Abstract
INTRODUCTION:
The diversity of cell types is a defining feature of the neuronal circuitry that makes up the areas and layers of the mammalian cortex. At a molecular level, the extent of this diversity is now better appreciated through recent efforts to census all potential cortical cell types through single-cell transcriptional profiling. Cortical populations can be hierarchically subdivided into multiple putative transcriptomic cell classes, subclasses, and types. This new catalog of neuronal subclasses and subtypes opens up new questions and avenues of investigation for how these cell types are collectively organized into circuits that function to process information and adapt to changes in experience.
RATIONALE:
We investigated the function of newly identified cell types in layers 2 or 3 (L2/3) of the primary somatosensory cortex, a region that integrates bottom-up sensory information with top-down internal representations. Current in vivo methods primarily allow cell types to be investigated one at a time and have limited ability to label cell types defined by combinations of expressed genes. To densely survey these cell types and investigate how they interact during task behavior, we developed a platform, Comprehensive Readout of Activity and Cell Type Markers (CRACK), that combines population calcium imaging with subsequent multiplexed fluorescent in situ hybridization. Multiplexed labeling of mRNA transcripts is critical to deciphering the identity of cell types defined by combinatorial patterns of gene expression.
RESULTS:
We profiled the functional responses of three excitatory cell types and eight inhibitory subclasses in L2/3 as mice performed a whisker-based tactile working memory task. Task-related properties of both excitatory and inhibitory neurons continue to differentiate as they are segregated into increasingly discrete molecular types. Our analysis revealed that the excitatory cell type, L2/3 intratelencephalic Baz1a (Baz1a), functions as a highly active detector of tactile features. Simultaneous imaging across identified cell types enabled measurements of functional connectivity between subpopulations. Functional connectivity analysis indicated that Baz1a neurons orchestrate local network activity patterns. We found that Baz1a neurons show strong functional connections with dendrite-targeting, somatostatin-expressing (Sst) inhibitory neurons. Trans-monosynaptic viral tracing confirmed that Baz1a neurons preferentially synapse onto Sst neurons. Baz1a neurons also show enrichment of select plasticity-related, immediate early genes, including Fos. To determine whether the expression pattern of immediate early genes is a stable property of Baz1a neurons and how this relates to neuronal plasticity, we tracked Fos expression and neuronal activity in mice subjected to whisker deprivation. We found that Baz1a neurons homeostatically adapt to sensory deprivation while stably maintaining Fos expression.
CONCLUSION:
These results demonstrate that Baz1a neurons are a component of a molecularly defined circuit motif that is capable of recruiting local circuits for sensory processing when salient features are encountered during behavior. This cell type also functions to preserve sensory representations during ongoing and altered sensory experience. This builds on our knowledge for how local circuits in somatosensory cortex are implemented to negotiate bottom-up and top-down information. The ability to map functional and transcriptional relationships across neuronal populations provides insight into how the organizing principles of the cortex give rise to the computations it performs.
Graphical Abstract
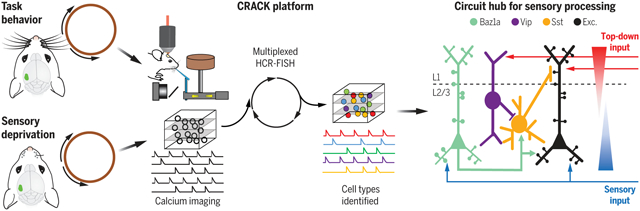
CRACK platform reveals a circuit hub for sensory processing. Functional profiling of molecularly defined cells was achieved with in vivo two-photon calcium imaging in L2/3 of the primary somatosensory cortex during task behavior or sensory deprivation followed by multiplexed fluorescent in situ hybridization. Excitatory Baz1a neurons form a connection motif capable of recruiting local circuits and preserving sensory representations during ongoing and altered sensory experience. HCR-FISH, hybridization chain reaction–fluorescence in situ hybridization; Vip, vasoactive intestinal peptide–expressing; Exc., excitatory.
Although single-cell transcriptomics of the neocortex has uncovered more than 300 putative cell types, whether this molecular classification predicts distinct functional roles is unclear. We combined two-photon calcium imaging with spatial transcriptomics to functionally and molecularly investigate cortical circuits. We characterized behavior-related responses across major neuronal subclasses in layers 2 or 3 of the primary somatosensory cortex as mice performed a tactile working memory task. We identified an excitatory intratelencephalic cell type, Baz1a, that exhibits high tactile feature selectivity. Baz1a neurons homeostatically maintain stimulus responsiveness during altered experience and show persistent enrichment of subsets of immediately early genes. Functional and anatomical connectivity reveals that Baz1a neurons residing in upper portions of layers 2 or 3 preferentially innervate somatostatin-expressing inhibitory neurons. This motif defines a circuit hub that orchestrates local sensory processing in superficial layers of the neocortex.
Cells of the neocortex can be defined on the basis of their molecular composition, the diversity of which is reflected in their transcriptome. The transcriptional profiles observed across this brain region indicate that cortical populations can be hierarchically subdivided into multiple putative transcriptomic cell classes [such as γ-aminobutyric acid (GABA)–ergic or glutamatergic], subclasses (such as GABAergic Pvalb), and types (such as GABAergic Pvalb Vipr2) (1, 2). Even within a single layer of one cortical area, transcriptional diversity remains high (3). This organization may have developmental origins (4, 5) or reflect anatomical specificity (6, 7) or physiological properties (8, 9). The extent to which this diversity relates to information encoding during goal-directed behavior is unclear. In superficial layers of the neocortex, excitatory layer-2 or –3 (L2/3) pyramidal neurons can be disinhibited by subclasses of inhibitory vasoactive intestinal peptide–expressing (Vip) neurons through subclasses of inhibitory somatostatin-expressing (Sst) neurons. The degree to which this motif is part of a larger circuit composed of other transcriptomic cell types is unclear.
The ability to link molecularly identified neurons with their function during behavior requires monitoring the activity of cell types in vivo. Traditional approaches to label cell types by use of transgenic lines or post hoc immunohistochemistry are limited to one to three molecular markers (10, 11). This has restricted investigations to classes of excitatory and inhibitory neurons to the broadest hierarchical levels of cell type diversity. Techniques for spatial transcriptional profiling increase the number of genes that can be simultaneously identified in tissue (12–16). Combinatorial expression patterns of multiple genes can then be used to define finer divisions in the transcriptomic taxonomy that correspond to more specific neuronal subclasses and types. Further, spatial profiling of gene expression in intact tissue readily enables dense multimodal registration of anatomical and functional measurements across neurons within a single sample (17). We developed a platform, Comprehensive Readout of Activity and Cell Type Markers (CRACK), that combines in vivo two-photon calcium imaging with post hoc multiplexed fluorescence in situ hybridization. Using this platform, we sought to determine whether finer divisions in the transcriptomic taxonomy (subclasses and types) exhibit distinct functional characteristics and connection motifs. We focused on newly identified cell types in L2/3 of the primary somatosensory cortex (S1), a region involved in processing and integrating tactile information with motor and associative input.
CRACK platform
The CRACK platform uses a multi-area two-photon microscope (18) configured to perform simultaneous population calcium imaging across multiple tissue depths, providing three-dimensional (3D) spatial information of neuron location for later post hoc identification (Fig. 1A, fig. S1, and movie S1). After functional in vivo experiments, tissue encompassing the imaged volume was sectioned parallel to the imaging plane. The tissue was embedded in hydrogel and cleared (19) to facilitate labeling of mRNA transcripts by using hybridization chain reaction–fluorescence in situ hybridization (HCR-FISH) (13) and confocal imaging. Because HCR-FISH is a DNA-based labeling strategy, probes for different mRNA transcripts were labeled, imaged, and then stripped by using deoxyribonuclease (DNase) across multiple rounds. To reidentify and register in vivo neurons across multiple rounds of HCR-FISH, we dedicated one imaging channel (561) to repeated labeling and imaging of transcripts of the red genetically encoded calcium indicator, RCaMP1.07, which we used for functional imaging (fig. S2 and supplementary text S1) (20). Other imaging channels were used for labeling cell type–specific markers (table S1).
Fig. 1. Multiplexed identification of transcriptomic cell subclasses and types in functionally imaged neurons.
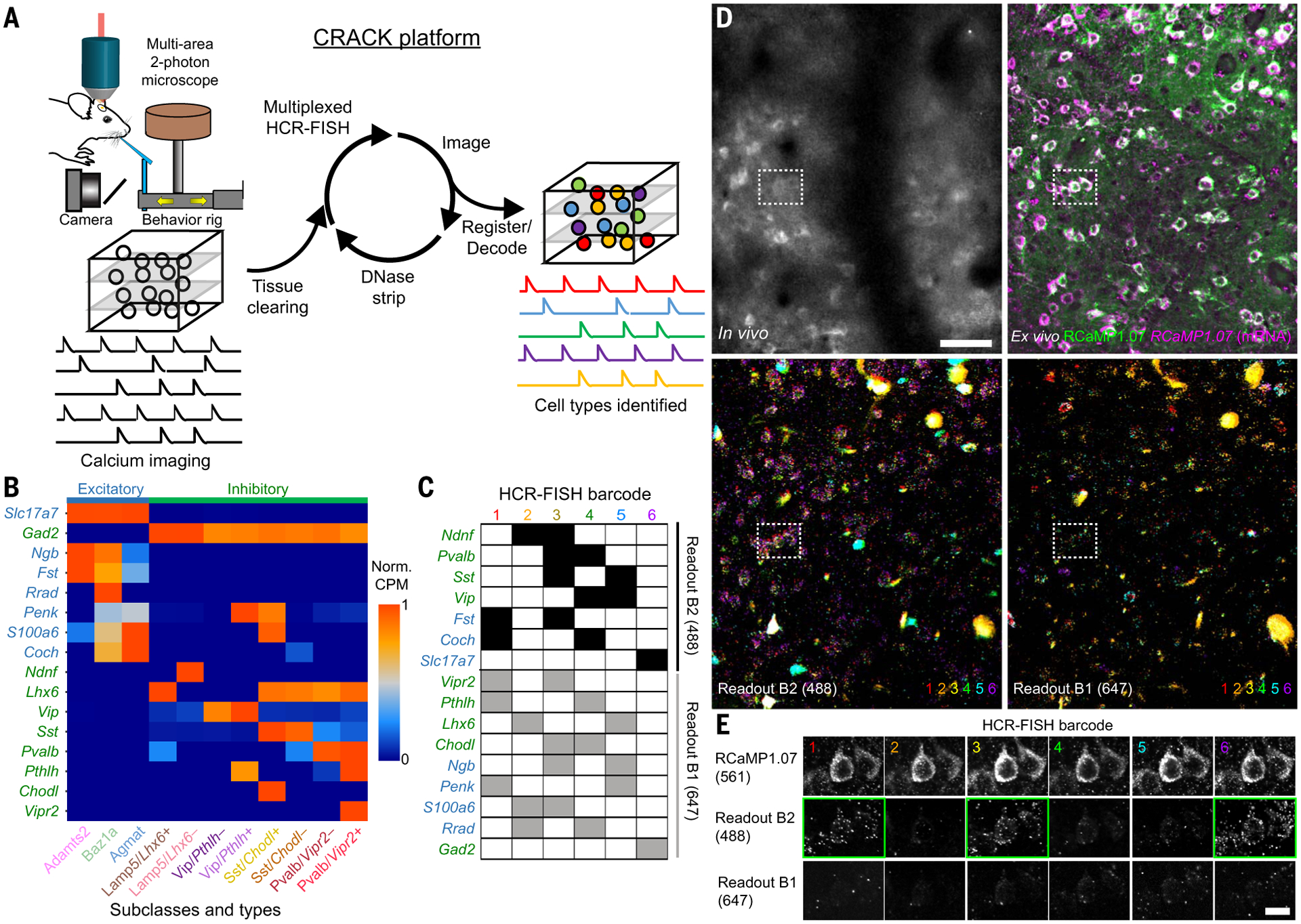
(A) Schematic of the CRACK platform. (B) Expression patterns of genes selected to identify L2/3 S1 excitatory (blue) and inhibitory (green) cell subclasses and types. (C) Barcode scheme for multiplexed HCR-FISH of selected genes. (D) Registration of in vivo calcium-imaged neurons to ex vivo tissue section across multiple rounds of HCR-FISH. (Top left) In vivo two-photon images of RCaMP1.07+ neurons. (Top right) Ex vivo confocal images of reidentified RCaMP1.07+ neurons showing endogeneous protein (green) followed by HCR-FISH staining transcripts (magenta). (Bottom) Overlays of (left) B2-488 and (right) B1-647 readout channels across all HCR-FISH barcode rounds. (E) Decoding of in vivo imaged neuron [(D), dotted rectangle] identified as an Adamts2 cell type expressing Fst and Slc17a7. Positive readouts are identified with green rectangles. Scale bars, (D) 50 μm; (E) 20 μm.
Although expression of a small number of genes can be detected through multiple rounds of sequential staining, a barcode readout scheme provides high read depth (100 to 1000 genes) in an error-robust manner. Using barcode readouts to decode arbitrary gene sets relies on single-molecule mRNA resolution, which is sensitive to image registration errors and has only been demonstrated in thin tissue sections (<40 μm) (12, 16). To obviate the need for single-molecule mRNA-resolution registration so that larger volumes of tissue (150 to 300 μm) could be imaged and analyzed, we programmed our barcode for cellular-resolution readout. This approach relies on prior knowledge of gene expression patterns so that binary decoding for each imaging channel and hybridization round could be programmed at cellular rather than mRNA resolution. This approach is highly compatible with identifying cell types defined by nonoverlapping gene expression patterns.
We analyzed single-cell RNA-sequencing (scRNA-seq) data from S1 that were acquired as part of a larger study of the molecular diversity of the isocortex (21). On the basis of combinatorial expression patterns, L2/3 intratelencephalic (IT) pyramidal neurons in S1 were observed to be segregated into three transcriptomic cell types: L2/3 IT Adamts2 (Adamts2), L2/3 IT Baz1a (Baz1a), and L2/3 IT Agmat (Agmat) (fig. S4). Excitatory neurons in L2/3 show both cell type–specific and area-specific gene expression patterns. When comparing S1 L2/3 cell types to those in the primary visual (V1) and anterior lateral motor (ALM) cortex, Baz1a and Agmat cells showed similarity to cell types identified in V1 and ALM, whereas Adamts2 cells were present in V1 but not ALM (1).
Inhibitory neuron cell types in S1 were shared with other cortical areas and found to be hierarchically organized. Although the major nonoverlapping inhibitory subclasses (Lamp5, Pvalb, Sst, and Vip) have each been investigated at the broadest level (22, 23), further subdivisions have not been investigated during task behavior. Thus, we selected gene markers that defined the next level of transcriptional subdivision (fig. S5). Lamp5 neurons were subdivided into two subclasses according to mutually exclusive expression of LIM homeobox 6 (Lhx6) or neuron-derived neurotrophic factor (Ndnf). Pvalb, Sst, or Vip neurons were subdivided according to expression of vasoactive intestinal peptide receptor 2 (Vipr2), chondrolectin (Chodl), or parathryroid hormone–like hormone (Pthlh), respectively. We devised a barcode scheme for detection of 16 mRNA species across six rounds of staining to resolve 11 transcriptomically defined cell populations (three excitatory types and eight inhibitory subclasses) (Fig. 1, B to E).
Task encoding across excitatory types
To identify functional differences between transcriptionally defined cell populations in L2/3 of S1, two-photon calcium imaging was carried out on expert wild-type mice (n = 7) performing a head-fixed whisker-based delayed nonmatch to sample (DNMS) task (fig. S6) (24). In this context-dependent sensory processing task, a motorized rotor is used to deflect multiple whiskers in either an anterior or posterior direction during an initial “sample” and a later “test” period, separated by a 2-s delay (Fig. 2A). During the delay period and the intertrial interval, the rotor was withdrawn to prevent whisker-rotor contact. Behavior was reported as “go/no go,” in which animals licked on “go” trials for a water reward (“hit”) when the presented sample and test stimulus were nonmatching and withheld licking on “no go” trials (“correct rejection”) when the presented sample and test stimulus were matching. High-speed videography was also performed to monitor whisking behavior.
Fig. 2. Task encoding across L2/3 excitatory cell types.
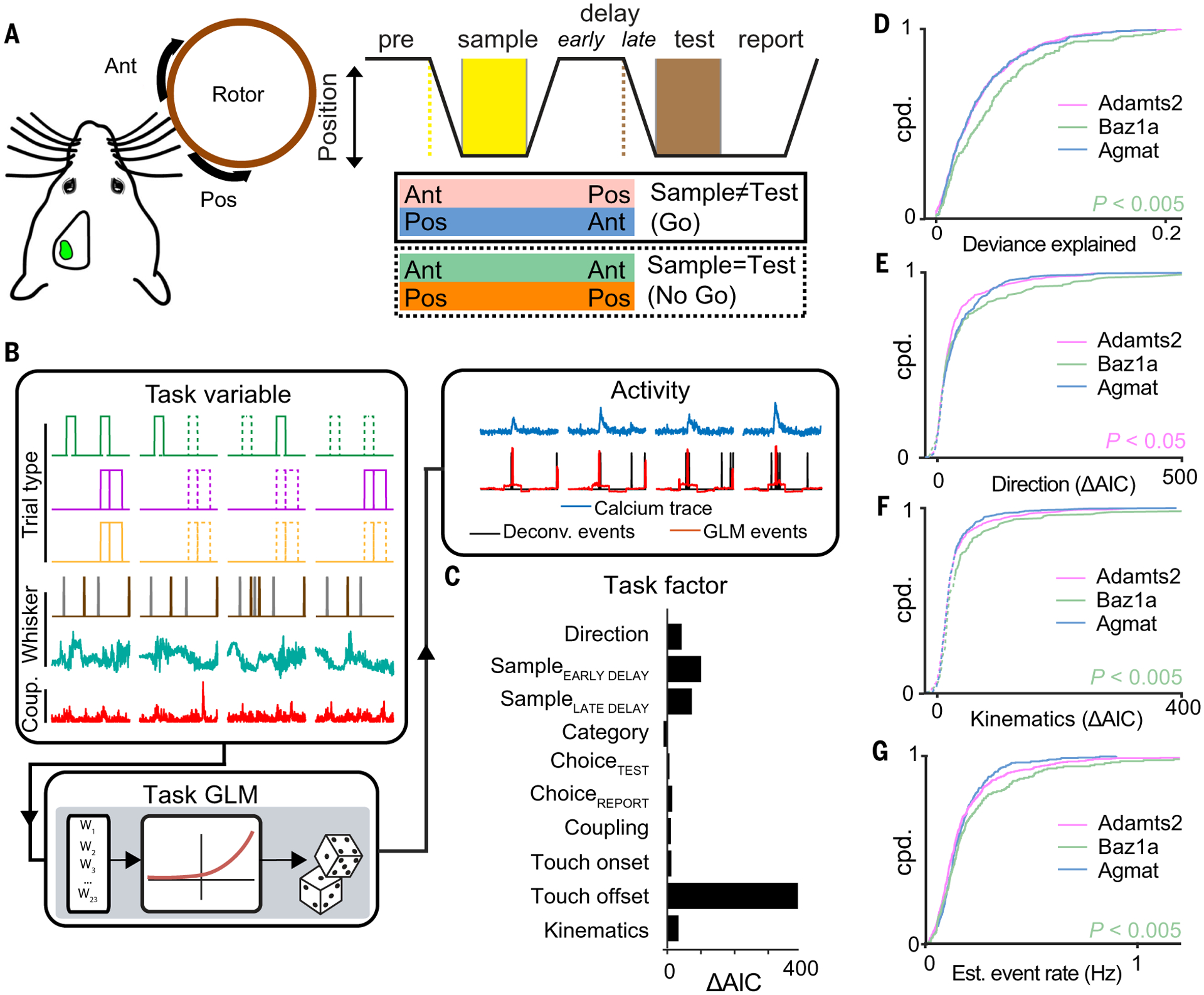
(A) Schematic of whisker-based delayed nonmatch to sample behavioral task. (B) Encoding of task-related activity in individual neurons using a GLM. (C) Encoding of task factors determined by comparing full and partial GLM fits (ΔAIC). (D to G) Cumulative probability distributions of (D) full model deviance explained, (E) encoding strength of stimulus direction, (F) encoding strength of whisker kinematics, and (G) estimated event rate across the three excitatory cell types. [(D) to (F)] Mann Whitney U test; (G) one-tailed Student’s t test. In (E) and (F), solid and dotted lines indicate significant (P < 0.01) and nonsignificant encoding strengths, respectively, by means of χ2 test. n = 1107 neurons from seven animals.
We previously reported diverse task-related responses in L2/3 of S1 during the DNMS task (24). To characterize task-related responses for each recorded cell in a more comprehensive manner, we fit a generalized linear model (GLM) to each neuron’s estimated calcium event activity against a range of “task variables” (Fig. 2B, figs. S9 and S10, and supplementary text S2) (25). Task variables representing a related feature were grouped into “task factors” (such as stimulus direction and trial category). The ability for a neuron to encode a particular task factor was determined by calculating the difference in the Akaike information criterion (ΔAIC) between a full model and a partial model that excludes task variables representing that task factor. A positive ΔAIC value indicates reduced fit quality from the full to the partial model, revealing that the excluded task factor in the partial model is an important contributor to the modeled neuron’s activity. Thus, we interpret significant, positive ΔAIC values to indicate neuronal encoding of the excluded task factor (Fig. 2C, fig. S11, and supplementary text S3). We analyzed 10 task-related factors. Six of the 10 task factors were defined by trial type information. This included information related to the direction of the task stimulus (direction), trial category defined by the combination of the sample and test stimulus (category), and the animal’s choice during the test (choiceTEST) and report period (choiceREPORT). Although our previous study found no evidence of sustained activity in S1 during the delay period (25), we included task factors that represent the sample stimulus at later points in the trial (sample encoded early in the delay period, sampleEARLY DELAY; sample information late in the delay period, sampleLATE DELAY). Another set of task factors describing whisker movement and tactile-object interactions were derived from video analysis of whisker tracking and included whisker-object touch onset (touch onset), whisker-object touch offset (touch offset), and whisker kinematics (kinematics). A final task factor was derived from the activity of all other simultaneously recorded neurons to assess the level of coupling the neuron had with overall network activity (coupling) (26). Overall, we identified neurons that were selective to a single or multiple task factors (fig. S12).
We first compared differences in task encoding across the three excitatory cell types. Baz1a neurons showed the best overall GLM fit (Fig. 2D) and more strongly encoded whisker kinematics compared with the other two excitatory cell types (P < 0.005, Mann Whitney U test) (Fig. 2F). By contrast, Adamts2 neurons more weakly encoded stimulus direction and touch offset (direction, P < 0.05; touch offset, P < 0.02; Mann Whitney U test) (Fig. 2E and fig. S14), whereas Agmat neurons more strongly encoded choiceREPORT (P < 0.05, Mann Whitney U test). Baz1a neurons also showed overall higher event rates (P < 0.005, one-tailed Student’s t test) and response reliability to sample and test stimuli (Fig. 2G and fig. S13). However, encoding of touch onset did not differ between excitatory cell types, suggesting that Baz1a neurons are more tuned to specific kinematic features rather than more sensitive to nonspecific tactile input (fig. S14).
Persistent stimulus activity and Fos expression in Baz1a neurons
Highly active, sensory-driven L2/3 S1 neurons exhibit high expression of the immediate early gene, Fos (27, 28). scRNA-seq analysis in naïve, untrained mice shows that whereas all cell types express some number of immediate early genes (IEGs), Baz1a neurons show consistent enrichment of Fos along with a subset of IEGs (Fig. 3A). Although Fos expression is dynamic and driven by experience-dependent plasticity (29), we speculated that Fos and other IEGs may be stably expressed in Baz1a neurons. To confirm this and address how it relates to neuronal function, we extended the CRACK platform to track Fos expression and stimulus-evoked activity during altered sensory experience using transgenic fosGFP mice (30) along with virally coexpressed Rcamp1.07 in S1 (n = 3) (Fig. 3, B and C). Ex vivo HCR-FISH confirmed that high fosGFP fluorescence corresponded with higher Fos mRNA and that green fluorescent protein (GFP) mRNA was also enriched in Baz1a neurons (Fig. 3D and fig. S18A). Although estimated event rates correlated with FosGFP expression levels in Adamts2 and Agmat neurons, they did not in Ba1za neurons, confirming that Fos does not necessarily reflect ongoing activity in this cell type (fig. S18B).
Fig. 3. Persistent IEG expression and homeostatic plasticity in Baz1a neurons.
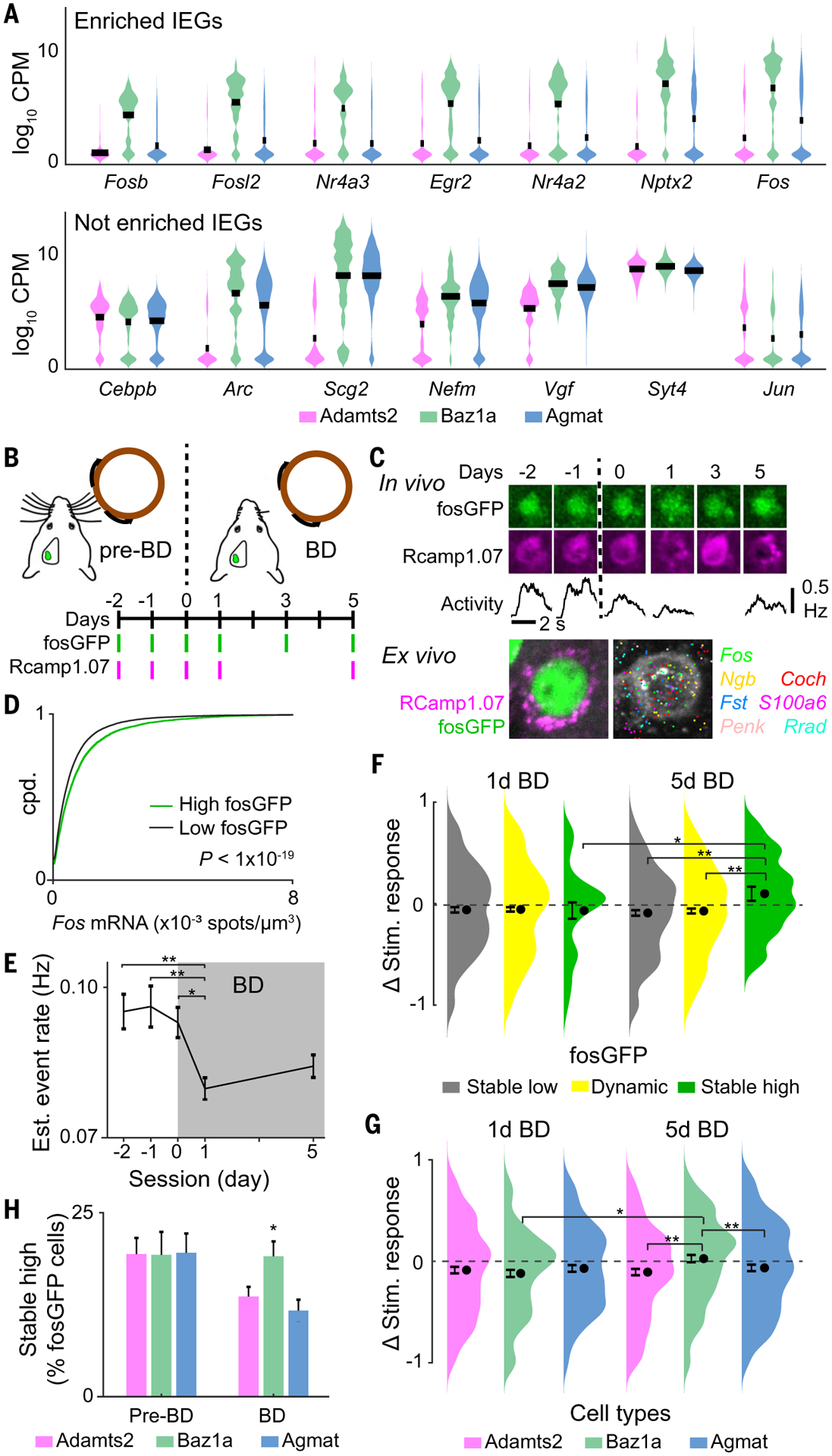
(A) Examples of selectively (top) enriched and (bottom) not enriched immediate early genes in Baz1a cells. (B) Time course of bilateral whisker deprivation (BD) experiment. (C) (Top) Example of Baz1a neuron with stable high fosGFP expression across in vivo imaging sessions. (Middle) Average stimulus responses during calcium imaging. (Bottom) Post hoc identification of neuron and HCR-FISH for select genes. (D) HCR-FISH Fos spot density in high (1.2-fold above background) and low fluorescent fosGFP cells (two-tailed Student’s t test). (E) Mean stimulus-evoked activity before and after BD across functionally imaged neurons (one-way ANOVA with post hoc multiple comparison test, n = 2569 cells from three animals). (F) Change in stimulus-evoked responses before BD versus at 1 day or 5 days BD across neurons with stable low, dynamic, and stable high fosGFP expression (two-tailed Student’s t test, n = 790 cells from three animals). (G) Change in stimulus-evoked responses before BD versus at 1 day or 5 days BD across excitatory cell types (χ2 test, n = 181 Adamts2, 136 Baz1a, and 153 Agmat cells from three animals). (H) Fraction of fosGFP neurons with stable high expression across all pre-BD sessions (days −2, −1, and 0) and across all BD sessions (days 1, 3, and 5) for excitatory cell types (two-tailed Student’s t test, n = 3753 cells from three animals). * P < 0.05, ** P < 0.005 in (E) to (G). Error bars = SEM; (H) SD from bootstrap analysis.
FosGFP and sensory-evoked calcium responses were tracked before and for 5 days after bilateral whisker deprivation (BD). During BD, the principal whisker corresponding to the imaged S1 barrel column was trimmed to a minimum length so that stimulus-evoked activity could still be tracked (fig. S17) (31). Overall, BD resulted in a decrease in stimulus-evoked activity after 1 day followed by a slow homeostatic compensation after 5 days [P < 0.0002, one-way analysis of variance (ANOVA), post hoc multiple comparison test] (Fig. 3E) (32). We first asked how fosGFP expression related to functional response changes during BD. Cells were divided into three groups according to fosGFP expression: (i) stable low fosGFP expression across all imaging sessions; (ii) stable high fosGFP expression across all imaging sessions, and (iii) dynamic fosGFP expression between at least two imaging sessions (fig. S19). All groups showed decreased stimulus-evoked activity after 1 day of BD. However, after 5 days, responses in stable low and dynamic fosGFP neurons remained depressed, whereas stable high fosGFP neurons exhibited an enhancement in sensory response magnitude and reliability compared with pre-BD conditions (P < 0.05, Student’s t test) (Fig. 3F and fig. S20).
Ex vivo cell type identification revealed that similar fractions of stable high fosGFP neurons were observed across all excitatory cell types before deprivation. However, during BD, there was increased fosGFP turnover in Adamts2 and Agmat neurons, whereas the fraction of stable high fosGFP cells remained unchanged in Baz1a neurons (P < 0.05, χ2 test) (Fig. 3H). Functionally, all three cell types showed reduced stimulus activity after 1 day BD, whereas only Baz1a neurons showed recovery after 5 days of BD (P < 0.005, Student’s t test) (Fig. 3G and fig. S20).
Task encoding in inhibitory subclasses and subdivisions
We next compared task encoding in three of the major subclasses of inhibitory neurons (Pvalb, Sst, and Vip). Lamp5 neurons were excluded from analysis because of their low numbers captured in the data set (table S2). Overall, Pvalb neurons exhibited the weakest coding of tactile-related features (Fig. 4, A to C, and fig. S15). However, the high firing rates of Pvalb neurons and associated difficulties in reliably inferring spiking-related calcium events in this subclass by calcium imaging may underestimate the strength of GLM-derived task responses (supplementary text S2) (11, 33). We therefore focused our analysis on Sst and Vip neurons.
Fig. 4. Task encoding across L2/3 inhibitory subclasses.
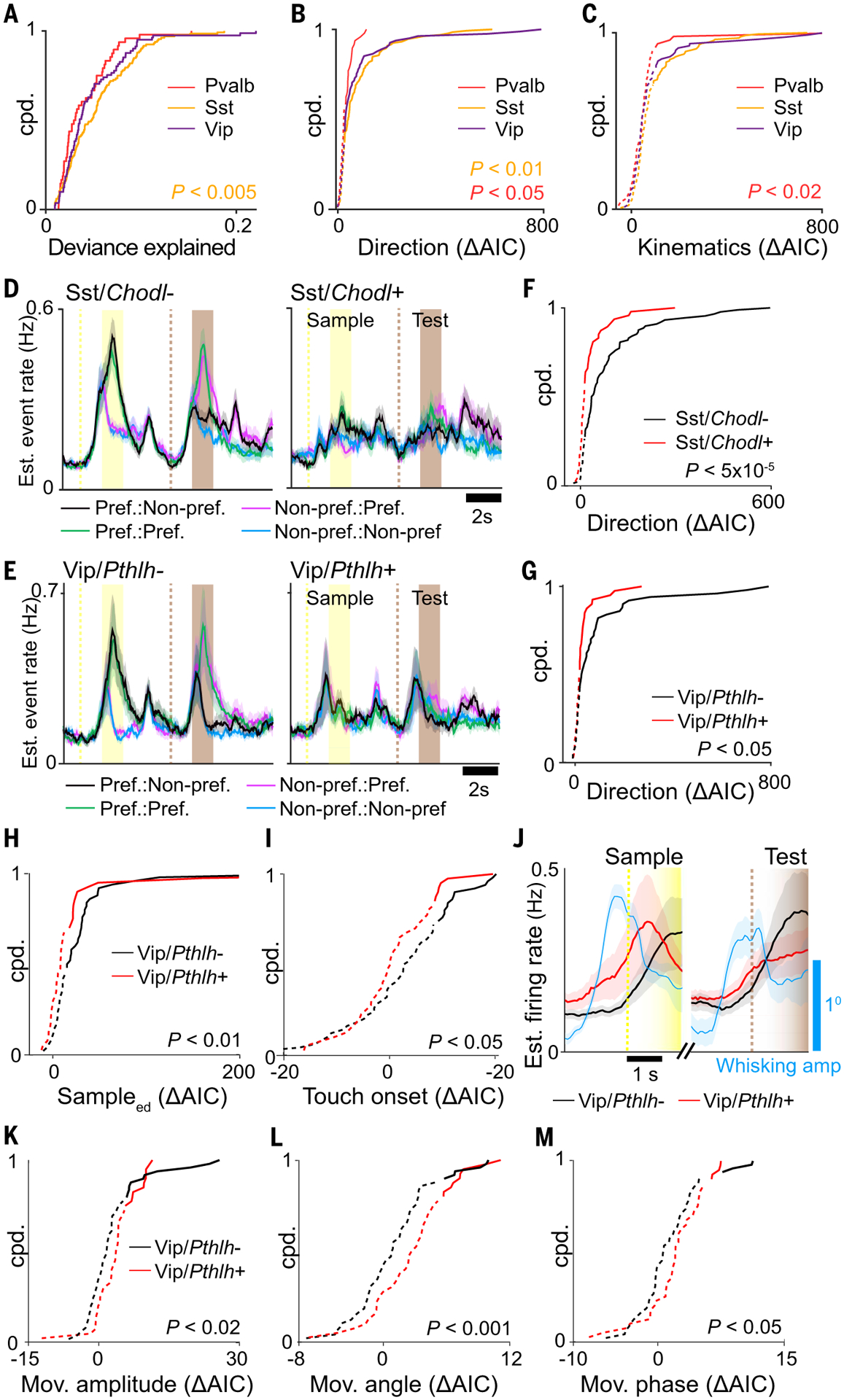
(A to C) Cumulative probability distributions for (A) full model deviance explained, (B) encoding strength of stimulus direction, and (C) encoding strength of whisker kinematics for three major inhibitory subclasses (Mann Whitney U Test). (D and E) Estimated event rate responses to preferred stimulus direction for (D) Sst subclasses and (E) Vip subclasses. (F and G) Cumulative probability distribution of ΔAIC for task factor encoding direction for (F) Sst subclasses and (G) Vip subclasses (Mann Whitney U Test). (H and I) Cumulative probability distribution of ΔAIC for (H) task factors encoding sampleEARLY DELAY and (I) touch onset for Vip subclasses (Mann Whitney U Test). (J) Estimated event rate for Vip subclasses along with mean whisking amplitude aligned to whisker-rotor touch onset preceding sample and test periods. (K to M) Cumulative probability distribution of ΔAIC for task factors encoding (K) free whisking amplitude, (L) angle, and (M) phase for Vip subclasses (Mann Whitney U Test). In (B), (C), (F) to (I), and (K) to (M), solid and dotted lines indicate significant (P < 0.01) and nonsignificant encoding strengths, respectively, by means of χ2 test. Shaded regions in (D) and (E) indicate SEM. n = 48 Pvalb cells, 47 Sst/Chodl+ cells, 88 Sst/Chodl− cells, 40 Vip/Pthlh+ cells, and 49 Vip/Pthlh− cells from seven animals.
We investigated whether more task-related differences emerge when inhibitory subclasses are further divided into finer transcriptomic subclasses or types. Among inhibitory subclasses, Sst showed the best overall GLM fit (P < 0.005, Mann-Whitney U test) (Fig. 4A) and strongly encoded stimulus direction (P < 0.05, Mann-Whitney U test) (Fig. 4B). We asked whether stimulus direction was encoded similarly in two subdivisions of Sst neurons. Sst/Chodl+ neurons express nitric oxide synthase (Nos1) (34), display long-range axonal projection patterns (35, 36), and are active during slow-wave sleep (37). During the DNMS task, Sst/Chodl+ encoded direction more weakly compared with Sst/Chodl− neurons (P < 5 × 10−5, Mann Whitney U test) (Fig. 4, D and F, and fig. S16A).
We next compared task differences between Vip/Pthlh+ and Vip/Pthlh− neurons (Fig. 4E). Vip neurons belonging to the Pthlh+ subdivision coexpress choline acetyltransferase (Chat) and calretinin, typically have bipolar morphologies, and preferentially target Sst neurons (36, 38–40). Vip neurons belonging to the Pthlh− subdivision coexpress synuclein gamma (Sncg) and cholecystokinin (Cck), have multipolar and basket cell morphologies, and preferentially target Pvalb neurons (36, 41). Vip/Pthlh− neurons more strongly encoded direction, sampleEARLY DELAY, and touch onset than did Vip/Pthlh+ neurons (direction, P < 0.05; sampleEARLY DELAY, P < 0.01; onset, P < 0.05, Mann-Whitney U test) (Fig. 4, G to I, and fig. S16B). Analysis of calcium events with respect to touch onset at the beginning of the sample and test period neurons showed elevated firing for Vip/Pthlh+ neurons preceding touch onset, which correlated with an anticipatory increase in whisking amplitude (Fig. 4J). This pretouch activity suggested that Vip/Pthlh+ neurons are driven by free whisking behavior. To disentangle movement-related from tactile-related whisker responses, we fit neuronal activity to a GLM with whisker kinematic variables using only time periods before touch onset during the prestimulus and delay period. Vip/Pthlh+ neurons more strongly encoded whisker amplitude, angle, and phase task factors during free whisking periods compared with those of Vip/Pthlh− neurons (amplitude, P < 0.02; angle, P < 0.001; phase, P < 0.05; Mann-Whitney U test) (Fig. 4, K to M).
Network interactions between major subclasses and types
The ability to simultaneously record across all identified cell subclasses and types enables a comprehensive characterization of cell type–specific network structures that underlie coding of task information. Non-negative matrix factorization across varying ranks captures population dynamics across distinct functional subpopulations (supplementary text S4) (26). Neurons that exhibit strong population coupling with increasing ranks suggest functional relationships with multiple subpopulations. Compared with other excitatory neurons, Baz1a neurons consistently showed higher coupling across ranks, indicating that they are highly integrated into the local L2/3 network (P < 0.02, F2,6, repeated measures ANOVA) (Fig. 5A). To investigate coupling between specific cell-type populations, we constructed a GLM that included all previously described task variables while subdividing the activity of other neurons into different “coupling factors” according to Adamts2, Baz1a, Agmat, Pvalb, Sst, and Vip transcriptomic populations (Fig. 5B). For a modeled neuron, the ΔAIC for each cell-type coupling factor constituted a measure of “functional connectivity” between that neuron and other simultaneously recorded cell types. Functional connections consist of positive and negative noise correlations that reflect either direct interactions or common input from nonrecorded neurons (fig. S21). From these measures, a directional weighted network graph can be constructed composed of the six subpopulations as nodes and functional connectivity as edges. To assess interactions between cell populations encoding different task factors, task-specific networks were generated by selecting for neurons with significant ΔAIC (P < 0.01, χ2 test) for a given task factor (Fig. 5C).
Fig. 5. Cell type functional connectivity across task networks.
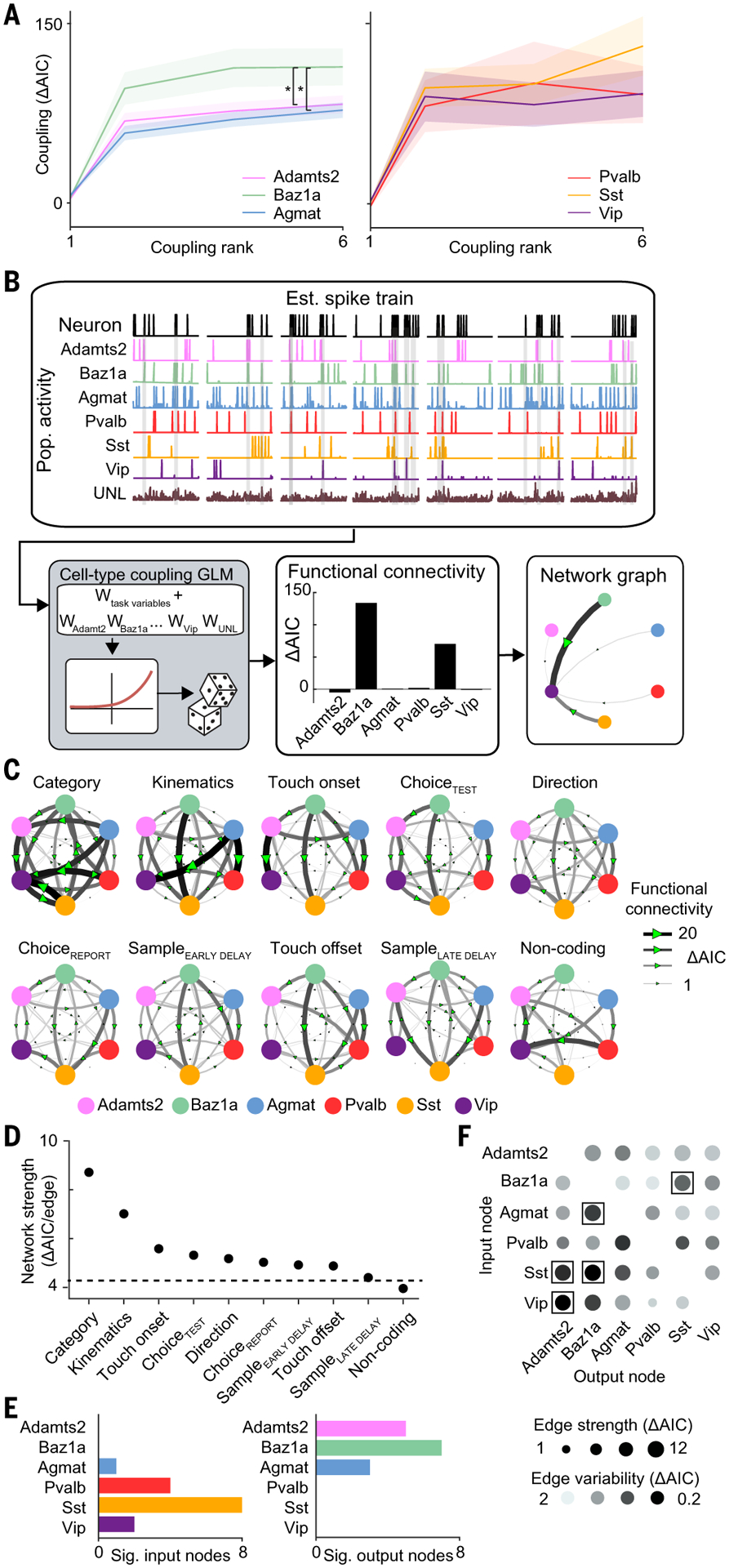
(A) Strength of coupling factor encoding across varying coupling ranks. *P < 0.02, repeated measures ANOVA test, F2,6. (B) Schematic of network analysis for example neuron. (C) Task-specific networks generated by selecting for neurons with significant encoding for a given task factor in the task GLM. Networks are sorted according to average edge strength. (D) Network strength across task networks. The dotted line indicates strength of shuffled network. (E) Cell type and subclass differences in number of significant (left) input and (right) output nodes across task networks. (F) Strength and variability of functional connectivity in network edges across task networks. Network edges with significantly high strength and low variability are indicated with a box. P < 0.05, permutation test. Shaded region in (A) indicates SEM. n = 1996 neurons, direction; 1374 neurons, sampleEARLY DELAY; 1076 neurons sampleLATE DELAY; 360 neurons, category; 623 neurons, choiceTEST; 830 neurons, choiceREPORT; 898 neurons, touch onset; 1033 neurons, touch offset; 864 neurons, kinematics; and 273 neurons, noncoding from seven animals.
We observed different network patterns across task factors. All task factor networks exhibited population-specific functional connection weights that were greater than chance (with the exception of the network that contained noncoding neurons, which exhibited random connection weights) (P < 0.05, bootstrap test) (Fig. 5D). Functional connectivity was strongest among neurons encoding category and whisker kinematics (fig. S22A). We further investigated the structure of these networks. For each cell-type node, we used the input edge strengths to determine how other cell populations influence the activity of the measured node and the output edge strengths to determine how the measured node influences the activity of other cell populations (Fig. 5E and fig. S22B). Inhibitory neurons were more likely than excitatory neurons to be influenced by network activity patterns. Sst neurons exhibited the highest input node strength across all task conditions (P < 0.05, bootstrap test). This is in line with evidence that suggests that Sst neurons follow local network activity (23). By contrast, excitatory neurons had a greater influence on other cell types, with Baz1a cells showing high output node strength in seven out of the nine task factor networks (P < 0.05, bootstrap test).
Given the differences in node strengths across task factor networks, we asked whether functional connectivity between any two subpopulations varied across task factor networks. High variability suggests that functional connections between cell types are dynamic and depend on the information being processed, whereas low variability suggests a stable motif that is intrinsic to the underlying circuitry. We measured the overall strength of each connection by calculating the mean edge weight across task factor networks. The stability of this connection was reported as the coefficient of variation of the edge weight across task factor networks (Fig. 5F). The majority of connections exhibited variability between task factor networks that were equivalent to chance levels, suggesting that connection strengths were dynamic and depend on the encoded task factor. However, a subset of connections [Adamts2→Vip, Adamts2→Sst, Agmat→Baz1a, and Baz1a↔Sst (output node→input node)] were consistently strong and stable across task factor networks, suggesting that they represent intrinsic functional motifs between specific cell populations (P < 0.05, permutation test).
Cell type–specific tracing confirms intrinsic functional connectivity
The observed functional connections that persisted across task networks could be explained by cell type–specific synaptic connections. Trans-monosynaptic rabies tracing enables input mapping to specific cell types but requires genetic access for conditional infection. Because transgenic lines for the three excitatory cell types are not available, we focused on input patterns to Sst and Vip inhibitory classes. Because Baz1a neurons showed stable functional connectivity with Sst neurons but not Vip neurons, we compared Baz1a synaptic connectivity between these two inhibitory classes. Using Sst-IRES-Cre (n = 4) and Vip-IRES-Cre (n = 4) mice (42), L2/3 Sst and Vip starter cells were first labeled by using a Cre-dependent adeno-associated virus (AAV) expressing TVA, CVS-N2cG, and dTomato, followed by delivery of the EnvA-pseudotyped CVS-N2c(ΔG) rabies virus expressing histone-GFP (Fig. 6, A and B) (43). We examined the sublaminar distribution of histone-GFP–positive inputs (nGFP+) to Sst and Vip cells across L2/3. Overall, Sst and Vip neurons received a greater number of inputs from cells located in deeper L2/3 (>200 μm below the pia mater) as compared with superficial L2/3. However, Sst neurons received more of their inputs from superficial L2/3 neurons compared with Vip neurons (Sst, 29.1 ± 2.7%; Vip, 21.2 ± 2.6%; mean ± SEM; P < 0.05, one-tailed Student’s t test) (Fig. 6C). We performed multiplexed HCR-FISH to identify cell type of nGFP+ input neurons. The overall density of Baz1a inputs was greater for Sst neurons as compared with Vip neurons (Sst, 12.8 ± 1.8%; Vip, 8.4 ± 1.2%; P < 0.05; one-tailed Student’s t test) (Fig. 6D). This difference was greatest among cells in superficial L2/3 (Sst, 22.1 ± 0.9%; Vip, 17.3 ± 2.0%; P < 0.05, one-tailed Student’s t test) (Fig. 6E).
Fig. 6. Upper layer Ba1za neurons target Sst neurons.
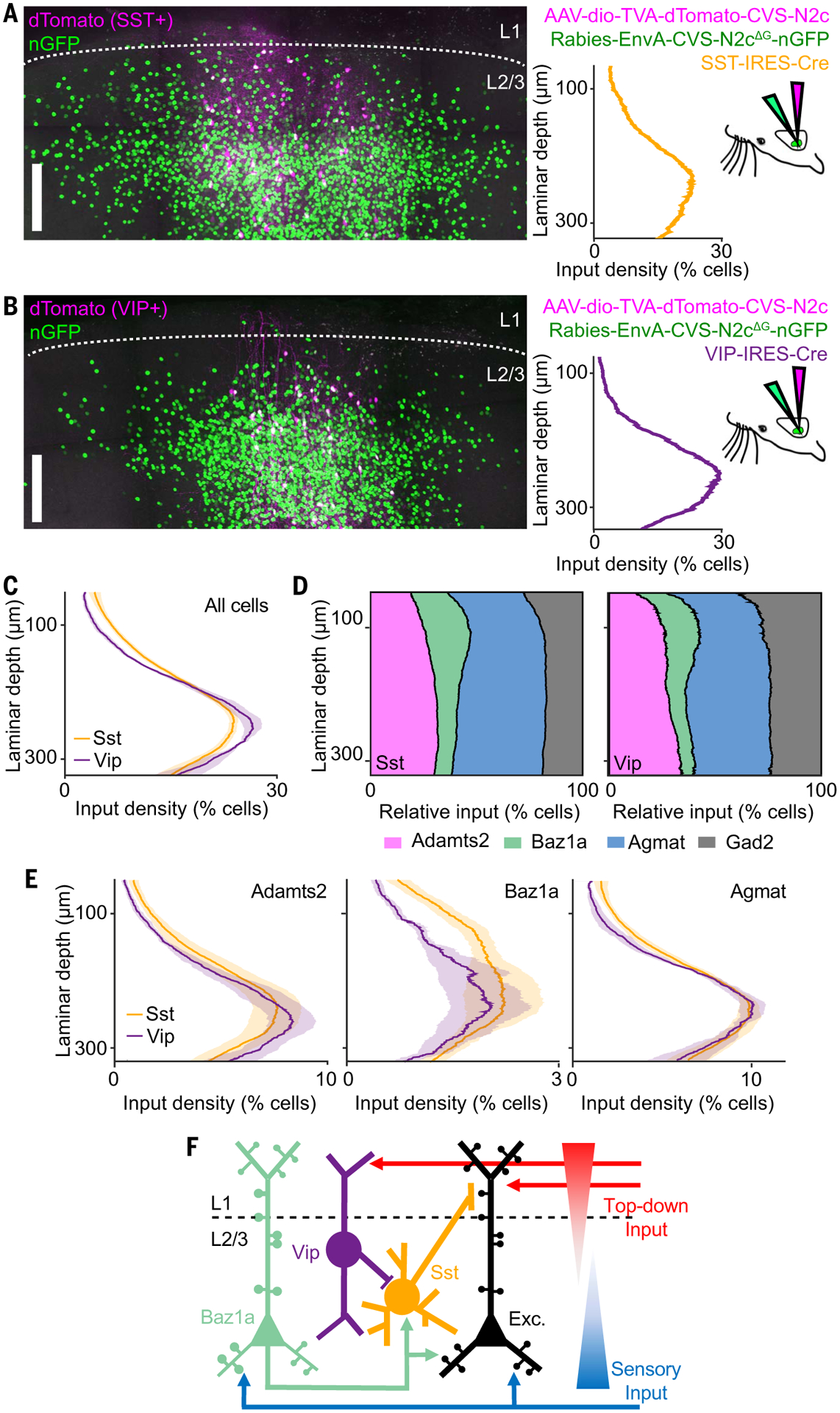
(A and B) Example of cell type–specific transmonosynaptic tracing in (A) Sst-IRES-Cre and (B) Vip-IRES-Cre mice. (Left) Confocal images of starter cells (magenta) and nGFP+ input neurons (green). (Right) Sublaminar distribution of input density from left images, along with injection scheme. (C) Average sublaminar somatic density distribution of inputs across L2/3 for Sst and VIP neurons. (D) Relative proportion of excitatory cell types and Gad2+ inhibitory neurons as a function of laminar depth for Sst and Vip input neurons. (E) Density of excitatory cell types as a function of laminar depth for Sst and Vip input neurons. (F) Circuit model of L2/3 illustrating cell type–specific connectivity between Vip, Sst, Baz1a, and other local excitatory neurons. Shaded regions in (C) and (E) indicate SEM. n = 4 Sst-IRES-Cre animals, 16 slices, 33,957 neurons; and 4 Vip-IRES-Cre animals, 14 slices, 35,926 neurons. Scale bars, 100 μm.
Discussion
We developed a platform to densely survey the functional and molecular properties of neuronal populations in vivo and applied it to study cell types in L2/3 of S1. We found evidence for increasing functional specialization; excitatory and inhibitory neurons are divided into more discrete subclasses and types. We focused on the role of Baz1a neurons in neocortical function. Enriched Fos expression suggests that Baz1a neurons are members of a previously described, highly interconnected FosGFP population that operates as a network hub in S1 (27). S1 is important for both tactile feature discrimination as well as sensorimotor integration for object localization (44). Given their highly selective stimulus responsiveness, Baz1a neurons are well poised to act as feature detectors and recruit local circuits for sensory processing. Superficial L2/3 pyramidal neurons are laminarly situated to integrate both top-down motor and associative signals arriving in L1 onto apical dendrites with bottom-up sensory signal arriving from L4 onto basal dendrites (Fig. 6F) (45, 46). Basal dendrites also contain highly recurrent, lateral connections between neighboring excitatory neurons (47). Integration of top-down signals and associative memory formation in L2/3 pyramidal neurons is mediated by Vip disinhibition (40, 48) through apical dendrite-targeting Sst neurons (49). We propose that excitatory connections from superficial Baz1a neurons onto Sst neurons can counteract Vip-mediated disinhibition so as to inhibit top-down inputs and bias synaptic integration in local L2/3 pyramidal neurons toward bottom-up and recurrent inputs on basal dendrites. Therefore, these circuit motifs operate complementary to one another, allowing S1 to shift between gating long-range feedback inputs and engaging feed-forward computations.
Baz1a neurons are also functionally distinct in their ability to adapt to altered sensory experience by homeostatically maintaining their response to tactile stimuli. Sensory deprivation transiently induces changes in IEG expression, resulting in experience-dependent plasticity (50). We speculate that stable expression of Fos and other select IEGs in Baz1a cells primes this cell type to adapt to changes in experience through molecular mechanisms that could modulate excitatory-inhibitory balance, synaptic scaling, or intrinsic excitability. This plasticity suggests that Baz1a neurons serve additional roles in preserving existing sensory representations in the face of novel experiences. The presence of cell types in V1 and ALM with similar expression profiles as that of Baz1a neurons suggests that homologous circuits with common functions may exist across neocortical areas.
Supplementary Material
ACKNOWLEDGMENTS
We thank O. Gonen, S. Kenyon, G. Shechter, N. Weston, and C. Xin for software development; A. Ahrens, G. House, K. Marmon, N. Josephs, D. Lee, and S. Wang for assistance in analysis; and M. Economo, D. Lee,B. Scott, and C. Habjan for comments on the manuscript.
Funding:
This work was supported by a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation, the Richard and Susan Smith Family Foundation, an Elizabeth and Stuart Pratt Career Development Award, the Whitehall Foundation, Harvard NeuroDiscovery Center, National Institutes of Health BRAIN Initiative Award (R01NS109965 to J.L.C. and U19MH114830 to H.Z.), National Institutes of Health New Innovator Award (DP2NS111134), and National Institutes of Health Ruth L. Kirschstein Predoctoral Individual National Research Service Award (F31NS111896) to C.C.
Footnotes
Competing interests: T.N.N. is currently employed at Cajal Neuroscience.
Data and materials availability:
scRNA-seq data are available to the public in the following repositories: https://assets.nemoarchive.org/dat-jb2f34y and https://assets.nemoarchive.org/dat-v39m5v1. Data and code related to the CRACK platform are available to the public at (51).
REFERENCES AND NOTES
- 1.Tasic B et al. , Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018).doi: 10.1038/s41586-018-0654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeisel A et al. , Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018). doi: 10.1016/j.cell.2018.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klingler E et al. , A translaminar genetic logic for the circuit identity of intracortically projecting neurons. Curr. Biol 29, 332–339.e5 (2019). doi: 10.1016/j.cub.2018.11.071 [DOI] [PubMed] [Google Scholar]
- 4.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H,Macklis JD, Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci 14, 755–769 (2013). doi: 10.1038/nrn3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim L, Mi D, Llorca A, Marín O, Development and functional diversification of cortical interneurons. Neuron 100, 294–313 (2018). doi: 10.1016/j.neuron.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorensen SA et al. , Correlated gene expression and target specificity demonstrate excitatory projection neuron diversity. Cereb. Cortex 25, 433–449 (2015). doi: 10.1093/cercor/bht243 [DOI] [PubMed] [Google Scholar]
- 7.Paul A et al. , Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell 171, 522–539.e20 (2017). doi: 10.1016/j.cell.2017.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kepecs A, Fishell G, Interneuron cell types are fit to function. Nature 505, 318–326 (2014). doi: 10.1038/nature12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris KD, Shepherd GM, The neocortical circuit: Themes and variations. Nat. Neurosci 18, 170–181 (2015). doi: 10.1038/nn.3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daigle TL et al. , A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality. Cell 174, 465–480.e22 (2018). doi: 10.1016/j.cell.2018.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan AG et al. , Distinct learning-induced changes in stimulus selectivity and interactions of GABAergic interneuron classes in visual cortex. Nat. Neurosci 21, 851–859 (2018).doi: 10.1038/s41593-018-0143-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X, RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).doi: 10.1126/science.aaa6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi HMT et al. , Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018). doi: 10.1242/dev.165753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah S et al. , Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development 143, 2862–2867 (2016).doi: 10.1242/dev.138560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L, Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361 (2014). doi: 10.1038/nmeth.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X et al. , Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018). doi: 10.1126/science.aat5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicovich PR et al. , Multimodal cell type correspondence by intersectional mFISH in intact tissues. bioRxiv [Preprint] 525451 (2019). doi: 10.1101/525451 [DOI] [Google Scholar]
- 18.Chen JL, Voigt FF, Javadzadeh M, Krueppel R,Helmchen F, Long-range population dynamics of anatomically defined neocortical networks. eLife 5, e14679 (2016).doi: 10.7554/eLife.14679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Treweek JB et al. , Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat. Protoc 10, 1860–1896 (2015). doi: 10.1038/nprot.2015.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohkura M, Sasaki T, Kobayashi C, Ikegaya Y, Nakai J, An improved genetically encoded red fluorescent Ca2+ indicator for detecting optically evoked action potentials. PLOS ONE 7, e39933 (2012). doi: 10.1371/journal.pone.0039933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao Z et al. , A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241.e26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abs E et al. , Learning-related plasticity in dendrite-targeting layer 1 interneurons. Neuron 100, 684–699.e6 (2018).doi: 10.1016/j.neuron.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J, Hu H, Agmon A, Svoboda K, Recruitment of GABAergic interneurons in the barrel cortex during active tactile behavior. Neuron 104, 412–427.e4 (2019). doi: 10.1016/j.neuron.2019.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Condylis C et al. , Context-dependent sensory processing across primary and secondary somatosensory cortex. Neuron 106, 515–525.e5 (2020). doi: 10.1016/j.neuron.2020.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pillow JW et al. , Spatio-temporal correlations and visual signalling in a complete neuronal population. Nature 454, 995–999 (2008). doi: 10.1038/nature07140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Runyan CA, Piasini E, Panzeri S, Harvey CD, Distinct timescales of population coding across cortex. Nature 548, 92–96 (2017). doi: 10.1038/nature23020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yassin L et al. , An embedded subnetwork of highly active neurons in the neocortex. Neuron 68, 1043–1050 (2010). doi: 10.1016/j.neuron.2010.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jouhanneau J-S et al. , Cortical fosGFP expression reveals broad receptive field excitatory neurons targeted by POm. Neuron 84, 1065–1078 (2014). doi: 10.1016/j.neuron.2014.10.014 [DOI] [PubMed] [Google Scholar]
- 29.Yap EL, Greenberg ME, Activity-regulated transcription: Bridging the gap between neural activity and behavior. Neuron 100, 330–348 (2018). doi: 10.1016/j.neuron.2018.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barth AL, Gerkin RC, Dean KL, Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. J. Neurosci 24, 6466–6475 (2004). doi: 10.1523/JNEUROSCI.4737-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margolis DJ et al. , Reorganization of cortical population activity imaged throughout long-term sensory deprivation. Nat. Neurosci 15, 1539–1546 (2012). doi: 10.1038/nn.3240 [DOI] [PubMed] [Google Scholar]
- 32.Gainey MA, Feldman DE, Multiple shared mechanisms for homeostatic plasticity in rodent somatosensory and visual cortex. Philos. Trans. R. Soc. London B Biol. Sci 372, 20160157 (2017). doi: 10.1098/rstb.2016.0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwan AC, Dan Y, Dissection of cortical microcircuits by single-neuron stimulation in vivo. Curr. Biol 22, 1459–1467 (2012). doi: 10.1016/j.cub.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tasic B et al. , Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci 19, 335–346 (2016). doi: 10.1038/nn.4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomioka R et al. , Demonstration of long-range GABAergic connections distributed throughout the mouse neocortex. Eur. J. Neurosci 21, 1587–1600 (2005). doi: 10.1111/j.1460-9568.2005.03989.x [DOI] [PubMed] [Google Scholar]
- 36.He M et al. , Strategies and Tools for Combinatorial Targeting of GABAergic Neurons in Mouse Cerebral Cortex. Neuron 91, 1228–1243 (2016). doi: 10.1016/j.neuron.2016.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerashchenko D et al. , Identification of a population of sleep-active cerebral cortex neurons. Proc. Natl. Acad. Sci. U.S.A 105, 10227–10232 (2008). doi: 10.1073/pnas.0803125105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dudai A et al. , Barrel cortex VIP/ChAT interneurons suppress sensory responses in vivo. PLOS Biol 18, e3000613 (2020). doi: 10.1371/journal.pbio.3000613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prönneke A et al. , Characterizing VIP neurons in the barrel cortex of VIPcre/tdTomato mice reveals layer-specific differences. Cereb. Cortex 25, 4854–4868 (2015).doi: 10.1093/cercor/bhv202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S, Kruglikov I, Huang ZJ, Fishell G, Rudy B, A disinhibitory circuit mediates motor integration in the somatosensory cortex. Nat. Neurosci 16, 1662–1670 (2013). doi: 10.1038/nn.3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hioki H et al. , Preferential inputs from cholecystokinin-positive neurons to the somatic compartment of parvalbumin-expressing neurons in the mouse primary somatosensory cortex. Brain Res 1695, 18–30 (2018). doi: 10.1016/j.brainres.2018.05.029 [DOI] [PubMed] [Google Scholar]
- 42.Taniguchi H et al. , A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013 (2011). doi: 10.1016/j.neuron.2011.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reardon TR et al. , Rabies virus CVS-N2c(DG) strain enhances retrograde synaptic transfer and neuronal viability. Neuron 89, 711–724 (2016). doi: 10.1016/j.neuron.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diamond ME, von Heimendahl M, Knutsen PM, Kleinfeld D,Ahissar E, ‘Where’ and ‘what’ in the whisker sensorimotor system. Nat. Rev. Neurosci 9, 601–612 (2008). doi: 10.1038/nrn2411 [DOI] [PubMed] [Google Scholar]
- 45.Xu NL et al. , Nonlinear dendritic integration of sensory and motor input during an active sensing task. Nature 492, 247–251 (2012). doi: 10.1038/nature11601 [DOI] [PubMed] [Google Scholar]
- 46.Doron G et al. , Perirhinal input to neocortical layer 1 controls learning. Science 370, eaaz3136 (2020). doi: 10.1126/science.aaz3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spruston N, Pyramidal neurons: Dendritic structure and synaptic integration. Nat. Rev. Neurosci 9, 206–221 (2008). doi: 10.1038/nrn2286 [DOI] [PubMed] [Google Scholar]
- 48.Williams LE, Holtmaat A, Higher-order thalamocortical inputs gate synaptic long-term potentiation via disinhibition. Neuron 101, 91–102.e4 (2019). doi: 10.1016/j.neuron.2018.10.049 [DOI] [PubMed] [Google Scholar]
- 49.Wang Y et al. , Anatomical, physiological and molecular properties of Martinotti cells in the somatosensory cortex of the juvenile rat. J. Physiol 561, 65–90 (2004). doi: 10.1113/jphysiol.2004.073353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loebrich S, Nedivi E, The function of activity-regulated genes in the nervous system. Physiol. Rev 89, 1079–1103 (2009). doi: 10.1152/physrev.00013.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Condylis C et al. , Dense functional and molecular readout of a circuit hub in sensory cortex. G-Node (2021); doi: 10.12751/g-node.7q0lz0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq data are available to the public in the following repositories: https://assets.nemoarchive.org/dat-jb2f34y and https://assets.nemoarchive.org/dat-v39m5v1. Data and code related to the CRACK platform are available to the public at (51).


