Abstract
Toll-like receptors (TLRs) recognize pathogen- and host-derived factors and control immune responses via the adaptor protein MyD88 and members of the interferon regulatory transcription factor (IRF) family. IRFs orchestrate key effector functions, including cytokine release, cell differentiation, and, under certain circumstances, inflammation pathology. Here, we show that IRF activity is generically controlled by the Src kinase family member LYN, which phosphorylates all TLR-induced IRFs at a conserved tyrosine residue, resulting in K48-linked polyubiquitination and proteasomal degradation of IRFs. We further show that LYN activity is controlled by the upstream kinase C-terminal Src kinase (CSK), whose activity, in turn, is controlled by the adaptor protein SPOP, which serves as molecular bridge to recruit CSK into the TLR signaling complex and to activate CSK catalytic activity. Consistently, deletion of SPOP or CSK results in increased LYN activity, LYN-directed IRF degradation, and inhibition of IRF transcriptional activity. Together, the data reveal a key regulatory mechanism for IRF family members controlling TLR biology.
TLRs use MyD88, SPOP, CSK, and LYN to control IRF activity via phosphorylation of a conserved tyrosine residue.
INTRODUCTION
Toll-like receptors (TLRs) are master regulators of inflammation, which is physiologically required to initiate immune responses. With the exception of TLR3, all TLRs initiate signaling via recruitment of the adaptor protein MyD88, followed by activation of defined signaling pathways, such as the nuclear factor κB (NF-κB), mitogen-activated protein kinase (MAPK), and interferon regulatory factor (IRF) pathway. The IRF family of transcription factors consists of nine members, which control various TLR effector functions ranging from the eponymous type I interferons (IFN-I) and cell differentiation to other key immune regulators, such as interleukin-12 (IL-12) (1–6). A role in TLR/MyD88-mediated gene regulation is particularly well documented for IRF1/5/7/8 controlling IFN-I expression, and IRF5 and IRF8 controlling IL-12p40 transcription (1–3, 7–12). While the critical contribution of these factors in immune regulation is clearly established, the underlying molecular mechanisms involved are only partially understood (13, 14). In particular, how IRFs are negatively regulated remains largely unknown and is the major focus of this study.
Because of the pivotal inflammatory function of TLRs, inadequate TLR activity promotes various inflammatory and autoimmune diseases, extending from bacterial sepsis and ischemia reperfusion injury to obesity-related metabolic inflammation and systemic lupus erythematosus (SLE) (15–19). Evidence for a pathogenic function of TLRs is particularly well supported in SLE with disease-associated polymorphisms in TLR signaling proteins, such as TLR7, IRAK1, IRF5, LYN, TNIP1 (ABIN1), and TNFAIP3 (A20) (20). Consistently, lupus-like disease symptoms in various mouse models are largely prevented by deletion of the TLR adaptor protein MyD88 (21–26). The role of IRFs in immune pathology, in turn, is particularly well documented for IRF5, with strongly ameliorated disease upon IRF5 deletion in all lupus models investigated so far (27–33). This includes the Lyn−/− model, which is particularly interesting as the Src kinase LYN was shown to interact with and control IRF5 activity. Given that all lupus models, including the LYN−/− model, appear to depend on MyD88 and IRF5, the data collectively indicate a SLE-linked pathogenicity axis constituted by MyD88, LYN, and IRF5. Still, it needs to be further investigated if these proteins act in one coherent signaling cascade that operates in the same cell type (23, 33, 34). LYN’s inhibitory function for IRF5 was attributed primarily to LYN’s capacity to bind IRF5, rather than its tyrosine kinase activity. This is unexpected in context of the established polymorphism found in C-terminal Src kinase (CSK), the master negative regulatory kinase of Src family kinases (SFKs). This SLE-linked polymorphism in CSK results in increased protein expression and reduced LYN kinase activity, which seems particularly compatible with a scenario where CSK controls LYN kinase activity and thus IRF5 activity (35).
Here, we used a proteomic approach to study the composition of the TLR/MyD88 signaling complex to identify proteins controlling signal transduction and gene regulation. This led to the identification of the adaptor protein speckle-type POZ protein (SPOP) as MyD88-binding protein. The function of SPOP as substrate recognition adaptor protein for the Cullin3-RING ubiquitin ligase (CRL3) is well established, and defined SPOP mutants cause cancer (36). Previous studies also described an interaction between MyD88 and SPOP, with SPOP controlling MyD88 protein levels in hematopoietic stem cells (HSCs) and diffuse large B cell lymphoma cells (37, 38). Our study reveals an additional function of SPOP, i.e., as adaptor protein for CSK, in mature immune cells. This function is critical for recruiting CSK into the TLR/MyD88 signaling complex where it is required to contain LYN activity. Detailed analyses using SPOP-deficient mice and biochemical assays reveal a critical role of SPOP, CSK, and LYN in IRF regulation, with LYN phosphorylating IRFs at a conserved tyrosine residue, resulting in polyubiquitination, proteasomal degradation, and inhibition of IRF transcriptional activity. Of note, these mechanisms are not restricted to IRF5 but appear to control all IRFs that are activated through MyD88, revealing a general mechanism of transcription factor regulation.
RESULTS
Identification of SPOP as MyD88-interacting protein
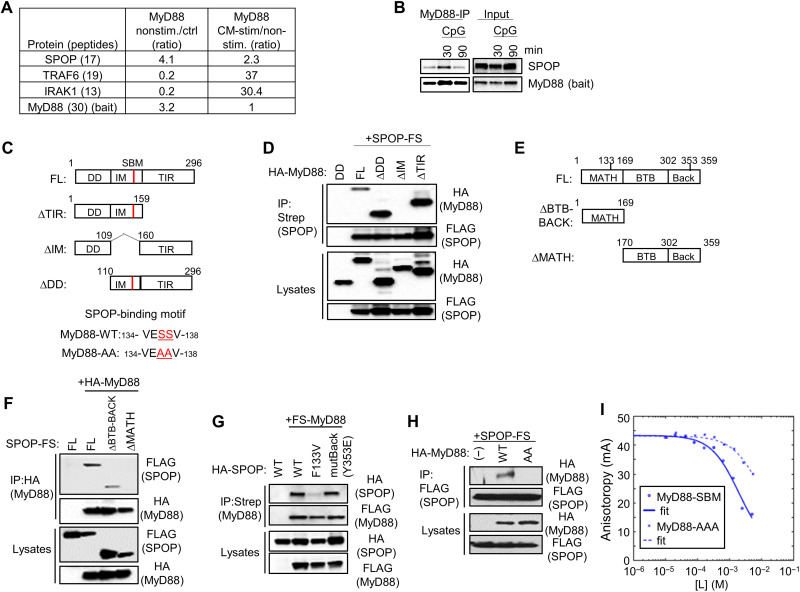
To identify MyD88-interacting proteins that control TLR signaling, we conducted proteomic experiments based on RAW264.7 cells expressing an epitope-tagged fusion protein of MyD88 and gyrase B (MyD88-GyrB). Using the GyrB-binding bivalent compound coumermycin A1 (CM), this protein allows GyrB-mediated inducible dimerization and formation of the MyD88 signaling complex, mimicking physiological TLR activation. Using this approach, components that are recruited into the activated signaling complex can be identified efficiently by quantitative mass spectrometry (MS) upon affinity purification (39, 40). Here, we used stable isotope labeling by amino acids in cell culture (SILAC) as a quantitative method in combination with MyD88-GyrB–expressing RAW264.7 macrophages (plus/minus CM), as well as RAW264.7 cells expressing GyrB alone as controls. While known components of the MyD88 complex, including TRAF6 and IRAK1, were primarily identified upon CM-induced MyD88 activation, SPOP was identified as constitutive, MyD88-interacting protein, whose abundance increased further upon CM activation (Fig. 1A). SPOP was originally identified as protein exhibiting a discrete speckled pattern in cell nuclei, and later found to serve as adaptor protein of CRL3, recruiting substrates to CRL3 for ubiquitination and subsequent proteasomal degradation (41–44). We confirmed interaction between MyD88 and SPOP by immunoprecipitation (IP)/immunoblotting (IB) with antibodies against endogenous proteins, showing constitutive interaction of the two proteins, which was further increased upon CpG-DNA (TLR9)–mediated cell activation (Fig. 1B). A previous large-scale yeast two-hybrid screen had identified interaction between MyD88 and SPOP, suggesting direct protein interaction, which was confirmed by mentioned recent publications (37, 38). To further confirm and characterize this interaction, we generated mutant proteins of MyD88 and SPOP and performed co-IP studies in human embryonic kidney (HEK) 293T cells (Fig. 1C). As shown in Fig. 1D, the full-length proteins interacted robustly, as expected. Mutant forms of MyD88 lacking either of the two structured globular domains of MyD88, i.e., the N-terminal death domain (DD) or the C-terminal Toll/IL-1 receptor homology domains (TIR), maintained SPOP binding, indicating that interaction is mediated by the so-called intermediate domain (IM) between DD and TIR. Deletion of IM prevented interaction, suggesting that SPOP binds to this region of MyD88 (Fig. 1D). Different structural domains of SPOP have been characterized, including an N-terminal MATH (meprin and TRAF homology) domain that mediates substrate recognition, as well as C-terminal BTB (broad-complex, tramtrack, and bric-à-brac) and BACK (BTB and C-terminal kelch) domains, which contribute to SPOP oligomerization and protein interaction with CUL3 (Fig. 1E) (45). While a mutant form of SPOP lacking BTB and BACK domains still interacted with MyD88, a MATH deletion mutant failed to bind MyD88, indicating that SPOP might use its substrate recognition site to mediate MyD88 binding (Fig. 1F). Consistent with this possibility, the oncogenic point mutation at F133 (F133V) within the MATH domain of SPOP, which impedes substrate recognition, displayed strongly reduced interaction with MyD88. A Y353E mutation in the BACK domain, which prevents SPOP oligomerization, remained without effect (Fig. 1G) (46). The IM of MyD88 contains a sequence with similarity to the defined SPOP-binding motif (SBM) Φ Π S S/T S/T (Fig. 1C) (45). Replacement of the two central serine residues with alanine residues (MyD88-AA) abrogated the protein interaction between MyD88 and SPOP in HEK293T cells, suggesting a critical contribution of this SBM for protein interaction (Fig. 1H). This interpretation was further supported by fluorescence anisotropy (FA) competition binding assays, in which a peptide containing the MyD88 SBM outcompeted the interaction between SPOP and a peptide from the phosphatase Puc containing the highest affinity SBM. Kd (dissociation constant) was ~0.3 mM and decreased ~23-fold upon mutation of key serine residues to alanine (MyD88-AA mutant; Fig. 1I and Table 1). These observations are also consistent with recent reports and show that MyD88 interacts directly with SPOP, using the substrate recognition cleft of SPOP and the corresponding SBM in the IM domain of MyD88 (37, 38).
Fig. 1. Identification of SPOP as MyD88-interacting protein.
(A) Quantitative MS analysis of proteins copurifying with MyD88. Numbers reflect ratios of proteins copurifying with nonstimulated MyD88-GyrB versus control protein (GyrB) (left) and CM-stimulated (dimerized) versus nonstimulated MyD88-GyrB (right). Numbers in brackets indicate number of unique peptides identified. (B) IP/IB analysis (left) of RAW264.7 cells with antibodies against endogenous proteins. IB of protein input (right) is shown for comparison. (C) Schematic representation of MyD88 constructs. MyD88-AA, MyD88 SS136/137AA. (D) IP/IB analysis of proteins transfected into HEK293T cells. HA, hemagglutinin. (E) Schematic representation of SPOP constructs. (F to H) IP/IB analysis of proteins exogenously expressed in HEK293T cells. (I) FA competition binding assay. Peptides containing the candidate SBM of MyD88 and the mutated sequence MyD88-AA compete with fluorescein-Puc91–106 for binding to the SPOP MATH domain in an FA competition assay. [L] is the peptide concentration. Continuous lines are nonlinear least-squares fits to a complete competitive binding model. See Table 1 for Kd values.
Table 1. Peptide affinities to the SPOP MATH domain.
| Motif | Residue no. | Sequence | Kd (mM)* |
| MyD88-SBM | 134–138 | VESSV | 0.83 ± 0.36 |
| MyD88-AA | 134–138 | VEAAV | 7.40 ± 3.48 |
| CSK-SBM | 338–342 | EASST | 0.45 ± 0.13 |
| CSK-AAA | 338–342 | EAAAA | 5.13 ± 1.36 |
*Errors represent SEs from four to five replicates from FA competition binding experiments.
SPOP controls expression of a select group of TLR target genes in myeloid cells
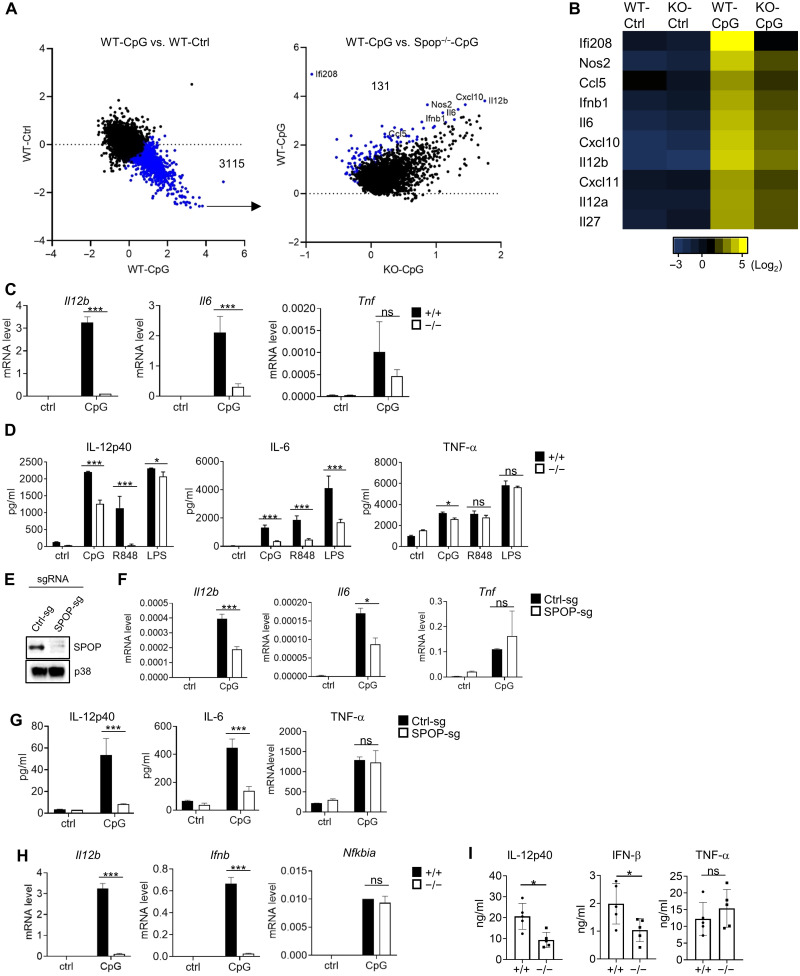
To investigate the function of SPOP in TLR biology, we established SPOP-deficient mice based on embryonic stem cells obtained from the knockout mouse phenotyping (KOMP) consortium (Spop Frt mice). Germline deletion of Spop results in embryonic or perinatal lethality; however, embryos harvested up to day E13.5 (embryonic day 13.5) showed the expected Mendelian ratio (47). We used fetal livers of E13.5 embryos to establish chimeric mice by adoptive transfer, whose bone marrow (BM) was used to generate dendritic cells (DCs). These BM-derived DCs (BMDCs) were stimulated with the TLR9 agonist CpG-DNA for 6 hours, and mRNA expression was analyzed by RNA sequencing (RNA-seq). A total of 3115 genes were up-regulated upon CpG-DNA stimulation in wild-type (wt) BMDC (Fig. 2A). Among those genes, 131 were significantly reduced in SPOP-deficient BMDC. These SPOP-dependent genes included well-characterized immune regulators, such as Nos2 [INOS (inducible nitric oxide synthase)], Ifnb1 (IFN-β), Il6 (IL-6), and IL12b (IL-12 p40) (Fig. 2B and table S1).
Fig. 2. Selective defect of TLR-induced genes in Spop−/− immune cells.
(A) RNA-seq–based gene expression analysis of BMDCs. Left: Treatment effect (4-hour CpG-DNA) of wt BMDC. A total of 3115 CpG-DNA–inducible transcripts (out of 21,267 transcripts) are highlighted in blue [fold change (FC) > 2, false discovery rate (FDR) < 0.05]. Right: Genotype effect on selected 3115 transcripts in wt and Spop−/− BMDC. A total of 131 transcripts were down-regulated in Spop−/− BMDC (red; FC > 4, FDR < 0.05). (B) Heatmap representation of selected transcripts from (A). (C) qPCR analysis of BMDC stimulated with CpG-DNA for 4 hours. ns, nonspecific. (D) Cytokine analysis [enzyme-linked immunosorbent assay (ELISA)] of BMDC stimulated with CpG-DNA, R848, or LPS for 6 hours. (E) IB analysis of RAW264.7 cells expressing CAS9 that were transduced with vectors encoding control (ctrl) and Spop-specific single-guide RNAs (sgRNAs). (F) qPCR analysis of RAW264.7 cells with SPOP deletion (SPOP-sg) or controls (Ctrl-sg) that were stimulated with CpG-DNA for 4 hours. (G) Cytokine analysis (ELISA) of Ctrl-sg– or SPOP-sgRNA–treated RAW264.7 cells that were stimulated with CpG-DNA for 6 hours. (H) qPCR analysis of plasmacytoid DC stimulated with CpG-DNA for 4 hours. (I) In vivo cytokine analysis (ELISA) upon R848 challenge for 90 min. Data represent means ± SD from three independent experiments. *P < 0.05, ***P < 0.005 are determined by two-way analysis of variance (ANOVA) with Sidak’s multiple comparison test (C, D, and F to I) and unpaired t test (I).
Deregulation of a select number of these genes was confirmed by quantitative polymerase chain reaction (qPCR), demonstrating significantly reduced expression of CpG-DNA–induced Il12b and Il6, while Tnfa expression was not affected (Fig. 2C). These changes in gene regulation were recapitulated on a protein level and were also apparent for other TLR ligands engaging MyD88-dependent signaling pathways, including R848 (TLR7) and lipopolysaccharide (LPS) (TLR4) (Fig. 2D). To exclude a potential influence by cell differentiation, we further recapitulated these findings using the macrophage cell line RAW264.7, where we deleted SPOP using CRISPR-CAS9 (Fig. 2E). Similar to primary cells, SPOP-deficient RAW264.7 cells exhibited a significant defect in CpG-DNA–mediated IL-12 and IL-6 expression, while TNF-α expression was not affected (Fig. 2, F and G, and fig. S1, A and B). Similar results were obtained from BM-derived plasmacytoid DC, where CpG-DNA–induced Il12b and Ifnb were strongly reduced in SPOP-deficient cells, while Ikba, an NF-κB target gene, was not affected (Fig. 2H). To confirm these findings in vivo, we established conditional SPOP-deficient mice containing Spop flanked by loxP sites (Spopf/f; see Materials and Methods). These mice were crossed with Vav-iCre mice to delete Spop in hematopoietic tissues (48). As expected, SPOP was efficiently deleted in splenocytes of Spopf/f Vav-iCre mice (fig. S1C). Consistent with the in vitro experiments, in vivo challenge of Spopf/f Vav-iCre mice with R848 resulted in significantly reduced serum levels of IL-12 p40 and IFN-β, while TNF-α was not affected (Fig. 2I).
As noted, two recent reports described interaction between MyD88 and SPOP. One was based on overexpression experiments suggesting SPOP/CUL3-mediated suppression of oncogenic MyD88 function in lymphoma cells (38). The other report suggested a CUL3-dependent function of SPOP specifically in HSC (37). SPOP deletion appeared to result in increased MyD88 levels in an HSC cell line (possibly owing to defective CUL3-mediated ubiquitination and protein degradation), resulting in increased neutrophilia upon challenge with the TLR3 agonist double-stranded RNA [poly I:C (Polyinosinic : polycytidylic acid)] (37). While described functions appeared restricted to specific cell types, both suggested a gain of function of MyD88 upon SPOP deletion, due to increased MyD88 protein levels. These observations are reminiscent of the established SPOP/CUL3 function in the androgen receptor (AR), estrogen receptor (ER), and MYC pathway, where loss of SPOP function results in increased expression of AR, ER, and MYC, ultimately driving oncogenicity (49–53). This phenotype is not observed in mature innate immune cells, where neither increased TLR-driven effector functions, such as IFN-I or IL-12 expression, nor increased MyD88 expression and enhanced signaling pathway activity, e.g., of the NF-κB and MAPK pathways, is observed upon SPOP deletion (Fig. 2 and see below). Together, results based on gene-deficient innate immune cells demonstrate that SPOP is required for activation of a subset of genes induced by TLRs that signal via MyD88.
SPOP controls IRF activation
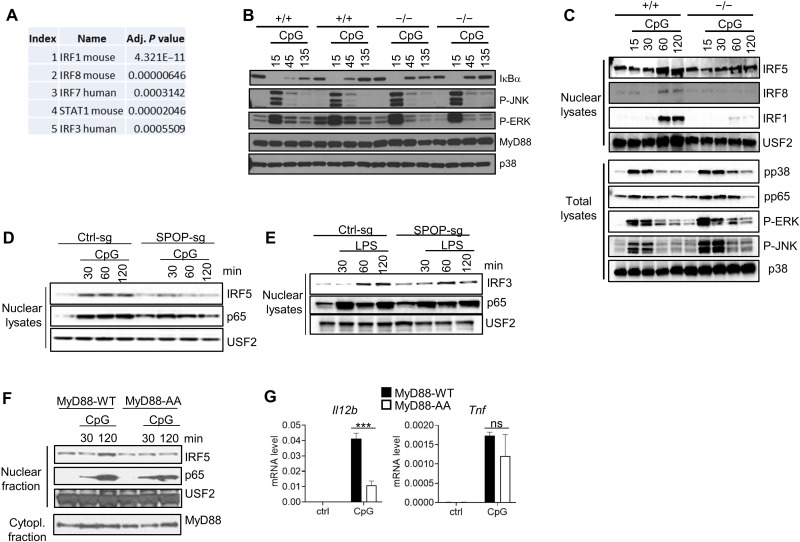
To delineate signaling pathways involved in SPOP-mediated gene regulation, we analyzed the RNA-seq dataset for transcription factors linked to genes deregulated in SPOP-deficient DC. The five top hits identified by transcriptional regulatory relationships unraveled by sentence-based text mining (TRRUST), a curated database of human and mouse transcriptional regulatory networks (available through Enrichr), included four IRF transcription factors (Fig. 3A) (54–56). As mentioned, various IRFs, including IRF1, IRF5, and IRF8, are known regulators of TLR-mediated gene transcription in myeloid cells, including BMDC and macrophages, and we analyzed their nuclear translocation along with other major TLR signaling pathways, i.e., the NF-κB and MAPK pathways. TLR9-mediated phosphorylation of RELA (p65) and the MAPKs c-Jun N-terminal kinase 1/2 (JNK1/2), p38, and extracellular signal–regulated kinase 1/2 (ERK1/2) was comparable in wt and Spop−/− BM-derived macrophages (BMMs) (Fig. 3B). As noted above, no differences in MyD88 expression, neither constitutively nor during TLR activation, were observed in various independently established cell populations (Fig. 3B). In contrast, nuclear translocation of IRF1, IRF5, and IRF8 was almost completely lost in SPOP-deficient cells (Fig. 3C). This defect was also apparent in SPOP-deficient RAW264.7 cells (Fig. 3D). To see whether this defect in IRF activation was specific for the MyD88 pathway, we stimulated cells with LPS, which activates IRF3 via the TRIF pathway (57). LPS-mediated nuclear translocation of IRF3 was largely unaffected in SPOP-deficient cells, suggesting that the TRIF pathway proceeds independent of SPOP (Fig. 3E). To confirm that the defined MyD88-SPOP interaction was required for IRF activation, we reconstituted Myd88−/− multipotent progenitor cells with MyD88-wt or the SBM-defective MyD88-AA mutant, followed by macrophage colony-stimulating factor (M-CSF)–driven cell differentiation into macrophages and stimulation with CpG-DNA. While MyD88-wt conferred responsiveness to CpG-DNA–mediated NF-κB-, IRF5-, Il12b, and Tnfa activation, MyD88-AA conferred similar responsiveness to NF-κB and Tnfα activation but failed to rescue IRF5 translocation and (largely) Il12b activation (Fig. 3, F and G). Together, these data are consistent with the interpretation that SPOP engagement via MyD88 is critical for IRF activity, while activation of the NF-κB and MAPK pathways proceeds largely independent of SPOP.
Fig. 3. SPOP controls TLR-induced IRF nuclear translocation and gene activation.
(A) RNA-seq/Enrichr/TTrust-based analysis of top 100 differentially regulated genes in CpG-DNA–induced wt and Spop−/− BMDC. The five top-scoring transcription factors are shown. (B) IB analysis of total lysates of BM-derived macrophages (BMMs) obtained from SPOP+/+ or SPOP−/− fetal liver chimeric mice. BMMs from two independent mice per genotype are shown. (C) IB analysis of nuclear extracts and total lysates from SPOP+/+ or SPOP−/− DCs derived from multipotent progenitor cells and stimulated with CpG-DNA. (D) IB analysis of nuclear extracts from Ctrl-sg or SPOP-sg RAW264.7 cells stimulated with CpG-DNA. (E) IB analysis of nuclear extracts from Ctrl-sg or SPOP-sg RAW264.7 cells stimulated with LPS. (F) IB analysis of nuclear and cytoplasmic fractions from CpG-DNA–stimulated macrophages derived from MyD88−/− multipotent progenitor cells that were reconstituted with wt or mutant MyD88 [MyD88-AA (SS136/137AA)]. (G) qPCR analysis of cells described in (F). Data represent mean ± SD from three independent experiments. ***P < 0.005 is determined by two-way ANOVA with Sidak’s multiple comparison test (B).
SPOP is required to counteract tyrosine phosphorylation of IRFs
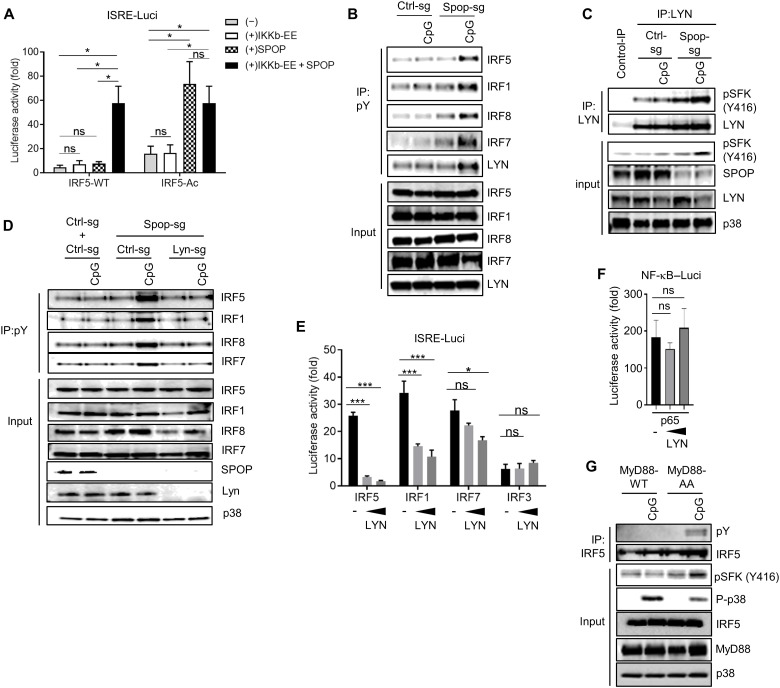
To delineate the molecular mechanism of IRF regulation, we focused our initial studies on IRF5, which can be studied well in macrophage cell lines, such as RAW264.7 cells. Although the regulation of IRF5 is only partially understood, two principal mechanisms were described in more detail, i.e., activation via IKKβ (inhibitor of NF-κB kinase β)–mediated serine phosphorylation and, less well characterized, inactivation via the Src kinase LYN (33, 58, 59). We first tested SPOP activity in the context of IKKβ-induced transcriptional IRF5 activation using luciferase reporter assays. While neither SPOP nor a constitutively active form of IKKβ (IKKβ-EE) alone induced significant IRF5 activity, coexpression of both proteins led to robust transcriptional activity of IRF5, supporting the role of SPOP in IRF activation (Fig. 4A). As expected, an IRF5 mutant with phospho-mimetic reconstitution of critical IKKβ-targeted serine sites (IRF5-Ac) showed increased basal activity in this reporter assay (Fig. 4A). SPOP enhanced activity of IRF5-Ac significantly, while coexpression of IKKβ-EE had no additional effect (Fig. 4A). While these data confirm the relevance of characterized serine residues as IKKβ-specific phosphorylation sites, the data also strongly suggest that SPOP controls IRF5 activity independent of IKKβ.
Fig. 4. LYN controls tyrosine phosphorylation and activity of IRFs.
(A) Luciferase reporter assay in HEK293T cells transfected with ISRE (IFN-stimulated response element) reporter vector, IRF5-WT, IRF5-S/D active mutant (S434,436,439,449D), SPOP, and constitutively active IKKβ (IKKβ-EE). (B) IP/IB analysis based on control (Ctrl-sg) and SPOP-deficient (Spop-sg) RAW264.7 cells stimulated with CpG-DNA for 1 hour. (C) IP/IB analysis based on control (Ctrl-sg) and SPOP-deficient (Spop-sg) RAW264.7 cells stimulated with CpG-DNA for 1 hour. (D) IP/IB analysis based on RAW264.7 cells with CRISPR-Cas9–mediated deletion of indicated genes. (E and F) Luciferase reporter assay in HEK293T cells transfected with ISRE reporter vector (E) or NF-κB reporter vector (F) plus IRF and LYN expression vectors (0.01 and 0.03 μg). Onefold luciferase activity corresponds to reporter activity without IRF. (G) IP/IB analysis of CpG-DNA–stimulated macrophages derived from MyD88−/− multipotent progenitor cells that were reconstituted with wt or mutant MyD88 [MyD88-AA (SS136/137AA)]. Data represent mean ± SD from three independent experiments. *P < 0.05, ***P < 0.005 are determined by two-way ANOVA with Sidak’s multiple comparison test (A, E, and F).
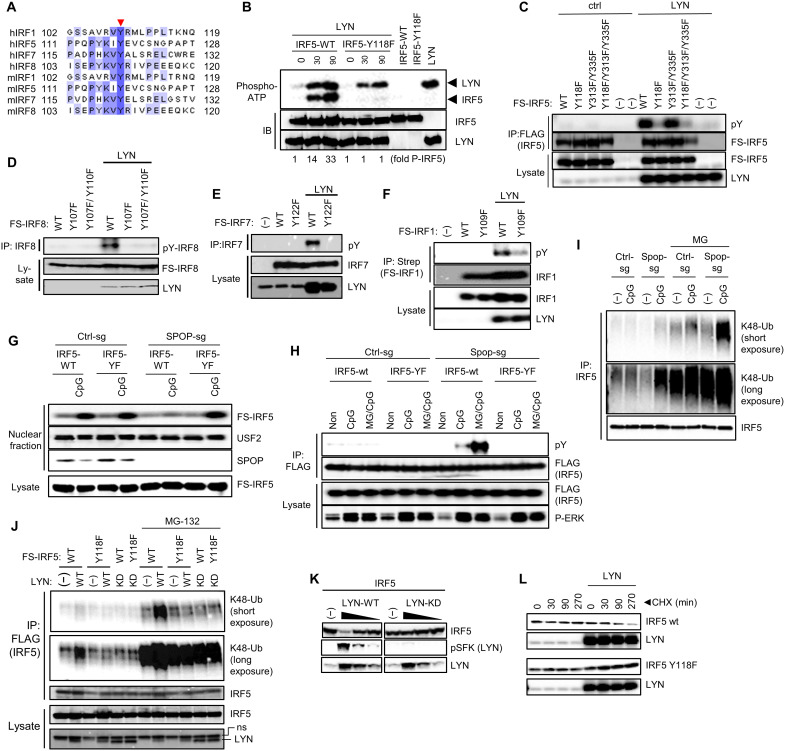
As mentioned, IRF5 was shown to be negatively regulated by the protein tyrosine kinase LYN. Protein interaction between LYN and IRF5, rather than LYN-mediated IRF5 phosphorylation, was proposed as primary mode of regulation (33). To see whether tyrosine phosphorylation was affected by SPOP, we stimulated SPOP-deficient RAW264.7 cells with CpG-DNA, followed by IP using phospho-tyrosine–specific antibodies and immunoblot analysis of different IRFs and LYN. As shown in Fig. 4B, SPOP-deficient cells showed strongly increased tyrosine phosphorylation of LYN, indicating increased kinase activity, as well as increased phosphorylation of IRF5. Of note, increased tyrosine phosphorylation was not limited to IRF5 but was also clearly observed for other IRFs that are activated through the MyD88 pathway, i.e., IRF1, IRF7, and IRF8 (Fig. 4B).
Similar to other Src family members, LYN activity is regulated by tyrosine phosphorylation of a conserved motif in the activation loop (Y397, corresponding to Y416 in c-Src) (60, 61). Consistent with the described results based on phospho-tyrosine IP experiments, CpG-DNA stimulation induced a distinct increase of LYN phosphorylation in SPOP-deficient cells (Fig. 4C). CRISPR-CAS9–mediated deletion of LYN in SPOP-deficient cells prevented the increased tyrosine phosphorylation of all IRFs analyzed, suggesting that LYN is critical for SPOP-mediated phosphorylation of various IRFs and, possibly, control of their transcriptional activity (Fig. 4D). Consistent with this idea, LYN-mediated repression of IRF transcriptional activity was observed for IRF5, IRF1, and IRF7 (Fig. 4E). IRF3 activity, which is physiologically induced by the TLR/TRIF pathway, or RELA-mediated NF-κB activity was not affected, demonstrating selectivity of LYN regulatory activity (Fig. 4, E and F). Of note, the described increase in tyrosine phosphorylation of IRF5 and LYN was also observed in cells expressing the SPOP binding–deficient MyD88-AA mutant, suggesting that MyD88-SPOP interaction is required to prevent tyrosine phosphorylation and functional repression (Fig. 4G). Together, the data show (i) that MyD88-SPOP interaction is required to counteract LYN activation and (ii) that increased LYN activity results in increased tyrosine phosphorylation of MyD88-dependent IRFs, whose phosphorylation correlates with reduced transcriptional activity.
LYN phosphorylates a conserved tyrosine residue in IRFs that controls nuclear translocation
Using in vitro phosphorylation assays and trypsin-based MS, two LYN phosphorylation sites in IRF5 were identified previously (Y313/Y335) (33). However, IRF5 variants with corresponding phenylalanine mutations did not show significantly reduced transcriptional activity in the presence of LYN, suggesting that LYN-IRF5 interaction, rather than IRF5 phosphorylation, might be the primary mode of inhibition. We took advantage of our observation that TLR-induced tyrosine phosphorylation of several IRFs is controlled by LYN. We thus aligned IRFs to identify possibly conserved tyrosine residues. Only one such tyrosine residue was identified located at the C terminus of the DNA binding domain of IRFs (Fig. 5A). Of note, this phosphorylation site in IRF5 appeared not amenable to trypsin-based MS due to the large fragment size of the resulting peptide (fig. S2). To identify LYN-targeted phosphorylation sites, we therefore conducted in vitro phosphorylation assays based on recombinant LYN and IRF5 and included GluC along with trypsin as enzymes for MS analysis (fig. S2). While various phosphorylated peptides were identified upon tryptic digest of IRF5, GluC digest of LYN-treated IRF5 led to identification of a peptide with an MS profile consistent with phosphorylation of either the conserved Y118 or, possibly, the adjacent Y115 (figs. S2 to S4). To differentiate between these possibilities and to confirm the critical pY residue in IRF5, we conducted in vitro kinase assays based on recombinant LYN and an IRF5 Y118F mutant protein (Fig. 5B). While wt IRF5 was efficiently phosphorylated by LYN (along with apparent LYN autophosphorylation), the Y118F mutation abrogated phosphorylation almost completely (Fig. 5B). To confirm and extend these findings, we conducted phosphorylation assays in HEK293T, including IRF5 variants with mutations in the previously identified phosphorylation sites (IRF5 Y313/335F). Similar to the in vitro kinase assay, wt IRF5 was efficiently phosphorylated by LYN, while phosphorylation of the Y118F mutant was almost completely abolished (Fig. 5C). The previously described IRF5 Y313/335F mutant did not show an apparent reduction in LYN-mediated phosphorylation, neither alone nor in combination with Y118F (Fig. 5C). Similar results were obtained for IRF8, IRF7, and IRF1, where mutation of the corresponding conserved tyrosines led to substantial decrease in LYN-mediated phosphorylation (Fig. 5, D to F). To address the functional impact of Y118 phosphorylation, we reconstituted wt and SPOP-deficient RAW264.7 cells with either IRF5 wt or IRF5 Y118F, followed by CpG-DNA treatment and nuclear translocation analysis. No impact of Y118 was observed in wt cells, while nuclear translocation of IRF5 wt was abolished in SPOP-deficient cells, as expected (Fig. 5G). However, the IRF5 Y118F mutant translocated comparably in control cells and SPOP-deficient cells, consistent with the interpretation that Y118 phosphorylation is the cause of failed nuclear translocation of wt IRF5 in SPOP-deficient cells (Fig. 5G). Similar data were obtained for IRF1 and IRF8 (figs. S5A and S6A). Together, these data suggest that the conserved tyrosine in IRFs represents the major phosphorylation site of LYN that controls IRF activation.
Fig. 5. LYN phosphorylates a conserved tyrosine in IRFs controlling K48 ubiquitination and stability.
(A) Sequence alignment of MyD88-induced IRFs. The conserved tyrosine is indicated by arrow. (B) In vitro LYN kinase assay using recombinant proteins expressed in HEK293T cells. Incorporation of P32 was analyzed by phosphor imaging. (C) IP/IB experiments using HEK293T cells and transfected proteins. (D to F) IP/IB experiments using HEK293T cells and transfected proteins. (G) Nuclear translocation assay of wt and SPOP-deficient RAW264.7 cells that were reconstituted retrovirally with IRF5 wt or IRF5 Y118F. (H) IP/IB analysis of IRF5 phosphorylation using cells described in (G). MG, MG-132. (I) IP/IB analysis of K48 ubiquitination of IRF5 in wt and SPOP-deficient RAW264.7 cells. (J) IP/IB analysis of K48 ubiquitination of IRF5 (I) using transfected HEK293T cells. (K) IB analysis of IRF5 and phospho-LYN upon overexpression of LYN wt or kinase-dead LYN [LYN-KD (D385G)]. (L) IB analysis of HEK293T cells that were transfected with IRF5 wt or IRF5 Y118 (L), followed by cycloheximide (CHX) treatment to block protein translation for indicated time points.
Tyrosine-phosphorylated IRF5 is targeted for K48-linked ubiquitination and proteasomal degradation
Given the correlation of IRF tyrosine phosphorylation and loss of nuclear translocation, we hypothesized that tyrosine phosphorylation of IRFs may serve as a signal for proteasomal protein degradation preventing nuclear translocation. Consistent with this hypothesis, treatment of RAW264.7 cells with the proteasome inhibitor MG132 strongly increased the amount of tyrosine-phosphorylated IRF5 in CpG-DNA–treated, SPOP-deficient cells (Fig. 5H). Likewise, the decoration of IRF5 with K48-linked polyubiquitin chains, targeting proteins for proteasomal degradation, was strongly increased in SPOP-deficient RAW264.7 cells treated with CpG-DNA and MG132 (Fig. 5I). Similar data were obtained for IRF8 (fig. S6B). K48 ubiquitination of IRF5 was recapitulated in HEK293T cells upon overexpression of LYN, where IRF5 ubiquitination depended on LYN kinase activity and the Y118 phosphorylation site in IRF5, as apparent from a kinase-deficient form of LYN and the Y118 mutation in IRF5, respectively (Fig. 5J). Similar results were obtained for IRF8 and IRF1, where LYN-induced IRF K48 ubiquitination was strongly increased in the presence of MG132 and dependent on the conserved tyrosine residues (figs. S5B and S6C). As expected, overexpression of LYN was accompanied by increased LYN autophosphorylation and loss of IRF5, while overexpression of the kinase-deficient form of LYN mediated neither autophosphorylation nor loss of IRF5 (Fig. 5K). To directly assess the impact of LYN activity on IRF stability, we treated cells for different periods of time with the translation inhibitor cycloheximide. As shown in Fig. 5L, LYN induced a clear decline of IRF5 protein levels, which was not observed for the IRF5 Y118F mutant. Similar observations were obtained for IRF8 and IRF1, where mutation of the conserved tyrosine rescued LYN-driven loss of IRFs (figs. S5C and S6D). In summary, the data show that LYN phosphorylates a conserved tyrosine in IRF proteins, as shown for IRF5, IRF1, IRF7, and IRF8, which triggers K48 ubiquitination and proteasomal degradation, thereby preventing nuclear translocation and thus activation of IRF proteins.
SPOP binds and recruits CSK to the activated MyD88 signaling complex
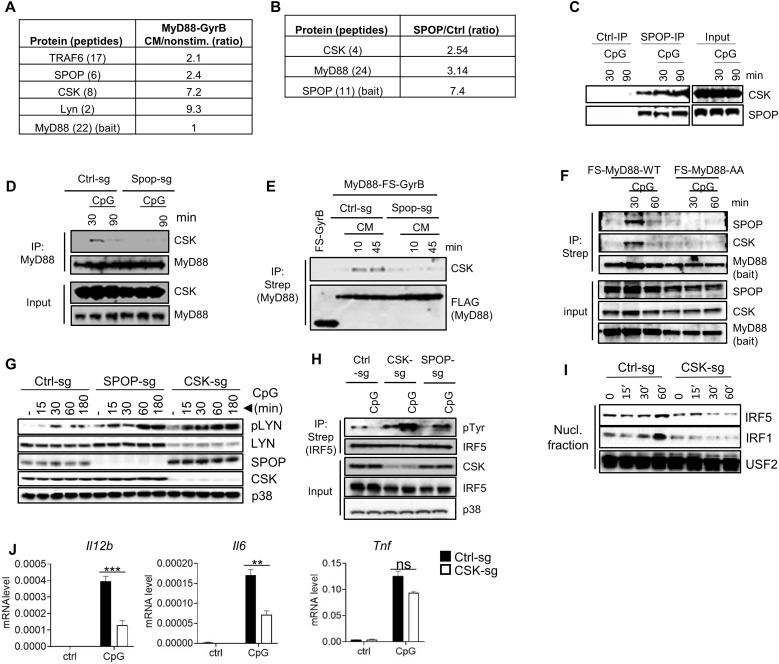
Given the deregulation of LYN activity in SPOP-deficient cells, a key question is how SPOP controls LYN. To identify potential candidate regulators, we performed up-scaled, quantitative MS experiments based on immunopurified MyD88 and SPOP. MyD88 interacted upon GyrB-driven dimerization induced by CM specifically with TRAF6, SPOP, and LYN, as expected (Fig. 6A). MyD88 also interacted with CSK, which was identified with eight unique peptides (Fig. 6A). SPOP, in turn, interacted constitutively with MyD88, as expected, but also with CSK, which was identified unequivocally with four unique peptides (Fig. 6B). We confirmed the SPOP-CSK interaction using antibodies against endogenous proteins, which demonstrated constitutive interaction that was slightly increased upon TLR activation (Fig. 6C). More detailed analysis using endogenous, as well as epitope-tagged MyD88 as bait during TLR stimulation in SPOP-deficient cells showed that CSK recruitment to MyD88 is dependent on SPOP (Fig. 6, D and E). This function of SPOP serving as adaptor protein for recruitment of CSK was further supported by experiments based on macrophages expressing wt or the SPOP binding–deficient MyD88-AA mutant, where both SPOP and CSK recruitment to MyD88 was abolished in the absence of SPOP (Fig. 6F).
Fig. 6. CSK interacts with SPOP and controls LYN activity and IRF activation.
(A) Quantitative MS analysis of proteins copurifying with dimerized MyD88. Numbers reflect protein ratios of CM-treated (dimerized) versus nonstimulated samples. Numbers in brackets indicate number of unique peptides identified. (B) Quantitative MS analysis of proteins copurifying with SPOP. Numbers reflect protein ratios of SPOP versus control protein (GyrB). (C) IP/IB analysis using isotype control antibodies or antibodies against endogenous proteins. (D) IP/IB analysis using control or SPOP-deficient RAW264.7 cells and antibodies against endogenous proteins. (E) IP/IB analysis using control or SPOP-deficient RAW264.7 cells expressing FS-MyD88-GyrB and antibodies against the epitope tag (Strep). (F) IP/IB analysis of macrophages derived from MyD88−/− multipotent progenitor cells that were reconstituted with wt or mutant MyD88 [MyD88-AA (SS136/137AA)]. (G) IB analysis of RAW264.7 cells with deletion of SPOP or CSK. (H) IP/IB analysis of RAW264.7 cells with deletion of SPOP or CSK. (I) Nuclear translocation assay of RAW264.7 cells with deletion of CSK. (J) qPCR analysis of RAW264.7 cells with deletion of CSK. Data represent mean ± SD from three independent experiments. **P < 0.01, ***P < 0.005 are determined by two-way ANOVA with Sidak’s multiple comparison test (J).
To evaluate the functional relevance of CSK in TLR-induced IRF activation, we deleted CSK in RAW264.7 cells and analyzed LYN activity in these cells in comparison to wt and SPOP-deficient cells during TLR activation. Similar to SPOP-deficient cells, LYN autophosphorylation was strongly increased in CSK-deficient cells (Fig. 6G). This increase in phosphorylation is particularly remarkable considering the relatively low expression levels of LYN in CSK-deficient cells, which is a known phenomenon in cells with reduced CSK activity (62, 63). Similar to LYN, tyrosine phosphorylation of IRF5 was strongly increased upon CSK deletion, and nuclear translocation of IRF5 and IRF1 was prevented during TLR activation (Fig. 6, H and I). Corresponding to these results and similar to SPOP-deficient cells, TLR-induced Il12b and Il6 mRNA expression was reduced in CSK-deficient cells, while Tnf expression was not significantly affected (Fig. 6J). Together, the data suggest that SPOP controls LYN activity and IRF phosphorylation via CSK, whose recruitment into the MyD88 signaling complex depends on SPOP.
SPOP interacts directly with CSK and triggers CSK activation
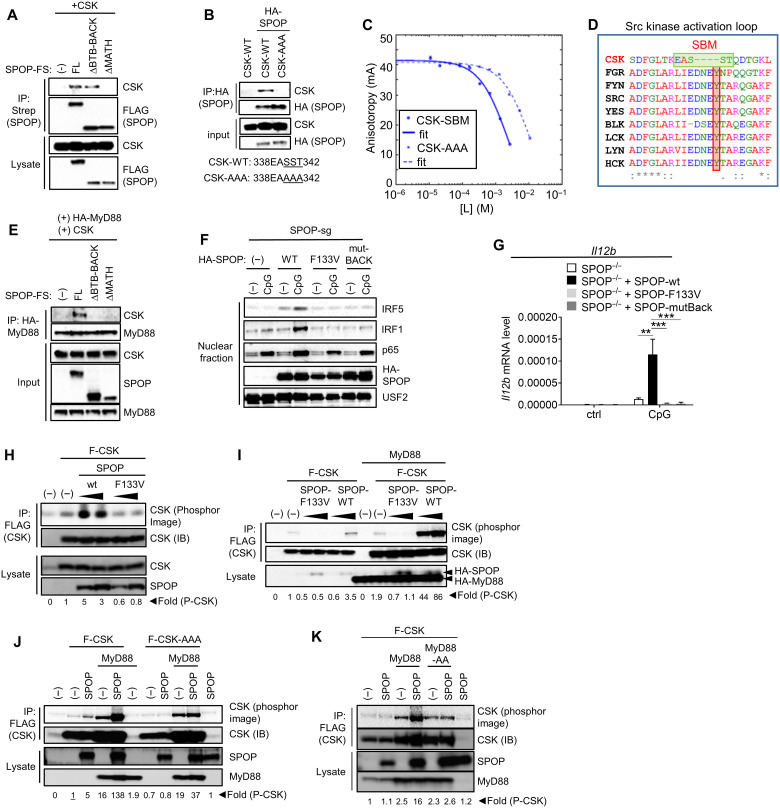
To further characterize SPOP-CSK interaction, we performed co-IP experiments in HEK293T cells, which demonstrated robust interaction of the two proteins (Fig. 7, A and B). SPOP deletion mutants showed that the MATH domain of SPOP was required for CSK interaction, suggesting that SPOP might bind CSK via its classic substrate-binding mode (Fig. 7A). Analyzing CSK computationally for SBMs, we found one motif that was located at a structurally exposed loop of CSK (fig. S7) (64). The functional relevance of this site was confirmed by mutagenesis experiments, where a CSK variant with replacement of the central part of the SBM with alanine residues failed to interact with SPOP (Fig. 7B). This interaction was further investigated by FA competition binding assays, where a CSK-derived peptide containing the described SBM outcompeted the interaction of SPOP and a Puc peptide with a Kd of 0.1 mM. Consistent with the co-IP experiments, the affinity of this interaction decreased ~40-fold upon replacement of key serine/threonine residues with alanine residues (Fig. 7C and Table 1). Of note, as illustrated by the alignment with other Src kinase family members, the SBM-containing loop represents the kinase activation loop, with the SBM virtually replacing the sequence containing the conserved tyrosine that controls catalytic activity in SFKs (Fig. 7D) (65). Hence, it appears that SPOP uses its substrate-binding MATH domain to bind CSK via a defined SBM in the activation loop of CSK. On the basis of SPOP’s described interaction with MyD88, the data also imply that SPOP serves as a molecular bridge between MyD88 and CSK. Given that SPOP engages with both MyD88 and CSK via its substrate-binding domain, this strongly suggests that dimer (or oligomer) formation of SPOP is required to allow for simultaneous binding of both target proteins. SPOP oligomerization is well characterized and depends on the synergistic dimerization of its BTB and BACK domains (45, 66). As demonstrated above, these domains are not required for MyD88 or CSK binding. To see whether they might be required for bridging MyD88 and CSK, we performed co-IP studies. While MyD88 did not pull down CSK when cotransfected in HEK293T cells, as expected, addition of full-length SPOP mediated interaction between the two proteins (Fig. 7E). In contrast, SPOP mutants lacking either the MATH or the BTB/BACK domains were unable to restore interaction (Fig. 7E). Consistent with these results, reconstitution of SPOP-deficient RAW264.7 cells with wt SPOP restored TLR-induced IRF5 and IRF1 nuclear translocation, while SPOP variants with loss-of-function mutation in the substrate-binding domain (F133V) or oligomerization-defective BACK mutation (Y353E) failed to do so. Consistently, wt SPOP restored TLR-induced Il12b expression, while both mutant forms of SPOP did not (Fig. 7G).
Fig. 7. SPOP acts as adaptor protein to recruit CSK into the TLR signaling complex.
(A and B) IP/IB analysis of HEK293T cells based on exogenously expressed proteins. The SBM in CSK and its mutation in CSK-AAA are indicated. (C) FA competition binding assay. Peptides containing the candidate SBM of CSK and the mutated sequence CSK-AAA compete with fluorescein-Puc91–106 for binding to the SPOP MATH domain. [L] is the peptide concentration. Continuous lines are nonlinear least-squares fits to a complete competitive binding model. See Table 1 for Kd values. (D) Clustal Omega–based sequence alignment of Src family kinases and CSK. The SBM and the conserved tyrosine are highlighted by green and red boxes, respectively. (E) IP/IB analysis of HEK293T cells based on exogenously expressed proteins. (F) Nuclear translocation assay of SPOP-deficient RAW264.7 cells that were reconstituted with indicated forms of SPOP. (G) qPCR analysis of cells described in (F) that were stimulated with CpG-DNA for 4 hours. (H to K) In vitro kinase assays based on proteins expressed in HEK293T cells. CSK-AAA, SST340/341/341AAA. Fold differences of CSK autophosphorylation as determined by phosphor image analysis. The activity used for reference (set as 1) is underlined. Data represent mean ± SD from three independent experiments. **P < 0.01, ***P < 0.005 are determined by two-way ANOVA with Sidak’s multiple comparison test (G).
As noted, the SBM in CSK is located in the activation loop. While the mechanism of CSK regulation remains only partially understood (see Discussion), it seemed an intriguing possibility that CSK activity is regulated by SPOP via this SBM. To investigate this idea, we performed in vitro kinase assays based on CSK that was isolated from HEK293T cells expressing either SPOP-wt or the SBM binding–deficient SPOP F133V mutant. SPOP-wt, but not the F133V mutant, induced clear CSK activity, reflected by increased autophosphorylation (Fig. 7H). Given that SPOP and CSK act downstream of MyD88, we also tested the influence of MyD88 on SPOP-mediated CSK activity. While MyD88 increased CSK expression levels (and thus kinase activity) to some extent, coexpression of SPOP, but not SPOP F133V, strongly increased CSK kinase activity (Fig. 7I). SPOP’s dependency on its substrate-binding site for CSK activation was mirrored by the SBM-variant CSK (CSK-AAA), whose protein levels were similarly up-regulated as CSK-wt upon MyD88 expression, but remained resistant to SPOP activation. Likewise, constitutive kinase activity was comparable between CSK-wt and CSK-AAA (Fig. 7J). The coactivating function of MyD88 depended on its SPOP-binding SBM, indicating that MyD88/SPOP complex formation is important for CSK activation (Fig. 7K).
Collectively, these data support the role of SPOP as adaptor protein recruiting CSK into the MyD88 complex. The data also support the idea that, in addition to its role as adaptor, SPOP activates CSK through interaction with the SBM in the CSK activation loop. The latter process is supported by MyD88/SPOP interaction, possibly serving as a molecular scaffold for orchestrating efficient CSK autophosphorylation. A schematic model based on observations described in this article is shown in fig. S8.
DISCUSSION
Data shown here reveal a key negative regulatory mechanism that controls activity of TLR/MyD88-induced IRF transcription factor family members, i.e., IRF1, IRF5, IRF7, and IRF8. This mechanism involves LYN-mediated phosphorylation of a conserved tyrosine residue found in all IRF proteins, which results in K48-linked polyubiquitination (via a so far uncharacterized E3 ubiquitin ligase) and proteasomal degradation. LYN activity is controlled by CSK, with SPOP serving as CSK adaptor protein (and possibly CSK activator) that is required to recruit CSK using defined SBMs in MyD88 and CSK. Mutation of these SBMs results in loss of CSK recruitment and function, increased LYN activity and IRF phosphorylation/degradation, and, ultimately, inhibition of transcriptional IRF activity (fig. S8). As noted, in the mature immune cells studied in this article, we did not notice changes in MyD88 protein levels or increased canonical MyD88 signaling, suggesting that CUL3-mediated regulation of MyD88 observed in HSC and cancer cells may be restricted to certain cell types (37, 38). Apart from fundamental implications for IRF-mediated gene regulation, signaling events described in our work provide a solid molecular framework for how TLRs promote inflammation, including pathological inflammation as observed in SLE.
As noted, our interpretations related to LYN as IRF-targeting kinase are in conflict with interpretations published in a previous report, favoring protein interaction rather than phosphorylation by LYN as primary IRF5 inhibitory mechanism (33). This interpretation was largely based on LYN overexpression and specific tyrosine mutants of IRF5 (IRF5 Y313F/Y335F), the latter of which showed little reduction in LYN-mediated phosphorylation in our experiments as well as the mentioned report. Hence, inferences based on this mutant are uncertain. In contrast, mutations of the conserved tyrosine (Y118 in IRF5) led to substantially reduced LYN-mediated IRF5 phosphorylation, K48 ubiquitination, and increased protein stability. The interpretation of results based on overexpressed LYN in HEK293T is likewise difficult, as forced expression might have biased physiological LYN/IRF5 interaction toward artificial inhibition. The Y118F modification of IRF5 rescued nuclear translocation in SPOP-deficient cells (in the presence of physiological LYN levels), strongly suggesting that LYN-mediated phosphorylation is the dominant mode of regulation, at least during TLR activation and in the cell types studied. Last, as noted above, this mechanism fits nicely to observations related to CSK polymorphisms found in SLE patients. Nevertheless, given the fact that the protein levels of LYN can vary, as demonstrated for CSK-deficient cells, it is possible that IRF5 binding through LYN contributes to inhibition under certain circumstances (62, 63). One aspect of LYN and IRFs, which has not been addressed so far, is, if interaction between these proteins controls LYN activation. Such mechanism has been described for many other SFK-activating proteins, whose binding via SH2/SH3 domains opens up the closed conformation of inactive Src family members, which are then further activated via dephosphorylation of their C-terminal tyrosine and consecutive autophosphorylation (67). Such scenario is consistent with our data, but further analyses related to the dynamics of LYN/IRF5 interaction and its impact on kinase activity are needed to address this question.
One piece of the puzzle missing is the postulated E3 ligase that recognizes phosphorylated IRFs and catalyzes their decoration with K48-linked polyubiquitin chains. We speculate that this E3 ligase belongs to one of the established pY-directed E3 ligases, and a possible candidate is c-CBL, which contains a conserved N-terminal pY-binding PTB domain used to bind activated protein tyrosine kinases and other pY-containing proteins (68–70). c-CBL was described as IRF8-binding and IRF8-ubiquitinating protein in macrophages treated with IFN-γ and LPS (71). Genetic deletion of c-CBL led to increased IRF8 stability and IL-12 production, consistent with (inverse) results obtained for SPOP/CSK-deficient cells. However, this paper focused on total IRF8 protein levels, not on nuclear translocation, which are likely controlled by IFN-γ and not TLR4/LPS. Hence, it is possible that the molecular mechanism of IRF8 degradation is different from the one controlled by TLR activation.
CSK is similar to SFKs with regard to the functional domain arrangement, including SH3, SH2, and kinase domains, but lacks the N-terminal acylation sites (targeting SFKs to the cell membrane), the autophosphorylation site in the activation loop (controlling kinase activity), and the C-terminal tyrosine site (controlling kinase inhibition). Because of these features and its apparent kinase activity ex vivo and in vitro, CSK was considered a constitutively active kinase, whose function is controlled primarily via adaptor proteins that recruit CSK to specific cellular localizations, such as lipid rafts and focal adhesions (67). Various such proteins are defined, which upon phosphorylation interact with CSK invariably through pY/SH2 interactions (72–78). Data shown here establish SPOP as adaptor protein that is essential to recruit CSK to the MyD88 signaling complex to facilitate LYN phosphorylation; however, the interaction with the CSK activation loop (not the SH2 domain) appears unique. This raises the question how binding and consecutive CSK recruitment is dynamically regulated during TLR activation. As shown here, MyD88-SPOP interaction is primarily constitutive, while SPOP-CSK interaction is induced upon TLR/MyD88 activation. Given that MyD88 oligomerizes during TLR activation, it seems possible that MyD88-mediated oligomerization of SPOP controls CSK binding, e.g., through increased avidity involving CSK self-association or additional SBMs in CSK. While our data establish the SBM in the CSK activation loop as critical binding site, they do not exclude the possibility that additional cooperative interaction surfaces exist. Alternatively, other proteins or protein modifications associated with activated MyD88 could contribute to SPOP-CSK binding. Obviously, more detailed studies, including structural analyses, will be required to differentiate between these possibilities.
A particularly interesting aspect of CSK regulation through SPOP is the apparent activation of CSK via the SBM motif in the activation loop. Although all evidence is based on overexpression of SPOP and MyD88 in HEK293T cells, its exquisite reliance on the SBM motifs in CSK and MyD88 indicates functional relevance. As mentioned, CSK is typically considered a constitutively active kinase; however, examples exist where protein interaction or phosphorylation increases kinase activity (79, 80). In case of the CSK-binding protein Pragmin, activation entails classic CSK-SH2 interaction with phosphorylated Pragmin, whose dimerization is critical for CSK activation (81). On the basis of the oligomerization potential of MyD88 and SPOP, it seems possible that induced proximity contributes to SPOP-mediated CSK activation. However, given that SPOP binds the activation loop of CSK, another possibility is that conformational changes of the activation loop increase kinase activity. This option seems particularly intriguing considering the unique feature of the CSK activation loop, which is short in comparison to all SFKs, hindering classic interactions of anchoring points that are usually found in active kinases (82). Protein interaction involving the activation loop of kinases has been described, including both phosphorylated and nonphosphorylated states, which at least in part contribute to kinase activation via tethering the activation loop in its active confirmation (83–85). Thus, we hypothesize that SPOP mediates such noncanonical anchoring of the CSK activation loop, ultimately increasing kinase activity. Again, structural analysis of the SPOP-CSK complex will likely provide an opportunity to differentiate between these possibilities. In summary, our work establishes a negative regulatory mode of TLR/MyD88-driven IRF activation with defined protein modifications and protein interactions, which are likely subject of physiological and pathological regulation and, possibly, can be targeted by therapeutic intervention.
MATERIALS AND METHODS
Study design
We initiated this project with a proteomic approach to study the composition of the TLR/MyD88 signaling complex to identify proteins controlling signal transduction and gene regulation. Upon identification of SPOP as previously unidentified component of the MyD88 signaling complex, we used Spop-deficient mice and biochemical assays to define the role of SPOP in innate immune cells. All experiments were repeated in at least two to three independent experiments. The number of replicates and statistical analyses performed to evaluate the significance of each experiment are described in the individual figure legends.
Reagents
CpG-DNA refers to the phosphorothioate backbone containing oligonucleotide 1668 (TCCATGACGTTCCTGATGCT) (TIB Molbiol). Other agonists used were LPS (Escherichia coli 0127:B8) (Sigma-Aldrich), CM (Sigma-Aldrich), and R848 (InvivoGen). Antibodies were sourced as follows: SPOP (ProteinTech); FLAG-M2 (Sigma-Aldrich); MyD88, IκBα, P-p38, P-p65, P-JNK, P-ERK, p38, IRF5, IRF1, IRF8, CSK, P-SFKs (Y416), LYN, P-Tyr1000, and K48-Ub (Cell Signaling Technology); hemagglutinin (HA) (3F10) (Sigma-Aldrich); IRF3, IRF7, p65, and USF2 (Santa Cruz Biotechnology); and secondary antibodies conjugated to horseradish peroxidase (HRP) [Amersham ECL Rabbit IgG, HRP-linked F(ab′)2 fragment from donkey and Amersham ECL Mouse IgG, HRP-linked F(ab’)2 fragment from sheep] (Cytiva). Chemiluminescent substrate was from Bio-Rad. Beads for IP were from Sigma-Aldrich (FLAG-M2 resin and anti-HA affinity matrix, clone 3F10), IBA Lifesciences (strep-XT beads), and Cell Signaling Technology (P-Tyr1000 sepharose beads and protein A agarose beads). Enzyme-linked immunosorbent assay (ELISA) kits were from eBioscience (TNF-α and IL-6) and R&D Systems (IL-12p40 and IFN-β). Luciferase assay system was from Promega. Lipofectamine 2000 was from Thermo Fisher Scientific. MG-132 and cycloheximide were from Sigma-Aldrich. Recombinant LYNa protein was from Carna Biosciences.
Plasmids
Expression plasmids were established by conventional molecular biology techniques and verified by Sanger DNA sequencing. Epitope tags consisted of tandem triple tags (HA, FLAG) and a fusion tag consisting of triple FLAG tag and StrepOne tag (FS). MyD88-FS-GyrB was expressed as fusion protein consisting of full-length mouse MyD88 and C-terminal FS tag and GyrB moiety using a pcDNA3-based vector with EF1α promoter. HA-tagged full-length mouse MyD88, ΔTIR (amino acids 1 to 159 of mouse MyD88), ΔDD (amino acids 110 to 296 of mouse MyD88), and deletion mutants Δ109–160 (ΔIM of mouse MyD88) were expressed as N-terminal HA-tagged proteins using an expression vector with cytomegalovirus (CMV) promoter. N-terminal triple HA-tagged full-length mouse MyD88-WT and MyD88-AA (S136/137A), N-terminal HA-tagged full-length mouse SPOP-WT and SPOP mutants (F133V or Y353E), and mouse LYN-WT and LYN-KD (D385G) were expressed using a lentiviral vector with ubiquitin promoter. C-terminal FS-tagged mouse SPOP, N-terminal FLAG-tagged Δ1–169 (ΔMATH of mouse SPOP), and C-terminal FS-tagged Δ170–374 (ΔBTB-Back of mouse SPOP) were expressed using a pcDNA3-based vector with EF1α promoter. N-terminal FS-tagged full-length mouse MyD88-WT and MyD88-AA (S136/137A), N-terminal FS-tagged mouse IRF5-WT and FS-tagged mouse IRF5-Active mutant (S434/436/439/445D), and N-terminal green fluorescent protein (GFP)–tagged mouse IRF7-WT and IRF7-Y122F were expressed using a murine stem cell virus (MSCV)–based retroviral vector. The cDNA of mouse IRF5 (RefSeq accession no.: XM_006505097.1) was obtained by reverse transcription PCR (RT-PCR) using mRNA from BMMs. N-terminal FS-tagged mouse IRF5-WT, IRF5-Y118F, and Y313/335F and N-terminal FS-tagged mouse IRF1-WT and IRF1-Y109F were expressed using a lentiviral vector with EF1α promoter. N-terminal FLAG-tagged mouse CSK-WT and CSK-SST340/341/342AAA were expressed using the vector pCMV3-N-FLAG (Sino Biological). pcDNA3-N-FLAG-hIRF8 was from Sino Biological. N-terminal FS-tagged human IRF8-WT and IRF8-Y110F were expressed using pcDNA3. pCMV3-N-FLAG-CSK was from Sino Biological. pEGFP-N1-human-LYN-GFP was obtained from Addgene (no. 35958). N-terminal HA-tagged human IKKβ-active mutant (S177/181E) was expressed using a vector containing a CMV promoter (pRC). Mutations were introduced using the Q5 Site-Directed Mutagenesis Kit according to the manufacturer’s instructions (New England Biolabs).
Mice, cell culture, and retrovirus generation
Spoptm1a(KOMP)Mbp were obtained from Knockout Mouse Project (www.komp.org). Mice carrying the targeted mutation were generated by injection of sperm into female C57BL/6J mice and crossing with C57BL/6J mice. For fetal liver transplantation, 2 × 106 fetal liver cells isolated from Spop+/+ or Spop−/− embryos on E13.5 were injected into lethally irradiated (950 cGy) C57BL/6 mice. Spopfl/fl mice were generated by crossing Spoptm1a(KOMP)Mbp mice with FLPeR (flipper) mice carrying the FLP recombinase gene targeted to the ROSA locus (B6.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/RainJ (86). Progeny carrying a Spop conditional allele (Spopfllfl) (as confirmed by PCR) was crossed with Vav-iCre mice (B6.Cg-Commd10Tg(Vav1-icre)A2Kio/J) to selectively abrogate SPOP expression in hematopoietic cells (48). All mouse studies were carried out in accordance with protocols approved by the Institutional Animal Care and Use Committee at University of Utah and St. Jude Children’s Research Hospital.
HEK293T cells were maintained in growth medium D-10 {Dulbecco’s modified Eagle’s medium (Life Technologies) containing phenol red, supplemented with 10% (v/v) fetal calf serum (FCS) (HyClone), 50 μM 2-mercaptoethanol, antibiotics [penicillin G (100 IU/ml) and streptomycin sulfate (100 IU/ml)], and pyruvate (1 mM)}. RAW264.7 cells were cultured in RP-10 {RPMI 1640 (Life Technologies), supplemented with 10% (v/v) FCS (HyClone), 50 μM 2-mercaptoethanol, and antibiotics [penicillin G (100 IU/ml) and streptomycin sulfate (100 IU/ml)]}. BMM and BMDC were generated by cultivating unfractionated BM cells (obtained from female C57BL/6 mice and corresponding wt control mice, as indicated in figure legends) for 6 days in D-10 supplemented with 30% L cell–conditioned medium (BMM) or 2% granulocyte-macrophage CSF (GM-CSF) conditioned medium (BMDC) (87).
Conditionally immortalized multipotent progenitor cells (referred to as Hoxb8-FL cells) were established as described (87). All cell culture was performed in RP-10. BM cells were partially purified using a Ficoll-Paque (GE Healthcare) gradient centrifugation step to removed neutrophils and red blood cells. The resulting cells were transduced using a retrovirus expressing a fusion protein of HOXB8 and the G400V variant of the estrogen-binding domain of the human ER (ERHBD), which renders the hormone-binding domain insensitive to physiological concentrations of estrogen or phenol red contained in growth medium (88). Transduced cells were expanded in RP-10 supplemented with 1 μM β-estradiol and 5% supernatant from an Flt3L-producing B16 melanoma cell line (provided by R. Steinman). For generation of Hoxb8-FL–derived macrophages, estrogen was removed and cells were differentiated for 7 days in RP-10 supplemented with 30% L cell–conditioned medium.
Replication-deficient lentivirus and MSCV were generated using Lipofectamine 2000 (Invitrogen)–based transient transfection of HEK293T cells using a four-plasmid system that was generously provided by I. Verma (for lentivirus) or an ecotropic, MSCV-based two-plasmid system. Cas9-mediated deletion of genes in RAW264.7 cells was done on the basis of cells transduced with the lentiviral CAS9 expression vector lentiCas9-Blast (Addgene, no. 52962). Single-guide RNAs (sgRNAs) were expressed by lentiviral delivery of specific sgRNA [mouse Spop genomic target sequence: CCTCCGGCAGAAATGTCGAG (SPOP-sg), GTGGTCCCGTTGCCGAGAGC (SPOP-sg2), mouse Csk genomic target sequence: GCAATACATTCTGTACCGGA, mouse Lyn genomic target sequence: AGAGATTATGACCCTATGCA] using the lentiviral vector lentiGuide-Puro (Addgene, no. 52963). Transduced cells were selected with puromycin (10 μg/ml) and used as polyclonal cell population.
Affinity purification and quantitative mass spectrometry
Following procedures previously described for SILAC (89), RAW264.7 cells expressing stably MyD88-FS-GyrB, SPOP-FS, or FS-GyrB were cultured in arginine- and lysine-free RP-10, supplemented with either l-arginine and l-lysine (light), l-arginine-HCl (13C6; CLM-2265 [R6]) and l-lysine-2HCl (4,4,5,5 D4; DLM-2640 [K4]) (medium), or l-arginine-HCl (13C6, 15N4; CLM-539 [R10]) and l-lysine-2HCl (13C6, 15N2; DLM-291 [K8]) (heavy) (Cambridge Isotope Labs). For complete incorporation of labeled amino acids, cells were passaged three times in SILAC medium over a period of 5 days. After cell treatment, the medium was replaced by ice-cold phosphate-buffered saline (PBS) and cells were collected by cell scraping and centrifugation. Cell pellets were incubated with lysis buffer [LB; 20 mM Hepes/NaOH (pH 7.5), 1.5 mM MgCl2, 150 mM NaCl, 1 mM EDTA, 10% glycerol, PhosSTOP (Roche), and cOmplete protease inhibitors (Roche)] and 0.5% NP-40 for 20 min. Samples were cleared by centrifugation and loaded five times over M2 FLAG-bead–containing columns. Unbound proteins were removed by washing column with LB plus 0.1% NP-40, and proteins were eluted at pH 3.5 in water supplemented with 100 mM glycine, 50 mM NaCl, 0.1% NP-40, and Roche complete protease inhibitors. The proteins were concentrated using spin columns with a 3-kDa cutoff [Amicon Ultra3K MWCO (EMD Millipore)] and dissolved in SDS–polyacrylamide gel electrophoresis (PAGE) loading buffer (Bio-Rad). The dissolved proteins derived from light, medium, and heavy conditions were combined, followed by separation on a 10% bis-tris gel (Bio-Rad) and staining with SYPRO Ruby protein stain (Sigma-Aldrich). The entire lane was cut into individual bands, digested using a proteolytic enzyme, and analyzed by liquid chromatography–MS/MS (LC-MS/MS) as described below, using a nanoACQUITY UPLC (Waters) coupled to an Orbitrap ELITE high-resolution mass spectrometer (Thermo Fisher Scientific).
Sample preparation and electrospray ionization LC
As described previously (89), the protein gel bands were reduced with dithiothreitol (DTT) and alkylated with iodoacetamide and then digested overnight with either trypsin (Promega) or GluC (Pierce) according to the manufacturer’s instructions. The resulting peptides from the digest were injected into the mass spectrometer using in-line chromatography with reversed-phase (C18) ultrahigh-pressure LC on a nanoACQUITY UPLC (Waters). The column used was a Waters BEHC18 with an inner diameter of 75 μm and a bed length of 10 cm. The particle size was 1.7 μm. Peptides were gradient eluted over a gradient [0 to 70% B for 60 min and 70 to 100% B for 10 min, where B was 70% (v/v) acetonitrile, 0.2% formic acid] using a flow rate of 250 nl/min into the high-resolution Orbitrap ELITE through a noncoated spray needle with voltage applied to the liquid junction.
MS/MS analysis with LTQ ELITE (Thermo Fisher Scientific) and database analysis
Following procedures previously described in (89), data-dependent scanning was incorporated to select the 20 most abundant ions (one microscan per spectrum; precursor isolation width, 2.0 Da; 35% collision energy; 10-ms ion activation; 15-s dynamic exclusion duration; 5-s repeat duration; and a repeat count of 1 from a full-scan mass spectrum at 60,000 resolution). MS/MS was performed in the ion trap by collision-activated dissociation. Database searches were performed using RAW files in combination with Andromeda search engine that is part of the MaxQuant software (version 1.1.1.32) developed at the Max Planck Institute. The SwissProt 2012_08 [537,505 sequences; 190,795,142 residues; taxonomy: Mus musculus (16,605 sequences)] database was used for peptide and protein identification. MaxQuant was also used to quantitate peptides and proteins and to provide ratios generated in Excel format. Protein assignments were made on the basis of both MS and MS/MS spectra, whereas peptide quantitation was based solely on MS data. The following residue modifications were allowed in the search: carbamidomethylation on cysteine (fixed modification), oxidation on methionine (variable modification), phosphorylation (variable modification) on serine, threonine, and tyrosine, and label:13C(6) on arginine, label:13C(10) on arginine, label:13C(4) on lysine, and label:13C(8) on lysine. The MS1 mass tolerance was set to 15 ppm (parts per million), the MS/MS tolerance was set to 0.5 Da, and protein false discovery rate (FDR) was set to 0.01. The identifications from the automated search were verified by manual inspection of the raw data.
FA competition binding assay
The MATH domain of SPOP (residues 28 to 166) was purified as previously described. All FA binding assays were performed in 20 mM tris (pH 7.6), 150 mM NaCl, 5 mM DTT, 0.01% Triton X-100, and bovine serum albumin (10 mg/ml) as previously described (90). Serial dilutions of each peptide were prepared in 384-well plates ranging from several millimolar to low micromolar concentrations. MATH domain and fPuc (with the sequence of Puc residues 91 to 106, Ac-ENLACDEVTSTTSSST-NH2, N-terminally fluorescently labeled) were added to final concentrations of 6 μM and 40 nM, respectively. Anisotropy was measured using a CLARIOstar plate reader (BMG LABTECH). Analysis was performed as described previously (91). For each FA assay, three independent experiments were performed and fit. Kd was reported as the average from the three fits, and the error was reported as the SD.
IP and nuclear protein extraction
IP studies were performed in HEK293T cells, RAW264.7 cells, and Hoxb8-FL–derived macrophages. HEK293T cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Cells were lysed for 20 min at 4°C in LB [20 mM Hepes/NaOH (pH 7.5), 1.5 mM MgCl2, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, and 10% glycerol] supplemented with cOmplete, EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich) and PhosSTOP (Sigma-Aldrich). After clearance of lysates by centrifugation (10 min, 20,817g, 4°C), lysates were subjected to IP using FLAG M2 resin (Sigma-Aldrich), anti–HA-matrix (clone 3F10, Sigma-Aldrich), strep-XT beads (IBA Lifesciences), or P-Tyr1000 sepharose beads (Cell Signaling Technology) for 1 hour at 4°C. For IP of endogenous proteins, protein extracts were incubated with antibodies for 4 hours, followed by protein A agarose beads (Cell Signaling Technology) or TrueBlot Anti-Rabbit Ig IP Agarose Beads (ROCKLAND) for 1 hour. Precipitated proteins were dissolved from beads by heating at 95°C for 5 min in SDS sample buffer (Bio-Rad), followed by centrifugation (3 min, 20,817g, room temperature) and analysis by IB. Nuclear and cytoplasmic proteins were extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific) according to the manufacturer’s instructions.
RNA analysis by RNA-seq and qPCR
Following the procedures previously described in (92), total RNA was isolated using TRIzol (Invitrogen) and quantified by measuring the optical density at 260 nm using a NanoDrop spectrophotometer. For RNA-seq, libraries were generated from ∼1 μg of total RNA using the Illumina TruSeq Stranded Total RNA Prep Kit with the Ribo-Zero Gold Human/Mouse/Rat Ribosomal Reduction Kit. Libraries were sequenced on an Illumina HiSeq 2500 Sequencing system using paired-end 100–base pair sequencing chemistry. For qPCR, cDNA was prepared from 1 μg of total RNA by reverse transcription using SuperScript II reverse transcriptase (Thermo Fisher Scientific) and oligo(dT) primers. Real-time PCR was performed using SYBR Green Master Mix (Thermo Fisher Scientific) and gene-specific primers in the 7300 Sequence Detector System (Thermo Fisher Scientific). Data were normalized by the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression in each individual sample. The genes and primer sequences used for qPCR are listed in Table 2.
Table 2. Primer sequences used for quantitative polymerase chain reaction.
| Gene | Sense | Antisense |
| Gapdh | 5′-GTGTTCCTACCCCCAATGTG-3′ | 5′-GGTCCTCAGTGTAGCCCAAG-3′ |
| Il6 | 5′-CTGATGCTGGTGACAACCAC-3′ | 5′-AGCCTCCGACTTGTGAAGTG-3′ |
| Il12b | 5′-AAACCAGACCCGCCCAAGAAC-3′ | 5′-AAAAAGCCAACCAAGCAGAAGACAG-3′ |
| Tnf | 5′-ACAGAAAGCATGATCCGCG-3′ | 5′-GCCCCCCATCTTTTGGG-3′ |
| Nfkbia | 5′-CTCACGGAGGACGGAGACTC-3′ | 5′-CTCTTCGTGGATGATTGCCA-3′ |
Protein expression and purification of IRF substrates for in vitro kinase assay
FS-IRF5, FS-IRF1, or FS-IRF8 proteins used for in vitro kinase assays were expressed in HEK293T cells by transient transfection using Lipofectamine 2000 (Thermo Fisher Scientific). Forty-eight hours after transfection, the cells were lysed using LB [20 mM Hepes/NaOH (pH 7.5), 1.5 mM MgCl2, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, and 10% glycerol] supplemented with complete protease inhibitors and phosphatase inhibitors cocktails (Sigma-Aldrich). After clearance by centrifugation (10 min, 20,817g, 4°C), lysates were subjected to column purification using strep-XT beads according to the manufacturer’s instructions (IBA Lifesciences). FS-tagged proteins were eluted with buffer BXT and concentrated using spin columns with a 3-kDa cutoff [Amicon Ultra3K MWCO (EMD Millipore)].
In vitro kinase assay
The protein tyrosine kinase activity of LYN was determined by measuring incorporation of P32 from [β-32P]ATP (adenosine triphosphate) into IRF substrates. Autophosphorylation activity of CSK was determined by measuring incorporation of P32 from [β-32P]ATP into CSK. For expression of CSK, HEK293T cells were cotransfected with FLAG-tagged CSK-WT or CSK-AA, HA-tagged MyD88-WT or MyD88-AA, and HA-tagged SPOP-WT/SPOP-F133V or SPOP-mutBack, as indicated in figures. Twenty-four hours after transfection, cells were lysed and FLAG-CSK was immunopurified with FLAG beads, followed by the in vitro kinase assay (CSK autophosphorylation). Kinase assays were carried out at 30°C in a 50-μl volume of kinase buffer [25 mM Hepes-NaOH (pH7.5), 5 mM MnCl2, 1 mM EDTA, 2 mM DTT, 0.01% Tween 20, PhosSTOP (Sigma-Aldrich), 50 μM cold ATP, and 10 μCi [γ-32P]ATP]. One microgram of LYN-KD and 0.5 μg of IRF substrates were used per sample. The reaction was terminated with 20 μl of 4× SDS sample buffer and heating at 95°C for 5 min. The protein solution was subjected to SDS-PAGE, immunoblotted onto nitrocellulose membranes, and analyzed by Phosphor Imager (Typhoon, GE Healthcare).
Analysis of protein stability
To analyze IRF protein stability, HEK293T cells were cotransfected with plasmids expressing FS-IRF5, FS-IRF1, or FS-IRF8 along with LYN using Lipofectamine 2000. Forty-eight hours after transfection, the cells were treated with cycloheximide (20 μg/ml) for time periods indicated in figures to inhibit protein synthesis. Cells were lysed using RIPA (radioimmunoprecipitation assay) buffer [50 mM tris-HCl (pH 8), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS (pH8.0)], and protein levels of IRF5, IRF1, and IRF8 were analyzed by IB.
In vivo ubiquitination assay
HEK293T cells were cotransfected with plasmids expressing FS-IRF5, FS-IRF1, or FS-IRF8 along with LYN using Lipofectamine 2000. Forty-eight hours after transfection, cells were treated with 10 μM MG132 for 4 hours. IRF-expressing RAW264.7 cells were treated with 10 μM MG132 for 1 hour followed by CpG-DNA stimulation (1 μM) for 1 hour. HEK293T and RAW264.7 cells were lysed in urea buffer [8 M urea, 20 mM Hepes/NaOH (pH 7.5), 1.5 mM MgCl2, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, and 10% glycerol] supplemented with complete protease and phosphatase inhibitors (Sigma-Aldrich) and 20 mM N-ethylmaleimide. After clearance by centrifugation (10 min, 20,817g, 4°C), lysates were subjected to IP using strep-XT beads for 4 hours at 4°C. Denatured IP samples were analyzed by IB using antibodies against K48-linked polyubiquitin.
Luciferase reporter assay
HEK293T cells were transfected with an ISRE (IFN-stimulated response element) reporter (firefly luciferase) vector containing a triple ISRE binding site and minimal promoter, or an NF-κB reporter vector containing a triple NF-κB binding site and a minimal promoter, Renilla luciferase vector (pRL-TK; Promega), and indicated expression plasmids using Lipofectamine 2000 (Thermo Fisher Scientific). Twenty-four hours after transfection, the cells were lysed in passive LB (Promega). Luciferase activity was determined using the dual luciferase kit (Promega), and firefly luciferase activity values were normalized to Renilla luciferase activity.
Analysis of in vivo cytokine levels
For analysis of R848-induced in vivo cytokine release, Spopfl/fl and Spopfl/fl-Vav-cre mice were intraperitoneally injected with 50 nmol R848 (in 0.1 ml of PBS) and serum samples were collected 1.5 hours later. Cytokine concentrations of IFN-β, TNF-α, IL-6, and IL-12p40 were determined by ELISA (IFN-β and IL-12p40 from R&D Systems and TNF-α and IL-6 from Thermo Fisher Scientific).
Statistical analyses
Statistical analyses were performed using GraphPad Prism 8 software. Pairwise comparisons were performed with two-tailed unpaired Student’s t test. Multiple comparisons were analyzed by two-way analysis of variance (ANOVA) with Sidak’s multiple comparison test followed by posttests as specified in figure legends.
Acknowledgments
Funding: H.H., V.R., and K.T. were supported by NIH grants AI145877 and AI139014, and T.M. was supported by NIH grant R01GM112846. This work was supported by the American Lebanese Syrian Associated Charities (ALSAC).
Author contributions: K.T. performed experiments and analyzed data. V.R. performed experiments. R.W. and J.K. established fetal liver chimeras. J.J.B. and T.M. performed fluorescence anisotropy competition binding assays, and B.K.L. performed bioinformatics analyses of RNA-seq experiments. A.M. and A.A.H. conducted mass spectrometry analyses. H.H. conceived and supervised the project, performed experiments, analyzed data, and wrote the manuscript. All authors edited the manuscript.
Competing interests: V.R. and H.H. are cofounders of Nanospot.ai, and T.M. is a consultant for Faze Therapeutics Inc. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The vectors used in the manuscript either are available through Addgene or can be provided by H.H. pending scientific review and a completed material transfer agreement. Requests for the vectors should be submitted to H.H.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Other Supplementary Material for this manuscript includes the following:
Table S1
REFERENCES AND NOTES
- 1.Miyamoto M., Fujita T., Kimura Y., Maruyama M., Harada H., Sudo Y., Miyata T., Taniguchi T., Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell 54, 903–913 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Scharton-Kersten T., Contursi C., Masumi A., Sher A., Ozato K., Interferon consensus sequence binding protein-deficient mice display impaired resistance to intracellular infection due to a primary defect in interleukin 12 p40 induction. J. Exp. Med. 186, 1523–1534 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giese N. A., Gabriele L., Doherty T. M., Klinman D. M., Tadesse-Heath L., Contursi C., Epstein S. L., Morse H. C. III, Interferon (IFN) consensus sequence-binding protein, a transcription factor of the IFN regulatory factor family, regulates immune responses in vivo through control of interleukin 12 expression. J. Exp. Med. 186, 1535–1546 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holtschke T., Lohler J., Kanno Y., Fehr T., Giese N., Rosenbauer F., Lou J., Knobeloch K. P., Gabriele L., Waring J. F., Bachmann M. F., Zinkernagel R. M., Morse H. C. III, Ozato K., Horak I., Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 87, 307–317 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Tamura T., Nagamura-Inoue T., Shmeltzer Z., Kuwata T., Ozato K., ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity 13, 155–165 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Tamura T., Yanai H., Savitsky D., Taniguchi T., The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 26, 535–584 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Matsuyama T., Kimura T., Kitagawa M., Pfeffer K., Kawakami T., Watanabe N., Kundig T. M., Amakawa R., Kishihara K., Wakeham A., Potter J., Furlonger C. L., Narendran A., Suzuki H., Ohashi P. S., Paige C. J., Taniguchi T., Mak T. W., Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell 75, 83–97 (1993). [PubMed] [Google Scholar]
- 8.Negishi H., Fujita Y., Yanai H., Sakaguchi S., Ouyang X., Shinohara M., Takayanagi H., Ohba Y., Taniguchi T., Honda K., Evidence for licensing of IFN-gamma-induced IFN regulatory factor 1 transcription factor by MyD88 in Toll-like receptor-dependent gene induction program. Proc. Natl. Acad. Sci. U.S.A. 103, 15136–15141 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takaoka A., Yanai H., Kondo S., Duncan G., Negishi H., Mizutani T., Kano S., Honda K., Ohba Y., Mak T. W., Taniguchi T., Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 434, 243–249 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Yanai H., Chen H. M., Inuzuka T., Kondo S., Mak T. W., Takaoka A., Honda K., Taniguchi T., Role of IFN regulatory factor 5 transcription factor in antiviral immunity and tumor suppression. Proc. Natl. Acad. Sci. U.S.A. 104, 3402–3407 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yasuda K., Nundel K., Watkins A. A., Dhawan T., Bonegio R. G., Ubellacker J. M., Marshak-Rothstein A., Rifkin I. R., Phenotype and function of B cells and dendritic cells from interferon regulatory factor 5-deficient mice with and without a mutation in DOCK2. Int. Immunol. 25, 295–306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tailor P., Tamura T., Kong H. J., Kubota T., Kubota M., Borghi P., Gabriele L., Ozato K., The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity 27, 228–239 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jefferies C. A., Regulating IRFs in IFN driven disease. Front. Immunol. 10, 325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu B. Q., Jin J., Li Y. Y., Ubiquitination modification: Critical regulation of IRF family stability and activity. Sci. China Life Sci. 64, 957–965 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B., Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science 282, 2085–2088 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Knapp S., Update on the role of Toll-like receptors during bacterial infections and sepsis. Wien. Med. Wochenschr. 160, 107–111 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Vilahur G., Badimon L., Ischemia/reperfusion activates myocardial innate immune response: The key role of the toll-like receptor. Front. Physiol. 5, 496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joosten L. A., Abdollahi-Roodsaz S., Dinarello C. A., O’Neill L., Netea M. G., Toll-like receptors and chronic inflammation in rheumatic diseases: New developments. Nat. Rev. Rheumatol. 12, 344–357 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Tall A. R., Yvan-Charvet L., Cholesterol, inflammation and innate immunity. Nat. Rev. Immunol. 15, 104–116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H. S., Bae S. C., What can we learn from genetic studies of systemic lupus erythematosus? Implications of genetic heterogeneity among populations in SLE. Lupus 19, 1452–1459 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Sadanaga A., Nakashima H., Akahoshi M., Masutani K., Miyake K., Igawa T., Sugiyama N., Niiro H., Harada M., Protection against autoimmune nephritis in MyD88-deficient MRL/lpr mice. Arthritis Rheum. 56, 1618–1628 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Nickerson K. M., Christensen S. R., Shupe J., Kashgarian M., Kim D., Elkon K., Shlomchik M. J., TLR9 regulates TLR7- and MyD88-dependent autoantibody production and disease in a murine model of lupus. J. Immunol. 184, 1840–1848 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamagna C., Scapini P., van Ziffle J. A., DeFranco A. L., Lowell C. A., Hyperactivated MyD88 signaling in dendritic cells, through specific deletion of Lyn kinase, causes severe autoimmunity and inflammation. Proc. Natl. Acad. Sci. U.S.A. 110, E3311–E3320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng Y., B cell responses to apoptotic cells in MFG-E8−/− mice. PLOS ONE 13, e0205172 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuriakose J., Redecke V., Guy C., Zhou J., Wu R., Ippagunta S. K., Tillman H., Walker P. D., Vogel P., Hacker H., Patrolling monocytes promote the pathogenesis of early lupus-like glomerulonephritis. J. Clin. Invest. 129, 2251–2265 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nanda S. K., Venigalla R. K., Ordureau A., Patterson-Kane J. C., Powell D. W., Toth R., Arthur J. S., Cohen P., Polyubiquitin binding to ABIN1 is required to prevent autoimmunity. J. Exp. Med. 208, 1215–1228 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richez C., Yasuda K., Bonegio R. G., Watkins A. A., Aprahamian T., Busto P., Richards R. J., Liu C. L., Cheung R., Utz P. J., Marshak-Rothstein A., Rifkin I. R., IFN regulatory factor 5 is required for disease development in the FcgammaRIIB−/-Yaa and FcgammaRIIB−/− mouse models of systemic lupus erythematosus. J. Immunol. 184, 796–806 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watkins A. A., Yasuda K., Wilson G. E., Aprahamian T., Xie Y., Maganto-Garcia E., Shukla P., Oberlander L., Laskow B., Menn-Josephy H., Wu Y., Duffau P., Fried S. K., Lichtman A. H., Bonegio R. G., Rifkin I. R., IRF5 deficiency ameliorates lupus but promotes atherosclerosis and metabolic dysfunction in a mouse model of lupus-associated atherosclerosis. J. Immunol. 194, 1467–1479 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yasuda K., Watkins A. A., Kochar G. S., Wilson G. E., Laskow B., Richez C., Bonegio R. G., Rifkin I. R., Interferon regulatory factor-5 deficiency ameliorates disease severity in the MRL/lpr mouse model of lupus in the absence of a mutation in DOCK2. PLOS ONE 9, e103478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savitsky D. A., Yanai H., Tamura T., Taniguchi T., Honda K., Contribution of IRF5 in B cells to the development of murine SLE-like disease through its transcriptional control of the IgG2a locus. Proc. Natl. Acad. Sci. U.S.A. 107, 10154–10159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y., Lee P. Y., Li Y., Liu C., Zhuang H., Han S., Nacionales D. C., Weinstein J., Mathews C. E., Moldawer L. L., Li S. W., Satoh M., Yang L. J., Reeves W. H., Pleiotropic IFN-dependent and -independent effects of IRF5 on the pathogenesis of experimental lupus. J. Immunol. 188, 4113–4121 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L., Feng D., Bi X., Stone R. C., Barnes B. J., Monocytes from Irf5−/− mice have an intrinsic defect in their response to pristane-induced lupus. J. Immunol. 189, 3741–3750 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ban T., Sato G. R., Nishiyama A., Akiyama A., Takasuna M., Umehara M., Suzuki S., Ichino M., Matsunaga S., Kimura A., Kimura Y., Yanai H., Miyashita S., Kuromitsu J., Tsukahara K., Yoshimatsu K., Endo I., Yamamoto T., Hirano H., Ryo A., Taniguchi T., Tamura T., Lyn kinase suppresses the transcriptional activity of IRF5 in the TLR-MyD88 pathway to restrain the development of autoimmunity. Immunity 45, 319–332 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Pellerin A., Yasuda K., Cohen-Bucay A., Sandra V., Shukla P., Jr B. K. H., Nundel K., Viglianti G. A., Xie Y., Klein U., Tan Y., Bonegio R. G., Rifkin I. R., Monoallelic IRF5 deficiency in B cells prevents murine lupus. JCI Insight 6, e141395 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manjarrez-Orduno N., Marasco E., Chung S. A., Katz M. S., Kiridly J. F., Simpfendorfer K. R., Freudenberg J., Ballard D. H., Nashi E., Hopkins T. J., Cunninghame Graham D. S., Lee A. T., Coenen M. J., Franke B., Swinkels D. W., Graham R. R., Kimberly R. P., Gaffney P. M., Vyse T. J., Behrens T. W., Criswell L. A., Diamond B., Gregersen P. K., CSK regulatory polymorphism is associated with systemic lupus erythematosus and influences B-cell signaling and activation. Nat. Genet. 44, 1227–1230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuneo M. J., Mittag T., The ubiquitin ligase adaptor SPOP in cancer. FEBS J. 286, 3946–3958 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guillamot M., Ouazia D., Dolgalev I., Yeung S. T., Kourtis N., Dai Y., Corrigan K., Zea-Redondo L., Saraf A., Florens L., Washburn M. P., Tikhonova A. N., Malumbres M., Gong Y., Tsirigos A., Park C., Barbieri C., Khanna K. M., Busino L., Aifantis I., The E3 ubiquitin ligase SPOP controls resolution of systemic inflammation by triggering MYD88 degradation. Nat. Immunol. 20, 1196–1207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin X., Shi Q., Li Q., Zhou L., Wang J., Jiang L., Zhao X., Feng K., Lin T., Lin Z., Zhuang H., Yang J., Hu C., Zhang L., Shen L., Lu Y., Zhu J., Wang H., Qi H., Meng X., Xi Y., Pan J., Fang S., Tian H., Zhou C., Zhang P., Gao K., Zhao S. M., Li Y., Gong Z., Wang C., CRL3-SPOP ubiquitin ligase complex suppresses the growth of diffuse large B-cell lymphoma by negatively regulating the MyD88/NF-κB signaling. Leukemia 34, 1305–1314 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Farrar M. A., Alberol-Ila J., Perlmutter R. M., Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature 383, 178–181 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Hacker H., Redecke V., Blagoev B., Kratchmarova I., Hsu L. C., Wang G. G., Kamps M. P., Raz E., Wagner H., Hacker G., Mann M., Karin M., Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 439, 204–207 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Nagai Y., Kojima T., Muro Y., Hachiya T., Nishizawa Y., Wakabayashi T., Hagiwara M., Identification of a novel nuclear speckle-type protein, SPOP. FEBS Lett. 418, 23–26 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Kwon J. E., La M., Oh K. H., Oh Y. M., Kim G. R., Seol J. H., Baek S. H., Chiba T., Tanaka K., Bang O. S., Joe C. O., Chung C. H., BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J. Biol. Chem. 281, 12664–12672 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Hernandez-Munoz I., Lund A. H., van der Stoop P., Boutsma E., Muijrers I., Verhoeven E., Nusinow D. A., Panning B., Marahrens Y., van Lohuizen M., Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl. Acad. Sci. U.S.A. 102, 7635–7640 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kent D., Bush E. W., Hooper J. E., Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development 133, 2001–2010 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Zhuang M., Calabrese M. F., Liu J., Waddell M. B., Nourse A., Hammel M., Miller D. J., Walden H., Duda D. M., Seyedin S. N., Hoggard T., Harper J. W., White K. P., Schulman B. A., Structures of SPOP-substrate complexes: Insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell 36, 39–50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barbieri C. E., Baca S. C., Lawrence M. S., Demichelis F., Blattner M., Theurillat J. P., White T. A., Stojanov P., Van Allen E., Stransky N., Nickerson E., Chae S. S., Boysen G., Auclair D., Onofrio R. C., Park K., Kitabayashi N., MacDonald T. Y., Sheikh K., Vuong T., Guiducci C., Cibulskis K., Sivachenko A., Carter S. L., Saksena G., Voet D., Hussain W. M., Ramos A. H., Winckler W., Redman M. C., Ardlie K., Tewari A. K., Mosquera J. M., Rupp N., Wild P. J., Moch H., Morrissey C., Nelson P. S., Kantoff P. W., Gabriel S. B., Golub T. R., Meyerson M., Lander E. S., Getz G., Rubin M. A., Garraway L. A., Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 44, 685–689 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai H., Liu A., Spop promotes skeletal development and homeostasis by positively regulating Ihh signaling. Proc. Natl. Acad. Sci. U.S.A. 113, 14751–14756 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Boer J., Williams A., Skavdis G., Harker N., Coles M., Tolaini M., Norton T., Williams K., Roderick K., Potocnik A. J., Kioussis D., Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33, 314–325 (2003). [DOI] [PubMed] [Google Scholar]
- 49.An J., Wang C., Deng Y., Yu L., Huang H., Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 6, 657–669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geng C., Rajapakshe K., Shah S. S., Shou J., Eedunuri V. K., Foley C., Fiskus W., Rajendran M., Chew S. A., Zimmermann M., Bond R., He B., Coarfa C., Mitsiades N., Androgen receptor is the key transcriptional mediator of the tumor suppressor SPOP in prostate cancer. Cancer Res. 74, 5631–5643 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang P., Gao K., Jin X., Ma J., Peng J., Wumaier R., Tang Y., Zhang Y., An J., Yan Q., Dong Y., Huang H., Yu L., Wang C., Endometrial cancer-associated mutants of SPOP are defective in regulating estrogen receptor-alpha protein turnover. Cell Death Dis. 6, e1687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li C., Ao J., Fu J., Lee D. F., Xu J., Lonard D., O’Malley B. W., Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene 30, 4350–4364 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geng C., Kaochar S., Li M., Rajapakshe K., Fiskus W., Dong J., Foley C., Dong B., Zhang L., Kwon O. J., Shah S. S., Bolaki M., Xin L., Ittmann M., O’Malley B. W., Coarfa C., Mitsiades N., SPOP regulates prostate epithelial cell proliferation and promotes ubiquitination and turnover of c-MYC oncoprotein. Oncogene 36, 4767–4777 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V., Clark N. R., Ma’ayan A., Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., Koplev S., Jenkins S. L., Jagodnik K. M., Lachmann A., McDermott M. G., Monteiro C. D., Gundersen G. W., Ma’ayan A., Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han H., Cho J. W., Lee S., Yun A., Kim H., Bae D., Yang S., Kim C. Y., Lee M., Kim E., Lee S., Kang B., Jeong D., Kim Y., Jeon H. N., Jung H., Nam S., Chung M., Kim J. H., Lee I., TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 46, D380–D386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S., Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301, 640–643 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Lopez-Pelaez M., Lamont D. J., Peggie M., Shpiro N., Gray N. S., Cohen P., Protein kinase IKKβ-catalyzed phosphorylation of IRF5 at Ser462 induces its dimerization and nuclear translocation in myeloid cells. Proc. Natl. Acad. Sci. U.S.A. 111, 17432–17437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ren J., Chen X., Chen Z. J., IKKβ is an IRF5 kinase that instigates inflammation. Proc. Natl. Acad. Sci. U.S.A. 111, 17438–17443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sotirellis N., Johnson T. M., Hibbs M. L., Stanley I. J., Stanley E., Dunn A. R., Cheng H. C., Autophosphorylation induces autoactivation and a decrease in the Src homology 2 domain accessibility of the Lyn protein kinase. J. Biol. Chem. 270, 29773–29780 (1995). [DOI] [PubMed] [Google Scholar]
- 61.Kmiecik T. E., Johnson P. J., Shalloway D., Regulation by the autophosphorylation site in overexpressed pp60c-src. Mol. Cell. Biol. 8, 4541–4546 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Freedman T. S., Tan Y. X., Skrzypczynska K. M., Manz B. N., Sjaastad F. V., Goodridge H. S., Lowell C. A., Weiss A., LynA regulates an inflammation-sensitive signaling checkpoint in macrophages. eLife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brian B. F. IV, Jolicoeur A. S., Guerrero C. R., Nunez M. G., Sychev Z. E., Hegre S. A., Saetrom P., Habib N., Drake J. M., Schwertfeger K. L., Freedman T. S., Unique-region phosphorylation targets LynA for rapid degradation, tuning its expression and signaling in myeloid cells. eLife 8, e46043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ogawa A., Takayama Y., Sakai H., Chong K. T., Takeuchi S., Nakagawa A., Nada S., Okada M., Tsukihara T., Structure of the carboxyl-terminal Src kinase, Csk. J. Biol. Chem. 277, 14351–14354 (2002). [DOI] [PubMed] [Google Scholar]
- 65.Madeira F., Park Y. M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A. R. N., Potter S. C., Finn R. D., Lopez R., The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marzahn M. R., Marada S., Lee J., Nourse A., Kenrick S., Zhao H., Ben-Nissan G., Kolaitis R. M., Peters J. L., Pounds S., Errington W. J., Prive G. G., Taylor J. P., Sharon M., Schuck P., Ogden S. K., Mittag T., Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 35, 1254–1275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okada M., Regulation of the SRC family kinases by Csk. Int. J. Biol. Sci. 8, 1385–1397 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cooper J. A., Kaneko T., Li S. S., Cell regulation by phosphotyrosine-targeted ubiquitin ligases. Mol. Cell. Biol. 35, 1886–1897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joazeiro C. A., Wing S. S., Huang H., Leverson J. D., Hunter T., Liu Y. C., The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286, 309–312 (1999). [DOI] [PubMed] [Google Scholar]
- 70.Zheng N., Wang P., Jeffrey P. D., Pavletich N. P., Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102, 533–539 (2000). [DOI] [PubMed] [Google Scholar]
- 71.Xiong H., Li H., Kong H. J., Chen Y., Zhao J., Xiong S., Huang B., Gu H., Mayer L., Ozato K., Unkeless J. C., Ubiquitin-dependent degradation of interferon regulatory factor-8 mediated by Cbl down-regulates interleukin-12 expression. J. Biol. Chem. 280, 23531–23539 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Sabe H., Hata A., Okada M., Nakagawa H., Hanafusa H., Analysis of the binding of the Src homology 2 domain of Csk to tyrosine-phosphorylated proteins in the suppression and mitotic activation of c-Src. Proc. Natl. Acad. Sci. U.S.A. 91, 3984–3988 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu J., Liu J., Ghirlando R., Saltiel A. R., Hubbard S. R., Structural basis for recruitment of the adaptor protein APS to the activated insulin receptor. Mol. Cell 12, 1379–1389 (2003). [DOI] [PubMed] [Google Scholar]
- 74.Saito K., Enya K., Oneyama C., Hikita T., Okada M., Proteomic identification of ZO-1/2 as a novel scaffold for Src/Csk regulatory circuit. Biochem. Biophys. Res. Commun. 366, 969–975 (2008). [DOI] [PubMed] [Google Scholar]
- 75.Baumeister U., Funke R., Ebnet K., Vorschmitt H., Koch S., Vestweber D., Association of Csk to VE-cadherin and inhibition of cell proliferation. EMBO J. 24, 1686–1695 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brdickova N., Brdicka T., Angelisova P., Horvath O., Spicka J., Hilgert I., Paces J., Simeoni L., Kliche S., Merten C., Schraven B., Horejsi V., LIME: A new membrane Raft-associated adaptor protein involved in CD4 and CD8 coreceptor signaling. J. Exp. Med. 198, 1453–1462 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cao H., Courchesne W. E., Mastick C. C., A phosphotyrosine-dependent protein interaction screen reveals a role for phosphorylation of caveolin-1 on tyrosine 14: Recruitment of C-terminal Src kinase. J. Biol. Chem. 277, 8771–8774 (2002). [DOI] [PubMed] [Google Scholar]
- 78.Kawabuchi M., Satomi Y., Takao T., Shimonishi Y., Nada S., Nagai K., Tarakhovsky A., Okada M., Transmembrane phosphoprotein Cbp regulates the activities of Src-family tyrosine kinases. Nature 404, 999–1003 (2000). [DOI] [PubMed] [Google Scholar]
- 79.Wong L., Lieser S. A., Miyashita O., Miller M., Tasken K., Onuchic J. N., Adams J. A., Woods V. L. Jr., Jennings P. A., Coupled motions in the SH2 and kinase domains of Csk control Src phosphorylation. J. Mol. Biol. 351, 131–143 (2005). [DOI] [PubMed] [Google Scholar]
- 80.Safari F., Murata-Kamiya N., Saito Y., Hatakeyama M., Mammalian Pragmin regulates Src family kinases via the Glu-Pro-Ile-Tyr-Ala (EPIYA) motif that is exploited by bacterial effectors. Proc. Natl. Acad. Sci. U.S.A. 108, 14938–14943 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lecointre C., Simon V., Kerneur C., Allemand F., Fournet A., Montarras I., Pons J. L., Gelin M., Brignatz C., Urbach S., Labesse G., Roche S., Dimerization of the Pragmin pseudo-kinase regulates protein tyrosine phosphorylation. Structure 26, 545–554.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 82.Levinson N. M., Seeliger M. A., Cole P. A., Kuriyan J., Structural basis for the recognition of c-Src by its inactivator Csk. Cell 134, 124–134 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Couture C., Deckert M., Williams S., Russo F. O., Altman A., Mustelin T., Identification of the site in the Syk protein tyrosine kinase that binds the SH2 domain of Lck. J. Biol. Chem. 271, 24294–24299 (1996). [DOI] [PubMed] [Google Scholar]
- 84.Tarricone C., Dhavan R., Peng J., Areces L. B., Tsai L. H., Musacchio A., Structure and regulation of the CDK5-p25nck5a complex. Mol. Cell 8, 657–669 (2001). [DOI] [PubMed] [Google Scholar]
- 85.Ahmed Z., Smith B. J., Kotani K., Wilden P., Pillay T. S., APS, an adapter protein with a PH and SH2 domain, is a substrate for the insulin receptor kinase. Biochem. J. 341 (Pt. 3), 665–668 (1999). [PMC free article] [PubMed] [Google Scholar]
- 86.Farley F. W., Soriano P., Steffen L. S., Dymecki S. M., Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28, 106–110 (2000). [PubMed] [Google Scholar]
- 87.Redecke V., Wu R., Zhou J., Finkelstein D., Chaturvedi V., High A. A., Hacker H., Hematopoietic progenitor cell lines with myeloid and lymphoid potential. Nat. Methods 10, 795–803 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tora L., Mullick A., Metzger D., Ponglikitmongkol M., Park I., Chambon P., The cloned human oestrogen receptor contains a mutation which alters its hormone binding properties. EMBO J. 8, 1981–1986 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ippagunta S. K., Pollock J. A., Sharma N., Lin W., Chen T., Tawaratsumida K., High A. A., Min J., Chen Y., Guy R. K., Redecke V., Katzenellenbogen J. A., Hacker H., Identification of Toll-like receptor signaling inhibitors based on selective activation of hierarchically acting signaling proteins. Sci. Signal. 11, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bouchard J. J., Otero J. H., Scott D. C., Szulc E., Martin E. W., Sabri N., Granata D., Marzahn M. R., Lindorff-Larsen K., Salvatella X., Schulman B. A., Mittag T., Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell 72, 19–36.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roehrl M. H., Wang J. Y., Wagner G., A general framework for development and data analysis of competitive high-throughput screens for small-molecule inhibitors of protein–protein interactions by fluorescence polarization. Biochemistry 43, 16056–16066 (2004). [DOI] [PubMed] [Google Scholar]
- 92.Ippagunta S. K., Gangwar R., Finkelstein D., Vogel P., Pelletier S., Gingras S., Redecke V., Hacker H., Keratinocytes contribute intrinsically to psoriasis upon loss of Tnip1 function. Proc. Natl. Acad. Sci. U.S.A. 113, E6162–E6171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Table S1