Abstract
In the process of structure-function studies on the class II molecule expressed in autoimmunity prone SJL mice, I-As, we discovered a disparity from the reported sequence of the MHC class II beta chain. The variant is localized at a highly conserved site of the beta chain, at residue 58. Our studies revealed that this single amino acid substitution of Pro for Ala at this residue, found in I-As, changes the structure of the MHC class II molecule, as evidenced by a loss of recognition by two monoclonal antibodies, and elements of MHC class II conformational stability identified through molecular dynamics simulation. Two other rare polymorphisms in I-As involved in hydrogen bonding potential between the alpha chain and the peptide main chain are located at the same end of the MHC class II binding pocket, studied in parallel may impact the consequences of the β chain variant. Despite striking changes in MHC class II structure, CD4 T cell recognition of influenza-derived peptides was preserved. These disparate findings were reconciled by discovering, through monoclonal antibody blocking approaches, that CD4 T cell recognition by I-As restricted CD4 T cells focused more on the region of MHC class II at the peptide’s amino terminus. These studies argue that the conformational variability or flexibility of the MHC class II molecule in that region of I-As select a CD4 T cell repertoire that deviates from the prototypical docking mode onto MHC class II peptide complexes. Overall, our results are consistent with the view that naturally occurring MHC class II molecules can possess polymorphisms that destabilize prototypical features of the MHC class II molecule but that can maintain T cell recognition of the MHC class II:peptide ligand via alternate docking modes
Keywords: CD4 T cells, MHC class Il structure, polymorphism, autoimmunity
1. Introduction
The proteins encoded by the major histocompatibility complex locus (MHC or H-2 in mice, HLA in humans) play a crucial role in the adaptive immune response. MHC class I molecules present antigenic or self-peptides to CD8+ T cells, while MHC class II molecules serve the same function for CD4+ T cells. The MHC locus is the most polymorphic among mammalian genes, a feature responsible for the ability of MHC to present a broad diversity of pathogen-derived peptides and ensure a protective immune response to pathogenic organisms (de Bakker and Raychaudhuri, 2012; Trowsdale and Parham, 2004). Paradoxically, this genetic variability is also often associated with disease, with one of the strongest associations being tissue-specific autoimmunity (Karami et al., 2019). Among the many genes that contribute to complex dysregulation in autoimmunity, the biggest influence is commonly linked to MHC class II genes (Hollenbach and Oksenberg, 2015; Jacobi et al., 2020; Joubert and Dalmau, 2019; Megiorni and Pizzuti, 2012; Pociot and Lernmark, 2016; Sospedra and Martin, 2016). This dominant role is likely due to the key regulatory role that MHC class II-restricted antigen presentation plays in many aspects of T cell-dependent immune responses. One of the best examples of this documented link between MHC class II and autoimmunity is the spontaneous model of insulin-dependent diabetes in non-obese diabetic (NOD) mice. These mice express the MHC class II allele I-Ag7, which shares a polymorphism with the diabetes-associated human allele HLA-DQ in the beta chain at the periphery of the peptide binding groove (Noble, 2015; Pociot and Lernmark, 2016; Todd et al., 1987). This loss of aspartic acid in the beta chain at amino acid Asp57 eliminates the potential for an inter-chain salt bridge with alpha Arg76. This polymorphism, like others linked to T cell-dependent autoimmunity in humans, is thought to control defective self-tolerance induction to self-peptides or acquisition of such peptides to initiate or propagate the autoimmune cascade. Other MHC class II genes in both mice and humans have been linked to other specific diseases (Hollenbach and Oksenberg, 2015; Karami et al., 2019; Megiorni and Pizzuti, 2012; Trowsdale and Knight, 2013).
We became intrigued with a particular MHC class II allele associated with autoimmunity, I-As, expressed in the SJL mouse, and associated with several autoimmune syndromes employed as models for multiple sclerosis (MS). These include experimental autoimmune encephalomyelitis (EAE) (Glatigny and Bettelli, 2018; Robinson et al., 2014), induced by vaccination with myelin basic protein or other central nervous system components, and Theiler’s murine encephalomyelitis virus (TMEV) (Miller et al., 2001; Oleszak et al., 2004; Tompkins et al., 2002), initiated by viral infection in the central nervous system. The I-As molecule was noted to possess an exceptionally rare set of polymorphisms in the alpha chain (His68Tyr and Asn69Thr) (Kalbus et al., 2001; Landais et al., 1985). In all other mouse I-A molecules sequenced to date, these two highly conserved residues (alpha His68 and alpha Asn69) constitute part of the general features of peptide:MHC class II interactions visualized in the early crystal structures of MHC class II molecules (Jardetzky et al., 1996; Stern et al., 1994). Our previous studies had revealed that these conserved amino acids at the periphery of the peptide binding pockets form hydrogen bonds with the peptide main chain and contribute to the kinetic stability of peptide binding to the MHC class II molecule (Bryant et al., 1999; McFarland et al., 2001; Sant et al., 1999). In the process of studying the impact of these polymorphisms in the I-As alpha chain, we discovered that the I-As beta chain possessed an unusual mutation that was not reported in the original sequencing of this gene (Estess et al., 1986). In the study we report here, we provide evidence for an associated conformational and physical impact on I-As based on serological reactivity of monoclonal antibodies, molecular modeling and CD4 T cell recognition. These studies suggest a generalized model for the role of MHC polymorphism in T cell recognition and defects in self-tolerance induction (Giarratana et al., 2007; J. A. Pearson et al., 2016).
2. Materials and Methods
2.1. Animals and infections.
Female B10.S (H-2s), SJL and C3H mice, purchased from Jackson laboratories, were maintained in a specific pathogen-free facility at the University of Rochester Medical Center, according to institutional guidelines. For influenza infections, mice were anaesthetized with avertin (2,2,2-Tribromoethanol) via intraperitoneal injection and infected intranasally with 5 × 104 50% egg infectious dose (EID50) of influenza A/New Caledonia/20/1999 (H1N1) virus. All experimental groups were age-matched and used between 8–16 weeks of age.
2.1.1. Ethics Statement.
Mice were maintained in a specific pathogen-free facility at the University of Rochester Medical Center according to institutional guidelines. All animal protocols used in this study adhere to the AAALAC International, the Animal Welfare Act and the PHS Guide, and were approved by the University of Rochester Committee on Animal Resources, Animal Welfare Assurance Number A3291–01. The protocol under which these studies were conducted was originally approved March 4, 2006 (protocol # 2006–030) and has been reviewed and re-approved every 36 months with the most recent review and approval December 29, 2020.
2.2. DNA-mediated transfection and cell maintenance.
Ltk− cells were transfected by calcium phosphate precipitation with cDNA genes encoding the WT I-As α and β chains (I-As (β-Ala58) or I-As (β-Pro58)) that had been cloned into the SV40 based vector pcEXV as we have previously described (Loss and Sant, 1993; Peterson and Miller, 1990), plus the neomycin resistance gene pSV2neo. Transfected, drug resistant cells were selected for MHC class II expression using mAb based sorting and cloned by limiting dilution to maintain homogeneous MHC class II expression. They were maintained in selection media and stained for MHC class II expression every 2–3 weeks, but were removed from selective drug for 2 d before T cell assays.
2.3. Monoclonal Antibody staining and flow cytometry.
Monoclonal antibodies (mAb) were produced from cultured hybridoma cells. Supernatant collected at high cell density was used at saturating doses (typically 1:4) in a 2-step staining procedure as previously described (Sant et al., 1987). Culture media or the I-E specific antibody, 14–4-4S (Ozato et al., 1980), plus a secondary Goat anti-mouse IgG were used as negative controls. Transfected cells were harvested from culture, washed, pelleted and resuspended in ice-cold PBS supplemented with 2.5% bovine calf serum and 0.1% sodium azide (PBS-2.5). Cells (5 X 105) were placed in round bottom 96-well plates with saturating amounts of mAb as culture supernatant, and incubated at 4°C for 45 min. The cells were washed twice with PBS-2.5 and incubated with a saturating concentration of FITC GAM Ig (Cappel Laboratories). Cells were incubated at 4°C for 45 min, washed and analyzed on an Accuri flow cytometer. Data were analyzed using histograms generated using Flowjo software (FlowJo, LLC), version 10. In these assays, more than 90% of cells were MHC class II positive and the data are represented as the mean fluorescent intensity (MFI) of the population.
2.4. Peptides.
17-mer peptides overlapping by 11 amino acids of the HA and NA from the H1N1 virus A/New Caledonia/20/99, and the NP from the H1N1virus A/New York/348/2003 were obtained from BEI Resources, ATCC.
2.5. EliSpot assays.
EliSpot assays were performed as described (Nayak et al., 2010; Richards et al., 2010). Briefly, 96-well filter plates (Millipore) were coated with rat anti-mouse IL-2 or IFN-γ (clone JES6–1A12 and clone AN-18, respectively, BD Biosciences), washed and blocked. CD4 T cells were co-cultured with APC and with the indicated peptide at a final concentration as indicated in the Figure for 18–20 h at 37°C and 5% CO2. For blocking experiments, 50μl of antibody supernatant 14.4.4S (I-As negative control), MKD6 (I-Ak negative control), 10.2–16 and 3F12 were added to the cultures, APC and antibody were incubated for 30 minutes at room temperature prior to the addition of CD4 T cells and peptides. Plates were washed and incubated with biotinylated rat anti-mouse IL-2 or IFN-γ (clone JES6–5H4, clone XMG1.2, BD Biosciences), and developed, dried and quantified with an Immunospot reader series 2A, using Immunospot software, version 3.2.
2.6. Molecular dynamics simulation.
Starting coordinates for I-Ak presenting a 13mer fragment of hen egg lysozyme were obtained from the Protein Data Bank, accession code 1IAK (Fremont et al., 1998). Three I-Ak variants were also simulated to probe differences in dynamics compared to I-As, for which there is no crystal structure: αHis68Tyr/Asn69Thr, βAla58Pro, and a combination of these. Mutations were introduced computationally from the wild type I-Ak starting coordinates utilizing Rosetta (Das and Baker, 2008) via the PyRosetta (Chaudhury et al., 2010) interface. After this, the site of mutation and three residues upstream and downstream were remodeled using cyclic coordinate descent (Canutescu and Dunbrack, 2003). Molecular dynamics simulations were performed as described previously (Devlin et al., 2020). Briefly, complexes were simulated using GPU-accelerated implementation of AMBER16, the ff14SB force field, and explicit SPC/E water. After minimization, simulations were heated to 300 K while restrained to initial coordinates. Restraints were relaxed over 20 ps after which systems were allowed to equilibrate with no restraints for 20 ns. Production trajectories were calculated for 500 ns each, conducted in triplicate for each variant. Hydrogen bond occupancy, secondary structure, RMS fluctuations, and coordinate RMS deviations were calculated from production trajectories using the cpptraj package. Occupancies and RMS fluctuations were averaged for the three triplicate simulations.
3. Results
The SJL mouse has been a widely used model for multiple sclerosis (MS) for a number of years. The neurological disorder can be initiated via priming with central nervous system (CNS) expressed antigens (Chastain et al., 2011; Kuchroo et al., 2002). As with other models of autoimmune diseases, like the prototypic non-obese diabetic (NOD) mouse, both MHC class II and non-MHC genes play a role in disease susceptibility (Anderson and Bluestone, 2005; Ridgway et al., 2008; Shapiro et al., 2021). In the process of structure-function studies on the MHC class II molecule expressed in SJL mice, I-As, we discovered a disparity from the reported sequence published by Estess in 1986 (Estess et al., 1986), studying the gene sequence of I-Aβs from the A.TH strain that expresses I-As. The original published sequence placed alanine at β58, the most common and highly conserved amino acid at this beta chain residue (Figure 1). This common allele sequence at β58 in I-As has been published numerous times in the literature (Acha-Orbea and McDevitt, 1987; Beck et al., 1987; Holmdahl et al., 1989; C. I. Pearson et al., 1999). Our sequencing of the beta chain from commercially purchased SJL mice and B10.S mice, which share the I-As molecule, revealed that the β chains in both mice possess proline instead of alanine at β58 (Figure 1). Sequencing of this gene in these common inbred strains of mice was performed several times with the same result (Supplemental Figure 1).
Figure 1. Sequence of common MHC class II I-A beta chain alleles.
Shown are the amino acid sequences of the beta chain alleles of commonly used mouse strains and common human alleles, as well as two alleles of I-A that have a polymorphism at position 58 (boxed). The original I-As sequence was reported as Ala, but our studies of inbred SJL and B10.S mouse strains that express I-As found a proline at beta 58. The adjacent beta Asp 57 forms a critical inter-chain salt bridge with alpha Arg 76, that is substituted for Ser in autoimmune prone NOD mice as well as other autoimmunity prone human alleles.
We were intrigued by this anomaly in I-As, in part because it was adjacent to the widely studied β57 residue that has been implicated in autoimmune diabetes (Todd et al., 1988). Also of interest was its location at the periphery of the peptide binding pocket of MHC class II molecules at the peptides carboxy terminus, across the MHC class II binding site from the other rare polymorphisms in the alpha chain (α68 and α69), discussed previously that we have found. To evaluate the consequences of the unusual amino acid variation in the I-As beta chain in expression and structure of I-As, stable transfectants were constructed with the reported sequence (β-Ala58) or the correct, mouse wild type strain-derived sequence (β-Pro58), with the wild type I-As alpha chain, all expressed as cDNA and introduced into MHC negative Ltk-fibroblasts, as we have previously described (Loss and Sant, 1993; Sant and Germain, 1989). Stable Ltk-transfectants expressing the WT I-Aα chain and either the I-As (β-Pro58) found naturally occurring in SJL and B10.S mice, or the mutated I-As (β-Ala58) at similar cell surface densities were then analyzed in detail. Cell surface expression of MHC class II molecules was monitored by flow cytometry with well-characterized anti-murine MHC class II molecules, including four monoclonal antibodies known to be reactive with I-As. Two of these I-As reactive mAb (3F12 and 3JP) were known to react with the I-A alpha chain and two (10.2–16 and MKS4) reacted with the beta chain. All antibodies tested, with their previously defined specificities, are shown in Table I. Conclusions about the sites of recognition for the mAb, cited in this paper, were inferred in published studies by testing spleen cells from I-A disparate mice, by mutagenesis of the alpha or beta chains or through genetic exchange of MHC class II gene segments followed by transfection. Antibodies previously defined to react with I-As included MKS4 which was elicited against I-As expressing splenocytes and recognizes an epitope on I-As, I-Au and I-Af (Kappler et al., 1981), that is not expressed by other common MHC class II molecules that have been tested, including I-Ak and I-Ad. 10.2–16 (Oi et al., 1978) is reactive to many class II molecules (I-As, I-Au, I-Af, I-Ag7, I-Ar and I-Ak), but fails to react with common I-Ad and I-Ab expressed in BALB/c and C57BL/6 mice, respectively. It is thought to recognize an epitope that is dependent on a 2-amino acid deletion in the beta chain at (β65 and β66) and requires Glu at position 69 (see Figure 1). 3F12 detects most class II molecules examined, including (I-Ab,k,r,s,q) but not I-Ad (Beck et al., 1986), and its epitope has been mapped to the alpha1 domain around residue 44 (Wei et al., 1994), while 3JP detects specificities associated with the alpha chain and is broadly reactive with many class II molecules, including I-Ak,r,s,q,b but not I-Ad (Braunstein et al., 1990; Landias et al., 1986). We included antibodies not previously identified to react with I-As because of the unpredictable nature of the consequences of the induced mutation and “correction” to the highly conserved Ala at β58.
Table I.
Monoclonal antibodies tested and their previously defined specificities.
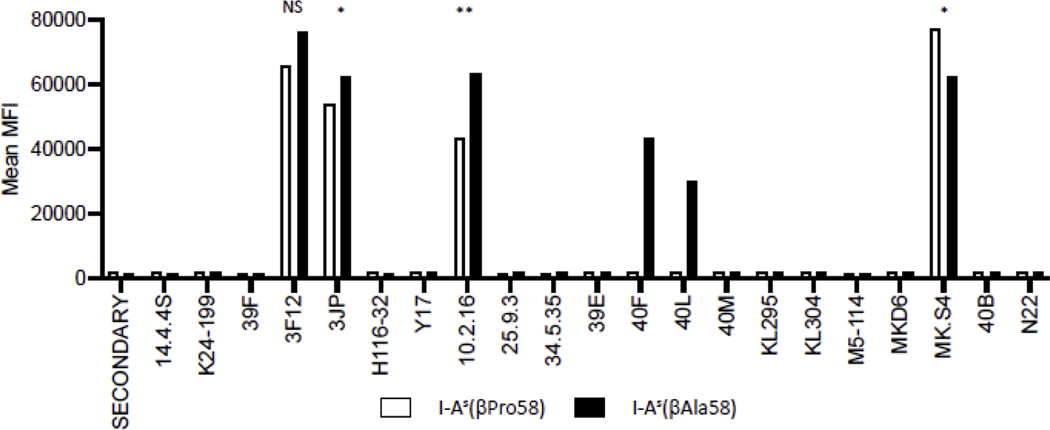
Figure 2 shows the results of flow cytometry experiments. Of the 20 mAb studied, four detected epitopes express by both I-As β-Pro58 or I-As β-Ala58 class II molecules. When additional mAb were tested, many detected neither class II molecule. Strikingly, however, two mAb (40F and 40L) that were previously shown to fail to react with I-As when spleen cells expressing this MHC class II allele have been tested (Pierres et al., 1981) and that has been assigned as beta chain specific, regained reactivity when cells expressing I-As β-Ala58 (Figure 2, black bars). This result suggests this single amino acid residue alters the properties of the beta chain, where Pro at amino acid 58, the natural amino acid in I-As, disrupts these epitopes. Thus, the replacement of Pro to Ala at β58 can be viewed as a gain in function change, restoring expression of a commonly expressed antibody epitope. This is also notable because other class II alleles including I-Ak, I-Af, and I-Au that share some sequence similarity with I-As but include β–Ala58 also react with 40F and 40L (Landias et al., 1986). The failure of reactivity of 40F and 40L to detect the I-As in splenocytes from mice and constructed APC reported in these studies is also notable because these antibodies were produced and characterized in 1981 (Pierres et al., 1981), indicating that this rare Pro at β58 has been expressed in I-As in the common SJL and B10.S inbred strain for at least 40 years.
Figure 2. Reactivity of APC expressing either I-As(βPro58) or its variant I-As(βAla58) with monoclonal antibodies (mAb).
Transfectants expressing either I-As(βPro58), white bars, or I-As(βAla58), black bars, were stained with the indicated mAb and then mean fluorescence intensity (MFI) of the population, which was >90% positive, was scored by flow cytometry in a two-step staining procedure. The MFI is indicated for each monoclonal antibody. Shown here is a representative figure. Each antibody was tested in at least two independent experiments. Statistical analysis was completed for mAb with reactivity to both MHC class II molecules, using an unpaired t-test.
Also of interest from the mAb and flow cytometry analyses were the results obtained with the widely used mAb 10.2–16. All murine MHC class II molecules that share the deletion at β65–66 segment react with 10.2–16 which is associated with the Ia.17 serological specificity. Early studies with 10.2–16 revealed positive but modestly diminished reactivity with I-As, relative to the common I-Ak and I-Af class II molecules (Oi et al., 1978). The “correction” of Proβ58 to Alaβ58 in the I-As transfectants studied here leads to a reproducible gain in reactivity with 10.2–16 that reached statistical significance (Figure 2 and Supplemental Figure 2). Collectively, these studies show that substitution of the rare polymorphism of β-Pro58 with the more common β-Ala58 in the I-As molecule leads to a complete “gain of function” phenotype with two monoclonal antibodies (40L and 40F) as well as a quantitative gain in reactivity with an additional mAb (10.2–16) an epitope localized to this region of the I-A beta chain.
To evaluate the conformational consequences of the novel βPro58 polymorphism in I-As, we structurally modeled the polymorphism and examined its impact via molecular dynamics simulations. Figure 3 illustrates the MHC class II structure. No crystal structure of I-As has been solved on which to base the modeling. Therefore, we used the crystallographic structure of the closely related and well-characterized I-Ak molecule (Fremont et al., 1998) as a reference structure (Beck et al., 1986; Braunstein et al., 1990). I-Ak shares many features of the beta chain with I-As, as well as some monoclonal antibody epitopes with I-As. The I-Ak MHC class II molecule, like I-As also shares a small deletion in the beta chain at amino acids 65 and 66 (see Figure 1), suggesting this to be the best structure to begin modeling with. The coordinates for I-Ak molecule complexed to HEL 46–61 peptide (Fremont et al., 1998) were used to produce three structural models representing I-As: one incorporating only the His68→Tyr and Asn69→Thr polymorphisms present in the I-As α chain, a second incorporating only the newly discovered Ala58→Pro in the β chain, and a third incorporating all three. These three models and wild type I-Ak were used as starting coordinates for 500 ns of molecular dynamics simulations, in explicit solvent replicated in triplicate for each protein variant. The simulations showed changes in dynamic character as discussed below, but comparisons among the three replicates demonstrated reproducibility and convergence (Supplemental Figure 3).
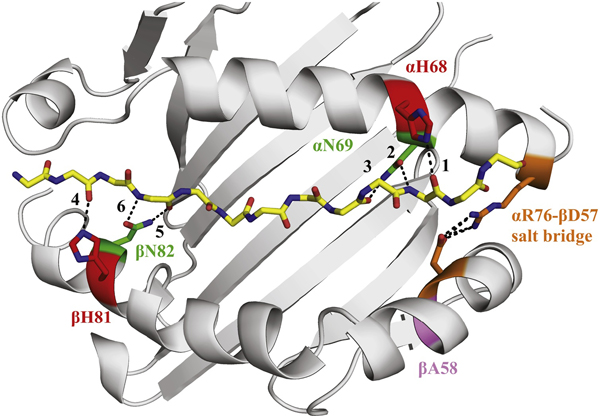
Figure 3. Structure of I-Ak presenting the HEL peptide highlighting key residues.
The structure of I-Ak presenting the HEL peptide was used for molecular dynamics simulations, with key positions highlighted. The polymorphic residues αH68 and αN69 form hydrogen bonds to the peptide near its C-terminus. The polymorphic site βA58 is within the shorter arm of the β helix, across the groove from αH68 and αN69. βH81 and βN82 are highlighted as control sites examined for hydrogen bond character. The αR76-βD57 salt-bridge is highlighted because of its known role in the stability of this region of class II. Hydrogen bonds between the peptide main chain and the MHC class II alpha helices that have been investigated in the simulations are numbered 1–6.
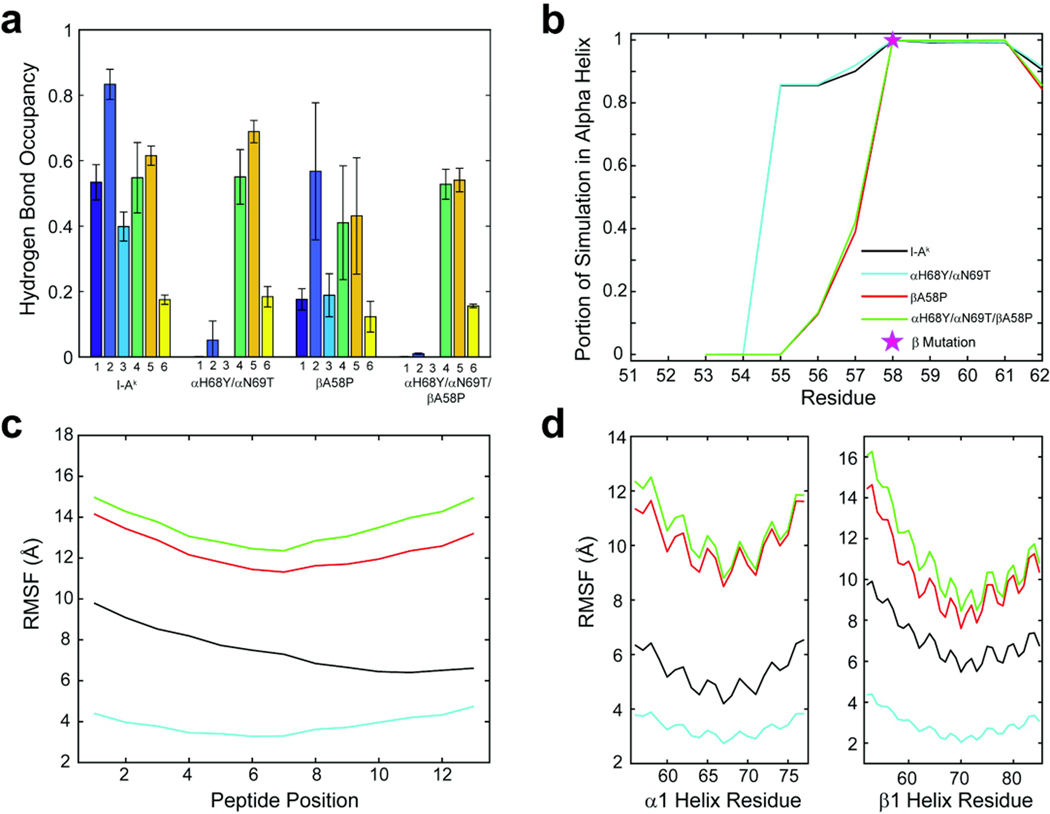
As noted, αHis68 and αAsn69 play a critical role in stabilizing presented peptides (McFarland et al., 2001; McFarland et al., 2005; Sant et al., 1999) via hydrogen bonds with the peptide backbone near the peptide C-terminus and are present in nearly all I-A complexes, except I-As (Landais et al., 1985) (hydrogen bonds labeled 1–3 in Fig. 3). To understand how the polymorphisms present in I-As might disrupt these interactions and how the unique features of I-As might interact with each other, we investigated the occupancy of these hydrogen bonds in the simulations. As expected, the alpha chain substitutions αH68Y and αN69T in I-As exhibited significantly decreased hydrogen bond occupancy (Figure 4). Hydrogen bonds from the beta chain to the N-terminal half of the peptide (hydrogen bonds 4–6 in Fig. 3) were largely unaffected, indicating the disruptions are localized around the polymorphic sites. Surprisingly, βA58P alone was associated with a significant decrease in C-terminal hydrogen bond occupancies. The combination of all three substitutions resulted in a total loss of the C-terminal hydrogen bonds in the simulations. To probe the effect of these substitutions on the conformational stability of the β1 helix, we investigated the secondary structure of this region around the β58 residue (Figure 5A). Consistent with proline’s role as a “helix breaker”, compared to I-Ak, the βA58P polymorphism eliminated helical character in residues 54–55 and reduced helical character in residues 56–57.
Figure 4. Polymorphisms that distinguish I-Ak from I-As impact peptide-protein hydrogen bonds and α helix character.
Average occupancy of hydrogen bonds 1–6 as indicated in Fig. 3 between residues α68 and α69, and the peptide backbone. Bars are shown as the mean occupancy over the course of simulations and error bars are the population standard deviation between the three replicate simulations for each variant. Hydrogen bonds are denoted by a number on the x-axis, which corresponds to the labels in Figure 3.
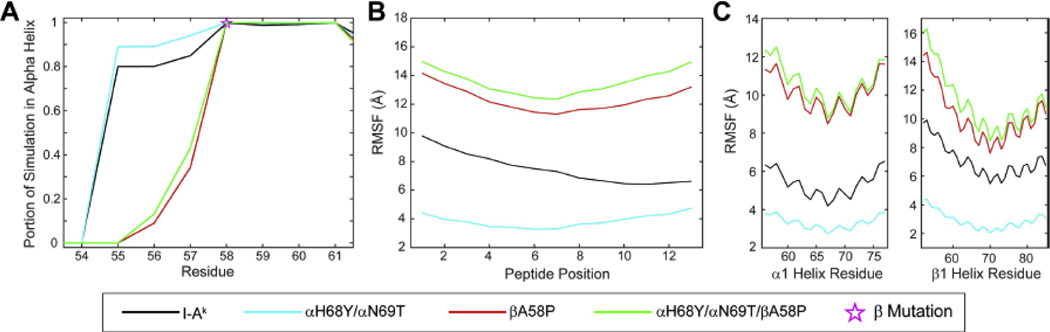
Figure 5. Peptide and protein fluctuations in the molecular dynamic simulations.
In panel A, the proportion of simulation that the β1 helix spent in alpha helical dihedral space. The β58 position is denoted by a pink star. The root mean square (RMS) fluctuations of the peptide calculated over the lengths of the simulations and shown in panel B and panel C illustrates the RMS fluctuations of select regions of the alpha1 and beta1 helices over the lengths of the simulations.
We next examined peptide and protein fluctuations in the molecular dynamics simulations by calculating root mean square (RMS) fluctuations for amino acid α carbons. Compared to I-Ak, the αH68Y and αN69T polymorphisms reduced fluctuations across the peptide and the α1 and β1 helices of the peptide binding groove (Figure 5B and C, Supplemental Figure 3), possibly due to a loss of correlated motion. Introduction of the βA58P polymorphisms, whether alone or in combination with αH68Y and αN69T, substantially increased fluctuations. Altogether, the modeling and simulations suggest that the three polymorphisms in I-As act to influence the conformational stability of the C-terminal half of the peptide as well as the adjacent alpha helix of the β chain. These differences could result in a different surface as “seen” by antibodies and T cell receptors and could significantly alter their recognition.
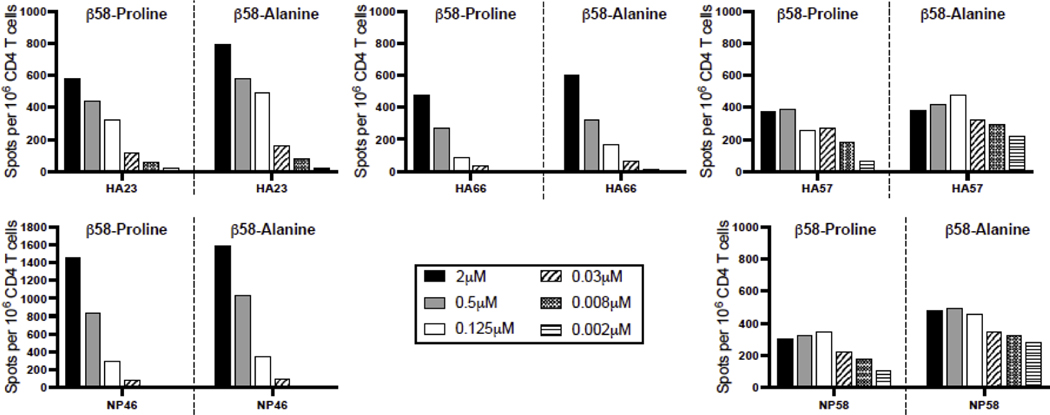
Because of the evidence for conformational and dynamic alterations that this single amino acid residue in the beta chain imparts, it was of interest to evaluate CD4 T cell recognition. The polyclonal CD4 T cell response to series of influenza-derived peptide epitopes using I-As βPro58 and I-AsβAla58 were examined. B10.S mice, expressing the I-As as its only MHC class II molecule, were infected with A/New Caledonia/20/99 influenza A virus, as we have previously described (Richards et al., 2009; Richards et al., 2018). CD4 T cells were assayed for recognition of the two alternative I-As molecules (I-As (β-Ala58) or I-As (β-Pro58)) by EliSpot assays using peptides previously identified by our laboratory as being immunodominant in H-2s mice (Knowlden and Sant, 2016; Nayak et al., 2010) and the pair of constructed antigen presenting cells that express the two alternative class II molecules that differ only at β58 used in the flow cytometry experiments. The results of these assays are shown in Figure 6 and Supplemental Figure 4. For the most part, intriguingly, there was little to no evidence of preferential recognition of the I-As (β-Pro58) relative to I-As (β-Ala58), despite the CD4 T cells being selected in vivo by the I-As (β-Pro58) MHC class II molecule. Even as the dose of peptide was reduced to suboptimal levels, both MHC class II molecules were able to capture the peptide epitopes and present them to the infection-elicited CD4 T cells. In fact, for some epitope specificities such as HA23 and NP46, there was a modest enhancement of recognition of the I-As (β-Ala58) class II molecule. Overall, these results suggest that the regions of the MHC class II protein that are relevant to peptide acquisition and TcR recognition elicited with the rare allelic variant proline at amino acid 58 in the beta chain are not disrupted by the alanine substitution.
Figure 6. CD4 T cell recognition of APC expressing either I-As(βPro58) or its variant I-As(βAla58).
B10.S mice were infected with A/New Caledonia/20/99 H1N1 influenza virus and at 10 days post infection CD4 T cells were isolated from spleen and tested for reactivity by production of IFNγ with the indicated influenza peptides, previously identified by our laboratory. Graded doses of the indicated peptide were added to the cultures and cytokine producing cells were enumerated using cytokine ELISPOT assays. Results are presented as the spots per million CD4 T cells with background subtracted. Each panel is a separate influenza peptide epitope with the response with APC I-As (βPro58) to the left and APC I-As(βAla58) to the right. Shown here is a representative figure. Each peptide was tested in at least two independent experiments. There are no statistically significant differences in CD4 T cell recognition of APC I-As (βPro58) and its variant I-As (βAla58).
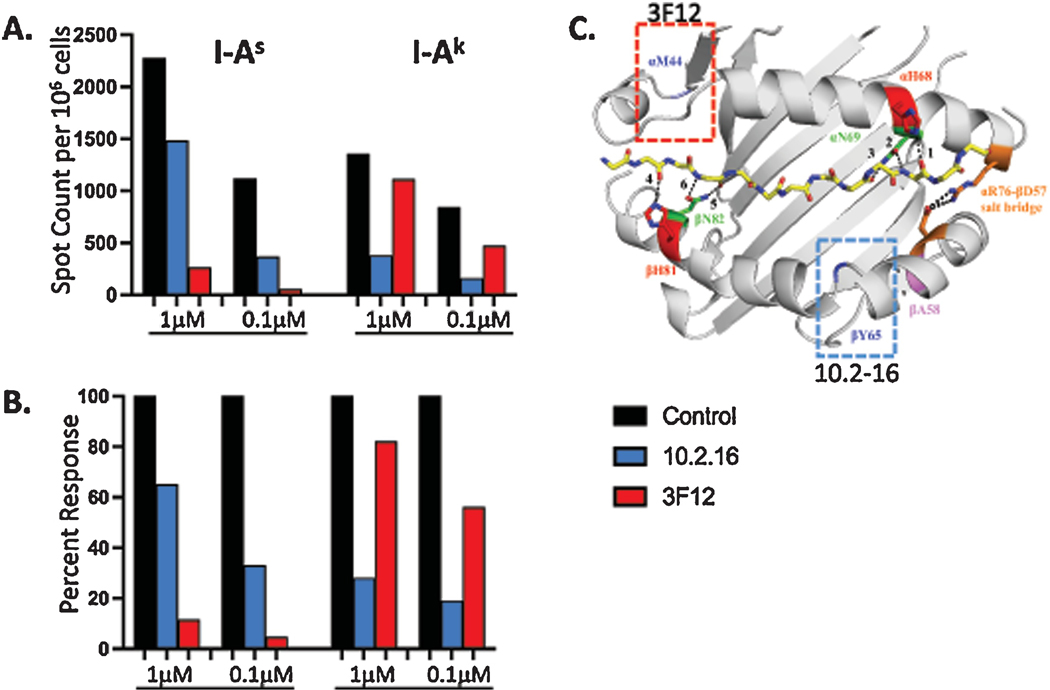
Based on the modest impact of the Ala>Pro substitution at β58 on CD4 T cell recognition, despite the evidence for dynamical changes in this region, we considered a model to explain this effect. We hypothesized that the rare polymorphisms (α-68,69 and β-58) that change the local structure of the MHC class II:peptide complex at the peptide carboxyterminal region have minimal impact on T cell recognition because of a preference in the manner in which the TcR repertoire engages I-As. Atypical TcR docking modes have been previously observed with some MHC molecules, particularly those associated with autoimmunity and unusual MHC sequences at the periphery of the MHC binding pocket (Deng and Mariuzza, 2007; Kerr et al., 2008; Li et al., 2005; Wucherpfennig et al., 2009). To evaluate this possibility, we performed antibody blocking experiments with antigen-specific CD4 T cells using two well characterized antibodies that detect epitopes at opposite ends of the binding pocket (see Figure 7C). 3F12 is an alpha chain-specific antibody thought to bind around residue 44 towards the peptide’s amino terminus (Wei et al., 1994). In contrast, 10.2–16 is a beta chain-specific antibody that recognizes an epitope associated with a 2-amino acid deletion in the beta chain at residues 65–66 expressed by the class II alleles I-Ak, I-Ag7, I-Ar, I-As and I-Au; the Ia17 antibody specificity (Braunstein et al., 1990; Landias et al., 1986). B10.S mice, expressing I-As and C3H mice that express the prototypical and extensively studied I-Ak molecule, were primed with influenza virus to initiate a broad CD4 T cell response. The elicited CD4 T cells were incubated with previously determined antigenic peptides (Nayak et al., 2010) in EliSpot assays, with inclusion of either 3F12, 10.2–16 or negative control antibodies in the culture. Shown in Figure 7 are the results of this experiment, where the residual response of the polyclonal response after blocking with the specific antibodies is shown as a sum of the total response (Fig. 7A) and as the fraction of that elicited with the negative control antibody (Fig. 7B). These data show that I-Ak specific responses are almost completely blocked by 10.2–16, in agreement with studies of others (Fathman and Kimoto, 1981; Naquet et al., 1983; Rosloniec et al., 1989). In contrast, total responses were only modestly blocked by 3F12. The opposite pattern was observed with I-As restricted CD4 T cells, where 3F12 was much more effective at blocking CD4 T cell recognition than the 10.2–16 mAb that recognizes the region of MHC class II in proximity to the variant beta chain residues. These results support the model that the structural features of I-As with a proline at the periphery of the binding pocket close to the peptide’s carboxy terminus, and perhaps with the alpha chain polymorphisms that destabilize hydrogen bonding interactions, may together disrupt prototypical TcR docking modes, and may instead recruit CD4 T cells with receptors that engage the more stable regions of this class II molecule located near the peptide amino terminus.
Figure 7. Identification of CD4 T cell recognition sites by sensitivity to blocking by monoclonal antibodies.
B10.S (I-As) and C3H (I-Ak) mice were infected with A/New Caledonia/20/99 H1N1 influenza virus, at day 12 post infection CD4 T cells were isolated and tested for reactivity in the presence of blocking antibodies by production of IL-2 when cultured with known influenza peptide epitopes presented by I-As or I-Ak, respectively, using EliSpot assays. Graded doses of peptide were added to the cultures and cytokine producing cells were enumerated by cytokine EliSpot assays, shown is the response of the peptides summed. Panel A illustrates the number of IL-2 producing cells per million CD4 T cells. Panel B indicates the percent of the total residual response in the presences of the indicated antibody, where the negative control antibody was set as 100 % of the response. Panel C is the structure of I-Ak presenting the HEL (46–61) peptide, highlighting key residues (as shown in Figure 3) with the additional highlights of αM44 and βY65 and the boxed regions where mAb 3F12 (red) and 10.2–16 (blue) have been mapped, respectively.
4. Discussion
Here we report the consequences of a rare, previously unsuspected allelic polymorphism in an MHC class II molecule (I-As) that is expressed in mice that are prone to autoimmunity. This β58A>P substitution is extremely unusual within murine I-A molecules as well as the human analog, HLA-DQ, which share high homology in this region of the beta chain. The amino acid segment (β56–60) PDAEY at the periphery of the alpha helical region that frames the peptide binding pocket is highly conserved in both human and mouse I-A/HLA-DQ as well as many I-E/HLA-DR molecules (see Figure 1). Our studies suggest that a single amino acid substitution of Pro for Ala at β58 found in I-As changes the structural and dynamic properties of class II molecules, as evidenced by a loss of recognition by two mAb antibodies, and elements in MHC class II conformational flexibility identified through molecular dynamics simulation in the region where the unique features of this MHC class II molecule were explored. Importantly, this substitution is adjacent to the amino acid (β-D57) that forms a critical salt bridge across the MHC class II binding pocket with alpha 76, suggesting that this region of MHC class II contributes to the conformational stability of the peptide binding pocket of MHC class II molecules. It should be noted that some HLA-DR alleles (e.g. DRB1*1101) as well as HLA-DP molecules alleles have substitution of a charged residue for Ala at β58 position.
In our search of all I-A alleles expressed in inbred mice, we found that one other class II molecule (I-Ar), expressed in B10.RIII mice, expresses Pro at β58. Interestingly, the B10.RIII mice, like the SJL strain of mice, are also used as a model for several autoimmune diseases, including experimental autoimmune uveoretinitis (Kerr et al., 2008; Xu et al., 1997), collagen-induced arthritis (Myers et al., 1995; Wooley et al., 1985), and chronic and relapsing experimental autoimmune encephalomyelitis (Jansson et al., 1995). In the latter autoimmune syndrome, disease can be initiated in both B10.RIII and SJL mice with MBP 89–101. Although there are a number of amino acid differences between these two MHC class II heterodimers, it is intriguing that they share this one rare β chain polymorphism and present the same peptide that is able to initiate an autoimmune syndrome.
Although we found evidence that this unusual polymorphism at the periphery of the MHC class II peptide binding pocket impacts the dynamic properties of the MHC class II molecule, there was little discernable impact on CD4 T cell recognition, with a modest trend of the CD4 T cells preferring the mutated I-As containing (β-Ala 58). These results were unexpected because the CD4 T cells derived from B10.S developed in the thymus, and thus “educated” on I-As containing the β-Pro58 and were activated in vivo after infection by this MHC class II molecule. Because there are many examples in the literature of single amino acid changes in MHC class II alpha or beta chains or the bound peptide altering CD4 T cell recognition (Fox et al., 1987; Liberman et al., 1998; Racioppi et al., 1991), this result was unexpected.
Based on the modest impact on CD4 T cells recognition, we considered a model to explain how a non-conservative polymorphism that changes local structure of the peptide binding pocket does not detectably affect T cell recognition. We speculated that this atypical class II molecule recruits CD4 T cells with TcR that dock towards the peptide amino terminus, distal to the change in structure. In support of this possibility was the finding that an antibody that binds at the peptides amino terminus (3F12) blocks T cell recognition of I-As restricted CD4 T cells elicited in B10.S mice. In contrast, a widely used antibody, (10.2–16), that typically blocks T cell recognition (Fathman and Kimoto, 1981; Naquet et al., 1983; Rosloniec et al., 1989) and detects a region of MHC class II molecules around β67–69, at the carboxyterminal end of the peptides, minimally blocked T cell recognition by I-As restricted CD4 T cells. This finding is consistent with the observations by others that some TcR:MHC-peptide complexes associated with autoimmunity possess unusual docking modes relative to most common diagonal mode of the TcR over the center of the MHC-peptide complex (Hahn et al., 2005; Li et al., 2005; Maynard et al., 2005). In these autoimmunity-related TcR:MHC-peptide complexes, the primary contacts are shifted away from the prototypical diagonal mode that focusses the CDR3 region centered at p5 of the MHC class II-bound peptide (Rossjohn et al., 2015). Instead, autoimmune associated TcR:MHC-peptide complexes, in both human and mice, are shifted to the amino terminus of the peptide, focusing on interactions between residues p2 and p3 of the peptide, with very few contacts at the carboxyterminus (Deng and Mariuzza, 2007; Li et al., 2005; Wucherpfennig et al., 2009). It has not been clear if these unusual docking orientations are a property of the autoimmune MHC class II molecule itself or are unique to particular self-peptide complexes.
Here we provide evidence that the same atypical docking mode is likely present in CD4 T cells specific for foreign, non-self, influenza-derived peptides. We speculate that in general, the antigen-elicited I-As-restricted CD4 T cells may primarily express T cell receptors that dock towards the amino terminus of the bound peptide because of the less stable structure and conformational flexibility near the proline near β58 that are combined with the alterations in hydrogen bonding potential between the peptide main chain and the MHC class II alpha chains in the same region. Under these conditions, the changes induced by the substitution of the rare β-Pro58 with the more common β-Ala58 may not disrupt CD4 TcR recognition. An intriguing question would be whether the lack of discrimination of these two class II molecules would be preserved with CD4 T cells selected with the prototypical substituted variant of I-As (Ala β58). If this model is correct, this variant and “corrected” MHC class II molecule, expressing the prototypical Ala residue at β58 would have permitted a more stable conformation of the MHC class II molecule and thus might promote the prototypical docking of the TcR. We also might find that the peptides selected for presentation and recruitment of CD4 T cells would be altered. Also, it is known that the conformational flexibility of MHC class II molecules, rather than specific sites on the MHC class II dimer are critical for the biological activity of HLA-DM (Yin et al., 2014). Thus, the greater dynamic flexibility of I-As may perturb DM-mediated selection of peptides on this MHCclass II molecule, a possibility we have not yet explored.
Overall, our results are consistent with the view that naturally occurring MHC class II molecules can possess amino acid polymorphisms that destabilize prototypical features of the MHC class II structure. The conformational stability of these MHC class II:peptide complexes may be associated with selective losses in tolerance, CD4 T cell dependent autoimmunity and atypical interactions with the polyclonal T cell repertoire. Although the impact of this feature has been most extensively discussed with regard to autoimmune diabetes (Ettinger et al., 1998; Miyadera et al., 2015), we suggest that it may extend to other MHC class II molecules that have polymorphisms that disrupt canonical aspects of MHC class II structure that contribute to the structural stability of the ligand that engages predictable clonotypic T cell receptors. Such MHC class II molecules, although perhaps at one time offering a selective advantage to the host against a pathogenic organism (Dean et al., 2002; Ghosh, 2008; Mangalam et al., 2013), may be paradoxically associated with less efficient tolerance induction and thus lead to a host predisposed towards autoimmune recognition by CD4 T cells. Interestingly, recent studies have shown that the orientation of the TcR on the MHC:peptide complex is critical for functional engagement of CD8 co-receptor (Zareie et al., 2021), which support the possibility that CD4 T cells whose TcR engages MHC class II:peptide in a atypical docking mode might lead to alterations in selection during development or recruitment during an immune response.
Supplementary Material
Highlights:
A rare and unreported mutation was discovered in I-As expressed autoimmunity prone SJL mice
A single amino acid polymorphism alters MHC class II structural features
CD4 T cell reactivity with I-As suggests TcR docking towards peptides’ amino terminus
Autoimmune T cells may share common features of T cell receptor recognition
Acknowledgements
This work was supported by Federal funds from the National Institute of Allergy and Infectious Diseases, National Institute of Health, Department of Health and Human Services, under CEIRS contract [HHSN272201400005C] and University of Rochester institutional funds to AJS and by Federal funds from the National Institute of General Medical Sciences, National Institute of Health award R35GM118166 to BMB. GK was supported by an Indiana CTSI pre-doctoral fellowship, supported by National Institutes of Health award UL1TR002529.
Footnotes
Declaration of Interests: None
Table References not included in the text (Bhattacharya et al., 1981; Hauptfeld-Dolejsek and Shreffler, 1989; Janeway et al., 1984; Koch et al., 1982; LaPan et al., 1991; Lerner et al., 1980; Melino et al., 1983; Metlay et al., 1990; Ozato and Sachs, 1981; Santambrogio et al., 1999; Wilson et al., 1998).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acha-Orbea H, McDevitt HO, 1987. The first external domain of the nonobese diabetic mouse class II I-A beta chain is unique. Proc Natl Acad Sci U S A 84, 2435–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MS, Bluestone JA, 2005. The NOD mouse: a model of immune dysregulation. Annu Rev Immunol 23, 447–485. [DOI] [PubMed] [Google Scholar]
- Beck BN, Buerstedde JM, Krco CJ, Nilson AE, Chase CG, McKean DJ, 1986. Characterization of cell lines expressing mutant I-Ab and I-Ak molecules allows the definition of distinct serologic epitopes on A alpha and A beta polypeptides. J Immunol 136, 2953–2961. [PubMed] [Google Scholar]
- Beck BN, Pease LR, Bell MP, Buerstedde JM, Nilson AE, Schlauder GG, McKean DJ, 1987. DNA sequence analysis of I-Ak beta mutants reveals serologically immunodominant region. J Exp Med 166, 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Dorf ME, Springer TA, 1981. A shared alloantigenic determinant on Ia antigens encoded by the I-A and I-E subregions: evidence for I region gene duplication. J Immunol 127, 2488–2495. [PubMed] [Google Scholar]
- Braunstein NS, Germain RN, Loney K, Berkowitz N, 1990. Structurally interdependent and independent regions of allelic polymorphism in class II MHC molecules. Implications for Ia function and evolution. J Immunol 145, 1635–1645. [PubMed] [Google Scholar]
- Bryant PW, Roos P, Ploegh HL, Sant AJ, 1999. Deviant trafficking of I-Ad mutant molecules is reflected in their peptide binding properties. Eur J Immunol 29, 2729–2739. [DOI] [PubMed] [Google Scholar]
- Canutescu AA, Dunbrack RL Jr., 2003. Cyclic coordinate descent: A robotics algorithm for protein loop closure. Protein Sci 12, 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain EM, Duncan DS, Rodgers JM, Miller SD, 2011. The role of antigen presenting cells in multiple sclerosis. Biochim Biophys Acta 1812, 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury S, Lyskov S, Gray JJ, 2010. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Baker D, 2008. Macromolecular modeling with rosetta. Annu Rev Biochem 77, 363–382. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Raychaudhuri S, 2012. Interrogating the major histocompatibility complex with high-throughput genomics. Hum Mol Genet 21, R29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Carrington M, O’Brien SJ, 2002. Balanced polymorphism selected by genetic versus infectious human disease. Annu Rev Genomics Hum Genet 3, 263–292. [DOI] [PubMed] [Google Scholar]
- Deng L, Mariuzza RA, 2007. Recognition of self-peptide-MHC complexes by autoimmune T-cell receptors. Trends Biochem Sci 32, 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin JR, Alonso JA, Ayres CM, Keller GLJ, Bobisse S, Vander Kooi CW, Coukos G, Gfeller D, Harari A, Baker BM, 2020. Structural dissimilarity from self drives neoepitope escape from immune tolerance. Nat Chem Biol 16, 1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estess P, Begovich AB, Koo M, Jones PP, McDevitt HO, 1986. Sequence analysis and structure-function correlations of murine q, k, u, s, and f haplotype I-A beta cDNA clones. Proc Natl Acad Sci U S A 83, 3594–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger RA, Liu AW, Nepom GT, Kwok WW, 1998. Exceptional stability of the HLA-DQA1*0102/DQB1*0602 alpha beta protein dimer, the class II MHC molecule associated with protection from insulin-dependent diabetes mellitus. J Immunol 161, 6439–6445. [PubMed] [Google Scholar]
- Fathman CG, Kimoto M, 1981. Studies Utilizing murine T cell clones: Ir genes, Ia antigens and MLR stimulating determinants. Immunol Rev 54, 57–79. [DOI] [PubMed] [Google Scholar]
- Fox BS, Chen C, Fraga E, French CA, Singh B, Schwartz RH, 1987. Functionally distinct agretopic and epitopic sites. Analysis of the dominant T cell determinant of moth and pigeon cytochromes c with the use of synthetic peptide antigens. J Immunol 139, 1578–1588. [PubMed] [Google Scholar]
- Fremont DH, Monnaie D, Nelson CA, Hendrickson WA, Unanue ER, 1998. Crystal structure of I-Ak in complex with a dominant epitope of lysozyme. Immunity 8, 305–317. [DOI] [PubMed] [Google Scholar]
- Ghosh K, 2008. Evolution and selection of human leukocyte antigen alleles by Plasmodium falciparum infection. Hum Immunol 69, 856–860. [DOI] [PubMed] [Google Scholar]
- Giarratana N, Penna G, Adorini L, 2007. Animal models of spontaneous autoimmune disease: type 1 diabetes in the nonobese diabetic mouse. Methods Mol Biol 380, 285–311. [DOI] [PubMed] [Google Scholar]
- Glatigny S, Bettelli E, 2018. Experimental Autoimmune Encephalomyelitis (EAE) as Animal Models of Multiple Sclerosis (MS). Cold Spring Harb Perspect Med 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW, 2005. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat Immunol 6, 490–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptfeld-Dolejsek V, Shreffler DC, 1989. Antigenic properties of 36 new H-2 congenic strains and 4 independently derived strains, W10LT, DDD, BZH, and FM. Immunogenetics 30, 132–136. [DOI] [PubMed] [Google Scholar]
- Hollenbach JA, Oksenberg JR, 2015. The immunogenetics of multiple sclerosis: A comprehensive review. J Autoimmun 64, 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmdahl R, Karlsson M, Andersson ME, Rask L, Andersson L, 1989. Localization of a critical restriction site on the I-A beta chain that determines susceptibility to collagen-induced arthritis in mice. Proc Natl Acad Sci U S A 86, 9475–9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi T, Massier L, Kloting N, Horn K, Schuch A, Ahnert P, Engel C, Loffler M, Burkhardt R, Thiery J, Tonjes A, Stumvoll M, Bluher M, Doxiadis I, Scholz M, Kovacs P, 2020. HLA Class II Allele Analyses Implicate Common Genetic Components in Type 1 and Non-Insulin-Treated Type 2 Diabetes. J Clin Endocrinol Metab 105. [DOI] [PubMed] [Google Scholar]
- Janeway CA Jr., Conrad PJ, Lerner EA, Babich J, Wettstein P, Murphy DB, 1984. Monoclonal antibodies specific for Ia glycoproteins raised by immunization with activated T cells: possible role of T cellbound Ia antigens as targets of immunoregulatory T cells. J Immunol 132, 662–667. [PubMed] [Google Scholar]
- Jansson L, Diener P, Engstrom A, Olsson T, Holmdahl R, 1995. Spreading of the immune response to different myelin basic protein peptides in chronic experimental autoimmune encephalomyelitis in B10.RIII mice. Eur J Immunol 25, 2195–2200. [DOI] [PubMed] [Google Scholar]
- Jardetzky TS, Brown JH, Gorga JC, Stern LJ, Urban RG, Strominger JL, Wiley DC, 1996. Crystallographic analysis of endogenous peptides associated with HLA-DR1 suggests a common, polyproline II-like conformation for bound peptides. Proc Natl Acad Sci U S A 93, 734–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert B, Dalmau J, 2019. The role of infections in autoimmune encephalitides. Rev Neurol (Paris) 175, 420–426. [DOI] [PubMed] [Google Scholar]
- Kalbus M, Fleckenstein BT, Offenhausser M, Bluggel M, Melms A, Meyer HE, Rammensee HG, Martin R, Jung G, Sommer N, 2001. Ligand motif of the autoimmune disease-associated mouse MHC class II molecule H2-A(s). Eur J Immunol 31, 551–562. [DOI] [PubMed] [Google Scholar]
- Kappler JW, Skidmore B, White J, Marrack P, 1981. Antigen-inducible, H-2-restricted, interleukin-2-producing T cell hybridomas. Lack of independent antigen and H-2 recognition. J Exp Med 153, 1198–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karami J, Aslani S, Jamshidi A, Garshasbi M, Mahmoudi M, 2019. Genetic implications in the pathogenesis of rheumatoid arthritis; an updated review. Gene 702, 8–16. [DOI] [PubMed] [Google Scholar]
- Kerr EC, Raveney BJ, Copland DA, Dick AD, Nicholson LB, 2008. Analysis of retinal cellular infiltrate in experimental autoimmune uveoretinitis reveals multiple regulatory cell populations. J Autoimmun 31, 354–361. [DOI] [PubMed] [Google Scholar]
- Knowlden ZA, Sant AJ, 2016. CD4 T cell epitope specificity determines follicular versus non-follicular helper differentiation in the polyclonal response to influenza infection or vaccination. Sci Rep 6, 28287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch N, Hammerling GJ, Tada N, Kimura S, Hammerling U, 1982. Cross-blocking studies with monoclonal antibodies against I-A molecules of haplotypes b, d and k. Eur J Immunol 12, 909–914. [DOI] [PubMed] [Google Scholar]
- Kuchroo VK, Anderson AC, Waldner H, Munder M, Bettelli E, Nicholson LB, 2002. T cell response in experimental autoimmune encephalomyelitis (EAE): role of self and cross-reactive antigens in shaping, tuning, and regulating the autopathogenic T cell repertoire. Annu Rev Immunol 20, 101–123. [DOI] [PubMed] [Google Scholar]
- Landais D, Matthes H, Benoist C, Mathis D, 1985. A molecular basis for the Ia.2 and Ia.19 antigenic determinants. Proc Natl Acad Sci U S A 82, 2930–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landias D, Beck BN, Buerstedde JM, Degraw S, Klein D, Koch N, Murphy D, Pierres M, Tada T, Yamamoto K, et al. , 1986. The assignment of chain specificities for anti-Ia monoclonal antibodies using L cell transfectants. J Immunol 137, 3002–3005. [PubMed] [Google Scholar]
- LaPan KE, Klapper DG, Frelinger JA, 1991. Production and characterization of a peptide specific, anti-major histocompatibility complex class II, monoclonal antibody. Mol Immunol 28, 499–504. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Matis LA, Janeway CA Jr., Jones PP, Schwartz RH, Murphy DB, 1980. Monoclonal antibody against an Ir gene product? J Exp Med 152, 1085–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Huang Y, Lue J, Quandt JA, Martin R, Mariuzza RA, 2005. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J 24, 2968–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman IS, Ivanov SN, Vinogradova TV, Avenirova EA, 1998. [Comparative study of peripheral circulation, lipid metabolism and hemocoagulation in families with atherosclerosis and diabetes mellitus]. Ter Arkh 70, 10–15. [PubMed] [Google Scholar]
- Loss GE Jr., Sant AJ, 1993. Invariant chain retains MHC class II molecules in the endocytic pathway. J Immunol 150, 3187–3197. [PubMed] [Google Scholar]
- Mangalam AK, Taneja V, David CS, 2013. HLA class II molecules influence susceptibility versus protection in inflammatory diseases by determining the cytokine profile. J Immunol 190, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard J, Petersson K, Wilson DH, Adams EJ, Blondelle SE, Boulanger MJ, Wilson DB, Garcia KC, 2005. Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: insights into MHC bias and antigen specificity. Immunity 22, 81–92. [DOI] [PubMed] [Google Scholar]
- McFarland BJ, Katz JF, Beeson C, Sant AJ, 2001. Energetic asymmetry among hydrogen bonds in MHC class II*peptide complexes. Proc Natl Acad Sci U S A 98, 9231–9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland BJ, Katz JF, Sant AJ, Beeson C, 2005. Energetics and cooperativity of the hydrogen bonding and anchor interactions that bind peptides to MHC class II protein. J Mol Biol 350, 170–183. [DOI] [PubMed] [Google Scholar]
- Megiorni F, Pizzuti A, 2012. HLA-DQA1 and HLA-DQB1 in Celiac disease predisposition: practical implications of the HLA molecular typing. J Biomed Sci 19, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melino MR, Epstein SL, Sachs DH, Hansen TH, 1983. Idiotypic and fluorometric analysis of the antibodies that distinguish the lesion of the I-A mutant B6.C-H-2bm12. J Immunol 131, 359–364. [PubMed] [Google Scholar]
- Metlay JP, Witmer-Pack MD, Agger R, Crowley MT, Lawless D, Steinman RM, 1990. The distinct leukocyte integrins of mouse spleen dendritic cells as identified with new hamster monoclonal antibodies. J Exp Med 171, 1753–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SD, Olson JK, Croxford JL, 2001. Multiple pathways to induction of virus-induced autoimmune demyelination: lessons from Theiler’s virus infection. J Autoimmun 16, 219–227. [DOI] [PubMed] [Google Scholar]
- Miyadera H, Ohashi J, Lernmark A, Kitamura T, Tokunaga K, 2015. Cell-surface MHC density profiling reveals instability of autoimmunity-associated HLA. J Clin Invest 125, 275–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers LK, Miyahara H, Terato K, Seyer JM, Stuart JM, Kang AH, 1995. Collageninduced arthritis in B10.RIII mice (H-2r): identification of an arthritogenic T-cell determinant. Immunology 84, 509–513. [PMC free article] [PubMed] [Google Scholar]
- Naquet P, Marchetto S, Pierres M, 1983. Dissection of the Poly(Glu60Ala30Tyr10) (GAT)-specific T-cell repertoire in H-2Ik mice. II. The use of monoclonal antibodies to study the recognition of Ia antigens by GAT-reactive T-cell clones. Immunogenetics 18, 559–574. [DOI] [PubMed] [Google Scholar]
- Nayak JL, Richards KA, Chaves FA, Sant AJ, 2010. Analyses of the specificity of CD4 T cells during the primary immune response to influenza virus reveals dramatic MHC-linked asymmetries in reactivity to individual viral proteins. Viral Immunol 23, 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble JA, 2015. Immunogenetics of type 1 diabetes: A comprehensive review. J Autoimmun 64, 101–112. [DOI] [PubMed] [Google Scholar]
- Oi VT, Jones PP, Goding JW, Herzenberg LA, Herzenberg LA, 1978. Properties of monoclonal antibodies to mouse Ig allotypes, H-2, and Ia antigens. Curr Top Microbiol Immunol 81, 115–120. [DOI] [PubMed] [Google Scholar]
- Oleszak EL, Chang JR, Friedman H, Katsetos CD, Platsoucas CD, 2004. Theiler’s virus infection: a model for multiple sclerosis. Clin Microbiol Rev 17, 174–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozato K, Mayer N, Sachs DH, 1980. Hybridoma cell lines secreting monoclonal antibodies to mouse H-2 and Ia antigens. J Immunol 124, 533–540. [PubMed] [Google Scholar]
- Ozato K, Sachs DH, 1981. Monoclonal antibodies to mouse MHC antigens. III. Hybridoma antibodies reacting to antigens of the H-2b haplotype reveal genetic control of isotype expression. J Immunol 126, 317–321. [PubMed] [Google Scholar]
- Pearson CI, Gautam AM, Rulifson IC, Liblau RS, McDevitt HO, 1999. A small number of residues in the class II molecule I-Au confer the ability to bind the myelin basic protein peptide Ac1–11. Proc Natl Acad Sci U S A 96, 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JA, Wong FS, Wen L, 2016. The importance of the Non Obese Diabetic (NOD) mouse model in autoimmune diabetes. J Autoimmun 66, 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M, Miller J, 1990. Invariant chain influences the immunological recognition of MHC class II molecules. Nature 345, 172–174. [DOI] [PubMed] [Google Scholar]
- Pierres M, Devaux C, Dosseto M, Marchetto S, 1981. Clonal analysis of B- and T-cell responses to Ia antigens. I. Topology of epitope regions on I-Ak and I-Ek molecules analyzed with 35 monoclonal alloantibodies. Immunogenetics 14, 481–495. [DOI] [PubMed] [Google Scholar]
- Pociot F, Lernmark A, 2016. Genetic risk factors for type 1 diabetes. Lancet 387, 2331–2339. [DOI] [PubMed] [Google Scholar]
- Racioppi L, Ronchese F, Schwartz RH, Germain RN, 1991. The molecular basis of class II MHC allelic control of T cell responses. J Immunol 147, 3718–3727. [PubMed] [Google Scholar]
- Richards KA, Chaves FA, Sant AJ, 2009. Infection of HLA-DR1 transgenic mice with a human isolate of influenza a virus (H1N1) primes a diverse CD4 T-cell repertoire that includes CD4 T cells with heterosubtypic cross-reactivity to avian (H5N1) influenza virus. J Virol 83, 6566–6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KA, DiPiazza AT, Rattan A, Knowlden ZAG, Yang H, Sant AJ, 2018. Diverse Epitope Specificity, Immunodominance Hierarchy, and Functional Avidity of Effector CD4 T Cells Established During Priming Is Maintained in Lung After Influenza A Virus Infection. Front Immunol 9, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KA, Topham D, Chaves FA, Sant AJ, 2010. Cutting edge: CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J Immunol 185, 4998–5002. [DOI] [PubMed] [Google Scholar]
- Ridgway WM, Peterson LB, Todd JA, Rainbow DB, Healy B, Burren OS, Wicker LS, 2008. Gene-gene interactions in the NOD mouse model of type 1 diabetes. Adv Immunol 100, 151–175. [DOI] [PubMed] [Google Scholar]
- Robinson AP, Harp CT, Noronha A, Miller SD, 2014. The experimental autoimmune encephalomyelitis (EAE) model of MS: utility for understanding disease pathophysiology and treatment. Handb Clin Neurol 122, 173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosloniec EF, Gay D, Freed JH, 1989. Epitopic analysis by anti-I-Ak monoclonal antibodies of I-Ak-restricted presentation of lysozyme peptides. J Immunol 142, 4176–4183. [PubMed] [Google Scholar]
- Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J, 2015. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol 33, 169–200. [DOI] [PubMed] [Google Scholar]
- Sant AJ, Beeson C, McFarland B, Cao J, Ceman S, Bryant PW, Wu S, 1999. Individual hydrogen bonds play a critical role in MHC class II: peptide interactions: implications for the dynamic aspects of class II trafficking and DM-mediated peptide exchange. Immunol Rev 172, 239–253. [DOI] [PubMed] [Google Scholar]
- Sant AJ, Braunstein NS, Germain RN, 1987. Predominant role of amino-terminal sequences in dictating efficiency of class II major histocompatibility complex alpha beta dimer expression. Proc Natl Acad Sci U S A 84, 8065–8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant AJ, Germain RN, 1989. Intracellular competition for component chains determines class II MHC cell surface phenotype. Cell 57, 797–805. [DOI] [PubMed] [Google Scholar]
- Santambrogio L, Sato AK, Fischer FR, Dorf ME, Stern LJ, 1999. Abundant empty class II MHC molecules on the surface of immature dendritic cells. Proc Natl Acad Sci U S A 96, 15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MR, Thirawatananond P, Peters L, Sharp RC, Ogundare S, Posgai AL, Perry DJ, Brusko TM, 2021. De-coding genetic risk variants in type 1 diabetes. Immunol Cell Biol. [DOI] [PMC free article] [PubMed]
- Sospedra M, Martin R, 2016. Immunology of Multiple Sclerosis. Semin Neurol 36, 115–127. [DOI] [PubMed] [Google Scholar]
- Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC, 1994. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature 368, 215–221. [DOI] [PubMed] [Google Scholar]
- Todd JA, Bell JI, McDevitt HO, 1987. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature 329, 599–604. [DOI] [PubMed] [Google Scholar]
- Todd JA, Bell JI, McDevitt HO, 1988. A molecular basis for genetic susceptibility to insulin-dependent diabetes mellitus. Trends Genet 4, 129–134. [DOI] [PubMed] [Google Scholar]
- Tompkins SM, Fuller KG, Miller SD, 2002. Theiler’s virus-mediated autoimmunity: local presentation of CNS antigens and epitope spreading. Ann N Y Acad Sci 958, 26–38. [PubMed] [Google Scholar]
- Trowsdale J, Knight JC, 2013. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet 14, 301–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J, Parham P, 2004. Mini-review: defense strategies and immunity-related genes. Eur J Immunol 34, 7–17. [DOI] [PubMed] [Google Scholar]
- Wei BY, Schreiber K, Buerstedde JM, Bell M, Nilson A, Huntoon C, Chase C, McKean DJ, 1994. Substitution of class II alpha chain polymorphic residues defines location of A alpha k serologic epitopes and alters association between alpha beta and Ii polypeptides. Int Immunol 6, 297–305. [DOI] [PubMed] [Google Scholar]
- Wilson NA, Wolf P, Ploegh H, Ignatowicz L, Kappler J, Marrack P, 1998. Invariant chain can bind MHC class II at a site other than the peptide binding groove. J Immunol 161, 4777–4784. [PubMed] [Google Scholar]
- Wooley PH, Luthra HS, Griffiths MM, Stuart JM, Huse A, David CS, 1985. Type II collagen-induced arthritis in mice. IV. Variations in immunogenetic regulation provide evidence for multiple arthritogenic epitopes on the collagen molecule. J Immunol 135, 2443–2451. [PubMed] [Google Scholar]
- Wucherpfennig KW, Call MJ, Deng L, Mariuzza R, 2009. Structural alterations in peptide-MHC recognition by self-reactive T cell receptors. Curr Opin Immunol 21, 590–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Rizzo LV, Silver PB, Caspi RR, 1997. Uveitogenicity is associated with a Th1-like lymphokine profile: cytokine-dependent modulation of early and committed effector T cells in experimental autoimmune uveitis. Cell Immunol 178, 69–78. [DOI] [PubMed] [Google Scholar]
- Yin L, Trenh P, Guce A, Wieczorek M, Lange S, Sticht J, Jiang W, Bylsma M, Mellins ED, Freund C, Stern LJ, 2014. Susceptibility to HLA-DM protein is determined by a dynamic conformation of major histocompatibility complex class II molecule bound with peptide. J Biol Chem 289, 23449–23464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zareie P, Szeto C, Farenc C, Gunasinghe SD, Kolawole EM, Nguyen A, Blyth C, Sng XYX, Li J, Jones CM, Fulcher AJ, Jacobs JR, Wei Q, Wojciech L, Petersen J, Gascoigne NRJ, Evavold BD, Gaus K, Gras S, Rossjohn J, La Gruta NL, 2021. Canonical T cell receptor docking on peptide-MHC is essential for T cell signaling. Science 372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.