Abstract
Background:
Genome-wide sequencing has emerged as a promising strategy for the timely diagnosis of rare diseases, but it is not yet available as a clinical test performed in Canadian diagnostic laboratories. We describe the protocol for evaluating a 2-year pilot project, Genome-wide Sequencing Ontario, to offer high-quality clinical genome-wide sequencing in Ontario, Canada.
Methods:
The Genome-wide Sequencing Ontario protocol was codesigned by the Ontario Ministry of Health, the Hospital for Sick Children in Toronto and the Children’s Hospital of Eastern Ontario in Ottawa. Enrolment of a prospective cohort of patients began on Apr. 1, 2021. Eligible cases with blood samples available for the index case and both parents (i.e., trios) are randomized to receive exome sequencing or genome sequencing. We will collect patient-level data and ascertain costs associated with the laboratory workflow for exome sequencing and genome sequencing. We will compare point estimates for the diagnostic utility and timeliness of exome sequencing and genome sequencing, and we will determine an incremental cost-effectiveness ratio (expressed as the incremental cost of genome sequencing versus exome sequencing per additional patient with a causal variant detected).
Interpretation:
Findings from this work will provide robust evidence for the diagnostic utility, cost-effectiveness and timeliness of exome sequencing and genome sequencing, and will be disseminated via academic publications and policy briefs. Findings will inform provincial and cross-provincial policy related to the long-term organization, delivery and reimbursement of clinical-grade genome diagnostics for rare disease.
An accurate diagnosis is an essential component of care for patients with rare diseases, enabling tailored patient management, cascade family testing, family planning and peer support.1 Exome sequencing (ES) has emerged as a diagnostic test in many jurisdictions to facilitate diagnoses for patients with a suspected rare genetic disease.2–4 Extending the capabilities of ES, genome sequencing (GS) offers coverage of the coding and noncoding regions of the genome, and improved detection of copy number and structural variants.5 The term genome-wide sequencing (GWS) has emerged to refer to both ES and GS. In addition to its diagnostic capabilities, GWS can also be used to identify secondary findings — variants that are unrelated to the indication for testing but are associated with medically actionable, often presymptomatic health risks.6,7
Despite the enhanced capabilities of GS compared to ES, a recent meta-analysis found that the diagnostic utility (i.e., rate of identifying causative genotypes in known disease genes) of GS and ES was not significantly different.8 However, for both ES and GS, the likelihood of diagnosis was significantly higher when sequencing was performed on the index case plus 2 biological relatives (usually parents; referred to as “trios”) compared to the index case alone (referred to as “singletons”).8 Although studies of the cost-effectiveness of ES and GS are emerging for a range of conditions,9–11 a recent systematic review of 36 studies concluded that the economic evidence base to support the use of ES and GS in the clinic is very limited and that robust comparative studies are urgently needed to support their translation into clinical practice.11
Alongside this growing evidence base, the Ontario Ministry of Health, which is responsible for administering the health care system for Ontario’s population of almost 15 million,12 has been instrumental in enabling access to clinical ES in Ontario since 2014 through its Out-of-Country/Out-of-Province (OOC/OOP) Prior Approval Program.13 This program is an exceptional access mechanism that allows the use of services outside the province or country when they are not available in the province and if specific criteria are met. These criteria are informed by input from relevant experts and stakeholders, and by professional best practice statements.
In Ontario, criteria for access to ES were based on a report for the Ontario Ministry of Health’s Genetic Testing Advisory Committee;14 this report was informed by the Canadian College of Medical Geneticists position statement on the clinical implementation of GWS.15 In 2020, in response to demand for local ES services, Ontario Health (an agency of the Ontario Ministry of Health) undertook a comprehensive health technology assessment of ES and GS for people with unexplained developmental disabilities or multiple congenital anomalies.16 Based on the evidence reviewed in that assessment, the Ontario Health Technology Advisory Committee recommended public funding for ES as a second-tier test for these individuals.9,16
As a result of policy efforts and the evolution in recent years of exceptional access programs in Ontario and other Canadian provinces, approximately 1500 Canadian patients with rare diseases receive access to clinical ES performed outside the country each year.17 Although the approach to date has enabled access to high-quality ES in an important subset of eligible patients, establishing local infrastructure is essential to enable the development of integrated laboratory services, a knowledge base that is locally representative and a performance measurement system that will guide technical and policy decisions related to the use of this technology over time.
We developed the Genome-wide Sequencing Ontario (GSO) pilot project, the objective of which is to evaluate the clinical performance and cost-effectiveness of ES and GS in Ontario. The project is intended to provide a high-quality, timely, cost-effective, equitable and sustainable service for residents of the province. GSO will provide evidence to inform longer-term decisions about technology adoption in Ontario and serve as a model for evidence development and clinical translation in other jurisdictions where clinical GWS policy is a priority. We describe a protocol that aims to evaluate the performance of this 2-year pilot project in Ontario.
Methods
Study design and setting
Informed by the strategies that have guided implementation efforts for ES and GS internationally,18,19 we have developed a 2-year, prospective, hybrid type 2 implementation-effectiveness study design, tailored to Ontario’s needs and resources.20 With a design that blends components of clinical effectiveness and implementation research, this approach enables more rapid translational gains, more effective implementation strategies and more useful information for decisionmakers.20 Although this project is not designed as a hypothesis-driven effectiveness trial, participants are randomized to receive ES or GS to mitigate bias in determining and comparing performance and implementation outcomes for these sequencing strategies.
GSO is cohosted by the genome diagnostics laboratories and divisions of clinical genetics at The Hospital for Sick Children (SickKids) in Toronto and the Children’s Hospital of Eastern Ontario (CHEO) in Ottawa. Although the project was developed to meet the real-world evidence needs of our end users at the Laboratory and Diagnostics Branch, Ontario Ministry of Health, these policy partners did not influence the design or the dissemination plan for the work.
We will use the Standards for Quality Improvement Reporting Excellence (SQUIRE) reporting guidelines checklist.21
Intervention
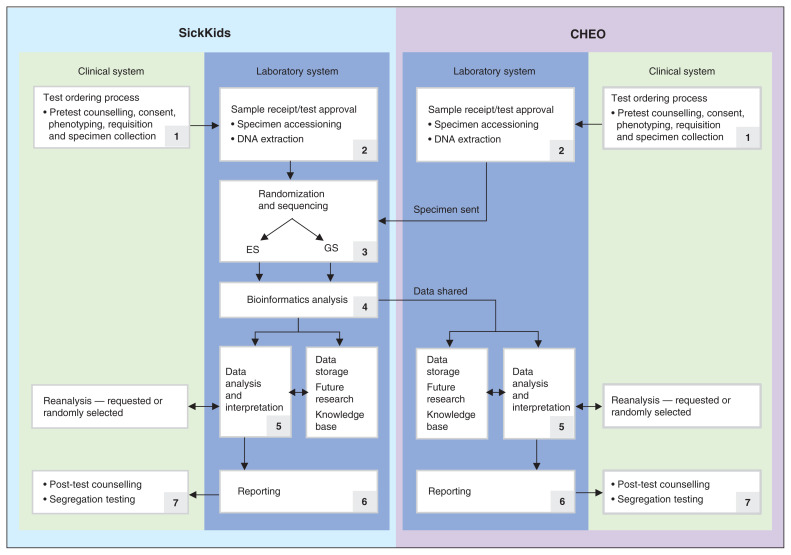
Routine (i.e., nonrapid) genome-wide sequencing — ES and GS — are performed on the Novaseq6000 sequencing platform at SickKids, followed by genome alignment and variant calling. Resulting variants undergo annotation, filtration and interpretation. A cloud-based platform facilitates data flow between laboratories, analysis and delivery, storage, and sharing (Figure 1). Harmonized interpretation and reporting protocols were developed by the SickKids and CHEO laboratories based on technical specifications from an expert working group established by the Ministry of Health Laboratories and Diagnostics Branch.22
Figure 1:
Genome-wide Sequencing Ontario harmonized, multi-institutional model for delivering genome-wide sequencing (GWS). CHEO = The Children’s Hospital of Eastern Ontario, GS = genome sequencing, ES = exome sequencing, SickKids = The Hospital for Sick Children.
Participants
The target population includes adults and children and aligns with the eligibility criteria for GWS that are in use by Ontario’s OOC/OOP Prior Approval Program, as established in the Genetic Testing Advisory Committee report14 and the Canadian College of Medical Geneticists position statement.15 Specifically, eligible patients include those for whom (a) a baseline genetics evaluation has been completed (e.g., phenotyping, family history, pretest genetic counselling and consent, and, where indicated, chromosome microarray and targeted testing, including biochemical testing); (b) a genetic etiology is the most likely explanation for the phenotype, as supported by a clinical presentation that includes any one of the following: moderate to severe developmental or functional impairment, multisystem involvement, a progressive clinical course that cannot be explained by another cause, or a differential diagnosis that includes 2 or more conditions that would require evaluation by separate gene panels; and (c) blood samples are available from the index patient and both parents (i.e., trios).
Ineligible patients include those for whom (a) the clinical presentation is limited to isolated mild intellectual disability or learning disabilities, nonsyndromic autism, isolated neurobehavioural disabilities (e.g., attention-deficit/hyperactivity disorder) or isolated neuropsychiatric conditions (e.g., schizophrenia); or (b) the phenotype is highly specific to a known genetic condition for which optimized genetic testing (e.g., a multigene panel) exists or is suggestive of an aneuploidy, a methylation defect or a trinucleotide repeat disorder.
As an implementation project, the sample size (n = 650) is based on OOC/OOP Program annual testing volumes for SickKids and CHEO.
Procedure
Patients are enrolled prospectively and complete a clinical consent form that is harmonized across sites to enable data-sharing between the SickKids and CHEO laboratories, optimizing the consistency of variant interpretation. After a genetics consultation during which ES or GS is deemed to be indicated, the requisition and sample collection are completed. All requisitions are reviewed by a genetic counsellor; if the patient meets the eligibility criteria, DNA is extracted and sent to the SickKids laboratory for sequencing.
Requisitions for testing of cases with uncertain eligibility are reviewed by a joint committee of clinical and laboratory experts. If eligibility criteria are not met, testing does not proceed, and a rejection report is issued to the ordering provider. For cases that are excluded, a physician can choose to appeal, whereupon the joint committee process would be used to evaluate any substantive additional clinical information or published evidence that is provided to support the case for testing. If preferred, ordering providers can submit a request for pre-approval before sample collection.
Randomization and matching
Patients are randomly assigned to ES or GS using a 1:1 ratio and an unblinded, stratified, permuted block randomization design. Cases are stratified by clinical site, phenotype and molecular testing history. Depending on the expected recruitment in each stratum, block sizes (i.e., the number of patients in each stratum) are 2, 4 or 6.
Outcomes
Three core dimensions on quality care will be ascertained: timeliness, diagnostic utility and cost-effectiveness.23 Timeliness represents the core implementation outcome, and diagnostic utility and cost-effectiveness represent core effectiveness outcomes. In addition to patient-level clinical characteristics (i.e., enrolment site, age, physical features or symptoms, ethnicity, consanguinity, molecular testing history and test urgency) and patient preferences related to research recontact and receipt of secondary findings, patient-level details are collected to facilitate the assessment of implementation and effectiveness outcomes.
Patient-level details relevant to the assessment of timeliness include the following: the date samples are received or accessioned; the date eligibility for ES or GS is confirmed; the date ES or GS is initiated and completed; the date the ES or GS result is reported; and the date segregation analysis is initiated and reported. Timeliness will be defined from a laboratory perspective and measured as the number of weeks elapsed from sample accessioning to laboratory reporting. It will be reported as the proportion of cases for whom laboratory turnaround time is less than 12 weeks.
Patient-level details relevant to the assessment of effectiveness in the form of diagnostic utility include the following: the number and laboratory interpretation of primary ES or GS variants identified (i.e., pathogenic variant, likely pathogenic variant, variant of uncertain clinical significance); the clinical interpretation of primary ES or GS variants identified (i.e., diagnostic, partially diagnostic, potentially diagnostic, nondiagnostic); and the number and interpretation of medically actionable secondary variants identified. Diagnostic utility will be defined as the rate of causative, pathogenic or likely pathogenic genotypes in known disease genes. It will be reported as the proportion of cases for whom diagnostic, partially diagnostic and medically actionable secondary variants are identified.
To facilitate the assessment of cost-effectiveness, costs for ES and GS will be determined by microcosting the laboratory workflow components for each sequencing approach.9,16,24,25 Data on the volume of resource use and unit prices for each workflow input will be obtained from laboratory protocols and managers. Cost-effectiveness will be reported as the incremental costs of local GS compared to ES for each additional patient with a molecular diagnosis achieved.
Data sources
Patient characteristics and preferences related to research recontact and receipt of secondary findings are ascertained from ES or GS requisition forms that are submitted to the laboratory. Laboratory process measures (i.e., dates) are ascertained from the SickKids and CHEO laboratory information systems, and the results of the ES or GS analyses are obtained from the harmonized, cloud-based genome analysis platform.
All data are entered into a centralized REDCap database. Data collection is facilitated by members of the study team and monitored monthly by a data audit team for accuracy and completion. Missing data identified through monthly quality checks will be obtained by the laboratory genetic counsellor from the ordering provider, technologist, genome analyst or director assigned to the case.
Statistical analysis
We will analyze our data using descriptive statistics (mean, median, range and standard deviation). Point estimates for diagnostic utility and timeliness will be compared statistically for ES and GS. We will examine the relationship between clinical characteristics (e.g., age, age of onset, sex, phenotype, prior testing history) and diagnostic yield using parametric or nonparametric univariate statistics as appropriate. If indicated, we will investigate explanatory variables of diagnostic utility using a regression model.
To facilitate the cost-effectiveness analysis, we will determine a cost per trio ES and cost per trio GS. Costs for each input related to the ES and GS laboratory workflows will be calculated by multiplying resource volume by unit price. For labour, the time in minutes for each task will be multiplied by wage rates. We will use the most recently available price units (2021). Otherwise, the prices as reported in previous microcosting studies24,25 will be assumed to be stable based on consultation with laboratory managers. The costs of laboratory workflow inputs for each sequencing approach will be summed to determine a per-sample cost. We will then undertake a cost-effectiveness analysis from an institutional payer perspective, in which the difference in costs between GS and ES will be calculated and divided by the difference in diagnostic yield to calculate an incremental cost-effectiveness ratio, expressed as the incremental cost of GS versus ES per additional patient detected with a pathogenic variant.
As an implementation project, this cost-effectiveness analysis is limited to laboratory costs and outcomes. Given the complexity of diagnostic outcomes and the heterogeneity of the patient sample, it will not be possible to model patients’ health states and the range of treatment options for a given diagnostic result to generate health benefits over a lifetime (such as quality-adjusted life-years). All costs will be reported in 2021 Canadian dollars.
Ethics approval
Individuals who undergo GWS as part of this protocol provide clinical consent for GWS and research consent through Clinical Trials Ontario [CTO-1577], the provincial body responsible for approving clinical trials and observational studies that involve 2 or more academic or health care institutions in Ontario.
Interpretation
As a robust comparison of diagnostic utility, cost-effectiveness and timeliness for clinical grade ES and GS, this study will be instrumental in guiding provincial policy and funding decisions related to their ongoing use in clinical care. We will also assess the relationship between key clinical characteristics (e.g., age, age of symptom onset, phenotype, prior testing history) and these performance outcomes to understand the relative performance of these technologies in different patient groups. In particular, our data will inform the development of diagnostic pathway guidelines that provide recommendations on the use of ES and GS for different clinical indications and when the pathway should include ES versus GS. Finally, having established a centralized sequencing–distributed interpretation model between 2 sites, data collected for this project will inform the feasibility and costs of scaling this model across the province.
In addition to disseminating our findings through academic publications and presentations, we will generate a report for the provincial policy partners who have been engaged in the design and implementation of this work. We will also develop educational materials for an expanding network of ordering providers to improve genomic literacy among non–genetics providers and generate lay summaries of our findings for patients with rare disease.
All patients are provided with the option to participate in future research as part of their clinical consent process. Patients who consent to recontact can enrol in additional research ethics board–approved studies linked to GSO, including the clinical reanalysis of genomic data and the patient and system impacts of medically actionable secondary findings. Findings from these studies will inform best practices and guidelines for clinical reanalysis and for managing secondary findings in a single-payer health care system. Participants in GSO will also be able to enrol in other rare disease research initiatives focused on gene discovery, characterizing natural history and trialling precision therapies.26
Moving beyond Ontario, we will engage our clinical, research, policy and funding partners to understand province-specific barriers and facilitators of ES and GS implementation. We will also strategize (as a community of practice invested in the equitable and sustainable delivery of high-quality genomic medicine) to develop concrete implementation structures and processes for Canadians.
Limitations
The primary limitations of this work relate to the inclusion of cases from only 2 clinical sites in Ontario, and to the restricted inclusion criteria. With an emphasis on pediatric patients with neurodevelopmental phenotypes, findings related to the performance of ES and GS may be limited in their generalizability to adult patients with rare disease and to those with non-neurodevelopmental phenotypes.
Conclusion
This protocol lays the foundation for the ongoing development and implementation of clinical ES and GS technologies. Going forward, our evaluation framework will not only serve to monitor performance and inform continuous system improvement, but also guide our evaluation of emerging-omic technologies and applications in the years to come.
Supplementary Material
Acknowledgements
The authors wish to acknowledge the support and guidance of Dr. Cynthia Ho and Ms. Neeta Sarta as representatives of the Laboratories and Diagnostics Branch, Ontario Ministry of Health, Toronto, Ontario.
Footnotes
Competing interests: The authorship group has received funding from the Ontario Ministry of Health and Genome Canada to examine the implementation of genome-wide sequencing in Ontario. Dimitri Stavropoulos has equity in PhenoTips. Wendy Ungar chairs the Ontario Genetics Advisory Committee, is a member of the Ontario Health Technology Assessment Committee, and receives funding from the Pharmaceutical Research and Manufacturers of America Foundation and honoraria from the Taiwan Center for Drug Evaluation. Robin Hayeems is a member of the Ontario Genetics Advisory Committee. Robin Hayeems, Martin Somerville and Kym Boycott are members of the Provincial Genetics Advisory Committee.
This article has been peer reviewed.
Contributors: Robin Hayeems, Christian Marshall, Meredith Gillespie, Anna Szuto, Caitlin Chisholm, Dimitri Stavropoulos, Viji Venkataramanan, Kate Tsiplova, Sarah Sawyer, Magda Price, Lynette Lau, Reem Khan, Whiwon Lee, Lijia Huang, Olga Jarinova, Wendy Ungar, Roberto Mendoza-Londono, Martin J. Somerville and Kym Boycott contributed to study design, data acquisition or data analysis planning. Robin Hayeems drafted the manuscript, and Christian Marshall, Meredith Gillespie, Anna Szuto, Caitlin Chisholm, Dimitri Stavropoulos, Viji Venkataramanan, Kate Tsiplova, Sarah Sawyer, Magda Price, Lynette Lau, Reem Khan, Whiwon Lee, Lijia Huang, Olga Jarinova, Wendy Ungar, Roberto Mendoza-Londono, Martin J. Somerville and Kym Boycott revised it critically for important content. All authors provided final approval of the version to be published and agree to act as guarantors of the work.
Funding: Funding was provided by the Government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-186).
Data sharing: As data collection is underway, data are not currently available to or accessible by others.
Supplemental information: For reviewer comments and the original submission of this manuscript, please see www.cmajopen.ca/content/10/2/E460/suppl/DC1.
References
- 1.Boycott KM, Vanstone M, Bulman DE, et al. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. 2013;14:681–91. doi: 10.1038/nrg3555. [DOI] [PubMed] [Google Scholar]
- 2.Stark Z, Tan TY, Chong B, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016;18:1090–6. doi: 10.1038/gim.2016.1. [DOI] [PubMed] [Google Scholar]
- 3.Meng L, Pammi M, Saronwala A, et al. Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr. 2017;171:e173438. doi: 10.1001/jamapediatrics.2017.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delaney SK, Hultner ML, Jacob HJ, et al. Toward clinical genomics in everyday medicine: perspectives and recommendations. Expert Rev Mol Diagn. 2016;16:521–32. doi: 10.1586/14737159.2016.1146593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lelieveld SH, Spielmann M, Mundlos S, et al. Comparison of exome and genome sequencing technologies for the complete capture of protein-coding regions. Hum Mutat. 2015;36:815–22. doi: 10.1002/humu.22813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller DT, Lee K, Chung WK, et al. ; ACMG Secondary Findings Working Group. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2021;23:1381–90. doi: 10.1038/s41436-021-01172-3. [DOI] [PubMed] [Google Scholar]
- 7.Cohn I, Paton TA, Marshall CR, et al. Genome sequencing as a platform for pharmacogenetic genotyping: a pediatric cohort study. NPJ Genom Med. 2017;2:19. doi: 10.1038/s41525-017-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark MM, Stark Z, Farnaes L, et al. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom Med. 2018;3:16. doi: 10.1038/s41525-018-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C, Vandersluis S, Holubowich C, et al. Cost-effectiveness of genome-wide sequencing for unexplained developmental disabilities and multiple congenital anomalies. Genet Med. 2021;23:451–60. doi: 10.1038/s41436-020-01012-w. [DOI] [PubMed] [Google Scholar]
- 10.Yuen T, Carter MT, Szatmari P, et al. Cost-effectiveness of genome and exome sequencing in children diagnosed with autism spectrum disorder. Appl Health Econ Health Policy. 2018;16:481–93. doi: 10.1007/s40258-018-0390-x. [DOI] [PubMed] [Google Scholar]
- 11.Schwarze K, Buchanan J, Taylor JC, et al. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018;20:1122–30. doi: 10.1038/gim.2017.247. [DOI] [PubMed] [Google Scholar]
- 12.Table 17-10-0009-01: Population estimates, quarterly. Ottawa: Statistics Canada; 2022. [accessed 2022 May 1]. Available: www150.statcan.gc.ca/t1/tbl1/en/tv.action?pid=1710000901. [Google Scholar]
- 13.OHIP Out of Country Prior Approval program. Toronto: Ontario Ministry of Health; [accessed 2021 Oct. 4]. modified 2018 Nov 13. Available: www.health.gov.on.ca/en/public/programs/ohip/outofcountry/prior_approval.aspx. [Google Scholar]
- 14.Use of genome-wide sequencing for undiagnosed rare genetic diseases in Ontario [final report] Toronto: Genetic Testing Advisory Committee, Ontario Ministry of Health; 2016. [accessed 2021 Oct. 4]. Available: www.health.gov.on.ca/en/pro/programs/gtac/docs/gtac_report_use_of_gws_for_undiagnosed_rare_genetic_diseases.pdf. [Google Scholar]
- 15.Boycott K, Hartley T, Adam S, et al. Canadian College of Medical Geneticists. The clinical application of genome-wide sequencing for monogenic diseases in Canada: position statement of the Canadian College of Medical Geneticists. J Med Genet. 2015;52:431–7. doi: 10.1136/jmedgenet-2015-103144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genome-wide sequencing for unexplained developmental disabilities or multiple congenital anomalies: a health technology assessment. Toronto: Ontario Health; 2020. [accessed 2021 Oct. 4]. Available: www.hqontario.ca/Portals/0/Documents/evidence/reports/hta-genome-wide-sequencing.pdf. [PMC free article] [PubMed] [Google Scholar]
- 17.Bell C. Pathway to the clinical implementation of genome-wide sequencing for rare disease. Ottawa: Genome Canada; 2016. [accessed 2021 Oct. 4]. Available: www.genomecanada.ca/sites/default/files/genomics_and_precision_health_forum_en.pdf. [Google Scholar]
- 18.Turnbull C, Scott RH, Thomas E, et al. 100 000 Genomes Project. The 100 000 Genomes Project: bringing whole genome sequencing to the NHS. BMJ. 2018;361:k1687. doi: 10.1136/bmj.k1687. [DOI] [PubMed] [Google Scholar]
- 19.Stark Z, Boughtwood T, Phillips P, et al. Australian genomics: a federated model for integrating genomics into healthcare. Am J Hum Genet. 2019;105:7–14. doi: 10.1016/j.ajhg.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curran GM, Bauer M, Mittman B, et al. Effectiveness-implementation hybrid designs: combining elements of clinical effectiveness and implementation research to enhance public health impact. Med Care. 2012;50:217–26. doi: 10.1097/MLR.0b013e3182408812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogrinc G, Davies L, Goodman D, et al. SQUIRE 2.0 (Standards for QUality Improvement Reporting Excellence): revised publication guildelines from a detailed consensus process. BMJ Qual Saf. 2016;25:986–92. doi: 10.1136/bmjqs-2015-004411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Technical Specifications Working Group for Clinical Genomic Testing in Ontario. Technical specifications for clinical exome and genome sequencing in constitutional disease testing in Ontario submitted to the Laboratories and Diagnostics Branch, Ontario Ministry of Health. Toronto: Laboratories and Diagnostics Branch, Ministry of Health; 2019. [Google Scholar]
- 23.Krahn M, Miller F, Bayoumi A, et al. Development of the Ontario Decision Framework: a values based framework for health technology assessment. Int J Technol Assess Health Care. 2018;34:290–9. doi: 10.1017/S0266462318000235. [DOI] [PubMed] [Google Scholar]
- 24.Tsiplova K, Zur RM, Marshall CR, et al. A microcosting and cost-consequence analysis of clinical genomic testing strategies in autism spectrum disorder. Genet Med. 2017;19:1268–75. doi: 10.1038/gim.2017.47. [DOI] [PubMed] [Google Scholar]
- 25.Jegathisawaran J, Tsiplova K, Hayeems R, et al. Determining accurate costs for genomic sequencing technologies — a necessary prerequisite. J Community Genet. 2020;11:235–8. doi: 10.1007/s12687-019-00442-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.All for one policy toolkit. Ottawa: Genome Canada; 2021. [accessed accessed 2021 Oct. 4]. Available: www.genomecanada.ca/en/all-one-policy-toolkit. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.