Abstract
Purpose:
Radiotherapy (RT) is a mainstay of cancer care and accumulating evidence suggests the potential for synergism with components of the immune response. However, little data describes the tumor immune contexture in relation to RT-sensitivity. To address this challenge, we employed the radiation sensitivity index (RSI) gene signature to estimate the RT-sensitivity of >10,000 primary tumors and characterized their immune microenvironments in relation to the RSI.
Material and Methods:
We analyzed gene expression profiles of 10,469 primary tumors (31 types) within a prospective tissue collection protocol. The RT-sensitivity of each tumor was estimated by the RSI and respective distributions were characterized. The tumor biology measured by the RSI was evaluated by differentially expressed genes (DEGs) combined with single sample gene set enrichment analysis (ssGSEA). Differences in the expression of immune regulatory molecules were assessed and deconvolution algorithms were used to estimate immune cell infiltrates in relation to the RSI. A subset (n=2,368) of tumors underwent DNA sequencing for mutational frequency characterization.
Results:
We identified a wide range of RSI values within and across various tumor types, with several demonstrating non-unimodal distributions (e.g. colon, renal, lung, prostate, esophagus, pancreas and PAM50 breast subtypes; p <0.05). Across all tumors types, stratifying RSI at a tumor type-specific median, identified 7,148 DEGs, of which 146 were coordinate in direction. Network topology analysis demonstrates RSI measures a coordinated STAT1, IRF1, and CCL4/MIP-1β transcriptional network. Tumors with an estimated high sensitivity to RT demonstrated distinct enrichment of interferon-associated signaling pathways and immune cell infiltrates (e.g. CD8+ T cells, activated natural killer cells, M1-macrophages; q < 0.05), which was in the context of diverse expression patterns of various immunoregulatory molecules.
Conclusion:
This analysis describes the immune microenvironments of patient tumors in relation to the RSI gene expression signature.
Introduction
There is an appreciable spectrum of in vitro sensitivity to radiation across various cancer cell types that is regulated by the underlying molecular repertoire1 and capacity to utilize available nutrients.2 However, in a tumor comprised of diverse cellular architecture, which is constantly evolving, the response to radiation is more complex. Despite acknowledged variations in radiosensitivity in the preclinical and clinical setting, the field of radiation oncology currently does not individualize radiation dose prescription based on the biology of a patient’s tumor.
To better understand the diversity of radiosensitivity in cancer and to identify conserved radiation response modifiers, we previously modeled the response of 48 genetically annotated human cancer cell lines to radiation. By integrating the basal transcriptome, TP53 and RAS isoform mutational status, tissue of origin, and clonogenic survival following a radiation dose of 2 Gy (SF2), we identified an interaction network of 474 genes, which includes regulators of DNA damage repair (DDR) (e.g. ATM, XRCC6), oxidative stress (e.g. PRDX1, TXN), as well as ten dominant signaling hubs: AR, JUN, STAT1, PKCB, RELA, ABL1, SUMO1, PAK2, HDAC1 and IRF1.3 From this network the radiosensitivity index (RSI) was derived by training a multi-gene algorithm to predict SF2 in the 48 cancer cell lines. Notably, the RSI is agnostic to cancer type and has been independently validated as a biomarker of radioresponsiveness across multiple human tumor types by several investigators.4–7 Also, the RSI has been proposed as a measurable tumor feature to guide radiotherapy dose selection in patients via integration into the genomically-adjusted radiation dose (GARD).8,9
The clinical utility of harnessing a patient’s own immune system has emerged as a new pillar of anti-tumor therapy and various strategies are being investigated to modulate the host immune response, including the use of radiotherapy.10,11 Indeed, emerging pre-clinical work has shown that irradiated tumors may act as adjuvants for the immune system, by facilitating local tumor control and by provoking regression of tumor deposits at distant sites via the abscopal effect.12 In addition to the potential systemic effects, radiation significantly alters the tumor antigen landscape, recruits anti-tumor T-cells, and instigates diverse inflammatory milieus.13 Still, the relationship between radiation and the immune response remains complex with contextual dependencies, and the optimal way to integrate radiation and immune-based therapies still remain uncertain.
To begin to address this question, we performed a systematic multi-tier analysis on a cohort of 10,469 transcriptionally profiled, prospectively collected tumor samples representing 31 tumor types. These analyses underscore the heterogeneity in tumor radiosensitivity and provide a framework to explore how this is related to individual tumor immune microenvironments.
Methods
Patient tumor cohorts.
Patients were consented to the Total Cancer Care™, a prospective tissue collection protocol at (IRB-approved, Liberty IRB #12.11.0023), a prospective tissue collection protocol at Moffitt Cancer Center and Research Institute.14 Pathology quality control evaluation of macrodissected tumors was performed, which included measurements of percent tumor, stroma and necrosis to ensure sample integrity. Tumor samples were profiled for gene expression using the custom Rosetta/Merck HuRSTA_2a520709 Affymetrix gene expression microarray platform (GEO: GPL15048). CEL files (Affymetrix GeneChip probe data file format) were normalized against the median CEL file using IRON,15 which yields Log2 intensity values per probeset. Principal component analysis (PCA) of all samples revealed the first component was highly correlated to RIN (RNA integrity number) value. A partial least squares (PLS) model was trained upon samples for which RIN values were available and this was used to re-estimate the quality of RNA of all samples; the first component was then removed to correct the signals for RNA quality. We identified 10,469 unique fresh frozen macrodissected primary tumors (see Supplemental Table 1) for the related analyses; patient demographics, including age at diagnosis and sex grouping for each tumor type are shown in Supplemental Table 2. Two publicly available datasets were used for additional analysis: breast cancer (GSE1456)16 and lung adenocarcinoma (GSE68465)17.
t-SNE:
Using the 10,469 samples, we first filtered the 60,607 probesets to those having a standard deviation (across the cohort) > 2, yielding 1,399 probesets. The NIPALS algorithm (R/nipals package v0.5) was used to compute the first 50 principal components of the expression data. Rtsne (v0.15) was used with perplexity=100 to generate a two-dimensional mapping from these principal components.
Calculation of RSI:
RSI was calculated as previously described3 by first rank-ordering the ten signature genes in each sample and then applying the linear equation (RSI) with predetermined coefficients. The following 10 probesets, corresponding to the originally defined HG-U133plus2 probesets, were used: merck-NM_007313_s_at (ABL1), merck-NM_000044_a_at (AR), merck-NM_004964_at (HDAC1), merck-NM_002198_at (IRF1), merck-NM_002228_at (JUN), merck-BQ646444_a_at (PAK2, CDK1), merck2-X06318_at (PRKCB), merck2-BC069248_at (RELA), merck-NM_139266_at (STAT1), merck-NM_001005782_s_at (SUMO1). RSI provides a continuous score, with a higher score estimating more radioresistance.
RSIlo vs. RSIhi.
For probesets that were differentially expressed (DE) between RSIlo and RSIhi within each tumor type, samples were ranked by RSI score and divided into RSIlo and RSIhi groups at the median RSI value. A probeset was determined to be DE within a tumor type if the following criteria were met: (i) probeset must not be annotated as antisense; (ii) one of the two group averages must be > 5; (iii) |log2 ratio| ≥ ~0.585 (1.5-fold); (iv) two-tailed t-test ≤ 0.01; and (v) Hellinger distance ≥ 0.25.
Violin Plots:
Plots were generated using R (3.6.0) using an in-house custom code (https://github.com/steveneschrich/densityplots). The violin plots represent the smoothed density with nrd0 bandwidth kernel.
ESTIMATE:
The ESTIMATE algorithm18 was used to estimate stromal and immune cell fractions, as well as tumor purity, from bulk gene expression of the primary tumor (Estimate R package v1.0.13). Gene expression probeset identifiers were mapped to Entrez GeneIDs and, as defined in the Estimate package, the data were filtered to only the common genes. A single probeset was selected per gene by choosing the probeset having the highest median expression across all tumors. This resulting dataset was used in the estimate function to produce immune scores for each tumor.
Immunophenotype:
Based on the characterization of immune escape-related genes described by Charoentong et al.,19 we extracted the gene expression corresponding to genes detailed in the three categories: Immunostimulator, Immunoinhibitor and Major histocompatibility complex (MHC). We calculated the log2 ratio as the difference in log2 expression of median RSIlo tumors vs median RSIhi tumors for each gene and tissue type combination. ComplexHeatmap20 version 2.8.0 was used to visualize these log2 ratios, using the column grouping from Charoentong et al., clustering genes within groups and row ordering (tumor type) from decreasing median immune score.
CIBERSORT:
The HuRSTA array was reduced to the LM22 signature genes defined by the CIBERSORT algorithm21 by choosing a representative probeset that detects the gene and has the highest median expression among matching probesets. The CIBERSORT web tool (https://cibersort.stanford.edu/index.php) was accessed on 2017–05-19 to generate signature scores (using quantile normalization).
Immune cell infiltrate normalized content abundance:
Immune cell infiltrate composition proportions from CIBERSORT were extracted in relative mode, where the proportions are relative to the total immune cell fraction of the tumor. To normalize the content across tumors, we scaled the ESTIMATE immune scores so the lowest immune score was 0 (rather than negative) and analyzed multiple immune cell infiltrate fractions by this adjusted immune score, as described previously.22
Immune cell infiltrate enrichment:
RSI was dichotomized into RSI_HIGH and RSI_LOW by tumor subtype using the median RSI for that type. Each immune cell type derived from CIBERSORT was likewise dichotomized using the same approach into immune cell type_HIGH and immune cell type_LOW. A Fisher exact test was performed for each comparison of dichotomized RSI and immune cell type counts to determine the independence of the variables. The q value package (2.16.0) was used to compute FDR q values to correct for multiple testing. The immune cell infiltrates were classified as composing adaptive and innate cell types based on current literature and PCA. The enrichment heatmap was generated using ComplexHeatmap (2.0.0).20 Each cell represents the proportion of samples with immune cell type_HIGH in either RSI_LO (top row) or RSI_HI (bottom row). The percentage of tumor types with a significant q value (q<0.05) of differences between immune cell type_HIGH in RSI_LOW vs. RSI_HIGH is noted.
ssGSEA:
Single-sample GSEA was used to evaluate the MSigDB Hallmarks Genesets.23 The GSVA24 package (R package GSVA, v1.32.0) was used to calculate ssGSEA scores. ssGSEA scores were combined by tumor type using the median score.
Metacore pathway.
Probesets that were identified as DE in ≥ 6 tumor types and that had no conflicting direction of change (197 probesets, 146 genes) were used as seeds for MetaCore (Clarivate Analytics) literature network generation. Networks were generated using the Build Network-Shortest Paths method with the following non-default settings: (i) do not show disconnected seed nodes; (ii) do not show shortest path edges only; and (iii) use only Phosphorylation, Dephosphorylation, and Transcription Regulation interaction types of known Activation / Inhibition status. The resulting interaction network was exported and further filtered to contain only nodes and edges consistent with the observed RSIlo vs. RSIhi directions of change, which were then visualized using Cytoscape v3.3.25 Major nodes were identified as nodes in the network with the largest degree, or the largest number of connected edges (incoming or outgoing).
Interferon-RSI-Immune Cell Type Comparison.
We performed two tests of proportions (RSI and IFNα) with specific Immune Cell (IC) hi/lo categorizations using Fisher exact tests, dichotomizing all variables at the median (e.g., RSI hi/lo, IFNα hi/lo and IC hi/lo). Multiple testing corrections were applied using p.adjust (FDR option). ComplexHeatmap was used with a custom code for showing significance (q < 0.05, Fisher Exact Test).
DNA sequencing and analysis
Sequencing analysis are described in detail as previous.26 Briefly, coding regions of 1,321 genes were targeted with SureSelect (Agilent Technologies, Santa Clara, CA) custom capture and sequenced on GAIIx sequencing instruments (Illumina, Inc., San Diego, CA) in paired-end 90bp configuration. Sequence reads were aligned to the reference human genome with the Burrows-Wheeler Aligner (BWA),27 and duplicate identification, insertion/deletion realignment, quality score recalibration, and variant identification were performed with PICARD (http://picard.sourceforge.net/) and the Genome Analysis ToolKit (GATK).28 Genotypes were determined with GATK UnifiedGenotyper and refined using Variant Quality Score Recalibration. Variants were filtered based on quality (highest sensitivity tranche were excluded) and population frequency (variant observed in 1000 Genomes Project or NHLBI Exome Sequencing Project or at >1% in 238 normal samples from our institutional cohort were excluded). Sequence variants were annotated to determine genic context (ie, non-synonymous, splicing) using ANNOVAR.29
Statistics:
Statistical analyses with Student’s t-test, Mann-Whitney U test, and linear regression were performed using R 3.6.0. The RSI distribution across all tumor types and for each specific tumor type was tested for being unimodal using Hartigan’s dip test (R package diptest, v0.75–7) and visualized using ggplot2 (3.2.1) and ggpubr (v0.2.3). RSI and Immune score were compared using Pearson’s (r) and Spearman’s (ρ) correlation and visualized using ggpubr (v0.2.3).
Results
Characterization of the spectrum of intrinsic radiation sensitivity within and across tumor types with the RSI.
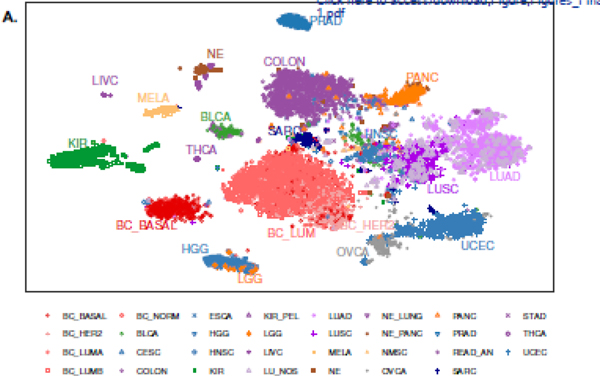
The ‘omics’ era offers many insights into tumor biology and provides a means to classify tumor types based on similar or dissimilar attributes. In this regard, we performed multi-dimensional reduction analysis with the t-distributed stochastic neighbor embedding (t-SNE) method on the bulk transcriptome of 10,469 non-metastatic, primary macrodissected tumors. We identified the vast majority of 31 individual tumor types cluster based on their underlying gene expression programs (Figure 1).
Figure 1. Tumor types cluster by transcriptional programs.
Multidimensional reduction analysis of transcription data from 10,469 tumors by the t-distributed stochastic neighbor embedding (t-SNE) method demonstrates distinct clustering of tumor types.
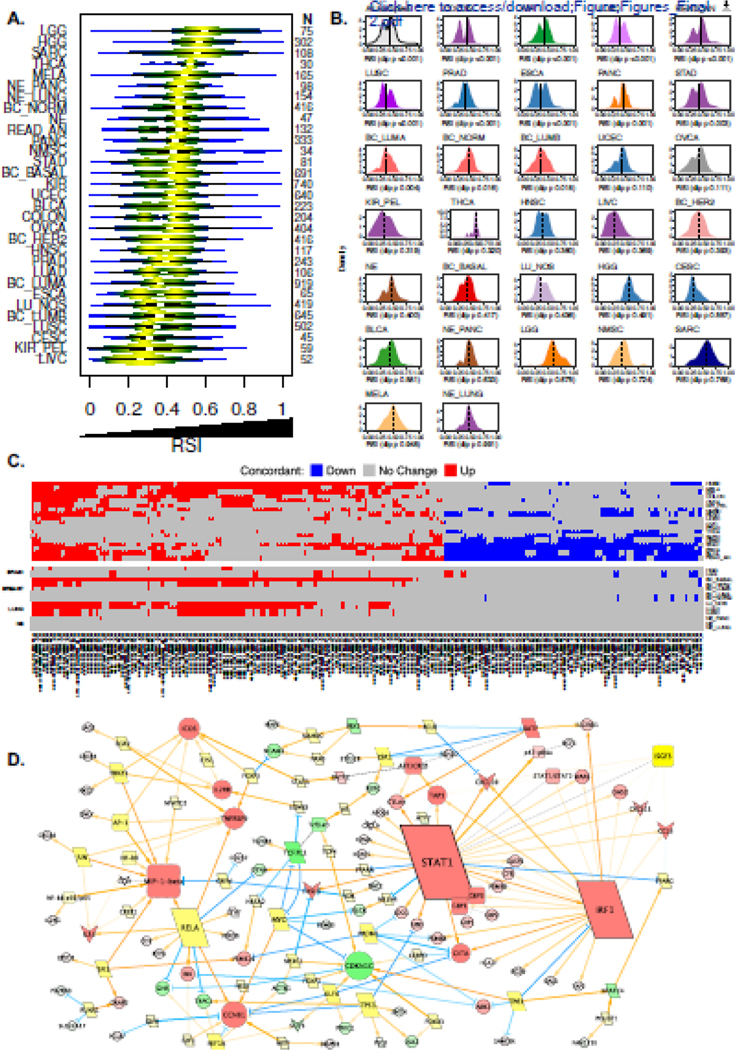
To estimate the radiosensitivity of human tumors, we used the cancer-type agnostic RSI signature. With this approach, we identified a spectrum of RSI values within and across the 31 tumor types (Figure 2a). Stratification of breast cancers into PAM50 subtypes revealed normallike tumors have a higher median RSI compared to luminal A and B types (P < 0.001). In contrast, no statistical differences in the median RSI are noted between the broad histologic subtypes of lung cancer (P = 0.096). Evaluation of interquartile range (IQR) ratios for the RSI reveal that tumors originating from the renal pelvis, liver, gastrointestinal tract, bladder and lung have the greatest dispersion (median ratio ≥ 1.5), whereas tumors considered more radioresistant (e.g. gliomas, sarcomas) have more uniformly distributed RSI values (Supplemental Table 3). As a whole, the RSI distribution among all tumor types is not unimodal based on Hartigan’s dip test statistic, which likely reflects the underlying diversity of tumor types grouped together. Separately, several tumor types display non-unimodal distributions in RSI, including kidney, colorectal, prostate, esophagus, pancreas, stomach, and lung and breast subtypes (Figure 2b). Importantly, the median RSI value (dashed line) varies across different tumor types, suggesting dichotomization into radiation sensitive (RSIlo) and resistant (RSIhi) tumors may differ based on tumor type.
Figure 2. Characterization of tumor radiosensitivity and coordinated biology estimated by the RSI.
(a) Violin plots depicting distribution of RSI values across 10,469 primary tumor samples representing 31 tumor types. (b) Hartigan’s dip statistic across 31 tumor types to evaluate for unimodal distributions of RSI. Analysis demonstrates that several tumor types do not have a unimodal distribution of RSI values, which suggests biologic heterogeneity. Dashed line on each density plot represents the tumor type-specific median RSI value. (c) Heatmap of differentially expressed probesets (n=209, representing 146 unique genes) between radiosensitive (RSIlo) and radioresistant (RSIhi) tumors across six or more of the 31 tumor types, which are concordant in direction of expression (up, red; down, blue; no change, gray). See Supplemental Table 5 for list of probesets. (d) Differentially expressed probesets (146 genes) from Figure 2c were used as seeds for network generation (see Methods). Genes upregulated (red), downregulated (green) in RSIlo versus RSIhi tumors are depicted; node size indicates degree of edges. Yellow nodes indicate bridging genes.
The RSI measures coordinated biology across several tumor types
In addition to heterogeneity in the RSI across and within tumor types, there are also overlapping RSI values (continuous scale) between tumor types. We hypothesized that comparable RSI values, despite different tumor types, may be related to similar underlying biology. To begin to explore the biologic discriminators, we evaluated differentially expressed genes (DEGs) between RSIlo and RSIhi tumors (based on tumor type median RSI) for each tumor type (Supplemental Table 4). Across all tumor types there were 7,184 DEGs between the RSI groups. Interestingly, prostate, neuroendocrine or non-PAM50 subtyped breast tumors have ≤ 2 RSI-influenced DEGs, whereas PAM50 breast tumor subtypes, except luminal variants, have >150 DEGs between the RSI groupings. Furthermore, neuroendocrine tumors stratified by site of origin unmasked additional RSI-related DEGs, underscoring the contribution of the microenvironment.
Next, we investigated whether the RSI is associated with a conserved transcriptional program in tumors. We identified 209 unique probesets (155 genes) that are differentially expressed between RSIlo and RSIhi tumors in at least six tumor types, and of these, 146 genes were concordant in their direction of expression (Figure 2c and Supplemental Table 5). Network topology analyses with the 146 conserved genes identified an expansive interacting network with STAT1, IRF1, and CCL4/MIP-1β as major upregulated nodes in RSIlo tumors (Figure 2d), suggesting these may act as central regulators of the biology measured by the RSI.
The RSI describes distinct tumor immune microenvironments
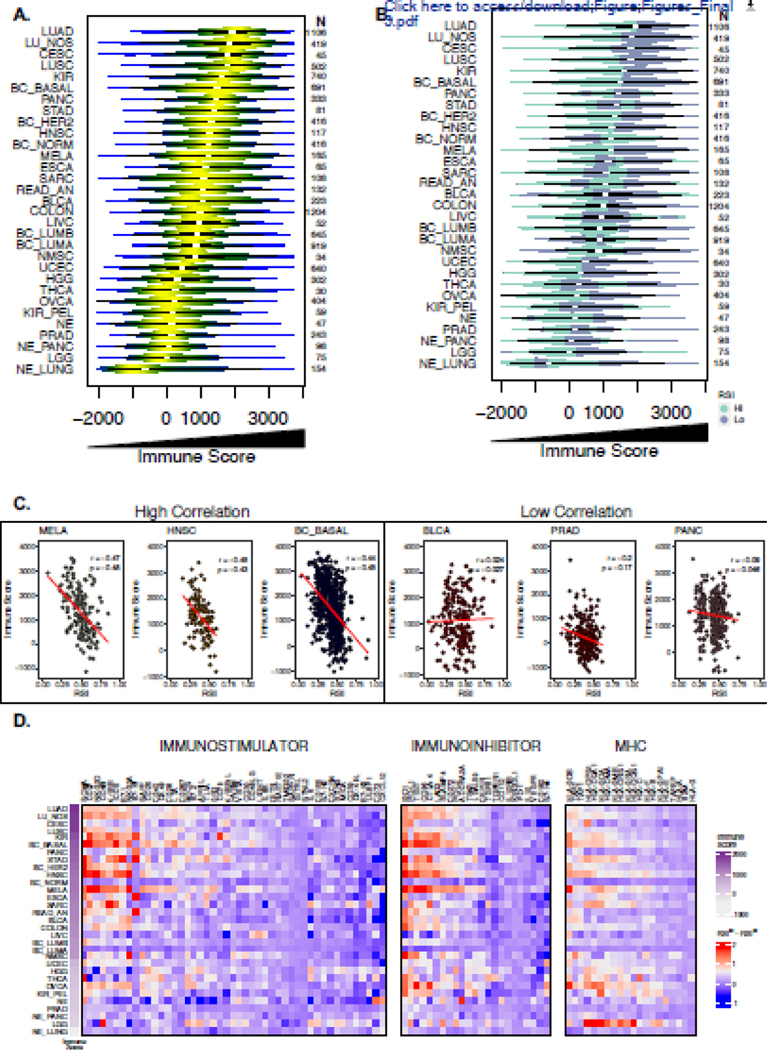
Prior data by Strom et al. hints at a relationship between the RSI and a 12-chemokine gene signature30, thus combining these findings with our network analysis suggests the measurement of RSI in tumors may be partially related to the immune microenvironment. To evaluate this relationship further, we inferred the presence of immune cell infiltrates in each tumor by the ESTIMATE algorithm. Similar to the variance observed with the RSI, ESTIMATE-derived approximations of immune cell infiltrates identified a wide range across and within tumor types (Figure 3a). Next, we again classified each tumor sample within a given tumor type as radiosensitive (RSIlo) or radioresistant (RSIhi) based on the median RSI value within a given tumor type. Integration of the RSI and immune score revealed most RSIlo tumors are characterized by increased immune cell infiltration across most tumor types (Figure 3b). However, among all tumors, we identified a weak negative correlation between the RSI and immune score when assessed as continuous variables (Pearson’s r = −0.28; Spearman’s ρ = 0.27, P <0.001). Also, the strength and direction of the correlation varied based on tumor type. Figure 3c highlights select tumor types with the highest (e.g. melanoma [MELA] r = - 0.47, ρ = - 0.48) and lowest correlations (e.g. pancreas [PANC] r = −0.09, ρ = −0.05) between the RSI and immune score; a full composite of correlations for all tumor types is shown in Supplemental Figure 1.
Figure 3. Evaluation of the association between immune cell infiltration, regulators of immune escape and the RSI.
(a) Violin plots depicting distribution of ESTIMATE-derived immune score values across 10,469 primary tumor samples representing 31 tumor types. (b) Integration of RSI and the immune score where radiosensitive (RSIlo; blue-green) radioresistant (RSIhi; purple) categorization is determined by the median RSI value within each tumor type. (c) Scatter plots of the RSI and immune score among high correlated and low correlated tumor types with Pearson’s correlation coefficient and Spearman’s ρ shown. (d) Heatmap of log2 gene expression differences of markers of immune escape between RSIlo and RSIhi tumors. Markers are categorized as defined by Charoentong et al. into immunostimulators, immunosuppressors and MHCs (class I, class II, non-classical) and genes were clustered within groups and row ordered (tumor type) from decreasing median immune score.
The above data suggests dichotomizing the RSI by tumor type-specific median values enriches for an interconnected STAT1, IRF1, and CCL4/MIP-1β signaling network which may instigate certain immune infiltrate patterns in RSIlo tumors. Various factors are known to regulate the tumor immune contexture, which may include immune checkpoint molecules major histocompatibility complex (MHC) repertoires, or mutational landscapes.31–33 To gain a better understanding of the immunologic environment in RSIlo vs RSIhi tumors, we first used the immunophenotype classification specified by Charoentong et al. to describe a catalog of molecules involved in tumor immune escape.19 Cluster analysis identified variation in expression differences in both immunostimulatory (e.g. ICOS, CD27, NKG2D, 4–1BB) and immunoinhibitory (e.g. IDO1, TIGIT, PD-L1, CTLA-4, LAG3) molecule classes across tumor types in relation with the RSI (Figure 3d). We also directly examined whether established immune checkpoint molecules (e.g. PD-L1, PD-1, CD47 and CTLA-4) and T cell receptor subunits (e.g. CD8A and CD8B) are associated with RSI (Supplemental Table 6, Supplemental Figure 2).
Previous investigations have identified certain mutational profiles may correlate with radiosensitivity1,34 and immune response, therefore we performed targeted sequencing of a subset of tumors (n=2,368) across all tumor types and found that mutation frequency (both non-synonymous and synonymous) only weakly correlated with the RSI (r = −0.07; P = 0.001), immune score (r = −0.01; P = 0.54) and individual immune cell subtypes (Supplemental Table 7).
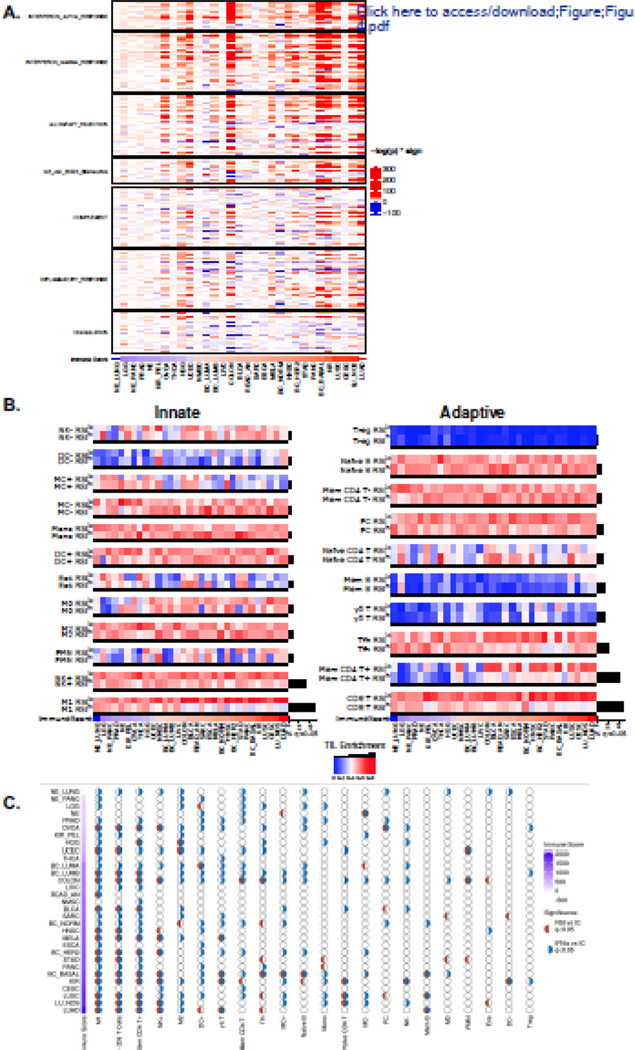
Due to the diversity in immune molecule expression differences between RSIlo and RSIhi tumors, we next employed single sample gene set enrichment analysis (ssGSEA) using the immune-related MSigDB hallmark gene sets to evaluate associated inflammatory pathways.23 This revealed that RSIlo vs RSIhi tumors are enriched in various inflammatory pathways, which were mostly manifest in tumor types classified as also having the highest immune scores. The predominant pathways enriched in RSIlo versus RSIhi tumors were the Type I (α) and II (γ) interferon (IFN) modules (Figure 4a).
Figure 4. The RSI describes distinct tumor immune microenvironments.
(a) RSIlo versus RSIhi tumors were compared with single sample gene set enrichment analysis (ssGSEA) using the MSigDB hallmark pathway genes related to immune signaling. Heatmap intensity depicts the -log p-value of the Fisher exact test multiplied by the directionality of expression difference; enriched in RSIlo (red) or RSIhi (blue). Tumor types are ordered by the immune score. (b) Tumor types are ordered by immune score and normalized CIBERSORT immune cell infiltrate estimates are compared within a given tumor type by dichotomizing at the median RSI value to identify RSIlo and RSIhi tumors. Immune cell infiltrates involved in the innate (left panel) and the adaptive (right panel) response. Hypothesis tests (Fisher’s exact test) in tumor types between RSIhi and RSIlo were performed; a black bar beneath the comparison denotes false-discovery rate adjusted q < 0.05. Black bars to the right of heatmap indicate proportion of significant tumor types for given immune cell infiltrate enrichment comparison between RSIlo and RSIhi tumors. (d) Associations of IFNα and the RSI with immune cell (IC) subtypes as independent or coordinated biology. Half-circles represent whether comparison within a tumor type and IC subtype are significant (q < 0.05); red (RSI vs IC), blue (IFNα vs IC). Empty circles imply no association between variables. Tumor types are ordered by immune score.
The ESTIMATE-derived immune score provides a generic readout of immune cell presence, and does not elucidate the composition or functional state of the immune infiltrate. To evaluate the immune repertoire further we utilized the CIBERSORT deconvolution algorithm to infer the presence of 22 distinct immune cell subtypes within each tumor. We found that immune cells involved in the adaptive or innate responses were differentially enriched between RSIlo and RSIhi tumors within each tumor type. A common pattern of enrichment was seen in RSIlo vs RSIhi tumors for CD4+ memory T cells (Mem CD4+), CD8+ T cells (CD8+ T), follicular helper T cells (Tfh), activated natural killer (NK) cells (NK+) and M1-polarized macrophages (M1) (Figure 4b; q-value < 0.05). This was further evaluated for NK+, CD8+ T and M1 cells as a continuous measure (Supplemental Figure 3). This additional level of immune cell characterization also demonstrates differences in immune cell subtypes between RSI groupings even in tumors with lower immune scores, suggesting these relationships may persist in both ‘hot’ and ‘cold’ immune microenvironments.
In certain tumor microenvironments IFN signaling may have opposing functions within different cellular compartments. For instance, type I IFN signaling in immune cells is beneficial for radiation-mediated tumor control,35 but may also create an immunosuppressive environment following radiotherapy.36 In tumor cells, intact type I IFN signaling has been shown to reduce the anti-tumor immune response following radiation,37 and in some cancer cell lines it reduces radiation-mediated DNA damage.38 Due to these paradoxical relationships, we further explored how IFNα and RSI were related to the presence of immune cell types. We found several tumor types had significant differences (q < 0.05) in both IFNα and RSI in relation to specific immune cell types, both independently and cooperatively (Figure 4c). For example, M1, CD8+ T and Mem CD4+ cells show cooperative behavior between IFNα and RSI in lung cancer types (LUSC, LU_NOS, and LUAD). We also found comparable trends in independent patient cohorts with breast cancer and lung adenocarcinoma (Supplemental Figure 4). Similarly, these observed relationships persisted despite the overall immune cell infiltrate level.
Discussion
In addition to broad categorization, tumor profiling has also provided a basis for targeted therapy selection (e.g., kinase inhibitors, immune checkpoint blockade) for several malignancies.39,40 In contrast, radiation dose selection is not currently informed by tumor biology, but is rather guided by clinicopathologic features and empirically derived tolerances of surrounding normal tissue. Further, although substantial experimental and clinical evidence suggest tumors in the same anatomic location and of similar histological subtype may vary in their response to radiation, uniform dosing schemes are generally employed in clinic. In this study we employed multi-tier computational analyses to characterize several important tumor features that may impact future efforts to personalize radiotherapy and strategically integrate immunotherapies.
First, we used the RSI to estimate radiosensitivity in over 10,000 primary tumor samples. Interestingly, though t-SNE analysis demonstrated clustering of tumor types by their transcriptomes, the RSI values varied considerably within and between these tumor types. This suggests composite transcriptomes may regulate higher order tumor type-specific biology, which has been previously described in subtype analyses,41–43 but only partially contribute to the dynamics of radiation response. Characterization of the RSI dispersion by IQR and dip statistics demonstrated that many clinically-defined radioresistant tumors (e.g. glioma, sarcoma, melanoma) have less dispersion and more unimodality. Though there may be variability of biology within these unimodal distributions, tumor types with greater dispersion and non-unimodal RSI distributions imply a higher probability of biologic subtypes with differential responses to radiation. To further underscore this diversity, visualization of the RSI distributions shows the most sensitive tumors in one tumor type overlap with the most resistant tumors of another type, and vice versa. Since the RSI is measured on a continuous scale and is a readout of a multi-gene interacting network, similar RSI values or ranges among tumors may imply comparable dominant underlying biology.
Cell autonomous radiosensitivity is influenced by complex interactions between intrinsic polygenic traits. This was elegantly demonstrated by in vitro studies in over 500 tumor cell lines where DDR and genomic stability emerged as major regulators of radiosensitivity at varying doses.1 However, tumor radiosensitivity is also governed by the other ‘hallmarks’ of cancer, including tumor microenvironment dynamics, utilization of nutrients and diverse cellular composites. Similar to these interconnected features, the RSI network also unifies various biologic components of DDR, stress adaptation and cell signaling. Notably, the 10 major genes of the RSI signature are represented by 5 transcription factor complexes, 3 kinases and 2 regulators of reversible post-translational modifications. Thus, the RSI represents an element of the radiation response, which may begin at the membrane-cytoplasmic interface and propagate to the nucleus to regulate broad transcription programs. This work demonstrates the RSI measures a coordinated biology characterized by the predominance of a STAT1, IRF1, and CCL4/MIP-1β signaling network in tumors classified as radiosensitive.
The connection of the RSI with an immune associated signaling network in patient tumors is supported by prior studies showing partial associations with a 12-chemokine panel30 as well as inflammatory subgroups.30,44 Though the measurement of the RSI may be related to immune signaling, correlations are weakly moderate, suggesting the RSI is not solely a measure of immune activity. The strength of this RSI-immune relationship varies by tumor type with only 20–30% of variance explained even in the highest correlated tumor types (e.g. thyroid, cervical, skin, head and neck).
To explore this further, we characterized components of the immune contexture in each tumor and evaluated how this was related to the RSI. When looking across all tumor types, integrating the ESTIMATE immune score with an approximation of radiation sensitivity by RSIlo and RSIhi groupings, we identified most tumor types with high immune cell presence are also characterized as RSIlo. This may suggest tumors with high immune cell content are more sensitive to radiation by the fact that a major compartment of RSIlo tumors are enriched with leukocytes, which are known to be highly radiosensitive. Although, emerging data demonstrates differential radiation sensitivity between lymphoid and myeloid lineages in patients45 and Arina et al. have elegantly demonstrated pre-existent resident T cells are resistant to radiation doses used in clinic.46 Our analysis of immune cell subtypes demonstrates that over a third of tumor types were enriched with CD8+ T, M1 and NK+ cells in RSIlo vs RSIhi tumors. Thus, it is possible the RSI metric may partly reflect the diversity in abundance of the radiation-sensitive CD8+ T cells vs -resistant macrophage lineages in patient tumors, which also varies on organ microenvironment.
Recent data support a connection between the DDR and antitumor immunity via conserved mechanisms that detect cytosolic nucleic acids to combat foreign pathogens. For example, cyclic GMP-AMP synthase (cGAS)/stimulator of IFN genes (STING) signaling, a major regulator of type I IFN production47, influences the radiation response48 and type I IFNs are known to stimulate both arms of the immune response49 and are essential for radiation efficacy.35,50 Although the signaling effectors MB21D1/cGAS and TMEM173/STING failed to meet our statistical criteria for identifying concordant DEGs in a prespecified number of tumor types between RSI groups, our analyses revealed that RSIlo vs RSIhi tumors are enriched in inflammatory signaling pathways (e.g. type I/II IFN). Associations between the RSI, immune cell infiltrates and IFN signaling were most evident in tumors with the highest levels of immune cell infiltration. We also demonstrate significant differences (q < 0.05) in both IFNα and RSI in relation to specific immune cell types, both independently and cooperatively, despite tumor type (Figure 4c); notably, this cooperative biology was most prominent in lung, kidney, melanoma, bladder and certain breast tumor subtypes. Although, despite predominant IFN signaling and abundant effector immune cells in RSIlo tumors, our analysis on immunoregulatory molecules (Figure 3d), suggests various immunostimulatory molecules remain suppressed at the transcript level. These types of analyses (e.g. IFNα vs immune cell type vs RSI) neglect known tumor and radiation dynamics. The importance of these dynamics are further highlighted by a recent study using agent-based modeling, which proposed certain starting tumor microenvironments are differentially primed to respond to radiation therapy.51 Specifically, this study suggested that fractionated radiation may shift the tumor to an anti-tumor or immunosuppressive ecosystem based on the initial proportions of resident immune effector and suppressor cells.
Others have found that IFNα and STAT1 signaling may lead to a radioresistant phenotype in some tumors.52,53 Though, it is important to note that IFN-STAT1 signaling has been shown to be context dependent,36 at times with opposing effects,54–56 and may be instigated by tumor-immune cell interactions or autonomously by tumors cells.57 In some tumors, type I IFNs are important for radiation efficacy,35 as is STAT1 signaling.58 Thus, IFN-STAT1 biology is diverse with end effects having a dependence on acute vs chronic stimulation.59 This complex relationship of IFN-STAT1 is reflected in our analyses where we identified diversity in immunoregulatory molecules and immune infiltrate composition despite predominating type I IFN signaling. Overall, this underscores the balanced immune stimulation-inhibition context in different tumor types, which may influence radiation response.
The RSI was originally derived from the NCI-60 cell line panel under uniform in vitro culture conditions by modeling the relationship between the basal molecular repertoire and clonogenic survival following a clinically relevant radiation dose.3 So, how does one explain how an experimental system devoid of immune cell activity identifies dominant immune signaling pathways in patient tumors? We hypothesize that promiscuous genomic instability60 and replication stress manifest in these cell lines may have produced danger associated molecular patterns (DAMPS; e.g., cytosolic DNA) provoking an IFN-based stress response57 which was identified by in vitro assays of radiosensitivity. The data currently supports a model where mislocalized nucleic acids instigate a ‘viral mimicry’ response in the context of radiation, leading to IFN signaling via shared STAT1-IRF1 transcriptional stress programs.
This study has several limitations. First, the measurement of radiation sensitivity is not static, but likely represents the dynamic continuum of cell proliferation-death cycles within an evolving tumor microenvironment. Thus, the RSI is a ‘snapshot’ of tumor biology which may continue to evolve during radiation treatment. Second, although analyzed tumors were prospectively collected, macrodissected, and underwent strenuous pathology quality control, there is still chance for sampling bias. Thus, our study did not address intratumor heterogeneity of the RSI, nor can it differentiate whether the inferred biology from gene expression is from tumor vs non-tumor cells. Future experiments with single-cell RNA sequencing (scRNA-seq) may help elucidate these differences. For example, Jang et al. used scRNA-seq in breast cancer cells and identified variation in RSI across individual tumor cells, which was associated with differences in mutation burden and alterations in DNA repair genes.61 Though, our analysis is based on bulk gene expression, the average biologic signal within the tumor is likely a major driver of the microenvironment cues. These findings are underscored by the recent work of Dai et al. which found a similar relationship between the RSI and immune response,44 which now combined with this work, represents >20,000 independent tumor samples.
This data suggests radiation sensitivity, estimated by the RSI, may be related to unique tumor immune microenvironments that may be leveraged with combinatorial approaches using immunotherapeutics. At present, there are limited tumor samples from patients treated with both radiation and immunotherapy, therefore future investigations retrieving these types of samples will be needed to assess this interaction formally. Last, inference of the tumor immune contexture by bulk gene expression analysis may not fully recapitulate the true immune microenvironment, thus will require further interrogation of patient tissues coupled with mechanistic studies.
At present, radiation therapy is not personalized according to a patient’s tumor biology. Great strides are currently being made in the field of radiation oncology to address this unmet need, as it is becoming more evident the molecular make-up and cellular composition of a tumor may influence the diversity of therapeutic responses seen in clinic. The genomically-adjusted radiation dose (GARD), is a measurable tumor feature that has been proposed to help inform radiation dose selection in patients.8,9 The RSI is a major input, and consequently, a readout of the GARD. Here we show the RSI describes heterogeneity in radiation sensitivity across various tumor types and measures a coordinated biology driven by a STAT1-IRF1 transcriptional program. This RSI transcriptional network supercluster infers certain immune cell types populate IFN-rich tumors which are classified as radiosensitive. Similar to other previously identified transcriptomic signatures,62 the RSI may represent a pan-cancer transcriptional network supercluster which links two assumed distinct tumor attributes, radiosensitivity and immune activity, into one coordinated biologic readout. The tumor immune contexture is increasingly being identified as a major rheostat of patient outcomes in relation to various anti-cancer therapies, thus a deeper understanding of the interconnectedness of these features is needed to optimize strategies combining radiation and immune-based therapies.
Supplementary Material
Pearson’s (r) and Spearman’s (ρ) correlation analysis for RSI and immune score among 31 individual tumor types.
The gene expression values of select immune markers (PD-L1, PD-1, CD47, CTLA-4, CD8A, CD8B) were compared among 31 tumor types by RSIlo and RSIhi groupings. Mann-Whitney U test was used to assess for significant differences.
CIBERSORT-derived estimates of CD8 T, NK+ and M1 cells were compared between RSIlo and RSIhi groupings in HNSC, BC_BASAL and MELA tumors. Groups were assessed for significance by the Mann-Whitney U test.
Publicly available breast cancer (GSE1456) and lung adenocarcinoma (GSE68465) patient cohorts with gene expression data were analyzed by: 1) the RSI, 2) ESTIMATE immune score, and 3) ssGSEA-derived MSigDB hallmark pathways (IFNα and IFNγ). Comparisons between RSIlo and RSIhi groupings were compared by the Mann-Whitney U test.
Supplemental Table 1: Nomenclature for tumor types in the cohort (n=31) and immune cell types (n=22).
Supplemental Table 2: Demographic summary of patient cohort by tumor type with evaluable demographic data (n=8,321). Shown are the total number of patients (N), RSI value (median, IQR), gender (N, percentage) and Age At Diagnosis (median, IQR).
Supplemental Table 3: Quartile estimates of RSI by tumor type with the ratio (Q3/Q1) of RSI.
Supplemental Table 4: Differentially Expressed Genes (DEGs) Between RSIlo and RSIhi Tumors
Supplemental Table 5: Differentially Expressed Genes Across at Least Six Tumor Types Between RSIlo and RSIhi Tumors
Supplemental Table 6: Association of the RSI with Immune-Regulatory Molecules (CD8A, CD8B, CD47, PD-L1, PD-1, CTLA-4) in RSIhi and RSIlo tumors, by tumor type. A t-test is used to evaluate the differences between RSIhi and RSIlo by tumor type/gene combination. Additionally, we accounted for the covariates of age and sex using linear regression.
Supplemental Table 7: Pearson’s Correlation Between Mutation Frequencies, the RSI, Immune Score and Immune Infiltrates
Acknowledgements:
We extend our sincere thanks to the Biostatistics and Informatics Core of H. Lee Moffitt Cancer Center and Research Institute.
Funding:
This work was supported by the NCI Cancer Center Support Grant P30-CA076292 to the Moffitt Cancer Center, by the Cortner-Couch Chair for Cancer Research of the University of the South Florida School of Medicine (J.L.C.), National Institutes of Health R21CA101355 (J.T-R.), De Bartolo Personalized Medicine Institute, the Florida Breast Cancer Foundation (H.E.), the State of Florida to the Florida Academic Cancer Centers Alliance and National Institutes of Health 1U01CA244100 (H.E.).
Footnotes
Conflict of Interest: J.T-R. and S.A.E report intellectual property (RSI) and stock in Cvergenx. J.K.T. consults and has ownership stake in Interpares Biomedicine. J.J.M has ownership interest (including patents) in Fulgent Genetics, Inc., Aleta Biotherapeutics, Inc., Cold Genesys, Inc., Myst Pharma, Inc., Verseau Therapeutics, Inc., and Tailored Therapeutics, Inc., and is a consultant/advisory board member for Celgene Corporation, ONCoPEP, Inc., Cold Genesys, Inc., Morphogenesis, Inc., Mersana Therapeutics, Inc., GammaDelta Therapeutics, Ltd., Myst Pharma, Inc., Tailored Therapeutics, Inc., Verseau Therapeutics, Inc., Iovance Biotherapeutics, Inc., Vault Pharma, Inc., Noble Life Sciences Partners, Fulgent Genetics, Inc., Orpheus Therapeutics, Inc., UbiVac, LLC, Vycellix, Inc., and Aleta Biotherapeutics, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability:
Research data are not available at this time.
References
- 1.Yard BD, Adams DJ, Chie EK, et al. A genetic basis for the variation in the vulnerability of cancer to DNA damage. Nat Commun. Apr 25 2016;7:11428. doi: 10.1038/ncomms11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang L, Wei F, Wu Y, et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J Exp Clin Cancer Res. Apr 23 2018;37(1):87. doi: 10.1186/s13046-018-0758-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eschrich S, Zhang H, Zhao H, et al. Systems biology modeling of the radiation sensitivity network: a biomarker discovery platform. Int J Radiat Oncol Biol Phys 2009;75:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sjostrom M, Staaf J, Eden P, et al. Identification and validation of single-sample breast cancer radiosensitivity gene expression predictors. Breast Cancer Res. Jul 4 2018;20(1):64. doi: 10.1186/s13058-018-0978-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grass GD, Mills MN, Scott JG, Eschrich S, Torres-Roca JF. Genomics and radiomics: Tools to see the unseen to personalize radiation therapy. Applied Radiation Oncology. 2019;8(4):9–22. [Google Scholar]
- 6.Khan MT, Yang L, More E, et al. Developing Tumor Radiosensitivity Signatures Using LncRNAs. Radiat Res. Apr 1 2021;195(4):324–333. doi: 10.1667/RADE-20-00157.1. [DOI] [PubMed] [Google Scholar]
- 7.Cavalieri S, Serafini MS, Carenzo A, et al. Clinical Validity of a Prognostic Gene Expression Cluster-Based Model in Human Papillomavirus-Positive Oropharyngeal Carcinoma. JCO Precis Oncol. 2021;5. doi: 10.1200/PO.21.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott JG, Berglund A, Schell MJ, et al. A genome-based model for adjusting radiotherapy dose (GARD): a retrospective, cohort-based study. Lancet Oncol. Feb 2017;18(2):202–211. doi: 10.1016/S1470-2045(16)30648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott JG, Sedor G, Ellsworth P, et al. Pan-cancer prediction of radiotherapy benefit using genomic-adjusted radiation dose (GARD): a cohort-based pooled analysis. Lancet Oncol. Aug 4 2021. doi: 10.1016/S1470-2045(21)00347-8. [DOI] [PubMed]
- 10.Herrera FG, Bourhis J, Coukos G. Radiotherapy combination opportunities leveraging immunity for the next oncology practice. CA Cancer J Clin. Jan 2017;67(1):65–85. doi: 10.3322/caac.21358. [DOI] [PubMed] [Google Scholar]
- 11.Weichselbaum RR, Liang H, Deng L, Fu YX. Radiotherapy and immunotherapy: a beneficial liaison? Nat Rev Clin Oncol. Jun 2017;14(6):365–379. doi: 10.1038/nrclinonc.2016.211. [DOI] [PubMed] [Google Scholar]
- 12.Ngwa W, Irabor OC, Schoenfeld JD, Hesser J, Demaria S, Formenti SC. Using immunotherapy to boost the abscopal effect. Nat Rev Cancer. May 2018;18(5):313–322. doi: 10.1038/nrc.2018.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demaria S, Guha C, Schoenfeld J, et al. Radiation dose and fraction in immunotherapy: one-size regimen does not fit all settings, so how does one choose? J Immunother Cancer. Apr 2021;9(4). doi: 10.1136/jitc-2020-002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenstermacher DA, Wenham RM, Rollison DE, Dalton WS. Implementing personalized medicine in a cancer center. Cancer J 2011;17:528–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welsh EA, Eschrich SA, Berglund AE, Fenstermacher DA. Iterative rank-order normalization of gene expression microarray data. BMC Bioinformatics. May 7 2013;14:153. doi: 10.1186/1471-2105-14-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pawitan Y, Bjohle J, Amler L, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7(6):R953–964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Director’s Challenge Consortium for the Molecular Classification of Lung A, Shedden K, Taylor JM, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. Aug 2008;14(8):822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. Jan 3 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 20.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. Sep 15 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 21.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. May 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grisaru-Tal S, Itan M, Grass DG, et al. Primary tumors from mucosal barrier organs drive unique eosinophil infiltration patterns and clinical associations. Oncoimmunology. Dec 30 2020;10(1):1859732. doi: 10.1080/2162402X.2020.1859732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. Dec 23 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. Jan 16 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. Nov 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teer JK, Zhang Y, Chen L, et al. Evaluating somatic tumor mutation detection without matched normal samples. Hum Genomics 2017;11:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. Jul 15 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. May 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. Sep 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strom T, Harrison LB, Giuliano AR, et al. Tumour radiosensitivity is associated with immune activation in solid tumours. Eur J Cancer. Oct 2017;84:304–314. doi: 10.1016/j.ejca.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wellenstein MD, de Visser KE. Cancer-Cell-Intrinsic Mechanisms Shaping the Tumor Immune Landscape. Immunity. Mar 20 2018;48(3):399–416. doi: 10.1016/j.immuni.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, Hogg GD, DeNardo DG. Rethinking immune checkpoint blockade: ‘Beyond the T cell’. J Immunother Cancer. Jan 2021;9(1). doi: 10.1136/jitc-2020-001460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity. Apr 17 2018;48(4):812–830 e814. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerns SL, Chuang KH, Hall W, et al. Radiation biology and oncology in the genomic era. Br J Radiol. Nov 2018;91(1091):20170949. doi: 10.1259/bjr.20170949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burnette BC, Liang H, Lee Y, et al. The efficacy of radiotherapy relies upon induction of type i interferon-dependent innate and adaptive immunity. Cancer Res. Apr 1 2011;71(7):2488–2496. doi: 10.1158/0008-5472.CAN-10-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khodarev NN, Minn AJ, Efimova EV, et al. Signal transducer and activator of transcription 1 regulates both cytotoxic and prosurvival functions in tumor cells. Cancer Res. Oct 1 2007;67(19):9214–9220. doi: 10.1158/0008-5472.CAN-07-1019. [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Cao Y, Markelc B, Kaeppler J, Vermeer JA, Muschel RJ. Type I IFN protects cancer cells from CD8+ T cell-mediated cytotoxicity after radiation. J Clin Invest. Oct 1 2019;129(10):4224–4238. doi: 10.1172/JCI127458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weichselbaum RR, Ishwaran H, Yoon T, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A. Nov 25 2008;105(47):18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doroshow JH, Kummar S. Translational research in oncology−−10 years of progress and future prospects. Nat Rev Clin Oncol. Nov 2014;11(11):649–662. doi: 10.1038/nrclinonc.2014.158. [DOI] [PubMed] [Google Scholar]
- 40.Sun S, Xu L, Zhang X, et al. Systematic Assessment of Transcriptomic Biomarkers for Immune Checkpoint Blockade Response in Cancer Immunotherapy. Cancers (Basel). Apr 1 2021;13(7). doi: 10.3390/cancers13071639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. Oct 4 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. Jan 19 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamoun A, de Reynies A, Allory Y, et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur Urol. Apr 2020;77(4):420–433. doi: 10.1016/j.eururo.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai YH, Wang YF, Shen PC, et al. Radiosensitivity index emerges as a potential biomarker for combined radiotherapy and immunotherapy. NPJ Genom Med. Jun 2 2021;6(1):40. doi: 10.1038/s41525-021-00200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heylmann D, Ponath V, Kindler T, Kaina B. Comparison of DNA repair and radiosensitivity of different blood cell populations. Sci Rep. Jan 28 2021;11(1):2478. doi: 10.1038/s41598-021-81058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arina A, Beckett M, Fernandez C, et al. Tumor-reprogrammed resident T cells resist radiation to control tumors. Nat Commun. Sep 2 2019;10(1):3959. doi: 10.1038/s41467019-11906-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng KW, Marshall EA, Bell JC, Lam WL. cGAS-STING and Cancer: Dichotomous Roles in Tumor Immunity and Development. Trends Immunol. Jan 2018;39(1):44–54. doi: 10.1016/j.it.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 48.Deng L, Liang H, Xu M, et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. Nov 20 2014;41(5):843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nat Rev Immunol. Jul 2015;15(7):405–414. doi: 10.1038/nri3845. [DOI] [PubMed] [Google Scholar]
- 50.Lim JY, Gerber SA, Murphy SP, Lord EM. Type I interferons induced by radiation therapy mediate recruitment and effector function of CD8(+) T cells. Cancer Immunol Immunother. Mar 2014;63(3):259–271. doi: 10.1007/s00262-013-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alfonso JCL, Grass GD, Welsh E, et al. Tumor-immune ecosystem dynamics define an individual Radiation Immune Score to predict pan-cancer radiocurability. Neoplasia. Nov 2021;23(11):1110–1122. doi: 10.1016/j.neo.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Post AEM, Smid M, Nagelkerke A, et al. Interferon-Stimulated Genes Are Involved in Cross-resistance to Radiotherapy in Tamoxifen-Resistant Breast Cancer. Clin Cancer Res. Jul 15 2018;24(14):3397–3408. doi: 10.1158/1078-0432.CCR-17-2551. [DOI] [PubMed] [Google Scholar]
- 53.Khodarev NN, Roizman B, Weichselbaum RR. Molecular pathways: interferon/stat1 pathway: role in the tumor resistance to genotoxic stress and aggressive growth. Clin Cancer Res. Jun 1 2012;18(11):3015–3021. doi: 10.1158/1078-0432.CCR-11-3225. [DOI] [PubMed] [Google Scholar]
- 54.Benci JL, Xu B, Qiu Y, et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell. Dec 1 2016;167(6):15401554 e1512. doi: 10.1016/j.cell.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee AJ, Ashkar AA. The Dual Nature of Type I and Type II Interferons. Front Immunol. 2018;9:2061. doi: 10.3389/fimmu.2018.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meissl K, Macho-Maschler S, Muller M, Strobl B. The good and the bad faces of STAT1 in solid tumours. Cytokine. Jan 2017;89:12–20. doi: 10.1016/j.cyto.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 57.Liu H, Golji J, Brodeur LK, et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat Med. Jan 2019;25(1):95–102. doi: 10.1038/s41591018-0302-5. [DOI] [PubMed] [Google Scholar]
- 58.Raspaglio G, Buttarelli M, Filippetti F, et al. Stat1 confers sensitivity to radiation in cervical cancer cells by controlling Parp1 levels: a new perspective for Parp1 inhibition. Cell Death Dis. Oct 12 2021;12(10):933. doi: 10.1038/s41419-021-04229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi LZ, Bonner JA. Bridging Radiotherapy to Immunotherapy: The IFN-JAK-STAT Axis. Int J Mol Sci. Nov 14 2021;22(22). doi: 10.3390/ijms222212295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reinhold WC, Erliandri I, Liu H, Zoppoli G, Pommier Y, Larionov V. Identification of a predominant co-regulation among kinetochore genes, prospective regulatory elements, and association with genomic instability. PLoS One. 2011;6(10):e25991. doi: 10.1371/journal.pone.0025991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jang BS, Han W, Kim IA. Tumor mutation burden, immune checkpoint crosstalk and radiosensitivity in single-cell RNA sequencing data of breast cancer. Radiother Oncol. Jan 2020;142:202–209. doi: 10.1016/j.radonc.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 62.Martinez E, Yoshihara K, Kim H, Mills GM, Trevino V, Verhaak RG. Comparison of gene expression patterns across 12 tumor types identifies a cancer supercluster characterized by TP53 mutations and cell cycle defects. Oncogene. May 21 2015;34(21):2732–2740. doi: 10.1038/onc.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pearson’s (r) and Spearman’s (ρ) correlation analysis for RSI and immune score among 31 individual tumor types.
The gene expression values of select immune markers (PD-L1, PD-1, CD47, CTLA-4, CD8A, CD8B) were compared among 31 tumor types by RSIlo and RSIhi groupings. Mann-Whitney U test was used to assess for significant differences.
CIBERSORT-derived estimates of CD8 T, NK+ and M1 cells were compared between RSIlo and RSIhi groupings in HNSC, BC_BASAL and MELA tumors. Groups were assessed for significance by the Mann-Whitney U test.
Publicly available breast cancer (GSE1456) and lung adenocarcinoma (GSE68465) patient cohorts with gene expression data were analyzed by: 1) the RSI, 2) ESTIMATE immune score, and 3) ssGSEA-derived MSigDB hallmark pathways (IFNα and IFNγ). Comparisons between RSIlo and RSIhi groupings were compared by the Mann-Whitney U test.
Supplemental Table 1: Nomenclature for tumor types in the cohort (n=31) and immune cell types (n=22).
Supplemental Table 2: Demographic summary of patient cohort by tumor type with evaluable demographic data (n=8,321). Shown are the total number of patients (N), RSI value (median, IQR), gender (N, percentage) and Age At Diagnosis (median, IQR).
Supplemental Table 3: Quartile estimates of RSI by tumor type with the ratio (Q3/Q1) of RSI.
Supplemental Table 4: Differentially Expressed Genes (DEGs) Between RSIlo and RSIhi Tumors
Supplemental Table 5: Differentially Expressed Genes Across at Least Six Tumor Types Between RSIlo and RSIhi Tumors
Supplemental Table 6: Association of the RSI with Immune-Regulatory Molecules (CD8A, CD8B, CD47, PD-L1, PD-1, CTLA-4) in RSIhi and RSIlo tumors, by tumor type. A t-test is used to evaluate the differences between RSIhi and RSIlo by tumor type/gene combination. Additionally, we accounted for the covariates of age and sex using linear regression.
Supplemental Table 7: Pearson’s Correlation Between Mutation Frequencies, the RSI, Immune Score and Immune Infiltrates
Data Availability Statement
Research data are not available at this time.