Abstract
Standardized monitoring of BCR::ABL1 mRNA levels is essential for the management of chronic myeloid leukemia (CML) patients. From 2016 to 2021 the European Treatment and Outcome Study for CML (EUTOS) explored the use of secondary, lyophilized cell-based BCR::ABL1 reference panels traceable to the World Health Organization primary reference material to standardize and validate local laboratory tests. Panels were used to assign and validate conversion factors (CFs) to the International Scale and assess the ability of laboratories to assess deep molecular response (DMR). The study also explored aspects of internal quality control. The percentage of EUTOS reference laboratories (n = 50) with CFs validated as optimal or satisfactory increased from 67.5% to 97.6% and 36.4% to 91.7% for ABL1 and GUSB, respectively, during the study period and 98% of laboratories were able to detect MR4.5 in most samples. Laboratories with unvalidated CFs had a higher coefficient of variation for BCR::ABL1IS and some laboratories had a limit of blank greater than zero which could affect the accurate reporting of DMR. Our study indicates that secondary reference panels can be used effectively to obtain and validate CFs in a manner equivalent to sample exchange and can also be used to monitor additional aspects of quality assurance.
Subject terms: Oncogenesis, Prognosis
Introduction
Molecular monitoring of chronic myeloid leukemia (CML) patients undergoing tyrosine kinase inhibitor (TKI) therapy provides important prognostic information for individual patients and is used to assess time-dependent treatment milestones, including early molecular response (EMR), major molecular response (MMR), and deep molecular response (DMR) [1, 2]. Molecular monitoring is usually performed using reverse transcriptase quantitative PCR (RT-qPCR), which estimates the number of copies of BCR::ABL1 mRNA relative to those of an internal reference gene, most commonly ABL1, GUSB, or BCR, thus controlling for variation in sample quality and quantity [3, 4]. Current guidelines specify that assay results should be expressed on the International Scale (IS) for BCR::ABL1 measurement, which is effectively the same as that used in the International Randomized Study of Interferon and STI571 (IRIS). On this scale, 100% BCR::ABL1IS corresponds to the IRIS standardized baseline derived from analysis of 30 pre-treatment chronic phase CML cases. [5] EMR is defined as ≤10% BCR::ABL1IS, MMR (also known as MR3, i.e., a molecular response of ≥3 logs below the standardized baseline) as ≤0.1% BCR::ABL1IS, and levels ≤0.01% (MR4) as DMR. [3] Testing laboratories derive results on the IS either by using commercially available kits or systems that have been calibrated to the World Health Organization (WHO) International Genetic Reference Panel for quantitation of BCR::ABL1 mRNA, or by using a laboratory-developed test (LDT) in conjunction with a laboratory-specific conversion factor (CF) to the IS derived by sample exchange [4, 6–12].
Sample exchange typically involves testing around 30 CML patient samples spanning the range from MR2-MR4.5 (i.e., 2–4.5 logs below the IRIS standardized baseline) in both an established reference laboratory and a test laboratory followed by calculation of the mean difference by Bland–Altman analysis. The CF is then defined as the multiplication factor needed to correct for the difference [13]. This process has enabled many laboratories with validated CFs to establish themselves as national or regional reference laboratories and then repeat the process of sample exchange, thus propagating CFs to local centers [14]. Although this has worked well for laboratories with tests that are stable over time, it is evident that the establishment and validation of CFs by sample exchange is time consuming, complex, expensive, and can be difficult for smaller laboratories to access [7, 15].
In 2010, the first International Genetic Reference Panel for quantitation of BCR::ABL1 mRNA was developed as a primary, WHO-accredited standard for IS calibration [8]. The panel is made of lyophilized K562 and HL60 cell line mixtures and therefore incorporates cellular RNA extraction into the IS calibration process. The panel includes four BCR::ABL1IS levels, with different values assigned to each depending on whether ABL1, BCR, or GUSB is used as a reference genes. To conserve this limited resource, the WHO panel is only available to manufacturers of BCR::ABL1 test kits and secondary standards [15]. In 2016, the first cell-based BCR::ABL1 secondary reference panel was produced. This is traceable to the WHO panel and has been produced using a similar format (lyophilized K562 and HL60 cell mixes) with the addition of a fifth sample corresponding to MR4.5. BCR::ABL1IS values were assigned to the secondary panel using reverse-transcription droplet digital PCR (RT-ddPCR) with reference to ABL1, BCR, and GUSB and the panel was successfully evaluated by 44 different BCR::ABL1 laboratories [12]. Recently this panel has been commercialized and is now available for laboratories using ABL1 as a reference gene (AcroMetrix™ BCR-ABL Panel, Thermo Fisher Scientific).
In addition to accurate measurement of detectable residual disease, it is also important to ensure that assays are sensitive enough to detect DMR on a routine basis. Many CML patients achieve sustained (>2 years) DMR on TKI therapy and around half remain in treatment-free remission (TFR) after stopping therapy [2, 16]. Standardization of molecular monitoring at deep levels of response is particularly important, not only to meet the recommended criteria for attempting TFR, but also to identify patients showing signs of molecular relapse, for whom DMR is usually regained after rapid resumption of treatment [17].
To maintain confidence in a CF, ensure that BCR::ABL1 and reference gene assays are stable over time, and monitor the ability of assays to detect DMR, testing laboratories need to perform rigorous internal quality control (IQC) and validate their CF regularly. IQC is important to monitor variation in assay performance over time and ensure that low level BCR::ABL1 detection is achieved consistently [18]. Branford et al. have recommended the analysis of high (c. 10% BCR::ABL1IS) and low (c. 0.1% BCR::ABL1IS) standards on a regular basis, and ideally on every run to check that BCR::ABL1 and reference gene assays are stable over time [19, 20].
Given the increased technical sensitivity required for low level BCR::ABL1 detection, a better understanding of the limits of BCR::ABL1 assay performance is crucial [21]. The limit of detection (LoD) and limit of quantification (LoQ) of a qPCR test is dependent on the background signal (the limit of blank; LoB), which ideally should be zero. Current BCR::ABL1 RT-qPCR molecular response (MR) guidelines assume that all laboratories are able to detect BCR::ABL1 with maximal efficiency [17], but this has never been formally tested and it is possible that differences in LoB and LoD for BCR::ABL1 assays between laboratories result in variation in the way that MR is reported [22].
From 2016 to 2021 the European Treatment and Outcomes Study (EUTOS) for CML has explored the use of the newly available cell-based secondary BCR::ABL1 reference panels to assign and validate CFs for testing laboratories. In addition, the ability of laboratories to detect MR4.5 reliably was assessed and approaches to IQC were explored. Here, we present the results of this study and EUTOS recommendations for ongoing standardization of molecular monitoring for CML using RT-qPCR.
Methods
Ability of laboratories to reliably detect MR4.5
From 2016–2021 three batches of nine samples were distributed annually (5 distributions) from the Wessex Regional Genetics Laboratory, Salisbury to EUTOS reference laboratories who agreed to participate (2016, n = 49; 2017, n = 48; 2019, n = 50; 2020, n = 49; 2021, n = 49). Three samples consisted of locally-prepared HL60/K562 cell line mixtures (5 × 105 cells/vial) at approximately 10%, 0.1% and 0.0032% (DMR cell line lysate) BCR::ABL1IS lysed in either Trizol (Thermo Fisher Scientific, Waltham, Massachusetts, USA), RLT (QIAGEN, Hilden, Germany) or Promega Homogenization Solution containing 1-Thioglycerol (Promega, Madison, Wisconsin, USA) according to the preferred RNA extraction method of each center. Plasmid DNA samples (ERMAD623 BCR-ABL pDNA calibrant, Sigma, St. Louis, Missouri, USA) were supplied as a mock “cDNA sample”. Each plasmid sample contained identical and precisely defined ABL1, GUSB, and BCR::ABL1 copy numbers [11] and were used to establish whether ABL1, GUSB, and BCR::ABL1 RT-qPCR assays were performing with equal efficiency. Plasmid samples with different copy numbers were provided for each annual round of testing. Secondary cell-based reference material panels were provided and were composed of five vials of lyophilized cells (HL60/K562) spanning the range 10% - 0.0032% BCR::ABL1IS and supplied by Novartis Pharmaceuticals Corporation (2016–2019) [12] or Thermo Fisher Scientific (AcroMetrix™ BCR-ABL Panel, 2020, 2021). Both secondary panels have BCR::ABL1IS values assigned for the reference gene ABL1 and the Novartis panel also had BCR::ABL1IS values assigned for the reference gene GUSB. To enable the AcroMetrix™ BCR-ABL Panel to be used to assign CFs to laboratories using GUSB as a reference gene, BCR::ABL1IS values were assigned to the batch by calibrating the reagents with the WHO panel at the laboratory in Wessex [8].
All samples were tested using RT-qPCR using standard laboratory protocols following the process shown in Supplementary Fig. 1. To monitor the quality of local routine samples, anonymized reference gene transcript copy numbers were collected for 50 local samples analysed at each laboratory over a 4-week audit period.
Derivation of conversion factors and monitoring stability over time
CFs were determined using laboratory BCR::ABL1 results from the secondary reference lyophilized cell line panels using the method described at https://www.nibsc.org/documents/ifu/09-138.pdf; (included in the Supplementary Information along with a CF calculation spreadsheet). The stability of laboratory CFs was assigned annually using the following criteria, which were based on the previously described definition of optimal performance (+/− 1.2 fold difference from reference method) [7], and the observed mean standard deviation in the initial international assessment of the freeze dried cell secondary reference panel (0.2 log/1.6 fold) [12].
Optimal (+/− 1.2 fold): Previous panel CF/New panel CF = 0.83–1.2
Satisfactory (+/− 1.6 fold): Previous panel CF/New panel CF = 0.63–1.58
Unvalidated: Previous panel CF/New panel CF < 0.63 or >1.58
The unvalidated category also included new laboratory assays where there was no existing CF for comparison.
Monitoring of internal quality control and correlation with stability of conversion factors
We aimed to measure variation in assay performance over time for individual laboratories and assess how this correlated with stability of CFs. We prepared high and low internal quality control standards by making mixtures of HL60 and K562 cell lines (see Supplementary Information) which were stored and distributed as lysates in either Trizol (Thermo Fisher Scientific), RLT (QIAGEN), or Homogenization Solution containing 1-Thioglycerol (Promega). These standards had BCR::ABL1IS values of approximately 5% (high level control) and 0.05% (low level control). Participants were asked to use their established protocols to extract RNA from both controls on a monthly basis, prepare two independent cDNA samples and test by RT-qPCR. Each laboratory submitted a minimum of 12 results from both high- and low-level controls over the 6-month period of the study. Data were submitted for reference gene transcript number, BCR::ABL1 transcript number, %BCR::ABL1/reference gene and BCR::ABL1IS for each IQC sample type. Six batches of high- and low-level control samples were distributed to 46 laboratories and 43 data sets were returned from 41 laboratories at the completion of the study (89%).
Limit of blank for BCR::ABL1 RT-qPCR assays
We aimed to determine the LoB for BCR::ABL1 RT-qPCR in a subset of experienced molecular monitoring laboratories (n = 12). The LoB is defined as the highest measurement result that is likely to be observed for a negative sample i.e., the likelihood of reporting a false positive BCR::ABL1 result at a defined probability (α). When α = 0.95, the likelihood of a true negative sample giving a result greater than zero (false positive result) is 5%. To determine the LoB, the Clinical and Laboratory Standards Institute guidelines recommend the following minimum requirements: test four negative samples, using two reagent lots of qPCR master mix, on one instrument, on three independent days, analysing two replicates per sample, generating 60 blank replicate results per reagent lot [21]. Prior to the study, a pre-study questionnaire was sent to all laboratories to determine sample requirements (lysis type for subsequent RNA extraction and volume). Fresh (<48 h), 4 ml non-leukemic peripheral blood samples (n = 360) were processed and pooled to generate BCR::ABL1 negative lysates with sufficient ABL1 copies (Trizol n = 4, RLT n = 4). BCR::ABL1 negative samples (n = 4) were provided to each participating laboratory. After local RNA extraction and cDNA synthesis, 18 RT-qPCR replicates (15x BCR::ABL1, 3x ABL1) were performed per sample, per reagent lot using their local standard protocols. Four BCR::ABL1 negative samples were analysed using two reagent lots of RT-qPCR master mix, on one instrument, on three independent days, analysing two replicates per sample. This generated 144 individual RT-qPCRs in total; 60 BCR::ABL1 and 12 ABL1 replicates for each reagent lot (Supplementary Fig. 2). To calculate the LoB for each reagent lot, the BCR::ABL1 copy number measurements of all samples were ranked in order from lowest to highest X(1), X(2), …, X(60). The rank position corresponding to the chosen value of α was calculated using the equation: “Rank position = 0.5 + (B x α)” where B is the number of replicates and α was 0.95. For most laboratories, the rank position was assigned as 57.5 (B = 60). The LoB was the highest measurement value of the sample at the given rank position across both lots.
Results
Ability of laboratories to detect MR4.5
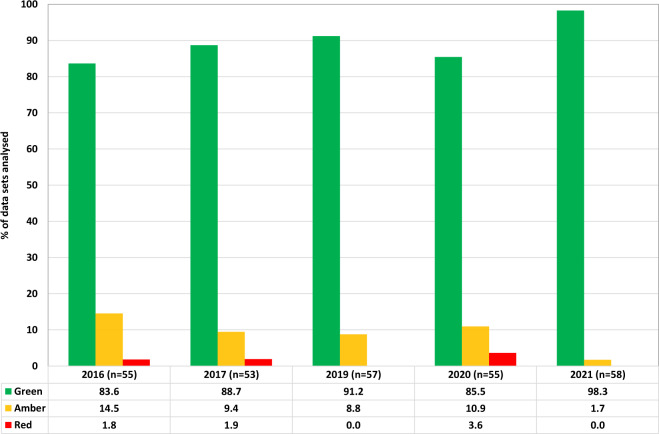
Analysis of information collected from participating EUTOS reference laboratories showed that there is substantial variation in the methodology used to perform molecular monitoring for CML. Laboratories used different RNA extraction methods, reference genes, PCR machines and RT-qPCR methods (Supplementary Table 1). To assess whether individual laboratories could reliably detect MR4.5, data from all test samples were analysed according to five categories of relevant technical measures: (i) median number of reference gene transcripts reported for cell line lysates, (ii) detection of BCR::ABL1 in the DMR cell line lysates and MR4.5 freeze dried reference panel samples, (iii) reference gene and BCR::ABL1 transcript numbers per µl cDNA, (iv) %BCR::ABL1/reference gene for cDNA sample and (v) quality of reference gene audit data. Each category was scored and arbitrarily weighted according to the perceived relevance of each component: cell line results and MR4.5 detection> reference gene copy number audit data >cDNA transcript values and cDNA ratio (see Supplementary Table 2 for more details). Combined scores were calculated and an overall laboratory score per reference gene was defined as green (detects MR4.5 in a high proportion of samples, combined score >80%), amber (detects MR4.5 in most samples, combined score >60%) or red (unable to detect MR4.5 in most samples, combined score <60%) as detailed in Supplementary Table 2. The number of data sets in each category, per year, per reference gene are shown in Fig. 1. Several laboratories submitted data for more than one reference gene or assay and therefore the number of data sets analysed is greater than the number of participating laboratories. The categories for cDNA transcript values and cDNA ratios were not scored during the 2021 round due to technical issues. Due to the small sample size and variability of assay conditions it was not possible to observe any significant differences in performance between platform or lysate type.
Fig. 1. Ability of laboratories to detect MR4.5.
Overall laboratory scores per reference gene were defined as green (detects MR4.5 in a high proportion of samples, combined score > 80%), amber (detects MR4.5 in most samples, combined score > 60%) or red (unable to detect MR4.5 in most samples, combined score < 60%). The bar charts show the number of data sets in each category for all laboratories. Several laboratories submitted data for more than one reference gene or assay and therefore the number of data sets analysed is greater than the number of participating laboratories.
Provision of conversion factors and monitoring stability over time
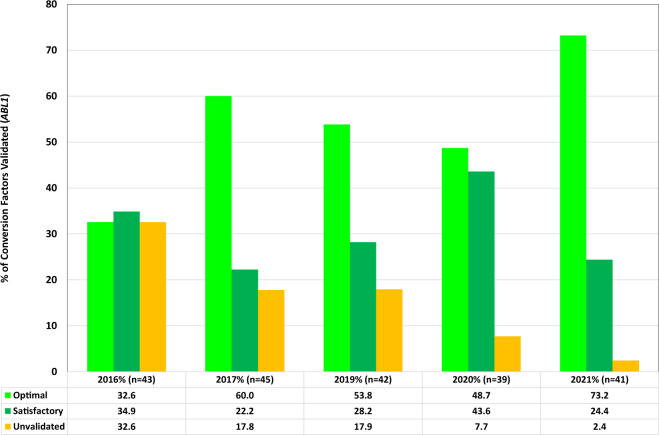
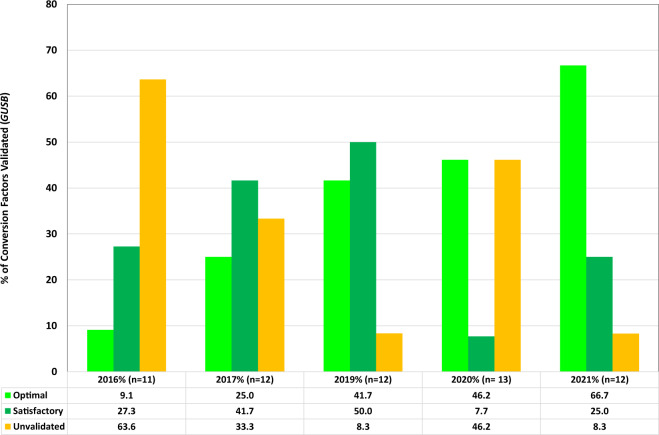
CFs were calculated and provided to laboratories on an annual basis for RT-qPCR assays using ABL1 and GUSB as reference genes. The stability of each CF was determined as either optimal, satisfactory or unvalidated by comparison with the previous year’s CF. At the start of the study, laboratories supplied the CF that they were currently using to report BCR::ABL1IS in their laboratory (n = 49). Where information was provided (n = 41), the laboratory-specific CFs had been obtained using sample exchange from 2014–2016 (93%) or 2012–2013 (7%). Figures 2 and 3 show the number of laboratories for each category, per year, for ABL1 and GUSB reference gene data sets, respectively. Several laboratories submitted data for more than one reference gene or assay and therefore the number of data sets analysed is greater than the number of participating laboratories. The mean, median, maximum, and minimum laboratory CFs for each reference gene per year are shown in Supplementary Table 3. The median CF value from data sets submitted over the course of the study were 0.604 for ABL1 (interquartile range (IQR) = 0.480–0.780, n = 213) and 1.576 for GUSB (IQR = 1.16–2.29, n = 70) (Supplementary Fig. 3, Supplementary Table 3). This compares to median CFs of 0.563 for ABL1 (IQR = 0.37–0.81, n = 245) and 0.960 for GUSB (IQR = 0.68–1.34, n = 44) for CFs derived by the EUTOS sample exchange program between 2006 and 2016.
Fig. 2. Stability of CFs for laboratories using ABL1 as a reference gene.
CFs were calculated and provided to laboratories on an annual basis. The stability of each CF was determined as either optimal (bright green), satisfactory (green) or unvalidated (amber) by comparison with the previous year’s value using the following criteria; Optimal (+/− 1.2 fold): Old CF/New CF = 0.83–1.2, Satisfactory (+/− 1.6 fold): Old CF/New CF = 0.63–1.58 or Unvalidated: Old CF/New CF < 0.63 or >1.58. The bars charts show the number of laboratories for each category, per year for ABL1 reference gene data sets. Several laboratories submitted data for more than one assay and therefore the number of data sets analysed may be greater than the number of participating laboratories.
Fig. 3. Stability of CFs for laboratories using GUSB as a reference gene.
CFs were calculated and provided to laboratories on an annual basis. The stability of each CF was determined as either optimal (bright green), satisfactory (green) or unvalidated (amber) by comparison with the previous year’s value using the following criteria; Optimal (+/− 1.2 fold): Old CF/New CF = 0.83–1.2, Satisfactory (+/− 1.6 fold): Old CF/New CF = 0.63–1.58 or Unvalidated: Old CF/New CF < 0.63 or >1.58. The bars charts show the number of laboratories for each category, per year for GUSB reference gene data sets. Several laboratories submitted data for more than one reference gene or assay and therefore the number of data sets analysed may be greater than the number of participating laboratories.
To assess whether the CFs were converting data to the IS reliably, the raw data (%BCR::ABL1/reference gene) from each laboratory were converted to the IS using the newly derived CF for the three test samples. For example, in the 2017 round, 72.3% of results were reported within 2-fold of the expected IS value when no conversion factor was applied. This increased to 95.5% of results when data were converted to BCR::ABL1IS using the newly derived CF (Supplementary Table 4). Similar results were seen for all rounds.
Use of internal quality control material
For the high and low-level standards, the CV was calculated for BCR::ABL1IS, total reference gene transcript values and BCR::ABL1 transcript values for each laboratory. The median, 1st quartile and 3rd quartile CVs for each laboratory and for each parameter are summarized in Table 1.
Table 1.
1st quartile, median, and 3rd quartile for the CV (%) values calculated per laboratory for BCR::ABL1IS, reference gene copy number, BCR::ABL1 copy number for the high and low standard.
| High Level IQC Sample CV (%) | Low Level IQC Sample CV (%) | ||
|---|---|---|---|
| BCR::ABL1IS | 1st quartile | 9.7 | 14.6 |
| Median | 14.3 | 21.1 | |
| 3rd quartile | 22.5 | 28.9 | |
| Reference gene copies | 1st quartile | 21.8 | 22.9 |
| Median | 28.2 | 28.2 | |
| 3rd quartile | 38.3 | 35.4 | |
| BCR::ABL1 copies | 1st quartile | 25.1 | 26.8 |
| Median | 31.0 | 33.3 | |
| 3rd quartile | 38.7 | 45.6 | |
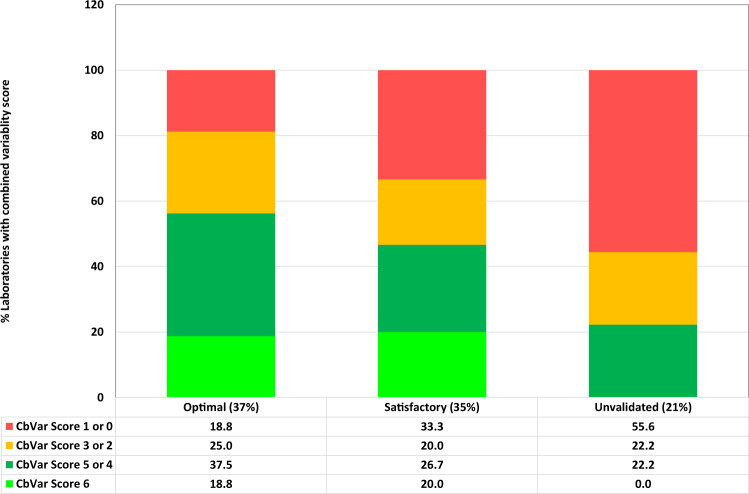
Overall, the degree of variability for BCR::ABL1IS was comparable to that seen in a previous study [20]. CVs for BCR::ABL1IS determination were used to assess how assay variability might correlate with CF status (optimal, satisfactory, or unvalidated) using data for 2019/2020 since this corresponded to the period when the variability data was collected. The stability of a CF is likely to be affected by variability in assessment of both the high and low standard and therefore we assigned a combined “variability score” using the following criteria:
3 points: CV < 1st quartile
2 points: CV between 1st quartile and median
1 point: CV between median and 3rd quartile
0 points: CV > 3rd quartile
Variability Score (CbVar) = score high level standard + score for low level standard.
The data obtained (Fig. 4) shows that 56% of laboratories with unvalidated conversion factors had red variability scores compared to only 19% of optimal laboratories. Overall, there is a clear relationship between variability and CF stability and therefore the BCR::ABL1IS CV of IQC samples is an important quality control metric for laboratories to record routinely.
Fig. 4. Use of IQC material to assess how CFs correlate with assay variability.
CVs for BCR::ABL1IS results from high and low level internal quality control material were used to assess how assay variability might correlate with CF status (optimal, 37% of laboratories who tested the internal quality control material; satisfactory, 35% of laboratories; unvalidated, 21% of laboratories). Combined variability scores for the high and low standards were assigned using the following criteria: 3 points: CV < 1st quartile, 2 points: CV between 1st quartile and median, 1 point: CV between median and 3rd quartile. 0 points: CV > 3rd quartile. The overall variability score (CbVar) was defined as the sum of the scores for the high and low level standards. The bar charts show the % of laboratories per CF status that had a combined variability scores of 6 (bright green). 4 or 5 (green), 2 or 3 (amber) or 1/0 (red).
Assessment of limit of blank for BCR::ABL1 detection
For 75% of laboratories (n = 9, Laboratories A - I) the likelihood of a true negative sample giving a result greater than zero (i.e., a false positive result) was 5% (Table 2). However, for 25% of laboratories (n = 3: laboratories J, K, & L) the likelihood of a true negative sample giving a result greater than zero ranged from 10–50% (Table 2), indicating a significant background of false positive results.
Table 2.
Limit of blank: data for the 12 participating laboratories.
| Lab | A | B | C | D | E | F | G | H | I | J | K | L |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Final BCR::ABL1 LoB (95%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.41 | 2.35 | 2.57 |
| Final BCR::ABL1 LoB (90%) | – | – | – | – | – | – | – | – | – | 0 | 0.6 | 2.17 |
| Final BCR::ABL1 LoB (85%) | – | – | – | – | – | – | – | – | – | – | 0 | 1.95 |
| Final BCR::ABL1 LoB (50%) | – | – | – | – | – | – | – | – | – | – | – | 0.79 |
| Total BCR::ABL1 replicates | 120 | 120 | 120 | 120 | 120 | 120 | 90 | 120 | 120 | 120 | 120 | 120 |
| No. of negative BCR::ABL1 replicates | 120 | 120 | 120 | 120 | 120 | 120 | 90 | 119 | 119 | 117 | 110 | 3 |
| % Negative BCR::ABL1 replicates | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99.2 | 99.2 | 97.5 | 91.7 | 2.5 |
| Max BCR::ABL1 copy number | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.44 | 2.15 | 2.67 | 2.9 | 4.25 |
Laboratories A–I have a likelihood of ≤5% that a true BCR::ABL1 negative sample will give a result greater than zero. Laboratories J, K, and L have a likelihood ranging from 10–50% (indicated in bold) that a true BCR::ABL1 negative sample will give a result greater than zero.
Discussion
MRD results directly impact treatment decisions in CML thus it is very important that the accuracy and precision of BCR::ABL1 assays are maintained across the entire measurement range, and that the sensitivity of the test is sufficient to measure DMR. It is well known that variability exists between RT-qPCR methods [23] and considerable work been undertaken to improve standardization of results for patients with detectable MRD [14, 24], but detailed assessment of the ability of laboratories to detect MR4.5 has not been undertaken. Furthermore, the “gold standard” methodology for deriving laboratory-specific CFs by sample exchange has proven to be unsustainable.
The EUTOS molecular standardization study indicates that secondary reference panels can be used effectively to obtain and validate IS CFs over time in a manner equivalent to sample exchange. They can also be used to monitor additional aspects of quality assurance. Over the period of the study, the percentage of laboratories with CFs validated as optimal or satisfactory increased from 67.5% (2016) to 97.6% (2021) and 36.4% (2016) to 91.7% (2021) for ABL1 and GUSB, respectively. The percentage of laboratories able to detect MR4.5 in most samples was high across all years with a median of 98.2% (range 96.4 to 100%).
The distribution of ABL1 CF values was similar to that observed by the EUTOS sample exchange program between 2006 and 2016. However, the distribution of GUSB CF values showed an approximate 1.64-fold increase compared to those obtained using sample exchange. It is unclear why this difference occurs, but if the assumption is made that the level of GUSB transcripts is 2.4 fold higher than ABL1, as shown previously in patient samples [17], then the CF values for GUSB assays would be expected to be correspondingly higher. This suggests that the CF values obtained from the cell line panels are valid (ABL1 median CF 0.604 vs GUSB median CF 1.576; 2.6 fold difference). In this study, laboratories using GUSB as a reference gene had a higher percentage of unvalidated CFs compared to ABL1 laboratories (2.4–32.6% ABL1 vs 8.3–63.6% GUSB). The GUSB assays also demonstrated a higher degree of variation (mean CbVar = 2) compared to ABL1 laboratories (mean CbVar = 3.14) when testing internal quality control material, suggesting that the GUSB assay may be more inherently variable. It should also be noted that the number of GUSB datasets was low for both studies and several GUSB laboratories reported technical difficulties using the lyophilized material, possibly due to inexperience in handling this material, resulting in low GUSB copy numbers.
Nevertheless, given the potential instability of GUSB assays observed in this study, we would suggest that laboratories using this reference gene should monitor the stability of their assays at least monthly using high- and low-level control samples. If instability is detected, the laboratory should consider switching to a validated ABL1 assay until investigations into the GUSB assay stability have been undertaken and successfully actioned. More data are required to fully investigate the use of the panels to derived CFs for GUSB assays. Unfortunately, the current AcroMetrix™ BCR-ABL Panel has not been calibrated to the primary WHO material for GUSB (or BCR) and therefore this panel cannot be used to directly derive CFs for this reference gene. Although other approaches may be possible, laboratories using GUSB (or BCR) as a reference gene will need to continue to perform sample exchange with a reference laboratory to derive a CF. Once a laboratory has established a CF, it should also be possible to revalidate that CF or derive a new CF using archived samples (e.g., lysates) with known IS values that span the range from MR1 to MR4.5 in a manner analogous to sample exchange with an external reference laboratory.
It is difficult to define exactly how frequently CFs should be revalidated, but we suggest it should be performed at least annually if ongoing IQC data demonstrates assay stability at high and low BCR::ABL1 values. If the newly derived CF is classified as optimal or satisfactory then it is acceptable to continue to use the original CF, although some centers may prefer to adopt the newly derived CF. However, when a newly derived CF is classified as unvalidated (and the assay has remained unchanged) further investigations should be considered to improve the assay stability. If the method or equipment is changed, or assay drift is noted though ongoing IQC then a new CF will need to be derived [7], although it is important to demonstrate first that any new assay is stable over time. Although we have demonstrated that commercially available secondary reagents can be used to derive a CF, it is important to note that this is not the only option, e.g., sample exchange with a validated laboratory remains an alternative approach, and laboratories may perform their own internal sample exchange, e.g., by comparing results from around 30 stored samples (ideally lysates) spanning 10% to DMR tested with the new method against results from the same samples with the previous, validated method.
For IQC procedures, it is recommended that laboratories attempt initially to optimize assays to decrease variability such that the CV for each category (BCR::ABL1IS, reference gene copy number, BCR::ABL1 copy number) are at least less than the 3rd quartile value obtained in this study (Table 1). Ideally, variability should be close to or lower than the median CV values (Table 1). Once assay variability is established in this range then the application of Westgard rules to accept or reject each run based on the performance of high and low controls (as recommended by Branford et al. [7, 19]) could be used to monitor assays on a regular basis (Supplementary Fig. 4). Laboratories may elect to use a lower standard e.g., 0.01% in addition to, or instead of, 0.1%. The exact level is not critical but we recommend that all laboratories regularly monitor the performance of their assays using at least two standards. Standards may be best prepared locally as lysates of cell line mixtures (see Supplementary Information), or may be purchased from commercial suppliers (e.g., the AcroMetrixTM BCR-ABL panel).
The use of high- and low-level standards can help monitor all the processes in the assay from RNA extraction through to RT-qPCR. Collecting data and monitoring the reference gene number, BCR::ABL1 copy number and %BCR::ABL/reference gene is an ideal way to observe if there are any immediate technical problems occurring with the assay as well as monitoring assay stability over time. Each parameter can provide different information e.g., the copy number information may be useful to determine variability in the cDNA synthesis. In this case, the BCR::ABL1IS may be unaffected but the copy numbers for the reference and target gene may be variable between runs, which may, in turn, affect the LoD. However, if the copy number of one gene is more variable than the other then this may indicate an issue with the RT-qPCR reagents or processes. This would likely affect the BCR::ABL1IS value obtained. For robust IQC, it is therefore recommended to record values for BCR::ABL1IS, reference gene copy number, BCR::ABL1 copy number and also the gradient of the plasmid standard curves and Cq values for each standards on every run. For laboratories using the ERM plasmid the Cq values for ABL1 and BCR::ABL1 should be comparable for each standard as the plasmid standard contains exactly the same number of BCR::ABL1 and ABL1 copies [11].
Laboratories should be aware of the variability of their assay and communicate this to clinical staff so that they are informed of the acceptable degree of variability of BCR::ABLIS values at critical clinical decision points. For example, a laboratory that has an optimal CV of 9.7% for a high level control sample could reproducibly report a 10% BCR::ABLIS sample in the range of 9.03–10.97% (this range is based on one standard deviation; some laboratories may prefer to use two standard deviations). In the case of a laboratory with an assay demonstrating high variability, e.g., a CV of 22.5%, the range for the same sample increases to 7.75–12.25%. For samples at MMR (0.1% BCR::ABLIS) the same laboratories would report a true MMR sample in the range 0.085–0.115% (CV 14.6%) and 0.071–0.129% (CV 28.9%) respectively.
Defining the LoB and LoD of quantitative assays is important for validation of molecular tests and is necessary for accreditation of a diagnostic test to ISO 15189 (2012). Our study provides a practical recommended protocol for determining the LoB for BCR::ABL1 RT qPCR testing, and we recommend that laboratories establish their LoB. A major challenge was the production of truly BCR::ABL1 negative samples. Initially, material was prepared from several BCR::ABL1 negative human cell lines from different sources but in our hands these showed very low level but reproducible amplification with BCR::ABL1 EAC RT-qPCR assays. Therefore, the use of cell line derived material for LoB studies is not recommended. Preparation of pooled blood samples from non-leukemic patients was time consuming however provided good quality material for the study. Using this material, we found that 25% of laboratories had a LoB greater than zero which may have implications for the accurate reporting of DMR, thus demonstrating the importance of establishing a LoB. Laboratories with poorly optimized assays may either fail to detect BCR::ABL1 and erroneously conclude that a patient had achieved DMR (variation in LoD) or exhibit a low level false positive rate and erroneously detect BCR::ABL1 (variation in LoB). Laboratory LoBs and LoDs have not been examined comprehensively to date because of a lack of suitable control reagents and agreed methodology.
In summary, we provide a number of recommendations for optimal monitoring of residual disease in CML by RT-qPCR, including establishment of laboratory-specific CFs and maintenance of reporting on the IS. We anticipate that these recommendations will further help to improve the quality of molecular monitoring for CML, with resulting benefits for patient management.
Supplementary information
Acknowledgements
We thank Dr. Peter Rigsby at the National Institute for Biological Standards and Control (NIBSC), UK for permission to reproduce document 09-138 in the Supplementary Material.
Author contributions
The 2016–2021 study was designed by HEW, MS, and NCPC with support from other members of the EUTOS molecular monitoring subcommittee (SS, TE, KMP, AH). Materials were prepared and distributed by HEW and MS; data analysis was performed by HEW. MM and CD provided information from previous EUTOS sample exchange rounds. All other authors represent individual EUTOS reference laboratories that analysed samples and returned data for central analysis.
Funding
The study was supported by Novartis through the European Treatment and Outcome Study (EUTOS) for CML.
Data availability
The datasets generated during the study are available from the corresponding author on reasonable request.
Competing interests
HW has received honoraria from Novartis. SS has received honoraria from Incyte, Novartis, Pfizer, Bristol Myers Squibb, and Roche and research support from Incyte, Novartis, and Bristol Myers Squibb. J-MC has received research support and honoraria from Incyte, and honoraria from Novartis and Cepheid. TL has received honoraria from Incyte, Novartis, Pfizer, Angelini, Bristol Myers Squibb, and research support from Incyte, Novartis, and Pfizer. PV has received honoraria from Astra-Zeneca, Eli Lilly, Gilead; GlaxoSmithKline, Novartis, Pfizer, Roche, Teva, and research support from Novartis and Pfizer. SM is employed by the MLL Munich Leukemia Laboratory GmbH, Munich. CD and MM are employed by the Institute for Hematology and Oncology (IHO GmbH), Mannheim. KMP has received honoraria from Angelini and Incyte. AH received research support from Novartis, BMS, Pfizer, and Incyte. NC has received research support and honoraria from Novartis, and honoraria from Incyte and Astellas.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-022-01607-z.
References
- 1.Deininger MW, Shah NP, Altman JK, Berman E, Bhatia R, Bhatnagar B, et al. Chronic myeloid leukemia, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2020;18:1385–415. doi: 10.6004/jnccn.2020.0047. [DOI] [PubMed] [Google Scholar]
- 2.Hochhaus A, Baccarani M, Silver RT, Schiffer C, Apperley JF, Cervantes F, et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia. 2020;34:966–84. doi: 10.1038/s41375-020-0776-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes T, Deininger M, Hochhaus A, Branford S, Radich J, Kaeda J, et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108:28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cross NC. Standardisation of molecular monitoring for chronic myeloid leukaemia. Best Pr Res Clin Haematol. 2009;22:355–65. doi: 10.1016/j.beha.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Branford S, Rudzki Z, Harper A, Grigg A, Taylor K, Durrant S, et al. Imatinib produces significantly superior molecular responses compared to interferon alfa plus cytarabine in patients with newly diagnosed chronic myeloid leukemia in chronic phase. Leukemia. 2003;17:2401–9. doi: 10.1038/sj.leu.2403158. [DOI] [PubMed] [Google Scholar]
- 6.Branford S, Cross NC, Hochhaus A, Radich J, Saglio G, Kaeda J, et al. Rationale for the recommendations for harmonizing current methodology for detecting BCR-ABL transcripts in patients with chronic myeloid leukaemia. Leukemia. 2006;20:1925–30. doi: 10.1038/sj.leu.2404388. [DOI] [PubMed] [Google Scholar]
- 7.Branford S, Fletcher L, Cross NC, Müller MC, Hochhaus A, Kim DW, et al. Desirable performance characteristics for BCR-ABL measurement on an international reporting scale to allow consistent interpretation of individual patient response and comparison of response rates between clinical trials. Blood. 2008;112:3330–8. doi: 10.1182/blood-2008-04-150680. [DOI] [PubMed] [Google Scholar]
- 8.White HE, Matejtschuk P, Rigsby P, Gabert J, Lin F, Lynn Wang Y, et al. Establishment of the first World Health Organization International Genetic Reference Panel for quantitation of BCR-ABL mRNA. Blood. 2010;116:e111–7. doi: 10.1182/blood-2010-06-291641. [DOI] [PubMed] [Google Scholar]
- 9.White HE, Hedges J, Bendit I, Branford S, Colomer D, Hochhaus A, et al. Establishment and validation of analytical reference panels for the standardization of quantitative BCR-ABL1 measurements on the international scale. Clin Chem. 2013;59:938–48. doi: 10.1373/clinchem.2012.196477. [DOI] [PubMed] [Google Scholar]
- 10.Mauté C, Nibourel O, Réa D, Coiteux V, Grardel N, Preudhomme C, et al. Calibration of BCR-ABL1 mRNA quantification methods using genetic reference materials is a valid strategy to report results on the international scale. Clin Biochem. 2014;47:1333–6. doi: 10.1016/j.clinbiochem.2014.05.067. [DOI] [PubMed] [Google Scholar]
- 11.White H, Deprez L, Corbisier P, Hall V, Lin F, Mazoua S, et al. A certified plasmid reference material for the standardisation of BCR-ABL1 mRNA quantification by real-time quantitative PCR. Leukemia. 2015;29:369–76. doi: 10.1038/leu.2014.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross NC, White HE, Ernst T, Welden L, Dietz C, Saglio G, et al. Development and evaluation of a secondary reference panel for BCR-ABL1 quantification on the International Scale. Leukemia. 2016;30:1844–52. doi: 10.1038/leu.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes TP, Kaeda J, Branford S, Rudzki Z, Hochhaus A, Hensley ML, et al. Frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukemia. N. Engl J Med. 2003;349:1423–32. doi: 10.1056/NEJMoa030513. [DOI] [PubMed] [Google Scholar]
- 14.Müller MC, Cross NC, Erben P, Schenk T, Hanfstein B, Ernst T, et al. Harmonization of molecular monitoring of CML therapy in Europe. Leukemia. 2009;23:1957–63. doi: 10.1038/leu.2009.168. [DOI] [PubMed] [Google Scholar]
- 15.Cross NC, Hochhaus A, Müller MC. Molecular monitoring of chronic myeloid leukemia: principles and interlaboratory standardization. Ann Hematol. 2015;94:S219–25. doi: 10.1007/s00277-015-2315-1. [DOI] [PubMed] [Google Scholar]
- 16.Mahon FX, Réa D, Guilhot J, Guilhot F, Huguet F, Nicolini F, et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010;11:1029–35. doi: 10.1016/S1470-2045(10)70233-3. [DOI] [PubMed] [Google Scholar]
- 17.Cross NC, White HE, Colomer D, Ehrencrona H, Foroni L, Gottardi E, et al. Laboratory recommendations for scoring deep molecular responses following treatment for chronic myeloid leukemia. Leukemia. 2015;29:999–1003. doi: 10.1038/leu.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spiess B, Naumann N, Galuschek N, Rinaldetti S, Kossak-Roth U, Tarnopolscaia I, et al. The benefit of quality control charts (QCC) for routine quantitative BCR-ABL1 monitoring in chronic myeloid leukemia. PLoS One. 2018;13:e0196326. doi: 10.1371/journal.pone.0196326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Branford S, Hughes T. Diagnosis and monitoring of chronic myeloid leukemia by qualitative and quantitative RT-PCR. Methods Mol Med. 2006;125:69–92. doi: 10.1385/1-59745-017-0:69. [DOI] [PubMed] [Google Scholar]
- 20.Branford S. Molecular monitoring in chronic myeloid leukemia-how low can you go? Hematol Am Soc Hematol Educ Program. 2016;2016:156–63. doi: 10.1182/asheducation-2016.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CSLI. Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures; Approved Guidelines - Second Edition. CLSI document EP17-A2. 2012: Clinical and Laboratory Standards Institute
- 22.White H, Salmon M, Bochicchio MT, Cayuela JM, Colomer D, Daraio F, et al. Variation in Limit of Blank for BCR-ABL1 Detection Between Laboratories Impacts on Scoring of Deep Molecular Response HemaSphere 2018: Abstract PF372.
- 23.Scott S, Travis D, Whitby L, Bainbridge J, Cross NCP, Barnett D. Measurement of BCR-ABL1 by RT-qPCR in chronic myeloid leukaemia: findings from an International EQA Programme. Br J Haematol. 2017;177:414–22. doi: 10.1111/bjh.14557. [DOI] [PubMed] [Google Scholar]
- 24.Müller MC, Erben P, Saglio G, Gottardi E, Nyvold CG, Schenk T, et al. Harmonization of BCR-ABL mRNA quantification using a uniform multifunctional control plasmid in 37 international laboratories. Leukemia. 2008;22:96–102. doi: 10.1038/sj.leu.2404983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the study are available from the corresponding author on reasonable request.