Abstract
Preclinical mechanistic studies have pointed towards RNA interference-mediated off-target effects as a major driver of hepatotoxicity for GalNAc–siRNA conjugates. Here, we demonstrate that a single glycol nucleic acid or 2′–5′-RNA modification can substantially reduce small interfering RNA (siRNA) seed-mediated binding to off-target transcripts while maintaining on-target activity. In siRNAs with established hepatotoxicity driven by off-target effects, these novel designs with seed-pairing destabilization, termed enhanced stabilization chemistry plus (ESC+), demonstrated a substantially improved therapeutic window in rats. In contrast, siRNAs thermally destabilized to a similar extent by the incorporation of multiple DNA nucleotides in the seed region showed little to no improvement in rat safety suggesting that factors in addition to global thermodynamics play a role in off-target mitigation. We utilized the ESC+ strategy to improve the safety of ALN-HBV, which exhibited dose-dependent, transient and asymptomatic alanine aminotransferase elevations in healthy volunteers. The redesigned ALN-HBV02 (VIR-2218) showed improved specificity with comparable on-target activity and the program was reintroduced into clinical development.
INTRODUCTION
RNA interference (RNAi)-based therapeutics harness a highly conserved natural mechanism whereby short, 20–25-nucleotide-long double-stranded small interfering RNAs (siRNAs) guide the sequence-specific downregulation of target messenger RNA (mRNA) transcripts via the RNA-induced silencing complex (RISC). Following many years of effort dedicated to advancing the underlying platform technologies including siRNA design, chemistry and delivery, RNAi therapeutics are emerging as a new class of medicines for the treatment of a wide variety of diseases across major therapeutic areas. The field has been galvanized by recent regulatory approvals of the first four RNAi therapeutics, ONPATTRO® (patisiran), GIVLAARI® (givosiran), OXLUMO® (lumasiran) and LEQVIO® (inclisiran). Inclisiran targets PCSK9 for the treatment of hypercholesteremia and is a prominent example of the technology rapidly expanding from orphan genetic indications to highly prevalent diseases.
The evolution of RNAi therapeutics has been driven by key advances in siRNA design and chemistry combined with delivery platforms suitable for clinical development. In particular, the use of a synthetic multivalent N-acetylgalactosamine (GalNAc) ligand, which enables the safe and efficient delivery of chemically modified siRNAs to hepatocytes in the liver, has transformed the field (1). This approach has now been widely adopted for different classes of investigational oligonucleotide therapeutics (2). Whereas the first generation of GalNAc–siRNA conjugates required relatively high doses to afford the desired in vivo activity (3), further refinement of the chemical modification pattern led to enhanced stabilization chemistry (ESC) designs with substantially improved potency and duration of effect (1,4,5).
While the overall safety profile for ESC GalNAc–siRNA conjugates in humans is very encouraging, the occurrence of dose-dependent, asymptomatic and transient liver function test (LFT) increases was observed in a small subset of our early clinical programs. The onset of these LFT increases largely coincided with the onset of maximum silencing activity. Since other development candidates with very similar siRNA chemistry and designs did not show LFT increases in humans, even at significantly higher doses, we hypothesized this to be a sequence-dependent phenomenon. This was supported by nonclinical observations where a subset of GalNAc–siRNA conjugates generally dropped out of the lead discovery process during exploratory safety studies in rats due to significant hepatotoxicity, albeit at higher, suprapharmacological doses.
Based on these observations, we launched a comprehensive mechanistic investigation to identify potential causes for the observed hepatotoxicity across species (6). We were able to rule out chemical toxicity and competition of GalNAc–siRNAs with endogenous microRNAs (miRNAs) for the RNAi pathway. Instead, all evidence pointed toward RNAi-mediated, hybridization-based off-target effects. We found that these off-target effects are largely driven by binding of the RISC-loaded siRNA to off-target transcripts mediated through base pairing between the seed region of the siRNA guide strand (nucleotides 2–8) and complementary site(s) in the 3′-untranslated region (3′-UTR) of mRNAs. This noncatalytic mechanism essentially mimics the post-transcriptional silencing by endogenous miRNAs and can lead to translational repression and/or mRNA destabilization at suprapharmacological levels of RISC-loaded siRNA (7–9), with reductions in mRNA levels accounting for most (66% to >90%) of the post-transcriptional repression mediated by mammalian miRNAs (10–13). A similar hybridization-based off-target mechanism was recently reported for antisense oligonucleotides (ASOs) whereby the toxicity of some high-affinity ASOs was associated with sequence-dependent RNase H-mediated cleavage of imperfectly matched transcripts (14). The fact that both siRNAs and ASOs can produce unwanted hybridization-based off-target effects, leading to potential toxicities in humans, highlights the need for robust screening methods, as well as both bioinformatics- and chemistry-based approaches for the design of safe therapeutics.
Previous reports have outlined the concept of using thermally destabilizing modifications in the seed region of the siRNA guide strand to enhance siRNA specificity by mitigating miRNA-like repression of off-targets while still allowing for productive full-length on-target recognition (15–21). However, to our knowledge, this approach has thus far only been demonstrated in vitro leaving the question of whether it could be translated into an improved therapeutic index in an in vivo setting. We recently reported that seed-pairing destabilization through a single nucleotide substitution in the seed region with the (S)-isomer of glycol nucleic acid (GNA) could improve the mRNA target specificity and abrogate the hepatotoxicity of a GalNAc–siRNA in rats (6). This followed another report that GNA could be tolerated at several positions of the siRNA guide strand in vitro and in vivo (22).
Here, we report on the further advancement of this approach and the generation of a new design with improved specificity, termed ESC+. The ESC+ design was found to be suitable for transition into clinical development and was applied to ALN-HBV, an investigational GalNAc–siRNA targeting the hepatitis B virus (HBV). ALN-HBV showed dose-dependent, asymptomatic and transient alanine aminotransferase (ALT) elevations in a phase 1 clinical study of healthy volunteers (manuscript submitted). We found that the ESC+ version of ALN-HBV, named ALN-HBV02, with a single GNA substitution at position 6 of the guide strand, and without any further changes in sequence or chemistry, minimizes off-target dysregulation without compromising on-target activity. ALN-HBV02, later termed VIR-2218, was reintroduced to preclinical and clinical development, thereby providing unique insight into the human translation of this new ESC+ design.
MATERIALS AND METHODS
Oligonucleotide synthesis
All oligonucleotides were synthesized on a MerMade 192, MerMade 12 or ÄKTA oligopilot 100 synthesizer according to previously published protocols (1,22). 2′-O-Methyl (2′-OMe), 2′-deoxy-2′-fluoro (2′-F), 2′-deoxy and 2′–5′-RNA (3′-OTBDMS-protected) phosphoramidites were purchased commercially. (S)-GNA phosphoramidites were synthesized according to previously published protocols (23–25). The identity and purity of all oligonucleotides in these studies (see Supplementary Table S1) were confirmed using ESI–LC/MS and IEX HPLC, respectively.
Thermal melting experiments
Melting studies were performed in 1 cm path length quartz cells on a Cary 300 UV–visible spectrophotometer equipped with a Peltier-thermostatted multicell holder. Each cuvette contained 800 μL of sample solution covered by 200 μL of light mineral oil. Samples were first annealed in the instrument by heating at a rate of 5°C/min from 25 to 95°C followed by cooling at the same rate from 95 to 25°C. After a waiting period of 3 min, melting curves were monitored at 260 nm with a heating rate of 1°C/min from 25 to 95°C. Melting temperatures (Tm) were calculated from the first derivatives of the heating curves and the reported values are the result of at least two independent measurements.
In vitro screening in the luciferase reporter assay
COS-7 cells were cultured at 37°C and 5% CO2 in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum. Cells were co-transfected in 96-well plates (15 000 cells/well) with 10 ng luciferase reporter plasmid and 0.64 pM to 50 nM siRNA in 5-fold dilutions using 2 μg/mL Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Cells were harvested at 48 h after transfection for the dual luciferase assay (Promega) according to the manufacturer’s instructions. The reporter plasmids were generated by cloning targeting sequences into the psiCHECK2 vector between Xho1 and Not1 restriction sites. The on-target reporter plasmid contained a single site with perfect complementarity to the guide strand in the 3′-UTR of Renilla luciferase. The off-target reporter plasmid contained four tandem seed-complementary sites separated by a 19-nucleotide spacer (5′-TAATATTACATAAATAAAA-3′) in the 3′-UTR of Renilla luciferase (18,26). Both plasmids co-expressed firefly luciferase as a transfection control. The specific insert sequences were as follows:
Ttr on-target: ATAAACAAGGTTTGACATCAATCTAGCTATATCTTTAAGAATGATAAACTAAACAGTGTTCTTGCTCTATAAGACATTGGTGAGGAAAAATCCTTTGGCCGTTTCCAAGATCTGACAGTGCA
Ttr off-target: ATAAACAAGGTTTGACATCAATCTAGCTATATCTTTAAGAATGATAAACTGCTCTATAATAATATTACATAAATAAAAGCTCTATAATAATATTACATAAATAAAAGCTCTATAATAATATTACATAAATAAAAGCTCTATAAGACATTGGTGAGGAAAAATCCTTTGGCCGTTTCCAAGATCTGACAGTGCA
Hao1 on-target: ATAAACAAGGTTTGACATCAATCTAGCTATATCTTTAAGAATGATAAACTTTGTCGATGACTTTCACATTCGACATTGGTGAGGAAAAATCCTTTGGCCGTTTCCAAGATCTGACAGTGCA
Hao1 off-target: ATAAACAAGGTTTGACATCAATCTAGCTATATCTTTAAGAATGATAAACTCATCGACAATAATATTACATAAATAAAACATCGACAATAATATTACATAAATAAAACATCGACAATAATATTACATAAATAAAACATCGACAAGACATTGGTGAGGAAAAATCCTTTGGCCGTTTCCAAGATCTGACAGTGCA
Care and use of laboratory animals
All studies were conducted by certified laboratory personnel using protocols consistent with local, state and federal regulations, as applicable, and experimental protocols were approved by the Institutional Animal Care and Use Committee at Alnylam Pharmaceuticals. All animals were acclimated in-house for 48 h prior to study start.
In vivo screening, serum protein quantification, liver mRNA quantification and liver siRNA quantification in mice
Female C57BL/6 mice ∼6–8 weeks of age were obtained from Charles River Laboratories and randomly assigned to each group. All dosing solutions were stored at 4°C until 1 h before the time of injection, when they were removed from storage and allowed to reach room temperature. Animals received a single subscapular subcutaneous injection of siRNA, 1× PBS or 0.9% NaCl (saline) at the indicated dose, prepared as an injection volume of 10 μL/g in PBS or 0.9% NaCl. At the indicated time pre- or post-dosing, blood was collected via retroorbital bleed. Serum samples were kept at room temperature for 1 h and then spun in a microcentrifuge at 21 000 × g at room temperature for 10 min and subsequently stored at −80°C until analysis. TTR serum protein levels were measured by ELISA (serum was diluted 1:4000 and used in a mouse prealbumin kit according to the manufacturer's instructions, ALPCO, 41-PALMS-E01). Animals were sacrificed at the days indicated in the figures, after which livers were harvested and snap frozen for further analysis.
For the extraction of RNA, powdered liver (∼10 mg) was resuspended in 500 μL QIAzol and a 5-mm steel grinding ball was added to each sample. Samples were homogenized at 25/s for 2 × 5 min at 4°C using a TissueLyser LT (Qiagen). Samples were incubated at room temperature for 5 min followed by addition of 100 μL chloroform. Samples were mixed by vigorously shaking the tubes, followed by a 10-min incubation at room temperature. Samples were spun at 12 000 × g for 15 min at 4°C and the supernatant was removed to a new tube and 1.5 volumes of 100% ethanol was added. Samples were then purified using an RNeasy Kit (Qiagen). Samples were eluted from RNeasy columns with 60 μL RNase-free water (Ambion) and quantified on a NanoDrop (Thermo Fisher Scientific). 1.5 μg of RNA was used to generate cDNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, 4368813). qPCR reactions were performed using gene-specific TaqMan assays for each target (Thermo Fisher, Mm00443267_m1 for Ttr and Mm00439249_m1 for Hao1) and mouse Gapdh as an endogenous control (Thermo Fisher, 4352339E). Real-time PCR was performed in a Roche LightCycler 480 using LightCycler 480 Probes Master Mix (Roche, 04707494001). Data were analyzed using the ΔΔCt method normalizing to control animals dosed with PBS or 0.9% NaCl alone.
Where applicable, cohorts of mice were sacrificed on various days post-dose, and livers were snap frozen in liquid nitrogen and ground into powder for further analysis. Total liver siRNA levels from powdered liver were quantified by TaqMan PCR (SL-RT qPCR) as previously described (27–29).
Rat toxicity studies
The test articles were diluted with 0.9% NaCl to achieve appropriate dosing concentrations and dosed subcutaneously on the upper back to male Sprague Dawley rats (6–8 weeks old) in a dose volume of 5 mL/kg with four to five animals per group. Whole venous blood was collected into serum separator tubes (BD Microtainer) and allowed to clot at room temperature for 30 min prior to centrifugation at 3000 rpm (1489 × g) for 10 min at 4°C. Serum was then aliquoted and stored at −80°C until analysis. Serum chemistries were analyzed using the AU400 chemistry analyzer (Beckman Coulter, Brea, CA, USA), with reagents provided by Beckman Coulter, Randox and Sekisui Diagnostics. Livers were fixed in 10% neutral buffered formalin for 72 h prior to routine processing using TissueTek VIP 6A1 (Sakura). Tissues were trimmed, embedded into paraffin blocks, sectioned at 4 µm, stained with hematoxylin and eosin (H&E) using TissueTek Prisma A1D (Sakura) and coverslipped using TissueTek Glass g2 (Sakura). Two sections were examined microscopically from each liver in an unblinded fashion, followed by blinded assessment to confirm subtle findings. The range of severity grade for each microscopic finding when present was graded on a four-point scale (with 1 = minimal, 2 = mild, 3 = moderate and 4 = marked).
Bioinformatics and RNA sequencing
RNA was extracted from primary rat hepatocytes transfected with siRNA as previously described (30). Rat livers collected at the end of the toxicity studies were snap frozen, powdered and RNA was extracted with the miRNeasy kit (Qiagen). cDNA libraries were prepared with the TruSeq Stranded Total RNA Library Prep Kit (Illumina) and sequenced on a NextSeq500 sequencer (Illumina), all according to manufacturers’ instructions. Raw RNA sequencing (RNA-seq) reads were filtered with minimal mean quality scores of 25 and minimal remaining length of 36, using the ea-utils software fastq-mcf (https://expressionanalysis.github.io/eautils/). Filtered reads were aligned to the Rattus norvegicus genome (Rnor_6.0) using STAR (ultrafast universal RNA-seq aligner) (31). Uniquely aligned reads were counted by featureCounts (32) with the minimum mapping quality score set to 10. Differential gene expression analysis was performed by the R package DESeq2 with the betaPrior parameter set to TRUE to shrink log2 fold-change estimates for noisy, low-count genes (32,33). Estimates of delta log2 fold change for seed-match categories in CDF plots are beta coefficients from a linear model regressing log2 fold change against binary seed-match classifications across genes. Asterisks following the delta estimates indicate the level of (two-tailed) statistical significance derived from the t-statistic associated with each beta coefficient, and follow typical convention (n.s. = not significant, *P < 0.05, **P < 0.01 and ***P < 0.001).
Pharmacodynamic evaluation of HBV-targeting siRNAs in an AAV-HBV mouse model
Pharmacology of HBV-targeting siRNAs was assayed in an adeno-associated virus 8 encoding the 1.3× HBV genome (genotype D, HBV-AAV8) mouse model. HBV-AAV8 (SignaGen Laboratories) was diluted in sterile 1× PBS to a final concentration of 2 × 1012 GC/mL. Male C57BL/6 mice 6–8 weeks of age were injected intravenously via lateral tail vein with 2 × 1011 GC/mouse in a fixed volume of 100 μL. siRNAs were diluted with sterile 1× PBS and administered with a variable volume of 10 μL/g. Blood that was collected at the days indicated via retroorbital bleed was allowed to clot for 30 min, and then was spun in a microcentrifuge for 10 min at 13 000 rpm and 4°C. HBV surface antigen protein levels were evaluated via ELISA (serum was diluted 1:2000 in 1× PBS and used in a human HBsAg kit, BioTang, Waltham, MA, USA) with only minor changes to the protocol. A human HBsAg protein standard (US Biologics, Memphis, TN, USA) was used to generate the standard curve.
Specificity evaluation of ALN-HBV01 and ALN-HBV02 by RNA-seq in HepG2.2.15 cells
HepG2.2.15 cells, a HepG2-derived cell line stably transfected with full-genome HBV, were diluted with culture media to a final concentration of 187 500 cells/mL, and 80 μL was pipetted into 96-well collagen-coated plates (BD BioCoat, cat #356407) to give a final concentration of 15 000 cells/well. RNAiMAX (Thermo Fisher, cat #13778150) was diluted with Opti-MEM (Thermo Fisher, cat #31985062) at a concentration of 0.3 μL RNAiMAX/10 μL Opti-MEM and incubated for 5 min at room temperature. After incubation, 10 μL/well was added to each well of 96-well collagen-coated plates (BD BioCoat, cat #356407), along with 10 μL/well of the appropriate dsRNA agent dilution (as appropriate in 1× PBS), mixed gently and incubated for 20 min at room temperature. Eighty microliters of prepared cell suspension was added to each well to give final cell density of 15 000 cells/well and final dsRNA agent concentrations of 10 nM. Cells were incubated in a 37°C incubator with 5% CO2 for 16–22 h. Cells were plated such that each experimental condition had 16 wells and the experiment was performed two times. The Thermo Fisher RNAqueous-96 Total RNA Isolation Kit was used to isolate RNA as per the protocol. cDNA library preparation, sequencing and read filtering were performed as mentioned earlier. Filtered reads were simultaneously aligned to the human (hgl9/GRCh37) and HBV (GenBank nucleotide ID: U95551; NCBI gene ID: 7276) genomes using STAR (ultrafast universal RNA-seq aligner) (31). Due to the circular structure of the HBV genome, 46 base pairs were repeated at the end of a linearized version of the HBV sequence to allow for reads to map at the break point. Counting of uniquely aligned reads and differential gene expression analysis was performed as mentioned earlier.
Statistical analysis
Differences between group means relative to control or parent siRNAs were evaluated for statistical significance using a one-way ANOVA in GraphPad Prism 8 and are indicated in each graph (*P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001). Only those that were statistically significant are indicated.
RESULTS
Positional effect of GNA on in vitro on- and off-target activities
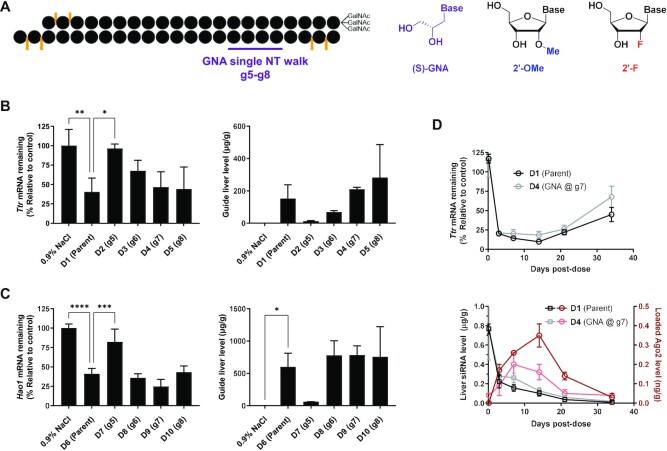
To evaluate the positional impact of GNA incorporation on the activity of GalNAc–siRNAs, a single GNA nucleotide was walked across the seed region of two separate GalNAc–siRNAs fully modified with 2′-OMe and 2′-F nucleotides, and terminal phosphorothioate linkages (Figure 1A and Table 1). For these initial studies and to expand on our previous work, we focused on two GalNAc–siRNAs targeting Ttr and Hao1 that we have previously reported to demonstrate seed-driven off-target hepatotoxicity in rats [siRNA-4 and siRNA-5, respectively, in ref. (6)]. For the siRNA targeting Ttr (D1), the pattern of 2′-OMe and 2′-F modifications was adjusted to make it similar to the lower 2′-F design of the Hao1 siRNA (D6). Following our previous work on the tolerability of GNA substitution across both strands of an siRNA (22), we utilized these two parent sequences to explore the positional effect of GNA incorporation at positions 5–8 of the guide strand (g5–g8) on both potency and specificity (Table 1 and Supplementary Table S1). The on- and off-target activities of each siRNA were evaluated in COS-7 cells expressing a luciferase reporter, which contained either a single fully complementary site to the guide sequence in its 3′-UTR (on-target reporter) or four tandem repeats of the sequence complementary to g2–g8 in the guide seed region (off-target reporter), respectively (18,26). The readout of the luciferase reporter assay is protein-based, thus capturing both potential mechanisms of miRNA-like activity: translational repression and mRNA destabilization. Minor reductions in on-target activity were observed for GNA substitution across most positions of both siRNAs except for position 8 in the sequence targeting Ttr (D5) exhibiting a 4-fold loss in potency relative to the parent D1.
Figure 1.
Evaluation of GNA-modified siRNAs in mice. (A) Structures of (S)-GNA, 2′-OMe and 2′-F nucleosides. Impact of GNA substitution at the specified position on activity (left) and liver guide strand concentration measured by RT-qPCR (right) in mice (n = 3) 7 days post-dose for Ttr-targeting (B) or Hao1-targeting (C) GalNAc–siRNAs dosed subcutaneously at either 0.5 mg/kg (Ttr) or 1.0 mg/kg (Hao1). Statistically significant differences relative to the parent siRNA are shown in each graph. (D) Impact of GNA substitution on Ttr mRNA knockdown in mice (n = 3) after a single subcutaneous dose of 0.75 mg/kg D1 or D4 and guide strand concentration in whole liver and Ago2 over time from the same study.
Table 1.
Evaluation of on- and off-target knockdown by GNA-modified GalNAc–siRNAs using a dual luciferase reporter assay
| Ttr | Hao1 | ||||||
|---|---|---|---|---|---|---|---|
| 5′-a•a•caguGuUCUugcucuauaaL-3′, 3′-u•u•uugucAcAagaacgaGaua•U•u-5′ | 5′-g•a•auguGaaAGucaucgacaaL-3′, 3′-g•u•cuuacAcUuucaGUaGcug•U•u-5′ | ||||||
| On-target IC50 (pM) | Off-target IC50 (nM) | Off-/on-target ratio | On-target IC50 (nM) | Off-target IC50 (nM) | Off-/on-target ratio | ||
| D1 (parent) | 2.16 | 0.07 | 31 | D6 (parent) | 0.42 | 2.48 | 6 |
| D2 (g5) | 4.19 | >50 | >11 945 | D7 (g5) | 0.64 | >50 | >78 |
| D3 (g6) | 4.55 | 3.21 | 705 | D8 (g6) | 0.67 | >50 | >75 |
| D4 (g7) | 5.40 | >50 | >9264 | D9 (g7) | 0.56 | >50 | >89 |
| D5 (g8) | 8.98 | 2.05 | 228 | D10 (g8) | 0.72 | 3.89 | 5 |
Italicized uppercase and lowercase letters represent 2′-F and 2′-OMe modifications, respectively, to adenosine, cytosine, guanosine and uridine. Phosphorothioate linkages are indicated by the ‘•’ symbol. ‘L’ represents the tri-N-acetylgalactosamine ligand. The top and bottom strands represent the passenger (5′→3′) and guide (3′→5′) strands of each siRNA, respectively. The underlined nucleotides in each guide strand represent those that were modified with GNA at the position specified in parentheses in the table.
On the other hand, GNA incorporation had a much more pronounced effect on the seed-mediated off-target activity that was both position- and sequence-dependent (Table 1). For the Ttr-targeting sequence, GNA at positions g5 (D2) and g7 (D4) was most effective at reducing the off-target reporter activity with a >500-fold improvement. For the sequence targeting Hao1, positions g5–g7 (D7–D9) strongly minimized the off-target activity with a >25-fold improvement observed relative to the parent. The ratio of in vitro off- to on-target activity provided a measure of the overall benefit of GNA substitution and allowed for a better comparison across compounds. For the Ttr-targeting siRNAs, the strongest effect was observed for GNA substitution at g5 (D2) or g7 (D4) with a >300-fold improvement in the ratio of off- to on-target activity compared to the parent. The impact of GNA substitution was overall less pronounced for the Hao1-targeting siRNAs with a >12-fold improvement for positions g5–g7 (D7–D9) and no positive impact for GNA substitution of position g8 (D10).
The presence of GNA can impact siRNA metabolic stability
The same set of GalNAc–siRNAs was used to evaluate activity in mice. For the series of siRNAs targeting Ttr, those modified with GNA at positions g7 (D4) or g8 (D5) exhibited a similar level of target mRNA knockdown to that of the parent siRNA (D1) 7 days post-dose, but those modified with GNA at g5 (D2) or g6 (D3) showed a significant reduction in activity (Figure 1B, left). The discrepancy between the in vivo and in vitro on-target activities suggested that additional factors, such as metabolic stability, may affect the observed in vivo activity. In fact, the total level of guide strand detected in liver by RT-qPCR correlated well with the in vivo efficacy as measured by mRNA target knockdown, illustrating the strong correlation between metabolic stability and in vivo activity (Figure 1B, right) (30). Similar observations were made for the series of GalNAc–siRNAs targeting Hao1 (Figure 1C). GNA-modified compounds D8–D10, which exhibited comparable efficacy to that of the parent (D6), were found to have similar tissue exposure levels. The poor knockdown observed with D7, on the other hand, is accompanied by a low level of guide strand in liver, again indicative of reduced metabolic stability.
To further evaluate the impact of GNA substitution on pharmacokinetics (PK) and pharmacodynamics (PD) of GalNAc–siRNA conjugates, the silencing activity, liver levels and Ago2 loading for the Ttr-targeting GNA-modified D4 and its parent D1 were followed over the course of 35 days after a single dose of 0.75 mg/kg in mice. The level of Ttr mRNA reduction and time–concentration profiles of the guide strand in the liver reflected the comparable PD and PK profiles, respectively, for both siRNAs over the course of the study (Figure 1D). Interestingly, the modification with GNA appeared to slightly impact the interaction with RISC as indicated by the ∼2-fold lower area under the curve for the Ago2-loaded GNA-modified guide strand compared to the parent (Figure 1D). However, it should be noted that quantification of GNA-modified test article can be confounded by a less reliable RT-qPCR amplification step (34). Taken together, these data confirm that GNA substitution is tolerated at certain positions in GalNAc–siRNAs with little impact on the PD profile in mice.
GNA improves the therapeutic index in rats
Based on the entirety of in vitro and in vivo data, GalNAc–siRNAs modified at position g7 with GNA (D4 and D9) were chosen for further evaluation alongside the parent ESC designs that had previously demonstrated significant off-target-driven hepatotoxicity in rats (6). GalNAc–siRNAs containing GNA at alternate positions, which demonstrated a loss of in vivo activity due to lower metabolic stability, a weak ability to suppress off-targets, or both, were not evaluated in rat toxicity studies since we expect their toxicity to be mitigated simply due to reduced exposure and RISC loading. The potency of the chosen compounds was assessed in a six-point dose response study ranging from 0.075 to 3 mg/kg in rats (Table 2). With an ED50 of 0.075 mg/kg, the GNA-modified D4 was found to be slightly less potent than the parent D1 (ED50 = 0.05 mg/kg; Supplementary Figure S1). For the siRNAs targeting Hao1, the parent D6 and its GNA-modified version D9 showed similar levels of knockdown across all doses tested, resulting in an ED50 of 0.3 mg/kg for both siRNAs (Supplementary Figure S2).
Table 2.
GalNAc–siRNAs used in rat toxicity studies
| siRNA duplex | Target mRNA | Passenger (5′–3′), guide (3′–5′) | Rat ED50 (mg/kg) | T m (°C) |
|---|---|---|---|---|
| D1 | Ttr | a•a•caguGuUCUugcucuauaaL, u•u•uugucAcAagaacgaGaua•U•u |
0.05 | 65.5 |
| D4 | Ttr | a•a•caguGuUCUugcucuauaaL, u•u•uugucAcAagaacgAGaua•U•u |
0.075 | 61.5 |
| D11 | Ttr | a•a•caguGuUCUugcucuauaaL, u•u•uugucAcAagaacgagata•U•u |
n.d. | 60.5 |
| D16 | Ttr | a•a•caguGuUCUugcucuauaaL, u•u•uugucAcAagaacg(A)Gaua•U•u |
n.d. | 61.5 |
| D6 | Hao1 | g•a•auguGaaAGucaucgacaaL, g•u•cuuacAcUuucaGUaGcug•U•u |
0.3 | 66.5 |
| D9 | Hao1 | g•a•auguGaaAGucaucgacaaL, g•u•cuuacAcUuucaGUAGcug•U•u |
0.3 | 64.8 |
| D12 | Hao1 | g•a•auguGaaAGucaucgacaaL, g•u•cuuacAcUuucagtagctg•U•u |
n.d. | 61.0 |
| D20 | Hao1 | g•a•auguGaaAGucaucgacaaL, g•u•cuuacAcUuucaGU(A)Gcug•U•u |
n.d. | 63.5 |
Italicized uppercase, lowercase, uppercase bold underlined, lowercase bold underlined and uppercase letters in parentheses represent 2′-F, 2′-OMe, (S)-GNA, 2′-deoxy and 2′–5′-linked ribose sugar modifications, respectively, to adenosine, cytosine, guanosine, uridine and thymidine. ‘L’ represents the tri-N-acetylgalactosamine ligand. Phosphorothioate linkages are indicated by the ‘•’ symbol. ED50 values were determined in rats; n.d. = not determined. All Tm values are the average of two independent measurements of the fully modified duplex at a concentration of 1 μM in 0.1× PBS.
To elucidate the effect of seed-pairing destabilization using multiple, individually less destabilizing modifications instead of a single GNA residue, these studies also included equipotent siRNAs D11 (Ttr) and D12 (Hao1) modified in the seed region with three DNA nucleotides. Seed destabilization using DNA is similar to an approach previously reported to mitigate off-target effects in vitro (18). The pattern of alternating 2′-OMe/DNA modification used in the seed region of D11 and D12 was one of several designs that showed in vitro activity equivalent to the parent (data not shown), a level of thermal destabilization comparable to GNA and sufficient metabolic stability to support in vivo evaluation. Analysis of guide strand metabolites by LC–MS and liver levels by RT-qPCR after a single dose in mice confirmed that the introduction of DNA into these GalNAc–siRNAs did not significantly impact overall metabolism (Supplementary Table S2 and Supplementary Figure S3) (30). GalNAc–siRNAs modified with several consecutive DNA nucleotides did not provide the necessary metabolic stability in vivo to warrant further evaluation (data not shown). Although the DNA-modified siRNAs D11 and D12 exhibited an equivalent or greater level of thermal destabilization compared to the GNA-modified D4 and D9, respectively, there was little to no reduction in off-target silencing compared to the parent siRNAs in the luciferase off-target reporter assay (Table 2 and Supplementary Figures S4–S7).
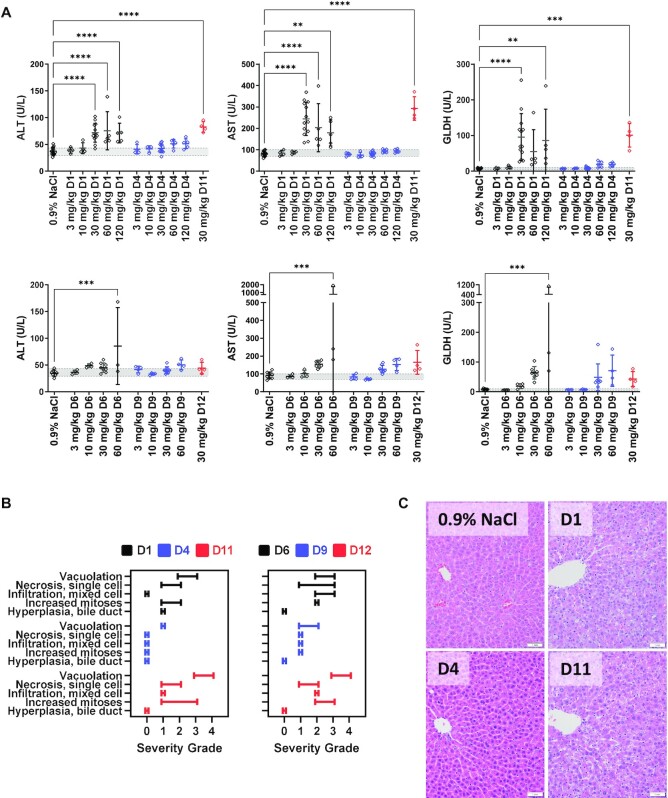
To assess the impact of GNA modification on the therapeutic index, GalNAc–siRNAs D1, D4, D6 and D9 were subsequently evaluated in an exploratory toxicity study in rats, with endpoints that included survival, serum chemistry and microscopic liver findings to assess the level of hepatotoxicity after three weekly doses of 3, 10, 30, 60 or 120 mg/kg. As expected, a dose-dependent elevation of liver enzymes was observed after repeated weekly administration of Ttr-targeting D1 with a 2–3-fold increase in ALT and aspartate aminotransferase (AST), and an 8–10-fold increase in glutamate dehydrogenase (GLDH) at the highest dose (Figure 2A). The greater sensitivity of GLDH compared to ALT/AST is consistent with the profile of our GalNAc–siRNA platform in rat toxicity studies, where GLDH was found to be most predictive for microscopic changes in the liver across 28 rat toxicity studies (35). The substitution of GNA in D4 led to a notable decrease or complete attenuation of liver ALT, AST and GLDH elevations across all doses tested. Similar trends were observed across a panel of three additional clinical chemistry parameters measured in this study (Supplementary Figures S8–S13 and Supplementary Table S3). In agreement with the observed lack of off-target mitigation in vitro, rats treated with a repeated weekly administration of DNA-modified D11 at 30 mg/kg demonstrated liver ALT elevations comparable to the parent D1 (Figure 2A, Supplementary Figures S8–S13 and Supplementary Table S4).
Figure 2.
Selected clinical pathology parameters and microscopic liver findings measured in rats (n = 4–5) following three weekly doses of parent, GNA- or DNA-modified GalNAc–siRNAs targeting Ttr or Hao1. (A) Measured liver enzymes from serum that was collected 24 h after final dose. Data were collected from three different studies; D1, D4, D6 and D9 were evaluated in a single study at 3, 10 and 30 mg/kg, and two subsequent studies evaluated D1 and D4 or D6 and D9 at 30, 60 and 120 mg/kg, and D11 or D12 at 30 mg/kg. Controls and the overlapping 30 mg/kg groups were combined and plotted above. (B) Summary of the range of microscopic liver findings based on severity grade. (C) Microscopic liver findings in rats following three weekly doses of 30 mg/kg with the indicated siRNAs targeting Ttr compared to the 0.9% NaCl control. Livers were collected 24 h post-final dose for analysis and sections were stained with H&E.
Increased ALT, AST and particularly GLDH levels were also observed with the Hao1-targeting siRNA D6. However, several unscheduled mortalities in the 60 and 120 mg/kg groups led to a more complicated analysis (Figure 2A, Supplementary Figures S8–S13 and Supplementary Table S5). In fact, all the rats dosed weekly with 120 mg/kg of D6 were found to be moribund or deceased 4–5 days after the second dose of siRNA and had to be sacrificed prior to completion of the study. However, all the animals dosed at 120 mg/kg with the GNA-modified D9 successfully completed the study. In addition, D9 demonstrated a significant reduction in liver AST and GLDH elevations at doses >60 mg/kg, with slight elevations relative to the control group at doses >10 mg/kg. Although all rats dosed weekly with 120 mg/kg of D9 successfully completed the study, they did show elevated liver enzymes at the end of the study suggesting that the single GNA modification reduced but did not completely attenuate hepatotoxicity. Again, in contrast to GNA-modified D9, rats treated with seed DNA-modified D12 at 30 mg/kg demonstrated a level of ALT, AST and GLDH elevations equivalent to that of the parent D6 (Figure 2A, Supplementary Figures S8–S13 and Supplementary Table S4).
Liver samples were harvested at the end of the study for microscopic evaluation (Figure 2B and C). Rats administered ≥30 mg/kg of the parent Ttr-targeting siRNA D1 had substantial hepatotoxicity consisting of minimal to mild single cell necrosis and/or coagulative necrosis, karyomegaly, mild to moderate hepatocellular degeneration, oval cell hyperplasia, increased mitoses and/or minimal to moderate hepatocellular vacuolation (Figure 2B and C, and Supplementary Table S6). A similar spectrum of microscopic findings was observed with the siRNA D11 modified with DNA in the seed (Figure 2B and C, and Supplementary Table S7). The addition of GNA to this sequence eliminated most of these microscopic findings apart from minimal vacuolation, single cell necrosis and/or increased mitoses up to the highest tested dose of 120 mg/kg. Thus, the addition of GNA resulted in an increase in the NOAEL (no observed adverse effect level) from 10 mg/kg with D1 to ≥120 mg/kg with D4.
Salient test article-related microscopic liver findings observed in rats administered ≥30 mg/kg of the parent Hao1-targeting siRNA D6 consisted of minimal to mild eosinophilic cytoplasmic change, minimal to moderate increased mitoses, single cell necrosis, hepatocellular vacuolation, hepatocellular hypertrophy, minimal to marked bile duct and/or oval cell hyperplasia, and moderate to marked necrosis (Figure 2B and Supplementary Table S8). The severity and incidence of these findings were reduced with the GNA-modified siRNA D9 across all dose levels, with generally minimal to mild findings at ≤60 mg/kg. However, at the highest dose of 120 mg/kg, the microscopic findings in the liver were generally noted at an increased incidence and/or severity. Taken together, the addition of GNA resulted in an increase in NOAEL from 10 mg/kg with D6 to 60 mg/kg with D9. The microscopic findings with D12 at 30 mg/kg were in line with the parent siRNA D6 indicating that this DNA-modified siRNA design did not improve safety (Figure 2B and Supplementary Table S7).
We calculated the therapeutic index in rats as the ratio of the NOAEL to the dose at which the siRNA demonstrated an ED50 for target knockdown (Supplementary Figure S14). The therapeutic index for the parent D1 targeting Ttr was improved by ≥8-fold through the single GNA substitution in D4. Similarly, the therapeutic index for the Hao1-targeting series improved by 6-fold for the GNA-modified D9.
GNA mitigates gene dysregulation in rat liver
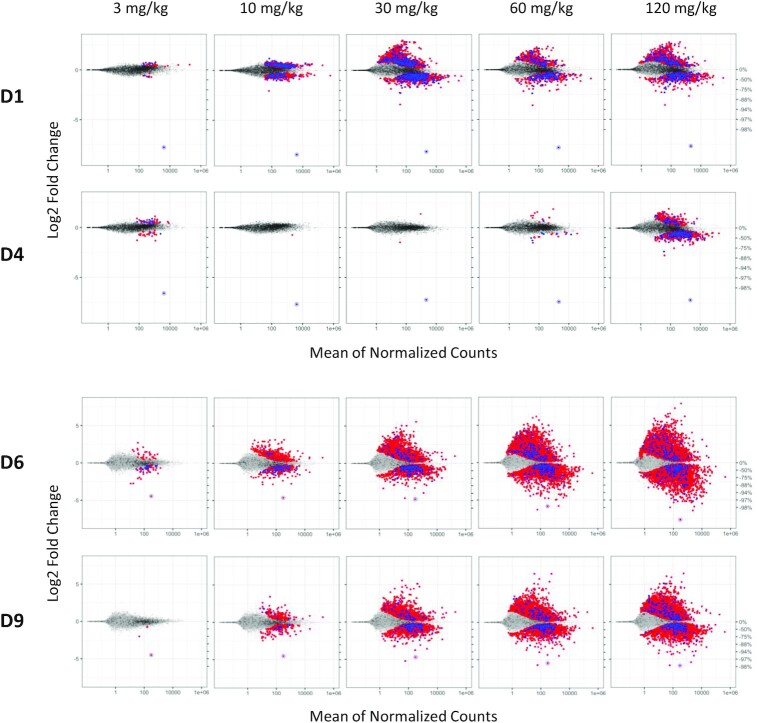
Livers from the rat toxicity studies were subsequently analyzed using RNA-seq to determine the level of transcriptional dysregulation due to the administration of each GalNAc–siRNA. While a transcriptome-based RNA-seq assay does not capture potential translational inhibition events resulting from off-target binding, mRNA destabilization has been demonstrated to be responsible for most of the observed miRNA-mediated decrease in protein production in mammalian cells (10–13). Hence, RNA-seq should provide a comprehensive measure of the off-target potential of a given siRNA. Weekly administration of Ttr-targeting D1 resulted in a dose-dependent increase of differentially expressed gene transcripts up to 30 mg/kg, at which point the number of differentially expressed genes appears to plateau (Figure 3, Supplementary Figure S15, and Supplementary Tables S9 and S10). The magnitude of cumulative distribution function (CDF) shift in the corresponding plots showed a dose-dependent increase across three of the four most common canonical seed matches (mer8, mer7-m8 and mer7-A1) up to 30 mg/kg ( Supplementary Figure S15, and Supplementary Tables S9 and S10) (36). In contrast, rats treated with the GNA-modified D4 showed little to no transcriptional dysregulation or substantial CDF shift up to and including 60 mg/kg, demonstrating that the incorporation of GNA was able to significantly mitigate off-target binding to all canonical seed sites. Consistent with the off-target reporter data, liver enzyme levels and histopathological findings, modification of the siRNA in the seed region with DNA (D11) had little to no impact on minimizing transcriptional dysregulation in vivo (Supplementary Figures S16 and S17).
Figure 3.
Transcriptional dysregulation in rat livers following three weekly doses of GalNAc–siRNAs targeting Ttr (D1, D4, top) or Hao1 (D6, D9, bottom). Frozen livers were collected 24 h after last dose for RNA-seq analysis. Log2 fold change plots (MA plot) represent the average signal from each cohort (n = 4–5). Dots represent individual rat gene transcripts, their average read count and the level of change in expression compared to the control group dosed with 0.9% NaCl. Whereas gray dots represent gene transcripts that were not determined to be differentially expressed after siRNA treatment relative to the control, the blue and red dots represent differentially expressed gene transcripts (false discovery rate < 0.05) with or without a canonical miRNA match (8mer, 7mer-m8, 7mer-A1 and mer6) to the guide seed region, respectively. On-target knockdown is represented by the circled dot.
Likewise, weekly administration of the Hao1-targeting D6 resulted in a strong and dose-dependent dysregulation of thousands of gene transcripts, many of which could not be attributed to one of the four most common canonical seed matches to the administered siRNA (Figure 3, Supplementary Figure S18 and Supplementary Table S9). Dysregulation of these gene transcripts without a canonical seed match is thought to be the result of secondary effects following the dysregulation of multiple primary (seed-matched) targets as well as other noncanonical matches to the parent siRNA, including those with complementarity to the supplemental region 3′ of the cleavage site (7,36). Although less pronounced than the Ttr-targeting series, liver samples from rats treated with the GNA-containing D9 showed a reduction in the number of dysregulated gene transcripts across all dose levels (Figure 3, Supplementary Figure S18, and Supplementary Tables S9 and S10). The reduced magnitude of CDF shift demonstrated a clear mitigation of off-targeting across all four canonical seed matches (mer8, mer7-m8, mer7-A1 and mer6) at doses up to and including 10 mg/kg. At higher doses, mitigation of these primary seed-mediated effects was less effective and overshadowed by secondary effects. As observed in the previous series, the seed DNA-containing D12 did not significantly alter the profile of differentially expressed gene transcripts compared to the parent D6 (Supplementary Figures S16 and S17).
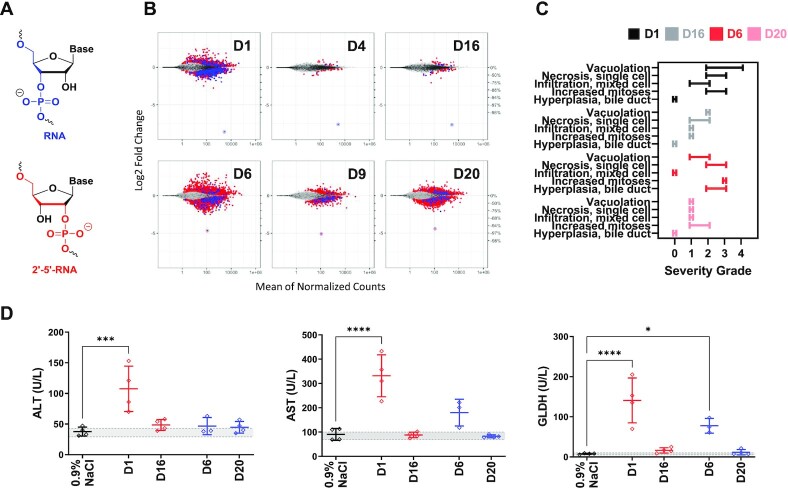
Expanding ESC+ designs with 2′–5′-RNA modification
To further expand our toolbox of ESC+ modifications and to explore the generalizability of our findings, we investigated 2′–5′-RNA linkages, which have been shown to thermally destabilize the formation of RNA duplexes (Figure 4A) (37–39). The pharmacodynamics of target silencing were evaluated in siRNAs containing a single 2′–5′-RNA linkage from positions g5–g8 in the same Ttr- and Hao1-targeting sequences that were utilized for GNA. For the series of siRNAs targeting Ttr, those modified with 2′–5′-RNA at position g7 (D16) or g8 (D17) showed activity and duration of silencing similar to that of the parent D1 through day 35 (Supplementary Figure S19). Although those siRNAs modified at g5 (D14) or g6 (D15) showed similar activity at Nadir, silencing activity recovered faster than the parent. A similar sequence-dependent activity was observed in the series of siRNAs targeting Hao1 where those modified with 2′–5′-RNA at g5, g6 or g7 (D18, D19 and D20, respectively) showed activity like the parent D6; however, the siRNA modified at g8 (D21) showed reduced activity (Supplementary Figure S20).
Figure 4.
Evaluation of 2′–5′-RNA-modified siRNAs in rats. (A) Structures of RNA and 2′–5′-RNA nucleotides. (B) Transcriptional dysregulation in primary rat hepatocytes following transfection of the specified siRNAs at a dose of 50 nM (n = 4). (C) Summary of the range of microscopic liver findings based on severity grade after three weekly doses of 30 mg/kg in rats (n = 4). (D) Measured liver enzymes from serum that was collected 24 h after final dose.
The ability of a single 2′–5′-RNA modification to mitigate seed-mediated off-targets of an siRNA was first evaluated using the off-target reporter assay. As with GNA, the effect was position- and sequence-dependent where the strongest mitigation of off-target silencing was observed with the siRNAs modified at position g7 with 2′–5′-RNA (D16 and D20; Supplementary Figures S21 and S22, respectively). Modification of g5, g6 or g8 had only modest effects on off-target mitigation in this reporter system. Interestingly, the incorporation of a single 2′–5′-RNA linkage in D16 and D20 led to a thermal destabilization of 3–4°C, similar to that of GNA, but less than the corresponding DNA designs (Table 2). We subsequently performed RNA-seq to further evaluate the effect of 2′–5′-RNA-modified siRNAs D16 and D20 on in vitro activity after transfection in primary rat hepatocytes (Figure 4B). The incorporation of 2′–5′-RNA in D16 and D20 led to a substantial reduction in off-targets and a significant improvement in the CDF plots in a fashion similar to, but to a lesser extent than, the corresponding GNA-modified siRNAs (D4 and D9; Figure 4B and Supplementary Figure S23).
These 2′–5′-RNA-modified GalNAc–siRNAs targeting Ttr or Hao1 were further evaluated in an exploratory rat toxicity study alongside the parent siRNAs at a dose of 30 mg/kg. Microscopic evaluation of the livers demonstrated a reduction in hepatotoxicity for both D16 and D20 relative to the parent siRNAs D1 and D6, respectively (Figure 4C). The increased levels of ALT, AST and GLDH observed in animals treated with the parent D1 were completely normalized with D16, which closely mimicked that of the saline control (Figure 4D). D20 targeting Hao1 also completely attenuated AST and GLDH elevations relative to the parent D6; however, inconsistent with our previous studies, ALT levels for animals treated with both Hao1-targeting siRNAs showed no difference to the control animals.
Application of ESC+ design to improve the specificity and safety of ALN-HBV
ALN-HBV is a GalNAc–siRNA conjugate that was designed to target all viral transcripts of the HBV for the treatment of chronic HBV infection, which can lead to cirrhosis, liver failure, hepatocellular carcinoma and death (40). This compound passed preclinical safety evaluations in both the rats and nonhuman primates. However, in a phase 1 single ascending dose study in healthy volunteers, transient, asymptomatic and dose-dependent increases in serum ALT levels ≥3 times the upper limit of normal were observed in 5 of the 18 subjects treated with ALN-HBV (manuscript submitted). The fact that such LFT increases were not observed in clinical studies with other development candidates of very similar chemistry and design pointed toward a sequence-dependent effect. Furthermore, the onset of liver enzyme elevations appeared to correlate with the expected onset of maximal PD activity and RISC loading 3–4 weeks post-dose. Taken together, we hypothesized that, in line with our preclinical studies, these findings may be due to RNAi-mediated miRNA-like off-target effects and that our ESC+ approach may be able to improve the therapeutic index in humans. Because of the imperfect transcriptome conservation between humans and standard toxicology species, safety testing of siRNA drug candidates in rodents may not fully predict the consequences of potential hybridization-based off-target activity in humans. Such effects may be better recapitulated in animal models expressing the human transcriptome, such as liver-humanized mice (manuscript submitted). However, our ESC+ approach employing thermally destabilizing seed modifications is expected to mitigate off-target binding in a species-agnostic manner.
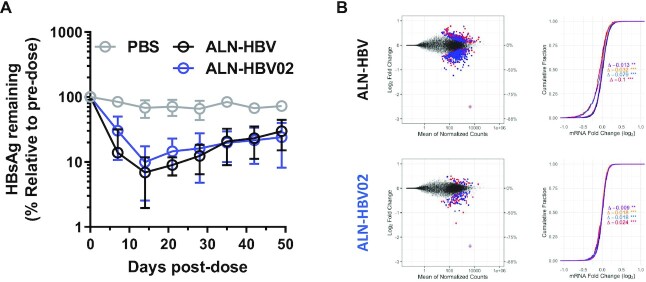
To assess whether the ESC+ approach could attenuate liver transaminase increases observed with ALN-HBV, we introduced GNA at g6 (ALN-HBV02) or g7 (D13) of the same siRNA sequence. On-target activity was evaluated by measuring serum HBsAg reduction in an HBV-AAV mouse model (Figure 5A). Target silencing with ALN-HBV and ALN-HBV02 after a single dose was found to be comparable over the course of 49 days, whereas the siRNA with substitution of GNA at g7 (D13) resulted in a loss of activity relative to the parent and was thereby deemed unsuitable for further development (Supplementary Figure S24). This finding highlights the sequence and position dependence of GNA substitution in designing ESC+ siRNAs; a balance between pharmacodynamics and off-target mitigation is necessary to develop therapeutically relevant siRNAs. In this sequence targeting HBV, only GNA substitution at g6 was found to have the optimal balance of on- and off-target activities, while in case of the Ttr- or Hao1-targeting sequences, only g7 or both g6 and g7 were found to be suitable, respectively. RNA-seq was used to evaluate the level of transcriptional dysregulation after transfection of these siRNAs in a HepG2-derived cell line stably transfected with the full genome of HBV. A significant reduction in the total number of differentially expressed gene transcripts and a shifted distribution with loss of seed-type statistical significance were observed with ALN-HBV02 relative to ALN-HBV in the log2 fold change (MA) and CDF plots, respectively (Figure 5B, and Supplementary Tables S11 and S12). ALN-HBV02 was advanced through preclinical development and evaluated as VIR-2218 in a phase 1/2 clinical trial in healthy volunteers or adult patients with chronic HBV infection (manuscript submitted).
Figure 5.
Evaluation of the efficacy and specificity of ALN-HBV02. (A) Pharmacodynamics after a single 1 mg/kg dose of ALN-HBV or ALN-HBV02 in mice transduced with HBV-AAV8. Serum HBsAg levels represent the average and error bars represent the standard deviation of all animals from a given cohort (n = 3), each normalized to individual pre-dose serum HBsAg levels. (B) Measurement of transcriptional dysregulation in HepG2.2.15 after transfection at 10 nM with ALN-HBV or ALN-HBV02 relative to a mock control. MA and CDF plots are shown on the left and right, respectively. In CDF plots, each colored line represents the impact of different types of seed matches on the cumulative dysregulation of gene transcripts: black = background; purple = mer6; yellow = mer7-A1; blue = mer7-m8; and red = mer8.
DISCUSSION
The development of RNAi therapeutics relies on the discovery of safe and effective siRNAs, which specifically target the desired transcript with minimal off-target effects. We have previously reported that RNAi-mediated off-target effects are largely responsible for the hepatotoxicity observed for a small subset of GalNAc–siRNAs across species and outlined a possible mitigation strategy using GNA to destabilize seed-mediated pairing to off-target transcripts. The present report describes the development of a novel siRNA design aimed to directly address these shortcomings through a systematic assessment of GNA incorporation with regard to siRNA on- and off-target activities, in vivo translation and therapeutic index in rats. This design, termed ESC+, was found to significantly reduce siRNA off-target effects and mitigate the safety findings without compromising on-target activity. To our knowledge, this constitutes the first report demonstrating the improvement of the therapeutic index of siRNAs in animals through a chemical modification pattern aimed toward a local destabilization of seed-mediated off-target engagement. Taken together, our investigations also point toward a remarkably negligible contribution of chemistry-based toxicities, i.e. non-hybridization-based side effects resulting from the accumulation of siRNA and/or metabolites in target tissues. This highlights the highly biocompatible nature of the chemistries used for GalNAc–siRNA conjugates.
A single incorporation of GNA in the guide strand between g5 and g8 in two model sequences only slightly impacted the in vitro on-target activity, while substantially suppressing off-target activity in a position-dependent manner. The lack of in vivo translation observed in mice for several GNA-modified siRNAs could be attributed to lower concentrations of intact siRNA in the liver. This suggests that, as was previously reported, the siRNA may be exposed to a more expedient nucleolytic degradation due to an increased fraying of the duplex in the vicinity of the strong thermal destabilization introduced by GNA modification (22,30). Nevertheless, we were able to identify at least one position for each sequence for which the in vivo efficacies measured by mRNA knockdown closely matched those of the parent siRNAs.
The work presented here highlights the sequence dependence of the ESC+ design requiring a somewhat empirical discovery process to arrive at lead candidates with the desired potency, specificity and in vivo translation. However, we also showed that this process follows a straightforward screening paradigm combining RNA-seq and reporter off-target assays with in vitro silencing for initial screening followed by in vivo pharmacology for a selected subset of compounds. When this strategy was applied to two model GalNAc–siRNAs with established off-target potential and hepatotoxicity, quantitative evaluation of potency and safety demonstrated a 6–8-fold improved therapeutic window in the rat based on survival, clinical pathology and liver histopathology. The incorporation of GNA at g7 was well tolerated in vivo with little to no loss in potency (ED50) and toxicity studies showed reduced mortality, reduced liver enzyme elevations, reduced microscopic findings and quieter off-target RNA-seq profiles in livers of animals treated with GNA-containing GalNAc–siRNAs. Remarkably for one of the sequences, the clinical pathology and RNA-seq profiles of the GNA-modified version (D4), in stark contrast to the parent siRNA (D1), were found to be very similar to the saline control group for doses up to 60 mg/kg. Our in vitro assays were able to predict the rank order and differences between the therapeutic window of the parent and GNA-modified siRNAs for each of the two sequences evaluated in these studies, albeit not quantitatively.
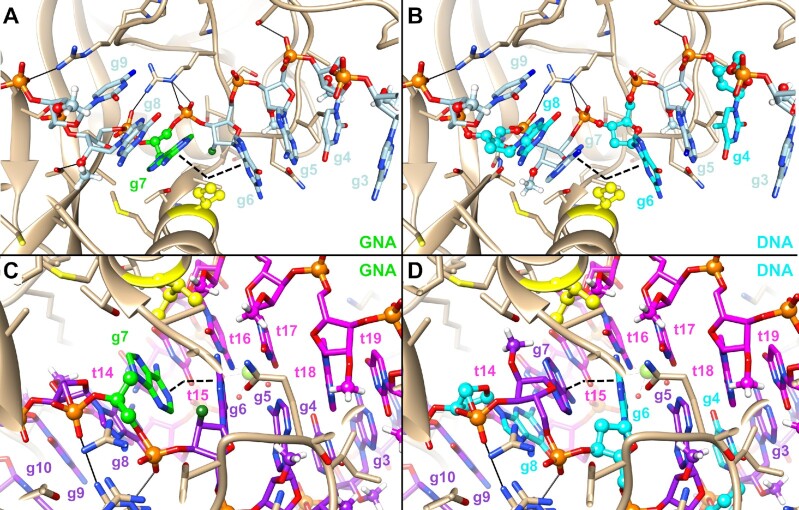
The results from the DNA- and 2′–5′-RNA-modified siRNAs demonstrated that destabilization of seed pairing measured in the context of free siRNA (not bound to RISC) is not sufficient to predict an improved safety of hepatotoxic siRNAs. Although the introduction of three interspersed DNA nucleotides in the seed region decreased the duplex thermal stability to a similar or greater extent than the incorporation of a single GNA or 2′–5′-RNA nucleotide, the DNA-modified siRNAs did not demonstrate a meaningful improvement in specificity or safety in any of our assays. In contrast, the siRNAs modified with a single GNA or 2′–5′-RNA demonstrated an improved specificity and safety across all assays. Intrigued by this result, we performed modeling of GNA- or DNA-modified guide strands into published structures of Ago2 in complex with guide only or Ago2 bound to a guide–target duplex (Figure 6). The models in Figure 6 show that the different nucleic acid modifications used in these designs (2′-OMe, 2′-F, DNA and GNA) are not expected to interfere strongly with binding of the guide strand to the protein, with the substituent at the 2′-position forming very few interactions with the Ago2 protein (Figure 6). This is consistent with previous reports that describe that the major interactions between Ago2 and RNA guide strand appear to be driven largely through contacts between the bridging phosphate groups and the protein backbone, with H-bond interactions to the 2′-OH groups playing only a minor role for guide strand recognition (41–43). Given that the global structure of Ago2 appears much less flexible than its corresponding guide counterpart (43), it is reasonable to assume that Ago2 can pre-organize the DNA nucleotides in the seed into an A-like conformation, thereby closely mimicking the global structure of the parent siRNAs in this study. Since it has also been reported that DNA nucleotides adopt an RNA-like C3′-endo/C2′-exo (North) conformation within the context of an RNA duplex (44–46), we hypothesize that the local structure of these DNA nucleotides in the siRNA duplex, and when bound as a guide strand by Ago2, closely resembles a pre-organized A-form conformation like RNA, 2′-OMe or 2′-F nucleotides in the parent siRNA.
Figure 6.
Modeling of modified guide strands in human Ago2. GNA-modified guide strand of D4 (A) or DNA-modified guide strand of D11 (B) modeled into the structure of Ago2–guide complex (PDB code 4F3T) (41). The angle of the kink in the guide strand introduced by Ile365 is indicated by the black dashed wedge. The same guide strands, now modeled into a structure of Ago2 in complex with both guide and target RNAs (PDB code 4W5T), are shown in panels (C) and (D) (7). A relax of the kink between g6 and g7 can be observed with a close to A-like conformation of the guide–target duplex.
Recognition of candidate sites by miRNA guide strands is proposed to propagate through an initial binding of guide nucleotides g2–g5 in which the Watson–Crick faces of the nucleobases are exposed to solvent in a pre-organized A-form conformation. The interaction between target RNA and guide strand is thereafter further stabilized through a conformational shift of helix 7 in the Argonaute protein, which relaxes a kink of the A-form structure between g6 and g7 caused by the insertion of an isoleucine residue on helix 7 (Supplementary Figure S25) (7,41,42). This, in turn, allows the guide strand to form a continuous A-form helix from g2 to g7, and also pre-organizes g13–g16 into the A-form to allow for potential pairing in the supplemental region (7). When Ago2 is bound only to guide RNA in the guide-only conformation, the P–P distances between g5 and g6, g6 and g7 (site of the induced kink), and g7 and g8 are 5.6, 5.5 and 6.5 Å, respectively, distinct from that of the average distance in A-form duplexes of 5.9 Å (Supplementary Figure S26). After the conformational shift of Ago2 to allow continuous binding from g2 to g7, the P–P distances of 5.9, 5.6 and 5.9 Å between g5 and g6, g6 and g7, and g7 and g8, respectively, more closely mimic those found in the average A-form duplex. Given the importance of this conformational shift that allows nucleotides g2–g7 to engage in productive miRNA–target recognition, the altered P–P distance introduced by GNA or 2′–5′-RNA would appear to favor the guide-only complex with Ago2 in the absence of full target complementarity, that is the conformation that precludes continuous seed-only binding. The incorporation of GNA at g7 would introduce flexibility and allow the guide strand to more easily adopt a shorter (22) g6–g7 P–P distance of 5.5 Å, potentially favoring the guide-only state and explaining the ability of GNA to mitigate seed-only binding. Similarly, the introduction of a 2′–5′-RNA linkage at g7 would lengthen (38) the P–P distance between g7 and g8, which could more easily adopt to the 6.5 Å observed in the guide-only structure, and may explain the strong preference for optimal off-target mitigation when this modification was incorporated at g7. On the other hand, DNA introduced in this context would be induced into an A-form and would not be proposed to strongly affect the P–P distances along the backbone of the guide strand relative to native RNA (44–46), and thereby would have little effect on mitigating an miRNA-like recognition of off-targets.
To address the question of whether this novel ESC+ design could help to improve the therapeutic index of clinically relevant siRNAs by reducing their propensity toward off-target effects, we applied this strategy to refine ALN-HBV, which was discontinued due to elevations in liver transaminases observed in several subjects in a phase 1 study in healthy volunteers. To improve the safety of this siRNA, we developed ALN-HBV02, the corresponding sequence-matched ESC+ version with a single chemistry change where the 2′-F modification at g6 was replaced with GNA. ALN-HBV02 showed a strong reduction in transcriptional dysregulation, while maintaining a similar PD profile to ALN-HBV in an AAV mouse model, providing support for the introduction of ALN-HBV02 into preclinical and clinical development. ALN-HBV02 demonstrated an improved hepatic safety profile and robust dose-dependent HBsAg in humans, suggesting an enhanced therapeutic window and providing the first human proof of concept for the ESC+ approach (manuscript submitted). This widened therapeutic window allows ALN-HBV02 to be dosed at higher levels to maximize and prolong anti-HBV knockdown, a critical feature for this antiviral strategy.
The present work in conjunction with the human safety data (manuscript submitted) highlights an example in which learnings from bedside were brought back to the bench to successfully address a specific safety issue. It also presents a rare opportunity for drug discovery to deconvolute the impact of a small but critical structural change on the performance of a given drug and its translation into humans. With several other ESC+ candidates already advancing through clinical development, we expect further human data confirming the improved specificity and safety of this novel design.
DATA AVAILABILITY
The RNA-seq data in this manuscript have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE184929.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all members of the RNA synthesis, in vitro screening, investigational toxicology, early development, RNA-seq and animal care facilities who helped support the execution of these studies.
Contributor Information
Mark K Schlegel, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Maja M Janas, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Yongfeng Jiang, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Joseph D Barry, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Wendell Davis, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Saket Agarwal, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Daniel Berman, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Christopher R Brown, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Adam Castoreno, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Sarah LeBlanc, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Abigail Liebow, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Tara Mayo, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Stuart Milstein, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Tuyen Nguyen, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Svetlana Shulga-Morskaya, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Sarah Hyde, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Sally Schofield, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
John Szeto, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Lauren Blair Woods, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Vedat O Yilmaz, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Muthiah Manoharan, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Martin Egli, Department of Biochemistry, School of Medicine, Vanderbilt University, Nashville, TN 37232, USA.
Klaus Charissé, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Laura Sepp-Lorenzino, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Patrick Haslett, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Kevin Fitzgerald, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Vasant Jadhav, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
Martin A Maier, Alnylam Pharmaceuticals, Inc., Cambridge, MA 02142, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Alnylam Pharmaceuticals. Funding for open access charge: Alnylam Pharmaceuticals.
Conflict of interest statement. With the exception of Martin Egli, all authors were employees of Alnylam Pharmaceuticals during the time this work was conducted.
REFERENCES
- 1. Nair J.K., Willoughby J.L.S., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel’in A.V., Milstein S.et al.. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014; 136:16958–16961. [DOI] [PubMed] [Google Scholar]
- 2. Prakash T.P., Graham M.J., Yu J., Carty R., Low A., Chappell A., Schmidt K., Zhao C., Aghajan M., Murray H.F.et al.. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 2014; 42:8796–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zimmermann T.S., Karsten V., Chan A., Chiesa J., Boyce M., Bettencourt B.R., Hutabarat R., Nochur S., Vaishnaw A., Gollob J.. Clinical proof of concept for a novel hepatocyte-targeting GalNAc–siRNA conjugate. Mol. Ther. 2017; 25:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Foster D.J., Brown C.R., Shaikh S., Trapp C., Schlegel M.K., Qian K., Sehgal A., Rajeev K.G., Jadhav V., Manoharan M.et al.. Advanced siRNA designs further improve in vivo performance of GalNAc–siRNA conjugates. Mol. Ther. 2018; 26:708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nair J.K., Attarwala H., Sehgal A., Wang Q., Aluri K., Zhang X., Gao M., Liu J., Indrakanti R., Schofield S.et al.. Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc–siRNA conjugates. Nucleic Acids Res. 2017; 45:10969–10977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Janas M.M., Schlegel M.K., Harbison C.E., Yilmaz V.O., Jiang Y., Parmar R., Zlatev I., Castoreno A., Xu H., Shulga-Morskaya S.et al.. Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat. Commun. 2018; 9:723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schirle N.T., Sheu-Gruttadauria J., MacRae I.J.. Structural basis for microRNA targeting. Science. 2014; 346:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jackson A.L., Burchard J., Schelter J., Chau B.N., Cleary M., Lim L., Linsley P.S.. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006; 12:1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Birmingham A., Anderson E.M., Reynolds A., Ilsley-Tyree D., Leake D., Fedorov Y., Baskerville S., Maksimova E., Robinson K., Karpilow J.et al.. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006; 3:199–204. [DOI] [PubMed] [Google Scholar]
- 10. Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P.. The impact of microRNAs on protein output. Nature. 2008; 455:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hendrickson D.G., Hogan D.J., McCullough H.L., Myers J.W., Herschlag D., Ferrell J.E., Brown P.O.. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009; 7:e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo H., Ingolia N.T., Weissman J.S., Bartel D.P.. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010; 466:835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eichhorn S.W., Guo H., McGeary S.E., Rodriguez-Mias R.A., Shin C., Baek D., Hsu S.H., Ghoshal K., Villén J., Bartel D.P.. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell. 2014; 56:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burel S.A., Hart C.E., Cauntay P., Hsiao J., Machemer T., Katz M., Watt A., Bui H.-H., Younis H., Sabripour M.et al.. Hepatotoxicity of high affinity gapmer antisense oligonucleotides is mediated by RNase H1 dependent promiscuous reduction of very long pre-mRNA transcripts. Nucleic Acids Res. 2015; 44:2093–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Laursen M.B., Pakula M.M., Gao S., Fluiter K., Mook O.R., Baas F., Langklaer N., Wengel S.L., Wengel J., Kjems J.et al.. Utilization of unlocked nucleic acid (UNA) to enhance siRNA performance in vitro and in vivo. Mol. Biosyst. 2010; 6:862–870. [DOI] [PubMed] [Google Scholar]
- 16. Mook O., Vreijling J., Wengel S.L., Wengel J., Zhou C., Chattopadhyaya J., Baas F., Fluiter K.. In vivo efficacy and off-target effects of locked nucleic acid (LNA) and unlocked nucleic acid (UNA) modified siRNA and small internally segmented interfering RNA (sisiRNA) in mice bearing human tumor xenografts. Artif. DNA PNA XNA. 2010; 1:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vaish N., Chen F., Seth S., Fosnaugh K., Liu Y., Adami R., Brown T., Chen Y., Harvie P., Johns R.et al.. Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res. 2011; 39:1823–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ui-Tei K., Naito Y., Zenno S., Nishi K., Yamato K., Takahashi F., Juni A., Saigo K.. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008; 36:2136–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee H.S., Seok H., Lee D.H., Ham J., Lee W., Youm E.M., Yoo J.S., Lee Y.S., Jang E.S., Chi S.W.. Abasic pivot substitution harnesses target specificity of RNA interference. Nat. Commun. 2015; 6:10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seok H., Jang E.S., Chi S.W.. Rationally designed siRNAs without miRNA-like off-target repression. BMB Rep. 2016; 49:135–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bramsen J.B., Pakula M.M., Hansen T.B., Bus C., Langkjaer N., Odadzic D., Smicius R., Wengel S.L., Chattopadhyaya J., Engels J.W.et al.. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010; 38:5761–5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schlegel M.K., Foster D.J., Kel’in A.V., Zlatev I., Bisbe A., Jayaraman M., Lackey J.G., Rajeev K.G., Charisse K., Harp J.et al.. Chirality dependent potency enhancement and structural impact of glycol nucleic acid modification on siRNA. J. Am. Chem. Soc. 2017; 139:8537–8546. [DOI] [PubMed] [Google Scholar]
- 23. Zhang L., Peritz A., Meggers E.. A simple glycol nucleic acid. J. Am. Chem. Soc. 2005; 127:4174–4175. [DOI] [PubMed] [Google Scholar]
- 24. Zhang L., Peritz A.E., Carroll P.J., Meggers E.. Synthesis of glycol nucleic acids. Synthesis. 2006; 4:645–653. [Google Scholar]
- 25. Schlegel M.K., Meggers E.. Improved phosphoramidite building blocks for the synthesis of the simplified nucleic acid GNA. J. Org. Chem. 2009; 74:4615–4618. [DOI] [PubMed] [Google Scholar]
- 26. Doench J.G., Petersen C.P., Sharp P.A.. siRNAs can function as miRNAs. Genes Dev. 2003; 17:438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R.et al.. Real-time quantification of microRNAs by stem–loop RT-PCR. Nucleic Acids Res. 2005; 33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pei Y., Hancock P.J., Zhang H., Bartz R., Cherrin C., Innocent N., Pomerantz C.J., Seitzer J., Koser M.L., Abrams M.T.et al.. Quantitative evaluation of siRNA delivery in vivo. RNA. 2010; 16:2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Elkayam E., Parmar R., Brown C.R., Willoughby J.L., Theile C.S., Manoharan M., Joshua-Tor L.. siRNA carrying an (E)-vinylphosphonate moiety at the 5′ end of the guide strand augments gene silencing by enhanced binding to human Argonaute-2. Nucleic Acids Res. 2016; 45:3528–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schlegel M.K., Matsuda S., Brown C.R., Harp J.M., Barry J.D., Berman D., Castoreno A., Schofield S., Szeto J., Manoharan M.et al.. Overcoming GNA/RNA base-pairing limitations using isonucleotides improves the pharmacodynamic activity of ESC+ GalNAc–siRNAs. Nucleic Acids Res. 2021; 49:10851–10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 33. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Castellanos-Rizaldos E., Brown C.R., Dennin S., Kim J., Gupta S., Najarian D., Gu Y., Aluri K., Enders J., Brown K.et al.. Reverse transcription quantitative polymerase chain reaction methods to support pharmacokinetics and drug mechanism of action to advance development of RNA interference therapeutics. Nucleic Acid Ther. 2020; 30:133–142. [DOI] [PubMed] [Google Scholar]
- 35. Janas M.M., Harbison C.E., Perry V.K., Carito B., Sutherland J.E., Vaishnaw A.K., Keirstead N.D., Warner G.. The nonclinical safety profile of GalNAc-conjugated RNAi therapeutics in subacute studies. Toxicol. Pathol. 2018; 46:735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McGeary S.E., Lin K.S., Shi C.Y., Pham T.M., Bisaria N., Kelley G.M., Bartel D.P.. The biochemical basis of microRNA targeting efficacy. Science. 2019; 366:eaav1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wasner M., Arion D., Borkow G., Noronha A., Uddin A.H., Parniak M.A., Damha M.J.. Physicochemical and biochemical properties of 2′,5′-linked RNA and 2′,5′-RNA:3′,5′-RNA “hybrid” duplexes. Biochemistry. 1998; 37:7478–7486. [DOI] [PubMed] [Google Scholar]
- 38. Sheng J., Li L., Engelhart A.E., Gan J., Wang J., Szostak J.W.. Structural insights into the effects of 2′–5′ linkages on the RNA duplex. Proc. Natl Acad. Sci. U.S.A. 2014; 111:3050–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Habibian M., Harikrishna S., Fakhoury J., Barton M., Ageely E.A., Cencic R., Fakih H.H., Katolik A., Takahashi M., Rossi J.et al.. Effect of 2–5′/3–5′ phosphodiester linkage heterogeneity on RNA interference. Nucleic Acids Res. 2020; 48:4643–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seto W.K., Lo Y.R., Pawlotsky J.M., Yuen M.F.. Chronic hepatitis B virus infection. Lancet. 2018; 392:2313–2324. [DOI] [PubMed] [Google Scholar]
- 41. Elkayam E., Kuhn C.-D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L.. The structure of human Argonaute-2 in complex with miR-20a. Cell. 2012; 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schirle N.T., MacRae I.J.. The crystal structure of human Argonaute2. Science. 2012; 336:1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schirle N.T., Kinberger G.A., Murray H.F., Lima W.F., Prakash T.P., MacRae I.J.. Structural analysis of human Argonaute-2 bound to a modified siRNA guide. J. Am. Chem. Soc. 2016; 138:8694–8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Egli M., Usman N., Rich A.. Conformational influence of the ribose 2′-hydroxyl group: crystal structures of DNA–RNA chimeric duplexes. Biochemistry. 1993; 32:3221–3237. [PubMed] [Google Scholar]
- 45. Wahl M.C., Sundaralingam M.. B-form to A-form conversion by a 3′-terminal ribose: crystal structure of the chimera d(CCACTAGTG)r(G). Nucleic Acids Res. 2000; 28:4356–4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kel’in A.V., Zlatev I., Harp J., Jayaraman M., Bisbe A., O'Shea J., Taneja N., Manoharan R.M., Khan S., Charisse K.et al.. Structural basis of duplex thermodynamic stability and enhanced nuclease resistance of 5′-C-methyl pyrimidine-modified oligonucleotides. J. Org. Chem. 2016; 81:2261–2279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data in this manuscript have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE184929.