Abstract
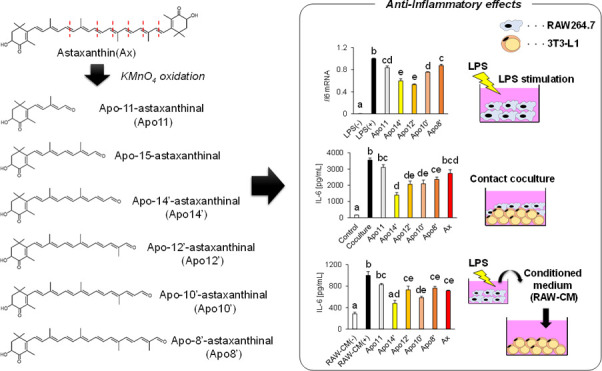
Apocarotenoids are carotenoid derivatives in which the polyene chain is cleaved via enzymatic or nonenzymatic action. They are found in animal tissues and carotenoid-containing foods. However, limited information on the biological functions of apocarotenoids is available. Here, we prepared apocarotenoids from astaxanthin via chemical oxidation and evaluated their anti-inflammatory action against macrophages and adipocytes. A series of astaxanthin-derived apoastaxanthinals, apo-11-, apo-15-, apo-14′-, apo-12′-, apo-10′-, and apo-8′-astaxanthinals, were successfully characterized by chromatography and spectroscopic analysis. The apoastaxanthinals inhibited inflammatory cytokine production and mRNA expression against lipopolysaccharide-stimulated RAW 264.7 macrophages. Apoastaxanthinals suppressed interleukin-6 overexpression in an in vitro model with macrophages and adipocytes in the following cultures: (1) contact coculture of 3T3-L1 adipocytes and RAW264.7 macrophages and (2) 3T3-L1 adipocytes in a RAW264.7-derived conditioned media. These results indicate that the apoastaxanthinals have the potential for regulation of adipose tissue inflammation observed in obesity.
1. Introduction
Astaxanthin is a red carotenoid found in several marine animals and microorganisms.1 It comprises a C40 polyene skeleton with 13 conjugated double bonds and 2 β-ionone rings substituted with hydroxy and keto groups. Previous reports indicate that astaxanthin possesses antioxidant and anticancer properties2,3 and could be used for lifestyle-related disease prevention4 and brain function improvement,5 which has led to an increased interest in its applications in food, animal feed, and nutraceutical and pharmaceutical products.
Apocarotenoids are cleavage products formed through enzymatic or nonenzymatic reactions.6 For example, retinoids, including vitamin A, are well-characterized apocarotenoids that play essential roles in many physiological processes including visual7 and immune systems.8 These compounds are produced through the central oxidative cleavage of provitamin A carotenoids by β-carotene 15,15′-oxygenase. In addition, β-carotene 9′,10′-oxygenase (BCO2) which has been identified in mammals, can also recognize both non-provitamin A and provitamin A carotenoids as substrates and produce asymmetric apocarotenoids.9
It is established that β-carotene- and lycopene-derived apocarotenoids, including β-apo-10′-carotenal and apo-12′-lycopenal, are found in human plasma.10,11 Furthermore, zeaxanthin- and lutein-derived apocarotenoids, including apo-10′-zeaxanthinal and ε-apo-12-luteinal, were also identified in human plasma12 and raw and processed foods.11,13 However, only a few studies on astaxanthin-derived apocarotenoids, including 3-hydroxy-4-oxo-β-ionone and 3-hydroxy-4-oxo-β-ionol, were reported in human plasma.14 Available information on the biological function of xanthophylls-derived apocarotenoids is also limited. On the other hand, it is known that β-carotene- and lycopene-derived apocarotenoids act as ligands for nuclear receptors including retinoid X receptor and retinoic acid receptor,15 inhibiting the proliferation of some cancer cells such as leukemia and prostate cancer cell lines.16 Further, apo-10′-lycopenoic acid inhibits liver and lung cancer cells by activating nuclear receptor peroxisome proliferator-activated receptor γ (PPARγ).17
Inflammation is associated with the incidence and development of various noncommunicable diseases (NCDs) including fatty liver disease and diabetes mellitus.18 Macrophages play an important role in regulating inflammation in the body.19 They are responsible for the host’s defense against infectious organisms and tissue homeostasis through the production of inflammatory cytokines and other mediators, including interleukin (IL), tumor necrosis factor-α, monocyte chemoattractant protein-1 (MCP-1), prostaglandins, and nitric oxide (NO).20,21 The dysregulation of these mediators causes chronic inflammatory disorders, leading to NCDs.19 In obese adipose tissues, activated macrophages stimulate adipocytes by secreting numerous inflammatory mediators. This exacerbates adipose tissue inflammation, which then leads to the development of insulin resistance and type-2 diabetes mellitus.22−25 Hence, it is important to regulate the overproduction of inflammatory factors to prevent NCDs. However, there is little information regarding regulatory effects of apocarotenoids against inflammation.
It was previously reported that astaxanthin inhibited NO and prostaglandin E2 by downregulating Nos2 and Ptgs2 mRNA expression in lipopolysaccharides (LPS)-activated RAW264.7 macrophages.26 Furthermore, it was demonstrated that astaxanthin suppressed inflammation in the adipose tissue of diet-induced obesity mouse model.27 However, the anti-inflammatory activity of astaxanthin-derived apocarotenoids has not been established.
In this study, we prepared astaxanthin-derived apocarotenoids through the oxidation of astaxanthin with potassium permanganate (KMnO4). The anti-inflammatory activity of the apoastaxanthinals was then investigated in activated RAW264.7 macrophages. In addition, using an obesity-induced inflammation in vitro model, we evaluated the inhibitory effect of apoastaxanthinals against inflammatory cytokine production induced through the interaction between 3T3-L1 adipocytes and RAW264.7 macrophages. This study demonstrated that the cleavage products derived from astaxanthin have a potential for prevention of inflammation-related NCDs.
2. Results
2.1. Preparation and Identification of Apocarotenoids Generated by Chemical Oxidation of Astaxanthin
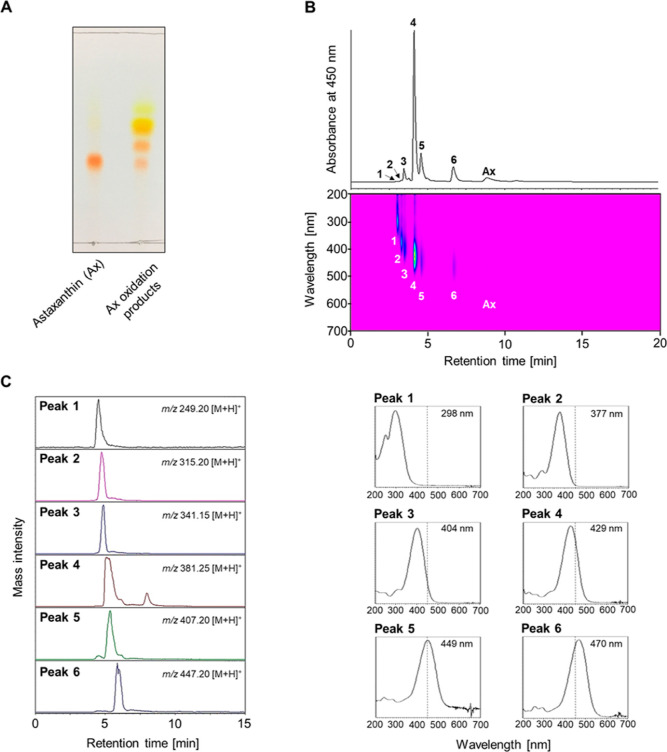
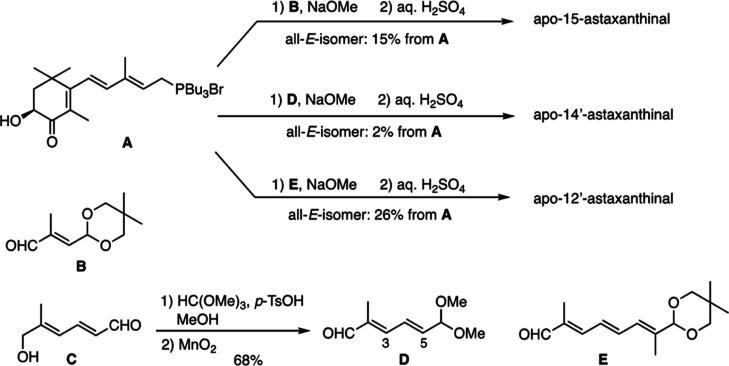
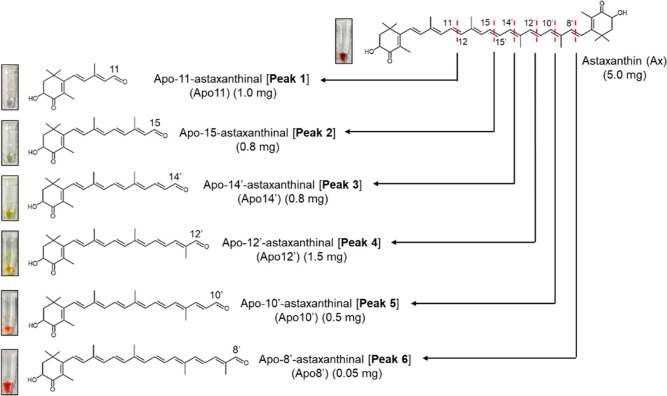
From the oxidation of astaxanthin with KMnO4, new spots of various colors were observed on the thin-layer chromatography (TLC) plate (Figure 1A). Subsequently, reverse-phase high-performance liquid chromatography (HPLC) equipped with a C30 column successfully separated astaxanthin and the six derivatives that had earlier retention time and shorter absorption maxima than the parent astaxanthin (Figure 1B). Furthermore, liquid chromatography–mass spectrometry (LC–MS) analysis revealed that these peaks had smaller molecular ions (protonated ions of peak 1–6; m/z 249.20, 315.20, 341.15, 381.25, 407.20, and 447.20) than astaxanthin (Figure 1C), indicating that the astaxanthin oxidation products had shorter chain lengths. To compare the oxidation products, we synthesized apoastaxanthinals (Scheme 1 and Supporting Information). According to the corresponding HPLC retention times and absorption spectra, peaks 2, 3, and 4 were identified as apo-15-astaxanthinal, apo-14′-astaxanthinal (apo14′), and apo-12′-apoastaxanthinal (apo12′), respectively. Peaks 1, 5, and 6 were further purified using HPLC and characterized by 1H NMR analysis (Table 1). The UV–vis, molecular ion, and 1H NMR data of peaks 1, 5, and 6 were identical to the previously reported data for apo-11-astaxanthinal (apo11), apo-10′-astaxanthinal (apo10′), and apo-8′-astaxanthinal (apo8′), respectively.28 Thus, six apoastaxanthinals were prepared by the oxidation of astaxanthin with KMnO4 (Figure 2).
Figure 1.
Identification of apoastaxanthinals generated by the oxidation of astaxanthin with potassium permanganate. (A) TLC analysis of astaxanthin and its oxidation products. (B) Reverse-phase HPLC equipped with C30-column was performed with methanol at a flow rate 1.0 mL/min. The products were monitored at wavelengths of 200 and 700 nm. Peak number described in the HPLC chromatogram corresponds to that on the photodiode array detector contour map (upper panel) and on the absorption spectra (lower panel). Absorption maxima are depicted within each figure, and black dotted lines are drawn at 450 nm. (C) LC–MS analysis equipped with C18-column was performed with methanol at a flow rate 0.1 mL/min. The peak number described in each mass chromatogram corresponds to that in (B).
Scheme 1. Synthesis of Apo-15-astaxanthinal, Apo-14′-astaxanthinal, and Apo-12′-astaxanthinal.
Table 1. 1H NMR Analysis of Peaks 1, 5, and 6 Generated by Astaxanthin Oxidationa.
| peak 1 | peak 5 | peak 6 | |
|---|---|---|---|
| position | δ (ppm) mult. J (Hz) | δ (ppm) mult. J (Hz) | δ (ppm) mult. J (Hz) |
| H-2α | 2.2 dd (13, 5) | 2.18 dd (13, 5) | 2.16 dd (13.5, 6) |
| H-2β | 1.88 dd (13, 13) | 1.84 dd (13, 13) | 1.82 dd (13.5, 13.5) |
| H-3 | 4.35 ddd (13, 5, 2) | 4.35 ddd (13, 5, 2) | 4.33 ddd (13.5, 6, 2) |
| H-7 | 6.7 d (17) | 6.27 d (17) | 6.23 d (15.5) |
| H-8 | 6.36 d (17) | 6.45 d (17) | 6.43 d (15.5) |
| H-10 | 6.03 d (7) | 6.32 d (11) | 6.3 d (11) |
| H-11 | 10.18 d (7) | 6.76 dd (15, 11) | 6.68 dd (15, 11) |
| H-12 | 6.49 d (15) | 6.45 d (15) | |
| H-14 | 6.36 d (11) | 6.33 d (11) | |
| H-15 | 6.88 dd (15, 11) | 6.77 dd (15, 11) | |
| H3-16 | 1.34 s | 1.35 s | 1.33 s |
| H3-17 | 1.19 s | 1.23 s | 1.21 s |
| H3-18 | 1.89 s | 1.96 s | 1.95 s |
| H3-19 | 2.35 s | 2.04 s | 2.02 s |
| H3-20 | 2.05 s | 2.02 s | |
| H-8′ | 9.46 s | ||
| H-10′ | 9.62 d (9) | 6.94 d (11) | |
| H-11′ | 6.23 dd (15, 9) | 6.69 dd (15, 11) | |
| H-12′ | 7.18 d (15) | 6.75 d (15) | |
| H-14′ | 6.64 d (11) | 6.45 d (11) | |
| H-15′ | 6.68 dd (15, 11) | 6.7 dd (15, 11) | |
| H3-19′ | 1.91 s | ||
| H3-20′ | 1.94 s | 2.02 s | |
| OH | 3.68 d (2) | 3.4 d (2) | 3.68 d (2) |
s, singlet; d, doublet; dd, doublet-of-doublets; ddd, doublet-of-doublets-of-doublets.
Figure 2.
Structure of the apoastaxanthinals derived from astaxanthin. The number between the brackets after each compound name corresponds to each peak described in Figure 1. The amount of astaxanthin used as a starting material and the yields of the six products are stated in milligrams. Pictures depict each apoastaxanthinal and astaxanthin dissolved in DMSO.
2.2. Cell Viability of RAW264.7 and 3T3-L1 Cells Treated with Apoastaxanthinals and Astaxanthin
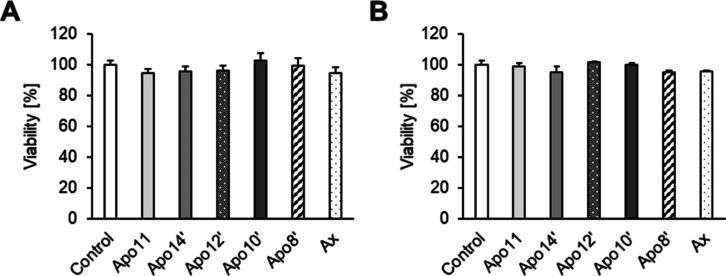
To evaluate the cell viability by WST-1 assay, RAW264.7 or differentiated 3T3-L1 cells were incubated in the presence of apoastaxanthinals or astaxanthin for 24 h. It was observed that the viability of the RAW264.7 and 3T3-L1 cells was not affected by the treatment (5 μM) with carotenoids (Figure 3).
Figure 3.
Cell viability of the macrophage-like RAW264.7 cells (A) and differentiated 3T3-L1 adipocytes (B) treated with apoastaxanthinals and astaxanthin. After incubation with each carotenoid (5 μM for 24 h), cell viability was determined using the WST-1 assay. Data are represented as the mean ± SEM [n = 5 (A) or n = 3 (B)]. Statistical analysis was performed using one-way ANOVA followed by the Tukey’s HSD test. Apo11: apo-11-astaxanthinal. Apo14′: apo-14′-astaxanthinal. Apo12′: apo-12′-astaxanthinal. Apo10′: apo-10′-astaxanthinal. Apo8′: apo-8′-astaxanthinal. Ax: astaxanthin.
2.3. Inhibitory Effect of Apo-12′-astaxanthinal on the Inflammatory Factor Expression in Activated RAW264.7 Macrophages
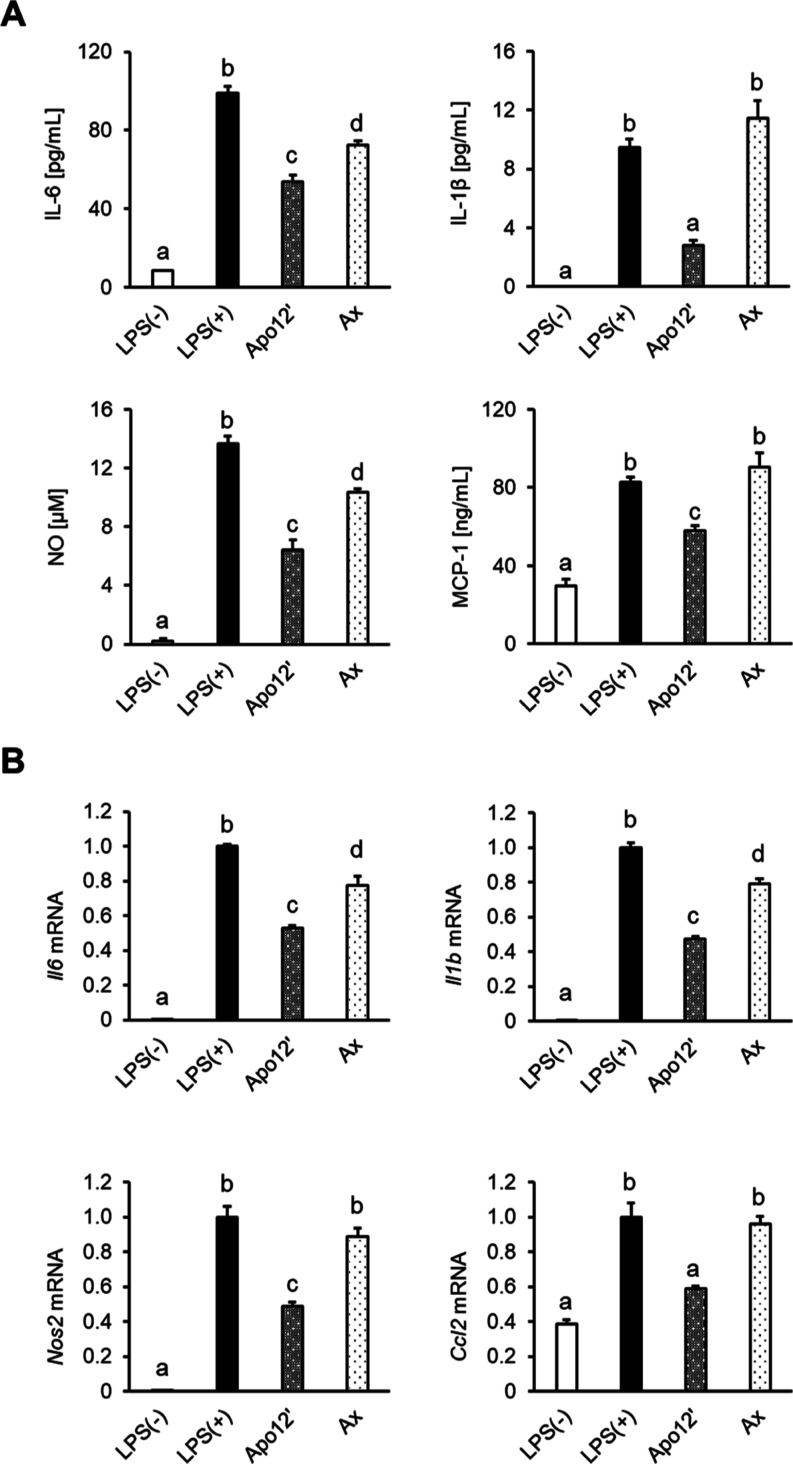
Anti-inflammatory activity of apo-12′-astaxanthinal was evaluated using LPS-activated RAW264.7 cells. In the culture media of the apo12′-treated cells, lower levels of inflammatory factors were observed than those in the LPS-treated [LPS (+)] and LPS and astaxanthin-treated (Ax) groups (Figure 4A). Further, treatment of apo12′ significantly downregulated mRNA expression of Il6, Il1b, Nos2, and Ccl2 than LPS (+) and Ax groups (Figure 4B). These results indicate that apo-12′-astaxanthinal is an effective anti-inflammatory agent against LPS-stimulated macrophages.
Figure 4.
Apo-12′-astaxanthinal significantly inhibited the expression of anti-inflammatory factors compared with parental astaxanthin in activated RAW264.7 macrophages. (A) Carotenoid-pretreated (5 μM for 2 h) cells are stimulated by LPS (100 ng/mL for 24 h) in the presence of carotenoids (5 μM). The levels of each inflammatory factor in the culture supernatant were determined by ELISA and Griess method. (B) Carotenoid-pretreated (5 μM for 2 h) cells are stimulated by LPS (100 ng/mL for 6 h). The levels of each inflammatory factor mRNA expression were determined by quantitative PCR method. Gapdh was used as the endogenous control. Data are represented as the mean ± SEM (n = 3) with different letters (p < 0.05). Apo12′: apo-12′-astaxanthinal. Ax: astaxanthin.
2.4. Downregulation of the Inflammatory Factor Gene Expression in Activated RAW264.7 Macrophages Treated with Apoastaxanthinals
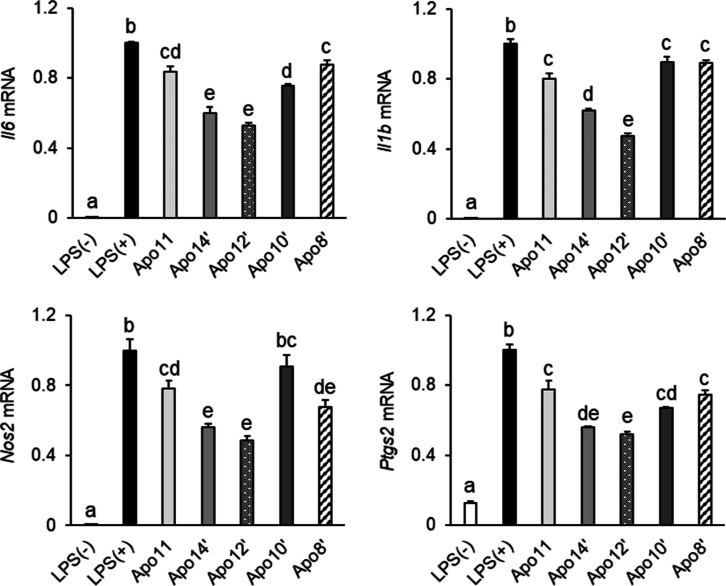
The anti-inflammatory activities of the apoastaxanthinals were compared in activated RAW264.7 cells. It was observed that the mRNA levels of Il6, Il1b, and Ptgs2 were downregulated by the treatment with the apoastaxanthinal derivatives, although those of Nos2 were not changed in the apo10′-treated cells (Figure 5). Among the apoastaxanthinal derivatives, apo14′ and apo12′ strongly downregulated the inflammatory factor mRNA expression. By contrast, the suppressive effects were comparable between apo11, apo10′, and apo8′ (Figure 5). These results indicated that the apoastaxanthinal derivatives downregulated the inflammatory factor mRNA expression in LPS-activated RAW264.7 and that the anti-inflammatory activity was dependent on their chain length.
Figure 5.
Apoastaxanthinals suppressed the mRNA expression of inflammatory factors in the activated RAW264.7 macrophages. Carotenoid-pretreated (5 μM for 2 h) cells are stimulated by LPS (100 ng/mL for 6 h) in the presence of carotenoids (5 μM). The levels of each inflammatory factor mRNA expression were determined by the quantitative PCR method. Gapdh was used as the endogenous control. Data are represented as the mean ± SEM (n = 3) with different letters (p < 0.05). Apo11: apo-11-astaxanthinal. Apo14′: apo-14′-astaxanthinal. Apo12′: apo-12′-astaxanthinal. Apo10′: apo-10′-astaxanthinal. Apo8′: apo-8′-astaxanthinal.
2.5. Apoastaxanthinals Inhibited IL-6 Production and mRNA Expression Induced by Interaction between Macrophages and Adipocytes
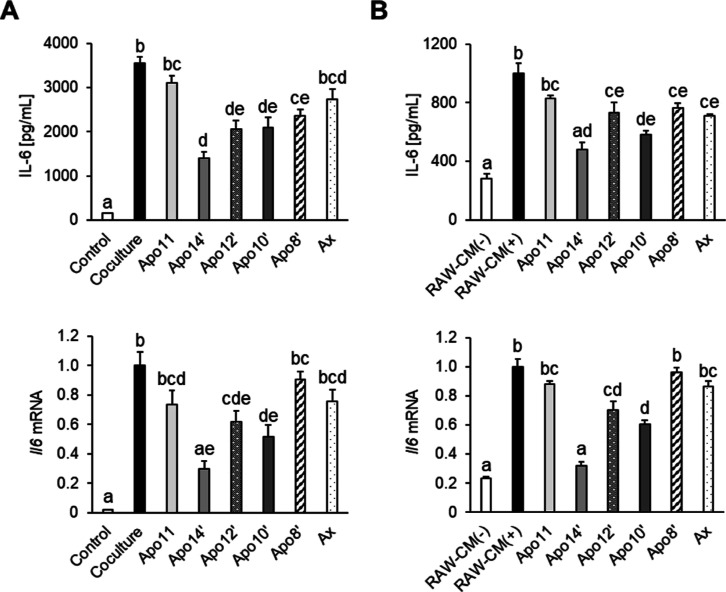
Dysregulation of macrophage and adipocyte interactions causes obesity-induced inflammation followed by insulin resistance. To ensure the suppressive effect against this interaction in vitro, we cocultured RAW264.7 with differentiated 3T3-L1 cells in the presence of apoastaxanthinals (Figure 6A). Compared to those in the coculture group, IL-6 protein levels in the culture media were reduced after treatment with the apoastaxanthinals, except for apo11′ (Figure 6A upper panel). In addition, Il6 mRNA expression was downregulated in apo12′- and apo10′-treated cells. In particular, its expression was the lowest in the apo14′-treated cells (Figure 6A lower panel). These results suggest that apoastaxanthinals attenuated IL-6 production induced by the coculture of 3T3-L1 adipocytes and RAW264.7 macrophages by downregulating its mRNA expression.
Figure 6.
Apoastaxanthinals and astaxanthin suppressed IL-6 production and mRNA expression induced by the interaction between RAW264.7 macrophages and 3T3-L1 adipocytes. (A) Contact coculture of RAW264.7 and 3T3-L1 cells (day 10) in the presence or absence of carotenoids (5 μM for 24 h). (B) Incubation of 3T3-L1 adipocytes in RAW264.7-derived conditioned media (RAW-CM). 3T3-L1 cells (day 10) were incubated in RAW-CM in the presence or absence of carotenoids (5 μM for 24 h). RAW-CM(−) represents the media collected from RAW264.7 cultured without LPS. Secreted IL-6 protein in the culture supernatant and Il6 mRNA levels were evaluated by ELISA and quantitative PCR methods, respectively. Actb was used as the endogenous control. Data are represented as the mean ± SEM (n = 3) with different letters (p < 0.05). Apo11: apo-11-astaxanthinal. Apo14′: apo-14′-astaxanthinal. Apo12′: apo-12′-astaxanthinal. Apo10′: apo-10′-astaxanthinal. Apo8′: apo-8′-astaxanthinal. Ax: astaxanthin.
To investigate the anti-inflammatory action of the apoastaxanthinals against 3T3-L1 adipocytes, the cells were incubated in RAW-CM recovered from the RAW264.7 cultures (Figure 6B). Although incubation with RAW-CM(+) significantly increased the IL-6 levels compared to that with RAW-CM(−), the addition of astaxanthin and apoastaxanthinals, except for apo11, significantly decreased the IL-6 levels (Figure 6B upper panel). In addition, apo14′, apo12′, and apo10′ significantly downregulated Il6 mRNA expression induced by RAW-CM(+) in 3T3-L1 cells (Figure 6B, lower panel). Notably, apo14′ attenuated both IL-6 protein and mRNA expression to the same level as that with RAW-CM(−) (Figure 6B). These results indicate that apoastaxanthinals, especially apo14′, can regulate IL-6 expression, which is induced by humoral factors secreted from LPS-stimulated RAW264.7 cells.
3. Discussion
It is established that the auto-oxidation of astaxanthin generates several carotenoids.28 To date, however, there have been few investigations on the preparation and biological activity of apoastaxanthinal derivatives. In this study, through the oxidation of astaxanthin with KMnO4, we synthesized and characterized six apoastaxanthinal derivatives, namely, apo-11-, apo-10′-, apo-8′-, apo-15-, apo-14′-, and apo-12′-apoastaxanthinal. We also evaluated their anti-inflammatory actions against activated RAW264.7 and 3T3-L1 cells. Although the reaction of astaxanthin with peroxynitrite generates apo-12′- and apo-10′-astaxanthinal,29 to our knowledge, this is the first report of apoastaxanthinal synthesis via astaxanthin oxidation with KMnO4 (Figure 2).
Analysis of the anti-inflammatory activity of the apoastaxanthinal derivatives on LPS-stimulated RAW264.7 cells revealed that apo12′ significantly inhibited the production of IL-6, MCP-1, IL-1β, and NO via downregulating their mRNA expression (Figure 4). Compared with astaxanthin, apo12′ potently suppressed these inflammatory factor expressions, suggesting that the cleavage products derived from astaxanthin augmented the anti-inflammatory activity on the LPS-stimulated macrophages (Figure 4). Notably, the downregulation of inflammatory factor gene expression was different among the apoastaxanthinal derivatives with different chain lengths (apo11, apo14′, apo12′, apo10′, and apo8′). Interestingly, apo14′ (the number of carbons; C22) and apo12′ (C25) displayed stronger suppressive properties compared with apo10′ (C27), apo8′ (C30), and apo11 (C15) in activated RAW264.7 cells (Figure 5). Apo-12′-lycopenal (C25), unlike apo-6′ (C32) and apo-8′-lycopenal (C30), specifically activated transcription mediated by PPARγ.30 PPARγ activation attenuated LPS-induced inflammation in RAW264.7 cells.31 Thus, the data obtained in this study suggest that the chain length of the apoastaxanthinal derivatives should be crucial for the anti-inflammatory effects.
Excessive and chronic production of inflammatory factors such as IL-6 from adipocytes is known to induce insulin resistance.24 Therefore, we investigated the inhibitory action of apoastaxanthinals against IL-6 production using two types of cell culture system: (1) the contact coculture of 3T3-L1 and RAW264.7 cells and (2) 3T3-L1 adipocytes in RAW-CM. The purpose of cell culture system 1 was to evaluate the regulatory effect on the direct interaction between macrophages and adipocytes. However, this system is unable to distinguish the effects of apoastaxanthinals on each cell. Therefore, we then perform cell culture system 2 to investigate the effect of apoastaxanthinals on adipocytes stimulated by macrophage-derived humoral factors associated with obesity-induced inflammation. Although the contact coculture demonstrated an increase in IL-6 production compared to the control group (RAW264.7 and 3T3-L1 alone), the apoastaxanthinals, except for apo11, significantly attenuated IL-6 production (Figure 6A). Apo14′ was the most potent at inhibiting IL-6 production by downregulating Il6 mRNA expression (Figure 6A). Further, mRNA expression of other inflammatory cytokines such as Il1b and Ccl2 were downregulated by apo14′ treatment (Supporting Information). On the other hand, RAW-CM(+) promoted the production of IL-6 in the 3T3-L1 cells compared to RAW-CM(−). This indicates that the activated macrophages also exacerbated inflammation in the adipocytes through the secretion of inflammatory humoral factors (Figure 6B). We also observed that astaxanthin and apoastaxanthinals, except for apo11, suppressed the IL-6 production in 3T3-L1 cells induced by RAW-CM(+). Notably, apo14′ strongly suppressed both the IL-6 production and mRNA expression when compared with other derivatives and astaxanthin. These results indicate that apo14′ significantly attenuated the inflammatory reaction in 3T3-L1 cells in addition to RAW264.7 cells, suggesting that the anti-inflammatory action of apoastaxanthinals requires a specific chain length.
Anti-inflammatory activity of apo14′ and apo12′ looks similar in LPS-stimulated RAW264.7 cells (Figure 5). However, apo14′ showed much more potent anti-inflammatory activity in systems using RAW264.7 and 3T3-L1 cells (Figure 6). This difference may be caused by PPARγ activation by the apoastaxanthinals on each cell. In addition to RAW264.7 macrophages,31 inflammation of 3T3-L1 adipocytes is suppressed by activation of PPARγ.32 Since adipocytes express more PPARγ proteins and genes than macrophages,33 apo14′ can exhibit potent anti-inflammatory activity in systems using RAW264.7 and 3T3-L1 cells. On the other hand, antioxidant enzymes associate with anti-inflammatory action by carotenoids.34 Lycopene-derived apocarotenoids strongly upregulate the gene and protein expression of antioxidant enzymes such as heme oxygenase 1 and NAD(P)H:quinone oxidoreductase 1 than lycopene.35 These enzyme expressions may be influenced by the treatment of apoastaxanthinals and astaxanthin in each cell. Since astaxanthin treatment did not increase the intracellular levels of apoastaxanthinals in both RAW264.7 and 3T3-L1 cells (data not shown), astaxanthin and apoastaxanthinals can exhibit anti-inflammatory effects via different mechanisms. Future work for the understanding of anti-inflammatory mechanism of apoastaxanthinals is expected.
Interestingly, apo10′ also exerted a potent anti-inflammatory activity than parent astaxanthin (Figure 6). Since BCO2 can produce apo-10′-carotenoids from parent xanthophylls,9 astaxanthin may be metabolized to apo10′ in the body. Recently, it has been reported that astaxanthin is accumulated in the liver of BCO2 knockout mouse fed astaxanthin,36 suggesting that astaxanthin is metabolized by BCO2 in the body. Given that astaxanthin suppresses inflammation in the adipose tissue of diet-induced obesity mouse model,27 apo10′ may be associated with this effect. Further investigation for the anti-inflammatory mechanism, metabolism, and distribution of apoastaxanthinals in the body is needed. To elucidate them, this study demonstrating the preparation of apoastaxanthinals and biological activity can be helpful.
4. Conclusions
We prepared and characterized six apoastaxanthinals from astaxanthin with KMnO4, namely, apo-11-, apo-15-, apo-14′-, apo-12′-, apo-10′-, and apo-8′-astaxanthinals. These apoastaxanthinals inhibited the LPS-induced mRNA expression of inflammatory cytokines and mediators in RAW264.7 macrophages. Furthermore, the apoastaxanthinals suppressed the overexpression of inflammatory cytokine IL-6 induced by the interaction of RAW264.7 and differentiated 3T3-L1 cells in an obesity-induced inflammation in vitro model. These results indicate, for the first time, the potential of astaxanthin-derived apoastaxanthinals as health beneficial compounds.
5. Experimental Section
5.1. Reagents and Chemicals
RAW264.7 macrophages and 3T3-L1 preadipocytes were purchased from the European Collection of Authenticated Cell Cultures (Salisbury, UK) and the American Type Culture Collection (Manassas, VA, USA), respectively. Fetal bovine serum (FBS) was purchased from Gibco (Grand Island, NY, USA). LPS from Escherichia coli O111:B4, isobutylmethylxanthine (IBMX), dexamethasone, insulin, N-[1-naphtyl]ethylenediamine dihydrochloride, and sulfanilamide were purchased from Sigma-Aldrich (St. Louis, MO, USA). Dulbecco’s modified Eagle’s medium (DMEM) was obtained from Nissui Pharmaceutical Co., Ltd. (Tokyo, Japan). Cetyltrimethylammonium bromide (CTAB) was obtained from Nacalai Tesque Inc. (Kyoto, Japan). Penicillin/streptomycin, RPMI1640, astaxanthin, and organic solvents were purchased from Fujifilm Wako Pure Chemical Co. Ltd. (Osaka, Japan).
5.2. Chemical Oxidation of Astaxanthin and Chromatography Analysis
Astaxanthin (5 mg) and CTAB (0.8 mg) were dissolved in 40 mL of chloroform, and then 10 mL of KMnO4 solution was added (180 mg/10 mL in distilled water). After 3 h of reaction at room temperature (20–25 °C), the oxidation products were separated using chloroform/methanol/distilled water (10:5:3, v/v/v). The organic layer was then collected, and the solvent was removed in vacuo. Astaxanthin oxidation was confirmed by TLC with RP-18 F254S plates (Merck Millipore, Burlington, MA, USA), performed using methanol. LC–MS was carried out using an LCMS-8040 (Shimadzu, Kyoto, Japan) spectrometer with an ODS-UG-3 (150 × 2.0 mm, Nomura Chemical Co., Inc., Aichi, Japan) column. The column temperature was set at 30 °C, and methanol was eluted as the mobile phase at a flow rate of 0.1 mL/min. A triple quadrupole mass spectrometer with electrospray ionization (positive ion mode) was used with a total ion scanning range of m/z 50–700 under the following conditions: nebulizer gas (N2, 2.0 L/min), drying gas (N2, 15.0 L/min), desolvation line temperature (250 °C), and heat block temperature (400 °C). Isolation of each astaxanthin oxidation product was conducted by HPLC with an SPD-M20A detector (Shimadzu) and a C30-UG-5 column (250 × 4.6 mm, Nomura Chemical Co.) eluted by methanol (1.0 mL/min flow rate).
5.3. Identification of Apo-11-astaxanthinal, Apo-10′-astaxanthinal, and Apo-8′-astaxanthinal Using 1H NMR Analysis
1H NMR (500 MHz) analysis, including 1H–1H COSY and NOESY, were conducted using a Varian UNITY INOVA 500 spectrometer in CDCl3.
5.4. Synthesis of Apo-15-astaxanthinal, Apo-14′-astaxanthinal, and Apo-12′-astaxanthinal
These apoastaxanthinals were chemically synthesized by Wittig condensation of previously reported phosphonium salt A(37) with acetal-aldehydes B,38D, and E(39) and subsequent acid-hydrolysis as shown in Scheme 1. Acetal-aldehyde D was prepared by dimethylacetalization of hydroxy-aldehyde C(40) and subsequent MnO2-oxidation. Experimental details are described in the Supporting Information.
5.5. Cell Culture
All cells used in this study were incubated in a humidified atmosphere of 95% air and 5% CO2 at 37 °C. RAW264.7 cells (passage number 15–25) were cultured in RPMI 1640 with 10% FBS containing 100 μg/mL streptomycin and 100 U/mL penicillin. Twenty-four hours preincubated RAW264.7 cells were treated with carotenoids (5 μM) dissolved in dimethyl sulfoxide (DMSO) and incubated for an additional 2 h. Control groups were treated with DMSO alone. To prevent cytotoxicity, DMSO was added to all culture media to 0.1%. LPS (100 ng/mL) was then added to the media, and the cells were stimulated for an additional 6 h (for mRNA expression analysis) or 24 h (for MCP-1, IL-6, IL-1β, and NO secretion analysis).
3T3-L1 preadipocytes (passage number 4) were cultured in DMEM with 10% FBS containing 100 μg/mL streptomycin and 100 U/mL penicillin. After reaching confluence (day 0), 3T3-L1 cells were incubated in fresh DMEM for another 2 days (day 0–2). To differentiate, the cells were replaced in fresh DMEM with 1 μM dexamethasone, 500 μM IBMX, and 10 μg/mL insulin and incubated for 2 days (day 2–4). On day 4, the media were replaced to fresh DMEM with 5 μg/mL insulin. The insulin-containing media were changed every 2 days. On day 10, the differentiated 3T3-L1 cells were used for further experiments (the picture of differentiated 3T3-L1 cells at day 10 in the Supporting Information).
5.6. Viability of RAW264.7 and 3T3-L1 Cells
RAW264.7 macrophages (2 × 104 cells/well) or differentiated 3T3-L1 cells were inoculated to 96-well culture plates and then treated with 5 μM of each carotenoid for 24 h. Then, WST-1 reagent (10 μL of each well) was added and incubated for an additional 4 h. The absorbance at 450 nm of each well was determined using a microplate reader (Molecular Devices, CA, USA). Media samples containing each carotenoid without cells were used as blanks.
5.7. Coculture of RAW264.7 and 3T3-L1 Cells
As previously described,23 RAW264.7 cells (1 × 105 cells/mL) were inoculated onto 3T3-L1 cells and incubated for an additional 24 h in the presence or absence of carotenoids without insulin. As a control, each cell, the number of which were equal to those in the contact system, was cultured separately and mixed after harvesting. The control group was treated with DMSO alone. To prevent cytotoxicity, DMSO was added to all culture media to 0.1%. Using a commercially available kit (Thermo Fisher Scientific, Frederick, MD, USA), the supernatant was subjected to enzyme-linked immunosorbent assay (ELISA). The adherent cells after removing the culture supernatant were subjected to mRNA expression analysis.
5.8. RAW-CM Preparation and Stimulation to Differentiated 3T3-L1 Adipocytes
RAW264.7-derived conditioned media (RAW-CM) were prepared following previous reports22,41 with a few modifications. In brief, RAW264.7 cells (5 × 104 cells/mL) in DMEM with 10% FBS were preincubated in 24-well plates for 48 h. The media was then replaced with DMEM with or without LPS (100 ng/mL). After 12 h of stimulation, the cells were then incubated in DMEM without LPS for an additional 12 h. The culture media were filtered using Millex-GP 0.22 μm (Merck Millipore, Burlington, MA, USA) and stored at −80 °C for further experiments (RAW-CM). The CM recovered from RAW264.7 cells treated with or without LPS was named RAW-CM(+) and RAW-CM(−), respectively. 3T3-L1 adipocytes were incubated in RAW-CM in the presence or absence of carotenoids for 24 h without insulin. The cells in the RAW-CM(−) group were treated with DMSO alone. To prevent cytotoxicity, DMSO was added to all culture media to 0.1%. After 24 h incubation in RAW-CM, the culture supernatant and the cells were subjected to ELISA and mRNA expression analysis, respectively.
5.9. Reverse Transcription Quantitative PCR
Total RNA was obtained using the QIAzol lysis reagent (Qiagen, Hilden, Germany). To synthesize cDNA from the total RNA, reverse transcription was performed using ReverTra Ace (Toyobo, Osaka, Japan) according to the manufacturer’s protocol. Gene expression level was quantified using GeneAce Probe qPCR Mix II (Nippon gene, Tokyo, Japan) on the StepOnePlus real-time PCR system (Applied Biosystems Japan Ltd., Tokyo, Japan). The PCR cycling condition was 50 °C for 2 min, 95 °C for 10 min, and 40 cycles of 95 °C for 30 s and 60 °C for 1 min. TaqMan Gene Expression Assays purchased from Thermo Fisher Scientific were follows: Nos2 (Mm00440502_m1), Il6 (Mm00446190_m1), Ccl2 (Mm00441242_m1), Ptgs2 (Mm00478374_m1), Il1b (Mm00434228_m1), Gapdh (Mm99999915_g1), and Actb (Mm00607939_s1). Relative quantification was performed using the standard curve method.42 The target quantity was divided by the endogenous (Gapdh or Actb) quantity to obtain a normalized target value.
5.10. Measurement of IL-6, IL-1β, MCP-1, and NO Levels in the Culture Supernatant
The levels of MCP-1, IL-6, and IL-1β in the culture supernatant were determined using a commercial ELISA kit. NO levels in the culture supernatant were determined using the Griess method.43 After LPS stimulation for 24 h, the culture media were collected and mixed with an equal amount of Griess reagent (0.1% N-[1-naphthyl]ethylenediamine dihydrochloride, 1% sulfanilamide, and 2.5% phosphoric acid in distilled water), and then the absorbance at 550 nm was determined. To calculate NO levels, a standard curve was created using NaNO2. For the blank sample, the media containing carotenoids without cells were used.
5.11. Statistics
The results are represented as the mean ± standard error of the mean (SEM). One-way ANOVA followed by the Tukey’s honest significant difference (HSD) test were used to evaluate the statistical difference (P < 0.05).
Acknowledgments
JSPS KAKENHI grant numbers 19J11147 and 18H02274 supported this work.
Glossary
Abbreviations Used
- LPS
lipopolysaccharide
- MCP-1
monocyte chemoattractant protein-1
- IL
interleukin
- Ptgs2
prostaglandin-endoperoxide synthase 2
- NO
nitric oxide
- Nos2
nitric oxide synthase 2
- Ccl2
C–C motif chemokine ligand 2
- RAW-CM
RAW264.7 derived conditioned media
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c01164.
Experimental details regarding synthesis of apo-15-astaxanthinal, (2E,4E)-6,6-dimethoxy-2-methylhexa-2,4-dienal, apo-14′-astaxanthinal, and apo-12′-astaxanthinal; pictures of differentiated 3T3-L1 adipocytes at day 10; and additional mRNA expression data of inflammatory cytokines in coculturing of RAW264.7 macrophages and 3T3-L1 adipocytes (PDF)
Author Contributions
N.T.: conceptualization, acquisition of data, funding acquisition, and writing of original draft. F.B.: conceptualization and editing of manuscript. Y.Y.: acquisition of data and editing of manuscript. T.M.: acquisition of data and editing of manuscript. K.M.: supervision and editing of manuscript. M.H.: conceptualization, funding acquisition, supervision, and editing of manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Higuera-Ciapara I.; Félix-Valenzuela L.; Goycoolea F. M. Astaxanthin: A review of its chemistry and applications. Crit. Rev. Food Sci. Nutr. 2006, 46, 185–196. 10.1080/10408690590957188. [DOI] [PubMed] [Google Scholar]
- Hussein G.; Sankawa U.; Goto H.; Matsumoto K.; Watanabe H. Astaxanthin, a carotenoid with potential in human health and nutrition. J. Nat. Prod. 2006, 69, 443–449. 10.1021/np050354+. [DOI] [PubMed] [Google Scholar]
- Guerin M.; Huntley M. E.; Olaizola M. Haematococcus astaxanthin: Applications for human health and nutrition. Trends Biotechnol. 2003, 21, 210–216. 10.1016/S0167-7799(03)00078-7. [DOI] [PubMed] [Google Scholar]
- Visioli F.; Artaria C. Astaxanthin in cardiovascular health and disease: Mechanisms of action, therapeutic merits, and knowledge gaps. Food Funct. 2017, 8, 39–63. 10.1039/c6fo01721e. [DOI] [PubMed] [Google Scholar]
- Grimmig B.; Kim S.-H.; Nash K.; Bickford P. C.; Douglas Shytle R. Neuroprotective mechanisms of astaxanthin: A potential therapeutic role in preserving cognitive function in age and neurodegeneration. GeroScience 2017, 39, 19–32. 10.1007/s11357-017-9958-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton G.Overview of carotenoid biosynthesis. In Carotenoids; Britton G., Liaaen-Jensen S., Pfander H., Eds.; Birkhäuser Verlag: Basel, Switzerland, 1998; Vol. 3, p 90. [Google Scholar]
- Palczewski K. G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 2006, 75, 743–767. 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephensen C. B. Vitamin A, infection, and immune function. Annu. Rev. Nutr. 2001, 21, 167–192. 10.1146/annurev.nutr.21.1.167. [DOI] [PubMed] [Google Scholar]
- Kiefer C.; Hessel S.; Lampert J. M.; Vogt K.; Lederer M. O.; Breithaupt D. E.; Von Lintig J. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J. Biol. Chem. 2001, 276, 14110–14116. 10.1074/jbc.M011510200. [DOI] [PubMed] [Google Scholar]
- Cooperstone J. L.; Novotny J. A.; Riedl K. M.; Cichon M. J.; Francis D. M.; Curley R. W. Jr.; Schwartz S. J.; Harrison E. H. Limited appearance of apocarotenoids is observed in plasma after consumption of tomato juices: A randomized human clinical trial. Am. J. Clin. Nutr. 2018, 108, 784–792. 10.1093/ajcn/nqy177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec R. E.; Riedl K. M.; Harrison E. H.; Curley R. W. Jr.; Hruszkewycz D. P.; Clinton S. K.; Schwartz S. J. Identification and quantification of apo-lycopenals in fruits, vegetables, and human plasma. J. Agric. Food Chem. 2010, 58, 3290–3296. 10.1021/jf100415z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoccali M.; Giuffrida D.; Salafia F.; Giofrè S. V.; Mondello L. Carotenoids and apocarotenoids determination in intact human blood samples by online supercritical fluid extraction-supercritical fluid chromatography-tandem mass spectrometry. Anal. Chim. Acta 2018, 1032, 40–47. 10.1016/j.aca.2018.06.022. [DOI] [PubMed] [Google Scholar]
- Fleshman M. K.; Lester G. E.; Riedl K. M.; Kopec R. E.; Narayanasamy S.; Curley R. W. Jr.; Schwartz S. J.; Harrison E. H. Carotene and novel apocarotenoid concentrations in orange-fleshed Cucumis melo melons: Determinations of β-carotene bioaccessibility and bioavailability. J. Agric. Food Chem. 2011, 59, 4448–4454. 10.1021/jf200416a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler A.; Liechti H.; Pichard L.; Wolz E.; Oesterhelt G.; Hayes A.; Maurel P. Metabolism and CYP-inducer properties of astaxanthin in man and primary human hepatocytes. Arch. Toxicol. 2002, 75, 665–675. 10.1007/s00204-001-0287-5. [DOI] [PubMed] [Google Scholar]
- Harrison E. H.; Dela Sena C.; Eroglu A.; Fleshman M. K. The formation, occurrence, and function of β-apocarotenoids: β-carotene metabolites that may modulate nuclear receptor signaling. Am. J. Clin. Nutr. 2012, 96, 1189S–1192S. 10.3945/ajcn.112.034843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharoni Y.; Linnewiel-Hermoni K.; Khanin M.; Salman H.; Veprik A.; Danilenko M.; Levy J. Carotenoids and apocarotenoids in cellular signaling related to cancer: A review. Mol. Nutr. Food Res. 2012, 56, 259–269. 10.1002/mnfr.201100311. [DOI] [PubMed] [Google Scholar]
- Cheng J.; Miao B.; Hu K.-Q.; Fu X.; Wang X.-D. Apo-10′-lycopenoic acid inhibits cancer cell migration and angiogenesis and induces peroxisome proliferator-activated receptor γ. J. Nutr. Biochem. 2018, 56, 26–34. 10.1016/j.jnutbio.2018.01.003. [DOI] [PubMed] [Google Scholar]
- Furman D.; Campisi J.; Verdin E.; Carrera-Bastos P.; Targ S.; Franceschi C.; Ferrucci L.; Gilroy D. W.; Fasano A.; Miller G. W.; Miller A. H.; Mantovani A.; Weyand C. M.; Barzilai N.; Goronzy J. J.; Rando T. A.; Effros R. B.; Lucia A.; Kleinstreuer N.; Slavich G. M. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019, 25, 1822–1832. 10.1038/s41591-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapouri-Moghaddam A.; Mohammadian S.; Vazini H.; Taghadosi M.; Esmaeili S. A.; Mardani F.; Seifi B.; Mohammadi A.; Afshari J. T.; Sahebkar A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018, 233, 6425–6440. 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- Fujiwara N.; Kobayashi K. Macrophages in inflammation. Curr. Drug Targets Inflamm. Allergy 2005, 4, 281–286. 10.2174/1568010054022024. [DOI] [PubMed] [Google Scholar]
- Oishi Y.; Manabe I. Macrophages in inflammation, repair and regeneration. Int. Immunol. 2018, 30, 511–528. 10.1093/intimm/dxy054. [DOI] [PubMed] [Google Scholar]
- Permana P. A.; Menge C.; Reaven P. D. Macrophage-secreted factors induce adipocyte inflammation and insulin resistance. Biochem. Biophys. Res. Commun. 2006, 341, 507–514. 10.1016/j.bbrc.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Suganami T.; Nishida J.; Ogawa Y. A paracrine loop between adipocytes and macrophages aggravates inflammatory changes: Role of free fatty acids and tumor necrosis factor α. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 2062–2068. 10.1161/01.ATV.0000183883.72263.13. [DOI] [PubMed] [Google Scholar]
- Burhans M. S.; Hagman D. K.; Kuzma J. N.; Schmidt K. A.; Kratz M. Contribution of adipose tissue inflammation to the development of type 2 diabetes mellitus. Compr. Physiol. 2018, 9, 1–58. 10.1002/cphy.c170040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilherme A.; Virbasius J. V.; Puri V.; Czech M. P. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat. Rev. Mol. Cell Biol. 2008, 9, 367–377. 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J.; Bai S. K.; Lee K. S.; Namkoong S.; Na H. J.; Ha K. S.; Han J. A.; Yim S. V.; Chang K.; Kwon Y. G.; Lee S. K.; Kim Y. M. Astaxanthin inhibits nitric oxide production and inflammatory gene expression by suppressing IκB kinase-dependent NF-κB activation. Mol. Cells 2003, 16, 97–105. [PubMed] [Google Scholar]
- Kim B.; Farruggia C.; Ku C. S.; Pham T. X.; Yang Y.; Bae M.; Wegner C. J.; Farrell N. J.; Harness E.; Park Y.-K.; Koo S. I.; Lee J.-Y. Astaxanthin inhibits inflammation and fibrosis in the liver and adipose tissue of mouse models of diet-induced obesity and nonalcoholic steatohepatitis. J. Nutr. Biochem. 2017, 43, 27–35. 10.1016/j.jnutbio.2016.01.006. [DOI] [PubMed] [Google Scholar]
- Etoh H.; Suhara M.; Tokuyama S.; Kato H.; Nakahigashi R.; Maejima Y.; Ishikura M.; Terada Y.; Maoka T. Auto-oxidation products of astaxanthin. J. Oleo Sci. 2012, 61, 17–21. 10.5650/jos.61.17. [DOI] [PubMed] [Google Scholar]
- Hayakawa T.; Kulkarni A.; Terada Y.; Maoka T.; Etoh H. Reaction of astaxanthin with peroxynitrite. Biosci. Biotechnol. Biochem. 2008, 72, 2716–2722. 10.1271/bbb.80358. [DOI] [PubMed] [Google Scholar]
- Takahashi S.; Waki N.; Mohri S.; Takahashi H.; Ara T.; Aizawa K.; Suganuma H.; Kawada T.; Goto T. Apo-12′-lycopenal, a lycopene metabolite, promotes adipocyte differentiation via peroxisome proliferator-activated receptor γ activation. J. Agric. Food Chem. 2018, 66, 13152–13161. 10.1021/acs.jafc.8b04736. [DOI] [PubMed] [Google Scholar]
- Takahashi N.; Kawada T.; Goto T.; Kim C.-S.; Taimatsu A.; Egawa K.; Yamamoto T.; Jisaka M.; Nishimura K.; Yokota K.; Yu R.; Fushiki T. Abietic acid activates peroxisome proliferator-activated receptor-γ (PPARγ) in RAW264.7 macrophages and 3T3-L1 adipocytes to regulate gene expression involved in inflammation and lipid metabolism. FEBS Lett 2003, 550, 190–194. 10.1016/S0014-5793(03)00859-7. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi K.; Kang M.-S.; Goto T.; Hirai S.; Ohyama K.; Kusudo T.; Yu R.; Yano M.; Sasaki T.; Takahashi N.; Kawada T. Citrus auraptene acts as an agonist for PPARs and enhances adiponectin production and MCP-1 reduction in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 2008, 366, 219–225. 10.1016/j.bbrc.2007.11.119. [DOI] [PubMed] [Google Scholar]
- Lefterova M. I.; Steger D. J.; Zhuo D.; Qatanani M.; Mullican S. E.; Tuteja G.; Manduchi E.; Grant G. R.; Lazar M. A. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol. Cell Biol. 2010, 30, 2078–2089. 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaulmann A.; Bohn T. Carotenoids, inflammation, and oxidative stress--implications of cellular signaling pathways and relation to chronic disease prevention. Nutr. Res. 2014, 34, 907–929. 10.1016/j.nutres.2014.07.010. [DOI] [PubMed] [Google Scholar]
- Yang C.-M.; Huang S.-M.; Liu C.-L.; Hu M.-L. Apo-8′-lycopenal induces expression of HO-1 and NQO-1 via the ERK/p38-Nrf2-ARE pathway in human HepG2 cells. J. Agric. Food Chem. 2012, 60, 1576–1585. 10.1021/jf204451n. [DOI] [PubMed] [Google Scholar]
- Wu L.; Lyu Y.; Srinivasagan R.; Wu J.; Ojo B.; Tang M.; El-Rassi G. D.; Metzinger K.; Smith B. J.; Lucas E. A.; Clarke S. L.; Chowanadisai W.; Shen X.; He H.; Conway T.; von Lintig J.; Lin D. Astaxanthin-shifted gut microbiota is associated with inflammation and metabolic homeostasis in mice. J. Nutr. 2020, 150, 2687–2698. 10.1093/jn/nxaa222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano Y.; Sato Y.; Watanabe Y.; Namikawa K.; Miki W.; Ito M. Carotenoids and related polyenes. Part 6. Stereoselective synthesis of astaxanthin analogues and their antioxidant activities. J. Chem. Soc. Perkin Trans. I 2001, 16, 1862–1869. 10.1039/b104338m. [DOI] [Google Scholar]
- Martin H.-D.; Jäger C.; Ruck C.; Schmidt M.; Walsh R.; Paust J. Anti- and prooxidant properties of carotenoids. J. Prakt. Chem. 1999, 341, 302–308. . [DOI] [Google Scholar]
- Bernhard K.; Englert G.; Mayer H.; Müller R. K.; Rüttimann A.; Vecchi M.; Widmer E.; Zell R. Synthese von optisch aktiven, natürlichen carotinoiden und strukturell verwandten naturprodukten. IX. Synthese von (3R)-hydroxyechinenon, (3R, 3′R)- und (3R, 3′S)-adonixanthin, (3R)-adonirubin, deren optischen antipoden und verwandten verbindungen. Helv. Chim. Acta 1981, 64, 2469–2484. 10.1002/hlca.19810640754. [DOI] [Google Scholar]
- Barrero A. F.; Herrador M. M.; Arteaga P.; Gil J.; González J.-A.; Alcalde E.; Cerdá-Olmedo E. New apocarotenoids and β-carotene cleavage in Blakeslea trispora. Org. Biomol. Chem. 2011, 9, 7190–7195. 10.1039/c1ob05712j. [DOI] [PubMed] [Google Scholar]
- Yang H.-E.; Li Y.; Nishimura A.; Jheng H.-F.; Yuliana A.; Kitano-Ohue R.; Nomura W.; Takahashi N.; Kim C.-S.; Yu R.; Kitamura N.; Park S.-B.; Kishino S.; Ogawa J.; Kawada T.; Goto T. Synthesized enone fatty acids resembling metabolites from gut microbiota suppress macrophage-mediated inflammation in adipocytes. Mol. Nutr. Food Res. 2017, 61, 1700064. 10.1002/mnfr.201700064. [DOI] [PubMed] [Google Scholar]
- Čikoš Š.; Bukovská A.; Koppel J. Relative quantification of mRNA: Comparison of methods currently used for real-time PCR data analysis. BMC Mol. Biol. 2007, 8, 113. 10.1186/1471-2199-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. K.; Cheon B. S.; Kim Y. H.; Kim S. Y.; Kim H. P. Effects of naturally occurring flavonoids on nitric oxide production in the macrophage cell line RAW 264.7 and their structure–activity relationships. Biochem. Pharmacol. 1999, 58, 759–765. 10.1016/S0006-2952(99)00160-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.