Abstract
RIPK1 is a master regulator of multiple cell death pathways, including apoptosis and necroptosis, and inflammation. Importantly, activation of RIPK1 has also been shown to promote the transcriptional induction of proinflammatory cytokines in cells undergoing necroptosis, in animal models of amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease (AD), and in human ALS and AD. Rare human genetic carriers of non-cleavable RIPK1 variants (D324V and D324H) exhibit distinct symptoms of recurrent fevers and increased transcription of proinflammatory cytokines. Multiple RIPK1 inhibitors have been advanced into human clinical trials as new therapeutics for human inflammatory and neurodegenerative diseases, such as ALS and AD. However, it is unclear whether and how RIPK1 kinase activity directly mediates inflammation independent of cell death as the nuclear function of RIPK1 has not yet been explored. Here we show that nuclear RIPK1 is physically associated with the BAF complex. Upon RIPK1 activation, the RIPK1/BAF complex is recruited by specific transcription factors to active enhancers and promoters marked by H3K4me1 and H3K27ac. Activated nuclear RIPK1 mediates the phosphorylation of SMARCC2, a key component of the BAF complex, to promote chromatin remodeling and the transcription of specific proinflammatory genes. Increased nuclear RIPK1 activation and RIPK1/BAF-mediated chromatin-remodeling activity were found in cells expressing non-cleavable RIPK1, and increased enrichment of activated RIPK1 on active enhancers and promoters was found in an animal model and human pathological samples of ALS. Our results suggest that RIPK1 kinase serves as a transcriptional coregulator in nucleus that can transmit extracellular stimuli to the BAF complex to modulate chromatin accessibility and directly regulate the transcription of specific genes involved in mediating inflammatory responses.
Subject terms: Mechanisms of disease, Cell signalling
Introduction
RIPK1 is a master regulator of inflammatory signaling as well as multiple cell death pathways, including apoptosis and necroptosis. Activation of RIPK1 kinase in TNFR1 signaling pathway can promote its interaction with FADD and caspase-8 to form complex IIa for mediating RIPK1-dependent apoptosis (RDA) or with RIPK3 to form complex IIb for promoting necroptosis.1 While the formation of complex IIa and complex IIb occurs in cytoplasm, the activation of RIPK1 has been shown to promote the transcriptional induction of proinflammatory cytokines in animal models of amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease (AD) and in human ALS and AD.2–4 Rare human genetic carriers of RIPK1 variants (D324V and D324H) which block the inactivating cleavage of RIPK1 mediated by caspase-8 exhibit distinct symptoms of recurrent fevers and increased transcription of proinflammatory cytokines.5,6 Multiple RIPK1 inhibitors have been advanced into human clinical trials as new therapeutics for human inflammatory and neurodegenerative diseases, including ALS and AD.1 However, it is unclear how the activation of RIPK1 may directly promote the transcription of proinflammatory cytokines independent of cell death as the nuclear function of RIPK1 has not been explored.
Mammalian genomic DNA is tightly packed into chromatins which must be modulated by ATP-dependent remodeling complexes in order to allow accessibility and transcription of specific genes.7,8 BRG1/BRM-associated factor (BAF) chromatin-remodeling complexes, the mammalian homolog of the SWI/SNF complex in yeast, are important regulators of gene expression and higher-order chromatin organization that modulate chromatin accessibility by displacing the nucleosomes in an ATP-dependent manner in response to developmental and environmental signals.9 In addition, the functional status of genomic regions is marked with differentially modified histones. The active enhancer (AE) regions are marked by high levels of histone H3 with monomethylated lysine 4 (H3K4me1) and with acetylated lysine 27 (H3K27ac).10,11 However, it remains unclear how cells selectively promote transcriptional induction of inflammatory factors by modulating chromatin accessibility in response to environmental signals.
Here we investigated the mechanism by which activated RIPK1 in the nucleus regulated the transcriptional induction of proinflammatory cytokines. Our study discovers a nuclear RIPK1/BAF complex that is involved in promoting the transcriptional induction of proinflammatory responses. We show that the RIPK1/BAF complex is recruited by specific transcriptional factors known to be involved in mediating inflammation to AEs and activated promoters (APs) marked by H3K4me1 and H3K27ac when RIPK1 is activated, and mediates the phosphorylation of SMARCC2 which in turn promotes chromatin remodeling and the transcription of specific genes involved in mediating inflammatory responses. Inhibition of RIPK1 kinase activity blocks the transcription of inflammatory factors by inactivating SMARCC2, a key component of the BAF complexes. Our results suggest that RIPK1 kinase serves an important signaling function in the nucleus by transmitting signals from extracellular stimuli to the BAF complex in order to regulate the chromatin accessibility and transcription of specific genes involved in mediating inflammatory responses.
Results
Activated RIPK1 is found in nucleus
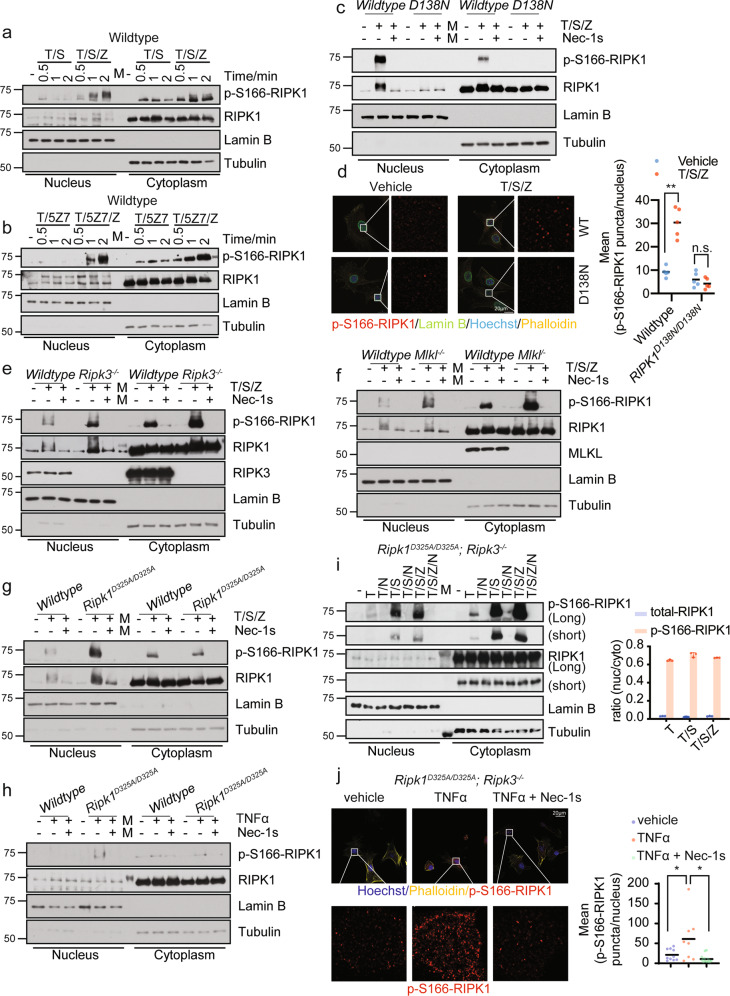
To explore the mechanism by which RIPK1 mediates the transcriptional induction of inflammatory responses, we first examined whether activated RIPK1 might be localized in the nucleus in apoptotic and necroptotic conditions that were known to involve the activation of RIPK1.12,13 In murine embryonic fibroblasts (MEFs) stimulated with TNFα/SM-164(T/S) or TNFα/5Z-7-oxozeaenol (T/5Z7) to promote RDA, or TNFα/SM-164/zVAD (T/S/Z) or TNFα/5Z7/zVAD (T/5Z7/Z) to promote necroptosis, we analyzed the presence of activated RIPK1 using p-S166-RIPK1 as a biomarker1 in the nuclear and cytosolic fractionations as RIPK1 is auto-phosphorylated at Ser 166 in response to stimuli causing RDA or necroptosis.12,14 We found that the activated RIPK1 in the nucleus was rapidly detectable with the same time course as that in cytoplasm after the stimuli (Fig. 1a, b). To confirm the signal of p-S166-RIPK1 detected in nucleus is specific, we analyzed the presence of activated RIPK1 in necroptotic cells in both wild-type and D138N kinase-dead knock-in MEFs in which the activation of RIPK1 was genetically blocked. We detected activated RIPK1 (p-S166-RIPK1+) in the nuclei of wild-type cells undergoing necroptosis by both fractionation experiment (Fig. 1c) and immunostaining (Fig. 1d). The nuclear p-S166-RIPK1 signal was specific as it was blocked by treatment with the RIPK1 kinase inhibitor R-7-Cl-O-necrostatin-1 (Nec-1s) (Fig. 1c) and genetic inactivation of RIPK1 with D138N mutation (Fig. 1c, d).
Fig. 1. Activated RIPK1 can be found in nucleus.
a, b Wild-type MEFs were treated as indicated and the fraction of nucleus or cytoplasm was analyzed by western blotting. 5 ng/mL mTNFα, 50 µM SM164, 100 nM (5Z)-7-oxozeaeno, 20 µM zVAD-fmk were used. mTNFα was added 0.5 h after the addition of other compounds and treated for the indicated period. c, e, f, g Wild-type, Ripk1D138N/D138N, Ripk3−/−, Mlkl−/−, or Ripk1D325A/D325A MEFs were pre-incubated with 50 µM SM164, 20 µM zVAD-fmk, with or without 10 µM Nec-1s for 0.5 h and then treated with 5 ng/mL mTNFα. The levels of p-S166-RIPK1, RIPK1, RIPK3, MLKL in the fractions of nucleus and cytoplasm were determined by western blotting. d Immunostaining of p-S166 RIPK1 (red) in wild-type and Ripk1D138N/D138N MEFs treated with 50 µM SM164, 20 µM zVAD-fmk for 0.5 h and then treated with 5 ng/mL mTNFα for 1 h. Representative images and enlarged nuclear staining of p-S166-RIPK1 are shown. Cell membranes were stained with Phalloidin (yellow). Nuclear membrane was stained with Lamin B (green). Nuclei were stained with Hoechst (blue). Quantification of the mean number of p-S166-RIPK1 puncta per nucleus in each field is shown on the right (n = 5). **P < 0.01, n.s., not significant. h Ripk1D325A/D325A MEFs were treated with or without 20 µM Nec-1s for 2 h followed by 20 ng/mL mTNFα for 1 h. The fraction of nucleus or cytoplasm was analyzed by western blotting. i Ripk1D325A/D325A;Ripk3−/− MEFs were treated with or without 20 µM Nec-1s for 2 h followed by 100 ng/mL mTNFα for 1 h or treated with 50 µM SM164, 20 µM zVAD-fmk for 0.5 h and then treated with 5 ng/mL mTNFα for 1 h. The fraction of nucleus or cytoplasm was analyzed by western blotting. Quantification of the ratio of nuclear/cytoplastic p-S166-RIPK1 or total RIPK1 were shown on the right. j Immunostaining of p-S166-RIPK1 in Ripk1D325A/D325A;Ripk3−/− MEFs treated with vehicle, or ± 20 µM Nec-1s for 0.5 h followed by 20 ng/mL mTNFα for 1 h. Representative images with enlarged nuclear staining of p-S166-RIPK1 and quantification of the mean number of p-S166-RIPK1 puncta per nucleus in each field are shown (n ≥ 8). *P < 0.05.
RIPK3 and MLKL are the downstream mediators that are activated by RIPK1 to mediate the execution of necroptosis.15 Interestingly, the presence of nuclear p-S166-RIPK1 was not affected by RIPK3 knockout or MLKL knockout which blocked cell death induced by necroptotic stimuli T/S/Z (Fig. 1e, f; Supplementary information, Fig. S1a). These results suggest that the nuclear localization of activated RIPK1 in necroptosis is not the consequence of cell death.
Blocking the caspase-8-mediated cleavage of murine RIPK1 by D325A knock-in mutation in mice models the human RIPK1 D324V or D324H mutation, which increases the activation of RIPK1.5,16,17 We next characterized the nuclear presence of activated RIPK1 by fractionation of lysates from Ripk1D325A/D325A MEFs treated with T/S/Z or TNFα. We detected a stronger nuclear p-S166-RIPK1 signal upon the treatment of T/S/Z in Ripk1D325A/D325A MEFs compared to wild-type MEFs (Fig. 1g). In addition, the nuclear p-S166-RIPK1 signal upon the treatment of TNFα in Ripk1D325A/D325A MEFs was detected while no RIPK1 activation in the nucleus of wild-type MEFs was detected (Fig. 1h). The p-S166-RIPK1 signal was specific as it was blocked by treatment with Nec-1s14 (Fig. 1g, h).
Treatment of Ripk1D325A/D325A;Ripk3−/− MEFs with TNFα for 1 h, 2 h, 4 h or 6 h is known to induce the transcription of proinflammatory cytokines independent of cell death.6 Notably, we detected activated RIPK1 in both nuclear and cytosolic fractions 1 h following addition of TNFα, which was inhibited by treatment with Nec-1s (Fig. 1i). In addition, the activation of RIPK1 was elevated in both the cytoplasm and nuclei of Ripk1D325A/D325A;Ripk3−/− MEFs undergoing RDA stimulated by T/S (Fig. 1i). The total amount of RIPK1 was higher in the cytoplasm than that in the nucleus, and the ratio of activated RIPK1 to total RIPK1 was also much higher in the nuclei of cells undergoing necroptosis or RDA than that in the cytoplasm of Ripk1D325A/D325A;Ripk3−/− MEFs. The presence of activated RIPK1 in the nucleus was further confirmed by immunostaining using an anti-p-S166-RIPK1 antibody in Ripk1D325A/D325A;Ripk3−/− MEFs treated with TNFα and the activation of RIPK1 was inhibited by treatment with Nec-1s (Fig. 1j). Thus, the nuclear localization of activated RIPK1 is an early event in TNFα-treated cells with a RIPK1 D325A mutation. The activation of RIPK1 was independent of cell death as increased p-S166-RIPK1 was also detected in both nuclei and cytoplasm of Ripk1D325A/D325A;Ripk3−/− MEFs stimulated by T/S/Z which could not induce necroptosis due to the RIPK3 deficiency16 (Fig. 1j).
We next examined the nuclear presence of activated RIPK1 in human cells. Human cancer cell lines frequently lose expression of RIPK3 which make them resistant to necroptosis induction.18,19 RIPK3-null cancer cell lines were treated with T/S/Z which could not induce cell death in these RIPK3-deficient cancer cells;18 nevertheless, the activated RIPK1 in nucleus was robustly detected following treatment with T/S/Z and the activation of RIPK1 was inhibited by treatment with Nec-1s (Supplementary information, Fig. S1b). We further examined the activated RIPK1 in human A375 cells treated with T/S/Z by immunostaining. Activated RIPK1 was also found in viable A375 cells treated with T/S/Z and the activation of RIPK1 was inhibited by treatment with Nec-1s (Supplementary information, Fig. S1b, c). Together, these results demonstrate the nuclear presence of activated RIPK1 when RIPK1 is activated by multiple conditions, which is independent of RIPK1’s role in activating cell death.
Nuclear RIPK1 is physically associated with the BAF complex and specific transcription regulators
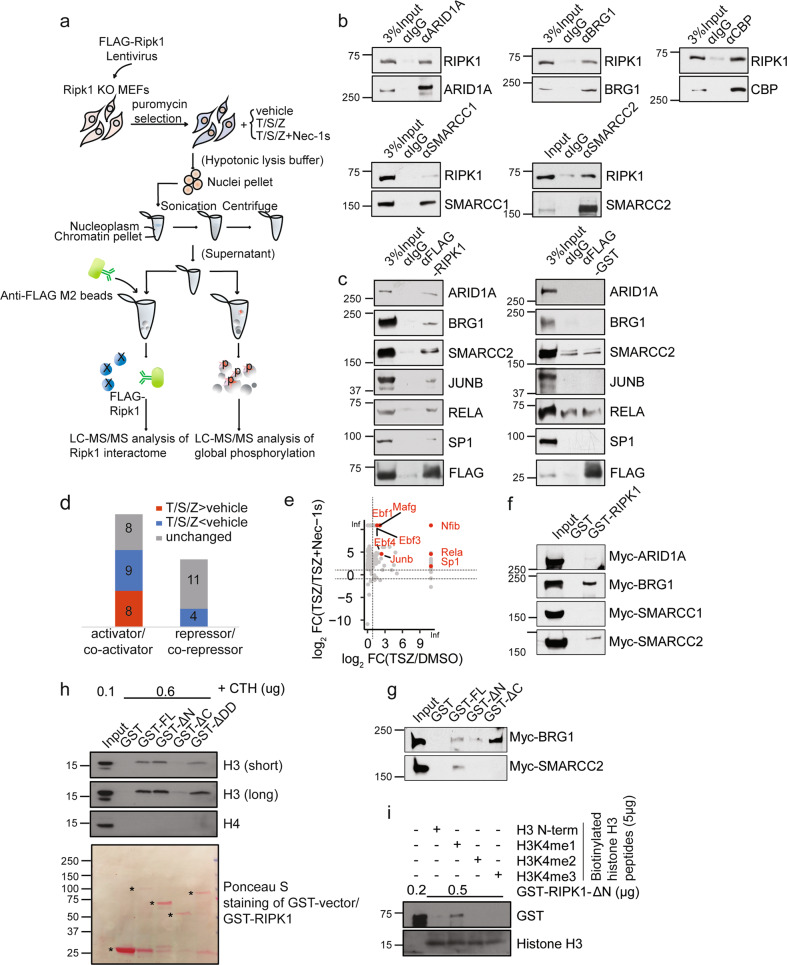
We next interrogated the nuclear interactome of RIPK1 in the nuclear fraction using affinity purification followed by quantitative mass spectrometry (Fig. 2a). We found that the nuclear RIPK1 was copurified with multiple transcription factors (SP1, JUNB, RELA and NFIB) (Supplementary information, Fig. S1d), co-regulators such as CREBBP (CBP) (Supplementary information, Table S1), histones (Supplementary information, Fig. S1e) and strikingly, most components of the BAF complex, including SMARCC1, SMARCA4 (BRG1), ACTL6A, SMARCC2, DPF2, SMARCE1, SMARCB1, and ARID1A (Supplementary information, Fig. S1d). Co-immunoprecipitation (Co-IP) was performed using antibodies against FLAG-tagged RIPK1 and endogenous RIPK1, validating the interactions of RIPK1 with identified transcription factors and most of the components of the BAF complex, including endogenous RELA, SP1, NFIB, ARID1A, BRG1, SMARCC1, SMARCC2, and CBP (Fig. 2b; Supplementary information, Fig. S1f, g). To further confirm the specificity of the interactions between RIPK1 and these proteins, all FLAG-RIPK1 IPs were eluted with FLAG peptide, and FLAG-glutathione S-transferase (GST) IPs were performed as negative control (Fig. 2c). Among the transcriptional regulators/coregulators detected in the mass spectrometry analysis of RIPK1 nuclear interactome, we found that the intensities of 8 transcriptional activators/coactivators (colored red in Fig. 2d) were higher in the RIPK1 interactome from MEFs treated with T/S/Z compared with the vehicle group, while the intensities of all transcriptional repressors/corepressors (colored blue or gray in Fig. 2d) were unchanged or lower. In addition, the binding intensities of all the 8 transcriptional activators/coactivators including JUNB, RELA, and SP1 decrease in the RIPK1 interactome from MEFs with addition of Nec-1s (Fig. 2e). These results demonstrate that activated RIPK1 in the nucleus preferentially binds to transcriptional activators/coactivators, suggesting that nuclear RIPK1 is involved in mediating transcriptional activation.
Fig. 2. RIPK1 is a transcriptional co-activator associated with multiple transcription factors and BAF complex.
a Schematic illustration of the quantitative mass spectrometry approach to identify the RIPK1 nuclear interactome and phosphoproteome. b Cell lysates from wild-type MEFs were immunoprecipitated with antibodies against the indicated proteins followed by western blotting with antibody against RIPK1. c Ripk1−/− MEFs were infected with FLAG-Ripk1 or FLAG-GST lentivirus. Nuclear lysates were immunoprecipitated with anti-FLAG M2 beads or IgG followed by protein G beads, eluted and followed by western blotting with antibodies against the indicated proteins. d The intensities of proteins detected in RIPK1 nuclear interactome under different treatments were compared and the RIPK1-interactive proteins were divided into three groups: T/S/Z > vehicle (red), T/S/Z < vehicle (blue) and unchanged (gray). Numbers of the transcriptional activators/co-activators and repressors/co-repressors from the three groups detected to interact with RIPK1 are shown. e Plot showing log2(TSZ/Vehicle) ratio of intensities detected in nuclear RIPK1 interactome on x axis and log2(TSZ/(TSZ + Nec-1s)) ratio of intensities on y axis. Active transcription factors which were detected to interact with activated RIPK1 are labeled. f The result of GST pull-down assays with Myc-ARID1A, Myc-BRG1, Myc-SMARCC1, Myc-SMARCC2 synthesized by in vitro transcription/translation, and GST, GST-RIPK1 purified from E. coli. g The result of GST pull-down assays with Myc-BRG1, Myc-SMARCC2 synthesized by in vitro transcription/translation, and GST, GST-RIPK1-full length (FL), GST-RIPK1-ΔN, GST-RIPK1-ΔC purified from E. coli. h In vitro pull-down of calf thymus histones (CTH) with indicated purified GST-fused FL or truncations of human RIPK1(ΔN, ΔC or ΔDD) was determined by western blotting with antibodies against histone H3 and H4. RIPK1-ΔDD, RIPK1 with the death domain deleted. i In vitro peptide pull-down assays. Biotinylated histone H3 peptides with mono-, di-, tri-methylation (5 µg), or without methylation on H3K4 were used to pull down purified GST-RIPK1-ΔN. The histone binding preferences were determined by western blotting with the antibody against GST tag.
To further characterize the physical association between RIPK1 and the BAF complex and gain insights into the molecular mechanism underlying the regulation on the interaction of these proteins, we purified GST-fused proteins of full-length or truncated RIPK1 (Supplementary information, Fig. S1h). GST pull-down experiments were then performed between GST-fused RIPK1 and in vitro transcribed/translated individual components of the BAF complex, including ARID1A, BRG1, SMARCC1, and SMARCC2. We found that RIPK1 could interact directly with BRG1 and SMARCC2 in vitro (Fig. 2f). GST pull-down experiments were performed between an irrelevant GST-3C and in vitro transcribed/translated BRG1 and SMARCC2 as a negative control (Supplementary information, Fig. S1i), indicating the interactions between BRG1, SMARCC2 and RIPK1 are specific. In addition, we found that the interaction between RIPK1 and BRG1 required the N-terminal fragment (1–289 amino acid (aa)) of RIPK1, whereas the interaction between RIPK1 and SMARCC2 required the full-length RIPK1 (Fig. 2g).
We then performed a GST pull-down assay between GST-fused RIPK1 proteins and histones in vitro to validate the interactions detected by mass spectrometry in Supplementary information, Fig. S1e. As the BAF complex has been shown to preferentially bind with the enhancers marked by H3K4me1 and remodel H3K4me1-modifed nucleosomes,20,21 we first checked the interaction between RIPK1 and histone H3 with histone H4 as a control. We found that RIPK1 preferentially bound to histone H3 (Supplementary information, Fig. S1j). Furthermore, we found that the intermediate domain (290–583 aa) of RIPK1 was required for the binding between RIPK1 and histone H3 (Fig. 2h; Supplementary information, Fig. S1h). To characterize the binding preference of RIPK1 with mono-, di-, tri-, or un-methylated H3K4, we performed in vitro peptide pull-down assays between the C-terminal fragment of RIPK1 (275–671 aa) and different biotinylated histone H3 peptides. The results showed that RIPK1 preferentially bound with monomethylated H3K4 (H3K4me1) (Fig. 2i), which is known as a histone mark linked with enhancers.10
Together, these results support the physical association of nuclear RIPK1 with transcription factors, the BAF complex and histone H3K4me1, which suggests that nuclear RIPK1 is directly involved in transcriptional regulation.
Activated nuclear RIPK1 promotes gene transcription in a cell death-independent manner
To explore the biological function of nuclear RIPK1, we used RNA sequencing (RNA-seq) to analyze the expression profile of immortalized Ripk1D325A/D325A;Ripk3−/− MEFs treated with vehicle, TNFα, or TNFα plus Nec-1s for 2 h, or 3 h. Gene Set Enrichment Analysis (GSEA) identified 71 genes involved in inflammatory responses (FDR q value = 0.0595, Enrichment score = 0.6234). These genes were shown enriched in the group treated with TNFα for 3 h compared to the other two groups (vehicle and TNFα plus Nec-1s) (Supplementary information, Fig. S2a). Real-time quantitative reverse transcription PCR (RT-qPCR) experiments performed in wild-type, Ripk1D138N/D138N, Ripk3−/−, Mlkl−/− MEFs with multiple treatments showed the transcriptional activation of target genes induced by RIPK1 independent of cell death (Supplementary information, Fig. S2b–d). Several genes involved in mediating inflammatory responses, such as Il6, Il1a, Cxcl1, Cxcl2, Tnfaip3, Csf1, Areg, and Ereg, were selected as representative target genes to validate the RNA-seq results by RT-qPCR in Ripk1D325A/D325A;Ripk3−/− MEFs treated with TNFα, or TNFα plus Nec-1s (Supplementary information, Fig. S2e). These results further support that activation of RIPK1 leads to the transcriptional induction of proinflammatory gene expression.
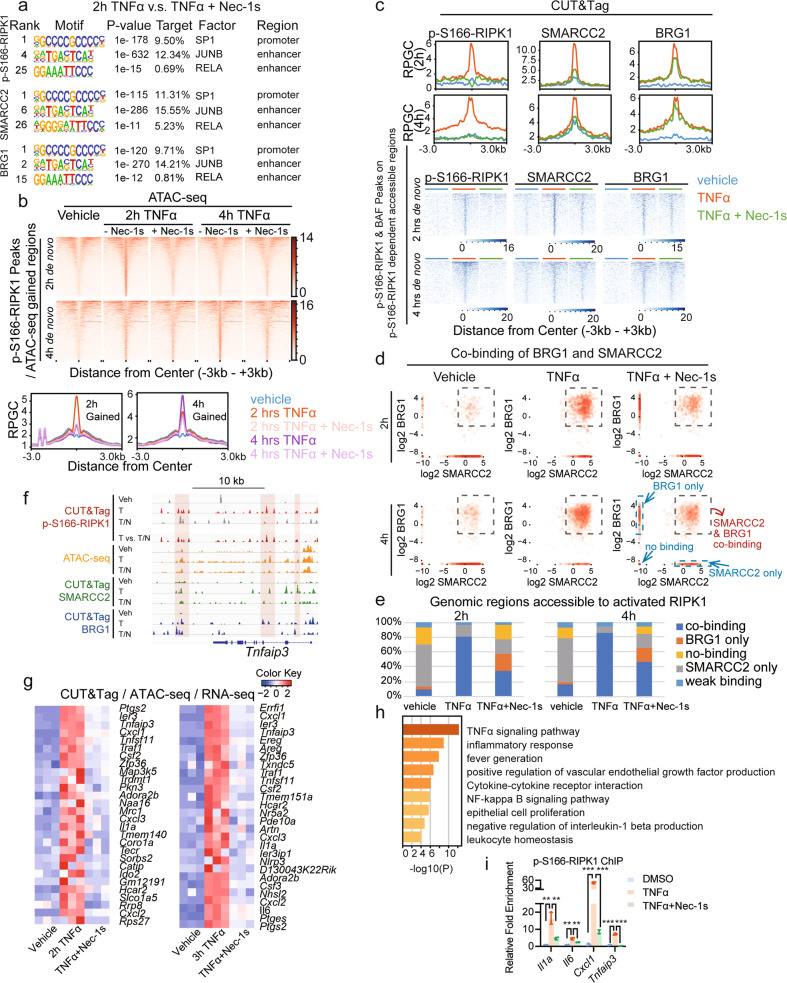
To define the motifs of transcription factors enriched in activated RIPK1-binding regions, we performed Cleavage Under Targets and Tagmentation (CUT&Tag)22,23 using the antibodies against p-S166-RIPK1 and conducted HOMER motif analysis of p-S166-RIPK1-binding peaks on promoters and enhancers. We found differential transcription factor-binding motifs in cells upon 2 h treatment with TNFα compared to those treated with TNFα plus Nec-1s. Representative binding motifs of transcription factors (SP1, JUNB and RELA) shown to interact with nuclear RIPK1 upon TSZ stimulation (Supplementary information, Fig. S1d) were found in the binding regions of p-S166-RIPK1, SMARCC2 and BRG1 (Fig. 3a), and additional transcription factor-binding motifs were listed in Supplementary information, Fig. S3a. In summary, these results suggest that activated nuclear RIPK1 is recruited by multiple transcription factors to their genomic targets.
Fig. 3. Inhibition of RIPK1 kinase activity disrupts genome-wide DNA accessibility by repressing the co-occupancy of SMARCC2 and BRG1.
a HOMER motif analysis of the binding peaks of p-S166-RIPK1, SMARCC2 or BRG1 on promoters or enhancers detected by CUT&Tag under the treatment of TNFα versus TNFα plus Nec-1s. b Heatmap (top) and summary plots (below) of ATAC-seq genomic accessibility reads under the indicated conditions over the de novo sites in Ripk1D325A/D325A;Ripk3−/− MEFs treated with TNFα for 2 h or 4 h. The de novo sites were calculated by the intersection of p-S166-RIPK1-binding peaks with the accessible genomic regions induced by TNFα. c The summary plots (top) and binding intensities (below) of p-S166-RIPK1, SMARCC2 and BRG1 under the treatment of vehicle, TNFα and TNFα plus Nec-1s were shown by CUT&Tag reads over the defined sites (See Supplementary information, Fig. S3c) in Ripk1D325A/D325A;Ripk3−/− MEFs treated with TNFα for 2 h (1110 sites) or 4 h (2159 sites). The defined sites were calculated by the intersection between p-S166-RIPK1/SMARCC2/BRG1 co-binding peaks under the treatment of TNFα and RIPK1 kinase activity-dependent accessible regions. The RIPK1 kinase activity-dependent accessible regions were defined as the accessible regions that induced by TNFα and became less accessible when adding Nec-1s. d Log2-transformed binding scores of SMARCC2 and BRG1 were plotted to show the co-occupancy of SMARCC2 and BRG1 under the treatment of vehicle, TNFα and TNFα plus Nec-1s over the interested genomic regions defined in Supplementary information, Fig. S3c. Each point represents a specific site with the binding of SMARCC2 (x value) and BRG1 (y value). The sites selected by the square are defined as SMARCC2/BRG1 co-binding sites, while the sites which are plotted on x axis are defined as sites bound with SMARCC2 only, and the ones on y axis are defined as sites bound with BRG1 only. The sites which are plotted on the minus infinity of x axis and y axis are defined as no-binding sites, and the rest are defined as weak binding sites. e Summary of the percentage changes of SMARCC2/BRG1 co-binding, SMARCC2 only, BRG1 only, no-binding and weak binding sites upon the treatment of vehicle, TNFα and TNFα plus Nec-1s in d. The genomic regions used here are defined in Supplementary information, Fig. S3c. f The ATAC-seq profiles (orange) and CUT&Tag profiles of p-S166-RIPK1 (red), SMARCC2 (green) and BRG1 (blue) genomic binding upon the treatment of vehicle, TNFα and TNFα plus Nec-1s on Tnfaip3 gene locus. g Heatmap showing the scaled normalized counts in RNA-seq data of the selected genes. These genes are not only induced by TNFα and repressed by TNFα plus Nec-1s at 2 h (42 genes) or 3 h (45 genes), but also the genomic targets of p-S166-RIPK1, SMARCC2 and BRG1 (in CUT&Tag) whose accessibility is induced by TNFα and disrupted by TNFα plus Nec-1s (in ATAC-seq). See also Supplementary information, Table S2. h Pathway analysis by Metascape for the selected target genes of p-S166-RIPK1, SMARCC2 and BRG1 in g. i p-S166-RIPK1 ChIP-qPCR in Ripk1D325A/D325A;Ripk3−/− MEFs treated with vehicle, TNFα and TNFα plus Nec-1s. For each position, ChIP binding normalized to input is shown as fold change to the condition treated with vehicle. n = 3, **P < 0.01, ***P < 0.001.
RIPK1 kinase modulates mSWI/SNF nucleosome-remodeling activity
We used assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) to characterize the impact of RIPK1 on chromatin accessibility in Ripk1D325A/D325A;Ripk3−/− MEFs treated with TNFα for 2 h or 4 h in the presence or absence of Nec-1s. We found that treatment with TNFα induced a significant enrichment of accessible chromatin regions that bound with activated RIPK1, which was reduced upon the treatment with Nec-1s (Fig. 3b; Supplementary information, Fig. S3b).
To assess the function of the RIPK1/BAF complex at a genome-wide level, we cross-analyzed the CUT&Tag data and the ATAC-seq data to study the distribution of activated RIPK1, SMARCC and BRG1 on chromatin accessible regions. We found specific sets of chromatin regions which were co-bound with p-S166-RIPK1, SMARCC2 and BRG1, and became accessible in cells treated with TNFα at both 2 h or 4 h and less accessible in cells treated with TNFα plus Nec-1s (1110 sites at 2 h, TNFα treatment; 2159 sites at 4 h, TNFα treatment) (Supplementary information, Fig. S3c). The genomic co-occupancy of activated RIPK1 and SMARCC2 to these chromatin regions was inhibited by Nec-1s after 2-h and 4-h treatment of TNFα plus Nec-1s compared to that after treatment of TNFα alone, while BRG1 co-occupancy was reduced at 2 h by Nec-1s (Fig. 3c).
In addition, we calculated the binding scores of SMARCC2 and BRG1 on these chromatin regions and plotted the log2 score of SMARCC2 (x axis) and BRG1 (y axis) of individual regions (Fig. 3d). We found a substantial reduction in the chromatin regions co-bound with SMARCC2 and BRG1 in cells treated with TNFα plus Nec-1s compared to that of TNFα only (Fig. 3d). We next summarized the percentage changes of chromatin regions with SMARCC2/BRG1 co-bound, BRG1 bound only, SMARCC2 bound only, weak binding, and no-binding in response to TNFα or TNFα plus Nec-1s treatment. Results showed higher TNFα-induced co-occupancy of SMARCC2 and BRG1 in chromatin regions that were accessible to activated RIPK1, which was diminished by Nec-1s (Fig. 3e). Notably, the DNA accessibility and co-occupancy of BRG1 and SMARCC2 at p-S166 RIPK1 binding sites are exemplified by Tnfaip3, Cxcl1, Il6, Il1a loci, which are representatives of several other loci (Fig. 3f; Supplementary information, Fig. S3e).
We then analyzed the chromatin occupancy of SMARCC2 and BRG1 in an alternative way by calculating the binding scores of SMARCC2 or BRG1 on defined regions (TNFα-inducible, p-S166-RIPK1-dependent accessible regions that are co-bound with activated RIPK1, SMARCC2 and BRG1) upon treatments of vehicle, TNFα, and TNFα plus Nec-1s. After the log2 ratio was plotted, we found that more than half of the 2 h, 4 h TNFα treatment-inducible (log2[TNF/Vehicle] > 1) SMARCC2-binding regions and 2 h TNFα treatment-inducible BRG1-binding regions showed significantly reduced occupancy when treated with Nec-1s (Supplementary information, Fig. S3d).
To characterize the target genes transcriptionally regulated by the RIPK1/BAF complex, we cross-analyzed the data of RNA-seq, ATAC-seq and CUT&Tag from Ripk1D325A/D325A;Ripk3−/− MEFs. The analysis of RNA-seq data identified 69 genes which showed at least 1.5-fold upregulation upon TNFα stimulation and at least 1.5-fold downregulation by the treatment with Nec-1s. We found that 43 of the 69 genes showed co-binding of p-S166-RIPK1/SMARCC2/BRG1 as detected by CUT&Tag and were among TNFα-induced accessible genomic regions which could be inhibited by treatment with Nec-1s as detected by ATAC-seq (Fig. 3g; Supplementary information, Table S2). These genes identified include Il6, Cxcl1–3, Il1a, Cxcl1–3, which were well-established RIPK1 kinase-regulated proinflammatory target genes.2–4 Strikingly, TNF signaling pathway, inflammatory responses and fever generation are the top 3 enriched pathways based on the annotation and calculation of the 43 overlapped target genes by Metascape24 (Fig. 3h). Validation for p-S166-RIPK1/SMARCC2/BRG1 targets by p-S166-RIPK1 ChIP-qPCR confirmed strong enrichment of activated RIPK1 on Il1a, Il6, Cxcl1, Tnfaip3 in Ripk1D325A/D325A;Ripk3−/− MEFs treated with TNFα and the enrichment was blocked by Nec-1s (Fig. 3i). These results demonstrate the pathological mechanism by which activated RIPK1 directly mediates the transcriptional induction of proinflammatory cytokines in patients with activating D324V and D324H RIPK1 mutations who suffer from an autoinflammatory disease with recurrent fevers, and strong RIPK1-dependent activation of inflammatory signaling pathways, and overproduction of inflammatory cytokines and chemokines.6 The congruence of three orthogonal, unbiased genome-wide analyses strongly support the role of RIPK1/BAF complex-regulated chromatin-remodeling activity in the activation of proinflammatory responses.
Regulation of BAF complex by RIPK1-mediated phosphorylation
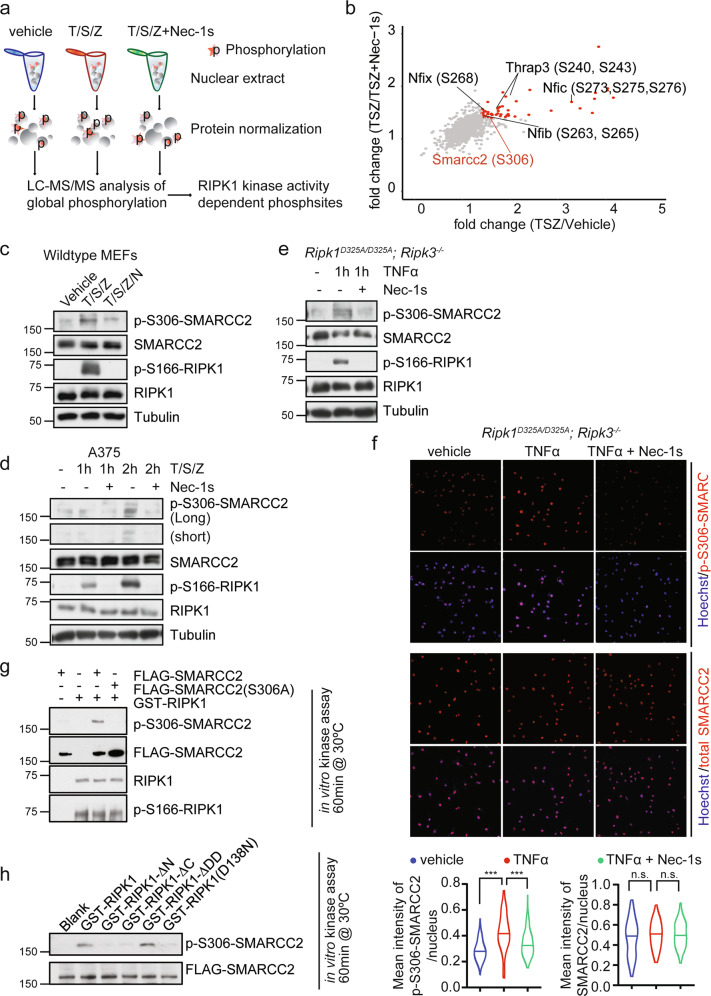
To further investigate the interaction between nuclear RIPK1 and its interacting partners, we next characterized the nuclear phosphoproteome in MEFs treated with T/S/Z or T/S/Z + Nec-1s using quantitative mass spectrometry (Figs. 2a, right and 4a). Nuclear extracts were isolated from WT MEFs treated with vehicle, T/S/Z or T/S/Z + Nec-1s for 1 h, a timepoint prior to necroptotic cell death occurring. The RIPK1 kinase-dependent phosphorylation of nuclear proteins was summarized (red, Fig. 4b; Supplementary information, Table S3). Intriguingly, this analysis identified RIPK1-dependent phosphorylation of SMARCC2, which was also found to be a nuclear RIPK1-interacting protein (Supplementary information, Fig. S1d and Table S1).
Fig. 4. Nuclear RIPK1 modulates nucleosome-remodeling activity by mediating the phosphorylation of SMARCC2.
a Schematic illustration of the quantitative mass spectrometry approach to identify the RIPK1 kinase activity-related phosphoproteome. MEFs were treated with vehicle, or 50 µM SM164, 20 µM zVAD-fmk, with or without 20 µM Nec-1s for 0.5 h followed by 5 ng/mL mTNFα for 1 h. b Plot showing log2(TSZ/Vehicle) ratio of phosphosites on x axis and log2(TSZ/(TSZ + Nec-1s)) ratio of phosphosites on y axis. Phosphosites (localization probability > 0.75) of transcriptional regulators/co-regulators which were detected to interact with activated RIPK1 are labeled. c MEFs were treated with vehicle, or 50 µM SM164, 20 µM zVAD-fmk, ± 20 µM Nec-1s for 0.5 h followed by 5 ng/mL mTNFα for 1 h. Cell lysates were immunoblotted with the indicated antibodies. d A375 cells were treated with vehicle, or 50 µM SM164, 20 µM zVAD-fmk, ± 20 µM Nec-1s for 0.5 h followed by 5 ng/mL hTNFα for 1 h. Cell lysates were immunoblotted with the indicated antibodies. e Ripk1D325A/D325A;Ripk3−/− MEFs were treated with vehicle, or ± 20 µM Nec-1s for 0.5 h followed by 100 ng/mL mTNFα for 1 h or 2 h and analyzed by immunoblotting. f Immunostaining of p-S306 SMARCC2, SMARCC2 in Ripk1D325A/D325A; Ripk3−/− MEFs treated with vehicle, or ± 20 µM Nec-1s for 0.5 h followed by 20 ng/mL mTNFα for 1 h. Representative images and quantification of the mean intensity of p-S306 SMARCC2 or SMARCC2 per nucleus are shown. ***P < 0.001, n.s., not significant. g In vitro kinase assay with GST-RIPK1 purified from E. coli and FLAG-SMARCC2 or FLAG-SMARCC2(S306A) purified from 293T cells. The reaction was incubated at 30 °C for 1 h in vitro with the presence of 200 µM ATP. h In vitro kinase assay with GST-RIPK1, GST-RIPK1-ΔN, GST-RIPK1-ΔC, GST-RIPK1-ΔDD, GST-RIPK1(D138N) purified from E. coli and FLAG-SMARCC2 purified from 293T cells. The reaction was incubated at 30 °C for 1 h in vitro with the presence of 200 µM ATP.
Considering that SMARCC2 binds to RIPK1 directly, we next explored the association between the phosphorylation of SMARCC2 and RIPK1 kinase activity. Cell lysates from wild-type MEFs treated with vehicle, T/S/Z, or T/S/Z plus Nec-1s for 1 h were analyzed by western blotting with an anti-p-S306 SMARCC2 antibody (Fig. 4c) to confirm the result from mass spectrometry. p-S306 SMARCC2 was detected in MEFs treated with T/S/Z, which was inhibited by the addition of Nec-1s. The p-S306 SMARCC2 was also detected in the human A375 cells treated with T/S/Z for 2 h and the signal was also inhibited by the addition of Nec-1s (Fig. 4d). We next confirmed that the levels of p-S306-SMARCC2 in Ripk1D325A/D325A;Ripk3−/− MEFs were induced by 1-h ΤΝFα treatment and inhibited by treatment with Nec-1s using western blotting (Fig. 4e) and immunostaining (Fig. 4f) with p-S306-SMARCC2 antibody. Furthermore, to test whether RIPK1 is able to phosphorylate Ser306-SMARCC2, we carried out in vitro kinase assay with GST-fused full-length RIPK1, truncated RIPK1 or D138N RIPK1 mutant and FLAG-purified wild-type SMARCC2 (FLAG-SMARCC2) or S306A-SMARCC2 mutant (FLAG-SMARCC2(S306A)). The result showed that Ser306-SMARCC2 can be directly phosphorylated by RIPK1 in vitro (Fig. 4g) and both the kinase domain and intermediate domain are necessary for the reaction (Fig. 4h).
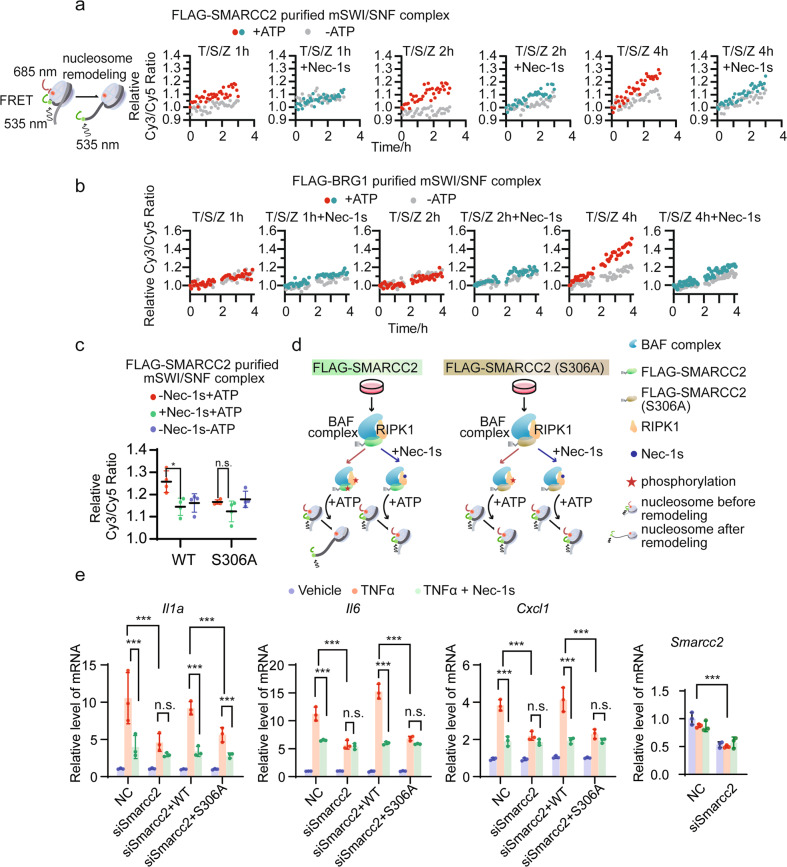
To explore the role of RIPK1 in the function of chromatin-remodeling complex, we performed a nucleosome remodeling assay with purified mSWI/SNF complex and mononucleosomes assembled from recombinant human Cy5-conjugated histones expressed in E. coli wrapped with a 207 base-pair 5’ Cy3-conjugated DNA sequence (Fig. 5a, left). As 293T cells are resistant to TSZ-induced necroptosis within 4 h (Supplementary information, Fig. S4b), we were able to purify RIPK1/mSWI/SNF complex using FLAG-SMARCC2 (referred to as FLAG-SMARCC2-purified complex hereafter, Fig. 5a) or FLAG-BRG1 (referred to as FLAG-BRG1-purified complex hereafter, Fig. 5b) from 293T cells treated with T/S/Z or T/S/Z plus Nec-1s. The major components of the purified complexes were confirmed by western blotting (Supplementary information, Fig. S4a). The nucleosome-remodeling activities were assessed using Cy3/Cy5 FRET and quantified as the Cy3/Cy5 ratio. Significant remodeling activities were detected from the reaction with the addition of the FLAG-SMARCC2-purified complex from 293T cells treated with T/S/Z without Nec-1s for 2 h and 4 h (Fig. 5a). In addition, the FLAG-BRG1-purified complex from 293T cells treated with T/S/Z for 4 h showed a significant remodeling activity, whereas the FLAG-BRG1-purified complex from cells treated with T/S/Z plus Nec-1s displayed dramatically decreased remodeling activity (Fig. 5b).
Fig. 5. Nuclear RIPK1 promotes transcription by mediating the phosphorylation of SMARCC2.
a Schematic diagram showing nucleosome remodeling assay with mononucleosome assembled from recombinant human Cy5-conjugated histones expressed in E. coli wrapped with a 207 base-pair 5′ Cy3-conjugated DNA sequence (left). The nucleosome-remodeling activity of FLAG-SMARCC2-purified mSWI/SNF complexes from 293T cells treated under indicated conditions are assessed using Cy3/Cy5 FRET with (red, green) or without (gray) the presence of 4 mM ATP. b The nucleosome-remodeling activity of FLAG-BRG1-purified mSWI/SNF complexes from 293T cells treated under indicated conditions are assessed using Cy3/Cy5 FRET with (red, green) or without (gray) the presence of 4 mM ATP. c The nucleosome-remodeling activity of FLAG-SMARCC2- or FLAG-SMARCC2(S306A)-purified mSWI/SNF complexes from 293T cells are assessed using Cy3/Cy5 FRET with the presence of 4 mM ATP (red), or with 4 mM ATP plus 20 µM Nec-1s (green), or without ATP (blue) (n = 3, means ± SD). *P < 0.05, n.s., not significant. d Schematic summary of results in c. e Ripk1D325A/D325A;Ripk3−/− MEFs were transfected with the Smarcc2 siRNA with or without the indicated plasmids for wild-type SMARCC2 or SMARCC2(S306A) expression. Cells were treated with the indicated conditions before subjected to RT-qPCR analyses of the expression levels of the indicated genes (n = 3, means ± SD). ***P < 0.001, n.s., not significant.
As these results suggest that RIPK1 activation is required in the RIPK1/mSWI/SNF complex for nucleosome remodeling in TNFα-treated cells, we then sought to determine whether the RIPK1 activation could affect nucleosome-remodeling activity via the phosphorylation of Ser306-SMARCC2. To this end, the complexes purified by FLAG-SMARCC2 and FLAG-SMARCC2(S306A) from 293T cells with or without Nec-1s treatment were added into the reaction of nucleosome remodeling assay, respectively. We found that only the reaction with the FLAG-SMARCC2-purified complex showed RIPK1-dependent remodeling activity upon the addition of ATP compared to the control group without the addition of ATP. Moreover, we observed a remodeling defect in the FLAG-SMARCC2(S306A)-purified complex in which SMARCC2 was not able to be phosphorylated by RIPK1 (Fig. 5c, d). Altogether, these results suggest that RIPK1 modulates nucleosome-remodeling activity by phosphorylating SMARCC2 at Ser306 site.
Given our observation that the in vitro nucleosome-remodeling activity of the RIPK1/mSWI/SNF complex requires the phosphorylation of S306-SMARCC2 mediated by activated nuclear RIPK1, we next examined if p-S306-SMARCC2 was necessary for the nuclear RIPK1/BAF complex to promote the transcription of its target genes. Ripk1D325A/D325A;Ripk3−/− MEFs were transfected with SMARCC2 siRNA to knockdown endogenous SMARCC2 and then complemented with wild-type SMARCC2 or S306A-SMARCC2 mutant expression plasmid; the cells were treated with vehicle, TNFα, or TNFα plus Nec-1s. The expression levels of three representative target genes (Il1a, Il6 and Cxcl1) selected from the 48 overlapping genes were measured by RT-qPCR. Results showed that the knockdown of SMARCC2 resulted in a diminishment of TNFα-induced, RIPK1 kinase activity-dependent transcription activation, which was rescued by adding back wild-type SMARCC2 but not S306A-SMARCC2 mutant (Fig. 5e), supporting the necessity of Ser306-SMARCC2 phosphorylation in transcription activation mediated by activated nuclear RIPK1.
RIPK1/BAF complex regulates inflammatory gene expression in ALS patients
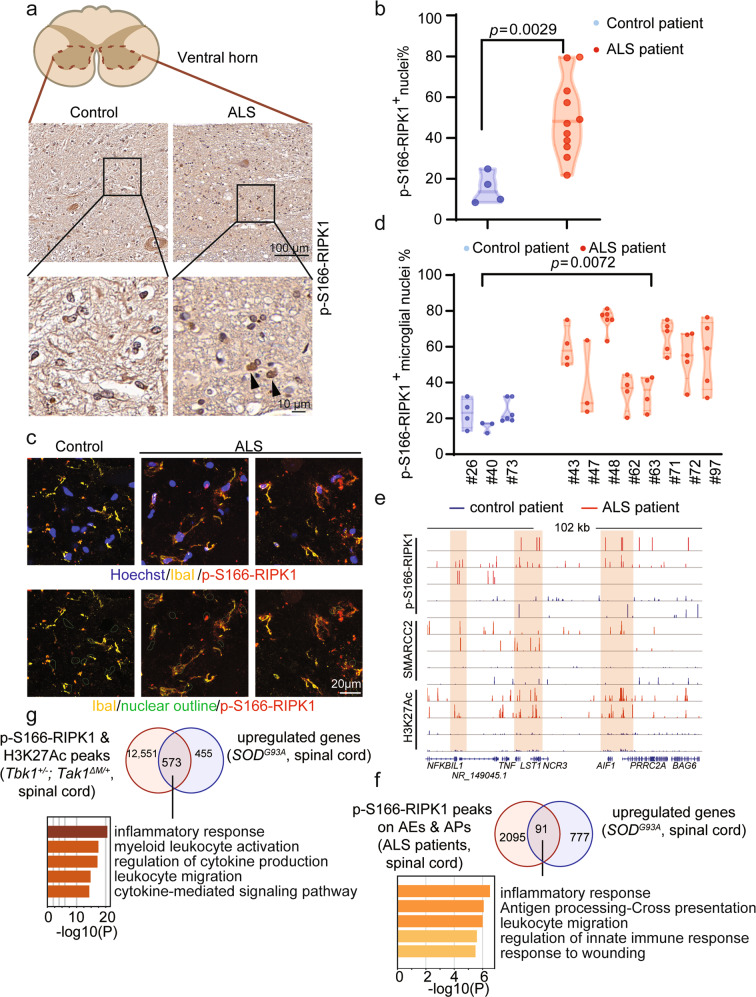
Given that the results described above demonstrated the role and mechanism of activated RIPK1 in the nuclei of wild-type and D325A-Ripk1 mutant MEFs in promoting chromatin remodeling to mediate the transcriptional induction of proinflammatory cytokines upon TNFα stimulation, we next examined the role of nuclear RIPK1/BAF complex in promoting the transcription of inflammatory genes in human pathological contexts. Since RIPK1-dependent transcriptional induction of proinflammatory cytokines has been found in the spinal cords of multiple ALS mouse models and the activation of RIPK1 has also been found in the lysates of spinal cords from the post-mortem pathological samples of ALS patients,2,25 we next examined the activated RIPK1 in the paraffin-embedded sections of upper spinal cord tissues from 11 ALS patients and age-matched control patients by immunohistochemistry (IHC) using p-S166-RIPK1 antibody (Supplementary information, Table S4). We found an increase in the number of nuclei with activated RIPK1 (p-S166-RIPK1+) staining in the ventral horns of spinal cords from ALS patients compared to that of age-matched controls (Fig. 6a; Supplementary information, Fig. S5a). An increase in the percentage of p-S166-RIPK1+ nuclei was found in the spinal cords from ALS patients (Fig. 6b). Further double immunostaining of p-S166-RIPK1 and microglial marker IBA1 suggests that at least a subset of cells with activated RIPK1 in the spinal cords of ALS patients were microglia, which were known to be increased in numbers26 (Fig. 6c, d).
Fig. 6. Genome-wide analysis of the transcriptional targets of RIPK1 and BAF complex in ALS patients and ALS mouse model.
a, b IHC staining of upper spinal cord sections from human patients with ALS (n = 11) and age-relevant controls (n = 4). Representative images (a) and the percentage of p-S166 RIPK1+ nuclei (b) in ventral horn are shown. c, d Immunostaining of upper spinal cord sections from human patients with ALS (n = 8) and age-relevant controls (n = 3) against p-S166 RIPK1 and microglia marker IbaI. Representative images (c), quantification of the percentage of p-S166-RIPK1+ microglial nuclei (d) in ventral horn are shown. e The CUT&Tag profiles of p-S166-RIPK1, SMARCC2 genomic binding and H3K27Ac in spinal cords from ALS patients (red) and control patients (blue) on the indicated gene loci. f Venn diagram shows the intersected genes calculated by the gene targets of activated RIPK1 on AEs and APs in the spinal cords from ALS patients and the upregulated genes in the spinal cords from SOD1G93A ALS mouse model from published RNA-seq datasets. 91 overlapped target genes were analyzed by Metascape for the enriched pathways. g Venn diagram shows the intersected genes calculated by the gene targets enriched with p-S166-RIPK1/H3K27Ac peaks in the spinal cords from Tbk1+/−; Tak1ΔM/+ mice and the upregulated genes in the spinal cords from SOD1G93A ALS mouse model from published RNA-seq datasets. 573 overlapped target genes were analyzed by Metascape for the enriched pathways.
To identify the genomic targets of nuclear RIPK1/BAF complex in ALS, nuclei were isolated from fresh-frozen post-mortem human spinal cords and subjected to CUT&Tag with antibodies against p-S166-RIPK1, SMARCC2, H3K27Ac, and H3K4me1. Differential peaks of H3K4me1, H3K27Ac modifications, and p-S166-RIPK1 binding in the spinal cords from ALS patients relative to those of the controls were called using the DiffBind package. Visualization of the peaks showed increased binding of p-S166-RIPK1, SMARCC2, and H3K27Ac on TNF gene locus in ALS group compared to those of controls (Fig. 6e). H3K27Ac is a marker for transcriptionally active chromatin regions.27,28 AEs were annotated by intersection of H3K27Ac- and H3K4me1-binding regions, while APs were annotated by H3K27Ac-binding regions 2 kb upstream from transcription starting sites. Intersected peaks of p-S166-RIPK1 on AEs or APs differentially enriched in ALS group were calculated, which identified a total of 2186 genes. Cross-analysis of CUT&Tag in the spinal cords from ALS patients and the published RNA-seq in the spinal cords from SOD1G93A ALS mouse model29 showed 91 overlapped target genes (mouse gene symbols were converted to human gene symbols before the comparison) (Supplementary information, Table S5). These genes were then identified as highly enriched in inflammatory responses using Metascape (Fig. 6f).
To further validate the transcriptional targets of nuclear RIPK1, we performed CUT&Tag in Tbk1+/−;Tak1ΔM/+ ALS mouse model25 with antibodies against p-S166-RIPK1, SMARCC2 and H3K27Ac. Intersection of specific peaks of p-S166-RIPK1 and H3K27Ac in the spinal cords from Tbk1+/−;Tak1ΔM/+ mice were defined as RIPK1-bound active regions to identify potential target genes. These genes were then intersected with the 1028 upregulated genes detected by the published RNA-seq of whole spinal cords from SOD1G93A ALS mouse model.29 The 573 overlapped genes (Supplementary information, Table S5) were then analyzed by Metascape, showing high enrichment in inflammatory responses (Fig. 6g). Collectively, the above observations revealed a critical role of nuclear RIPK1 in the progression of inflammatory diseases by modulating the remodeling activity of the BAF complex via the phosphorylation of SMARCC2 to promote the transcription of downstream pro-inflammatory genes.
Discussion
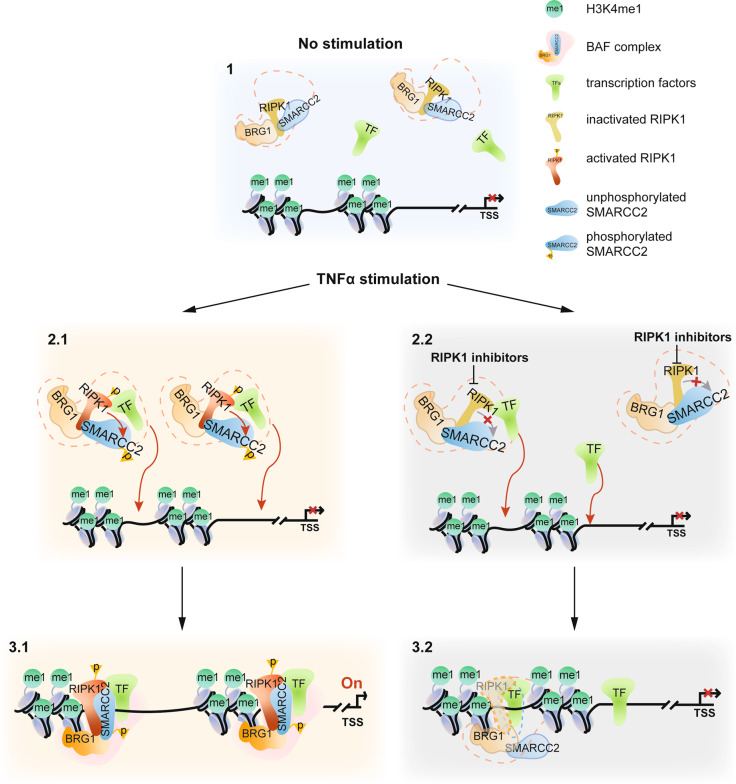
Chromatin accessibility, defined as the degree that specific regulators of gene transcription are allowed to interact with genomic DNA in cells, is a critical regulatory mechanism that is continuously and dynamically modulated to control transcription factor binding and remodeler-mediated nucleosomal removal to promote transcription in response to different stimuli under physiological and pathological conditions.30 In this regard, the role of TNFα as a potent inducer of transcriptional induction of proinflammatory factors has been well-established. The importance of TNFα in promoting inflammatory responses in human diseases has been underscored by the success of anti-TNF therapy.31 However, it remains unclear how TNFα stimulation or inflammation in general leads to chromatin remodeling to promote the expression of specific proinflammatory genes. Our results described above suggest that RIPK1 interacts with the BAF complex, and upon its activation induced by TNFα signaling is recruited by multiple active transcription factors to specific genome regions to modulate the accessibility of these specific genomic DNA sites to promote the transcription of proinflammatory genes (Fig. 7). The BAF complex is known to regulate nucleosome positioning and recruit transcriptional machinery.32,33 Our results suggest that in cells stimulated by TNFα, transcription factors bind to their target enhancers and recruit RIPK1/BAF complex to the genomic target sites where RIPK1 binds to H3K4me1 directly. Activated RIPK1 mediates the phosphorylation of SMARCC2, a subunit important in promoting the BAF complex-medicated chromatin remodeling.34 As a result, the enhancer DNA accessibility is increased, and the transcription of target genes is activated. Inhibition of RIPK1 suppresses the recruitment of RIPK1/BAF complex by transcription factors and the phosphorylation of SMARCC2, which disrupts the co-occupancy of SMARCC2 and BRG1 on the target sites and ultimately suppresses the transcription of proinflammatory factors by reducing the enhancer DNA accessibility (Fig. 7). Our study demonstrates the role of the RIPK1/BAF complex in promoting inflammatory responses in ALS patients who carry genetic variants of RIPK1 (D324V and D324H) that block the cleavage by caspase-8. Mutations in the various subunits of the BAF complex have been found to be responsible for BAFopathies, a heterogeneous group of disorders including mental retardation, autism, schizophrenia, and ALS.35,36 Our results suggest that dysregulated RIPK1-mediated inflammatory response might also contribute to the pathology of BAFopathies.
Fig. 7. A model of RIPK1 kinase activity-dependent transcriptional activation by regulating chromatin-modeling activity.
RIPK1 interacts with BRG1 and SMARCC2 directly. Upon TNFα stimulation, transcription factors recruit RIPK1/BAF complex to the genomic target sites where RIPK1 is able to bind H3K4me1 directly. Activated RIPK1 mediates the phosphorylation of SMARCC2, which is important in promoting the BAF complex-medicated chromatin remodeling. As a result, the DNA accessibility of the genomic target sites is increased, and the transcription of target genes is activated. With inhibition of RIPK1 kinase activity, less RIPK1/BAF complex is recruited by specific transcription factors to the genomic targets. In addition, inhibition of RIPK1 kinase activity also suppresses the phosphorylation of SMARCC2, which disrupts the co-occupancy of SMARCC2 and BRG1 on the target sites and the chromatin-remodeling activity of the BAF complex, and as a result, suppresses the transcription of proinflammatory factors by reducing the DNA accessibility on the genomic regions.
Our study identified the role of RIPK1-mediated phosphorylation of S306 SMARCC2 in promoting chromatin remodeling to regulate inflammatory responses. p-S302, p-S304 and p-S306 are a set of closely positioned phosphorylation sites on SMARCC2. In our analysis, p-S306 is regulated by RIPK1, while p-S302 and p-S304 were unaffected. The phosphorylation of S302/S304 in SMARCC2 have been found in several global phosphoproteome studies, including mouse tissues37 and in association with neural differentiation.34 In addition, the phosphorylation of S302/S304/S306 was found in cancer cells treated with tyrosine kinase inhibitor Dasatinib.34 It is possible that S302/S304/S306 in SMARCC2 represents an important regulatory module whose phosphorylation controls the chromatin-remodeling activity of the BAF complex downstream of different signaling events-triggered activation.
Transcriptional induction of proinflammatory factors is a key aspect of inflammation in different tissues and contexts. We show that RIPK1 is recruited by multiple transcription factors involved in inflammatory responses. The roles of different transcriptional activators/coactivators in mediating inflammatory responses have been well-documented in the literature. In particular, NF-κB has a well-established role in mediating the transcription of inflammatory factors in different inflammatory diseases.38 The activation of NF-κB and Sp1 has been suggested to mediate inflammation in microvascular endothelial cells.39 Binding of NF-κB with super enhancers is involved in mediating transcriptional induction of inflammatory response in vascular endothelial cells and atherogenesis.40 In addition, JUNB is a key transcriptional modulator of macrophage-mediated inflammatory response41 and inflammation of the skin in animal model of psoriasis.42 Transcriptional activity of Sp1 may be involved in mediating neurodegeneration in Huntington’s disease.43,44 However, it is unclear how cells coordinately regulate the activation of these different transcriptional factors to promote inflammation. Our results suggest that RIPK1/BAF complex is a key regulator of chromatin remodeling that promotes the chromatin accessibility to diverse array of transcriptional factors to mediate the transcriptional induction of inflammatory genes.
Increased number and activation of microglia is a hallmark of neurodegenerative diseases such as ALS.26 Activation of NF-κB has been proposed to mediate inflammation to promote the death of motor neurons in ALS.45–47 Activation of RIPK1 in microglia upon loss of Optineurin in mice, a genetic model of ALS, leads to transcriptional induction of proinflammatory factors with enrichment for Sp1 target genes.2 While excessive activation of inflammation has a well-recognized role in human diseases, attempts to inhibit the transcription of proinflammatory factors for the treatment of human inflammatory diseases have been met with the technical challenge of blocking transcriptional factors which are difficult to target by traditional small-molecule approaches. Additionally, targeting NF-κB-activating proteins such as TAK1 and IKKs results in significant on-target toxicities which is at least in part due to the roles of TAK1 and IKKs in performing inhibitory phosphorylation on RIPK1.48,49 Since multiple small-molecule inhibitors of RIPK1 kinase have been shown to be safe in humans,1 our study suggests that inhibition of RIPK1 kinase provides an exciting opportunity to reduce the transcriptional induction of proinflammatory factors by modulating BAF complex-mediated chromatin-remodeling activity for the treatment of human inflammatory and neurodegenerative diseases.
Materials and methods
Cell lines and cell culture
All cells were cultured at 37 °C with 5% CO2. All cell lines and primary cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Cat# 11965) with 10% (vol/vol) fetal bovine serum (FBS; Thermo Fisher Scientific, Cat# 10082–147).
Generation and immortalization of MEFs
Immortalized MEFs were generated as previously described.25 Briefly, MEFs were isolated from E11–E13 embryos and were cultured in high glucose DMEM supplemented with 15% FBS, non-essential amino acids, sodium pyruvate, penicillin, streptomycin and amphotericin B. Primary MEFs were immortalized by transfection with SV40 large T antigen-expressing plasmid (Addgene 22298) at passages 4–6.
Animals
Tbk1+/− mice and Tak1fl/fl mice50,51 were used in this study. Lyz2Cre/Cre mice were from The Jackson Laboratory (Cat# 004781). Tbk1+/−;Tak1ΔM/+ mice and Tbk1+/−;Tak1ΔM/+;Ripk1D138N/+ mice were generated as previously described.25 All animals were maintained in a pathogen-free environment, and experiments on mice were conducted according to the protocols approved by the Harvard Medical School Animal Care Committee.
Human tissues
The research involving human pathological samples was approved by the Institutional Review Board (IRB) of Harvard University. Frozen tissues from the spinal cord of patients with ALS and control subjects without ALS were obtained from Dr. John Ravits at UCSD. Paraffin-embedded tissue slides were obtained from Dr. John Ravits at UCSD and Dr. Matthew Frosch of Harvard Neuropathology Services. More information is shown in Supplementary information, Table S4.
Plasmid construction
For protein purification, cDNA encoding full-length hRIPK1 (1−671 aa) and truncated mRIPK1 (275−671, 1−289, 1−583 aa) were cloned into pGEX-4T-3 plasmid for E. coli expression. cDNA encoding MYC-tagged ARID1A, BRG1, SMARCC1 and SMARCC2 were cloned into pGBKT7 plasmid for in vitro transcription/translation of the proteins. SMARCC2 S306A mutant and hRIPK1 D138N mutant were generated using MutExpress II mutagenesis kit (Vazyme Biotech). All plasmids were verified by DNA sequencing and the details of the plasmids sequences are available upon request.
Nuclear/Cytoplastic fractionation
Nuclear/Cytoplastic fractionation was done according to a previous protocol.52
Mass spectrometry
FLAG-RIPK1 reconstituted Ripk1−/− MEFs were treated with vehicle or 50 µM SM164, 20 µM zVAD-fmk, with or without 10 µM Nec-1s for 0.5 h and then treated with 5 ng/mL mTNFα for 1 h. The nuclear proteins were extracted. RIPK1-binding proteins were co-immunoprecipitated using FLAG-M2 beads and trypsin digested on beads. The resulting peptides were desalted and directly analyzed on the Q Exactive HF mass spectrometer (Thermo Scientific) for the detection of RIPK1 interactome. The peptides from the nuclear extraction were subjected to the enrichment of phosphorylated peptides using TiO2. The enriched phosphorylated peptides were analyzed on the Q Exactive HF mass spectrometer (Thermo Scientific). The identification and quantification of RIPK1-binding proteins and phosphorylated peptides were done by MaxQuant.53 The tandem mass spectra were searched against UniProt human protein database together with a set of commonly observed contaminants. The precursor and the fragment mass tolerance were set as 20 ppm. The cysteine carbamidomethylating was set as a static modification, and the methionine oxidation as well as serine, threonine and tyrosine phosphorylation were set as variable modifications. The FDR at peptide spectrum match level and protein level were controlled below 1%. Proteins in the results were annotated to transcription factors and transcriptional coregulators with the Gene Ontology terms.
IP and western blotting
IP and western blotting were performed according to the procedures described previously.54 Immunoprecipitated complexes were eluted from FLAG-M2 beads in FLAG IP.
RT-qPCR
RT-qPCR was performed the same as previously described.55
Protein expression and purification
Recombinant GST-hRIPK1 (1−671, 275−671, 1−289, 1−583 aa) protein fragments were expressed in E. coli BL21(DE3) after induction with 0.1 mM IPTG overnight at 16 °C. Bacteria were harvested by centrifugation and cell pellet was lysed in PBS containing 1% Triton X-100 and 5 mM DTT with mild sonication at 4 °C for 15 min. The lysates were cleared by centrifugation. GST-tagged RIPK1 fragments were incubated with GST agarose (GE Healthcare) at room temperature for 1 h and washed with wash buffer (0.5% NP40 in PBS) for five times.
GST pull-down assays
GST pull-down assays were done as previously described.54 Briefly, the GST fusion proteins with glutathione-Sepharose beads was incubated with in vitro transcribed/translated products by TnT® Quick Coupled Transcription/Translation System (Promega) in binding buffer (0.8% BSA in PBS) at 4 °C for 2 h in the presence of the protease inhibitor mixture. The beads were washed with wash buffer 1 (0.5% NP40 in PBS) for two times, wash buffer 2 (0.5% NP40, 300 mM KCl in PBS) for one time, and wash buffer 1 for two times. The beads were then resuspended in 2× SDS-PAGE loading buffer and detected by western blotting.
Histone binding
Recombinant full-length or truncated hRIPK1-GST fusion proteins (0.5 μg) were incubated with 0.6 μg of native calf thymus histones (Sigma, # H9250) in binding buffer (50 mM Tris-HCl, pH 7.5, 1 M NaCl, 1% Nonidet P-40, 0.5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF) plus protease inhibitors (Roche)) at 4 °C for 4 h. Protein complexes were pulled down with glutathione beads, washed 5 times with the binding buffer, and subjected to western blotting using anti-H3 and anti-H4 antibodies.
Histone peptide pull-down
Histone peptide pull-down assays were performed according to a previous protocol.56 Unmodified H3 N-terminal peptides (# SKU12–0001) and all modified H3 peptides (# SKU12–0007, 12–0008, 12–0009) were purchased from Epicypher. 0.5 μg purified GST-RIPK1-ΔN was incubated with 5 μg of biotinylated histone peptides in 100 μL of binding buffer (50 mM Tris-HCl, pH7.5, 150 mM NaCl, 0.05% NP-40, 0.3 mg/mL BSA plus protease inhibitors) overnight at 4 °C. Protein−peptide complexes were pulled down with streptavidin beads (Millipore), washed 5 times with the binding buffer, and subjected to western blotting using anti-GST antibody.
EpiDyne-FRET nucleosome remodeling assay
EpiDyne-FRET nucleosome substrates (# SKU 16–4201) were purchased from Epicypher and the assays were performed according to standard protocol. Briefly, 1 µL of FLAG-SMARCC2, SMARCC2 S306A mutant or BRG1-purified BAF complex was incubated with 20 nM EpiDyne-FRET substrates at room temperature in a final volume of 10 µL of remodeling buffer (20 mM Tris-HCl, pH 7.5, 50 mM KCl, 3 mM MgCl2, 100 µg/mL BSA). The reaction was initiated by adding ATP (2 mM) and measured by 384-well plate reader capable of Cy3/Cy5 detection (Perkin Elmer EnVision). Data are shown as the ratio of Cy3 and Cy5 emission signals at each time point.
In vitro kinase assay
Flag-tagged SMARCC2 wild type or S306A mutant were immunoprecipitated from HEK293T cells stably expressing Flag-SMARCC2 or S306A mutant and washed with high-salt lysis buffer (5% glycerol, 0.5% NP-40, 500 mM NaCl and 50 mM Tris-HCl (pH7.4)) for 5 times and then with water for 3 additional times. Beads were eluted with Flag peptides after the final wash step. In vitro kinase assays were performed with 0.2 μg GST-fused full-length hRIPK1, truncated hRIPK1 or kinase-dead hRIPK1 D138N mutant purified from E. coli and 1 μg Flag-tagged SMARCC2 or SMARCC2 S306A mutant in 20 μL 1× kinase buffer (CST, # 9802S) with 200 μM ATP. Kinase reactions were performed for 60 min at 30 °C and quenched by the addition of 5× sample loading buffer and boiling at 100 °C for 10 min.
CUT&Tag analysis
CUTANATM pAG-Tn5 (# SKU15–1017) were purchased from Epicypher. CUT&Tag experiments were performed with 20,000–50,000 nuclei from Ripk1D325A/D325A;Ripk3−/− MEFs or spinal cords according to a previous protocol22,23 and the protocol from Epicypher. The size distribution and concentration of libraries were determined by capillary electrophoresis using an Agilent Bioanalyzer. Barcoded libraries were mixed to achieve equal representation. After mixing, SPRI bead cleanup was performed to remove residual PCR primers. Paired-end 41 × 41 bp sequencing was performed on the barcoded libraries following the manufacturer’s instructions of NextSeq500. Paired-end reads were aligned using Bowtie2 (v2.3.4.3)57 with parameters: --local --very-sensitive --no-unal --no-mixed --no-discordant --phred33 -I 10 -X 700. Peaks were called by SEACR peak caller.58 Bedtools (v2.27.1)59 was used to find intersected or unique peaks among groups. Motif analysis was done by HOMER (v4.10.3).60 Average scores for each of the files in every genomic region were computed and heatmaps were generated using deepTools (v3.0.2).61 Peaks were visualized by IGV.62
RNA-seq
RNA was extracted from wild-type or Ripk1D325A/D325A;Ripk3−/− MEFs using Quick-RNA MiniPrep kit (Zymo Research) with the in-column DNase treatment. Libraries were prepared and sequenced by the Bauer Core facility at Harvard University. Each library was prepared using KAPA RNA HyperPrep Kits. Libraries were sequenced on an Illumina NextSeq500 instrument with paired 38 bp reads. Reads were aligned to the mm9 UCSC mouse transcriptome with Salmon63 and expression values were determined with DESeq2.64
ATAC-seq
ATAC-seq was performed according to a previous protocol.65,66 The size distribution and concentration of libraries were determined by capillary electrophoresis using an Agilent Bioanalyzer. Paired-end 41 × 41 bp sequencing were performed on the barcoded libraries following the manufacturer’s instructions of NextSeq500 to access open vs closed chromatin regions. Data were processed following ATAC-seq guidelines by Harvard FAS informatics (https://informatics.fas.harvard.edu/atac-seq-guidelines.html). Paired-end reads were aligned using Bowtie2 (v2.3.4.3) with parameters: --very-sensitive -k 10. Peaks were called by Genrich. Bedtools (v2.27.1) was used to find intersected or unique peaks among groups. Heatmaps were generated using deepTools (v3.0.2). Peaks were visualized by IGV.62
ChIP-qPCR
ChIP experiments were performed according to the procedure described previously.67 DNA was purified with the QIAquick PCR Purification Kit.
Nucleus isolation from spinal cords
Nucleus isolation was performed according to a previous protocol.68 Frozen human spinal cords or fresh mouse spinal cords were homogenized in 5 mL Dulbecco’s phosphate buffered saline (PBS; Corning) on ice. Homogenates were washed 2 times with NF1 buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 5 mM MgCl2, 0.1 M sucrose, 0.5% Triton X-100). Homogenates were incubated on ice in 5 mL NF1 for 30 min. Nuclei were extracted from homogenates using a 7 mL WheatonTM Dounce Tissue Grinder (DWK Life Sciences, # 357542) and passed through a 70 μm strainer. Homogenates were underlaid with a 1.2 M sucrose cushion and centrifuged at 4000× g for 30 min. Pelleted nuclei were washed with NF1 buffer at 1600× g for 5 min. Nuclei were washed with Nuclear Extraction (NE) Buffer (20 mM HEPES-KOH, pH 7.9, 10 mM KCl, 0.1% Triton X-100, 20% Glycerol, 0.5 mM Spermidine, 1× Roche cOmpleteTM, Mini, EDTA-free Protease Inhibitor) for the following CUT&Tag analysis. Nuclei were stored at –80 °C prior to library preparation.
IHC
Cells were fixed in 4% paraformaldehyde, permeabilized with 1% Triton X-100, and incubated with appropriate primary antibodies followed by addition of rhodamine-conjugated secondary antibodies. For immunostaining of human tissues, frozen spinal cords were sectioned on the cryostat and then fixed with ice-cold acetone. Tissue sections were mounted and blocked with 10% normal goat serum and 1% BSA and then incubated with primary antibodies at 4 °C overnight and secondary antibodies at room temperature for 1 h. Four to six images per sample were randomly selected from ventral horn of spinal cords. All images were collected with a Nikon Ti-E confocal microscope equipped with A1R scan head with spectral detector and resonant scanners; images were acquired with Nikon Elements Software (v4.11). For IHC analysis, paraffin-embedded sections were subjected to antigen unmasking with citrate-based solution (Vector Laboratories, # H-3300, pH 6.0) and incubated with primary antibodies overnight at 4 °C in a humidified chamber, followed by incubation with the HRP-conjugated secondary antibodies. The staining was completed with standard DAB protocols (Vector Laboratories, # MP-7800). The IHC images were scanned by iHisto Inc.
Quantification of IHC
CellProfiler (v3.1.8)69 was used for the quantifications of microglial nuclei co-localized with p-S166-RIPK1 in immunostaining. IBA1+ cells were segmented in each section as microglial cells and Hoechst+IBA1+ signals were segmented as microglial nuclei. p-S166-RIPK1+ microglial nuclei were then identified based on the fluorescence intensity. p-S166-RIPK1+ microglial nuclei and total microglial nuclei were counted, and the ratio was calculated. For IHC staining with paraffin-embedded slides, color deconvolution in ImageJ was used to separate the hematoxylin counterstaining and p-S166-RIPK1 IHC staining. Total nuclei were counted. p-S166-RIPK1-stained nuclei with mean intensity greater than 100 were considered as p-S166-RIPK1+ nuclei. The percentages of p-S166-RIPK1+ nuclei were calculated and compared between ALS and control groups. IHC results were confirmed by a second blinded researcher.
Antibodies
Antibodies of RIPK1 (CST, # 3493), p-S166-RIPK1 (Arigo, # ARG66476), p-S166-RIPK1 (CST, # 44590), LaminB (Santa Cruz, # sc-374015), Tubulin (TransGen, # I11107), RIPK3 (CST, # 95702), MLKL (CST, # 37705), ARID1A (CST, # 12354S), BRG1 (CST, # 49360S), SMARCC1 (CST, # 11956), SMARCC2 (CST, # 12760S), Myc (Sigma, # M4493), RELA (CST, # 8242), SP1 (Santa Cruz, # sc-17824), NFIB (Bethyl Laboratories, # A303–565A-T), SMARCB1 (CST, # 91735), CBP (CST, # 7389S), H3 (CST, # 9715), H4 (CST, # 2935), H3K4me1 (Thermo Fisher, # 710795), H3K27Ac (Sigma, # 07–360), GST (Santa Cruz, # sc-138), p-S306-SMARCC2 (developed by CST), Flag (CST, # 2368), IBA1 (Wako, # 019–19741) were used.
The antibody of p-306-SMARCC2 was developed in collaboration with Cell Signaling Technology. The antibody was generated by immunizing rabbits with a synthetic phosphopeptide corresponding to residues surrounding Ser306 of human SMARCC2. The antibody was purified by peptide affinity chromatography.
Quantification and statistical analysis
Multiple comparisons were statistically evaluated using a two-tailed Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. represents no significant difference unless further specified. Differences were considered to be significant if P value < 0.05. Results are presented as means ± SD unless further specified. Pairwise comparisons between two groups were performed using the Student’s t-test. For multiple comparisons with the three treatments, we performed one-way ANOVA. Analysis was performed blinded with respect to mouse genotype and treatment condition.
Supplementary information
Acknowledgements
We thank Dr. Kevin Struhl for critical reading of this manuscript. We thank Dr. Jianke Zhang of Thomas Jefferson University, for Ripk1D325A/D325A/Ripk3−/− MEFs, Dr. Matthew Frosch of Harvard Neuropathology Services for providing post-mortem human spinal cord pathological samples, Dr. Jennifer Walters at the Harvard Medical School Nikon microscope facility for assistance with fluorescence microscopy, Dr. Gary Kasof of Cell Signaling Technology for developing p-S306 SMARCC2 antibody and Prof. Jiahuai Han of Xiamen University for providing Ripk1D325A/D325A MEFs during revision. This work was supported in part by the National Natural Science Foundation of China (82188101, 21837004 and 91849204), and the National Natural Science Youth Foundation of China (31701210).
Author contributions
J.Y. and W.L. contributed to the conception of this project. J.Y. supervised the work. W.L. designed and performed experiments and interpreted data. B.S., C.Z., H.W., M.-M.Z., H.Z., M.G.N., D.X., V.J.M., Z.H. made contributions to the specific data acquisition. J.R. provided the patient samples. J.Y. and W.L. drafted and edited the paper. L.M. and J.R. edited the paper.
Data availability
ATAC-seq, RNA-seq and CUT&Tag data have been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/) with an accession number GSE179018. All the codes used in the analysis are available upon request.
Competing interests
J.Y. is a consultant for Sanofi. The other authors declare that they have no competing financial interests.
Contributor Information
Wanjin Li, Email: wanjinli@sioc.ac.cn.
Junying Yuan, Email: junying_yuan@sioc.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-022-00673-3.
References
- 1.Mifflin L, Ofengeim D, Yuan J. Receptor-interacting protein kinase 1 (RIPK1) as a therapeutic target. Nat. Rev. Drug Discov. 2020;19:553–571. doi: 10.1038/s41573-020-0071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito Y, et al. RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science. 2016;353:603–608. doi: 10.1126/science.aaf6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ofengeim D, et al. RIPK1 mediates a disease-associated microglial response in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2017;114:E8788–E8797. doi: 10.1073/pnas.1714175114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu K, et al. Necroptosis promotes cell-autonomous activation of proinflammatory cytokine gene expression. Cell Death Dis. 2018;9:500. doi: 10.1038/s41419-018-0524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lalaoui N, et al. Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease. Nature. 2020;577:103–108. doi: 10.1038/s41586-019-1828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao P, et al. A dominant autoinflammatory disease caused by non-cleavable variants of RIPK1. Nature. 2020;577:109–114. doi: 10.1038/s41586-019-1830-y. [DOI] [PubMed] [Google Scholar]
- 7.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sokpor G, Xie Y, Rosenbusch J, Tuoc T. Chromatin remodeling BAF (SWI/SNF) complexes in neural development and disorders. Front. Mol. Neurosci. 2017;10:243. doi: 10.3389/fnmol.2017.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 11.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ofengeim D, et al. Activation of necroptosis in multiple sclerosis. Cell Rep. 2015;10:1836–1849. doi: 10.1016/j.celrep.2015.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, et al. Ubiquitination of RIPK1 regulates its activation mediated by TNFR1 and TLRs signaling in distinct manners. Nat. Commun. 2020;11:6364. doi: 10.1038/s41467-020-19935-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Degterev A, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallach D, Kang TB, Dillon CP, Green DR. Programmed necrosis in inflammation: Toward identification of the effector molecules. Science. 2016;352:aaf2154. doi: 10.1126/science.aaf2154. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Dowling JP, Zhang J. RIPK1 can mediate apoptosis in addition to necroptosis during embryonic development. Cell Death Dis. 2019;10:245. doi: 10.1038/s41419-019-1490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newton K, et al. Cleavage of RIPK1 by caspase-8 is crucial for limiting apoptosis and necroptosis. Nature. 2019;574:428–431. doi: 10.1038/s41586-019-1548-x. [DOI] [PubMed] [Google Scholar]
- 18.Najafov A, et al. BRAF and AXL oncogenes drive RIPK3 expression loss in cancer. PLoS Biol. 2018;16:e2005756. doi: 10.1371/journal.pbio.2005756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Najafov A, Chen H, Yuan J. Necroptosis and cancer. Trends Cancer. 2017;3:294–301. doi: 10.1016/j.trecan.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Local A, et al. Identification of H3K4me1-associated proteins at mammalian enhancers. Nat. Genet. 2018;50:73–82. doi: 10.1038/s41588-017-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henikoff S, Henikoff JG, Kaya-Okur HS, Ahmad K. Efficient chromatin accessibility mapping in situ by nucleosome-tethered tagmentation. Elife. 2020;9:e63274. doi: 10.7554/eLife.63274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaya-Okur HS, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019;10:1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu D, et al. TBK1 suppresses RIPK1-driven apoptosis and inflammation during development and in aging. Cell. 2018;174:1477–1491.e19. doi: 10.1016/j.cell.2018.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song F, Chiang P, Wang J, Ravits J, Loeb JA. Aberrant neuregulin 1 signaling in amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 2012;71:104–115. doi: 10.1097/NEN.0b013e3182423c43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng D, et al. The genome-wide transcriptional regulatory landscape of ecdysone in the silkworm. Epigenet. Chromatin. 2018;11:48. doi: 10.1186/s13072-018-0216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrera-Uribe J, et al. Changes in H3K27ac at gene regulatory regions in porcine alveolar macrophages following LPS or polyIC exposure. Front. Genet. 2020;11:817. doi: 10.3389/fgene.2020.00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiu IM, et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 2013;4:385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iurlaro M, et al. Mammalian SWI/SNF continuously restores local accessibility to chromatin. Nat. Genet. 2021;53:279–287. doi: 10.1038/s41588-020-00768-w. [DOI] [PubMed] [Google Scholar]
- 31.Monaco C, Nanchahal J, Taylor P, Feldmann M. Anti-TNF therapy: past, present and future. Int. Immunol. 2015;27:55–62. doi: 10.1093/intimm/dxu102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bao X, et al. A novel ATAC-seq approach reveals lineage-specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biol. 2015;16:284. doi: 10.1186/s13059-015-0840-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 34.Ochoa D, et al. The functional landscape of the human phosphoproteome. Nat. Biotechnol. 2020;38:365–373. doi: 10.1038/s41587-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chesi A, et al. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat. Neurosci. 2013;16:851–855. doi: 10.1038/nn.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machol K, et al. Expanding the spectrum of BAF-related disorders: de novo variants in SMARCC2 cause a syndrome with intellectual disability and developmental delay. Am. J. Hum. Genet. 2019;104:164–178. doi: 10.1016/j.ajhg.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huttlin EL, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tak PP, Firestein GS. NF-kappaB: a key role in inflammatory diseases. J. Clin. Invest. 2001;107:7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiuza C, et al. Inflammation-promoting activity of HMGB1 on human microvascular endothelial cells. Blood. 2003;101:2652–2660. doi: 10.1182/blood-2002-05-1300. [DOI] [PubMed] [Google Scholar]
- 40.Brown JD, et al. NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fontana MF, et al. JUNB is a key transcriptional modulator of macrophage activation. J. Immunol. 2015;194:177–186. doi: 10.4049/jimmunol.1401595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schonthaler HB, Guinea-Viniegra J, Wagner EF. Targeting inflammation by modulating the Jun/AP-1 pathway. Ann. Rheum. Dis. 2011;70:i109–i112. doi: 10.1136/ard.2010.140533. [DOI] [PubMed] [Google Scholar]
- 43.Qiu Z, et al. Sp1 is up-regulated in cellular and transgenic models of Huntington disease, and its reduction is neuroprotective. J. Biol. Chem. 2006;281:16672–16680. doi: 10.1074/jbc.M511648200. [DOI] [PubMed] [Google Scholar]
- 44.Ravache M, Weber C, Merienne K, Trottier Y. Transcriptional activation of REST by Sp1 in Huntington’s disease models. PLoS One. 2010;5:e14311. doi: 10.1371/journal.pone.0014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swarup V, et al. Deregulation of TDP-43 in amyotrophic lateral sclerosis triggers nuclear factor kappaB-mediated pathogenic pathways. J. Exp. Med. 2011;208:2429–2447. doi: 10.1084/jem.20111313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frakes AE, et al. Microglia induce motor neuron death via the classical NF-kappaB pathway in amyotrophic lateral sclerosis. Neuron. 2014;81:1009–1023. doi: 10.1016/j.neuron.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikiz B, et al. The regulatory machinery of neurodegeneration in in vitro models of amyotrophic lateral sclerosis. Cell Rep. 2015;12:335–345. doi: 10.1016/j.celrep.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geng J, et al. Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat. Commun. 2017;8:359. doi: 10.1038/s41467-017-00406-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dondelinger Y, et al. Serine 25 phosphorylation inhibits RIPK1 kinase-dependent cell death in models of infection and inflammation. Nat. Commun. 2019;10:1729. doi: 10.1038/s41467-019-09690-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 51.Sato S, et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 2005;6:1087–1095. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- 52.Gagnon KT, Li L, Janowski BA, Corey DR. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat. Protoc. 2014;9:2045–2060. doi: 10.1038/nprot.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 54.Si W, et al. Dysfunction of the reciprocal feedback loop between GATA3- and ZEB2-nucleated repression programs contributes to breast cancer metastasis. Cancer Cell. 2015;27:822–836. doi: 10.1016/j.ccell.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 55.Li W, et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J. Clin. Invest. 2017;127:3421–3440. doi: 10.1172/JCI94233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwase S, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 57.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meers MP, Tenenbaum D, Henikoff S. Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling. Epigenet. Chromatin. 2019;12:42. doi: 10.1186/s13072-019-0287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramirez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, et al. Corepressor protein CDYL functions as a molecular bridge between polycomb repressor complex 2 and repressive chromatin mark trimethylated histone lysine 27. J. Biol. Chem. 2011;286:42414–42425. doi: 10.1074/jbc.M111.271064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nott A, et al. Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science. 2019;366:1134–1139. doi: 10.1126/science.aay0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McQuin C, et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018;16:e2005970. doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ATAC-seq, RNA-seq and CUT&Tag data have been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/) with an accession number GSE179018. All the codes used in the analysis are available upon request.