Abstract
The first member and eponym of the rhodopsin family was identified in the 1930s as the visual pigment of the rod photoreceptor cell in the animal retina. It was found to be a membrane protein, owing its photosensitivity to the presence of a covalently bound chromophoric group. This group, derived from vitamin A, was appropriately dubbed retinal. In the 1970s a microbial counterpart of this species was discovered in an archaeon, being a membrane protein also harbouring retinal as a chromophore, and named bacteriorhodopsin. Since their discovery a photogenic panorama unfolded, where up to date new members and subspecies with a variety of light-driven functionality have been added to this family. The animal branch, meanwhile categorized as type-2 rhodopsins, turned out to form a large subclass in the superfamily of G protein-coupled receptors and are essential to multiple elements of light-dependent animal sensory physiology. The microbial branch, the type-1 rhodopsins, largely function as light-driven ion pumps or channels, but also contain sensory-active and enzyme-sustaining subspecies. In this review we will follow the development of this exciting membrane protein panorama in a representative number of highlights and will present a prospect of their extraordinary future potential.
Keywords: membrane protein, photoreceptor, retinal protein, visual pigments, optogenetics, ion pumps, microbial, eukaryotic
Introduction
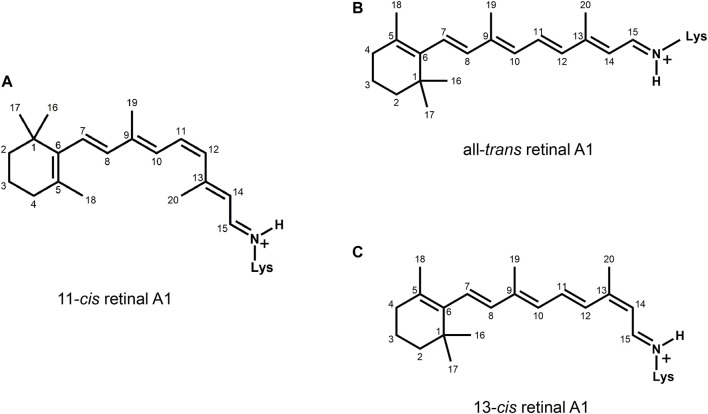
The first member and eponym of the rhodopsin family was identified in the 1930s as the visual pigment of the rod photoreceptor cell in the animal retina (Tansley, 1931; Wald, 1935). It turned out to be a membrane protein, owing its photosensitivity to the presence of a covalently bound chromophoric group. This photosensitive group, derived from vitamin A, was appropriately coined retinene, later officially renamed as retinal (Wald, 1935; Morton and Goodwin, 1944; Morton and Pitt, 1957). The visual pigments harboured a special conformer of this polyene compound, in casu the 11-cis configuration (Hubbard and Wald, 1952; Hubbard et al., 1971) (Figure 1). Upon photo-activation the chromophore was converted into the all-trans configuration, which triggered a sequel of conformational changes in the protein, leading to its active state (Wald, 1953; Morton and Pitt, 1957; Dartnall, 1962a). Eventually the chromophore was released as all-trans retinal (Dartnall, 1962c; Wald, 1968; Hubbard et al., 1971; Bridges, 1972). Surprisingly, in the 1970s a microbial counterpart of this protein was discovered in the archaeon Halobacterium salinarum (at the time referred to as Halobacterium halobium), which also harboured retinal as a chromophore, and was named bacteriorhodopsin (Oesterhelt and Stoeckenius, 1971). This membrane protein, however, contained the all-trans configuration, which upon photo-activation was converted into the 13-cis configuration (Smith et al., 1985; Oesterhelt, 1998). The resulting active state of the protein in this case thermally decayed in a sequel of steps whereby the chromophore eventually was thermally re-isomerized into the all-trans configuration returning to the original starting state (Oesterhelt, 1998; Lanyi, 2004).
FIGURE 1.
Chemical structures of the most common chromophore configurations in the rhodopsin families. The type-2 pigments contain an 11-cis, 15-anti retinylidene Schiff base of retinal A1 (A) in the “dark state” (or “ground state” in photophysical terminology), which is photo-excited into the all-trans configuration. The Type-1 pigments contain the all-trans configuration (B) in the “dark state.” This is photo-excited into the 13-cis, 15-anti configuration (C), which thermally relaxes and re-isomerizes, returning to the ground state. The ring-polyene chain orientation is different for type-2 (6-s-cis) and type-1 (6-s-trans) rhodopsins.
Since their discovery a photogenic panorama unfolded, where up to date new members and subspecies with a variety of light-driven functionality have been added to these families. The animal branch, categorized as type-2 rhodopsins, turned out to form part of the major subclass in the superfamily of G protein-coupled receptors (Bennett et al., 1982; Kühn, 1984; Crescitelli, 1991; Hargrave and McDowell, 1992). Currently they have diversified into at least eleven groups (Opn1–Opn9, R-group, Cn-group) most of which are essential to multiple elements of light-dependent animal sensory physiology. Depending on the animal species, they can be located in multiple tissues next to the eye (Terakita, 2005; Davies et al., 2015). Meanwhile, the microbial branch was named as type-1 rhodopsins, which largely function as light-driven ion pumps or channels, but also contain sensory-active and enzyme-sustaining subspecies (Oesterhelt, 1998; Spudich et al., 2000; Ernst et al., 2014; Leung and Montell, 2017; Nagata and Inoue, 2022). The most recent addition to the microbial rhodopsins is the heliorhodopsin family, which is remarkably different from the type-1 family in their inverted orientation in the membrane, with the N-terminal now residing in the intracellular compartment (Pushkarev et al., 2018; Shihoya et al., 2019; Kovalev et al., 2020b; Rozenberg et al., 2021; Chazan et al., 2022). The physiological function of this new family has not become very clear as of yet.
In this review we follow the historical development of this exciting membrane protein panorama in a representative number of highlights and present a prospect of their extraordinary future potential. We broadly outline their functional diversity and physiological relevance, as a comprehensive description is outside the scope of this review. A large number of excellent reviews on the rhodopsin families have been published, many of which we have referred to where appropriate, along with the most relevant early and recent papers. We refrain from presenting many molecular details, and therefore we refer to the following more recent reviews (DeGrip and Rothschild, 2000; Hofmann, 2000; Spudich et al., 2000; Hofmann et al., 2009; Yizhar et al., 2011; Palczewski and Orban, 2013; Ernst et al., 2014; Imamoto and Shichida, 2014; Inoue et al., 2014; Deisseroth, 2015; Hofmann and Palczewski, 2015; Brown and Ernst, 2017; Bando et al., 2019; El Khatib and Atamian, 2019; Dowling, 2020; Kandori, 2020; Kwon et al., 2020; Baillie et al., 2021; Moraes et al., 2021; Rozenberg et al., 2021; Bondar, 2022; Broser, 2022; Brown, 2022; Khelashvili and Menon, 2022; Nagata and Inoue, 2022).
This review presents a historical perspective and is therefore organized according to the landmark discoveries or progress in the field. In the following sections, we first discuss milestone studies and the common elements of the type-2 and type-1 rhodopsins, followed by individual subsections presenting typical elements for the type-2 and type-1 family, respectively. For the interested reader, we have compiled additional relevant citations in tables accompanying every section.
Discovery
The discovery and identification of rhodopsins was governed by their spectral properties. Since they all absorb photons in the visible spectrum, careful visual observations were the cornerstone for these early studies.
Type-2 Family
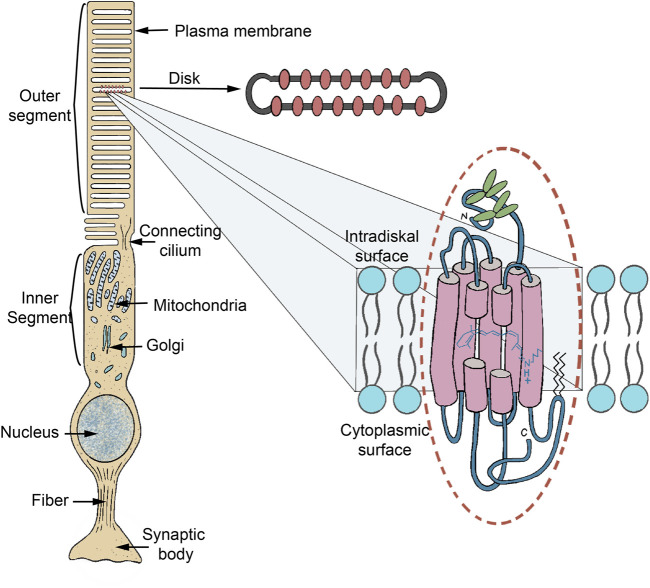
Rhodopsin, the founding father of the type-2 family was first identified as the visual pigment of the rod photoreceptor cell. In the 19th century, groundbreaking research on vision by Müller, Boll and Kühne led to the visual perception, that light capture occurred in the distal part of the human retina (Figure 2), in particular the outer segments of the photoreceptor cells (Müller, 1855; Boll, 1877; Ewald and Kühne, 1878). The typical red color of this tissue disappeared upon illumination, which was termed “bleaching,” and could to some extent be regenerated upon subsequent dark adaptation of the isolated eyecup. As of the 1930s it became apparent that a membrane-bound protein in the rod photoreceptor cell was responsible for the red color (Tansley, 1931; Bliss, 1948; Wald, 1953). This protein was named rhodopsin, after the ancient Greek words ροδεοσ (rhodeos, rose-coloured) and οψισ (opsis, which appropriately can be translated as sight or eyes). It was found to owe its spectral properties to a covalently bound cofactor, eventually named retinal (Wald, 1935, 1953, 1968; Hubbard et al., 1971). Subsequently, it was discovered that the cone photoreceptors in the vertebrate retina harboured closely related visual pigments (Morton and Pitt, 1957; Dartnall, 1962c; Mustafi et al., 2009). Thereafter, it became known that the invertebrate retina applied structurally very similar, but photochemically slightly differently operating visual pigments (Hara et al., 1967; Suzuki et al., 1993; Gärtner, 2000). Similar “bi-stable” pigments in fact are also active in the vertebrate retina, like the well-known melanopsins (Provencio et al., 1998; Kumbalasiri and Provencio, 2005). Another highlight was the growing insight that the visual pigments form part of the superfamily of G protein-coupled receptors (Kühn, 1984; Hargrave and McDowell, 1992; Palczewski and Orban, 2013). As a matter of fact, rhodopsin is the cornerstone of the major subfamily in this widespread receptor family.
FIGURE 2.
Schematic of a vertebrate rod photoreceptor cell (scotopic vision), zooming in on the location of the rod visual pigment rhodopsin. The rod outer segment (ROS), a ciliary outgrowth, is densely filled with isolated flattened vesicles (discs) which contain rhodopsin as the major (ca 90% w/w) membrane protein. The vertebrate visual pigments are therefore also designated as “ciliary rhodopsins.” Other disc membrane proteins are involved in signal propagation, stabilization of the disc shape and communication with the plasma membrane (PM). The phospholipids in the disc membrane have an exceptionally high content (ca 40%) of highly unsaturated fatty acids (22:6∞3) (Daemen, 1973). The discs are continuously generated at the base of the ROS as invaginations of the PM, then are nipped off and move upwards. After 7–10 days they reach the top of the ROS, which is pinched off in a circadian rhythm and degraded in the adjacent retinal pigment epithelium (RPE) (Young, 1976). The vertebrate cone photoreceptor (photopic vision) is organized in a similar fashion, except that the “discs” remain continuous with the PM as invaginations and are not pinched off. The organization of invertebrate visual photoreceptors is roughly similar, but the photoreceptive membranes are organized as numerous microvilli in rhabdomeric structures (Warrant and McIintyre, 1993) and their rhodopsins are also designated as rhabdomeric visual pigments. Only the classical visual pigments (Opn1, Opn2 and R-gene families) are organized in these specialized cellular outgrowths. All other type-2 and all type-1 pigments are targeted to the PM or an eyespot and form only a small part (up to several percent) of that membrane protein population.
Type-1 Family
In the early 1970s, fascinated by the dark-purple colonies of the salt lake thriving archaeon Halobacterium salinarum, Oesterhelt reported the surprising discovery that an intrinsic membrane protein was dominating purple patches in the cellular membrane of this archaeon and also harboured retinal as the chromophoric cofactor (Oesterhelt and Stoeckenius, 1971; Oesterhelt and Hess, 1973). At that time archaea were considered a subfamily of bacteria, and Oesterhelt coined the name bacteriorhodopsin (BR). Surprisingly, it was discovered that bacteriorhodopsin functions as a light-driven outward-directed proton pump, creating a proton-motive force enabling the cellular ATP-synthase complex to supply the cell with metabolic energy in the form of ATP (Oesterhelt et al., 1991). While bacteriorhodopsin is the dominant photoreceptor in Halobacterium salinarum, this archaeon eventually turned out to harbour several related photosensitive proteins, both with ion transport and sensory functions (Oesterhelt, 1998). Since the 1990s this field exploded, with more strains, including eukaryotic organisms like algae and fungi, and other functionalities being revealed every year (Béjà et al., 2000; Spudich et al., 2000; Brown, 2004; Rozenberg et al., 2021; Broser, 2022; Nagata and Inoue, 2022). More recently even viral rhodopsins have been discovered (Philosof and Béjà, 2013; Bratanov et al., 2019; Zabelskii et al., 2020). The overall structure and photochemistry of these pigments are very similar, and they are now considered to be a primary factor in marine phototrophy and solar energy conversion (Kirchman and Hanson, 2013; Gómez-Consarnau et al., 2019).
Spectral and Structural Properties, and Solubilization
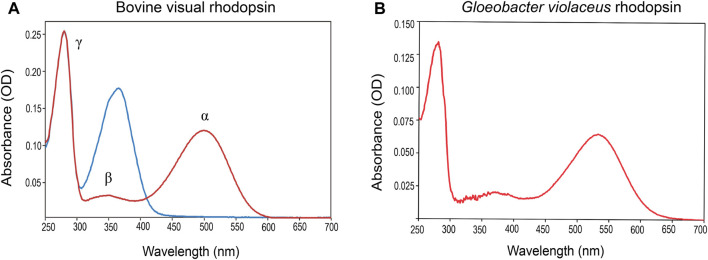
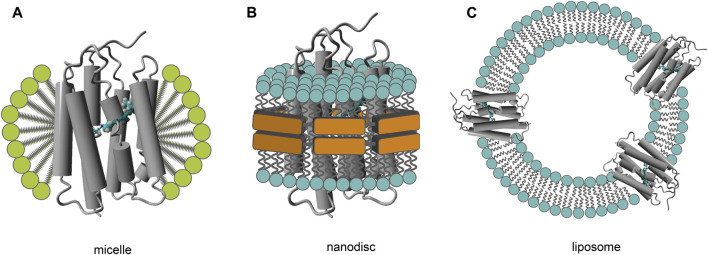
The spectral properties of all rhodopsins were discovered by visual observation, thanks to their absorbance of photons in the visible spectrum (350–750 nm). Accurate recording of their absorbance spectra was complicated in the spectrophotometers available at that time, due to the intense scattering of light by the rhodopsin containing membrane fragments isolated from host cells. Strong chemical reagents or alkaline conditions could dissolve these fragments, but with concomitant denaturation of the proteins and loss of their native spectral properties (bleaching). In the 1950s synthetic surface-active agents, termed detergents, became available, that were able to solubilize these membrane proteins in smaller mixed detergent-lipid-protein micelles, which strongly reduced light scattering (Hallett et al., 1991). Strong detergents like SDS still led to denaturation and release of retinal, but milder detergents were developed to avoid rapid partial unfolding at lab temperature or below. Accurate recording of absorbance spectra could then be established in detergent solutions. If some scattering still remained, or other visible light material interfered, difference spectroscopy was established by recording spectra before and after illumination in the presence of hydroxylamine and taking a difference spectrum. Hydroxylamine captures the released retinal as retinaloxime, which absorbs outside the main absorbance band of most rhodopsins (Wald and Brown, 1953; Hubbard et al., 1971; Kropf, 1975). This usually provides an accurate profile of the main absorbance band or at least the absorbance maximum. (Figure 3). A more recent and elegant approach is to insert a membrane protein into small nanodiscs (Civjan et al., 2003; Borch and Hamann, 2009; Ritchie et al., 2009) (Figure 4). This also strongly reduces light scattering and has the important advantage of embedding the protein in the more stabilizing lipid bilayer environment (Banerjee et al., 2008; Tsukamoto et al., 2011; Zhou and Cross, 2013; Ganapathy et al., 2020). Nanodiscs can be generated using either lipoproteins and membrane scaffold protein derivatives (MSPs) or small synthetic polymers of the amphipol or styrene-maleic acid copolymer family (SMAs) (Knowles et al., 2009; Popot et al., 2011; Hoi et al., 2021). For MSPs usually a brief detergent solubilization step is still required, while SMAs can extract the protein directly from the membrane, but have a smaller pH-profile (Shirzad-Wasei et al., 2015; Dörr et al., 2016; Kopf et al., 2020; Ueta et al., 2020).
FIGURE 3.
Typical dark state absorbance spectra (red curves) of a purified type-2 (A) and type-1 (B) pigment. Both spectra exhibit a major peak (α-band) and a small satellite (β-band), both originating in the chromophore, and a γ-band near 280 nm, mainly originating in protein residues. The α-band derives from the whole conjugated polyene system (S0-S1) (cf. Figure 1), while the β-band derives from a smaller segment, and its intensity also depends on the torsion in the polyene chain. Upon short illumination of the monostable type-2 pigment (A) in the presence of hydroxylamine, the liberated retinal is converted into retinaloxime (blue curve). The Meta state of bistable type-2 pigments, also reacts with hydroxylamine generating retinaloxime, but usually quite slowly. Short illumination of type-1 pigments (B) in the presence of hydroxylamine hardly affects the photocycle and the return to the ground state. However, upon prolonged illumination hydroxylamine will slowly attack photo-intermediates, mainly M and N, releasing retinaloxime.
FIGURE 4.
Membrane mimics for purified membrane proteins. Schematics of the micellar (A), nanodisc (B) and vesicular (C; proteoliposome) organization are displayed. Note that the relative dimensions are not to scale: diameters vary from 10–50 nm for the micelles and nanodiscs, and from 100 nm up to 10 µm for the liposomes. Several amphipatic components functioning as bilayer-stabilizing agents in the nanodiscs have been generated (MSP derivatives from lipoproteins, synthetic amphipols and SMAs, respectively), and are still under further development. Purification of the protein in a detergent environment generates the classical micellar state (A). Because the thermal stability of membrane proteins in the micelles is generally reduced, often (phospho)lipids are added (bicelles). Alternatively, membrane proteins can be transferred into the bilayer membrane of a nanodisc (B) or liposome (C). Membrane proteins can be directly (amphipol or SMA nanodiscs) or under very brief detergent exposure (MSP nanodiscs) transferred from the native membrane into nanodiscs, and the classical purification techniques can be applied upon the resulting nanodisc population. Liposomes offer a broader selection for the lipid population, and are used in vectorial transport studies and in AFM, FTIR and solid-state NMR spectroscopy. However, they are less suitable in optical spectroscopy because of their larger dimension, resulting in strong light scattering.
The spectral profile of rhodopsins in the visible and near-UV region is very similar (Figure 3). It consists of the most red-shifted main absorbance band or α-band, a smaller β-band, both originating in the bound retinal, and the γ-band near 280 nm, that largely originates in the aromatic residues of the protein part termed “opsin.” In all rhodopsins retinal is covalently linked to a lysine residue in the seventh transmembrane segment (TM7) via a Schiff base (Figure 1) which is mostly protonated. The α-band is strongly red-shifted from the absorbance band of free retinal (maximum around 380 nm). This unusual polar grouping in the middle of a membrane protein is stabilized by the negatively charged “counterion complex,” containing one, two or occasionally three protein residues (mostly Glu/Asp, sometimes Lys or in anion pumps a Cl − ion) in a H-bonded network with nearby residues and bound water molecules (Lanyi, 2004; Ernst et al., 2014; Gerwert et al., 2014; Nomura et al., 2018). Thus, the excitation energy in this retinylidene moiety is strongly reduced, compared to free retinal, which results in a red-shift of the absorbance profile. The magnitude of the red-shift strongly depends on the structure of the H-bonded network and counterion complex involving variable electrostatic interactions with the protonated Schiff base and to a lesser extent on the properties of protein residues in the opsin binding pocket (Lesca et al., 2018; Nikolaev et al., 2020; Shen et al., 2021; Shtyrov et al., 2021; Church et al., 2022a). By modifying these elements Nature created the spectacular broad variance in the spectral profile of rhodopsins, allowing them to cover the entire visible region.
The three-dimensional (3-D) structure of rhodopsins has been extensively investigated by classical electron diffraction on 2-D crystals and X-ray crystallography on large 3-D crystals, by solid-state NMR spectroscopy on membrane fragments and more recently by X-ray free electron lasers (XFEL) on small crystals (Schertler and Hargrave, 1995; Palczewski, 2012; Ladizhansky, 2017; Smith, 2021). Cryo-electron microscopy (Cryo-EM) has been traditionally performed on micellar solutions, but has also evolved to include nanodiscs (Maeda et al., 1991; Bertazolli-Filho et al., 2001; Hasegawa et al., 2018; Zhao et al., 2019; Zhang M. et al., 2021). Only solid-state NMR can be directly applied to membrane suspensions, but overall, there is quite good agreement between the various approaches. The overall structure is quite similar for all rhodopsin families, with the main scaffold consisting of seven closely packed transmembrane α-helices, which creates a tightly fitting binding pocket lined by a lysine residue to covalently bind retinal (Figure 2). The protein N- and C-terminal stretch reside at the extracellular and intracellular side of the membrane, respectively, except for the heliorhodopsin family where this sidedness is reverted (Pushkarev et al., 2018). However, the packing of the α-helices, the size of the loops connecting the α-helices and of the N-terminal and C-terminal stretches outside the membrane differ significantly between the type-1 and type-2 families.
Type-2 Family
Most type-2 rhodopsins, and in particular cone visual pigments and invertebrate pigments are very sensitive to at least partial denaturation upon solubilization in detergent solution (Bliss, 1948; Kropf, 1982; Okano et al., 1989). While commercial detergents like Triton X-100, CTAB, LDAO and Emulphogene BC-720 could dissolve the vertebrate rod pigment rhodopsin into mixed micelles with none or only very slow loss of spectral properties at room temperature, for most other pigments only the very mild agent digitonin could be applied (Tansley, 1931; Knudsen and Hubbell, 1978; Okano et al., 1989; Hofmann and Palczewski, 2015). This natural compound, a steroidal glycone extracted from Digitalis purpurea, however has the disadvantage that its commercial preparations were quite expensive and did vary in composition and aqueous solubility (Bridges, 1977). Major progress was attained in the 1970s upon development of the alkylsaccharide detergents 1-O-n-β-D-octylglucoside (octylglucoside, OG), nonylglucoside (NG) and dodecylmaltoside (DDM) (Stubbs et al., 1976; DeGrip and Bovee-Geurts, 1979). DDM in particular turned out to maintain thermal stability and spectral and photochemical properties of rhodopsin almost as well as digitonin (DeGrip, 1982; VanAken et al., 1986). Additionally, DDM is well accessible and affordable through organic synthesis, and has therefore become the most popular detergent in the membrane protein field. Also, in case a protein purified in DDM needs to be reconstituted in a lipid bilayer for certain applications (nanodisc or proteoliposome, Figure 4), DDM can be easily extracted via cyclodextrin inclusion (DeGrip et al., 1998). More recently, a large number of novel detergents based upon the structural principle of DDM have been developed, some of which provide better thermal stability or better crystallization conditions for selected membrane proteins than DDM, but all requiring more complex synthesis (Hussain et al., 2016; Nguyen et al., 2018; Ehsan et al., 2020; Urner et al., 2020).
The absorbance band profiles of type-2 rhodopsins are quite similar (Figure 3), but the position of the α-band varies strongly for the visual pigments. The vertebrate rod photoreceptor pigment rhodopsin has quite a broad range in its absorbance maximum (Rh1 subset, 440–520 nm), with fresh-water animals slightly red-shifted and marine animals blue-shifted depending on the depth of their habitat (Locket, 1977; Luk et al., 2016; Musilova et al., 2019). Vertebrate cone pigments cover the entire visible spectrum, and can be divided into four subsets, the long-wavelength (LWS, absorbance maximum range 520–640 nm), green (Rh2, 460–530 nm), blue (SWS2, 400–470 nm), and UV (SWS1, 350–450 nm) sensitive pigments (Crescitelli, 1991; Yokoyama and Yokoyama, 2000; Imamoto and Shichida, 2014). This classification is not only based upon spectral sensitivity, but also upon sequence similarity (Nathans, 1987; Hunt and Collin, 2014; Jacobs, 2018; El Khatib and Atamian, 2019). Invertebrate visual pigments are more scattered over the visible region and can range from 340 nm up to 600 nm (Gärtner, 2000; Katz and Minke, 2009; Tsukamoto and Terakita, 2010). Non-visual animal rhodopsins are scattered over the 340–550 nm region (Leung and Montell, 2017; Pérez J. H. et al., 2019; Moraes et al., 2021).
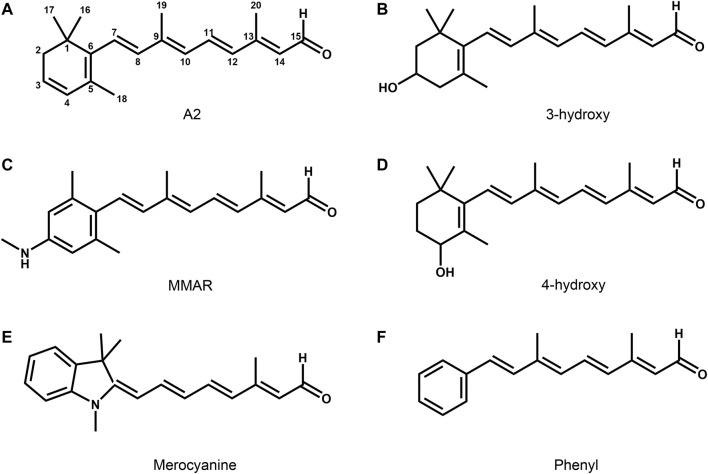
The spectral properties of the type-2 rhodopsins depend on the 11-cis configuration of the retinylidene chromophore. Next to the standard retinal (retinal A1, Figure 1), several natural modifications occur (analogs). In fresh-water and coastal vertebrates 11-cis 3-dehydroretinal (retinal A2) has been observed (Figure 5) (Bridges, 1972; Yoshizawa, 1984; Imai et al., 1999). The longer conjugated chain red-shifts the absorbance maximum by 20–40 nm in rod pigments and up to 70 nm in cone pigments, as compared to retinal A1, to compensate for the lower blue light intensity in their habitat (Dartnall, 1962c; Hubbard et al., 1971). These “A2-rhodopsins” are also referred to as porphyropsins. In insects and some other invertebrates, 11-cis 3-hydroxy- and 4-hydroxyretinals have been detected (Figure 5) (Vogt and Kirschfeld, 1984; Matsui et al., 1988; Seki and Vogt, 1998). These modifications blue-shift the absorbance maximum by 20–40 nm, as compared to retinal A1 (Sekharan et al., 2011).
FIGURE 5.
Some uncommon retinal analogs occurring as natural chromophores or in engineered pigment analogs. 3, 4-didehydroretinal (retinal A2, (A)) red-shifts the rhodopsin spectrum relative to A1, and is mostly found in fish and amphibian visual pigments. 3-hydroxy- (B) and 4-hydroxy- (D) retinal A1 induce a blue-shift relative to A1 and are found in the visual pigments of insects and deep-sea shrimps, respectively. Phenylretinal (F), MMAR (C) and the merocyanine derivative (E) are synthetic analogs, that, respectively, induce a blue-shift (F) and the largest red-shifts, observed so far ((C,E); see text). All these analogs bind to the lysine residue in the native opsin binding pocket with a protonated Schiff base.
While such natural modifications are exploited to modulate the spectral position of a rhodopsin, the most effective approaches to shift the absorbance spectrum of the rhodopsin chromophore away from that of free retinal (380 nm) are protonation of the Schiff base and mutation of selected opsin residues lining the retinal binding pocket. For instance, only the Schiff base in UV absorbing rhodopsins, which absorb in the 350–380 nm region, is not protonated, while in all other classes it is protonated (Kusnetzow et al., 2004; Imamoto and Shichida, 2014). The large variation in the spectral properties in the latter classes is mainly due to the combined inductive effect of opsin binding pocket residues, in combination with H-bonding networks involving water molecules. On top of that, some vertebrate LWS visual pigments have developed a unique mutation (Glu197->His) creating a chloride binding site that effectuates a further 20–30 nm red-shift (Wang et al., 1993).
With respect to structural biology, bovine rod rhodopsin was a forerunner among all animal intrinsic membrane proteins, presenting the first detailed 3-D structure via X-ray crystallography in 2000, with many more to follow (Palczewski et al., 2000; Li et al., 2004; Okada et al., 2004). The seven transmembrane α-helical scaffold surrounding an accessible cofactor binding pocket proved to be the general motif for the entire G protein-coupled receptor family (Figure 6) (Sanchez-Reyes et al., 2017). This feat has stimulated advances in many other research fields, including drug design in the pharmaceutical sciences, study of protein structure-function correlations, and membrane protein-lipid interactions, both from experimental, theoretical and in-silico standpoints. Several natural factors concurred to enable this important step forward. First of all, rod rhodopsin is one of the few intrinsic membrane proteins that is available in relatively large quantities in domesticated animals, the most used being cattle (up to 1 mg of rhodopsin per eye), bullfrogs (up to 100 μg per eye) and chick (up to 100 μg LWS cone pigment per eye) (DeGrip et al., 1980; Toba and Hanawa, 1985; Yoshizawa and Kuwata, 1991). After enucleation and proper dark adaptation of the eyes, intact rod or cone outer segments (ROS or COS) can be easily isolated in a dark room under dim red light (>650 nm) that will not activate and bleach the pigment (Figure 3). Further, in dark-adapted ROS, rhodopsin makes up about 85% of the total protein content (DeGrip et al., 1980). Eventually, dark-adapted bovine retinae even became commercially available (Hormel Co., Austin, Minnesota, United States). Finally, bovine rod rhodopsin was found to be relatively resistant to destabilization by detergents as compared to most other visual pigments, allowing extensive purification. Likewise, it proved to be sufficiently stable in less mild but more crystal-production-favoring small detergents like OG and NG to facilitate crystallization trials (Palczewski et al., 2000; Park et al., 2008).
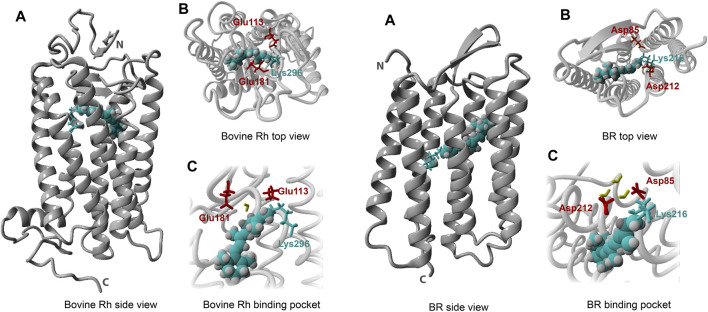
FIGURE 6.
Comparison of structural features of the type-2 and type-1 pigment archetypes bovine rod rhodopsin (left section) and bacteriorhodopsin (right section), respectively. Full crystal structures are presented in (A) and (A) (pdb 1U19 and 5ZIN), a top view is shown in (B) and (B) and a binding pocket exposure in (C) and (C), respectively. The retinylidene chromophore (cyan) is represented as space-filling spheres, and the retinal binding lysine residue (cyan) is presented as sticks. The two protein residues displayed (red) contribute to the counterion complex stabilizing the pronated Schiff base. The two crystal structures share the seven α-helical transmembrane segment bundle, but the packing of the helices, the location and assembly of the binding pocket and the structure of the chromophore are clearly different.
The bovine rhodopsin amino acid sequence was established thanks to heroic protein sequencing efforts (Abdulaev et al., 1982; Hargrave et al., 1983). Over time, sequence information became available more easily via genome mining and c-DNA-sequencing. Thus, it came out that most invertebrate visual pigments are similar in size to the vertebrate pigments (36–42 kD), but mollusc pigments are significantly larger (46–55 kD), because of the presence of a much longer C-terminal (Ovchinnikov et al., 1988b; Gärtner, 2000). This additional stretch is unique in having an insertion of up to eleven copies of a peculiar pentapeptide sequence (Pro-Pro-Gln-Gly-Tyr), which probably helps in immobilization of the protein in the microvillar membrane (Ryba et al., 1993; Gärtner, 2000). Longer C-terminal stretches are also found in non-visual rhodopsins. For instance, the VA-opsin and melanopsin family also show this feature, except that the pentapeptide insertion does not occur. Here the extra sequence probably has a function in complex regulation of signal processing and desensitization (Valdez-Lopez et al., 2020; Contreras et al., 2021). Some VA-opsins and melanopsins are even produced in two or more splicing isoforms, with longer and shorter C-terminals (Davies et al., 2010).
While squid provides fair quantities of visual pigment, the first complete 3-D crystal structures only became available since 2008, both because of the much lower stability of the pigments in detergent solution and since crystallization could only be achieved after proteolytic removal of most of the long C-terminal (Murakami and Kouyama, 2008; Shimamura et al., 2008). The overall fold of the seven-transmembrane α-helical scaffold is quite similar to bovine rhodopsin, but the structure of the long C-terminal could not be determined, of course. The position of the retinal chromophore is slightly different, since the Glu residue functioning as the direct counterion for the protonated Schiff base is displaced from the site in the vertebrate pigments (Terakita et al., 2004). The first crystal structure of an arthropod rhodopsin (jumping spider) was only recently published in 2019, and again shows the familiar seven α-helical fold with overall high similarity with the squid structure (Varma et al., 2019). So far, crystal structures of non-visual rhodopsins have not been reported.
The crystal unit cell of bovine rod rhodopsin contains a dimer, but its interaction pattern is very different from the natural one (Fotiadis et al., 2006; Palczewski, 2006). In fact, rhodopsin is equally active as a monomer, and the organization in the ROS disc membranes is still debated (monomer, dimer, longer stretches?) (Fotiadis et al., 2004; Chabre and LeMaire, 2005; Mishra et al., 2016; Zhang et al., 2016; Feldman et al., 2019; Zhao et al., 2019). Invertebrate visual rhodopsins are probably rigidly immobilized in their native membrane, which allows to discern the polarization plane of the incoming light (Gärtner, 2000; Stavenga et al., 2000).
The crystal structures are essential to resolve the protein fold of the rhodopsins and have confirmed several conjectures of the binding pocket. Biochemical, vibrational (resonance Raman and FTIR spectroscopy) and solid-state NMR studies already produced very strong evidence that it indeed harboured the 11-cis configuration of retinal (Groenendijk et al., 1980; Mathies et al., 1987; Lugtenburg et al., 1988; DeGrip and Rothschild, 2000; Mathies and Lugtenburg, 2000). Surely enough, this configuration best fitted the non-protein electronic density in the binding pocket. The same is true for the covalent binding of retinal to a lysine residue, for which the above-mentioned techniques also already provided a wealth of evidence (Bownds, 1967; DeGrip et al., 1973; Creemers et al., 1999; Mathies and Lugtenburg, 2000). However, to firmly establish protonation of the Schiff base the resolution of the crystal structures is not high enough. Instead, the evidence produced by vibrational and NMR spectroscopy is very convincing and in fact was later underpinned by quantum-chemical computation (Palings et al., 1987; Herzfeld and Lansing, 2002; Gascón et al., 2005; Tastan et al., 2014).
Type-1 Family
The sensitivity to detergent action also varies strongly between microbial rhodopsins. For instance, while bacteriorhodopsin (BR) is quite stable in OG, Triton X-100 and dodecylphosphocholine (DPC) even as a monomer, the rhodopsin proton pump from the cyanobacterium Gloeobacter violaceus (GR) strongly prefers DDM and is very unstable in DPC (Dencher and Heyn, 1978; Brouillette et al., 1989; Ganapathy et al., 2020). In general, OG and DDM are the preferred agents for solubilization of type-1 rhodopsins.
The spectral range of type-1 rhodopsins (360–690 nm) is comparable to that of type-2. There is less evidence for a clear relation to activity or habitat, an exception being the proton pump proteorhodopsin, which exhibits a blue-shift in deeper marine environments (Béjà et al., 2001; Bielawski et al., 2004). In a major distinction from type-2, microbial rhodopsins invariably exploit retinal A1 in the all-trans configuration as the basis for their light absorbance. Here, as well, a plethora of experimental evidence has demonstrated retinal binding to a lysine residue via a protonated Schiff base (Haupts et al., 1999).
Advanced angular electron diffraction studies on 2-D BR crystals in membrane patches already afforded a first glimpse into the organization of the helical transmembrane segments of type-1 rhodopsins (Henderson and Unwin, 1975; Mitra et al., 1993; Grigorieff et al., 1996; Heymann et al., 1997; Mitsuoka et al., 1999). The first 3-D crystal structures were reported for BR from 1997 onwards, and at a very high resolution slightly before that of bovine rhodopsin (Luecke et al., 1999; Pebay-Peyroula et al., 2000). This progress was aided by its high stability in detergent solutions and the relatively simple isolation from its native source. Bacteriorhodopsin is organized in large singular patches in the cellular membrane of Halobacterium salinarum, which can visually be observed and separated from other membrane fragments quite easily (Oesterhelt and Stoeckenius, 1971). In addition, type-1 rhodopsins complete a full photocycle (see below) and after photo-activation do not release the retinal, but thermally return to the ground state. This obviates the complexity of using dark rooms and shielding all experimental manipulations from room light exposure. Meanwhile, quite a number of crystal structures have been resolved for various classes of type-1 rhodopsins (Table 1). The most recent high resolution 3-D structures actually capitalized on the fantastic progress in cryo-EM (Hirschi et al., 2021; Kishi et al., 2022).
TABLE 1.
Selected additional citations for the section “Spectral and structural properties and solubilization”.
The available type-1 3-D structures show high similarity in protein fold and retinal pocket location. The basic seven α-helical transmembrane organization is comparable to that of type-2 (Figure 6), but for type-1 the helical packing is somewhat different and more compact. The loop segments connecting the helices are generally shorter and the retinal pocket is positioned differently to accommodate the longer all-trans chromophore instead of the curved 11-cis one (Figures 1, 6). Aspects of the binding pocket (retinal isomer and binding to a lysine residue via a Schiff base) again were in line with a wealth of evidence generated by biochemical and spectroscopic techniques (Lanyi, 2004). A recent XFEL study of the bacteriorhodopsin photocycle achieved a very high structural (ca 1.5 Å) and temporal (femtosecond) resolution and produced evidence for protonation of the Schiff base (Nogly et al., 2018). Also in the type-1 case the evidence generated by biophysical techniques like vibrational, EPR and NMR spectroscopy and by quantum-chemical computation is most convincing (Ernst et al., 2014; Brown and Ernst, 2017; Ryazantsev et al., 2019; Nagata and Inoue, 2022).
A conspicuous feature of most type-1 rhodopsins is that they organize in homo-oligomers, whether observed in the native membrane or in host cells. The most common arrangement for bacterial and archaeal rhodopsins are trimers or pentamers, though occasionally hexamers do occur as well (Hussain et al., 2015; Shibata et al., 2018; Kao et al., 2019). Circular dichroism spectroscopy provides evidence for exciton coupling between the chromophores (Cassim, 1992; Fujimoto and Inoue, 2020; Fujimoto, 2021). For eukaryotic type-1 rhodopsins, homo-dimeric as well as hetero-dimeric complexes are observed (Mukherjee et al., 2019; Govorunova et al., 2021; Broser, 2022). Isolated type-1 monomers are also functionally active, indicating that the oligomeric assembly probably affords optimal packing and mutual stabilization, and/or the opportunity to modulate monomer activity by inter-subunit interplay (Iizuka et al., 2019).
A novel feature was discovered in the enzyme-rhodopsins i.e. an additional transmembrane segment at the N-terminal (TM8), which functions as a connector with the cognate soluble enzyme domain and seems to be essential for modulating its activity (Ikuta et al., 2020; Tsunoda et al., 2021).
Interestingly, several thermostable microbial rhodopsins have been discovered. The crystal structure of the highly thermophilic rhodopsin (TR) from Thermus thermophilus was resolved to be very similar to that of the much less thermally stable xanthorhodopsin (XR) from Salinibacter ruber, including the binding crevice for the carotenoid antenna (Tsukamoto et al., 2016). Likewise, the crystal structure of the thermostable rhodopsin proton-pump from Rubrobacter xylanophilus (RxR) is very similar to that of bacteriorhodopsin (Hayashi et al., 2020). An unusually widely stable proton pump (pH, detergent, temperature), named Tara76 rhodopsin, was isolated from uncultured bacteria (Shim et al., 2021). Such data shed new light on the design options to increase thermal and environmental stability without a significant sacrifice in dynamics and activity (Hayashi et al., 2020).
Additional selected references relevant for this section have been compiled in Table 1.
Functional Diversity, Phylogeny
It was relatively simple in the old days. On one hand, we knew of animal rhodopsins, being G protein-coupled receptors, very nicely developed and evolved into a set of proteins allowing photopic vision (color discrimination) and a single class for extremely sensitive scotopic vision (black-and-white). On the other hand, another class of retinal-proteins had evolved in archaea to exploit solar energy for active transport of protons and chloride ions. However, with the awakening of the genome era, this view became totally obsolete. While the notion that the type-1 and type-2 families probably do not have a common ancestor and have little overlap in physiological function was consistent, over time many new members were discovered and their classification revised (Porter et al., 2012; Yee et al., 2013; Zabelskii et al., 2021). In hindsight, it was to be expected that ahead of the large carotenoid and chlorophyll dependent protein complexes in the photosynthetic reaction centers, Nature would have taken advantage of the abundance of solar energy making maximal use of this fantastic toolbox of retinal-proteins, that are relatively simply to bioproduce and adapt.
It is likely that many products of this toolbox are yet to be discovered, but already the genetic and functional diversity is so vast and complex, that we provide a very general overview below and mostly refer to selected reviews.
Type-2 Family
The animal rhodopsins have meanwhile been classified in at least nine gene families (Opn1–Opn9) and two separate sets with some members still awaiting further assignment (Table 2). Physiological function and tissue distribution show incredible diversity (Janssen et al., 2003; Leung and Montell, 2017; Liebert et al., 2021; Moraes et al., 2021; Calligaro et al., 2022). The classical visual pigments come within Opn1 (cone pigments) and Opn2 (rod pigments). Pigments discovered later in the vertebrate retina, such as melanopsin, VA-opsin or neuropsin, peropsin (RRH) and RGR fall under Opn4, unclassified and Opn5, respectively (Table 2). The common thread still is primary signal transduction via at least one of the available G-protein species (Gt, Go, Gi, Gq, and Gs), with cross-activation, modulation or desensitization via a variety of other mediators. However, RGR and its mollusc counterpart retinochrome are exceptional in this context, since they act as photo-isomerases, binding all-trans retinal in the dark state, and releasing 11-cis retinal after photo-activation as a supply for regeneration of visual opsins (Hara et al., 1967; Pepe and Cugnoli, 1992; Zhang et al., 2019; Choi et al., 2021; Vöcking et al., 2021). Another remarkable subset are Opn5L, peropsin and Opn7 members, which also bind all-trans retinal in the dark state, but that seems to be the active state binding the G protein. Upon illumination they generate the 11-cis chromophore, which represents the resting state that in the case of Opn5L members may even thermally revert to the active state (Nagata et al., 2018; Yamashita, 2020; Karapinar et al., 2021; Sakai et al., 2022). An even more surprising observation is that some type-2 pigments may be involved in recognizing temperature differences or mechanical changes, or function as chemosensors or tumorigenic elements, possibly even without requiring their retinal cofactor (Shen et al., 2011; Park et al., 2013; Baker et al., 2015; Pérez-Cerezales et al., 2015; Leung et al., 2020; Xu et al., 2020; Córdova et al., 2021; Moraes et al., 2021).
TABLE 2.
Current classification of type-2 rhodopsins.
Spectral range and mono/bistability not always exclusive within a group and very limited known for Opn6-Opn9 and Cn-opsins.
Cone pigments are mainly involved in color (photopic) vision, rod pigments in (scotopic) dim-light vision. In mammals melanopsins are important for pupillary contraction and circadian regulation. Retinochromes (R-opsins) and peropsins and RGRs (Opn5) have photoisomerase activity (all-trans → 11-cis).
The Opn5L group (Sato et al., 2018b; Yamashita, 2020) may have been classified wrongly, since they clade within the Opn6-9 framework.
Type-1 Family
Type-1 rhodopsins have been identified in archaea and eubacteria, including cyanobacteria, as well as in unicellular eukaryotes (algae, fungi, yeast) and more recently also in choanoflagellates and viruses (Lamarche et al., 2017; Bratanov et al., 2019; Zabelskii et al., 2020; Rozenberg et al., 2021; Govorunova et al., 2022b; Nagata and Inoue, 2022). Most of these pigments function as light-driven ion transporters or ion channels (Figure 7). The newly discovered xenorhodopsins and schizorhodopsins are exceptional as they perform inward-directed proton transport (Inoue et al., 2018; Inoue et al., 2020; Weissbecker et al., 2021; Brown, 2022). However, some type-1 rhodopsins display a photosensory function (sensory rhodopsins) and signal via a cognate transducer protein, which is totally different functionally and structurally from the animal G-proteins (Bogomolni and Spudich, 1991; Krah et al., 1994; Deininger et al., 1995). Overall, type-1 pigments are the dominant contributors to marine phototrophy (Casey et al., 2017; Larkum et al., 2018; Gómez-Consarnau et al., 2019). In addition, eukaryotic type-1 rhodopsins have been discovered which are intracellularly fused to an enzymatic domain and mediate light-driven enzyme activation (guanylyl cyclase, phosphodiesterase) or inhibition (guanylyl cyclase, based upon histidine kinase activity) (Avelar et al., 2014; Lamarche et al., 2017; Luck et al., 2019; Mukherjee et al., 2019; Tsunoda et al., 2021; Broser, 2022; Tian et al., 2022). These pigments have been termed as enzyme-rhodopsins.
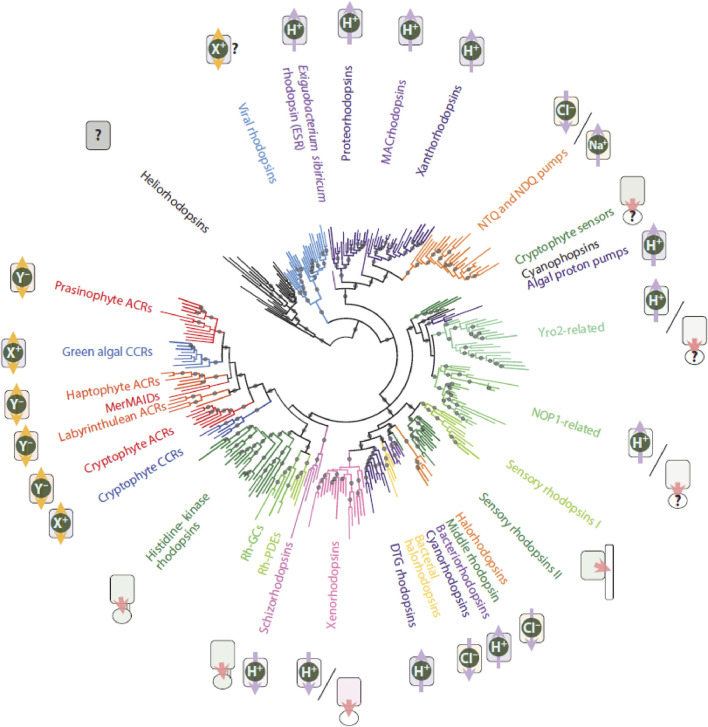
FIGURE 7.
Global phylogeny of type-1 pigments illustrating their formidable diversification. The figure was modified with consent from Rozenberg et al., 2021. We refer to the original paper for the construction of the tree and for all abbreviations. Purple arrows represent active outward (away from center) and inward ion transport, respectively. Orange arrows represent ion channels. Pink arrows represent enzyme-rhodopsins (fused enzyme domains) and sensory rhodopsins (detachable transducers). For further details of the various classes we refer to recent literature (Hasemi et al., 2016; Govorunova et al., 2016; id-, 2022; Nakajima et al., 2018; Oppermann et al., 2019; Kovalev et al., 2020a; Rozenberg et al., 2021).
The overall structure and photochemistry of all type-1 rhodopsins present a very similar pattern, though the sequence identity can be as low as 12%, and the kinetics of the photocycle can vary up to at least thousand-fold. The most recent addition, the heliorhodopsins, are not very different in their protein fold from e.g. BR in spite of a very low sequence identity (<10%) (Shihoya et al., 2019). Considering their inverted insertion into the membrane, very long photocycle and so far unknown functionality, they probably are better classified separately as type-3 rhodopsins (Tanaka et al., 2020; Chazan et al., 2022).
Heterologous Expression and Purification
The congruent broad heterogeneity in the rhodopsin superfamily offers a fascinating spectrum for mechanistic studies as well as biomimetic adaptation and application. However, mechanistic studies still require large quantities of relatively pure material (at least several mg). With the exception of some visual pigments and archaeal rhodopsins, such quantities are not available from native sources. Besides, purifying minor quantities of rhodopsins out of a large excess of cellular membrane proteins turned out to be a “hell of a job” (Dartnall, 1962b; Hubbard et al., 1971). Furthermore, in-depth mechanistic studies and biomimetic applications need the ability to make modifications biosynthetically at the protein residue level, and synthetically at the chromophore level. And even when in silico molecular dynamics and quantum chemical computation would have reached the time-scale of protein conformational changes (femtoseconds to seconds range) and the native accuracy, then still experimental verification is in order. Experimentally modifying rhodopsins in the native organism was completely out of hand at the time, except for some limited success with bacteriorhodopsin mutants in Halobacterium salinarum which still did not solve the quantity requirement (Krebs et al., 1993). Hence, the search for suitable heterologous expression hosts started in the 1980s, and over time it became obvious that the eukaryotic rhodopsins required quite a different perspective.
With the start of the genome era, recombinant DNA technology (genome mining, DNA and c-DNA sequence information and comparison, DNA sequence modification) became accessible and have now become common experimental tools (Khorana, 1979; Khorana et al., 1987). Likewise, total synthesis of retinal isomers and a plethora of derivatives has improved significantly (Dawadi and Lugtenburg, 2010; Liu and Liu, 2011; Álvarez et al., 2014; El-Tahawy et al., 2020).
Type-2 Family
Type-2 rhodopsins can undergo a variety of posttranslational modifications (disulfide-bridge formation, N- and O-glycosylation, methylation, acetylation, myristylation, palmitoylation, phosphorylation), most of which are not properly executed by the bacterial or archaeal biosynthetic machinery (Table 3). Expression of bovine rhodopsin in bacteria and even yeast did not yield promising results (Mollaaghababa et al., 1996; Abdulaev and Ridge, 2000). Hence, for optimal heterologous expression a eukaryotic cell type had to be selected as a host. Attempts have been made to express type-2 pigments and related receptors in the eye of whole organisms (mouse, Xenopus) and in Caenorhabditis elegans using viral vectors or transgenic animals, but this gave relatively low yields or even led to retinal degeneration (Zhang et al., 2005; Salom et al., 2008; Cao et al., 2012; Salom et al., 2012). Eventually, the best results with sufficient posttranslational modification and targeting to the plasma membrane were obtained in some mammalian cell lines using plasmid transfection (COS, HEK, Neuroblastoma cell lines), in insect cell lines using baculoviral infection (Spodoptera Sf9 and Sf12 and Trichoplusia “High-Five”) and in Xenopus oocytes (Oprian et al., 1987; Janssen et al., 1988; Khorana et al., 1988; Karnik et al., 1993; Kazmi et al., 1996). The highest expression levels of functional pigments, with addition of 11-cis retinal during culture or after isolation of the cells, were obtained in suspension culture of insect cells or specially adapted HEK293 cells, with yields up to 130 nmol/L, equivalent to ca 5 mg bovine rhodopsin per liter (Klaassen and DeGrip, 2000; Reeves et al., 2002). Even then the pigment accounts for maximally 5 percent of the total cellular membrane protein, and further purification is inevitable. Eventually, gene manipulation lent a helping hand and it has now become common practice to add a small sequence tag to the pigment c-DNA, encoding a short peptide sequence to easily identify and purify the expressed pigment. Two approaches have become the most popular in the type-2 rhodopsin field. One exploited the availability of a monoclonal antibody against the C-terminal octapeptide of bovine rhodopsin (Molday, 1989). This allows for highly selective immuno-affinity purification using a suitable detergent like DDM for solubilization (Ridge et al., 1995). By adding to or replacing the native C-terminal with this octapeptide, the resulting tagged protein can be comfortably isolated. The second approach involved extending the C-terminal with six to ten histidine residues (His6-tag to His10-tag), which upon solubilization with a suitable detergent allows metal affinity purification over a matrix containing immobilized Ni2+ or Co2+ complexes (Janknecht et al., 1991; Janssen et al., 1995). Both approaches are very effective with hardly any perturbation of expression level and functionality of the pigment (Reeves et al., 1999; Bosman et al., 2003). Nevertheless, if necessary, a short target peptide sequence for a selective proteolytic enzyme can be introduced in front of the purification tag to remove it after purification (Sarramegna et al., 2006). Most Opn1 and Opn2 pigments can be satisfactorily purified by either procedure (Vissers and DeGrip, 1996; Shirzad-Wasei and DeGrip, 2016; Katayama et al., 2017; Katayama et al., 2019). Some pigments from the other subsets have been difficult to solubilize or are too unstable in detergent solution to survive purification. The alternative option then is to transfer the protein into the stabilizing lipid environment of nanodiscs (Figure 4), which requires hardly any detergent (amphipol or SMA-type) or very brief exposure to a suitable mild detergent (MSP-type). Exploiting the sequence tag on the incorporated protein, the protein-nanodisc unit is then easily purified again by affinity chromatography (Shirzad-Wasei et al., 2015; Cai et al., 2017; Ganapathy et al., 2020).
TABLE 3.
Selected additional citations for the section “Heterelogous expression and purification”.
The opportunity to modify, bio-generate and purify type-2 pigments in sufficient quantities has given a tremendous boost to all mechanistic and functional studies. Analysis of the native proteins or binding pocket mutants, often in combination with 2H-, 13C- or F-labeling and/or chemical modification of retinal and/or with 15N- and/or 13C-labeling of protein residues or inserting modified amino acids, has provided a wealth of data, underpinning, extending and refining the information obtained from crystal structures (see next sections). Groundbreaking details of dark state structures have been excavated by biochemical (e.g. limited proteolysis, selective chemical modification, selective deuteration, atomic force microscopy, cryo-EM) and biophysical studies (e.g. FTIR and resonance Raman spectroscopy, solid-state NMR spectroscopy, EPR spectroscopy) (Table 3). This also fueled a large body of theoretical and in-silico efforts (molecular dynamics, quantum-chemical calculation and modeling) (Ryazantsev et al., 2019; Pedraza-González et al., 2020; Fujimoto, 2021; Mroginski et al., 2021; Church et al., 2022b). As a result of all these exertions, it has already been possible to construct a highly detailed picture of the dark state of bovine rhodopsin.
Type-1 Family
For the archaeal and bacterial type-1 rhodopsins, a heterologous expression host was more easily identified. Escherichia coli strains had already been developed for uncomplicated suspension culture, high productivity, low proteolytic activity and easy transformation. Plasmids with inducible promoters became available, and were further engineered with specific features, like producing the necessary enzymatic machinery to generate all-trans retinal from its precursor β-carotene (Kim et al., 2008). Nevertheless, in most cases just supplementing the cell culture with all-trans retinal together with inducing opsin expression or even after membrane isolation was sufficient to produce the full equivalent of the corresponding rhodopsin (Spudich et al., 2000; Ganapathy et al., 2015). In this way yields up to 20 mg/L have been reported (Ganapathy et al., 2015; Song et al., 2020). For some archaeal pigments, this straightforward approach only gave low yields and had to be adapted e.g. for bacteriorhodopsin itself (Bratanov et al., 2015; Tu et al., 2018). On the other hand, heterologous expression was more problematic for the eukaryotic type-1 rhodopsins, again because of their more complex posttranslational modification profile. Channelrhodopsins are commonly produced in yeast (Pichia pistoris), but successful production of eukaryotic type-1 pigments in insect and mammalian cell lines, Caenorhabditis elegans and Xenopus oocytes is also reported (Nagel et al., 2003; Bruun et al., 2015; Govorunova et al., 2017). An interesting new approach is using the trypanosome Leishmania tarentolae for over-expression (Volkov et al., 2017). For optogenetic applications (see below), functional production and targeting in a mammalian context is imperative, and often requires insertion of trafficking or targeting signals and/or sequence optimization to mammalian genetic code preferences.
The C-terminal His-tag has become the most popular option for purification of archaeal and eubacterial rhodopsins. For eukaryotic type-1 rhodopsins, several tags are used, including the His-tag, although the latter may sometimes interfere with particular electrophysiological or enzymatic analyses (Govorunova et al., 2021; Rozenberg et al., 2021; Tsunoda et al., 2021; Govorunova et al., 2022b).
Thanks to the powerful combination of the recombinant DNA toolbox with heterologous expression and purification making sufficient protein material available, an astounding repertoire of structural and functional data has also become available for the type-1 rhodopsins (Table 3). As a result, bacteriorhodopsin has become the best studied and fathomed membrane protein, with unprecedented insight into its structure and function (Ernst et al., 2014; Larkum et al., 2018; Nogly et al., 2018; Weinert et al., 2019). Next to that, the type-1 community has delivered prospects for a wealth of biotechnological and biomimical applications, far beyond any prognosis (see below).
Photochemical Properties
The initial rapid steps after photoactivation of type-1 and type-2 rhodopsins are quite comparable (Figure 8). Ultrafast photoisomerization of the chromophore leads to the first stable photoproduct within ps. This conversion is extremely efficient with quantum yields between 0.6 and 0.7 for type-2 pigments and varying between 0.3 and 0.7 for type-1 pigments and very low energy loss through fluorescence (Gozem et al., 2017). Often, this red-shifted photoproduct then thermally relaxes via spectrally distinguishable photo-intermediates within ms to a blue-shifted M(eta) intermediate, where the chromophore-binding Schiff base has become deprotonated through transfer of the proton to the direct counterion (Nakagawa et al., 1999; Hofmann, 2000; Tsukamoto and Terakita, 2010; Ernst et al., 2014; Govorunova et al., 2017). This explains the large blue-shift. In some type-1 pigments, a deprotonated M state is not formed, however a protonated L-like equivalent is observed (Spudich et al., 2014; Govorunova et al., 2017; Engelhard et al., 2018). The M or its L-like equivalent intermediate is the active state of the pigment, where the conformational changes in the protein evoke the subsequent cognate activity (grouping with cognate G protein or transducer, opening up an ion channel or vectorial ion pathway, regulating the enzymatic domain, etc.) (Table 4). At the M or L-like stage the type-2 and type-1 pathways take completely different directions.
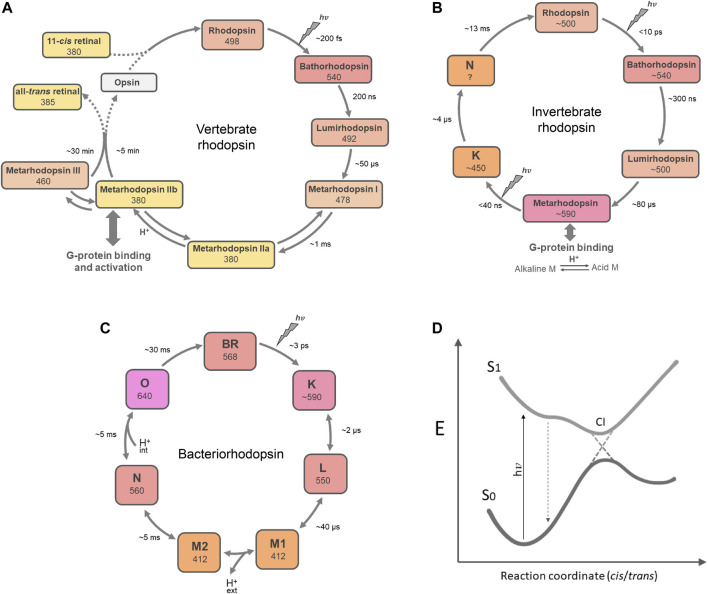
FIGURE 8.
Global presentation of the predominant photochemical pathways in the rhodopsin families. (A) Bovine rod rhodopsin as the archetype of the monostable type-2 pigments, (B) squid/fly visual pigment chimera, typical for the bistable type-2 pigments, and (C) bacteriorhodopsin (BR) as a prototype for the type-1 pigments. The “dark state” 11-cis, 15-anti chromophore configuration in type-2 pigments is photo-excited into all-trans. The all-trans chromophore configuration in type-1 pigments is photo-excited into 13-cis, 15-anti, which thermally relaxes, eventually returning to the ground state. The early photo-intermediates still contain a protonated Schiff base and relax thermally to deprotonated Meta II or M states. In proton pumps like BR this is accompanied by opening up proton pathways in the protein, while in most type-2 pigments binding of a G protein is initiated. At this stage, the pathways divert, as further explained in the text. Of course, here are exceptions: some type-2 pigments contain all-trans in the “dark state,” which is photoexcited into 11-cis, either to change activity or to release 11-cis retinal for regeneration of visual opsins (Table 2). Some type-1 pigments can also photo-generate 9-cis or 11-cis states with deviating photocycles and/or functions. (D) Simplified schematic of a conical intersection where the excited chromophore at the S1 energy surface can cross over to the S0 energy surface of the photoproduct. The S1 surface can contain thermal transitions, and in type-1 pigments the kinetics to reach and cross-over at the conical intersection also depend on the pKa of the direct counterion to the Schiff base (Chang et al., 2022).
TABLE 4.
Selected additional citations for the section “Photochemical properties”.
Type-2 Family
For the type-2 rhodopsins ultrarapid spectroscopy data are limited, and mainly available for Opn1, Opn2, and R-type pigments (Shichida et al., 1978; Shichida, 1990; Schoenlein et al., 1991; Vought et al., 2000; Imamoto and Shichida, 2014; Schnedermann et al., 2018). Generally speaking, two schemes have been identified: Monostable pigments eventually release all-trans retinal (all Opn1 and Opn2 rhodopsins, Figure 8A) following which the opsins require supplementation with retinal re-isomerized elsewhere to regenerate the original “dark” state. Bistable pigments (most other type-2 pigments investigated, Table 2) progress until a stable M-intermediate is reached (all-trans chromophore), that requires photo-isomerization to return to the original “dark” state (11-cis chromophore) (Figure 8B) (Hillman et al., 1983; Gärtner, 2000; Stavenga et al., 2000).
The photochemical profile of the monostable bovine rod rhodopsin has been explored in great detail. The native pigment and a variety of isotopically labeled and/or mutant pigments have been investigated by femtosecond optical spectroscopy and vibrational and NMR spectroscopy. These studies have revealed intimate details on the kinetics, conformational changes in the chromophore and surrounding H-bonded networks with constrained water molecules, protein-chromophore interplay and Schiff base (de)protonation (Table 4). Overall protein conformational changes have been elucidated by fluorescence, ESR and NMR spectroscopy and TR-WAXS (DeGrip et al., 1999; Kusnetzow et al., 2006; Alexiev and Farrens, 2014; Malmerberg et al., 2015; Van Eps et al., 2017; Smith, 2021). Crystal structures have been resolved for all photo-intermediates and present a broad structural basis (Table 4). The power of theoretical and quantum-chemical calculations has grown immensely, laying a strong foundation for electronic and energetic elements of the process, in particular (Schapiro et al., 2011; Gozem et al., 2017; Schnedermann et al., 2018; Agathangelou et al., 2021; Nikolaev et al., 2021).
A very effective combination of selectively labeled chromophore with femtosecond spectroscopy and advanced quantum chemical computation resolved many remaining issues in the photoisomerization process of bovine rhodopsin (Schnedermann et al., 2018). The global picture has arisen that after photo-excitation of the chromophore into the Franck-Condon state it rapidly relaxes along a barrierless trajectory on the potential surface to a minimal energy conical intersection (Figure 8D). Here, productive resonance of the electronic wave packet at the excited state potential surface with torsional and HOOP vibrational modes in the twisted C10-C13 segment of the 11-cis chromophore, can prime very effective cross-over to a ground state energy surface, generating a hot all-transoid state (photorhodopsin) within tens of fs (Johnson et al., 2015). This relaxes thermally in about 200 fs into the photoproduct bathorhodopsin, which contains a still highly twisted all-trans chromophore, but is stable below 130 K (Yoshizawa and Wald, 1963). In free retinal, photoexcitation results in formation of several isomers (predominantly all-trans, 13-cis, 9-cis, and 11-cis), but in rhodopsins this conversion is remarkably selective from 11-cis to all-trans. This is clearly facilitated by the constraints of the binding site and the twist in the C10-C13 segment of the chromophore (Bismuth et al., 2007; Weingart, 2007; Schnedermann et al., 2018).
At room temperature, the ca 35 kcal of excitation energy stored in bathorhodopsin (Cooper, 1979) drives further relaxation via several intermediates until the metarhodopsin IIa-IIb equilibrium is reached within ms. This relaxation process subtly rearranges chromophore, protein residues and H-bonded networks up to the metarhodopsin stage, where the Schiff base transfers its proton, the counterion and another Glu at the intracellular side of the protein become protonated and an interhelical activity switch reshuffles helical segments to open up binding residues for the G-protein (Hofmann, 2000; Vogel et al., 2007; Vogel et al., 2008; Pope et al., 2020). The chromophore is subsequently slowly released via hydrolysis of the Schiff base to generate the nearly inactive apoprotein opsin (Wald, 1953; Rothschild et al., 1987; Jastrzebska et al., 2011). In vivo the active state is rapidly inactivated through phosphorylation and arrestin binding, however, which blocks activation of the G protein (Ranganathan and Stevens, 1995).
The photochemical profile of other monostable pigments (human rod rhodopsin, several cone pigments) has been investigated to much less depth, but is quite comparable to the bovine rod pigment (Barry and Mathies, 1987; Kusnetzow et al., 2001; Hofmann and Palczewski, 2015; Kazmin et al., 2015). However, the kinetics differ somewhat. For instance, the investigated cone pigments show more rapid kinetics in most steps (Imai et al., 1997; Vissers et al., 1998; Chen et al., 2012; Sato et al., 2012). Ultra-violet absorbing cone pigments may be more complex, as photoisomerization is accompanied by protonation of the Schiff base (Kusnetzow et al., 2004; Mooney et al., 2012).
The photochemical profile of bistable pigments, investigated thus far (squid, octopus and some insect pigments), follow a scheme similar to the monostable pigments up through formation of the M-intermediate and with comparable kinetics (Figure 8B) (Gärtner, 2000; Stavenga et al., 2000; Vought et al., 2000; Murakami and Kouyama, 2015). It is reported that in cephalopods the M-intermediate in fact forms a pH-dependent equilibrium between a protonated (acid M) and a deprotonated state (alkaline M). This involves the Schiff base of the chromophore, and the alkaline M is strongly blue-shifted (Liang et al., 1994; Vought et al., 2000). Photo-reisomerization of the M state to the original “dark” state is again quite efficient with a quantum yield around 0.4 (Stavenga et al., 2000).
Type-1 Family
The “dark” state of type-1 pigments contains a chromophore with the all-trans, 15-syn configuration (Figure 1). Rapid spectroscopy has been performed on quite a number of type-1 pigments, and the global scheme is quite similar to that of bacteriorhodopsin (Figure 8C). However, the kinetics of the slower steps (M and subsequent ones) and thereby the overall cycle time can vary considerably from ms up to minutes (Rozenberg et al., 2021; Tsunoda et al., 2021; Broser, 2022; Nagata and Inoue, 2022).
Out of all rhodopsins the photochemistry of BR is understood in most detail (Wickstrand et al., 2015; Nango et al., 2016). Femtosecond XFEL crystallography has even revealed very early responses to photoexcitation of the chromophore (Nogly et al., 2018). The adjacent protein residues and water molecules already react to the charge delocalization in the excited chromophore before the isomerization is initiated (Tahara et al., 2019a). During the isomerization process more of the protein environment becomes involved while the chromophore rapidly relaxes along a 2-state trajectory on the excited state potential surface to a conical intersection, where it effectively crosses in ca 500 fs over to a ground state energy surface into a “hot” transient hybrid state (J) and then relaxes thermally in about 3 ps into the photoproduct K, which contains a still significantly twisted 13-cis, 15-anti chromophore, but is stable below 150 K (Lanyi, 2004). Here, a major driving force is the elongation of the C13-C14 bond in the excited state in combination with electrostatic re-arrangement and weakening of the hydrated H-bonded network in the Schiff base region. At room temperature, the ca 15 kcal of excitation energy stored in K (this can be higher in sensory rhodopsins) (Birge et al., 1991; Govorunova et al., 2017; Rozenberg et al., 2021) drives further relaxation via the spectrally distinguishable L intermediate until the M states are reached in ca 50 µs. This relaxation process again subtly re-arranges chromophore, protein helices and H-bonded networks up to the M states, where the Schiff base transfers its proton via a water molecule to the counterion and the hydrated H-bonded network opens up a proton gateway to the extracellular membrane surface. The M-states thermally decay via several intermediates in tens of ms to the BR ground state, during which the Schiff base is reprotonated via proton transfer from residue Asp96, a proton is taken up from the intracellular surface and the chromophore is re-isomerized to the all-trans, 15-syn configuration. In fact, all-trans is the most stable configuration for free retinal (Ganapathy and Liu, 1992). Nevertheless, in some archaeal rhodopsins including BR the chromophore slowly enters an all-trans, 15-anti ↔ 13-cis, 15-syn equilibrium when stored in the dark (dark adaptation). The latter chromophore is photo-excited in the light and via a separate non-productive photocycle rerouted to the ground state BR (Smith et al., 1989; Oesterhelt et al., 1991). In channelrhodopsins the opposite phenomenon is observed, where prolonged illumination reduces the activity, since an equilibrium between pigments with an all-trans, 15-anti and a 13-cis, 15-syn chromophore configuration is generated (light-adaptation with partial desensitization) (Bruun et al., 2015; Kuhne et al., 2019; Rozenberg et al., 2021; Govorunova et al., 2022b).
Using serial synchrotron crystallography, the slower conformational changes from 5 to ca 40 ms were recorded in the BR photocycle and involve small α-helical rearrangements, chromophore re-isomerization and proton uptake, ending in formation of the ground state (Weinert et al., 2019). A very recent study using advanced high-resolution atomic force spectroscopy at the single-molecule level investigated the BR photocycle after M formation (Perrino et al., 2021). It was concluded that a cytoplasmic gate for proton uptake opens up at about 3 ms after photo-excitation lasting for about 14 ms. Surprisingly, this same study observes a “black-out period” of tens of ms before a recycled ground state can be photo-reactivated. This uncovers a very interesting new phenomenon reminiscent of comparable nonresponsive states in animal voltage-regulated channels (Armstrong, 1992). Meanwhile, XFEL studies have also been performed on other ion pumps and channels. A femtosecond XFEL study of the sodium-pumping rhodopsin from Krokinobacter eikastus (KR2) again observed photo-isomerization of the chromophore to start in the femtosecond range and completed within 2 ps (Skopintsev et al., 2020). Changes in the local structure of the binding site and early conformational changes in the protein backbone are observed in the early nanosecond range. Further subtle rearrangements result in Schiff base deprotonation in µs and in the early ms range a gate opens up and transient binding of a Na + ion in the vicinity of the Schiff base is observed with release within 20 ms. A femtosecond XFEL study of the chloride pump from the flavobacterium Nonlabens marinus follows the conformational adaptations between 1 and 100 ps after photo-excitation (Yun et al., 2021). It shows the final rearrangements of the chromophore to the 13-cis configuration within 50 ps, together with the dynamics of the hydrated H-bonded network and deformations in the local α-helical elements. Following chromophore isomerization the chloride ion first dissociates from the protonated Schiff base and then starts to diffuse away. Additional molecular details of the interactions and trajectory of the chloride ion are provided by recent ps up to ms studies using time-resolved serial crystallography in combination with spectral and theoretical analysis (Hosaka et al., 2022; Mous et al., 2022). An XFEL study of the channelrhodopsin chimera C1C2, that photochemically behaves like ChR1, investigated the photo-induced conformational changes from 1 µs to 4 ms (Oda et al., 2021). Photo-isomerization induces a kink in the chromophore structure, triggering shifts in the retinal binding lysine residue and TM7, starting at around 1 µs and increasing during formation of the M-state up to 4 ms. This induces small lateral shifts of the chromophore and in TM7 and TM3 at around 50 µs. It is postulated that these rearrangements forebode the subsequent opening of the gates in the cation channel pore, although these were not observed in the crystal. The XFEL and serial crystallography studies beautifully illustrate the powerful but subtle design and the broad potential of the photo-driven nanomachinery. Less detailed studies basically show a similar pattern (Table 4). Subtle differences in early kinetics and conformational adaptation in chromophore and adjacent protein elements following photo-excitation are observed in the ultrarapid studies. A cautious interpretation could be that the structure of the hydrated H-bonding network in the complex counterion is an important roadmap for the light-triggered protein activity, which also depends on the pKa of the direct counterion (Hontani et al., 2017b; Oda et al., 2021; Chang et al., 2022).
In this context it should be realized that crystal structures have their limitations (García-Nafría and Tate, 2020; Guo, 2020). Detergent exposure may affect elements of the protein structure, and the crystal will certainly constrain larger conformational alterations in the protein, which may occur in the slower phase of the photocycle (Weinert et al., 2019; Oda et al., 2021; Govorunova et al., 2022b). Hence, it would be preferable to study the slower photocycle phases with experimental approaches that can handle membrane-bound systems as shown in Figure 4, like time-resolved AFM, cryo-EM and vibrational spectroscopy.
The general scheme for the photocycle of BR (Figure 8C) also holds for other type-1 pigments, though the kinetics after M formation can vary significantly (Wand et al., 2013; Tahara et al., 2015; Han et al., 2020; Smitienko et al., 2021). The decay is much slower for sensory rhodopsins, enzyme-rhodopsins and heliorhodopsins, possibly since longer interaction with their cognate partner is required for regulated signal transduction. In fact, some sensory rhodopsins and enzyme-rhodopsins exhibit a bistable photocycle (Kawanabe et al., 2007; Broser et al., 2020) and proton transfer to the counterion may not occur (Bergo et al., 2006).
Bioengineering
This section samples the impressive expansion in the field of rhodopsins bioengineered by creative exploitation of their design principles. Often, similar strategies are utilized for both type-1 and type-2 pigments, and therefore they are clustered together in the following subsections.
Shifts in Spectral And/or Functional Properties
Chromophore
Very early on in the 1960s, it was realized that the beautiful design and versatility of rhodopsins could be studied and exploited by modifying the chromophore and changing the spectral properties (Blatz et al., 1969; Kropf et al., 1973). Since protein modeling was not really established at that time, this led to a surge of trial-and-error synthetic efforts to test a large number of retinal analogs on their ability to incorporate into the binding site and to modulate spectral and/or functional properties (Balogh-Nair and Nakanishi, 1982; Derguini and Nakanishi, 1986; Liu and Asato, 1990; Crouch et al., 2002). Initially, this was mainly performed on bovine rod rhodopsin and bacteriorhodopsin, which were easily isolated in sufficient quantities. In this way both bathochromic and hypsochromic spectral shifts up to ca 80 nm could be realized, frequently with retardation of photo-kinetics or total loss of function. For instance, using “locked” retinals (blocking functional photo-transformations) it was confirmed that the photo-isomerization process was essential for the functionality and that the ring-polyene chain connection was 6-s-cis in type-2 rhodopsins and 6-s-trans in type-1 pigments (Figure 1) (Crouch et al., 1984; Fukada et al., 1984; Harbinson et al., 1985; van der Steen et al., 1986; DeGrip et al., 1990; Bhattacharya et al., 1992a; Ganapathy et al., 2015). Also, the remarkable observation was made with bovine opsin, that next to the 11-cis and 9-cis retinal, also the 7-cis, 7, 9-dicis, and 7, 9, 13-tricis retinal isomers could form a functional pigment, inducing a 40–50 nm blue-shift but reducing thermostability (DeGrip et al., 1976; Liu et al., 1984). In general, it turned out that the bovine opsin binding pocket could better accommodate more voluminous modifications than the bacterio-opsin pocket, suggesting a more constrained character for the latter one. This was later validated in 3-D structures, but other type-1 pigments or photo-intermediates can be less selective (Popp et al., 1993; Inoue et al., 2012; Mori et al., 2013). New analogs are still frequently generated, in particular because recombinant production of mutated opsins modifies the binding pocket constraints. In addition, protein modeling has become more straightforward and for optogenetics larger spectral shifts and other functionalities like higher photosensitivity or higher fluorescence yields are in demand (see below).
Protein Joins In
Once recombinant DNA technology allowed the production of functional opsins in heterologous hosts, one could use this technology to adapt the intrinsic potential of opsins to one’s need and design. Combining synthetic retinal design with recombinant DNA opsin modification opened up a marvelous toolbox to investigate the structure and functional mechanism of rhodopsins as well as to probe new functionalities and applications. This trend is evolving more and more rapidly. Initially, binding site residues were modified to probe their contribution to the packing, stabilization and spectral tuning of the chromophore (Khorana et al., 1987; Sakmar et al., 1989; Nathans, 1992; Sakmar and Fahmy, 1995; Giesbers et al., 2007). A salient example is the accumulating evidence for type-1 pigments, that three positions around the retinal chromophore, corresponding to L93, P186 and Ala215 in BR, function as natural spectral-tuning modules that systematically shift the absorbance spectrum of the chromophore without affecting molecular function (Table 5). This further inspired detailed analysis with modified and/or labeled retinals (13C, 2H, F) and protein residues (13C, 15N, F, azido, spin labels) using fluorescence, vibrational, EPR and NMR spectroscopy (Table 5). This profited from as well as steered development of sophisticated theoretical and in-silico procedures, like DFT, QM/MM and molecular dynamics (Altun et al., 2008; Collette et al., 2018; Del Carmen Marín et al., 2019b; Dokukina et al., 2019; Pieri et al., 2019; Shao et al., 2020; Nikolaev et al., 2021; Scholz and Neugebauer, 2021; Shen et al., 2021; Pedraza-González et al., 2022). All these elements have already profoundly deepened our insight into the structure and mechanism of bovine rod rhodopsin and bacteriorhodopsin, the frontrunners of type-2 and -1, respectively. However, more members are following up. Some recent conspicuous examples are mentioned in the next section.
TABLE 5.
Selected additional citations for the section “Bioengineering”.
Conversion
The manipulations described in the previous subsection frequently revealed surprising conversions in activity profile, exemplifying the versatile design principle of the rhodopsins (Kaneko et al., 2017). An interesting example is presented by the Nonlabens marinus inward chloride pump NMR-3 and the Krokinobacter eikastus sodium exporter KR2 (Hosaka et al., 2016; Yun et al., 2020). With only 35% sequence identity, the crystal structures are remarkably similar, but the gating residues for Cl− and Na+ are located at the opposite site of the membrane (Kato et al., 2015a; Hosaka et al., 2016; Kovalev et al., 2019; Yun et al., 2020). Another example is the huge mutagenesis effort that converted a thermophilic rhodopsin into the best thermally stable rhodopsin available to date, while retaining pump activity (Yasuda et al., 2022). On the other hand, selective mutations in the opsin could convert BR into an inward chloride pump, the sodium pump KR2 into a selective light-driven cation channel, the proton pumps Archaerhodopsin-3 (AR3) and Coccomyxa subellipsoidea rhodopsin (CsR) into light-driven proton channels, and the proton pump GR from Gloeobacter violaceus into a fluorescent chloride sensor (Sasaki et al., 1995; Brown et al., 1996; Inoue et al., 2015; 2016; Fudim et al., 2019; Vogt et al., 2019; Tutol et al., 2021). Alternatively, a cyanobacterial chloride pump could be converted into a proton pump (Hasemi et al., 2016; Kikukawa, 2021). Novel retinal A1 and A2 analogs with an elongated polyene chain (10 instead of 9 carbons) still could incorporate into the binding pocket of the ReaChR channelrhodopsin inducing red-shifts up to ca 30 nm (Okitsu et al., 2020). However, when tested upon AR3, one A2 analog induced a 41 nm blue-shift and again converted it into a light-driven proton channel (Takayama et al., 2018). Another novel retinal analog (MMAR, Figure 5.) smoothly incorporated into the binding pocket of the proton pump Green Proteorhodopsin (GPR), inducing a 47 nm red-shift, but when combined with a Phe →Ser mutation near the binding pocket, an unprecedented 200 nm red-shift was observed (Ganapathy et al., 2017). This retinal analog not only maintains some pump activity under near-infrared illumination (700–900 nm region; NIR), but also induces strong fluorescence emission in the NIR, probably emitted in the first picoseconds after excitation (Hontani et al., 2018; Mei et al., 2018; 2020). Proton-pumping rhodopsins in several eubacteria (XR, GR and TR) harbor a carotenoid derivative (salinixanthin) close enough to act as an antenna and transfer electronic excitation to the retinal (Balashov et al., 2010; Imasheva et al., 2011; Misra et al., 2019; Jana et al., 2020). This combination significantly broadens the spectral sensitivity of the rhodopsins for blue wavelengths, and the carotenoid binding option can also be introduced into other pigments (Anashkin et al., 2018). Attempts have also been made to generate chimeric pigments with combined functionality. The earliest example was a BR mutant containing loops of rod rhodopsin being able to weakly activate the G protein (Geiser et al., 2006). This concept in BR was further developed (Sasaki et al., 2014; Kaneko et al., 2017; Yoshida et al., 2017) and also found wider application in other rhodopsins (Kaneko et al., 2017). Chimeras could be produced between type-1 and type-2 pigments, often with shared properties and variable potential for G protein activation (Kojima et al., 1996; Geiser et al., 2006; Airan et al., 2009; Nakatsuma et al., 2011; Sasaki et al., 2014; Bedbrook et al., 2017a; Kaneko et al., 2017; Hickey et al., 2021). A remarkable example is that the C1C2 chimera could be crystallized and a high-resolution crystal structure obtained long before its “parent” channelrhodopsins ChR1 and ChR2, (Kato et al., 2012). In a sequel, new chimeric channelrhodopsins with better performance were generated using structure-guided recombination (Bedbrook et al., 2017a). The chimeric concept has also resulted in type-2 recombinants with variable success (Kojima et al., 1996; Giesbers et al., 2007; Hickey et al., 2021).
These selected examples, along with some more references collected in Table 5, already give an impression of the fabulous potential and prospects of the rhodopsin clan. The most impressive flux, however, is noticeable in the optogenetics field.
Optogenetics
Neuronal activity and circuitry are of the essence for multicellular life. Much effort is dedicated to studying activity regulation and circuitry in complex tissues like the brain. This used to be a highly challenging electrophysiological operation, requiring invasive electrodes and precise surgical location. Once it was realized, that rhodopsins could be properly expressed in animal tissues with genetic targeting to specific neurons using selective promoters, it became possible to monitor and regulate neuronal activity by light using endogenously expressed rhodopsins (Boyden et al., 2005). This led to an explosion of research activity in a new field, coined optogenetics (Deisseroth, 2010, 2015; Rost et al., 2017; Kandori, 2020; Friedman, 2021). Initially, only type-1 rhodopsins were considered, since ion fluxes can directly modulate neuronal activity. Also, all-trans retinal is intrinsically available in animal cells and type-1 pigments complete a full photocycle.
In a first breakthrough, a cation-selective channelrhodopsin originally identified in Chlamydomonas reinhardtii termed ChR2 (Nagel et al., 2003) was exploited. ChR2 was shown to elicit action potentials in cultured neurons upon illumination (Boyden et al., 2005; Cardin et al., 2010; Klapoetke et al., 2014; Berndt et al., 2016; Deisseroth and Hegemann, 2017). This domain rapidly expanded into ion pumps, which can activate or silence neuronal activity (Chow et al., 2010). Simultaneously, pigments were modified to change spectral range, increase current output, alter photo- and response kinetics, improve membrane targeting, etc. (Lin et al., 2013; Kushibiki et al., 2014; Kato et al., 2015b; Brinks et al., 2016; Govorunova et al., 2017; Cho et al., 2019; Krol et al., 2019; Kojima et al., 2020b; Gong et al., 2020). Eventually, enzyme-rhodopsins as well as bistable type-2 pigments also entered the field, being able to modulate cellular metabolic processes up to gene expression (Mukherjee et al., 2019; Karapinar et al., 2021; Mahn et al., 2021; Rodgers et al., 2021; Tsunoda et al., 2021; Vierock et al., 2021). Bistable type-1 and -2 pigments allow further control, since their activity is triggered by illumination, but ends near the M(eta) stage, which can be photoreversed by illumination in another spectral range (Sheves and Friedman, 1986; Koyanagi and Terakita, 2014; Mederos et al., 2019; Eickelbeck et al., 2020).
A second breakthrough came with the discovery that the intensity of the fluorescence emission of the proton pumps GPR and AR3, be it quite weak, is modulated by the membrane potential (Kralj et al., 2011; Saint Clair et al., 2012b; Kralj et al., 2012). This triggered another burst of research dedicated to improve the voltage sensing of these pumps (minimizing pump activity, shifting spectral range, improving quantum yield, voltage sensing potential, temporal resolution, etc.) by a range of technologies like directed and scanning mutagenesis, multidimensional directed evolution, library screening and machine learning (McIsaac et al., 2014; Engqvist et al., 2015; McIsaac et al., 2015; Abdelfattah et al., 2016; Karasuyama et al., 2018; Kojima et al., 2020a). This was initially mostly performed on AR3, generating a whole family of mutants with different response characteristics (Quasar1 to 3, pa-Quasar3, Novarch, Archon1 and 2, Arch-EEN, Quasar6, Somarchon to name a few) (Piatkevich et al., 2019; Chien et al., 2021). The fluorescence of these voltage sensors most likely originates in late-stage photo-intermediates (Maclaurin et al., 2013). The introduced mutations may even result in a complex bistable photo-equilibrium between a fluorescent and a non-fluorescent state (Mei et al., 2021; Penzkofer et al., 2021). Meanwhile a host of additional voltage sensors have been developed. Next to optimized rhodopsins and chimeric rhodopsin fusions, fusion proteins of light-sensitive opsin cores with other fluorophores, often GFP derivatives or synthetic dyes, and of other voltage sensors with fluorescent rhodopsins have become popular (Bando et al., 2019; Kannan et al., 2019; Lee et al., 2019; Berglund et al., 2020; Zhang X. M. et al., 2021).
Further control has been sought by combining optogenetics with classical electrophysiology (electro-optogenetics) or combining voltage sensors and neuronal activators and/or silencers both based on rhodopsins (all-optical electrophysiology) (Hochbaum et al., 2014; Afshar Saber et al., 2018; Sridharan et al., 2022). In the latter case, it is important to separate the spectral sensitivities to allow selective control and avoid optical cross-talk. In addition, much effort has been put into shifting the spectral range of the optogenetic tools and sensors as far as possible into the NIR, since NIR radiation penetrates much further into the mammalian brain (up to cm compared to several mm for e.g. blue-green light) (Larkum et al., 2018; Govorunova et al., 2020; Broser, 2022). For this purpose, mutagenesis of far-red absorbing rhodopsins like Crimson and CrimsonSA would be a good starting point (Oda et al., 2018). Another option is the novel channelrhodopsin ChRmine, which has quite unusual properties, including a trimeric structure similar to BR (Marshel et al., 2019; Kishi et al., 2022). A very fascinating example is NeoR, a subunit in the heterodimeric rhodopsin-cyclase from the fungus Rhizoclosmatium globosum. NeoR is quite exceptional, as it harbors three carboxyl residues near the chromophore and has an absorbance maximum at 690 nm with strong fluorescence emission at 707 nm (Broser, 2022). Other gateways could include special optical technologies or local NIR-converting nanoparticles and two-photon spectroscopy, which are more complicated (Sneskov et al., 2013; Chen, 2019; Matarèse et al., 2019; Yu et al., 2019; Adesnik and Abdeladim, 2021; Lehtinen et al., 2022), or designing special retinal analogs. The latter was quite successful, shifting absorbance maxima up to ca 750 nm with fluorescent emission around 800 nm using merocyanine analogs or MMAR (Figure 5) (Derguini et al., 1983; Hoischen et al., 1997; Liu and Asato, 2003; Herwig et al., 2017; Hontani et al., 2018; Mei et al., 2020). The strong red-shift in these analog chromophores, as well as in the A1 chromophore in NeoR is contributed to extensive delocalization of the positive charge from the protonated Schiff base over the polyene element (Figures 1, 5) (Liu and Asato, 2003; Lutnaes et al., 2004; Ganapathy et al., 2019; Broser, 2022). This will strongly reduce the energy gap between the ground and first excited state. Incorporation of retinal A2 into NeoR-opsin already effectuates a further 69 nm red-shift (Broser et al., 2020). Hence, it would be very interesting to investigate whether the combination of NeoR-opsin or mutants with bathochromic analogs like MMAR would even further red-shift the absorbance band and increase the gap with the emission band. Optogenetic application, however, requires invasive administration of the retinal analog and may need transient depletion of the endogenous A1.
So far, the field of optogenetics has progressed spectacularly, from neuron and brain slice cultures, up to intact animals including insects, C. elegans, mice and macaques (Bi et al., 2006; Flytzanis et al., 2014; Inagaki et al., 2014; AzimiHashemi et al., 2019; Babl et al., 2019; Piatkevich et al., 2019; Gong et al., 2020; Wagner et al., 2021; Wright et al., 2021) and is being extended to human disease models (Wright et al., 2017; Williams et al., 2019; Córdova et al., 2021; Fougère et al., 2021; Lindner et al., 2021). Future prospects will be touched upon in the next section.
Cell Factories
While rhodopsins drive important physiological processes in prokaryotes and eukaryotes, and can contribute significantly to the energy requirement of their hosts, implementing this into biotechnological resources like cell factories has not yet developed very far (Walter et al., 2010). E. coli can profit from expression of a rhodopsin proton pump (Martinez et al., 2007; Choi et al., 2014; Na et al., 2015; Wang et al., 2015; Kim et al., 2017; Song et al., 2020). However, the extent to which this can for instance support production of useful consumables or commodity chemicals needs to be established. Cyanobacteria like Synechocystis sp. PCC6803 and Synechococcus already exploit chlorophyll-based oxidative photosynthesis to gather solar energy and are under intense investigation as cellular factories (Wijffels et al., 2013; Angermayr et al., 2015; Du et al., 2018; Knoot et al., 2018; Carpine et al., 2020). They do not have an endogenous opsin, but do produce all-trans retinal and can serve as a heterologous host for expression of rhodopsin proton pumps (Chen et al., 2016b; Chen et al., 2017; Chen et al., 2019a). Expression of these pumps was considered as a potential extra energy source, but the contribution of these pumps towards cellular energy production appeared to be limited (Chen et al., 2019b). This may be due to the metabolic constraint of proton fluxes, and/or to the chlorophylls and carotenoids absorbing much of the incoming radiation up to ca 650 nm (the PAR region). Attempts to express the GPR F234S mutant in combination with the retinal analog MMAR were successful in generating a proton pump absorbing in the 700–800 nm range, outside the PAR region. However, this still did not generate sufficient additional energy due to the lower pump activity of this mutant and failed to sustain bacterial growth under NIR illumination (Chen et al., 2018).
Prospects
A major asset of the rhodopsin family is the impressive versatility of the design principle: a relatively simple photosensitive ligand, constrained to allow selective photoisomerization with a high quantum yield, triggering subtle but effective conformational changes in the protein opening up specific binding sites or ion transport pathways.
Genome mining will undoubtedly discover new type-1 and type-2 or related variants, especially considering the still vast reservoir of unexplored microbial and invertebrate life forms. For instance, the apparent non-photic activity of (rhod)opsins in certain physiological conditions (thermo-, mechano- or chemo-sensing) may add a new chapter to this family saga (Leung and Montell, 2017; Katana et al., 2019; Baden et al., 2020; Fleming et al., 2020; Hasegawa et al., 2020; Mei et al., 2020; Zabelskii et al., 2021; Feuda et al., 2022). Next to that, insight into the effect of pathological mutations will become an ever more important asset in medical diagnostics and potential treatment. This has already been widely explored in the case of rod rhodopsin and retina-degenerative diseases, (Athanasiou et al., 2018). Expression and functional and structural characterization of new (rhod)opsins or mutants still involves an elaborate effort, but this may be considerably mitigated soon.
The phenomenal progress in artificial intelligence and machine learning already culminated in the design of software packages like RoseTTAFold and Alphafold, that are quite successful in predicting the protein fold from the primary sequence (Humphreys et al., 2021; Jumper et al., 2021). Considering the respectable number of crystal structures for type-1 pigments and G protein-coupled receptors already obtained, this in silico approach will be of invaluable help to close in on the 3-D structures of rhodopsin sequences identified to date, as well as those yet to be identified. A similar track is conceivable for the assessment of spectral and functional properties. Experimental analyses, in combination with in-silico techniques like DFT, machine learning and quantum-chemical computing already made big strides in establishing the contribution of individual opsin residues and water molecules to the spectral tuning of rhodopsins (Kato et al., 2015; Bedbrook et al., 2017b; Karasuyama et al., 2018; Bedbrook et al., 2019; Nikolaev et al., 2020; Inoue et al., 2021; Yang et al., 2022). However, this approach always requires 3-D information. It would be very desirable to build in additional functionalities, e.g. to predict an approximate absorbance maximum, into the sequence-to-structure software packages. This could then be easily expanded towards predicting the effect of mutations and the fit and effects of retinal derivatives or even more distant chromophores. Suggestions for functionality (specific pump or channel, enzymatic domains, thermal stability) probably could also be in reach, though mechanistic details (photoisomerization process, quantum yield of isomerization or fluorescence emission, early conformational changes) may be aiming too high.
Such developments will be a goldmine for optogenetics. Rapid prediction of spectral and functional properties and optimal targeting of desired mutants would be very valuable. Likewise, assessment of new constructs like chimeric pigments, fused monomers, oligomeric assemblies, enzyme activating pigments, new signaling partners and the like can be set up in silico and will require much less experimental justification (Sasaki et al., 2014; Abdelfattah et al., 2020). This would undoubtedly be accompanied by further physiological expansion of optogenetic tools. A wider spectral range of neuronal activity modulators and voltage sensors together with improved optics will increase the scope for (all)-optical electrophysiological characterization of neural circuitry, also lending insight into neuronal function (and dysfunction) in the brain (Villette et al., 2019; Guimarães Backhaus et al., 2021; Sharma et al., 2021; Zou et al., 2021; Prakash et al., 2022; Sridharan et al., 2022; Tan et al., 2022). Other important medical targets may also arise using optogenetics to correct physiological defects and address pathological conditions, where first steps have already been taken (Braun et al., 1995; Deubner et al., 2019; Shen et al., 2020; Acharya et al., 2021; Cokic et al., 2021; Kathe et al., 2021; Gilhooley et al., 2022; Sun et al., 2022).
Several concepts to utilize rhodopsins in bioelectronic and biomimic nanotechnology have already been attempted, but did not yet really come to maturation (Khodonov et al., 2000; Kuang et al., 2014; Hirschi et al., 2019; Aprahamian, 2020; Shim et al., 2021). With the rapidly growing insight in the structural and mechanistic potential of the rhodopsin pigments, this is expected to change at short notice. So far, electro-optical phenomena have been investigated in 2D crystals, lipid films and other matrices (Oesterhelt et al., 1991; Miyasaka et al., 1992; Hong, 1994; Wagner et al., 2013; Zhao et al., 2015; Ji et al., 2017; Gruber et al., 2022). With help of the above mentioned software packages, the design of specific constructs with high performance and stability under the system’s conditions will be facilitated.
This would also be the case for application in cell factories. The most interesting and rewarding application in this respect is the notion of “synergistic photosynthesis,” the combination of chlorophyll-based oxidative photosynthesis with retinal-based phototrophy, using high-performance rhodopsin proton pumps absorbing in the NIR (Chen et al., 2016a; Chen et al., 2019b). This will also require adaptation of proton regulation in the host cell or introduction of special cellular organelles containing the pump and an ATP-synthase. In eukaryotic cells like algae or fungi, targeting of a proton pump to mitochondria to increase ATP levels for production of commodity chemicals under selected conditions can be further developed (Hoffmann et al., 1994; Hara et al., 2013; Tkatch et al., 2017; Imai et al., 2019; Berry and Wojtovich, 2020). In general, designing highly active ion pumps absorbing in the 700–800 nm region, i.e. outside the PAR region, is essential for productive “synergistic photosynthesis.” Again, artificial intelligence can be a decisive factor here.
Epilogue
In roughly 10 years, the rhodopsin field has reached a century’s worth of experimental investigation. In this review, we have mainly touched upon the surface of the phenomenal development in this field, somewhat like molecular force microscopy. In the coming 10 years we expect its expansion to continue and to eventually require an at least ten-volume book series for full documentation. By that time, we will hopefully have a better understanding of how a selection of twenty amino acids can lead a membrane protein domain of 300–400 amino acids surrounding a small chromophoric group to such mechanistic versatility.
Acknowledgments
WdeG acknowledges the many Master’s and PhD students, postdocs, visiting researchers and collaborating colleagues for their contribution to the work in his research labs. Special thanks go to Petra Bovee-Geurts and Jenny van Oostrum (Radboudumc) for their long-term technical assistance and to Ken Rothschild (Boston University), Johan Lugtenburg and Huub de Groot (Leiden University) and Giel Bosman (Radboudumc) for a lasting amalgamation of science and friendship. The writing of this review was financially supported by Leiden University, Delft University and Radboudumc.
Author Contributions
WdeG conceptualized and wrote the first draft of the manuscript. SG elaborated on sections of the manuscript and prepared the figures. Both authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AFM, Atomic force microscopy; AR3, Archaerhodopsin-3; BR, Bacteriorhodopsin; C1C2, chimera between channelrhodopsin-1 and -2; Cryo-EM, Cryo-electron microscopy; CTAB, Cetyltrimethylammonium bromide; DDM, Dodecylmaltoside; DFT, Density functional theory; DPC, Dodecylphosphocholine; EPR, Electroparamagnetic resonance; FTIR, Fourier-transform infra-red; GR, Gloeobacter violaceus rhodopsin; LDAO, Lauryldimethylaminoxide; MSP, Membrane scaffold protein; NG, Nonylglucoside; OG, octylglucoside; PAR, Photosynthetically active region; PM, plasma membrane; RGR, RPE-retinal G protein-coupled receptor; RPE, Retinal Pigment Epithelium; SMA, Styrene-maleic-acid-copolymer; TR, Thermophilic rhodopsin; TR-WAXS, Time-resolved wide-angle X-ray scattering; XFEL, X-ray free electron laser.
References
- Abdelfattah A. S., Farhi S. L., Zhao Y., Brinks D., Zou P., Ruangkittisakul A., et al. (2016). A Bright and Fast Red Fluorescent Protein Voltage Indicator that Reports Neuronal Activity in Organotypic Brain Slices. J. Neurosci. 36, 2458–2472. 10.1523/jneurosci.3484-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelfattah A. S., Valenti R., Zheng J., Wong A., Chuong A. S., Hasseman J. P., et al. (2020). A General Approach to Engineer Positive-Going eFRET Voltage Indicators. Nat. Commun. 11, 3444–34413448. 10.1038/s41467-020-17322-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdulaev N. G., Artamonov I. D., Bogachuk A. S., Feigina M. Y., Kostina M. B., Kudelin A. B., et al. (1982). Structure of Light-Activated Proteins - Visual Rhodopsin. Biochem. Int. 5, 693–703. [Google Scholar]
- Abdulaev N. G., Ridge K. D. (2000). Heterologous Expression of Bovine Opsin in Pichia pastoris . Meth. Enzymol. 315, 3–11. 10.1016/s0076-6879(00)15831-8 [DOI] [PubMed] [Google Scholar]
- Acharya A. R., Vandekerckhove B., Larsen L. E., Delbeke J., Wadman W. J., Vonck K., et al. (2021). In Vivo blue Light Illumination for Optogenetic Inhibition: Effect on Local Temperature and Excitability of the Rat hippocampus. J. Neural Eng. 18, 066038–066031. 10.1088/1741-2552/ac3ef4 [DOI] [PubMed] [Google Scholar]
- Adesnik H., Abdeladim L. (2021). Probing Neural Codes with Two-Photon Holographic Optogenetics. Nat. Neurosci. 24, 1356–1366. 10.1038/s41593-021-00902-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshar Saber W., Gasparoli F. M., Dirks M. G., Gunn-Moore F. J., Antkowiak M. (2018). All-Optical Assay to Study Biological Neural Networks. Front. Neurosci. 12, 451–451412. 10.3389/fnins.2018.00451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agathangelou D., Orozco-Gonzalez Y., Del Carmen Marín M., Roy P. P., Brazard J., Kandori H., et al. (2018). Effect of Point Mutations on the Ultrafast Photo-Isomerization of Anabaena Sensory Rhodopsin. Faraday Discuss. in the press. 10.1039/c7fd00200a [DOI] [PubMed] [Google Scholar]
- Agathangelou D., Roy P. P., Del Carmen Marín M., Ferré N., Olivucci M., Buckup T., et al. (2021). Sub-picosecond C=C Bond Photo-Isomerization: Evidence for the Role of Excited State Mixing. Comptes Rendus Phys. 22, 1–28. 10.5802/crphys.41 [DOI] [Google Scholar]
- Aguilà M., Toledo D., Morillo M., Dominguez M., Vaz B., Álvarez R., et al. (2009). Structural Coupling of 11-Cis-7-Methyl-Retinal and Amino Acids at the Ligand Binding Pocket of Rhodopsin. Photochem. Photobiol. 85, 485–493. 10.1111/j.1751-1097.2009.00535.x [DOI] [PubMed] [Google Scholar]
- Airan R. D., Thompson K. R., Fenno L. E., Bernstein H., Deisseroth K. (2009). Temporally Precise In Vivo Control of Intracellular Signalling. Nature 458, 1025–1029. 10.1038/nature07926 [DOI] [PubMed] [Google Scholar]
- Alabugin A. (2019). Near-IR Photochemistry for Biology: Exploiting the Optical Window of Tissue. Photochem. Photobiol. 95, 722–732. 10.1111/php.13068 [DOI] [PubMed] [Google Scholar]
- Alexander N. S., Katayama K., Sun W. Y., Salom D., Gulati S., Zhang J. Y., et al. (2017). Complex Binding Pathways Determine the Regeneration of Mammalian Green Cone Opsin with a Locked Retinal Analogue. J. Biol. Chem. 292, 10983–10997. 10.1074/jbc.m117.780478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiev U., Farrens D. L. (2014). Fluorescence Spectroscopy of Rhodopsins: Insights and Approaches. Biochimica Biophysica Acta-Bioenergetics 1837, 694–709. 10.1016/j.bbabio.2013.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiev U., Mollaaghababa R., Khorana H. G., Heyn M. P. (2000). Evidence for Long Range Allosteric Interactions between the Extracellular and Cytoplasmic Parts of Bacteriorhodopsin from the Mutant R82A and its Second Site Revertant R82A/G231C. J. Biol. Chem. 275, 13431–13440. 10.1074/jbc.275.18.13431 [DOI] [PubMed] [Google Scholar]
- Alexiev U., Rimke I., Pöhlmann T. (2003). Elucidation of the Nature of the Conformational Changes of the EF-Interhelical Loop in Bacteriorhodopsin and of the Helix VIII on the Cytoplasmic Surface of Bovine Rhodopsin: A Time-Resolved Fluorescence Depolarization Study. J. Mol. Biol. 328, 705–719. 10.1016/s0022-2836(03)00326-7 [DOI] [PubMed] [Google Scholar]
- Alfonsa H., Lakey J. H., Lightowlers R. N., Trevelyan A. J. (2016). Cl-out Is a Novel Cooperative Optogenetic Tool for Extruding Chloride from Neurons. Nat. Commun. 7, 13495–13499. 10.1038/ncomms13495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbach C. A., Kusnetzow A. K., Ernst O. P., Hofmann K. P., Hubbell W. L. (2008). High-resolution Distance Mapping in Rhodopsin Reveals the Pattern of Helix Movement Due to Activation. Proc. Natl. Acad. Sci. U. S. A. 105, 7439–7444. 10.1073/pnas.0802515105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun A., Yokoyama S., Morokuma K. (2008). Spectral Tuning in Visual Pigments: An ONIOM(QM : MM) Study on Bovine Rhodopsin and its Mutants. J. Phys. Chem. B 112, 6814–6827. 10.1021/jp709730b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez R., Vaz B., Gronemeyer H., De Lera A. R. (2014). Functions, Therapeutic Applications, and Synthesis of Retinoids and Carotenoids. Chem. Rev. 114, 1–125. 10.1021/cr400126u [DOI] [PubMed] [Google Scholar]
- Amsden J. J., Kralj J. M., Chieffo L. R., Wang X. H., Erramilli S., Spudich E. N., et al. (2007). Subpicosecond Protein Backbone Changes Detected during the Green-Absorbing Proteorhodopsin Primary Photoreaction. J. Phys. Chem. B 111, 11824–11831. 10.1021/jp073490r [DOI] [PubMed] [Google Scholar]
- Anashkin V. A., Bertsova Y. V., Mamedov A. M., Mamedov M. D., Arutyunyan A. M., Baykov A. A., et al. (2018). Engineering a Carotenoid-Binding Site in Dokdonia Sp PRO95 Na+-Translocating Rhodopsin by a Single Amino Acid Substitution. Photosynth. Res. 136, 161–169. 10.1007/s11120-017-0453-0 [DOI] [PubMed] [Google Scholar]
- Angel T. E., Gupta S., Jastrzebska B., Palczewski K., Chance M. R. (2009). Structural Waters Define a Functional Channel Mediating Activation of the GPCR, Rhodopsin. Proc. Natl. Acad. Sci. U. S. A. 106, 14367–14372. 10.1073/pnas.0901074106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermayr S. A., Rovira A. G., Hellingwerf K. J. (2015). Metabolic Engineering of Cyanobacteria for the Synthesis of Commodity Products. Trends Biotechnol. 33, 352–361. 10.1016/j.tibtech.2015.03.009 [DOI] [PubMed] [Google Scholar]
- Aprahamian I. (2020). The Future of Molecular Machines. Acs Central Sci. 6, 347–358. 10.1021/acscentsci.0c00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong C. M. (1992). Voltage-dependent Ion Channels and Their Gating. Physiol. Rev. 72, S5–S13. 10.1152/physrev.1992.72.suppl_4.s5 [DOI] [PubMed] [Google Scholar]
- Arnaboldi M., Motto M. G., Tsujimoto K., Balogh-Nair V., Nakanishi K. (1979). Hydroretinals and Hydrorhodopsins. J. Am. Chem. Soc. 101, 7082–7084. 10.1021/ja00517a059 [DOI] [Google Scholar]
- Asenjo A. B., Rim J., Oprian D. D. (1994). Molecular Determinants of Human Red/green Color Discrimination. Neuron 12, 1131–1138. 10.1016/0896-6273(94)90320-4 [DOI] [PubMed] [Google Scholar]
- Asido M., Kar R. K., Kriebel C. N., Braun M., Glaubitz C., Schapiro I., et al. (2021). Transient Near-UV Absorption of the Light-Driven Sodium Pump Krokinobacter Eikastus Rhodopsin 2: A Spectroscopic Marker for Retinal Configuration. J. Phys. Chem. Lett. 12, 6284–6291. 10.1021/acs.jpclett.1c01436 [DOI] [PubMed] [Google Scholar]
- Astakhova L. A., Novoselov A. D., Ermolaeva M. E., Firsov M. L., Rotov A. Y. (2021). Phototransduction in Anuran Green Rods: Origins of Extra-sensitivity. Int. J. Mol. Sci. 22, 13400. 10.3390/ijms222413400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou D., Aguila M., Bellingham J., Li W. W., Mcculley C., Reeves P. J., et al. (2018). The Molecular and Cellular Basis of Rhodopsin Retinitis Pigmentosa Reveals Potential Strategies for Therapy. Prog. Retin. Eye Res. 62, 1–23. 10.1016/j.preteyeres.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avelar G. M., Schumacher R. I., Zaini P. A., Leonard G., Richards T. A., Gomes S. L. (2014). A Rhodopsin-Guanylyl Cyclase Gene Fusion Functions in Visual Perception in a Fungus. Curr. Biol. 24, 1234–1240. 10.1016/j.cub.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axford D., Judge P. J., Bada Juarez J. F., Kwan T. O. C., Birch J., Vinals J., et al. (2022). Two States of a Light-Sensitive Membrane Protein Captured at Room Temperature Using Thin-Film Sample Mounts. Acta Crystallogr. Sect. D. Struct. Biol. 78, 52–58. 10.1107/s2059798321011220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azadi-Chegeni F., Schiphorst C., Pandit A. (2018). In Vivo NMR as a Tool for Probing Molecular Structure and Dynamics in Intact Chlamydomonas Reinhardtii Cells. Photosynth. Res. 135, 227–237. 10.1007/s11120-017-0412-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimihashemi N., Bergs A. C. F., Schüler C., Scheiwe A. R., Costa W. S., Bach M., et al. (2019). Rhodopsin-based Voltage Imaging Tools for Use in Muscles and Neurons of Caenorhabditis elegans . Proc. Natl. Acad. Sci. U. S. A. 116, 17051–17060. 10.1073/pnas.1902443116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimihashemi N., Erbguth K., Vogt A., Riemensperger T., Rauch E., Woodmansee D., et al. (2014). Synthetic Retinal Analogues Modify the Spectral and Kinetic Characteristics of Microbial Rhodopsin Optogenetic Tools. Nat. Commun. 5, 5810. 10.1038/ncomms6810 [DOI] [PubMed] [Google Scholar]
- Babl S. S., Rummell B. P., Sigurdsson T. (2019). The Spatial Extent of Optogenetic Silencing in Transgenic Mice Expressing Channelrhodopsin in Inhibitory Interneurons. Cell. Rep. 29, 1381–1395. 10.1016/j.celrep.2019.09.049 [DOI] [PubMed] [Google Scholar]
- Bada Juarez J. F., Judge P. J., Adam S., Axford D., Vinals J., Birch J., et al. (2021). Structures of the Archaerhodopsin-3 Transporter Reveal that Disordering of Internal Water Networks Underpins Receptor Sensitization. Nat. Commun. 12, 629. 10.1038/s41467-020-20596-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden T., Euler T., Berens P. (2020). Understanding the Retinal Basis of Vision across Species. Nat. Rev. Neurosci. 21, 5–20. 10.1038/s41583-019-0242-1 [DOI] [PubMed] [Google Scholar]
- Bagley K. A., Eisenstein L., Ebrey T. G., Tsuda M. (1989). A Comparative Study of the Infrared Difference Spectra for octopus and Bovine Rhodopsins and Their Bathorhodopsin Photointermediates. Biochemistry-USA 28, 3366–3373. 10.1021/bi00434a036 [DOI] [PubMed] [Google Scholar]
- Baillie J. S., Stoyek M. R., Quinn T. A. (2021). Seeing the Light: The Use of Zebrafish for Optogenetic Studies of the Heart. Front. physiology 12, 748570. 10.3389/fphys.2021.748570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker G. E., De Grip W. J., Turton M., Wagner H.-J., Foster R. G., Douglas R. H. (2015). Light Sensitivity in a Vertebrate Mechanoreceptor? J. Exp. Biol. 218, 2826–2829. 10.1242/jeb.125203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashov S. P., Imasheva E. S., Choi A. R., Jung K.-H., Liaaen-Jensen S., Lanyi J. K. (2010). Reconstitution of Gloeobacter Rhodopsin with Echinenone: Role of the 4-keto Group. Biochemistry 49, 9792–9799. 10.1021/bi1014166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balogh-Nair V., Nakanishi K. (1982). Synthetic Analogs of Retinal, Bacteriorhodopsin and Bovine Rhodopsin. Meth. Enzymol. 88, 496–506. 10.1016/0076-6879(82)88067-1 [DOI] [Google Scholar]
- Bamann C., Bamberg E., Wachtveitl J., Glaubitz C. (2014). Proteorhodopsin. Biochimica Biophysica Acta-Bioenergetics 1837, 614–625. 10.1016/j.bbabio.2013.09.010 [DOI] [PubMed] [Google Scholar]
- Bando Y., Grimm C., Cornejo V. H., Yuste R. (2019). Genetic Voltage Indicators. BMC Biol. 17, 71–7112. 10.1186/s12915-019-0682-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Huber T., Sakmar T. P. (2008). Rapid Incorporation of Functional Rhodopsin into Nanoscale Apolipoprotein Bound Bilayer (NABB) Particles. J. Mol. Biol. 377, 1067–1081. 10.1016/j.jmb.2008.01.066 [DOI] [PubMed] [Google Scholar]
- Banskota S., Raguram A., Suh S., Du S. W., Davis J. R., Choi E. H., et al. (2022). Engineered Virus-like Particles for Efficient Invivo Delivery of Therapeutic Proteins. Cell. 185. 10.1016/j.cell.2021.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry B., Mathies R. A. (1987). Raman Microscope Studies on the Primary Photochemistry of Vertebrate Visual Pigments with Absorption Maxima from 430 to 502 Nm. Biochemistry 26, 59–64. 10.1021/bi00375a009 [DOI] [PubMed] [Google Scholar]
- Bayburt T. H., Grinkova Y. V., Sligar S. G. (2006). Assembly of Single Bacteriorhodopsin Trimers in Bilayer Nanodiscs. Archives Biochem. Biophysics 450, 215–222. 10.1016/j.abb.2006.03.013 [DOI] [PubMed] [Google Scholar]
- Bayburt T. H., Vishnivetskiy S. A., Mclean M. A., Morizumi T., Huang C.-C., Tesmer J. J. G., et al. (2011). Monomeric Rhodopsin Is Sufficient for Normal Rhodopsin Kinase (GRK1) Phosphorylation and Arrestin-1 Binding. J. Biol. Chem. 286, 1420–1428. 10.1074/jbc.m110.151043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar H., Fields A. P., Kralj J. M., Spudich J. L., Rothschild K. J., Cohen A. E. (2012). Ultrasensitive Measurements of Microbial Rhodopsin Photocycles Using Photochromic FRET. Photochem. Photobiol. 88, 90–97. 10.1111/j.1751-1097.2011.01011.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker-Baldus J., Bamann C., Saxena K., Gustmann H., Brown L. J., Brown R. C. D., et al. (2015). Enlightening the Photoactive Site of Channelrhodopsin-2 by DNP-Enhanced Solid-State NMR Spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 112, 9896–9901. 10.1073/pnas.1507713112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook C. N., Rice A. J., Yang K. K., Ding X. Z., Chen S. Y., Leproust E. M., et al. (2017a). Structure-guided SCHEMA Recombination Generates Diverse Chimeric Channelrhodopsins. Proc. Natl. Acad. Sci. U. S. A. 114, E2624–E2633. 10.1073/pnas.1700269114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook C. N., Yang K. K., Rice A. J., Gradinaru V., Arnold F. H. (2017b). Machine Learning to Design Integral Membrane Channelrhodopsins for Efficient Eukaryotic Expression and Plasma Membrane Localization. Plos Comput. Biol. 13, 1005786. 10.1371/journal.pcbi.1005786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook C. N., Yang K. K., Robinson J. E., Mackey E. D., Gradinaru V., Arnold F. H. (2019). Machine Learning-Guided Channelrhodopsin Engineering Enables Minimally Invasive Optogenetics. Nat. Methods 16, 1176–1184. 10.1038/s41592-019-0583-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béjà O., Aravind L., Koonin E. V., Suzuki M. T., Hadd A., Nguyen L. P., et al. (2000). Bacterial Rhodopsin: Evidence for a New Type of Phototrophy in the Sea. Science 289, 1902–1906. 10.1126/science.289.5486.1902 [DOI] [PubMed] [Google Scholar]
- Béjà O., Spudich E. N., Spudich J. L., Leclerc M., Delong E. F. (2001). Proteorhodopsin Phototrophy in the Ocean. Nature 411, 786–789. 10.1038/35081051 [DOI] [PubMed] [Google Scholar]
- Belrhali H., Nollert P., Royant A., Menzel C., Rosenbusch J. P., Landau E. M., et al. (1999). Protein, Lipid and Water Organization in Bacteriorhodopsin Crystals: A Molecular View of the Purple Membrane at 1.9 Angstrom Resolution. Struct. Fold. Des. 7, 909–917. 10.1016/s0969-2126(99)80118-x [DOI] [PubMed] [Google Scholar]
- Bennett N., Michel-Villaz M., Kühn H. (1982). Light-induced Interaction between Rhodopsin and the GTP-Binding Protein: Metarhodopsin-II Is the Major Photoproduct Involved. Eur. J. Biochem. 127, 97–103. 10.1111/j.1432-1033.1982.tb06842.x [DOI] [PubMed] [Google Scholar]
- Berglund K., Clissold K., Li H. F. E., Wen L., Park S. Y., Gleixner J., et al. (2016). Luminopsins Integrate Opto- and Chemogenetics by Using Physical and Biological Light Sources for Opsin Activation. Proc. Natl. Acad. Sci. U. S. A. 113, E358–E367. 10.1073/pnas.1510899113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund K., Fernandez A. M., Gutekunst C. a. N., Hochgeschwender U., Gross R. E. (2020). Step-function Luminopsins for Bimodal Prolonged Neuromodulation. J. Neurosci. Res. 98, 422–436. 10.1002/jnr.24424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo V. B., Ntefidou M., Trivedi V. D., Amsden J. J., Kralj J. M., Rothschild K. J., et al. (2006). Conformational Changes in the Photocycle of Anabaena Sensory Rhodopsin - Absence Of the Schiff Base Counterion Protonation Signal . J. Biol. Chem. 281, 15208–15214. 10.1074/jbc.m600033200 [DOI] [PubMed] [Google Scholar]
- Berndt A., Lee S. Y., Ramakrishnan C., Deisseroth K. (2014). Structure-Guided Transformation of Channelrhodopsin into a Light-Activated Chloride Channel. Science 344, 420–424. 10.1126/science.1252367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt A., Lee S. Y., Wietek J., Ramakrishnan C., Steinberg E. E., Rashid A. J., et al. (2016). Structural Foundations of Optogenetics: Determinants of Channelrhodopsin Ion Selectivity. Proc. Natl. Acad. Sci. U. S. A. 113, 822–829. 10.1073/pnas.1523341113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry B. J., Wojtovich A. P. (2020). Mitochondrial Light Switches: Optogenetic Approaches to Control Metabolism. FEBS J. 287, 4544–4556. 10.1111/febs.15424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M. H., Holt A., Salari A., Veit J., Visel M., Levitz J., et al. (2019). Restoration of High-Sensitivity and Adapting Vision with a Cone Opsin. Nat. Commun. 10, 1221. 10.1038/s41467-019-09124-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertazolli-Filho R., Ghosh S., Huang W. H., Wollmann G., Coca-Prados M. (2001). Molecular Evidence that Human Ocular Ciliary Epithelium Expresses Components Involved in Phototransduction. Biochem. Biophysical Res. Commun. 284, 317–325. 10.1006/bbrc.2001.4970 [DOI] [PubMed] [Google Scholar]
- Besaw J. E., Ou W. L., Morizumi T., Eger B. T., Sanchez Vasquez J. D., Chu J. H. Y., et al. (2020). The Crystal Structures of a Chloride-Pumping Microbial Rhodopsin and its Proton-Pumping Mutant Illuminate Proton Transfer Determinants. J. Biol. Chem. 295, 14793–14804. 10.1074/jbc.ra120.014118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S., Hall S. E., Vaidehi N. (2008). Agonist-induced Conformational Changes in Bovine Rhodopsin: Insight into Activation of G-Protein-Coupled Receptors. J. Mol. Biol. 382, 539–555. 10.1016/j.jmb.2008.06.084 [DOI] [PubMed] [Google Scholar]
- Bhattacharya S. S., Marti T., Otto H., Heyn M. P., Khorana H. G. (1992a). A Bacteriorhodopsin Analog Reconstituted with a Nonisomerizable 13-trans Retinal Derivative Displays Light Insensitivity. J. Biol. Chem. 267, 6757–6762. 10.1016/s0021-9258(19)50490-2 [DOI] [PubMed] [Google Scholar]
- Bhattacharya S. S., Ridge K. D., Knox B. E., Khorana H. G. (1992b). Light-stable Rhodopsin. 1. A Rhodopsin Analog Reconstituted with a Nonisomerizable 11-cis Retinal Derivative. J. Biol. Chem. 267, 6763–6769. 10.1016/s0021-9258(19)50491-4 [DOI] [PubMed] [Google Scholar]
- Bi A. D., Cui J. J., Ma Y.-P., Olshevskaya E. V., Pu M. L., Dizhoor A. M., et al. (2006). Ectopic Expression of a Microbial-type Rhodopsin Restores Visual Responses in Mice with Photoreceptor Degeneration. Neuron 50, 23–33. 10.1016/j.neuron.2006.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibow S. (2019). Opportunities and Challenges of Backbone, Sidechain, and RDC Experiments to Study Membrane Protein Dynamics in a Detergent-free Lipid Environment Using Solution State NMR. Front. Mol. Biosci. 6, 103. 10.3389/fmolb.2019.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickelmann C., Morrow J. M., Du J., Schott R. K., Van Hazel I., Lim S., et al. (2015). The Molecular Origin and Evolution of Dim-Light Vision in Mammals. Evolution 69, 2995–3003. 10.1111/evo.12794 [DOI] [PubMed] [Google Scholar]
- Bielawski J. P., Dunn K. A., Sabehi G., Béjà O. (2004). Darwinian Adaptation of Proteorhodopsin to Different Light Intensities in the Marine Environment. Proc. Natl. Acad. Sci. U. S. A. 101, 14824–14829. 10.1073/pnas.0403999101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birge R. R., Cooper T. M., Lawrence A. F., Masthay M. B., Zhang C.-F., Zidovetzki R. (1991). Revised Assignment of Energy Storage in the Primary Photochemical Event in Bacteriorhodopsin. J. Am. Chem. Soc. 113, 4327–4328. 10.1021/ja00011a043 [DOI] [Google Scholar]
- Bismuth O., Friedman N., Sheves M., Ruhman S. (2007). Photochemical Dynamics of All-Trans Retinal Protonated Schiff-Base in Solution: Excitation Wavelength Dependence. Chem. Phys. 341, 267–275. 10.1016/j.chemphys.2007.06.052 [DOI] [Google Scholar]
- Blackshaw S., Snyder S. H. (1999). Encephalopsin: A Novel Mammalian Extraretinal Opsin Discretely Localized in the Brain. J. Neurosci. 19, 3681–3690. 10.1523/jneurosci.19-10-03681.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw S., Snyder S. H. (1997). Parapinopsin, a Novel Catfish Opsin Localized to the Parapineal Organ, Defines a New Gene Family. J. Neurosci. 17, 8083–8092. 10.1523/jneurosci.17-21-08083.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship E., Vahedi-Faridi A., Lodowski D. T. (2015). The High-Resolution Structure of Activated Opsin Reveals a Conserved Solvent Network in the Transmembrane Region Essential for Activation. Structure 23, 2358–2364. 10.1016/j.str.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatz P. E., Lin M., Balasubramaniyan P., Balasubramaniyan V., Dewhurst P. B. (1969). A New Series of Synthetic Visual Pigments from Cattle Opsin and Homologs of Retinal. J. Am. Chem. Soc. 91, 5930–5931. 10.1021/ja01049a069 [DOI] [PubMed] [Google Scholar]
- Bliss A. F. (1948). The Absorption Spectra of Visual Purple of the Squid and its Bleaching Products. J. Biol. Chem. 176, 563–569. 10.1016/s0021-9258(19)52673-4 [DOI] [PubMed] [Google Scholar]
- Bogomolni R. A., Spudich J. L. (1991). Archaebacterial Rhodopsins: Sensory and Energy Transducing Membrane Proteins. Mod. Cell. Biol. 10, 233–255. [Google Scholar]
- Boll F. (1877). Zur Anatomie und Physiologie der Retina. Arch. Anat. Physiol. 2, 175–286. 10.1007/BF02962033 [DOI] [Google Scholar]
- Bondar A.-N. (2022). Mechanisms of Long-Distance Allosteric Couplings in Proton-Binding Membrane Transporters. Adv. protein Chem. Struct. Biol. 128, 199–239. 10.1016/bs.apcsb.2021.09.002 [DOI] [PubMed] [Google Scholar]
- Borch J., Hamann T. (2009). The Nanodisc: A Novel Tool for Membrane Protein Studies. Biol. Chem. 390, 805–814. 10.1515/BC.2009.091 [DOI] [PubMed] [Google Scholar]
- Borgia A., Borgia M. B., Bugge K., Kissling V. M., Heidarsson P. O., Fernandes C. B., et al. (2018). Extreme Disorder in an Ultrahigh-Affinity Protein Complex. Nature 555, 61–66. 10.1038/nature25762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosman G. J. C. G. M., Vanoostrum J., Breikers G., Bovee-Geurts P. H. M., Klaassen C. H. W., DeGrip W. J. (2003). Functional Expression of His-Tagged Rhodopsin in Sf9 Insect Cells. Meth. Mol. Biol. 228, 73–86. 10.1385/1-59259-400-X:73 [DOI] [PubMed] [Google Scholar]
- Bovee-Geurts P. H. M., Fernández Fernández I., Liu R. S. H., Mathies R. A., Lugtenburg J., DeGrip W. J. (2009). Fluoro Derivatives of Retinal Illuminate the Decisive Role of the C12-H Element in Photoisomerization and Rhodopsin Activation. J. Am. Chem. Soc. 131, 17933–17942. 10.1021/ja907577p [DOI] [PubMed] [Google Scholar]
- Bovee-Geurts P. H. M., Lugtenburg J., DeGrip W. J. (2017). Coupled HOOP Signature Correlates with Quantum Yield of Isorhodopsin and Analog Pigments. Biochimica Biophysica Acta-Bioenergetics 1858, 118–125. 10.1016/j.bbabio.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Bownds M. D. (1967). Site of Attachment of Retinal in Rhodopsin. Nature 216, 1178–1181. 10.1038/2161178a0 [DOI] [PubMed] [Google Scholar]
- Boyden E. S., Zhang F., Bamberg E., Nagel G., Deisseroth K. (2005). Millisecond-timescale, Genetically Targeted Optical Control of Neural Activity. Nat. Neurosci. 8, 1263–1268. 10.1038/nn1525 [DOI] [PubMed] [Google Scholar]
- Bratanov D., Balandin T., Round E., Shevchenko V., Gushchin I., Polovinkin V., et al. (2015). An Approach to Heterologous Expression of Membrane Proteins. The Case of Bacteriorhodopsin. PLoS ONE 10, e0128390. 10.1371/journal.pone.0128390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratanov D., Kovalev K., Machtens J.-P., Astashkin R., Chizhov I., Soloviov D., et al. (2019). Unique Structure and Function of Viral Rhodopsins. Nat. Commun. 10, 4939. 10.1038/s41467-019-12718-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun R. D., Linsenmeier R. A., Goldstick T. K. (1995). Oxygen Consumption in the Inner and Outer Retina of the Cat. Investig. Ophthalmol. Vis. Sci. 36, 542–554. [PubMed] [Google Scholar]
- Bridges C. D. B. (1977). Method for Preparing Stable Digitonin Solutions for Visual Pigment Extraction. Vis. Res. 17, 301–302. 10.1016/0042-6989(77)90095-5 [DOI] [PubMed] [Google Scholar]
- Bridges C. D. B. (1972). “The Rhodopsin-Porphyropsin Visual System,” in Photochemistry of Vision. Editor Dartnall H. J. A. (Berlin: Springer-Verlag; ), 417–480. 10.1007/978-3-642-65066-6_11 [DOI] [Google Scholar]
- Brinkmann A., Sternberg U., Bovee-Geurts P. H. M., Fernández Fernández I., Lugtenburg J., Kentgens A. P. M., et al. (2018). Insight into the Chromophore of Rhodopsin and its Meta-II Photointermediate by 19F Solid-State NMR and Chemical Shift Tensor Calculations. Phys. Chem. Chem. Phys. 20, 30174–30188. 10.1039/c8cp05886e [DOI] [PubMed] [Google Scholar]
- Brinks D., Adam Y., Kheifets S., Cohen A. E. (2016). Painting with Rainbows: Patterning Light in Space, Time, and Wavelength for Multiphoton Optogenetic Sensing and Control. Accounts Chem. Res. 49, 2518–2526. 10.1021/acs.accounts.6b00415 [DOI] [PubMed] [Google Scholar]
- Broecker J., Eger B. T., Ernst O. P. (2017). Crystallogenesis of Membrane Proteins Mediated by Polymer-Bounded Lipid Nanodiscs. Structure 25, 384–392. 10.1016/j.str.2016.12.004 [DOI] [PubMed] [Google Scholar]
- Broser M. (2022). Far-Red Absorbing Rhodopsins, Insights from Heterodimeric Rhodopsin-Cyclases. Front. Mol. Biosci. 8, 806922. 10.3389/fmolb.2021.806922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broser M., Spreen A., Konold P. E., Peter E., Adam S., Borin V., et al. (2020). NeoR, a Near-Infrared Absorbing Rhodopsin. Nat. Commun. 11, 5682. 10.1038/s41467-020-19375-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillette C. G., Mcmichens R. B., Stern L. J., Khorana H. G. (1989). Structure and Thermal-Stability of Monomeric Bacteriorhodopsin in Mixed Phospholipid Detergent Micelles. Proteins-Structure Funct. Genet. 5, 38–46. 10.1002/prot.340050106 [DOI] [PubMed] [Google Scholar]
- Brown J., Behnam R., Coddington L., Tervo D. G. R., Martin K., Proskurin M., et al. (2018). Expanding the Optogenetics Toolkit by Topological Inversion of Rhodopsins. Cell. 175, 1131–1140. 10.1016/j.cell.2018.09.026 [DOI] [PubMed] [Google Scholar]
- Brown L. S., Ernst O. P. (2017). Recent Advances in Biophysical Studies of Rhodopsins - Oligomerization, Folding, and Structure. Biochimica Biophysica Acta-Proteins Proteomics 1865, 1512–1521. 10.1016/j.bbapap.2017.08.007 [DOI] [PubMed] [Google Scholar]
- Brown L. S. (2004). Fungal Rhodopsins and Opsin-Related Proteins: Eukaryotic Homologues of Bacteriorhodopsin with Unknown Functions. Photochem. Photobiological Sci. 3, 555–565. 10.1039/b315527g [DOI] [PubMed] [Google Scholar]
- Brown L. S., Ladizhansky V. (2015). Membrane Proteins in Their Native Habitat as Seen by Solid-State NMR Spectroscopy. Protein Sci. 24, 1333–1346. 10.1002/pro.2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L. S. (2022). Light-driven Proton Transfers and Proton Transport by Microbial Rhodopsins - A Biophysical Perspective. Biochimica Biophysica Acta-Biomembranes 1864, 183867. 10.1016/j.bbamem.2022.183867 [DOI] [PubMed] [Google Scholar]
- Brown L. S., Needleman R., Lanyi J. K. (1996). Interaction of Proton and Chloride Transfer Pathways in Recombinant Bacteriorhodopsin with Chloride Transport Activity: Implications for the Chloride Translocation Mechanism. Biochemistry 35, 16048–16054. 10.1021/bi9622938 [DOI] [PubMed] [Google Scholar]
- Bruun S., Stoeppler D., Keidel A., Kuhlmann U., Luck M., Diehl A., et al. (2015). Light-dark Adaptation of Channelrhodopsin Involves Photoconversion between the All-Trans and 13-cis Retinal Isomers. Biochemistry 54, 5389–5400. 10.1021/acs.biochem.5b00597 [DOI] [PubMed] [Google Scholar]
- Buda F., Keijer T., Ganapathy S., De Grip W. J. (2017). A Quantum-Mechanical Study of the Binding Pocket of Proteorhodopsin: Absorption and Vibrational Spectra Modulated by Analogue Chromophores. Photochem. Photobiol. 93, 1399–1406. 10.1111/php.12800 [DOI] [PubMed] [Google Scholar]
- Buhrke D., Hildebrandt P. (2020). Probing Structure and Reaction Dynamics of Proteins Using Time-Resolved Resonance Raman Spectroscopy. Chem. Rev. 120, 3577–3630. 10.1021/acs.chemrev.9b00429 [DOI] [PubMed] [Google Scholar]
- Butt H.-J. (1990). Quantum Efficiency of Native and Mutant Bacteriorhodopsin Obtained from Blue Light Induced Relaxation Experiments. Eur. Biophys. J. 19, 31–39. 10.1007/bf00223571 [DOI] [Google Scholar]
- Caffrey M. (2003). Membrane Protein Crystallization. J. Struct. Biol. 142, 108–132. 10.1016/s1047-8477(03)00043-1 [DOI] [PubMed] [Google Scholar]
- Cai Y. Y., Liu Y. T., Culhane K. J., Devree B. T., Yang Y., Sunahara R. K., et al. (2017). Purification of Family B G Protein-Coupled Receptors Using Nanodiscs: Application to Human Glucagon-like Peptide-1 Receptor. PLoS ONE 12, 0179568. 10.1371/journal.pone.0179568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calligaro H., Dkhissi-Benyahya O., Panda S. (2022). Ocular and Extraocular Roles of Neuropsin in Vertebrates. Trends Neurosci. 1776, 1–12. 10.1016/j.tins.2021.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P. X., Sun W., Kramp K., Zheng M., Salom D., Jastrzebska B., et al. (2012). Light-sensitive Coupling of Rhodopsin and Melanopsin to Gi/o and Gq Signal Transduction in Caenorhabditis elegans . FASEB J. 26, 480–491. 10.1096/fj.11-197798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin J. A., Carlén M., Meletis K., Knoblich U., Zhang F., Deisseroth K., et al. (2010). Targeted Optogenetic Stimulation and Recording of Neurons In Vivo Using Cell-type-specific Expression of Channelrhodopsin-2. Nat. Protoc. 5, 247–254. 10.1038/nprot.2009.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpine R., Olivieri G., Hellingwerf K. J., Pollio A., Marzocchella A. (2020). Industrial Production of Poly-Beta-Hydroxybutyrate from CO2: Can Cyanobacteria Meet This Challenge? Processes 8, 323–321323. [Google Scholar]
- Carravetta M., Zhao X., Johannessen O. G., Lai W. C., Verhoeven M. A., Bovee-Geurts P. H. M., et al. (2004). Protein-induced Bonding Perturbation of the Rhodopsin Chromophore Detected by Double-Quantum Solid-State NMR. J. Am. Chem. Soc. 126, 3948–3953. 10.1021/ja039390q [DOI] [PubMed] [Google Scholar]
- Casey J. R., Ferrón S., Karl D. M. (2017). Light-Enhanced Microbial Organic Carbon Yield. Front. Microbiol. 8, 2157. 10.3389/fmicb.2017.02157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassim J. Y. (1992). Unique Biphasic Band Shape of the Visible Circular Dichroism of Bacteriorhodopsin in Purple Membrane. Excitons, Multiple Transitions or Protein Heterogeneity? Biophys. J. 63, 1432–1442. 10.1016/s0006-3495(92)81701-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabre M., Lemaire M. (2005). Monomeric G-Protein-Coupled Receptor as a Functional Unit. Biochemistry 44, 9395–9403. 10.1021/bi050720o [DOI] [PubMed] [Google Scholar]
- Chan T., Lee M., Sakmar T. P. (1992). Introduction of Hydroxyl-Bearing Amino Acids Causes Bathochromic Spectral Shifts in Rhodopsin - Amino Acid Substitutions Responsible for Red-Green Color Pigment Spectral Tuning. J. Biol. Chem. 267, 9478–9480. 10.1016/s0021-9258(19)50115-6 [DOI] [PubMed] [Google Scholar]
- Chang C.-F., Kuramochi H., Singh M., Abe-Yoshizumi R., Tsukuda T., Kandori H., et al. (2022). A Unified View on Varied Ultrafast Dynamics of the Primary Process in Microbial Rhodopsins. Angew. Chem. Int. Ed. 61, e202111930. 10.1002/anie.202111930 [DOI] [PubMed] [Google Scholar]
- Chang C. F., Kuramochi H., Singh M., Abe-Yoshizumi R., Tsukuda T., Kandori H., et al. (2019). Acid-base Equilibrium of the Chromophore Counterion Results in Distinct Photoisomerization Reactivity in the Primary Event of Proteorhodopsin. Phys. Chem. Chem. Phys. 21, 25728–25734. 10.1039/c9cp04991f [DOI] [PubMed] [Google Scholar]
- Charvolin D., Perez J.-B., Rouvière F., Giusti F., Bazzacco P., Abdine A., et al. (2009). The Use of Amphipols as Universal Molecular Adapters to Immobilize Membrane Proteins onto Solid Supports. Proc. Natl. Acad. Sci. U. S. A. 106, 405–410. 10.1073/pnas.0807132106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla U., Perera S. M. D. C., Fried S. D. E., Eitel A. R., Mertz B., Weerasinghe N., et al. (2021). Activation of the G-Protein-Coupled Receptor Rhodopsin by Water. Angew. Chemie-International Ed. 60, 2288–2295. 10.1002/anie.202003342 [DOI] [PubMed] [Google Scholar]
- Chazan A., Rozenberg A., Mannen K., Nagata T., Tahan R., Yaish S., et al. (2022). Diverse Heliorhodopsins Detected via Functional Metagenomics in Freshwater Actinobacteria, Chloroflexi and Archaea . Environ. Microbiol. 2022, 15890. 10.1111/1462-2920.15890 [DOI] [PubMed] [Google Scholar]
- Chen G.-Q., Gouaux J. E. (1996). Overexpression of Bacterio-Opsin in Escherichia coli as a Water-Soluble Fusion to Maltose Binding Protein: Efficient Regeneration of the Fusion Protein and Selective Cleavage with Trypsin. Protein Sci. 5, 456–467. 10.1002/pro.5560050307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.-H., Kuemmel C., Birge R. R., Knox B. E. (2012). Rapid Release of Retinal from a Cone Visual Pigment Following Photoactivation. Biochemistry 51, 4117–4125. 10.1021/bi201522h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Arents J. C., Schuurmans J. M., Ganapathy S., De Grip W. J., Cheregi O., et al. (2019b). Combining Retinal-Based and Chlorophyll-Based (Oxygenic) Photosynthesis: Proteorhodopsin Expression Increases Growth Rate and Fitness of a ΔPSI Strain of Synechocystis Sp. PCC6803. Metab. Eng. 52, 68–76. 10.1016/j.ymben.2018.11.002 [DOI] [PubMed] [Google Scholar]
- Chen Q., Arents J., Ganapathy S., DeGrip W. J., Hellingwerf K. J. (2017). Functional Expression of Gloeobacter Rhodopsin in Synechocystis Sp PCC6803. Photochem. Photobiol. 93, 772–781. 10.1111/php.12745 [DOI] [PubMed] [Google Scholar]
- Chen Q., Arents J., Schuurmans J. M., Ganapathy S., De Grip W. J., Cheregi O., et al. (2019a). Functional Expression of Gloeobacter Rhodopsin in PSI-Less Synechocystis Sp. PCC6803. Front. Bioeng. Biotechnol. 7, 67. 10.3389/fbioe.2019.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Montesarchio D., Hellingwerf K. J. (2016a). “‘Direct Conversion’: Artificial Photosynthesis with Cyanobacteria,” in Artificial Photosynthesis. Editor Bruno R., 43–61. 10.1016/bs.abr.2016.03.001 [DOI] [Google Scholar]
- Chen Q., Van Der Steen J. B., Arents J. C., Hartog A. F., Ganapathy S., De Grip W. J., et al. (2018). Deletion of Sll1541 in Synechocystis Sp Strain PCC 6803 Allows Formation of a Far-Red-Shifted Holo-Proteorhodopsin In Vivo . Appl. Environ. Microbiol. 84, e024351–0241714. 10.1128/AEM.02435-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Van Der Steen J. B., Dekker H. L., Ganapathy S., De Grip W. J., Hellingwerf K. J. (2016b). Expression of Holo-Proteorhodopsin in Synechocystis Sp PCC 6803. Metab. Eng. 35, 83–94. 10.1016/j.ymben.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Chen S. (2019). Optical Modulation Goes Deep in the Brain. Science 365, 456–457. 10.1126/science.aay4350 [DOI] [PubMed] [Google Scholar]
- Chien M.-P., Brinks D., Testa-Silva G., Tian H., Brooks F. P. I., Adam Y., et al. (2021). Photoactivated Voltage Imaging in Tissue with an Archaerhodopsin-Derived Reporter. Sci. Adv. 7, eabe3216. 10.1126/sciadv.abe3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhov I., Chernavskii D. S., Engelhard M., Mueller K.-H., Zubov B. V., Hess B. (1996). Spectrally Silent Transitions in the Bacteriorhodopsin Photocycle. Biophysical J. 71, 2329–2345. 10.1016/s0006-3495(96)79475-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. K., Park D., Yang A. M., Chen F., Chuong A. S., Klapoetke N. C., et al. (2019). Multidimensional Screening Yields Channelrhodopsin Variants Having Improved Photocurrent and Order-Of-Magnitude Reductions in Calcium and Proton Currents. J. Biol. Chem. 294, 3806–3821. 10.1074/jbc.ra118.006996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe H.-W., Kim Y. J., Park J. H., Morizumi T., Pai E. F., Krauß N., et al. (2011). Crystal Structure of Metarhodopsin II. Nature 471, 651–655. 10.1038/nature09789 [DOI] [PubMed] [Google Scholar]
- Choi A. R., Shi L. C., Brown L. S., Jung K.-H. (2014). Cyanobacterial Light-Driven Proton Pump, Gloeobacter Rhodopsin: Complementarity between Rhodopsin-Based Energy Production and Photosynthesis. PLoS ONE 9, e110643. 10.1371/journal.pone.0110643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E. H., Daruwalla A., Suh S., Leinonen H., Palczewski K. (2021). Retinoids in the Visual Cycle: Role of the Retinal G Protein-Coupled Receptor. J. Lipid Res. 62, 1000–1040. 10.1194/jlr.tr120000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow B. Y., Han X., Dobry A. S., Qian X. F., Chuong A. S., Li M. J., et al. (2010). High-performance Genetically Targetable Optical Neural Silencing by Light-Driven Proton Pumps. Nature 463, 98–102. 10.1038/nature08652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuon K., Kim S. Y., Meas S., Shim J.-G., Cho S.-G., Kang K.-W., et al. (2021). Assembly of Natively Synthesized Dual Chromophores into Functional Actinorhodopsin. Front. Microbiol. 12, 652328. 10.3389/fmicb.2021.652328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church J. R., Amoyal G. S., Borin V. A., Adam S., Olsen J. M. H., Schapiro I. (2022). Deciphering the Spectral Tuning Mechanism in Proteorhodopsin: The Dominant Role of Electrostatics Instead of Chromophore Geometry. Chem. - A Eur. J. 28, e202200139. 10.1002/chem.202200139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church J. R., Haugaard Olsen J. M., Schapiro I. (2022b). The Impact of Retinal Configuration on the Protein-Chromophore Interactions in Bistable Jumping Spider Rhodopsin-1. Molecules 27, 71. 10.3390/molecules27010071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civjan N. R., Bayburt T. H., Schuler M. A., Sligar S. G. (2003). Direct Solubilization of Heterologously Expressed Membrane Proteins by Incorporation into Nanoscale Lipid Bilayers. BioTechniques 35, 556–563. 10.2144/03353rr02 [DOI] [PubMed] [Google Scholar]
- Cokic M., Bruegmann T., Sasse P., Malan D. (2021). Optogenetic Stimulation of Gi Signaling Enables Instantaneous Modulation of Cardiomyocyte Pacemaking. Front. physiology 12, 768495. 10.3389/fphys.2021.768495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collette F., Renger T., Müh F., Schmidt Am Busch M. (2018). Red/Green Color Tuning of Visual Rhodopsins: Electrostatic Theory Provides a Quantitative Explanation. J. Phys. Chem. B 122, 4828–4837. 10.1021/acs.jpcb.8b02702 [DOI] [PubMed] [Google Scholar]
- Concistrè M., Gansmüller A., Mclean N., Johannessen O. G., Marín-Montesinos I., Bovee-Geurts P. H. M., et al. (2008). Double-quantum 13C Nuclear Magnetic Resonance of Bathorhodopsin, the First Photointermediate in Mammalian Vision. J. Am. Chem. Soc. 130, 10490–10491. 10.1021/ja803801u [DOI] [PubMed] [Google Scholar]
- Contreras E., Nobleman A. P., Robinson P. R., Schmidt T. M. (2021). Melanopsin Phototransduction: beyond Canonical Cascades. J. Exp. Biol. 224, 224–221214. 10.1242/jeb.226522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A. (1979). Energetics of Rhodopsin and Isorhodopsin. FEBS Lett. 100, 382–384. 10.1016/0014-5793(79)80375-0 [DOI] [PubMed] [Google Scholar]
- Copits B. A., Gowrishankar R., O'neill P. R., Li J.-N., Girven K. S., Yoo J. J., et al. (2021). A Photoswitchable GPCR-Based Opsin for Presynaptic Inhibition. Neuron 109, 1791–1809. 10.1016/j.neuron.2021.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Córdova C., Lozano C., Rodríguez B., Marchant I., Zúñiga R., Ochova P., et al. (2021). Optogenetic Control of Cancer Cell Survival in ChR2-Transfected HeLa Cells. Int. J. Exp. pathology 102, 242. 10.1111/iep.12426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creemers A. F. L., Kiihne S. R., Bovee-Geurts P. H. M., DeGrip W. J., Lugtenburg J., De Groot H. J. M. (2002). 1H and 13C MAS NMR Evidence for Pronounced Ligand-Protein Interactions Involving the Ionone Ring of the Retinylidene Chromophore in Rhodopsin. Proc. Nat. Acad. Sci. U. S. A. 99, 9101–9106. 10.1073/pnas.112677599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creemers A. F. L., Klaassen C. H. W., Bovee-Geurts P. H. M., Kelle R., Kragl U., Raap J., et al. (1999). 15N Solid State NMR Evidence for a Compex Schiff Base Counterion in the Visual G Protein-Coupled Receptor Rhodopsin. Biochemistry-USA 38, 7195–7199. 10.1021/bi9830157 [DOI] [PubMed] [Google Scholar]
- Crescitelli F. (1991). The Natural History of Visual Pigments: 1990. Prog. Retin. Res. 11, 1–32. 10.1111/j.1749-6632.1958.tb39548.x [DOI] [Google Scholar]
- Crouch R. K., Kefalov V. J., Gärtner W., Cornwall M. C. (2002). Use of Retinal Analogues for the Study of Visual Pigment Function. Meth. Enzymol. 343, 29–48. 10.1016/s0076-6879(02)43126-6 [DOI] [PubMed] [Google Scholar]
- Crouch R. K., Nodes B. R., Perlman J. I., Pepperberg D. R., Akita H., Nakanishi K. (1984). Cycloheptatrienylidene Analog of 11-cis Retinal. Formation of Pigment in Photoreceptor Membranes. Investig. Ophthalmol. Vis. Sci. 25, 419–428. [PubMed] [Google Scholar]
- Daemen F. J. M. (1973). Vertebrate Rod Outer Segment Membranes. Biochimica Biophysica Acta 300, 255–288. 10.1016/0304-4157(73)90006-3 [DOI] [PubMed] [Google Scholar]
- Dartnall H. J. A. (1962a). “The Chemical Structure and Photochemistry of the Visual Pigments,” in The Visual Process. Editor Davson H.. 1 ed (New York, U.S.A. Academic Press; ), 427–471. [Google Scholar]
- Dartnall H. J. A. (1962b). “The Identity and Distribution of Visual Pigments in the Animal Kingdom,” in The Visual Process. Editor Davson H. (New York, U.S.A. Academic Press; ), 367–426. [Google Scholar]
- Dartnall H. J. A. (1962c). “The Properties of Visual Pigments in Photoreceptors,” in The Visual Process. Editor Davson H.. 1 ed (New York, U.S.A. Academic Press; ), 473–533. [Google Scholar]
- Davidson F. F., Loewen P. C., Khorana H. G. (1994). Structure and Function in Rhodopsin: Replacement by Alanine of Cysteine Residues 110 and 187, Components of a Conserved Disulfide Bond in Rhodopsin, Affects the Light-Activated Metarhodopsin II State. Proc. Nat. Acad. Sci. U. S. A. 91, 4029–4033. 10.1073/pnas.91.9.4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A., Gowen B. E., Krebs A. M., Schertler G. F. X., Saibil H. R. (2001). Three-dimensional Structure of an Invertebrate Rhodopsin and Basis for Ordered Alignment in the Photoreceptor Membrane. J. Mol. Biol. 314, 455–463. 10.1006/jmbi.2001.5167 [DOI] [PubMed] [Google Scholar]
- Davies A., Schertler G. F. X., Gowen B. E., Saibil H. R. (1996). Projection Structure of an Invertebrate Rhodopsin. J. Struct. Biol. 117, 36–44. 10.1006/jsbi.1996.0067 [DOI] [PubMed] [Google Scholar]
- Davies W. I. L., Collin S. P., Hunt D. M. (2012). Molecular Ecology and Adaptation of Visual Photopigments in Craniates. Mol. Ecol. 21, 3121–3158. 10.1111/j.1365-294x.2012.05617.x [DOI] [PubMed] [Google Scholar]
- Davies W. I. L., Hankins M. W., Foster R. G. (2010). Vertebrate Ancient Opsin and Melanopsin: Divergent Irradiance Detectors. Photochem. Photobiological Sci. 9, 1444–1457. 10.1039/c0pp00203h [DOI] [PubMed] [Google Scholar]
- Davies W. I. L., Sghari S., Upton B. A., Nord C., Hahn M., Ahlgren U., et al. (2021). Distinct Opsin 3 (Opn3) Expression in the Developing Nervous System during Mammalian Embryogenesis. eNeuro 8, 0141–0121. 10.1523/eneuro.0141-21.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies W. I. L., Tamai T. K., Zheng L., Fu J. K., Rihel J., Foster R. G., et al. (2015). An Extended Family of Novel Vertebrate Photopigments Is Widely Expressed and Displays a Diversity of Function. Genome Res. 25, 1666–1679. 10.1101/gr.189886.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawadi P. B. S., Lugtenburg J. (2010). Synthesis and Use of Stable Isotope Enriched Retinals in the Field of Vitamin A. Molecules 15, 1825–1872. 10.3390/molecules15031825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva S. R., Barnard A. R., Hughes S., Tam S. K. E., Martin C., Singh M. S., et al. (2017). Long-term Restoration of Visual Function in End-Stage Retinal Degeneration Using Subretinal Human Melanopsin Gene Therapy. Proc. Natl. Acad. Sci. U. S. A. 114, 11211–11216. 10.1073/pnas.1701589114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrip W. J., Bonting S. L., Daemen F. J. M. (1973). The Binding Site of Retinaldehyde in Cattle Rhodopsin. Biochimica Biophysica Acta 303, 189–193. 10.1016/0005-2795(73)90162-1 [DOI] [PubMed] [Google Scholar]
- DeGrip W. J., Bovee-Geurts P. H. M. (1979). Synthesis and Properties of Alkylglucosides with Mild Detergent Action: Improved Synthesis and Purification of β-1-octyl, -nonyl- and -Decyl-Glucose. Synthesis of β-1-undecylglucose and β-1-dodecylmaltose. Chem. Phys. Lipids 23, 321–335. 10.1016/0009-3084(79)90010-0 [DOI] [Google Scholar]
- DeGrip W. J., Bovee-Geurts P. H. M., Van Der Hoef I., Lugtenburg J. (2007). 7, 8-Dihydro-Retinals Outperform the Native Retinals in Conferring Photosensitivity to Visual Opsin. jacs 129, 13265–13269. 10.1021/ja074937c [DOI] [PubMed] [Google Scholar]
- DeGrip W. J., Bovee-Geurts P. H. M., Wang Y.-J., Verhoeven M. A., Lugtenburg J. (2011). Cyclopropyl and Isopropyl Derivatives of 11-cis and 9-cis Retinals at C-9 and C-13: Subtle Steric Differences with Major Effects on Ligand Efficacy in Rhodopsin. J. Nat. Prod. 74, 383–390. 10.1021/np100744v [DOI] [PubMed] [Google Scholar]
- DeGrip W. J., Daemen F. J. M., Bonting S. L. (1980). Isolation and Purification of Bovine Rhodopsin. Meth. Enzymol. 67, 301–320. 10.1016/s0076-6879(80)67038-4 [DOI] [PubMed] [Google Scholar]
- DeGrip W. J., DeLange F., Klaassen C. H. W., Verdegem P. J. E., Wallace-Williams S. E., Creemers A. F. L., et al. (1999). “Photoactivation of Rhodopsin: Interplay between Protein and Chromophore,” in Rhodopsins and Phototransduction. Editor Goode J. A. (Chichester, UK: John Wiley & Sons; ), 102–118. [DOI] [PubMed] [Google Scholar]
- DeGrip W. J., Gillespie J., Rothschild K. J. (1985). Carboxyl Group Involvement in the Meta I and Meta II Stages in Rhodopsin Bleaching. A Fourier Transform Infra-red Spectroscopic Study. Biochim. Biophys. Acta 809, 97–106. 10.1016/0005-2728(85)90172-0 [DOI] [PubMed] [Google Scholar]
- DeGrip W. J., Gray D., Gillespie J., Bovee-Geurts P. H. M., Vandenberg E. M. M., Lugtenburg J., et al. (1988). Photoexcitation of Rhodopsin: Conformation Changes in the Chromophore, Protein and Associated Lipid, as Determined by FTIR Difference Spectroscopy. Photochem. Photobiol. 48, 497–504. 10.1111/j.1751-1097.1988.tb02852.x [DOI] [PubMed] [Google Scholar]
- DeGrip W. J., Liu R. S. H., Ramamurthy V., Asato A. E. (1976). Rhodopsin Analogues from Highly Hindered 7-cis Isomers of Retinal. Nature 262, 416–418. 10.1038/262416a0 [DOI] [PubMed] [Google Scholar]
- DeGrip W. J., Rothschild K. J. (2000). “Structure and Mechanism of Vertebrate Visual Pigments,” in Molecular Mechanisms in Visual Transduction. Editors Stavenga D. G., DeGrip W. J., PughJr. E. N. (Amsterdam, Netherlands: Elsevier Science Pub.), 1–54. 10.1016/s1383-8121(00)80004-4 [DOI] [Google Scholar]
- DeGrip W. J. (1982). Thermal Stability of Rhodopsin and Opsin in Some Novel Detergents. Meth. Enzymol. 81, 256–265. 10.1016/s0076-6879(82)81040-9 [DOI] [PubMed] [Google Scholar]
- DeGrip W. J., VanOostrum J., Bovee-Geurts P. H. M. (1998). Selective Detergent-Extraction from Mixed Detergent/lipid/protein Micelles, Using Cyclodextrin Inclusion Compounds: A Novel Generic Approach for the Preparation of Proteoliposomes. Biochem. J. 330, 667–674. 10.1042/bj3300667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrip W. J., VanOostrum J., Bovee-Geurts P. H. M., Van Der Steen R., Van Amsterdam L. J. P., Groesbeek M., et al. (1990). 10, 20-Methanorhodopsins: (7E, 9E, 13E)-10, 20-methanorhodopsin and (7E, 9Z, 13Z)-10, 20-methanorhodopsin - 11-Cis-Locked Rhodopsin Analog Pigments with Unusual Thermal and Photo-Stability. Eur. J. Biochem. 191, 211–220. 10.1111/j.1432-1033.1990.tb19112.x [DOI] [PubMed] [Google Scholar]
- Deininger W., Kröger P., Hegemann U., Lottspeich F., Hegemann P. (1995). Chlamyrhodopsin Represents a New Type of Sensory Photoreceptor. EMBO J. 14, 5849–5858. 10.1002/j.1460-2075.1995.tb00273.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K. (2010). Controlling the Brain with Light. Sci. Am. 303, 48–55. 10.1038/scientificamerican1110-48 [DOI] [PubMed] [Google Scholar]
- Deisseroth K., Hegemann P. (2017). The Form and Function of Channelrhodopsin. Science 357, eaan5544. 10.1126/science.aan5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K. (2015). Optogenetics: 10 Years of Microbial Opsins in Neuroscience. Nat. Neurosci. 18, 1213–1225. 10.1038/nn.4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Carmen Marín M., Agathangelou D., Orozco-Gonzalez Y., Valentini A., Kato Y., Abe-Yoshizumi R., et al. (2019a). Fluorescence Enhancement of a Microbial Rhodopsin via Electronic Reprogramming. J. Am. Chem. Soc. 141, 262–271. 10.1021/jacs.8b09311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Carmen Marín M., De Vico L., Dong S. J. S., Gagliardi L., Truhlar D. G., Olivucci M. (2019b). Assessment of MC-PDFT Excitation Energies for a Set of QM/MM Models of Rhodopsins. J. Chem. Theory Comput. 15, 1915–1923. 10.1021/acs.jctc.8b01069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLange F., Bovee-Geurts P. H. M., Pistorius A. M. A., Rothschild K. J., DeGrip W. J. (1999). Probing Intramolecular Orientations in Rhodopsin and Metarhodopsin II by Polarized Infrared Difference Spectroscopy. Biochemistry-USA 38, 13200–13209. 10.1021/bi9909501 [DOI] [PubMed] [Google Scholar]
- DeLange F., Bovee-Geurts P. H. M., Vanoostrum J., Portier M. D., Verdegem P. J. E., Lugtenburg J., et al. (1998a). An Additional Methyl Group at the 10-position of Retinal Dramatically Slows Down the Kinetics of the Rhodopsin Photocascade. Biochemistry-USA 37, 1411–1420. 10.1021/bi972397y [DOI] [PubMed] [Google Scholar]
- DeLange F., Klaassen C. H. W., Wallace-Williams S. E., Bovee-Geurts P. H. M., Liu X.-M., DeGrip W. J., et al. (1998b). Tyrosine Structural Changes Detected during the Photoactivation of Rhodopsin. J. Biol. Chem. 273, 23735–23739. 10.1074/jbc.273.37.23735 [DOI] [PubMed] [Google Scholar]
- DeLange F., Merkx M., Bovee-Geurts P. H. M., Pistorius A. M. A., DeGrip W. J. (1997). Modulation of the Metarhodopsin I/metarhodopsin II Equilibrium of Bovine Rhodopsin by Ionic Strength - Evidence for a Surface Charge Effect. Eur. J. Biochem. 243, 174–180. 10.1111/j.1432-1033.1997.0174a.x [DOI] [PubMed] [Google Scholar]
- Demoulin B., Maiuri M., Berbasova T., Geiger J. H., Borhan B., Garavelli M., et al. (2021). Control of Protonated Schiff Base Excited State Decay within Visual Protein Mimics: A Unified Model for Retinal Chromophores. Chemistry-A Eur. J. 27, 16389. 10.1002/chem.202102383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dencher N. A., Heyn M. P. (1978). Formation and Properties of Bacteriorhodopsin Monomers in the Nonionic Detergents Octyl-β-D-Glucoside and Triton X-100. FEBS Lett. 96, 322–326. 10.1016/0014-5793(78)80427-x [DOI] [PubMed] [Google Scholar]
- Derguini F., Caldwell C. G., Motto M. G., Balogh-Nair V., Nakanishi K. (1983). Bacteriorhodopsins Containing Cyanine Dye Chromophores - Support for the External Point-Charge Model. J. Am. Chem. Soc. 105, 646–648. 10.1021/ja00341a068 [DOI] [Google Scholar]
- Derguini F., Nakanishi K. (1986). Synthetic Rhodopsin Analogs. Photobiochem. Photobiophys. 13, 259–283. [Google Scholar]
- Deubner J., Coulon P., Diester I. (2019). Optogenetic Approaches to Study the Mammalian Brain. Curr. Opin. Struct. Biol. 57, 157–163. 10.1016/j.sbi.2019.04.003 [DOI] [PubMed] [Google Scholar]
- Devine E. L., Oprian D. D., Theobald D. L. (2013). Relocating the Active-Site Lysine in Rhodopsin and Implications for Evolution of Retinylidene Proteins. Proc. Natl. Acad. Sci. U. S. A. 110, 13351–13355. 10.1073/pnas.1306826110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X. Y., Sun C., Cui H. L., Chen S. J., Gao Y. J., Yang Y. A., et al. (2018). Functional Roles of Tyrosine 185 during the Bacteriorhodopsin Photocycle as Revealed by In Situ Spectroscopic Studies. Biochimica Biophysica Acta-Bioenergetics 1859, 1006–1014. 10.1016/j.bbabio.2018.05.011 [DOI] [PubMed] [Google Scholar]
- Dokukina I., Nenov A., Garavelli M., Marian C. M., Weingart O. (2019). QM/MM Photodynamics of Retinal in the Channelrhodopsin Chimera C1C2 with OM3/MRCI. ChemPhotoChem 3, 107–116. 10.1002/cptc.201800185 [DOI] [Google Scholar]
- Döring C. C., Kumar S., Tumu S. C., Kourtesis I., Hausen H. (2020). The Visual Pigment Xenopsin Is Widespread in Protostome Eyes and Impacts the View on Eye Evolution. Elife 9, e55193. 10.7554/eLife.55193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr J. M., Scheidelaar S., Koorengevel M. C., Dominguez J. J., Schäfer M., Van Walree C. A., et al. (2016). The Styrene-Maleic Acid Copolymer: A Versatile Tool in Membrane Research. Eur. Biophysics J. Biophysics Lett. 45, 3–21. 10.1007/s00249-015-1093-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling J. E. (2020). Vitamin A: its Many Roles - from Vision and Synaptic Plasticity to Infant Mortality. J. Comp. Physiology a-Neuroethology Sens. Neural Behav. Physiology 206, 389–399. 10.1007/s00359-020-01403-z [DOI] [PubMed] [Google Scholar]
- Du W., Caicedo Burbano P., Hellingwerf K. J., Branco Dos Santos F. (2018). “Challenges in the Application of Synthetic Biology towards Synthesis of Commodity Products by Cyanobacteria via “Direct Conversion”,” in Synthetic Biology of Cyanobacteria. Editors Zhang W., Song X. (Gateway East, Singapore: Springer Nature Singapore Pte Ltd.), 3–26. 10.1007/978-981-13-0854-3_1 [DOI] [PubMed] [Google Scholar]
- Duda M., Domagalik A., Orlowska-Feuer P., Krzysztynska-Kuleta O., Beldzik E., Smyk M. K., et al. (2020). Melanopsin: From a Small Molecule to Brain Functions. Neurosci. Biobehav. Rev. 113, 190–203. 10.1016/j.neubiorev.2020.03.012 [DOI] [PubMed] [Google Scholar]
- Dunham T. D., Farrens D. L. (1999). Conformational Changes in Rhodopsin - Movement of Helix F Detected by Site-specific Chemical Labeling and Fluorescence Spectroscopy. J. Biol. Chem. 274, 1683–1690. 10.1074/jbc.274.3.1683 [DOI] [PubMed] [Google Scholar]
- Ebrey T. G., Koutalos Y. (2001). Vertebrate Photoreceptors. Prog. Retin. Eye Res. 20, 49–94. 10.1016/s1350-9462(00)00014-8 [DOI] [PubMed] [Google Scholar]
- Ehsan M., Katsube S., Cecchetti C., Du Y., Mortensen J. S., Wang H. Q., et al. (2020). New Malonate-Derived Tetraglucoside Detergents for Membrane Protein Stability. ACS Chem. Biol. 15, 1697–1707. 10.1021/acschembio.0c00316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickelbeck D., Rudack T., Tennigkeit S. A., Surdin T., Karapinar R., Schwitalla J. C., et al. (2020). Lamprey Parapinopsin ("UVLamP"): a Bistable UV-Sensitive Optogenetic Switch for Ultrafast Control of GPCR Pathways. ChemBioChem 21, 612–617. 10.1002/cbic.201900485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M., Goncalves J. A., Ahuja S., Kirkup C., Hirshfeld A., Simmerling C., et al. (2012). Structural Transitions of Transmembrane Helix 6 in the Formation of Metarhodopsin I. J. Phys. Chem. B 116, 10477–10489. 10.1021/jp3019183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M., Ying W. W., Reeves P. J., Khorana H. G., Smith S. O. (2002). Magic Angle Spinning Nuclear Magnetic Resonance of Isotopically Labeled Rhodopsin. Meth. Enzymol. 343, 212–222. 10.1016/s0076-6879(02)43137-0 [DOI] [PubMed] [Google Scholar]
- El Khatib S., Atamian A. (2019). Evolution of Color Vision in Vertebrates. Delhi, India: Akinik Publications, 19–42. [Google Scholar]
- El-Tahawy M. M. T., Conti I., Bonfanti M., Nenov A., Garavelli M. (2020). Tailoring Spectral and Photochemical Properties of Bioinspired Retinal Mimics by In Silico Engineering. Angew. Chemie-International Ed. 59, 20619–20627. 10.1002/anie.202008644 [DOI] [PubMed] [Google Scholar]
- Engel A., Gaub H. E. (2008). Structure and Mechanics of Membrane Proteins. Annu. Rev. Biochem. 77, 127–148. 10.1146/annurev.biochem.77.062706.154450 [DOI] [PubMed] [Google Scholar]
- Engelhard C., Chizhov I., Sieber F., Engelhard M. (2018). Microbial Halorhodopsins: Light-Driven Chloride Pumps. Chem. Rev. 118, 10629–10645. 10.1021/acs.chemrev.7b00715 [DOI] [PubMed] [Google Scholar]
- Engqvist M. K. M., Mcisaac R. S., Dollinger P., Flytzanis N. C., Abrams M., Schor S., et al. (2015). Directed Evolution of Gloeobacter Violaceus Rhodopsin Spectral Properties. J. Mol. Biol. 427, 205–220. 10.1016/j.jmb.2014.06.015 [DOI] [PubMed] [Google Scholar]
- Erbguth K., Prigge M., Schneider F., Hegemann P., Gottschalk A. (2012). Bimodal Activation of Different Neuron Classes with the Spectrally Red-Shifted Channelrhodopsin Chimera C1V1 in Caenorhabditis elegans . PLoS ONE 7, e46827. 10.1371/journal.pone.0046827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst O. P., Lodowski D. T., Elstner M., Hegemann P., Brown L. S., Kandori H. (2014). Microbial and Animal Rhodopsins: Structures, Functions, and Molecular Mechanisms. Chem. Rev. 114, 126–163. 10.1021/cr4003769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald A., Kühne W. (1878). “Untersuchungen über den Sehpurpur,” in Untersuchungen aus dem Physiologischen Institute der Universität Heidelberg. Editor Kühne W., 248–290. [Google Scholar]
- Farrens D. L. (2010). What Site-Directed Labeling Studies Tell Us about the Mechanism of Rhodopsin Activation and G-Protein Binding. Photochem. Photobiological Sci. 9, 1466–1474. 10.1039/c0pp00283f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman T. B., Ivankov O. I., Kuklin A. I., Murugova T. N., Yakovleva M. A., Smitienko O. A., et al. (2019). Small-angle Neutron and X-Ray Scattering Analysis of the Supramolecular Organization of Rhodopsin in Photoreceptor Membrane. Biochimica Biophysica Acta-Biomembranes 1861, 183000. 10.1016/j.bbamem.2019.05.022 [DOI] [PubMed] [Google Scholar]
- Feng J., Brown M. F., Mertz B. (2015). Retinal Flip in Rhodopsin Activation? Biophysical J. 108, 2767–2770. 10.1016/j.bpj.2015.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Mertz B. (2015). Proteorhodopsin Activation Is Modulated by Dynamic Changes in Internal Hydration. Biochemistry 54, 7132–7141. 10.1021/acs.biochem.5b00932 [DOI] [PubMed] [Google Scholar]
- Feng S., Powell S. M., Wilson R., Bowman J. P. (2013). Light-stimulated Growth of Proteorhodopsin-Bearing Sea-Ice Psychrophile Psychroflexus Torquis Is Salinity Dependent. ISME J. 7, 2206–2213. 10.1038/ismej.2013.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Verdegem P. J. E., Lee Y. K., Sandström D., Edén M., Bovee-Geurts P. H. M., et al. (1997). Direct Determination of a Molecular Torsional Angle in the Membrane Protein Rhodopsin by Solid-State NMR. J. Am. Chem. Soc. 119, 6853–6857. 10.1021/ja970710d [DOI] [Google Scholar]
- Feuda R., Menon A. K., Göpfert M. C. (2022). Rethinking Opsins. Mol. Biol. Evol. 39, msac033. 10.1093/molbev/msac033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuda R., Rota-Stabelli O., Oakley T. H., Pisani D. (2014). The Comb Jelly Opsins and the Origins of Animal Phototransduction. Genome Biol. Evol. 6, 1964–1971. 10.1093/gbe/evu154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer P., Mukherjee S., Peter E., Broser M., Bartl F., Hegemann P. (2021). The Inner Mechanics of Rhodopsin Guanylyl Cyclase during cGMP-Formation Revealed by Real-Time FTIR Spectroscopy. eLife 10, e71384. 10.7554/eLife.71384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming J. F., Feuda R., Roberts N. W., Pisani D. (2020). A Novel Approach to Investigate the Effect of Tree Reconstruction Artifacts in Single-Gene Analysis Clarifies Opsin Evolution in Nonbilaterian Metazoans. Genome Biol. Evol. 12, 3906–3916. 10.1093/gbe/evaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flytzanis N. C., Bedbrook C. N., Chiu H., Engqvist M. K. M., Xiao C., Chan K. Y., et al. (2014). Archaerhodopsin Variants with Enhanced Voltage-Sensitive Fluorescence in Mammalian and Caenorhabditis elegans Neurons. Nat. Commun. 5, 4894. 10.1038/ncomms5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R. G., Hankins M. W. (2002). Non-rod, Non-cone Photoreception in the Vertebrates. Prog. Retin. Eye Res. 21, 507–527. 10.1016/s1350-9462(02)00036-8 [DOI] [PubMed] [Google Scholar]
- Fotiadis D., Jastrzebska B., Philippsen A., Müller D. J., Palczewski K., Engel A. (2006). Structure of the Rhodopsin Dimer: A Working Model for G-Protein-Coupled Receptors. Curr. Opin. Struct. Biol. 16, 252–259. 10.1016/j.sbi.2006.03.013 [DOI] [PubMed] [Google Scholar]
- Fotiadis D., Liang Y., Filipek S., Saperstein D. A., Engel A., Palczewski K. (2004). The G Protein-Coupled Receptor Rhodopsin in the Native Membrane. FEBS Lett. 564, 281–288. 10.1016/s0014-5793(04)00194-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fougère M., Van Der Zouwen C. I., Boutin J. A., Neszvecsko K., Sarret P., Ryczko D. (2021). Optogenetic Stimulation of Glutamatergic Neurons in the Cuneiform Nucleus Controls Locomotion in a Mouse Model of Parkinson's Disease. Proc. Natl. Acad. Sci. U. S. A. 118, e2110934118. 10.1073/pnas.2110934118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M., Carlson D. B., Hunter M. S., Williams G. J., Messerschmidt M., Zatsepin N. A., et al. (2014). Femtosecond X-Ray Diffraction from Two-Dimensional Protein Crystals. IUCrJ 1, 95–100. 10.1107/s2052252514001444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfeld J., Löving R., Armache J.-P., Sonnen A. F.-P., Guettou F., Moberg P., et al. (2016). A Saposin-Lipoprotein Nanoparticle System for Membrane Proteins. Nat. Methods 13, 345–351. 10.1038/nmeth.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. M. (2021). How the Discovery of Microbial Opsins Led to the Development of Optogenetics. Cell. 184, 5266–5270. 10.1016/j.cell.2021.08.022 [DOI] [PubMed] [Google Scholar]
- Friedman N., Sheves M., Ottolenghi M. (1989). Model Systems for Rhodopsins: The Photolysis of Protonated Retinal Schiff-Bases, Cyanine Dye, and Artificial Cyanine-Bacteriorhodopsin. J. Am. Chem. Soc. 111, 3203–3211. 10.1021/ja00191a015 [DOI] [Google Scholar]
- Friedrich D., Perodeau J., Nieuwkoop A. J., Oschkinat H. (2020). MAS NMR Detection of Hydrogen Bonds for Protein Secondary Structure Characterization. J. Biomol. NMR 74, 247–256. 10.1007/s10858-020-00307-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudim R., Szczepek M., Vierock J., Vogt A., Schmidt A., Kleinau G., et al. (2019). Design of a Light-Gated Proton Channel Based on the Crystal Structure of Coccomyxa Rhodopsin. Sci. Signal. 12, eaav4203. 10.1126/scisignal.aav4203 [DOI] [PubMed] [Google Scholar]
- Fujimoto K. J. (2021). Electronic Couplings and Electrostatic Interactions behind the Light Absorption of Retinal Proteins. Front. Mol. Biosci. 8, 752700. 10.3389/fmolb.2021.752700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto K. J., Hayashi S., Hasegawa J., Nakatsuji H. (2007). Theoretical Studies on the Color-Tuning Mechanism in Retinal Proteins. J. Chem. Theory Comput. 3, 605–618. 10.1021/ct6002687 [DOI] [PubMed] [Google Scholar]
- Fujimoto K. J., Inoue K. (2020). Excitonic Coupling Effect on the Circular Dichroism Spectrum of Sodium-Pumping Rhodopsin KR2. J. Chem. Phys. 153, 04510. 10.1063/5.0013642 [DOI] [PubMed] [Google Scholar]
- Fujita S., Endo T., Ju J.-M., Kean E. L., Kobata A. (1994). Structural Studies of the N-Linked Sugar Chains of Human Rhodopsin. Glycobiology 4, 633–640. 10.1093/glycob/4.5.633 [DOI] [PubMed] [Google Scholar]
- Fujiyabu C., Sato K., Nishio Y., Imamoto Y., Ohuchi H., Shichida Y., T. (2022). Amino Acid Residue at Position 188 Determines the UV-Sensitive Bistable Property of Vertebrate Non-visual Opsin Opn5. Commun. Biol. 5, 63. 10.1038/s42003-022-03010-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada Y., Shichida Y., Yoshizawa T., Ito M., Kodama A., Tsukida K. (1984). Studies on Structure and Function of Rhodopsin by Use of Cyclopentatrienylidene 11-Cis-Locked-Rhodopsin. Biochemistry 23, 5826–5832. 10.1021/bi00319a023 [DOI] [PubMed] [Google Scholar]
- Furuse M., Tamogami J., Hosaka T., Kikukawa T., Shinya N., Hato M., et al. (2015). Structural Basis for the Slow Photocycle and Late Proton Release in Acetabularia Rhodopsin I from the Marine Plant Acetabularia Acetabulum . Acta Crystallogr. Sect. F-Structural Biol. D71, 2203–2216. 10.1107/s1399004715015722 [DOI] [PubMed] [Google Scholar]
- Furutani Y., Kandori H., Shichida Y. (2003). Structural Changes in Lumirhodopsin and Metarhodopsin I Studied by Their Photoreactions at 77 K. Biochemistry 42, 8494–8500. 10.1021/bi034438y [DOI] [PubMed] [Google Scholar]
- Furutani Y., Terakita A., Shichida Y., Kandori H. (2005). FTIR Studies of the Photoactivation Processes in Squid Retinochrome. Biochemistry 44, 7988–7997. 10.1021/bi050219w [DOI] [PubMed] [Google Scholar]
- Ganapathy S., Bécheau O., Venselaar H., Frölich S., Van Der Steen J. B., Chen Q., et al. (2015). Modulation of Spectral Properties and Pump Activity of Proteorhodopsins by Retinal Analogues. Biochem. J. 467, 333–343. 10.1042/bj20141210 [DOI] [PubMed] [Google Scholar]
- Ganapathy S., Kratz S., Chen Q., Hellingwerf K. J., De Groot H. J. M., Rothschild K. J., et al. (2019). Redshifted and Near-Infrared Active Analog Pigments Based upon Archaerhodopsin-3. Photochem. Photobiol. 95, 959–968. 10.1111/php.13093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathy S., Liu R. S. H. (1992). Photoisomerization of Sixteen Isomers of Retinal. Initial Product Distribution in Direct and Sensitized Irradiation. Photochem. Photobiol. 56, 959–964. 10.1111/j.1751-1097.1992.tb09718.x [DOI] [Google Scholar]
- Ganapathy S., Opdam L., Hontani Y., Frehan S., Chen Q., Hellingwerf K. J., et al. (2020). Membrane Matters: The Impact of a Nanodisc-Bilayer or a Detergent Microenvironment on the Properties of Two Eubacterial Rhodopsins. Biochimica Biophysica Acta-Biomembranes 1862, 183113. 10.1016/j.bbamem.2019.183113 [DOI] [PubMed] [Google Scholar]
- Ganapathy S., Venselaar H., Chen Q., De Groot H. J. M., Hellingwerf K. J., De Grip W. J. (2017). Retinal-based Proton Pumping in the Near Infrared. J. Am. Chem. Soc. 139, 2338–2344. 10.1021/jacs.6b11366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Nafría J., Tate C. G. (2020). Cryo-Electron Microscopy: Moving beyond X-Ray Crystal Structures for Drug Receptors and Drug Development. Annu. Rev. Pharmacol. Toxicol. 60, 51–71. [DOI] [PubMed] [Google Scholar]
- Garczarek F., Gerwert K. (2006). Functional Waters in Intraprotein Proton Transfer Monitored by FTIR Difference Spectroscopy. Nature 439, 109–112. 10.1038/nature04231 [DOI] [PubMed] [Google Scholar]
- Gärtner W. (2000). “Invertebrate Visual Pigments,” in Molecular Mechanisms in Visual Transduction. Editors Stavenga D. G., DeGrip W. J., PughJr. E. N. (Amsterdam, Netherlands: Elsevier Science Pub.), 298–388. [Google Scholar]
- Gärtner W., Ullrich D., Vogt K. (1991). Quantum Yield of CHAPSO-Solubilized Rhodopsin and 3-Hydroxy-Retinal Containing Bovine Opsin. Photochem. Photobiol. 54, 1047–1055. [DOI] [PubMed] [Google Scholar]
- Gascón J. A., Sproviero E. M., Batista V. S. (2005). QM/MM Study of the NMR Spectroscopy of the Retinyl Chromophore in Visual Rhodopsin. J. Chem. Theory Comput. 1, 674–685. 10.1021/ct0500850 [DOI] [PubMed] [Google Scholar]
- Geiser A. H., Sievert M. K., Guo L. W., Grant J. E., Krebs M. P., Fotiadis D., et al. (2006). Bacteriorhodopsin Chimeras Containing the Third Cytoplasmic Loop of Bovine Rhodopsin Activate Transducin for GTP/GDP Exchange. Protein Sci. 15, 1679–1690. 10.1110/ps.062192306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard E., Mutt E., Nagata T., Koyanagi M., Flock T., Lesca E., et al. (2018). Convergent Evolution of Tertiary Structure in Rhodopsin Visual Proteins from Vertebrates and Box Jellyfish. Proc. Natl. Acad. Sci. U. S. A. 115, 6201–6206. 10.1073/pnas.1721333115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwert K., Freier E., Wolf S. (2014). The Role of Protein-Bound Water Molecules in Microbial Rhodopsins. Biochimica Biophysica Acta-Bioenergetics 1837, 606–613. 10.1016/j.bbabio.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Ghanbarpour A., Nairat M., Nosrati M., Santos E. M., Vasileiou C., Dantus M., et al. (2019). Mimicking Microbial Rhodopsin Isomerization in a Single Crystal. J. Am. Chem. Soc. 141, 1735–1741. 10.1021/jacs.8b12493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson S. K., Parkes J. H., Liebman P. A. (1999). Phosphorylation Alters the pH-dependent Active State Equilibrium of Rhodopsin by Modulating the Membrane Surface Potential. Biochemistry-USA 38, 11103–11114. 10.1021/bi990411w [DOI] [PubMed] [Google Scholar]
- Giesbers M. E., Bosman G. J. C. G. M., Bovee-Geurts P. H. M., DeGrip W. J. (2007). Introduction of a Rod Aromatic Cluster Does Not Improve the Structural Stability of the Human Green Cone Pigment. J. Struct. Biol. 159, 222–227. 10.1016/j.jsb.2007.01.010 [DOI] [PubMed] [Google Scholar]
- Giesbers M. E., Shirzad-Wasei N., Bosman G. J. C. G. M., DeGrip W. J. (2008). Functional Expression, Targeting and Ca2+ Signaling of a Mouse Melanopsin-eYFP Fusion Protein in a Retinal Pigment Epithelium Cell Line. Photochem. Photobiol. 84, 990–995. 10.1111/j.1751-1097.2008.00347.x [DOI] [PubMed] [Google Scholar]
- Gilhooley M. J., Lindner M., Palumaa T., Hughes S., Peirson S. N., Hankins M. W. (2022). A Systematic Comparison of Optogenetic Approaches to Visual Restoration. Mol. Ther. Methods & Clin. Dev. 25, 111. 10.1016/j.omtm.2022.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Consarnau L., Raven J. A., Levine N. M., Cutter L. S., Wang D. L., Seegers B., et al. (2019). Microbial Rhodopsins Are Major Contributors to the Solar Energy Captured in the Sea. Sci. Adv. 5, eaaw8855. 10.1126/sciadv.aaw8855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X., Mendoza-Halliday D., Ting J. T., Kaiser T., Sun X. Y., Bastos A. M., et al. (2020). An Ultra-sensitive Step-Function Opsin for Minimally Invasive Optogenetic Stimulation in Mice and Macaques. Neuron 107, 38–51. 10.1016/j.neuron.2020.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova E. G., Gou Y. Y., Sineshchekov O. A., Li H., Wang Y. M., Brown L. S., et al. (2022a). Kalium Rhodopsins: Natural Light-Gated Potassium Channels. bioRxiv. 10.1101/2021.09.17.460684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova E. G., Sineshchekov O. A., Li H., Spudich J. L. (2017). Microbial Rhodopsins: Diversity, Mechanisms, and Optogenetic Applications. Annu. Rev. Biochem. 86, 845–872. 10.1146/annurev-biochem-101910-144233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova E. G., Sineshchekov O. A., Li H., Wang Y., Brown L. S., Palmateer A., et al. (2021). Cation and Anion Channelrhodopsins: Sequence Motifs and Taxonomic Distribution. mBio 12, e0165621. 10.1128/mbio.01656-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova E. G., Sineshchekov O. A., Li H., Wang Y. M., Brown L. S., Spudich J. L. (2020). RubyACRs, Nonalgal Anion Channelrhodopsins with Highly Red-Shifted Absorption. Proc. Natl. Acad. Sci. U. S. A. 117, 22833–22840. 10.1073/pnas.2005981117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova E. G., Sineshchekov O. A., Spudich J. L. (2022b). Emerging Diversity of Channelrhodopsins and Their Structure-Function Relationships. Front. Cell. Neurosci. 15, 800313. 10.3389/fncel.2021.800313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova E. G., Sineshchekov O. A., Spudich J. L. (2016). Structurally Distinct Cation Channelrhodopsins from Cryptophyte Algae. Biophysical J. 110, 2302–2304. 10.1016/j.bpj.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozem S., Luk H. L., Schapiro I., Olivucci M. (2017). Theory and Simulation of the Ultrafast Double-Bond Lsomerization of Biological Chromophores. Chem. Rev. 117, 13502–13565. 10.1021/acs.chemrev.7b00177 [DOI] [PubMed] [Google Scholar]
- Griffiths J. M., Bennett A. E., Engelhard M., Siebert F., Raap J., Lugtenburg J., et al. (2000). Structural Investigation of the Active Site in Bacteriorhodopsin: Geometric Constraints on the Roles of Asp-85 and Asp-212 in the Proton-Pumping Mechanism from Solid-State NMR. Biochemistry 39, 362–371. 10.1021/bi991106d [DOI] [PubMed] [Google Scholar]
- Grigorieff N., Ceska T. A., Downing K. H., Baldwin J. M., Henderson R. A. (1996). Electron-crystallographic Refinement of the Structure of Bacteriorhodopsin. J. Mol. Biol. 259, 393–421. 10.1006/jmbi.1996.0328 [DOI] [PubMed] [Google Scholar]
- Grime R. L., Logan R. T., Nestorow S. A., Sridhar P., Edwards P. C., Tate C. G., et al. (2021). Differences in SMA-like Polymer Architecture Dictate the Conformational Changes Exhibited by the Membrane Protein Rhodopsin Encapsulated in Lipid Nano-Particles. Nanoscale 13, 13519–13528. 10.1039/d1nr02419a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenendijk G. W. T., DeGrip W. J., Daemen F. J. M. (1980). Quantitative Determination of Retinals with Complete Retention of Their Geometric Configuration. Biochim. Biophys. Acta 617, 430–438. 10.1016/0005-2760(80)90009-0 [DOI] [PubMed] [Google Scholar]
- Grote M., Engelhard M., Hegemann P. (2014). Of Ion Pumps, Sensors and Channels - Perspectives on Microbial Rhodopsins between Science and History. Biochimica Biophysica Acta-Bioenergetics 1837, 533–545. 10.1016/j.bbabio.2013.08.006 [DOI] [PubMed] [Google Scholar]
- Gruber E., Kabylda A. M., Brøndsted Nielsen M., Rasmussen A. P., Teiwes R., Kusochek P. A., et al. (2022). Light Driven Ultrafast Bioinspired Molecular Motors: Steering and Accelerating Photoisomerization Dynamics of Retinal. J. Am. Chem. Soc. 144, 69–73. 10.1021/jacs.1c10752 [DOI] [PubMed] [Google Scholar]
- Guimarães Backhaus R., Fu T., Backhaus H., Stroh A. (2021). Pipeline for 2-photon All-Optical Physiology in Mouse: From Viral Titration and Optical Window Implantation to Binarization of Calcium Transients. Star. Protoc. 2, 101010. 10.1016/j.xpro.2021.101010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati S., Jastrzebska B., Banerjee S., Placeres A. L., Miszta P., Gao S. Q., et al. (2017). Photocyclic Behavior of Rhodopsin Induced by an Atypical Isomerization Mechanism. Proc. Natl. Acad. Sci. U. S. A. 114, E2608–E2615. 10.1073/pnas.1617446114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Wu Y., Gong Z., Chen X., Cao F., Kala S., et al. (2022). Photonic Nanojet-Mediated Optogenetics. Adv. Sci. 9, e2104140. 10.1002/advs.202104140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y. (2020). Be Cautious with Crystal Structures of Membrane Proteins or Complexes Prepared in Detergents. Crystals 10, 86. 10.3390/cryst10020086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S., Freedman M. S., Bellingham J., Inglis S. L., Poopalasundaram S., Soni B. G., et al. (2001). Characterization of a Novel Human Opsin Gene with Wide Tissue Expression and Identification of Embedded and Flanking Genes on Chromosome 1q43. Genomics 72, 203–208. 10.1006/geno.2001.6469 [DOI] [PubMed] [Google Scholar]
- Hallett F. R., Watton J., Krygsman P. (1991). Vesicle Sizing. Number Distributions by Dynamic Light Scattering. Biophys. J. 59, 357–362. 10.1016/s0006-3495(91)82229-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Kim S.-H., Cho J. C., Song J., Bleckner G., Jung K.-H. (2020). Photochemical Characterization of Flavobacterial Rhodopsin: The Importance of the Helix E Region for Heat Stability. Biochimica Biophysica Acta-Bioenergetics 1861, 148092. 10.1016/j.bbabio.2019.148092 [DOI] [PubMed] [Google Scholar]
- Hanai S., Katayama K., Imai H., Kandori H. (2021). Light-induced Difference FTIR Spectroscopy of Primate Blue-Sensitive Visual Pigment at 163 K. Biophysics Physicobiology 18, 40–49. 10.2142/biophysico.bppb-v18.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K. Y., Wada T., Kino K., Asahi T., Sawamura N. (2013). Construction of Photoenergetic Mitochondria in Cultured Mammalian Cells. Sci. Rep. 3, 1635. 10.1038/srep01635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Hara R., Takeuchi J. (1967). Vision in Octopus and Squid. Nature 214, 572–575. 10.1038/214572a0 [DOI] [PubMed] [Google Scholar]
- Harada Y., Senda T., Sakamoto T., Takamoto K., Ishibashi T. (1994). Expression of octopus Rhodopsin in Escherichia coli . J. Biochem. Tokyo 115, 66–75. 10.1093/oxfordjournals.jbchem.a124307 [DOI] [PubMed] [Google Scholar]
- Harbison G. S., Smith S. O., Pardoen J. A., Courtin J. M. L., Lugtenburg J., Herzfeld J., et al. (1985). Solid-state 13C NMR Detection of a Perturbed 6-S-Trans Chromophore in Bacteriorhodopsin. Biochemistry 24, 6955–6962. 10.1021/bi00345a031 [DOI] [PubMed] [Google Scholar]
- Hargrave P. A., Mcdowell J. H., Curtis D. R., Wang J. K., Juszcak E., Fong S.-L., et al. (1983). The Structure of Bovine Rhodopsin. Biophys. Struct. Mech. 9, 235–244. 10.1007/bf00535659 [DOI] [PubMed] [Google Scholar]
- Hargrave P. A., Mcdowell J. H. (1992). Rhodopsin and Phototransduction - A Model System for G-Protein-Linked Receptors. FASEB J. 6, 2323–2331. 10.1096/fasebj.6.6.1544542 [DOI] [PubMed] [Google Scholar]
- Hargrave P. A. (1982). Rhodopsin Chemistry, Structure and Topography. Prog. Retin. Res. 1, 2–51. 10.1016/0278-4327(82)90003-7 [DOI] [Google Scholar]
- Hargrave P. A. (1977). The Amino-Terminal Tryptic Peptide of Bovine Rhodopsin. A Glycopeptide Containing Two Sites of Oligosaccharide Attachment. Biochim. Biophys. Acta 492, 83–94. 10.1016/0005-2795(77)90216-1 [DOI] [PubMed] [Google Scholar]
- Haris P. I., Robillard G. T., Vandijk A. A., Chapman D. (1992). Potential of 13C and 15N Labeling for Studying Protein-Protein Interactions Using Fourier Transform Infrared Spectroscopy. Biochemistry-USA 31, 6279–6284. 10.1021/bi00142a016 [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Hosaka T., Kojima K., Nishimura Y., Nakajima Y., Kimura-Someya T., et al. (2020). A Unique Clade of Light-Driven Proton-Pumping Rhodopsins Evolved in the Cyanobacterial Lineage. Sci. Rep. 10, 16752. 10.1038/s41598-020-73606-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa N., Miki K., Takeda K. (2018). X-ray Structure Analysis of Bacteriorhodopsin at 1.3 Ä Resolution. Sci. Rep. 8, 13123. 10.1038/s41598-018-31370-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasemi T., Kikukawa T., Kamo N., Demura M. (2016). Characterization of a Cyanobacterial Chloride-Pumping Rhodopsin and its Conversion into a Proton Pump. J. Biol. Chem. 291, 355–362. 10.1074/jbc.m115.688614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S., Takeuchi H., Nakagawa M., Tsuda M. (1996). Ultraviolet Resonance Raman Evidence for the Absence of Tyrosinate in octopus Rhodopsin and the Participation of Trp Residues in the Transition to Acid Metarhodopsin. FEBS Lett. 398, 239–242. 10.1016/s0014-5793(96)01250-1 [DOI] [PubMed] [Google Scholar]
- Haupts U., Tittor J., Oesterhelt D. (1999). Closing in on Bacteriorhodopsin: Progress in Understanding the Molecule. Annu. Rev. Biophys. Biomol. Struct. 28, 367–399. 10.1146/annurev.biophys.28.1.367 [DOI] [PubMed] [Google Scholar]
- Havelka W. A., Henderson R. A., Oesterhelt D. (1995). Three-dimensional Structure of Halorhodopsin at 7 Å Resolution. J. Mol. Biol. 247, 726–738. 10.1016/s0022-2836(05)80151-2 [DOI] [PubMed] [Google Scholar]
- Hayashi M., Kojima K., Sudo Y., Yamashita A. (2021). An Optogenetic Assay Method for Electrogenic Transporters Using Escherichia coli Co-expressing Light-Driven Proton Pump. Protein Sci. 30, 2161–2169. 10.1002/pro.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Tajkhorshid E., Pebay-Peyroula E., Royant A., Landau E. M., Navarro J., et al. (2001). Structural Determinants of Spectral Tuning in Retinal Proteins-Bacteriorhodopsin vs Sensory Rhodopsin II. J. Phys. Chem. B 105, 10124–10131. 10.1021/jp011362b [DOI] [Google Scholar]
- Hayashi T., Yasuda S., Suzuki K., Akiyama T., Kanehara K., Kojima K., et al. (2020). How Does a Microbial Rhodopsin RxR Realize its Exceptionally High Thermostability with the Proton-Pumping Function Being Retained? J. Phys. Chem. B 124, 990–1000. 10.1021/acs.jpcb.9b10700 [DOI] [PubMed] [Google Scholar]
- Heath G. R., Kots E., Robertson J. L., Lansky S., Khelashvili G., Weinstein H., et al. (2021). Localization Atomic Force Microscopy. Nature 594, 385–390. 10.1038/s41586-021-03551-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Unwin P. N. T. (1975). Three-dimensional Model of Purple Membrane Obtained by Electron-Microscopy. Nature 257, 28–32. 10.1038/257028a0 [DOI] [PubMed] [Google Scholar]
- Herwig L., Rice A. J., Bedbrook C. N., Zhang R. J. K., Lignell A., Cahn J. K. B., et al. (2017). Directed Evolution of a Bright Near-Infrared Fluorescent Rhodopsin Using a Synthetic Chromophore. Cell. Chem. Biol. 24, 415–425. 10.1016/j.chembiol.2017.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzfeld J., Lansing J. C. (2002). Magnetic Resonance Studies of the Bacteriorhodopsin Pump Cycle. Annu. Rev. Biophysics Biomol. Struct. 31, 73–95. 10.1146/annurev.biophys.31.082901.134233 [DOI] [PubMed] [Google Scholar]
- Heymann J. B., Müller D. J., Mitsuoka K., Engel A. (1997). Electron and Atomic Force Microscopy of Membrane Proteins. Curr. Opin. Struct. Biol. 7, 543–549. 10.1016/s0959-440x(97)80120-0 [DOI] [PubMed] [Google Scholar]
- Hickey D. G., Davies W. I. L., Hughes S., Rodgers J., Thavanesan N., Maclaren R. E., et al. (2021). Chimeric Human Opsins as Optogenetic Light Sensitisers. J. Exp. Biol. 224, 240580. 10.1242/jeb.240580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi A., Shihoya W., Konno M., Ikuta T., Kandori H., Inoue K., et al. (2021). Crystal Structure of Schizorhodopsin Reveals Mechanism of Inward Proton Pumping. Proc. Natl. Acad. Sci. U. S. A. 118, 2016328118. 10.1073/pnas.2016328118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand P. W., Scheerer P., Park J. H., Choe H.-W., Piechnick R., Ernst O. P., et al. (2009). A Ligand Channel through the G Protein Coupled Receptor Opsin. PLoS ONE 4, e4382. 10.1371/journal.pone.0004382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt V., Polakowski F., Büldt G. (1991). Purple Fission Yeast: Overexpression and Processing of the Pigment Bacteriorhodopsin in Schizosaccharomyces pombe . Photochem. Photobiol. 54, 1009–1016. 10.1111/j.1751-1097.1991.tb02123.x [DOI] [Google Scholar]
- Hillman P., Hochstein S., Minke B. (1983). Transduction in Invertebrate Photoreceptors - Role of Pigment Bistability. Physiol. Rev. 63, 668–772. 10.1152/physrev.1983.63.2.668 [DOI] [PubMed] [Google Scholar]
- Hirano T., Fujioka N., Imai H., Kandori H., Wada A., Ito M., et al. (2006). Assignment of the Vibrational Modes of the Chromophores of Iodopsin and Bathoiodopsin: Low-Temperature Fourier Transform Infrared Spectroscopy of 13C and 2H-Labeled Iodopsins. Biochemistry 45, 1285–1294. 10.1021/bi0517077 [DOI] [PubMed] [Google Scholar]
- Hirschi S., Fischer N., Kalbermatter D., Laskowski P. R., Ucurum Z., Müller D. J., et al. (2019). Design and Assembly of a Chemically Switchable and Fluorescently Traceable Light-Driven Proton Pump System for Bionanotechnological Applications. Sci. Rep. 9, 1046. 10.1038/s41598-018-37260-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi S., Kalbermatter D., Ucurum Z., Lemmin T., Fotiadis D. (2021). Cryo-EM Structure and Dynamics of the Green-Light Absorbing Proteorhodopsin. Nat. Commun. 12, 4107. 10.1038/s41467-021-24429-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochbaum D. R., Zhao Y., Farhi S. L., Klapoetke N. C., Werley C. A., Kapoor V., et al. (2014). All-optical Electrophysiology in Mammalian Neurons Using Engineered Microbial Rhodopsins. Nat. Methods 11, 825–833. 10.1038/nmeth.3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A., Hildebrandt V., Heberle J., Büldt G. (1994). Photoactive Mitochondria: In Vivo Transfer of a Light-Driven Proton Pump into the Inner Mitochondrial Membrane of Schizosaccharomyces pombe . Proc. Nat. Acad. Sci. U. S. A. 91, 9367–9371. 10.1073/pnas.91.20.9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K. P. (2000). “Late Photoproducts and Signaling States of Bovine Rhodopsin,” in Molecular Mechanisms in Visual Transduction. Editors Stavenga D. G., DeGrip W. J., PughJr. E. N. (Amsterdam, Netherlands: Elsevier Science Pub.), 91–142. 10.1016/s1383-8121(00)80006-8 [DOI] [Google Scholar]
- Hofmann K. P., Scheerer P., Hildebrand P. W., Choe H. W., Park J. H., Heck M., et al. (2009). A G Protein-Coupled Receptor at Work: the Rhodopsin Model. Trends Biochem. Sci. 34, 540–552. 10.1016/j.tibs.2009.07.005 [DOI] [PubMed] [Google Scholar]
- Hofmann L., Palczewski K. (2015). Advances in Understanding the Molecular Basis of the First Steps in Color Vision. Prog. Retin. Eye Res. 49, 46–66. 10.1016/j.preteyeres.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoi K. K., Bada Juarez J. F., Judge P. J., Yen H.-Y., Wu D., Vinals J., et al. (2021). Detergent-free Lipodisq Nanoparticles Facilitate High-Resolution Mass Spectrometry of Folded Integral Membrane Proteins. Nano Lett. 21, 2824–2831. 10.1021/acs.nanolett.0c04911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoischen D., Steinmüller S., Gärtner W., Buss V., Martin H.-D. (1997). Merocyanines as Extremely Bathochromically Absorbing Chromophores in the Halobacterial Membrane Protein Bacteriorhodopsin. Angew. Chem. Int. Ed. 36, 1630–1633. [Google Scholar]
- Hong F. T. (1994). “Retinal Proteins in Photovoltaic Devices,” in Molecular and Biomolecular Electronics. Editor Birge R. R. (Washington, DC, USA: American Chemical Society; ), 1–27. [Google Scholar]
- Hontani Y., Broser M., Luck M., Weissenborn J., Kloz M., Hegemann P., et al. (2020). Dual Photoisomerization on Distinct Potential Energy Surfaces in a UV-Absorbing Rhodopsin. J. Am. Chem. Soc. 142, 11464–11473. 10.1021/jacs.0c03229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hontani Y., Broser M., Silapetere A., Krause B. S., Hegemann P., Kennis J. T. M. (2017a). The Femtosecond-To-Second Photochemistry of Red-Shifted Fast-Closing Anion Channelrhodopsin PsACR1. Phys. Chem. Chem. Phys. 19, 30402–30409. 10.1039/c7cp06414d [DOI] [PubMed] [Google Scholar]
- Hontani Y., Ganapathy S., Frehan S., Kloz M., De Grip W. J., Kennis J. T. M. (2019). Photoreaction Dynamics of Red-Shifting Retinal Analogues Reconstituted in Proteorhodopsin. J. Phys. Chem. B 123, 4242–4250. 10.1021/acs.jpcb.9b01136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hontani Y., Ganapathy S., Frehan S., Kloz M., De Grip W. J., Kennis J. T. M. (2018). Strong pH-dependent Near-Infrared Fluorescence in a Microbial Rhodopsin Reconstituted with a Red-Shifting Retinal Analogue. J. Phys. Chem. Lett. 9, 6469–6474. 10.1021/acs.jpclett.8b02780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hontani Y., Marazzi M., Stehfest K., Mathes T., Van Stokkum I. H. M., Elstner M., et al. (2017b). Reaction Dynamics of the Chimeric Channelrhodopsin C1C2. Sci. Rep. 7, 7217. 10.1038/s41598-017-07363-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope A. J., Partridge J. C., Dulai K. S., Hunt D. M. (1997). Mechanisms of Wavelength Tuning in the Rod Opsins of Deep-Sea Fishes. Proc. R. Soc. B-Biological Sci. 264, 155–163. 10.1098/rspb.1997.0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornak V., Ahuja S., Eilers M., Goncalves J. A., Sheves M., Reeves P. J., et al. (2010). Light Activation of Rhodopsin: Insights from Molecular Dynamics Simulations Guided by Solid-State NMR Distance Restraints. J. Mol. Biol. 396, 510–527. 10.1016/j.jmb.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka T., Nomura T., Kubo M., Nakane T., Fangjia L., Sekine S.-I., et al. (2022). Conformational Alterations in Unidirectional Ion Transport of a Light-Driven Chloride Pump Revealed Using X-Ray Free Electron Lasers. Proc. Natl. Acad. Sci. U. S. A. 119, e2117433119. 10.1073/pnas.2117433119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka T., Yoshizawa S., Nakajima Y., Ohsawa N., Hato M., Delong E. F., et al. (2016). Structural Mechanism for Light-Driven Transport by a New Type of Chloride Ion Pump, Nonlabens Marinus Rhodopsin-3. J. Biol. Chem. 291, 17488–17495. 10.1074/jbc.m116.728220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. G. G., Sun B. Q. Q., Bizounok M., Hatcher M. E., Lansing J. C., Raap J., et al. (1998). Early and Late M Intermediates in the Bacteriorhodopsin Photocycle: A Solid-State NMR Study. Biochemistry-USA 37, 8088–8096. 10.1021/bi973168e [DOI] [PubMed] [Google Scholar]
- Huang L., Deng H., Koutalos Y., Ebrey T. G., Groesbeek M., Lugtenburg J., et al. (1997). A Resonance Raman Study of the C=C Stretch Modes in Bovine and octopus Visual Pigments with Isotopically Labeled Retinal Chromophores. Photochem. Photobiol. 66, 747–754. 10.1111/j.1751-1097.1997.tb03219.x [DOI] [PubMed] [Google Scholar]
- Hubbard R., Brown P. K., Bownds M. D. (1971). Methodology of Vitamin A and Visual Pigments. Meth. Enzymol. 18C, 615–653. 10.1016/s0076-6879(71)18045-7 [DOI] [Google Scholar]
- Hubbard R., Wald G. (1952). Cis-trans Isomers of Vitamin A and Retinene in the Rhodopsin System. J. General Physiology 36, 269–315. 10.1085/jgp.36.2.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell W. L., Altenbach C., Hubbell C. M., Khorana H. G. (2003). Rhodopsin Structure, Dynamics, and Activation: A Perspective from Crystallography, Site-Directed Spin Labeling, Sulfhydryl Reactivity, and Disulfide Cross-Linking. Adv. Protein Chem. 63, 243–290. 10.1016/s0065-3233(03)63010-x [DOI] [PubMed] [Google Scholar]
- Humphreys I. R., Pei J. M., Baek M., Krishnakumar A., Anishchenko I., Ovchinnikov S., et al. (2021). Computed Structures of Core Eukaryotic Protein Complexes. Science 374, 1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt D. M., Collin S. P. (2014). “The Evolution of Photoreceptors and Visual Photopigments in Vertebrates,” in Evolution of Visual and Non-visual Pigments. Editor Hunt D. M. (New York: Springer Science+Business Media New York; ), 163–217. 10.1007/978-1-4614-4355-1_6 [DOI] [Google Scholar]
- Hunt D. M., Dulai K. S., Partridge J. C., Cottrill P., Bowmaker J. K. (2001). The Molecular Basis for Spectral Tuning of Rod Visual Pigments in Deep-Sea Fish. J. Exp. Biol. 204, 3333–3344. 10.1242/jeb.204.19.3333 [DOI] [PubMed] [Google Scholar]
- Hussain H., Du Y., Scull N. J., Mortensen J. S., Tarrasch J., Bae H. E., et al. (2016). Accessible Mannitol-Based Amphiphiles (MNAs) for Membrane Protein Solubilisation and Stabilisation. Chemistry-A Eur. J. 22, 7068–7073. 10.1002/chem.201600533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S., Kinnebrew M., Schonenbach N. S., Aye E., Han S. G. (2015). Functional Consequences of the Oligomeric Assembly of Proteorhodopsin. J. Mol. Biol. 427, 1278–1290. 10.1016/j.jmb.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwa J., Reeves P. J., Klein-Seetharaman J., Davidson F. F., Khorana H. G. (1999). Structure and Function in Rhodopsin: Further Elucidation of the Role of the Intradiscal Cysteines, Cys-110, -185, and -187, in Rhodopsin Folding and Function. Proc. Nat. Acad. Sci. U. S. A. 96, 1932–1935. 10.1073/pnas.96.5.1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka A., Kajimoto K., Fujisawa T., Tsukamoto T., Aizawa T., Kamo N., et al. (2019). Functional Importance of the Oligomer Formation of the Cyanobacterial H+ Pump Gloeobacter Rhodopsin. Sci. Rep. 9, 10711. 10.1038/s41598-019-47178-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda D., Furutani Y., Kandori H. (2007). FTIR Study of the Retinal Schiff Base and Internal Water Molecules of Proteorhodopsin. Biochemistry 46, 5365–5373. 10.1021/bi700143g [DOI] [PubMed] [Google Scholar]
- Ikuta T., Shihoya W., Sugiura M., Yoshida K., Watari M., Tokano T., et al. (2020). Structural Insights into the Mechanism of Rhodopsin Phosphodiesterase. Nat. Commun. 11, 5605. 10.1038/s41467-020-19376-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H., Hirano T., Terakita A., Shichida Y., Muthyala R. S., Chen R.-L., et al. (1999). Probing for the Threshold Energy for Visual Transduction: Red-Shifted Visual Pigment Analogs from 3-Methoxy-3-Dehydroretinal and Related Compounds. Photochem. Photobiol. 70, 111–115. 10.1111/j.1751-1097.1999.tb01956.x [DOI] [PubMed] [Google Scholar]
- Imai H., Imamoto Y., Yoshizawa T., Shichida Y. (1995). Difference in Molecular Properties between Chicken Green and Rhodopsin as Related to the Functional Difference between Cone and Rod Photoreceptor Cells. Biochemistry-USA 34, 10525–10531. 10.1021/bi00033a026 [DOI] [PubMed] [Google Scholar]
- Imai H., Terakita A., Tachibanaki S., Imamoto Y., Yoshizawa T., Shichida Y. (1997). Photochemical and Biochemical Properties of Chicken Blue- Sensitive Cone Visual Pigment. Biochemistry-USA 36, 12773–12779. 10.1021/bi970809x [DOI] [PubMed] [Google Scholar]
- Imai Y., Inoshita T., Meng H. R., Shiba-Fukushima K., Hara K. Y., Sawamura N., et al. (2019). Light-driven Activation of Mitochondrial Proton-Motive Force Improves Motor Behaviors in a Drosophila Model of Parkinson's Disease. Commun. Biol. 2, 424. 10.1038/s42003-019-0674-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto Y., Kandori H., Okano T., Fukada Y., Shichida Y., Yoshizawa T. (1989). Effect of Chloride Ion on the Thermal Decay Process of the Batho Intermediate of Iodopsin at Low Temperature. Biochemistry 28, 9412–9416. 10.1021/bi00450a025 [DOI] [PubMed] [Google Scholar]
- Imamoto Y., Shichida Y. (2014). Cone Visual Pigments. Biochimica Biophysica Acta-Bioenergetics 1837, 664–673. 10.1016/j.bbabio.2013.08.009 [DOI] [PubMed] [Google Scholar]
- Imamoto Y., Yoshizawa T., Shichida Y. (1996). Chromophore Configuration of Iodopsin and its Photoproducts Formed at Low Temperatures. Biochemistry 35, 14599–14607. 10.1021/bi9614850 [DOI] [PubMed] [Google Scholar]
- Imasheva E. S., Balashov S. P., Wang J. M., Lanyi J. K. (2011). Removal and Reconstitution of the Carotenoid Antenna of Xanthorhodopsin. J. Membr. Biol. 239, 95–104. 10.1007/s00232-010-9322-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki H. K., Jung Y., Hoopfer E. D., Wong A. M., Mishra N., Lin J. Y., et al. (2014). Optogenetic Control of Drosophila Using a Red-Shifted Channelrhodopsin Reveals Experience-dependent Influences on Courtship. Nat. Methods 11, 325–U311. 10.1038/nmeth.2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Del Carmen Marin M., Tomida S., Nakamura R., Nakajima Y., Olivucci M., et al. (2019). Red-shifting Mutation of Light-Driven Sodium-Pump Rhodopsin. Nat. Commun. 10, 1993. 10.1038/s41467-019-10000-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Karasuyama M., Nakamura R., Konno M., Yamada D., Mannen K., et al. (2021). Exploration of Natural Red-Shifted Rhodopsins Using a Machine Learning-Based Bayesian Experimental Design. Commun. Biol. 4, 362. 10.1038/s42003-021-01878-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Nomura Y., Kandori H. (2016). Asymmetric Functional Conversion of Eubacterial Light-Driven Ion Pumps. J. Biol. Chem. 291, 9883–9893. 10.1074/jbc.m116.716498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Reissig L., Sakai M., Kobayashi S., Homma M., Fujii M., et al. (2012). Absorption Spectra and Photochemical Reactions in a Unique Photoactive Protein, Middle Rhodopsin MR. J. Phys. Chem. B 116, 5888–5899. 10.1021/jp302357m [DOI] [PubMed] [Google Scholar]
- Inoue K., Sasaki J., Morisaki M., Tokunaga F., Terazima M. (2004). Time-resolved Detection of Sensory Rhodopsin II-Transducer Interaction. Biophysical J. 87, 2587–2597. 10.1529/biophysj.104.043521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Sudo Y., Homma M., Kandori H. (2011). Spectrally Silent Intermediates during the Photochemical Reactions of Salinibacter Sensory Rhodopsin I. J. Phys. Chem. B 115, 4500–4508. 10.1021/jp2000706 [DOI] [PubMed] [Google Scholar]
- Inoue K., Tahara S., Kato Y., Takeuchi S., Tahara T., Kandori H. (2018). Spectroscopic Study of Proton-Transfer Mechanism of Inward Proton-Pump Rhodopsin, Parvularcula Oceani Xenorhodopsin. J. Phys. Chem. B 122, 6453–6461. 10.1021/acs.jpcb.8b01279 [DOI] [PubMed] [Google Scholar]
- Inoue K., Tsukamoto T., Shimono K., Suzuki Y., Miyauchi S., Hayashi S., et al. (2015). Converting a Light-Driven Proton Pump into a Light-Gated Proton Channel. J. Am. Chem. Soc. 137, 3291–3299. 10.1021/ja511788f [DOI] [PubMed] [Google Scholar]
- Inoue K., Tsukamoto T., Sudo Y. (2014). Molecular and Evolutionary Aspects of Microbial Sensory Rhodopsins. Biochimica Biophysica Acta-Bioenergetics 1837, 562–577. 10.1016/j.bbabio.2013.05.005 [DOI] [PubMed] [Google Scholar]
- Inoue K., Tsunoda S. P., Singh M., Tomida S., Hososhima S., Konno M., et al. (2020). Schizorhodopsins: A Family of Rhodopsins from Asgard Archaea that Function as Light-Driven Inward H+ Pumps. Sci. Adv. 6, 2441. 10.1126/sciadv.aaz2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Iwaki M., Sugita S., Abe-Yoshizumi R., Iwata T., Inoue K., et al. (2018). Unique Hydrogen Bonds in Membrane Protein Monitored by Whole Mid-IR ATR Spectroscopy in Aqueous Solution. J. Phys. Chem. B 122, 165–170. 10.1021/acs.jpcb.7b11064 [DOI] [PubMed] [Google Scholar]
- Iwasa T., Colmenares L. U., Hirata K., Arime Y., Nakagawa M., Kikkawa S., et al. (1998). 19F-NMR and UV-Vis Absorption Spectroscopic Studies of Fluorinated octopus Rhodopsin and its Photoproducts. J. Phys. Chem. A 102, 5602–5610. 10.1021/jp9802477 [DOI] [Google Scholar]
- Iyer E. S. S., Misra R., Maity A., Liubashevski O., Sudo Y., Sheves M., et al. (2016). Temperature Independence of Ultrafast Photoisomerization in Thermophilic Rhodopsin: Assessment versus Other Microbial Proton Pumps. J. Am. Chem. Soc. 138, 12401–12407. 10.1021/jacs.6b05002 [DOI] [PubMed] [Google Scholar]
- Jacobs G. H. (2018). Photopigments and the Dimensionality of Animal Color Vision. Neurosci. Biobehav. Rev. 86, 108–130. 10.1016/j.neubiorev.2017.12.006 [DOI] [PubMed] [Google Scholar]
- Jäger S., Lewis J. W., Zvyaga T. A., Szundi I., Sakmar T. P., Kliger D. S. (1997). Chromophore Structural Changes in Rhodopsin from Nanoseconds to Microseconds Following Pigment Photolysis. Proc. Nat. Acad. Sci. U. S. A. 94, 8557–8562. 10.1073/pnas.94.16.8557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S., Jung K. H., Sheves M. (2020). The Chirality Origin of Retinal-Carotenoid Complex in Gloeobacter Rhodopsin: a Temperature-dependent Excitonic Coupling. Sci. Rep. 10, 13992. 10.1038/s41598-020-70697-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht R., Demartynoff G., Lou J., Hipskind R. A., Nordheim A., Stunnenberg H. G. (1991). Rapid and Efficient Purification of Native Histidine-Tagged Protein Expressed by Recombinant Vaccinia Virus. Proc. Nat. Acad. Sci. U. S. A. 88, 8972–8976. 10.1073/pnas.88.20.8972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen J. J. M., Bovee-Geurts P. H. M., Merkx M., DeGrip W. J. (1995). Histidine Tagging Both Allows Convenient Single-step Purification of Bovine Rhodopsin and Exerts Ionic Strength-dependent Effects on its Photochemistry. J. Biol. Chem. 270, 11222–11229. 10.1074/jbc.270.19.11222 [DOI] [PubMed] [Google Scholar]
- Janssen J. J. M., Mulder W. R., DeCaluwé G. L. J., Vlak J. M., DeGrip W. J. (1991). In Vitro expression of Bovine Opsin Using Recombinant Baculovirus: The Role of Glutamic Acid (134) in Opsin Biosynthesis and Glycosylation. Biochim. Biophys. Acta 1089, 68–76. 10.1016/0167-4781(91)90086-2 [DOI] [PubMed] [Google Scholar]
- Janssen J. J. M., VandeVen W. J. M., VanGroningen-Luyben W. a. H. M., Roosien J., Vlak J. M., DeGrip W. J. (1988). Synthesis of Functional Bovine Opsin in Insect Cells under Control of the Baculovirus Polyhedrin Promotor. Mol. Biol. Rep. 13, 65–71. 10.1007/bf00539052 [DOI] [PubMed] [Google Scholar]
- Janssen J. W. H., David-Gray Z. K., Bovee-Geurts P. H. M., Nevo E., Foster R. G., DeGrip W. J. (2003). A Green Cone-like Pigment in the 'blind' Mole-Rat Spalax Ehrenbergi: Functional Expression and Photochemical Characterization. Photochem. Photobiol. Sci. 2, 1287–1291. 10.1039/b300059c [DOI] [PubMed] [Google Scholar]
- Jastrzebska B., Palczewski K., Golczak M. (2011). Role of Bulk Water in Hydrolysis of the Rhodopsin Chromophore. J. Biol. Chem. 286, 18930–18937. 10.1074/jbc.m111.234583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L. L., Ma B. F., Meng Q., Li L. J., Liu K., Chen D. L. (2017). Detergent-resistant Oligomeric Leptosphaeria Rhodopsin Is a Promising Bio-Nanomaterial and an Alternative to Bacteriorhodopsin. Biochem. Biophysical Res. Commun. 493, 352–357. 10.1016/j.bbrc.2017.09.018 [DOI] [PubMed] [Google Scholar]
- Jiang M. S., Pandey S., Fong H. K. W. (1993). An Opsin Homologue in the Retina and Pigment Epithelium. Investig. Ophthalmol. Vis. Sci. 34, 3669–3678. [PubMed] [Google Scholar]
- Johnson P. J. M., Halpin A., Morizumi T., Brown L. S., Prokhorenko V. I., Ernst O. P., et al. (2014). The Photocycle and Ultrafast Vibrational Dynamics of Bacteriorhodopsin in Lipid Nanodiscs. Phys. Chem. Chem. Phys. 16, 21310–21320. 10.1039/c4cp01826e [DOI] [PubMed] [Google Scholar]
- Johnson P. J. M., Halpin A., Morizumi T., Prokhorenko V. I., Ernst O. P., Miller R. J. D. (2015). Local Vibrational Coherences Drive the Primary Photochemistry of Vision. Nat. Chem. 7, 980–986. 10.1038/nchem.2398 [DOI] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly Accurate Protein Structure Prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun N. Y., Cardin J. A. (2020). Activation of Distinct Channelrhodopsin Variants Engages Different Patterns of Network Activity. Eneuro 7, 0222–0218. 10.1523/ENEURO.0222-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahremany S., Sander C. L., Tochtrop G. P., Kubas A., Palczewski K. (2019). Z-isomerization of Retinoids through Combination of Monochromatic Photoisomerization and Metal Catalysis. Org. Biomol. Chem. 17, 8125–8139. 10.1039/c9ob01645g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandori H., Inoue K., Tsunoda S. P. (2018). Light-Driven Sodium-Pumping Rhodopsin: A New Concept of Active Transport. Chem. Rev. 118, 10646–10658. 10.1021/acs.chemrev.7b00548 [DOI] [PubMed] [Google Scholar]
- Kandori H. (2020). Retinal Proteins: Photochemistry and Optogenetics. Bull. Chem. Soc. Jpn. 93, 76–85. 10.1246/bcsj.20190292 [DOI] [Google Scholar]
- Kanehara K., Yoshizawa S., Tsukamoto T., Sudo Y. (2017). A Phylogenetically Distinctive and Extremely Heat Stable Light-Driven Proton Pump from the Eubacterium Rubrobacter Xylanophilus DSM 9941T . Sci. Rep. 7, 44427. 10.1038/srep44427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko A., Inoue K., Kojima K., Kandori H., Sudo Y. (2017). Conversion of Microbial Rhodopsins: Insights into Functionally Essential Elements and Rational Protein Engineering. Biophys. Rev. 9, 861–876. 10.1007/s12551-017-0335-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan M., Vasan G., Pieribone V. A. (2019). Optimizing Strategies for Developing Genetically Encoded Voltage Indicators. Front. Cell. Neurosci. 13, 53. 10.3389/fncel.2019.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao Y.-M., Cheng C.-H., Syue M.-L., Huang H.-Y., Chen I.-C., Yu T.-Y., et al. (2019). Photochemistry of Bacteriorhodopsin with Various Oligomeric Statuses in Controlled Membrane Mimicking Environments: A Spectroscopic Study from Femtoseconds to Milliseconds. J. Phys. Chem. B 123, 2032–2039. 10.1021/acs.jpcb.9b01224 [DOI] [PubMed] [Google Scholar]
- Karapinar R., Schwitalla J. C., Eickelbeck D., Pakusch J., Mücher B., Grömmke M., et al. (2021). Reverse Optogenetics of G Protein Signaling by Zebrafish Non-visual Opsin Opn7b for Synchronization of Neuronal Networks. Nat. Commun. 12, 4488. 10.1038/s41467-021-24718-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasuyama M., Inoue K., Nakamura R., Kandori H., Takeuchi I. (2018). Understanding Colour Tuning Rules and Predicting Absorption Wavelengths of Microbial Rhodopsins by Data-Driven Machine-Learning Approach. Sci. Rep. 8, 15580–15511. 10.1038/s41598-018-33984-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik S. S., Ridge K. D., Bhattacharya S. S., Khorana H. G. (1993). Palmitoylation of Bovine Opsin and its Cysteine Mutants in COS Cells. Proc. Nat. Acad. Sci. U. S. A. 90, 40–44. 10.1073/pnas.90.1.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik S. S., Sakmar T. P., Chen H.-B., Khorana H. G. (1988). Cysteine Residues 110 and 187 Are Essential for the Formation of Correct Structure in Bovine Rhodopsin. Proc. Nat. Acad. Sci. U. S. A. 85, 8459–8463. 10.1073/pnas.85.22.8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katana R., Guan C. L., Zanini D., Larsen M. E., Giraldo D., Geurten B. R. H., et al. (2019). Chromophore-Independent Roles of Opsin Apoproteins in Drosophila Mechanoreceptors. Curr. Biol. 29, 2961–2969. 10.1016/j.cub.2019.07.036 [DOI] [PubMed] [Google Scholar]
- Katanosaka K., Tokunaga F., Kawamura S., Ozaki K. (1998). N-linked Glycosylation of Drosophila Rhodopsin Occurs Exclusively in the Amino-Terminal Domain and Functions in Rhodopsin Maturation. FEBS Lett. 424, 149–154. 10.1016/s0014-5793(98)00160-4 [DOI] [PubMed] [Google Scholar]
- Kataoka C., Inoue K., Katayama K., Béjà O., Kandori H. (2019). Unique Photochemistry Observed in a New Microbial Rhodopsin. J. Phys. Chem. Lett. 10, 5117–5121. 10.1021/acs.jpclett.9b01957 [DOI] [PubMed] [Google Scholar]
- Katayama K., Furutani Y., Imai H., Kandori H. (2012). Protein-bound Water Molecules in Primate Red- and Green-Sensitive Visual Pigments. Biochemistry 51, 1126–1133. 10.1021/bi201676y [DOI] [PubMed] [Google Scholar]
- Katayama K., Gulati S., Ortega J. T., Alexander N. S., Sun W. Y., Shenouda M. M., et al. (2019). Specificity of the Chromophore-Binding Site in Human Cone Opsins. J. Biol. Chem. 294, 6082–6093. 10.1074/jbc.ra119.007587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama K., Nonaka Y., Tsutsui K., Imai H., Kandori H. (2017). Spectral Tuning Mechanism of Primate Blue-Sensitive Visual Pigment Elucidated by FTIR Spectroscopy. Sci. Rep. 7, 4904. 10.1038/s41598-017-05177-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathe C., Michoud F., Schönle P., Rowald A., Brun N., Ravier J., et al. (2021). Wireless Closed-Loop Optogenetics across the Entire Dorsoventral Spinal Cord in Mice. Nat. Biotechnol. 40, 198. 10.1038/s41587-021-01019-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H. E., Inoue K., Abe-Yoshizumi R., Kato Y., Ono H., Konno M., et al. (2015). Structural Basis for Na+ Transport Mechanism by a Light-Driven Na+ Pump. Nature 521, 48–53. 10.1038/nature14322 [DOI] [PubMed] [Google Scholar]
- Kato H. E., Kamiya M., Sugo S., Ito J., Taniguchi R., Orito A., et al. (2015). Atomistic Design of Microbial Opsin-Based Blue-Shifted Optogenetics Tools. Nat. Commun. 6, 7177. 10.1038/ncomms8177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H. E., Zhang F., Yizhar O., Ramakrishnan C., Nishizawa T., Hirata K., et al. (2012). Crystal Structure of the Channelrhodopsin Light-Gated Cation Channel. Nature 482, 369–374. 10.1038/nature10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz B., Minke B. (2009). Drosophila Photoreceptors and Signaling Mechanisms. Front. Cell. Neurosci. 3, 2. 10.3389/neuro.03.002.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann J. C. D., Krause B. S., Adam S., Ritter E., Schapiro I., Hegemann P., et al. (2020). Modulation of Light Energy Transfer from Chromophore to Protein in the Channelrhodopsin ReaChR. Biophysical J. 119, 705–716. 10.1016/j.bpj.2020.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J., Kriebel C. N., Eberhardt P., Jakdetchai O., Leeder A. J., Weber I., et al. (2019). Solid-state NMR Analysis of the Sodium Pump Krokinobacter Rhodopsin 2 and its H30A Mutant. J. Struct. Biol. 206, 55–65. 10.1016/j.jsb.2018.06.001 [DOI] [PubMed] [Google Scholar]
- Kaushal S., Ridge K. D., Khorana H. G. (1994). Structure and Function in Rhodopsin: The Role of Asparagine-Linked Glycosylation. Proc. Nat. Acad. Sci. U. S. A. 91, 4024–4028. 10.1073/pnas.91.9.4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura I., Seki H., Tajima S., Makino Y., Shigeta A., Okitsu T., et al. (2021). Structure of a Retinal Chromophore of Dark-Adapted Middle Rhodopsin as Studied by Solid-State Nuclear Magnetic Resonance Spectroscopy. Biophysics Physicobiology 18, 177–185. 10.2142/biophysico.bppb-v18.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura S., Gerstung M., Colozo A. T., Helenius J., Maeda A., Beerenwinkel N., et al. (2013). Kinetic, Energetic, and Mechanical Differences between Dark-State Rhodopsin and Opsin. Structure 21, 426–437. 10.1016/j.str.2013.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanabe A., Furutani Y., Jung K.-H., Kandori H. (2007). Photochromism of Anabaena Sensory Rhodopsin. J. Am. Chem. Soc. 129, 8644–8649. 10.1021/ja072085a [DOI] [PubMed] [Google Scholar]
- Kawasaki Y., Konno M., Inoue K. (2021). Thermostable Light-Driven Inward Proton Pump Rhodopsins. Chem. Phys. Lett. 779, 138868. 10.1016/j.cplett.2021.138868 [DOI] [Google Scholar]
- Kazmi M. A., Dubin R. A., Oddoux C., Ostrer H. (1996). High-level Inducible Expression of Visual Pigments in Transfected Cells. BioTechniques 21, 304–311. 10.2144/96212rr05 [DOI] [PubMed] [Google Scholar]
- Kazmin R., Rose A., Szczepek M., Elgeti M., Ritter E., Piechnick R., et al. (2015). The Activation Pathway of Human Rhodopsin in Comparison to Bovine Rhodopsin. J. Biol. Chem. 290, 20117–20127. 10.1074/jbc.m115.652172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelashvili G., Menon A. K. (2022). Phospholipid Scrambling by G Protein-Coupled Receptors. Annu. Rev. Biophysics 51, 39–61. 10.1146/annurev-biophys-090821-083030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodonov A. A., Shevyakov S. V., Mironova E. V., Shvets V. I., Alexeeva S. G., Demina O. V., et al. (2000). Bacteriorhodopsin Analogs, Bearing Modified Chromophore as a Basis for the Photochromic Materials. Mol. Cryst. Liq. Cryst. 345, 641–646. 10.1080/10587250008023938 [DOI] [Google Scholar]
- Khorana H. G., Braiman M. S., Chao B. H., Doi T., Flitsch S. L., Gilles-Gonzalez M. A., et al. (1987). Site-specific Mutagenesis in Structure - Function Studies of Bacteriorhodopsin. Chem. Scr. 27B, 137–147. [Google Scholar]
- Khorana H. G., Knox B. E., Nasi E., Swanson R., Thompson D. A. (1988). Expression of a Bovine Rhodopsin Gene in Xenopus Oocytes: Demonstration of Light-dependent Ionic Currents. Proc. Nat. Acad. Sci. U. S. A. 85, 7917–7921. 10.1073/pnas.85.21.7917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorana H. G. (1979). Total Synthesis of a Gene. Science 203, 614–625. 10.1126/science.366749 [DOI] [PubMed] [Google Scholar]
- Kikukawa T. (2021). Unique Cl- Pump Rhodopsin with Close Similarity to H+ Pump Rhodopsin. Biophysics Physicobiology 18, 317–326. 10.2142/biophysico.bppb-v18.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-J., Kwon Y. D., Lee S. Y., Kim P. (2012). An Engineered Escherichia coli Having a High Intracellular Level of ATP and Enhanced Recombinant Protein Production. Appl. Microbiol. Biotechnol. 94, 1079–1086. 10.1007/s00253-011-3779-0 [DOI] [PubMed] [Google Scholar]
- Kim H. A., Kim H. J., Park J., Choi A. R., Heo K., Jeong H., et al. (2017). An Evolutionary Optimization of a Rhodopsin-Based Phototrophic Metabolism in Escherichia coli . Microb. Cell. Factories 16, 111. 10.1186/s12934-017-0725-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-Y., Waschuk S. A., Brown L. S., Jung K.-H. (2008). Screening and Characterization of Proteorhodopsin Color-Tuning Mutations in Escherichia coli with Endogenous Retinal Synthesis. Biochimica Biophysica Acta-Bioenergetics 1777, 504–513. 10.1016/j.bbabio.2008.03.010 [DOI] [PubMed] [Google Scholar]
- Kimura Y., Vassylyev D. G., Miyazawa A., Kidera A., Matsushima M., Mitsuoka K., et al. (1997). High Resolution Structure of Bacteriorhodopsin Determined by Electron Crystallography. Photochem. Photobiol. 66, 764–767. 10.1111/j.1751-1097.1997.tb03221.x [DOI] [Google Scholar]
- Kirchman D. L., Hanson T. E. (2013). Bioenergetics of Photoheterotrophic Bacteria in the Oceans. Environ. Microbiol. Rep. 5, 188–199. 10.1111/j.1758-2229.2012.00367.x [DOI] [PubMed] [Google Scholar]
- Kishi K. E., Kim Y. S., Fukuda M., Inoue M., Kusakizako T., Wang P. Y., et al. (2022). Structural Basis for Channel Conduction in the Pump-like Channelrhodopsin ChRmine. Cell. 185, 1–18. 10.1016/j.cell.2022.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen C. H. W., DeGrip W. J. (2000). Baculovirus Expression System for Expression and Characterization of Functional Recombinant Visual Pigments. Meth. Enzymol. 315, 12–29. 10.1016/s0076-6879(00)15832-x [DOI] [PubMed] [Google Scholar]
- Klapoetke N. C., Murata Y., Kim S. S., Pulver S. R., Birdsey-Benson A., Cho Y. K., et al. (2014). Independent Optical Excitation of Distinct Neural Populations. Nat. Methods 11, 338–U333. 10.1038/nmeth.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klyszejko A. L., Shastri S., Mari S. A., Grubmüller H., Müller D. J., Glaubitz C. (2008). Folding and Assembly of Proteorhodopsin. J. Mol. Biol. 376, 35–41. 10.1016/j.jmb.2007.11.030 [DOI] [PubMed] [Google Scholar]
- Knoot C. J., Ungerer J., Wangikar P. P., Pakrasi H. B. (2018). Cyanobacteria: Promising Biocatalysts for Sustainable Chemical Production. J. Biol. Chem. 293, 5044–5052. 10.1074/jbc.r117.815886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles T. J., Finka R., Smith C., Lin Y.-P., Dafforn T., Overduin M. (2009). Membrane Proteins Solubilized Intact in Lipid Containing Nanoparticles Bounded by Styrene Maleic Acid Copolymer. J. Am. Chem. Soc. 131, 7484–7485. 10.1021/ja810046q [DOI] [PubMed] [Google Scholar]
- Knudsen P., Hubbell W. L. (1978). Stability of Rhodopsin in Detergent Solutions. Membr. Biochem. 1, 297–322. 10.3109/09687687809063853 [DOI] [PubMed] [Google Scholar]
- Kochendoerfer G. G., Lin S. W., Sakmar T. P., Mathies R. A. (1999). How Color Visual Pigments Are Tuned. Trends biochem. Sci. 24, 300–305. 10.1016/s0968-0004(99)01432-2 [DOI] [PubMed] [Google Scholar]
- Kojima D., Imai H., Okano T., Fukada Y., Crescitelli F., Yoshizawa T., et al. (1995). Purification and Low Temperature Spectroscopy of Gecko Visual Pigments Green and Blue. Biochemistry-USA 34, 1096–1106. 10.1021/bi00003a047 [DOI] [PubMed] [Google Scholar]
- Kojima D., Oura T., Hisatomi O., Tokunaga F., Fukada Y., Yoshizawa T., et al. (1996). Molecular Properties of Chimerical Mutants of Gecko Blue and Bovine Rhodopsin. Biochemistry 35, 2625–2629. 10.1021/bi9511548 [DOI] [PubMed] [Google Scholar]
- Kojima D., Terakita A., Ishikawa T., Tsukahara Y., Maeda A., Shichida Y. (1997). A Novel Go-Mediated Phototransduction Cascade in Scallop Visual Cells. J. Biol. Chem. 272, 22979–22982. 10.1074/jbc.272.37.22979 [DOI] [PubMed] [Google Scholar]
- Kojima K., Kurihara R., Sakamoto M., Takanashi T., Kuramochi H., Zhang X. M., et al. (2020a). Comparative Studies of the Fluorescence Properties of Microbial Rhodopsins: Spontaneous Emission versus Photointermediate Fluorescence. J. Phys. Chem. B 124, 7361–7367. 10.1021/acs.jpcb.0c06560 [DOI] [PubMed] [Google Scholar]
- Kojima K., Miyoshi N., Shibukawa A., Chowdhury S., Tsujimura M., Noji T., et al. (2020c). Green-sensitive, Long-Lived, Step-Functional Anion Channelrhodopsin-2 Variant as a High-Potential Neural Silencing Tool. J. Phys. Chem. Lett. 11, 6214–6218. 10.1021/acs.jpclett.0c01406 [DOI] [PubMed] [Google Scholar]
- Kojima K., Shibukawa A., Sudo Y. (2020b). The Unlimited Potential of Microbial Rhodopsins as Optical Tools. Biochemistry 59, 218–229. 10.1021/acs.biochem.9b00768 [DOI] [PubMed] [Google Scholar]
- Kojima K., Ueta T., Noji T., Saito K., Kanehara K., Yoshizawa S., et al. (2020d). Vectorial Proton Transport Mechanism of RxR, a Phylogenetically Distinct and Thermally Stable Microbial Rhodopsin. Sci. Rep. 10, 282. 10.1038/s41598-019-57122-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno M., Inoue K., Kandori H. (2021). Ion Transport Activity Assay for Microbial Rhodopsin Expressed in Escherichia coli Cells. Bio-Protocol 11, 4115. 10.21769/bioprotoc.4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf A. H., Dörr J. M., Koorengevel M. C., Antoniciello F., Jahn H., Killian J. A. (2020). Factors Influencing the Solubilization of Membrane Proteins from Escherichia coli Membranes by Styrene-Maleic Acid Copolymers. Biochimica Biophysica Acta-Biomembranes 1862, 183125. 10.1016/j.bbamem.2019.183125 [DOI] [PubMed] [Google Scholar]
- Kovalev K., Astashkin R., Gushchin I., Orekhov P., Volkov D., Zinovev E. V., et al. (2020a). Molecular Mechanism of Light-Driven Sodium Pumping. Nat. Commun. 11, 21371. 10.1038/s41467-020-16032-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalev K., Polovinkin V., Gushchin I., Alekseev A., Shevchenko V., Borshchevskiy V., et al. (2019). Structure and Mechanisms of Sodium-Pumping KR2 Rhodopsin. Sci. Adv. 5, eaav2671. 10.1126/sciadv.aav2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalev K., Volkov D., Astashkin R., Alekseev A., Gushchin I., Haro-Moreno J. M., et al. (2020b). High-resolution Structural Insights into the Heliorhodopsin Family. Proc. Natl. Acad. Sci. U. S. A. 117, 4131–4141. 10.1073/pnas.1915888117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi M., Terakita A. (2014). Diversity of Animal Opsin-Based Pigments and Their Optogenetic Potential. Biochimica Biophysica Acta-Bioenergetics 1837, 710–716. 10.1016/j.bbabio.2013.09.003 [DOI] [PubMed] [Google Scholar]
- Kraack J. P., Buckup T., Motzkus M. (2011). Vibrational Analysis of Excited and Ground Electronic States of All-Trans Retinal Protonated Schiff-Bases. Phys. Chem. Chem. Phys. 13, 21402–21410. 10.1039/c1cp22245g [DOI] [PubMed] [Google Scholar]
- Krah M., Marwan W., Verméglio A., Oesterhelt D. (1994). Phototaxis of Halobacterium Salinarium Requires a Signalling Complex of Sensory Rhodopsin I and its Methyl-Accepting Transducer HtrI. EMBO J. 13, 2150–2155. 10.1002/j.1460-2075.1994.tb06491.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj J. M., Douglass A. D., Hochbaum D. R., Maclaurin D., Cohen A. E. (2012). Optical Recording of Action Potentials in Mammalian Neurons Using a Microbial Rhodopsin. Nat. Methods 9, 90–95. 10.1038/nmeth.1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj J. M., Hochbaum D. R., Douglass A. D., Cohen A. E. (2011). Electrical Spiking in Escherichia coli Probed with a Fluorescent Voltage-Indicating Protein. Science 333, 345–348. 10.1126/science.1204763 [DOI] [PubMed] [Google Scholar]
- Krebs A., Edwards P. C., Villa C., Li J.-D., Schertler G. F. X. (2003). The Three-Dimensional Structure of Bovine Rhodopsin Determined by Electron Cryomicroscopy. J. Biol. Chem. 278, 50217–50225. 10.1074/jbc.M307995200 [DOI] [PubMed] [Google Scholar]
- Krebs M. P., Mollaaghababa R., Khorana H. G. (1993). Gene Replacement in Halobacterium Halobium and Expression of Bacteriorhodopsin Mutants. Proc. Nat. Acad. Sci. U. S. A. 90, 1987–1991. 10.1073/pnas.90.5.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol A., Lopez-Huerta V. G., Corey T. E. C., Deisseroth K., Ting J. T., Feng G. P. (2019). Two eARCHT3.0 Lines for Optogenetic Silencing of Dopaminergic and Serotonergic Neurons. Front. Neural Circuits 13, 4. 10.3389/fncir.2019.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropf A. (1982). A New Detergent for the Study of Visual Pigments. Vis. Res. 22, 495–497. 10.1016/0042-6989(82)90199-7 [DOI] [PubMed] [Google Scholar]
- Kropf A. (1975). The Nature of the Chromophore-Protein Interaction in Visual Pigments as Studied by Visual Pigment Analogues. Abstr. Annu. Meet. Biophysical Soc. Jpn. 31, 281–282. [Google Scholar]
- Kropf A., Whittenberger B. P., Goff S. P., Waggoner A. S. (1973). The Spectral Properties of Some Visual Pigment Analogs. Exp. Eye Res. 17, 591–606. 10.1016/0014-4835(73)90088-2 [DOI] [PubMed] [Google Scholar]
- Kuang L. J., Fernandes D. A., O'halloran M., Zheng W., Jiang Y. J., Ladizhansky V., et al. (2014). Frozen" Block Copolymer Nanomembranes with Light-Driven Proton Pumping Performance. ACS Nano 8, 537–545. 10.1021/nn4059852 [DOI] [PubMed] [Google Scholar]
- Kühn H. (1984). Interactions between Photoexcited Rhodopsin and Light-Activated Enzymes in Rods. Prog. Retin. Res. 3, 123–156. [Google Scholar]
- Kuhne J., Eisenhauer K., Ritter E., Hegemann P., Gerwert K., Bartl F. J. (2015). Early Formation of the Ion-Conducting Pore in Channelrhodopsin-2. Angew. Chemie-International Ed. 54, 4953–4957. 10.1002/anie.201410180 [DOI] [PubMed] [Google Scholar]
- Kuhne J., Vierock J., Tennigkeit S. A., Dreier M.-A., Wietek J., Petersen D., et al. (2019). Unifying Photocycle Model for Light Adaptation and Temporal Evolution of Cation Conductance in Channelrhodopsin-2. Proc. Natl. Acad. Sci. U. S. A. 116, 9380–9389. 10.1073/pnas.1818707116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni R. U., Miller E. W. (2017). Voltage Imaging: Pitfalls and Potential. Biochemistry 56, 5171–5177. 10.1021/acs.biochem.7b00490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbalasiri T., Provencio I. (2005). Melanopsin and Other Novel Mammalian Opsins. Exp. Eye Res. 81, 368–375. 10.1016/j.exer.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Kushibiki T., Okawa S., Hirasawa T., Ishihara M. (2014). Optogenetics: Novel Tools for Controlling Mammalian Cell Functions with Light. Int. J. Photoenergy 2014, 895039. 10.1155/2014/895039 [DOI] [Google Scholar]
- Kusnetzow A. K., Altenbach C., Hubbell W. L. (2006). Conformational States and Dynamics of Rhodopsin in Micelles and Bilayers. Biochemistry-USA 45, 5538–5550. 10.1021/bi060101v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnetzow A. K., Dukkipati A., Babu K. R., Ramos L., Knox B. E., Birge R. R. (2004). Vertebrate Ultraviolet Visual Pigments: Protonation of the Retinylidene Schiff Base and a Counterion Switch during Photoactivation. Proc. Natl. Acad. Sci. U. S. A. 101, 941–946. 10.1073/pnas.0305206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnetzow A. K., Dukkipati A., Babu K. R., Singh D., Vought B. W., Knox B. E., et al. (2001). The Photobleaching Sequence of a Short-Wavelength Visual Pigment. Biochemistry 40, 7832–7844. 10.1021/bi010387y [DOI] [PubMed] [Google Scholar]
- Kwon S.-K., Jun S.-H., Kim J. F. (2020). Omega Rhodopsins: A Versatile Class of Microbial Rhodopsins. J. Microbiol. Biotechnol. 30, 633–641. 10.4014/jmb.1912.12010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y. M., Patra A. K., Chiura H. X., Kim S.-J. (2019). Production of Extracellular Vesicles with Light-Induced Proton Pump Activity by Proteorhodopsin-Containing Marine Bacteria. MicrobiologyOpen 8, e808. 10.1002/mbo3.808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladizhansky V. (2017). “Advances in Solid-State NMR Studies of Microbial Rhodopsins,” in Modern Magnetic Resonance. Editor Webb G. A. (New York City: Springer International Publishing AG; ), 1–19. 10.1007/978-3-319-28275-6_65-1 [DOI] [Google Scholar]
- Lamarche L. B., Kumar R. P., Trieu M. M., Devine E. L., Cohen-Abeles L. E., Theobald D. L., et al. (2017). Purification and Characterization of RhoPDE, a Retinylidene/Phosphodiesterase Fusion Protein and Potential Optogenetic Tool from the Choanoflagellate Salpingoeca Rosetta . Biochemistry 56, 5812–5822. 10.1021/acs.biochem.7b00519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y., Wang Y., Lu H. (2020). Opsin 3 Is a Key Regulator of Ultraviolet A-Induced Photoageing in Human Dermal Fibroblast Cells. Br. J. Dermatology 182, 1228–1244. 10.1111/bjd.18410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang-Hinrichs C., Queck I., Büldt G., Stahl U., Hildebrandt V. (1994). The Archaebacterial Membrane Protein Bacterio-Opsin Is Expressed and N-Terminally Processed in the Yeast Saccharomyces cerevisiae . Mol. Gen. Genet. 244, 183–188. 10.1007/bf00283521 [DOI] [PubMed] [Google Scholar]
- Lanyi J. K. (2004). Bacteriorhodopsin. Annu. Rev. Physiology 66, 665–688. 10.1146/annurev.physiol.66.032102.150049 [DOI] [PubMed] [Google Scholar]
- Larkum A. W. D., Ritchie R. J., Raven J. A. (2018). Living off the Sun: Chlorophylls, Bacteriochlorophylls and Rhodopsins. Photosynthetica 56, 11–43. 10.1007/s11099-018-0792-x [DOI] [Google Scholar]
- Lavington S., Watts A. (2020). Lipid Nanoparticle Technologies for the Study of G Protein-Coupled Receptors in Lipid Environments. Biophys. Rev. 12, 1287–1302. 10.1007/s12551-020-00775-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Huang K.-C., Mei G. X., Mamaeva N., DeGrip W. J., Rothschild K. J., et al. (2019). “Pre-resonance Stimulated Raman Scattering Spectroscopy and Imaging of Membrane Potential Using Near-Infrared Rhodopsins,” in Multiphoton Microscopy in the Biomedical Sciences. Editors Periasamy M., So P. T. C., König K. (Bellingham, U.S.A: SPIE; ), 81. 10.1117/12.2506833 [DOI] [Google Scholar]
- Lee S., Ghosh S., Jana S., Robertson N., Tate C. G., Vaidehi N. (2020). How Do Branched Detergents Stabilize GPCRs in Micelles? Biochemistry 59, 2125–2134. 10.1021/acs.biochem.0c00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen K., Nokia M. S., Takala H. (2022). Red Light Optogenetics in Neuroscience. Front. Cell. Neurosci. 15, 778900. 10.3389/fncel.2021.778900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesca E., Panneels V., Schertler G. F. X. (2018). The Role of Water Molecules in Phototransduction of Retinal Proteins and G Protein-Coupled Receptors. Faraday Discuss. 207, 27–37. 10.1039/c7fd00207f [DOI] [PubMed] [Google Scholar]
- Leung N. Y., Montell C. (2017). Unconventional Roles of Opsins. Annu. Rev. Cell. Dev. Biol. 33, 241–264. 10.1146/annurev-cellbio-100616-060432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung N. Y., Thakur D. P., Gurav A. S., Kim S. H., Di Pizio A., Niv M. Y., et al. (2020). Functions of Opsins in Drosophila Taste. Curr. Biol. 30, 1367–1379. 10.1016/j.cub.2020.01.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. W., Hug S. J., Wallace-Williams S. E., Kliger D. S. (1990). Direct Evidence for an Equilibrium between Early Photolysis Intermediates of Rhodopsin. J. Am. Chem. Soc. 112, 6711–6712. 10.1021/ja00174a040 [DOI] [Google Scholar]
- Lewis J. W., Van Kuijk F. J. G. M., Carruthers J. A., Kliger D. S. (1997). Metarhodopsin III Formation and Decay Kinetics: Comparison of Bovine and Human Rhodopsin. Vis. Res. 37, 1–8. 10.1016/s0042-6989(96)00138-1 [DOI] [PubMed] [Google Scholar]
- Li H., Huang C.-Y., Govorunova E. G., Schafer C. T., Sineshchekov O. A., Wang M. T., et al. (2019). Crystal Structure of a Natural Light-Gated Anion Channelrhodopsin. eLife 8, e41741. 10.7554/eLife.41741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Huang C.-Y., Govorunova E. G., Sineshchekov O. A., Yi A., Rothschild K. J., et al. (2021). The Crystal Structure of Bromide-Bound GtACR1 Reveals a Pre-activated State in the Transmembrane Anion Tunnel. Elife 10, 65903. 10.7554/elife.65903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Edwards P. C., Burghammer M., Villa C., Schertler G. F. X. (2004). Structure of Bovine Rhodopsin in a Trigonal Crystal Form. J. Mol. Biol. 343, 1409–1438. 10.1016/j.jmb.2004.08.090 [DOI] [PubMed] [Google Scholar]
- Li L. Z., Lu L. H., Ren Y., Tang G., Zhao Y., Cai X., et al. (2022). Colocalized, Bidirectional Optogenetic Modulations in Freely Behaving Mice with a Wireless Dual-Color Optoelectronic Probe. Nat. Commun. 13, 839. 10.1038/s41467-022-28539-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Steinberg G., Livnah N., Sheves M., Ebrey T. G., Tsuda M. (1994). The pKa of the Protonated Schiff Bases of Gecko Cone and octopus Visual Pigments. Biophys. J. 67, 848–854. 10.1016/s0006-3495(94)80544-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichty J. J., Malecki J. L., Agnew H. D., Michelson-Horowitz D. J., Tan S. (2005). Comparison of Affinity Tags for Protein Purification. Protein Expr. Purif. 41, 98–105. 10.1016/j.pep.2005.01.019 [DOI] [PubMed] [Google Scholar]
- Liebel M., Schnedermann C., Bassolino G., Taylor G., Watts A., Kukura P. (2014). Direct Observation of the Coherent Nuclear Response after the Absorption of a Photon. Phys. Rev. Lett. 112, 238301. 10.1103/PhysRevLett.112.238301 [DOI] [PubMed] [Google Scholar]
- Liebert A., Pang V., Bicknell B., Mclachlan C., Mitrofanis J., Kiat H. (2021). A Perspective on the Potential of Opsins as an Integral Mechanism of Photobiomodulation: It's Not Just the Eyes. Photobiomodul Photomed. Laser Surg. 40, 123–135. 10.1089/photob.2021.0106 [DOI] [PubMed] [Google Scholar]
- Lin J. Y., Knutsen P. M., Muller A., Kleinfeld D., Tsien R. Y. (2013). ReaChR: a Red-Shifted Variant of Channelrhodopsin Enables Deep Transcranial Optogenetic Excitation. Nat. Neurosci. 16, 1499–1508. 10.1038/nn.3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. W., Kochendoerfer G. G., Carroll H. S., Wang D., Mathies R. A., Sakmar T. P. (1998). Mechanisms of Spectral Tuning in Blue Cone Visual Pigments - Visible and Raman Spectroscopy of Blue-Shifted Rhodopsin Mutants. J. Biol. Chem. 273, 24583–24591. 10.1074/jbc.273.38.24583 [DOI] [PubMed] [Google Scholar]
- Lin S. W., Sakmar T. P., Franke R. R., Khorana H. G., Mathies R. A. (1992). Resonance Raman Microprobe Spectroscopy of Rhodopsin Mutants: Effect of Substitutions in the Third Transmembrane Helix. Biochemistry-USA 31, 5105–5111. 10.1021/bi00137a003 [DOI] [PubMed] [Google Scholar]
- Lincereghino J., Cregg J. M. (2000). Heterologous Protein Expression in the Methylotrophic Yeast Pichia pastoris . FEMS Microbiol. Rev. 24, 45–66. 10.1111/j.1574-6976.2000.tb00532.x [DOI] [PubMed] [Google Scholar]
- Lindner M., Gilhooley M. J., Peirson S. N., Hughes S., Hankins M. W. (2021). The Functional Characteristics of Optogenetic Gene Therapy for Vision Restoration. Cell. Mol. Life Sci. 78, 1597–1613. 10.1007/s00018-020-03597-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips D., Schuurmans J. M., Branco Dos Santos F., Hellingwerf K. J. (2018). Many Ways towards 'solar Fuel': Quantitative Analysis of the Most Promising Strategies and the Main Challenges during Scale-Up. Energy & Environ. Sci. 11, 10–22. 10.1039/c7ee02212c [DOI] [Google Scholar]
- Liu R. S. H., Asato A. E. (1990). “The Binding Site of Opsin Based on Analog Studies with Isomeric, Fluorinated, Alkylated, and Other Modified Retinals,” in Chemistry and Biology of Synthetic Retinoids. Editors Dawson M. I., Okamura W. H. (Boca Raton, Fl, U.S.A. CRC Press; ), 52–75. [Google Scholar]
- Liu R. S. H., Asato A. E. (2003). Tuning the Color and Excited State Properties of the Azulenic Chromophore: NIR Absorbing Pigments and Materials. J. Photochem. Photobiol. C Photochem. Rev. 4, 179–194. 10.1016/j.jphotochemrev.2003.09.001 [DOI] [Google Scholar]
- Liu R. S. H., Liu J. (2011). Fluorinated Retinoids and Carotenoids. J. Nat. Prod. 74, 512–517. 10.1021/np1006394 [DOI] [PubMed] [Google Scholar]
- Liu R. S. H., Matsumoto H., Kini A., Asato A. E., Denny M., Kropf A., et al. (1984). Seven New Hindered Isomeric Rhodopsins - A Reexamination of the Stereospecificity of the Binding-Site of Bovine Opsin. Tetrahedron 40, 473–482. 10.1016/s0040-4020(01)88435-0 [DOI] [Google Scholar]
- Liu Y., Cui Y. M., Chi H., Xia Y., Liu H. N., Rossiter S. J., et al. (2019). Scotopic Rod Vision in Tetrapods Arose from Multiple Early Adaptive Shifts in the Rate of Retinal Release. Proc. Natl. Acad. Sci. U. S. A. 116, 12627–12628. 10.1073/pnas.1900481116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Z., Zhang W., Du X. X., Liu Y. X., Qu J. B., Liu X. B., et al. (2021). Genome‐wide Identification of Nonvisual Opsin Family Reveals Amplification of RPE‐retinal G Protein Receptor Gene (RGR) and Offers Novel Insights into Functions of RGR(s) in Paralichthys olivaceus (Paralichthyidae, Teleostei). J. Exp. Zoology Part B Mol. Dev. Evol. 334, 25–36. 10.1002/jez.b.22914 [DOI] [PubMed] [Google Scholar]
- Locket N. A. (1977). “Adaptations to the Deep-Sea Environment,” in The Visual System in Vertebrates. Editor Crescitelli F. (Berlin: Springer-Verlag; ), 67–192. [Google Scholar]
- López S., Rodriguez V., Montenegro J., Saá C., Alvarez R., López C. S., et al. (2005). Synthesis of N-Heteroaryl Retinals and Their Artificial Bacteriorhodopsins. ChemBioChem 6, 2078–2087. 10.1002/cbic.200500148 [DOI] [PubMed] [Google Scholar]
- Lórenz-Fonfría V. A., Bamann C., Resler T., Schlesinger R., Bamberg E., Heberle J. (2015a). Temporal Evolution of Helix Hydration in a Light-Gated Ion Channel Correlates with Ion Conductance. Proc. Natl. Acad. Sci. U. S. A. 112, E5796–E5804. 10.1073/pnas.1511462112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lórenz-Fonfría V. A., Kandori H. (2009). Spectroscopic and Kinetic Evidence on How Bacteriorhodopsin Accomplishes Vectorial Proton Transport under Functional Conditions. J. Am. Chem. Soc. 131, 5891–5901. 10.1021/ja900334c [DOI] [PubMed] [Google Scholar]
- Lórenz-Fonfría V. A., Schultz B.-J., Resler T., Schlesinger R., Bamann C., Bamberg E., et al. (2015b). Pre-gating Conformational Changes in the ChETA Variant of Channelrhodopsin-2 Monitored by Nanosecond IR Spectroscopy. J. Am. Chem. Soc. 137, 1850–1861. 10.1021/ja5108595 [DOI] [PubMed] [Google Scholar]
- Lórenz-Fonfría V. A., Yagi K., Ito S., Kandori H. (2021). Retinal Vibrations in Bacteriorhodopsin Are Mechanically Harmonic but Electrically Anharmonic: Evidence from Overtone and Combination Bands. Front. Mol. Biosci. 8, 749261. 10.3389/fmolb.2021.749261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Zhou X. E., Gao X., Wang N., Xia R. X., Xu Z. M., et al. (2020). Crystal Structure of Heliorhodopsin 48C12. Cell. Res. 30, 88–90. 10.1038/s41422-019-0266-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck M., Velázquez Escobar F., Glass K., Sabotke M.-I., Hagedorn R., Corellou F., et al. (2019). Photoreactions of the Histidine Kinase Rhodopsin Ot-HKR from the Marine Picoalga Ostreococcus Tauri . Biochemistry 58, 1878–1891. 10.1021/acs.biochem.8b01200 [DOI] [PubMed] [Google Scholar]
- Luecke H., Schobert B., Richter H.-T., Cartailler J.-P., Lanyi J. K. (1999). Structure of Bacteriorhodopsin at 1.55 Å Resolution. J. Mol. Biol. 291, 899–911. 10.1006/jmbi.1999.3027 [DOI] [PubMed] [Google Scholar]
- Luecke H., Schobert B., Stagno J., Imasheva E. S., Wang J. M., Balashov S. P., et al. (2008). Crystallographic Structure of Xanthorhodopsin, the Light-Driven Proton Pump with a Dual Chromophore. Proc. Natl. Acad. Sci. U. S. A. 105, 16561–16565. 10.1073/pnas.0807162105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenburg J., Creemers A. F. L., Verhoeven M. A., Van Wijk A. a. C., Verdegem P. J. E., Monnee M. C. F., et al. (1999). Synthesis of 13C-Labeled Carotenoids and Retinoids. Pure Appl. Chem. 71, 2245–2251. 10.1351/pac199971122245 [DOI] [Google Scholar]
- Lugtenburg J., Mathies R. A., Griffin R. G., Herzfeld J. (1988). Structure and Function of Rhodopsins from Solid State NMR and Resonance Raman Spectroscopy of Isotopic Retinal Derivatives. Trends biochem. Sci. 13, 388–393. 10.1016/0968-0004(88)90181-8 [DOI] [PubMed] [Google Scholar]
- Luk H. L., Bhattacharyya N., Montisci F., Morrow J. M., Melaccio F., Wada A., et al. (2016). Modulation of Thermal Noise and Spectral Sensitivity in Lake Baikal Cottoid Fish Rhodopsins. Sci. Rep. 6, 38425. 10.1038/srep38425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutnaes B. F., Kildahl-Andersen G., Krane J., Liaaen-Jensen S. (2004). Delocalized Carotenoid Cations in Relation to the Soliton Model. J. Am. Chem. Soc. 126, 8981–8990. 10.1021/ja0492541 [DOI] [PubMed] [Google Scholar]
- Ma J.-X., Kono M., Xu L., Das J., Ryan J. C., Hazard E. S., Iii, et al. (2001). Salamander UV Cone Pigment: Sequence, Expression, and Spectral Properties. Vis. Neurosci. 18, 393–399. 10.1017/s0952523801183057 [DOI] [PubMed] [Google Scholar]
- Maclaurin D., Venkatachalam V., Lee H., Cohen A. E. (2013). Mechanism of Voltage-Sensitive Fluorescence in a Microbial Rhodopsin. Proc. Natl. Acad. Sci. U. S. A. 110, 5939–5944. 10.1073/pnas.1215595110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A., Sasaki J., Pfefferle J. M., Shichida Y., Yoshizawa T. (1991). Fourier Transform Infrared Spectral Studies on the Schiff Base Mode of All-Trans Bacteriorhodopsin and its Photointermediates-K and Photointermediates-L. Photochem. Photobiol. 54, 911–921. 10.1111/j.1751-1097.1991.tb02111.x [DOI] [Google Scholar]
- Maeda T., Imanishi Y., Palczewski K. (2003). Rhodopsin Phosphorylation: 30 Years Later. Prog. Retin. Eye Res. 22, 417–434. 10.1016/s1350-9462(03)00017-x [DOI] [PubMed] [Google Scholar]
- Mahn M., Saraf-Sinik I., Patil P., Pulin M., Bitton E., Karalis N., et al. (2021). Efficient Optogenetic Silencing of Neurotransmitter Release with a Mosquito Rhodopsin. Neuron 109, 1621–1635. 10.1016/j.neuron.2021.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmerberg E., Bovee-Geurts P. H. M., Katona G., Deupi X., Arnlund D., Wickstrand C., et al. (2015). Conformational Activation of Visual Rhodopsin in Native Disc Membranes. Sci. Signal. 8, ra26. 10.1126/scisignal.2005646 [DOI] [PubMed] [Google Scholar]
- Maly T., Debelouchina G. T., Bajaj V. S., Hu K.-N., Joo C.-G., Mak-Jurkauskas M. L., et al. (2008). Dynamic Nuclear Polarization at High Magnetic Fields. J. Chem. Phys. 128, 052211. 10.1063/1.2833582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J. F., Aladin V., Jin X. S., Leeder A. J., Brown L. J., Brown R. C. D., et al. (2019). Exploring Protein Structures by DNP-Enhanced Methyl Solid-State NMR Spectroscopy. J. Am. Chem. Soc. 141, 19888–19901. 10.1021/jacs.9b11195 [DOI] [PubMed] [Google Scholar]
- Mao J. F., Do N.-N., Scholz F., Reggie L., Mehler M., Lakatos A., et al. (2014). Structural Basis of the Green-Blue Color Switching in Proteorhodopsin as Determined by NMR Spectroscopy. J. Am. Chem. Soc. 136, 17578–17590. 10.1021/ja5097946 [DOI] [PubMed] [Google Scholar]
- Marrero H., Rothschild K. J. (1987). Conformational Changes in Bacteriorhodopsin Studied by Infrared Attenuated Total Reflection. Biophys. J. 52, 629–635. 10.1016/s0006-3495(87)83254-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshel J. H., Kim Y. S., Machado T. A., Quirin S., Benson B., Kadmon J., et al. (2019). Cortical Layer-specific Critical Dynamics Triggering Perception. Science 365, eaaw5202. 10.1126/science.aaw5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A., Bradley A. S., Waldbauer J. R., Summons R. E., Delong E. F. (2007). Proteorhodopsin Photosystem Gene Expression Enables Photophosphorylation in a Heterologous Host. Proc. Natl. Acad. Sci. U. S. A. 104, 5590–5595. 10.1073/pnas.0611470104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S., Morita E. H., Tasumi M., Iwasa T., Tsuda M. (1993). Infrared Studies of octopus Rhodopsin - Existence of a Long- Lived Intermediate and the States of the Carboxylic Group of Asp- 81 in Rhodopsin and its Photoproducts. FEBS Lett. 317, 223–227. 10.1016/0014-5793(93)81280-d [DOI] [PubMed] [Google Scholar]
- Matarèse B. F. E., Feyen P. L. C., De Mello J. C., Benfenati F. (2019). Sub-millisecond Control of Neuronal Firing by Organic Light-Emitting Diodes. Front. Bioeng. Biotechnol. 7, 278. 10.3389/fbioe.2019.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathies R. A., Lugtenburg J. (2000). “The Primary Photoreaction of Rhodopsin,” in Molecular Mechanisms in Visual Transduction. Editors Stavenga D. G., DeGrip W. J., PughJr. E. N. (Amsterdam, Netherlands: Elsevier Science Pub.), 55–90. 10.1016/s1383-8121(00)80005-6 [DOI] [Google Scholar]
- Mathies R. A., Smith S. O., Palings I. (1987). “Determination of Retinal Chromophore Structure in Rhodopsins,” in Resonance Raman Spectra of Polyenes and Aromatics. Editor Spiro T. G. (New York: John Wiley & Sons; ), 59–108. [Google Scholar]
- Matsui S., Seidou M., Uchiyama I., Sekiya N., Hiraki K., Yoshihara K., et al. (1988). 4-Hydroxyretinal, a New Visual Pigment Chromophore Found in the Bioluminescent Squid, Watasenia Scintillans . Biochimica Biophysica Acta 966, 370–374. 10.1016/0304-4165(88)90087-6 [DOI] [PubMed] [Google Scholar]
- Mccamant D. W., Kukura P., Mathies R. A. (2005). Femtosecond Stimulated Raman Study of Excited-State Evolution in Bacteriorhodopsin. J. Phys. Chem. B 109, 10449–10457. 10.1021/jp050095x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdermott A. E. (2009). Structure and Dynamics of Membrane Proteins by Magic Angle Spinning Solid-State NMR. Annu. Rev. Biophysics 38, 385–403. 10.1146/annurev.biophys.050708.133719 [DOI] [PubMed] [Google Scholar]
- Mcisaac R. S., Bedbrook C. N., Arnold F. H. (2015). Recent Advances in Engineering Microbial Rhodopsins for Optogenetics. Curr. Opin. Struct. Biol. 33, 8–15. 10.1016/j.sbi.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcisaac R. S., Engqvist M. K. M., Wannier T., Rosenthal A. Z., Herwig L., Flytzanis N. C., et al. (2014). Directed Evolution of a Far-Red Fluorescent Rhodopsin. Proc. Natl. Acad. Sci. U. S. A. 111, 13034–13039. 10.1073/pnas.1413987111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckee A. G., Kuntz C. P., Ortega J. T., Woods H., Most V., Roushar F. J., et al. (2021). Systematic Profiling of Temperature- and Retinal-Sensitive Rhodopsin Variants by Deep Mutational Scanning. J. Biol. Chem. 2021, 101359. 10.1016/j.jbc.2021.101359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mederos S., Hernández-Vivanco A., Ramírez-Franco J., Martín-Fernández M., Navarrete M., Yang A., et al. (2019). Melanopsin for Precise Optogenetic Activation of Astrocyte-Neuron Networks. Glia 67, 915–934. 10.1002/glia.23580 [DOI] [PubMed] [Google Scholar]
- Mei G. X., Cavini C. M., Mamaeva N., Wang P., DeGrip W. J., Rothschild K. J. (2021). Optical Switching between Long-Lived States of Opsin Transmembrane Voltage Sensors. Photochem. Photobiol. 97, 1001–1015. 10.1111/php.13428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei G. X., Mamaeva N., Ganapathy S., Wang P., DeGrip W. J., Rothschild K. J. (2020). Analog Retinal Redshifts Visible Absorption of QuasAr Transmembrane Voltage Sensors into Near-Infrared. Photochem. Photobiol. 96, 55–66. 10.1111/php.13169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei G. X., Mamaeva N., Ganapathy S., Wang P., DeGrip W. J., Rothschild K. J. (2018). Raman Spectroscopy of a Near Infrared Absorbing Proteorhodopsin: Similarities to the Bacteriorhodopsin O Photointermediate. Plos One 13, e0209506. 10.1371/journal.pone.0209506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melaccio F., Del Carmen Marín M., Valentini A., Montisci F., Rinaldi S., Cherubini M., et al. (2016). Toward Automatic Rhodopsin Modeling as a Tool for High-Throughput Computational Photobiology. J. Chem. Theory Comput. 12, 6020–6034. 10.1021/acs.jctc.6b00367 [DOI] [PubMed] [Google Scholar]
- Melyan Z., Tarttelin E. E., Bellingham J., Lucas R. J., Hankins M. W. (2005). Addition of Human Melanopsin Renders Mammalian Cells Photoresponsive. Nature 433, 741–745. 10.1038/nature03344 [DOI] [PubMed] [Google Scholar]
- Milosevic M. M., Jang J., Mckimm E. J., Zhu M. H., Antic S. D. (2020). In Vitro Testing of Voltage Indicators: Archon1, ArcLightD, ASAP1, ASAP2s, ASAP3b, Bongwoori-Pos6, BeRST1, FlicR1, and Chi-VSFP-Butterfly. eNeuro 7, 0060. 10.1523/eneuro.0060-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A. K., Gragg M., Stoneman M. R., Biener G., Oliver J. A., Miszta P., et al. (2016). Quaternary Structures of Opsin in Live Cells Revealed by FRET Spectrometry. Biochem. J. 473, 3819–3836. 10.1042/bcj20160422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra R., Eliash T., Sudo Y., Sheves M. (2019). Retinal-Salinixanthin Interactions in a Thermophilic Rhodopsin. J. Phys. Chem. B 123, 10–20. 10.1021/acs.jpcb.8b06795 [DOI] [PubMed] [Google Scholar]
- Mitra A. K., Miercke L. J. W., Turner G. J., Shand R. F., Betlach M. C., Stroud R. M. (1993). Two-dimensional Crystallization of Escherichia Coli-Expressed Bacteriorhodopsin and its D96N Variant: High Resolution Structural Studies in Projection. Biophys. J. 65, 1295–1306. 10.1016/s0006-3495(93)81169-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuoka K., Hirai T., Murata K., Miyazawa A., Kidera A., Kimura Y., et al. (1999). The Structure of Bacteriorhodopsin at 3.0 Å Resolution Based on Electron Crystallography: Implication of the Charge Distribution. J. Mol. Biol. 286, 861–882. 10.1006/jmbi.1998.2529 [DOI] [PubMed] [Google Scholar]
- Miyasaka T., Koyama K., Itoh I. (1992). Quantum Conversion and Image Detection by a Bacteriorhodopsin- Based Artificial Photoreceptor. Science 255, 342–344. 10.1126/science.255.5042.342 [DOI] [PubMed] [Google Scholar]
- Molday R. S. (1989). Monoclonal Antibodies to Rhodopsin and Other Proteins of Rod Outer Segments. Prog. Retin. Res. 8, 173–209. [Google Scholar]
- Mollaaghababa R., Davidson F. F., Kaiser C., Khorana H. G. (1996). Structure and Function in Rhodopsin: Expression of Functional Mammalian Opsin in Saccharomyces cerevisiae . Proc. Nat. Acad. Sci. U. S. A. 93, 11482–11486. 10.1073/pnas.93.21.11482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollevanger L. C. P. J., Kentgens A. P. M., Pardoen J. A., Courtin J. M. L., Veeman W. S., Lugtenburg J., et al. (1987). High-resolution Solid-State 13C-NMR Study of Carbons C-5 and C-12 of the Chromophore of Bovine Rhodopsin: Evidence for a 6-S-Cis Conformation with Negative-Charge Perturbation Near C-12. Eur. J. Biochem. 163, 9–14. 10.1111/j.1432-1033.1987.tb10729.x [DOI] [PubMed] [Google Scholar]
- Mooney V. L., Szundi I., Lewis J. W., Yan E. C. Y., Kliger D. S. (2012). Schiff Base Protonation Changes in Siberian Hamster Ultraviolet Cone Pigment Photointermediates. Biochemistry 51, 2630–2637. 10.1021/bi300157r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes M. N., Monteiro De Assis L. V., Provencio I., De Lauro Castrucci A. M. (2021). Opsins outside the Eye and the Skin: a More Complex Scenario Than Originally Thought for a Classical Light Sensor. Cell. Tissue Res. 385, 519–538. 10.1007/s00441-021-03500-0 [DOI] [PubMed] [Google Scholar]
- Morello J.-P., Bouvier M. (1996). Palmitoylation: A Post-translational Modification that Regulates Signalling from G Protein-Coupled Receptors. Biochem. Cell. Biol. 74, 449–457. 10.1139/o96-049 [DOI] [PubMed] [Google Scholar]
- Mori A., Yagasaki J., Homma M., Reissig L., Sudo Y. (2013). Investigation of the Chromophore Binding Cavity in the 11-cis Acceptable Microbial Rhodopsin MR. Chem. Phys. 419, 23–29. 10.1016/j.chemphys.2012.11.020 [DOI] [Google Scholar]
- Morizumi T., Ou W.-L., Van Eps N., Inoue K., Kandori H., Brown L. S., et al. (2019). X-ray Crystallographic Structure and Oligomerization of Gloeobacter Rhodopsin. Sci. Rep. 9, 11283–1128111215. 10.1038/s41598-019-47445-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton R. A., Goodwin T. W. (1944). Preparation of Retinene In Vitro . Nature 153, 405–406. 10.1038/153405a0 [DOI] [Google Scholar]
- Morton R. A., Pitt G. A. J. (1957). Visual Pigments. Fortschritte Chem. Org. Naturst. 14, 244–316. 10.1007/978-3-7091-7164-6_6 [DOI] [PubMed] [Google Scholar]
- Mous S., Gotthard G., Ehrenberg D., Sen S., Weinert T., Johnson P. J. M., et al. (2022). Dynamics and Mechanism of a Light-Driven Chloride Pump. Science 375, 845. 10.1126/science.abj6663 [DOI] [PubMed] [Google Scholar]
- Moutsaki P., Whitmore D. H., Bellingham J., Sakamoto K., David-Gray Z. K., Foster R. G. (2003). Teleost Multiple Tissue (Tmt) Opsin: A Candidate Photopigment Regulating the Peripheral Clocks of Zebrafish? Mol. Brain Res. 112, 135–145. 10.1016/s0169-328x(03)00059-7 [DOI] [PubMed] [Google Scholar]
- Mroginski M.-A., Adam S., Amoyal G. S., Barnoy A., Bondar A.-N., Borin V. A., et al. (2021). Frontiers in Multiscale Modeling of Photoreceptor Proteins. Photochem. Photobiol. 97, 243–269. 10.1111/php.13372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Hegemann P., Broser M. (2019). Enzymerhodopsins: Novel Photoregulated Catalysts for Optogenetics. Curr. Opin. Struct. Biol. 57, 118–126. 10.1016/j.sbi.2019.02.003 [DOI] [PubMed] [Google Scholar]
- Müller D. J., Kessler M., Oesterhelt F., Möller C., Oesterhelt D., Gaub H. (2002). Stability of Bacteriorhodopsin α-helices and Loops Analyzed by Single-Molecule Force Spectroscopy. Biophysical J. 83, 3578–3588. 10.1016/S0006-3495(02)75358-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H. (1855). Über die entoptische Wahrnemung der Netzhautgefässe, insbesondere als Beweismittel für die Lichtperception durch die nach hinten gelegene Netzhautelemente. Verhandlungen. Physikalisch-Medizinische Gesellschaft Würzburg 5, 411–447. [Google Scholar]
- Müller M. (1992). Proteolysis in Protein Import and Export: Signal Peptide Processing in Eu- and Prokaryotes. Experientia 48, 118–129. [DOI] [PubMed] [Google Scholar]
- Munro R. A., De Vlugt J., Ward M. E., Kim S. Y., Lee K. A., Jung K.-H., et al. (2019). Biosynthetic Production of Fully Carbon-13 Labeled Retinal in E. coli for Structural and Functional Studies of Rhodopsins. J. Biomol. NMR 73, 49–58. 10.1007/s10858-019-00225-9 [DOI] [PubMed] [Google Scholar]
- Muradin-Szweykowska M., Peters A. J. M., Lugtenburg J. (1984). The Interaction of Bacterioopsin with 11, 14-bridged Retinals - the Synthesis of 13-demethyl-11, 14-Imino-Retinal, 13-Demethyl-N-Methyl-11, 14-imino, 13-demethyl-11, 14-Thio-Retinal, 13-demethyl-11, 14-Etheno-Retinal, 11, 14-Imino-Retinal and Their Binding with Bacterioopsin. Recl. Des. Trav. Chim. Des. Pays-Bas 103, 105–109. [Google Scholar]
- Murakami M., Kouyama T. (2008). Crystal Structure of Squid Rhodopsin. Nature 453, 363–U333. 10.1038/nature06925 [DOI] [PubMed] [Google Scholar]
- Murakami M., Kouyama T. (2011). Crystallographic Analysis of the Primary Photochemical Reaction of Squid Rhodopsin. J. Mol. Biol. 413, 615–627. 10.1016/j.jmb.2011.08.044 [DOI] [PubMed] [Google Scholar]
- Murakami M., Kouyama T. (2015). Crystallographic Study of the LUMI Intermediate of Squid Rhodopsin. PLoS ONE 10, e0126970. 10.1371/journal.pone.0126970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musilova Z., Cortesi F., Matschiner M., Davies W. I. L., Patel J. S., Stieb S. M., et al. (2019). Vision Using Multiple Distinct Rod Opsins in Deep-Sea Fishes. Science 364, 588–592. 10.1126/science.aav4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musio C., Santillo S., Taddei-Ferretti C., Robles L. J., Vismara R., Barsanti L., et al. (2001). First Identification and Localization of a Visual Pigment in Hydra (Cnidaria, Hydrozoa). J. Comp. Physiology A - Sens. Neural Behav. Physiology 187, 79–81. 10.1007/s003590100180 [DOI] [PubMed] [Google Scholar]
- Mustafi D., Engel A. H., Palczewski K. (2009). Structure of Cone Photoreceptors. Prog. Retin. Eye Res. 28, 289–302. 10.1016/j.preteyeres.2009.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Y.-A., Lee J.-Y., Bang W.-J., Lee H. J., Choi S.-I., Kwon S.-K., et al. (2015). Growth Retardation of Escherichia coli by Artificial Increase of Intracellular ATP. J. Industrial Microbiol. Biotechnol. 42, 915–924. 10.1007/s10295-015-1609-6 [DOI] [PubMed] [Google Scholar]
- Nagata T., Inoue K. (2022). Rhodopsins at a Glance. J. Cell. Sci. 134, jcs258989. 10.1242/jcs.258989 [DOI] [PubMed] [Google Scholar]
- Nagata T., Koyanagi M., Lucas R., Terakita A. (2018). An All-Trans-Retinal-Binding Opsin Peropsin as a Potential Dark-Active and Light-Inactivated G Protein-Coupled Receptor. Sci. Rep. 8, 3535. 10.1038/s41598-018-21946-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T., Koyanagi M., Tsukamoto H., Mutt E., Schertler G. F. X., Deupi X., et al. (2019). The Counterion-Retinylidene Schiff Base Interaction of an Invertebrate Rhodopsin Rearranges upon Light Activation. Commun. Biol. 2, 180–181189. 10.1038/s42003-019-0409-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G., Szellas T., Huhn W., Kateriya S., Adeishvili N., Berthold P., et al. (2003). Channelrhodopsin-2, a Directly Light-Gated Cation-Selective Membrane Channel. Proc. Natl. Acad. Sci. U. S. A. 100, 13940–13945. 10.1073/pnas.1936192100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito A., Makino Y., Shigeta A., Kawamura I. (2019). Photoreaction Pathways and Photointermediates of Retinal-Binding Photoreceptor Proteins as Revealed by In Situ Photoirradiation Solid-State NMR Spectroscopy. Biophys. Rev. 11, 167–181. 10.1007/s12551-019-00501-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M., Iwasa T., Kikkawa S., Takao T., Shimonishi Y., Tsuda M. (1997). Identification of Two Palmitoyl Groups in octopus Rhodopsin. Photochem. Photobiol. 65, 185–189. 10.1111/j.1751-1097.1997.tb01897.x [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Iwasa T., Kikkawa S., Tsuda M., Ebrey T. G. (1999). How Vertebrate and Invertebrate Visual Pigments Differ in Their Mechanism of Photoactivation. Proc. Nat. Acad. Sci. U. S. A. 96, 6189–6192. 10.1073/pnas.96.11.6189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y., Pedraza-González L., Barneschi L., Inoue K., Olivucci M., Kandori H. (2021). Pro219 Is an Electrostatic Color Determinant in the Light-Driven Sodium Pump KR2. Commun. Biol. 4, 1185. 10.1038/s42003-021-02684-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y., Tsukamoto T., Kumagai Y., Ogura Y., Hayashi T., Song J., et al. (2018). Presence of a Haloarchaeal Halorhodopsin-like Cl- Pump in Marine Bacteria. Microbes Environ. 33, 89–97. 10.1264/jsme2.me17197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi H., Okada T. (2006). Crystallographic Analysis of Primary Visual Photochemistry. Angew. Chem. Int. Ed. 45, 4270–4273. 10.1002/anie.200600595 [DOI] [PubMed] [Google Scholar]
- Nakamura A., Kojima D., Imai H., Terakita A., Okano T., Shichida Y., et al. (1999). Chimeric Nature of Pinopsin between Rod and Cone Visual Pigments. Biochemistry-USA 38, 14738–14745. 10.1021/bi9913496 [DOI] [PubMed] [Google Scholar]
- Nakao S., Kojima K., Sudo Y. (2021). Microbial Rhodopsins as Multi-Functional Photoreactive Membrane Proteins for Optogenetics. Biol. Pharm. Bull. 44, 1357–1363. 10.1248/bpb.b21-00544 [DOI] [PubMed] [Google Scholar]
- Nakao S., Kojima K., Sudo Y. (2022). Phototriggered Apoptotic Cell Death (PTA) Using the Light-Driven Outward Proton Pump Rhodopsin Archaerhodopsin-3. J. Am. Chem. Soc. 144, 3771. 10.1021/jacs.1c12608 [DOI] [PubMed] [Google Scholar]
- Nakatsuma A., Yamashita T., Sasaki K., Kawanabe A., Inoue K., Furutani Y., et al. (2011). Chimeric Microbial Rhodopsins Containing the Third Cytoplasmic Loop of Bovine Rhodopsin. Biophysical J. 100, 1874–1882. 10.1016/j.bpj.2011.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T. A., Khorana H. G. (1991). Mapping of the Amino Acids in Membrane-Embedded Helices that Interact with the Retinal Chromophore in Bovine Rhodopsin. J. Biol. Chem. 266, 4269–4275. 10.1016/s0021-9258(20)64317-4 [DOI] [PubMed] [Google Scholar]
- Nango E., Royant A., Kubo M., Nakane T., Wickstrand C., Kimura T., et al. (2016). A Three-Dimensional Movie of Structural Changes in Bacteriorhodopsin. Science 354, 1552–1557. 10.1126/science.aah3497 [DOI] [PubMed] [Google Scholar]
- Nathans J. (1987). Molecular Biology of Visual Pigments. Annu. Rev. Neurosci. 10, 163–194. 10.1146/annurev.ne.10.030187.001115 [DOI] [PubMed] [Google Scholar]
- Nathans J. (1992). Rhodopsin - Structure, Function, and Genetics. Biochemistry 31, 4923–4931. 10.1021/bi00136a001 [DOI] [PubMed] [Google Scholar]
- Neitz M., Neitz J. (1998). “Molecular Genetics and the Biological Basis of Color Vision,” in Color Vision - Perspectives from Different Disciplines. Editors Backhaus W., Kliegl R. (Berlin, Germany: Walter de Gruyter & Co.), 101–119. [Google Scholar]
- Neumann K., Verhoefen M.-K., Weber I., Glaubitz C., Wachtveitl J. (2008). Initial Reaction Dynamics of Proteorhodopsin Observed by Femtosecond Infrared and Visible Spectroscopy. Biophysical J. 94, 4796–4807. 10.1529/biophysj.107.125484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neutze R., Brändén G., Schertler G. F. X. (2015). Membrane Protein Structural Biology Using X-Ray Free Electron Lasers. Curr. Opin. Struct. Biol. 33, 115–125. 10.1016/j.sbi.2015.08.006 [DOI] [PubMed] [Google Scholar]
- Nguyen K.-A., Peuchmaur M., Magnard S., Haudecoeur R., Boyère C., Mounien S., et al. (2018). Glycosyl-Substituted Dicarboxylates as Detergents for the Extraction, Overstabilization, and Crystallization of Membrane Proteins. Angew. Chemie-International Ed. 57, 2948–2952. 10.1002/anie.201713395 [DOI] [PubMed] [Google Scholar]
- Nielsen M. B. (2009). Model Systems for Understanding Absorption Tuning by Opsin Proteins. Chem. Soc. Rev. 38, 913–924. 10.1039/b802068j [DOI] [PubMed] [Google Scholar]
- Nikolaev D. M., Manathunga M., Orozco-Gonzalez Y., Shtyrov A. A., Omar Guerrero Martínez Y., Gozem S., et al. (2021). Free Energy Computation for an Isomerizing Chromophore in a Molecular Cavity via the Average Solvent Electrostatic Configuration Model: Applications in Rhodopsin and Rhodopsin-Mimicking Systems. J. Chem. Theory Comput. 17, 5885–5895. 10.1021/acs.jctc.1c00221 [DOI] [PubMed] [Google Scholar]
- Nikolaev D. M., Shtyrov A. A., Mereshchenko A. S., Panov M. S., Tveryanovich Y. S., Ryazantsev M. N. (2020). An Assessment of Water Placement Algorithms in Quantum Mechanics/molecular Mechanics Modeling: the Case of Rhodopsins' First Spectral Absorption Band Maxima. Phys. Chem. Chem. Phys. 22, 18114–18123. 10.1039/d0cp02638g [DOI] [PubMed] [Google Scholar]
- Nikolaev D. M., Shtyrov A. A., Panov M. S., Jamal A., Chakchir O. B., Kochemirovsky V. A., et al. (2018). A Comparative Study of Modern Homology Modeling Algorithms for Rhodopsin Structure Prediction. ACS Omega 3, 7555–7566. 10.1021/acsomega.8b00721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogly P., Weinert T., James D., Carbajo S., Ozerov D., Furrer A., et al. (2018). Retinal Isomerization in Bacteriorhodopsin Captured by a Femtosecond X-Ray Laser. Science 361, 145–151. 10.1126/science.aat0094 [DOI] [PubMed] [Google Scholar]
- Nomura Y., Ito S., Teranishi M., Ono H., Inoue K., Kandori H. (2018). Low-temperature FTIR Spectroscopy Provides Evidence for Protein-Bound Water Molecules in Eubacterial Light-Driven Ion Pumpsl. Phys. Chem. Chem. Phys. 20, 3165–3171. 10.1039/c7cp05674e [DOI] [PubMed] [Google Scholar]
- Nonaka Y., Hanai S., Katayama K., Imai H., Kandori H. (2020). Unique Retinal Binding Pocket of Primate Blue-Sensitive Visual Pigment. Biochemistry 59, 2602–2607. 10.1021/acs.biochem.0c00394 [DOI] [PubMed] [Google Scholar]
- O'tousa J. E. (1992). Requirement of N-Linked Glycosylation Site in Drosophila Rhodopsin. Vis. Neurosci. 8, 385–390. 10.1017/s0952523800004910 [DOI] [PubMed] [Google Scholar]
- Oda K., Nomura T., Nakane T., Yamashita K., Inoue K., Ito S., et al. (2021). Time-resolved Serial Femtosecond Crystallography Reveals Early Structural Changes in Channelrhodopsin. eLife 10, 62389. 10.7554/eLife.62389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K., Vierock J., Oishi S., Rodriguez-Rozada S., Taniguchi R., Yamashita K., et al. (2018). Crystal Structure of the Red Light-Activated Channelrhodopsin Chrimson. Nat. Commun. 9, 3949. 10.1038/s41467-018-06421-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D., Bräuchle C., Hampp N. (1991). Bacteriorhodopsin - A Biological Material for Information Processing. Quart. Rev. Biophys. 24, 425–478. 10.1017/s0033583500003863 [DOI] [PubMed] [Google Scholar]
- Oesterhelt D., Hess B. (1973). Reversible Photolysis of Purple Complex in Purple Membrane of Halobacterium Halobium . Eur. J. Biochem. 37, 316–326. 10.1111/j.1432-1033.1973.tb02990.x [DOI] [PubMed] [Google Scholar]
- Oesterhelt D., Stoeckenius W. (1971). Rhodopsin-like Protein from the Purple Membrane of Halobacterium Halobium . Nat. New Biol. 233, 149–152. 10.1038/newbio233149a0 [DOI] [PubMed] [Google Scholar]
- Oesterhelt D. (1998). The Structure and Mechanism of the Family of Retinal Proteins from Halophilic Archaea. Curr. Opin. Struct. Biol. 8, 489–500. 10.1016/s0959-440x(98)80128-0 [DOI] [PubMed] [Google Scholar]
- Ogonah O., Shuler M. L., Granados R. R. (1991). Protein Production (β-Galactosidase) from a Baculovirus Vector in Spodoptera Frugiperda and Trichpolusia Ni Cells in Suspension Culture. Biotechnol. Lett. 13, 265–270. 10.1007/bf01041482 [DOI] [Google Scholar]
- Ogren J. I., Yi A., Mamaev S., Li H., Lugtenburg J., DeGrip W. J., et al. (2015). Comparison of the Structural Changes Occurring during the Primary Phototransition of Two Different Channelrhodopsins from Chlamydomonas Algae. Biochemistry 54, 377–388. 10.1021/bi501243y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T., Sugihara M., Bondar A.-N., Elstner M., Entel P., Buss V. (2004). The Retinal Conformation and its Environment in Rhodopsin in Light of a New 2.2 Å Crystal Structure. J. Mol. Biol. 342, 571–583. 10.1016/j.jmb.2004.07.044 [DOI] [PubMed] [Google Scholar]
- Okano T., Fukada Y., Artamonov I. D., Yoshizawa T. (1989). Purification of Cone Visual Pigments from Chicken Retina. Biochemistry-USA 28, 8848–8856. 10.1021/bi00448a025 [DOI] [PubMed] [Google Scholar]
- Okano T., Yoshizawa T., Fukada Y. (1994). Pinopsin Is a Chicken Pineal Photoreceptive Molecule. Nature 372, 94–97. 10.1038/372094a0 [DOI] [PubMed] [Google Scholar]
- Okitsu T., Yamano Y., Shen Y. C., Sasaki T., Kobayashi Y., Morisawa S., et al. (2020). Synthesis of One Double Bond-Inserted Retinal Analogs and Their Binding Experiments with Opsins: Preparation of Novel Red-Shifted Channelrhodopsin Variants. Chem. Pharm. Bull. 68, 265–272. 10.1248/cpb.c19-01005 [DOI] [PubMed] [Google Scholar]
- Olinski L. E., Lin E. M., Oancea E. (2020). Illuminating Insights into Opsin 3 Function in the Skin. Adv. Biol. Regul. 75, 100668. 10.1016/j.jbior.2019.100668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppermann J., Fischer P., Silapetere A., Liepe B., Rodriguez-Rozada S., Flores-Uribe J., et al. (2019). MerMAIDs: a Family of Metagenomically Discovered Marine Anion-Conducting and Intensely Desensitizing Channelrhodopsins. Nat. Commun. 10, 3315. 10.1038/s41467-019-11322-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprian D. D., Asenjo A. B., Lee N., Pelletier S. L. (1991). Design, Chemical Synthesis, and Expression of Genes for the Three Human Color Vision Pigments. Biochemistry 30, 11367–11372. 10.1021/bi00112a002 [DOI] [PubMed] [Google Scholar]
- Oprian D. D., Molday R. S., Kaufman R. J., Khorana H. G. (1987). Expression of a Synthetic Bovine Rhodopsin Gene in Monkey Kidney Cells. Proc. Nat. Acad. Sci. U. S. A. 84, 8874–8878. 10.1073/pnas.84.24.8874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega J. T., Jastrzebska B. (2019). The Retinoid and Non-retinoid Ligands of the Rod Visual G Protein-Coupled Receptor. Int. J. Mol. Sci. 20, 6218. 10.3390/ijms20246218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Abdulaev N. G., Bogachuk A. S. (1988a). Two Adjacent Cysteine Residues in the C-Terminal Cytoplasmic Fragment of Bovine Rhodopsin Are Palmitylated. FEBS Lett. 230, 1–5. 10.1016/0014-5793(88)80628-8 [DOI] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Abdulaev N. G., Zolotarev A. S., Artamonov I. D., Bespalov I. A., Dergachev A. E., et al. (1988b). Octopus Rhodopsin - Amino Acid Sequence Deduced from C-DNA. FEBS Lett. 232, 69–72. 10.1016/0014-5793(88)80388-0 [DOI] [PubMed] [Google Scholar]
- Owen S. F., Liu M. H., Kreitzer A. C. (2019). Thermal Constraints on In Vivo Optogenetic Manipulations. Nat. Neurosci. 22, 1061–1065. 10.1038/s41593-019-0422-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki Y., Kawashima T., Abe-Yoshizumi R., Kandori H. (2014). A Color-Determining Amino Acid Residue of Proteorhodopsin. Biochemistry 53, 6032–6040. 10.1021/bi500842w [DOI] [PubMed] [Google Scholar]
- Palczewski K. (2012). Chemistry and Biology of Vision. J. Biol. Chem. 287, 1612–1619. 10.1074/jbc.r111.301150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K. (2006). G Protein-Coupled Receptor Rhodopsin. Annu. Rev. Biochem. 75, 743–767. 10.1146/annurev.biochem.75.103004.142743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., et al. (2000). Crystal Structure of Rhodopsin: A G Protein-Coupled Receptor. Science 289, 739–745. 10.1126/science.289.5480.739 [DOI] [PubMed] [Google Scholar]
- Palczewski K., Orban T. (2013). From Atomic Structures to Neuronal Functions of G Protein-Coupled Receptors. Annu. Rev. Neurosci. 36, 139–164. 10.1146/annurev-neuro-062012-170313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palings I., Pardoen J. A., Vandenberg E. M. M., Winkel C., Lugtenburg J., Mathies R. A. (1987). Assignment of Fingerprint Vibrations in the Resonance Raman Spectra of Rhodopsin, Isorhodopsin, and Bathorhodopsin: Implications for Chromophore Structure and Environment. Biochemistry-USA 26, 2544–2556. 10.1021/bi00383a021 [DOI] [PubMed] [Google Scholar]
- Panda S., Nayak S. K., Campo B., Walker J. R., Hogenesch J. B., Jegla T. (2005). Illumination of the Melanopsin Signaling Pathway. Science 307, 600–604. 10.1126/science.1105121 [DOI] [PubMed] [Google Scholar]
- Panda S., Sato T. K., De Lauro Castrucci A. M., DeGrip W. J., Rollag M. D., Hogenesch J. B., et al. (2002). Melanopsin (Opn4) Is Required for Circadian Phase Shifting under Low Light Conditions. Science 298, 2213–2216. 10.1126/science.1076848 [DOI] [PubMed] [Google Scholar]
- Pande C., Pande A., Yue K. T., Callender R. H., Ebrey T. G., Tsuda M. (1987). Resonance Raman Spectroscopy of octopus Rhodopsin and its Photoproducts. Biochemistry 26, 4941–4947. 10.1021/bi00390a009 [DOI] [PubMed] [Google Scholar]
- Panneels V., Wu W. T., Tsai C.-J., Nogly P., Rheinberger J., Jaeger K., et al. (2015). Time-resolved Structural Studies with Serial Crystallography: A New Light on Retinal Proteins. Struct. Dyn. 2, 041718. 10.1063/1.4922774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzer S., Zhang C., Konte T., Bräuer C., Diemar A., Yogendran P., et al. (2021). Modified Rhodopsins from Aureobasidium Pullulans Excel with Very High Proton-Transport Rates. Front. Mol. Biosci. 8, 750528. 10.3389/fmolb.2021.750528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Morizumi T., Li Y. F., Hong J. E., Pai E. F., Hofmann K. P., et al. (2013). Opsin, a Structural Model for Olfactory Receptors? Angew. Chemie-International Ed. 52, 11021–11024. 10.1002/anie.201302374 [DOI] [PubMed] [Google Scholar]
- Park J. H., Scheerer P., Hofmann K. P., Choe H.-W., Ernst O. P. (2008). Crystal Structure of the Ligand-free G-Protein-Coupled Receptor Opsin. Nature 454, 183–187. 10.1038/nature07063 [DOI] [PubMed] [Google Scholar]
- Park P. S.-H., Sapra K. T., Jastrzebska B., Maeda T., Maeda A., Pulawski W., et al. (2009). Modulation of Molecular Interactions and Function by Rhodopsin Palmitylation. Biochemistry 48, 4294–4304. 10.1021/bi900417b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passamaneck Y. J., Furchheim N., Hejnol A., Martindale M. Q., Lüter C. (2011). Ciliary Photoreceptors in the Cerebral Eyes of a Protostome Larva. EvoDevo 2, 6. 10.1186/2041-9139-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J. S., Brown C. J., Ytreberg F. M., Stenkamp D. L. (2018). Predicting Peak Spectral Sensitivities of Vertebrate Cone Visual Pigments Using Atomistic Molecular Simulations. Plos Comput. Biol. 14, e1005974. 10.1371/journal.pcbi.1005974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarchi T., Shen A., He W., Baikoghli M., Cheng R. H., Xiang Y. K., et al. (2018). Nanodelivery of a Functional Membrane Receptor to Manipulate Cellular Phenotype. Sci. Rep. 8, 3556. 10.1038/s41598-018-21863-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebay-Peyroula E., Neutze R., Landau E. M. (2000). Lipidic Cubic Phase Crystallization of Bacteriorhodopsin and Cryotrapping of Intermediates: Towards Resolving a Revolving Photocycle. Biochimica Biophysica Acta-Bioenergetics 1460, 119–132. 10.1016/s0005-2728(00)00134-1 [DOI] [PubMed] [Google Scholar]
- Pediani J. D., Ward R. J., Marsango S., Milligan G. (2018). Spatial Intensity Distribution Analysis: Studies of G Protein-Coupled Receptor Oligomerisation. Trends Pharmacol. Sci. 39, 175–186. 10.1016/j.tips.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza-González L., Barneschi L., Padula D., De Vico L., Olivucci M. (2022). Evolution of the Automatic Rhodopsin Modeling (ARM) Protocol. Top. Curr. Chem. 380 (21), 21–48. 10.1007/s41061-022-00374-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza-González L., Del Carmen Marín M., Jorge A. N., Ruck T. D., Yang X. C., Valentini A., et al. (2020). Web-ARM: A Web-Based Interface for the Automatic Construction of QM/MM Models of Rhodopsins. J. Chem. Inf. Model. 60, 1481–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirson S. N., Bovee-Geurts P. H. M., Lupi D., Jeffery G., DeGrip W. J., Foster R. G. (2004). Expression of the Candidate Circadian Photopigment Melanopsin (Opn4) in the Mouse Retinal Pigment Epithelium. Mol. Brain Res. 123, 132–135. 10.1016/j.molbrainres.2004.01.007 [DOI] [PubMed] [Google Scholar]
- Penzkofer A., Silapetere A., Hegemann P. (2021). Photocycle Dynamics of the Archaerhodopsin 3 Based Fluorescent Voltage Sensor Archon2. J. Photochem. Photobiol. B-Biology 225, 112331. 10.1016/j.jphotobiol.2021.112331 [DOI] [PubMed] [Google Scholar]
- Pepe I. M., Cugnoli C. (1992). Retinal Photoisomerase - Role in Invertebrate Visual Cells. J. Photochem. Photobiol. B-Biol 13, 5–17. 10.1016/1011-1344(92)80035-t [DOI] [PubMed] [Google Scholar]
- Pérez A. A., Chen Q., Pineda Hernández H., Branco Dos Santos F., Hellingwerf K. J. (2019a). On the Use of Oxygenic Photosynthesis for the Sustainable Production of Commodity Chemicals. Physiol. Plant. 166, 413–427. 10.1111/ppl.12946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez J. H., Tolla E., Dunn I. C., Meddle S. L., Stevenson T. J. (2019b). A Comparative Perspective on Extra-retinal Photoreception. Trends Endocrinol. Metabolism 30, 39–53. 10.1016/j.tem.2018.10.005 [DOI] [PubMed] [Google Scholar]
- Pérez-Cerezales S., Boryshpolets S., Afanzar O., Brandis A., Nevo R., Kiss V., et al. (2015). Involvement of Opsins in Mammalian Sperm Thermotaxis. Sci. Rep. 5, 16146. 10.1038/srep16146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrino A. P., Miyagi A., Scheuring S. (2021). Single Molecule Kinetics of Bacteriorhodopsin by HS-AFM. Nat. Commun. 12, 7225. 10.1038/s41467-021-27580-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters L. D. M., Kussmann J., Ochsenfeld C. (2020). Combining Graphics Processing Units, Simplified Time-dependent Density Functional Theory, and Finite-Difference Couplings to Accelerate Nonadiabatic Molecular Dynamics. J. Phys. Chem. Lett. 11, 3955–3961. 10.1021/acs.jpclett.0c00320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philosof A., Béjà O. (2013). Bacterial, Archaeal and Viral-like Rhodopsins from the Red Sea. Environ. Microbiol. Rep. 5, 475–482. 10.1111/1758-2229.12037 [DOI] [PubMed] [Google Scholar]
- Piatkevich K. D., Bensussen S., Tseng H.-A., Shroff S. N., Lopez-Huerta V. G., Park D., et al. (2019). Population Imaging of Neural Activity in Awake Behaving Mice. Nature 574, 413–417. 10.1038/s41586-019-1641-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkevich K. D., Jung E. E., Straub C., Linghu C. G., Park D., Suk H.-J., et al. (2018). A Robotic Multidimensional Directed Evolution Approach Applied to Fluorescent Voltage Reporters. Nat. Chem. Biol. 14, 352–360. 10.1038/s41589-018-0004-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechnick R., Ritter E., Hildebrand P. W., Ernst O. P., Scheerer P., Hofmann K.-P., et al. (2012). Effect of Channel Mutations on the Uptake and Release of the Retinal Ligand in Opsin. Proc. Natl. Acad. Sci. U. S. A. 109, 5247–5252. 10.1073/pnas.1117268109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieri E., Ledentu V., Sahlin M., Dehez F., Olivucci M., Ferré N. (2019). CpHMD-Then-QM/MM Identification of the Amino Acids Responsible for the Anabaena Sensory Rhodopsin pH-dependent Electronic Absorption Spectrum. J. Chem. Theory Comput. 15, 4535–4546. 10.1021/acs.jctc.9b00221 [DOI] [PubMed] [Google Scholar]
- Pinhassi J., Delong E. F., Béjà O., González J. M., Pedrós-Alió C. (2016). Marine Bacterial and Archaeal Ion-Pumping Rhodopsins: Genetic Diversity, Physiology, and Ecology. Microbiol. Mol. Biol. Rev. 80, 929–953. 10.1128/mmbr.00003-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachetzki D. C., Fong C. R., Oakley T. H. (2012). Cnidocyte Discharge Is Regulated by Light and Opsin-Mediated Phototransduction. BMC Biol. 10, 17. 10.1186/1741-7007-10-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchard N., Point E., Dahmane T., Giusti F., Renault M., Le Bon C., et al. (2014). The Use of Amphipols for Solution NMR Studies of Membrane Proteins: Advantages and Constraints as Compared to Other Solubilizing Media. J. Membr. Biol. 247, 827–842. 10.1007/s00232-014-9654-z [DOI] [PubMed] [Google Scholar]
- Poddar H., Heyes D. J., Schiro G., Weik M., Leys D., Scrutton N. S. (2022). A Guide to Time-Resolved Structural Analysis of Light-Activated Proteins. FEBS J. 289, 576–595. 10.1111/febs.15880 [DOI] [PubMed] [Google Scholar]
- Polito R., Temperini M. E., Ritter E., Puskar L., Schade U., Broser M., et al. (2021). Conformational Changes of a Membrane Protein Determined by Infrared Difference Spectroscopy beyond the Diffraction Limit. Phys. Rev. Appl. 16, 014048. 10.1103/physrevapplied.16.014048 [DOI] [Google Scholar]
- Polli D., Rivalta I., Nenov A., Weingart O., Garavelli M., Cerullo G. (2015). Tracking the Primary Photoconversion Events in Rhodopsins by Ultrafast Optical Spectroscopy. Photochem. Photobiological Sci. 14, 213–228. 10.1039/c4pp00370e [DOI] [PubMed] [Google Scholar]
- Pope A. L., Sanchez-Reyes O. B., South K., Zaitseva E., Ziliox M., Vogel R., et al. (2020). A Conserved Proline Hinge Mediates Helix Dynamics and Activation of Rhodopsin. Structure 28, 1004–1013. 10.1016/j.str.2020.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popot J.-L., Althoff T., Bagnard D., Banères J.-L., Bazzacco P., Billon-Denis E., et al. (2011). Amphipols from A to Z. Annu. Rev. Biophysics 40, 379–408. 10.1146/annurev-biophys-042910-155219 [DOI] [PubMed] [Google Scholar]
- Popp A., Wolperdinger M., Hampp N., Bräuchle C., Oesterhelt D. (1993). Photochemical Conversion of the O-Intermediate to 9-Cis-Retinal- Containing Products in Bacteriorhodopsin Films. Biophys. J. 65, 1449–1459. 10.1016/s0006-3495(93)81214-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M. L., Blasic J. R., Jr., Bok M. J., Cameron E. G., Pringle T., Cronin T. W., et al. (2012). Shedding New Light on Opsin Evolution. Proc. R. Soc. B-Biological Sci. 279, 3–14. 10.1098/rspb.2011.1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash M., Murphy J., St Laurent R., Friedman N., Crespo E. L., Bjorefeldt A., et al. (2022). Selective Control of Synaptically-Connected Circuit Elements by All-Optical Synapses. Commun. Biol. 5, 33. 10.1038/s42003-021-02981-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencio I., Jiang G., DeGrip W. J., Hayes W. P., Rollag M. D. (1998). Melanopsin: An Opsin in Melanophores, Brain and Eye. Proc. Nat. Acad. Sci. U. S. A. 95, 340–345. 10.1073/pnas.95.1.340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencio I., Rodriguez I. R., Jiang G., Hayes W. P., Moreira E. F., Rollag M. D. (2000). A Novel Human Opsin in the Inner Retina. J. Neurosci. 20, 600–605. 10.1523/jneurosci.20-02-00600.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkarev A., Inoue K., Larom S., Flores-Uribe J., Singh M., Konno M., et al. (2018). A Distinct Abundant Group of Microbial Rhodopsins Discovered Using Functional Metagenomics. Nature 558, 595–599. 10.1038/s41586-018-0225-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X. D., Kumbalasiri T., Carlson S. M., Wong K. Y., Krishna V. R., Provencio I., et al. (2005). Induction of Photosensitivity by Heterologous Expression of Melanopsin. Nature 433, 745–749. 10.1038/nature03345 [DOI] [PubMed] [Google Scholar]
- Radlwimmer F. B., Yokoyama S. (1997). Cloning and Expression of the Red Visual Pigment Gene of Goat (Capra hircus). Gene 198, 211–215. 10.1016/s0378-1119(97)00316-8 [DOI] [PubMed] [Google Scholar]
- Ranganathan R., Stevens C. F. (1995). Arrestin Binding Determines the Rate of Inactivation of the G Protein-Coupled Receptor Rhodopsin In Vivo . Cell. 81, 841–848. 10.1016/0092-8674(95)90004-7 [DOI] [PubMed] [Google Scholar]
- Rath P., Decaluwé G. L. J., Bovee-Geurts P. H. M., DeGrip W. J., Rothschild K. J. (1993). Fourier Transform Infrared Difference Spectroscopy of Rhodopsin Mutants: Light Activation of Rhodopsin Causes Hydrogen-Bonding Changes in Residue Aspartic Acid-83 during Meta II Formation. Biochemistry 32, 10277–10282. 10.1021/bi00090a001 [DOI] [PubMed] [Google Scholar]
- Rath P., DeLange F., DeGrip W. J., Rothschild K. J. (1998). Hydrogen Bonding Changes of Internal Water Molecules in Rhodopsin during Metarhodopsin I and Metarhodopsin II Formation. Biochem. J. 329, 713–717. 10.1042/bj3290713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlinson K. A., Lapraz F., Ballister E. R., Terasaki M., Rodgers J., Mcdowell R. J., et al. (2019). Extraocular, Rod-like Photoreceptors in a Flatworm Express Xenopsin Photopigment. eLife 8, e45465. 10.7554/eLife.45465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P. J., Kim J.-M., Khorana H. G. (2002). Structure and Function in Rhodopsin: A Tetracycline-Inducible System in Stable Mammalian Cell Lines for High-Level Expression of Opsin Mutants. Proc. Natl. Acad. Sci. U. S. A. 99, 13413–13418. 10.1073/pnas.212519199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P. J., Klein-Seetharaman J., Getmanova E. V., Eilers M., Loewen M. C., Smith S. O., et al. (1999). Expression and Purification of Rhodopsin and its Mutants from Stable Mammalian Cell Lines: Application to NMR Studies. Biochem. Soc. Trans. 27, 950–955. 10.1042/bst0270950 [DOI] [PubMed] [Google Scholar]
- Regan C. M., DeGrip W. J., Daemen F. J. M., Bonting S. L. (1978). Sulfhydryl Group Reactivity as a Probe of Transient Protein Conformational Changes during Rhodopsin Photolysis. Biochim. Biophys. Acta 537, 145–152. 10.1016/0005-2795(78)90609-8 [DOI] [PubMed] [Google Scholar]
- Ren Z., Ren P. X., Balusu R., Yang X. J. (2016). Transmembrane Helices Tilt, Bend, Slide, Torque, and Unwind between Functional States of Rhodopsin. Sci. Rep. 6, 34129. 10.1038/srep34129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge K. D., Abdulaev N. G. (2000). Folding and Assembly of Rhodopsin from Expressed Fragments. Meth. Enzymol. 315, 59–70. 10.1016/s0076-6879(00)15834-3 [DOI] [PubMed] [Google Scholar]
- Ridge K. D., Lu Z. J., Liu X.-M., Khorana H. G. (1995). Structure and Function in Rhodopsin. Separation and Characterization of the Correctly Folded and Misfolded Opsins Produced on Expression of an Opsin Mutant Gene Containing Only the Native Intradiscal Cysteine Codons. Biochemistry-USA 34, 3261–3267. 10.1021/bi00010a016 [DOI] [PubMed] [Google Scholar]
- Ritchie T. K., Grinkova Y. V., Bayburt T. H., Denisov I. G., Zolnerciks J. K., Atkins W. M., et al. (2009). Reconstitution of Membrane Proteins in Phospholipid Bilayer Nanodiscs. Methods Enzym. 464, 211–231. 10.1016/s0076-6879(09)64011-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter E., Zimmermann K., Heck M., Hofmann K. P., Bartl F. J. (2004). Transition of Rhodopsin into the Active Metarhodopsin II State Opens a New Light-Induced Pathway Linked to Schiff Base Isomerization. J. Biol. Chem. 279, 48102–48111. 10.1074/jbc.m406857200 [DOI] [PubMed] [Google Scholar]
- Rodgers J., Bano-Otalora B., Belle M. D. C., Paul S., Hughes R., Wright P., et al. (2021). Using a Bistable Animal Opsin for Switchable and Scalable Optogenetic Inhibition of Neurons. Embo Rep. 22, 51866. 10.15252/embr.202051866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödig C., Chizhov I., Weidlich O., Siebert F. (1999). Time-resolved Step-Scan Fourier Transform Infrared Spectroscopy Reveals Differences between Early and Late M Intermediates of Bacteriorhodopsin. Biophys. J. 76, 2687–2701. 10.1016/S0006-3495(99)77421-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer B., Goletz P. W., Znoiko S. L., Ablonczy Z., Ma J.-X., Redmond T. M., et al. (2003). Correlation of Regenerable Opsin with Rod ERG Signal in RPE65(-/-) Mice during Development and Aging. Investigative Ophthalmol. Vis. Sci. 44, 310–315. 10.1167/iovs.02-0567 [DOI] [PubMed] [Google Scholar]
- Rost B. R., Schneider-Warme F., Schmitz D., Hegemann P. (2017). Optogenetic Tools for Subcellular Applications in Neuroscience. Neuron 96, 572–603. 10.1016/j.neuron.2017.09.047 [DOI] [PubMed] [Google Scholar]
- Rothschild K. J., Andrew J. R., DeGrip W. J., Stanley H. E. (1976). Opsin Structure Probed by Raman Spectroscopy of Photoreceptor Membranes. Science 191, 1176–1178. 10.1126/science.1257742 [DOI] [PubMed] [Google Scholar]
- Rothschild K. J., Cantore W. A., Marrero H. (1983). Fourier Transform Infrared Difference Spectra of Intermediates in Rhodopsin Bleaching. Science 219, 1333–1335. 10.1126/science.6828860 [DOI] [PubMed] [Google Scholar]
- Rothschild K. J., DeGrip W. J., Sanches R. (1980). Fourier Transform Infrared Study of Photoreceptor Membrane. I. Group Assignments Based on Rhodopsin Delipidation and Reconstitution. Biochim. Biophys. Acta 596, 338–351. 10.1016/0005-2736(80)90121-2 [DOI] [PubMed] [Google Scholar]
- Rothschild K. J., Gillespie J., DeGrip W. J. (1987). Evidence for Rhodopsin Refolding during the Decay of Meta II. Biophys. J. 51, 345–350. 10.1016/s0006-3495(87)83341-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild K. J., Marrero H., Braiman M. S., Mathies R. A. (1984). Primary Photochemistry of Bacteriorhodopsin - Comparison of Fourier-Transform Infrared Difference Spectra with Resonance Raman-Spectra. Photochem. Photobiol. 40, 675–679. 10.1111/j.1751-1097.1984.tb05359.x [DOI] [PubMed] [Google Scholar]
- Rothschild K. J., Marrero H. (1982). Infrared Evidence that the Schiff Base of Bacteriorhodopsin Is Protonated: bR570 and K Intermediates. Proc. Nat. Acad. Sci. U. S. A. 79, 4045–4049. 10.1073/pnas.79.13.4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild K. J. (2016). The Early Development and Application of FTIR Difference Spectroscopy to Membrane Proteins: A Personal Perspective. Biomed. Spectrosc. Imaging 5, 231–267. 10.3233/bsi-160148 [DOI] [Google Scholar]
- Rothschild K. J., Zagaeski M., Cantore W. A. (1981). Conformational Changes of Bacteriorhodopsin Detected by Fourier Transform Infrared Difference Spectroscopy. Biochem. Biophys. Res. Commun. 103, 483–489. 10.1016/0006-291x(81)90478-2 [DOI] [PubMed] [Google Scholar]
- Rousso I., Gat Y., Lewis A., Sheves M., Ottolenghi M. (1998). Effective Light-Induced Hydroxylamine Reactions Occur with C-13 = C-14 Nonisomerizable Bacteriorhodopsin Pigments. Biophysical J. 75, 413–417. 10.1016/s0006-3495(98)77526-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P. P., Kato Y., Abe-Yoshizumi R., Pieri E., Ferré N., Kandori H., et al. (2018). Mapping the Ultrafast Vibrational Dynamics of All-Trans and 13-cis Retinal Isomerization in Anabaena Sensory Rhodopsin. Phys. Chem. Chem. Phys. 20, 30159–30173. 10.1039/c8cp05469j [DOI] [PubMed] [Google Scholar]
- Royant A., Nollert P., Edman K., Neutze R., Landau E. M., Pebay-Peyroula E., et al. (2001). X-ray Structure of Sensory Rhodopsin II at 2.1-Å Resolution. Proc. Natl. Acad. Sci. U. S. A. 98, 10131–10136. 10.1073/pnas.181203898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenberg A., Inoue K., Kandori H., Béjà O. (2021). Microbial Rhodopsins: The Last Two Decades. Annu. Rev. Microbiol. 75, 427–447. 10.1146/annurev-micro-031721-020452 [DOI] [PubMed] [Google Scholar]
- Rupenyan A., Van Stokkum I. H. M., Arents J. C., Van Grondelle R., Hellingwerf K. J., Groot M. L. (2008). Characterization of the Primary Photochemistry of Proteorhodopsin with Femtosecond Spectroscopy. Biophysical J. 94, 4020–4030. 10.1529/biophysj.107.121376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupenyan A., Van Stokkum I. H. M., Arents J. C., Van Grondelle R., Hellingwerf K. J., Groot M. L. (2009). Reaction Pathways of Photoexcited Retinal in Proteorhodopsin Studied by Pump-Dump-Probe Spectroscopy. J. Phys. Chem. B 113, 16251–16256. 10.1021/jp9065289 [DOI] [PubMed] [Google Scholar]
- Ruprecht J. J., Mielke T., Vogel R., Villa C., Schertler G. F. X. (2004). Electron Crystallography Reveals the Structure of Metarhodopsin I. EMBO J. 23, 3609–3620. 10.1038/sj.emboj.7600374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryazantsev M. N., Nikolaev D. M., Struts A. V., Brown M. F. (2019). Quantum Mechanical and Molecular Mechanics Modeling of Membrane-Embedded Rhodopsins. J. Membr. Biol. 252, 425–449. 10.1007/s00232-019-00095-0 [DOI] [PubMed] [Google Scholar]
- Ryba N. J. P., Hoon M. A., Findlay J. B. C., Saibil H. R., Wilkinson J. R., Heimburg T., et al. (1993). Rhodopsin Mobility, Structure, and Lipid-Protein Interaction in Squid Photoreceptor Membranes. Biochemistry 32, 3298–3305. 10.1021/bi00064a012 [DOI] [PubMed] [Google Scholar]
- Sadaf A., Cho H. C., Byrne B., Chae P. S. (2015). Amphipathic Agents for Membrane Protein Study. Meth. Enzymol. 557, 57–94. 10.1016/bs.mie.2014.12.021 [DOI] [PubMed] [Google Scholar]
- Saint Clair E. C., Ogren J. I., Mamaev S., Kralj J. M., Rothschild K. J. (2012a). Conformational Changes in the Archaerhodopsin-3 Proton Pump: Detection of Conserved Strongly Hydrogen Bonded Water Networks. J. Biol. Phys. 38, 153–168. 10.1007/s10867-011-9246-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint Clair E. C., Ogren J. I., Mamaev S., Russano D., Kralj J. M., Rothschild K. J. (2012b). Near-IR Resonance Raman Spectroscopy of Archaerhodopsin 3: Effects of Transmembrane Potential. J. Phys. Chem. B 116, 14592–14601. 10.1021/jp309996a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K., Imamoto Y., Su C.-Y., Tsukamoto H., Yamashita T., Terakita A., et al. (2012). Photochemical Nature of Parietopsin. Biochemistry 51, 1933–1941. 10.1021/bi2018283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K., Shichida Y., Imamoto Y., Yamashita T. (2022). Creation of Photocyclic Vertebrate Rhodopsin by Single Amino Acid Substitution. eLife 11, 75979. 10.7554/elife.75979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakmar T. P., Fahmy K. (1995). Properties and Photoactivity of Rhodopsin Mutants. Isr. J. Chem. 35, 325–337. 10.1002/ijch.199500034 [DOI] [Google Scholar]
- Sakmar T. P., Franke R. R., Khorana H. G. (1989). Glutamic Acid-113 Serves as the Retinylidene Schiff Base Counterion in Bovine Rhodopsin. Proc. Nat. Acad. Sci. U. S. A. 86, 8309–8313. 10.1073/pnas.86.21.8309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakmar T. P., Menon S. T., Marin E. P., Awad E. S. (2002). Rhodopsin: Insights from Recent Structural Studies. Annu. Rev. Biophysics Biomol. Struct. 31, 443–484. 10.1146/annurev.biophys.31.082901.134348 [DOI] [PubMed] [Google Scholar]
- Salcedo E., Huber A., Henrich S., Chadwell L. V., Chou W. H., Paulsen R., et al. (1999). Blue- and Green-Absorbing Visual Pigments of Drosophila: Ectopic Expression and Physiological Characterization of the R8 Photoreceptor Cell-specific Rh5 and Rh6 Rhodopsins. J. Neurosci. 19, 10716–10726. 10.1523/jneurosci.19-24-10716.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salom D., Cao P. X., Sun W. Y., Kramp K., Jastrzebska B., Jin H., et al. (2012). Heterologous Expression of Functional G-Protein-Coupled Receptors in Caenorhabditis elegans . FASEB J. 26, 492–502. 10.1096/fj.11-197780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salom D., Jin H., Gerken T. A., Yu C., Huang L., Palczewski K. (2019). Human Red and Green Cone Opsins Are O-Glycosylated at an N-Terminal Ser/Thr-Rich Domain Conserved in Vertebrates. J. Biol. Chem. 294, 8123–8133. 10.1074/jbc.ra118.006835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salom D., Wu N., Sun W. Y., Dong Z., Palczewski K., Jordan S., et al. (2008). Heterologous Expression and Purification of the Serotonin Type 4 Receptor from Transgenic Mouse Retina. Biochemistry 47, 13296–13307. 10.1021/bi8018527 [DOI] [PubMed] [Google Scholar]
- Sanchez-Reyes O. B., Cooke A. L. G., Tranter D. B., Rashid D., Eilers M., Reeves P. J., et al. (2017). G Protein-Coupled Receptors Contain Two Conserved Packing Clusters. Biophysical J. 112, 2315–2326. 10.1016/j.bpj.2017.04.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardet C., Tardieu A., Luzzati V. (1976). Shape and Size of Bovine Rhodopsin: A Small-Angle X-Ray-Scattering Study of A Rhodopsin-Detergent Complex. J. Mol. Biol. 105, 383–407. 10.1016/0022-2836(76)90100-5 [DOI] [PubMed] [Google Scholar]
- Sarramegna V., Muller I., Milon A., Talmont F. (2006). Recombinant G Protein-Coupled Receptors from Expression to Renaturation: A Challenge towards Structure. Cell. Mol. Life Sci. 63, 1149–1164. 10.1007/s00018-005-5557-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki J., Brown L. S., Chon Y.-S., Kandori H., Maeda A., Needleman R., et al. (1995). Conversion of Bacteriorhodopsin into a Chloride Ion Pump. Science 269, 73–75. 10.1126/science.7604281 [DOI] [PubMed] [Google Scholar]
- Sasaki J., Takahashi H., Furutani Y., Kandori H., Spudich J. L. (2011). Sensory Rhodopsin-I as a Bidirectional Switch: Opposite Conformational Changes from the Same Photoisomerization. Biophysical J. 100, 2178–2183. 10.1016/j.bpj.2011.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Yamashita T., Yoshida K., Inoue K., Shichida Y., Kandori H. (2014). Chimeric Proton-Pumping Rhodopsins Containing the Cytoplasmic Loop of Bovine Rhodopsin. PLoS ONE 9, e91323. 10.1371/journal.pone.0091323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Yamashita T., Imamoto Y., Shichida Y. (2012). Comparative Studies on the Late Bleaching Processes of Four Kinds of Cone Visual Pigments and Rod Visual Pigment. Biochemistry 51, 4300–4308. 10.1021/bi3000885 [DOI] [PubMed] [Google Scholar]
- Sato K., Yamashita T., Kojima K., Sakai K., Matsutani Y., Yanagawa M., et al. (2018a). Pinopsin Evolved as the Ancestral Dim-Light Visual Opsin in Vertebrates. Commun. Biol. 1, 156. 10.1038/s42003-018-0164-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Yamashita T., Ohuchi H., Shichida Y. (2011). Vertebrate Ancient-Long Opsin Has Molecular Properties Intermediate between Those of Vertebrate and Invertebrate Visual Pigments. Biochemistry 50, 10484–10490. 10.1021/bi201212z [DOI] [PubMed] [Google Scholar]
- Sato K., Yamashita T., Ohuchi H., Takeuchi A., Gotoh H., Ono K., et al. (2018b). Opn5L1 Is a Retinal Receptor that Behaves as a Reverse and Self-Regenerating Photoreceptor. Nat. Commun. 9, 125. 10.1038/s41467-018-03603-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer C. T., Farrens D. L. (2015). Conformational Selection and Equilibrium Governs the Ability of Retinals to Bind Opsin. J. Biol. Chem. 290, 4304–4318. 10.1074/jbc.m114.603134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer G., Shastri S., Verhoefen M.-K., Vogel V., Glaubitz C., Wachtveitl J., et al. (2009). Characterizing the Structure and Photocycle of PR 2D Crystals with CD and FTIR Spectroscopy. Photochem. Photobiol. 85, 529–534. 10.1111/j.1751-1097.2008.00491.x [DOI] [PubMed] [Google Scholar]
- Schapiro I., Ruhman S. (2014). Ultrafast Photochemistry of Anabaena Sensory Rhodopsin: Experiment and Theory. Biochimica Biophysica Acta-Bioenergetics 1837, 589–597. 10.1016/j.bbabio.2013.09.014 [DOI] [PubMed] [Google Scholar]
- Schapiro I., Ryazantsev M. N., Frutos L. M., Ferré N., Lindh R., Olivucci M. (2011). The Ultrafast Photoisomerizations of Rhodopsin and Bathorhodopsin Are Modulated by Bond Length Alternation and HOOP Driven Electronic Effects. J. Am. Chem. Soc. 133, 3354–3364. 10.1021/ja1056196 [DOI] [PubMed] [Google Scholar]
- Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauß N., Choe H.-W., et al. (2008). Crystal Structure of Opsin in its G-Protein-Interacting Conformation. Nature 455, 497–502. 10.1038/nature07330 [DOI] [PubMed] [Google Scholar]
- Schertler G. F. X., Hargrave P. A. (1995). Projection Structure of Frog Rhodopsin in Two Crystal Forms. Proc. Nat. Acad. Sci. U. S. A. 92, 11578–11582. 10.1073/pnas.92.25.11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertler G. F. X. (2005). Structure of Rhodopsin and the Metarhodopsin I Photointermediate. Curr. Opin. Struct. Biol. 15, 408–415. 10.1016/j.sbi.2005.07.010 [DOI] [PubMed] [Google Scholar]
- Schey K. L., Papac D. I., Knapp D. R., Crouch R. K. (1992). Matrix-assisted Laser Desorption Mass Spectrometry of Rhodopsin and Bacteriorhodopsin. Biophys. J. 63, 1240–1243. 10.1016/s0006-3495(92)81699-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlinkmann K. M., Plückthun A. (2013). Directed Evolution of G-Protein-Coupled Receptors for High Functional Expression and Detergent Stability. Meth. Enzymol. 520, 67–97. 10.1016/b978-0-12-391861-1.00004-6 [DOI] [PubMed] [Google Scholar]
- Schnedermann C., Muders V., Ehrenberg D., Schlesinger R., Kukura P., Heberle J. (2016). Vibronic Dynamics of the Ultrafast All-Trans to 13-cis Photoisomerization of Retinal in Channelrhodopsin-1. J. Am. Chem. Soc. 138, 4757–4762. 10.1021/jacs.5b12251 [DOI] [PubMed] [Google Scholar]
- Schnedermann C., Yang X., Liebel M., Spillane K. M., Lugtenburg J., Fernández I., et al. (2018). Evidence for a Vibrational Phase-dependent Isotope Effect on the Photochemistry of Vision. Nat. Chem. 10, 449–455. 10.1038/s41557-018-0014-y [DOI] [PubMed] [Google Scholar]
- Schobert B., Cupp-Vickery J., Hornak V., Smith S. O., Lanyi J. K. (2002). Crystallographic Structure of the K Intermediate of Bacteriorhodopsin: Conservation of Free Energy after Photoisomerization of the Retinal. J. Mol. Biol. 321, 715–726. 10.1016/s0022-2836(02)00681-2 [DOI] [PubMed] [Google Scholar]
- Schoenlein R. W., Peteanu L. A., Mathies R. A., Shank C. V. (1991). The First Step in Vision: Femtosecond Isomerization of Rhodopsin. Science 254, 412–415. 10.1126/science.1925597 [DOI] [PubMed] [Google Scholar]
- Scholz L., Neugebauer J. (2021). Protein Response Effects on Cofactor Excitation Energies from First Principles: Augmenting Subsystem Time-dependent Density-Functional Theory with Many-Body Expansion Techniques. J. Chem. Theory Comput. 17, 6105–6121. 10.1021/acs.jctc.1c00551 [DOI] [PubMed] [Google Scholar]
- Schreiber M., Sugihara M., Okada T., Buss V. (2006). Quantum Mechanical Studies on the Crystallographic Model of Bathorhodopsin. Angew. Chem. Int. Ed. 45, 4274–4277. 10.1002/anie.200600585 [DOI] [PubMed] [Google Scholar]
- Sekharan S., Yokoyama S., Morokuma K. (2011). Quantum Mechanical/Molecular Mechanical Structure, Enantioselectivity, and Spectroscopy of Hydroxyretinals and Insights into the Evolution of Color Vision in Small White Butterflies. J. Phys. Chem. B 115, 15380–15388. 10.1021/jp208107r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T., Isono K., Ozaki K., Tsukahara Y., Shibata-Katsuta Y., Ito M., et al. (1998). The Metabolic Pathway of Visual Pigment Chromophore Formation in Drosophila melanogaster - All-Trans (3S)-3-Hydroxyretinal Is Formed from All-Trans Retinal via (3R)-3-Hydroxyretinal in the Dark. Eur. J. Biochem. 257, 522–527. 10.1046/j.1432-1327.1998.2570522.x [DOI] [PubMed] [Google Scholar]
- Seki T., Vogt K. (1998). Evolutionary Aspects of the Diversity of Visual Pigment Chromophores in the Class Insecta. Comp. Biochem. Physiol. B 119, 53–64. 10.1016/s0305-0491(97)00322-2 [DOI] [Google Scholar]
- Shao Y. H., Mei Y., Sundholm D., Kaila V. R. I. (2020). Benchmarking the Performance of Time-dependent Density Functional Theory Methods on Biochromophores. J. Chem. Theory Comput. 16, 587–600. 10.1021/acs.jctc.9b00823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Jäckel Z., Schneider A., Paul O., Diester I., Ruther P. (2021). Multifunctional Optrode for Opsin Delivery, Optical Stimulation, and Electrophysiological Recordings in Freely Moving Rats. J. neural Eng. 18, 066013. 10.1088/1741-2552/ac3206 [DOI] [PubMed] [Google Scholar]
- Shen C., Jin X., Glover W. J., He X. (2021). Accurate Prediction of Absorption Spectral Shifts of Proteorhodopsin Using a Fragment-Based Quantum Mechanical Method. Molecules 26, 4486. 10.3390/molecules26154486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W. L., Kwon Y., Adegbola A. A., Luo J. J., Chess A., Montell C. (2011). Function of Rhodopsin in Temperature Discrimination in Drosophila . Science 331, 1333–1336. 10.1126/science.1198904 [DOI] [PubMed] [Google Scholar]
- Shen Y., Campbell R. E., Cote D. C., Paquet M. E. (2020). Challenges for Therapeutic Applications of Opsin-Based Optogenetic Tools in Humans. Front. Neural Circuits 14, 41. 10.3389/fncir.2020.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheves M., Friedman N. (1986). Influence of External Negative Charges on the Absorption Maxima of Symmetrical Cyanines. A Study with Model Compounds and Artificial Bacteriorhodopsin Pigments. Angewandte Chemie-International Ed. Engl. 25, 284–286. 10.1002/anie.198602841 [DOI] [Google Scholar]
- Shi L. C., Ahmed M. a. M., Zhang W. R., Whited G., Brown L. S., Ladizhansky V. (2009). Three-dimensional Solid-State NMR Study of a Seven-Helical Integral Membrane Proton Pump-Structural Insights. J. Mol. Biol. 386, 1078–1093. 10.1016/j.jmb.2009.01.011 [DOI] [PubMed] [Google Scholar]
- Shibata M., Inoue K., Ikeda K., Konno M., Singh M., Kataoka C., et al. (2018). Oligomeric States of Microbial Rhodopsins Determined by High-Speed Atomic Force Microscopy and Circular Dichroic Spectroscopy. Sci. Rep. 8, 8262. 10.1038/s41598-018-26606-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichida Y., Kobayashi T., Ohtani H., Yoshizawa T., Nagakura S. (1978). Picosecond Laser Photolysis of Squid Rhodopsin at Room and Low-Temperatures. Photochem. Photobiol. 27, 335–341. 10.1111/j.1751-1097.1978.tb07609.x [DOI] [Google Scholar]
- Shichida Y. (1986). Primary Intermediates of Photobleaching of Rhodopsin. Photobiochem. Photobiophys. 13, 287–307. [Google Scholar]
- Shichida Y. (1990). Ultra-fast Laser Spectroscopy of Visual Pigments. Photochem. Photobiol. 52, 1179–1185. 10.1111/j.1751-1097.1990.tb08456.x [DOI] [PubMed] [Google Scholar]
- Shigeta A., Ito S., Inoue K., Okitsu T., Wada A., Kandori H., et al. (2017). Solid-State Nuclear Magnetic Resonance Structural Study of the Retinal-Binding Pocket in Sodium Ion Pump Rhodopsin. Biochemistry 56, 543–550. 10.1021/acs.biochem.6b00999 [DOI] [PubMed] [Google Scholar]
- Shihoya W., Inoue K., Singh M., Konno M., Hososhima S., Yamashita K., et al. (2019). Crystal Structure of Heliorhodopsin. Nature 574, 132–136. 10.1038/s41586-019-1604-6 [DOI] [PubMed] [Google Scholar]
- Shim J.-G., Kang N.-R., Chuon K., Cho S.-G., Meas S., Jung K.-H. (2022). Mutational Analyses Identify a Single Amino Acid Critical for Color Tuning in Proteorhodopsins. FEBS Lett. 596, 784. 10.1002/1873-3468.14297 [DOI] [PubMed] [Google Scholar]
- Shim J.-G., Soum V., Kang K.-W., Chuon K., Cho S.-G., Kim J.-H., et al. (2021). Discovery of a Microbial Rhodopsin that Is the Most Stable in Extreme Environments. iScience 24, 102620. 10.1016/j.isci.2021.102620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura T., Hiraki K., Takahashi N., Hori T., Ago H., Masuda K., et al. (2008). Crystal Structure of Squid Rhodopsin with Intracellularly Extended Cytoplasmic Region. J. Biol. Chem. 283, 17753–17756. 10.1074/jbc.c800040200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono K., Ikeura Y., Sudo Y., Iwamoto M., Kamo N. (2001). Environment Around the Chromophore in Pharaonis Phoborhodopsin: Mutation Analysis of the Retinal Binding Site. Biochimica Biophysica Acta-Biomembranes 1515, 92–100. 10.1016/s0005-2736(01)00394-7 [DOI] [PubMed] [Google Scholar]
- Shirzad-Wasei N., DeGrip W. J. (2016). Heterologous Expression of Melanopsin: Present, Problems and Prospects. Prog. Retin. Eye Res. 52, 1–21. 10.1016/j.preteyeres.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Shirzad-Wasei N., Van Oostrum J., Bovee-Geurts P. H. M., Kusters L. J. A., Bosman G. J. C. G. M., DeGrip W. J. (2015). Rapid Transfer of Overexpressed Integral Membrane Protein from the Host Membrane into Soluble Lipid Nanodiscs without Previous Purification. Biol. Chem. 396, 903–915. 10.1515/hsz-2015-0100 [DOI] [PubMed] [Google Scholar]
- Shirzad-Wasei N., Van Oostrum J., Bovee-Geurts P. H. M., Wasserman M., Bosman G. J. C. G. M., DeGrip W. J. (2013). Large Scale Expression and Purification of Mouse Melanopsin-L in the Baculovirus Expression System. Protein Expr. Purif. 91, 134–146. 10.1016/j.pep.2013.07.010 [DOI] [PubMed] [Google Scholar]
- Shtyrov A. A., Nikolaev D. M., Mironov V. N., Vasin A. V., Panov M. S., Tveryanovich Y. S., et al. (2021). Simple Models to Study Spectral Properties of Microbial and Animal Rhodopsins: Evaluation of the Electrostatic Effect of Charged and Polar Residues on the First Absorption Band Maxima. Int. J. Mol. Sci. 22, 3029. 10.3390/ijms22063029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sineshchekov O. A., Govorunova E. G., Wang J. H., Spudich J. L. (2012). Enhancement of Long-Wavelength Sensitivity of Optogenetic Microbial Rhodopsins by 3, 4-dehydroretinal. Biochemistry 51, 4499–4506. 10.1021/bi2018859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Inoue K., Pushkarev A., Béjà O., Kandori H. (2018). Mutation Study of Heliorhodopsin 48C12. Biochemistry 57, 5041–5049. 10.1021/acs.biochem.8b00637 [DOI] [PubMed] [Google Scholar]
- Skopintsev P., Ehrenberg D., Weinert T., James D., Kar R. K., Johnson P. J. M., et al. (2020). Femtosecond-to-millisecond Structural Changes in a Light-Driven Sodium Pump. Nature 583, 314–322. 10.1038/s41586-020-2307-8 [DOI] [PubMed] [Google Scholar]
- Smith S. O., Aschheim K., Groesbeek M. (1996). Magic Angle Spinning NMR Spectroscopy of Membrane Proteins. Quart. Rev. Biophys. 29, 395–449. 10.1017/s0033583500005898 [DOI] [PubMed] [Google Scholar]
- Smith S. O., De Groot H. J. M., Gebhard R., Courtin J. M. L., Lugtenburg J., Herzfeld J., et al. (1989). Structure and Protein Environment of the Retinal Chromophore in Light-Adapted and Dark-Adapted Bacteriorhodopsin Studied by Solid-State NMR. Biochemistry 28, 8897–8904. 10.1021/bi00448a032 [DOI] [PubMed] [Google Scholar]
- Smith S. O., De Groot H. J. M., Gebhard R., Lugtenburg J. (1992). Magic Angle Spinning NMR Studies on the Metarhodopsin II Intermediate of Bovine Rhodopsin: Evidence for an Unprotonated Schiff Base. Photochem. Photobiol. 56, 1035–1039. 10.1111/j.1751-1097.1992.tb09726.x [DOI] [PubMed] [Google Scholar]
- Smith S. O. (2021). Deconstructing the Transmembrane Core of Class A G Protein-Coupled Receptors. Trends Biochem. Sci. 46, 1017. 10.1016/j.tibs.2021.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. O., Myers A. B., Mathies R. A., Pardoen J. A., Winkel C., Vandenberg E. M. M., et al. (1985). Vibrational Analysis of the All-Trans Retinal Protonated Schiff Base. Biophys. J. 47, 653–664. 10.1016/s0006-3495(85)83961-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. O. (2010). Structure and Activation of the Visual Pigment Rhodopsin. Annu. Rev. Biophysics 39, 309–328. 10.1146/annurev-biophys-101209-104901 [DOI] [PubMed] [Google Scholar]
- Smitienko O. A., Feldman T. B., Petrovskaya L. E., Nekrasova O. V., Yakovleva M. A., Shelaev I. V., et al. (2021). Comparative Femtosecond Spectroscopy of Primary Photoreactions of Exiguobacterium Sibiricum Rhodopsin and Halobacterium Salinarum Bacteriorhodopsin. J. Phys. Chem. B 125, 995–1008. 10.1021/acs.jpcb.0c07763 [DOI] [PubMed] [Google Scholar]
- Smitienko O. A., Nekrasova O. V., Kudriavtsev A. V., Yakovleva M. A., Shelaev I. V., Gostev F. E., et al. (2017). Femtosecond and Picosecond Dynamics of Recombinant Bacteriorhodopsin Primary Reactions Compared to the Native Protein in Trimeric and Monomeric Forms. Biochemistry-Moscow 82, 490–500. 10.1134/S0006297917040113 [DOI] [PubMed] [Google Scholar]
- Sneskov K., Olsen J. M. H., Schwabe T., Hãttig C., Christiansen O., Kongsted J. (2013). Computational Screening of One- and Two-Photon Spectrally Tuned Channelrhodopsin Mutants. Phys. Chem. Chem. Phys. 15, 7567–7576. 10.1039/c3cp44350g [DOI] [PubMed] [Google Scholar]
- Sonar S., Liu X.-M., Lee C.-P., Coleman M., He Y.-W., Pelletier S., et al. (1995). Site-directed Isotope Labeling and FT-IR Spectroscopy: The Tyr 185/Pro 186 Peptide Bond of Bacteriorhodopsin Is Perturbed during the Primary Photoreaction. J. Am. Chem. Soc. 117, 11614–11615. 10.1021/ja00151a041 [DOI] [Google Scholar]
- Song Y. Z., Cartron M. L., Jackson P. J., Davison P. A., Dickman M. J., Zhu D., et al. (2020). Proteorhodopsin Overproduction Enhances the Long-Term Viability of Escherichia coli . Appl. Environ. Microbiol. 86, 02087–02019. 10.1128/AEM.02087-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni B. G., Foster R. G. (1997). A Novel and Ancient Vertebrate Opsin. FEBS Lett. 406, 279–283. 10.1016/s0014-5793(97)00287-1 [DOI] [PubMed] [Google Scholar]
- Spooner P. J. R., Sharples J. M., Goodall S. C., Bovee-Geurts P. H. M., Verhoeven M. A., Lugtenburg J., et al. (2004). The Ring of the Rhodopsin Chromophore in a Hydrophobic Activation Switch within the Binding Pocket. J. Mol. Biol. 343, 719–730. 10.1016/j.jmb.2004.08.049 [DOI] [PubMed] [Google Scholar]
- Spudich J. L., Sineshchekov O. A., Govorunova E. G. (2014). Mechanism Divergence in Microbial Rhodopsins. Biochimica Biophysica Acta-Bioenergetics 1837, 546–552. 10.1016/j.bbabio.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L., Yang C.-S., Jung K.-H., Spudich E. N. (2000). Retinylidene Proteins: Structures and Functions from Archaea to Humans. Annu. Rev. Cell. Dev. Biol. 16, 365–392. 10.1146/annurev.cellbio.16.1.365 [DOI] [PubMed] [Google Scholar]
- Sridharan S., Gajowa M. A., Ogando M. B., Jagadisan U. K., Abdeladim L., Sadahiro M., et al. (2022). High-performance Microbial Opsins for Spatially and Temporally Precise Perturbations of Large Neuronal Networks. Neuron 110, 1–17. 10.1016/j.neuron.2022.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Ramon E., Cordomí A., Garriga P. (2014). Binding Specificity of Retinal Analogs to Photoactivated Visual Pigments Suggest Mechanism for Fine-Tuning GPCR-Ligand Interactions. Chem. Biol. 21, 369–378. 10.1016/j.chembiol.2014.01.006 [DOI] [PubMed] [Google Scholar]
- Standfuss J., Xie G. F., Edwards P. C., Burghammer M., Oprian D. D., Schertler G. F. X. (2007). Crystal Structure of a Thermally Stable Rhodopsin Mutant. J. Mol. Biol. 372, 1179–1188. 10.1016/j.jmb.2007.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavenga D. G., Oberwinkler J., Postma M. (2000). “Modeling Primary Visual Processes in Insect Photoreceptors,” in Molecular Mechanisms in Visual Transduction. Editors Stavenga D. G., DeGrip W. J., PughJr. E. N. (Amsterdam, Netherlands: Elsevier Science Pub.), 527–574. 10.1016/s1383-8121(00)80013-5 [DOI] [Google Scholar]
- Steinhoff H.-J., Mollaaghababa R., Altenbach C. A., Khorana H. G., Hubbell W. L. (1995). Site Directed Spin Labeling Studies of Structure and Dynamics in Bacteriorhodopsin. Biophys. Chem. 56, 89–94. 10.1016/0301-4622(95)00019-t [DOI] [PubMed] [Google Scholar]
- Stenkamp R. E. (2008). Alternative Models for Two Crystal Structures of Bovine Rhodopsin. Acta Crystallogr. d-biol. Cryst. 64, 902–904. 10.1107/s0907444908017162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward L. E., Chamberlin A. R. (1998). Protein Engineering with Nonstandard Amino Acids. Meth. Mol. Biol. 77, 325–354. 10.1385/0-89603-397-X:325 [DOI] [PubMed] [Google Scholar]
- Struts A. V., Salgado G. F. J., Tanaka K., Krane S., Nakanishi K., Brown M. F. (2007). Structural Analysis and Dynamics of Retinal Chromophore in Dark and Metal States of Rhodopsin from 2H NMR of Aligned Membranes. J. Mol. Biol. 372, 50–66. 10.1016/j.jmb.2007.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs G. W., Smith H. G., Litman B. J. (1976). Alkyl Glucosides as Effective Solubilizing Agents for Bovine Rhodopsin - A Comparison with Several Commonly Used Detergents. Biochim. Biophys. Acta 426, 46–56. 10.1016/0005-2736(76)90428-4 [DOI] [PubMed] [Google Scholar]
- Su C.-Y., Luo D.-G., Terakita A., Shichida Y., Liao H.-W., Kazmi M. A., et al. (2006). Parietal-eye Phototransduction Components and Their Potential Evolutionary Implications. Science 311, 1617–1621. 10.1126/science.1123802 [DOI] [PubMed] [Google Scholar]
- Subramaniam S., Lindahl I., Bullough P. A., Faruqi A. R., Tittor J., Oesterhelt D., et al. (1999). Protein Conformational Changes in the Bacteriorhodopsin Photocycle. J. Mol. Biol. 287, 145–161. 10.1006/jmbi.1999.2589 [DOI] [PubMed] [Google Scholar]
- Sudo Y., Ihara K., Kobayashi S., Suzuki D., Irieda H., Kikukawa T., et al. (2011). A Microbial Rhodopsin with a Unique Retinal Composition Shows Both Sensory Rhodopsin II and Bacteriorhodopsin-like Properties. J. Biol. Chem. 286, 5967–5976. 10.1074/jbc.m110.190058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo Y., Okazaki A., Ono H., Yagasaki J., Sugo S., Kamiya M., et al. (2013). A Blue-Shifted Light-Driven Proton Pump for Neural Silencing. J. Biol. Chem. 288, 20624–20632. 10.1074/jbc.m113.475533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto T., Katayama K., Kandori H. (2021). Role of Thr82 for the Unique Photochemistry of TAT Rhodopsin. Biophysics Physicobiology 18, 108–115. 10.2142/biophysico.bppb-v18.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Gilbert D. J., Copeland N. G., Jenkins N. A., Nathans J. (1997). Peropsin, a Novel Visual Pigment-like Protein Located in the Apical Microvilli of the Retinal Pigment Epithelium. Proc. Nat. Acad. Sci. U. S. A. 94, 9893–9898. 10.1073/pnas.94.18.9893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li M. J., Cao S., Xu Y., Wu P., Xu S., et al. (2022). Optogenetics for Understanding and Treating Brain Injury: Advances in the Field and Future Prospects. Int. J. Mol. Sci. 23, 1800. 10.3390/ijms23031800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki E., Katayama E., Hirosawa K. (1993). Structure of Photoreceptive Membranes of Drosophila Compound Eyes as Studied by Quick-Freezing Electron Microscopy. J. Electron Microsc. 42, 178–184. [PubMed] [Google Scholar]
- Suzuki K., Del Carmen Marín M., Konno M., Bagherzadeh R., Murata T., Inoue K. (2022). Structural Characterization of Proton-Pumping Rhodopsin Lacking a Cytoplasmic Proton Donor Residue by X-Ray Crystallography. J. Biol. Chem. 298, 101722. 10.1016/j.jbc.2022.101722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara S., Kuramochi H., Takeuchi S., Tahara T. (2019a). Protein Dynamics Preceding Photoisomerization of the Retinal Chromophore in Bacteriorhodopsin Revealed by Deep-UV Femtosecond Stimulated Raman Spectroscopy. J. Phys. Chem. Lett. 10, 5422–5427. 10.1021/acs.jpclett.9b02283 [DOI] [PubMed] [Google Scholar]
- Tahara S., Singh M., Kuramochi H., Shihoya W., Inoue K., Nureki O., et al. (2019b). Ultrafast Dynamics of Heliorhodopsins. J. Phys. Chem. B 123, 2507–2512. 10.1021/acs.jpcb.9b00887 [DOI] [PubMed] [Google Scholar]
- Tahara S., Takeuchi S., Abe-Yoshizumi R., Inoue K., Ohtani H., Kandori H., et al. (2015). Ultrafast Photoreaction Dynamics of a Light-Driven Sodium-Ion-Pumping Retinal Protein from Krokinobacter Eikastus Revealed by Femtosecond Time-Resolved Absorption Spectroscopy. J. Phys. Chem. Lett. 6, 4481–4486. 10.1021/acs.jpclett.5b01994 [DOI] [PubMed] [Google Scholar]
- Takayama R., Kaneko A., Okitsu T., Tsunoda S. P., Shimono K., Mizuno M., et al. (2018). Production of a Light-Gated Proton Channel by Replacing the Retinal Chromophore with its Synthetic Vinylene Derivative. J. Phys. Chem. Lett. 9, 2857–2862. 10.1021/acs.jpclett.8b00879 [DOI] [PubMed] [Google Scholar]
- Tam B. M., Moritz O. L. (2009). The Role of Rhodopsin Glycosylation in Protein Folding, Trafficking, and Light-Sensitive Retinal Degeneration. J. Neurosci. 29, 15145–15154. 10.1523/jneurosci.4259-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P., He L., Huang Y., Zhou Y. (2022). Optophysiology: Illuminating Cell Physiology with Optogenetics. Physiol. Rev. 102, 1263. 10.1152/physrev.00021.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Singh M., Shihoya W., Yamashita K., Kandori H., Nureki O. (2020). Structural Basis for Unique Color Tuning Mechanism in Heliorhodopsin. Biochem. Biophysical Res. Commun. 533, 262–267. 10.1016/j.bbrc.2020.06.124 [DOI] [PubMed] [Google Scholar]
- Tansley K. (1931). The Regeneration of Visual Purple: its Relation to Dark Adaptation and Night Blindness. J. Physiology 71, 442–458. 10.1113/jphysiol.1931.sp002749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarttelin E. E., Bellingham J., Hankins M. W., Foster R. G., Lucas R. J. (2003). Neuropsin (Opn5): A Novel Opsin Identified in Mammalian Neural Tissue. FEBS Lett. 554, 410–416. 10.1016/s0014-5793(03)01212-2 [DOI] [PubMed] [Google Scholar]
- Tarttelin E. E., Fransen M. P., Edwards P. C., Hankins M. W., Schertler G. F. X., Vogel R., et al. (2011). Adaptation of Pineal Expressed Teleost Exo-Rod Opsin to Non-image Forming Photoreception through Enhanced Meta II Decay. Cell. Mol. Life Sci. 68, 3713–3723. 10.1007/s00018-011-0665-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tastan O., Dutta A., Booth P. J., Klein-Seetharaman J. (2014). Retinal Proteins as Model Systems for Membrane Protein Folding. Biochimica Biophysica Acta-Bioenergetics 1837, 656–663. 10.1016/j.bbabio.2013.11.021 [DOI] [PubMed] [Google Scholar]
- Tavanti F., Tozzini V. (2014). A Multi-Scale-Multi-Stable Model for the Rhodopsin Photocycle. Molecules 19, 14961–14978. 10.3390/molecules190914961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakita A., Koyanagi M., Tsukamoto H., Yamashita T., Miyata T., Shichida Y. (2004). Counterion Displacement in the Molecular Evolution of the Rhodopsin Family. Nat. Struct. Mol. Biol. 11, 284–289. 10.1038/nsmb731 [DOI] [PubMed] [Google Scholar]
- Terakita A. (2005). The Opsins. Genome Biol. 6, 213. 10.1186/gb-2005-6-3-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Yang S., Nagel G., Gao S. Q. (2022). Characterization and Modification of Light-Sensitive Phosphodiesterases from Choanoflagellates. Biomolecules 12, 88. 10.3390/biom12010088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkatch T., Greotti E., Baranauskas G., Pendin D., Roy S., Nita L. I., et al. (2017). Optogenetic Control of Mitochondrial Metabolism and Ca2+ Signaling by Mitochondria-Targeted Opsins. Proc. Natl. Acad. Sci. U. S. A. 114, E5167–E5176. 10.1073/pnas.1703623114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba Y., Hanawa I. (1985). Photoreceptor Sensitivity as a Function of Rhodopsin Content in the Isolated Bullfrog Retina. Jpn. J. Physiology 35, 483–494. 10.2170/jjphysiol.35.483 [DOI] [PubMed] [Google Scholar]
- Tomida S., Ito S., Mato T., Furutani Y., Inoue K., Kandori H. (2020). Infrared Spectroscopic Analysis on Structural Changes Around the Protonated Schiff Base upon Retinal Isomerization in Light-Driven Sodium Pump KR2. Biochimica Biophysica Acta-Bioenergetics 1861, 148190. 10.1016/j.bbabio.2020.148190 [DOI] [PubMed] [Google Scholar]
- Tomida S., Kitagawa S., Kandori H., Furutani Y. (2021). Inverse Hydrogen-Bonding Change between the Protonated Retinal Schiff Base and Water Molecules upon Photoisomerization in Heliorhodopsin 48C12. J. Phys. Chem. B 125, 8331–8341. 10.1021/acs.jpcb.1c01907 [DOI] [PubMed] [Google Scholar]
- Tomobe K., Yamamoto E., Kholmurodov K., Yasuoka K. (2017). Water Permeation through the Internal Water Pathway in Activated GPCR Rhodopsin. PLoS ONE 12, e0176876. 10.1371/journal.pone.0176876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townson S. M., Chang B. S. W., Salcedo E., Chadwell L. V., Pierce N. E., Britt S. G. (1998). Honeybee Blue-And Ultraviolet-Sensitive Opsins: Cloning, Heterologous Expression in Drosophila, and Physiological Characterization. J. Neurosci. 18, 2412–2422. 10.1523/jneurosci.18-07-02412.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribet C., Audebert R., Popot J.-L. (1996). Amphipols: Polymers that Keep Membrane Proteins Soluble in Aqueous Solutions. Proc. Nat. Acad. Sci. U. S. A. 93, 15047–15050. 10.1073/pnas.93.26.15047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.-J., Marino J., Adaixo R., Pamulal F., Muehle J., Maeda S., et al. (2019). Cryo-EM Structure of the Rhodopsin-Gαι-Βγ Complex Reveals Binding of the Rhodopsin C-Terminal Tail to the Gβ Subunit. Elife 8, 46041. 10.7554/elife.46041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura M., Ishikita H. (2020). Insights into the Protein Functions and Absorption Wavelengths of Microbial Rhodopsins. J. Phys. Chem. B 124, 11819–11826. 10.1021/acs.jpcb.0c08910 [DOI] [PubMed] [Google Scholar]
- Tsujimura M., Noji T., Saito K., Kojima K., Sudo Y., Ishikita H. (2021). Mechanism of Absorption Wavelength Shifts in Anion Channelrhodopsin-1 Mutants. Biochimica Biophysica Acta-Bioenergetics 1862, 148349. 10.1016/j.bbabio.2020.148349 [DOI] [PubMed] [Google Scholar]
- Tsukamoto H., Szundi I., Lewis J. W., Farrens D. L., Kliger D. S. (2011). Rhodopsin in Nanodiscs Has Native Membrane-like Photointermediates. Biochemistry 50, 5086–5091. 10.1021/bi200391a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto H., Terakita A. (2010). Diversity and Functional Properties of Bistable Pigments. Photochem. Photobiological Sci. 9, 1435–1443. 10.1039/c0pp00168f [DOI] [PubMed] [Google Scholar]
- Tsukamoto T., Mizutani K., Hasegawa T., Takahashi M., Honda N., Hashimoto N., et al. (2016). X-ray Crystallographic Structure of Thermophilic Rhodopsin - Implications For High Thermal Stability and Optogenetic Function . J. Biol. Chem. 291, 12223–12232. 10.1074/jbc.m116.719815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda S. P., Ewers D., Gazzarrini S., Moroni A., Gradmann D., Hegemann P. (2006). H+-pumping Rhodopsin from the Marine Alga Acetabularia . Biophysical J. 91, 1471–1479. 10.1529/biophysj.106.086421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda S. P., Sugiura M., Kandori H. (2021). “Molecular Properties and Optogenetic Applications of Enzymerhodopsins,” in Optogenetics: Light-Sensing Proteins and Their Applications in Neuroscience and beyond. Editors Yawo H., Kandori H., Koizumi A., Kageyama R.. 2nd ed (Singapore: Springer; ), 153–165. 10.1007/978-981-15-8763-4_9 [DOI] [PubMed] [Google Scholar]
- Tu C.-H., Yi H.-P., Hsieh S.-Y., Lin H.-S., Yang C.-S. (2018). Overexpression of Different Types of Microbial Rhodopsins with a Highly Expressible Bacteriorhodopsin from Haloarcula Marismortui as a Single Protein in E. coli . Sci. Rep. 8, 14026. 10.1038/s41598-018-32399-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutol J. N., Lee J., Chi H. C., Faizuddin F. N., Abeyrathna S. S., Zhou Q., et al. (2021). A Single Point Mutation Converts a Proton-Pumping Rhodopsin into a Red-Shifted, Turn-On Fluorescent Sensor for Chloride. Chem. Sci. 12, 5655–5663. 10.1039/d0sc06061e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta T., Kojima K., Hino T., Shibata M., Nagano S., Sudo Y. (2020). Applicability of Styrene-Maleic Acid Copolymer for Two Microbial Rhodopsins, RxR and HsSRI. Biophysical J. 119, 1760–1770. 10.1016/j.bpj.2020.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urner L. H., Liko I., Yen H.-Y., Hoi K.-K., Bolla J. R., Gault J., et al. (2020). Modular Detergents Tailor the Purification and Structural Analysis of Membrane Proteins Including G Protein-Coupled Receptors. Nat. Commun. 11, 564. 10.1038/s41467-020-14424-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez-Lopez J. C., Petr S. T., Donohue M. P., Bailey R. J., Gebreeziabher M., Cameron E. G., et al. (2020). The C-Terminus and Third Cytoplasmic Loop Cooperatively Activate Mouse Melanopsin Phototransduction. Biophysical J. 119, 389–401. 10.1016/j.bpj.2020.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Steen R., Biesheuvel P. L., Mathies R. A., Lugtenburg J. (1986). Retinal Analogs with Locked 6-7 Conformations Show that Bacteriorhodopsin Requires the 6-S-Trans Conformation of the Chromophore. J. Am. Chem. Soc. 108, 6410–6411. 10.1021/ja00280a060 [DOI] [Google Scholar]
- Van Eps N., Caro L. N., Morizumi T., Kusnetzow A. K., Szczepek M., Hofmann K. P., et al. (2017). Conformational Equilibria of Light-Activated Rhodopsin in Nanodiscs. Proc. Natl. Acad. Sci. U. S. A. 114, E3268–E3275. 10.1073/pnas.1620405114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanAken T., Foxall-Vanaken S., Castleman S., Ferguson-Miller S. (1986). Alkyl Glycoside Detergents: Synthesis and Applications to the Study of Membrane Proteins. Methods Enzym. 125, 27–35. 10.1016/s0076-6879(86)25005-3 [DOI] [PubMed] [Google Scholar]
- Varma N., Mutt E., Mühle J., Panneels V., Terakita A., Deupi X., et al. (2019). Crystal Structure of Jumping Spider Rhodopsin-1 as a Light Sensitive GPCR. Proc. Natl. Acad. Sci. U. S. A. 116, 14547–14556. 10.1073/pnas.1902192116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdegem P. J. E., Bovee-Geurts P. H. M., DeGrip W. J., Lugtenburg J., De Groot H. J. M. (1999). Retinylidene Ligand Structure in Bovine Rhodopsin, Metarhodopsin I, and 10-methylrhodopsin from Internuclear Distance Measurements Using 13C-Labeling and 1-D Rotational Resonance MAS NMR. Biochemistry-USA 38, 11316–11324. 10.1021/bi983014e [DOI] [PubMed] [Google Scholar]
- Verhoefen M.-K., Schäfer G., Shastri S., Weber I., Glaubitz C., Mäntele W., et al. (2011). Low Temperature FTIR Spectroscopy Provides New Insights in the pH-dependent Proton Pathway of Proteorhodopsin. Biochimica Biophysica Acta-Bioenergetics 1807, 1583–1590. 10.1016/j.bbabio.2011.09.001 [DOI] [PubMed] [Google Scholar]
- Verhoeven M. A., Bovee-Geurts P. H. M., De Groot H. J. M., Lugtenburg J., DeGrip W. J. (2006). Methyl Substituents at the 11- or 12-position of Retinal Profoundly and Differentially Affect Photochemistry and Signalling Activity of Rhodopsin. J. Mol. Biol. 363, 98–113. 10.1016/j.jmb.2006.07.039 [DOI] [PubMed] [Google Scholar]
- Verhoeven M. A., Creemers A. F. L., Bovee-Geurts P. H. M., DeGrip W. J., Lugtenburg J., De Groot H. J. M. (2001). Ultra-high-field MAS NMR Assay of a Multispin Labeled Ligand Bound to its G-Protein Receptor Target in the Natural Membrane Environment: Electronic Structure of the Retinylidene Chromophore in Rhodopsin. Biochemistry 40, 3282–3288. 10.1021/bi0023798 [DOI] [PubMed] [Google Scholar]
- Vierock J., Rodriguez-Rozada S., Dieter A., Pieper F., Sims R., Tenedini F., et al. (2021). BiPOLES Is an Optogenetic Tool Developed for Bidirectional Dual-Color Control of Neurons. Nat. Commun. 12, 4527. 10.1038/s41467-021-24759-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villette V., Chavarha M., Dimov I. K., Bradley J., Pradhan L., Mathieu B., et al. (2019). Ultrafast Two-Photon Imaging of a High-Gain Voltage Indicator in Awake Behaving Mice. Cell. 179, 1590–1609. 10.1016/j.cell.2019.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers P. M. a. M., Bovee-Geurts P. H. M., Portier M. D., Klaassen C. H. W., DeGrip W. J. (1998). Large-scale Production and Purification of the Human Green Cone Pigment: Characterization of Late Photo-Intermediates. Biochem. J. 330, 1201–1208. 10.1042/bj3301201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers P. M. a. M., DeGrip W. J. (1996). Functional Expression of Human Cone Pigments Using Recombinant Baculovirus: Compatibility with Histidine Tagging and Evidence for N-Glycosylation. FEBS Lett. 396, 26–30. 10.1016/0014-5793(96)01064-2 [DOI] [PubMed] [Google Scholar]
- Vlasov A. V., Maliar N. L., Bazhenov S. V., Nikelshparg E. I., Brazhe N. A., Vlasova A. D., et al. (2020). Raman Scattering: From Structural Biology to Medical Applications. Crystals 10, 38–3149. 10.3390/cryst10010038 [DOI] [Google Scholar]
- Vöcking O., Leclère L., Hausen H. (2021). The Rhodopsin-Retinochrome System for Retinal Re-isomerization Predates the Origin of Cephalopod Eyes. BMC Ecol. Evol. 21, 1–141. 10.1186/s12862-021-01939-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel R., Mahalingam M., Lüdeke S., Huber T., Siebert F., Sakmar T. P. (2008). Functional Role of the "Ionic Lock" - an Interhelical Hydrogen-Bond Network in Family a Heptahelical Receptors. J. Mol. Biol. 380, 648–655. 10.1016/j.jmb.2008.05.022 [DOI] [PubMed] [Google Scholar]
- Vogel R., Sakmar T. P., Sheves M., Siebert F. (2007). Coupling of Protonation Switches during Rhodopsin Activation. Photochem. Photobiol. 83, 286–292. 10.1562/2006-06-19-ir-937 [DOI] [PubMed] [Google Scholar]
- Vogeley L., Sineshchekov O. A., Trivedi V. D., Sasaki J., Spudich J. L., Luecke H. (2004). Anabaena Sensory Rhodopsin: A Photochromic Color Sensor at 2.0 Å. Science 306, 1390–1393. 10.1126/science.1103943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt A., Guo Y., Tsunoda S. P., Kateriya S., Elstner M., Hegemann P. (2015). Conversion of a Light-Driven Proton Pump into a Light-Gated Ion Channel. Sci. Rep. 5, 16450. 10.1038/srep16450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt A., Silapetere A., Grimm C., Heiser F., Möller M. A., Hegemann P. (2019). Engineered Passive Potassium Conductance in the KR2 Sodium Pump. Biophysical J. 116, 1941–1951. 10.1016/j.bpj.2019.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt K., Kirschfeld K. (1984). Chemical Identity of the Chromophores of Fly Visual Pigment. Naturwissenschaften 71, 211–213. 10.1007/bf00490436 [DOI] [Google Scholar]
- Volkov O., Kovalev K., Polovinkin V., Borshchevskiy V., Bamann C., Astashkin R., et al. (2017). Structural Insights into Ion Conduction by Channelrhodopsin 2. Science 358, eaan8862. 10.1126/science.aan8862 [DOI] [PubMed] [Google Scholar]
- Vought B. W., Dukkipati A., Max M., Knox B. E., Birge R. R. (1999). Photochemistry of the Primary Event in Short-Wavelength Visual Opsins at Low Temperature. Biochemistry-USA 38, 11287–11297. 10.1021/bi990968b [DOI] [PubMed] [Google Scholar]
- Vought B. W., Salcedo E., Chadwell L. V., Britt S. G., Birge R. R., Knox B. E. (2000). Characterization of the Primary Photointermediates of Drosophila Rhodopsin. Biochemistry 39, 14128–14137. 10.1021/bi001135k [DOI] [PubMed] [Google Scholar]
- Wada A., Fujioka N., Tanaka Y., Ito M. (2000). A Highly Stereoselective Synthesis of 11Z-Retinal Using Tricarbonyliron Complex. J. Org. Chem. 65, 2438–2443. 10.1021/jo9916030 [DOI] [PubMed] [Google Scholar]
- Wada T., Shimono K., Kikukawa T., Hato M., Shinya N., Kim S.-Y., et al. (2011). Crystal Structure of the Eukaryotic Light-Driven Proton-Pumping Rhodopsin, Acetabularia Rhodopsin II, from Marine Alga. J. Mol. Biol. 411, 986–998. 10.1016/j.jmb.2011.06.028 [DOI] [PubMed] [Google Scholar]
- Wagner M. J., Savall J., Hernandez O., Mel G., Inan H., Rumyantsev O., et al. (2021). A Neural Circuit State Change Underlying Skilled Movements. Cell. 184, 3731–3747. 10.1016/j.cell.2021.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner N. L., Greco J. A., Ranaghan M. J., Birge R. R. (2013). Directed Evolution of Bacteriorhodopsin for Applications in Bioelectronics. J. R. Soc. Interface 10, 201301971. 10.1098/rsif.2013.0197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald G., Brown P. K. (1953). The Molar Extinction of Rhodopsin. J. General Physiology 37, 189–200. 10.1085/jgp.37.2.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald G. (1935). Carotenoids and the Visual Cycle. J. General Physiology 19, 351–371. 10.1085/jgp.19.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald G. (1953). The Biochemistry of Vision. Annu. Rev. Biochem. 22, 497–526. 10.1146/annurev.bi.22.070153.002433 [DOI] [PubMed] [Google Scholar]
- Wald G. (1968). The Molecular Basis of Visual Excitation. Nature 219, 800–807. 10.1038/219800a0 [DOI] [PubMed] [Google Scholar]
- Walter J. M., Greenfield D., Liphardt J. (2010). Potential of Light-Harvesting Proton Pumps for Bioenergy Applications. Curr. Opin. Biotechnol. 21, 265–270. 10.1016/j.copbio.2010.03.007 [DOI] [PubMed] [Google Scholar]
- Wand A., Gdor I., Zhu J. Y., Sheves M., Ruhman S. (2013). Shedding New Light on Retinal Protein Photochemistry. Annu. Rev. Phys. Chem. 64, 437–458. 10.1146/annurev-physchem-040412-110148 [DOI] [PubMed] [Google Scholar]
- Wang N., Wang M. T., Gao Y. Y., Ran T. T., Lan Y. L., Wang J., et al. (2012). Crystallization and Preliminary X-Ray Crystallographic Analysis of a Blue-Light-Absorbing Proteorhodopsin. Acta Crystallogr. Sect. F-Structural Biol. Cryst. Commun. 68, 281–283. 10.1107/s1744309111043612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. L., Munro R. A., Shi L. C., Kawamura I., Okitsu T., Wada A., et al. (2013). Solid-state NMR Spectroscopy Structure Determination of a Lipid-Embedded Heptahelical Membrane Protein. Nat. Methods 10, 1007–1012. 10.1038/nmeth.2635 [DOI] [PubMed] [Google Scholar]
- Wang Y.-J., Bovee-Geurts P. H. M., Lugtenburg J., DeGrip W. J. (2004). Constraints of the 9-methyl Group Binding Pocket of the Rhodopsin Chromophore Probed by 9-halogeno Substitution. Biochemistry 43, 14802–14810. 10.1021/bi048404h [DOI] [PubMed] [Google Scholar]
- Wang Y., Li Y., Xu T., Shi Z. Y., Wu Q. (2015). Experimental Evidence for Growth Advantage and Metabolic Shift Stimulated by Photophosphorylation of Proteorhodopsin Expressed in Escherichia coli at Anaerobic Condition. Biotechnol. Bioeng. 112, 947–956. 10.1002/bit.25504 [DOI] [PubMed] [Google Scholar]
- Wang Z. Y., Asenjo A. B., Oprian D. D. (1993). Identification of the Cl-Bbinding Site in the Human Red and Green Color Vision Pigments. Biochemistry-USA 32, 2125–2130. 10.1021/bi00060a001 [DOI] [PubMed] [Google Scholar]
- Warrant E. J., Locket N. A. (2004). Vision in the Deep Sea. Biol. Rev. 79, 671–712. 10.1017/s1464793103006420 [DOI] [PubMed] [Google Scholar]
- Warrant E. J., Mciintyre P. D. (1993). Arthropod Eye Design and the Physical Limits to Spatial Resolving Power. Prog. Neurobiol. 40, 413–461. 10.1016/0301-0082(93)90017-m [DOI] [PubMed] [Google Scholar]
- Watari M., Ikuta T., Yamada D., Shihoya W., Yoshida K., Tsunoda S. P., et al. (2019). Spectroscopic Study of the Transmembrane Domain of a Rhodopsin-Phosphodiesterase Fusion Protein from a Unicellular Eukaryote. J. Biol. Chem. 294, 3432–3443. 10.1074/jbc.ra118.006277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T., Skopintsev P., James D., Dworkowski F., Panepucci E., Kekilli D., et al. (2019). Proton Uptake Mechanism in Bacteriorhodopsin Captured by Serial Synchrotron Crystallography. Science 365, 61–65. 10.1126/science.aaw8634 [DOI] [PubMed] [Google Scholar]
- Weingart O. (2007). The Twisted C11=C12 Bond of the Rhodopsin Chromophore - A Photochemical Hot Spot. J. Am. Chem. Soc. 129, 10618–10619. 10.1021/ja071793t [DOI] [PubMed] [Google Scholar]
- Weissbecker J., Boumrifak C., Breyer M., Wiessalla T., Shevchenko V., Mager T., et al. (2021). The Voltage Dependent Sidedness of the Reprotonation of the Retinal Schiff Base Determines the Unique Inward Pumping of Xenorhodopsin. Angew. Chemie-International Ed. 60, 23010–23017. 10.1002/anie.202103882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner K., Lehner I., Dhiman H. K., Richter C., Glaubitz C., Schwalbe H., et al. (2007). Combined Solid State and Solution NMR Studies of α, ε-15N Labeled Bovine Rhodopsin. J. Biomol. NMR 37, 303–312. 10.1007/s10858-007-9143-0 [DOI] [PubMed] [Google Scholar]
- Wickstrand C., Dods R., Royant A., Neutze R. (2015). Bacteriorhodopsin: Would the Real Structural Intermediates Please Stand up? Biochimica Biophysica Acta-General Subj. 1850, 536–553. 10.1016/j.bbagen.2014.05.021 [DOI] [PubMed] [Google Scholar]
- Wietek J., Beltramo R., Scanziani M., Hegemann P., Oertner T. G., Wiegert J. S. (2015). An Improved Chloride-Conducting Channelrhodopsin for Light-Induced Inhibition of Neuronal Activity In Vivo . Sci. Rep. 5, 14807. 10.1038/srep14807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijffels R. H., Kruse O., Hellingwerf K. J. (2013). Potential of Industrial Biotechnology with Cyanobacteria and Eukaryotic Microalgae. Curr. Opin. Biotechnol. 24, 405–413. 10.1016/j.copbio.2013.04.004 [DOI] [PubMed] [Google Scholar]
- Williams R. H., Tsunematsu T., Thomas A. M., Bogyo K., Yamanaka A., Kilduff T. S. (2019). Transgenic Archaerhodopsin-3 Expression in Hypocretin/Orexin Neurons Engenders Cellular Dysfunction and Features of Type 2 Narcolepsy. J. Neurosci. 39, 9435–9452. 10.1523/jneurosci.0311-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P., Rodgers J., Wynne J., Bishop P. N., Lucas R. J., Milosavljevic N. (2021). Viral Transduction of Human Rod Opsin or Channelrhodopsin Variants to Mouse on Bipolar Cells Does Not Impact Retinal Anatomy or Cause Measurable Death in the Targeted Cells. Int. J. Mol. Sci. 22, 13111. 10.3390/ijms222313111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright W., Gajjeraman S., Batabyal S., Pradhan S., Bhattacharya S., Mahapatra V., et al. (2017). Restoring Vision in Mice with Retinal Degeneration Using Multicharacteristic Opsin. Neurophotonics 4, 041505. 10.1117/1.nph.4.4.049801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Wang R. X., Yang Y. F., Xu T. Y., Li Y., Xu J., et al. (2020). Expression of OPN3 in Lung Adenocarcinoma Promotes Epithelial-Mesenchymal Transition and Tumor Metastasis. Thorac. Cancer 11, 286–294. 10.1111/1759-7714.13254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. X., Peng L. X., Wang S. C., Wang A. Q., Ma R. R., Zhou Y., et al. (2018). Hybrid Indicators for Fast and Sensitive Voltage Imaging. Angew. Chemie-International Ed. 57, 3949–3953. 10.1002/anie.201712614 [DOI] [PubMed] [Google Scholar]
- Yaguchi M., Jia X., Schlesinger R., Jiang X., Ataka K., Heberle J. (2022). Near-Infrared Activation of Sensory Rhodopsin II Mediated by NIR-To-Blue Upconversion Nanoparticles. Front. Mol. Biosci. 8, 782688. 10.3389/fmolb.2021.782688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalouz S., Senjean B., Günther J., Buda F., O'brien T. E., Visscher L. (2021). A State-Averaged Orbital-Optimized Hybrid Quantum-Classical Algorithm for a Democratic Description of Ground and Excited States. Quantum Sci. Technol. 6, 024004. 10.1088/2058-9565/abd334 [DOI] [Google Scholar]
- Yamashita T., Ohuchi H., Tomonari S., Ikeda K., Sakai K., Shichida Y. (2010). Opn5 Is a UV-Sensitive Bistable Pigment that Couples with Gi Subtype of G Protein. Proc. Natl. Acad. Sci. U. S. A. 107, 22084–22089. 10.1073/pnas.1012498107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T., Ono K., Ohuchi H., Yumoto A., Gotoh H., Tomonari S., et al. (2014). Evolution of Mammalian Opn5 as a Specialized UV-Absorbing Pigment by a Single Amino Acid Mutation. J. Biol. Chem. 289, 3991–4000. 10.1074/jbc.m113.514075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T. (2020). Unexpected Molecular Diversity of Vertebrate Nonvisual Opsin Opn5. Biophys. Rev. 12, 333–338. 10.1007/s12551-020-00654-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan E. C. Y., Ganim Z., Kazmi M. A., Chang B. S. W., Sakmar T. P., Mathies R. A. (2004). Resonance Raman Analysis of the Mechanism of Energy Storage and Chromophore Distortion in the Primary Visual Photoproduct. Biochemistry 43, 10867–10876. 10.1021/bi0400148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Manathunga M., Gozem S., Léonard J., Andruniów T., Olivucci M. (2022). Quantum-classical Simulations of Rhodopsin Reveal Excited-State Population Splitting and its Effects on Quantum Efficiency. Nat. Chem. 14, 441. 10.1038/s41557-022-00892-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S. I., Akiyama T., Kojima K., Ueta T., Hayashi T., Ogasawara S., et al. (2022). Development of an Outward Proton Pumping Rhodopsin with a New Record in Thermostability by Means of Amino Acid Mutations. J. Phys. Chem. B 126, 1004. 10.1021/acs.jpcb.1c08684 [DOI] [PubMed] [Google Scholar]
- Ye S. X., Huber T., Vogel R., Sakmar T. P. (2009). FTIR Analysis of GPCR Activation Using Azido Probes. Nat. Chem. Biol. 5, 397–399. 10.1038/nchembio.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S. X., Zaitseva E., Caltabiano G., Schertler G. F. X., Sakmar T. P., Deupi X., et al. (2010). Tracking G-Protein-Coupled Receptor Activation Using Genetically Encoded Infrared Probes. Nature 464, 1386–1U14. 10.1038/nature08948 [DOI] [PubMed] [Google Scholar]
- Yee D. C., Shlykov M. A., Västermark A., Reddy V. S., Arora S., Sun E. I., et al. (2013). The Transporter-Opsin-G Protein-Coupled Receptor (TOG) Superfamily. FEBS J. 280, 5780–5800. 10.1111/febs.12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh V., Lee T.-Y., Chen C.-W., Kuo P.-C., Shiue J., Chu L.-K., et al. (2018). Highly Efficient Transfer of 7TM Membrane Protein from Native Membrane to Covalently Circularized Nanodisc. Sci. Rep. 8, 13501. 10.1038/s41598-018-31925-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi A., Li H., Mamaeva N., De Cordoba R. E. F., Lugtenburg J., DeGrip W. J., et al. (2017). Structural Changes in an Anion Channelrhodopsin: Formation of the K and L Intermediates at 80 K. Biochemistry 56, 2197–2208. 10.1021/acs.biochem.7b00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi A., Mamaeva N., Li H., Spudich J. L., Rothschild K. J. (2016). Resonance Raman Study of an Anion Channelrhodopsin: Effects of Mutations Near the Retinylidene Schiff Base. Biochemistry 55, 2371–2380. 10.1021/acs.biochem.6b00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O., Fenno L. E., Davidson T. J., Mogri M., Deisseroth K. (2011). Optogenetics in Neural Systems. Neuron 71, 9–34. 10.1016/j.neuron.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Yokoyama S. (1995). Amino Acid Replacements and Wavelength Absorption of Visual Pigments in Vertebrates. Mol. Biol. Evol. 12, 53–61. 10.1093/oxfordjournals.molbev.a040190 [DOI] [PubMed] [Google Scholar]
- Yokoyama S. (2000). Molecular Evolution of Vertebrate Visual Pigments. Prog. Retin. Eye Res. 19, 385–419. 10.1016/s1350-9462(00)00002-1 [DOI] [PubMed] [Google Scholar]
- Yokoyama S., Yokoyama R. (2000). “Comparative Molecular Biology of Visual Pigments,” in Molecular Mechanisms in Visual Transduction. Editors Stavenga D. G., DeGrip W. J., PughJr. E. N. (Amsterdam, Netherlands: Elsevier Science Pub.), 257–296. 10.1016/s1383-8121(00)80009-3 [DOI] [Google Scholar]
- Yoshida K., Yamashita T., Sasaki K., Inoue K., Shichida Y., Kandori H. (2017). Chimeric Microbial Rhodopsins for Optical Actvation of Gs-Proteins. Biophysics physicobiology 14, 183–190. 10.2142/biophysico.14.0_183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T., Kandori H. (1991a). Primary Photochemical Events in the Rhodopsin Molecule. Prog. Retin. Res. 11, 33–55. 10.1016/0278-4327(91)90023-u [DOI] [Google Scholar]
- Yoshizawa T., Kuwata O. (1991b). Iodopsin, a Red-Sensitive Cone Visual Pigment in the Chicken Retina. Photochem. Photobiol. 54, 1061–1070. 10.1111/j.1751-1097.1991.tb02130.x [DOI] [PubMed] [Google Scholar]
- Yoshizawa T. (1984). Photophysiological Functions of Visual Pigments. Adv. Biophysics 17, 5–67. 10.1016/0065-227x(84)90024-8 [DOI] [PubMed] [Google Scholar]
- Yoshizawa T., Wald G. (1967). Photochemistry of Iodopsin. Nature 214, 566–571. 10.1038/214566a0 [DOI] [PubMed] [Google Scholar]
- Yoshizawa T., Wald G. (1963). Pre-lumirhodopsin and the Bleaching of Visual Pigments. Nature 197, 1279–1286. 10.1038/1971279a0 [DOI] [PubMed] [Google Scholar]
- Young R. W. (1976). Visual Cells and the Concept of Renewal. Investig. Ophthalmol. Vis. Sci. 15, 700–725. [PubMed] [Google Scholar]
- Yu H., Siewny M. G. W., Edwards D. T., Sanders A. W., Perkins T. T. (2017). Hidden Dynamics in the Unfolding of Individual Bacteriorhodopsin Proteins. Science 355, 945–949. 10.1126/science.aah7124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N., Huang L., Zhou Y. B., Xue T., Chen Z. G., Han G. (2019). Near-Infrared-Light Activatable Nanoparticles for Deep-Tissue-Penetrating Wireless Optogenetics. Adv. Healthc. Mater. 8, 1801132. 10.1002/adhm.201801132 [DOI] [PubMed] [Google Scholar]
- Yu S. M., Mcquade D. T., Quinn M. A., Hackenberger C. P. R., Krebs M. P., Polans A. S., et al. (2000). An Improved Tripod Amphiphile for Membrane Protein Solubilization. Protein Sci. 9, 2518–2527. 10.1110/ps.9.12.2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J.-H., Li X. X., Park J.-H., Wang Y., Ohki M., Jin Z. Y., et al. (2019). Non-cryogenic Structure of a Chloride Pump Provides Crucial Clues to Temperature-dependent Channel Transport Efficiency. J. Biol. Chem. 294, 794–804. 10.1074/jbc.ra118.004038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J.-H., Li X. X., Yue J. N., Park J.-H., Jin Z. Y., Li C. F., et al. (2021). Early-stage Dynamics of Chloride Ion-Pumping Rhodopsin Revealed by a Femtosecond X-Ray Laser. Proc. Natl. Acad. Sci. U. S. A. 118, 2020486118. 10.1073/pnas.2020486118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J.-H., Ohki M., Park J.-H., Ishimoto N., Sato-Tomita A., Lee W., et al. (2020). Pumping Mechanism of NM-R3, a Light-Driven Bacterial Chloride Importer in the Rhodopsin Family. Sci. Adv. 6, eaay204. 10.1126/sciadv.aay2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabelskii D., Alekseev A., Kovalev K., Rankovic V., Balandin T., Soloviov D., et al. (2020). Viral Rhodopsins 1 Are an Unique Family of Light-Gated Cation Channels. Nat. Commun. 11, 5707. 10.1038/s41467-020-19457-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabelskii D., Dmitrieva N., Volkov O., Shevchenko V., Kovalev K., Balandin T., et al. (2021). Structure-based Insights into Evolution of Rhodopsins. Commun. Biol. 4, 821. 10.1038/s42003-021-02326-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Y., Choi E. H., Tworak A., Salom D., Leinonen H., Sander C. L., et al. (2019). Photic Generation of 11-Cis-Retinal in Bovine Retinal Pigment Epithelium. J. Biol. Chem. 294, 19137–19154. 10.1074/jbc.ra119.011169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Salom D., He J. H., Okun A., Ballesteros J., Palczewski K., et al. (2005). Expression of Functional G Protein-Coupled Receptors in Photoreceptors of Transgenic Xenopus laevis . Biochemistry 44, 14509–14518. 10.1021/bi051386z [DOI] [PubMed] [Google Scholar]
- Zhang L., Wang K., Ning S., Pedersen P. A., Duelli A. S., Gourdon P. E. (2022). Isolation and Crystallization of the D156C Form of Optogenetic ChR2. Cells 11, 895. 10.3390/cells11050895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Gui M., Wang Z.-F., Gorgulla C., Yu J. J., Wu H., et al. (2021a). Cryo-EM Structure of an Activated GPCR-G Protein Complex in Lipid Nanodiscs. Nat. Struct. Mol. Biol. 28, 258–267. 10.1038/s41594-020-00554-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. S., Zheng S. N., Sun J. H., Zeng X. X., Duan Y. K., Luan G. D., et al. (2021). Rapidly Improving High Light and High Temperature Tolerances of Cyanobacterial Cell Factories through the Convenient Introduction of an AtpA-C252f Mutation. Front. Microbiol. 12, 647164. 10.3389/fmicb.2021.647164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Cao L.-H., Kumar S., Enemchukwu N. O., Zhang N., Lambert A., et al. (2016). Dimerization of Visual Pigments In Vivo . Proc. Natl. Acad. Sci. U. S. A. 113, 9093–9098. 10.1073/pnas.1609018113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. M., Yokoyama T., Sakamoto M. (2021b). Imaging Voltage with Microbial Rhodopsins. Front. Mol. Biosci. 8, 738829. 10.3389/fmolb.2021.738829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Iwasa T., Tsuda M., Kobata A., Takasaki S. (1997). A Novel Monoantennary Complex-type Sugar Chain Found in octopus Rhodopsin: Occurrence of the Galβ1->3Fuc Group Linked to the Proxiranal N-Acetylglucosamine Residue of the Trimannosyl Core. Glycobiology 7, 1153–1158. 10.1093/glycob/7.8.1153 [DOI] [PubMed] [Google Scholar]
- Zhao D. Y., Pöge M., Morizumi T., Gulati S., Van Eps N., Zhang J. Y., et al. (2019). Cryo-EM Structure of the Native Rhodopsin Dimer in Nanodiscs. J. Biol. Chem. 294, 14215–14230. 10.1074/jbc.ra119.010089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z. L., Wang P., Xu X. L., Sheves M., Jin Y. D. (2015). Bacteriorhodopsin/Ag Nanoparticle-Based Hybrid Nano-Bio Electrocatalyst for Efficient and Robust H2 Evolution from Water. J. Am. Chem. Soc. 137, 2840–2843. 10.1021/jacs.5b00200 [DOI] [PubMed] [Google Scholar]
- Zhou H.-X., Cross T. A. (2013). Influences of Membrane Mimetic Environments on Membrane Protein Structures. Annu. Rev. Biophysics 42, 361–392. 10.1146/annurev-biophys-083012-130326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Ding M. Q., Duan X. D., Konrad K. R., Nagel G., Gao S. Q. (2021). Extending the Anion Channelrhodopsin-Based Toolbox for Plant Optogenetics. Membranes 11, 287–281212. 10.3390/membranes11040287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Tian H. H., Guan S. L., Ding J. F., Gao L., Wang J. F., et al. (2021). Self-assembled Multifunctional Neural Probes for Precise Integration of Optogenetics and Electrophysiology. Nat. Commun. 12, 5871–58715879. 10.1038/s41467-021-26168-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zundel G. (1988). “Hydrogen-bond Systems as Proton Wires Formed by Side Chains of Proteins and by Side Chains and Phosphates,” in Transport through Membranes: Carriers, Channels and Pumps. Editor Pullman A. (Dordrecht, Netherlands: Kluwer Academic Publishers; ), 409–420. 10.1007/978-94-009-3075-9_27 [DOI] [Google Scholar]