Abstract
Circular dichroism (CD) spectroscopy is widely used to characterize the secondary structure composition of proteins. To derive accurate and detailed structural information from the CD spectra, we have developed the Beta Structure Selection (BeStSel) method (PNAS, 112, E3095), which can handle the spectral diversity of β-structured proteins. The BeStSel webserver provides this method with useful accessories to the community with the main goal to analyze single or multiple protein CD spectra. Uniquely, BeStSel provides information on eight secondary structure components including parallel β-structure and antiparallel β-sheets with three different groups of twist. It overperforms any available method in accuracy and information content, moreover, it is capable of predicting the protein fold down to the topology/homology level of the CATH classification. A new module of the webserver helps to distinguish intrinsically disordered proteins by their CD spectrum. Secondary structure calculation for uploaded PDB files will help the experimental verification of protein MD and in silico modelling using CD spectroscopy. The server also calculates extinction coefficients from the primary sequence for CD users to determine the accurate protein concentrations which is a prerequisite for reliable secondary structure determination. The BeStSel server can be freely accessed at https://bestsel.elte.hu.
Graphical Abstract
Graphical Abstract.
Functions of the BeStSel web server for the analysis of protein CD spectra.
INTRODUCTION
The far-UV circular dichroism (CD) spectrum of a protein is characteristic to the secondary structure composition and is widely used to investigate protein structure. Although it does not provide site-specific structural information, CD spectroscopy is useful when a fast, inexpensive technique is needed or the application of high-resolution techniques (X-ray or NMR) is problematic. Applications cover all areas of protein science. CD spectroscopy can be used to verify the correct fold of recombinant proteins, study the effect of environmental conditions (pH, ionic strength, additives, crowding) and protein modifications (e.g. mutations and post-translational modifications) on the structure and stability. Moreover, CD spectroscopy is a suitable technique for the experimental verification of the structural information predicted by bioinformatics tools that make use of the ever-growing protein databases.
The instrumentation of CD spectroscopy is well developed and benchtop instruments are routinely used. Synchrotron radiation (SR) CD is also available with a broad wavelength range and can be used for high quality and special applications (1). A central question in protein CD spectroscopy is the spectral contribution of the various secondary structural elements. Numerous algorithms were developed in the last decades to gain information on the secondary structure from the CD spectra, however, accurate structure estimation was mostly limited to α-helical proteins because of the large spectral diversity of β-structured proteins (2,3). We have shown that the parallel-antiparallel orientation and the twist of the β-sheets account for this spectral diversity and developed the Beta Structure Selection (BeStSel) method for the accurate secondary structure estimation from protein CD spectra (4). BeStSel provides detailed structural information distinguishing eight structural components and overperforms any other method in accuracy. Moreover, it is capable of fold prediction down to the topology/homology level of CATH protein fold classification (5). The BeStSel webserver provides free access to the method for the scientific community and enables the fast, easy and accurate analysis of CD spectra. Here, we introduce the recent developments and present status of the BeStSel webserver.
MATERIALS AND METHODS (WEBSERVER DESCRIPTION)
Secondary structure components of BeStSel and the twist of β-sheets
Eight secondary structure components are defined in BeStSel based on the Dictionary of Secondary Structure of Proteins (DSSP) (6,7). Residues assigned to α-helix by DSSP are divided into two groups, regular, and distorted, as the middle part of α-helices (Helix1) and two-two residues at the ends of α-helices (Helix2), respectively. Residues assigned to β-strands by DSSP are considered for the four β-sheet groups of BeStSel, (i) parallel β-sheet and antiparallel β-sheet of three different twists: (ii) left-hand twisted (Anti1), (iii) relaxed (slightly right-hand twisted, Anti2), and (iiii) right-hand twisted (Anti3) (4). Turn is defined identically to that in DSSP (7). All other elements including missing residues are assigned to ‘Others’.
Optimization of BeStSel basis spectra
BeStSel uses precalculated, fixed basis spectra corresponding to the eight structural components to determine the secondary structure composition of proteins. These basis spectra are optimized using a reference CD spectrum set of 73 proteins with known 3D-structures (4). An independent set of β-structure rich proteins or proteins with rare structural composition was also used as test. The optimization procedure to get the basis spectra sets is described in detail by Micsonai et al. (4). The entire optimization process was executed separately for the different wavelength ranges now offered from 175–250 nm to 200–250 nm at 5 nm steps. For each range, there are eight sets of the eight basis spectra, and each set is optimized to be the most accurate for one of the secondary structure components. The fraction of that component is taken from the fitting with the linear combination of the corresponding eight spectra to the CD spectrum of the unknown protein and thus eight fittings will provide the fractions of the eight secondary structure components.
Protein fold prediction by BeStSel
Protein folds can be characterized by specific secondary structure patterns. The eight secondary structure components of BeStSel provide sufficient structural information to predict the protein fold. The β-sheet composition, including the parallel-antiparallel β-sheets and the level of twist in the antiparallel β-sheets adequately characterize the diverse folds with β-structures, while the two α-helix components reveal the number and average length of α-helices in the proteins (4). In the BeStSel package, the CATH protein fold classification is used (8) distinguishing hierarchical levels of protein fold from Class to Architecture, Topology, Homology (superfamily) and further levels. The advantage of using CATH is that most of the protein domains in the PDB are classified and the database is continuously maintained. Every single protein structure of the PDB can be represented as a point in the eight-dimensional secondary structural space of BeStSel and, in turn, the result of the BeStSel analysis of the CD spectrum can also be projected to this space. To find the fold of a protein, we search for points representing PDB structures that have similar secondary structure composition based on their Euclidean distance and then we determine their fold classification. However, the regions that different protein folds occupy in the eight-dimensional secondary structure space can be overlapping and it might be challenging to find the correct fold. The BeStSel package offers various different methods for fold recognition. A simple method performs a search on the entire PDB database for 20 structures closest to the target structure in Euclidean distance in the eight dimensional secondary structure space. The CATH classification of the corresponding structures are listed. In the case of multidomain proteins, it might be difficult to pinpoint the correct fold. To predict the fold of single domain proteins, we use a reference database containing a non-redundant (95% maximal sequence identity) single domain reference subset of CATH 4.3 (5). The corresponding secondary structure compositions are calculated from the PDB structures of the domains.
This subset contains 61 932 single domains covering the five fold classes, 43 architectures, 1467 topologies and 6540 homologies. Three prediction methods were constructed. (i) Search for the closest structures in the Euclidean space. This is useful for structures lying in a rarely populated part of the fold space. (ii) Search for all the chains that lie within the expectable error of BeStSel secondary structure determination, more exactly, within a distance of 1.5 × RMSD of BeStSel's average performance on SP175 reference set. The hits are sorted out for classes, architectures and topologies. The result table shows the frequencies and percentages of the different groups in the CATH categories. In dense regions, hundreds of structures can be found within the expected error of BeStSel, and the closest ones are not necessarily the correct ones (4). Usually, this method provides the highest reliability. (iii) The weighted k nearest neighbors’ (WKNN) (9) method predicts the Class, Architecture, Topology and Homology of the protein. In each layer, the predicted categories are ordered by WKNN score (10).
Disordered-ordered binary classification
262 ordered or disordered protein CD spectra were collected from the PCDDB (11), were the results of our own measurements, or were collected from the literature (12). The classification method uses the k nearest neighbor model with cosine distance function (13) using CD data at three wavelengths (197-206-233nm, or 212-217-225 nm). Disordered-ordered classification is based on the analysis of the 10 nearest neighbors in the reference set.
The operation of the BeStSel web server
The BeStSel web server is freely accessible. A detailed guide is provided in the tutorial file, which can be downloaded from the website in pdf format. The homepage also provides short explanations and tips for users. Error messages explain if input data format is not suitable. Warning messages draw the attention to possible problems like abnormal spectral amplitudes, which can be a result of improper data normalization or CD unit choice.
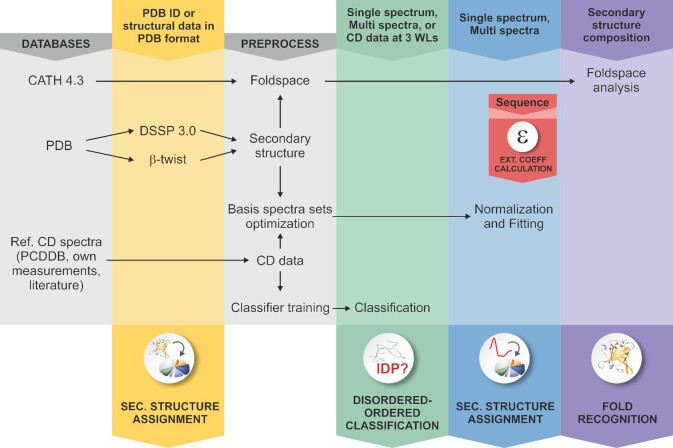
The server provides 8 program modules: Single spectrum analysis, Multiple spectra analysis, Fold recognition, Secondary structure decomposition for 3D-structures, Calculation of extinction coefficients from the primary sequence, Disordered-ordered binary classification, a searchable collection of publications using CD spectroscopy with BeStSel analysis, and a Guide to CD spectroscopy and data analysis helping CD users. A schematic diagram shows the modules and function of the web server in Figure 1.
Figure 1.
Schematic representation of the BeStSel server. Block diagram shows the modules and function of the BeStSel package. Arrows indicate the input and output data. From a single CD spectrum the secondary structure contents are estimated and then, based on these, the protein fold can be predicted. A series of CD spectra as input can be evaluated at once to get the secondary structure contents. Users can provide arbitrary secondary structure contents and carry out the fold prediction for that secondary structure composition. Users can also enter PDB IDs or upload structure files in PDB format as input to find the corresponding secondary structure contents and fold classification. Based on the CD data, a binary disordered-ordered classification can be carried out. To aid correct concentration determination, extinction coefficients at 205 and 214 nm can be calculated from the primary sequence of the protein (15,16).
In Single spectrum analysis, a CD spectrum can be uploaded and analyzed by the BeStSel method for secondary structure content. Data can be copied to the text window in the form of two columns or can be uploaded from txt files. The program automatically recognizes the file headers and in case of data pitch different from 1 nm, sorts out and uses integer nm data for analysis. Measurement files of various instruments saved in text format are handled properly by the server. Input units can be Δϵ (M–1cm–1), [Θ] (mean residue ellipticity in deg cm2 dmol–1) or measured ellipticity in mdeg units. In the latter case, concentration, residue number and pathlength data should be given and the server will normalize the data to Δϵ. After clicking on the submit button, a Data examination window appears to verify that the data was uploaded correctly. With one more click, the secondary structure contents are calculated using the eight secondary structure components and presented in the form of a graphical output together with the spectral fitting with RMSD and normalized RMSD (NRMSD) data. At first, fitting is carried out for the widest wavelength range. The user then can change the lower wavelength limit at 5 nm increments and recalculate the secondary structures. Details of the output image can be configured at the bottom of the page and the image can be redrawn. Alternatively, fitting results can be saved as .txt or .csv files for further data processing or figure preparation. Secondary structure contents can be recalculated by rescaling the spectrum with a chosen factor. The ‘Best factor’ function makes multiple recalculations with scaling factors between 0.5 and 2. This might help to examine the dependence of the fitting results on the CD amplitude and might help to find possible errors in concentration determination or normalization. However, the factor with the lowest NRMSD should not be taken as correction for the normalized spectrum when used in the 190–250 or 200–250 nm range. The correct concentration determination is essential for accurate analysis.
Protein fold prediction based on the secondary structure content can be initiated by one click. Four different types of analyses are carried out as described in Materials and Methods. The Fold recognition module can be used separately from CD spectrum analysis to predict the protein fold by manually entering the eight secondary structure contents and the chain length. The output of fold prediction is a list of the highest ranked 1, 5, 10 and 15 CATH classes, architectures, topologies and homologies, respectively. We recommend the WKNN method for structural studies to discover the fold of model structures or structures originated from the PDB.
Multiple spectra analysis will analyze a series of CD spectra simultaneously which might be helpful when multiple spectra are collected as a function of ligand or denaturant concentration, temperature, time, etc. Data table can be copied to the text window or can be opened from a text file. Input units are the same as in Single spectrum analysis. After ‘Data examination’ secondary structure contents are calculated by a single click and presented either as a graphical output or can be saved in .txt or .csv files for the convenience of the users. Wavelength range and scaling factor can be set for recalculation the same way as in Single spectrum analysis.
The Secondary structure from PDB files module calculates the eight BeStSel components and for comparison, DSSP (7) and SELCON3 (14) composition for 3D structures. For structures deposited in the PDB the input is the four-letter PDB ID and the program will provide the CATH information as well, if it exists. Structural files in PDB format (max. 20 MB) can also be uploaded to the server and the calculation will be carried out automatically. Both graphical and text outputs are available. This module is especially useful for experimental verification of MD results or in silico models by CD spectroscopy, making the structural information comparable (see in Results and Discussion).
The Extinction coefficient calculation module provides the extinction coefficients of proteins and peptides at 214 nm (15) and 205 nm (16), based on their primary sequence and number of disulfide bridges. The amino acid sequence should be entered or copied to the text window. The extinction coefficients can be used for concentration determination directly on the CD sample.
The Disordered-ordered classification module analyses far-UV CD data to identify disordered structures. The classification is based on CD data at three wavelengths (197-206-233 nm or 212-217-225 nm triplet). Data can be copied into the text window. The first column contains the wavelength values and the other columns contain the corresponding spectral data. Entire spectrum, series of spectra, or CD data only at the necessary wavelengths, all will be accepted and handled properly. The output is a table containing the CD data at the wavelength triplet used for the classification and the predicted results.
The Guide to CD and data analysis opens a separate window with practical and important considerations for CD spectroscopy measurements.
Cited by… opens a separate window and provides a database of scientific articles that used CD spectroscopy with BeStSel analysis. Article identifiers and keywords are provided, and the collection is searchable showing useful examples of using CD spectroscopy and BeStSel for the convenience of the users.
RESULTS AND DISCUSSION
Performance
Whereas the instrumentation of CD spectroscopy is well developed, there is a great need to efficiently extract the structural information buried in the CD spectra. By developing the BeStSel method, we could solve a general problem of structure determination by CD spectroscopy: the diversity of β-structures. By distinguishing parallel and antiparallel β-structures and three different twists of antiparallel β-sheets and including two α-helix components, the overall eight components of BeStSel provide a more accurate structural estimation for any secondary structure component than any previous method (4). A great advantage of the method is that it can be used for a reliable structure estimation of β-sheet-rich proteins, including membrane proteins, protein aggregates and amyloid fibrils. Moreover, BeStSel provides extra structural information, which is sufficient for protein fold prediction down to the topology/homology levels of CATH fold classification. In the present upgrade, BeStSel basis spectra were re-optimized by using the DSSP 3.0 algorithm to assign the major secondary structure components in the reference database. In the earlier version of DSSP, residues in π-helices might have been erroneously assigned to α-helix. Moreover, the BeStSel basis spectra were calculated for new wavelength ranges, now available at 5 nm increments starting from 175 nm. Supplementary Table S1 shows the performance compared to the previous version of BeStSel on the reference database. Overall, the accuracy of the method is improved, e.g. in the 190–250 nm wavelength range, the RMSD for α-helix estimation decreased from 0.052 to 0.042 (Supplementary Table S1). Performance was also tested on an independent set of β-sheet rich or rare protein structures and compared to other available methods for secondary structure estimation (Supplementary Table S2). Calculated to a common basis of helix, antiparallel β, parallel β, overall β-sheet and ‘turn + others’ structures, the RMSDs for secondary structure estimation were proved to be 0.034, 0.049, 0.037, 0.035 and 0.038 for BeStSel, while the other methods provided RMSDs in the ranges 0.083–0.26, 0.12–0.214, 0.076–0.198, 0.068–0.23 and 0.074–0.232, respectively. None of the previous methods performed evenly for the different secondary structure components.
The Fold prediction module was upgraded from CATH 4.2 (17) to using CATH 4.3 (5) data, resulting in a significant increase in the number of protein folds at all levels of classification. Supplementary Table S3 shows the theoretical reliability of fold prediction on the domains of CATH 4.3 (5) as secondary structure inputs in a 5-fold cross-validated manner.
A new function at the webserver is the Disordered-ordered binary classification of proteins based on their CD spectra (12). Such classifier, using experimental data has not been available yet and is highly needed by the community studying intrinsically disordered proteins (IDPs). It can be used for simple and fast experimental verification for a variety of bioinformatics tools identifying IDPs and can also facilitate the growth of experimental data in IDP databases, such as DisProt (18). The method uses the k nearest neighbors mathematical model with cosine-distance function on CD data at three wavelengths (13). Using the 197-206-233 nm wavelength triplet, the estimation error is 4.7, 1.7 and 3.9% on ordered, disordered proteins and in overall error, respectively, on the dataset of CD spectra with 190 nm wavelength cut-off (12). For a cut-off at 200 nm, using the 212–217–225 nm wavelength triplet, the error of classification is 3.3, 7.5 and 4.6%, respectively.
The functionality of the BeStSel webserver is compared to the other available online tools in Table 1. Besides its superior accuracy and the more detailed secondary structure information, the BeStSel webserver has an intelligent interface and provides useful functions for CD spectroscopy users.
Table 1.
Comparison of the functionality of the BeStSel webserver to other available online services for secondary structure estimation from the CD spectra of proteins
| BeStSel | Dichroweb(46) | CAPITO(47) | K2D2(48) | K2D3(49) | |
|---|---|---|---|---|---|
| Access without registration | • | • | • | • | |
| Text input | • | • | • | • | |
| File input | • | • | • | ||
| Input unit selection | • | • | • | ||
| Auto normalization from mdeg | • | • | |||
| Change wavelength range without resubmission | • | ||||
| Best factor | • | ||||
| Download fitted spectrum | • | •a | • | • | |
| Different reference sets | • | ||||
| Different algorithms | •b | ||||
| Multiple spectra analysis | • | • | |||
| Decompose parallel/antiparallel β-sheet and antiparallel β-sheet twist | • | ||||
| Fold recognition | • | ||||
| Disordered-ordered classification | • | •c | |||
| PDB file analysis | • | •d | |||
| Extinction coefficient calculation | • | ||||
| Similarity analysis | • |
aNot for VARSLC; bFor comparison of the performance of various algorithms to BeStSel, see Micsonai et al. (4) and Supplementary Table S1 and S2; cPlot only; dVia 2Struct server (50).
Applications
Application of CD spectroscopy in combination with BeStSel analysis covers all areas of protein science. BeStSel has been used in over 1,000 scientific studies since its first publication in 2015 (4). We made a searchable database of these works at the webserver, providing valuable examples for users. The broad applicability of BeStSel is represented by the vast variation of studies conducted using the algorithm. Some notable examples are presented below.
An increasing number of users are applying the method to investigate the effects of nanoparticle–protein interactions on protein structure. Barbalinardo et al. (19) report that lysozyme amyloid fibrils are less cytotoxic in the presence of gold nanoparticles than fibrils alone. They investigated the conformational changes of fibrils caused by gold nanoparticles. Brito et al. (20) showed that stability and gain of specific activity of bromelain protein complexes can be improved by immobilizing bromelain on gold nanoparticles. By applying CD spectroscopy, the authors observed structural changes in proteins upon binding to nanoparticles. Barbir et al. (21) studied the effects of silver nanoparticles on the structure of plasma transport proteins by CD.
There are several cases where BeStSel was applied in SARS-CoV-2-related experiments. Mycroft-West et al. (22) found that heparin alters the conformation of the SARS-CoV-2 spike protein and inhibits infection. Van Oosten et al. (23) developed a virus-like particle (VLP)-based vaccine for SARS-CoV-2 using the baculovirus—insect cell expression system. They used secondary structure analysis to compare the recombinant and the wild-type spike protein. A number of further studies involve the application of BeStSel for demonstrating that recombinant proteins have the correct secondary structure (24–27). Another purpose BeStSel is often used for is investigating the structure of antibodies (28–30).
Numerous works addressed the structural changes and β-sheet conversion or formation upon protein aggregation and amyloid formation. Kazman et al. (31) studied antibody light chain amyloid formation and explored the process of β-sheet transition from antiparallel to parallel in oligomers. Kaur et al. (32) revealed that the CarD transcription regulator from M. tuberculosis has a tendency to form amyloid-like fibrils and undergoes reversible thermal folding in solution. Do et al. (33) followed the aggregation of the functional amyloid CRES (cystatin-related epididymal spermatogenic) and pointed out that its amyloid form is rich in antiparallel β-sheets instead of the more common parallel β-sheets. Amodeo et al. (34) discovered that the c subunit of the ATP synthase is amyloidogenic and spontaneously folds into β-sheets.
There are also studies about characterizing individual proteins of interest: Bowen et al. (35) microbially produced high-performance titin polymers and used CD to examine the secondary structure and fold of the purified polymers. Balacescu et al. (36) investigated the structural behavior of apomyoglobin under different denaturing conditions. Ji et al. (37) designed a self-assembling drug delivery system and examined its target protein by CD spectroscopy.
Recently we predicted the structure of α-synuclein, an IDP associated with Parkinson's disease, by AlphaFold2 (38), which predicted 64% α-helix content. Experimental verification by CD spectroscopy and BeStSel analysis showed no α-helix content under physiological conditions. However, in the presence of 30% TFE, 47% α-helix was observed (12).
Case studies
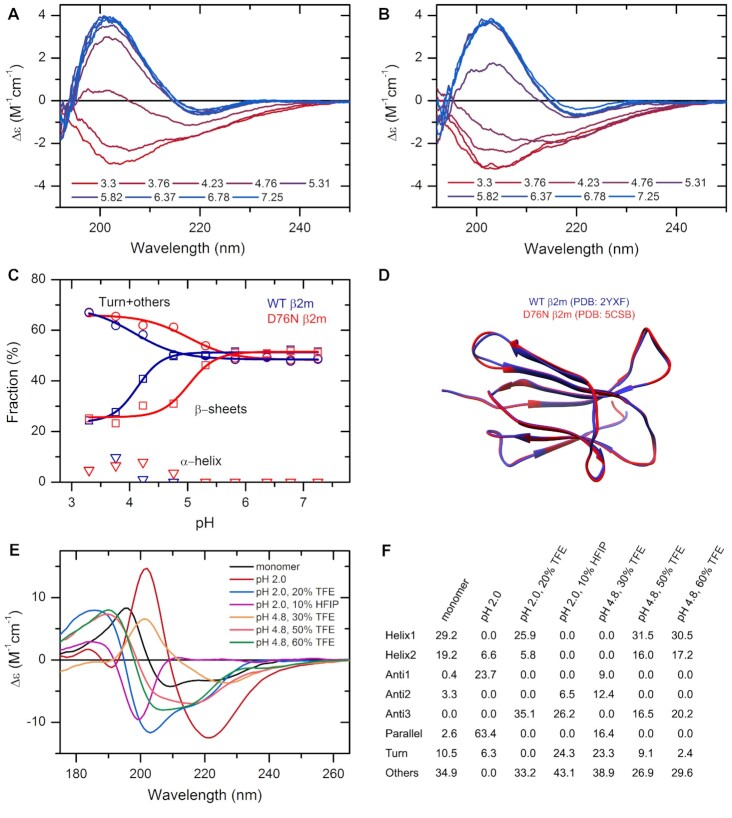
β2-Microglobulin (β2m) is the light chain of the major histocompatibility complex I. Dissociating from the complex, the protein circulates in monomeric form in the blood and is associated with dialysis related amyloidosis in long-term haemodialysis patients. In 2012, a variant of the protein carrying a D76N point mutation was discovered causing a hereditary systemic amyloidosis with pathophysiology strikingly different from that of the wild-type protein (39,40). The first investigations found that the mutant exhibits a high-resolution structure almost identical to that of the wild-type protein (Figure 2D) and its unique behavior can be discovered by using a rather complex methodology. Here, using CD spectroscopy and BeStSel analysis we show that the mutant protein exhibits increased sensitivity to a decrease in pH and below pH 6 it tends to unfold and loose its β-structured native state, which might facilitate its amyloid aggregation. The wild-type protein is more resistant to low pH as shown in Figure 2A–C.
Figure 2.
The effect of environmental conditions on the protein structure studied by CD spectroscopy and analyzed by the BeStSel web server. (A, B) CD spectra of wild type (A) and D76N mutant β2m (B) were recorded at various pH values in 10 mM Na-citrate buffer at 37°C. (C) Secondary structure contents provided by BeStSel were added up as α-helix, β-sheet and turn + others. The mutant protein (red) is more sensitive to pH drop than the wild-type one (blue) and starts to loose its β-structure below pH 6.0. CD measurements were carried out on a benchtop Jasco J-1500 spectropolarimeter (1 mm pathlength, 50 nm/min scan rate, 2 sec response time, 1 nm bandwidth, accumulation: 6). (D) The two β2m variants exhibit very similar high-resolution native structure making difficult to explain the difference in the pathology. (E) Insulin can exhibit various conformations depending on the solution conditions. Its native, monomeric state is α-helical. Under nonnative conditions, such as low pH and the presence of alcohols, it forms β-structured aggregates with different β-sheet compositions as shown in (F). At pH 2.0, it forms amyloid fibrils with characteristic parallel β-sheets. These spectra were collected by SRCD at DISCO beamline in SOLEIL Synchrotron, France.
CD spectroscopy has a great advantage in characterizing the conformation of proteins as a function of environmental conditions. High-resolution techniques cannot handle large number of samples, while in silico methods often cannot address environmental parameters properly. We studied the structure of human insulin at different pH values in the presence of additives. As revealed by the CD spectra, under native conditions insulin exhibits α-helical structure. At low pH and in the presence of TFE or HFIP, insulin forms oligomers and amyloid fibrils with various different secondary structure composition (Figure 2E, F and (41)).
NEW FEATURES
After the first release in 2015 (4), the next larger update of BeStSel was introduced in 2018 when fold prediction was improved on the basis of the CATH 4.2 and the WKNN search engine was built-in (10). The current version of the webserver uses updated background databases. The DSSP algorithm was replaced by the 3.0 version, which solved issues with assigning residues in π-helices as α-helix. The BeStSel basis spectra were re-optimized on the new assignations resulting in improved accuracy (Supplementary Tables S1-S2). Moreover, wavelength ranges for CD spectrum analysis are now available at 5 nm increments. Fold prediction was further enhanced by processing the CATH 4.3 data. The background databases are up-to-date, 184 307 PDB structures are now recognized for secondary structure analysis and fold classification. The updated single domain dataset used as a basis for fold prediction contains 61932 single domains based on CATH 4.3 covering 43 architectures, 1467 topologies and 6540 homologies. A recent addition is the binary classification of ordered-disordered structures based on the CD spectra, which helps the experimental identification of intrinsically disordered proteins (IDPs) and the verification of the results of bioinformatics tools.
One of the new features is that users can upload any 3D structures in PDB format and have the eight secondary structure components of BeStSel calculated along with DSSP and SELCON components for comparison. This is crucial for the experimental verification of MD simulation results or in silico models, such as AlphaFold2 (42) structures, making the structural comparison with the results of CD spectrum analysis possible. Another useful accessory is the extinction coefficient calculator from the amino acid sequence based on the works of Kuipers et al. (15) and Anthis et al. (16) for direct concentration determination of CD samples.
The detailed, downloadable tutorial has been updated and further improved. Information and help is provided throughout the use of the webserver.
LIMITATIONS AND FURTHER DEVELOPMENTS
The eight secondary structure components of BeStSel do not account for polyproline-II helix which is characteristic of collagen-like structures, different type of turns that are often the main structural components of short peptides, 310-helices, which appear in higher amounts in some globular proteins, and thus, analysis for such structures is not adequate. BeStSel does not treat aromatic contributions (other algorithms neither do), which might affect the results in the case of high number of aromatic residues.
For highly disordered proteins, some part of the disordered structure is counted as highly right-twisted antiparallel β-sheet (Anti3) because of the spectral similarities (4,10). When a protein or peptide is not expected to have globular structure, and the secondary structure estimation provides high Anti3 component with no or very low Anti2 content, Anti3 might be considered as disordered and added to the ‘Others’ component.
BeStSel has an advantage over the previously available methods that it is capable of estimating the β-sheet-rich structure of protein aggregates and amyloid fibrils (4). However, in case of such samples, spectral artifacts caused by differential light scattering, precipitation, or linear dichroism might affect or obstruct the accurate secondary structure analysis. Therefore, it is essential to make sure that the sample measured is a transparent, homogenous solution without large insoluble precipitates (4,43). An indication of light scattering might be when after a proper baseline subtraction there is a substantial remaining signal in the 250–260 nm wavelength region (be sure it is not nucleic acid contamination). Light scattering effects can be decreased by making the size of the aggregates or amyloid fibrils smaller by applying a slight ultrasonication on the sample and placing the cuvette close to the detector. Precipitation makes the sample inhomogeneous and results in absorption flattening (distortion and shrinking of the CD signal) (4,43,44), which makes the quantitative structure analysis impossible. Amyloid fibrils might become oriented in the cell causing linear dichroism effects (45), which can be detected by rotating the cell in the instrument.
One of the main goals for the future is to significantly increase the number of reference proteins and further improve the accuracy on β-structured proteins and IDPs.
CONCLUSIONS
The BeStSel web server provides the BeStSel method for the community to analyze protein CD spectra for secondary structure composition and protein fold prediction. The eight secondary structure components give detailed structural information from the CD spectra including the β-structure composition (orientation and twist). The method has an accuracy superior to any previously available method on any type of secondary structures. It is especially usable for β-sheet-rich proteins, protein aggregates and membrane proteins. Single and multiple CD spectra can be analyzed and the protein fold can be predicted with a few clicks. Adjustable wavelength ranges, scaling of spectra, links to corresponding PDB structures make the site a swiss-knife for CD users. A new module of the webserver helps to distinguish intrinsically disordered proteins by their CD spectrum. Secondary structure calculation for uploaded PDB files will help the experimental verification of protein MD and in silico modelling using CD spectroscopy. The server is capable of high-throughput calculations and makes the methodology of protein CD spectroscopy complete with accurate analyses at any field of protein science, structural biochemistry, biotechnology and pharmaceutical industry.
DATA AVAILABILITY
The datasets generated for this study are available on request to the corresponding author. The BeStSel web server is freely accessible at https://bestsel.elte.hu.
Supplementary Material
Contributor Information
András Micsonai, ELTE NAP Neuroimmunology Research Group, Department of Biochemistry, Institute of Biology, ELTE Eötvös Loránd University, Budapest H-1117, Hungary.
Éva Moussong, ELTE NAP Neuroimmunology Research Group, Department of Biochemistry, Institute of Biology, ELTE Eötvös Loránd University, Budapest H-1117, Hungary.
Frank Wien, Synchrotron SOLEIL, Gif-sur-Yvette 91192, France.
Eszter Boros, Department of Biochemistry, Institute of Biology, ELTE Eötvös Loránd University, Budapest H-1117, Hungary.
Henrietta Vadászi, ELTE NAP Neuroimmunology Research Group, Department of Biochemistry, Institute of Biology, ELTE Eötvös Loránd University, Budapest H-1117, Hungary.
Nikoletta Murvai, Department of Biochemistry, Institute of Biology, ELTE Eötvös Loránd University, Budapest H-1117, Hungary; Institute of Enzymology, Research Centre for Natural Sciences, Budapest H-1117, Hungary.
Young-Ho Lee, Research Center of Bioconvergence Analysis, Korea Basic Science Institute (KBSI), Ochang 28119, Republic of Korea; Bio-Analytical Science, University of Science and Technology (UST), Daejeon 34113, Republic of Korea; Graduate School of Analytical Science and Technology (GRAST), Chungnam National University (CNU), Daejeon 34134, Republic of Korea.
Tamás Molnár, ELTE NAP Neuroimmunology Research Group, Department of Biochemistry, Institute of Biology, ELTE Eötvös Loránd University, Budapest H-1117, Hungary.
Matthieu Réfrégiers, Synchrotron SOLEIL, Gif-sur-Yvette 91192, France; Centre de Biophysique Moléculaire, CNRS UPR4301, Orléans, France.
Yuji Goto, Global Center for Medical Engineering and Informatics, Osaka University, Osaka 565-0871, Japan.
Ágnes Tantos, Institute of Enzymology, Research Centre for Natural Sciences, Budapest H-1117, Hungary.
József Kardos, ELTE NAP Neuroimmunology Research Group, Department of Biochemistry, Institute of Biology, ELTE Eötvös Loránd University, Budapest H-1117, Hungary.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research, Development and Innovation Fund of Hungary [K120391, K138937, K125340, PD135510, 2017-1.2.1-NKP-2017-00002]; International Collaboration [2019-2.1.11-TÉT-2019-00079, 2018-2.1.17-TÉT-KR-2018-00008, 2019-2.1.6-NEMZ_KI-2019-00012, 2019-2.1.11-TÉT-2020-00101]; SOLEIL Synchrotron, France [20181890, 20191810, 20200751]; Institute for Protein Research, Osaka University; Japan Society for the Promotion of Science, Core-to-Core Program A (Advanced Research Networks to Y.G.). Funding for open access charge: National Research, Development and Innovation Fund of Hungary [K120391, K138937, PD 135510].
Conflict of interest statement. None declared.
REFERENCES
- 1. Wallace B.A. Synchrotron radiation circular-dichroism spectroscopy as a tool for investigating protein structures. J. Synchrotron Radiat. 2000; 7:289–295. [DOI] [PubMed] [Google Scholar]
- 2. Greenfield N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006; 1:2876–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khrapunov S. Circular dichroism spectroscopy has intrinsic limitations for protein secondary structure analysis. Anal. Biochem. 2009; 389:174–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Micsonai A., Wien F., Kernya L., Lee Y.H., Goto Y., Refregiers M., Kardos J.. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3095–E3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sillitoe I., Bordin N., Dawson N., Waman V.P., Ashford P., Scholes H.M., Pang C.S.M., Woodridge L., Rauer C., Sen N.et al.. CATH: increased structural coverage of functional space. Nucleic Acids Res. 2021; 49:D266–D273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cooley R.B., Arp D.J., Karplus P.A.. Evolutionary origin of a secondary structure: pi-helices as cryptic but widespread insertional variations of alpha-helices that enhance protein functionality. J. Mol. Biol. 2010; 404:232–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kabsch W., Sander C.. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983; 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 8. Orengo C.A., Michie A.D., Jones S., Jones D.T., Swindells M.B., Thornton J.M.. CATH–a hierarchic classification of protein domain structures. Structure. 1997; 5:1093–1108. [DOI] [PubMed] [Google Scholar]
- 9. Dudani S.A. The distance-weighted k-nearest-neighbor rule. IEEE Trans. Syst. Man Cybern. 1976; SMC-6:325–327. [Google Scholar]
- 10. Micsonai A., Wien F., Bulyaki E., Kun J., Moussong E., Lee Y.H., Goto Y., Refregiers M., Kardos J.. BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018; 46:W315–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Whitmore L., Woollett B., Miles A.J., Klose D.P., Janes R.W., Wallace B.A.. PCDDB: the protein circular dichroism data bank, a repository for circular dichroism spectral and metadata. Nucleic Acids Res. 2011; 39:D480–D486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Micsonai A., Moussong E., Murvai N., Tantos A., Toke O., Réfrégiers M., Wien F., Kardos J.. Disordered-ordered protein binary classification by circular dichroism spectroscopy. Front. Mol. Biosci. 2022; 863141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Manning C.D., Raghavan P., Schütze H.. Introduction to Information Retrieval. 2008; Cambridge: Cambridge University Press. [Google Scholar]
- 14. Sreerama N., Venyaminov S.Y., Woody R.W.. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999; 8:370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuipers B.J., Gruppen H.. Prediction of molar extinction coefficients of proteins and peptides using UV absorption of the constituent amino acids at 214 nm to enable quantitative reverse phase high-performance liquid chromatography-mass spectrometry analysis. J. Agric. Food Chem. 2007; 55:5445–5451. [DOI] [PubMed] [Google Scholar]
- 16. Anthis N.J., Clore G.M.. Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm. Protein Sci. 2013; 22:851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sillitoe I., Lewis T.E., Cuff A., Das S., Ashford P., Dawson N.L., Furnham N., Laskowski R.A., Lee D., Lees J.G.et al.. CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 2015; 43:D376–D381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Quaglia F., Meszaros B., Salladini E., Hatos A., Pancsa R., Chemes L.B., Pajkos M., Lazar T., Pena-Diaz S., Santos J.et al.. DisProt in 2022: improved quality and accessibility of protein intrinsic disorder annotation. Nucleic Acids Res. 2021; 50:D480–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barbalinardo M., Antosova A., Gambucci M., Bednarikova Z., Albonetti C., Valle F., Sassi P., Latterini L., Gazova Z., Bystrenova E.. Effect of metallic nanoparticles on amyloid fibrils and their influence to neural cell toxicity. Nano Res. 2020; 13:1081–1089. [Google Scholar]
- 20. Brito A.M.M., Oliveira V., Icimoto M.Y., Nantes-Cardoso I.L.. Collagenase activity of bromelain immobilized at gold nanoparticle interfaces for therapeutic applications. Pharmaceutics. 2021; 11:810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barbir R., Capjak I., Crnkovic T., Debeljak Z., Domazet Jurasin D., Curlin M., Sinko G., Weitner T., Vinkovic Vrcek I.. Interaction of silver nanoparticles with plasma transport proteins: a systematic study on impacts of particle size, shape and surface functionalization. Chem. Biol. Interact. 2021; 335:109364. [DOI] [PubMed] [Google Scholar]
- 22. Mycroft-West C.J., Su D., Pagani I., Rudd T.R., Elli S., Gandhi N.S., Guimond S.E., Miller G.J., Meneghetti M.C.Z., Nader H.B.et al.. Heparin inhibits cellular invasion by SARS-CoV-2: structural dependence of the interaction of the spike S1 receptor-binding domain with heparin. Thromb. Haemost. 2020; 120:1700–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Oosten L., Altenburg J.J., Fougeroux C., Geertsema C., van den End F., Evers W.A.C., Westphal A.H., Lindhoud S., van den Berg W., Swarts D.C.et al.. Two-Component nanoparticle vaccine displaying glycosylated spike S1 domain induces neutralizing antibody response against SARS-CoV-2 variants. mBio. 2021; 12:e0181321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kibria M.G., Fukutani A., Akazawa-Ogawa Y., Hagihara Y., Kuroda Y.. Anti-EGFR VHH antibody under thermal stress is better solubilized with a lysine than with an arginine SEP tag. Biomolecules. 2021; 11:810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brindha S., Kibria M.G., Saotome T., Unzai S., Kuroda Y.. EGFR extracellular domain III expressed in escherichia coli with SEP tag shows improved biophysical and functional properties and generate anti-sera inhibiting cancer cell growth. Biochem. Biophys. Res. Commun. 2021; 555:121–127. [DOI] [PubMed] [Google Scholar]
- 26. Bortnov V., Tonelli M., Lee W., Lin Z., Annis D.S., Demerdash O.N., Bateman A., Mitchell J.C., Ge Y., Markley J.L.et al.. Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum. Nat. Commun. 2019; 10:5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Anathy V., Lahue K.G., Chapman D.G., Chia S.B., Casey D.T., Aboushousha R., van der Velden J.L.J., Elko E., Hoffman S.M., McMillan D.H.et al.. Reducing protein oxidation reverses lung fibrosis. Nat. Med. 2018; 24:1128–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eliseev I.E., Ukrainskaya V.M., Yudenko A.N., Mikushina A.D., Shmakov S.V., Afremova A.I., Ekimova V.M., Vronskaia A.A., Knyazev N.A., Shamova O.V.. Targeting erbb3 receptor in cancer with inhibitory antibodies from llama. Biomedicines. 2021; 9:1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dash R., Rathore A.S.. Freeze thaw and lyophilization induced alteration in mAb therapeutics: trastuzumab as a case study. J. Pharm. Biomed. Anal. 2021; 201:114122. [DOI] [PubMed] [Google Scholar]
- 30. Karch C.P., Bai H., Torres O.B., Tucker C.A., Michael N.L., Matyas G.R., Rolland M., Burkhard P., Beck Z.. Design and characterization of a self-assembling protein nanoparticle displaying HIV-1 env V1V2 loop in a native-like trimeric conformation as vaccine antigen. Nanomedicine. 2019; 16:206–216. [DOI] [PubMed] [Google Scholar]
- 31. Kazman P., Absmeier R.M., Engelhardt H., Buchner J.. Dissection of the amyloid formation pathway in AL amyloidosis. Nat. Commun. 2021; 12:6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaur G., Kaundal S., Kapoor S., Grimes J.M., Huiskonen J.T., Thakur K.G.. Mycobacterium tuberculosis CarD, an essential global transcriptional regulator forms amyloid-like fibrils. Sci. Rep. 2018; 8:10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Do H.Q., Hewetson A., Myers C., Khan N.H., Hastert M.C., F M.H., Latham M.P., Wylie B.J., Sutton R.B., Cornwall G.A. The functional mammalian CRES (Cystatin-Related epididymal spermatogenic) amyloid is antiparallel beta-Sheet rich and forms a metastable oligomer during assembly. Sci. Rep. 2019; 9:9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amodeo G.F., Lee B.Y., Krilyuk N., Filice C.T., Valyuk D., Otzen D.E., Noskov S., Leonenko Z., Pavlov E.V.. C subunit of the ATP synthase is an amyloidogenic calcium dependent channel-forming peptide with possible implications in mitochondrial permeability transition. Sci. Rep. 2021; 11:8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bowen C.H., Sargent C.J., Wang A., Zhu Y., Chang X., Li J., Mu X., Galazka J.M., Jun Y.S., Keten S.et al.. Microbial production of megadalton titin yields fibers with advantageous mechanical properties. Nat. Commun. 2021; 12:5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Balacescu L., Schrader T.E., Radulescu A., Zolnierczuk P., Holderer O., Pasini S., Fitter J., Stadler A.M.. Transition between protein-like and polymer-like dynamic behavior: internal friction in unfolded apomyoglobin depends on denaturing conditions. Sci. Rep. 2020; 10:1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ji T., Li Y., Deng X., Rwei A.Y., Offen A., Hall S., Zhang W., Zhao C., Mehta M., Kohane D.S.. Delivery of local anaesthetics by a self-assembled supramolecular system mimicking their interactions with a sodium channel. Nat. Biomed. Eng. 2021; 5:1099–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A.et al.. Highly accurate protein structure prediction with alphafold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bulyaki E., Kun J., Molnar T., Papp A., Micsonai A., Vadaszi H., Marialigeti B., Kovacs A.I., Gellen G., Yamaguchi K.et al.. Pathogenic D76N variant of beta2-Microglobulin: synergy of diverse effects in both the native and amyloid states. Biology (Basel). 2021; 10:1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Valleix S., Gillmore J.D., Bridoux F., Mangione P.P., Dogan A., Nedelec B., Boimard M., Touchard G., Goujon J.M., Lacombe C.et al.. Hereditary systemic amyloidosis due to asp76asn variant beta2-microglobulin. N. Engl. J. Med. 2012; 366:2276–2283. [DOI] [PubMed] [Google Scholar]
- 41. Muta H., Lee Y.H., Kardos J., Lin Y., Yagi H., Goto Y.. Supersaturation-limited amyloid fibrillation of insulin revealed by ultrasonication. J. Biol. Chem. 2014; 289:18228–18238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Zidek A., Bridgland A., Cowie A., Meyer C., Laydon A.et al.. Highly accurate protein structure prediction for the human proteome. Nature. 2021; 596:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Micsonai A., Bulyaki E., Kardos J.. BeStSel: from secondary structure analysis to protein fold prediction by circular dichroism spectroscopy. Methods Mol. Biol. 2021; 2199:175–189. [DOI] [PubMed] [Google Scholar]
- 44. Wallace B.A., Teeters C.L.. Differential absorption flattening optical effects are significant in the circular dichroism spectra of large membrane fragments. Biochemistry. 1987; 26:65–70. [DOI] [PubMed] [Google Scholar]
- 45. Wallace B.A. Protein characterisation by synchrotron radiation circular dichroism spectroscopy. Q. Rev. Biophys. 2009; 42:317–370. [DOI] [PubMed] [Google Scholar]
- 46. Lobley A., Whitmore L., Wallace B.A.. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002; 18:211–212. [DOI] [PubMed] [Google Scholar]
- 47. Wiedemann C., Bellstedt P., Gorlach M.. CAPITO–a web server-based analysis and plotting tool for circular dichroism data. Bioinformatics. 2013; 29:1750–1757. [DOI] [PubMed] [Google Scholar]
- 48. Perez-Iratxeta C., Andrade-Navarro M.A.. K2D2: estimation of protein secondary structure from circular dichroism spectra. BMC Struct. Biol. 2008; 8:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Louis-Jeune C., Andrade-Navarro M.A., Perez-Iratxeta C.. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins. 2012; 80:374–381. [DOI] [PubMed] [Google Scholar]
- 50. Klose D.P., Wallace B.A., Janes R.W.. 2Struc: the secondary structure server. Bioinformatics. 2010; 26:2624–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author. The BeStSel web server is freely accessible at https://bestsel.elte.hu.