Abstract
Objectives
Although myocardial fibrosis is a common pathophysiological process associated with many heart diseases, the molecular mechanisms regulating the development of fibrosis have not been fully determined. Recently, single cell RNA sequencing (scRNA-seq) analysis has been used to examine cellular fate and function during cellular differentiation and has contributed to elucidating the mechanisms of various diseases. The main purpose of this study was to characterize the fate of cardiac fibroblasts (CFs) and the dynamic gene expression patterns in a model of cardiac pressure overload using scRNA-seq analysis.
Methods
The public scRNA-seq dataset of the transverse aortic coarctation (TAC) model in mice was downloaded from the GEO database, GSE155882. First, we performed quality control, dimensionality reduction, clustering, and annotation of the data through the Seurat R package (v4.0.5). Then, we constructed the pseudotime trajectory of cell development and identified key regulatory genes using the Monocle R package (v2.22.0). Different cell fates and groups were fully characterized by Gene Set Enrichment Analysis (GSEA) analysis and Transcription factor (TF) activity analysis. Finally, we used Cytoscape (3.9.1) to extensively examine the gene regulatory network related to cell fate.
Results
Pseudotime analysis showed that CFs differentiated into two distinct cell fates, one of which produced activated myofibroblasts, and the other which produced protective cells that were associated with reduced fibrosis levels, increased antioxidative stress responses, and the ability to promote angiogenesis. In the TAC model, activated CFs were significantly upregulated, while protective cells were downregulated. Treatment with the bromodomain inhibitor JQ1 reversed this change and improved fibrosis. Analysis of dynamic gene expression revealed that Gpx3 was significantly upregulated during cell differentiation into protective cells. Gpx3 expression was affected by JQ1 treatment. Furthermore, Gpx3 expression levels were negatively correlated with the different levels of fibrosis observed in the various treatment groups. Finally, we found that transcription factors Jun, Fos, Atf3, and Egr1 were upregulated in protective cells, especially Egr1 was predicted to be involved in the regulation of genes related to antioxidant stress and angiogenesis, suggesting a role in promoting differentiation into this cell phenotype.
Conclusions
The scRNA-seq analysis was used to characterize the dynamic changes associated with fibroblast differentiation and identified Gpx3 as a factor that might be involved in the regulation of myocardial fibrosis under cardiac pressure overload. These findings will help to further understanding of the mechanism of fibrosis and provide potential intervention targets.
1. Introduction
The heart compensates for increased workload through structural remodeling, which can ultimately lead to heart failure [1]. Myocardial fibrosis, as a form of cardiac remodeling, is a common process in many pathophysiologies, such as hypertension [2], myocardial infarction [3], and valvular disease [4]. In response to injury, myocardial fibroblasts show abnormal activation and secrete excessive extracellular matrix, resulting in chamber wall stiffness and diastolic dysfunction [5]. Many studies have revealed potential mechanisms associated with myocardial fibrosis. Myocardial fibrosis is the result of crosstalk between various types of cells, such as macrophages, which can secret pro- and antifibrotic factors [6], regulatory T cells, which can participate in regulating fibrosis by inhibiting excessive inflammation [7], and endothelial cells, which can secrete factors that interfere with the process of fibrosis [8]. Moreover, inflammatory reactions [9], oxidative stress [10], energy metabolism [11], apoptosis [12], and other mechanisms all play a role in the regulation of fibrosis. However, despite recent scientific advances, the current antifibrotic treatment options are still limited [13]. Therefore, exploring the mechanism of myocardial fibrosis and developing new therapeutic interventions are critical.
In recent years, single cell RNA sequencing (scRNA-seq) has become a standard tool for medical research that has been used in many fields [14]. Single cell sequencing allows for analysis of cell heterogeneity, intercellular communication, and cell developmental trajectories, which are important in the study of tissue development and pathology [15]. Several therapeutic targets for myocardial fibrosis, such as Ddah1, Meox1, and Ctrhc1, have been identified by single cell sequencing analysis and have provided novel theoretical targets for the treatment of myocardial fibrosis [16–18].
The purpose of this study was to explore the heterogeneity and activation of myocardial fibroblasts in a stress overload mouse model using scRNA-seq analysis. Our aim was to clarify some of the mechanisms of fibrosis and explore potential targets for treating myocardial fibrosis.
2. Materials and Methods
2.1. Analysis and Integration of scRNA-seq Data
The datasets analyzed in this study were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). First, we filtered the presumed doublet cells through DoubletFinder (v2.0.3) [19]. R package Seurat (v4.0.5) [20] was used for data integration, cell filtration, normalization, clustering, and UMAP or tSNE dimensional reduction. Harmony (v0.1.0) [21] was used to remove the batch effect between groups. Overall, we excluded cells with nFeatures < 200, nFeatures > 3,000, and percent.mt > 10% for initial quality control. After clustering, the cell clusters with the most significant differences in nFeatures and percent.mt were further excluded.
2.2. Pseudotime Trajectory Analysis
Monocle2 was used for pseudotime analysis [22]. Cell progression genes were defined based on differential gene expression between Seurat clusters. “BEAM_res” function was used for analysis of gene dynamics during cell differentiation.
2.3. Gene Ontology (GO) Enrichment Analysis
Enriched GO terms in differentially expressed genes were identified using the clusterprofileR package (v0.5.0) [23] with the default parameters.
2.4. Hub Gene Analysis
First, differential genes were introduced into the STRING v11 database (https://string-db.org/) to obtain the PPI network. Then, hub genes were analyzed via Cytoscape v3.9.1 [24]. Notably, the hub genes were ranked by “degree.”
2.5. Gene Set Enrichment Analysis (GSEA)
The GSEABase (v1.56.0) package (http://www.bioconductor.org/packages/release/bioc/html/GSEABase.html) was used to complete GO and KEGG enrichment analysis of marker genes in all groups.
2.6. Transcription Factor Activity Evaluation and Transcription Factor Target Overrepresentation Analysis
The transcription factor activity of different cell states was predicted using the DoRothEA (v1.6.0) package [25] with default parameters. The transcription factors of gene sets related to angiogenesis and antioxidant stress were predicted by ChEA3 [26], and the gene sets were submitted online (https://maayanlab.cloud/chea3/). By comparing multiple libraries and scoring, the top 15 transcription factors were finally obtained by sorting with “mean rank.”
2.7. Receptor Ligand Interaction Analysis
We evaluated intercellular receptor ligand signals by NicheNet [27], fibroblasts were used as sender and endothelial cells as receiver, and the other parameters were default.
3. Results and Discussion
3.1. The scRNA-seq Analysis of Cardiac Fibroblasts (CFs) in a Transverse Aortic Coarctation (TAC) Model
Alexanian et al. [17] established TAC and Sham models in mice and administered JQ1 [28], a small molecule BET bromodomain inhibitor, 18 days later. Mice were treated with JQ1 for one month, then JQ1 treatment was stopped in one cohort of mice for 14 days. Finally, the heart samples (n = 2) of four groups were harvested for single cell sequencing (Figure 1(a)). First, we carried out quality control on the data and identified a total of 58,175 cells; after quality control, the data had no significant batch effect, and the nFeatures and percen.mt of the data were within a reasonable range (Supplementary Figure 1(a, b)). After dimensionality reduction and clustering, these cells were annotated according to marker genes (Supplementary Table 1). A total of 14 cell subtypes were obtained (Figure 1(b)), in which activated CFs expressed high levels of profibrotic factors such as Postn, Sparc, and Cilp [29–31]. Ly6a+ CFs expressed high levels of the stem cell marker Ly6a [32] and fibrotic inhibitor Pi16 [33]. Another group of CFs (G0s2+ CFs) showed high expression of G0s2, which blocks lipolysis and inhibits the cell cycle [34], as well as expressing the Antioxidant stress-related gene Gpx3 [35] and cardioprotective gene Adm [36] (Supplementary Figure 1(c)).
Figure 1.
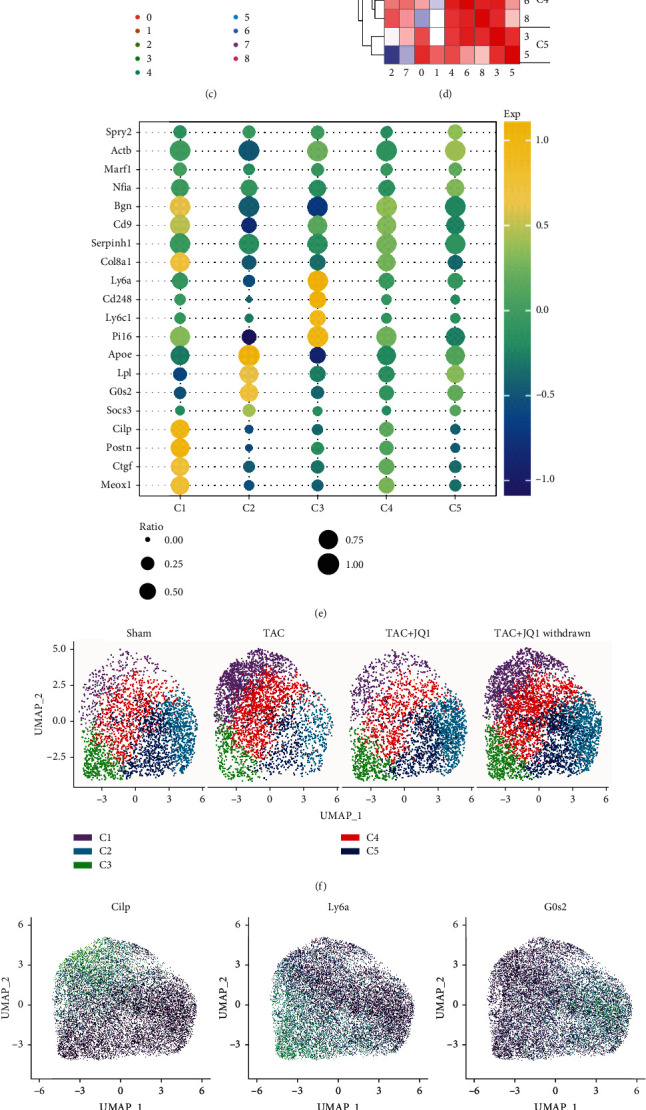
Overview of the fibroblast populations profiled in healthy and nonhealthy hearts. (a) Grouping and interventions in the single cell datasets. (b) The tSNE plot of nonmyocardial cells in 4 groups. (c) UMAP plot of CFs in four groups. A total of nine cell clusters were obtained. (d) Correlation heat map between clusters. The cell clusters were renamed C1 − C5 based on their correlation. (e) Relative expression levels of the marker genes for C1 − C5 shown by a bubble plot. (f) Unsupervised clustering of CFs visualized by groups. (g) FeaturePlot of some representative marker genes. (h) The proportion of C1 − C5 in each group.
To further explore CF heterogeneity in cardiac fibrosis, we defined additional CF populations using unsupervised graph-based clustering and UMAP. In total, nine cell clusters were identified in the whole dataset (Figure 1(c)). We extracted the top marker genes with the largest standard deviation of each cell cluster to calculate the correlation. Based on this, we reclassified these nine cell clusters into five CF subgroups, C1-C5 (Figure 1(d)). C1 and C4 displayed high expression levels of profibrotic genes such as Postn, Ctgf, and Meox1, indicating that these cell groups had a stronger fibrotic phenotype. C2 overexpressed Apoe, G0s2, and Lpl, which are thought to be related to fat metabolism, while C3 overexpressed Ly6a, Pi16, and Cd248. Gene expression levels in the C4 and C5 subgroups were not as high as those observed in the C1-C3 subgroups and were therefore considered to be cell subsets in a transition state (Figures 1(e) and 1(g)). Although we did not observe significant batch effects, there were significant differences in cell distribution among the different groups (Figure 1(f)). As shown in Figure 1(h), the number of cells in C1 and C4 was significantly increased in the TAC group, decreased after treatment with JQ1, and partially attenuated by JQ1 withdrawal. Since changes in the number of cells in the C2, C3, and C5 subgroups were the opposite of those found in C1 and C4, we concluded that these two types of cells played different roles in CFs under pressure overload.
3.2. JQ1 Treatment Attenuates Oxidative Stress and Fibrosis in CFs
Since JQ1 can improve fibrosis and enhance cardiac function, examining the characteristics of the four treatment groups may further our understanding of the mechanisms of fibrosis. We analyzed the marker genes of each group and displayed the data by volcano plot. The TAC group showed high expression of profibrotic-related genes such as Ctgf, Cilp, and Thbs1, as well as high expression of Actn1 and Csrp2, which are associated with actin contraction (Figure 2(b)). After JQ1 treatment, the fibrotic-related genes were significantly downregulated, while upregulation of the heart protective genes Eef2 [37] and Dusp1 [38] was observed (Figure 2(c)). To some extent, JQ1 treatment restored the gene expression pattern in the TAC group to that observed in the Sham group (Figure 2(a)). Withdrawal of JQ1 resulted in downregulation of the protective and fibrogenic genes, which might explain lower levels of heart protection in the JQ1 withdrawal group compared to the JQ1 treatment group (Figure 2(d)). Next, we obtained the marker genes of each group with the parameters of “min.pct =0.01, logfc.threshold = 0” and sorted them by “log2FC.” Then, we performed GO and KEGG enrichment analysis of the marker genes for each group and displayed them with radar plot (Supplementary Table 2). GO terms related to fibrosis, such as “Supramolecular Fiber Organization” and “Cell Cell Adhesion,” were significantly upregulated in the TAC group, but improved after JQ1 treatment. In addition, we found that JQ1 significantly improved oxidative stress, and no significant differences in the enrichment level of the GO term “Negative Regulation Of Response To Oxidative Stress” were observed between the TAC + JQ1 and Sham groups (Figure 2(e)). Similar results were found by KEGG enrichment analysis. For example, the gene set “Drug Metabolism Cytochrome P450” related to antioxidative stress and including genes such as Gstm1, Gsta3, and Gpx3 was found to be upregulated in the JQ1 treatment group compared to the TAC group.
Figure 2.
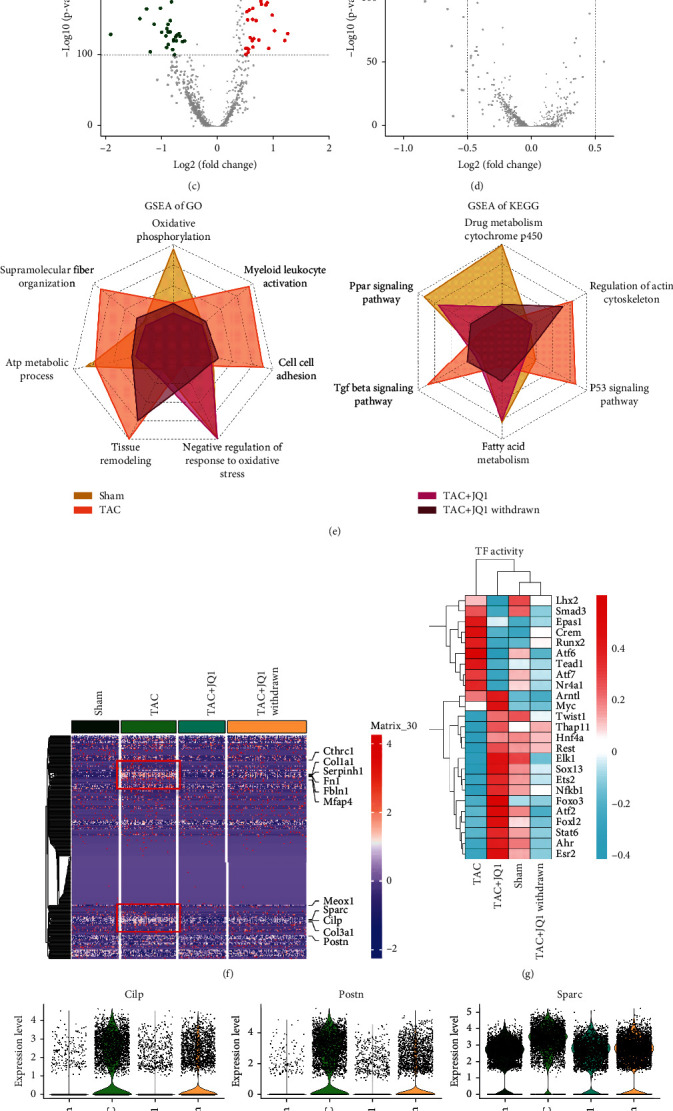
Characteristics of CFs in each treatment group. (a)–(d) Volcano plot showing the marker genes of each group. Marker genes with p value < 1e100 and |log2fc| > 0.5 are labelled. (e) GSEA enrichment analysis of GO and KEGG terms in each group is displayed by radar plot. (f) Heat map of genes related to fibrosis. (g) Heat map showing the relative levels of transcription factor activity in each group. (h) Volcano plot showing the expression of representative profibrotic genes in each experimental group.
Next, we studied the effect of JQ1 therapy on fibrosis. We obtained a gene list of fibrosis-related GO terms (Supplementary Table 3) and used heat map to compare their expression in the four groups. We found that fibrosis-related genes in the TAC group were significantly upregulated compared to those in the Sham group, and that JQ1 treatment reversed this outcome, while withdrawal of JQ1 weakened the JQ1-induced improvement in fibrosis (Figure 2(f)). The expression of representative fibrogenic genes is shown in Figure 2(h) and is consistent with the heat map data. Because transcription factors regulate gene expression, they play an important role in the development of diseases. Thus, we analyzed the activity of transcription factors in each group. Our data revealed significant upregulation of Atf6, Runx2, Crem, and Tead1 in the TAC group, with Atf6 and Runx2 associated with oxidative stress [39, 40], while Crem and Tead1 were involved in promoting fibrosis [41, 42]. Transcription factor activity levels in the TAC + JQ1 and Sham groups were very similar, with Atf2, Elk1, Ets1, and Esr1 associated with inhibition of oxidative stress [43–45]. In the TAC + JQ1 withdrawal group, both protective and harmful transcription factor activities were impaired (Figure 2(g)).
3.3. Differential Fates of CF Differentiation
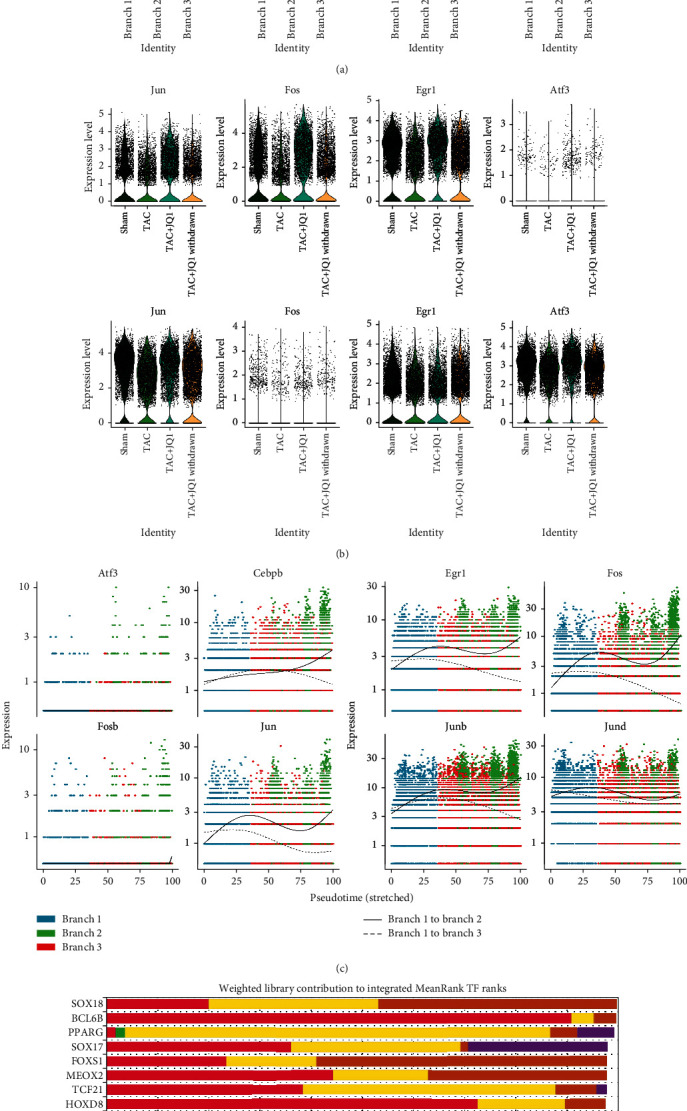
Our data indicated that the four CF treatment groups had different fibrotic phenotypes and biological functions. Next, we examined the differentiation trajectories of CFs and compared the differences between the groups through pseudotime analysis. In the cell trajectory, we identified three main branches, which we named branches 1 − 3 (Figure 3(c)). As a putative starting point of development, branch 1 was mainly composed of C4, which highly expressed the stem cell marker, Ly6a. Two distinct differentiation fates were derived from branch 2. Branch 2 was mainly composed of C2 and C5, while branch 3 was mainly composed of C1 and C4, which exhibited high expression levels of fibrogenic genes (Figures 3(a) and 3(b)). We evaluated the proportion of branches in each group and found that the TAC group was dominated by branch 3, suggesting that branch 3 was closely related to the fibrotic phenotype. In contrast, the JQ1 treatment group showed a significant reduction in the proportion of branch 3, even larger than that observed in the Sham group (Figure 3(c)). A higher proportion of branch 2 was observed in the JQ1 treatment group relative to the Sham group, suggesting that JQ1 treatment altered the fate of CFs.
Figure 3.
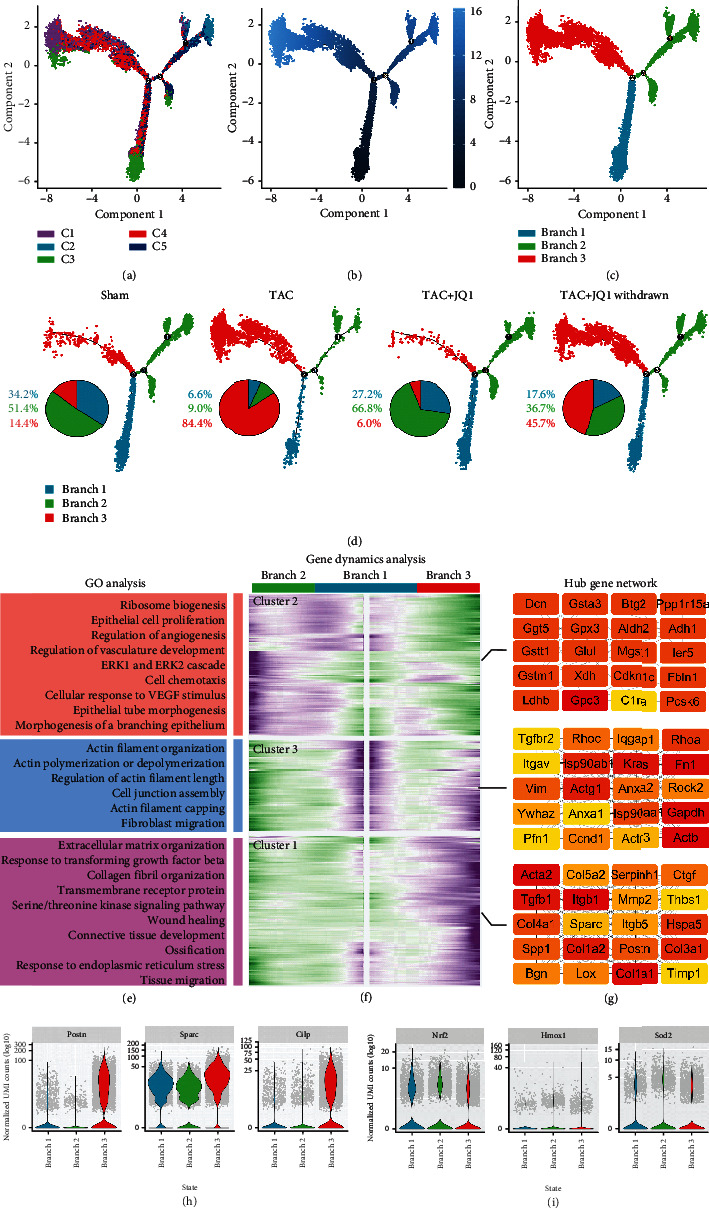
Simulation of the developmental trajectory of CFs and analysis of gene expression patterns inferred by Monocle2. (a) Pseudotime trajectory analysis of fibroblasts, using significantly regulated genes between C1 and C5. (b) Time series inference of cell trajectories. The color gradient indicates pseudotime progression. (c) Cell trajectories were renamed according to three main branches. (d) Cell trajectories of each group. Pie chart showing the proportion of cells in each branch of each group. (e) GO biological process terms enriched in each gene cluster. (f) Heat map hierarchical clustering showing differentially expressed genes together with the pseudotime curve. Color key from green to purple indicates relative expression levels from low to high. (g) Gene network displaying the top 20 hub genes from each cluster. (h, i) Expression of representative profibrotic genes and antioxidative stress genes in each branch.
To explore the mechanism of CF differentiation, we evaluated the gene dynamics of two different cell fates. Genes were divided into three clusters according to their expression patterns: genes of cluster 2 were gradually upregulated in branch 2, while genes of cluster 1 and cluster 3 were gradually upregulated in branch 3 (Figure 3(e)). Genes of cluster 2 were found to be mainly enriched in angiogenesis and ribosome composition (Supplementary Table 4). We further analyzed its PPI network using Cytoscape and sorting genes by “Eccentricity.” Of the selected top 20 genes, Gstt1, Gpx3, Gstm1, Gpc3, and Ggt5 were widely considered to be related to antioxidative stress. The gene function of cluster 3 was mainly enriched in actin filament organization, and its top 20 genes (sorted by degree) included Actb, Actg1, and Actr3. The top 20 genes (sorted by degree) of cluster 1 included Acta2, Tgfb1, Postn, Ctgf, and Col1a1, which are well-established fibrogenic genes. The GO enrichment analysis data were consistent with these findings. The expression of genes related to fibrosis and oxidative stress in each branch was evaluated by vlnplot. We found that branch 2 had reduced expression of fibrosis-related genes and increased expression of genes associated with antioxidative stress (Figures 3(f) and 3(g)).
3.4. Gpx3 May Be Involved in Regulating the Differentiation of CFs
According to the results described above, genes of cluster2 that were upregulated in branch 2 were associated with angiogenesis and antioxidative stress. In order to further explore its specific mechanism, we selected the top 100 genes in the PPI network of cluster 2 and classified each gene according to reports in the literature (Figure 4(a)). Genes labelled blue are related to ribosomal synthesis, while yellow-labelled genes may be involved in the negative regulation of fibrosis. The dark-green-labelled genes encode transcription factors that play a protective role in a variety of diseases. In addition, gene sets labelled red such as Gpx3, Ggt5, and Gstm1 were considered to be associated with antioxidative stress, and the differential expression of these genes in the cell trajectories was evaluated further. The expression of Gpx3, Gstm1, Ggt5, and Gsta3 showed different trends in the two cell fates, in contrast to Postn, Sparc, and Ctgf expressions (Figure 4(b)). Compared with other red-labelled genes, the expression of Gpx3 in the cell track changed more significantly (Supplementary Figure 2(a)) and compared its expression pattern with Postn and Col1a1. Gpx3 was mainly expressed in branch 2, while Postn and Col1a1 were predominantly expressed in branch 3. Furthermore, Gpx3 was downregulated in the TAC group and upregulated after treatment with JQ1, whereas withdrawal of JQ1 weakened the JQ1-induced upregulation of Gpx3. Meanwhile, the expression trend of the profibrotic gene Postn and Col1a1 was the opposite of Gpx3 (Supplementary Figure 2(b)). These data suggested that Gpx3 may inhibit CFs from differentiating into branch 3 and may promote differentiation into branch 2. In addition, we found that Apoe, Lpl, and Socs3 were also highly expressed in branch 2, suggesting that there might be metabolic differences between the branches. Thus, we evaluated the expression of metabolism-related genes in each group and found that the fatty acid metabolism levels of branch 2 were significantly increased, while the glycolysis and mitochondrial respiration levels were decreased, especially mitochondrial respiration. Of note, branch 3 had the highest glycolysis level among all branches (Figure 4(c)). We also evaluated the levels of pentose phosphate pathway, glycogen synthesis, glycogen decomposition, and gluconeogenesis, and the results showed no significant difference (Supplementary Figure 3(a)).
Figure 4.
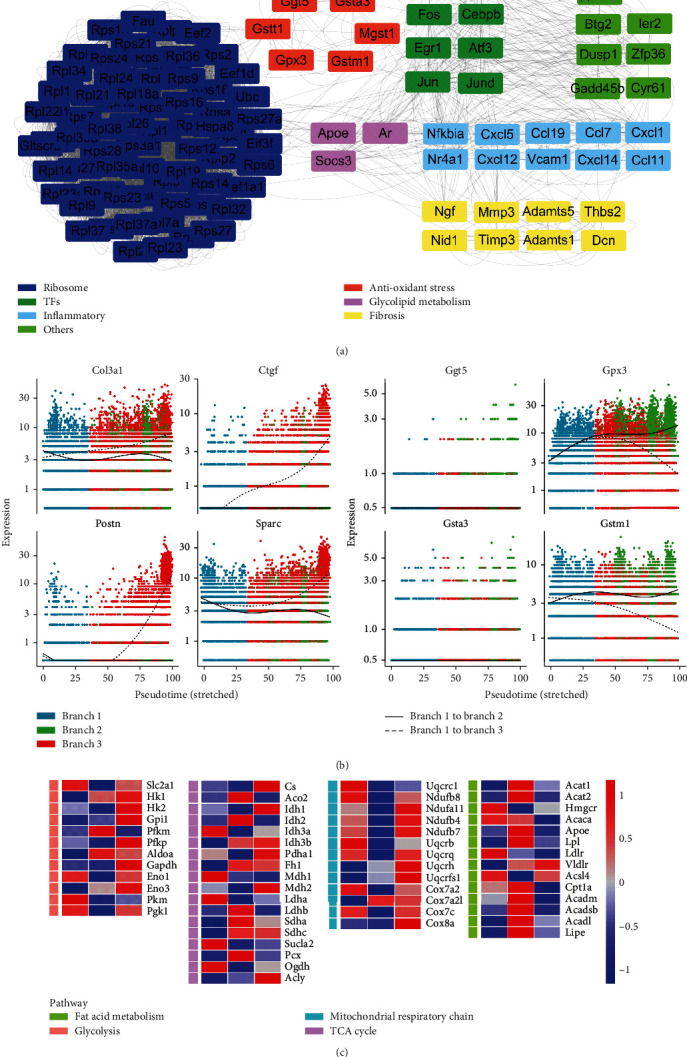
Examination of the function of upregulated genes in branch 2. (a) The top 100 hub gene network in gene cluster1 (described in Figure 2(f)), showing genes that were upregulated in branch 2. The function of these genes was classified and color-coded based on reports in the literature. (b) Differences in expression of Gpx3, Gstm1, Gsta3, Ggt5, and fibroblast-promoting genes in the different cell trajectories. (c) Heat map showing the relative expression levels of key genes related to fatty acid (FA) signals, tricarboxylic acid (TCA) cycle, glycolytic pathway, and mitochondrial respiration in each branch.
3.5. CFs of Branch 2 Are Involved in Promoting Angiogenesis
It is interesting to note that, as shown in Figure 3(e), genes that were gradually upregulated in branch 2 were mainly associated with angiogenesis. Because CFs are unable to promote the sprouting of endothelial cells directly, we assumed that CFs promote angiogenesis by secreting proteins. Thus, we downloaded 3970 genes encoding secretory proteins (Supplementary Table 5), as well as angiogenesis-promoting genes (Supplementary Table 6), and intersected them with the marker genes of each branch. We found that four marker genes in branch 1 encoded angiogenic secretory proteins, compared to five in branch 3, and 12 in branch 2 (Figures 5(a)–5(c)). Figure 5(d) shows the expression of these 12 genes through vlnplot. Our data indicated that branch 2 had the largest number of genes encoding proteins that promote angiogenesis compared with the other two branches, which was consistent with the results described above. Next, we selected CFs and endothelial cell subsets and mapped the branch labels of the trajectory of CFs into these cells (Supplementary Figure 3(b)). Then, the NicheNet package was used to explore the ligand-receptor (L-R) signal communication between the three branches of CFs and endothelial cells. Trf and Lamb1 were found to be highly expressed in branch 2, while Manf and Bdnf were highly expressed in branch 3. Based on the L-R pairs shown in Figure 5(g), we identified the corresponding receptors. Itga6 was identified as the receptor of Lamb1 and was found to be highly expressed in capillary endothelial cells, while Fzd4 was the receptor of Trf, which was shown to be enriched in arterial endothelial cells. Both receptors have been shown to be associated with angiogenesis. F11r, the receptor of Bdnf, was expressed in arterial endothelial cells and has been reported to be associated with atherosclerosis. Sec63 was found to be the receptor of Manf and was widely expressed in CFs of branch 3.
Figure 5.
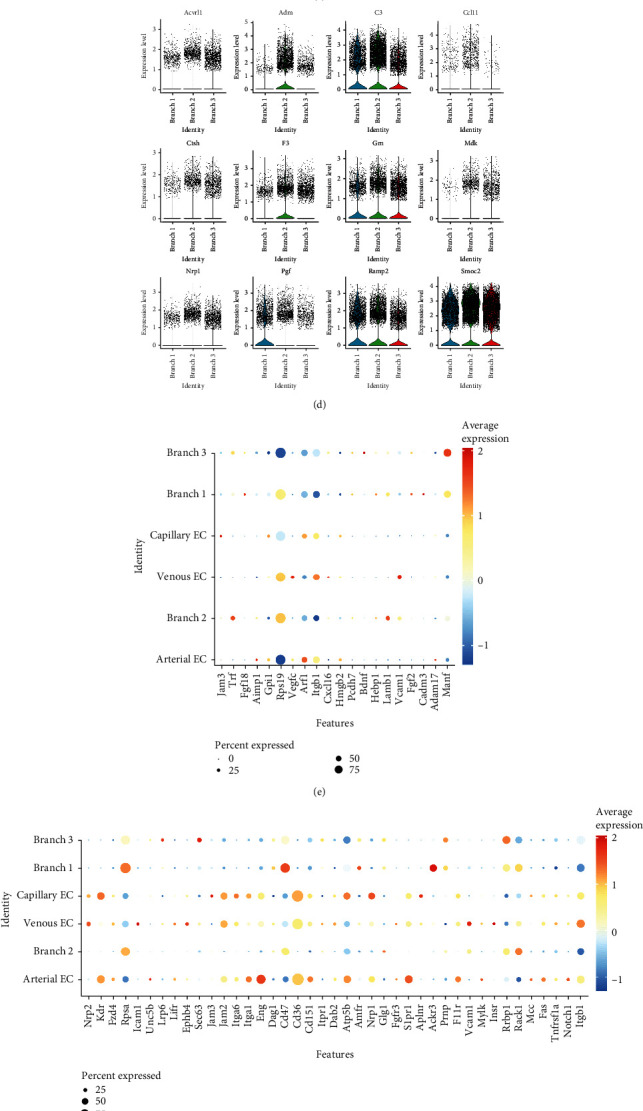
CFs participate in angiogenesis. (a)–(c) The secretory protein gene set and the angiogenic gene set were, respectively, intersected with the marker genes of each branch. Four overlapping genes were found in branch 1, 12 in branch 2, and five in branch 3. (d) Vlnplot showing the expression of 12 genes encoding angiogenesis-promoting secretory proteins in branch 2. (e) Top ligands in each cell type obtained by NicheNet analysis. (f) Top receptors in each cell type. (g) L-R pairs between cell types.
3.6. Egr1 May Be Involved in Reprogramming the Fate of CFs
In the gene network of branch 2, multiple genes encoding transcription factors were upregulated and served as the core of the network (Figure 4(a)). Transcription factors play an important role in the pathophysiological process of disease, and thus, in-depth examination of these transcription factors will improve our understanding of fibroblast differentiation. With the exception of Atf3 and Fosb, these transcription factors were widely expressed in CFs and were significantly upregulated in branch 2 (Figure 6(a)). The expression levels of these transcription factors were found to be decreased in the TAC group, but increased after JQ1 treatment, and decreased after JQ1 withdrawal. These findings were consistent with the Gpx3 data, but in contrast to the Postn data (Figure 6(b)). Similarly, the changing trends of these transcription factors on cell trajectories were consistent with Gpx3 but opposite to Postn (Figure 6(c)). Since the expression levels of these transcription factors were consistent with those of Gpx3, Gstm1, and genes encoding angiogenesis-promoting secretory proteins, we further predicted the transcription factors of these genes by ChEA3. Egr1 was identified from the top 15 predicted transcription factors and was also found to be highly expressed in branch 2. Based on the above results, we speculated that Egr1 may be an upstream transcription factor that has a role in improving oxidative stress and promoting angiogenesis.
Figure 6.
Comprehensive characterization of Clec3b expression in fibroblasts. (a) Expression of the transcription factors Jun, Fos, Fosb, Junb, Jund, Cebpd, Atf3, and Egr1 in each branch are displayed by vlnplot. (b) Expression of these eight transcription factors in each treatment group. (c) Expression of these 8 transcription factors in each cell trajectory. (d) Top 15 transcription factors predicted by ChEA3, sorted by average integrated ranks across the libraries.
4. Discussion
This study fully characterized the heterogeneity of CFs in a mouse TAC model, as well as the cell development trajectory through scRNA-seq analysis. Based on scRNA-seq analysis, we propose for the first time that there are two distinct cell differentiation fates for CFs under pressure overload. Fibroblasts expressing the stem cell marker Ly6a served as the starting point of differentiation (branch 1), from which two types of cell differentiation trajectories were derived, one of which was involved in the promotion of fibrosis (branch 3) and the other, which was involved in antioxidative stress and angiogenesis (branch 2). In the nonstress state, CFs are mainly differentiated into branch 2. However, under pressure overload conditions, CFs increased their differentiation into branch 3, which promoted fibrosis. Treatment with JQ1 significantly restored the fate of branch 2 and significantly alleviated fibrosis levels, which proved to be the powerful therapeutic effect of JQ1.
GSEA analysis revealed that the PPAR signaling pathway in the TAC heart was significantly downregulated compared with the Sham group. PPARs belong to the nuclear hormone receptor superfamily [46, 47]. PPARγ expression is closely associated with myocardial fibrosis [48, 49], due to its involvement in regulating inflammation, energy metabolism, and oxidative stress [50–53]. PPAR agonists are expected to be beneficial in antimyocardial fibrosis therapy [54]. Furthermore, CFs in branch 2 were found to express high levels of Apoe, Lpl, and Socs3, which are related to lipid metabolism. To evaluate metabolic differences in fibroblast differentiation, we evaluated the expression of key genes of fatty acid metabolism, glycolysis, the TCA cycle, and mitochondrial respiratory chain in cell development trajectories. We found that during the differentiation of fibroblasts into nonprotective branch 3, glycolysis levels gradually increased and were accompanied by a reduction in fatty acid metabolism. Thus, the balance of normal cardiac energy substrates was dysregulated, and so, under pressure overload, fibroblasts tend to be fueled by the glycolytic pathway. Many studies have reported that excessive glycolysis is involved in the process of organ fibrosis. Thus, inhibition of glycolysis can inhibit the activation of fibroblasts and improve organ fibrosis [55–58]. This change of energy metabolism may be an adaptive measure under hypoxia, as cells tend to undergo glycolysis in the absence of oxygen [59, 60]. Thus, we speculate that restoring normal levels of fatty acid metabolism may be a potential treatment for fibrosis.
Next, we assessed the level of oxidative stress in fibroblasts in each group. We found that the ability to resist oxidative stress was significantly impaired in the TAC group. Oxidative stress also plays an important role in the differentiation of fibroblasts. CFs that differentiated into branch 2 exhibited higher antioxidative stress abilities. However, differentiation into branch 3 was accompanied by a reduction in antioxidative stress ability. JQ1 treatment promoted the differentiation of cells into branch 2 and significantly restored the expression levels of antioxidative stress-related genes in fibroblasts, especially Gpx3. However, the withdrawal of JQ1 partially weakened this protective effect. These results indicated that oxidative stress was involved in the regulation of fibrosis, while JQ1 was found to have an anti-fibrotic role, at least in part through the regulation of oxidative stress [61–64]. Notably, Sdha and Sdhc, key genes involved in mediating succinic acid metabolism, were significantly reduced in fibroblasts from the TAC heart. However, previous studies have reported that Sdh-mediated succinic acid oxidation drives mitochondria to produce ROS, which is involved in the promotion of oxidative stress injury [65, 66]. This may be the compensation mechanism of energy metabolism reprogramming to cope with oxidative stress [67–70].
In addition, we observed an interesting phenotype. We found that, compared with other branches, branch 2 had a higher ability to promote angiogenesis, due to increased production of secreted proteins that promote angiogenesis, such as C3, Mdk, Pgf, and Ramp2 [71–73]. We analyzed the L-R signals between CFs of the three branches and endothelial cells via NicheNet and obtained similar results. We identified Trf-Fzd4 and Lamb1-Itga6 as the L-R pairs between branch 2 and ECs. Studies have shown that Fzd4, as a receptor of Wnt, participates in promoting angiogenesis [73], while Itga6 promotes endothelial morphogenesis through regulation of Cxcr4 [74]. Bdnf-F11r was identified as a L-R pair between branch 3 and endothelial cells. A role for F11r in promoting atherosclerosis has previously been reported [75]. Our results indicated that branch 2, as one of the potential fates of fibroblast differentiation, has an increased ability to promote angiogenesis. The concept of fibroblasts having a role in the promotion of angiogenesis is not new. Fibroblasts reportedly exhibit characteristics associated with promoting angiogenesis on the third day after myocardial infarction but have an antiangiogenesis role on day 7 [76]. This mechanism also plays an important role in tumor angiogenesis [77]. Our results not only support this concept but also clearly show that CFs that promote angiogenesis constitute one of the branches of CF fate. At the same time, this branch has a strong antioxidative stress capability, as well as increased fatty acid metabolism.
Finally, we evaluated the expression of transcription factors such as Jund, Fos, Atf3, and Egr1 in branch 2. Expression of these transcription factors was reduced in the TAC group, but improved following JQ1 treatment. Studies have shown that these transcription factors play a beneficial role in many pathophysiological processes. Overexpression of Atf, for example, can alleviate ischemia-reperfusion injury in the heart by reducing oxidative stress [78]. Indeed, Atf3 may be a target of Nrf2 and may therefore be involved in the regulation of oxidative stress [78]. JunD, a member of the AP-1 transcription factor family, plays a protective role in diabetic cardiomyopathy through the regulation of antioxidative stress [79]. Similarly, Cebpd has been shown to be involved in inhibiting oxidative stress and maintaining mitochondrial function [80, 81]. Here, we predicted the transcription factors of angiogenesis-promoting genes including Gpx3, Gstm1, and Gstt1. Among the predicted transcription factors, Egr1, which was also enriched in branch 2, was of particular interest. Multiple studies have reported that Egr1 promotes collagen synthesis and fibrosis [82]. Egr1 has also been shown to lead to upregulation of oxidative stress levels and has been associated with a variety of adverse outcomes, such as myocardial fibrosis, renal fibrosis, pulmonary hypertension, and arrhythmia after myocardial infarction [83–86]. However, in a model of myocardial ischemia-reperfusion injury, remote ischemic preconditioning was found to improve myocardial ischemia-reperfusion injury, through the protective role of Egr1 against apoptosis via the Jak-Stat pathway [87]. In most studies, Egr1 has been examined in the context of an acute pathological model. However, in the cardiac stress overload model, Egr1 was been studied in the context of myocardial hypertrophy. To date, there are no reports describing the expression and function of Egr1 in CFs in a chronic TAC model. However, Cupesi et al. reported that in a TAC model, Egr1 was upregulated after week 1, but downregulated after week 3 [88]. Based on our scRNA-seq analysis of the TAC model, we found that Egr1 expression levels were downregulated in fibroblasts in the TAC model. However, JQ1 treatment led to upregulation of Egr1 expression levels, which was accompanied by improved fibrosis. Thus, we propose that Egr1 expressed by fibroblasts may play a protective role in the model of chronic cardiac pressure overload. Our results show that the function of Egr1 may be inconsistent with most literature reports, which is worthy of further exploration. Finally, this study was carried out based on the analysis of single-cell sequencing data and has not been verified by experiments, which is the deficiency of this study.
5. Conclusions
This study found that there were two distinct cell fates in CFs. One fate may be related to angiogenesis and antioxidative stress, accompanied by metabolic reprogramming. Furthermore, Gpx3 and Egr1 may be involved in promoting the differentiation of CFs into a protective state.
Acknowledgments
Heartfelt thanks are due to Michael Alexanian and his team for selflessly sharing their valuable data, which has furthered our understanding of myocardial fibrosis.
Contributor Information
Dongying Zhang, Email: zhangdongying@cqmu.edu.cn.
Gang Li, Email: ligang@cqmu.edu.cn.
Data Availability
The dataset of the transverse aortic coarctation (TAC) model in mice was downloaded from the GSE155882 dataset.
Additional Points
Code Availability. Codes for R analysis of data presented in this study are available on GitHub (https://github.com/Leeguoxi/scRNA_TAC).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Guoxing Li and Yuhong Qin contributed equally and co-first authors.
Supplementary Materials
Supplementary Figure 1: (a) the vlnplot shows the nFeatures and percent.mt of all cell subsets. (b) Unsupervised clustering of all cells visualized by groups, which showed that there was no obvious batch effect. (c) FeaturePlot showed the markers of fibroblast. Supplementary Figure 2: (a) the expression of Gpx3, Gstm1, Ggt5, Gsta3, Mgst1, and Gstt1 in three branches of the trajectory. (b) Comparative analysis of expression patterns of Gpx3, Col1a1, and Postn. Supplementary Figure 3: (a) heat map of genes related to glycogen synthesis, glycogen decomposition, and pentose phosphate pathway. (b) We renamed the fibroblasts with branch labels and then selected subgroups for reclustering before performing receptor ligand interaction analysis. Supplementary Table 1: top 20 markers of all subsets. Supplementary Table 2: GSEA results of KEGG and GO. Supplementary Table 3: list of fibrogenic genes. Supplementary Table 4: GO analysis of three branches. Supplementary Table 5: list of gene encoding secreted protein. Supplementary Table 6: list of angiogenesis genes.
References
- 1.Alam P., Maliken B. D., Jones S. M., et al. Cardiac remodeling and repair: recent approaches, advancements, and future perspective. International Journal of Molecular Sciences . 2021;22(23):p. 13104. doi: 10.3390/ijms222313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nwabuo C. C., Vasan R. S. Pathophysiology of hypertensive heart disease: beyond left ventricular hypertrophy. Current Hypertension Reports . 2020;22(2):p. 11. doi: 10.1007/s11906-020-1017-9. [DOI] [PubMed] [Google Scholar]
- 3.Varasteh Z., Weber W. A., Rischpler C. Nuclear molecular imaging of cardiac remodeling after myocardial infarction. Pharmaceuticals . 2022;15(2):p. 183. doi: 10.3390/ph15020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debonnaire P., Delgado V., Bax J. J. Potential role of fibrosis imaging in severe valvular heart disease. Heart . 2015;101(5):397–407. doi: 10.1136/heartjnl-2013-304679. [DOI] [PubMed] [Google Scholar]
- 5.Travers J. G., Kamal F. A., Robbins J., Yutzey K. E., Blaxall B. C. Cardiac fibrosis: the fibroblast awakens. Circulation Research . 2016;118(6):1021–1040. doi: 10.1161/CIRCRESAHA.115.306565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiskirchen R., Weiskirchen S., Tacke F. Organ and tissue fibrosis: molecular signals, cellular mechanisms and translational implications. Molecular Aspects of Medicine . 2019;65:2–15. doi: 10.1016/j.mam.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Sun K., Li Y. Y., Jin J. A double-edged sword of immuno-microenvironment in cardiac homeostasis and injury repair. Signal Transduction and Targeted Therapy . 2021;6(1):p. 79. doi: 10.1038/s41392-020-00455-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Widyantoro B., Emoto N., Nakayama K., et al. Endothelial cell-derived endothelin-1 promotes cardiac fibrosis in diabetic hearts through stimulation of endothelial-to-mesenchymal transition. Circulation . 2010;121(22):2407–2418. doi: 10.1161/CIRCULATIONAHA.110.938217. [DOI] [PubMed] [Google Scholar]
- 9.Prabhu S. D., Frangogiannis N. G. The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circulation Research . 2016;119(1):91–112. doi: 10.1161/CIRCRESAHA.116.303577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pagan L. U., Gomes M. J., Gatto M., Mota G. A. F., Okoshi K., Okoshi M. P. The role of oxidative stress in the aging heart. Antioxidants . 2022;11(2):p. 336. doi: 10.3390/antiox11020336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibb A. A., Lazaropoulos M. P., Elrod J. W. Myofibroblasts and fibrosis: mitochondrial and metabolic control of cellular differentiation. Circulation Research . 2020;127(3):427–447. doi: 10.1161/CIRCRESAHA.120.316958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin W., Cao L., Massey I. Y. Role of PI3K/Akt signaling pathway in cardiac fibrosis. Molecular and Cellular Biochemistry . 2021;476(11):4045–4059. doi: 10.1007/s11010-021-04219-w. [DOI] [PubMed] [Google Scholar]
- 13.López B., Ravassa S., Moreno M. U., et al. Diffuse myocardial fibrosis: mechanisms, diagnosis and therapeutic approaches. Nature Reviews Cardiology . 2021;18(7):479–498. doi: 10.1038/s41569-020-00504-1. [DOI] [PubMed] [Google Scholar]
- 14.Adey A. C. Tagmentation-based single-cell genomics. Genome Research . 2021;31(10):1693–1705. doi: 10.1101/gr.275223.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Litvinukova M., Talavera-López C., Maatz H., et al. Cells of the adult human heart. Nature . 2020;588(7838):466–472. doi: 10.1038/s41586-020-2797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz-Villalba A., Romero J. P., Hernández S. C., et al. Single-cell RNA sequencing analysis reveals a crucial role for CTHRC1 (collagen triple helix repeat containing 1) cardiac fibroblasts after myocardial infarction. Circulation . 2020;142(19):1831–1847. doi: 10.1161/CIRCULATIONAHA.119.044557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexanian M., Przytycki P. F., Micheletti R., et al. A transcriptional switch governs fibroblast activation in heart disease. Nature . 2021;595(7867):438–443. doi: 10.1038/s41586-021-03674-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhuang L., Lu L., Zhang R., Chen K., Yan X. Comprehensive integration of single-cell transcriptional profiling reveals the heterogeneities of non-cardiomyocytes in healthy and ischemic hearts. Frontiers in Cardiovascular Medicine . 2020;7:p. 615161. doi: 10.3389/fcvm.2020.615161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGinnis C. S., Murrow L. M., Gartner Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Systems . 2019;8(4):329–337.e4. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satija R., Farrell J. A., Gennert D., Schier A. F., Regev A. Spatial reconstruction of single-cell gene expression data. Nature Biotechnology . 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korsunsky I., Millard N., Fan J., et al. Fast, sensitive and accurate integration of single-cell data with harmony. Nature Methods . 2019;16(12):1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu X., Mao Q., Tang Y., et al. Reversed graph embedding resolves complex single-cell trajectories. Nature Methods . 2017;14(10):979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu G., Wang L. G., Han Y., He Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS . 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research . 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holland C. H., Tanevski J., Perales-Patón J., et al. Robustness and applicability of transcription factor and pathway analysis tools on single-cell RNA-seq data. Genome Biology . 2020;21(1):p. 36. doi: 10.1186/s13059-020-1949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keenan A. B., Torre D., Lachmann A., et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Research . 2019;47(W1):W212–W224. doi: 10.1093/nar/gkz446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Browaeys R., Saelens W., Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nature Methods . 2020;17(2):159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 28.Filippakopoulos P., Qi J., Picaud S., et al. Selective inhibition of BET bromodomains. Nature . 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oka T., Xu J., Kaiser R. A., et al. Genetic manipulation of periostin expression reveals a role in cardiac hypertrophy and ventricular remodeling. Circulation Research . 2007;101(3):313–321. doi: 10.1161/CIRCRESAHA.107.149047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding W., Pu W., Jiang S., et al. Evaluation of the antifibrotic potency by knocking down SPARC, CCR2 and SMAD3. eBioMedicine . 2018;38:238–247. doi: 10.1016/j.ebiom.2018.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q. J., He Y., Li Y., et al. Matricellular protein Cilp1 promotes myocardial fibrosis in response to myocardial infarction. Circulation Research . 2021;129(11):1021–1035. doi: 10.1161/CIRCRESAHA.121.319482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinto A. R., Ilinykh A., Ivey M. J., et al. Revisiting cardiac cellular composition. Circulation Research . 2016;118(3):400–409. doi: 10.1161/CIRCRESAHA.115.307778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng M., Yang S., Ji Y., et al. Overexpression of peptidase inhibitor 16 attenuates angiotensin II–induced cardiac fibrosis via regulating HDAC1 of cardiac fibroblasts. Journal of Cellular and Molecular Medicine . 2020;24(9):5249–5259. doi: 10.1111/jcmm.15178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Hou Y., Zhang W., et al. Lipolytic inhibitor G0S2 modulates glioma stem-like cell radiation response. Journal of Experimental & Clinical Cancer Research . 2019;38(1):p. 147. doi: 10.1186/s13046-019-1151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohamed M. M., Sabet S., Peng D. F., Nouh M. A., el-Shinawi M., el-Rifai W. Promoter hypermethylation and suppression of glutathione peroxidase 3 are associated with inflammatory breast carcinogenesis. Oxidative Medicine and Cellular Longevity . 2014;2014:9. doi: 10.1155/2014/787195.787195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimosawa T., Shibagaki Y., Ishibashi K., et al. Adrenomedullin, an endogenous peptide, counteracts cardiovascular damage. Circulation . 2002;105(1):106–111. doi: 10.1161/hc0102.101399. [DOI] [PubMed] [Google Scholar]
- 37.Pires Da Silva J., Monceaux K., Guilbert A., et al. SIRT1 protects the heart from ER stress-induced injury by promoting eEF2K/eEF2-dependent autophagy. Cell . 2020;9(2):p. 426. doi: 10.3390/cells9020426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin Q., Li R., Hu N., et al. DUSP1 alleviates cardiac ischemia/reperfusion injury by suppressing the Mff- required mitochondrial fission and Bnip3-related mitophagy via the JNK pathways. Redox Biology . 2018;14:576–587. doi: 10.1016/j.redox.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin J. K., Blackwood E. A., Azizi K., et al. ATF6 decreases myocardial ischemia/reperfusion damage and links ER stress and oxidative stress signaling pathways in the heart. Circulation Research . 2017;120(5):862–875. doi: 10.1161/CIRCRESAHA.116.310266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raaz U., Schellinger I. N., Chernogubova E., et al. Transcription factor Runx2 promotes aortic fibrosis and stiffness in type 2 diabetes mellitus. Circulation Research . 2015;117(6):513–524. doi: 10.1161/CIRCRESAHA.115.306341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewin G., Matus M., Basu A., et al. Critical role of transcription factor cyclic AMP response element modulator in beta1-adrenoceptor-mediated cardiac dysfunction. Circulation . 2009;119(1):79–88. doi: 10.1161/CIRCULATIONAHA.108.786533. [DOI] [PubMed] [Google Scholar]
- 42.Bugg D., Bretherton R., Kim P., et al. Infarct collagen topography regulates fibroblast fate via p38-yes-associated protein transcriptional enhanced associate domain signals. Circulation Research . 2020;127(10):1306–1322. doi: 10.1161/CIRCRESAHA.119.316162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan N. M., Ahmad I., Haqqi T. M. Nrf2/ARE pathway attenuates oxidative and apoptotic response in human osteoarthritis chondrocytes by activating ERK1/2/ELK1-P70S6K-P90RSK signaling axis. Free Radical Biology & Medicine . 2018;116:159–171. doi: 10.1016/j.freeradbiomed.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menazza S., Sun J., Appachi S., et al. Non-nuclear estrogen receptor alpha activation in endothelium reduces cardiac ischemia-reperfusion injury in mice. Journal of Molecular and Cellular Cardiology . 2017;107:41–51. doi: 10.1016/j.yjmcc.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H. Y., Yu L. F., Zhou T. G., et al. Lipopolysaccharide-stimulated bone marrow mesenchymal stem cells-derived exosomes inhibit H2O2-induced cardiomyocyte inflammation and oxidative stress via regulating miR-181a-5p/ATF2 axis. European Review for Medical and Pharmacological Sciences . 2020;24(19):10069–10077. doi: 10.26355/eurrev_202010_23224. [DOI] [PubMed] [Google Scholar]
- 46.Janani C., Ranjitha Kumari B. D. PPAR gamma gene--a review. Diabetes and Metabolic Syndrome: Clinical Research and Reviews . 2015;9(1):46–50. doi: 10.1016/j.dsx.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 47.Hassan F. U., Nadeem A., Li Z., et al. Role of peroxisome proliferator-activated receptors (PPARs) in energy homeostasis of dairy animals: exploiting their modulation through nutrigenomic interventions. International Journal of Molecular Sciences . 2021;22(22):p. 12463. doi: 10.3390/ijms222212463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azhar S. Peroxisome proliferator-activated receptors, metabolic syndrome and cardiovascular disease. Future Cardiology . 2010;6(5):657–691. doi: 10.2217/fca.10.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu Q., Hu S., Guo P., et al. PPAR-γ with its anti-fibrotic action could serve as an effective therapeutic target in T-2 toxin-induced cardiac fibrosis of rats. Food and Chemical Toxicology . 2021;152:p. 112183. doi: 10.1016/j.fct.2021.112183. [DOI] [PubMed] [Google Scholar]
- 50.Lei W., Li X., Li L., et al. Compound Danshen dripping pill ameliorates post ischemic myocardial inflammation through synergistically regulating MAPK, PI3K/AKT and PPAR signaling pathways. Journal of Ethnopharmacology . 2021;281:p. 114438. doi: 10.1016/j.jep.2021.114438. [DOI] [PubMed] [Google Scholar]
- 51.Wagner K. D., Wagner N. PPARs and myocardial infarction. International Journal of Molecular Sciences . 2020;21(24):p. 9436. doi: 10.3390/ijms21249436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen B., Huang S., Su Y., et al. Macrophage Smad3 protects the infarcted heart, stimulating phagocytosis and regulating inflammation. Circulation Research . 2019;125(1):55–70. doi: 10.1161/CIRCRESAHA.119.315069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garg S., Khan S. I., Malhotra R. K., et al. The molecular mechanism involved in cardioprotection by the dietary flavonoid fisetin as an agonist of PPAR-γ in a murine model of myocardial infarction. Archives of Biochemistry and Biophysics . 2020;694:p. 108572. doi: 10.1016/j.abb.2020.108572. [DOI] [PubMed] [Google Scholar]
- 54.Lu Q., Guo P., Guo J., et al. Targeting peroxisome proliferator-activated receptors: a new strategy for the treatment of cardiac fibrosis. Pharmacology & Therapeutics . 2021;219:p. 107702. doi: 10.1016/j.pharmthera.2020.107702. [DOI] [PubMed] [Google Scholar]
- 55.Xie N., Tan Z., Banerjee S., et al. Glycolytic reprogramming in myofibroblast differentiation and lung fibrosis. American Journal of Respiratory and Critical Care Medicine . 2015;192(12):1462–1474. doi: 10.1164/rccm.201504-0780OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barcena-Varela M., Paish H., Alvarez L., et al. Epigenetic mechanisms and metabolic reprogramming in fibrogenesis: dual targeting of G9a and DNMT1 for the inhibition of liver fibrosis. Gut . 2021;70(2):388–400. doi: 10.1136/gutjnl-2019-320205. [DOI] [PubMed] [Google Scholar]
- 57.Li Q., Li C., Elnwasany A., et al. PKM1 exerts critical roles in cardiac remodeling under pressure overload in the heart. Circulation . 2021;144(9):712–727. doi: 10.1161/CIRCULATIONAHA.121.054885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Z. T., Gao Q. Y., Wu M. X., et al. Glycolysis inhibition alleviates cardiac fibrosis after myocardial infarction by suppressing cardiac fibroblast activation. Frontiers in Cardiovascular Medicine . 2021;8:p. 701745. doi: 10.3389/fcvm.2021.701745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang M., Xie X., Cao F., Wang Y. Mitochondrial metabolism in myocardial remodeling and mechanical unloading: implications for ischemic heart disease. Frontiers in Cardiovascular Medicine . 2021;8:p. 789267. doi: 10.3389/fcvm.2021.789267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng T. C., Philip J. L., Tabima D. M., Hacker T. A., Chesler N. C. Multiscale structure-function relationships in right ventricular failure due to pressure overload. American Journal of Physiology. Heart and Circulatory Physiology . 2018;315(3):H699–H708. doi: 10.1152/ajpheart.00047.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han X., Ding C., Sang X. N., et al. Targeting Sirtuin1 to treat aging-related tissue fibrosis: from prevention to therapy. Pharmacology & Therapeutics . 2022;229:p. 107983. doi: 10.1016/j.pharmthera.2021.107983. [DOI] [PubMed] [Google Scholar]
- 62.Yao R., Cao Y., Wang C., et al. Taohuajing reduces oxidative stress and inflammation in diabetic cardiomyopathy through the sirtuin 1/nucleotide-binding oligomerization domain-like receptor protein 3 pathway. BMC Complementary Medicine and Therapies . 2021;21(1):p. 78. doi: 10.1186/s12906-021-03218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purnomo Y., Piccart Y., Coenen T., Prihadi J., Lijnen P. Oxidative stress and transforming growth factor-β1-induced cardiac fibrosis. Cardiovascular & Hematological Disorders Drug Targets . 2013;13(2):165–172. doi: 10.2174/1871529X11313020010. [DOI] [PubMed] [Google Scholar]
- 64.Liu H., Wang L., Weng X., et al. Inhibition of Brd4 alleviates renal ischemia/reperfusion injury-induced apoptosis and endoplasmic reticulum stress by blocking FoxO4-mediated oxidative stress. Redox Biology . 2019;24:p. 101195. doi: 10.1016/j.redox.2019.101195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mills E. L., Kelly B., Logan A., et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell . 2016;167(2):457–470.e13. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prag H. A., Pala L., Kula-Alwar D., et al. Ester prodrugs of malonate with enhanced intracellular delivery protect against cardiac ischemia-reperfusion injury in vivo. Cardiovascular Drugs and Therapy . 2022;36(1):1–13. doi: 10.1007/s10557-020-07033-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tin A., Scharpf R., Estrella M. M., et al. The loss ofGSTM1 associates with kidney failure and heart failure. Journal of the American Society of Nephrology . 2017;28(11):3345–3352. doi: 10.1681/ASN.2017030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song Y., Shan Z., Liu X., et al. An updated meta-analysis showed smoking modify the association of GSTM1null genotype on the risk of coronary heart disease. Bioscience Reports . 2021;41(2) doi: 10.1042/BSR20200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cresci M., Foffa I., Ait-Ali L., et al. Maternal and paternal environmental risk factors, metabolizing GSTM1 and GSTT1 polymorphisms, and congenital heart disease. The American Journal of Cardiology . 2011;108(11):1625–1631. doi: 10.1016/j.amjcard.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 70.Olson E., Pravenec M., Landa V., et al. Transgenic overexpression of glutathione S-transferase μ-type 1 reduces hypertension and oxidative stress in the stroke-prone spontaneously hypertensive rat. Journal of Hypertension . 2019;37(5):985–996. doi: 10.1097/HJH.0000000000001960. [DOI] [PubMed] [Google Scholar]
- 71.Lu J., Liu Q. H., Wang F., et al. Exosomal miR-9 inhibits angiogenesis by targeting MDK and regulating PDK/AKT pathway in nasopharyngeal carcinoma. Journal of Experimental & Clinical Cancer Research . 2018;37(1):p. 147. doi: 10.1186/s13046-018-0814-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Balser C., Wolf A., Herb M., Langmann T. Co-inhibition of PGF and VEGF blocks their expression in mononuclear phagocytes and limits neovascularization and leakage in the murine retina. Journal of Neuroinflammation . 2019;16(1):p. 26. doi: 10.1186/s12974-019-1419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lai C. H., Chen A. T., Burns A. B., et al. RAMP2-AS1 regulates endothelial homeostasis and aging. Frontiers in Cell and Development Biology . 2021;9:p. 635307. doi: 10.3389/fcell.2021.635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu H., Pumiglia K., LaFlamme S. E. Laminin-511 and α6 integrins regulate the expression of CXCR4 to promote endothelial morphogenesis. Journal of Cell Science . 2020;133(11) doi: 10.1242/jcs.246595. [DOI] [PubMed] [Google Scholar]
- 75.Babinska A., Clement C. C., Przygodzki T., et al. A peptide antagonist of F11R/JAM-A reduces plaque formation and prolongs survival in an animal model of atherosclerosis. Atherosclerosis . 2019;284:92–101. doi: 10.1016/j.atherosclerosis.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 76.Mouton A. J., Ma Y., Rivera Gonzalez O. J., et al. Fibroblast polarization over the myocardial infarction time continuum shifts roles from inflammation to angiogenesis. Basic Research in Cardiology . 2019;114(2):p. 6. doi: 10.1007/s00395-019-0715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Unterleuthner D., Neuhold P., Schwarz K., et al. Cancer-associated fibroblast-derived WNT2 increases tumor angiogenesis in colon cancer. Angiogenesis . 2020;23(2):159–177. doi: 10.1007/s10456-019-09688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Y., Hu Y., Xiong J., Zeng X. Overexpression of activating transcription factor 3 alleviates cardiac microvascular ischemia/reperfusion injury in rats. Frontiers in Pharmacology . 2021;12:p. 598959. doi: 10.3389/fphar.2021.598959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hussain S., Khan A. W., Akhmedov A., et al. Hyperglycemia induces myocardial dysfunction via epigenetic regulation of JunD. Circulation Research . 2020;127(10):1261–1273. doi: 10.1161/CIRCRESAHA.120.317132. [DOI] [PubMed] [Google Scholar]
- 80.Banerjee S., Aykin-Burns N., Krager K. J., et al. Loss of C/EBPδ enhances IR-induced cell death by promoting oxidative stress and mitochondrial dysfunction. Free Radical Biology & Medicine . 2016;99:296–307. doi: 10.1016/j.freeradbiomed.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Banerjee S., Alexander T., Majumdar D., et al. Loss of C/EBPδ exacerbates radiation-induced cognitive decline in aged mice due to impaired oxidative stress response. International Journal of Molecular Sciences . 2019;20(4):p. 885. doi: 10.3390/ijms20040885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li J., Gong L., Zhang R., et al. Fibroblast growth factor 21 inhibited inflammation and fibrosis after myocardial infarction via EGR1. European Journal of Pharmacology . 2021;910:p. 174470. doi: 10.1016/j.ejphar.2021.174470. [DOI] [PubMed] [Google Scholar]
- 83.Li J., Xu C., Liu Y., et al. Fibroblast growth factor 21 inhibited ischemic arrhythmias via targeting miR-143/EGR1 axis. Basic Research in Cardiology . 2020;115(2):p. 9. doi: 10.1007/s00395-019-0768-4. [DOI] [PubMed] [Google Scholar]
- 84.Ai K., Li X., Zhang P., et al. Genetic or siRNA inhibition of MBD2 attenuates the UUO- and I/R-induced renal fibrosis via downregulation of EGR1. Molecular Therapy--Nucleic Acids . 2022;28:77–86. doi: 10.1016/j.omtn.2022.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van der Feen D. E., Dickinson M. G., Bartelds B., et al. Egr-1 identifies neointimal remodeling and relates to progression in human pulmonary arterial hypertension. The Journal of Heart and Lung Transplantation . 2016;35(4):481–490. doi: 10.1016/j.healun.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 86.Marques F. Z., Nelson E., Chu P. Y., et al. High-fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation . 2017;135(10):964–977. doi: 10.1161/CIRCULATIONAHA.116.024545. [DOI] [PubMed] [Google Scholar]
- 87.Zhang S., Tao X., Cao Q., et al. lnc003875/miR-363/EGR1 regulatory network in the carcinoma -associated fibroblasts controls the angiogenesis of human placental site trophoblastic tumor (PSTT) Experimental Cell Research . 2020;387(2):p. 111783. doi: 10.1016/j.yexcr.2019.111783. [DOI] [PubMed] [Google Scholar]
- 88.Cupesi M., Yoshioka J., Gannon J., Kudinova A., Stewart C. L., Lammerding J. Attenuated hypertrophic response to pressure overload in a lamin A/C haploinsufficiency mouse. Journal of Molecular and Cellular Cardiology . 2010;48(6):1290–1297. doi: 10.1016/j.yjmcc.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: (a) the vlnplot shows the nFeatures and percent.mt of all cell subsets. (b) Unsupervised clustering of all cells visualized by groups, which showed that there was no obvious batch effect. (c) FeaturePlot showed the markers of fibroblast. Supplementary Figure 2: (a) the expression of Gpx3, Gstm1, Ggt5, Gsta3, Mgst1, and Gstt1 in three branches of the trajectory. (b) Comparative analysis of expression patterns of Gpx3, Col1a1, and Postn. Supplementary Figure 3: (a) heat map of genes related to glycogen synthesis, glycogen decomposition, and pentose phosphate pathway. (b) We renamed the fibroblasts with branch labels and then selected subgroups for reclustering before performing receptor ligand interaction analysis. Supplementary Table 1: top 20 markers of all subsets. Supplementary Table 2: GSEA results of KEGG and GO. Supplementary Table 3: list of fibrogenic genes. Supplementary Table 4: GO analysis of three branches. Supplementary Table 5: list of gene encoding secreted protein. Supplementary Table 6: list of angiogenesis genes.
Data Availability Statement
The dataset of the transverse aortic coarctation (TAC) model in mice was downloaded from the GSE155882 dataset.


