Abstract
Pre-exposing (priming) plants to mild, non-lethal elevated temperature improves their tolerance to a later higher-temperature stress (triggering stimulus), which is of great ecological importance. ‘Thermomemory’ is maintaining this tolerance for an extended period of time. NAM/ATAF1/2/CUC2 (NAC) proteins are plant-specific transcription factors (TFs) that modulate responses to abiotic stresses, including heat stress (HS). Here, we investigated the potential role of NACs for thermomemory. We determined the expression of 104 Arabidopsis NAC genes after priming and triggering heat stimuli, and found ATAF1 expression is strongly induced right after priming and declines below control levels thereafter during thermorecovery. Knockout mutants of ATAF1 show better thermomemory than wild type, revealing a negative regulatory role. Differential expression analyses of RNA-seq data from ATAF1 overexpressor, ataf1 mutant and wild-type plants after heat priming revealed five genes that might be priming-associated direct targets of ATAF1: AT2G31260 (ATG9), AT2G41640 (GT61), AT3G44990 (XTH31), AT4G27720 and AT3G23540. Based on co-expression analyses applied to the aforementioned RNA-seq profiles, we identified ANAC055 to be transcriptionally co-regulated with ATAF1. Like ataf1, anac055 mutants show improved thermomemory, revealing a potential co-control of both NAC TFs over thermomemory. Our data reveals a core importance of two NAC transcription factors, ATAF1 and ANAC055, for thermomemory.
Subject terms: Plant sciences, Plant stress responses, Heat
Introduction
Temperatures higher than the plants’ adaptive thresholds cause heat stress (HS), which diminishes their growth, survival and productivity. Plants, by nature, have the ability to tolerate a certain degree of high temperature above their ambient temperature. This is known as ‘basal thermotolerance’. In addition to basal thermotolerance, plants also have the ability to acquire thermotolerance if they are pre-exposed to mild non-lethal higher temperatures, in a process called ‘heat priming’1–4. This heat priming induces not only an immediate response, but also leads to molecular and metabolic changes that persist for some time in the absence of the stress and that allow plants to respond more effectively to a second HS event. The second stress is known as the ‘triggering stimulus’3,4. The time period between priming and triggering stimuli is called the ‘memory phase’, during which stress memory can form and consolidate. Thermomemory refers to the maintenance of some, but not all, HS-induced changes, which ‘primes’, or prepares plants to respond more rapidly and strongly if such stress recurs before the memory fades. Experimental evidence indicates that thermomemory can range from several hours to days4–6 or even generations7. Thermomemory appears to be regulated, at least in part, by a set of genes different from those acting in basal thermotolerance and acquired thermotolerance1,4,5,8–11.
HS priming involves the activation of heat shock transcription factors (HSFs) that induce the expression of heat shock proteins (HSPs) which help to protect cellular proteins from denaturation and contribute to the repair or removal of misfolded proteins12,13. Several HSFs have been shown to be involved in the HS response9,14–16. In particular, class HSFA1s are considered as ‘master regulators’ of the HS response16, regulating the expression of several other transcription factor (TF) genes, including DEHYDRATION RESPONSIVE ELEMENT BINDING PROTEIN 2A (DREB2A), HSFA2, HSFA7a, HSFBs, and MULTIPROTEIN BRIDGING FACTOR 1C (MBF1C)17. While immediate responses to HS are relatively well studied, the molecular and physiological processes underlying thermomemory in plants are still not well understood. One of the possible mechanisms involves the accumulation of TFs in their inactive state after a priming stimulus (during the memory phase), and their activation upon experiencing the triggering stimulus18,19. For example, heat shock factor A2 (HSFA2) has been identified as a key component in the establishment of thermomemory in primed plants9. HSFA2 induces the expression of HEAT STRESS-ASSOCIATED 32 (HSA32), which was found to be required specifically for the maintenance of thermomemory and to participate in the maintenance of cellular homeostasis during high temperature stress8.
NAM/ATAF1/2/CUC2 (NAC) is a family of plant-specific TFs that have an important role in the response to different biotic and abiotic stresses, including water deficit, salinity stress and temperature stress20–22. Several NACs have been reported to be involved in the basal HS response in Arabidopsis thaliana and crop plants23–27. For example, overexpression of ANAC019 improved the thermotolerance of Arabidopsis, presumably by regulating the expression of HSFA1s and other HSFs including HSFBs25. The Arabidopsis membrane-associated NAC transcription factor gene NTL4 has also been found to be responsive to heat stress, and ntl4 mutants exhibited a higher cell viability and less H2O2 accumulation than WT, under HS27. In rice (Oryza sativa), expression of SNAC3 is induced by several stresses, including heat, and its overexpression enhances tolerance to high temperature, drought and oxidative stress26. The expression of the Triticum aestivum (wheat) NAC transcription factor TaNAC2L is also induced by high temperature, and its overexpression in Arabidopsis improves its acquired heat tolerance via regulating expression of its heat stress-related genes23. However, there is limited evidence on the involvement of NAC TFs in regulating thermomemory. Arabidopsis JUNGBRUNNEN1 (JUB1) is the only NAC TF that has so far been reported to regulate thermomemory. JUB1 expression is induced by HS and its expression pattern during the memory phase is similar to the expression pattern of other thermomemory associated genes, such as HSFA224. Overexpression of JUB1 improves the thermomemory of Arabidopsis seedlings24.
In this study, we set out to systematically identify NAC TFs responsive to thermomemory in Arabidopsis. We analyzed the expression patterns of nearly all (104) Arabidopsis NACs after priming HS, during the thermomemory phase, and after the triggering stimulus. One NAC identified in our screen is ARABIDOPSIS TRANSCRIPTION ACTIVATOR FACTOR 1 (ATAF1; also called ANAC002). ATAF1 was previously identified to be involved in the gene regulatory networks of senescence, drought and sugar signaling28–31. We found that overexpression of ATAF1 in transgenic Arabidopsis plants limits thermomemory capacity, while knocking out ATAF1 profoundly enhances thermomemory and survival of plants exposed to a more severe HS. To identify genes regulated by ATAF1 in thermomemory, we employed RNA-seq to compare the heat responses in wild-type plants and mutants with altered ATAF1 levels (overexpressors and knockout mutants). We found that ATAF1 is co-expressed with ANAC055. With respect to thermomemory, the ataf1/anac055 double-mutant displayed a phenotype similar to that of the single knock-out mutants ataf1 and anac055, suggesting that the two TFs cooperatively regulate thermomemory.
Results
Identification of priming-responsive and memory-associated NAC transcription factors
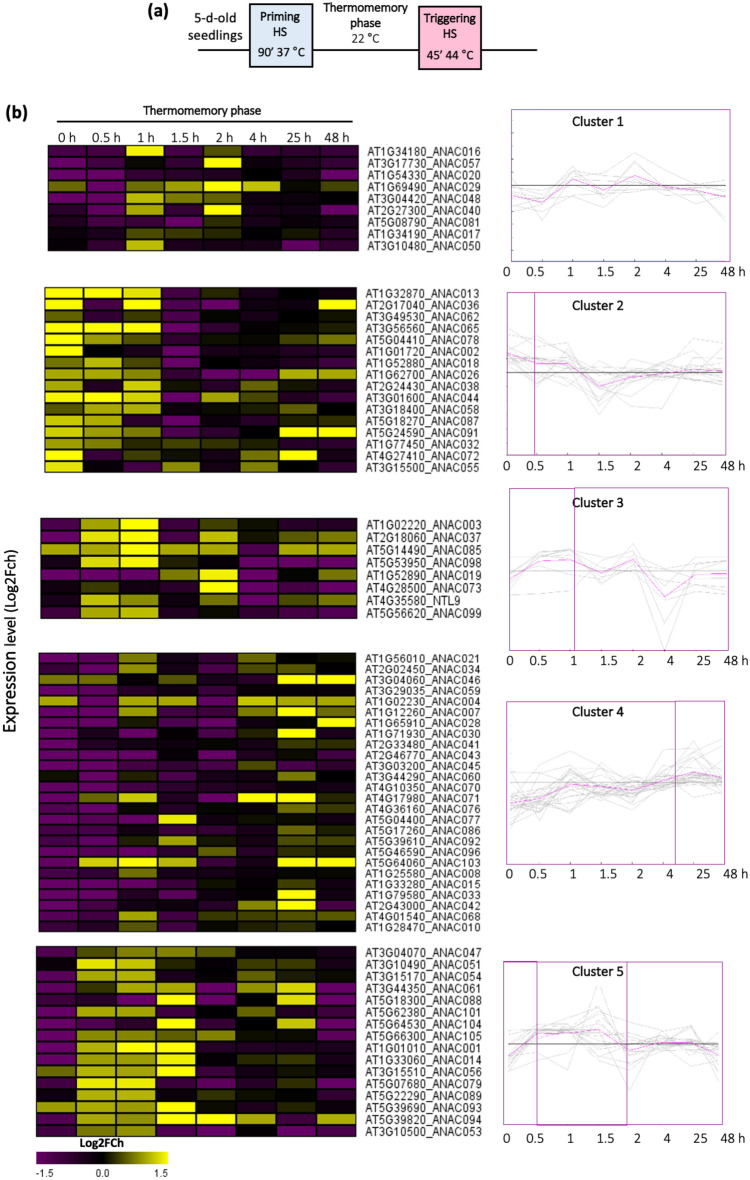
To investigate the expression pattern of NAC TFs in response to HS priming and during the memory phase, we treated Arabidopsis seedlings with a mild HS (priming) before exposing them to a severe HS (triggering stimulus). We collected samples at multiple time points after priming (up to 48 h into the thermomemory phase) and tested the expression of 104 NAC genes; seedlings kept in unprimed conditions were used as controls (Fig. 1).
Figure 1.
Expression of NAC transcription factors during the memory phase, after heat priming. (a) Schematic representation of the heat stress (HS) regime applied to asses HS memory. (b) Heat map and k-means clustering (k = 5 clusters) of NACs, based on their change in expression (HS samples compared to unstressed controls) during the memory phase at the time points indicated in the representation in (a). The color indicates the expression ratio as log2-fold change for the different time points, where ‘yellow’ denotes an increased expression and ‘purple’ a reduced expression. The list of genes in each cluster and their expression values are shown in Supplementary Table S1.
Considering a 1.5-fold change as cut-off, 75 of the NACs were differentially expressed upon priming compared to control, while expression of 29 NAC genes was undetectable in the whole-seedling samples. The cut-off threshold was chosen considering that even moderate changes in TF expression levels can elicit strong downstream responses32. The expression of some NACs (including e.g., ANAC013 and ANAC029/AtNAP) in response to HS priming was similar to that reported in a previous study by Kilian et al.30. NACs were grouped in clusters based on their expression pattern using k-means clustering. Changes in the expression of many NACs was already detectable immediately after the priming treatment (Fig. 1, Supplementary Table S1). Three clusters comprising 33 genes in total (clusters 1, 2 and 3) appeared upregulated during early timepoints, and largely unchanged during later timepoints (Fig. 1), while 26 genes (in cluster 4) were initially strongly downregulated, and then strongly upregulated during later time points (Fig. 1). For the genes in cluster 5 we observed a strong downregulation at time point 0, which is immediately after completion of the 90 min HS, followed by a strong upregulation wthin the time frame of up to 2 h after the HS, and then a moderate to unchanged expression (Fig. 1). Taken together, the NAC TFs analyzed exhibited different expression profiles during HS priming and memory, suggesting different roles in the response to HS.
The effect of heat priming and triggering on the expression of NAC transcription factors
Pre-exposure of plants to a moderate stress (priming) alters their response to upcoming severe temperature stress. We tested this, at the molecular level, by investigating the transcriptional response of NAC TFs. We tested whether or not heat-priming affects the expression of NACs after the triggering HS. To this end, we determined the expression of NAC TFs after the triggering stimulus (T plants) and compared it with the expression in primed and triggered plants (P + T), using a 1.5-fold change cut-off. To identify specific expression patterns between P + T and T plants, a k-means clustering approach was applied, using 10 clusters (Fig. 2, Supplementary Table S2). We noticed that expression of most NAC TFs was induced in primed + triggered plants, compared with triggered-only plants. Only few genes (cluster 9) showed an opposite expression pattern, being mostly repressed in primed + triggered plants and induced in triggered-only plants (Fig. 2). ATAF1 displayed a particularly interesting pattern: its expression remained almost unchanged in plants after priming + triggering (P + T), and expression was considerably induced in triggered-only conditions (without prior priming) (Fig. 2, cluster 9).
Figure 2.
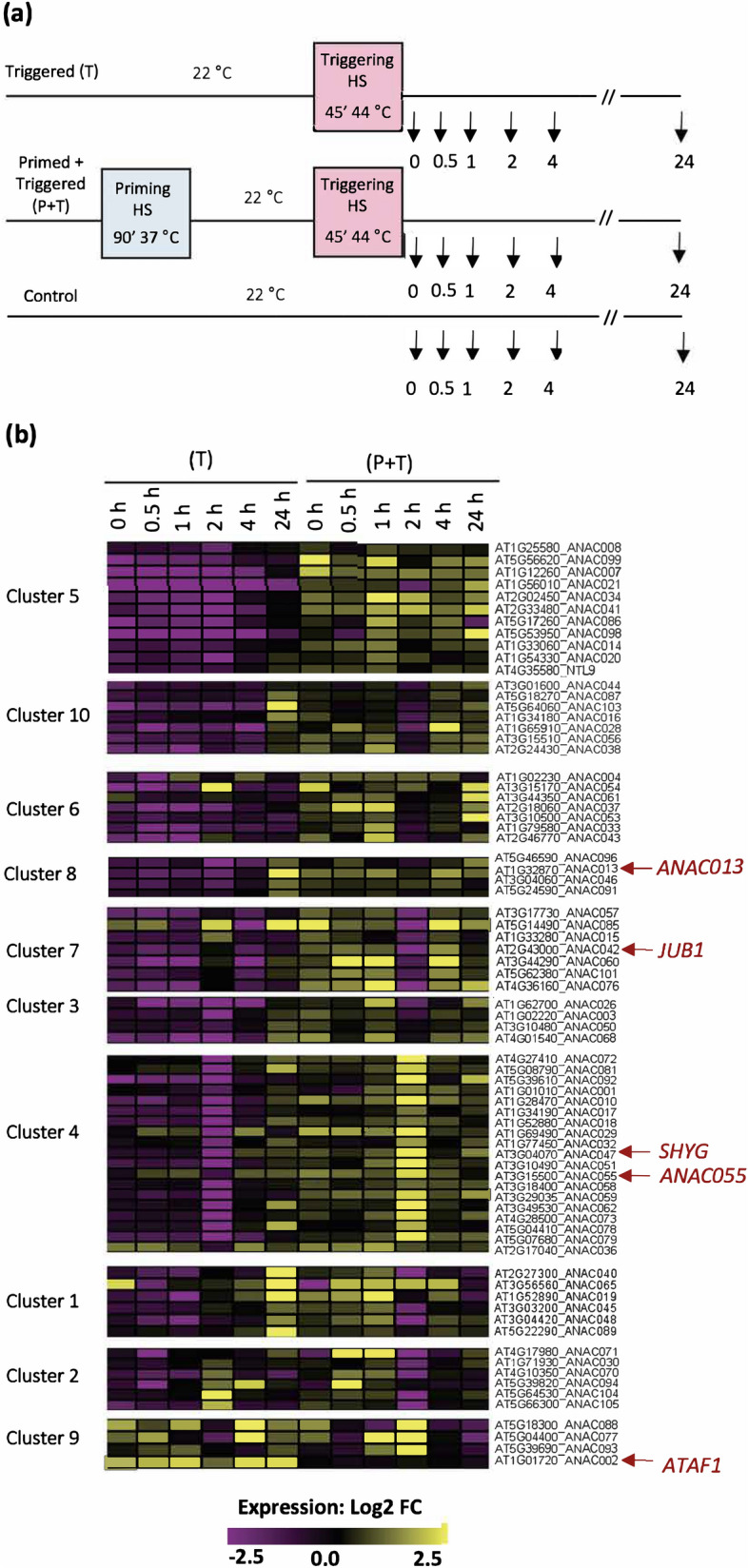
Expression of NACs after a triggering stimulus. (a) Schematic representation of the time points used for expression analysis of NACs in triggered plants (T), and in plants that were primed before triggering (P + T). In triggered (T) plants, the expression was calculated as a ratio of HS samples compared to unstressed controls. In primed and triggered (P + T) plants, the expression was calculated as primed and triggered (P + T), compared to triggered only (T). (b) K-means clustering of NAC expression profiles of primed and triggered (P + T) and triggered (T) plants after the triggering stimulus (k = 10 clusters). The expression of genes in each cluster is displayed as a heat map. The color indicates the expression as log2-fold change for the different time points, where yellow denotes an increased expression and purple a reduced expression. The list of genes in each cluster and their expression values are shown in Supplementary Table S2.
To test the hypothesis that ATAF1 is functionally involved in thermotolerance, additional physiological experiments were performed on ataf1 mutants and plants overexpressing ATAF1. Two other genes from clusters 4 (ANAC047/SHYG, AT3G04070) and 8 (ANAC013, AT1G32870) were included for comparison.
Functional analysis of selected NACs for thermomemory
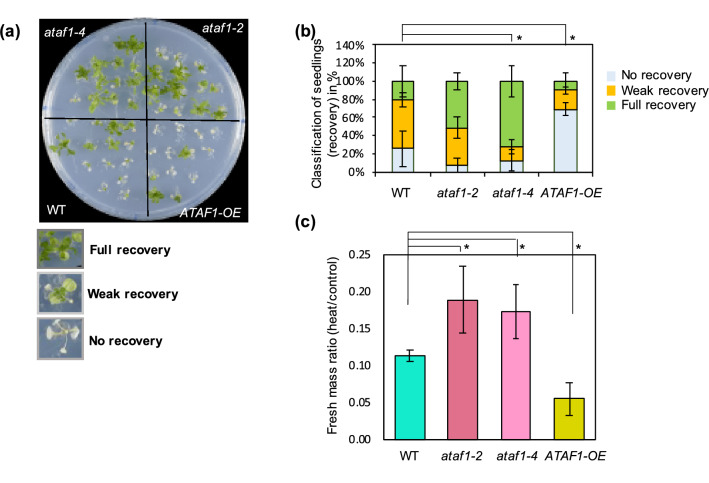
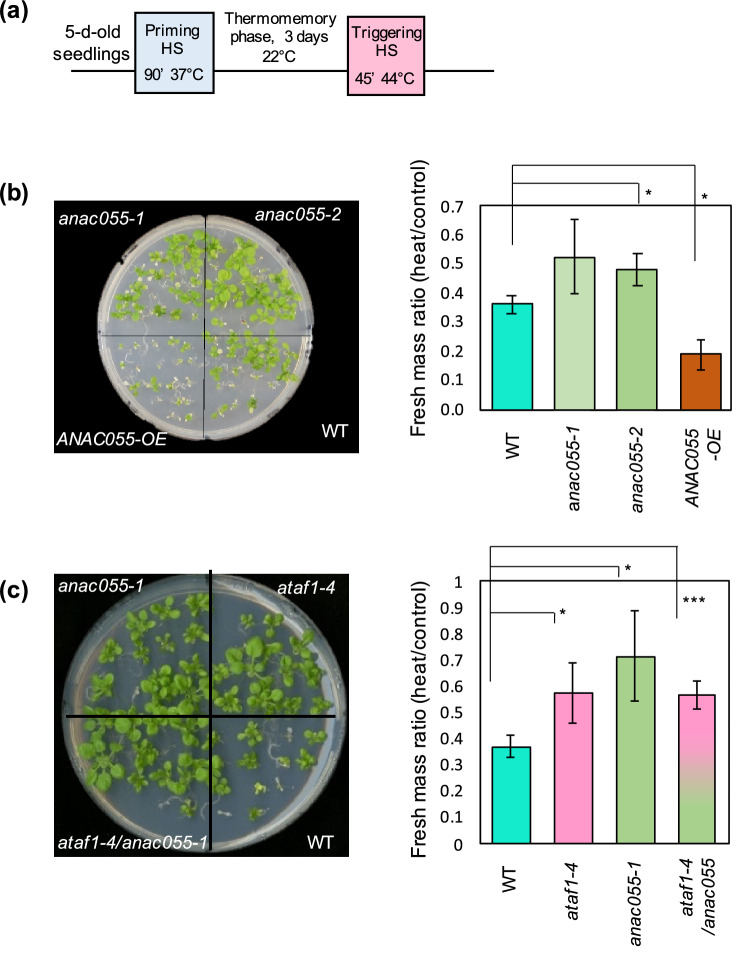
To investigate the impact of the selected NAC genes on thermomemory, we assessed their functional involvement in the process by analyzing the phenotype of overexpressor and knockout (or knockdown) mutants in response to a triggering HS given 3 days after heat priming (3 days of thermomemory). As shown in Supplementary Fig. S1, ANAC013 and SHYG overexpressors, shyg knockout and anac013 knockdown lines were not significantly different from wild type when compared in the thermomemory assay. However, ATAF1 appeared to be involved in thermomemory. A phenotypic analysis of ATAF1 transgenic plants for thermomemory showed a strong phenotype, where the ataf1–2 and ataf1–4 mutants had a significantly higher survival rate and fresh mass compared with WT plants, while plants overexpressing ATAF1 (called ATAF1-OE in the following) exhibited a severely reduced thermomemory (Fig. 3a–c). When the ataf1–4 mutant was transformed with an ATAF1 allele (pATAF1::ATAF1-GFP) which expresses an ATAF1-GFP fusion from the native ATAF1 promoter, thermomemory was restored to wild-type response (Supplementary Fig. S2). Taken together, these results suggest a negative regulatory role of ATAF1 in thermomemory.
Figure 3.
Improved thermomemory in ataf1 mutants. (a) Thermomemory phenotype of ATAF1 transgenic plants. Seedlings of ATAF1-OE, ataf1-2, ataf1–4, and WT were exposed to the HS regime schematically shown in Fig. 1a. Photos were taken 14 days after the second HS (triggering stimulus). The phenotype of one representative replicate of at least three independent biological replicates is shown. (b,c) Quantification of the results shown in (a). (b) Percentage of seedlings in different phenotypic classes; the thermotolerance phenotype was classified into three classes depending of the extent of plant recovery. The phenotype classes are illustrated in panel (a). (c) Seedling fresh mass in HS-primed and triggered plants compared to unstressed control plants. Error bars represent the standard deviation, calculated from three biological replicates; each replicate is the average mass of 13 seedlings. Significant differences between transgenic and WT plants were calculated using Student’s t-test; *indicates P-value < 0.05.
Potential target genes of ATAF1
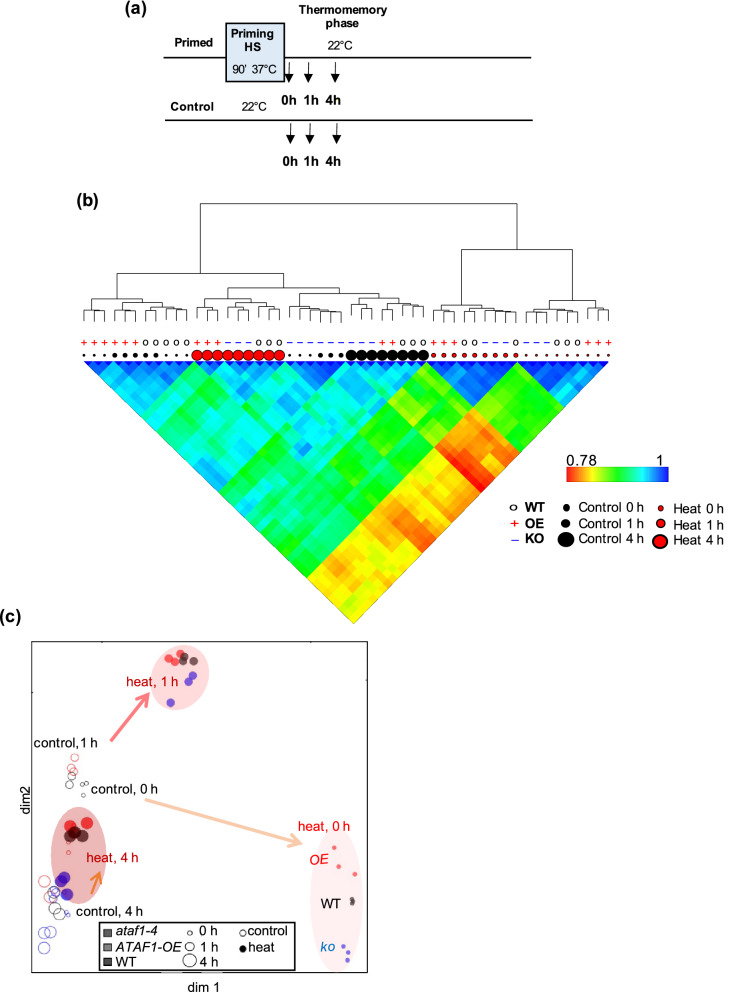
As our data suggested that ATAF1 is a negative regulator of thermomemory we sought to understand the role of ATAF1 in response to a heat priming stimulus by identifying its regulated target genes. To this end, WT, ATAF1-OE and ataf1–4 mutant seedlings were treated with the priming stimulus (37 °C for 90 min), and samples were then collected at three time points (0 h, 1 h and 4 h; Fig. 4a) after the heat priming for gene expression profiling by RNA-seq. To eliminate the potential influence from other environmental factors, control samples (unprimed control) were collected at the same time points as the heat-treated samples.
Figure 4.
Clustering of RNA-seq data. (a) Schematic presentation of the time points used for expression analysis by RNA-seq. (b) Top: Hierarchical clustering of the samples according to their expression patterns. “+” denotes ATAF1-OE lines; “0” denotes wild-type Col-0; and “−” denotes ataf1–4. The sizes of the circles indicate the time elapsed since HS (0, 1, and 4 h after heat stress). The black and red colors indicate whether the plants underwent control or heat temperature treatment for 90 min. Legend: colored heat map of Pearson’s coefficients of expression profiles between all possible pairs of the samples. A partial rainbow color scheme, from red to blue, indicates the range of correlation values. (c) Multidimensional scaling (MDS) plot. Overall similarities and differences between samples, based on a multidimensional scaling analysis. Briefly, the Euclidean distance between all samples was calculated according to the expression levels of all genes, and represented in a two-dimensional space.
To understand the relationship between samples in the RNA-seq, a hierarchical clustering analysis was performed and a heat map of the Pearson’s correlation coefficients of the expression profiles between all possible pairs of the samples was established (Fig. 4b). We observed a clustering between biological replicates, suggesting a high reproducibility of the experiment. The heat map also showed a strong separation between samples based on HS conditions, and by genotype (Fig. 4b). The overall similarities and differences between samples were confirmed by multidimensional scaling (MDS) (Fig. 4c), supporting a stronger separation by condition (heat and control) than by genotype. We also noticed a similarity of samples under control temperature and those after 4 h of heat stress, suggesting a rapid recovery of most changes in gene expression. Of note, the expression of ATAF1 in WT seedlings during the memory phase, as revealed by RNA-seq, followed the pattern determined by qRT-PCR (Supplementary Fig. S3).
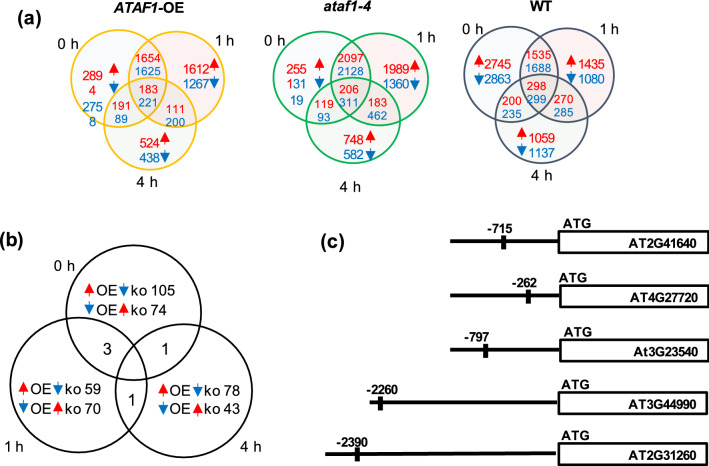
For all three genotypes, we found the highest number of differentially expressed genes (DEGs) at time point 0, right at the end of the 90-min heat priming treatment (Fig. 5a). To identify priming HS-associated genes, we used the same criteria that were previously used by Sedaghatmehr et al.4. We investigated genes whose expression was induced after priming and remained high at all examined time points in the memory phase, as well as genes whose expression was down-regulated and remained low during the memory phase (Fig. 5a). Among the DEGs are thermomemory-associated HSPs4 whose expression in response to heat priming was similarly induced in ATAF1-OE, ataf1–4 mutant and WT, such as HSP22, HSP21, HSP17.4 and HSP18.2 (Supplementary Table S3). This finding corroborates the conclusion that these HSPs contribute to a general heat-response, which is common to all three genotypes.
Figure 5.
Global transcriptomic changes of thermomemory-associated genes potentially regulated by ATAF1. (a) Venn diagrams of heat-induced and heat-repressed genes, after heat priming (heat relative to control) in WT, ataf1–4 mutant, and ATAF1-OE transgenic plants. (b) Venn diagram of genes upregulated in ATAF1-OE and downregulated in ataf1 mutant, and vice versa, compared to WT, in response to a priming HS, after 0 h, 1 h or 4 h. (c) Schematic representation of the position of ATAF1-binding sites in the upstream regions of genes identified in (b).
To identify differential responses of ATAF1-OE and ataf1–4 mutant plants to heat priming, we analysed the RNA-seq data for genes upregulated in ATAF1-OE, compared to WT, but downregulated in ataf1–4 mutant plants, and vice versa (Fig. 5b). Next, to identify potential direct target genes of ATAF1, we analyzed the promoters of differentially expressed genes for either the presence of the ATAF1 binding site (reported by Garapati et al.31) or for binding by ATAF1 as determined by DNA affinity purification sequencing (DAP-seq) experiments33. The analysis was performed for each time point after the heat priming (0 h, 1 h and 4 h). In total, we identified 64 genes as likely direct ATAF1 targets (Supplementary Table S4). We then refined our analysis by searching for those potential target genes that are commonly regulated by ATAF1 in all three, or two, time points after heat priming. Five genes, i.e., AT2G31260 (ATG9), AT2G41640 (GT61), AT3G44990 (XTH31), AT4G27720 and AT3G23540 were commonly regulated at two of the three timepoints, suggesting they might be priming-associated direct targets of ATAF1 (Fig. 5c); no gene was regulated at all three time points. ATG9 is an autophagy gene34 and experimental evidence indicates that autophagy plays a role in the heat stress response35,36. Its involvement in thermomemory is yet to be confirmed. The two genes GT61 and XTH31 are related to cell wall biosynthesis and expansion. Glycosyltransferase 61 (GT61) belongs to the glycosyltransferase (GT) family. GT proteins have diverse functions in plants, but most of them are likely involved in the biosynthesis of polysaccharides and glycoproteins in the cell wall. In grasses, including rice (Oryza sativa) and wheat (Triticum aestivum), GT61 family enzymes are involved in the synthesis of xylans, one of the main components of the cell wall37–39. GT61 has not yet been shown to be involved in heat stress priming or tolerance. XTH31 belongs to the xyloglucan endotransglucosylase/hydrolase (XTH) family. Generally, members of the XTH family are involved in cell wall remodeling, expansion and morphogenesis suggesting a potential role in stress responses40. In Arabidopsis, XTH31 is involved in regulating cell wall xyloglucan content41. The xth31 loss-of-function mutant has a reduced sensitivity to ABA, and seeds germinate faster than those of the WT41,42. Transgenic soybean (Glycine max) plants overexpressing XTH31 from Arabidopsis display enhanced tolerance to flooding along with more adventitious roots and longer primary roots43. The two genes AT4G27720 and AT3G23540 are not well characterized. AT4G27720 is annotated to encode a major facilitator superfamily protein with a molybdate ion transporter function, while AT3G23540 is annotated to encode a protein of the alpha/beta-hydrolase superfamily. The α/β-hydrolase enzymes are involved in various processes, including biosynthesis, metabolism, signal transduction, gene regulation44, and in the plants’ response and tolerance to salinity stress45.
Co-regulatory network of ATAF1 reveals potential co-regulation with ANAC055
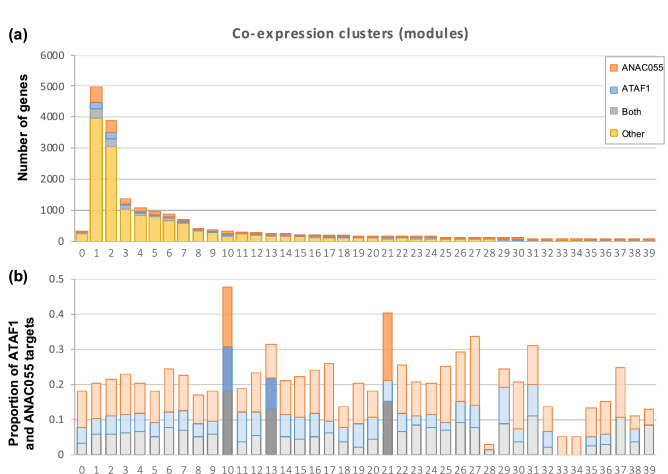
Genes with similar expression patterns often share similar functions and are potentially regulated by the same transcription factors. In order to identify genes co-regulated with ATAF1 in response to heat priming, a co-expression analysis was performed using the transcriptomic data of ATAF1-OE, ataf1–4 mutant and WT plants, under control condition and upon exposure to heat. A weighted correlation network analysis was used to find modules of highly correlated genes. This method is commonly used to investigate relationships between genes and to identify gene candidates for further analysis46. The co-expression analysis revealed 40 modules in our data set (Fig. 6a). These modules contained groups of genes expressed in a similar way. It is likely that genes regulated directly by ATAF1, in response to heat, display similar expression patterns; hence, we took publicly available data on genes that have already been identified to be potentially targeted by ATAF1 (in general, not just by heat stress) and queried if these genes were present in our co-expression modules. The publicly available data of the potential direct targets were generated using a DAP-seq assay33. Putative target genes were designated as having a DAP-seq peak within the first 1000 bp upstream of the transcription start site. Thus, we compared the list of genes that are proposed direct targets of ATAF1 (from O’Malley et al.33) with the genes that are clustered in the co-expression analysis; we noticed that the potential targets of ATAF1 were overrepresented in co-expression clusters 10 and 13 (Fig. 6b). These clusters needed further investigation because they likely contained direct target genes of ATAF1, in response to heat stress. The clusters were further interrogated with the proposed TF targets identified using the DAP-seq assay developed by O’Malley et al.33. We found that three of the clusters (10, 13, 21) showed significant enrichment for genes targeted by both ATAF1 and ANAC055, suggesting that the genes in these clusters might be co-regulated by the two NACs (Fig. 6b).
Figure 6.
Potential targets of ATAF1 in response to heat stress. (a) The bar graph shows the number of genes in each co-expression cluster. A clustering analysis was performed by weighted gene correlation network analysis, which resulted in 40 clusters of genes co-expressed together. (b) The bar graph shows the proportion of ATAF1 and ANAC055 targets in each cluster, including genes targeted by both; clusters with significant enrichment of ATAF1 and ANAC055 target genes are shaded (P < < 0.001); only two clusters (10 and 13) are highly enriched with ATAF1 targets; clusters 10 and 21 are significantly enriched for ANAC055, and clusters 10, 13 and 21 are enriched for genes that are targeted by both transcription factors.
ATAF1 and ANAC055 appear to have a common role in thermomemory
Our co-expression analysis showed that ATAF1 and ANAC055 had a large number of shared co-expressed targets in response to HS (Supplementary Table S5). We therefore tested if ANAC055 is also functionally involved in regulating thermomemory. To this end, we assessed the thermomemory phenotype of transgenic lines with altered expression of ANAC055 (Fig. 7a,b). Two anac055 mutants (anac055-1 and anac055-2) were tested, and both showed a substantial increase in fresh mass ratio, survival, and chlorophyll content compared with WT, while plants overexpressing ANAC055 showed a decrease in those parameters (Fig. 7b, Supplementary Fig. S4). These results indicate a negative role for ANAC055 in the regulation of thermomemory, similar to that of ATAF1. To test whether or not ATAF1 and ANAC055 worked in a fully or partially redundant manner, we generated an ataf1/anac055 double mutant and exposed seedlings to HS and compared their phenotypes with the phenotypes of the corresponding single-gene mutants. The fresh mass ratio of the ataf1/anac055 double-mutant was higher than that of the WT, but not significantly different from that of the single-gene knockout mutants ataf1–4 and anac055-1 (Fig. 7c). This result indicates that ATAF1 and ANAC055 require each others function to control thermomemory.
Figure 7.
Thermomemory is improved in the ataf1–4/anac055 double-knockout mutant compared to WT. (a) Schematic representation of HS regime. (b) Thermomemory phenotype of ANAC055-OE, anac055-1 and anac055-2 mutants, and WT plants. (c) Thermomemory phenotype of ATAF1-OE, ataf1–4 mutant, ataf1–4/anac055 double-mutant, and WT plants. Seedlings were exposed to the HS regime shown in (a). Left panels: seedlings photographed 14 days after exposure to the triggering heat stress. The phenotype of one representative replicate of at least three (b) or five (c) independent biological replicates is shown. Right panels: seedling fresh mass upon heat stress, compared to control plants (no heat stress). Error bars represent standard deviation, which was calculated from three biological replicates, where each replicate was the average mass of 18–19 seedlings. Significant differences between transgenic lines and WT plants were calculated using Student’s t-test; * if P < 0.05; *** if P < 0.001.
Discussion
HS is one of the major abiotic stresses negatively affecting plant growth and reproduction globally. The impact of HS on crop production increases with climate change. Prior exposure to mild HS (heat priming) enables plants to acquire and maintain tolerance to HS by establishing a cellular memory of a prior HS (thermomemory)1–4. Here, we studied the role of NAC TFs in thermomemory. We found that several NACs substantially responded at their expression level after heat stress and that the changes in expression persisted during the memory phase. These findings are similar to those reported for the expression pattern of other thermomemory-associated genes during the memory phase (such as HSFA2, HSA32, and JUB1)8,9,24.
We then analyzed the expression of NAC TFs after the triggering stimulus (second heat stress) to investigate if their expression was affected by a priming treatment. Our data show that NAC expression was altered after the triggering stimulus, indicating the presence of transcriptional memory that controls their expression. A similar expression pattern has previously been reported for drought memory, where the expression of a subset of DEGs is altered after a second drought stress. Ding et al.47 classified the memory genes into four distinct groups, each showing different expression patterns: genes with increased (or decreased) expression levels after a first stress, and a further increased (or decreased) level after a second stress, which they, respectively, denoted as (+/+, or −/−); ‘revised memory genes’ that are induced after a first stress and then decreased after the second stress (+/−), or the opposite (−/+). Many NACs showed an expression pattern similar to those reported by Ding et al.47.
We found that ATAF1 (ANAC002) expression increases rapidly right after the priming HS, and then decreases to below control levels during the memory phase (Fig. 1; cluster 2). We also found that ataf1 knockout mutants were more tolerant to heat stress than the wild type, revealing a negative regulatory role of ATAF1 in HS memory. ATAF1 was previously identified as an important regulator of senescence and the response to drought stress29,31,48. It has been well established that ATAF1 promotes senescence by directly regulating the expression of two TFs involved in senescence and chloroplast maintenance: ANAC092 (ORE1) and GOLDEN2-LIKE1 (GLK1)31. ATAF1 also regulates drought response by regulating the expression of 9-CIS-EPOXYCAROTENOID DIOXYGENASE 3 (NCED3), which encodes a key enzyme of ABA biosynthesis31,49.
We used RNA-seq analysis to identifying early transcriptional changes controlled by ATAF1. Several mechanisms (genes) have been previously identified to be involved in basal heat tolerance: they include ROS detoxification, lipid signaling, and activation of heat shock proteins and heat shock factors8–10,27,50. Our data found similar responses in ATAF1-OE, ataf1–4 mutant and WT plants, where these mechanisms were similarly activated.
Co-expression analysis has been established as a powerful tool to identify TF target genes51,52. For instance, Jensen et al.49 performed a co-expression analysis across publicly available microarray datasets and identified 25 genes co-expressed with ATAF1. The promoter regions of these ATAF1 co-expressors were significantly enriched for ATAF1 binding sites. We performed a co-expression analysis and combined this information with publicly available data from a DAP-seq approach33. We found that four co-expression clusters were enriched with predicted ATAF1 target genes; and those were also enriched for target genes of another NAC TF, ANAC055. We found many target genes commonly regulated by ATAF1 and ANAC055, suggesting at least partial overlap between the two TFs with respect to their regulatory capacity. Often, TFs act in a redundant manner. For example, the transcription factors WRKY11 and WRKY17 operate as negative regulators of basic resistance to Pseudomonas syringae pv. tomato (Pst)50. The loss of function of WRKY11 increases the resistance toward Pst. An expression analysis of selected genes in single and double mutants revealed that both TFs modulate transcriptional changes in response to pathogen infection; however, each one of them acted either specifically or in a partly redundant manner, depending on the specific target gene50. In our work, we identified ATAF1 and ANAC055 as negative regulators of thermotolerance. Phenotypic and gene expression data of ataf1 and anac055 single and double mutants showed a partial regulatory redundancy between ATAF1 and ANAC055.
Our results suggest that ATAF1 and ANAC055 work both as negative regulators of thermomemory because (i) the single ataf1 and anac055 mutants as well as the double-mutant plants have a similar phenotype in thermomemory, (ii) they appear to be co-expressed, and (iii) their potential target genes overlap. Further protein-interaction studies are needed to test if ATAF1 and ANAC055 also interact physically, for instance as heterodimers or if they are only connected via their gene-regulatory networks. This interaction needs to be investigated in a temporal and spatial context to evaluate its relevance for thermomemory.
It has recently been suggested that the heat-inducible transcription factor HSFA2 is involved in thermomemory, particularly regulating HS memory genes in the shoot apical meristem (SAM), suggesting that the establishment of thermomemory involves organ-specific mechanisms53,54. Our results show that in ataf1–4 lines, a higher proportion of plants displayed full recovery, i.e. seedlings and cotyledons remained green compared to WT plants. In plants that only partially recovered, cotyledons remained bleached and the developing true leaves emerged green (Supplementary Fig. S4). The green emerging leaves suggest that mechanisms that protect the SAM, where new leaves develop, play a role in thermomemory.
Most of the current understanding of thermotolerance and thermopriming is based on Arabidopsis seedlings. However, there have been some limited studies performed to assess the thermopriming in crop plants. For instance, heat priming of winter wheat during the stem-elongation, bolting and anthesis stages significantly enhanced grain-yield under heat stress during the grain filling phase55. Another mechanism for improving thermotolerance may be through root endophytes which appear to enhance thermotolerance of crops by impacting the expression of thermomemory associated gene HSFA256. Our study shows the importance of two genes, ATAF1 and ANAC055, as negative regulators of thermomemory in seedlings; it remains to be tested if later growth or seed yield are also modified under recursive HS. At least for seedlings tested here, a biomass penalty under non-stress conditions was not observed. Once a better understanding of thermomemory is established in model organisms and crops, genome editing technology could be employed to enhance crop performance in the field in anticipated future climatic conditions.
Materials and methods
Plant materials
Arabidopsis thaliana ecotype Col-0 was used as wild type and obtained from the European Arabidopsis Stock Center (NASC) (http://arabidopsis.info/). The ATAF1-OE, ataf1-2 (SALK-057618) and ataf1–4 (GABI-Kat GK565H08) seeds were described previously in Garapati et al.31. Seeds of the T-DNA insertion lines of ANAC055 (anac055-1; SALK_014331 and anac055-2; SALK_011069) were obtained from The European Arabidopsis Stock Centre (NASC) seed collection (http://arabidopsis.info/). The ANAC047 transgenic lines, i.e., ANAC047-OE (35S::ANAC047-GFP) and the two knockout lines (T-DNA insertion lines shyg-1/SALK-066615 and shyg-2/GABI-Kat GK-343D11) were previously described by Rauf et al.57. Seeds of ANAC013-OE and knockdown (anac013-kd) lines were provided by Prof. Frank van Breusegem (Ghent University, Ghent, Belgium) and described in De Clercq et al.58.
Plasmid construction and plant transformation
For 35S::ANAC055 (ANAC055-OE), the ANAC055 open reading frame was amplified by PCR from Arabidopsis Col-0 leaf cDNA and inserted into pUni/V5-His-TOPO (Invitrogen). The cDNA was then cloned via added PmeI-PacI sites into a modified pGreen0229-35S plant transformation vector59. The resulting pGreen0229-35S::ANAC055 vector was electroporated into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis wild type via floral dip. To construct pATAF1::ATAF1-GFP, a genomic fragment encompassing the 1.5-kb ATAF1 promoter and the ATAF1 coding region (without the translation stop codon) was inserted upstream of the green fluorescence protein coding sequence (in-frame fusion) in plasmid pK7FWG2,060. The cloning replaced the CaMV 35S promoter present in the plasmid with the ATAF1 promoter. The ataf1 knockout complementation line was generated by transforming the ataf1–4 mutant31 with the pATAF1::ATAF1-GFP construct.
Double mutant
The ataf1/anac055 double-knockout mutant was generated by crossing the homozygous ataf1–4 mutant with the homozygous anac055-1 mutant. Plants from the F2 generation were PCR-screened to select homozygous double-mutant plants. The list of primers used for genotyping is given in Supplementary Table S6.
Plant growth conditions
Arabidopsis seeds were surface-sterilized by washing them with 70% ethanol for 2 min; the ethanol solution was then replaced with 20% sodium hypochlorite and seeds were incubated in the solution for 20 min, with continuous shaking. Subsequently, seeds were washed four times with sterile water and left to dry on sterilized filter paper, in a laminar clean bench. Approximately equal numbers of seeds (around 500 seeds) were germinated on small petri dishes containing half-strength Murashige and Skoog (MS) medium supplemented with 1% (w/v) sucrose. For stratification, plates were kept in a dark cold room (8 °C) for 2 days before placing them in a growth chamber (photoperiod 16 h/8 h: light/dark at 22.5 °C, under 120 μmol m−2 s−1 photon irradiance). Five-day-old seedlings were used for HS assays.
Heat stress treatment
For NAC expression profiling, 5-day-old Arabidopsis seedlings were exposed to a HS regime. Plants were primed with mild heat stress at 37 °C for 90 min in a growth chamber, and subsequently returned back to normal growth condition to recover for 2 days to assess their thermomemory. Plants were then challenged with a triggering HS of 44 °C for 45 min. Arabidopsis seedlings were collected during the memory phase, and after the triggering stimulus, at the time points indicated in Fig. 1. Whole seedlings collected from one single plate were considered as one biological replicate. Control samples (without treatment) were collected at the same time to reduce circadian effects. Three biological replicates per treatment, per time point, and per genotype were analysed. Samples were directly frozen in liquid nitrogen, and stored at − 80 °C until further analysis.
Chlorophyll content
The Chlorophyll content was measure using the protocol of Hiscox and Israelstam61. Frozen seedling of ten were immersed in 0.5 mL preheated dimethylsulfoxid (DMSO, 65 °C) and incubated at 65 °C for 20 min. For the measurement the extract was diluted 1:10 or 1:100 with DMSO. Absorbance was recorded at 663 nm and 645 nm, respectively, for chlorophyll a and b. The following formula62 was used to calculate the concentration of chlorophyll a, b and total chlorophyll as mg g−1 fresh mass (FM):
Data was normalized to the respective control of each genotype.
RNA isolation, cDNA synthesis and qRT-PCR for NAC expression profiling
For expression profiling of NAC genes, total RNA was extracted using Trizol (Ambion, Life Technologies), followed by purification of RNA using columns of the PureLink RNA Mini kit (Ambion, Life Technologies) according to the manufacturer’s instructions. RNA concentration was measured using a NanoDrop spectrophotometer (Thermo Scientific); genomic DNA was digested using the TURBO DNA-free Kit (Ambion) according to the manufacturer’s instructions. The absence of genomic DNA contamination was confirmed by qRT-PCR, using intron-specific primers of control gene AT5G65080 (for primer sequences, see Supplementary Table S6). cDNA was synthesized from 4 µg high-quality RNA, using the Revert Aid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific), following the instructions of the manufacturer.
The qRT-PCR reaction was performed using SYBR Green master mix (Applied Biosystems): cDNA 0.5 μL, SYBR Green master mix (2×) 2.5 μL, forward and reverse primer mix (each 0.5 μM) 2 μL. The reactions were incubated in an ABI PRISM 7900 HT sequence-detection system (Applied Biosystems) using the following protocol: temperature was first set at 95 °C and kept constant for 10 min, and then underwent a series of 40 cycles of 95 °C for 15 s and 60 °C for 1 min. ACTIN2 (AT3G18780) was used as the reference gene for data analysis. The primer platform of the 104 Arabidopsis NAC TF genes was established using the QuantPrime tool63. Primer sequences are given in Supplementary Table S6. After 40 reaction cycles, a melting curve was generated by increasing the temperature from 60 to 95 °C, with a ramp speed of 1.9 °C min−1, to verify the amplification of the desired products. The efficiency of the reaction was estimated using LinRegPCR software, as described by Caldana et al.64.
Data were analyzed using the comparative Ct method, as described in Kamranfar et al.65. Briefly, the delta Ct value (ΔCt) was calculated by normalizing each Ct value with the Ct value of the reference gene ACTIN2. Then, the level of gene expression was expressed as the difference between an arbitrary value of 40 and the ΔCt value (a high 40-ΔCt value indicates a high gene expression level). A threshold of 1.5-fold change was used to select differentially expressed genes.
RNA isolation and RNA-seq
For the RNA-seq analysis, 5-day-old Arabidopsis seedlings of the three genotypes were used: WT, ataf1–4 mutant, and ATAF1-OE. After treatment with the heat priming temperature (37 °C for 90 min), seedlings were harvested at time points 0 h, 1 h and 4 h during the memory phase.
The transcriptomes of the three genotypes WT, ataf1–4 and ATAF1-OE was analyzed using RNA-seq technology. Total RNA was isolated from 5-day-old Arabidopsis seedlings of the three genotypes. Whole seedlings collected from each plate (pool of at least 10 seedlings) were considered as one biological replicate. Three biological replicates per treatment, per time point, and per genotype, were used, which made 72 samples in total. RNA was isolated using Direct-zol RNA MiniPrep Plus (Zymo Research, CA, USA) according to the manufacturer’s instructions. Quality was assessed using a Bioanalyzer (Agilent Technologies, Waldbrann, Germany), and quantity was determined using Qubit (Invitrogen, Life Technologies). High-quality RNA (3 µg) was used to prepare libraries for sequencing. RNA-sequencing 150-bp paired-end (PE) libraries were prepared from mRNA-enriched samples, using the NEB Next Ultra DNA Library Prep Kit for Illumina sequencing (New England Biolabs), according to the manufacturers’ instructions. A total of 12 samples were pooled in one lane of the flow cell. RNA was sequenced, using an Illumina HiSeq4000 machine, at KAUST Core Labs.
RNA-seq data analysis
HISAT was used to align the RNA-seq reads onto the genome of Arabidopsis thaliana (TAIR10)66. Only sequencing reads that mapped uniquely on the genome were retained. Htseq-count was used to count the number of sequencing reads of the transcripts67. VOOM, a transformation method that allows the use of RNA-seq data for the linear model framework, was used to calculate the transcript abundance, and quantile normalization was used to normalize the abundance distribution between samples68. EdgeR and LIMMA69,70 were used as a framework for calculating the differentially expressed genes. R, a general statistical environment, was used for data management, analyses, and generating figures71.
The customized linear contrast was set up to calculate the differential expression between genotypes, or temperatures. More specifically, second-order interaction effects were calculated using the contrast framework70. For example, a calculation could be made to quantitatively assess the changes in gene expression under heat, in comparison with the control for ataf1 and WT: (heatataf1 − controlataf1) − (heatWT − controlWT). All statistical significance cut-offs were adjusted for multiple testing by controlling the False Discovery Rate (Benjamini-Hochberg) at 0.0572 using “decideTests” (…, method = “global”, adjust.method = “BH”, p.value = “0.05”, lfc = 0.
For all genes and samples, a weighted gene correlation network analysis was performed to identify groups of co-expressed genes46, using default parameters. The soft thresholding power parameter was set at 6, which is the value, based on graphical examination, that the optimal network conditions are met (maximum scale-free fit index and minimum mean connectivity). For multi-dimensional scaling, the cmscale function in R was used.
Transcription factor binding sites (TFBS) acquired via DAP-seq as reported by O’Malley et al.33 were extracted from the publicly available plant cistrome database. DAP-seq binding motif peaks were extracted in narrowPeak format and mapped to the Arabidopsis genome. Distance from peak location to closest transcription start site (TSS) was then computed via the bedtools ‘closest’ command73, where TSS locations were taken from the TAIR 10 annotation file74. Putative ATAF1 and ANAC055 target genes were defined as having at least one DAP-seq peak within the first 1000 bp upstream of the respective TSSs. Statistical analysis for enrichment of target genes within the co-expression clusters was performed in R, using a hypergeometric test for overrepresentation71.
Legislation statement
The experimental research and field studies on plant materials comply with relevant institutional, national, and international guidelines and legislation.
Supplementary Information
Acknowledgements
The research was supported by funding from King Abdullah University of Science and Technology (KAUST), Saudi Arabia. NOA received support through a L’Oréal-UNESCO For Women in Science 2014 Middle East Fellowship. SMS received funding from the Ministry for Science, Research and Art of Baden-Wuerttemberg, Germany (Az: 75533-30-20/1). BMR thanks the University of Potsdam, and SB thanks the Max Planck Institute of Molecular Plant Physiology for financial support. BMR thanks the European Union’s Horizon 2020 Research and Innovation Programme, project PlantaSYST (SGA-CSA No. 739582 under FPA No. 664620) for funding. TDW thanks the International Max Planck Research School ‘Primary Metabolism and Plant Growth’, Potsdam, for financial support. The authors thank Prof. Frank van Breusegem (Ghent University, Ghent, Belgium) for providing seeds of ANAC013-OE and knockdown (anac013-kd) lines.
Author contributions
S.B., B.M.R., S.M.S. and M.T. designed the research. N.O.A. performed the experiments and acquired the experimental data. S.L.O. performed some of the phenotyping and determined chlorophyll levels. N.O.A., Y.H.W., T.D.W., S.B., and S.M.S. performed data analyses. I.K. gnerated the complementation line of ataf1–4. A.D.A. supervised qRT-PCR experiments. All authors contributed to the interpretation of the data. S.B., B.M.R., and M.T. acquired funding. N.O.A., S.B., and S.M.S. drafted the manuscript, which was finalized by S.B., S.M.S., and B.M.R. All authors reviewed and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
RNA sequencing data are available from the NCBI Bioproject database under ID SUB8978006 (https://ngdc.cncb.ac.cn/search/?dbId=gsa&q=SUB8978006). All other data generated or analysed during this study are included in this published article and its Supplementary Information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Salma Balazadeh, Email: s.balazadeh@biology.leidenuniv.nl.
Sandra M. Schmöckel, Email: sandra.schmoeckel@uni-hohenheim.de
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-14429-x.
References
- 1.Yeh CH, Kaplinsky NJ, Hu C, Charng YY. Some like it hot, some like it warm: Phenotyping to explore thermotolerance diversity. Plant Sci. 2012;195:10–23. doi: 10.1016/j.plantsci.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balmer A, Pastor V, Gamir J, Flors V, Mauch-Mani B. The 'prime-ome': Towards a holistic approach to priming. Trends Plant Sci. 2015;20:443–452. doi: 10.1016/j.tplants.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Hilker M, et al. Priming and memory of stress responses in organisms lacking a nervous system. Biol. Rev. Camb. Philos. Soc. 2016;91:1118–1133. doi: 10.1111/brv.12215. [DOI] [PubMed] [Google Scholar]
- 4.Sedaghatmehr M, Mueller-Roeber B, Balazadeh S. The plastid metalloprotease FtsH6 and small heat shock protein HSP21 jointly regulate thermomemory in Arabidopsis. Nat. Commun. 2016;7:12439. doi: 10.1038/ncomms12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stief A, et al. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell. 2014;26:1792–1807. doi: 10.1105/tpc.114.123851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma M, Banday ZZ, Shukla BN, Laxmi A. Glucose-regulated HLP1 acts as a key molecule in governing thermomemory. Plant Physiol. 2019;180:1081. doi: 10.1104/pp.18.01371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, et al. An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 2019;29:379–390. doi: 10.1038/s41422-019-0145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charng YY, Liu HC, Liu NY, Hsu FC, Ko SS. Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol. 2006;140:1297–1305. doi: 10.1104/pp.105.074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charng YY, et al. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007;143:251–262. doi: 10.1104/pp.106.091322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meiri D, Breiman A. Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 2009;59:387–399. doi: 10.1111/j.1365-313X.2009.03878.x. [DOI] [PubMed] [Google Scholar]
- 11.Brzezinka K, et al. Arabidopsis FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling. Elife. 2016;5:e17061. doi: 10.7554/eLife.17061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haslbeck M, Vierling E. A first line of stress defense: Small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 2015;427:1537–1548. doi: 10.1016/j.jmb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finka A, Sharma SK, Goloubinoff P. Multi-layered molecular mechanisms of polypeptide holding, unfolding and disaggregation by HSP70/HSP110 chaperones. Front. Mol. Biosci. 2015;2:29. doi: 10.3389/fmolb.2015.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda M, Mitsuda N, Ohme-Takagi M. Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 2011;157:1243–1254. doi: 10.1104/pp.111.179036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schramm F, et al. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008;53:264–274. doi: 10.1111/j.1365-313X.2007.03334.x. [DOI] [PubMed] [Google Scholar]
- 16.Liu HC, Liao HT, Charng YY. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011;34:738–751. doi: 10.1111/j.1365-3040.2011.02278.x. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida T, et al. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genom. 2011;286:321–332. doi: 10.1007/s00438-011-0647-7. [DOI] [PubMed] [Google Scholar]
- 18.Bruce TJ, Matthes MC, Napier JA, Pickett JA. Stressful, “memories” of plants: Evidence and possible mechanisms. Plant Sci. 2007;173:603–608. doi: 10.1016/j.plantsci.2007.09.002. [DOI] [Google Scholar]
- 19.Conrath U. Systemic acquired resistance. Plant Signal Behav. 2006;1:179–184. doi: 10.4161/psb.1.4.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nuruzzaman M, Sharoni AM, Kikuchi S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013 doi: 10.3389/fmicb.2013.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marques DN, dos Reis SP, de Souza CRB. Plant NAC transcription factors responsive to abiotic stresses. Plant Gene. 2017;11:170–179. doi: 10.1016/j.plgene.2017.06.003. [DOI] [Google Scholar]
- 22.Yoon Y, et al. The role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy. 2020 doi: 10.3390/agronomy10060788. [DOI] [Google Scholar]
- 23.Guo W, et al. The wheat NAC transcription factor TaNAC2L is regulated at the transcriptional and post-translational levels and promotes heat stress tolerance in transgenic Arabidopsis. PLoS ONE. 2015;10:e0135667. doi: 10.1371/journal.pone.0135667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shahnejat-Bushehri S, Mueller-Roeber B, Balazadeh S. Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal Behav. 2012;7:1518–1521. doi: 10.4161/psb.22092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan Q, Yue X, Zeng H, Zhu J. The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress–responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell. 2014;26:438–453. doi: 10.1105/tpc.113.118927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang Y, et al. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2015;66:6803–6817. doi: 10.1093/jxb/erv386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, et al. The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Sci. 2014;227:76–83. doi: 10.1016/j.plantsci.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Garapati P, et al. Transcription factor Arabidopsis activating factor1 integrates carbon starvation responses with trehalose metabolism. Plant Physiol. 2015;169:379–390. doi: 10.1104/pp.15.00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y, et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009;19:1279–1290. doi: 10.1038/cr.2009.108. [DOI] [PubMed] [Google Scholar]
- 30.Kilian J, et al. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 31.Garapati P, Xue G-P, Munné-Bosch S, Balazadeh S. Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 2015;168:1122–1139. doi: 10.1104/pp.15.00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim MJ, Ruzicka D, Shin R, Schachtman DP. The Arabidopsis AP2/ERF transcription factor RAP2.11 modulates plant response to low-potassium conditions. Mol. Plant. 2012;5:1042–1057. doi: 10.1093/mp/sss003. [DOI] [PubMed] [Google Scholar]
- 33.O’Malley RC, et al. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016;165:1280–1292. doi: 10.1016/j.cell.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Y, Klionsky DJ. Autophagic membrane delivery through ATG9. Cell Res. 2017;27:161–162. doi: 10.1038/cr.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J, Wang J, Yu J-Q, Chen Z. Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 2014 doi: 10.3389/fpls.2014.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gou W, et al. Autophagy in plant: A new orchestrator in the regulation of the phytohormones homeostasis. Int. J. Mol. Sci. 2019 doi: 10.3390/ijms20122900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiniquy D, et al. XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc. Nat. Acad. Sci. 2012;109:17117. doi: 10.1073/pnas.1202079109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders N, et al. Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc. Nat. Acad. Sci. 2012;109:989. doi: 10.1073/pnas.1115858109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell RAC, Dupree P, Shewry PR. A novel bioinformatics approach identifies candidate genes for the synthesis and feruloylation of arabinoxylan. Plant Physiol. 2007;144:43. doi: 10.1104/pp.106.094995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iurlaro A, et al. Drought and heat differentially affect XTH expression and XET activity and action in 3-day-old seedlings of durum wheat cultivars with different stress susceptibility. Front. Plant Sci. 2016;7:1686–1686. doi: 10.3389/fpls.2016.01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu XF, et al. XTH31, encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis. Plant Cell. 2012;24:4731. doi: 10.1105/tpc.112.106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Endo A, et al. Tissue-specific transcriptome analysis reveals cell wall metabolism, flavonol biosynthesis and defense responses are activated in the endosperm of germinating Arabidopsis thaliana Seeds. Plant Cell Physiol. 2011;53:16–27. doi: 10.1093/pcp/pcr171. [DOI] [PubMed] [Google Scholar]
- 43.Song L, Valliyodan B, Prince S, Wan J, Nguyen H. Characterization of the XTH gene family: New insight to the roles in soybean flooding tolerance. Int. J. Mol. Sci. 2018;19:2705. doi: 10.3390/ijms19092705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lord CC, Thomas G, Brown JM. Mammalian alpha beta hydrolase domain (ABHD) proteins: Lipid metabolizing enzymes at the interface of cell signaling and energy metabolism. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2013;1831:792–802. doi: 10.1016/j.bbalip.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu D, et al. A novel α/β-hydrolase gene IbMas enhances salt tolerance in transgenic sweetpotato. PLoS ONE. 2014;9:e115128. doi: 10.1371/journal.pone.0115128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding Y, Fromm M, Avramova Z. Multiple exposures to drought train transcriptional responses in Arabidopsis. Nat. Commun. 2012;3:740. doi: 10.1038/ncomms1732. [DOI] [PubMed] [Google Scholar]
- 48.Lu PL, et al. A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol. Biol. 2007;63:289–305. doi: 10.1007/s11103-006-9089-8. [DOI] [PubMed] [Google Scholar]
- 49.Jensen MK, et al. ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thalian. FEBS Openbio. 2013;3:321–327. doi: 10.1016/j.fob.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Journot-Catalino N, Somssich IE, Roby D, Kroj T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell. 2006;18:3289–3302. doi: 10.1105/tpc.106.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Truman W, Glazebrook J. Co-expression analysis identifies putative targets for CBP60g and SARD1 regulation. BMC Plant Biol. 2012;12:216. doi: 10.1186/1471-2229-12-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y. Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 2009;150:535. doi: 10.1104/pp.109.136028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olas JJ, et al. Primary carbohydrate metabolism genes participate in heat-stress memory at the shoot apical meristem of Arabidopsis thaliana. Mol. Plant. 2021;14:1508–1524. doi: 10.1016/j.molp.2021.05.024. [DOI] [PubMed] [Google Scholar]
- 54.Friedrich T, et al. Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in Arabidopsis. Nat. Commun. 2021;12:3426. doi: 10.1038/s41467-021-23786-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan Y, et al. Heat priming during early reproductive stages enhances thermo-tolerance to post-anthesis heat stress via improving photosynthesis and plant productivity in winter wheat (Triticum aestivum L.) Front Plant Sci. 2018 doi: 10.3389/fpls.2018.00805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shekhawat K, et al. Root endophyte induced plant thermotolerance by constitutive chromatin modification at heat stress memory gene loci. EMBO Rep. 2021;22:e51049. doi: 10.15252/embr.202051049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rauf M, et al. NAC transcription factor speedy hyponastic growth regulates flooding-induced leaf movement in Arabidopsis. Plant Cell. 2013;25:4941–4955. doi: 10.1105/tpc.113.117861. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.De Clercq I, et al. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell. 2013;25:3472–3490. doi: 10.1105/tpc.113.117168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skirycz A, et al. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 2006;47:10–24. doi: 10.1111/j.1365-313X.2006.02767.x. [DOI] [PubMed] [Google Scholar]
- 60.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 61.Hiscox JD, Israelstam GF. A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 1979;57:1332–1334. doi: 10.1139/b79-163. [DOI] [Google Scholar]
- 62.Arnon DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B. QuantPrime a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinform. 2008;9:465. doi: 10.1186/1471-2105-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caldana C, Scheible WR, Mueller-Roeber B, Ruzicic S. A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods. 2007;3:7. doi: 10.1186/1746-4811-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamranfar I, et al. Transcription factor RD26 is a key regulator of metabolic reprogramming during dark-induced senescence. New Phytol. 2018;218:1543–1557. doi: 10.1111/nph.15127. [DOI] [PubMed] [Google Scholar]
- 66.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anders S, Pyl PT, Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gentleman R, editor. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; 2005. pp. 397–420. [Google Scholar]
- 71.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2013. [Google Scholar]
- 72.Benjamini Y, Hochberg Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. B Met. 1995;57:289–300. [Google Scholar]
- 73.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berardini TZ, et al. The arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis J. Genet. Dev. 2015;53:474–485. doi: 10.1002/dvg.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data are available from the NCBI Bioproject database under ID SUB8978006 (https://ngdc.cncb.ac.cn/search/?dbId=gsa&q=SUB8978006). All other data generated or analysed during this study are included in this published article and its Supplementary Information files.