2019;15:205‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mattsson N, Insel PS, Donohue M, et al. Independent information from cerebrospinal fluid amyloid‐beta and florbetapir imaging in Alzheimer's disease. Brain. 2015;138(Pt 3), 772‐783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reimand J, de Wilde A, Teunissen CE, et al. PET and CSF amyloid‐β status are differently predicted by patient features: information from discordant cases. Alzheimers Res Ther. 2019;11:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jack Clifford R., Bennett David A., Blennow Kaj, Carrillo Maria C., Feldman Howard H., Frisoni Giovanni B., Hampel Harald, Jagust William J., Johnson Keith A., Knopman David S., Petersen Ronald C., Scheltens Philip, Sperling Reisa A., Dubois Bruno (2016) A/T/N: An unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology, 87 (5), 539–547. 10.1212/wnl.0000000000002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jack CR, Bennett DA, Blennow K, et al. NIA‐AA Research Framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14:535‐562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bertens D, Tijms BM, Scheltens P, Teunissen CE, Visser PJ. Unbiased estimates of cerebrospinal fluid β‐amyloid 1‐42 cutoffs in a large memory clinic population. Alzheimers Res Ther. 2017;9:8. [[Internet]]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jack Clifford R., Bennett David A., Blennow Kaj, Carrillo Maria C., Feldman Howard H., Frisoni Giovanni B., Hampel Harald, Jagust William J., Johnson Keith A., Knopman David S., Petersen Ronald C., Scheltens Philip, Sperling Reisa A., Dubois Bruno (2016) A/T/N: An unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology, 87 (5), 539–547. 10.1212/wnl.0000000000002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garrett Stephanie L., McDaniel Darius, Obideen Malik, Trammell Antoine R., Shaw Leslie M., Goldstein Felicia C., Hajjar Ihab (2019) Racial Disparity in Cerebrospinal Fluid Amyloid and Tau Biomarkers and Associated Cutoffs for Mild Cognitive Impairment. JAMA Network Open, 2 (12), e1917363 10.1001/jamanetworkopen.2019.17363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


