Abstract
Bacteriophages (phages) are selective viral predators of bacteria. Abundant and ubiquitous in nature, phages can be used to treat bacterial infections (phage therapy), including refractory infections and those resistant to antibiotics. However, despite an abundance of anecdotal evidence of efficacy, significant hurdles remain before routine implementation of phage therapy into medical practice, including a dearth of robust clinical trial data. Phage–bacterium interactions are complex and diverse, characterized by co‐evolution trajectories that are significantly influenced by the environments in which they occur (mammalian body sites, water, soil, etc.). An understanding of the molecular mechanisms underpinning these dynamics is essential for successful clinical translation. This review aims to cover key aspects of bacterium–phage interactions that affect bacterial killing by describing the most relevant published literature and detailing the current knowledge gaps most likely to influence therapeutic success.
Keywords: antimicrobials, bacteriophages, phage therapy, phage–bacterium dynamics
Subject Categories: Microbiology, Virology & Host Pathogen Interaction
This comprehensive review discusses bacterium–phage interactions, antimicrobial potential of phages and research challenges that impact realization of the successful bacteriophage therapy.

Glossary
- Bacteriophages
Bacteriophages, or phages, are viruses that specifically and selectively infect bacteria
- Biofilm
Surface‐attached, structured community of microorganisms embedded in a self‐produced extracellular matrix (polysaccharides, DNA, water)
- Enzybiotics
Phage‐derived antibacterial enzymes with therapeutic potential. Depolymerases catalyse the hydrolysis of the capsule polysaccharide of Gram‐negative bacteria, while lysins (also endolysins or murein hydrolases) are hydrolytic enzymes capable of cleaving the cell wall (peptidoglycan) of both Gram‐negative and Gram‐positive species
- L‐forms
Cell wall‐deficient bacteria resistant to supra‐therapeutic concentrations of cell wall targeting antibiotics
- Lysogenic conversion
Phage–bacterium interaction in which a prophage encodes proteins that enhance bacterial fitness or virulence
- Lysogeny or lysogenic cycle
Phage life cycle in which the viral genome stably integrates in the bacterial chromosome, replicating with it
- Lytic infection and productive lysis
Infecting phages replicate their genome and assemble new viral particles (virions) by hijacking host resources. Phage‐directed cell lysis then releases this viral progeny ready to infect new cells, in an exponential growth cycle (productive lysis) limited only by availability of bacterial prey and their response/s to phage attack
- Obligate lytic phages
Phages that cannot undergo lysogeny. Preferred for therapeutic applications
- Phage adsorption
Molecular interactions between phage proteins and specific bacterial receptors that bind the phage to the bacterial cell surface allowing for infection (phage genome release into the cytosol) to occur
- Phage cocktail
Combination of multiple phages for therapeutic application. Phages in a cocktail ideally act synergistically against a bacterial target and limit the development of phage‐resistant variants. Cocktails combining phages with different host specificity allow for broader therapeutic targeting
- Phage therapy
Medical use of phages as antimicrobials for treatment of bacterial infections
- Pseudolysogeny
Phage–bacterium interaction in which the phage genome resides within the host cell without chromosomal integration, in an unstable, inactive state
- Temperate phages
Phages capable of undergoing lysogeny. These may lie “dormant” within a living bacterial cell while integrated into the host chromosome as “prophages”, but have the potential to enter a lytic infection cycle under certain conditions (e.g. host cell stress). Temperate phages are less preferred for therapy
- Transduction
Phage‐mediated horizontal transmission of genetic information from one bacterial cell to another, as opposed to genetic inheritance through reproduction (“vertical transmission”). Mainly associated with the lysogenic life cycle
Brief introduction to phage therapy
With the discovery of antibiotics and the development of vaccines, the 20th century saw an unprecedented steady decline in mortality attributable to bacterial infections (Armstrong et al, 1999). This progress built on advances in microbiology and sanitation in the 1880s led by Louis Pasteur and Ignaz Semmelweis (Cavaillon & Chrétien, 2019). In the late 1910s, following initial work by Ernest Hankin and Frederick Twort, Felix D’Herelle identified viruses that specifically and selectively kill bacteria, naming them bacteriophages (phages) [from “bacterium” + “phagêin” (Greek, to eat)], and immediately recognized their potential as antimicrobial agents (Sulakvelidze et al, 2001; Kutter & Sulakvelidze, 2004; Wittebole et al, 2014). In the following decades, however, the development of phage‐based therapy was hampered by a poor understanding of phage biology, some early clinical failures and the meteoric rise of antibiotics (Sulakvelidze et al, 2001; Wittebole et al, 2014; Rohwer & Segall, 2015).
Regrettably, the use (and misuse) of antibiotics has since led to the emergence of globally disseminated bacterial pathogens that are resistant to last‐line treatments, and antibiotic resistance now poses a significant global health and economic burden (Fair & Tor, 2014; O'Neill, 2016; WHO, 2017; Baker et al, 2018). As investment in the discovery and production of new antibiotics dwindles, the development of alternative antimicrobial therapies, including revaluation of phage therapy, is a primary goal (Moelling et al, 2018; Rohde et al, 2018; Petrovic Fabijan et al, 2020a).
In parts of eastern Europe (e.g. Georgia, Poland and Russia), phages have been in routine medical practice for over 70 years and this experience provides a rich source of empirical data (Sulakvelidze et al, 2001; Stone, 2002; Rohwer & Segall, 2015; Górski et al, 2020). Several reviews of recent progress in the development of phage therapy cover preclinical experimentation in animal models, compassionate use in critically ill humans and a few clinical trials (Wittebole et al, 2014; McCallin & Brüssow, 2017; Gordillo Altamirano & Barr, 2019; Nale & Clokie, 2021; Pirnay & Kutter, 2021). Most of the cited studies attest to the safety of phage therapy, but clinical effectiveness has not yet been conclusively demonstrated (McCallin & Brüssow, 2017; Gordillo Altamirano & Barr, 2019; Pirnay & Kutter, 2021). In addition, the results of experimentation in small animal models do not consistently translate into clinical success (Wittebole et al, 2014; Nale & Clokie, 2021), just as in vitro phage activity often fails to correlate with in vivo efficacy (Melo et al, 2020a). These inconsistencies complicate the design of clinical protocols, undermine confidence in phage application and hinder progress towards clinical implementation.
While the number of completely sequenced phage genomes has doubled in the last 5 years (Fig 1) (Cook et al, 2021), these represent a minuscule fraction of the prokaryotic virosphere, estimated to exceed 1031 particles (Hatfull, 2015). Phages are found in all bacterial habitats (Kutter & Sulakvelidze, 2004; Clokie et al, 2011) and are a key driving force of microbial ecology and evolution (Dion et al, 2020). Tailed double‐stranded DNA phages (order Caudovirales) constitute the largest group described to date (Clokie et al, 2011) and are easily isolated with simple techniques from diverse environmental sources (Ackermann, 1998). Tailed phages have high target specificity, which can be redirected by forced evolution or genetic engineering (Pires et al, 2016a; Burrowes et al, 2019), and are the only phage type to have been trialled in therapy so far (Ackermann, 1998; Kutter & Sulakvelidze, 2004).
Figure 1. Phage whole genome sequencing.
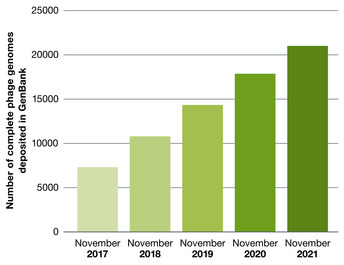
Number of complete phage genomes deposited in GenBank in the past 5 years (with permission from Cook et al, 2021).
Phages eliminate target bacteria by bursting bacterial cells (lysis) within minutes of infection (Kutter & Sulakvelidze, 2004), thereby releasing newly formed phage particles (virions) that go on to infect new host cells in a self‐perpetuating cycle (Kutter & Sulakvelidze, 2004; Kortright et al, 2019). Crucially, phage activity is unaffected by antibiotic resistance.
The highly specific virus–host pairing is central to microbial population dynamics and is deeply connected to environmental conditions and ecological niches. In therapeutic applications, the risk of undesirable adaptive outcomes of the phage–bacterium interplay (e.g. resistance development in bacteria) is pragmatically addressed by the use of combinations of multiple phages (cocktails) with differing adaptive strategies (Chan & Abedon, 2012; Chan et al, 2013; Pirnay et al, 2018; Rohde et al, 2018). Combining phages into therapeutic cocktails (as opposed to monophage therapy), broadening their utility and commercialization potential, requires a clear understanding of phage–phage and phage–bacterium dynamics (Chan & Abedon, 2012; Schmerer et al, 2014; Gordillo Altamirano & Barr, 2019; Venturini et al, 2019a; Pirnay, 2020; Haines et al, 2021).
The key mechanisms that may allow prediction of in vivo pharmacokinetics and dynamics linked to therapeutic outcome have not yet been fully elucidated. Here, we provide an overview of the biological processes linked to phages’ antimicrobial potential and highlight some of the research challenges that remain.
Phage infection
Infection cycles
Phages depend on their bacterial hosts for survival and multiplication, but bacterial growth rates can fluctuate significantly even in nutritious habitats. Doubling times for wild‐type Escherichia coli laboratory strains in optimal conditions are approximately 20 min (Gibson et al, 2018), while those measured in the mammalian gut can range from 40 min to 140 h (Abedon, 1989; Poulsen et al, 1995). Although one infective phage particle may yield as many as 20,000 new virions per infected bacterial cell in optimal conditions (Zinder, 1980), bacteria rarely encounter such habitats in nature and phages that would ordinarily propagate exponentially may fail to do so when bacterial growth is limited (e.g. by nutritional stress) (Lourenço et al, 2020; Attrill et al, 2021).
In exponentially growing bacteria, phages replicate typically via either a lytic or a lysogenic cycle (Fig 2A). Phage therapy traditionally uses “virulent” or “obligate lytic” phages (lytic cycle only) that lyse bacteria immediately upon infection in preference to “temperate” phages, which undergo a lysogenic cycle, integrating their genome into the bacterial host chromosome and replicating passively with it as “prophages” (Fig 2A–C) (Lamont et al, 1989; Howard‐Varona et al, 2017; Li et al, 2020). Therapeutic use of temperate phages risks transfer of genes (“transduction”) that may enhance bacterial fitness or virulence (e.g. toxins) or confer antibiotic resistance to the bacterial host (Brussow et al, 2004). This is known as “lysogenic conversion”, a process by which important pathogens have acquired cardinal virulence factors (e.g. Corynebacterium diphtheriae carrying the siphovirus β‐phage that encodes the diphtheria toxin Tox (Holmes, 2000) or enterohaemorrhagic E. coli with the lambdoid phage encoding Shiga toxins (Schmidt, 2001)). Stable chromosomal integration is mainly a function of the phage itself (Brussow et al, 2004; Fortier & Sekulovic, 2013; Argov et al, 2019; Petrovic Fabijan et al, 2021) but also depends on host conditions; when these change (e.g. nutritional stress or DNA damage), prophages may excise from the chromosome and enter a lytic cycle that leads to bacterial cell death (Banks et al, 2003; Nanda et al, 2014; Balasubramanian et al, 2019; Chatterjee & Duerkop, 2019; Benler & Koonin, 2020; Filipiak et al, 2020). Importantly, quorum‐sensing mechanisms and communication via signalling molecules are also increasingly implicated in phage–bacterium interactions, including switching between lytic and lysogenic lifestyles (León & Bastías, 2015; Silpe & Bassler, 2019).
Figure 2. Phage replicative cycles.
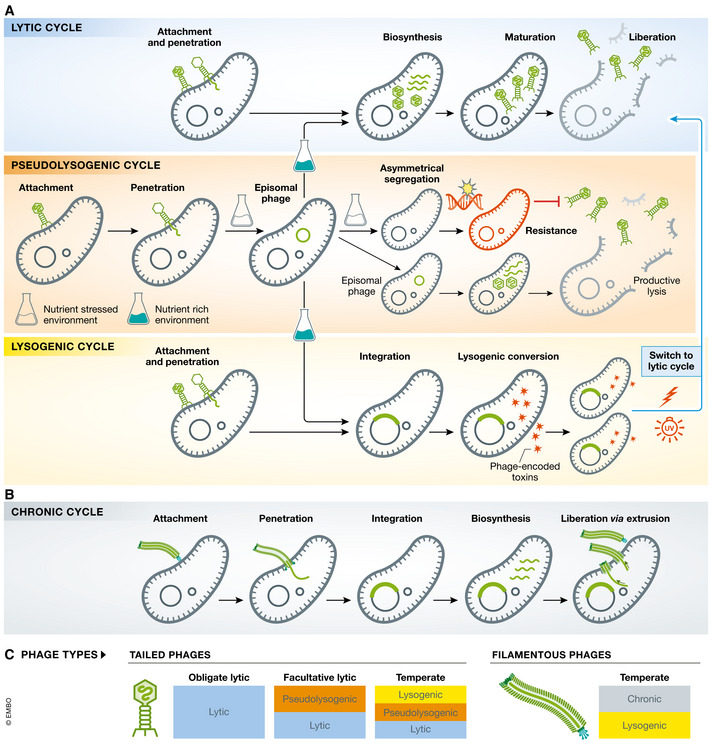
(A) Modes of phage infection characteristic of tailed phages: (i) lytic cycle—phage replication immediately follows infection, with assembly and release of virions leading to cell lysis. Each virion is free to start a new lytic cycle leading to a burst of “productive” infection; (ii) lysogenic cycle—phages can integrate into the bacterial chromosome and replicate with it as prophages, until a lytic cycle is triggered; and (iii) pseudolysogeny—phage genomes persist in a episomal state within the host cell before resolving into a lytic or lysogenic cycle. Episomal phages typically segregate asymmetrically during cell division, while a small fraction undergoes a productive lytic cycle (scavenger response) favouring development of phage‐resistant bacterial subpopulations. (B) Chronic infection cycles are characteristic of “filamentous phages” (family Inoviridae) that continuously produce progeny released by extrusion without cell death/lysis. (C) Phage types by replication cycle: tailed phages that always lyse bacteria upon infection are “virulent” or “obligate lytic”, while “facultative lytic” phages may also undergo pseudolysogeny. “Temperate” phages may have a lysogenic or pseudolysogenic lifestyle until triggered to enter a lytic cycle, typically when the host bacteria experience stress conditions. Filamentous phages typically follow a chronic productive cycle, though some have the capacity to also enter a lysogenic cycle.
Chronic infection is a distinct replication cycle characteristic of “filamentous” phages belonging to the family Inoviridae (single‐stranded DNA phages; order Tubulavirales) (Fig 2B) (Secor et al, 2020; Mantynen et al, 2021). Unlike lytic and lysogenic cycles, chronic infection leads to continuous virion production without lysis of the bacterial cell (Loh et al, 2019). Filamentous phages are well suited for the horizontal exchange of DNA and many encode important virulence factors. The best understood filamentous phages involved in lysogenic conversion of their hosts are those that infect Vibrio cholerae (e.g. CTXø, encoding the cholera toxin) (Waldor & Mekalanos, 1996; Karaolis et al, 1999) and Pf phages that infect Pseudomonas aeruginosa promoting biofilm production in infected bacteria (Secor et al, 2015). Filamentous phages are considered unsuitable for therapy.
While lytic and lysogenic lifestyles and their impact on therapeutic outcome have been extensively reviewed (Sulakvelidze et al, 2001), the impact of pseudolysogeny has not yet been defined. This additional phage infection mode (Fig 2A), which some propose should be defined altogether as a separate cycle (Mantynen et al, 2021), was first recognized in the early 1960s (Los & Wegrzyn, 2012), but as yet there is no unanimous definition for this phenomenon and its molecular bases remain largely unexplored. Pseudolysogeny has been defined as a “phage carrier” state (Ripp & Miller, 1997) or, perhaps more accurately, as “stalled phage development” (Los & Wegrzyn, 2012). In pseudolysogeny, neither multiplication nor synchronized replication of the phage occurs within the host cell, but when conditions allow, the phage enters either a “normal” lysogenic cycle or a lytic cycle.
Pseudolysogeny has been observed primarily in Gram‐negative species, generally when bacterial growth was limited (Los et al, 2003; Cenens et al, 2013; Latino et al, 2016), suggesting a role in long‐term phage survival in unfavourable conditions, perhaps by providing many of the advantages of the lysogenic state while avoiding chromosomal integration. Lytic phages are recognized by their efficient killing activity in vitro (high lytic efficacy) and the absence of classic lysogeny genes (integrases, repressor genes, etc.), but there is no established genetic marker of pseudolysogenic capacity, as it is not usually a feature of exponentially growing bacteria. Replication of obligate lytic T4‐like phages is completely inhibited in nutrient‐stressed E. coli, but it has been reported that under the same conditions, a T4rI mutant (defective in the function of the holin inhibitor) keeps producing viable virions (Los et al, 2003). Bryan et al (2016) showed that T4 phages efficiently bind to and infect, but fail to successfully lyse, E. coli in the stationary phase. Under nutrient‐limiting conditions in vitro, the lytic cycle still occurs in a small subpopulation of infected bacterial cells (“scavenger response”), fully resuming in the rest of the population only upon nutrient addition with restoration of logarithmic growth. P. aeruginosa and Yersinia enterocolitica can support pseudolysogenic infection with apparently obligate lytic myoviruses or podoviruses that provide bacteria with immunity from further phage infection (superinfection exclusion) (Latino et al, 2016; León‐Velarde et al, 2016). Thus, it seems that not all virulent phages are truly obligate lytic viruses or, at least, that a replication pause may occur in the lytic cycle. This provides advantages for both the virus and the parasitized host cell, especially when the host bacterial population is stressed, by preventing extinction of vulnerable bacterial population on which the predating virus is dependent.
Temperate phages may also enter a pseudolysogenic state in bacteria that are stressed or starved: the podovirus P22 can stably persist in episomal form in Salmonella cells, asymmetrically segregating upon cell division (Cenens et al, 2013). This is linked to the specific phage‐mediated and targeted depression of the host dgo operon via the pid phage gene (Cenens et al, 2013) and suggests some advantages of pseudolysogeny even in phages that have developed the capacity to integrate, perhaps as a more agile response to bacterial population stress. Other temperate phages, variably defined as “phage‐like plasmids” (Pfeifer et al, 2021) or “phagemids” (Kittleson et al, 2012), are found in the host as extra‐chromosomal elements that encode partitioning systems (Salje, 2010) and replicate within the cell cycle. In the well‐studied P1 E. coli myovirus (Lobocka et al, 2004) and its many variants (Walker & Anderson, 1970; Rosner, 1972; Venturini et al, 2019b), ATP‐dependent post‐segregational killing promotes symmetrical distribution of phage episomes via common plasmid partitioning and maintenance mechanisms (Lobocka et al, 2004).
Although much remains to be investigated, it seems plausible for pseudolysogeny to represent a route to both short‐ and long‐term phage survival through (i) physical protection from harsh environmental conditions outside the host (e.g. UV‐light, pH and temperature can drastically reduce the half‐life of virions) (Jonczyk et al, 2011), and (ii) hibernation (replication pause) in unfavourable conditions that threaten the host population (e.g. stationary phase or persister populations) (Bryan et al, 2016).
A better understanding of the diversity and genetic regulation of phage life cycles is paramount for successful therapeutic applications. Future progress will likely benefit from “multiomics” approaches and investigation of these complex phenomena at a single‐cell level (Dang & Sullivan, 2014; Skurnik, 2022). Genetic engineering approaches may also prove useful for redirecting phage lifestyles to suit therapeutic goals (e.g. enhance lysis by elimination of lysogeny genes in temperate phages (Dedrick et al, 2019)).
Multiplicity of infection and the concept of phage dosing
Self‐amplification through progressive productive infection is a unique distinction between phages and traditional (drug) antibiotics with important clinical implications (Levin & Bull, 2004). Phage growth parameters such as adsorption rate, latent period (duration of infection cycle from replication to virion assembly) and burst size (number of released virions per lysed cell) are commonly used to quantify productive lytic infection in vitro (Levin & Bull, 2004; Dennehy & Abedon, 2021). These parameters are specific to each phage and can vary considerably, and as such have been the focus of theoretical studies attempting to model lysis outcomes of bacterium–phage pairs to inform therapeutic strategies (Bull et al, 2004; Levin & Bull, 2004; Wang, 2006; Heineman & Bull, 2007).
Modelling of in vivo dynamics, even for the simplest phage–bacterium interaction, must consider the availability of resources to bacterial prey populations (Weitz et al, 2013), other mobile genetic elements (Harrison et al, 2017), community effects (bystander microflora) (Blazanin & Turner, 2021) and the spatial structures at the site where predator and prey meet (Lourenço et al, 2020; Attrill et al, 2021). Bacterial density directly affects adsorption rate and phage replication duration, as well as opportunities for further viral propagation. If target bacteria are slow‐growing and sparsely separated, the productive exponential infection may not proceed (Payne & Jansen, 2001; Kasman et al, 2002; Levin & Bull, 2004; Heineman & Bull, 2007; Abedon, 2011).
Multiplicity of infection (MOI) is the term used to indicate the ratio of phages to bacteria in in vitro testing and is often applied in vivo as a “dosing” concept. A MOI of > 10 may be more advantageous in murine sepsis models (Yuan et al, 2019; Hesse et al, 2020), and this has been used as a target for human dosing (Khatami et al, 2021), but this extrapolation is problematic because not all phages administered reach their target and not all phages that adsorb to a bacterial cell will infect it (Attachment mechanisms and receptor specificity) (Abedon, 2016). Direct measurement of phage and bacterial densities in vivo is not practical except for urine (Abedon, 1989; Khawaldeh et al, 2011; Dąbrowska & Abedon, 2019) or blood (Petrovic Fabijan et al, 2020b) so that even once the target MOI is defined and the amplification process can be monitored, these samples of convenience can only serve as surrogates for the site of infection in tissues. Therapy with antibiotic drugs leads to relatively predictable relationships between tissue and blood concentrations, which can be determined and used to optimize dosing. Evidence of phage amplification derived from samples of convenience might become a useful surrogate for successful delivery to site. However, in vivo amplification appears to subside quickly, likely due to both therapeutic “success” (i.e. elimination of prey populations) and host control of the administered therapeutic virus by innate and acquired immune responses (The eukaryotic host: phage‐induced immune responses). The pharmacodynamics and pharmacokinetics of phage therapy are also subject to variable and possibly virus‐specific tissue penetration (Górski et al, 2015; Dąbrowska & Abedon, 2019). Careful monitoring of clinical sites and samples in the course of carefully structured therapeutic regimens will therefore be extremely important to lasting and robust therapeutic application (Abedon, 2011).
Attachment mechanisms and receptor specificity
Phage adsorption to the bacterial cell is a first and crucial step in the infection process (Bertozzi Silva et al, 2016; Letarov & Kulikov, 2017). For “best” phage therapy (optimal lytic efficiency = optimal bactericidal activity), the majority of virus–bacterium contacts should lead to productive infection (Multiplicity of infection and the concept of phage dosing), making the molecular interactions at the bacterial cell surface a key aspect of therapeutic success (Nobrega et al, 2018). Membrane‐embedded proteins are common phage receptors, but phage access to these receptors is highly regulated by various protective glycan structures such as peptidoglycan, capsule or lipopolysaccharide (LPS) found on bacterial envelopes.
Phage tail machines as sophisticated infection devices
Although phage–bacterium interactions via capsid proteins have been described (Casjens & Molineux, 2012), adsorption to the bacterial cell envelope is most commonly mediated by sophisticated phage tail machines that specifically recognize diverse bacterial cell surface structures and are implicated in other important infection‐aiding processes (Chua et al, 1999; Letarov & Kulikov, 2017; Nobrega et al, 2018) (Fig 3). Three tail morphologies are known: short non‐contractile tails in the Podoviridae; long non‐contractile tails in the Siphoviridae; and long contractile tails in the Myoviridae (Ackermann & Prangishvili, 2012) (Fig 3A). Tailed phages have evolved to deliver much larger genomes to their hosts than non‐tailed phages (Davidson et al, 2012) and are highly specialized in overcoming the protective layers of Gram‐negative and Gram‐positive bacterial envelope architectures (Fig 3B and C). For host recognition, tailed phages use fibres, longitudinal, multimeric protein assemblies, or shorter and more compact protein oligomers termed spikes.
Figure 3. Bacterial envelopes and receptors for tailed phages.
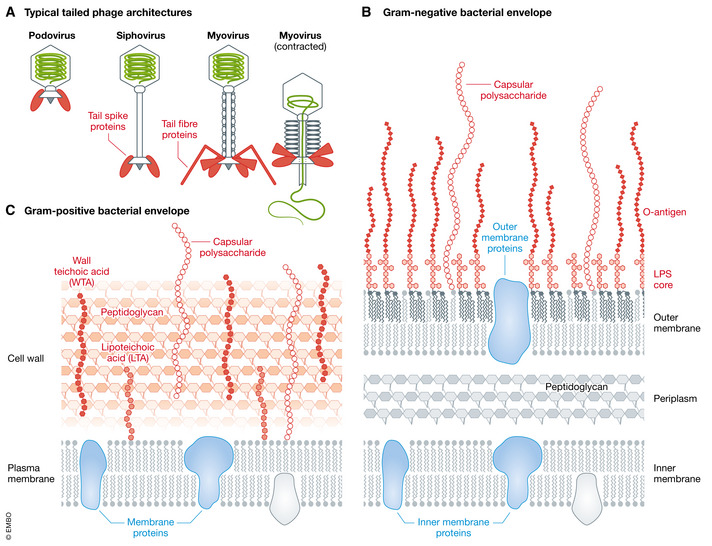
(A) Schematic overview of the three typical tailed phage architectures. Fibre and spike components in the baseplate that interact with bacterial envelope glycans are shown in red. (B) Gram‐negative bacterial envelope. (C) Gram‐positive bacterial envelope with glycan (red) and protein (blue) phage receptors.
Surface attachment and infection
Phage recognition of bacterial cell surface receptors is a well‐orchestrated process comprising several individual sequential steps (Broeker & Barbirz, 2017; Broeker et al, 2017). The diversity of bacterial cell envelopes (Fig 3) has required tailed phages to develop different strategies to initiate infection. This initial and often reversible interaction with the primary receptors precedes subsequent “secondary receptor” binding, which leads to changes in the tail machine that are irreversible (Casjens & Molineux, 2012). Phages preferentially encounter as primary receptors all the exposed surface structures on host bacteria, i.e. capsule, exopolysaccharide, peptidoglycan or teichoic acids (Dunne et al, 2018), and in Gram‐negative target also LPS (Broeker & Barbirz, 2017). Flagella and adhesins may also serve as primary receptors for some phages (Esteves et al, 2021; Montemayor et al, 2021). Many primary receptors are distal to the cell surface, and phages employ diverse active mechanisms to approach the bacterial membrane. “Flagellotropic” phages, for example, ride on flagella towards the host surface, harnessing bacterial motility for infection progression (Tittes et al, 2021), and many tailed phages produce tailborne depolymerases to specifically destroy the polysaccharide‐based glycan protective shields (Knecht et al, 2020). Many of the initial fibre‐ or spike‐mediated receptor interactions are reversible, which allows phage particles to dissociate from the cell surface until they reach a site for irreversible attachment.
Irreversible attachment to these secondary receptors can initiate a cascade of steps that lead to permanent conformational rearrangements in the phage tail assembly (Taylor et al, 2018), priming the phage for DNA release. Conserved transmembrane proteins (e.g. transporters and channels), efflux pumps and pilus portals often serve as secondary receptors (Bertozzi Silva et al, 2016), with their extracellularly exposed parts providing highly specific phage attachment sites with numerous opportunities for bacterial adaptation to halt the phage infection cycle, for example by mutation of outer membrane extracellular loops (Porcek & Parent, 2015; Rocker et al, 2020). As shown for purified outer membrane proteins (Chiaruttini et al, 2010; Evilevitch, 2018), binding to secondary receptors can trigger the phage molecular machine for DNA release in vitro, thus rendering phage particles non‐infectious. Gram‐negative host‐specific phages incubated with protein‐free LPS preparations typically lose their infectivity as contact with these receptor molecules induces particle opening and DNA loss (Jesaitis & Goebel, 1955; Lindberg, 1973; Andres et al, 2012; Broeker et al, 2019). How entirely protein‐free LPS preparations trigger DNA release in the absence of a host cell remains to be elucidated (Andres et al, 2010; Broeker & Barbirz, 2017). Cryotomography studies of phages attached to bacteria have revealed the formation of channel‐like structures spanning the envelope that ensure the integrity of the cell surface during phage genome transfer into the cytosol. However, the molecular composition of these channels is not yet fully understood (Hu et al, 2015; Farley et al, 2017; Wang et al, 2019); in some cases, phages seem to extend their tails to reach the interior of the cell, while in others, phage‐synthesized ejection proteins recruit other protein components from the bacterial envelope to facilitate DNA movement.
Adsorption regulation—the unique role of bacterial surface glycans
In bacterial ecosystems, regulation of interactions with predatory viruses takes place both at extracellular and at intracellular levels (Hampton et al, 2020). As phage receptors, surface glycans (Fig 3) modulate phage entry and are important in evolutionary adaptations to phage infection (Phage–bacterium co‐adaptation). Bacterial cell surface glycans also face the human immune system and are often described as important participants in so‐called pathogen‐associated molecular patterns (PAMPs). Changes induced by phages thus affect the innate immune response to pathogens (The eukaryotic host: phage‐induced immune responses), and phage‐encoded enzymes that remove protective glycan layers (e.g. depolymerases), exposing underlying PAMPs at the envelope (Majkowska‐Skrobek et al, 2018; Liu et al, 2020; Volozhantsev et al, 2020), may directly enhance clearance of bacteria by the innate immune system (Oliveira et al, 2019a).
In the presence of actively infecting phages, bacteria may alter surface glycan structures through transcriptional control of glycosyltransferases. This “phase variation” is achieved by altered glycan composition or LPS chain length or even by complete abrogation of the assembly of protective capsule or O‐antigens (Huan et al, 1997; Seed et al, 2012; Cai et al, 2019; de Sousa et al, 2020; Whitfield et al, 2020). Similarly, prophages may alter bacterial surface glycan composition via glycosylation or acetylation to exclude other phages from infection (Cenens et al, 2015; Schmidt et al, 2016; Teh et al, 2020).
Phages bind bacterial surface glycans using specific tail proteins (Broeker et al, 2017; Nobrega et al, 2018; Knecht et al, 2020). Many host adsorption proteins are depolymerases that facilitate surface access through O‐antigen or capsular layers, this being an essential step for infection by some phages (Broeker & Barbirz, 2017). The glycan adsorptive capacity of these tail proteins also enables phages to persist in glycan‐rich niches, for example by binding heparan sulphates of mucins in the mammalian gut (Green et al, 2021). Phage glycan depolymerases can strip off glycan coats without initiating cell rupture, thereby avoiding critical concentrations of microbial cell envelope fragments that may drive a damaging immune response in clinical sepsis (Ryu et al, 2017). LPS‐mediated sepsis and septic shock are primary drivers of mortality in Gram‐negative infection (Opal et al, 1999), and several studies have shown that pretreatment with phage depolymerases to degrade O‐antigen polysaccharides reduces pro‐inflammatory responses and protects mice from lethal sepsis (Liu et al, 2019; Oliveira et al, 2019b; Chen et al, 2020).
Outer membrane vesicles (OMVs) also play a unique role in controlling phages as they can effectively trap them, preventing host infection (Schwechheimer & Kuehn, 2015; Reyes‐Robles et al, 2018), as shown for Salmonella phage P22 where only few phages eject their DNA into the OMV lumen, with the majority of particles stalling at the membrane (Stephan et al, 2020).
The specificity of these attachment mechanisms limits phage clinical range, when compared to traditional antibiotics with broad‐spectrum activity against multiple bacterial species. This potential therapeutic limitation is mainly being obviated by the use of phage cocktails, but it can also be addressed via natural phage “training” to broaden host range by successive passage (Yu et al, 2015; Burrowes et al, 2019) or by formal synthetic biology approaches (Chen et al, 2019; Dedrick et al, 2019). The use of enzybiotics (depolymerases or endolysins) is also being considered (Pires et al, 2016b; Olsen et al, 2018). Phage endolysins attack the peptidoglycan layer of Gram‐positive and Gram‐negative envelopes (Fig 3), thus acting less specifically than depolymerases (Broendum et al, 2018; Sao‐Jose, 2018; De Maesschalck et al, 2020; Mondal et al, 2020; Chen et al, 2021; Linden et al, 2021; Murray et al, 2021). Importantly, the development of bacterial resistance to externally applied endolysins is unlikely as these enzymes target cellular structures essential for bacterial survival (Roach & Donovan, 2015). However, all the outlined approaches crucially require the maintenance and accessibility of well‐curated and diverse phage banks, which are still scarce (Nagel et al, 2022).
Phage–bacterium co‐adaptation
The interaction between phages and bacteria is a major contributor to the diversity and evolution of microbial populations, involving fine‐tuned, complex co‐adaptation dynamics, with bacteria trying to minimize susceptibility to phage infection as phages strive to retain or regain it (Díaz‐Muñoz & Koskella, 2014; Koskella & Brockhurst, 2014; Seed et al, 2014). Bacterial adaptations are not without cost, and both mathematical models and experimental observations suggest that bacterial resistance to phage can be overcome (Levin & Bull, 2004), but the development of bacterial phage resistance in vivo has not been yet systematically researched (Hesse et al, 2020; Gordillo Altamirano et al, 2021; Salazar et al, 2021).
Alteration of cell surface phage receptors (“adsorption resistance”, through modification or masking or by synthesis of competitive inhibitors; Attachment mechanisms and receptor specificity) is arguably the most common adaptive response to phage predation; CRISPR/Cas may be a close second (Doron et al, 2018; Ofir & Sorek, 2018; Alseth et al, 2019; Rostøl & Marraffini, 2019; Hampton et al, 2020). Bacterial susceptibility to phages may be modulated by horizontal exchange of receptors mediated by OMVs (Tzipilevich et al, 2017) or more often by genetic modification of cell surface structures targeted by phages, which may affect both pathogenic potential and overall survival of target bacteria (Verma et al, 2009; Capparelli et al, 2010; Chan et al, 2016; Markwitz et al, 2021). The resulting fitness cost can increase bacterial vulnerability to both the immune system and antibiotics (León & Bastías, 2015).
Attempts to use phages to clear Klebsiella pneumoniae and Acinetobacter baumannii infection in vivo have resulted in phage‐resistant capsular mutants that appear to be less virulent and more susceptible to antibiotics (Verma et al, 2009; Gordillo Altamirano et al, 2021), and therefore easier to eliminate. E. coli responds to phage challenge by modification of LPS biosynthesis with concomitant fitness loss and attenuation in a murine model of systemic infection (Salazar et al, 2021). In K. pneumoniae, mutations in the porin OmpK36 lead to increased antibiotic resistance and are poorly tolerated in vivo (reduced growth rates) (Fajardo‐Lubian et al, 2019), while in Shigella flexneri Omp‐targeting phages have been shown to lead to resistant mutants incapable of intracellular spread (Kortright et al, 2022). As such, Omp‐specific phages, for example, might have particular value in managing these pathogens. Phage‐insensitive variants appear to be rarely isolated after phage administration in the clinic, suggesting that the many varied outcomes predicted and observed in vitro may be transient in vivo, with few phage‐resistant subtypes (“fittest” mutants) actually able to succeed in nature (Bohannan & Lenski, 2000; León & Bastías, 2015; Hernandez & Koskella, 2019; Aslam et al, 2020; Petrovic Fabijan et al, 2020b).
Conversely, phages may counterevolve to regain their infectivity by modification of their own host attachment receptors (tails), resulting in host range expansion (Salazar et al, 2021). In therapy, the use of cocktails of multiple phages acting in synergy (to optimize lysis of target bacteria) has been shown to both broaden target range and minimize the occurrence of phage resistance (Abedon et al, 2021a). While the development of cross‐resistance is also a possibility (Wright et al, 2018), mixtures of phages with different receptor specificities are expected to exert multiple simultaneous selective pressures on the target host (Schmerer et al, 2014) that come at increased costs to bacterial fitness. Carefully “tailored” phage combinations using original and “evolved” phages against the one host have been shown to successfully target both the wild‐type strain and its variants (Yu et al, 2018; Aslam et al, 2020; Abedon et al, 2021a; Salazar et al, 2021).
Phage attack can affect antibiotic susceptibility in target bacteria (Ryan et al, 2012; Segall et al, 2019; Gordillo Altamirano et al, 2021), and the careful use of antibiotic–phage combinations may also be useful in limiting the development of bacterial variants resistant to both (Gu Liu et al, 2020 et al, 2020; Gordillo Altamirano et al, 2021). As outlined in several recent exhaustive reviews (Segall et al, 2019; Tagliaferri et al, 2019; Morrisette et al, 2020; Li et al, 2021), phage–antibiotic synergy (PAS) has been successfully demonstrated in both Gram‐positive and Gram‐negative bacteria, though many studies have focused on E. coli and P. aeruginosa (Comeau et al, 2007; Allen et al, 2017; Chaudhry et al, 2017; Gu Liu et al, 2020), and may have important clinical implications. However, synergy is not the only outcome of simultaneous exposure to phages and antibiotics with addition, neutrality and antagonism also possible.
The effects of phage–antibiotic combinations on target bacteria depend on many factors including the specific antibiotic tested (results obtained with one antibiotic are not always replicated with another antibiotic of the same class), the testing conditions (e.g. type of media, bacterial growth (planktonic cells versus biofilm), in vitro versus in vivo conditions), phage type (even very closely related phages can give different outcomes), and timing of administration (e.g. simultaneous or sequential) (Segall et al, 2019; Tagliaferri et al, 2019; Morrisette et al, 2020; Li et al, 2021). Only recently, Gu Liu et al (2020) presented the first in‐depth analysis of the mechanisms underlying the efficacy of phage–antibiotic combinations against a highly virulent E. coli ST131 strain. Their work clearly demonstrates the complexity of these interactions and the urgent need for applying this type of comprehensive approach to other bacterial species and antibiotic–phage combinations for a clear understanding of possible outcomes to guide clinical application.
Phage–bacterium co‐adaptation is predicted to drive a stalemate that favours bacterial survival in nature (Bohannan & Lenski, 2000; Koskella & Brockhurst, 2014; Fernández et al, 2018; Makalatia et al, 2021), and successful therapy requires us to contrive situations in which natural balances are tipped in favour of the phage (Levin & Bull, 2004), the specifics of which will depend on the interacting phage–bacterium pair and their immediate environment. Phage‐resistant variants arising in vivo can be problematic (Schooley et al, 2017), but phage‐resistant bacteria are sometimes less virulent (Olszak et al, 2019) or less antibiotic‐resistant (Oechslin, 2018) than their parent (Ryan et al, 2012; Chaudhry et al, 2017). A detailed understanding of receptor specificities (Bertozzi Silva et al, 2016) and co‐adaptation trajectories both in vitro and in vivo (Doron et al, 2018; Makalatia et al, 2021) must be developed in order to inform new mathematical models and “artificial intelligence” (AI) solutions (Schmerer et al, 2014; Cowley et al, 2018; Hesse et al, 2020; Pirnay, 2020; Haines et al, 2021; Maffei et al, 2021) to help deconvolute these natural biological and evolutionary complexities.
Bacterial targets
Reduced growth states: stationary phase bacteria and L‐forms
Bacterial pathogens have evolved to defend themselves effectively against commonly encountered stressors in the mammalian host (e.g. oxidative, nutritional and antibiotic). Given the ubiquity of phages in nature and the aeons of co‐evolution with bacteria, an array of finely tuned and well‐established defences against phage attack are also to be expected. The physiological state of the bacterial host population is an important determinant of phage replication (Infection cycles), and the exponential growth conditions used for antibiotic and phage susceptibility testing in diagnostic laboratories are probably rare in nature, with “stationary phase” growth being common in chronic and relapsing infections (Gefen et al, 2014) (Fig 4).
Figure 4. Escherichia coli growth states.
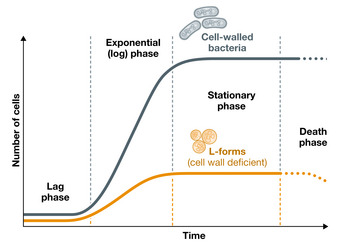
Growth in optimal conditions (37°C; rich medium) of wild‐type E. coli (blue curve) and its L‐forms (cell wall‐deficient variants; orange curve). At reaching stationary phase, bacterial metabolic activity and growth are slowed to near nil.
The impact of bacterial stress on the lytic/pseudolysogenic pathways may be therapeutically important. Phages that ordinarily pseudolysogenize stressed bacteria (Bryan et al, 2016) may be poor choices for the management of some infections. Cell wall‐deficient “L‐forms” are more metabolically active and faster growing than stationary phase‐walled cells (Mercier et al, 2014; Mickiewicz et al, 2019) but divide more slowly than exponential phase bacteria (Fig 4), using a primitive mechanism that is independent of essential elements of binary fission (e.g. FtsZ) (Leaver et al, 2009). L‐forms can be induced by innate immune effectors, such as lysozyme, and by exposure to cell wall targeting antibiotics (e.g. β‐lactams), to which they are completely resistant. This is important because cell wall targeting antibiotics are the mainstay of modern infection therapy (Care, 2021) and because biofilms (Special states: biofilms) and multi‐drug‐resistant infections, against which such antibiotics often fail, are key indications for phage therapy. Therefore, targeting L‐forms with phages may be an important therapeutic option. However, L‐form susceptibility to phages has not yet been well characterized except for a few reports, suggesting that the capacity for efficient lysis of L‐forms is retained at least by some phages (Kawacka et al, 2020).
Special states: intracellular pathogens
Certain bacterial pathogens responsible for high rates of infection and mortality (GBD Tuberculosis, 2018; Khalil et al, 2018; GBD Non‐Typhoidal Salmonella, 2019; GBD Antimicrobial Resistance, 2022) routinely replicate inside human cells including professional phagocytes such as monocyte‐derived macrophages (Ogawa & Sasakawa, 2006) (Fig 5A). These bacteria are protected from the immune system and from bactericidal agents in their intracellular niches. In addition, intracellular bacteria can take advantage of the biology of the host cell to disseminate to tissues beyond the site of infection. Most antibiotics commonly used in medicine do not penetrate mammalian cells efficiently and are therefore ineffective against intracellular pathogens (Abed & Couvreur, 2014; Kamaruzzaman et al, 2017). The few exceptions (e.g. quinolones, macrolides and tetracyclines) (Carryn et al, 2003; Kamaruzzaman et al, 2017) are widely used orally, and resistance to these is rising in target pathogens (WHO, 2017). Phages could therefore be of value for the treatment of intracellular infections.
Figure 5. Intracellular lifestyles of bacterial pathogens and barriers in the treatment of intracellular infections.
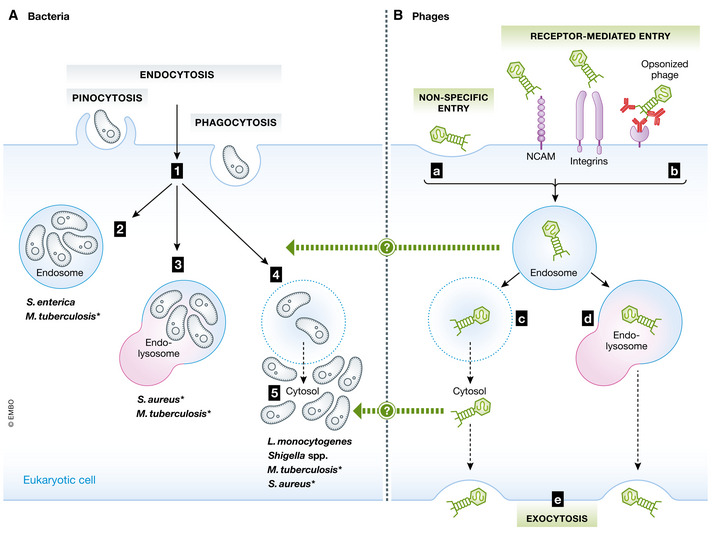
(A) Intracellular bacteria penetrate mammalian cells by endocytosis, phagocytosis or pinocytosis (1), and reside inside different subcellular compartments: the endosome (2), the endolysosome (3; formed by fusion of the endosome with a lysosome) or the cytosol (5), after escaping endosome inclusion (4, light‐blue dotted ring) (Cossart & Sansonetti, 2004; Ray et al, 2009). (B) Free phages must cross the eukaryotic cell membrane by non‐specific (a) or receptor‐mediated (b) entry. Endocytosed phages may then escape the vacuole (c, light‐blue dotted ring) or remain in the endosome (blue ring) with eventual fusion to a lysosome (d). After cytosolic release (c) or lysosomal fusion (d), viable phages may be released from the mammalian cell via exocytosis (e). * indicates bacteria that can replicate in more than one host cell compartment. The exact details of how phages reach their intracellular targets are still largely unknown (?).
The first evidence of phages crossing the eukaryotic cell barrier dates back more than 50 years (Monsur et al, 1970), and it is known that these viruses can penetrate human cells and even enter their nucleus (Nieth et al, 2015; Lehti et al, 2017; Nguyen et al, 2017; Zhang et al, 2017; Sweere et al, 2019). Phages may enter the eukaryotic cell non‐specifically by phagocytosis or pinocytosis, or through receptor‐mediated entry by binding specifically to cell surface structures like the neural cell adhesion molecule (NCAM; a major polysialic acid carrier that mimics bacterial receptors) or to cell surface integrins, or by antibody‐mediated uptake when phages are opsonized by circulating immunoglobulins (Bodner et al, 2021; Goswami et al, 2021) (Fig 5B).
Phages have been detected in early endosomes, endolysosomes and the Golgi apparatus (Nieth et al, 2015; Lehti et al, 2017; Zhang et al, 2017; Moller‐Olsen et al, 2018), and can escape eukaryotic vacuoles to reach bacteria replicating in the cytosol (Nieth et al, 2015). Phage–bacterium interactions in subcellular compartments are expected to be heavily modulated by the host eukaryotic cell, potentially in ways that alter phage infectivity or bacterial susceptibility as bacteria respond to intracellular stress (e.g. low pH, reactive oxygen species and antimicrobial peptides) and to nutrient depravation. Phages can certainly kill intracellular bacteria (Zhang et al, 2017; Moller‐Olsen et al, 2018), but further investigation of how phages reach their intracellular targets will be essential for designing successful therapeutic protocols.
Special states: biofilms
In many natural niches, including human body sites (e.g. respiratory and urinary tract), both Gram‐positive and Gram‐negative bacteria live in complex sessile biofilm communities (Hall‐Stoodley et al, 2004), often polymicrobial. Bacterial biofilms are common in chronic and persistent infections (Bjarnsholt, 2013) and on abiotic surfaces such as medical devices (prosthetic joints, catheters, heart valves) (Donlan, 2001; Petrovic Fabijan et al, 2019). Diverse components (Smirnova et al, 2010) make up an extracellular matrix in which bacteria are embedded, which gives stability and strength to the growing biofilm (Flemming & Wingender, 2010). Biofilm formation and maturation are guided by the coordinated activity of embedded bacteria, regulated by refined quorum‐sensing mechanisms in response to population density variation (Parsek & Greenberg, 2005; Nadell et al, 2008). Bacteria in a biofilm exhibit different metabolic activity and physiological state depending on their position in the biofilm and on the age of the biofilm (Stewart & Franklin, 2008). Antibiotics are often ineffective against biofilm‐mediated infections as bacteria are physically protected from external agents and more tolerant to antimicrobial challenge due to their modified metabolism and often reduced growth states (Lebeaux et al, 2014; Yan & Bassler, 2019).
The finger‐like bacterial fimbriae and other adhesins that are important in biofilm initiation (Déziel et al, 2001; Pohlschroder & Esquivel, 2015; Maldarelli et al, 2016; Delerue & Ramamurthi, 2021) are also common phage receptors (Phage tail machines as sophisticated infection devices). Phages have proven useful against bacteria in biofilms (Abedon, 2019; Patey et al, 2019; Melo et al, 2020b; Petrovic Fabijan et al, 2021), but the study of these systems is difficult (Abedon et al, 2021b; Pires et al, 2021). Although phages often exhibit potent in vitro activity against bacteria in biofilms, effective biofilm eradication may require combination strategies (Verma et al, 2009; Seth et al, 2013; Tkhilaishvili et al, 2018; Henriksen et al, 2019; Morris et al, 2019), with failures attributed to difficulties in accessing target cells and the development of phage‐resistant subpopulations.
The biofilm matrix shields bacteria from phage attack by trapping phage particles and preventing diffusion (Sutherland et al, 2004; González et al, 2018; Dunsing et al, 2019; Melo et al, 2020b), and phage size and concentration have been shown to differentially impact biofilm disruption ability (González et al, 2018). A biofilm can protect phages from the eukaryotic immune system, and these trapped viruses may in turn limit biofilm growth (Simmons et al, 2018; Hansen et al, 2019; Bond et al, 2021) so that in a stabilized biofilm, bacteria and phages may coexist in dynamic equilibrium (Fernández et al, 2018; Hansen et al, 2019; Pires et al, 2021). Bacteria may produce matrix‐degrading substances when challenged with phages (Alcock & Palmer, 2021; de Cássia Oliveira et al, 2021) and can also secrete phage‐inactivating substances (Pires et al, 2021). E. coli can halt phage invasion of mature biofilms through expression of curli fibres that affect biofilm architecture, hinder phage diffusion and physically protect the bacterial cell surface (Price & Chapman, 2018; Vidakovic et al, 2018; Bond et al, 2021). Also relevant when considering phage therapy for chronic infections (Pires et al, 2017) is the fact that older biofilms are often characterized by thicker matrix and by subpopulations of bacteria that are less metabolically active (Testa et al, 2019), these two factors alone mitigating the potential impact of phage therapeutic intervention.
Phage‐produced lysins and depolymerases (Attachment mechanisms and receptor specificity) are less sensitive to biofilm heterogeneity, bacterial metabolic state and physical barriers and may have a role in matrix degradation (Olsen et al, 2018; Wu et al, 2019; Rakov et al, 2021; Shahed‐Al‐Mahmud et al, 2021). Delivery of phages or their derived enzymes together with antibiotics and/or disinfectants may be synergistic, with disruption of the extracellular matrix by phage enzymes and/or chemical antimicrobials expected to allow better access to subsequent antibiotics and phages (Chan & Abedon, 2015; Ferriol‐González & Domingo‐Calap, 2020).
Bacteria in biofilms use much the same adaptation mechanisms as free‐living bacteria (Phage–bacterium co‐adaptation) (Azeredo et al, 2021). Added protection derived from the population density in biofilms comes from quorum‐sensing signalling to manage receptor modulation (Moreau et al, 2017; Azeredo et al, 2021; León‐Félix & Villicaña, 2021), e.g. in E. coli (Høyland‐Kroghsbo et al, 2013) and P. aeruginosa (Høyland‐Kroghsbo et al, 2017; Broniewski et al, 2021), and through modification of bacterial physiology (Qin et al, 2017).
The eukaryotic host: phage‐induced immune responses
The natural immunogenicity of phages may result in both an innate immune response (Petrovic Fabijan et al, 2020b; Khatami et al, 2021) and an adaptive immune response (e.g. phage‐specific antibodies) to viral nucleic acids (DNA or RNA) and proteins (capsid and tail) (Gonzalez‐Mora et al, 2020). The sustained phage viraemia arising from therapeutic infusion (Dąbrowska & Abedon, 2019; Petrovic Fabijan et al, 2020b) does not seem to present a safety risk but may be associated with modulation of the human immune response (Górski et al, 2017b; Petrovic Fabijan et al, 2020b; Khatami et al, 2021) by mechanisms that are as yet unclear. This topic has been well reviewed (Popescu et al, 2021), but key aspects to highlight include the following:
Phagocytosis
Non‐specific phagocytosis of viral particles may play a major role in the rapid clearance or neutralization of phages through the mammalian host reticuloendothelial system (Merril et al, 1996) and promote the presentation of antigens to T cells for the development of specific or adaptive immune response against phages themselves (Dąbrowska & Abedon, 2019). Phage binding may also facilitate phagocytosis of bacteria by macrophages or dendritic cells. Early studies (D'Herelle, 1923; Nelson, 1928) showed that phage‐resistant bacteria are protected from this effect, and it has been suggested that this “opsonization” process may be important for the eradication of pathogenic bacteria in vivo (Górski et al, 2017b) and may explain observations of reduced phage efficacy in neutropenic hosts (Roach et al, 2017).
Inflammation
Minor pro‐inflammatory responses ex vivo (Van Belleghem et al, 2017) and in treated patients (Khatami et al, 2021) have been attributed to LPS release into the system following bacterial lysis. However, the use of highly purified therapeutic phage preparations has not been associated with significant inflammatory responses (Górski et al, 2012; Krut & Bekeredjian‐Ding, 2018) so it is thought that contaminating endotoxins in early therapeutic phage preparations may have been primarily responsible for activation of Toll‐like receptor (TLR) signalling pathways and early reports of post‐infusion fevers (D'Herelle, 1930; Hashiguchi et al, 2010; Krut & Bekeredjian‐Ding, 2018).
Anti‐inflammatory immune response
Highly purified (“GMP‐grade”) phage preparations may induce the expression of key anti‐inflammatory genes, including IL‐1RA and IL‐10 family cytokines (Van Belleghem et al, 2017). An apparent anti‐inflammatory profile has been demonstrated both in vivo (Van Belleghem et al, 2017) and in vitro (Dhungana et al, 2021) and observed in critically ill patients with infective endocarditis and sepsis receiving adjunct phage therapy (Petrovic Fabijan et al, 2020b; Khatami et al, 2021). Other studies have shown a significant decrease in C‐reactive protein values, erythrocyte sedimentation rates and white cell counts in patients treated with phage (Miedzybrodzki et al, 2009), although these could equally be simple responses to reduced bacterial burden. It is conceivable that phages evolved to attack human colonizers and pathogens might also be able to survive attack by the immune system, and while the immunomodulatory and anti‐inflammatory mechanisms remain unclear, some studies suggest that phage interaction with immune cells may also be directly implicated (Górski et al, 2017a; Sweere et al, 2019).
Antiviral immune response
This has been well described in filamentous phages (Sweere et al, 2019). Pf phages can trigger maladaptive innate viral responses via TLR3 and interferon‐β production, and inhibition of TNF and phagocytosis, impairing bacterial clearance. It remains unclear, however, whether widely used therapeutic tailed phages can trigger similar antiviral responses.
Adaptive humoral immune response
Due to their immunogenic nature, phages can induce a strong humoral response (phage‐neutralizing IgG, IgM and, to a lesser extent, IgA antibodies), which can impact phage bioavailability in vivo and potentially hamper therapeutic success. The timing and strength of the humoral antiphage immune response mainly depend on phage immunogenic properties based on different structural protein composition (e.g. capsid proteins are known to be highly antigenic, for example the major capsid protein and outer capsid protein (Hoc) in T4‐like phages (Dąbrowska et al, 2014)), but are also affected by the route of administration, dose and the patient's immune status (Zaczek et al, 2016; Lusiak‐Szelachowska et al, 2017). Previous reports indicated that orally administered phages induce no or very weak humoral response in healthy volunteers (Sarker et al, 2012). In contrast, intravenously administered phages induce a strong humoral response, which usually arises within 10 days of phage therapy initiation (Pescovitz et al, 2011; Lusiak‐Szelachowska et al, 2014; Petrovic Fabijan et al, 2020b), with strong IgM induction in the first days of therapy, and high IgG levels recorded between 7 and 14 days. While earlier studies from the Hirszfeld Institute for Experimental Therapy (Poland) and the Eliava Institute (Georgia) showed no significant correlation between clinical outcome and level of antiphage antibodies (Lusiak‐Szelachowska et al, 2014), recent reports indicate that robust antibody response against certain phage types may limit phage efficacy in vivo and lead to therapeutic failure (Dedrick et al, 2021). Although our understanding of the influence of the humoral immune response on phage bioavailability and therapeutic success is limited, genetic engineering approaches (e.g. modification of phage capsid proteins) may prove key to overcoming these immunogenicity barriers (Hodyra‐Stefaniak et al, 2020).
Phages that have evolved to protect their prey populations by down‐regulating the host immune response may prove to be difficult choices in therapy. Conversely, phage‐mediated immunomodulation may be a good therapeutic trade‐off in severe sepsis where attenuation of a lethal cytokine‐mediated inflammatory response may be the most important therapeutic goal.
Concluding remarks
In this review, we sought to highlight the main areas of phage and bacterial biology that may directly relate to therapeutic outcome and in need of further investigation (Table 1).
Table 1.
Key biological aspects in phage–bacterium interaction that may affect clinical outcomes.
| Biological mechanism | Biological role | Desired properties for therapy | Implications for therapy | Focus for improvement of clinical outcomes |
|---|---|---|---|---|
| Phage attachment | Infectivity (lytic activity) | High lytic activity: large burst size | Dosing and timing of administration | Diverse banks of characterized phages; genome engineering |
| Receptor specificity | Infectivity (lytic activity; host range) | Defined host range | Targeting; clinical spectrum of activity (target bacteria); resistance | Personalized therapy; curated phage/bacteria banks; AI/machine learning approaches; phage cocktails; phage “training”; genome engineering |
| Phage life cycle | Infectivity (lytic activity); transduction | High lytic activity; low transduction rates | Bacterial killing efficiency; transmission of virulence/resistance | Phage genomics; curated phage banks; genome engineering |
| Bacterial cell physiological state/ density | Niche colonization and invasion | High lytic activity; high penetration | Dosing and timing of administration; phage/antibiotic synergy; target diseases | Smart delivery |
| Bacterial lifestyle | Communal (biofilms); intracellular | High penetration | Penetration (target availability); clinical spectrum of activity (type of disease) | Smart delivery |
| Co‐adaptation | Microbial evolution | Poor ability to elicit resistance; stable high infectivity | Resistance development | Phage–phage and phage–antibiotic synergy |
However, bringing phages into the pharmacopoeia requires attention to several other areas that we have not fully discussed. The limited host range of most therapeutic phages means that this precision therapy needs well‐curated and accessible phage sources, which is a biobanking and information management challenge (Nagel et al, 2022). The prioritization of target infections is key in determining the content and purpose of such collections and will vary with the intended use and the balance of research and commercial sustainability agendas (commercial priorities in sustainable phage production will differ from research priorities).
Modification of phages to enhance their therapeutic potential (Pires et al, 2016; Brown et al, 2017; Chen et al, 2019; Kilcher & Loessner, 2019; Monteiro et al, 2019) is complicated by the presence of large proportions of uncharacterized genetic material (“dark matter”) in phage genomes, which must be experimentally addressed (Hatfull & Hendrix, 2011; Wittebole et al, 2014; Hatfull, 2015; Philipson et al, 2018; Moreno‐Gallego & Reyes, 2021).
The complexities of variable penetration into eukaryotic cells, tissue layers and mammalian host compartments such as the gut have also not been addressed in this review, but readers are referred to others for this important topic (Barr et al, 2015; Dąbrowska & Abedon, 2019; Hofer, 2019; Huh et al, 2019). We have also set aside the difficulties of production and manufacturing protocols for GMP‐grade phage preparations: safe phage therapy involves not only quality processing but also the careful selection of suitable production hosts to ensure efficiency and avoid inadvertent gene transduction. The ideal phages for formulation into therapy must not only behave predictably in complex microbial niches but must also be readily purified and stable in storage (Merabishvili et al, 2018; Moelling et al, 2018; Rohde et al, 2018; Pirnay et al, 2019). The safety of phages for compassionate use means that there may be some opportunities to “learn as we go”, but we must now proceed with eyes wide open, and we must be guided as much as possible by the basic physiology of the main actors, the phages and their bacterial hosts.
Author contributions
Carola Venturini: Conceptualization; Visualization; Writing—original draft; Writing—review & editing. Aleksandra Petrovic Fabijan: Conceptualization; Visualization; Writing—original draft; Writing—review & editing. Alicia Fajardo Lubian: Conceptualization; Visualization; Writing—original draft; Writing—review & editing. Stefanie Barbirz: Conceptualization; Visualization; Writing—original draft; Writing—review & editing. Jonathan Iredell: Conceptualization; Writing—review & editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
CV, JI, APF, AFL and SB conceptualized and wrote the manuscript. CV coordinated the preparation of the manuscript. CV and JI made final edits to the manuscript.
Pending issues
Limited well‐curated and accessible phage biobanks
Narrow host range
Exclusive reliability on obligate lytic phages
Occurrence of phage‐resistant bacterial mutants
Priority types of infection targeted
Application of phage cocktails vs monophage therapy
Therapeutic phage monitoring, dosing and administration protocols
Formulation and stabilization of phage therapeutics
Regulatory and intellectual property protection
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
For more information
Online links to relevant sources
International Society for Viruses of Microorganisms (ISVM) (international non‐profit organization dedicated to the advancement of the science and utility of the viruses of microorganisms, including archaeal viruses, bacteriophages and the viruses of microbial eukaryotes)—http://www.isvm.org/
Phage Directory (curated database of phage laboratories, phages and host strains to advance research and phage therapy)—https://phage.directory/
Phages for Human Applications Group Europe (international non‐profit organization to support phage research and phage therapy in Europe)—P.H.A.G.E. vzw ‐ Home (p‐h‐a‐g‐e.org)
Phage Australia (Australian national network of phage researchers and clinician‐scientists to professionalize phage therapy)—https://phageaustralia.org/
Center for Phage Biology and Therapy at Yale (newly established centre to advance phage biology and develop phage therapy into a safe, effective, scientifically sound and rational approach to infection control)—http://www.yalephagecenter.com/
Centre on Innovative Phage Applications and Therapeutics (first dedicated phage therapy centre in North America)—Center for Innovative Phage Applications and Therapeutics (ucsd.edu)
Acknowledgements
CV holds an SSVS Postdoctoral Research Associate position funded through the Mabs Melville Bequest (University of Sydney, Sydney, Australia). APF and AFL are supported by Office for Health and Medical Research (New South Wales, Australia) Phage Therapy Fellowships. JI is supported by a National Health Medical Research Council (Australian Government) Investigator Grant (Iredell_APP1197534). SB is funded by the German Science Foundation (BA4046/4‐1, SPP 2330 New Concepts in Prokaryotic Virus‐host Interactions). The authors thank Dr. Andrew Millard and Mr. Ryan Cook from Millard’s laboratory at Warwick Medical School (University of Warwick, UK) for providing us with an updated number of complete phage genomes deposited in GenBank since 2017.
EMBO Mol Med (2022) 14: e12435
See the Glossary for abbreviations used in this article.
Contributor Information
Carola Venturini, Email: carola.venturini@sydney.edu.au.
Jonathan Iredell, Email: jonathan.iredell@sydney.edu.au.
References
- Abed N, Couvreur P (2014) Nanocarriers for antibiotics: a promising solution to treat intracellular bacterial infections. Int J Antimicrob Agents 43: 485–496 [DOI] [PubMed] [Google Scholar]
- Abedon ST (1989) Selection for bacteriophage latent period length by bacterial density: a theoretical examination. Microb Ecol 18: 79–88 [DOI] [PubMed] [Google Scholar]
- Abedon ST (2011) Phage therapy pharmacology: calculating phage dosing. Adv Appl Microbiol 77: 1–40 [DOI] [PubMed] [Google Scholar]
- Abedon ST (2016) Phage therapy dosing: the problem(s) with multiplicity of infection (MOI). Bacteriophage 6: e1220348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abedon ST (2019) Use of phage therapy to treat long‐standing, persistent, or chronic bacterial infections. Adv Drug Deliv Rev 145: 18–39 [DOI] [PubMed] [Google Scholar]
- Abedon ST, Danis‐Wlodarczyk KM, Wozniak DJ (2021a) Phage cocktail development for bacteriophage therapy: toward improving spectrum of activity breadth and depth. Pharmaceuticals 14: 1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abedon ST, Danis‐Wlodarczyk KM, Wozniak DJ, Sullivan MB (2021b) Improving phage‐biofilm in vitro experimentation. Viruses 13: 1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann H‐W (1998) Tailed bacteriophages: the order caudovirales. Adv Virus Res 51: 135–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann HW, Prangishvili D (2012) Prokaryote viruses studied by electron microscopy. Arch Virol 157: 1843–1849 [DOI] [PubMed] [Google Scholar]
- Alcock F, Palmer T (2021) Activation of a bacterial killing machine. PLoS Genet 17: e1009261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RC, Pfrunder‐Cardozo KR, Meinel D, Egli A, Hall AR (2017) Associations among antibiotic and phage resistance phenotypes in natural and clinical Escherichia coli isolates. MBio 8: e01341‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseth EO, Pursey E, Luján AM, McLeod I, Rollie C, Westra ER (2019) Bacterial biodiversity drives the evolution of CRISPR‐based phage resistance. Nature 574: 549–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres D, Hanke C, Baxa U, Seul A, Barbirz S, Seckler R (2010) Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro . J Biol Chem 285: 36768–36775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres D, Roske Y, Doering C, Heinemann U, Seckler R, Barbirz S (2012) Tail morphology controls DNA release in two Salmonella phages with one lipopolysaccharide receptor recognition system. Mol Microbiol 83: 1244–1253 [DOI] [PubMed] [Google Scholar]
- Argov T, Sapir SR, Pasechnek A, Azulay G, Stadnyuk O, Rabinovich L, Sigal N, Borovok I, Herskovits AA (2019) Coordination of cohabiting phage elements supports bacteria‐phage cooperation. Nat Commun 10: 5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong GL, Conn LA, Pinner RW (1999) Trends in infectious disease mortality in the United States during the 20th century. JAMA 281: 61–66 [DOI] [PubMed] [Google Scholar]
- Aslam S, Lampley E, Wooten D, Karris M, Benson C, Strathdee S, Schooley RT (2020) Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug‐resistant bacterial infections at a single center in the United States. Open Forum Infect Dis 7: ofaa389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attrill EL, Claydon R, Łapińska U, Recker M, Meaden S, Brown AT, Westra ER, Harding SV, Pagliara S (2021) Individual bacteria in structured environments rely on phenotypic resistance to phage. PLoS Biol 19: e3001406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azeredo J, García P, Drulis‐Kawa Z (2021) Targeting biofilms using phages and their enzymes. Curr Opin Biotechnol 68: 251–261 [DOI] [PubMed] [Google Scholar]
- Baker S, Thomson N, Weill F‐X, Holt KE (2018) Genomic insights into the emergence and spread of antimicrobial‐resistant bacterial pathogens. Science 360: 733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Osburne MS, BrinJones H, Tai AK, Leong JM (2019) Prophage induction, but not production of phage particles, is required for lethal disease in a microbiome‐replete murine model of enterohemorrhagic E. coli infection. PLoS Pathog 15: e1007494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks DJ, Lei BF, Musser JM (2003) Prophage induction and expression of prophage‐encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect Immun 71: 7079–7086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr JJ, Auro R, Sam‐Soon N, Kassegne S, Peters G, Bonilla N, Hatay M, Mourtada S, Bailey B, Youle M et al (2015) Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters. Proc Natl Acad Sci USA 112: 13675–13680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benler S, Koonin EV (2020) Phage lysis‐lysogeny switches and programmed cell death: danse macabre. BioEssays 42: 2000114 [DOI] [PubMed] [Google Scholar]
- Bertozzi Silva J, Storms Z, Sauvageau D (2016) Host receptors for bacteriophage adsorption. FEMS Microbiol Lett 363: fnw002 [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T (2013) The role of bacterial biofilms in chronic infections. APMIS 121: 1–58 [DOI] [PubMed] [Google Scholar]
- Blazanin M, Turner PE (2021) Community context matters for bacteria‐phage ecology and evolution. ISME J 15: 3119–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodner K, Melkonian AL, Covert MW (2021) The enemy of my enemy: new insights regarding bacteriophage‐mammalian cell interactions. Trends Microbiol 29: 528–541 [DOI] [PubMed] [Google Scholar]
- Bohannan BJ, Lenski RE (2000) Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol Lett 3: 362–377 [Google Scholar]
- Bond MC, Vidakovic L, Singh PK, Drescher K, Nadell CD (2021) Matrix‐trapped viruses can prevent invasion of bacterial biofilms by colonizing cells. eLife 10: e65355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeker NK, Andres D, Kang Y, Gohlke U, Schmidt A, Kunstmann S, Santer M, Barbirz S (2017) Complex carbohydrate recognition by proteins: fundamental insights from bacteriophage cell adhesion systems. Perspect Sci (Neth) 11: 45–52 [Google Scholar]
- Broeker NK, Barbirz S (2017) Not a barrier but a key: how bacteriophages exploit host's O‐antigen as an essential receptor to initiate infection. Mol Microbiol 105: 353–357 [DOI] [PubMed] [Google Scholar]
- Broeker NK, Roske Y, Valleriani A, Stephan MS, Andres D, Koetz J, Heinemann U, Barbirz S (2019) Time‐resolved DNA release from an O‐antigen‐specific Salmonella bacteriophage with a contractile tail. J Biol Chem 294: 11751–11761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broendum SS, Buckle AM, McGowan S (2018) Catalytic diversity and cell wall binding repeats in the phage‐encoded endolysins. Mol Microbiol 110: 879–896 [DOI] [PubMed] [Google Scholar]
- Broniewski JM, Chisnall MAW, Høyland‐Kroghsbo NM, Buckling A, Westra ER (2021) The effect of quorum sensing inhibitors on the evolution of CRISPR‐based phage immunity in Pseudomonas aeruginosa . ISME J 15: 2465–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R, Lengeling A, Wang B (2017) Phage engineering: how advances in molecular biology and synthetic biology are being utilized to enhance the therapeutic potential of bacteriophages. Quant Biol 5: 42–54 [Google Scholar]
- Brussow H, Canchaya C, Hardt WD (2004) Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68: 560–602, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan D, El‐Shibiny A, Hobbs Z, Porter J, Kutter EM (2016) Bacteriophage T4 infection of stationary phase E. coli: life after log from a phage perspective. Front Microbiol 7: 1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull J, Pfennig DW, Wang N (2004) Genetic details, optimization and phage life histories. Trends Ecol Evol 19: 76–82 [DOI] [PubMed] [Google Scholar]
- Burrowes BH, Molineux IJ, Fralick JA (2019) Directed in vitro evolution of therapeutic bacteriophages: the Appelmans protocol. Viruses 11: 241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R, Wang G, Le S, Wu M, Cheng M, Guo Z, Ji Y, Xi H, Zhao C, Wang X et al (2019) Three capsular polysaccharide synthesis‐related glucosyltransferases, GT‐1, GT‐2 and WcaJ, are associated with virulence and phage sensitivity of Klebsiella pneumoniae . Front Microbiol 10: 1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capparelli R, Nocerino N, Lanzetta R, Silipo A, Amoresano A, Giangrande C, Becker K, Blaiotta G, Evidente A, Cimmino A (2010) Bacteriophage‐resistant Staphylococcus aureus mutant confers broad immunity against staphylococcal infection in mice. PLoS One 5: e11720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care ACoSaQiH (2021) AURA 2021: fourth Australian report on antimicrobial use and resistance in human health. [Google Scholar]
- Carryn S, Van Bambeke F, Mingeot‐Leclercq MP, Tulkens PM (2003) Activity of beta‐lactams (ampicillin, meropenem), gentamicin, azithromycin and moxifloxacin against intracellular Listeria monocytogenes in a 24 h THP‐1 human macrophage model. J Antimicrob Chemother 51: 1051–1052 [DOI] [PubMed] [Google Scholar]
- Casjens SR, Molineux IJ (2012) Short noncontractile tail machines: adsorption and DNA delivery by Podoviruses. In Viral mol machines, Rossmann MG, Rao VB (eds), pp 143–179. New York, NY: Springer; [DOI] [PubMed] [Google Scholar]
- de Cássia Oliveira V, Steixner S, do Nascimento C, Pagnano VO, Silva‐Lovato CH, Paranhos HFO, Wilflingseder D, Coraça‐Huber D, Watanabe E (2021) Expression of virulence factors by Pseudomonas aeruginosa biofilm after bacteriophage infection. Microb Pathog 154: 104834 [DOI] [PubMed] [Google Scholar]
- Cavaillon J‐M, Chrétien F (2019) From septicemia to sepsis 3.0—from Ignaz Semmelweis to Louis Pasteur. Genes Immun 20: 371–382 [DOI] [PubMed] [Google Scholar]
- Cenens W, Makumi A, Govers SK, Lavigne R, Aertsen A (2015) Viral transmission dynamics at single‐cell resolution reveal transiently immune subpopulations caused by a carrier state association. PLoS Genet 11: e1005770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenens W, Mebrhatu MT, Makumi A, Ceyssens PJ, Lavigne R, Van Houdt R, Taddei F, Aertsen A (2013) Expression of a novel P22 ORFan gene reveals the phage carrier state in Salmonella typhimurium . PLoS Genet 9: e1003269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan BK, Abedon ST (2012) Phage therapy pharmacology: phage cocktails. Adv App Microbiol 78: 1–23 [DOI] [PubMed] [Google Scholar]
- Chan BK, Abedon ST, Loc‐Carrillo C (2013) Phage cocktails and the future of phage therapy. Future Microbiol 8: 769–783 [DOI] [PubMed] [Google Scholar]
- Chan BK, Abedon ST (2015) Bacteriophages and their enzymes in biofilm control. Curr Pharm Des 21: 85–99 [DOI] [PubMed] [Google Scholar]
- Chan BK, Sistrom M, Wertz JE, Kortright KE, Narayan D, Turner PE (2016) Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa . Sci Rep 6: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Duerkop BA (2019) Sugar and fatty acids accelerate prophage induction. Cell Host Microbe 25: 175–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry WN, Concepcion‐Acevedo J, Park T, Andleeb S, Bull JJ, Levin BR (2017) Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS One 12: e0168615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liu M, Zhang PF, Leung SSY, Xia J (2021) Membrane‐permeable antibacterial enzyme against multidrug‐resistant Acinetobacter baumannii . ACS Infect Dis 7: 2192–2204 [DOI] [PubMed] [Google Scholar]
- Chen Y, Batra H, Dong J, Chen C, Rao VB, Tao P (2019) Genetic engineering of bacteriophages against infectious diseases. Front Microbiol 10: 954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YB, Li XM, Wang S, Guan LY, Li XX, Hu DY, Gao DY, Song JY, Chen HC, Qian P (2020) A novel tail‐associated O91‐specific polysaccharide depolymerase from a podophage reveals lytic efficacy of shiga toxin‐producing Escherichia coli . Appl Environ Microbiol 86: e00145‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini N, de Frutos M, Augarde E, Boulanger P, Letellier L, Viasnoff V (2010) Is the in vitro ejection of bacteriophage DNA quasistatic? A bulk to single virus study. Biophys J 99: 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua JE, Manning PA, Morona R (1999) The Shigella flexneri bacteriophage Sf6 tailspike protein (TSP)/endorhamnosidase is related to the bacteriophage P22 TSP and has a motif common to exo‐and endoglycanases, and C‐5 epimerases. Microbiology 145: 1649–1659 [DOI] [PubMed] [Google Scholar]
- Clokie MRJ, Millard AD, Letarov AV, Heaphy S (2011) Phages in nature. Bacteriophage 1: 31–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau AM, Tetart F, Trojet SN, Prere MF, Krisch HM (2007) Phage‐antibiotic synergy (PAS): beta‐lactam and quinolone antibiotics stimulate virulent phage growth. PLoS One 2: e799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook R, Brown N, Redgwell T, Rihtman B, Barnes M, Clokie M, Stekel DJ, Hobman J, Jones MA, Millard A (2021) Infrastructure for a Phage Reference database: identification of large‐scale biases in the current collection of cultured phage genomes. PHAGE 2: 214–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P, Sansonetti PJ (2004) Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304: 242–248 [DOI] [PubMed] [Google Scholar]
- Cowley LA, Low AS, Pickard D, Boinett CJ, Dallman TJ, Day M, Perry N, Gally DL, Parkhill J, Jenkins C (2018) Transposon insertion sequencing elucidates novel gene involvement in susceptibility and resistance to phages T4 and T7 in Escherichia coli O157. MBio 9: e00705‐18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dąbrowska K, Abedon ST (2019) Pharmacologically aware phage therapy: pharmacodynamic and pharmacokinetic obstacles to phage antibacterial action in animal and human bodies. Microbiol Mol Biol Rev 83: e00012‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dąbrowska K, Miernikiewicz P, Piotrowicz A, Hodyra K, Owczarek B, Lecion D, Kazmierczak Z, Letarov A, Gorski A (2014) Immunogenicity studies of proteins forming the T4 phage head surface. J Virol 88: 12551–12557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang VT, Sullivan MB (2014) Emerging methods to study bacteriophage infection at the single‐cell level. Front Microbiol 5: 724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AR, Cardarelli L, Pell LG, Radford DR, Maxwell KL (2012) Long noncontractile tail machines of bacteriophages. Viral Mol Mach 726: 115–142 [DOI] [PubMed] [Google Scholar]
- De Maesschalck V, Gutierrez D, Paeshuyse J, Lavigne R, Briers Y (2020) Advanced engineering of third‐generation lysins and formulation strategies for clinical applications. Crit Rev Microbiol 46: 548–564 [DOI] [PubMed] [Google Scholar]
- Dedrick RM, Freeman KG, Nguyen JA, Bahadirli‐Talbott A, Smith BE, Wu AE, Ong AS, Lin CT, Ruppel LC, Parrish NM et al (2021) Potent antibody‐mediated neutralization limits bacteriophage treatment of a pulmonary Mycobacterium abscessus infection. Nat Med 27: 1357–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick RM, Guerrero‐Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs‐Sera D, Schooley RT et al (2019) Engineered bacteriophages for treatment of a patient with a disseminated drug‐resistant Mycobacterium abscessus . Nat Med 25: 730–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delerue T, Ramamurthi KS (2021) How bacteria block their own biofilms. J Biol Chem 296: 100392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennehy JJ, Abedon ST (2021) Phage infection and lysis. In Bacteriophages, Harper DR, Abedon ST, Burrowes BH, McConville ML (eds), pp 341–383. Cham: Springer; [Google Scholar]
- Déziel E, Comeau Y, Villemur R (2001) Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J Bacteriol 183: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Herelle F (1923) The bacteriophage: its role in immunity. Am J Public Health 13: 406–408 [Google Scholar]
- D'Herelle F (1930) Studies upon asiatic cholera. Yale J Biol Med 1: 195–219 [PMC free article] [PubMed] [Google Scholar]
- Dhungana G, Nepal R, Regmi M, Malla R (2021) Pharmacokinetics and pharmacodynamics of a novel virulent Klebsiella phage Kp_Pokalde_002 in a mouse model. Front Cell Infect Microbiol 11, 684704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz‐Muñoz SL, Koskella B (2014) Bacteria–phage interactions in natural environments. Adv App Microbiol 89: 135–183 [DOI] [PubMed] [Google Scholar]
- Dion MB, Oechslin F, Moineau S (2020) Phage diversity, genomics and phylogeny. Nat Rev Microbiol 18: 125–138 [DOI] [PubMed] [Google Scholar]
- Donlan RM (2001) Biofilms and device‐associated infections. J Emerg Infect Dis 7: 277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, Sorek R (2018) Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359: eaar4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne M, Hupfeld M, Klumpp J, Loessner MJ (2018) Molecular basis of bacterial host interactions by Gram‐positive targeting bacteriophages. Viruses 10: 397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunsing V, Irmscher T, Barbirz S, Chiantia S (2019) Purely polysaccharide‐based biofilm matrix provides size‐selective diffusion barriers for nanoparticles and bacteriophages. Biomacromol 20: 3842–3854 [DOI] [PubMed] [Google Scholar]
- Esteves NC, Porwollik S, McClelland M, Scharf BE (2021) The multidrug efflux system AcrABZ‐TolC is essential for infection of Salmonella typhimurium by the flagellum‐dependent bacteriophage Chi. J Virol 95: e00394‐21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evilevitch A (2018) The mobility of packaged phage genome controls ejection dynamics. eLife 7: e37345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair RJ, Tor Y (2014) Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem 6: 25–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo‐Lubian A, Ben Zakour NL, Agyekum A, Qi Q, Iredell JR (2019) Host adaptation and convergent evolution increases antibiotic resistance without loss of virulence in a major human pathogen. PLoS Pathog 15: e1007218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley MM, Tu JG, Kearns DB, Molineux IJ, Liu J (2017) Ultrastructural analysis of bacteriophage Phi 29 during infection of Bacillus subtilis . J Struct Biol 197: 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L, Rodríguez A, García P (2018) Phage or foe: an insight into the impact of viral predation on microbial communities. ISME J 12: 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferriol‐González C, Domingo‐Calap P (2020) Phages for biofilm removal. Antibiotics 9: 268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipiak M, Los JM, Los M (2020) Efficiency of induction of shiga‐toxin lambdoid prophages in Escherichia coli due to oxidative and antibiotic stress depends on the combination of prophage and the bacterial strain. J Appl Genet 61: 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H‐C, Wingender J (2010) The biofilm matrix. Nat Rev Microbiol 8: 623–633 [DOI] [PubMed] [Google Scholar]
- Fortier LC, Sekulovic O (2013) Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 4: 354–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD Antimicrobial Resistance C (2022) Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399: 629–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD Non‐Typhoidal Salmonella IDC (2019) The global burden of non‐typhoidal Salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis 19: 1312–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD Tuberculosis C (2018) Global, regional, and national burden of tuberculosis, 1990–2016: results from the Global Burden of Diseases, Injuries, and Risk Factors 2016 Study. Lancet Infect Dis 18: 1329–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefen O, Fridman O, Ronin I, Balaban NQ (2014) Direct observation of single stationary‐phase bacteria reveals a surprisingly long period of constant protein production activity. Proc Natl Acad Sci USA 111: 556–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson B, Wilson DJ, Feil E, Eyre‐Walker A (2018) The distribution of bacterial doubling times in the wild. Proc Biol Sci 285: 20180789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González S, Fernández L, Gutiérrez D, Campelo AB, Rodríguez A, García P (2018) Analysis of different parameters affecting diffusion, propagation and survival of staphylophages in bacterial biofilms. Front Microbiol 9: 2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Mora A, Hernandez‐Perez J, Iqbal HMN, Rito‐Palomares M, Benavides J (2020) Bacteriophage‐based vaccines: a potent approach for antigen delivery. Vaccines 8: 504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordillo Altamirano FL, Barr JJ (2019) Phage therapy in the postantibiotic era. Clin Microbiol Rev 32: e00066‐18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordillo Altamirano F, Forsyth JH, Patwa R, Kostoulias X, Trim M, Subedi D, Archer SK, Morris FC, Oliveira C, Kielty L et al (2021) Bacteriophage‐resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat Microbiol 6: 157–161 [DOI] [PubMed] [Google Scholar]
- Górski A, Dąbrowska K, Hodyra‐Stefaniak K, Borysowski J, Międzybrodzki R, Weber‐Dąbrowska B (2015) Phages targeting infected tissues: novel approach to phage therapy. Future Microbiol 10: 199–204 [DOI] [PubMed] [Google Scholar]
- Górski A, Dabrowska K, Miedzybrodzki R, Weber‐Dabrowska B, Lusiak‐Szelachowska M, Jonczyk‐Matysiak E, Borysowski J (2017a) Phages and immunomodulation. Future Microbiol 12: 905–914 [DOI] [PubMed] [Google Scholar]
- Górski A, Jonczyk‐Matysiak E, Lusiak‐Szelachowska M, Miedzybrodzki R, Weber‐Dabrowska B, Borysowski J (2017b) The Potential of phage therapy in sepsis. Front Immunol 8: 1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górski A, Miedzybrodzki R, Borysowski J, Dabrowska K, Wierzbicki P, Ohams M, Korczak‐Kowalska G, Olszowska‐Zaremba N, Lusiak‐Szelachowska M, Klak M et al (2012) Phage as a modulator of immune responses: practical implications for phage therapy. Adv Virus Res 83: 41–71 [DOI] [PubMed] [Google Scholar]
- Górski A, Międzybrodzki R, Węgrzyn G, Jończyk‐Matysiak E, Borysowski J, Weber‐Dąbrowska B (2020) Phage therapy: current status and perspectives. Med Res Rev 40: 459–463 [DOI] [PubMed] [Google Scholar]
- Goswami A, Sharma PR, Agarwal R (2021) Combatting intracellular pathogens using bacteriophage delivery. Crit Rev Microbiol 47: 461–478 [DOI] [PubMed] [Google Scholar]
- Green SI, Gu Liu C, Yu X, Gibson S, Salmen W, Rajan A, Carter HE, Clark JR, Song X, Ramig RF et al (2021) Targeting of mammalian glycans enhances phage predation in the gastrointestinal tract. MBio 12: 3356–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Liu C, Green SI, Min L, Clark JR, Salazar KC, Terwilliger AL, Kaplan HB, Trautner BW, Ramig RF, Maresso AW (2020) Phage‐antibiotic synergy is driven by a unique combination of antibacterial mechanism of action and stoichiometry. MBio 11: e01462‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines ME, Hodges FE, Nale JY, Mahony J, Van Sinderen D, Kaczorowska J, Alrashid B, Akter M, Brown N, Sauvageau D (2021) Analysis of selection methods to develop novel phage therapy cocktails against antimicrobial resistant clinical isolates of bacteria. Front Microbiol 12: 564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall‐Stoodley L, Costerton JW, Stoodley P (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2: 95–108 [DOI] [PubMed] [Google Scholar]
- Hampton HG, Watson BN, Fineran PC (2020) The arms race between bacteria and their phage foes. Nature 577: 327–336 [DOI] [PubMed] [Google Scholar]
- Hansen MF, Svenningsen SL, Røder HL, Middelboe M, Burmølle M (2019) Big impact of the tiny: bacteriophage–bacteria interactions in biofilms. Trends Microbiol 27: 739–752 [DOI] [PubMed] [Google Scholar]
- Harrison E, Hall JP, Paterson S, Spiers AJ, Brockhurst MA (2017) Conflicting selection alters the trajectory of molecular evolution in a tripartite bacteria–plasmid–phage interaction. Mol Ecol 26: 2757–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi S, Yamaguchi Y, Takeuchi O, Akira S, Sugimura K (2010) Immunological basis of M13 phage vaccine: regulation under MyD88 and TLR9 signaling. Biochem Biophys Res Commun 402: 19–22 [DOI] [PubMed] [Google Scholar]
- Hatfull GF (2015) Dark matter of the biosphere: the amazing world of bacteriophage diversity. J Virol 89: 8107–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF, Hendrix RW (2011) Bacteriophages and their genomes. Curr Opin Virol 1: 298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineman RH, Bull JJ (2007) Testing optimality with experimental evolution: lysis time in a bacteriophage. Evolution 61: 1695–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen K, Rørbo N, Rybtke ML, Martinet MG, Tolker‐Nielsen T, Høiby N, Middelboe M, Ciofu O (2019) P. aeruginosa flow‐cell biofilms are enhanced by repeated phage treatments but can be eradicated by phage‐ciprofloxacin combination. Pathog Dis 77: ftz011 [DOI] [PubMed] [Google Scholar]
- Hernandez CA, Koskella B (2019) Phage resistance evolution in vitro is not reflective of in vivo outcome in a plant‐bacteria‐phage system. Evolution 73: 2461–2475 [DOI] [PubMed] [Google Scholar]
- Hesse S, Rajaure M, Wall E, Johnson J, Bliskovsky V, Gottesman S, Adhya S (2020) Phage resistance in multidrug‐resistant Klebsiella pneumoniae ST258 evolves via diverse mutations that culminate in impaired adsorption. MBio 11: e02530‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodyra‐Stefaniak K, Kaźmierczak Z, Majewska J, Sillankorva S, Miernikiewicz P, Międzybrodzki R, Górski A, Azeredo J, Lavigne R, Lecion D et al (2020) Natural and induced antibodies against phages in humans: induction kinetics and immunogenicity for structural proteins of PB1‐related phages. PHAGE 1: 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer U (2019) Pairing phages with their hosts in the human gut. Nat Rev Microbiol 17: 589 [DOI] [PubMed] [Google Scholar]
- Holmes RK (2000) Biology and molecular epidemiology of diphtheria toxin and the tox gene. J Infect Dis 181: S156–167 [DOI] [PubMed] [Google Scholar]
- Howard‐Varona C, Hargreaves KR, Abedon ST, Sullivan MB (2017) Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J 11: 1511–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høyland‐Kroghsbo NM, Maerkedahl RB, Svenningsen SL (2013) A quorum‐sensing‐induced bacteriophage defense mechanism. MBio 4: e00362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høyland‐Kroghsbo NM, Paczkowski J, Mukherjee S, Broniewski J, Westra E, Bondy‐Denomy J, Bassler BL (2017) Quorum sensing controls the Pseudomonas aeruginosa CRISPR‐Cas adaptive immune system. Proc Natl Acad Sci USA 114: 131–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Margolin W, Molineux IJ, Liu J (2015) Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc Natl Acad Sci USA 112: E4919–E4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan PT, Bastin DA, Whittle BL, Lindberg AA, Verma NK (1997) Molecular characterization of the genes involved in O‐antigen modification, attachment, integration and excision in Shigella flexneri bacteriophage SfV. Gene 195: 217–227 [DOI] [PubMed] [Google Scholar]
- Huh H, Wong S, St Jean J, Slavcev R (2019) Bacteriophage interactions with mammalian tissue: therapeutic applications. Adv Drug Deliv Rev 145: 4–17 [DOI] [PubMed] [Google Scholar]
- Jesaitis MA, Goebel WF (1955) Lysis of T4 phage by the specific lipocarbohydrate of phase II Shigella sonnei . J Exp Med 102: 733–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonczyk E, Klak M, Miedzybrodzki R, Gorski A (2011) The influence of external factors on bacteriophages. Folia Microbiol 56: 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaruzzaman NF, Kendall S, Good L (2017) Targeting the hard to reach: challenges and novel strategies in the treatment of intracellular bacterial infections. Br J Pharmacol 174: 2225–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaolis DK, Somara S, Maneval DR, Johnson JA, Kaper JB (1999) A bacteriophage encoding a pathogenicity island, a type‐IV pilus and a phage receptor in cholera bacteria. Nature 399: 375–379 [DOI] [PubMed] [Google Scholar]
- Kasman LM, Kasman A, Westwater C, Dolan J, Schmidt MG, Norris JS (2002) Overcoming the phage replication threshold: a mathematical model with implications for phage therapy. J Virol 76: 5557–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawacka I, Olejnik‐Schmidt A, Schmidt M, Sip A (2020) Effectiveness of phage‐based inhibition of Listeria monocytogenes in food products and food processing environments. Microorganisms 8: 1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil IA, Troeger C, Blacker BF, Rao PC, Brown A, Atherly DE, Brewer TG, Engmann CM, Houpt ER, Kang G et al (2018) Morbidity and mortality due to Shigella and enterotoxigenic Escherichia coli diarrhoea: the global burden of disease study 1990–2016. Lancet Infect Dis 18: 1229–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatami A, Lin RC, Petrovic‐Fabijan A, Alkalay‐Oren S, Almuzam S, Britton PN, Brownstein MJ, Dao Q, Fackler J, Hazan R (2021) Bacterial lysis, autophagy and innate immune responses during adjunctive phage therapy in a child. EMBO Mol Med 13: e13936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khawaldeh A, Morales S, Dillon B, Alavidze Z, Ginn AN, Thomas L, Chapman SJ, Dublanchet A, Smithyman A, Iredell JR (2011) Bacteriophage therapy for refractory Pseudomonas aeruginosa urinary tract infection. J Med Microbiol 60: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Kilcher S, Loessner MJ (2019) Engineering bacteriophages as versatile biologics. Trends Microbiol 27: 355–367 [DOI] [PubMed] [Google Scholar]
- Kittleson JT, DeLoache W, Cheng HY, Anderson JC (2012) Scalable plasmid transfer using engineered P1‐based phagemids. ACS Synth Biol 1: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht LE, Veljkovic M, Fieseler L (2020) Diversity and function of phage encoded depolymerases. Front Microbiol 10: 2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortright KE, Chan BK, Koff JL, Turner PE (2019) Phage therapy: a renewed approach to combat antibiotic‐resistant bacteria. Cell Host Microbe 25: 219–232 [DOI] [PubMed] [Google Scholar]
- Kortright KE, Done RE, Chan BK, Souza V, Turner PE (2022) Selection for phage resistance reduces virulence of Shigella flexneri . Appl Environ Microbiol 88: e0151421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B, Brockhurst MA (2014) Bacteria–phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev 38: 916–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krut O, Bekeredjian‐Ding I (2018) Contribution of the immune response to phage therapy. J Immunol 200: 3037–3044 [DOI] [PubMed] [Google Scholar]
- Kutter E, Sulakvelidze A (2004) Bacteriophages: biology and applications. Boca Raton, FL: CRC Press; [Google Scholar]
- Lamont I, Brumby AM, Egan JB (1989) UV induction of coliphage 186: prophage induction as an SOS function. Proc Natl Acad Sci USA 86: 5492–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latino L, Midoux C, Hauck Y, Vergnaud G, Pourcel C (2016) Pseudolysogeny and sequential mutations build multiresistance to virulent bacteriophages in Pseudomonas aeruginosa . Microbiol 162: 748–763 [DOI] [PubMed] [Google Scholar]
- Leaver M, Dominguez‐Cuevas P, Coxhead JM, Daniel RA, Errington J (2009) Life without a wall or division machine in Bacillus subtilis . Nature 460: 538 [DOI] [PubMed] [Google Scholar]
- Lebeaux D, Ghigo J‐M, Beloin C (2014) Biofilm‐related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78: 510–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti TA, Pajunen MI, Skog MS, Finne J (2017) Internalization of a polysialic acid‐binding Escherichia coli bacteriophage into eukaryotic neuroblastoma cells. Nat Commun 8: 1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León M, Bastías R (2015) Virulence reduction in bacteriophage resistant bacteria. Front Microbiol 6: 343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León‐Félix J, Villicaña C (2021) The impact of quorum sensing on the modulation of phage‐host interactions. J Bacteriol 203: e00687‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León‐Velarde CG, Happonen L, Pajunen M, Leskinen K, Kropinski AM, Mattinen L, Rajtor M, Zur J, Smith D, Chen S et al (2016) Yersinia enterocolitica‐specific infection by bacteriophages TG1 and ϕR1‐RT is dependent on temperature‐regulated expression of the phage host receptor OmpF. Appl Environ Microbiol 82: 5340–5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letarov AV, Kulikov EE (2017) Adsorption of bacteriophages on bacterial cells. Biochemistry 82: 1632–1658 [DOI] [PubMed] [Google Scholar]
- Levin BR, Bull JJ (2004) Population and evolutionary dynamics of phage therapy. Nat Rev Microbiol 2: 166–173 [DOI] [PubMed] [Google Scholar]
- Li GL, Cortez MH, Dushoff J, Weitz JS (2020) When to be temperate: on the fitness benefits of lysis vs. lysogeny. Virus Evol 6: veaa042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, He Y, Wang Z, Wei J, Hu T, Si J, Tao G, Zhang L, Xie L, Abdalla AE et al (2021) A combination therapy of phages and antibiotics: two is better than one. Int J Biol Sci 17: 3573–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg AA (1973) Bacteriophage receptors. Annu Rev Microbiol 27: 205–241 [DOI] [PubMed] [Google Scholar]
- Linden SB, Alreja AB, Nelson DC (2021) Application of bacteriophage‐derived endolysins to combat streptococcal disease: current state and perspectives. Curr Opin Biotechnol 68: 213–220 [DOI] [PubMed] [Google Scholar]
- Liu YN, Leung SSY, Guo YT, Zhao LL, Jiang N, Mi LY, Li PY, Wang C, Qin YH, Mi ZQ et al (2019) The capsule depolymerase Dpo48 rescues Galleria mellonella and mice from Acinetobacter baumannii systemic infections. Front Microbiol 10: 545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YN, Leung SSY, Huang Y, Guo YT, Jiang N, Li PY, Chen JC, Wang RT, Bai CQ, Mi ZQ et al (2020) Identification of two depolymerases from phage IME205 and their antivirulent functions on K47 capsule of Klebsiella pneumoniae . Front Microbiol 11: 218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobocka MB, Rose DJ, Plunkett G 3rd, Rusin M, Samojedny A, Lehnherr H, Yarmolinsky MB, Blattner FR (2004) Genome of bacteriophage P1. J Bacteriol 186: 7032–7068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh B, Kuhn A, Leptihn S (2019) The fascinating biology behind phage display: filamentous phage assembly. Mol Microbiol 111: 1132–1138 [DOI] [PubMed] [Google Scholar]
- Los M, Wegrzyn G, Neubauer P (2003) A role for bacteriophage T4 rI gene function in the control of phage development during pseudolysogeny and in slowly growing host cells. Res Microbiol 154: 547–552 [DOI] [PubMed] [Google Scholar]
- Los M, Wegrzyn G (2012) Pseudolysogeny. Adv Virus Res 82: 339–349 [DOI] [PubMed] [Google Scholar]
- Lourenço M, Chaffringeon L, Lamy‐Besnier Q, Pedron T, Campagne P, Eberl C, Bérard M, Stecher B, Debarbieux L, De Sordi L (2020) The spatial heterogeneity of the gut limits predation and fosters coexistence of bacteria and bacteriophages. Cell Host Microbe 28: 390–401 [DOI] [PubMed] [Google Scholar]
- Lusiak‐Szelachowska M, Zaczek M, Weber‐Dabrowska B, Miedzybrodzki R, Klak M, Fortuna W, Letkiewicz S, Rogoz P, Szufnarowski K, Jonczyk‐Matysiak E et al (2014) Phage neutralization by sera of patients receiving phage therapy. Viral Immunol 27: 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusiak‐Szelachowska M, Zaczek M, Weber‐Dabrowska B, Miedzybrodzki R, Letkiewicz S, Fortuna W, Rogoz P, Szufnarowski K, Jonczyk‐Matysiak E, Olchawa E et al (2017) Antiphage activity of sera during phage therapy in relation to its outcome. Future Microbiol 12: 109–117 [DOI] [PubMed] [Google Scholar]
- Maffei E, Shaidullina A, Burkolter M, Heyer Y, Estermann F, Druelle V, Sauer P, Willi L, Michaelis S, Hilbi H (2021) Systematic exploration of Escherichia coli phage–host interactions with the BASEL phage collection. PLoS Biol 19: e3001424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majkowska‐Skrobek G, Latka A, Berisio R, Squeglia F, Maciejewska B, Briers Y, Drulis‐Kawa Z (2018) Phage‐borne depolymerases decrease Klebsiella pneumoniae resistance to innate defense mechanisms. Front Microbiol 9: 2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalatia K, Kakabadze E, Bakuradze N, Grdzelishvili N, Stamp B, Herman E, Tapinos A, Coffey A, Lee D, Papadopoulos NG (2021) Investigation of Salmonella phage–bacteria infection profiles: network structure reveals a gradient of target‐range from generalist to specialist phage clones in nested subsets. Viruses 13: 1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli GA, Piepenbrink KH, Scott AJ, Freiberg JA, Song Y, Achermann Y, Ernst RK, Shirtliff ME, Sundberg EJ, Donnenberg MS et al (2016) Type IV pili promote early biofilm formation by Clostridium difficile . Pathog Dis 74: ftw061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantynen S, Laanto E, Oksanen HM, Poranen MM, Diaz‐Munoz SL (2021) Black box of phage‐bacterium interactions: exploring alternative phage infection strategies. Open Biol 11: 210188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwitz P, Olszak T, Gula G, Kowalska M, Arabski M, Drulis‐Kawa Z (2021) Emerging phage resistance in Pseudomonas aeruginosa PAO1 is accompanied by an enhanced heterogeneity and reduced virulence. Viruses 13: 1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallin S, Brüssow H (2017) Clinical trials of bacteriophage therapeutics. In Bacteriophages, Harper D, Abedon S, Burrowes B, McConville M (eds), pp 1–29. Cham: Springer; [Google Scholar]
- Melo LD, Oliveira H, Pires DP, Dabrowska K, Azeredo J (2020a) Phage therapy efficacy: a review of the last 10 years of preclinical studies. Crit Rev Microbiol 46: 78–99 [DOI] [PubMed] [Google Scholar]
- Melo LD, Pinto G, Oliveira F, Vilas‐Boas D, Almeida C, Sillankorva S, Cerca N, Azeredo J (2020b) The protective effect of Staphylococcus epidermidis biofilm matrix against phage predation. Viruses 12: 1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabishvili M, Pirnay J‐P, De Vos D (2018) Guidelines to compose an ideal bacteriophage cocktail. In Bacteriophage therapy, Azeredo J, Sillankorva S (eds), pp 99–110. New York, NY: Springer; [DOI] [PubMed] [Google Scholar]
- Mercier R, Kawai Y, Errington J (2014) General principles for the formation and proliferation of a wall‐free (L‐form) state in bacteria. eLife 3: e04629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merril CR, Biswas B, Carlton R, Jensen NC, Creed GJ, Zullo S, Adhya S (1996) Long‐circulating bacteriophage as antibacterial agents. Proc Natl Acad Sci USA 93: 3188–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickiewicz KM, Kawai Y, Drage L, Gomes MC, Davison F, Pickard R, Hall J, Mostowy S, Aldridge PD, Errington J (2019) Possible role of L‐form switching in recurrent urinary tract infection. Nat Commun 10: 4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedzybrodzki R, Fortuna W, Weber‐Dabrowska B, Gorski A (2009) A retrospective analysis of changes in inflammatory markers in patients treated with bacterial viruses. Clin Exp Med 9: 303–312 [DOI] [PubMed] [Google Scholar]
- Moelling K, Broecker F, Willy C (2018) A wake‐up call: we need phage therapy now. Viruses 10: 688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller‐Olsen C, Ho SFS, Shukla RD, Feher T, Sagona AP (2018) Engineered K1F bacteriophages kill intracellular Escherichia coli K1 in human epithelial cells. Sci Rep 8: 17559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal SI, Draper LA, Ross RP, Hill C (2020) Bacteriophage endolysins as a potential weapon to combat Clostridioides difficile infection. Gut Microb 12: 1813533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsur KA, Rahman MA, Huq F, Islam MN, Northrup RS, Hirschhorn N (1970) Effect of massive doses of bacteriophage on excretion of vibrios, duration of diarrhoea and output of stools in acute cases of cholera. Bull World Health Organ 42: 723–732 [PMC free article] [PubMed] [Google Scholar]
- Monteiro R, Pires DP, Costa AR, Azeredo J (2019) Phage therapy: going temperate? Trends Microbiol 27: 368–378 [DOI] [PubMed] [Google Scholar]
- Montemayor EJ, Ploscariu NT, Sanchez JC, Parrell D, Dillard RS, Shebelut CW, Ke ZL, Guerrero‐Ferreira RC, Wright ER (2021) Flagellar structures from the bacterium Caulobacter crescentus and implications for phage phi CbK predation of multi‐flagellin bacteria. J Bacteriol 203: e00399‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P, Diggle SP, Friman VP (2017) Bacterial cell‐to‐cell signaling promotes the evolution of resistance to parasitic bacteriophages. Ecol Evol 7: 1936–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno‐Gallego JL, Reyes A (2021) Informative regions in viral genomes. Viruses 13: 1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JL, Letson HL, Elliott L, Grant AL, Wilkinson M, Hazratwala K, McEwen P (2019) Evaluation of bacteriophage as an adjunct therapy for treatment of peri‐prosthetic joint infection caused by Staphylococcus aureus . PLoS One 14: e0226574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisette T, Kebriaei R, Lev KL, Morales S, Rybak MJ (2020) Bacteriophage therapeutics: a primer for clinicians on phage‐antibiotic combinations. Pharmacotherapy 40: 153–168 [DOI] [PubMed] [Google Scholar]
- Murray E, Draper LA, Ross RP, Hill C (2021) The advantages and challenges of using endolysins in a clinical setting. Viruses 13: 680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Xavier JB, Levin SA, Foster KR (2008) The evolution of quorum sensing in bacterial biofilms. PLoS Biol 6: e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel T, Musila L, Muthoni M, Nikolich M, Nakavuma JL, Clokie MR (2022) Phage banks as potential tools to rapidly and cost‐effectively manage antimicrobial resistance in the developing world. Curr Opin Virol 53: 101208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nale JY, Clokie MR (2021) Preclinical data and safety assessment of phage therapy in humans. Curr Opin Biotechnol 68: 310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda AM, Heyer A, Kramer C, Grunberger A, Kohlheyer D, Frunzke J (2014) Analysis of SOS‐induced spontaneous prophage induction in Corynebacterium glutamicum at the single‐cell level. J Bacteriol 196: 180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AR (1928) The effect of bacteriophage upon the phenomena of leukocytosis and phagocytosis. J Immunol 15: 43–64 [Google Scholar]
- Nguyen S, Baker K, Padman BS, Patwa R, Dunstan RA, Weston TA, Schlosser K, Bailey B, Lithgow T, Lazarou M et al (2017) Bacteriophage transcytosis provides a mechanism to cross epithelial cell layers. MBio 8: e01874‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieth A, Verseux C, Barnert S, Suss R, Romer W (2015) A first step toward liposome‐mediated intracellular bacteriophage therapy. Expert Opin Drug Deliv 12: 1411–1424 [DOI] [PubMed] [Google Scholar]
- Nobrega FL, Vlot M, de Jonge PA, Dreesens LL, Beaumont HJE, Lavigne R, Duthil BE, Brouns SJJ (2018) Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol 16: 760–773 [DOI] [PubMed] [Google Scholar]
- Oechslin F (2018) Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 10: 351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir G, Sorek R (2018) Contemporary phage biology: from classic models to new insights. Cell 172: 1260–1270 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Sasakawa C (2006) Intracellular survival of Shigella . Cell Microbiol 8: 177–184 [DOI] [PubMed] [Google Scholar]
- Oliveira H, Melo LDR, Santos SB (2019a) Bacteriophage proteins as antimicrobials to combat antibiotic resistance. In Antibiotic drug resistance, Capelo‐Martínez JL, Igrejas G (eds), pp. 343–406. Hoboken, NJ: John Wiley & Sons, Inc. [Google Scholar]
- Oliveira H, Mendes A, Fraga AG, Ferreira A, Pimenta AI, Mil‐Homens D, Fialho AM, Pedrosa J, Azeredo J (2019b) K2 capsule depolymerase is highly stable, is refractory to resistance, and protects larvae and mice from Acinetobacter baumannii sepsis. App Environ Microb 85: e00934‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen N, Thiran E, Hasler T, Vanzieleghem T, Belibasakis GN, Mahillon J, Loessner MJ, Schmelcher M (2018) Synergistic removal of static and dynamic Staphylococcus aureus biofilms by combined treatment with a bacteriophage endolysin and a polysaccharide depolymerase. Viruses 10: 438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T, Danis‐Wlodarczyk K, Arabski M, Gula G, Maciejewska B, Wasik S, Lood C, Higgins G, Harvey BJ, Lavigne R (2019) Pseudomonas aeruginosa PA5oct jumbo phage impacts planktonic and biofilm population and reduces its host virulence. Viruses 11: 1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill J (2016) Review on antimicrobial resistance: tackling drug‐resistant infections globally: final report and recommendations. [Google Scholar]
- Opal SM, Scannon PJ, Vincent J‐L, White M, Carroll SF, Palardy JE, Parejo NA, Pribble JP, Lemke JH (1999) Relationship between plasma levels of lipopolysaccharide (LPS) and LPS‐binding protein in patients with severe sepsis and septic shock. J Infect Dis 180: 1584–1589 [DOI] [PubMed] [Google Scholar]
- Parsek MR, Greenberg E (2005) Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol 13: 27–33 [DOI] [PubMed] [Google Scholar]
- Patey O, McCallin S, Mazure H, Liddle M, Smithyman A, Dublanchet A (2019) Clinical indications and compassionate use of phage therapy: personal experience and literature review with a focus on osteoarticular infections. Viruses 11: 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RJ, Jansen VA (2001) Understanding bacteriophage therapy as a density‐dependent kinetic process. J Theor Biol 208: 37–48 [DOI] [PubMed] [Google Scholar]
- Pescovitz MD, Torgerson TR, Ochs HD, Ocheltree E, McGee P, Krause‐Steinrauf H, Lachin JM, Canniff J, Greenbaum C, Herold KC et al (2011) Effect of rituximab on human in vivo antibody immune responses. J Allergy Clin Immunol 128: 1295–1302 e1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic Fabijan A, Aleksic Sabo V, Gavric D, Doffkay Z, Rakhely G, Knezevic P (2021) Are Bordetella bronchiseptica siphoviruses (Genus Vojvodinavirus) appropriate for phage therapy‐bacterial allies or foes? Viruses 13: 1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic Fabijan A, Ben Zakour NL, Ho J, Lin RCY, Iredell J, Westmead Bacteriophage Therapy T , AmpliPhi Biosciences C (2019) Polyclonal Staphylococcus aureus bacteremia. Ann Intern Med 171: 940–941 [DOI] [PubMed] [Google Scholar]
- Petrovic Fabijan A, Khalid A, Maddocks S, Ho J, Gilbey T, Sandaradura I, Lin RC, Ben Zakour N, Venturini C, Bowring B (2020a) Phage therapy for severe bacterial infections: a narrative review. Med J Aust 212: 279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic Fabijan A, Lin RCY, Ho J, Maddocks S, Ben Zakour NL, Iredell JR, Westmead Bacteriophage Therapy T (2020b) Safety of bacteriophage therapy in severe Staphylococcus aureus infection. Nat Microbiol 5: 465–472 [DOI] [PubMed] [Google Scholar]
- Pfeifer E, de Sousa JAM, Touchon M, Rocha EPC (2021) Bacteria have numerous distinctive groups of phage‐plasmids with conserved phage and variable plasmid gene repertoires. Nucleic Acids Res 49: 2655–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipson CW, Voegtly LJ, Lueder MR, Long KA, Rice GK, Frey KG, Biswas B, Cer RZ, Hamilton T, Bishop‐Lilly KA (2018) Characterizing phage genomes for therapeutic applications. Viruses 10: 188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires DP, Cleto S, Sillankorva S, Azeredo J, Lu TK (2016a) Genetically engineered phages: a review of advances over the last decade. Microbiol Mol Biol Rev 80: 523–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires DP, Melo LD, Boas DV, Sillankorva S, Azeredo J (2017) Phage therapy as an alternative or complementary strategy to prevent and control biofilm‐related infections. Curr Opin Microbiol 39: 48–56 [DOI] [PubMed] [Google Scholar]
- Pires DP, Melo LD, Azeredo J (2021) Understanding the complex phage‐host interactions in biofilm communities. Annu Rev Virol 8: 73–94 [DOI] [PubMed] [Google Scholar]
- Pires DP, Oliveira H, Melo LDR, Sillankorva S, Azeredo J (2016b) Bacteriophage‐encoded depolymerases: their diversity and biotechnological applications. Appl Microbiol Biotechnol 100: 2141–2151 [DOI] [PubMed] [Google Scholar]
- Pirnay J‐P (2020) Phage therapy in the year 2035. Front Microbiol 11: 1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirnay J‐P, De Vos D, Verbeken G (2019) Clinical application of bacteriophages in Europe. Microbiol Aust 40: 8–15 [Google Scholar]
- Pirnay J‐P, Kutter E (2021) Bacteriophages: it's a medicine, Jim, but not as we know it. Lancet Infect Dis 21: 309–311 [DOI] [PubMed] [Google Scholar]
- Pirnay J‐P, Verbeken G, Ceyssens P‐J, Huys I, De Vos D, Ameloot C, Fauconnier A (2018) The magistral phage. Viruses 10: 64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlschroder M, Esquivel RN (2015) Archaeal type IV pili and their involvement in biofilm formation. Front Microbiol 6: 190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu M, Van Belleghem JD, Khosravi A, Bollyky PL (2021) Bacteriophages and the immune system. Annu Rev Virol 8: 415–435 [DOI] [PubMed] [Google Scholar]
- Porcek NB, Parent KN (2015) Key residues of S. flexneri OmpA mediate infection by bacteriophage Sf6. J Mol Biol 427: 1964–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen LK, Licht TR, Rang C, Krogfelt KA, Molin S (1995) Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin‐treated mice. J Bacteriol 177: 5840–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JE, Chapman MR (2018) Phaged and confused by biofilm matrix. Nat Microbiol 3: 2–3 [DOI] [PubMed] [Google Scholar]
- Qin X, Sun Q, Yang B, Pan X, He Y, Yang H (2017) Quorum sensing influences phage infection efficiency via affecting cell population and physiological state. J Basic Microbiol 57: 162–170 [DOI] [PubMed] [Google Scholar]
- Rakov C, Ben Porat S, Alkalay‐Oren S, Yerushalmy O, Abdalrhman M, Gronovich N, Huang LN, Pride D, Coppenhagen‐Glazer S, Nir‐Paz R et al (2021) Targeting biofilm of MDR Providencia stuartii by phages using a catheter model. Antibiotics 10: 375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Marteyn B, Sansonetti PJ, Tang CM (2009) Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol 7: 333–340 [DOI] [PubMed] [Google Scholar]
- Reyes‐Robles T, Dillard RS, Cairns LS, Silva‐Valenzuela CA, Housman M, Ali A, Wright ER, Camilli A (2018) Vibrio cholerae outer membrane vesicles inhibit bacteriophage infection. J Bacteriol 200: e00792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripp S, Miller RV (1997) The role of pseudolysogeny in bacteriophage‐host interactions in a natural freshwater environment. Microbiology 143: 2065–2070 [DOI] [PubMed] [Google Scholar]
- Roach DR, Donovan DM (2015) Antimicrobial bacteriophage‐derived proteins and therapeutic applications. Bacteriophage 5: e1062590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach DR, Leung CY, Henry M, Morello E, Singh D, Di Santo JP, Weitz JS, Debarbieux L (2017) Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe 22: 38–47 [DOI] [PubMed] [Google Scholar]
- Rocker A, Lacey JA, Belousoff MJ, Wilksch JJ, Strugnell RA, Davies MR, Lithgow T (2020) Global trends in proteome remodeling of the outer membrane modulate antimicrobial permeability in Klebsiella pneumoniae. MBio 11: e00603‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde C, Resch G, Pirnay J‐P, Blasdel BG, Debarbieux L, Gelman D, Górski A, Hazan R, Huys I, Kakabadze E (2018) Expert opinion on three phage therapy related topics: bacterial phage resistance, phage training and prophages in bacterial production strains. Viruses 10: 178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F, Segall AM (2015) A century of phage lessons. Nature 528: 46–47 [DOI] [PubMed] [Google Scholar]
- Rosner JL (1972) Formation, induction, and curing of bacteriophage P1 lysogens. Virology 48: 679–689 [DOI] [PubMed] [Google Scholar]
- Rostøl JT, Marraffini L (2019) (Ph)ighting phages: how bacteria resist their parasites. Cell Host Microbe 25: 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan EM, Alkawareek MY, Donnelly RF, Gilmore BF (2012) Synergistic phage‐antibiotic combinations for the control of Escherichia coli biofilms in vitro . FEMS Immunol Med Microbiol 65: 395–398 [DOI] [PubMed] [Google Scholar]
- Ryu JK, Kim SJ, Rah SH, Kang JI, Jung HE, Lee D, Lee HK, Lee JO, Park BS, Yoon TY et al (2017) Reconstruction of LPS transfer cascade reveals structural determinants within LBP, CD14, and TLR4‐MD2 for efficient LPS recognition and transfer. Immunity 46: 38–50 [DOI] [PubMed] [Google Scholar]
- Salazar KC, Ma L, Green SI, Zulk JJ, Trautner BW, Ramig RF, Clark JR, Terwilliger AL, Maresso AW (2021) Antiviral resistance and phage counter adaptation to antibiotic‐resistant extraintestinal pathogenic Escherichia coli . MBio 12: e00211‐21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje J (2010) Plasmid segregation: how to survive as an extra piece of DNA. Crit Rev Biochem Mol Biol 45: 296–317 [DOI] [PubMed] [Google Scholar]
- Sao‐Jose C (2018) Engineering of phage‐derived lytic enzymes: improving their potential as antimicrobials. Antibiotics 7: 29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker SA, McCallin S, Barretto C, Berger B, Pittet A‐C, Sultana S, Krause L, Huq S, Bibiloni R, Bruttin A (2012) Oral T4‐like phage cocktail application to healthy adult volunteers from Bangladesh. Virology 434: 222–232 [DOI] [PubMed] [Google Scholar]
- Schmerer M, Molineux IJ, Bull JJ (2014) Synergy as a rationale for phage therapy using phage cocktails. PeerJ 2: e590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Rabsch W, Broeker NK, Barbirz S (2016) Bacteriophage tailspike protein based assay to monitor phase variable glucosylations in Salmonella O‐antigens. BMC Microbiol 16: 207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H (2001) Shiga‐toxin‐converting bacteriophages. Res Microbiol 152: 687–695 [DOI] [PubMed] [Google Scholar]
- Schooley RT, Biswas B, Gill JJ, Hernandez‐Morales A, Lancaster J, Lessor L, Barr JJ, Reed SL, Rohwer F, Benler S et al (2017) Development and use of personalized bacteriophage‐based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother 61: e00954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Kuehn MJ (2015) Outer‐membrane vesicles from Gram‐negative bacteria: biogenesis and functions. Nat Rev Microbiol 13: 605–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secor PR, Burgener EB, Kinnersley M, Jennings LK, Roman‐Cruz V, Popescu M, Van Belleghem JD, Haddock N, Copeland C, Michaels LA et al (2020) Pf bacteriophage and their impact on Pseudomonas virulence, mammalian immunity, and chronic infections. Front Immunol 11: 244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secor PR, Sweere JM, Michaels LA, Malkovskiy AV, Lazzareschi D, Katznelson E, Rajadas J, Birnbaum ME, Arrigoni A, Braun KR et al (2015) Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe 18: 549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed KD, Faruque SM, Mekalanos JJ, Calderwood SB, Qadri F, Camilli A (2012) Phase variable O antigen biosynthetic genes control expression of the major protective antigen and bacteriophage receptor in Vibrio cholerae O1. PLoS Pathog 8: e1002917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed KD, Yen M, Shapiro BJ, Hilaire IJ, Charles RC, Teng JE, Ivers LC, Boncy J, Harris JB, Camilli A (2014) Evolutionary consequences of intra‐patient phage predation on microbial populations. eLife 3: e03497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall AM, Roach DR, Strathdee SA (2019) Stronger together? Perspectives on phage‐antibiotic synergy in clinical applications of phage therapy. Curr Opin Microbiol 51: 46–50 [DOI] [PubMed] [Google Scholar]
- Seth AK, Geringer MR, Nguyen KT, Agnew SP, Dumanian Z, Galiano RD, Leung KP, Mustoe TA, Hong SJ (2013) Bacteriophage therapy for Staphylococcus aureus biofilm–infected wounds: a new approach to chronic wound care. J Plast Reconstr Aesthet Surg 131: 225–234 [DOI] [PubMed] [Google Scholar]
- Shahed‐Al‐Mahmud M, Roy R, Sugiokto FG, Islam MN, Lin MD, Lin LC, Lin NT (2021) Phage phi AB6‐borne depolymerase combats Acinetobacter baumannii biofilm formation and infection. Antibiotics 10: 279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silpe JE, Bassler BL (2019) A host‐produced quorum‐sensing autoinducer controls a phage lysis‐lysogeny decision. Cell 176: 268–280 e213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons M, Drescher K, Nadell CD, Bucci V (2018) Phage mobility is a core determinant of phage–bacteria coexistence in biofilms. ISME J 12: 531–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurnik M (2022) Editorial of Viruses special issue on phage–host interactions 2021. Viruses 14: 236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova T, Didenko L, Azizbekyan R, Romanova YM (2010) Structural and functional characteristics of bacterial biofilms. Microbiol 79: 413–423 [Google Scholar]
- de Sousa JAM, Buffet A, Haudiquet M, Rocha EPC, Rendueles O (2020) Modular prophage interactions driven by capsule serotype select for capsule loss under phage predation. ISME J 14: 2980–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan MS, Broeker NK, Saragliadis A, Roos N, Linke D, Barbirz S (2020) In vitro analysis of O‐antigen specific bacteriophage P22 inactivation by Salmonella outer membrane vesicles. Front Microbiol 11: 510638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Franklin MJ (2008) Physiological heterogeneity in biofilms. Nat Rev Microbiol 6: 199–210 [DOI] [PubMed] [Google Scholar]
- Stone R (2002) Bacteriophage therapy. Stalin's forgotten cure. Science 298: 728–731 [DOI] [PubMed] [Google Scholar]
- Sulakvelidze A, Alavidze Z, Morris JG Jr (2001) Bacteriophage therapy. Antimicrob Agents Chemother 45: 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland IW, Hughes KA, Skillman LC, Tait K (2004) The interaction of phage and biofilms. FEMS Microbiol Lett 232: 1–6 [DOI] [PubMed] [Google Scholar]
- Sweere JM, Van Belleghem JD, Ishak H, Bach MS, Popescu M, Sunkari V, Kaber G, Manasherob R, Suh GA, Cao X (2019) Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 363: eaat9691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliaferri TL, Jansen M, Horz HP (2019) Fighting pathogenic bacteria on two fronts: phages and antibiotics as combined strategy. Front Cell Infect Microbiol 9: 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NMI, van Raaij MJ, Leiman PG (2018) Contractile injection systems of bacteriophages and related systems. Mol Microbiol 108: 6–15 [DOI] [PubMed] [Google Scholar]
- Teh MY, Furevi A, Widmalm G, Morona R (2020) Influence of Shigella flexneri 2a O antigen acetylation on its bacteriophage Sf6 receptor activity and bacterial interaction with human cells. J Bacteriol 202: e00363‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa S, Berger S, Piccardi P, Oechslin F, Resch G, Mitri S (2019) Spatial structure affects phage efficacy in infecting dual‐strain biofilms of Pseudomonas aeruginosa . Commun Biol 2: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittes C, Schwarzer S, Quax TEF (2021) Viral hijack of filamentous surface structures in archaea and bacteria. Viruses 13: 164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkhilaishvili T, Lombardi L, Klatt A‐B, Trampuz A, Di Luca M (2018) Bacteriophage Sb‐1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus . Int J Antimicrob Agents 52: 842–853 [DOI] [PubMed] [Google Scholar]
- Tzipilevich E, Habusha M, Ben‐Yehuda S (2017) Acquisition of phage sensitivity by bacteria through exchange of phage receptors. Cell 168: 186–199 [DOI] [PubMed] [Google Scholar]
- Van Belleghem JD, Clement F, Merabishvili M, Lavigne R, Vaneechoutte M (2017) Pro‐ and anti‐inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Sci Rep 7: 8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini C, Fabjian AP, Lin RC (2019a) Bacteriophage therapy for severe infections. Microbiol Aust 40: 20–23 [Google Scholar]
- Venturini C, Zingali T, Wyrsch ER, Bowring B, Iredell J, Partridge SR, Djordjevic SP (2019b) Diversity of P1 phage‐like elements in multidrug resistant Escherichia coli . Sci Rep 9: 18861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V, Harjai K, Chhibber S (2009) Restricting ciprofloxacin‐induced resistant variant formation in biofilm of Klebsiella pneumoniae B5055 by complementary bacteriophage treatment. J Antimicrob Chemother 64: 1212–1218 [DOI] [PubMed] [Google Scholar]
- Vidakovic L, Singh PK, Hartmann R, Nadell CD, Drescher K (2018) Dynamic biofilm architecture confers individual and collective mechanisms of viral protection. Nat Microbiol 3: 26–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volozhantsev NV, Shpirt AM, Borzilov AI, Komisarova EV, Krasilnikova VM, Shashkov AS, Verevkin VV, Knirel YA (2020) Characterization and therapeutic potential of bacteriophage‐encoded polysaccharide depolymerases with beta‐galactosidase activity against Klebsiella pneumoniae K57 capsular type. Antibiotics 9: 732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor MK, Mekalanos JJ (1996) Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272: 1910–1914 [DOI] [PubMed] [Google Scholar]
- Walker DH Jr, Anderson TF (1970) Morphological variants of coliphage P1. J Virol 5: 765–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Tu JG, Liu J, Molineux IJ (2019) Structural dynamics of bacteriophage P22 infection initiation revealed by cryo‐electron tomography. Nat Microbiol 4: 1049–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I‐N (2006) Lysis timing and bacteriophage fitness. Genetics 172: 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz JS, Poisot T, Meyer JR, Flores CO, Valverde S, Sullivan MB, Hochberg ME (2013) Phage–bacteria infection networks. Trends Microbiol 21: 82–91 [DOI] [PubMed] [Google Scholar]
- Whitfield C, Williams DM, Kelly SD (2020) Lipopolysaccharide O‐antigens‐bacterial glycans made to measure. J Biol Chem 295: 10593–10609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2017) Global priority list of antibiotic‐resistant bacteria to guide research, discovery and development of new antibiotics. Who 2017: 1–7 [Google Scholar]
- Wittebole X, De Roock S, Opal SM (2014) A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 5: 226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RC, Friman V‐P, Smith MC, Brockhurst MA (2018) Cross‐resistance is modular in bacteria–phage interactions. PLoS Biol 16: e2006057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Wang R, Xu M, Liu Y, Zhu X, Qiu J, Liu Q, He P, Li Q (2019) A novel polysaccharide depolymerase encoded by the phage SH‐KP152226 confers specific activity against multidrug‐resistant Klebsiella pneumoniae via biofilm degradation. Front Microbiol 10: 2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Bassler BL (2019) Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Wang S, Guo Z, Liu H, Sun D, Yan G, Hu D, Du C, Feng X, Han W (2018) A guard‐killer phage cocktail effectively lyses the host and inhibits the development of phage‐resistant strains of Escherichia coli . App Microbiol Biotechnol 102: 971–983 [DOI] [PubMed] [Google Scholar]
- Yu P, Mathieu J, Li M, Dai Z, Alvarez PJ (2015) Isolation of polyvalent bacteriophages by sequential multiple‐host approaches. Appl Environ Microbiol 82: 808–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Wang L, Li X, Tan D, Cong C, Xu Y (2019) Efficacy of a phage cocktail in controlling phage resistance development in multidrug resistant Acinetobacter baumannii . Virus Res 272: 197734 [DOI] [PubMed] [Google Scholar]
- Zaczek M, Lusiak‐Szelachowska M, Jonczyk‐Matysiak E, Weber‐Dabrowska B, Miedzybrodzki R, Owczarek B, Kopciuch A, Fortuna W, Rogoz P, Gorski A (2016) Antibody production in response to staphylococcal MS‐1 phage cocktail in patients undergoing phage therapy. Front Microbiol 7: 1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Sun L, Wei R, Gao Q, He T, Xu C, Liu X, Wang R (2017) Intracellular Staphylococcus aureus control by virulent bacteriophages within MAC‐T bovine mammary epithelial cells. Antimicrob Agents Chemother 61: e01990‐16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder ND (1980) Portraits of viruses: RNA phage. Intervirology 13: 257–270 [DOI] [PubMed] [Google Scholar]


