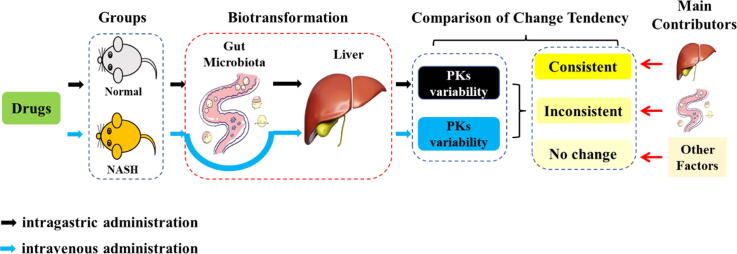
Graphical abstract
Keywords: Pharmacokinetic variability, Gut microbiota, Host Cyp450s, Non-alcoholic steatohepatitis, Personalized medicine
Highlights
-
•
Drugs’ pharmacokinetics were changed in NASH disease.
-
•
A systematical research on cocktail drugs in NASH.
-
•
Gut microbiota can bio-transform some drugs in vitro, and the metabolic rate was different in NASH.
-
•
The gut microbiota and the host co-contributed the pharmacokinetic variability of drugs in NASH.
-
•
The degree of influence on pharmacokinetic variability varies from drug to drug.
Abstract
Introduction
Pharmacokinetic variability in disease state is common in clinical practice, but its underlying mechanism remains unclear. Recently, gut microbiota has been considered to be pharmacokinetically equivalent to the host liver. Although some studies have explored the roles of gut microbiota and host Cyp450s in drug pharmacokinetics, few have explored their effects on pharmacokinetic variability, especially in disease states.
Objectives
In this study, we aim to investigate the effects of gut microbiota and host Cyp450s on pharmacokinetic variability in mice with non-alcoholic steatohepatitis (NASH), and to elucidate the contribution of gut microbiota and host Cyp450s to pharmacokinetic variability in this setting.
Methods
The pharmacokinetic variability of mice with NASH was explored under intragastric and intravenous administrations of a cocktail mixture of omeprazole, phenacetin, midazolam, tolbutamide, chlorzoxazone, and metoprolol, after which the results were compared with those obtained from the control group. Thereafter, the pharmacokinetic variabilities of all drugs and their relations to the changes in gut microbiota and host Cyp450s were compared and analyzed.
Results
The exposures of all drugs, except metoprolol, significantly increased in the NASH group under intragastric administration. However, no significant increase in the exposure of all drugs, except tolbutamide, was observed in the NASH group under intravenous administration. The pharmacokinetic variabilities of phenacetin, midazolam, omeprazole, and chlorzoxazone were mainly associated with decreased elimination activity in the gut microbiota. By contrast, the pharmacokinetic variability of tolbutamide was mainly related to the change in the host Cyp2c65. Notably, gut microbiota and host Cyp450s exerted minimal effects on the pharmacokinetic variability of metoprolol.
Conclusion
Gut microbiota and host Cyp450s co-contribute to the pharmacokinetic variability in mice with NASH, and the degree of contribution varies from drug to drug. The present findings provide new insights into the explanation of pharmacokinetic variability in disease states.
Introduction
Pharmacokinetic variability is common in clinical practice and in personalized medicine, constituting a major challenge in drug therapy, especially for drugs with narrow therapeutic indexes [1]. Pharmacokinetics refers to the relationship between the plasma profile of a drug and its dose. Theoretically and ideally, the plasma profiles of drugs could be similar in patients receiving the same dose regimen. However, in reality, even if the same drug dose is administered, there is always a significant difference in the plasma profiles between patients; this is known as pharmacokinetic variability [2]. Generally, pharmacokinetic variability is influenced by a variety of factors in vivo or in vitro, such as the physiological status of the body, gender, age, diseases, and external stimuli [3]. Among these factors, liver function is the main cause of pharmacokinetic variability. Drug-metabolizing enzymes and transporters are distributed in the liver, and nearly all clinical drugs are metabolized through oxidation, reduction, hydrolysis, conjugation, and/or biliary excretion [4], [5], [6], [7]. Thus, the activities of liver Cyp450s directly affect the metabolism of drugs in the body, which is the primary factor considered for pharmacokinetic variability in clinical medicine.
In recent years, studies on gut microbiota have increased in number, becoming a hot topic in the field. The gut microbiota plays a vital role in many processes, including the metabolism of polysaccharides, production of essential vitamins, development and differentiation of the host’s intestinal epithelium and immune systems, and maintenance of tissue homeostasis [8], [9], [10]. In addition, commensal gut microbiota densities exceed 108 cells/mL in the small intestine and 1011 cells/mL in the large intestine [11]. The gut microbiota has 150-fold more coding genes than the human genome and includes a rich repository of enzymes with the potential to metabolize drugs [12]. In addition to the liver drug-metabolizing enzymes of the host, the gut microbiota is another important factor affecting pharmacokinetic variability [13]. The gut microbiota can affect the bioavailability of drugs through bacterial metabolism, bacterial transport, drug-metabolizing enzymes, and regulation of intestinal transport proteins [14]. Many studies have reported that the gut microbiota participates in the pharmacokinetics of most drugs and regulates their pharmacological and toxicological effects [15], [16]. Thus, its influencing ability is equivalent to that of the host liver and can be regarded as the “second liver” [17].
Although some studies have explored the roles of host Cyp450s and gut microbiota in drug pharmacokinetics, few have explored their effects on pharmacokinetic variability, especially in disease states [18], [19], [20], [21], [22], [23]. Non-alcoholic fatty liver disease (NAFLD) is a chronic liver disease that is the manifestation of metabolic syndrome in the liver, including simple steatosis, necrotizing inflammation, fibrosis, and cirrhosis [24], [25], [26]. Non-alcoholic steatohepatitis (NASH) is a necrotic inflammatory reaction in NAFLD, which is the main cause of liver cirrhosis and liver-related death [27]. NASH is also associated with metabolic risk factors, such as diabetes, dyslipidemia, obesity, atherosclerosis and, in some cases, genetic predisposition [28], [29]. Epidemiological studies have shown that the global prevalence of NAFLD is approximately 4%–46%, whereas that of NASH is approximately 3%–5%. In particular, the prevalence of NAFLD in China is as high as 32.9%, with 20%–40% of patients progressing to NASH [30], [31]. Thus, it has seriously endangered the health of the public, bringing about changes in people’s lifestyles and diets [32]. The drug treatment of NASH is mainly based on the pathogenesis to select appropriate drugs, including insulin sensitizers, antioxidants, PPAR agonists, anti-diabetic drugs, hypertensive drugs, and liver-protective drugs [33], [34]. NASH medications have many types, and the use of drugs in combined disease state complicates the medication situation [35], [36]. Furthermore, NASH is closely related to the gut microbiota. One of the mechanisms of NASH is related to the lipopolysaccharide (LPS or endotoxin) derived from the gut microbiota via the dysfunctional gut barrier to the portal vein and liver, thereby inducing an inflammatory response through the activation of inflammatory cells in the liver [37], [38]. The gut microbiota in disease state is obviously different from that in healthy state, and the effect of gut microbiota on pharmacokinetic variabilities can be imagined. Although several studies have reported pharmacokinetic variabilities in NASH [39], the underlying mechanism remains unclear.
The host liver is the primary site of drug metabolism, and the gut microbiota is referred to as the “second liver” [40]. Therefore, the pharmacokinetic variability for the individualized administration of NASH based on the influences of the host Cyp450s and the gut microbiota can be identified from the perspectives of safety and efficacy. However, a systematic research focusing on the effects of host Cyp450s and gut microbiota on the pharmacokinetic variability of NASH is lacking. Therefore, in this study, liver Cyp450 enzyme probe drugs, including omeprazole, phenacetin, midazolam, tolbutamide, chlorzoxazone, and metoprolol, were selected to examine the effects of gut microbiota and host Cyp450s on the pharmacokinetic variabilities in mice with NASH. It should be emphasized that although these drugs are not directly used to treat NASH, they may be co-administered in corresponding comorbidities such as digestive ulcer, hyperglycemia, and hypertension, among others, during the therapy of NASH. In addition, they are the classical probe drug combinations commonly used in pharmacological studies to evaluate the activities of Cyp450 enzymes, including Cyp2c29, Cyp1a2, Cyp3a11, Cyp2c65, Cyp2e1, and Cyp2d22 [41], [42]. Furthermore, these six drug metabolism enzymes metabolize almost 90% of the clinically used drugs [43]. Given the foregoing reasons, the current cocktail approach could fully reflect the metabolic status of the host Cyp450s or other co-influencing factors such as gut microbiota, as well as provide a scientific basis for the individual medication of NASH in clinical settings.
Materials and methods
Chemicals and materials
Phenacetin (99.9%, T0778), acetaminophen (99.9%, T0065), tolbutamide (99.7%, T1054), metoprolol (99.8%, T0487), and chlorzoxazone (99.7%, T1650) were purchased from Target Molecule Corp. (Massachusetts, USA); omeprazole (>99%, MB1692-S) from Dalian Meilun Biotech Co., Ltd. (Dalian, China); midazolam (5 mg/mL, H10980025) from Jiangsu Enhua Pharmaceutical Co., Ltd. (Xuzhou, China); 4ʹ-hydroxytolbutamide (100%, H969850), 5ʹ-hydroxyomepreole (95%, H948110), αʹ-hydroxymetoprolol (100%, H948390), and 6ʹ-hydroxychlorzoxazone (100%, H825120) from Toronto Research Chemicals (Toronto, Canada); 1ʹ-hydroxymidazolam (95%, 10385-1) from Cayman Chemicals (Michigan, USA); and lamotrigine (100775-200401) from the National Institutes for Food and Drug Control (Beijing, China). Glycyrrhetic acid (>98%, B20412) was obtained from Yuanye Biotechnology Co., Ltd. (Shanghai, China) and used as internal standard (IS). HPLC-grade acetonitrile and methanol were purchased from Merck Company (Darmstadt, Germany); formic acid from CNW Technologies (Dusseldorf, Germany); phosphate buffer saline from Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China); ReadiUse NADPH Regenerating Kit from Wuhan PrimeTox Bio-pharma Technology Co., Ltd. (Wuhan, China); and deionized water from a Millipore Milli-Q Plus system (Bedford, USA).
NASH mouse model
Ethics statement
The C57BL/6J mice were purchased from Hunan SJA Laboratory Animal Co., Ltd. (Changsha, China). The animals were maintained in natural light–dark cycle conditions with controlled temperature (22 ± 2 °C) and given food and drinking water freely. All mice were housed at the Laboratory Animal Research Center of Central South University. All experimental procedures have been approved ethically by the Administration Committee of Experimental Animals of Central South University (No. 2021sydw0052, Changsha, China).
Construction of NASH mouse model
Thirty-two male C57BL/6J (25 ± 3 g) mice were randomly divided into a control group (n = 16) and a NASH model group (n = 16). The mice in the control group were fed with methionine- and choline-supplement (MCS) diet, whereas the mice in the NASH group were fed with methionine- and choline-deficient (MCD) diet for five continuous weeks. The day after the NASH modeling experiment, individual mouse fecal specimens were collected for 16S rRNA gene sequencing analysis. After the collection of pharmacokinetic blood samples, the mice were euthanized using carbon dioxide.
Biochemical assay
Up to 200 μL of mouse serum was sent to the Laboratory Department of Xiangya Hospital (Hunan, China) for the detection of liver function indexes and lipid profiles using a fully automatic biochemical analyzer (AU5821, Beckman, USA).
Histopathological assay
The right liver lobes were removed, fixed in 4% paraformaldehyde, dehydrated, and then embedded in paraffin. Hematoxylin and eosin (H&E) staining was performed in a routine histopathological examination. The degree of liver steatosis was examined under a 200X and a 400X light microscope and then photographed.
16S rRNA sequencing for gut microbiota
The compositions of gut microbiota in the control and NASH groups were determined via 16S rRNA genes analysis. DNA was isolated from the frozen fecal samples using the E.Z.N.A.® Stool DNA Kit (Omega Bio-tek, Norcross, GA, USA) in accordance with the manufacturer’s protocols. Then, PCR amplification of the V4 hypervariable regions of the bacterial 16S rRNA gene was performed using universal primers incorporating the FLX Titanium adaptors and a barcode sequence. Amplicons were extracted from 2% agarose gels and purified using the GeneJETTM Gel Extraction Kit (Thermo Scientific, Massachusetts, USA). Sequencing libraries were generated using the Ion Plus Fragment Library Kit 48 rxns (Thermo Scientific, Massachusetts, USA) in accordance with the manufacturer’s recommendations. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific, Massachusetts, USA). The library was sequenced on an Ion S5TM XL platform, and 400 bp/600 bp single-end reads were generated. Sequence analysis was performed using Uparse software. Sequences with ≥ 97% similarity were assigned to the same operational taxonomic units (OTUs). The representative sequence for each OTU was screened for further annotation.
Pharmacokinetic study in mice with NASH
Pharmacokinetic study
The control and NASH groups (n = 8) were all administered a mixture of probe drugs including phenacetin (10 mg/kg), omeprazole (10 mg/kg), metoprolol (10 mg/kg), midazolam (10 mg/kg), tolbutamide (1 mg/kg), and chlorzoxazone (1 mg/kg) through intragastric administration to investigate the influence of host Cyp450s and gut microbiota. Capillary microsampling was performed at 0, 0.08, 0.17, 0.25, 0.33, 0.5, 0.75, 1, 1.5, 2, 4, 6, 8, and 12 h, respectively. About 30 μL of caudal vein blood samples were collected at each time point. To examine the influence of host Cyp450s, probe drugs including phenacetin (5 mg/kg), omeprazole (5 mg/kg), metoprolol (5 mg/kg), midazolam (5 mg/kg), tolbutamide (1 mg/kg), and chlorzoxazone (1 mg/kg) were also intravenously administered to the control and NASH groups (n = 8 each group), respectively. The blood samples were collected according to the above steps and stored in a −80 °C refrigerator for ultra-high performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) analysis.
Blood sample preparation
The mouse blood samples (10 μL) were mixed with 100 μL acetonitrile and 20 μL IS solution containing lamotrigine (10.2 ng/mL) and glycyrrhetinic acid (23.3 ng/mL), which were vortex-mixed for 5 min and centrifuged at 20,379 g (4 °C) for 10 min to remove the precipitated proteins. Next, 50 μL of the supernatant was transferred to another clean tube containing 200 μL of solvents A and B (v/v, 1:1). The mixture was vortexed for 2 min and re-centrifuged at 20,379 g (4 °C) for another 5 min. Finally, 100 μL of supernatant was transferred to a sample bottle for UPLC-MS/MS detection.
UPLC-MS/MS method
The AB Sciex Qtrap 6500+ UPLC-MS/MS system (AB Sciex, Massachusetts, USA) was used to determine the concentration of the probe drugs. Targeted analytes were separated using a Phenomenex Luna C18 100 A column (150 mm × 2.0 mm, 5 μm, Torrance, CA, USA) at a flow rate of 0.35 mL/min. The mobile phase consisted of solvent A (deionized water with 0.1% formic acid) and solvent B (acetonitrile). The gradient elution program was as follows: 0–0.2 min (10% B); 0.2–3.5 min (10%–90% B); 3.5–4.5 min (90% B); 4.5–4.6 min (90%–10% B); 4.6–8 min (10% B). The autosampler temperature was maintained at 15 °C and the injection volume was set at 2 μL. The mass spectrometer was operated in negative or positive ion mode through multiple-reaction monitoring. The detailed optimized MS/MS parameters are presented in Table 1. Data acquisition was performed with the Analyst Software Version 1.6.3 (AB Sciex, Massachusetts, USA).
Table 1.
Detailed UPLC-MS/MS parameters for the determination of probe drugs, their main metabolites, and internal standard.
| Targeted analytes | Precursor → production (m/z) | ESI model (Positive/Negative) | CE(V) | DP(V) | CXP(V) | EP(V) |
|---|---|---|---|---|---|---|
| Omeprazole | 346.1 → 329.0 | Positive | 10 | 30 | 14 | 10 |
| Phenacetin | 180.1 → 110.1 | Positive | 28 | 70 | 13 | 10 |
| Midazolam | 326.1 → 291.2 | Positive | 38 | 120 | 15 | 10 |
| Tolbutamide | 269.1 → 170.1 | Negative | –22 | −55 | −20 | −10 |
| Chlrozoxazone | 168.1 → 132.0 | Negative | −28 | −80 | −15 | −10 |
| Metoprolol | 268.1 → 133.1 | Positive | 34 | 90 | 16 | 10 |
| 5-OH-Omeprazole | 362.1 → 214.2 | Positive | 17 | 45 | 11 | 10 |
| Acetaminophen | 152.1 → 110.1 | Positive | 22.5 | 70 | 13 | 10 |
| OH-Midazolam | 342.1 → 324.1 | Positive | 30 | 50 | 10 | 10 |
| OH-Tolbutamide | 285.1 → 186.1 | Negative | −25 | −60 | −9 | −10 |
| 6-OH-Chlorzoxazone | 183.9 → 120.0 | Negative | −25 | −50 | −12 | −10 |
| α-OH-Metoprolol | 284.0 → 74.1 | Positive | 26 | 90 | 11 | 10 |
| Lamotrigine (IS) | 256.2 → 145.2 | Positive | 45 | 150 | 10 | 10 |
| Glycyrrhetic (IS) | 469.3 → 355.3 | Negative | −59 | −158 | −20 | −10 |
Note: ESI: electrospray ion source; V: voltage; CE: collision energy; DP: declustering potential; CXP: cell exit potential; EP: entrance potential; IS: internal standard.
Determination of host Cyp450 activities and expressions
Liver microsomal enzyme activity
Mouse liver microsomes from each group were prepared through differential centrifugation as previously described [44]. Incubation reactions of the experimental and control groups were carried out in parallel. All microsomal incubations were performed in a final incubation volume of 100 μL. The reaction medium contained 49 μL of NADPH regenerating reagent (pH 7.4), 50 μL of mouse liver microsomes (0.5 mg/mL), and 1 μL cocktail of substrates (0.705 μg/mL phenacetin, 6.48 μg/mL omeprazole, 2.5 μg/mL midazolam, 1.225 μg/mL tolbutamide, 1.675 μg/mL metoprolol, and 2.405 μg/mL chlorzoxazone). The mixture was incubated at 37 °C. The incubation time for each probe drug was as follows: 1.5 min for midazolam; 10 min for phenacetin, omeprazole, and chlorzoxazone; 15 min for metoprolol; and 20 min for tolbutamide. The enzymatic reaction was stopped by adding 400 μL of cold acetonitrile including IS. The mixture was centrifuged at 20,379 g for 5 min. Then, 200 μL of the supernatant was transferred into a sample bottle for the determination of metabolites through UPLC-MS/MS analysis.
Western blotting
Total protein was extracted from the control and NASH mouse liver with RIPA buffer and PMSF. Protein content was determined using a BCA protein assay reagent. Samples with 30 µg total protein each were separated on 10% SDS PAGE gels and transferred onto polyvinylidene difluoride membranes. The blots were probed with rabbit anti-Cyp1a2 (Proteintech Group, Rosemont, USA), anti-Cyp3a4 (Proteintech Group, Rosemont, USA), anti-Cyp2c19 (Abcam, Cambridge, UK), anti-Cyp2c9 (Bioworld, Bloomington, USA), anti-Cyp2d6 (Proteintech Group, Rosemont, USA), mouse anti-Cyp2e1 (Abcam, Cambridge, UK), and anti-tubulin (Zhengeng, China). Signals were visualized by enhanced chemiluminescence detection.
Detection of biotransformation ability of gut microbiota
Each fecal sample (about 1 g) was fully suspended in sterile physiological saline (4 mL) and centrifuged at 1699 g for 10 min. The supernatant was obtained and mixed evenly with the anaerobic culture solution (GAM broth, 1:9). After incubation for 24 h, the gut microbiota work solution was obtained. The biotransformation of the probe drugs by the gut microbiota was performed in an incubation system containing 1 mL of the gut microbiota work solution and 5 μL of the probe drug stock solution (1.996 μg/mL omeprazole, 1.835 μg/mL phenacetin, 2.604 μg/mL midazolam, 1.973 μg/mL tolbutamide, 8.016 μg/mL chlorzoxazone, and 5.583 μg/mL metoprolol). The incubation system was anaerobically incubated at 37 °C for 72 h and stopped with the addition of 400 μL cold acetonitrile including IS in a 100-μL incubation system. The samples were centrifuged at 20,379 g for 10 min and detected by UPLC-MS/MS.
Statistical analysis
The pharmacokinetic parameters of the probe drugs were analyzed through non-compartmental assessment of the data with the Drug and Statistical Software Version 3.2.2 (Clinical Drug Evaluation Center, Wannan Medical College, Anhui, China). The pharmacokinetic variabilities of the probe drugs and the relation between the gut microbiota and the host Cyp450 enzyme were compared and analyzed. Statistical analyses were performed using an unpaired Student’s t test in SPSS (IBM, v. 20.0) software. A Chi-square test was applied for nonparametric statistics and significant differences were analyzed using two-tailed tests (p < 0.05).
Results
Characteristics of host Cyp450s and gut microbiota in mice with NASH
NASH mouse model induced by MCD diet
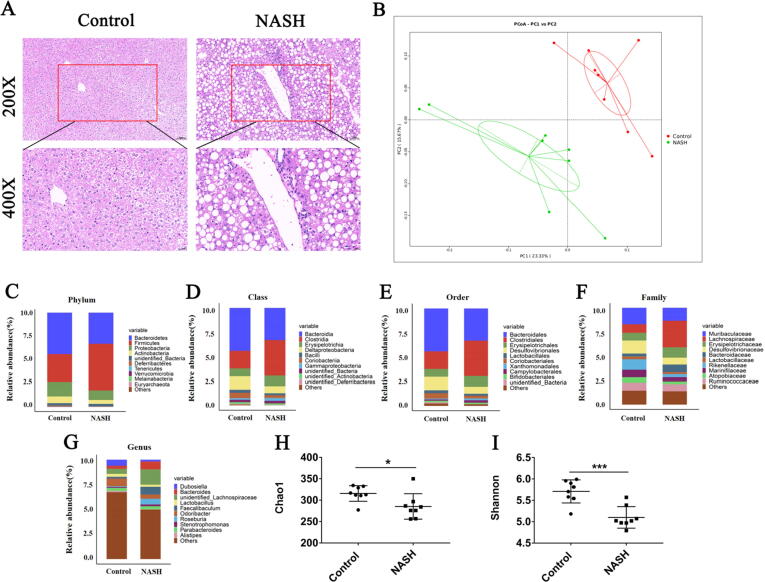
The liver function indexes, including direct bilirubin (DBIL), total bilirubin (TBIL), albumin (ALB), alkaline phosphatase (ALP), γ-glutamyl transpeptidase (γ-GT), triglyceride (TG), high-density lipid-cholesterol (HDL-c), and malondialdehyde (MDA), were significantly higher in the MCD-induced NASH group than in the control group, whereas the low-density lipoprotein cholesterol (LDL-c) level was significantly lower in the MCD-induced NASH group than in the control group (Table 2). Compared with the control group, H&E staining of liver tissues in the NASH group showed obvious bullae steatosis with different sizes of lipid droplets (Fig. 1A). These phenomena are consistent with the characteristics of mice with MCD-induced NASH, indicating the success of the mouse model.
Table 2.
Comparison of biochemical indexes between control and NASH mice.
| Variables | Control group | NASH group | Change trend |
|---|---|---|---|
| ALT (U/L) | 303.624 ± 156.477 | 405.847 ± 132.862 | − |
| DBIL (μmol/L) | 17.134 ± 3.128 | 25.320 ± 4.083*** | ↑ |
| TBIL (μmol/L) | 25.092 ± 5.175 | 41.306 ± 4.649*** | ↑ |
| ALB (g/L) | 38.330 ± 4.513 | 49.912 ± 2.199*** | ↑ |
| ALP (U/L) | 103.984 ± 10.823 | 171.207 ± 26.146*** | ↑ |
| γ-GT (U/L) | 4.849 ± 4.063 | 10.108 ± 2.220** | ↑ |
| TG (mmol/L) | 1.761 ± 0.307 | 2.445 ± 0.373** | ↑ |
| TC (mmol/L) | 3.727 ± 0.395 | 3.345 ± 0.354 | − |
| HDL-c (mmol/L) | 0.449 ± 0.137 | 0.775 ± 0.214** | ↑ |
| LDL-c (mmol/L) | 1.497 ± 0.270 | 1.092 ± 0.282* | ↓ |
| MDA (μmol/g) | 6.906 ± 1.964 | 9.938 ± 4.057 | − |
Note: ALT: alanine transaminase; DBIL: direct bilirubin; TBIL: total bilirubin; ALB: albumin; ALP: alkaline phosphatase; γ-GT: gamma-glutamyl transpeptidase; TG: triglyceride; TC: total cholesterol; HDL-c: high-density lipoprotein cholesterol; LDL-c: low-density lipoprotein cholesterol; MDA: malondialdehyde. Data are mean ± SD, n = 8; *p < 0.05, **p < 0.01, ***p < 0.001 versus control group. −: not significant. ↑: significantly increased. ↓: significantly decreased.
Fig. 1.
Comparison of gut microbiota between control and NASH mice. (A): H&E staining for liver tissues (scale bar: 50 μM for the top and 20 μM for the bottom row); (B): Unweighted UniFrac principal coordinate analysis; (C–G): Most abundant taxa at the phylum (C), class (D), order (E), family (F), and genus (G) levels; (H–I): Chao1 (H) and Shannon (I) alpha diversity metrics were reduced in NASH mice. Data are mean ± SD, n = 8; *p < 0.05, **p < 0.01, ***p < 0.001 versus control group.
Changes of Cyp450 enzyme activity and expression in mice with NASH
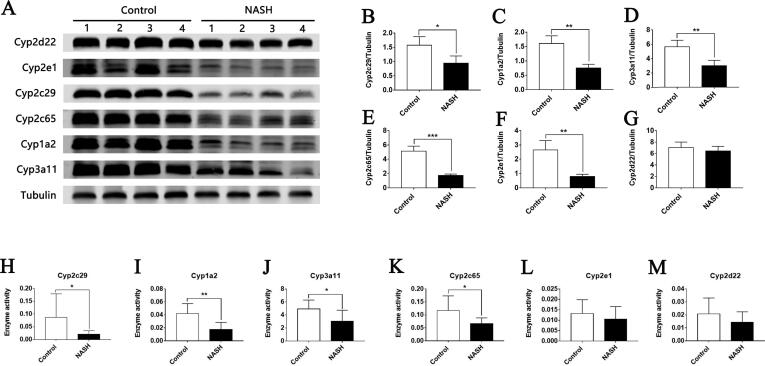
Compared with the control group, the expression levels of Cyp1a2, Cyp2c65, Cyp3a11, Cyp2c29, and Cyp2e1 in the NASH group were significantly downregulated (Fig. 2A–G). Meanwhile, the activities of Cyp1a2, Cyp2c65, Cyp3a11, and Cyp2c29 in the NASH group significantly reduced by 44.33%, 51.14%, 34.02%, and 24.12%, respectively (Fig. 2H–M). The activities of Cyp2e1 and Cyp2d22 in the NASH group exhibited downward trends, but the differences were not statistically significant. Results showed that the enzyme activity and expression of mice with NASH induced by MCD diet were lower than those of normal mice.
Fig. 2.
Comparison of host Cyp450s enzymes between control and NASH mice. (A) Western blot showing the expression of Cyp450 enzymes in the liver. Tubulin was used as a loading control. (B–G) Densitometric quantification for Western blot. (H–M) Activities of Cyp450 enzymes in liver microsome. Data are mean ± SD; *p < 0.05, **p < 0.01, ***p < 0.001 versus control group.
Changes of gut microbiota in mice with NASH
The changes of gut microbiota in the control and NASH groups were analyzed by 16S rRNA gene sequencing. Similar sequences (≥97%) were divided into OTUs. Species annotation was conducted according to the 16S rRNA gene database. Significant separation was observed between the control and NASH groups (Fig. 1B). Sequence alignment was performed using an RDP classifier to compare the differences in gut microbiota between the two groups at different classification levels. We selected the top 10 species with the largest abundance at each classification level (Phylum, Class, Order, Family, Genus) in each group. Compared with the control group, the proportion of species in the NASH group at each classification level changed (Fig. 1C–G). The alpha diversity analysis index of different samples under the threshold of 97% agreement was counted. The Chao1 index was used to estimate the total number of species contained in the community and the Shannon index represents the total number of classifications in the sample. Results showed that the Chao1 and Shannon indices of the NASH group were significantly lower than those of the control group (Fig. 1H, I), indicating that the gut microbiota composition of the mice with NASH induced by MCD diet decreased in varying degrees.
Pharmacokinetic variability co-influenced by host Cyp450s and gut microbiota in mice with NASH
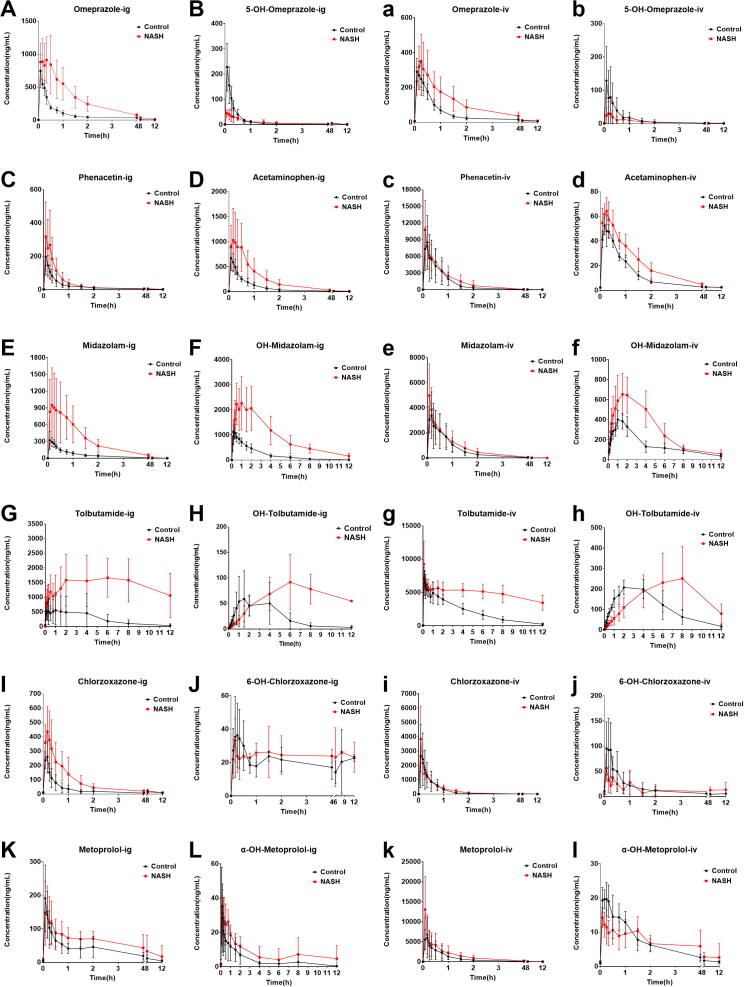
The exposure of the probe drugs, except metoprolol, was significantly higher in the NASH group than in the control group after intragastric administration. The pharmacokinetic parameters, including AUC and Cmax, of the probe drugs changed in different degrees. Moreover, their respective metabolites showed significant changes. The details are described as follows.
Omeprazole
The exposure of omeprazole in the NASH group was significantly higher than that in the control group, and AUC(0-12h) and AUC(0-infinity) significantly increased. Meanwhile, CLz/F significantly decreased. Its metabolite 5-OH-omeprazole showed an obvious opposite change compared with the parent drug (Table 3 and Fig. 3A, B).
Table 3.
Pharmacokinetic parameters of analytes between control and NASH mice by intragastric administration.
| Analytes | Groups | AUC(0-12h), ng·h/mL | AUC(0-infinity), ng·h/mL | Cmax, ng/mL | Tmax, h | t1/2, h | CLz/F, L/h/kg |
|---|---|---|---|---|---|---|---|
| Omeprazole | Control | 525.43 ± 67.62 | 549.67 ± 59.45 | 758.00 ± 242.89 | 0.11 ± 0.04 | 4.33 ± 2.78 | 18,362.04 ± 1,836.04 |
| NASH | 1765.71 ± 684.29*** | 1821.70 ± 731.27*** | 965.67 ± 340.37 | 0.22 ± 0.16 | 2.67 ± 1.07 | 7116.28 ± 5250.54*** | |
| 5-OH-omeprazole | Control | 73.49 ± 28.59 | 74.04 ± 28.25 | 229.98 ± 93.95 | 0.10 ± 0.03 | 0.51 ± 0.31 | 89248.71 ± 124294.64 |
| NASH | 36.46 ± 11.66* | 57.26 ± 28.94 | 53.00 ± 16.46*** | 0.15 ± 0.10 | 1.50 ± 0.73* | 215121.12 ± 104444.70 | |
| Phenacetin | Control | 134.92 ± 42.61 | 143.64 ± 41.11 | 199.33 ± 106.23 | 0.10 ± 0.03 | 4.59 ± 1.87 | 75382.89 ± 24126.47 |
| NASH | 231.43 ± 126.05* | 314.64 ± 197.88* | 423.95 ± 319.05* | 0.10 ± 0.03 | 7.78 ± 3.88 | 42948.82 ± 23087.23** | |
| Acetaminophen | Control | 609.00 ± 215.27 | 1291.43 ± 679.24 | 712.14 ± 243.35 | 0.11 ± 0.04 | 6.23 ± 3.88 | 14547.78 ± 3328.31 |
| NASH | 1277.79 ± 677.38* | 1291.43 ± 679.24 | 1042.67 ± 623.48 | 0.28 ± 0.25 | 1.72 ± 1.04* | 9374.47 ± 3743.18* | |
| Midazolam | Control | 302.17 ± 58.88 | 303.44 ± 58.44 | 358.29 ± 135.28 | 0.13 ± 0.07 | 2.13 ± 0.66 | 34263.46 ± 8162.03 |
| NASH | 1518.64 ± 790.20*** | 1521.71 ± 791.59*** | 988.17 ± 655.44** | 0.24 ± 0.145 | 1.54 ± 0.31 | 11891.56 ± 14110.88** | |
| OH-midazolam | Control | 2561.26 ± 558.16 | 2574.50 ± 556.76 | 1123.71 ± 225.50 | 0.37 ± 0.28 | 1.48 ± 0.35 | 4032.33 ± 805.79 |
| NASH | 10885.25 ± 4549.00*** | 11510.02 ± 5062.40*** | 2347.17 ± 962.87** | 0.96 ± 0.40** | 2.61 ± 0.71** | 1289.74 ± 1256.38*** | |
| Tolbutamide | Control | 3069.03 ± 3378.87 | 3123.13 ± 3422.15 | 669.43 ± 508.15 | 0.98 ± 1.45 | 1.78 ± 0.47 | 742.03 ± 557.94 |
| NASH | 21686.36 ± 15503.09** | 56355.58 ± 49559.98* | 2471.67 ± 2023.48* | 5.67 ± 1.51*** | 28.07 ± 24.80** | 33.46 ± 24.09** | |
| OH-Tolbutamide | Control | 324.32 ± 258.43 | 329.04 ± 260.28 | 56.17 ± 28.47 | 2.14 ± 0.90 | 1.66 ± 0.74 | 3024.55 ± 4818.47 |
| NASH | 1339.71 ± 1043.23* | 1630.81 ± 1040.30* | 159.04 ± 91.64* | 11.33 ± 1.63*** | 10.94 ± 8.48* | 251.87 ± 300.30 | |
| Chlorzoxazone | Control | 250.06 ± 88.36 | 271.46 ± 99.36 | 276.37 ± 105.55 | 0.14 ± 0.04 | 4.30 ± 2.23 | 3.99 ± 0.98 |
| NASH | 602.00 ± 267.84** | 629.87 ± 251.86** | 571.87 ± 299.33* | 0.19 ± 0.09 | 3.44 ± 2.74 | 1.88 ± 0.88*** | |
| 6-OH-chlorzoxazone | Control | 224.10 ± 35.78 | 510.70 ± 602.95 | 64.96 ± 27.38 | 0.26 ± 0.12 | 29.58 ± 48.45 | 2.92 ± 1.86 |
| NASH | 266.55 ± 114.43 | 484.46 ± 348.08 | 49.313 ± 24.289 | 3.10 ± 3.58 | 10.30 ± 7.93 | 3.48 ± 2.66 | |
| Metoprolol | Control | 317.71 ± 128.62 | 328.84 ± 127.43 | 216.40 ± 93.22 | 0.12 ± 0.09 | 2.52 ± 0.69 | 3.41 ± 1.20 |
| NASH | 413.45 ± 218.46 | 453.42 ± 298.21 | 189.13 ± 88.97 | 0.27 ± 0.32 | 2.60 ± 1.44 | 2.92 ± 1.59 | |
| α-OH-metoprolol | Control | 59.11 ± 21.21 | 61.32 ± 21.90 | 47.40 ± 17.72 | 2.77 ± 1.51 | 0.14 ± 0.09 | 2.77 ± 1.51 |
| NASH | 68.28 ± 40.72 | 70.39 ± 43.97 | 37.77 ± 11.34 | 1.70 ± 1.01 | 0.23 ± 0.23 | 1.70 ± 1.01 |
Note: AUC(0–12 h): area under the plasma concentration–time curve from 0 to 12 h; AUC(0-infinity): area under the plasma concentration–time curve from 0 to infinity h; Cmax: maximal concentration; t1/2: eliminate half-life; Tmax: time to peak concentration; CLz/F: blood clearance. Data are mean ± SD, n = 8; *p < 0.05, **p < 0.01, ***p < 0.001 versus control group.
Fig. 3.
Comparison of mean plasma concentration–time curves of probe drugs and metabolites by intragastric and intravenous administration between control and NASH mice. (A–L) Probe drugs intragastrically administered. (a–l) Probe drugs intravenously administered. Data are mean ± SD, n = 8.
Phenacetin
The exposure of phenacetin in the NASH group was significantly higher than that in the control group, and AUC(0-12h), AUC(0-infinity), and Cmax significantly increased. Meanwhile, CLz/F significantly decreased. The exposure of acetaminophen was also significantly higher in the NASH group than in the control group (Table 3 and Fig. 3C, D).
Midazolam
The exposure of midazolam in the NASH group was significantly higher than that in the control group, and AUC(0-12h), AUC(0-infinity), and Cmax significantly increased. Meanwhile, CLz/F significantly decreased. The exposure of OH-midazolam was also significantly higher in the NASH group than in the control group (Table 3 and Fig. 3E, F).
Tolbutamide
The exposure of tolbutamide in the NASH group was significantly higher than that in the control group, and AUC(0-12h), AUC(0-infinity), Cmax, Tmax, and t1/2 significantly increased. Meanwhile, CLz/F significantly decreased. The exposure of OH-tolbutamide was also significantly higher in the NASH group than in the control group (Table 3 and Fig. 3G, H).
Chlorzoxazone
The exposure of chlorzoxazone in the NASH group was significantly higher than that in the control group, and AUC(0-12h), AUC(0-infinity), and Cmax significantly increased. Meanwhile, CLz/F significantly decreased. However, the exposure of 6-OH-chlorzoxazone showed no obvious changes in the two groups (Table 3 and Fig. 3I, J).
Metoprolol
No significant changes in metoprolol and α-OH-metoprolol exposures were found in the two groups. The parameters, including the AUC(0-12h), AUC(0-infinity), Cmax, Tmax, t1/2, and CLz/F, of metoprolol and α-OH-metoprolol showed no statistically significant differences between the NASH and control groups (Table 3 and Fig. 3K, L).
Pharmacokinetic variability mainly influenced by the host Cyp450s in mice with NASH
Compared with that of the control group, the pharmacokinetics of the NASH group during intravenous administration showed no significant differences, except for higher exposures of tolbutamide, OH-tolbutamide, and OH-midazolam.
No significant changes were noted in the exposure and pharmacokinetic parameters of omeprazole, phenacetin, midazolam, chlorzoxazone, and metoprolol or their metabolites (Table 4 and Fig. 3a–e, i–l). However, the exposure of tolbutamide in the NASH group was significantly higher than that in the control group, and the AUC(0-12h), AUC(0-infinity), and Tmax of tolbutamide were significantly higher than those in the control group. Meanwhile, CLz/F significantly decreased (Table 4 and Fig. 3g). The pharmacokinetic parameters, including the AUC and Cmax, of OH-tolbutamide and OH-midazolam also significantly increased (Table 4 and Fig. 3f, h).
Table 4.
Pharmacokinetic parameters of probe drugs between control and NASH mice by intravenous administration.
| Analytes | Groups | AUC(0-12h), ng·h/mL | AUC(0-infinity), ng·h/mL | Cmax, ng/mL | Tmax, h | t1/2, h | CLz/F, L/h/kg |
|---|---|---|---|---|---|---|---|
| Omeprazole | Control | 335.64 ± 61.51 | 436.49 ± 128.46 | 307.00 ± 60.89 | 0.17 ± 0.11 | 9.21 ± 8.38 | 10.91 ± 4.41 |
| NASH | 586.75 ± 178.81** | 654.77 ± 150.47 | 294.57 ± 95.30 | 0.24 ± 0.09 | 5.89 ± 5.47 | 8.02 ± 1.98 | |
| 5-OH-omeprazole | Control | 77.67 ± 63.23 | 87.05 ± 66.82 | 113.15 ± 105.36 | 0.12 ± 0.07 | 9.36 ± 4.21 | 116.94 ± 119.32 |
| NASH | 33.53 ± 21.78 | 43.26 ± 22.92 | 21.54 ± 20.58 | 0.28 ± 0.25 | 9.72 ± 3.13 | 144.01 ± 67.17 | |
| Phenacetin | Control | 7017.39 ± 3514.16 | 7190.82 ± 3361.43 | 9648.33 ± 4219.11 | 0.11 ± 0.04 | 4.76 ± 3.48 | 0.90 ± 0.59 |
| NASH | 7714.58 ± 3764.24 | 7857.32 ± 3965.15 | 10555.71 ± 5268.20 | 0.15 ± 0.12 | 3.26 ± 1.85 | 0.80 ± 0.42 | |
| Acetaminophen | Control | 81.98 ± 9.56 | 156.44 ± 65.85 | 52.93 ± 7.734 | 0.21 ± 0.07 | 29.92 ± 11.59 | 37.72 ± 16.85 |
| NASH | 128.52 ± 42.13** | 168.30 ± 38.61 | 64.46 ± 13.84 | 0.25 ± 0.13 | 12.46 ± 12.23 | 31.02 ± 6.58 | |
| Midazolam | Control | 3203.76 ± 1650.28 | 3206.56 ± 1649.65 | 3594.29 ± 1708.41 | 0.11 ± 0.04 | 0.81 ± 0.21 | 1.97 ± 0.99 |
| NASH | 4749.76 ± 2566.02 | 4755.99 ± 2567.31 | 5352.50 ± 2465.10 | 0.12 ± 0.09 | 1.21 ± 0.45 | 1.31 ± 0.61 | |
| OH-midazolam | Control | 1836.54 ± 490.21 | 2130.84 ± 824.13 | 428.86 ± 86.04 | 1.32 ± 0.59 | 3.81 ± 1.72 | 2.55 ± 0.64 |
| NASH | 3561.41 ± 1190.62** | 3857.57 ± 1372.45* | 694.88 ± 189.24** | 1.50 ± 0.38 | 2.84 ± 1.59 | 1.45 ± 0.54** | |
| Tolbutamide | Control | 25583.23 ± 6932.28 | 26403.97 ± 7552.17 | 7807.14 ± 1459.56 | 0.36 ± 0.44 | 2.10 ± 0.36 | 0.20 ± 0.05 |
| NASH | 58276.69 ± 11536.71*** | 129682.45 ± 63485.35*** | 9076.25 ± 3163.69 | 0.16 ± 0.10 | 12.96 ± 8.82** | 0.05 ± 0.03*** | |
| OH-Tolbutamide | Control | 1329.61 ± 330.20 | 1414.25 ± 333.57 | 227.00 ± 35.12 | 2.92 ± 1.20 | 2.64 ± 1.89 | 0.75 ± 0.21 |
| NASH | 2658.69 ± 1322.89* | 3218.168 ± 1371.781* | 398.50 ± 180.36* | 8.00 ± 2.53** | 3.99 ± 2.47 | 0.38 ± 0.20* | |
| Chlorzoxazone | Control | 1694.57 ± 1024.63 | 1700.78 ± 1018.04 | 3318.00 ± 2218.99 | 0.14 ± 0.10 | 1.07 ± 1.83 | 4.09 ± 2.50 |
| NASH | 1714.34 ± 580.13 | 1757.71 ± 610.534 | 3081.43 ± 1836.52 | 0.14 ± 0.11 | 1.15 ± 0.68 | 3.12 ± 0.95 | |
| 6-OH-chlorzoxazone | Control | 123.34 ± 45.67 | 229.62 ± 195.23 | 117.40 ± 49.59 | 0.17 ± 0.10 | 12.94 ± 18.21 | 32.43 ± 18.09 |
| NASH | 119.76 ± 33.28 | 270.50 ± 236.25 | 58.43 ± 22.65 | 0.25 ± 0.34 | 31.13 ± 43.61 | 26.46 ± 15.86 | |
| Metoprolol | Control | 3949.38 ± 1727.20 | 3975.79 ± 1719.27 | 5518.33 ± 2476.02 | 0.08 ± 0.00 | 2.02 ± 1.30 | 1.52 ± 0.75 |
| NASH | 8342.53 ± 4802.86 | 8391.23 ± 4795.22 | 11991.43 ± 8308.01 | 0.12 ± 0.09 | 1.83 ± 0.73 | 0.81 ± 0.50 | |
| α-OH-metoprolol | Control | 47.31 ± 8.80 | 76.52 ± 56.13 | 21.36 ± 3.42 | 0.21 ± 0.15 | 5.55 ± 4.57 | 84.64 ± 34.11 |
| NASH | 64.10 ± 28.85 | 128.15 ± 191.56 | 14.95 ± 4.53 | 1.03 ± 1.33 | 6.34 ± 8.22 | 77.34 ± 35.26 |
Note: AUC(0-12h): area under the plasma concentration–time curve from 0 to 12 h; AUC(0-infinity): area under the plasma concentration–time curve from 0 to infinity h; Cmax: maximal concentration; t1/2: eliminate half-life; Tmax: time to peak concentration; CLz/F: blood clearance. Data are mean ± SD, n = 8; *p < 0.05, **p < 0.01, ***p < 0.001 versus control group.
Pharmacokinetic variability related to host Cyp450s and gut microbiota alterations in mice with NASH
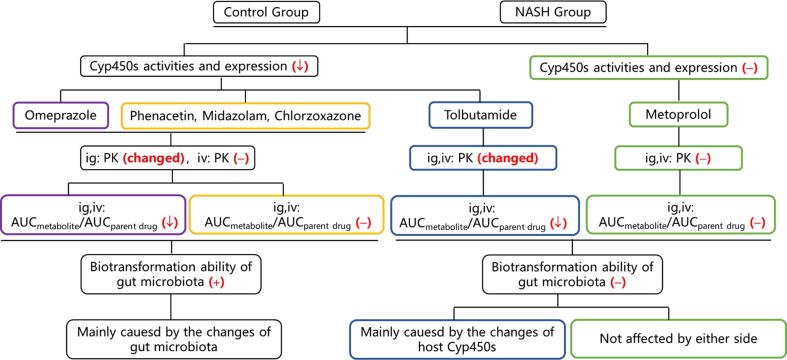
The ratio of AUCmetabolite to AUCparent drug can reflect the activity of the corresponding Cyp450 enzyme in vivo. Combined with the results of Cyp450 enzyme activity and protein expression in vitro, the relationship between the host Cyp450 enzyme and the pharmacokinetic variability in mice with NASH could be analyzed (Table 5). Compared with that in the control group, Cyp2c65 in the NASH group showed reduced activity after intragastric and intravenous administrations, which is consistent with the reduced enzyme activity and protein expression in vitro. This result suggests that the pharmacokinetic variability of tolbutamide was mainly caused by the changes in host Cyp450 enzymes. The activities of Cyp1a2, Cyp3a11, and Cyp2e1 enzymes did not change in vivo but decreased in vitro, suggesting that the pharmacokinetic variabilities of phenacetin, midazolam, and chlorzoxazone were affected by the changes in the gut microbiota in NASH. The activity of Cyp2d22 changed neither in vivo nor in vitro, indicating that the metabolism of metoprolol was hardly affected by the host Cyp450s and gut microbiota. Although the activity of Cyp2c29 significantly decreased in vivo and in vitro, no significant difference was observed in the pharmacokinetics of intravenous administration, suggesting that the pharmacokinetic variability of omeprazole was caused by other influencing factors.
Table 5.
Comparison of activities and expressions of Cyp450 enzymes determined through different methods.
| Cyp450s | AUCmetabolite/AUCparent drug by intragastric administration (in vivo) |
AUCmetabolite/AUCparent drug by intravenous administration (in vivo) |
Incubation by liver microsomes (in vitro) |
Protein expression (in vitro) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | NASH | Change trend | Control | NASH | Change trend | Control | NASH | Control | NASH | |
| Cyp2c29 | 0.138 ± 0.046 | 0.031 ± 0.022*** | ↓ | 0.241 ± 0.210 | 0.063 ± 0.034* | ↓ | − | ↓ | − | ↓ |
| Cyp1a2 | 4.612 ± 1.017 | 5.075 ± 1.245 | − | 0.015 ± 0.008 | 0.019 ± 0.009 | − | − | ↓ | − | ↓ |
| Cyp3a11 | 8.540 ± 1.215 | 7.950 ± 1.945 | − | 0.697 ± 0.331 | 0.956 ± 0.582 | − | − | ↓ | − | ↓ |
| Cyp2c65 | 0.137 ± 0.056 | 0.065 ± 0.027** | ↓ | 0.071 ± 0.035 | 0.034 ± 0.019* | ↓ | − | ↓ | − | ↓ |
| Cyp2e1 | 0.986 ± 0.327 | 0.632 ± 0.527 | − | 0.093 ± 0.039 | 0.090 ± 0.055 | − | − | − | − | ↓ |
| Cyp2d22 | 0.175 ± 0.090 | 0.167 ± 0.033 | − | 0.013 ± 0.005 | 0.009 ± 0.007 | − | − | − | − | − |
Note: Data are mean ± SD; *p < 0.05, **p < 0.01, ***p < 0.001 versus control group. −: not significant. ↓: significantly decreased.
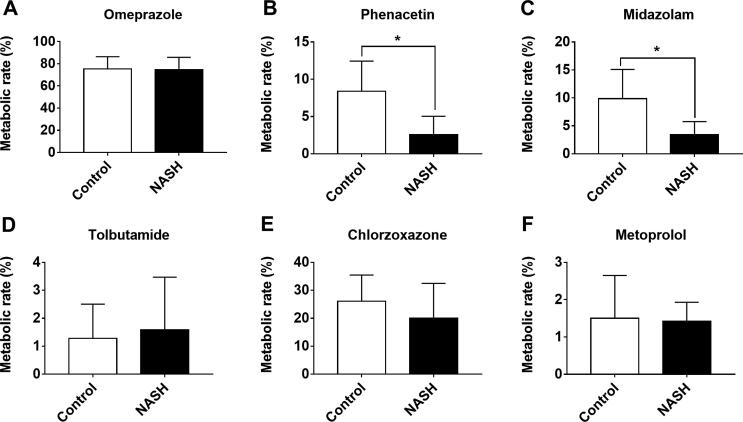
Considering the influence of the gut microbiota, we analyzed its biotransformation ability with regard to the probe drugs. The results showed that gut microbiota did not metabolize tolbutamide and metoprolol but metabolized omeprazole, phenacetin, midazolam, and chlorzoxazone. The metabolism rates of omeprazole, phenacetin (p < 0.05), midazolam (p < 0.05), and chlorzoxazone were lower in the NASH group compared with the control group (Fig. 4).
Fig. 4.
Comparison of biotransformation of probe drugs by gut microbiota between control and NASH mice. Data are mean ± SD, n = 8; *p < 0.05 versus control group.
In sum, the higher exposures of omeprazole, phenacetin, midazolam, and chlorzoxazone in the NASH group than in the control group were mainly due to changes in the gut microbiota. The pharmacokinetic variability of tolbutamide was mainly caused by the changes in host Cyp450 enzymes. However, the pharmacokinetic variability of metoprolol was not affected by either factor. The results are summarized in Table 6 and the detailed logical judgment process is shown in Fig. 5.
Table 6.
Pharmacokinetic variabilities of NASH group compared with control group and judgment of related influencing factors.
| Drugs | PK changes (ig) |
PK changes (iv) |
Cyp450s changes |
AUCmetabolite/AUCparent drug |
Gut microbiota |
Main influencing factors | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Parent drug | Metabolite | Parent drug | Metabolite | Activity | Expression | ig | iv | Biotransformation | Host/gut microbiota | |
| Omeprazole | AUC↑, CLz/F↓ |
AUC↓, Cmax ↓, t1/2↓ | AUC(0-12h)↑ | − | Cyp2c29↓ | Cyp2c29↓ | ↓ | ↓ | Yes | gut microbiota |
| Phenacetin | AUC↑, Cmax ↑,CLz/F↓ | AUC↑, t1/2 ↓, CLz/F↓ | − | AUC(0-12h)↑ | Cyp1a2↓ | Cyp1a2↓ | − | − | Yes | gut microbiota |
| Midazolam | AUC↑, Cmax ↑,CLz/F↓ | AUC↑, Cmax ↑, Tmax ↑, t1/2↑, CLz/F↓ |
− | AUC↑, Cmax ↑,CLz/F↓ | Cyp3a11↓ | Cyp3a11↓ | − | − | Yes | gut microbiota |
| Chlorzoxazone | AUC↑, Cmax ↑,CLz/F↓ | − | − | − | Cyp2e1− | Cyp2e1↓ | − | − | Yes | gut microbiota |
| Tolbutamide | AUC↑, Cmax ↑, Tmax ↑ t1/2↑, CLz/F↓ |
AUC↑, Cmax ↑, Tmax ↑, t1/2↑ | AUC↑, t1/2↑,CLz/F↓ | AUC↑, Cmax ↑, Tmax ↑,CLz/F↓ | Cyp2c65↓ | Cyp2c65↓ | ↓ | ↓ | No | host |
| Metoprolol | − | − | − | − | Cyp2d22− | Cyp2d22− | − | − | No | Neither |
Note: The results in the table are from the NASH group vs. the control group in mice. −: not significant. ↑: significantly increased. ↓: significantly decreased. ig: intragastric administration. iv: intravenous administration. Based on the results of Cyp450 enzymes in vitro as the standard, combined with the results of Cyp450 enzymes in vivo, their relationships can be roughly divided into three categories: 1) if the results were consistent, the pharmacokinetic variabilities were mainly related to the host alteration; 2) if the results were inconsistent, the pharmacokinetic variabilities were mainly related to the gut microbiota; and 3) if the results did not change, the pharmacokinetics were less affected by both factors.
Fig. 5.
Detailed logical judgment process of the main influencing factors of pharmacokinetic variabilities. The results in the figure are from the NASH group vs. the control group in mice; −: no change or no having; ↓: decrease; +: having; ig: intragastric administration. iv: intravenous administration.
Discussion
The host Cyp450 enzymes and the gut microbiota are important factors affecting pharmacokinetics. Diseases such as NASH usually affect the activity of host Cyp450s and the abundance or composition of gut microbiota [45], [46], thus affecting pharmacokinetic variability. The activity and expression of the main host Cyp450s (Cyp2c29, Cyp1a2, Cyp3a11, Cyp2c65, and Cyp2e1) involved in the drug metabolism of omeprazole, phenacetin, midazolam, tolbutamide, and chlorzoxazone were significantly lower in the NASH group than in the control group. Previous studies support our findings. To elucidate, Li et al. reported that the activity and expression of Cyp450 enzymes, including Cyp1a2, Cyp3a11, and Cyp2c29, decreased in the NASH group induced by MCD diet [47], [48]. It is worth mentioning that the host Cyp450s involved in those studies were considered individually, whereas six host Cyp450s were simultaneously investigated in our study, which are responsible for the metabolism of over 90% of clinically used drugs [43]. The category and quantity of the gut microbiota in the NASH group significantly reduced, and the composition and abundance were significantly different from those in the control group. Meanwhile, NASH significantly decreased the alpha diversity index (Chao1 and Shannon) of the gut microbiota. Some previous studies support our results. For instance, Ye et al. found that MCD diet increased Firmicutes population but decreased Proteobacteria and Bacteroidetes populations [49]. Kai et al. found a decrease in the alpha diversity index (Chao1) in the NASH group [50]. However, our study was more comprehensive and systematic because we considered the host CYP450 enzymes and the gut microbiota.
Changes in the host Cyp450 activity and gut microbiota caused by NASH might give rise to pharmacokinetic variability. Therefore, the same drug under different administration routes may have different pharmacokinetic variabilities because of the different effects of the host Cyp450s and gut microbiota under different drug administration pathways. Generally, pharmacokinetics is influenced by the host Cyp450s and the gut microbiota under intragastric administration. By contrast, drugs bypass the intestine and directly enter the bloodstream under intravenous administration, thereby avoiding intestinal absorption. In this case, pharmacokinetics is mainly affected by the host [51]. Based on the above logic, under intragastric administration, the exposure and pharmacokinetic parameters of omeprazole, phenacetin, midazolam, tolbutamide, and chlorzoxazone were considerably higher in the NASH group than in the control group. For their main metabolites, the pharmacokinetics of 5-OH-omeprazole, acetaminophen, OH-midazolam, and OH-tolbutamide also significantly changed. However, the pharmacokinetics of metoprolol and α-OH-metoprolol showed no significant change. Under intravenous administration, the exposure and pharmacokinetic parameters of tolbutamide, OH-toluamide, and OH-midazolam in the NASH group were much higher than those in the control group, whereas those of omeprazole, phenacetin, midazolam, chlorzoxazone, metoprolol, and their metabolites did not obviously vary between the two groups. Comparing the pharmacokinetics results between these two administration routes, we found that the pharmacokinetics of omeprazole, phenacetin, midazolam, chlorzoxazone, and their metabolites in the NASH group significantly changed under intragastric administration but not under intravenous administration. The pharmacokinetics of tolbutamide and its metabolite (OH-tolbutamide) significantly changed under the two administration routes. The pharmacokinetics of metoprolol and its metabolite (α-OH-metoprolol) showed no significant changes between the two administration routes. Considering these results, we speculated that the pharmacokinetic variabilities of omeprazole, phenacetin, midazolam, and chlorzoxazone in the NASH group are mainly caused by changes in the gut microbiota, while the pharmacokinetic variability of tolbutamide may be mainly due to changes in the host Cyp450 enzymes. Meanwhile, the pharmacokinetic variability of metoprolol was neither affected by the host Cyp450s nor by the gut microbiota.
We further analyzed the different measurement values of host Cyp450 activities between the groups to explain the relation of the pharmacokinetic variability of these drugs with the changes in the host Cyp450s and gut microbiota from another side. Previous studies reported that the ratio of AUCmetabolite to AUCparent drug of the probe drugs can reflect the activity of the corresponding Cyp450 enzymes in vivo [52]. However, these ratios via intragastric or intravenous administration were inevitably affected by the host factors and gut microbiota. By contrast, determination of the host Cyp450 activity by in vitro human liver microsomal incubation was relatively accurate. Therefore, using the results of Cyp450 enzymes in vitro as a reference, we can divide their relationships into three types to compare the differences between the results of Cyp450 enzyme activity determined in vivo. First, if the enzyme activities in vitro and in vivo showed the same trend, the pharmacokinetic variability in NASH mainly caused the change in host Cyp450s. Second, if the enzyme activities were inconsistent in vitro and in vivo, the pharmacokinetic variability in NASH was mainly caused by the change in gut microbiota. Third, if no difference existed between these enzyme activities, then the pharmacokinetics variability was less affected by both factors. In the present study, the activity of Cyp2c65 in vivo was similar to that in vitro, suggesting that the pharmacokinetic variability of tolbutamide was mainly caused by changes in the host Cyp450s. Although the activity of Cyp2c29 significantly decreased in vivo and in vitro, no significant difference existed in the pharmacokinetics of intravenous administration, suggesting that the pharmacokinetic variability of omeprazole was related to the changes in gut microbiota. The in vivo and in vitro activities of Cyp1a2, Cyp3a11, and Cyp2e1 were inconsistent, indicating that the pharmacokinetic variabilities of phenacetin, midazolam, and chlorzoxazone were mainly related to the changes in gut microbiota. In addition, the biotransformation experiments showed that gut microbiota can metabolize omeprazole, phenacetin, midazolam, and chlorzoxazone. The metabolic rates of the normal control group were higher than those of the NASH group, which supports our inference from the other side. The activity of Cyp2d22 did not change, indicating that metoprolol metabolism was not affected by either factor.
Regarding the study of pharmacokinetic variability under NASH, only morphine was reported. The Cmax and AUC of morphine-3-glucuronic acid and morphine-6-glucuronic acid significantly increased in patients with NASH, and the changes are related to the increased expression of the transporter multidrug resistance-associated protein 3 (MRP3) [39]. Meanwhile, the influence of the host Cyp450s and gut microbiota was not considered in the study. For other studies, the analysis of the influencing factors of pharmacokinetic variability was mostly based on single host Cyp450s or the gut microbiota. Few studies simultaneously considered these two factors.
The cocktail probe is superior to the single-probe approach in terms of efficiency, in saving mice and costs, and in terms of obtaining more information from the same procedure. This study mainly referred to the previously published Cyp450 enzyme probe cocktail method [53], [54], [55]. The most common probes for Cyp1a2 are phenacetin and caffeine; those for Cyp3a11 are midazolam and dapsone; those for Cyp2c65 are tolbutamide and warfarin; those for Cyp2c29 are omeprazole and mephenytoin; that for Cyp2e1 is chlorzoxazone; and those for Cyp2d22 are metoprolol and dextromethorphan. Based on a group of probe drugs commonly used by researchers [56], phenacetin, midazolam, omeprazole, tolbutamide, chlorzoxazone, and metoprolol were selected as the present probes. Notably, the potential interaction between probe drugs is one of the most frequent concerns [57]. However, some experiments already demonstrated that no metabolic interaction occurs when these drugs are administered simultaneously in relatively low dosages [58]. The interactions between the probes can be reduced or controlled by regulating the drug doses. Many previous studies used the cocktail method to determine the activity of Cyp450 enzymes. Similarly, our selection of the probe drugs was based on a combination that had been used frequently in past studies. Thus, the drug metabolic interactions were controllable.
Some limitations need to be noted. Intravenous administration was used rather than gavage in germ-free or pseudo-germ-free mice to rule out the influence of gut microbiota. Germ-free or pseudo-germ-free mice are usually used to study the gut microbiota [59], but these animals require special breeding conditions or continuous antibiotic administration, respectively. Germ-free mice are ideal models but are very expensive [60]. For pseudo-germ-free mice, there might be concerns about drug–drug interactions between the probes and the antibiotics. Thus, intravenous administration effectively meets the needs of the present research. Furthermore, although intravenous administration is more convenient and quicker and not more expensive than sterile mice, the influence of enterohepatic circulation on the pharmacokinetic variability cannot be completely avoided; the same holds true for germ-free mice despite the absence of gut microbiota. In general, enterohepatic circulation may prolong the pharmacological effect of certain drugs and drug metabolites. Of particular importance is the potential amplifying effect of enterohepatic variability in defining differences in the bioavailability, apparent volume of distribution, and clearance of a given compound. In plasma concentration–time curves, enterohepatic circulation is often associated with multiple peaks and long apparent half-lives, which are more pronounced in drugs excreted through the gut [61]. Genetic abnormalities, disease states, orally administered adsorbents, and certain co-administered drugs all affect enterohepatic circulation. The gut microbiota is also a newly discovered factor affecting enterohepatic circulation [61], [62]. The six probe drugs used in the present study are not excreted mainly through the gut and there is a relative dearth of information about the influence of enterohepatic circulation on these drugs, so we speculated that enterohepatic circulation might not contribute much to these drugs’ pharmacokinetic variabilities in NASH mice. Even so, these phenomena need to be validated progressively in a single drug in germ-free or pseudo-germ-free mice or further in a clinical disease state to provide better clinical advice.
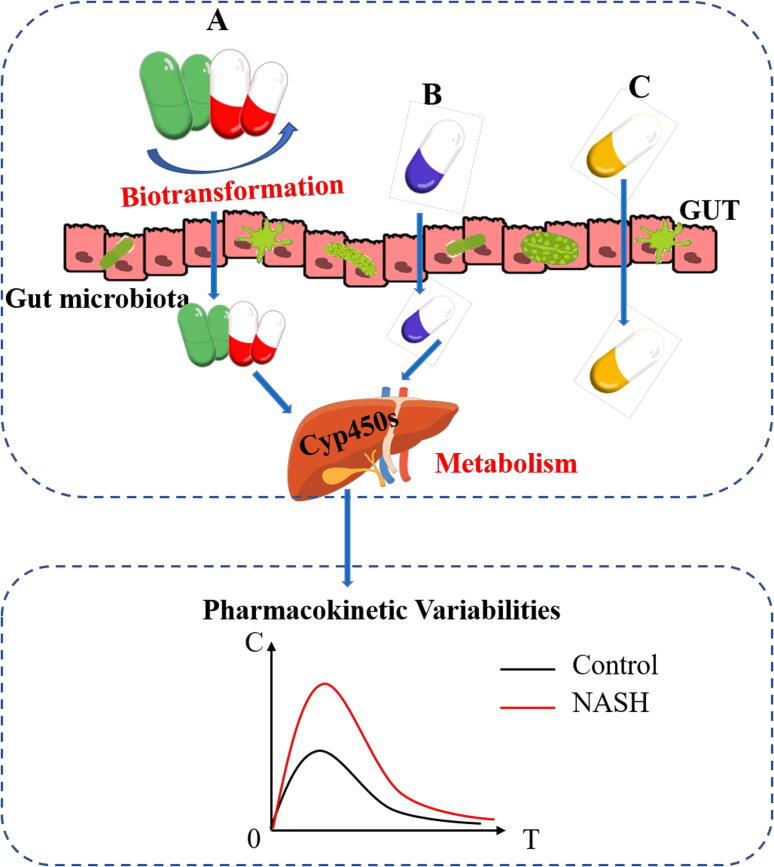
In summary, we observed the changes in host Cyp450 enzymes and gut microbiota that lead to pharmacokinetic variability in mice with NASH. Results show that the degree of their respective influence varies from drug to drug. NASH is a common type of disease or a risk factor for many metabolic diseases, such as hypertension and diabetes [63]. The medications given to patients with NASH or those with NASH symptoms are often complex in clinical settings. Therefore, the dosages of most drugs need to be adjusted according to their pharmacokinetic variabilities in vivo in order to meet their therapeutic concentration or to avoid toxicity. The factors leading to the pharmacokinetic variability of individual drugs are often different. Thus, the influence of the host or the gut microbiota in accordance with different medications should be analyzed. The pharmacokinetic variability of individual drugs requires specific analyses and cannot be generalized (Fig. 6). In the future, with the development and popularization of microbial sequencing and high-throughput sequencing technology, the influence of the host and gut microbiota should be considered during the individualized treatment of NASH and its complications.
Fig. 6.
Gut microbiota and host Cyp450s co-contribute to pharmacokinetic variabilities in NASH mice. The pharmacokinetic variabilities of some drugs are mainly caused by alterations in the gut microbiota (A), some may be mainly due to changes in the host Cyp450 enzymes (B), while others are not affected either by the host Cyp450 enzyme or by the gut microbiota (C).
Conclusion
The exposures of most drugs were increased and the pharmacokinetic variabilities of most drugs were found in mice with NASH. The gut microbiota and the host co-influenced the pharmacokinetic variability of drugs in mice with NASH, with the degree of contribution varying from drug to drug. Given these findings, the influence of the host and the gut microbiota should be simultaneously considered in personalized medicine.
Author contributions
Jing Guo and Yao Chen conceived of and designed the experiments and data analyses, as well as contributed to the writing and editing of the manuscript. Jing Guo, Ying Xu, Li-jie Chen, and Song-xia Zhang performed the experiments. Jing Guo, Xiao-ping Chen, Zhi-rong Tan, and Yao Chen contributed to the discussions. Yu-ligh Liou contributed to the English writing, grammar revision, and language polishing of the manuscript during its preparation. Hong-hao Zhou, Wei Zhang, and Yao Chen provided the reagents, materials, analysis tools, and experiment platforms that were crucial for the completion of the work.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Scientific Foundation of China (Grants 81974513 and 81302850) and the National Science and Technology Plan of China (Grant 2017ZX09304014). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. Thanks to Kang He, Li Li, Yi-cheng Wang, Tai Rao, Ying Guo, and Jing-bo Peng from the Drug Analysis Center of Xiangya Hospital, Central South University for providing technology assistance.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Porayette P., Flockhart D., Gupta S.K. One size fits one: pharmacogenetics in gastroenterology. Clin Gastroenterol Hepatol. 2014;12(4):565–570. doi: 10.1016/j.cgh.2014.01.035. [DOI] [PubMed] [Google Scholar]
- 2.Lin J.H. Pharmacokinetic and pharmacodynamic variability: a daunting challenge in drug therapy. Curr Drug Metab. 2007:109–136. doi: 10.2174/138920007779816002. [DOI] [PubMed] [Google Scholar]
- 3.Sun S., Wang Y., Wu A., Ding Z., Liu X. Influence factors of the pharmacokinetics of herbal resourced compounds in clinical practice. Evid Based Complement Alternat Med. 2019;2019:1–16. doi: 10.1155/2019/1983780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kulkarni NM, Malampati S, Mahat MY, Chandrasekaran S, Raghul J, Khan AA et al. Altered pharmacokinetics of rosiglitazone in a mouse model of non-alcoholic fatty liver disease. Drug Metab Pers Ther 2016;165–71. doi: 10.1515/dmpt-2016-0008. [DOI] [PubMed]
- 5.ter Heine R, Binkhorst L, de Graan AJ, de Bruijn P, Beijnen JH, Mathijssen RH, et al. Population pharmacokinetic modelling to assess the impact of CYP2D6 and CYP3A metabolic phenotypes on the pharmacokinetics of tamoxifen and endoxifen, Br J Clin Pharmacol. 2014;572-86. doi: 10.1111/bcp.12388. [DOI] [PMC free article] [PubMed]
- 6.Shu Y., Brown C., Castro R.A., Shi R.J., Lin E.T., Owen R.P., et al. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin Pharmacol Ther. 2008;83(2):273–280. doi: 10.1038/sj.clpt.6100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koukoula M, Dotsikas Y, Molou E, Schulpis KH, Thodi G, Chatzidaki M et al. Study of the effect of CYP2C19 polymorphisms on omeprazole pharmacokinetics by utilizing validated LC-MS/MS and Real Time-PCR methods. J Chromatogr B Analyt Technol Biomed Life Sci. 2017;173–179. doi:10.1016/j.jchromb.2016.06.046. [DOI] [PubMed]
- 8.Choi M.S., Yu J.S., Yoo H.H., Kim D.-H. The role of gut microbiota in the pharmacokinetics of antihypertensive drugs. Pharmacol Res. 2018;130:164–171. doi: 10.1016/j.phrs.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 9.Sommer F., Bäckhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 10.Neish A.S. Preimmune recognition and response to microbial metabolites. Physiology (Bethesda) 2021;36(2):94–101. doi: 10.1152/physiol.00023.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body, PLoS Biol 2016;e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed]
- 12.Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R., Goodman A.L. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science. 2019;363(6427) doi: 10.1126/science:aat9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarmusch A.K., Vrbanac A., Momper J.D., Ma J.D., Alhaja M., Liyanage M., et al. Enhanced characterization of drug metabolism and the influence of the intestinal microbiome: a pharmacokinetic, microbiome, and untargeted metabolomics study. Clin Transl Sci. 2020;13(5):972–984. doi: 10.1111/cts.v13.510.1111/cts.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson CH, Patterson AD, Idle JR, Gonzalez FJ. Xenobiotic metabolomics: major impact on the metabolome, Annu Rev Pharmacol Toxicol 2012;37–56. doi: 10.1146/annurev-pharmtox-010611-134748. [DOI] [PMC free article] [PubMed]
- 15.Jeong H.G., Kang M.J., Kim H.G., Oh D.G., Kim J.S., Lee S.K., et al. Role of intestinal microflora in xenobiotic-induced toxicity. Mol Nutr Food Res. 2013;57(1):84–99. doi: 10.1002/mnfr.v57.110.1002/mnfr.201200461. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X., Han Y., Huang W., Jin M., Gao Z. The influence of the gut microbiota on the bioavailability of oral drugs. Acta Pharm Sin B. 2021;11(7):1789–1812. doi: 10.1016/j.apsb:2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gill S.R., Pop M., DeBoy R.T., Eckburg P.B., Turnbaugh P.J., Samuel B.S., et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. doi: 10.1126/science:1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noh K., Kang Y.R., Nepal M.R., Shakya R., Kang M.J., Kang W., et al. Impact of gut microbiota on drug metabolism: an update for safe and effective use of drugs. Arch Pharm Res. 2017;40(12):1345–1355. doi: 10.1007/s12272-017-0986-y. [DOI] [PubMed] [Google Scholar]
- 19.Kang M.J., Ko G.S., Oh D.G., Kim J.S., Noh K., Kang W., et al. Role of metabolism by intestinal microbiota in pharmacokinetics of oral baicalin. Arch Pharm Res. 2014;37(3):371–378. doi: 10.1007/s12272-013-0179-2. [DOI] [PubMed] [Google Scholar]
- 20.Matuskova Z, Anzenbacherova E, Vecera R, Tlaskalova-Hogenova H, Kolar M, Anzenbacher P. Administration of a probiotic can change drug pharmacokinetics: effect of E. coli Nissle 1917 on amidarone absorption in rats, PLoS One. 2014;e87150. doi:10.1371/journal.pone.0087150. [DOI] [PMC free article] [PubMed]
- 21.Mallory E.K., Acharya A., Rensi S.E., Turnbaugh P.J., Bright R.A., Altman R.B. Chemical reaction vector embeddings: towards predicting drug metabolism in the human gut microbiome. Pac Symp Biocomput. 2018;56–67 [PMC free article] [PubMed] [Google Scholar]
- 22.Lindenbaum J., Rund D.G., Butler V.P., Tse-Eng D., Saha J.R. Inactivation of digoxin by the gut flora: reversal by antibiotic therapy. N Engl J Med. 1981;305(14):789–794. doi: 10.1056/NEJM198110013051403. [DOI] [PubMed] [Google Scholar]
- 23.Haiser H.J., Seim K.L., Balskus E.P., Turnbaugh P.J. Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes. 2014;5(2):233–238. doi: 10.4161/gmic.27915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24(7):908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue F., Xia K.e., Wei L., Xing L., Wu S., Shi Y., et al. Effects of constant light exposure on sphingolipidomics and progression of NASH in high-fat-fed rats. J Gastroenterol Hepatol. 2020;35(11):1978–1989. doi: 10.1111/jgh.v35.1110.1111/jgh.15005. [DOI] [PubMed] [Google Scholar]
- 26.Reimer K.C., Wree A., Roderburg C., Tacke F. New drugs for NAFLD: lessons from basic models to the clinic. Hepatol Int. 2020;14(1):8–23. doi: 10.1007/s12072-019-10001-4. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki A, Diehl AM. Nonalcoholic Steatohepatitis, Annu Rev Med.(2017) 85-98. doi:10.1146/annurev-med-051215-031109. [DOI] [PubMed]
- 28.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K et al. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology, Gastroenterology 2012;1592–609. doi: 10.1053/j.gastro.2012.04.001. [DOI] [PubMed]
- 29.Jiang M., Li C., Liu Q., Wang A., Lei M. Inhibiting ceramide synthesis attenuates hepatic steatosis and fibrosis in rats with non-alcoholic fatty liver disease. Front Endocrinol (Lausanne) 2019:665. doi: 10.3389/fendo.2019.00665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherif Z.A., Saeed A., Ghavimi S., Nouraie S.-M., Laiyemo A.O., Brim H., et al. Global epidemiology of nonalcoholic fatty liver disease and perspectives on US minority populations. Dig Dis Sci. 2016;61(5):1214–1225. doi: 10.1007/s10620-016-4143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J, Zhou F, Wang W, Zhang XJ, Ji YX, Zhang P et al. Epidemiological Features of NAFLD From 1999 to 2018 in China, Hepatology 2020;1851–1864. doi: 10.1002/hep.31150. [DOI] [PubMed]
- 32.Chen L.-J., Guo J., Zhang S.-X., Xu Y., Zhao Q., Zhang W., et al. Sirtuin3 rs28365927 functional variant confers to the high risk of non-alcoholic fatty liver disease in Chinese Han population. Lipids Health Dis. 2021;20(1) doi: 10.1186/s12944-021-01520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sumida Y., Yoneda M. Current and future pharmacological therapies for NAFLD/NASH. J Gastroenterol. 2018;53(3):362–376. doi: 10.1007/s00535-017-1415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konerman M.A., Jones J.C., Harrison S.A. Pharmacotherapy for NASH: current and emerging. J Hepatol. 2018;68(2):362–375. doi: 10.1016/j.jhep.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 35.Cobbina E., Akhlaghi F. Non-alcoholic fatty liver disease (NAFLD) - pathogenesis, classification, and effect on drug metabolizing enzymes and transporters. Drug Metab Rev. 2017;49(2):197–211. doi: 10.1080/03602532.2017.1293683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machado M.V., Diehl A.M. Pathogenesis of nonalcoholic steatohepatitis. Gastroenterology. 2016;150(8):1769–1777. doi: 10.1053/j.gastro.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brandl K., Schnabl B. Intestinal microbiota and nonalcoholic steatohepatitis. Curr Opin Gastroenterol. 2017:128–133. doi: 10.1097/mog.0000000000000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolodziejczyk A.A., Zheng D., Shibolet O., Elinav E. The role of the microbiome in NAFLD and NASH. EMBO Mol Med. 2019 doi: 10.15252/emmm.201809302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferslew B.C., Johnston C.K., Tsakalozou E., Bridges A.S., Paine M.F., Jia W., et al. Altered morphine glucuronide and bile acid disposition in patients with nonalcoholic steatohepatitis. Clin Pharmacol Ther. 2015;97(4):419–427. doi: 10.1002/cpt.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albillos A., de Gottardi A., Rescigno M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J Hepatol. 2020;72(3):558–577. doi: 10.1016/j.jhep.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Qin C.-Z., Ren X., Tan Z.-R., Chen Y., Yin J.-Y., Yu J., et al. A high-throughput inhibition screening of major human cytochrome P450 enzymes using an in vitro cocktail and liquid chromatography-tandem mass spectrometry. Biomed Chromatogr. 2014;28(2):197–203. doi: 10.1002/bmc.3003. [DOI] [PubMed] [Google Scholar]
- 42.Chen X.P., Tan Z.R., Huang S.L., Huang Z., Ou-Yang D.S., Zhou H.H. Isozyme-specific induction of low-dose aspirin on cytochrome P450 in healthy subjects. Clin Pharmacol Ther. 2003:264–271. doi: 10.1067/mcp.2003.14. [DOI] [PubMed] [Google Scholar]
- 43.Meyer U.A. Cytochrome P450 enzymes. Drug Metabol Drug Interact. 2012:1–2. doi: 10.1515/dmdi-2012-0002. [DOI] [PubMed] [Google Scholar]
- 44.Spaggiari D., Geiser L., Daali Y., Rudaz S. Phenotyping of CYP450 in human liver microsomes using the cocktail approach. Anal Bioanal Chem. 2014;406(20):4875–4887. doi: 10.1007/s00216-014-7915-4. [DOI] [PubMed] [Google Scholar]
- 45.de Faria Ghetti F., Oliveira D.G., de Oliveira J.M., de Castro Ferreira L.E.V.V., Cesar D.E., Moreira A.P.B. Influence of gut microbiota on the development and progression of nonalcoholic steatohepatitis. Eur J Nutr. 2018;57(3):861–876. doi: 10.1007/s00394-017-1524-x. [DOI] [PubMed] [Google Scholar]
- 46.Gómez-Lechón M.J., Jover R., Donato M.T. Cytochrome p450 and steatosis. Curr Drug Metab. 2009:692–699. doi: 10.2174/138920009789895543. [DOI] [PubMed] [Google Scholar]
- 47.Na A.-Y., Jo J.J., Kwon O.K., Shrestha R., Cho P.J., Kim K.M., et al. Investigation of nonalcoholic fatty liver disease-induced drug metabolism by comparative global toxicoproteomics. Toxicol Appl Pharmacol. 2018;352:28–37. doi: 10.1016/j.taap.2018.05.021. [DOI] [PubMed] [Google Scholar]
- 48.Li H., Clarke J.D., Dzierlenga A.L., Bear J., Goedken M.J., Cherrington N.J. In vivo cytochrome P450 activity alterations in diabetic nonalcoholic steatohepatitis mice. J Biochem Mol Toxicol. 2017;31(2):e21840. doi: 10.1002/jbt.2017.31.issue-210.1002/jbt.21840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye J.-Z., Li Y.-T., Wu W.-R., Shi D., Fang D.-Q., Yang L.-Y., et al. Dynamic alterations in the gut microbiota and metabolome during the development of methionine-choline-deficient diet-induced nonalcoholic steatohepatitis. World J Gastroenterol. 2018;24(23):2468–2481. doi: 10.3748/wjg.v24.i23.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider K., Mohs A., Kilic K., Candels L., Elfers C., Bennek E., et al. Intestinal microbiota protects against MCD diet-induced steatohepatitis. Int J Mol Sci. 2019;20(2):308. doi: 10.3390/ijms20020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Tong Q., Shou J.-W., Zhao Z.-X., Li X.-Y., Zhang X.-F., et al. Gut microbiota-mediated personalized treatment of hyperlipidemia using berberine. Theranostics. 2017;7(9):2443–2451. doi: 10.7150/thno.18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohamed M.-E., Feng T., Enejosa J.V., Fisniku O., Othman A.A. Effects of upadacitinib coadministration on the pharmacokinetics of sensitive Cytochrome P450 probe substrates: a study with the modified cooperstown 5+1 cocktail. J Clin Pharmacol. 2020;60(1):86–95. doi: 10.1002/jcph.v60.110.1002/jcph.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun J., Lu Y., Li Y., Pan J., Liu C., Gong Z., et al. Influence of Shenxiong glucose injection on the activities of Six CYP isozymes and metabolism of warfarin in rats assessed using probe cocktail and pharmacokinetic approaches. Molecules. 2017;22(11):1994. doi: 10.3390/molecules22111994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turpault S, Brian W, Van Horn R, Santoni A, Poitiers F, Donazzolo Y et al. Pharmacokinetic assessment of a five-probe cocktail for CYPs 1A2, 2C9, 2C19, 2D6 and 3A, Br J Clin Pharmacol 2009;928–35. doi:10.1111/j.1365-2125.2009.03548.x. [DOI] [PMC free article] [PubMed]
- 55.Cheng C., Qian J., Wang Z., Li W., Huang C., Chen M., et al. Influences of corydalis decumbens on the activities of CYP450 enzymes in rats with a cocktail approach. Biomed Res Int. 2019;2019:1–9. doi: 10.1155/2019/9614781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin Y., Wei Y., Hu X., Wu M., Ying X., Ding M. Influences of oldenlandia diffusa on the CYP450 activities in rats using a cocktail method by UHPLC-MS/MS. Biochem Res Int. 2018;2018:1–6. doi: 10.1155/2018/1467143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paolini M, Biagi GL, Bauer C, Cantelli-Forti G. Cocktail strategy: complications and limitations, J Clin Pharmacol 1993;1011–2. doi:10.1002/j.1552-4604.1993.tb01936.x. [DOI] [PubMed]
- 58.Frye R.F., Matzke G.R., Adedoyin A., Porter J.A., Branch R.A. Validation of the five-drug “Pittsburgh cocktail” approach for assessment of selective regulation of drug-metabolizing enzymes. Clin Pharmacol Ther. 1997;62(4):365–376. doi: 10.1016/S0009-9236(97)90114-4. [DOI] [PubMed] [Google Scholar]
- 59.Fan L, Zhao X, Tong Q, Zhou X, Chen J, Xiong W et al. Interactions of Dihydromyricetin, a Flavonoid from Vine Tea (Ampelopsis grossedentata) with Gut Microbiota, J Food Sci 2018;1444–1453. doi: 10.1111/1750-3841.14128. [DOI] [PubMed]
- 60.Lange ME, Uwiera RRE, Inglis GD. Housing gnotobiotic mice in conventional animal facilities. Curr Protoc Mouse Biol 2019;e59. doi: 10.1002/cpmo.59. [DOI] [PubMed]
- 61.Roberts M.S., Magnusson B.M., Burczynski F.J., Weiss M. Enterohepatic circulation: physiological, pharmacokinetic and clinical implications. Clin Pharmacokinet. 2002;41(10):751–790. doi: 10.2165/00003088-200241100-00005. [DOI] [PubMed] [Google Scholar]
- 62.Orme M.L'E., Back D.J. Factors affecting the enterohepatic circulation of oral contraceptive steroids. Am J Obstet Gynecol. 1990;163(6):2146–2152. doi: 10.1016/0002-9378(90)90555-L. [DOI] [PubMed] [Google Scholar]
- 63.Byrne CD, Targher G. NAFLD: a multisystem disease, J Hepatol 2015;S47-64. doi: 10.1016/j.jhep.2014.12.012. [DOI] [PubMed]