Abstract
The selection of the best-fitting substitution model of molecular evolution is a traditional step for phylogenetic inferences, including ancestral sequence reconstruction (ASR). However, a few recent studies suggested that applying this procedure does not affect the accuracy of phylogenetic tree reconstruction. Here, we revisited this debate topic by analyzing the influence of selection among substitution models of protein evolution, with focus on exchangeability matrices, on the accuracy of ASR using simulated and real data. We found that the selected best-fitting substitution model produces the most accurate ancestral sequences, especially if the data present large genetic diversity. Indeed, ancestral sequences reconstructed under substitution models with similar exchangeability matrices were similar, suggesting that if the selected best-fitting model cannot be used for the reconstruction, applying a model similar to the selected one is preferred. We conclude that selecting among substitution models of protein evolution is recommended for reconstructing accurate ancestral sequences.
Keywords: substitution models of protein evolution, substitution model selection, molecular evolution, ancestral sequence reconstruction, phylogenetics, protein evolution
Introduction
Ancestral sequence reconstruction (ASR) constitutes a powerful framework in evolutionary biology with a variety of applications (Liberles 2007; Selberg et al. 2021). For example, it has been used to develop vaccines based on centralized (ancestral) sequences (Kothe et al. 2006; Arenas and Posada 2010a) and to understand the stability and functional properties of diverse paleoenzymes such as thioredoxins (Perez-Jimenez et al. 2011), beta-lactamases (Risso et al. 2013), RuBisCO (Shih et al. 2016), or alcohol dehydrogenases (Thomson et al. 2005), among others (Merkl and Sterner 2016). These molecular reconstructions are not only of interest to evolutionary researchers, they can also present useful applications in industrial processes (Thomson et al. 2005) due to the biological and physicochemical properties (i.e., high thermostability) of the resurrected enzymes (Trudeau et al. 2016).
As for other phylogenetic analyses, probabilistic ASR methods (i.e., maximum-likelihood) require the specification of a substitution model of molecular evolution (Yang 2006). At the protein level, the substitution model includes the rates of change among the 20 amino acids (exchangeability matrix) and the equilibrium amino acid frequencies (Arenas 2015). Traditionally, the reconstruction of ancestral protein sequences is based on empirical substitution models of evolution (Thorne 2000; Arenas 2015). Despite the serve limits of these substitution models (i.e., all sites evolve under the same rates of change among amino acids, which is highly unrealistic; Echave et al. 2016), their mathematical simplicity (i.e., assuming site-independent evolution simplifies the likelihood function; Yang 2006) favored their establishment in phylogenetics. Empirical substitution models of evolution have been developed for diverse taxonomic, species, and protein groups (i.e., nuclear and mitochondrial proteins; Thorne 2000; Arenas 2015). Thus, nearly 100 empirical substitution models of protein evolution are currently available, many have been recently developed (Ng et al. 2000; Le et al. 2017; Le and Vinh 2020; Del Amparo and Arenas 2022) and still require efforts for their implementation in analytical phylogenetic frameworks (i.e., to perform substitution model selection and phylogenetic tree reconstruction, among others). Despite some data, the selection of an empirical substitution model is straightforward (i.e., when there is a substitution model biologically related to the study protein data), and in other times, this selection is unclear and traditionally requires the selection of a best-fitting substitution model (among the currently available substitution models) using likelihood-based methods (Yang et al. 1994; Zhang and Nei 1997; Zhang 1999; Minin et al. 2003; Lemmon et al. 2004). However, a few recent studies found that the selection of the best-fitting substitution model of protein evolution may not be mandatory for phylogenetic tree reconstruction (Abadi et al. 2019; Spielman and Shapiro 2020; Tao et al. 2020), although other studies suggested the opposite (Posada 2001; Minin et al. 2003; Dornburg et al. 2019). This debate brings the question of whether the selection among substitution models of protein evolution affects the accuracy of protein ASR, a relevant issue already mentioned in Pupko et al. (2007). For this application, a few studies suggested that substitution model selection does not affect ASR (Williams et al. 2006; Abadi et al. 2019), but they focused on ASR under substitution models of DNA evolution (Abadi et al. 2019) or ignored the influence of varying the amino acid exchangeability matrix (Williams et al. 2006). Here, we revisited this topic by the evaluation of the influence of selection among empirical substitution models of protein evolution, with focus on the exchangeability matrix, on the accuracy of ASR using simulated and real data. Overall, we found that the accuracy of the reconstructed ancestral sequences enhances the application of the selected best-fitting substitution model, especially in data presenting large genetic diversity.
Results
Evaluation of Substitution Model Selection for ASR Based on Simulated Protein Data
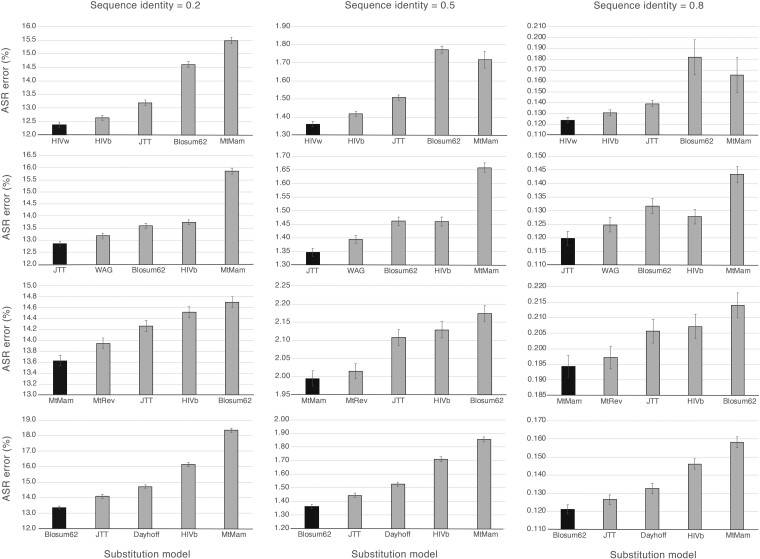
We evaluated the error of reconstructing ancestral sequences under diverse substitution models using computer simulations. We found that ancestral sequences inferred under the true (simulated) substitution model are more similar to the true ancestral sequences than ancestral sequences inferred under any other substitution model (fig. 1 and supplementary fig. S2, Supplementary Material online). In addition, we found that ancestral sequences reconstructed under a substitution model with an exchangeability matrix similar to that of the true substitution model are more accurate (in terms of similarity with the true ancestral sequences) than ancestral sequences reconstructed under a substitution model with exchangeability matrix far from that of the true substitution model (fig. 1 and supplementary fig. S2, Supplementary Material online). Interestingly, we also found that the sequence identity of the data affects the influence of the substitution model selection on the reconstructed ancestral sequences (fig. 1 and supplementary fig. S2, Supplementary Material online). In particular, the ASR from data with low-sequence identity (large genetic diversity) was more sensible to the selection of the substitution model than the ASR from data with high-sequence identity. Finally, we found that increasing the number of sequences (while maintaining genetic diversity) of the data qualitatively produced similar ASR error (compare fig. 1 and supplementary fig. S2, Supplementary Material online).
FIG. 1.
Influence of substitution model selection on ancestral sequence reconstruction using simulated data. Distances between true ancestral sequences and ancestral sequences reconstructed under true (black bars) and other substitution models (gray bars; including from the left to the right a model that is similar, intermediate, and far from the true model). The distances are shown in percentage. The study is based on 1,000 simulated data sets of 50 protein sequences with sequence identity 0.2 (large genetic diversity; plots on the left), 0.5 (intermediate genetic diversity; middle plots), and 0.8 (low genetic diversity; plots on the right). Error bars indicate 95% confidence intervals. The same results showing ASR error (y-axis) from zero are presented in supplementary figure S3, Supplementary Material online.
Evaluation of Substitution Model Selection for ASR Based on Real Protein Families
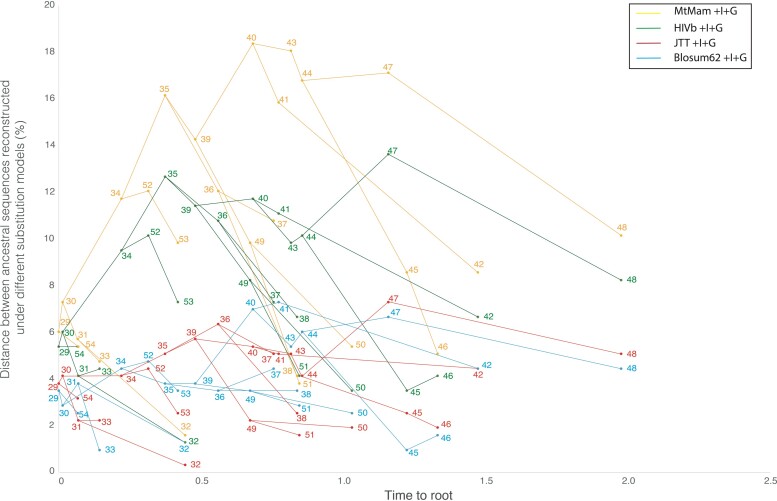
The studied real protein families showed that ancestral sequences reconstructed under different substitution models differ (fig. 2 and supplementary fig. S5, Supplementary Material online). In agreement with the results from simulated data, the distance between ancestral sequences reconstructed under different substitution models increases with the distance between the exchangeability matrices of the corresponding models and this can be observed at every internal node (fig. 2 and supplementary fig. S5, Supplementary Material online). Interestingly, the distance between ancestral sequences reconstructed under the best-fitting substitution model and ancestral sequences reconstructed under any other substitution model overall increased, going backwards in time with a maximum divergence near the center of tree (Deng et al. 2010), although with some fluctuations over time (fig. 2 and supplementary fig. S5, Supplementary Material online). This finding indicates that the ASR error caused by applying a substitution model that poorly fits with the data can affect the reconstructed sequences at all the internal nodes, and especially sequences belonging to internal nodes that are at a greater distance from the tip nodes.
FIG. 2.
Influence of substitution model selection on ancestral sequence reconstruction of the TRXB protein family. The figure shows the distance between ancestral sequences reconstructed under the best-fitting substitution model (LG + I + G) and other substitution models (MtMam + I + G, HIVb + I + G, JTT + I + G, and Blosum62 + I + G; shown with different colors) at every internal node and as a function of the time to root. The distances are shown in percentage. Note that all the nodes shown in the figure are internal nodes, the tip nodes are excluded because their sequences are given (thus, they are not reconstructed).
Concerning the number of CTL epitopes detected in the inferred ancestral sequences of the HIV-1 env data, we found that it varies depending on the substitution model applied in the ASR (supplementary tables S1 and S2, Supplementary Material online). Interestingly and in line with our previous findings, ancestral sequences reconstructed under substitution models with similar exchangeability matrices displayed a similar number of predicted epitopes (supplementary tables S1 and S2, Supplementary Material online).
Discussion
Selecting a best-fitting (in terms of likelihood) substitution model of evolution, among the available set of substitution models, and applying this model for probabilistic phylogenetic reconstruction is a well-established methodology. It is based on the natural reasoning of the phenotypic consequences caused by amino acid substitution events (i.e., >50 years ago Zuckerkandl and Pauling (1965) indicated that “it is the type rather than number of amino acid substitutions that is decisive”) and was supported by multiple likelihood-based phylogenetic studies for >20 years (Yang et al. 1994; Zhang and Nei 1997; Zhang 1999; Minin et al. 2003; Lemmon et al. 2004). However, a few recent works suggested that substitution model selection has little effect on phylogenetic tree reconstruction (Abadi et al. 2019; Spielman and Shapiro 2020; Tao et al. 2020) leading to a debate topic in the field. With regard to ASR, a study suggested that the selection among substitution models of DNA evolution does not influence nucleotide ASR (Abadi et al. 2019) and others investigated the influence of substitution rate variation among sites (Yang 1994; but under the same exchangeability matrix) on protein ASR (Pupko et al. 2002; Williams et al. 2006) or did not quantify the protein ASR error with computer simulations (Moshe and Pupko 2019). At the DNA level, we believe that the effect of substitution model selection on the accuracy of phylogenetic reconstructions could be reduced due to its lower number of character states compared with amino acids. Evaluating the influence of substitution rate variation among sites with a fixed exchangeability matrix is indeed relevant but still does not inform about the phylogenetic consequences of selection among substitution models of protein evolution considering different exchangeability matrices. Note that the currently available substitution models of protein evolution present differing empirical exchangeability matrices that are required to mimic diverse evolutionary patterns observed in nature (supplementary fig. S1, Supplementary Material online; Thorne 2000; Arenas 2015). In order to clarify this aspect, here, we revisited this topic to find that the selection among substitution models of protein evolution, with different exchangeability matrices, can seriously affect the reconstruction of ancestral sequences.
We simulated protein sequences to quantify the distance between ancestral sequences inferred under diverse substitution models, including the true substitution model, and we found that applying the true substitution model produces the most accurate ancestral sequences (compared with ancestral sequences reconstructed under other substitution models). Interestingly, we found that substitution models with exchangeability matrices similar to the exchangeability matrix of the true substitution model led to more accurate ancestral sequences than substitution models with exchangeability matrices far from that of the true substitution model. In practice, this suggests that if the best-fitting substitution model is not available to perform the ASR (i.e., it is not implemented in the ASR evolutionary framework), applying a substitution model with an exchangeability matrix as similar as possible to the exchangeability matrix of the best-fitting substitution model is recommended. Next, we found that the influence of the substitution model on ASR is affected by the genetic diversity of the study data. In particular, data with large genetic diversity produce ancestral sequences more influenced by the applied substitution model. Note that data with large genetic diversity accumulated multiple substitution events during their evolutionary histories and thus involve a more intense contribution of the substitution model in the likelihood function of probabilistic ASR methods (Yang 2006). Indeed, any substitution model produces error and thus applying more frequently a substitution model can increase error, especially (as we demonstrate in this study) if the applied model does not fit well with the studied data.
The ASR of real protein families performed under different substitution models showed that the reconstructed ancestral sequences (and also their biological properties in terms of number of CTL epitopes) differ depending on the applied substitution model. In particular, more distant substitution models produced more different ancestral sequences (with more different biological properties), in agreement with the results from the simulated data. Again, we found that using substitution models as similar as possible to the selected best-fitting substitution model is recommended when the best-fitting substitution model cannot be used for the ASR for any reason. The influence of the substitution model of protein evolution on ASR affects all the reconstructed ancestral sequences, but especially those belonging to the most internal nodes where the ASR method has to make more complicated decisions due to the long distance from the extant sequences. Despite some studies suggested that the selection of the best-fitting substitution model of evolution may not be a mandatory task for phylogenetic tree reconstruction (see Introduction), here we clearly found that substitution model selection is highly recommended for the reconstruction of ancestral proteins.
As indicated in the Introduction, protein ASR is frequently applied in diverse fields such as paleoenzymology and biotechnology (e.g., Perez-Jimenez et al. 2011; Holinski et al. 2017) and the reconstructed molecules should be as realistic as possible to display reliable biological properties. The observed patterns of amino acid substitution are the consequence of diverse selection constrains (i.e., selection on the protein function and stability; Lorenzo-Redondo et al. 2014; Arenas et al. 2016; Duchene et al. 2016; Echave et al. 2016; Kirchner et al. 2017; Geoghegan and Holmes 2018; Jimenez-Santos et al. 2018; Moshe and Pupko 2019) that can differ among taxonomic levels (Duchene et al. 2016; Chang et al. 2020), protein families (Rios et al. 2015; Del Amparo and Arenas 2022), and even within protein families (Del Amparo and Arenas 2022). Here we show that these different selection processes, mimicked with different substitution models, should be taken into consideration to more accurately reconstruct ancestral proteins.
Materials and Methods
Analysis of the Influence of Substitution Model Selection on ASR Using Simulated Protein Data
We simulated data to evaluate the distance between ancestral sequences reconstructed under the true (simulated) substitution model and ancestral sequences reconstructed under other (close or far from the true model; supplementary fig. S1, Supplementary Material online) substitution models. First, we simulated phylogenetic trees with random topologies using the function rtree implemented in the library ape of R (Paradis et al. 2004). Next, for each simulated tree, we simulated protein sequence evolution (we assumed a sequence length of 250 amino acids) under a particular substitution model with the function simSeq implemented in the phangorn library of R (Schliep 2011). We applied the HIVw (Nickle et al. 2007), JTT (Jones et al. 1992), Blosum62 (Henikoff and Henikoff 1992), and MtMam (Yang et al. 1998) substitution models in the simulations (true models) to include representative models of viral, nuclear, and mitochondrial proteins. We evaluated the influence of substitution model selection on ASR in six evolutionary scenarios of simulated data with variable number of protein sequences (50 and 100) and sequence identity (pairwise sequence comparisons, 0.2, 0.5, and 0.8). For each evolutionary scenario, we simulated a total of 1,000 multiple sequence alignments. As a control check, we applied ProtTest3 (Darriba et al. 2011) to verify that the true substitution models are selected as the best-fitting substitution models from the simulated data. Next, for each simulated data set, we reconstructed its ancestral sequences using the simulated phylogenetic tree (thus, avoiding potential biases from phylogenetic tree reconstruction) with the ML ASR method implemented in the function ancestral.pml of the phangorn library of R. The ASR was performed under diverse substitution models that included the true model and other models that are close and far from the true models. In particular, data simulated under the HIVw substitution model were evaluated with the HIVw (true), HIVb (close to the true; Nickle et al. 2007), JTT (intermediate), Blosum62, and MtMam (far from the true) substitution models; data simulated under the JTT substitution model were evaluated with the JTT (true), WAG (close; Whelan and Goldman 2001), HIVb (intermediate), Blosum62, and MtMam (far) substitution models; data simulated under the Blosum62 substitution model were evaluated with the Blosum62 (true), JTT (close), Dayhoff (intermediate), HIVb, and MtMam (far) substitution models; and data simulated under the MtMam substitution model were evaluated with the MtMam (true), MtRev (close; Adachi and Hasegawa 1996), JTT (intermediate), Blosum62, and HIVb (far) substitution models. Finally, we calculated the distance between simulated (true) ancestral sequences and ancestral sequences reconstructed under each substitution model. This distance is the sequence dissimilarity calculated as the proportion of different amino acid states (comparing every site) between the sequences.
Analysis of the Influence of Substitution Model Selection on ASR Using Real Protein Families
We analyzed the prokaryotic protein families D-ala D-ala ligases and Thioredoxins I (TRXB; table 1) as illustrative real examples. These protein families, available from the PFAM database (table 1), include a putative group of homologs from many bacterial species (Bastolla et al. 2004) with extant sequences longer than 200 amino acids that allow well-supported phylogenetic reconstructions (Arenas et al. 2017; Arenas and Bastolla 2020) and also have been previously analyzed with ASR (Perez-Jimenez et al. 2011; Meziane-Cherif et al. 2012; Ingles-Prieto et al. 2013). We realigned the sequences with MAFFT (Katoh and Standley 2013) as a prudent procedure. Next, we identified the best-fitting substitution model of protein evolution with ProtTest3 (table 1) and inferred an ML phylogenetic tree with RAxML-NG (Kozlov et al. 2019) under the best-fitting substitution model. We reconstructed the ancestral sequences under the best-fitting substitution model and other substitution models with close and distant exchangeability matrices (i.e., JTT, HIVb, Blosum62, and MtMam). Finally, for every internal node of the phylogenetic tree, we evaluated the distance between the ancestral sequences reconstructed under the best-fitting substitution model and the ancestral sequences reconstructed under every other substitution model.
Table 1.
Empirical Protein Families.
| Protein Family | PFAM Code | Number of Sequences | Sequence Length | Sequence Identity | Best-fitting Substitution Model |
|---|---|---|---|---|---|
| D-ala D-ala ligases (DDL) | PF07478 | 42 | 399 | 0.40 | LG + I + G |
| Thioredoxins I (TRXB) | PF00070 | 28 | 375 | 0.46 | LG + I + G |
Note.—For each data set, the table includes name of the protein family, PFAM code, number of sequences, sequence length (number of amino acids), sequence identity (ranging from 0 to 1), and the best-fitting substitution model selected with ProtTest3.
In order to provide an illustration of the biological consequences of substitution model selection in ASR, we also evaluated the predicted number of CTL epitopes in the ancestral sequences (reconstructed under different substitution models) of two alignments of the HIV-1 env region obtained from Arenas and Posada (2010b). Note that ancestral HIV-1 envelope proteins were widely used to design centralized vaccines against this virus (Nickle et al. 2003) and the accuracy of ASR can be crucial to obtain ancestral sequences with realistic immunological properties (Arenas and Posada 2010a). The first data set was a HIV-1 group M reference alignment with an outgroup from the Los Alamos HIV sequence database (41 sequences, 758 amino acids). The second data set included subtype B viruses and an outgroup (38 sequences, 810 amino acids; Doria-Rose et al. 2005). For each data set, we identified the best-fitting substitution model and inferred an ML tree under the best-fitting substitution model. Next, we reconstructed the ancestral sequences under the best-fitting substitution model and other, similar, and different substitution models. Then, we scanned the root ancestral sequences for known CTL epitopes with MHCPred (Guan et al. 2003).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank Centro de Supercomputación de Galicia (CESGA) for the computer resources. This work was supported by the Spanish Ministry of Economy and Competitiveness (grant numbers RYC-2015-18241 and PID2019-107931GA-I00). Funding for open access charge: Universidade de Vigo/CISUG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Roberto Del Amparo, CINBIO, Universidade de Vigo, Vigo, Spain; Departamento de Bioquímica, Xenética e Immunoloxía, Universidade de Vigo, Vigo, Spain.
Miguel Arenas, CINBIO, Universidade de Vigo, Vigo, Spain; Departamento de Bioquímica, Xenética e Immunoloxía, Universidade de Vigo, Vigo, Spain; Galicia Sur Health Research Institute (IIS Galicia Sur), Vigo, Spain.
Author Contributions
M.A. conceived the study. R.D.A performed the computer simulations and analyzed the empirical and simulated data. M.A. and R.D.A. wrote and revised the manuscript.
Data Availability
The empirical and simulated data are available at Zenodo repository from the URL https://doi.org/10.5281/zenodo.6412799.
References
- Abadi S, Azouri D, Pupko T, Mayrose I. 2019. Model selection may not be a mandatory step for phylogeny reconstruction. Nat Commun. 10:934. 10.1038/s41467-019-08822-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi J, Hasegawa M. 1996. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J Mol Evol. 42:459–468. 10.1007/BF02498640 [DOI] [PubMed] [Google Scholar]
- Arenas M. 2015. Trends in substitution models of molecular evolution. Front Genet. 6:319. 10.3389/fgene.2015.00319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas M, Bastolla U. 2020. ProtASR2: ancestral reconstruction of protein sequences accounting for folding stability. Methods Ecol Evol. 11:248–257. 10.1111/2041-210X.13341 [DOI] [Google Scholar]
- Arenas M, Lorenzo-Redondo R, Lopez-Galindez C. 2016. Influence of mutation and recombination on HIV-1 in vitro fitness recovery. Mol Phylogenet Evol. 94:264–270. 10.1016/j.ympev.2015.09.001 [DOI] [PubMed] [Google Scholar]
- Arenas M, Posada D. 2010a. Computational design of centralized HIV-1 genes. Curr HIV Res. 8:613–621. 10.2174/157016210794088263 [DOI] [PubMed] [Google Scholar]
- Arenas M, Posada D. 2010b. The effect of recombination on the reconstruction of ancestral sequences. Genetics 184:1133–1139. 10.1534/genetics.109.113423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas M, Weber CC, Liberles DA, Bastolla U. 2017. ProtASR: an evolutionary framework for ancestral protein reconstruction with selection on folding stability. Syst Biol. 66:60. [DOI] [PubMed] [Google Scholar]
- Bastolla U, Moya A, Viguera E, van Ham RCHJ . 2004. Genomic determinants of protein folding thermodynamics in prokaryotic organisms. J Mol Biol. 343:1451–1466. 10.1016/j.jmb.2004.08.086 [DOI] [PubMed] [Google Scholar]
- Chang H, Nie Y, Zhang N, Zhang X, Sun H, Mao Y, Qiu Z, Huang Y. 2020. MtOrt: an empirical mitochondrial amino acid substitution model for evolutionary studies of Orthoptera insects. BMC Evol Biol. 20:57. 10.1186/s12862-020-01623-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. 10.1093/bioinformatics/btr088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Amparo R, Arenas M. 2022. HIV protease and integrase empirical substitution models of evolution: protein-specific models outperform generalist models. Genes 13(1):61. 10.3390/genes13010061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Maust B, Nickle D, Learn G, Liu Y, Heath L, Kosakovsky Pond S, Mullins J. 2010. DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. Biotechniques 48:405–408. 10.2144/000113370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Learn GH, Rodrigo AG, Nickle DC, Li F, Mahalanabis M, Hensel MT, McLaughlin S, Edmonson PF, Montefiori D, et al. 2005. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J Virol. 79:11214–11224. 10.1128/JVI.79.17.11214-11224.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornburg A, Su Z, Townsend JP. 2019. Optimal rates for phylogenetic inference and experimental design in the era of genome-scale data sets. Syst Biol. 68:145–156. 10.1093/sysbio/syy047 [DOI] [PubMed] [Google Scholar]
- Duchene S, Di Giallonardo F, Holmes EC. 2016. Substitution model adequacy and assessing the reliability of estimates of virus evolutionary rates and time scales. Mol Biol Evol. 33:255–267. 10.1093/molbev/msv207 [DOI] [PubMed] [Google Scholar]
- Echave J, Spielman SJ, Wilke CO. 2016. Causes of evolutionary rate variation among protein sites. Nat Rev Genet. 17:109–121. 10.1038/nrg.2015.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JL, Holmes EC. 2018. The phylogenomics of evolving virus virulence. Nat Rev Genet. 19:756–769. 10.1038/s41576-018-0055-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P, Doytchinova IA, Zygouri C, Flower DR. 2003. MHCPred: a server for quantitative prediction of peptide-MHC binding. Nucleic Acids Res. 31:3621–3624. 10.1093/nar/gkg510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. 1992. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 89:10915–10919. 10.1073/pnas.89.22.10915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holinski A, Heyn K, Merkl R, Sterner R. 2017. Combining ancestral sequence reconstruction with protein design to identify an interface hotspot in a key metabolic enzyme complex. Proteins 85:312–321. 10.1002/prot.25225 [DOI] [PubMed] [Google Scholar]
- Ingles-Prieto A, Ibarra-Molero B, Delgado-Delgado A, Perez-Jimenez R, Fernandez JM, Gaucher EA, Sanchez-Ruiz JM, Gavira JA. 2013. Conservation of protein structure over four billion years. Struct Lond Engl. 1993(21):1690–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Santos MJ, Arenas M, Bastolla U. 2018. Influence of mutation bias and hydrophobicity on the substitution rates and sequence entropies of protein evolution. PeerJ 6:e5549. 10.7717/peerj.5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 8:275–282. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner S, Cai Z, Rauscher R, Kastelic N, Anding M, Czech A, Kleizen B, Ostedgaard LS, Braakman I, Sheppard DN, et al. 2017. Alteration of protein function by a silent polymorphism linked to tRNA abundance. PLoS Biol. 15:e2000779. 10.1371/journal.pbio.2000779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe DL, Li Y, Decker JM, Bibollet-Ruche F, Zammit KP, Salazar MG, Chen Y, Weng Z, Weaver EA, Gao F, et al. 2006. Ancestral and consensus envelope immunogens for HIV-1 subtype C. Virology 352:438–449. 10.1016/j.virol.2006.05.011 [DOI] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35:4453–4455. 10.1093/bioinformatics/btz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le VS, Dang CC, Le QS. 2017. Improved mitochondrial amino acid substitution models for metazoan evolutionary studies. BMC Evol Biol. 17:136. 10.1186/s12862-017-0987-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TK, Vinh LS. 2020. FLAVI: an amino acid substitution model for flaviviruses. J Mol Evol. 88:445–452. 10.1007/s00239-020-09943-3 [DOI] [PubMed] [Google Scholar]
- Lemmon AR, Moriarty EC. 2004. The importance of proper model assumption in Bayesian phylogenetics. Syst Biol. 53:265–277. 10.1080/10635150490423520 [DOI] [PubMed] [Google Scholar]
- Liberles DA. 2007. Ancestral sequence reconstruction. Oxford, United Kingdom. Oxford University Press. [Google Scholar]
- Lorenzo-Redondo R, Delgado S, Moran F, Lopez-Galindez C. 2014. Realistic three dimensional fitness landscapes generated by self organizing maps for the analysis of experimental HIV-1 evolution. PLoS One. 9:e88579. 10.1371/journal.pone.0088579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkl R, Sterner R. 2016. Ancestral protein reconstruction: techniques and applications. Biol Chem. 397:1–21. 10.1515/hsz-2015-0158 [DOI] [PubMed] [Google Scholar]
- Meziane-Cherif D, Saul FA, Haouz A, Courvalin P. 2012. Structural and functional characterization of VanG D-Ala:D-Ser ligase associated with vancomycin resistance in Enterococcus faecalis. J Biol Chem. 287:37583–37592. 10.1074/jbc.M112.405522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin V, Abdo Z, Joyce P, Sullivan J. 2003. Performance-based selection of likelihood models for phylogeny estimation. Syst Biol. 52:674–683. 10.1080/10635150390235494 [DOI] [PubMed] [Google Scholar]
- Moshe A, Pupko T. 2019. Ancestral sequence reconstruction: accounting for structural information by averaging over replacement matrices. Bioinformatics 35:2562–2568. 10.1093/bioinformatics/bty1031 [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff JG, Henikoff S. 2000. PHAT: a transmembrane-specific substitution matrix. Bioinformatics 16:760–766. 10.1093/bioinformatics/16.9.760 [DOI] [PubMed] [Google Scholar]
- Nickle DC, Heath L, Jensen MA, Gilbert PB, Mullins JI, Kosakovsky Pond SL. 2007. HIV-specific probabilistic models of protein evolution. PLoS One. 2:e503. 10.1371/journal.pone.0000503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickle DC, Jensen MA, Gottlieb GS, Shriner D, Learn GH, Rodrigo AG, Mullins JI. 2003. Consensus and ancestral state HIV vaccines. Science 299:1515–1518. 10.1126/science.299.5612.1515c [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- Perez-Jimenez R, Ingles-Prieto A, Zhao ZM, Sanchez-Romero I, Alegre-Cebollada J, Kosuri P, Garcia-Manyes S, Kappock TJ, Tanokura M, Holmgren A, et al. 2011. Single-molecule paleoenzymology probes the chemistry of resurrected enzymes. Nat Struct Mol Biol. 18:592–596. 10.1038/nsmb.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. 2001. The effect of branch length variation on the selection of models of molecular evolution. J Mol Evol. 52:434–444. 10.1007/s002390010173 [DOI] [PubMed] [Google Scholar]
- Pupko T, Doron-Faigenboim A, Liberles DA, Cannarozzi GM. 2007. Probabilistic models and their impact on the accuracy of reconstructed ancestral protein sequences. In: Liberkes D, editor. Ancestral sequence reconstruction. Oxford: Oxford University Press. Available from: https://oxford.universitypressscholarship.com/10.1093/acprof:oso/9780199299188.001.0001/acprof-9780199299188-chapter-4 [Google Scholar]
- Pupko T, Pe’er I, Hasegawa M, Graur D, Friedman N. 2002. A branch-and-bound algorithm for the inference of ancestral amino-acid sequences when the replacement rate varies among sites: application to the evolution of five gene families. Bioinformatics 18:1116–1123. 10.1093/bioinformatics/18.8.1116 [DOI] [PubMed] [Google Scholar]
- Rios S, Fernandez MF, Caltabiano G, Campillo M, Pardo L, Gonzalez A. 2015. GPCRtm: an amino acid substitution matrix for the transmembrane region of class A G protein-coupled receptors. BMC Bioinformatics. 16:206. 10.1186/s12859-015-0639-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM. 2013. Hyperstability and substrate promiscuity in laboratory resurrections of precambrian β-lactamases. J Am Chem Soc. 135:2899–2902. 10.1021/ja311630a [DOI] [PubMed] [Google Scholar]
- Schliep KP. 2011. Phangorn: phylogenetic analysis in R. Bioinformatics 27:592–593. 10.1093/bioinformatics/btq706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selberg AGA, Gaucher EA, Liberles DA. 2021. Ancestral sequence reconstruction: from chemical paleogenetics to maximum likelihood algorithms and beyond. J Mol Evol. 89:157–164. 10.1007/s00239-021-09993-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih PM, Occhialini A, Cameron JC, Andralojc PJ, Parry MAJ, Kerfeld CA. 2016. Biochemical characterization of predicted Precambrian RuBisCO. Nat Commun. 7:10382. 10.1038/ncomms10382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman SJ. 2020. Relative model fit does not predict topological accuracy in single-gene protein phylogenetics. Mol Biol Evol. 37:2110–2123. 10.1093/molbev/msaa075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q, Barba-Montoya J, Huuki LA, Durnan MK, Kumar S. 2020. Relative efficiencies of simple and complex substitution models in estimating divergence times in phylogenomics. Mol Biol Evol. 37:1819–1831. 10.1093/molbev/msaa049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Gaucher EA, Burgan MF, De Kee DW, Li T, Aris JP, Benner SA. 2005. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet. 37:630–635. 10.1038/ng1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne JL. 2000. Models of protein sequence evolution and their applications. Curr Opin Genet Dev. 10:602–605. [DOI] [PubMed] [Google Scholar]
- Trudeau DL, Kaltenbach M, Tawfik DS. 2016. On the potential origins of the high stability of reconstructed ancestral proteins. Mol Biol Evol. 33:2633–2641. 10.1093/molbev/msw138 [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 18:691–699. 10.1093/oxfordjournals.molbev.a003851 [DOI] [PubMed] [Google Scholar]
- Williams PD, Pollock DD, Blackburne BP, Goldstein RA. 2006. Assessing the accuracy of ancestral protein reconstruction methods. PLoS Comput Biol. 2:e69. 10.1371/journal.pcbi.0020069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 39:306–314. 10.1007/BF00160154 [DOI] [PubMed] [Google Scholar]
- Yang Z. 2006. Computational molecular evolution. Oxford, United Kingdom: Oxford University Press. [Google Scholar]
- Yang Z, Goldman N, Friday A. 1994. Comparison of models for nucleotide substitution used in maximum-likelihood phylogenetic estimation. Mol Biol Evol. 11:316–324. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Hasegawa M. 1998. Models of amino acid substitution and applications to mitochondrial protein evolution. Mol Biol Evol. 15:1600–1611. 10.1093/oxfordjournals.molbev.a025888 [DOI] [PubMed] [Google Scholar]
- Zhang J. 1999. Performance of likelihood ratio tests of evolutionary hypotheses under inadequate substitution models. Mol Biol Evol. 16:868–875. 10.1093/oxfordjournals.molbev.a026171 [DOI] [PubMed] [Google Scholar]
- Zhang J, Nei M. 1997. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J Mol Evol. 44(Suppl. 1):S139–S146. 10.1007/PL00000067 [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. New York: Academic Press. p. 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The empirical and simulated data are available at Zenodo repository from the URL https://doi.org/10.5281/zenodo.6412799.