Abstract
A highly integrated, morphologically diverse bacterial community is associated with the dorsal surface of Alvinella pompejana, a polychaetous annelid that inhabits active high-temperature deep-sea hydrothermal vent sites along the East Pacific Rise (EPR). Analysis of a previously prepared bacterial 16S ribosomal DNA (rDNA) library identified a spirochete most closely related to an endosymbiont of the oligochete Olavius loisae. This spirochete phylotype (spirochete A) comprised only 2.2% of the 16S rDNA clone library but appeared to be much more dominant when the same sample was analyzed by denaturing gradient gel electrophoresis (DGGE) and the terminal restriction fragment length polymorphism procedure (12 to 18%). PCR amplification of the community with spirochete-specific primers used in conjunction with DGGE analysis identified two spirochete phylotypes. The first spirochete was identical to spirochete A but was present in only one A. pompejana specimen. The second spirochete (spirochete B) was 84.5% similar to spirochete A and, more interestingly, was present in the epibiont communities of all of the A. pompejana specimens sampled throughout the geographic range of the worm (13°N to 32°S along the EPR). The sequence variation of the spirochete B phylotype was less than 3% for the range of A. pompejana specimens tested, suggesting that a single spirochete species was present in the A. pompejana epibiotic community. Additional analysis of the environments surrounding the worm revealed that spirochetes are a ubiquitous component of high-temperature vents and may play an important role in this unique ecosystem.
Symbiotic relationships between invertebrates and bacteria are a prominent feature in marine systems. Investigations of these marine symbiotic relationships have been spurred on by the discovery of their dominance in invertebrates from extreme environments, such as hydrothermal vents. One of the first symbioses described in hydrothermal vent environments was that of the endosymbiotic chemoautotrophic sulfur-oxidizing bacteria associated with the deep-sea hydrothermal vent giant tubeworm, Riftia pachyptila (8). While these bacteria have not been cultured, their role in the symbiosis has been determined in detail by using a variety of physiological and molecular methods (16, 22). Other hydrothermal vent symbiotic associations that have been described include chemoautotrophic endosymbionts associated with other vestimentiferans and bivalves, as well as the episymbionts associated with the shrimp Rimicaris exoculata and the polychaetes Alvinella pompejana and Alvinella caudata (5, 7, 13, 15, 19, 28).
A. pompejana is considered to be the most thermotolerant and eurythermal metazoan yet described (6, 9). It forms tubes and colonizes the walls of high-temperature chimneys (40 to 105°C) at vent sites along the East Pacific Rise (EPR) from 32°S to 21°N latitude (11; C. A. Di Meo, G. Luther, and S. C. Cary, unpublished data). A. pompejana is characterized by a dense, specific epibiotic microflora associated with the dorsal integument of the worm (5, 12). Although electron microscopy studies of the A. pompejana epibionts suggest that they are highly diverse, two bacterial morphotypes, a filamentous sheathed form and a rod-shaped form, appear to predominate (17, 18). The filamentous form is integrated into specialized expansions of the intersegmentary parts, while the rod-shaped form appears to be less abundant but evenly distributed on the dorsal surface (17, 18). The role of these epibionts in the symbiotic association with A. pompejana is unclear. Ribulose-1,5-biphosphate carboxylase/oxygenase (RuBisCO), 14C labeling, and bicarbonate uptake assays have not implicated the dominant filamentous morphotype in autotrophic CO2 fixation (1, 2). However, it has been suggested that the symbionts may provide a food source for the worm (5), as hypothesized for the ectosymbionts of R. exoculata (28, 31), detoxify the immediate environment surrounding the worm, or possibly provide thermal insulation (11, 29).
In previous investigations, a combination of restriction fragment length polymorphism (RFLP) in conjunction with sequence analysis of a 16S ribosomal DNA (rDNA) clone library and in situ hybridizations performed with clone-specific oligonucleotides demonstrated that the A. pompejana community is dominated by 4 families (of 32 families) that form a tight clade within the epsilon subdivision of the Proteobacteria, at least 2 of which are made up of filamentous organisms (5, 19). In the present study, a novel spirochete phylotype was discovered after further screening of the same 16S rDNA clone library. Spirochete A appeared to be much more dominant in the community when two separate DNA fingerprinting methods were used. However, spirochete A was not found in other A. pompejana specimens when spirochete-specific primers were used. We also demonstrated that a second, unique spirochete (spirochete B), while accounting for only a small percentage of the community, was a consistent component of the A. pompejana epibiont population throughout the geographic range of the worm.
MATERIALS AND METHODS
Specimen collection and isolation of the DNAs of the A. pompejana epibionts.
A. pompejana specimens were collected from five geographically distinct hydrothermal vent sites along the EPR. A single A. pompejana specimen (APG1B) was collected from the Elsa vent site, (latitude, 13°N) during the joint French U.S. Hydrothermal Environment Research Observatory expeditions in 1991. Other A. pompejana samples were collected at latitudes of 32°S and 18°S (dives 3340 and 3333, respectively; Southern EPR cruise, December 1998 to January 1999, voyage 3, leg 30, R/V Atlantis), 9°N (dives 3317 and 3308, November 1998, voyage 3, leg 29, R/V Atlantis), and 13°N (dives 2874 and 2875, November 1994, cruise 131, leg 25, R/V Atlantis; dives AM-01 and AM-08, June 1999, AMISTAD cruise, L'Atalante) (Table 1). After the worms were washed three times in sterile seawater, bacterial filaments were carefully cut from the dorsal surface of each worm, homogenized in a lysis buffer (5 M guanidinium isothiocyanate, 50 mM Tris [pH 7.4], 25 mM EDTA, 0.8% 2-mercaptoethanol), frozen at −80°C on the ship, and stored until extractions were performed in the laboratory. Total genomic DNAs were isolated from the frozen lysed samples by using an IsoQuick extraction method according to the instructions of the manufacturer (ORCA Research, Bothwell, Wash.). The extraction efficiency of the method was comparable to those of other DNA extraction methods (5; data not shown).
TABLE 1.
Spirochete sequence similarities for various A. pompejana epibiont communities and environmental samples
| Sample (dive) | Geographic location (latitude, longitude) | Vent site | Year | % Sequence similarity of major DGGE banda |
|---|---|---|---|---|
| AP32 (3340) | 31°50′S, 111°W′ | Snow Ghosts | 1999 | 100 |
| AP18 (3333) | 18°24′S, 113°24′W | Sojourn | 1998 | 97.8 |
| AP9N (3317) | 9°51′N, 104°18′W | Robin's nest | 1998 | 98.4 |
| AP9N (3308) | 9°51′N, 104°18′W | M | 1998 | 98.4 |
| APG1B | 12°59′N, 103°56′W | Elsa | 1991 | 84.5 (98.4)b |
| AP13N (2874) | 12°48.8′N, 103°56.4′W | Totem | 1994 | 98.4 |
| AP13N (AM-01) | 12°48.7′N, 103°56.4′W | Grandbonum | 1999 | 98.4 |
| AP13N (AM-08) | 12°49.84′N, 103°56.8′W | PP55 | 1999 | 98.4 |
| Chim (2864) | 9°49.1N, 104°17.3′W | Pillar | 1994 | 89.6 |
| APTube (2857) | 12°59′N, 103°56′W | Elsa | 1994 | 75.3 |
Values were based on a DNA distance analysis (similarity to sample AP32 from dive 3340).
The value in parentheses is the percent similarity for a minor DGGE band.
Sequencing and phylogenetic analysis of epibionts.
A 16S rDNA clone (APG10), representing an RFLP family (3 of 139 clones screened) in a previously prepared 16S rDNA library (19), was bidirectionally sequenced at the 5′ end by using primers 27F and 519R (25) and a Perkin-Elmer Big Dye terminator sequencing kit (Applied Biosystems Inc., Foster City, Calif.). The reaction mixtures were ethanol precipitated and sequenced with an ABI 310 automated sequencer (ABI, Foster City, Calif.). The resulting sequences were edited and aligned with their complementary strands by using AutoAssembler DNA sequence assembly software (ABI).
Community structure analysis.
Denaturing gradient gel electrophoresis (DGGE) was performed as described by Muyzer et al. (25), with some modifications. Briefly, 25 ng of isolated genomic DNA or 2.5 ng of plasmid DNA from the clone described above (unless otherwise noted) was used for PCR amplification of the V3 region of 16S rDNA with the following primers: 338F/GC clamp and 519R (25) or 804R (5′-CTACCAGGGTATCTAATCC-3′; universal bacterial primer designed from a Ribosomal Database Project II (RDP II) alignment of the 16S rDNA of all prokaryotes in the database). Triplicate PCR were performed in 50-μl mixtures which contained each primer at a concentration of 0.1 μM, 1.25 U of Taq polymerase (Promega, Madison, Wis.), 2.5 U of Pfu polymerase (Stratagene, La Jolla, Calif.), 1× Promega Taq PCR buffer, 2 mM MgCl2, and each deoxyribonucleoside triphosphate at a concentration of 200 μM. Samples were pooled, and the DNA was precipitated and quantified by densitometry by using an AlphaImager 2000 document and analysis system, version 3.3b (Alpha Innotech Corporation, Calif.).
Amplification products (approximately 250 ng for the plasmid clones and approximately 1,500 ng for the genomic DNA products) were electrophoresed through a 25 to 55% denaturing gradient, 6 or 8% acrylamide gel (100% denaturant was 7 M urea plus 40% deionized formamide) for 5 h at a constant voltage of 130 V and a temperature of 60°C by using a Dcode universal mutation detection system (Bio-Rad, Hercules, Calif.). DNA bands were visualized with a UV transilluminator and were photographed by using the AlphaImager system (Alpha Innotech).
Individual separated DGGE fragments were stabbed with an aerosol-free pipette tip, and each resulting gel piece was resuspended in 20 μl of sterile water. One-half of the sample was used in a 50-μl PCR mixture as described above. Amplification products were checked for purity by DGGE of a portion of the sample. If necessary, a second round of stabbing and amplification was performed. Excess primers were removed from PCR products by passage through a Qiagen PCR purification column (Qiagen, Valencia, Calif.) according to the manufacturer's instructions and were quantified by UV spectrophotometry. Approximately 40-ng portions of purified products were used in 10-μl Big Dye terminator sequencing reaction mixtures (ABI) along with either forward primer 338F (3) or reverse primer 519R. Sequence analysis of the resulting fragments was performed as described above.
Terminal RFLP (T-RFLP) analysis was performed as described by Liu et al. (23), with several modifications. Briefly, triplicate 50-μl PCR mixtures were amplified by using fluorescently labeled primers 27f-HEX and 926r-TET and the cycling conditions described previously except that 50 ng of genomic DNA was added to each reaction mixture, the annealing temperature was changed to 54°C, and the number of cycles was reduced from 35 to 30. Fifty nanograms of each pooled amplification product was digested with 5 U of CfoI, HaeIII, or AluI (Promega) in a final volume of 10 μl for 6 h at 37°C. These enzymes were chosen because of their frequent cutting potential (23). Two microliters of each digest was resuspended in 20 μl of formamide, and 0.5 μl of a 0-500 TAMRA size standard (ABI) was added to each tube. Denatured samples were electrophoresed with a model 310 automated sequencer (ABI). Individual T-RFLP fragments were visualized, and the sizes and integrated areas under the peaks were automatically determined with the ABI 310 GeneScan software (GeneScan, version 2.1).
Molecular analyses of spirochete members.
To amplify the DNAs of the majority of spirochetes in a mixed microbial community, a spirochete-specific forward primer, Spi33F (5′GGCGGCGCGTWTTAAG-3′), was designed based on a survey of the GenBank database and a previously published phylogenetic analysis of the spirochetes (26). This forward primer was used in PCR amplifications with reverse primer 519R or 926R under the following conditions: 94°C for 1 min, 68°C for 1 min (56°C with the 926R primer), and 72°C for 1 min for 35 cycles. Three sets of DNA extraction mixtures were amplified: (i) mixtures from the A. pompejana specimens mentioned above, (ii) mixtures from a bacterial film present on a sample of chimney (dive 2864, Pillar site, 9°N, voyage 131, leg 25, November 1994), and (iii) mixtures from the outsides of A. pompejana tubes (sample 137, dive 2857, 13°N, Elsa; sample 150, dive 2859, 9°N, X5; both from the cruise mentioned above). The negative 16S rDNA controls used were from representatives of the alpha, beta, gamma, delta, and epsilon subdivisions of the Proteobacteria, as well as the cytophaga group (data not shown). RFLPs were determined by digestion of each amplification product as described previously (19).
A 0.5-μl portion of each spirochete-specific amplified product (from the preparations described above) was used in a second PCR performed with primers 338F/GC clamp and 519R and was subjected to DGGE analysis as described above. The bands were stabbed, amplified, sequenced, and phylogenetically grouped as described above. DGGE analysis with primers 338F/GC clamp and 804R was also performed directly with the A. pompejana and environmental DNAs described above.
Nucleotide sequence accession numbers.
The consensus nucleotide sequence of 16S rDNA clone APG10 has been deposited in the GenBank database (4) under accession number AF180309. The nucleotide sequences of the amplification products from DGGE fragments have been deposited in the GenBank database under accession numbers AF180311 to AF180319. The nucleotide sequences of spirochete-specific amplified products have been deposited in the GenBank database under accession numbers AY007429 to AY007433 and AF300986.
RESULTS
Community analysis of A. pompejana epibionts.
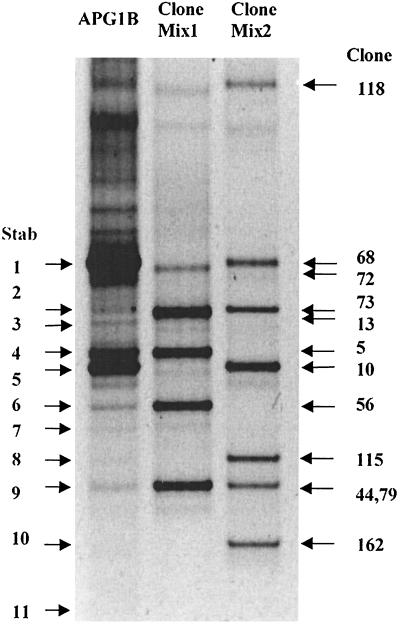
Previous molecular characterization of the epibiont community associated with A. pompejana demonstrated a dominance of phylotypes belonging to the epsilon subdivision of the Proteobacteria (5, 19). Molecular characterization of the minor members of a community in many cases may reveal important information regarding a symbiotic association, especially when diverse bacteria are found in the association, as in A. pompejana (19). Therefore, in this study, we focused on a distinct microbe present in the A. pompejana epibiont community. A clone from a previously prepared 16S rDNA library (19) (APG10 [= spirochete A]; frequency in 16S rDNA library, 2.2%) was characterized, and this clone phylogenetically grouped with the Spirochaeta species (sequence similarity with Olavius loisae endosymbiont, 86.5%). Two techniques (DGGE and T-RFLP analysis) were used to confirm the clone dominance predicted from the 16S rDNA library analysis. More than 11 fragments were identified from a short, amplified region of the 16S rDNA from the alvinellid community (APG1B) DNA by using DGGE (Fig. 1). DGGE analysis confirmed that the majority of the epibionts were members of the epsilon subdivision of the Proteobacteria; 10 of the 11 bands in Fig. 1 were from members of this group, compared to 9 of 11 clones from the 16S rDNA library. The total frequency of members of the epsilon subdivision in the library was, therefore, at least 80% (not all of the clone families have been characterized). DGGE analysis also indicated that one of the bands (Fig. 1, band 5: corresponding to spirochete A) was present at a significantly higher level (15.5%) than in the 16S rDNA library (2.2%). Similar relative frequencies were obtained in T-RFLP analyses with another set of primers, which confirmed the DGGE data. The relative frequencies of the spirochete A 5′-terminal restriction fragment (T-RF) obtained with AluI, CfoI, and HaeIII, were 18, 12, and 15%, respectively. Frequencies were determined by comparing the relative peak areas of T-RF with the sum of all T-RF areas by using the Gene Scan software.
FIG. 1.
DGGE gel comparing a portion of the 16S rDNA from APG1B (uncloned) to 16S rDNA library clones. Triplicate PCR mixtures obtained with APG1B DNA were pooled and ethanol precipitated, and approximately 1.5 μg was loaded into the first lane. Mixtures of amplified products of other clones were loaded into the second lane (clones 72, 5A, 13B, 44B, 56B) (Clone Mix1) and third lane (clones 118, 68, 73, 10, 115, 79, 162) (Clone Mix2).
Spirochetes in the Alvinella community.
To determine if spirochete A (= APG10) was a consistent member of the epibiotic community associated with A. pompejana, additional alvinellid and environmental samples (A. pompejana tubes, chimney) were screened. This was achieved in two ways: (i) 16S rDNA RFLP analysis after amplification with spirochete-specific primers and (ii) DGGE and sequence analysis after amplification with the spirochete-specific primer or a universal bacterial primer set.
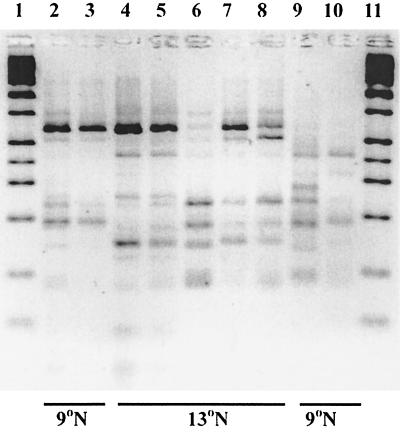
Spirochete-specific amplification products from seven A. pompejana worms and two tubes, as well as a chimney sample taken from a different vent site, were screened by RFLP analysis (Fig. 2). Digestion of the PCR products obtained with primers 33F and 926R with MboI and HaeIII revealed that each of the samples had a dominant phylotype, as demonstrated by the sum obtained for darkest bands present in each lane. For instance, in Fig. 2, lane 1, the prominent bands corresponded to approximately 190 and 700 bp, and the sum represented the full-length product. There also was evidence of minor members, as indicated by the fainter bands present in each of the lanes. Interestingly, only the amplified DNAs from latitude 13°N had minor RFLP bands identical to the bands obtained for spirochete A, which was also from latitude 13°N (Fig. 2, lanes 4 through 8).
FIG. 2.
RFLP analysis of the 16S rDNA gene of various microbial communities after amplification with spirochete-specific primers and digestion with MboI and HaeIII. Lanes 1 and 11, molecular weight marker; lane 2, A. pompejana community from dive 3317; lane 3, community from dive 3308; lane 4, community from dive 2874; lane 5, community from dive 2875; lane 6, APG1B; lane 7, community from dive AM-08; lane 8, community from dive AM-01; lane 9, bacterial film on chimney; lane 10, community from outside A. pompejana tube in dive 2859. Geographic locations are indicated at the bottom.
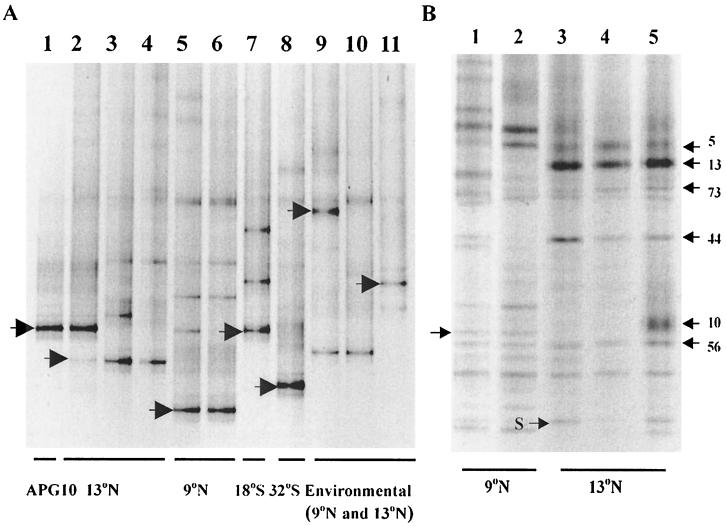
To determine if the spirochete association was a ubiquitous trait found in all A. pompejana epibiont communities, we screened A. pompejana samples from a large geographic range along the EPR and several other environmental samples. Two sets of primers were used in PCR amplifications, and the amplicons from the second set were subjected to DGGE analysis (Fig. 3A). The first primer set (a spirochete primer and a universal primer) amplified a large segment of the 16S rDNA gene. Amplicons obtained with the first set were used as templates for the second set of primers (bacterium-specific primers used to amplify the V3 region of the 16S rDNA gene). The majority of the geographically diverse epibiont communities produced one major spirochete-specific band (indicated by the arrows in Fig. 3A) and several minor bands, confirming the results obtained in the RFLP analysis. The major bands resulting from amplifications of the bacterial communities from different worms taken from the same latitude (e.g., 13°N) (Fig. 3A, lanes 3 and 4) along the EPR were identical, even though in some cases the alvinellids were collected from different vent sites and in different years (Table 1). The one exception to this was the APG1B sample obtained at latitude 13°N (Fig. 3A, lane 2), from which the original 16S rDNA library was prepared. The major species in this case corresponded to spirochete A (clone APG10) identified in the 16S rDNA library and DGGE analyses described above. However, a minor spirochete-specific band (Fig. 3A, lane 2, arrow) was present in the amplified sample that migrated just like the other latitude 13°N samples (Fig. 3A, lanes 3 and 4).
FIG. 3.
DGGE gels containing the 16S rDNA gene from various microbial communities. (A) Gel after two rounds of amplification, the first with a spirochete-specific and bacterial primer set. Lane 1, APG10 plasmid control; lane 2, APG1B; lane 3, community from dive 2874; lane 4, community from dive AM-08; lane 5, community from dive 3317; lane 6, community from dive 3308; lane 7, community from dive 3333; lane 8, community from dive 3340; lane 9, A. pompejana tube community from dive 2857; lane 10, A. pompejana tube community from dive 2859; lane 11, chimney sample community from dive 2864. (B) Gel after amplification of the 16S rDNA gene (E. coli positions 338 to 804). Lane 1, A. pompejana community from dive 3317; lane 2, community from dive 3308; lane 3, community from dive 2874; lane 4, community from dive 2875; lane 5, APG1B. The arrows indicate the bands sequenced for further analysis. Geographic locations are indicated at the bottom.
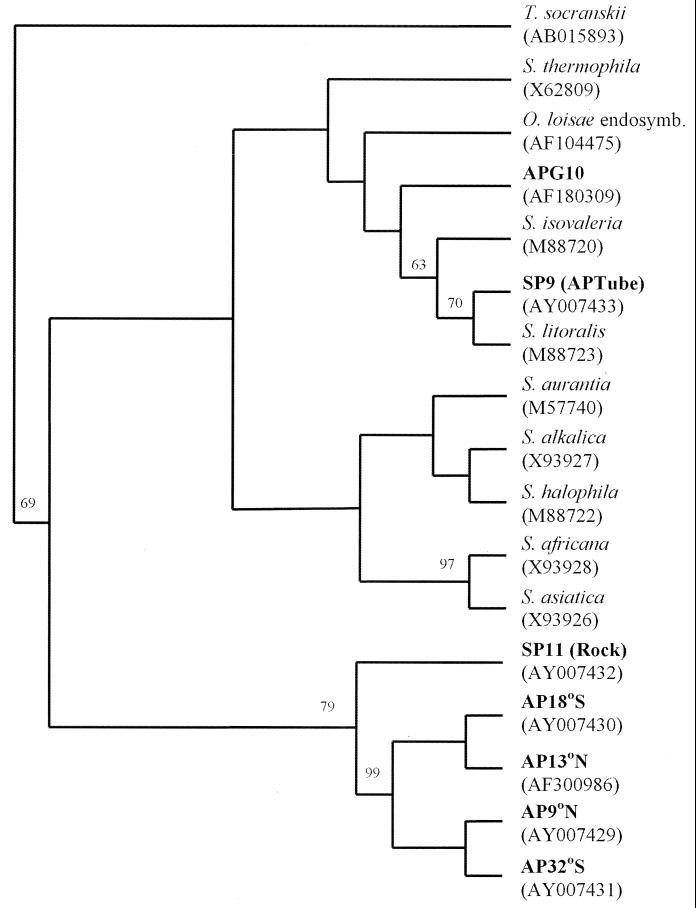
DNA distance analysis of sequences obtained from all the major bands (and the minor band in the APG1B sample) indicated that closely related spirochetes were present in all the A. pompejana epibiont communities tested (Table 1). The levels of DNA similarity between spirochetes obtained from A. pompejana were greater than 97.8%, except for the value for clone APG10 from APG1B, which was 84.5% similar to AP32. Since the phylotypes were more than 97% identical, they were considered members of a single species (spirochete B). Phylogenetic analysis of the 16S rDNA gene from several spirochetes demonstrated that all of the hydrothermal vent spirochetes characterized in this study cluster within the genus Spirochaeta (Fig. 4). Three distinct phylogenetic clusters were observed, and one of these groups contained all but one of the spirochetes associated with A. pompejana. A second cluster included spirochete A (= APG10) as identified by 16S rDNA library screening and DGGE analysis and the endosymbiont from O. loisae (14). The third group contained the spirochete from an A. pompejana tube and Spirochaeta litoralis. Values from phylogenetic bootstrap analyses of the sequences obtained directly from the DGGE bands (amplified region from position 338 to position 519 of the Escherichia coli 16S rDNA gene) were low, most likely due to the small fragment available for sequencing. Since the bootstrap values were low, the tree topologies were confirmed by obtaining nearly identical trees with the maximum-likelihood and neighbor-joining algorithms (data not shown).
FIG. 4.
Consensus bootstrapped cladogram (100 replicates) depicting the ancestral relationships indicated. The cladogram was calculated by using the DNA parsimony algorithm (PHYLIP, version 3.5c). The analysis was based on the region corresponding to bases 338 to 519 of the E. coli 16S rDNA gene, excluding insertions, deletions, and ambiguous bases. Various members of the genus Spirochaeta were included (GenBank accession numbers in parentheses). Treponema socranskii was used as the outgroup species.
In order to compare the relative levels of the two unique spirochetes obtained from A. pompejana, the relative frequencies of both were estimated by DGGE analysis. Several different bands were observed on a DGGE gel prepared from bacterial 16S rDNA products (region corresponding to 16S rDNA of E. coli at positions 338 to 804) amplified from alvinellid epibiont samples obtained from the 9°N and 13°N sites (Fig. 3B). The calculated relative frequency of spirochete A (arrow indicating APG10 in Fig. 3B, lane 5) as determined by densitometry was 15.5%. Sequence analysis of a similarly migrating band (arrow in lane 1) demonstrated that it did not correspond to a spirochete; however, sequence analysis of 20 additional bands revealed a band corresponding to spirochete B (arrow S in lane 3). The relative frequency of this band was approximately 1%, as determined by densitometry. All of the other bands were identical or closely related to APG5A, APG13B, APG44B, or APG56B, the four clones identified in a previous investigation (19; data not shown).
DISCUSSION
A. pompejana is characterized by a dense coating of epibionts whose role in the symbiotic association remains unknown. A previous study of the complexity of this association revealed the prevalence of two filamentous epsilon-proteobacterial phylotypes as determined by both 16S rDNA library screening and in situ hybridization, confirming previous morphological evidence of dominant filamentous symbionts (5, 19). Although these two closely related bacterial phylotypes dominate the community, there is morphological and molecular evidence of a more diverse assemblage of bacteria on the dorsal surface of A. pompejana (5, 10, 17–19, 29). In this investigation, besides confirming the diversity of members of the epsilon subdivision of the Proteobacteria by DGGE analysis, we determined the presence of spirochetes in the A. pomepjana community and found that one of these spirochetes is a minor, but consistent member.
Spirochete B was detected in all A. pompejana epibiont communities surveyed (at latitudes from 13°N to 32°S along the EPR), although it was a minor member of the community as determined by DGGE analysis, accounting for (∼1%) of the population. More importantly, its close phylogenetic relationship throughout almost the entire geographic range of A. pompejana (more than 97% similarity) and the fact that identical spirochetes were found at the same vent sites in different years suggest that this spirochete may be involved in an integrated symbiosis with A. pompejana. The fidelity of this relationship has yet to be determined. None of the other spirochetes detected were consistently present in the A. pompejana epibiont community, and most were found in the hydrothermal vent environment separate from the worm.
The genus Spirochaeta is comprised of obligately and facultatively chemoheterotrophic anaerobes that are generally free-living freshwater and marine species (26). However, Dubilier et al. demonstrated the presence of three symbionts associated with cuticular invaginations of O. loisae, an annelid inhabiting marine sediments, one of which is a member of the Spirochaeta family (14). Until our study, only one spirochete had been discovered in a hydrothermal vent environment (21). Although this spirochete was not phylogenetically characterized, its growth characteristics were consistent with those of other anaerobic chemoheterotrophic Spirochaeta strains, the majority of which ferment available carbohydrates to produce acetate, CO2, and H2 (21). Therefore, the metabolism of the A. pompejana spirochete epibiont most likely is fermentative, and the spirochete probably uses available carbohydrates in the mucus surrounding the worm and potentially supplies carbon sources and electron donors to the other epibionts (members of the epsilon subdivision of the Proteobacteria).
The A. pompejana epibiont community described in this study and many other symbiotic and nonsymbiotic communities have been characterized by using PCR. The drawbacks to PCR-based analyses of microbial communities have been reviewed at length (33). Two of the major concerns lie with PCR bias and different resolution capabilities of the methods. A determination of whether a member is considered dominant or minor in a community by PCR-based techniques may be questioned if only a single parameter is used for analysis. We felt that this study was a unique opportunity to characterize the spirochetes in a microbial community by using three separate, PCR-based methods. For each analysis (16S rDNA library screening, DGGE, and T-RFLP analysis) we used different primers, cycling conditions, and resolution techniques. DGGE and T-RFLP analyses, in which 16S rDNA amplification products that were approximately 171 and 918 bp long, respectively, were used, resulted in similar relative frequencies of spirochete A. The frequency of the corresponding clone was significantly different in the 16S rDNA library, where the entire 16S rDNA gene had been amplified (19). Alignment of the sequence of the 3′ primer used for the 16S rDNA library with the 10 sequences most similar to spirochete A (= APG10) demonstrated that 7 of 10 members of the spirochete family had four or more mismatches in the 3′ end of the primer sequence. Thus, many of the spirochetes could not be amplified representatively with this primer and were underrepresented in the full-length 16S rDNA community analysis. Like the results of other investigators, our results indicate that primer bias may be extremely important in a comparison of the members of a community (27, 30, 34).
There have been only a few previous molecular biology-based studies that described the microbial diversity observed at hydrothermal vents, and in none of them were spirochetes found (20, 24, 32). This may have been due to absence of the organisms or, as suggested by this study, to the choice of primers used for PCR amplification. During this investigation, we discovered several novel spirochetes not only in the A. pompejana epibiont community but also in the surrounding tubes and chimney samples from other hydrothermal vent environments. Because of the difficulty of isolating and cultivating spirochetes, detailed molecular analysis of similar environments with spirochete-specific primers would most likely reveal other, previously unknown members of the spirochete division. We are currently screening other hydrothermal vent habitats with such primers to identify additional novel spirochetes that are most likely important components of the microbial communities associated with deep-sea hydrothermal vent environments.
Unlike many other symbiotic associations in marine systems, the bacterial community associated with A. pompejana is morphologically and phylogenetically diverse. A group of closely related members of the epsilon subdivision of the Proteobacteria dominates the community (19; this study), while a smaller percentage of the A. pompejana epibiont community members from a wide geographic range studied in this work consists of more diverse members of the epsilon subdivision of the Proteobacteria and of a closely related Spirochaeta phylogenetic cluster. The role of the spirochetes remains unclear, but our results support the hypothesis that some of the epibionts of A. pompejana are chemoheterotrophic. Further research is currently being performed in our laboratory to characterize the mixture of A. pompejana epibionts collected from geographically, thermally, and chemically diverse environments and to decipher their metabolic capabilities in an effort to discover their role in this integrated episymbiosis.
ACKNOWLEDGMENTS
This research was supported by grants to S.C.C. from the National Science Foundation (grant OCE-9314595), the LEXEN initiative (grant OPP-9907666) and the Delaware Sea Grant Program (grant R/B37).
We thank K. Coyne, M. Cottrell, and C. Di Meo for helpful discussions and for critically reviewing the manuscript. We thank the captain and crew of the R/V Atlantis and especially the DSV Alvin pilots for their critical roles in the collection of specimens. We thank D. Prieur for the invitation to participate on the AMISTAD cruise and C. Jeanthon and the pilots and crew of L'Atalante and DSV Nautile for their assistance. We also thank R. Vrijenhoek for providing access to samples collected during the Southern EPR cruise.
REFERENCES
- 1.Alayse-Danet A M, Desbruyères D, Gaill F. The possible nutritional or detoxification role of the epibiotic bacteria of alvinellid polychaetes: review of the current data. Symbiosis. 1987;4:51–62. [Google Scholar]
- 2.Alayse-Danet A M, Gaill F, Desbruyères D. In situ bicarbonate uptake by bacteria-Alvinella associations. Mar Ecol (Pubbl Stn Zool Napoli I) 1986;7:233–240. [Google Scholar]
- 3.Amann R I, Krumholz L, Stahl D A. Fluorescent oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson D A, Karsch-Mizrachi I, Lipman D J, Ostell J, Rapp B A, Wheeler D L. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cary S C, Cottrell M T, Stein J L, Camacho F, Desbruyères D. Molecular identification and localization of a filamentous symbiotic bacteria associated with the hydrothermal vent annelid, Alvinella pompejana. Appl Environ Microbiol. 1997;63:1124–1130. doi: 10.1128/aem.63.3.1124-1130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cary S C, Shank T, Stein J. Worms bask in extreme temperatures. Nature. 1998;391:545–546. [Google Scholar]
- 7.Cavanaugh C M. Microbiol symbiosis: patterns of diversity in the marine environment. Am Zool. 1994;34:79–89. [Google Scholar]
- 8.Cavanaugh C M, Jones M L, Jannasch H W, Waterbury J B. Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: possible chemoautotrophic symbionts. Science. 1981;213:340–342. doi: 10.1126/science.213.4505.340. [DOI] [PubMed] [Google Scholar]
- 9.Chevaldonné P, Desbruyères D, Childress J J. Some like it hot, some like it hotter. Nature. 1992;359:593–594. [Google Scholar]
- 10.Cottrell M T, Cary S C. Diversity of dissimilatory bisulfite reductase genes of bacteria associated with the deep-sea hydrothermal vent polychaete annelid Alvinella pompejana. Appl Environ Microbiol. 1999;65:1127–1132. doi: 10.1128/aem.65.3.1127-1132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desbruyères D, Chevaldonné P, Alayse A-M, Jollivet D, Lallier F H, Jouin-Toulmond C, Zal F, Sarradin P-M, Cosson R, Caprais J-C, Arndt C, O'Brien J, Guezennec J, Hourdez S, Riso R, Gaill F, Laubier L, Toulmond A. Biology and ecology of the “Pompeii worm” (Alvinella pompejana Desbruyères and Laubier), a normal dweller of an extreme deep-sea environment: a synthesis of current knowledge and recent developments. Deep-Sea Res Part II. 1998;45:383–422. [Google Scholar]
- 12.Desbruyères D, Gaill F, Laubier L, Fouquet Y. Polychaetous annelids from hydrothermal vent ecosystems: an ecological overview. Bull Biol Soc Wash. 1985;6:103–116. [Google Scholar]
- 13.Distel D L, Lee H K, Cavanaugh C M. Intracellular coexistence of methano- and thioautotrophic bacteria in a hydrothermal vent mussel. Proc Natl Acad Sci USA. 1995;92:9598–9602. doi: 10.1073/pnas.92.21.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubilier N, Amann R, Erseus C, Muyzer G, Park S Y, Giere O, Cavanaugh C M. Phylogenetic diversity of bacterial endosymbionts in the gutless marine oligochete Olavius loisae (Annelida) Mar Ecol Prog Ser. 1999;178:271–280. [Google Scholar]
- 15.Feldman R A, Black M B, Cary C S, Lutz R A, Vrijenhoek R C. Molecular phylogenetics of bacterial endosymbionts and their vestimentiferan hosts. Mol Mar Biol Biotechnol. 1997;6:268–277. [PubMed] [Google Scholar]
- 16.Fisher C R, Childress J J, Minnich E. Autotrophic carbon fixation by the chemoautotrophic symbionts of Riftia pachyptila. Biol Bull (Woods Hole) 1989;177:372–385. [Google Scholar]
- 17.Gaill F, Desbruyères D, Prieur D. Bacterial communities associated with “Pompeii worms” from the East Pacific Rise hydrothermal vents: SEM, TEM observations. Microb Ecol. 1987;13:129–139. doi: 10.1007/BF02011249. [DOI] [PubMed] [Google Scholar]
- 18.Gaill F, Desbruyères D, Prieur D, Gourret J P. Mise en évidence de communautés bactériennes épibiontes du “ver de Pompéi.”. C R Acad Sci Paris Ser III. 1984;298:553–558. [Google Scholar]
- 19.Haddad M A, Camacho F, Durand P, Cary S C. Phylogenetic characterization of the epibiotic bacteria associated with the hydrothermal vent polychaete Alvinella pompejana. Appl Environ Microbiol. 1995;61:1679–1687. doi: 10.1128/aem.61.5.1679-1687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmsen H J M, Prieur D, Jeanthon C. Distribution of microorganisms in deep-sea hydrothermal vent chimneys investigated by whole-cell hybridization and enrichment culture of thermophilic subpopulations. Appl Environ Microbiol. 1997;63:2876–2883. doi: 10.1128/aem.63.7.2876-2883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harwood C S, Jannasch H W, Canale-Parola E. Anaerobic spirochete from a deep-sea hydrothermal vent. Appl Environ Microbiol. 1982;44:234–237. doi: 10.1128/aem.44.1.234-237.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laue B E, Nelson D C. Characterization of the gene encoding the autotrophic ATP sulfurylase from the bacterial endosymbiont of the hydrothermal vent tubeworm Riftia pachyptila. J Bacteriol. 1994;176:3723–3729. doi: 10.1128/jb.176.12.3723-3729.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moyer C L, Dobbs F C, Karl D M. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1995;61:1555–1562. doi: 10.1128/aem.61.4.1555-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paster B J, Dewhirst F E, Weisburg W G, Tordoff L A, Fraser G J, Hespell R B, Stanton T B, Zablen L, Mandelco L, Woese C R. Phylogenetic analysis of the spirochetes. J Bacteriol. 1991;173:6101–6109. doi: 10.1128/jb.173.19.6101-6109.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polz M F, Cavanaugh C M. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polz M F, Cavanaugh C M. Dominance of one bacterial phylotype at a Mid-Atlantic Ridge hydrothermal vent site. Proc Natl Acad Sci USA. 1995;92:7232–7236. doi: 10.1073/pnas.92.16.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prieur D, Jeanthon C. Preliminary study of heterotrophic bacteria isolated from two deep sea hydrothermal vent invertebrates: Alvinella pompejana (polychaete) and Bathymodiolus thermophilus (bivalve) Symbiosis. 1987;4:87–98. [Google Scholar]
- 30.Rainey F, Ward N, Sly L, Stackebrandt E. Dependence on the taxon composition of clone libraries for PCR amplified, naturally occurring 16S rDNA, on the primer pair and the cloning system used. Experientia. 1994;50:796–797. [Google Scholar]
- 31.Rieley G, Van Dover C L, Hedrick D B, Eglinton G. Trophic ecology of Rimicaris exoculata: a combined lipid abundance stable isotope approach. Mar Biol (New York) 1999;133:495–499. [Google Scholar]
- 32.Sievert S M, Brinkhoff T, Muyzer G, Ziebis V, Kuever J. Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island (Greece) Appl Environ Microbiol. 1999;65:3834–3842. doi: 10.1128/aem.65.9.3834-3842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wintzingerode F V, Gb̈el U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 34.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]