Summary
It is widely accepted that adipose-derived regenerative cells (ADRCs) can differentiate into mesodermal lineage cells. However, reprogramming adult ADRCs into mature cardiomyocytes is challenging. We investigated the induction of myocardial differentiation in ADRCs via direct reprogramming using lentiviral gene transfer. First, we identified candidate transcriptional factors by performing RNA sequencing and ultimately confirmed that the combination of six unique factors (Baf60c, Gata4, Gata6, Klf15, Mef2a, and Myocd) could efficiently express enhanced green fluorescent protein (GFP) in ADRCs isolated from adult alpha-myosin heavy chain promoter-driven GFP transgenic mice. The GFP-positive ADRCs induced by six factors (6F-ADRCs) expressed multiple cardiac genes and revealed cardiac differentiation in bioinformatic analysis. Moreover, injection of 6F-ADRCs into acute myocardial infarcted tissues in vivo resulted in the improvement of survival rate, fractional shortening, and reduction of infarction scar area. This study provides an alternative method for direct reprogramming of adult ADRCs into cardiomyocytes.
Subject areas: Cell biology, Stem cells research, Molecular biology
Graphical abstract
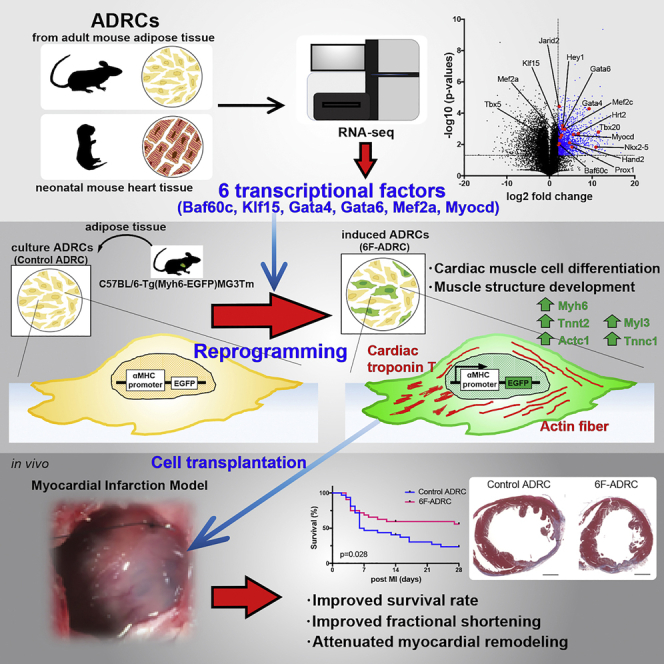
Highlights
-
•
Six transcriptional factors can differentiate ADRCs (6F-ADRCs) into cardiomyocytes
-
•
Bioinformatic analysis shows that 6F-ADRCs express multiple cardiac genes
-
•
Gene Ontology analysis indicates 6F-ADRCs toward cardiac differentiation
-
•
6F-ADRCs have potential for cardiac regeneration in in vivo transplantation model
Cell biology, Stem cells research, Molecular biology
Introduction
The mortality rate in patients with severe heart disease remains high. Ischemic heart disease is a leading cause of death (Puymirat et al., 2017). The common pathology of severe heart disease is extensive loss of functional cardiomyocytes with poor regenerative potential (Bergmann et al., 2015). Over recent decades, various types of cardiac regeneration strategies have been tested to improve cardiac function (Menasché et al., 2008; Menasche, 2009; Ptaszek et al., 2012; Zimmet et al., 2012; Kawamura et al., 2012; Eschenhagen et al., 2017). Among these strategies, cell-based therapy is at the center of regenerative medicine. Adipose-derived regenerative cells (ADRCs), which are a promising source for cell-based therapy, have been tested in various organs, including the heart (Zuk et al., 2001, 2002; Dai et al., 2016; Ma et al., 2017).
Although the sample sizes were small, several clinical trials demonstrated that the implantation of ADRCs after myocardial ischemia, when compared with placebo (Meliga et al., 2007; Houtgraaf et al., 2012; Perin et al., 2014), improved the clinical manifestation. In the mouse or rat myocardial infarction (MI) model, it has been reported that the paracrine effect of angiogenesis-promoting factors vascular endothelial growth factor (VEGF) and basic fibroblast growth factor (bFGF) from the transplanted ADRCs promoted angiogenesis, reduced the MI size, suppressed fibrosis, and improved the survival rate and cardiac function (Ii et al., 2011; Ishii et al., 2014; Gautam et al., 2015; Eguchi et al., 2019). Another example is that Neuregulin-1, an epidermal growth factor acting through receptor ErbB tyrosine kinases (Fuller et al., 2008), is also secreted by ADRCs, inducing cardiac repair (Bersell et al., 2009). We previously reported that stromal cell-derived factor 1 (SDF-1), a chemokine that directs the migration of different cell types, is secreted by implanted ADRCs, promoting the recovery of hindlimb ischemia by directing the migration of recipient cells (Kondo et al., 2009). In addition, almost no engraftment of ADRCs has been observed for several weeks post-implantation. This evidence indicates that the favorable effects of implanted ADRCs are not sustainable and are achieved by the paracrine effect of the cells and not by the differentiation of implanted ADRCs into new functional cardiomyocytes.
Here, we hypothesized that cardiac lineage-induced ADRCs could be engrafted for a long time to improve cardiac function and prognosis post MI. It has been previously reported that cardiac myofibroblast could be directly transdifferentiated into a cardiomyocyte-like cell by three defined factors: Gata4, Mef2c, Tbx5 (GMT) (Ieda, M. et al., 2010). In that report, up to 20% of myofibroblasts could be transdifferentiated into cardiomyocytes; however, it remains to be tested whether adult ADRCs can be transdifferentiated into cardiomyocytes by defined factors.
In the present study, we examined the profile of transcriptional factors expressed in the e11.5 embryonic heart to identify key molecules that transdifferentiate ADRCs into cardiomyocytes. We found that, among the highly expressed genes in the embryonic heart, a combination of six transcriptional factors can efficiently induce the cardiomyocyte phenotype in ADRCs. We also found that cardiac lineage-induced ADRCs can be engrafted for a longer period while maintaining the characteristics of cardiomyocytes, which improves cardiac function post MI in vivo.
Results
Candidate transcriptional factors for direct reprogramming of ADRCs to cardiomyocytes are identified by RNA sequencing analysis
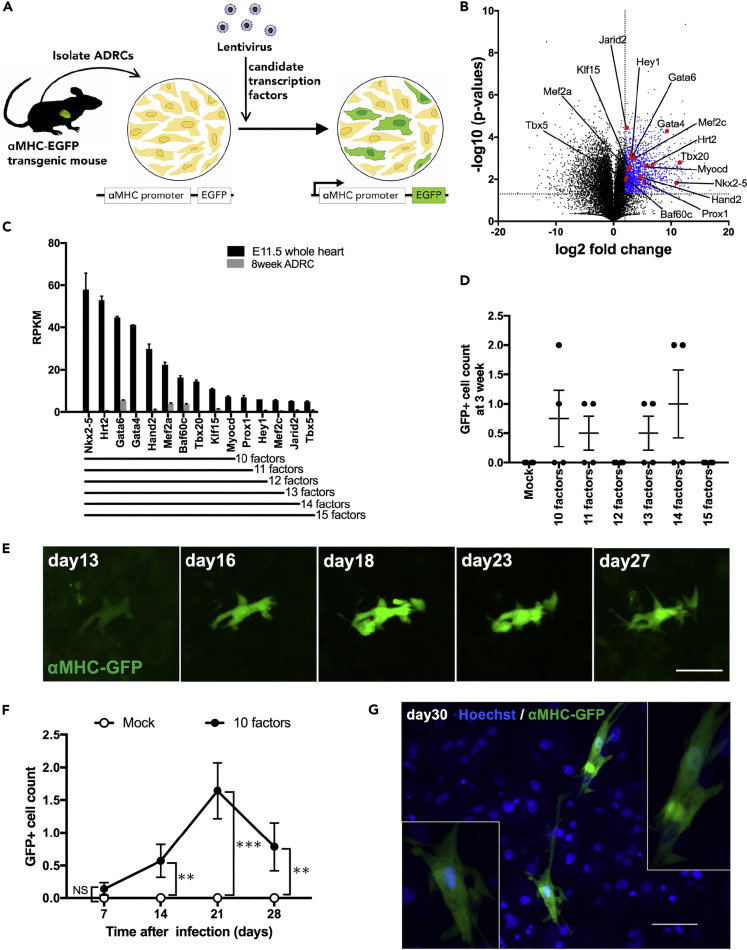
To visualize the promoter activation of myosin heavy chain 6 (αMHC), which is a marker of cell transdifferentiation into mature cardiomyocytes, we utilized the transgenic mice (C57BL/6-Tg (Myh6-EGFP) MG3Tm; MG3) carrying enhanced green fluorescent protein (αMHC-GFP) tagged to the αMHC promoter (Figure S1). Using lentiviral vectors, we introduced target genes into ADRCs isolated from adult MG3 adipose tissue (Figure 1A). The combination of three transcriptional factors, Gata4, Mef2c, and Tbx5 (GMT), can successfully help to transdifferentiate cardiac fibroblasts into cardiomyocyte-like cells (Ieda, M. et al., 2010). We tried this combination in ADRC; however, no green fluorescent protein (GFP)-positive cells were observed by either fluorescence microscopy or flow cytometry (Figures S2B and S2C). Therefore, using next-generation sequencing, we performed transcriptome analysis of adult mouse ADRCs and e11.5 embryonic mouse hearts to find unique or new combinations of transcriptional factors for transdifferentiating ADRCs into cardiomyocytes. Among the significantly and abundantly expressed genes in the e11.5 mouse heart, we identified 15 myocardium-specific transcription factors by annotation analysis (Figures 1B and 1C). In contrast, these 15 factors were poorly expressed in the adult ADRCs (Figure 1D). Notably, GMT transcriptional factors were included in 15 factors (Figure 1C). To identify the essential factors for reprogramming ADRCs toward cardiomyocytes, we removed the less expressed factors from the initial 15. We found that transduction of ten factors (Nkx2-5, Hrt2, Gata6, Gata4, Hand2, Mef2a, Baf60c, Tbx20, Klf15, and Myocd) into ADRCs (10F-ADRCs) could induce αMHC-GFP (Figure 1E), even though the induction efficiency was still extremely limited after three weeks of culture (Figure 1D). The number of GFP-positive ADRCs introduced by the ten factors gradually increased, peaking after 3 weeks (Figure 1F); however, contracting or striated cells were not observed microscopically (Figure 1G).
Figure 1.
Searching for factors inducing direct reprogramming of ADRCs to cardiomyocytes by performing RNA sequencing
(A) Scheme of strategy for exploring candidate factors.
(B) A volcano plot for the identification of candidate factors comparing ADRC versus embryonic heart (e11.5). The plot displays the log2-fold change in the expression for all detected RNAs. Transcripts with log2 fold change >4 and p value <0.05 are highlighted in blue.
(C) Fifteen myocardium-specific transcriptional factors, listed in descending order of their expression in whole embryonic heart (n = 2).
(D) Count of GFP+ADRCs transfected with factors 10-15 each. Cells were counted 3 weeks after viral infection (n = 4).
(E) Fluorescence microscopy of GFP+ADRCs transfected with 10 factors. The same cell was observed for 1 month (scale bar, 50 μm).
(F) Changes in the number of GFP+ cells after the transfection of 10 factors (n = 14). Cells were counted weekly (n = 14). (G) A representative image of GFP+ ADRC 30 days after transfection with 10 factors. Scale bar, 100 μM.
Data are represented as mean ± SEM (C, D, F). Statistical significance was determined with the Student’s t-test between two groups (F). ∗∗p < 0.05; ∗∗∗p < 0.01.
Six transcriptional factors are essential for inducing cardiomyocyte-specific genes on ADRCs
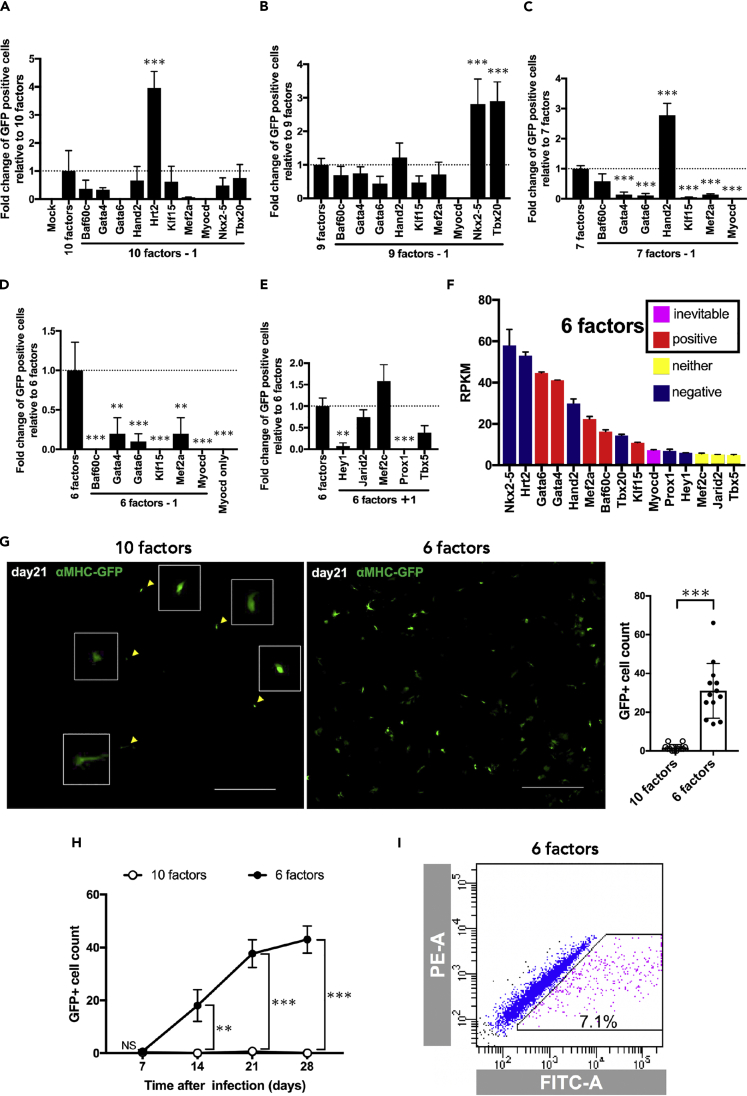
To identify the ideal combination of the ten factors, we removed each factor one by one and found that, if Hrt2 (Hey2) was removed from the initial ten factors, the efficiency of GFP expression was significantly increased (Figure 2A), indicating that Hrt2 is an inhibitory factor. Nkx2-5, Tbx20, and Hand2 were, in the same manner as Hrt2, identified as negative factors and were excluded from the combination of essential factors (Figures 2B and 2C). Importantly, when Myocd was excluded, no GFP+ cells were observed (Figures 2A–2C), indicating that Myocd is an indispensable factor (Figure 2F) (Iwafuchi-Doi and Zaret, 2014). None of the remaining five transcription factors (Gata6, Gata4, Mef2a, Baf60c, and Klf15) could be excluded (Figure 2D). We tested the 11th to 15th transcriptional factors (Prox1, Mef2c, Jarid2, and Tbx5) to determine whether they could induce more GFP+ cells in the absence of the already excluded negative factors (Hrt2, Nkx2-5, Tbx20, Hand2); however, none of them could effectively induce GFP+ cells (Figure 2E). As a result, we identified six factors (Gata6, Gata4, Mef2a, Baf60c, Myocd, and Klf15) essential for reprogramming ADRCs toward cardiomyocytes (Figure 2F). The combination of these six factors dramatically increased αMHC-GFP expression (Figure 2G). GFP+ADRCs were gradually visualized after 1 week of induction; afterward, the number linearly increased for 3 weeks, followed by a gradual increase thereafter. (Figure 2H). However, no beating colonies were observed. Using fluorescence-activated cell sorting (FACS) analysis, 5.033 ± 0.943% (n = 6) of GFP+ADRCs were detected after 3 weeks of transduction (Figure 2I, see also Figures S2B and S2C).
Figure 2.
The combination of six unique factors (Baf60c, Gata4, Gata6, Klf15, Mef2a, and Myocd) could efficiently express αMHC-GFP in ADRCs
(A–D) Impact of substruction of single factor from the combination of 10 factors (A), 9 factors (B), 7 factors (C), and 6 factors (D). GFP+ cells were counted 21 days after transfection. One-way ANOVA was used to examine statistical significance and an adjusted p value < 0.05 was considered significant. (A) n = 4, (B) n = 3, (C) n = 4, (D) n = 3. (E) Impact of factor no 11 to 15 in addition to six factors. GFP+ cells were counted 21 days after transfection (n = 3).
(F) Summarized characteristics of the 15 factors used for inducing GFP+ADRCs. (G) Representative images (left) of GFP+ cells induced by 10 factors and 6 factors 21 days after transfection. The number of GFR+ cells is dramatically increased by six factors (right). Scale bars, 1,000 μm (n = 13). (H) Transitions of GFP+ cells induced by 10 factors (open circle) and 6 factors (filled circle) (n = 3 each). (I) A representative FACS plot of αMHC-GFP+ADRCs 3 weeks after the transfection of six factors.
Data are represented as mean ± SEM (A-H). Statistical significance was determined with the ordinary one-way ANOVA with Dunnett's multiple comparisons test (A-E) or the Student’s t-test between two groups (G, H). ∗∗p < 0.05; ∗∗∗p < 0.01.
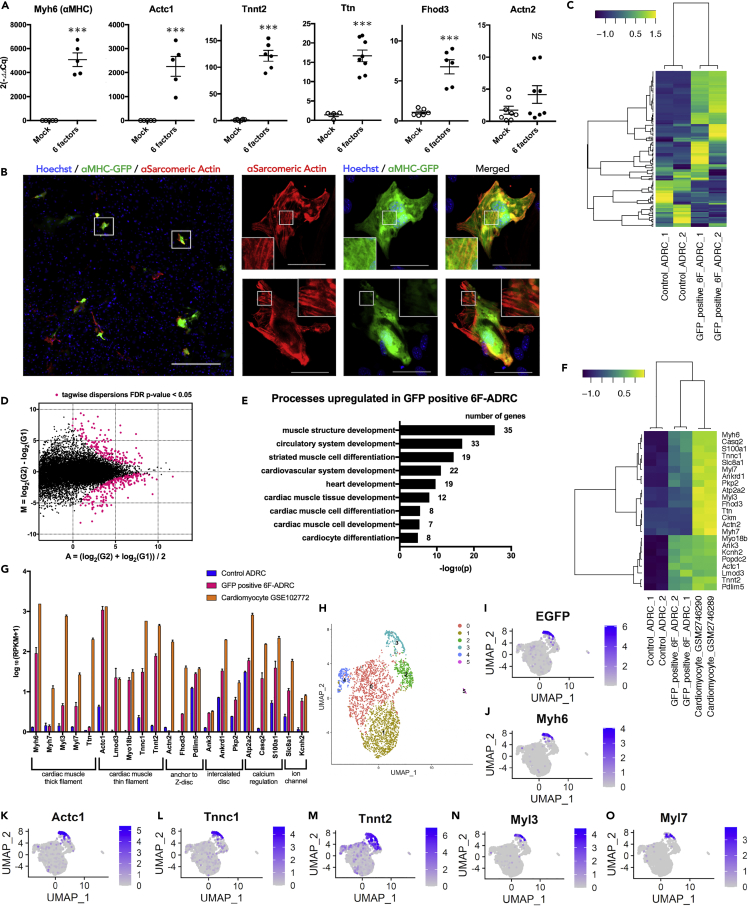
ADRCs induced with six factors express multiple myocardial genes and have the characteristics of cardiac differentiation accessed by bioinformatics analysis
We performed qPCR for cardiomyocyte-specific genes to determine whether the six factors could successfully induce the cardiomyocyte lineage in ADRCs. We found that several cardiomyocyte-specific genes, such as Myh6 (α-myosin heavy chain), Actc1 (cardiac α-actin), and Tnnt2 (Troponin T2, Cardiac Type), were significantly upregulated in the ADRCs induced by six factors (6F-ADRCs) compared with the mock control (Figure 3A).
Figure 3.
ADRCs induced with six factors express multiple myocardial genes and have the characteristics of cardiac differentiation accessed by bioinformatics analysis
(A) qPCR analysis of cardiac marker genes in 6F-ADRCs and mock induced ADRCs 3 weeks after transfection. Myh6 (n = 5), Actc1 (n = 5), Tnnt2 (n = 6), Ttn (n = 8), Fhod3 (n = 6), Actn2 (n = 8) (B) Representative images of GFP+ADRCs immunostaining for α-sarcomeric actin after induction of six factors. The secondary antibody was labeled with Alexa 594 (red). Scale bars, 500 μm (left panel) and 50 μm (right panels) (C) Heatmap image of RNA sequencing analysis illustrating selected cardiac genes among uninduced ADRCs and GFP+ 6F-ADRCs (FACS sorted 3 weeks after transfection). Log10(RPKM+1) values obtained from RNA-seq are shown (n = 2). Lists of genes are shown in Table S5.
(D) MA plot of differentially regulated genes between GFP+ 6F-ADRCs (G2) and uninduced ADRCs (G1). M=log2(G2)-log2(G1), A=(log2(G2)+log2(G1))/2. Strawberry colored plots show differentially expressed genes (DEGs) with tagwise dispersions FDR p value cutoff <0.05. Lists of DEGs are shown in Table S6.
(E) Gene ontology analysis for significantly upregulated genes (RNA-seq analysis) in GFP+ 6F-ADRCs. Lists of genes are shown in Figure S4 and Table S7.
(F) Heatmap image of RNA-seq analysis illustrating 23 cardiac genes among uninduced ADRCs, GFP+ 6F-ADRCs, and 12.9 weeks C57BL/6J adult ventricular cardiomyocytes (GSM2746289, GSM2746290).
(G) RNA-seq-based 21 cardiac genes expression levels among uninduced ADRCs, GFP+ 6F-ADRCs, and adult ventricular cardiomyocytes (GSM2746289, GSM2746290). Log10(RPKM+1) values obtained from RNA-seq are shown (n = 2).
(H) Uniform manifold approximation and projection (UMAP) clustering of six factors ADRCs 3 weeks after transfection, mixed with uninduced ADRCs. FeaturePlots of each gene expression showing (I) EGFP, (J) Myh6, (K) Actc1, (L) Tnnc1, (M) Tnnt2, (N) Myl3, (O) Myl7 expression.
Data are represented as mean ± SEM (A, G). Statistical significance was determined with the Student’s t-test between two groups (A). ∗∗∗p < 0.01.
The expression of Tnnt2, which is specifically expressed on thin filaments and regulates muscle contraction in response to alterations in intracellular calcium ion concentration, was significantly elevated in 6F-ADRCs compared with the control. Genes involved in sarcomere formation and Z body formation, such as Ttn (Titin) and Fhod3 (Formin Homology 2 Domain Containing 3), were also significantly upregulated (Figure 3A). On the other hand, the expression of Actn2 (actinin α2), which helps to anchor the myofibrillar actin filament, was not increased compared with that in the mock control (far right in Figure 3A). Immunocytochemical staining showed that most of the GFP+ADRCs induced by the six factors were co-positive with sarcomeric α-actin; however, consistent with the qPCR results, co-staining with α-actinin was not observed (Figure 3B). Although GFP+ADRC showed a fibrous conformation that was evident with sarcomeric α-actin staining (Figure 3B), a well-organized striated structure was not observed. Similar results were obtained for immunostaining of cardiac troponin T for GFP+ADRC (Figure S3). For comprehensive characterization of GFP+ADRCs, we next performed RNA sequencing (RNA-seq) on GFP+ADRCs collected 3 weeks after transduction by FACS on cultured ADRCs without gene transfer as a negative control (Control ADRCs). Heatmap images of selected cardiac-related genes illustrated that there were differentially expressed genes among GFP+ADRCs and the negative control (Figure 3C and Table S5, n = 2 in each group). We identified 414 genes that were differentially expressed between GFP+ADRCs and the negative control, with a false discovery rate (FDR) < 0.05 (Figure 3D and Table S6). Using the differentially expressed genes, we performed gene ontology (GO) term analysis based on the MGI_ID (Mouse Genome Informatics: http://www.informatics.jax.org/) of 160 genes significantly expressed in GFP+ADRCs (Figure S4 and Table S7). Processes that were upregulated in the GFP+ADRC group included muscle tissue/structure development and cardiac muscle cell development/differentiation, which indicate cardiomyocyte characteristics (Figure 3E). This analysis showed that a variety of cardiac genes, including Myh6, Actc1, and Tnnt2, were highly expressed in GFP+ADRCs, thus validating the qPCR results (Figures 3A, 3F, and 3G). The sarcomeric genes Myh6, Actc1, and Tnnt2 showed similar expression when compared with 12.9 weeks C57BL/6J adult ventricular cardiomyocytes (GSM2746289, GSM2746290, Chevalier et al., 2018) by RNA-seq (Figures 3F and 3G). To explore GFP+ADRCs at the single-cell level, we performed single-cell RNA sequence analysis (scRNA-seq) of 6F-ADRCs induced 3 weeks previously. After setting the thresholds, we retained the high-quality data of 2,911 cells in the reference. UMAP cells showed the presence of an EGFP expression cluster, and this distribution was identical to that of Myh6 (Figures 3H–3J). The distributions of Actc1, Tnnt2, and Tnnc1 (Troponin C Type 1), which constitute cardiac muscle thin filaments, were also almost identical to that of EGFP cluster (Figures 3K, 3L, and 3M). Interestingly, Myl3 (Myosin Light Chain 3, also known as MLC1v), which is a regionalized ventricle marker expressed after heart looping, was distributed in the same position as the EGFP cluster. However, Myl7 (Myosin Light Chain 7, also known as MLC2a), which is a pan-myocardial marker indicating the onset of cardiac differentiation, was distributed around the EGFP cluster (Figures 3N and 3O) (England and Loughna, 2013). The overexpressed six factors also showed accumulation around the EGFP cluster (Figures S5A–S5F). These scRNA-seq results were consistent with the data of RNA-seq given above, which showed that GFP+ADRCs expressed multiple cardiac genes simultaneously on a cell-by-cell basis. Taken together, GFP+ADRCs, although not identical, have similar characteristics to cardiomyocytes in the pattern of gene expression but are structurally premature compared with cardiomyocytes.
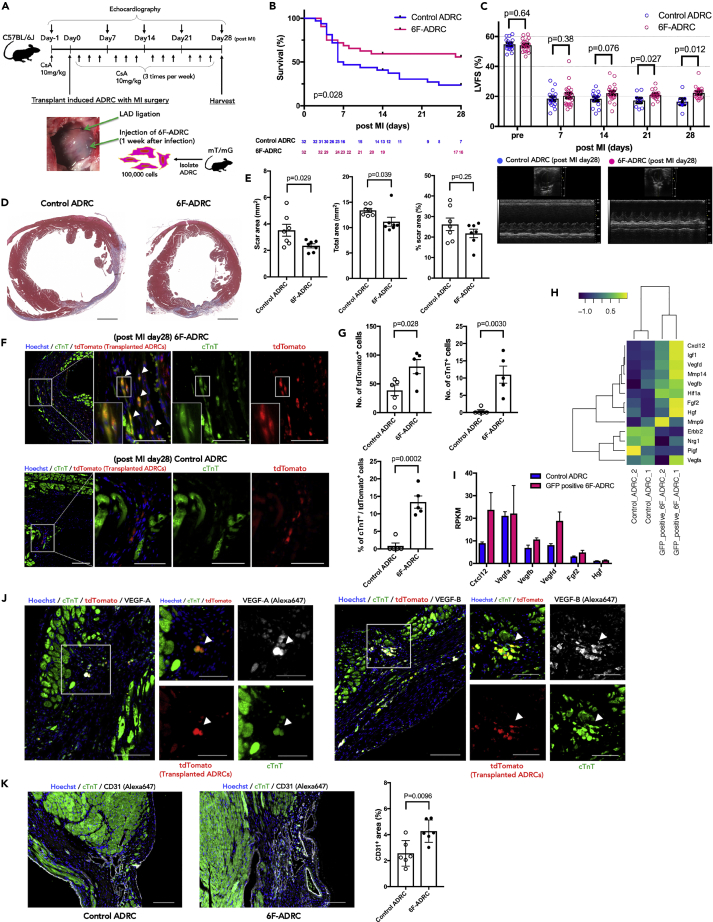
Transplantation of 6F-ADRCs into acute myocardial infarcted tissues improves chronic cardiac function in vivo
To investigate whether 6F-ADRCs transplantation might contribute to cardiac functional recovery by its direct or indirect effect on the acute phase of MI, we harvested 6F-ADRCs 1 week after viral induction and injected them into acute MI mouse hearts, whose left anterior descending (LAD) artery was ligated before cell transplantation. ADRCs from B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (mT/mG) mice (Muzumdar et al., 2007) were used to identify transplanted ADRCs by immunostaining with red fluorescent protein (RFP)-tagged antibodies. After 28 days of observation post operation, the surviving mice were euthanized for sample collection (Figure 4A). The survival rate was improved in the 6F-ADRCs implanted group compared with that in the non-induced ADRCs (Control ADRCs) group (Figure 4B, p = 0.028, with log rank [Mantel-Cox] testing). Echocardiography performed at 7, 14, 21, and 28 days after cell transplantation showed that the difference in left ventricular fractional shortening (LVFS) between 6F-ADRCs treated and control ADRCs treated groups gradually became clearer over time (Figure 4C, LVFS on Day 7, 20.45 ± 1.623 vs. 18.44 ± 1.452 mm, p = 0.38; LVFS on Day 14, 22.18 ± 1.391 vs. 18.32 ± 1.511 mm, p = 0.076) and was statistically significant at Days 21 and 28 (LVFS on Day 21, 20.97 ± 0.7922 vs 17.25 ± 1.595%, p = 0.027; LVFS on Day 28, 22.39 ± 1.127 vs. 16.48 ± 2.026%, p = 0.012). Cross-sectional tissue examination showed that the scar area and total left ventricle (LV) area were significantly reduced in the 6F-ADRCs group than the control ADRCs group, resulting in no significant difference in the percentage of scar area between the groups (Figures 4D and 4E). Accordingly, these results indicate that transplantation of 6F-ADRCs into acute MI model mice improved myocardial function and attenuated myocardial remodeling.
Figure 4.
Transplantation of 6F-ADRCs into acute myocardial infarcted tissues improves chronic cardiac function in vivo
(A) Experimental scheme using 8-week-old C57Bl/6J mice subjected to intracardiac injection of 6F-ADRCs. ADRCs were harvested from mT/mG mice so that the implanted ADRCs could be identified. As the treatment group, 6F-ADRCs 1 week after viral induction were injected (6F-ADRC group). As the control group, ADRCs without viral induction were injected (Control ADRC group).
(B) Survival plots up to 28 days after coronary artery ligation followed by the implantation of ADRCs (n = 22–32). Survival curves were analyzed using Kaplan–Meier estimators and log-rank (Mantel-Cox) tests.
(C) Echocardiogram analysis of left ventricular fractional shortening.
(D) Representative images of Masson’s trichrome staining were captured 4 weeks after transplantation. Scale bars, 1,000 μm. (E) Transplantation of 6F-ADRCs decreases the scar area (left) and cross-sectional area (middle) of the heart after MI. Relatively, the percent scar area was not significantly different (right).
(F) Transplanted ADRCs labeled with tdTomato (enhanced with immunostaining of anti-RFP/Alexa 647) from mT/mG mice were identified in the border area 4 weeks after acute MI. Immunostaining showed that double-positive cells of tdTomato and cTnT (with Alexa 488) were frequently observed in the 6F-ADRCs group compared with the uninduced ADRCs group. Scale bars, 100 μm (low magnified images) and 50 μm (high magnified images) (G) The number of dtTomato-positive cells representing implanted ADRCs significantly increased in the 6F-ADRC group compared with the Control ADRC group (up right). Almost no cTnT-positive implanted cell was observed in Control ADRC group (up left); however, 13.36 ± 1.754% of implanted cells were positive for cTnT in the 6F-ADRC group (bottom) (n = 5).
(H) Heatmap image of RNA-seq analysis illustrating 13 genes related to angiogenic paracrine factors among uninduced control ADRCs and GFP+ 6F-ADRCs. Log10(RPKM+1) values obtained from RNA-seq are shown (n = 2).
(I) RNA-seq-based angiogenic paracrine signals including VEGF-A and VEGF-B expression levels of GFP+ 6F-ADRCs relative to uninduced ADRCs. RPKM values obtained from RNA-seq are shown (n = 2).
(J) Transplanted 6F-ADRCs labeled with tdTomato and stained with cTnT were stained with VEGF-A and VEGF-B in the border area 4 weeks after acute MI surgery. The left row shows VEGF-A and the right row shows VEGF-B staining. Scale bars, 100 μm (low magnified images) and 50 μm (high magnified images). (K) CD31 immunofluorescence staining of section border zone of acute MI scar area, and CD31+ fraction area, compared with Control ADRCs treatment group and 6F-ADRCs treatment group (n = 6). Scale bars, 100 μm.
Data are represented as mean ± SEM (C, E, G, I, K). Statistical significance was determined with the Student’s t-test between two groups (C, E, G, K).
It has been previously reported that transplanted ADRCs were not frequently observed in heart samples 1 month after MI surgery (Ii, M. et al., 2011) or were transdifferentiated into endothelial cells (Cao et al., 2005; Martínez-Estrada et al., 2005; Ishii et al., 2014; Gautam et al., 2015). To clarify whether implanted 6F-ADRCs settled in cardiac tissue while maintaining the characteristics of cardiomyocytes, we observed the peri-infarcted area by histochemical staining. Implanted 6F-ADRCs were clearly distinguished as RFP-positive cells in the recipient tissue. We identified RFP+ cells co-stained with cardiac troponin T (cTnT) around the border zone of MI where 6F-ADRCs were implanted, indicating that the transplanted cells survived around the MI lesion with cardiomyocyte characteristics (Figure 4F). The number of ADRCs that survived 4 weeks after implantation, identified as RFP+ cells, increased significantly in the 6F-ADRCs group compared with the control ADRCs group. In addition, the cTnT-positive rate in RFP-positive cells showed a clear increase in the 6F-ADRCs group compared with the control ADRCs group (Figure 4G). RNA-seq data showed that 6F-ADRCs expressed no less level of multiple paracrine factors, such as vascular endothelial growth factor, than that of ADRCs without induction suggesting that 6F-ADRCs have considerably enhanced angiogenic effects via secretion of angiogenic cytokine (Figures 4H and 4I). The expression levels of VEGF-A and VEGF-B were confirmed by qPCR and were comparable in the GFP+ 6F-ADRCs group and the mock control ADRCs group (Figure S6). Fluorescence immunostaining was used to identify the cells expressing VEGF-A and VEGF-B, respectively, among the transplanted cTnT expressing 6F-ADRCs (Figure 4J). Specifically, VEGF-B promotes cardiac neovascularization after MI and improves cardiac tissue remodeling and function (Räsänen et al., 2021). We found that angiogenesis, which is thought to develop in response to paracrine factors secreted by transplanted cells at the site of acute MI, was significantly increased with 6F-ADRCs treatment group around the infarcted area (Figure 4K). Collectively, 6F-ADRCs remained viable with the characteristics of cardiomyocytes and the ability of angiogenic factor secretion for over 4 weeks, which promoted angiogenesis in the border zone of the acute MI area, thereby improving cardiac function and mortality after MI.
Discussion
Zuk et al. reported that a putative stem cell population isolated from adipose stroma could differentiate into adipogenic, osteogenic, chondrogenic, and myogenic lineages and express multiple surface markers similar to those detected on mesenchymal stem cells (MSCs) (Zuk et al., 2001, 2002). Because it is easy to access and obtain adipose-derived MSCs, a synonym for ADRCs, they have been widely applied in tissue engineering and clinical trials (Meliga et al., 2007; Valina et al., 2007; Katagiri et al., 2020). A successful phase II clinical trial (Houtgraaf et al., 2012; Perin et al., 2014) involving a double-blind placebo control study was conducted, in which adipose-derived stem cells (ADSCs), a synonym of ADRCs, or placebo was administered in cardiac ischemia by intramyocardial injection. In that study, patients administered ADSCs showed improvement in clinical manifestations, including NYHA and CCS classification, and a reduction in 12-month hospitalization rate compared with placebo; however, no significant difference was seen in the left ventricular ejection fraction between the groups (Henry et al., 2017). Numerous animal studies have explored the mechanisms underlying ADRC therapy. According to these findings, bilateral mechanistic insights have been made: a paracrine effect by which tissue survival is increased or apoptosis is prevented (Ii et al., 2011; Ishii et al., 2014; Gautam et al., 2015; Eguchi et al., 2019) and direct transdifferentiation of implanted ADRCs into cardiomyocytes. While a limited number of reports have concluded that ADRCs spontaneously differentiate into beating cardiomyocytes with minimal supplementation of growth factors (Planat-Bénard et al., 2004; Jumabay et al., 2010), in a vast majority of studies, the cardioprotective and proangiogenic effects of ADRCs have been attributed to the secretion of cytokines and growth factors such as insulin-like growth factor-1 (IGF-1) or the VEGF family (Ii, M. et al., 2011, Yang et al., 2013; Ishii et al., 2014; Gautam et al., 2015; Räsänen et al., 2021). Previously, we reported that, although ADRCs did not differentiate into myocytes in hindlimb ischemia, implanted ADRCs induced host cell migration by organizing neovascularization through SDF-1 secretion (Kondo et al., 2009). Accordingly, if a fraction of ADRCs can be differentiated into cardiomyocytes, which aids systolic function, the therapeutic effect could also be synergistically improved.
We showed that a combination of six transcriptional factors, Gata6, Gata4, Mef2a, Baf60c, Klf15, and Myocd, can efficiently induce cardiac genes similar to cardiomyocytes in adult ADRCs. The 6F-ADRCs have a limited ability to proliferate and do not form massive colonies. If implanted in MI, 6F-ADRCs can survive for an extended period, expressing cardiomyocyte-specific genes, promoting angiogenesis, and contributing to the improvement of cardiac function. Among the six factors, Myocd was characterized as an inevitable factor that did not induce GFP+ADRCs, either alone or when deleted, and induced cardiac differentiation in ADRCs (Figures 2A–2D). We also found that several transcriptional factors expressed in the e11.5 embryonic heart, such as Nkx2-5, Hrt2, and Hand2, adversely inhibited transdifferentiation of ADRCs into cardiomyocytes (Figures 2A–2C). We then analyzed the expression of the six factors exogenously expressed in ADRCs using scRNA-seq. Although there were differences in the distribution of expression among the factors, common high accumulations, which were consistent with the EGFP cluster, were observed for each of the six factors (Figures S5A–S5F). While the gene expression pattern of 6F-ADRCs was similar to that of cardiomyocytes, self-beating cells were not found in in vitro settings, even 4 weeks post induction. In addition, no mature sarcomere formation was observed in immunostaining of 6F-ADRC (Figure 3B). One of the problems would be that some genes required for sarcomere organization in cardiomyocytes were missing. For example, alpha actinin (Actn2) that is required for sarcomere assembly did not show a significant increase in expression (far right in Figure 3A). The next step would require further widespread expression including sarcomere-related genes.
In the current study, we isolated ADRCs from an αMHC-EGFP reporter mouse expressing GFP, specifically in cardiomyocytes. This system has been extensively used in a series of reprogramming studies as an indicator of transdifferentiation of fibroblast to cardiomyocyte (Ieda et al., 2010; Srivastava and Yu, 2015). We also took advantage of this system and found that six transcriptional factors could induce approximately 2–7% GFP in ADRCs (Figure S2C); however, we could not find mature cardiomyocytes. This is because several sarcomere genes, such as α-actinin, which are localized to the Z-disc and promote the elongation and maturation of actin filaments (Choi et al., 2008), were not detected in 6F-ADRCs (far right in Figure 3A). Wang et al. reported that the stoichiometry of the transcriptional factors or different isoforms largely influences the efficiency and quality of induced cardiac myocyte reprogramming (Wang et al., 2015, 2020). They showed that the combination of a higher level of Mef2c and a lower level of Gata4 and Tbx5 could induce 10-fold more mature cardiomyocytes than the number induced by the same level of these factors alone. Thus, it may be necessary to consider stoichiometry when inducing the six factors in ADRCs.
Despite the favorable effects of their implantation in MI, ADRCs can neither be engrafted for a long period of time nor develop into mature cardiomyocytes (Cai et al., 2009; Ii et al., 2011; Yang et al., 2013; Ishii et al., 2014; Gautam et al., 2015). Vagnozzi et al. suggested that the favorable effect of stem cell therapy is based on innate immune responses of the host heart and not on myocardial differentiation or paracrine effects of implanted cells (Vagnozzi et al., 2020). Accordingly, they advocated that the cell type of implantation has little to do with the therapeutic effect of stem cell therapy. In contrast to their results, 6F-ADRC showed favorable effects compared with uninduced ADRCs, indicating that the therapeutic effects differ among implanted cell types. One potential explanation for this discrepancy is that 6F-ADRCs may exert favorable effects in the relatively late phase of cardiac injury since the graft survival rate of 6F-ADRCs is better than that of uninduced ADRCs.
Generally, the engraftment capacity of derived cardiomyocytes varies with the differentiation stage for induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs), and mature differentiated iPSC-CMs are optimal for engraftment in mouse heart (Funakoshi et al., 2016). Our histological analysis showed that 6F-ADRCs were superior to non-induced ADRCs in terms of the efficacy of engraftment (Figure 3G). Multiple cardiac genes are expressed in 6F-ADRCs, and their cardiac properties may increase the efficiency of engraftment. Our data indicated that implantation of 6F-ADRCs improved cardiac function compared with non-induced ADRCs, which was statistically evident after 4 weeks of implantation. One possible mechanism is that, in addition to the direct effect of differentiation into cardiomyocytes, 6F-ADRCs survived for a long time by secreting certain factors that inhibit collagen generation and the activity of matrix metalloproteases, thus preventing LV remodeling after MI (Lindsey, 2018). We showed that multiple paracrine factors of VEGF-A/VEGF-B were expressed in engrafted 6F-ADRCs at the border zone of the MI area (Figure 4J) and promoted angiogenesis 4 weeks after transplantation (Figure 4K).
In conclusion, six transcription factors can transdifferentiate adult ADRCs into cardiac lineage cells. If implanted in MI, 6F-ADRCs engrafted around the MI border area could stay there for a longer period of time while maintaining the characteristics of cardiomyocytes, which may prevent LV remodeling and impairment of cardiac function. Further refinement is required to induce adult ADRCs into mature cardiomyocytes.
Limitations of the study
First, the detailed cellular and molecular mechanisms by which the six transcriptional factors reprogram ADRCs toward cardiomyocytes need further investigation. Second, although six factors reprogrammed ADRCs toward cardiomyocytes in this study, some cardiac genes (e.g., alpha actinin and beta cardiac myosin) that require sarcomere assembly (Chopra et al., 2018) did not show a significant increase in expression. It is possible that, if we overcome this problem, we could obtain contracting, mature cardiomyocytes. The next step would require further widespread expression of these myocardial component genes. Finally, as we used only male mice in in vivo studies, sex differences should be considered in future studies, as females may have a different response than those observed here. This is a limitation of the experimental model.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-alpha sarcomeric actin | Sigma Aldrich | Cat# A2172; RRID: AB_476695 |
| Mouse anti-Cardiac Troponin T | Abcam | Cat# ab10214; RRID: AB_2206574 |
| Mouse anti-Cardiac Troponin T | Abcam | Cat# ab8295; RRID: AB_306445 |
| Rabbit anti-RFP | Medical & Biological Laboratories | Cat# PM005; RRID: AB_591279 |
| Rat anti-RFP | ChromoTek | Cat# 5F8; RRID: AB_2336064 |
| Rabbit anti-VEGFA | Abcam | Cat# ab39250; RRID: AB_778794 |
| Rabbit anti-VEGFB | Abcam | Cat# ab185696 |
| Goat anti-CD31/PECAM-1 | Bio-Techne | Cat# AF3628; RRID: AB_2161028 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat# A11001; RRID: AB_2534069 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 546 | Invitrogen | Cat# A11081; RRID: AB_141738 |
| Goat anti-Mouse IgM (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Invitrogen | Cat# A21044; RRID: AB_2535713 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Invitrogen | Cat# A11005; RRID: AB_2534073 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen | Cat# A21244; RRID: AB_2535812 |
| Donkey anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Abcam | Cat# ab150105; RRID: AB_2732856 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen | Cat# A21447; RRID: AB_141844 |
| Bacterial and virus strains | ||
| pCSⅡ-CMV-MCS-IRES2-mRFP1 | RIKEN BioResource Reserch Center | Cat# RDB07918 |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s Modified Eagle’s Medium - low glucose | Sigma Aldrich | Cat# D6046-500ML |
| D-MEM(High Glucose) with L-Glutamine, Phenol Red, Sodium Pyruvate and 1,500mg/l Sodium Bicarbonate | FUJIFILM | Cat# 049-32645 |
| MEM Non-essential Amino Acids Solution (×100) | FUJIFILM | Cat# 139-15651 |
| Opti-MEM™ I Reduced Serum Medium | Gibco | Cat# 31985-062 |
| TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) | Takara Bio | Cat# RR820A |
| PrimeSTAR® Max DNA Polymerase | Takara Bio | Cat# R045A |
| In-Fusion® HD Cloning Kit w/Cloning Enhancer | Clontech | Cat# 639633 |
| Lipofectamine™ 2000 Transfection Reagent | Invitrogen | Cat# 11668019 |
| Collagenase TypeⅠ | FUJIFILM | Cat# 037-17603 |
| Blasticidin S Hydrochloride | FUJIFILM | Cat# 029-18701 |
| Cyclosporin A | FUJIFILM | Cat# 031-24931 |
| 7-AAD | BD Pharmingen | Cat# 559925 |
| CellCover | ANACYTE Laboratories | Cat# 800-125 |
| Critical commercial assays | ||
| miRNeasy Micro Kit (50) | QIAGEN | Cat# 217084 |
| TURBO DNA-free™ Kit | Invitrogen | Cat# AM1907 |
| PrimeScript™ RT reagent Kit (Perfect Real Time) | Takara Bio | Cat# RR037A |
| QIAquick Gel Extraction Kit (50) | QIAGEN | Cat# 28704 |
| QIAprep Spin Miniprep Kit (50) | QIAGEN | Cat# 27104 |
| Plasmid Midi Kit (100) | QIAGEN | Cat# 12145 |
| Lenti-X™ GoStix™ Plus | Takara Bio | Cat# 631280 |
| NEBNext Poly(A) mRNA Magnetic Isolation Module | New England Biolabs | Cat# E7490 |
| NEBNext rRNA Depletion Kit (Human/Mouse/Rat) | New England Biolabs | Cat# E6310 |
| NEBNext Ultra II RNA Library Prep Kit for illumina | New England Biolabs | Cat# E7770 |
| Chromium Next GEM Single Cell 3ʹ Reagent Kits v3.1 | 10X Genomics, Inc | Cat# PN-1000121 |
| Deposited data | ||
| Bulk transcriptome data (RNA-Seq) | This study | GEO: GSE205165 |
| Bulk transcriptome data (RNA-Seq) | This study | GEO: GSE205166 |
| Single cell transcriptome data (scRNA-Seq) | This study | GEO: GSE205168 |
| Experimental models: Cell lines | ||
| Human: Platinum-GP (Plat-GP) | Cell Biolabs, Inc. | Cat# RV-103 |
| Experimental models: organisms/strains | ||
| Mouse: C57BL6/J | Charles River Laboratories | RRID:IMSR_JAX:000664 |
| Mouse: C57BL/6-Tg(Myh6-EGFP)MG3Tm;MG3 | JCRB Laboratory | https://animal.nibiohn.go.jp/e_mg3.html |
| Mouse: B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | Muzumdaret al., 2007 | RRID:IMSR_JAX:007676 |
| Oligonucleotides | ||
| Primers for In-fusion cloning and qPCR | See Table S1A and S1B | N/A |
| Recombinant DNA | ||
| Plasmid: pCMV-R8.74 | Addgene | Cat# 22036 |
| Plasmid: pCMV-VSV-G | Addgene | Cat# 8454; RRID:Addgene_8454 |
| Plasmid: pAdVAntage | Promega | Cat# E1711 |
| Software and algorithms | ||
| BZ-X analyzer software | Kyence | https://www.keyence.co.jp/support/bio/analyzer/ |
| CFX ManagerTM Software Security Edition (v2.0) | Bio-Rad | https://www.bio-rad.com/ja-jp/product/cfx96-touch-real-time-pcr-detection-system?ID=LJB1YU15 |
| BD FACS Software | BD Biosciences | https://www.bdbiosciences.com/ja-jp/products/instruments/flow-cytometers/research-cell-sorters/bd-facsaria-iii |
| CLC Genomics Workbench (v21.0.5) | CLC bio/QIAGEN | https://digitalinsights.qiagen.com/ja/qiagen-clc-genomics-workbench/ |
| GradhPad Prism 7 (v 7.0d) | GradhPad | https://www.graphpad.com/scientific-software/prism/ |
| R Statistical Computing software (v4.1.1) | The R Foundation | https://www.r-project.org |
| DAVID Bioinformatics Resources 6.8 | Laboratory of Human Retrovirology and Immunoinformatics (LHRI) | https://david.ncifcrf.gov |
| CountessTM II FL | Invitrogen | Cat# AMQAF1000 |
| Cell Ranger (v6.1.2) | 10x Genomics | https://support.10xgenomics.com |
| Seurat (v4.0.6) | Hao et al., 2021 | https://satijalab.org/seurat/ |
| ImageJ (1.53a) | the Research Services Branch (RSB) of the National Institute of Mental Health (NIMH), part of the National Institutes of Health (NIH) | https://imagej.nih.gov/ij/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kazumasa Unno (kunno@med.nagoya-u.ac.jp)
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Mouse model
All protocols of animal care and animal use in this study were approved by the Animal Ethics Review Board of Nagoya University School of Medicine (Center for Animal Research and Education, Nagoya University: https://www.care.nagoya-u.ac.jp/statute.html). Wild-type C57BL/6J mice (Charles River Laboratories, Wilmington, MA, USA) were used. αMHC-GFP transgenic mice (C57BL/6-Tg (Myh6-EGFP) MG3Tm; MG3, Figure S1) were used as reporter mice to detect the αMHC (Myh6) promoter activation in vitro. The B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,−EGFP)Luo/J (mT/mG) reporter mice (Muzumdar, M. D. et al., 2007) were used for transplantation studies in vivo. Anesthesia was maintained by administering medetomidine hydrochloride (0.3 mg/kg), midazolam (4 mg/kg), and butorphanol tartrate (5 mg/kg) into the abdominal cavity before the surgical procedure. The C57BL/6-Tg (Myh6-EGFP) MG3Tm mice were deposited by Dr. Takayuki Morisaki, and provided from Laboratory Animal Resource Bank at NIBIOHN, Japan.
Isolation of mouse ADRCs
ADRCs were isolated from the inguinal and axillary fat pads of mice after sufficient anesthesia. The adipose tissues were washed with phosphate buffered saline (PBS), chopped fully with scissors, and then treated with 5 ml of collagenase type 1 (FUJIFILM, Cat# 037-17603) solution; subsequently, 2 mg/ml Hank's balanced salt solution was added after passing through a 0.22 μm mesh filter. Enzyme treatment was performed with gentle rocking at 37°C for 30 to 45 min. The digested tissues were filtered through a 100 μm mesh filter and centrifuged at 500 × g for 5 min. The supernatant was removed and the pellet was stirred with 10 ml Dulbecco’s modified Eagle medium (DMEM; Sigma Aldrich, Cat# D6046-500ML) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. The suspension was plated in 10 cm dishes (passage 0), and the medium was exchanged the day after isolating the ADRC. ADRCs were cultured under 5% CO2 at 37°C conditions. The medium was changed every 2–3 days. After achieving the desired confluency of 80–90%, ADRCs were harvested by trypsinization for 5 min at 37°C and pipetting to passage 1.
Method details
RNA extraction
Each tissue or cell sample was added to the QIAzol Lysis Reagent (QIAGEN, Cat# 79,306). Total RNA was extracted using the miRNeasy Micro Kit (QIAGEN, Cat# 217,084) and the TURBO DNA-freeTM Kit (Thermo Fisher Scientific, Cat# AM1907) to remove contaminating DNA.
Lentivirus cloning and preparation
We amplified the coding regions of candidate genes by PCR with PrimeSTAR (Takara Bio, Cat# R045A) using template complementary DNA (cDNA) isolated from fetal or neonatal C57BL/6 mouse hearts (The primers are listed in Table S1.). These inserts were cloned into the multicloning site of the pCSⅡ-CMV-MCS-IRES-mRFP1 vector (RIKEN BioResource Research Center, Cat# RDB07918) using an In-Fusion PCR cloning kit (Clontech, Cat# 639633). Before transduction, Plat-GP cells (Cell Biolabs, Cat# RV-103) were seeded at 6 × 106 cells per 10 cm2 of the dish in DMEM high glucose (FUJIFILM, Cat# 049-32645) supplemented with 10% FBS and 1% MEM non-essential amino acid solution (FUJIFILM, Cat# 139-15651). The following day, pCSII-based lentivirus vectors were induced into Plat-GP cells using lipofectamine 2000 (Invitrogen, Cat# 11668019). A mass of 8 μg pCSⅡ-CMV-MCS-IRES-mRFP1, 16 μg pCMV-R8.74 (Addgene, Cat# 22036), 4 μg pCMV-VSV-G (Addgene, Cat# 8454), and 2 μg pAdVAntage (Promega, Cat# E1711) were diluted in 500 μl of Opti-MEM (Gibco, Cat# 31985-062). A quantity of 60 μl of lipofectamine 2000 was gently mixed with 500 μl of Opti-MEM and incubated for 5 min. Thereafter, it was mixed with the plasmid, left for 20 min, and evenly added dropwise to the Plat-GP cells, followed by incubation (37 °C 5% CO2). The supernatant was replaced after approximately 10 h. The virus solution was harvested twice, approximately 48 h and 72 h after transfection, centrifuged, and filtered (0.22 μm). The virus solutions were stored at −80°C. The vector plasmid of CSII-CMV-MCS-IRES2-mRFP1 was developed by the late Dr. Hiroyuki Miyoshi and provided from RIKEN BioResource Research Center, Japan.
ADRC culture and lentiviral transfection
ADRCs used for gene transfer were isolated from MG3 mice (aged 12–24 weeks). After reaching approximately 80-90% confluency at passage 1, ADRCs were harvested and plated on 24 well plates at 30,000 cells/well. The next day, each viral solution was diluted with DMEM (with 10% FBS and 1% penicillin/streptomycin), and 10 μg/ml polybrene was added to the cells. The viral titer was confirmed using Lenti-XTM GoStixTM Plus (Cat #631280; Takara Bio), and the amount of each virus solution was adjusted to MOI=2. The mock cells were infected with a control lentivirus that did not contain the inserted target genes. The plates were incubated for 24 h. The medium was changed every 2–3 days.
Quantitative evaluation of α-MHC-GFP using a microscope
Keyence BZ-X710 was used to observe αMHC-EGFP-positive ADRCs. High-resolution images were captured under a GFP filter using the CFI Plan Apo at 4x magnification. BZ-X analyzer software (Keyence) was used to connect 3×3 adjacent images (image joints). The hybrid cell count of the BZ-X analyzer software was used for the quantitative evaluation of GFP-positive cells. All shooting conditions and thresholds for quantitative evaluation were unified.
Quantitative PCR (qPCR) reaction
Extracted RNA (500 ng of extracted RNA was converted to cDNA using the PrimeScriptTM RT Reagent Kit (Takara Bio, Cat# RR037A). Real-time qPCR was performed using SYBR Premix Ex TaqTM II (Takara Bio, Cat# RR820A) on a CFX96 Real-Time PCR Detection System with CFX ManagerTM Software Security Edition (Bio-Rad) to determine the cycle number (Cq). The 2-ΔΔCq method was used to determine gene expression changes, using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a housekeeping gene. Primer information and amplification conditions are listed in Tables S2–S4.
Immunocytochemistry
Cells were cultured under the appropriate conditions on a glass slide and fixed with 4% paraformaldehyde for 30 min, followed by several PBS washes. They were permeabilized with ice-cold methanol for 30 min at -20 °C for alpha sarcomeric actin staining or with 0.5% Triton X for 15 min on ice for cardiac troponin T staining, followed by several rinses with PBS. The cells were then incubated with a blocking solution containing PBS and 1% BSA for 1 h at room temperature. After blocking, cells were incubated with primary mouse anti-alpha sarcomeric actin (Abcam, Cat# A2172, RRID: AB_476695, 1:1000) for 2 h at 37 °C, or with primary mouse anti-cardiac troponin T (Abcam, Cat# ab8295, RRID: AB_306445, 1:100) overnight at 4 °C. Samples were rinsed and then incubated with goat anti-mouse IgM (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 (Thermo Fisher Scientific, Cat# A21044, RRID: AB_2535713, 1:500), or goat anti-mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 (Thermo Fisher Scientific, Cat# A11005, RRID: AB_2534073, 1:500) for 2 h at 37 °C. After several washes, nuclei were counterstained with Hoechst@33342 Stain (Invitrogen, Cat# H3570, 1:1000) for 10 min at room temperature. Finally, the cells were washed thoroughly and sealed with VECTASHIELD (Vector Laboratories, Cat# H-1000). Serial image sections with a step size of 0.1-0.3 μm thickness were collected using a fluorescence microscope BZ-X710 and BZ-X Analyzer.
Fluorescence-activated cell sorting (FACS) Analysis and Sorting
Cells were harvested by trypsinization for 5 min at 37 °C and stained with 7-AAD solution (BD Pharmingen, Cat# 559925, 1:200 with DMEM) for 10 min at room temperature to exclude dead cells. Cells were fixed with CellCover (AL Anacyte Laboratories, Cat# 800-125) and sorted using FACS Aria 2 (BD Biosciences) (Figure S2A).
RNA-sequencing analysis
RNA was isolated as previously described. RNA quality was assessed using the Agilent TapeStation system with a high-sensitivity RNA ScreenTape (Agilent). RNA of 20-50 ng was used to prepare the sequencing libraries. Complementary DNA synthesis and library preparation were performed using NEBNext Poly(A) mRNA Magnetic Isolation Module (NEW ENGLAND Biolabs, Cat# E7490) and NEBNext Ultra II RNA Library Prep Kit for Illumina (NEW ENGLAND Biolabs, Cat# E7770) to identify candidate transcriptional factors according to the manufacturer’s instructions. The NEBNext rRNA Depletion Kit (Human/Mouse/Rat) (NEW ENGLAND Biolabs, Cat# E6310) and NEBNext Ultra II RNA Library Prep Kit for Illumina (NEW ENGLAND Biolabs, Cat# E7770) were used for GFP+ADRC sequencing. Sequencing was performed using a HiSeq2500 next-generation sequencing platform (Illumina). The sequenced raw data analysis was performed using the CLC Genomics Workbench (CLC bio/QIAGEN, Version: 21.0.5), including the trimming operation, quality assessment, gene symbol annotation, and count data normalization. As a positive control for gene expression, we downloaded the dataset GSE102772, in which 12.9 weeks C57BL/6J adult ventricular cardiomyocyte gene expression data (GSM2746289, GSM2746290) were included (Chevalier et al., 2018). Mouse genome reference sequences and annotation files were downloaded from the iGenome portal (https://support.illumina.com/sequencing/sequencing_software/igenome.html). A volcano plot was created using GraphPad Prism 7, in which differentially expressed genes were selected using a t-test, p-value cut-off < 0.05, and fold change > 4. The MA plot created on GradhPad Prism 7 showed differentially expressed genes, which were selected by tagwise dispersions with false discovery rate (FDR) p-value cut-off < 0.05, among the samples using the CLC Genomics Workbench. Heat maps were generated using heatmap.3 in the R package and GraphPad Prism 7. GO terms of biological processes were analyzed using the DAVID Bioinformatics Resources 6.8.
Single-cell RNA sequencing (scRNA-seq)
ADRCs were introduced from MG3 transgenic mice as previously described (please see methods above). After three weeks of transduction with the six factors, the ADRCs were harvested. Transduced ADRCs were counted in 9,000 cells using CountessTM II FL (Invitrogen). As a negative control, 1,000 ADRCs without lentiviral induction were mixed with the sample. Single cells were encapsulated by emulsion beads using 10X GemCode technology running a Chromium Controller (10X Genomics). The Chromium Next GEM Single Cell 3’ Reagent kit v3.1 (10X Genomics, Cat# PN-1000121) was used to label each cell and individual transcript with a unique molecular identifier (UMI), and cDNA libraries were generated according to the manufacturer’s instructions. The libraries were sequenced on a HiSeq X Ten Illumina platform. Sequencing results were demultiplexed and converted into the FASTQ format using the Illumina package bcl2fastq. Alignment, barcode processing, and UMI counting were performed using 10x Cell Ranger (version 6.1.2) with a 10mm-based default reference. Downstream work was predominantly performed using the R package Seurat (version 4.0.6) (Hao et al., 2021). Cells with less than 200 features, less than 2,000 or more than 75,000 transcripts, and more than 30% mitochondrial reads were excluded. Expression counts were normalized using Seurat’s NormalizeData function and variable features were selected by FindVariableFeatures (nFeatures = 2,000, selection.method=‘vst’). The data were scaled using Seurat’s ScaleData function. The main cell-type clusters were identified using Louvain clustering with a resolution parameter of 0.2, utilizing the top six principal components. Dimensionality reduction plots are based on the uniform manifold approximation and projection (UMAP) using Seurat’s RunUMAP function. Doublets were identified using DoubletFinder (version 2.0.3) and were subsequently removed. A total of 2,911 cells were used for further analysis. Marker genes were visualized using Seurat’s feature plot function.
Mouse model of myocardial infarction and ADRC transplantation
MI model mice were created by permanent ligation of the left anterior descending (LAD) coronary artery in eight-week-old wild-type C57BL/6 mice. Anesthesia was maintained by administering anesthetics into the abdominal cavity. Permanent ligation was performed around the LAD coronary artery 2–3 mm from its origin using an 8-0 silk suture. ADRCs for transplantation were isolated from mT/mG mice and the transplanted ADRCs were visualized on a fluorescence microscope. ADRCs in the treatment group were harvested one week after transduction of the six factors. As a negative control, the ADRCs were prepared without viral transduction. The total ADRCs for each cell were calculated as 1×105 cells and suspended in 10 μl DMEM (1×105 cells / 10 μl). Immediately after MI injury, ADRC solutions were injected at 5–10 sites (1-2 μl per site) in the anterior and posterior infarct border zones of the post-MI myocardium using a Hamilton syringe with a 30-gauge needle. For the MI + ADRCs with no induced factor groups, the same volume of concentrated ADRCs with DMEM was intramyocardially injected at the same sites of the infarct border zones. The induced or uninduced ADRCs to be transplanted were prepared by another experimenter, and surgery and echocardiographic evaluations were performed in a blind condition. For C57BL/6 mice, the immunosuppressant drug cyclosporin A (FUJIFILM, Cat# 031-24931) was administered three times a week (10 mg/kg, i.p. every 2-3 days) to achieve equivalent allograft transplantation throughout the duration of the study (Diehl et al., 2017). Transthoracic echocardiography was performed at baseline and every week after ADRCs transplantation using a Vevo 1100 with a 40-MHz probe (FUJIFILM). Cardiac function, including left ventricular dimensions at end diastole (LVDd), end systole (LVDs), and fractional shortening (FS), was measured using the M-mode. Hearts were collected on the 28th postoperative day.
Immunohistochemistry
The hearts of MI-induced mice were harvested 28 days after surgery. The hearts were fixed in 4% PFA/PBS for 24 h and embedded in paraffin. Heart sections of 4 μm-thickness were deparaffinized with xylene and ethanol, followed by blocking with 1% BSA for 10 min. For double staining of cardiac troponin T and tdTomato, primary anti-RFP antibody (Medical & Biological Laboratories, Cat# PM005, RRID: AB_591279, 1:2000) was first mounted on the sections overnight at 4 °C. Sections were rinsed and incubated with goat anti-rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (Thermo Fisher Scientific, Cat# A21244, RRID: AB_2535812, 1:500) for 1.5 h at 37 °C. The sections were again fixed with 4% PFA/PBS for 10 min, exposed to citrate buffer (2.4 g/l sodium citrate, 0.35 g/l citric acid), and boiled for 10 min in a microwave for antigen retrieval. Then for double staining, after blocking with 1% BSA for 10 min, primary mouse anti-cardiac troponin T antibody (Abcam, Cat# ab10214, RRID: AB_2206574, 1:600) was mounted on the sections overnight at 4 °C. Sections were rinsed and incubated with goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (Thermo Fisher Scientific, Cat# A11001; RRID: AB_2534069, 1:500) for 1.5 h at 37 °C. Triple staining for cardiac troponin T, tdTomato, and VEGF-A (or VEGF-B) was performed as follows. The heart sections were deparaffinized with xylene and ethanol, followed by blocking with 1% BSA for 10 min. For the retrieval process, the sections were exposed to citrate buffer and boiled for 10 min in a microwave. The primary antibodies used were as follows: rat anti-RFP (ChromoTek, Cat# 5F8, RRID: AB_2336064, 1:1000), rabbit anti-VEGF-A (Abcam, Cat# ab39250, RRID: AB_778794, 1:200) (or VEGF-B (Abcam, Cat# ab185696, 1:200)), and mouse anti-cardiac troponin T (Abcam, Cat# ab10214, RRID: AB_2206574, 1:600). The secondary antibodies used were Alexa Fluor 488 (Thermo Fisher Scientific, Cat# A11001; RRID: AB_2534069, 1:500), Alexa Fluor 546 (Thermo Fisher Scientific, Cat# A11081, RRID: AB_141738, 1:500), and Alexa Fluor 647 (Thermo Fisher Scientific, Cat# A21244, RRID: AB_2535812, 1:500). Incubation with antibodies was carried out at 4 °C overnight for primary antibodies and at 37 °C for 1.5 h for secondary antibodies. Double staining with CD31 and cardiac troponin T was performed, as previously described. The primary antibodies used were goat anti-CD31/PECAM-1 (BioTechne, Cat# AF3628, RRID: AB_2161028, 5 μg/mL) and mouse anti-cardiac troponin T (Abcam, Cat# ab10214, RRID: AB_2206574, 1:600). The secondary antibodies used were Alexa Fluor 488 (Thermo Fisher Scientific, Cat# ab150105; RRID: AB_2732856, 1:500) and Alexa Fluor 647 (Thermo Fisher Scientific, Cat# A21447; RRID: AB_141844, 1:500). After several washes, the samples were incubated with Hoechst@33342 stain (1:1000) for 10 min at room temperature for nuclear staining. Samples were enclosed with VECTASHIELD, and observations were performed using BZ-X710, as in immunocytochemistry. Serial image sections with a step size of 0.3-0.5 μm thickness were collected using the fluorescence microscope BZ-X710 and BZ-X Analyzer (Keyence). High-resolution images were taken under GFP, Texas Red, and Cy5 filters using the CFI Plan Apo at 20x magnification. For quantitative evaluation of tdTomato and cTnT positive cells, or CD31+ area, ImageJ software (version 1.53a) was used. All shooting conditions and thresholds for quantitative evaluation were combined.
Quantification and statistical analysis
Statistical analysis
Statistical analyses were performed using GraphPad Prism 7 software (GraphPad Software, La Jolla, CA, USA). Data are expressed as mean ± standard error of the mean (SEM). Statistical significance was examined using the Student’s t-test between two groups or ANOVA between more than two groups. Survival curves were analyzed using Kaplan–Meier estimators and log-rank (Mantel-Cox) tests. Significance was defined as p < 0.05 (∗, p < 0.1; ∗∗, p < 0.05; ∗∗∗, p < 0.01).
Acknowledgments
We wish to thank Yoko Inoue for her surgical assistance. We wish to thank Hidetaka Suga (Department of Endocrinology and Diabetes, Nagoya University Graduate School of Medicine, Nagoya, Japan) for the technical support. We wish to thank Manabu Wakamatsu (Department of Pediatrics, Nagoya University Graduate School of Medicine, Nagoya, Japan) for the experimental and technical support with single-cell transcriptomic analyses. We wish to thank Hiroyuki Miyoshi (RIKEN, Japan) and Dider Trono (Ecole Polytechnique Federale de Lausanne, Switzerland) for plasmids. We wish to thank Takayuki Morisaki (Tokyo University, Japan) for MG3 mice. This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Science, and Technology of Japan awarded to K.U. (17K09573), K. Kondo (17K09574), K. Kato (20K17145), and T.M. (21H02911, 18H02805). This work was supported in part by grants from JST PRESTO (grant number JPMJPR19H5) to Y. Sato. This work was supported in part by JST CREST (grant number JPMJCR19H4).
Author contributions
K.U., K. Kondo, R.S., and T.M. designed the study and reviewed the data. S.N., K.U., Y.O., K. Kato, and R.S. wrote the paper. K.U., K. Kondo, Y. Shimizu, and T.M. contributed to study conception and design. S.N., Y. Sato, and R.H. performed the lentivirus experiments and analyzed the data. S.N., K. Kato, Y.O., Y. Sato, and Y.T. performed the bioinformatics analysis. S.N., K.U., K. Kato, and Y.F. performed the histological experiments and analysis.
Declaration of interests
All authors declare no competing interests.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104651.
Supplemental information
Data and code availability
This paper does not report original code. RNA-seq and Single-cell RNA-seq data have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
References
- Bergmann O., Zdunek S., Felker A., Salehpour M., Alkass K., Bernard S., Sjostrom S., Szewczykowska M., Jackowska T., dos Remedios C., et al. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- Bersell K., Arab S., Haring B., Kühn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- Cai L., Johnstone B.H., Cook T.G., Tan J., Fishbein M.C., Chen P.S., March K.L. IFATS collection: human adipose tissue-derived stem cells induce angiogenesis and nerve sprouting following myocardial infarction, in conjunction with potent preservation of cardiac function. Stem Cell. 2009;27:230–237. doi: 10.1634/stemcells.2008-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Sun Z., Liao L., Meng Y., Han Q., Zhao R.C. Human adipose tissue-derived stem cells differentiate into endothelial cells in vitro and improve postnatal neovascularization in vivo. Biochem Bioph Res Co. 2005;332:370–379. doi: 10.1016/j.bbrc.2005.04.135. [DOI] [PubMed] [Google Scholar]
- Chevalier M., Vermij S.H., Wyler K., Gillet L., Keller I., Abriel H. Transcriptomic analyses of murine ventricular cardiomyocytes. Sci. Data. 2018;5 doi: 10.1038/sdata.2018.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.K., Vicente-Manzanares M., Zareno J., Whitmore L.A., Mogilner A., Horwitz A.R. Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra A., Kutys M.L., Zhang K., Polacheck W.J., Sheng C.C., Luu R.J., Eyckmans J., Hinson J.T., Seidman J.G., Seidman C.E., Chen C.S. Force generation via β-cardiac myosin, titin, and α-actinin drives cardiac sarcomere assembly from cell-matrix adhesions. Dev. Cell. 2018;44:87–96.e5. doi: 10.1016/j.devcel.2017.12.012. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai R., Wang Z., Samanipour R., Koo K.i., Kim K. Adipose-derived stem cells for tissue engineering and regenerative medicine applications. Stem Cells Int. 2016;2016:1–19. doi: 10.1155/2016/6737345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl R., Ferrara F., Müller C., Dreyer A.Y., McLeod D.D., Fricke S., Boltze J. Immunosuppression for in vivo research: state-of-the-art protocols and experimental approaches. Cell. Mol. Immunol. 2017;14:146–179. doi: 10.1038/cmi.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi S., Takefuji M., Sakaguchi T., Ishihama S., Mori Y., Tsuda T., Takikawa T., Yoshida T., Ohashi K., Shimizu Y., et al. Cardiomyocytes capture stem cell-derived, anti-apoptotic microRNA-214 via clathrin-mediated endocytosis in acute myocardial infarction. J. Biol. Chem. 2019;294:11665–11674. doi: 10.1074/jbc.ra119.007537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England J., Loughna S. Heavy and light roles: myosin in the morphogenesis of the heart. Cell. Mol. Life Sci. 2013;70:1221–1239. doi: 10.1007/s00018-012-1131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenhagen T., Bolli R., Braun T., Field L.J., Fleischmann B.K., Frisén J., Giacca M., Hare J.M., Houser S., Lee R.T., et al. Cardiomyocyte regeneration. Circulation. 2017;136:680–686. doi: 10.1161/circulationaha.117.029343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller S.J., Sivarajah K., Sugden P.H. ErbB receptors, their ligands, and the consequences of their activation and inhibition in the myocardium. J. Mol. Cell. Cardiol. 2008;44:831–854. doi: 10.1016/j.yjmcc.2008.02.278. [DOI] [PubMed] [Google Scholar]
- Funakoshi S., Miki K., Takaki T., Okubo C., Hatani T., Chonabayashi K., Nishikawa M., Takei I., Oishi A., Narita M., et al. Enhanced engraftment, proliferation, and therapeutic potential in heart using optimized human iPSC-derived cardiomyocytes. Sci Rep-Uk. 2016;6 doi: 10.1038/srep19111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam M., Fujita D., Kimura K., Ichikawa H., Izawa A., Hirose M., Kashihara T., Yamada M., Takahashi M., Ikeda U., Shiba Y. Transplantation of adipose tissue-derived stem cells improves cardiac contractile function and electrical stability in a rat myocardial infarction model. J. Mol. Cell. Cardiol. 2015;81:139–149. doi: 10.1016/j.yjmcc.2015.02.012. [DOI] [PubMed] [Google Scholar]
- Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T.D., Pepine C.J., Lambert C.R., Traverse J.H., Schatz R., Costa M., Povsic T.J., David Anderson R., Willerson J.T., Kesten S., Perin E.C. The Athena trials: autologous adipose-derived regenerative cells for refractory chronic myocardial ischemia with left ventricular dysfunction. Catheter Cardio Inte. 2017;89:169–177. doi: 10.1002/ccd.26601. [DOI] [PubMed] [Google Scholar]
- Houtgraaf J.H., den Dekker W.K., van Dalen B.M., Springeling T., de Jong R., van Geuns R.J., Geleijnse M.L., Fernandez-Aviles F., Zijlsta F., Serruys P.W., Duckers H.J. First experience in humans using adipose tissue–derived regenerative cells in the treatment of patients with ST-segment elevation myocardial infarction. J. Am. Coll. Cardiol. 2012;59:539–540. doi: 10.1016/j.jacc.2011.09.065. [DOI] [PubMed] [Google Scholar]
- Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ii M., Horii M., Yokoyama A., Shoji T., Mifune Y., Kawamoto A., Asahi M., Asahara T. Synergistic effect of adipose-derived stem cell therapy and bone marrow progenitor recruitment in ischemic heart. Lab. Invest. 2011;91:539–552. doi: 10.1038/labinvest.2010.191. [DOI] [PubMed] [Google Scholar]
- Ishii M., Shibata R., Shimizu Y., Yamamoto T., Kondo K., Inoue Y., Ouchi N., Tanigawa T., Kanemura N., Ito A., et al. Multilayered adipose-derived regenerative cell sheets created by a novel magnetite tissue engineering method for myocardial infarction. Int. J. Cardiol. 2014;175:545–553. doi: 10.1016/j.ijcard.2014.06.034. [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M., Zaret K.S. Pioneer transcription factors in cell reprogramming. Gene Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumabay M., Zhang R., Yao Y., Goldhaber J.I., Boström K.I. Spontaneously beating cardiomyocytes derived from white mature adipocytes. Cardiovasc. Res. 2010;85:17–27. doi: 10.1093/cvr/cvp267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T., Kondo K., Shibata R., Hayashida R., Shintani S., Yamaguchi S., Shimizu Y., Unno K., Kikuchi R., Kodama A., et al. Therapeutic angiogenesis using autologous adipose-derived regenerative cells in patients with critical limb ischaemia in Japan: a clinical pilot study. Sci Rep-Uk. 2020;10 doi: 10.1038/s41598-020-73096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura M., Miyagawa S., Miki K., Saito A., Fukushima S., Higuchi T., Kawamura T., Kuratani T., Daimon T., Shimizu T., et al. Feasibility, safety, and therapeutic efficacy of human induced pluripotent stem cell-derived cardiomyocyte sheets in a porcine ischemic cardiomyopathy model. Circulation. 2012;126:S29–S37. doi: 10.1161/circulationaha.111.084343. [DOI] [PubMed] [Google Scholar]
- Kondo K., Shintani S., Shibata R., Murakami H., Murakami R., Imaizumi M., Kitagawa Y., Murohara T. Implantation of adipose-derived regenerative cells enhances ischemia-induced angiogenesis. Arteriosclerosis Thrombosis Vasc Biology. 2009;29:61–66. doi: 10.1161/atvbaha.108.166496. [DOI] [PubMed] [Google Scholar]
- Lindsey M.L. Assigning matrix metalloproteinase roles in ischaemic cardiac remodelling. Nat. Rev. Cardiol. 2018;15:471–479. doi: 10.1038/s41569-018-0022-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Sun J., Zhao Z., Lei W., Chen Y., Wang X., Yang J., Shen Z. A brief review: adipose-derived stem cells and their therapeutic potential in cardiovascular diseases. Stem Cell Res. Ther. 2017;8:124. doi: 10.1186/s13287-017-0585-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Estrada O.M., Muñoz-Santos Y., Julve J., Reina M., Vilaró S. Human adipose tissue as a source of Flk-1+ cells: new method of differentiation and expansion. Cardiovasc. Res. 2005;65:328–333. doi: 10.1016/j.cardiores.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Meliga E., Strem B.M., Duckers H.J., Serruys P.W. Adipose-derived cells. Cell Transplant. 2007;16:963–970. doi: 10.3727/096368907783338190. [DOI] [PubMed] [Google Scholar]
- Menasche P. Cell-based therapy for heart disease: a clinically oriented perspective. Mol. Ther. 2009;17:758–766. doi: 10.1038/mt.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menasché P., Alfieri O., Janssens S., McKenna W., Reichenspurner H., Trinquart L., Vilquin J.-T., Marolleau J.-P., Seymour B., Larghero J., et al. The myoblast autologous grafting in ischemic cardiomyopathy (MAGIC) trial. Circulation. 2008;117:1189–1200. doi: 10.1161/circulationaha.107.734103. [DOI] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Perin E.C., Sanz-Ruiz R., Sánchez P.L., Lasso J., Pérez-Cano R., Alonso-Farto J.C., Pérez-David E., Fernández-Santos M.E., Serruys P.W., Duckers H.J., et al. Adipose-derived regenerative cells in patients with ischemic cardiomyopathy: the PRECISE Trial. Am. Heart J. 2014;168:88–95.e2. doi: 10.1016/j.ahj.2014.03.022. e2. [DOI] [PubMed] [Google Scholar]
- Planat-Bénard V., Menard C., André M., Puceat M., Perez A., Garcia-Verdugo J.M., Pénicaud L., Casteilla L. Spontaneous cardiomyocyte differentiation from adipose tissue stroma cells. Circ. Res. 2004;94:223–229. doi: 10.1161/01.res.0000109792.43271.47. [DOI] [PubMed] [Google Scholar]
- Ptaszek L.M., Mansour M., Ruskin J.N., Chien K.R. Towards regenerative therapy for cardiac disease. Lancet. 2012;379:933–942. doi: 10.1016/s0140-6736(12)60075-0. [DOI] [PubMed] [Google Scholar]
- Puymirat E., Simon T., Cayla G., Cottin Y., Elbaz M., Coste P., Lemesle G., Motreff P., Popovic B., Khalife K., et al. Acute myocardial infarction. Circulation. 2017;136:1908–1919. doi: 10.1161/circulationaha.117.030798. –1919. [DOI] [PubMed] [Google Scholar]
- Räsänen M., Sultan I., Paech J., Hemanthakumar K.A., Yu W., He L., Tang J., Sun Y., Hlushchuk R., Huan X., et al. VEGF-B promotes endocardium-derived coronary vessel development and cardiac regeneration. Circulation. 2021;143:65–77. doi: 10.1161/circulationaha.120.050635. [DOI] [PubMed] [Google Scholar]
- Srivastava D., Yu P. Recent advances in direct cardiac reprogramming. Curr. Opin. Genet. Dev. 2015;34:77–81. doi: 10.1016/j.gde.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnozzi R.J., Maillet M., Sargent M.A., Khalil H., Johansen A.K.Z., Schwanekamp J.A., York A.J., Huang V., Nahrendorf M., Sadayappan S., Molkentin J.D. An acute immune response underlies the benefit of cardiac stem cell therapy. Nature. 2020;577:405–409. doi: 10.1038/s41586-019-1802-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valina C., Pinkernell K., Song Y.H., Bai X., Sadat S., Campeau R.J., Le Jemtel T.H., Alt E. Intracoronary administration of autologous adipose tissue-derived stem cells improves left ventricular function, perfusion, and remodelling after acute myocardial infarction. Eur. Heart J. 2007;28:2667–2677. doi: 10.1093/eurheartj/ehm426. [DOI] [PubMed] [Google Scholar]
- Wang L., Liu Z., Yin C., Asfour H., Chen O., Li Y., Bursac N., Liu J., Qian L. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ. Res. 2015;116:237–244. doi: 10.1161/circresaha.116.305547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Huang P., Near D., Ravi K., Xu Y., Liu J., Qian L. Isoform specific effects of Mef2C during direct cardiac reprogramming. Cells. 2020;9:268. doi: 10.3390/cells9020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Wang W., Li L., Peng Y., Chen P., Huang H., Guo Y., Xia X., Wang Y., Wang H., et al. The relative contribution of paracine effect versus direct differentiation on adipose-derived stem cell transplantation mediated cardiac repair. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmet H., Porapakkham P., Porapakkham P., Sata Y., Haas S.J., Itescu S., Forbes A., Krum H. Short- and long-term outcomes of intracoronary and endogenously mobilized bone marrow stem cells in the treatment of ST-segment elevation myocardial infarction: a meta-analysis of randomized control trials. Eur. J. Heart Fail. 2012;14:91–105. doi: 10.1093/eurjhf/hfr148. [DOI] [PubMed] [Google Scholar]
- Zuk P.A., Zhu M., Ashjian P., De Ugarte D.A., Huang J.I., Mizuno H., Alfonso Z.C., Fraser J.K., Benhaim P., Hedrick M.H. Human adipose tissue is a source of multipotent stem cells. Mol. Biol. Cell. 2002;13:4279–4295. doi: 10.1091/mbc.e02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk P.A., Zhu M., Mizuno H., Huang J., Futrell J.W., Katz A.J., Benhaim P., Lorenz H.P., Hedrick M.H. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper does not report original code. RNA-seq and Single-cell RNA-seq data have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.