Abstract
We describe single-component optogenetic probes whose activation dynamics depend on both light and temperature. We used the BcLOV4 photoreceptor to stimulate Ras and PI3K signaling in mammalian cells, allowing activation over a large dynamic range with low basal levels. Surprisingly, we found that BcLOV4 membrane translocation dynamics could be tuned by both light and temperature such that membrane localization spontaneously decayed at elevated temperatures despite constant illumination. Quantitative modeling predicted BcLOV4 activation dynamics across a range of light and temperature inputs and thus provides an experimental roadmap for BcLOV4-based probes. BcLOV4 drove strong and stable signal activation in both zebrafish and fly cells, and thermal inactivation provided a means to multiplex distinct blue-light sensitive tools in individual mammalian cells. BcLOV4 is thus a versatile photosensor with unique light and temperature sensitivity that enables straightforward generation of broadly applicable optogenetic tools.
Introduction
Optogenetic probes permit light-induced control of intracellular biochemistry. Such probes are typically engineered from proteins that evolved to respond to their host’s environmental conditions, i.e. to its light status1,2, but in some cases also to temperature3-5. Light-responsive actuators now exist for control of protein dimerization6-10, allostery11,12, oligomerization13, ion transport14, and membrane recruitment15,16, providing an extensive toolset for precise manipulation of an array of biological processes, including cell signaling.
Ras and phosphatidyl inositol-3-kinase (PI3K) are signaling regulators that together control essential cell processes including transcription, translation, growth, survival, proliferation and migration17-20. Optogenetic control of these two pathways has enabled recent discoveries of how their spatiotemporal dynamics regulate cell and tissue growth, form, and disease21-24. Currently, optogenetic activation of Ras or PI3K is achieved through membrane recruitment of signaling effectors via light-induced protein heterodimerization25,26. However, this approach is limited by the necessity for two distinct proteins, which can require stoichiometric tuning of both components to permit signaling through a large dynamic range with minimal elevated basal signaling27. While stoichiometric tuning is feasible in single cells, it is more challenging in tissues and organisms.
Single-component membrane translocation was recently described using the BcLOV4 photoreceptor, which translocates from the cytoplasm to membrane phospholipids under blue light in mammalian cells15 (Figure 1A). BcLOV4 has already served as a modular technology for light-induced activation of the Rho GTPases Rac128, RhoA29, Cdc4230, suggesting that BcLOV4 may be adapted to regulate many additional pathways.
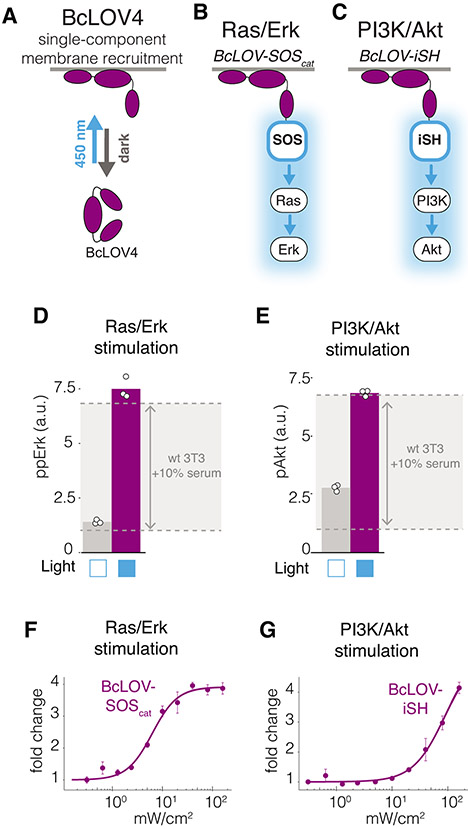
Figure 1. Single-component BcLOV4 fusions allow control of Ras and PI3K signaling.
A) BcLOV4 binds the cell membrane when exposed to blue light. The three BcLOV4 domains represent the LOV, DUF, and RGS domains, as previously described15 B) Light-induced membrane recruitment of BcLOV4 fused to the SOScat catalytic domain will induce Ras/Erk signaling. C) Analogous recruitment of the iSH domain will induce PI3K/Akt signaling. D,E) Five minutes of blue light stimulation (160 mW/cm2 at 20% duty cycle) increases intracellular ppErk levels in cells that express BcLOV-SOScat (D) and increases pAkt in cells that express BcLOV-iSH (E). Grey zone indicates the change in ppErk or pAkt in wild-type cells that were stimulated with 10% calf serum for 10 minutes. Data represent means of three biologically independent replicates, each representing the mean signal intensity from ~2000-4000 single cells. F,G) Light intensity dose-response of (F) ppErk fold-change induction in BcLOV-SOScat-expressing cells or (G) pAkt fold-change induction in BcLOV-iSH cells at 100% duty cycle after 5 minutes of illumination. Data represent means +/− SD of three biologicially independent replicates, each representing the mean signal intensity from ~300-500 single cells. All stimulation in A-G was achieved using the optoPlate-96. All stimulation and environmental conditions for all figures can be found in Supplementary Table 1.
In this work, our initial goal was to generate and characterize BcLOV4-based probes for Ras or PI3K activation. Surprisingly, we discovered that BcLOV4 translocation and signal activation respond not only to blue light but also to temperature, such that under sustained, long-term stimulation, BcLOV4 becomes inactivated and dissociates from the membrane as a function of increased temperature and light intensity. Through systematic characterization, we developed and validated a quantitative model that predicted BcLOV4 and downstream signaling dynamics as a function of light and temperature, providing a roadmap for BcLOV4 usage over a range of experimental conditions, particularly during long time course experiments. We demonstrate the broad applicability and stable activation of our probes in zebrafish embryos and Drosophila Schneider 2 (S2) cells, which operate at low temperatures (22-30 °C). Finally, we demonstrate that temperature inactivation of BcLOV4 can be leveraged to allow multiplexing of blue-sensitive optogenetic probes in individual mammalian cells.
Results
Engineering control of Ras and PI3K signaling with BcLOV4
To generate an actuator of Ras/Erk signaling, we fused BcLOV4 to the catalytic domain of the Ras guanine nucleotide exchange factor Son of Sevenless 2 (SOScat), which activates Ras upon recruitment to the membrane (Figure 1B)26. We generated an analogous probe to control PI3K signaling by replacing the SOScat domain with the inter-SH2 domain of the p85 subunit (iSH) (Figure 1C)25,31. We refer to these probes as BcLOV-SOScat and BcLOV-iSH, respectively.
To test probe activity, we illuminated NIH 3T3 cells that stably expressed either BcLOV-SOScat or BcLOV-iSH, and we quantified levels of phospho-Erk (ppErk) or phosho-Akt (pAkt) using immunofluorescence imaging (Supplementary Figure 1). In the absence of blue light, BcLOV-SOScat cells exhibited low basal levels of ppErk, similar to wild-type (wt) cells. Upon illumination, ppErk levels rose dramatically and reached their peak within 5 minutes (Supplementary Figure 2), reaching levels comparable to wt cells stimulated with 10% serum (Figure 1D). BcLOV-iSH expressing cells also induced strong levels of pAkt, although basal activation in the absence of light was somewhat higher relative to wt cells (Figure 1E). These results suggest that our BcLOV-derived probes stimulate physiologically relevant levels of pathway activation while minimally disrupting endogenous cell physiology in the dark state.
To better characterize light-induced stimulation of BcLOV-SOScat and BcLOV-iSH, we measured the dose-response of signal activation as a function of light intensity. We found that BcLOV-SOScat achieves half-maximal pathway stimulation with 7 mW/cm2 of blue light and saturates near 40 mW/cm2 with a 4-fold signal induction (Figure 1F). Conversely, we estimate that BcLOV-iSH reaches half-maximal signal induction with 80 mW/cm2, though we did not reach saturation at the highest levels of stimulation (4-fold at 160 mW/cm2) (Figure 1G). Taken together, given the large fold-change induction, minimal basal level of stimulation, and single-component nature, BcLOV4-based probes offer several beneficial qualities for optogenetic stimulation of Ras and PI3K signaling.
Optogenetic activation decays during extended stimulation.
We next asked how BcLOV4-based probes regulate signaling through time. We used recently-described illumination devices for microwell plates (the optoPlate-96) to perform time course stimulation experiments27. After stimulation, cells were immunostained and quantified to assess pathway activity (Figure 2A).
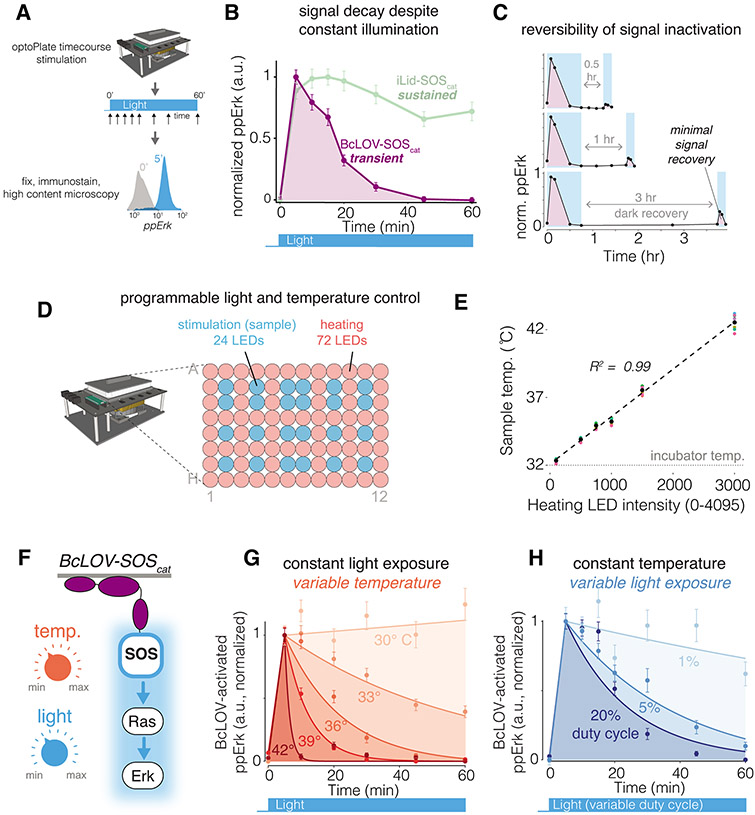
Figure 2. BcLOV-induced signaling dynamics depend on experimental temperature and light exposure.
A) Schematic of experimental protocol. B) Sustained stimulation of BcLOV-SOScat (160 mW/cm2 at 20% duty cycle) reveals that ppErk signal decays rapidly after an initial signal increase, whereas activation of iLID-SOScat remains sustained. C) Recovery (dark) periods of up to 3 hr after signal decay do not permit recovery of activatible BcLOV-SOScat, suggesting that BcLOV4 inactivation is effectively irreversible. D) BcLOV-SOScat signal dynamics were examined under variable light and temperature conditions. E) Schematic for how the optoPlate-96 was repurposed to allow independent control of experimental light and temperature conditions. For more information, see Supplementary Figure 7. F) Steady-state sample temperature was a linear function of the intensity of the 72 heater LEDs (precise intensity-temperature relationship should be determined empirically for each individual optoPlate). See Methods. G) At a given light exposure level (here, 160 mW/cm2 at 20% duty cycle), BcLOV-SOScat signal decays more rapidly at higher temperatures. H) At a given temperature (here, 36°C), BcLOV-SOScat decay increases with increased light exposure (variable duty cycles of 160 mW/cm2 light). Data points in B/C/G/H represent the mean +/− SEM of ~1000-4000 individual cells. Traces in G/H are exponential decay functions fit to data points at each temperature and duty cycle, as described in the Methods section. Data are normalized between the min and max of each trace. Normalization was performed separately for each temperature and duty cycle to highlight the change in the rate of BcLOV-SOScat inactivation rather than absolute signal. Absolute signal traces can be found in Supplementary Figure 8.
We were surprised to find that, despite constant stimulation for 60 minutes, cells that expressed BcLOV-SOScat showed an initial increase followed by rapid and complete decay of ppErk (Figure 2B, Supplementary Figure 3). To determine if transient signaling was caused by BcLOV4 or alternative mechanisms (e.g. negative feedback within the Ras/Erk pathway), we compared activation dynamics to those achieved with an orthogonal optogenetic system, the commonly-used blue-light inducible iLID/sspB (nano) heterodimerizing pair6, which can be adapted to recruit SOScat to the membrane (iLID-SOScat23, (Supplementary Figure 4). In contrast to BcLOV-SOScat, iLID-SOScat produced stable signaling under identical illumination conditions (see Supplementary Table 1 for detailed experimental conditions for all experiments), indicating that Ras/Erk signal decay was a feature of BcLOV4 stimulation. Similarly, sustained illumination of BcLOV-iSH cells led to an initial increase followed by rapid decrease of pathway activity, whereas stimulation with an analogous iLID-based probe (iLID-iSH, Supplementary Figure 4) resulted in sustained activity (Supplementary Figure 5A,B). These results further indicate that transient activation dynamics were a function of BcLOV4 activation and not the pathway under study.
To understand the nature of signal decay, we performed a series of experiments using the BcLOV-SOScat probe. We first asked whether BcLOV-SOScat inactivation could be reversed after removal of the light stimulus. We stimulated cells with blue light until the signal decayed, withdrew light for either 0.5, 1, or 3 hr, and then restimulated for 10 minutes (Figure 2C). As before, the initial 45 minutes of blue light led to a pulse of ppErk signal. However, only ~10% of the original signal could be obtained upon restimulation even after 3 hrs of light withdrawal. This small amount of reactivation was partially a result of new protein production rather than reversal of inactivated BcLOV-SOScat (Supplementary Figure 6). These results suggest that BcLOV4 can undergo a spontaneous transition into an uncharacterized, long-lived inactivated state.
We next asked whether certain experimental parameters could modulate the observed signal decay rate. We noticed that pathway decay kinetics could change as a function of the illumination settings of individual experiments. Because higher light intensity can also cause heating of the sample27, we tested the effects of both temperature and light on BcLOV-SOScat stimulation kinetics. To perform optogenetic time course experiments at specific temperatures, we adapted the optoPlate-96 to precisely control both illumination as well as sample temperature (Figure 2D,E, Supplementary Figure 7). Briefly, we decoupled sample heating from the illumination profile by designating 24 LED positions for light stimulation and using the remaining 72 LED positions as heating elements (Figure 2D). Illumination of the 72 “heater” LEDs over 3000 intensity levels correlated linearly with sample temperature in the wells above the 24 “stimulation” LEDs (R2 = 0.99) (Figure 2E).
Strikingly, we observed that BcLOV-SOScat decay kinetics were strongly correlated to sample temperature, where signal decay was faster at higher temperatures (Figure 2F,G, Supplementary Figure 8A). At the extremes, ppErk decayed with a half-life of ~6 minutes at 42 °C, while decay was minimal at 30 °C. We observed strong ppErk signal induction at all temperatures tested, suggesting that temperature only impacted BcLOV4 function in its lit, activated form. (Supplementary Figure 8A). The BcLOV-iSH probe was also temperature-sensitive (Supplementary Figure 8B). By contrast, iLID-SOS- and iLID-iSH-induced signaling was sustained at both high and low temperatures (Supplementary Figure 8C,D), further indicating that temperature sensitivity is a property of BcLOV4 control. Finally, we used a live cell reporter of Erk activity (ErkKTR32) as an orthogonal readout to verify that, in contrast to temperature-regulated inactivation, optogenetic inactivation (removal of blue light) was reversible over multiple illumination cycles at both high (37 °C) and low (30 °C) temperatures (Supplementary Figure 9).
We also observed that BcLOV-SOScat signal decay rate was dependent on light intensity, where higher intensity led to rapid decay, while lower intensity led to more sustained stimulation (Figure 2H, Supplementary Figure 8E). Low light achieved sustained signaling at the expense of signal amplitude (Supplementary Figure 8E), although we note that this tradeoff will be specific to the pathway under study. For example, in a previous report, comparable sparse illumination conditions (1.6% duty cycle) yielded saturating activation levels of BcLOV-regulated Rho GTPase signaling28,29.
Because signal decay was observed with both the BcLOV-SOScat and BcLOV-iSH probes but not with analogous iLID-based probes (Figure 2B, Supplementary Figure 8C), we suspected that the observed light- and temperature-dependent decay kinetics were a property of the BcLOV4 photosensor itself. We thus quantified membrane translocation of BcLOV-mCherry under various light and temperature conditions (Figure 3A, Supplementary Figure 10). In accordance with our signaling results, sustained illumination resulted in sustained membrane localization at low temperatures (25 °C) but only transient localization at 37 °C, with inactivated BcLOV4 returning to the cytoplasm after ~30 min (Figures 3 B,C, Supplementary Figure 10, Supplementary Movie 1). Similarly, when we varied light intensity at a constant temperature, increased intensity (duty cycle) increased the decay rate of membrane translocation (Figure 3D). Decay of membrane fluorescence was not due to BcLOV4 degradation (Supplementary Figure 11). As before, we observed initial membrane recruitment at all temperatures (Supplementary Figure 10), and BcLOV4 inactivation was irreversible for at least 3 hrs (Supplementary Figure 12). Together, these data demonstrate that BcLOV4 acts as not only a photosensor but also a temperature sensor.
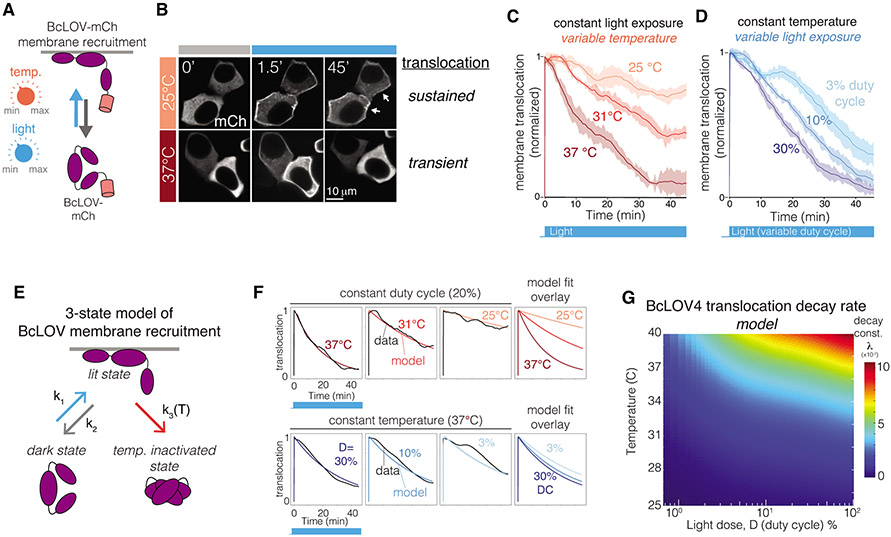
Figure 3. BcLOV4 membrane translocation dynamics depend on temperature and light exposure.
A) BcLOV-mCh membrane recruitment was quantified at various temperatures and light exposures using live cell imaging. B) Representative images of membrane recruitment at low and high temperatures. Activation at 25 °C permitted sustained membrane recruitment while recruitment at 37 °C was transient (stimulation performed at 1.45 W/cm2 and 3% duty cycle). Image brightness was adjusted at each time point for clarity to account for photobleaching. C) Quantification of membrane recruitment at various temperatures (1.45 W/cm2 at 3% duty cycle) reveals a temperature-dependent decay of membrane translocation. D) Quantification of membrane recruitment at various light exposures (at 36 °C and 1.45 W/cm2) shows light-dependent decay of BcLOV-mCh translocation. Each trace is the mean membrane fluorescence +/− SEM of three biologically independent samples, with each replicate representing the mean of ~100 cells. See Supplemental Figure 10 for unnormalized traces and quantification workflow. E) Schematic of a 3-state model of BcLOV4 membrane translocation. F) Fitting the model to live-cell translocation data provides parameter values for k1, k2, and k3(T). G) Heat map depicts the decay rate of BcLOV4 membrane localization as a function of temperature and light exposure. Decay rates were calculated by simulating sustained illumination over a range of duty cycles and temperatures and fitting the modeled decay rate to a single exponential decay. Color indicates the decay constant λ (1 divided by the time to reach 37% of maximum signal). Larger λ indicates faster decay. See Supplementary Figure 10, 13, and Methods for imaging and model details.
Modeling dependence of BcLOV on temperature and light
To explain and predict BcLOV-mCh translocation dynamics, we developed a computational model. We reasoned that, in addition to the dark and lit states, a third state of BcLOV4 could account for our observations of light- and temperature-dependent decay kinetics (Figure 3E). This third state, which we call the temperature-inactivated (TI) state, is only accessible from the lit (membrane-bound) state, and transition to the TI state is irreversible on the timescales we consider (Figure 2C, Supplementary Figures 6,12). We predicted that the rate of entry into the TI state (k3) would increase as a function of temperature. Thus, because more light increases the amount of BcLOV4 in the lit state, and higher temperature increases the transition rate from the lit to the TI state, our model could explain why both increased light and temperature can increase the decay rate of BcLOV4 membrane translocation. For details on model development, see Figure 3E-G, Supplementary Figure 10, and Methods.
We parameterized our model by fitting values for k1, k2, and k3 to live-cell data of BcLOV-mCh translocation dynamics over a range of temperatures (25-40 °C) and light exposures (1.1%, 3.3%, 10% duty cycle) (Figure 3E,F and Supplementary Figures 10,13). Across experimental conditions, we obtained consistent values for k1 and k2 (see Methods), which correspond closely to reported values of BcLOV4 membrane translocation and dissociation (half-times of ~1 second for association and ~1 minute for dissociation)15. As expected, k3 showed a strong exponential dependence on temperature (Supplementary Figure 13). We then used our parameterized model to generate a 2D heatmap of predicted BcLOV4 decay rate as a function of temperature and light dose during sustained stimulation (Figure 3G). This heatmap represents a systematic roadmap of translocation dynamics that can be used to predict the behavior of BcLOV4-based probes.
To validate our model and predict signaling dynamics downstream of BcLOV-SOScat stimulation, we integrated our model of membrane translocation with a model of Ras/Erk signal transmission (Figure 4A). We modeled the Ras/Erk pathway with a transfer function that represents signal transmission from membrane-localized SOScat to Erk phosphorylation as a dynamic filter. Previous work defined the Ras/Erk module as a second-order low pass filter (LPF) with a 2 mHz cutoff frequency26. However, this previous work applied SOScat membrane localization as an input and measured nuclear localization of fluorescently-tagged Erk2 as the output, whereas in our case the output was cytoplasmic levels of endogenous ppErk. Thus, to choose the most appropriate model, we performed dynamic stimulation of BcLOV-SOScat and fitted either first- or second-order low-pass filter models to the data. We found that signal transmission was best modeled by a first-order LPF with a cutoff frequency of 2 mHz (Figure 4B,C). Because first order filters transmit fast signal dynamics more efficiently than analogous second-order filters (Figure 4B), our results suggest that the Ras/Erk pathway can transmit fast signal fluctuations (< ~4 min) more effectively than previously measured26. Our results may differ from previous measurements because of additional biochemical steps required to transduce phosphorylation into nuclear translocation of fluorescently-tagged Erk, compared to the direct observation of Erk phosphorylation in our work. Notably, our model captured both the fast timescale dynamics of ppErk fluctuations, as well as the slow timescale decay of ppErk due to progressive BcLOV4 inactivation (Figure 4C).
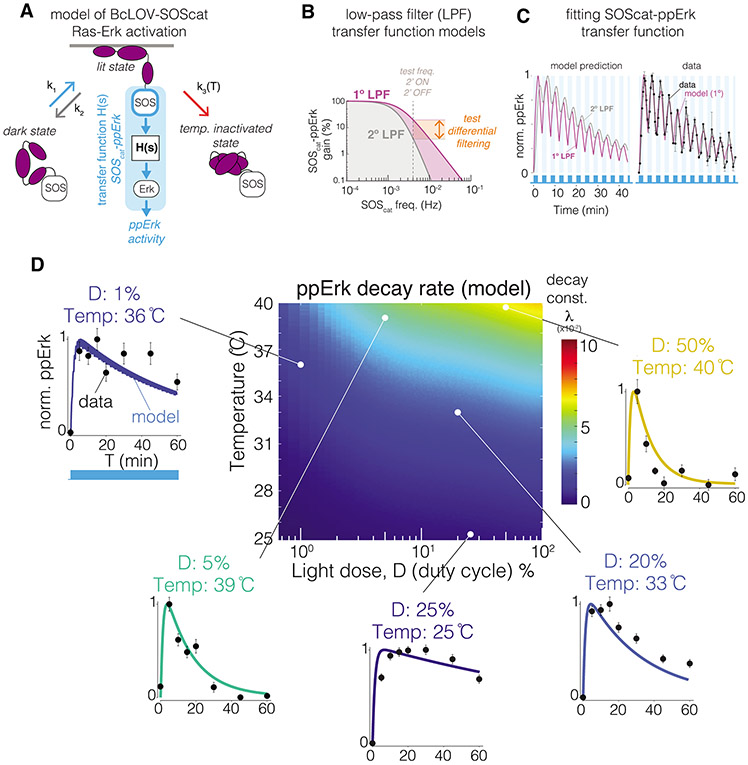
Figure 4. Modeling predicts BcLOV-SOScat-induced ppErk dynamics and reveals dynamic filtering properties of Ras/Erk signaling.
A) A model of ppErk activation was developed by integrating the BcLOV4 membrane translocation model with a transfer function model that describes the input/output response of SOScat membrane localization (input) to ppErk activation (output). B) Filtering properties of a 1° vs 2° low pass filter (LPF). 1° LPFs attenuate high frequency inputs less than 2° low pass filters. C) A 1° LPF with 2 mHz cutoff frequency best describes ppErk dynamics when stimulated with fast 2’ON/2’OFF BcLOV-SOScat oscillations. Data points are the mean +/− SD of three biologically independent replicates, with each replicate representing the mean of ~1000-2000 cells. D) Heat map depicts the predicted Erk activation dynamics resulting from BcLOV4 membrane translocation dynamics over the indicated light and temperature conditions. Plots show model predictions of Erk activation at the indicated experimental conditions, and data points show experimental results. Datapoints represent means +/− SEM of ~1000-4000 cells. Unnormalized plots are presented in Supplementary Figure 14. See Methods for model details.
Our integrated model of membrane translocation and Erk activation predicted how specific light and temperature inputs shape BcLOV-SOScat-induced ppErk dynamics. We used this model to generate a heatmap of ppErk decay rate as a function of temperature and light dose during constant illumination (Figure 4D). To validate our model, we performed stimulation time course experiments at temperature and light conditions that were sampled from regions of our heatmap with diverse decay rates. Measured ppErk decay rates matched closely to the rates predicted by our model over all experimental conditions tested (Figure 4D, Supplementary Figure 14). Phototoxicity was not observed under the illumination conditions used in this study (Supplementary Figure 15). Together, our data and models comprehensively describe how BcLOV4 and optogenetic probes thereof will behave as a function of light and temperature condition. We note, however, that decay rates will likely vary between pathways due to pathway-specific biochemisty and must thus be determined empirically.
BcLOV4-based signal activation in model organisms
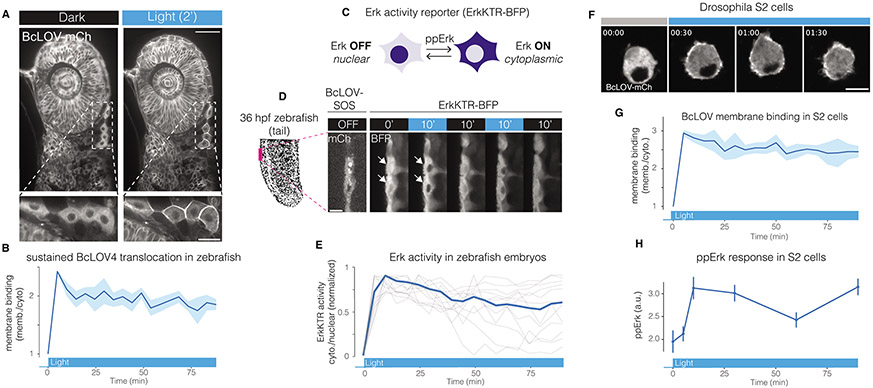
The single-component nature and low rate of spontaneous decay at < 30 °C position BcLOV4-based tools as highly suited for experiments in tissues and model organisms that operate at lower temperatures. We thus tested performance of BcLOV4 and BcLOV-SOScat in both zebrafish embryos and Drosophila S2 cells. BcLOV-mCh expressed well in zebrafish embryos and, upon illumination, rapidly translocated to the membrane in all cells (Figure 5A, Supplementary Movie 2). Membrane translocation was sustained through 90 min of illumination, as expected from our experiments in mammalian cells (Figure 5B). To determine whether BcLOV-SOScat could stimulate Ras signaling in zebrafish, we co-expressed BcLOV-SOScat with the ErkKTR reporter, which has previously been used in zebrafish (Figure 5C)33. In cells that co-expressed BcLOV-SOScat and ErkKTR-BFP, we observed rapid and reversible ErkKTR translocation that could be stimulated over multiple cycles (Figure 5D, Supplementary Movie 3), consistent with our data from mammalian cells. Notably, over 90 min of sustained stimulation, Erk activity remained high, demonstrating that both BcLOV-SOScat translocation and signal activation could be maintained (Figure 5E). We also expressed BcLOV4 probes in Drosophila S2 cells as an orthogonal model system that grows at temperatures permissive to stable BcLOV4 translocation, and we again observed sustained BcLOV-mCh membrane localization through 90 min of blue light illumination (Figure 5F,G). Furthermore, expression and sustained stimulation of BcLOV-SOScat allowed sustained activation of ppErk (Figure 5H). Together, these data show that BcLOV4-based probes can serve as simple and sensitive optogenetic probes across diverse cells, tissues, and organisms of study.
Figure 5. BcLOV4 and BcLOV-SOScat in zebrafish embryos and Drosophila cells.
A) Blue-light induced membrane translocation of BcLOV-mCherry in a zebrafish embryo (24 hours post fertilization [hpf]). Scale bar = 50 μm, inset scale bar = 20 μm. B) BcLOV-mCh translocation is sustained over 90 min in zebrafish embryos. Data represent mean +/− SD of 10 cells. C) Schematic of ErkKTR activity. ErkKTR is nuclear when Erk signaling is off and translocates to the cytoplasm when Erk is activated. D) The Ras/Erk pathway can be reversibly stimulated over multiple cycles in zebrafish embryos (24 hpf) that co-express express BcLOV-SOScat and ErkKTR-BFP, as measured by ErkKTR-BFP translocation. White arrows highlight nuclei where ErkKTR translocation is evident. Scale bar = 10 μm. E) Sustained illumination of BcLOV-SOScat permits sustained elevated Erk activity in 24 hpf zebrafish embryos. Plot shows ErkKTR cytoplasmic/nuclear ratios of 12 single cells (light grey; blue trace represents mean) measured over two experiments. Trajectories are normalized between 0 and 1 to permit comparison between experiments. For (A-E) stimulation was performed using 1.45 W/cm2 488 nm light at 1.5% duty cycle. F) BcLOV-mCh membrane translocation in Drosophila S2 cells stimulated with blue light (1.45 W/cm2 at 3% duty cycle) for 90 min. Scale bar = 10 μm. G) Quantification of (F) shows sustained membrane translocation in S2 cells. Data represent mean +/− SD of 10 cells. H) Sustained stimulation of BcLOV-SOScat in S2 cells shows sustained elevated ppErk levels over 90 min, measured by immunofluorescence. Data is the mean +/− SD of three biologically independent samples, with each replicate representing the mean of ~100-200 cells.. Stimulation was performed at (160 mW/cm2 at 20% duty cycle). All experiments shown in Figure 5 were performed at room temperature.
Optogenetic multiplexing using BcLOV4
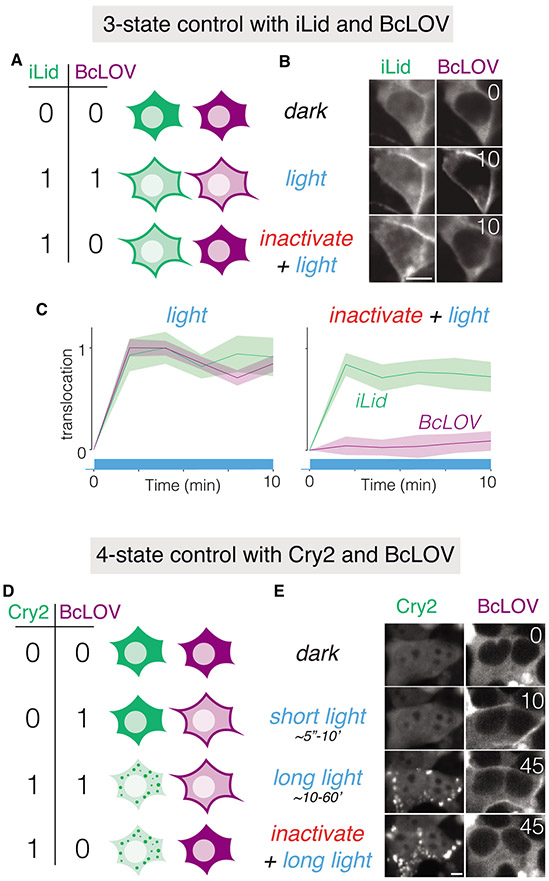
Finally, we reasoned that the unique light- and temperature-responsiveness of BcLOV4 could be leveraged as a novel control mode to regulate multiple optogenetic proteins in single cells. Currently, such multiplexing can be achieved using optogenetic probes with different activation spectra (e.g. blue and red-absorbing). However, there is a relative lack of optogenetic proteins that respond to red-shifted (non-blue) light, and the absorption spectra of these few probes can also reach into the 400-500 nm (blue) range, thus challenging orthogonal multiplexing with blue-sensitive probes27,34. An arguably simpler approach would be to multiplex control of distinct blue-sensitive probes. To demonstrate how temperature regulation of BcLOV4 enables such multiplexing, we co-expressed BcLOV4 with one of two blue light-sensitive tools: an iLID/sspB membrane binding system (Supplementary Figure 4) or Cryptochrome 2(Cry2), which forms large aggregates when activated by blue light13. Coexpression of BcLOV4-mCherry and iLID/sspB-GFP allowed control of 3 activation states: none (dark), both (blue light), or iLID-ONLY, achieved by sequential heat inactivation of BcLOV4 and subsequent light stimulation of iLID (Figure 6 A-C).
Figure 6. BcLOV4 temperature sensitivity enables orthogonal multiplexing of multiple blue-light sensitive tools in single cells.
A) Schematic of how coexpression of BcLOV4 and iLID/sspB can allow blue light control of 3 separate cell states. B) Coexpression in of BcLOV-mCh and iLid/sspB-GFP in HEK 293T cells demonstrates 3-state control using blue light with or without temperature inactivation. Light stimulation was performed at 37 °C using 1 s of blue light (1.45 W/cm2) every 30 seconds for 10 minutes in the presence or absence of prior BcLOV inactivation. Prior inactivation was achieved by illuminating with these same light settings for 1 hour. C) Quantification of light-induced membrane binding (activation) of BcLOV4 and iLID in the absence (left) or presence (right) of prior heat inactivation. Traces are the normalized mean +/− SEM of three biologically independent replicates, with each replicate representing the mean of ~100 cells. D) Schematic of how co-expression of BcLOV4 and Cry2 can allow blue-light control of 4 separate cell states. E) Co-expression of BcLOV-GFP and Cry2-mCh in HEK 293T cells demonstrates 4-state control. “Short light” (10 min) and “long light” (45 min) exposure were both achieved using 100 ms of light (1.45 W/cm2) every 30 s at 30 °C. To achieve the Cry2-ONLY state (bottom row), images were acquired after BcLOV was inactivated using 1 s of blue light (1.45 W/cm2) every 30 seconds for 45 min at 37 °C. Scale bars = 10 μM. See Methods and Supplementary Table 1 for full illumination conditions. Time is given in minutes. The multiplexing experiments depicted are representative of two independent experiments for each pair of optogenetic proteins.
Co-expression of BcLOV4-GFP with Cry2-mCherry allowed control of all 4 possible activation states (Figure 6D,E). In this arrangement, the BcLOV-ONLY state can be achieved because of differential ON-kinetics of fast BcLOV translocation vs. the slower formation of large Cry2 clusters. We note that although the Cry2-mCherry displayed in Figure 6E was imaged at 45 minutes, the time at which clusters appears is highly variable (~minutes to tens of minutes) and depends on Cry2 concentration. Nevertheless, because Cry2 clustering kinetics are slower than BcLOV translocation (~seconds), they permit a BcLOV-ONLY state at relatively short timescales.
Discussion
We describe the application of BcLOV4 membrane translocation to generate single-component probes for optical control over Ras/Erk or PI3K signaling. We characterized these probes in mammalian cells and we found that the BcLOV4-based probes can provide signaling through a large, physiologically relevant dynamic range with low basal signaling and high photosensitivity. In addition, BcLOV4-based probes are single-protein systems, eliminating the need for stoichiometric tuning of analogous multi-component tools. Such tuning can be difficult in model organisms like Drosophila and zebrafish, in which we show that BcLOV4-based probes function well. More generally, our work adds to the growing library of BcLOV4-based optogenetic signaling tools28-30, highlighting BcLOV4 as a modular optogenetic actuator of effector/membrane interaction to regulate signaling across biological models, including yeast, flies, zebrafish, and mammalian cells15,28.
We discovered that BcLOV4 is a temperature sensor in addition to its known role as a photosensor. While temperature-dependence has been observed in certain photosensors and optogenetic probes, this dependence mostly manifests as decreased protein stability or photoreactivity at elevated temperatures3,10,35-39. By contrast, BcLOV4 folds and translocates rapidly when exposed to light at all temperatures, but then, under sustained illumination, enters a long-lived inactive state and reverts to the cytoplasm at a rate that increases with both temperature and light dose (Figure 2,3, Supplementary Figure 10). This behavior is consistent with a temperature-dependent photoinactivation where, once BcLOV4 is at the membrane in its active state, elevated temperatures accelerate its transition into a state that is incompatible with membrane binding. Although the structural details of such inactivation remain unknown, light-induced oxidation40 and thermal denaturation during the photocycle41 have both been observed in blue light photosensors and could conceivably play a role in BcLOV4 temperature inactivation.
We developed quantitative models of BcLOV4 membrane translocation and signal activation to predict activity as a function of light and temperature. We found that sustained, whole-cell illumination will result in sustained translocation only under low light exposure or low temperature. These conditions may explain why BcLOV4 inactivation was not previously noticed, as its use to date has been performed at either ≤ 30 °C or over short time periods, necessitated only sparse illumination (~1% duty cycle), or used subcellular regions of stimulation, which preserves unstimulated, activatible BcLOV4 outside the region of illumination15,28,29.
In addition to shaping long-term activation dynamics, BcLOV4 temperature sensitivity can be leveraged to allow multiplexing of blue-light sensitive tools in single cells, allowing control of 3 or 4 distinct cell states using a single blue light channel. Our approach is complementary to a recent report wherein distinct transcriptional targets were activated using blue light with different temporal patterns42. Our method provides similar capability but at the post-translational level, for example for the study of how multiple signals (e.g. Ras and PI3K) are integrated in single cells. We note that when attempting 4-state control (with Cry2), the duration of the BcLOV4-ONLY state can be altered by tuning the ability of Cry2 to form large clusters, either by changing Cry2 concentration or through the use of Cry2 variants that change its propensity for cluster formation43.
Combined with previous work, our studies provide a roadmap for how to use BcLOV4-based optogenetic tools. BcLOV4 membrane recruitment can be faithfully and precisely controlled over short durations (< ~30 min) across temperatures but requires low temperatures or sparse illumination for sustained (> 30 min) stimulation. Specific BcLOV4 translocation dynamics over a range of light and temperature conditions can be predicted using our 3-state model (Figure 3E-G). Although our work predicts a temperature-inactivated state of BcLOV4, future studies will be required to understand the molecular basis for this temperature sensitivity. Such studies will inform protein engineering efforts to modulate BcLOV4 temperature responsiveness for enhanced optical and thermal control across biological systems.
Methods
Cell Culture
Lenti-X HEK 293T cells were maintained in 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S) in DMEM. NIH 3T3 cells were maintained in 10% calf serum and 1% P/S in DMEM. All cells were cultured in standard cell culture incubators at 37 °C and 5% CO2. Drosophila Schneider 2 (S2) cells were maintained in Schneider’s Drosophila Medium with 10% heat-inactivated Fetal Bovine Serum at room temperature. All cell lines were purchased commercially (Lenti-X HEK 293T: Takarabio 632180; NIH 3T3: ATCC CRL-1658; S2: ThermoFisher R69007). Cell lines were not verified after purchase. Cells were not cultured in proximity to commonly misidentified cell lines.
Plasmid design and assembly
Constructs for stable transduction in mammalian cells were cloned into the pHR lentiviral backbone with an SFFV promoter driving the gene of interest. The pHR backbone was linearized using MluI and NotI restriction sites. BcLOV4, iLID, BFP, SOScat, and iSH coding DNA fragments were generated via PCR and inserted into the pHR backbone via HiFi cloning mix (New England Biolabs). For expression in Drosophila S2 cells, BcLOV-mCherry, BcLOV-iSH, and BcLOV-SOScat were amplified and inserted into the pbphi-nanos promoter-αTubulin 3’UTR vector44 between the NheI and BamHI restriction sites. The resulting vectors were digested with NotI and XhoI to replace the nanos protomer with the metallothionein promoter (pMt)45, which was synthesized by gBlocks gene fragments (Integrated DNA Technologies). The pMt promoter permits inducible expression in the presence of heavy metals, e.g. copper. For zebrafish mRNA expression experiments, BcLOV-mCherry, BcLOV-SOScat and ERK-KTR-BFP (adapted from Regot et al32) were amplified with primers containing att sites for Gateway cloning. PCR amplicons were transferred into pDONR221 plasmids and sequence verified. Gateway cloning was used to transfer each insert into pCSDest plasmids 46.
Plasmid transfection.
HEK 293Ts were transfected using the following calcium phosphate method: Per 1 mL of media of the cell culture to be transfected, 50 μL of 2x HeBS28,29 buffer, 1 μg of each DNA construct, and H2O up to 94 μL was mixed. 6 μL of 2.5mM CaCl2 was added after mixing of initial components, incubated for 1:45 minutes at room temperature, and added directly to cell culture. S2 cells were transfected with Lipofectamine 3000 reagent (ThermoFisher) following the manufacturer’s protocol. Transfection mixture contained 10ng/μL of DNA, 1.5% Lipofectamine 3000 reagent, and 2% P3000 reagent, and was brought up to volume with Opti-MEM (ThermoFisher). Transfection mix was incubated for 15 min at room temperature and was then added directly to the S2 cells. 100 μL of transfection mix per 1 mL of cell culture media was used. The transfected cells were imaged 72 hr after the transfection (24 hr after promoter induction).
Lentiviral packaging and cell line generation
Lentivirus was packaged by cotransfecting the pHR transfer vector, pCMV-dR8.91, and pMD2.G (Addgene #12259) into Lenti-X HEK293T. Briefly, cells were seeded one day prior to transfection at a concentration of 350,000 cells/mL in a 6-well plate. Plasmids were transfected using the calcium phosphate method. Media was removed one day post-transfection and replaced with fresh media. Two days post-transfection, media containing virus was collected and centrifuged at 800 x g for 3 minutes. The supernatant was passed through a 0.45 μm filter. 500 μL of filtered virus solution was added to 100,000 NIH3T3 cells seeded in a 6-well plate. Cells were expanded over multiple passages, and successfully transduced cells were enriched through fluorescence activated cell sorting (BD FACS Aria II) (see gating strategy in Supplementary Figure 16).
Zebrafish maintenance and mRNA injection
For mRNA generation, pCSDest BcLOV-SOScat, pCSDest BcLOV-mCherry and pCSDest ERK-KTR-BFP were digested with NotI. mRNA was generated using the SP6 mMessage Machine kit (Invitrogen) according to the manufacturer’s specifications. 400 pg of BcLOV-SOScat or BcLOV-mCherry were injected. For the KTR construct, we injected 100 pg. For double injections, mRNAs were mixed prior to injection. Embryos were derived by natural spawning in the morning of injection and injected with the desired construct(s). Imaging was performed at 24 hours post fertilization after embedding the embryos in 1% low melting point agarose in a glass bottom dish. Animal protocols (#806819) were approved by the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC). Wildtype fish of the AB strain were used for experiments at the indicated time points of development. The sex of the animals cannot be determined at the embryonic stage.
Preparation of cells for plate-based experiments
96- or 384-well plates were seeded with cells, as previously described27. Briefly, wells were coated with 50uL of MilliporeSigma™ Chemicon™ Human Plasma Fibronectin Purified Protein fibronectin solution diluted 100x in PBS and were incubated at 37 °C for 30 min. NIH 3T3 cells were seeded in 96/384-well format at a density of 3500/1000 cells/well in 100/50 μL respectively and were spun down at 100 x g for 1 minute. After 24 hr, cells were starved by performing 7 80% washes with starvation media (DMEM + 1% P/S). Experiments were performed after 3 hr of starvation.
Optogenetic stimulation
The optoPlate-96 was used for optogenetic stimulation of individual wells in microwell plates 27. A single-color optoPlate was configured with two blue LEDs for maximum dynamic range of blue light intensity. Arduino IDE (version 1.8) was used to program the Arduino Micro found on the optoPlate-96. A low-profile (9 mm tall) well-plate adapter was used for experiments where we simultaneously stimulated and modulated sample temperature. A tall adapter was used for experiments in 384-well plates, as recommended27. Stimulation time courses were performed by assigning timepoints to individual wells. Wells corresponding to different time points were started sequentially, such that all wells could be fixed simultaneously at the end of each experiment. For live cell imaging experiments, the 488 nm laser was used to stimulate BcLOV4 membrane translocation.
Temperature-controlled optoPlate experiments.
Control of sample temperature leveraged the fact that the optoPlate generates heat when operated under conditions that draw large amounts of current27. To independently control a sample’s illumination conditions and its temperature, we designated 24 LEDs as “stimulation” LEDs and repurposed the remaining 72 LEDs as “heater” LEDs (for details see Figure 2 and Supplementary Figure 7). Constant illumination of the 72 heater LEDs at varying intensities permitted linear and uniform control of temperature in the 24 sample wells. Experiments were performed with the heatsink fan operating at maximum speed. To perform temperature-controlled experiments, the cell culture incubator temperature was reduced to 25 °C or 30 °C and optoPlate heating was used to increase temperature according to the relationship described in Figure 2F. Sample plates were first equilibrated to the lower incubator temperature for 2 hrs, and then were equilibrated on the optoPlate to achieve the desired increased temperature for 1.5 hrs before the illumination program began.
Immunofluorescence staining
Immediately following the completion of a stimulation protocol, 16% paraformaldehyde (PFA) was added to each well to a final concentration of 4%, and cells were incubated in PFA in the dark for 10 min. Cells were then permeabilized with 100/50 μL (for 96/384-well plates) with phosphate buffered saline (PBS) + 0.1% Triton-X for 10 min. Cells were then further permeabilized with ice cold methanol for 10 min. After permeabilization, cells were blocked with 1% BSA at room temperature for 30 min. Primary antibody was diluted in PBS + 1% BSA according to the manufacturer's recommendation for immunofluorescence (phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204), Cell Signaling #4370, 1:400 dilution; phospho-Akt (Ser473), Cell Signaling Technologies #9271, 1:800 dilution). 96/384-well wells were incubated with 50/25 μL of antibody dilution for 2 hr at room temperature (RT). Samples were incubated at room temperature in primary antibody (for two hours, after which primary antibody was removed and samples underwent five washes in PBS + 0.1% TWEEN-20 (PBS-T). Cells were then incubated with secondary antibody (Jackson Immunoresearch Alexa Fluor® 488 AffiniPure Goat Anti-Rabbit IgG (H+L)) and DAPI (ThermoFisher, #D1306, 300 nM) in PBS-T + 0.1% BSA for 1 hour at RT. Secondary antibody was removed, samples underwent 5 washes with PBS-T. Samples were imaged in PBS-T.
Imaging
Live cell imaging.
Live-cell imaging was performed using a Nikon Ti2-E microscope equipped with a Yokagawa CSU-W1 spinning disk, 405/488/561/640 nm laser lines, an sCMOS camera (Photometrics), a motorized stage, and an environmental chamber (Okolabs). HEK 293Ts expressing BcLOV-mCh were imaged with a 20X objective at variable temperatures and 5% CO2. Cells were incubated at the desired temperature for 2 hours before imaging to ensure cells equilibrated at the desired temperature. Temperatures were verified by using the temperature sensor shown in Supplementary Figure 7A. The temperature probes were submerged in PBS in wells of a 96-well plate, and the probe/plate apparatus was placed on the microscope stage inside the environmental chamber to record temperatures during imaging. BcLOV4 was stimulated using a 488nm laser. Zebrafish and Drosophila S2 cells were imaged at RT using a 40X oil immersion objective.
High content imaging.
Fixed samples were imaged using a Nikon Ti2E epifluorescence microscope equipped with DAPI/FITC/Texas Red/Cy5 filter cubes, a SOLA SEII 365 LED light source, and motorized stage. High content imaging was performed using the Nikon Elements AR software. Image focus was ensured using image-based focusing in the DAPI channel.
Image processing and analysis
Immunofluorescence quantification.
Images were processed using Cell Profiler47. Cells were segmented using the DAPI channel, and cytoplasm was identified using a 5 pixel ring around the nucleus. Nuclear and cytoplasmic fluorescence values were then exported and analyzed using R (https://cran.r-project.org/) and R-Studio (https://rstudio.com/). Data was processed and visualized using the dplyr48 and ggplot249 packages. For Figures 2G and H, exponential decay functions of the form a * e−b*x were fit to data points in each condition in order to visualize the rate of decay in signaling. Curves were fit using the MATLAB R2020a cftool program.
Membrane recruitment.
Membrane localization was quantified using the iLastik machine learning software50. Briefly, iLastik was used to identify pixels that correspond to the plasma membrane based on user annotations of images of cells that expressed iRFP-CAAX protein, which localizes to the plasma membrane. The resulting image masks were imported into Cell Profiler and were used to quantify the amount of BcLOV4 membrane localization within the same frame. Total BcLOV4, membrane-localized BcLOV4, and total iRFP-CAAX intensity was recorded and further processed in R. Bleaching was corrected by dividing the total intensity of masked mCh images by the total intensity of mCh in the original unmasked image. This method assumed that loss of fluorescence was not due to degradation, which we empirically confirmed (Supplementary Figure 11). Zebrafish and Drosophila BcLOV4 membrane recruitment in Figure 5 was performed by manually comparing the pixel intensity of membrane and cytoplasmic intensities in ≥10 cells.
Modeling
The 3-parameter model found in Figure 3 was based on the following observations: 1) The rate of transition from the temperature inactivated (TI) to the dark state can be approximated as 0 because BcLOV4 inactivation is effectively irreversible (Figure 2C). 2) The rate of BcLOV4 transition from the dark state directly to the TI state can be approximated as 0 because BcLOV4 could be strongly activated even when pre-incubated for 2 hours before stimulation across a wide range of experimental temperatures, with no obvious correlation between temperature and signal strength (Supplementary Figure 8). 3) The rate of BcLOV4 transition from the TI state to the lit state is 0 because temperature-inactivated membrane recruitment and signal activation decay to zero, whereas a non-zero reversion to the lit state would result in a non-zero equilibrium between lit and inactivated state.
Under these assumptions, the following equations were used to model BcLOV4 activity. These equations allow only for bi-directional BcLOV4 movement between the dark and lit state and irreversible movement from the lit state to the temperature inactivated state.
| (eq. 1) |
| (eq. 2) |
| (eq. 3) |
D = Dark State BcLOV4
L = Lit State BcLOV4
TI = Temperature Inactivated State BcLOV4
k1 = Dark to Lit state transition rate
k2 = Lit to Dark state transition rate
k3 = Lit to Temperature Inactivated state transition rate
This system of equations was implemented in MATLAB and was solved numerically using the Euler method. The rate constants k1, k2, and k3 were found by fitting the model to live cell imaging data of BcLOV4 membrane translocation through minimization of mean squared error (MSE)(Figure 3F). Fitting was performed using a custom script that iteratively calculated vertical R-squared across a coarse-grained range of parameter values, and then testing a more finely-resolved set of values centered on the best fitting value chosen from the previous iteration. Under the assumption that:
| (eq. 4) |
k1 and k2 were found by setting k3 = 0 and fitting observed BcLOV4 translocation kinetics over short (1 minute) time periods, yielding the following rate constants:
k3 was then determined by keeping k1 and k2 constant and fitting k3 to observed decay rates at each temperature.
The transfer function model of SOScat-to-ppErk transmission was implemented using the image processing toolbox in MATLAB. Based on previous work26, we hypothesized that the transfer function could be modeled as a 1° or 2° low-pass filter (LPF):
| (eq. 5) |
| (eq. 6) |
where H(s) is the ratio between system input and output, s is the complex variable (frequency), and ω0 is the cutoff frequency. To discriminate between these potential models, we measured ppErk in cells when stimulated by a dynamic 2 min ON/2 min OFF pulse train of light, which corresponds to a frequency that should be ~90% suppressed by the second order LPF, but substantially less-suppressed (~40%) by a first order LPF (Figure 4B). We integrated the 1° or 2° LPF models into our model of BcLOV4 translocation, and we fit these integrated models to the data as described above. We found that dynamic Erk stimulation through BcLOV4 translocation was best described by a 1° LPF with a 2 mHz cutoff frequency.
For all experiments related to modeling, illumination duty cycle was used to modulate the intensity of BcLOV4 stimulation. Duty cycle parameters were limited to patterns where the OFF period was < ~1 min to ensure that BcLOV4 membrane recruitment was maintained at intermediate levels, as determined by its measured inactivation kinetics15.
Multiplexing Experiments
HEK 293T cells were seeded in a 96-well plate and were co-transfected with 100 ng each of the following plasmids. BcLOV/iLID multiplexing: BcLOV-mCh, Sspb-GFP-P2A-iLID-CAAX, and iRFP-CAAX; BcLOV/Cry2 multiplexing: BcLOV-mCh, Cry2(PHR)-GFP13. Images were acquired 24 hours post-transfection using confocal microscopy. For BcLOV/iLID multiplexing, light stimulation was performed at 37 °C using 1 s of blue light (1.45 W/cm2) every 30 seconds for 10 min in the presence or absence of prior BcLOV inactivation. Inactivation was achieved using these same light settings for 1 hour. For BcLOV/Cry2 multiplexing, “short light” (10 min) and “long light” (45 min) exposure was achieved using 100 ms of light (1.45 W/cm2) every 30 s at 30 °C. BcLOV inactivation was achieved using 1 s of blue light (1.45 W/cm2) every 30 seconds for 45 min. The Cry2-ONLY state was imaged after 45 min of light inactivation.
Supplementary Material
Acknowledgements.
The authors thank Dr. Christoph Seiler and the CHOP Aquatic Zebrafish Core for assistance in injecting and mounting zebrafish embryos, Daniel Wu and Dr. David Chenoweth for assistance with TIRF microscopy, and Drs. Heath Johnson and Jared Toettcher (Princeton) for providing Drosophila S2 cells. This work was supported by the National Institutes of Health (R35GM138211, R21GM132831 for L.J.B, R35GM133425 for H.D. and B.L., R01NS101106 for E.E.B and B.Y.C. and R01HL152086 to A.F.S.) and the National Science Foundation (Graduate Research Fellowship Program to W.B., CAREER MCB1652003 for E.E.B. and B.Y.C.).
Footnotes
Competing Interests Statement
The authors declare no competing interests.
Code availability
Matlab code used to fit data and model BcLOV4 activation dynamics can be found at the following link: https://drive.google.com/drive/folders/1h0eDceDplxYUguSpUNyg5CHA4uudZO39?usp=sharing
Data Availability
All raw data used to generate the figures can be found at the following link: https://drive.google.com/drive/folders/1h0eDceDplxYUguSpUNyg5CHA4uudZO39?usp=sharing
References
- 1.Christie JM, Salomon M, Nozue K, Wada M & Briggs WR LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. U. S. A 96, 8779–8783 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwerdtfeger C & Linden H VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 22, 4846–4855 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dietler J et al. A Light-Oxygen-Voltage Receptor Integrates Light and Temperature. J. Mol. Biol 433, 167107 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Hunt SM, Elvin M, Crosthwaite SK & Heintzen C The PAS/LOV protein VIVID controls temperature compensation of circadian clock phase and development in Neurospora crassa. Genes Dev. 21, 1964–1974 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutiérrez-Medina B & Candia CNH Aggregation kinetics of the protein photoreceptor Vivid. Biochim. Biophys. Acta: Proteins Proteomics 140620 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Guntas G et al. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. U. S. A 112, 112–117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennedy MJ et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimizu-Sato S, Huq E, Tepperman JM & Quail PH A light-switchable gene promoter system. Nat. Biotechnol 20, 1041–1044 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Strickland D et al. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods 9, 379–384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawano F, Suzuki H, Furuya A & Sato M Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat. Commun 6, 6256 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Zhou XX, Chung HK, Lam AJ & Lin MZ Optical control of protein activity by fluorescent protein domains. Science 338, 810–814 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu YI et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS & Schaffer DV Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10, 249–252 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Boyden ES, Zhang F, Bamberg E, Nagel G & Deisseroth K Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci 8, 1263–1268 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Glantz ST et al. Directly light-regulated binding of RGS-LOV photoreceptors to anionic membrane phospholipids. Proc. Natl. Acad. Sci. U. S. A 115, E7720–E7727 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L et al. Optical control of membrane tethering and interorganellar communication at nanoscales. Chem. Sci 8, 5275–5281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCubrey JA et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta 1773, 1263–1284 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang F et al. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: potential targeting for therapeutic intervention. Leukemia 17, 1263–1293 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Vanhaesebroeck B, Stephens L & Hawkins P PI3K signalling: the path to discovery and understanding. Nat. Rev. Mol. Cell Biol 13, 195–203 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Yang J et al. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol. Cancer 18, 26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bugaj LJ et al. Cancer mutations and targeted drugs can disrupt dynamic signal encoding by the Ras-Erk pathway. Science 361, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hino N et al. ERK-Mediated Mechanochemical Waves Direct Collective Cell Polarization. Dev. Cell 53, 646–660.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Johnson HE et al. The Spatiotemporal Limits of Developmental Erk Signaling. Dev. Cell 40, 185–192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson HE & Toettcher JE Signaling Dynamics Control Cell Fate in the Early Drosophila Embryo. Dev. Cell 48, 361–370.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toettcher JE, Gong D, Lim WA & Weiner OD Light-based feedback for controlling intracellular signaling dynamics. Nat. Methods 8, 837–839 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toettcher JE, Weiner OD & Lim WA Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bugaj LJ & Lim WA High-throughput multicolor optogenetics in microwell plates. Nat. Protoc 14, 2205–2228 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Berlew EE, Kuznetsov IA, Yamada K, Bugaj LJ & Chow BY Optogenetic Rac1 engineered from membrane lipid-binding RGS-LOV for inducible lamellipodia formation. Photochem. Photobiol. Sci 19, 353–361 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berlew EE et al. Single-component optogenetic tools for inducible RhoA GTPase signaling. Advanced Biology 2100810 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hannanta-Anan P, Glantz ST & Chow BY Optically inducible membrane recruitment and signaling systems. Curr. Opin. Struct. Biol 57, 84–92 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suh B-C, Inoue T, Meyer T & Hille B Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science 314, 1454–1457 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Regot S, Hughey JJ, Bajar BT, Carrasco S & Covert MW High-Sensitivity Measurements of Multiple Kinase Activities in Live Single Cells. Cell 157, 1724–1734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayr V, Sturtzel C, Stadler M, Grissenberger S & Distel M Fast Dynamic in vivo Monitoring of Erk Activity at Single Cell Resolution in DREKA Zebrafish. Front Cell Dev Biol 6, 111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adrian M, Nijenhuis W, Hoogstraaten RI, Willems J & Kapitein LC A Phytochrome-Derived Photoswitch for Intracellular Transport. ACS Synth. Biol 6, 1248–1256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golic AE et al. BlsA Is a Low to Moderate Temperature Blue Light Photoreceptor in the Human Pathogen Acinetobacter baumannii. Front. Microbiol 10, 1925 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakasone Y, Ono T-A, Ishii A, Masuda S & Terazima M Temperature-sensitive reaction of a photosensor protein YcgF: possibility of a role of temperature sensor. Biochemistry 49, 2288–2296 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Nakasone Y et al. Stability of dimer and domain-domain interaction of Arabidopsis phototropin 1 LOV2. J. Mol. Biol 383, 904–913 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Richter F et al. Engineering of temperature- and light-switchable Cas9 variants. Nucleic Acids Res. 44, 10003–10014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benedetti L et al. Optimized Vivid-derived Magnets photodimerizers for subcellular optogenetics. Cold Spring Harbor Laboratory 2020.08.28.272807 (2020) doi: 10.1101/2020.08.28.272807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hernández-Candia CN, Casas-Flores S & Gutiérrez-Medina B Light induces oxidative damage and protein stability in the fungal photoreceptor Vivid. PLoS One 13, e0201028 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Brederode ME, Hoff WD, Van Stokkum IH, Groot ML & Hellingwerf KJ Protein folding thermodynamics applied to the photocycle of the photoactive yellow protein. Biophys. J 71, 365–380 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benzinger D, Ovinnikov S & Khammash M Synthetic gene networks recapitulate dynamic signal decoding and differential gene expression. bioRxiv 2021.01.07.425755 (2021) doi: 10.1101/2021.01.07.425755. [DOI] [PubMed] [Google Scholar]
- 43.Duan L et al. Understanding CRY2 interactions for optical control of intracellular signaling. Nat. Commun 8, 547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 44.Fukaya T, Lim B & Levine M Enhancer Control of Transcriptional Bursting. Cell 166, 358–368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santos MG, Jorge SAC, Brillet K & Pereira CA Improving heterologous protein expression in transfected Drosophila S2 cells as assessed by EGFP expression. Cytotechnology 54, 15–24 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villefranc JA, Amigo J & Lawson ND Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn 236, 3077–3087 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamprecht MR, Sabatini DM & Carpenter AE CellProfiler™: free, versatile software for automated biological image analysis. Biotechniques 42, 71–75 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Wickham H, François R, Henry L & Müller K dplyr: a grammar of data manipulation. R package version 0.8. 0.1 Retrieved January 13, 2020 (2019). [Google Scholar]
- 49.Wickham H ggplot2: ggplot2. Wiley Interdiscip. Rev. Comput. Stat 3, 180–185 (2011). [Google Scholar]
- 50.Berg S et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data used to generate the figures can be found at the following link: https://drive.google.com/drive/folders/1h0eDceDplxYUguSpUNyg5CHA4uudZO39?usp=sharing