Abstract
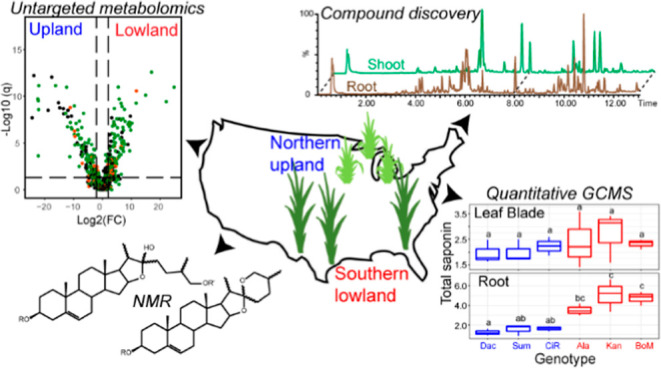
Switchgrass (Panicum virgatum L.) is a bioenergy crop that grows productively on lands not suitable for food production and is an excellent target for low-pesticide input biomass production. We hypothesize that resistance to insect pests and microbial pathogens is influenced by low-molecular-weight compounds known as specialized metabolites. We employed untargeted liquid chromatography–mass spectrometry, quantitative gas chromatography–mass spectrometry (GC–MS), and nuclear magnetic resonance spectroscopy to identify differences in switchgrass ecotype metabolomes. This analysis revealed striking differences between upland and lowland switchgrass metabolomes as well as distinct developmental profiles. Terpenoid- and polyphenol-derived specialized metabolites were identified, including steroidal saponins, di- and sesqui-terpenoids, and flavonoids. The saponins are particularly abundant in switchgrass extracts and have diverse aglycone cores and sugar moieties. We report seven structurally distinct steroidal saponin classes with unique steroidal cores and glycosylated at one or two positions. Quantitative GC–MS revealed differences in total saponin concentrations in the leaf blade, leaf sheath, stem, rhizome, and root (2.3 ± 0.10, 0.5 ± 0.01, 2.5 ± 0.5, 3.0 ± 0.7, and 0.3 ± 0.01 μg/mg of dw, respectively). The quantitative data also demonstrated that saponin concentrations are higher in roots of lowland (ranging from 3.0 to 6.6 μg/mg of dw) than in upland (from 0.9 to 1.9 μg/mg of dw) ecotype plants, suggesting ecotypic-specific biosynthesis and/or biological functions. These results enable future testing of these specialized metabolites on biotic and abiotic stress tolerance and can provide information on the development of low-input bioenergy crops.
Keywords: switchgrass, sustainability, biofuels, biomass, ecotype, metabolomics, NMR, specialized metabolite, diosgenin, saponin
Introduction
Development of environmentally sustainable and economical production of transportation fuels and industrial feedstocks using plant biomass is an important goal for the bioeconomy. Dedicated energy crops that are productive with low or no chemical fertilizers and pesticides on land that is unsuitable for food and fiber crops have received much attention.1 This requires development of plants with a suite of “ideal” traits,2 including perennial life cycle, rapid growth under conditions of low soil fertility and water content, as well as being resilient to pests and pathogens.
Plants are master biochemists, producing a wide variety of general and specialized metabolites adapted to their ecological niches.3 The structurally diverse tissue- and clade-specific specialized metabolites play varied roles in how plants cope with biotic and abiotic stresses, both by reducing deleterious impacts and promoting beneficial interactions. For instance, glucosinolates produced by crucifers such as mustard, cabbage, and horseradish mediate interactions with insect herbivores,4 and flavonoids that induce the rhizobial lipochitooligosaccharides (“nod factors”) initiate the rhizobium–legume nitrogen fixation symbiosis.5 The root-accumulating avenacin triterpene saponins are well documented to protect oat plants (Avena spp.) from the fungal pathogen-induced “take-all” disease,6−8 contributing to oat productivity. Modifying plant-specialized metabolism is an attractive target for bioengineering or trait breeding to create low-input bioenergy crops that can thrive on “marginal” lands unsuitable for food and fiber crops.
Although hundreds of thousands of specialized metabolites are estimated to be produced by plants,3 there are reasons why this number is almost certainly an underestimate. First, these metabolites are taxonomically restricted, often showing interspecies or even intraspecies variation;3 thus, any sampled species, ecotype, or cultivar will have a small subset of the overall plant kingdom’s phenotypic diversity. Second, specialized metabolites tend to be produced in a subset of cell or tissue types in any plant species analyzed; thus, cataloging the metabolic potential of even a single species requires extraction of multiple tissues over the plant’s development. Third, accumulation of these metabolites can be impacted by growth conditions and induced by abiotic or biotic stress.9 Finally, identification and structural characterization of newly discovered metabolites require specialized capabilities, typically a combination of mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy analysis.10
The North American native perennial switchgrass (Panicum virgatum L.) has the potential to be cultivated as a low-input bioenergy crop for growing on nonagricultural land.11 The two principal ecotypes of switchgrass are phenotypically distinct, with variation in flowering time, plant size, physiology, and disease resistance. Upland ecotypes exhibit robust freezing tolerance but produce relatively low biomass yield in part due to early flowering.12−15 Plants of lowland ecotype typically found in riparian areas produce large amounts of biomass and are more flooding- and heat-tolerant, pathogen-resistant, and nutrient-use-efficient than those of the upland ecotype.12,16,17 However, these lowland ecotypes do not perform well in northern areas, largely due to cold intolerance.
Although morphological and physiological properties associated with the adaptive divergence of upland and lowland switchgrass have been intensively studied, the specialized metabolite diversity and their ecotypic differences remain underexplored, partially due to the technical challenges mentioned above. Lee et al.18 detected large amounts of diosgenin-derived steroidal saponins from aerial tissues of four different switchgrass cultivars. Diosgenin is synthesized from cholesterol and cyclized and oxidized through several spontaneous steps and enzymatic reactions catalyzed by cytochrome P450 enzymes (CYP450s).19 It is the backbone of spirostanol-type steroidal saponins that are important defensive compounds with documented antimicrobial and antiherbivory activities.20−22 These natural compounds also have pharmaceutical value. Diosgenin has been used as a major precursor for synthesizing steroidal drugs including hormonal contraceptives and corticosteroid anti-inflammatory agents.20 Beyond steroidal saponins, quercetin-derived flavonoids23 and biotic/abiotic stress-elicited C10–C20 terpenes24,25 were also identified in switchgrass. A comprehensive metabolomics survey will be beneficial to understand the natural product diversity in this important bioenergy crop.
In this study, we developed and deployed approaches to compare metabolomes of three upland and three lowland switchgrass cultivars by untargeted liquid chromatography–MS (LC–MS), targeted gas chromatography (GC)–MS, and NMR. The two ecotypes were documented to have distinct metabolomes, especially in the rhizome and root. We identified seven structurally distinct classes of diosgenin-derived steroidal saponins and cataloged a variety of flavonoid glycosides and di- and sesqui-terpenoids. Steroidal saponins were notably abundant, accounting for more than 30% total ion counts (averaging 5 μg/mg dry tissue weight) in lowland roots from the reproductive developmental stage. Furthermore, ecotype- and/or tissue-type-specific accumulations were observed for individual saponin classes as well as total saponins. Our study provides a comprehensive analysis of the specialized metabolites produced by different switchgrass cultivars and sets the stage for developing dedicated bioenergy crops with varied plant and microbiome traits.
Results
Metabolome Comparisons between Tissue Types, Developmental Stages, and Genotypes
LC–MS was used to develop an overview comparison of the metabolomes of upland and lowland ecotypes in different tissue types and developmental stages. Tissue extracts in 80% methanol were prepared from a sample panel (Figure S1B) containing three upland (Dacotah, Summer, and Cave-in-Rock) and three lowland (Alamo, Kanlow, and BoMaster) cultivars grown from seeds in a controlled environment (Materials and Methods). Shoot, rhizome, and root tissues were analyzed from plants at three developmental stages—vegetative, the transition between vegetative and reproductive, and early reproductive (Figure S1A).26 In total, 4,668 distinct metabolite features were identified from the positive ionization mode data set; these were annotated as retention time/mass-to-charge ratio pairs and include multiple in-source fragments from single analytes. Downstream statistical analyses focused on the 2586 of 4,668 features whose maximum abundance among the biological samples was ≥500 (Table S1).
We employed two complementary approaches to annotate the features by the metabolite class. First, relative mass defect (RMD) filtering27 was used to guide assignments of all metabolite signals to putative chemical classes (Materials and Methods). As a result, 42 and 15% of the 2586 features in the data set were tentatively annotated as terpenoid glycosides and polyphenol-derived metabolites, respectively (Figure 1A and Table S1 columns E and F), with RMD scores distinguishing classes based on the fractional hydrogen content in each measured ion. We then searched the molecular ion mass-to-charge ratios (m/z) and associated fragment ion information in available online mass spectral databases (Materials and Methods) and found strong matches to 169 previously characterized metabolites (level-2 non-novel metabolite identifications,28Table S1 column KL–KT).
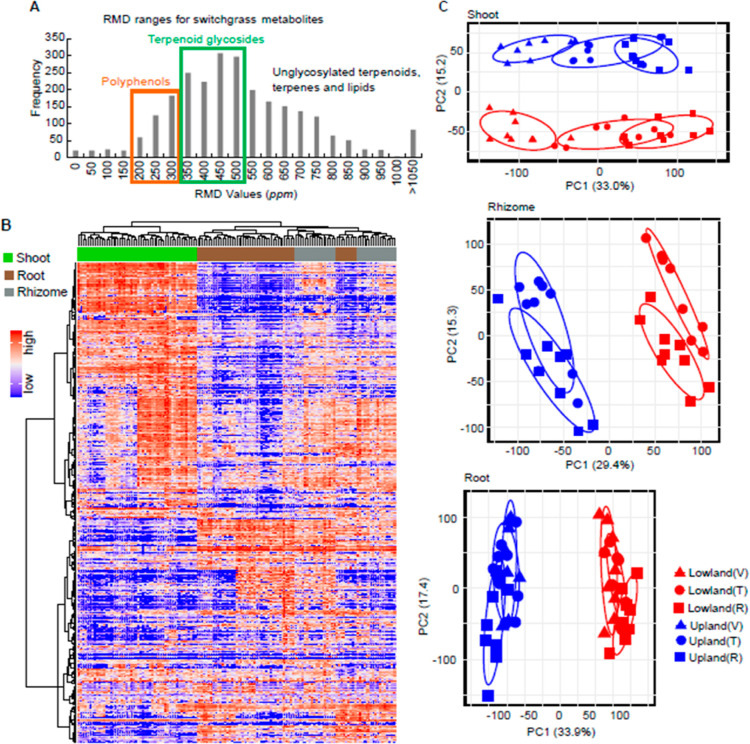
Figure 1.
Untargeted metabolome profiling for switchgrass. (A) Histogram of RMD values for the total 2586 features detected in this study by LC–MS in the positive ion mode. The green and orange rectangles highlight regions corresponding to the ranges of the RMD values anticipated for terpenoid glycosides and polyphenols, respectively. (B) Metabolome of the six switchgrass cultivars, three tissue types, and three developmental stages shown by a heatmap with HCA. The row and column clusters symbolize the 2586 features and 48 sample groups (containing 139 individual samples), respectively. The values representing the metabolite abundances that were used to make the heatmap were scaled to a range from 0 (the lowest abundance) to 1 (the highest abundance). (C) PCA score plots for the switchgrass shoot, rhizome, and root metabolite profiles (n = 8 for “upland vegetative” and “lowland reproductive”; n = 9 for all the other groups). The percentage of explained variation is shown on the x- and y-axes. V, vegetative phase; T, transition phase; and R, reproductive phase.
The untargeted metabolome data provide a comprehensive view of specialized metabolite variation among the samples, and broad patterns of variation were revealed using hierarchical clustering analysis (HCA, Figure 1B). The aerial (shoot) and subterranean tissue (root and rhizome) metabolites differed noticeably, consistent with the hypothesis that there are fundamental dissimilarities between the above- and below-ground tissue metabolomes. In contrast, the rhizome and root tissues were more similar to each other. Differences in metabolomes of each of the three tissue types across switchgrass cultivars (genotypes) were investigated using principal component analysis (PCA). The shoot metabolite profiles clustered into distinct groups in the PCA score plot, corresponding with the upland (blue) and lowland (red) switchgrass ecotypes (Figure 1C, top panel). Separation of the metabolite profiles was especially clear for different developmental stages (developmental stages are differentiated by the symbol shape and “V, T, and R”, standing for the “vegetative, transition, and reproductive phase”, respectively, in Figure 1C, top panel). The PCAs also showed clear-cut differences in metabolite profiles between the upland and lowland genotypes in both the rhizomes and roots (Figure 1C, middle and bottom panels, respectively). The developmental stage-associated variance in metabolite profiles of these two subterranean tissues was less apparent compared to that in the aerial tissue. Taken together, the metabolite PCAs revealed distinct patterns between upland and lowland switchgrass cultivars of the three tissues across the three developmental stages.
Upland and Lowland Ecotypes Have Strikingly Distinct Metabolomes
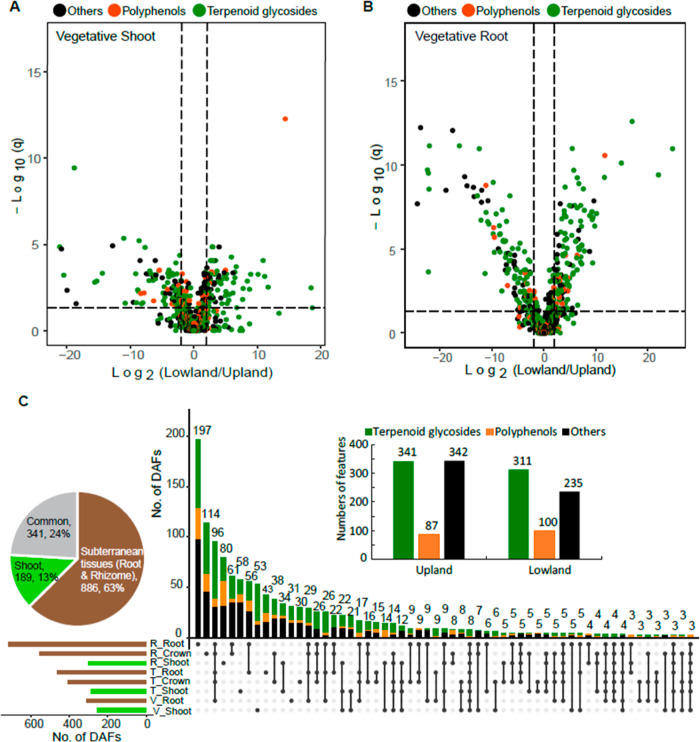
We next focused on each tissue type x developmental stage combination and identified the metabolite features that differentially accumulated in either upland or lowland ecotypes. Surprisingly, 25% (256 of 1035, Table 1) of the features detected in extracts of the vegetative-stage tillers predominantly accumulated in one or the other switchgrass ecotype. Such features were termed ecotype “differentially accumulated features” (DAFs). Specifically, there are 126 upland-enriched and 130 lowland-enriched DAFs in extracts of the vegetative-stage tillers (Figure 2A and Table 1). Analysis of vegetative-stage roots further revealed a total of 879 features with 35% (310, Table 1) meeting the ecotype DAF statistical threshold. Of these 310 DAFs, similar numbers of features were found to be either upland- (149) or lowland- (161) enriched (Figure 2B and Table 1). DAFs were also identified for the other developmental stage x tissue type combinations (Figure S2 and Table 1).
Table 1. Numbers of Total Detected Features and DAFs in Each of the Eight Developmental Stage x Tissue Type Sample Classes.
| no.
of DAFsb |
||||
|---|---|---|---|---|
| developmentalstage xtissue type | no. of total detected featuresa | upland-enriched | lowland-enriched | total |
| vegetative-stagex shoot | 1035 | 126 | 130 | 256 |
| vegetative-stagex root | 879 | 149 | 161 | 310 |
| transition-stagex shoot | 1037 | 155 | 133 | 288 |
| transition-stagex rhizome | 1114 | 268 | 139 | 407 |
| transition-stagex root | 965 | 226 | 235 | 461 |
| reproductive-stagex shoot | 1612 | 100 | 200 | 300 |
| reproductive-stagex rhizome | 1378 | 324 | 229 | 553 |
| reproductive-stagex root | 1327 | 408 | 309 | 717 |
Sum of the numbers in this column is larger than the numbers of the total detected features (2586) as some features were counted multiple times in several sample classes.
Sum of the numbers in the column of total is larger than the total detected unique DAFs (1416) as some DAFs were counted multiple times. The detailed information regarding these DAFs can be found in Tables S2–S9. Criteria for the DAFs: false discovery rate (FDR) adjusted p ≤ 0.05; fold changes ≥2.
Figure 2.
DAFs were identified between the upland and lowland ecotypes. Significance analysis (cutoff threshold: FDR adjusted p ≤ 0.05; fold changes ≥2) was performed to screen for the DAFs between the upland and lowland switchgrass ecotypes (n = 8 or 9) in various developmental stage x tissue type samples. Results of the analyses for (A) vegetative-stage shoots and (B) vegetative-stage roots are shown here using volcano plots. Putative terpenoid glycosides, polyphenols, and metabolites from the other categories are classified using RMD filtering and the results color-coded. (C) Upset plot of the 1416 unique (non-overlapping) ecotype DAFs across the 8 developmental stage x tissue type sample groups. The rows and columns in the Upset plot represent the sets (the eight sample groups in this case) and their intersections, respectively. For each set that is part of a given intersection, a black filled circle is placed in the corresponding matrix cell; otherwise, a light-gray circle is shown. The numbers of DAFs in the intersections are shown as a bar chart above the matrix. The horizontal bar chart to the left of the matrix shows the size of each set. The inserted bar plot shows that upland and lowland ecotypes accumulated similar numbers of the predominant DAFs likely terpenoid glycosides (green) and polyphenols (orange). The inserted pie chart summarizes percentages of the DAFs contributed by aerial (shoot) vs subterranean (root/rhizome) tissues. V, vegetative phase; T, transition phase; and R, reproductive phase.
Altogether, 1416 unique ecotype DAFs were identified for the eight tissue type x developmental stage combinations included in this study (Figure 2C and Tables S2–S9), accounting for approximately half of the features in the full data set. Based on RMD filtering, 46 and 13% of the DAFs were predicted to be terpenoid glycosides and polyphenol-derived metabolites, respectively (green and orange dots in Figures 2A-,B and S2, respectively). The numbers of the DAFs preferentially accumulated in upland and lowland ecotypes are equivalent to each other (Figure 2C inset: barplot). Furthermore, 63% of the DAFs were found in subterranean tissues, while only 13% were unique to the aerial tissues. The remaining 24% were detected in both above- and below-ground tissues (Figure 2C inset: pie chart). These results reveal that switchgrass subterranean tissues are the major sources of the observed specialized metabolic ecotypic diversity.
Detailed Analysis of Metabolite Diversity Using MS/MS
The observation that terpenoid and polyphenol metabolite classes are highly represented in DAFs led us to use high-resolution LC–MS/MS to characterize these features in more detail. In total, we tentatively identified 72 saponins, 10 diterpenoids, 4 sesquiterpenoids, and 7 flavonoid glycosides (confidence level 2,28Figure 3A and Tables S11–S14) from the 6 switchgrass cultivars. The identified saponins (Table S14) vary in their precursor masses and retention times (RTs) due to differences in both aglycones and sugar moieties. Assignments of multiple fragment ions generated by collision-induced dissociation resulting from the losses of sugar units allowed for annotation of the conjugated monosaccharides as well as the aglycones. For 44 (out of the 72) saponins, the presence of a signal at m/z 415 was diagnostic of the diosgenin aglycone.29 Fragmentations of 20 saponins yielded an aglycone fragment ion at m/z 431, while further loss of 18 Da (H2O) resulted in m/z 413; this suggests that this core has an additional oxygen atom compared to diosgenin. For three saponins, the presence of a signal at m/z 417 was indicative of a tigogenin aglycone.30 Finally, larger aglycone fragments at m/z 457 and 473 were observed for six and nine saponins, respectively. Elemental composition suggested that these 2 aglycones both have 29 carbons and differed by 1 oxygen, as predicted for nortriterpenoids. Moreover, the LC–MS spectra for about two-thirds of the 72 saponins displayed an abundant [M + H–H2O]+ ion, characteristic of furostanol saponins, which contain a C-22 labile hydroxyl group due to the hemiketal structure. The furostanol saponins are often glycosylated at their sidechains (also shown by our NMR results below, Figure 4B–H). Therefore, based on the type of aglycone and sidechain glycosylation, we grouped the 72 identified saponins into 7 classes: D-415-SCG, D-415, D-431-SCG, D-431, D-417-SCG, D-457, and D-473 (“D” indicates a diosgenin-derived aglycone; the numerical number reflects the m/z value of the aglycone fragment ion detected by positive-mode MS; “SCG” indicates “sidechain glycosylation”).
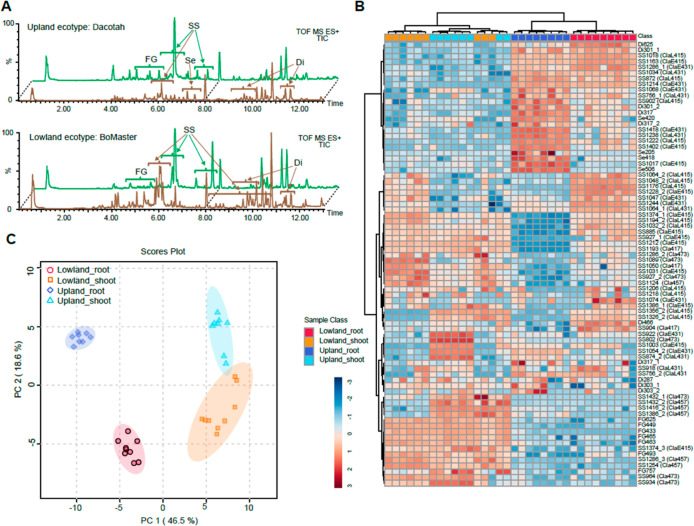
Figure 3.
Upland and lowland ecotypes are distinct in specialized metabolite profiles. (A) TICs for upland Dacotah and lowland BoMaster shoot (green) and root (brown) extracts. Areas where the specialized metabolites were identified are labeled by the short names. FG, flavonoid glycoside; ESS, early-eluting steroidal saponin; LSS, late-eluting steroidal saponin; TS, triterpenoid saponin; Di, diterpenoid; and Se, sesquiterpenoid. (B) Heatmap showing relative abundances of the specialized metabolites (vertical axis) across the biological samples (horizontal axis). Classifications of the saponins are shown in the parentheses. Relative metabolite abundances were log10-scaled to a range between −3 (lowest) and 3 (highest). Clustering method/distance: Ward/Euclidean. (C) PCA score plot showing distinct separations of the specialized metabolite profiles among the upland shoot, upland root, lowland shoot, and lowland root.
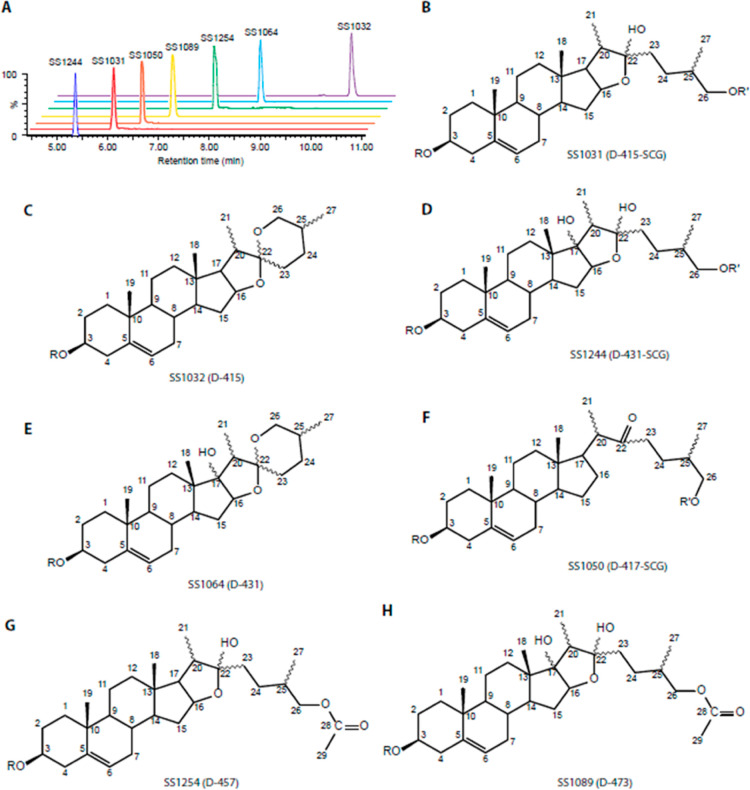
Figure 4.
Chemical structures of the saponins identified in switchgrass. (A) Seven purified saponins, representing the seven switchgrass saponin classes, have distinct RTs between 5 and 11 min. (B–H) Structures and numbering of the aglycones for the saponins SS1031, SS1032, SS1244, SS1064, SS1050, SS1254, and SS1089. Classifications of the saponins are shown in the parentheses. R and R′ indicate the position of the sugar moiety at C-3 and C-26 (on the side chain), respectively.
PCA (Figure 3C) using all the identified specialized metabolite features as the loadings (Figures 3B and S5) revealed that the ecotype plays an important role in the separation of the metabolite profiles even when multiple tissue types are included. Specifically, flavonoid glycosides predominately accumulated in shoots, while diterpenoids and sesquiterpenoids preferentially accumulated in roots (Figures 3B and S5A); together, these features separated the tissue types on the PCA (Figure 3C). Notably, all four sesquiterpenoids (Table S12) were exclusively detected in upland ecotype roots, >1000-fold higher than they were found in lowland roots (Table S10). In contrast, a diterpenoid glycoside, Di466 (Table S13), was abundant in lowland roots but nearly undetectable in upland root samples (also a >1000-fold accumulation difference, Table S10). Hence, such features also contribute to separation of the ecotypes on the PCA (Figure 3C).
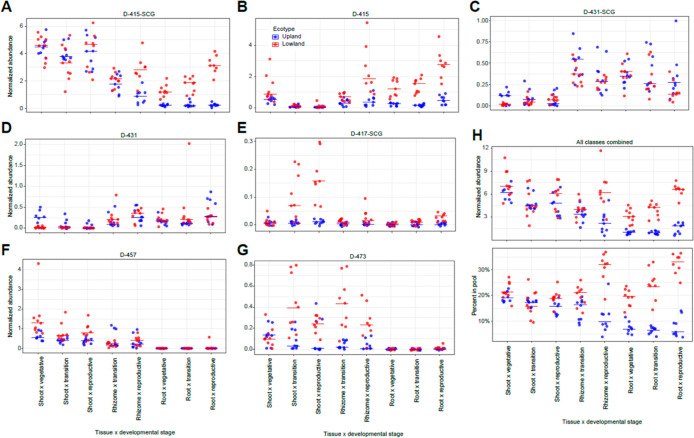
Accumulation patterns of the saponins (Figures 3B and S5B) were relatively complex compared with the C15 and C20 terpenes. Five general patterns were observed: (1) D-415 saponins preferentially accumulated in lowland ecotype roots; (2) D-431-SCG and D-431 saponins preferentially accumulated in root tissues, but accumulation was not ecotype-specific; (3) D-457 and D-473 saponins predominately accumulated in shoots with no apparent ecotype specificity; (4) D-417-SCG saponins preferentially were found in lowland shoots; and (5) D-415-SCG saponins showed neither strong tissue- nor ecotype-specific accumulation.
NMR Characterization of Saponins
To unequivocally determine the structures for the switchgrass saponins, we selected seven saponins that represent unique classes (Figure 4A) for high-performance (HP) LC purification: SS1031, SS1032, SS1244, SS1064, SS1050, SS1254, and SS1089 (“SS” abbreviation for steroidal saponins). Their molecular formulas were proposed based upon positive ionization mode UPLC-high-resolution MS/MS analyses (Figure S6–S12; see Materials and Methods for details). NMR spectra (1H, DEPTQ, HSQC, COSY, HMBC, and TOCSY) were generated for them (Tables S15–S21 and Figures S13–S61). The proton resonances determined from 1H, correlated spectroscopy (COSY), and total correlation spectroscopy (TOCSY) spectra in all samples fell into three distinct chemical shift regions: 0.8–2.6 ppm from aglycone backbone hydrogens; 3.2–4.2 ppm from sugar hydrogens; and sugar ring anomeric and aglycone olefinic hydrogens from 4.2–5.6 ppm. The heteronuclear single quantum coherence (HSQC), heteronuclear multiple bond correlation (HMBC), and distortionless enhancement by polarization transfer with retention of quaternary nuclei (DEPTQ) spectra were used to assign chemical shifts of the saponin core carbons, beginning with those downfield-shifted carbons due to direct connections with oxygens or double bonds. When combined with 1H–1H couplings established from COSY and TOCSY spectra, we could assign each aglycone position. Positions of sugar moiety substitutions were determined from HMBC spectra, based on 1H–13C correlations separated by two or three bonds. All seven saponins were glycosylated at the C-3 hydroxyl group, while three were also glycosylated at the C-26 position (side-chain glycosylation). We also observed an olefinic hydrogen from HSQC and 1H, indicating a double bond between the C-5 and C-6, for all seven saponins; thus, it rules out the possibility that SS1050 contained a tigogenin core. Collectively, these data identified SS1031 (Figure 4B) as the previously characterized switchgrass saponin, protodioscin, with sidechain glycosylation18 and SS1032 (Figure 4C) as a steroidal saponin derived from a diosgenin with the characteristic core spiroketal moiety but no sidechain glycosylation. These two saponins are likely biosynthetically related as the final six-member heterocyclic ring closure forming the spiroketal structure in diosgenin is spontaneous in planta.19 Similarly, SS1244 (Figure 4D) and SS1064 (Figure 4E) are related saponins that are derived from diosgenin: the former with side chain glycosylation and the latter without. Both share an additional tertiary hydroxyl group on C-17. The relatively low-abundance SS1050 (Figure 4F) has a cholesterol-like C27 aglycone with only four rings, side chain glycosylation, and a C-22 ketone group (characterized by a chemical shift of 215.6 ppm for the carbonyl carbon).
A unique feature of the diosgenin-derived saponins SS1254 (Figure 4G) and SS1089 (Figure 4H) is that they appear to be acetylated—rather than glycosylated—at C-26. This is consistent with the observed C29 aglycone mass of 456 Da (42 Da larger than that of diosgenin, which is 414 Da) and 472 Da (42 Da larger than the 430 Da of oxydiosgenin), respectively. This structural feature was revealed by HMBC data where the C-28 carbonyl carbon at 171.0 ppm is only correlated with the C-26 methylene and C-29 methyl protons. SS1089 has an additional tertiary hydroxyl group on the C-17 position compared to SS1254.
Differential Saponin Accumulation between Upland and Lowland Revealed by LC– and GC–MS Analysis
Documenting the differential accumulations of the different saponin classes is expected to help with identification of their biosynthetic enzymes and regulatory genes. As an approach to quantifying differential accumulation of the saponins that is orthogonal to PCA loading plots (Figure S5B) and HCA (Figure 3B), we compared cumulative ion counts for each saponin class between the ecotypes across different tissue x developmental stage sample groups. The D-415-SCG and D-415 were the dominant saponin forms in terms of their accumulative ion counts and were approximately 1 order of magnitude higher than the other classes. The lowland ecotypes accumulated much higher levels of these saponins than did the upland in root tissues, and this was especially notable during the reproductive stage (Figure 5A,B). The two C-17 hydroxylated saponin classes, D-431-SCG and D-431, had higher accumulations in below-ground tissues, without apparent accumulation differences between the two ecotypes (Figure 5C,D). In contrast, the two acetylated sidechain saponin forms (D-457 and D-473) were found to have much higher accumulations in above-ground tissue than in below-ground tissues (Figure 5F,G). The only saponin form not oxidized at the C-16 position (D-417-SCG, Figure 5E) showed a tendency toward higher accumulations in lowland shoots at later developmental stages. For the total (summed) saponins, lowland roots accumulated more than upland roots at all three developmental stages. In contrast, shoots of the two ecotypes accumulated comparable amounts of total saponins across development stages (Figure 5H upper panel). The saponins represented about 20 to 35% versus 5 to 10% of the total ion counts in lowland and upland roots, respectively (Figure 5H lower panel), indicating the contribution of these specialized metabolites in defining distinct phenotypes between ecotypes.
Figure 5.
Relative quantification of individual saponin classes and total saponins. (A–G) Accumulative ion counts for each individual saponin classes and (H, lower panel) the total saponins (all classes combined) from the eight tissue type x developmental stage sample classes measured by positive-mode LC–MS. (H, lower panel) Percentages of the saponins in the total ion pools. Normalized abundances (y-axis) are calculated as (ion intensity of the feature/ion intensity of the internal standard). For all panels, the horizontal bars represent median values (n = 8 or 9). Red, lowland ecotype; blue, upland ecotype.
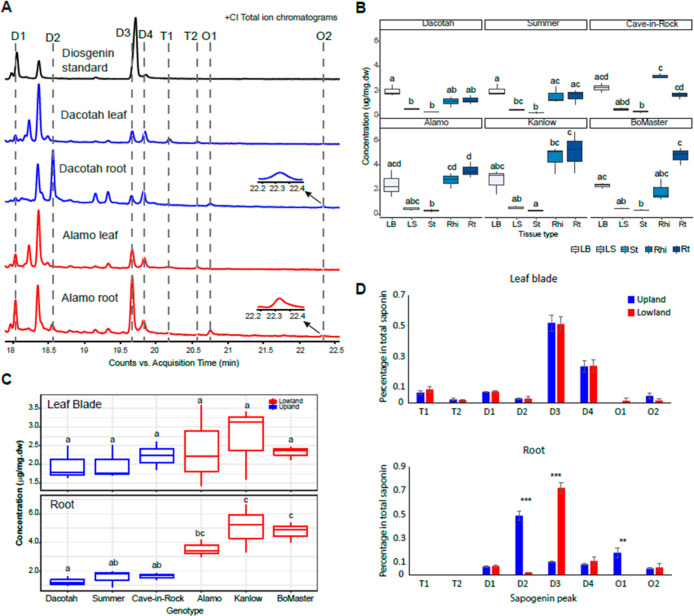
Although this LC–MS approach is excellent for documenting the diversity of saponin types, it is not ideal for quantification because of the uneven ionization efficiencies of the early- versus late-eluting analytes (caused by the changing ratio of the water and organic mobile phase during chromatography). To more accurately determine total saponin concentrations in different tissues and cultivars, a GC–MS-based quantification method was developed to quantify the sapogenins after hydrolytic removal of sugars through the comparison with an authentic diosgenin standard (Materials and Methods). As a result, we identified six diosgenin-derived steroidal sapogenin peaks and two peaks annotated as triterpenes (Figure 6A and Table 2). The triterpene peaks were only present in leaf blade samples and might arise from acetylated steroids if the hydrolytic removal of acetates was incomplete.
Figure 6.
Sapogenin aglycone peaks identified in switchgrass extracts are quantified by GC–MS. (A) +(CI) GC–MS TICs of the diosgenin standard (black), Dacotah leaf and root (blue), and Alamo leaf root (red). The identified sapogenin aglycone peaks in the switchgrass and standard samples are indicated and aligned by the dashed lines. Zoomed-in views for the peak O2 in the Dacotah root and Alamo root are indicated by the arrows. (B) Comparison of the total sapogenin concentrations among the five tissue types for each switchgrass cultivar (Kruskal–Wallis test: p = 0.012, 0.026, 0.009, 0.024, 0.017, and 0.011 for Dacotach, Summer, Cave-in-Rock, Alamo, Kanlow, and BoMaster, respectively). LB, leaf blade; LS, leaf sheath, St, stem; Rhi, rhizome; and Rt, root. (C) Comparison of the total sapogenin among the six switchgrass cultivars in the leaf blade (Kruskal–Wallis test: p = 0.766) and root (Kruskal–Wallis test, p = 0.016). Different lower-case letters on top of the boxes designate statistically different means (post-hoc test: Dunn’s test). (D) Ratio of the individual sapogenins in the leaf blade (upper) and root (lower) of upland and lowland ecotypes. Heights of the bars reflect the means of the nine replicates (three cultivars × three replicates) for each ecotype; error bars show the standard error of the mean; ** standards for 0.001 ≤ p ≤ 0.01 and *** standards for p < 0.001 (one-tailed t-test).
Table 2. TMS-Derivatized Sapogenin Aglycones Identified in Switchgrass Extracts by CI GC–MS Analysisa.
| peak | retention index (RI)b | key ion annotations (m/z)c | abundant fragment ion m/zd | formula of the neutral molecule | mass of the neutral molecule (Da) | annotation of the neutral moleculee |
|---|---|---|---|---|---|---|
| D1 | 3114 | 397 [M + H]+ | 139, 282 | C27H40O2 | 396 | anhydro-diosgenin |
| D2 | 3194 | 397 [M + H–TMS–OH]+, 471 [M–CH3]+, 485 [M – H]+, 487 [M + H]+ | 139, 282 | C30H50O3Si | 486 | diosgenin (TMS-derivatized) |
| D3 | 3339 | 397 [M + H–TMS–OH]+, 471 [M–CH3]+, 485 [M – H]+, 487 [M + H]+ | 139, 187, 282 | C30H50O3Si | 486 | diosgenin (TMS-derivatized) |
| D4 | 3361 | 397 [M + H–TMS–OH]+, 471 [M–CH3]+, 485 [M – H]+, 487 [M + H]+ | 139, 187, 282 | C30H50O3Si | 486 | diosgenin (TMS-derivatized) |
| T1 | 3399 | 409 [M + H–TMS–OH]+, 483 [M–CH3]+, 497 [M – H]+ | 218, 203, 190 | C33H58OSi | 498 | β-amyrin (TMS-derivatized) |
| T2 | 3439 | 409 [M + H–TMS–OH]+, 483 [M–CH3]+, 497 [M – H]+ | 218, 203, 190 | C33H58OSi | 498 | β-amyrin (TMS-derivatized) |
| O1 | 3462 | 395 [M + H–TMS–OH–H2O]+, 413[M + H–TMS–H2O]+, 487 [M–CH3]+, 501 [M – H]+, 503 [M + H]+ | 139, 187, 298 | C30H50O4Si | 502 | oxydiosgenin (TMS-derivatized) |
| O2 | 3620 | 395 [M + H–TMS–2H2O]+, 413[M + H–TMS–OH]+, 487 [M–CH3]+, 501 [M – H]+, 503 [M + H]+ | 139, 187, 298 | C30H50O4Si | 502 | oxydiosgenin (TMS-derivatized) |
The TMS-derivatized molecules are considered as the molecular ions in each case here. TMS, trimethylsilyl group [Si(CH3)3].
The RIs were calculated using a homologous series of n-alkane standards with the same GC–MS method as employed for the samples (Figure S4A).
The m/z information for the key ion annotations was obtained by (CI) GC–MS (Figure S62).
The m/z information for the fragment ions was obtained by electron ionization (EI) GC–MS (Figure S4B).
The annotations were made based on the pseudomolecular ions and fragment ion information from (EI) MS (Figure S4B) by comparing with the diosgenin standard and/or searching against the National Institute of Standards and Technology (NIST) NIST17 GC–MS library (www.chemdata.nist.gov) for the best matching compounds.
The total saponin (all eight peaks summed) concentrations were first compared across tissue types for each switchgrass cultivar (Figure 6B and Table S22). For every cultivar, saponins were detectable in all five analyzed tissue types and were higher in the leaf blade, rhizome, and root than they were in the leaf sheath and shaved stem. The two upland cultivars Dacotah and Summer have the highest saponin concentration in leaf blades, whereas all three lowland cultivars accumulate the highest saponin level in roots. The third upland cultivar, Cave-in-Rock, however, accumulates the highest total saponin in rhizomes. We then compared the total saponins in leaf blades and roots across the six cultivars. The total saponin concentrations in lowland roots are uniformly higher than those in roots of the three upland cultivars, with Kanlow and BoMaster showing statistical significance (p < 0.05, Kruskal–Wallis test). In comparison, leaf blade total saponins revealed no ecotype-related statistical difference (Figure 6C). Moreover, different sapogenin compositions were also found between upland and lowland roots but not leaf blades (Figure 6D). This was due to the differentiated accumulations of two diosgenin isomers, D2 and D3, and one oxydiosgenin isomer, O1, identified only in roots. Taken together, quantitative analysis of sugar-free sapogenins supports the results of LC–MS analysis showing strong genetic difference in root saponin accumulation. Considering the high abundances of the saponins, water solubilities of these molecules, and the documented bioactivities to the microbes,31 the differential accumulation in roots might play a role in shaping the ecotype-specific rhizosphere microbiomes in switchgrass.
Discussion
Plants produce a plethora of structurally diverse specialized metabolites that serve roles ranging from attracting beneficial organisms to combating deleterious biotic and abiotic agents. However, domestication and improvement of crops typically lead to reduced amounts and types of these advantageous metabolites. While restoring these beneficial traits to existing crops is generally not feasible, development of new food, fuel, and fiber crops can be done in such a way to maintain or enhance existing metabolic variation, potentially limiting the need for toxic pesticides.
Switchgrass is a compelling example of a low-fertilizer and low-pesticide cellulosic bioenergy crop in the USA.11 Switchgrass populations (ecotypes) are found from the northern Midwest to Texas and along the East Coast; unlike the relatively narrow genetic and phenotypic variation of traditional crops, there is abundant genetic variation across these populations. Striking differences in numerous plant traits were documented between populations of the upland and lowland ecotypes,32 including biomass production,33 rust pathogen (Uromyces graminicola) resistance,34 and tolerance to low-nitrogen, drought, and freezing conditions.14,35,36 These documented phenotypic and physiological differences encouraged us to compare the upland and lowland switchgrass metabolomes and catalog the ecotype-specific specialized metabolites. In addition to being of interest for pathway discovery and as tools to understand the genetic architecture of switchgrass, such information should be valuable for breeding switchgrass varieties that are highly productive with no or low pesticide inputs.
Analysis of six switchgrass accessions representing both upland and lowland ecotypes of eight specific tissue type x developmental stage sample classes identified a remarkable amount of metabolite variation. In fact, 1416 ecotype DAFs (Tables 1 and S2–S9) account for half of the metabolite features detected in this study. Many of these differences were quite large: there were 157 DAFs showing >1000-fold accumulation difference between the two ecotypes in at least 1 of the 8 sample classes (Table S10). In contrast, published metabolomics analyses for maize showed that metabolite profiles of the six genetically defined genome-wide association study populations failed to separate in PCA, even when the metabolite data were independently analyzed within the same tissue type.37
Based on RMD filtering,27 46 and 13% of the switchgrass ecotype-specific DAFs are proposed to be terpenoid glycosides and polyphenol-derived metabolites, respectively. Relatively few switchgrass terpenoid- and polyphenol-specialized metabolites were identified in the past, including leaf steroidal saponins,18,38 diterpenoid-derived antimicrobial phytoalexins,39 and quercetin-based flavonoids.23 Our identification of more than 1000 unannotated DAFs suggests the possibility of many more switchgrass metabolites to be characterized and underlines the complexity of analysis of complex mixtures of structurally diverse small molecules. By generating MS/MS spectra beginning with the most abundant metabolites, we tentatively identified close to 100 specialized metabolites. In contrast to the flavonoid glycosides (Table S11), the diterpenoids (Table S12), sesquiterpenoids (Table S13), and most of the steroidal saponins (Table S14) did not match known compounds in MS/MS databases (Materials and Methods): this led us to subject representative saponins to NMR analysis.
Saponins stand out in this study as highly abundant and differentially accumulating metabolites, exhibiting diversity in their cores and glycosylation positions and types. This structural diversity indicates tissue- and/or genotype-specific activities of as yet uncharacterized CYP450s, UDP-glycosyltransferases, and other tailoring enzymes (e.g., acyltransferases). This conclusion is supported by the seven distinct sapogenin aglycone structures elucidated based upon LC–MS and NMR data and their accumulation patterns in switchgrass. These include diosgenin cores with the characteristic 5,6-spiroketal moiety (Figure 4C) and a diosgenin core that presumably is derived from a metabolic intermediate,19 in which the sidechain is stabilized by glycosylation and thus prevented from the spontaneous cyclization to form the final pyranosidic ring (Figure 4B); this metabolite was previously identified in switchgrass aerial tissues.18 Besides glycosylation, the sidechain also can be stabilized by acetylation resulting in the C29 aglycones (Figure 4G,H). Predominant accumulation of the sidechain-acetylated saponins in shoots implies involvement of one or more tissue-specific acyltransferase activities in switchgrass saponin biosynthesis. Likewise, diosgenin C-17 hydroxylation (Figure 4D,E,H) was found for the saponins preferentially accumulated in root tissues, suggesting a root-specific CYP450 activity. We also characterized a saponin core that is not oxidized at the C-16 position (Figure 4F), which might derive from a precursor or side product of the diosgenin biosynthetic pathway.19 Saponins with this core were seen across the cultivars analyzed but were more abundant in lowland shoots especially at a later developmental stage. This indicates that the switchgrass CYP450 responsible for the cholesterol C-16 oxidation is possibly regulated in an ecotype x tissue x development manner in switchgrass.
The switchgrass saponins we characterized were either singly glycosylated at C-3 (Figure 4C,E,G,H) or glycosylated at both C-3 and on the C-26 sidechain (Figure 4B,D,F). Glycosylation diversity also comes from variation in the conjugating saccharide types, with one to six monosaccharides observed (Table S14). MS neutral mass losses corresponding to anhydrous glucose/galactose, rhamnose, and xylose were observed for these saponins, and some monosaccharides were also acetylated (Table S14). Published studies indicated that the conjugated sugar moieties impact saponin bioactivities; for example, the oat avenacoside steroidal saponins are activated by deglycosylation upon leaf damage or pathogen attack,40,41 while antimicrobial activities of other saponins seem to rely on glycosylation.31 The purified switchgrass saponins with distinct glycosylation patterns provide good opportunities to perform structure–function analyses using in vitro microbial bioassays.
The total saponin levels in roots of three lowland switchgrass cultivars, Alamo, Kanlow, and BoMaster, are 3.5, 5.1, and 4.7 μg/mg dw, respectively, which are higher than those of the three upland cultivars, Dacotah, Summer, and Cave-in-Rock, at 1.2, 1.5, and 1.6 μg/mg dw, respectively (Figure 6C lower panel and Table S22). The root saponin contents in the lowland cultivars are close to those observed in legume roots: for example, the Medicago truncatula(42) and two Medicago sativa cultivars, Radius43 and Kleszczewska,44 contain 5.9, 5.0, and 9.3 μg/mg dw saponins in their roots, respectively.
Information about accession and ecotypic differences in the saponin content could provide tools for improvement of switchgrass biomass. For example, avenacins have well documented protective roles protecting oat roots from the fungal “take-all” disease.6−8 Saponin root differential accumulation among switchgrass ecotypes suggests that they might be attractive targets for breeding cultivars with an increased ability to improve yield by modulating the microbiome structure and function.45−47 In contrast, breeding for low leaf saponins might produce varieties with biomass that is efficiently converted into fuel by avoiding accumulation of toxins that interfere with growth of processing microbes. Taken together, our results provide opportunities to identify targets for producing switchgrass varieties with improved plant/microbiome traits and increased biomass yield or biofuel conversion at lower economic and environmental costs.
Materials and Methods
Plant Material
The six switchgrass cultivars used in this study were the upland ecotypes Dacotah, Summer, and Cave-in-Rock and lowland ecotypes Alamo, Kanlow, and BoMaster. The seeds were ordered from Native Connections (http://nativeconnections.net, Three Rivers, MI). The plants were grown under controlled growth conditions: temperature was set at 27 °C with 16 h light (500 μE m–2s–1) per day and relative humidity set to 53%. Seeds were sown directly in a 1:1 mixture of sand and vermiculite, watered twice a week with deionized water, and fertilized once every 2 weeks using half-strength Hoagland’s solution.48
For untargeted LC–MS analysis, plant tissues were harvested at 1, 2, and 3 months after imbibition, corresponding to the vegetative, transition, and early reproductive developmental stages, respectively.26 Roots, rhizomes, and shoots (Figure S1A) were collected separately for all the switchgrass cultivars. For example, one sample represented a specific cultivar (genotype) x developmental stage x tissue type combination (Figure S1B). There were three biological replicates from three independent plants with two exceptions: two samples each from two independent plants for vegetative-phase Cave-in-Rock samples and early reproductive-phase Alamo samples (due to sample loss). For the GC–MS quantification of sapogenins, samples were only collected from the 3 month old (early reproductive phase) plants. All samples were immediately frozen in liquid nitrogen and stored at −80 °C until extraction.
Metabolite Extraction
All chemicals were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. The samples were frozen in liquid nitrogen and powdered using 15mL polycarbonate grind vial sets (OPS Diagnostics, Lebanon, NJ) on a MiniG high-throughput homogenizer (SPEX SamplePrep, Metuchen, NJ). 500 mg of each sample was extracted at 4 °C overnight (14–16 h) in 5 mL of 80% methanol containing the 1 μM telmisartan internal standard. Extracts were centrifuged at 4000 g for 20 min at room temperature to remove solids. The supernatant from each sample was transferred to an HPLC vial and stored at −80 °C prior to LC–MS analysis. For GC–MS, 50 mg of the lyophilized sample was extracted in 1 mL of 80% methanol following the workflow described for LC–MS sample preparation above, as described by Tzin et al.49
UPLC–ESI-QToF-MS Analysis
Reversed-phase UPLC–positive mode electrospray ionization-quadrupole time-of-flight MS (UPLC–(+)ESI-QToF-MS) analyses were performed with a Waters ACQUITY UPLC system coupled to a Waters Xevo G2-XS QToF mass spectrometer (Waters, Milford, MA). The chromatographic separations were performed using a reversed-phase, UPLC BEH C18, 2.1 mm × 150 mm, 1.7 μm column (Waters) with a flow rate of 0.4 mL/min. The mobile phase consisted of solvent A (10 mM ammonium formate/water) and solvent B (100% acetonitrile). The column oven was maintained at 40 °C. Separations were achieved utilizing a 20 min method, injecting 10 μL of the extract and using the following method (%A/%B): 0–1.0 min hold (99/1), linear gradient to 15 min (1/99), hold (1/99) until 18 min, returning at 18.01 min (99/1), and holding until 20 min. The Xevo G2-XS QToF mass spectrometer was operated using the following static instrument parameters: desolvation temperature of 350 °C; desolvation gas flow rate at 600 L/h; capillary voltage of 3.0 kV; and cone voltage of 30 V. Mass spectra were acquired in the continuum mode over m/z 50 to 1500 using data-independent acquisition (DIA, MSE) or data-dependent MS/MS acquisition (DDA), with collision potential scanned between 20 and 80 V for the higher-energy function for DIA (and 20–60 V for DDA). The DDA mode automatically selected the three most abundant molecular ions to pass through the mass filter for fragmentation analysis at each scan. The MS system was calibrated using sodium formate, and leucine enkephalin was used as the lock mass compound, but automated mass correction was not applied during DIA data acquisition. QC and reference samples were analyzed every 20 injections to evaluate the stability of the LC–MS system.
Data Processing and Metabolite Mining for the Untargeted Metabolomic Analysis
Acquired raw MS data were processed using the Progenesis QI software package (v.3.0, Waters, Milford, MA) using RT alignment, lock mass correction, peak detection, adduct grouping, and deconvolution. The identified compounds were defined by the RT and m/z information, and we also refer to these as features. The parameters used with Progenesis processing were as follows: sensitivity for peak picking, default; minimum chromatographic peak width, 0.15 min; and RT range, 0.3–15.5 min. Intensities (ion abundances) of all the detected features were normalized to the internal standard, telmisartan, before downstream statistical analyses. Online databases—including KEGG,50 MassBank,51 PubChem,52 and MetaboLights53—were used to provide annotations to the features based on a 10 ppm precursor tolerance, 95% isotope similarity, and 10 ppm theoretical fragmentation pattern matching with fragment tolerance.
The complementary method RMD filtering27 was used to assign chemical classes for the features. Briefly, an RMD value of each feature was calculated in ppm as (mass defect/measured monoisotopic mass) × 106. This value reflects the fractional hydrogen content of a feature and provides an estimate of the relative reduced states of carbons in the metabolite precursor of that feature. For example, in this study, we defined terpenoid glycosides (RMD of 350–550 ppm) or phenolics (RMD of 200–350 ppm) using this method. Features with RMD >1200 ppm are likely contaminants (e.g., inorganic salts) in the MS system.
The DDA was carried out for a set of pooled samples to generate positive-mode MS/MS spectra for the abundant ions. Specialized metabolite discovery was performed by mining the DDA data and beginning with the most abundant metabolites. Characteristic precursor/fragment ions and RMD were used to assign metabolites to a particular chemical class (e.g., flavonoid glycoside, diterpenoid, sesquiterpenoid, and saponin).
Saponin Purification
The switchgrass (Kanlow) plants were grown in a growth chamber using the conditions described in the Plant Material section. About 150–200 g of fresh root or shoot tissues from fully matured plants (3 months post-germination) was harvested. The tissues were ground into powders with liquid nitrogen and placed in a 2 L beaker. 1.5–2 L of 80% methanol (in water) was added, and the mixture was incubated for 48 h at 4 °C. The mixture was centrifuged at 4000g for 15 min to remove insoluble debris. The supernatant volume was reduced under vacuum using a rotary evaporator, followed by evaporation to dryness using a SpeedVac vacuum concentrator.
The residue was redissolved in 100 mL of water. The liquid–liquid phase partitioning was carried out against first hexane and then ethyl acetate, with a 1:1 ratio, to remove the non-polar interfering metabolites. The resultant water phase was then loaded onto a 35 cc C18 SPE cartridge (Waters). The cartridge was washed three times each using 0, 10, 20, and 50% methanol (in water) to remove the polar interfering metabolites. The cartridge was eluted three times each using 70, 80, and 90% methanol (in water) to obtain the saponin-enriched fractions (eluates). The solvent was evaporated to dryness under vacuum using SpeedVac, and the residue was redissolved in 8 mL of 80% methanol (in water). The insoluble residue was removed by centrifugation at 4000g for 5 min at 25 °C. Supernatants were transferred to autosampler vials.
Purification was carried out using a Waters 2795 pump/autosampler connected with an LKB Superrac 2211 fraction collector and a Waters Symmetry C18 HPLC column (100 Å, 5 μm, 4.6 mm × 150 mm). The mobile phase consisted of 0.15% formic acid in water, pH 2.8 (solvent A) and acetonitrile (solvent B). The linear gradient elution used to purify saponins SS1244, SS1031, SS1064, and SS1032 from root tissues was 1% B at 0 min, 30% B at 1.01 min and linearly increased to 40% B at 7 min, 50% B at 7.01 min and linearly increased to 70% B at 15 min, and 99% B at 15.01 min and held at 99% B between 15.01 and 18 min. A slightly modified linear gradient elution was used to purify saponins SS1050, SS1098, and SS1254 from shoot tissues, 1% B at 0 min, 30% B at 1.01 min and linear-increased to 40% B at 8 min, 50% B at 8.01 min and linear-increased to 60% B at 15 min, and 99% B at 15.01 min and held at 99% B between 15.01 and 18 min. The solvent flow rate was 1.5 mL/min, and the column temperature was 40 °C. The eluate was collected every 10 s as one fraction for each injection, using an injection volume of 100 μL. The HPLC fractions containing the seven targeted saponins were estimated to be >75% pure based on LC–MS analysis results. Fractions of adequate purity for the same saponins were pooled.
NMR Spectroscopy
NMR spectra of purified saponin samples were acquired using a Bruker Ascend 600 MHz spectrometer (Bruker Biospin, Germany) operating at 600.13 MHz for proton and equipped with an inverse 1.7 mm TCI micro-cryoprobe and a SampleJet auto-sampler unit. For 1H NMR spectra, solvent suppression with a shaped pulse program (Wetdc) was used with a scan number of 16 at temperature 298 K with pulse width 10.75 μs and power 0.2 W. Acquisition time for each scan was 0.681 s with a delay time of 3 s for a spectral width of 20 ppm. For locking the magnetic field, CD3OD was used as the solvent, and 1H spectra were calibrated using the residual solvent peaks. Baseline and phase correction was performed manually using Bruker TopSpin 3.5.6 software. Subsequently, 2D NMR spectra were collected using the default Bruker pulse program cosygpmfppqf for COSY (ns = 16, sw = 13 for F1 and F2), hsqcedetgpsp.3 for HSQC (ns = 64, sw = 13 for F2 and 220 for F1), hmbcgpndqf for HMBC (ns = 128, sw = 13 for F1 and F2), and mlevphpr.2 for TOCSY (ns = 64, sw = 13 for F1 and F2) with pulse width 10.75 μs and power 0.2 W for 1H and pulse width 12 μs and power 68 W for 13 C. DEPT-Q spectra were collected using a Bruker Avance III 800 MHz spectrometer equipped with a 5 mm TCI cryoprobe. Data were obtained using the deptsp135 pulse program with 3072 scans for an sw 222 ppm and 0.734 s acquisition time for each scan with pulse width 14 μs and power 107.15 W for 13 C. Finally, the data were visualized using both Topspin 3.5.6 and MestReNova software for peak assignments.
Analysis of Switchgrass Sapogenins by Acid Hydrolysis, Derivatization, and GC–MS
To analyze sapogenins, acid hydrolysis was carried out according to a previously published protocol.54 In brief, 300 μL of the switchgrass extract, 200 μL of distilled water, and 100 μL of 12 M hydrochloric acid (MilliporeSigma, Burlington, MA) were mixed in a polypropylene microcentrifuge tube and incubated at 85 °C for 2 h. The samples were cooled and evaporated to dryness under vacuum with the temperature ≤40 °C. The resultant pellet was dissolved in 500 μL of distilled water and extracted with 500 μL of ethyl acetate for phase partition. After this, 300 μL of the ethyl acetate layer was transferred to a new microcentrifuge tube and evaporated to dryness under vacuum at room temperature. The dry residue was dissolved in 100 μL of N-methyl-N-(trimethylsilyl)trifluoroacetamide, derivatized overnight at 60 °C and analyzed using a 30 m VF5 column (Agilent Technologies, Santa Clara, CA; 0.25 mm ID, 0.25 μm film thickness) coupled to an Agilent 5975 single quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA) operated using 70 eV electron ionization (EI). Then, the same set of samples was analyzed on an Agilent 7010B triple quadrupole mass spectrometer using the same column for chemical ionization (CI). The MS scanning range was m/z 80–800. Splitless sample injection was used, with helium as carrier gas at a constant flow of 1 mL/min and the inlet and transfer line held at 280 °C. The GC temperature program was as follows: held at 50 °C for 1 min; ramped at 30 °C/min to 200 °C; and ramped at 10 °C/min to 320 °C and held for 10 min.
Absolute and relative quantification of the sapogenins was performed using commercially available diosgenin (∼95%, Sigma-Aldrich) as an external standard. Serially diluted standards (6–192 μg/mL dissolved in 80% ethanol) were pre-treated in the same way as the other samples; after hydrochloric acid hydrolysis, four target peaks were identified that are derived from the commercial diosgenin. They were termed diosgenin standard (DS) 1–4. DS4, eluting at 21.3 min, could be detected by EI GC–MS at high standard concentrations (Figure S3A). All target peaks were combined when plotted against the standard’s concentrations to generate a six-point response curve. Duplicate technical replicate analyses were done for each standard sample used to generate the standard response curve, which was linear (r2 > 0.97, Figure S3B) and was used to calculate relative concentrations for sapogenins detected in switchgrass extracts. Quantifications were based on the peak areas calculated from total ion chromatograms (TICs) for standards, diosgenin-type sapogenins, and the unknown sapogenins with chemical structures similar to that of diosgenin. Six individual plants were harvested for each switchgrass genome type and pooled into three groups of two individual plants. Pooling permitted collection of enough tissue to perform separate analysis of the leaf blade, leaf sheath, stem, rhizome, and root to overcome the issue of the limited amount of plant tissue.
Statistical Analysis
To visualize the metabolomic variation in tissue types, developmental stages, and ecotypes of switchgrass, HCA and PCA were performed using the R (v. 3.5.1) and MetaboAnalyst 5.0 online tool platform.55 Signals were normalized to internal standard area and tissue mass, log-transformed, and scaled using Pareto scaling prior to these analyses. To assess the relationship among samples and among features, hierarchal clustering with Euclidean distance as the similarity measure and Ward.D2 as the clustering algorithm was used. The relationship results were visualized in the form of dendrograms on the heatmap. Significance analyses were carried out using the Progenesis QI software (Waters) to identify the DAFs between the upland and lowland ecotypes. The cutoff threshold of the significance analyses was FDR-adjusted Student’s t-test p ≤ 0.05 and fold change ≥ 2. Results of the analyses were visualized using volcano plots. To examine statistical differences in the sapogenin concentrations among samples, the Kruskal–Wallis test and post-hoc Dunn’s tests were performed in R. p ≤ 0.05 was considered statistically significant.
Acknowledgments
We thank Gregory Bonito (Michigan State University) for providing the switchgrass seeds used in this study and helpful advice on the manuscript and Anthony Schilmiller and Cassandra Johnny (MSU Mass Spectrometry and Metabolomics Core) for LC–MS- and GC–MS analysis-related technical support. This material is based upon work supported by the Great Lakes Bioenergy Research Center, U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, under award No. DE-SC0018409. A.D.J. acknowledges support from the USDA National Institute of Food and Agriculture, Hatch project MICL02474.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jafc.2c01306.
Method for sample collection and the sample panel; significance analysis for the DAFs; six-point instrument response curve for GC–MS quantification; (EI) GC–MS spectral information for sapogenin aglycones; biplot of the PCA; LC–MS/MS spectral information for saponins; NMR spectral information for saponins; and (CI) GC–MS spectral information for sapogenin aglycones (PDF)
LC–MS metabolomic data sets; NMR chemical shift data for saponins; saponin contents in different tissues of the six switchgrass cultivars in this study; and metabolite reporting checklist (XLSX)
Author Contributions
X.L.: conceived and performed research, created figures and tables, and wrote the manuscript. S.J.S.: generated NMR data at the University of Missouri. L.W.S.: oversaw NMR experiments at the University of Missouri. A.D.J.: conceived of experimental approaches, provided technical input, reviewed data, and edited the manuscript. R.L.L.: conceived of experimental approaches and wrote and edited the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Sands R. D.; Malcolm S. A.; Suttles S. A.; Marshall E. Dedicated Energy Crops and Competition for Agricultural Land. Agric. Econ. 2017, 10.22004/ag.econ.252445. [DOI] [Google Scholar]; (https://ageconsearch.umn.edu/record/252445)
- Jiao Y.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- Pichersky E.; Lewinsohn E. Convergent Evolution in Plant Specialized Metabolism. Annu. Rev. Plant Biol. 2011, 62, 549–566. 10.1146/annurev-arplant-042110-103814. [DOI] [PubMed] [Google Scholar]
- Hartmann T. From waste products to ecochemicals: Fifty years research of plant secondary metabolism. Phytochemistry 2007, 68, 2831–2846. 10.1016/j.phytochem.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Poole P.; Ramachandran V.; Terpolilli J. Rhizobia: from saprophytes to endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291–303. 10.1038/nrmicro.2017.171. [DOI] [PubMed] [Google Scholar]
- Bowyer P.; Clarke B. R.; Lunness P.; Daniels M. J.; Osbourn A. E. Host Range of a Plant Pathogenic Fungus Determined by a Saponin Detoxifying Enzyme. Science 1995, 267, 371. 10.1126/science.7824933. [DOI] [PubMed] [Google Scholar]
- Burkhardt H. J.; Maizel J. V.; Mitchell H. K. Avenacin, an Antimicrobial Substance Isolated from Avena sativa. II. Structure*. Biochemistry 1964, 3, 426–431. 10.1021/bi00891a021. [DOI] [PubMed] [Google Scholar]
- Papadopoulou K.; Melton R. E.; Leggett M.; Daniels M. J.; Osbourn A. E. Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 12923–12928. 10.1073/pnas.96.22.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A.; Ziegler J.; Vogt T.. Specialized Plant Metabolites: Diversity and Biosynthesis. In Ecological Biochemistry; Krauss G.-J., Nies D. H., Eds.; 14–37; Wiley-VCH Verlag GmbH & Co. KGaA, 2014. [Google Scholar]
- Last R. L.; Jones A. D.; Shachar-Hill Y. Towards the plant metabolome and beyond. Nat. Rev. Mol. Cell Biol. 2007, 8, 167–174. 10.1038/nrm2098. [DOI] [PubMed] [Google Scholar]
- Sanderson M. A.; Adler P. R.; Boateng A. A.; Casler M. D.; Sarath G. Switchgrass as a biofuels feedstock in the USA. Can. J. Plant Sci. 2006, 86, 1315–1325. 10.4141/p06-136. [DOI] [Google Scholar]
- Casler M. D.; Vogel K. P.; Harrison M. Switchgrass Germplasm Resources. Crop Sci. 2015, 55, 2463–2478. 10.2135/cropsci2015.02.0076. [DOI] [Google Scholar]
- Kiniry J. R.; et al. Perennial Biomass Grasses and the Mason–Dixon Line: Comparative Productivity across Latitudes in the Southern Great Plains. BioEnergy Res. 2013, 6, 276–291. 10.1007/s12155-012-9254-7. [DOI] [Google Scholar]
- Nielsen E. L. Polyploidy and Winter Survival in Panicum virgatum L.1. Agron. J. 1947, 39, 822–827. 10.2134/agronj1947.00021962003900090010x. [DOI] [Google Scholar]
- Sage R. F.; de Melo Peixoto M.; Friesen P.; Deen B. C4 bioenergy crops for cool climates, with special emphasis on perennial C4 grasses. J. Exp. Bot. 2015, 66, 4195–4212. 10.1093/jxb/erv123. [DOI] [PubMed] [Google Scholar]
- Aspinwall M. J.; et al. Genotypic variation in traits linked to climate and aboveground productivity in a widespread C4 grass: evidence for a functional trait syndrome. New Phytol. 2013, 199, 966–980. 10.1111/nph.12341. [DOI] [PubMed] [Google Scholar]
- Uppalapati S. R.; et al. Characterization of the Rust Fungus, Puccinia emaculata, and Evaluation of Genetic Variability for Rust Resistance in Switchgrass Populations. BioEnergy Res. 2013, 6, 458–468. 10.1007/s12155-012-9263-6. [DOI] [Google Scholar]
- Lee S. T.; et al. Isolation, Characterization, and Quantification of Steroidal Saponins in Switchgrass (Panicum virgatum L.). J. Agric. Food Chem. 2009, 57, 2599–2604. 10.1021/jf803907y. [DOI] [PubMed] [Google Scholar]
- Christ B.; et al. Repeated evolution of cytochrome P450-mediated spiroketal steroid biosynthesis in plants. Nat. Commun. 2019, 10, 3206. 10.1038/s41467-019-11286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesus M.; Martins A. P.; Gallardo E.; Silvestre S. Diosgenin: Recent Highlights on Pharmacology and Analytical Methodology. J. Anal. Methods Chem. 2016, 2016, 4156293. 10.1155/2016/4156293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K.; Gadewar M.; Tahilyani V.; Patel D. K. A review on pharmacological and analytical aspects of diosmetin: a concise report. Chin. J. Integr. Med. 2013, 19, 792–800. 10.1007/s11655-013-1595-3. [DOI] [PubMed] [Google Scholar]
- Udalova Z. V.; Zinov’eva S. V.; Vasil’eva I. S.; Paseshnichenko V. A. Correlation between the Structure of Plant Steroids and Their Effects on Phytoparasitic Nematodes. Appl. Biochem. Microbiol. 2004, 40, 93. 10.1023/B:ABIM.0000010362.79928.77. [DOI] [Google Scholar]
- Uppugundla N.; et al. Switchgrass Water Extracts: Extraction, Separation and Biological Activity of Rutin and Quercitrin. J. Agric. Food Chem. 2009, 57, 7763–7770. 10.1021/jf900998q. [DOI] [PubMed] [Google Scholar]
- Muchlinski A.; et al. Biosynthesis and Emission of Stress-Induced Volatile Terpenes in Roots and Leaves of Switchgrass (Panicum virgatum L.). Front. Plant Sci. 2019, 10, 1144. 10.3389/fpls.2019.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelot K. A.; et al. Functional Diversity of Diterpene Synthases in the Biofuel Crop Switchgrass. Plant Physiol. 2018, 178, 54–71. 10.1104/pp.18.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin C. F.; et al. Standardization of Switchgrass Sample Collection for Cell Wall and Biomass Trait Analysis. Bioenergy Res. 2013, 6, 755–762. 10.1007/s12155-012-9292-1. [DOI] [Google Scholar]
- Ekanayaka E. A. P.; Celiz M. D.; Jones A. D. Relative Mass Defect Filtering of Mass Spectra: A Path to Discovery of Plant Specialized Metabolites. Plant Physiol. 2015, 167, 1221. 10.1104/pp.114.251165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner L. W.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. T.; Stegelmeier B. L.; Gardner D. R.; Vogel K. P. The isolation and identification of steroidal sapogenins in switchgrass. J. Nat. Toxins 2001, 10, 273–281. [PubMed] [Google Scholar]
- De Combarieu E.; Fuzzati N.; Lovati M.; Mercalli E. Furostanol saponins from Tribulus terrestris. Fitoterapia 2003, 74, 583–591. 10.1016/s0367-326x(03)00152-7. [DOI] [PubMed] [Google Scholar]
- Moses T.; Papadopoulou K. K.; Osbourn A. Metabolic and functional diversity of saponins, biosynthetic intermediates and semi-synthetic derivatives. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 439–462. 10.3109/10409238.2014.953628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D. B.; et al. Adaptations between ecotypes and along environmental gradients in Panicum virgatum. Am. Nat. 2014, 183, 682–692. 10.1086/675760. [DOI] [PubMed] [Google Scholar]
- Wullschleger S. D.; Davis E. B.; Borsuk M. E.; Gunderson C. A.; Lynd L. R. Biomass Production in Switchgrass across the United States: Database Description and Determinants of Yield. Agron. J. 2010, 102, 1158–1168. 10.2134/agronj2010.0087. [DOI] [Google Scholar]
- Cornelius D. R.; Johnston C. O. Differences in Plant Type and Reaction to Rust among Several Collections of Panicum Virgatum L.1. Agron. J. 1941, 33, 115–124. 10.2134/agronj1941.00021962003300020003x. [DOI] [Google Scholar]
- Stroup J.; Sanderson M. A.; Muir J. P.; McFarland M. J.; Reed R. L. Comparison of growth and performance in upland and lowland switchgrass types to water and nitrogen stress. Bioresour. Technol. 2003, 86, 65–72. 10.1016/s0960-8524(02)00102-5. [DOI] [PubMed] [Google Scholar]
- Yang J.; et al. Natural Variation for Nutrient Use and Remobilization Efficiencies in Switchgrass. Bioenergy Res. 2009, 2, 257–266. 10.1007/s12155-009-9055-9. [DOI] [Google Scholar]
- Zhou S.; et al. Metabolome-Scale Genome-Wide Association Studies Reveal Chemical Diversity and Genetic Control of Maize Specialized Metabolites. Plant Cell 2019, 31, 937. 10.1105/tpc.18.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoli J. R.; Reid R. L.; Belesky D. P. Photosensitization in Lambs Grazing Switchgrass. Agron. J. 1992, 84, 1077–1080. 10.2134/agronj1992.00021962008400060033x. [DOI] [Google Scholar]
- Murphy K. M.; Zerbe P. Specialized diterpenoid metabolism in monocot crops: Biosynthesis and chemical diversity. Phytochemistry 2020, 172, 112289. 10.1016/j.phytochem.2020.112289. [DOI] [PubMed] [Google Scholar]
- Gus-Mayer S.; Brunner H.; Schneider-Poetsch H. r. A. W.; Rüdiger W. Avenacosidase from oat: purification, sequence analysis and biochemical characterization of a new member of the BGA family of beta-glucosidases. Plant Mol. Biol. 1994, 26, 909–921. 10.1007/bf00028858. [DOI] [PubMed] [Google Scholar]
- Nisius A. The stromacentre inAvena plastids: an aggregation ofβ-glucosidase responsible for the activation of oat-leaf saponins. Planta 1988, 173, 474–481. 10.1007/bf00958960. [DOI] [PubMed] [Google Scholar]
- Huhman D. V.; Berhow M. A.; Sumner L. W. Quantification of Saponins in Aerial and Subterranean Tissues of Medicago truncatula. J. Agric. Food Chem. 2005, 53, 1914–1920. 10.1021/jf0482663. [DOI] [PubMed] [Google Scholar]
- Bialy Z.; Jurzysta M.; Oleszek W.; Piacente S.; Pizza C. Saponins in alfalfa (Medicago sativa L.) root and their structural elucidation. J. Agric. Food Chem. 1999, 47, 3185–3192. 10.1021/jf9901237. [DOI] [PubMed] [Google Scholar]
- Oleszek W.; Jurzysta M.; Price K. R.; Fenwick G. R. High-performance liquid chromatography of alfalfa root saponins. J. Chromatogr. 1990, 519, 109–116. 10.1016/0021-9673(90)85139-m. [DOI] [Google Scholar]
- Hestrin R.; Lee M. R.; Whitaker B. K.; Pett-Ridge J. The Switchgrass Microbiome: A Review of Structure, Function, and Taxonomic Distribution. Phytobiomes J. 2021, 5, 14–28. 10.1094/pbiomes-04-20-0029-fi. [DOI] [Google Scholar]
- Liu T.-Y.; et al. Drought stress and plant ecotype drive microbiome recruitment in switchgrass rhizosheath. J. Integr. Plant Biol. 2021, 63, 1753–1774. 10.1111/jipb.13154. [DOI] [PubMed] [Google Scholar]
- Singer E.; Bonnette J.; Kenaley S. C.; Woyke T.; Juenger T. E. Plant compartment and genetic variation drive microbiome composition in switchgrass roots. Environ. Microbiol. Rep. 2019, 11, 185–195. 10.1111/1758-2229.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D. R.; Dennis R.; Arnon D. I.; Daniel I.. The Water-Culture Method for Growing Plants without Soil; College of Agriculture, University of California: Berkeley, California, 1950. [Google Scholar]
- Tzin V.; et al. Integrated metabolomics identifies CYP72A67 and CYP72A68 oxidases in the biosynthesis of Medicago truncatula oleanate sapogenins. Metabolomics 2019, 15, 85. 10.1007/s11306-019-1542-1. [DOI] [PubMed] [Google Scholar]
- Kanehisa M.; Furumichi M.; Sato Y.; Ishiguro-Watanabe M.; Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai H.; et al. MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- Kim S.; et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug K.; et al. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020, 48, D440–D444. 10.1093/nar/gkz1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiełbasa A.; Krakowska A.; Rafińska K.; Buszewski B. Isolation and determination of saponin hydrolysis products from Medicago sativa using supercritical fluid extraction, solid-phase extraction and liquid chromatography with evaporative light scattering detection. J. Sep. Sci. 2019, 42, 465–474. 10.1002/jssc.201800994. [DOI] [PubMed] [Google Scholar]
- Pang Z.; et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.