Graphical abstract
Keywords: Neuroblastoma, Network simulation, Chromosomal abnormalities, Inferred karyotype
Abstract
Neuroblastoma (NB) is the most common extracranial solid tumor in children. Although only a few recurrent somatic mutations have been identified, chromosomal abnormalities, including the loss of heterozygosity (LOH) at the chromosome 1p and gains of chromosome 17q, are often seen in the high-risk cases. The biological basis and evolutionary forces that drive such genetic abnormalities remain enigmatic. Here, we conceptualize the Gene Utility Model (GUM) that seeks to identify genes driving biological signaling via their collective gene utilities and apply it to understand the impact of those differentially utilized genes on constraining the evolution of NB karyotypes. By employing a computational process-guided flow algorithm to model gene utility in protein–protein networks that built based on transcriptomic data, we conducted several pairwise comparative analyses to uncover genes with differential utilities in stage 4 NBs with distinct classification. We then constructed a utility karyotype by mapping these differentially utilized genes to their respective chromosomal loci. Intriguingly, hotspots of the utility karyotype, to certain extent, can consistently recapitulate the major chromosomal abnormalities of NBs and also provides clues to yet identified predisposition sites. Hence, our study not only provides a new look, from a gene utility perspective, into the known chromosomal abnormalities detected by integrative genomic sequencing efforts, but also offers new insights into the etiology of NB and provides a framework to facilitate the identification of novel therapeutic targets for this devastating childhood cancer.
1. Introduction
Understanding the etiology and finding key therapeutic targets for childhood cancers is a challenging endeavor, as these tumors often harbor fewer somatic mutations than adult cancers [1], [2]. Neuroblastoma (NB) is a rare solid tumor derived from peripheral sympathetic nervous system. As a devastating childhood cancer, only a few recurrent mutations have been identified in the past decade [3]. ALK is known as the most frequently mutated gene in NB, whose mutations only accounts for only ∼ 12% of sporadic tumors [4], [5], [6]. Nevertheless, advanced stage NBs do exhibit typical segmental chromosomal aberrancies that are associated with poor prognosis [8], in particular loss of heterozygosity (LOH) at chromosome 1p or 11q, and segmental gain of chromosome 17q [9]. Some of these chromosomal abnormalities appear to be significantly associated with MYCN amplification, high risk status and poor disease outcome [10], [11]. As a hallmark of high-risk NB, MYCN amplification accounts for ∼ 20% of total cases and is associated with advanced stage disease and poor diagnosis [12], [13], [14]. However, the biological basis for the association of MYCN amplification with those specific chromosomal aberrancies, such as 1p loss or 17q gain, remains enigmatic. Although loss of ARID1A (located in the 1p36 deleted region [15]), or overexpression of BIRC5 (located in the commonly gained 17q segment [16], [17] have been recently shown to synergize with MYCN to promote NB tumorigenesis [18], [19], limited number of genes within these altered chromosomal regions were found to be bona fide tumor suppressors or oncogenes or to cooperate with MYCN to contribute to NB pathogenesis. Hence, gain or loss-of-function of individual oncogenes or tumor suppressor genes cannot fully address the association of unique patterns of segmental chromosomal aberrancies to the advanced stage NB.
Since genes do not act alone and alterations at the chromosomal level could provide a more effective means for cancer cells to gain growth and/or survival advantage, a computational model from a systems biology perspective, which can explain the contribution of comprehensive abnormalities in chromosomes to the pathogenesis of NB, would be appealing. However, the conventional systems biology approaches are largely focused on the analyses of genetic mutations and differential gene expression, which fail to address the molecular mechanisms underlying the establishment of common chromosomal aberrations. Here, we proposed Gene Utility Model (GUM) and hypothesized that the utility of gene, i.e., the involvement of a gene in conveying intracellular signals to execute certain cellular process, could be the key factor in determining the molecular pathogenesis of NB.
With this motivation, we employed a context-dependent systems biology approach that utilizes a process-guided flow algorithm [20] on the transcriptomic data retrieved from advanced stage NBs to model gene utility. We inferred the information flow that goes through a gene in a biological network to detect genes with dramatic changes in their utilities. In particular in NB we focused in pairwise comparations of different groups of tumors stratified by their risk or status of MYCN amplification. Then, through mapping these differentially utilized genes to their respective chromosomal loci, we built a “utility karyotype”. Intriguingly, our study revealed that our utility karyotype, to a certain extent, can consistently recapitulate previously reported chromosomal abnormalities (segmental gains or loss) in the advanced stage NBs. This indicates that gene utility could be a critical biological variable that can acts as an evolutionary pressure contributing to the selection of chromosomal aberrations during NB tumorigenesis. Besides genes with conserved alterations in their utilities and genomic occurrences, significant number of genes with dramatic differential utilities were also found in the chromosomal regions that are lack of any known genetic alterations. Interestingly, certain chromosomal loci also have enriched differentially utilized genes, forming the “utility hotspots”. Taken together, GUM provides a new avenue for us to better understand the etiology of NB that harbors low somatic mutations and to uncover novel therapeutic candidates, such as those genes with significant changes in their utilities under different disease context, that could easily be missed from past integrative genomic analyses.
2. Materials and methods
2.1. Data
The transcriptomic data used in this study was downloaded from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE49710. Raw expression profiles were background corrected and summarized into probeset values. Probesets mapped to the same gene were averaged and each array was normalized by dividing each expression value by the total expression value in the respective array as described in our previous work [20], [21]. Information for accession number and annotation with respect to MYCN amplification status, disease stage, and NB risk for samples used for pairwise comparisons and fold change of gene expression in this study is provided in Data S1 and Data S2, respectively.
2.2. Protein-protein interaction network
We downloaded the iRefIndex version 14.0 and constructed protein–protein interaction (PPI) network using all interactions available as described in our previous study [20]. We removed self-loops and multiple edges, creating a final iRefIndex PPI network containing 15,608 proteins and 180,044 interactions. Our network is available for download as an R object from our website.
2.3. Construction of context-specific weighted PPI network
Context-specific weighted PPI networks with respect to advanced stage high-risk and nonhigh-risk NBs were constructed using PPI network described above with a co-expression network generated from transcriptomic data corresponding to each of these NB states. Briefly, Pearson correlation coefficients (PCC) were computed across samples with respect to each NB state for each interaction pair available in the PPI network. The absolute values of the PCC were used as interaction weights for PPI network specific to each NB state context.
2.4. The design of the process-guided flow algorithm
The basic concept of the process-guided flow algorithm is adopted from flow algorithms broadly used in engineering field such as simulating traffic flow. In our case, information flow in a gene is analogous to a flow in traffic or electric current with least resistance. Therefore, the process-guided flow algorithm is formulated as a minimum-cost flow optimization problem where the edge capacities of interacting protein pairs are defined as the absolute value of the PCC in the context-specific weighted PPI network and the cost as −log(PCC). PCC is used to capture phenotype-specific information as co-expression is a good indicator of genes cooperating in the same or related pathways. In brief, the process-guided flow algorithm takes a weighted PPI network G(V, E) as input (where V is vertices, i.e., genes and E is edges, i.e., interaction between genes) with a list of proteins P ⊂ V to be used as sources, and a list of proteins to be used as targets T ⊂ V. Each edge is defined by a capacity and cost. Negative flow and flow higher than the edge capacity are not allowed. Also, each node (vertice) has to satisfy the local equilibrium condition, i.e., the inflow must be equal to the outflow at every node, except at the source and sink.
The uniqueness of the process-guided flow algorithm is that it only uses one parameter, that is, the size of the functional neighborhood (SFN) associated with a particular gene that find paths between sources and sinks. Here, SFN is defined as the biological processes (in GO term) associated with the interacting partners of a gene whose edge capacities (i.e., flow capacities that pass through interacting protein pairs) are higher than a threshold (SFN = 0.95 in all calculations presented here). Since SFN controls the number of first-neighbor (directly interacting partners) of the genes, the higher the SFN value, more stringent the path selection and therefore leads to reduced number of paths. If there is a shared GO term between two interacting proteins, the shared GO term will be included in the flow paths. If there is no shared GO term, GO terms of their highly correlated interacting partners (with high edge capacities) will be used. Further details of process-guided flow algorithm can be found in our previous publication [20].
2.5. Identification of source genes
We deemed over-expressed genes as functional relevant candidates that convey key signaling events in disease development. However, over-expressed genes are not identified by fold change method by averaging expression levels across samples. Instead, we used template matching method [22] to identify genes that are preferentially expressed at higher levels across most samples in a given NB state but lower expression levels in another NB state during a pairwise comparison. Top 0.5% of genes with the highest template-matching scores were selected as sources that serve as input to indicate where flows begin in process-guided flow algorithm.
2.6. Assignment of sinks (targets)
Sinks are genes in context-specific weighted PPI network where flows end. In current study, by default we defined sinks as genes involved in transcriptional regulation which include the following three GO terms: GO:0003676 (nucleic acid binding), GO:0006355 (regulation of transcription, DNA-templated) and GO:0008134 (transcription factor binding).
2.7. Identification of differentially utilized genes
We selected the top 30% of the absolute differential gene utility values as the cuff off to select differential utility genes (see utility distribution plot in Data S3). Thus, a gene is defined as high differentially utilized if its differential gene utility for a pairwise comparison ≥ 0.8. Likewise, a gene is defined as low differentially utilized if its differential gene utility for a pairwise comparison ≤ -0.8. These genes comprised top 30% of absolute differential gene utilities (see utility distribution plot in Data S3).
2.8. Construction of differentially utilized karyotypes
Differential utilities of cytogenetic sub-bands (e.g., 1p36.1 and 1p36.2) are the basic components that constitute the utility karyotype. To recognize a utility band at least two differentially utilized genes showing a consistent utility directionality (i.e., high or low differential utilities) is required.
In the case where a cytogenetic band harbors mixed gene utilities (i.e., with both high and low utilized genes), at least two extra genes with same utility directionality (e.g., high utility) than opposite utility directionality (e.g., low utility) should be present for a given cytogenetic band to be considered as a differentially utilized band. Positioning these utility bands onto their respective chromosomal sites gives rise to utility karyotype (Fig. 1).
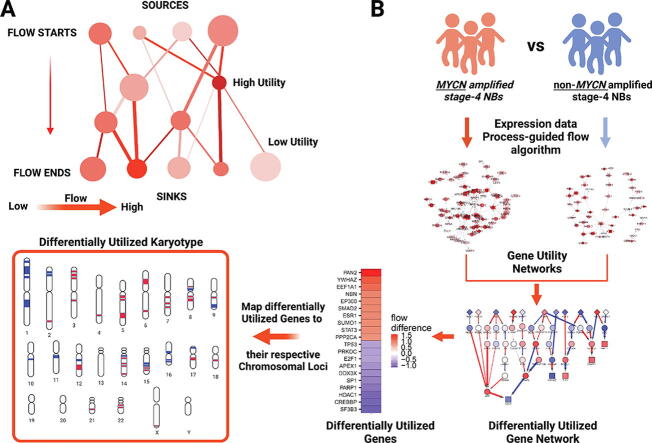
Fig. 1.
Gene Utility conceptual model and computational pipeline. (A). Gene Utility Model (GUM) for interacting proteins. Gene utility reflects the contribution of a gene to cellular signaling processes. Genes (nodes) with high information flows in a disease network are deemed highly utilized genes although they might not be expressed at high levels (small circles). A flow algorithm is used in a disease network to simulate gene utility. Information flows (i.e., signal transduction) begin at the sources and end at in sinks, and their propagation from one protein to another occurs through existing interactions (edges). Thicker edges indicate larger signal transduction capacity. The extent of signal transduction (flow) mediated by a gene is deemed as its utility. (B) Overview of the process-guided flow algorithm to compute signal transduction capacity for each gene (i.e., gene utility). In this example, we used transcriptomic data from MYCN amplified stage-4 neuroblastoma (NB) and non-MYCN amplified stage-4 NBs to perform a pairwise comparison and build two networks. Over-expressed genes between these two disease states are used as sources, where flow starts. Next, a network of differentially utilized genes is computed by subtracting gene utility profiles of the two conditions. Genes with significant differential utilities (|utility| ≥ 0.8) in this pairwise comparison are further mapped to their respective chromosomal loci to generate a differential utility karyotype. (C) Schematic illustration of advance NB states considered in this study for pairwise comparisons. Figure created with BioRender.
2.9. Over-representation analyses (ORA)
ORA were performed using gene list and query at WebGestalt [23]. Human genome was used as reference and statistical significance was computed based on Bonferroni correction. Pathways from KEGG and REACTOME databases with at least 5 genes were used for ORA. Processes with false discovery rate (FDR < 0.05) are deemed statistically significant in over-representation.
3. Results
3.1. Gene utility model and the construction of utility karyotype
In living cells, the extent of functional interactions between gene products (proteins and RNAs) are dependent on their molecular identities, subcellular localizations, and relative abundance, which collectively make up the molecular constituents (called cellular context hereafter) of a given cellular state [24]. Under a specific cellular context, molecular signals are conveyed via protein interactions. Depending on the abundance of proteins and extent of interactions in the protein–protein interaction (PPI) network, the unique phenotypic properties of a cellular state will be established. If a protein is actively and heavily involved in signaling transductions, it is considered highly utilized. An analogy to gene utility is the flow of vehicles in traffic, i.e., the busier a road is, the more important role it plays in commuting, albeit it could be a narrow road (analogous to low levels of gene expression). Thus, a gene received high information flow is deemed to carry high gene utility. The extent of utility of a gene is not necessarily correlated with its expression levels, but rather dependent on the levels of interactions, the numbers and availabilities of its interacting partners in a contain cellular or disease context (Fig. 1A).
Here, we proposed Gene Utility Model (GUM) to understand the importance of genes under specific cellular contexts. GUM states that it is the utility of genes that provides selective pressure for the survival and fitness of cancer cells. Using GUM, it is possible to construct an “utility karyotype” by mapping differentially utilized genes to their respective chromosomal loci. Further, GUM predicts that the resulting utility karyotype can recapitulate, to a certain extent, the chromosomal aberrancies observed in NB.
We have previously developed a network biology tool, NetDecoder, that uses process-guided flow algorithm to reconstruct profiles of network-based, context-specific information flow in pairwise phenotypic comparative analyses [20]. Process-guided flow algorithm is based on the principle that proteins which share similar functions or in the same pathways tend to interact together to transduce signaling information and cooperatively execute a defined process under evolutionary pressure [25]. Briefly, information flows begin with a number of differentially expressed genes, defined as source genes; relayed via interactions among proteins with shared function; and ended with a panel of transcription regulatory genes, defined as sinks (Fig. 1A). To build a GUM, we first generated protein–protein interaction (PPI) networks weighted based on the specific context, by computing Pearson Correlation Coefficients (PCCs) of genes and proteins across neuroblastoma samples within the same classification. Next, the absolute PCC values were used as weights to represent the capacities and costs of flow interacting gene pairs (analogous to width of a road in traffic). Under a specific context of GUM, we defined genes with relatively higher information flows as highly utilized genes and vice versa. Differentially utilized gene network was then computed by subtracting information flows between two different contexts, where genes with dramatically increased or decreased utilities were identified. The subsequent mapping of these differentially utilized genes to their respective chromosomal loci was conducted to generate differentially utilized karyotype for a given pairwise comparison (Fig. 1B).
3.2. High MYCN utilities are seen in MYCN-amplified NBs, while low TP53 utilities are detected in all high-risk NBs
In this study, we are interested in identifying differentially utilized genes associated to stage 4 high-risk NBs or NBs with MYCN amplification (Fig. 1C). The following three pairwise comparisons of the publicly available NB transcriptomic data (GSE49710) were applied, including i) stage-4 NBs with MYCN amplification (M+, n = 65) against all the rest stage-4 NBs (M−, n = 116), ii) stage-4 NBs with MYCN amplification (M+R+, n = 65) against stage-4 high-risk NBs without MYCN amplification (M−R+, n = 83), and iii) stage-4 MYCN non-amplified high-risk NBs (M−R+, n = 83) against all the rest stage-4 MYCN non-amplified NBs (M−R−, n = 33) (Data S1). For a given pairwise comparison, we first computed differential gene expression (Data S2) and used template matching method [22] to select genes with differential expression levels across samples for a given group as sources. For example, for M+ vs. M− pairwise comparison, genes with higher expression levels across samples in M+ tumors were selected as sources. The transcriptional regulatory genes were used as sinks. Then, gene utility networks with respect to each aforementioned group were generated. Differentially utilized genes from a pairwise comparison were finally identified by subtracting utilities of each individual genes involved in the gene utility networks (Fig. 1B and Data S2).
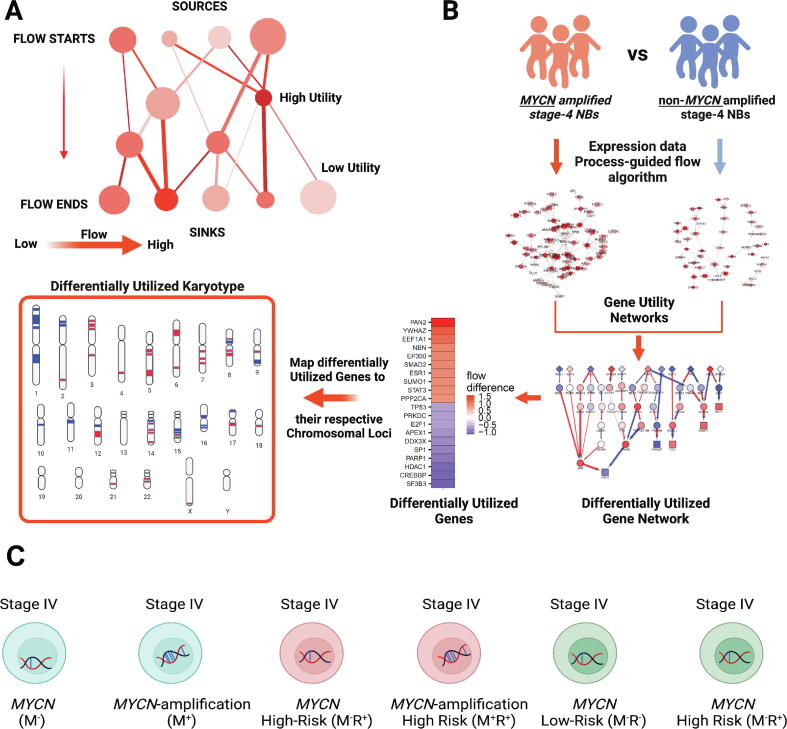
We first examined the expression levels and utilities of MYCN, MYC, and TP53 across these three pairwise comparisons (Fig. 2). As expected, we detected significant high expression levels of MYCN in stage-4 NBs with MYCN amplification (Fig. 2A left panel). This is also consistent with the higher utilities of MYCN in both M+ vs. M− and M+R+ vs. M−R+ comparisons (Fig. 2A right panel), supporting the critical role of MYCN in the tumorigenesis of high-risk NBs. Interestingly, the expression of MYCN is inversely correlated with that of MYC (Fig. 2B left panel) as well as their gene utilities (Fig. 2B right panel). We did not detect flow to MYCN in M−R+ vs. M−R− comparison (Fig. 2A right panel), suggesting less important role of MYCN is playing in the pathogenesis stage-4 NB without its amplification. Intriguingly, we found low utilities of TP53 in all three comparisons (Fig. 2C right panel), albeit its significant overexpression in MYCN-amplified NBs (Fig. 2C left panel). Low utilities of TP53 in high-risk NBs regardless of the MYCN amplification status suggest TP53 indeed functions as a tumor suppressor in high-risk NBs. Our finding, therefore, provides a new evidence on the tumor suppressor role of TP53, which is through its reduced utilities in high-risk NBs, other than the inactivation mutations as detected in many adult cancers [26].
Fig. 2.
Differential gene expression and utility profiles of MYCN, MYC, and TP53 across three pairwise comparisons with respect MYCN amplification status and risk. Relative gene expression (in log2 scale) and utilities for (A) MYCN, (B) MYC and (C) TP53. M+ vs. M−: stage-4 MYCN amplified state against stage-4 non-MYCN amplified state; M+R+ vs. M−R+: stage-4 MYCN amplified high-risk state against stage-4 non-MYCN amplified high-risk state; M−R+ vs. M−R−: stage-4 non-MYCN amplified high-risk state against stage-4 non-MYCN amplified nonhigh-risk state.
3.3. Utility karyotype reveals the association of karyotypic utility bands with MYCN amplification and risk status
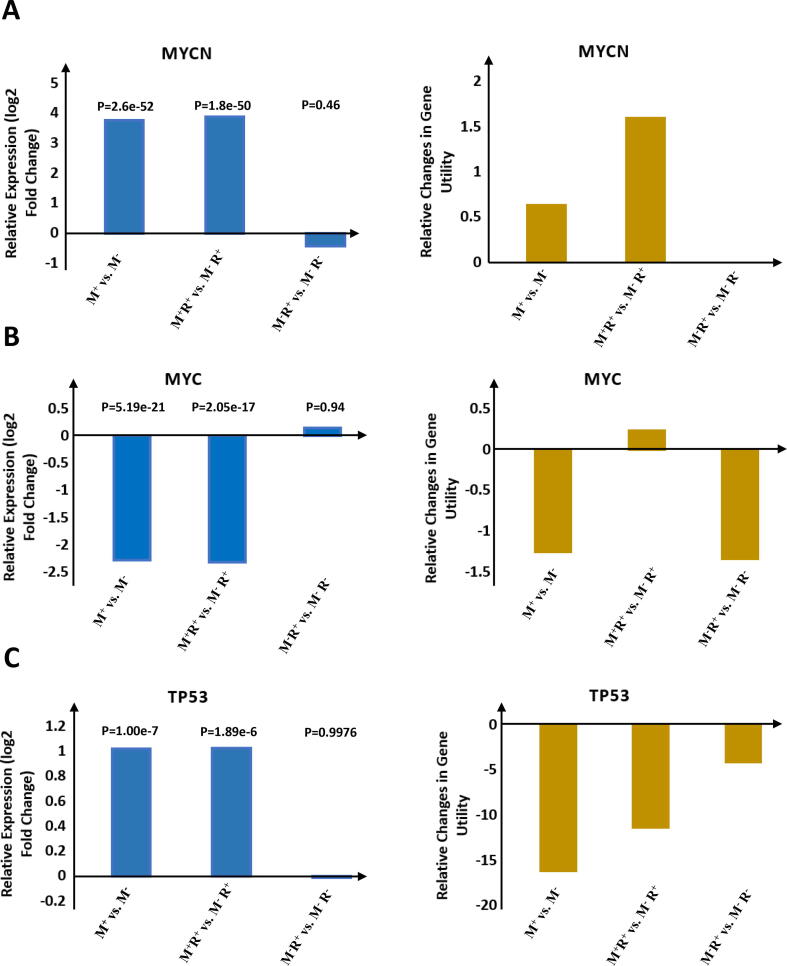
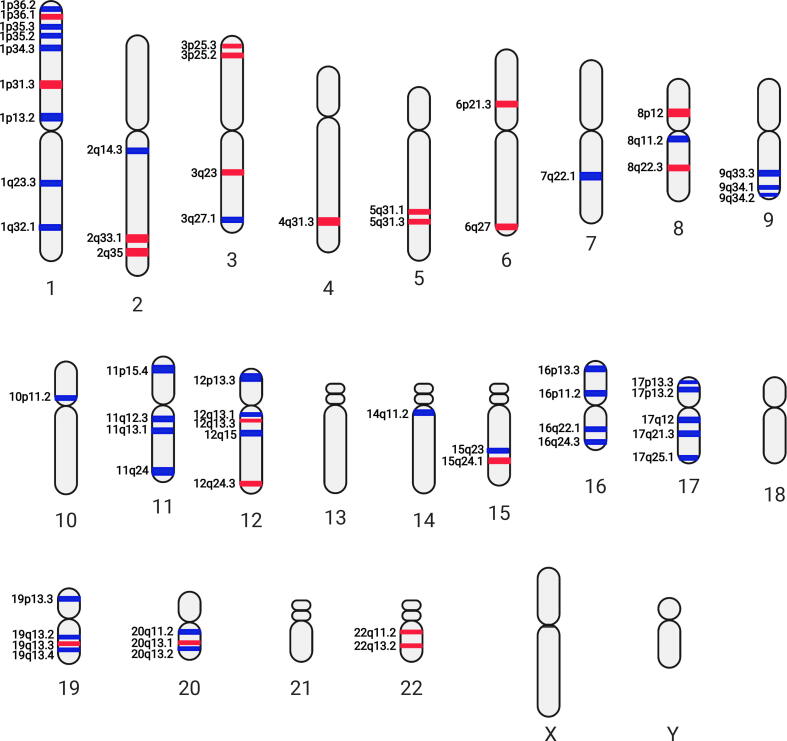
We next asked whether there are patterns of gene utility that could explain the NB etiology when mapping those differentially utilized genes to their respective chromosomal sites. We therefore constructed differentially utilized karyotype for each pairwise comparison by mapping genes with dramatically increased utilities (differential gene utility ≥ 0.8) or decreased utilities (differential gene utility ≤ -0.8) to their respective chromosomal loci (Data S3). The chromosomal region containing at least two outstanding gain or reduced utilized genes over all differentially utilized genes residing in the same cytogenetic sub-band was defined as a “karyotypic utility band”. The resulting differentially utilized karyotypes with respect to M+ vs. M−, M+R+ vs. M−R+, and M−R+ vs. M−R− are presented in Fig. 3, Fig. 4, Fig. 5, respectively.
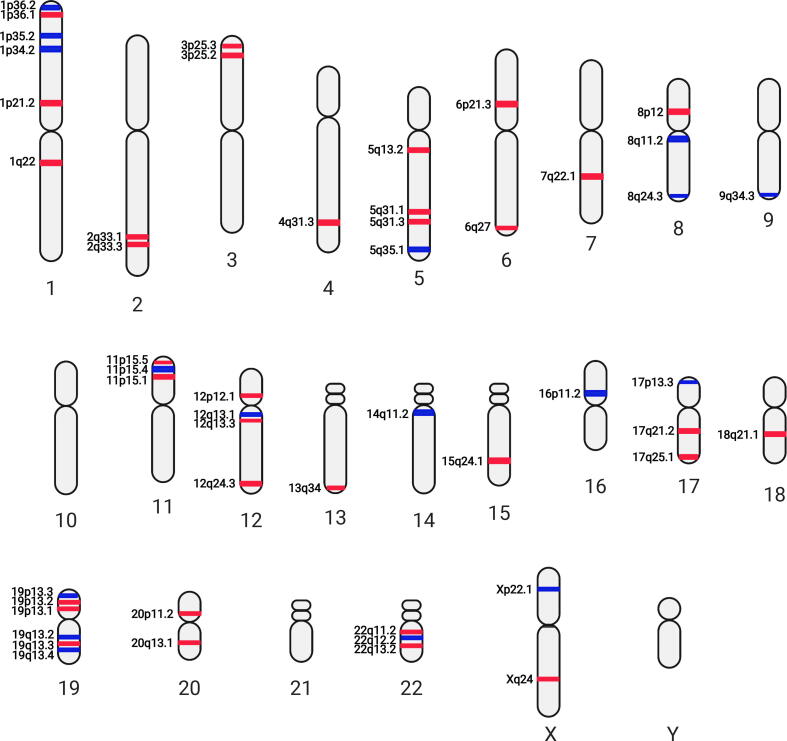
Fig. 3.
Differentially utilized karyotype with respect to stage-4 MYCN amplified state against stage-4 non-MYCN amplified state (M+ vs. M−) pairwise comparison. Red bands: high differentially utilized; blue bands: low differentially utilized. Figure created with BioRender. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
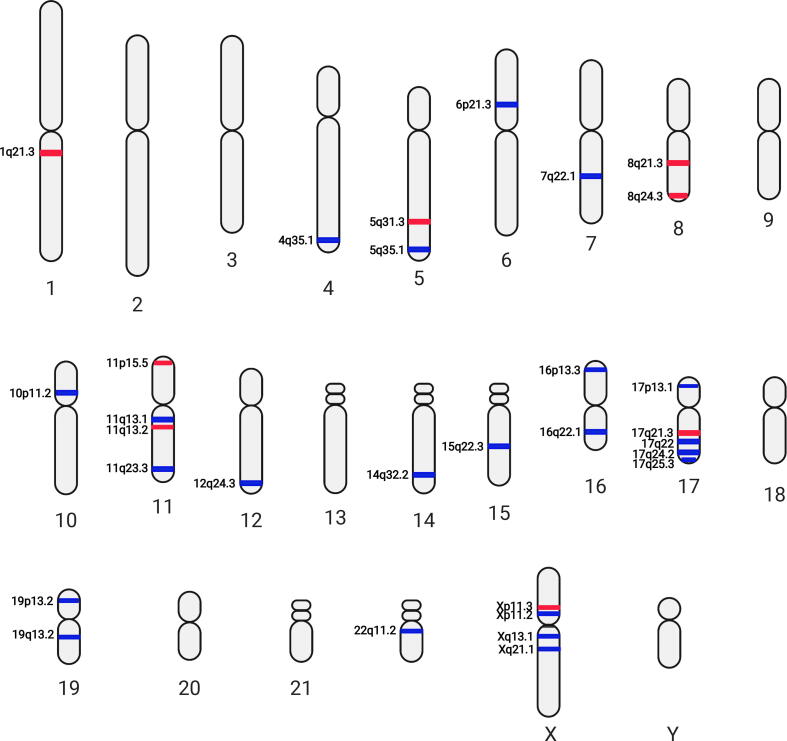
Fig. 4.
Differentially utilized karyotype with respect to stage-4 MYCN amplified high-risk state against stage-4 non-MYCN amplified high-risk state (M+R+ vs. M−R+) pairwise comparison. Red bands: high differentially utilized; blue bands: low differentially utilized. Figure created with BioRender. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Differentially utilized karyotype with respect to stage-4 non-MYCN amplified high-risk state against stage-4 non-MYCN amplified nonhigh-risk state (M−R+ vs. M−R−) pairwise comparison. Red bands: high differentially utilized; blue bands: low differentially utilized. Figure created with BioRender. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In this study, we found a couple of chromosomes, such as 1, 2, 3, 8, 11, 12, 16, and 17 contain more than 2 karyotypic utility bands in respect to MYCN amplification status (M+ vs. M−, or M+R+ vs. M−R+, Fig. 3, Fig. 4). Intriguingly, certain karyotypic utility patterns are well overlapped with the chromosomal aberrancies frequently detected in NBs with MYCN amplification, e.g., low utilities at 1p34 and 1p35 corresponding to the 1p deletion and high utilities at 17q21 and 17q25 corresponding to the 17q gain (Fig. 4). In contrast largely distinct karyotypic utility patterns were observed in the M−R+ vs. M−R− comparison (Fig. 5). These findings suggest that the inferred karyotypic utility bands with respect to the M+ vs. M− or M+R+ vs. M−R+ (Fig. 3, Fig. 4) are most likely associated to MYCN amplification.
3.4. Gene clusters with distinct utility profiles can imply the risk and etiology of NB
To determine whether karyotypic utility patterns can explain NB etiology, we first examined the genes with distinct utility profiles across all pairwise comparisons. We grouped genes with high or low differential utilities (|gene utility| ≥ 0.8) into 6 clusters according to their utility profiles (Data S3 and Data S4): (i) Genes with low differential utilities in both M+ vs. M− and M+R+ vs. M−R+ comparisons (the light blue cluster, n = 80 genes). Genes in this cluster correspond to those with decreased utilities in MYCN amplified NBs regardless of their risk status. (ii) Genes with high differential utilities in both M+ vs. M− and M+R+ vs. M−R+ comparisons (the dark yellow cluster, n = 119 genes). Genes in this cluster correspond to those with increased utilities in MYCN amplified NBs regardless of the risk status. (iii) Genes with low differential utilities in both M+ vs. M− and M+R+ vs. M−R+ comparisons but with high differential utilities in M−R+ vs. M−R− comparison (the green cluster, n = 26 genes). Genes in this cluster correspond to those with decreased utilities in MYCN amplified NBs but seems highly required for non-MYCN amplified high-risk NBs. (iv) Genes with high differential utilities in both M+ vs. M− and M+R+ vs. M−R+ comparisons but with low differential utilities in M−R+ vs. M−R− (the orange cluster, n = 25 genes). Genes in this cluster correspond to those with increased utilities in MYCN amplified cases but seems less involved in non-MYCN amplified high-risk NBs. (v) Genes with low differential utilities in all comparisons (the dark grey cluster, n = 15 genes). And (vi) Genes with high differential utilities in all comparisons (the pink cluster, n = 11 genes).
Genes of the dark grey [low utilities, (v)] or pink [high utilities, (vii)] clusters are of particular interest, because their utilities are associated with high-risk status regardless of the MYCN amplification. Genes in the dark grey cluster could most likely play tumor suppressing role, while genes in the pink cluster could be critical for maintaining the aggressiveness of high-risk NBs. Consistent with this hypothesis, TP53 was found in the grey cluster (Data S4). Another gene of interest in this cluster is RXRA (retinoid X receptor-α). It has been shown that retinoic acid plays a major role in the development of nervous system during embryogenesis and it can induce differentiation of neuroblastoma cells [27]. As an important receptor of retinoic acid, we observed the consistent association of decreased utility of RXRA with increased risk of NB (Data S4). Besides RXRA, SMAD3 was also appeared in this dark grey cluster (Data S4). An earlier study has shown that SMAD4, a close family member of SMAD3, inhibits the growth, invasion, and metastasis of NB cells both in vitro and in vivo [28]. In addition, our over-representation pathway analysis of dark grey cluster showed that genes involved in cell cycle process are significantly enriched (Data S4), suggesting their roles as negative regulators of cell cycle during NB pathogenesis.
On the other hand, genes in pink cluster are enriched with ribosomal genes (Data S4), suggesting the importance of risk-conferring roles of ribosomal processes in advanced stage NBs. Of particular interest is a recent study showing that inhibition of ribosome biogenesis suppresses the growth of MYCN-amplified neuroblastoma [29]. Our data suggest targeting ribosomal genes in pink cluster, such as RPL6, RPL9, RPS20, and RPS3A, might inhibit progression of high-risk NBs, regardless the presence of MYCN amplification. Interestingly, we found pathways related to SLITs and ROBOs are enriched in light blue, dark yellow, and pink clusters (Data S4). SLIT-ROBO is known to regulate diverse axon guidance [30] that had been shown to be involved in NB development [31]. Therefore, our data further suggest the contribution of proteasomal and ribosomal genes to NB pathogenesis and the genes utilities along with different utility clusters can consistently recapitulate early findings resulted from experimental models.
3.5. Gene utility might contribute to the established patterns of chromosomal aberrancies in advanced stage NBs
We next reason that if gene utility is an important criterion contributing to evolutionary selective forces shaping chromosomal aberrancies in NB, then the inferred utility karyotype should be able to recapitulate and explain to a certain extent known chromosomal aberrancies. We therefore compiled findings from reported cytogenetic studies on chromosomal aberrancies found in NB (Data S5) to examine to what extent the constructed utility karyotypes recapitulate observed pattern of chromosomal aberrancies.
Chromosomal aberrations of 1p are frequently reported in NB, especially LOH at 1p34-36 have been found to be associated with high-risk NB harboring MYCN amplification [32], [33], [34]. Indeed, both M+ vs. M− and M+R+ vs. M−R+ pairwise comparisons indicate low gene utilities at 1p34 and 1p35 (Fig. 3, Fig. 4), suggesting the association of low gene utilities in these chromosomal regions with MYCN amplification. For example, NFYC, γ subunit of nuclear transcription factor Y (a light blue cluster gene) located at 1p34, has low utilities in both M+ vs. M− and M+R+ vs. M−R+ pairwise comparisons (Data S3). A novel splice variant of NFYA, α subunit of nuclear transcription factor Y, has been identified recently in NB [35], suggesting a potential involvement of nuclear transcription factor Y in NB pathogenesis. We found 4 green cluster genes, including RBBP4, EIF3I, HDAC1, and RPA2, at 1p35 showed low utilities in MYCN amplified NBs but high utilities in non-MYCN amplified high-risk NBs (Data S3). An earlier study indicates that selective HDAC1/2 inhibitors may induce differentiation in NB cell lines [36]. Thus, our data suggest that inhibition of HDAC1 might be more effective to non-MYCN amplified high-risk NBs due to its relatively increased utility in these tumors. Nevertheless, the roles of low utilities or deletions of the other 3 green cluster genes in MYCN amplified cases remain to be elucidated.
Interestingly, we also found succinate dehydrogenase complex iron sulfur subunit B (SDHB), located at 1p36.1 in an orange cluster, shows high gene utilities in both M+ vs. M− and M+R+ vs. M−R+ comparisons (Data S3). Since 1p36 is a region with frequent loss of heterozygosity (LOH) in high-risk NBs, high utilities of SDHB in MYCN amplified NBs seems enigmatic. We reason that SDHB plays important roles in respiratory chain, its total impairment can cause metabolic and functional derangement in NB cells. Therefore, there is no homozygous loss of 1p in NBs and high utility of SDHB could be a compensatory effect to sustain NB cell survival. In addition, we hypothesize that heterozygous loss of one of copy of genes located at 1p36 can confer survival advantages to MYCN amplified NB cells. A recent study showed 1p36 deletions affect ARID1A, a tumor suppressor candidate associated to MYCN amplification [15]. Our utility models did not capture significant utility changes for ARID1A. But we detected low gene utilities for TARDBP, a TAR DNA-binding protein located at 1p36.2 (in the light blue cluster) in MYCN amplified NBs (Data S3). We suspect low utilities and heterozygous loss of this gene might be related to the reduced production of TDP-43 to avoid cytotoxic effect of its accumulation to neuron cells [37] including NB cells [38]. Interestingly, a TARDBP associated gene, amyloid β precursor protein (APP, located at 21q21), showed high gene utilities in MYCN amplified but low utility in non-MYCN amplified NBs (Data S3), although there is no obvious utility karyotype band formed at 21q21. Our data, therefore, suggest low utilities of genes located at 1p34 and 1p35 correspond to LOH in MYCN amplified NBs, whereas high utilities of genes located at 1p36.1 with respect to the 1p36 deletion could be due to their functional compensation, such as critical roles of SDHB in respiratory chain and TARDBP in reducing cytotoxicity.
Segmental gain of 17q is another frequently reported region associated with high-risk NBs. High correlation of gain at 17q21-qter with MYCN amplification is a well-known hallmark for high-risk NB and associated with poor outcome of NB patients [11]. Intriguingly, our constructed utility karyotypes with respect to M+R+ vs. M−R+ pairwise comparisons indeed indicate high gene utilities at 17q21.2 and 17q25.1 (Fig. 4, Data S4). We found STAT3 (an orange cluster gene) and RARA (dark yellow cluster genes) at 17q21.2 both show high utilities in MYCN amplified NBs. Although gains of 17q are also reported in non-MYCN amplified NBs and we detected high utilities at 17q21.3 in the M−R+ vs. M−R− comparison (Fig. 5), our gene utility models suggests that certain genes located at 17q might play more important roles in MYCN amplified states since their gene utilities are dramatically increased. Of particular interest is STAT3 which exhibits high gene utilities in MYCN amplified NBs but low utility in non-MYCN amplified cases (Data S3). This data is consistent with earlier report showing that STAT3 activity is required for the regulation of MYCN expression [39].
LOH on chromosome 14q has been shown to be associated with stage 4 NBs without MYCN amplification [10], [40], [41]. Our GUM consistently detected the low utilities at 14q32.2 for M−R+ vs. M−R− comparison (Fig. 5). We found poly(A) polymerase α (PAPOLA), located at 14q32.2 in the orange cluster, exhibited with high utilities in MYCN-amplified NBs but low utility in non-MYCN amplified high-risk NBs (Data S3). An early study indicated that induction of axon formation in neuroblastoma cells requires increased activity of poly(A) polymerase [42]. Therefore, the GUM provides a new light to explain LOH of chromosome 14q in tumors without MYCN amplification.
It has been reported the LOH at 16p in familial NB and at 16p12-13 in sporadic cases [43]. Further examination indicated LOH at 16p12-13 contained predisposition gene that might contribute to the pathogenesis of NB [44]. Our results showed low gene utilities at 16p11.2 and 16p13.3 for M+ vs. M− (Fig. 3) and at 16p11.2 for M+R+ vs. M−R+ (Fig. 4). Low gene utilities at these chromosomal sites might strengthen the loss of function effect of genes in MYCN amplified NBs. We found UBE2I, located at 16p13.3 in the grey cluster, exhibited low gene utilities in all high-risk NBs regardless of MYCN amplification status. Similarly, KCTD5 located at 16p13.3 in the light blue cluster, showed low gene utilities in MYCN amplified NBs (Data S3), suggesting their potential tumor suppressor roles in NB.
Deletion at chromosome 3p is frequently found in non-MYCN amplified NBs but rarely detected in MYCN-amplified NBs8. We did not observe dramatically reduced utilities at 3p for M−R+ vs. M−R− comparison (Fig. 5). However, we detected high utilities at 3p25.2 and 3p25.3 for both M+ vs. M− (Fig. 3) and M+R+ vs. M−R+ (Fig. 4), suggesting increased gene utilities at 3p25 might be resulted from the reduced utilities of these genes in MYCN non-amplified NBs.
In comparison to other cancers with childhood onset, NB shares overlapping but also unique chromosomal properties. Retinoblastoma, a disease that is similar to neuroblastoma in age of onset and occasional gene mutation profiles, is typically associated with deletions in the chromosome 13q14 with secondary aberrations in 1q, 6p/q, and additional deletions within chromosome 13 [45]. Wilms tumor is associated 11p13 and also paired with 11p15 aberrations, which has been linked to NB through other disorders as both conditions are correlated to Beckwith-Wiedemann Syndrome [46]. Acute myeloid leukemia has a poor prognosis that shares similar alterations at 11q23 with high-risk NBs, likely indicating presence of critical genes contribute to both cancer [47]. However, none of these diseases show co-occurred chromosomal aberrations such as LOH at 1p or gain at 17q, together with MYCN amplification. Hence, our model for utility karyotypes, to certain extent, can recapitulate chromosomal aberrancies that are particularly important to NBs.
3.6. Utility karyotype suggests novel predisposition chromosomal sites
We next examined whether utility karyotypes can recognize novel predisposition sites. Studies by Diskin et al on rare variants in TP53 and copy number variations identified susceptible sites at 17p13 and 1q21, respectively [48], [49]. Our recent study identified deficiency of GAS7 at 17p13.1 promotes MYCN-driven metastasis [50]. Although we did not detect flows in GAS7 in the current study (Data S2), we found low gene utilities at 17p13.2 and 17p13.3 for M+ vs. M− (Fig. 3), at 17p13.3 for M+R+ vs. M−R+ (Fig. 4), and at 17p13.1 for M−R+ vs. M−R− (Fig. 5). Beside tumor suppressor gene TP53, 17p13 also contains differentially utilized genes in MYCN-amplified NBs, such as PRPF8 (a light blue cluster gene) and RPA1 (a green cluster gene) (Data S3), suggesting that genes located at 17p13 might play predisposition role in high-risk NBs. Our study also identified high utilities at 1q21.3 for M−R+ vs. M−R− (Fig. 5), which might harbor predisposition genes associated with non-MYCN amplified NBs. In addition, previous integrative genomics study had identified that LMO1, located at 11p15, is an important NB susceptibility gene [51]. Although we did not detect the information flow to LMO1 in GUM, the constructed utility karyotypes for both M+ vs. M− and M+R+ vs. M−R+ pairwise comparisons indeed indicate the involvement of 11p15 in high-risk NBs (Fig. 3, Fig. 4).
Our constructed utility karyotypes also suggest the importance of X chromosome in NB etiology. We found Xp22.1 and Xq24 (Fig. 4), Xp11.2, Xp11.3, Xq13.1 and Xq21.1 (Fig. 5) are plausible predisposition sites responsible for NB development. In particular, ATRX, located at Xq21.1, showed opposite utility changes in high-risk NBs with and without MYCN amplification (Data S3). It is known that inactivation mutations in ATRX and MYCN amplification are mutually exclusive [52]. Interestingly, we detected higher ATRX utilities in MYCN-amplified NBs, which is most likely resulted from the reduced utility of ATRX in non-MYCN amplified NBs (Data S3). Besides, we also identified low differential gene utilities at Xq13 and Xq21 in high-risk NBs without MYCN amplification (Fig. 5). A study by Satge et al revealed that patients with Turner Syndrome, where one X chromosome is completely or partially missing in females, developed NB [53]. They also noted that patients with Klinefelter syndrome harboring XXY karyotypes or triple-X syndrome (XXX karyotype) developed neuroblastic tumors [53]. Another study by Parodi et al showed that loss of whole chromosome X is associated to poor prognosis of NB [54], which further supports the potential protective role of chromosome X in NB tumorigenesis.
3.7. A gene utility-based evolutionary selection model for the development and survival of NB cells
One of the hallmarks of cancer cells is that they undergo high rate of glycolysis resulting in a surplus of lactate production regardless of oxygen supply [55]. Excess lactate is then converted back to glucose in liver, leading to a net ATP consumption that can lead to body wasting, such as cachexia in cancer patients [56]. Hence, cancer development is energetic costly. Energetic expenditure is therefore a non-genetic evolutionary force to select cancer cells with genomic architectures that facilitate their fitness and survival. A proper channeling molecular resources that involves coordinated information flow throughout protein–protein interaction network and the optimized utilization of genes in a given context could help cancer cells better meet the energetic demand for their survival. We, therefore, propose a hypothetical model to explain how gene utility contributes to the selection of karyotypes for NB development (Fig. 6).
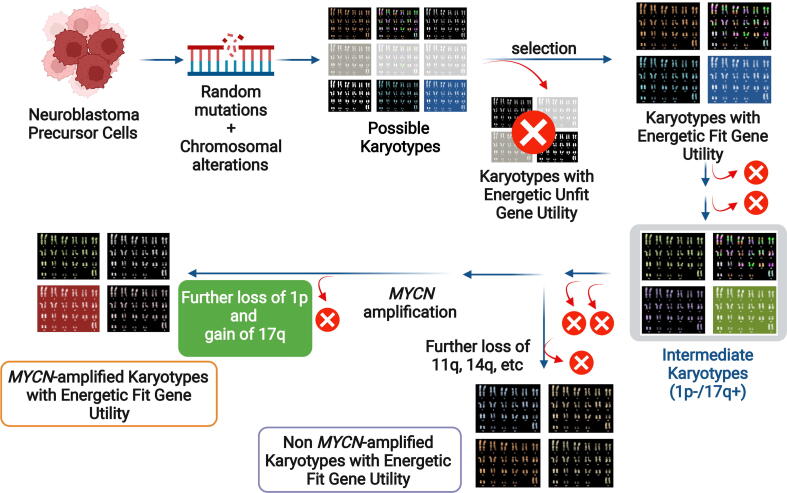
Fig. 6.
A scheme for evolutionary selection of NB karyotypes and associated outcomes based on gene utility and energetic cost concepts. Early random somatic mutational and chromosomal abnormality events occurring on neuroblastoma precursor cells can lead to a variety of karyotypes. A set of these karyotypes harbor gene utilities that provide an energetically fit advantage and promote survival of tumor cells. Other rounds of somatic mutations and acquisition of other chromosomal aberrations and selection events lead to intermediate karyotypes, which show a partial deletion at 1p and/or gain of 17q in the majority of cases. In this intermediate stage, NB cells can either acquire MYCN amplification, followed by 1p deletion and 17q addition to reach a more favorable gene utility state which promotes cell survival. Alternatively, tumor cells with a loss of 11q and/or 14q divert from the evolutionary course of NB development to reach a MYCN amplification independent state. x: energetically unfit karyotypes. Figure created with BioRender.
In our model, neuroblastoma precursor cells acquire random mutations and chromosomal aberrancies. Through cycles of selection, clones with mutations and abnormal chromosomal architectures exhibit gene utility profiles that fail to meet energetic demand for survival are weeded out. However, a set of abnormal chromosomal architectures show certain survival advantage by utility profiles can favor energetic fitness. This process is in part supported by the limited number of mutations detected in NB. Further, we observed several genes that promote cell cycle progression, such as PCNA, DNA response, such as NBN, and protein synthesis, such as RPL6 and RPL9. All these are pink cluster genes with high gene utilities in high-risk NBs.
Once energetically fit, karyotypes are established in early stages. Evolutionary forces will continue to impact their selection, leading to intermediate karyotypes, such as partial 1p deletion and/or 17q gain with energetically fit gene utilities. This assumption is supported by the fact that MYCN amplification is often found to be accompanied by either 1p loss [57]or 17q gain [11], or both. However, both LOH at 1p and gain at 17q can be observed in the absence of MYCN amplification [58]. Furthermore, gain of 17q is strongly linked to both 1p deletion and MYCN amplification [11], suggesting that either 1p deletion and/or 17q gain precedes MYCN amplification. Such randomly acquired genomic instability leads to the presence of karyotypes with both energetically fit and unfit gene utility. For example, the gain of 17q21-qter is strongly associated with tumor progression [11]. Further accumulation of segmental alterations had been suggested to linked to NB progression [59]. As chromosomes become more unstable with the advancement of the disease and in the extreme but rare cases, chromothripsis that involves massive chromosomal rearrangements can occur [7]. During NB evolutionary course, acquisition of MYCN amplification via circular recombination [60] might offer additional survival advantages to certain tumor cells. Finally, post-MYCN amplification selection for cells carrying larger deletion 1p and gain of 17q can have a utility fitness advantage that foster high-risk tumors harboring MYCN-amplified karyotypes. Alternatively, the acquisition of other chromosomal aberrancies such as deletion at 11q and 14q can also provide an alternative gene utility setting that confers cancer cell survival without MYCN amplification. Earlier works and current analysis suggest mutually exclusive gene utility profiles between tumors with and without MYCN amplification, for instance the inactivation and utility of ATRX. Hence, we hypothesize that there exist certain chromosomal aberrancies at late intermediate stage (probably after the acquisition of 1p deletion and 17q amplification) that dictate the evolutionary trajectories, and therefore the acquisition of MYCN amplification could offer better fitness to cancer cells. Chromosomal aberrancies that dictate the bifurcation and determination of process in “karyotypic” fates into MYCN amplification remain to be elucidated.
4. Discussion
NB is a childhood cancer with high metastatic rate and motility. However, its etiology is still not well understood. Since current genomics approaches largely relied on genetic mutations as hallmarks to benchmark the importance of genes for disease etiology, low number of somatic mutations in sporadic high-risk NB renders this disease “a tough nut to crack” [61]. Although with less recurrent somatic mutations, advanced stage NBs do show conserved patterns of chromosomal alterations. But the evolutionary forces and biological basis behind these conserved patterns of chromosomal aberrations remains elusive. For example, limited tumor suppressor genes or oncogenes were identified in the chromosome arms with deletion or gain/amplification. Thus, we reason that current studies based primarily on genetic alterations could not provide a complete understanding of the molecular bases underlying NB pathogenesis.
In this study, we introduce a new conceptual framework, called GUM, to understand disease etiology from the perspective of how a gene is utilized in a biological network. In brief, gene utility concerns how a gene is used in the context of myriad of molecular interactions in biological network to influence the functionality of a biological process. Unlike gene expression that is dependent on whether the promoter and enhancer regions of a gene is accessible by the transcription factor that regulates its expression, gene utility is an emergent property in that the extent of utility of a gene is dependent on cellular context that includes how a gene interacts with its interacting counterparts in PPI network as determined by their functionality shaped by evolution and their expression levels (or protein levels).
With this concept, we employed process-guided flow algorithm to model genome-wide information flow to represent gene utility corresponds to NB with specific classification. We then perform pairwise comparisons to identified differentially utilized genes and construct differentially utilized karyotypes by mapping these genes to their respective chromosomal loci. Our study shows distinct utility karyotypic profiles for high-risk NBs with or without MYCN amplification and the advanced stage NBs with different risk stratification. Further analyses indeed show the constructed utility karyotypes recapitulate the chromosome segmental changes associated with MYCN amplification, such as low differential utility bands at 1p and high differential utility band at 17q in MYCN-amplified NBs. Furthermore, the constructed utility karyotypes also recapitulate chromosomal aberrancies that are unique to NB without MYCN amplification, such as LOH at 14q and 22q. In addition, differential utility bands at the constructed utility karyotypes suggest novel predisposition sites, such as high tendency in loss of 17p, and the potential protective roles of X chromosome on high-risk NB.
Hence, our study shows that gene utility is a useful conceptual framework, which provides insightful explanation to the biological basis of the chromosomal abnormalities observed in advanced stage NBs. We further propose gene utilities can be influenced by energetic expenditure that act together as non-genetic factors to constrain the evolutionary forces to impact tumorigenesis. Our work, therefore, unveils hidden information encoded within the whole transcriptome via computational methods, such as process-guided flow algorithm. The decoded biological knowledge not only recovers findings from previous cytogenetic studies, but also provides further insights on novel utility karyotype hotspots. Inferred karyotypes can be investigated for their association to predisposition risk in certain diseases, and leveraged to identify gene utility hotspots and selection of new targets that are amenable to drug interventions.
Despite the robustness of our GUM, this computational approach, like many other network biology methods, is subjected to a number of limitations related to the completeness of PPI network and the size of samples for the analysis. In particular, many genes, such as non-coding RNAs, are not included in the PPI network. For instance, genes encoding miR34-a at 1p36 and miR-125b at 11q24 are frequently deleted in NB [62], [63], [64], [65], but cannot to be captured in our analyses. Furthermore, it is known that genetic mutations can cause rewiring of PPI network in cancer [66], [67]. However, the rewired PPI networks, especially in NB, is largely unknown, which might lead to incomplete recapitulation of the key findings from previous genomic analyses. For instance, we failed to identify low utility at chromosome 3p where deletions were found in NBs without MYCN amplification [10]. In addition, we could not capture all the information flow through genes that are known to be involved in NB pathogenesis, such as GAS7 [50], which could be due to the limited numbers of source genes selected for the pairwise analyses in GUM. Moreover, current study cannot address chromothripsis, a massive chromosomal rearrangement event throughout whole genome in cancer cells [68], [69], or epigenetic regulation, including the chromatin remodeling that affects three-dimensional organization of chromatin and gene expression.
Nonetheless, our gene utility study sheds a new light on the understanding of biological basis that drives neuroblastoma development and provides insightful explanation on the relative conserved patterns of chromosomal aberrancies that contribute to the pathogenesis of NBs. Importantly, GUM can facility the identification of novel therapeutic targets based on their differential utilities but not expression levels or genomic alterations, which opens a new avenue for developing effective targeted therapy for childhood cancer where somatic mutations are scarce.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
Funding: This work was supported by a grant R01 CA240323 (S. Z.) from the National Cancer Institute, United States; a grant W81XWH-17-1-0498 (S.Z.) from the United States Department of Defense (DoD), United States; a V Scholar award (D2018-005) from the V Foundation for Cancer Research (S. Z.), United States; and a grant support from the Mayo Clinic DERIVE Office and Mayo Center for Biomedical Discovery (S. Z.), United States. A grant P30CA015083 from the Mayo Clinic Cancer Center, Mayo Center for Biomedical Discovery and Center for Individualized Medicine (H. L and S. Z.), United States; grants support from the Glenn Foundation for Medical Research, and the David F. and Margaret T. Grohne Cancer Immunology and Immunotherapy Program (H. L.), United States; and grants R01AG056318 and P50CA136393 (H. L.) from the NIH, United States.
Author's contributions: C.Y.U., H.L., and S.Z. contributed to the conception and design of the study. C.Y.U., T.M.L., C.Z., C.C., K.S.Y. H.L. and S.Z. contributed to the acquisition of data, C.Y.U., T.M.L., C.Z., C.C., K.S.Y. H.L. and S.Z. contributed to the analysis and interpretation of data. C.Y.U., T.M.L., C.Z., C.C., K.S.Y. H.L. and S.Z. drafted the manuscript. S.Z. supervised the study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.06.024.
Contributor Information
Hu Li, Email: li.hu@mayo.edu.
Shizhen Zhu, Email: zhu.shizhen@mayo.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Vogelstein B., et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grobner S.N., et al. The landscape of genomic alterations across childhood cancers. Nature. 2018;555:321–327. doi: 10.1038/nature25480. [DOI] [PubMed] [Google Scholar]
- 3.Pugh T.J., et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosse Y.P., et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George R.E., et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y., et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 7.Molenaar J.J., et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 8.Brady S.W., et al. Pan-neuroblastoma analysis reveals age- and signature-associated driver alterations. Nat Commun. 2020;11:5183. doi: 10.1038/s41467-020-18987-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maris J.M. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vandesompele J., et al. Genetic heterogeneity of neuroblastoma studied by comparative genomic hybridization. Genes Chromosomes Cancer. 1998;23:141–152. doi: 10.1002/(sici)1098-2264(199810)23:2<141::aid-gcc7>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Bown N., et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med. 1999;340:1954–1961. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- 12.Brodeur G.M., Seeger R.C., Schwab M., Varmus H.E., Bishop J.M. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 13.Seeger R.C., et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 14.Schwab M., et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305:245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Lopez, J. et al. Large 1p36 Deletions Affecting Arid1a Locus Facilitate Mycn-Driven Oncogenesis in Neuroblastoma. Cell Rep30, 454-464 e455, doi:10.1016/j.celrep.2019.12.048 (2020). [DOI] [PMC free article] [PubMed]
- 16.Adida C., Berrebi D., Peuchmaur M., Reyes-Mugica M., Altieri D.C. Anti-apoptosis gene, survivin, and prognosis of neuroblastoma. Lancet. 1998;351:882–883. doi: 10.1016/S0140-6736(05)70294-4. [DOI] [PubMed] [Google Scholar]
- 17.Islam A., et al. High expression of Survivin, mapped to 17q25, is significantly associated with poor prognostic factors and promotes cell survival in human neuroblastoma. Oncogene. 2000;19:617–623. doi: 10.1038/sj.onc.1203358. [DOI] [PubMed] [Google Scholar]
- 18.Shi, H. et al. ARID1A loss in neuroblastoma promotes the adrenergic-to-mesenchymal transition by regulating enhancer-mediated gene expression. Sci Adv6, eaaz3440, doi:10.1126/sciadv.aaz3440 (2020). [DOI] [PMC free article] [PubMed]
- 19.Decaesteker B., et al. From DNA Copy Number Gains and Tumor Dependencies to Novel Therapeutic Targets for High-Risk Neuroblastoma. J Pers Med. 2021;11 doi: 10.3390/jpm11121286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.da Rocha E.L., Ung C.Y., McGehee C.D., Correia C., Li H. NetDecoder: a network biology platform that decodes context-specific biological networks and gene activities. Nucleic Acids Res. 2016;44:e100. doi: 10.1093/nar/gkw166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghanat Bari M., Ung C.Y., Zhang C., Zhu S., Li H. Machine Learning-Assisted Network Inference Approach to Identify a New Class of Genes that Coordinate the Functionality of Cancer Networks. Sci Rep. 2017;7:6993. doi: 10.1038/s41598-017-07481-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pavlidis, P. & Noble, W. S. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol2, RESEARCH0042, doi:10.1186/gb-2001-2-10-research0042 (2001). [DOI] [PMC free article] [PubMed]
- 23.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh J.A., et al. Protein complexes are under evolutionary selection to assemble via ordered pathways. Cell. 2013;153:461–470. doi: 10.1016/j.cell.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivlin N., Brosh R., Oren M., Rotter V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer. 2011;2:466–474. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carpentier A., et al. Distinct sensitivity of neuroblastoma cells for retinoid receptor agonists: evidence for functional receptor heterodimers. Oncogene. 1997;15:1805–1813. doi: 10.1038/sj.onc.1201335. [DOI] [PubMed] [Google Scholar]
- 28.Qu H., et al. Smad4 suppresses the tumorigenesis and aggressiveness of neuroblastoma through repressing the expression of heparanase. Sci Rep. 2016;6:32628. doi: 10.1038/srep32628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hald O.H., et al. Inhibitors of ribosome biogenesis repress the growth of MYCN-amplified neuroblastoma. Oncogene. 2019;38:2800–2813. doi: 10.1038/s41388-018-0611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blockus H., Chedotal A. The multifaceted roles of Slits and Robos in cortical circuits: from proliferation to axon guidance and neurological diseases. Curr Opin Neurobiol. 2014;27:82–88. doi: 10.1016/j.conb.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Li Y., et al. Genomic analysis-integrated whole-exome sequencing of neuroblastomas identifies genetic mutations in axon guidance pathway. Oncotarget. 2017;8:56684–56697. doi: 10.18632/oncotarget.18079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biegel J.A., et al. Constitutional 1p36 deletion in a child with neuroblastoma. Am J Hum Genet. 1993;52:176–182. [PMC free article] [PubMed] [Google Scholar]
- 33.White P.S., et al. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–2694. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- 34.White P.S., et al. Detailed molecular analysis of 1p36 in neuroblastoma. Med Pediatr Oncol. 2001;36:37–41. doi: 10.1002/1096-911X(20010101)36:1<37::AID-MPO1010>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 35.Cappabianca L., et al. Discovery, characterization and potential roles of a novel NF-YAx splice variant in human neuroblastoma. J Exp Clin Cancer Res. 2019;38:482. doi: 10.1186/s13046-019-1481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frumm S.M., et al. Selective HDAC1/HDAC2 inhibitors induce neuroblastoma differentiation. Chem Biol. 2013;20:713–725. doi: 10.1016/j.chembiol.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gendron T.F., Rademakers R., Petrucelli L. TARDBP mutation analysis in TDP-43 proteinopathies and deciphering the toxicity of mutant TDP-43. J Alzheimers Dis. 2013;33(Suppl 1):S35–S45. doi: 10.3233/JAD-2012-129036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russo A., et al. Increased cytoplasmic TDP-43 reduces global protein synthesis by interacting with RACK1 on polyribosomes. Hum Mol Genet. 2017;26:1407–1418. doi: 10.1093/hmg/ddx035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sattu K., et al. Phosphoproteomic analysis of anaplastic lymphoma kinase (ALK) downstream signaling pathways identifies signal transducer and activator of transcription 3 as a functional target of activated ALK in neuroblastoma cells. FEBS J. 2013;280:5269–5282. doi: 10.1111/febs.12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki T., et al. Frequent loss of heterozygosity on chromosome 14q in neuroblastoma. Cancer Res. 1989;49:1095–1098. [PubMed] [Google Scholar]
- 41.Thompson P.M., et al. Loss of heterozygosity for chromosome 14q in neuroblastoma. Med Pediatr Oncol. 2001;36:28–31. doi: 10.1002/1096-911X(20010101)36:1<28::AID-MPO1008>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 42.Simantov R., Sachs L. Induction of polyadenylate polymerase and differentiation in neuroblastoma cells. Eur J Biochem. 1975;55:9–14. doi: 10.1111/j.1432-1033.1975.tb02132.x. [DOI] [PubMed] [Google Scholar]
- 43.Furuta S., Ohira M., Machida T., Hamano S., Nakagawara A. Analysis of loss of heterozygosity at 16p12-p13 (familial neuroblastoma locus) in 470 neuroblastomas including both sporadic and mass screening tumors. Med Pediatr Oncol. 2000;35:531–533. doi: 10.1002/1096-911x(20001201)35:6<531::aid-mpo6>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 44.Maris J.M., et al. Evidence for a hereditary neuroblastoma predisposition locus at chromosome 16p12-13. Cancer Res. 2002;62:6651–6658. [PubMed] [Google Scholar]
- 45.Potluri V.R., Helson L., Ellsworth R.M., Reid T., Gilbert F. Chromosomal abnormalities in human retinoblastoma. A review Cancer. 1986;58:663–671. doi: 10.1002/1097-0142(19860801)58:3<663::aid-cncr2820580311>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 46.Scott R.H., Stiller C.A., Walker L., Rahman N. Syndromes and constitutional chromosomal abnormalities associated with Wilms tumour. J Med Genet. 2006;43:705–715. doi: 10.1136/jmg.2006.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raimondi S.C., et al. Chromosomal abnormalities in 478 children with acute myeloid leukemia: clinical characteristics and treatment outcome in a cooperative pediatric oncology group study-POG 8821. Blood. 1999;94:3707–3716. [PubMed] [Google Scholar]
- 48.Diskin, S. J. et al. Rare variants in TP53 and susceptibility to neuroblastoma. J Natl Cancer Inst106, dju047, doi:10.1093/jnci/dju047 (2014). [DOI] [PMC free article] [PubMed]
- 49.Diskin S.J., et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong Z., et al. GAS7 Deficiency Promotes Metastasis in MYCN-Driven Neuroblastoma. Cancer Res. 2021;81:2995–3007. doi: 10.1158/0008-5472.CAN-20-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang K., et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–220. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeineldin M., et al. MYCN amplification and ATRX mutations are incompatible in neuroblastoma. Nat Commun. 2020;11:913. doi: 10.1038/s41467-020-14682-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satge D., et al. Abnormal constitutional karyotypes in patients with neuroblastoma: a report of four new cases and review of 47 others in the literature. Cancer Genet Cytogenet. 2003;147:89–98. doi: 10.1016/s0165-4608(03)00203-6. [DOI] [PubMed] [Google Scholar]
- 54.Parodi S., et al. Loss of whole chromosome X predicts prognosis of neuroblastoma patients with numerical genomic profile. Pediatr Blood Cancer. 2019;66:e27635. doi: 10.1002/pbc.27635. [DOI] [PubMed] [Google Scholar]
- 55.Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science324, 1029-1033, doi:10.1126/science.1160809 (2009). [DOI] [PMC free article] [PubMed]
- 56.Fearon K.C., Glass D.J., Guttridge D.C. Cancer cachexia: mediators, signaling, and metabolic pathways. CellMetab. 2012;16:153–166. doi: 10.1016/j.cmet.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 57.Fong C.T., et al. Loss of heterozygosity for the short arm of chromosome 1 in human neuroblastomas: correlation with N-myc amplification. Proc Natl Acad Sci U S A. 1989;86:3753–3757. doi: 10.1073/pnas.86.10.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caron H. Allelic loss of chromosome 1 and additional chromosome 17 material are both unfavourable prognostic markers in neuroblastoma. Med Pediatr Oncol. 1995;24:215–221. doi: 10.1002/mpo.2950240402. [DOI] [PubMed] [Google Scholar]
- 59.Schleiermacher G., et al. Accumulation of segmental alterations determines progression in neuroblastoma. J Clin Oncol. 2010;28:3122–3130. doi: 10.1200/JCO.2009.26.7955. [DOI] [PubMed] [Google Scholar]
- 60.Rosswog C., et al. Chromothripsis followed by circular recombination drives oncogene amplification in human cancer. Nat Genet. 2021;53:1673–1685. doi: 10.1038/s41588-021-00951-7. [DOI] [PubMed] [Google Scholar]
- 61.Speleman F., Park J.R., Henderson T.O. Neuroblastoma: A Tough Nut to Crack. Am Soc Clin Oncol Educ Book. 2016;35:e548–e557. doi: 10.14694/EDBK_159169. [DOI] [PubMed] [Google Scholar]
- 62.Tivnan A., et al. MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer. 2011;11:33. doi: 10.1186/1471-2407-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le M.T., et al. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol. 2009;29:5290–5305. doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cole K.A., et al. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res. 2008;6:735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Welch C., Chen Y., Stallings R.L. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 66.Bandyopadhyay S., et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim, M. et al. A protein interaction landscape of breast cancer. Science374, eabf3066, doi:10.1126/science.abf3066 (2021). [DOI] [PMC free article] [PubMed]
- 68.Stephens P.J., et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Forment J.V., Kaidi A., Jackson S.P. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.