Abstract
The TP63 is an indispensable transcription factor for development and homeostasis of epithelia and its derived glandular tissue. It is also involved in female germline cell quality control, muscle and thymus development. It is expressed as multiple isoforms transcribed by two independent promoters, in addition to alternative splicing occurring at the mRNA 3′-UTR. Expression of the TP63 gene, specifically the amino-deleted p63 isoform, ΔNp63, is required to regulate numerous biological activities, including lineage specification, self-renewal capacity of epithelial stem cells, proliferation/expansion of basal keratinocytes, differentiation of stratified epithelia. In cancer, ΔNp63 is implicated in squamous cancers pathogenesis of different origin including skin, head and neck and lung and in sustaining self-renewal of cancer stem cells. How this transcription factor can control such a diverse set of biological pathways is central to the understanding of the molecular mechanisms through which p63 acquires oncogenic activity, profoundly changing its down-stream transcriptional signature. Here, we highlight how different proteins interacting with p63 allow it to regulate the transcription of several central genes. The interacting proteins include transcription factors/regulators, epigenetic modifiers, and post-transcriptional modifiers. Moreover, as p63 depends on its interactome, we discuss the hypothesis to target the protein interactors to directly affect p63 oncogenic activities and p63-related diseases.
Keywords: cancer, epithelial cells, keratinocytes, p63
Introduction
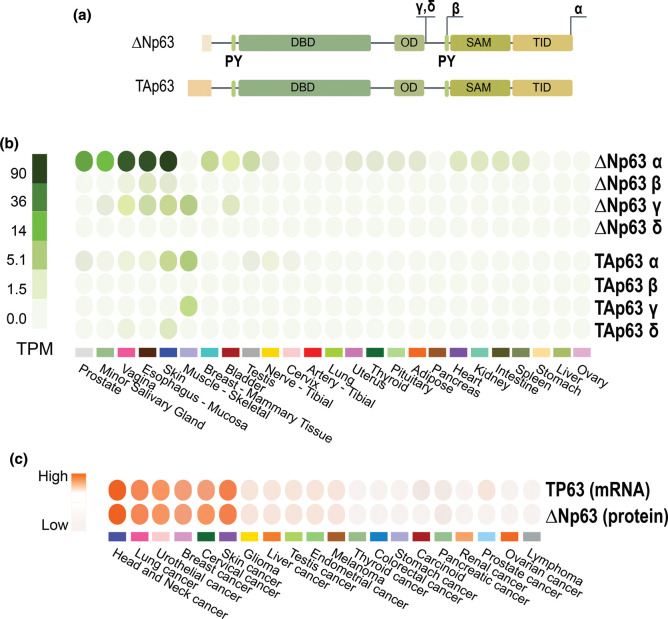
The transcription factor p63, a member of the p53 family of transcription factors [1–5], is a master regulator of gene transcription in epithelial development and homeostasis [6–10], as shown by the p63 knock-out phenotype. It is expressed as different isoforms because of the usage of two different promoters. One promoter precedes the first exon, while the second lies in the third intron Figure 1; by specific usage of these two distinct promoters, TP63 transcription generates two N-terminally different variants, named TAp63 and ΔNp63. Both transcripts undergo alternative splicing, giving rise to C-terminus variants ɑ, β and γ, so that in total, one gene encodes for at least six proteins (TAp63ɑ, TAp63β, TAp63γ, ΔNp63ɑ, ΔNp63β and ΔNp63γ) [8, 11, 12]. Additional isoforms (TAp63δ and ΔNp63δ) have been described in specific biological contests, Figure 1a [13]. p63 proteins are structurally organized in domains, all isoforms having a DNA-binding domain (DBD) and an oligomerization domain (OD). While the TAp63 isoforms contain an N-terminus acidic transactivation domain (TA), the ΔNp63 ones maintain the transactivating capability due to the presence of 14 unique residues [14]. Furthermore, both isoforms show an additional TA2 domain localized in the C-terminus [15]. The complexity of the proteins arises also from the C-terminus, in fact, the ɑ isoforms, but not the β and γ, contain a Sterile alpha-motif (SAM) domain and a Trans-Inhibitory domain (TID). These modules function as a protein–protein interaction domain, and as a regulatory module of TAp63ɑ transcriptional activity, respectively [16]. In addition, two proline-rich domains (PPxY) termed PY motifs are also involved in protein–protein interactions [17]. The TID domain and the PY motif undergo extensive post-translational modifications including sumoylation and ubiquitination [17].
Figure 1. The TP63 gene architecture and protein domains.
(a) p63 protein isoforms and organization. TP63 encodes for two main protein isoforms, TAp63 and ΔNp63, thanks to the alternative promoters (P1 and P2). Alternative splicing events at the C-terminus generate other variants: α, β, and γ. Exons skipping event produces variants δ. p63 protein isoforms are organized in different domains: TA, transactivating domain (absent from the ΔNp63 isoforms); DBD, DNA-binding domain; OD, oligomerization domain; SAM, sterile alpha motif; TID, trans-inhibitory domain. PY protein–protein interacting motifs are also indicated. (b) TAp63 and ΔNp63 isoforms expression level in different normal tissues (GTex portal). (c) TP63 mRNA and ΔNp63 protein expression in several tumors tissue (The Human Protein Atlas), see also (https://www.proteinatlas.org/ENSG00000073282-TP63).
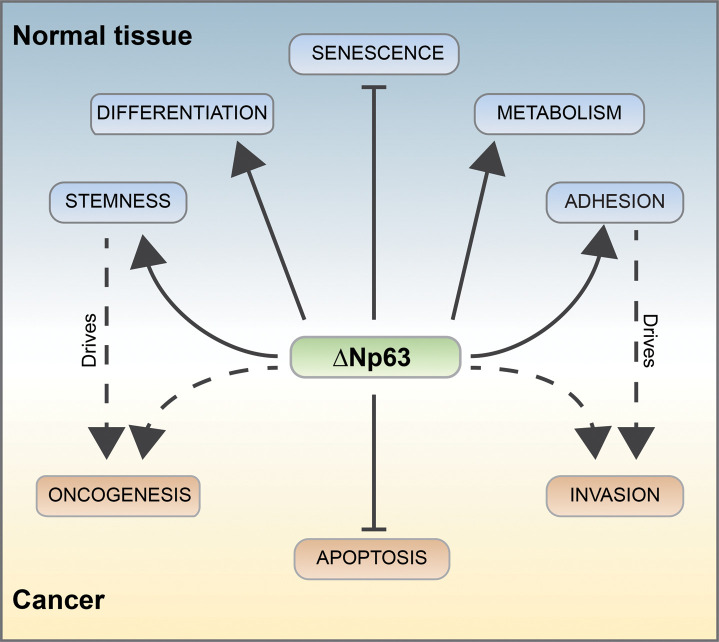
TAp63 and ΔNp63 isoforms are typically differentially expressed in tissues (Figure 1b,c) [18]. The complexity of the p63 transcripts and protein isoforms leads scientists to predict the functional complexity of the gene. The existence of several p63 isoforms expressed in different tissues, with distinct, often opposing functions, allows p63 to have a wide array of effects (Figure 2). Indeed, we still need to define the molecular mechanisms regulating p63 transcriptional signature in different biological contexts. The TAp63 isoforms bind canonical p53-responsive elements (RE) on DNA and can activate the transcription of several p53 target genes. Therefore, similarly to p53, TAp63 acts as a tumor suppressor, inducing cell cycle arrest and apoptosis [11, 19–22]. The TAp63 isoforms are detectable at basal level in skeletal muscles and testis (Figure 1b). In addition, TAp63 isoform is expressed in resting oocytes, and it is post-translationally activated in response to DNA damaging agents [18, 23]. In oocytes, TAp63α is activated by CHK2, CK1 and c-Abl phosphorylation in response to DNA damage [24, 25], leading to Puma and Noxa expression [26, 27] and cell death. This apoptotic role of p63, specifically TAp63 isoform, is also conserved during neuronal development, together with p53, it can regulate neuronal death [28, 29]. The ΔNp63 isoforms present a specific p63-RE. ΔNp63 protein is mainly involved in maintaining self-renewing capacity of progenitor cells in different epithelia, including epidermis and thymic epithelial cells [7, 30–32]. The epidermis is characterized by a high turnover, that is regenerated by mitotically active keratinocytes in the inner basal layer. Basal keratinocytes migrate to the outer cornified layer, giving rise to the terminally differentiated keratinized compartment [33–36]. The renewal capacity of the epidermis is due to the presence of epidermal stem cells and transient amplifying (TA) cells in the basal layer, expressing high levels of ΔNp63. ΔNp63 is highly expressed also in the proliferative compartments of glandular and simple epithelia [7, 37–39]. In the thymus ΔNp63, controlling the expression of Jag2 and EgfR2-IIIb, is essential for thymus development controlling thymic stem cell maintenance [7, 32].At a functional level, ΔNp63 directly regulates the expression of many genes, thereby affecting several biological processes. ΔNp63 transcriptional signature is crucial to control: (i) morphogenesis and differentiation; (ii) fate specification: (iii) proliferation; (iv) senescence; (v) cell–cell and cell–matrix interactions; (vi) cell metabolism (Figure 2) [40–42]. ΔNp63 has a crucial role in embryonic development and morphogenesis, as demonstrated by the phenotype of both the global p63 knockout and the selective ΔNp63 knockouts [32, 43, 6 44]. p63 knockout embryos manifest by E10.5 an array of severe alterations affecting epithelial-derived body parts, including skin, limbs, hair, teeth, hard palate, salivary and mammary glands, and the urogenital tract. Not last p63 affects the mitochondrial metabolism [45–47], which redox regulation [48–50] is crucial to determine cell fate [51, 52].
Figure 2. p63-dependent pathways.
p63 can regulate several biochemical pathways relevant for normal epithelia tissue homeostasis and tumorigenesis. ΔNp63 controls the formation and the homeostasis of the epidermis by regulating epithelial stemness, differentiation, metabolism, and adhesion. These pathways, when deregulated, could also participate in tumor formation and progression, even though additional interactors may well direct the transcription activity towards distinct promoters. p63 also negatively regulates senescence in normal tissue [53, 54] and counteract apoptotic response in cancer cells.
ΔNp63 regulates keratinocyte proliferation, differentiation and homeostasis, by directly transactivating a plethora of target genes, for example KRT14, PERP, EVPK, BPAG, CDKN1A, ITGA3, ITGB4, JAG1, and JAG2 [8]. In keratinocytes, p63 also regulates the chromatin landscape by directly transcribing chromatin-modulating factors that allow the opening of the chromatin. For instance, in epithelial progenitor cells, ΔNp63 directly controls the transcription of BRG1, the catalytic subunit the ATP-dependent chromatin remodeler BAF (SWI/SNF) that allows nucleosome remodeling in the region of the epidermal differentiation complex (EDC), a keratinocytes lineage-specific gene cluster [55–57]. ΔNp63 also directly transcribes the chromobox protein homolog 4 (Cbx4), a transcriptional repressor required to inhibit the expression of non-epidermal genes during differentiation and in stem cells [58, 59].
Mutations of TP63 cause developmental disorders, including ectrodactyly–ectodermal dysplasia–cleftlip/palate (EEC) syndrome, ankyloblepharon–ectodermal defects–cleft lip/palate (AEC) syndrome and split-hand/foot malformation-IV syndrome (SHFM) [38]. These mutations are localized in the DBD, the SAM domain and the TID, respectively [60]. TP63 is also linked to cancer.
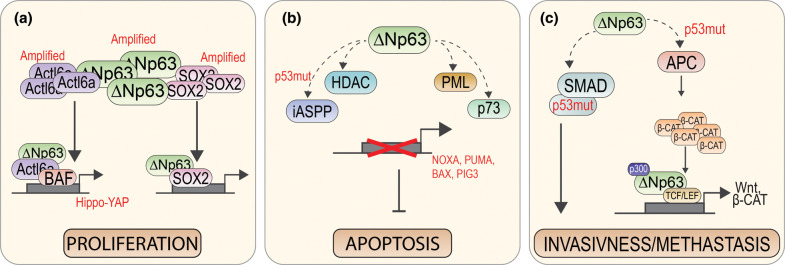
TP63 is rarely mutated in human cancers [61], thereby excluding it as a classic tumor suppressor gene. Yet, in specific poorly differentiated carcinomas, including squamous cell carcinomas (SCCs), ΔNp63 is overexpressed and/or amplified [62–64], conferring a proliferative advantage to cancer cells (Figures 1c, 3). SCC of the Head and Neck (HNSCC) and Skin Squamous Cell Carcinoma (sSCC) are the sixth most common cancers worldwide [65–69]. Tumors originate from squamous epithelia and/or mucosa in different anatomical sites. Whole-exome sequencing data, while confirming mutations in known genes (TP53, CDKN2A, PTEN, PIK3CA and HRAS), also identified 30% of the cases harboring mutations/amplifications in genes important for squamous differentiation (including NOTCH and TP63), indicating that epithelial differentiation deregulation strongly participate to SCC carcinogenesis [70, 71, 72 57]. ΔNp63 expression has been also described as an indicator of poor prognosis for several different tumor types [73]. ΔNp63 can engage specific oncogenic programs related to apoptosis resistance [74] and promotion of anchorage-dependent and-independent growth, mobility, and invasion in HNSCC [42, 64] (Figure 3a). Interestingly, when overexpressed in epithelial cancer cells, ΔNp63, and its interactors, acts as an oncogene, profoundly changing their down-stream transcriptional signature [64]. The mechanisms driving ΔNp63-selective transcription in cancer cells as well as its contribution to the regulation of the epigenetic landscape in tumors have not been fully investigated yet. In this review, we will focus on p63α isoform, we describe how multiple proteins that interact with ΔNp63α enable it to regulate the transcription of several genes in different biological contexts. Specifically, we will highlight transcription factors/epigenetic complexes and post-transcriptional modifiers that are crucial to uncover ΔNp63α functions in keratinocytes and SCCs. The list of selected interactors is shown in Table 1. Figure 4 shows the molecular determinants of p63 that are responsible for these interactions.
Figure 3. p63 regulated pathway in cancer.
(a) In SCC p63 is co-amplified with ACTL6a [64]. Their interaction maintains the proliferative state of the tumor cells together with other subunits of the BAF-complex [75]. The p63-ACTL6a interaction is also important in the regulation of the Hippo-YAP signaling pathway in cancer [64, 75, 76]. Similarly, the amplification of SOX2 in SCC results in the interaction with p63 and the proliferation regulation of cancer cell, among other co-regulated genes [77]. (b) To allow the cancer cell survival p63 can inhibit the proliferation program through its interactors. The p63 interaction with HDAC PML and p73 inhibits the transcription of pro-apoptotic gene, such as BAX and PUMA [78–82]. The interaction of p63 with iASPP leads to apoptosis inhibition only when p53 is mutated [83]. (c) The interaction of p63 with APC complex is also mediated by the p53 mutation. Under this condition, the β-catenin accumulation occurs in the nucleus leading to the aberrant regulation of the Wnt pathway, also mediated by p63-TCF/LEF interaction [84, 85]. Moreover, the p63-p300 contributes to the β-catenin accumulation in the nucleus [86–88]. Lastly the p63/SMAD/mutant-p53 complex formation is observed in cancer and leads to the down-regulation of metastasis suppressor genes [89–91].
Table 1. ΔNp63 and TAp63 interactors.
| Interactor | p63 isoform | p63 domain | Normal tissue/cell cancer type | References |
|---|---|---|---|---|
| Transcription factors/regulators | ||||
| p73 | ΔNp63α | OD | HNSCC | [78, 82] |
| p53 tumor derived mutants | ΔNp63α/γ | DBD | H-Ker; cancer cell lines | [89, 90, 93] |
| ASPP1/2iASPP | ΔNp63γ | DBD | c-SCLC | [83, 94] |
| SMAD2/3 | ΔNp63α | Not specified | BRCA | [90] |
| c-REL | ΔNP63α | C-terminus | H-Ker; HNSCC | [95, 168] |
| PML | TA/ΔNp63α | Not specified | H-Ker | [79] |
| β-CATENIN/GSK3β/PP2A | ΔNp63α | C-terminus/Not specified/N-terminus | HNSCC | [84, 85] |
| SOX2 | ΔNp63α | Not specified | LSCC | [77] |
| SETX | TA/ΔNp63α | C-terminus | H-Ker; HNSCC | [96] |
| NRF2 | ΔNp63α | Not specified | H-Ker | [97] |
| ABPP1 | TA/ΔNp63α | SAM | H-Ker | [98] |
| Chromatin modifiers | ||||
| BAF complex | ΔNp63α | C-terminus | H-Ker | [75] |
| ACTL6a | ΔNp63α | C-terminus | SCC; H-Ker | [64, 76] |
| KMT2D | ΔNp63α/β/γ | DBD | H-Ker | [99] |
| Dnmt3a | ΔNp63α | Not specified | H-Ker | [100] |
| p300 | ΔNP63α/γ; TAp63γ | N-terminus; C-terminus | SCC, OS | [86–88] |
| BRDT | ΔNp63α | Not specified | SCC | [101] |
| HDAC1/2 | ΔNp63α | TID | NSkin, SCC | [80, 81] |
| SRCAP | ΔNp63α | Not specified | SCC | [76] |
| CBX4 | ΔNp63α | Not specified | Nskin | [102] |
| SETDB1 | ΔNp63α | C-terminus | BRCA | [103] |
| Post-translation modifiers | ||||
| Ubiquitination | ||||
| MDM2; Fbw7 | TA/ΔNp63α | SAM | OS | [104, 169] |
| WWP1 | TA/ΔNp63α | PY | Bepithelium, CRC | [105] |
| PIR2/Rnf144B | ΔNp63α | Not specified | H-Ker; SCC | [105] |
| PIRH2 | TA/ΔNp63α | Not specified | H-Ker | [105] |
| PIN1 | TA/ΔNp63α | T538P | Nskin, SCC | [106] |
| ITCH | TA/ΔNp63α | Between PY and SAM | H-Ker | [107] |
| CHIP | TA/ΔNp63 | SAM | SCC | [108] |
| FBXO3 | ΔNp63α | Not specified | BRCA | [170] |
| Tmub1 | ΔNp63 | Not specified | HCC | [171] |
| SCF βTrCp1 | TA/ΔNp63γ | N/C-terminus | c-SCLC, H-Ker | [172] |
| Phosphorylation | ||||
| IKK(b) | ΔNp63α; TAp63γ | N-terminus PY | c-SCLC, H-Ker | [109, 173] |
| HIPK2 | ΔNp63α | C-terminus (T397) | H-Ker, tumor cell lines | [110] |
| APC/C complex | ΔNp63α | D-box (Degradation box) of the SAM | SCC, H-Ker | [174] |
| ATM; CDK2; p70s6K | ΔNp63α | On specific phosphorylation sites | SCC | [111] |
| p38 | ΔNp63α | C-tertminal (aa 435–408) | H-Ker | [112] |
| CRBN | TA/ΔNp63α | SAM (S509) | H-Ker | [175] |
| RACK1; SFN | ΔNp63α | C-terminus | SCC | [176] |
| Cdk12 | ΔNp63α | DBD | H-Ker | [177] |
| SMG1 | ΔNp63α | C-terminus | H-Ker | [177] |
| Sumoylation | ||||
| UBC9; SUMO1 | TA/ΔNp63α | SAM | H-Ker | [113] |
Abbreviation: OS, Osteosarcoma; LSCC, Lung squamous cell carcinoma; SCC, squamous cell carcinoma; HNSCC, Head&Neck squamous cell carcinoma; BRCA, Breast cancer; CRC, Colon rectal cancer; Nskin, Normal skin; c-SCLC, small cell lung cancer; Bepithelium, Breast epithelium; H-Ker, human keratinocytes; HCC, Hepatocellular carcinoma. The list of interactors was obtained using web databases (https://string-db.org/; https://www.ebi.ac.uk/intact/). The interactors were restricted based on strong biochemical experiments (endoguenous/semiendogenous co-immunoprecipitation, co-localization, mass-spectrometry analysis of pool-down experiments) present in literature, supporting a direct interaction in human cancer cell lines or tumour samples.
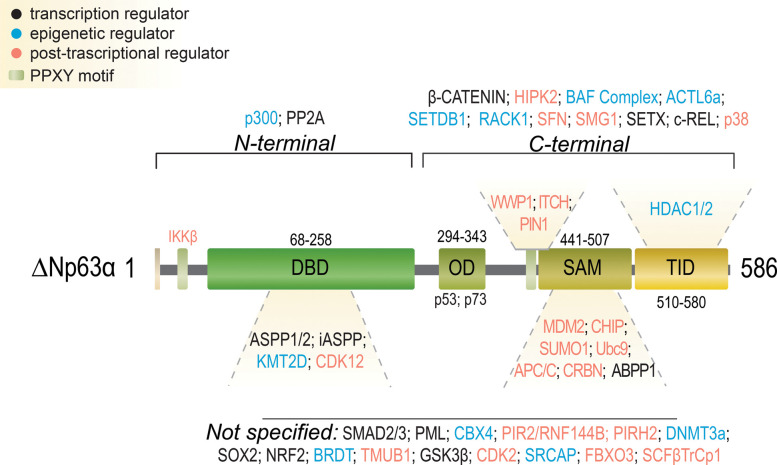
Figure 4. p63 interacting proteins.
Detailed representation of p63 protein structure. The p63 interacting domain with the different interactor is also indicated. The different types of interactors are indicated by color code. Transcription regulators: black; epigenetic regulators: blue; post-transcriptional regulators: orange (for further details and references, see Table 1). The PPXXY motifs are indicated in light green.
Exploiting the p63 interactome in epithelial development and cancer
P63 interacts with transcriptional factors and co-regulators
Chromatin immunoprecipitation-sequencing experiments demonstrated that the p63 role is mainly related to enhancer-mediated transactivation. As matter of fact, the majority of the p63-binding sites and p63-bound open chromatin regions in keratinocytes are active enhancers, while only a small number of p63-bound regions are promoters [75, 114–116]. Beside coordinating chromatin-remodeling enzymes and nucleosome modifiers (see section below) at the enhancer sites, ΔNp63 also directly interacts with other transcriptional factors and regulators, resulting in mutual regulation of their transcriptional activities (see Table 1 and Figure 4 for details).
In cancer cells, p63 interacts also with its own family members, including p73 and mutant-p53 (Figure 3b,c). The TAp73 isoform regulates apoptosis and cell cycle via binding p53 RE on DNA to transactivate specific target genes [117, 118]. Giving their shared homology in the OD, it is expected that they could interact, forming hetero tetramers [89, 119]. In HNSCC, ΔNp63α interacts with TAp73β and inhibits TAp73 pro-apoptotic activity (Figure 3b). ΔNp63α overexpression in HNSCC cells leads to cell survival, and its down-regulation results in pro-apoptotic gene induction (PUMA and NOXA) thanks to the release of the inhibition on TAp73β [78]. Co-transcriptional regulators of the p53 family can also act on p63. ASPP1 and ASSP2, for instance, are apoptosis regulators binding the p53 DBD. This interaction enhances p53 transactivation function on apoptotic genes, such as BAX and PIG3 [120]. ASPP1 and ASPP2 can also interact with p63 and p73, stimulating their transactivation function specifically on the promoters of BAX, PIG3, and PUMA but not on those of MDM2 or p21, although the mechanism of selectivity is not known [94]. The other member of the ASPP family, iASPP, inhibitor of p53 activity, also interacts with p63 in normal keratinocytes [121] to allow proper proliferation and differentiation programmes. iASPP participates in the p63-mediated epithelial integrity program by regulating the expression of genes controlling cell adhesion. In cutaneous SCC, iASPP and p63 autoregulatory feedback loop controls cell migration facilitating proliferation and blocking epithelial-mesenchymal transition [122]. In other cancers, the p63/p73 interaction with iASPP leads to the inhibition of p53-independent apoptosis, suggesting a possible rescue by down-regulation of iASPP, and the consequent apoptosis activation mediated by p63/p73, in cancers where p53 is mutated (Figure 3b) [83, 122].
In normal epithelial cells, p63 is important to sustain proliferation and epithelial specification, However, p63 opposes TGF-β induced metastasis in breast cancer cells. From a mechanistic point of view, TGFβ acts in concert with mutant-p53 and oncogenic Ras to promote the formation of a mutant-p53/p63 protein complex in which SMADs proteins are the crucial components [90]. When p63 is sequestered in the ternary complex, its binding to DNA is impaired and its functions are antagonized, leading to the down-regulation of two metastasis suppressor genes, Sharp-1 and Cyclin G2[90]. In several HNSCC cell lines, the NF-kB subunit c-REL together with TAp73, mutant-p53, and ΔNp63α forms a nuclear complex. Upon TNFα stimulation, c-REL interacts with ΔNp63α, and dissociates TAp73 and ΔNp63α. Therefore, TNFα or the c-REL level affects ΔNp63α/TAp73 interaction and transcription of different p63 targets, like p21, WAF1, NOXA, and PUMA [95].
The promyelocytic leukemia protein (PML) that functions via its association with PML-nuclear bodies (PML-NBs) can act as a transcriptional cofactor and is another regulator of the p53 family members. PML plays a role in a wide range of crucial cellular processes, including transcriptional regulation, tumor suppression, apoptosis, senescence, DNA damage response, as well as in viral defense [95, 123]. It has been previously shown that both p53 and p73 physically interact with PML through their DBDs [3, 95, 123, 124]. PML physically interacts with TAp63α and ΔNp63α isoforms as well. p63 is confined into the PML nuclear-bodies (PML-NBs) in vivo, and PML manages p63 transcriptional activity (Figure 3b). This interaction increases the levels of p63 in cultured cells and enhances its capability to transactivate the p21, GADD45 and BAX promoters [79].
ΔNp63 also modulates the Wnt/β-Catenin signal transduction pathway that plays an important role in controlling cell proliferation in tumor initiation and development [125, 126]. Upon Wnt activation, the β-Catenin accumulation in the nucleus leads to Tcf/Lef-responsive genes expression, responsible for increasing cell proliferation and de-differentiation [127]. Lack of Wnt, instead, leads to the interaction of β-Catenin with glycogen synthase kinase 3β (GSK3β), Axin, adenomatous polyposis coli (APC) and protein phosphatase 2A (PP2A). GSK3β phosphorylates β-Catenin and targets it to the proteasome [128]. By two hybrid assays, it has been shown that ΔNp63 is directly associated with B56a, a regulatory subunit of PP2A and APC complex. Under normal conditions, p53 mediates ΔNp63 and β-catenin degradation. In SCC, instead, mutant-p53 causes APC complex and ΔNp63 release, leading to β-catenin aberrant accumulation in the nucleus (Figure 3c) [84, 85, 129].
In SCC, ΔNp63 co-operates with the transcription factor SOX2. SOX2 is involved in the maintenance of pluripotency and stemness [130]. These two factors physically interact and co-occupy several genetic loci in SCC. Together, they regulate the transcription of many target genes, including the transcription factor ETV4, a gene generally amplified in SCC tumors (Figure 3a) [77, 130].
During keratinocyte differentiation, ΔNp63 associates with Senataxin (SETX), a R-loop-resolving enzyme. Both ΔNp63 and SETX physically bind the p63 DNA-binding motif present in early epidermal differentiation genes, in order to facilitate R-loop removal and allow efficient transcriptional termination. Interestingly, SETX expression is deregulated in cutaneous SCC, indicating that ΔNp63 and SETX interaction is important in skin pathologies [96]. To sustain proliferation and wound repair in keratinocytes, ΔNp63 interacts with nuclear factor erythroid 2-related factor 2 (NFE2L2; NRF2). Chromatin immunoprecipitation and sequencing data demonstrated the binding of human NRF2 and p63 at a genome-wide level to both enhancers and promoters of selected genes [97]. This opens the discussion on the epigenetic regulation of p63, see also below. Interestingly, p63 controls the stemness potential of the lung regeneration [131]; indeed following a necrotizing degeneration from H1N1 viral infection, Distal Airway Stem Cells (DASC) expressing Trp63 (p63) and keratin 5, called DASC(p63/Krt5) seems to be crucial for the repair and tissue regeneration [131]. This occurs also in normal lung homeostasis, and the presence of pre-existing DASC(p63/Krt5) cells is pivotal to allow regeneration and prevent fibrotic lesions which would reduce oxygen exchange, as seen in Chronic Obstructive Pulmonary Disease, COPD. Here, an epigenetic regulation on Trp63 predisposes to an inflammatory response, driving the transcription complex towards pro-inflammatory and pro-fibrotic programs able to trigger immune infiltrations of neutrophils, T-cells, NK cells and macrophages [132].
Finally, p63 also participates in splicing regulation by interacting with the splicing machinery. Both TAp63 and ΔNp63 isoforms bind apobec-1-binding protein-1 (ABBP1), a component of the RNA processing machinery. This interaction causes a shift in the alternative splicing of FGFR-2, a gene implicated in epithelial differentiation. In AEC syndrome, mutations of p63 in the SAM domain affect the RNA processing, since p63 is enabled to bind ABBP1 [97, 98].
Altogether these studies, although they report only selected p63 interactors, consistently highlight that p63 co-operates with specific transcription factors and regulators during keratinocyte maturation and in different phases of tumor development.
P63 co-ordinates chromatin-remodeling enzymes and nucleosome modifiers
Several studies implicated p63 as a crucial signaling node during epidermal differentiation and stem cell maintenance, through the control of the chromatin state [133–135]. As a matter of fact, several epigenetic determinants are physically associated with ΔNp63 (Table 1, Figure 4).
In keratinocytes, an important role is played by the BAF (SWI/SNF) complex, an ATP-dependent nucleosome modifier, that is important to open the chromatin and to allow the expression of differentiation genes [136]. Interestingly, ATAC-Seq, ChIP-seq and co-immunoprecipitation experiments demonstrated that p63 physically interacts with the BAF complex, and their cooperative fashion acts to control keratinocyte-specific open chromatin structure to allow epidermal differentiation [75]. These data showed that high levels of p63 and ACTL6A are necessary to suppress differentiation and promote the epithelial progenitor state in normal epidermis. The association of p63 with BAF also occurs in HNSCCs, where the ACTL6A is a central oncogenic driver, commonly found amplified at genomic level or overexpressed [137]. Interestingly, both ACTL6A and TP63 are co-amplified in HNSCC, they are also physically co-associated and co-expressed together with other BAF complex subunits co-regulating a key set of relevant targets, including genes encoding components of the Hippo-YAP signaling pathway (Figure 3a) [64]. These findings indicated that p63/BAF function together in a common pathway to counteract differentiation both in normal epithelial progenitor cells and in tumor cells [57, 64].
Another ΔNp63 interactor required for proper epithelial stem cell maintenance and differentiation is the epigenetic regulator KMT2D (MLL4). KMT2D is the most frequently mutated gene in several cancer types, including cutaneous SCC and basaliomas [99]. KMT2D is a histone methyltransferase that deposits the monomethylation (H3K4me1) to the histone H3 Lys 4 (H3K4), facilitates enhancers activation and transcription factor binding [99]. In proliferating keratinocytes, KMT2D interacts with p63 at enhancer sites, controlling the expression of genes involved in epithelial development, adhesion, and differentiation. KMT2D participates in the p63-mediated transcription of genes typically expressed in the self-renewing and proliferating compartment, including integrins, laminins as well as key regulators of proper epidermal differentiation (retinoid and vitamin D) [99]. It is not known at present, whether this interaction is maintained or lost in tumors.
DNA methylation is also an important epigenetic modification to control tissue homeostasis and renewal [138]. In epithelial progenitor cells, the de novo DNA methyltransferases DNMT3a and DNMT3b are expressed at high levels. DNMT3a increases during differentiation while DNMT3b decreases [139]. DNMT3a is highly associated with enhancer regions and physically interacts with ΔNp63 to maintain high levels of DNA hydroxymethylation in the enhancer region. This interaction, through a mechanism involving the DNA methylase Tet2, allows enhancer activation [100]. It has been estimated that ∼50% of the DNMT3a-associated enhancers were bound by p63 in epithelial progenitor cells, and specifically regulate the expression of genes involved in keratinocyte proliferation [100].
The histone acetyltransferase p300 (cAMP response element-binding protein (CBP) or p300) is a transcriptional co-activator known to regulate both p53 and p73 [100, 140–143]. It has been demonstrated that p300 acetylates also non-histone proteins, including different p63 isoforms. p300 enhances p63γ-mediated transcription of p21, consequently leading to G1 arrest [86]. TAp63α also interacts with the Taz2 domain of p300. This interaction inhibits the DNA binding activity of TAp63 and its association with the transcriptional machinery [87]. A direct and specific interaction has been also described between the ΔNp63α C-terminus and p300. This association correlates with p63 function on the β-catenin promoter activation, in HNSCC (Figure 3c) [88]. In esophageal SCC, ΔNp63 physically interacts and co-operates with a member of the bromodomain testis-specific protein (BRDT), to control the expression of a subset of ΔNp63-specific genes related to the squamous cancer cell phenotype. The abnormal expression of BRDT enhances the cancer cells dependencies of ΔNp63α [101].
ΔNp63-mediated target gene repression has been described to be mediated by direct interaction with different chromatin modifiers. In HNSCC, ΔNp63α-HDAC1 and/or -HDAC2 physical interaction mediates transcriptional repression of genes involved in controlling cancer cell proliferation and survival (Figure 3b) [80, 81]. Chromatin immunoprecipitation experiment showed that endogenous p63 binds the PUMA promoter together with HDAC1. This interaction is necessary for SCC cell survival, leading to the repression of apoptosis [80]. In normal skin the p63-HDAC interaction leads to a reduction in proliferation without apoptosis activation [81]. Interestingly, the HDAC null mice phenotypically resemble p63 null mice, supporting the hypothesis that HDAC1/2 directly mediates the repression of many p63-dependent genes [81]. Another mechanism of ΔNp63α transcriptional repression in HNSCC is mediated by its interaction with a subunit of the SRCAP chromatin remodeling complex. This causes an histone exchange from H2A to H2A.Z. H2A.Z acts as a transcriptional repressor when localized near the transcription start sites [81, 144]. Thanks to H2A.Z deposition, several pro-proliferation ΔNp63α target genes are repressed [76]. In thymic epithelial cells (TEC) ΔNp63α also plays an important role in maintaining a high proliferative potential [32]. As a matter of fact, p63 null mice present several defects in TEC proliferation and expansion [7]. Cbx4, a subunit of the polycomb repressive complex 1 (PRC1), and ΔNp63α physically interact and act in the same pathway to stimulate the proliferative and self-renewal capacity of thymic epithelial stem cells [102]. In keratinocytes, Cbx4 represses non-epidermal lineage gene expression interacting with PRC1 [59]. Thus, p63 and Cbx4 represses the non-epidermal transcription program during keratinocyte proliferation via PRC1-mediated transcriptional repression. Interestingly, Cbx4 is also frequently amplified in SCCs [59, 145], indicating that the interaction between ΔNp63 and Cbx4 could also contribute to SCC formation. Finally, in breast cancer cells, SETDB1, a histone lysine methyltransferase, part of the SET (Suppression of variegation, Enhancer of zeste, Trithorax)-domain containing enzymes, specifically interacts with the ΔNp63α isoform [103, 146].
Altogether these studies demonstrated that during keratinocytes differentiation and tumor formation, several epigenetic modulators, including chromatin-remodeling complexes and enzymes, are involved in p63-mediated epigenomic reprogramming. In normal progenitor cells and in keratinocytes, p63 expression is required for establishing an epidermal-specific chromatin structure [75, 116, 147]. On the other hand, as a pro-survival oncogene in SCCs, ΔNp63α is utilized by tumor cells to establish squamous-specific chromatin architecture, which fosters tumor development [148].
Post-translational regulators/interactors controlling ΔNp63 protein levels
A third category of interactors is represented by enzymes that post-translationally modify p63, including kinases, E3 ubiquitin ligases (E3s) and SUMO ligases (Table 1, Figure 4). Indeed, similarly to p53, p63 regulation is mostly accomplished at post-translational level [149]. Here, we will review the contribution of phosphorylation and ubiquitination to the modulation of both p63 protein stability and transcriptional function. These two modifications are undoubtedly those that more broadly impact on p63.
Phosphorylation of p63 occurs predominantly in response to genotoxic stress. Coherently, p63 plays an instrumental role in cellular stress responses including those elicited by DNA damaging insults. ΔNp63 protein is a target for direct phosphorylation by ATM, CDK2 and p70s6K kinases following DNA damage exposure of HNSCC cells [111]. The authors demonstrated that ΔNp63 is sequentially phosphorylated by ATM, CDK2 and p70s6K kinases, ultimately leading to its proteasomal-dependent degradation, though the specific E3 involved in this mechanism has not been identified yet. Since ΔNp63 transcriptionally activates ATM, this would establish a feedback regulatory mechanism similar to the p53/MDM2 loop to control ΔNp63 protein level.
Similarly, phosphorylation of ΔNp63 at threonine residue 397 by Homeodomain-interacting protein kinase 2 (HIPK2) promotes its proteasomal proteolysis in response to different genotoxic stress and increases the susceptibility of p53-defective cells to chemotherapeutic drugs [110]. The Authors demonstrated that a non-phosphorylatable ΔNp63-T397A mutant is resistant to DNA damage-dependent, HIPK2-induced degradation, and confers resistance of cancer cells to doxorubicin.
An alternative molecular mechanism underlying the regulation of ΔNp63 protein stability in response to chemotherapeutic agents is mediated by IkappaB kinase (IKK). IKK is a multi-subunit protein kinase consisting of two catalytic subunits, IKKα and IKKβ, and a regulatory subunit, IKKγ. Chemotherapy-induced activation of IKKβ promotes ΔNp63 degradation leading to enhanced transactivation of p53 target genes and increased susceptibility to DNA damage-induced cell death [109].
Another kinase involved in p63 modification is p38, which is activated upon UVB irradiation, and regulates ΔNp63 functions by direct phosphorylation in keratinocytes. This modification acts as a signal for ΔNp63 to detach from the promoters of p53-regulated genes involved in DNA damage-induced apoptosis and allows p53 to eliminate potentially harmful cells. Of note, phosphorylation of ΔNp63 by p38 is a specific mechanism that is detected only in response to UV irradiation [112]. Additional kinases, CHK2, Abl, phosphorylate TAp63 isoform specifically during oocyte maturation and DNA-damage [25, 26, 150].
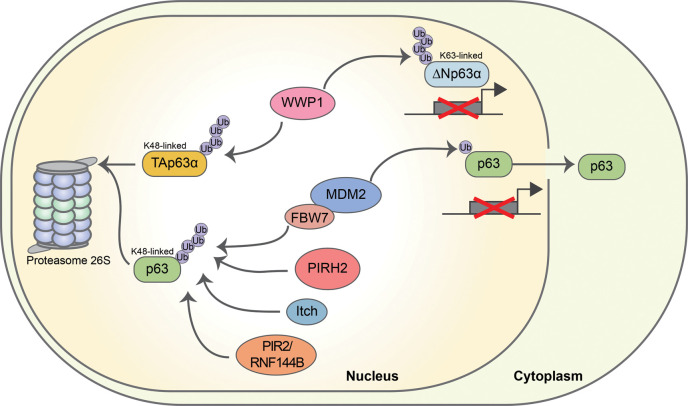
The most important mechanism of p63 degradation is the proteasome-dependent pathway. TAp63 proteins are highly labile, while ΔNp63 proteins are much more stable (Figure 5).
Figure 5. E3s involved in the regulation of p63 stabilization and transcriptional functions.
Several E3s, including ITCH, WWP1, PIRH2, PIR2/Rnf144B and MDM2 acting in concert with FBXW7, determine the ubiquitin-dependent degradation of p63 — through the formation of K48 polyubiquitin chains [92, 93, 105, 107]. WWP1 is also able to inhibit p63 transcriptional activity through K63 polyubiquitination. The mono-ubiquitination mediated by MDM2 allows p63 to translocate into the cytoplasm, ultimately inhibiting its transcriptional activity.
Several E3s, belonging to either the RING-finger-type or the HECT-type subfamilies, have been implicated in controlling p63 protein stability or function. A major role in the regulation of p63 stability in physiological conditions is executed by the HECT-type E3 ITCH [151–153] that interacts with the proline-rich motif (PPXY) present in the α and β isoforms of p63 through its WW domains [47, 107, 154]. ITCH targets endogenous p63 for proteasomal degradation in keratinocytes by modifying two lysine residues (K193 and K194), whose mutations have been associated with the SHFM-4 syndrome. Hence, these mutations likely act by altering the stability of p63a/b isoforms during development. Overall, these findings emphasize a relevant role for the ITCH/p63 functional interaction in the regulation of epithelial homeostasis (Figure 5).
The PPXY motif of p63 is characterized by the nearby presence of a (T/S) P motif, which is a recognition site for the WW domain present in the prolyl-isomerase Pin1. Phosphorylation of the threonine residue and proline cis/trans isomerization by Pin1 of this (T/S) P motif is a crucial regulatory event that results in an increase in the ITCH-WW-p63 complex stability [154].
Another HECT-type E3 involved in p63 regulation is WW domain-containing E3 ubiquitin protein ligase 1 (WWP1) [155, 156]. Modification of p63 by WWP1 may have distinct outcomes depending on the nature of the assembled ubiquitin chains (Figure 5). Attachment of K63-polyubiquitin chains increases ΔNp63-dependent transcriptional activation and its pro-survival behavior [156]. On the contrary, modification of TAp63 by K48-linked chains promotes its proteasomal degradation in colon cancer cells, conferring resistance to DNA damage-induced cell death [155, 156]. In both cases, WWP1 contributes to the oncogenic activity of p63 in cancer cells.
Pin1 has been reported to interfere with the WWP1-WW-p63- association by directly binding to the threonine residue located in (T/S) P motif. This interaction results in the inhibition of p63 proteasomal degradation by WWP1 and, hence, promotes ΔNp63-induced cell proliferation [106].
The RING-finger type MDM2 (murine double minute-2), the primary regulator of p53/p73 protein stability [157, 158], also binds p63 though through weaker interactions. The formation of the MDM2-p63 complex seems to have contradictory effects on p63 function. Mono-ubiquitination of p63 by MDM2 enhances its nuclear export, thus negatively affecting its transactivation potential, with no effect on its degradation [159]. Another study reported that MDM2 increases p63 protein stability resulting in increased transcriptional activity (Figure 5) [160].
Although Mdm2 is incapable of directly targeting p63 for proteolysis, it can co-operate with the F-Box protein Fbw7 to poly-ubiquitinate ΔNp63 and target it for proteasomal degradation [104]. The Authors reported that MDM2 binds ΔNp63 in the nucleus promoting its translocation to the cytoplasm, where p63 is targeted for degradation by the Fbw7 E3-ubiquitin ligase. Fbw7 recognizes a phosphodegron located in the SAM domain and containing a threonine residue that has to be phosphorylated by glycogen synthase kinase 3 in order for Fbw7 to associate with p63 and promote its degradation. The MDM2/Fbw7 cooperation is functionally relevant during keratinocyte differentiation when ΔNp63 needs to be degraded (Figure 5) [104].
PIRH2 (p53-induced protein with a RING-H2 domain), a monomeric RING-finger-type E3, plays a physiologically important role in keratinocyte differentiation through the ubiquitin-mediated proteasomal degradation of p63. Binding of PIRH2 to a yet not identified structural motif of p63 leads to its ubiquitination and subsequent reduction in protein half-life of p63 in keratinocytes [161]. The PIRH2/p63 functional axis becomes relevant also in a cancer context. Indeed, arsenic trioxide treatment of cancer cells leads to Pirh2 promoter transactivation, which in turn decreases the stability of ΔNp63 reducing its pro-oncogenic activities [162].
The RING-finger E3 PIR2/Rnf144B is transcriptionally induced by ΔNp63 in primary keratinocytes, where it functions as a negative regulator of both p21 and p63 to control cell fate by sustaining proliferation and differentiation of epidermal cells [105]. Indeed, the Authors also reported that PIR2/Rnf14 is capable of ubiquitinating p63 to induce its proteasomal degradation, thus generating an autoregulatory loop, like the p53–MDM2 feedback circuit [105]. High expression of both ΔNp63 and PIR2/Rnf14 in squamous cell carcinomas (SCCs) suggests that the ΔNp63-PIR2/Rnf14 axis may be not operative in these tumors resulting in the protection of ΔNp63 degradation, which ultimately leads to ΔNp63 accumulation. These findings indicate that the p63/PIR2/p21 axis has a major role in regulating the homeostasis of stratified epithelia.
CHIP (C-terminus of Hsc-70 interacting protein), an E3 containing a U-box domain, also promotes p63 degradation via the ubiquitin-proteasome pathway [108]. Here, the authors reported that Hsp70 plays a key role in regulating CHIP-mediated p63 ubiquitination. In cancer cells, it has been found that Hsp70 overexpression increases TAp63 ubiquitination and reduces TAp63 protein levels, while prevents ΔNp63 ubiquitination and causes its accumulation. This regulatory mechanism by Hsp70 ultimately contributes to cancer cell migration and invasion.
ΔNp63 protein levels are strictly regulated by conjugation with SUMO. Sumoylation of ΔNp63 occurs at lysine residue K637 located in the SAM domain. Sumoylation represents a prerequisite for efficient ΔNp63α poly-ubiquitylation [163, 164]. Interestingly, p63 mutants having alteration in their sumoylation capacity are found in the human hereditary congenital SHFM, EEC and LMS (Limb Mammary Syndrome) [113].
Concluding remarks
Here, we highlight the complexity of ΔNp63α interactome in keratinocyte proliferation/differentiation and in squamous cell carcinomas. Squamous cell carcinoma remains an unresolved challenge to human health due to its aggressiveness and lack of specific therapeutic options. These tumors are highly dependent on ΔNp63α. The presence of p63 in chromatin-remodeling complexes provides an opportunity to target SCCs overexpressing ΔNp63α via epigenetic drugs. To note, many of p63 interactors, such as KMT2D and SWI/SNF, are frequently mutated in SCC [165–167], further supporting the relevance that the normal p63-mediated transcriptional program in cells must be perturbed to allow SCC initiation and development. Fully understanding the ΔNp63α-interactors pattern in SCCs may be a valuable strategy to enable the development of novel therapeutic approaches. As a matter of fact, the abrogation of ΔNp63α interaction with interactors/co-activators could broadly affect ΔNp63-dependent transcription. On the other hand, since phosphorylation and ubiquitination are undoubtedly the modifications that more broadly impact on p63 protein stability and transcriptional function, it is conceivable that small molecules, by modulating the activity of these enzymes, could be explored as therapeutic options in SCCs. Additional studies are necessary to fully understand the complexity of the interactions between epigenetic regulators and ΔNp63α.
Abbreviations
- ABBP1
apobec-1-binding protein-1
- APC
adenomatous polyposis coli
- BRDT
bromodomain testis-specific protein
- DASC
distal airway stem cells
- DBD
DNA-binding domain
- HIPK2
homeodomain-interacting protein kinase 2
- IKK
IkappaB kinase
- OD
oligomerization domain
- PML
promyelocytic leukemia protein
- RE
responsive elements
- SAM
Sterile alpha-motif
- SCCs
squamous cell carcinomas
- SETX
Senataxin
- SHFM
split-hand/foot malformation-IV syndrome
- TA
transactivation domain
- TID
trans-inhibitory domain
- WWP1
WW domain-containing E3 ubiquitin protein ligase 1
Competing Interests
G.M. is a member of the Editorial Board of Biochemical Journal. The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was mainly supported by AIRC IG Grant 22206 to EC. Partially supported by the Italian Ministry of Health, IDI-IRCCS (RC to EC, CRI-NMSC, RF to E.D. and E.C.), and by AIRC IG Grant 23232 to F.B. and to G.M. (IG#20473; 2018-2022). Work has been also supported by Regione Lazio through LazioInnova Progetto Gruppo di Ricerca n 33 & 55-2021-T0002E0001.
CRediT Author Contribution
Gerry Melino: Resources, Data curation, Supervision, Funding acquisition, Validation, Writing — original draft, Writing — review and editing. Rosalba Pecorari: Data curation, Formal analysis, Validation, Investigation, Writing — original draft, Writing — review and editing. Francesca Bernassola: Data curation, Writing — original draft, Writing — review and editing. Eleonora Candi: Conceptualization, Resources, Data curation, Supervision, Validation, Writing — original draft, Project administration, Writing — review and editing.
References
- 1.De Laurenzi, V. and Melino, G. (2000) Evolution of functions within the p53/p63/p73 family. Ann. N. Y. Acad. Sci. 926, 90–100 10.1111/j.1749-6632.2000.tb05602.x [DOI] [PubMed] [Google Scholar]
- 2.Candi, E., Agostini, M., Melino, G. and Bernassola, F. (2014) How the TP53 family proteins TP63 and TP73 contribute to tumorigenesis: regulators and effectors. Hum. Mutat. 35, 702–714 10.1002/humu.22523 [DOI] [PubMed] [Google Scholar]
- 3.Bernassola, F., Salomoni, P., Oberst, A., Di Como, C.J., Pagano, M., Melino, G.et al. (2004) Ubiquitin-dependent degradation of p73 is inhibited by PML. J. Exp. Med. 199, 1545–1557 10.1084/jem.20031943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amelio, I. and Melino, G. (2015) The p53 family and the hypoxia-inducible factors (HIFs): determinants of cancer progression. Trends Biochem. Sci. 40, 425–434 10.1016/j.tibs.2015.04.007 [DOI] [PubMed] [Google Scholar]
- 5.Tomasini, R., Tsuchihara, K., Tsuda, C., Lau, S.K., Wilhelm, M., Rufini, A.et al. (2009) TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc. Natl Acad. Sci. U.S.A. 106, 797–802 10.1073/pnas.0812096106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang, A., Schweitzer, R., Sun, D., Kaghad, M., Walker, N., Bronson, R.T.et al. (1999) P63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714–718 10.1038/19539 [DOI] [PubMed] [Google Scholar]
- 7.Senoo, M., Pinto, F., Crum, C.P. and McKeon, F. (2007) P63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129, 523–536 10.1016/j.cell.2007.02.045 [DOI] [PubMed] [Google Scholar]
- 8.Candi, E., Cipollone, R., Rivetti di Val Cervo, P., Gonfloni, S., Melino, G. and Knight, R. (2008) P63 in epithelial development. Cell. Mol. Life Sci. 65, 3126–3133 10.1007/s00018-008-8119-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shalom-Feuerstein, R., Lena, A.M., Zhou, H., De La Forest Divonne, S., Van Bokhoven, H., Candi, E.et al. (2011) Δnp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 18, 887–896 10.1038/cdd.2010.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Candi, E., Terrinoni, A., Rufini, A., Chikh, A., Lena, A.M., Suzuki, Y.et al. (2006) P63 is upstream of IKK alpha in epidermal development. J. Cell Sci. 119, 4617–4622 10.1242/jcs.03265 [DOI] [PubMed] [Google Scholar]
- 11.Yang, A., Kaghad, M., Wang, Y., Gillett, E., Fleming, M.D., Dötsch, V.et al. (1998) P63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2, 305–316 10.1016/S1097-2765(00)80275-0 [DOI] [PubMed] [Google Scholar]
- 12.Lena, A.M., Duca, S., Novelli, F., Melino, S., Annicchiarico-Petruzzelli, M., Melino, G.et al. (2015) Amino-terminal residues of ΔNp63, mutated in ectodermal dysplasia, are required for its transcriptional activity. Biochem. Biophys. Res. Commun. 467, 434–440 10.1016/j.bbrc.2015.09.111 [DOI] [PubMed] [Google Scholar]
- 13.Mangiulli, M., Valletti, A., Caratozzolo, M.F., Tullo, A., Sbisà, E., Pesole, G.et al. (2009) Identification and functional characterization of two new transcriptional variants of the human p63 gene. Nucleic Acids Res. 37, 6092–6104 10.1093/nar/gkp674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helton, E.S., Zhu, J. and Chen, X. (2006) The unique NH2-terminally deleted (DeltaN) residues, the PXXP motif, and the PPXY motif are required for the transcriptional activity of the deltaN variant of p63. J. Biol. Chem. 281, 2533–2542 10.1074/jbc.M507964200 [DOI] [PubMed] [Google Scholar]
- 15.Duijf, P.H.G. (2002) Gain-of-function mutation in ADULT syndrome reveals the presence of a second transactivation domain in p63. Hum. Mol. Genet. 11, 799–804 10.1093/hmg/11.7.799 [DOI] [PubMed] [Google Scholar]
- 16.Ghioni, P., Bolognese, F., Duijf, P.H.G., Van Bokhoven, H., Mantovani, R. and Guerrini, L. (2002) Complex transcriptional effects of p63 isoforms: identification of novel activation and repression domains. Mol. Cell. Biol. 22, 8659–8668 10.1128/MCB.22.24.8659-8668.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collavin, L., Lunardi, A. and Del Sal, G. (2010) p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death Differ. 17, 901–911 10.1038/cdd.2010.35 [DOI] [PubMed] [Google Scholar]
- 18.Su, X., Chakravarti, D. and Flores, E.R. (2013) P63 steps into the limelight: crucial roles in the suppression of tumorigenesis and metastasis. Nat. Rev. Cancer 13, 136–143 10.1038/nrc3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flores, E.R., Tsai, K.Y., Crowley, D., Sengupta, S., Yang, A., McKeon, F.et al. (2002) P63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416, 560–564 10.1038/416560a [DOI] [PubMed] [Google Scholar]
- 20.Guo, X., Keyes, W.M., Papazoglu, C., Zuber, J., Li, W., Lowe, S.W.et al. (2009) TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat. Cell Biol. 11, 1451–1457 10.1038/ncb1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suh, E.-K., Yang, A., Kettenbach, A., Bamberger, C., Michaelis, A.H., Zhu, Z.et al. (2006) P63 protects the female germ line during meiotic arrest. Nature 444, 624–628 10.1038/nature05337 [DOI] [PubMed] [Google Scholar]
- 22.Flores, E.R., Sengupta, S., Miller, J.B., Newman, J.J., Bronson, R., Crowley, D.et al. (2005) Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7, 363–373 10.1016/j.ccr.2005.02.019 [DOI] [PubMed] [Google Scholar]
- 23.Candi, E., Melino, G., Tóth, A. and Dötsch, V. (2021) Mechanisms of quality control differ in male and female germ cells. Cell Death Differ. 28, 2300–2302 10.1038/s41418-021-00818-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan, M., Luong, P., Hudson, C., Gudmundsdottir, K. and Basu, S. (2010) c-Abl phosphorylation of ΔNp63α is critical for cell viability. Cell Death Dis. 1, e16 10.1038/cddis.2009.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonfloni, S., Di Tella, L., Caldarola, S., Cannata, S.M., Klinger, F.G., Di Bartolomeo, C.et al. (2009) Inhibition of the c-Abl-TAp63 pathway protects mouse oocytes from chemotherapy-induced death. Nat. Med. 15, 1179–1185 10.1038/nm.2033 [DOI] [PubMed] [Google Scholar]
- 26.Lena, A.M., Rossi, V., Osterburg, S., Smirnov, A., Osterburg, C., Tuppi, M.et al. (2021) The p63 C-terminus is essential for murine oocyte integrity. Nat. Commun. 12, 383 10.1038/s41467-020-20669-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gebel, J., Tuppi, M., Sänger, N., Schumacher, B. and Dötsch, V. (2020) DNA damaged induced cell death in oocytes. Molecules 25, 5714 10.3390/molecules25235714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancino, G.I., Yiu, A.P., Fatt, M.P., Dugani, C.B., Flores, E.R., Frankland, P.W.et al. (2013) P63 regulates adult neural precursor and newly born neuron survival to control hippocampal-dependent behavior. J. Neurosci. 33, 12569–12585 10.1523/JNEUROSCI.1251-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs, W.B., Govoni, G., Ho, D., Atwal, J.K., Barnabe-Heider, F., Keyes, W.M.et al. (2005) P63 is an essential proapoptotic protein during neural development. Neuron 48, 743–756 10.1016/j.neuron.2005.10.027 [DOI] [PubMed] [Google Scholar]
- 30.Xin, L., Lukacs, R.U., Lawson, D.A., Cheng, D. and Witte, O.N. (2007) Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells 25, 2760–2769 10.1634/stemcells.2007-0355 [DOI] [PubMed] [Google Scholar]
- 31.Chakrabarti, R., Wei, Y., Hwang, J., Hang, X., Andres Blanco, M., Choudhury, A.et al. (2014) Δnp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat. Cell Biol. 16, 1004–1015, 1–13 10.1038/ncb3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Candi, E., Rufini, A., Terrinoni, A., Giamboi-Miraglia, A., Lena, A.M., Mantovani, R.et al. (2007) Deltanp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc. Natl Acad. Sci. U.S.A. 104, 11999–12004 10.1073/pnas.0703458104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lena, A.M., Shalom-Feuerstein, R., Rivetti di Val Cervo, P., Aberdam, D., Knight, R.A., Melino, G.et al. (2008) miR-203 represses “stemness” by repressing DeltaNp63. Cell Death Differ. 15, 1187–1195 10.1038/cdd.2008.69 [DOI] [PubMed] [Google Scholar]
- 34.Candi, E., Schmidt, R. and Melino, G. (2005) The cornified envelope: a model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 6, 328–340 10.1038/nrm1619 [DOI] [PubMed] [Google Scholar]
- 35.Aberdam, D., Candi, E., Knight, R.A. and Melino, G. (2008) miRNAs, “stemness” and skin. Trends Biochem. Sci. 33, 583–591 10.1016/j.tibs.2008.09.002 [DOI] [PubMed] [Google Scholar]
- 36.Steinert, P.M., Candi, E., Tarcsa, E., Marekov, L.N., Sette, M., Paci, M.et al. (1999) Transglutaminase crosslinking and structural studies of the human small proline rich 3 protein. Cell Death Differ. 6, 916–930 10.1038/sj.cdd.4400568 [DOI] [PubMed] [Google Scholar]
- 37.Chakrabarti, R., Sinha, S. and Kang, Y. (2016) Abstract B21: DNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signaling. Mol. Cancer Res. 14, B1 10.1158/1557-3125.DEVBIOLCA15-B21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Celli, J., Duijf, P., Hamel, B.C., Bamshad, M., Kramer, B., Smits, A.P.et al. (1999) Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 99, 143–153 10.1016/S0092-8674(00)81646-3 [DOI] [PubMed] [Google Scholar]
- 39.Mills, A.A., Zheng, B., Wang, X.J., Vogel, H., Roop, D.R. and Bradley, A. (1999) P63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713 10.1038/19531 [DOI] [PubMed] [Google Scholar]
- 40.Candi, E., Smirnov, A., Panatta, E., Lena, A.M., Novelli, F., Mancini, M.et al. (2017) Metabolic pathways regulated by p63. Biochem. Biophys. Res. Commun. 482, 440–444 10.1016/j.bbrc.2016.10.094 [DOI] [PubMed] [Google Scholar]
- 41.Dotsch, V., Bernassola, F., Coutandin, D., Candi, E. and Melino, G. (2010) P63 and p73, the ancestors of p53. Cold Spring Harb. Perspect. Biol. 2, a004887 10.1101/cshperspect.a004887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Compagnone, M., Gatti, V., Presutti, D., Ruberti, G., Fierro, C., Markert, E.K.et al. (2017) ΔNp63-mediated regulation of hyaluronic acid metabolism and signaling supports HNSCC tumorigenesis. Proc. Natl Acad. Sci. U.S.A. 114, 13254–13259 10.1073/pnas.1711777114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vanbokhoven, H., Melino, G., Candi, E. and Declercq, W. (2011) P63, a story of mice and men. J. Invest. Dermatol. 131, 1196–1207 10.1038/jid.2011.84 [DOI] [PubMed] [Google Scholar]
- 44.Candi, E., Rufini, A., Terrinoni, A., Dinsdale, D., Ranalli, M., Paradisi, A.et al. (2006) Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ. 13, 1037–1047 10.1038/sj.cdd.4401926 [DOI] [PubMed] [Google Scholar]
- 45.Viticchiè, G., Agostini, M., Lena, A.M., Mancini, M., Zhou, H., Zolla, L.et al. (2015) P63 supports aerobic respiration through hexokinase II. Proc. Natl Acad. Sci. U.S.A. 112, 11577–11582 10.1073/pnas.1508871112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Annicchiarico-Petruzzelli, M., Di Daniele, N. and Candi, E. (2015) ßnp63 controls cellular redox status. Oncoscience 2, 661–662 10.18632/oncoscience.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bellomaria, A., Barbato, G., Melino, G., Paci, M. and Melino, S. (2012) Recognition mechanism of p63 by the E3 ligase itch: novel strategy in the study and inhibition of this interaction. Cell Cycle 11, 3638–3648 10.4161/cc.21918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bucciarelli, T., Sacchetta, P., Pennelli, A., Cornelio, L., Romagnoli, R., Melino, S.et al. (1999) Characterization of toad liver glutathione transferase. Biochim. Biophys. Acta 1431, 189–198 10.1016/S0167-4838(99)00036-9 [DOI] [PubMed] [Google Scholar]
- 49.Vitali, A., Botta, B., Delle Monache, G., Zappitelli, S., Ricciardi, P., Melino, S.et al. (1998) Purification and partial characterization of a peroxidase from plant cell cultures of cassia didymobotrya and biotransformation studies. Biochem. J. 331, 513–519 10.1042/bj3310513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nepravishta, R., Sabelli, R., Iorio, E., Micheli, L., Paci, M. and Melino, S. (2012) Oxidative species and S-glutathionyl conjugates in the apoptosis induction by allyl thiosulfate. FEBS J. 279, 154–167 10.1111/j.1742-4658.2011.08407.x [DOI] [PubMed] [Google Scholar]
- 51.Gallo, M., Paludi, D., Cicero, D.O., Chiovitti, K., Millo, E., Salis, A.et al. (2005) Identification of a conserved N-capping box important for the structural autonomy of the prion alpha 3-helix: the disease associated D202N mutation destabilizes the helical conformation. Int. J. Immunopathol. Pharmacol. 18, 95–112 10.1177/039463200501800111 [DOI] [PubMed] [Google Scholar]
- 52.Melino, S., Nepravishta, R., Bellomaria, A., Di Marco, S. and Paci, M. (2009) Nucleic acid binding of the RTN1-C C-terminal region: toward the functional role of a reticulon protein. Biochemistry 48, 242–253 10.1021/bi801407w [DOI] [PubMed] [Google Scholar]
- 53.Rivetti di Val Cervo, P., Lena, A.M., Nicoloso, M., Rossi, S., Mancini, M., Zhou, H.et al. (2012) p63-microRNA feedback in keratinocyte senescence. Proc. Natl Acad. Sci. U.S.A. 109, 1133–1138 10.1073/pnas.1112257109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Notari, M., Hu, Y., Koch, S., Lu, M., Ratnayaka, I., Zhong, S.et al. (2011) Inhibitor of apoptosis-stimulating protein of p53 (iASPP) prevents senescence and is required for epithelial stratification. Proc. Natl Acad. Sci. U.S.A. 108, 16645–16650 10.1073/pnas.1102292108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Botchkarev, V.A. (2015) Epigenetic regulation of epidermal development and keratinocyte differentiation. J. Investig. Dermatol. Symp. Proc. 17, 18–19 10.1038/jidsymp.2015.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Panatta, E., Lena, A.M., Mancini, M., Smirnov, A., Marini, A., Delli Ponti, R.et al. (2020) Long non-coding RNA uc.291 controls epithelial differentiation by interfering with the ACTL6A/BAF complex. EMBO Rep. 21, e46734 10.15252/embr.201846734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mancini, M., Cappello, A., Pecorari, R., Lena, A.M., Montanaro, M., Fania, L.et al. (2021) Involvement of transcribed lncRNA uc.291 and SWI/SNF complex in cutaneous squamous cell carcinoma. Discov. Oncol. 12, 14 10.1007/s12672-021-00409-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luis, N.M., Morey, L., Mejetta, S., Pascual, G., Janich, P., Kuebler, B.et al. (2011) Regulation of human epidermal stem cell proliferation and senescence requires polycomb- dependent and -independent functions of Cbx4. Cell Stem Cell 9, 233–246 10.1016/j.stem.2011.07.013 [DOI] [PubMed] [Google Scholar]
- 59.Mardaryev, A.N., Liu, B., Rapisarda, V., Poterlowicz, K., Malashchuk, I., Rudolf, J.et al. (2016) Cbx4 maintains the epithelial lineage identity and cell proliferation in the developing stratified epithelium. J. Cell Biol. 212, 77–89 10.1083/jcb.201506065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ianakiev, P., Kilpatrick, M.W., Toudjarska, I., Basel, D., Beighton, P. and Tsipouras, P. (2000) Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am. J. Hum. Genet. 67, 59–66 10.1086/302972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Melino, G., Lu, X., Gasco, M., Crook, T. and Knight, R.A. (2003) Functional regulation of p73 and p63: development and cancer. Trends Biochem. Sci. 28, 663–670 10.1016/j.tibs.2003.10.004 [DOI] [PubMed] [Google Scholar]
- 62.Candi, E., Dinsdale, D., Rufini, A., Salomoni, P., Knight, R.A., Mueller, M.et al. (2007) TAp63 and ΔNp63 in cancer and epidermal development. Cell Cycle 6, 274–285 10.4161/cc.6.3.3797 [DOI] [PubMed] [Google Scholar]
- 63.Ramsey, M.R., Wilson, C., Ory, B., Rothenberg, S.M., Faquin, W., Mills, A.A.et al. (2013) FGFR2 signaling underlies p63 oncogenic function in squamous cell carcinoma. J. Clin. Invest. 123, 3525–3538 10.1172/JCI68899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saladi, S.V., Ross, K., Karaayvaz, M., Tata, P.R., Mou, H., Rajagopal, J.et al. (2017) ACTL6A is co-amplified with p63 in squamous cell carcinoma to drive YAP activation, regenerative proliferation, and poor prognosis. Cancer Cell 31, 35–49 10.1016/j.ccell.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reddy, P., Yao, M. and Patel, M. (2021) Investigative landscape in advanced non-melanoma skin cancers. Curr. Treat. Options Oncol. 22, 56 10.1007/s11864-021-00853-0 [DOI] [PubMed] [Google Scholar]
- 66.Park, J.-O., Nam, I.-C., Kim, C.-S., Park, S.-J., Lee, D.-H., Kim, H.-B.et al. (2022) Sex differences in the prevalence of head and neck cancers: A 10-Year follow-Up study of 10 million healthy people. Cancers 14, 2521 10.3390/cancers14102521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siegel, R.L., Miller, K.D., Fuchs, H.E. and Jemal, A. (2021) Cancer statistics, 2021. CA Cancer J. Clin. 71, 7–33 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 68.Fania, L., Samela, T., Moretta, G., Ricci, F., Dellambra, E., Mancini, M.et al. (2021) Attitudes among dermatologists regarding non-melanoma skin cancer treatment options. Discov. Oncol. 12, 31 10.1007/s12672-021-00421-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ragaini, B.S., Blizzard, L., Newman, L., Stokes, B., Albion, T. and Venn, A. (2021) Temporal trends in the incidence rates of keratinocyte carcinomas from 1978 to 2018 in Tasmania, Australia: a population-based study. Discov. Oncol. 12, 30 10.1007/s12672-021-00426-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dotto, G.P. and Rustgi, A.K. (2016) Squamous cell cancers: a unified perspective on biology and genetics. Cancer Cell 29, 622–637 10.1016/j.ccell.2016.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Campbell, J.D., Yau, C., Bowlby, R., Liu, Y., Brennan, K., Fan, H.et al. (2018) Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep. 23, 194–212.e6 10.1016/j.celrep.2018.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stransky, N., Egloff, A.M., Tward, A.D., Kostic, A.D., Cibulskis, K., Sivachenko, A.et al. (2011) The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garcia, S., Dalès, J.-P., Charafe-Jauffret, E., Carpentier-Meunier, S., Andrac-Meyer, L., Jacquemier, J.et al. (2007) Poor prognosis in breast carcinomas correlates with increased expression of targetable CD146 and c-Met and with proteomic basal-like phenotype. Hum. Pathol. 38, 830–841 10.1016/j.humpath.2006.11.015 [DOI] [PubMed] [Google Scholar]
- 74.Yang, A., Zhu, Z., Kapranov, P., McKeon, F., Church, G.M., Gingeras, T.R.et al. (2006) Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell 24, 593–602 10.1016/j.molcel.2006.10.018 [DOI] [PubMed] [Google Scholar]
- 75.Bao, X., Rubin, A.J., Qu, K., Zhang, J., Giresi, P.G., Chang, H.Y.et al. (2015) A novel ATAC-seq approach reveals lineage-specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biol. 16, 284 10.1186/s13059-015-0840-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gallant-Behm, C.L., Ramsey, M.R., Bensard, C.L., Nojek, I., Tran, J., Liu, M.et al. (2012) Δnp63α represses anti-proliferative genes via H2A.Z deposition. Genes Dev. 26, 2325–2336 10.1101/gad.198069.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watanabe, H., Ma, Q., Peng, S., Adelmant, G., Swain, D., Song, W.et al. (2014) SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J. Clin. Invest. 124, 1636–1645 10.1172/JCI71545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rocco, J.W., Leong, C.-O., Kuperwasser, N., DeYoung, M.P. and Ellisen, L.W. (2006) P63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell 9, 45–56 10.1016/j.ccr.2005.12.013 [DOI] [PubMed] [Google Scholar]
- 79.Bernassola, F., Oberst, A., Melino, G. and Pandolfi, P.P. (2005) The promyelocytic leukaemia protein tumour suppressor functions as a transcriptional regulator of p63. Oncogene 24, 6982–6986 10.1038/sj.onc.1208843 [DOI] [PubMed] [Google Scholar]
- 80.Ramsey, M.R., He, L., Forster, N., Ory, B. and Ellisen, L.W. (2011) Physical association of HDAC1 and HDAC2 with p63 mediates transcriptional repression and tumor maintenance in squamous cell carcinoma. Cancer Res. 71, 4373–4379 10.1158/0008-5472.CAN-11-0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.LeBoeuf, M., Terrell, A., Trivedi, S., Sinha, S., Epstein, J.A., Olson, E.N.et al. (2010) Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev. Cell 19, 807–818 10.1016/j.devcel.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davison, T.S., Vagner, C., Kaghad, M., Ayed, A., Caput, D. and Arrowsmith, C.H. (1999) P73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J. Biol. Chem. 274, 18709–18714 10.1074/jbc.274.26.18709 [DOI] [PubMed] [Google Scholar]
- 83.Cai, Y., Qiu, S., Gao, X., Gu, S.-Z. and Liu, Z.-J. (2012) iASPP inhibits p53-independent apoptosis by inhibiting transcriptional activity of p63/p73 on promoters of proapoptotic genes. Apoptosis 17, 777–783 10.1007/s10495-012-0728-z [DOI] [PubMed] [Google Scholar]
- 84.Patturajan, M., Nomoto, S., Sommer, M., Fomenkov, A., Hibi, K., Zangen, R.et al. (2002) Deltanp63 induces beta-catenin nuclear accumulation and signaling. Cancer Cell 1, 369–379 10.1016/S1535-6108(02)00057-0 [DOI] [PubMed] [Google Scholar]
- 85.Drewelus, I., Göpfert, C., Hippel, C., Dickmanns, A., Damianitsch, K., Pieler, T.et al. (2010) P63 antagonizes Wnt-induced transcription. Cell Cycle 9, 580–587 10.4161/cc.9.3.10593 [DOI] [PubMed] [Google Scholar]
- 86.MacPartlin, M., Zeng, S., Lee, H., Stauffer, D., Jin, Y., Thayer, M.et al. (2005) P300 regulates p63 transcriptional activity. J. Biol. Chem. 280, 30604–30610 10.1074/jbc.M503352200 [DOI] [PubMed] [Google Scholar]
- 87.Krauskopf, K., Gebel, J., Kazemi, S., Tuppi, M., Löhr, F., Schäfer, B.et al. (2018) Regulation of the activity in the p53 family depends on the organization of the transactivation domain. Structure 26, 1091–1100.e4 10.1016/j.str.2018.05.013 [DOI] [PubMed] [Google Scholar]
- 88.Katoh, I., Maehata, Y., Moriishi, K., Hata, R.-I. and Kurata, S.-I. (2019) C-terminal α domain of p63 binds to p300 to coactivate β-catenin. Neoplasia 21, 494–503 10.1016/j.neo.2019.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gaiddon, C., Lokshin, M., Ahn, J., Zhang, T. and Prives, C. (2001) A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 21, 1874–1887 10.1128/MCB.21.5.1874-1887.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Adorno, M., Cordenonsi, M., Montagner, M., Dupont, S., Wong, C., Hann, B.et al. (2009) A mutant-p53/Smad complex opposes p63 to empower TGFβ-induced metastasis. Cell 137, 87–98 10.1016/j.cell.2009.01.039 [DOI] [PubMed] [Google Scholar]
- 91.Strano, S., Fontemaggi, G., Costanzo, A., Rizzo, M.G., Monti, O., Baccarini, A.et al. (2002) Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J. Biol. Chem. 277, 18817–18826 10.1074/jbc.M201405200 [DOI] [PubMed] [Google Scholar]
- 92.Bellomaria, A., Barbato, G., Melino, G., Paci, M. and Melino, S. (2010) Recognition of p63 by the E3 ligase ITCH: Effect of an ectodermal dysplasia mutant. Cell Cycle 9, 3730–3739 10.4161/cc.9.18.12933 [DOI] [PubMed] [Google Scholar]
- 93.Oberst, A., Malatesta, M., Aqeilan, R.I., Rossi, M., Salomoni, P., Murillas, R.et al. (2007) The Nedd4-binding partner 1 (N4BP1) protein is an inhibitor of the E3 ligase Itch. Proc. Natl Acad. Sci. U.S.A. 104, 11280–11285 10.1073/pnas.0701773104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bergamaschi, D., Samuels, Y., Jin, B., Duraisingham, S., Crook, T. and Lu, X. (2004) ASPP1 and ASPP2: common activators of p53 family members. Mol. Cell. Biol. 24, 1341–1350 10.1128/MCB.24.3.1341-1350.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu, H., Yang, X., Duggal, P., Allen, C.T., Yan, B., Cohen, J.et al. (2011) TNF-α promotes c-REL/ΔNp63α interaction and TAp73 dissociation from key genes that mediate growth arrest and apoptosis in head and neck cancer. Cancer Res. 71, 6867–6877 10.1158/0008-5472.CAN-11-2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gatti, V., Fierro, C., Compagnone, M., La Banca, V., Mauriello, A., Montanaro, M.et al. (2022) ΔNp63-Senataxin circuit controls keratinocyte differentiation by promoting the transcriptional termination of epidermal genes. Proc. Natl Acad. Sci. U.S.A. 119, e2104718119 10.1073/pnas.2104718119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kurinna, S., Seltmann, K., Bachmann, A.L., Schwendimann, A., Thiagarajan, L., Hennig, P.et al. (2021) Interaction of the NRF2 and p63 transcription factors promotes keratinocyte proliferation in the epidermis. Nucleic Acids Res. 49, 3748–3763 10.1093/nar/gkab167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fomenkov, A., Huang, Y.-P., Topaloglu, O., Brechman, A., Osada, M., Fomenkova, T.et al. (2003) P63 alpha mutations lead to aberrant splicing of keratinocyte growth factor receptor in the Hay-Wells syndrome. J. Biol. Chem. 278, 23906–23914 10.1074/jbc.M300746200 [DOI] [PubMed] [Google Scholar]
- 99.Lin-Shiao, E., Lan, Y., Coradin, M., Anderson, A., Donahue, G., Simpson, C.L.et al. (2018) KMT2D regulates p63 target enhancers to coordinate epithelial homeostasis. Genes Dev. 32, 181–193 10.1101/gad.306241.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rinaldi, L., Datta, D., Serrat, J., Morey, L., Solanas, G., Avgustinova, A.et al. (2016) Dnmt3a and Dnmt3b associate with enhancers to regulate human epidermal stem cell homeostasis. Cell Stem Cell 19, 491–501 10.1016/j.stem.2016.06.020 [DOI] [PubMed] [Google Scholar]
- 101.Wang, X., Kutschat, A.P., Yamada, M., Prokakis, E., Böttcher, P., Tanaka, K.et al. (2021) Bromodomain protein BRDT directs ΔNp63 function and super-enhancer activity in a subset of esophageal squamous cell carcinomas. Cell Death Differ. 28, 2207–2220 10.1038/s41418-021-00751-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu, B., Liu, Y.-F., Du, Y.-R., Mardaryev, A.N., Yang, W., Chen, H.et al. (2013) Cbx4 regulates the proliferation of thymic epithelial cells and thymus function. Development 140, 780–788 10.1242/dev.085035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Regina, C., Compagnone, M., Peschiaroli, A., Lena, A.M., Melino, G. and Candi, E. (2016) Δnp63α modulates histone methyl transferase SETDB1 to transcriptionally repress target genes in cancers. Cell Death Discov. 2, 16015 10.1038/cddiscovery.2016.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Galli, F., Rossi, M., D'Alessandra, Y., De Simone, M., Lopardo, T., Haupt, Y.et al. (2010) MDM2 and Fbw7 cooperate to induce p63 protein degradation following DNA damage and cell differentiation. J. Cell Sci. 123, 2423–2433 10.1242/jcs.061010 [DOI] [PubMed] [Google Scholar]
- 105.Conforti, F., Yang, A.L., Piro, M.C., Mellone, M., Terrinoni, A., Candi, E.et al. (2013) PIR2/Rnf144B regulates epithelial homeostasis by mediating degradation of p21WAF1 and p63. Oncogene 32, 4758–4765 10.1038/onc.2012.497 [DOI] [PubMed] [Google Scholar]
- 106.Li, C., Chang, D.L., Yang, Z., Qi, J., Liu, R., He, H.et al. (2013) Pin1 modulates p63α protein stability in regulation of cell survival, proliferation and tumor formation. Cell Death Dis. 4, e943 10.1038/cddis.2013.468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rossi, M., Aqeilan, R.I., Neale, M., Candi, E., Salomoni, P., Knight, R.A.et al. (2006) The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc. Natl Acad. Sci. U.S.A. 103, 12753–12758 10.1073/pnas.0603449103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu, H.H., Wang, B., Armstrong, S.R., Abuetabh, Y., Leng, S., Roa, W.H.Y.et al. (2021) Hsp70 acts as a fine-switch that controls E3 ligase CHIP-mediated TAp63 and ΔNp63 ubiquitination and degradation. Nucleic Acids Res. 49, 2740–2758 10.1093/nar/gkab081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chatterjee, A., Sen, T., Chang, X. and Sidransky, D. (2010) Yes-associated protein 1 regulates the stability of ΔNp63α. Cell Cycle 9, 162–167 10.4161/cc.9.1.10321 [DOI] [PubMed] [Google Scholar]
- 110.Lazzari, C., Prodosmo, A., Siepi, F., Rinaldo, C., Galli, F., Gentileschi, M.et al. (2011) HIPK2 phosphorylates ΔNp63α and promotes its degradation in response to DNA damage. Oncogene 30, 4802–4813 10.1038/onc.2011.182 [DOI] [PubMed] [Google Scholar]
- 111.Huang, Y., Sen, T., Nagpal, J., Upadhyay, S., Trink, B., Ratovitski, E.et al. (2008) ATM kinase is a master switch for the ΔNp63α phosphorylation/degradation in human head and neck squamous cell carcinoma cells upon DNA damage. Cell Cycle 7, 2846–2855 10.4161/cc.7.18.6627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Papoutsaki, M., Moretti, F., Lanza, M., Marinari, B., Sartorelli, V., Guerrini, L.et al. (2005) A p38-dependent pathway regulates DeltaNp63 DNA binding to p53-dependent promoters in UV-induced apoptosis of keratinocytes. Oncogene 24, 6970–6975 10.1038/sj.onc.1208835 [DOI] [PubMed] [Google Scholar]
- 113.Huang, Y.-P., Wu, G., Guo, Z., Osada, M., Fomenkov, T., Park, H.L.et al. (2004) Altered sumoylation of p63alpha contributes to the split-hand/foot malformation phenotype. Cell Cycle 3, 1587–1596 10.4161/cc.3.12.1290 [DOI] [PubMed] [Google Scholar]
- 114.Kouwenhoven, E.N., Oti, M., Niehues, H., van Heeringen, S.J., Schalkwijk, J., Stunnenberg, H.G.et al. (2015) Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep. 16, 863–878 10.15252/embr.201439941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sethi, I., Sinha, S. and Buck, M.J. (2014) Role of chromatin and transcriptional co-regulators in mediating p63-genome interactions in keratinocytes. BMC Genomics 15, 1042 10.1186/1471-2164-15-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Qu, J., Yi, G. and Zhou, H. (2019) P63 cooperates with CTCF to modulate chromatin architecture in skin keratinocytes. Epigenetics Chromatin 12, 31 10.1186/s13072-019-0280-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Melino, G., De Laurenzi, V. and Vousden, K.H. (2002) P73: friend or foe in tumorigenesis. Nat. Rev. Cancer 2, 605–615 10.1038/nrc861 [DOI] [PubMed] [Google Scholar]
- 118.Ramadan, S., Terrinoni, A., Catani, M.V., Sayan, A.E., Knight, R.A., Mueller, M.et al. (2005) P73 induces apoptosis by different mechanisms. Biochem. Biophys. Res. Commun. 331, 713–717 10.1016/j.bbrc.2005.03.156 [DOI] [PubMed] [Google Scholar]
- 119.Chan, W.M., Siu, W.Y., Lau, A. and Poon, R.Y.C. (2004) How many mutant p53 molecules are needed to inactivate a tetramer? Mol. Cell. Biol. 24, 3536–3551 10.1128/MCB.24.8.3536-3551.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Samuels-Lev, Y., O'Connor, D.J., Bergamaschi, D., Trigiante, G., Hsieh, J.-K., Zhong, S.et al. (2001) ASPP proteins specifically stimulate the apoptotic function of p53. Mol. Cell 8, 781–794 10.1016/S1097-2765(01)00367-7 [DOI] [PubMed] [Google Scholar]
- 121.Chikh, A., Matin, R.N.H., Senatore, V., Hufbauer, M., Lavery, D., Raimondi, C.et al. (2011) iASPP/p63 autoregulatory feedback loop is required for the homeostasis of stratified epithelia. EMBO J. 30, 4261–4273 10.1038/emboj.2011.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Robinson, D.J., Patel, A., Purdie, K.J., Wang, J., Rizvi, H., Hufbauer, M.et al. (2019) Epigenetic regulation of iASPP-p63 feedback loop in cutaneous squamous cell carcinoma. J. Invest. Dermatol. 139, 1658–1671.e8 10.1016/j.jid.2019.01.020 [DOI] [PubMed] [Google Scholar]
- 123.Lallemand-Breitenbach, V. and de Thé, H. (2018) PML nuclear bodies: from architecture to function. Curr. Opin. Cell Biol. 52, 154–161 10.1016/j.ceb.2018.03.011 [DOI] [PubMed] [Google Scholar]
- 124.Guo, A., Salomoni, P., Luo, J., Shih, A., Zhong, S., Gu, W.et al. (2000) The function of PML in p53-dependent apoptosis. Nat. Cell Biol. 2, 730–736 10.1038/35036365 [DOI] [PubMed] [Google Scholar]
- 125.Yu, J. and Virshup, D.M. (2014) Updating the Wnt pathways. Biosci. Rep. 34, e00142 10.1042/BSR20140119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhan, T., Rindtorff, N. and Boutros, M. (2017) Wnt signaling in cancer. Oncogene 36, 1461–1473 10.1038/onc.2016.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bryja, V., Červenka, I. and Čajánek, L. (2017) The connections of Wnt pathway components with cell cycle and centrosome: side effects or a hidden logic? Crit. Rev. Biochem. Mol. Biol. 52, 614–637 10.1080/10409238.2017.1350135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Clevers, H. and Nusse, R. (2012) Wnt/β-Catenin signaling and disease. Cell 149, 1192–1205 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 129.Katoh, I., Fukunishi, N., Fujimuro, M., Kasai, H., Moriishi, K., Hata, R.-I.et al. (2016) Repression of Wnt/β-catenin response elements by p63 (TP63). Cell Cycle 15, 699–710 10.1080/15384101.2016.1148837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Takahashi, K. and Yamanaka, S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 131.Zuo, W., Zhang, T., Wu, D.Z., Guan, S.P., Liew, A.-A., Yamamoto, Y.et al. (2015) P63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature 517, 616–620 10.1038/nature13903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rao, W., Wang, S., Duleba, M., Niroula, S., Goller, K., Xie, J.et al. (2020) Regenerative metaplastic clones in COPD lung drive inflammation and fibrosis. Cell 181, 848–864.e18 10.1016/j.cell.2020.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yi, M., Tan, Y., Wang, L., Cai, J., Li, X., Zeng, Z.et al. (2020) TP63 links chromatin remodeling and enhancer reprogramming to epidermal differentiation and squamous cell carcinoma development. Cell. Mol. Life Sci. 77, 4325–4346 10.1007/s00018-020-03539-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Melino, G., Memmi, E.M., Pelicci, P.G. and Bernassola, F. (2015) Maintaining epithelial stemness with p63. Sci. Signal. 8, re9 10.1126/scisignal.aaa1033 [DOI] [PubMed] [Google Scholar]
- 135.Avgustinova, A. and Benitah, S.A. (2016) Epigenetic control of adult stem cell function. Nat. Rev. Mol. Cell Biol. 17, 643–658 10.1038/nrm.2016.76 [DOI] [PubMed] [Google Scholar]
- 136.Bao, X., Tang, J., Lopez-Pajares, V., Tao, S., Qu, K., Crabtree, G.R.et al. (2013) ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell 12, 193–203 10.1016/j.stem.2012.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.National Cancer Institute. (2020) The Cancer Genome Atlas. Definitions, National Cancer Institute [Google Scholar]
- 138.Rinaldi, L. and Benitah, S.A. (2015) Epigenetic regulation of adult stem cell function. FEBS J. 282, 1589–1604 10.1111/febs.12946 [DOI] [PubMed] [Google Scholar]
- 139.Sen, G.L., Reuter, J.A., Webster, D.E., Zhu, L. and Khavari, P.A. (2010) DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 463, 563–567 10.1038/nature08683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Avantaggiati, M.L., Ogryzko, V., Gardner, K., Giordano, A., Levine, A.S. and Kelly, K. (1997) Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89, 1175–1184 10.1016/S0092-8674(00)80304-9 [DOI] [PubMed] [Google Scholar]
- 141.Gu, W. and Roeder, R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 10.1016/S0092-8674(00)80521-8 [DOI] [PubMed] [Google Scholar]
- 142.Lill, N.L., Grossman, S.R., Ginsberg, D., DeCaprio, J. and Livingston, D.M. (1997) Binding and modulation of p53 by p300/CBP coactivators. Nature 387, 823–827 10.1038/42981 [DOI] [PubMed] [Google Scholar]
- 143.Zeng, X., Li, X., Miller, A., Yuan, Z., Yuan, W., Kwok, R.P.et al. (2000) The N-terminal domain of p73 interacts with the CH1 domain of p300/CREB binding protein and mediates transcriptional activation and apoptosis. Mol. Cell. Biol. 20, 1299–1310 10.1128/MCB.20.4.1299-1310.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Marques, M., Laflamme, L., Gervais, A.L. and Gaudreau, L. (2010) Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics 5, 267–272 10.4161/epi.5.4.11520 [DOI] [PubMed] [Google Scholar]
- 145.Zhang, L., Zhou, Y., Cheng, C., Cui, H., Cheng, L., Kong, P.et al. (2015) Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am. J. Hum. Genet. 96, 597–611 10.1016/j.ajhg.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Serber, Z., Lai, H.C., Yang, A., Ou, H.D., Sigal, M.S., Kelly, A.E.et al. (2002) A C-terminal inhibitory domain controls the activity of p63 by an intramolecular mechanism. Mol. Cell. Biol. 22, 8601–8611 10.1128/MCB.22.24.8601-8611.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lin-Shiao, E., Lan, Y., Welzenbach, J., Alexander, K.A., Zhang, Z., Knapp, M.et al. (2019) P63 establishes epithelial enhancers at critical craniofacial development genes. Sci. Adv. 5, eaaw0946 10.1126/sciadv.aaw0946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jiang, Y., Jiang, Y.-Y., Xie, J.-J., Mayakonda, A., Hazawa, M., Chen, L.et al. (2018) Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat. Commun. 9, 3619 10.1038/s41467-018-06081-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Browne, G., Cipollone, R., Lena, A.M., Serra, V., Zhou, H., van Bokhoven, H.et al. (2011) Differential altered stability and transcriptional activity of ΔNp63 mutants in distinct ectodermal dysplasias. J. Cell Sci. 124, 2200–2207 10.1242/jcs.079327 [DOI] [PubMed] [Google Scholar]
- 150.Tuppi, M., Kehrloesser, S., Coutandin, D.W., Rossi, V., Luh, L.M., Strubel, A.et al. (2018) Oocyte DNA damage quality control requires consecutive interplay of CHK2 and CK1 to activate p63. Nat. Struct. Mol. Biol. 25, 261–269 10.1038/s41594-018-0035-7 [DOI] [PubMed] [Google Scholar]
- 151.Bernassola, F., Karin, M., Ciechanover, A. and Melino, G. (2008) The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14, 10–21 10.1016/j.ccr.2008.06.001 [DOI] [PubMed] [Google Scholar]
- 152.Melino, G., Gallagher, E., Aqeilan, R.I., Knight, R., Peschiaroli, A., Rossi, M.et al. (2008) Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ. 15, 1103–1112 10.1038/cdd.2008.60 [DOI] [PubMed] [Google Scholar]
- 153.Delvecchio, V.S., Fierro, C., Giovannini, S., Melino, G. and Bernassola, F. (2021) Emerging roles of the HECT-type E3 ubiquitin ligases in hematological malignancies. Discov. Oncol. 12, 39 10.1007/s12672-021-00435-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Melino, S., Bellomaria, A., Nepravishta, R., Paci, M. and Melino, G. (2014) P63 threonine phosphorylation signals the interaction with the WW domain of the E3 ligase Itch. Cell Cycle 13, 3207–3217 10.4161/15384101.2014.951285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li, Y., Zhou, Z. and Chen, C. (2008) WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ. 15, 1941–1951 10.1038/cdd.2008.134 [DOI] [PubMed] [Google Scholar]
- 156.Peschiaroli, A., Scialpi, F., Bernassola, F., El Sherbini, E.S. and Melino, G. (2010) The E3 ubiquitin ligase WWP1 regulates ΔNp63-dependent transcription through Lys63 linkages. Biochem. Biophys. Res. Commun. 402, 425–430 10.1016/j.bbrc.2010.10.050 [DOI] [PubMed] [Google Scholar]
- 157.Busuttil, V., Droin, N., McCormick, L., Bernassola, F., Candi, E., Melino, G.et al. (2010) NF-kappaB inhibits T-cell activation-induced, p73-dependent cell death by induction of MDM2. Proc. Natl Acad. Sci. U.S.A. 107, 18061–18066 10.1073/pnas.1006163107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Khoo, K.H., Verma, C.S. and Lane, D.P. (2014) Drugging the p53 pathway: understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 13, 217–236 10.1038/nrd4236 [DOI] [PubMed] [Google Scholar]
- 159.Kadakia, M., Slader, C. and Berberich, S.J. (2001) Regulation of p63 function by Mdm2 and MdmX. DNA Cell Biol. 20, 321–330 10.1089/10445490152122433 [DOI] [PubMed] [Google Scholar]
- 160.Calabrò, V., Mansueto, G., Parisi, T., Vivo, M., Calogero, R.A. and La Mantia, G. (2002) The human MDM2 oncoprotein increases the transcriptional activity and the protein level of the p53 homolog p63. J. Biol. Chem. 277, 2674–2681 10.1074/jbc.M107173200 [DOI] [PubMed] [Google Scholar]
- 161.Jung, Y.-S., Qian, Y., Yan, W. and Chen, X. (2013) Pirh2 E3 ubiquitin ligase modulates keratinocyte differentiation through p63. J. Invest. Dermatol. 133, 1178–1187 10.1038/jid.2012.466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yan, W., Chen, X., Zhang, Y., Zhang, J., Jung, Y.-S. and Chen, X. (2013) Arsenic suppresses cell survival via Pirh2-mediated proteasomal degradation of ΔNp63 protein. J. Biol. Chem. 288, 2907–2913 10.1074/jbc.M112.428607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Straub, W.E., Weber, T.A., Schäfer, B., Candi, E., Durst, F., Ou, H.D.et al. (2010) The C-terminus of p63 contains multiple regulatory elements with different functions. Cell Death Dis. 1, e5 10.1038/cddis.2009.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ranieri, M., Vivo, M., De Simone, M., Guerrini, L., Pollice, A., La Mantia, G.et al. (2018) Sumoylation and ubiquitylation crosstalk in the control of ΔNp63α protein stability. Gene 645, 34–40 10.1016/j.gene.2017.12.018 [DOI] [PubMed] [Google Scholar]
- 165.Li, X., Tian, D., Guo, Y., Qiu, S., Xu, Z., Deng, W.et al. (2020) Genomic characterization of a newly established esophageal squamous cell carcinoma cell line from China and published esophageal squamous cell carcinoma cell lines. Cancer Cell Int. 20, 184 10.1186/s12935-020-01268-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chang, C.-Y., Shipony, Z., Lin, S.G., Kuo, A., Xiong, X., Loh, K.M.et al. (2021) Increased ACTL6A occupancy within mSWI/SNF chromatin remodelers drives human squamous cell carcinoma. Mol. Cell 81, 4964–4978.e8 10.1016/j.molcel.2021.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gao, Y.-B., Chen, Z.-L., Li, J.-G., Hu, X.-D., Shi, X.-J., Sun, Z.-M.et al. (2014) Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 46, 1097–1102 10.1038/ng.3076 [DOI] [PubMed] [Google Scholar]
- 168.King, K.E., Ponnamperuma, R.M., Allen, C., Lu, H., Duggal, P., Chen, Z.et al. (2008) The p53 homologue DeltaNp63alpha interacts with the nuclear factor-kappaB pathway to modulate epithelial cell growth. Cancer Res. 68, 5122–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Stindt, M.H., Muller, P.A.J., Ludwig, R.L., Kehrloesser, S., Dötsch, V. and Vousden, K.H.. ( 2015) Functional interplay between MDM2, p63/p73 and mutant p53. Oncogene 34, 4300–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Niu, M., He, Y., Xu, J., Ding, L., He, T., Yi, Y.et al. (2021) Noncanonical TGF-β signaling leads to FBXO3-mediated degradation of ΔNp63α promoting breast cancer metastasis and poor clinical prognosis. PLoS Biol. 19, e3001113. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 171.Fu, H., Zhang, Y., Chen, J., Zhou, B., Chen, G. and Chen, P. (2020) Tmub1 Suppresses Hepatocellular Carcinoma by Promoting the Ubiquitination of ΔNp63 Isoforms. Mol. Ther. Oncolytics. 18, 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gallegos, J.R., Litersky, J., Lee, H., Sun, Y., Nakayama, K., Nakayama, K.et al. (2008) SCF TrCP1 activates and ubiquitylates TAp63gamma. J. Biol. Chem. 283, 66–75 [DOI] [PubMed] [Google Scholar]
- 173.MacPartlin, M., Zeng, S.X. and Lu, H. (2008) Phosphorylation and stabilization of TAp63gamma by IkappaB kinase-beta. J. Biol. Chem. 283, 15754–15761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rokudai, S., Li, Y., Otaka, Y., Fujieda, M., Owens, D.M., Christiano, A.M.et al. (2018) STXBP4 regulates APC/C-mediated p63 turnover and drives squamous cell carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 115, E4806–E4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Asatsuma-Okumura, T., Ando, H., De Simone, M., Yamamoto, J., Sato, T., Shimizu, N.et al. (2019) p63 is a cereblon substrate involved in thalidomide teratogenicity. Nat. Chem. Biol. 15, 1077–1084 [DOI] [PubMed] [Google Scholar]
- 176.Fomenkov, A., Zangen, R., Huang, Y.-P., Osada, M., Guo, Z., Fomenkov, T.et al. (2004) RACK1 and stratifin target DeltaNp63alpha for a proteasome degradation in head and neck squamous cell carcinoma cells upon DNA damage. Cell Cycle 3, 1285–1295 [DOI] [PubMed] [Google Scholar]
- 177.Bamberger, C., Pankow, S. and Yates, J.R. (2021) SMG1 and CDK12 Link ΔNp63α Phosphorylation to RNA Surveillance in Keratinocytes. J. Proteome Res. [DOI] [PMC free article] [PubMed] [Google Scholar]